Abstract
Treacher Collins syndrome (TCS) is a severe congenital disorder characterized by craniofacial malformations, including cleft palate, hypoplasia of the facial bones, downward slanting of the palpebral fissures and malformation of the external and middle ear. Worldwide, 90% of cases of TCS are caused by mutations in the TCOF1 gene, which are inherited via an autosomal dominant pattern, while <2% cases are caused by POLR1D and POLR1C genes, which are inherited via autosomal dominant and autosomal recessive patterns, respectively. The present study describes the clinical findings and molecular diagnostics of a Chinese family with TCS. TCS was diagnosed in a 9-year-old female Chinese proband and her mother, while no craniofacial abnormalities were apparent in other family members. Exons of the TCOF1 gene and segregation analysis were used to examine causative mutations using the Sanger sequencing approach. A single novel heterozygous mutation in TCOF1 exon 3 splicing site c.165-1G>A was detected in the proband. Furthermore, the same mutation was identified in her mother, but not in other family members. These results suggest that c.165-1G>A is a novel heterozygous mutation of the TCOF1 gene that caused the development of TCS in the proband and her mother. The TCOF1 mutation that was identified in proband was inherited from her mother and so can be considered as de novo mutation.
Keywords: Treacher Collins syndrome, TCOF1, mutation
Introduction
Treacher Collins syndrome (TCS) is a severe congenital disorder characterized by craniofacial malformation that occurs in 1 out of 50,000 live births (1–4). The condition was first described by Thompson in 1846, however TCS was named after E. Treacher Collins, who described the essential components of the condition in 1900 (5). The first extensive review of the condition was performed by Franceschetti and Klein in 1949, who used the term mandibulofacial dysostosis (MFD) to describe relative clinical features (6–8). The major characteristics of TCS include cleft palate, hypoplasia of the facial bones, the mandible and zygomatic complex, downward slanting of the palpebral fissures and malformation of the external and middle ear (9–12). Patients affected by TCS require management strategies or treatment plans for hearing, respiration, a variety of malformations, bad overall quality of life and mental health, which describes a great burden to individuals, families and society (13).
TCOF1 mutations occur in >90% of cases of TCS and are inherited via an autosomal dominant pattern (12). Over the past decade, two additional gene mutations have been reported in <2% of TCS patients: POLR1D, which is autosomal dominant, and POLR1C, which is inherited via autosomal recessive pattern (14–17). Furthermore, >60% of patients with TCS have no previous family history of the disease and TCS arises as the result of de novo mutations (6,17). Affected individuals may transmit the defect to each child with a 50% probability according to Mendelian laws of genetics, which emphasizes the importance of genetic counseling to affected individuals (5). In the present study, the clinical findings and molecular diagnosis of a Chinese family with TCS are reported.
Materials and methods
Patients
A 9-year-old Chinese girl (the proband; Fig. 1) and her 40-year-old mother (Fig. 2) from Jiangsu, China, were diagnosed with TCS based on a physical examination in December 2013 at the Department of Otolaryngology Head and Neck Surgery, Chinese PLA 97th Hospital (Beijing, China). No craniofacial abnormalities were apparent in the proband's father, sister or maternal grandfather, grandmother, aunts or uncle.
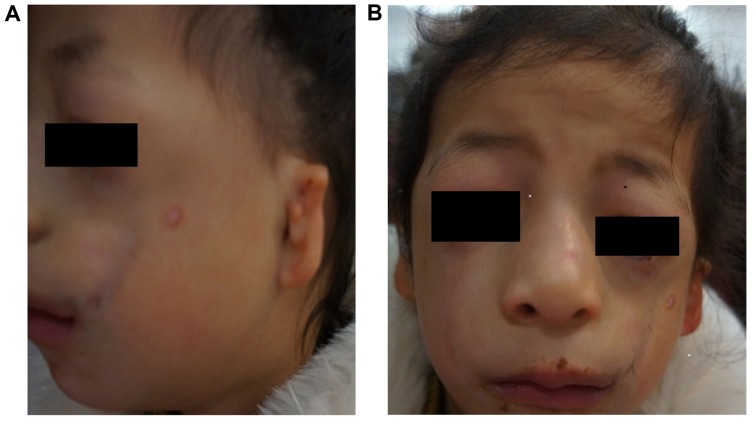
Figure 1.
(A) Facial and (B) ear malformations observed in the proband. Microtia, narrowed nasal cavity, downward slanting of the eyelids, hypoplasia of the zygomatic bone, mandibular hypoplasia and coloboma with eyelashes absent in the medial part of the eyelids were observed.
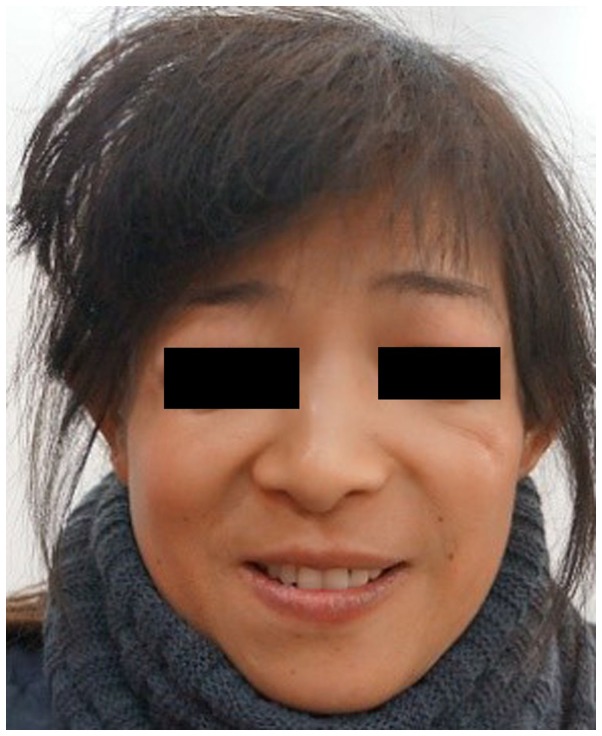
Figure 2.
Facial features of the mother. Narrowed nasal cavity, downward slanting of the eyelids, hypoplasia of the zygomatic bone, mandibular hypoplasia and coloboma with eyelashes absent in the medial part of the eyelids were observed.
Based on the physical examination of the proband, the following clinical features were observed: Conductive hearing loss, microtia, hypoplasia of the middle ear with concomitant atresia of the external auditory canal, narrow nasal cavity, soft palate drooping downward, downward slanting of the eyelids, hypoplasia of the zygomatic bone, mandibular hypoplasia, coloboma with eyelashes absent in the medial part of the eyelids and funnel chest. Similar features, but with a lower level of clinical severity, assessed by slight deformity, including mandibular and zygomatic hypoplasia; partial absence of lower eyelashes; and coloboma of the lower lateral eyelid, were detected in her mother. In addition, the proband and her mother used hearing aids and were undergoing speech therapy. The family pedigree is presented in Fig. 3.
Figure 3.
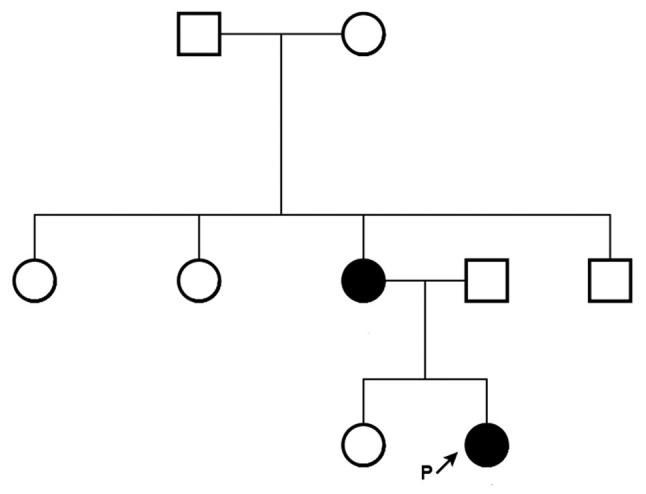
Family pedigree. Squares indicate males, circles indicate females. Black shading indicates a patient with Treacher Collins syndrome.
The present study was approved by the General Hospital of PLA Clinical Human Research Ethics Committee (Beijing, China). All participants provided written informed consent and all procedures complied with the Declaration of Helsinki.
Sanger sequencing
Following the physical examination, peripheral blood samples (3–5 ml) were collected from the elbow vein of the subjects, including the proband and the sister, father, mother, maternal grandfather and maternal grandmother. Blood samples were stored at 4°C for DNA extraction within 3 days or at −80°C for a longer storage times. DNA was extracted from peripheral blood samples using the AxyPrep DNA blood Midi kit (Axygen; Corning Incorporated, Corning, NY, USA) according to the manufacturer's protocol. All TCOF1 exons were sequenced to identify the causative mutation in the proband. A total of 27 exons of TCOF1 were amplified using polymerase chain reaction (PCR) under optimal conditions. Specific primers were designed using Primer3-v.0.4.0 online software (http://bioinfo.ut.ee/) (Table I).
Table I.
Self-designed primers used for TCOF1 gene amplification and sequencing.
| Exon | Direction | Sequence (5′-3′) | Length (bp) |
|---|---|---|---|
| 1 | Forward | GAAAGAGGAGCCGGAAGTGT | 397 |
| Reverse | ACTGAAGTCGCAGTGGGAAG | ||
| 2 | Forward | CCTCTTCTGAACCACCTGTCTA | 233 |
| Reverse | CTTATTCCAAGCTGAGTTGCTT | ||
| 3 | Forward | TGCCTATACTGTGTTTTCACCA | 392 |
| Reverse | CCCAGGGTCTTTTAGGTCTTCT | ||
| 4 | Forward | ACAGAGCTCATTCCTGCAAGTC | 381 |
| Reverse | TAAGATCCCACAACTGGTGACA | ||
| 5 | Forward | TTGAAGGGGTAGTTTACCCAAA | 275 |
| Reverse | CCCTCGTCTAGGTGATGAGAAA | ||
| 6 | Forward | CTTTGATGAGCAGCTGGTTT | 228 |
| Reverse | AGGTTCCTGGAAGGGTTAGAG | ||
| 6A | Forward | AAGCCTTGTGTACTTTTCTGGA | 486 |
| Reverse | AGAGGTGCTCATGGCAGAGT | ||
| 7 | Forward | GTGTGGCCAAAGTATCAGTCAA | 497 |
| Reverse | ACACAGTGAGAGGGGAGTAAGG | ||
| 8 | Forward | GGACTTGTTCTCCCACTCTGG | 500 |
| Reverse | GAAACAGGATGAGGGGAGAG | ||
| 9 | Forward | GAATCGGACAGTGAGGAGGAG | 455 |
| Reverse | GGAAAAGTCAAAACCACAGGAG | ||
| 10 | Forward | CCTGTGGTTTTGACTTTTCCTC | 500 |
| Reverse | GAGATACACAGGATCGGGAGAG | ||
| 11 | Forward | ACCTCACACTGGGACTCTGTCT | 480 |
| Reverse | GGAATTTTCAGAGCTGGTTTTG | ||
| 12 | Forward | ATGGACAACTCGGAGAGCAG | 587 |
| Reverse | GACAAGGGGAAGAGAGGTGTC | ||
| 13 | Forward | GTGAGGCCTGTGTTTTCTGG | 400 |
| Reverse | CTGAGGCTTCTGCACACCTG | ||
| 14 | Forward | CTCAGGTTCACACGCCTATTG | 399 |
| Reverse | CCCCACTATGGCACAACTCT | ||
| 15 | Forward | GGTAGAGAGGAGGACCAGTCAC | 485 |
| Reverse | AGCTCTGATCTGGTGGGTCTT | ||
| 16 | Forward | TAACACCTTTGCCACATCCA | 388 |
| Reverse | GCCTCCCAAAGTGCTAGGAT | ||
| 16A | Forward | CCGACCACGTGCTTATCC | 246 |
| Reverse | ATGGCGAGATTTTCCCTATG | ||
| 17 | Forward | GTGGACCCTTTGCCTTGTAA | 373 |
| Reverse | ACTCAGCCAGTGTCCTGTCC | ||
| 18 | Forward | GCTCTAGATCACCAGCACAGG | 393 |
| Reverse | TAGGAGGCCAGAAAGCCTCT | ||
| 19 | Forward | CAGTTTTGCCCCTTTGACTG | 359 |
| Reverse | CAAACCAAGTGCAGAGGTCA | ||
| 20 | Forward | CATGTGTGCCCCATCTAACA | 474 |
| Reverse | TACAGGTGGGGAAACTGAGG | ||
| 21 | Forward | GTGAGGGACCTGCAGAGAGA | 270 |
| Reverse | CTGAGGGATCGGGTAGACAG | ||
| 22 | Forward | AGGGCAGGGTGATCCTAGAG | 298 |
| Reverse | CTGTTTTAGGGGACAACATGC | ||
| 23 | Forward | ATTGGTGGAAAGGTGTGAGC | 837 |
| Reverse | AGGAATGAGACCAGGTGCTG | ||
| 24 | Forward | CTGGGATTGCAGGAATGAAC | 398 |
| Reverse | GGTGTGTCACAACCCCTGAC | ||
| 25 | Forward | CGCTGCAGACCCAGTATCTA | 250 |
| Reverse | TCAGGTCTGCCTGGCTCTCT |
PCR amplification was performed in a 25 µl reaction volume using the Taq PCR Master mix (Biomed Gene Technology Co., Ltd., Beijing, China) according to the manufacturer's protocol. Thermocycling conditions were as follows: Denaturation at 95°C for 5 min followed by 14 cycles of 95°C for 30 sec, annealing at 60°C for 30 sec and extension at 72°C for 45 sec, followed by 21 cycles at 95°C for 30 sec, annealing at 56°C for 30 sec and extension at 72°C for 45 sec. The reaction was completed with a final extension step at 72°C for 7 min. PCR products were purified and sequenced. Sequencing was performed according to the manufacture's protocol using an ABI PRISM Big Dye Terminator v3.1 Cycle Sequencing-ready Reaction kit (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA) with an ABI 3130 Genetic analyzer (Applied Biosystems; Thermo Fisher Scientific, Inc.).
Mutational analysis
Sanger sequencing reads were mapped to a human genome reference sequence (hg19; University of California, Santa Cruz, CA, USA) using the Mutation Surveyor (v.5.0.2; SoftGenetics, LLC, State College, PA, USA). Consequently, the frequency of each variant was obtained from dbSNP database (version 132, http://www.ncbi.nlm.nih.gov/snp/); all variants with a frequency >1% were filtered. Finally, the possible causative variants were predicted using the combined annotation dependent depletion tool (http://cadd.gs.washington.edu/) for missense variants. To validate candidate mutations, phenotype-genotype co-segregation analysis was performed in family members, including the proband and the sister, father, mother, maternal grandfather and maternal grandmother, using the Sanger sequencing results (Figs. 3 and 4).
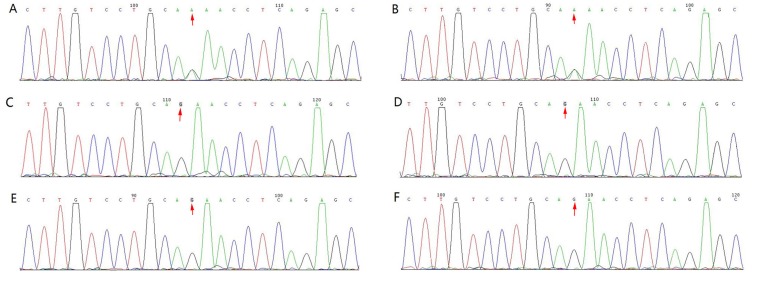
Figure 4.
Pedigree and sequence analysis of TCOF1 mutations in (A) P and her (B) mother, (C) father, (D) sister, (E) grandfather and (F) grandmother. The results of Sanger sequencing demonstrate heterozygous mutations in P and her mother. P, proband.
Results
Clinical features
Prominent TCS features were observed in the proband (Fig. 1). Furthermore, similar features with lower clinical severity were observed in her mother, including conductive hearing loss and deformities in face and ear (Fig. 2).
Sequencing and diagnosis
One novel heterozygous mutation was identified in the TCOF1 exon 3 splicing site c.165-1G>A (Fig. 4) of the proband and her mother. Furthermore, segregation analysis confirmed the co-segregation of this TCOF1 exon 3 coding region mutation (Fig. 4). No apparent craniofacial abnormalities were observed in the other family members assessed. In addition, their hearing was normal and there were no changes in this coding region post-sequencing (Fig. 4).
Discussion
TCS is associated with autosomal dominant mutations in the TCOF1 gene, which is located on chromosome 5q32-q33 (6,10,18). TCOF1 encodes a 144 kDa nucleolar phosphoprotein, Treacle, which serves a role in ribosomal DNA gene transcription via interacting with upstream binding factors and RNA polymerase I in the nucleolus (2). TCOF1 is broadly expressed throughout the embryo, with particularly strong activity in the neuroepithelium where it serves an essential role in cell survival (2,11,19). Treacle participates in ribosome biogenesis by controlling preribosomal (pre-r)RNA synthesis or by pre-rRNA processing (8,12). Dixon et al (11) suggested that general cranio skeletal hypoplasia observed in individuals with TCS is caused by a deficiency in neural crest cells, rather than neural crest cell migration defect. TCOF1/treacle is essential for neural crest cell formation, neuroepithelial survival and neural crest cell proliferation (11,20). However, the biological role of Treacle remains to be fully elucidated.
It was previously believed that the TCOF1 gene included 25 exons, 49–561 bp in length (21). In 2004, two additional exons were discovered by So et al (22): 6A, 231 bp, situated between exons 6 and 7; and 16A, 108 bp, localized between exons 16 and 17 (21). Pathogenic mutations in the TCOF1 gene are spread throughout its coding region and typically comprise point mutations or small frame shift deletions and insertions, the majority of which are family-specific (23). The existence of mutational hot spots in TCOF1 has been suggested, indicating that exons 23 and 24 are responsible for ~1/3 of all known pathogenic changes (23). The majority of mutations responsible for TCS are localized in exons, mainly in the hot spots of exons 10, 15, 16, 23 and 24 (21,24). The TCOF1 gene mutations include missense, nonsense, small deletions and duplications. The most common classes of TCOF1 alleles are small deletions (60%) and duplications (25%), resulting in frame shift (17).
Multiple exons have been identified within the TCOF1 gene and different splicing patterns result in several variants of the mutant gene. So far, >200 mutations have been identified (2). Combined analysis of the variants and clinical features has not revealed a clear association between genotype and phenotype (2). Spontaneous mutations can occur first in the proband or be inherited from a parent. There is no gender predilection and mutations can be splice site, nonsense or deletion variants. All mutations lead to the insertion of a premature termination codon (2,17,25).
In the present study, the proband was diagnosed based on physical examination; and TCS was subsequently confirmed using molecular analysis. Briefly, one novel heterozygous mutation in the TCOF1 gene was identified. To the best of our knowledge, this mutation has not previously been reported in TCS. The Deafness Variation Database (http://deafnessvariationdatabase.org) and Exome Aggregation Consortium (http://exac.broadinstitute.org) databases were also searched and the mutation was not listed. As such, the mutation described in the present study, chr5:149743675:G>A; NM_000356.3:c.165-1G>A, appears to be a novel TCOF1 mutation. The mutation is localized in exon 3 of the TCOF1 gene and is a splice site mutation that can lead to exon 3 being skipped in the matured mRNA. This results in the formation of a truncated TCOF1 protein with loss of function.
The phenotypic variability observed in TCS makes diagnosis challenging. Although there is no clear explanation yet, genotype and phenotype discordance have been suggested by some studies. In some individuals, phenotypic expression of TCS is so mild that it is near impossible to diagnose based on physical examination alone (26). In contrast, some patients may succumb to respiratory distress soon after birth due to the severity of TCS symptoms (26). More than 60% of patients with TCS have no positive family history and the condition is thought to arise from de novo mutations (5). At present, the reason for the differences in TCS presentation remains unknown. The wide variation in symptoms has been attributed to modifier genes, epigenetic factors and the role of wild-type alleles (26–28). Some studies have demonstrated that patient phenotype is not dependent on the type or localization of the mutation responsible (21,23). Identical mutations in the same family may cause variable expressivity in different individuals, variable expressivity of TCOF1 mutations may not be a consequence of mutational heterogeneity and phenotypic expression of this disorder may be modified by combined effect of genetic, environmental and stochastic factors, while there are indications of increased severity over generations phenotypic expression can not modified by the gender of the parent (29,30). In the present study, with the exception of her mother, the proband's family did not have any features of TCS and no mutations were identified in the same region. Since the novel TCOF1 gene mutation was detected in the same region in the mother as in the proband, it follows that this mutation initially occurred in the mother and was inherited by the proband with increased severity.
In conclusion, in the present study a novel pathogenic mutation of the TCOF1 gene was identified in a proband with TCS and her mother. The results suggest that this mutation could be passed on to the next generation in an autosomal dominant manner. As a result, the progeny of the proband and her mother have a 50% risk of suffering from TCS and therefore require genetic counseling.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- MD
mandibulofacial dysostosis
- TCS
Treacher Collins syndrome
Funding
The present study was supported by the National Natural Science foundation of China (grant no. 81030017).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
HY and DH contributed to the conception and design of the study. ZY, YL, YW, XZ, HD, JC performed the experiments. ZY and YL analyzed the data. ZY prepared the manuscript. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
The present study was approved by the General Hospital of PLA Clinical Human Research Ethics Committee (Beijing, China). All participants provided written informed consent.
Patient consent for publications
All participants provided written informed consent.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Rosa F, Coutinho MB, Ferreira JP, Sousa CA. Ear malformations, hearing loss and hearing rehabilitation in children with Treacher Collins syndrome. Acta Otorrinolaringol Esp. 2016;67:142–147. doi: 10.1016/j.otoeng.2016.05.001. (In English, Spanish) [DOI] [PubMed] [Google Scholar]
- 2.Sakai D, Dixon J, Achilleos A, Dixon M, Trainor PA. Prevention of Treacher Collins syndrome craniofacial anomalies in mouse models via maternal antioxidant supplementation. Nat Commun. 2016;7:10328. doi: 10.1038/ncomms10328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luquetti DV, Hing AV, Rieder MJ, Nickerson DA, Turner EH, Smith J, Park S, Cunningham ML. ‘Mandibulofacial dysostosis with microcephaly’ caused by EFTUD2 mutations: Expanding the phenotype. Am J Med Genet A. 2013;161A:1–113. doi: 10.1002/ajmg.a.35696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vincent M, Collet C, Verloes A, Lambert L, Herlin C, Blanchet C, Sanchez E, Drunat S, Vigneron J, Laplanche JL, et al. Large deletions encompassing the TCOF1 and CAMK2A genes are responsible for Treacher Collins syndrome with intellectual disability. Eur J Hum Genet. 2014;22:52–56. doi: 10.1038/ejhg.2013.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mohan RP, Verma S, Agarwal N, Singh U. Treacher Collins syndrome: A case report. BMJ Case Rep 2013. 2013;pii doi: 10.1136/bcr-2013-009341. bcr2013009341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sakai D, Trainor PA. Treacher Collins syndrome: Unmasking the role of Tcof1/treacle. Int J Biochem Cell Biol. 2009;41:1229–1232. doi: 10.1016/j.biocel.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mehrotra D, Hasan M, Pandey R, Kumar S. Clinical spectrum of Treacher Collins syndrome. J Oral Biol Craniofac Res. 2011;1:36–40. doi: 10.1016/S2212-4268(11)60009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Renju R, Varma BR, Kumar SJ, Kumaran P. Mandibulofacial dysostosis (Treacher Collins syndrome): A case report and review of literature. Contemp Clin Dent. 2014;5:532–534. doi: 10.4103/0976-237X.142826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shetty SB, Thomas A, Pidamale R. Treacher Collins syndrome: A case report and a brief review on diagnostic aids. Int J Clin Pediatr Dent. 2011;4:235–239. doi: 10.5005/jp-journals-10005-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andrade EC, Junior VS, Didoni AL, Freitas PZ, Carneiro AF, Yoshimoto FR. Treacher Collins Syndrome with choanal atresia: A case report and review of disease features. Braz J Otorhinolaryngol. 2005;71:107–110. doi: 10.1016/S1808-8694(15)31296-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dixon J, Edwards SJ, Anderson I, Brass A, Scambler PJ, Dixon MJ. Identification of the complete coding sequence and genomic organization of the Treacher Collins syndrome gene. Genome Res. 1997;7:223–234. doi: 10.1101/gr.7.3.223. [DOI] [PubMed] [Google Scholar]
- 12.Weiner AM, Scampoli NL, Calcaterra NB. Fishing the molecular bases of Treacher Collins syndrome. PLoS One. 2012;7:e29574. doi: 10.1371/journal.pone.0029574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geirdal AØ, Saltnes SS, Storhaug K, Åsten P, Nordgarden H, Jensen JL. Living with orofacial conditions: Psychological distress and quality of life in adults affected with Treacher Collins syndrome, cherubism, or oligodontia/ectodermal dysplasia-a comparative study. Qual Life Res. 2015;24:927–35. doi: 10.1007/s11136-014-0826-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schaefer E, Collet C, Genevieve D, Vincent M, Lohmann DR, Sanchez E, Bolender C, Eliot MM, Nürnberg G, Passos-Bueno MR, et al. Autosomal recessive POLR1D mutation with decrease of TCOF1 mRNA is responsible for Treacher Collins syndrome. Genet Med. 2014;16:720–724. doi: 10.1038/gim.2014.12. [DOI] [PubMed] [Google Scholar]
- 15.Weaver KN, Watt KE, Hufnagel RB, Navajas Acedo J, Linscott LL, Sund KL, Bender PL, König R, Lourenco CM, Hehr U, et al. Acrofacial dysostosis, Cincinnati type, a mandibulofacial dysostosis syndrome with limb anomalies, is caused by POLR1A dysfunction. Am J Hum Genet. 2015;96:765–774. doi: 10.1016/j.ajhg.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noack Watt KE, Achilleos A, Neben CL, Merrill AE, Trainor PA. The roles of RNA polymerase I and III subunits Polr1c and Polr1d in craniofacial development and in Zebrafish models of Treacher Collins syndrome. PLoS Genet. 2016;12:e1006187. doi: 10.1371/journal.pgen.1006187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conte C, D'Apice MR, Rinaldi F, Gambardella S, Sangiuolo F, Novelli G. Novel mutations of TCOF1 gene in European patients with Treacher Collins syndrome. BMC Med Genet. 2011;12:125. doi: 10.1186/1471-2350-12-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Positional cloning of a gene involved in the pathogenesis of Treacher Collins syndrome The Treacher Collins syndrome collaborative group. Nat Genet. 1996;12:130–136. doi: 10.1038/ng0296-130. [DOI] [PubMed] [Google Scholar]
- 19.Chang CC, Steinbacher DM. Treacher Collins syndrome. Semin Plast Surg. 2012;26:83–90. doi: 10.1055/s-0032-1320066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dixon J, Jones NC, Sandell LL, Jayasinghe SM, Crane J, Rey JP, Dixon MJ, Trainor PA. Tcof1/Treacle is required for neural crest cell formation and proliferation deficiencies that cause craniofacial abnormalities. Proc Natl Acad Sci USA. 2006;103:13403–13408. doi: 10.1073/pnas.0603730103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marszalek-Kruk BA, Wojcicki P, Smigiel R, Trzeciak WH. Novel insertion in exon 5 of the TCOF1 gene in twin sisters with Treacher Collins syndrome. J Appl Genet. 2012;53:279–282. doi: 10.1007/s13353-012-0091-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.So RB, Gonzales B, Henning D, Dixon J, Dixon MJ, Valdez BC. Another face of the Treacher Collins syndrome (TCOF1) gene: Identification of additional exons. Gene. 2004;328:49–57. doi: 10.1016/j.gene.2003.11.027. [DOI] [PubMed] [Google Scholar]
- 23.Splendore A, Jabs EW, Felix TM, Passos-Bueno MR. Parental origin of mutations in sporadic cases of Treacher Collins syndrome. Eur J Hum Genet. 2003;11:718–722. doi: 10.1038/sj.ejhg.5201029. [DOI] [PubMed] [Google Scholar]
- 24.Splendore A, Silva EO, Alonso LG, Richieri-Costa A, Alonso N, Rosa A, Carakushanky G, Cavalcanti DP, Brunoni D, Passos-Bueno MR. High mutation detection rate in TCOF1 among Treacher Collins syndrome patients reveals clustering of mutations and 16 novel pathogenic changes. Hum Mutat. 2000;16:315–322. doi: 10.1002/1098-1004(200010)16:4<315::AID-HUMU4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 25.Trainor PA. Craniofacial birth defects: The role of neural crest cells in the etiology and pathogenesis of Treacher Collins syndrome and the potential for prevention. Am J Med Genet A. 2010;152A:1–2994. doi: 10.1002/ajmg.a.33454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ulusal S, Gurkan H, Vatansever U, Kurkcu K, Tozkir H, Acunas B. A case of Treacher Collins syndrome. Balkan J Med Genet. 2013;16:77–80. doi: 10.2478/bjmg-2013-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horiuchi K, Ariga T, Fujioka H, Kawashima K, Yamamoto Y, Igawa H, Sugihara T, Sakiyama Y. Mutational analysis of the TCOF1 gene in 11 Japanese patients with Treacher Collins Syndrome and mechanism of mutagenesis. Am J Med Genet A. 2005;134:363–367. doi: 10.1002/ajmg.a.30357. [DOI] [PubMed] [Google Scholar]
- 28.Schlump JU, Stein A, Hehr U, Karen T, Möller-Hartmann C, Elcioglu NH, Bogdanova N, Woike HF, Lohmann DR, Felderhoff-Mueser U, et al. Treacher Collins syndrome: Clinical implications for the paediatrician-a new mutation in a severely affected newborn and comparison with three further patients with the same mutation, and review of the literature. Eur J Pediatr. 2012;171:1611–1618. doi: 10.1007/s00431-012-1776-7. [DOI] [PubMed] [Google Scholar]
- 29.Geirdal AO, Saltnes SS, Storhaug K, Asten P, Nordgarden H, Jensen JL. Living with orofacial conditions: Psychological distress and quality of life in adults affected with Treacher Collins syndrome, cherubism, or oligodontia/ectodermal dysplasia-a comparative study. Qual Life Res. 2015;24:927–935. doi: 10.1007/s11136-014-0826-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teber OA, Gillessen-Kaesbach G, Fischer S, Böhringer S, Albrecht B, Albert A, Arslan-Kirchner M, Haan E, Hagedorn-Greiwe M, Hammans C, et al. Genotyping in 46 patients with tentative diagnosis of Treacher Collins syndrome revealed unexpected phenotypic variation. Eur J Hum Genet. 2004;12:879–890. doi: 10.1038/sj.ejhg.5201260. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.