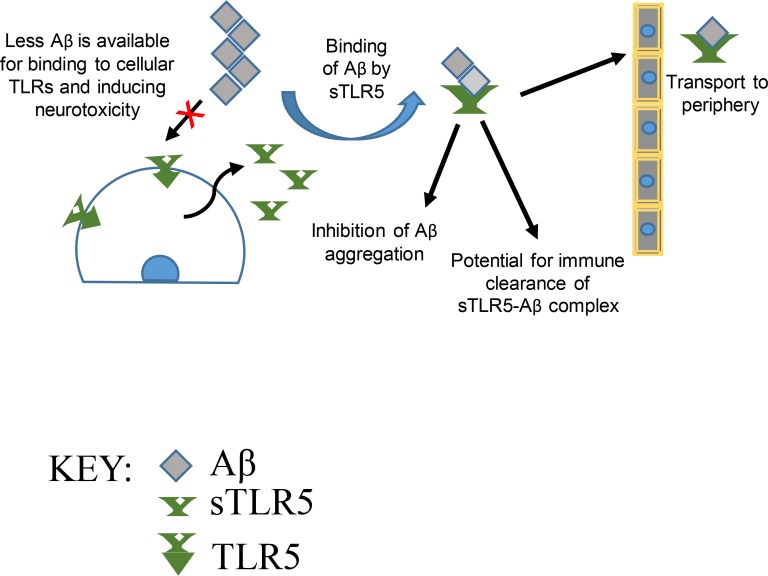
Chakrabarty et al. show that human TLR5 ectodomain reduces amyloid β (Aβ) plaques by direct interaction with Aβ, demonstrating the feasibility of such immune decoy receptor strategies as potential biotherapies in Alzheimer’s disease.
Abstract
There is considerable interest in harnessing innate immunity to treat Alzheimer’s disease (AD). Here, we explore whether a decoy receptor strategy using the ectodomain of select TLRs has therapeutic potential in AD. AAV-mediated expression of human TLR5 ectodomain (sTLR5) alone or fused to human IgG4 Fc (sTLR5Fc) results in robust attenuation of amyloid β (Aβ) accumulation in a mouse model of Alzheimer-type Aβ pathology. sTLR5Fc binds to oligomeric and fibrillar Aβ with high affinity, forms complexes with Aβ, and blocks Aβ toxicity. Oligomeric and fibrillar Aβ modulates flagellin-mediated activation of human TLR5 but does not, by itself, activate TLR5 signaling. Genetic analysis shows that rare protein coding variants in human TLR5 may be associated with a reduced risk of AD. Further, transcriptome analysis shows altered TLR gene expression in human AD. Collectively, our data suggest that TLR5 decoy receptor–based biologics represent a novel and safe Aβ-selective class of biotherapy in AD.
Graphical Abstract
Introduction
An invariant feature of the pathological cascade in Alzheimer’s disease (AD) is reactive gliosis, reflecting underlying alterations in the innate immune activation state. Association of single-nucleotide polymorphisms (SNPs) and functional variants in immune genes with AD demonstrates an important role for innate immunity in AD (International Genomics of Alzheimer’s Disease Consortium (IGAP), 2015; Karch and Goate, 2015; Saykin et al., 2015). Experimental studies in transgenic mouse models with AD-like pathologies demonstrate that manipulating innate immune pathways can have positive or negative effects on AD proteostasis, cognition, and neurodegeneration (Heneka et al., 2015; Heppner et al., 2015), a phenomenon we have collectively termed immunoproteostasis (Chakrabarty et al., 2015). These data suggest the potential of targeting immunoproteostasis for therapeutic benefit in AD.
TLRs are pattern recognition receptors of the innate immune system that are activated by pathogen- (PAMP) or damage-associated molecular patterns (DAMPs; Kawai and Akira, 2011). Engagement of TLRs can lead to a wide spectrum of outcomes: neuronal injury under chronic inflammatory conditions, but also functional recovery following nerve injury or ischemia (Rivest, 2009). AD-associated amyloid β (Aβ) aggregates appear to be DAMPs and can interact with and activate endogenous pattern recognition receptors, including a complex of TLR2, CD14, and TLR4, resulting in chronic inflammatory activation (Liu et al., 2005; Reed-Geaghan et al., 2009; Stewart et al., 2010). On the other hand, select TLRs may regulate Aβ clearance, as TLR4−/− microglia are less proficient in Aβ uptake, and bitransgenic TLR4−/−/amyloid precursor protein (APP) mice show increased Aβ plaques (Tahara et al., 2006). Similarly, stimulation of endogenous TLR4 or TLR9 activity using specific ligands reduces Aβ burden and attenuates AD-related pathology (Herber et al., 2004; Michaud et al., 2013). Thus, these two actions of TLRs (ligand binding and downstream immune signaling) might be predicted to have opposing effects on Aβ pathology. Here, we examined whether we could use the soluble ectodomains of various TLRs as immune decoy receptors to alter phenotypes in an APP transgenic model (TgCRND8; Janus et al., 2000) of AD-like Aβ deposition without activating chronic inflammation. Toward that end, we explored (1) the patterns of TLR expression in the human brain; (2) the efficacy of select soluble TLRs (sTLRs) in modulating Aβ burden and behavioral impairment in the TgCRND8 mouse model of AD; and (3) association of TLR5 haplotypes with AD risk.
Results and discussion
RNA sequencing (RNAseq) shows TLR upregulation in AD patient brains
Previous studies have suggested that select TLRs may regulate immunoproteostasis in AD models (Rivest, 2009); however, TLR expression networks have not been systematically studied in human AD. To determine whether TLR levels are differentially altered in AD patients (Table S1), we performed differential gene expression (DEG) analysis of TLRs in two brain regions of human AD and control brains: the temporal cortex (TCX), which is directly affected, and the cerebellum (CER), which is generally spared in AD (Tables S2 and S3). Using a “simple model” (see Materials and methods), we found that six TLR genes (1, 2, 4, 5, 6, and 8) were significantly upregulated in the TCX of AD patients (q < 0.05), two (TLRs 3 and 7) had suggestive DEGs, and one (TLR10) was unaffected at 0.05 < q < 0.1. TLR9 was unique in that it was significantly downregulated in AD compared with controls. In the CER, only TLRs 1, 4, and 6 were significantly upregulated in AD patients, with TLR8 showing suggestive upregulation. We subsequently reanalyzed the DEG using a more stringent “comprehensive model” (see Materials and methods), which adjusts for cell-specific gene expression changes. The comprehensive model shows that TLR-related DEG changes are mostly abolished after accounting for microglia-specific (CD68) gene expression changes, suggesting that the DEG changes were mostly attributable to increased microgliosis in AD patients.
We also determined coexpression patterns of TLR family genes using pairwise correlations of gene levels measured in TCX or CER of AD and control subjects. After adjustment of gene levels using all covariates from the comprehensive model, we found that brain expression levels of select TLRs have strong positive correlations in AD patients and controls for both TCX and CER regions (Fig. S1 and Tables S4, S5, S6, and S7). TLRs 1, 6, and 7 have the largest number of significant positive correlations across all diagnostic and brain region groups, followed by TLRs 5, 8, and 10, whereas TLRs 2, 3, 4, and 9 have the least number of positive correlations. Thus, expression levels of multiple TLRs, including TLR5, are correlated in both TCX and CER. Overall, our RNAseq data reveal that TLR transcript level correlations reflect both alterations in microglial numbers and intrinsic regulatory networks in AD.
Multiple decoy TLRs affect plaque burden in TgCRND8 mice
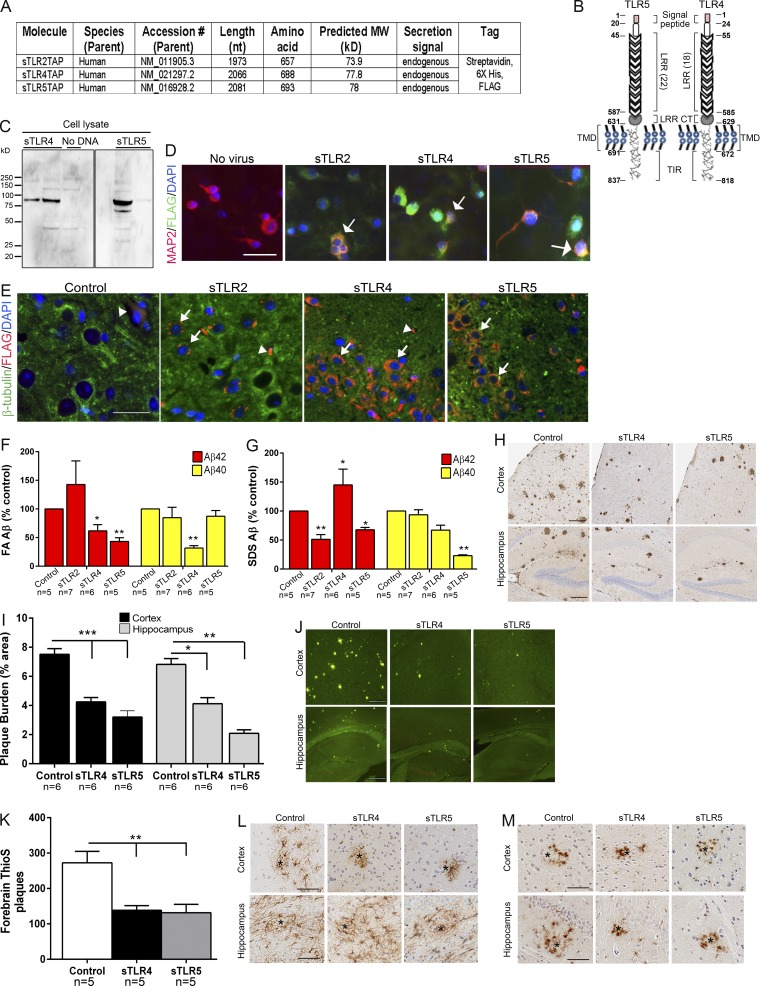
Our DEG data show that several TLRs are associated with AD neuropathology, characterized by Aβ plaques and neurofibrillary tangles, but the functional significance remains unclear. Based on previous data that Aβ interacts with select TLRs (Liu et al., 2005; Tahara et al., 2006), we hypothesized that soluble ectodomains of select human TLRs could be used as decoy receptors to bind and alter Aβ and AD-like phenotypes in an APP transgenic model (TgCRND8; Janus et al., 2000). We expected that ectodomains of TLRs (sTLRs) may scavenge Aβ or Aβ aggregates and, further, that these sTLRs might act as decoy nonsignaling receptors by being unable to initiate downstream immune signaling in the brain (Liew et al., 2005). Given that these two actions could have opposing effects on Aβ accumulation that would be challenging to evaluate in vitro, we performed neonatal somatic brain transgenesis studies using our recombinant adeno-associated virus (rAAV) vector toolkit (Chakrabarty et al., 2013). We generated rAAV2 plasmid constructs for chicken β-actin promoter–driven ectodomains of TLRs 2, 4, and 5 with a C terminus molecular tag consisting of FLAG, streptavidin, and 6X-histidine (sTLRs 2, 4, and 5; Fig. 1, A and B). We packaged these constructs in rAAV1 capsid, validated expression in cell culture and in mouse brains (Fig. 1, C–E), and delivered these to TgCRND8 mice and their wild-type littermates via intracerebroventricular injections on neonatal day P0 (Chakrabarty et al., 2010). Cohorts of mice were also injected with rAAV2/1-EGFP, which serve as a well-validated control for these studies (Chakrabarty et al., 2010, 2015). Although there was no previous data to suggest that a TLR5-based decoy receptor might interact with Aβ, we included it in these studies, as TLR5’s only known ligand, flagellin, is a fibrillar protein that can form β-sheet fibrils resembling Aβ (Hakalehto et al., 1997; Caspar, 2009). Mice were aged to 5 mo followed by examination of amyloid loads. sTLR5 expression robustly decreased formic acid and SDS-extractable insoluble Aβ42 levels (formic acid, ↓56.8%, P < 0.01; SDS, ↓49%, P < 0.01), amyloid plaque burden (hippocampus, ↓69%, P < 0.001; cortex, ↓49%, P < 0.001), and forebrain cored plaques (↓51.7%, P < 0.001; Fig. 1, F–K). sTLR4 expression also resulted in robust attenuation of formic acid–associated Aβ42 and Aβ40 levels, Aβ plaque burden, and cored plaques (Fig. 1, F–K). sTLR2 expression did not affect formic acid–associated Aβ42 and Aβ40 levels (Fig. 1 F), and we did not further examine Aβ plaque burden in these mice.
Figure 1.
Expression of sTLR4 and sTLR5 reduces Aβ plaques in TgCRND8 mice. (A) Table depicting the details of sequences used for generating rAAV2 constructs used in the study. MW, molecular weight. (B) Linear diagram of human full-length TLR5 molecule with four major domains: secretion signal peptide, leucine repeat–rich (LRR) domain, LRR C terminus (LRR CT), transmembrane domain (TMD), and Toll/IL-1 receptor (TIR) domain. The numbers depict the amino acid corresponding to each of the domains. (C) sTLR protein was detected with anti-FLAG antibody in HEK cell lysates. (D) Immunofluorescence analysis from primary neuroglial cultures transduced with rAAV2/1-sTLR shows sTLR expression (Alexa Fluor 488 nm) in MAP2-immunopositive (Alexa Fluor 594 nm) neurons (arrows). Immunofluorescence was used to confirm expression of these rAAV constructs in 6-mo-old CRND8 mouse forebrains. (E) sTLR expression (FLAG antibody, Alexa Fluor 594 nm) could be detected in β-tubulin III–immunopositive neurons (Alexa Fluor 488 nm; arrows). Arrowheads mark putative blood vessel staining. (D and E) DAPI marks nuclei. Scale bars, 50 µm (D); 25 µm (E). (F–M) Neonatal CRND8 mice were injected with AAV2/1-sTLR or AAV2/1-EGFP (control) in the cerebral ventricles and analyzed after 5 mo for biochemical Aβ levels (F and G) and Aβ plaques (H–K). (F and G) Biochemical analyses showed that sTLR4 and sTLR5 tandem affinity purification reduces formic acid– and SDS-extractable insoluble Aβ42 levels. Data (normalized to percentage of control mice in each cohort) represent mean ± SEM; n = 5–7/group. (H–K) Both anti-Aβ mAb 33.1.1 (H and I) and thioflavin S (ThioS) staining (J and K) depict reduced Aβ deposition in sTLR4- and sTLR5-expressing mice compared with controls. (L and M) Representative GFAP (L) and Iba-1 (M) immunoreactivity in AAV-expressing mice. Scale bars, 125 µm (H and J); 100 µm (L and M). n = 5–6/group. Data represent mean ± SEM. ***P < 0.001, **P < 0.01, *P < 0.05; one-way ANOVA.
Fc-tagged sTLR5 acts as a biotherapy in two independent cohorts of TgCRND8
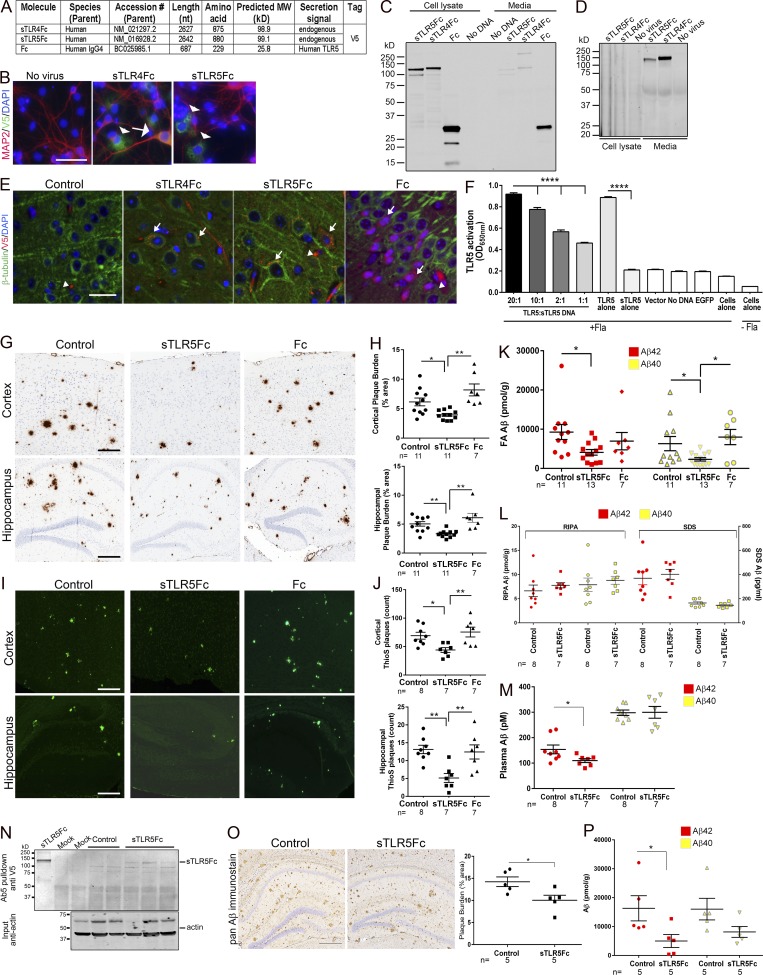
To further investigate whether sTLRs can be developed into immunobiotherapeutics, we created functionalized versions of sTLR4 and sTLR5 by adding human IgG4 Fc domain to the C terminus of the sTLR (Fig. 2 A; Huang, 2009; Czajkowsky et al., 2012). In addition to rAAV2/1-EGFP, for these experiments, we used rAAV2/1-(IgG4)Fc (“Fc”) as additional controls. These constructs produced secreted sTLRs in transiently transfected human embryonic kidney (HEK) cells and in rAAV2/1-transduced primary neuroglial cultures and are expressed in mouse brains following AAV delivery (Fig. 2, A–E). Notably, sTLR5Fc was able to functionally compete with full-length TLR5 in flagellin-dependent TLR5 activation assay (Fig. 2 F). rAAV-sTLR5Fc, rAAV-EGFP, and rAAV-Fc were delivered into neonatal TgCRND8 mice via intracerebroventricular injections and aged to 6 mo. Following behavioral studies, mice were euthanized and brain amyloid loads were assessed. Expression of sTLR5Fc could be detected in the Tris-buffered saline–soluble lysate in the brains of TgCRND8 mice (Fig. S2 A). Expression of sTLR5Fc resulted in significantly reduced Aβ plaques (cortex, ↓35.6%; hippocampus, ↓33.4%) and thioflavin S–cored plaques (cortex, ↓36.3%; hippocampus, ↓60.8%) compared with control mice (Fig. 2, G–J). Biochemical analyses showed robust reductions in insoluble Aβ42 (↓55.8%) and Aβ40 (↓63.3%) in sTLR5Fc mice (Fig. 2 K). Radioimmunoprecipitation assay (RIPA)– and SDS-associated soluble Aβ, which collectively comprise <1–3% of total Aβ levels in TgCRND8 mice at this age, was unaltered (Fig. 2 L). Aβ plaque burden, the number of cored plaques, and Aβ biochemistry were unaltered between control mice and Fc-expressing mice (Fig. 2, G–L). Plasma Aβ42 (but not Aβ40) levels were also lowered in the sTLR5Fc mice compared with control EGFP (enhanced GFP) mice (↓28%, P < 0.05; Fig. 2 M). A significant increase in C99/Aβ trimer levels in both the RIPA and SDS fractions and a slight trend toward increased monomeric Aβ in the RIPA fraction of sTLR5Fc mice forebrain were noted (Fig. S2, B–D). No significant changes in APP or APP C-terminal fragments (CTFs) were detected in the sTLR5 cohort compared with control mice (Fig. S2, E and F). Using the anti–Aβ1-16 antibody Ab5, sTR5Fc could be immunoprecipitated from the Tris-buffered saline brain extracts of sTLR5Fc-expressing APP mice, suggesting that sTLR5Fc forms a complex with Aβ in vivo (Fig. 2 N). We could also detect sTLR5Fc in the plasma of injected mice, suggesting that sTLR5Fc expressed in the brain is transported to the periphery (Fig. S2 G). Using a second independent cohort of neonatal CRND8 mice injected with a different rAAV2/1-sTLR5Fc preparation, we further confirmed that sTLR5Fc expression resulted in decreased Aβ plaque burden, thioflavin S–reactive cored plaques (Fig. S2, H–K), and attenuated formic acid–associated Aβ42 levels (Fig. S2, L and M).
Figure 2.
sTLR5Fc reduces Aβ plaques in TgCRND8 mice. (A–F) Validation of Fc-tagged decoy TLR4 and TLR5 constructs. (A) Table depicting the details of sequences used for generating Fc-tagged rAAV2 constructs. (B) Immunofluorescence analysis from primary neuroglial cultures transduced with rAAV2/1-sTLR-Fc shows sTLR expression (Alexa Fluor 488 nm) in MAP2-immunopositive (Alexa Fluor 594 nm) neurons (arrow) or in nonneuronal cells (arrowheads). (C) sTLRFc was detected with anti-V5 antibody in HEK cell lysates following transient transfection. (D) Immunoblotting shows that most of the detectable Fc-tagged sTLR proteins are secreted into the media of neuroglial cultures. (E) Immunofluorescence was used to confirm expression of these Fc-tagged rAAV constructs in 6-mo-old CRND8 mouse forebrains. sTLRFc expression (V5 antibody, Alexa Fluor 594 nm) could be detected in β-tubulin III–immunopositive neurons (Alexa Fluor 488 nm; arrows). Arrowheads mark putative blood vessel staining. (B and E) DAPI marks nuclei. Scale bars, 50 µm (B); 25 µm (E). (F) Transfection of full-length TLR5 in HEK-Blue-Null1 cells results in flagellin-induced TLR5 activation (detected using alkaline phosphatase), which is abolished in the presence of sTLR5Fc. Increasing the ratio of full-length TLR5 to sTLR5Fc (ratio: 20:1, 10:1, 2:1, and 1:1) DNA during transfection into HEK-Blue-Null1 cells increases production of alkaline phosphatase, showing that sTLR5Fc can compete with TLR5 binding to exogenous flagellin. Transfection with sTLR5Fc, vector, or EGFP alone does not lead to flagellin-induced activation of alkaline phosphatase production. Data represent mean ± SEM. ****P < 0.0001. (G–P) Neonatal CRND8 mice were injected with AAV2/1-sTLRFc, AAV2/1-Fc, or AAV2/1-EGFP (control) in the cerebral ventricles and analyzed after 6 mo. Aβ staining protocols (anti-Aβ mAb 33.1.1, G and H; and thioflavin S [ThioS], I and J) depict reduced Aβ deposition in sTLR5Fc-expressing mice compared with controls. Scale bars, 125 µm (cortex and hippocampus). n = 7–11/group. Data represent mean ± SEM. **P < 0.01, *P < 0.05; one-way ANOVA. Aβ levels in the forebrain were analyzed following serial extraction in RIPA, SDS, and formic acid. (K) Both formic acid–extracted Aβ42 and Aβ40 levels are reduced in sTLR5Fc-expressing mice. (L) There was no change in RIPA- and SDS-extracted Aβ42 and Aβ40. n = 7–13 mice/group. (M) Plasma Aβ42 levels (but not Aβ40 levels) were reduced in sTLR5Fc-expressing mice compared with controls. *P < 0.05; unpaired two-tailed Student’s t test. (N) sTLR5Fc protein was immunoprecipitated with anti–Aβ1-16 (Ab5) antibody from Tris-buffered saline–soluble brain fractions and detected using anti-V5 antibody. “Mock” represents a sham immunoprecipitation assay with no input, while purified recombinant sTLR5Fc is depicted in the left lane. A fraction of the input material was separated on a polyacrylamide gel and immunoblotted for β-actin protein to determine protein loading across the lanes. (O) 6-mo-old CRND8 mice were injected in the hippocampus with AAV2/1-sTLR5Fc or AAV2/1-EGFP (control) and analyzed at 9 mo. Representative images of the hippocampal plaque pathology (stained with mAb 33.1.1) is shown. Scale bar, 125 μm. (O and P) Both hippocampal plaque burden (O) and formic acid–extractable insoluble Aβ42 levels (P) show significant reduction in mice expressing sTLR5Fc compared with EGFP. Data represent mean ± SEM. (P) A nonsignificant reduction of formic acid–extractable Aβ40 levels is noticed in the sTLR5Fc-injected mice. n = 5 mice/group; *P < 0.05; unpaired two-tailed Student’s t test.
We next tested the therapeutic potential of sTLR5Fc by delivering AAV-sTLR5Fc in the hippocampus of plaque-depositing CRND8 mice. Following intrahippocampal injections in 9-mo-old TgCRND8 mice, we observed that sTLR5Fc expression decreased biochemical Aβ42 levels (↓67%, P < 0.05) and Aβ plaque burden (↓26%, P < 0.05) after 3 mo (Fig. 2, O and P).
sTLR5Fc attenuates expression of AD-associated genes but does not alter behavior
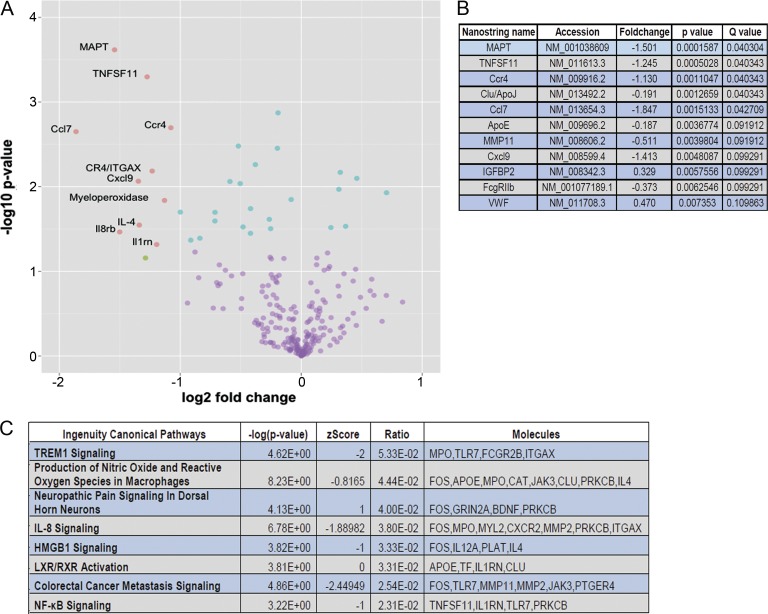
We next conducted a focused analysis of transcriptomic changes in the sTLR5Fc- and Fc-expressing mice using a custom “Neurodegeneration” NanoString array (Chakrabarty et al., 2015). This array consists of immune and select nonimmune genes that have been implicated in AD-type neurodegeneration (Matarin et al., 2015). Of 240 genes tested in this array, 2 genes were upregulated and 9 genes were downregulated in sTLR5Fc mice compared with Fc-expressing mice (q < 0.1; Fig. 3, A and B; and Table S8). Notable among the downregulated genes are cytokines and chemokines (CCl7, Cxcl9, and TNFSF11), receptors (FcγRIIb, Ccr4, clusterin, and APOE), MMP11, and microtubule-associated protein tau (MAPT). Two genes, IGFBP2 and VWF, were upregulated (P < 0.005; Fig. 3, A and B). We used differential rank conservation (DIRAC; Eddy et al., 2010) analysis to determine networks where the relative expression values of their components were most different in sTLR5Fc mice compared with Fc control cohorts. DIRAC-based analysis showed that the networks with the most significantly altered transcriptomes in the sTLR5Fc mice corresponded to Trem1 signaling, IL-8 signaling, HMGB1, and NF-κB signaling (Fig. 3 C). Since MAPT, which is a risk factor in tauopathies (Ghetti et al., 2015), was shown to be downregulated by NanoString, we tested whether endogenous tau protein was altered. We did not find any significant changes in tau protein levels in sTLR5Fc-expressing mice (Fig. S2, N and O).
Figure 3.
Transcriptome characterization of sTLR5Fc-expressing TgCRND8 mice. (A) Volcano plot depicts differentially expressed genes in sTLR5Fc- and Fc-expressing TgCRND8 mice as analyzed by NanoString Neurodegeneration custom array. Blue dots indicate q < 0.1; green dots indicate fold change values, FC > 2; and pink dots represent statistically significant changes with FC > 2 and q < 0.1. (B) Representative rank-ordered list of genes that are differentially downregulated in TgCRND8 mice is tabulated. All differentially expressed genes (q < 0.1) were submitted to ingenuity pathway analysis for canonical network analysis. (C) These pathways were analyzed by DIRAC and presented as rank-ordered network alterations. n = 5–6 mice/group.
We analyzed whether sTLR5Fc expression altered microgliosis and astrocytosis in CRND8 mice brains. sTLR5Fc-expressing mice showed a trend toward reduced glial fibrillary-associated protein (GFAP; marker of astrocyte) and cd11b (marker of activated microglia) levels by immunoblotting compared with both EGFP- and Fc-expressing control mice (Fig. S3, A–E). Notably, there was an overall reduction in microglia and possibly cells of myeloid lineage in sTLR5Fc mice as indicated by significantly reduced cd68 levels, though reductions in cd11b and GFAP levels did not reach significance (Fig. S3, A–E).
Next, we tested fear-conditioning memory (Chakrabarty et al., 2015) and burrowing activity (Deacon, 2012) in these mice. There was a trend toward improved context memory in sTLR5Fc mice (Fig. S3 F) and burrowing activity (Fig. S3 G), but tone fear memory was not altered (Fig. S3 F). We analyzed various postsynaptic and presynaptic proteins in these mice: PSD95, spinophilin, synapsin1, and synaptophysin (Fig. S3, H–L). While sTLR5Fc expression did not alter the levels of PSD95, spinophilin, and synapsin1 compared with EGFP-expressing controls, we observed an insignificant decrease in synaptophysin levels in the sTLR5Fc-expressing mice (Fig. S3 K). Interestingly, Fc expression augmented synapsin1 expression but had no significant effect on the other synaptic proteins analyzed (Fig. S3 J).
sTLR4Fc does not alter plaque pathology in TgCRND8 mice
In contrast to the effects of rAAV2/1-sTLR5Fc and rAAV2/1-sTLR4, rAAV2/1-sTLR4Fc expression did not affect Aβ plaque pathology following neonatal intracerebroventricular delivery into TgCRND8 litters. We did not detect any changes in Aβ plaque burden (Fig. S4 A), insoluble Aβ levels (Fig. S4 B), levels of APP or CTFs, and astrogliosis (GFAP level; Fig. S4 C). No changes were seen in contextual or tone fear–conditioning behavior in sTLR4Fc mice (Fig. S4 D).
TLR5 ectodomain binds Aβ and attenuates Aβ toxicity in neurons
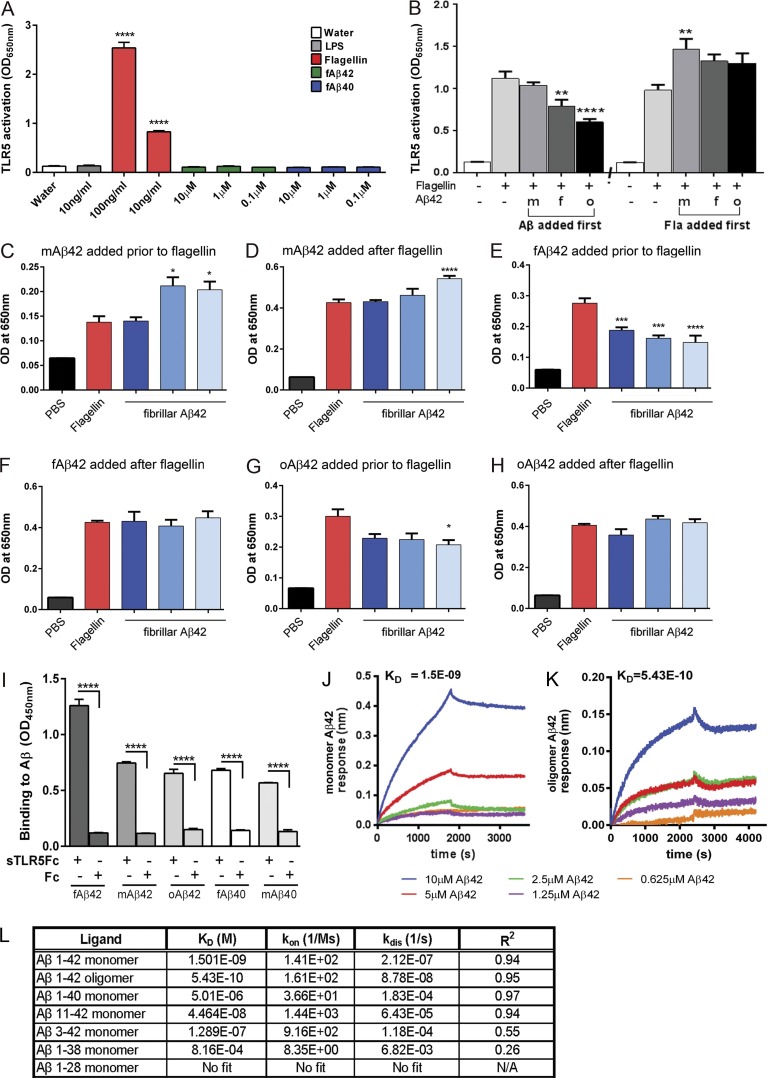
We explored if the mechanism by which sTLR5Fc leads to Aβ plaque attenuation is by direct binding and sequestration of Aβ or by indirect pathways. Initial experiments showed that neither aggregated Aβ42 nor Aβ40 acted as canonical ligands for TLR5, as these do not activate TLR5-dependent signaling in a cell-based reporter assay (Fig. 4 A). Aβ42 could, however, partially inhibit flagellin-dependent canonical TLR5 signaling if added to the cells before the addition of flagellin (Fig. 4 B). This antagonistic effect was most apparent with oligomeric Aβ42 compared with monomeric Aβ42 or fibrillar Aβ42 (Fig. 4 B). In contrast, monomeric Aβ42 showed a synergistic effect in flagellin-dependent TLR5 activation when added to the HEK-Blue-TLR5 after the cells were exposed to flagellin (Fig. 4 B). Overall, this suggests that Aβ may be an allosteric modulator of TLR5 activity by possibly interacting with the receptor. To understand this phenomenon, we next explored whether this interaction is dependent on the dose of the different forms of Aβ42. We observed that prior addition of different doses of fibrillar and oligomeric Aβ42 partially inhibited flagellin-induced TLR5 signaling, whereas adding these Aβ42 preparations to the cells after flagellin addition did not significantly affect TLR5 signaling (Fig. 4, E–H). Monomeric Aβ42, on the other hand, did not affect flagellin-induced TLR5 signaling when added before flagellin. However, when added in high concentrations (10 µM) after addition of flagellin, monomeric Aβ42 could enhance TLR5 signaling (Fig. 4, C and D). These observations confirm that varying the amount of Aβ or the specific form of Aβ could modulate TLR5 activation.
Figure 4.
sTLR5Fc modulates flagellin-mediated TLR5 activation and binds Aβ with high specificity. (A) HEK-Blue-TLR5 cells were exposed to either flagellin or increasing concentrations of fAβ42 and fAβ40. Following stimulation with 100 ng/ml flagellin, NF-κB/AP-1–driven production of alkaline phosphatase was detected in the medium, but the addition of different concentrations of fAβ42, fAβ40, or LPS (TLR4 agonist) did not elicit this response. ****P < 0.0001 compared with water control; one-way ANOVA. n = 3 replicates. (B) Application of Aβ42 (f, fibrillar; m, monomer; and o, oligomer) modulated the ability of flagellin to activate the TLR5-signaling cascade depending on whether it was added before or after the addition of flagellin to the HEK-TLR5 cells. **P < 0.01 and ****P < 0.0001 compared with flagellin treatment; one-way ANOVA. n = 3 replicates. (C–H) The interactions between Aβ42 species and TLR5 leading to modulation of flagellin-dependent TLR5 activation is dose dependent (C and D, monomeric Aβ42; E and F, fibrillar Aβ42; G and H, oligomeric Aβ42). *P < 0.05, ***P < 0.001. (I–L) ELISA-based assay (I) and biolayer interferometry (J–L) confirm that sTLR5Fc interacts with different forms of Aβ42 and Aβ40. (I) ****P < 0.0001; one-way ANOVA. n = 3 replicates. (J and K) Representative fitted curve showing binding affinity of monomeric Aβ42 (J) and oligomeric Aβ42 (K) to sTLR5Fc using biolayer interferometry. Under identical conditions, Fc (either recombinant or produced from Chinese hamster ovary clonal cell line) does not bind any Aβ species. (L) Binding affinities of different Aβ species to sTLR5Fc are tabulated. KD, affinity constant; kon, association rate; kdis, dissociation rate; R, correlation coefficient; N/A, not applicable. Data are representative of three replicate experiments. Data represent mean ± SEM.
Using well-characterized Aβ preparations and recombinant sTLR5Fc and Fc proteins (Fig. S5, A–G), we performed ELISA and biolayer interferometry assays to examine direct interactions between sTLR and Aβ. Direct ELISA binding assay using varying doses of sTLR5Fc protein showed that recombinant sTLR5Fc bound strongly and specifically to fibrillar Aβ42 and, to a lesser extent, to other forms of Aβ42 and Aβ40 (Fig. 4 I and Fig. S5, H and I). Under similar conditions, recombinant Fc does not interact with these Aβ species (Fig. 4 I). To better quantitate the affinities of sTLR5Fc to Aβ, we next used biolayer interferometry assays (Wilson et al., 2010). Real-time kinetic data confirmed that sTLR5Fc specifically bound to Aβ42, with higher affinities for oligomeric versus monomer and higher affinities for Aβ42 relative to Aβ40 (Fig. 4, J–L; and Fig. S5 J). Interestingly, Aβ species truncated at the C terminus (for example, Aβ1–16 and Aβ1–28) did not bind sTLR5Fc (Fig. 4 L). Two independent N-terminal–specific Aβ1–16 antibodies (4G8 and Ab5) could detect the sTLR5Fc-Aβ42 complex bound to the interferometry sensor, while an end-specific (Aβ35–42) antibody was unable to recognize this complex (Fig. S5 K), suggesting that the C terminus of Aβ42 may be a key part of the interaction domain. Neither Fc nor recombinant sTLR4Fc showed any binding to Aβ42 under these experimental conditions (Fig. S5 J). Overall, using both the ELISA and biolayer interferometry assays, we demonstrate a preferential binding of sTLR5Fc to Aβ42 aggregates.
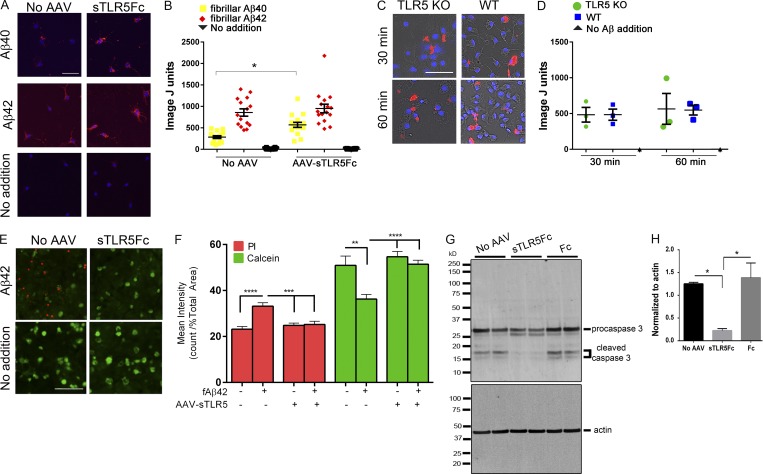
To explore how TLR5 or its decoy receptor could affect microglial Aβ uptake, we performed two experiments: in the first one, we investigated whether the presence of medium conditioned with sTLR5Fc would alter microglial uptake of fluorescently labeled fibrillar Aβ40 or fibrillar Aβ42 by wild-type microglia. We found that the presence of sTLR5Fc resulted in increased uptake of fibrillar Aβ40 preparations by wild-type microglia (Fig. 5, A and B). This suggests that a complex of sTLR and fibrillar Aβ40 may be internalized more efficiently by wild-type microglia. In a second experiment, we tested whether TLR5−/− microglia shows altered fibrillar Aβ40 uptake. Using an experimental paradigm that was used in the previous experiments, we additionally found that there was no difference in fibrillar Aβ uptake between the wild-type and TLR5−/− microglia following incubation for either 30 min or 60 min (Fig. S5, C and D).
Figure 5.
sTLR5Fc facilitates uptake of fibrillar Aβ42 and prevents Aβ-induced cell death in primary neuroglia. (A and B) Wild-type primary mouse microglia were grown on an astrocyte layer, which was transduced with rAAV2/1-sTLR5Fc, leading to expression and extrusion of sTLR5Fc in the growth medium. Microglial uptake of fibrillized Alexa Fluor 555-nm–labeled Aβ40 or Aβ42 was done in the presence of sTLR5Fc-conditioned medium or control medium not containing sTLR5Fc. Images were analyzed using ImageJ. Individual fields of view, containing at least five microglia from two experimental replicates, were imaged and analyzed. *P < 0.05; one-way ANOVA. (C and D) Primary microglia derived from wild-type and TLR5 KO mice were tested for uptake of fibrillized Alexa Fluor 555-nm–labeled Aβ following incubation for either 30 min or 60 min. n = 3 replicates, each containing at least 100 microglia. The y axis, denoted as ImageJ units, corresponds to Aβ fluorescence units normalized to DAPI cell count in both panels. (A and C) Scale bars, 50 µm. (E–H) Primary neuroglial cultures were transduced with rAAV2/1-sTLR5Fc or control (no virus added) for 4 d followed by exposure to 0.5 µM fAβ42 for 24 h. (E and F) Cell death was assayed by adding a mix of ethidium homodimer-1 and calcein-AM; dead cells are marked by red fluorescence of ethidium homodimer-1, while surviving cells are indicated by green fluorescence of calcein-AM. Scale bar, 100 µm. (F) At least 10–12 independent fields of view from two biological replicates were analyzed by a blinded experimenter using ImageJ. Data are representative of two independent experiments. **P < 0.01, ***P < 0.005, ****P < 0.001; one-way ANOVA. (G) Representative immunoblot showing AAV-sTLR5Fc, but not AAV-Fc, attenuates fAβ42-induced caspase 3 cleavage. (H) Quantitative densitometric analysis of cleaved caspase 3 (doublet at 17–20 kD) is depicted following normalization to actin levels. n = 2/experiment. *P < 0.05; one-way ANOVA. Data represent mean ± SEM.
We next investigated whether the interaction of Aβ with sTLR5Fc altered its toxicity in primary mouse neuroglial cultures. We transduced wild-type mouse primary neuroglial cultures with rAAV2/1-sTLR5Fc, and after 7 d we exposed these cultures to fAβ42 for 24 h. Treatment with fAβ42 resulted in increased cell death (decreased calcein and increased ethidium homodimer uptake; Decherchi et al., 1997). sTLR5Fc transduction before fibrillar Aβ42 addition efficiently blocked Aβ-induced neurotoxicity (Fig. 5, E and F). Fibrillar Aβ42 induces activation of different classes of caspases, including caspase 3, resulting in apoptosis of primary cultures (Mattson et al., 1998). Pre-exposure of primary neuroglial cultures to sTLR5Fc, but not Fc, attenuated fibrillar Aβ42-induced pro-caspase3 cleavage (Fig. 5, G and H).
Association of common, protein-altering TLR5 variants with AD
Previous studies have reported that protein-altering genetic variants in the TLR5 gene are associated with human disease. A common nonsense variant in the human TLR5 gene (rs5744168), which has a minor allele frequency of 5.6%, has been proposed to be a risk factor for Legionnaire’s disease (Hawn et al., 2003) and conversely to confer resistance to neonatal sepsis (Abu-Maziad et al., 2010). This variant results in a TLR5 variant (p.Arg392X) that encodes a secreted TLR5 protein (1–392) that is truncated relative to the sTLR5 ectodomain (1–636) used in our study. This variant and another common variant (p.F616L) have been shown to blunt the NF-κB responses downstream of TLR5 following flagellin challenge (Grossman et al., 2013).
To explore whether these or other protein-altering variants in the TLR5 gene alter the risk of AD, we analyzed six protein-altering variants with minor allele frequencies of 1.0% or more in a case-control series with 2,418 AD and 3,469 nondemented control subjects (Table S9). These six variants form four haplotypes (Table S10). The most common haplotype (H1) has a frequency of 55.5% and referent alleles at all six loci. H2 (36.4%), which contains the alternative allele of rs5744174, encodes a protein with one amino acid substitution (p.F616L). H3 (5.9%), which contains the alternative alleles of rs45528236, rs5744168, and rs5744174, encodes a truncated protein with one amino acid substitution (p.Q181K-R392X). H4 (1.0%), which contains the alternative alleles of rs5744177, rs5744175, rs5744174, and rs5744171, encodes a protein with four amino acid substitutions (p.L478I-F616L-I644F-D846G).
Each variant was analyzed by univariate logistic regression using an additive model with series, age, sex, and APOE dosage as covariates (Table S11). The most common SNP (rs5744174) encoding p.F616L, which occurs on haplotypes H2, H3, and H4, showed suggestive (P = 0.084) association with a reduced risk of AD (odds ratio [OR] = 0.92, 0.84–1.01). rs5744177, rs5744175, and rs5744171 almost always occur together, and they tag the H4 haplotype encoding p.L478I-F616L-I644F-D846G. These SNPs showed significant association (P < 0.02) with reduced risk of AD (identical ORs = 0.55). The two remaining SNPs (rs5744168 and rs45528236), which tag the H3 haplotype encoding a truncated (p.Q181K-R392X) protein, showed no evidence of association (P values were 0.33 for rs5744168 and 0.35 for rs45528236).
Joint analysis of the four TLR5 haplotypes by multivariate logistic regression with series, age, sex, APOE ε2 dosage, and APOE ε4 dosage as covariates and H1 as the referent yielded a global P value of 0.012 for haplotypic association (Table S10). This significant association was driven primarily by H4 (p.L478I-F616L-I644F-D846G), which showed significant (P = 0.0083) association with a reduced risk of AD (OR = 0.53, 0.33–0.85). There was also some contribution from H2 (TLR5 p.F616L), which showed suggestive (P = 0.067) association with a reduced risk of AD (OR = 0.91, 0.82–1.01). H3 (TLR5 p.Q181K-R392X) showed no evidence of association with AD (P = 0.57; OR = 1.06, 0.87–1.29). These results provide suggestive evidence that common protein-altering variants in TLR5 may be associated with a reduced risk of AD.
In summary, here we demonstrate that sTLR5/sTLR5Fc acts as a novel anti-Aβ biotherapeutic agent in mice in both preventive and therapeutic paradigms. The effect sizes of sTLR5/sTLR5Fc for reducing biochemical amyloid loads as reported here are among the strongest reported to date in the TgCRND8 model of Aβ deposition (Janus et al., 2000; Levites et al., 2006a,b). The efficacy for reducing amyloid loads and blocking toxicity can be attributed to the direct binding of TLR5 ectodomain to Aβ. Notably, the strength of this binding was similar to many of the high-affinity monoclonal anti-Aβ antibodies used in current AD immunotherapies (Levites et al., 2006a; Adolfsson et al., 2012; Demattos et al., 2012; Miles et al., 2013; Wisniewski and Goñi, 2015; Sevigny et al., 2016). Further sTLR5Fc blocks Aβ toxicity in primary neuroglial cultures, and sTLR5Fc directly facilitates microglial uptake of Aβ in vitro. These data reveal an unexpected binding activity of the TLR5 ectodomain to Aβ that can potentially be harnessed for therapeutic benefit in AD. Indeed, the only previously reported ligand of TLR5 is the bacterial protein flagellin (Hayashi et al., 2001). However, as opposed to flagellin, which is a TLR5 agonist, Aβ appears to be an allosteric modulator of TLR5 function and is incapable of signaling through the TLR5 receptor by itself.
RNAseq of human AD brains showed that levels of multiple TLRs are highly correlated, suggestive of common regulatory and effector pathways. Using the in situ hybridization data from the Allen Brain Atlas, we found that at the resting healthy state, there is very little expression of TLR5 detectable in a mouse brain or the human brain (Lein et al., 2007; Hawrylycz et al., 2012). Combined with comprehensive RNAseq model analysis, this suggests that TLR5 levels may increase in neuropathologically affected brain regions in AD, likely secondary to microglial activation. Given that secondary factors such as the neuropathological burden may induce TLRs in the AD brain, we can speculate that using a decoy receptor, such as sTLR5, could have additional neuroprotective properties, as it does protect from Aβ neurotoxicity in culture. TLR decoys can have potential usage for blocking excessive TLR signaling as well as recognizing DAMPs such as Aβ and other neurotoxic entities in AD and related dementias. Since the mouse model used in this study is primarily a model for Alzheimer-type Aβ plaque deposition, the major effects observed in our study can be directly attributed to sTLR5 binding to Aβ and its sequestration. Though this mouse model is well recognized as a primary model for Alzheimer-type Aβ plaque deposition, it does not recapitulate the entire Alzheimer neurodegenerative cascade. Therefore, the potential of sTLR5 in dampening immune activation and related neurotoxic pathways needs to be further explored in multiple models of AD. Overall, given there is precedence of sTLRs as a preclinical therapeutic in dampening systemic inflammation (Liew et al., 2005; Henrick et al., 2016), the significance of our study lies in the fact that this decoy receptor paradigm may have similar effects in AD or related disorders.
Genetic analysis of protein-coding variants of TLR5 provides compelling, though preliminary, evidence that the H4 haplotype lowers AD risk. Notably, the H3 haplotype that produces a secreted TLR5 ectodomain (a C-terminal truncated form of the entire TLR5 ectodomain used here) showed no evidence of association with AD, supporting the potential safety of using the sTLR5 decoy for AD; however, no firm conclusion should be drawn until the strength and significance of the association are assessed in a large, independent, case-controlled series.
An important aspect to consider in anti-Aβ therapeutics that have immunomodulatory functions are the inherent safety issues; immunomodulatory therapies could have significant side effects that might limit their use in an aged population (Sperling et al., 2012). Our strategy harnesses a naturally occurring phenomenon for many innate immune receptors, including select TLRs, whereby ectodomain shedding of the ligand binding domain results in a decoy receptor (Liew et al., 2005). Compared with pharmacological TLR inhibitors or antagonists (Kanzler et al., 2007), this approach utilizes the essential beneficial component of innate immune activation, i.e., direct binding and scavenging Aβ, as opposed to altering intracellular signaling of these receptors once engaged to their cognate DAMPs.
Clearly one of the limitations of peripherally delivered AD immunobiotherapies is limited brain bioavailability; for example, brain levels of peripherally delivered antibodies are typically ∼0.1% of plasma levels (Levites et al., 2006c; Atwal et al., 2011; Golde et al., 2011). Therefore, direct central nervous system delivery of a biological therapy may have several advantages, including (1) limiting peripheral side effects; (2) reducing potential for immune responses against the biologic; and (3) enhancing efficacy by increasing exposure in the target organ (Levites et al., 2006b; Thakker et al., 2009). Consistent with other studies, the presence of sTLR5Fc in plasma in our study suggests an efficient efflux pathway for biologics delivered directly to the brain, though the mechanism for this efflux remains uncertain (Abuqayyas and Balthasar, 2013). Thus, biological agents expressed in, or delivered directly to, the brain can engage toxic factors and chaperone them into the periphery where they can be “detoxified” either by clearance or simple dilution.
sTLR5Fc expression reduced overall microglial burden (cd68) and also resulted in a downward trend on astrocytosis (Gfap) and levels of activated microglia (cd11b). Similarly, a focused NanoString array indicated downregulation of a select group of immune regulatory molecules. Though MAPT RNA was downregulated, protein levels remained unchanged. In a well-controlled behavioral study, we observed only a suggestive trend toward improved cognition in the fear context test and in burrowing behavior in the sTLR5Fc cohort. Because TgCRND8 is a very aggressive early-onset model of amyloid deposition, even large reductions in amyloid may be insufficient to revert behavioral impairments at 6 mo of age. Alternatively, both control and sTLR5Fc cohorts could have reached a ceiling performance in the fear-conditioning test, while the lower burrowing activity of TgCRND8 mice might reflect broader effects of APP overexpression on complex species-specific home cage behavior. More importantly, there was no evidence for adverse effects in nontransgenic mice, strongly suggesting that sTLR5Fc or Fc itself does not impair memory functions. Most of the synaptic markers that we investigated were not altered in the sTLR5Fc-expressing animals, while one in particular, synaptophysin, was slightly reduced in the sTLR5Fc cohort compared with controls. It is possible that this reduction in synaptophysin (also observed in the Fc cohort) may partially explain why the reduction in Aβ plaques was not fully recapitulated in improved cognition.
Since we designed the studies to assess the comparative efficacies of sTLR4 and sTLR5 decoy receptors in parallel, for complete transparency, we have included the disparate data of the sTLR4 and sTLR4Fc cohorts. Additional studies will be needed to understand why sTLR4 decoy strategy reduced amyloid loads, whereas sTLR4Fc did not. Though we cannot rule out that the original sTLR4 observation could have been a chance occurrence, it is also possible that Fc functionalization of the sTLR4 decoy altered it in some way to make it less effective.
In summary, our experiments represent a paradigm shift from previous studies by our laboratory and others in the area of AD immunobiotherapy (Golde et al., 2011; Reiman, 2016). As opposed to using highly engineered antigen-specific antibodies targeting Aβ, Aβ aggregates, or both, sTLR5/STLRFc can potentially be used as a decoy receptor to therapeutically target amyloid and amyloid-like aggregates. By directly interacting with Aβ and attenuating Aβ levels in vivo, the functionalized TLR5 decoy receptor represents a novel and potentially safe class of immunomodulatory agents for AD.
Materials and methods
Materials used in the study
All reagents, materials, mouse models, and software used in the study are described in Table 1.
Table 1. Description of reagents used in this study.
| Reagent | Source | Description |
|---|---|---|
| Antibodies | ||
| Anti-Aβ 33.1.1 | Dr. Todd E. Golde | PMID: 19825975 |
| Anti-Aβ Ab5 | Dr. Todd E. Golde | PMID: 19825975 |
| Anti-Aβ 2.1.3 | Dr. Todd E. Golde | PMID: 19825975 |
| Anti-Aβ 13.1.1 | Dr. Todd E. Golde | PMID: 19825975 |
| Anti-APP CT20 | Dr. Todd E. Golde | PMID: 19825975 |
| Total Tau | Dr. Benoit I. Giasson | PMID: 28760159 |
| Cd11b | Abcam | Cat # ab133357 |
| Iba-1 | Wako | Cat # 01919741 |
| GFAP | Cell Signaling | Cat # 3670 |
| GFAP | Dako | Cat # Z0334 |
| Anti-Aβ 82E1 | IBL | Cat # 10323 |
| PSD95 | Cell Signaling | Cat # 3409 |
| Spinophilin | Cell Signaling | Cat # 14136 |
| Synapsin 1 | Cell Signaling | Cat # 5297 |
| Synaptophysin | AbCam | Cat # ab32127 |
| Cleaved caspase 3 | Cell Signaling | Cat # 9661 |
| β-actin | Sigma | Cat # A5441 |
| V5 | Life Technologies | Cat # R96025 |
| FLAG | Sigma | Cat # F1804 |
| β-tubulin | Sigma | Cat # T2200 |
| HRP-labeled secondary antibodies (ImmPress) | Vector Biolabs | Cat # MP7401; MP7402 |
| Alexa Fluor–labeled secondary antibodies | Thermo Fisher | Cat # A-11001; A-11008 |
| Bacterial and virus strains | ||
| Not applicable | ||
| Biological samples | ||
| Adeno-associated viruses serotype 1 | Dr. Todd E. Golde | Made according to PMID: 23825679 |
| Chemicals, peptides, and recombinant proteins | ||
| Aβ42, Aβ40, fluorescent Aβ40 | Anaspec | Cat # AS-24224; AS-24236; AS-60492-01 |
| Flagellin | Invivogen | Cat # tlrl-epstfla |
| Critical commercial assays | ||
| Human IgG Fc biosensor | Fortebio, Pall Life Sciences | Not applicable |
| HEK-Blue Detection | Invivogen | Cat # hb-det3 |
| NanoString custom array | NanoString | PMID: 25619653 |
| LIVE/DEAD Viability/Cytotoxicity Kit for mammalian cells | Thermo Fisher | Cat # L3224 |
| Deposited data | ||
| RNAseq data | https://www.synapse.org/#!Synapse:syn2580853/wiki/66722 | |
| Experimental models: cell lines | ||
| HEK-Blue hTLR5 cells | Invivogen | Not applicable |
| Chinese hamster ovary | ATCC | Not applicable |
| HEK 293 T | ATCC | Not applicable |
| Experimental models: organisms/strains | ||
| B6/C3H | Envigo | Not applicable |
| TgCRND8 | Dr. Todd E. Golde | Originally obtained from the Tanz Centre for Research in Neurodegenerative Diseases, Toronto, Canada |
| TLR5 KO | Jax Labs | Stock # 008377 |
| Oligonucleotides | ||
| Not applicable | ||
| Recombinant DNA | ||
| TLR and sTLR constructs (pAG3 or pAAV constructs) | Dr. Todd E. Golde | Fig. S2 |
| Software and algorithms | ||
| NanoStringNorm R package | NanoString | PMID: 22513995 |
| DIRAC | Dr. Nathan D. Price | PMID: 20523739 |
| Bioconductor package CQN | Dr. Nilüfer Ertekin-Taner | PMID: 22285995 |
| Corrplot R-package | Dr. Nilüfer Ertekin-Taner | https://github.com/taiyun/corrplot |
| Imagescope Pixel Count Program | Aperio/Leica Biosystems | Not applicable |
Animal models and AAV2/1 injection
All animal procedures were approved by the Institutional Animal Care and Use Committee at the University of Florida, Gainesville. TgCRND8 mice were maintained in-house by breeding APP transgenic males (carrying the wild-type retinal degeneration gene) with C57B6/C3H F1 females (Envigo; Janus et al., 2000). These mice have florid AD-type Aβ plaque pathology in their forebrains starting around 3 mo of age. No sex difference in either Aβ load or cognitive parameters has been noted in this transgenic line (Hanna et al., 2012; Chakrabarty et al., 2015). rAAV2/1 were generated as described previously (Chakrabarty et al., 2013). Neonatal intracerebroventricular and adult hippocampal injections with 1–2 × 1010 genome particles of AAV2/1 were performed as described before (Chakrabarty et al., 2015). For neonatal sTLR experiments, neonatal CRND8 mice were injected and analyzed at 5 mo of age. The cohort sizes were control (5F, 1M), sTLR2 (3F, 3M), sTLR4 (3F, 2M), and sTLR5 (3F, 2M; Fig. 1). For neonatal sTLR5Fc experiments, CRND8 mice were injected with AAV2/1-sTLR5Fc (7F, 6M), AAV2/1-Fc (3F, 4M), or control (6F, 5M) in the cerebral ventricles and aged for 6 mo (Fig. 2 and Fig. S2). For the adult hippocampal cohort, we used rAAV2/1-sTLR5Fc (4F, 1M) or EGFP (3F, 2M; Fig. 2). We conducted post-hoc secondary analysis and determined there were no gender-dependent effects on formic acid–associated Aβ42 values or Aβ plaque burden in the sTLR5Fc cohort (P = 0.7081 and P = 0.6712, respectively, two-tailed Student’s t test).
Preparation of Aβ and TLR5 activation assay
1,1,1,3,3,3-hexafluoro-2-propanol (Sigma)–treated Aβ1-40 and Aβ1-42 (Anaspec) were solubilized in DMSO (Sigma). Aggregation was performed in 10 mM HCl, whereas oligomerization was performed in phenol red–free Ham’s F12 media as described earlier (Stine et al., 2003). TLR5 activation assay was performed with Aβ according to the manufacturer’s instructions. Briefly, HEK-Blue hTLR5 cells (Invivogen), suspended in DMEM and 10% heat-inactivated FBS, were seeded at 40% in 180 µl/well in 96-well tissue culture–treated plates (Costar) 24 h before assay. Flagellin (Invivogen) or Aβ, prepared in endotoxin-free water, was added in duplicate, and cells were incubated at 37°C for 16 h. Alkaline phosphatase secreted to the medium, the measure of TLR5 signaling, was detected using Quanti-Blue detection reagent (Invivogen) by measuring the optical density at 650 nM.
Recombinant protein purification from Chinese hamster ovary clonal cell line and binding to Aβ
Stable cell lines were made from sTLR4Fc, sTLR5Fc, or Fc construct in pAG3 vector and maintained in 5% ultra-low IgG FBS and zeocin (Life Technologies). Media from cell lines were mixed with ProteinA Sepharose beads (Thermo Fisher) at 4°C for 16 h at a ratio of 1 µl bead slurry/1 ml culture media. Recombinant sTLR was eluted from the beads with two bed volumes of 100 mM glycine, pH 2.7, followed by neutralization with 8% (vol/vol) 1 M Tris, pH 8.8. For biolayer interferometry, purified samples were allowed to bind to human IgG Fc biosensors on Octet Red (Fortebio, Pall Life Sciences), followed by binding to sTLR protein diluted in PBS. Baseline alignment with buffer and association and disassociation kinetics were determined using Fortebio Data Analysis software. For determination of binding by ELISA, 1 μg/ml of Aβ was coated onto Immulon 4BX plates (Thermo Fisher) in 100 mM sodium carbonate buffer overnight. Plates were blocked in BlockAce (AbD Serotec) and allowed to bind to Fc-tagged proteins at 4°C overnight. Detection was performed using HRP-conjugated donkey anti-human IgG (1:5,000; Jackson ImmunoResearch).
Immunoblotting and Aβ ELISA of mouse brains
CRND8 mice brains were sagittally dissected, and the left hemisphere was used for protein extraction using a sequential extraction protocol of RIPA buffer, 2% SDS, and 70% formic acid followed by ELISA using Aβ end-specific antibodies, as previously described (Chakrabarty et al., 2015). All Aβ ELISA assays were done with experimental and control samples belonging to the individual cohort (sTLR cohort, Fig. 1; sTLR5Fc cohort 1, Fig. 2; sTLR5Fc cohort 2, Fig. S2; sTLR4Fc cohort 1, Fig. S4). ELISA results have been represented either as absolute values, or, when multiple experimental cohorts are described simultaneously, the values are represented as percent control (corresponding to each cohort). For immunoblotting, proteins were separated in 4–12% Bis-Tris gels, and the following antibodies were used: anti-Aβ 82E1 (1:500; IBL), anti-Aβ 4G8 (1:2,000; BioLegend), V5 (1:2,000; Invitrogen), PSD95 (1:500; Cell Signaling), synapsin 1 (1:500; Cell Signaling), synaptophysin (1:500; AbCam), spinophilin (1:500; Cell Signaling), cd11b (1:200; Novus), CD68 (1:500; Thermo Fisher), GFAP (1:1,000; Dako), Actin AC-15 (1:1,000, Sigma), total tau (1:1,000; B.I. Giasson), and anti-APP CT20 (1:1,000; T.E. Golde). Detection was performed using SuperSignal West Femto Chemiluminescent Substrate (Thermo Scientific) or near-infrared detection (Li-Cor). Relative band intensity was quantified using ImageJ software (National Institutes of Health).
Immunohistochemical imaging and image processing
Immunohistochemical or immunofluorescent staining on formalin-fixed, paraffin-embedded brains was done using pan-Aβ antibody 33.1.1 (1:1,500; T.E. Golde), biotinylated Ab5 antibody (Aβ 1–15; 1:500; T.E. Golde), Iba-1 (1:1,000; Wako), and GFAP (1:500; Chemicon). Antigen retrieval was done using steam for 20 min, except for biotinylated Ab5 antibody, which required additional treatment in 70% formic acid for 5 min at room temperature. Detection was performed using ImmPress reagents (Vector Laboratories). Aβ plaque burden was calculated using the Positive Pixel Count program (Aperio) by averaging two to three sections per sample, 30 µm apart. 1% thioflavin S (Sigma) staining was done using established protocols. Immunohistochemically and fluorescent-stained sections were captured using the Scanscope XT or FL image scanner (Aperio) and analyzed using the ImageScope program. Thioflavin S plaques were counted by a blinded observer.
Primary neuroglial cultures
Primary neuroglia was prepared from cerebral cortices of wild-type mice as described earlier (Li et al., 2015). Briefly, primary cultures were maintained in Neurobasal medium (Invitrogen) containing NeuroCult SM1 Neuronal Supplement and L-glutamine (Stem Cell Technologies) for 6 d with two changes of cell growth media, followed by transduction with 2 × 108 virions of rAAVs. Cells were harvested after 5 d and analyzed by immunoblotting using anti-cleaved caspase 3 (1:500; Cell Signaling), anti-V5 (1:1,000; Invitrogen), and anti-actin (1:1,000; Sigma) antibodies. Live cultures were used for performing live/dead viability assay (Thermo Fisher) using the manufacturer’s instructions, images were captured using the EVOS FL Imaging System (Thermo Fisher), and fluorescence intensity was analyzed by ImageJ. Immunofluorescence was done using the primary antibodies β-tubulin (1:1,000; Sigma), MAP2 (1:1,000; AbCaM), FLAG M2 (1:1,000; Sigma), and V5 (1:1,000; Invitrogen) and detected with Alexa Fluor–labeled secondary antibodies (Invitrogen).
Ex vivo microglia culture, Aβ phagocytosis assay, and data analysis
Alexa Fluor 555-nm–labeled Aβ (Anaspec) was solubilized in DMSO, diluted in sterile PBS to a concentration of 100 µM, and fibrillized at 37°C for 2 d. Microglial cultures were obtained from either wild-type B6/C3H (Envigo) or TLR5 KO (stock no. 008377; Jackson Labs) pups on day P2, as described earlier (Chakrabarty et al., 2010). Microglia were maintained on an astrocyte bed in DMEM containing 5–10% FBS for at least 7–10 d. For phagocytosis assays, microglia were shaken off at 37°C, plated on coated chamber slides (Lab-tek CC2), and allowed to settle overnight. The following morning, 2.5 µM fibrillized Aβ was sonicated and added to the microglia and incubated at 37°C for the indicated times. All images were processed using Fiji (ImageJ v1.51s; Schneider et al., 2012). For Fig. 5, A and B, images were captured using an Olympus BX60 microscope, and the following normalization was done. Scale was removed, and pixel measurements were used. Images were first processed by splitting channels into RGB stack, subtracting background from the red slice, running “sharpen” on the red slice, and setting an autothreshold using the intermodes algorithm with dark background. This threshold was then converted to mask. Then the “analyze particles” tool was used with size = 1–1,000 and circularity = 0.0–1.00. Results were then sent to the summary window. For the blue slice, the intermodes algorithm was used to threshold the image, a mask was created, and analyze particles was run with size = 250–100,000 and circularity = 0.0–1.00. Results were sent to the summary window. Aβ fluorescence was quantified by taking the total red area and dividing it by the number of microglia, as determined by the blue count. Results were then plotted in GraphPad (v5.04) and analyzed using one-way ANOVA with Bonferroni post test. For Fig. 5, C and D, images of the whole well were captured using a Keyence BZ-X710 microscope, and the following normalization was used. Images first had scale removed, and pixel measurements were used. Images in the blue channel were thresholded using Color Thresholder (version 2.0.0-rc-64/1.51s) with hue = 0–255, saturation = 0–255, and brightness = 126–255. A mask was then created from thresholding results. Analyze particles was then used with size = 250–100,000 and circularity = 0.0–1.00. Results were sent to the summary window. Images in the red channel were thresholded with hue = 0–255, saturation = 0–255, and brightness = 127–255. A mask was then created from thresholding results. Analyze particles was then used with size = 50–100,000 and circularity = 0.0–1.00. Results were sent to summary window. R (version 3.4.2; R Core Team, 2017) was then used in order to process the summary results. The count and area parameters from each well were then added together in order to calculate the parameters from the entire well. Normalized expression was then calculated by dividing the Aβ area by the microglia count, as determined by DAPI count.
Behavioral assays: Contextual fear conditioning
This was done as described previously in Chakrabarty et al. (2015) and Hanna et al. (2012). In the contextual fear-conditioning test, a mouse learned the association between characteristics (size, texture of walls, floor, etc.) of a training chamber that represents an initially neutral conditional stimulus and an aversive, brief electric foot shock (unconditional stimulus) that takes place in the training context. In the training session, a mouse was allowed to explore a training chamber for 120 s, with a coterminating 2-s foot shock immediately following a 30-s tone. The mouse recovered for 60 s and then received another 2-s foot shock following a 30-s tone (80 dB). The final post unconditional stimulus period was 60 s. For the context test session, on day 3, the mice were exposed to the same training context and tested for freezing response at the presentation of the tone (no shock was applied). For the tone test session, on day 4, the mouse was placed in a modified chamber; during the first 180 s, the mouse was allowed to explore the new environment; during the second 180 s, the tone was delivered, and percent freezing during both 180-s periods was recorded. The activity and freezing response of mice were recorded online by an image analysis system (FreezeFrame; Actimetrics), and the freezing was presented as a percent change with respect to any freezing exhibited during training. No shock was applied during either of the test days.
Burrowing activity test
Rodent species typical behavior, such as burrowing, can be considered as equivalent to activities of daily living and complement tests of spatial learning and memory (Deacon, 2006). During training sessions, a burrow (a 200-mm-long, 60-mm-diameter plastic pipe, slightly elevated at the open end) filled with food pellets was put into the cage where the animals were housed together to acclimate mice to using burrows. For the test, individual mice were transferred to a new cage with a container or burrow filled with 200 g of food pellets and a nestlet. Food and water were available ad libitum. The amount of pellets displaced from the burrow over the next 2 h was measured.
RNA preparation and NanoString data analysis
RNA from cryopulverized mouse cortices was prepared using Trizol reagent (Invitrogen) and purified using PureLink RNA Mini kit (Thermo Fisher) following DNase treatment (n = 5–6/group). 100 ng of RNA was used for NanoString assays using the custom-made Neurodegeneration array described earlier (Chakrabarty et al., 2015). The NanoStringNorm R package (Waggott et al., 2012) was used to normalize nCounter data from all mouse experiments. All samples were normalized collectively to minimize batch effects between experimental groups. Specifically, we normalized the raw nCounter data using the exogenous control probes common to all samples: positive controls, negative controls, and a defined list of housekeeping genes. NanoStringNorm scales endogenous gene expression based on levels (counts) of positive controls and housekeeping genes in each sample; by contrast, negative controls are used to determine a cutoff for detection of genes, in which any values below the threshold are set to zero. Normalization was done by following the recommended commands: CodeCount: “geo.mean”; Background: “mean.2sd”; and SampleContent: “housekeeping.geo.mean.” One sample from the sTLR5Fc transgenic group was removed, as it was identified as having abnormal values relative to the other samples.
Human RNAseq and data analysis
All autopsied late-onset AD subjects met neuropathologic criteria for definite AD according to NINCDS-ADRDA criteria (McKhann et al., 1984). Control subjects had a Braak neurofibrillary tangle stage of III or less and CERAD54 neuritic and cortical plaque densities of 0 (none) or 1 (sparse) and lacked any of the following pathological diagnoses: AD, Parkinson’s disease, dementia with Lewy bodies, vascular dementia, progressive supranuclear palsy, motor neuron disease, corticobasal syndrome, Pick’s disease, Huntington’s disease, frontotemporal lobar degeneration, hippocampal sclerosis, or dementia lacking distinctive histology. Within this cohort, all AD subjects were from the Mayo Clinic Brain Bank. 34 control CER and 31 control TCX samples were from the Mayo Clinic Brain Bank, and the remaining control tissue was from the Banner Sun Health Institute. All subjects were North American Caucasians. Late-onset AD subjects had ages at death ≥60; however, a more relaxed lower age cutoff of ≥50 was applied for normal controls to achieve sample sizes similar to that of AD subjects. No upper age limit was imposed on this cohort; however, when subjects had ages at death of ≥90, their ages were recorded as “90_or_above” and shown as “90” in Table S1 to protect patient confidentiality. As expected, there was a greater frequency of APOE4-positive subjects in the AD group compared with control subjects. AD subjects had a greater female sex frequency (57%) than controls (49%). RNA integrity number (RIN) for all samples was selected to be ≥5.0. Control samples had slightly lower RINs than AD samples due to limitations in availability of samples in this diagnostic category.
Brain samples for the human RNAseq study underwent RNA extractions via the Trizol/chloroform/ethanol method, followed by DNase and cleanup of RNA using Qiagen RNeasy Mini Kit and Qiagen RNase-Free DNase Set (Allen et al., 2016). The quantity and quality of the RNA samples were determined by the Agilent 2100 Bioanalyzer using the Agilent RNA 6000 Nano Chip (Agilent Technologies).
Human RNA samples were randomized by age, sex, APOE genotype, and RIN before transfer to the Mayo Clinic Medical Genome Facility Gene Expression Core and Sequencing Core for library preparation and RNAseq. CER and TCX samples underwent RNAseq at different times, but all samples for a single brain region were measured at the same time. The TruSeq RNA Sample Prep Kit (Illumina) was used for library preparation for all samples. The library concentration and size distribution were determined on an Agilent Bioanalyzer DNA 1000 chip. All samples were run in triplicate using barcoding (three samples per flowcell lane) and underwent 101-bp, paired-end sequencing. RNAseq measures were processed through the Mayo Clinic MAP-RSeq pipeline (Kalari et al., 2014) as previously described (Allen et al., 2016). Raw read counts were normalized using conditional quantile normalization (Bioconductor package CQN; Hansen et al., 2012) accounting for sequencing depth, gene length, and GC content.
Using RNAseq, we measured levels of TLR family genes in the TCX from 84 patients with a neuropathologic diagnosis of AD and 80 elderly control subjects who lacked any neurodegenerative pathology, and cerebellar tissue from 86 AD and 80 control subjects (Table S1). All subjects in the human RNAseq also underwent genome-wide genotyping with the Illumina Omni 2.5 Beadchips. These samples had quality control using both the RNAseq and genotype data. For the TCX dataset, four AD patients and four control subjects were excluded from analysis: two AD subjects with sex mismatch based on Y chromosome RNAseq and GWAS data; two AD and two control subjects who were heterozygosity outliers based on GWAS data; one control who was a third-degree relative of another subject; and one control subject who was a population outlier based on Eigenstrat (Price et al., 2006). For the CER datasets, six AD patients and four control subjects were excluded from analysis: three AD patients for mismatched gender identification and three AD patients for being outliers in heterozygosity analysis; one control based on low mapped reads; two controls who were heterozygosity outliers; and one control with a third-degree relation. Using normalized gene expression levels of the post–quality control subjects and Pearson pairwise correlation analyses, each TLR level was correlated with every other TLR, separately in the AD and control subjects and TCX and CER. These analyses were performed using residual gene expression levels obtained after accounting for key biological and technical covariates (age at death, gender, RIN, tissue source, and RNAseq flowcell), as well as brain cell–type markers for five cell-specific genes (CD68, microglia; CD34, endothelial; OLIG2, oligodendroglia; GFAP, astrocyte; and ENO2, neuron) to account for cell number changes that occur with AD neuropathology. DEG analyses were performed using multivariable linear regression with normalized gene levels as the outcome and diagnosis as the independent variable. These analyses were done under both a simple and comprehensive model. The simple model includes age at death, gender, RIN, tissue source, and RNAseq flowcell as covariates. The comprehensive model includes these and also five cell type markers to account for cell number differences between AD and control tissue. False discovery rate–based q values taking into account all tested genes (n = 55,221 for comprehensive model in TCX and n = 55,751 for CER) were used to assess the significance of DEGs. Correlation plots for Fig. S1 were generated using the “corrplot” R-package (Wei and Simko, 2016). All analyses were done with R-code. Human RNAseq data have been deposited in the Accelerating Medicines Partnership-AD knowledge portal (https://www.nia.nih.gov/alzheimers/amp-ad-target-discovery-and-preclinical-validation-project).
Genetic analysis on human brain samples
Demographic information on the late-onset AD patients and nondemented control subjects that were analyzed is shown in Table S9. Written informed consent and institutional review board approval were obtained for all individuals who participated in this study. The Mayo case-control series consisted of European Americans ascertained at the Mayo Clinic Jacksonville, Mayo Clinic Rochester, and the Mayo Clinic autopsy-confirmed samples. All subjects in the Mayo clinical case-control series were diagnosed by a neurologist at the Mayo Clinic in Jacksonville, FL, or Rochester, MN. The neurologist confirmed a Clinical Dementia Rating score of 0 for all Jacksonville and Rochester subjects enrolled as controls; cases had diagnoses of possible or probable AD made according to NINCDS-ADRDA criteria (McKhann et al., 1984). In the autopsy-confirmed series, all brains were evaluated by Dr. Dennis Dickson and came from the brain bank he maintains at the Mayo Clinic in Jacksonville, FL. In the autopsy series, the diagnosis of definite AD was also made according to NINCDS-ADRDA criteria. Only samples with an age at diagnosis >60 yr, with sex and APOE covariates (E2, E3, and E4 alleles) available, were included in this study. Genotyping was performed using Sequenom’s MassArray iPLEX technology. Sequenom’s Typer Analyzer 4.0 was used to conduct off-machine processing and genotype calling. Analysis of control subjects using PLINK showed that all variants were in Hardy–Weinberg equilibrium (Purcell et al., 2007). Haplotypes were identified using PLINK. Analysis of single variants and haplotypes was performed by logistic regression using an additive model adjusting for sex and age at diagnosis, APOE E2 dosage, and APOE E4 dosage.
Statistical analysis
One-way ANOVA with Tukey’s multiple comparison test was used for statistical comparison unless otherwise stated (SigmaStat 3.0 version). For t tests with multiple groups, multiple comparison test parameters were applied controlling for a false discovery rate of 5%. For t tests with two experimental groups, an unpaired Student’s t test was performed. Graphical analyses were done using Prism 5 (GraphPad Software), and final images were created using Photoshop CS2 (Adobe).
Online supplemental material
Fig. S1 shows TLR coexpression network in human AD. Fig. S2 shows that sTLR5Fc alters Aβ plaques but does not affect APP levels. Fig. S3 shows that sTLR5Fc expression in transgenic CRND8 mice alters immune activation but does not alter cognitive impairment. Fig. S4 shows that rAAV2/1-sTLR4Fc does not alter Aβ plaque deposition. Fig. S5 delineates the binding parameters of recombinant sTLR5Fc to Aβ. Table S1 tabulates human subjects used in TLR coexpression analysis. Table S2 shows differential expression of TLR genes in human TCX. Table S3 shows differential expression of TLR genes in the human CER. Table S4 shows TLR family gene expression correlations in TCX of AD subjects. Table S5 shows TLR family gene expression correlations in TCX of control subjects. Table S6 shows TLR family gene expression correlations in CER of AD subjects. Table S7 shows TLR family gene expression correlations in CER of control subjects. Table S8 tabulates DEG of TgCRND8 mice expressing sTLR5Fc compared with control mice. Table S9 tabulates human subjects used for analysis of protein-coding TLR variants in AD and controls. Table S10 describes association of protein-coding variants of TLR5 with risk for AD. Table S11 shows analysis of TLR5-associated SNPs in human brain.
Supplementary Material
Acknowledgments
We thank Thomas G. Beach (Banner Sun Health Institute, Sun City, AZ) for sharing human tissue through the Sun Health Research Institute Brain and Body Donation Program. P.E. Cruz thanks Shruti Desai for help in cloning rAAV constructs.
The Brain and Body Donation Program is supported by the National Institute of Neurological Disorders and Stroke (U24 NS072026, National Brain and Tissue Resource for Parkinson’s Disease and Related Disorders), the National Institute on Aging (P30 AG19610, Arizona Alzheimer’s Disease Core Center), the Arizona Department of Health Services (contract 211002, Arizona Alzheimer’s Research Center), the Arizona Biomedical Research Commission (contracts 4001, 0011, 05-901, and 1001 to the Arizona Parkinson’s Disease Consortium), and the Michael J. Fox Foundation for Parkinson’s Research. This work was supported by the National Institutes of Health/National Institute on Aging (U01 AG046139 to T.E. Golde, N.D. Price, and N. Ertekin-Taner and R01 AG018454 to T.E. Golde) and a BrightFocus Foundation Fellowship (P. Chakrabarty).
T.E. Golde and P. Chakrabarty are coinventors on a patent for the use of sTLRs as disease-modifying therapies for neurodegenerative proteinopathies (patent no. PCT/US2014/029202). The other authors declare no competing financial interests.
Author contributions: P. Chakrabarty, A. Li, E.J. Koller, and M.R. Strickland performed immunoblots and immunohistochemistry; P. Chakrabarty and X. Liu performed mouse experiments; T.B. Ladd performed protein purification, binding experiments, and immunoprecipitation; P. Chakrabarty and P.E. Cruz cloned recombinant constructs; B.D. Moore and C. Janus performed behavioral experiments; C. Ceballos-Diaz performed NanoString hybridization; C. Ceballos-Diaz, M. Yaroshenko, and M.R. Strickland performed experiments with primary cultures; J.D. Burgess, M. Allen, X. Wang, C. Younkin, and N. Ertekin-Taner performed human RNAseq data analysis; D.W. Dickson provided human brain tissue; C.C. Funk, H.-D. Li, and N.D. Price performed NanoString R and DIRAC analysis on mouse brains; A.M. Rosario prepared viruses; J. Reddy, B. Lohrer, L. Mehrke, C. Medway, and S.G. Younkin analyzed protein-coding variants of TLR5 in human brain; B.I. Giasson generated anti–total tau antibody; and P. Chakrabarty and T.E. Golde planned and executed all experiments and wrote the manuscript.
References
- Abu-Maziad A., Schaa K., Bell E.F., Dagle J.M., Cooper M., Marazita M.L., and Murray J.C.. 2010. Role of polymorphic variants as genetic modulators of infection in neonatal sepsis. Pediatr. Res. 68:323–329. 10.1203/PDR.0b013e3181e6a068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abuqayyas L., and Balthasar J.P.. 2013. Investigation of the role of FcγR and FcRn in mAb distribution to the brain. Mol. Pharm. 10:1505–1513. 10.1021/mp300214k [DOI] [PubMed] [Google Scholar]
- Adolfsson O., Pihlgren M., Toni N., Varisco Y., Buccarello A.L., Antoniello K., Lohmann S., Piorkowska K., Gafner V., Atwal J.K., et al. 2012. An effector-reduced anti-β-amyloid (Aβ) antibody with unique aβ binding properties promotes neuroprotection and glial engulfment of Aβ. J. Neurosci. 32:9677–9689. 10.1523/JNEUROSCI.4742-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen M., Burgess J.D., Ballard T., Serie D., Wang X., Younkin C.S., Sun Z., Kouri N., Baheti S., Wang C., et al. 2016. Gene expression, methylation and neuropathology correlations at progressive supranuclear palsy risk loci. Acta Neuropathol. 132:197–211. 10.1007/s00401-016-1576-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwal J.K., Chen Y., Chiu C., Mortensen D.L., Meilandt W.J., Liu Y., Heise C.E., Hoyte K., Luk W., Lu Y., et al. 2011. A therapeutic antibody targeting BACE1 inhibits amyloid-β production in vivo. Sci. Transl. Med. 3:84ra43 10.1126/scitranslmed.3002254 [DOI] [PubMed] [Google Scholar]
- Caspar D.L. 2009. Inconvenient facts about pathological amyloid fibrils. Proc. Natl. Acad. Sci. USA. 106:20555–20556. 10.1073/pnas.0910978107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty P., Jansen-West K., Beccard A., Ceballos-Diaz C., Levites Y., Verbeeck C., Zubair A.C., Dickson D., Golde T.E., and Das P.. 2010. Massive gliosis induced by interleukin-6 suppresses Abeta deposition in vivo: evidence against inflammation as a driving force for amyloid deposition. FASEB J. 24:548–559. 10.1096/fj.09-141754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty P., Rosario A., Cruz P., Siemienski Z., Ceballos-Diaz C., Crosby K., Jansen K., Borchelt D.R., Kim J.Y., Jankowsky J.L., et al. 2013. Capsid serotype and timing of injection determines AAV transduction in the neonatal mice brain. PLoS One. 8:e67680 10.1371/journal.pone.0067680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty P., Li A., Ceballos-Diaz C., Eddy J.A., Funk C.C., Moore B., DiNunno N., Rosario A.M., Cruz P.E., Verbeeck C., et al. 2015. IL-10 alters immunoproteostasis in APP mice, increasing plaque burden and worsening cognitive behavior. Neuron. 85:519–533. 10.1016/j.neuron.2014.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czajkowsky D.M., Hu J., Shao Z., and Pleass R.J.. 2012. Fc-fusion proteins: new developments and future perspectives. EMBO Mol. Med. 4:1015–1028. 10.1002/emmm.201201379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deacon R.M. 2006. Burrowing in rodents: a sensitive method for detecting behavioral dysfunction. Nat. Protoc. 1:118–121. 10.1038/nprot.2006.19 [DOI] [PubMed] [Google Scholar]
- Deacon R. 2012. Assessing burrowing, nest construction, and hoarding in mice. J. Vis. Exp. 59:e2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decherchi P., Cochard P., and Gauthier P.. 1997. Dual staining assessment of Schwann cell viability within whole peripheral nerves using calcein-AM and ethidium homodimer. J. Neurosci. Methods. 71:205–213. 10.1016/S0165-0270(96)00146-X [DOI] [PubMed] [Google Scholar]
- Demattos R.B., Lu J., Tang Y., Racke M.M., Delong C.A., Tzaferis J.A., Hole J.T., Forster B.M., McDonnell P.C., Liu F., et al. 2012. A plaque-specific antibody clears existing β-amyloid plaques in Alzheimer’s disease mice. Neuron. 76:908–920. 10.1016/j.neuron.2012.10.029 [DOI] [PubMed] [Google Scholar]
- Eddy J.A., Hood L., Price N.D., and Geman D.. 2010. Identifying tightly regulated and variably expressed networks by Differential Rank Conservation (DIRAC). PLOS Comput. Biol. 6:e1000792 10.1371/journal.pcbi.1000792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghetti B., Oblak A.L., Boeve B.F., Johnson K.A., Dickerson B.C., and Goedert M.. 2015. Invited review: Frontotemporal dementia caused by microtubule-associated protein tau gene (MAPT) mutations: a chameleon for neuropathology and neuroimaging. Neuropathol. Appl. Neurobiol. 41:24–46. 10.1111/nan.12213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golde T.E., Schneider L.S., and Koo E.H.. 2011. Anti-aβ therapeutics in Alzheimer’s disease: the need for a paradigm shift. Neuron. 69:203–213. 10.1016/j.neuron.2011.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman S.R., Andersen K.G., Shlyakhter I., Tabrizi S., Winnicki S., Yen A., Park D.J., Griesemer D., Karlsson E.K., Wong S.H., et al. 1000 Genomes Project . 2013. Identifying recent adaptations in large-scale genomic data. Cell. 152:703–713. 10.1016/j.cell.2013.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakalehto E., Santa H., Vepsäläinen J., Laatikainen R., and Finne J.. 1997. Identification of a common structural motif in the disordered N-terminal region of bacterial flagellins--evidence for a new class of fibril-forming peptides. Eur. J. Biochem. 250:19–29. 10.1111/j.1432-1033.1997.00019.x [DOI] [PubMed] [Google Scholar]
- Hanna A., Iremonger K., Das P., Dickson D., Golde T., and Janus C.. 2012. Age-related increase in amyloid plaque burden is associated with impairment in conditioned fear memory in CRND8 mouse model of amyloidosis. Alzheimers Res. Ther. 4:21 10.1186/alzrt124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen K.D., Irizarry R.A., and Wu Z.. 2012. Removing technical variability in RNA-seq data using conditional quantile normalization. Biostatistics. 13:204–216. 10.1093/biostatistics/kxr054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawn T.R., Verbon A., Lettinga K.D., Zhao L.P., Li S.S., Laws R.J., Skerrett S.J., Beutler B., Schroeder L., Nachman A., et al. 2003. A common dominant TLR5 stop codon polymorphism abolishes flagellin signaling and is associated with susceptibility to legionnaires’ disease. J. Exp. Med. 198:1563–1572. 10.1084/jem.20031220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawrylycz M.J., Lein E.S., Guillozet-Bongaarts A.L., Shen E.H., Ng L., Miller J.A., van de Lagemaat L.N., Smith K.A., Ebbert A., Riley Z.L., et al. 2012. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 489:391–399. 10.1038/nature11405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi F., Smith K.D., Ozinsky A., Hawn T.R., Yi E.C., Goodlett D.R., Eng J.K., Akira S., Underhill D.M., and Aderem A.. 2001. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 410:1099–1103. 10.1038/35074106 [DOI] [PubMed] [Google Scholar]
- Heneka M.T., Golenbock D.T., and Latz E.. 2015. Innate immunity in Alzheimer’s disease. Nat. Immunol. 16:229–236. 10.1038/ni.3102 [DOI] [PubMed] [Google Scholar]
- Henrick B.M., Yao X.D., Taha A.Y., German J.B., and Rosenthal K.L.. 2016. Insights into Soluble Toll-Like Receptor 2 as a Downregulator of Virally Induced Inflammation. Front. Immunol. 7:291 10.3389/fimmu.2016.00291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heppner F.L., Ransohoff R.M., and Becher B.. 2015. Immune attack: the role of inflammation in Alzheimer disease. Nat. Rev. Neurosci. 16:358–372. 10.1038/nrn3880 [DOI] [PubMed] [Google Scholar]
- Herber D.L., Roth L.M., Wilson D., Wilson N., Mason J.E., Morgan D., and Gordon M.N.. 2004. Time-dependent reduction in Abeta levels after intracranial LPS administration in APP transgenic mice. Exp. Neurol. 190:245–253. 10.1016/j.expneurol.2004.07.007 [DOI] [PubMed] [Google Scholar]
- Huang C. 2009. Receptor-Fc fusion therapeutics, traps, and MIMETIBODY technology. Curr. Opin. Biotechnol. 20:692–699. 10.1016/j.copbio.2009.10.010 [DOI] [PubMed] [Google Scholar]
- International Genomics of Alzheimer’s Disease Consortium (IGAP) 2015. Convergent genetic and expression data implicate immunity in Alzheimer’s disease. Alzheimers Dement. 11:658–671. 10.1016/j.jalz.2014.05.1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janus C., Pearson J., McLaurin J., Mathews P.M., Jiang Y., Schmidt S.D., Chishti M.A., Horne P., Heslin D., French J., et al. 2000. A beta peptide immunization reduces behavioural impairment and plaques in a model of Alzheimer’s disease. Nature. 408:979–982. 10.1038/35050110 [DOI] [PubMed] [Google Scholar]
- Kalari K.R., Nair A.A., Bhavsar J.D., O’Brien D.R., Davila J.I., Bockol M.A., Nie J., Tang X., Baheti S., Doughty J.B., et al. 2014. MAP-RSeq: Mayo Analysis Pipeline for RNA sequencing. BMC Bioinformatics. 15:224 10.1186/1471-2105-15-224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzler H., Barrat F.J., Hessel E.M., and Coffman R.L.. 2007. Therapeutic targeting of innate immunity with Toll-like receptor agonists and antagonists. Nat. Med. 13:552–559. 10.1038/nm1589 [DOI] [PubMed] [Google Scholar]
- Karch C.M., and Goate A.M.. 2015. Alzheimer’s disease risk genes and mechanisms of disease pathogenesis. Biol. Psychiatry. 77:43–51. 10.1016/j.biopsych.2014.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T., and Akira S.. 2011. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 34:637–650. 10.1016/j.immuni.2011.05.006 [DOI] [PubMed] [Google Scholar]
- Lein E.S., Hawrylycz M.J., Ao N., Ayres M., Bensinger A., Bernard A., Boe A.F., Boguski M.S., Brockway K.S., Byrnes E.J., et al. 2007. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 445:168–176. 10.1038/nature05453 [DOI] [PubMed] [Google Scholar]
- Levites Y., Das P., Price R.W., Rochette M.J., Kostura L.A., McGowan E.M., Murphy M.P., and Golde T.E.. 2006a Anti-Abeta42- and anti-Abeta40-specific mAbs attenuate amyloid deposition in an Alzheimer disease mouse model. J. Clin. Invest. 116:193–201. 10.1172/JCI25410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levites Y., Jansen K., Smithson L.A., Dakin R., Holloway V.M., Das P., and Golde T.E.. 2006b Intracranial adeno-associated virus-mediated delivery of anti-pan amyloid beta, amyloid beta40, and amyloid beta42 single-chain variable fragments attenuates plaque pathology in amyloid precursor protein mice. J. Neurosci. 26:11923–11928. 10.1523/JNEUROSCI.2795-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levites Y., Smithson L.A., Price R.W., Dakin R.S., Yuan B., Sierks M.R., Kim J., McGowan E., Reed D.K., Rosenberry T.L., et al. 2006c Insights into the mechanisms of action of anti-Abeta antibodies in Alzheimer’s disease mouse models. FASEB J. 20:2576–2578. 10.1096/fj.06-6463fje [DOI] [PubMed] [Google Scholar]
- Li A., Ceballos-Diaz C., DiNunno N., Levites Y., Cruz P.E., Lewis J., Golde T.E., and Chakrabarty P.. 2015. IFN-γ promotes τ phosphorylation without affecting mature tangles. FASEB J. 29:4384–4398. 10.1096/fj.15-275834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew F.Y., Xu D., Brint E.K., and O’Neill L.A.. 2005. Negative regulation of toll-like receptor-mediated immune responses. Nat. Rev. Immunol. 5:446–458. 10.1038/nri1630 [DOI] [PubMed] [Google Scholar]
- Liu Y., Walter S., Stagi M., Cherny D., Letiembre M., Schulz-Schaeffer W., Heine H., Penke B., Neumann H., and Fassbender K.. 2005. LPS receptor (CD14): a receptor for phagocytosis of Alzheimer’s amyloid peptide. Brain. 128:1778–1789. 10.1093/brain/awh531 [DOI] [PubMed] [Google Scholar]
- Matarin M., Salih D.A., Yasvoina M., Cummings D.M., Guelfi S., Liu W., Nahaboo Solim M.A., Moens T.G., Paublete R.M., Ali S.S., et al. 2015. A genome-wide gene-expression analysis and database in transgenic mice during development of amyloid or tau pathology. Cell Reports. 10:633–644. 10.1016/j.celrep.2014.12.041 [DOI] [PubMed] [Google Scholar]
- Mattson M.P., Partin J., and Begley J.G.. 1998. Amyloid beta-peptide induces apoptosis-related events in synapses and dendrites. Brain Res. 807:167–176. 10.1016/S0006-8993(98)00763-X [DOI] [PubMed] [Google Scholar]
- McKhann G., Drachman D., Folstein M., Katzman R., Price D., and Stadlan E.M.. 1984. Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology. 34:939–944. 10.1212/WNL.34.7.939 [DOI] [PubMed] [Google Scholar]
- Michaud J.P., Hallé M., Lampron A., Thériault P., Préfontaine P., Filali M., Tribout-Jover P., Lanteigne A.M., Jodoin R., Cluff C., et al. 2013. Toll-like receptor 4 stimulation with the detoxified ligand monophosphoryl lipid A improves Alzheimer’s disease-related pathology. Proc. Natl. Acad. Sci. USA. 110:1941–1946. 10.1073/pnas.1215165110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles L.A., Crespi G.A., Doughty L., and Parker M.W.. 2013. Bapineuzumab captures the N-terminus of the Alzheimer’s disease amyloid-beta peptide in a helical conformation. Sci. Rep. 3:1302 10.1038/srep01302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., and Reich D.. 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38:904–909. 10.1038/ng1847 [DOI] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., and Sham P.C.. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81:559–575. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team 2017. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Available at http://www.r-project.org/.
- Reed-Geaghan E.G., Savage J.C., Hise A.G., and Landreth G.E.. 2009. CD14 and toll-like receptors 2 and 4 are required for fibrillar Abeta-stimulated microglial activation. J. Neurosci. 29:11982–11992. 10.1523/JNEUROSCI.3158-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiman E.M. 2016. Alzheimer’s disease: Attack on amyloid-β protein. Nature. 537:36–37. 10.1038/537036a [DOI] [PubMed] [Google Scholar]
- Rivest S. 2009. Regulation of innate immune responses in the brain. Nat. Rev. Immunol. 9:429–439. 10.1038/nri2565 [DOI] [PubMed] [Google Scholar]
- Saykin A.J., Shen L., Yao X., Kim S., Nho K., Risacher S.L., Ramanan V.K., Foroud T.M., Faber K.M., Sarwar N., et al. Alzheimer’s Disease Neuroimaging Initiative . 2015. Genetic studies of quantitative MCI and AD phenotypes in ADNI: Progress, opportunities, and plans. Alzheimers Dement. 11:792–814. 10.1016/j.jalz.2015.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., and Eliceiri K.W.. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9:671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevigny J., Chiao P., Bussière T., Weinreb P.H., Williams L., Maier M., Dunstan R., Salloway S., Chen T., Ling Y., et al. 2016. The antibody aducanumab reduces Aβ plaques in Alzheimer’s disease. Nature. 537:50–56. 10.1038/nature19323 [DOI] [PubMed] [Google Scholar]
- Sperling R., Salloway S., Brooks D.J., Tampieri D., Barakos J., Fox N.C., Raskind M., Sabbagh M., Honig L.S., Porsteinsson A.P., et al. 2012. Amyloid-related imaging abnormalities in patients with Alzheimer’s disease treated with bapineuzumab: a retrospective analysis. Lancet Neurol. 11:241–249. 10.1016/S1474-4422(12)70015-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart C.R., Stuart L.M., Wilkinson K., van Gils J.M., Deng J., Halle A., Rayner K.J., Boyer L., Zhong R., Frazier W.A., et al. 2010. CD36 ligands promote sterile inflammation through assembly of a Toll-like receptor 4 and 6 heterodimer. Nat. Immunol. 11:155–161. 10.1038/ni.1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stine W.B. Jr., Dahlgren K.N., Krafft G.A., and LaDu M.J.. 2003. In vitro characterization of conditions for amyloid-beta peptide oligomerization and fibrillogenesis. J. Biol. Chem. 278:11612–11622. 10.1074/jbc.M210207200 [DOI] [PubMed] [Google Scholar]
- Tahara K., Kim H.D., Jin J.J., Maxwell J.A., Li L., and Fukuchi K.. 2006. Role of toll-like receptor signalling in Abeta uptake and clearance. Brain. 129:3006–3019. 10.1093/brain/awl249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakker D.R., Weatherspoon M.R., Harrison J., Keene T.E., Lane D.S., Kaemmerer W.F., Stewart G.R., and Shafer L.L.. 2009. Intracerebroventricular amyloid-beta antibodies reduce cerebral amyloid angiopathy and associated micro-hemorrhages in aged Tg2576 mice. Proc. Natl. Acad. Sci. USA. 106:4501–4506. 10.1073/pnas.0813404106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waggott D., Chu K., Yin S., Wouters B.G., Liu F.F., and Boutros P.C.. 2012. NanoStringNorm: an extensible R package for the pre-processing of NanoString mRNA and miRNA data. Bioinformatics. 28:1546–1548. 10.1093/bioinformatics/bts188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei T., and Simko V.. 2016. Corrplot. Available at: https://github.com/taiyun/corrplot (accessed August 29, 2017).
- Wilson J.L., Scott I.M., and McMurry J.L.. 2010. Optical biosensing: Kinetics of protein A-IGG binding using biolayer interferometry. Biochem. Mol. Biol. Educ. 38:400–407. 10.1002/bmb.20442 [DOI] [PubMed] [Google Scholar]
- Wisniewski T., and Goñi F.. 2015. Immunotherapeutic approaches for Alzheimer’s disease. Neuron. 85:1162–1176. 10.1016/j.neuron.2014.12.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.