Abstract
MicroRNA (miR)-936 has been reported to inhibit the cell cycle and glioma cell proliferation. However, the roles of miR-936 in other human tumors remain largely unknown. In the present study, it was indicated that miR-936 was significantly downregulated in non-small cell lung cancer (NSCLC) tissues compared with adjacent normal tissues by reverse transcription-quantitative polymerase chain reaction (RT-qPCR). Results also indicated that miR-936 was downregulated in NSCLC cell lines compared with 16HBE cells. Furthermore, it was demonstrated that overexpression of miR-936 significantly inhibited the proliferation, cell cycle progression and invasion of NSCLC cells. Notably, E2F2 was identified as a target gene of miR-936 in NSCLC cells. The results indicated that E2F2 was upregulated in NSCLC tissues and cell lines, and its expression was negatively correlated with that of miR-936 in NSCLC tissues. Overexpression of miR-936 significantly reduced the protein expression levels of E2F2 in NSCLC cells. Furthermore, restoration of E2F2 rescued the proliferation and invasion of NSCLC cells transfected with miR-936 mimics. To the best of our knowledge, the present findings demonstrated for the first time that miR-936 suppressed NSCLC progression by directly targeting E2F2.
Keywords: microRNA-936, non-small cell lung cancer, proliferation, invasion, E2F2
Introduction
Lung cancer is one of the leading factors of cancer-associated fatality around the world (1). Every year, >300,000 new patients are diagnosed with lung cancer in the United States (2). A complexity of factors and signaling pathways contribute to the development and progression of lung cancer, including environmental pollution (3). Among all lung cancer cases, non-small cell lung cancer (NSCLC) accounts for ~85% (4). Although advances have been made on lung cancer intervention, the outcomes of patients remain very poor. The five-year survival rate of patients with NSCLC remains low (5). Therefore, to improve the outcome of NSCLC the identification of novel biomarkers for NSCLC diagnosis and prognosis and the development of effective therapeutic targets are urgently required.
MicroRNAs (miRs) are a class of endogenous noncoding RNAs with a length of 19–22 nucleotides that are widely expressed in almost all cell types (6). miRs are able to regulate gene expression by binding to the complementary sequence of the 3′-untranslated region (3′-UTR) of target mRNAs (7). An increasing number of studies indicate that miRs are critical for the regulation of nearly all physiological processes, including the cell cycle, proliferation, migration, invasion and differentiation of cells (8). Therefore, abnormal expression of miRs typically results in occurrence of human cancer, including NSCLC (5). For example, upregulation of miR-383 has been demonstrated to inhibit the proliferation, migration and invasion of colon cancer cells (9). A previous study also revealed that miR-185 was downregulated and suppressed pancreatic cell proliferation by targeting transcriptional coactivator with PDZ-binding motif in pancreatic cancer (10). Furthermore, several miRs are reported to be promising predictors for cancer diagnosis and prognosis (11–13). Thus, investigation into the roles and functional mechanisms of miRs is crucial to NSCLC therapy.
miR-936 is located at chromosome 10q25.1 and has a length of 22 nucleotides (14). A previous study indicated that miR-936 inhibits glioma cell proliferation by targeting CKS1 (15). However, the function of miR-936 in other cancer types remains unclear.
The present study investigated the expression of miR-936 in NSCLC tissues and cell lines compared with normal tissues or cells. Furthermore, the associated mechanism of miR-936 in NSCLC progression was assessed and its effect on the proliferation, cell cycle and invasion of NSCLC cells was determined.
Materials and methods
Clinical specimens
The procedures in the present study were approved by the Ethics Committee on Human Experimentation of The Second Hospital of Anhui Medical University (Hefei, China) and written informed consent was also obtained from each patient. Samples of primary cancer tissues (n=35; male:female ratio, 26:9; age range, 39–68 years; median age, 55 years) and adjacent normal tissues (n=13) were obtained from patients who had undergone surgery at The Second Hospital of Anhui Medical University from January 2014 to December 2016. The patients who received chemotherapy and radiotherapy were excluded. The clinical pathological parameters are listed in Table I. All samples were rapidly placed in liquid nitrogen and stored at −80°C until use.
Table I.
Association of the expression of miR-936 with clinicopathological features.
| Clinicopathological parameters | miR-936 high (n=17) | miR-936 low (n=18) | P-valuea |
|---|---|---|---|
| Age (years) | 0.443 | ||
| ≤50 | 12 | 15 | |
| >50 | 5 | 3 | |
| Tumor stage | 0.044 | ||
| I/II | 11 | 5 | |
| III/IV | 6 | 13 | |
| Tumor size (cm) | 0.035 | ||
| ≤4 | 14 | 8 | |
| >4 | 3 | 10 | |
| Lymph node metastasis | 0.028 | ||
| Negative | 8 | 2 | |
| Positive | 9 | 16 |
Chi-square test was used to calculate P-values.
Cell culture and transfection
Lung adenocarcinoma cell lines (A549, SPC-A1, H1299, and H23) and the bronchial epithelial cell line (16HBE) were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA) and were cultured in Dulbecco's modified Eagle medium (DMEM, Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) with 10% fetal bovine serum (FBS, Gibco; Thermo Fisher Scientific, Inc.) at 37°C in an atmosphere containing 5% CO2. miR-936 mimic (5′-ACAGUAGAGGGAGGAAUCGCAG-3′) and scramble mimic (5′-ACAUCUGCGUAAGAUUCGAGUCUA-3′) were synthesized by the Shanghai GenePharma (Shanghai, China). An E2F2 coding sequence was constructed into a pcDNA3 vector (Addgene, Inc., Cambridge, MA, USA) to overexpress E2F2. miR-936 mimics (100 nM), scramble (100 nM) and pcDNA3-E2F2 vectors were transfected into A549 cells using the Lipofectamine 2000 Kit (Invitrogen, Thermo Fisher Scientific, Inc.) according to the manufacturer's information. A549 cells were collected 48 h following transfection.
Cell proliferation assay
Cell proliferation was measured using the Cell Counting Kit-8 (CCK-8, Beyotime Institute of Biotechnology, Jiangsu, China) according to the manufacturer's instructions. A549 cells were seeded into 96-well plates at a density of 200 cells/well. At A549 cells were cultured at 37°C for 0, 24, 48, or 72 h, following which, 10 µl of CCK-8 reagent was added to each well and the samples were incubated at 25°C for 1 h further. The absorbance of each well was measured at 450 nm using a microplate reader (Thermo Fisher Scientific, Inc.).
Transwell invasion assay
The invasion ability of A549 cells transfected with miR-936 mimics and NC was analyzed using Transwell chambers with an 8-µm pore polycarbonate membrane (EMD Millipore, Billerica, MA, USA). The Transwell chambers were pre-coated with Matrigel (BD Biosciences, San Jose, CA, USA). A total of 2×104 transfected cells in 100 µl serum-free DMEM medium were placed into the upper chambers. A volume of 500 µl DMEM medium supplemented with 20% FBS was added into the lower chambers. Subsequent to incubation at 37°C for 24 h, cells that did not invade through the pores were carefully wiped away with cotton wool. Subsequently, the inserts were fixed with 100% methanol for 10 min, and stained with 0.5% crystal violet (Beyotime Institute of Biotechnology) for 30 min at 25°C and imaged with an inverted light microscope (IX71; Olympus Corporation, Tokyo, Japan) at a magnification of ×200.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cultured cell lines or NSCLC tissues and clinical specimens using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). cDNA was synthesized from isolated RNA using a TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems; Thermo Fisher Scientific, Inc.). qPCR was performed with a Taqman MicroRNA Assay Kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) on an ABI7500 PCR detection system. U6 small nuclear RNA was used as an internal control. The thermocycling conditions were as follows: Denaturation at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 sec and elongation at 60°C for 1 min. All reactions were run in triplicates and the relative expression of miR-936 to U6 was calculated. The fold change of miR-936 in NSCLC relative to the adjacent noncancerous lung tissues was determined by the 2−ΔΔCq method (16). The primer sequences were as follows: miR-936 forward, 5′-AACGAGACGACGACAGAC-3′ and reverse, 5′-ACAGTAGAGGGAGGAATCGCAG-3′; U6 forward, 5′-AACGAGACGACGACAGAC-3′ and reverse, 5′-GCAAATTCGTGAAGCGTTCCATA-3′; E2F2 forward, 5′-GTCTCCGCCGAGCTTGAGG-3′ and reverse, 5′-GAGCAGAGAGCAGCGCTTAG-3′; GAPDH forward, 5′-ATGTTGCAACCGGGAAGGAA-3′ and reverse, 5′-AGGAAAAGCATCACCCGGAG-3′.
Bioinformatics analysis
The Target Scan tool (http://www.targetscan.org/index.html) was used to predict the potential targets of miR-936.
Western blot analysis
A549 cells were lyzed using radio immunoprecipitation buffer (Thermo Fisher Scientific, Inc.) and total protein lysates were obtained. Protein concentration was determined using a BCA assay. Total protein (20 µg) was separated in 4–20% SDS-PAGE gels and transferred to polyvinylidene fluoride membranes at 4°C. Membranes were then blocked at 25°C for 1 h with 5–10% milk/Tris-buffered saline with Tween-20 and were incubated with primary antibodies at 4°C overnight. These antibodies included: E2F2 (1:1,000; cat. no. sc-9967), PCNA (1:2,000; cat. no. sc-56), Cyclin D1 (1:1,000; cat. no. sc-4074) and GAPDH (1:2,000; cat. no. sc-365062). All antibodies were sourced from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA). Membranes were incubated with the goat anti-mouse horseradish perxoidase-conjugated secondary antibody (1:2,000; cat. no. sc-2005; Santa Cruz Biotechnology, Inc.). Protein signals on membranes were detected using ECL reagents (GE Healthcare, Chicago, IL, USA).
Cell cycle analysis
Cells were harvested, washed twice with ice-cold PBS and fixed in 70% ethanol for 24 h at 4°C. Cells were then washed three times with ice cold PBS and incubated with 1 mg/ml RNase A (R6148; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for 30 min at 37°C. Subsequently, cells were stained at 25°C for 10 min with 50 µg/ml propidium iodide (BD Bioscience, Franklin Lakes, NJ, USA) in 0.5% Tween-20 with PBS and subjected to analysis of cell cycle distribution using a BD FACScan flow cytometer (Becton Dickinson) coupled with Cell Quest acquisition and analysis programs (version 2; BD Bioscience).
Luciferase reporter assay
The 3′-UTR region of E2F2 containing the binding site of miR-936 was constructed into the pGL3 vector (Promega Corporation, Madison, WI, USA). For the luciferase reporter assay, A549 cells were plated in a 96-well plate and incubated at 37°C for 24 h. Subsequently, the cells were transfected with miR-936 mimics or Scramble, and pGL3-E2F2-3′-UTR or pGL3-E2F2-3′-UTR-MUT plasmids using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. A total of 24 h later, luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega Corporation). Luciferase activity was normalized to Renilla luciferase activity.
Statistical analysis
All statistical analyses were performed using SPSS 20.0 (IBM Corp., Armonk, NY, USA) and GraphPad Prism (version 6; GraphPad Software, Inc., La Jolla, CA, USA). The Student's t-test and one-way analysis of variance followed by Tukey's post hoc test were used to analyze two or multiple groups, respectively, for statistical significance. Pearson correlation coefficient analysis was used to determine the correlations. The association between the expression of miR-936 and clinicopathological features was analyzed using χ2 test. P<0.05 was considered to indicate a statistically significant difference.
Results
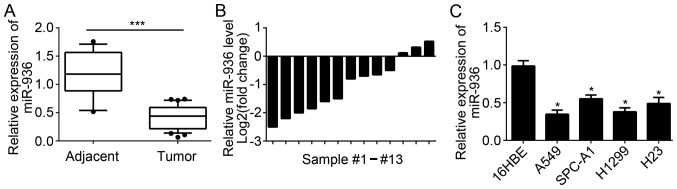
miR-936 expression is downregulated in NSCLC tissues
A total of 35 NSCLC tissues and 13 adjacent normal tissues were collected. RT-qPCR was used to measure the expression levels of miR-936 in these tissues. Results indicated that miR-936 expression was significantly downregulated in 35 NSCLC tissues compared with 13 normal tissues (Fig. 1A). Furthermore, the expression of miR-936 in 13 pairs of NSCLC tissues and matched adjacent normal tissues was analyzed. The results indicated that miR-936 expression was also downregulated in the majority of the 13 NSCLC tissues compared to matched normal tissues (Fig. 1B). Consistently, significant downregulation of miR-936 expression was also observed in NSCLC cell lines compared with 16HBE cells (Fig. 1C). These data demonstrated that miR-936 was downregulated in NSCLC tissues and cells, which implied miR-936 may be involved in the progression of NSCLC.
Figure 1.
miR-936 expression was downregulated in NSCLC tissues. (A) miR-936 expression levels were determined by RT-qPCR in NSCLC tissues (n=35) and adjacent normal tissues (n=13). (B) Relative expression levels of miR-936 in 13 NSCLC tissues and 13 matched adjacent normal tissues were measured by RT-qPCR. (C) Relative expression levels of miR-936 in NSCLC cell lines and the bronchial epithelial cell line (16HBE) were analyzed by RT-qPCR. *P<0.05 and ***P<0.001 vs. the control group. NSCLC, non-small cell carcinoma; miR, microRNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
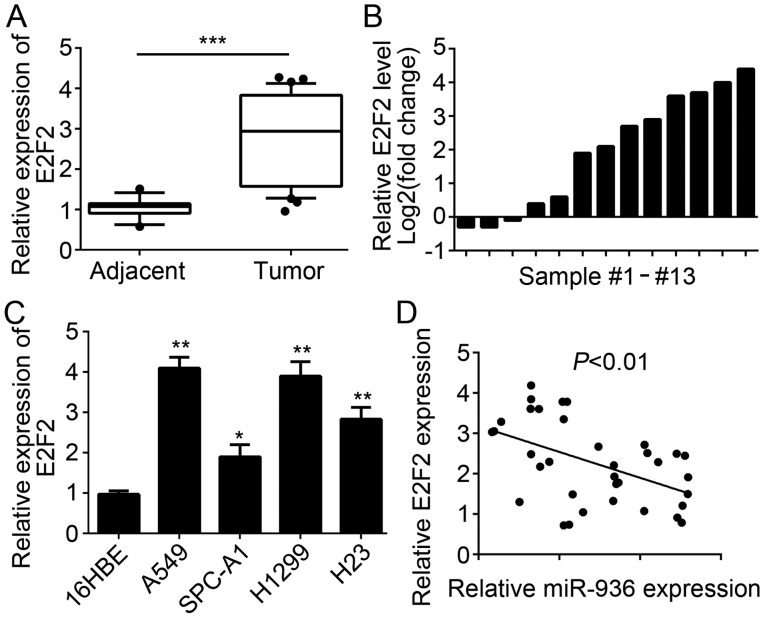
E2F2 expression is upregulated in NSCLC tissues
The E2F2 expression level in NSCLC tissues and cell lines was also examined. RT-qPCR analysis results indicated that E2F2 was significantly upregulated in NSCLC tissues compared with the adjacent normal tissues (Fig. 2A). Similarly, E2F2 expression was upregulated in the majority of paired NSCLC tissues compared with 13 matched normal tissues (Fig. 2B). Furthermore, the expression patterns of E2F2 in NSCLC cell lines were determined and the results indicated that E2F2 was significantly upregulated in NSCLC cell lines compared with 16HBE cells (Fig. 2C). Additionally, miR-936 expression level was inversely correlated with that of E2F2 in NSCLC tissues (Fig. 2D).
Figure 2.
E2F2 expression was upregulated in NSCLC tissues. (A) E2F2 expression levels were determined by RT-qPCR in NSCLC tissues (n=35) and adjacent normal tissues (n=13). (B) Relative expression levels of E2F2 in 13 NSCLC tissues and 13 matched adjacent normal tissues were measured by RT-qPCR. (C) Relative expression levels of E2F2 in NSCLC cell lines and the bronchial epithelial cell line (16HBE) were analyzed by RT-qPCR. (D) miR-936 expression level was reversely correlated with the expression level of E2F2 in the NSCLC tissues. *P<0.05, **P<0.01 and ***P<0.001 vs. the control group. NSCLC, non-small cell carcinoma; miR, microRNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
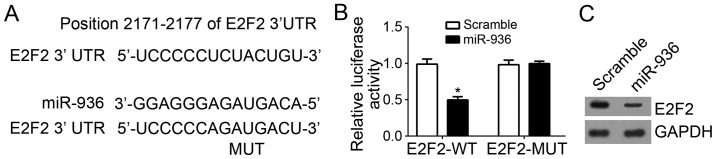
E2F2 is a direct target gene of miR-936 in NSCLC cells
The binding site between miR-936 and E2F2 3′-UTR was identified using Target Scan (http://www.targetscan.org/index.html) (Fig. 3A). To further confirm their interaction, luciferase reporter assays were performed, which revealed that overexpression of miR-936 significantly inhibited the luciferase activity in A549 cells (Fig. 3B). Furthermore, the protein expression level of E2F2 in A549 cells was also downregulated after transfection with miR-936 mimics compared with scramble control (Fig. 3C).
Figure 3.
E2F2 was a direct target gene of miR-936 in the non-small cell carcinoma cells. (A) The binding site between miR-936 and E2F2 3′-UTR was indicated. (B) miR-936 overexpression decreased the luciferase activity when this reporter gene included the wild-type E2F2 3′-UTR in A549 cell. There was no change in the luciferase activity when this reporter gene included the mutated E2F2 3′-UTR. (C) Overexpression of miR-936 suppressed the E2F2 protein expression levels in the A549 cells. *P<0.05 vs. the control group. miR, microRNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; UTR, untranslated region; WT, wild-type; MUT, mutant.
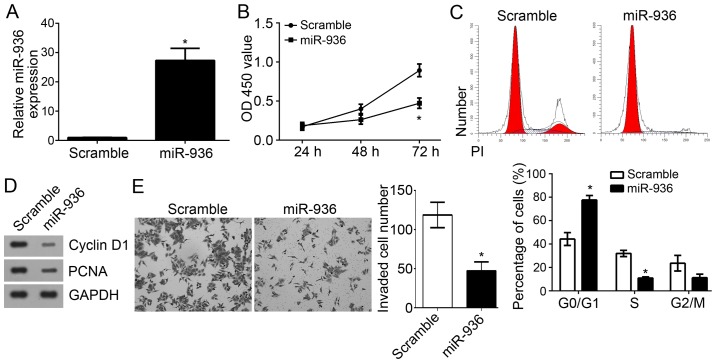
miR-936 overexpression suppresses the proliferation, cell cycle and invasion of NSCLC cells
To further explore the functions of miR-936 in NSCLC, miR-936 was overexpressed by transfection with miR-936 mimics in A549 cells. RT-qPCR analysis indicated that miR-936 was effectively and significantly upregulated in A549 cells transfected with miR-936 mimics (Fig. 4A). CCK8 assays were used to determine the proliferation of A549 cells. Notably, overexpression of miR-936 significantly inhibited the proliferation of A549 cells (Fig. 4B). As the cell cycle is directly linked to proliferation (17), the effect of miR-936 on the cell cycle was analyzed. Results indicated that overexpression of miR-936 significantly reduced the cell percentage in S phase but increased the cells in G0/G1 phase (Fig. 4C). Furthermore, lower protein expression levels of proliferating cell nuclear antigen (PCNA) and Cyclin D1 were observed in A549 cells transfected with miR-936 mimics compared with the scramble control group (Fig. 4D). Notably, Transwell invasion assays revealed that overexpression of miR-936 significantly suppressed the invaded A549 cell number (Fig. 4E). Taken together, these findings demonstrated that miR-936 may serve as a tumor suppressor by inhibiting cell proliferation, the cell cycle and invasion.
Figure 4.
miR-936 overexpression suppressed the proliferation, cell cycle and invasion of non-small cell carcinoma cells. (A) Reverse transcription analysis indicated that miR-936 was significantly upregulated in A549 cells transfected with miR-936 mimics compared with the scramble control group. (B) The Cell Counting Kit-8 assay indicated that miR-936 overexpression inhibited the proliferation of A549 cells. (C) Cell cycle distribution was determined in A549 cells by flow cytometry. (D) Protein expression levels of PCNA and Cyclin D1 in A549 cells were determined by western blot analysis. (E) Transwell assays were utilized to measure the invasion of A549 cells (magnification, ×100). *P<0.05 vs. the control group. PCNA, proliferating cell nuclear antigen; miR, micro RNA.
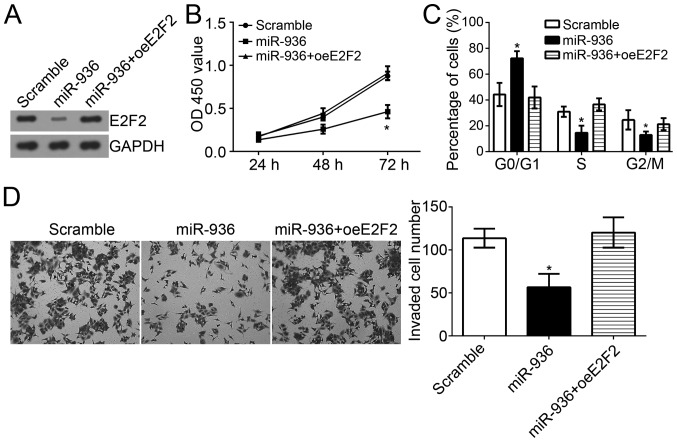
miR-936 suppresses NSCLC cell proliferation, the cell cycle and invasion by regulating E2F2 expression
E2F2 was identified as a target gene of miR-936 in NSCLC and E2F2 expression was indicated to be upregulated in NSCLC tissues in previous results. To investigate whether miR-936 regulates NSCLC progression by suppression of E2F2 expression, the protein expression level of E2F2 was restored in A549 cells transfected with miR-936 mimics. Western blot analysis indicated that E2F2 expression was markedly upregulated in A549 cells (Fig. 5A). CCK8 assays were performed to evaluate cell proliferation. The results indicated that miR-936 overexpression significantly suppressed the proliferation of A549 cells, whereas restoration of E2F2 inhibited this effect (Fig. 5B). Furthermore, overexpression of miR-936 significantly arrested the cell cycle, whereas overexpression of E2F2 in miR-936-overexpressing A549 cells reversed this effect (Fig. 5C). Transwell assays were performed to assess cell invasion. The results revealed that overexpression of miR-936 significantly suppressed cell invasion, whereas restoration of E2F2 expression enhanced the invasion of A549 cells transfected with miR-936 mimics (Fig. 5D). Taken together, these data suggested that miR-936 suppressed the proliferation, cell cycle and invasion of NSCLC cells by directly regulating E2F2 expression.
Figure 5.
miR-936 suppressed the NSCLC cell proliferation, cell cycle and invasion by regulating E2F2 expression. (A) Western blot analysis revealed that E2F2 expression was significantly rescued in A549 cells transfected with miR-936 mimics. (B) CCK8 assay results indicated that overexpression of E2F2 promoted the proliferation of A549 cells treated with miR-936 mimics. (C) Cell cycle progression was rescued by E2F2 overexpression in A549 cells transfected with miR-936 mimics. (D) Transwell assays were used to analyze the invasion of A549 cells (magnification, ×100). *P<0.05 vs. the control group.
Discussion
NSCLC is a common malignant human cancer that is associated with a large number of cancer-associated fatalities globally (1). However, the underlying molecular mechanism of NSCLC occurrence and progression remains largely unknown. Thus, there is an urgent requirement to identify the key molecules and signaling pathways involved in NSCLC. In the present study, the expression of miR-936 was significantly downregulated in NSCLC tissues compared with adjacent normal tissues. Consistently, the expression of miR-936 was also significantly downregulated in NSCLC cell lines compared with 16HBE cells. Functional experiments suggested that overexpression of miR-936 significantly inhibited the proliferation, cell cycle and invasion of A549 cells. Mechanistically, the present study also indicated that E2F2 was a direct target of miR-936 in NSCLC cells and that overexpression of miR-936 significantly inhibited the protein expression of E2F2 in A549 cells. Notably, a reverse correlation between the expression of miR-936 and E2F2 in NSCLC tissues was also demonstrated. Furthermore, E2F2 was significantly upregulated in the NSCLC tissues and cell lines compared with normal tissues or the 16HBE cell line. In addition, the restoration of E2F2 in miR-936-overexpressing A549 cells enhanced the proliferation, restored the cell cycle and promoted cell invasion. Taken together, the present study demonstrated that miR-936 acted as a tumor suppressor in NSCLC through targeting E2F2.
Accumulating studies have indicated that dysregulated expression of miRs is a characteristic of nearly all types of human malignancy, including stomach adenocarcinoma (18), gastric cancer (19), esophageal squamous cell carcinoma (20), head and neck squamous cell carcinoma (21), cervical cancer (22), breast cancer (23), liver cancer (24) and NSCLC (25). miRs have been reported to serve as oncogenes or tumor suppressors to regulate cancer cell proliferation and migration. For instance, Zhang et al (26) reported that miR-630 promotes cell proliferation and inhibits apoptosis in the HCT116 human colorectal cancer cell line. Furthermore, Liu et al (27) revealed that miR-1297 contributes to tumor growth of human breast cancer by targeting PTEN/phosphoinositide 3-kinase/AKT signaling. Previous evidence has also indicated that miR-936 were downregulated in glioma specimens and that cell cycle arrest was induced, which targeted CKS1 (15). However, the functions of miR-936 in other cancer types, including NSCLC, have not been investigated. In the present study, it was demonstrated that miR-936 was significantly downregulated in NSCLC tissues and cell lines, which indicated that miR-936 may be involved in the development and progression of NSCLC.
Identification of miR-936 target genes is crucial for understanding its molecular mechanism in NSCLC carcinogenesis and for the development of effective therapeutic targets. Through bioinformatics analysis, it was identified that E2F2 was a target gene of miR-936 in NSCLC cells. Notably, E2F2 is a E2F member, which represent a family of transcription factors regulating a myriad of biological processes (28). Previous findings have indicated that E2F2 is an oncogene in some types of cancer, including lung cancer (28), osteosarcoma (29), melanoma (30), ovarian cancer (31) and glioma (32). In addition, Wang et al (33) reported that E2F2 was inhibited by miR-31 and contributed to the malignance of gastric cancer. However, how E2F2 expression is regulated in NSCLC requires further investigation. In the present study, it was demonstrated that miR-936 targeted E2F2 and suppressed the expression of E2F2 in NSCLC cells. Furthermore, E2F2 was upregulated in NSCLC tissues and restoration of E2F2 restored the proliferation, cell cycle and invasion of miR-936-overexpressing A549 cells. These data indicated that E2F2 was downregulated by miR-936 and acted as an oncogene in NSCLC.
In conclusion, the present study demonstrated the key role of miR-936 in NSCLC cells. It was also demonstrated that miR-936 suppressed the proliferation, cell cycle and invasion of NSCLC cells by targeting E2F2. The findings suggested that the miR-936/E2F2 axis may serve as a potential therapeutic target for NSCLC treatment.
Acknowledgements
Not applicable.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
XZ and HT initiated, designed the present study, analyzed, interpreted the results and wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
For the use of human samples, the protocol for the present study was approved by the Institutional Ethics Committee of The Second Hospital of Anhui Medical University and all enrolled patients signed a written informed consent document.
Patient consent for publication
All patients within this study provide consent for the publication of their data.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 3.Yang X, Chen BB, Zhang MH, Wang XR. MicroRNA-126 inhibits the proliferation of lung cancer cell line A549. Asian Pac J Trop Med. 2015;8:239–242. doi: 10.1016/S1995-7645(14)60323-0. [DOI] [PubMed] [Google Scholar]
- 4.Peters S, Adjei AA, Gridelli C, Reck M, Kerr K, Felip E. ESMO Guidelines Working Group: Metastatic non-small-cell lung cancer (NSCLC): ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2012;23(Suppl 7):vii56–vii64. doi: 10.1093/annonc/mds226. [DOI] [PubMed] [Google Scholar]
- 5.Yang S, Zhang Y, Zhao X, Wang J, Shang J. microRNA-361 targets Wilms' tumor 1 to inhibit the growth, migration and invasion of non-small-cell lung cancer cells. Mol Med Rep. 2016;14:5415–5421. doi: 10.3892/mmr.2016.5858. [DOI] [PubMed] [Google Scholar]
- 6.Chen C, Zhao Z, Liu Y, Mu D. microRNA-99a is downregulated and promotes proliferation, migration and invasion in non-small cell lung cancer A549 and H1299 cells. Oncol Lett. 2015;9:1128–1134. doi: 10.3892/ol.2015.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang H, Zhang H, Hu X, Li W. Knockdown of long non-coding RNA XIST inhibits cell viability and invasion by regulating miR-137/PXN axis in non-small cell lung cancer. Int J Biol Macromol. 2018;111:623–631. doi: 10.1016/j.ijbiomac.2018.01.022. [DOI] [PubMed] [Google Scholar]
- 8.Lei T, Zhu Y, Jiang C, Wang Y, Fu J, Fan Z, Qin H. MicroRNA-320 was downregulated in non-small cell lung cancer and inhibited cell proliferation, migration and invasion by targeting fatty acid synthase. Mol Med Rep. 2016;14:1255–1262. doi: 10.3892/mmr.2016.5370. [DOI] [PubMed] [Google Scholar]
- 9.Cui Y, Chen LG, Yao HB, Zhang J, Ding KF. Upregulation of microRNA-383 inhibits the proliferation, migration and invasion of colon cancer cells. Oncol Lett. 2018;15:1184–1190. doi: 10.3892/ol.2017.7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xia D, Li X, Niu Q, Liu X, Xu W, Ma C, Gu H, Liu Z, Shi L, Tian X, et al. MicroRNA-185 suppresses pancreatic cell proliferation by targeting transcriptional coactivator with PDZ-binding motif in pancreatic cancer. Exp Ther Med. 2018;15:657–666. doi: 10.3892/etm.2017.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 13.Anninos P, Chatzimichael A, Adamopoulos A, Kotini A, Tsagas N. A combined study of MEG and pico-Tesla TMS on children with autism disorder. J Integra Neurosci. 2016;15:497–513. doi: 10.1142/S0219635216500278. [DOI] [PubMed] [Google Scholar]
- 14.Olbromski M, Grzegrzolka J, Jankowska-Konsur A, Witkiewicz W, Podhorska-Okolow M, Dziegiel P. MicroRNAs modulate the expression of the SOX18 transcript in lung squamous cell carcinoma. Oncol Rep. 2016;36:2884–2892. doi: 10.3892/or.2016.5102. [DOI] [PubMed] [Google Scholar]
- 15.Wang D, Zhi T, Xu X, Bao Z, Fan L, Li Z, Ji J, Liu N. MicroRNA-936 induces cell cycle arrest and inhibits glioma cell proliferation by targeting CKS1. Am J Cancer Res. 2017;7:2131–2143. [PMC free article] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Wang D, Cao Q, Qu M, Xiao Z, Zhang M, Di S. MicroRNA-616 promotes the growth and metastasis of non-small cell lung cancer by targeting SOX7. Oncol Rep. 2017;38:2078–2086. doi: 10.3892/or.2017.5854. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Liu GW, Qin ZM, Shen QH. An ensemble method integrated with miRNA expression data for predicting miRNA targets in stomach adenocarcinoma. Cancer Biomark. 2017;20:617–625. doi: 10.3233/CBM-170595. [DOI] [PubMed] [Google Scholar]
- 19.Liu S, Suo J, Wang C, Sun X, Wang D, He L, Zhang Y, Li W. Prognostic significance of low miR-144 expression in gastric cancer. Cancer Biomark. 2017;20:547–552. doi: 10.3233/CBM-170351. [DOI] [PubMed] [Google Scholar]
- 20.Mei LL, Qiu YT, Huang MB, Wang WJ, Bai J, Shi ZZ. miR-99a suppresses proliferation, migration and invasion of esophageal squamous cell carcinoma cells through inhibiting the IGF1R signaling pathway. Cancer Biomark. 2017;20:527–537. doi: 10.3233/CBM-170345. [DOI] [PubMed] [Google Scholar]
- 21.Krishnan AR, Zheng H, Kwok JG, Qu Y, Zou AE, Korrapati A, Li PX, Califano JA, Hovell MF, Wang-Rodriguez J, Ongkeko WM. A comprehensive study of smoking-specific microRNA alterations in head and neck squamous cell carcinoma. Oral Oncol. 2017;72:56–64. doi: 10.1016/j.oraloncology.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ou L, Wang D, Zhang H, Yu Q, Hua F. Decreased expression of miR-138-5p by LncRNA H19 in cervical cancer promotes tumor proliferation. Oncol Res. 2017 Aug 10; doi: 10.3727/096504017X15017209042610. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong Y, Liu Y, Jiang A, Li R, Yin M, Wang Y. MicroRNA-335 suppresses the proliferation, migration, and invasion of breast cancer cells by targeting EphA4. Mol Cell Biochem. 2018;439:95–104. doi: 10.1007/s11010-017-3139-1. [DOI] [PubMed] [Google Scholar]
- 24.Yang L, Peng F, Qin J, Zhou H, Wang B. Downregulation of microRNA-196a inhibits human liver cancer cell proliferation and invasion by targeting FOXO1. Oncol Rep. 2017;38:2148–2154. doi: 10.3892/or.2017.5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ling DJ, Chen ZS, Zhang YD, Liao QD, Feng JX, Zhang XY, Shi TS. MicroRNA-145 inhibits lung cancer cell metastasis. Mol Med Rep. 2015;11:3108–3114. doi: 10.3892/mmr.2014.3036. [DOI] [PubMed] [Google Scholar]
- 26.Zhang L, Feng G, Zhang X, Ding Y, Wang X. microRNA630 promotes cell proliferation and inhibits apoptosis in the HCT116 human colorectal cancer cell line. Mol Med Rep. 2017;16:4843–4848. doi: 10.3892/mmr.2017.7159. [DOI] [PubMed] [Google Scholar]
- 27.Liu C, Liu Z, Li X, Tang X, He J, Lu S. MicroRNA-1297 contributes to tumor growth of human breast cancer by targeting PTEN/PI3K/AKT signaling. Oncol Rep. 2017;38:2435–2443. doi: 10.3892/or.2017.5884. [DOI] [PubMed] [Google Scholar]
- 28.Feliciano A, Garcia-Mayea Y, Jubierre L, Mir C, Hummel M, Castellvi J, Hernández-Losa J, Paciucci R, Sansano I, Sun Y, et al. miR-99a reveals two novel oncogenic proteins E2F2 and EMR2 and represses stemness in lung cancer. Cell Death Dis. 2017;8:e3141. doi: 10.1038/cddis.2017.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tao T, Shen Q, Luo J, Xu Y, Liang W. MicroRNA-125a Regulates cell proliferation via directly targeting E2F2 in osteosarcoma. Cell Physiol Biochem. 2017;43:768–774. doi: 10.1159/000481560. [DOI] [PubMed] [Google Scholar]
- 30.Zhao H, Tang W, Chen X, Wang S, Wang X, Xu H, Li L. The NAMPT/E2F2/SIRT1 axis promotes proliferation and inhibits p53-dependent apoptosis in human melanoma cells. Biochem Biophys Res Commun. 2017;493:77–84. doi: 10.1016/j.cbpb.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 31.Xie L, Li T, Yang LH. E2F2 induces MCM4, CCNE2 and WHSC1 upregulation in ovarian cancer and predicts poor overall survival. Eur Rev Med Pharmacol Sci. 2017;21:2150–2156. [PubMed] [Google Scholar]
- 32.Song H, Zhang Y, Liu N, Zhang D, Wan C, Zhao S, Kong Y, Yuan L. Let-7b inhibits the malignant behavior of glioma cells and glioma stem-like cells via downregulation of E2F2. J Physiol Biochem. 2016;72:733–744. doi: 10.1007/s13105-016-0512-6. [DOI] [PubMed] [Google Scholar]
- 33.Wang H, Zhang X, Liu Y, Ni Z, Lin Y, Duan Z, Shi Y, Wang G, Li F. Downregulated miR-31 level associates with poor prognosis of gastric cancer and its restoration suppresses tumor cell malignant phenotypes by inhibiting E2F2. Oncotarget. 2016;7:36577–36589. doi: 10.18632/oncotarget.9288. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.