Abstract
Osteoarthritis (OA) is the most common type of arthritis, observed mainly in the population aged >65 years. However, the mechanism underlying the development and progression of OA has remained largely elusive. The present study aimed to identify differentially expressed mRNAs and lncRNAs in OA. By analyzing the GSE48556 and GSE82107 datasets, a total of 202 up- and 434 downregulated mRNAs were identified in OA. Gene ontology and Kyoto Encyclopedia of Genes and Genomes pathway analysis indicated that differently expressed genes were mainly involved in regulating antigen processing and presentation, interspecies interaction between organisms, immune response, transcription and signal transduction. In addition, a series of long non-coding (lnc)RNAs were differently expressed in OA. To provide novel data on the molecular mechanisms and functional roles of these lncRNAs in OA, a co-expression analysis was performed, which revealed that the dysregulated lncRNAs were associated with transcription, signal transduction, immune response and cell adhesion. In addition, certain key genes in protein-protein interaction networks were identified. The present study provided useful information for exploring potential candidate biomarkers for the diagnosis and prognosis of OA, as well as novel drug targets.
Keywords: differentially expressed gene, long non-coding RNA, osteoarthritis, protein-protein interaction network, expression profiling
Introduction
Osteoarthritis (OA), the most common type of arthritis, reduces the quality of life of affected individuals and is most common in the population aged >65 years (1). Although previous studies made efforts to develop therapeutic methods and explore the underlying mechanisms of OA, they remain far from being completely elucidated; however, it has been revealed that inflammation is involved in the development and progression of OA (2). Protein-protein interaction (PPI) network analysis is a powerful tool to explore the pathological mechanisms of human diseases (3). Furthermore, microarray analysis has been widely applied to monitor gene expression in OA. Thus, combining PPI and public microarray data will help to identify novel pathways regulating OA progression.
Long non-coding RNAs (lncRNAs) are a major class of ncRNAs with a length of >200 nucleotides (4). lncRNAs have important roles in regulating a vast variety of biological processes at the transcriptional, post-transcriptional and post-translational level. Recently, several studies have indicated that lncRNAs may act as contributors to OA. Certain lncRNAs, including ubiquitin-fold modifier conjugating enzyme 1 (UFC1) (5), thymosin-β-4 (6), growth arrest-specific 5 (GAS5) (7) act as natural microRNA sponges in human OA. A well-known lncRNA, HOX transcript antisense intergenic RNA (HOTAIR) (8), was reported to participate in interleukin-1β-induced matrix metalloproteinase overexpression and chondrocyte apoptosis in temporomandibular joint OA. However, the functional roles of most lncRNAs in OA have remained largely elusive.
In the present study, two public datasets were analyzed to identify differentially expressed lncRNAs and mRNAs in OA. Next, gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were performed to explore the potential roles of differentially expressed genes (DEGs). A series of lncRNAs that were differently expressed in OA were identified. To provide novel information on the molecular mechanisms and functional roles of lncRNAs in OA, a co-expression analysis was also performed. The present study provided useful information for exploring potential candidate biomarkers for the diagnosis and prognosis of RA, as well as drug targets.
Materials and methods
Microarray data and data pre-processing
The microarray datasets GSE48556 (9) and GSE82107 (10) were downloaded from the National Centre for Biotechnology Information (NCBI) Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/). A total of 139 samples were included in GSE48556. Of these, 106 biopsies were obtained from OA patients and 33 biopsies were obtained from healthy controls. A total of 17 samples (10 from OA patients and 7 from healthy controls) were included in the GSE82107 dataset. mRNAs with fold changes ≥2 and P<0.05 were considered to be significantly differentially expressed.
GO and KEGG pathway analysis
To identify the functions of DEGs in OA, a GO function enrichment analysis was performed. KEGG pathway enrichment analysis was also performed to identify pathways enriched in OA using the Molecule Annotation System (http://bioinfo.capitalbio.com/mas3/). The P-value was calculated by hypergeometric distribution and a pathway with P<0.05 was considered as significant.
LncRNA classification pipeline
LncRNA expression patterns were assessed in the microarray dataset GSE82107. The following criteria were used to identify the unique probe sets for lncRNAs from the Affymetrix array. Refseq IDs labelled as ‘NR_’ which is indicative of ncRNA in the Refseq database, were retained (11). Finally, 2,448 annotated lncRNA transcripts with corresponding Affymetrix probe IDs were obtained. lncRNAs with fold changes ≥2 and P<0.05 were selected as significantly differentially expressed lncRNAs.
Identification of lncRNA-associated PPI modules
The Search Tool for the Retrieval of Interacting Genes/ Proteins (STRING) online software (https://string-db.org) was used to assess the interactions. The interactions of the proteins encoded by the differentially expressed genes were searched using STRING online software, and the combined score of >0.4 was used as the cut-off criterion. The PPI network was visualized using Cytoscape software (http://www.cytoscape.org).
Co-expression network construction and analysis
In the present study, the Pearson correlation coefficient of DEG-lncRNA pairs was calculated according to their expression values. The co-expressed DEG-lncRNA pairs with an absolute value of the Pearson correlation coefficient of ≥0.8 were selected and the co-expression network was established by using Cytoscape software. The Cytoscape MCODE plug-in (version 3.4.0) was applied to search for clustered sub-networks of highly connected nodes from the co-expression network. The resulting network was subjected to module analyses with the Plugin MCODE with the following default parameters: Degree cut-off, ≥3; and nodes with edges, ≥3-core).
Statistical analysis
Numerical data are presented as the mean ± standard deviation of at least three determinations. Statistical comparisons between groups of normalized data were performed using the t-test or Mann-Whitney U-test according to the test conditions. P<0.05 was considered to indicate a statistically significant difference with a 95% confidence level. R (version 3.2.4; http://www.r-project.org/) was used to perform all analyses.
Results
Identification of DEGs in OA
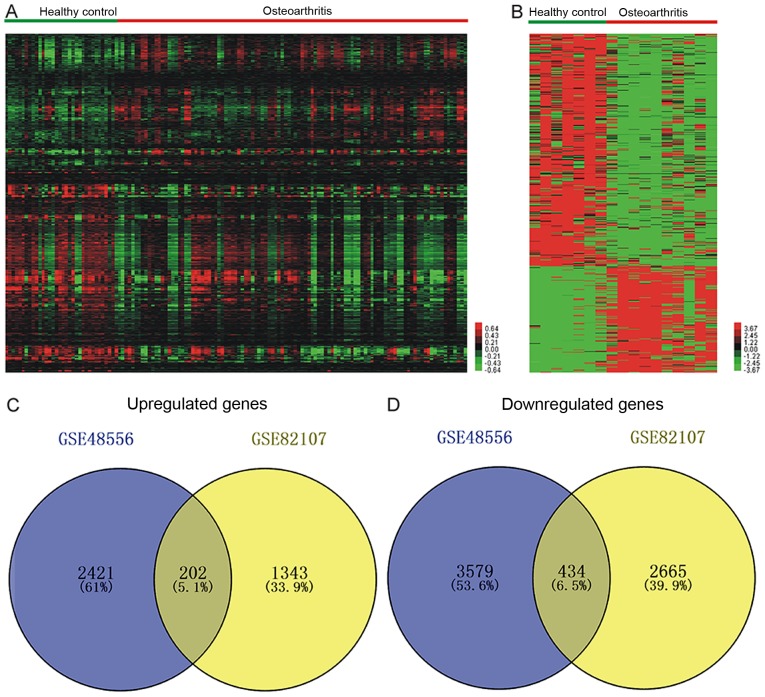
To identify the significantly differentially expressed mRNAs between OA patients vs. healthy controls, two publicly available gene expression datasets, GSE48556 and GSE82107, were analyzed. A total of 2,623 up- and 4,013 downregulated mRNAs were identified in GSE48556 (Fig. 1A), and from the GSE82107 dataset, 1,545 up- and 3,099 downregulated mRNAs were obtained (Fig. 1B). Integrated analysis of these two datasets revealed that the two datasets had 202 up- and 434 downregulated mRNAs in common (Fig 1C and D). The top 10 up- and downregulated mRNAs are listed in Table I.
Figure 1.
Clustering analysis of differentially expressed mRNAs between osteoarthritis patients and healthy controls in the datasets (A) GSE48556 and (B) GSE82107. Integrated analysis of (C) upregulated and (D) downregulated mRNAs.
Table I.
Top 10 up- and downregulated mRNAs in the GSE82107 dataset.
| Gene symbol | P-value | Ave (control) | Ave (OA) |
|---|---|---|---|
| Downregulated genes | |||
| GPD1 | 0.04825352 | 104.04 | 11.85 |
| MYF6 | 0.046950299 | 529.61 | 60.34 |
| PCDH15 | 0.018945043 | 7.96 | 0.93 |
| KIAA1257 | 0.00647357 | 12.73 | 1.49 |
| ARG1 | 0.03656324 | 10.41 | 1.26 |
| LOC101927734 | 0.035854747 | 26.83 | 3.45 |
| LOC101929609 | 0.0011542 | 13.47 | 1.88 |
| SHC3 | 0.001053533 | 28.54 | 4.02 |
| KIAA1661 | 0.035300865 | 11.66 | 1.69 |
| FBXL21 | 0.011937398 | 9.66 | 1.41 |
| Upregulated genes | |||
| CYR61 | 0.030911933 | 307.24 | 1482.31 |
| DLX1 | 0.031583337 | 1.67 | 8.47 |
| LRRC15 | 0.049914446 | 145.54 | 747.21 |
| SOCS3 | 0.014017918 | 159.34 | 850.29 |
| VMO1 | 0.022699188 | 4.83 | 25.96 |
| SELO | 0.001439415 | 2.16 | 11.88 |
| HORMAD1 | 0.025736549 | 1.53 | 8.46 |
| RARRES1 | 0.045620106 | 14.71 | 108.20 |
| STMN2 | 0.020694784 | 5.23 | 39.04 |
| GJB2 | 0.02794137 | 99.03 | 746.29 |
OA, osteoarthritis; Ave, average value; mRNA, messenger RNA.
GO and KEGG analysis of differentially expressed mRNAs
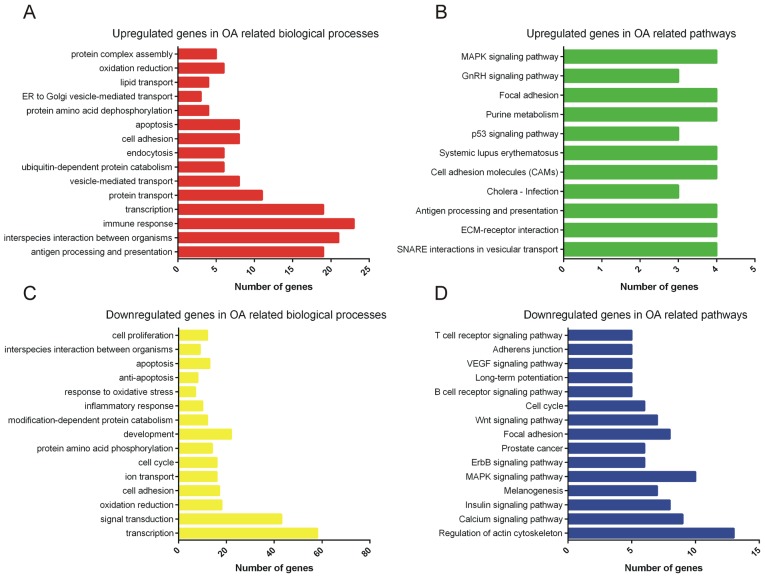
Next, the differentially expressed mRNAs were subjected to GO and KEGG analyses (Fig. 2). GO analysis indicated that the upregulated genes were mainly involved in regulating antigen processing and presentation, interspecies interaction between organisms, immune response, transcription, protein transport, vesicle-mediated transport, ubiquitin-dependent protein catabolism, endocytosis, cell adhesion and apoptosis (Fig. 2A). Furthermore, the downregulated genes were mainly enriched in categories associated with transcription, signal transduction, oxidation/reduction, cell adhesion, ion transport, cell cycle, protein amino acid phosphorylation, development, modification-dependent protein catabolism and inflammatory response (Fig. 2B). The above pathways may therefore participate in regulating the progression of OA.
Figure 2.
GO and KEGG pathway analysis of differentially expressed mRNAs. (A) GO analysis and (B) KEGG analysis of upregulated mRNAs in OA-associated biological processes and pathways, respectively. (C) GO analysis and (D) KEGG analysis of downregulated mRNAs in OA-associated biological processes and pathways, respectively. GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; OA, osteoarthritis; ER, endoplasmic reticulum; ECM, extracellular matrix; VEGF, vascular endothelial growth factor; MAPK, mitogen-activated protein kinase; GnRH, gonadotropin-releasing hormone; SNARE, soluble NSF attachment protein receptor.
KEGG pathway analysis revealed that upregulated genes were primarily enriched in pathways associated with the p53, gonadotropin-releasing hormone and mitogen-associated protein kinase (MAPK) signaling pathways (Fig. 2C). Downregulated genes were mainly associated with calcium, insulin, MAPK, ErbB, Wnt, B-cell receptor signaling pathway and vascular endothelial growth factor (VEGF) signaling pathways (Fig. 2D).
PPI network construction
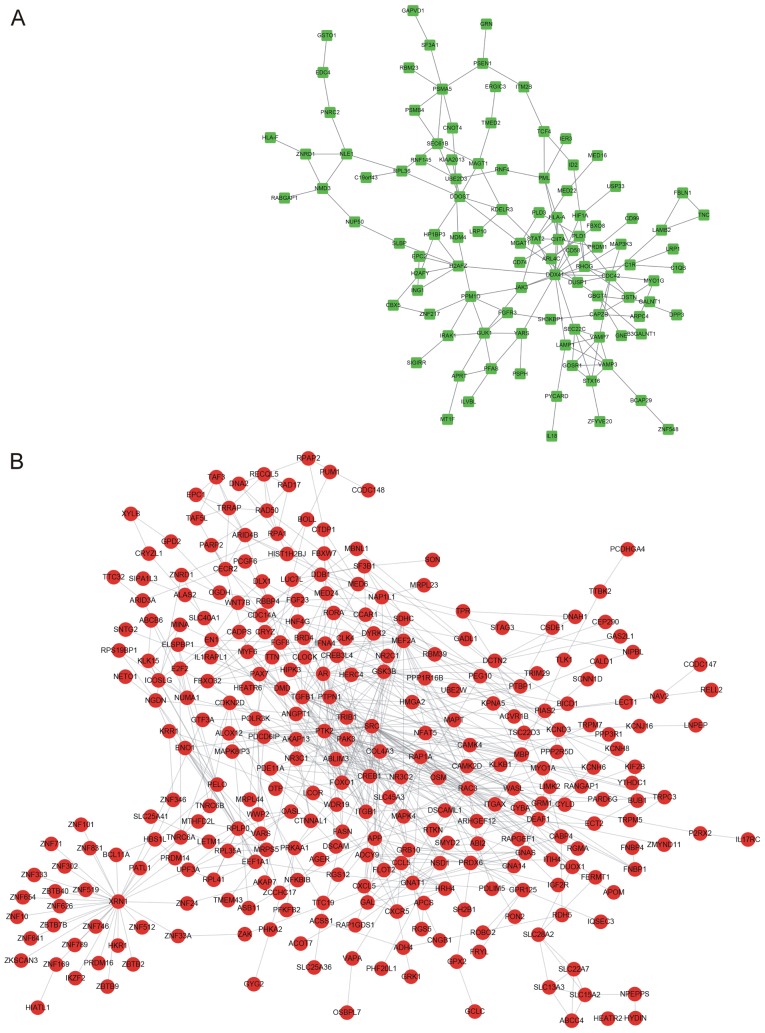
The PPI networks constructed for the up- and downregulated genes are presented in Fig. 3. The PPI network for the upregulated genes contained 78 nodes and 158 edges, and the hub nodes with the highest connectivity degree were DEAD box helicase 41 (DDX41; connectivity degree=13) and cell division cycle 41 (CDC42; connectivity degree=6; Fig. 3A). The PPI network for the downregulated genes contained 219 nodes and 656 edges, and the hub nodes with the highest connectivity degree were cyclic AMP responsive element binding protein 1 (CREB; connectivity degree=28), SH2 domain containing (SRC; connectivity degree=27) and androgen receptor (AR; d connectivity degree=18; Fig. 3B).
Figure 3.
Protein-protein interaction networks of (A) upregulated and (B) downregulated genes in osteoarthritis patients.
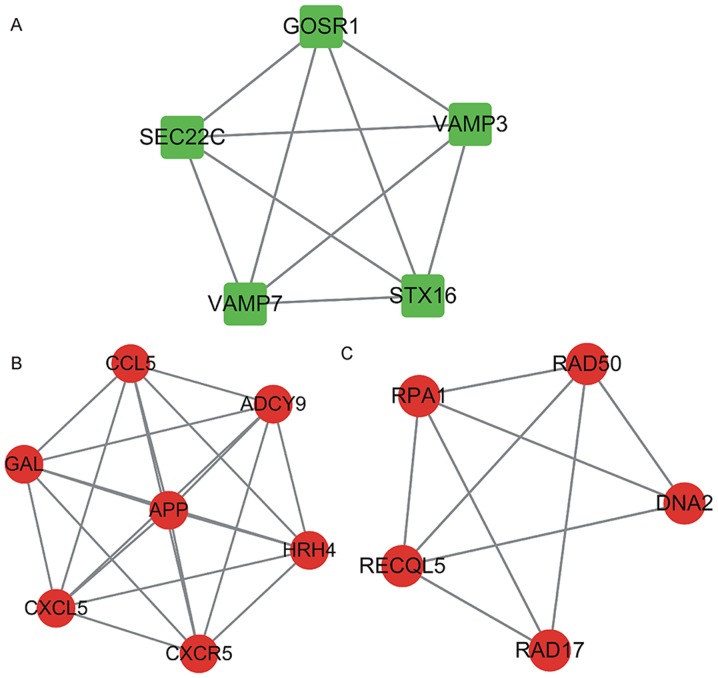
Furthermore, a module analysis of the network was performed using the MCODE plugin (degree cut-off, ≥3; the nodes with edges, ≥3-core). In the respective PPI networks, a total of 5 up-regulated proteins, including golgi SNAP receptor complex member 1 (GOSR1), SEC22 homolog C vesicle trafficking protein (SEC22C), vesicle associated membrane protein 7 (VAMP7), syntaxin 16 (STX16) and vesicle associated membrane protein 3 (VAMP3; Fig. 4A) and 12 down-regulated proteins including, C-C motif chemokine ligand 5 (CCL5), galanin and GMAP prepropeptide (GAL), C-X-C motif chemokine ligand 5 (CXCL5), amyloid beta precursor protein (APP), adenylate cyclase 9 (ADCY9), histamine receptor H4 (HRH4), C-X-C motif chemokine receptor 5 (CXCR5), replication protein A1 (RPA1), RAD50 double strand break repair protein (RAD50), DNA replication helicase/nuclease 2 (DNA2), RAD17 checkpoint clamp loader component (RAD17) and RecQ like helicase 5 (RECQL5; Fig. 4B and C) were identified as key proteins/genes.
Figure 4.
Hub modules from the PPI network in osteoarthritis patients were constructed. (A) A total of five upregulated proteins constituted a hub PPI module: GOSR1, SEC22C, VAMP7, STX16) and VAMP3. (B and C) Twelve downregulated proteins constituted 2 hub PPI modules: CCL5, GAL, CXCL5, APP, ADCY9, HRH4, CXCR5, RPA1, RAD50, DNA2, RAD17 and RECQL5. PPI, protein-protein interaction. GOSR1, golgi SNAP receptor complex member 1; SEC22C, SEC22 homolog C vesicle trafficking protein; VAMP7, vesicle associated membrane protein 7; STX16, syntaxin 16; VAMP3, vesicle associated membrane protein 3; CCL5, C-C motif chemokine ligand 5; GAL, galanin and GMAP prepropeptide; CXCL5, C-X-C motif chemokine ligand 5; APP, amyloid beta precursor protein; ADCY9, adenylate cyclase 9; HRH4, histamine receptor H4; CXCR5, C-X-C motif chemokine receptor 5; RPA1, replication protein A1; RAD50, RAD50 double strand break repair protein; DNA2, DNA replication helicase/nuclease 2; RAD17, RAD17 checkpoint clamp loader component; RECQL5, RecQ like helicase 5.
Co-expression network analysis of differently expressed lncRNAs in OA
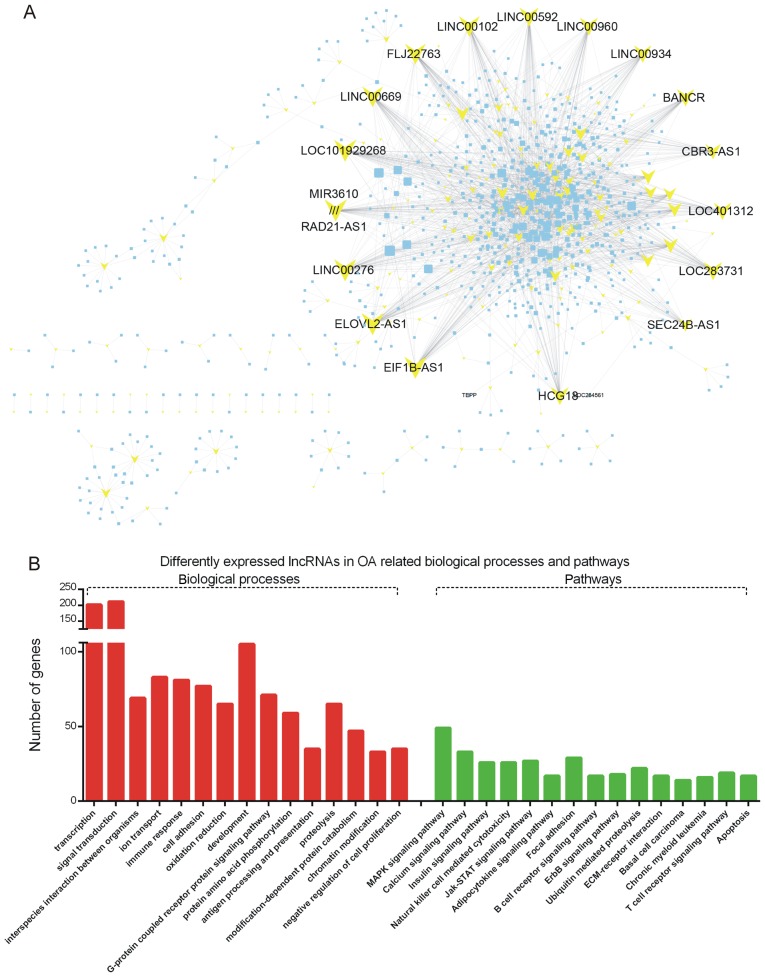
Furthermore, the differently expressed lncRNAs were identified from the GSE82107 dataset. A total of 25 up- and 326 downregulated lncRNAs were identified in OA. The top 10 up- and down-regulated lncRNAs are presented in Table II. To predict the potential functional roles of these lncRNAs, the Pearson correlation coefficient for lncRNA-DEG pairs was first calculated according to their expression value. The co-expressed DEG-lncRNA pairs with an absolute value of their Pearson correlation coefficient of ≥0.8 were selected. As presented in Fig. 5A, the network included 348 differentially expressed lncRNAs and 2,883 DEGs (Fig. 5A).
Table II.
Top 10 up- and downregulated lncRNAs in the GSE82107 dataset.
| Gene symbol | P-value | Ave (control) | Ave (OA) |
|---|---|---|---|
| Downregulated lncRNA | |||
| LEMD1-AS1 | 2.68×10−3 | 7.39 | 1.06 |
| NAV2-AS5 | 1.28×10−2 | 9.59 | 1.4 |
| LOC101928107 | 4.46×10−2 | 6.70 | 1.07 |
| LOC286083 | 7.45×10−3 | 10.49 | 1.82 |
| LOC340107 | 3.90×10−3 | 9.66 | 1.72 |
| GRIK1-AS2 | 3.08×10−3 | 13.34 | 2.39 |
| LINC00551 | 1.88×10−2 | 5.94 | 1.07 |
| LOC401134 | 1.19×10−2 | 9.70 | 1.81 |
| SNORA74A | 2.98×10−2 | 7.26 | 1.41 |
| CBR3-AS1 | 5.04×10−3 | 14.81 | 2.89 |
| Upregulated lncRNA | |||
| FLJ32255 | 5.84×10−4 | 21.84 | 46.97 |
| LINC01094 | 2.11×10−2 | 48.00 | 114.5 |
| NPHP3-AS1 | 2.75×10−2 | 5.61 | 13.8 |
| LINC00957 | 9.84×10−3 | 32.21 | 86.68 |
| LINC00963 | 3.02×10−2 | 3.30 | 9.47 |
| FLJ27354 | 3.85×10−3 | 4.54 | 14.76 |
| LOC101927720 | 1.08×10−2 | 4.77 | 15.67 |
| SIGLEC16 | 1.09×10−2 | 6.30 | 20.8 |
| LOC100506119 | 4.60×10−2 | 4.43 | 16.33 |
| LOC101060091 | 1.75×10−2 | 1.56 | 7.68 |
OA, osteoarthritis; Ave, average value; lncRNA, long non-coding RNA.
Figure 5.
(A) Differentially expressed gene-lncRNA co-expression pairs with an absolute value of the Pearson correlation coefficient of ≥0.9 were selected. (B) Gene ontology and Kyoto Encyclopedia of Genes and Genomes pathway analysis of lncRNAs based on the set of co-expressed mRNAs. OA, osteoarthritis; lncRNA, long non-coding RNA; ER, endoplasmic reticulum; ECM, extracellular matrix; MAPK, mitogen-activated protein kinase; Jak, Janus kinase; STAT, signal transducer and activator of transcription; LINC00592, long intergenic non-protein coding RNA 592; LINC00960, long intergenic non-protein coding RNA 960; LINC00934, long intergenic non-protein coding RNA 934; BANCR, BRAF-activated non-protein coding RNA; CBR3-AS1, CBR3 antisense RNA 1; SEC24B-AS1, SEC24B antisense RNA 1; HCG18, HLA complex group 18 (non-protein coding); EIF1B-AS1, EIF1B antisense RNA 1; ELOVL2-AS1, ELOVL2 antisense RNA 1; LINC00276, long intergenic non-protein coding RNA 276; RAD21-AS1, RAD21 antisense RNA 1; MIR3610, microRNA 3610; LINC00669, MIR924 host gene; FLJ2276, chromosome 3 open reading frame 85; LINC00102, long intergenic non-protein coding RNA 102.
To predict the functions of the differentially expressed lncRNAs, GO and KEGG pathway analysis was performed for each given lncRNA by using the respective set of co-expressed mRNAs. According to the KEGG pathway analysis, the dysregulated lncRNAs were primarily enriched in pathways associated with the MAPK, calcium and insulin pathways, natural killer cell-mediated cytotoxicity, and with the Janus kinase/signal transducer and activator of transcription, adipocytokine, B-cell receptor and ErbB signaling pathways (Fig. 5B). GO analysis revealed that the dysregulated lncRNAs were associated with transcription, signal transduction, interspecies interaction between organisms, ion transport, immune response, cell adhesion, oxidation/reduction, development, G-protein-coupled receptor protein signaling pathway and protein amino acid phosphorylation (Fig. 5B).
Identification of key lncRNAs in OA
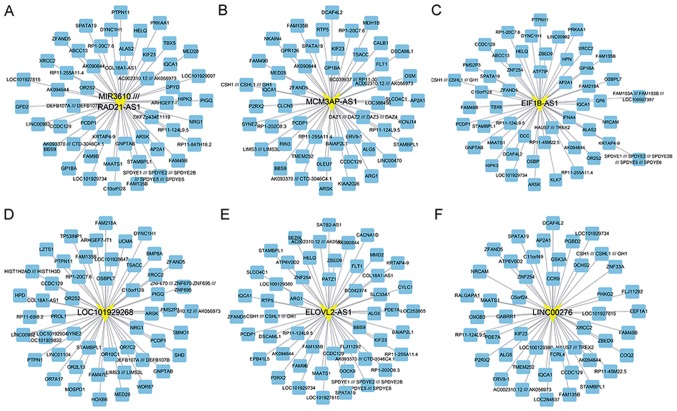
In the present study, a total of lncRNAs, including EIF1B antisense RNA 1 (EIF1B-AS1), ELOVL2 antisense RNA 1 (ELOVL2-AS1), long intergenic non-protein coding RNA 276 (LINC00276), LOC101929268, MCM3AP antisense RNA 1 (MCM3AP-AS1) and RAD21 antisense RNA 1 (RAD21-AS1) were identified to be co-expressed with an absolute value of the Pearson correlation coefficient of ≥0.8. These lncRNAs were identified as key lncRNAs in OA. The co-expression network of each key lncRNAs is presented in Fig. 6.
Figure 6.
Co-expression networks for key long non-coding RNAs in osteoarthritis. The co-expression networks were constructed for (A) RAD21-AS1, (B) MCM3AP-AS1, (C) EIF1B-AS1, (D) LOC101929268, (E) ELOVL2-AS1 and (F) LINC00276. RAD21-AS1, RAD21 antisense RNA 1; MCM3AP-AS1, MCM3AP antisense RNA 1; EIF1B-AS1, EIF1B antisense RNA 1; ELOVL2-AS1, ELOVL2 antisense RNA 1; LINC00276, long intergenic non-protein coding RNA 276.
Discussion
OA has become one of the most frequent chronic diseases, which is considered synonymous with aging (12). Elucidation of the mechanisms underlying the development and progression of OA is urgently required in order to identify novel biomarkers for the diagnosis and prognosis of OA patients, and for the development of drug targets. In the present study, the GSE48556 and GSE82107 datasets were analyzed to identify differentially expressed mRNAs and lncRNAs in OA patients. A total of 202 up- and 434 downregulated mRNAs were obtained in OA. GO analysis indicated that the upregulated genes were mainly involved in regulating antigen processing and presentation, interspecies interaction between organisms, immune response and transcription, while the downregulated genes were mainly enriched in transcription, signal transduction, oxidation reduction and cell adhesion. KEGG pathway analysis revealed that the upregulated genes were primarily enriched in pathways associated with the p53, GnRH and MAPK signaling pathways, while the downregulated genes were mainly associated with calcium, insulin, MAPK, ErbB, Wnt, B-cell receptor and VEGF signaling pathways. The GO terms and KEGG pathways that the dysregulated genes were enriched in provide an insight in the pathways associated with OA.
PPI network analysis is a useful tool to explore the pathological mechanisms of human diseases. For instance, Chen et al (13) identified DEGs in salivary adenoid cystic carcinoma cells. In the present study, PPI networks for up- and downregulated genes in OA were constructed. The PPI network for the upregulated genes contained 78 nodes and 158 edges, and the hub nodes with the highest connectivity degree were DDX41 (degree=13) and CDC42 (degree=6). The PPI network for the downregulated genes contained 219 nodes and 656 edges, and the hub nodes with the highest connectivity degree were CREB1 (degree=28), SRC (degree=27) and AR (degree=18). Of note, it was observed that the PPI for the down-regulated genes included hub genes involved in various hormone-associated pathways, suggesting that hormones are involved in the development and progression of OA. Furthermore, a module analysis of the network was performed using the MCODE plugin. A total of upregulated proteins (GOSR1, SEC22C, VAMP7, STX16 and VAMP3) and 12 downregulated proteins (CCL5, GAL, CXCL5, APP, ADCY9, HRH4, CXCR5, RPA1, RAD50, DNA2, RAD17 and RECQL5) were identified as key genes in the PPI network for the up- and downregulated genes, respectively.
Emerging studies have indicated lncRNAs have regulatory roles in various physiological processes and are deregulated in certain human diseases, including prostate cancer (14), breast cancer (15), colon cancer (16,17) and OA (5,18). A number of lncRNAs, including ubiquitin-fold modifier conjugating enzyme 1 (5), HOX transcript antisense RNA (8), maternally expressed 3 (non-protein coding) (19), growth arrest specific 5 (non-protein coding) (7) and imprinted maternally expressed transcript (non-protein coding) (20), were reported to be significantly associated with the progression of OA. However, the functional roles of most lncRNAs in OA remain largely elusive. In the present study, the lncRNA expression patterns in OA were identified from the GSE82107 dataset. A total of 25 up- and 326 downregulated lncRNAs were identified in OA. A total of 6 lncRNAs (EIF1B-AS1, ELOVL2-AS1, LINC00276, LOC101929268, MCM3AP-AS1 and RAD21-AS1) were identified as key lncRNAs, based on an absolute value of the Pearson correlation coefficient of ≥0.8. Combined with co-expression, GO analysis and KEGG analysis, it was identified that the dysregulated lncRNAs were primarily enriched in pathways associated with the MAPK, calcium and insulin signaling pathways. GO analysis revealed that the dysregulated lncRNAs were associated with transcription, signal transduction, interspecies interaction between organisms, ion transport, immune response, cell adhesion, oxidation reduction, development, G-protein-coupled receptor protein signaling pathway and protein amino acid phosphorylation.
In conclusion, in the present study, two public datasets were analyzed to identify differentially expressed lncRNAs and mRNAs in OA. GO and KEGG pathway analysis indicated that DEGs were mainly involved in regulating antigen processing and presentation, interspecies interaction between organisms, immune response, transcription and signal transduction. A series of differently expressed lncRNAs in OA was also identified. To provide novel information on the molecular mechanisms and functional roles of lncRNAs in OA, a co-expression analysis was performed, which key provided several key lncRNAs in OA. The present study provided useful information to explore potential candidate biomarkers for the diagnosis and prognosis of OA patients, and to identify novel drug targets.
Acknowledgements
Not applicable.
Funding
This work was supported by grants from Tongji Hospital of Shanghai (grant nos. KPB1608 and HBRC1607) and the Sailing Program from the Shanghai Science and Technology Commission (grant no. 16YF1410300).
Availability of data and materials
The analyzed data sets generated during the study are available from GEO database. The accession number of dataset GSE48556 is GDS5363 (https://www.ncbi.nlm.nih.gov/sites/GDSbrowser?acc=GDS5363) and GSE82107 for GSE82107 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE82107).
Authors' contributions
YQS made substantial contributions to the conception and design of the study. LC and JCZ made substantial contributions to the acquisition of data and drafted the manuscript. YQZ was involved in the analysis and interpretation of data. ZTR made substantial contributions to the analysis of data and revised the manuscript critically for important intellectual content. All authors read and approved the final manuscript.
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Cho HJ, Morey V, Kang JY, Kim KW, Kim TK. Prevalence and risk factors of spine, Shoulder, Hand, Hip, and Knee osteoarthritis in community-dwelling koreans older than age 65 years. Clin Orthop Relat Res. 2015;473:3307–3314. doi: 10.1007/s11999-015-4450-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greene MA, Loeser RF. Aging-related inflammation in osteoarthritis. Osteoarthritis Cartilage. 2015;23:1966–1971. doi: 10.1016/j.joca.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kong B, Yang T, Chen L, Kuang YQ, Gu JW, Xia X, Cheng L, Zhang JH. Protein-protein interaction network analysis and gene set enrichment analysis in epilepsy patients with brain cancer. J Clin Neurosci. 2014;21:316–319. doi: 10.1016/j.jocn.2013.06.026. [DOI] [PubMed] [Google Scholar]
- 4.Imamura K, Akimitsu N. Long non-coding RNAs involved in immune responses. Front Immunol. 2014;5:573. doi: 10.3389/fimmu.2014.00573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang G, Wu Y, Xu D, Yan X. Long noncoding RNA UFC1 promotes proliferation of chondrocyte in osteoarthritis by acting as a sponge for miR-34a. DNA Cell Biol. 2016;35:691–695. doi: 10.1089/dna.2016.3397. [DOI] [PubMed] [Google Scholar]
- 6.Liu Q, Hu X, Zhang X, Dai L, Duan X, Zhou C, Ao Y. The TMSB4 pseudogene lncRNA functions as a competing endogenous RNA to promote cartilage degradation in human osteoarthritis. Mol Ther. 2016;24:1726–1733. doi: 10.1038/mt.2016.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song J, Ahn C, Chun CH, Jin EJ. A long non-coding RNA, GAS5, plays a critical role in the regulation of miR-21 during osteoarthritis. J Orthop Res. 2014;32:1628–1635. doi: 10.1002/jor.22718. [DOI] [PubMed] [Google Scholar]
- 8.Zhang C, Wang P, Jiang P, Lv Y, Dong C, Dai X, Tan L, Wang Z. Upregulation of lncRNA HOTAIR contributes to IL-1β-induced MMP overexpression and chondrocytes apoptosis in temporomandibular joint osteoarthritis. Gene. 2016;586:248–253. doi: 10.1016/j.gene.2016.04.016. [DOI] [PubMed] [Google Scholar]
- 9.Zhang L, Ma D, Li X, Deng C, Shi Q, You X, Leng X, Li M, Tang F, Zhang F, Li Y. Gene expression profiles of peripheral blood mononuclear cells in primary biliary cirrhosis. Clin Exp Med. 2014;14:409–416. doi: 10.1007/s10238-013-0253-6. [DOI] [PubMed] [Google Scholar]
- 10.Broeren MG, de Vries M, Bennink MB, van Lent PL, van der Kraan PM, Koenders MI, Thurlings RM, van de Loo FA. Functional tissue analysis reveals successful cryopreservation of human osteoarthritic synovium. PLoS One. 2016;11:e0167076. doi: 10.1371/journal.pone.0167076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X, Sun S, Pu JK, Tsang AC, Lee D, Man VO, Lui WM, Wong ST, Leung GK. Long non-coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiol Dis. 2012;48:1–8. doi: 10.1016/j.nbd.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 12.Laulan J, Marteau E, Bacle G. Wrist osteoarthritis. Orthop Traumatol Surg Res. 2015;101(Suppl 1):S1–S9. doi: 10.1016/j.otsr.2014.06.025. [DOI] [PubMed] [Google Scholar]
- 13.Chen W, Liu BY, Zhang X, Zhao XG, Cao G, Dong Z, Zhang SL. Identification of differentially expressed genes in salivary adenoid cystic carcinoma cells associated with metastasis. Arch Med Sci. 2016;12:881–888. doi: 10.5114/aoms.2016.60973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wan X, Huang W, Yang S, Zhang Y, Pu H, Fu F, Huang Y, Wu H, Li T, Li Y. Identification of androgen-responsive lncRNAs as diagnostic and prognostic markers for prostate cancer. Oncotarget. 2016;7:60503–60518. doi: 10.18632/oncotarget.11391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi SJ, Wang LJ, Yu B, Li YH, Jin Y, Bai XZ. LncRNA-ATB promotes trastuzumab resistance and invasion-metastasis cascade in breast cancer. Oncotarget. 2015;6:11652–11663. doi: 10.18632/oncotarget.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yue B, Qiu S, Zhao S, Liu C, Zhang D, Yu F, Peng Z, Yan D. LncRNA-ATB mediated E-cadherin repression promotes the progression of colon cancer and predicts poor prognosis. J Gastroenterol Hepatol. 2016;31:595–603. doi: 10.1111/jgh.13206. [DOI] [PubMed] [Google Scholar]
- 17.He X, Tan X, Wang X, Jin H, Liu L, Ma L, Yu H, Fan Z. C-Myc-activated long noncoding RNA CCAT1 promotes colon cancer cell proliferation and invasion. Tumour Biol. 2014;35:12181–12188. doi: 10.1007/s13277-014-2526-4. [DOI] [PubMed] [Google Scholar]
- 18.Fu M, Huang G, Zhang Z, Liu J, Zhang Z, Huang Z, Yu B, Meng F. Expression profile of long noncoding RNAs in cartilage from knee osteoarthritis patients. Osteoarthritis Cartilage. 2015;23:423–432. doi: 10.1016/j.joca.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 19.Su W, Xie W, Shang Q, Su B. The long noncoding RNA MEG3 is downregulated and inversely associated with vegf levels in osteoarthritis. Biomed Res Int. 2015;2015:356893. doi: 10.1155/2015/356893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steck E, Boeuf S, Gabler J, Werth N, Schnatzer P, Diederichs S, Richter W. Regulation of H19 and its encoded microRNA-675 in osteoarthritis and under anabolic and catabolic in vitro conditions. J Mol Med (Berl) 2012;90:1185–1195. doi: 10.1007/s00109-012-0895-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed data sets generated during the study are available from GEO database. The accession number of dataset GSE48556 is GDS5363 (https://www.ncbi.nlm.nih.gov/sites/GDSbrowser?acc=GDS5363) and GSE82107 for GSE82107 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE82107).