Martini-Stoica et al. demonstrate that the TFEB-mediated lysosomal pathway in astrocytes is increased in tauopathy and plays a functional role in modulating extracellular tau and the propagation of neuronal tau pathology in a mouse model of tau spreading.
Abstract
The progression of tau pathology in Alzheimer’s disease follows a stereotyped pattern, and recent evidence suggests a role of synaptic connections in this process. Astrocytes are well positioned at the neuronal synapse to capture and degrade extracellular tau as it transits the synapse and hence could potentially have the ability to inhibit tau spreading and delay disease progression. Our study shows increased expression and activity of Transcription Factor EB (TFEB), a master regulator of lysosomal biogenesis, in response to tau pathology in both human brains with dementia and transgenic mouse models. Exogenous TFEB expression in primary astrocytes enhances tau fibril uptake and lysosomal activity, while TFEB knockout has the reverse effect. In vivo, induced TFEB expression in astrocytes reduces pathology in the hippocampus of PS19 tauopathy mice, as well as prominently attenuates tau spreading from the ipsilateral to the contralateral hippocampus in a mouse model of tau spreading. Our study suggests that astrocytic TFEB plays a functional role in modulating extracellular tau and the propagation of neuronal tau pathology in tauopathies such as Alzheimer’s disease.
Introduction
Tauopathies include a broad range of neurological diseases that are characterized by intracellular inclusions of aberrant tau protein. In Alzheimer’s disease, the most common tauopathy, intraneuronal neurofibrillary tangles (NFTs) composed of hyperphosphorylated tau accumulate and are strongly correlated with progressive cognitive decline and neurodegeneration (Giannakopoulos et al., 2003). Evidence from postmortem Alzheimer’s disease brains suggests that NFT pathology progresses in a hierarchical, stereotyped pattern beginning in the transentorhinal cortex and eventually spreading to synaptically connected brain regions such as the hippocampus and, later, the cortex (Braak and Braak, 1995). This pathological spreading pattern is the basis of Braak staging in Alzheimer’s disease and is associated with changes in memory and other cognitive domains (Grober et al., 1999). However, the precise mechanism of the tau spreading pattern is unknown. Mounting evidence suggests tau aggregates can cross cell membranes and seed tau pathology followed by subsequent spreading of pathology to other cells, resulting in the cell-to-cell transfer or prion-like propagation of tau pathology (Frost et al., 2009; Guo and Lee, 2011). This theory of tau spreading suggests a crucial role of extracellular tau species in disease progression. Lending further support to this mechanism are studies where the intracranial injection of synthetic tau fibrils or tauopathy mouse brain extract in transgenic tau mice resulted in the induction of NFT-like pathology and spreading of pathology to synaptically connected brain regions (Clavaguera et al., 2009; Iba et al., 2013). Moreover, studies have shown increased CSF tau levels in Alzheimer’s disease, as well as detected tau in the brain interstitial fluid via in vivo microdialysis in a tauopathy mouse model (Riemenschneider et al., 2003; Yamada et al., 2011). Given the proposed role of extracellular tau in the pathogenesis of Alzheimer’s disease, targeting this species of tau could be relevant to halting disease progression.
Evidence suggests that tau spreading between neurons occurs trans-synaptically (Wu et al., 2016). While neurons would seem to be the main players in this process, astroglia may be uniquely poised to modulate this process given their close physical proximity to neurons and formation of “tripartite” synapses consisting of astroglial projections and neuronal pre- and post-synaptic terminals (Halassa et al., 2007; Perea et al., 2009). Astrocytes are also the most abundant cell type in the brain and play many complex roles in the central nervous system (CNS; such as modulating synapse formation, maintaining neuronal homeostasis through metabolic support, and comprising part of the blood–brain barrier (Barres, 2008). Astroglia also react to neuronal injury and neurodegenerative conditions such as Alzheimer’s disease with changes in morphology, gene expression, and function (Pekny et al., 2016). In Alzheimer’s disease, reactive astrocytes have a prominent role in neuroinflammation through the release of pro-inflammatory mediators and cytotoxic molecules, which is thought to exacerbate pathology (Heneka et al., 2015). However, astrocytes also express genes involved in phagocytosis, which may attenuate pathology through uptake and clearance of protein aggregates (Cahoy et al., 2008). This phagocytic capacity has been demonstrated with respect to Aβ in several studies (Wyss-Coray et al., 2003; Basak et al., 2012; Li et al., 2014; Xiao et al., 2014), but little is known about the ability of astrocytes to modulate tau pathology.
In a transgenic model of tau spreading, astrocytes take up hyperphosphorylated tau as synapses degenerate (de Calignon et al., 2012). Tau inclusions are also present in astroglia in several tauopathies, including progressive supranuclear palsy (PSP), corticobasal degeneration (CBD), and Pick’s disease (Chin and Goldman, 1996). In addition, glial fibrillary tangles composed of hyperphosphorylated and aggregated tau accumulate in the medial temporal lobe of older individuals in what are called thorn-shaped astrocytes (Schultz et al., 2004). More recently, neuropathologists have sought to more systematically characterize the accumulation of abnormally phosphorylated tau in astrocytes, broadly referring to this phenomenon as aging-related tau astrogliopathy (ARTAG; Kovacs et al., 2016, 2017a,b). ARTAG may be a benign part of normal aging, but may also be a sinister harbinger of the development of a primary tauopathy, either resulting in pathological tau inclusions in astrocytes or indicating astrocyte involvement in early stages of trans-synaptic tau spreading (Kovacs et al., 2017b). Recent findings also suggest the possibility of neuron to astrocyte or even glia to glia tau spreading mechanisms in certain tauopathies such as PSP and CBD (Narasimhan et al., 2017). How and why these glial aggregates accumulate (tau uptake versus aberrant/ectopic tau expression) is still a mystery, but a possible explanation lies in the impairment of one of the main mechanisms of bulk protein degradation, the autophagy lysosomal pathway. Defects in the lysosomal pathway can occur with aging (Kato et al., 1998; Cuervo and Dice, 2000; Wolfe et al., 2013) or due to familial Alzheimer’s disease–related loss of presenilin (Lee et al., 2010, 2015; Coen et al., 2012). Furthermore, lysosomal dysfunction alone results in the accumulation of aberrant tau (Oyama et al., 1998; Nixon, 2004). One possible approach to combat the accumulation of tau is to enhance the degradative capacity of the lysosomal pathway. Transcription Factor EB (TFEB) is a ubiquitously expressed transcription factor that stimulates lysosomal biogenesis and autophagy when activated, resulting in the breakdown of proteins and lipids for nutrients (Sardiello et al., 2009; Settembre et al., 2011). TFEB also promotes clearance of aberrant storage material in lysosomal storage disorders (Medina et al., 2011; Song et al., 2013; Spampanato et al., 2013; Palmieri et al., 2017). CNS expression of TFEB results in clearance of aberrant tau species in tau transgenic mice, though this was primarily attributed to an intraneuronal/cell autonomous mechanism (Polito et al., 2014). Here, we demonstrate a functional role for astroglial TFEB in stimulating the uptake and clearance of aberrant extracellular tau thereby preventing neuronal spreading of tau pathology in a mouse model of tauopathy.
Results
TFEB activity is enhanced with clinical disease progression and tauopathy
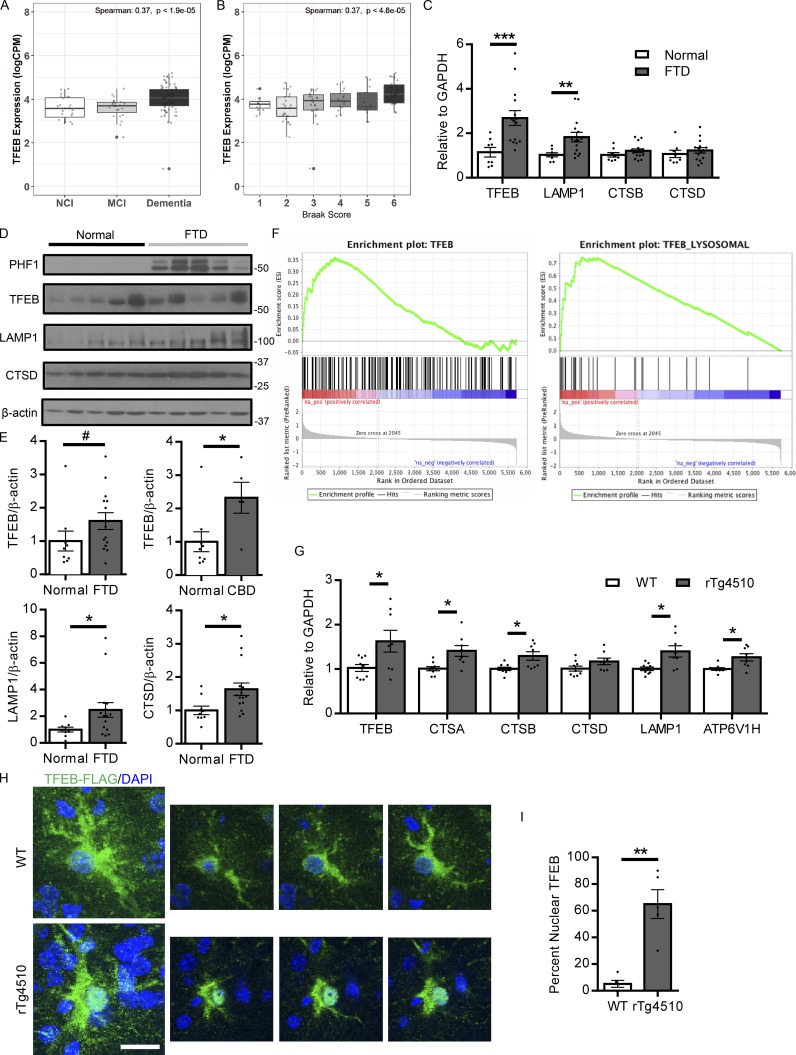
TFEB activation has been linked to lysosomal stress and the accumulation of aberrant protein aggregates, indicated by TFEB’s nuclear localization in lysosomal storage disorders (Sardiello et al., 2009; Song et al., 2013). To determine if a similar phenomenon occurs in the protein aggregopathies of Alzheimer’s disease and other tauopathies, the status of TFEB activation in relation to clinical disease progression was analyzed (Mount Sinai School of Medicine [MSSM], Accelerating Medicines Partnership-Alzheimer’s disease [AMP-AD] RNA-seq project, Synapse ID: syn8484987). Transcriptional levels of TFEB and several of its well-known lysosomal targets were stratified by no dementia, mild cognitive impairment, and dementia, showing that these transcript levels positively correlate with cognitive decline (Fig. 1 A and Fig. S1, A–C). Further, transcriptional levels of these same genes were also positively correlated with increasing Braak score, indicating a relationship between the TFEB lysosomal pathway and worsening pathology (Fig. 1 B and Fig. S1, D–F). In addition, human brains diagnosed with frontotemporal dementia (FTD) demonstrated an increase in transcriptional levels of TFEB and TFEB lysosomal target gene LAMP1 (Fig. 1 C). On the protein level, TFEB lysosomal target genes LAMP1 and Cathepsin D (CTSD) are also increased in the FTD frontal cortex relative to normal subjects, and TFEB protein levels trended increased with statistical significance in the CBD group (Fig. 1, D and E). In the rTg4510 mouse model of tauopathy (Ramsden et al., 2005; SantaCruz et al., 2005), gene set enrichment analysis (GSEA) of microarray data revealed a similar enrichment of TFEB’s transcriptional profile when comparing 4-mo-old transgenic mice with widespread NFTs to wild-type mice, indicating TFEB’s activation with tau pathology (Fig. 1 F). Examining the protein levels in another tauopathy mouse model, hippocampal lysates from PS19 mice (Yoshiyama et al., 2007) at 10 mo of age showed increases in TFEB and lysosomal proteins LAMP1 and CTSD compared with wild-type mice (Fig. S1, G and H). With TFEB protein expression detected in total brain lysate, we also confirmed its expression at the protein level in specific CNS cell types including astrocytes, neurons, and microglia isolated for primary culture (Fig. S1 I). Since multiple cell types in the CNS express TFEB, we isolated astrocytes from the forebrain of the rTg4510 mice via FACS and performed quantitative RT-PCR (qRT-PCR) to understand the astroglial contribution to TFEB activation in tauopathy (Swartzlander et al., 2018). Transcriptomic analysis of these isolated astrocytes revealed an increase in TFEB transcripts, as well as transcripts of several of its well-known target genes in the rTg4510 relative to wild-type controls (Fig. 1 G). This cell type specific data suggests that TFEB is activated in astrocytes in response to tauopathy. In addition, TFEB’s subcellular localization in astrocytes was analyzed via FLAG immunostaining of AAV-GFAP-TFEB3×FLAG–injected mice. Two groups of mice, wild-type and rTg4510 mice were injected intracerebroventricularly (ICV) at post-natal day 3 (P3) with an Adeno-associated virus (AAV)–expressing a human TFEB-FLAG fusion protein under a GFAP promoter for astrocyte specific expression. Quantitation of TFEB-3×FLAG nuclear localization, a marker of TFEB activation, in these mice revealed a significant increase in TFEB nuclear localization with tau pathology (Fig. 1, H and I). A similar increase in astrocyte TFEB nuclear localization was also observed in tau spreading mice (Fig. S1, J and K). Overall, these findings indicate an activation of TFEB relative to both cognitive decline in humans and tau pathology in mouse models. In particular, this activation is present in astrocytes, suggesting that astroglial TFEB may be part of a compensatory/adaptive response to cellular stress induced by aberrant protein aggregation.
Figure 1.
TFEB activity is enhanced in response to AD/tauopathy disease progression. (A) Human brain TFEB transcript levels stratified by clinical assessment of cognition from the MSSM cohorts in the AMP-AD reprocessed RNA-seq project (parahippocampal gyrus; n = 130). The three dementia groups are defined based on CERAD score (CDR): CDR = 0 is NCI, CDR = 0.5 is MCI, CDR ≥1 is Dementia. Performed once. (B) Human brain TFEB transcript levels stratified by Braak score (1–6) from the MSSM cohorts in AMP-AD reprocessed RNA-seq project (parahippocampal gyrus; n = 130). Spearman’s correlation coefficient is computed between the expression and dementia group status (A) or Braak score (B) and P value show the significance of the observed correlation computed using the asymptotic t approximation algorithm. Performed once. (C) qRT-PCR analysis of the transcriptional levels of TFEB and its lysosomal target genes in the frontal cortex of FTD or normal human subjects. n = 15 FTD, 8 normal cases. Student’s t test, experiments technically replicated twice. (D and E) Representative Western blot with quantitation (E) of TFEB and its lysosomal targets LAMP1 and CTSD, as well as phospho tau levels indicated by phospho tau antibody PHF1 in normal versus FTD human subjects. n = 15 FTD, 9 normal cases. #P = 0.14. Student’s t test; experiment technically replicated twice. (F) GSEA of transcriptomic changes in the brains of 4-mo-old wild-type versus rTg4510 mice. The left panel shows enrichment of TFEB transcriptional targets (ES: 0.36, P value: 0.0096), while the right panel demonstrates enrichment of TFEB lysosomal transcriptional targets (ES: 0.72; P value: <0.001). n = 4/group. Performed once. (G) qRT-PCR of TFEB regulated lysosomal genes in astrocytes from 4.5-mo-old wild-type versus rTg4510 mice. n = 8/group. Student’s t test; pooled from three independent experiments. CTSA, Cathepsin A; CTSB, Cathepsin B. (H and I) Representative confocal images of FLAG (green) and DAPI (blue) staining in the forebrain of P3 AAV-GFAP-TFEB injected wild-type or rTg4510 mice at 4 mo of age with quantification (I). Bar, 10 µm. n = 5/group with 5 images/animal; Student’s t test, representative of two independently performed experiments. Molecular mass indicated in kilodaltons. Error bars designate SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
TFEB enhances lysosomal biogenesis and stimulates uptake of preformed fibrils of tau (pffs) in primary astrocytes
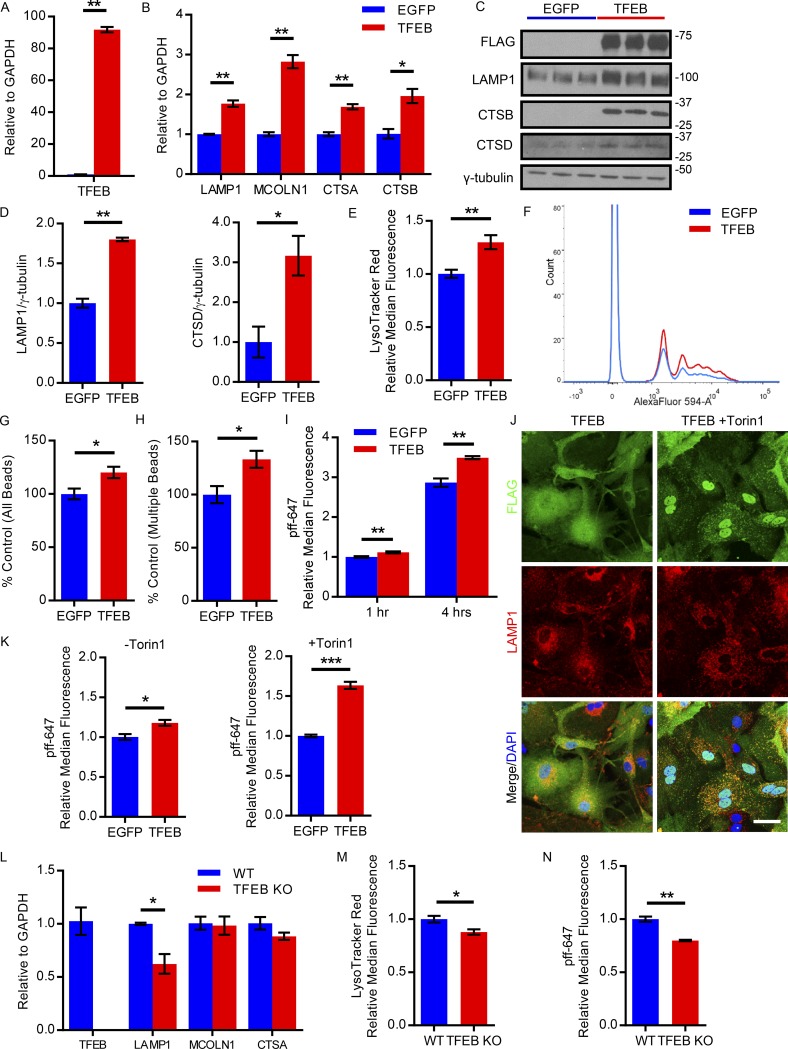
To examine TFEB’s regulation of known lysosomal targets in astrocytes, we cultured primary astrocytes from wild-type mice (C57/BL6) and used an AAV delivery approach to overexpress TFEB or EGFP as described above (Fig. 2 A). As expected, the expression of several TFEB lysosomal target genes was significantly up-regulated in TFEB-transduced astrocytes, including lysosomal marker LAMP1 and lysosomal proteases, cathepsins A and B (Fig. 2 B). Likewise, protein expression of lysosomal TFEB targets LAMP1, CTSB, and the mature form of CTSD were increased (Fig. 2, C and D). To determine if TFEB overexpression increases overall lysosomal biogenesis, we stained AAV-transduced primary astrocytes with LysoTracker red dye and demonstrated an increase in the abundance of acidic organelles such as lysosomes and late endosomes in TFEB-transduced astrocytes (Fig. 2 E). In sum, these results demonstrate that TFEB regulates the lysosomal pathway in primary astrocytes.
Figure 2.
TFEB enhances lysosomal biogenesis and uptake of tau fibrils in primary astrocytes. (A and B) qRT-PCR analysis of the transcriptional levels of TFEB and its lysosomal target genes (B) in primary astrocytes transduced with AAV-GFAP-EGFP or AAV-GFAP-TFEB, the latter expressed as a 3×FLAG fusion protein. n = 3/group; Student’s t test, representative of three independently performed experiments. (C and D) Western blot with quantitation (D) of TFEB lysosomal targets in primary astrocytes transduced with AAV-GFAP-EGFP or AAV-GFAP-TFEB. TFEB expression was detected by an anti-FLAG antibody. n = 3/group; Student’s t test, representative of three independently performed experiments. (E) Flow cytometry analysis of LysoTracker red staining in primary astrocytes transduced with AAV-GFAP-EGFP or AAV-GFAP-TFEB. n = 4/group; Student’s t test, representative of three independently performed experiments. (F) Representative tracing flow cytometry analysis of red fluorescent latex bead uptake in primary astrocytes transduced with AAV-GFAP-EGFP or AAV-GFAP-TFEB following 1-h incubation. n = 4/group; representative of three independently performed experiments. (G and H) Flow cytometry analysis of red fluorescent bead uptake in primary astrocytes transduced with AAV-GFAP-EGFP or AAV-GFAP-TFEB showing total number of cells taking up beads and total number of cells taking up multiple beads (H). n = 4/group; Student’s t test, representative of three independently performed experiments. (I) Flow cytometry analysis of AlexaFluor-647–conjugated tau fibril uptake in primary astrocytes transduced with AAV-GFAP-EGFP or AAV-GFAP-TFEB following 1 or 4 h of fibril incubation. n = 4/group; Student’s t test, representative of three independently performed experiments. (J) Representative confocal images of TFEB (FLAG, green), DAPI (blue), and LAMP1 (red) costaining in basal conditions (TFEB) versus 1-h treatment with 250 nM Torin1 (TFEB + Torin1). Bar, 20 µm. n = 4/group, representative of two independently performed experiments. (K) Flow cytometry analysis of dye-conjugated tau fibril uptake in primary astrocytes transduced with AAV-GFAP-EGFP or AAV-GFAP-TFEB following 1-h incubation with fibrils in basal conditions (−Torin1) or 3 h of 250 nM Torin1 treatment (+Torin1). n = 4/group; Student’s t test, representative of two independently performed experiments. (L) qRT-PCR analysis of the transcriptional levels of TFEB and its lysosomal target genes in primary astrocytes from nestin-cre tcfeb knockout (TFEB KO) or wild-type pups. (n = 4/group; Student’s t test, representative of two independently performed experiments). (M) Flow cytometry analysis of LysoTracker red staining in primary astrocytes from TFEB KO or wild-type pups. n = 4/group; Student’s t test, representative of two independently performed experiments. (N) Flow cytometry analysis of AlexaFluor-647–conjugated tau fibril uptake in primary astrocytes from TFEB KO or wild-type pups following 3 h of fibril incubation. (n = 4/group; Student’s t test, representative of two independently performed experiments). Molecular mass indicated in kilodaltons. Error bars designate SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To test TFEB’s role in phagocytosis in primary astrocytes, we used flow cytometry to analyze the uptake of red fluorescent latex beads in conditions of AAV-mediated TFEB overexpression versus EGFP transduced controls. TFEB not only increased the proportion of cells taking up beads, but also increased the proportion of cells taking up multiple beads, suggesting an enhanced phagocytic capacity (Fig. 2, F–H). Given that TFEB stimulates uptake of a nonspecific substance such as latex beads in astrocytes, we proceeded to test if this increased phagocytic capacity could also apply to the uptake of pffs. Synthetic tau fibrils of the truncated form of human tau with the P301L mutation (K18 P301L) were generated as previously described (Li and Lee, 2006; Guo and Lee, 2011) and dye-conjugated to AlexaFluor-647 for ease of detection. The dye-conjugated pffs were then incubated with primary astrocytes transduced with TFEB or EGFP, followed by trypsinization to remove cell surface–bound pffs. The suspended live cells were then subject to flow cytometry analysis to detect the cellular fluorescent signal from internalized dye-conjugated pffs. Our results demonstrate a statistically significant increase of 12% in the uptake of pffs in TFEB transduced astrocytes relative to control at the 1-h time point, with a 22% relative increase in uptake in the TFEB transduced cells at 4 h (Fig. 2 I). This suggests that not only does TFEB increase pff uptake, it also increases the rate of uptake. While TFEB overexpression in astrocytes resulted in a significant increase in pff uptake, the magnitude of the change was rather modest. One possible explanation for these small but significant changes is inefficient activation (i.e., reduced nuclear localization) of TFEB in our AAV-transduced primary astrocyte culture. Using FLAG immunostaining for the transduced FLAG-tagged TFEB construct, confocal microscopy revealed that under basal primary astrocyte culture conditions, TFEB was predominantly cytoplasmic in its inactive state (Fig. 2 J). To drive TFEB nuclear localization and recapitulate the activation status observed in tauopathy, primary astrocytes were treated with the mTORC1 inhibitor Torin1. mTORC1 phosphorylates TFEB, which sequesters the transcription factor in the cytoplasm (Peña-Llopis et al., 2011; Martina et al., 2012; Roczniak-Ferguson et al., 2012; Settembre et al., 2012). Inhibiting mTORC1 blocks the phosphorylation of TFEB, allowing unphosphorylated TFEB to translocate to the nucleus and become transcriptionally active (Settembre et al., 2012). Confocal microscopy of FLAG immunostaining for FLAG-tagged TFEB demonstrates the nuclear localization of TFEB following Torin1 treatment as shown by colabeling with DAPI (Fig. 2 J). Additional LAMP1 staining also reveals TFEB’s colocalization with the lysosome with Torin1 treatment, an indication of TFEB temporarily trapped on the lysosomal surface due to its interaction with the recently inactivated mTORC1 (Fig. 2 J; Settembre et al., 2012). Given that Torin1 treatment leads to increased nuclear localization of TFEB relative to basal conditions, we tested whether Torin1 would further increase the uptake of pffs in TFEB transduced primary astrocytes. Primary astrocytes were subjected to 3 h of Torin1 treatment or basal conditions before 1-h incubation with dye-conjugated pffs. Flow cytometry revealed that Torin1 treatment of TFEB transduced astrocytes increased dye-conjugated pff uptake 63% relative to EGFP transduced controls as shown by median fluorescence intensity, while under basal conditions, the TFEB overexpressing astrocytes increased uptake just 18% relative to EGFP expressing controls (Fig. 2 K). Thus, TFEB enhances phagocytic pathways in astrocytes, in particular increasing the uptake of pffs.
To contrast and complement the TFEB overexpression studies, we cultured primary astrocytes from nestin-cre tcfeb knockout (TFEB KO) pups. qRT-PCR demonstrated undetectable transcript levels of TFEB as well as a reduction in LAMP1 mRNA in TFEB KO astrocytes compared with littermate controls (Fig. 2 L). In addition, the TFEB KO astrocyte culture demonstrated a modest reduction in LysoTracker Red staining, indicating a reduction in lysosomes and/or lysosomal acidity (Fig. 2 M). Recapitulating the uptake assay with dye-conjugated pffs in TFEB KO astrocytes, we observed a modest reduction pff uptake compared with primary astrocytes from littermate controls (Fig. 2 N). These findings indicate that endogenous TFEB regulates cellular uptake and provide additional support for a functional role of TFEB in astroglial uptake of tau.
TFEB enhances uptake of pffs via macropinocytosis and trafficking to the lysosome in astrocytes
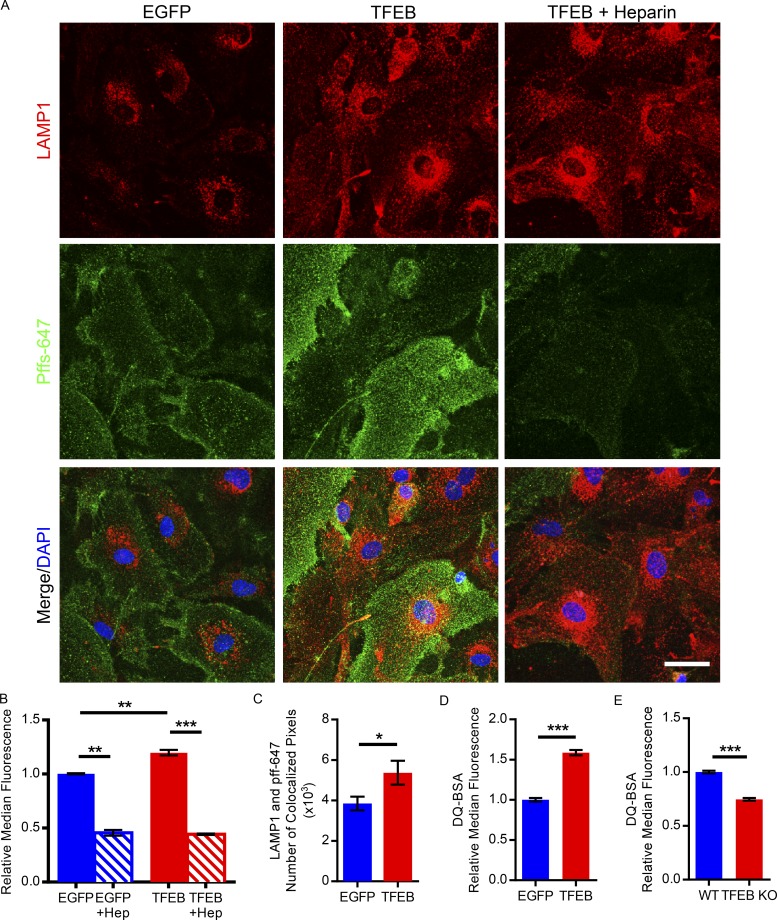
We next sought to determine the mechanism by which TFEB enhances astroglial uptake of pffs and the subcellular localization of phagocytosed pffs. To study pff handling in astrocytes, primary astrocyte cultures were incubated with dye-conjugated pffs and underwent immunostaining with LAMP1 antibody. Confocal microscopy revealed an expected increase in lysosomal marker LAMP1 staining, as well as an increase in dye-conjugated fibril fluorescence signal in TFEB transduced astrocytes (Fig. 3 A). Enhanced colocalization of dye-conjugated pffs and LAMP1 is observed in the TFEB transduced astrocytes, which is quantified based on number of colocalized pixels (Fig. 3, A and C). These findings indicate TFEB increases the incidence of phagocytosed pffs inside the lysosome. In addition, we also sought to demonstrate that TFEB enhances both uptake and degradation of phagocytosed proteins. Dye-quenched BSA (DQ-BSA) or boron-dipyrromethene dye–conjugated BSA is taken up by macropinocytosis and fluoresces following proteolysis in lysosomes. Flow cytometry analysis of primary astrocytes incubated with DQ-BSA revealed an ∼60% increase in median fluorescence for TFEB-transduced astrocytes, indicating that TFEB enhances uptake and proteolysis of DQ-BSA (Fig. 3 D). In contrast, TFEB KO astrocytes had an ∼25% decrease in median fluorescence following a 3-h incubation with DQ-BSA (Fig. 3 E). In sum, these data suggest that TFEB stimulates uptake and trafficking of pffs to the lysosome for degradation.
Figure 3.
TFEB enhances uptake of tau fibrils via macropinocytosis and trafficking to the lysosome in primary astrocytes. (A) Representative confocal images of AlexaFluor-647–conjugated tau fibrils (green) and LAMP1 costaining (red) in primary astrocytes transduced with AAV-GFAP-EGFP or AAV-GFAP-TFEB. Also depicted are primary astrocytes transduced with AAV-GFAP-TFEB treated with 100 µg/ml; 18 U/ml heparin. Bar, 20 µm. (n = 3/group; representative of two independently performed experiments). (B) Flow cytometry analysis of dye-conjugated tau fibril uptake in primary astrocytes transduced with AAV-GFAP-EGFP or AAV-GFAP-TFEB following 1-h incubation with or without heparin (100 µg/ml; 18 U/ml) treatment in each group. n = 4/group; Student’s t test, representative of two independently performed experiments. (C) Analysis of colocalization based on number of colocalized pixels of LAMP1 and AlexaFluor-647–conjugated tau fibrils in primary astrocytes transduced with AAV-GFAP-EGFP or AAV-GFAP-TFEB. n = 12 confocal images/group; Student’s t test, representative of two independently performed experiments. (D) Flow cytometry analysis of DQ-BSA uptake and cleavage in the lysosomal acidic compartment of primary astrocytes transduced with AAV-GFAP-EGFP or AAV-GFAP-TFEB following three hours of incubation with DQ-BSA. n = 4/group; Student’s t test, representative of two independently performed experiments. (E) Flow cytometry analysis of DQ-BSA uptake and cleavage in the lysosomal acidic compartment of primary astrocytes from TFEB KO or wild-type pups following 3 h of incubation with DQ-BSA. n = 4/group; Student’s t test, representative of two independently performed experiments. Error bars designate SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Previous studies indicate pff uptake in the cell-to-cell transfer process occurs via macropinocytosis through interaction with heparan sulfate proteoglycans (HSPGs) on the cell surface (Holmes et al., 2013). Heparin, a pharmacologic agent used as an anticoagulant, is in the same family as HSPGs and is structurally similar to this cell surface moiety. Thus, heparin can be used to competitively inhibit pff binding to HSPGs and prevent cellular uptake via macropinocytosis. Confocal microscopy revealed that heparin treatment reduces the fluorescence signal of pffs in primary astrocytes transduced with TFEB (Fig. 3 A). In addition, flow cytometry analysis demonstrated that heparin reduces dye-conjugated pff uptake in both TFEB and EGFP transduced astrocytes, suggesting that macropinocytosis is responsible for astroglial pff uptake (Fig. 3 B). Overall, this data indicates TFEB enhances astroglial uptake of pffs via macropinocytosis for subsequent delivery to the lysosome for degradation.
AAV-GFAP-TFEB specifically targets astrocytes and enhances lysosomal gene expression in vivo
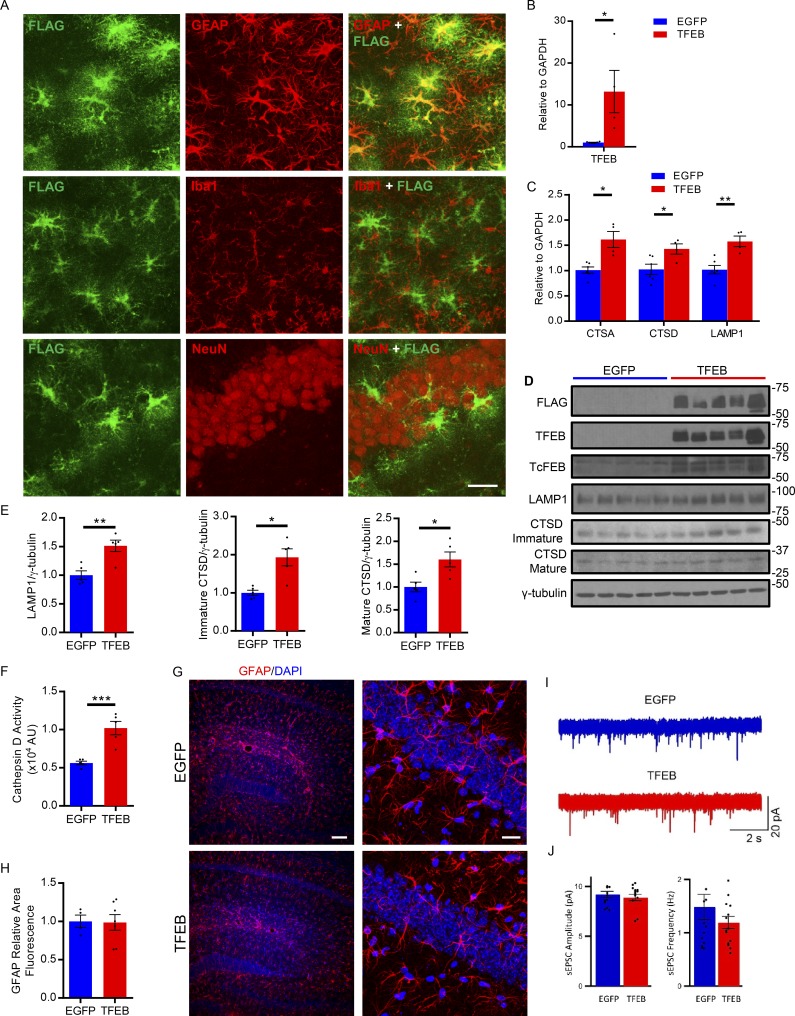
Given that TFEB enhances lysosomal biogenesis and uptake of extracellular pffs in vitro, we next sought to determine whether astroglial TFEB impacts tau pathology and tau spreading in vivo. To test this potential role of astroglial TFEB, we used an AAV-mediated gene delivery strategy of AAV-GFAP-TFEB (or AAV-GFAP-EGFP as a control) described above. Previous studies have shown that the GFAP promoter primarily targets astrocytes (von Jonquieres et al., 2013; Xiao et al., 2014). The AAV was injected ICV into both lateral ventricles of mouse pups at P3. Pups were injected at P3 based on a prior study indicating the AAV serotype 2/8 preferentially infects astrocytes at this age (Chakrabarty et al., 2013). ICV AAV injections at this age also lead to widespread expression in the cortex and hippocampus (Fig. S2, A and B). 4 mo after injection, the mice were assessed for expression patterns of the AAV delivered TFEB. Confocal microscopy of costaining for FLAG-tagged TFEB and astrocytic marker GFAP in the hippocampus showed AAV-mediated expression of TFEB occurred in astrocytes as signified by the colocalization of the two antibodies (Fig. 4 A). Quantification of GFAP/FLAG colocalization revealed that 44.4 ± 3.8% (average ± SEM, n = 4) of GFAP-positive astrocytes were also FLAG-positive in the hippocampus following P3 injections. To demonstrate that AAV-mediated TFEB expression was astrocyte specific, costaining of FLAG with Iba1 and NeuN, a microglial marker and neuronal marker, respectively, is also shown, with no colocalization between FLAG and neuronal or microglial markers present (Fig. 4 A). This evidence suggests that this AAV-mediated gene delivery predominantly affects astrocytes. Having shown the targeting specificity of this gene delivery strategy, we next sought to confirm the activity of exogenous astroglial TFEB in vivo. Hippocampal brain extract from TFEB-injected mice exhibited an ∼13-fold increase in TFEB transcripts as well as significant increases in the transcript levels of TFEB target genes compared with EGFP-injected littermates (Fig. 4, B and C). On the protein level, LAMP1 and both immature and mature forms of CTSD were significantly increased in the hippocampus of TFEB-injected mice (Fig. 4, D and E). In addition, protein homogenate from the hippocampus of TFEB-injected mice had increased CTSD enzyme activity relative to EGFP-injected littermates (Fig. 4 F). Thus, our AAV gene delivery strategy is astrocyte specific and up-regulates lysosomal target genes in vivo.
Figure 4.
AAV-GFAP-TFEB specifically targets astrocytes and enhances lysosomal gene expression in vivo. (A) Representative confocal images demonstrating expression of TFEB with GFAP, Iba-1, and NeuN (red; bottom) in the hippocampus of wild-type mice injected ICV at P3 with AAV-GFAP-TFEB viral particles. Bar, 20 µm. n = 3, representative of two independently performed experiments. (B and C) qRT-PCR analysis of TFEB and its lysosomal target genes (C) from P3-injected AAV-GFAP-EGFP or AAV-GFAP-TFEB wild-type hippocampus samples at 3 mo of age. n = 6/group; Student’s t test, representative of two independently performed experiments. (D and E) Western blot with quantitation (E) of TFEB lysosomal targets LAMP1 and CTSD from the hippocampus of P3-injected AAV-GFAP-EGFP or AAV-GFAP-TFEB wild-type mice at 3 mo of age. n = 5/group; Student’s t test, representative of two independently performed experiments. (F) CTSD enzyme activity assay from hippocampal homogenate of P3-injected AAV-GFAP-EGFP or AAV-GFAP-TFEB wild-type mice at 3 mo of age. n = 5/group; Student’s t test, representative of two independently performed experiments. (G) Representative fluorescent confocal images of GFAP immunostaining (red) of the hippocampus in 7–9-mo-old wild-type mice P3-injected with AAV-GFAP-EGFP or AAV-GFAP-TFEB at low (left) and high (right) magnification. Bars, 100 µm and 20 µm, respectively. n = 5–7; representative of two independently performed experiments. (H) Area fluorescence quantitation of GFAP immunostaining in the hippocampus of 7–9-mo-old wild-type mice. n = 5 EGFP/7 TFEB; Student’s t test, representative of two independently performed experiments. (I) sEPSC sample traces from one granule neuron in P3-injected AAV-GFAP-EGFP or AAV-GFAP-TFEB wild-type mice at 9 mo of age. (J) Mean sEPSC amplitude and frequency showed no difference between P3-injected AAV-GFAP-EGFP (n = 14 neurons/3 mice) or AAV-GFAP-TFEB (n = 16 neurons/3 mice) wild-type mice at 9 mo of age (Student’s t test; representative of two independently performed experiments). Molecular mass indicated in kilodaltons. Error bars designate SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Having established that astrocytic TFEB has a role in tau handling through lysosomal and cellular uptake pathways, we sought to investigate whether astroglial TFEB overexpression affects additional pathways that may impact tau pathology/tau spreading. In particular, we studied the effect on general astrocyte properties and neuronal activity. To determine if AAV-mediated astroglial TFEB expression influenced astrocyte morphology or reactivity, P3-injected wild-type mice underwent immunostaining with reactive astrocyte marker GFAP. Representative fluorescent confocal images of GFAP staining in the hippocampus show no obvious morphological changes in astrocytes between groups (Fig. 4 G), which is validated by quantitation of area fluorescence of GFAP immunostaining (Fig. 4 H). In addition, we used electrophysiology to study neuronal activity in the context of astroglial TFEB overexpression. Previous work indicates that neuronal activity influences the rate of synaptic tau release, which may impact tau spreading (Wu et al., 2016), and astrocytes can impact neuronal signaling through lysosomal exocytosis of ATP (Zhang et al., 2007; Lalo et al., 2016). Since TFEB can enhance lysosomal exocytosis, we sought to determine if astroglial TFEB alters neuronal activity. From acute hippocampal slices of 9-mo-old P3-injected mice with AAV-GFAP-EGFP or AAV-GFAP-TFEB, spontaneous excitatory post-synaptic currents (sEPSCs) were recorded from granule neurons in the dentate gyrus. sEPSCs events are generated by both action potential dependent neurotransmitter release, as well as action potential independent neurotransmitter release, and therefore can represent ongoing neuronal activity in the hippocampal circuitry. Neither sEPSCs amplitude nor frequency show statistically significant differences between AAV-GFAP-EGFP and AAV-GFAP-TFEB mice (Fig. 4, I and J). Thus, AAV-mediated astroglial TFEB expression does not impact astrocyte reactivity/morphology or neuronal activity in wild-type mice, excluding these possible indirect effects on tau release.
Astroglial TFEB does not impact tau pathology in rTg4510 mice
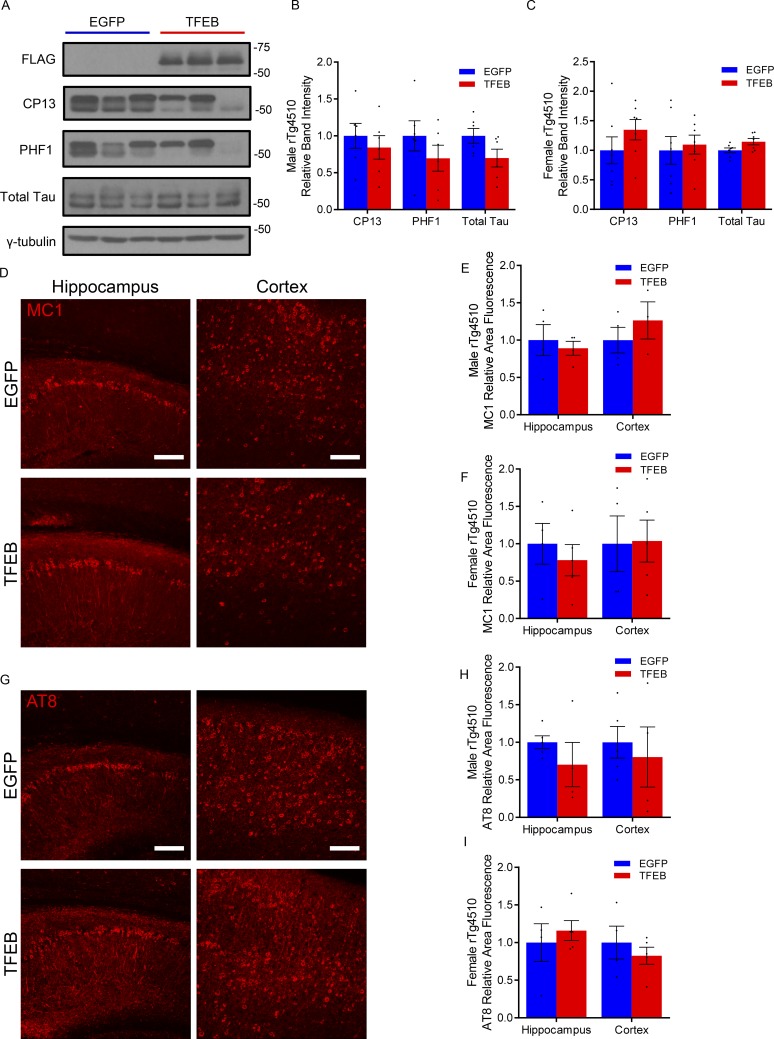
After validating our AAV-mediated gene delivery technique for astroglial TFEB expression in vivo, we next focused on the impact of astroglial TFEB on tau pathology in the rTg4510 tauopathy mouse model. Using P3 ICV AAV injections of GFAP-TFEB or GFAP-EGFP in rTg4510 mice, tau pathology was analyzed biochemically and histologically at 4 mo of age. Immunoblotting for total tau and phospho-tau antibodies, PHF1 and CP13, demonstrated no statistical difference in hippocampal brain lysate from TFEB- versus EGFP-injected rTg4510 male or female mice (Fig. 5, A–C). Similarly, immunofluorescence staining for an early pathological conformation of tau using the MC1 antibody showed no significant difference in area fluorescence in the hippocampus or cortex of TFEB and EGFP groups, nor did staining with phospho-tau antibody AT8 (Fig. 5, D–I). Female mice also showed no difference in area fluorescence using the same antibodies for pathological tau (Fig. 5, F and I). In addition, immunofluorescence staining for astro- and microgliosis markers GFAP and Iba1, respectively, showed no change in area fluorescence in TFEB- or EGFP-injected male mice (Fig. S3). Thus, tau pathology and gliosis in the rTg4510 is not impacted by astroglial TFEB in the cortex or hippocampus at 4 mo of age.
Figure 5.
Astroglial TFEB does not impact tau pathology in rTg4510 mice. (A and B) Representative Western blot with quantitation (B) of total and phosphorylated tau species using antibodies CP13 and PHF1 from the hippocampus of male 4-mo-old rTg4510 mice injected ICV with AAV-GFAP-EGFP or AAV-GFAP-TFEB at P3. n = 6, 7/group; Student’s t test, representative of two independently performed experiments. (C) Quantitation of Western blot for female mice. n = 6/group; Student’s t test, representative of two independently performed experiments. (D–G) Representative confocal images of MC1 and AT8 (E) immunostaining with quantitation (F and G) from the cortex and hippocampus of AAV-GFAP-EGFP– or AAV-GFAP-TFEB–injected rTg4510 mice. Bar, 75 µm. n = 4/group; Student’s t test, representative of two independently performed experiments. (H and I) Quantitation of MC1 and AT8 (I) immunostaining for female mice. n = 4/group; Student’s t test, representative of two independently performed experiments. Molecular mass indicated in kilodaltons. Error bars indicate SEM.
Astroglial TFEB reduces tau pathology in PS19 mice
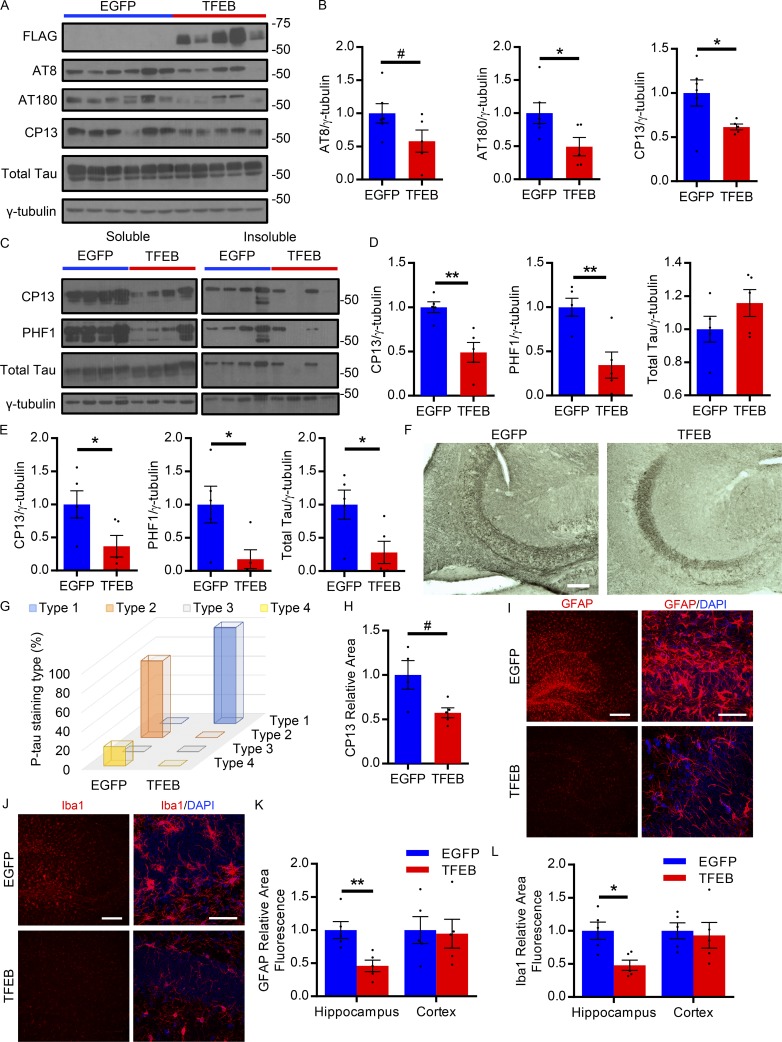
While astroglial TFEB overexpression did not impact tau pathology in our study of the aggressive rTg4510 tauopathy mouse model, we sought to determine if it may affect pathology in a slower progressing model such as the PS19. Unlike the rapid progression of tau pathology in the rTg4510 mice, the PS19 have a slower onset of pathology, with MC1 and AT8 immunofluorescence staining in the neuronal cell body reliably present at 6–8 mo and robust at 9 mo of age (Yoshiyama et al., 2007; Holmes et al., 2014). Using a similar experimental design described for the rTg4510 mice, PS19 pups were injected ICV with AAV-GFAP-TFEB or AAV-GFAP-EGFP at P3 and analyzed at 9 mo of age. Tau pathology was analyzed biochemically and histologically, as was reactive gliosis. Total hippocampal brain lysates indicated on Western blot a reduction in phospho-tau species recognized by AT8 (trending), AT180, and CP13 antibodies in the TFEB-injected mice (Fig. 6, A and B). Following fractionation, immunoblotting for phospho and total tau species in the soluble pool similarly showed a significant reduction in phospho-tau species recognized by CP13 and PHF1 antibodies, with no change in total tau (Fig. 6, C and D). In the insoluble fraction, phospho- and total tau species were all reduced in the TFEB mice, as demonstrated by Western blot (Fig. 6, C and E). Immunostaining with phospho-tau antibody CP13 revealed a distinct difference in staining pattern when comparing the TFEB- and EGFP-injected mice (Fig. 6 F and Fig. S3 G). In particular, the majority of 9-mo-old PS19 mice injected with AAV-EGFP had abundant cell body CP13 staining in the CA3 region of the hippocampus (Fig. 6 F). In contrast, cell body staining was absent in the CA3 region of AAV-TFEB–injected mice with instead prominent mossy fiber staining present (Fig. 6 F). The staining patterns described correspond well with those reported in the PS19 mice studied in Shi et al. (2017). Accordingly, the prominent mossy fiber staining in the CA3 of the TFEB-injected mice indicate a type 1 pattern, associated with earlier stages of pathology, while the cell body staining in the CA3 region and the dense staining over the entire hippocampus of the EGFP-injected mice indicate the more severe type 2 and type 4 pattern of pathology, respectively. The distribution of the CP13 staining patterns indicated patterns associated with later stage pathology were more abundant in EGFP-injected mice, while earlier stage, less severe phospho-tau staining patterns were present in the TFEB-injected mice (Fig. 6 G). With respect to phospho-tau staining in the cortex of these mice, a trending reduction in the area of CP13 staining is observed in the TFEB-injected group (Fig. 6 H). In line with a reduction in tau pathology, gliosis was also reduced in the hippocampus though not in the cortex, as shown by GFAP and Iba1 immunostaining (Fig. 6, I–L). Overall, these results show that astroglial TFEB overexpression reduces tau pathology and gliosis in the hippocampus of PS19 tauopathy mice.
Figure 6.
Astroglial TFEB reduces tau pathology in the hippocampus of PS19 mice. (A and B) Western blot with quantification (B) of total and phosphorylated tau species using antibodies AT8, AT180, and CP13 from the hippocampus of 9-mo-old PS19 mice injected ICV with AAV-GFAP-EGFP or AAV-GFAP-TFEB at P3. n = 6, 5/group. #, P = 0.09; Student’s t test, representative of two independently performed experiments. (C–E) Western blot with quantification of total and phosphorylated tau species using antibodies CP13 and PHF1 in soluble (D) and insoluble (E) fractions from the hippocampus of 9-mo-old AAV-injected PS19 mice. n = 5/group; Student’s t test, experiment technically replicated twice. (F) Representative images of CP13 immunostaining in the CA3 of the hippocampus in 9-mo-old AAV-injected PS19 mice. Bar, 100 µm. n = 5/group; representative of two independently performed experiments. (G) Distribution of the four phospho-tau staining types in 9-mo-old AAV-injected PS19 mice. n = 5/group; representative of two independently performed experiments. (H) Quantification of CP13 immunostaining area in the cortex of 9-mo-old AAV-injected PS19 mice. n = 5/group. #, P = 0.07; Student’s t test, representative of two independently performed experiments. (I and J) Representative fluorescent confocal images of GFAP (I) and Iba1 (J) immunostaining (red) of the dentate gyrus of 9-mo-old AAV-injected PS19 mice at low (left) and high (right) magnification. Bars, 200 µm and 50 µm, respectively. n = 5/group, representative of two independently performed experiments. (K and L) Area fluorescence quantitation of GFAP (K) and Iba1 (L) immunostaining of 9-mo-old AAV-injected PS19 mice. n = 5/group; Student’s t test, representative of two independently performed experiments. Molecular mass indicated in kilodaltons. Error bars designate SEM. *, P < 0.05; **, P < 0.01.
Astroglial TFEB reduces tau spreading in PS19 mice
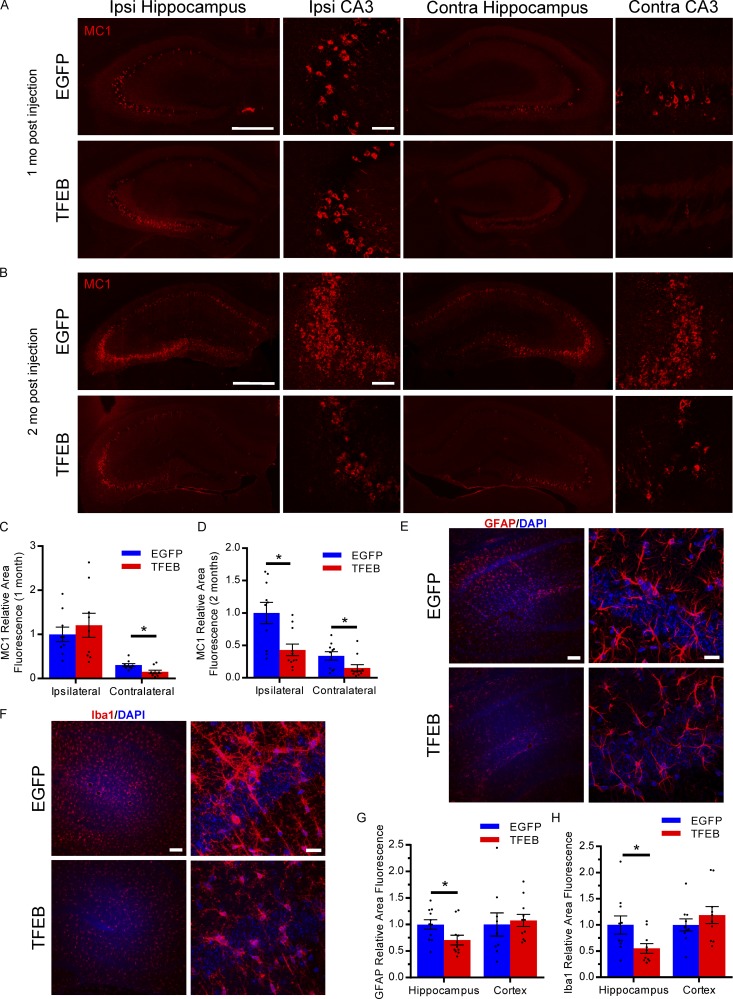
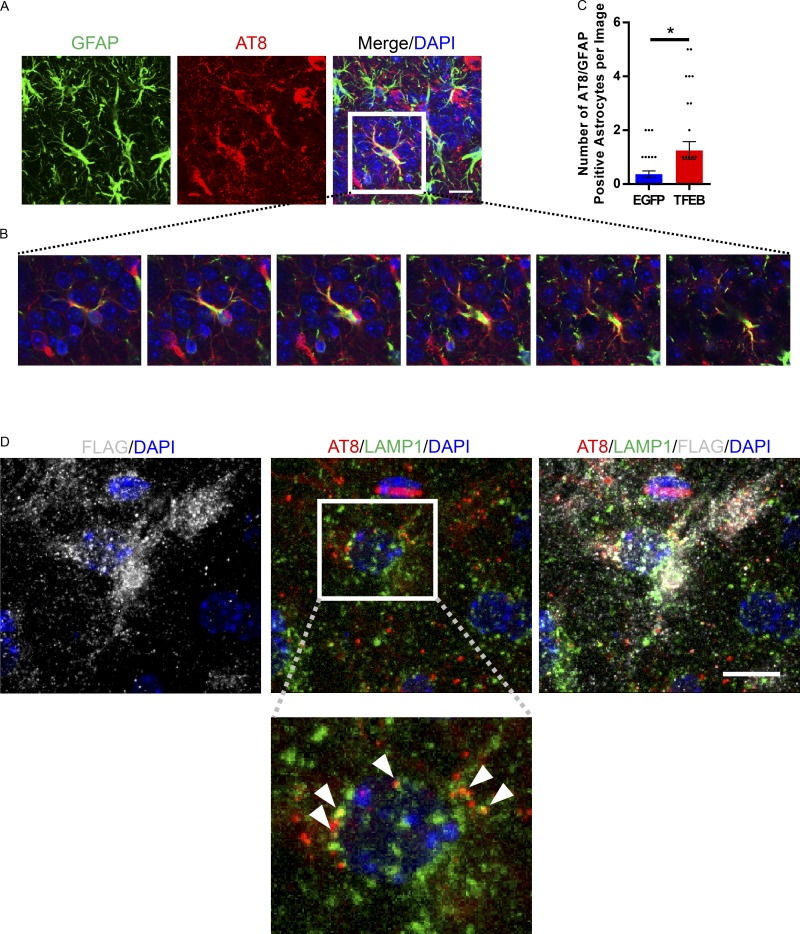
Since astroglial TFEB overexpression reduces overall pathology in a slower progressing transgenic mouse model, we sought to determine if it may also affect tau spreading. To assess the role of astroglial TFEB in tau spreading, we designed an in vivo tau spreading assay based on a previously published model of pff induced NFT transmission in mice (Iba et al., 2013). In this case, pffs were delivered via stereotaxic injection into the left hippocampus of 3-mo-old PS19 tauopathy mice. This resulted in a reliable tau spreading model with ipsilateral hippocampal pathology within 1–2 wk (Iba et al., 2013) and contralateral pathology starting at 3 wk (Fig. S4, A and B). As mentioned before, the PS19 have a slower onset of pathology, with MC1 and AT8 immunofluorescence staining reliably present at 6–8 mo and robust at 9 mo of age (Fig. S4 C; Yoshiyama et al., 2007; Holmes et al., 2014). Thus, the MC1 and phospho-tau staining in the tau spreading assay should be exclusively pff-induced tau pathology up to 2 mo after fibril injection (5 mo of age; Fig. S4 C). To test the impact of astroglial TFEB, PS19 mice underwent P3 ICV injections of AAV-GFAP-TFEB or EGFP as described for the rTg4510 mice. At 3 mo of age these mice were subjected to stereotaxic injections of tau pffs in the left hippocampus (Fig. S5 A; Lein et al., 2007). At 1 mo after pff injections, we observed MC1 staining in the ipsilateral hippocampus, with less MC1 positivity in the contralateral hippocampus for both TFEB and EGFP groups (Fig. 7 A). Quantifying the area fluorescence of MC1 staining for both hippocampi from sections representing the entire volume of hippocampus showed that astroglial TFEB had no impact on MC1 staining on the ipsilateral side, but significantly reduced the area of MC1 staining on the contralateral side (Fig. 7, A and C). Similar results were observed with immunofluorescence using phospho-tau antibody AT8 (Fig. S5, B and E). Progressing from 1 mo to 2 mo after injection resulted in overall worsening pathology in both groups. Interestingly, at the 2-mo post-pff injection time point, similar measurements showed a reduction in MC1 staining area in both the ipsilateral and contralateral hippocampi in the TFEB groups compared with EGFP controls (Fig. 7 B, D). Again, similar results are shown with AT8 antibody staining at 2 mo after pff injection (Fig. S5, C and F). In addition, X-34 staining, which labels β sheet structures in tangle-like pathology, was reduced in the TFEB-injected mice 2 mo after pff injection on both the ipsilateral and contralateral side (Fig. S5, D and G). These findings indicate that while astroglial TFEB does not seem to impact the initial seeding of tau pathology in the ipsilateral hippocampus, tau spreading to the contralateral hippocampus is reduced at early time points, with both hippocampi demonstrating reduced tau pathology at the later time point. Correlated with a reduction in tau pathology at 2 mo after pff injection, TFEB-injected mice also have reduced gliosis in the hippocampus, as shown by reduced area fluorescence when staining for reactive astrocytes marker GFAP (Fig. 7, E and G) and microglial marker Iba1 (Fig. 7, F and H). In addition to a reduction in tau pathology/tau spreading, the TFEB-injected mice also demonstrate an increased incidence of astrocytes with AT8 positive phospho-tau (Fig. 8, A–C). AT8-positive tau was found to colocalize with lysosomal marker LAMP1 in AAV-TFEB transduced hippocampal astrocytes (Fig. 8 D). Overall, the results from the tau spreading model demonstrate that astroglial TFEB reduces tau spreading and reactive gliosis and also increases astroglial engulfment of phospho-tau in vivo.
Figure 7.
Astroglial TFEB reduces tau spreading in PS19 mice. (A and B) Representative fluorescent images of MC1 immunostaining (red) of the ipsilateral and contralateral hippocampus of PS19 mice with P3 ICV injection of AAV-GFAP-EGFP or AAV-GFAP-TFEB at 1 mo (A) and 2 mo (B) after unilateral tau fibril injection. Bars: 500 µm for whole hippocampus; 40 µm for CA3. n = 7–11/group; representative of two independently performed experiments for 1 mo; 2 mo technically replicated twice. (C and D) Area fluorescence quantitation of MC1 immunostaining of the ipsilateral and contralateral hippocampus of PS19 mice 1 mo (C) and 2 mo (D) after tau fibril injection. n = 7–11/group; Student’s t test, representative of two independently performed experiments for 1 mo; 2 mo technically replicated twice. (E and F) Representative fluorescent confocal images of GFAP (E) and Iba1 (F) immunostaining (red) of the hippocampus of PS19 mice 2 mo after tau fibril injection at low (left) and high (right) magnification. Bars, 100 µm and 20 µm, respectively. n = 10/group; technically replicated twice. (G and H) Area fluorescence quantitation of GFAP (G) and Iba1 (H) immunostaining of PS19 mice 2 mo after tau fibril injection. (n = 10/group; Student’s t test, technically replicated twice). Error bars designate SEM. *, P < 0.05.
Figure 8.
Astroglial TFEB increases phospho tau present in astrocytes in a tau spreading mouse model. (A) Representative confocal image of AT8 (red), GFAP (green), and DAPI (blue) staining from the contralateral CA3 hippocampus of AAV-GFAP-TFEB–injected tau spreading mouse 2 mo after tau fibril injection. Bar, 10 µm. n = 4/group; representative of two independently performed experiments. (B) Z-stack confocal images from merged image shown in A. (C) Quantification of the number of AT8 positive astrocytes in the hippocampus of the tau spreading mouse model injected ICV at P3 with AAV-GFAP-EGFP or AAV-GFAP-TFEB. n = 40 sections/group, 4 mice/group; Student’s t test, representative of two independently performed experiments. (D) Representative confocal images of AT8 (red), LAMP1 (green), FLAG (gray), and DAPI (blue) staining from the hippocampus of AAV-GFAP-TFEB–injected tau spreading mouse 2 mo after tau fibril injection. Arrows indicate colocalization of AT8 and LAMP1 within the FLAG-positive cell. Bar, 10 µm. n = 4/group; representative of two independently performed experiments. Error bars designate SEM. *, P < 0.05.
Discussion
TFEB activation is a known consequence of lysosomal stress/dysfunction as demonstrated in lysosomal storage disorders and pharmacologic inhibition of lysosomal acidification (Sardiello et al., 2009; Song et al., 2013). Whether due to normal aging, the lysosomal stress of accumulating protein aggregates, or Alzheimer’s disease–related mutations causing inadequate lysosomal acidification, lysosomal dysfunction has long been implicated in Alzheimer’s disease (Cuervo and Dice, 2000; Lee et al., 2010, 2015; Coen et al., 2012; Wolfe et al., 2013). Focusing specifically on astrocytes, we see evidence of TFEB activation in tauopathies: (1) sorted astrocytes from whole brain demonstrate transcriptional up-regulation of TFEB and several of its lysosomal target genes, and (2) TFEB nuclear localization in astrocytes is increased. Since astrocytes are not the predominant cell type harboring undegraded protein aggregates in Alzheimer’s disease or frontotemporal dementia, it is unlikely that this activation is mediated by lysosomal stress due to astroglial protein accumulation. Instead, the astrocyte is intimately associated with the neuronal synapse, both anatomically and metabolically, making it uniquely poised to modulate or respond to neuronally secreted tau and its trans-synaptic spreading (Wu et al., 2016). Astroglial TFEB activation could be due to several factors, including the presence of aberrant extracellular tau, as well as signals from a stressed neuron, or secreted inflammatory molecules (Pastore et al., 2016). The precise mechanism is unknown and warrants further investigation. Despite this activation of TFEB in tauopathy, the response is insufficient to reverse or even halt disease progression. Our findings suggest that enhancing the lysosomal pathway in astrocytes through TFEB overexpression can mitigate disease progression and reduce tau spreading.
Our study demonstrates that astroglial TFEB overexpression prominently affects tau spreading, but has mixed results on tau pathology (rTg4510 vs. PS19 mice). Analysis of postmortem human brains suggest a stereotyped spatial progression of tau pathology via synaptic connections (Braak and Braak, 1995). The study of animal and cellular models of this process indicate a mechanism of trans-cellular tau pathology propagation, where aberrant tau produced in one cell is released extracellularly and enters another adjacent cell to seed tau aggregation (Clavaguera et al., 2009; Frost et al., 2009; de Calignon et al., 2012; Kfoury et al., 2012; Liu et al., 2012; Iba et al., 2013; Wu et al., 2016). Similar to other tau spreading models, the model used in our study relies on this noncell autonomous mechanism of tau pathology progression with extracellular aberrant tau serving in a crucial role. In contrast, the importance of extracellular tau in the pathogenesis of the rTg4510 or PS19 tau transgenic mice is not well-established. Possible explanations for the lack of efficacy of astroglial TFEB in the rTg4510 mice are the predominance of a cell autonomous mechanism in the pathogenesis of neuropathology or the model’s severity and rapid progression. However, given that astroglial TFEB overexpression seems to be more effective at reducing tau pathology in the less severe, slower progressing PS19 mice, the rTg4510 mice may have reached a disease stage too advanced to detect a difference between TFEB- and EGFP-injected groups. Others have similarly had success in targeting extracellular tau in PS19 tau transgenic mice. For example, passive tau immunization with antibodies designed to target extracellular tau effectively reduced tau pathology in the PS19 (Yanamandra et al., 2013, 2015, 2017). While these antibodies have been shown to target exogenously administered extracellular tau, mechanisms targeting intracellular tau cannot be ruled out and may be responsible in part for reducing pathology (Sigurdsson, 2009; Yanamandra et al., 2013). Nonetheless, the passive immunization studies and our work indicate that (1) extracellular tau may play a role in disease pathogenesis in transgenic mice as well as tau spreading mice, and (2) this implicit targeting of extracellular tau can be an effective method to halt the progression of tau pathology. However, as with the passive immunization, additional mechanisms independent of the direct targeting of extracellular tau may also factor into the observed effect. Astroglial TFEB overexpression, in addition to increasing cellular uptake and clearance of extracellular tau seeds, may also indirectly impact disease pathogenesis based on interactions with neurons, modulation of neuroinflammation, or other important disease modifying pathways, which are yet to be reported in this particular context.
In terms of glial involvement in modulating tau spreading, initial studies implicate microglia in this process. Microglia are thought to engulf tau aggregates bound to anti-tau antibodies in passive immunization studies (Funk et al., 2015; Yanamandra et al., 2015), while another study postulates microglia may facilitate tau spreading via exosome secretion (Asai et al., 2015). However, little is known about astroglial involvement in tau spreading. Though microglia are typically thought of as the professional phagocytes of the CNS, astrocytes are also phagocytic cells (Cahoy et al., 2008). In fact, numerous studies have emphasized the phagocytic capacity of astrocytes in the uptake and clearance of Aβ (Wyss-Coray et al., 2003; Basak et al., 2012; Li et al., 2014). Furthermore, a prior study demonstrated targeted astroglial TFEB expression reduced plaque pathology in an Alzheimer’s disease mouse model through enhanced uptake and lysosomal clearance pathways (Xiao et al., 2014). With respect to tau, we observe that astroglial TFEB expression increases macropinocytosis of pffs, followed by enhanced delivery of engulfed tau to the lysosome for degradation. In vivo, the increased presence of phospho-tau in TFEB overexpressing astrocytes suggests that TFEB-induced astrocytic engulfment and clearance may be responsible for the reduction in tau spreading and modulation of overall tau pathology. However, whether or not the direct mechanism investigated in vitro is solely responsible for our in vivo findings or if indirect mechanisms may also contribute remains to be established. One possible alternative mechanism investigated is whether modifying astrocytes via TFEB overexpression would impact synaptic activity. Tau release has been linked to neuronal activity (Pooler et al., 2013; Yamada et al., 2014; Wu et al., 2016), and astrocytes are intimately associated with the synapse, even becoming functionally part of the synapse. Despite this potential link, our findings indicate that TFEB overexpression in astrocytes does not modulate neuronal activity or impact trans-synaptic tau spreading via this particular mechanism.
In terms of our in vivo findings of reduced tau spreading with astroglial TFEB overexpression, one must consider the limitations of the in vivo tau spreading assay. While the propagation of tau pathology from the ipsilateral pff injection site to the contralateral side is meant to mimic progression of tau pathology that serves as the basis of Braak staging, the injection of micrograms of pffs into the hippocampus is far from being a physiological event. However, the initial impact of astroglial TFEB seems to be on contralateral pathology, rather than initial seeding on the ipsilateral side. Thus, astroglial TFEB is not directly impacting the injected pff seeding ability, but rather limiting the propagation process further downstream. Furthermore, the AT8 positive tau observed in astrocytes in our spreading model results from human tau expressed in the transgenic mice, not the injected tau pffs which do not contain this phospho-tau epitope. This impact on tau spreading eventually results in an attenuation of overall pathology at 2 mo after injection, impacting the ipsilateral and contralateral side. Another complicating matter in this particular spreading assay is that while the pattern of pathology suggests tau spreading to synaptically connected regions, trans-synaptic tau spreading is unlikely to be the sole mechanism of spreading in this model. Tau spreading via direct physical connections or through the extracellular space without specifically involving synapses cannot be ruled out (Gousset et al., 2009; Yamada et al., 2011; Asai et al., 2015). Which particular mechanism(s) of tau propagation astroglial TFEB predominantly impacts warrants further study. Nonetheless, while the in vivo tau spreading assay is artificial in nature, the propagation of pathology from the ipsilateral to contralateral side provides an experimental model in which to test interventions that can impact the spatial progression of tau pathology that is observed in Alzheimer’s disease and correlated with cognitive decline (Grober et al., 1999; Giannakopoulos et al., 2003).
Overall, our work supports the premise that TFEB overexpression in astrocytes reduces tau spreading, possibly through targeting aberrant extracellular tau for uptake and degradation. In terms of physiological relevance, evidence from whole brain and sorted astrocytes suggests the endogenous TFEB lysosomal system represents an adaptive response to tau pathology. Although we demonstrate a functional role of astroglial TFEB overexpression in modulating tau pathology and spreading, future in vivo studies using astroglial TFEB loss-of-function mutants are necessary to fully understand the physiological role of astroglial TFEB in this process. Other cell-type specific TFEB knockouts, particularly in neurons and microglia, are also warranted to gain a comprehensive assessment of the role of TFEB in tau handling. Nonetheless, our findings in astroglial TFEB overexpression describe a novel role for astrocytes in modulating tau pathology and open new avenues for leveraging the lysosomal TFEB pathway for therapeutic development.
Materials and methods
Experimental design
The aim of this study was to determine the impact of astroglial TFEB on tau pathology. We studied the effects on two aspects of tauopathy, general tau pathology and tau spreading, using the rTg4510 and PS19 transgenic mouse models, respectively. All control mice were littermates to experimental mice. Astroglial TFEB overexpression was achieved via ICV injections of AAV at P3. The PS19 mice additionally underwent unilateral hippocampal injections of pffs at 3 mo of age to create the tau spreading model. Mice were randomly assigned to TFEB or EGFP treatment groups. Both sexes were analyzed. Animals were housed in groups of five animals maximum per cage. Investigators were blinded to study group identity during experiments, data collection, and analysis. In vitro experiments were repeated a minimum of two to three times, while in vivo experiments were either repeated, analyzed at different time points with similar results, or corroborating experiments were performed with separate cohorts of mice. Sample size was determined based on experience with similar studies (Polito et al., 2014) and/or power analysis described in Statistics.
Human tauopathy samples
Postmortem brain tissue (mid frontal cortex) from subjects with FTD (including PSP, CBD, and Pick’s disease) and normal subjects were provided in the form of frozen blocks by Center for Neurodegenerative Disease Research at the University of Pennsylvania (demographic information listed below). Brains were considered normal based on the following criteria: exclusion of schizophrenia and fetal cases, confirmed Braak stage = 1 or 0 and CERAD = 0. Brains were designated as FTD (including CBD, PSP, and Pick’s disease) based on the following criteria: exclude secondary pathology, exclude CERAD >0, medial frontal cortex tau 3+ or 2+, and confirmed sporadic disease. Demographic data are included in Table S1. Each brain was homogenized in 1 ml RIPA (∼1:10 wt/vol) with protease and phosphatase inhibitor (Roche). For RNA extraction, frozen tissue was homogenized in TRIzol using the RNeasy Mini kit (Qiagen).
Animals
All protocols involving mice were approved by the Institutional Animal Care and Use Committee of Baylor College of Medicine. rTg4510 mice were obtained from Jackson Labs and maintained by crossing the transactivator line CaMKIIα (129S6 background) with the tau responder line (FVB background; Ramsden et al., 2005; SantaCruz et al., 2005). Wild-type littermates were used as controls. PS19 mice were obtained from Jackson Labs (Yoshiyama et al., 2007). Heterozygotes were bred to B6C3F1/J wild-type mice to maintain the line. The conditional tcfeb-flox mice were a generous gift from A. Ballabio (Telethon Institutes of Genetics and Medicine, Naples, Italy; Settembre et al., 2012) and crossed to nestin-Cre mice from Jackson Labs. Control mice were littermate tcfeb-flox homozygotes without the cre.
Antibodies
CP13, PHF1, and MC1 antibodies were generous gifts from Peter Davies (Albert Einstein College of Medicine, Bronx, NY). All other antibodies were purchased from commercial sources described in Table S2.
In vivo astrocyte isolation
Astrocytes were isolated from 4.5-mo-old wild-type and rTg4510 (n = 9) mice as described previously (Swartzlander et al., 2018). In brief, mice were anesthetized and perfused with ice-cold saline. The forebrain was dissected and then finely minced using a razor blade. The minced forebrain was resuspended in HBSS and enzymatically digested in a solution containing papain (LK003178; Worthington Biochemical Corporation) and DNase. The suspension was then triturated with a glass Pasteur pipette to mechanically dissociate the minced forebrain. Cellular debris and myelin were removed by filtration through a 40-µm filter and centrifugation in 21% isotonic percoll resulting in a pellet of single cells. To isolate astrocytes from the dissociated singles cells, the cells were first blocked in HBSS with 0.5% BSA and mouse Fc Block (BD Biosciences) before staining for viability with LIVE/DEAD blue (L23105; Thermo Fisher) and with the fluorescent antibodies CD45 and ACSA-2. Approximately 200,000 live ACSA-2 positive, CD45 negative astrocytes were isolated per brain through FACS using a BD FACSAria II.
GSEA
GSEA was performed as previously described (Subramanian et al., 2005). Microarray data were obtained as described in our previous work (Polito et al., 2014) and is available in the NCBI’s Gene Expression Omnibus database (accession no.: GSE53480). The cumulative distribution function was constructed by performing 1,000 random gene set membership assignments. A nominal P value of <0.01 and an FDR of <10% were used to assess the significance of the enrichment score (ES). The lists that define TFEB targets and TFEB lysosomal targets were obtained from previous studies (Sardiello et al., 2009; Palmieri et al., 2011).
Human transcriptomic data
The human reprocessed RNA-seq data from the parahippocampal gyrus was generated by the MSSM and available on Synapse as part of the AMP-AD consortium (Synapse ID: syn8484987, download date March 27, 2017 and March 29, 2017). Samples with an RNA integrity number (RIN) >6 were included in the analysis. The cohort was divided in to groups based on cognition and Braak staging. For cognition, there was no cognitive impairment (NCI; n = 23; RIN = 7.3 ± 0.1), mild cognitive impairment (MCI; n = 23; RIN = 7.2 ± 0.1), and dementia (n = 84; RIN = 7.1 ± 0.1). The breakdown for Braak stages 1–6 were as follows: 1 (n = 12; RIN = 7.3 ± 0.1), 2 (n = 25; RIN = 7.0 ± 0.1), 3 (n = 25; RIN = 7.1 ± 0.1), 4 (n = 15; RIN = 7.1 ± 0.2), 5 (n = 13; RIN = 7.9 ± 0.2), and 6 (n = 27; RIN = 6.9 ± 0.1). For the entire cohort (n = 130) analyzed, the average RIN score is 7.1 ± 0.1. Statistics on genes of interest are described in the Statistics section.
Primary astrocyte culture
Murine cortices and hippocampi were isolated from P2 pups for astrocyte culture. The forebrain was dissected in ice-cold dissection medium (HBSS with 10 mM Hepes, 0.6% glucose, and 1% vol/vol Pen/Strep) and finely minced. The tissue was digested in 2.5% trypsin at 37°C for 15 min, followed by the addition of trypsin inhibitor (1 mg/ml) and DNase. The tissue was then centrifuged at 1,000 g for 5 min, triturated, and resuspended in astrocyte culture media (DMEM + 10% FBS). Cells were plated on poly-d-lysine (PDL)–coated T75 flasks at 50,000 cells/cm2. The mixed glial culture was allowed to reach confluency before shaking at 250 rpm overnight at 37°C to remove unwanted cells types (microglia, oligodendrocytes, neurons, and fibroblasts). After a 24-h recovery in fresh media, astrocytes were trypsinized in 0.25% trypsin/EDTA and replated onto appropriate PDL-coated vessels for experiments. Media was changed every 3–5 d.
Primary microglia culture
The same procedure as described for primary astrocyte culture was followed with noted exceptions. Confluent mixed glial cultures were shaken for 2 h at 250 rpm at 37°C. The T-75 flasks were then tapped vigorously 10–15 times on the bench top to loosen microglia growing on top of the astrocytes. The media along with floating microglia were collected from the flask and centrifuged for 5 min at 1,000 g. The cell pellet from one T-75 flask was resuspended in 3 ml of astrocyte culture media and plated in 3 wells of a PDL coated 12-well plate. The media was changed 24 h later. After 48–72 h in culture, microglia were collected for experiments.
Primary neuronal culture
P0 pups were used for neuronal culture. The dissection procedure for primary astrocyte culture was followed with the noted exception of using neuronal culture media (neurobasal media + B-27 supplement + pen/strep + l-glutamine) instead of astrocyte media. Media on primary neurons was changed after 24 h. Weekly, fresh media of half the total volume of the well was added. Neurons were harvested at 18 days in vitro for experiments.
Plasmid construction and AAV transduction
Human TFEB coding sequence containing a C-terminal 3×FLAG tag was cloned into pAAV-GFAP-EGFP by replacing the EGFP sequence. Both pAAV-GFAP-TFEB3×FLAG and pAAV-GFAP-EGFP vectors were used to generate AAV 2/8 by the Gene Vector Core (Baylor College of Medicine). Serotype 8 AAVs were prepared by the transfection of three plasmids (transfer vector, p5E18-VD2/8 Rep-Cap plasmid, and pAdΔF6 helper plasmid) using iMFectin Poly DNA Transfection Reagent (GenDEPOT). AAV purification was performed by the method developed by Ayuso et al., with modifications (Ayuso et al., 2010). AAVs were dialyzed against phosphate buffered saline without Mg2+ and Ca2+, and the titer was determined by real time PCR as described previously (Ljungberg et al., 2012).
For AAV transduction in primary astrocytes, AAV was added to reduced serum medium (DMEM + 2% FBS) at 500,000 multiplicity of infection in half the usual volume for the particular surface area. Following 4–6 h of incubation, equal amount of fresh astrocyte medium (DMEM + 10% FBS) was added to the culture. Media was then changed 48 h after the addition of AAV. Astrocytes were cultured an additional 5–7 d before performing experiments to reach optimal TFEB or EGFP expression levels. Transduction efficiency was observed at >80% for these in vitro studies.
RNA extraction, reverse transcription, and qRT-PCR
Total RNA was extracted from primary astrocytes using an RNeasy Mini kit (Qiagen), and cDNA was synthesized from 500 ng total RNA using SuperScript III First-Strand Synthesis System (Invitrogen). For hippocampal brain samples, TRIzol reagent (Invitrogen) was used to extract total RNA and cDNA was synthesized from 2 µg total RNA. cDNA was diluted to 2 ng/µL and 4 µl were added to 10 µl 2× FastStart Universal SYBR Green PCR Master (Roche). Each sample was run in triplicate using a StepOnePlus Real-Time PCR System (Applied Biosystems). Ct values were normalized to the housekeeping gene GAPDH, which was amplified in parallel. The 2−ΔΔCT method was used to calculate relative gene expression levels. The following qRT-PCR primers were used: TFEB, 5′- TCAGAAGCGAGAGCTAACAGAT-3′ and 5′-TGTGATTGTCTTTCTTCTGCCG-3′; LAMP1, 5′-CAGCACTCTTTGAGGTGAAAAAC-3′ and 5′-ACGATCTGAGAACCATTCGCA-3′; MCOLN1 5′-CTGACCCCCAATCCTGGGTAT-3′ and 5′-GGCCCGGAACTTGTCACAT-3′; CTSA, 5′-CCCTCTTTCCGGCAATACTCC-3′ and 5′-CGGGGCTGTTCTTTGGGTC-3′; CTSB, 5′-TCCTTGATCCTTCTTTCTTGCC-3′ and 5′-ACAGTGCCACACAGCTTCTTC-3′; CTSD, 5′-GCTTCCGGTCTTTGACAACCT-3′ and 5′-CACCAAGCATTAGTTCTCCTCC-3′; ATP6V1H, 5′-GGATGCTGCTGTCCCAACTAA-3′ and 5′-TCTCTTGCTTGTCCTCGGAAC-3′; GAPDH, 5′-AACTTTGGCATTGTGGAAGG-3′ and 5′-ACACATTGGGGGTAGGAACA-3′; human TFEB, 5′-CCAGAAGCGAGAGCTCACAGAT-3′ and 5′-TGTGATTGTCTTTCTTCTGCCG-3′; human LAMP1, 5′-ACGTTACAGCGTCCAGCTCAT-3′ and 5′-TCTTTGGAGCTCGCATTGG-3′; human CTSB, 5′-AGTGGAGAATGGCACACCCTA-3′ and 5′-AAGAAGCCATTGTCACCCCA-3′; human CTSD, 5′-AACTGCTGGACATCGCTTGCT-3′ and 5′-CATTCTTCACGTAGGTGCTGGA-3′; human GAPDH, 5′-TGCACCACCAACTGCTTAGC-3′ and 5′-GGCATGGACTGTGGTCATGAG-3′.
Immunoblotting
For Western blot, cells, forebrain, or dissected hippocampus were lysed in radioimmunoprecipitation assay buffer (TBS with 1% NP-40, 1% sodium deoxycholic acid, 0.1% sodium dodecyl sulfate, and protease/phosphatase inhibitor cocktails [Roche]). Lysates were sonicated six pulses at 50% duty cycle and incubated on ice for 30 min. Samples were then centrifuged at 20,000 g for 20 min. Supernatants were collected and quantified using a Pierce BCA Protein Assay kit (Thermo Fisher). Lysates were incubated 7 min at 90°C in sample loading buffer. 15-µg protein samples were loaded onto 12% SDS-PAGE gels and then transferred to nitrocellulose membranes (Bio-Rad). Membranes were blocked in 5% nonfat milk in PBS + 0.1% Tween 20 (PBS-T). Blots were probed with primary antibody, washed with PBS-T, and then probed with the appropriate HRP-conjugated secondary antibody, followed by additional washes. The signal was developed with Pierce ECL Western Blotting Substrate (Thermo Fisher) and detected using x-ray films. Band intensity was quantified using ImageJ software (National Institutes of Health) and normalized to the loading control (γ-tubulin or β-actin). The loading control normalized samples were then normalized to the average of either wild-type or EGFP samples, so that wild-type or EGFP samples would average to 1. The values reported are relative to the control (either EGFP or wild type).
Soluble/insoluble fractionation
The hippocampus was dissected from 9-mo-old PS19 mice injected with AAV at P3. The hippocampus was weighed for each of the mice and recorded to later use to normalize the loading of the soluble fraction. The tissue was suspended in 350 µl of TBS with 10% sucrose + protease/phosphatase inhibitor cocktails and disrupted using a handheld homogenizer. The mixture was centrifuged at 800 g for 5 min at 4°C. Pellets were resuspended in the same volume of the above buffer and centrifuged again under the same conditions. Supernatants from the two steps were pooled together and 700 µl of the solution from each mouse underwent ultracentrifugation at 100,000 g for 1 h at 4°C. The supernatants from ultracentrifugation were collected as the soluble fractions. Pellets from the ultracentrifugation were resuspended in 700 µl of 1% sarkosyl in TBS with protease/phosphatase inhibitor cocktails, sonicated, incubated in shaking for 30 min, and ultracentrifuged again at 100,000 g for 1 h at 4°C. Pellets from the second ultracentrifugation step were resuspended in 150 µl of 1% SDS, sonicated and kept as the insoluble fraction. The soluble fraction loading volume was normalized based on the weight of the hippocampus. The insoluble fraction was loaded in equal volumes for all samples. 30 µl of insoluble fractions per gel were added to 4× sample loading buffer and heated at 95°C for 7 min. Samples were subjected to SDS-PAGE and Western blot analysis.
Flow cytometry
Primary astrocytes were incubated with 100 nM Lysotracker red (Life Technologies) for 1 h before trypsinization and harvested for flow cytometry. Flow cytometry analysis was completed in the Cytometry and Cell Sorting Core (Baylor College of Medicine) on an LSR Fortessa (BD Biosciences). BD FACSDiva 8.0.1 software was used to analyze 20,000 events/sample. Primary astrocytes were also incubated with 1 µg/ml DQ Red BSA (Life Technologies) for 3 h followed by flow cytometry analysis.
Fluorescent bead uptake assay
Primary astrocytes were incubated with red fluorescent latex beads, amine-modified polystyrene of 1.0 µm mean particle size (Sigma) for 1 h in starvation medium (HBSS with Ca2+ and Mg2+ + 10 mM Hepes + 1% vol/vol Pen/Strep) at 0.01% (vol/vol). Cells were then washed three times with ice-cold PBS to remove beads on the cell surface. Astrocytes were then trypsinized and subjected to flow cytometry analysis (20,000 events/sample).
Tau fibril preparation
Recombinant tau fibrils or pffs were prepared as described previously (Li and Lee, 2006; Xu et al., 2016). In brief, truncated human tau containing only four microtubule-binding repeats (4R) with a 5′ Myc tag and P301L MAPT mutation (5′Myc-K18/P301L) was cloned into pRK172 for bacterial expression. The protein was purified and underwent in vitro fibrillization by mixing recombinant tau with heparin. Fibrillization was confirmed using the thioflavin T fluorescence assay, sedimentation test, and cryo-electron microscopy. For cryo-electron microscopy, 5 µl of K18PL pffs was added to carbon coated grids. Pffs were imaged on a Jeol JEM2100 laB6 transmission electron microscope at 15,000× magnification. Following confirmation, the pffs were frozen as single use aliquots (40 µM) at −80°C. Before use in experiments, frozen aliquots were thawed followed by sonication of 30 brief pulses. The sonicated tau fibrils were then dye conjugated to AlexaFluor-647 using an AlexaFluor-647 Protein Labeling kit (Life Technologies). The dye-conjugated pffs were eluted with PBS and collected for further experiments. Dye conjugation of tau fibrils was confirmed via Western blot for Myc and total tau.
Tau fibril uptake assay
Primary astrocytes were incubated in starvation medium 1 h before the addition of AlexaFluor-647 pffs (500 nM). Cells were incubated with dye-conjugated pffs for 1 or 4 h, then washed with ice-cold PBS three times before trypsinization for the removal of cell surface associated pffs and detachment of cells. Harvested cells were subjected to flow cytometry analysis (20,000 events/sample). For heparin treatment, primary astrocytes received 100 µg/ml heparin (Sigma) along with the dye-conjugated pffs in starvation medium for 1 h before washout, trypsinization, and flow cytometry analysis.
For Torin1 experiments, astrocytes were treated with 250 nM Torin1 in astrocyte culture medium for 3 h prior to addition of pffs (as described above). Astrocytes were incubated with Torin1 and pffs for 1 h before washout, trypsinization, and harvest for flow cytometry.
Immunofluorescence
Primary astrocytes grown on coverslips were fixed in 4% paraformaldehyde (PFA) for 20 min at room temperature after multiple washes with ice-cold PBS. Following fixation, coverslips were gently washed with PBS. Coverslips were then incubated in blocking buffer (PBS + 2% donkey serum + 0.1% Triton X-100) for 1 h at room temperature. After blocking, coverslips were incubated with primary antibodies (FLAG, 1:1,000; LAMP1, 1:50) overnight in blocking buffer at 4°C. Coverslips were then washed in PBS followed by incubation with secondary antibodies (anti-mouse IgG AlexaFluor-488; anti-rat IgG AlexaFluor-555 [Invitrogen]) for 2 h in blocking buffer at room temperature. Coverslips were then washed in PBS and mounted using DAPI containing mounting media. Cells were imaged by confocal microscopy (Leica TCS SPE). For colocalization analysis, Coloc2 plugin from ImageJ was used to calculate number of colocalized pixels of LAMP1 and pffs-647. Fields of view were randomly selected at 40× for a total of 12 images per group, and the number of colocalized pixels were averaged for each group.
Mice were transcardially perfused with saline followed by 4% PFA, and the brains were collected and post-fixed overnight in 4% PFA followed by dehydration in 30% sucrose. The brains were then sectioned on a sliding microtome in 30-µm increments. The sections underwent extensive washing in TBS before addition of primary antibody (FLAG, 1:250; MC1, 1:500; AT8, 1:500; Iba1, 1:1,000; NeuN, 1:1,000; and GFAP, 1:1,000) in blocking buffer overnight at 4°C. Sections were then washed in TBS followed by incubation with secondary fluorescence antibodies for 2 h at room temperature. After a final wash in TBS, floating sections were mounted on gelatin-coated glass slides and allowed to dry before adding mounting medium containing DAPI. Sections were imaged by confocal microscopy (Leica TCS SPE). The Leica TCS SPE is equipped with a 405-, 488-, 561-, and 635-nm laser lines and HC PL APO 10×/0.40, HC PL APO 20×/0.70 CS, ACS APO 40×/1.15 OIL CS, ACS APO 63×/1.30 OIL CS objectives.
X-34 staining
The X-34 staining protocol was adapted from a previous publication (Styren et al., 2000). Free floating sections cut at 30-µm thickness were incubated in a 40% ethanol staining solution containing 10 µM X-34 (SML1954; Sigma) for 10 min. Excess X-34 was rinsed off with PBS, and sections were mounted on microscope slides with Fluoromount-G (00-4958-02; Thermo Fisher).
Immunohistochemistry
Free-floating tissue sections were incubated at 4°C overnight in a solution containing CP13 antibody at a 1:250 dilution, 2% donkey serum, and 0.4% Triton X-100 in PBS. The primary solution was washed off with three washes of PBS before incubating in a 1:1,000 dilution of biotinylated anti-mouse IgG (BA-2000; Vector) in PBS for 2 h at room temperature. Sections were washed again and incubated in the Vectastain ABC kit (PK-6100; Vector) following their protocol for 2 h at room temperature. The sections were then washed twice with PBS and twice more with 0.1 M sodium acetate before being submerged in the staining solution for 60 s. The chemical staining solution consists of 4 ml distilled water, 5 ml of 5% nickel ammonium sulfate, 10 mg DAB (D8001; Sigma), and 5 µl of 30% hydrogen peroxide. The reaction was quenched in 0.1 M sodium acetate before washing in PBS and mounting on slides with mounting media.
Immunofluorescence quantification
TFEB nuclear localization was calculated based on counting instances of DAPI and FLAG staining colocalization and dividing by total number of FLAG-positive cells per confocal image, averaged for each animal (five images per animal, n = 5). Colocalization was determined based on multiple z-stack slices (20 slices per 30-µm section). Random areas of the cortex and hippocampus were selected for imaging at 40×.
Number of astrocytes containing tau in EGFP and TFEB tau spreading mice were calculated based on counting number of astrocytes with colocalization of GFAP and AT8 staining. Colocalization was determined based on confocal microscropy of multiple z-stack slices (30 slices per 30-µm section). Random areas of the hippocampus were selected for imaging at 40× (40 images per group, n = 4).
For calculating area fluorescence of GFAP, Iba1, MC1, and AT8 antibody staining, the slide containing representative slices of the entire mouse brain was scanned on an EVOS fluorescence microscope. Area fluorescence in specific brain regions was calculated after thresholding to eliminate background and nonspecific staining using ImageJ. Area fluorescence of GFAP, Iba1, MC1, or AT8 staining in the cortex or hippocampus was averaged across all consistently represented sections for each animal to signify the relative pathology or gliosis within the entire volume of the brain region analyzed. The hippocampus and cortex were thresholded separately.
In vivo gene delivery
Pups at P3 were anesthetized via hypothermia before bilateral ICV injections. Anesthetized pups were injected free-hand with 1010 viral particles per side of AAV2/8-GFAP-TFEB or AAV2/8-GFAP-EGFP using at 28-gauge needle attached to a Hamilton syringe.
CTSD enzyme activity assay
CTSD activity was measured using a CTSD enzyme assay kit (Ab65302; Abcam). 25 µg of protein homogenate was adjusted to 50 µl per well with cell lysis buffer (provided in the kit) for experimental samples. 50 µl of reaction buffer (provided in the kit) per reaction well was added to the protein homogenate. 50 µl of reaction buffer with 50 µl lysis buffer per reaction well without protein homogenate was used as a negative control or blank. Finally, 2 µl substrate (provided in the kit) was added to each reaction well including the blanks and excitation/emission (Ex/Em) = 328/460 was measured using a Synergy 2 Multi-Detection Microplate Reader and samples were left to incubate at 37°C in dark, before taking the reading for the next time point. Fluorescence was measured in technical triplicates from n = 5 samples per genotype at the indicated times. Appropriate calculations to measure first-order rate constant (k) of enzymatic reaction were obtained upon subtracting blank fluorescence units from the values of the experimental wells.
Electrophysiology
In brief, adult 9-mo-old mice were anesthetized with isoflurane and decapitated. The brain was rapidly removed and placed into ice-cold modified NMDG solution (composition in mM: 110 NMDG, 110 HCl, 3 KCl, 10 MgCl2 6*H2O, 1.1 NaH2PO4 H2O, 0.5 CaCl2 dihydrate, 25 glucose, 3 pyruvic acid, 10 ascorbic acid, 25 NaHCO3, and 305–310 mOsm; Ting et al., 2014; Miller et al., 2017). 350-µm horizontal hippocampal sections were harvested using Leica VT1200S vibratome. Immediately after slicing, slices were allowed to recover in oxygenated (95% oxygen and 5% CO2) NMDG for 30 min at 34°C, followed by another 30-min incubation in bicarbonate buffered ACSF (composition in mM: 124 NaCl, 4 KCl, 26 NaHCO3, 1.26 NaH2PO4, 1.3 MgCl2, and 2 CaCl2) at room temperature before recording. ACSF solutions were bubbled with 95% O2/5% CO2 at all times to maintain consistent oxygenation and pH.
To record sEPSCs, pipettes were filled with a cesium-substituted intracellular solution (mM): 10 CsCl, 105 CsMeSO3, 0.5 ATP, 0.3 GTP, 10 Hepes, 5 glucose, 2 MgCl2, and 1 EGTA. Neurons were held at the measured GABA reversal potential (−65 mV), corrected for liquid junction potential (Chen et al., 2016). No drugs were included in the bath. Whole cell patch clamp recordings were acquired (MultiClamp 700B; Axon Instruments) and digitized sampled at 20 kHZ (Digidata 1550; Axon Instruments), filtered at 2 kHz and acquired with pClamp software (Axon Instruments). Pipette resistances ranged from 4 to 7 MΩ Access resistance ranged from 8 to 20 MΩ and was monitored for consistency; for analyzed cells, change was never >20%. Recordings were discarded if the initial leak current was >200 pA or changed by >20% over the course of recording. Off-line analysis was performed using Clampfit.
Tau spreading assay
PS19 pups underwent bilateral ICV injections at P3 with AAV2/8-GFAP-TFEB or AAV2/8-GFAP-EGFP as described above. In brief, 3-mo-old PS19 mice were deeply anesthetized with 2% isoflurane and placed in a Kopf stereotaxic device (Model 940) to secure the temporal bone. A microinjection pipette (Nanoject II; Drummond Scientific) was used to aseptically inject 2.0 µl of pffs into the left hippocampus (stereotaxic coordinates from bregma: −2.5 mm posterior, −2.0 mm lateral, and −1.8 mm ventral) at a flow rate of 50 nanoliters every 6 s. The needle was pulled vertically in 0.6-mm increments during removal to reduce off target tau seeding in the cortex due to capillary action from needle withdrawal.
Statistics
All data are presented as ±SEM. Power analysis was performed using a confidence interval of α = 0.05. Pairwise comparisons were analyzed using a two-tailed Student’s t test. Human transcriptomic data were analyzed using Spearman’s correlation coefficient and significance was computed using algorithm AS 89 via the asymptotic t approximation implemented by the stats package in R. P values <0.05 were considered statistically significant (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Online supplemental material
Fig. S1 shows the correlation of additional lysosomal TFEB target transcripts with cognitive decline and Braak staging, as well as TFEB nuclear localization in the tau spreading mouse model. Fig. S2 shows widespread astroglial expression of AAV-GFAP-TFEB/EGFP in the mouse brain. Fig. S3 shows immunostaining and quantification of gliosis in the rTg4510 mice. Fig. S4 shows phospho tau immunostaining validating the time points chosen for the tau spreading model. Fig. S5 shows AT8 immunostaining and X-34 staining with quantification in the tau spreading model. Table S1 lists human brain demographic data. Table S2 lists antibodies used in experiments.
Supplementary Material
Acknowledgments
We are grateful to P. Davies for the gift of PHF1, CP13, and MC1 antibodies. We thank N. Timchenko for assisting with protein purification. This project was supported by core services at Baylor College of Medicine, including the Cytometry and Cell Sorting Core with funding from the National Institutes of Health (grants P30 AI036211, P30 CA125123, and S10 RR024574) and the assistance of J. Sederstrom, the Gene Vector Core with assistance from K. Oka and S. Deng, and the Protein and Monoclonal Antibody Production Core with assistance from K. Phillips and M. Lee (NIH funding: grant P30 CA125123). We appreciate the generosity of Wah Chiu and the technical assistance of Patrick Mitchell in generating cryo-electron microscopy images of tau pffs. We thank members of the Zheng laboratory for insightful discussions.
This study was supported by grants from the NIH (R01 NS093652, R01 AG020670, and RF1 AG054111 to H. Zheng; ADCC AG010124 and FTD PPG AG17586 to J.Q. Trojanowski and V.M.Y. Lee; and F30 AG050412 to H. Martini-Stoica) and Wyncote Foundation (FTD Center) to J.Q. Trojanowski and V.M.Y. Lee.
The authors declare no competing financial interests.
Author contributions: H. Martini-Stoica designed, performed, and analyzed all experiments with the noted exceptions. A.L. Cole provided technical support in immunostaining and performed stereotaxic injections. D.B. Swartzlander assisted with flow cytometry and performed astrocyte isolation for FACS. F. Chen performed electrophysiology experiments. Y.-W. Wan analyzed MSSM data. M. Sardiello performed GSEA. L. Bajaj performed the cathepsin D enzyme activity assay. D.A. Bader performed immunoblotting of endogenous TFEB in primary cultures and provided critical editing of the manuscript. J.Q. Trojanowski provided human FTD samples and provided input on the writing of the manuscript. V.M.Y. Lee provided K18PL constructs and protocols in addition to providing input on the writing of the manuscript. Z. Liu provided guidance/assistance on bioinformatics. H. Martini-Stoica and H. Zheng conceived of the project. H. Martini-Stoica wrote the paper with H. Zheng’s input. All authors critically reviewed the paper.
References
- Asai H., Ikezu S., Tsunoda S., Medalla M., Luebke J., Haydar T., Wolozin B., Butovsky O., Kügler S., and Ikezu T.. 2015. Depletion of microglia and inhibition of exosome synthesis halt tau propagation. Nat. Neurosci. 18:1584–1593. 10.1038/nn.4132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayuso E., Mingozzi F., Montane J., Leon X., Anguela X.M., Haurigot V., Edmonson S.A., Africa L., Zhou S., High K.A., et al. 2010. High AAV vector purity results in serotype- and tissue-independent enhancement of transduction efficiency. Gene Ther. 17:503–510. 10.1038/gt.2009.157 [DOI] [PubMed] [Google Scholar]
- Barres B.A. 2008. The mystery and magic of glia: a perspective on their roles in health and disease. Neuron. 60:430–440. 10.1016/j.neuron.2008.10.013 [DOI] [PubMed] [Google Scholar]
- Basak J.M., Verghese P.B., Yoon H., Kim J., and Holtzman D.M.. 2012. Low-density lipoprotein receptor represents an apolipoprotein E-independent pathway of Aβ uptake and degradation by astrocytes. J. Biol. Chem. 287:13959–13971. 10.1074/jbc.M111.288746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak H., and Braak E.. 1995. Staging of Alzheimer’s disease-related neurofibrillary changes. Neurobiol. Aging. 16:271–278, discussion :278–284. 10.1016/0197-4580(95)00021-6 [DOI] [PubMed] [Google Scholar]
- Cahoy J.D., Emery B., Kaushal A., Foo L.C., Zamanian J.L., Christopherson K.S., Xing Y., Lubischer J.L., Krieg P.A., Krupenko S.A., et al. 2008. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci. 28:264–278. 10.1523/JNEUROSCI.4178-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarty P., Rosario A., Cruz P., Siemienski Z., Ceballos-Diaz C., Crosby K., Jansen K., Borchelt D.R., Kim J.Y., Jankowsky J.L., et al. 2013. Capsid serotype and timing of injection determines AAV transduction in the neonatal mice brain. PLoS One. 8:e67680 10.1371/journal.pone.0067680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Moran J.T., Zhang Y., Ates K.M., Yu D., Schrader L.A., Das P.M., Jones F.E., and Hall B.J.. 2016. The transcription factor NeuroD2 coordinates synaptic innervation and cell intrinsic properties to control excitability of cortical pyramidal neurons. J. Physiol. 594:3729–3744. 10.1113/JP271953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin S.S.-M., and Goldman J.E.. 1996. Glial inclusions in CNS degenerative diseases. J. Neuropathol. Exp. Neurol. 55:499–508. 10.1097/00005072-199605000-00001 [DOI] [PubMed] [Google Scholar]
- Clavaguera F., Bolmont T., Crowther R.A., Abramowski D., Frank S., Probst A., Fraser G., Stalder A.K., Beibel M., Staufenbiel M., et al. 2009. Transmission and spreading of tauopathy in transgenic mouse brain. Nat. Cell Biol. 11:909–913. 10.1038/ncb1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coen K., Flannagan R.S., Baron S., Carraro-Lacroix L.R., Wang D., Vermeire W., Michiels C., Munck S., Baert V., Sugita S., et al. 2012. Lysosomal calcium homeostasis defects, not proton pump defects, cause endo-lysosomal dysfunction in PSEN-deficient cells. J. Cell Biol. 198:23–35. 10.1083/jcb.201201076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo A.M., and Dice J.F.. 2000. When lysosomes get old. Exp. Gerontol. 35:119–131. 10.1016/S0531-5565(00)00075-9 [DOI] [PubMed] [Google Scholar]
- de Calignon A., Polydoro M., Suárez-Calvet M., William C., Adamowicz D.H., Kopeikina K.J., Pitstick R., Sahara N., Ashe K.H., Carlson G.A., et al. 2012. Propagation of tau pathology in a model of early Alzheimer’s disease. Neuron. 73:685–697. 10.1016/j.neuron.2011.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost B., Jacks R.L., and Diamond M.I.. 2009. Propagation of tau misfolding from the outside to the inside of a cell. J. Biol. Chem. 284:12845–12852. 10.1074/jbc.M808759200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk K.E., Mirbaha H., Jiang H., Holtzman D.M., and Diamond M.I.. 2015. Distinct Therapeutic Mechanisms of Tau Antibodies: Promoting Microglial Clearance Versus Blocking Neuronal Uptake. J. Biol. Chem. 290:21652–21662. 10.1074/jbc.M115.657924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannakopoulos P., Herrmann F.R., Bussière T., Bouras C., Kövari E., Perl D.P., Morrison J.H., Gold G., and Hof P.R.. 2003. Tangle and neuron numbers, but not amyloid load, predict cognitive status in Alzheimer’s disease. Neurology. 60:1495–1500. 10.1212/01.WNL.0000063311.58879.01 [DOI] [PubMed] [Google Scholar]
- Gousset K., Schiff E., Langevin C., Marijanovic Z., Caputo A., Browman D.T., Chenouard N., de Chaumont F., Martino A., Enninga J., et al. 2009. Prions hijack tunnelling nanotubes for intercellular spread. Nat. Cell Biol. 11:328–336. 10.1038/ncb1841 [DOI] [PubMed] [Google Scholar]
- Grober E., Dickson D., Sliwinski M.J., Buschke H., Katz M., Crystal H., and Lipton R.B.. 1999. Memory and mental status correlates of modified Braak staging. Neurobiol. Aging. 20:573–579. 10.1016/S0197-4580(99)00063-9 [DOI] [PubMed] [Google Scholar]
- Guo J.L., and Lee V.M.. 2011. Seeding of normal Tau by pathological Tau conformers drives pathogenesis of Alzheimer-like tangles. J. Biol. Chem. 286:15317–15331. 10.1074/jbc.M110.209296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halassa M.M., Fellin T., and Haydon P.G.. 2007. The tripartite synapse: roles for gliotransmission in health and disease. Trends Mol. Med. 13:54–63. 10.1016/j.molmed.2006.12.005 [DOI] [PubMed] [Google Scholar]
- Heneka M.T., Carson M.J., El Khoury J., Landreth G.E., Brosseron F., Feinstein D.L., Jacobs A.H., Wyss-Coray T., Vitorica J., Ransohoff R.M., et al. 2015. Neuroinflammation in Alzheimer’s disease. Lancet Neurol. 14:388–405. 10.1016/S1474-4422(15)70016-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes B.B., DeVos S.L., Kfoury N., Li M., Jacks R., Yanamandra K., Ouidja M.O., Brodsky F.M., Marasa J., Bagchi D.P., et al. 2013. Heparan sulfate proteoglycans mediate internalization and propagation of specific proteopathic seeds. Proc. Natl. Acad. Sci. USA. 110:E3138–E3147. 10.1073/pnas.1301440110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes B.B., Furman J.L., Mahan T.E., Yamasaki T.R., Mirbaha H., Eades W.C., Belaygorod L., Cairns N.J., Holtzman D.M., and Diamond M.I.. 2014. Proteopathic tau seeding predicts tauopathy in vivo. Proc. Natl. Acad. Sci. USA. 111:E4376–E4385. 10.1073/pnas.1411649111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iba M., Guo J.L., McBride J.D., Zhang B., Trojanowski J.Q., and Lee V.M.. 2013. Synthetic tau fibrils mediate transmission of neurofibrillary tangles in a transgenic mouse model of Alzheimer’s-like tauopathy. J. Neurosci. 33:1024–1037. 10.1523/JNEUROSCI.2642-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y., Maruyama W., Naoi M., Hashizume Y., and Osawa T.. 1998. Immunohistochemical detection of dityrosine in lipofuscin pigments in the aged human brain. FEBS Lett. 439:231–234. 10.1016/S0014-5793(98)01372-6 [DOI] [PubMed] [Google Scholar]
- Kfoury N., Holmes B.B., Jiang H., Holtzman D.M., and Diamond M.I.. 2012. Trans-cellular propagation of Tau aggregation by fibrillar species. J. Biol. Chem. 287:19440–19451. 10.1074/jbc.M112.346072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs G.G., Ferrer I., Grinberg L.T., Alafuzoff I., Attems J., Budka H., Cairns N.J., Crary J.F., Duyckaerts C., Ghetti B., et al. 2016. Aging-related tau astrogliopathy (ARTAG): harmonized evaluation strategy. Acta Neuropathol. 131:87–102. 10.1007/s00401-015-1509-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs G.G., Lee V.M., and Trojanowski J.Q.. 2017a Protein astrogliopathies in human neurodegenerative diseases and aging. Brain Pathol. 27:675–690. 10.1111/bpa.12536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs G.G., Robinson J.L., Xie S.X., Lee E.B., Grossman M., Wolk D.A., Irwin D.J., Weintraub D., Kim C.F., Schuck T., et al. 2017b Evaluating the Patterns of Aging-Related Tau Astrogliopathy Unravels Novel Insights Into Brain Aging and Neurodegenerative Diseases. J. Neuropathol. Exp. Neurol. 76:270–288. 10.1093/jnen/nlx007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalo U., Palygin O., Verkhratsky A., Grant S.G., and Pankratov Y.. 2016. ATP from synaptic terminals and astrocytes regulates NMDA receptors and synaptic plasticity through PSD-95 multi-protein complex. Sci. Rep. 6:33609 10.1038/srep33609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Yu W.H., Kumar A., Lee S., Mohan P.S., Peterhoff C.M., Wolfe D.M., Martinez-Vicente M., Massey A.C., Sovak G., et al. 2010. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell. 141:1146–1158. 10.1016/j.cell.2010.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., McBrayer M.K., Wolfe D.M., Haslett L.J., Kumar A., Sato Y., Lie P.P., Mohan P., Coffey E.E., Kompella U., et al. 2015. Presenilin 1 Maintains Lysosomal Ca(2+) Homeostasis via TRPML1 by Regulating vATPase-Mediated Lysosome Acidification. Cell Reports. 12:1430–1444. 10.1016/j.celrep.2015.07.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein E.S., Hawrylycz M.J., Ao N., Ayres M., Bensinger A., Bernard A., Boe A.F., Boguski M.S., Brockway K.S., Byrnes E.J., et al. 2007. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 445:168–176. 10.1038/nature05453 [DOI] [PubMed] [Google Scholar]
- Li W., and Lee V.M.. 2006. Characterization of two VQIXXK motifs for tau fibrillization in vitro. Biochemistry. 45:15692–15701. 10.1021/bi061422+ [DOI] [PubMed] [Google Scholar]
- Li Y., Cheng D., Cheng R., Zhu X., Wan T., Liu J., and Zhang R.. 2014. Mechanisms of U87 astrocytoma cell uptake and trafficking of monomeric versus protofibril Alzheimer’s disease amyloid-β proteins. PLoS One. 9:e99939 10.1371/journal.pone.0099939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Drouet V., Wu J.W., Witter M.P., Small S.A., Clelland C., and Duff K.. 2012. Trans-synaptic spread of tau pathology in vivo. PLoS One. 7:e31302 10.1371/journal.pone.0031302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljungberg M.C., Ali Y.O., Zhu J., Wu C.S., Oka K., Zhai R.G., and Lu H.C.. 2012. CREB-activity and nmnat2 transcription are down-regulated prior to neurodegeneration, while NMNAT2 over-expression is neuroprotective, in a mouse model of human tauopathy. Hum. Mol. Genet. 21:251–267. 10.1093/hmg/ddr492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina J.A., Chen Y., Gucek M., and Puertollano R.. 2012. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 8:903–914. 10.4161/auto.19653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina D.L., Fraldi A., Bouche V., Annunziata F., Mansueto G., Spampanato C., Puri C., Pignata A., Martina J.A., Sardiello M., et al. 2011. Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev. Cell. 21:421–430. 10.1016/j.devcel.2011.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller O.H., Bruns A., Ben Ammar I., Mueggler T., and Hall B.J.. 2017. Synaptic Regulation of a Thalamocortical Circuit Controls Depression-Related Behavior. Cell Reports. 20:1867–1880. 10.1016/j.celrep.2017.08.002 [DOI] [PubMed] [Google Scholar]
- Narasimhan S., Guo J.L., Changolkar L., Stieber A., McBride J.D., Silva L.V., He Z., Zhang B., Gathagan R.J., Trojanowski J.Q., and Lee V.M.Y.. 2017. Pathological Tau Strains from Human Brains Recapitulate the Diversity of Tauopathies in Nontransgenic Mouse Brain. J. Neurosci. 37:11406–11423. 10.1523/JNEUROSCI.1230-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon R.A. 2004. Niemann-Pick Type C disease and Alzheimer’s disease: the APP-endosome connection fattens up. Am. J. Pathol. 164:757–761. 10.1016/S0002-9440(10)63163-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama F., Murakami N., and Ihara Y.. 1998. Chloroquine myopathy suggests that tau is degraded in lysosomes: implication for the formation of paired helical filaments in Alzheimer’s disease. Neurosci. Res. 31:1–8. 10.1016/S0168-0102(98)00020-0 [DOI] [PubMed] [Google Scholar]
- Palmieri M., Impey S., Kang H., di Ronza A., Pelz C., Sardiello M., and Ballabio A.. 2011. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum. Mol. Genet. 20:3852–3866. 10.1093/hmg/ddr306 [DOI] [PubMed] [Google Scholar]
- Palmieri M., Pal R., Nelvagal H.R., Lotfi P., Stinnett G.R., Seymour M.L., Chaudhury A., Bajaj L., Bondar V.V., Bremner L., et al. 2017. mTORC1-independent TFEB activation via Akt inhibition promotes cellular clearance in neurodegenerative storage diseases. Nat. Commun. 8:14338 10.1038/ncomms14338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastore N., Brady O.A., Diab H.I., Martina J.A., Sun L., Huynh T., Lim J.A., Zare H., Raben N., Ballabio A., and Puertollano R.. 2016. TFEB and TFE3 cooperate in the regulation of the innate immune response in activated macrophages. Autophagy. 12:1240–1258. 10.1080/15548627.2016.1179405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekny M., Pekna M., Messing A., Steinhäuser C., Lee J.-M., Parpura V., Hol E.M., Sofroniew M.V., and Verkhratsky A.. 2016. Astrocytes: a central element in neurological diseases. Acta Neuropathol. 131:323–345. 10.1007/s00401-015-1513-1 [DOI] [PubMed] [Google Scholar]
- Peña-Llopis S., Vega-Rubin-de-Celis S., Schwartz J.C., Wolff N.C., Tran T.A., Zou L., Xie X.J., Corey D.R., and Brugarolas J.. 2011. Regulation of TFEB and V-ATPases by mTORC1. EMBO J. 30:3242–3258. 10.1038/emboj.2011.257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perea G., Navarrete M., and Araque A.. 2009. Tripartite synapses: astrocytes process and control synaptic information. Trends Neurosci. 32:421–431. 10.1016/j.tins.2009.05.001 [DOI] [PubMed] [Google Scholar]
- Polito V.A., Li H., Martini-Stoica H., Wang B., Yang L., Xu Y., Swartzlander D.B., Palmieri M., di Ronza A., Lee V.M., et al. 2014. Selective clearance of aberrant tau proteins and rescue of neurotoxicity by transcription factor EB. EMBO Mol. Med. 6:1142–1160. 10.15252/emmm.201303671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pooler A.M., Phillips E.C., Lau D.H., Noble W., and Hanger D.P.. 2013. Physiological release of endogenous tau is stimulated by neuronal activity. EMBO Rep. 14:389–394. 10.1038/embor.2013.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsden M., Kotilinek L., Forster C., Paulson J., McGowan E., SantaCruz K., Guimaraes A., Yue M., Lewis J., Carlson G., et al. 2005. Age-dependent neurofibrillary tangle formation, neuron loss, and memory impairment in a mouse model of human tauopathy (P301L). J. Neurosci. 25:10637–10647. 10.1523/JNEUROSCI.3279-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemenschneider M., Wagenpfeil S., Vanderstichele H., Otto M., Wiltfang J., Kretzschmar H., Vanmechelen E., Förstl H., and Kurz A.. 2003. Phospho-tau/total tau ratio in cerebrospinal fluid discriminates Creutzfeldt-Jakob disease from other dementias. Mol. Psychiatry. 8:343–347. 10.1038/sj.mp.4001220 [DOI] [PubMed] [Google Scholar]
- Roczniak-Ferguson A., Petit C.S., Froehlich F., Qian S., Ky J., Angarola B., Walther T.C., and Ferguson S.M.. 2012. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci. Signal. 5:ra42 10.1126/scisignal.2002790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SantaCruz K., Lewis J., Spires T., Paulson J., Kotilinek L., Ingelsson M., Guimaraes A., DeTure M., Ramsden M., McGowan E., et al. 2005. Tau suppression in a neurodegenerative mouse model improves memory function. Science. 309:476–481. 10.1126/science.1113694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello M., Palmieri M., di Ronza A., Medina D.L., Valenza M., Gennarino V.A., Di Malta C., Donaudy F., Embrione V., Polishchuk R.S., et al. 2009. A gene network regulating lysosomal biogenesis and function. Science. 325:473–477. 10.1126/science.1174447 [DOI] [PubMed] [Google Scholar]
- Schultz C., Ghebremedhin E., Del Tredici K., Rüb U., and Braak H.. 2004. High prevalence of thorn-shaped astrocytes in the aged human medial temporal lobe. Neurobiol. Aging. 25:397–405. 10.1016/S0197-4580(03)00113-1 [DOI] [PubMed] [Google Scholar]
- Settembre C., Di Malta C., Polito V.A., Garcia Arencibia M., Vetrini F., Erdin S., Erdin S.U., Huynh T., Medina D., Colella P., et al. 2011. TFEB links autophagy to lysosomal biogenesis. Science. 332:1429–1433. 10.1126/science.1204592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C., Zoncu R., Medina D.L., Vetrini F., Erdin S., Erdin S., Huynh T., Ferron M., Karsenty G., Vellard M.C., et al. 2012. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 31:1095–1108. 10.1038/emboj.2012.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Yamada K., Liddelow S.A., Smith S.T., Zhao L., Luo W., Tsai R.M., Spina S., Grinberg L.T., Rojas J.C., et al. Alzheimer’s Disease Neuroimaging Initiative . 2017. ApoE4 markedly exacerbates tau-mediated neurodegeneration in a mouse model of tauopathy. Nature. 549:523–527. 10.1038/nature24016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdsson E.M. 2009. Tau-focused immunotherapy for Alzheimer’s disease and related tauopathies. Curr. Alzheimer Res. 6:446–450. 10.2174/156720509789207930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Wang F., Savini M., Ake A., di Ronza A., Sardiello M., and Segatori L.. 2013. TFEB regulates lysosomal proteostasis. Hum. Mol. Genet. 22:1994–2009. 10.1093/hmg/ddt052 [DOI] [PubMed] [Google Scholar]
- Spampanato C., Feeney E., Li L., Cardone M., Lim J.A., Annunziata F., Zare H., Polishchuk R., Puertollano R., Parenti G., et al. 2013. Transcription factor EB (TFEB) is a new therapeutic target for Pompe disease. EMBO Mol. Med. 5:691–706. 10.1002/emmm.201202176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styren S.D., Hamilton R.L., Styren G.C., and Klunk W.E.. 2000. X-34, a fluorescent derivative of Congo red: a novel histochemical stain for Alzheimer’s disease pathology. J. Histochem. Cytochem. 48:1223–1232. 10.1177/002215540004800906 [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartzlander D.B., Propson N.E., Roy E.R., Saito T., Saido T., Wang B., and Zheng H.. 2018. Concurrent cell type-specific isolation and profiling of mouse brains in inflammation and Alzheimer’s disease. JCI Insight. 3:e121109 10.1172/jci.insight.121109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting J.T., Daigle T.L., Chen Q., and Feng G.. 2014. Acute brain slice methods for adult and aging animals: application of targeted patch clamp analysis and optogenetics. Methods Mol. Biol. 1183:221–242. 10.1007/978-1-4939-1096-0_14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Jonquieres G., Mersmann N., Klugmann C.B., Harasta A.E., Lutz B., Teahan O., Housley G.D., Fröhlich D., Krämer-Albers E.-M., and Klugmann M.. 2013. Glial promoter selectivity following AAV-delivery to the immature brain. PLoS One. 8:e65646 10.1371/journal.pone.0065646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe D.M., Lee J.H., Kumar A., Lee S., Orenstein S.J., and Nixon R.A.. 2013. Autophagy failure in Alzheimer’s disease and the role of defective lysosomal acidification. Eur. J. Neurosci. 37:1949–1961. 10.1111/ejn.12169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.W., Hussaini S.A., Bastille I.M., Rodriguez G.A., Mrejeru A., Rilett K., Sanders D.W., Cook C., Fu H., Boonen R.A., et al. 2016. Neuronal activity enhances tau propagation and tau pathology in vivo. Nat. Neurosci. 19:1085–1092. 10.1038/nn.4328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss-Coray T., Loike J.D., Brionne T.C., Lu E., Anankov R., Yan F., Silverstein S.C., and Husemann J.. 2003. Adult mouse astrocytes degrade amyloid-beta in vitro and in situ. Nat. Med. 9:453–457. 10.1038/nm838 [DOI] [PubMed] [Google Scholar]
- Xiao Q., Yan P., Ma X., Liu H., Perez R., Zhu A., Gonzales E., Burchett J.M., Schuler D.R., Cirrito J.R., et al. 2014. Enhancing astrocytic lysosome biogenesis facilitates Aβ clearance and attenuates amyloid plaque pathogenesis. J. Neurosci. 34:9607–9620. 10.1523/JNEUROSCI.3788-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Martini-Stoica H., and Zheng H.. 2016. A seeding based cellular assay of tauopathy. Mol. Neurodegener. 11:32 10.1186/s13024-016-0100-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., Cirrito J.R., Stewart F.R., Jiang H., Finn M.B., Holmes B.B., Binder L.I., Mandelkow E.M., Diamond M.I., Lee V.M., and Holtzman D.M.. 2011. In vivo microdialysis reveals age-dependent decrease of brain interstitial fluid tau levels in P301S human tau transgenic mice. J. Neurosci. 31:13110–13117. 10.1523/JNEUROSCI.2569-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., Holth J.K., Liao F., Stewart F.R., Mahan T.E., Jiang H., Cirrito J.R., Patel T.K., Hochgräfe K., Mandelkow E.M., and Holtzman D.M.. 2014. Neuronal activity regulates extracellular tau in vivo. J. Exp. Med. 211:387–393. 10.1084/jem.20131685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanamandra K., Kfoury N., Jiang H., Mahan T.E., Ma S., Maloney S.E., Wozniak D.F., Diamond M.I., and Holtzman D.M.. 2013. Anti-tau antibodies that block tau aggregate seeding in vitro markedly decrease pathology and improve cognition in vivo. Neuron. 80:402–414. 10.1016/j.neuron.2013.07.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanamandra K., Jiang H., Mahan T.E., Maloney S.E., Wozniak D.F., Diamond M.I., and Holtzman D.M.. 2015. Anti-tau antibody reduces insoluble tau and decreases brain atrophy. Ann. Clin. Transl. Neurol. 2:278–288. 10.1002/acn3.176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanamandra K., Patel T.K., Jiang H., Schindler S., Ulrich J.D., Boxer A.L., Miller B.L., Kerwin D.R., Gallardo G., Stewart F., et al. 2017. Anti-tau antibody administration increases plasma tau in transgenic mice and patients with tauopathy. Sci. Transl. Med. 9:eeal2029 10.1126/scitranslmed.aal2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiyama Y., Higuchi M., Zhang B., Huang S.M., Iwata N., Saido T.C., Maeda J., Suhara T., Trojanowski J.Q., and Lee V.M.. 2007. Synapse loss and microglial activation precede tangles in a P301S tauopathy mouse model. Neuron. 53:337–351. 10.1016/j.neuron.2007.01.010 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Chen G., Zhou W., Song A., Xu T., Luo Q., Wang W., Gu X.S., and Duan S.. 2007. Regulated ATP release from astrocytes through lysosome exocytosis. Nat. Cell Biol. 9:945–953. 10.1038/ncb1620 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.