Abstract
Two variant cohesin complexes containing SMC1, SMC3, RAD21 and either STAG/SA1 or SA2 are present in all cell types. We report here their genomic distribution and their specific contributions to genome organization in human cells. While both variants are found at CTCF sites, a fraction of cohesin-SA2 localizes to enhancers lacking CTCF, is linked to tissue-specific transcription and cannot be replaced by cohesin-SA1 when SA2 is absent, a condition observed in several tumours. Downregulation of either variant has different consequences for gene expression and genome architecture. Our results suggest that cohesin-SA1 preferentially contributes to the stabilization of TAD boundaries together with CTCF, while cohesin-SA2 promotes cell type-specific contacts between enhancers and promoters independently of CTCF. Loss of SA2 rewires local chromatin contacts and alters gene expression. These findings provide insights on how cohesin mediates chromosome folding and establish a novel framework to address the consequences of cohesin mutations in cancer.
In addition to mediating sister chromatid cohesion, cohesin contributes to the spatial organization of the genome in chromatin loops and topologically associated domains (TADs)1–5. In vertebrate somatic cells, cohesin complexes carry one of two versions of the SA subunit, namely SA1 or SA2, which are encoded by the STAG1 and STAG2 genes, respectively6. Studies in human and mouse cells indicate that cohesin-SA1 and cohesin-SA2 are specifically required for telomere and centromere cohesion, respectively7,8. Nevertheless, cohesion provided by either variant complex is sufficient to allow cell proliferation9. Whether the two variants have specific roles in genome organization and gene regulation is unclear, although murine STAG1 null embryos die before birth and show altered cohesin distribution and gene expression10. Importantly, loss of function mutations in the STAG2 gene have been identified in several human cancers including bladder cancer, Ewing sarcoma and acute myeloid leukaemia11. While cohesin-SA1 is sufficient to carry out the essential functions of cohesin in STAG2 deficient cancer cells9 it may not be able to compensate for other non-essential cohesin-SA2 functions. Consistent with this idea, current evidence suggests that the contribution of cohesin dysfunction to tumourigenesis is not related to cohesion defects or genome instability12–14 but to altered gene regulation15,16.
How cohesin affects gene expression remains poorly understood. Analysis of cohesin distribution in mammalian cells shows a large overlap with the sites occupy by the architectural protein CTCF 17–19. Cohesin and CTCF are present at the boundaries of Topologically Associated Domains (TADs), submegabase regions identified in whole-genome chromatin conformation capture experiments (Hi-C) that encompass DNA sequences interacting more frequently with sequences inside than outside the domain5,20. TADs are thought to regulate transcription by facilitating interactions between enhancers and promoters present in the same TAD while preventing interactions between elements from different TADs. Deletion of CTCF sites at TAD boundaries changes local topology and affects gene expression21,22. A model for TAD generation proposes that after loading, cohesin extrudes DNA to generate progressively longer chromatid loops until it dissociates from chromatin by the action of Wapl or until it reaches an obstacle such as CTCF bound to chromatin where it gets stalled 23–26. Cohesin and CTCF are also found inside TADs and contribute to cell type specific sub-TAD organization3. Moreover, cohesin non-CTCF sites have also been identified in which the complex occupies regions bound by tissue-specific transcription factors or transcriptional regulators such as Mediator27–29. In most of these studies, the potential differences between the two variant cohesin complexes were not addressed. We therefore set out to analyse the distribution of cohesin-SA1 and cohesin-SA2 in non-tumour human cells as well as the consequences of their specific downregulation in gene expression and chromatin architecture. Our results reveal important differences between the behaviour of the two complexes in the sites they occupy, the dynamics of their chromatin association, their interaction partners and, as a consequence, their contribution to 3D genome organization.
Cohesin non-CTCF sites carry SA2 and are present at enhancers
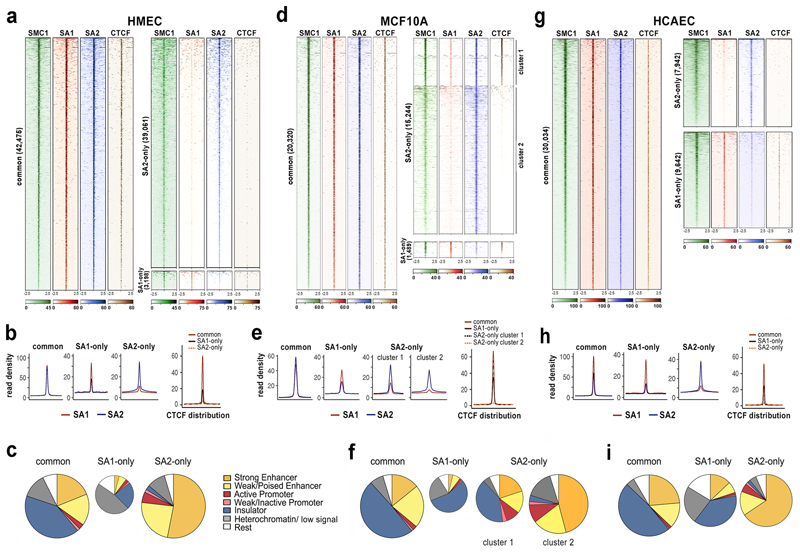
To characterize the specific roles of cohesin-SA1 and cohesin-SA2 in chromatin architecture we selected a primary cell line with comparable levels of the two variant complexes, human mammary epithelial cells (HMEC, Supplementary Fig.1). We first analysed the genomic distribution of SMC1, SA1 and SA2 by chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) using custom-made, validated antibodies and high depth sequencing (about 100 million reads) to ensure whole genome coverage (Supplementary Dataset 2). Reads were aligned to the reference genome and peaks were called using MACS2 (FDR <0.01). Overlaps between the peaks obtained in the SA1 and SA2 immunoprecipitates defined three categories: common, SA2-only and SA1-only positions. Common cohesin positions (42,475) were occupied by either variant complex and colocalized with CTCF (Fig. 1a). They were featured by high cohesin occupancy and similar read density for SA1 and SA2 (Fig. 1b). In contrast, most SA2-only cohesin positions (39,061) had none or very little CTCF and a lower read density. The fraction of SA1-only positions was small (3,198), contained some SA2 and CTCF (Fig. 1a, lower right; Fig. 1b). Analysis of the distribution of these cohesin binding sites in chromatin states defined by ChromHMM in HMECs30 revealed that most SA2-only cohesin positions (77%) were in enhancers, particularly active ones (Fig. 1c). The distribution of the common positions was very different, with only 35% present in enhancers while another 41% were in insulators defined by the sole presence of CTCF. Some SA1-only positions were in insulators (23%) and enhancers (10%) but most were present in a chromatin state designated as “heterochromatin, low signal” 30. Motif discovery analysis showed that both common and SA1-only positions were significantly enriched in CTCF binding motif while SA2-only positions were populated by recognition motifs of several transcription factors other than CTCF (Supplementary Fig. 2a).
Fig. 1. A large fraction of cohesin-SA2 localizes to enhancers independently of CTCF.
a, Analysis of ChIP-seq read distribution for SA1, SA2, SMC1 and CTCF around common, cohesin SA1-only and cohesin SA2-only positions within a 5-kb window in HMECs. b, Average read density plots for SA1 (red) and SA2 (blue) distribution in common, SA1-only and SA2-only positions as well as for CTCF (separate plot on the right). c, Pie charts showing the distribution of cohesin positions in chromatin states defined in HMECs. d-f, same as a-c in MCF10A cells. g-i, same as a-c in HCAECs. CTCF datasets are from ENCODE (Supplementary Dataset 2).
We validated the above findings in MCF10A cells, a non-tumourigenic epithelial breast cell line that, unlike HMECs, can be easily grown and transfected for functional analyses (Fig. 1d,e). Common positions had similar average read density for SA1 and SA2 and overlapped with CTCF. Among SA2-only positions assigned by peak calling, read distribution heatmaps distinguished two clusters (Fig. 1d). While cohesin positions in both clusters were enriched in SA2, those in cluster 1 contained some SA1 and CTCF (Fig. 1d) and its distribution among chromatin states was not very different to that of common and SA1-only positions (Fig. 1e). The larger cluster 2, in contrast, grouped true SA2-only positions: as in HMECs, these positions lacked CTCF and were enriched in enhancers and depleted in insulators compared with common and SA1-only positions (Fig. 1e,f). Cohesin-SA2 may have partners other than CTCF at enhancers and promoters, most likely transcription factors. Consistent with this possibility, proteomic analyses of immunoprecipitates obtained with SA1 and SA2 antibodies from MCF10A cell extracts identified several transcriptional regulators interacting with SA2 and not SA1, including Zmym2 and YAP1 (Supplementary Dataset 3). Zmym2 acts as a corepressor in association with the CoREST complex while YAP1 is a coactivator. ChIP-seq analyses for Zmym2 (this study) and activated YAP131 confirmed the presence of SA2, and not SA1 or CTCF, at their binding sites in MCF10A cells (Supplementary Fig. 2b). We conclude that cohesin can be found at CTCF sites and non-CTCF sites and in the latter, cohesin-SA2 is the predominant variant. These cohesin-SA2 non-CTCF positions are enriched in cis-regulatory elements co-occupied by transcriptional regulators.
Cohesin-SA2 is linked to tissue-specific transcription
We determined the distribution of cohesin-SA1 and cohesin-SA2 in a third cell line, this time of different embryonic origin, human cardiac endothelial cells (HCAEC). Here the number of SA1-only and SA2-only positions was similar (Fig. 1g). Read density profile plots for SA1 and SA2 in common positions suggested that the SA2 ChIP had been less efficient in these cells (Fig. 1h). We suspect that this has two consequences: (1) SA2-only positions with low cohesin occupancy go undetected and (2) a fraction of the positions assigned as SA1-only by peak calling are in fact common positions. In any case, as in the other two cell lines, common and most SA1-only positions overlapped with CTCF, while SA2-only positions lacked CTCF. The distribution of SA1-only positions among chromatin states was close to that of common positions, with a prevalence in insulators whereas SA2-only positions were enriched in enhancers, as previously described for epithelial cells (Fig. 1i).
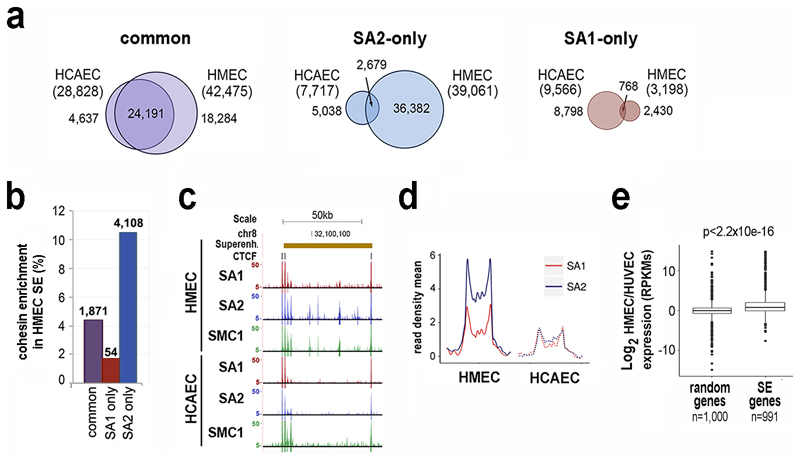
We observed that a large fraction of common positions was conserved between the epithelial and endothelial cells, while SA1-only and SA2-only positions were not (Fig. 2a). Moreover, cohesin-SA2 only sites in HMECs were particularly enriched in super-enhancers defined in the same cell line, which control cell identity genes32 (Fig. 2b,c). Genome-wide, SA2 signals were enriched above SA1 signals in active super-enhancers (Fig. 2d) and the loss of SA2 at these super-enhancers in HCAECs correlated with decreased expression of its associated genes (Fig. 2e).
Fig. 2. Cohesin SA2-only positions are enriched in cell-type specific super-enhancers.
a, Venn diagrams showing overlap of cohesin binding sites between HMECs and HCAECs. Common positions are clearly more conserved. b, Cohesin enrichment in super-enhancers (SE) defined in HMECs. c, Example of cohesin distribution in HMEC and HCAEC cells within a HMEC-specific SE. d, Plot shows SA1 and SA2 enrichment in HMEC and HCAEC cells along HMEC SE. e, Boxplot comparing changes in expression between random genes and genes associated with HMEC-specific SE32. Boxes represent interquartile range (IQR), midline represent the median, whiskers are 1.5xIQR and individual points are outliers. Statistical significance was calculated with a Wilcoxon signed-rank test.
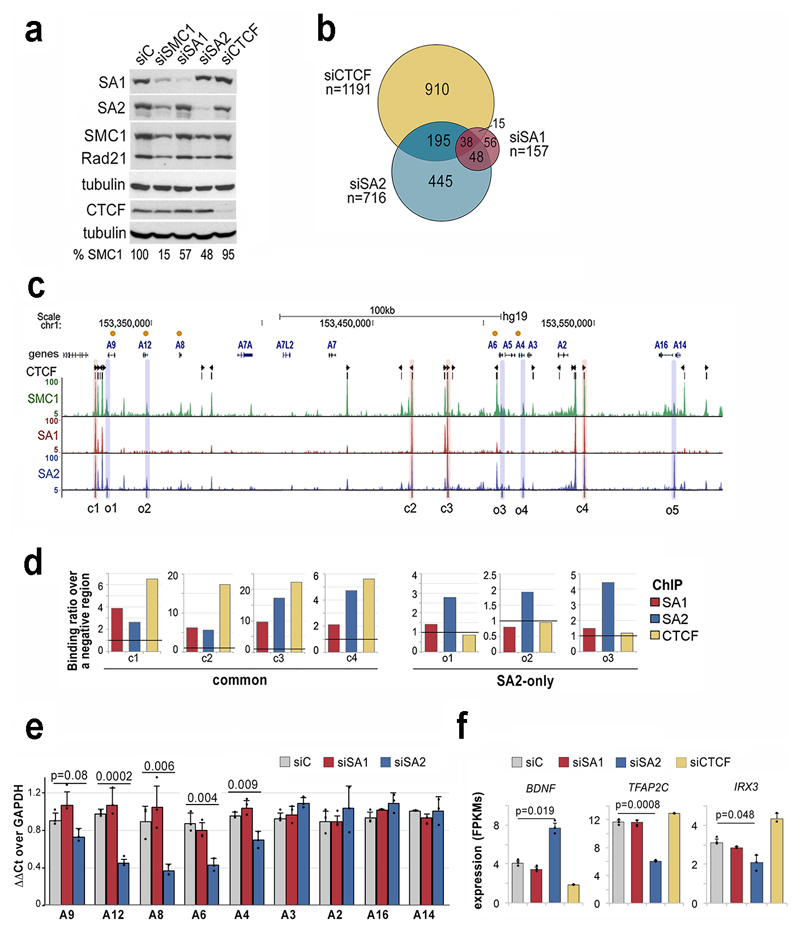
To further understand the effect of each cohesin variant on gene regulation, we transfected MCF10A cells with siRNAs against cohesin subunits and, for comparison, siRNAs against CTCF and SMC1. Comparable extent of depletion of SA1 or SA2 left similar amounts of cohesin (SMC1) in the cells (Fig. 3a). Using a stringent criterion on RNA sequencing (RNA-seq) data analysis, 157 and 716 differentially expressed genes (DEGs) were identified in cells treated with SA1 and SA2 siRNAs, respectively (Fig. 3b and Supplementary Datasets 4-6). Out of 630 genes deregulated only after SA2 depletion, 445 were not affected by CTCF knock down confirming a CTCF independent role for SA2 in the control of gene expression. Among genes deregulated in siSA2-treated cells there were several members of the S100 family of calcium binding proteins which are located in a 300-kb long gene cluster in chromosome 1 (Supplementary Dataset 5 and Fig. 3c, orange dots). This region contains strong common cohesin peaks as well as less prominent cohesin-SA2-only binding sites at the promoters of the deregulated genes (Fig. 3c). We used this locus to validate ChIP-seq data by ChIP-qPCR (Fig. 3d) and RNA-seq data by qRT-PCR (Fig. 3e). Other genes whose expression was affected by SA2 downregulation were BDNF (Brain Derived Neurotrophic Factor), a known target of CoREST in non-neuronal cells33 and two of the top ten core transcription factors proposed to control cell identity in mammary gland cells34, Irx3 and Tfap2c (Fig. 3f). Gene set enrichment analyses also revealed aberrant upregulation of pathways specific of the hematopoietic system and the nervous system in MCF10A cells after SA2 siRNA treatment (Supplementary Fig. 3). Taken together with the preferential enrichment of cohesin-SA2 at super-enhancers, these evidences support a contribution of cohesin-SA2 to tissue-specific gene expression.
Fig. 3. SA2-specific changes in transcription are related to cell identity.
a, Levels of cohesin and CTCF after siRNA transfection in MCF10A cells (uncropped blot images are shown in Supplementary Dataset 1). b, Venn diagram showing the overlap between genes deregulated after downregulation of SA1, SA2 or CTCF compared to mock transfected cells (FDR<0.05, log2fold change<-0.5 or >0.5 and FPKM>3 in at least one condition). b, UCSC browser image of the S100A gene cluster showing genes (orange dots indicate those deregulated in SA2 depleted cells), CTCF peaks (and motif orientation), and genomic distribution of SMC1, SA1 and SA2 in MCF10A cells. Positions corresponding to common (c) and SA2-only (o) cohesin binding sites used in ChIP-qPCR analyses are shadowed in red and blue, respectively. d, ChIP-qPCR validation of SA1, SA2 and CTCF binding to (c) and (o) positions. e-f, Gene expression levels of S100A genes (e) and cell-type specific transcription factors (f) in control, siSA1, siSA2 and siCTCF conditions (mean and SD from three independent experiments). Student's T test was used to assess statistical significance.
Different dynamic behaviour of cohesin-SA1 and cohesin-SA2
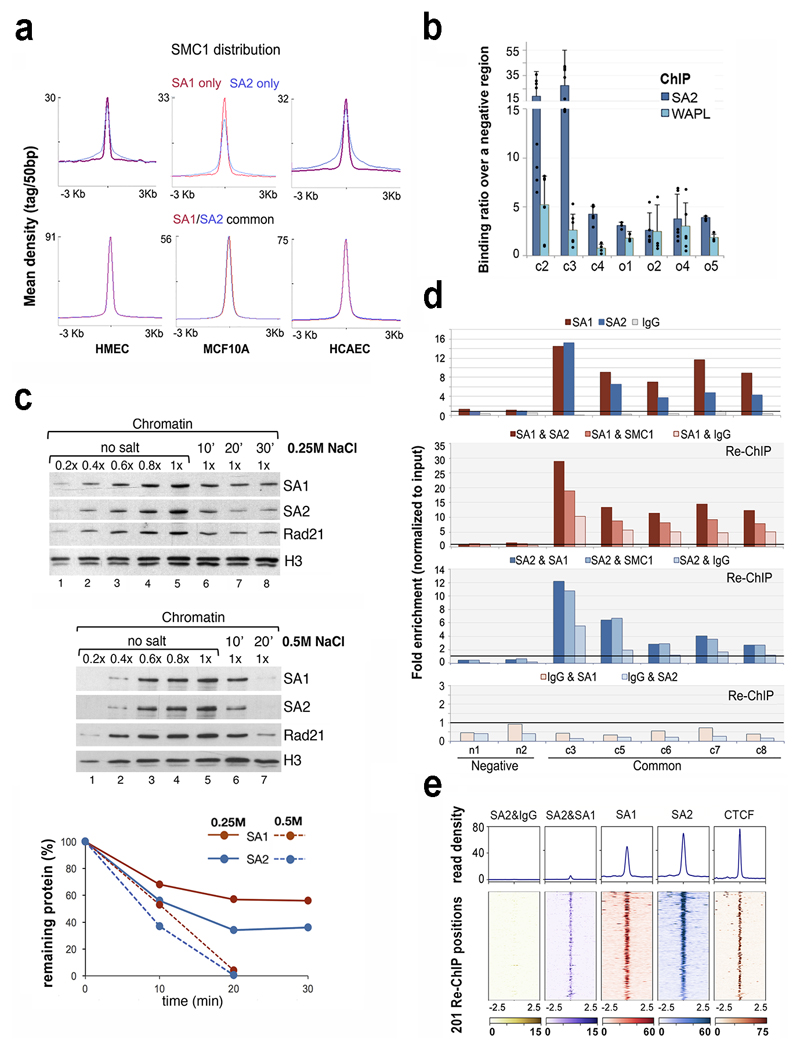
ChIP-seq read density plots of SMC1 distribution around common and cohesin-SA1-only positions produce sharp and narrow profiles in all three cell lines analysed, while for SA2-only positions the profiles are broader (Fig. 4a). These data suggest that cohesin-SA2 present at these positions is more dynamic. Consistent with this possibility, quantitative ChIP-qPCR analyses showed that cohesin-SA2 complexes at common positions are less likely to associate with Wapl, a factor that dissociates cohesin from chromatin35, compared to those present at SA2-only positions (Fig. 4b). Moreover, Wapl removal in HAP1 cells23 increases SMC1 occupancy more in cohesin-non CTCF sites, most likely bound by cohesin-SA2, than in cohesin-CTCF sites (Supplementary Fig. 4). There is also more Wapl in SA2 immunoprecipitates than in SA1 (highlighted in Supplementary Dataset 3).
Fig. 4. Different behaviour of cohesin in common and SA2-only positions.
a, Cohesin SMC1 distribution in HMECs, MCF10A and HCAECs. Maximum mean tag density is indicated on the Y axis. b, SA2 and WAPL binding to the indicated cohesin positions from the S100 locus. Bars represent the mean of at least three independent experiments performed in triplicates; error bar=SD. c, Chromatin bound cohesin was determined upon 0.25M (mid panel) and 0.5M (lower panel) NaCl treatment at different time points. Quantification is shown at the bottom. Uncropped blot images are shown in Supplementary Dataset 1. d-e, The simultaneous presence of at least two cohesin complexes at a given position within the same cell was assayed by Re-ChIP-qPCR (d), and confirmed by Re-ChIP-seq (e). Chromatin eluted from the first ChIP with SA1 or SA2 was incubated with SA2 and SA1 antibodies, respectively, as well as SMC1 and IgG as positive and negative controls. Lower panel, Re-ChIP of chromatin eluted from IgG beads with SA1 and SA2. Positions c3-c8 are "common" cohesin binding sites; n1 and n2, are negative regions. All the positions captured by Re-ChIP-seq (e) correspond to common sites in MCF10A cells.
To further test our hypothesis that cohesin-SA2 is more dynamic than cohesin-SA1, we performed a salt extraction experiment. The chromatin fraction of MCF10A cells was treated with 0.25M or 0.5M NaCl for 10 or 20 min and the amount of each variant remaining on chromatin was assessed by immunoblotting. We found that SA2 was more sensitive to the salt than SA1, as seen at all time points in the lower salt treatment. At higher salt the enhanced sensitivity of SA2 could be seen at the shorter time point. (Fig. 4c). We conclude that the association of cohesin-SA2 with chromatin is less tight or, in other words, more dynamic, than the association of cohesin-SA1.
Both cohesin-SA1 and cohesin-SA2 can be found at common cohesin binding sites. This may be because cells in a population can have either variant complex or because both complexes can coexist at a given position within a cell. Re-ChIP experiments with SA1 and SA2 antibodies revealed that at least two independent cohesin rings can coexist in the same genomic position in the same cell (Fig. 4 d,e). We speculate that stacking at CTCF-bound sites may contribute to stabilize cohesin binding by preventing access of Wapl to cohesin. Alternatively, CTCF itself may stop cohesin progression36 and at the same time prevent its dissociation by a yet unclear mechanism.
Cohesin-SA1 cannot occupy SA2-only sites
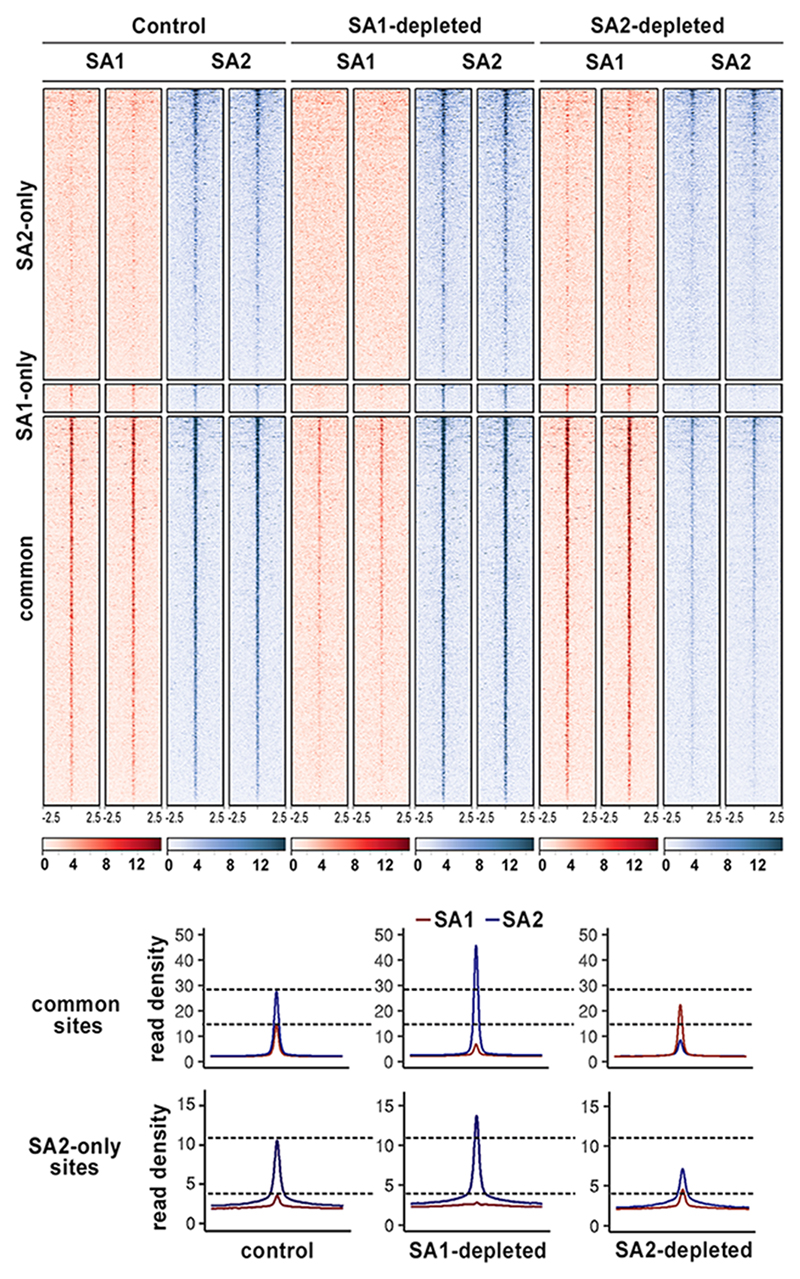
Next, we asked how cohesin distribution changes upon depletion of SA1 or SA2. Calibrated ChIP-seq analyses with SA1 and SA2 antibodies were performed in cells mock depleted or depleted of SA1 or SA2. In SA1 depleted cells there was little cohesin-SA1 left at any position while the presence of cohesin-SA2 increased both at common and SA2-only sites compared to mock transfected cells, and even at SA1-only sites (Fig. 5). It is likely that these SA1-only sites, defined based on peak calling (Fig. 1), are in fact common positions in which SA2 is immunoprecipitated less efficiently. Importantly, cohesin-SA1 could not occupy cohesin-SA2 only sites in SA2 depleted cells, and instead accumulated further at common positions. We conclude that both cohesin-SA1 and cohesin-SA2 can reach common binding sites independently of each other. Thus, in the absence of one variant, the other could in principle compensate for its loss at these CTCF bound sites. In contrast, cohesin-SA1 cannot occupy cohesin-SA2 only positions when SA2 is missing.
Fig. 5. Cohesin-SA1 cannot occupy SA2-only sites.
Read heatmap (up) and read density plots (down) showing SA1 and SA2 distribution around cohesin positions defined in Fig. 1d in control cells and cells treated with siRNA against SA1 or SA2. Two independent replicates were performed per condition. Read density plots were built merging the reads from the two replicates.
Cohesin-SA1 and cohesin-SA2 make different contributions to genome architecture
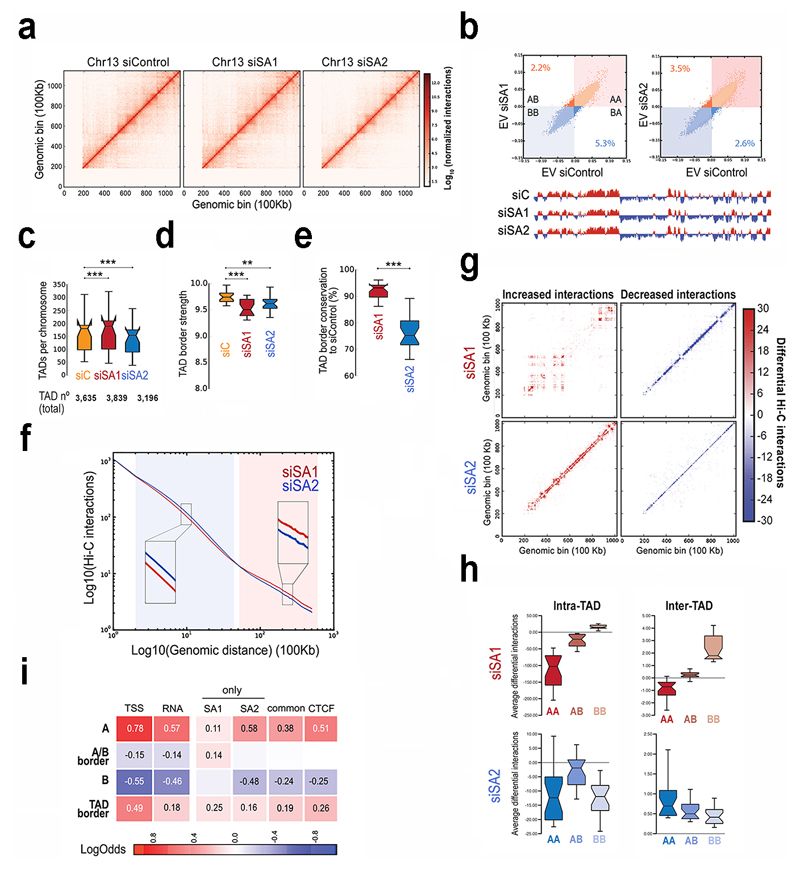
To address the consequences of SA1 or SA2 depletion for genome architecture, we carried out Hi-C experiments in MCF10A cells depleted from SA1 or SA2 (Fig. 6a, Supplementary Fig. 5a,b and Supplementary Dataset 7). Identity of active (A) and repressive (B) compartments37 was mostly preserved (Fig. 6b and Supplementary Fig. 5c). TAD number increased in 204 TADs by SA1 depletion but decreased in 439 TADs by SA2 depletion (Fig. 6c). TAD border strength was lessened, particularly in SA1 depleted cells (Fig. 6d) whereas TAD border conservation diminished by 25% after SA2 depletion (Fig. 6e and Supplementary Fig. 5d). We therefore suggest that some TADs might arise in a CTCF independent manner and instead depend on the interaction of cohesin-SA2 with different transcriptional regulators. Although we could not test this idea with the current resolution of our Hi-C analyses, it agrees with recent data showing that around 20% of TAD borders are maintained after acute elimination of CTCF in mouse ES cells38 as well as with high-resolution Hi-C maps from the same cells revealing a set of TAD boundaries featured by the presence of cohesin and active marks but no CTCF39.
Fig. 6. Distinct contribution of cohesin-SA1 and cohesin-SA2 to genome architecture.
a, Vanilla normalized Hi-C matrices for chr13 at 100Kb resolution in MCF10A cells. b, Scatter plot of eigenvectors of the intra-chromosomal interaction matrices indicated. Numbers within the plot show the % of bins that change compartment. First eigenvector for chr13 at 100Kb resolution is shown below. c-e, Boxplots showing number of TADs per chromosome (c), TAD border strength (d) and TAD border conservation (e). The box extends from the lower to upper quartile values of the data, with a line at the median. Notches represent the confidence interval around the median. f, Hi-C interactions as a function of genomic distance averaged across the genome for a maximum distance of 50 Mb. g, Overall genome-wide increased and decreased interactions between siControl and siSA1 or siSA2 in chr15. h, Effect of the SA1 or SA2 depletion in differential inter- and intra-TAD interactions in different compartments. Boxplots are for the chromosome average values. i, Enrichment of SA1-only, SA2-only and common sites in AB compartments, A/B borders and TAD borders. Squares with numbers are significant (Fisher exact test, p-values < 0.001).
Analysis of the genomic interactions as a function of the genomic distance further evidenced specific contributions of the two cohesin variants to chromatin architecture (Fig. 6f and Supplementary Fig. 5e). Loss of SA2 increased mid-range contacts (0.1 -1.3 Mb) while loss of cohesin-SA1 increased long-range (>1.4 Mb) contacts. These distinct effects were also evident in matrices representing separately gained and lost interactions for each condition compared to control cells (Fig. 6g and Supplementary Fig. 6). SA1 depletion increased very long-range interactions, most of them located within the B compartment, while mid-range interactions within the A compartment were lost (Fig. 6g and 6h, top). One possible interpretation of these data is that SA1 depletion results in a more "relaxed" A compartment, which is compensated by increased compaction of the B compartment. In contrast, SA2 depletion increased inter-TAD mid-range contacts, mostly within the A compartment (Fig. 6g and 6h, bottom), at least in part due to loss of TAD borders. SA2 depletion also decreased short-range intra-TAD contacts that could correspond to enhancer-enhancer or enhancer-promoter interactions, given the prevalence of SA2-only positions in these elements. Finally, the specific enrichment of cohesin-SA1 only positions in A/B borders (Fig. 6i) prompt us to speculate that cohesin-SA1 might play a unique role in modulating A/B compartment identity. However, visual examination of the Hi-C matrices and the resulting Eigen values used in compartment analyses did not reveal compartment switches in any condition (Fig. 6b, lower part).
To interpret our results, we propose that cohesin-SA1 would have a more structural role in genome organization supporting TAD/subTAD formation together with CTCF, while cohesin-SA2 would be more critical for functional intra-TAD contacts together with transcriptional regulators. In the absence of cohesin-SA1, cohesin-SA2 can still cooperate with CTCF in genome organization although border strength is decreased and the A compartment is loosened. In the absence of cohesin-SA2, short-range intra-TAD contacts decrease while new contacts are formed between neighbouring TADs and these changes have more noticeable consequences for gene expression.
Discussion
Recent studies depleting cohesin or CTCF in different cellular systems have led to the conclusion that TADs and compartments arise independently38,40–42. TADs would depend on cohesin and CTCF, whereas genomic compartmentalization would rely mostly on epigenetic features regardless of chromatin contacts. While TAD boundaries are largely invariant across cell types5, the specific interactions within TADs may not43. Moreover, single-cell Hi-C experiments imply a certain degree of stochasticity in the TAD boundary definition among cells in the population44,45. Our results show for the first time that the two variant cohesin complexes have non-redundant functions in genome organization. Upon downregulation of one or the other, the changes that we observe are, not surprisingly, different from those observed after removal of all cohesin40–42.The amount of total cohesin present on chromatin in siSA1 and siSA2 treated cells is very similar, while the relative abundance of each variant changes dramatically bringing about the changes in cohesin distribution, chromatin contacts and gene expression reported above.
Previous analyses have shown that cohesin colocalizes with transcription factors independently of CTCF and thereby contributes to mediate tissue-specific transcription29. We here show that cohesin-SA2 is the prevalent variant at cohesin-non CTCF sites and confirm that these SA2-only sites tend to be tissue-specific and are enriched at enhancers and super-enhancers. Importantly, cohesin-SA1 cannot replace cohesin-SA2 at non-CTCF sites. The mechanisms that position SA1 and SA2-containing complexes remain to be identified. The two SA subunits are highly similar, with over 70% sequence identity along the central part of the protein. The homology decreases in the N- and C-terminal regions and, for instance, SA1 but not SA2 interacts with telomeric protein TRF1 through its amino terminus46. In the same way, SA2 may interact with certain transcriptional regulators through its unique regions. Alternatively, chromatin loops between enhancer and promoters and between CTCF sites may arise by distinct mechanisms, the latter being possibly loop extrusion, and the two SA subunits may be preferentially used for one or the other. In this regard, it is interesting to note recent in vitro data proposing that establishment of DNA-DNA interactions by a cohesin ring already embracing dsDNA requires the second DNA molecule to be ssDNA47 and that purified SA2 binds ssDNA better than SA148. One could envision cohesin-SA2 interacting with eRNA to stabilize an enhancer-promoter loop49.
Somatic mutations in STAG2 have been reported in multiple human cancers, most prominently bladder, Ewing sarcoma and myeloid malignancies11. The presence of cohesin-SA1 allows STAG2 deficient cancer cells to survive by ensuring sufficient cohesion between the sister chromatids9. However, cohesin-SA1 cannot occupy SA2-only sites involved in enhancer-promoter interactions and, as a consequence, expression of some key genes may be altered. Recent studies have shown that elimination of all chromatin loops mediated by cohesin has little impact on steady-state transcription40,42. Yet, cohesin may be most relevant for transcriptional responses induced upon differentiation or lineage commitment50 and when deregulated may contribute to tumourigenesis, as shown for haematopoietic stem and progenitor cells51.
Online Methods
Cell lines
Human primary cell lines were purchased from Lonza and cultured according to the manufacturer's recommendations. NHA (Normal Human Astrocytes, CC-2565) were grown in ABM basal medium (CC-3187) supplemented with AGM Bulletkit (CC-4123); SKMC (Skeletal Muscle Cells, CC-2561) were cultured in SkBM basal medium (CC-3161) supplemented with SkGM Bulletkit (CC-4139); NHBE (Normal Human Bronchial Epithelial Cells, CC-2540) were cultured in BEBM basal medium (CC-3171) supplemented with BEGM Bulletkit (CC-4175); HCAEC (Coronary Artery Endothelial Cells, CC-2585) were grown in EBM2 basal medium (CC-3156) supplemented EGM2-MV Bulletkit (CC-4147); NHEK (Normal Human Epidermal Keratinocytes, #00192627) were grown in KBM-Gold basal medium (#00192151) supplemented with KGM-Gold Bulletkit (#00192060). HMEC (Normal Mammary Epithelial Cells, CC-2551) were cultured in MEBM basal medium (CC-3171) supplemented with MEGM Bulletkit (CC-3150). NHOst (Normal Human Osteoblasts) were grown in OBM basal medium (CC-3208) supplemented with OGM Bulletkit (CC-3207). PrEC (Prostate Epithelial Cells, CC-2555) were cultured with PrEBM basal medium (CC-3165) supplemented with PrEGM Bulletkit (CC-3166). HUVEC (Human Umbilical Vein Endothelial Cells, CC-2517) were grown in EBM basal medium (CC-3121) supplemented with EGM Bulletkit (CC-3124). MCF10A cells (a gift from M. Quintela, CNIO) were grown in DMEM/F12 (#31330038, Thermofisher) supplemented with 20ng/ml of EGF, 0.5mg/ml hydrocortisone, 100ng/ml of cholera toxin, 10mg/ml of insulin and 5% of horse serum.
Antibodies
A rabbit polyclonal antibody recognizing human Wapl was generated using a recombinant C-terminal fragment of the protein (352 amino acids long) obtained by PCR amplification of full length hWapl cDNA [a gift from T. Hirano (RIKEN, Japan)]. A rat monoclonal antibody was raised against the N-terminal region of mouse SA1 and used for western blotting. Additional custom made antibodies have been previously described: SA1, SA2 and SMC18, RAD2152, Zmym253 [a gift from H. Yu (UT Southwestern, US). Commercial antibodies were CTCF (07-729, Millipore), tubulin (DM1A, Sigma), histone H3 (Abcam AB1791).
Quantitative immunoblotting in whole cell extracts and chromatin fractions
Cells were collected by trypsinization, counted, resuspended in SDS-PAGE loading buffer at 107 cells/ml, sonicated and boiled. Equal volumes were separated by SDS-PAGE and analyzed by immunoblotting. Chromatin fractionation was performed as described54 and fractions were run on SDS gels alongside increasing amounts of recombinant proteins corresponding to C-terminal fragments of human SA1 and SA2 to estimate the amount of each variant subunit10. To assess the strength of chromatin association of cohesin variants, chromatin fractions were treated with modified buffer A (10mM HEPES, 1.5mM MgCl2, 0.34M sucrose, 10% glycerol, 1mM DTT and protease inhibitors) containing 0.25 or 0.5M NaCl for 10, 20 or 30 min on ice. Solubilized proteins were separated from insoluble chromatin by low speed centrifugation (4 min at 1,700 x g) and the latter analyzed by immunoblotting.
siRNA
_MCF10A cells were transfected with 50 nM onTARGETplus SMARTpool siRNAs (Dharmacon L-010638, L-021351, L-006833 and L-020165 for SA1, SA2, SMC1 and CTCF, respectively) using DharmaFECT reagent 1. Transfection efficiency was first estimated by qRT-PCR 24 h after transfection, and typically reached more than 90% downregulation (data not shown). Cells were taken at 72h and protein levels assessed by immunoblot.
ChIP sequencing and analysis
Chromatin immunoprecipitation (ChIP) was performed as described34, with some modifications. Confluent cells were cross-linked with 1% formaldehyde added to the media for 15 minutes at RT. After quenching with 0.125M Glycine, fixed cells were washed twice with PBS containing 1μM PMSF and protease inhibitors, pelleted and lysed in lysis buffer (1%SDS, 10mM EDTA, 50mM Tris-HCl pH 8.1) at 2x107 cells/ml. 107 cells equivalent to 40-50 μg of chromatin were used per immunoprecipitation reaction with 25 μg of antibody. Sonication was performed with a Covaris system (shearing time 30 min, 20% duty cycle, intensity 6, 200 cycles per burst and 30 s per cycle) in a minimum volume of 2 ml. For calibrated ChIP-seq in siC, siSA1 and siSA2-treated MCF10A cells, 20% of chromatin from mouse ES cells was added to the human chromatin. We doubled the amount of antibody used for the immunoprecipitations in order to reduce differences on antibody saturation among conditions. ChIP-seq profiles for each antibody were multiplied by the occupancy ratio (OR) = (WmIPh)/(WhIPm), where Wh and IPh are the number of reads mapped to the human genome from input (W) and immunoprecipitated fractions (IP) and Wm and IPm are reads mapped to the mouse genome from input and IP fractions55.
From 6 to 10 ng of immunoprecipitated chromatin (as quantitated by fluorometry) were electrophoresed on an agarose gel and independent sample-specific fractions of 100–200 bp were taken. Adapter-ligated library was completed by limited-cycle PCR with Illumina PE primers (11 to 13 cycles). DNA libraries were applied to an Illumina flow cell for cluster generation and sequenced on the Illumina Genome Analyzer IIx (GAIIx). Image analysis was performed with Illumina Real Time Analysis software (RTA1.8).
Alignment of 50-bp (76-bp for calibrated ChIP samples) long sequences to the reference genome (GRCh37/hg19, February 2009) was performed using 'BWA and Bowtie2'56 under default settings. Duplicates were removed using Picardtools (version 1.60) and peak calling was carried out using MACS2 (version 2.1.1.20160309) setting a q value (FDR) to 0.05 or 0.01 (SMC1, SA1, SA2 in HMEC) and using the '--extsize' argument with the values obtained in the 'macs2 predictd' step57. All comparisons used the input tracks as "control", and each one of the datasets as "treatment".
Common, SA1-only and SA2-only positions were defined using BEDtools v2.26 with a minimum of 1 nt overlap. Common positions were defined in two steps: 1) overlap between SMC1 and SA1 bed files was performed using '-wa -wb' argument and the positions obtained were concatenated and sorted using 'cat' and 'sort -k1,1 -k2,2n' commands. The output was merged using 'bedtools merge' function and considered as one dataset; 2) this was overlapped with the SA2 dataset as above. SA1-only and SA2-only positions are those where SA1 or SA2 do not overlap among each other.
Mean read density profiles and read density heatmaps for different chromatin binding proteins were generated with deepTools 2.058 BAM files of processed reads and plotting them around peak summits of SA1 or SA2 only or common positions.
For Motif discovery analysis, whole sequences of cohesin positions were extracted and used for motif enrichment analysis using MEME-ChIP from MEME59. Default parameters were used except for the following ones: -ccut 0, -meme-mod anr, -meme-minw: 6, -meme-maxw: 50, -nmeme: 600, -meme-nmotifs: 10, -meme-maxsize: 200,000.
Enrichment of cohesin positions (SA1 and SA2-only and common) at HMEC and HCAEC chromatin states30 was defined using 'intersect' function from BEDtools utilities (v2.26) with a minimum of 1nt overlap. The analysis was performed making sure that one position does not belong to two different chromatin states.
To analyse cohesin distribution along super-enhancers, ChIP-seq reads from SA1 and SA2 in HMECs and HCAECs were plotted along HMEC super-enhancers32 using the "scale-regions" parameter from deepTools to adjust all the super-enhancers to a predefined size and applying a local regression (LOESS) to smooth the read signals.
ChIP-qPCR and Re-ChIP
ChIP-qPCR on immunoprecipitated chromatin was performed using the SYBR Green PCR Master Mix and an ABI Prism® 7900HT instrument (Applied Biosystems®). Primers were designed using OligoPerfect Designer™ (Invitrogen) and reactions were performed in triplicate. Chromosome coordinates of the validated peaks and the corresponding primers are listed in Supplementary Table 1. The relative amount of each amplified fragment was normalized with respect to the amplification obtained from input DNA using the ΔΔCt method and represented as indicated in the corresponding figure legends.
ReChIP experiment was performed with the Re-ChIP-IT kit (#53016, Active Motif) according to the manufacturer's protocol. Briefly, MCF10A cells were fixed, lysed and sonicated as described in the ChIP protocol. Fifty μg of chromatin were incubated with 20 μg of the first antibody (SA1, SA2 or IgG) in presence of magnetic beads, washed, eluted and further incubated with 5 μg of the second antibody (SA1, SA2, SMC1 or IgG). Eluted chromatin was analyzed by quantitative PCR. 1 ng of immunoprecipitated chromatin from two conditions, SA2 ChIP followed by IgG ReChIP and SA2 ChIP followed by SA1 ReChIP, was used to prepare libraries for Re-ChIP sequencing. Libraries were prepared with 18 PCR cycles. Peaks were called in SA2-SA1 ReChIP upon normalization with SA2-IgG ReChIP signals.
Quantitative RT-PCR and RNA-sequencing
cDNAs were prepared using the Superscript II reverse transcriptase (Invitrogen) from total RNA (RNeasy Mini Kit, Qiagen) and qRT-PCR analyses were performed using the SYBR Green PCR Master Mix and an ABI Prism® 7900HT instrument (Applied Biosystems®). Primers (Supplementary Table 1) were designed using OligoPerfect Designer™ (Invitrogen). Reactions were performed in triplicate. Quantifications were normalized to endogenous GAPDH, using the ΔΔCt method.
For RNA-seq libraries (three replicates for condition), polyA+RNA was purified with the Dynabeads mRNA purification kit (Invitrogen) from DNaseI-treated total RNA, randomly fragmented, converted to cDNA and processed through subsequent enzymatic treatments of end-repair, dA-tailing, and ligation to adapters as in Illumina's "TruSeq RNA Sample Preparation Guide" (Part # 15008136 Rev. A). Adapter-ligated library was completed by limited-cycle PCR with Illumina PE primers (8 cycles). The resulting purified cDNA library was applied to an Illumina flow cell for cluster generation (TruSeq cluster generation kit v5) and sequenced on the Genome Analyzer IIx with SBS TruSeq v5 reagents by following manufacturer's protocols. Fastq files with 50-nt single-end sequenced reads were quality-checked with FastQC (S. Andrews, http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and aligned to the human genome (GRCh37/hg19) with Nextpresso (http://bioinfo.cnio.es/nextpresso/) executing TopHat-2.0.0 using Bowtie 0.12.7 and Samtools 0.1.16 allowing two mismatches and five multi-hits. Transcript assembly, estimation of their abundances and differential expression were calculated with Cufflinks 1.3.0 using th6e mouse genome annotation data set GRCm37.v65 from Ensembl. To account for multiple hypotheses testing, the estimated significance level (p value) was adjusted using Benjamini-Hochberg False Discovery Rate (FDR) correction. For differential expression, FDR<0.05, log2fold change<-0.5 or >0.5 and fpkm>3 in at least one of the two conditions compared was required.
GSEAPreranked was used to perform a gene set enrichment analysis60. We used the RNA-seq gene list ranked by statistic, setting ‘gene set’ as the permutation method and we run it with 1000 permutations.
Hi-C
MCF10A cells were arrested in G1 by means of high confluency culture (150,000 cells per cm2). Hi-C was performed as described42 using MboI enzyme. Two library replicates per condition were sequenced (>200 million reads each, Supplementary Dataset 7). Data were processed using TADbit61 for read quality control, read mapping, interaction detection, interaction filtering, and matrix normalization. First, the reads were checked using an implemented FastQC protocol in TADbit. This allowed discarding problematic samples and detect systematic artefacts. Then, we used a fragment-based strategy in TADbit for mapping the remaining reads to the reference human genome (GRCh38). The mapping strategy resulted in about 80% of reads mapped uniquely to the genome. Next, we filtered non-informative contacts between two reads, including self-circles, dangling-ends, errors, random breaks or duplicates. The final interaction matrices resulted in 272 to 303 millions of valid interactions per experimental condition (Supplementary Dataset 7). These valid interactions were then used to generate genome-wide interaction maps at 100 Kb and 40 Kb to segment the genome into the so-called A/B compartments, Topologically Associating Domains (TADs), and produce differential interaction maps.
A/B compartments were calculated using vanilla normalized and decay corrected matrices as implemented in TADbit. Briefly, compartments are detected by calculating the first component of a PCA of chromosome-wide matrices and assigning A compartments to genomic bin with positive PCA1 values and high genes density (Fig. 6b). Conversely, B compartments are assigned to genomic bin with negative PCA1 values and low genes density. TADs were identified using 40 Kb resolution vanilla normalized and decay corrected matrices as input to the TAD detection algorithm implemented in TADbit. TAD border localization as well as strength was calculated and used to identify conserved borders and their strength (Fig. 6c-e). A border was considered conserved between siControl and siSA1 or siSA2 experiments if it was localized within +/- 2 two bins in both experiments. Boxplots were generated with the Python plotting library Matplotlib. Raw matrices normalized by coverage (that is, all three experiments were scaled to have the same number of final valid interactions) at 100Kb resolution were also used for studying Hi-C interactions as function of genomic distance. This genomic decay was obtained per chromosome to a maximum genomic distance of 50Mb and then averaged to obtain a genome-wide curve in siSA1 and siSA2 experiments (Fig. 6f). The same 100Kb matrices were used to determine differential Hi-C interactions between siControl and siSA1 or siSA2 experiments (Fig. 6g). These differential interactions maps were then used to assess the chromosome average differential interaction as a function of compartment localization and separating then in intra- and inter-TAD (Fig. 6h). Finally, the enrichment or depletion of genes (represented by their TSS), RNA (based on RNA-seq data), and CTCF and cohesin binding sites (SA1-only, SA2 and common) was analyzed by a log odds analysis of observing such features in genomic bins belonging to A, B compartments, A/B borders or TAD borders (Fig. 6i). The log odds distributions were assessed for their distribution being statistically different than zero as for a Fisher exact test (p-value <0.005). TADbit used for Hi-C analyses is freely available as a github repository at https://github.com/3DGenomes/tadbit.
Immunoprecipitation and LC-MS/MS analysis
Whole cell extracts from MCF10A cells were prepared by lysis on ice for 30 min in TBS supplemented with 0.5% NP-40, 0.5mM DTT, 0.1mM PMSF and 1X complete protease inhibitor cocktail (Roche) followed by sonication. NaCl was added to 0.3M and the extract rotated for 30 min at 4ºC. After centrifugation, the soluble fraction was recovered and diluted to bring the extract back to 0.1M NaCl and 10% glycerol was added. Antibodies were cross-linked to protein A Pureproteome magnetic beads (Millipore) at 1 mg/ml (SA1, SA2 and IgG as control) and incubated with extracts overnight at 4ºC. The beads were washed 6 times with 20 vol of lysis buffer and proteins were eluted in two consecutive steps in 2 vol of elution buffer (8M urea, 100mM Tris-HCl pH 8) by shaking for 10 min. Samples were digested by standard Filter Aided Sample Preparation (FASP)62. Proteins were reduced with 10mM DTT, alkylated with 50mM IAA for 20 min in the dark and digested with 1:50 Lys-C (Wako) for 4 h. Samples were diluted in 50mM ammonium bicarbonate and digested with 1:100 Trypsin (Promega) overnight at 37ºC. Resulting peptides were desalted using a Sep-Pak C18 cartridge for SPE (Waters Corp.), vacuum-dried and resuspended in 0.5% FA. Immunoprecipitates were analysed using a nanoLC Ultra system (Eksigent, Dublin, CA) coupled with a LTQ-Orbitrap Velos instrument (Thermo) via nanoESI (ProxeonBiosystem, Waltham, MA). Two technical replicates were performed. Raw data were analysed using MaxQuant1.5.3.3063 with Andromeda64 as the search engine against UniProtKB/Swiss-Prot (20,584 sequences). Peptides were filtered at 1% FDR. For protein assessment (FDR <1%), at least one unique peptide was required for both identification and quantification. Other parameters were set as default. The resulting “proteingroup.txt” file was loaded in Perseus65 (v1.5.1.6). Missing values were imputed from a normal distribution. A two-sample Student’s T-Test (one side) was used corrected for multiple testing using a permutation-based approach.
Supplementary Material
Acknowledgments
We thank Y. Cuartero and J. Quilez (4D Genome-CRG) for technical help with Hi-C experiments; D. Rico (Newcastle University), F.X. Real (CNIO) and M. Manzanares (CNIC) for comments to the manuscript; T. Hirano (RIKEN) and H. Yu (UT Southwestern) for reagents. This work has been supported by the Spanish Ministry of Economy and Competitiveness and FEDER funds (grants BFU2013-48481-R and BFU2016-79841-R to AL; BFU2013-47736-P to MAM-R; BES-2014-069166 fellowship to MDK; Centro de Excelencia Severo Ochoa SEV-2015-0510 to CNIO and SEV-2012-0208 to CRG); the European Research Council (FP7/2010-2015, ERC grant agreement 609989) and the EU Horizon 2020 research and innovation programme (agreement 676556) to MAM-R; CERCA Programme/Generalitat de Catalunya to MAM-R; La Caixa Foundation (PhD fellowship to AK).
Footnotes
Data availability. ChIP-seq, RNA-seq and Hi-C data from this study have been submitted to GEO database (GSE101921). Additional source data are available upon reasonable request.
Author contributions: AC and AK performed most experiments with technical help from MR-C; MDK performed immunoprecipitation and salt extraction experiments; FLD and AK performed Hi-C; GG-L analysed RNA-seq data; AK and DG-L analysed ChIP-seq data; MAM-R analysed Hi-C data; AC and AL planned the project and wrote the manuscript with contributions from all the authors.
Competing interests: The authors declare no competing interests.
References
- 1.Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu Rev Genet. 2009;43:525–558. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- 2.Hadjur S, et al. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–413. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Phillips-Cremins JE, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zuin J, et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci U S A. 2014;111:996–1001. doi: 10.1073/pnas.1317788111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Losada A, Yokochi T, Kobayashi R, Hirano T. Identification and characterization of SA/Scc3p subunits in the Xenopus and human cohesin complexes. J Cell Biol. 2000;150:405–416. doi: 10.1083/jcb.150.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Canudas S, Smith S. Differential regulation of telomere and centromere cohesion by the Scc3 homologues SA1 and SA2, respectively, in human cells. J Cell Biol. 2009;187:165–173. doi: 10.1083/jcb.200903096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Remeseiro S, et al. Cohesin-SA1 deficiency drives aneuploidy and tumourigenesis in mice due to impaired replication of telomeres. EMBO J. 2012;31:2076–2089. doi: 10.1038/emboj.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Lelij P, et al. Synthetic lethality between the cohesin subunits STAG1 and STAG2 in diverse cancer contexts. Elife. 2017;6 doi: 10.7554/eLife.26980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuadrado A, Remeseiro S, Gomez-Lopez G, Pisano DG, Losada A. The specific contributions of cohesin-SA1 to cohesion and gene expression: Implications for cancer and development. Cell Cycle. 2012;11:2233–2238. doi: 10.4161/cc.20318. [DOI] [PubMed] [Google Scholar]
- 11.De Koninck M, Losada A. Cohesin mutations in cancer. Cold Spring Harb Perspect Med. 2016;6 doi: 10.1101/cshperspect.a026476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balbas-Martinez C, et al. Recurrent inactivation of STAG2 in bladder cancer is not associated with aneuploidy. Nat Genet. 2013;45:1464–1469. doi: 10.1038/ng.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Solomon DA, et al. Frequent truncating mutations of STAG2 in bladder cancer. Nat Genet. 2013;45:1428–1430. doi: 10.1038/ng.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kon A, et al. Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nat Genet. 2013;45:1232–1237. doi: 10.1038/ng.2731. [DOI] [PubMed] [Google Scholar]
- 15.Viny AD, et al. Dose-dependent role of the cohesin complex in normal and malignant hematopoiesis. J Exp Med. 2015;212:1819–1832. doi: 10.1084/jem.20151317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corces MR, Corces VG. The three-dimensional cancer genome. Current Opinion in Genetics and Development. 2016;36:1–7. doi: 10.1016/j.gde.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wendt KS, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 18.Parelho V, et al. Cohesins Functionally Associate with CTCF on Mammalian Chromosome Arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 19.Rubio ED, et al. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci U S A. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nora EP, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo Y, et al. CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell. 2015;162:900–910. doi: 10.1016/j.cell.2015.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Narendra V, et al. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science (80-. ) 2015;347:1017–1021. doi: 10.1126/science.1262088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haarhuis JHI, et al. The Cohesin Release Factor WAPL Restricts Chromatin Loop Extension. Cell. 2017;169:693–707.e14. doi: 10.1016/j.cell.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanborn AL, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci U S A. 2015;112:E6456–65. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fudenberg G, et al. Formation of Chromosomal Domains by Loop Extrusion. Cell Rep. 2016;15:2038–2049. doi: 10.1016/j.celrep.2016.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Wit E, et al. CTCF Binding Polarity Determines Chromatin Looping. Mol Cell. 2015;60:676–684. doi: 10.1016/j.molcel.2015.09.023. [DOI] [PubMed] [Google Scholar]
- 27.Kagey MH, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt D, et al. A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res. 2010;20:578–588. doi: 10.1101/gr.100479.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Faure AJ, et al. Cohesin regulates tissue-specific expression by stabilising highly occupied cis-regulatory modules. Genome Res. 2012;22:2163–2175. doi: 10.1101/gr.136507.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jang W, Kim T, Koo JS, Kim S, Lim D. Mechanical cue-induced YAP instructs Skp2-dependent cell cycle exit and oncogenic signaling. EMBO J. 2017 doi: 10.15252/embj.201696089. e201696089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 34.D’Alessio AC, et al. A systematic approach to identify candidate transcription factors that control cell identity. Stem Cell Reports. 2015;5:763–775. doi: 10.1016/j.stemcr.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tedeschi A, et al. Wapl is an essential regulator of chromatin structure and chromosome segregation. Nature. 2013;501:564–568. doi: 10.1038/nature12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davidson IF, et al. Rapid movement and transcriptional re-localization of human cohesin on DNA. EMBO J. 2016;35:2671–2685. doi: 10.15252/embj.201695402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science (80-. ) 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nora EP, et al. Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell. 2017;169:930–944.e22. doi: 10.1016/j.cell.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bonev B, et al. Multiscale 3D Genome Rewiring during Mouse Article Multiscale 3D Genome Rewiring during Mouse Neural Development. Cell. 2017;171:557.e1–557.e24. doi: 10.1016/j.cell.2017.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rao SSP, et al. Cohesin Loss Eliminates All Loop Domains. Cell. 2017;171:305–320.e24. doi: 10.1016/j.cell.2017.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wutz G, et al. Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J. 2017;36:3573–3599. doi: 10.15252/embj.201798004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwarzer W, et al. Two independent modes of chromatin organization revealed by cohesin removal. Nature. 2017;551:51–56. doi: 10.1038/nature24281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dixon JR, et al. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518:331–336. doi: 10.1038/nature14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Flyamer IM, et al. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature. 2017;544:110–114. doi: 10.1038/nature21711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stevens TJ, et al. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature. 2017;544:59–64. doi: 10.1038/nature21429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Canudas S, et al. Protein requirements for sister telomere association in human cells. EMBO J. 2007;26:4867–4878. doi: 10.1038/sj.emboj.7601903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murayama Y, Samora CP, Kurokawa Y, Iwasaki H, Uhlmann F. Establishment of DNA-DNA Interactions by the Cohesin Ring. Cell. 2018;172:465–477.e15. doi: 10.1016/j.cell.2017.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Countryman P, et al. Cohesin SA2 is a sequence-independent DNA-binding protein that recognizes DNA replication and repair intermediates. J Biol Chem. 2018;293:1054–1069. doi: 10.1074/jbc.M117.806406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li W, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498:516–20. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ing-Simmons E, et al. Spatial enhancer clustering and regulation of enhancer-proximal genes by cohesin. Genome Res. 2015;25:504–13. doi: 10.1101/gr.184986.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mullenders J, et al. Cohesin loss alters adult hematopoietic stem cell homeostasis, leading to myeloproliferative neoplasms. J Exp Med. 2015;212:1833–1850. doi: 10.1084/jem.20151323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carretero M, Ruiz-Torres M, Rodriguez-Corsino M, Barthelemy I, Losada A. Pds5B is required for cohesion establishment and Aurora B accumulation at centromeres. EMBO J. 2013;32:2938–2949. doi: 10.1038/emboj.2013.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gocke CB, Yu H. ZNF198 stabilizes the LSD1-CoREST-HDAC1 complex on chromatin through its MYM-type zinc fingers. PLoS One. 2008;3 doi: 10.1371/journal.pone.0003255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mendez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu B, et al. Biological chromodynamics: A general method for measuring protein occupancy across the genome by calibrating ChIP-seq. Nucleic Acids Res. 2015;43 doi: 10.1093/nar/gkv670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ramírez F, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bailey TL, et al. MEME Suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009;37 doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. GSEA-P: A desktop application for gene set enrichment analysis. Bioinformatics. 2007;23:3251–3253. doi: 10.1093/bioinformatics/btm369. [DOI] [PubMed] [Google Scholar]
- 61.Serra F, et al. Automatic analysis and 3D-modelling of Hi-C data using TADbit reveals structural features of the fly chromatin colors. PLoS Comput Biol. 2017;13 doi: 10.1371/journal.pcbi.1005665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wisniewski JR, Zougman A, Nagaraj N, Mann M, Wi JR. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:377–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 63.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 64.Cox J, et al. Andromeda: A peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 65.Tyanova S, et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nature Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.