Abstract
During viral infection, the innate immune RIG-I like receptors (RLRs) recognize viral double stranded RNA (dsRNA) and trigger filament assembly of the adaptor protein Mitochondrial Anti-viral Signaling protein (MAVS). The MAVS filament then activates anti-viral signaling events including the up-regulation of type I interferon expression. In recent years, much insight has been gained into how RLRs recognize dsRNA, but the precise mechanism of how activated RLRs stimulate MAVS filament formation remains less understood. In this chapter, we describe an in vitro reconstitution assay that we have previously developed to study the RLR-catalyzed filament assembly of MAVS. We provide technical guidance for purifying the caspase activation recruitment domain (CARD) of MAVS (MAVSCARD) as a functional monomer and also preformed filament seed. We also describe the methods to monitor the monomer-to-filament transition of MAVSCARD upon stimulation. This protocol provides a minimalist approach to studying RLR signaling events and can potentially be applied to elucidate signaling mechanisms of other innate immune receptors, such as Toll-like receptors and inflammasomes, that involve higher order assemblies of CARDs or related domains for their downstream signal activation.
Keywords: RIG-I, MDA5, MAVS, Caspase activation recruitment domain (CARD), Filament formation, IFNα/β signaling pathway, Protein refolding, Filament assembly, Innate immune signaling
1. Introduction
Effective immune defense against microbial infection is dependent upon efficient detection of Pathogen Associated Molecular Pattern (PAMP) by the Pattern Recognition Receptors (PRRs). Retinoic acid-inducible gene 1 (RIG-I) and its paralog, Melanoma Differentiation-Associated protein 5 (MDA5) compose one such family of PRRs. They are broadly expressed, cytoplasmic proteins and are responsible for detection of double-stranded RNA (dsRNA) generated by a wide range of viruses [1–3]. Upon recognition of viral dsRNA, RIG-I/MDA5 activate the type I interferon signaling pathway through the common adaptor molecule, Mitochondrial Anti-viral Signaling protein (MAVS, also known as IPS-1, Cardif, and VISA) [4–7]. Despite the shared downstream pathway, RIG-I and MDA5 play nonredundant roles in antiviral immunity by recognizing largely distinct groups of viruses [8–10].
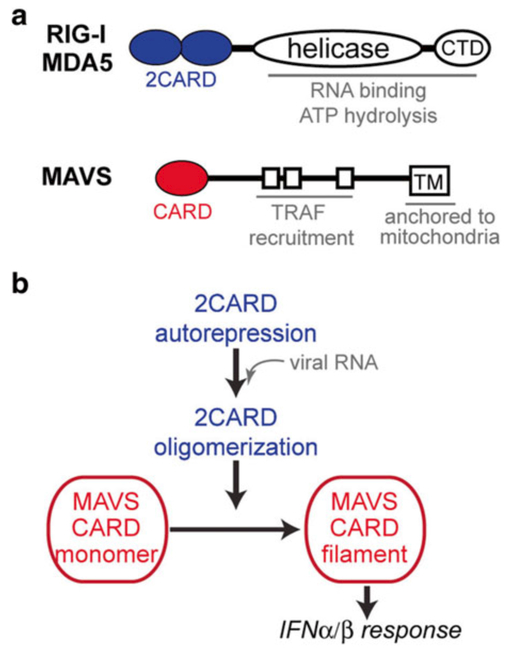
Over the last several years, we and others have investigated the structural and biochemical mechanisms by which these two receptors recognize different types of viral RNAs and activate the IFN signaling pathway [11–14]. RIG-I and MDA5 commonly contain the N-terminal tandem caspase activation recruitment domain (2CARD), the central DExD/H motif helicase domain, and the C-terminal domain (CTD) (Fig. 1a) [15]. 2CARD is responsible for interaction with MAVS and IFN signaling, whereas the helicase domain and CTD together function as an RNA recognition unit (Fig. 1a) [16, 17]. The crystal structure of ligand-free, full-length RIG-I suggested that 2CARD is masked by the protein structure in the absence of RNA, but is released upon dsRNA interaction (Fig. 1b) [13]. The exposed 2CARD then assembles into a helical oligomer [18], aided by high local concentration of 2CARD within the oligomeric assembly of RIG-I and/or bridged by K63-linked polyubiquitin chains [19, 20]. The assembled oligomer of 2CARD then associates with the CARD domain of MAVS (MAVSCARD, Fig. 1b) and nucleates MAVS filament formation (Fig. 1b) by extending the helical trajectory predefined by the 2CARD oligomer [21]. The MAVS filament in turn recruits further downstream signaling molecules, such as TRAF2, 5, and 6, to activate the type I interferon signaling pathway [22].
Fig. 1.
(a) Domain architectures of RIG-I/MDA5 and MAVS. This article focuses on 2CARD of RIG-I/MDA5 and CARD of MAVS. (b) Schematic of the signal activation processes of RIG-I/MDA5. Upon viral RNA recognition, 2CARDs of RIG-I/MDA5 are released from auto-repression and undergo homo-oligomerization. The oligomeric 2CARD then nucleates a MAVS CARD filament for activation of the IFNα/β signaling pathway
One of the challenges in the field for some time has been the lack of in vitro biochemical assays to analyze the signaling activities of RIG-I and MDA5. Interactions between RIG-I/MDA5 2CARD and MAVSCARD were often difficult to measure with confidence, possibly indicating the transient nature of their interactions or sufficiency of a few interactions for signal amplification. In our effort to understand the signal activation processes of RIG-I and MDA5, our laboratory has developed an assay (namely MAVS activation assay) that allows one to monitor the transition of MAVSCARD from monomeric to filamentous state in response to RIG-I/MDA5. This assay recapitulates many features of the cellular signaling processes of RIG-I/MDA5, for example the requirement for K63-linked polyubiquitin for isolated 2CARD [19] and alleviation of such requirement in pre-oligomerized full-length RIG-I [18]. Furthermore, this assay enabled detailed mechanistic analysis of the signal activation processes of RIG-I/MDA5 at the level of molecular structure and biochemistry [19, 21]. It should be noted that an analogous cell-free assay has been developed [23]. The advantage of our assay is that our system is reconstituted entirely from purified components (i.e., receptor, ligand, and adaptor), which greatly simplifies data interpretation and allows more detailed investigation of the molecular events.
We here describe our reconstitution of the MAVS activation assay. This assay utilizes a MAVS construct containing just its CARD domain (residues 1–97) fused to a SNAP tag (MAVSCARD-SNAP) [24]. The advantage of this fusion construct is that it has superior solubility over the construct having just the CARD domain, and also allows specific fluorescent labeling via the SNAP tag. MAVSCARD-SNAP oligomerizes into a filamentous structure with prion-like characteristics similar to those observed with full-length MAVS or MAVSCARD [11, 22]. We provide detailed guidelines on how to purify functional MAVSCARD-SNAP in its filamentous and monomeric forms. We also describe how to monitor monomer-to-filament transition of MAVSCARD-SNAP using native gel electrophoretic migration shift assay (EMSA).
2. Materials
2.1. Protein Purification
pET47b expression vector encoding MAVS CARD (residues 1–97) with an N-terminal His-tag and a C-terminal SNAP-tag (MAVSCARD-SNAP) (see Note 1).
LB broth and agar plate containing 30 μg/ml kanamycin.
BL21(DE3) competent cells.
0.5 M Isopropyl β-d-l-thiogalactopyranoside (IPTG).
2.8 L fernbach baffled flasks.
A temperature-controlled shaking incubator for 2.8 L flasks that can be set to 20–37 °C.
A spectrophotometer and cuvette that can measure absorbance at 600 nm.
A high-speed centrifuge (e.g., Sorvall refrigerated centrifuge).
Lysis buffer: 20 mM Tris, pH 7.5, 250 mM NaCl, 20 mM imidazole, pH 7.5, 0.05 % CHAPS, 10 % glycerol.
Emulsiflex (Avestin) for cell lysis.
Nickel-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen).
Gravity flow column for nickel-affinity protein purification.
Ni-NTA wash buffer: 20 mM Tris, pH 7.5, 250 mM NaCl, 40 mM imidazole, pH 7.5, 0.05 % CHAPS, 10 % glycerol.
Ni-NTA elution buffer: 20 mM Tris, pH 7.5, 250 mM NaCl, 300 mM imidazole, pH 7.5, 0.05 % CHAPS, 10 % glycerol.
HRV 3C protease (GE Healthcare or prepared in-house [25]).
Dialysis membrane with a 12,000–14,000 MWCO and clips (Spectrum Labs).
3C protease digestion buffer: 20 mM Tris, pH 7.5, 150 mM NaCl.
CARD storage buffer: 20 mM Tris, pH 7.5, 150 mM NaCl, 0.5 mM EDTA.
7 M Guanidinium chloride (GndCl, CalBiochem).
2.2. Preparation of Monomeric MAVS
Magnetic stir bar.
Stir plate.
A temperature-controlled shaking incubator that can be set to 37 °C.
Dialysis membrane with a 12,000–14,000 MWCO and clips (Spectrum Labs).
MAVSCARD-SNAP refolding buffer: 20 mM Tris, pH 7.5, 500 mM NaCl, 0.5 mM EDTA.
β-Mercaptoethanol (BME, MP biomedicals).
0.1 μm syringe filter (Millex).
5 ml disposable syringe.
2.3. MAVS Activation Assay
Benzylguanosine-conjugated Alexa647 (BG-Alexa-647 surface labeling fluorophore, New England Biolabs).
RIG-I truncation variant containing only 2CARD (expression and purification of which is described [19]).
Unanchored K63-linked polyubiquitin chains (production of which is described in [19]).
CARD storage buffer: 20 mM Tris pH 7.5, 150 mM NaCl, 5 mM EDTA.
6× native gel loading dye: 70 % sucrose with 0.0025 % of bromophenol blue and xylene cyanol.
SYBR-Gold dye (Life Technologies) for nucleic acid staining.
Krypton (Thermo Scientific) for protein staining.
3–12 % Bis-Tris-Tricine native gel (Novex by Life Technologies).
Bis-Tris-Tricine running buffer: 50 mM BisTris, 50 mM Tricine.
FLA-9000 laser gel image scanner (GE Healthcare) or equivalent machine capable of scanning multiple fluorophore channels (488 nm, 555 nm and 647 nm).
3. Methods
3.1. Protein Purification
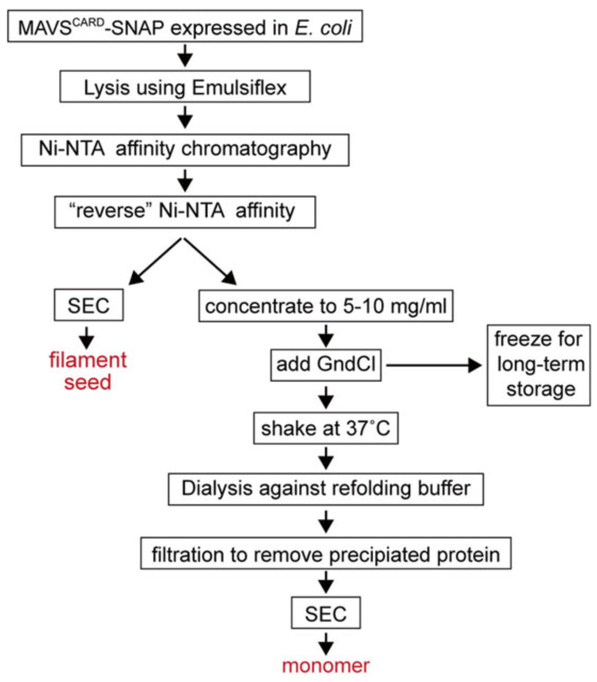
MAVSCARD-SNAP purified from E. coli is in the form of short filaments, which we refer to as filament seeds. The following protocol describes the purification strategies for MAVSCARD-SNAP filament seeds (see Fig. 2).
Fig. 2.
Schematic of the purification strategies for the MAVSCARD-SNAP filament seed and monomer
Transform BL21(DE3) cells with pET47b encoding His-tagged MAVSCARD-SNAP and plate transformed cells onto an LB agar+kanamycin plate. Let the cells grow overnight at 37 °C.
Pick a colony from the LB agar plate and inoculate 5–20 ml of LB broth + kanamycin. Shake the starter culture at 37 °C overnight.
Transfer 5–10 ml of starter culture to 1 L of LB broth + kanamycin in a 2.8 L fernbach flask. Shake at ~200 rpm at 37 °C for a few hours. Monitor cell growth by measuring the OD600.
When the OD600 of the culture reaches 0.4, lower the incubator temperature to 20 °C. Induce with 0.4 mM IPTG, when the OD600 reaches 0.6. Shake the induced culture overnight.
All subsequent steps are performed at 4–8 °C unless otherwise stated. Harvest the cells by spinning down the culture at 4500 × g for 10 min, and resuspend with 25 ml lysis buffer per liter of culture.
Cells are lysed using an Emulsiflex (see Note 2).
Once cells are sufficiently lysed, spin the cell lysate at 38,000 × g for 30 min.
Apply the cleared lysate onto pre-equilibrated Ni-NTA gravity flow column (5–10 ml agarose per liter of culture). To enhance MAVSCARD-SNAP enrichment on Ni-NTA agarose, incubate lysate with the Ni-NTA column for at least 10 min before starting the column flow. Batch Ni-NTA binding can work as well.
Wash the Ni-NTA column with 20 column volumes of Ni-NTA wash buffer.
First elute MAVSCARD-SNAP with a half column volume of Ni-NTA elution buffer. This fraction will most likely contain contaminants and should not be pooled with the second Ni-NTA elution.
Elute MAVSCARD-SNAP with 2 column volumes of Ni-NTA elution buffer.
Cleave off the His-tag using HRV 3C protease (50 μg per 1 mg of MAVSCARD-SNAP) while dialyzing against 3C protease digestion buffer overnight.
Remove the His-tag by applying the 3C protease digestion onto pre-equilibrated Ni-NTA beads. MAVSCARD-SNAP will be in the unbound fraction and a subsequent wash using half column volume of the CARD storage buffer. MAVSCARD-SNAP should be of >95 % purity as analyzed by SDS-PAGE.
At this point, MAVSCARD-SNAP is a filamentous seed. A few milliliters of MAVSCARD-SNAP seed (~1 mg/ml) can be stored at 4 °C for a month or frozen at −20 °C for longer periods.
3.2. Preperation of Monomeric MAVS
Chemical denaturation of MAVSCARD-SNAP filament seeds with GndCl followed by refolding yields monomeric MAVSCARD-SNAP (see Note 3).
After purification of the MAVSCARD-SNAP filament seed (Subheading 3.1), concentrate it to 5–10 mg/ml (see Note 4).
Denature MAVSCARD-SNAP by adding 6× volumes of 7 M GndCl. Vortex to mix at room temperature.
Make 0.5–1.5 ml aliquots of denatured MAVSCARD-SNAP. These aliquots can be snap frozen in liquid nitrogen and stored at −80 °C for 2 months, or can be directly used for preparation of monomeric MAVSCARD-SNAP as in the following steps.
Thaw frozen denatured MAVSCARD-SNAP samples by shaking at ~200 rpm for 30 min at 37 °C (see Note 5).
While protein is being thawed, prepare ice-cold refolding buffer (at least 50× volume of the denatured MAVSCARD-SNAP).
To the thawed MAVSCARD-SNAP, directly add BME to a final concentration of 100 mM and transfer the MAVSCARD-SNAP + BME mixture into dialysis tubing (see Note 6).
Place the dialysis setup into ice-cold refolding buffer (see Note 7).
Supplement refolding buffer with BME to a final concentration of 20 mM.
Dialyze MAVSCARD-SNAP for 45–60 min at 4 °C (see Note 8). Be sure to have the dialysis setup constantly spinning at ~60 rpm.
Recover the refolded MAVSCARD-SNAP. There will be some precipitated protein after dialysis. Spin down the precipitated protein at >12,000 × g for 1 min.
Syringe filter (0.1 μm) the cleared MAVSCARD-SNAP sample, but be careful not to introduce any air bubbles. Expect to recover ~50 % of the protein after filtration.
3.3. MAVS Activation Assay
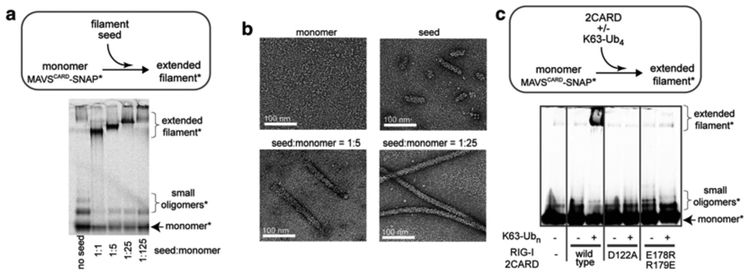
Since monomeric MAVSCARD-SNAP will spontaneously oligomerize over the course of several hours, the filament formation assay should be performed as quickly as possible. Each batch of monomeric MAVSCARD-SNAP needs to be tested for proper refolding. In our lab, we use at least two criteria to confirm functionality of monomeric MAVSCARD-SNAP. That is, correctly refolded MAVSCARD-SNAP should (1) efficiently extend the filament seeds (Fig. 3a, b) and (2) form de novo filaments upon addition of RIG-I with K63-linked polyubiquitin chains (Fig. 3c). Below, we describe both strategies.
Fig. 3.
(a) Monomer-to-filament transition of MAVSCARD-SNAP* upon addition of the filament seed. Asterisk indicates fluorescent labeling of the SNAP tag. The filament seed is nonfluorescent. (b) Representative electron micrographs of filaments extended in (a). (c) Monomer-to-filament transition of MAVSCARD-SNAP* upon addition of RIG-I 2CARD in the presence or absence of K63-linked polyubiquitin (K63-Ubn). MAVS stimulatory activities of wild-type 2CARD and oligomerization-deficient mutants (D122A and E178R/R179E) were compared. Figure images in (a) and (b) were adopted from [11], and the image in (c) from [19]
Add BG-Alexa-647 dye (1 μM, final) to monomeric MAVSCARD-SNAP (20 μM or 0.6 mg/ml). Incubate the reaction on ice for 10 min (see Note 9). Monomeric MAVSCARD-SNAP can be purified by Size Exclusion Chromatography (SEC) to remove free dye. Alternatively, the labeling reaction can be quenched by addition of an excess amount of free benzylguanosine. Labeled MAVSCARD-SNAP will be referred to as MAVSCARD-SNAP*. All reactions with MAVSCARD-SNAP* will be performed using CARD storage buffer (see Note 10).
If MAVSCARD-SNAP* was properly refolded, it should not form filaments on its own within ~2 h post-refolding. To test this, incubate 10 μl of 10 μM MAVSCARD-SNAP* at 22 °C for 15–30 min.
For filament extension, add the MAVSCARD-SNAP seed (prepared in Subheading 3.1) to monomeric MAVSCARD-SNAP* (10 μM, final) (prepared in Subheading 3.2), and incubate at 22 °C for 15–30 min. We typically use the seed:monomer mass ratio of 25:1, but different ratios can be used to obtain filaments with different lengths (see Fig. 3b).
For filament induction by RIG-I, add RIG-I 2CARD (10 μM, final) and unanchored K63-linked polyubiquitin chains (80 μg/ml, final) to MAVSCARD-SNAP* (10 μM, final) and incubate at 22 °C for 15–30 min.
To reactions prepared in steps 2–4 (10 μl, add 2 μl of 6× native gel loading dye.
Immediately load samples into a 3–12 % Bis-Tris-Tricine native gel (see Note 11). Run the native gel in Bis-Tris-Tricine running buffer at 200 V for 70–80 min at 4 °C or until the dye front runs off the gel.
Scan the gel using the Alexa647 fluorescence with a gel scanner FLA-9000. We obtain optimal visualization using the 100 μm resolution and 300–600 V laser sensitivity settings (see Note 12).
4. Notes
-
The plasmid encoding the CARD domain (residues 1–97) of MAVS (UnitProtKB: Q7Z434.2) fused to the SNAP tag (version 1.0, New England Biolabs) was generated by inserting MAVS CARD between the BamHI and EcoRI restriction sites and SNAP between the EcoRI and XhoI restriction sites in pET47b. The amino acid sequence of the resultant fusion protein, His-tagged MAVSCARD-SNAP, is
MAHHHHHHSAALEVLFQGPGYQDPMPFAEDKTYKYICRNFSNFCNVDVVEILPYLPCLTARDQDRLRATCTLSGNRDTLWHLFNTLQRRPGWVEYFIAALRGCELVDLADEVASVYQSYQPEFMDKDCEMKRTTLDSPLGKLELSGCEQGLHEIKLLGKGTSAADAVEVPAPAAVLGGPEPLMQATAWLNAYFHQPEAIEEFPVPALHHPVFQQESFTRQVLWKLLKVVKFGEVISYQQLAALAGNPAATAAVKTALSGNPVPILIPCHRVVSSSGAVGGYEGGLAVKEWLLAHEGHRLVNRVWDLQV
Other mechanical devices can be used to lyse cells, but avoid using a sonicator as it may increase fragmentation of filament seeds.
The refolded monomer displays the same three dimensional structure [21] as the one without refolding [26].
We found that the concentration of MAVSCARD-SNAP (5–10 mg/ml) at this step is important. At higher concentrations, MAVSCARD-SNAP does not refold as well and leads to heavy precipitation.
We found that incubating the denatured MAVSCARD-SNAP at 37 °C before refolding is critical for obtaining correctly refolded monomeric MAVSCARD-SNAP.
Since MAVSCARD-SNAP has multiple cysteines, it is important to keep high concentrations of BME to maintain a reducing environment during refolding. Add fresh BME to the refolding buffer just before its use. Tris(2-carboxyethyl)phosphine (TCEP) can be used as an alternative reducing agent.
It is important to use ice-cold buffer for optimal refolding of MAVSCARD-SNAP.
If the refolding buffer is not compatible with a downstream assay, you must first refold the protein in the refolding buffer for 30 min and then perform a secondary dialysis using your desired buffer. We have extensively optimized the refolding buffer and do not recommend altering the composition of this buffer.
The labeling reaction is not sensitive to buffer conditions and can also be performed at 22 °C.
We found that MAVSCARD-SNAP* filament formation (in particular seed extension) is not sensitive to salt concentration of the reaction buffer. You may alter these reaction conditions to suit your desired experimental setup.
While other native gel buffer systems may be used, we found that the best results were obtained with Bis-Tris-Tricine native gels.
While the native gel assay provides a convenient method to examine filament formation of MAVS CARD, it has its limitations. First, it cannot distinguish between the filament of MAVSCARD-SNAP and nonfilamentous aggregate. In addition, very long filaments can fail to enter the gel, although disappearance of monomeric MAVSCARD-SNAP can be analyzed instead. We recommend using negative stain electron microscopy as a complementary method to verify filament formation.
References
- 1.Takeuchi O, Akira S (2008) MDA5/RIG-I and virus recognition. Curr Opin Immunol 20(1):17–22. doi: 10.1016/j.coi.2008.01.002 [DOI] [PubMed] [Google Scholar]
- 2.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T (2004) The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol 5(7):730–737. doi: 10.1038/ni1087 [DOI] [PubMed] [Google Scholar]
- 3.O’Neill LA, Bowie AG (2010) Sensing and signaling in antiviral innate immunity. Curr Biol 20(7): R328–R333. doi: 10.1016/j.cub.2010.01.044 [DOI] [PubMed] [Google Scholar]
- 4.Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB (2005) VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell 19(6): 727–740. doi: 10.1016/j.molcel.2005.08.014 [DOI] [PubMed] [Google Scholar]
- 5.Seth RB, Sun L, Ea CK, Chen ZJ (2005) Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that acti-vates NF-kappaB and IRF 3. Cell 122(5):669–682. doi: 10.1016/j.cell.2005.08.012 [DOI] [PubMed] [Google Scholar]
- 6.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J (2005) Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437(7062):1167–1172. doi: 10.1038/nature04193 [DOI] [PubMed] [Google Scholar]
- 7.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S (2005) IPS-1, an adaptor triggering RIG-I-and Mda5-mediated type I interferon induc-tion. Nat Immunol 6(10):981–988. doi: 10.1038/ni1243 [DOI] [PubMed] [Google Scholar]
- 8.Loo YM, Fornek J, Crochet N, Bajwa G, Perwitasari O, Martinez-Sobrido L, Akira S, Gill MA, Garcia-Sastre A, Katze MG, Gale M Jr (2008) Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol 82(1):335–345. doi: 10.1128/JVI.01080-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S (2006) Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441(7089):101–105. doi: 10.1038/nature04734 [DOI] [PubMed] [Google Scholar]
- 10.Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, Endres S, Hartmann G (2006) 5′-Triphosphate RNA is the ligand for RIG-I. Science 314(5801):994–997. doi: 10.1126/science.1132505 [DOI] [PubMed] [Google Scholar]
- 11.Wu B, Peisley A, Richards C, Yao H, Zeng X, Lin C, Chu F, Walz T, Hur S (2013) Structural basis for dsRNA recognition, fi la-ment formation, and antiviral signal activa-tion by MDA5. Cell 152(1–2):276–289. doi: 10.1016/j.cell.2012.11.048, S0092–8674(12)01436–5 [pii] [DOI] [PubMed] [Google Scholar]
- 12.Luo D, Ding SC, Vela A, Kohlway A, Lindenbach BD, Pyle AM (2011) Structural insights into RNA recognition by RIG-I. Cell 147(2):409–422. doi: 10.1016/j.cell.2011.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, Gerlier D, Cusack S (2011) Structural basis for the activa-tion of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell 147(2):423–435. doi: 10.1016/j.cell.2011.09.039 [DOI] [PubMed] [Google Scholar]
- 14.Jiang F, Ramanathan A, Miller MT, Tang GQ, Gale M Jr, Patel SS, Marcotrigiano J (2011) Structural basis of RNA recognition and activation by innate immune receptor RIGI. Nature 479(7373):423–427. doi: 10.1038/nature10537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoneyama M, Fujita T (2008) Structural mechanism of RNA recognition by the RIG-I-like receptors. Immunity 29(2):178–181 [DOI] [PubMed] [Google Scholar]
- 16.Kato H, Takahasi K, Fujita T (2011) RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immunol Rev 243:91–98 [DOI] [PubMed] [Google Scholar]
- 17.Wilkins C, Gale M Jr (2010) Recognition of viruses by cytoplasmic sensors. Curr Opin Immunol 22:41–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peisley A, Wu B, Yao H, Walz T, Hur S (2013) RIG-I forms signaling-competent fi laments in an ATP-dependent, ubiquitin-independent manner. Mol Cell 51:573–583 [DOI] [PubMed] [Google Scholar]
- 19.Peisley A, Wu B, Xu H, Chen ZJ, Hur S (2014) Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature 509(7498):110–114. doi: 10.1038/nature13140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang X, Kinch LN, Brautigam CA, Chen X, Du F, Grishin NV, Chen ZJ (2012) Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity 36(6):959–973. doi: 10.1016/j.immuni.2012.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu B, Peisley A, Tetrault D, Li Z, Egelman EH, Magor KE, Walz T, Penczek PA, Hur S (2014) Molecular imprinting as a signal-activation mechanism of the viral RNA sensor RIG-I. Mol Cell. doi: 10.1016/j.molcel.2014.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ (2011) MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell 146(3):448–461.doi: 10.1016/j.cell.2011.06.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeng W, Sun L, Jiang X, Hou F, Adhikari A, Xu M, Chen ZJ (2010) Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell 141:315–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, Johnsson K (2003) A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat Biotechnol 21(1):86–89. doi: 10.1038/nbt765 [DOI] [PubMed] [Google Scholar]
- 25.Alexandrov A, Dutta K, Pascal SM (2001) MBP fusion protein with a viral protease cleavage site: one-step cleavage/purification of insoluble proteins. Biotechniques 30:1194–1198 [DOI] [PubMed] [Google Scholar]
- 26.Potter JA, Randall RE, Taylor GL (2008) Crystal structure of human IPS-1/MAVS/VISA/Cardif caspase activation recruitment domain. BMC Struct Biol 8:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]