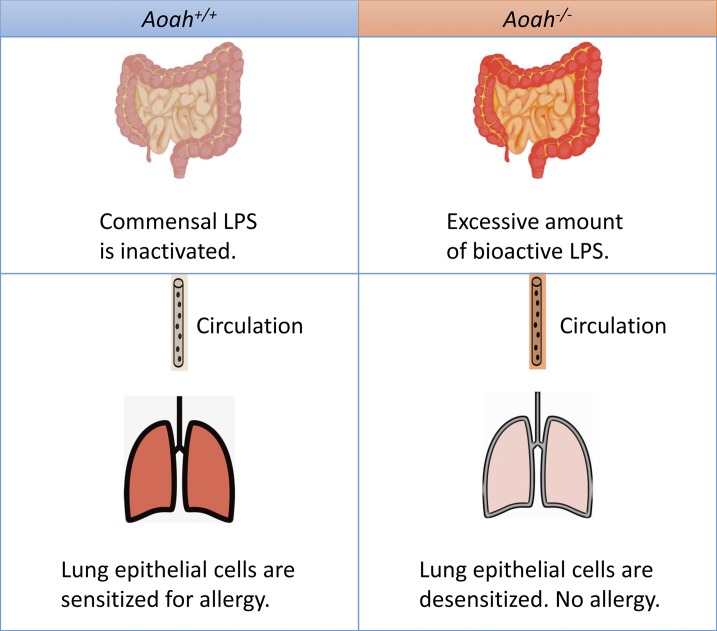
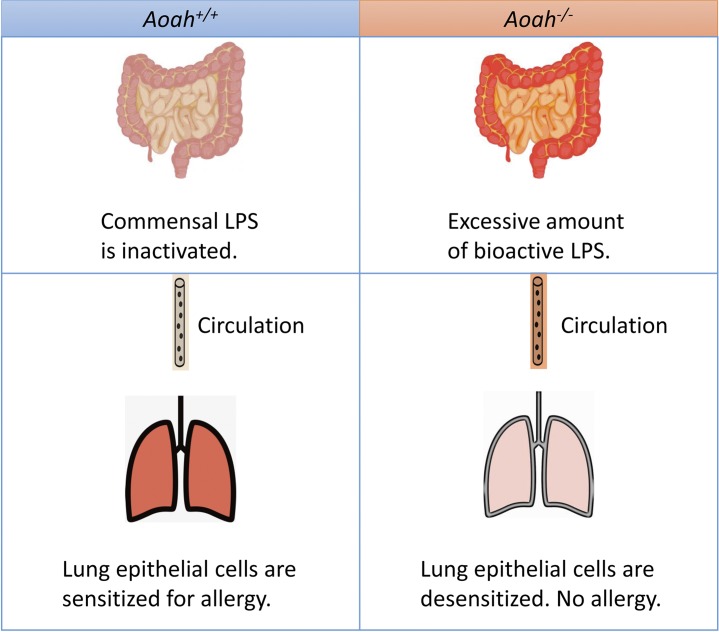
This study provides strong evidence that intestinal commensal LPS desensitizes lung epithelial cells and therefore diminishes allergic responses to inhaled allergens. A host lipase, acyloxyacyl hydrolase (AOAH), prevents the desensitization by inactivating commensal LPS.
Abstract
Allergic asthma is a chronic inflammatory disease primarily mediated by Th2 immune mechanisms. Numerous studies have suggested that early life exposure to lipopolysaccharide (LPS) is negatively associated with allergic asthma. One proposed mechanism invokes desensitization of lung epithelial cells by LPS. We report here that acyloxyacyl hydrolase (AOAH), a host lipase that degrades and inactivates LPS, renders mice more susceptible to house dust mite (HDM)–induced allergic asthma. Lung epithelial cells from Aoah−/− mice are refractory to HDM stimulation, decreasing dendritic cell activation and Th2 responses. Antibiotic treatment that diminished commensal LPS-producing bacteria normalized Aoah−/− responses to HDM, while giving LPS intrarectally ameliorated asthma. Aoah−/− mouse feces, plasma, and lungs contained more bioactive LPS than did those of Aoah+/+ mice. By inactivating commensal LPS, AOAH thus prevents desensitization of lung epithelial cells. An enzyme that prevents severe lung inflammation/injury in Gram-negative bacterial pneumonia has the seemingly paradoxical effect of predisposing to a Th2-mediated airway disease.
Graphical Abstract
Introduction
Allergic asthma is a common disease worldwide and recently its prevalence has increased dramatically in developed societies (Lambrecht and Hammad, 2015). In response to environmental allergens, such as house dust mite (HDM), cockroach, fungal spores, or pollen, the conducting airways develop chronic inflammation. Allergic asthma is most often a Th2-dominant inflammatory process although Th1/Th17 immunity also contributes to certain forms of asthma (Lambrecht and Hammad, 2015).
Airway epithelial cells, the first site of allergen exposure, orchestrate Th2 responses (Lambrecht and Hammad, 2012, 2014a). Expressing pattern recognition receptors, epithelial cells respond to microbe-associated molecular patterns (MAMPs) to release CCL-20, GM-CSF, MCP-1, IL-33, IL-25, and TSLP to recruit and activate dendritic cells (DCs) for Th2 polarization (Hammad and Lambrecht, 2008; Hammad et al., 2009; Lambrecht and Hammad, 2012, 2014a). Conventional CD11c+ CD11b+ CD103− lung resident DCs are able to acquire allergens and migrate to draining lymph nodes to induce allergen specific Th2 responses (sensitization phase), while monocyte-derived CD11b+ CD64+ FcεRI+ DCs are mainly responsible for effector T cell recruitment to the lung and their activation (challenge phase; van Rijt et al., 2005; Hammad et al., 2010; Plantinga et al., 2013). Epithelial cell–derived IL-33, IL-25, and TSLP also activate group 2 innate lymphoid cells (ILC-2) to promote both innate allergic inflammation and the adaptive Th2 response (Halim et al., 2014, 2016; Martinez-Gonzalez et al., 2015). Effector T cells release Th2 cytokines, such as IL-4, IL-5, IL-13, and IL-9, to drive allergen-specific IgE production, lung eosinophilia, mucus overproduction, and bronchial hyper-reactivity, which are characteristic features of allergic asthma (Locksley, 2010; Islam and Luster, 2012; Lambrecht and Hammad, 2015).
Genes, microbiota, and environmental factors all contribute to an individual’s susceptibility to allergic asthma. Epidemiological studies have suggested that high microbial exposures in traditional dairy farms may provide protection against allergic asthma (Braun-Fahrländer et al., 2002; von Mutius and Vercelli, 2010; Ege et al., 2011). Gram-negative bacterial membrane LPS (endotoxin), the most potent MAMP molecule that stimulates host cells via the MD2-TLR4 complex, has been present at high levels in air samples collected from farms, and the amount of LPS exposure has negatively correlated with the risk of developing asthma (Braun-Fahrländer et al., 2002; Doreswamy and Peden, 2011; Stein et al., 2016). Recently, Schuijs et al. have shown that chronic exposure to LPS up-regulates the negative cell signaling regulator A20 in airway epithelial cells, thereby reducing epithelial cytokine/chemokine responses to allergens, suppressing DC recruitment and Th2 immunity (Schuijs et al., 2015). Their findings suggest that airway exposure to LPS or farm dust increases the threshold for allergen recognition by epithelial cells, thus suppressing allergic responses (Schuijs et al., 2015). In addition to environmental MAMP exposure, intestinal microbiota also modulate allergic asthma (Russell et al., 2012, 2013; Arrieta et al., 2015), and a low abundance of intestinal LPS has been linked to a high risk of airway inflammation (Arrieta et al., 2015). Therefore, both environmental and commensal LPS may play a profound role in regulating allergic asthma (Huang and Boushey, 2015). To date, whether modulating LPS bioactivity may influence susceptibility to allergy has not been reported.
LPS can be deacylated and inactivated by a unique host enzyme, acyloxyacyl hydrolase (AOAH; Munford et al., 2009). AOAH specifically removes secondary fatty acyl chains from the lipid A moiety of LPS (Munford and Hall, 1986). The deacylated LPS not only has diminished bioactivity but may also serve as a LPS antagonist (Kitchens et al., 1992). Expressed mainly in phagocytes and NK cells, AOAH is known to limit LPS-induced B cell activation, polyclonal antibody production (Lu et al., 2005; Lu and Munford, 2011), and hepatomegaly (Shao et al., 2007, 2011). We have recently shown that AOAH-dependent LPS inactivation is required for the resolution of lung inflammation/injury induced by Gram-negative bacteria (Zou et al., 2017). Importantly, AOAH promotes recovery from endotoxin tolerance, a transient refractory state or cellular reprogramming that can follow LPS exposure (Biswas and Lopez-Collazo, 2009), allowing macrophages or DCs to respond to a subsequent LPS challenge (Lu et al., 2008, 2013; Janelsins et al., 2014). There are reports that intronic AOAH single nucleotide polymorphisms are associated with asthma (Barnes et al., 2006; Ferreira et al., 2017) or rhinosinusitis (Zhang et al., 2012), yet whether the enzyme promotes or ameliorates allergic airway responses has not been known.
We hypothesized that AOAH regulates allergic responses by modulating LPS bioactivity. We used an HDM-induced mouse asthma model to study allergic responses in Aoah−/− and Aoah+/+ mice. As one of the commonest aeroallergens, HDMs and their fecal pellets contain MAMP molecules including endotoxin, chitin, and β-glucans (Gregory and Lloyd, 2011); HDM also has Der p 2, which amplifies TLR4 signaling by mimicking MD-2 (Trompette et al., 2009). Notably, innate stimulation is required for Th2 allergic responses to inhaled allergens (Eisenbarth et al., 2002; Tan et al., 2010), and stimulation of TLR4 on lung structural cells, most likely epithelial cells, is necessary and sufficient to activate DCs and to promote allergic responses to HDM extract (Hammad et al., 2009). LPS, Der p 2, and other MAMPs in HDM may synergistically stimulate TLR4 to activate airway epithelial cells (Hammad et al., 2009). In this study, we found that Aoah−/− mice are resistant to HDM-induced allergic asthma, as manifested by reduced Th2 responses, defective DC recruitment and activation, diminished ILC-2 responses as well as refractory epithelial cell responses. We also provide evidence that intestinal commensal-derived LPS may desensitize Aoah−/− lung epithelial cells. Thus, AOAH may promote allergic asthma by inactivating commensal LPS and preventing airway epithelial cell desensitization. We show that a host-derived LPS-degrading lipase plays a previously unappreciated role in modulating allergic asthma.
Results
AOAH promotes HDM-induced allergic asthma
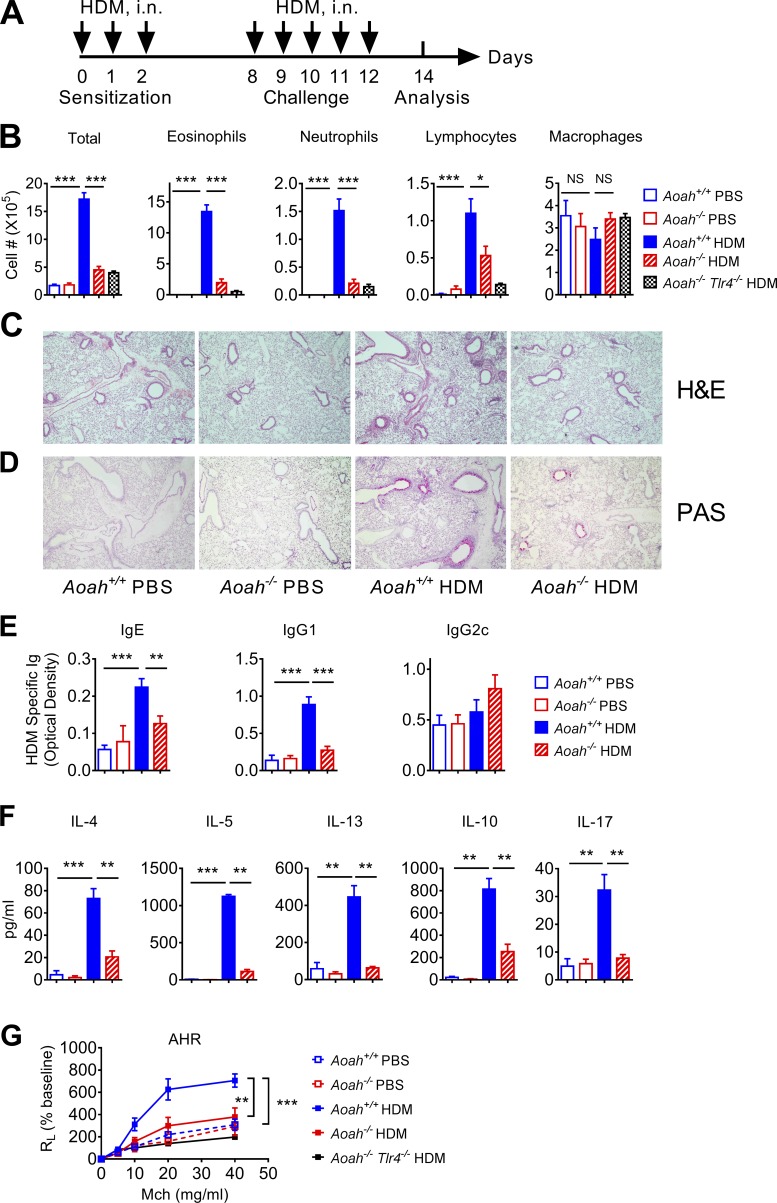
To find out the effects that AOAH may have on allergic asthma, we used an HDM-induced asthma model (Fig. 1 A). On day 14, bronchoalveolar lavage fluid (BALF) from HDM-treated Aoah+/+ mice had more eosinophils, neutrophils, and lymphocytes than did BALF from PBS-treated control mice, demonstrating that HDM induced allergic inflammation in Aoah+/+ airways (Fig. 1 B). In contrast, Aoah−/− mice had significantly reduced numbers of eosinophils, neutrophils, and lymphocytes in their airways (Fig. 1 B). Aoah−/−Tlr4−/− mice also had diminished eosinophilic airway inflammation (Fig. 1 B). In keeping with these findings, the lung histological studies also showed that HDM exposure induced much smaller peri-bronchial and peri-vascular leukocyte infiltrates in Aoah−/− mouse lungs (Fig. 1 C). PAS staining showed diminished goblet cell hyperplasia and mucus production in Aoah−/− airways (Fig. 1 D). We measured HDM-specific IgE, IgG1 (Th2 antibodies), and IgG2c (a Th1 antibody in C57BL/6J mice) in mouse sera and found that Aoah−/− mice had decreased levels of anti-HDM IgE and IgG1, but similar levels of anti-HDM IgG2c compared with Aoah+/+ mice, suggesting defective Th2 responses in Aoah−/− mice (Fig. 1 E). To further measure HDM-specific Th2 responses, we restimulated cells from the mediastinal lymph nodes (MLNs) with HDM allergen in vitro and found that MLN cells from HDM-treated Aoah−/− mice had significantly less Th2 cytokine (IL-4, IL-5, and IL-13) secretion than did MLN cells from Aoah+/+ mice. Notably, IL-10 and IL-17 production was also reduced in Aoah−/− MLN cells (Fig. 1 F). We also found reduced IL-4, IL-5, and IL-13 in Aoah−/− BALF (Fig. S1). When we measured methacholine-induced airway hyperresponsiveness (AHR), HDM-treated Aoah−/− mice had significantly reduced AHR compared with Aoah+/+ mice (Fig. 1 G). Collectively, in response to HDM stimulation, the presence of AOAH allowed higher Th2 responses, greater eosinophilic lung inflammation, more mucosal overproduction, and AHR, strong evidence that AOAH promotes HDM-induced allergic asthma.
Figure 1.
Aoah−/− mice have reduced HDM-induced allergic asthma. (A) Mice were sensitized with 10 µg HDM on days 0, 1, and 2 and challenged with 10 µg HDM daily from days 8–12. Mice were euthanized on day 14 for analysis. Control mice received PBS i.n. for sensitization and challenge. (B) Cells in BALF were analyzed following cytospin and Wright-Giemsa staining. Data were combined from three experiments. n = 9. (C) Paraformaldehyde-fixed lungs were cut and stained with H&E. n = 3–4. (D) Lung sections were stained with periodic acid-Schiff (PAS) to measure goblet cell hyperplasia and mucosal secretion in airways. n = 3–4. (E) HDM-specific IgE, IgG1, and IgG2c levels were measured in mouse sera using ELISA. Data were combined from three experiments. n = 9. (F) MLNs were harvested and single-cell suspensions were prepared. MLN cells were restimulated with medium containing 30 µg/ml HDM for 5 d. The culture media were used for ELISA assays. Data were combined from two experiments. n = 7–8. (G) In response to increasing doses of methacholine, the lung resistance was measured as described in Materials and methods. Data were combined from two experiments. n = 5–6. (B–F) Mann-Whitney test was used. (G) Two-way ANOVA was used. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
AOAH promotes lung dendritic cell and ILC-2 responses upon HDM stimulation
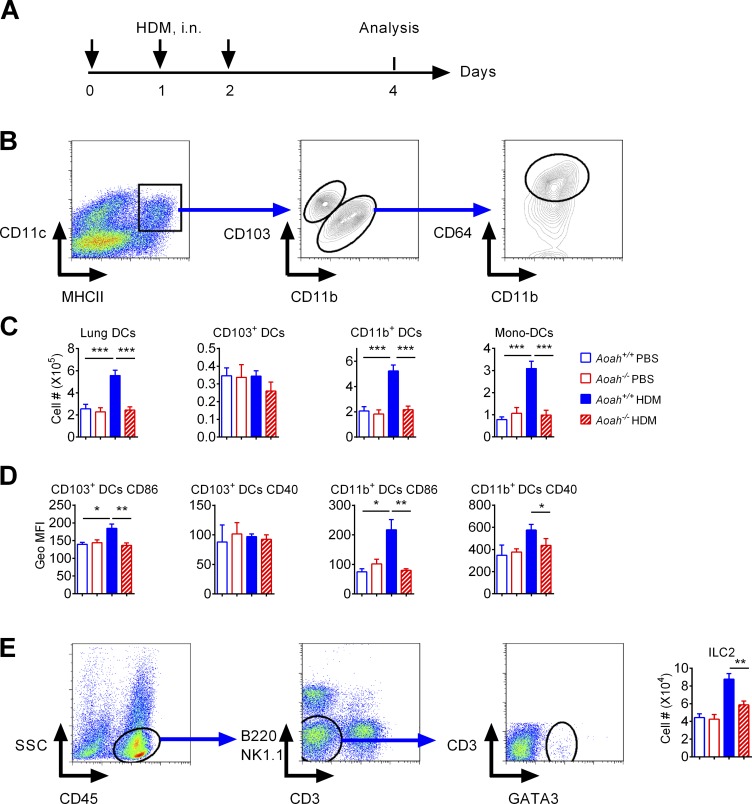
In response to allergen stimulation, lung DCs present allergens to naive T cells to initiate Th2 responses (van Rijt et al., 2005; Hammad et al., 2010; Plantinga et al., 2013). As Aoah−/− mice had defective Th2 responses to HDM, we wondered whether DC activation was impaired during the sensitization phase. Aoah+/+ or Aoah−/− mice were sensitized with HDM and their lung DC numbers and activation status were tested (Fig. 2 A). DCs were identified as CD11chi MHCIIhi Autofluorescencelo cells and they were further segregated as CD103+ DCs or CD11b+ DCs (Fig. 2 B). CD11b+ DCs included monocyte-derived DCs (CD64+) or conventional CD11b+ DCs (CD64−; Fig. 2 B; Plantinga et al., 2013). After HDM sensitization, Aoah+/+ lungs had significantly more DCs than did PBS-instilled control mice; the number of CD103+ DC did not increase while CD11b+ DC numbers significantly increased, largely due to greater abundance of monocyte-derived DCs (Fig. 2 C). CD11b+ DCs are known to be more important than CD103+ DCs in HDM-induced allergic responses (Plantinga et al., 2013). Aoah−/− mouse lung CD11b+ DCs, which included monocyte-derived DCs (CD11b+ CD64+), did not increase with HDM stimulation (Fig. 2 C), suggesting defective DC recruitment in Aoah−/− lungs. To confirm that Aoah−/− lungs have reduced monocyte-derived DCs after HDM sensitization, we labeled monocytes by injecting fluorescent microspheres (beads) i.v. (Tacke et al., 2006) and measured bead+ (monocyte-derived) DCs in the lung after HDM treatment (Fig. S2 A). Fluorescent beads labeled ∼3% of monocytes; Aoah+/+ and Aoah−/− monocytes were labeled with similar efficiency (Fig. S2, B and C). With HDM treatment, bead+ DCs were recovered in the lung, mainly in the CD11b+ DC population (Fig. S2 D). There were significantly fewer bead+ DCs in Aoah−/− lungs than in Aoah+/+ lungs (Fig. S2 E), consistent with the observation that Aoah−/− lungs had fewer monocyte-derived DCs (Fig. 2 C). When measuring DC activation markers, we found that CD103+ DCs from Aoah−/− mice had reduced CD86, and CD11b+ DCs from Aoah−/− mice had reduced CD86 and CD40, compared with their counterparts from Aoah+/+ mice (Fig. 2 D), suggesting that DCs from Aoah−/− mice were less activated after HDM exposure. As ILC-2 are known to be activated at early stage of sensitization upstream of DCs (Halim et al., 2014; Martinez-Gonzalez et al., 2015), we further checked ILC-2 responses to HDM in the lungs. The lung ILC-2 were identified as SSClo, CD45+, CD3−, B220−, NK1.1−, and GATA3+ cells (Fig. 2 E). ILC-2 numbers also decreased in HDM-treated Aoah−/− mouse lungs after HDM stimulation (Fig. 2 E). Collectively, after exposure to HDM, AOAH promotes ILC-2 and DC activation, in keeping with robust Th2 responses.
Figure 2.
Aoah−/− mouse lungs have diminished DC and ILC-2 responses to HDM. (A) Mice were sensitized with HDM on days 0, 1, and 2 and analyzed on day 4. (B) Gating strategy to identify DCs and DC subsets in mouse lungs. (C) Lung DCs and DC subset numbers in Aoah+/+ and Aoah−/− lungs. Data were combined from three experiments. n = 10–12. (D) DC activation markers in Aoah+/+ and Aoah−/− lungs. Data were combined from two experiments. n = 5–6. (E) Lung ILC-2 gating strategy and Aoah−/− mouse lungs had reduced ILC-2 numbers compared with Aoah+/+ mouse lungs after HDM exposure. Data were combined from two experiments. n = 5–6. (C–E) Mann-Whitney test was used. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
AOAH promotes airway epithelial recognition of allergens
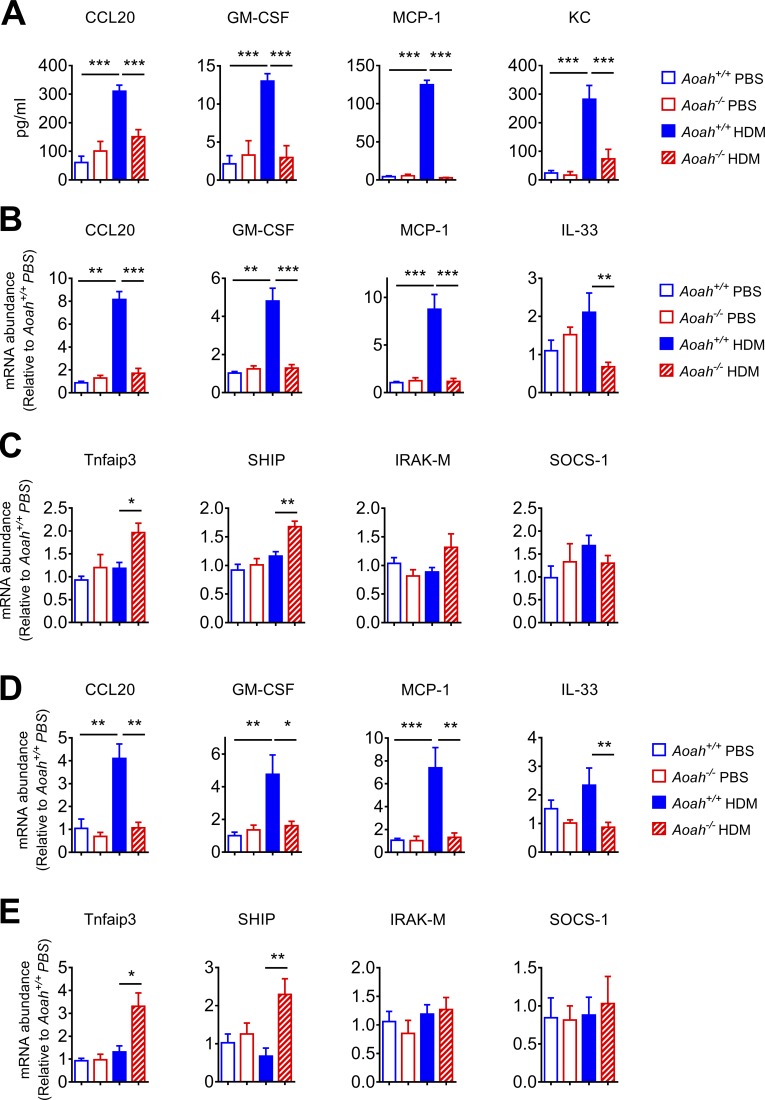
Upon allergen stimulation, lung epithelial cells secret pro-allergic cytokines or chemokines to drive ILC-2 and DC activation and therefore Th2 immune responses (Hammad and Lambrecht, 2008; Hammad et al., 2009; Lambrecht and Hammad, 2012; Schuijs et al., 2015). To test whether lung epithelial cells from Aoah−/− mice are hypo-responsive to allergens, we instilled 10 µg HDM extract (which contained ∼1 endotoxin unit [EU]) i.n. and measured the BALF concentrations of CCL20, GM-CSF, MCP-1, and KC, cytokines, or chemokines known to be predominantly produced by lung epithelial cells and to be critical for pulmonary allergic responses (Hammad et al., 2009; Schuijs et al., 2015). Aoah−/− mice had significantly lower epithelial cell cytokine/chemokine secretion (Fig. 3 A), suggesting that lung epithelial cells from Aoah−/− mice are refractory to HDM stimulation. Lung homogenates from Aoah−/− mice also had reduced CCL20, GM-CSF, MCP-1 and IL-33 mRNA abundance (Fig. 3 B). The expression of Tnfaip3 (encoding A20) and SHIP, two negative regulators of TLR signaling, was up-regulated in HDM exposed Aoah−/− mouse lungs, suggesting that they may contribute to the decreased responses (Fig. 3 C). However, the expression levels of two other negative regulators, IRAK-M and SOCS-1, were similar in Aoah+/+ and Aoah−/− lungs (Fig. 3 C). To further explore the cytokine/chemokine responses in epithelial cells, we instilled HDM i.n. and then sorted CD45−CD326+ lung epithelial cells and found that HDM-exposed epithelial cells from Aoah−/− mice had decreased CCL20, GM-CSF, MCP-1, and IL-33 mRNA expression and elevated Tnfaip3 and SHIP compared with their counterparts in Aoah+/+ mice (Fig. 3, D and E). Thus, in response to HDM exposure, AOAH promotes lung epithelial cells to produce pro-allergic CCL20, GM-CSF, MCP-1, and IL-33, in keeping with the evidence that Aoah+/+ lungs had vigorous DC and ILC-2 responses upon HDM stimulation (Fig. 2).
Figure 3.
Lung epithelial cells from Aoah−/− mice have defective innate responses to HDM. (A) Aoah+/+ or Aoah−/− mice were instilled i.n. with HDM and 5 h later, cytokine/chemokine levels were measured in BALF using ELISA. Data were combined from three experiments. n = 6–9. (B) 5 h after HDM instillation, cytokine/chemokine mRNA were measured in lung homogenates using qPCR. Data were combined from two experiments. n = 6–8. (C) TLR signaling negative regulators Tnfaip3, SHIP, IRAK-M, and SOCS-1 mRNA levels were measured in lung homogenates. Data were combined from two experiments. n = 6–8. (D) 5 h after instillation of HDM, lung epithelial cells were sorted by using flow cytometry, and cytokine/chemokine mRNA were measured using qPCR. Data were combined from two experiments. n = 6–8. (E) Tnfaip3, SHIP, IRAK-M, and SOCS-1 mRNA levels were measured in sorted lung epithelial cells. Data were combined from two experiments. n = 6–8. (A–E) Mann-Whitney test was used. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
AOAH lowers the allergen recognition threshold of airway epithelial cells
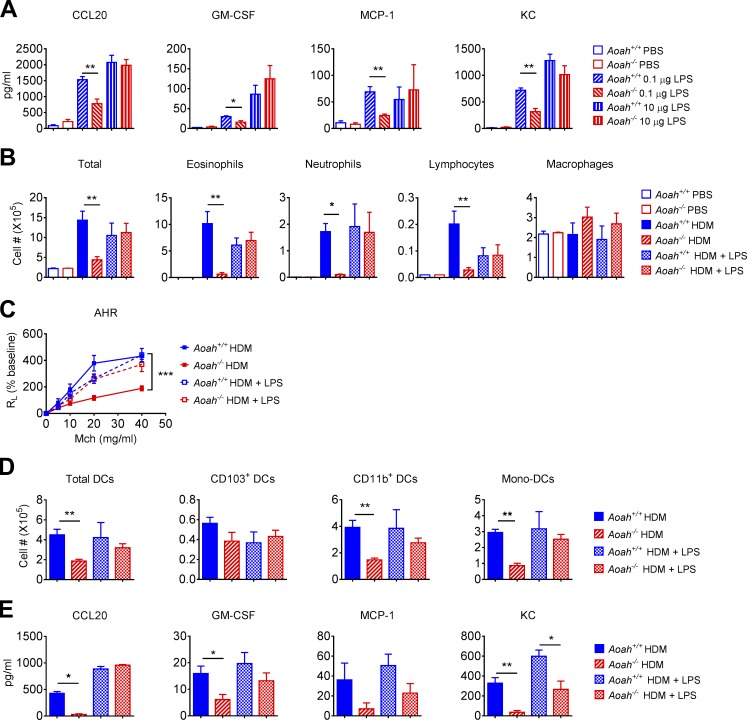
Lung epithelial cells set the allergen recognition threshold (Hammad and Lambrecht, 2008; Hammad et al., 2009; Lambrecht and Hammad, 2012; Schuijs et al., 2015). To test whether Aoah−/− lung epithelial cells have an increased stimulation threshold, we first instilled low or high doses of LPS i.n. and measured several innate pro-allergic responses. Aoah−/− mice had significantly lower epithelial cytokine/chemokine secretion in response to low-dose LPS instillation (0.1 µg, i.n.), while the responses to a high dose of LPS (10 µg) were similar in the two strains of mice (Fig. 4 A), suggesting that Aoah−/− lung epithelial cells have a higher threshold for MAMP recognition.
Figure 4.
Lung epithelial cells from Aoah−/− mice have elevated allergen recognition threshold. (A) Mice were instilled with 0.1 µg or 10 µg LPS, i.n., and 5 h later, the concentrations of inflammatory cytokines/chemokines in BALF were determined using ELISA. Data were combined from three experiments. n = 8. (B) Aoah+/+ and Aoah−/− mice were sensitized by 10 µg HDM or 10 µg HDM plus 0.1 µg LPS, and challenged with 10 µg HDM only. On day 14, cells in BALF were analyzed following cytospin and Wright-Giemsa staining. Data were combined from two experiments. n = 6. (C) AHR was measured on day 14. Data were combined from two experiments. n = 5–7. (D) Aoah+/+ and Aoah−/− mice were instilled with 10 µg HDM only or 10 µg HDM plus 0.1 µg LPS on days 0, 1 and 2, and on day 4 the lung DCs were analyzed by using flow cytometry. n = 5–6. (E) Aoah+/+ and Aoah−/− mice were instilled with 10 µg HDM only or 10 µg HDM plus 0.1 µg LPS, and the BALF was collected after 5 h cytokines/chemokines in BALF were determined using ELISA. n = 5–6. (A, B, D, and E) Mann-Whitney test was used. (C) Two-way ANOVA was used. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
We next added a small amount of LPS (0.1 µg) to HDM extract during the sensitization phase and asked whether Aoah−/− epithelial responses can be normalized. Aoah−/− mice then had Aoah+/+-like airway eosinophilic inflammation, AHR, and lung DC and epithelial responses (Fig. 4, B–E). Altogether, in this asthma model, AOAH lowers the threshold for allergen recognition by lung epithelial cells.
Aoah−/− mice have normal airway allergic responses when sensitization bypasses lung epithelial cells
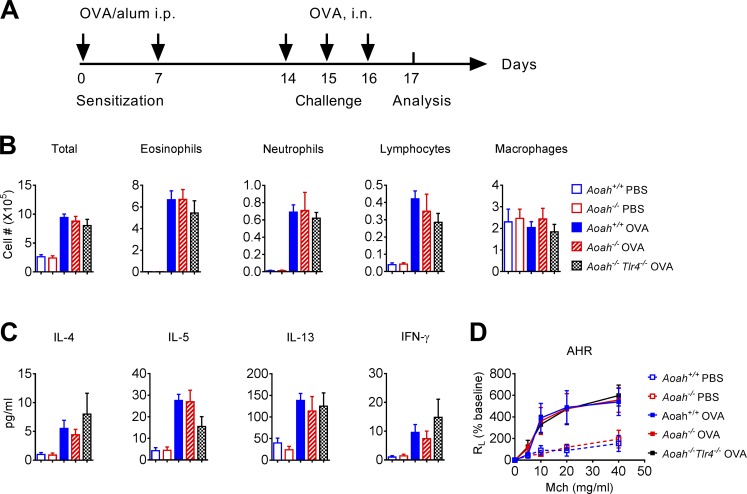
To further show that the reduced allergic response to HDM in Aoah−/− mice is due to hypo-responsive lung epithelial cells to allergens and is not caused by other unknown effects that AOAH might have on hypersensitivity, we used a traditional OVA/aluminum hydroxide (Alum)–induced allergic asthma model in which sensitization bypasses the lung epithelium and TLR4 is not required for generating Th2 responses to the OVA allergen (Fig. 5 A; Eisenbarth et al., 2002). Aoah+/+, Aoah−/−, and Aoah−/−Tlr4−/− mice had similar eosinophil, neutrophil, and lymphocyte infiltration (Fig. 5 B), similar airway Th2 and Th1 cytokine secretion (Fig. 5 C) and comparable AHR (Fig. 5 D). These results indicate that Aoah−/− mice are not intrinsically incapable of mounting Th2 responses.
Figure 5.
Aoah−/− mice have normal airway allergic responses when sensitization bypasses lung epithelial cells. (A) Mice were sensitized by injecting OVA/Alum i.p. on days 0 and 7, then challenged with OVA i.n. on day 14–16 and analyzed on day 17. (B) The cell differential counts in BALF from Aoah+/+, Aoah−/−, and Aoah−/−Tlr4−/− mice. Data were combined from two experiments. n = 6. (C) Th1/Th2 cytokines in BALF from Aoah+/+ Aoah−/−, and Aoah−/−Tlr4−/−mice. Data were combined from two experiments. n = 6. (D) AHR were measured on day 17. n = 5–6. (B and D) Mann-Whitney test was used. (D) Two-way ANOVA was used.
Preexposure to LPS in airway or circulation inhibits HDM-induced allergic asthma
As Aoah−/− lung epithelial cells are refractory to stimulation by a low dose of LPS or HDM and we know that LPS exposure leads to prolonged tolerance in Aoah−/− mouse peritoneal macrophages and colonic DCs (Lu et al., 2008, 2013; Janelsins et al., 2014), we wondered whether LPS in the airway or circulating blood induces lung epithelial desensitization in Aoah−/− mice. We first tested whether preexposure to LPS in airways inhibited allergic responses in Aoah+/+ mice. On days 6, 4, and 2 before HDM exposure, we instilled 1 µg LPS i.n. (Fig. S3 A) and found that preexposure to LPS dramatically inhibited airway eosinophilic inflammation (Fig. S3 B) and Th2 responses (Fig. S3 C), consistent with previous studies (Schuijs et al., 2015). Because LPS is ubiquitous in the environment and the lung microbiome has been found to contain Gammaproteobacteria, many of which make TLR4-stimulating LPS (Gollwitzer et al., 2014; Huang and Boushey, 2015; Munford, 2016), we then tested whether there was more bioactive LPS in Aoah−/− mouse airways. By using a cell-based endotoxin detection method, we could detect a small amount of TLR4-stimulating activity in cell-free BALF; yet, there was no difference between Aoah−/− and Aoah+/+ mouse BALF (Fig. S3 D), suggesting that Aoah−/− mouse lung epithelial desensitization was not caused by LPS in the airway.
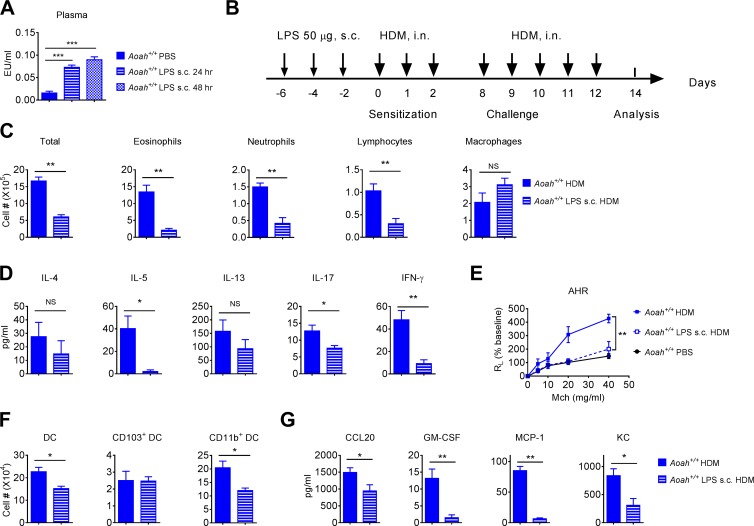
We then asked whether systemic LPS may inhibit HDM-induced allergic reactions. LPS injected s.c. into a mouse drains slowly and continuously for weeks to regional lymph nodes; it reaches the circulation via the thoracic duct and slowly appears in the liver and other organs (Lu and Munford, 2011). A low amount of LPS (∼0.12% to 0.84% of the injected LPS) is present in the circulation for weeks following s.c. injection (Lu and Munford, 2011). We injected 50 µg LPS s.c. in Aoah+/+ mice. 24 and 48 h later, we found significantly increased TLR4-stimulating activity in mouse plasma (Fig. 6 A). When LPS was administered s.c. 6, 4, and 2 d before HDM treatment (Fig. 6 B), the airway eosinophilic inflammation, Th2 responses, and AHR dramatically decreased (Fig. 6, C–E). The DC and lung epithelial responses were also diminished (Fig. 6, F and G). When we decreased the s.c. injection LPS dose to 5 µg, there was little inhibition, possibly because the LPS was inactivated in the injection site and draining lymph nodes before it could reach the circulation (Lu and Munford, 2011). Collectively, these results suggest that circulating LPS can also inhibit airway allergic responses.
Figure 6.
Preexposure to LPS injected s.c. inhibits allergic asthma. (A) Aoah+/+ mice were injected with 50 µg of LPS s.c. 24 and 48 h later, TLR4-stimulating activities were measured in their plasma. Data were combined from two experiments. n = 12. (B) Aoah+/+ mice were treated with LPS s.c. 6, 4, and 2 d before HDM sensitization. (C) Cells in BALF were analyzed on day 14. Data were combined from two experiments. n = 6. (D) BALF cytokine levels were measured on day 14 using ELISA. Data were combined from two experiments. n = 6. (E) AHR was measured on day 14. n = 4. (F) Mice were pretreated with LPS s.c. and then were sensitized with HDM on days 0, 1, and 2. Lung DC numbers were measured on day 4. n = 5. (G) Mice were pretreated with LPS s.c. and then were instilled with HDM; 5 h later, their BALF cytokines were measured. n = 5. (A, C, D, F, and G) Mann-Whitney test was used. (E) Two-way ANOVA was used. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
AOAH prevents airway epithelial desensitization induced by intestinal commensal Gram-negative bacteria
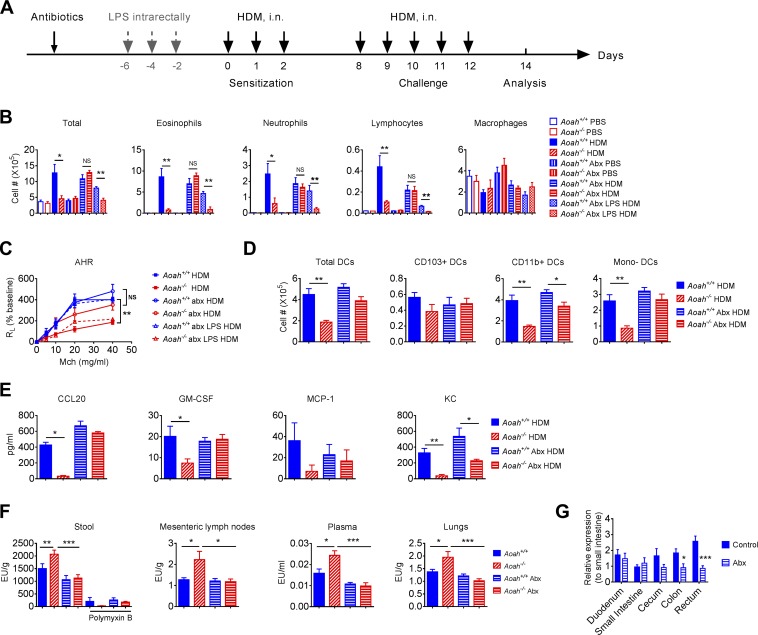
We next asked whether intestinal commensal Gram-negative bacterial LPS contribute to Aoah−/− airway epithelial desensitization. We surmised that in Aoah−/− mice, more gut-derived LPS might translocate and traffic to the lung without being inactivated. We found that cohoused Aoah+/+ and Aoah−/− mice had a similar intestinal microbiome (Fig. S4, A–E). The Enterobacteriaceae family of the Gammaproteobacteria class are more likely to produce LPS that can stimulate MD2-TLR4 and be inactivated by AOAH than are Bacteroides species (Erridge, 2010; Erridge et al., 2010; Munford, 2016; Vatanen et al., 2016; Faraj et al., 2017). No Gammaproteobacteria were detected in the microbiome analysis, probably due to their low relative abundance. We cultured Aoah+/+ and Aoah−/− mouse stools on MacConkey plates, which primarily allow the growth of aerobic Gram-negative bacteria such as Enterobacteriaceae, and we found similar numbers CFUs (Fig. S4 F). We also isolated stool DNA and quantitated total bacteria, Enterobacteriaceae, Bacteroidaceae, Lactobacillaceae, and Clostridiaceae families by using quantitative PCR (qPCR). No difference was observed between Aoah+/+ and Aoah−/− mouse stool (Fig. S4 G). We then added neomycin and metronidazole to the drinking water for 3 wk before sensitization to deplete commensal aerobic and anaerobic Gram-negative bacteria (Fig. 7 A). Antibiotic-treated Aoah−/− mice regained airway eosinophil inflammation, AHR, lung DC activation, and lung epithelial responses to HDM (Fig. 7, B–E). Importantly, when we gave LPS intrarectally to antibiotic-treated mice, the airway inflammation and AHR in Aoah−/− mice were reduced again (Fig. 7, B and C), strongly suggesting that LPS produced in the intestine can prevent allergic responses. When only neomycin was used, the allergic responses in Aoah−/− mice were also normalized, suggesting that LPS derived from aerobic Enterobacteriaceae inhibits airway allergy (Fig. S5, A and B). We then found that indeed Aoah−/− feces contained more TLR4-stimulating activity than did Aoah+/+ feces and that antibiotic treatment could reduce this activity to similar levels in the feces of the two strains of mice (Fig. 7 F). The TLR4-stimulating activity in stools was inhibited by polymyxin B, supporting that LPS was the major TLR4 stimulant (Fig. 7 F). The intestinal contents of Aoah−/− mouse colon and rectum, which contain high densities of commensal bacteria, had elevated TLR4-stimulating activities (Fig. S5 C). In addition, we found that Aoah−/− mouse mesenteric lymph nodes, plasma, and lung homogenates contained more TLR4-stimulants, which could also be reduced by antibiotic treatment (Fig. 7 F). We measured AOAH expression in the intestine by using qPCR and found that AOAH mRNA was expressed throughout the intestine and peaked in the rectum (Fig. 7 G). Neomycin treatment reduced AOAH expression, suggesting that commensal Gram-negative bacteria induce AOAH expression in the colon and rectum (Fig. 7 G). As expected, the CD45+ cells in the large intestine, including macrophages and DCs express high levels of AOAH mRNA (Fig. S5D). To find out if the diminished allergic responses in Aoah−/− mice are mediated by LPS but not by other MAMPs, we first normalized the allergic responses in Aoah−/− mice by antibiotic treatment, and then injected 5 µg LPS or 7 × 108 heat-inactivated Gram-positive bacteria, Streptococcus pneumoniae, s.c. before HDM exposure. LPS injection induced protection in Aoah−/− mice but not in Aoah+/+ mice, while Streptococcus pneumoniae had similar protective effects in both strains of mice, suggesting that AOAH specifically modulates LPS-mediated protection (Fig. S5 E). Regulatory T cells (T reg cells) can be induced to inhibit allergic responses (Lewkowich et al., 2005; Duan et al., 2011; Soroosh et al., 2013), while LPS releases airway tolerance by suppressing T reg cells (Duan et al., 2008). We found that Aoah+/+ and Aoah−/− mice had similar pulmonary T reg cell populations with or without HDM treatment, suggesting that T reg cells may not account for the hypo-responsiveness to HDM in Aoah−/− mice (Fig. S5F). Collectively, these data all support the hypothesis that intestinal microbiota-derived, stimulatory LPS diminishes lung allergic responses in Aoah−/− mice.
Figure 7.
Intestinal commensal Gram-negative bacterial LPS dampens airway allergic responses to HDM in Aoah−/− mice. (A) Antibiotics were added to sterile drinking water for 3 wk before HDM treatment. In some groups, after 2 wk of antibiotic treatment, 50 µg LPS in 200 µl PBS was administered intrarectally every other day thrice before HDM treatment. In all experiments, water containing antibiotics was fed till the end of experiments. (B) On day 14, BALF cells were analyzed. Data were combined from three experiments. n = 6–9. Abx, antibiotics. (C) AHR was measured on day 14. Data were combined from two experiments. n = 3–7. (D) On day 0, 1 and 2, mice were sensitized with HDM. On day 4, mouse lung DCs were analyzed using flow cytometry. n = 5–6. (E) Mice were instilled with HDM i.n. and BALF was obtained 5 h later. Cytokines/chemokines were measured using ELISA. n = 5–6. (F) Heated feces suspension from Aoah+/+ and Aoah−/− mice treated with and without antibiotics were tested for TLR4-stimulating activities. In some wells, 20 µg/ml polymyxin B was added. Data were combined from four experiments. n = 16. Mesenteric lymph node homogenates were heated and tested for TLR4 stimulants. n = 12–18. Diluted and heated plasma from Aoah+/+ and Aoah−/− mice were tested for TLR4-stimulating activity. n = 12–18. Lung homogenates were heated and tested for TLR4-stimulants. n = 12–18. (H) AOAH mRNA levels were measured in different intestinal segments. Data were combined from four experiments. n = 8–14. As experiments in D and E were done simultaneously with those in Fig. 4 (D and E), they shared the same Aoah+/+ HDM and Aoah−/− HDM groups. (B and D–G) Mann-Whitney test was used. (C) Two-way ANOVA was used. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Discussion
Allergen-induced Th2 immunity underlies most allergic asthma. Both genetic and environmental factors are involved in the induction or modulation of Th2 responses. LPS or endotoxin is a common environmental pollutant, a constituent of many foodstuffs, and it also is produced by Gram-negative bacteria in the intestinal microbiota (Doreswamy and Peden, 2011; Erridge, 2011a,b; Huang and Boushey, 2015). Early exposure to a LPS-rich environment has been reported to provide protection against asthma (Braun-Fahrländer et al., 2002; Stein et al., 2016). One proposed protective mechanism is that LPS exposure induces the barrier epithelial cell desensitization, and therefore, the downstream DC and Th2 responses are reduced (Schuijs et al., 2015). In this study, we found that mice lacking a host LPS-degrading enzyme, AOAH, were resistant to HDM-induced allergic asthma. Compared with Aoah+/+ control mice, Aoah−/− mice had reduced eosinophilic airway inflammation, diminished airway goblet cell hyperplasia, and mucus production, as well as decreased airway hyper-reactivity. We further found that Aoah−/− lung epithelial cells were refractory to a low dose of LPS or HDM stimulation, which led to defective ILC-2 and DC activation as well as reduced Th2 responses. When a small amount of LPS was added to HDM, the epithelial activation threshold was exceeded, and DC and Th2 activation were restored. When the allergen sensitization bypassed lung epithelial cells, in contrast, Aoah−/− mice had normal allergic responses. In addition, we found that preexposure to circulating LPS tolerized lung epithelial cells; that antibiotic treatment that decreased intestinal commensal LPS restored Aoah−/− lung epithelial cell responses to HDM; and that instilling LPS intrarectally to antibiotic-treated mice restored tolerance. Finally, we showed that Aoah−/− feces, mesenteric lymph nodes, blood, and lungs contained more bioactive endotoxin than did the same samples from their Aoah+/+ counterparts. We conclude that AOAH sensitizes lung epithelial cells for allergic responses by decreasing the bioactivity of intestinal commensal LPS (Fig. 8). Thus, not only environmental LPS, but also LPS within the body may desensitize lung epithelial cells to prevent allergic asthma.
Figure 8.
Graphic summary. When AOAH is present, commensal LPS is inactivated and lung epithelial cells remain sensitized for allergen stimulation. When AOAH is missing, commensal LPS cannot be inactivated and excessive amount of bioactive LPS is present in the gut. LPS translocates and reaches the lung via circulation, desensitizing epithelial cells. Allergic responses are diminished.
Previously, we showed that when very low levels of bioactive LPS were present in the extracellular environment in vivo, Aoah−/− peritoneal macrophages remained desensitized (tolerant) for months (Lu et al., 2008, 2013). Similarly, Janelsins et al. found that commensal LPS diminished the ability of Aoah−/− colonic DCs to induce Th17 immunity (Janelsins et al., 2014). In this study, we extended the tolerance induced by prolonged exposure to LPS to include epithelial cells. Our data strongly suggest that lung epithelial cells from Aoah−/− mice, which may be constantly exposed to LPS derived from intestinal commensal Gram-negative bacteria, became tolerant or cross-tolerant to LPS or other MAMPs in HDM extract and therefore prevented downstream allergic responses. Notably, when we added LPS to HDM, the epithelial responses were restored. Thus, lung epithelial cells set the activation threshold for allergen stimulation (Schuijs et al., 2015) and AOAH tunes the threshold.
It has been shown previously that intestinal commensal bacteria regulate allergic asthma (Herbst et al., 2011; Hill et al., 2012; Budden et al., 2017). Arrieta et al., reported that the abundance of four bacteria was significantly decreased in the intestines of children at risk of asthma. One of them was Veillonella, a Gram-negative anaerobe that has an Escherichia coli–like lipid A that may stimulate TLR4 (Matera et al., 2009; Arrieta et al., 2015). Furthermore, the LPS biosynthesis pathway is altered in the microbiota of atopy/wheeze patients so that a decreased amount of LPS was found in their feces (Arrieta et al., 2015). These results provide evidence that the presence of bioactive LPS in the gut increases protection from allergic asthma. Intestinal commensal LPS may enter the circulation via lymphatics or the portal system (Munford, 2016; Faraj et al., 2017) to directly inhibit lung epithelial cells. In our experimental setting, inhaled LPS or LPS in food (irradiated) and water (autoclaved) may have played a minor role as we detected similar TLR4-stimulating activity in Aoah+/+ and Aoah−/− mouse BALF; antibiotic treatment almost completely restored allergic responses in Aoah−/− mice. Recently, it has been shown that the lung also has commensal bacteria, but its microbiota is of low abundance and probably transient (Budden et al., 2017; Huffnagle et al., 2017). We were unable to isolate sufficient bacterial DNA from mouse lungs to perform microbiome analysis. As we found that intestinal and circulating TLR4-stimulating activity negatively correlated with lung allergic responses, intestinal commensal Gram-negative bacteria may be the major source of LPS that modulates lung epithelial sensitivity.
Exposure to airway LPS increased Tnfaip3 expression in epithelial cells, and their activation threshold increased; when Tnfaip3 gene was specifically deleted in epithelial cells, the protective effects of LPS preexposure were largely lost (Schuijs et al., 2015). We found that the basal levels of Tnfaip3 expression did not differ between Aoah−/− and Aoah+/+ epithelial cells, while after HDM exposure, epithelial cells from Aoah−/−mice up-regulated Tnfaip3 mRNA, which may have inhibited their innate responses. In addition, another negative regulator (SHIP) was also induced by HDM, while IRAK-M and SOCS-1 expression was unchanged in Aoah−/− epithelium. These results are different from tolerant Aoah−/− peritoneal macrophages, which had increased IRAK-M expression (Lu et al., 2013). Furthermore, unlike tolerant macrophages, which are even refractory to high concentrations of LPS, we found that adding a small amount of LPS to HDM could restore lung epithelial responsiveness in Aoah−/− mice, suggesting that different mechanisms underlie the reprogramming of peritoneal macrophages and epithelial cells. In addition to the negative cell signaling regulators, epigenetic changes in epithelial cells may also contribute to the tolerance of Aoah−/− epithelium (Foster et al., 2007; Neagos et al., 2015). This possibility awaits further investigation.
Many studies have suggested that preexposure to LPS promotes Th1 responses and therefore prevents a Th2 bias to allergens (Kim et al., 2007; Doreswamy and Peden, 2011; Daan de Boer et al., 2013). We found that in Aoah−/− mice, the Th1 or Th17 responses did not increase and sometimes even decreased, suggesting that the decreased Th2 responses are not caused by Th1 bias and instead are promoted by a general inhibitory mechanism. It has been shown previously that T reg cells inhibit allergic responses, while LPS antagonizes the suppression (Lewkowich et al., 2005; Duan et al., 2008, 2011; Soroosh et al., 2013). We did not observe significant differences in T reg cell populations between Aoah+/+ and Aoah−/− mouse lungs with and without HDM stimulation. In addition, we found that mediastinal lymph node cells from Aoah−/− mouse secreted less IL-10 when restimulated with HDM in vitro. These results suggest that reduced HDM-induced allergic responses in Aoah−/− are not caused by enhanced T reg cell function.
We found that in Aoah+/+ mice, AOAH mRNA was expressed in the macrophages and DCs of the large intestine and induced by commensal Gram-negative bacteria. Janelsins et al. also showed that DCs from colonic lamina propria had much greater AOAH mRNA abundance and enzymatic activity than DCs from small intestine, lymph nodes, spleen, and lung; TLR4 signaling and intestinal commensal Gram-negative bacteria stimulated AOAH expression in colonic DCs (Janelsins et al., 2014). These DCs and macrophages may take up intestinal commensal Gram-negative bacteria and deacylate LPS (Katz et al., 1999; Lu et al., 2003). LPS may be further deacylated in the liver and the lung, as we have shown previously that Kupffer cells and liver DCs, lung macrophages, and neutrophils express AOAH (Shao et al., 2007, 2011; Zou et al., 2017). When AOAH is missing, the internalized LPS cannot be inactivated and bioactive LPS can be released into extracellular fluid (Lu et al., 2013). As AOAH can also be secreted, commensal LPS may be deacylated in the intestinal lumen (Staab et al., 1994; Gioannini and Weiss, 2007). In addition to AOAH, intestinal alkaline phosphatases, produced in the duodenum, have been reported to inactivate LPS in the gut and decrease serum endotoxin levels (Kaliannan et al., 2013). Our findings suggest that these phosphatases are unable to fully inactivate intestinal LPS in Aoah−/− mice.
Another host MAMP-degrading enzyme can also modulate allergic asthma. A chitin-degrading enzyme, acidic mammalian chitinase (AMCase), promotes allergic asthma partially through the IL-13 activation pathway (Zhu et al., 2004). Blocking or inhibiting AMCase caused a marked decrease in Th2 responses in an OVA-induced allergic asthma model (Zhu et al., 2004). In contrast, other studies have suggested that AMCase plays a protective role (Seibold et al., 2009; Van Dyken et al., 2011). These observations may have differed because the allergens and the time and routes of allergen exposure were different. These and our findings all demonstrate that MAMP-degrading enzymes can play a critical role in modulating pulmonary inflammatory responses.
In summary, our study highlights the prominent role that AOAH plays in sensitizing lung epithelial cells by degrading/inactivating intestinal commensal LPS, thereby increasing susceptibility to allergic asthma. Notably, AOAH also promotes the resolution of acute lung injury induced by Gram-negative bacteria (Zou et al., 2017). A gene that has been highly conserved through evolution (Munford and Varley, 2006) seems to regulate the counter-balance between infection and allergy.
Materials and methods
Mice
C57BL/6J Aoah+/+, Aoah−/− and Tlr4−/−Aoah−/− mice were obtained from the National Institutes of Health, Bethesda, MD (R.S. Munford). The generation of Aoah−/− mice had been described (Lu et al., 2003). The mutated Aoah gene had been backcrossed to C57BL/6J mice for at least 10 generations. Tlr4−/−Aoah−/− mice were produced by crossing Aoah−/− and Tlr4−/− mice. Female littermates of 6–10 wk of age from Aoah+/− parents were used and they were cohoused throughout the experiments. All mice were housed under specific pathogen-free conditions in Fudan University, the Department of Laboratory Animal Science, and studied using protocols approved by the Institutional Animal Care and Use Committee (IACUC) of Fudan University. All protocols adhered to the Guide for the Care and Use of Laboratory Animals.
Reagents
HDM extract was obtained from the Greer laboratories. The allergens were extracted from whole body crust of Dermatophagoides pteronyssinus, which contained 100 EU endotoxin/mg. E. coli 0111:B4 LPS was obtained from Sigma. Low endotoxin OVA (endotoxin level <1 EU/mg) was from Chondrex, Inc. Alum, which contained 40 mg/ml Al(OH)3, was from Thermo-Fisher. Anti-mouse antibodies used for flow cytometry were CD11c (clone N418, eBioscience), I-A/I-E (Clone M5/114.152, Biolegend), CD11b (clone M1/70, BD), CD103 (clone M290, BD), CD64 (clone X54-5/7.1, BD), CD86 (clone GC1, BD), CD40 (clone 3/23, BD), CD45 (clone 30-F11, Biolegend), CD3 (clone 17A2, BD), B220 (clone RA3-6B2, eBioscience), NK1.1 (clone PK136, BD), GATA3 (clone L50-823, BD), and CD326 (clone G8.8, Biolegend). ELISA kits IL-4, IL-5, IL-10, IFN-γ, and MCP-1 were from BD; IL-13, KC, CCL20, GM-CSF, and IL-17 kits were from R&D Systems.
Mouse asthma models
The HDM-induced allergic asthma method was modified from a previous study (Daan de Boer et al., 2013). Aoah+/+, Aoah−/−, and Tlr4−/−Aoah−/− mice were anesthetized i.p. with 0.5% pentobarbital sodium (50 µg/g body weight) and instilled i.n. with 10 µg HDM in 40 µl PBS daily on days 0, 1, and 2 (sensitization phase). From days 8–12, mice were challenged daily with 10 µg HDM in 40 µl PBS (challenge phase); control mice were instilled with PBS i.n. during both the sensitization and challenge phases. On day 14, mice were euthanized for analysis (Fig. 1 A). In some experiments, 10 µg HDM mixed with 0.1 µg LPS was instilled i.n. for sensitization.
For OVA/Alum-induced allergic asthma, mice were injected i.p. with a mixture of 50 µl of 1 mg/ml OVA (low endotoxin) and 50 µl of 40 mg/ml of Alum on days 0 and 7 (sensitization). From days 14–16, mice were anesthetized and instilled i.n. with 40 µl of 1.25 mg/ml OVA (total 50 µg) daily. 1 d after the last instillation, mice were euthanized for analysis (Fig. 5 A).
Analysis of BALF and mediastinal lymph nodes
Mice were anesthetized and exsanguinated by cutting the inferior vena cava. Bronchoalveolar lavage was performed using 1 ml of PBS containing 5 mM EDTA five times. The recovered BALF was combined and centrifuged at 700 g for 5 min at 4°C. The cell-free supernatant was used for cytokine or chemokine quantitation using ELISA kits. The cell pellets were resuspended in PBS and total cell numbers were counted using Cellometer (Nexcelom). After cytospin and Wright-Giemsa staining, macrophages, eosinophils, neutrophils, and lymphocytes were differentially counted. 300 cells were counted for each sample and the total numbers of each cell type in BALF were calculated.
Measurement of AHR
In the HDM-induced allergic asthma model, mice were anesthetized on day 14, the trachea was intubated, and the lungs were mechanically ventilated. Lung resistance (RL) was measured after increasing doses (5–40 mg/ml) of aerosolized methacholine (Sigma) was applied (Buxco Electronics). In the OVA-induced asthma model, mice were tested on day 17.
qPCR
RNA from lungs or sorted lung epithelial cells was purified using a RNA isolation kit (Tiangen) and then reversely transcribed (Tiangen). DNA from feces was isolated by using a QIAamp Fast DNA Stool Mini kit (QIAGEN). cDNA or bacterial DNA was analyzed by qPCR using SYBR green reagents (Tiangen) with the ABI 7500 system. The primers used for RT-PCR were listed in Table 1. Actin (lung samples) was used as the internal control and the relative gene expression levels were calculated by the ΔΔCt quantification method. Relative bacterial 16S rDNA abundance were calculated by the ΔCt quantification method and related to total DNA amount isolated from stool samples.
Table 1. Primers used for qPCR.
| Gene symbols | Primer sequence |
|---|---|
| mActin | Sense: 5′-GGCTGTATTCCCCTCCATCG-3′; antisense: 5′-CCAGTTGGTAACAATGCCATGT-3′ |
| mAOAH | Sense: 5′-GTTTTCCCAACGCTGCGGGG-3′; antisense: 5′-TGGCCTTCTGCCCGGGTACA-3′ |
| mCCL20 | Sense: 5′-TCCAGAGCTATTGTGGGTTTCA-3; antisense: 5′-CTGAGGAGGTTCACAGCCCTT-3′ |
| mGM-CSF | Sense: 5′-TCAAAGAAGCCCTGAACCTC-3′; Antisense: 5′-AAATTGCCCCGTAGACCCTGCT-3′ |
| mMCP-1 | Sense: 5′-GGCTCAGCCAGATGCAGTTAA-3′; antisense: 5′-CCTACTCATTGGGATCATCTTGCT-3′ |
| mIL-33 | Sense: 5′-ACTATGAGTCTCCCTGTCCTG-3′; antisense: 5′-ACGTCACCCCTTTGAAGC-3′ |
| mA20 | Sense: 5′-CTCAGAACCAGAGATTCCATGAAG-3′; antisense: 5′-ACCTGTGTAGTTCGAGGCATGTC-3′ |
| mSHIP | Sense: 5′- TCAGCCATATCTGCACTGACAAC-3′; antisense: 5′-ACTCCCACTGCTCCCTTGTTT-3′ |
| mIRAK-M | Sense: 5′-TCCCACCTGAGGTGAAGCAT-3′; antisense: 5′-TGTGACATTGGCTGGTTCCA-3′ |
| mSOCS-1 | Sense: 5′- CCGTGGGTCGCGAGAAC-3′; antisense: 5′-AGGAACTCAGGTAGTCACGGAGTA-3′ |
| Enterobacteriaceae | Sense: 5′-CATTGACGTTACCCGCAGAAGAAGC-3′; antisense: 5′-CTCTAC-GAGACTCAAGCTTGC-3′ |
| Bacteroidaceae | Sense: 5′-CCAATGTGGGGGACCTTC-3′; antisense: 5′-AACGCTAGCTACAGGCTT-3′ |
| Lactobacillaceae | Sense: 5′-AGCAGTAGGGAATCTTCC-3′; antisense: 5′-CGCCACTGGTGTTCYTCCATATA-3′ |
| Clostridiaceae | Sense: 5′-TTAACACAA-TAAGTWATCCACCTGG-3′; antisense: 5′-ACCTTCCTCCGTTTTGTCAAC-3′ |
| Specific primers for bacteria | Sense: 5′-GTGCCAG-CMGCCGCGGTAA-3′; antisense: 5′-GACTACCAGG-GTATCTAAT-3′ |
Preparing single-cell suspensions from the lungs and intestines for flow cytometric analysis and sorting
To measure immune cells in the lungs, the lungs were perfused first and instilled with 1 ml digestion buffer (RPMI 1640 containing 1 mg/ml collagenase IV [Sigma] and 5 U/ml DNase I [Sigma]). After the lungs were excised, cut into ∼1-mm3 pieces, and incubated in 2 ml digestion buffer at 37°C with shaking at 220 rpm for 1 h, the digested tissues were filtered through a 70-mm cell strainer, and red blood cells were lysed using ammonium-chloride-potassium lysis buffer (eBioscience). Cells were collected by centrifugation and counted. For FACS analysis, cells were incubated with Fc blocking antibody (purified anti-mouse CD16/32, BioLegend) to prevent binding of nonspecific FcγRIII/II and then incubated with fluorochrome-conjugated antibodies for 1 h on ice. To stain intracellular GATA3, cells were fixed and permeablized with Fix/Perm solution/kit (BD) before anti-GATA3 antibody was added. After washing, the samples were analyzed by using CyAn-ADP (Beckman Counter) and data were processed using Flow Jo software (TreeStar).
To sort lung epithelial cells, lung single-cell suspensions were stained with anti-mouse CD45 antibody and CD326 antibody and sorted on MoFlo XDP (Beckman Counter). After sorting, the purity of CD45− CD326+ lung epithelial cells was >85%.
Similarly, small and large intestines were minced after the lumen contents were washed and then digested with 1.5 mg/ml collagenase IV and 5 U/ml DNase I for 15 min while shaking at 220 rpm. After washing, the single-cell suspension was sorted using CD45 antibody-conjugated magnetic microbeads. The CD45+ cells were then sorted using FACS to obtain I-A+ CD11c+CD11bdull DCs and I-A+ CD11cdull CD11b+ macrophages.
ELISA for HDM-specific antibodies
In the model of HDM-induced allergic asthma, mice were bled on day 14. Sera were collected for HDM-specific antibody measurement. ELISA plates (Costar) were coated with 5 µg/ml HDM extract in PBS at 4°C for 18 h. Detection antibodies were goat anti-mouse IgE (Southern Biotechnology), IgG1 (Santa Cruz Biotechnology) and IgG2c (Novus Biologicals, Inc.) conjugated with HRP. HRP substrate TMB (BD) was used. Plates were read at a wavelength of 450 nm (Tecan). We used OD values to represent the abundance of HDM-specific IgE, IgG1, and IgG2c.
Histology
After lungs had been excised and fixed in 4% paraformaldehyde, they were sectioned and stained with H&E or periodic acid-Schiff. The samples were examined for inflammatory cell infiltration, tissue damage, goblet cell metaplasia, and mucus production by using a Nikon E200 microscope.
Antibiotic treatment
To deplete commensal Gram-negative bacteria, 1 g/liter of neomycin sulfate (Sigma) and metronidazole (BioVision) each were added to the drinking water. In some experiments, only 1 g/liter of neomycin was added. The water containing antibiotics was fed for 3 wk before HDM treatment. The feces was checked for aerobic Gram-negative bacteria by culturing on MacConkey agar plates (Oxoid). The feeding was continued until the end of experiments. In some experiments, 6, 4, and 2 d before HDM sensitization, 50 µg LPS was administered intrarectally in 100 µl PBS.
Measurement of TLR4-stimulating activities in mouse feces, lymph nodes, plasma, and lungs
Fresh feces were collected and resuspended in endotoxin-free PBS (0.1 g/ml) and centrifuged at 800 g for 5 min. The supernatant was heated at 70°C for 10 min. Mice were bled from the tail vein; 3 µl of 0.5 M EDTA was used as an anti-coagulant. Plasma was diluted 1:5 in RPMI medium and heated at 70°C for 10 min. Mesenteric lymph nodes and lungs were homogenized in PBS and centrifuged. The supernatant was heated at 70°C for 10 min. After heating, all the samples were centrifuged again, and the supernatant was collected for TLR4-stimulating activity by using a cell-based colorimetric endotoxin detection kit (HEK-Blue LPS Detection Kit2, Invivogen). In brief, diluted samples were added to human embryonic kidney (HEK-293) cells that express hTLR4 and an NF-κB–inducible secreted embryonic alkaline phosphatase reporter gene. After 18 h incubation, cell culture media were applied to QUANTI-Blue medium to measure alkaline phosphatase activity. A preparation of E. coli 055:B5 LPS, standardized according to FDA-approved control standard endotoxin and included in the kit, was used to quantitate TLR4-stimulating activity. Plates were read at a wavelength of 655 nm (Tecan). The detection limit was 0.008 EU/ml.
Diversity analysis of intestinal microbiota
Fresh stool samples were collected from cohoused Aoah+/+ and Aoah−/− littermates. Microbial DNA was extracted using the E.Z.N.A. soil DNA kit (Omega Bio-tek) according to manufacturer’s protocols. The V3-V4 hypervariable regions of the bacteria 16S rRNA gene were amplified with primers 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) by thermocycler PCR system (ABI). Purified PCR products were sequenced on an Illumina MiSeq platform (Majorbio Bio-Pharm Technology Co. Ltd.). Raw fastq files were demultiplexed, quality-filtered by Trimmomatic, and merged by FLASH. Operational taxonomic units (OTUs) were clustered with 97% similarity cutoff using UPARSE (version 7.1). The taxonomy of each 16S rRNA gene sequence was analyzed by RDP Classifier algorithm against the Silva (SSU123) 16S rRNA database (Wang et al., 2007) and the confidence threshold was set at 70%. We obtained 654,401 high-quality sequences with an average of 40,900 reads/sample. A total of 286 OTUs were obtained.
Statistical analysis
Data were presented as mean ± SE. Differences between groups were analyzed by Mann-Whitney U tests for unpaired data using PRISM (GraphPad). Two-way ANOVA was used in AHR experiments. The statistical significance was set at P < 0.05.
Primers
Primers are listed in Table 1.
Online supplemental material
Fig. S1 shows that Aoah−/− mice have reduced airway Th2 cytokine secretion in response to HDM exposure. Fig. S2 shows that Aoah−/− mouse lungs have reduced numbers of monocyte-derived DCs upon HDM exposure. Fig. S3 shows that preexposure to LPS in airways inhibits allergic asthma. Fig. S4 shows that cohoused Aoah+/+ and Aoah−/− mice have similar intestinal microbiomes. Fig. S5 shows that Aoah−/− mice treated with neomycin have normal eosinophilic airway inflammation; AOAH regulates commensal LPS bioactivity in the large intestine.
Supplementary Material
Acknowledgments
We thank Dr. Robert Munford for critiquing and improving the manuscript.
This work was supported by grants from the National Natural Science Foundation of China (31570910, 31770993, and 91742104 to M. Lu) and a grant from the Shanghai Committee of Science and Technology (14ZR1401700 to M. Lu).
The authors declare no competing financial interests.
Author contributions M. Lu and G. Qian designed the experiments and wrote the paper. G. Qian and M. Lu analyzed the data. G. Qian, W. Jiang, B. Zou, X. Cheng, and J. Feng conducted the experiments. C. Niu helped with the microbiome analysis. Y. Chu, R. He, T. Chu, and J. Gu provided advice and some reagents.
References
- Arrieta M.C., Stiemsma L.T., Dimitriu P.A., Thorson L., Russell S., Yurist-Doutsch S., Kuzeljevic B., Gold M.J., Britton H.M., Lefebvre D.L., et al. CHILD Study Investigators . 2015. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 7:307ra152 10.1126/scitranslmed.aab2271 [DOI] [PubMed] [Google Scholar]
- Barnes K.C., Grant A., Gao P., Baltadjieva D., Berg T., Chi P., Zhang S., Zambelli-Weiner A., Ehrlich E., Zardkoohi O., et al. . 2006. Polymorphisms in the novel gene acyloxyacyl hydroxylase (AOAH) are associated with asthma and associated phenotypes. J. Allergy Clin. Immunol. 118:70–77. 10.1016/j.jaci.2006.03.036 [DOI] [PubMed] [Google Scholar]
- Biswas S.K., and Lopez-Collazo E.. 2009. Endotoxin tolerance: new mechanisms, molecules and clinical significance. Trends Immunol. 30:475–487. 10.1016/j.it.2009.07.009 [DOI] [PubMed] [Google Scholar]
- Braun-Fahrländer C., Riedler J., Herz U., Eder W., Waser M., Grize L., Maisch S., Carr D., Gerlach F., Bufe A., et al. Allergy and Endotoxin Study Team . 2002. Environmental exposure to endotoxin and its relation to asthma in school-age children. N. Engl. J. Med. 347:869–877. 10.1056/NEJMoa020057 [DOI] [PubMed] [Google Scholar]
- Budden K.F., Gellatly S.L., Wood D.L., Cooper M.A., Morrison M., Hugenholtz P., and Hansbro P.M.. 2017. Emerging pathogenic links between microbiota and the gut-lung axis. Nat. Rev. Microbiol. 15:55–63. 10.1038/nrmicro.2016.142 [DOI] [PubMed] [Google Scholar]
- Daan de Boer J., Roelofs J.J., de Vos A.F., de Beer R., Schouten M., Hommes T.J., Hoogendijk A.J., de Boer O.J., Stroo I., van der Zee J.S., et al. . 2013. Lipopolysaccharide inhibits Th2 lung inflammation induced by house dust mite allergens in mice. Am. J. Respir. Cell Mol. Biol. 48:382–389. 10.1165/rcmb.2012-0331OC [DOI] [PubMed] [Google Scholar]
- Doreswamy V., and Peden D.B.. 2011. Modulation of asthma by endotoxin. Clin. Exp. Allergy. 41:9–19. 10.1111/j.1365-2222.2010.03628.x [DOI] [PubMed] [Google Scholar]
- Duan W., So T., and Croft M.. 2008. Antagonism of airway tolerance by endotoxin/lipopolysaccharide through promoting OX40L and suppressing antigen-specific Foxp3+ T regulatory cells. J. Immunol. 181:8650–8659. 10.4049/jimmunol.181.12.8650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan W., So T., Mehta A.K., Choi H., and Croft M.. 2011. Inducible CD4+LAP+Foxp3- regulatory T cells suppress allergic inflammation. J. Immunol. 187:6499–6507. 10.4049/jimmunol.1101398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ege M.J., Mayer M., Normand A.C., Genuneit J., Cookson W.O., Braun-Fahrländer C., Heederik D., Piarroux R., and von Mutius E.. GABRIELA Transregio 22 Study Group . 2011. Exposure to environmental microorganisms and childhood asthma. N. Engl. J. Med. 364:701–709. 10.1056/NEJMoa1007302 [DOI] [PubMed] [Google Scholar]
- Eisenbarth S.C., Piggott D.A., Huleatt J.W., Visintin I., Herrick C.A., and Bottomly K.. 2002. Lipopolysaccharide-enhanced, toll-like receptor 4-dependent T helper cell type 2 responses to inhaled antigen. J. Exp. Med. 196:1645–1651. 10.1084/jem.20021340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erridge C. 2010. Endogenous ligands of TLR2 and TLR4: agonists or assistants? J. Leukoc. Biol. 87:989–999. 10.1189/jlb.1209775 [DOI] [PubMed] [Google Scholar]
- Erridge C. 2011a Accumulation of stimulants of Toll-like receptor (TLR)-2 and TLR4 in meat products stored at 5 °C. J. Food Sci. 76:H72–H79. 10.1111/j.1750-3841.2010.02018.x [DOI] [PubMed] [Google Scholar]
- Erridge C. 2011b The capacity of foodstuffs to induce innate immune activation of human monocytes in vitro is dependent on food content of stimulants of Toll-like receptors 2 and 4. Br. J. Nutr. 105:15–23. 10.1017/S0007114510003004 [DOI] [PubMed] [Google Scholar]
- Erridge C., Duncan S.H., Bereswill S., and Heimesaat M.M.. 2010. The induction of colitis and ileitis in mice is associated with marked increases in intestinal concentrations of stimulants of TLRs 2, 4, and 5. PLoS One. 5:e9125 10.1371/journal.pone.0009125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraj T.A., McLaughlin C.L., and Erridge C.. 2017. Host defenses against metabolic endotoxaemia and their impact on lipopolysaccharide detection. Int. Rev. Immunol. 36:125–144. 10.1080/08830185.2017.1280483 [DOI] [PubMed] [Google Scholar]
- Ferreira M.A., Jansen R., Willemsen G., Penninx B., Bain L.M., Vicente C.T., Revez J.A., Matheson M.C., Hui J., Tung J.Y., et al. Australian Asthma Genetics Consortium Collaborators . 2017. Gene-based analysis of regulatory variants identifies 4 putative novel asthma risk genes related to nucleotide synthesis and signaling. J. Allergy Clin. Immunol. 139:1148–1157. 10.1016/j.jaci.2016.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster S.L., Hargreaves D.C., and Medzhitov R.. 2007. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 447:972–978. 10.1038/nature05836 [DOI] [PubMed] [Google Scholar]
- Gioannini T.L., and Weiss J.P.. 2007. Regulation of interactions of Gram-negative bacterial endotoxins with mammalian cells. Immunol. Res. 39:249–260. 10.1007/s12026-007-0069-0 [DOI] [PubMed] [Google Scholar]
- Gollwitzer E.S., Saglani S., Trompette A., Yadava K., Sherburn R., McCoy K.D., Nicod L.P., Lloyd C.M., and Marsland B.J.. 2014. Lung microbiota promotes tolerance to allergens in neonates via PD-L1. Nat. Med. 20:642–647. 10.1038/nm.3568 [DOI] [PubMed] [Google Scholar]
- Gregory L.G., and Lloyd C.M.. 2011. Orchestrating house dust mite-associated allergy in the lung. Trends Immunol. 32:402–411. 10.1016/j.it.2011.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halim T.Y., Steer C.A., Mathä L., Gold M.J., Martinez-Gonzalez I., McNagny K.M., McKenzie A.N., and Takei F.. 2014. Group 2 innate lymphoid cells are critical for the initiation of adaptive T helper 2 cell-mediated allergic lung inflammation. Immunity. 40:425–435. 10.1016/j.immuni.2014.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halim T.Y., Hwang Y.Y., Scanlon S.T., Zaghouani H., Garbi N., Fallon P.G., and McKenzie A.N.. 2016. Group 2 innate lymphoid cells license dendritic cells to potentiate memory TH2 cell responses. Nat. Immunol. 17:57–64. 10.1038/ni.3294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammad H., and Lambrecht B.N.. 2008. Dendritic cells and epithelial cells: linking innate and adaptive immunity in asthma. Nat. Rev. Immunol. 8:193–204. 10.1038/nri2275 [DOI] [PubMed] [Google Scholar]
- Hammad H., Chieppa M., Perros F., Willart M.A., Germain R.N., and Lambrecht B.N.. 2009. House dust mite allergen induces asthma via Toll-like receptor 4 triggering of airway structural cells. Nat. Med. 15:410–416. 10.1038/nm.1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammad H., Plantinga M., Deswarte K., Pouliot P., Willart M.A., Kool M., Muskens F., and Lambrecht B.N.. 2010. Inflammatory dendritic cells--not basophils--are necessary and sufficient for induction of Th2 immunity to inhaled house dust mite allergen. J. Exp. Med. 207:2097–2111. 10.1084/jem.20101563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst T., Sichelstiel A., Schär C., Yadava K., Bürki K., Cahenzli J., McCoy K., Marsland B.J., and Harris N.L.. 2011. Dysregulation of allergic airway inflammation in the absence of microbial colonization. Am. J. Respir. Crit. Care Med. 184:198–205. 10.1164/rccm.201010-1574OC [DOI] [PubMed] [Google Scholar]
- Hill D.A., Siracusa M.C., Abt M.C., Kim B.S., Kobuley D., Kubo M., Kambayashi T., Larosa D.F., Renner E.D., Orange J.S., et al. . 2012. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nat. Med. 18:538–546. 10.1038/nm.2657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.J., and Boushey H.A.. 2015. The microbiome in asthma. J. Allergy Clin. Immunol. 135:25–30. 10.1016/j.jaci.2014.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffnagle G.B., Dickson R.P., and Lukacs N.W.. 2017. The respiratory tract microbiome and lung inflammation: a two-way street. Mucosal Immunol. 10:299–306. 10.1038/mi.2016.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam S.A., and Luster A.D.. 2012. T cell homing to epithelial barriers in allergic disease. Nat. Med. 18:705–715. 10.1038/nm.2760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janelsins B.M., Lu M., and Datta S.K.. 2014. Altered inactivation of commensal LPS due to acyloxyacyl hydrolase deficiency in colonic dendritic cells impairs mucosal Th17 immunity. Proc. Natl. Acad. Sci. USA. 111:373–378. 10.1073/pnas.1311987111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaliannan K., Hamarneh S.R., Economopoulos K.P., Nasrin Alam S., Moaven O., Patel P., Malo N.S., Ray M., Abtahi S.M., Muhammad N., et al. . 2013. Intestinal alkaline phosphatase prevents metabolic syndrome in mice. Proc. Natl. Acad. Sci. USA. 110:7003–7008. 10.1073/pnas.1220180110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz S.S., Weinrauch Y., Munford R.S., Elsbach P., and Weiss J.. 1999. Deacylation of lipopolysaccharide in whole Escherichia coli during destruction by cellular and extracellular components of a rabbit peritoneal inflammatory exudate. J. Biol. Chem. 274:36579–36584. 10.1074/jbc.274.51.36579 [DOI] [PubMed] [Google Scholar]
- Kim Y.K., Oh S.Y., Jeon S.G., Park H.W., Lee S.Y., Chun E.Y., Bang B., Lee H.S., Oh M.H., Kim Y.S., et al. . 2007. Airway exposure levels of lipopolysaccharide determine type 1 versus type 2 experimental asthma. J. Immunol. 178:5375–5382. 10.4049/jimmunol.178.8.5375 [DOI] [PubMed] [Google Scholar]
- Kitchens R.L., Ulevitch R.J., and Munford R.S.. 1992. Lipopolysaccharide (LPS) partial structures inhibit responses to LPS in a human macrophage cell line without inhibiting LPS uptake by a CD14-mediated pathway. J. Exp. Med. 176:485–494. 10.1084/jem.176.2.485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrecht B.N., and Hammad H.. 2012. The airway epithelium in asthma. Nat. Med. 18:684–692. 10.1038/nm.2737 [DOI] [PubMed] [Google Scholar]
- Lambrecht B.N., and Hammad H.. 2014a Allergens and the airway epithelium response: gateway to allergic sensitization. J. Allergy Clin. Immunol. 134:499–507. 10.1016/j.jaci.2014.06.036 [DOI] [PubMed] [Google Scholar]
- Lambrecht B.N., and Hammad H.. 2015. The immunology of asthma. Nat. Immunol. 16:45–56. 10.1038/ni.3049 [DOI] [PubMed] [Google Scholar]
- Lewkowich I.P., Herman N.S., Schleifer K.W., Dance M.P., Chen B.L., Dienger K.M., Sproles A.A., Shah J.S., Köhl J., Belkaid Y., and Wills-Karp M.. 2005. CD4+CD25+ T cells protect against experimentally induced asthma and alter pulmonary dendritic cell phenotype and function. J. Exp. Med. 202:1549–1561. 10.1084/jem.20051506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locksley R.M. 2010. Asthma and allergic inflammation. Cell. 140:777–783. 10.1016/j.cell.2010.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M., and Munford R.S.. 2011. The transport and inactivation kinetics of bacterial lipopolysaccharide influence its immunological potency in vivo. J. Immunol. 187:3314–3320. 10.4049/jimmunol.1004087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M., Zhang M., Kitchens R.L., Fosmire S., Takashima A., and Munford R.S.. 2003. Stimulus-dependent deacylation of bacterial lipopolysaccharide by dendritic cells. J. Exp. Med. 197:1745–1754. 10.1084/jem.20030420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M., Zhang M., Takashima A., Weiss J., Apicella M.A., Li X.H., Yuan D., and Munford R.S.. 2005. Lipopolysaccharide deacylation by an endogenous lipase controls innate antibody responses to Gram-negative bacteria. Nat. Immunol. 6:989–994. 10.1038/ni1246 [DOI] [PubMed] [Google Scholar]
- Lu M., Varley A.W., Ohta S., Hardwick J., and Munford R.S.. 2008. Host inactivation of bacterial lipopolysaccharide prevents prolonged tolerance following gram-negative bacterial infection. Cell Host Microbe. 4:293–302. 10.1016/j.chom.2008.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M., Varley A.W., and Munford R.S.. 2013. Persistently active microbial molecules prolong innate immune tolerance in vivo. PLoS Pathog. 9:e1003339 10.1371/journal.ppat.1003339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Gonzalez I., Steer C.A., and Takei F.. 2015. Lung ILC2s link innate and adaptive responses in allergic inflammation. Trends Immunol. 36:189–195. 10.1016/j.it.2015.01.005 [DOI] [PubMed] [Google Scholar]
- Matera G., Muto V., Vinci M., Zicca E., Abdollahi-Roodsaz S., van de Veerdonk F.L., Kullberg B.J., Liberto M.C., van der Meer J.W., Focà A., et al. . 2009. Receptor recognition of and immune intracellular pathways for Veillonella parvula lipopolysaccharide. Clin. Vaccine Immunol. 16:1804–1809. 10.1128/CVI.00310-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munford R.S. 2016. Endotoxemia-menace, marker, or mistake? J. Leukoc. Biol. 100:687–698. 10.1189/jlb.3RU0316-151R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munford R.S., and Hall C.L.. 1986. Detoxification of bacterial lipopolysaccharides (endotoxins) by a human neutrophil enzyme. Science. 234:203–205. 10.1126/science.3529396 [DOI] [PubMed] [Google Scholar]
- Munford R.S., and Varley A.W.. 2006. Shield as signal: lipopolysaccharides and the evolution of immunity to gram-negative bacteria. PLoS Pathog. 2:e67 10.1371/journal.ppat.0020067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munford R., Lu M., and Varley A.. 2009. Chapter 2: Kill the bacteria...and also their messengers? Adv. Immunol. 103:29–48. 10.1016/S0065-2776(09)03002-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neagos J., Standiford T.J., Newstead M.W., Zeng X., Huang S.K., and Ballinger M.N.. 2015. Epigenetic Regulation of Tolerance to Toll-Like Receptor Ligands in Alveolar Epithelial Cells. Am. J. Respir. Cell Mol. Biol. 53:872–881. 10.1165/rcmb.2015-0057OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plantinga M., Guilliams M., Vanheerswynghels M., Deswarte K., Branco-Madeira F., Toussaint W., Vanhoutte L., Neyt K., Killeen N., Malissen B., et al. . 2013. Conventional and monocyte-derived CD11b(+) dendritic cells initiate and maintain T helper 2 cell-mediated immunity to house dust mite allergen. Immunity. 38:322–335. 10.1016/j.immuni.2012.10.016 [DOI] [PubMed] [Google Scholar]
- Russell S.L., Gold M.J., Hartmann M., Willing B.P., Thorson L., Wlodarska M., Gill N., Blanchet M.R., Mohn W.W., McNagny K.M., and Finlay B.B.. 2012. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 13:440–447. 10.1038/embor.2012.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell S.L., Gold M.J., Willing B.P., Thorson L., McNagny K.M., and Finlay B.B.. 2013. Perinatal antibiotic treatment affects murine microbiota, immune responses and allergic asthma. Gut Microbes. 4:158–164. 10.4161/gmic.23567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuijs M.J., Willart M.A., Vergote K., Gras D., Deswarte K., Ege M.J., Madeira F.B., Beyaert R., van Loo G., Bracher F., et al. . 2015. Farm dust and endotoxin protect against allergy through A20 induction in lung epithelial cells. Science. 349:1106–1110. 10.1126/science.aac6623 [DOI] [PubMed] [Google Scholar]
- Seibold M.A., Reese T.A., Choudhry S., Salam M.T., Beckman K., Eng C., Atakilit A., Meade K., Lenoir M., Watson H.G., et al. . 2009. Differential enzymatic activity of common haplotypic versions of the human acidic Mammalian chitinase protein. J. Biol. Chem. 284:19650–19658. 10.1074/jbc.M109.012443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao B., Lu M., Katz S.C., Varley A.W., Hardwick J., Rogers T.E., Ojogun N., Rockey D.C., Dematteo R.P., and Munford R.S.. 2007. A host lipase detoxifies bacterial lipopolysaccharides in the liver and spleen. J. Biol. Chem. 282:13726–13735. 10.1074/jbc.M609462200 [DOI] [PubMed] [Google Scholar]
- Shao B., Kitchens R.L., Munford R.S., Rogers T.E., Rockey D.C., and Varley A.W.. 2011. Prolonged hepatomegaly in mice that cannot inactivate bacterial endotoxin. Hepatology. 54:1051–1062. 10.1002/hep.24488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soroosh P., Doherty T.A., Duan W., Mehta A.K., Choi H., Adams Y.F., Mikulski Z., Khorram N., Rosenthal P., Broide D.H., and Croft M.. 2013. Lung-resident tissue macrophages generate Foxp3+ regulatory T cells and promote airway tolerance. J. Exp. Med. 210:775–788. 10.1084/jem.20121849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staab J.F., Ginkel D.L., Rosenberg G.B., and Munford R.S.. 1994. A saposin-like domain influences the intracellular localization, stability, and catalytic activity of human acyloxyacyl hydrolase. J. Biol. Chem. 269:23736–23742. [PubMed] [Google Scholar]
- Stein M.M., Hrusch C.L., Gozdz J., Igartua C., Pivniouk V., Murray S.E., Ledford J.G., Marques Dos Santos M., Anderson R.L., Metwali N., et al. . 2016. Innate Immunity and Asthma Risk in Amish and Hutterite Farm Children. N. Engl. J. Med. 375:411–421. 10.1056/NEJMoa1508749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacke F., Ginhoux F., Jakubzick C., van Rooijen N., Merad M., and Randolph G.J.. 2006. Immature monocytes acquire antigens from other cells in the bone marrow and present them to T cells after maturing in the periphery. J. Exp. Med. 203:583–597. 10.1084/jem.20052119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan A.M., Chen H.C., Pochard P., Eisenbarth S.C., Herrick C.A., and Bottomly H.K.. 2010. TLR4 signaling in stromal cells is critical for the initiation of allergic Th2 responses to inhaled antigen. J. Immunol. 184:3535–3544. 10.4049/jimmunol.0900340 [DOI] [PubMed] [Google Scholar]
- Trompette A., Divanovic S., Visintin A., Blanchard C., Hegde R.S., Madan R., Thorne P.S., Wills-Karp M., Gioannini T.L., Weiss J.P., and Karp C.L.. 2009. Allergenicity resulting from functional mimicry of a Toll-like receptor complex protein. Nature. 457:585–588. 10.1038/nature07548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyken S.J., Garcia D., Porter P., Huang X., Quinlan P.J., Blanc P.D., Corry D.B., and Locksley R.M.. 2011. Fungal chitin from asthma-associated home environments induces eosinophilic lung infiltration. J. Immunol. 187:2261–2267. 10.4049/jimmunol.1100972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rijt L.S., Jung S., Kleinjan A., Vos N., Willart M., Duez C., Hoogsteden H.C., and Lambrecht B.N.. 2005. In vivo depletion of lung CD11c+ dendritic cells during allergen challenge abrogates the characteristic features of asthma. J. Exp. Med. 201:981–991. 10.1084/jem.20042311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatanen T., Kostic A.D., d’Hennezel E., Siljander H., Franzosa E.A., Yassour M., Kolde R., Vlamakis H., Arthur T.D., Hämäläinen A.M., et al. DIABIMMUNE Study Group . 2016. Variation in Microbiome LPS Immunogenicity Contributes to Autoimmunity in Humans. Cell. 165:1551 10.1016/j.cell.2016.05.056 [DOI] [PubMed] [Google Scholar]
- von Mutius E., and Vercelli D.. 2010. Farm living: effects on childhood asthma and allergy. Nat. Rev. Immunol. 10:861–868. 10.1038/nri2871 [DOI] [PubMed] [Google Scholar]
- Wang Q., Garrity G.M., Tiedje J.M., and Cole J.R.. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267. 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Endam L.M., Filali-Mouhim A., Zhao L., Desrosiers M., Han D., and Zhang L.. 2012. Polymorphisms in RYBP and AOAH genes are associated with chronic rhinosinusitis in a Chinese population: a replication study. PLoS One. 7:e39247 10.1371/journal.pone.0039247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Zheng T., Homer R.J., Kim Y.K., Chen N.Y., Cohn L., Hamid Q., and Elias J.A.. 2004. Acidic mammalian chitinase in asthmatic Th2 inflammation and IL-13 pathway activation. Science. 304:1678–1682. 10.1126/science.1095336 [DOI] [PubMed] [Google Scholar]
- Zou B., Jiang W., Han H., Li J., Mao W., Tang Z., Yang Q., Qian G., Qian J., Zeng W., et al. . 2017. Acyloxyacyl hydrolase promotes the resolution of lipopolysaccharide-induced acute lung injury. PLoS Pathog. 13:e1006436 10.1371/journal.ppat.1006436 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.