Abstract
NLRC3, a member of the NLR family, has been reported as a negative regulator of inflammatory signaling pathways in innate immune cells. However, the direct role of NLRC3 in modulation of CD4+ T-cell responses in infectious diseases has not been studied. In the present study, we showed that NLRC3 plays an intrinsic role by suppressing the CD4+ T cell phenotype in lung and spleen, including differentiation, activation, and proliferation. NLRC3 deficiency in CD4+ T cells enhanced the protective immune response against Mycobacterium tuberculosis infection. Finally, we demonstrated that NLRC3 deficiency promoted the activation, proliferation, and cytokine production of CD4+ T cells via negatively regulating the NF-κB and MEK-ERK signaling pathways. This study reveals a critical role of NLRC3 as a direct regulator of the adaptive immune response and its protective effects on immunity during M. tuberculosis infection. Our findings also suggested that NLRC3 serves as a potential target for therapeutic intervention against tuberculosis.
Author summary
Accumulating evidence that NLRs play an intrinsic roles in regulating T cell responses in lungs and lymphoid tissues, however, to our known, contribution for NLRs modulation of T-cell responses directly in infectious diseases has not been studied. NLRC3, a member of the NLR family, has been reported as a negative regulator of inflammatory signaling pathways in innate immune cells. The direct role of NLRC3 in the modulation of CD4+ T-cell responses in infectious diseases has not been studied. In the present study, we focused on elucidating the roles of NLRC3 in shaping CD4+ T cell responses in vivo and in vitro, in particular during M. tuberculosis infection. We found that NLRC3 plays an intrinsic l roles by suppressing the CD4+ T cell phenotype in lungs and lymphoid tissues, including Th1 and Th17 differentiation, activation, and proliferation. NLRC3 deficiency in CD4+ T cells enhanced the protective immune response against M. tuberculosis infection. Finally, we demonstrated that NLRC3 deficiency promoted the activation, proliferation, and cytokine production of CD4+ T cells via negatively regulating the NF-κB and MEK-ERK signaling pathways. This study reveals a critical role of NLRC3 as a direct regulator of the adaptive immune response and its protective effects on immunity during M. tuberculosis infection. Our findings also suggested that NLRC3 serves as a potential target for therapeutic intervention against tuberculosis.
Introduction
Nucleotide-binding oligomerization domain-like receptors (NLRs) belong to a large family of cytoplasmic sensors that act in response to host perturbation by infectious agents or cellular stress [1,2]. NLRs participate in a very diverse range of biological functions by regulating innate and acquired immune responses, and thus, contribute to immunity against infectious diseases. Some NLRs, such as NLRP3, NLRP7, and NLRC4, have been reported to promote the production of the proinflammatory cytokines, pro-IL-1β and pro-IL-18, via inflammasomes [3–5]. Other NLRs, such as NOD1, NOD2, NLRC5, and CIITA, are known to activate nuclear factor-κB (NF-κB), mitogen-activated protein kinases (MAPKs), and interferon (IFN) regulatory factors (IRFs) to stimulate innate immunity [6,7]. On the contrary, NLRP4, NLRP6, NLRP12, and NLRX1 have been demonstrated as negative regulators of inflammation [8–10]. In recent years, the roles of NLRs in regulating T cell responses have increasingly received research attention. Surprisingly, NLRP12 was found to act as a vital negative regulator of T-cell-mediated immunity and to influence NF-κB regulation and IL-4 production [11]. NOD1 and NOD2 promote the positive maturation of CD8 single-positive thymocytes in a thymocyte-intrinsic manner [12]. NLRP3 expression in CD4+ T cells was found to be required for completion of the inflammasome-mediated differentiation of T helper type 1 (Th1) and Th2 cells [13,14]. However, the exact roles of various NLR family members in regulating adaptive immune responses remain unclear.
NLRC3, an intracellular member of NLR family, has been reported to be expressed by various immune cell populations, including macrophages, epithelial cell and T cells and suppress inflammatory signaling pathways in innate immune cells [15–20]. These pathways include the NF-κB and STING pathways, which are exploited by bacteria or viruses to evade the host immune response and to promote survival [15,16]. A previous study showed that NLRC3 was significantly upregulated in T lymphocytes and suggested that NLRC3 suppresses T cell activation [21]. However, the exact role of NLRC3 and the mechanisms underlying its effects on CD4+ T cell activation and differentiation remain unclear, particularly during in vivo pathogen infection.
Mycobacterium tuberculosis (M. tuberculosis) is the primary causal pathogen of tuberculosis and has infected one-third of the world's population, making it a serious threat to human health. Various strategies, including improved diagnosis of active disease, drug therapy, and new vaccines, are required for effective management of tuberculosis infections [22,23]. Therefore, elucidating the immunoregulatory mechanisms underlying M. tuberculosis infection is an important step for the development of novel therapeutic approaches and for improving the success of vaccination strategies [24]. Previous studies have demonstrated that host resistance to M. tuberculosis infection requires the coordinated actions of the innate and adaptive immune cells [25]. Activation of bacilli-laden macrophages by cytokines secreted by effector CD4+ T cells, including IFN-γ, IL-17, TNF-α, and GM-CSF, is the key step for the immunoregulation of M. tuberculosis infection [26–28]. However, M. tuberculosis can evade immune responses by suppressing the activation of CD4+ T cells through factors, such as PD-1, CTLA4, and Tim-3 [29,30]. Pattern recognition receptors (PRRs) regulate innate and adaptive immune responses during M. tuberculosis infection via Toll-like receptors, C-type lectin receptors, and NLRs [31,32]. Therefore, there is a need to elucidate the mechanisms that lead to abnormalities in PRR signaling pathways that can influence disease pathogenesis during M. tuberculosis infection.
The present study focused on investigating the molecular mechanisms underlying NLRC3 regulation of CD4+ T-cell effector functions during M. tuberculosis infection. Our results revealed that negative regulation of CD4+ T cell activation by NLRC3 is mediated by the NF-κB and ERK signaling pathways both in vitro and in vivo. Furthermore, NLRC3 regulates negatively CD4+ T cell responses in lungs and lymphoid tissues, including differentiation and proliferation, which in turn suppresses the innate immune responses and promotes M. tuberculosis survival. NLRC3 was found to mediate immune evasion by M. tuberculosis in vivo. The above findings provided new insights showing that the suppression of NLRs inhibits the immune response to M. tuberculosis by controlling CD4+ T-cell immunity.
Results
NLRC3–deficient environment impacts CD4+ T cell phenotype during M. tuberculosis infection
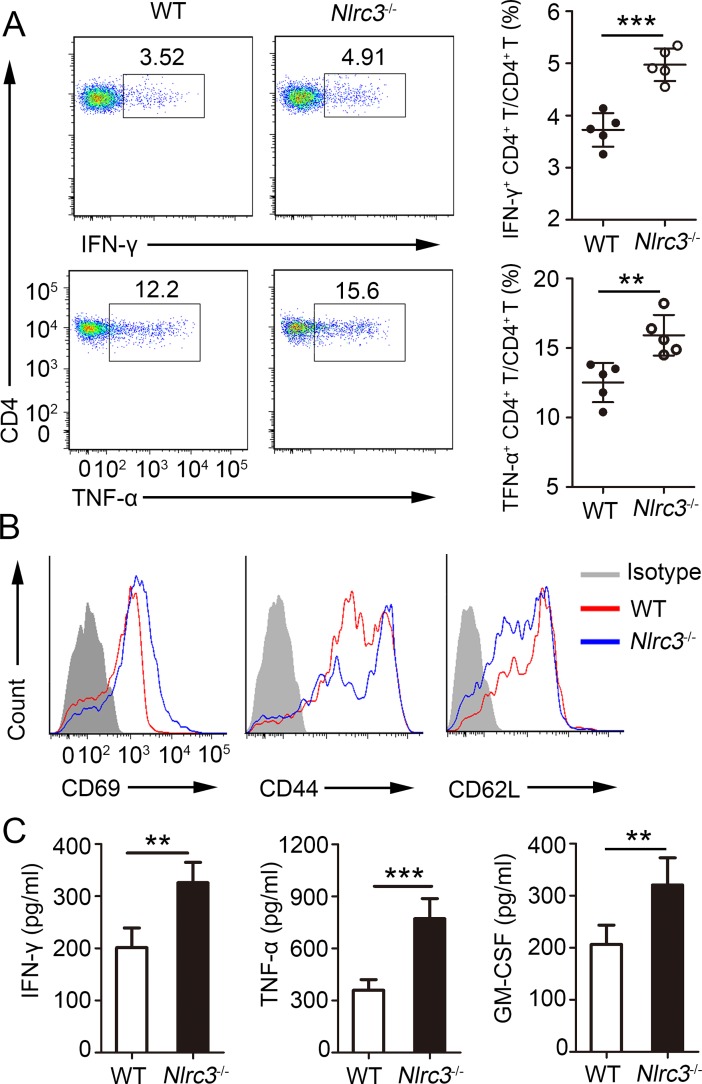
NLRC3 has emerged as a negative regulator of inflammatory signaling and is known to suppress the innate immune response during pathogen infection [16]. However, the ability of NLRC3 to modulate CD4+ T cell responses and the mechanisms by which NLRC3-mediated control of T cells can affect infectious disease progression remains poorly defined. Thus, to elucidate the effects of the NLRC3 knockdown on the CD4+ T cell phenotype during M. tuberculosis infection, we infected wild-type (WT) and NLRC3–deficient (Nlrc3-/-) mice with M. tuberculosis. CD4+ T cells isolated from the lungs of Nlrc3-/- mice showed stronger expression of intracellular IFN-γ, TNF-α and IL-17A after restimulation than that showed by CD4+ T cells isolated from lungs of WT mice (Fig 1A, S1A Fig) at 3 weeks post-infection (w.p.i.). However, NLRC3 knockdown did not affect Treg cells (CD25+ Foxp3+) (S1B Fig). Activation analysis revealed that Nlrc3-/- CD4+ T cells showed upregulation of CD44 and CD69 expression and downregulation of CD62L expression relative to that by WT CD4+ T cells (Fig 1B, S1C Fig). Consistent with these, the lungs of Nlrc3-/- mice contained higher levels of IFN-γ and TNF-α than WT mice (Fig 1C). GM-CSF, a kind of main CD4+ T cell-derived cytokines that activates human and murine macrophages to inhibit intracellular M. tuberculosis growth [27], was also increased in lungs of Nlrc3-/- mice (Fig 1C). Likewise, splenocytes isolated from Nlrc3-/- mice with M. tuberculosis at 3 w.p.i. were also found to secrete increased amounts of IFN-γ and TNF-α after stimulation with ESAT-6 peptide (S1D Fig). Taken together, the above results demonstrate that NLRC3 is a negative regulator of CD4+ T cell activation during M. tuberculosis infection.
Fig 1. NLRC3 deficiency promotes activation of CD4+ T in M. tuberculosis infection.
WT and Nlrc3-/- mice were infected with M. tuberculosis and mice were harvested at 3 weeks post-infection (w.p.i.). (A) Lung cells were restimulated with M. tuberculosis lysate directly ex vivo and the intracellular production of IFN-γ and TNF-α by CD4+ T cells was determined. Pooled data are presented in the right panel. (B) Expression of activation markers by lung CD4+ T cells. (C) Concentration of IFN-γ, TNF-α and GM-CSF in lungs (homogenized in 2 ml PBS and 0.05% Tween 80) as detected by ELISA. Data shown in (A, C) are the mean ±SD. **P < 0.01 and ***P < 0.001. Data are representative of three independent experiments with similar results.
Nlrc3-/- mice are protected from M. tuberculosis infection
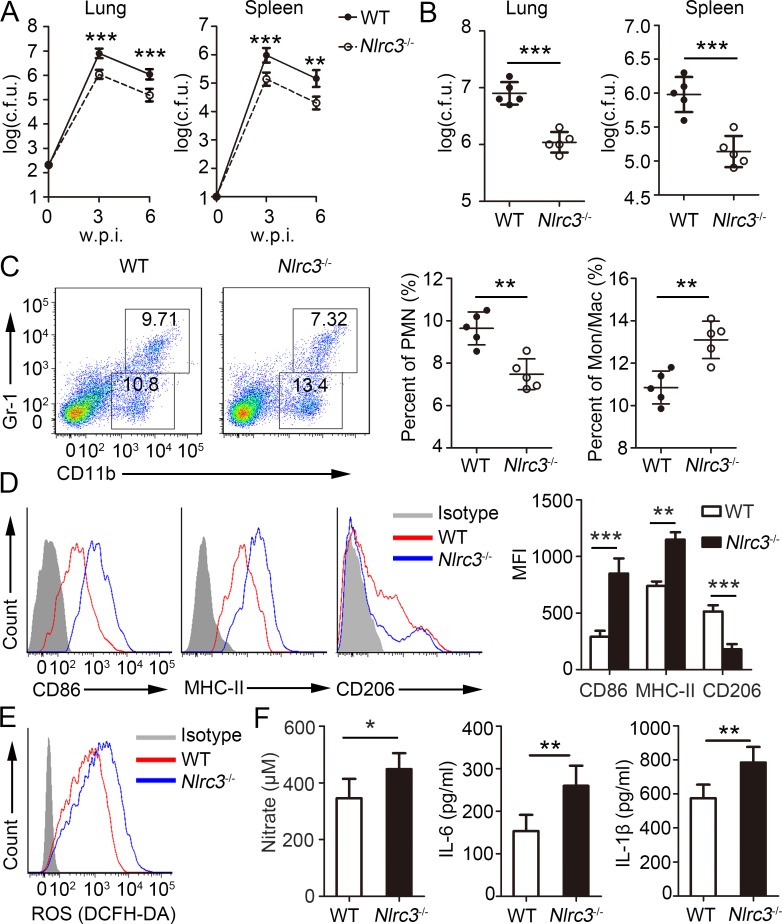
CD4+ T cells have been demonstrated to enhance the ability of macrophages to eliminate M. tuberculosis via the production of proinflammatory cytokines [25,33]. Given that CD4+ T cells showed enhanced activation phenotype under NLRC3-deficient conditions, we investigated the role of NLRC3 in the infectious disease. Previous studies showed that WT mice exposed to aerosol inoculation of M. tuberculosis showed improved control of bacterial burden and higher survival following CD4+ T recruitment into the lungs [34]. Consistent with the above findings, both WT and Nlrc3-/- mice showed no significant weight loss (S2A Fig) and mortality (S2B Fig). However, bacterial titers in the lungs and spleens of Nlrc3-/- mice were significantly lower than those of WT mice at 1, 3 and 6 w.p.i. (S2C Fig, Fig 2A and 2B). The bacterial burden in lungs from Nlrc3-/- mice was reduced by about 80% relate to WT mice (Fig 2B). To assess the lung-infiltrating innate cells at 1 w.p.i., we found that proportions and numbers of monocyte-macrophages (CD11b+ Gr-1-) and polymorph nuclear neutrophils (PMN) (CD11b+ Gr-1+) had no difference between Nlrc3-/- mice and WT mice (S2D and S2E Fig). The expression levels of surface marker CD86, MHC-II and CD206 on monocyte-macrophages were the similar between Nlrc3-/- mice and WT mice at 1 w.p.i. (S2F Fig). The IL-6 levels in the lung homogenates of Nlrc3-/- mice were significantly higher than those of WT mice, while IL-1β levels had no difference at 1 w.p.i. (S2G Fig). Moreover, we assessed the lung-infiltrating innate cells at 3 w.p.i., and found that total number of lung-infiltrating cells had no difference between Nlrc3-/- mice and WT mice via histological observation and inflammatory cell count identified by flow cytometry (S3A And S3B Fig). However, Nlrc3-/- mice contained significantly higher proportions and numbers of monocyte-macrophages, but lower proportions and numbers of PMN, relative to those in WT mice (Fig 2C, S3B Fig). Polarization of CD4+ T cells is known to influence macrophage differentiation in vivo [35]. We further assessed the effects of NLRC3 deficiency on macrophage differentiation. Surface marker expression analysis revealed that lung-infiltrating monocyte-macrophages in Nlrc3-/- mice showed upregulation of CD86 and MHC-II expression and downregulation of CD206 expression relative to those of WT mice (Fig 2D), which suggested that macrophages tend to differentiate into classically activated macrophages under NLRC3-deficient conditions [36]. Consistent with the above results, macrophages collected from Nlrc3-/- mice showed higher intracellular ROS levels than those collected from WT mice (Fig 2E). Nitrate, IL-6, and IL-1β levels in the lung homogenates of Nlrc3-/- mice were significantly higher relative to those of WT mice (Fig 2F). Together, the above results indicate that NLRC3-deficient mice showed stronger immune responses.
Fig 2. Nlrc3-/- mice are protected from M. tuberculosis infection.
WT and Nlrc3-/- mice infected with approximately 200 colony-forming units (c.f.u.) of M. tuberculosis were monitored. (A) Bacterial burdens were determined after infection at 3 and 6 w.p.i.. (B) Bacterial burdens were determined after infection at 3 w.p.i.. (C) Frequencies of lung-infiltrating cells that are neutrophils (CD11b+ Gr-1+) or monocyte-macrophages (CD11b+ Gr-1-) at 3 w.p.i.. (D) Expressions of CD86, MHC-II and CD206 were detected on monocyte-macrophages (CD11b+ Gr-1-) via flow cytometry at 3 w.p.i.. (E) ROS production by monocyte-macrophages (CD11b+ Gr-1-) were detected assessed as mean fluorescence intensity (MFI) of intracellular CFDA. (F) Concentrations of nitrate were measured by nitrate reductase assay and concentrations of IL-6 and IL-1β in lungs (homogenized in 2 ml PBS and 0.05% Tween 80) were detected by ELISA at 3 w.p.i.. Data shown in are the mean ±SD. *P < 0.05, **P < 0.01 and ***P < 0.001. Data are representative of three independent experiments with similar results.
NRLC3 directly suppresses CD4+ T cell activation in vitro
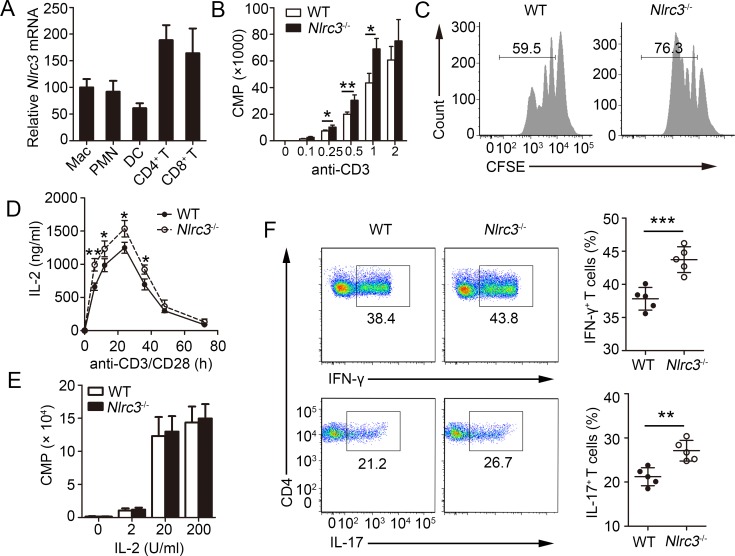
To elucidate how NLRC3-deficient environment impacts CD4+ T cell phenotype, first we detected T cell development in Nlrc3-/- mice. NLRC3 deficiency did not result in an obvious defect in thymic development (S4A and S4B Fig). However, the spleens had higher number of CD4+ T cells and CD8+ T cells (S4C and S4D Fig). Furthermore, CD4+ T cells from spleens of Nlrc3-/- mice produced greater amounts of intracellular IFN-γ and IL-17 (S4E Fig). These results indicate that NLRC3 is dispensable for thymic development but suppresses the CD4+ T cell functions in spleens in naive mice.
Next, we determined whether NLRC3 modulated CD4+ T development indirectly through antigen-presenting cells or directly. First, we evaluated expression of NLRC3 in various immune cell populations, and found that Nlrc3 mRNA could be detected in various immune cell populations, especially the highest expression among T cells (Fig 3A). These results were consistent with previous report [21]. Thus, we hypothesized that is a direct regulator of CD4+ T cells. To confirm this hypothesis, naïve CD4+ T cells from WT and Nlrc3-/- mice were activated in vitro. CD4+ T cell thymidine incorporation assays were conducted and found that purified Nlrc3-/- CD4+ T cells displayed increased thymidine incorporation relative to WT CD4+ T cells (Fig 3B). Likewise, Nlrc3-/- CD4+ T cells showed enhanced proliferation based on CFSE dye dilution (Fig 3C). IL-2 is a key cytokine that affects CD4+ T proliferation. Thus, we wondered whether the increased IL-2 production caused enhanced proliferation. To investigate this, we analyze the concentration of IL-2 in culture supernatant of CD4+ T with anti-CD3 and anti-CD28 antibodies. CD4+ T cells from Nlrc3-/- mice produced more IL-2 than did WT CD4+ T cells (Fig 3D). Furthermore, we stimulated CD4+ T cells with anti-CD3 and anti-CD28, then washed the cells and recultured them with exogenous IL-2. The proliferation would be no markedly difference between WT and Nlrc3-/- CD4+ T (Fig 3E). Our previous results showed that NLRC3-deficient environment impacts CD4+ T cell phenotype in vivo. We tested whether NLRC3-deficient in CD4+ T had the similar effect in vitro. The results showed that Nlrc3-/- CD4+ T cells had higher intracellular levels of IFN-γ and IL-17, produced from in vitro polarized Th cells, than those in WT CD4+ T cells (Fig 3F). On the contrary, WT and Nlrc3-/- CD4+ T cells showed no significant differences in intracellular IL-4 production (S5 Fig). Overall, these data indicate that NLRC3 suppresses activation and functions of CD4+ T directly.
Fig 3. T cell activation in vitro requires NLRC3.
(A) Relative expression of Nlrc3 in purified macrophages (CD11b+ Gr-1-), dendritic cells (CD11c+ MHCIIhi), polymorphonuclear leukocytes (PMNs) (CD11b+ Gr-1+), CD4+ T cells (CD3+ CD4+) and CD8+ T cells (CD3+ CD8+). (B) Purified naïve T cells isolated from WT and Nlrc3-/- mice were stimulated with plate bound anti-CD3 (increasing concentrations) and anti-CD28 (1 μg/ml) for 48 hr and the incorporation of thymidine was measured during the final 8 hr. (C) Purified WT and Nlrc3-/- naïve CD4+ T cells were labeled with CFSE and stimulated with anti-CD3 (1 μg/ml) and anti-CD28 (1 μg/ml) for 3 d. (D) Concentrations of IL-2 in supernatants of purified WT and Nlrc3-/- naïve CD4+ T cells stimulated for 0–80 h with anti-CD3 (1 μg/ml) and anti-CD28 (1 μg/ml) were detected by ELISA. (E) Thymidine incorporation in purified WT and Nlrc3-/- naive CD4+ T cells first primed with anti-CD3 and CD28 and then cultured with various concentrations of IL-2. (F) Purified WT and Nlrc3-/- naïve CD4+ T cells were polarized in Th1 or Th17 culture conditions for 4 days. Data shown in (B, C, E, F) are the mean ±SD. *P < 0.05 and **P < 0.01. Data are representative of three independent experiments with similar results.
NRLC3 directly suppresses CD4+ T cell activation in vivo
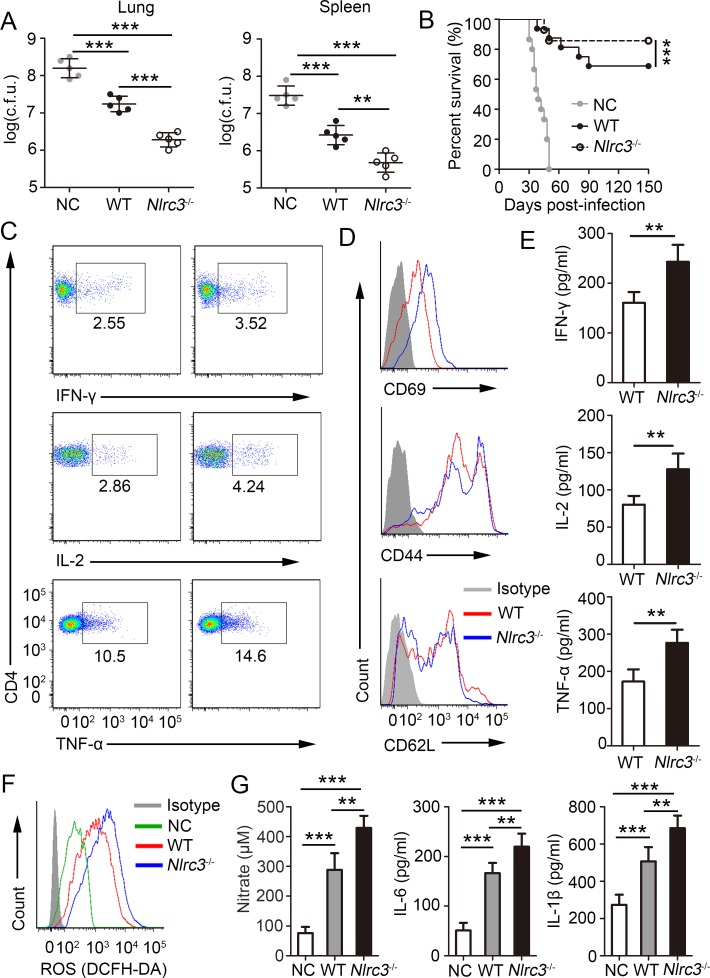
To further confirm the role of NLRC3 in the regulation of CD4+ T cells, competitive adoptive CD4+ T cell transfer assays were carried out. For these experiments, Rag2-/- recipient mice received 1:1 CD45.1+ WT and CD45.2+ Nlrc3-/- naïve CD4+ T cells and were subsequently infected with M. tuberculosis. We found that there was no significant difference in percentage of CD4+ T between CD45.1+ WT and CD45.2+ Nlrc3-/- CD4+ T in the same environment (Fig 4A) at 3 w.p.i. However, the percentages of CD4+ T cells producing IFN-γ were higher in CD45.2+ Nlrc3-/- CD4+ T cells than in CD45.1+ WT CD4+ T cells (Fig 4A). Furthermore, CD 69 expression was found to be upregulated in CD45.2+ Nlrc3-/- CD4+ T cells (Fig 4B). Consistent with the in vitro results, Nlrc3-/- CD4+ T cells showed higher IL-2 production after stimulation with M. tuberculosis lysate, thereby demonstrating the enhanced ability of CD45.2+ Nlrc3-/- CD4+ T cells to produce IL-2 in vivo (Fig 4C). These results establish an intrinsic role for NLRC3 as a negative regulator of CD4+ T cell activation during M. tuberculosis infection.
Fig 4. NLRC3 deficiency promotes differentiation of CD4+ T and IL-2 production in vivo.
Naïve CD4+ T cells from CD45.1+ WT and CD45.2+ Nlrc3-/- mice were mixed at a 1:1 ratio and competitively transferred via tail vein injection into Rag2-/- recipient mice. Then recipient mice were infected with M. tuberculosis and were harvested at 3w.p.i.. (A) Lung cells were restimulated with M. tuberculosis lysate directly ex vivo and the intracellular production of IFN-γ by CD4+ T cells was determined. Representative FACs plots depicting gating of CD4+ T cells are shown. Gating strategies to evaluate cytokine production by WT and Nlrc3-/- CD4+ T cells are provided. Numbers in the quadrants indicate the percent cells in each. Pooled data are presented in the right panel. (B) Expression of CD69 by lung CD4+ T cells. (C) Expression of IL-2 by lung CD4+ T cells. Pooled data are presented in the right panel. Data shown in (A, C) are the mean ±SD. *P < 0.05 and **P < 0.01. Data are representative of three independent experiments with similar results.
NLRC3 deficiency in CD4+ T cells promotes antibacterial immune responses
To define the role of NLRC3-deficiency of CD4+ T cells on M. tuberculosis infection, Rag2-/- mice were injected with naïve CD4+ T cells from WT or Nlrc3-/- mice and were subsequently infected with M. tuberculosis. We found that bacterial titres in the lungs and spleens of Rag2-/- mice injected with the Nlrc3-/- CD4+ T cells were significantly lower than those in mice injected with WT CD4+ T cells at 3 w.p.i. (Fig 5A). The survival of Rag2-/- mice given Nlrc3-/- CD4+ T cells was higher than that of Rag2-/- mice given WT CD4+ T (Fig 5B). Furthermore, to detect activation and development of CD4+ T, we found that CD4+ T cells isolated from lungs of Rag2-/- mice given Nlrc3-/- CD4+ T expressed higher levels of intracellular IFN-γ, IL-2 and TNF-α after restimulation than those given WT CD4+T (Fig 5C, S6A Fig). CD4+ T cells isolated from the lungs of Rag2-/- mice injected with Nlrc3-/- CD4+ T cells showed stronger expression of CD69 and CD44, but weaker expression of CD62L, relative to those isolated from the lungs of Rag2-/- mice injected with WT CD4+ T cells (Fig 5D, S6B Fig). Similarly, the lungs of Rag2-/- mice injected with Nlrc3-/- CD4+ T cells showed higher production of IFN-γ, IL-2 and TNF-α relative to those of WT mice (Fig 5E). In addition, CD4+ T cell counts in the draining lymph nodes (DLN), spleens, and lungs of Rag2-/- mice injected with Nlrc3-/- CD4+ T were higher than those of Rag2-/- mice injected with WT CD4+ T cells (S6C Fig). We next evaluated lung-infiltrating innate cells, and results revealed that the lungs of Rag2-/- mice injected with Nlrc3-/- CD4+ T cells had significantly higher percentages of monocyte-macrophages, but lower percentages of PMNs, relative to those in the lungs of Rag2-/- mice injected with WT CD4+ T cells (S7 Fig). Intracellular ROS in monocyte-macrophages (Fig 5F) and concentrations of nitrate, IL-6 and IL-1β in lung homogenate (Fig 5G) were all increased in Rag2-/- mice given Nlrc3-/- CD4+ T relative to those given WT CD4+ T. Together, these data indicate that absence of NLRC3 on CD4+ T cells promotes the antibacterial immune response of the body by regulating CD4+ T cell activation and further regulating the innate immune response.
Fig 5. NLRC3 deficiency promotes the antibacterial ability in vivo via regulating CD4+ T cells.
Purified WT or Nlrc3-/- naïve CD4+ T cells were adoptively transferred into Rag2-/- mice. Then recipient mice were infected with M. tuberculosis and parts of mice were harvested at 3w.p.i.. (A) Bacterial burdens were determined in lungs and spleen at 3w.p.i.. (B) Survival of mice every other day from 0–150 day post-infection (d.p.i.). (C) Lung cells were restimulated with M. tuberculosis lysate directly ex vivo and the intracellular production of IFN-γ, IL-2, and TNF-α by CD4+ T cells was determined. (D) Expression of activation markers by lung CD4+ T cells. (E) Concentration of IFN-γ, IL-2 and TNF-α in lungs (homogenized in 2 ml PBS and 0.05% Tween 80) as detected by ELISA. (F) ROS production by monocyte-macrophages (CD11b+ Gr-1-) were detected assessed as mean fluorescence intensity (MFI) of intracellular CFDA. (G) Concentrations of nitrate were measured by nitrate reductase assay and concentrations of IL-6 and IL-1β in lungs (homogenized in 2 ml PBS and 0.05% Tween 80) were detected by ELISA. Data shown in (A, E, G) are the mean ±SD. *P < 0.05, **P < 0.01 and ***P < 0.001. Data are representative of three independent experiments with similar results.
NLRC3 negatively regulates CD4+ T cell activation via NF-κB and ERK signaling
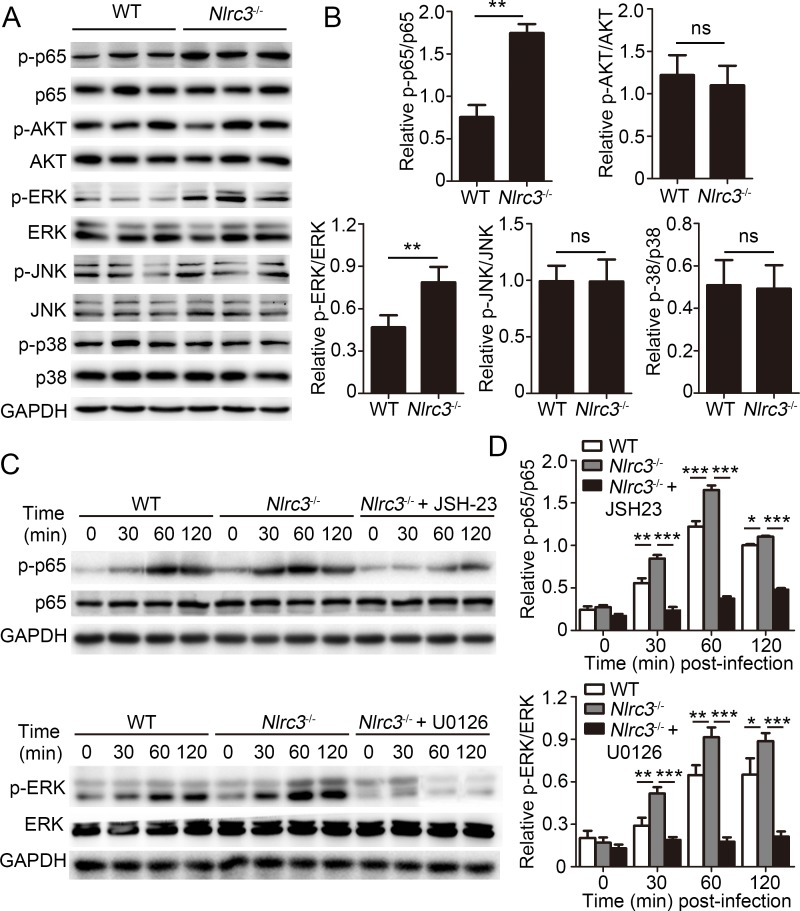
The specific molecular pathways that are controlled by NLRC3 in CD4+ T cells remain unclear. First, our results showed that transfer of Nlrc3-/- CD4+ T cells resulted in enhanced phosphorylation of NF-κB p65 and ERK (Fig 6A and 6B) in lungs of recipient mice. While the phosphorylation of AKT, JNK and p38 was no difference between two groups (Fig 6A and 6B). These results suggested that increased phosphorylation of NF-κB p65 and ERK caused by NLRC3 deficiency of CD4+ T might contribute to improved antibacterial ability of the body. To determine whether NLRC3 suppressed activation of CD4+ T via mediating regulation of NF-κB p65 and ERK activation, we evaluated the signaling in purified CD4+ T cells. As expected, the results demonstrated that stimulation of Nlrc3-/- CD4+ T cells with anti-CD3 and anti-CD28 led to increased phosphorylation of NF-κB p65 and ERK relative to WT CD4+ T cells (Fig 6C and 6D). Then we used NF-κB-inhibitor JSH-23 and MEK1/2-inhibitor U0126 to inhibit those pathway signals respectively, and observed phosphorylation of these proteins was decreased (Fig 6C and 6D).
Fig 6. NLRC3 negatively regulates NF-κB and ERK Signaling in CD4+ T cells.
(A-B) Purified WT or Nlrc3-/- naïve CD4+ T cells were adoptively transferred into Rag2-/- mice. Then recipient mice were infected with M. tuberculosis and were harvested at 3w.p.i.. Lungs were collected. (A) Immunoblot analysis of lung lysates. Each lane represents an individual mouse. (B) Densitometry quantification of band intensity for A. (C-D) Purified WT and Nlrc3-/- CD4+ T cells were stimulated with anti-CD3 (1 μg/ml) plus anti-CD28 (1 μg/ml) in the presence or absence of the NF-κB inhibitor JSH-23 (20 μM) or MEK1/2-inhibitor U0126 (40 μM). (C) Lysates were probed for total and phosphorylated p65 (p-p65), p65, p-ERK, ERK and GAPDH. (D) Densitometry quantification of band intensity for C. Data shown in (B and D) are the mean ±SD. *P < 0.05, **P < 0.01 and ***P < 0.001. Data are representative of three independent experiments with similar results.
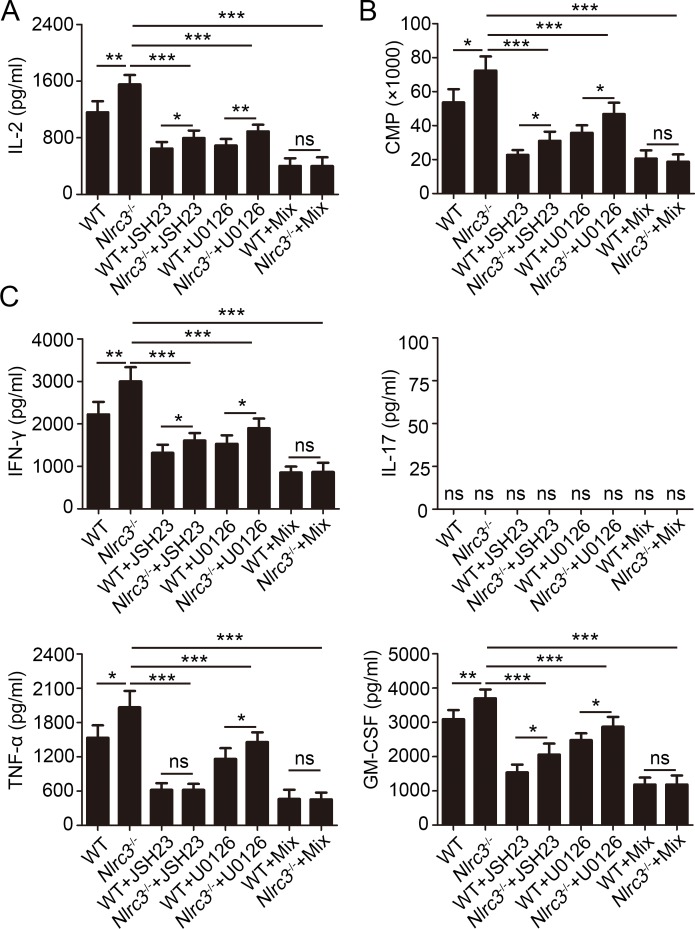
Next we examined the effect of inhibiting the phosphorylation of these proteins on activation of CD4+ T. Activation analysis revealed that Nlrc3-/- CD4+ T cells showed upregulation CD69 expression after stimulated with anti-CD3 and anti-CD28 relative to that by WT CD4+ T cells, while CD69 expression level would have no difference between the Nlrc3-/- CD4+ T and WT CD4+ T cells with mix of JSH-23 and U0126 (S8 Fig). Likewise, we found that mix of JSH-23 and U0126 suppressed increased IL-2 expression of Nlrc3-/- CD4+ T cells stimulated with anti-CD3 and anti-CD28 (Fig 7A), but one of them alone could not limit completely increased IL-2 expression caused by NLRC3- deficiency on CD4+ T (Fig 7A). Likewise, pharmacological inhibition of NF-κB and ERK treatments lowered the enhanced proliferation capacity exhibited by Nlrc3-/- CD4+ T (Fig 7B). Furthermore, pharmacological inhibitions of NF-κB and ERK were found to reduce IFN-γ, TNF-α and GM-CSF secretion by Nlrc3-/- CD4+ T cells (Fig 7C). However, we did not detect the IL-17 secretion by CD4+ T cells stimulated only with anti-CD3 and anti-CD28 (Fig 7C). In total, NLRC3 negatively regulates activation of CD4+ T via mediating NF-κB and ERK signaling pathways.
Fig 7. NLRC3 suppresses activation of CD4+ T cells via negatively regulating NF-κB and ERK Signaling.
Purified WT and Nlrc3-/- CD4+ T cells were stimulated for 48 hr with anti-CD3 (1 μg/ml) plus anti-CD28 (1 μg/ml) in the presence or absence of the NF-κB inhibitor JSH-23 (20 μM), MEK1/2-inhibitor U0126 (40 μM) or the mix of the two. (A) Concentrations of IL-2 in supernatants were detected by ELISA. (B) The incorporation of thymidine was measured during the final 8 hr. (C) Concentrations of IFN-γ, IL-17, TNF-α and GM-CSF in supernatants were detected by ELISA. Data shown are the mean ±SD. *P < 0.05, **P < 0.01 and ***P < 0.001. Data are representative of three independent experiments with similar results.
Discussion
Accumulating evidence that NLRs play an intrinsic role in regulating T cell responses lymphoid tissues [11,13]. However, to our known, contribution for NLRs modulation of T-cell responses directly in infectious diseases has not been studied. In the present study, we focused on elucidating the roles of NLRC3 in shaping CD4+ T cell responses in vivo and in vitro, in particular during M. tuberculosis infection. We found that NLRC3 played an intrinsic role in regulating negatively CD4+ T cell responses in lungs and lymphoid tissues, including differentiation and proliferation, which in turn suppresses the innate immune responses and promotes M. tuberculosis survival.
The current study provides a previously unidentified role that NLRC3 regulates Th1 and Th17 differentiation of CD4+ T during M. tuberculosis infection. NLRC3-deficient CD4+ T showed upregulated expression of Th1 and Th17-related cytokines, such as IFN-γ, IL-17, TNF-α and GM-CSF. These cytokines play vital role in activating macrophages to kill intracellular bacteria [24]. However, previous study showed that M. tuberculosis could induce immune evasion via T cell-costimulatory molecules, such as PD-1, CTLA4, and Tim-3 on T cells, to induce impairment of T-cell immunity [29,37,38]. The role of NLRC3 in inducing impairment of T-cell immunity is similar with T cell-costimulatory molecules. Thus, it might be an important strategy for M. tuberculosis to induce immune evasion via negative regulator of PRRs family suppressing adaptive immunity.
Following M. tuberculosis infection, phagocytic cells including macrophages and neutrophils will be recruited to the lung and form protective immune responses [39,40]. However, excessive PMN recruitment will cause damaging immunity and uncontrolled tissue damage [28]. NLRC3 has been described as a negative regulator of inflammatory signaling. Thus, it is worrying whether NLRC3-deficiency in body will lead to excessive inflammatory response. However, the results were almost completely opposite that PMN recruitment is reduced in NLRC3 deficient mice. Previous study showed that FN-γ directly inhibits neutrophil accumulation in lung and impairs neutrophil survival during M. tuberculosis infection [41]. Increased IFN-γ expression might contribute to the reduced PMN recruitment in NLRC3-deficient mice during M. tuberculosis infection.
Previous studies revealed that NLRC3 suppresses NF-κB signaling in macrophages and T cells [16,21]. We likewise observed that NLRC3-deficience in CD4+ T cells resulted in increased NF-κB p65 phosphorylation in CD4+ T cells. In addition, enhanced ERK phosphorylation was observed in NLRC3-deficient CD4+ T cells, which has not been reported in previous studies. Pharmacological inhibition of NF-κB or MEK-ERK signaling alone could not retune enhanced proliferation and cytokine production of CD4+ T cells induced by NLRC3-deficience. Thus, NLRC3 might regulate negatively activation of CD4+ T via suppressing both NF-κB and MEK-ERK signaling pathways. Previous study showed that NLRC3 inhibited activation of NF-κB by interacting with the adaptor TRAF6 to attenuate Lys63-linked ubiquitination of TRAF6 [15], but it is unclear whether NLRC3 regulates NF-κB signaling pathway via the same mechanism. The mechanism of NLRC3 regulating ERK signaling pathway remain to be investigated. Further studies are required to clarify these regulatory mechanisms. NLRC3 is a typical intracellular member of NLR family. However, the typical ligands of NLRC3 are still unclear. To study whether NLRC3 regulates NF-κB and ERK signal pathways via recognizing certain ligands will be very meaningful to enhance the understanding of its function.
In summary, we identified that NLRC3 regulated negatively CD4+ T cells directly. Loss of NLRC3 in CD4+ T cells enhanced the protective immune response against M. tuberculosis infection. Finally, we demonstrated that NLRC3-dificience in CD4+ T cells promoted the activation, proliferation and cytokine production of CD4+ T cell via negatively regulating NF-κB and MEK-ERK signaling pathways. Our study highlighted the critical role of NLRC3 in the regulation of the adaptive immune response and protective immunity during M. tuberculosis infection. Our findings also suggested that NLRC3 serves as a potential target for the development of therapeutic intervention against tuberculosis.
Materials and methods
Ethics statement
All animal experiments in this study were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All experimental protocols were reviewed and approved by the Medical Ethics Board and the Biosafety Management Committee of Southern Medical University (approval number SMU-L2015123).
Mice
C57BL/6 mice were purchased from the Lab Animal Center of Southern Medicine University (Guangzhou, China). NLRC3-deficient (Nlrc3-/-) mice were built by Shanghai Research Center for Model Organisms (Shanghai, China). CD45.1+ and Rag2-/- mice on a C57BL/6 background were purchased from Nanjing Biomedical Research Institute (Nanjing, China). All mice were maintained in the Lab Animal Center of Southern Medicine University under specific pathogen-free conditions.
M. tuberculosis infection of mice
6~8-week old female and male mice were exposed to 1 × 107 colony-forming units (CFU) of M. tuberculosis H37Rv (ATCC 27294, the same below) in an Inhalation Exposure System (Glas-Col, USA), which delivers ~ 200 bacteria to the lung per animal. At 24 h after infection, the bacterial titres in the lungs (left lobe) of at least two mice were determined to confirm the dose of M. tuberculosis H37Rv inoculation. Bacterial burden was determined by plating serial dilutions of lung (left lobe) and spleen homogenates onto 7H10 agar plates (BD Difco, USA) supplemented with 10% OADC. Plates were incubated at 37°C in 5% CO2 for 4 weeks before counting colonies.
Lymphocyte isolation
A single-cell suspension was prepared from the spleen or lymph node by passing the organ through a 70-μm nylon cell strainer, followed by treatment with red blood cell lysis buffer. Lung cell suspensions (right lobe) were prepared by perfusing cold saline containing heparin through the heart, removed, and sectioned in ice-cold medium. Dissected lung tissue was incubated in 0.7 mg/ml collagenase IV and 30 μg/ml DNase (Sangon Biotech (Shanghai), China) at 37°C for 30 min. Digested lungs were disrupted by passage through a 70-μm nylon cell strainer, treated with red blood cell lysis buffer, and processed over a 40:80% Percoll (GE Healthcare) gradient. The resulting cell suspension was washed and counted.
FACS analysis
For intracellular cytokine detection, isolated cells were cultured in 20 μg/ml of M. tuberculosis lysate or PMA/ionomycin for 1.5 h before 10 μg/ml Brefeldin A (eBioscience, USA) was added to the culture for 3.5 h more. For surface staining, lymphocytes were harvested, washed and stained for 30 min on ice with mixtures of fluorescently conjugated mAbs or isotype-matched controls. mAbs of mice were as follows: APC-Cy7-anti-CD3, PE-Cy7-anti-CD4, APC-anti-CD8a, PE-anti-CD45.1, Alexa Fluor 700-anti-CD45.2, PE-anti-CD25, PerCP-cy5.5-anti-CD69, PE-anti-CD44, FITC-anti-CD62L, PE-cy7-anti-Gr-1, PE-anti-CD11b, APC-ant-CD86, Alexa Fluor 700-anti-MHC-II and PerCP-cy5.5-anti-CD206 (eBioscience). For intracellular staining, the cells were incubated 20 min in IC Fixation buffer (eBioscience), followed by permeabilization buffer (eBioscience) and 1 h of incubation with appropriate mAbs of mice: PE-anti-IFN-γ, FITC-anti-IL-17, PerCP-cy5.5-anti-IL-4, FITC-anti-IL-2 and FITC-anti-Foxp3. Cell phenotype was analyzed by flow cytometry on a flow cytometer (BD LSR II) (BD Biosciences, USA). Data were acquired as the fraction of labeled cells within a live-cell gate and analyzed using FlowJo software (Tree Star). All gates were set on the basis of isotype-matched control antibodies.
Enzyme-linked immunosorbent assay (ELISA)
Lungs (right lobe) were homogenized in 2 ml PBS + 0.05% Tween 80. Homogenized tissue supernatants were filtered (0.22 μ m). Cell culture supernatants were collected. Cytokine production was measured by enzyme-linked immunosorbent assay of mouse IFN-γ, TNF-α, GM-CSF, nitrate, IL-6, IL-1β or IL-2 (ExCell Bio, China) according to the manufacturer’s protocol.
Nlrc3 expression
Macrophages (Gr-1- CD11b+), polymorphonuclear leukocytes (PMNs) (Gr-1+ CD11b+), dendritic cells (CD11c+ MHC-IIhi), CD4+ T cells (CD3+ CD4+) and CD8+ T cells (CD3+ CD8+) were purified by flow cytometry sorting and total RNA was isolated with Trizol (Invitrogen, USA) according to the manufacturer’s instructions. 1 mg of RNA was reverse transcribed to cDNA with random RNAspecific primers using the high-capacity cDNA reverse transcription kit (Applied Biosystems). Transcript amounts of Nlrc3 and Gapdh were analyzed with SYBR-Green (Applied Biosystems) according to the manufacturer’s recommendations. The primer sequences used for PCR are in S1 Table.
Thymidine incorporation assay
Splenocytes and LNs were harvested from WT and Nlrc3-/-mice. Naïve WT and Nlrc3-/- CD4+ T cells (CD3+ CD4+ CD44lo CD62Lhi) were purified by FACs sorting. Purified naive T cells were stimulated with increasing concentrations of plate bound anti-CD3 (eBioscience) and anti-CD28 (eBioscience) in triplicate wells for 48 hr. During the last 8 hr of stimulation, T cells were pulsed with [3H]thymidine and the amount of incorporated [3H]thymidine was measured as counts per minute (cpm). For signal pathway-inhibition studies, the NF-κB-inhibitor JSH-23 (20μM) and MEK1/2-inhibitor U0126 (40μM) (Selleck, USA) was added into culture media.
CFSE CD4+ T cell proliferation assay
Splenocytes and LNs were harvested from WT and Nlrc3-/-mice. Naïve WT and Nlrc3-/- CD4+ T cells (CD3+ CD4+ CD44lo CD62Lhi) were purified by FACs sorting. Purified T cells were labeled with 2.5 μM CFSE and then 5 × 104 T cells/well were stimulated with anti-CD3 (1.0 μg/ml) and anti-CD28 (1.0 μg/ml). T cells were cultured for 72 hrs and proliferation was determined by Flow cytometry analysis of CFSE dilution.
Th cells polarization
For Th cell polarization, splenocytes and LNs were harvested from WT and Nlrc3-/-mice. Naïve WT and Nlrc3-/- CD4+ T cells (CD3+ CD4+ CD44lo CD62Lhi) were purified by FACs sorting. Cells were cultured in RPMI 1640 medium (Thermo Fisher Scientific, USA) with plate-bound anti-CD3 and anti-CD28 antibodies in the presence of cytokines (R&D, USA) and neutralizing antibodies (R&D) as follows. Th1 conditions: IFN-γ (200 ng/ml), IL-12 (2 ng/ml), anti-IL-4 (5 mg/ml). Th2 conditions: IL-4 (10 ng/ml) and anti-IFN-γ (5 mg/ml). Th17 conditions: TGF-β3 (2ng/ml), IL-6 (25 ng/ml), IL-1β (10 ng/ml), IL-23 (10 ng/ml), anti-IL-4 (5 mg/ml) and anti-IFN-γ (5 mg/ml).
CD4+ T cells adoptive transfer
Splenocytes and LNs were harvested from WT and Nlrc3-/-mice. Naïve WT and Nlrc3-/- CD4+ T cells (CD3+ CD4+ CD44lo CD62Lhi) were purified by FACs sorting. Cells were counted and then (1x106 cells) adoptively transferred into Rag2-/- recipient mice via tail vein injection. These recipient mice were infected with M. tuberculosis after one day.
Competitive T cell adoptive transfer
Splenocytes and LNs were harvested from WT (CD45.1+) and Nlrc3-/- (CD45.2+) mice. Naïve WT and Nlrc3-/- CD4+ T cells (CD3+ CD4+ CD44lo CD62Lhi) were purified by FACs sorting and mixed at a 1:1 ratio. Mixed 1:1 naïve T cells (1x106 total cells) were then adoptively transferred into Rag2-/- recipient mice via tail vein injection. These recipient mice were infected with M. tuberculosis after one day. Lungs were harvested on day 21 post adoptive transfer and homeostatic expansion was evaluated using congenic CD45 markers and flow cytometry.
Western blotting
Tissue or cells were washed three times with ice-cold PBS and then lysed in lysis buffer containing 1 mM phenylmethylsulfonyl fluoride, 1% (vol/vol) protease inhibitor cocktail (Sigma Aldrich, USA), and 1 mM DTT. Equal amounts (20 mg) of cell lysates were resolved using 8–15% polyacrylamide gels transferred to PVDF membrane. Membranes were blocked in 5% non-fat dry milk in PBST and incubated overnight with the respective primary antibodies at 4°C. The membranes were incubated at room temperature for 1 h with appropriate HRP-conjugated secondary antibodies and visualized with Plus-ECL (PerkinElmer, CA) according to the manufacturer’s protocol.
Statistics
All experiments were performed at least twice. When shown, multiple samples represent biological (not technical) replicates of mice randomly sorted into each experimental group. No blinding was performed during animal experiments. Determination of statistical differences was performed with Prism 5 (Graphpad Software, Inc.) using unpaired two-tailed t-tests (to compare two groups with similar variances), or one-way ANOVA with Bonferonni’s multiple comparison test (to compare more than two groups).
Supporting information
NLRC3 deficiency promotes activation of CD4+ T in M. tuberculosis infection. WT and Nlrc3-/- mice were infected with M. tuberculosis and mice were harvested at 3 weeks post-infection (w.p.i.). (A) Lung cells were restimulated with M. tuberculosis lysate directly ex vivo and the intracellular production of IL-17 by CD4+ T cells was determined. Pooled data are presented in the right panel. (B) Expression of CD25 and Foxp3 were detected on CD4+ T cells from lungs. Pooled data are presented in the right panel. (C) Expression of activation markers were assessed as mean fluorescence intensity (MFI) on lung CD4+ T cells. (D) Splenocytes were stimulated for 48 hr with ESAT-6 peptide (5μg/ml) and cytokine production was measured by ELISA. Data shown are the mean ±SD. **P < 0.01 and ***P < 0.001. Data are representative of three independent experiments with similar results.
(TIF)
NLRC3 deficiency had no effect on weight and survival of mice with M. tuberculosis infection. Mice infected with approximately 200 colony-forming units of M. tuberculosis were monitored at various days post infection. (A) Weight change. (B) Survival every day from 0–200 day post-infection. Data shown are the mean ±SD. (C) Bacterial burdens were determined after infection at 1w.p.i.. (D) Frequencies of lung-infiltrating cells that are neutrophils (CD11b+ Gr-1+) or monocyte-macrophages (CD11b+ Gr-1-) at 1 w.p.i.. (E) Numbers of lung-infiltrating cells were counted at 1 w.p.i. (F) Expressions of CD86, MHC-II and CD206 were detected on monocyte-macrophages (CD11b+ Gr-1-) via flow cytometry at 1 w.p.i.. (G) Concentrations of IL-6 and IL-1β in lungs (homogenized in 2 ml PBS and 0.05% Tween 80) were detected by ELISA at 1 w.p.i.. Data shown are the mean ±SD. **P < 0.01. Data are representative of three independent experiments with similar results.
(TIF)
WT and Nlrc3-/- mice infected with approximately 200 colony-forming units of M. tuberculosis were monitored. (A) H&E-stained lung sections derived from two representative mice in each group of mice 3 w.p.i.. The magnification is shown at the right of each image. (B) Numbers of lung-infiltrating cells were counted at. Data shown are the mean ±SD. *P < 0.05 and **P < 0.01. Data are representative of three independent experiments with similar results.
(TIF)
NLRC3 does not affect thymic development but does influence mature CD4+ T cells. (A) Representative expression of CD4 and CD8 by WT and Nlrc3-/- thymocytes. Pooled data are presented in the right panel. DN, double negative (CD4- CD8-); DP, double positive (CD4+ CD8+); CD4, CD4 single positive (CD4+ CD8-); CD8, CD8 single positive (CD4- CD8+). (B) Total numbers of thymocytes in each stage of thymic development. (C) Representative expression of splenic CD4+ and CD8+ T cells from WT and Nlrc3-/- mice. Pooled data are presented in the right panel. (D) Enumeration of splenic CD4+ and CD8+ T cells from WT and Nlrc3-/- mice. (E) Production of IFN-γ and IL-17 by CD4+ T cells following stimulation with PMA/ionomycin. Pooled data are presented in the right panel. Data shown are the mean ±SD. *P < 0.05 and **P < 0.01. Data are representative of three independent experiments with similar results.
(TIF)
NLRC3 does not affect differentiation of Th2. Purified WT and Nlrc3-/- naïve CD4+ T cells were polarized in Th2 culture conditions for 4 days. Data shown are the mean ±SD. Data are representative of three independent experiments with similar results.
(TIF)
NLRC3 deficiency of CD4+ T promotes the antibacterial ability in vivo. Purified WT or Nlrc3-/- naïve CD4+ T cells were adoptively transferred into Rag2-/- mice. Then recipient mice were infected with M. tuberculosis and parts of mice were harvested at 3w.p.i.. (A) Lung cells were restimulated with M. tuberculosis lysate directly ex vivo and the intracellular production of IFN-γ, IL-2, and TNF-α by CD4+ T cells was determined. Pooled data are presented. (B) Mean fluorescence intensity (MFI) of activation markers by lung CD4+ T cells. (C) Enumeration of CD4+ cells in draining lymph nodes (DLNs), spleens and lungs. Data shown are the mean ±SD. **P < 0.01 and ***P < 0.001. Data are representative of three independent experiments with similar results.
(TIF)
NLRC3 deficiency of CD4+ T affected infiltration of myeloid cells to lung. Purified WT or Nlrc3-/- naïve CD4+ T cells were adoptively transferred into Rag2-/- mice. Then recipient mice were infected with M. tuberculosis and parts of mice were harvested at 3w.p.i.. Frequencies of lung-infiltrating cells that are neutrophils (CD11b+ Gr-1+) or monocyte-macrophages (CD11b+ Gr-1-). Pooled data are presented in the right panel. Data shown are the mean ±SD. **P < 0.01. Data are representative of three independent experiments with similar results.
(TIF)
NLRC3 suppresses activation of CD4+ T cells via negatively regulating NF-κB and ERK Signaling. Purified WT and Nlrc3-/- CD4+ T cells were stimulated for 48 hr with anti-CD3 (1 μg/ml) plus anti-CD28 (1 μg/ml) in the presence or absence of the NF-κB inhibitor JSH-23 (20 μM), MEK1/2-inhibitor U0126 (40 μM) or the mix of the two. And then expression of CD69 were detected. *P < 0.05 and **P < 0.01. Data are representative of three independent experiments with similar results.
(TIF)
(XLSX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
National Science and Technology Key Projects on Major Infectious Diseases (2017ZX10201301-008 LM), National Natural Science Foundation of China (81772150, 81571951, LM) and National Natural Science Foundation of China (81641062, SH) played role in the study design; Natural Science Foundation of Guangdong Province (2016A030311001, LM) and Natural Science Foundation of Guangdong Province (2017A030310268, SH), Science and Technology Project of Guangdong Province (2017A020212007, LM), Science and Technology Project of Guangzhou (201707010215, LM) and Medical Scientific Research Foundation of Guangdong Province (A2017073, SH) played role in data collection and analysis.
References
- 1.Yeretssian G (2012) Effector functions of NLRs in the intestine: innate sensing, cell death, and disease. Immunol Res 54: 25–36. 10.1007/s12026-012-8317-3 [DOI] [PubMed] [Google Scholar]
- 2.Kufer TA, Sansonetti PJ (2011) NLR functions beyond pathogen recognition. Nat Immunol 12: 121–128. 10.1038/ni.1985 [DOI] [PubMed] [Google Scholar]
- 3.Van Gorp H, Kuchmiy A, Van Hauwermeiren F, Lamkanfi M (2014) NOD-like receptors interfacing the immune and reproductive systems. FEBS J 281: 4568–4582. 10.1111/febs.13014 [DOI] [PubMed] [Google Scholar]
- 4.Kanneganti TD, Lamkanfi M, Nunez G (2007) Intracellular NOD-like receptors in host defense and disease. Immunity 27: 549–559. 10.1016/j.immuni.2007.10.002 [DOI] [PubMed] [Google Scholar]
- 5.Rathinam VA, Vanaja SK, Fitzgerald KA (2012) Regulation of inflammasome signaling. Nat Immunol 13: 333–342. 10.1038/ni.2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geddes K, Magalhaes JG, Girardin SE (2009) Unleashing the therapeutic potential of NOD-like receptors. Nat Rev Drug Discov 8: 465–479. 10.1038/nrd2783 [DOI] [PubMed] [Google Scholar]
- 7.Griebel T, Maekawa T, Parker JE (2014) NOD-like receptor cooperativity in effector-triggered immunity. Trends Immunol 35: 562–570. 10.1016/j.it.2014.09.005 [DOI] [PubMed] [Google Scholar]
- 8.Zhong Y, Kinio A, Saleh M (2013) Functions of NOD-Like Receptors in Human Diseases. Front Immunol 4: 333 10.3389/fimmu.2013.00333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anand PK, Malireddi RK, Lukens JR, Vogel P, Bertin J, et al. (2012) NLRP6 negatively regulates innate immunity and host defence against bacterial pathogens. Nature 488: 389–393. 10.1038/nature11250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui J, Li Y, Zhu L, Liu D, Songyang Z, et al. (2012) NLRP4 negatively regulates type I interferon signaling by targeting the kinase TBK1 for degradation via the ubiquitin ligase DTX4. Nat Immunol 13: 387–395. 10.1038/ni.2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lukens JR, Gurung P, Shaw PJ, Barr MJ, Zaki MH, et al. (2015) The NLRP12 Sensor Negatively Regulates Autoinflammatory Disease by Modulating Interleukin-4 Production in T Cells. Immunity 42: 654–664. 10.1016/j.immuni.2015.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinic M, Caminschi I, O'Keeffe M, Thinnes T, Grumont R, et al. (2017) The Bacterial Peptidoglycan-Sensing Molecules NOD1 and NOD2 Promote CD8(+) Thymocyte Selection. J Immunol 198: 2649–2660. 10.4049/jimmunol.1601462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bruchard M, Rebe C, Derangere V, Togbe D, Ryffel B, et al. (2015) The receptor NLRP3 is a transcriptional regulator of TH2 differentiation. Nat Immunol 16: 859–870. 10.1038/ni.3202 [DOI] [PubMed] [Google Scholar]
- 14.Arbore G, West EE, Spolski R, Robertson AAB, Klos A, et al. (2016) T helper 1 immunity requires complement-driven NLRP3 inflammasome activity in CD4(+) T cells. Science 352: aad1210 10.1126/science.aad1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schneider M, Zimmermann AG, Roberts RA, Zhang L, Swanson KV, et al. (2012) The innate immune sensor NLRC3 attenuates Toll-like receptor signaling via modification of the signaling adaptor TRAF6 and transcription factor NF-kappaB. Nat Immunol 13: 823–831. 10.1038/ni.2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang L, Mo J, Swanson KV, Wen H, Petrucelli A, et al. (2014) NLRC3, a member of the NLR family of proteins, is a negative regulator of innate immune signaling induced by the DNA sensor STING. Immunity 40: 329–341. 10.1016/j.immuni.2014.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karki R, Man SM, Malireddi RK, Kesavardhana S, Zhu Q, et al. (2016) NLRC3 is an inhibitory sensor of PI3K-mTOR pathways in cancer. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tocker AM, Durocher E, Jacob KD, Trieschman KE, Talento SM, et al. (2017) The Scaffolding Protein IQGAP1 Interacts with NLRC3 and Inhibits Type I IFN Production. J Immunol 199: 2896–2909. 10.4049/jimmunol.1601370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eren E, Berber M, Ozoren N (2017) NLRC3 protein inhibits inflammation by disrupting NALP3 inflammasome assembly via competition with the adaptor protein ASC for pro-caspase-1 binding. J Biol Chem 292: 12691–12701. 10.1074/jbc.M116.769695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo L, Kong Q, Dong Z, Dong W, Fu X, et al. (2017) NLRC3 promotes host resistance against Pseudomonas aeruginosa-induced keratitis by promoting the degradation of IRAK1. Int J Mol Med 40: 898–906. 10.3892/ijmm.2017.3077 [DOI] [PubMed] [Google Scholar]
- 21.Conti BJ, Davis BK, Zhang J, O'Connor W Jr., Williams KL, et al. (2005) CATERPILLER 16.2 (CLR16.2), a novel NBD/LRR family member that negatively regulates T cell function. J Biol Chem 280: 18375–18385. 10.1074/jbc.M413169200 [DOI] [PubMed] [Google Scholar]
- 22.Dye C, Glaziou P, Floyd K, Raviglione M (2013) Prospects for tuberculosis elimination. Annu Rev Public Health 34: 271–286. 10.1146/annurev-publhealth-031912-114431 [DOI] [PubMed] [Google Scholar]
- 23.Lienhardt C, Glaziou P, Uplekar M, Lonnroth K, Getahun H, et al. (2012) Global tuberculosis control: lessons learnt and future prospects. Nat Rev Microbiol 10: 407–416. 10.1038/nrmicro2797 [DOI] [PubMed] [Google Scholar]
- 24.Mortaz E, Adcock IM, Tabarsi P, Masjedi MR, Mansouri D, et al. (2015) Interaction of Pattern Recognition Receptors with Mycobacterium Tuberculosis. J Clin Immunol 35: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayer-Barber KD, Barber DL (2015) Innate and Adaptive Cellular Immune Responses to Mycobacterium tuberculosis Infection. Cold Spring Harb Perspect Med 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orme IM, Robinson RT, Cooper AM (2015) The balance between protective and pathogenic immune responses in the TB-infected lung. Nat Immunol 16: 57–63. 10.1038/ni.3048 [DOI] [PubMed] [Google Scholar]
- 27.Rothchild AC, Stowell B, Goyal G, Nunes-Alves C, Yang Q, et al. (2017) Role of Granulocyte-Macrophage Colony-Stimulating Factor Production by T Cells during Mycobacterium tuberculosis Infection. MBio 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Torrado E, Cooper AM (2010) IL-17 and Th17 cells in tuberculosis. Cytokine Growth Factor Rev 21: 455–462. 10.1016/j.cytogfr.2010.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Zhong H, Xie X, Chen CY, Huang D, et al. (2015) Long noncoding RNA derived from CD244 signaling epigenetically controls CD8+ T-cell immune responses in tuberculosis infection. Proc Natl Acad Sci U S A 112: E3883–3892. 10.1073/pnas.1501662112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldberg MF, Saini NK, Porcelli SA (2014) Evasion of Innate and Adaptive Immunity by Mycobacterium tuberculosis. Microbiol Spectr 2. [DOI] [PubMed] [Google Scholar]
- 31.Mishra A, Akhtar S, Jagannath C, Khan A (2017) Pattern recognition receptors and coordinated cellular pathways involved in tuberculosis immunopathogenesis: Emerging concepts and perspectives. Mol Immunol 87: 240–248. 10.1016/j.molimm.2017.05.001 [DOI] [PubMed] [Google Scholar]
- 32.Pahari S, Kaur G, Aqdas M, Negi S, Chatterjee D, et al. (2017) Bolstering Immunity through Pattern Recognition Receptors: A Unique Approach to Control Tuberculosis. Front Immunol 8: 906 10.3389/fimmu.2017.00906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brighenti S, Ordway DJ (2016) Regulation of Immunity to Tuberculosis. Microbiol Spectr 4. [DOI] [PubMed] [Google Scholar]
- 34.Kimmey JM, Huynh JP, Weiss LA, Park S, Kambal A, et al. (2015) Unique role for ATG5 in neutrophil-mediated immunopathology during M. tuberculosis infection. Nature 528: 565–569. 10.1038/nature16451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muraille E, Leo O, Moser M (2014) TH1/TH2 paradigm extended: macrophage polarization as an unappreciated pathogen-driven escape mechanism? Front Immunol 5: 603 10.3389/fimmu.2014.00603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang N, Liang H, Zen K (2014) Molecular mechanisms that influence the macrophage m1-m2 polarization balance. Front Immunol 5: 614 10.3389/fimmu.2014.00614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakai S, Kauffman KD, Sallin MA, Sharpe AH, Young HA, et al. (2016) CD4 T Cell-Derived IFN-gamma Plays a Minimal Role in Control of Pulmonary Mycobacterium tuberculosis Infection and Must Be Actively Repressed by PD-1 to Prevent Lethal Disease. PLoS Pathog 12: e1005667 10.1371/journal.ppat.1005667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roberts T, Beyers N, Aguirre A, Walzl G (2007) Immunosuppression during active tuberculosis is characterized by decreased interferon- gamma production and CD25 expression with elevated forkhead box P3, transforming growth factor- beta, and interleukin-4 mRNA levels. J Infect Dis 195: 870–878. 10.1086/511277 [DOI] [PubMed] [Google Scholar]
- 39.Natarajan K, Kundu M, Sharma P, Basu J (2011) Innate immune responses to M. tuberculosis infection. Tuberculosis (Edinb) 91: 427–431. [DOI] [PubMed] [Google Scholar]
- 40.Hu S, He W, Du X, Yang J, Wen Q, et al. (2017) IL-17 Production of Neutrophils Enhances Antibacteria Ability but Promotes Arthritis Development During Mycobacterium tuberculosis Infection. EBioMedicine 23: 88–99. 10.1016/j.ebiom.2017.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nandi B, Behar SM (2011) Regulation of neutrophils by interferon-gamma limits lung inflammation during tuberculosis infection. J Exp Med 208: 2251–2262. 10.1084/jem.20110919 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
NLRC3 deficiency promotes activation of CD4+ T in M. tuberculosis infection. WT and Nlrc3-/- mice were infected with M. tuberculosis and mice were harvested at 3 weeks post-infection (w.p.i.). (A) Lung cells were restimulated with M. tuberculosis lysate directly ex vivo and the intracellular production of IL-17 by CD4+ T cells was determined. Pooled data are presented in the right panel. (B) Expression of CD25 and Foxp3 were detected on CD4+ T cells from lungs. Pooled data are presented in the right panel. (C) Expression of activation markers were assessed as mean fluorescence intensity (MFI) on lung CD4+ T cells. (D) Splenocytes were stimulated for 48 hr with ESAT-6 peptide (5μg/ml) and cytokine production was measured by ELISA. Data shown are the mean ±SD. **P < 0.01 and ***P < 0.001. Data are representative of three independent experiments with similar results.
(TIF)
NLRC3 deficiency had no effect on weight and survival of mice with M. tuberculosis infection. Mice infected with approximately 200 colony-forming units of M. tuberculosis were monitored at various days post infection. (A) Weight change. (B) Survival every day from 0–200 day post-infection. Data shown are the mean ±SD. (C) Bacterial burdens were determined after infection at 1w.p.i.. (D) Frequencies of lung-infiltrating cells that are neutrophils (CD11b+ Gr-1+) or monocyte-macrophages (CD11b+ Gr-1-) at 1 w.p.i.. (E) Numbers of lung-infiltrating cells were counted at 1 w.p.i. (F) Expressions of CD86, MHC-II and CD206 were detected on monocyte-macrophages (CD11b+ Gr-1-) via flow cytometry at 1 w.p.i.. (G) Concentrations of IL-6 and IL-1β in lungs (homogenized in 2 ml PBS and 0.05% Tween 80) were detected by ELISA at 1 w.p.i.. Data shown are the mean ±SD. **P < 0.01. Data are representative of three independent experiments with similar results.
(TIF)
WT and Nlrc3-/- mice infected with approximately 200 colony-forming units of M. tuberculosis were monitored. (A) H&E-stained lung sections derived from two representative mice in each group of mice 3 w.p.i.. The magnification is shown at the right of each image. (B) Numbers of lung-infiltrating cells were counted at. Data shown are the mean ±SD. *P < 0.05 and **P < 0.01. Data are representative of three independent experiments with similar results.
(TIF)
NLRC3 does not affect thymic development but does influence mature CD4+ T cells. (A) Representative expression of CD4 and CD8 by WT and Nlrc3-/- thymocytes. Pooled data are presented in the right panel. DN, double negative (CD4- CD8-); DP, double positive (CD4+ CD8+); CD4, CD4 single positive (CD4+ CD8-); CD8, CD8 single positive (CD4- CD8+). (B) Total numbers of thymocytes in each stage of thymic development. (C) Representative expression of splenic CD4+ and CD8+ T cells from WT and Nlrc3-/- mice. Pooled data are presented in the right panel. (D) Enumeration of splenic CD4+ and CD8+ T cells from WT and Nlrc3-/- mice. (E) Production of IFN-γ and IL-17 by CD4+ T cells following stimulation with PMA/ionomycin. Pooled data are presented in the right panel. Data shown are the mean ±SD. *P < 0.05 and **P < 0.01. Data are representative of three independent experiments with similar results.
(TIF)
NLRC3 does not affect differentiation of Th2. Purified WT and Nlrc3-/- naïve CD4+ T cells were polarized in Th2 culture conditions for 4 days. Data shown are the mean ±SD. Data are representative of three independent experiments with similar results.
(TIF)
NLRC3 deficiency of CD4+ T promotes the antibacterial ability in vivo. Purified WT or Nlrc3-/- naïve CD4+ T cells were adoptively transferred into Rag2-/- mice. Then recipient mice were infected with M. tuberculosis and parts of mice were harvested at 3w.p.i.. (A) Lung cells were restimulated with M. tuberculosis lysate directly ex vivo and the intracellular production of IFN-γ, IL-2, and TNF-α by CD4+ T cells was determined. Pooled data are presented. (B) Mean fluorescence intensity (MFI) of activation markers by lung CD4+ T cells. (C) Enumeration of CD4+ cells in draining lymph nodes (DLNs), spleens and lungs. Data shown are the mean ±SD. **P < 0.01 and ***P < 0.001. Data are representative of three independent experiments with similar results.
(TIF)
NLRC3 deficiency of CD4+ T affected infiltration of myeloid cells to lung. Purified WT or Nlrc3-/- naïve CD4+ T cells were adoptively transferred into Rag2-/- mice. Then recipient mice were infected with M. tuberculosis and parts of mice were harvested at 3w.p.i.. Frequencies of lung-infiltrating cells that are neutrophils (CD11b+ Gr-1+) or monocyte-macrophages (CD11b+ Gr-1-). Pooled data are presented in the right panel. Data shown are the mean ±SD. **P < 0.01. Data are representative of three independent experiments with similar results.
(TIF)
NLRC3 suppresses activation of CD4+ T cells via negatively regulating NF-κB and ERK Signaling. Purified WT and Nlrc3-/- CD4+ T cells were stimulated for 48 hr with anti-CD3 (1 μg/ml) plus anti-CD28 (1 μg/ml) in the presence or absence of the NF-κB inhibitor JSH-23 (20 μM), MEK1/2-inhibitor U0126 (40 μM) or the mix of the two. And then expression of CD69 were detected. *P < 0.05 and **P < 0.01. Data are representative of three independent experiments with similar results.
(TIF)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.