de Jong et al. show that loss-of-function mutations in CIB1 are responsible for epidermodysplasia verruciformis, a cutaneous disease caused by human β-papillomavirus (β-HPV) infections, and that CIB1 forms a complex with EVER proteins, acting as a restriction factor against HPV.
Abstract
Patients with epidermodysplasia verruciformis (EV) and biallelic null mutations of TMC6 (encoding EVER1) or TMC8 (EVER2) are selectively prone to disseminated skin lesions due to keratinocyte-tropic human β-papillomaviruses (β-HPVs), which lack E5 and E8. We describe EV patients homozygous for null mutations of the CIB1 gene encoding calcium- and integrin-binding protein-1 (CIB1). CIB1 is strongly expressed in the skin and cultured keratinocytes of controls but not in those of patients. CIB1 forms a complex with EVER1 and EVER2, and CIB1 proteins are not expressed in EVER1- or EVER2-deficient cells. The known functions of EVER1 and EVER2 in human keratinocytes are not dependent on CIB1, and CIB1 deficiency does not impair keratinocyte adhesion or migration. In keratinocytes, the CIB1 protein interacts with the HPV E5 and E8 proteins encoded by α-HPV16 and γ-HPV4, respectively, suggesting that this protein acts as a restriction factor against HPVs. Collectively, these findings suggest that the disruption of CIB1–EVER1–EVER2-dependent keratinocyte-intrinsic immunity underlies the selective susceptibility to β-HPVs of EV patients.
Introduction
Epidermodysplasia verruciformis (EV; OMIM ID 226400) is a rare Mendelian genodermatosis. EV patients are highly and selectively susceptible to skin diseases due to cutaneous human papillomaviruses (HPVs) of the β genus (Orth, 2006, 2008; de Jong et al., 2018). They are otherwise healthy and normally resistant to other microorganisms including other viruses and skin-tropic pathogens and even all other cutaneous and mucosal HPVs. Early in childhood, these patients present with persistent, disseminated, flat warts and pityriasis versicolor–like lesions of the skin that are induced by β-HPVs. Some patients develop nonmelanoma skin cancer, particularly on areas of the body exposed to the sun. By contrast, β-HPV infection is widespread and asymptomatic in the general population. EV is transmitted as an autosomal recessive (AR) trait in most families but was shown to be X-linked recessive in one family (Androphy et al., 1985). Biallelic null mutations of either TMC6 or TMC8 encoding EVER1 and EVER2, respectively, account for about half the patients and families displaying EV (Ramoz et al., 2002; Burger and Itin, 2014; Imahorn et al., 2017; de Jong et al., 2018). These genes are widely expressed throughout the body, including in leukocytes, but patients with null mutations display no consistent abnormalities of the development or function of any subset of leukocytes (Lazarczyk et al., 2012; Crequer et al., 2013). EVER1 or EVER2 deficiency in keratinocytes, which would normally express both proteins and are the natural and exclusive host cells of β-HPVs, has thus been proposed as the cellular basis of the disease (Orth, 2006, 2008). The exceedingly narrow infectious phenotype and the lack of detectable leukocyte abnormalities prevented EV from being recognized as a primary immunodeficiency until the discovery of genetic etiologies in 2002 (Ramoz et al., 2002; Notarangelo et al., 2004; Casanova, 2015a,b). However, EV was shown to be an inborn error underlying viral lesions between 1922 and 1946 by the works of Wilhelm Lutz and Edward Cockayne (Lewandowsky and Lutz, 1922; Cockayne, 1933; Lutz, 1946), before the first descriptions of congenital neutropenia by Ralph Kostmann and inherited agammaglobulinemia by Ogden Bruton (Kostmann, 1950; Bruton, 1952).
Patients with an “atypical” form of inherited EV have recently been described (de Jong et al., 2018). These patients suffer from primary immunodeficiencies due to profound T cell defects caused by inactivating biallelic mutations of STK4 (Crequer et al., 2012a), RHOH (Crequer et al., 2012b), CORO1A (Stray-Pedersen et al., 2014), TPP2 (Stepensky et al., 2015), DCLRE1C (Tahiat et al., 2016), LCK (Li et al., 2016), RASGRP1 (Platt et al., 2017), or DOCK8 (Sanal et al., 2012; Liu et al., 2017). Other patients with atypical EV have T cell deficits of unknown genetic etiology (Azzimonti et al., 2005; Borgogna et al., 2014; Landini et al., 2014). In all these patients, persistent infection with β-HPVs causes skin lesions identical to those of patients with classic EV, but in a context of broader infectious manifestations, the breadth and severity of which depend on the mutated gene and the nature of the T cell deficit. Indeed, patients with inherited T cell deficiencies typically suffer from various viral, bacterial, fungal, and parasitic infections, including many infections of the skin and viral infections in particular (Notarangelo et al., 2004; Fischer, 2015). These patients are also prone to various autoimmune and, more rarely, tumoral manifestations. An additional role of these gene products in keratinocytes has not been formally excluded, but the T cell deficit common to all these patients strongly suggests that full T cell development and function are required for protective immunity to β-HPVs. Intriguingly, not all T cell deficits seem to confer a predisposition to β-HPV–driven lesions, and not all patients with such deficits display lesions of this type. Finally, β-HPV–induced skin lesions resembling typical EV have also been reported in a third group of patients who are otherwise healthy years after successful allogeneic hematopoietic stem cell transplantation (HSCT) for severe combined immunodeficiency due to mutations of IL2RG and JAK3 (Laffort et al., 2004). As both these genes are also normally expressed in keratinocytes, this third type of EV might be caused by a persistent deficiency of their products in keratinocytes that is not corrected by allogeneic HSCT. The products of these genes may be physiologically connected with EVER1 and EVER2 in keratinocytes.
The molecular basis of “typical” EV remains elusive. One plausible hypothesis is that β-HPVs are defective for a growth-promoting function encoded by the E5 gene, located between the E2-L2 genes of cutaneous α-HPVs, or the E8 gene, located in the E6 region of γ-, κ-, and μ-papillomaviruses, and that EVER1/2 deficiency may compensate for the missing viral function (Bravo and Alonso, 2004; Nonnenmacher et al., 2006; Orth, 2006, 2008). Indeed, it has been shown in vivo that cottontail rabbit papilloma virus (CRPV)-E8 is essential for the development of lesions in rabbits (Hu et al., 2002; Nonnenmacher et al., 2006). EVER1 and EVER2 belong to a larger family of transmembrane channel–like (TMC) proteins (Keresztes et al., 2003; Kurima et al., 2003). TMC1 and TMC2 are components of the sensory transduction ion channel in inner ear hair cells (Kawashima et al., 2011; Pan et al., 2013; Kurima et al., 2015). Both TMC6 and TMC8 are broadly expressed in human tissues, but expression levels are generally lower for TMC8 (www.proteinatlas.org). No knockout mice have been reported for either of these genes. It has been suggested that EVER1 and EVER2 control intracellular zinc homeostasis in human leukocytes and keratinocytes through interaction with the ER-resident zinc transporter ZnT1 (Lazarczyk et al., 2008, 2009, 2012). HPV16-E5α (E5α being encoded by mucosal and E5β by cutaneous HPVs) has been shown to interact with EVER1/2 (Lazarczyk et al., 2008), and both HPV16-E5α and E8 from CRPV interact with ZnT1 (Nonnenmacher et al., 2006; Lazarczyk et al., 2008). It has also been suggested that EVER2 modulates TNF receptor signaling in human keratinocytes (Gaud et al., 2013; Vuillier et al., 2014). The pathogenesis of EV-like lesions in patients with IL2RG and JAK3 mutations years after successful HSCT also remains unknown. Overall, the pathogenesis of β-HPV–driven lesions in EVER1- and EVER2-deficient patients and the mechanisms by which human keratinocytes normally control β-HPVs remain unexplained. We investigated patients with typical EV but no mutations of TMC6 or TMC8. We hypothesized that the discovery of novel genetic etiologies of EV might open up new avenues of investigation into the pathogenesis of EV and the interaction between human keratinocytes and β-HPVs.
Results
Clinical features of EV patients
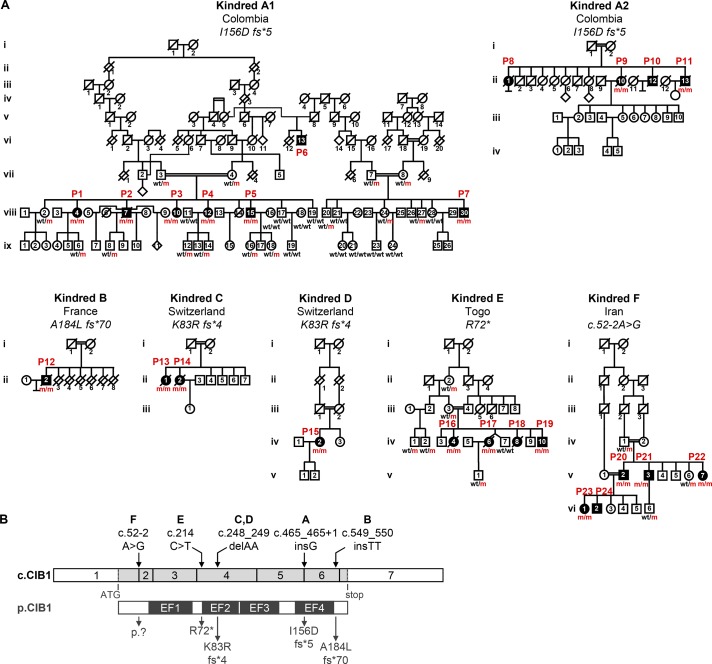
We investigated 24 individuals with unexplained typical EV (P1–P24) from six families originating from and living in five different countries (Colombia, France, Iran, Switzerland, and Togo; Fig. 1 A). None of the patients carried mutations in the exons and flanking intron regions of TMC6 or TMC8. The cohort was clinically homogeneous, with disseminated β-HPV–positive skin lesions at various sites on the body beginning in childhood or early adulthood (Table 1 and Fig. S1). β-HPV5 and/or -HPV8 genotypes have been identified in the lesions of all patients tested (12 of 24). Cutaneous squamous cell carcinoma (SCC) developed in 14 of the 24 patients, mostly on sun-exposed areas of the skin (Table 1). 12 of the 24 patients died: two from disseminated skin cancer (at the ages of 71 yr for P13 and 21 yr for P16) and 10 from causes unrelated to EV (aged 47–91 yr). None of the patients displayed any other unusually severe infectious diseases despite exposure to a large number of different infectious agents in urban or rural areas of countries as diverse as Colombia, France, Iran, Switzerland, and Togo. The clinical features of all patients other than kindred F from Iran have been described in detail elsewhere with the confirmation of β-HPV genotype and/or typical histological signs of β-HPV infection (Lutz, 1946; Rueda and Rodriguez, 1976; Kienzler et al., 1979; Kremsdorf et al., 1984; Deau et al., 1991, 1993; Rueda, 1993; Saka et al., 2009; Arnold et al., 2011; Imahorn et al., 2017). Case studies for all 24 patients are listed in Materials and methods. P13 and P14 were among the first EV patients to be documented by Wilhelm Lutz in 1946 (Lutz, 1946). Overall, these patients were clinically and virologically indistinguishable from patients with biallelic truncating mutations of TMC6 or TMC8, with an AR but unexplained inheritance of lesions due to β-HPVs.
Figure 1.
Identification of homozygous mutations affecting human CIB1 in a cohort of 24 EV patients. (A) Pedigrees of six kindreds affected by EV. Familial segregation of homozygous CIB1 mutations (m/m) in six consanguineous families indicating an AR pattern of inheritance with complete clinical penetrance. (B) Graphical representation of the CIB1 cDNA exon (c.CIB1) and protein (p.CIB1) structure with presentation of the EF-hand domains. The arrows at the top indicate the location of the cDNA positions affected by the CIB1 mutations found in the families, whereas those at the bottom indicate their consequences at protein level. *, stop codon; del, deletion; ins, insertion; fs, frameshift.
Table 1. Comparison of the clinical phenotypes of the 24 EV patients described in this study.
| Patient | Case | Sex | Age (yr) | Age onset (yr)/SCC | HPV type | Skin lesions (location) | SCC | Reference |
|---|---|---|---|---|---|---|---|---|
| P1 | A1.viii.4 | F | 50 | 12/no | Unknown | Head and hands | No | Rueda (1993) |
| P2 | A1.viii.7 | M | 47* | 6/18 | 8, 17, 20 | Head, neck, trunk, hands, legs, and feet | Forehead | Rueda (1993), case 11 |
| P3 | A1.viii.10 | F | 46 | 7/13 | 8, 20 | Head, trunk, legs, arms, and hands | Forehead, lips | Rueda (1993), case 12 |
| P4 | A1.viii.12 | F | 44 | 7/no | 8 | Head and trunk | No | Rueda (1993) |
| P5 | A1.viii.15 | M | 37 | 5/17 | 8, 20 | Head, trunk, hands, and upper legs | Forehead | Rueda (1993), case 13 |
| P6 | A1.vi.13 | M | 47* | 1/15 | Unknown | Head, trunk, hands, and legs | Nose, trunk | Rueda (1993), case 6 |
| P7 | A1.viii.30 | M | 47 | 5/10 | 5, 17, 36 | Head, trunk, arms, hands, and legs | Canthus, nose | Rueda (1993), case 7 |
| P8 | A2.ii.1 | F | 78* | 1/47 | 5, 8, 20, 24 | Head, trunk, and extremities | Forehead | Rueda (1993) and Rueda and Rodriguez (1976), case 1 both papers |
| P9 | A2.ii.10 | F | Unknown | 5/38 | 8, 9 | Head, trunk, and extremities | - | Rueda (1993) and Rueda and Rodriguez (1976), case 2 both papers |
| P10 | A2.ii.12 | M | 48* | 1/26 | Unknown | Head, trunk, and extremities | Forehead, lips | Rueda (1993) and Rueda and Rodriguez (1976), case 3 both papers |
| P11 | A2.ii.13 | M | 47* | 3/27 | 5, 8, 20 | Head, trunk, and extremities | Forehead | Rueda (1993) and Rueda and Rodriguez (1976), case 4 both papers |
| P12 | B.ii.2 | M | 72* | 23/43 | 8, 15 | Head, trunk, arms, hands, and legs | Forehead, check, ear | Kienzler et al. (1979) |
| P13 | C.ii.1 | F | 71* | Childhood/35 | 5 | Head, neck, hands, and legs | Scalp, nose | Lutz (1946) and Arnold et al. (2011), case 1 both papers |
| P14 | C.ii.2 | F | 91* | <7/52 | 5 | Head, neck, and hands | Nose, hand, neck, forehead, scalp | Lutz (1946) and Arnold et al. (2011), case 2 both papers |
| P15 | D.iv.2 | F | 59 | Childhood/54 | 5 | Head, neck, arms, hand, knee, and lower leg | Forehead, nose | Imahorn et al. (2017) |
| P16 | E.iv.4 | F | 21* | 7/unknown | Unknown | Face and trunk | Multiple (sun-exposed skin) | Saka et al. (2009) |
| P17 | E.iv.6 | F | Unknown* | 5/unknown | Unknown | Face and trunk | Unknown | Saka et al. (2009) |
| P18 | E.iv.8 | F | Unknown* | Unknown/unknown | Unknown | Unknown | Unknown | Saka et al. (2009) |
| P19 | E.iv.10 | M | 14 | 2/unknown | Unknown | Face and trunk | Unknown | Saka et al. (2009) |
| P20 | F.v.2 | M | 47 | 3/46 | Unknown | Head, neck, trunk, and upper and lower limb | Forehead (basosquamous carcinoma) | This study |
| P21 | F.v.3 | M | 44 | 2/no | Unknown | Head, neck, trunk, and upper and lower limb | - | This study |
| P22 | F.v.7 | F | 30 | 5/no | Unknown | Head, neck, trunk, and upper and lower limb | - | This study |
| P23 | F.vi.1 | F | 22 | 1/no | Unknown | Head, neck, trunk, and upper and lower limb | - | This study |
| P24 | F.vi.2 | M | 21 | 4/no | Unknown | Head, neck, trunk, and upper and lower limb | - | This study |
Information collected from listed references or oral communication with the treating physicians. Please see Fig. 1 A for individual deidentifiers. F, female; M, male; ages of death are marked with asterisks.
Immunological features of EV patients
Immunophenotyping of the patients tested (P3, P5, P15, P16, and P17) revealed normal numbers and compartmentalization of circulating T cells, B cells, and NK cells (Table S1) as in patients carrying homozygous inactivating mutations of TMC8 (Crequer et al., 2013). Furthermore, T cell function after CD3 stimulation (P3 and P5; Table S2) and B cell function as measured by antibody responses to common DNA and RNA viruses (P1–P5; Table S3) were normal. This finding is also consistent with those for patients with EVER2 deficiency (Crequer et al., 2013) but not for patients with RHOH, MST1, CORO1A, ARTEMIS, RASGRP, DOCK8, and TPP2 deficiencies who suffer from CD4+ T cell lymphopenia and various degrees of impairment of circulating T cell response to CD3 stimulation (Crequer et al., 2012a,b; Sanal et al., 2012; Stray-Pedersen et al., 2014; Stepensky et al., 2015; Tahiat et al., 2016; Liu et al., 2017; Platt et al., 2017). Finally, more detailed analyses of skin-homing T cell populations (CLA+, CCR10+, CLA+CCR4+, and CLA+CCR10+ subsets) revealed no frequency abnormalities in the five patients tested (P1–P4 and P15; Table S4), again contrasting with the smaller sizes of these subsets within the CD4+ compartment in RHOH-deficient patients (Table S4; Crequer et al., 2012b). These skin-homing subsets even displayed mild expansion in three EVER2-deficient patients (Crequer et al., 2013). Thus, none of the patients tested displayed any detectable lymphocyte abnormalities, suggesting that they suffer from a new AR genetic etiology of EV affecting keratinocyte-intrinsic immunity as in EVER1- and EVER2-deficient patients.
Identification of a 2.4-Mb chromosomal region by genome-wide linkage (GWL)
We used GWL analyses to search for genetic etiologies of EV in this cohort. Kindreds A, D, E, and F were known to be consanguineous, and consanguinity was suspected for kindreds B and C. We confirmed that all patients were born to consanguineous parents (Table S5) by showing that their inbreeding coefficients, as estimated with FSuite (Gazal et al., 2014) and genotyping data obtained with the Affymetrix Genome-wide Human Single Nucleotide Polymorphism (SNP) or Illumina Infinum Global Screening array, were high. GWL analysis was performed by homozygosity mapping on 22 members of two branches (termed A1 and A2; Fig. 1 A) of the extended kindred A, P12 from kindred B, P14 from kindred C, P15 from kindred D, six members of kindred E, and four members of kindred F in two generations. A highly significant linkage peak was found on chromosome 15, with a logarithm of odds (LOD) score of 16.7 for a 2.4-Mb interval (Fig. S2, A and B), strongly suggesting genetic homogeneity across these kindreds. We then searched for allelic homogeneity by looking at the haplotypes common to the various families. Kindred A is an extended family with two branches: A1 and A2 (Fig. 1 A). These two branches originate from the same region of Colombia and are homozygous for the same 3.5-Mb haplotype encompassing the linked interval, consistent with a common ancestral mutation. Kindreds C and D were not known to be related, but both are from Switzerland, albeit from different cantons. These two families share the same short homozygous 1.74-Mb haplotype encompassing the linked interval, suggesting a common ancestral mutation for these two Swiss families. The shared haplotype of kindreds C and D is part of the common haplotype of the Colombian patients. These findings reduce the interval of interest on chromosome 15 to 1.74 Mb between positions 89.8 and 91.5 Mb. Overall, these results strongly suggest that the 24 patients from these six families have the same AR genetic etiology of EV, different from EVER1 and EVER2 deficiency, as the corresponding genes TCM6 and TCM8, which are organized in tandem, are absent from this interval.
Identification of bi-allelic mutations of CIB1
We then performed whole-exome sequencing (WES) on the probands from kindreds A–C and E (P7, P12, P14, and P19) and four patients from kindred F (P20–P23; Figs. 1 A and S2 C). This analysis identified CIB1 (encoding calcium- and integrin-binding protein 1; CIB1) as the only gene carrying a very rare homozygous nonsynonymous coding mutation (minor allele frequency [MAF] < 0.001) in all patients tested. CIB1 is located within the 1.74-Mb interval detected by GWL and homozygosity mapping, which contains 33 other protein-coding genes and nine RNA genes. In particular, no other gene within the interval of interest on chromosome 15 was homozygous for rare nonsynonymous coding variants common to all patients. The coding exons of the 42 genes were all fully covered by WES. Sanger sequencing of genomic DNA confirmed that all 24 patients from the six kindreds were homozygous for CIB1 mutations (m/m), whereas the 34 unaffected family members tested were either heterozygous (wt/m) or homozygous for the WT (wt/wt) allele (Figs. 1 A and S2 D). The mutations were frameshift insertions (P1–P11, kindred A: c.465_465+1insG; P12, kindred B: c.549_550insTT) and deletions (P13–15, kindred C and D: c.248_249delAA) predicted to cause premature stop codons (P1–P11: p.I156Dfs*5; P13–15: p.K83Rfs*4) or a stop loss (P12: p.A184Lfs*70), a nonsense allele (P16–P19, kindred E: c.214C>T, p.R72*), and an essential splice site mutation (P20–P24, kindred F: c.52-2A>G; Fig. 1 B) predicted by the Genscan algorithm (Burge and Karlin, 1997) to eliminate the acceptor site of exon 2 and to activate a cryptic acceptor site in intron 1. This would result in the retention of 319 bp of intron 1 and the creation of a 47-aa protein product from exon 1 (aa 1–17) and part of intron 1 followed by a stop codon (aa 18–47). All 24 patients from these six kindreds were homozygous for very rare mutations of CIB1 and had no other homozygous genetic lesion in common. Moreover, the segregation of the mutant alleles suggested an AR trait with complete penetrance as in families with EVER1 or EVER2 deficiency.
The CIB1 mutations are predicted to be deleterious
The frameshift indels and splice-site mutations were not found in the public databases Genome Aggregation Database (gnomAD; Lek et al., 2016), Bravo (https://bravo.sph.umich.edu/freeze5/hg38/), or the Greater Middle East variome project (http://igm.ucsd.edu/gme/; Scott et al., 2016), and they were also absent from our own in-house database of >4,500 exomes. The nonsense R72* variant (dbSNP accession number rs143773090) was found in four heterozygous individuals from the total of 276,936 individuals for whom data were available in the gnomAD, indicating an allele frequency of <1.4 × 10−5, consistent with the very low prevalence of EV. Furthermore, no homozygous CIB1 nonsense or frameshift mutations were present in these public or in-house databases. The gene-damage index (Itan et al., 2015) of CIB1 is very low (1.131). Mutations of CIB1 predicted to be deleterious, with a combined annotation-dependent depletion (CADD) score above the CIB1-specific mutation significance cutoff of 2.3 (Kircher et al., 2014; Itan et al., 2016), are very uncommon in the general population, particularly in the homozygous state (only three missense mutations, each with a MAF <0.0027). The CADD scores of the CIB1 variants from the EV patients (35 for the insertions, 27.7 for the deletion, 24.8 for the essential splice, and 40 for the nonsense variants) were well above the mutation significance cutoff. These genetic findings strongly suggest that these CIB1 mutations, which are very rare or private to the six kindreds, are deleterious. Their strict unique recessive linkage with EV unambiguously demonstrates that homozygosity for deleterious CIB1 alleles is a new AR genetic etiology of EV.
Lack of CIB1 expression in patients’ cells
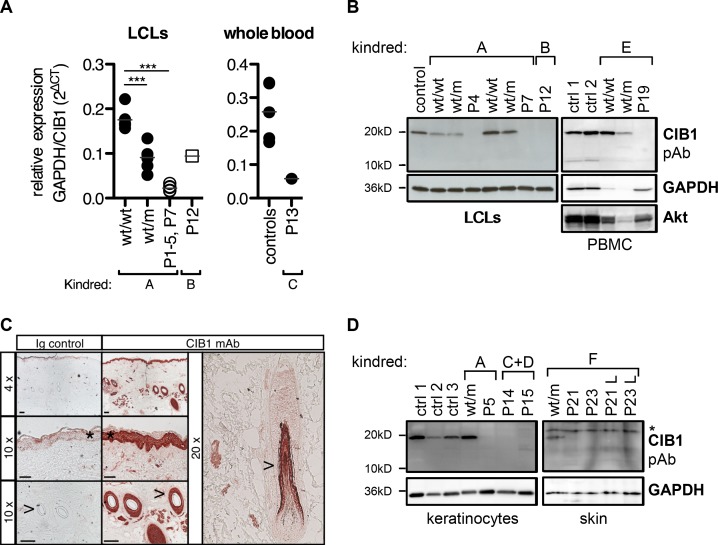
The CIB1 protein is ubiquitous throughout the body and was first identified as an intracellular regulator of integrin signaling in platelets on the basis of its calcium-binding EF-hand domains (Fig. 1 B). Human CIB1 has since emerged as a regulator of diverse cellular processes including migration, adhesion, proliferation, and cell death/survival (Leisner et al., 2016; Wang et al., 2017). We first assessed the biochemical impact of the mutations by measuring endogenous CIB1 mRNA and protein levels in leukocytes. Lymphoblastoid cell lines (LCLs) from P1–P5 and P7 had much lower levels of CIB1 mRNA than those from WT family members (wt/wt), whereas heterozygous (wt/m) family members had intermediate RNA levels (Fig. 2 A). The stop-loss mutation in P12 is predicted to lead to an elongated transcript. The levels of CIB1 mRNA in LCLs from P12 were higher than those for kindred A but much lower than those for WT controls (Fig. 2 A). P13 also had very low CIB1 mRNA levels in whole blood (Fig. 2 A), potentially reflecting nonsense-mediated mRNA decay due to the presence of premature stop codons. An antibody raised against an N-terminal epitope of CIB1 located upstream from all the premature stop codons seen in the patients detected no endogenous CIB1 protein in LCLs or peripheral blood mononuclear cells (PBMCs) from patients from kindreds A, B, and E, whereas CIB1 protein was detected in unrelated controls and healthy family members (whether WT homozygous or heterozygous for the mutant alleles; Fig. 2 B). Thus, all the CIB1 mutations tested greatly decreased mRNA levels and resulted in a complete absence of CIB1 protein from the cells tested. These data indicate that the patients had AR complete CIB1 deficiency.
Figure 2.
CIB1 levels in patient-derived cells and healthy skin biopsy specimens. (A) CIB1 mRNA levels were assessed by RT-qPCR in patient-derived LCLs for kindred A and P12 and in whole blood from P13. Each symbol represents the mean of three independent measurements per cell line/donor. Statistical significance (***, P < 0.001) was assessed in a one-way ANOVA followed by Dunnett’s multiple comparison test for kindred A. Levels were 47% lower than the corresponding control mean in P12 and 80% lower in P13. (B) CIB1 protein was detected in LCLs and PBMCs with a polyclonal antibody directed against the N terminus of CIB1. GAPDH and Akt served as loading controls. Cellular material from kindreds A and B was analyzed three times. PBMCs from kindred E were analyzed once. (C) CIB1 protein level in skin biopsy specimens from healthy donors were measured by IHC with a monoclonal antibody directed against CIB1. An isotype Ig was included as a control. These results are representative of two experiments, each performed on two patients. *, epidermis; >, hair shaft. Bars: (top) 100 μm; (middle and bottom) 50 μm. (D) CIB1 protein was detected in primary keratinocytes from three healthy controls (ctrl 1–3), kindreds A, C, and D, or in whole-skin lysates from kindred F. L, lesion. These results are representative of three and one independent experiments, respectively. Asterisk indicates a nonspecific band.
CIB1 is expressed in keratinocytes
The ubiquitous pattern of expression and the pleiotropic functions assigned to CIB1 are not easy to reconcile with the very narrow clinical phenotype of EV patients. β-HPVs have a strict tropism for the epidermis, and hair follicle stem cells probably serve as a reservoir for HPV persistence (Orth, 2008). We therefore assessed CIB1 protein levels by immunohistochemistry (IHC) in skin biopsy specimens from healthy controls. A strong signal for CIB1 was obtained for the epidermis and hair follicles (Fig. 2 C). We then assessed CIB1 protein levels in cultured primary keratinocytes from three healthy controls, one heterozygous healthy individual from kindred A (wt/m), and three patients from kindreds A, C, and D. CIB1 protein levels were high in control and heterozygous individuals, whereas this protein was not detected in the patients (Fig. 2 D). We also assessed CIB1 protein levels in whole-skin biopsy lysates from a heterozygous individual of kindred F (wt/m) and in lysates from both lesional and nonlesional skin biopsy specimens from his relatives P21 and P23. CIB1 protein was detected in the lysate from the heterozygous individual but not in lesional and nonlesional lysates from the patients (Fig. 2 D). Thus, CIB1 is clearly abundantly expressed in keratinocytes, the natural host cells of β-HPVs, and the patients’ skin and cultured keratinocytes display no detectable CIB1 expression. Finally, we showed that mRNA levels for CIB1, but also for TMC6 and TMC8, were not increased by type I IFN treatment in keratinocytes (Fig. S3), consistent with the lack of β-HPV lesions in patients with inborn errors of type I IFN immunity (Dupuis et al., 2001, 2003; Ciancanelli et al., 2015; Kreins et al., 2015; Lamborn et al., 2017). Collectively, these findings suggest that AR CIB1 deficiency may underlie EV by disrupting keratinocyte-intrinsic type I IFN-independent immunity to β-HPVs.
Lack of CIB1 expression in EVER1- and EVER2-deficient cells
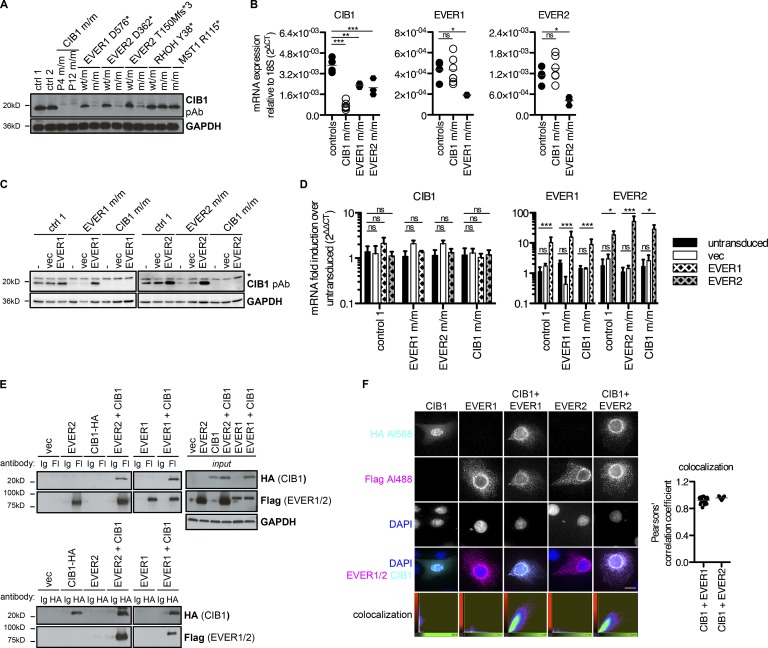
The clinical and virological phenotypes of EVER1-, EVER2-, and CIB1-deficient patients are indistinguishable. We therefore analyzed CIB1 levels in LCLs derived from patients with EVER1 or EVER2 deficiency. Strikingly, individuals carrying inactivating mutations of either EVER1 (D576*; Ramoz et al., 2002) or EVER2 (D362* [Ramoz et al., 2002] or T150Mfs*3 [Crequer et al., 2013]) had very low levels of CIB1 protein, whereas their heterozygous relatives had normal CIB1 protein levels (Fig. 3 A). In EVER1- and EVER2-deficient LCLs, CIB1 mRNA levels were ∼50% lower than in control LCLs (Fig. 3 B). EVER1 and EVER2 are therefore required for the correct expression of CIB1 protein. We detected normal EVER1 and EVER2 mRNA levels in CIB1-deficient LCLs (Fig. 3 B), but a lack of reliable antibodies against EVER proteins made it impossible for us to assess the levels of the corresponding proteins. We also tested LCLs from patients with T cell defects associated with persistent cutaneous β-HPV infections due to RHOH or MST1 (encoded by STK4) deficiency (Crequer et al., 2012a,b). CIB1 levels were normal in cells from these patients (Fig. 3 A). This dichotomy mirrors the lymphocyte frequency data (Tables S1 and S4) and suggests that isolated β-HPV–induced disease in patients with typical EV is caused by a common EVER1/EVER2- and CIB1-dependent mechanism different from the T cell–dependent mechanisms of disease in patients with atypical EV as well as infectious phenotypes including but not limited to β-HPV–induced lesions.
Figure 3.
CIB1 forms a complex with EVER1 and EVER2. (A) CIB1 protein levels in LCLs derived from patients with loss-of-function mutations of EVER1 (D576*) and EVER2 (D362* or T150Mfs*3), and comparison with those in heterozygous carriers, positive controls (ctrl 1 and 2), P4 and P11, and patients with RHOH and MST1 deficiencies. (B) CIB1, EVER1, and EVER2 mRNA levels were assessed by RT-qPCR in controls (n = 4), CIB1 m/m (P1–P6 and P12), EVER1 m/m, and EVER2 m/m (n = 3 each) LCLs. Each symbol represents the mean of one cell line measured in three independent experiments. Statistical significance was assessed by one-way ANOVA followed by Dunnett’s multiple comparison tests relative to healthy controls. (C) CIB1 protein levels in LCLs derived from patients with loss-of-function mutations of EVER1 (D576*), EVER2 (T150Mfs*3), and a healthy control after reconstitution with WT EVER1 or EVER2 by retroviral transduction and stable selection. Asterisk indicates a nonspecific band. (D) CIB1, EVER1, and EVER2 mRNA levels were measured by RT-qPCR in LCLs derived from patients with loss-of-function mutations of EVER1 (D576*), EVER2 (T150Mfs*3), or CIB1 and a healthy control after reconstitution with WT EVER1 or EVER2 by retroviral transduction and stable selection. The data were first normalized against RNaseP as a housekeeping gene and then against an appropriate untransduced parental cell line by the ΔΔCt method. Statistical significance was assessed by one-way ANOVA followed by Dunnett’s multiple comparison test relative to the corresponding untransduced control. (B and D) ns, P > 0.05; *, P < 0.05; **, P < 0.01; ***, P < 0.001. (E) HEK293T cells were transfected with plasmids encoding CIB1-HA, FLAG-EVER1, and FLAG-EVER2 either separately or together; 24 h after transfection, samples were subjected to immunoprecipitation with FLAG (Fl)- or HA-specific antibodies (+). Samples incubated with nonspecific IgG served as specificity controls. Western blots were performed to detect coimmunoprecipitated CIB1-HA and FLAG-EVER1 or FLAG-EVER2. The immunoprecipitation of EVER1/2 and CIB1 was confirmed by reincubation with antibodies specific for FLAG and HA, respectively. The presence of all proteins was checked by Western blotting of an input sample taken before immunoprecipitation. GAPDH served as a loading control. vec, vector. (F) Healthy control keratinocytes were transfected with plasmids encoding CIB1-HA, FLAG-EVER1, and FLAG-EVER2 either alone or in combination; 24 h after transfection, cells were subjected to immunofluorescence imaging with Alexa Fluor 568–HA and Alexa Fluor 488–FLAG antibody combinations. DAPI was used for counterstaining. Colocalization was assessed by calculating Pearson’s correlation coefficient with Imaris software. Bar, 13 µm. The results shown are representative of three independent experiments.
CIB1, EVER1, and EVER2 form a complex
These findings suggest that CIB1 protein stability might be controlled posttranscriptionally by the EVER1 and EVER2 proteins. Consistent with this hypothesis, the stable overexpression of EVER1 in EVER1-deficient LCLs or of EVER2 in EVER2-deficient LCLs rescued CIB1 protein (Fig. 3 C) but not mRNA levels (Fig. 3 D). As a control, the overproduction of either EVER protein in CIB1-deficient LCLs did not rescue CIB1 expression (Fig. 3, C and D). In the absence of robust antibodies against the EVER1 and EVER2 proteins, overexpression was confirmed by assessing mRNA levels (Fig. 3 D). These data suggest that EVER1 and EVER2 regulate CIB1 expression posttranslationally. We therefore hypothesized that the CIB1, EVER1, and EVER2 proteins form a multimeric complex that controls CIB1 protein stability. Reciprocal coimmunoprecipitation experiments in HEK293T cells showed that CIB1 interacted with both EVER1 and EVER2 (Fig. 3 E). Subsequent immunofluorescence analysis in primary keratinocytes indicated that overproduced CIB1 was mostly localized in the nucleus. However, on coexpression with EVER1 or EVER2, CIB1 was localized in perinuclear and cytoplasmic web-like structures that strongly colocalized with EVER1 or EVER2 as demonstrated quantitatively by the calculation of Pearson’s correlation coefficient (Fig. 3 F). Furthermore, mass spectrometry on CIB1 immunopurified after its ectopic expression in HEK293T cells identified endogenous EVER1 as the highest-ranked interactor of CIB1 (Fig. S4 A). EVER2 was not detected probably because its mRNA was barely detectable in HEK293T cells at levels lower than those of EVER1 by a factor of at least 60 (Fig. S4, B–D). Collectively, these data strongly suggest that a multimeric complex of CIB1 and EVER1/EVER2 proteins is required for CIB1 protein stability. They connect CIB1 directly with the previously identified EV-causing proteins EVER1 and EVER2.
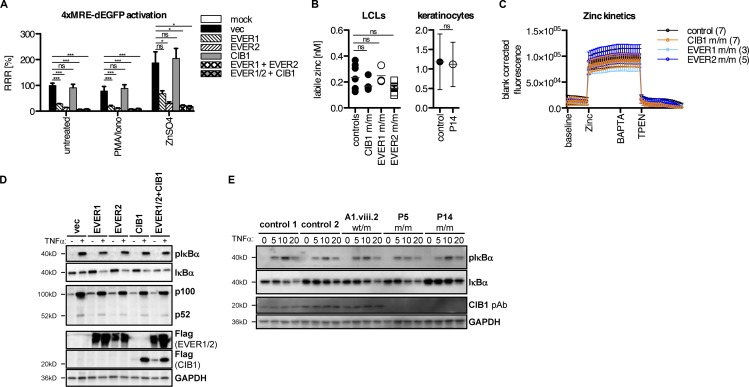
CIB1 does not affect intracellular free zinc and NF-κB signaling
The interaction between CIB1 and EVER1/2 led us to investigate the contribution of CIB1 to the known functions of EVER1 and EVER2 (Lazarczyk et al., 2008; Vuillier et al., 2014). EVER1 and EVER2 have been reported to control intracellular zinc homeostasis by binding to the zinc transporter ZnT1, increasing its activity to import zinc into the ER storage compartment, thereby controlling free intracellular zinc levels (Lazarczyk et al., 2008). We overproduced EVER1 and EVER2 in HEK293T cells; both proteins repressed the basal and zinc-induced transcription of a GFP reporter construct via the metal-response element (MRE; Günther et al., 2012; Fig. 4 A), consistent with previous findings (Lazarczyk et al., 2008). However, CIB1 overexpression had no effect on MRE activity, and no additional effect was observed when CIB1 was coexpressed with EVER1 or EVER2 (Fig. 4 A), consistent with the lack of colocalization of CIB1 and ZnT1 (Fig. S4 E). Moreover, our findings confirmed the interaction between the overproduced EVER1/2 and ZnT1 proteins (Lazarczyk et al., 2008), but we detected no interaction between CIB1 and ZnT1 in the same conditions (not depicted). At odds with this previous study (Lazarczyk et al., 2012), the absolute amounts of free zinc, determined with the fluorescent zinc reporter Fluozin-3, were unaffected in LCLs from patients with EVER1 or EVER2 deficiency (Fig. 4 B). Consistent with these findings, P1–P5, P7, and P12 also had normal free zinc levels in LCLs, and P14 had normal amounts of zinc in primary keratinocytes (Fig. 4 B). Furthermore, all cell lines had zinc flux kinetics in the control range (Fig. 4 C). No statistically significant effect of genotype was detected in a two-way repeated-measures ANOVA. Thus, the overexpression of EVER1 and EVER2 modulated zinc-dependent transcription, whereas that of CIB1 did not, and endogenous EVER1/2 and CIB1 deficiencies had no overt effect on zinc homeostasis, implying that the mechanism underlying EV is zinc independent. Finally, an analysis of TNFα-dependent canonical and noncanonical NF-κB activation upon overexpression of EVER1, EVER2, CIB1, or a combination of these genes in HEK293T cells revealed no differences relative to an empty vector–transfected control (Fig. 4 D), at odds with previous studies (Gaud et al., 2013; Vuillier et al., 2014). Moreover, CIB1-deficient keratinocytes from P5 and P14 displayed normal NF-κB activation in response to TNFα, as shown by assessments of IκBα phosphorylation (Fig. 4 E). This finding is consistent with the fact that none of the many known inborn errors of NF-κB underlie skin lesions caused by β-HPVs (Zhang et al., 2017). Thus, studies of the known functions of EVER1 and EVER2 identified no phenotype common to EVER1-, EVER2-, and CIB1-deficient cells. As the three genetic deficiencies are AR with complete penetrance and underlie strict virological and clinical phenocopies and the three proteins are required for the stability of an oligomeric complex, these findings suggest that the major mechanism of EV pathogenesis is probably unrelated to zinc metabolism and NF-κB activation.
Figure 4.
Analysis of zinc signaling/levels and NF-kB activation in the presence and absence of CIB1. (A) HEK293T cells were transfected with plasmids encoding CIB1, EVER1, and EVER2 either alone or in combination and with a 4×MRE-dependent EGFP reporter construct. After 24 h, cells were stimulated overnight with PMA/ionomycin (10 ng/ml and 50 ng/ml, respectively) or zinc sulfate (ZnSO4; 100 µM). The next day, cells were stained with 1 µg/ml DAPI to exclude dead cells, and GFP fluorescence was determined with an LSRII flow cytometer. The RRR with the value for vector-transfected cells was set at 100%. Statistical significance was assessed by one-way ANOVA followed by Dunnett’s multiple comparison test relative to the appropriate vector-transfected control (ns, P > 0.05; *, P < 0.05; ***, P < 0.001; n = 3). (B) Flow cytometric quantification of absolute amounts of labile zinc in LCLs derived from healthy controls, EVER1-, EVER2-, or CIB1-deficient patients, or in keratinocytes from P14 with 1 µM FluoZin-3 as described by Haase et al. (2006). Statistical significance was assessed by one-way ANOVA followed by Dunnett’s multiple comparison test relative to the healthy controls (n = 3). (C) Kinetics of zinc flux in LCLs derived from healthy controls and EVER1-, EVER2-, or CIB1-deficient patients. Cells were loaded with 1 µM FluoZin-3 for 30 min. Fluorimetric measurement were performed on a Victor microplate reader. Baseline fluorescence was recorded every minute for 10 min. Cells were then loaded with 100 µM ZnSO4 and recorded for 15 min. The specificity of the zinc signal was confirmed by adding the calcium-specific chelator BAPTA before the quenching of the signal with the zinc-specific chelator TPEN and recording for 20 min. No significant effect of genotype was detected in two-way repeat-measures ANOVA (n = 3). (D) HEK293T cells were transfected with plasmids encoding CIB1-FLAG, FLAG-EVER1, and EVER2 either alone or in combination. Cells were stimulated with 50 ng/ml TNFα 6 h after transfection, incubated overnight, and then harvested and processed for Western blotting. Membranes were probed for the canonical NF-κB component (p)IκBa and the noncanonical NF-κB component p100/p52. Expression of the constructs used for transfection was verified by incubation with a FLAG-specific antibody. GAPDH served as a loading control (n = 3). (E) Primary keratinocytes from unrelated donors (controls 1 and 2), a healthy family member from kindred A1 carrying the mutation in a heterozygous state (A1.viii.2), and one patient each from kindreds A1 and C were stimulated with 10 ng/ml TNFα for 5, 10, or 20 min and then harvested and processed for Western blotting. Membranes were probed for the canonical NF-κB component (p)IκBa. CIB1 levels were assessed with a polyclonal antibody. GAPDH served as a loading control. vec, empty vector (n = 3).
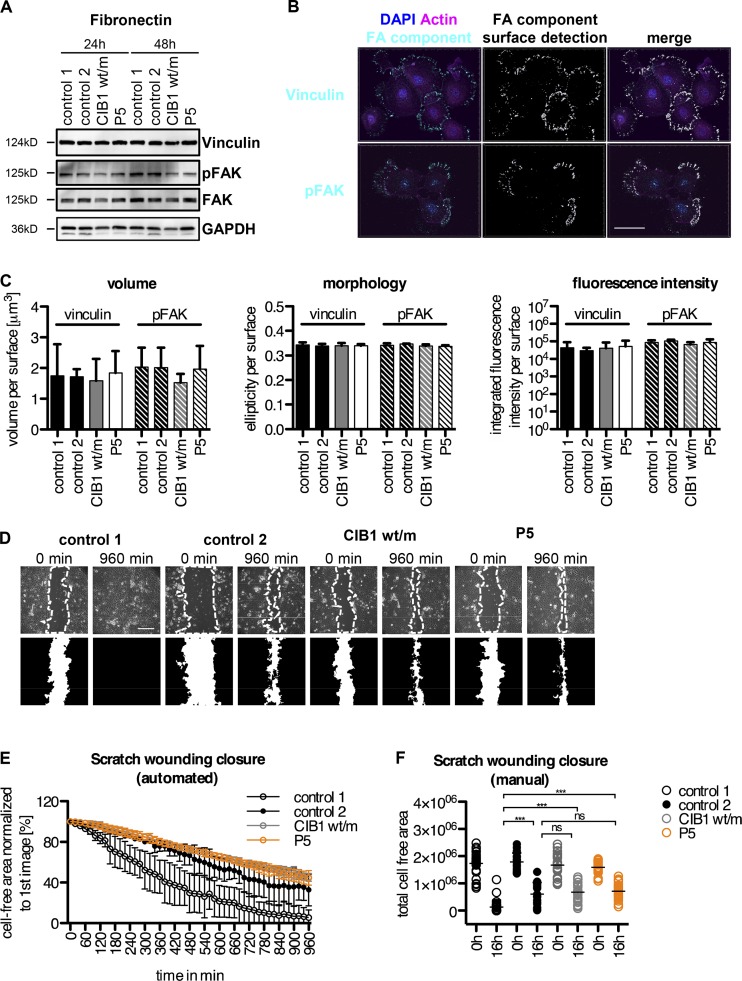
CIB1 deficiency does not affect keratinocyte adhesion and migration
We then focused our attention on the known functions of CIB1. None of the four phenotypes of CIB1-deficient mice were seen in our patients, who had no cardiac, vascular, hemostasis, or fertility abnormalities (Yuan et al., 2006; Zayed et al., 2007; Naik et al., 2009; Heineke et al., 2010). CIB1 has not been studied in human or mouse keratinocytes. However, it has been reported to localize to focal adhesions and to modulate cell migration (Leisner et al., 2016). The keratinocytes of P5 contained normal levels of the focal adhesion proteins vinculin and focal adhesion kinase (FAK) and displayed normal FAK activation, as shown by Western blot analyses of phosphorylation (pFAK), relative to two unrelated healthy controls and a heterozygous family member (Fig. 5 A). We assessed the formation of focal adhesions by performing immunofluorescence staining for vinculin- and pFAK-positive focal adhesion structures in CIB1-deficient keratinocytes. Automated surface detection and quantification showed these adhesions to be of normal size, fluorescence intensity, and morphology, similar to those of healthy controls and heterozygous relatives (Fig. 5, B and C). Consistent with these results, CIB1 deficiency had no detectable effect on the ability of keratinocytes to migrate in scratch-wounding assays, although the results obtained were at the lower end of the control range (Fig. 5, D, E, and F). Finally, we compared the transcriptomes of WT keratinocytes and CIB1-deficient keratinocytes and of CIB1-deficient keratinocytes and GST- or WT-CIB1–transduced CIB1-deficient keratinocytes. We identified only 40 genes displaying at least a twofold down- or up-regulation. These genes included CIB1 but not EVER1 or EVER2, consistent with our quantitative PCR (qPCR) data. No enrichment in a specific pathway was detected among these genes (Table S6). Thus, the lack of CIB1 caused no overt cellular phenotype related or unrelated to the known functions of EVER1, EVER2, or CIB1 in LCLs or primary keratinocytes. These data are consistent with the very narrow clinical phenotype of EV patients, who are highly and selectively vulnerable to β-HPV but otherwise healthy. However, they provide no insight into the potential mechanism underlying β-HPV–driven lesions as even keratinocytes displayed no detectable phenotype.
Figure 5.
Focal adhesion formation and scratch wounding in CIB1-deficient keratinocytes. (A) Primary keratinocytes from two donors, a healthy member of kindred A (A1.viii.2; CIB1 wt/m), and P5 were allowed to adhere to fibronectin-coated plates (10 µg/ml) for the times indicated and were then harvested and processed for Western blotting. The membranes were probed for the focal adhesion (FA) components vinculin and (p)FAK. GAPDH served as a loading control. (B and C) Primary keratinocytes from two donors, a healthy member of kindred A (A1.viii.2), and P5 were allowed to adhere to fibronectin-coated coverslips (10 µg/ml) for 24 h. Cells were fixed and stained with vinculin- and pFAK-specific antibodies. Z stacks were acquired with a confocal microscope. (B) Representative image for automated surface detection for control 1. Bar, 40 µm. (C) The volume (size), fluorescence intensity, and morphology of the vinculin- and pFAK-positive structures were automatically determined with Imaris. Values were normalized against the total number of surfaces detected. Statistical significance was assessed in one-way ANOVA followed by Dunnett’s multiple comparison test relative to control 1. No significant differences were detected for any of the parameters tested (P > 0.05). (D–F) Primary keratinocytes from two donors, a healthy member of kindred A (A1.viii.2; CIB1 wt/m), and P5 were allowed to adhere to fibronectin-coated plates (1 µg/ml) overnight. Scratch wounds were created with a pipette tip, and wound closure was monitored by live-cell microscopy every 20 min for 16 h. Bar, 283 µm. Wound closure was then quantified automatically with the MiToBo plugin in ImageJ with measurement of the cell-free area normalized against the starting point (set to 100%; D and E). Statistical significance was assessed by two-way repeated-measures ANOVA. Differences between controls 1 and 2, CIB1 wt/m, and P5 were statistically significant. Differences between control 2, CIB1 wt/m, and P5 were not significant. (F) For confirmation of the accuracy of the results obtained with the plugin, the total cell-free area at the start and end points was determined manually. The data shown are the means of two (controls 1 and 2) or three (CIB1 wt/m and P5) independent experiments with at least six data points acquired per set. Statistical significance was assessed by one-way ANOVA followed by Dunnett’s multiple comparison test relative to controls 1 or 2. (ns, P > 0.05; ***, P < 0.001).
CIB1, EVER1, and EVER2 interact with the HPV E5 and E8 proteins
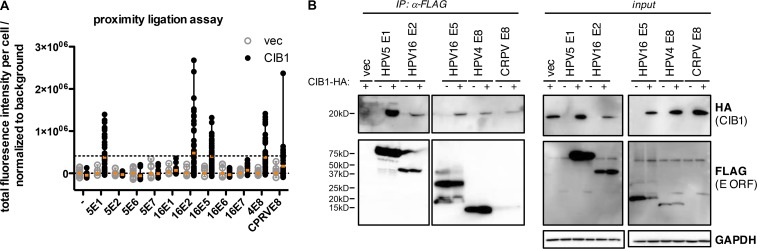
These results suggest that CIB1-deficient keratinocytes may display their EV-causing phenotype only in the presence of β-HPV infection. Antiviral restriction factors are commonly targeted and antagonized by viral proteins (Duggal and Emerman, 2012). The specific feature of the β-HPVs underlying EV lesions is their lack of the E5 and E8 open reading frames (ORFs), rendering them defective in healthy individuals (Orth, 2006). The E8 ORFs from μ-HPV1, γ-HPV4, and κ-CRPV are structurally related to the E5 ORF, and the corresponding proteins function as growth-promoting factors in E5-deficient κ-papillomaviruses (Orth, 2008). Indeed, it has been shown that CRPV-E8 is crucial in vivo (Hu et al., 2002; Nonnenmacher et al., 2006). E5 and E8 are both hydrophobic transmembrane proteins with weak transforming activity in vitro (Nonnenmacher et al., 2006). As EV patients are prone only to cutaneous disease caused by β-HPVs, we hypothesized that the EVER1–EVER2–CIB1 complex interacts with E8 and E5 produced early in infection. We considered γ-HPV4 E8 and κ-CRPV E8, all β-HPV5 early-expressed ORFs (E1, E2, E6, and E7), and as controls, all α-HPV16 early-expressed ORFs (E1, E2, E5α, E6, and E7). We assessed the ability of the corresponding proteins to interact with CIB1 in the human keratinocyte cell line HaCaT in Duolink proximity-ligation assays (PLAs) by fluorescence microscopy followed by the automated surface detection and quantification of total fluorescence intensity. This approach can be used to detect proteins lying in close proximity and therefore likely to form a complex. This approach revealed that CIB1 was indeed targeted by HPV4 E8, CRPV E8, and HPV16 E5 but also by HPV5 E1 and HPV16 E2 and not by any of the other six E proteins from these two HPVs (Fig. 6 A). We then confirmed these interactions by coimmunoprecipitation/immunoblotting in HaCaT cells (Fig. 6 B). Collectively, these experiments showed that CIB1 interacts with both the γ-HPV4 E8 and α-HPV16 E5 proteins. These findings are consistent with a previous study showing that EVER proteins interact with HPV16 E5 (Lazarczyk et al., 2008). The interaction between β-HPV5 E1 and HPV16 E2 and CIB1 but not between HPV5-E2 and CIB1 suggests that the CIB1–EVER1–EVER2 complex or at least CIB1 might be involved against other cutaneous or mucosal HPVs in patients without EV. All these observations provide a plausible mechanism of disease in patients with EV. β-HPVs lacking the E5 and E8 ORFs cannot overcome the CIB1–EVER1–EVER2 complex, which probably operates as a restriction factor in keratinocytes (Fig. 7). Conversely, in patients lacking EVER1, EVER2, or CIB1, the lack of this restriction factor allows β-HPV–driven lesions to develop.
Figure 6.
PLA and coimmunoprecipitation in HaCaT. (A) HaCaT cells were transfected with plasmids encoding FLAG-HPV5 E1, E2, E6, and E7, FLAG-HPV16 E1, E2, E5, E6, and E7, FLAG-HPV4 E8, FLAG–CRPV E8, and CIB1-HA alone or in combination. The day after transfection, samples were plated on microscopy slides, allowed to adhere, fixed in acetone, permeabilized, and subjected to Duolink PLAs with rabbit-HA– and mouse-FLAG–specific antibodies. Z stacks were acquired with a widefield microscope, and PLA-positive sites (defined as structures >0.35 µM2) were scored with Imaris software for 15–50 cells per condition. These pooled results were obtained in two independent experiments. The orange bars indicate the mean. (B) HaCaT cells were transfected with plasmids encoding CIB1-HA and the FLAG-tagged HPV E ORFs scoring positive in the PLA in A. 1 d after transfection, samples were subjected to immunoprecipitation (IP) with FLAG-specific antibodies. Western blots were performed to detect coimmunoprecipitated HPV5 E1, HPV16 E2, E5, HPV4 E8, and CRPV E8. The immunoprecipitation of CIB1 was confirmed by reincubation with a FLAG-specific antibody. The presence of all proteins was checked by Western blotting analysis on an input sample taken before immunoprecipitation. GAPDH served as a loading control (n = 3). vec, vector.
Figure 7.
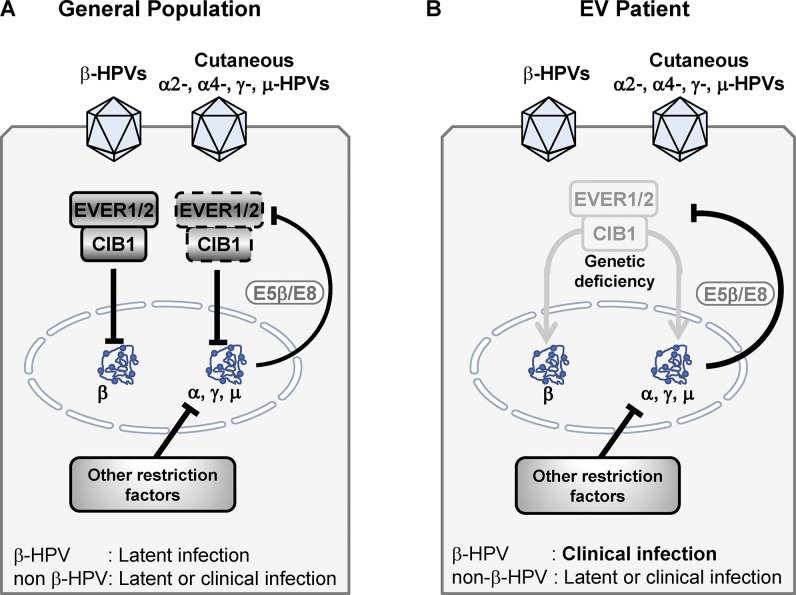
Mechanism underlying EV. (A) In the general population, the CIB1–EVER1–EVER2 complex restricts the transcription of minichromosome of β-HPV, leading to the absence of clinical manifestation. The proteins E5 and E8 expressed by the other cutaneous HPVs (α2-, α4-, γ-, and μ-HPVs) are able to antagonize the CIB1–EVER1–EVER2 complex. However, additional restriction factors are probably contributing to the absence of HPV lesions in the vast majority of people. (B) In EV patients, the lack of CIB1–EVER1–EVER2 permits the transcription of β-HPV minichromosome, which leads to the development of EV lesions on the skin. However, the probable presence of additional restriction factors against cutaneous HPVs other than β-HPVs accounts for their normal control, which does not differ from the general population.
Discussion
We report AR complete CIB1 deficiency as a new genetic cause of EV. We identified two frameshift insertions (I156Dfs*5 and A184Lfs*70), one frameshift deletion (K83Rfs*4 in two kindreds), one nonsense mutation (R72*), and one splice mutation (c.52-2A>G) in 24 patients from six families from five ethnically and ecologically diverse countries: Colombia, France, Iran, Switzerland, and Togo. None of the heterozygous relatives of the patients show any clinical signs of EV, whereas all homozygous individuals suffer from EV. The familial segregation of the mutant CIB1 alleles is consistent with an AR trait displaying complete clinical penetrance, but CIB1 deficiency is associated with somewhat variable expressivity, as also reported for the deficiencies in families with EVER1 and EVER2 mutations (Orth, 2006, 2008; Burger and Itin, 2014). For example, in kindred A, the phenotype ranged from very mild manifestations in P1 and P4 to more aggressive disease with early cancer development in siblings P2, P3, and P5. Nevertheless, in all patients, CIB1 deficiency led to the development of EV from the age of 23 yr onward (the age at which clinical penetrance can be said to be complete in this cohort). Finally, none of these patients suffered from any other unusually severe infection, like patients with EVER1 or EVER2 deficiencies and unlike those with atypical EV and T cell deficits. AR CIB1, EVER1, and EVER2 deficiencies are therefore indistinguishable clinical and virological phenocopies, consistent with a typical form of EV.
CIB1 has been reported to have pleiotropic functions both in vitro in cell culture models and in vivo in knockout mice (Leisner et al., 2016). Mice lacking Cib1 have vascular, cardiac, and hemorrhagic phenotypes, and their males are sterile (Yuan et al., 2006; Naik et al., 2009). By contrast, none of these phenotypes were observed in patients, indicating that CIB1 is not essential for these functions in humans. Low CIB1 mRNA levels are unlikely to be causally associated with oligoasthenozoospermia-related sterility (Sun et al., 2014) as a complete absence of the CIB1 protein had no effect on male fertility in our cohort. We know that this observation is robust because we have diagnosed 24 CIB1-deficient patients from six kindreds originating from very diverse ethnicities. Our findings suggest that the only nonredundant function of CIB1 in humans in vivo is the control of β-HPVs in keratinocytes. Cib1-deficient mice have not been challenged with infectious agents including skin-tropic viruses. The only known mouse-tropic papillomavirus, Mus musculus papillomavirus type 1 (MuPV1; Ingle et al., 2011), does not belong to the β genus (Joh et al., 2011).
Our identification of AR CIB1 deficiency establishes a third genetic etiology of typical EV after AR EVER1 and EVER2 deficiencies. Six different homozygous and two compound heterozygous mutations of TCM6 have been described in 14 patients from eight kindreds (Ramoz et al., 2002; Tate et al., 2004; Zuo et al., 2006; Aochi et al., 2007; Gober et al., 2007; Youssefian et al., 2018), and 11 homozygous mutations of TCM8 have been described in 17 patients from 11 kindreds (Ramoz et al., 2002; Sun et al., 2005; Berthelot et al., 2007; Rady et al., 2007; Landini et al., 2012; Burger and Itin, 2014; Miyauchi et al., 2016; Imahorn et al., 2017; de Jong et al., 2018; Youssefian et al., 2018). All these mutations are loss of function. The identification of five homozygous loss-of-function alleles of CIB1 markedly increases the number of EV cases for which explanations have been found at the genetic and molecular levels. Moreover, we identified a molecular connection between CIB1, EVER1, and EVER2 as shown by the very low levels of CIB1 proteins in cells from patients with EVER1 or EVER2 deficiency. This is reminiscent of the interaction between TMC1, TMC2, and CIB2 in inner-ear hair cells, which is required for normal sensory transduction (Giese et al., 2017). Our observation provides a unifying cellular phenotype common to all known genetic etiologies of typical EV. CIB1 protein profiling could therefore be used as a simple laboratory test for screening patients with suspected EV, making it possible to target sequencing efforts on CIB1, TMC6, and TMC8 in patients with low levels of CIB1 protein.
The clinical characteristics of EVER1- and EVER2-deficient patients (EV in the absence of other clinical manifestations; Orth, 2006, 2008; Burger and Itin, 2014) and of the patients with CIB1 deficiency described in this study are identical and markedly different from those of patients with atypical EV due to CD4+ T cell lymphopenia caused by RHOH, MST1 (encoded by STK4), CORO1A, ARTEMIS (encoded by DCLRE1C), DOCK8, RASGRP1, LCK, and TPP2 deficiencies. In such patients, persistent β-HPV–induced lesions occur in the context of a much broader infectious, autoimmune, and tumoral phenotype (Crequer et al., 2012a,b; Sanal et al., 2012; Stray-Pedersen et al., 2014; Stepensky et al., 2015; Li et al., 2016; Tahiat et al., 2016; Liu et al., 2017; Platt et al., 2017). Consistent with this observation, the CIB1-deficient patients tested in this study had no detectable T-lymphocyte defects in terms of either cell numbers or functions. The clinical and immunological dichotomy between typical and atypical EV is reflected at the cellular level by the profile of CIB1 expression profile, with very low levels of CIB1 in patients with EVER1 and EVER2 deficiencies but not in patients with RHOH and MST1 deficiencies. These findings support the notion that patients with typical EV have intact adaptive immunity and suffer from an inborn error of cell-intrinsic immunity, resulting in an inability to control β-HPV–driven keratinocyte proliferation.
Inherited CIB1 deficiency is not associated with any detectable, constitutive phenotype in keratinocytes in terms of the transcriptome, cell growth, migration, or adhesion. CIB1 deficiency also has no impact on zinc homeostasis and NF-κB activation, suggesting that EVER proteins have other functions shared by CIB1 that are involved in the pathogenesis of EV. We reveal in this study a phenotype of CIB1, EVER1, and EVER2 deficiencies that is conditional on infection with β-HPVs. Our findings suggest that the CIB1–EVER1–EVER2 complex interacts in vivo with both cutaneous γ-HPV4 E8 and κ-CRPV E8 as well as with α-HPV16 E5. In this light, it is plausible that β-HPVs that lack both the E5 and E8 ORFs cannot overcome the CIB1–EVER1–EVER2 complex, unlike α-, γ-, and μ-HPVs and CRPV, which cause disease more widely in the general population (Fig. 7). Both E5 and E8 are hydrophobic and transmembrane proteins with weak transforming activity in vitro but potent growth-promoting activity in vivo (Hu et al., 2002; García-Vallvé et al., 2005; Nonnenmacher et al., 2006; Maufort et al., 2007; Orth, 2008; Wechsler et al., 2018). Conversely, the E5 and E8 proteins target and antagonize the CIB1–EVER1–EVER2 complex, thereby contributing to virulence to α-, γ-, and μ-HPVs, which cause cutaneous warts in the general population. By contrast, β-HPVs are capable of causing disease only in EV patients lacking the CIB1–EVER1–EVER2 complex. The interaction of β-HPV5 E1 and α-HPV16 E2 with CIB1 is probably not sufficient to antagonize the EVER–CIB1 complex, yet it suggests that this complex might be involved in the pathogenesis of the disease caused by various HPVs. However, EV patients are apparently not more prone to develop common HPV warts, suggesting that immunity against cutaneous non–β-HPVs relies on mechanisms other than the CIB1–EVER1–EVER2 complex (Fig. 7). We have established that EV is caused by mutations of the genes encoding the proteins of the CIB1–EVER1–EVER2 complex, which acts as a restriction factor for HPVs in keratinocytes through interaction with two viral E proteins, E8 and E5, which are absent from β-HPVs.
The CIB1–EVER1–EVER2 complex fulfills the five criteria commonly used to define restriction factors active against viruses (Duggal and Emerman, 2012). First, the lack of CIB1, EVER1, or EVER2 is clinically associated with EV, lesions of which are caused by β-HPVs. Second, patients lacking the CIB1–EVER1–EVER2 complex display no other detectable cellular or clinical phenotype. Third, the CIB1–EVER1–EVER2 complex operates in keratinocytes, the host cells of β-HPVs. Fourth, E5 and E8 are viral virulence genes expressed by cutaneous α-, γ-, and μ-HPVs but not β-HPVs, and the products of these genes target the CIB1–EVER1–EVER2 complex. Fifth, human CIB1 belongs to a multigene family with four members. We do not provide experimental evidence for the control of β-HPVs by the CIB1–EVER1–EVER2 complex in keratinocytes in vitro as this work would have gone well beyond the scope of this study given the technical difficulties involved. Nevertheless, we provide the first in vivo evidence of a restriction factor operating in humans. Other restriction factors in humans have been validated in vitro or in animal models (Kluge et al., 2015; Subramanian et al., 2018). The pathogenesis of the β-HPV–driven skin lesions characteristic of EV in patients with AR CIB1, EVER1, or EVER2 deficiency involves the disruption of skin-restricted, cell-autonomous, nonhematopoietic, keratinocyte-intrinsic, IFN-independent restriction factor (the CIB1–EVER1–EVER2 complex)–mediated immunity against a specific set of defective viruses (β-HPVs, which lack E5 and E8).
Materials and methods
Patients and case studies
The clinical characteristics of all patients are summarized in Table 1. The experiments involving human subjects were conducted in accordance with local, national, and international regulations and were approved by the French Ethics Committee, the French National Agency for the Safety of Medicines and Health Products, and the French Ministry of Research (protocol C10-13) as well as The Rockefeller University Institutional Review Board (protocol JCA-0700). Informed consent was obtained from all patients or their family members included in this study.
Kindred A (branches A1 and A2; P1–11)
Branches A1 and A2 of kindred A originate from the same region of Colombia, and affected individuals in these two branches of the family are homozygous for the same 3-Mb haplotype encompassing a disease-linked interval on chromosome 15 that harbors CIB1, consistent with a common ancestor for these two families.
Kindred A, branch A1 (Figs. 1 A and S1; Tables 1, S1 A, S2, S3, S4, and S5)
P1–P5 were born to second-degree cousins (Fig. 1 A). The consanguinity of this family was confirmed by the high inbreeding coefficient determined by SNP 6.0 array analysis (Table S5). Neither of the parents displayed any signs of cutaneous HPV infection or EV. The clinical characteristics of P2, P3, and P5 have been reported previously (Rueda, 1993). All five patients mounted normal antibody responses to various DNA viruses (HSV-1/2, varicella zoster virus, CMV, EBV, and hepatitis B) and RNA viruses (hepatitis A, hepatitis C, measles, rubella, and mumps) as shown by comparison with 12 healthy family members (Table S3). Furthermore, P1–P4 were extensively phenotyped for skin-homing total CD3+, CD4+, and CD8+ T cell subsets (CLA+, CLA+CCR4+, CLA+CCR10+, and CCR10+), and no alterations were detected relative to five unrelated healthy donors and nine healthy family members (Table S4).
P1 (A1.viii.4; Fig. 1 A) has a very mild EV phenotype. She first developed persistent flat warts on the back of her hands at the age of 12 yr. At the age of 15 yr, she developed warts on her face. She is not currently followed by a dermatologist.
P2 (A1.viii.7; Fig. 1 A) presented with brownish macules on his hands at the age of 6 yr. He was formally diagnosed at the age of 13 yr. The hyperpigmented lesions progressively spread, forming flat warts on his breast, neck, forehead, legs, and feet (Fig. S1 A). The lesions on his forehead were hypochromic, and the lesions on his trunk were pityriasis versicolor–like. He developed an invasive ulcerating SCC on the right-hand side of his forehead, which was surgically removed. He was treated by cryotherapy and with topical 5-fluorouracil. Restriction enzyme analysis and Southern blot hybridization showed that P2 was carrying HPV8, HPV17, and HPV20. P2 also suffered from chronic hepatitis C virus–induced hepatitis and died from hepatic carcinoma at the age of 47 yr. This patient was described as case 11 by Rueda (1993).
P3 (A1.viii.10; Fig. 1 A) presented with flat warts at the age of 7 yr. She was formally diagnosed at the age of 13 yr. She displayed reddish and hyperpigmented flat warts on her forehead, trunk, arms, hands, and legs. She developed three SCCs on the forehead and lips after the age of 13 yr. Restriction enzyme analysis and Southern blot hybridization showed that P3 carried HPV8 and HPV20. This patient was reported as case 12 in a previous study (Rueda, 1993). She displays no immunological abnormalities of T cells, B cells, NK cells, or monocytes (Table S1 A), and her T cell proliferation after anti-CD3 antibody stimulation was normal (Table S2).
P4 (A1.viii.12; Fig. 1 A) presented with flat warts at the age of 7 yr and was formally diagnosed at the age of 22 yr. Her lesions are located on the head and trunk. She has not yet developed SCCs. She was shown to be HPV8 positive by restriction enzyme analysis and Southern blot hybridization.
P5 (A1.viii.15; Fig. 1 A) presented with flat warts on his forehead at the age of 5 yr. The lesions spread to his hands, trunk, and upper legs over time. He was formally diagnosed with EV at the age of 7 yr. He developed an SCC on his forehead at the age of 31 yr, which was surgically removed. He is currently being treated by cryotherapy and protection against sun exposure. Restriction enzyme and Southern blot analyses showed that he carried HPV8 and HPV20. This patient was described as case 13 in a previous study (Rueda, 1993). He displays no immunological abnormalities of T cells, B cells, NK cells, or monocytes (Table S1 A), and his T cell proliferation in response to anti-CD3 antibody stimulation was normal (Table S2).
P6 (A1.iv.13; Fig. 1 A) presented with diffuse flat warts at the age of 1 yr. He was formally diagnosed with EV at the age of 29 yr. He had numerous lesions over his entire body except for the scalp, palms, soles of his feet, and mucous membranes. The backs of his hands and his arms and legs were particularly severely affected. Erythematous squamous and pigmented lesions evolved to Bowen’s disease and recurrent invasive SCC after the age of 15 yr. Histologically, these lesions consisted of large cells with an extensive clear cytoplasm and vacuolar nuclei in both the stratum spinosum and granular layer of the epidermis, a feature pathognomonic for EV. In semithin sections, inclusion bodies, also typical of EV, were identified. This patient died at the age of 47 yr due to multiple aggressive SCCs. He was reported as case 6 in a previous study (Rueda, 1993). Unfortunately, this patient died >35 yr ago, precluding genetic analysis.
P7 (A1.viii.30; Fig. 1 A), belonging to another branch of kindred A1, displayed consanguinity because one of his parents was the offspring of second cousins. Familial consanguinity was confirmed by a high inbreeding coefficient determined by SNP 6.0 array analysis (Table S5). From the age of 5 yr, P7 presented eruptions of pityriasis versicolor–like and scaly erythematous lesions on the face and neck, which spread as flat warts to his trunk, forearms, legs, and the backs of his hands. He developed infiltrative lesions, histologically defined as Bowen’s disease, at the left medial canthus and on the nose. P7 was treated surgically for Bowen’s disease and then topically with retinoic acid and 5-fluoruracil. P7 tested positive for HPV5, HPV17, and HPV36 by restriction enzyme analysis and Southern blot hybridization. This patient was described as case 7 in a previous study (Rueda, 1993).
Kindred A, branch A2 (Fig. 1 A and Tables 1 and S5)
P8–P11 were born to healthy parents. Familial consanguinity was suspected and confirmed on the basis of a high inbreeding coefficient in P11 as determined by 250K array analysis (Table S5). The clinical characteristics of P8–P11 have been described previously (Rueda and Rodriguez, 1976; Rueda, 1993). All patients had normal karyotypes and serum Ig levels.
P8 (A2.ii.1; Fig. 1 A) first presented with EV at the age of 1 yr, but she was not formally diagnosed until the age of 59 yr. She presented with flat warts on her head, trunk, and extremities. She developed SCC on her forehead at the age of 47 yr. Restriction enzyme analysis and Southern blot hybridization demonstrated the presence of HPV5, HPV8, HPV20, and HPV24 in this patient. She died at the age of 78 yr. This patient was described as case 1 in previous studies (Rueda and Rodriguez, 1976; Rueda, 1993). HPV5 was cloned from benign and malignant lesions from P8 and characterized (Deau et al., 1993). HPV20 was originally cloned from a benign lesion from P8 (Kremsdorf et al., 1984).
P9 (A2.ii.10; Fig. 1 A) first presented with EV at the age of 5 yr, but she was not formally diagnosed until the age of 40 yr. She presented with flat warts on her head, trunk, and extremities. SCC development has never been reported for this patient. Restriction enzyme analysis and Southern blot hybridization confirmed that this patient was carrying HPV8. This patient was described as case 2 in previous studies (Rueda and Rodriguez, 1976; Rueda, 1993). HPV5 from P9 was cloned and characterized as case B by Deau et al. (1991) and case 4 by Deau et al. (1993).
P10 (A2.ii.12; Fig. 1 A) first presented with EV at the age of 1 yr. He was not formally diagnosed until the age of 40 yr. He presented with flat warts on his head, trunk, and extremities. He developed SCC on his forehead and lips after the age of 26 yr, and he died at the age of 48 yr. This patient was described as case 3 in two previous studies (Rueda and Rodriguez, 1976; Rueda, 1993).
P11 (A2.ii.13) first presented with EV at the age of 3 yr but was not formally diagnosed until the age of 36 yr. He presented with flat warts on his head, trunk, and extremities. He developed invasive SCC and basal cell carcinoma on his forehead after the age of 27 yr. He died at the age of 48 yr. Restriction enzyme analysis and Southern blot hybridization confirmed that this patient was carrying HPV5, HPV8, and HPV20. He was described as case 4 in two previous studies (Rueda and Rodriguez, 1976; Rueda, 1993).
Kindred B (P12; Fig. 1 A and Tables 1 and S5)
P12 (B.ii.2; Fig. 1 A) was previously described in a case study (Kienzler et al., 1979). He was born to French parents in 1933. Potential parental consanguinity was investigated by calculating an inbreeding coefficient with FSuite software (Gazal et al., 2014). This analysis indicated that his parents were first cousins (Table S5). P12 presented with recurrent polymorphic skin lesions typical of EV from the age of 23 yr onwards. He developed several SCCs on the forehead, cheek, and ear ∼20 yr after the onset of the disease. The viral etiology of his disease was confirmed by the identification of HPV8 by restriction enzyme analysis and by Southern blotting. P12 had normal levels of Ig, complement components, and cellular immunity as shown by measurements of dinitrochlorobenzene sensitization, rosette assays, leukocyte migration, and lymphocyte proliferation. P12 died from natural causes at the age of 72 yr.
Kindred C (P13 and P14; Fig. 1 A and Tables 1 and S5)
P13 (C.ii.1; Fig. 1 A) and P14 (C.ii.2; Fig. 1 A) were among the first EV patients documented by Lutz, who was the first to describe this disease and followed these two sisters from 1929/1930 onwards (Lutz, 1946). Both patients were also described in a more recent study (Arnold et al., 2011). They were born to Swiss parents who were shown to be consanguineous by inbreeding coefficient analyses (Table S5). P13 was born in 1912. She developed flat wart-like lesions on her neck, face, and on the backs of her hands in childhood, and these warts persisted throughout her life. After the age of 35 yr, she developed several actinic keratoses and SCC. She died of locally invasive SCC with regional lymph node metastasis at the age of 71 yr. P14 was born in 1923 and developed symptoms of EV in early childhood. Persistent lesions spread from the neck and hands to the patient’s legs. After the age of 52 yr, P14 developed several actinic keratoses, SCCs, and basal cell carcinomas that were clinically well managed by a combination of surgery and topical treatment. P14 died from natural causes at the age of 91 yr. HPV5 was identified by PCR amplification from specimens obtained from these two patients.
Kindred D (P15; Figs. 1 A and S1; Tables 1, S1 B, S4, and S5)
P15 (D.iv.2; Fig. 1 A) was born to consanguineous parents from Switzerland, and her phenotype was recently described in a case study (Imahorn et al., 2017). Familial consanguinity was confirmed by a high inbreeding coefficient determined with an SNP 6.0 array (Table S5). P15 developed first lesions during early childhood (Fig. S1 B). She was treated for several carcinomas in situ (Bowen type) and SCCs from the age of 54 yr onwards. HPV5 was detected by PCR amplification and Sanger sequencing. This patient has no immunological abnormalities of T cells, B cells, or NK cells (Table S1 B). She was also extensively phenotyped for skin-homing total CD3+, CD4+, and CD8+ cells and T cell subsets (CLA+, CLA+CCR4+, CLA+CCR10+, and CCR10+), none of which displayed any marked difference relative to five unrelated healthy donors (Table S4).
Kindred E (P16–P19; Figs. 1 A and S1 C; Tables 1 and S5)
P16 (E.iv.4; Fig. 1 A), P17 (E.iv.6; Fig. 1 A), and P19 (E.iv.10; Fig. 1 A) were first described in a case study (Saka et al., 2009). In brief, P16–P19 were born to consanguineous parents from Togo. Familial consanguinity was confirmed on the basis of a high inbreeding coefficient in an SNP 6.0 array analysis (Table S5). P16 first developed papules on her face at the age of 7 yr, and these lesions subsequently disseminated over her entire body. She was diagnosed at the age of 15 yr and died at the age of 21 yr from a pulmonary metastasis secondary to the development of multiple SCCs on areas of the skin exposed to the sun. P17 first developed papules on her face at the age of 5 yr, and these lesions subsequently disseminated over her entire body. P18 was not included in the initial case study. Only limited information about her clinical characteristics is available. She died in 2013 after an episode of fever. P19 is currently 14 yr of age and was first presented with disseminated skin lesions at the age of 2 yr (Fig. S1 C). P16, P17, and P19 had normal blood counts and renal and hepatic functions and were confirmed to be HIV negative. Histological analysis confirmed the presence of the pathognomonic cytopathic effect in a lesional skin biopsy. It was not possible to type the causal HPV due to infrastructure-related difficulties.
Kindred F (P20–P24; Figs. 1 A, S1 D, and S2; Tables 1 and S5)
Two patients, a 47-yr-old man and his 22-yr-old daughter, were referred to the Dermatology Department of the Tehran University of Medical Sciences for the evaluation of a chronic condition that had begun during childhood. Similar presentations were observed in other family members, with five in total all born to consanguineous parents, suggesting the presence of an AR Mendelian disorder in the extended family. All patients were healthy at birth, but they developed cutaneous lesions resembling widespread flat warts and thin plaques from the ages of 1–3 yr onwards. Multiple hyperpigmented plaques were observed on the skin of the male patient. These plaques had an occasionally verrucous surface and displayed ulceration and scaling with raised pearly borders. Three excisional biopsies of the ulcerated plaques revealed the presence of islands of infiltrative malignant cells in the dermis with palisading basaloid cells and polygonal squamous differentiation with areas of keratinization and intercellular bridges consistent with basosquamous carcinoma. The patient also had numerous slightly raised brown papules and plaques on the face, neck, arms, chest, and back of the hands, some coalescing into larger plaques with a papillomatous surface. Microscopic evaluations of these lesions revealed hyperkeratosis and acanthosis, with keratinocytes displaying coarse keratohyaline granules, perinuclear halos, and a blue-gray pallor characteristic of EV. A jaw x-ray (orthopantogram) was performed to evaluate jaw keratocysts and other abnormalities suggestive of nevoid basal cell carcinoma syndrome, which were found to be within normal limits. The index case and one of his affected siblings were positive for HBsAg, and both reported long-term hepatitis B infection and liver cirrhosis. The female patient had numerous erythematous or hyperpigmented papules with a flat surface, minimal scaling, and irregular borders on the face, neck, hands, and arms. Flat pink macules and larger patches with fine scales were present on the chest and upper arms. Biopsy of the lesions revealed pathological features typical of EV warts with minimal papillomatosis and hyperkeratosis, viral cytopathic effects, and a blue-gray hue.
Genome-wide linkage analysis and inbreeding coefficient determination
Genome-wide linkage analysis was performed essentially as previously described (Grant et al., 2011) but with minor modifications. 22 members of kindred A, P14, P15, and six members of kindred E were genotyped with the Affymetrix Genome-wide Human SNP array 6.0. P10 was genotyped with the Affymetrix Genome-wide Human Mapping 250K, and P11 was genotyped with the Affymetrix CytoscanHD array. Four members of family F were genotyped with the Illumina Infinum Global Screening Array (642,824 SNP markers) using 200 ng genomic DNA in accordance with the manufacturer’s instructions (Illumina). Genotypes were called with the Power Tools Software Package (Affymetrix), and Chromosome Analysis Suite Software was used for P11. SNPs presenting more than one Mendelian inconsistency were discarded. SNPs were further filtered with population-based filters with PLINK software (Purcell et al., 2007) according to the ethnicity of the kindred. A subset of markers common to the three arrays and optimized to decrease their linkage disequilibrium (LD) was used for parametric multipoint linkage analyses in MERLIN (Abecasis et al., 2002) for kindreds A1, A2, B, D, E, and F, considering founders to be second-degree relatives and assuming AR inheritance with complete penetrance and a deleterious allele frequency of 10−4. The family founders and unrelated individuals from HapMap CEU, MEX, and YRI were used to estimate allele frequencies for kindreds B and D, A1 and A2, and E, respectively, and to define linkage clusters with an r2 threshold of 0.4. For kindred F, we estimated allele frequencies from an unrelated Iranian cohort of 82 healthy individuals.
We estimated the inbreeding coefficient F by Markov processes to model homozygous states throughout the genome according to the FEstim method (Leutenegger et al., 2003). We used the FEstim_SUBS method to minimize LD between SNPs as recommended in a previous study (Gazal et al., 2014) for the random extraction of sparse markers at 0.5-cm intervals to create 100 submaps. This strategy does not require the estimation of LD scores for the data, and F is estimated by calculating the median value of the estimates obtained from the different maps. FSuite v1.0.3 software was used to calculate FEstim and to infer the degree of parental consanguinity for each individual from the genome-wide human SNP array data (Gazal et al., 2014).
WES
The method used for WES has been described previously (Bolze et al., 2010; Byun et al., 2010). Briefly, genomic DNA extracted from the patients’ blood cells or LCLs was sheared with a Covaris S2 Ultrasonicator. An adapter-ligated library was prepared with the Paired-End Sample Prep kit V1 (Illumina). Exome capture was performed with the SureSelect Human All Exon kit (50- or 71-Mb version; Agilent Technologies). Sequencing was performed on an Illumina Genome Analyzer IIx (Illumina) generating 72- or 100-base reads. For kindred F, DNA was extracted from peripheral blood samples by the salting out method. DNA concentration was measured with a Qubit 2.0 fluorimeter (Thermo Fisher Scientific). WES was performed for probands and affected family members. Exons and flanking intronic regions of ∼20,000 genes (45 Mb; 98% of consensus coding sequence exons) were captured with the Illumina TruSeq Exome Enrichment Kit and sequenced on a NextSeq 500 instrument (Illumina). We used a BWA-MEM algorithm (Li and Durbin, 2009) to align the sequences with the reference human genome sequence (hg19build). Downstream processing was performed with the Genome Analysis Toolkit (GATK; McKenna et al., 2010), SAMtools (Li et al., 2009), and Picard Tools (http://broadinstitute.github.io/picard/). Substitution calls were made with GATK UnifiedGenotyper, whereas indel calls were made with SomaticIndelDetector V2. All calls with a read coverage of <2× and a Phredscaled SNP quality of <20 were filtered out. Single-nucleotide variants were filtered on the basis of dbSNP135 (http://www.ncbi.nlm.nih.gov/SNP/) and 1000Genomes (http://www.internationalgenome.org) data. All variants were annotated with ANNOVAR (Wang et al., 2010). The sequence data are available from the Sequence Read Archive website under accession number SRP151153. All variant calls with a variant quality <200, depth of coverage <5, and mapping quality <40 were filtered out. Only exonic and splice variants were retained, and synonymous and heterozygous variants were subsequently removed. Single-nucleotide variants with a MAF of >0.1% in public databases were then excluded, given the rarity of EV. This process identified 26–56 variants per patient.
Sanger sequencing of genomic DNA
Genomic DNA samples from patients and their relatives were used as a template for the amplification of 300–600-bp regions encompassing the mutation by PCR with site-specific oligonucleotides. Amplicons were sequenced with BigDye Terminator technology on an ABI 3730 DNA sequencer. Lasergene Suite and SnapGene were used for sequence analysis (DNASTAR and SnapGene).
IHC
Briefly, skin punch biopsy specimens (6 mm in diameter) were obtained from waste skin removed from individuals undergoing abdominoplasty (two individuals). Biopsy specimens were frozen in optimal cutting temperature compound (Sakura) and stored at −80°C. Tissue sections were air dried, fixed with acetone, blocked by incubation with 10% normal serum, and stained with the anti-CIB1 monoclonal antibody UN-2 (Naik et al., 1997) or an IgG1-κ isotype control (Sigma-Aldrich) at a concentration of 13 µg/ml overnight at 4°C. Tissue sections were thoroughly washed and incubated with biotin-labeled horse anti-mouse secondary antibody (Vector Laboratories) for 30 min at room temperature. The slides were then treated with 0.3% H2O2 and washed, and the staining signal was amplified with avidin–biotin complex Vectastain (Vector Laboratories), activated with 0.015% H2O2, and developed with a 5% solution of the chromogen 3-amino-9-ethylcarbazole (Sigma-Aldrich) for ∼20 min. IHC was performed on specimens from four healthy donors.
RNA sequencing
Total RNA was isolated from primary keratinocytes from P5 and P14, two healthy donors (control 1 and control 2), and cells from P5 stably expressing GST as an irrelevant protein or complemented with an untagged CIB1 construct. Total cellular mRNA was prepared for sequencing with the NextSeq 500/550 High Output v2 kit. We generated 75-bp single reads on a NextSeq 500. Reads were mapped onto the reference genome with STAR v2.5.3a (Dobin et al., 2013). A genome index specific to our data was first created, and the single-end reads were then aligned in a two-pass strategy. In this strategy, new splicing junctions are first detected and used for the final mapping. Read counts were obtained for each gene with HTSeq v. 0.9.1 (Anders et al., 2015). Gene expression levels were estimated with Transcripts Per Kilobase Million (TPM). TPM takes the size of each sample library and gene lengths into account to assess expression levels (Conesa et al., 2016). Statistical analysis was performed with R v.3.2.3 (The R Foundation, 2015). Gene expression profiles are represented as the log(fold change) in expression values between two different groups.
RT-qPCR
Human tissue
RNA was isolated from LCLs, primary keratinocytes, and HEK293T cells with the RNeasy mini kit and from whole blood with the PaxGene blood RNA kit (QIAGEN). Reverse transcription was performed with the SuperScript III first-strand synthesis system and oligo(dT) primers according to the manufacturer’s protocol (Thermo Fisher Scientific). RT-qPCR was then performed with TaqMan assays using exon-spanning probes for CIB1 (Hs01089679_m1), EVER1/TMC6 (Hs00273164_m1), or EVER2/TMC8 (Hs00380060_m1; Thermo Fisher Scientific) with normalization against GAPDH (4310884E), 18S (Hs99999901_s1), or RNaseP (4403326) as housekeeping genes by the ΔCT method. Data are represented as fold changes and were analyzed with the ΔΔCT method and comparison with an untreated control sample. The data shown are the means of three experiments.
Cell lines and primary cell isolation
HEK 293T cells were obtained from ATCC (CRL-3216). PBMCs were isolated by Ficoll-Hypaque density centrifugation (Amersham Pharmacia Biotech) from cytopheresis or whole-blood samples obtained from healthy volunteers and patients, respectively. EBV-immortalized LCLs (EBV-B cells) were obtained as previously described (Dupuis et al., 2001). Primary keratinocyte isolation: Punch biopsy specimens from the inner upper arm were washed three times with serum-free DMEM supplemented with 50 µg/ml gentamycin. They were then treated with dispase (354235; BD) at 4°C overnight. The following day, dermis and epidermis were separated manually and treated with Accutase (A6964; Sigma-Aldrich). The epidermis was then cut into pieces and wounded to obtain basal keratinocytes, which were cultured as described below.
Cell culture and transfection
HEK293T and HaCaT cells were maintained in DMEM supplemented with 10% FBS. LCLs were maintained in RPMI 1640 supplemented with 10% FBS. Primary keratinocytes were maintained on mitomycin-C–inactivated (10 µg/ml; 3 h) MEF 3T3-J2 feeder cells in complete Green medium (DMEM/Ham’s F12 in a 2:1 ratio supplemented with 10% FBS, 180 nM adenine, 10 ng/ml EGF, 0.4 µg/ml hydrocortisone, 8.47 ng/ml cholera toxin, 5 µg/ml insulin, 1.36 ng/ml triiodothyronine, and 10 µM ROCK inhibitor Y-27632). Transfections were performed with XtremeGENE 9 (Roche), FuGENE6 (Promega), Lipofectamine 2000 (Thermo Fisher Scientific), or Lipofectamine LTX (Thermo Fisher Scientific) in accordance with the manufacturer’s instructions.
Plasmids
Myc/DDK-tagged pCMV6-entry expression plasmids for EVER1/TMC6, EVER2/TMC8, CIB1, HPV5 E1, E2, E6, and E7 as well as HPV16 E1, E2, E5, E6, and E7 were obtained from Origene. An HA-tagged CIB1 expression construct was generated by inserting the CIB1 expression cassette from pCMV6-entry into pcDNA3.1 between the BamHI and EcoRI sites. CIB1 was amplified by PCR with the primers 5′-ATTAGGATCCGCCACCATGGGGGGCTCGGGCAGTCGC-3′ and 5′-TCGAGAATTCTCAAGCGTAATCTGGAACATCGTATGGGTACAGGACAATCTTAAAGGAGC-3′. DDK-tagged pcDNA3.1+ expression constructs for CRPV E8 and HPV4 E8 were generated by inserting the E8 ORF (NCBI Reference Sequence NC_001541) amplified from the CRPV reference genome with 5′-ATTAGGATCCGCCACCATGgactacaaggacgacgatgacaagGGACCTGCAGAGACTGC-3′ and 5′-tcgagaattcTTAATCTTCTTCCGCAAACTG-3′ primers between the BamHI and EcoRI sites and restriction cloning the purified synthesized codon-optimized HPV4 (gi|9626597) E8 ORF (from IDT) using the BamHI and EcoRI sites. Lowercase letters denote restriction sites.
We generated the pLZRS-IRES-ΔNGFR (de Paus et al., 2013; Martinez-Barricarte et al., 2016) expression plasmids for EVER1/TMC6 and EVER2/TMC8 with the cold fusion cloning kit (System Bioscience) in accordance with the manufacturer’s instructions after linearization with EcoRI. The Myc/DDK-tagged expression cassettes were amplified by PCR from the abovementioned pCMV6 entry clones with the following primers: EVER1, 5′-ATTTAAATTCGAATTATGGCCCAGCCACTGGC-3′ and 5′-AGGCCTGCAGGAATTTTAAACCTTATCGTCGTCATCCTTGTAATCCAGGATATCATT-3′; and EVER2, 5′-ATTTAAATTCGAATTATGCTGCTGCCGCGG-3′ and 5′-AGGCCTGCAGGAATTTTAAACCTTATCGTCGTCATCCTTGTAATCCAGGATATCATT-3′. The integrity of all expression cassettes was confirmed by Sanger sequencing.
Retrovirus production and transduction
Retroviruses containing EVER1- or EVER2-encoding or empty vectors were produced with the pLZRS-IRES-ΔNGFR constructs described above in Phoenix A cells as previously described (Martinez-Barricarte et al., 2016). Freshly harvested retroviruses were used to infect LCLs from a healthy WT control and EV patients with EVER1 D576* m/m, EVER2 T150M fs*3, or CIB1 I156D fs*5. Cells were grown and purified as previously described (Martinez-Barricarte et al., 2016) but in RPMI 1640 supplemented with 10% FBS as the culture medium. Flow cytometry confirmed that the transduced and selected populations were >95% pure.
Immunoblotting and immunoprecipitation
Cells were lysed by incubation in radioimmunoprecipitation assay (RIPA) buffer supplemented with the inhibitors aprotinin, NaF, N-ethylmaleimide, Na3VO4, and leupeptin for 30 min on ice, and the resulting lysate was then cleared. For immunoprecipitation, 10% of the lysate was set aside as the input control. Cleared lysates were incubated with 1 µg of the indicated antibodies (FLAG, HA, GFP, or an Ig control) at 4°C on an overhead tumbler for 2–16 h. Protein A–coupled magnetic Dynabeads (50 µl from a stock solution; Thermo Fisher Scientific) were added, and antibody-bound protein was captured from the lysate by incubation for 1 h at 4°C on an overhead rotator. Beads were washed three to five times with 200 µl lysis buffer, resuspended in 100 µl lysis buffer, and transferred to new reaction tubes to prevent the coelution of protein bound nonspecifically to the wall of the tube. The beads were resuspended in 25–50 µl 2× Laemmli buffer and boiled at 95°C for 5 min. Samples were subjected to standard SDS-PAGE in Mini-Protean TGX 4–20% gels (Bio-Rad Laboratories), and the resulting bands were transferred onto polyvinylidene difluoride membrane by semidry transfer for 30–40 min at 200 mA and 25 V per gel. The primary antibodies used were specific for CIB1 (raised against amino acids 24–43 [9, 11]), the HA epitope (HA.11-16B12; Covance), the FLAG epitope (M2; HRP coupled or unlabeled; Sigma-Aldrich), IkBa (L35A5), pIkBa (5A5), and p100/p52 (18D10; Cell Signaling Technology), GAPDH HRP-coupled (FL-335), pFAK (2D11), and FAK (D-1; Santa Cruz Biotechnology, Inc.), vinculin (VIN-11-5; Sigma-Aldrich), GFP (JL-8; Takara Bio Inc.), and actin (137CT26.1.1; Abgent). We used normal rabbit and mouse IgG (Santa Cruz Biotechnology, Inc.) as immunoprecipitation controls. The secondary antibodies used were HRP-coupled anti-mouse IgG and anti-rabbit IgG (GE Healthcare) or anti-chicken IgY (Jackson ImmunoResearch Laboratories, Inc.). Antibody binding was detected by enhanced chemiluminescence with hyperfilm ECL or an RGB 600 Imager (GE Healthcare). The data shown are representative of at least three independent experiments unless too little material was available. See the figure legends for details.
Immunofluorescence, PLA, and microscopy
Primary keratinocytes were plated on poly-L-lysine–coated coverslips and transfected as indicated above. The cells were fixed by incubation with 4% PFA 24 h after transfection, permeabilized, and blocked by incubation in 0.2% Triton X-100, 3% BSA, and 10% normal serum, and then they were incubated overnight with the primary antibody. The antibodies used were directed against the FLAG (anti-DDK; Origene) and HA epitopes (Sigma-Aldrich) and were used at a dilution of 1:500. The secondary antibodies used were Alexa Fluor 488– or 568–conjugated goat anti-mouse or anti-rabbit IgG (Thermo Fisher Scientific) at a dilution of 1:500. Cells were counterstained with DAPI (0.5 µg/ml). Coverslips were mounted on slides in Prolong Antifade Gold (Thermo Fisher Scientific). Images were acquired with a DeltaVision Image Restoration microscope (Applied Precision Ltd.) using a 60× oil-immersion objective and 0.24-µm steps. Images were deconvoluted with SoftWoRx software. Deconvoluted z stacks were subjected to automated image and colocalization analysis with Imaris software (Bitplane). Representative data from three independent experiments are shown. For focal adhesion analysis, primary keratinocytes were plated on fibronectin-coated (10 µg/ml; Corning) coverslips, to which they were allowed to adhere for 24 h. Cells were fixed with 4% PFA, permeabilized with 0.3% Triton X-100, blocked in 5% normal serum and 0.3% Triton X-100, and incubated overnight with the primary antibody. The antibodies used were directed against vinculin (VIN-11-5; Sigma-Aldrich) and pFAK (44-624G; Thermo Fisher Scientific), and they were used at a dilution of 1:500. The secondary antibodies used were Alexa Fluor 488–conjugated goat anti-mouse or anti-rabbit IgG at a dilution of 1:500–1,000 (Thermo Fisher Scientific). Cells were counterstained with Alexa Fluor 568–phalloidin (1 U phallotoxin/slide; Thermo Fisher Scientific) and DAPI (0.5 µg/ml). Images were acquired with an AxioObserver.Z1 epifluorescence microscope (ZEISS) equipped with an ORCA-ER camera (Hamamatsu Photonics) and an ApoTome.2 (ZEISS) with a 40× 1.4 NA oil immersion objective in 0.24-µm steps with Zen software (ZEISS). Z stacks were subjected to automated image analysis and surface detection with Imaris software. Focal adhesions were defined as vinculin- or pFAK-stained surfaces with a volume >0.002 mm3 within the lower half of the z stack. The parameters analyzed included volume, fluorescence intensity, and morphology (measured as ellipticity). Values were normalized relative to the total number of surfaces detected to obtain a mean per detected surface structure. The data shown are the mean and SD of three experiments with eight data points per set. For PLAs, HaCaT cells were transfected with CIB1-HA or the indicated myc-DDK–tagged HPV5 and HPV16 E ORF plasmids alone or in combination. Cells were transferred to poly-L-lysine–coated coverslips after 24 h. They were allowed to adhere for another 24 h and were then fixed in ice-cold acetone. Duolink PLA (Sigma-Aldrich) was performed in accordance with the manufacturer’s instructions with anti-rabbit HA (Sigma-Aldrich) and anti-mouse DDK (Origene) antibodies at a dilution of 1:400 and then were incubated overnight at 4°C with the cells. Duolink In Situ Detection Reagents Red was used for visualization.
Reporter gene assay
HEK293T cells were cotransfected with the indicated plasmids and pMRE-GFP, a GFP reporter construct driven by four copies of a MRE 5′-TGCACTC-3′ (provided by W. Schaffner, University of Zurich, Zurich, Switzerland). This element is activated by the zinc-dependent metal transcription factor 1 (MTF-1). After 24 h, cells were stimulated with PMA/ionomycin (10 ng/ml and 50 ng/ml; Sigma-Aldrich) or ZnSO4 (100 µM) overnight. The next day, cells were stained with 1 µg/ml DAPI to exclude dead cells, and fluorescence was acquired with an LSRII flow cytometer (BD). Results are shown as the relative response ratio (RRR) calculated as RRR= 100% × (sample − negative control)/(positive control − negative control). Untreated mock-transfected cells served as the negative control, and untreated vector-transfected cells served as the positive control. The data shown are the mean and SD of three independent experiments.
Determination of absolute free zinc levels and zinc kinetics
Labile zinc levels in LCLs and keratinocytes were determined by flow cytometry as previously described (Haase et al., 2006). Briefly, cells were loaded with the zinc-specific fluorescent probe FluoZin-3 (1 µM; Thermo Fisher Scientific) for 30 min at room temperature. Samples were either left untreated (F sample) or were treated with ZnSO4 and the zinc ionophore pyrithione/2-mercaptopyridine N-oxide (referred to as Fmax; 100 µM and 50 µM; Sigma-Aldrich) or the zinc chelator TPEN (Fmin; 100 µM; Sigma-Aldrich). All samples were stained with DAPI (0.5 µg/ml) to exclude dead cells. Fluorescence was recorded on an LSRII flow cytometer. The concentration of intracellular labile zinc was calculated from the mean fluorescence according to the formula [Zn] = KD × ([F − Fmin]/[Fmax − F]). The dissociation constant of the FluoZin-3/zinc complex is 15 nM. The data shown are the means of three independent experiments for LCLs and the mean and SD of three independent experiments for keratinocytes. For zinc kinetic measurements, LCLs were stained with FluoZin-3 (1 µM; 30 min at room temperature) in HBSS (Thermo Fisher Scientific) and then allowed to rest for 60 min at room temperature. Fluorescence was recorded on a VICTOR plate reader (PerkinElmer). Baseline fluorescence was monitored every 60 s for 10 min. Cells were then sequentially loaded with ZnSO4 (100 µM), the calcium chelator BAPTA (25 µM; Sigma-Aldrich), and the zinc-specific chelator TPEN (200 µM). In each set of conditions, fluorescence was determined every 60 s for 10–20 min. BAPTA treatment ensured the specificity of the zinc signal. Unstained cells were used to correct values for background fluorescence. The data shown are the mean and SD for three independent experiments.
Flow cytometry/immunophenotyping
Whole blood was either processed directly or used for the isolation of PBMCs on a Ficoll density gradient. The PBMCs obtained were then either processed directly or frozen for future use. We lysed the red blood cells by incubating 100 µl fresh whole blood with fluorophore-conjugated antibodies for 20 min and then with FACS lysing solution (BD) for 10 min at room temperature. The antibodies used for staining were CD45-FITC, CD3-PE-Cy7, CD19-PE-Cy7, CD4–Pacific Blue, CD14-V450, CD8-PE-Cy5, CD8-V450, or CD16-CD56-PE, CD45RA-FITC, CCR7-PE, and CD3-APC. Fluorescence was measured on a FACSCanto II flow cytometer (BD). Frozen PBMCs were thawed, washed, and incubated with Zombie UV viability dye (BioLegend) and then with fluorophore-conjugated antibodies in brilliant stain buffer (BD) for 30 min. The antibodies used were CD45RO-BV421, CD4-BV605, CCR4-BV421, CLA–Alexa Fluor 647, CD45RA–Alexa Fluor 488, CCR7–Alexa Fluor 647, CD8-PE-Cy7, HLA-DR-FITC, CD16-APC-Cy7, CD20-PE (BioLegend), CD3-PerCP-Cy5.5, CCR10-BB515, and CD56-APC (BD). Fluorescence was measured on an LSRII flow cytometer. Data were analyzed with FlowJo software (v.9.8.3; Tree Star). Whole-blood experiments were performed for two patients (P3 and P5). Frozen PMBCs from healthy donors (n = 5), WT family members (n = 5), healthy heterozygous family members (n = 4), and patients (n = 5; P1–P4 and P15) were used. The data were obtained in a single–time point experiment.
T cell proliferation assay
Freshly isolated PBMCs were loaded with carboxyfluorescein succinimidyl ester (5 µM; Invitrogen) for 5 min, washed, and plated at a density of 1 × 105 cells/ml in RPMI 1640/10% FBS supplemented with 2% penicillin/streptomycin on plates coated with anti-CD3 antibody (5, 2.5, or 1.25 µg/ml; clone UCHT1). Cells were then stimulated with anti-CD28 antibody (1 µg/ml; clone ANC28/5D10) for 3 d at 37°C in a humidified incubator under an atmosphere containing 5% CO2. Cells were harvested and processed for flow cytometry by incubation with an APC-conjugated anti-CD3 antibody in flow cytometry buffer for 20 min. Fluorescence was measured on a FACSCanto II flow cytometer. The data were analyzed with FlowJo software using the proliferation analysis tool. The parameters displayed included proliferation index (the total number of divisions divided by the number of cells entering division), division index (the mean number of cell divisions undergone by a cell from the original population), and percent division (precursor frequency). The data shown were obtained in a single–time point experiment on specimens from two patients (P3 and P5) and two controls processed in parallel.
Scratch wound healing assays
Primary keratinocytes were plated on fibronectin-coated (10 µg/ml; Corning) glass-bottomed six-well culture plates (MatTek). The cells were allowed to adhere to the plates overnight, and the resulting monolayer was then scratched with a pipette tip. Wound closure was monitored every 20 min for 16 h with an inverted Andor-driven spinning-disk confocal system equipped with a Zyla 4.2 (Andor Technology) camera in brightfield conditions with a 10× 0.3 NA air objective. During imaging, cells were maintained at 37°C under an atmosphere containing 5% CO2. The images were subjected to automated analysis with the MiToBo plugin in ImageJ (National Institutes of Health) to determine the cell-free area, which was normalized relative to the initial cell-free area and expressed as a percentage. The start and endpoint images were also manually analyzed by measuring the total cell-free area with the freehand tool. The data shown are the means for two to three experiments with at least six data points per experiment.
Large-scale protein purification and mass spectrometry analysis
For identification of the cellular partners interacting with CIB1, we transfected HEK239T cells (∼107 cells per 10-cm dish) in 10 dishes with 12 µg plasmid encoding CIB1-FLAG or ORF75-FLAG (control) in the presence of GenJet transfection reagent (SignaGen). 36 h later, the cells were lysed with RIPA buffer supplemented with protease inhibitor cocktail and centrifuged at 13,000 rpm for 20 min at 4°C. Lysates were precleared with 100 µl Sepharose 4B (Sigma-Aldrich) to remove cell debris and then were mixed with a ∼50% slurry of anti-FLAG M2 Affinity Gel (Sigma-Aldrich) and incubated for 4 h at 4°C. The beads were thoroughly washed in lysis buffer, and the bound proteins were eluted by heating samples in 2× Laemmli SDS sample buffer for 5 min at 95°C before separation by electrophoresis on a NuPAGE 4–12% Bis-Tris gradient gel (Thermo Fisher Scientific). The coimmunoprecipitated proteins were stained by incubation with colloidal Coomassie solution (0.02% Coomassie Brilliant Blue G-250, 5% aluminum sulfate-(14–18)-hydrate, 10% ethanol, and 8% orthophosphoric acid [85%]; Kang et al., 2002) at room temperature overnight. Bands specifically present in the CIB1-FLAG sample but absent from the ORF75-FLAG sample were excised and analyzed by ion-trap mass spectrometry at the Taplin Biological Mass Spectrometry Facility (Harvard Medical School, Boston, MA).
Gene ontology analysis
Gene ontology analysis was performed according to the PANTHER classification system release 20170413 (Mi et al., 2013) with the overrepresentation test and the Reactome version 58 (release 20161207) against the Homo sapiens database. Bonferroni correction for multiple testing was applied. 239 mapped hits were run against 21,042 database entries. The data are represented as fold enrichments, with P values <0.05 considered significant.
Statistical analyses
Statistical analyses were performed with Prism 5.0 (GraphPad Software).
Online supplemental material
Fig. S1 depicts photographs of the clinical EV manifestation of the studied patients. Fig. S2 depicts the results of GWL analysis and whole-exome filtering performed on the studied patient cohort and shows chromatographs obtained by Sanger sequencing that confirmed the mutations identified in CIB1. Fig. S3 shows qPCR expression data of EV-related genes after interferon treatment of primary human control keratinocytes. Fig. S4 shows the results obtained by performing mass spectrometry after immunoprecipitation of CIB1 expressed in HEK293T cells as well as immunofluorescent microscopy and colocalization analysis of CIB1 and ZnT1 in primary human control keratinocytes. Table S1 shows the results obtained by immunophenotyping for the studied patients using flow cytometry. Table S2 shows the T cell proliferation. Table S3 shows the viral serologies. Table S4 shows the skin-homing T cell subpopulations. Table S5 shows the inbreeding coefficient. Table S6 shows the transcriptomic analysis.
Supplementary Material
Acknowledgments
We thank the patients and their families for their collaboration, both branches of the Laboratory of Human Genetics of Infectious Diseases for helpful discussions and support, J. Bustamante for translations and discussions, J.A. Alvarez and Y. Lopez for sample collection and flow cytometry support, B. Vieco and M. Velazquez from the Dermatology section and Pathology Department of the University of Antioquia for their assistance, and Y. Nemirovskaya, E. Anderson, M. Woollett, L. Amar, C. Patissier, and D. Papandrea for administrative support. We wish to dedicate this study to the memory of Professor Luis-Alfredo Rueda and Professor Pierre Agache for their invaluable collaboration with one of us (G. Orth). We would like to thank the gnomAD and the groups that provided exome and genome variant data to this resource. We thank Ross Tomaino (Harvard Medical School, Boston, MA) for mass spectrometry analysis.
The Laboratory of Human Genetics of Infectious Diseases is supported by grants from the National Institutes of Health (grant 5 R21 AI107508-02) and the French Cancer Institute (grant 2013-1-PL BIO-11-1). L.V. Parise and T.M. Leisner are supported by National Institutes of Health grant R41 CA200189. S.J. de Jong was supported by the German Research Foundation (Jo 1151-1), funds from the Women & Science Fellowship program at The Rockefeller University, and the National Institutes of Health Clinical and Translational Science Award program (UL1 TR001866). J.L. Franco and A.A. Arias were supported by the Fundación Diana Garcia de Olarte FIP and Colciencias Programa de Intercambio de investigadores e innovadores Colombia-Francia (ECOS-NORD/COLCIENCIAS/MEN/ICETEX; 619-2013 and Colciencias contract 713-2016 code 111574455633).
T.M. Leisner and L.V. Parise are cofounders of Reveris Therapeutics LLC. The authors declare no additional competing financial interests.
Author contributions: S.J. de Jong designed, performed, and interpreted most of the experiments and wrote the original manuscript. A. Créquer, E. Imahorn, H. Vahidnezhad, L. Youssefian, A.H. Saeidian, and A. d’Amico performed exome analysis. I. Matos helped with scratch wound healing experiments. D. Hum, A.A. Arias, and L. Lorenzo helped for constructs and patients’ material. C.Q.F. Wang performed IHC experiments. A.A. Arias, J.L. Franco, and J.G. Markle helped for flow cytometry experiments. F. Jabot-Hanin, E. Patin, and M. Bouaziz helped for linkage and RNA sequencing analyses. N. Portilla Maya, X. Rueda Cadena, B. Saka, N. Aghazadeh, and S. Zeinali, were responsible for medical care of patients. F. Full, A. Ensser, T.M. Leisner, L.V. Parise, J.G. Krueger, L. Laimins, L. Abel, E. Fuchs, J. Uitto, and B. Burger provided expertise and methodology. G. Orth, E. Jouanguy, and J.-L. Casanova conceptualized the study, supported design of experiments, analysis, and interpretation of data, and wrote the original manuscript. All coauthors reviewed and edited the manuscript.
References
- Abecasis, G.R., Cherny S.S., Cookson W.O., and Cardon L.R.. 2002. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 30:97–101. 10.1038/ng786 [DOI] [PubMed] [Google Scholar]
- Anders, S., Pyl P.T., and Huber W.. 2015. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 31:166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Androphy, E.J., Dvoretzky I., and Lowy D.R.. 1985. X-linked inheritance of epidermodysplasia verruciformis. Genetic and virologic studies of a kindred. Arch. Dermatol. 121:864–868. 10.1001/archderm.1985.01660070054014 [DOI] [PubMed] [Google Scholar]
- Aochi, S., Nakanishi G., Suzuki N., Setsu N., Suzuki D., Aya K., and Iwatsuki K.. 2007. A novel homozygous mutation of the EVER1/TMC6 gene in a Japanese patient with epidermodysplasia verruciformis. Br. J. Dermatol. 157:1265–1266. 10.1111/j.1365-2133.2007.08206.x [DOI] [PubMed] [Google Scholar]
- Arnold, A.W., Burger B., Kump E., Rufle A., Tyring S.K., Kempf W., Häusermann P., and Itin P.H.. 2011. Homozygosity for the c.917A→T (p.N306l) polymorphism in the EVER2/TMC8 gene of two sisters with epidermodysplasia verruciformis Lewandowsky-Lutz originally described by Wilhelm Lutz. Dermatology (Basel). 222:81–86. 10.1159/000322536 [DOI] [PubMed] [Google Scholar]
- Azzimonti, B., Mondini M., De Andrea M., Gioia D., Dianzani U., Mesturini R., Leigheb G., Tiberio R., Landolfo S., and Gariglio M.. 2005. CD8+ T-cell lymphocytopenia and lack of EVER mutations in a patient with clinically and virologically typical epidermodysplasia verruciformis. Arch. Dermatol. 141:1323–1325. [DOI] [PubMed] [Google Scholar]
- Berthelot, C., Dickerson M.C., Rady P., He Q., Niroomand F., Tyring S.K., and Pandya A.G.. 2007. Treatment of a patient with epidermodysplasia verruciformis carrying a novel EVER2 mutation with imiquimod. J. Am. Acad. Dermatol. 56:882–886. 10.1016/j.jaad.2007.01.036 [DOI] [PubMed] [Google Scholar]
- Bolze, A., Byun M., McDonald D., Morgan N.V., Abhyankar A., Premkumar L., Puel A., Bacon C.M., Rieux-Laucat F., Pang K., et al. 2010. Whole-exome-sequencing-based discovery of human FADD deficiency. Am. J. Hum. Genet. 87:873–881. 10.1016/j.ajhg.2010.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgogna, C., Landini M.M., Lanfredini S., Doorbar J., Bouwes Bavinck J.N., Quint K.D., de Koning M.N., Genders R.E., and Gariglio M.. 2014. Characterization of skin lesions induced by skin-tropic α- and β-papillomaviruses in a patient with epidermodysplasia verruciformis. Br. J. Dermatol. 171:1550–1554. 10.1111/bjd.13156 [DOI] [PubMed] [Google Scholar]
- Bravo, I.G., and Alonso A.. 2004. Mucosal human papillomaviruses encode four different E5 proteins whose chemistry and phylogeny correlate with malignant or benign growth. J. Virol. 78:13613–13626. 10.1128/JVI.78.24.13613-13626.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruton, O.C. 1952. Agammaglobulinemia. Pediatrics. 9:722–728. [PubMed] [Google Scholar]
- Burge, C., and Karlin S.. 1997. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268:78–94. [DOI] [PubMed] [Google Scholar]
- Burger, B., and Itin P.H.. 2014. Epidermodysplasia verruciformis. Curr. Probl. Dermatol. 45:123–131. 10.1159/000356068 [DOI] [PubMed] [Google Scholar]
- Byun, M., Abhyankar A., Lelarge V., Plancoulaine S., Palanduz A., Telhan L., Boisson B., Picard C., Dewell S., Zhao C., et al. 2010. Whole-exome sequencing-based discovery of STIM1 deficiency in a child with fatal classic Kaposi sarcoma. J. Exp. Med. 207:2307–2312. 10.1084/jem.20101597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova, J.L. 2015a. Human genetic basis of interindividual variability in the course of infection. Proc. Natl. Acad. Sci. USA. 112:E7118–E7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova, J.L. 2015b. Severe infectious diseases of childhood as monogenic inborn errors of immunity. Proc. Natl. Acad. Sci. USA. 112:E7128–E7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciancanelli, M.J., Huang S.X., Luthra P., Garner H., Itan Y., Volpi S., Lafaille F.G., Trouillet C., Schmolke M., Albrecht R.A., et al. 2015. Life-threatening influenza and impaired interferon amplification in human IRF7 deficiency. Science. 348:448–453. 10.1126/science.aaa1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockayne, E.A. 1933. Inherited Abnormalities of the Skin and Its Appendages. Oxford University Press, London. 156 pp. [Google Scholar]
- Conesa, A., Madrigal P., Tarazona S., Gomez-Cabrero D., Cervera A., McPherson A., Szcześniak M.W., Gaffney D.J., Elo L.L., Zhang X., and Mortazavi A.. 2016. A survey of best practices for RNA-seq data analysis. Genome Biol. 17:13. 10.1186/s13059-016-0881-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crequer, A., Picard C., Patin E., D’Amico A., Abhyankar A., Munzer M., Debré M., Zhang S.Y., de Saint-Basile G., Fischer A., et al. 2012a. Inherited MST1 deficiency underlies susceptibility to EV-HPV infections. PLoS One. 7:e44010. 10.1371/journal.pone.0044010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crequer, A., Troeger A., Patin E., Ma C.S., Picard C., Pedergnana V., Fieschi C., Lim A., Abhyankar A., Gineau L., et al. 2012b. Human RHOH deficiency causes T cell defects and susceptibility to EV-HPV infections. J. Clin. Invest. 122:3239–3247. 10.1172/JCI62949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crequer, A., Picard C., Pedergnana V., Lim A., Zhang S.Y., Abel L., Majewski S., Casanova J.L., Jablonska S., Orth G., and Jouanguy E.. 2013. EVER2 deficiency is associated with mild T-cell abnormalities. J. Clin. Immunol. 33:14–21. 10.1007/s10875-012-9749-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deau, M.C., Favre M., and Orth G.. 1991. Genetic heterogeneity among human papillomaviruses (HPV) associated with epidermodysplasia verruciformis: evidence for multiple allelic forms of HPV5 and HPV8 E6 genes. Virology. 184:492–503. 10.1016/0042-6822(91)90419-C [DOI] [PubMed] [Google Scholar]
- Deau, M.C., Favre M., Jablonska S., Rueda L.A., and Orth G.. 1993. Genetic heterogeneity of oncogenic human papillomavirus type 5 (HPV5) and phylogeny of HPV5 variants associated with epidermodysplasia verruciformis. J. Clin. Microbiol. 31:2918–2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong, S.J., Imahorn E., Itin P., Orth G., Jouanguy E., Casanova J.-L., and Burger B.. 2018. Epidermodysplasia verruciformis: inborn errors of immunity to human beta-papillomaviruses. Front. Microbiol. Virol. 9:1222. 10.3389/fmicb.2018.01222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Paus, R.A., Geilenkirchen M.A., van Riet S., van Dissel J.T., and van de Vosse E.. 2013. Differential expression and function of human IL-12Rβ2 polymorphic variants. Mol. Immunol. 56:380–389. 10.1016/j.molimm.2013.07.002 [DOI] [PubMed] [Google Scholar]
- Dobin, A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggal, N.K., and Emerman M.. 2012. Evolutionary conflicts between viruses and restriction factors shape immunity. Nat. Rev. Immunol. 12:687–695. 10.1038/nri3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis, S., Dargemont C., Fieschi C., Thomassin N., Rosenzweig S., Harris J., Holland S.M., Schreiber R.D., and Casanova J.L.. 2001. Impairment of mycobacterial but not viral immunity by a germline human STAT1 mutation. Science. 293:300–303. 10.1126/science.1061154 [DOI] [PubMed] [Google Scholar]
- Dupuis, S., Jouanguy E., Al-Hajjar S., Fieschi C., Al-Mohsen I.Z., Al-Jumaah S., Yang K., Chapgier A., Eidenschenk C., Eid P., et al. 2003. Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat. Genet. 33:388–391. 10.1038/ng1097 [DOI] [PubMed] [Google Scholar]
- Fischer, A. 2015. Recent advances in understanding the pathophysiology of primary T cell immunodeficiencies. Trends Mol. Med. 21:408–416. 10.1016/j.molmed.2015.04.002 [DOI] [PubMed] [Google Scholar]
- García-Vallvé, S., Alonso A., and Bravo I.G.. 2005. Papillomaviruses: different genes have different histories. Trends Microbiol. 13:514–521. 10.1016/j.tim.2005.09.003 [DOI] [PubMed] [Google Scholar]
- Gaud, G., Guillemot D., Jacob Y., Favre M., and Vuillier F.. 2013. EVER2 protein binds TRADD to promote TNF-α-induced apoptosis. Cell Death Dis. 4:e499. 10.1038/cddis.2013.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazal, S., Sahbatou M., Babron M.C., Génin E., and Leutenegger A.L.. 2014. FSuite: exploiting inbreeding in dense SNP chip and exome data. Bioinformatics. 30:1940–1941. 10.1093/bioinformatics/btu149 [DOI] [PubMed] [Google Scholar]
- Giese, A.P.J., Tang Y.Q., Sinha G.P., Bowl M.R., Goldring A.C., Parker A., Freeman M.J., Brown S.D.M., Riazuddin S., Fettiplace R., et al. 2017. CIB2 interacts with TMC1 and TMC2 and is essential for mechanotransduction in auditory hair cells. Nat. Commun. 8:43. 10.1038/s41467-017-00061-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gober, M.D., Rady P.L., He Q., Tucker S.B., Tyring S.K., and Gaspari A.A.. 2007. Novel homozygous frameshift mutation of EVER1 gene in an epidermodysplasia verruciformis patient. J. Invest. Dermatol. 127:817–820. 10.1038/sj.jid.5700641 [DOI] [PubMed] [Google Scholar]
- Grant, A.V., Boisson-Dupuis S., Herquelot E., de Beaucoudrey L., Filipe-Santos O., Nolan D.K., Feinberg J., Boland A., Al-Muhsen S., Sanal O., et al. 2011. Accounting for genetic heterogeneity in homozygosity mapping: application to Mendelian susceptibility to mycobacterial disease. J. Med. Genet. 48:567–571. 10.1136/jmg.2011.089128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günther, V., Davis A.M., Georgiev O., and Schaffner W.. 2012. A conserved cysteine cluster, essential for transcriptional activity, mediates homodimerization of human metal-responsive transcription factor-1 (MTF-1). Biochim. Biophys. Acta. 1823:476–483. 10.1016/j.bbamcr.2011.10.006 [DOI] [PubMed] [Google Scholar]
- Haase, H., Hebel S., Engelhardt G., and Rink L.. 2006. Flow cytometric measurement of labile zinc in peripheral blood mononuclear cells. Anal. Biochem. 352:222–230. 10.1016/j.ab.2006.02.009 [DOI] [PubMed] [Google Scholar]
- Heineke, J., Auger-Messier M., Correll R.N., Xu J., Benard M.J., Yuan W., Drexler H., Parise L.V., and Molkentin J.D.. 2010. CIB1 is a regulator of pathological cardiac hypertrophy. Nat. Med. 16:872–879. 10.1038/nm.2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, J., Han R., Cladel N.M., Pickel M.D., and Christensen N.D.. 2002. Intracutaneous DNA vaccination with the E8 gene of cottontail rabbit papillomavirus induces protective immunity against virus challenge in rabbits. J. Virol. 76:6453–6459. 10.1128/JVI.76.13.6453-6459.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imahorn, E., Yüksel Z., Spoerri I., Gürel G., Imhof C., Saraçoğlu Z.N., Koku Aksu A.E., Rady P.L., Tyring S.K., Kempf W., et al. 2017. Novel TMC8 splice site mutation in epidermodysplasia verruciformis and review of HPV infections in patients with the disease. J. Eur. Acad. Dermatol. Venereol. 31:1722–1726. 10.1111/jdv.14431 [DOI] [PubMed] [Google Scholar]
- Ingle, A., Ghim S., Joh J., Chepkoech I., Bennett Jenson A., and Sundberg J.P.. 2011. Novel laboratory mouse papillomavirus (MusPV) infection. Vet. Pathol. 48:500–505. 10.1177/0300985810377186 [DOI] [PubMed] [Google Scholar]
- Itan, Y., Shang L., Boisson B., Patin E., Bolze A., Moncada-Vélez M., Scott E., Ciancanelli M.J., Lafaille F.G., Markle J.G., et al. 2015. The human gene damage index as a gene-level approach to prioritizing exome variants. Proc. Natl. Acad. Sci. USA. 112:13615–13620. 10.1073/pnas.1518646112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itan, Y., Shang L., Boisson B., Ciancanelli M.J., Markle J.G., Martinez-Barricarte R., Scott E., Shah I., Stenson P.D., Gleeson J., et al. 2016. The mutation significance cutoff: gene-level thresholds for variant predictions. Nat. Methods. 13:109–110. 10.1038/nmeth.3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joh, J., Jenson A.B., King W., Proctor M., Ingle A., Sundberg J.P., and Ghim S.J.. 2011. Genomic analysis of the first laboratory-mouse papillomavirus. J. Gen. Virol. 92:692–698. 10.1099/vir.0.026138-0 [DOI] [PubMed] [Google Scholar]
- Kang D.-H., Gho Y.-S., Suh M.-K., and Kang C.-H.. 2002. Highly Sensitive and Fast Protein Detection with Coomassie Brilliant Blue in Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis. Bull. Korean Chem. Soc. 11:1511–1512. [Google Scholar]
- Kawashima, Y., Géléoc G.S., Kurima K., Labay V., Lelli A., Asai Y., Makishima T., Wu D.K., Della Santina C.C., Holt J.R., and Griffith A.J.. 2011. Mechanotransduction in mouse inner ear hair cells requires transmembrane channel-like genes. J. Clin. Invest. 121:4796–4809. 10.1172/JCI60405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keresztes, G., Mutai H., and Heller S.. 2003. TMC and EVER genes belong to a larger novel family, the TMC gene family encoding transmembrane proteins. BMC Genomics. 4:24. 10.1186/1471-2164-4-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kienzler, J.L., Laurent R., Coppey J., Favre M., Orth G., Coupez L., and Agache P.. 1979. [Epidermodysplasia verruciformis. A case report including ultrastructural, virological and photobiological investigations (author’s transl)]. Ann. Dermatol. Venereol. 106:549–563 (Epidermodysplasia verruciformis. A case report including ultrastructural, virological and photobiological investigations (author's transl)). [PubMed] [Google Scholar]
- Kircher, M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., and Shendure J.. 2014. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46:310–315. 10.1038/ng.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluge, S.F., Sauter D., and Kirchhoff F.. 2015. SnapShot: antiviral restriction factors. Cell. 163:774–774. [DOI] [PubMed] [Google Scholar]
- Kostmann, R. 1950. Hereditär reticulos - en ny systemsjukdom. Svenska Läkartdin. 47:2861–2868. [Google Scholar]
- Kreins, A.Y., Ciancanelli M.J., Okada S., Kong X.F., Ramírez-Alejo N., Kilic S.S., El Baghdadi J., Nonoyama S., Mahdaviani S.A., Ailal F., et al. 2015. Human TYK2 deficiency: Mycobacterial and viral infections without hyper-IgE syndrome. J. Exp. Med. 212:1641–1662. 10.1084/jem.20140280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremsdorf, D., Favre M., Jablonska S., Obalek S., Rueda L.A., Lutzner M.A., Blanchet-Bardon C., Van Voorst Vader P.C., and Orth G.. 1984. Molecular cloning and characterization of the genomes of nine newly recognized human papillomavirus types associated with epidermodysplasia verruciformis. J. Virol. 52:1013–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurima, K., Yang Y., Sorber K., and Griffith A.J.. 2003. Characterization of the transmembrane channel-like (TMC) gene family: functional clues from hearing loss and epidermodysplasia verruciformis. Genomics. 82:300–308. 10.1016/S0888-7543(03)00154-X [DOI] [PubMed] [Google Scholar]
- Kurima, K., Ebrahim S., Pan B., Sedlacek M., Sengupta P., Millis B.A., Cui R., Nakanishi H., Fujikawa T., Kawashima Y., et al. 2015. TMC1 and TMC2 Localize at the Site of Mechanotransduction in Mammalian Inner Ear Hair Cell Stereocilia. Cell Reports. 12:1606–1617. 10.1016/j.celrep.2015.07.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laffort, C., Le Deist F., Favre M., Caillat-Zucman S., Radford-Weiss I., Debré M., Fraitag S., Blanche S., Cavazzana-Calvo M., de Saint Basile G., et al. 2004. Severe cutaneous papillomavirus disease after haemopoietic stem-cell transplantation in patients with severe combined immune deficiency caused by common gammac cytokine receptor subunit or JAK-3 deficiency. Lancet. 363:2051–2054. 10.1016/S0140-6736(04)16457-X [DOI] [PubMed] [Google Scholar]
- Lamborn, I.T., Jing H., Zhang Y., Drutman S.B., Abbott J.K., Munir S., Bade S., Murdock H.M., Santos C.P., Brock L.G., et al. 2017. Recurrent rhinovirus infections in a child with inherited MDA5 deficiency. J. Exp. Med. 214:1949–1972. 10.1084/jem.20161759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landini, M.M., Zavattaro E., Borgogna C., Azzimonti B., De Andrea M., Colombo E., Marenco F., Amantea A., Landolfo S., and Gariglio M.. 2012. Lack of EVER2 protein in two epidermodysplasia verruciformis patients with skin cancer presenting previously unreported homozygous genetic deletions in the EVER2 gene. J. Invest. Dermatol. 132:1305–1308. 10.1038/jid.2011.399 [DOI] [PubMed] [Google Scholar]
- Landini, M.M., Borgogna C., Peretti A., Colombo E., Zavattaro E., Boldorini R., Miglio U., Doorbar J., Ravanini P., Kumar R., et al. 2014. alpha- and beta-papillomavirus infection in a young patient with an unclassified primary T-cell immunodeficiency and multiple mucosal and cutaneous lesions. J. Am. Acad. Dermatol. 71:108–115. [DOI] [PubMed] [Google Scholar]
- Lazarczyk, M., Pons C., Mendoza J.A., Cassonnet P., Jacob Y., and Favre M.. 2008. Regulation of cellular zinc balance as a potential mechanism of EVER-mediated protection against pathogenesis by cutaneous oncogenic human papillomaviruses. J. Exp. Med. 205:35–42. 10.1084/jem.20071311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarczyk, M., Cassonnet P., Pons C., Jacob Y., and Favre M.. 2009. The EVER proteins as a natural barrier against papillomaviruses: a new insight into the pathogenesis of human papillomavirus infections. Microbiol. Mol. Biol. Rev. 73:348–370. 10.1128/MMBR.00033-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarczyk, M., Dalard C., Hayder M., Dupre L., Pignolet B., Majewski S., Vuillier F., Favre M., and Liblau R.S.. 2012. EVER proteins, key elements of the natural anti-human papillomavirus barrier, are regulated upon T-cell activation. PLoS One. 7:e39995. 10.1371/journal.pone.0039995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leisner, T.M., Freeman T.C., Black J.L., and Parise L.V.. 2016. CIB1: a small protein with big ambitions. FASEB J. 30:2640–2650. 10.1096/fj.201500073R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek, M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., et al. Exome Aggregation Consortium . 2016. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 536:285–291. 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leutenegger, A.L., Prum B., Génin E., Verny C., Lemainque A., Clerget-Darpoux F., and Thompson E.A.. 2003. Estimation of the inbreeding coefficient through use of genomic data. Am. J. Hum. Genet. 73:516–523. 10.1086/378207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandowsky, F., and Lutz W.. 1922. Ein Fall einer bisher nicht beschriebenen Hauterkrankung (Epidermodysplasia verruciformis). Arch. Dermatol. Syph. 141:193–203. 10.1007/BF01938833 [DOI] [Google Scholar]
- Li, H., and Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25:1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., and Durbin R.. 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25:2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S.L., Duo L.N., Wang H.J., Dai W., Zhou E.H., Xu Y.N., Zhao T., Xiao Y.Y., Xia L., Yang Z.H., et al. 2016. Identification of LCK mutation in a family with atypical epidermodysplasia verruciformis with T-cell defects and virus-induced squamous cell carcinoma. Br. J. Dermatol. 175:1204–1209. 10.1111/bjd.14679 [DOI] [PubMed] [Google Scholar]
- Liu, Y.Q., Zhang G.L., Mo X.H., Wang B., Wu F., Chen J., Luo H., Zhu L.D., Xu M.Y., Zhou Q., et al. 2017. A novel homozygous DOCK8 mutation associated with unusual coexistence of gross molluscum contagiosum and epidermodysplasia verruciformis in a DOCK8 deficiency patient. J. Eur. Acad. Dermatol. Venereol. 31:e504–e505. 10.1111/jdv.14344 [DOI] [PubMed] [Google Scholar]
- Lutz, W. 1946. A propos de l’epidermodysplasie verruciforme. Dermatologica. 92:30–43. 10.1159/000255805 [DOI] [PubMed] [Google Scholar]
- Martinez-Barricarte, R., de Jong S.J., Markle J., de Paus R., Boisson-Dupuis S., Bustamante J., van de Vosse E., Fleckenstein B., and Casanova J.L.. 2016. Transduction of Herpesvirus saimiri-Transformed T Cells with Exogenous Genes of Interest. Curr. Protoc. Immunol. 115. 10.1002/cpim.15 [DOI] [PubMed] [Google Scholar]
- Maufort, J.P., Williams S.M., Pitot H.C., and Lambert P.F.. 2007. Human papillomavirus 16 E5 oncogene contributes to two stages of skin carcinogenesis. Cancer Res. 67:6106–6112. 10.1158/0008-5472.CAN-07-0921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna, A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., and DePristo M.A.. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi, H., Muruganujan A., and Thomas P.D.. 2013. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 41(D1):D377–D386. 10.1093/nar/gks1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyauchi, T., Nomura T., Suzuki S., Takeda M., Shinkuma S., Arita K., Fujita Y., and Shimizu H.. 2016. Genetic analysis of a novel splice-site mutation in TMC8 reveals the in vivo importance of the transmembrane channel-like domain of TMC8. Br. J. Dermatol. 175:803–806. 10.1111/bjd.14569 [DOI] [PubMed] [Google Scholar]
- Naik, M.U., Nigam A., Manrai P., Millili P., Czymmek K., Sullivan M., and Naik U.P.. 2009. CIB1 deficiency results in impaired thrombosis: the potential role of CIB1 in outside-in signaling through integrin alpha IIb beta 3. J. Thromb. Haemost. 7:1906–1914. 10.1111/j.1538-7836.2009.03581.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik, U.P., Patel P.M., and Parise L.V.. 1997. Identification of a novel calcium-binding protein that interacts with the integrin alphaIIb cytoplasmic domain. J. Biol. Chem. 272:4651–4654. 10.1074/jbc.272.8.4651 [DOI] [PubMed] [Google Scholar]
- Nonnenmacher, M., Salmon J., Jacob Y., Orth G., and Breitburd F.. 2006. Cottontail rabbit papillomavirus E8 protein is essential for wart formation and provides new insights into viral pathogenesis. J. Virol. 80:4890–4900. 10.1128/JVI.80.10.4890-4900.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notarangelo, L., Casanova J.L., Fischer A., Puck J., Rosen F., Seger R., and Geha R.. International Union of Immunological Societies Primary Immunodeficiency diseases classification committee . 2004. Primary immunodeficiency diseases: an update. J. Allergy Clin. Immunol. 114:677–687. 10.1016/j.jaci.2004.06.044 [DOI] [PubMed] [Google Scholar]
- Orth, G. 2006. Genetics of epidermodysplasia verruciformis: Insights into host defense against papillomaviruses. Semin. Immunol. 18:362–374. 10.1016/j.smim.2006.07.008 [DOI] [PubMed] [Google Scholar]
- Orth, G. 2008. Host defenses against human papillomaviruses: lessons from epidermodysplasia verruciformis. Curr. Top. Microbiol. Immunol. 321:59–83. [DOI] [PubMed] [Google Scholar]
- Pan, B., Géléoc G.S., Asai Y., Horwitz G.C., Kurima K., Ishikawa K., Kawashima Y., Griffith A.J., and Holt J.R.. 2013. TMC1 and TMC2 are components of the mechanotransduction channel in hair cells of the mammalian inner ear. Neuron. 79:504–515. 10.1016/j.neuron.2013.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt, C.D., Fried A.J., Hoyos-Bachiloglu R., Usmani G.N., Schmidt B., Whangbo J., Chiarle R., Chou J., and Geha R.S.. 2017. Combined immunodeficiency with EBV positive B cell lymphoma and epidermodysplasia verruciformis due to a novel homozygous mutation in RASGRP1. Clin. Immunol. 183:142–144. 10.1016/j.clim.2017.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell, S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., and Sham P.C.. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81:559–575. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rady, P.L., De Oliveira W.R., He Q., Festa C., Rivitti E.A., Tucker S.B., and Tyring S.K.. 2007. Novel homozygous nonsense TMC8 mutation detected in patients with epidermodysplasia verruciformis from a Brazilian family. Br. J. Dermatol. 157:831–833. 10.1111/j.1365-2133.2007.08123.x [DOI] [PubMed] [Google Scholar]
- Ramoz, N., Rueda L.A., Bouadjar B., Montoya L.S., Orth G., and Favre M.. 2002. Mutations in two adjacent novel genes are associated with epidermodysplasia verruciformis. Nat. Genet. 32:579–581. 10.1038/ng1044 [DOI] [PubMed] [Google Scholar]
- Rueda, L.A.1993. Specific cytopathic effects of human papillomavirus in epidermodysplasia verruciformis. In Dermatology, progress & perspectives: the proceedings of the 18th World Congress of Dermatology, held in New York, June 12-18, 1992. W.H.C. Burgdorf, and S.I. Katz, editors. Parthenon Pub. Group, New York. 226-229. [Google Scholar]
- Rueda, L.A., and Rodriguez G.. 1976. Verrugas humanas por virus papova. Med. Cutan. Ibero Lat. Am. 2:113–136. [PubMed] [Google Scholar]
- Saka, B., Mouhari-Touré A., Kombaté K., Pitché P., and Tchangaï-Walla K.. 2009. Scattered papules in three Togolese children from a consanguineous marrige: epidermodysplasia verruciformis. Med. Trop. (Mars.). 69:293–294. [PubMed] [Google Scholar]
- Sanal, O., Jing H., Ozgur T., Ayvaz D., Strauss-Albee D.M., Ersoy-Evans S., Tezcan I., Turkkani G., Matthews H.F., Haliloglu G., et al. 2012. Additional diverse findings expand the clinical presentation of DOCK8 deficiency. J. Clin. Immunol. 32:698–708. 10.1007/s10875-012-9664-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, E.M., Halees A., Itan Y., Spencer E.G., He Y., Azab M.A., Gabriel S.B., Belkadi A., Boisson B., Abel L., et al. Greater Middle East Variome Consortium . 2016. Characterization of Greater Middle Eastern genetic variation for enhanced disease gene discovery. Nat. Genet. 48:1071–1076. 10.1038/ng.3592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepensky, P., Rensing-Ehl A., Gather R., Revel-Vilk S., Fischer U., Nabhani S., Beier F., Brümmendorf T.H., Fuchs S., Zenke S., et al. 2015. Early-onset Evans syndrome, immunodeficiency, and premature immunosenescence associated with tripeptidyl-peptidase II deficiency. Blood. 125:753–761. 10.1182/blood-2014-08-593202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stray-Pedersen, A., Jouanguy E., Crequer A., Bertuch A.A., Brown B.S., Jhangiani S.N., Muzny D.M., Gambin T., Sorte H., Sasa G., et al. 2014. Compound heterozygous CORO1A mutations in siblings with a mucocutaneous-immunodeficiency syndrome of epidermodysplasia verruciformis-HPV, molluscum contagiosum and granulomatous tuberculoid leprosy. J. Clin. Immunol. 34:871–890. 10.1007/s10875-014-0074-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, G., Kuzmanovic T., Zhang Y., Peter C.B., Veleeparambil M., Chakravarti R., Sen G.C., and Chattopadhyay S.. 2018. A new mechanism of interferon’s antiviral action: Induction of autophagy, essential for paramyxovirus replication, is inhibited by the interferon stimulated gene, TDRD7. PLoS Pathog. 14:e1006877. 10.1371/journal.ppat.1006877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, W., Guan Q., Wen J., Zhang Q., Yang W., Zhang B., Cui W., Zou Z., and Yu Y.. 2014. Calcium- and integrin-binding protein-1 is down-regulated in the sperm of patients with oligoasthenozoospermia: CIB1 expression in patients with oligoasthenozoospermia. J. Assist. Reprod. Genet. 31:541–547. 10.1007/s10815-014-0177-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, X.K., Chen J.F., and Xu A.E.. 2005. A homozygous nonsense mutation in the EVER2 gene leads to epidermodysplasia verruciformis. Clin. Exp. Dermatol. 30:573–574. 10.1111/j.1365-2230.2005.01858.x [DOI] [PubMed] [Google Scholar]
- Tahiat, A., Badran Y.R., Chou J., Cangemi B., Lefranc G., Labgaa Z.M., Oussalam S., Kaddouri-Slimani A., Belarbi A., Bendissari-Bouzid K., et al. 2016. Epidermodysplasia verruciformis as a manifestation of ARTEMIS deficiency in a young adult. J. Allergy Clin. Immunol. [DOI] [PubMed] [Google Scholar]
- Tate, G., Suzuki T., Kishimoto K., and Mitsuya T.. 2004. Novel mutations of EVER1/TMC6 gene in a Japanese patient with epidermodysplasia verruciformis. J. Hum. Genet. 49:223–225. 10.1007/s10038-004-0135-6 [DOI] [PubMed] [Google Scholar]
- The R Foundation . 2015. The R Project for Statistical Computing. https://www.r-project.org/
- Vuillier, F., Gaud G., Guillemot D., Commere P.H., Pons C., and Favre M.. 2014. Loss of the HPV-infection resistance EVER2 protein impairs NF-kappaB signaling pathways in keratinocytes. PLoS One. 9:e89479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, K., Li M., and Hakonarson H.. 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., Peng X., Zhang X., Xu H., Lu C., Liu L., Song J., and Zhang Y.. 2017. The Emerging Roles of CIB1 in Cancer. Cell. Physiol. Biochem. 43:1413–1424. [DOI] [PubMed] [Google Scholar]
- Wechsler, E.I., Tugizov S., Herrera R., Da Costa M., and Palefsky J.M.. 2018. E5 can be expressed in anal cancer and leads to epidermal growth factor receptor-induced invasion in a human papillomavirus 16-transformed anal epithelial cell line. J. Gen. Virol. 99:631–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssefian, L., Vahidnezhad H., Mahmoudi H.R., Saeidian A.H., Daneshpazhooh M., Kamyab-Hesari K., Zeinali S., de Jong S.J., Orth G., Blanchet-Bardon C., et al. 2018. Epidermodysplasia verruciformis: Extensive genetic heterogeneity and novel EVER1 and EVER2 mutations revealed by genome-wide analysis. J. Invest. Dermatol. In press. [DOI] [PubMed] [Google Scholar]
- Yuan, W., Leisner T.M., McFadden A.W., Clark S., Hiller S., Maeda N., O’Brien D.A., and Parise L.V.. 2006. CIB1 is essential for mouse spermatogenesis. Mol. Cell. Biol. 26:8507–8514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zayed, M.A., Yuan W., Leisner T.M., Chalothorn D., McFadden A.W., Schaller M.D., Hartnett M.E., Faber J.E., and Parise L.V.. 2007. CIB1 regulates endothelial cells and ischemia-induced pathological and adaptive angiogenesis. Circ. Res. 101:1185–1193. [DOI] [PubMed] [Google Scholar]
- Zhang, Q., Lenardo M.J., and Baltimore D.. 2017. 30 Years of NF-kappaB: A Blossoming of Relevance to Human Pathobiology. Cell. 168:37–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo, Y.G., Ma D., Zhang Y., Qiao J., and Wang B.. 2006. Identification of a novel mutation and a genetic polymorphism of EVER1 gene in two families with epidermodysplasia verruciformis. J. Dermatol. Sci. 44:153–159. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.