Abstract
Leishmania infantum is one of the causative agents of visceral leishmaniasis (VL), a widespread, life-threatening disease. This parasite is responsible for the majority of human VL cases in Brazil, the Middle East, China, Central Asia and the Mediterranean basin. Its main reservoir are domestic dogs which, similar to human patients, may develop severe visceral disease and die if not treated. The drug allopurinol is used for the long-term maintenance of dogs with canine leishmaniasis. Following our report of allopurinol resistance in treated relapsed dogs, we investigated the mechanisms and markers of resistance to this drug. Whole genome sequencing (WGS) of clinical resistant and susceptible strains, and laboratory induced resistant parasites, was carried out in order to detect genetic changes associated with resistance. Significant gene copy number variation (CNV) was found between resistant and susceptible isolates at several loci, including a locus on chromosome 30 containing the genes LinJ.30.3550 through LinJ.30.3580. A reduction in copy number for LinJ.30.3560, encoding the S-adenosylmethionine synthetase (METK) gene, was found in two resistant clinical isolates and four induced resistant clonal strains. Using quantitative real time PCR, this reduction in METK copy number was also found in three additional resistant clinical isolates. Furthermore, inhibition of S-adenosylmethionine synthetase encoded by the METK gene in allopurinol susceptible strains resulted in increased allopurinol resistance, confirming its role in resistance to allopurinol. In conclusion, this study identified genetic changes associated with L. infantum resistance to allopurinol and the reduction in METK copy number identified may serve as a marker for resistance in dogs, and reduced protein activity correlated with increased allopurinol resistance.
Keywords: Leishmania infantum, Dogs, Allopurinol, Drug resistance, S-adenosylmethionine synthetase
Graphical abstract
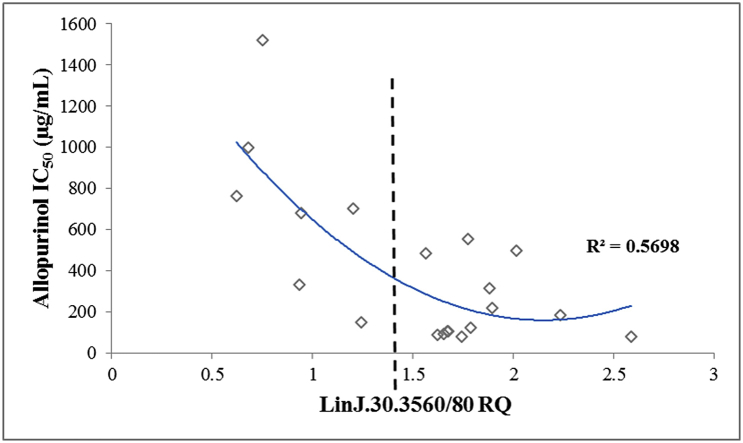
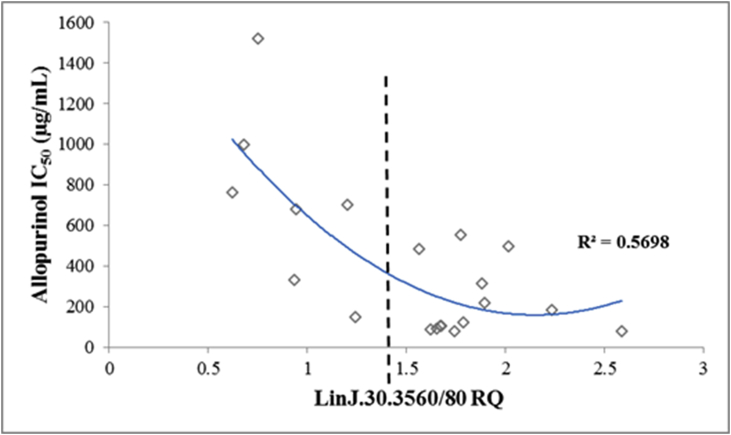
Distribution of relative quantity values (RQ) of gene LinJ.30.3560 from the 20 L. infantum clinical strains in relation to their allopurinol IC50.
Highlights
-
•
Allopurinol resistance was previously described in L. infantum isolated from dogs.
-
•
This study aimed at defining the genetic differences between susceptible and resistant strains.
-
•
Gene and chromosome copy numbers differed between susceptible and resistant L. infantum strains.
-
•
Decrease in METK gene copies was associated with increased allopurinol resistance.
-
•
Inhibition of the enzyme encoded by METK increased allopurinol resistance.
1. Introduction
Visceral leishmaniasis (VL) is a debilitating, life threatening disease affecting hundreds of thousands of people annually (Alvar et al., 2012). According to recent WHO estimations, 556 million people are exposed to VL in 12 high-burden countries, with 300,000 cases and 20,000 deaths worldwide annually. The causative agents of this disease are L. infantum in South America, the Mediterranean basin, and parts of Europe, North Africa, and Asia; and L. donovani in East Africa and the Indian subcontinent (World Health Organization, 2010; Alvar et al., 2012; Gardoni, 2013). A limited number of drugs are available to treat VL, mainly pentavalent antimonials, miltefosine, and amphotericin B in human patients (Freitas-Junior et al., 2012; Monge-Maillo and López-Vélez, 2013). Antimonials, miltefosine and allopurinol are the drugs of choice for dogs, which are the main reservoir for L. infantum (Solano-Gallego et al., 2011). Resistance to antileishmanial drugs is well documented in humans (Croft et al., 2006; Berg et al., 2013; Leprohon et al., 2014), but poorly studied in dogs. We have recently described resistance to allopurinol in L. infantum isolated from treated dogs with disease relapse (Yasur-Landau et al., 2016). Such resistant parasite strains may promote the spread of infection between dogs, and from dogs to humans (Quinnell and Courtenay, 2009; Yasur-Landau et al., 2016).
Allopurinol is a purine analog with anti-leishmanial activity (Pfaller and Marr, 1974). It is rarely prescribed for human VL (Mishra et al., 2007; Freitas-Junior et al., 2012; Monge-Maillo and López-Vélez, 2013), but extensively used for long term control of canine L. infantum infection, either by itself or in combination with meglumine antimoniate or miltefosine (Solano-Gallego et al., 2011). Based on the induction of allopurinol resistance in-vitro in susceptible L. infantum isolates from dogs in a previous study, we concluded that a genetic basis for resistance against this drug exists (Yasur-Landau et al., 2017). In the current study, whole genome sequencing was used to detect differences in the genomes of allopurinol-susceptible and resistant clones, and isolates from dogs. The resistant isolates were obtained from naturally-infected clinically relapsed dogs receiving allopurinol maintenance therapy, as well as strains induced in-vitro under drug pressure. Consistent phenotype-related changes between allopurinol susceptible and resistant strains were found in ploidy and gene copy numbers. Based on these results, we chose to focus on the role of the enzyme S-adenosylmethionine synthetase in allopurinol resistance.
2. Materials and methods
2.1. Allopurinol-susceptible and resistant strains
Twenty L. infantum strains were isolated from dogs showing clinical signs of leishmaniasis (Solano-Gallego et al., 2011; Yasur-Landau et al., 2016), 15 of which were isolated prior to any drug treatment (non-treated group – NT) and 5 after disease relapse during allopurinol treatment (treated relapsed group – TR). Each isolate was identified as L. infantum using a real time PCR assay amplifying a part of the parasite's internal transcribed spacer (ITS-1), followed by DNA sequencing (el Tai et al., 2000). Strains from the TR group were allopurinol resistant, showing an average IC50 value 4 folds higher compared to that of the NT group (Yasur-Landau et al., 2016). Strains NT1-10 as well as TR1-4 were previously described in detail (Yasur-Landau et al., 2016). Two of the susceptible strains (NT4 and NT5) from non-treated dogs were used in the induction of allopurinol resistance (termed NT4.L and NT5.L), achieved by culturing parasites in increasing concentrations of allopurinol over a course of 5 months and producing allopurinol resistant strains, as previously described (Yasur-Landau et al., 2017). Ten susceptible and 10 resistant strains representing 5 different sampling points in each of the two resistance induction experiments were used in the following experiments, as well as 6 clonal strains derived from the induced cultures of NT4.L and NT5.L on days 28, 104 and 28, 86, respectively (Supplementary figure 1). The latter 6 clonal strains, two resistant and one susceptible each from NT4.L and NT5.L were also submitted for whole genome sequencing (WGS), alongside strains NT10 and NT4, TR2 and TR4, each pair representing the susceptible (low IC50) and resistant (high IC50) phenotype, respectively. Information on all strains used in this study is included in supplementary Table 1. Clones were produced using the hanging drop method, as previously described (Evans and Smith, 1986; Yasur-Landau et al., 2016). Allopurinol susceptibility was determined using a viability assay and expressed as IC50 using Prism 5 software (GraphPad Software, San Diego, CA), as previously described (Yasur-Landau et al., 2016).
2.2. DNA extraction and WGS
DNA for WGS was extracted from mid-log phase promastigotes of each of the 10 strains and prepared for whole genome sequencing as previously described (Yasur-Landau et al., 2017). Sequencing was done using 100 bases paired ends reads, on an Ilumina HiSeq2000 platform. Clonal strains NT4.L.r1, NT4.L.r2, NT4.L.s, NT5.L.r1, NT5.L.r2 and NT5.L.s were sequenced at the DNA LandMarks Inc. Laboratory (St.-Jean-sur-Richelieu, Canada), and submitted to GenBank™ under the accession numbers SAMN09079779-84. Isolates MCAN/IL/2012/NT16, MCAN/IL/2009/TR2, MCAN/IL/2011/NT10 and MCAN/IL/2011/TR4 were sequenced at the Wellcome Trust Sanger Institute, UK, and submitted by the Sanger Institute to the European Nucleotide Archive with accession numbers SAMEA1708595-8, respectively.
Raw reads cleaning, trimming and mapping were also done as previously described, using the L. infantum JPCM5 genome, chromosomes 1–36 as reference (European nucleotide archive, BioProject PRJNA12658, FR796433 - FR796468). The soap.coverage (version 2.7.7) of the SOAP Short Oligonucleotide Analysis Package (https://www.ncbi.nlm.nih.gov/pubmed/18227114) was used for depth estimation of each chromosome in each sample.
2.3. Data analysis
Analysis of ploidy (number of copies of whole chromosomes) was done for each sequenced strain essentially as described by Rogers et al. (2011). Briefly, for each strain, the averaged median read depth of chromosomes 30, 34, 35 and 36 was set to 2 assuming a diploid arrangement, and median readings for each of the other chromosomes were normalized accordingly. Information including coding sequences, gene annotations and protein products of L. infantum was obtained from the TriTrypDB website (Aslett et al., 2010). To determine genomic loci amplification (copy number variation, CNV), the Bioconductor cn.MOPS software version 1.6.7 (Klambauer et al., 2012) was used in the R environment with default settings, WL = 2500 and minWidth = 4.
2.4. Quantitative PCR assays of genes LinJ.30.3560 and LinJ.36.0790
Following the analysis of CNV's in the WGS data, two loci showing a CNV with possible connection to allopurinol resistance were further studied. CNV in loci containing genes LinJ.30.3550-3580 and LinJ.36.0790 were further determined using quantitative real time PCR (qPCR) with primers designed to amplify parts of the LinJ.30.3560 and LinJ.36.0790 genes. Primers were designed using the NCBI/Primer-BLAST website (http://www.ncbi-.nlm.nih.gov/tools/primer-blast/). DNA of clones NT5.L.r1 and NT4.L.s was used as reference (RQ = 1) for LinJ.30.3560 and LinJ.36.0790, respectively. Forward and reverse primers for the GAPDH control gene (do Monte-Neto et al., 2011) were used to amplify a 226 base fragment of LinJ.30.2990 and LinJ.30.3000. According to the L. infantum JPCM5 reference genome there are 2 copies of GAPDH, and no evidence for a change in its copy number was found in the WGS data. Therefore, GAPDH was used as a reference gene for quantitation. Primer sequences are presented in supplementary Table 2. PCR reactions were done in a total volume of 20 μL each, including; 10 μL Fast SYBR Green Master Mix (x2) (Applied Biosystems, Foster City, CA), 2 ng DNA, ultra-pure water and a final concentration of 250 nM of the forward and reverse primers. The StepOnePlus real-time PCR thermal cycler (Applied Biosystems, Foster City, CA) was used with the following thermal profile; initial denaturation for 20 s at 95 °C, followed by denaturation for 3 s at 95 °C and annealing for 30 s at 59 °C for 40 cycles. Amplicons were subsequently subjected to a melt step with the temperature raised to 95 °C for 15 s and then lowered to 60 °C for 1 min. The temperature was then raised to 95 °C at a rate of 0.3 °C per second. Amplification, melt profiles and amplicon relative quantity (RQ) based on analysis of the ΔΔCT values, were analyzed using the StepOne V2.2.2 software (Applied Biosystems, Foster City, CA). Each reaction was repeated twice and an average RQ was calculated.
2.5. S-adenosylmethionine synthetase inhibition experiments
S-adenosylmethionine synthetase is an enzyme encoded by the two copies (LinJ.30.3560/80) of the METK gene, present in one of the two loci showing possible connection to allopurinol resistance. Its effect on allopurinol resistance was studied using an enzyme inhibition assay. Cycloleucine (1-Aminocyclopentanecarboxylic acid) is an inhibitor of the enzyme S-adenosylmethionine synthetase in bacteria, yeast, rat and human cells in culture (Lombardini and Talalay, 1970; Zhuge and Cederbaum, 2007; Jani et al., 2009). The effect of S-adenosylmethionine synthetase inhibition on the parasite susceptibility to allopurinol was determined in four strains with different initial allopurinol IC50. For each strain, 3 separate allopurinol IC50 tests were conducted as described above, but in addition to supplementation of the parasite cultures with allopurinol, cycloleucine was added at 12.5, 25, or 50 mg/mL. Cycloleucine was added at the start of each assay to all the test wells. Each set of experiments was repeated twice, and for each strain allopurinol IC50 was plotted against the various cycloleucine concentrations. In order to rule out the possibility that results seen in these experiments were solely attributed to a generic inhibitory effect of cycloleucine on parasite growth, regardless of the specific effect on METK, experiments were also repeated using amphotericin B at concentrations of 0, 0.125, 0.25 and 0.5 μM instead of cycloleucine. Allopurinol, cycloleucine and amphotericin B were purchased from Sigma-Aldrich, St. Louis, MO.
2.6. Statistical analysis
Allopurinol IC50 values of L. infantum strains were compared using the Tukey HSD test. Ploidy patterns of sequenced clones and strains were analyzed by measuring the Pearson correlation values followed by hierarchical clustering by r-values. Relative quantity (RQ) of LinJ.30.3560/30.3580 in parasite strains was compared between two categories (RQ<1.5 and RQ>1.5) using the Mann-Whitney Test. For all other analyses, a two-tailed t-test was applied. Analysis was done using the JMP12 software (SAS Institute Inc. Cary, NC), P values smaller than 0.05 were considered significant.
3. Results
3.1. Whole genome sequencing
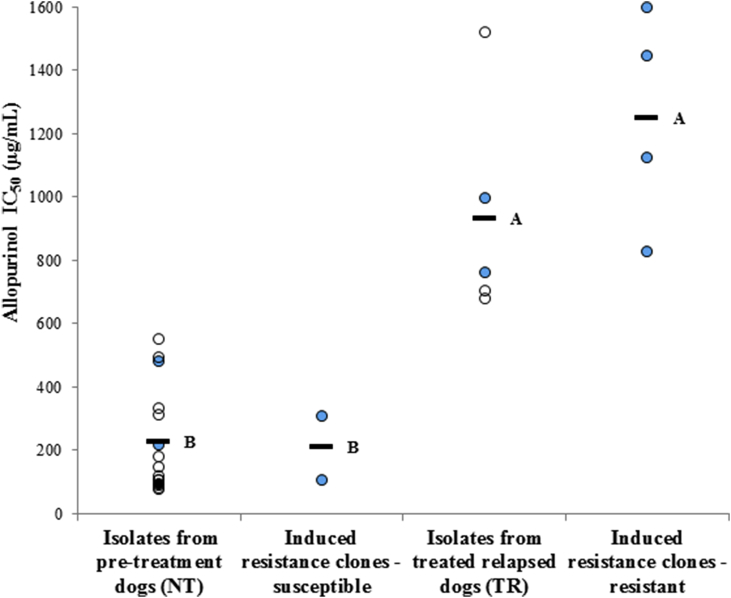
Six clonal strains, three from the NT4.L line and three from the NT5.L line were chosen for WGS. Each triplet included one clone from an initial stage during the induction of resistance with low/intermediate IC50, and two clones with high IC50, from a later stage of the resistance induction (Supplementary figure 1). Thus, parasites in each triplet possessed a similar genetic background and were exposed to comparable conditions, differing only by the time period and intensity of allopurinol pressure applied. Together with the four clinical isolate strains that were also sequenced, these strains were chosen to phenotypically represent low, medium and high allopurinol IC50 values compared to the 20 naturally-infected dog strains evaluated (Fig. 1).
Fig. 1.
Allopurinol susceptibilities of naturally-infected dog L. infantum strains (NT and TR groups, total n = 20), susceptible and resistant clonal strains (n = 2 and n = 4, respectively) used in this study. Bars indicate group IC50 averages; 226 and 932 μg/mL for clinical isolate strains (obtained from dogs pre-treatment - NT and following relapse - TR, respectively); 208 and 1249 μg/mL (susceptible and resistant clones, respectively). Values indicated by distinct letters differ significantly (Tukey HSD test, P < 0.05). Filled blue circles indicate strains and clones for which whole genome sequencing was done. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
WGS resulted in cleaned paired-end reads of 14-38 × 106 reads per sample, high mapped rate of 98.13–98.82% and coverage of × 85- × 218. Total number of raw reads obtained by the sequencing of each isolate, as well as number of cleaned reads, percent of reads mapped to the reference genome, and average median read depth are described in supplementary Table 3. WGS data was used to detect changes in copy numbers of whole chromosomes and specific loci.
3.2. Whole chromosome copy number variation
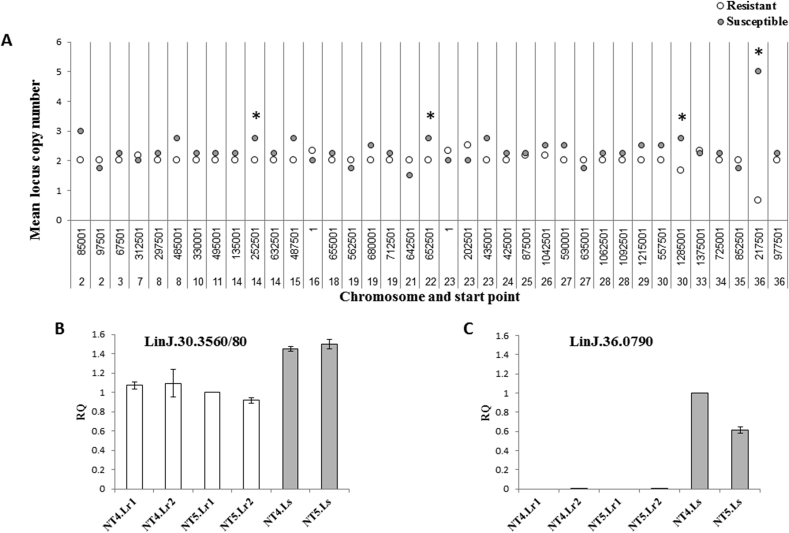
Complete results of ploidy estimation for the sequenced clones are presented in supplementary Table 4. Polyploidy was seen in all clones in chromosomes 8, 9 and 31, while diploidy was conserved in chromosomes 19, 21, 24, 27, 28, 30, 34, 35 and 36 (Fig. 2A). Analysis of hierarchical clustering of the ploidy patterns for all 36 chromosomes showed that the 4 resistant clones grouped closely together and separated from all the remaining strains and clones analyzed by WGS (Fig. 2B). For chromosomes 8, 15 and 33, resistant clones displayed a higher ploidy compared to their respective susceptible clones (T-test, P = 0.006, P = 0.02, P < 0.001, respectively). However, no such differences were found when comparing the resistant and susceptible clinical strains.
Fig. 2.
Analysis of whole chromosome copy number variation for the 10 sequenced L. infantum strains. A. Estimated normalized ploidy of all chromosomes for all sequenced strains and clones. Different colors represent ploidy levels (copy number of respective chromosomes). B. Hierarchical clustering of correlations in ploidy patterns of all chromosomes for all tested clones and strains (Pearson correlation, colors represent r values). The highest correlation was found between the 4 resistant clonal strains. Pearson correlations and hierarchical cluster were performed using R scripts using the heatmap.2 function of the gplots package (Warnes GR, Bolker B, Bonebakker L, Gentleman R, Liaw WHA, Lumley T et al., gplots: various R programming tools for plotting data. [Online]. (2012). Available at: http://CRAN.R-project.org/package=gplots [15 April 2015].). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. Genomic loci copy number variation analysis
The analysis preformed on the WGS data of the 10 sequenced strains using cn.MOPS produced 37 unique loci in which CNV were detected (supplementary Table 5). An increase in the number of copies (CNV>2) was the most prevalent variation and found mainly in susceptible strains, although reduction (CNV<2) or even an apparent deletion of a locus (CNV = 0) were also found in various strains during the analysis. When comparing the mean copy numbers for detected CNV's of the resistant strains to that of susceptible ones, several CNV's differed significantly (Fig. 3A). Two of these loci, containing genes LinJ.30.3550 through LinJ.30.3580 (chromosome 30, starting point 1285001) and LinJ.36.0790 (chromosome 36, starting point 217501), were further studied. For the CNV containing genes LinJ.30.3550-3580, cn.MOPS analysis demonstrated a 1.5–3 folds reduction in copy numbers in both resistant clones and resistant clinical strains compared to their allopurinol susceptible counterparts. For the CNV containing gene LinJ.36.0790, a complete deletion of the loci was found in all resistant clonal strains although this was not seen in the resistant clinical strains. Quantitative PCR done on the 6 clonal strains showed that for gene LinJ.30.3560 resistant clones had an average relative quantity (RQ) value of 1.02 ± 0.08 while the average for susceptible ones was 1.48 ± 0.04. (t-test, P = 0.017). These numbers are equivalent to 2 and 3 copies respectively of the LinJ.30.3560 gene per diploid genome, which match the numbers found by the cn.MOPS analysis and more importantly, suggest a deletion of one copy of this locus in the resistant clones. For gene LinJ.36.0790, the average RQ value of 0.001 ± 0.001 was found for the resistant clones compared to 0.81 ± 0.27 in susceptible clones (t-test, P = 0.024), thus confirming the full deletion of this locus as first seen in the cn.MOPS analysis. Comparison of RQ values for individual clones in the two loci is presented in Fig. 3B and C.
Fig. 3.
Analysis of genomic loci copy number variations (CNV's). A. Comparison of mean CNV number of the 10 sequenced resistant or susceptible strains at each locus in which CNV was detected by cn.MOPS. Each bar represents a different locus, recognized by the chromosome it is in and its start position within the chromosome. * Significant difference in means between resistant and sensitive strains (t-test, P = 0.05, 0.05, 0.01 and 0.02 for loci in chromosome 14, 22, 30 and 36, respectively). B, C. Quantitative real time PCR results of genes LinJ.30.3560 and LinJ.36.0790 for the 6 clonal strains. Results expressed as relative quantity values (RQ), verifying the cn.MOPS analysis results. Empty bars – resistant clones, Filled bars – susceptible clones. NT5.L.r1 was used as reference (RQ = 1) for LinJ.30.3560, and NT4.L.s for LinJ.36.0790.
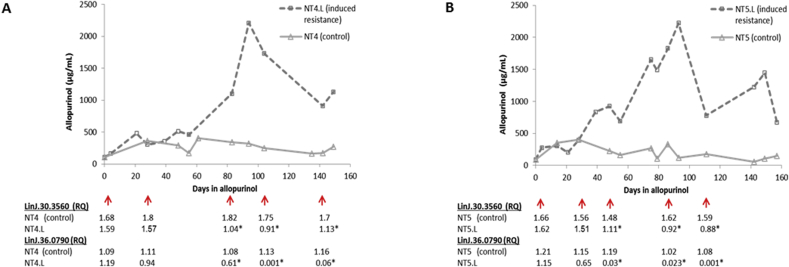
Subsequently, cryopreserved parasite strain samples from five different time points during the in vitro resistance induction experiments (NT4.L and NT5.L) were revived and tested for their allopurinol IC50 and RQ values of the two genes of interest. Firstly, for both LinJ.30.3560 and Linj.36.0790, decrease in RQ values accompanied the increase in IC50 in the induced resistant parasites (Fig. 4). Secondly, in these induced resistant strains, RQ values for the LinJ.30.3560 gene were 1.5 times lower or more compared to values seen for allopurinol susceptible control strains. This number translates to a deletion of one copy of this gene in the resistant strains compared to susceptible ones (in a diploid genome), and matches what was previously seen in the WGS data. Lastly, qPCR was performed for the 2 genes on all 20 clinical strains (supplementary Table 6). A moderate negative linear (R2 = 0.489) and quadratic (R2 = 0.57) correlation was found between RQ and IC50 values of each strain for LinJ.30.3560 (Fig. 5). However, when the strains were divided into those with RQ < 1.5 or ≥1.5 (equivalent to <3 or ≥3 gene copies), the former group presented a significantly higher IC50 values (Mann-Whitney Test, P < 0.001), and only two clinical strains presented a susceptible phenotype together with an RQ value below 1.5. Therefore, 3 copies (RQ ≥ 1.5) of the LinJ.30.3560 gene resulted in a susceptible phenotype in induced clones, induced isolates and the clinical isolates, while less than 3 copies (RQ < 1.5) resulted in resistant parasites. For gene Linj.36.0790, no correlation between IC50 and RQ was found among the 20 clinical isolates (R2 < 0.15), nor was any significant difference found in RQ between the 2 phenotypes.
Fig. 4.
Change in copy numbers of genes LinJ.30.3560 and LinJ.36.0790 relative to allopurinol IC50 during induction of resistance in two clinical L. infantum strains (Yasur-Landau et al., 2017). Cryopreserved promastigotes from several time points in this process were revived and tested. NT4.L and NT5.L are the strains under drug pressure, while NT4 and NT5 are non-treated controls of the same strains. Red arrows indicate time points at which strains were tested by qPCR. Results expressed as relative quantity values (RQ), for each of the tested genes. * Significant difference between induced isolate and control for respective time points (t-test, P < 0.05).
Fig. 5.
Distribution of relative quantity values (RQ) of gene LinJ.30.3560 from the 20 clinical strains in relation to their allopurinol IC50. Negative quadratic (R2 = 0.57) correlation was found between RQ and IC50 values of each strain for LinJ.30.3560. Moreover, average IC50 of strains with RQ < 1.5 was significantly higher compared to strains showing RQ > 1.5 (Mann-Whitney Test, P < 0.001).
3.4. S-adenosylmethionine synthetase inhibition
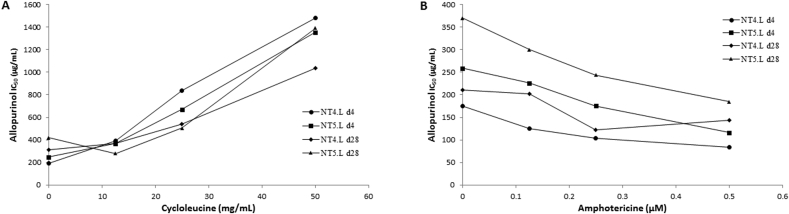
S-adenosylmethionine synthetase (METK), encoded by LinJ.30.3560, is a highly conserved, key enzyme in the synthesis of S-adenosylmethionine (AdoMet), which is a cofactor involved in various biological functions (Reguera et al., 2007; Grillo and Colombatto, 2008). Cycloleucine is a cyclic amino acid which inhibits METK by acting as a l-methionine analog (Lombardini and Talalay, 1970). Allopurinol, on the other hand, is a purine nucleobase analog (Pfaller and Marr, 1974; Balaña-Fouce et al., 1998), and is unlikely to inhibit S-adenosylmethionine synthetase by the same mechanism as cycloleucine. Since lower METK gene copy number was found to be associated with resistance to allopurinol, it was interesting to see whether inhibition of METK activity by cycloleucine would result in increased allopurinol IC50 values in drug sensitive strains. Indeed, a strong positive correlation in susceptible isolates, between the added cycloleucine concentrations and allopurinol IC50 values was found for 4 strains, with R2 values ranging between 0.8 and 0.98 (Fig. 6A). In contrast, when amphotericin B was used in place of cycloleucine, a negative correlation was found with R2 ranging between 0.56 and 0.98 (Fig. 6B).
Fig. 6.
Effect of cycloleucine (A) or amphotericin B (B) on allopurinol IC50 of susceptible L. infantum strains. Increase in cycloleucine concentrations resulted in higher allopurinol IC50, while amphotericin B caused an opposite effect.
4. Discussion
Domestic dogs are considered to be the main reservoir for L. infantum (Alvar et al., 2012; Ready, 2014) and allopurinol is currently the drug of choice for long-term control of this infection in dogs (Solano-Gallego et al., 2011). Thus, widespread resistance of L. infantum to allopurinol may have serious veterinary and public health implications. Following our report of allopurinol resistance in naturally-infected dogs (Yasur-Landau et al., 2016) and the ability to experimentally induce allopurinol resistance in L. infantum in-vitro (Yasur-Landau et al., 2017), we proceeded to look for genetic changes in resistant parasite strains, as a step in the search for markers and mechanisms of resistance to this drug.
By investigating both experimentally-induced and naturally-occurring resistant parasite strains we aimed to increase the likelihood of detecting resistance related modifications, existing or evolving naturally in these strains. Differences were first detected between induced-resistant clonal strains with varying drug susceptibilities, and then examined in clinical strains isolated from dogs.
Aneuploidy is a well-recognized phenomenon in cultured Leishmania spp., and ploidy patterns of the strains studied here, regardless of their allopurinol susceptibility and origin, indeed reflected the genus's “mosaic aneuploidy” (Mannaert et al., 2012; Sterkers et al., 2012). Moreover, chromosome duplication was also shown to be associated with drug resistance in Leishmania species from humans (Ubeda et al., 2008; Leprohon et al., 2009; Downing et al., 2011; Brotherton et al., 2013). In the present study, resistant clonal strains were distinguishable in their ploidy pattern from susceptible ones, including their own allopurinol-susceptible close ancestry, suggesting a role for chromosome duplication in resistance. More specifically, an increase of 1.5–2 folds in copy numbers of chromosomes 8, 15 and 33 was found in resistant clonal isolates compared to the respective susceptible clonal isolates. Since a large cohort of genes are duplicated in each event of whole chromosome duplication, it is a challenging task to determine which are directly involved in adaptation to the stressor, and which are merely “duplicated by proxy”. The fact that these changes were found only in the in-vitro induced resistance strains, subjected to drug pressure for a period of 5 months, suggests that chromosome duplication may not be a long-term adaptation to stressors, perhaps due to its high energetic or fitness cost, as suggested previously (Ubeda et al., 2008).
Specific loci CNV's have also been suggested to possess adaptive value and take part in drug resistance formation in Leishmania (Ubeda et al., 2008; Leprohon et al., 2009; Downing et al., 2011; Brotherton et al., 2013; Ritt et al., 2013). Of the 37 unique CNV's detected, we chose to focus on two shared by several strains, and therefore less likely to result from a random event. The first CNV was a deletion of the locus containing the LinJ.36.0790 gene, a hypothetical conserved protein, containing zinc-finger domains (Aslett et al., 2010). The deletion was consistent among the 4 resistant clonal strains, highlighted by the estimated high number of copies in the susceptible clonal strains. Since this deletion was not shared by the 20 clinical isolates, this finding may represent a unique adaptation to in-vitro selection under drug pressure when strains were cultured as promastigotes. In contrast, the decreased copy number of the locus containing genes LinJ.30.3550 through LinJ.30.3580 was consistent across sequencing data of all in vitro clonal as well as clinical resistant strains. By following this CNV during induction of drug resistance in two separate experiments, a reduction in copy number could be seen over a time frame of several months, persisting for at least several months, and matching the shift to a resistant phenotype. A lower copy number of this locus was also found in additional resistant clinical strains by qPCR. This locus contains two genes, each with two copies, LinJ.30.3550/70 and METK (LinJ.30.3560/80). Arrangement, transcriptional regulation and expression have been studied for this gene cluster in L. infantum by García-Estrada et al. (2003, 2007, 2008), establishing among other findings that both genes are expressed throughout the parasite's life cycle, with possible stage-specific post transcriptional regulation of their level. The METK gene encodes S-adenosylmethionine synthetase, a key enzyme in the synthesis of S-adenosylmethionine (AdoMet), a molecule involved in critical cellular functions such as trans-methylation, trans-sulphuration and polyamine synthesis in L. infantum (Reguera et al., 2002, 2007). The LinJ.30.3550/70 gene encodes the LORIEN protein. The role of this protein is uncertain, but it contains a SP-RING/Miz zinc-finger motif, suggesting that it is related to the Small Ubiquitin related Modifier (SUMO) conjugation pathway, promoting post translational regulation of target proteins (García-Estrada et al., 2008).
Focusing on this locus as a possible key player in allopurinol resistance of L. infantum, we performed experiments with the S-adenosylmethionine synthetase inhibitor cycloleucine to evaluate its influence on the allopurinol IC50 of L. infantum in culture. Increasing concentrations of cycloleucine resulted in higher allopurinol IC50 for all 4 susceptible strains tested. This finding is in line with the CNV results, suggesting that decreased levels of functional S-adenosylmethionine synthetase reduce allopurinol's inhibitory effect on the parasite. Amphotericin B, which exerts an inhibitory effect on Leishmania through its influence on membrane sterols (Saha et al., 1986), had an opposite effect on allopurinol IC50. The latter finding, suggests that results found with cycloleucine were not caused by a generic response to additional stress or to a mere growth inhibition by a second drug used in combination with allopurinol. Overexpression of S-adenosylmethionine in a resistant isolate together with a corresponding reduction in allopurinol IC50, will provide strong supporting evidence regarding the role of this enzyme in allopurinol susceptibility. However, our attempts to overexpress this gene using a Leishmania expression vector were so far unsuccessful, and confirmation by this technique awaits further studies. Together, these results demonstrate that the enzyme S-adenosylmethionine synthetase has an important role in allopurinol resistance, and raise new ideas concerning the mechanism by which allopurinol acts against this parasite. Since allopurinol can be incorporated into energetic nucleotides such as ATP (Balaña-Fouce et al., 1998; Mishra et al., 2007) it may be that such allopurinol containing nucleotides inhibit S-adenosylmethionine synthetase or are utilized by it, producing faulty products, which in turn inhibit the parasite's growth. Down-regulation of S-adenosylmethionine synthetase in resistant strains may reduce the levels of such faulty products. Increased transport of exogenous AdoMet via an alternative route that has been identified in Leishmania might compensate for the lower levels of de novo synthesis (Dridi et al., 2010). Another possible explanation is that allopurinol inhibits enzymes involved in the metabolism of AdoMet working downstream to S-adenosylmethionine synthetase, and causing an increase in AdoMet levels with deleterious effect on the parasite via hypermethylation. A decrease in S-adenosylmethionine synthetase levels or activity may counter this effect, thereby causing drug resistance. A similar mechanism was described for S-adenosylmethionine synthetase and the drug dl-α-difluoromethylornithine (DFMO). DFMO caused a sharp increase in AdoMet levels in susceptible Trypanosoma brucei rhodesiense isolates, while in resistant ones both S-adenosylmethionine synthetase activity and AdoMet levels were much lower (Bacchi et al., 1993). The fact that two susceptible clinical strains in our study also demonstrated a reduced copy number for the METK gene could be because resistance may be multifactorial and associated with other mechanisms. Even though decrease in the METK gene copy number was the most prominent change detected, additional genetic modulations may also be involved. Interestingly, we have not found any significant evidence for resistance-related genetic changes in the phosphoribosyl transferases genes, even though allopurinol is thought to be an inhibitor of these enzymes (Mishra et al., 2007), nor in any nucleobase or drug transporter genes.
In conclusion, although this study identified several chromosomes and genes with possible involvement in allopurinol resistance of L. infantum, the most remarkable finding was a decrease in the METK gene copy numbers in resistant parasite strains, suggesting that allopurinol resistance does not arise exclusively via effects on purine metabolism as might have been expected based on its presumed mode of action, but through impact on AdoMet related pathways. Focusing research on protein production levels and the activity of identified targets in resistant and susceptible parasite strains could help to determine their significance in allopurinol resistance in future studies.
Ethical statement
The animal care protocol used in this study was approved by the Hebrew University's Institutional Animal Care and Use Committee (IACUC); approval no. MD-08-11476-2, following the USA NIH guidelines.
Acknowledgments
We would like to thank Dr. James Cotton and Mrs. Mandy Sanders of the parasite genomics group, Sanger Institute, UK, for generating and submitting the whole genome sequencing data for 4 of the L. infantum strains (TR4, TR2, NT16, and NT10). We would also like to thank Dr. Christine Petersen from the University of Iowa, USA, for facilitating this cooperation. This study was partially supported by the Hebrew University Research Center for Sustainable Animal Husbandry and Health [grant no. 0366196]. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ijpddr.2018.08.002.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
Note: Nucleotide sequence data reported in this paper are available in GenBank™ under the accession numbers SAMN09079779-84 (isolates NT4.L.r1, NT4.L.r2, NT4.L.s, NT5.L.r1, NT5.L.r2, NT5.L.s) and EMBL under the accession numbers SAMEA1708595-8 (isolates NT16, TR2, NT10 and TR4).
References
- Alvar J., Vélez I.D., Bern C., Herrero M., Desjeux P., Cano J., Jannin J., den Boer M., WHO Leishmaniasis Control Team Leishmaniasis worldwide and global estimates of its incidence. PLoS One. 2012;7 doi: 10.1371/journal.pone.0035671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslett M., Aurrecoechea C., Berriman M., Brestelli J., Brunk B.P., Carrington M., Depledge D.P., Fischer S., Gajria B., Gao X., Gardner M.J., Gingle A., Grant G., Harb O.S., Heiges M., Hertz-Fowler C., Houston R., Innamorato F., Iodice J., Kissinger J.C., Kraemer E., Li W., Logan F.J., Miller J.A., Mitra S., Myler P.J., Nayak V., Pennington C., Phan I., Pinney D.F., Ramasamy G., Rogers M.B., Roos D.S., Ross C., Sivam D., Smith D.F., Srinivasamoorthy G., Stoeckert C.J., Jr., Subramanian S., Thibodeau R., Tivey A., Treatman C., Velarde G., Wang H. TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 2010;38(Database issue):D457–D462. doi: 10.1093/nar/gkp851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacchi C.J., Garofalo J., Ciminelli M., Rattendi D., Goldberg B., McCann P.P., Yarlett N. Resistance to DL-alpha-difluoromethylornithine by clinical isolates of Trypanosoma brucei rhodesiense. Role of S-adenosylmethionine. Biochem. Pharmacol. 1993;46(3):471–481. doi: 10.1016/0006-2952(93)90524-z. 3. [DOI] [PubMed] [Google Scholar]
- Balaña-Fouce R., Requera R.M., Cubría J.C., Ordóñez D. The pharmacology of leishmaniasis. Gen. Pharmacol. 1998;30:435–443. doi: 10.1016/s0306-3623(97)00268-1. [DOI] [PubMed] [Google Scholar]
- Berg M., Mannaert A., Vanaerschot M., Van Der Auwera G., Dujardin J.C. (Post-) Genomic approaches to tackle drug resistance in Leishmania. Parasitology. 2013;140(12):1492–1505. doi: 10.1017/S0031182013000140. [DOI] [PubMed] [Google Scholar]
- Brotherton M.C., Bourassa S., Leprohon P., Légaré D., Poirier G.G., Droit A., Ouellette M. Proteomic and genomic analyses of antimony resistant Leishmania infantum mutant. PLoS One. 2013;8(11) doi: 10.1371/journal.pone.0081899. 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft S.L., Sundar S., Fairlamb A.H. Drug resistance in leishmaniasis. Clin. Microbiol. Rev. 2006;19:111–126. doi: 10.1128/CMR.19.1.111-126.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- do Monte-Neto R.L., Coelho A.C., Raymond F., Légaré D., Corbeil J., Melo M.N., Frézard F., Ouellette M. Gene expression profiling and molecular characterization of antimony resistance in Leishmania amazonensis. PLoS Neglected Trop. Dis. 2011;5:e1167. doi: 10.1371/journal.pntd.0001167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downing T., Imamura H., Decuypere S., Clark T.G., Coombs G.H., Cotton J.A., Hilley J.D., de Doncker S., Maes I., Mottram J.C., Quail M.A., Rijal S., Sanders M., Schönian G., Stark O., Sundar S., Vanaerschot M., Hertz-Fowler C., Dujardin J.C., Berriman M. Whole genome sequencing of multiple Leishmania donovani clinical isolates provides insights into population structure and mechanisms of drug resistance. Genome Res. 2011;21(12):2143–2156. doi: 10.1101/gr.123430.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dridi L., Ahmed Ouameur A., Ouellette M. High affinity S-Adenosylmethionine plasma membrane transporter of Leishmania is a member of the folate biopterin transporter (FBT) family. J. Biol. Chem. 2010;285(26):19767–19775. doi: 10.1074/jbc.M110.114520. 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el Tai N.O., Osman O.F., el Fari M., Presber W., Schönian G. Genetic heterogeneity of ribosomal internal transcribed spacer in clinical samples of Leishmania donovani spotted on filter paper as revealed by single-strand conformation polymorphisms and sequencing. Trans. R. Soc. Trop. Med. Hyg. 2000;94:575–579. doi: 10.1016/s0035-9203(00)90093-2. [DOI] [PubMed] [Google Scholar]
- Evans D.A., Smith V. A simple method for cloning leishmanial promastigotes. Z. Parasitenkd. 1986;72:573–576. doi: 10.1007/BF00925476. [DOI] [PubMed] [Google Scholar]
- Freitas-Junior L.H., Chatelain E., Kim H.A., Siqueira-Neto J.L. Visceral leishmaniasis treatment: what do we have, what do we need and how to deliver it? Int J Parasitol Drugs Drug Resist. 2012;28(2):11–19. doi: 10.1016/j.ijpddr.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Estrada C., Reguera R.M., Villa H., Requena J.M., Müller S., Pérez-Pertejo Y., Balaña-Fouce R., Ordóñez D. Identification of a gene in Leishmania infantum encoding a protein that contains a SP-RING/MIZ zinc finger domain. Biochim. Biophys. Acta. 2003;1629(1–3):44–52. doi: 10.1016/j.bbaexp.2003.07.001. 1. [DOI] [PubMed] [Google Scholar]
- García-Estrada C., Pérez-Pertejo Y., Ordóñez D., Balaña-Fouce R., Reguera R.M. Analysis of genetic elements regulating the methionine adenosyltransferase gene in Leishmania infantum. Gene. 2007;389(2):163–173. doi: 10.1016/j.gene.2006.11.003. 15. [DOI] [PubMed] [Google Scholar]
- García-Estrada C., Pérez-Pertejo Y., Ordóñez D., Balaña-Fouce R., Reguera R.M. Characterization of the 5' region of the Leishmania infantum LORIEN/MAT2 gene cluster and role of LORIEN flanking regions in post-transcriptional regulation. Biochimie. 2008;90(9):1325–1336. doi: 10.1016/j.biochi.2008.03.007. [DOI] [PubMed] [Google Scholar]
- Gardoni L. Epidemiological surveillance of leishmaniasis in the European Union: operational and research challenges. Euro Surveill. 2013;18:20539. doi: 10.2807/1560-7917.es2013.18.30.20539. [DOI] [PubMed] [Google Scholar]
- Grillo M.A., Colombatto S. S-adenosylmethionine and its products. Amino Acids. 2008;34(2):187–193. doi: 10.1007/s00726-007-0500-9. [DOI] [PubMed] [Google Scholar]
- Jani T.S., Gobejishvili L., Hote P.T., Barve A.S., Joshi-Barve S., Kharebava G., Suttles J., Chen T., McClain C.J., Barve S. Inhibition of methionine adenosyltransferase II induces FasL expression, Fas-DISC formation and caspase-8-dependent apoptotic death in T leukemic cells. Cell Res. 2009;19:358–369. doi: 10.1038/cr.2008.314. [DOI] [PubMed] [Google Scholar]
- Klambauer G., Schwarzbauer K., Mayr A., Clevert D.A., Mitterecker A., Bodenhofer U., Hochreiter S. cn.MOPS: mixture of Poissons for discovering copy number variations in next-generation sequencing data with a low false discovery rate. Nucleic Acids Res. 2012;40:e69. doi: 10.1093/nar/gks003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leprohon P., Légaré D., Raymond F., Madore E., Hardiman G., Corbeil J., Ouellette M. Gene expression modulation is associated with gene amplification, supernumerary chromosomes and chromosome loss in antimony-resistant Leishmania infantum. Nucleic Acids Res. 2009;37(5):1387–1399. doi: 10.1093/nar/gkn1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leprohon P., Fernandez-Prada C., Gazanion É., Monte-Neto R., Ouellette M. Drug resistance analysis by next generation sequencing in Leishmania. Int J Parasitol Drugs Drug Resist. 2014;22(5):26–35. doi: 10.1016/j.ijpddr.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardini J.B., Talalay P. Formation, functions and regulatory importance of S-adenosyl-L-methionine. Adv. Enzym. Regul. 1970;9:349–384. doi: 10.1016/s0065-2571(71)80054-7. [DOI] [PubMed] [Google Scholar]
- Mannaert A., Downing T., Imamura H., Dujardin J.C. Adaptive mechanisms in pathogens: universal aneuploidy in Leishmania. Trends Parasitol. 2012;28(9):370–376. doi: 10.1016/j.pt.2012.06.003. [DOI] [PubMed] [Google Scholar]
- Mishra J., Saxena A., Singh S. Chemotherapy of leishmaniasis: past, present and future. Curr. Med. Chem. 2007;14:1153–1169. doi: 10.2174/092986707780362862. [DOI] [PubMed] [Google Scholar]
- Monge-Maillo B., López-Vélez R.E. Therapeutic options for visceral leishmaniasis. Drugs. 2013;73:1863–1888. doi: 10.1007/s40265-013-0133-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaller M.A., Marr J.J. Antileishmanial effect of allopurinol. Antimicrob. Agents Chemother. 1974;5:469–472. doi: 10.1128/aac.5.5.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinnell R.J., Courtenay O. Transmission, reservoir hosts and control of zoonotic visceral leishmaniasis. Parasitology. 2009;136:1915–1934. doi: 10.1017/S0031182009991156. [DOI] [PubMed] [Google Scholar]
- Ready P.D. Epidemiology of visceral leishmaniasis. Clin. Epidemiol. 2014;6:147–154. doi: 10.2147/CLEP.S44267. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguera R.M., Balaña-Fouce R., Pérez-Pertejo Y., Fernández F.J., García-Estrada C., Cubría J.C., Ordóñez C., Ordóñez D. Cloning expression and characterization of methionine adenosyltransferase in Leishmania infantum promastigotes. J. Biol. Chem. 2002;277(5):3158–3167. doi: 10.1074/jbc.M105512200. 2002 Feb 1. [DOI] [PubMed] [Google Scholar]
- Reguera R.M., Redondo C.M., Pérez-Pertejo Y., Balaña-Fouce R. S-Adenosylmethionine in protozoan parasites: functions, synthesis and regulation. Mol. Biochem. Parasitol. 2007;152(1):1–10. doi: 10.1016/j.molbiopara.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Ritt J.F., Raymond F., Leprohon P., Légaré D., Corbeil J., Ouellette M. Gene amplification and point mutations in pyrimidine metabolic genes in 5-fluorouracil resistant Leishmania infantum. PLoS Neglected Trop. Dis. 2013;7(11) doi: 10.1371/journal.pntd.0002564. e2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers M.B., Hilley J.D., Dickens N.J., Wilkes J., Bates P.A., Depledge D.P., Harris D., Her Y., Herzyk P., Imamura H., Otto T.D., Sanders M., Seeger K., Dujardin J.C., Berriman M., Smith D.F., Hertz-Fowler C., Mottram J.C. Chromosome and gene copy number variation allow major structural change between species and strains of Leishmania. Genome Res. 2011;21:2129–2142. doi: 10.1101/gr.122945.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A.K., Mukherjee T., Bhaduri A. Mechanism of action of amphotericin B on Leishmania donovani promastigotes. Mol. Biochem. Parasitol. 1986;19(3):195–200. doi: 10.1016/0166-6851(86)90001-0. [DOI] [PubMed] [Google Scholar]
- Solano-Gallego L., Miró G., Koutinas A., Cardoso L., Pennisi M.G., Ferrer L., The LeishVet Group LeishVet guidelines for the practical management of canine leishmaniosis. Parasites Vectors. 2011;4:86. doi: 10.1186/1756-3305-4-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterkers Y., Lachaud L., Bourgeois N., Crobu L., Bastien P., Pagès M. Novel insights into genome plasticity in Eukaryotes: mosaic aneuploidy in Leishmania. Mol. Microbiol. 2012;86(1):15–23. doi: 10.1111/j.1365-2958.2012.08185.x. [DOI] [PubMed] [Google Scholar]
- Ubeda J.M., Légaré D., Raymond F., Ouameur A.A., Boisvert S., Rigault P., Corbeil J., Tremblay M.J., Olivier M., Papadopoulou B., Ouellette M. Modulation of gene expression in drug resistant Leishmania is associated with gene amplification, gene deletion and chromosome aneuploidy. Genome Biol. 2008;9:R11. doi: 10.1186/gb-2008-9-7-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization Control of the Leishmaniases, report of a WHO expert committee. World Health Organ Tech Rep Ser. 2010;949:1–186. [Google Scholar]
- Yasur-Landau D., Jaffe C.L., David L., Baneth G. Allopurinol resistance in Leishmania infantum from dogs with disease relapse. PLoS Neglected Trop. Dis. 2016;10(1) doi: 10.1371/journal.pntd.0004341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasur-Landau D., Jaffe C.L., Doron-Faigenboim A., David L., Baneth G. Induction of allopurinol resistance in Leishmania infantum isolated from dogs. PLoS Neglected Trop. Dis. 2017;11(9) doi: 10.1371/journal.pntd.0005910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuge J., Cederbaum A.I. Depletion of S-adenosyl-l-methionine with cycloleucine potentiates cytochrome P450 2E1 toxicity in primary rat hepatocytes. Arch. Biochem. Biophys. 2007;466:177–185. doi: 10.1016/j.abb.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.