B cell–intrinsic activated CARD11 (aCARD11) expression promotes rapid formation and premature collapse of the germinal center while enhancing terminal differentiation due to heightened NF-κB and mTORC1 signaling.
Abstract
Activating mutations in the adapter protein CARD11 associated with diffuse large B cell lymphomas (DLBCLs) are predicted to arise during germinal center (GC) responses, leading to inappropriate activation of NF-κB signaling. Here, we modeled the B cell–intrinsic impact of the L251P activating mutation in CARD11 (aCARD11) on the GC response. Global B cell aCARD11 expression led to a modest increase in splenic B cells and a severe reduction in B1 B cell numbers, respectively. Following T cell–dependent immunization, aCARD11 cells exhibited increased rates of GC formation, resolution, and differentiation. Restriction of aCARD11 to GC B cells similarly altered the GC response and B cell differentiation. In this model, aCARD11 promoted dark zone skewing along with increased cycling, AID levels, and class switch recombination. Furthermore, aCard11 GC B cells displayed increased biomass and mTORC1 signaling, suggesting a novel strategy for targeting aCARD11-driven DLBCL. While aCARD11 potently impacts GC responses, the rapid GC contraction suggests it requires collaboration with events that limit terminal differentiation to promote lymphoma.
Introduction
Diffuse large B cell lymphoma (DLBCL) is the most common type of non-Hodgkin’s lymphoma (Pasqualucci and Zhang, 2016). While nearly half of DLBCLs are curable with current treatment, the activated B cell–like (ABC) subtype has an inferior prognosis (Lenz et al., 2008; Staudt, 2010; Shaffer et al., 2012). ABC-DLBCL is derived from germinal center (GC) B cells that have acquired progressive oncogenic hits (Staudt, 2010; Rui et al., 2011; Shaffer et al., 2012). In normal B cells, B cell receptor (BCR) engagement induces phosphorylation of the molecular scaffold CARD11, leading to conformational changes that promote assembly of a CARD11, Bcl10, MALT1 (CBM) signalosome (Sommer et al., 2005), which is required for NF-κB and JNK signaling and B cell proliferation, survival, and differentiation (Vallabhapurapu and Karin, 2009). Activating mutations in CARD11 (referred to hereafter as aCARD11) occur in 10% of ABC-DLBCLs (Lenz et al., 2008). Importantly, while aCARD11-expressing DLBCLs rely on constitutive NF-κB signals for survival (Ngo et al., 2006), additional aberrant signals are also likely required for tumor growth. Thus, a better understanding of how aCARD11 alters GC biology may inform the design of future therapies.
An initial in vivo analysis of aCARD11 variants demonstrated that oncogenic mutations altered the response of self-reactive B cells, promoting proliferation and autoantibody production upon exposure to self-antigen (Jeelall et al., 2012). In that study, DLBCL-derived aCARD11 mutants were introduced ex vivo (using retroviral gene delivery) into murine B cells following in vivo antigen-priming. Adoptive transfer of these cells into Rag1−/− recipients expressing the self-antigen led to broken tolerance and aberrant proliferation, plasmacytic differentiation, and autoantibody secretion. The impact of aCARD11 on T cell–dependent (TD) responses and the GC reaction were not addressed in this study.
A DLBCL-associated mutation resulting in an isoleucine insertion, CARD11-L225LI, is the most potent known NF-κB activating mutation (Lenz et al., 2008). In a B cell–intrinsic CARD11-L225LI mouse model, pups succumbed to early postnatal lethality resulting from aggressive B cell lymphoproliferation. Within 5 d after birth, transgenic mice displayed histopathological features of high-grade lymphoma, with blastoid cells infiltrating solid organs and bone marrow (BM). B cells isolated from transgenic mice exhibited elevated NF-κB and JNK activity compared with controls. This phenotype was abrogated by intercross with either Bcl10−/− or MALT1−/− mice, demonstrating that disruption of the CBM complex resolves aberrant NF-κB activation (Knies et al., 2015). While this study showed that a single mutation in CARD11 can yield a disease phenotype mirroring lymphoma, whether other CARD11 mutants—that result in a spectrum of NF-κB activity (Lenz et al., 2008)—will behave similarly is unknown. Also, as these animals succumbed to disease immediately after birth, this model was unable to provide insight into how aCARD11 mutants affect a GC response.
As the activating, somatic mutations in CARD11 that lead to DLBCL are predicted to occur during the B cell GC response, GC-specific analyses are likely to improve understanding of DLBCL biology. To evaluate the impact of aCARD11 on the GC response, we developed a transgenic model allowing inducible expression of aCARD11 (mouse CARD11-L251P) that mimics an analogous mutation identified in human DLBCL (L244P; Lenz et al., 2008). This construct was introduced in association with a downstream T2A-linked GFP marker into the endogenous Rosa26 locus. Crossing this strain to various B cell–intrinsic Cre-bearing strains gives rise to GFP+ cells coexpressing aCARD11. Importantly, this model was designed to facilitate aCARD11 expression levels similar to that observed in heterozygotes that develop DLBCL. Further, this specific mutant activates NF-κB to a lesser extent than the previously modeled L225LI mutation (Lenz et al., 2008; Knies et al., 2015) and was anticipated to permit a relatively normal B cell developmental program wherein animals would establish a mature peripheral B cell compartment. Using several B cell–intrinsic models, we assessed both how this mutation modulates B cell development and, most importantly, its impact within a GC response. Our findings indicate that cells expressing aCARD11 display enhanced NF-κB and mTORC1 signaling, significantly altered GC kinetics, cell fate determination, and class switch recombination (CSR) during the primary response to TD antigens.
Results
aCARD11 promotes follicular over marginal zone (MZ) B cell development and alters the B1 compartment
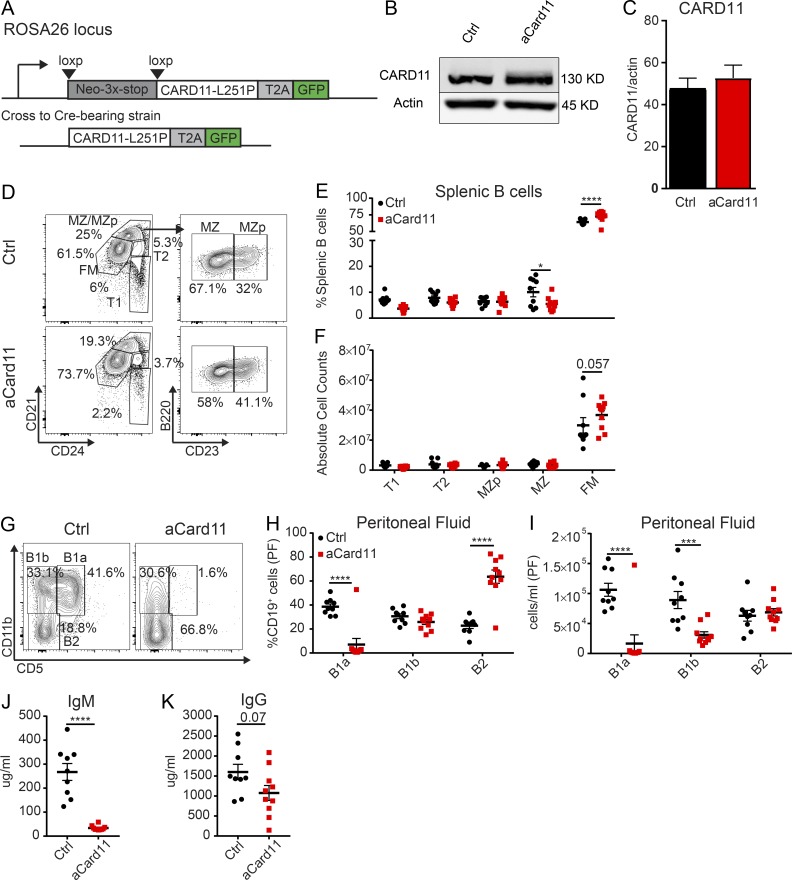
To better understand how aCARD11 impacts B cell biology, we developed an inducible model for expressing CARD11 with a L251P coding change (equivalent to human CARD11-L244P; Lenz et al., 2008) with a cis-linked GFP reporter knocked into the endogenous Rosa26 locus (Fig. 1 A). For the initial characterization of B cell maturation, ROSA-aCard11 animals were crossed to the Mb1-Cre strain (referred to as “Mb1-aCard11”) (Hobeika et al., 2006), which initiates Cre recombinase expression at the BM pro-B cell stage. Importantly, total CARD11 protein levels, in naive splenic B cells in Mb1-aCard11 mice, were minimally changed relative to control animals (Fig. 1, B and C). While unlikely, inefficient cleavage of the T2A fusion protein could impact mutant CARD11 protein levels and contribute to our observations of no difference in total CARD11 protein. Alternatively, the mutant CARD11 protein may be expressed at lower levels than the endogenous, WT protein, as it has been shown that activated forms of CARD11 have decreased stability at steady-state (Moreno-García et al., 2010). GFP expression was detected in pro–B cells (B220+IgM−CD43+; ∼30–50%) and in all later developmental stages (Fig. S1, A and B; >90% GFP+ starting at the small pre–B cell stage). There was no significant impact of aCARD11 on B cell subsets in the BM (Fig. S1 D). GFP expression in the periphery was restricted to cells of the B lineage (Fig. S1, A and C).
Figure 1.
B lineage aCARD11 expression leads to modest alterations in mature B2 B cells but near absence of B1 B cells. (A) Schematic showing activated mouse CARD11 (CARD11-L251P) with a self-cleaving T2A-linked GFP reporter in the Rosa26 locus. (B) Representative immunoblot of CARD11 levels in naive B cells in control (Ctrl) and Mb1-Cre-aCard11 mice. (C) Quantification of CARD11 levels showing data from two independent experiments (bar graphs depict mean + SEM; n = 5 Ctrl mice and 7 Mb1-aCard11 mice). (D) Representative flow plots of splenic B cell subsets with gating schema to identify B cell subsets: early transitional (T1; B220+CD24+CD21lo), late transitional (T2; B220+CD24+CD21int), MZ precursor (MZp; B220+CD24+CD21hiCD23+), MZ (B220+CD24+CD21hiCD23−), and FM (B220+CD24intCD21int) B cells. (E and F) Frequency (E; MZ: P = 0.04; FM: P < 0.0001) and absolute number (F; FM: 0.057) of B cell subsets. Black: Ctrl mice, animals lacking expression of the aCard11 transgene. Red: Mb1-aCard11 mice, homozygous for the aCard11 transgene and heterozygous for Mb1-Cre. (G) Representative flow plots of B cell subsets in PF: B1a (CD19+CD11b+CD5+), B1b (CD19+CD11b+CD5−), and B2 (CD19+CD11b−CD5−). (H) Frequency (B1a: P < 0.0001; B2: P < 0.0001) and (I) absolute number (B1a: P < 0.0001; B1b: P = 0.0008) of peritoneal B cell subsets (per milliliter of PF). Significance defined as P ≤ 0.05 by two-way ANOVA. (J) IgM (P < 0.001) and (K) IgG serum titers (P = 0.07). (J and K) Significance defined as P ≤ 0.05 by unpaired t test. (D–K) Data are from two independent experiments (n = 9 Ctrl mice and 10 Mb1-aCard11 mice at ∼12 wk of age). (E, F, and H–K) For summary graphs, lines and error bars represent mean ± SEM. *, P < 0.05; ****, P < 0.0001.
Despite expression of aCARD11 in all peripheral B cells, we did not observe striking differences in spleen weight or in absolute counts or frequency within the T or myeloid compartments (Fig. S1, E–H). Characterization of splenic B cells revealed a slight increase in total cell numbers (Fig. S1 H), decreased spontaneous GC B cells (Fig. S1 I), increased follicular mature (FM) B cells (percentage and absolute B cell counts), and a decreased percentage of MZ B cells (Fig. 1, D–F). There was also no significant B cell expansion evident in the spleens of older animals (>30 wk, data not shown). Characterization of B cell subsets in the peritoneal cavity revealed a near absence of B1a B cells (CD19+CD11b+CD5+) and a marked reduction of B1b B cells (CD19+CD11b+CD5−; Fig. 1, G–I). Mb1-aCard11 animals also exhibited decreased total IgM with a trend toward decreased total IgG, despite equivalent levels of IgG1 and IgG2c (Fig. 1, J and K; Fig. S1, J and K). The decrease in basal IgM production likely reflected absence of the B1a compartment, a major source of natural IgM (Baumgarth, 2016). Together, these data demonstrate establishment of a murine model, expressing an inducible, oncogenic CARD11 variant at near endogenous levels, leading to modest changes in B2 B cell development and marked reduction in B1 B cell compartment in the absence of immunization.
aCARD11 promotes more rapid formation and contraction of the GC response
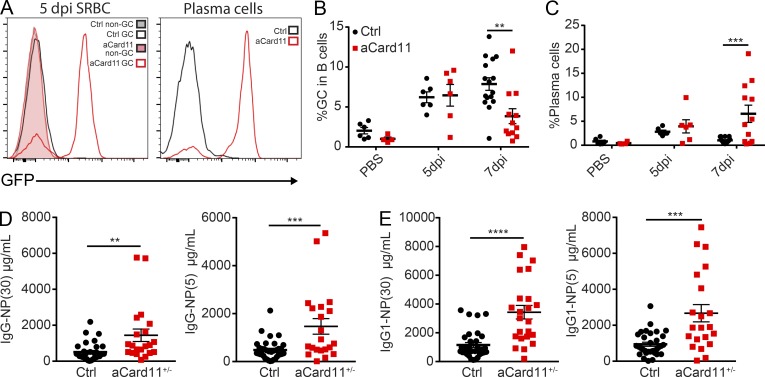
Previous work showed that overexpression of aCARD11 variants in mature mouse B cells promoted plasmacytic differentiation in response to self-antigen exposure (Jeelall et al., 2012). However, the impact of activating mutations on a GC reaction has not been assessed. To determine how aCARD11 impacts the response to TD antigens, we first immunized control and aCard11 mice with sheep RBCs (SRBCs) and evaluated the kinetics of the GC response. We hypothesized that aCARD11 would result in a larger and more prolonged GC response due to increased NF-κB signaling. Contrary to this prediction, we observed an altered kinetic in immunized Mb1-aCard11 vs. control mice, with the GC response reaching an earlier peak (Fig. 2, A–C, day 5 after immunization) followed by GC contraction as early as day 7 (Fig. 2, B and C). In contrast, the GC response in control animals peaked between days 7 and 10. Consistent with our flow-based data, immunofluorescence studies revealed larger and more abundant GCs in the spleen at day 5 in immunized Mb1-aCard11 mice (Fig. 2, D and E; and Fig. S1 L). Correlating with the more rapid GC contraction in Mb1-aCard11 mice, we observed an earlier expansion of the splenic plasma cell (PC; B220−CD138+) compartment at day 7 relative to controls (Fig. 2, F–H). Further, in contrast to controls, the splenic PC compartment was nearly absent by day 10 in Mb1-aCard11, without evidence for increased PC migration to the BM (data not shown). While serum IgM antibody titers increased with a similar kinetic after immunization in Mb1-aCard11 and control animals, total IgM levels were reduced at baseline (Figs. 1 J and 2 I) and remained lower at all time points compared with controls (Fig. 2 I). In contrast, total IgG, IgG1, and IgG2C levels were not significantly different in Mb1-aCard11 vs. control animals (Fig. 2, J and K; and data not shown). Together, these findings support a model wherein aCARD11 enhances GC generation and coordinately promotes terminal differentiation into short-lived PCs.
Figure 2.
B cell–intrinsic aCARD11 expression alters the kinetics of the GC response. Ctrl (Cre-negative littermates and Mb1Cre/+ mice) and Mb1-aCard11 mice (homozygous for aCard11 transgene) were immunized as indicated with either PBS or SRBCs. (A) Representative flow plots of splenic GC (B220+CD95hiCD38lo) B cells at 5 d postimmunization (dpi). Top: Ctrl; bottom: Mb1-aCard11 mice. (B) Frequency (5 dpi: P < 0.001; 7 dpi: P = 0.002; and 10 dpi: P < 0.001) and (C) absolute number of GC B cells after immunization at indicated time points (5 dpi: P < 0.001; 7 dpi: P = 0.08; and 10 dpi: P = 0.05). (D) Representative splenic tissue sections in Ctrl (littermate controls) and Mb1-aCard11 mice at 5 dpi. Blue: anti-CD3; red: anti-B220; and green: PNA. Scale bar, 1 mm. (E) Number of GCs (PNA+ foci) normalized to total area (square millimeters) of splenic tissue section (P = 0.01 calculated by unpaired t test). (D and E) Data are representative of five Ctrl and five Mb1-aCard11 mice 5 dpi with SRBCs from three independent experiments. For bar graph, lines represent mean + SEM. (F) Representative flow plots of PCs (B220−CD138+) at 5 dpi. Top: Ctrl; bottom: Mb1-aCard11 mice. (G) Frequency (7 dpi: P < 0.0001) and (H) absolute number of PCs after immunization (7 dpi: P < 0.0001). (I) IgM (PBS; 5 and 10 dpi: P < 0.0001; 7 dpi: P = 0.003; and 21 dpi: P = 0.0007), (J) IgG, and (K) IgG1 serum titers at 5, 7, 10, and 21 dpi. (B, C, and G–K) Significance defined as P ≤ 0.05 by two-way ANOVA. Data shown in A–C and F–H include PBS immunization: 13 Ctrl mice, 12 Mb1-aCard11 (12 expts) mice; SRBC immunization, day 5: 15 Ctrl mice, 15 Mb1-aCard11 (4 expts) mice; day 7: 10 Ctrl mice, 10 Mb1-aCard11 (4 expts) mice; day 10: five Ctrl mice, six Mb1-aCard11 (two expts) mice; day 21: seven Ctrl mice, seven Mb1-aCard11 (two expts) mice. Data shown in I–K include PBS injection: 18 Ctrl mice, 15 Mb1-aCard11 (12 expts); SRBC injection, day 5: 29 Ctrl mice, 26 Mb1-aCard11 (12 expts) mice; day 7: seven Ctrl mice, nine Mb1-aCard11 (three expts) mice; day 10: eight Ctrl mice, nine Mb1-aCard11 (four expts) mice; day 21: seven Ctrl mice, seven Mb1-aCard11 (two expts) mice. For summary graphs, lines and error bars represent mean ± SEM. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001.
GC B cell–intrinsic aCARD11 expression is sufficient to alter the kinetics of the GC response
The altered kinetics of TD GC response in Mb1-aCard11 mice might be caused by the increased development of FM B cells and/or preactivation of B cells in this strain. To specifically assess the impact of aCARD11 within an ongoing GC response, we crossed ROSA-aCard11 mice with the Cγ1-Cre strain. In the resulting model (Cγ1-aCard11), Cre expression is induced by transcription of the IgG1 constant region gene (Cγ1; Casola et al., 2006), limiting transgene expression to a subset of B cells that have class switched. Cγ1-aCard11 mice lacked GFP expression within the splenic non-GC B cells (Fig. 3 A). Unlike Mb1-aCard11 animals, Cγ1-aCard11 mice showed no difference in peritoneal B cell subsets (Fig. S2, A–C) and had comparable serum IgM and IgG titers at baseline (Fig. S2, D and E). Upon immunization with SRBCs, GFP expression was readily detected in splenic GC B cells and PCs (Fig. 3 A, left and right, respectively), consistent with Cγ1-Cre induction in response to immunization. Immunization with SRBCs led to robust GC response in both control and Cγ1-aCard11 mice. However, at day 7, Cγ1-aCard11 mice exhibited a more rapid contraction of the GC response relative to that observed in controls (Fig. 3 B). In parallel with more rapid GC contraction, we observed an increase in the PC compartment at 7 d after immunization in Cγ1-aCard11 mice relative to control mice (Fig. 3 C). While these findings mirrored the rapid contraction of the GC and earlier PC production present in Mb1-aCard11 mice, the Cγ1-aCard11 model did not reproduce the early peak in the GC, likely due to the requirement for class switching to trigger initial aCARD11 expression.
Figure 3.
GC B cell–intrinsic aCARD11 expression induces rapid GC peak and contraction with increased terminal differentiation. (A) Left: GFP expression in Ctrl (Cre-negative littermates) and aCard11+/+ mice crossed with the Cγ1-Cre strain showing non-GC (B220+CD95loCD38hi) and GC (B220+CD95hiCD38lo) B cells at 5 d postimmunization (dpi) with SRBCs. Filled histogram: non-GC (gray, Ctrl; pink, Cγ1-aCard11); open histogram: GC (black, Ctrl; red, Cγ1-aCard11). Right: GFP expression in PCs (B220−CD138+). (B) Percentage of splenic GC B cells in PBS controls and at 5 and 7 dpi with SRBC (P = 0.001 at 7 dpi). (C) Percentage of splenic PCs after immunization (P < 0.001 at 7 dpi). To measure antigen-specific antibody response, Cγ1-aCard11+/− and control mice (Cre-negative littermates and Cγ1Cre/+) were injected with PBS or 50 μg NP-CGG injected i.p. For B and C, significance was calculated by two-way ANOVA. Data represent six Ctrl mice, four Cγ1-aCard11 PBS-injected mice; SRBC injected, day 5: six Ctrl mice, six Cγ1-aCard11 (three expts) mice; day 7: 16 Ctrl mice, 12 Cγ1-aCard11 (6 expts) mice. (D) Low- (NP-30; P = 0.006) and high-affinity (NP-5; P = 0.0006) NP-specific IgG serum titers. (E) Low- (P < 0.0001) and high-affinity (P = 0.0001) NP-specific IgG1 serum titers. (D and E) Significance calculated by one-way ANOVA. Data are representative of 33 Ctrl mice and 21 Cγ1-aCard11+/− mice 12 dpi with NP-CGG from seven independent experiments. For summary graphs, lines and error bars represent mean ± SEM. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
To determine whether the PCs generated in aCard11 mice were capable of producing antigen-specific antibodies, we immunized Cγ1-aCard11 and control mice with the TD antigen NP-CGG in order to track the NP-specific antibody response. Despite the absence of GC B cells at 12 d after immunization, Cγ1-aCard11 mice exhibited increased serum titers for both high- and low-affinity NP-specific total IgG and IgG1 (Fig. 3, D and E). Together, these data suggest that aCARD11 promotes rapid terminal differentiation of B cells into PCs, including antigen-specific PCs, following antigen encounter. These findings also demonstrate that GC B cell–intrinsic expression of aCARD11 is sufficient to alter the kinetic of the GC response to TD antigens.
aCARD11 enhances cycling in vivo and skews GC polarization
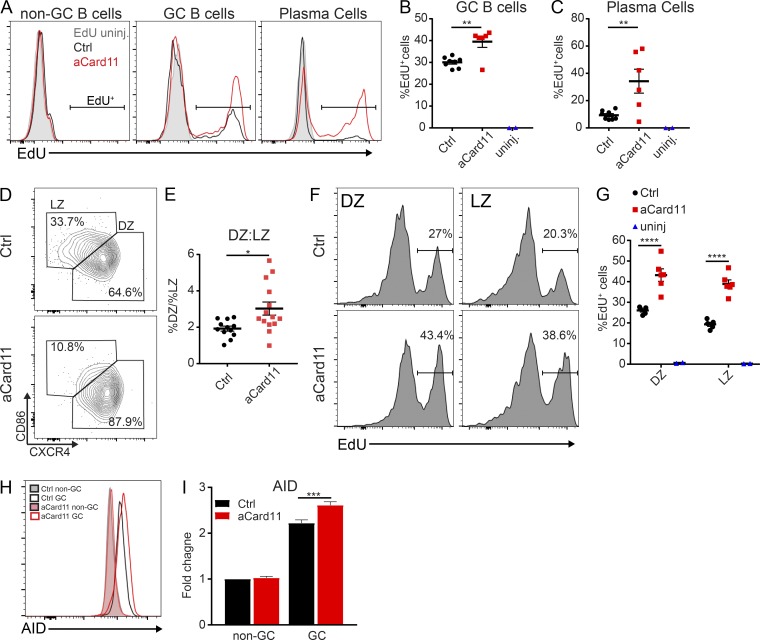
We next tested whether the earlier peak of the GC response in Mb1-aCard11 animals reflected enhanced cycling driven by aCARD11. Previous studies have shown that activating mutations in CARD11 enhance proliferation in vitro (Snow et al., 2012) and can switch the response to self-antigen recognition in vivo from antigen-induced cell death to proliferation (Jeelall et al., 2012). To determine whether GC B cells expressing aCARD11 exhibit higher rates of proliferation, we immunized control and Mb1-aCard11 mice with SRBCs and, at 1 h before sacrifice at the peak of the GC response (day 5 after immunization), mice were injected intraperitoneally with the thymidine analogue, ethynyl deoxyuridine (EdU). We observed a significant increase in the frequency of EdU+ cells within both the GC and PC compartments in Mb1-aCard11 compared with control mice (Fig. 4, A–C). In contrast, we observed no difference in EdU incorporation in pro/pre–B cells (IgM−B220+; Fig. S3, A and B) and no difference in EdU incorporation in non-GC B cells (Fig. S3 C, B220+CD95loCD38+). Thus, the increased cycling observed in aCARD11 GCs and PCs is unlikely the result of changes before immunization or GC entry. Together, these data demonstrate that aCARD11 significantly enhances both GC B cell and plasmablast (PB) proliferation.
Figure 4.
aCARD11 promotes GC B cell cycling and DZ skewing. (A–C, F, and G) Ctrl and Mb1-aCard11 mice were immunized with SRBCs and sacrificed on day 5 after immunization for analysis of B cell populations. (A–C) 1 mg EdU was injected i.p. at 1 h before sacrifice. (A) Representative histograms showing the relative proportion of EdU+ cells in the non-GC (B220+CD95loCD38hi), GC (B220+CD95hiCD38lo), and PC (B220−CD138+) compartments. (B) Percentage of EdU+ cells in GC B cells (P = 0.002). (C) Percentage of EdU+ cells in PCs (P = 0.007). Blue triangles, Ctrl animals without EdU labeling, one per experiment. (A–C) Data are representative of two independent experiments using eight Ctrl mice and six Mb1-aCard11 (EdU-injected) and two untreated animals; significance was calculated by Student’s unpaired t test. (D) Representative flow plots of DZ (CXCR4+CD86−) vs. LZ (CXCR4−CD86+) GC B cells at 5 d after immunization. (E) Ratio of DZ to LZ cells within the GC at 5 d after immunization (P = 0.013). (D and E) Data are representative of three independent experiments, with 13 Ctrl and 14 Mb1-aCard11 mice; significance was calculated by Student’s unpaired t test. (F) Representative histograms showing the relative proportion of EdU+ cells in DZ (left, CD19+CD95hiCD38loCXCR4+CD86−) and LZ (right, CD19+CD95hiCD38loCXCR4−CD86+) GC B cells in littermate controls (top) and Mb1-aCard11 (bottom) mice. (G) Percentage of EdU+ cells in the DZ (P = 0.0004) and LZ (P = 0.004) within the GC B cell population. Black circles, littermate controls; red squares, Mb1-aCard11, and blue triangles, animals without EdU labeling (one per experiment). (F and G) Data are representative of two independent experiments with five Ctrl mice and six Mb1-aCard11 (EdU-injected) and two untreated animals; significance was calculated by two-way ANOVA. (H) AID expression in non-GC (B220+CD95−PNA−) and GC (B220+CD95hiPNAhi) B cells at 5 d after immunization. Filled histograms—non-GC B cells: gray, Ctrl; pink, Mb1-aCard11. Open histograms—GC B cells: black, Ctrl; red, Mb1-aCard11. (I) Fold change in median fluorescent intensity for AID over Ctrl non-GC B cells (P = 0.001). (H and I) Data are representative of three independent experiments with 13 Ctrl and 13 Mb1-aCard11 mice; significance was calculated by two-way ANOVA. For summary graphs, lines represent mean ± SEM. For bars graphs, lines represent mean + SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Next, we assessed whether the enhanced cell cycling in aCARD11 GC B cells correlated with alterations in the polarization of the GC response. The GC is polarized into two zones: light zone (LZ) and dark zone (DZ). The DZ is the proliferative compartment within the GC (Mesin et al., 2016). Using the SRBC immunization model, we observed an ∼2:1 ratio of DZ (CD86loCXCR4hi) to LZ (CD86hiCXCR4lo) GC B cells in control mice at day 5 after immunization. In Mb1-aCard11 mice, GC B cells exhibited exaggerated DZ skewing yielding an ∼3:1 DZ/LZ ratio (Fig. 4, D and E). One possible explanation for this finding includes an increase in naive B cells that enter the GC response (perhaps due to a lower activation threshold). In contrast to this idea, however, we also observed an increased frequency of BrdU+ GC B cells (Fig. S3 D) and exaggerated DZ skewing (∼3:1 ratio; Fig. S3 E) in the Cγ1-aCard11. We queried whether the enhanced cycling we observed was restricted to the DZ, as that could account for the exaggerated skewing. Rather than a majority of EdU+ cells restricted to the DZ, we observed enhanced cycling in both DZ and LZ cells at similar proportions within Mb1-aCard11 mice compared with controls (Fig. 4, F and G). This finding implies that aCARD11 causes dysregulated cycling throughout the GC.
We also assessed the transcriptional program of GC B cells in the Mb1-aCard11 model. Activation-induced cytidine deaminase (AID) promotes both somatic hypermutation and CSR in GC B cells, and DZ B cells have increased AID expression compared with LZ B cells (Victora et al., 2012). Mb1-aCard11 GC B cells (B220+CD95hiPNAhi) expressed greater levels of AID compared with control GC B cells at the peak of the GC response (Fig. 4, H and I), consistent with skewing of the GC toward a DZ phenotype. AID expression is regulated by FOXO1, a critical transcriptional regulator of the DZ program. FOXO1 deletion in GC B cells results in a loss of the DZ (Dominguez-Sola et al., 2015). Surprisingly, we observed decreased expression of FOXO1 within total GC B cells (B220+CD95hiPNAhi) in Mb1-aCard11 mice at the peak of the GC response (Fig. S3, F and G). Decreased FOXO1 may indicate enhanced phosphoinositide 3-kinase (PI3K) signaling since Akt, downstream of PI3K, phosphorylates FOXO1, resulting in nuclear export and increased proteasomal degradation (Hay, 2011). However, GC B cell restricted expression of constitutively active PI3K results in a loss of the DZ, reminiscent of GC B cell–intrinsic deletion of FOXO1 (Dominguez-Sola et al., 2015; Sander et al., 2015). In summary, expression of aCARD11 in GC B cells increases their proliferation and promotes a DZ phenotype.
GC B cells expressing aCARD11 demonstrate elevated mTORC1 signaling
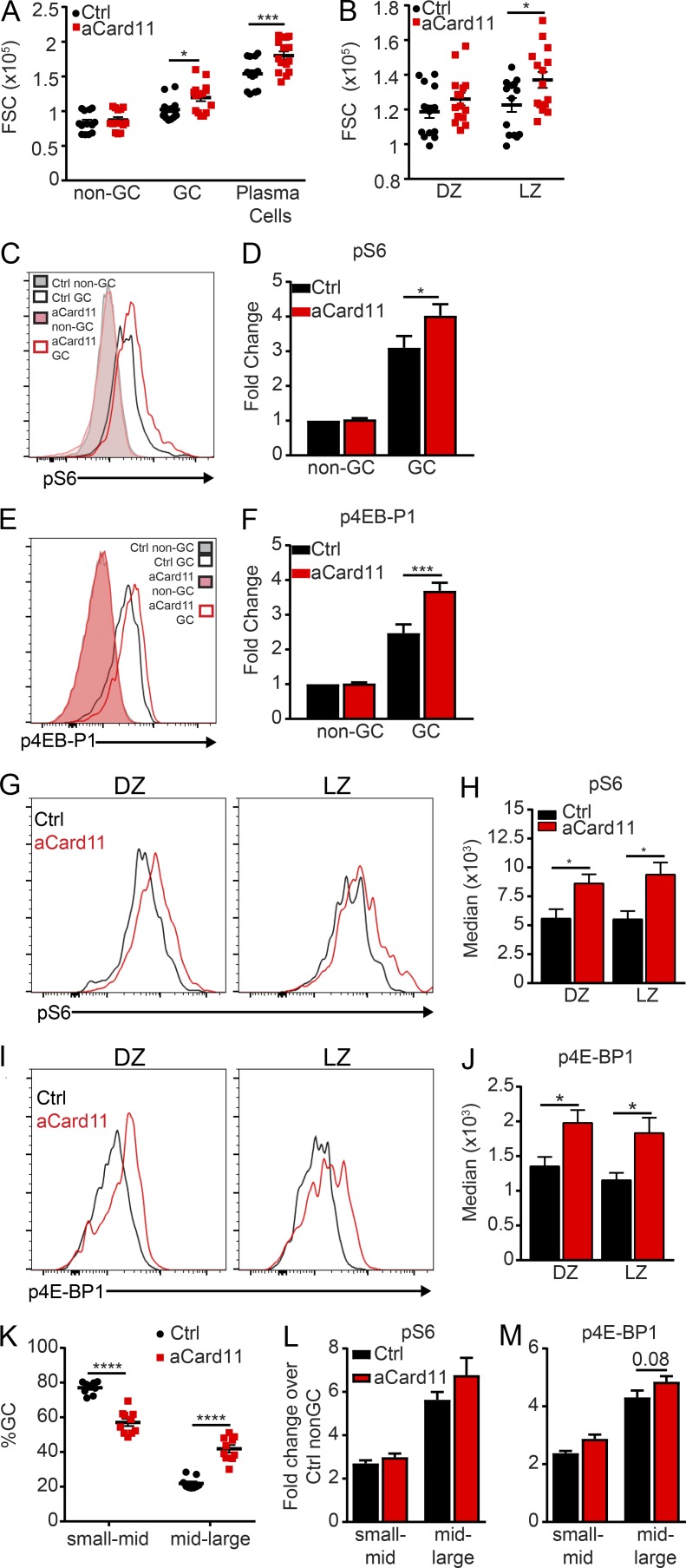
GC-specific, hyperactivation of mTOR results in DZ skewing and larger GC cell size (Ersching et al., 2017), reminiscent of the phenotype in the Mb1- and Cγ1-aCard11 models. mTORC1 signaling, triggered in response to positive selection within the LZ, yields increased biomass that permits GC B cells to undergo subsequent rounds of proliferation within the DZ. mTORC1 hyperactivation has also been shown to drive PC development when TSC1 is deleted in mature B cells (Benhamron et al., 2015). GC B cells are more metabolically active, correlating with increased mTORC1 activity, cell size, glucose uptake, and mitochondrial biomass (Jellusova et al., 2017). Using forward scatter (FSC) as a measure of cell size, we observed a significant increase in the size of GC B cells and PCs in both Mb1-aCard11 (Fig. 5 A) and Cγ1-aCard11 (Fig. S4, A and C) mice compared with controls (at day 5 after immunization with SRBCs). Further dissection of the GC into DZ vs. LZ cells showed that aCARD11-expressing cells in the DZ trended toward a larger FSC than their control counterparts, while LZ cells were significantly larger than control LZ cells (Figs. 5 B and S4 B).
Figure 5.
GC B cells expressing aCARD11 exhibit increased mTORC1 signaling. Ctrl and Mb1-aCard11 mice were immunized with SRBCs and sacrificed on day 5 after immunization for analysis of B cell populations. (A) Median FSC of non-GC B cells (B220+CD95loCD38hi), GC B cells (B220+CD95hiCD38lo; P = 0.04) and PCs (B220−CD138+; P = 0.0006). (B) Median FSC of DZ (CXCR4+CD86−) and LZ (CXCR4−CD86+; P = 0.03) GC B cells. (A and B) Data are representative of four independent experiments with 15 Ctrl mice and 15 Mb1-aCard11 mice injected with SRBCs. Statistics were calculated by two-way ANOVA. (C) pS6 in non-GC (B220+CD95loCD38hi) and GC (B220+CD95hiCD38lo) B cells. Filled histograms: non-GC B cells (gray, Ctrl; pink, Mb1-aCard11). Open histograms: GC B cells (black, Ctrl; and red, Mb1-aCard11). (D) Fold change in pS6 median flourescent intensity over Ctrl non-GC B cells (P = 0.044). Data are representative of two independent experiments; n = 8 Ctrl and n = 8 Mb1-aCard11 mice. Significance was calculated by two-way ANOVA. (E) p4E-BP1 in non-GC and GC B cells. Filled histograms: non-GC B cells (gray, Ctrl; pink, Mb1-aCARD11). Open histograms: GC B cells (black, Ctrl; red, Mb1-aCARD11). (F) Fold change in p4E-BP1 median flourescent intensity over Ctrl non-GC B cells (P = 0.0004). Data are representative of two independent experiments; n = 7 Ctrl and n = 6 Mb1-aCard11 mice. Significance was calculated by two-way ANOVA. (G) Representative histogram overlays pS6 within the DZ (left, CD19+CD95hiCD38loCXCR4+CD86−) and LZ (right, CD19+CD95hiCD38loCXCR4−CD86+) in Ctrl (black) and Mb1-aCard11 (red) mice. (H) Median fluorescent intensity of pS6 in DZ (P = 0.045) and LZ (P = 0.01) cells in Ctrl mice and Mb1-aCard11 mice. (I) Representative histogram overlays of p4E-BP1 in DZ and LZ in Ctrl (black) and Mb1-aCard11 (red) mice. (J) Median fluorescent intensity of p4E-BP1 in DZ (P = 0.048) and LZ (P = 0.03) cells in Ctrl mice and Mb1-aCard11 mice. (G–J) Data are representative of two independent experiments with five Ctrl mice and seven Mb1-aCard11 mice. Significance was calculated by two-way ANOVA. (K–M) GC B cells from immunized Mb1-aCARD11 vs. control mice were separated based upon cell size: (1) small- to mid-sized cells (side scatter [SSC] < 1.5 × 105 and FSC < 1.5 × 105) and (2) mid-large–sized cells (SSC > 1.5 × 105 and FSC >1.5 × 105). (K) Proportion of GC B cells in small-mid–sized gate (P < 0.0001) and the mid-large cell–sized gate (P < 0.0001). (L) Fold change in pS6 median fluorescent intensity over control non-GC B cells. (K and L) Data are representative of three independent experiments with 9 Ctrl mice and 10 Mb1-aCard11 mice injected with SRBCs. (M) Fold change in p4E-BP1 median fluorescent intensity over control non-GC B cells (P = 0.08). Data are representative of two independent experiments with seven Ctrl mice and six Mb1-aCard11 mice injected with SRBCs. Significance was calculated by two-way ANOVA. For summary graphs, lines represent mean ± SEM. For bar graphs, lines represent mean + SEM. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001.
To explicitly determine if mTORC1 signaling is increased in this strain, we quantified phosphorylation of ribosomal protein S6 and eukaryotic translation initiation factor 4E-binding protein 1 (4E-BP1), major downstream targets of the mTORC1 pathway (Iwata et al., 2017). Using flow cytometry, we observed a significant increase in pS6 and p4E-BP1 in GC B cells (B220+CD95hiCD38lo) in Mb1-aCard11 mice compared with controls (Fig. 5, C–F). Immunoblotting of total splenic B cells (including naive and GC B cells, PBs, and PCs) from immunized mice further supported our findings, with evidence for increased pS6 in Mb1-aCard11 mice (Fig. S4, D and E). As anticipated, we also observed a trend toward a decrease in total basal IκBα in unimmunized Mb1-aCard11 B cells compared with controls (Mb1Cre/+; Fig. S4, D and E), consistent with enhanced NF-κB activation (Lenz et al., 2008). Segregation of GC B cells into DZ vs. LZ cells demonstrated increased pS6 and p4E-BP1 in both populations in Mb1-aCard11 mice compared with controls (Fig. 5, G–J). Thus, the enhanced mTORC1 signature observed is not solely due to increased DZ skewing in Mb1-aCard11 mice. The increased pS6 and p4E-BP1 in the aCard11 GC B cells is potentially a result of their increased biomass. Separation of the GC B cells into two populations, differentiated by size, demonstrated an increased proportion of cells with increased biomass in the GC of Mb1-aCard11 mice compared with controls (Fig. 5 K). However, pS6 and p4E-BP1 levels were not significantly different in small-mid or mid-large populations of GC B cells from Mb1-aCard11 vs. control mice (Fig. 5, L and M). Thus, it remains unclear whether the increased biomass precedes elevated mTORC1 signaling or, conversely, is a result of enhanced mTORC1 signaling. mTORC1 signaling may also be impacted by increased responsiveness to T follicular helper cells, as aCARD11-expressing GC B cells exhibited increased CD40 surface expression and a trend for diminished MHC surface levels (Fig. S4, F and G). Together, these data indicate that aCARD11 expression enhances NF-κB signals and, directly or indirectly, potentiates mTORC1 signaling in response to TD antigens.
Cells overexpressing aCARD11 manifest an NF-κB–independent elevation in mTORC1 signaling
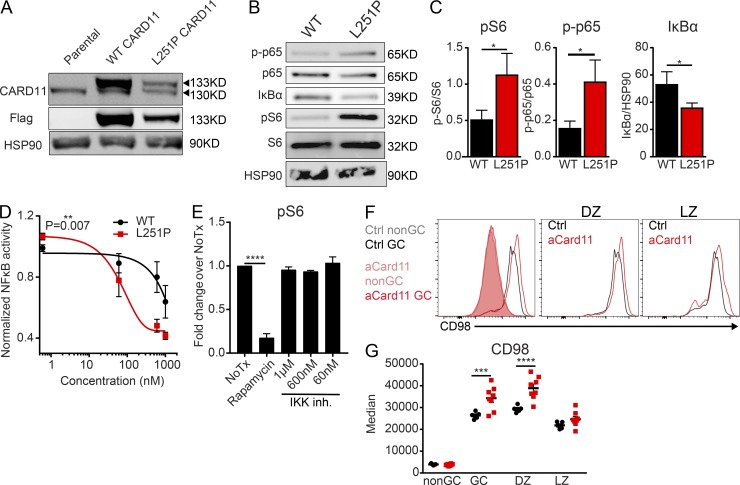
To further assess the impact of aCARD11 on signaling, we next analyzed basal signaling differences in a GC B cell–like (GCB)-DLBCL line, OCI-Ly7, whose survival is not dependent on CARD11. This GCB-DLBCL line was transduced with lentiviral vectors expressing either flag-tagged WT CARD11 or CARD11-L251P and a cis-linked GFP marker (Fig. 6 A). Using cells closely matched for GFP expression and viral copy number, we observed increased pS6 in aCARD11-expressing cells (Fig. 6, B and C). Of note, consistent with increased turnover of aCARD11 (Moreno-García et al., 2010), CARD11 protein levels were significantly reduced in CARD11-L251P transduced samples (data not shown). Consistent with this finding, aCARD11 cells exhibited enhanced NF-κB signaling as assessed by using an NF-κB reporter and by immunoblotting, demonstrating increased phospho-p65 and decreased total IκBα (Fig. 6, B–D). To determine whether the enhanced mTORC1 signaling characteristic of CARD11-L251P–expressing cells is dependent on NF-κB signaling, CARD11-L251P–expressing cells were treated with the IKK-2 inhibitor, MLN120B (Hideshima et al., 2006). While MLN120B treatment significantly reduced NF-κB signaling, measured by diminished NF-κB–driven luciferase production (Fig. 6 D), we observed no difference in pS6 levels after treatment (Fig. 6 E), unlike treatment with rapamycin. Our findings demonstrate that enhanced mTORC1 signaling in CARD11-L251P expressing cells is NF-κB–independent; however, whether mTORC1 signaling is directly or indirectly driven by oncogenic CARD11 remains unclear.
Figure 6.
DLBCL B cells expressing aCARD11 exhibit increased pS6 independent of NF-kB signaling. The GCB-DLBCL line, OCI-Ly7, was transduced with lentiviral vectors encoding murine, flag-tagged WT CARD11 or CARD11-L251P, and a cis-linked GFP reporter. (A) Immunoblot showing expression of flag-tagged CARD11 in transduced cell populations. (B) Representative immunoblot of basal signaling in WT or CARD11-L251P cells. (C) Left: Quantification of signaling by using ImageJ of pS6 normalized to total S6 (P = 0.017). Middle: p-p65 normalized to total p65 (P = 0.031). Right: Total IκBα normalized to HSP90 (P = 0.041). Combined data are from three experiments; significance was calculated by Student’s paired t test. Data in A–C are representative of seven biological replicates. (D) NF-κB activity (luciferase) normalized to viability (Cell Titer Glo) after treatment with different doses of IKK-2 inhibitor for 16 h. Area under the curve is significantly different between WT and CARD11-L251P (P = 0.007), calculated by Student’s paired t test. Data represent three independent experiments. (E) Fold change in pS6 median fluorescent intensity of CARD11-L251P expressing OCI-Ly7 cells normalized to untreated cells (P < 0.0001). Data are representative of three independent experiments with three biological replicates. Significance was calculated by one-way ANOVA. (F) Representative histogram overlays of CD98 expression on (left) non-GC B cells (filled histograms: gray, Ctrl; pink, Mb1-aCard11, CD19+CD95loCD38hi) and GC B cells (open histograms: black, Ctrl; red, Mb1-aCard11, CD19+CD95hiCD38lo); (middle) DZ (CD19+CD95hiCD38loCXCR4+CD86−) and (right) LZ (right, CD19+CD95hiCD38loCXCR4−CD86+) GC B cells in Ctrl (black) and Mb1-aCard11 (red) mice. (G) Median fluorescent intensity of CD98 on non-GC, GC (P = 0.0008), DZ (P < 0.0001), and LZ cells in Ctrl mice and Mb1-aCard11 mice. (F and G) Data are representative of two independent experiments with five Ctrl mice and seven Mb1-aCard11 mice 5 dpi with SRBCs. Significance was calculated by two-way ANOVA. For summary graphs, lines represent mean ± SEM. For bar graphs, lines represent mean + SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Activation of mTORC1 requires mTORC1 recruitment to the lysosome, which is induced by amino acid sensing. Patients with hypomorphic, dominant-negative mutations in CARD11 display decreased pS6, as well as diminished expression of amino acid transporters (Ma et al., 2017). CD98 is a heterodimer composed of a heavy chain (Slc3a2) covalently linked to one of several light chains (e.g., LAT1/Slc7a5 or LAT2/Slc7a8), which function as amino acid transporters (Cantor et al., 2009). Aside from amino acid transport, CD98 plays a role in cell adhesion, growth, and survival (Cantor and Ginsberg, 2012). To determine whether CD98 expression is altered in aCARD11-expressing cells, we analyzed GC B cells in control and Mb1-aCard11 mice at 5 d after immunization with SRBCs. We observed a significant increase in surface CD98 expression on aCard11 GC B cells compared with controls (Fig. 6, F and G). Analysis of DZ and LZ cells demonstrated that the DZ cells display greater expression of CD98 (Fig. 6, F and G). Increased expression of CD98 in DZ cells in Mb1-aCard11 mice may support the observed increase in cycling, allowing GC B cells to better compete in the setting of limited nutrient availability. However, our data cannot distinguish whether the increase in CD98 facilitates or merely reflects the observed exaggerated DZ skewing.
aCARD11 signaling likely enhances CSR
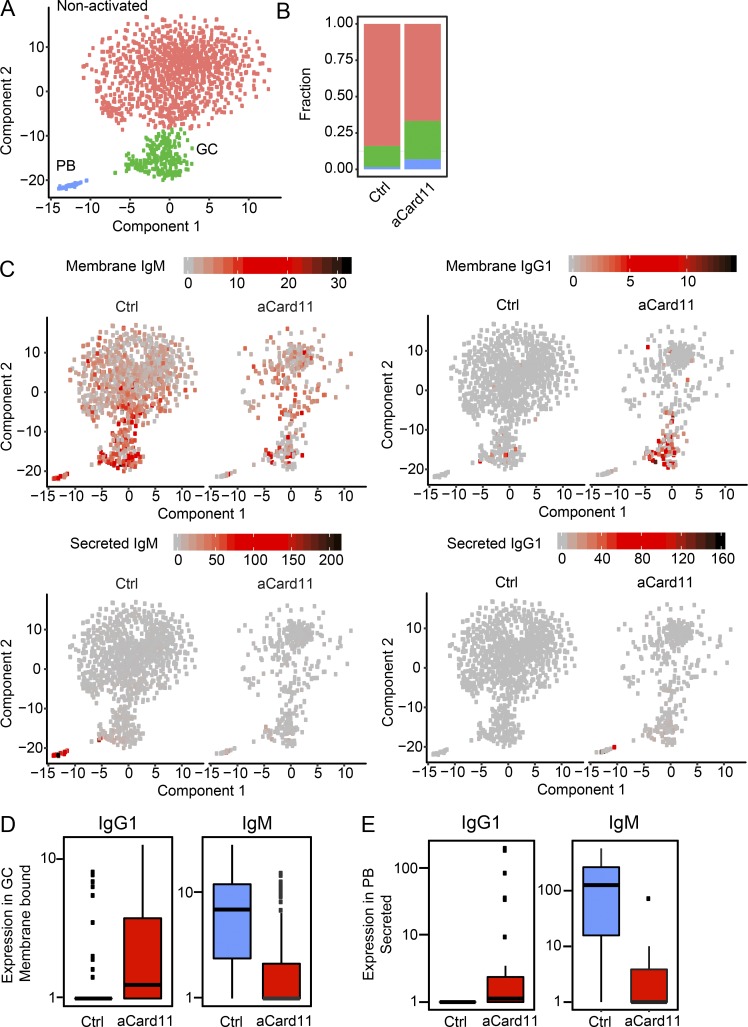
Pharmacological inhibition and murine models of B cell–intrinsic deletion of Rictor or Raptor demonstrate the importance of the mTORC1/pS6 axis in promoting CSR. Treatment of B cells with ATP-competitive mTOR kinase inhibitors that targeted both mTORC1/2 or conditional deletion of Rictor, a key component of the mTORC2 complex, resulted in enhanced CSR. In contrast, inhibition of mTORC1 by rapamycin or B cell–specific deletion of Raptor, required for mTORC1 function, reduced CSR and PB formation (Limon et al., 2014). Mass cytometry has demonstrated that GC B cells with the highest levels of pS6 also express the highest levels of IgG1 (Jellusova et al., 2017). These data support a model wherein cells with increased mTORC1 activity are selected to undergo CSR. Our findings indicate that aCARD11 alters the kinetic of the GC response and GC B cells expressing aCARD11 display enhanced mTORC1 signaling. We used single-cell RNA sequencing (scRNA-seq) to interrogate the impact of aCARD11 on the transcriptome of B cells in response to immunization. Total B cells and PCs were isolated from Mb1-aCard11 and control (Mb1Cre/+) mice at day 5 after immunization with SRBCs and processed for scRNA-seq by using the Chromium product suite by 10× Genomics (Zheng et al., 2017). Cell Ranger (10× Genomics) and Monocle 2 (Qiu et al., 2017) were used for data analysis. We obtained single-cell expression profiles for a total of 1,954 cells. Unsupervised clustering was performed to identify and remove non–B lineage cells (Fig. S5 A), as well as to separate B cells into nonactivated vs. GC B cells and PBs (Fig. 7 A). The GC cluster was characterized by high transcript levels of AID (Fig. S5 B) and genes associated with cell cycle progression and DNA-repair pathways previously shown to be up-regulated in GC B cells (Mesin et al., 2016). The PB cluster was characterized by high transcript levels of Xbp1 (Fig. S5 C), a transcription factor specifically up-regulated in antibody-secreting cells (ASCs; Tellier et al., 2016). Consistent with our flow and immunohistochemistry findings, Mb1-aCard11 mice were found to have increased GC and PB compartments (Fig. 7 B). Differential gene expression analysis between control and aCARD11 cells revealed membrane-bound and secreted BCR isotypes among the most significantly differentially expressed genes within the GC and PB compartments. The GC B cells in the aCARD11 samples showed an increase in membrane-bound IgG1 transcripts with a concomitant decrease in membrane-bound IgM, compared with controls (Fig. 7, C [top] and D). Similarly, within the PB cluster, aCARD11 cells exhibited increased secreted IgG1 transcripts and decreased secreted IgM compared with control cells (Fig. 7, C [bottom] and E). aCARD11 GC B cells also showed increased secreted IgG1 transcript levels, suggesting that they are on their way to becoming PBs, as well as decreased membrane bound IgM in the PB compartment (Fig. S5, D and E).
Figure 7.
aCARD11 promotes Ig class switching in GC B cells and generation of IgG1-secreting PCs. Ctrl (Mb1Cre/+) and Mb1-aCARD11 mice were immunized with SRBCs and sacrificed on day 5 after immunization for scRNA sequence analysis (see Materials and methods). (A) A t-SNE plot of 1,581 single B cells reveals three clusters: red, nonactivated; green, GC; and blue, PBs. (B) Fraction of B cell clusters in A within Ctrl and Mb1-aCard11 samples. (C) t-SNE plots of B cells (as shown in A) illustrating gene expression. Left: Ctrl cells; right: Mb1-aCard11. Depicted are transcript levels of membrane-bound IgM (upper left), secreted IgM (lower left), membrane-bound IgG1 (upper right), and secreted IgG1 (lower right). (D) Expression levels of membrane-bound IgG1 (left; P = 3.10−39) and IgM (right; P = 1.6 × 10−74) transcripts in GC B cells. (E) Expression levels of secreted IgG1 (left; P = 2.4 × 10−58) and IgM (right; P = 1.7 × 10−143) transcripts in GC B cells. (D and E) Blue, Ctrl; red, Mb1-aCard11. P values were calculated by using the likelihood ratio test included within Monocle’s differential-GeneTest function. Data shown are from one of two representative experiments. Graphs are box plots showing lower (first) and upper (third) quartiles with the line indicating the median. Lines extending from box plots indicate ±1.5 interquartile range (IQR). Individual data points are outlying points beyond ±1.5 IQR.
To determine if CSR is altered at the protein level, we also performed FACS analysis of PCs at day 5 after immunization in Mb1-aCard11 and control mice to assess intracellular Ig levels and the antibody isotype being produced. Compared with control cells, PCs in Mb1-aCard11 mice exhibited an increased proportion of in IgM−IgG1− PCs (B220−CD138+) following immunization (Fig. S5, F and G). These “double-negative” PCs have likely switched to an IgG isotype other than IgG1, as there is a significant decrease in IgG1+ PCs as well (Fig. 5, F and G). These findings, while not definitive, are consistent with a model whereby aCARD11 expression increases CSR during a TD immune response.
Discussion
CARD11 plays a pivotal role in B and T cell function through regulation of the antigen receptor (AgR)–NF-κB signaling axis. Expanding work has identified both germline and somatic, loss-of-function (LOF) and gain-of-function (GOF) variants in CARD11, providing insight into human immune development and function. Germline GOF CARD11 mutations lead to an immune dysregulation disorder referred to as B cell expansion with NF-κB and T cell anergy (BENTA; Snow et al., 2012; Brohl et al., 2015). These patients have defects in humoral responses to some vaccines, with decreased switched and memory B cells as well as T cell hyporesponsiveness. In contrast, somatic CARD11 GOF mutations are enriched in ABC-DLBCL, a post-GC malignancy exhibiting mutated Ig V genes and a gene expression signature that includes PB genes (Shaffer et al., 2012). Previous murine models of Card11 GOF variants used pan-B lineage transgenic expression (Knies et al., 2015) or lentiviral expression and adoptive transfer (Jeelall et al., 2012) to assess the impact on developing and mature B cells but did not assess the role of these variants in TD immune responses. In this study, we characterized the impact of an activating mutation in CARD11 on B cell development and, most notably, within the GC response. We show that B cell–intrinsic expression of aCARD11 markedly alters the innate B cell compartment while exerting only limited impact on the naive B2 B cell compartments. B cell–intrinsic aCARD11 expression substantially impacts GC dynamics leading to more rapid GC formation and resolution with enhanced terminal differentiation. aCARD11-expressing GC B cells exhibit increased cycling and DZ skewing. Surprisingly, in addition to increased NF-κB signals, aCARD11 GC B cells exhibit enhanced mTORC1 signaling. Consistent with these findings, a DLBCL cell line overexpressing CARD11-L251P demonstrated enhanced NF-κB and mTORC1 signaling in the absence of stimulation. Thus, aCARD11 potentially modulates multiple critical signaling pathways that uniquely alter the GC response.
Expression of aCARD11 during B cell development exerted only minimal impact on BM and splenic B2 B cell development but lead to an unexpected impact on the innate B cell compartment. aCARD11 expression had no appreciable impact on BM B cell development or the number or proportion of splenic transitional B cells. Mb1-aCard11 mice, however, exhibited a modest increase in the proportion and number of FM B cells, a change that accounted for a moderate increase in total B2 cell numbers. In parallel, these animals exhibited a decreased frequency of MZ B cells. BENTA patients exhibit polyclonal expansion of immature, transitional, and naive mature B cells (Arjunaraja et al., 2017). In contrast, aCARD11 expression led to loss of the peritoneal B1 compartment with near absence of the B1a B cell subset and marked reduction in B1b B cell numbers. Loss of these innate-like B cell populations correlated with decreased serum IgM in unimmunized Mb1-aCard11 mice. While not answered in our studies, a range of mechanisms might account for these findings. One possibility is that aCARD11 promotes terminal differentiation of B1 B cells, similar to the enhanced PC response observed following TD immunization (see below). However, the combined reduction of both B1 B cells and IgM argues against this idea. Alternatively, aCARD11 signals may impact B lineage fate at an early stage of B1 B cell development or limit resting and/or activated B1 cell survival. Notably, previous studies in Card11−/− and Bcl10−/− mice (Egawa et al., 2003; Xue et al., 2003) reported defects in both the MZ and B1 compartments, suggesting that signals mediated by the CBM complex are required for development and/or maintenance of these populations. The parallel findings in Card11 LOF vs. GOF mice suggest that precise tuning of one or more CARD11-dependent signals (NF-κB, JNK, and/or mTORC1; see also below) is required for B1 development or maintenance and that perturbation in either direction promotes loss of innate B cells. Of note, consistent with our data, IgM levels are consistently reduced in human subjects with BENTA (Snow et al., 2012; Brohl et al., 2015). BENTA patients exhibit impaired responses to T cell–independent polysaccharide vaccines, and low serum titers to some T cell–dependent viral vaccines have been observed in some patients (Arjunaraja et al., 2017). These defects in T cell–independent humoral responses parallel our findings of diminished innate B cell populations and decreased IgM titers in the Mb1-aCard11 mice. The similarities between our mouse model and the BENTA phenotype may indicate that aCARD11 expression would yield dampened T cell–independent humoral responses in our mouse model and will be a focus of future studies. While the B1 B cell compartment has not been evaluated in BENTA, this may be a fruitful area for future study. Defining CARD11-dependent signals essential for B1 development or survival is likely to provide insight into the regulation of this important population.
Using two distinct, inducible B cell-intrinsic models, we show that expression of the CARD11-L251P is sufficient to alter GC dynamics. Upon immunization, aCARD11 expression promoted both accelerated GC formation and resolution. At the peak of the GC response, aCard11 mice exhibited exaggerated DZ skewing compared with controls. This change correlated with enhanced cycling and increased AID expression, increased cell biomass, enhanced pS6 and p4E-BP1 levels, and decreased FOXO1 levels, illustrating a unique impact of aCARD11 on B cell–intrinsic signaling. Importantly, nearly all of these observations were recapitulated in a model in which aCARD11 expression was restricted to GC B cells (Cγ1-Cre), indicating that these changes are not due to a developmental impact of aCARD11 in the Mb1-cre model. GOF mutations in CARD11 yield a spectrum of N-FκB activation (Lenz et al., 2008) that supports survival, proliferation, and terminal differentiation. In contrast to previous work with retroviral overexpression of aCARD11 mutants in primary murine B cells (Jeelall et al., 2012), CARD11 expression was comparable to endogenous levels in our knock-in model. Further, while CARD11-L251P mediates less robust constitutive NF-κB activation compared with the L225LI mutation modeled in previous studies (Lenz et al., 2008; Jeelall et al., 2012; Knies et al., 2015), our combined data demonstrate that B cell–intrinsic, endogenous level CARD11-L251P expression (as would occur following mutation in somatic cells) is sufficient to drive enhanced cycling of GC DZ cells.
In addition to its impact on DZ cells, aCARD11-expressing LZ cells are likely more responsive to AgR and, potentially, to costimulatory or cytokine signals that mediate activation and promote differentiation, GC exit, and GC dissolution. NF-κB signaling in B cells promotes IRF4 and Blimp1 expression, leading to terminal differentiation. Consistent with this idea, both B cell–intrinsic GC models revealed a major impact on the rate of PC generation in response to immunization. Cγ1-aCard11 and Mb1-aCard11 animals exhibited approximately sixfold and eightfold increases in splenic PCs, respectively. PCs in the Mb1-aCard11 model exhibited enhanced cycling, and cell biomass was increased in both models at the peak of the GC response. Cγ1-aCard11 animals also exhibited increased antigen-specific IgG and IgG1 at 12 dpi. These observations are consistent with previous work showing that GOF mutations in CARD11 promote proliferation and plasmacytic differentiation (Jeelall et al., 2012). Similarly, expression of CARD11-L225LI using a similar Rosa26 transgenic model (Knies et al., 2015) lead to diffuse organ infiltration of plasmacytic cells and early mortality. In contrast to aCARD11, B cells expressing constitutively active IKKβ exhibit increased proliferation in vitro but fail to persist in vivo in the presence of self-antigen. Similarly, GC B cell–restricted expression of constitutively active IKK2 (IKKCA; Calado et al., 2010) did not significantly alter the GC response, antibody levels, or PC formation. The phenotypic differences observed across the aCARD11 vs. the IKKCA models supports the idea that aCARD11 can influence B cell fate via both NF-κB–dependent and –independent signals.
Consistent with the role for aCARD11 in enhanced NF-κB signaling (Lenz et al., 2008; Jeelall et al., 2012; Knies et al., 2015), CARD11-L251P–expressing murine B cells (Mb1-aCard11 model) and DLBCL lines exhibited reduced IκBα levels. Importantly, in the context of the GC, we demonstrate that cells expressing aCARD11 have additional alterations in signals critical for GC function including, most notably, mTORC1. Our findings demonstrate that CARD11-L251P may directly or indirectly promote enhanced mTORC1 signaling, as indicated by increased DZ skewing, biomass, and pS6 and p4E-BP1 levels in murine GC B cells, and elevated basal pS6 in a CARD11-L251P–expressing GCB-DLBCL cell line. mTORC1 signals play a critical role in GC biology. B cell–intrinsic deletion of Raptor, a key adaptor for the mTORC1 complex, blocks PC differentiation and antibody secretion (Jones et al., 2016). Similarly, hypomorphic mTOR variants inhibit humoral responses to antigen (Zhang et al., 2013). Raptor deletion at the peak of the GC or treatment with rapamycin after GC initiation results in premature collapse of the GC (Jones et al., 2016). In contrast, hyperactive mTORC1 skews GC B cells toward a DZ phenotype (Ersching et al., 2017). In adoptive transfer models, however, mTORC1 hyperactive GC B cells are outcompeted at later time points, demonstrating the importance of coordinate regulation of this pathway. We specifically observed that increased cell biomass and pS6 and p4E-BP1 levels were in aCARD11 GC B cells and PCs following immunization (and not in non-GC B cells). The GC comprises a hypoxic, nutrient-deprived microenvironment due, in part, to the rapid proliferation of activated B cells (Cho et al., 2016; Jellusova et al., 2017). The altered GC response in the animal models described here suggests that aCARD11 may be particularly effective in promoting GC expansion in this “stressful” environment. While not tested here, aCARD11 may also facilitate this process by lowering the BCR activation threshold, allowing for a more efficient positive selection by T follicular helper cells or follicular dendritic cell–delivered antigen in the LZ. In this context, aCARD11-expressing B cells might also gain increased sensitivity to coreceptor signals and amino acid sensing pathways that mediate mTORC1 activation.
How CARD11 links AgR signaling to mTORC1 remains unclear. Our findings suggest there is a link between CARD11 and mTORC1 signaling in GC B cells, an idea that is consistent with, and expands upon, earlier in vitro studies in T cell lines and primary T cells. Studies in CD4+ T cells support the idea that IKK activity is not required for CARD11-dependent mTORC1 induction (Hamilton et al., 2014; Nakaya et al., 2014). Work using CARD11-deficient Jurkat T cells and mouse CD4+ T cells reported decreased pS6 levels in response to CD3/CD28 stimulation (Hamilton et al., 2014). One model proposes that CARD11 complexes with MALT1 to activate mTORC1, independent of Bcl10. Consistent with this idea, the paracaspase activity of MALT1 was required for mTORC1 induction, whereas blocking Bcl-10 or IKK2 had no impact on pS6 levels after CD3/CD28 stimulation. In contrast, another study showed a partial impairment in mTORC1 in Bcl10-deficient naive CD4+ T cells after TCR/CD28 stimulation (Nakaya et al., 2014), perhaps reflecting differences in culture or stimulation conditions (Shi and Sun, 2015). Our studies of CARD11-L251P–expressing DLBCL cell lines support a model in which mTORC1 signaling is independent of NF-κB signaling. It has been postulated that CARD11 modulates a novel signaling axis involved in amino acid sensing that cooperates with PI3K/AKT to activate mTORC1. Diminished pS6 correlates with decreased expression of ASCT2 and CD98 in CD4+T cells from patients with dominant-negative CARD11 mutations (Ma et al., 2017). Conversely, our data suggest that aCARD11 enhances this signaling axis and plays a pivotal role in B cells, particularly during a GC response. While Slc1a5−/− (ASCT2−/−) mice do not exhibit defects in TD GC responses (Masle-Farquhar et al., 2017), B cell–intrinsic deletion of CD98 heavy chain (Slc3a5flox/floxCD19Cre/+) impairs antibody responses and PC formation (Cantor et al., 2009). CD98 functions in both integrin signaling and amino acid transport (Fenczik et al., 2001; Nicklin et al., 2009). However, the impact of these distinct functions in vivo has not been deconvoluted. CD98 (Slc3a2-Slc7a5 heterodimer) acts as a bidirectional transporter, which exports intracellular L-glutamine in exchange for the import of L-leucine and activates mTORC1 (Nicklin et al., 2009). CD98 is up-regulated upon B cell activation and confers a selective advantage to GC B cells (Cantor et al., 2009). The increase in CD98 expression on aCARD11 GC B cells suggests that they are likely more responsive to amino acid sensing, resulting in heightened mTORC1 signals. Alternatively, aCARD11 expression could sensitize cells to integrin-mediated signals, which would support enhanced cycling and survival. Thus, while the precise link(s) between CARD11 and mTORC1 in B cells remains to be elucidated, our data in association with previous findings in T cells, strongly support a model in which altered mTORC1 signaling, due to enhanced amino acid sensing and/or NF-κB–independent mTORC1 activation downstream of CARD11, functions in conjunction with enhanced NF-κB signals to alter the GC response.
Multiple types of B cell malignancies that arise from GC and post-GC B cells exhibit GOF mutations in CARD11, including diffuse large-, MZ-, mantle cell-, and follicular-B cell lymphomas and, less commonly, Waldenstrom macroglobulinemia, lymphoplasmacytic lymphoma, and primary central nervous system lymphoma (Shaffer et al., 2012; Wu et al., 2016; Arjunaraja et al., 2017; Onaindia et al., 2017; van den Brand et al., 2017). aCARD11 GC B cells exhibited increased AID expression, increased expression of switched transcripts, and an increase in the secreted form of switched transcripts within PBs by single-stranded RNA sequencing analysis. While Mb1-aCard11 and control mice displayed equivalent IgG and IgG1 levels after SRBC immunization, PCs in aCARD11 animals were predominantly IgM−IgG1− at day 5 after immunization, consistent with increased class switching. Previous work indicates that CSR to IgG1 drives GC B cells toward a PC fate (Gitlin et al., 2016), suggesting that enhanced class switching in aCARD11-expressing cells may similarly increase PC differentiation. Both enhanced CSR and PC differentiation may be due to enhanced mTORC1 signaling in aCARD11-expressing GC B cells, as mTORC1 promotes both CSR and PC differentiation (Limon et al., 2014; Tamahara et al., 2017). These events likely function in concert with additional mutations acquired during the GC response to promote lymphomagenesis in GC-derived B cells. ABC-DLBCLs express high levels of AID and heavily mutated IgH genes, and a requirement for AID activity during oncogenesis has been demonstrated in multiple models (Kotani et al., 2007; Pasqualucci et al., 2008). In our aCard11 mice, overcoming the premature collapse of the GC may be required to allow additional oncogenic hits to be acquired, promoting lymphomagenesis. DLBCLs exhibit inactivation of BLIMP1/PRDM1 at levels >50% (Mandelbaum et al., 2010), thus modeling aCARD11 signaling in the context of a block in terminal differentiation to study the impact of these coordinating oncogenic events in promoting lymphomagenesis is a line of inquiry we are currently pursuing.
In summary, we demonstrate that aCARD11 expression markedly alters GC dynamics and cell fate decisions. Disease-associated, activating CARD11 mutations have previously been shown to exhibit a spectrum of NF-κB activation. Our data build upon those findings, indicating that aCARD11 may potentiate mTORC1 signaling. Many ABC-DLBCLs (∼50%) are refractory to current therapeutic regimens, and it has been shown that follicular lymphomas (Bartlett et al., 2018) and mantle cell lymphomas expressing aCARD11 are resistant to ibrutinib treatment (Wu et al., 2016; Xu et al., 2017). Our combined findings suggest that aCARD11 contributes to resistance in aggressive lymphomas via enhancement of these distinct signaling programs and that mTORC1 inhibitors should be explored as a potential treatment option in tumors expressing aCARD11.
Materials and methods
Mice
1× Flag-tagged murine CARD11-L251P was knocked into the ROSA26 locus, with an upstream loxp-flanked 3×-stop codon and a downstream self-cleaving T2A linked GFP reporter. These mice are called activated Card11 (aCard11) mice throughout this article. The aCard11 mice were maintained by crossing to C57BL/6 mice and have been backcrossed five times. aCard11 mice were crossed to Mb1-Cre (Hobeika et al., 2006) and Cγ1-Cre (Casola et al., 2006) mice, which are on a fully C57BL/6 background. All mice were bred and maintained in a specific pathogen-free animal facility, and studies were performed in accordance with the Institutional Animal Care and Use Committee of Seattle Children’s Research Institute. Mb1Cre mice were provided by M. Reth (Max Planck Institute of Immunobiology, Freiburg, Germany).
Immunizations
12–15-wk-old mice were i.p. immunized with 200 μl of 20% by volume SRBCs (Colorado Serum Co.) diluted in 1× PBS and sacrificed 5, 7, 10, and 21 d after immunization. 12–23-wk-old mice were i.p. immunized with 50 μg of NP-CGG (Ratio>40; Biosearch Technologies) precipitated in alum and sacrificed at 12 d after immunization.
Reagents and antibodies
The LIVE/DEAD Fixable Near-IR Dead Cell Stain Kit (Invitrogen) was used according to the manufacturer’s instructions. Antimurine antibodies used in the study include those from eBioscience: CD38 (90), Bcl-6 (BCL-DWN), AID (mAID-2), IRF4 (3E4), IgM (II/41), CD8 (53–6.7), Gr-1/Ly-6G (RB6-8C5), and SA-e450; from Southern Biotech: IgD (11–26), BP-1 (FG35.4), and λ (JC5-1); from Biolegend: B220 (RA3-6B2), CD19 (6D5), CD24 (M1/69), CD21 (7E9 and 7G6), CD23 (B3B4), BP-1 (6C3), and CD4 (GK1.5); from BD Biosciences: CD43 (S7), CD21 (7G6) CD19 (1D3), CD138 (281–2), CD95/Fas (Jo2), IgG1 (A85-1), κ (187.1), CXCR4 (2B11), CD86 (GL1), CD11b (M1/70), CD5 (53–7.3), Blimp1 (5E7), and SA-PE; from Life Technologies: CD3 (500A2), SA-AF700, and SA-APC; from Vector Laboratories: PNA; and Cell Signaling Technology: FOXO1 (C29H4), p4E-BP1 (Thr37/46, 236B4), and p-S6 (Ser235/236, D57.2.2E). IKK-2 inhibitor MLN120B was purchased from Sigma Aldrich.
Spleen immunofluorescence staining
Mouse spleens were embedded in optimal cutting temperature compound and frozen over dry ice. 10-µm sections were cut on a cryostat, mounted on Superfrost Excell slides (Thermo Fisher Scientific), and fixed in −20°C acetone for 20 min. After rehydration in staining buffer (PBS, 1% goat serum, 1% BSA, 0.1% Tween-20), slides were stained with B220-PE (RA3-6B2; BD Biosciences), CD3-APC (500A2; Life Technologies) and PNA-FITC (Vector Laboratories). Images were acquired and analyzed by using a Leica DM6000B microscope at 10× magnification, a Leica DFL300 FX camera, and Leica Application Suite Advanced Fluorescence software. Analysis was performed by using ImageJ software.
Flow cytometric analysis
Single-cell suspensions for spleen, BM, and peritoneal fluid (PF) were obtained as previously described (Becker-Herman et al., 2011) and incubated with fluorescence-labeled Abs for 15 min at 4°C. Transcription factor labeling was performed by using the True-Nuclear Transcription Factor Buffer Set (Biolegend). Surface Abs were stained for 20 min at 4°C followed by incubation with a transcription factor fixation/permeabilization solution (TrueNuclear) for 35 min at room temperature (RT). Transcription factor Abs were then stained for 35 min at RT in a permeabilization buffer. For intracellular staining, cells were first fixed for 30 min at 4°C with fixation/permeabilization solution (BD Biosciences), followed by staining with intracellular Abs for 30 min at 4°C in fixation/permeabilization buffer (BD Biosciences).
All flow cytometric, EdU, BrdU, and phosphoflow data were collected on an LSRII (BD) and analyzed by using FlowJo software (TreeStar).
Cell cycle analysis using in vivo EdU labeling
EdU (Thermo Fisher Scientific) was dissolved in sterile 1× PBS or DMSO, and 1 mg in a volume of 100 μl was injected i.p. 1 h before sacrifice. EdU detection was performed with the Click-iT Plus EdU Pacific Blue and Alexa Fluor 647 Flow Cytometry Assay Kits (Thermo Fisher Scientific). Cells were stained for surface markers for 15 min at 4°C, followed by fixation and incubation with the Click-iT Plus reaction cocktail according to the manufacturer’s instructions.
Cell cycle analysis using in vivo BrdU labeling
For in vivo labeling of cycling cells, mice were i.p. injected with 1 mg in a volume of 100 μl BrdU 24 h before sacrifice. Both spleens and BM were collected, and cells were surface stained, fixed and permeabilized, and treated with DNase for 1 h at 37°C and stained with anti-BrdU FITC (BD Biosciences) immediately before FACS analysis.
Phospho-flow analysis
1e6 cells were fixed with Cytofix/Cytoperm solution (BD Biosciences) for 20 min at RT in the dark. Cells were resuspended in 90% methanol and stored at −20°C for a minimum of 30 min. Cells were stained with surface and phospho-site antibodies for 45 min at RT in the dark. For phosphorylation site staining in the DZ and LZ: cells were fixed as described above, then permeabilized in 90% methanol for 30mins at -20°C, followed by incubation with antibodies for surface markers for 45 min at RT in the dark, and a subsequent 45 min incubation with streptavidin and phospho-site antibodies at RT in the dark.
ELISA
96-well Nunc-Immuno MaxiSorp plates (Thermo Fisher Scientific) were precoated overnight at 4°C with 2 μg/ml anti-IgM, anti-IgG, anti-IgG1, and anti-IgG2C (Southern Biotech) or 50 µg/ml NP(5)-BSA and NP(30)-BSA (Biosearch Technologies). Plates were blocked for 1 h with 2% BSA in PBS before addition of diluted serum (1:6,250, 1:31,250, 1:156,250) for 2 h at RT or overnight at 4°C. Specific antibodies were detected by using goat anti–mouse IgM, IgG, IgG1, IgG3, or rat anti-IgG2C–horseradish peroxidase (1:2,000 dilution; Southern Biotech), and peroxidase reactions were developed by using OptEIA TMB substrate (BD Biosciences) and stopped with sulfuric acid. Absorbance at 450 nm was read by using a SpectraMax 190 microplate reader (Molecular Devices). 450-nm absorbance was corrected by subtraction of 570-nm absorbance.
Immunoblot analysis
Primary murine naive B cells were purified by CD43 microbead depletion (Miltenyi Biotec). For analysis of immunized animals, total B lineage cells were isolated with a Pan B Cell Isolation Kit II (StemCell). Cell pellets were lysed in 1× RIPA lysis buffer (1% NP-40, 0.25% sodium deoxycholate, 0.1% SDS, 150 mM NaCl, 20 mM Hepes, pH 7.5, 1 mM β-glycerophosphate, 1 mM NaF, 10 mM Na-pyrophosphate, 1 mM Na-orthovanadate, and a Pierce protease inhibitor tablet) on ice for 10 min. Lysates were cleared by centrifugation, and supernatant was collected. The protein concentration of cleared supernatants was determined by bicinchoninic acid analysis. 15 μg of sample was run on 4–12% Bis-Tris NuPAGE gels (Invitrogen) in 1× MOPs buffer (Invitrogen). Protein was transferred onto nitrocellulose in 1× Transfer buffer (Invitrogen) with 10% methanol. Nonspecific binding was prevented by incubation with Odyssey LI-COR Blocking Buffer. All primary antibodies were incubated for a minimum of 12 h at 4°C except actin, which was incubated for 1 h at RT. Primary anti–mouse antibodies used for immunoblot analysis included those from Cell Signaling Technology: CARD11 (1D12), HSP90, p-p65 (Ser536, 93H1), p65 (C-20), FOXO1 (D7C1H), p-S6 (Ser235/236), S6 (54D2), and IκBα (5A5); from Santa Cruz Biotechnology: p65; and from Sigma Aldrich: actin (Rb polyclonal). Secondary antibodies were incubated with membranes at 1:10,000 in 1× TBST for 30 min at RT in the dark. Membranes were imaged with Odyssey Infrared Imaging System (LI-COR Biotechnology). Immunoblot quantification was performed by using ImageJ software.
Guassia luciferase and Cell Titer Glo assay
0.106 transduced OCI-Ly7 cells in 100 μl media (IMDM, 10% FBS and BME) were cultured under the following conditions: DMSO 1:1,000, MLN120B (60 nM, 600 nM, and 1 μM), or 50 nM Rapamycin. After 16 h, 20 µl of culture supernatant was transferred into a 96-well opaque plate (Corning) with 10 µl Gluc reagent cocktail (1:100 of BioLux Gluc Substrate diluted into BioLux GLuc assay buffer; New England BioLabs) added and incubated for 45 s at RT before luminescence was recorded. The remaining 80 µl culture volume was transferred to an opaque 96-well flat-bottom plate with 10 µl CellTiter-Glo reagent (Promega). Cells were incubated with CellTiter-Glo for 15 min at RT in the dark before luminescence was recorded.
scRNA-seq
Single-cell suspensions were prepared from spleens harvested from mice 5 d after immunization with SRBCs. B lineage cells were enriched by using the EasySep Mouse B Cell Isolation Kit (StemCell). Single-cell cDNA libraries were generated by using the Chromium product suite by 10× Genomics (Zheng et al., 2017) and sequenced by using an Illumina NextSeq 500. Cell Ranger (by 10× Genomics) was used for sample demultiplexing, barcode processing, and single-cell 3′ gene counting, revealing a total cell count of 1,954. Monocle 2 was used for downstream analysis as described previously (Qiu et al., 2017) and in the Monocle vignette. Only cells with a total unique molecular identifier count of between 600 and 12,500 were included in the analysis (n = 1,945). Cells were clustered by using t-SNE, and the major cell types were identified by using canonical marker genes. Non–B cell clusters were completely removed, and in addition, only CD79a-positive cells were retained for downstream analysis (n = 1,581). Clustering of B cells only was again performed by using t-SNE. Differential gene expression analysis between WT and aCard111 cells was performed by using the differential-GeneTest function included within Monocle.
Accession no.
Single-cell RNA sequencing data were deposited to the Gene Expression Omnibus under accession no. GSE117895.
Statistical analysis
Statistical differences were determined using Prism (GraphPad Software). Significant P values are indicated in figures for the following ranges: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. In all summary figures, a single data point represents an individual mouse, and bars indicate the mean ± SEM. In bar graphs, bars indicate the mean + SEM.
Online supplemental material
Fig. S1 shows reporter expression tracking expression of aCARD11 in BM B cell subsets and splenic immune cell lineages, expression of aCARD11 has no significant impact on BM B cell subsets or splenic distribution of immune cells, Mb1-aCard11 mice have reduced spontaneous GC B cells under homeostatic conditions and no difference in serum IgG1 and IgG2C titers, and splenic GC structures in Mb1-aCard11 mice at 5 d after immunization with SRBCs are significantly larger (area) compared with GCs in controls. Fig. S2 shows no phenotypic difference in peritoneal B cell subsets in control and Cγ1-aCard11 mice, and no difference in serum IgM and total IgG titers under homeostatic conditions as well. Fig. S3 shows no difference in cycling of BM B cell subsets or non-GC (B220+CD95loCD38hi) splenic B cells with enhanced cycling in GC (B220+CD95hiCD38lo) and splenic PCs (PCs, B220−CD138+) in Mb1-aCard11 mice compared with controls 5 d after immunization with SRBCs, cycling of GC B cells is increased in Cγ1-aCard11 mice at 5 d after immunization with SRBCs, with exaggerated DZ skewing compared with controls, and GC B cells in Mb1-aCard11 mice have decreased total FOXO1 expression compared with control GC B cells 5 d after immunization with SRBCs. Fig. S4 shows a trend toward increased biomass of total GC B cells in Cγ1-aCard11 mice, with significant differences in cell size when separated into DZ and LZ compared with controls, PCs also demonstrated increased biomass compared with controls, immunoblotting of total B lineage in the spleen of Mb1-aCard11 vs. controls shows trends toward increased pS6 and decreased total FOXO1 and IκBα, and increased surface expression of CD40 on aCARD11-expressing GC B cells with decreased MHC II expression compared with controls 5 d after immunization with SRBCs. Fig. S5 shows separation of non-B cells from cells of B lineage in t-SNE plots with further subsetting of GC B cells by highest expression of AID (Aicda) and PBs by highest expression of XBP1 (Xbp1), expression of secreted isoform of IgG1 transcript in GC B cells is increased in Mb1-aCard11 mice while secreted IgM is decreased, membrane-bound IgG1 transcript is increased in PBs of Mb1-aCard11 mice while membrane-bound IgM is decreased, and intracellular staining of the BCR shows an increased proportion of IgM−IgG1− PCs in Mb1-aCard11 mice compared with controls 5 d after immunization with SRBCs.
Supplementary Material
Acknowledgments
We thank Martha Metzler, Eric Allenspach, and Shaun Jackson for helpful comments and critical review of the manuscript. We thank Iana Meitlis (Seattle Children's Research Institute, Seattle, WA) for reagents.
This work was supported by the National Institutes of Health under award numbers T32AI106677 (to M.N. Wray-Dutra), DP3-DK097672 and DP3-DK111802 (to D.J. Rawlings), R01CA201135 (to R.G. James), and DP2HD088158 and U54 DK107979 (to C. Trapnell). Additional support was provided by a Cancer Research Institute Training Grant (to M.N. Wray-Dutra); an Early Postdoctoral Mobility Fellowship from the Swiss National Science Foundation and the Forschungsfonds der Universität Basel (to R. Chawla); the W. M. Keck Foundation, the Dale F. Frey Award for Breakthrough Scientists, an Alfred P. Sloan Foundation Research Fellowship (to C. Trapnell); the Children's Guild Association Endowed Chair in Pediatric Immunology and the Benaroya Family Gift Fund (D.J. Rawlings). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The authors declare no competing financial interests.
Author contributions: M.W. Wray-Dutra, R. Chawla, K.R. Thomas, B.J. Seymore, and T. Arkatkar designed, performed, and interpreted the experiments. K.M. Sommer and D.J. Rawlings designed the murine model. R.G. James, C. Trapnell, and D.J. Rawlings designed the overall study and interpreted data. M.N. Wray-Dutra generated figures. M.N. Wray-Dutra, R. Chawla, R.G. James, and D.J. Rawlings wrote the manuscript.
References
- Arjunaraja S., Nosé B.D., Sukumar G., Lott N.M., Dalgard C.L., and Snow A.L.. 2017. Intrinsic plasma cell differentiation defects in B cell expansion with NF-κB and T cell anergy patient b cells. Front. Immunol. 8:913 10.3389/fimmu.2017.00913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett N.L., Costello B.A., LaPlant B.R., Ansell S.M., Kuruvilla J.G., Reeder C.B., Thye L.S., Anderson D.M., Krysiak K., Ramirez C., et al. . 2018. Single-agent ibrutinib in relapsed or refractory follicular lymphoma: A phase 2 consortium trial. Blood. 131:182–190. 10.1182/blood-2017-09-804641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgarth N. 2016. B-1 cell heterogeneity and the regulation of natural and antigen-induced IgM production. Front. Immunol. 7:324 10.3389/fimmu.2016.00324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker-Herman S., Meyer-Bahlburg A., Schwartz M.A., Jackson S.W., Hudkins K.L., Liu C., Sather B.D., Khim S., Liggitt D., Song W., et al. . 2011. WASp-deficient B cells play a critical, cell-intrinsic role in triggering autoimmunity. J. Exp. Med. 208:2033–2042. 10.1084/jem.20110200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamron S., Pattanayak S.P., Berger M., and Tirosh B.. 2015. mTOR activation promotes plasma cell differentiation and bypasses XBP-1 for immunoglobulin secretion. Mol. Cell. Biol. 35:153–166. 10.1128/MCB.01187-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohl A.S., Stinson J.R., Su H.C., Badgett T., Jennings C.D., Sukumar G., Sindiri S., Wang W., Kardava L., Moir S., et al. . 2015. Germline CARD11 mutation in a patient with severe congenital B cell lymphocytosis. J. Clin. Immunol. 35:32–46. 10.1007/s10875-014-0106-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calado D.P., Zhang B., Srinivasan L., Sasaki Y., Seagal J., Unitt C., Rodig S., Kutok J., Tarakhovsky A., Schmidt-Supprian M., and Rajewsky K.. 2010. Constitutive canonical NF-κB activation cooperates with disruption of BLIMP1 in the pathogenesis of activated B cell-like diffuse large cell lymphoma. Cancer Cell. 18:580–589. 10.1016/j.ccr.2010.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor J.M., and Ginsberg M.H.. 2012. CD98 at the crossroads of adaptive immunity and cancer. J. Cell Sci. 125:1373–1382. 10.1242/jcs.096040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor J., Browne C.D., Ruppert R., Féral C.C., Fässler R., Rickert R.C., and Ginsberg M.H.. 2009. CD98hc facilitates B cell proliferation and adaptive humoral immunity. Nat. Immunol. 10:412–419. 10.1038/ni.1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casola S., Cattoretti G., Uyttersprot N., Koralov S.B., Seagal J., Hao Z., Waisman A., Egert A., Ghitza D., and Rajewsky K.. 2006. Tracking germinal center B cells expressing germ-line immunoglobulin gamma1 transcripts by conditional gene targeting. Proc. Natl. Acad. Sci. USA. 103:7396–7401. 10.1073/pnas.0602353103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S.H., Raybuck A.L., Stengel K., Wei M., Beck T.C., Volanakis E., Thomas J.W., Hiebert S., Haase V.H., and Boothby M.R.. 2016. Germinal centre hypoxia and regulation of antibody qualities by a hypoxia response system. Nature. 537:234–238. 10.1038/nature19334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Sola D., Kung J., Holmes A.B., Wells V.A., Mo T., Basso K., and Dalla-Favera R.. 2015. The FOXO1 transcription factor instructs the germinal center dark zone program. Immunity. 43:1064–1074. 10.1016/j.immuni.2015.10.015 [DOI] [PubMed] [Google Scholar]
- Egawa T., Albrecht B., Favier B., Sunshine M.J., Mirchandani K., O’Brien W., Thome M., and Littman D.R.. 2003. Requirement for CARMA1 in antigen receptor-induced NF-kappa B activation and lymphocyte proliferation. Curr. Biol. 13:1252–1258. 10.1016/S0960-9822(03)00491-3 [DOI] [PubMed] [Google Scholar]
- Ersching J., Efeyan A., Mesin L., Jacobsen J.T., Pasqual G., Grabiner B.C., Dominguez-Sola D., Sabatini D.M., and Victora G.D.. 2017. Germinal center selection and affinity maturation require dynamic regulation of mTORC1 kinase. Immunity. 46:1045–1058.e6. 10.1016/j.immuni.2017.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenczik C.A., Zent R., Dellos M., Calderwood D.A., Satriano J., Kelly C., and Ginsberg M.H.. 2001. Distinct domains of CD98hc regulate integrins and amino acid transport. J. Biol. Chem. 276:8746–8752. 10.1074/jbc.M011239200 [DOI] [PubMed] [Google Scholar]
- Gitlin A.D., von Boehmer L., Gazumyan A., Shulman Z., Oliveira T.Y., and Nussenzweig M.C.. 2016. Independent roles of switching and hypermutation in the development and persistence of B lymphocyte memory. Immunity. 44:769–781. 10.1016/j.immuni.2016.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton K.S., Phong B., Corey C., Cheng J., Gorentla B., Zhong X., Shiva S., and Kane L.P.. 2014. T cell receptor-dependent activation of mTOR signaling in T cells is mediated by Carma1 and MALT1, but not Bcl10. Sci. Signal. 7:ra55 10.1126/scisignal.2005169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay N. 2011. Interplay between FOXO, TOR, and Akt. Biochim. Biophys. Acta. 1813:1965–1970. 10.1016/j.bbamcr.2011.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hideshima T., Neri P., Tassone P., Yasui H., Ishitsuka K., Raje N., Chauhan D., Podar K., Mitsiades C., Dang L., et al. . 2006. MLN120B, a novel IkappaB kinase beta inhibitor, blocks multiple myeloma cell growth in vitro and in vivo. Clin. Cancer Res. 12:5887–5894. 10.1158/1078-0432.CCR-05-2501 [DOI] [PubMed] [Google Scholar]
- Hobeika E., Thiemann S., Storch B., Jumaa H., Nielsen P.J., Pelanda R., and Reth M.. 2006. Testing gene function early in the B cell lineage in mb1-cre mice. Proc. Natl. Acad. Sci. USA. 103:13789–13794. 10.1073/pnas.0605944103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata T.N., Ramírez-Komo J.A., Park H., and Iritani B.M.. 2017. Control of B lymphocyte development and functions by the mTOR signaling pathways. Cytokine Growth Factor Rev. 35:47–62. 10.1016/j.cytogfr.2017.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeelall Y.S., Wang J.Q., Law H.D., Domaschenz H., Fung H.K., Kallies A., Nutt S.L., Goodnow C.C., and Horikawa K.. 2012. Human lymphoma mutations reveal CARD11 as the switch between self-antigen-induced B cell death or proliferation and autoantibody production. J. Exp. Med. 209:1907–1917. 10.1084/jem.20112744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jellusova J., Cato M.H., Apgar J.R., Ramezani-Rad P., Leung C.R., Chen C., Richardson A.D., Conner E.M., Benschop R.J., Woodgett J.R., and Rickert R.C.. 2017. Gsk3 is a metabolic checkpoint regulator in B cells. Nat. Immunol. 18:303–312. 10.1038/ni.3664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.D., Gaudette B.T., Wilmore J.R., Chernova I., Bortnick A., Weiss B.M., and Allman D.. 2016. mTOR has distinct functions in generating versus sustaining humoral immunity. J. Clin. Invest. 126:4250–4261. 10.1172/JCI86504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knies N., Alankus B., Weilemann A., Tzankov A., Brunner K., Ruff T., Kremer M., Keller U.B., Lenz G., and Ruland J.. 2015. Lymphomagenic CARD11/BCL10/MALT1 signaling drives malignant B-cell proliferation via cooperative NF-κB and JNK activation. Proc. Natl. Acad. Sci. USA. 112:E7230–E7238. 10.1073/pnas.1507459112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani A., Kakazu N., Tsuruyama T., Okazaki I.M., Muramatsu M., Kinoshita K., Nagaoka H., Yabe D., and Honjo T.. 2007. Activation-induced cytidine deaminase (AID) promotes B cell lymphomagenesis in Emu-cmyc transgenic mice. Proc. Natl. Acad. Sci. USA. 104:1616–1620. 10.1073/pnas.0610732104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz G., Davis R.E., Ngo V.N., Lam L., George T.C., Wright G.W., Dave S.S., Zhao H., Xu W., Rosenwald A., et al. . 2008. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 319:1676–1679. 10.1126/science.1153629 [DOI] [PubMed] [Google Scholar]
- Limon J.J., So L., Jellbauer S., Chiu H., Corado J., Sykes S.M., Raffatellu M., and Fruman D.A.. 2014. mTOR kinase inhibitors promote antibody class switching via mTORC2 inhibition. Proc. Natl. Acad. Sci. USA. 111:E5076–E5085. 10.1073/pnas.1407104111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C.A., Stinson J.R., Zhang Y., Abbott J.K., Weinreich M.A., Hauk P.J., Reynolds P.R., Lyons J.J., Nelson C.G., Ruffo E., et al. . 2017. Germline hypomorphic CARD11 mutations in severe atopic disease. Nat. Genet. 49:1192–1201. 10.1038/ng.3898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelbaum J., Bhagat G., Tang H., Mo T., Brahmachary M., Shen Q., Chadburn A., Rajewsky K., Tarakhovsky A., Pasqualucci L., and Dalla-Favera R.. 2010. BLIMP1 is a tumor suppressor gene frequently disrupted in activated B cell-like diffuse large B cell lymphoma. Cancer Cell. 18:568–579. 10.1016/j.ccr.2010.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masle-Farquhar E., Bröer A., Yabas M., Enders A., and Bröer S.. 2017. ASCT2 (SLC1A5)-deficient mice have normal B-cell development, proliferation, and antibody production. Front. Immunol. 8:549 10.3389/fimmu.2017.00549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesin L., Ersching J., and Victora G.D.. 2016. Germinal center B cell dynamics. Immunity. 45:471–482. 10.1016/j.immuni.2016.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-García M.E., Sommer K., Shinohara H., Bandaranayake A.D., Kurosaki T., and Rawlings D.J.. 2010. MAGUK-controlled ubiquitination of CARMA1 modulates lymphocyte NF-kappaB activity. Mol. Cell. Biol. 30:922–934. 10.1128/MCB.01129-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya M., Xiao Y., Zhou X., Chang J.H., Chang M., Cheng X., Blonska M., Lin X., and Sun S.C.. 2014. Inflammatory T cell responses rely on amino acid transporter ASCT2 facilitation of glutamine uptake and mTORC1 kinase activation. Immunity. 40:692–705. 10.1016/j.immuni.2014.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo V.N., Davis R.E., Lamy L., Yu X., Zhao H., Lenz G., Lam L.T., Dave S., Yang L., Powell J., and Staudt L.M.. 2006. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 441:106–110. 10.1038/nature04687 [DOI] [PubMed] [Google Scholar]
- Nicklin P., Bergman P., Zhang B., Triantafellow E., Wang H., Nyfeler B., Yang H., Hild M., Kung C., Wilson C., et al. . 2009. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 136:521–534. 10.1016/j.cell.2008.11.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onaindia A., Medeiros L.J., and Patel K.P.. 2017. Clinical utility of recently identified diagnostic, prognostic, and predictive molecular biomarkers in mature B-cell neoplasms. Mod. Pathol. 30:1338–1366. 10.1038/modpathol.2017.58 [DOI] [PubMed] [Google Scholar]
- Pasqualucci L., and Zhang B.. 2016. Genetic drivers of NF-κB deregulation in diffuse large B-cell lymphoma. Semin. Cancer Biol. 39:26–31. 10.1016/j.semcancer.2016.08.001 [DOI] [PubMed] [Google Scholar]
- Pasqualucci L., Bhagat G., Jankovic M., Compagno M., Smith P., Muramatsu M., Honjo T., Morse H.C. III, Nussenzweig M.C., and Dalla-Favera R.. 2008. AID is required for germinal center-derived lymphomagenesis. Nat. Genet. 40:108–112. 10.1038/ng.2007.35 [DOI] [PubMed] [Google Scholar]
- Qiu X., Mao Q., Tang Y., Wang L., Chawla R., Pliner H.A., and Trapnell C.. 2017. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods. 14:979–982. 10.1038/nmeth.4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui L., Schmitz R., Ceribelli M., and Staudt L.M.. 2011. Malignant pirates of the immune system. Nat. Immunol. 12:933–940. 10.1038/ni.2094 [DOI] [PubMed] [Google Scholar]
- Sander S., Chu V.T., Yasuda T., Franklin A., Graf R., Calado D.P., Li S., Imami K., Selbach M., Di Virgilio M., et al. . 2015. PI3 Kinase and FOXO1 transcription factor activity differentially control B cells in the germinal center light and dark zones. Immunity. 43:1075–1086. 10.1016/j.immuni.2015.10.021 [DOI] [PubMed] [Google Scholar]
- Shaffer A.L. III, Young R.M., and Staudt L.M.. 2012. Pathogenesis of human B cell lymphomas. Annu. Rev. Immunol. 30:565–610. 10.1146/annurev-immunol-020711-075027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J.H., and Sun S.C.. 2015. TCR signaling to NF-κB and mTORC1: Expanding roles of the CARMA1 complex. Mol. Immunol. 68(2, 2 Pt C):546–557. 10.1016/j.molimm.2015.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow A.L., Xiao W., Stinson J.R., Lu W., Chaigne-Delalande B., Zheng L., Pittaluga S., Matthews H.F., Schmitz R., Jhavar S., et al. . 2012. Congenital B cell lymphocytosis explained by novel germline CARD11 mutations. J. Exp. Med. 209:2247–2261. 10.1084/jem.20120831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer K., Guo B., Pomerantz J.L., Bandaranayake A.D., Moreno-García M.E., Ovechkina Y.L., and Rawlings D.J.. 2005. Phosphorylation of the CARMA1 linker controls NF-kappaB activation. Immunity. 23:561–574. 10.1016/j.immuni.2005.09.014 [DOI] [PubMed] [Google Scholar]
- Staudt L.M. 2010. Oncogenic activation of NF-kappaB. Cold Spring Harb. Perspect. Biol. 2:a000109 10.1101/cshperspect.a000109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamahara T., Ochiai K., Muto A., Kato Y., Sax N., Matsumoto M., Koseki T., and Igarashi K.. 2017. The mTOR-Bach2 cascade controls cell cycle and class switch recombination during B cell differentiation. Mol. Cell. Biol. 37:e00418-17 10.1128/MCB.00418-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellier J., Shi W., Minnich M., Liao Y., Crawford S., Smyth G.K., Kallies A., Busslinger M., and Nutt S.L.. 2016. Blimp-1 controls plasma cell function through the regulation of immunoglobulin secretion and the unfolded protein response. Nat. Immunol. 17:323–330. 10.1038/ni.3348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallabhapurapu S., and Karin M.. 2009. Regulation and function of NF-kappaB transcription factors in the immune system. Annu. Rev. Immunol. 27:693–733. 10.1146/annurev.immunol.021908.132641 [DOI] [PubMed] [Google Scholar]
- van den Brand M., Rijntjes J., Hebeda K.M., Menting L., Bregitha C.V., Stevens W.B., van der Velden W.J., Tops B.B., van Krieken J.H., and Groenen P.J.. 2017. Recurrent mutations in genes involved in nuclear factor-κB signalling in nodal marginal zone lymphoma-diagnostic and therapeutic implications. Histopathology. 70:174–184. 10.1111/his.13015 [DOI] [PubMed] [Google Scholar]
- Victora G.D., Dominguez-Sola D., Holmes A.B., Deroubaix S., Dalla-Favera R., and Nussenzweig M.C.. 2012. Identification of human germinal center light and dark zone cells and their relationship to human B-cell lymphomas. Blood. 120:2240–2248. 10.1182/blood-2012-03-415380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., de Miranda N.F., Chen L., Wasik A.M., Mansouri L., Jurczak W., Galazka K., Dlugosz-Danecka M., Machaczka M., Zhang H., et al. . 2016. Genetic heterogeneity in primary and relapsed mantle cell lymphomas: Impact of recurrent CARD11 mutations. Oncotarget. 7:38180–38190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Tsakmaklis N., Yang G., Chen J.G., Liu X., Demos M., Kofides A., Patterson C.J., Meid K., Gustine J., et al. . 2017. Acquired mutations associated with ibrutinib resistance in Waldenström macroglobulinemia. Blood. 129:2519–2525. 10.1182/blood-2017-01-761726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L., Morris S.W., Orihuela C., Tuomanen E., Cui X., Wen R., and Wang D.. 2003. Defective development and function of Bcl10-deficient follicular, marginal zone and B1 B cells. Nat. Immunol. 4:857–865. 10.1038/ni963 [DOI] [PubMed] [Google Scholar]
- Zhang S., Pruitt M., Tran D., Du Bois W., Zhang K., Patel R., Hoover S., Simpson R.M., Simmons J., Gary J., et al. . 2013. B cell-specific deficiencies in mTOR limit humoral immune responses. J. Immunol. 191:1692–1703. 10.4049/jimmunol.1201767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G.X., Terry J.M., Belgrader P., Ryvkin P., Bent Z.W., Wilson R., Ziraldo S.B., Wheeler T.D., McDermott G.P., Zhu J., et al. . 2017. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8:14049 10.1038/ncomms14049 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.