Abstract
CLARITY is a novel tissue clearing technique that transforms intact biological tissues into a nanoporous hydrogel-tissue hybrid, preserving anatomical structures, proteins and nucleic acids. The hydrogel-based structure is transparent after the removal of lipids and permits several rounds of immunostaining and imaging. This technique provides an ideal way for researchers to examine the central nervous system (i.e., mouse brain and spinal cord) intact. CLARITY was selected as one of ten breakthroughs in 2013 by Science. However, the original CLARITY technique still has severe technical limitations which impede its application in wider fields. Therefore, many modified clearing methods based on CLARITY have emerged. As all CLARITY-based tissue clearing techniques involve similar procedures, the present review attempted to divide these methods into individual procedures in order to provide new ways to test and combine tissue clearing methods. Furthermore, the combination of clearing methods could help to determine the optimal method for clearing and imaging large samples.
Keywords: tissue clearing, CLARITY, immunohistochemistry, tissue imaging
1. Introduction
Biological tissues are very complex and intrinsically three-dimensional. Since most biological tissues are not transparent, and imaging processes are limited by light scatter, imaging deep within the tissues is problematic (1–3). Scientists have made multiple attempts to reduce the impacts of light scatter, including the development of various light microscopes (such as confocal/multi-photon, light sheet, volume-illumination and Bessel-beam illumination microscopes) (1,4), as well as tissue clearing techniques (Scale (5), ScaleS (6), iDISCO (7), 3DISCO (8,9), SeeDB (10), ClearT (11), CUBIC (12), FRUIT (13), CLARITY (14)). Tissue clearing techniques preserve both the molecular and structural information with minimal disassembly of the sample. Combined with labeling approaches, this method enables the integration of molecular, cellular and systems biology across different scales (2). Thus, the increasing interest for tissue clearing techniques is driving the development of many new techniques from laboratories around the world (2,15).
Clear Lipid-exchanged Acrylamide-hybridized Rigid Imaging/Immunostaining/In situ hybridization-compatible Tissue-hYdrogel, or CLARITY (14), is a novel tissue clearing technique proposed in 2013. By infusing hydrogel monomers (acrylamide and bis-acrylamide), formaldehyde and thermally triggered initiators into tissues at 4°C, this technique transforms tissues into a hydrogel-hybridized form after heat initiation. The 3D hydrogel network is relatively stable, optically transparent and macromolecular-permeable (Fig. 1). Biological molecules are combined with the hydrogel network except lipids. Thus, proteins and nucleic acids are preserved after removal of lipids by detergent. CLARITY was selected as one of ten notable breakthroughs in 2013 by Science. As reported, CLARITY leaves tissues sufficiently sturdy to repeatedly infiltrate with different labels, unlike other methods, which render see-through brains too fragile (16). Therefore, it could be sped up by many fold tasks such as counting all the neurons in a given brain region.
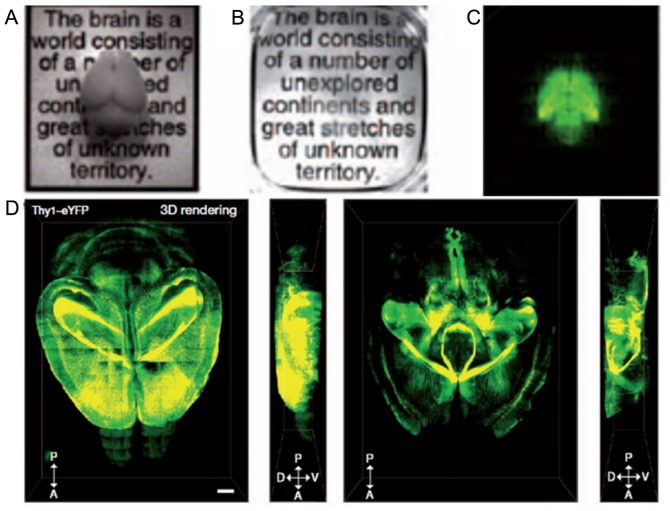
Figure 1.
Intact mouse brain imaging. This picture is cited from the article of Chung et al (14). (A) Intact mouse brain before CLARITY. (B) Intact mouse brain after CLARITY. (C) Fluorescence image of clarified mouse brain. (D) 3D rendering of clarified brain image. Dorsal, left, ventral and right side view of fluorescence 3D image. Scale bar, 1 mm.
The original CLARITY technique includes the following steps: Fixation of tissues; hydrogel monomer infusion; hydrogel-tissue hybridization; lipid extraction; molecular labeling (if needed); and refractive index matching and imaging (Fig. 2) (14,17). Although it may seem difficult to implement CLARITY effectively due to the complexity of the tissue clearing process, this technique has several advantages over other techniques. CLARITY can make tissues highly transparent without severe tissue deformation (tissue expansion occurs during lipid removal but is reversible after immersing in refractive index (RI) matching solutions), and it preserves both anatomical and molecular information well (only ~8% protein loss was observed) (14,18). In addition, as proteins and nucleic acids are bound to the hydrogel mesh with chemical tethering, it permits several rounds of molecular labeling and elution. If the sample has endogenous transgenic expression of fluorophores, it's also compatible with molecular labeling of other targets (14,17,19). After tissue clearing, the samples can be stored for weeks to months (17,20). In the years following its invention, CLARITY and its improved techniques have been widely used in many fields, especially neuroscience. Furthermore, this technique seems compatible with a variety of tissues and organs, such as brain, bone, pancreas, liver, spleen, kidney, lung, heart, intestine and even the whole body (21–25). It has also been reported that CLARITY is suitable for the 3D molecular imaging of plant organs, termed PEA-CLARITY (26).
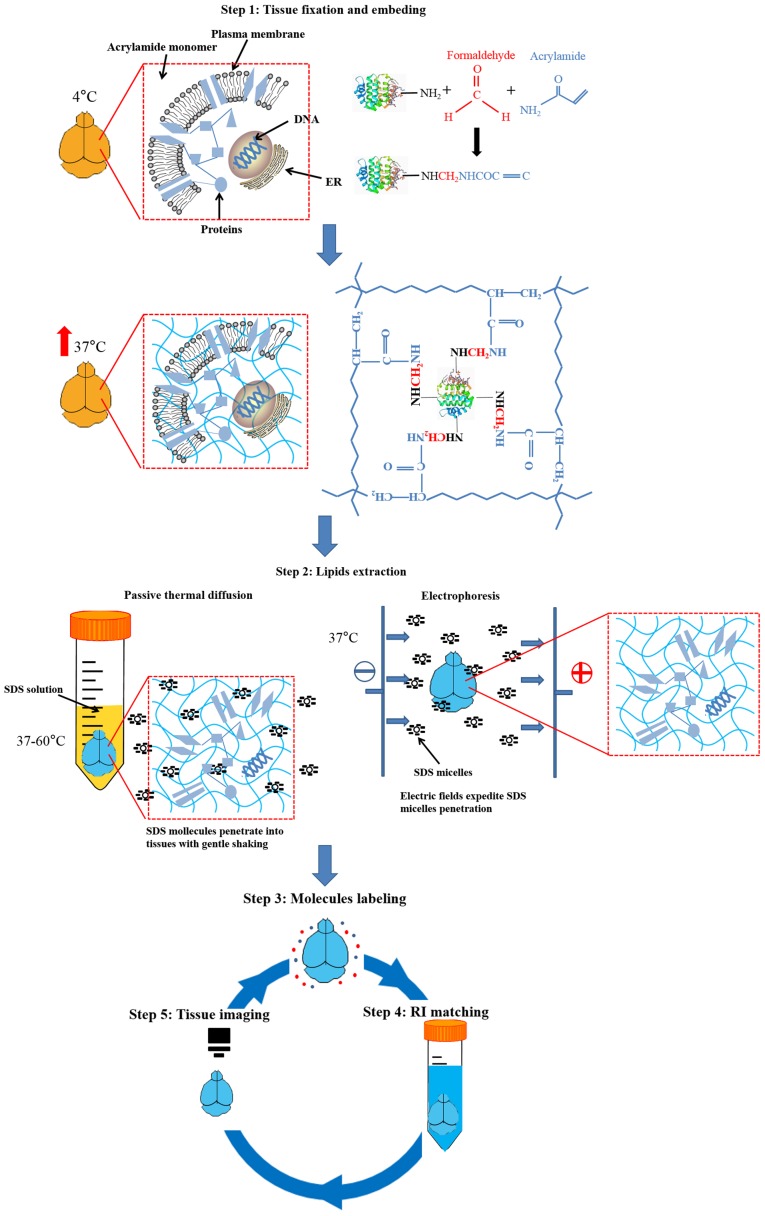
Figure 2.
Procedures of CLARITY. Step 1: Tissues are fixed with formaldehyde and acrylamide monomers, during which covalent links are formed between the natural molecules and monomers. Then, acrylamide monomers polymerize into a nanoporous hydrogel mesh with thermal initiation. Step 2: Lipids of tissue are eluted by an SDS detergent, with electric fields or gentle shaking applied. Tissue structure and biomolecules are preserved. Step 3: Cleared tissues may be stained with different labels. If the tissues have an endogenous transgenic expression of fluorophores, proceed directly to step 4. Step 4: Labeled tissues are incubated in RI-matching solutions to achieve homogeneity. Step 5: Tissues of interest become optically transparent and enable examination with microscopes.
However, there are several limitations and challenges facing CLARITY. For example, too much time is required from tissue clearing to the acquisition of imaging data. Also, electrophoretic tissue clearing (ETC) requires a custom-designed chamber and continuous exchange of detergent (SDS solution) (17). Moreover, during electrophoresis, the temperature of the solution increases gradually requiring temperature control or the sample will become brown due to the heat (14,17). In addition, the RI-matching solution (Focus Clear, Cell Explorer Labs) in the original protocols is not commonly affordable (17). These problems make the implementation of this technique difficult. Therefore, many groups are attempting to simplify and improve CLARITY. Many other clearing techniques based on hydrogel-tissue hybridization have emerged since the introduction of CLARITY (such as PACT, PARS, ACT-PRESTO and MAP) (18,27,28).
As all CLARITY-based tissue clearing techniques involve similar procedures, in this study, we introduce improvements for the CLARITY technique by dividing its component techniques into five implementation procedures: i) tissue fixation and embedding; ii) lipid extraction (including ETC and passive thermal diffusion); iii) molecule labeling; iv) refractive index matching; and v) tissue imaging. Thus, we provide a new perspective on this technique allowing for the integration of different methods. Additionally, this approach helps to develop a method for large-scale sample clearing.
2. Improvements for CLARITY technique
Tissue fixation and embedding
The first step in CLARITY is tissue fixation. In the original CLARITY protocol, animals were transcardially perfused with HM solution (including 4% PFA, 4% acrylamide, 0.05% bis-acrylamide and 0.25% VA-044 initiator in PBS) after removing blood. Next, the tissue is incubated in HM solution at 4°C for 2–3 days followed by hydrogel polymerization with heating (14). In a separate protocol, the incubation time was shortened to 1 day (17).
PFA, acrylamide, proteins and nucleic acids chemically bond together to form a hydrogel mesh. This binding preserves intrinsic molecules (proteins and nucleic acids) and permits the penetration of labeling dyes. However, the speed of clearing and the immunolabeling process depend on the pore size of hydrogel mesh. High concentrations of PFA and acrylamide result in a dense hydrogel mesh. This mesh creates a stable hydrogel-tissue hybridization and reduced the loss of proteins. However, a dense hydrogel mesh slows both tissue clearing and antibody perfusion (17,21,28,29). Therefore, many researchers have adjusted the mixture ratio for different tissues and clearing approaches to maintain a balance between hydrogel rigidity and porosity with minimal protein loss and a faster speed for clearing and immunolabeling (19).
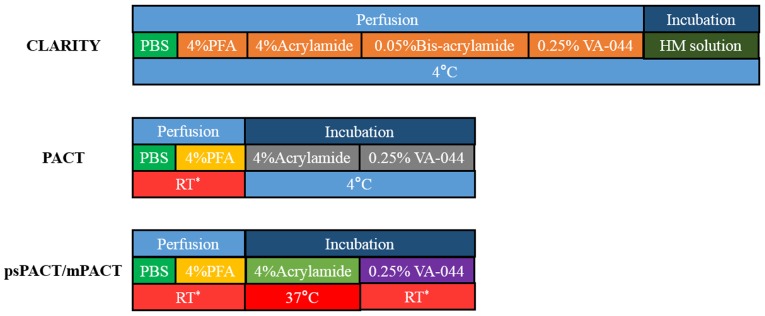
In ETC, the original solution (4% PFA, 4% acrylamide, 0.05% bis-acrylamide) is the most common ratio to date. Epp et al systematically researched the influence of different ETC temperatures and hydrogel composition on tissue transparency and expansion using an intact mouse brain (21). These researchers recommended a 4% formaldehyde and 4% acrylamide mixture for examining endogenous fluorescent markers and a composition of 3% acrylamide and 3% formaldehyde for immunohistochemistry. For passive CLARITY, the process of tissue fixation is the same as ETC in the original article (17). Next, Yang et al proposed PACT, which adopted a different perfusion method (28). Compared with the passive CLARITY protocol, PACT fixes tissues with 4% PFA followed by incubation at 4°C overnight in the hydrogel monomer solution (A4P0, 4% acrylamide in PBS supplemented with VA-044 initiator). These researchers also demonstrated the structural integrity of tissues throughout PACT processing using 1-mm-thick Thy1-eYFP tissue sections. The pore of the hydrogel mesh in tissues processing with PACT is larger than that observed with HM solution because of the obstruction of lipid bilayer when incubating in A4P0 (23). Thus, tissues fixed with A4P0 would be more easily deformed compared with those in original protocol. However, this provides advantages in certain dense tissues, such as bone (PACT-deCAL) (23,25). This perfusion method for PACT is also used in an improved ETC technique termed ACT-PRESTO (18). Lee et al optimized the original PACT procedure, creating psPACT (process-separated PACT) and mPACT (modified PACT: psPACT with added α-thioglycerol) (30). In psPACT, fixed tissues were incubated in A4P0 (4% acrylamide in PBS without VA-044) at 37°C for 24 h (the original protocol incubated tissues with A4P0 within VA-044 initiator and stored tissues at 4°C). Next, tissues were covered with 0.25% VA-044 at room temperature for 6 to 24 h. The authors compared psPACT, mPACT with PACT in the rat central nervous system (spinal cord and brain), as well as in other internal organs. These researchers concluded that this tissue fixation method could accelerate tissue clearing, with mPACT being faster than psPACT. We compare these different fixation methods in Fig. 3.
Figure 3.
Comparison of three main tissue fixation methods. For each method, there are three rows of colored boxes. The first row shows the tissue handling methods with fixation solutions; the second row displays the ingredients used for tissue fixation; the third row presents the proposed working temperature. Additionally, the second row also utilizes different colored boxes to delineate between the different ingredients used during tissue fixation. If the boxes are the same color, these ingredients are mixed together. The HM solution consists of the ingredients in orange box directly before it. RT, room temperature
We have observed that there is no obvious limitation to the perfusion method used in different tissue clearing approaches (ETC vs. passive CLARITY). The most important challenge is to shorten the time of tissue clearing by adjusting the pore diameter of hydrogel mesh using different concentrations of ingredients and perfusion methods while keeping tissue structure stable. Passive CLARITY and PACT requires more time to extract lipids from tissue-hydrogel hybrids with shaking with a higher temperature than would be used in ETC when handling relatively large tissues (ETC adopted 37°C; the original passive CLARITY protocol recommended 37°C-60°C; PACT first adopted 37°C; another study concluded 42°C-47°C is a better temperature range for PACT) (17,28,31). Thus, the requirement for tissue stability in PACT is higher than that of ETC, and the concentration of acrylamide and PFA is of vital importance when implementing passive CLARITY and PACT.
The following step after tissue fixation is hydrogel polymerization. In the original protocol, nitrogen and vacuum pump are used to extract and replace air in the tissue container (as oxygen impedes hydrogel formation) (14). Notably, oxygen must be completely removed; otherwise, hydrogel formation in the inner parts of tissue would be incomplete and result in the emergence of cavities after lipids extraction. It was also reported that adding a thick layer of mineral oil over the top of the hydrogel solution before polymerization could minimize the exposure of samples to the air (32).
Lipid extraction
Since membrane lipids are the main cause of light diffraction, lipid extraction becomes a key process in CLARITY (1,20,33). Tissue transparency primarily depends on the degree of lipid elution. In general, there are two methods for lipid extraction: passive thermal diffusion and electrophoresis. Passive thermal diffusion is a way to transport SDS micelles into tissues via heat-induced diffusion. And electrophoresis applies electric fields to accelerate the penetrating of SDS micelles. All modified CLARITY techniques elute membrane lipids based on these two methods (PACT, PARS, ETC, ACT-PRESTO- and stochastic electrotransport) (14,17,18,14). In this study, we compared strengths and weaknesses of these two methods in Table I. Next, we reviewed the modified CLARITY techniques based on the two methods.
Table I.
Comparison of passive thermal diffusion and electrophoresis lipids extraction.
| Methods | Principle | Elution time | Complexity of implementation | Refresh SDS buffer | Volume of SDS buffer | Circulation |
|---|---|---|---|---|---|---|
| Passive thermal diffusion | Transporting surfactant micelles into tissues via heat-induced diffusion | Weeks to months | Easy | Occasionally | Small (several ten milliliters) | No |
| Electrophoresis | Transporting surfactant micelles into tissues via electric fields | Several days | Difficult | Frequently | Large (several liters for each circulation) | Yes (to control the temperature) |
Passive thermal diffusion
Regarding passive thermal diffusion in-passive CLARITY, tissues are incubated in 4% SDS at pH 8.5 with gentle shaking at 37°C with an increased temperature of up to 60°C for faster clearing (17). Because of dense hydrogel mesh structure and low efficiency of SDS transportation, this method is time-consuming and may not reach a high degree of tissue transparency for larger tissues, except at higher temperatures (34). However, higher temperatures may cause potential tissue deformation and damage to natural molecules. Therefore, in order to accelerate SDS transportation and to keep tissue structure stable, Yang et al proposed new methods, termed PACT and PARS (23,28). PACT adopted 8% SDS buffer at pH 8.5 and samples are gently shaking at 37°C for tissue clearing. These researchers emphasized that only an 8% SDS concentration could achieve uniform tissue clearing throughout the entire 3-mm block, while the SDS concentration in ETC and most other modified techniques is 4% (14,17). PARS is a method for whole-body clearing and labeling (23,28). After fixing tissues with PFA and A4P0, tissues of interest are not removed from animal's body and the clearing process is achieved in vivo. PARS utilized the intact vasculature to deliver and circulate the SDS buffer. For different tissues, different parts of vasculature could be selected. For brain or spinal cord clearing, PACT regents could be delivered with a subdural cannula inserted above the region of interest, and the buffer circulation would be similar to that of cerebrospinal fluid (28). The principle of PARS is also based on passive diffusion. The difference is that PARS increased the contact area between the clearing buffer and tissues using the intrinsic vasculature; therefore, the tissue clearing speed becomes faster. Additionally, PARS minimizes tissue expansion during clearing, as the skull and other physiologic structures limit the available space. However, implementation of PARS is more complicated than PACT (a cannula needs to be set and kept stable as tissue vasculature is easily destroyed and obstructed during buffer circulation). Thus, the application of PARS is limited. After that, several modified methods based on PACT emerged. For mPACT, the author added 0.5% α-thioglycerol to 8% SDS buffer for lipid extraction with gentle shaking, and it showed faster clearing speed than PACT. The use of α-thioglycerol was also reported in other articles to avoid browning and autofluorescence accumulation as a result of the Maillard reaction (10,18). The α-thioglycerol works via the sulfhydryl, suggesting that other chemical materials with sulfhydryl, such as β-mercaptoethanol, may also work (10). It has also been reported that 0–50 mM sodium sulfite could exhibit an anti-browning effect in other tissue clearing methods, such as SWITCH (35). Lee et al also combined mPACT and PARS as a whole body perfusion method (PARS-mPACT) (30). PARS-mPACT fixed tissues via the transcardial perfusion of separated mPACT regents. Next, polymerized tissues were isolated and passively cleared according to the mPACT protocol. However, PARS-mPACT and mPACT showed no differences in tissue transparency. Another method, named CLARITY2, adopted a different way to accelerate tissue clearing (36). After tissue fixation and hydrogel polymerization, brain tissue was cut to 1–1.5-mm-thick coronal slices and underwent passive tissue clearing. This approach enabled us to accelerate and simplify the clearing, staining and imaging steps compared with original protocol. However, CLARITY2 did not avoid damaging the tissues and is similar to a traditional tissue slice. However, this technique is still an ideal time-saving method for methodological studies (32,37,38). In addition, it's reported that with pancreatic lipase breaking down lipid droplets, lipid-rich tissues were better cleared when performing passive clearing (39).
And there were no obvious tradeoffs in the quality or characteristics of cleared tissues with the SDS buffer at different pH, except for a slight enhancement in the rate of clearance at a more alkaline pH (23). Thus PACT, PARS and other passive CLARITY techniques could dissolve SDS using 1× PBS at pH 7.5 for convenience, adjusting the SDS concentration according to the needs of the study to maintain a balance between tissue integrity and transparency.
Electrophoresis
Electrophoresis is faster than passive thermal diffusion, as electric fields accelerate the transport of SDS. First, the ETC system is composed of fourelements: i) A chamber for containing samples and electrodes; ii) a power supply; iii) a circulator to control flow rate and buffer temperature, which is combined with the chamber; and iv) a filter to remove byproducts.
In the original protocol, a range of conditions (10–60 V) were applied across the tissue at 37–50°C for several days with clearing solution circulating through ETC chamber (14). However, the high temperature and voltage could cause a higher risk of tissue deformation, as well as epitope and fluorescence loss (17,22). Therefore, the voltage and circulation temperature should be precisely controlled to produce better outcomes. It was also recommended that ETC be completed at 25 V and 37°C for an adult mouse brain (17). The limitations to the application of high electric fields in the ETC system was compounded by the fact that the electrical properties in different regions of a tissue are not homogeneous, leading to regions with concentrated electric fields. Thus, high electric fields may cause deformation of these regions. This suggests that electrophoresis is ineffective for promoting the transportation of SDS into large, dense samples only if high electric fields are used. Stochastic electrotransport, created by Kim et al could promote the migration of freely moving molecules with high electromobility while suppressing the displacement of endogenous biological molecules with low electromobility within the sample. This technique may be an ideal way to solve this high-voltage ETC problem (22). Compared with the stationary samples and electrodes in original ETC chamber, the author implemented stochastic electrotransport by creating a continuously rotating sample chamber with respect to two parallel electrodes (22). Next, these researchers made a contrast among mouse brains cleared by stochastic electrotransport, static electrophoresis and thermal diffusion. The mouse brain cleared with stochastic electrotransport was remarkably transparent within 3 days, while others did not show the same results (22). Lee et al created a modified clearing method termed ACT (18). ACT alternated the platinum wires to platinum plate to generate a dense regular current in the ETC chamber and adopted a two-step fixation protocol as mentioned in PACT. Therefore, ACT could clear tissues faster than ETC (2 h of ACT could achieve complete optical transparency in 1-mm brain sections and 15 h was sufficient to clear entire adult rat brain) (18). However, this method has a high risk of tissue deformation because of the strong electric fields generated and relatively unstable tissue-hydrogel hybridization structure compared to HM solution fixed tissues.
Moreover, modifications to ETC were proposed via changing the parameters of ETC (current or temperature) to achieve better results. One study reported that different optimized current of ETC for various organs (including brain, pancreas, kidney, liver, intestine and lung) (24), demonstrating that the current and voltage of electrophoresis should correspond with the needs of the study. Epp et al tested effects of different ETC temperatures on tissue transparency and proposed a combined 37/55°C clearing protocol, in which they ran ETC at 20 V for the first 4 days at 37°C and increased the temperature to 55°C on the final day (21). Using this method, tissues showed better transparency and structural integrity than those produced by running ETC at only 37°C or 55°C.
There are additional problems when repeating the original protocol. First, the by-products formed during electrophoresis would discolor tissues and colored particles may deposit on the surface of tissues (24). To solve this problem, the author in stochastic electrotransport used a temperature controller to maintain SDS buffer temperature at ~15°C to prevent browning tissues due to heat. Apart from that step, nanoporous membranes were used to contain samples and mechanically divide the circulating solution into an inner portion and outer portion (22). The lower concentrations in outer portion were designed to slow down electro-oxidation of the surfactant molecules (22). Second, the continuously emerging bubbles during electrophoresis could interrupt electrophoresis if the ETC chamber was filled (14,17). In ACT (18), the author designed a long ETC chamber, which allowed bubbles float to the top where they were removed through the top outlet. Bastrup and Larsen (29) also designed an adjustable ETC chamber to fit variable tissue sizes.
Molecules labeling
In general, there are three methods to visualization interest molecules: i) Genetic introduction of fluorescent markers; ii) in vivo labeling of cells or regions with viruses or chemicals; and iii) chemical/antibody staining (40). For the first two methods, fluorescence may quench throughout clearing process and require amplification (2,19). Therefore, chemical/antibody staining is an alternative way to label cleared tissues. With this method, multiple rounds of staining [three rounds of staining with up to 4 channels were reported (14)] are possible without damaging the preserved structures as proteins and nucleic acids are chemically bound to the hydrogel mesh (19). The clearing solution SDS buffer can be used to wash antibodies and other molecular labels out of the hydrogel-tissue hybridization in preparation for another round of staining.
In the original protocol, tissues were immunostained by incubating them at 37°C with a high concentration of primary and secondary antibodies (dilution, 1:50-1:100) and gentle shaking. Itrequires between 2 days and 2 weeks, depending on the size of the tissue sample (14). The principle of this immunostaining method is the same with passive tissue clearing, expediting the penetration of antibodies by passive thermal diffusion. Some other methods to provide external forces for accelerating the transportation and penetration of antibodies were applied to labeling tissues with a large volume. Similar to lipid extraction, the improvement of immunostaining is also based on two aspects: passive diffusion and electrophoresis (41).
Passive diffusion labeling
Many labeling methods are based on passive diffusion as it's simple to implement. It has been reported that PARS was suitable for immunostaining and that all immunohistochemical solutions were delivered through the PARS circulation system (including blocking reagent, antibodies or fluorescently labeled molecules and wash buffers) (23,28). This delivery is target-specific and uniformly distributed throughout organs with a low background (28). However, similar to original protocol, it cannot expedite the labeling process (23). To solve this problem, another method termed PRESTO (pressure related efficient and stable transfer of macromolecules into organs) emerged (18). This method was composed of c-PRESTO (centrifugal PRESTO) and s-PRESTO (syringe PRESTO). The c-PRESTO technique applied a centrifugal force with a standard table top centrifuge (600 rcf) to expedite antibody diffusion. For example, kidney tissues incubated for 3 h with antibodies showed labeled structures 10–30 µm deep, whereas for normal tissues, 3 h was sufficient to label structures 120 µm deep (18). The s-PRESTO created a convection flow with a syringe pump, which infused labeling reagents into the specimen. This technique showed a labeling depth that was four times greater than that obtained in original passive labeling (41).
Almost all cleared tissues are applied a fluorescence-based phenotype to label and image interest tissues. But one study used a colorimetric, non-fluorescent method based on the conversion of horseradish peroxidase to diaminobenzidine to label PACT cleared tissues (42). However, this technique was only applied to 50 and 100 µm sections of adult mouse brain tissue; therefore, it did not contain the anatomical structure information of an intact brain. Furthermore, non-fluorescence labeling cannot achieve 3D reconstruction, limiting further applications in tissueimaging.
Electrophoresis labeling
As antibodies are charged molecules in solution with certain pH, applying external electric fields can expedite the transportation of them into tissues. A study by Li et al applied a simple constant electric field across a 500-µm-thick brain section, and it decreased the delivery time of antibodies by more than 800-fold over simple diffusion without incurring structural damage (43). In this study, the 500-µm-thick brain section was stained in only 30 min at an external voltage of 25 V. However, when applying this to larger tissues (whole brains or animal bodies), heat damage caused by electrodes may occur necessitating buffer circulation to control the temperature. To ensure the concentration of dyes, the staining solution and circulated buffer should be separated. Similar to lipid extraction with ETC, static electrophoresis resulted in substantial tissue damage in large-volume sample, as it needed a longer time for electrophoresis. This problem was solved with stochastic electrotransport (22). Similar to the device for clearing in stochastic electrotransport, antibodies were confined inside of the sample chamber with nanoporous membranes and PBS was circulated in the outer chamber to control the temperature.
Non-protein molecules labeling
However, these techniques have been poorly explored with RNA studies. Yang et al first used single-molecule fluorescence in situ hybridization (sm-FISH) to detect single RNA molecules in 100-µm-thick PACT-processed mouse brain sections (28). When applying a single-molecule hybridization chain reaction (smHCR), single mRNAs could be detected within 500-µm-thick PACT-processed brain slices (44). Additionally, EDC-CLARITY used EDC (1-Ethyl-3-3-dimethyl-aminopropyl carbodiimide) to link 5′-phosphate groups with surrounding amine-containing proteins to stably retain RNAs in clarified tissues (45). With the hairpin chain reaction (HCR) amplification system, it presented validation for selecting microRNAs, cell-type markers and immediate-early genes.
CLARITY achieved tissue transparency by eluting membrane lipids and lipophilic dyes. Therefore, lipophilic fluorescent dyes, such as DiI, which stains cellular membranes, is unable to be applied with CLARITY (46,47). A study by Jensen and Berg altered the molecular structure of the dye to adhere to both membranes and proteins such that the dye remained in the tissue after tissue clearing (47). These researchers tested three Dil-analogue dyes, CM-DiI, SP-DiI and FM 1-43FX in PACT-processed spinal cords of adult rats and mice. All three dyes remained in the tissue after lipid-clearing, but CM-DiI had the sharpest and FM 1-43FX had the strongest fluorescent signals. This modification provided a new way to label neurons (retrograde or anterograde) and mark the position of extracellular electrodes after electrophysiology.
Refractive index matching
Before imaging, cleared tissues need to be incubated in solutions that match the average RI of the tissue (~1.46) to achieve RI homogenization. Afterward, tissues would be optically transparent and photon scattering from both the excitation light and the emitted fluorescence signal would be reduced. In theory, the RI of each solution can be matched to the cleared tissue or microscope objective by adjusting concentration of the main ingredient. However, the anti-swelling of cleared tissues should also be considered an important factor affecting the solution. In this study, we contrasted several solutions used in CLARITY-based techniques.
In original protocol, the author applied FocusClear (CelExplorer Labs) and 85–87% glycerol to match RI (14). These reagents both promoted tissue transparency, but the best results were achieved with FocusClear. However, FocusClear is expensive and not suitable for long-term storage (long-term storage resuls in the formation of an irreversible white precipitate) (14,17). Yang et al used a solution termed RIMS (refractive index matching solution) to achieve tissue transparency (28). It contains 88% Histodenz in 0.02 M phosphate buffer with an RI of 1.47. RIMS is stable with long-term storage and preserves fluorescent markers over months. Their group had also derived sRIMS, containing 70% sorbitol in 0.02 M phosphate buffer and cRIMS, containing 88% histodenz in 0.005 M phosphate buffer (23). Next, one study found that 63% 2,2′-thiodiethanol (TDE) in PBS was more suitable for RI matching in CLARITY cleared tissues (48). A different solutuion, 80% Nycodenz solution (nRIMS), was applied with mPACT (30). In ACT-PRESTO, a CUBIC-mount solution (50% Sucrose, 25% urea and 25% N, N, N', N'-tetrakis (2-hydroxypropyl) ethylenediamine) was used to match the RI of the tissues (18). The comparison of these RI-matching solutions can be seen in Table II. However, in a study by Poguzhelskaya et al imaging without an RI-matching solution but in PBST was also possible (36).
Table II.
Comparison of different RI matching solutions.
| Solution | Focus clear | RIMS | sRIMS | cRIMS | nRIMS | THE | CUBIC-mount solution |
|---|---|---|---|---|---|---|---|
| Main ingredients | Unknown | 88% Histodenz in 0.02 M phosphate buffer | 70% sorbitol in 0.02 M phosphate buffer | 88% histodenz in 0.005 M phosphate buffer | 80% Nycodenz (TDE) in PBS | 63% 2, 2′-thiodiethanol | 50% Sucrose, 25% urea and 25% N, N, N', N'- tetrakis (2-hydroxypropyl) ethylenediamine |
Although all of these solutions were intended to match the RI of cleared tissues, for different samples, it may still fail to match the RI well after tissue clearing, as various elements differ in inner tissues. For example, a study by Liu et al reported that 47% TDE in phosphate buffer (not PBS) was more suitable for human brain tissues (37). Therefore, if the concentrations reported in articles do not make the interest tissue ideally transparent, different concentrations and solvents (PBS, phosphate buffer or normal saline) should be tested. Moreover, solutions with a high concentration could reduce tissue swelling. When imaging without an RI-matching solution but in simple purified water or PBST, the tissue will remain swollen, and fail to get the structural information. Therefore, RI-matching and anti-swelling should both be considered when choosing RI-matching solutions.
Tissue imaging
Finding an optimized, high-resolution deep-imaging approach for large-scale samples is still a major challenge. Furthermore, imaging a 3D volume is a time-consuming task. Therefore, when selecting a microscope and objective, imaging time, resolution and sample size should all be taken into account. In this study, we compared different microscopes and objectives applied to cleared samples.
There are three primary microscopes used for three-dimensional fluorescence imaging: i) The standard confocal microscope; ii) the two-photon microscope; and iii) the light sheet microscope. The standard confocal microscope achieves optical sectioning by using a pinhole in front of the photomultiplier tubes and scans samples point to point. Therefore, this microscope's processing time is notably slow, particularly in large samples, and long-time imaging can lead to photobleaching (20,49). A two photon microscope results in lower photobleaching of fluorophores and provides greater imaging depth than a confocal microscope, but point-by-point scanning is still slow. For the light sheet microscope, it uses fast sCMOS or CCD cameras to image a selectively illuminated focal plane and provides a high imaging speed (2–3 orders of magnitude faster than point-scanning methods), high signal-to-noise ratio and low levels of photobleaching (17,50). Tomer et al developed SPED (SPherical-aberration-assisted Extended Depth-of-field) light sheet microscopy, which combined an extension of the depth-of-field with the optical sectioning of a light sheet microscope, thereby eliminating the need to physically scan detection objectives for volumetric imaging (51). It enabled the scanning of thousands of volumes per-second, limited only by the camera acquisition rate.
The key microscope objective parameters are working distance (WD), numerical aperture (NA) and RI. WD is the distance between the objective lens and the focal plane. A long WD avoids the sample directly contacting with lens and ensures imaging of large samples (52). The NA of an objective relates to the collection of emitted signal, and higher NA enables higher resolution (17). The RI of the cleared tissues is ~1.46; therefore, it is better to choose objectives designed with an RI near 1.46 when using a non-optimized objective.
3. Challenges and opportunities
Though many efforts have been spared on the improvement for CLARITY, the primary issues still remain. First, we acknowledge that the implementation of CLARITY has limitations due to the factors mentioned above. A more simplified protocol is needed and the processing time should be shortened for examining samples more quickly.
The largest challenge for tissue clearing, as with other tissue clearing techniques, is the clearing of large-volume samples, such as a monkey brain (3). Though tissue clearing techniques are widely used in neuroscience, information acquired in this way concerning the human brain is still limited. Therefore, animals similar to humans are ideal models for studying the human brain. Therefore, integration of current techniques or the development of new methods for clearing large samples are of vital importance. Furthermore, in terms of imaging, large sample is not feasible due to the limitations of microscopes and data analysis. Thus, long WD, high NA microscopes and fast data analysis software are also in demand.
However, there are several advantages from other clearing and labeling techniques from which we can learn. SWITCH (System-Wide control of Interaction Time and kinetics of Chemicals) produces tissue-glutaraldehyde hybrids which are heat- and chemical-resistant and permit multiple rounds (>20) of labeling (35). When applying antibody labeling, SWITCH enables scalable and uniform labeling by controlling probe-target binding kinetics with a low concentration (0.5 to 1.0 mM) SDS. Therefore, it may also be accessible to control the binding kinetics in CLARITY in order to achieve homogeneity and more rounds of labeling. In CLARITY, we use RI-matching solutions to resist tissue swelling and maintain a normal size (17). In contrast, a method called MAP (magnified analysis of the proteome), which is also based on hydrogel-tissue hybridization, linearly expands entire organs four-fold while preserving their overall architecture and three-dimensional proteome organization (27). Similarly, Chen et al reported a method termed expansion microscopy (ExM) that can linearly expand tissues ~4.5-fold to enable imaging with conventional confocal microscopes to examine the subcellular architecture (53). Then, their group developed iterative expansion microscopy (iExM), in which a sample can be expanded ~20-fold. These methods provide us with new ways to utilize tissue expansion to study subcellular architecture, and even for expediting antibodies penetration during labeling (expanded tissues have bigger pore size of hydrogel network).
4. Conclusions
Compared with other tissue clearing techniques, CLARITY is more compatible with different samples and provides tissues with higher transparency, despite having several limitations. However, CLARITY still faces many challenges. High resolution and multi-level imaging of large-scale samples require the integration of different clearing methods and imaging techniques. Additionally, massive volumetric data sets acquired from samples with a larger volume present a challenge for data analysis. However, with future studies focused on these problems, tissue clearing can open the door to many new discoveries and provide a new view of the inner interactions among cells, organs and systems. The combination of existing clearing methods and development of new techniques may help us to address these challenges.
Acknowledgements
Not applicable.
Funding
This work was supported b the National Natural Science Foundation of China (61471367).
Availability of data and materials
Not applicable.
Authors' contributions
HD was a major contributor in writing the manuscript. PH wrote the ‘molecular labeling’ part of the manuscript and designed the illustrations. BZ contributed to the conception that divides different methods into five procedures and made the tables. QL designed the study and made important revisions to the manuscript. All authors contributed to assess and revise the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflict of interest.
References
- 1.Zhu X, Xia Y, Wang X, Si K, Gong W. Optical brain imaging: A powerful tool for neuroscience. Neurosci Bull. 2017;33:95–102. doi: 10.1007/s12264-016-0053-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seo J, Choe M, Kim SY. Clearing and labeling techniques for large-scale biological tissues. Mol Cells. 2016;39:439–446. doi: 10.14348/molcells.2016.0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richardson DS, Lichtman JW. Clarifying tissue clearing. Cell. 2015;162:246–257. doi: 10.1016/j.cell.2015.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Planchon TA, Gao L, Milkie DE, Davidson MW, Galbraith JA, Galbraith CG, Betzig E. Rapid three-dimensional isotropic imaging of living cells using Bessel beam plane illumination. Nat Methods. 2011;8:417–423. doi: 10.1038/nmeth.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hama H, Kurokawa H, Kawano H, Ando R, Shimogori T, Noda H, Fukami K, Sakaue-Sawano A, Miyawaki A. Scale: A chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat Neurosci. 2011;14:1481–1488. doi: 10.1038/nn.2928. [DOI] [PubMed] [Google Scholar]
- 6.Hama H, Hioki H, Namiki K, Hoshida T, Kurokawa H, Ishidate F, Kaneko T, Akagi T, Saito T, Saido T, Miyawaki A. ScaleS: An optical clearing palette for biological imaging. Nat Neurosci. 2015;18:1518–1529. doi: 10.1038/nn.4107. [DOI] [PubMed] [Google Scholar]
- 7.Renier N, Wu Z, Simon DJ, Yang J, Ariel P, Tessier-Lavigne M. iDISCO: A simple, rapid method to immunolabel large tissue samples for volume imaging. Cell. 2014;159:896–910. doi: 10.1016/j.cell.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 8.Ertürk A, Bradke F. High-resolution imaging of entire organs by 3-dimensional imaging of solvent cleared organs (3DISCO) Exp Neurol. 2013;242:57–64. doi: 10.1016/j.expneurol.2012.10.018. [DOI] [PubMed] [Google Scholar]
- 9.Ertürk A, Becker K, Jährling N, Mauch CP, Hojer CD, Egen JG, Hellal F, Bradke F, Sheng M, Dodt HU. Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat Protoc. 2012;7:1983–1995. doi: 10.1038/nprot.2012.119. [DOI] [PubMed] [Google Scholar]
- 10.Ke MT, Fujimoto S, Imai T. SeeDB: A simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat Neurosci. 2013;16:1154–1161. doi: 10.1038/nn.3447. [DOI] [PubMed] [Google Scholar]
- 11.Kuwajima T, Sitko AA, Bhansali P, Jurgens C, Guido W, Mason C. ClearT: A detergent- and solvent-free clearing method for neuronal and non-neuronal tissue. Development. 2013;140:1364–1368. doi: 10.1242/dev.091844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Susaki EA, Tainaka K, Perrin D, Kishino F, Tawara T, Watanabe TM, Yokoyama C, Onoe H, Eguchi M, Yamaguchi S, et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell. 2014;157:726–739. doi: 10.1016/j.cell.2014.03.042. [DOI] [PubMed] [Google Scholar]
- 13.Hou B, Zhang D, Zhao S, Wei M, Yang Z, Wang S, Wang J, Zhang X, Liu B, Fan L, et al. Scalable and DiI-compatible optical clearance of the mammalian brain. Front Neuroanat. 2015;9:19. doi: 10.3389/fnana.2015.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chung K, Wallace J, Kim SY, Kalyanasundaram S, Andalman AS, Davidson TJ, Mirzabekov JJ, Zalocusky KA, Mattis J, Denisin AK, et al. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332–337. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vigouroux RJ, Belle M, Chédotal A. Neuroscience in the third dimension: Shedding new light on the brain with tissue clearing. Mol Brain. 2017;10:33. doi: 10.1186/s13041-017-0314-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.2013 runners-up. CLARITY makes it perfectly clear. Science. 2013;342:1434–1435. doi: 10.1126/science.342.6165.1434-b. [DOI] [PubMed] [Google Scholar]
- 17.Tomer R, Ye L, Hsueh B, Deisseroth K. Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat Protoc. 2014;9:1682–1697. doi: 10.1038/nprot.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee E, Choi J, Jo Y, Kim JY, Jang YJ, Lee HM, Kim SY, Lee HJ, Cho K, Jung N, et al. ACT-PRESTO: Rapid and consistent tissue clearing and labeling method for 3-dimensional (3D) imaging. Sci Rep. 2016;6:18631. doi: 10.1038/srep18631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jensen KHR, Berg RW. Advances and perspectives in tissue clearing using CLARITY. J Chem Neuroanat. 2017;86:19–34. doi: 10.1016/j.jchemneu.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 20.Kim SY, Chung K, Deisseroth K. Light microscopy mapping of connections in the intact brain. Trends Cogn Sci. 2013;17:596–599. doi: 10.1016/j.tics.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 21.Epp JR, Niibori Y, Hsiang Liz HL, Mercaldo V, Deisseroth K, Josselyn SA, Frankland PW. Optimization of CLARITY for clearing whole-brain and other intact organs. eNeuro. 2015;2 doi: 10.1523/ENEURO.0022-15.2015. ENEURO. 0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim SY, Cho JH, Murray E, Bakh N, Choi H, Ohn K, Ruelas L, Hubbert A, McCue M, Vassallo SL, et al. Stochastic electrotransport selectively enhances the transport of highly electromobile molecules. Proc Natl Acad Sci USA. 2015;112:E6274–E6283. doi: 10.1073/pnas.1510133112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Treweek JB, Chan KY, Flytzanis NC, Yang B, Deverman BE, Greenbaum A, Lignell A, Xiao C, Cai L, Ladinsky MS, et al. Whole-body tissue stabilization and selective extractions via tissue-hydrogel hybrids for high-resolution intact circuit mapping and phenotyping. Nat Protoc. 2015;10:1860–1896. doi: 10.1038/nprot.2015.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee H, Park JH, Seo I, Park SH, Kim S. Improved application of the electrophoretic tissue clearing technology, CLARITY, to intact solid organs including brain, pancreas, liver, kidney, lung and intestine. Bmc Dev Biol. 2014;14:48. doi: 10.1186/s12861-014-0048-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greenbaum A, Chan KY, Dobreva T, Brown D, Balani DH, Boyce R, Kronenberg HM, McBride HJ, Gradinaru V. Bone CLARITY: Clearing, imaging and computational analysis of osteoprogenitors within intact bone marrow. Sci Transl Med. 2017;9:eeah6518. doi: 10.1126/scitranslmed.aah6518. [DOI] [PubMed] [Google Scholar]
- 26.Palmer WM1, Martin AP, Flynn JR, Reed SL, White RG, Furbank RT, Grof CP. PEA-CLARITY: 3D molecular imaging of whole plant organs. Sci Rep-Uk. 2015;5:13492. doi: 10.1038/srep13492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ku T, Swaney J, Park JY, Albanese A, Murray E, Cho JH, Park YG, Mangena V, Chen J, Chung K. Multiplexed and scalable super-resolution imaging of three-dimensional protein localization in size-adjustable tissues. Nat Biotechnol. 2016;34:973–981. doi: 10.1038/nbt.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang B, Treweek JB, Kulkarni RP, Deverman BE, Chen CK, Lubeck E, Shah S, Cai L, Gradinaru V. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell. 2014;158:945–958. doi: 10.1016/j.cell.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bastrup J, Larsen PH. Optimized CLARITY technique detects reduced parvalbumin density in a genetic model of schizophrenia. J Neurosci Meth. 2017;283:23–32. doi: 10.1016/j.jneumeth.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 30.Lee M, Seo JM, Park HS, Cho YE. Optimization of the optical transparency of rodent tissues by modified PACT-based passive clearing. Exp Mol Med. 2016;48:e274. doi: 10.1038/emm.2016.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu T, Qi Y, Zhu J, Xu J, Gong H, Luo Q, Zhu D. Elevated-temperature-induced acceleration of PACT clearing process of mouse brain tissue. Sci Rep. 2017;7:38848. doi: 10.1038/srep38848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng H, Rinaman L. Simplified CLARITY for visualizing immunofluorescence labeling in the developing rat brain. Brain Struct Funct. 2016;221:2375–2383. doi: 10.1007/s00429-015-1020-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yushchenko DA, Schultz C. Tissue clearing for optical anatomy. Angew Chem Int Ed. 2013;52:10949–10951. doi: 10.1002/anie.201306039. [DOI] [PubMed] [Google Scholar]
- 34.El-Sherbiny IM, Yacoub MH. Hydrogel scaffolds for tissue engineering: Progress and challenges. Glob Cardiol Sci Pract. 2013;2013:316–342. doi: 10.5339/gcsp.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murray E, Cho JH, Goodwin D, Ku T, Swaney J, Kim SY, Choi H, Park YG, Park JY, Hubbert A, et al. Simple, scalable proteomic imaging for high-dimensional profiling of intact systems. Cell. 2015;163:1500–1514. doi: 10.1016/j.cell.2015.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poguzhelskaya E, Artamonov D, Bolshakova A, Vlasova O, Bezprozvanny I. Simplified method to perform CLARITY imaging. Mol Neurodegener. 2014;9:19. doi: 10.1186/1750-1326-9-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu AK, Hurry ME, Ng OT, DeFelice J, Lai HM, Pearce RK, Wong GT, Chang RC, Gentleman SM. Bringing CLARITY to the human brain: Visualization of Lewy pathology in three dimensions. Neuropath Appl Neuro. 2016;42:573–587. doi: 10.1111/nan.12293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Magliaro C, Callara AL, Mattei G, Morcinelli M, Viaggi C, Vaglini F, Ahluwalia A. Clarifying CLARITY: Quantitative optimization of the diffusion based delipidation protocol for genetically labeled tissue. Front Neurosci. 2016;10:179. doi: 10.3389/fnins.2016.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lai M, Li X, Li J, Hu Y, Czajkowsky DM, Shao Z. Improved clearing of lipid droplet-rich tissues for three-dimensional structural elucidation. Acta Bioch Bioph Sin. 2017;49:465–467. doi: 10.1093/abbs/gmx018. [DOI] [PubMed] [Google Scholar]
- 40.Susaki EA, Ueda HR. Whole-body and whole-organ clearing and imaging techniques with single-cell resolution: Toward organism-level systems biology in mammals. Cell Chem Biol. 2016;23:137–157. doi: 10.1016/j.chembiol.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 41.Tainaka K, Kuno A, Kubota SI, Murakami T, Ueda HR. Chemical principles in tissue clearing and staining protocols for whole-body cell profiling. Annu Rev Cell Dev Biol. 2016;32:713–741. doi: 10.1146/annurev-cellbio-111315-125001. [DOI] [PubMed] [Google Scholar]
- 42.Sung K, Ding Y, Ma J, Chen H, Huang V, Cheng M, Yang CF, Kim JT, Eguchi D, Di Carlo D, et al. Simplified three-dimensional tissue clearing and incorporation of colorimetric phenotyping. Sci Rep. 2016;6:30736. doi: 10.1038/srep30736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li J, Czajkowsky DM, Li X, Shao Z. Fast immuno-labeling by electrophoretically driven infiltration for intact tissue imaging. Sci Rep. 2015;5:10640. doi: 10.1038/srep10640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shah S, Lubeck E, Schwarzkopf M, He TF, Greenbaum A, Sohn CH, Lignell A, Choi HM, Gradinaru V, Pierce NA, Cai L. Single-molecule RNA detection at depth by hybridization chain reaction and tissue hydrogel embedding and clearing. Development. 2016;143:2862–2867. doi: 10.1242/dev.138560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sylwestrak EL, Rajasethupathy P, Wright MA, Jaffe A, Deisseroth K. Multiplexed intact-tissue transcriptional analysis at cellular resolution. Cell. 2016;164:792–804. doi: 10.1016/j.cell.2016.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lai HM, Ng WL, Gentleman SM, Wu W. Chemical probes for visualizing intact animal and human brain tissue. Cell Chem Biol. 2017;24:659–672. doi: 10.1016/j.chembiol.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 47.Jensen KH, Berg RW. CLARITY-compatible lipophilic dyes for electrode marking and neuronal tracing. Sci Rep. 2016;6:32674. doi: 10.1038/srep32674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Costantini I, Ghobril JP, Di Giovanna AP, Mascaro Allegra AL, Silvestri L, Müllenbroich MC, Onofri L, Conti V, Vanzi F, Sacconi L, et al. A versatile clearing agent for multi-modal brain imaging. Sci Rep. 2015;5:9808. doi: 10.1038/srep09808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ji N, Freeman J, Smith SL. Technologies for imaging neural activity in large volumes. Nat Neurosci. 2016;19:1154–1164. doi: 10.1038/nn.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krzic U, Gunther S, Saunders TE, Streichan SJ, Hufnagel L. Multiview light-sheet microscope for rapid in toto imaging. Nat Methods. 2012;9:730–733. doi: 10.1038/nmeth.2064. [DOI] [PubMed] [Google Scholar]
- 51.Tomer R, Lovett-Barron M, Kauvar I, Andalman A, Burns VM, Sankaran S, Grosenick L, Broxton M, Yang S, Deisseroth K. SPED light sheet microscopy: Fast mapping of biological system structure and function. Cell. 2015;163:1796–1806. doi: 10.1016/j.cell.2015.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Keller PJ, Ahrens MB. Visualizing whole-brain activity and development at the single-cell level using light-sheet microscopy. Neuron. 2015;85:462–483. doi: 10.1016/j.neuron.2014.12.039. [DOI] [PubMed] [Google Scholar]
- 53.Chen F, Tillberg PW, Boyden ES. Expansion microscopy. Science. 2015;347:543–548. doi: 10.1126/science.1260088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.