Abstract
Cervical cancer (CC) is one of the most malignant tumors that affect women. Recent studies have reported that microRNAs (miRs) serve important roles in CC. The aim of the present study was to investigate the role of miR-218 in CC and to verify its underlying mechanism. The results of reverse transcription-quantitative polymerase chain reaction (RT-qPCR) revealed that miR-218 was dramatically downregulated in CC tissues and cell lines. Furthermore, the expression of Gli3 and Ki67 was measured using RT-qPCR and the results revealed that levels of these proteins were negatively correlated with miR-218 in CC tissues. The protein expression levels were determined by western blotting. Then SiHa cell line was used to investigate the mechanism of CC. Following cell transfection, cell apoptosis and cycle analyses were performed using the flow cytometry. The results revealed that miR-218 overexpression significantly inhibited cell proliferation, apoptosis and cell cycle. Additionally, luciferase reporter assay revealed that Gli3 may be a novel and direct target of miR-218 in CC. Taken together, the results of the present study suggest that miR-218 overexpression or Gli3 knockdown may have potential as therapeutic strategies for the treatment of CC.
Keywords: microRNA, microRNA-218, cervical cancer, Gli3
Introduction
Cervical cancer (CC) is the fourth most common cancer affecting women worldwide, with an annual mortality of ~270,000 (1,2). Standard therapies for CC, including radiotherapy, chemotherapy and surgical intervention, are often less effective for patients in the advanced stages of disease (3). The mechanisms of CC remain unclear and gaining a better understanding of tumor progression and identifying novel biomarkers for the disease is of great importance.
MicroRNAs (miRNAs or miRs) are a group of small non-coding RNAs (~22–25 nucleotides in length) that negatively regulate their target genes via binding to complementary sequences of the 3′-untranslated region (3′-UTR) (4). A number of studies have revealed that miRNAs serve important roles in the development of CC and are associated with tumor suppressing and oncogenic activities (5). miR-218 is a miRNA that has a tumor-suppressing effect in a number of cancers (6–8). The precursors of miR-218 are mir-218-1 and mir-218-2, which are located in the Slit2 and Slit3 introns and can be expressed together with Slit2 and Slit3 (6–7,9). Expression of the miR-218 precursors is downregulated in several human malignant tumors and has a significant tumor suppressive effect (7,10–12).
miR-218 has been reported to be downregulated in tumor tissues and sera of patients with CC (12). Additionally, miR-218 downregulation is associated with tumor invasion (13). A number of studies have reported that the expression of miR-218, which serves a tumor-suppressive role in CC, is affected by human papillomavirus (HPV) infection (14–19). A previous study on HPV, a major carcinogenic risk factor for CC, reported that the expression of miR-218 and its host gene tumor suppressor gene SLIT2 was lower in HPV-16 and −18 positive cells compared with normal cervical tissue and HPV negative cells (20). It was suggested that the expression of miR-218 in CC tissues was lower compared with normal cervical tissues and that there are certain strains of HPV that are associated with a greater risk of CC, may result in further downregulation (20). miR-218 is downregulated in a number of malignant tumors, including lung cancer, breast cancer, thyroid cancer, mesenteric adenocarcinoma, colon cancer, gastric cancer, prostate cancer, glioblastoma and bladder cancer (21). Low miR-218 expression was demonstrated to be associated with tumor cell invasion and migration, and poor prognosis (7,22,23). Furthermore, miR-218 expression is lower in malignant tumors compared with benign tumors (24). Low miR-218 expression is associated with tumor invasion, migration and poor prognosis (7,21–24). Together, these studies suggest that miR-218 serves an important role in the development of tumors. Although these studies link miR-218 with tumor metastasis, the specific mechanisms of miR-218 in tumors remain unclear and require further study. The aim of the present study was to investigate the role and mechanism of miR-218 in the metastasis of CC.
Gli3 serves a key and complex role in the occurrence and development of tumors, participating in multi-gene, multi-step regulatory processes (25). Bai et al (25) reported that Gli3 full-length protein (Gli3-FL) was found in the cytoplasm and Gli3 terminal removal protein (Gli3-TR) was found in the nucleus of tumor epithelial cells in small cell lung cancer. Gli3-FL and Gli3-TR are highly expressed in lung cancer; Gli3-FL is not associated with clinic pathological parameters and survival rate, while Gli3-TR is associated with lymph node metastasis and overall survival. Gli3-TR, together with tumor differentiation and disease stage, can be used as independent prognostic indicators in patients with lung cancer.
In the present study, Gli3 was confirmed as a potential target gene of miR-218 via a series of experiments. The regulatory mechanisms of miR-218 and Gli3 in CC were also investigated and it was demonstrated that miR-218 regulates the proliferation and apoptosis of CC cells by targeting to Gli3 in vitro. These results provide a theoretical basis for miR-218 as a target for CC progression and treatment
Materials and methods
Clinical samples
A total of 112 CC tissues and corresponding adjacent normal tissues were obtained from patients with cervical cancer (age range, 25–55 years old), who had not undergone radiotherapy or chemotherapy, in the First Affiliated Hospital of Bengbu Medical College (Bengbu, China) from 2014 to 2016. All tissue samples were immediately frozen in liquid nitrogen and stored at −80°C until use. All participants provided signed statements of informed consent according to the principles of the Declaration of Helsinki. The study protocol was approved by the Ethics Committee of the First Affiliated Hospital of Bengbu Medical College.
Cell lines and transfection
A human cervical epithelial cell line (HCvEpCs) and Hela, SiHa and C33A CC cell lines were obtained from the American Type Culture Collection (Manassas, VA, USA). Cells were grown in Dulbecco's Modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) at 37°C in an atmosphere containing 5% CO2. SiHa cells were seeded in 96-well plates at a density of 2×105 cells/well and grown to 70% confluence. Cells were then transfected with negative control miRNA (miR-NC), miR-218 mimics or miR-218 inhibitor using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Subsequently, the transfected cells were incubated for a further 72 h. The miR-218 sequences transfected were 5′-TGCATGGTTAGATCAAGCACAAGGG-3′ and 5′-CTCGTCTTATTTCCGTGACTGTTTT-3′.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
RT-qPCR was performed to measure the expression of miR-218 in CC cell lines and frozen tissue specimens. miRNAs were isolated using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.) and the concentration of total RNA was measured. Reverse transcription of the extracted RNA was completed by the two-step method using a SuperScript™ RT-PCR System with Platinum™ Taq DNA Polymerase kit (Fermentas; Thermo Fisher Scientific, Inc.). The reaction conditions were as follows: 70°C for 10 min, 4°C for 2 min, 42°C for 60 min, 70°C for 10 min and put temporary on hold at −80°C. The expression of miR-218, Ki67 and Gli3 was measured using the TaqMan miRNA assay kit (Invitrogen; Thermo Fisher Scientific, Inc.). The PCR thermocycling conditions were as follows: 97°C for 5 min, followed by 35 cycles at 95°C for 30 sec, 65°C for 30 sec and 73°C for 1 min, 73°C for 10 min, 4°C forever. PCR primers used were as follows: Gli3, forward 5′-TGGTTACATGGAGCCCCACTA-3′ and revers 5′-GAATCGGAGATGGATCGTAATGG-3′; KI67, forward 5′-GCCTGCTCGACCCTACAGA-3′ and reverse 5′-GCTTGTCAACTGCGGTTGC-3′; cleaved-caspase-3, forward 5′-AGAGGGGATCGTTGTAGAAGTC-3′ and reverse 5′-ACAGTCCAGTTCTGTACCACG-3′; cleaved-poly ADP-ribose polymerase (PARP), forward 5′-TGGAAAAGTCCCACACTGGTA-3′ and reverse 5′-AAGCTCAGAGAACCCATCCAC-3′; cyclin B1, forward 5′-AATAAGGCGAAGATCAACATGGC-3′ and reverse 5′-TTTGTTACCAATGTCCCCAAGAG-3′; cyclin D1, forward 5′-CAATGACCCCGCACGATTTC-3′ and reverse 5′-CATGGAGGGCGGATTGGAA-3′; and GAPDH, forward 5′-CTGGGCTACACTGAGCACC-3′ and reverse 5′-AAGTGGTCGTTGAGGGCAATG-3′. The relative expression of miR-218, Ki67, Gli3, cleaved-caspase-3, cleaved-PARP, cyclin B1 and cyclin D1 was calculated. The 2−ΔΔCq method was used for relative quantification (26). U6 was used as an endogenous control for miRNA quantification and GAPDH was used as an internal reference.
Cell proliferation analysis
A Cell Counting Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan) assay was performed to assess the proliferative ability of SiHa cells. At 24, 48 and 72 h following transfection, 2×103 cells/well were seeded into 96-well plates and 10 µl of CCK-8 solution was added to assess cell viability according to the manufacturer's protocol. The optical density (OD) was measured at 450 nm using a plate reader.
Apoptosis analysis
To elucidate the mechanisms of miR-218 in CC cells, a flow cytometry assay was performed to measure apoptosis. Harvested cells were incubated with Annexin V-phycoerythrin (PE)/7-amino-actinomycin D (7-AAD; KGA1015; Nanjing KeyGen Biotech Co., Ltd., Nanjing, China) for 15 min at room temperature and samples were analyzed using FACSCalibur flow cytometry (BD Biosciences, Franklin Lakes, NJ, USA).
Cell cycle analysis
Cell cycle distribution was assessed using flow cytometry. Following transfection, SiHa cells were harvested by trypsinization and washed twice with ice-cold PBS, following which they were fixed with 70% ethanol at 4°C overnight. Fixed cells were rehydrated in PBS and stained with Annexin V-PE/7-AAD for 15 min at room temperature in the dark and analyzed using FACSCalibur. Subsequently, the percentage of cells in the G1, S and G2 phase was measured using PV ELITE software (version 18.0; Integraph Corporation, Madison, AL, USA).
Dual luciferase reporter assay
A luciferase reporter assay was performed using a Dual-Luciferase Reporter Assay reagent (Promega, Madison, WI, USA) according to the manufacturer's instructions. A total of 3.5×104 SiHa cells/well were seeded in 24-well plates overnight and pGL3-Gli3-3′UTR-wild type (WT) or pGL3-Gli3-3′UTR-mutant (mut) and miR-218 or miR-control vectors were co-transfected into the cells using Lipofectamine® 2000. Three independent experiments were performed and data are presented as the mean ± standard deviation.
Bioinformatics analysis
Bioinformatics analysis with TargetScan (version 6.2; http://genes.mit.edu/targetscan) was conducted to predict its target gene.
Western blotting
Western blotting was performed to measure the expression of Gli3, cleaved-caspase-3 and cleaved-PARP proteins. SiHa cells were homogenized in lysis buffer (Beyotime Institute of Biotechnology, Haimen, China) and the total protein was extracted by centrifuged at 16,000 × g at 4°C for 5 min. The proteins (20 µg/lane) were quantified according to the protocol of the BCA kit (cat. no. AR0146; Wuhan Boshide Biological Engineering Co. Ltd., Wuhan, China), separated by 10% SDS-PAGE and transferred onto a polyvinylidene fluoride membrane. Membranes were subsequently incubated with primary antibodies against Gli3 (1:1,000, PA5-19822; Invitrogen, Thermo Fisher Scientific, Inc.), cleaved-caspase-3 (1:500; ab49822) cleaved-PARP (1:1,000; ab32064), cyclin B1 (1:500; ab72), cyclin D1 (1:10,000; ab134175; all Abcam, Cambridge, UK) and GAPDH (1:1,000 dilution; MA5-15738; Invitrogen; Thermo Fisher Scientific, Inc.) for 1 h at room temperature. GAPDH was the internal control. Membranes were then incubated with horseradish peroxidase-conjugated anti-mouse (1:1,000; ab131368) and anti-rabbit (1:1,000; ab191866; Abcam) antibodies at room temperature for 2 h. Finally, membranes were incubated with BeyoECL Plus (Beyotime Institute of Biotechnology) and the detected using Chemi Doc™ XRS+ imaging system (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Statistical analysis
All statistical analyses were performed using GraphPad Prism 6.0 software (GraphPad Software, Inc., La Jolla, CA, USA). Differences between two groups were analyzed using a Student's t-test. Differences between multiple groups were assessed using one-way analysis of variance followed by the Dunnett's post-hoc test. The correlation between miR-218 expression and other genes mRNA expression levels was examined using Pearson's correlation analysis. Each experiment was repeated ≥3 times. Values were presented as the mean ± standard deviation and P<0.05 was considered to indicate a statistically significant difference.
Results
miR-218 expression is negatively correlated with Ki67 and Gli3 in CC tissues
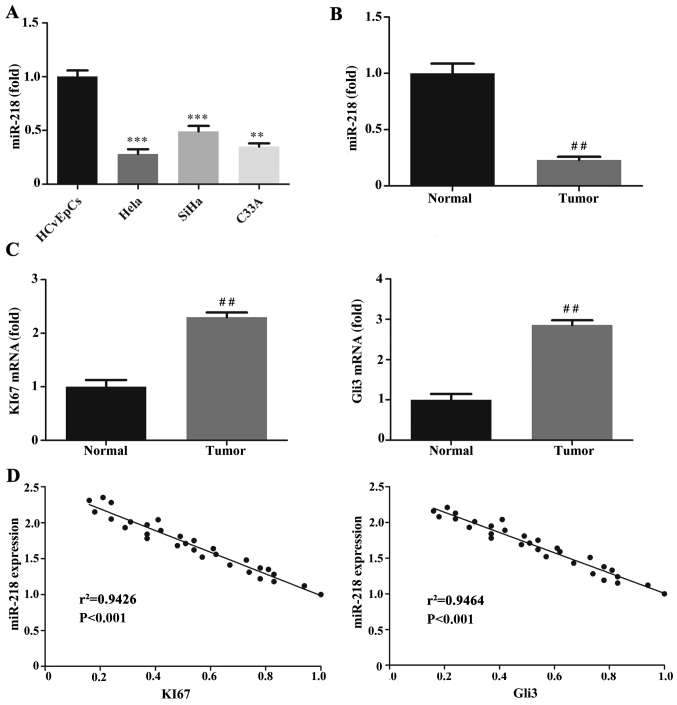
The expression of miR-218 was decreased in the CC cell lines and tissues compared with the HCvEpCs and normal tissues, respectively (Fig. 1A and B). Ki67 and Gli3 were also downregulated in CC tissues compared with normal tissues (Fig. 1C). The correlation between miR-218 and Ki67 and Gli3 expression was assessed using Pearson's correlation analysis and the results revealed that miR-218 was negatively correlated with Ki67 and Gli3 (Fig. 1D).
Figure 1.
Expression of miR-218, KI67 and Gli3 mRNA. RT-qPCR was performed to quantify miR-218 expression in (A) HCvEpCs, Hela, SiHa and C33A cells and (B) CC tumor and normal cervical tissues. (C) RT-qPCR was performed to measure KI67 and Gli3 mRNA expression in CC tumor and normal tissues. (D) Pearson's correlation analysis of miR-218 expression with KI67 or Gli3 mRNA in 112 cervical cancer tissues. KI67, r2=0.9426; Gli3, r2=0.9464. **P<0.01, ***P<0.001 vs. HCvEpCs ##P<0.01 vs. Normal. miR, microRNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; HCvEpCs, normal human cervical cells; CC, cervical cancer.
miR-218 suppresses CC proliferation, cell cycle progression and apoptosis in vitro
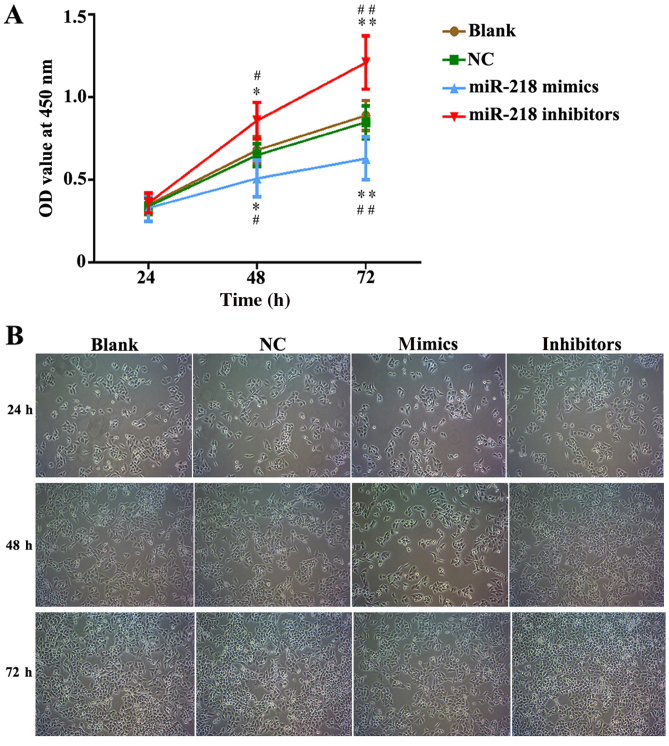
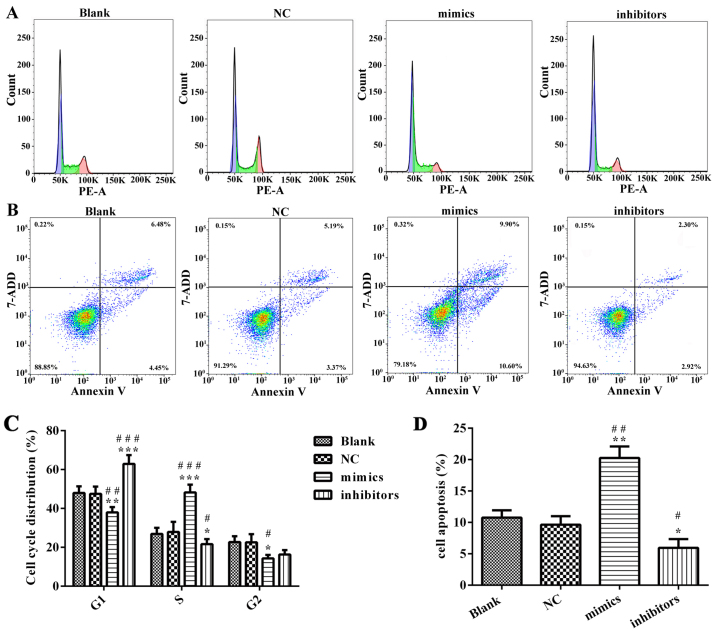
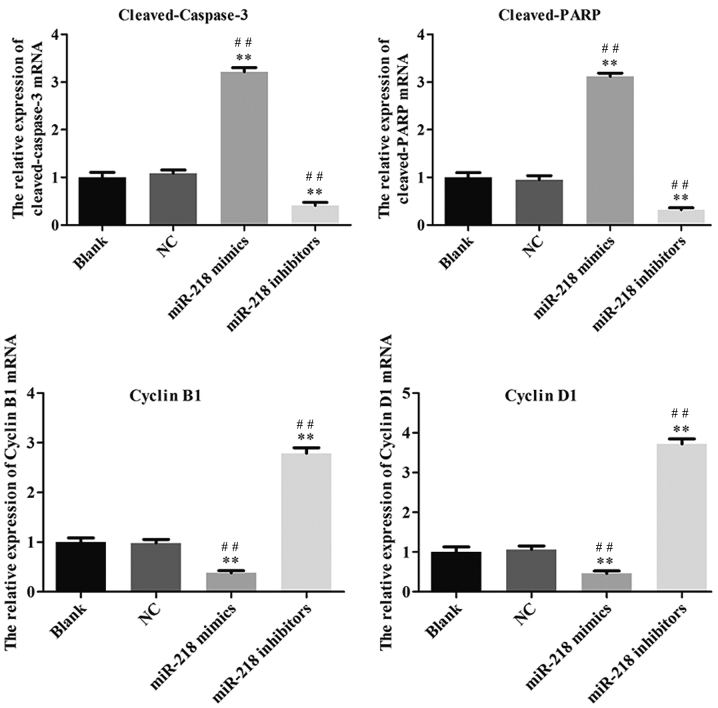
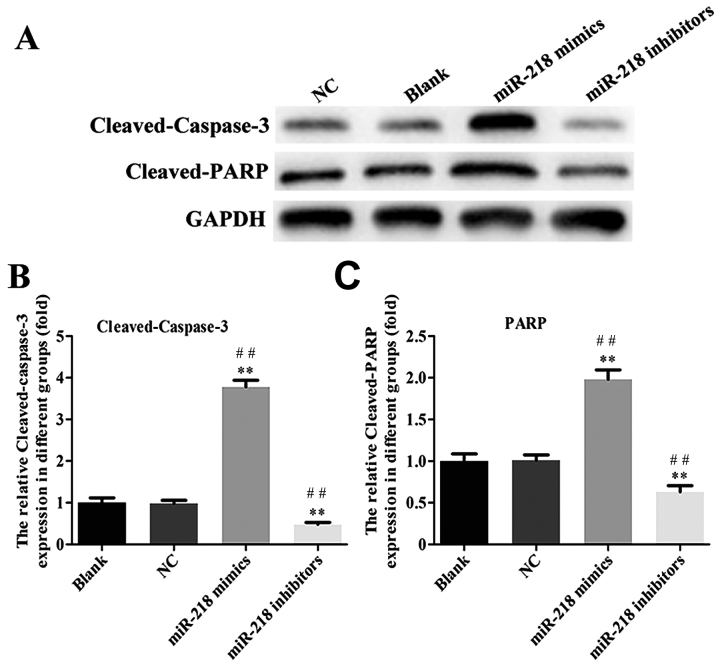
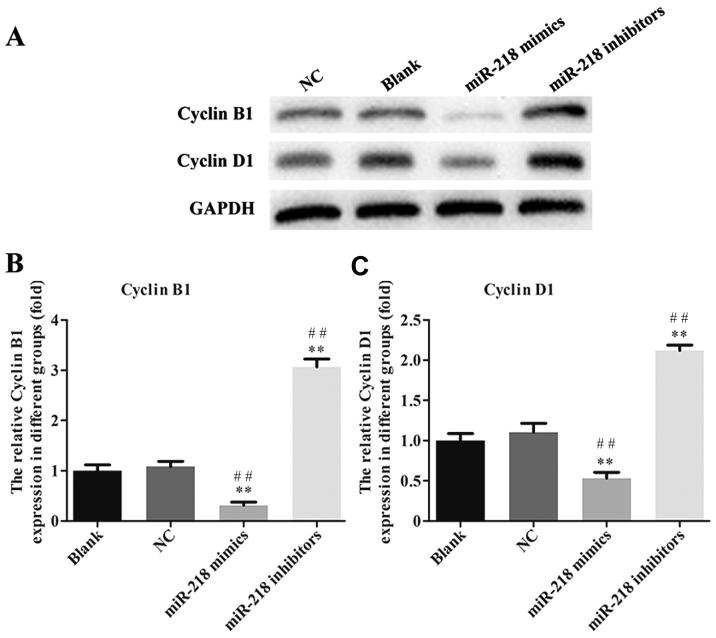
In order to explore the potential role of miR-218 in the pathogenesis of CC, SiHa cells were transfected with miR-218 mimics, NC vectors and inhibitors. The results of a cell viability assay revealed that miR-218 overexpression dramatically suppressed the proliferation of SiHa cells compared with the miR-NC group, while proliferation was significantly suppressed in the inhibitors group (Fig. 2A). Cell morphology was affected as presented in Fig. 2B; the number of cell invasions decreased in miR-218 mimics group and increased in the miR-218 inhibitors group compared with the NC group. Furthermore, the results of flow cytometry revealed that the percentage of cells in the G0/G1 phase was increased in the miR-218 mimics group and decreased in the miR-218 inhibitors group (Fig. 3A). Furthermore, the rate of apoptosis was increased in the miR-218 mimics group, while apoptosis was decreased in the inhibitors group (Fig. 3B). The quantified flow cytometry results demonstrated that a significantly greater number of cells treated with the inhibitor were in the G1 phase compared with the NC and Blank groups (Fig. 3C) and that apoptosis was significantly increased in the mimc group and decreased in the inhibitor group compared with the NC and Blank groups (Fig. 3D). The expression of cleaved-caspase-3, cleaved-PARP, cyclin and cyclin D1 was assessed in transfected SiHa cells. PCR results confirmed that the expression of cleaved-caspase-3 and cleaved-PARP was increased while the expression of cyclin B1 and cyclin D1 was decreased in the miR-218-mimics group at the mRNA (Fig. 4) and protein (Figs. 5 and 6) level. The opposite was observed in the miR-218 inhibitors group (Figs. 4–6).
Figure 2.
miR-218 suppresses cervical cancer proliferation in vitro. (A) Cell Counting Kit-8 analysis was performed for SiHa cells 48 and 72 h following transfection with miR-218 mimics or inhibitors. (B) SiHa cell morphology was assessed at 24, 48 and 72 h following transfection. *P<0.05 and **P<0.01 vs. NC. #P<0.05 and ##P<0.01 vs. Blank. Magnification, ×200. miR, microRNA; NC, negative control; OD, optical density.
Figure 3.
miR-218 suppresses cervical cancer cell cycle progression and apoptosis in vitro. (A) After SiHa cells were transfected with miR-218 inhibitors or mimics, flow cytometry was performed to test the cell cycle progression. (B) Apoptosis was evaluated using flow cytometry. (C) Quantification of cell cycle distribution, including G1-phase, G0-phase and S-phase. (D) Quantification of cell apoptosis. *P<0.05, **P<0.01, ***P<0.001 vs. NC. #P<0.05, ##P<0.01, ###P<0.001 vs. Blank. miR, microRNA; NC, negative control; PE, phycoerythrin; 7-AAD, 7-aminonactinomycin D.
Figure 4.
Reverse transcription-quantitative polymerase chain reaction was performed to assess the expression of cleaved-caspase-3, cleaved-PARP, cyclin B1 and cyclin D1 mRNA in SiHa cells. **P<0.01 vs. NC and ##P<0.01 vs. Blank. PARP, poly ADP-ribose polymerase; NC, negative control; miR, microRNA.
Figure 5.
(A) Western blotting was performed and the expression of (B) cleaved-caspase-3 and (C) cleaved-PARP protein was quantified. **P<0.01 vs. NC and ##P<0.01 vs. Blank. PARP, poly ADP-ribose polymerase; NC, negative control; miR, microRNA.
Figure 6.
(A) Western blotting was performed and the expression of (B) cyclin B1 and (C) cyclin-D1 protein was quantified. **P<0.01 vs. NC and ##P<0.01 vs. Blank. NC, negative control; miR, microRNA.
Gli3 is an important target of miR-218 in CC cells
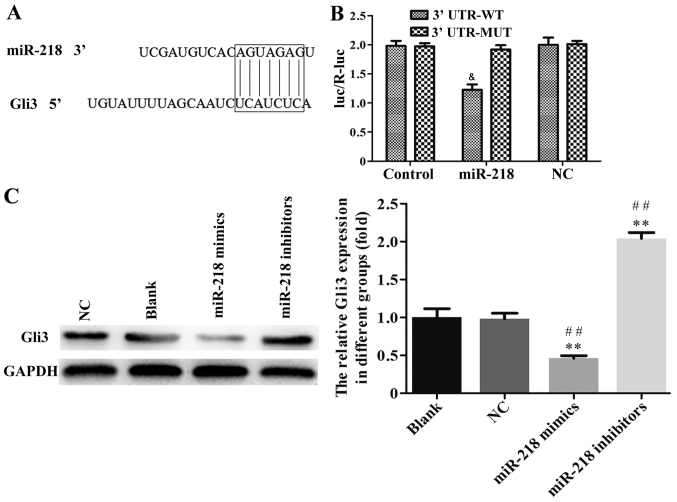
To further investigate the underlying mechanism of miR-218 in the progression of CC, TargetScan 6.2 was used to predict its target gene. As shown in the Fig. 7A, Gli3 may be a target of miR-218. The results of the luciferase activity assay indicated that miR-218 overexpression suppressed the activity of Gli3 WT-3′-UTR in SiHa cells compared with NC group (Fig. 7B). The association between miR-218 and Gli3 expression was further confirmed by western blotting and it was demonstrated that the expression of Gli3 protein was significantly downregulated in the miR-218 mimics group and upregulated in the miR-218 inhibitor group compared with the NC group (Fig. 7C).
Figure 7.
Gli3 is direct downstream target of miR-218. (A) The putative binding sites of miR-218 in the 3′-UTR regions of Gli3. (B) SiHa cells were co-transfected with miR-218 mimics or NC mimics with a luc-Gli3-WT or luc-Gli3-MUT and a dual luciferase activity assay was performed. &P<0.05 vs. miR-218 in the 3′-UTR-MUT group. (C) Western blotting was performed to measure the expression of Gli3 protein in SiHa cells following transfection with miR-218 mimics or inhibitors. **P<0.01 vs. NC and ##P<0.01 vs. Blank. miR, microRNA; UTR, untranslated region; NC, negative control; luc-Gli3-WT, luciferase vector containing the wild-type Gli3 3′-UTR; luc-Gli3-MUT, luciferase vector containing the mutant Gli3 3′-UTR.
Discussion
CC is one of the most prevalent and deadly malignancies in women worldwide (27,28). A World Health Organization report revealed that >270,000 women succumbed to CC in 2014 and >85% of cases occur in less developed regions (28). The morbidity and mortality of CC are high (29) and the age of onset is a decreasing (30,31). In the past few decades, great progress has been made researching treatments for CC (32,33). However, there remains a challenge to improve the survival of CC patients so far.
miRNAs have been reported to serve a role in the initiation, progression and metastasis of CC via regulating cancer cell proliferation, apoptosis, cell cycle arrest, migration and invasion (34,35). As a member of the miRNA family, miR-218 is correlated with tumor progression and poor prognosis (36,37). It has been reported that miR-218 binds to its target mRNA in order to inhibit or promote the formation and development of tumors (38). In previous studies, miR-218 downregulation was identified in the tumor tissues and sera of patients with CC (39). However, it remains unclear whether miR-218 expression is downregulated in CC cells.
The aim of the present study was to investigate the pivotal roles of miR-218 in the progression of CC. The expression of miR-218 in CC tissues and cells was examined and it was demonstrated that miR-218 was significantly decreased in CC tumor tissues compared with adjacent normal tissues. These results are consistent with those observed in CC and normal cervical cell lines. The expression of Ki67 mRNA in CC tissues was demonstrated to be increased and a negative correlation was observed between Ki67 and miR-218. The present study also revealed that, compared with adjacent tissues, Ki67 expression was upregulated in CC tissues, suggesting that miR-218 was negatively correlated with the proliferation marker Ki67. Following transfection with miR-218 mimics, the proliferation ability of CC cells was suppressed, suggesting that miR-218 inhibits the proliferation of CC cells and its downregulation serves a role in the development and progression of CC. Gli3 was predicted and confirmed as a gene target of miR-218 and its expression in CC tissues and transfected CC cell lines was assessed. A previous study indicated that the downregulated expression of Glli3 attenuates pancreatic cancer cell activity and proliferation (40). Gli3 is expressed in a subset of colon cancers and associated with the degree of tumor differentiation (40). Gli3 signaling significantly enhances the tumorigenicity of colon cancer (41). Kang et al (42) reported that, compared with normal tissues, Gli3 is upregulated in colon cancer tissues and Gli3 downregulation inhibits cancer cell proliferation. The downregulated of Gli3 expression was also reported to enhance the sensitivity of colon cancer cells to 5-FU (42). Together, these results suggest that the role of Gli3 in tumor tissue is complex. In the present study, Gli3 mRNA and protein expression decreased dramatically and was inversely correlated with levels of miR-218 in CC tissues. It was demonstrated that miR-218 could inhibit the proliferation, apoptosis and cell cycle progression of CC cells via suppressing Gli3 expression. The present study revealed that miR-218 mimic transfection in CC cells promotes apoptosis and enhances caspase-3 activity. These results suggest that miR-218 is able to inhibit cell growth and regulate tumor progression by enhancing caspase-3 activity and promoting apoptosis in CC. Similarly, it was also demonstrated that miR-218, which is downregulated in CC, inhibits cell growth by blocking the transition from G1 to S phase. These results provide a novel insight into the pathogenesis and potential treatment targets of CC and may have clinical implications.
In summary, miR-218 overexpression in CC cells and tissues can inhibit cancer progression by blocking transition from G0/G1 to S phase, promoting apoptosis, increasing the activity of proteins associated with cell apoptosis and decreasing the expression of cell cycle-associated proteins. The results of the present study suggest that miR-218 serves an important role in the pathogenesis of CC and provide a theoretical basis for the development of targeted therapy for CC.
Acknowledgements
The authors would like to thank the Department of Gynecology, First Affiliated Hospital of Bengbu Medical College.
Funding
Not applicable.
Availability of data and materials
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.
Authors' contributions
JZ contributed to writing the manuscript, study conception and design and the acquisition and analysis of data. SL contributed to study conception and design as well as revising and approving the final version of the manuscript. YL drafted the manuscript and analyzed data. HL collected and interpreted data and revised the final manuscript. YZ contributed to study conception, data analysis and interpretation and drafting the manuscript. QZ contributed to study conception and design, acquisition of data and drafting the manuscript. All authors have read and approved the final version of this manuscript to be published.
Ethics approval and consent to participate
All experimental protocols were performed in accordance with the principles of the Declaration of Helsinki and were approved by the Clinical Research Ethics Committee of the First Affiliated Hospital of Bengbu Medical College (Anhui, China). Prior to enrolment, written informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Lahue BJ, Baginska E, Li SS, Parisi M. Health technology assessment on cervical cancer screening 2000–2014. Int J Technol Assess Health Care. 2015;31:171–180. doi: 10.1017/S0266462315000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Subramanian S, Trogdon J, Ekwueme DU, Gardner JG, Whitmire JT, Rao C. Cost of cervical cancer treatment: Implications for providing coverage to low-income women under the Medicaid expansion for cancer care. Womens Health Issues. 2010;20:400–405. doi: 10.1016/j.whi.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 4.Arbyn M, Castellsague X, de Sanjose S, Bruni L, Saraiya M, Bray F, Ferlay J. Worldwide burden of cervical cancer in 2008. Ann Oncol. 2011;22:2675–2686. doi: 10.1093/annonc/mdr015. [DOI] [PubMed] [Google Scholar]
- 5.Gómez-Gómez Y, Organista-Nava J, Gariglio P. Deregulation of the miRNAs expression in cervical cancer: Human papillomavirus implications. Biomed Res Int 2013. 2013:407052. doi: 10.1155/2013/407052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fish JE, Wythe JD, Xiao T, Bruneau BG, Stainier DY, Srivastava D, Woo S. A Slit/miR-218/Robo regulatory loop is required during heart tube formation in zebrafish. Development. 2011;138:1409–1419. doi: 10.1242/dev.060046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alajez NM, Lenarduzzi M, Ito E, Hui AB, Shi W, Bruce J, Yue S, Huang SH, Xu W, Waldron J, et al. miR-218 suppresses nasopharyngeal cancer progression through downregulation of surviving and SLIT2-ROBO1 pathway. Cancer Res. 2011;71:2381–2391. doi: 10.1158/0008-5472.CAN-10-2754. [DOI] [PubMed] [Google Scholar]
- 8.Small EM, Sutherland LB, Rajagopalan KN, Wang S, Olson EN. MicroRNA-218 regulates vascular patterning by modulation of Slit-Robo signaling. Circ Res. 2010;107:1336–1344. doi: 10.1161/CIRCRESAHA.110.227926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao H, Anand AR, Ganju RK. Slit2-Robo4 pathway modulates lipopolysaccharide-induced endothelial inflammation and its expression is dysregulated during endotoxemia. J Immunol. 2014;192:385–393. doi: 10.4049/jimmunol.1302021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dickinson RE, Dallol A, Bieche I, Krex D, Morton D, Maher ER, Latif F. Epigenetic inactivation of SLIT3 and SLIT1 genes in human cancers. Br J Cancer. 2004;91:2071–2078. doi: 10.1038/sj.bjc.6602222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Narayan G, Goparaju C, Arias-Pulido H, Kaufmann AM, Schneider A, Dürst M, Mansukhani M, Pothuri B, Murty VV. Promoter hypermethylation-mediated inactivation of multiple Slit-Robo pathway genes in cervical cancer progression. Mol Cancer. 2006;5:16. doi: 10.1186/1476-4598-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dallol A, Morton D, Maher ER, Latif F. SLIT2 axon guidance molecule is frequently inactivated in colorectal cancer and suppresses growth of colorectal carcinoma cells. Cancer Res. 2003;63:1054–1058. [PubMed] [Google Scholar]
- 13.Xin SY, Feng XS, Zhou LQ, Sun JJ, Gao XL, Yao GL. Reduced expression of circulating microRNA-218 in gastric cancer and correlation with tumor invasion and prognosis. World J Gastroenterol. 2014;20:6906–6911. doi: 10.3748/wjg.v20.i22.6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding H, Wu YL, Wang YX, Zhu FF. Characterization of the MicroRNA expression profile of cervical squamous cell carcinoma metastases. Asian Pac J Cancer Prev. 2014;15:1675–1679. doi: 10.7314/APJCP.2014.15.4.1675. [DOI] [PubMed] [Google Scholar]
- 15.Zheng ZM, Wang X. Regulation of cellular miRNA expression by human papillomaviruses. Biochim Biophys Acta. 2011;1809:668–677. doi: 10.1016/j.bbagrm.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rao Q, Shen Q, Zhou H, Peng Y, Li J, Lin Z. Aberrant microRNA expression in human cervical carcinomas. Med Oncol. 2012;29:1242–1248. doi: 10.1007/s12032-011-9830-2. [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto N, Kinoshita T, Nohata N, Itesako T, Yoshino H, Enokida H, Nakagawa M, Shozu M, Seki N. Tumor suppressive microRNA-218 inhibits cancer cell migration and invasion by targeting focal adhesion pathways in cervical squamous cell carcinoma. Int J Oncol. 2013;42:1523–1532. doi: 10.3892/ijo.2013.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee H, Kim KR, Cho NH, Hong SR, Jeong H, Kwon SY, Park KH, An HJ, Kim TH, Kim I, et al. MicroRNA expression profiling and Notch1 and Notch2 expression in minimal deviation adenocarcinoma of uterine cervix. World J Surg Oncol. 2014;12:334. doi: 10.1186/1477-7819-12-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Banno K, Iida M, Yanokura M, Kisu I, Iwata T, Tominaga E, Tanaka K, Aoki D. MicroRNA in cervical cancer: OncomiRs and tumor suppressor miRs in diagnosis and treatment. ScientificWorldJournal. 2014;2014:178075. doi: 10.1155/2014/178075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez I, Gardiner AS, Board KF, Monzon FA, Edwards RP, Khan SA. Human papillomavirus type 16 reduces the expression of microRNA-218 in cervical carcinoma cells. Oncogene. 2008;27:2575–2582. doi: 10.1038/sj.onc.1210919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uesugi A, Kozaki K, Tsuruta T, Furuta M, Morita K, Imoto I, Omura K, Inazawa J. The tumor suppressive microRNA miR-218 targets the mTOR component rictor and inhibits AKT phosphorylation in oral cancer. Cancer Res. 2011;71:5765–5778. doi: 10.1158/0008-5472.CAN-11-0368. [DOI] [PubMed] [Google Scholar]
- 22.He H, Di Y, Liang M, Yang F, Yao L, Hao S, Li J, Jiang Y, Jin C, Fu D. The microRNA-218 and ROBO-1 signaling axis correlates with the lymphatic metastasis of pancreatic cancer. Oncol Rep. 2013;30:651–658. doi: 10.3892/or.2013.2516. [DOI] [PubMed] [Google Scholar]
- 23.Yang M, Liu R, Sheng J, Liao J, Wang Y, Pan E, Guo W, Pu Y, Yin L. Differential expression profiles of microRNAs as potential biomarkers for the early diagnosis of esophageal squamous cell carcinoma. Oncol Rep. 2013;29:169–176. doi: 10.3892/or.2012.2105. [DOI] [PubMed] [Google Scholar]
- 24.Tu K, Li C, Zheng X, Yang W, Yao Y, Liu Q. Prognostic significance of miR-218 in human hepatocellular carcinoma and its role in cell growth. Oncol Rep. 2014;32:1571–1577. doi: 10.3892/or.2014.3386. [DOI] [PubMed] [Google Scholar]
- 25.Bai XY, Lin JY, Zhang XC, Xie Z, Yan HH, Chen ZH, Xu CR, An SJ, Sheng GM, Wu YL. High expression of truncated GLI3 is associated with poor overall survival in patients with non-small cell lung cancer. Cancer Biomark. 2013;13:37–47. doi: 10.3233/CBM-130312. [DOI] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancerincidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 28.Dappa E, Elger T, Hasenburg A, Düber C, Battista MJ, Hötker AM. The value of advanced MRI techniques in the assessment of cervical cancer: A review. Insights Imaging. 2017;8:471–481. doi: 10.1007/s13244-017-0567-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang HY, Kim G, Cho H, Kim S, Lee D, Park S, Park KH, Lee H. Diagnostic performance of HPV E6/E7, hTERT, and Ki67 mRNA RT-qPCR assays on formalin-fixed paraffin-embedded cervical tissue specimens from women with cervical cancer. Exp Mol Pathol. 2015;98:510–516. doi: 10.1016/j.yexmp.2015.03.036. [DOI] [PubMed] [Google Scholar]
- 30.Arbyn M, Castellsagué X, de Sanjosé S, Bruni L, Saraiya M, Bray F, Ferlay J. Worldwide burden of cervical cancer in 2008. Ann Oncol. 2011;22:2675–2686. doi: 10.1093/annonc/mdr015. [DOI] [PubMed] [Google Scholar]
- 31.Luo Q, Zhang S, Wei H, Pang X, Zhang H. Roles of Foxp3 in the occurrence and development of cervical cancer. Int J Clin Exp Pathol. 2015;8:8717–8730. [PMC free article] [PubMed] [Google Scholar]
- 32.Burd EM. Human papillomavirus and cervical cancer. Clin Microbiol Rev. 2003;16:1–17. doi: 10.1128/CMR.16.1.1-17.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nour NM. Cervical cancer: A preventable death. Rev Obstet Gynecol. 2009;2:240–244. [PMC free article] [PubMed] [Google Scholar]
- 34.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:1–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 35.Kogo R, How C, Chaudary N, Bruce J, Shi W, Hill RP, Zahedi P, Yip KW, Liu FF. The microRNA-218~survivin axis regulates migration, invasion, and lymph node metastasis in cervical cancer. Oncotarget. 2015;6:1090–1100. doi: 10.18632/oncotarget.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu J, Wang Y, Dong R, Huang X, Ding S, Qiu H. Circulating MicroRNA-218 was reduced in cervical cancer and correlated with tumor invasion. J Cancer Res Clin Oncol. 2012;138:671–674. doi: 10.1007/s00432-012-1147-9. [DOI] [PubMed] [Google Scholar]
- 37.Yuan W, Xiaoyun H, Haifeng Q, Jing L, Weixu H, Ruofan D, Jinjin Y, Zongji S. MicroRNA-218 enhances the radiosensitivity of human cervical cancer via promoting radiation induced apoptosis. Int J Med Sci. 2014;11:691–696. doi: 10.7150/ijms.8880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hartono D, Lioe B, Zhang Y, Li B, Yu J. Impacts of particulate matter (PM2.5) on the behavior of freshwater snail Parafossarulus striatulus. Sci Rep. 2017;7:644. doi: 10.1038/s41598-017-00449-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang BB, Liu SY, Zhang YU, Wei LQ, Mao XL, Wang J, Li LI, Lu ZX. microRNA-218 expression and its association with the clinicopathological characteristics of patients with cervical cancer. Exp Ther Med. 2015;10:269–274. doi: 10.3892/etm.2015.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steg A, Amm HM, Novak Z, Frost AR, Johnson MR. Gli3 mediates cell survival and sensitivity to cyclopamine in pancreaticcancer. Cancer Biol Ther. 2010;10:893–902. doi: 10.4161/cbt.10.9.13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iwasaki H, Nakano K, Shinkai K, Kunisawa Y, Hirahashi M, Oda Y, Onishi H, Katano M. Hedgehog Gli3 activator signal augments tumorigenicity of colorectal cancer via upregulation of adherence-related genes. Cancer Sci. 2013;104:328–336. doi: 10.1111/cas.12073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang HN, Oh SC, Kim JS, Yoo YA. Abrogation of Gli3 expression suppresses the growth of colon cancercells via activation of p53. Exp Cell Res. 2012;318:539–549. doi: 10.1016/j.yexcr.2011.12.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.