Using genetic, pharmacological and animal model approaches, we elucidate a novel human mutation in a G protein coupled receptor that impairs receptor oligomerization and trafficking leading to fatal, non-immune hydrops fetalis associated with arrested lymphatic development.
Abstract
We report the first case of nonimmune hydrops fetalis (NIHF) associated with a recessive, in-frame deletion of V205 in the G protein–coupled receptor, Calcitonin Receptor-Like Receptor (hCALCRL). Homozygosity results in fetal demise from hydrops fetalis, while heterozygosity in females is associated with spontaneous miscarriage and subfertility. Using molecular dynamic modeling and in vitro biochemical assays, we show that the hCLR(V205del) mutant results in misfolding of the first extracellular loop, reducing association with its requisite receptor chaperone, receptor activity modifying protein (RAMP), translocation to the plasma membrane and signaling. Using three independent genetic mouse models we establish that the adrenomedullin–CLR–RAMP2 axis is both necessary and sufficient for driving lymphatic vascular proliferation. Genetic ablation of either lymphatic endothelial Calcrl or nonendothelial Ramp2 leads to severe NIHF with embryonic demise and placental pathologies, similar to that observed in humans. Our results highlight a novel candidate gene for human congenital NIHF and provide structure–function insights of this signaling axis for human physiology.
Introduction
In early stages of intrauterine development, changes in fluid distribution and equilibrium are not well tolerated (Brace, 1989; Brace and Valenzuela, 1990). Impaired lymphatic drainage in adults often presents with chronic swelling or lymphedema, but in the fetus it can develop into a more severe and lethal condition known as hydrops fetalis (HF; Apkon, 1995; Bukowski and Saade, 2000; Randenberg, 2010a,b). The term HF refers to excessive fluid accumulation in more than two fetal extra vascular compartments and body cavities and is characterized by generalized skin thickness of >5 mm, placental enlargement and edema, pericardial or pleural effusion, or ascites (Bellini et al., 2009, 2015; Bellini and Hennekam, 2012). HF is further subdivided into two categories, Rhesus iso-immunization or immune HF, which is common in developing countries, and nonimmune HF (NIHF). In Westernized countries, effective Rhesus (Rh) alloimmunization during pregnancy has reduced the number immune related HF cases to <10%, such that NIHF now accounts for 85–90% of all hydrops cases. The etiology of NIHF is further subdivided into 14 classification groups based on the cause of the disorder (Randenberg, 2010a,b). Within these subgroups, generalized lymphatic dysplasia (GLD) is the third leading cause of NIHF, following cardiovascular and idiopathic disorders, the latter of which account for 20% of all known NIHF cases.
In an attempt to understand the underlying causes of NIHF, Connell et al. developed a new classification system as a clinical and research tool to further delineate primary lymphedema conditions and facilitate the identification of new causative genes linked to GLD (Connell et al., 2013). Hennekam Syndrome (OMIM no. 235510) was the first inherited form of GLD described and is caused by loss-of-function mutations in the collagen and calcium binding EGF domains 1 (CCBE1) gene in an autosomal recessive manner (Alders et al., 2009; Connell et al., 2010). Consistent with the prevalent role of the CCBE1–VEGF-C–VEGFR3 signaling axis in lymphangiogenesis, genes encoding modifiers of this pathway have also been described in a limited number of families with Hennekam syndrome, such as atypical cadherin 4, FAT4 (Alders et al., 2014), and ADAM metallopeptidase with thrombospondin motif, ADAMTS3 (Brouillard et al., 2017). More recently, biallelic mutations in piezo-type mechanosensitive ion channel component 1 (PIEZO1) and heterozygous inactivating mutations in the Eph receptor B4 (EPHB4) have also been identified as causative of GLD in humans (Fotiou et al., 2015; Lukacs et al., 2015; Martin-Almedina et al., 2016). These new discoveries suggest that GLD is genetically heterogeneous and provides precedent for the identification of novel genetic pathways.
Calcitonin receptor-like receptor (CALCRL, gene; CLR, protein) is a G protein–coupled receptor (GPCR) that forms an active signaling complex for either adrenomedullin (AM) peptide or calcitonin gene related peptide (CGRP), depending on the receptor’s interaction with a family of receptor activity-modifying proteins (RAMPs 1–3) (McLatchie et al., 1998; Kamitani et al., 1999; Hay et al., 2006; Woolley and Conner, 2013). Coexpression and direct protein–protein interaction of a RAMP with CLR is required for their forward translocation from the cytoplasm to the plasma membrane. Moreover, the RAMP moiety also determines ligand binding specificity, such that an interaction between CLR and RAMP2 or RAMP3 leads to the formation of a receptor complex that preferentially binds AM (AM1 and AM2 receptors, respectively), while a CLR–RAMP1 complex creates the active form of the CGRP-binding receptor. Selective antagonists and neutralizing antibodies that exploit this unique paradigm of GPCR selectivity and function have very recently shown great promise in phase III clinical trials for the therapeutic treatment of migraine (Goadsby et al., 2017; Silberstein et al., 2017). Yet, there are currently no known conditions or diseases associated with loss-of-function of these genes and signaling pathways in humans.
We and others have used genetic approaches in mice to elucidate the developmental and physiological functions of this complex GPCR signaling pathway in vivo (Kamitani et al., 1999; Dackor et al., 2006; Fritz-Six et al., 2008; Ichikawa-Shindo et al., 2008; Kadmiel et al., 2011; Koyama et al., 2013). Although genetic loss of murine Ramp1, Ramp3, or CalcA/B is compatible with long-term survival (Dackor et al., 2007), global deletion of the genes encoding either AM, CLR, or RAMP2 results in embryonic lethality at mid-gestation characterized by marked HF caused by a developmental arrest of lymphangiogenesis (Caron and Smithies, 2001; Dackor et al., 2006, 2007; Fritz-Six et al., 2008; Ichikawa-Shindo et al., 2008). Additionally, female mice that are haploinsufficient for Adm, Calcrl, or Ramp2 exhibit significant subfertility associated with abnormal uterine receptivity, placental defects and fetal growth restriction, which are maternally linked phenotypes largely independent of the genotype of the fetus (Li et al., 2006, 2008).
Here, we identify a novel recessive form of GLD that presents with NIHF and fetal demise in the homozygous state, as well as accompanying clinical subfertility in women heterozygous for a loss-of-function mutation in the CALCRL gene. Using molecular dynamic modeling, newly developed biochemistry approaches, and in vivo conditional genetic models, we further reveal that the in-frame deletion variant in the CALCRL gene reduces the translocation of the active CLR–RAMP receptor complex to the plasma membrane, resulting in decreased receptor function, which in complementary genetic mouse models, underscores the cell autonomous requirement of the CLR–RAMP2–AM signaling axis within lymphatic endothelial cells.
Results
Clinical assessment of a family presenting with subfertility and hydropic fetuses
The proband, a 22-yr-old asymptomatic woman G4 P1211 (Fig. 1 A, III.4), with normal liver enzymes and no signs of diabetes mellitus or hypertension, was referred to the genetics clinic after experiencing three pregnancies resulting in intrauterine fetal demise. Her first pregnancy resulted in a healthy baby girl (IV.1). The second pregnancy (Fig. 1 A, IV.2) was complicated by HF and mild polyhydramnios, which were noticed during second trimester ultrasounds. The first ultrasound during the 22nd week of gestation was inconclusive due to fetal positioning. However, the second ultrasound during the 27th week of gestation (26 wk + 3 d), which exhibited normal radiological parameters, confirmed the presence of HF, with a biparietal diameter measurement equivalent to 40 wk + 5 d and a head circumference equivalent to 33 wk + 6 d. Consistent with the diagnostic criteria for HF (Apkon, 1995; Bellini et al., 2006, 2009; Bellini and Hennekam, 2012), significant interstitial fluid was noted in more than two extravascular compartments, including marked pleural effusion indicated by reduced lung size, marked skin edema, and pericardial effusion. The pericardial effusion was also associated with impaired cardiac outflow. The pregnancy continued to 29 wk, after which the fetal movement completely ceased.
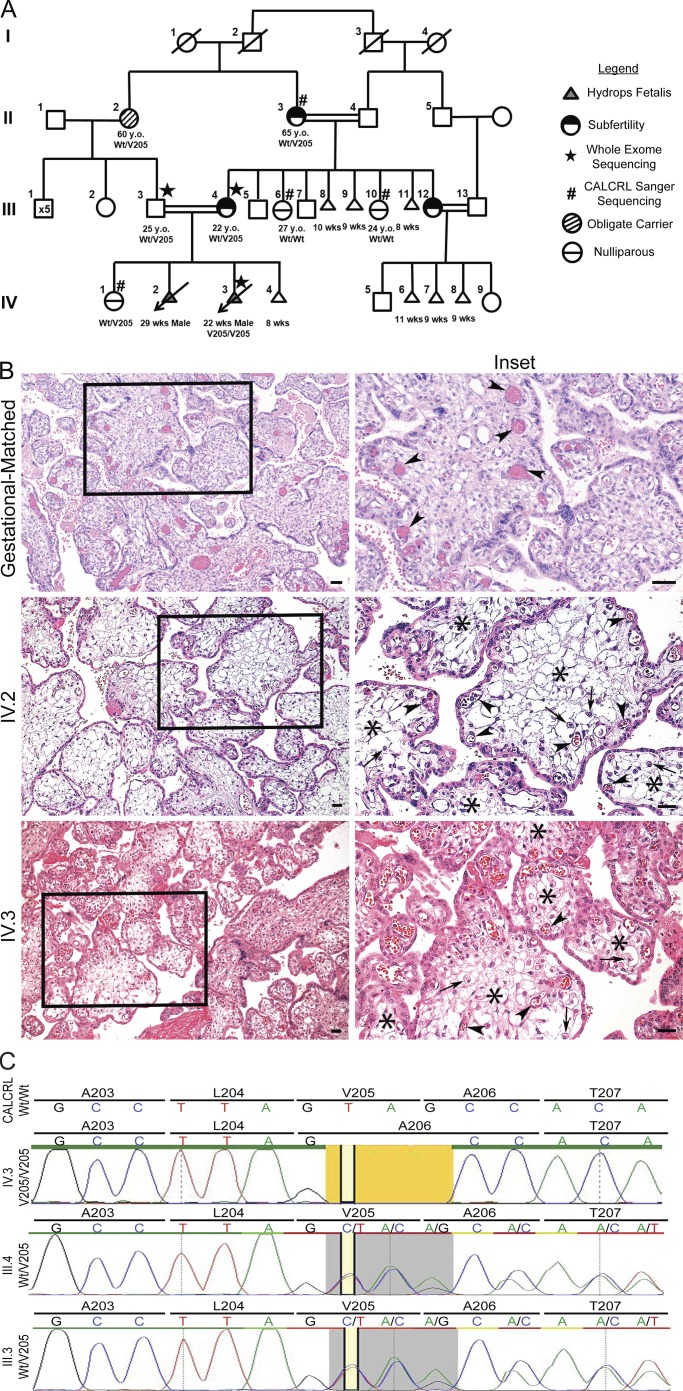
Figure 1.
Familial pedigree, placental histology and sequencing traces of family with NIHF. (A) Whole exome sequencing identified mutations in subject IV.3 in hCALCRL following the elimination of all previously implicated candidate genes. Parents, III.3 and III.4, and maternal grandmother were confirmed as heterozygous carriers of the hCALCRL(V205del) variant. Family history gives rise to two phenotypes dependent on haplotype: subfertility and nonimmune HF. (B) Placental histology from affected homozygous fetuses (IV.2 and IV.3) compared with a gestational-matched, normal placenta. The right column represents enlargements of boxed areas in the left column. Arrowheads indicate fetal vessels. In the case of the normal placenta, these vessels are completely filled with fetal erythrocytes and distributed both at the periphery and closer to the central villus core. In the case of the two affected placentas, the fetal vessels are more commonly found near the villus periphery, are compressed, and contain fewer erythrocytes. Arrows indicate presence of scattered Hofbauer cells. Asterisks indicate regions of severe edema within the chorionic villi of the affected placentas, a finding not seen in the normal placenta. (C) Whole exome sequencing traces for IV.3, III.4, and III.3 showing the amino acid residue valine 205 deletion (yellow). In individuals III.4 and III.3, the heterozygous allele (gray) is represented by the double peak.
During the proband’s third pregnancy (Fig. 1 A, IV.3), two ultrasounds were performed in the clinic. The first was at 12 wk of gestation and was determined to be normal. The second was at 21 wk and showed normal cardiac outflow with no major anomalies. However, at 22 wk of gestation, the proband noted poor intrauterine movements, and an ultrasound revealed mild polyhydramnios with significant presentation of HF. Due to the previous history of intrauterine fetal demise, blood was collected from the fetus for genetic testing.
The proband’s fourth pregnancy (Fig. 1 A, IV.4) resulted in spontaneous miscarriage at 8 wk of gestation when she presented with sudden onset of vaginal bleeding. No fetal or placental tissue was obtained from IV.4.
Placental pathology consistent with HF
Placental pathology was performed on IV.2 and IV.3 with findings consistent with the clinical presentation of HF. The umbilical cord of IV.2 appeared normal, with a trivascular structure consisting of two arteries and one vein. In addition, the fetal membranes were translucent and negative for acute chorioamnionitis. However, the singleton aborted placenta was edematous and large for the gestational age, weighing 1,113 g. Compared with an unaffected, gestationally matched, 24-wk placenta (Fig. 1 B, top panels), the placenta of IV.2 showed diffuse edematous chorionic villi, with severe villus edema and scattered Hofbauer cells (Fig. 1 B, middle panels). Furthermore, the fetal vessels appeared abnormally shaped and compressed, likely due to the encroaching villus edema.
Similar to IV.2, the IV.3 singleton placenta was large for gestation age, weighing 484 g. The umbilical cord was trivascular with no noticeable acute arteritis or phlebitis. The placenta was negative for villitis and viral cytopathic effects, with translucent membranes negative for acute chorioamnionitis. However, histopathological evaluation revealed edematous chorionic villi, an increased number of Hofbaeur cells and chorangiosis (Fig. 1 B, bottom panels). Once again, some of the fetal vessels appeared multi-lobular in shape and compressed to the outer rims of the chorionic villi, likely due to encroaching villus edema. Collectively, these placental findings are consistent with the fetal presentation of NIHF.
Exome sequencing to identify causal CALCRL (V205del) variant
The family history of consanguinity (Fig. 1 A) indicated that the fetal demise and resulting subfertility could be related to a homozygous variant. To address this hypothesis, exome sequencing was performed on blood samples collected from III.4, III.3, and IV.3. We first searched for potentially damaging variants in known genes recently implicated with NIHF with GLD, including PIEZO1 (Fotiou et al., 2015) and EPHB4 (Martin-Almedina et al., 2016), as well as CCBE1 (Hennekam Syndrome; Alders et al., 2009; Connell et al., 2010) and genes associated with known lymphatic malformations, including FLT4 (Joukov et al., 1996; Achen et al., 1998; Gordon et al., 2013), FOXC2 (Fang et al., 2000; Sabine et al., 2012), and SOX18 (François et al., 2008). In all instances, with an average coverage of 10 × 117.5 in all regions, we found these genes to be normal. However, exome sequencing analysis led to the detection of a previously unreported homozygous variant in exon 9 of the CALCRL gene: NM_005795.5: c.614_616del (p.Val205del), resulting in an in-frame deletion of amino acid valine 205 within the encoded protein, referred to as CLR (Fig. 1 C), hereafter referred to as V205del.
To date, this variant is not present in the Genome Aggregation Database, Exome Aggregation Consortium (ExAC), Exome Sequencing Project, or the 1000 Genomes browser. While the proband (III.4) and her unaffected daughter (IV.1) were heterozygous for the mutation, the affected fetus (IV.3) was found to be homozygous (Fig. 1 C). The V205del variant was confirmed by Sanger sequencing and further testing was performed in other surviving adult members of the pedigree. Importantly, the proband’s husband (III.3), who is also a first-cousin, is heterozygous for the V205del variant (Fig. 1 C), thus corroborating homozygous inheritance in IV.3. The proband’s mother, II.3, also experienced multiple pregnancy losses, and is heterozygous for the CALCRL V205del variant. Two of the proband’s sisters (III.6 and III.10) consented to Sanger-based genetic testing and did not carry the V205del variant. Another sister (III.12), who has also experienced significant early pregnancy losses and is married to a first cousin from the same familial pedigree (Fig. 1 A), declined genetic testing.
Individuals III.4, III.3, II.3, and IV.1 were also heterozygous, and fetus IV.3 was homozygous, for a variant in the ASAH1 gene: c.1045C>G (p.Arg349Gly), which is present in the ExAC South Asian and European (non-Finnish) populations. Biallelic loss-of-function mutations in ASAH1 are associated with Farber lipogranulomatosis (OMIM no. 228000), a lysosomal storage disorder that typically manifests during infancy, or in more severe cases, with perinatal lethality or antenatal HF (Kattner et al., 1997). However, we noted no clinical manifestation or symptoms indicative of this condition in IV.3 or in other nongenotyped family members within this four-generation pedigree. Altogether, the family pedigree, genetic inheritance pattern and clinical phenotype support the identification of mutations in the CALCRL gene as a novel causative pathway of human NIHF.
Genetic conservation of CALCRL and residue V205
The human CALCRL gene exhibits relatively few loss-of-function variants in the general population, as indicated by a high “probability of loss-of-function intolerance” (pLI) score (0.93) derived from the number of loss-of-function variants observed in the ExAC database compared with the expected number based on the size of the gene (16.9 Kb) (Lek et al., 2016). The V205del variant itself is absent from the Genome Aggregation Database, which contains genome-scale sequence data on nearly 140,000 individuals. In addition, this finding is highly consistent with the impaired subfertility of the heterozygous female carriers (patients II.3 and III.4), suggesting that their low fecundity would be expected to contribute fewer loss-of-function variants to the population at large.
Consistently, we identified 67 conserved orthologues of the hCALCRL gene and used multiple sequence alignment (Jalview and Clustal Omega; Sievers et al., 2011; McWilliam et al., 2013; Li et al., 2015) to establish the relative conservation of amino acid V205. We found that V205 is fully conserved across all species, except insects, being present in 62 of the 67 species represented. In four of the five species where the residue is not present (Lesser Hedgehog, Mouse Lemur, Sloth, and Wallaby), there is no equivalent residue, perhaps due to lack of sequencing coverage. A subset of the 62 species alignment is represented in Fig. S1, and reveals stringent conservation of the V205 residue, even within a core of amino acids that displays significant sequence degeneracy among fishes.
Molecular modeling of Human Calcitonin Receptor-Like Receptor (hCLR) and hCLR(V205del)
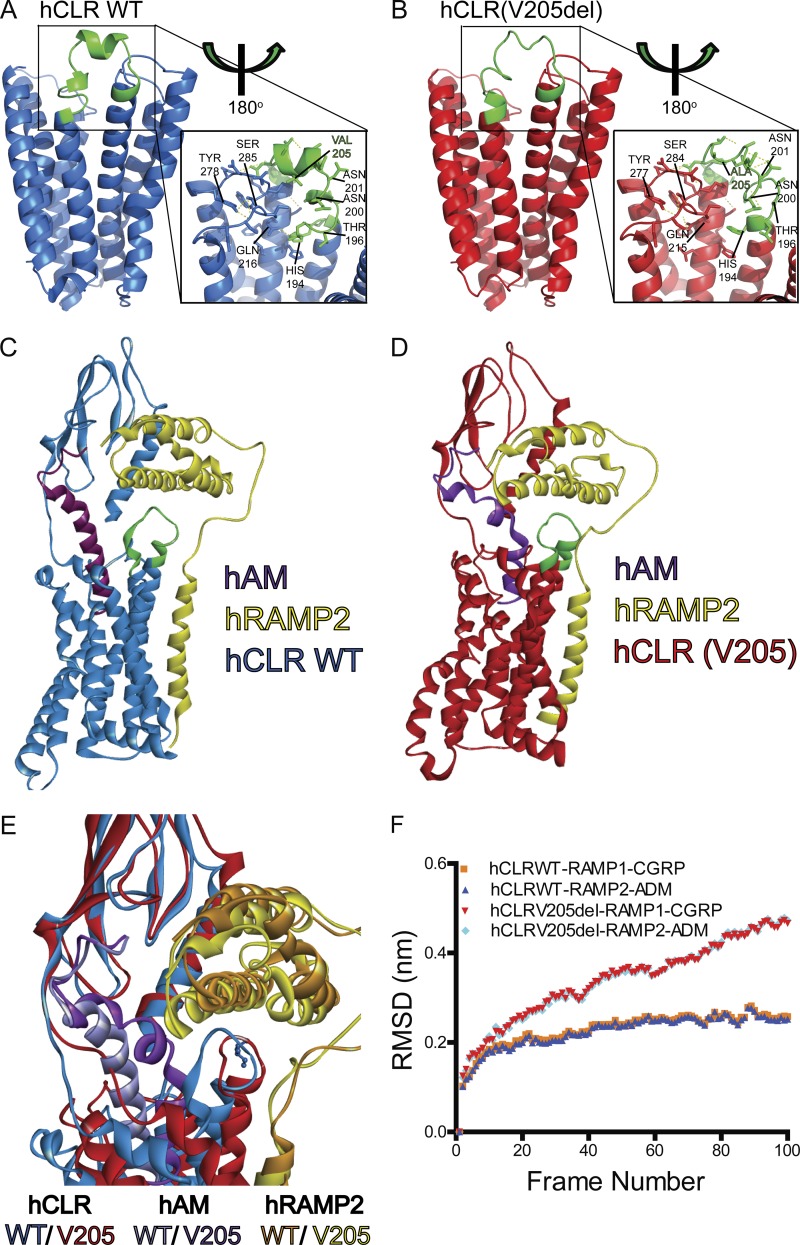
To visualize the consequences of V205 deletion on the hCLR structure, we generated homology models of the hCLR (Fig. 2 A, blue) and hCLR(V205del) (Fig. 2 B, red) structures using SWISS-MODEL (Arnold et al., 2006; Guex et al., 2009; Kiefer et al., 2009; Biasini et al., 2014), built on the structure of the corticotropin-releasing factor receptor 1 as a GPCR family B member template (Hollenstein et al., 2013). Notably, deletion of V205 is predicted to cause loss of α-helical character in extracellular loop 1 (ECL1) and gain of an extra turn in the extracellular-facing helix of transmembrane 2 (TM2, green). These changes are accompanied by predicted loss or gain of hydrogen bonding. For example, His194 loses the ability to interact with Gln215 and Thr196 undergoes a large shift in position. Asn200 and Asn201 gain and lose H-bonding contacts, respectively, and the shift of Ala206 to Ala205 contributes to the loss of α-helical character in ECL1. Finally, distal residues found in extracellular loop 2 (ECL2), Tyr277, and Ser285 lose the ability to interact. These changes in residues in ECL1 are predicted to interfere with ligand binding in the receptor’s allosteric site resulting in a loss of potency. Importantly, previous alanine mutagenesis studies have shown that many of these affected residues are important for receptor oligomerization with RAMPs and/or ligand binding (Barwell et al., 2011, 2012; Woolley and Conner, 2013; Watkins et al., 2016).
Figure 2.
Predicted structural changes and molecular dynamic modeling of wild-type and mutant hCLR and hCLR:RAMP2 complexes. (A and B) Predicted structural conformations of the hCLR and hCLR(V205del) by homology modeling using SWISS-MODEL. Cartoon views of the receptors predict structural changes centered on the site of mutation in extra cellular loop 1 (green). Inset shows predicted interactions involving side-chains and key residues are labeled to highlight changes in hydrogen bonding (H194/194, N200/200, N201/201, Q216/215, Y278/277, and S285/284), large residue positional changes (T196), and the site of deletion (V205 and A205). (C and D) Snap-shots of CLR–RAMP2–AM complexes after replica Monte Carlo simulations with either wild-type (C) or V205del (D) CLR. (E) Overlay of the RAMP/ECL1 interface of wild-type CLR–RAMP2–AM (blue) and CLR(V205del)–RAMP2–AM (red) complexes after REMC simulations. (F) Changes in root mean square deviation (RSMD) of wild-type and V205del CLR in complex with both RAMP1 and CGRP or RAMP2 and AM during REMC simulations.
Therefore, we next developed dynamic molecular modeling of wild-type hCLR (Fig. 2 C) or hCLR(V205del) (Fig. 2 D) in complex with hRAMP2 (yellow), which forms a functional receptor for hAM ligand (purple), using ROSETTA (Barth et al., 2007) and MODELLER (Šali and Blundell, 1993). The hCLR(V205del) complex reveals a shortened distance between hRAMP2 helix 2 and the hAM binding site, ultimately distorting the binding pocket and displacing the N’ terminus of hRAMP2. Furthermore, the repositioning of hRAMP2 helix 2 causes a knock-on effect on hCLR(V205del), distorting TM1, TM6, and TM7, which are essential for presenting the receptor ligand binding pocket. Consistent with these molecular modeling predictions, replica exchange Monte Carlo (REMC) molecular dynamic simulations (Fig. 2 E and Video 1) confirmed a distorted hCLR(V205)–hRAMP1 and –hRAMP2 interaction, with a significantly increased root mean square deviation for the hCGRP and hAM ligand docking (Fig. 2 F). Collectively, these structural modeling approaches predict that the mutant hCLR(V205del) protein is likely to display ineffective binding to both hRAMPs and ligands.
hCLR(V205del) abrogates cell signaling and RAMP-mediated, cell surface receptor expression
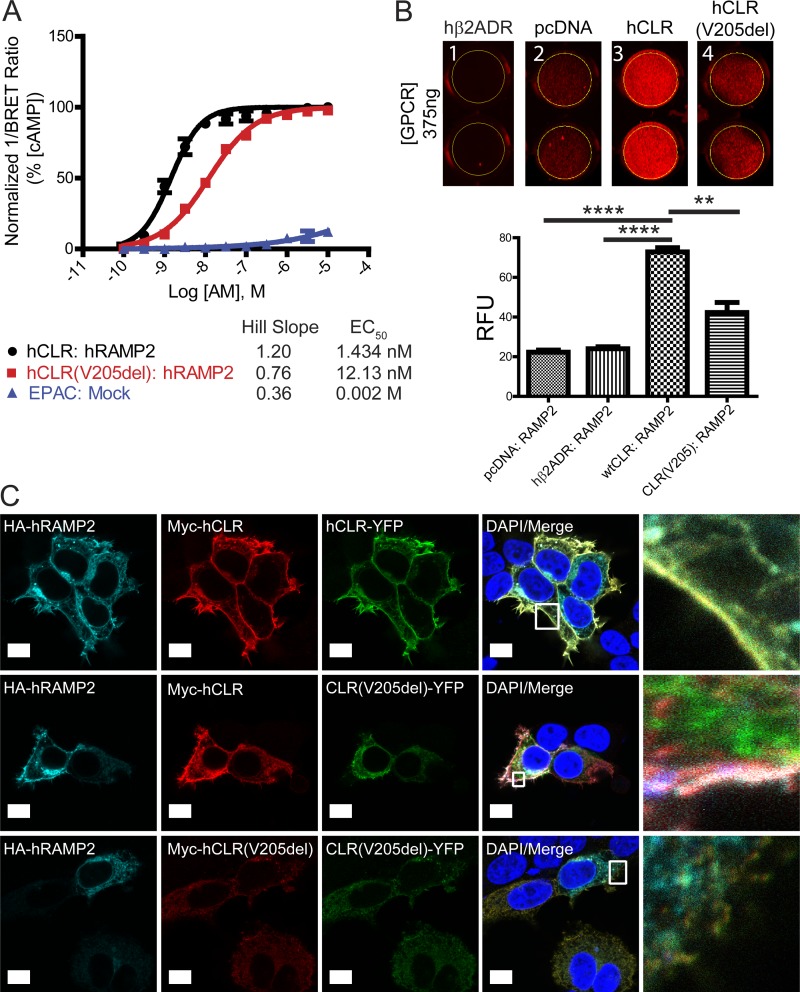
To directly test the signaling and receptor oligomerization competency of hCLR(V205del), we developed and used several orthogonal cell-based assays. First, using a bioluminescence resonance energy transfer (BRET)–based cAMP biosensor (Barak et al., 2008; Salahpour et al., 2012) we found that dose-dependent stimulation with the cognate peptide ligand, AM, robustly stimulated cAMP production in hCLR:hRAMP2-expressing HEK293T cells with maximal efficacy and nanomolar potency (Half maximal effective concentration, EC50 = 1.434 nM), as expected based on previous studies (Hay et al., 2003; Kuwasako et al., 2011; Fig. 3 A). However, the mutant hCLR(V205del) receptor exhibited a reduction in cAMP production represented by the statistically significant loss of potency (EC50 = 12.13 nM; Fig. 3 A). This loss of signaling by the hCLR(V205del) receptor implies that the V205 residue is critical for either ligand-mediated receptor activation, or for proper receptor cell surface localization, or both.
Figure 3.
hCLR (V205del) exhibits impaired signaling and plasma membrane localization. (A) Detection of cAMP production with the EPAC biosensor after 45 min of AM stimulation of hCLR:hRAMP2 or hCLR(V205del):hRAMP2. n = 5 independent experiments, in duplicate. (B) Infrared 24-well plate imaging of transiently transfected HEK293T cells with MarsCy1(FAP)-tagged hRAMP2. Cells transfected with 375 ng of pcDNA3.1, hβ2ADR, hCLR, and hCLR (V205del) were cotransfected with FAP-tagged RAMP2 and cells were stained with the MarsCy1 and membrane impermeant fluorogen SCi1. Compare cell surface expression to the pcDNA negative control. SCi1 stained cells were scanned at 700 nm (red, SCi1). n = 5 independent experiments, in duplicate. (C) Confocal microscopy of HEK293T cells expressing HA-hRAMP2 (cyan), Myc-hCLR (red), and hCLR-YFP (green). The top row shows the colocalization of all three wild-type proteins at the plasma membrane resulting in yellow-cyan color. The middle row shows the changes in protein distribution with coexpression of hCLR (V205)-YFP protein: wild-type HA-RAMP2 and Myc-hCLR colocalize to the plasma membrane (white merge), while the distribution of the hCLR (V205)-YFP protein (green) remains largely intracellular. The bottom row shows complete absence of all three proteins at the plasma membrane when both mutant forms of the hCLR(V205) receptor are expressed. Far right column shows the boxed regions at higher magnification. n = 2 independent experiments, with analysis of >50 cells per condition. Bar, 10 µm. **, P < 0.01; ****, P < 0.0001.
Next, to distinguish between these possibilities, we adapted an infrared fluorogen activating protein (IRFAP) assay (Fig. S2, A and B; Szent-Gyorgyi et al., 2008; Fisher et al., 2014; Snyder et al., 2015, 2017) to screen for cellular cotranslocation of hCLR–hRAMP heterodimer complexes to the cell surface, exploiting the fact that the forward trafficking of either hCLR or hRAMPs from the Golgi to the plasma membrane are mutually interdependent (McLatchie et al., 1998; Hay et al., 2006). As expected, the interaction of hCLR:FAP-hRAMP2 was robustly detected at 375 ng (Fig. 3 B, column 3). Conversely, the hCLR(V205del) variant exhibited diminished levels of chaperone ability for FAP-hRAMP2, resulting in a statistically significant decrease of FAP signal detected (Fig. 3 B, column 4). The intensity of pcDNA3.1 (column 2) or the non-RAMP interacting GPCR, β2-adrenergic receptor (column 1), are shown as background controls.
Confocal microscopy of HEK293T cells expressing fluorescently or epitope-tagged versions of the proteins corroborated these findings. In brief, cells were transfected to express 3xHA-hRAMP2 protein (cyan) in the presence of Myc-hCLR or Myc-hCLR(V205del) (red) and hCLR-YFP or hCLR(V205del)-YFP (green) to evaluate the subcellular localization of individual oligomer complexes (Fig. 3 C). In the 3xHA-RAMP2:Myc-hCLR:hCLR-YFP cotransfection, cells displayed robust colocalization of all three wild-type proteins at the plasma membrane, as indicated by the yellow-merged staining (top row, right column). However, the hCLR(V205del)-YFP variant shows reduced levels at the plasma membrane and remains largely intracellular (middle row, green). Importantly, under this condition, which mimics the phenotypic condition of individuals who are heterozygous for the hCLR(V205del) variant, the expression of the mutant hCLR(V205del) receptor does not invoke dominant negative effects on the forward trafficking of the wild-type hRAMP2 and hCLR to the plasma membrane (middle row, right column, white merge). Finally, none of the proteins traffic to the plasma membrane when both Myc-hCLR(V205del) and hCLR(V205del)-YFP are coexpressed with 3xHA-hRAMP2 (bottom row).
hCLR(V205del) fails to form oligomers with hRAMPs
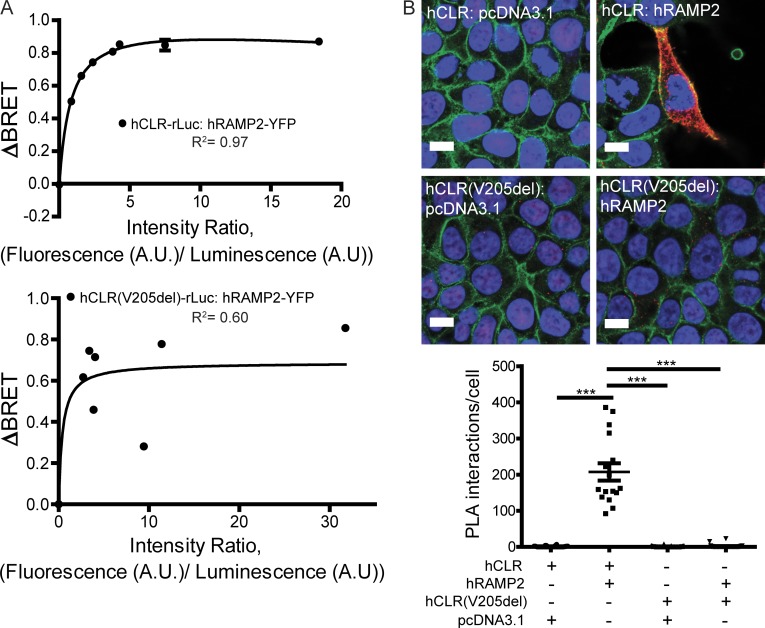
Since the effective signaling and plasma membrane localization of hCLR is fully dependent on RAMP protein interaction, we reasoned that the impaired functions of the hCLR(V205del) variant in the above assays might be caused principally by its inability to interact with RAMPs. To test this hypothesis, we used two different assays for protein–protein interaction. First, using BRET assays, we consistently observed robust and characteristic rectangular hyperbola saturation isotherms with wild-type hCLR-rLuc and hRAMP2-YFP, with a Bmax of 1.03, and a disassociation constant of Kd= 0.83 (Fig. 4 A, top). On the other hand, the hCLR(V205del) mutant resulted in a reduction of BRET signal indicated by a Bmax of 0.68, but interacted with a similar disassociation constant of Kd = 0.42 (Fig. 4 A, bottom). The biggest difference between the two saturation experiments was in the R2 or goodness of fit, where hCLR exhibited a 0.97 R2 and hCLR(V205) exhibited a 0.60 R2. Second, we used in situ proximity ligation assay (PLA; Chauhan et al., 2015), which detects protein–protein interactions at single molecule resolution within living cells. HEK293T cells transiently cotransfected with hCLR and hRAMP2 exhibited markedly robust PLA signal (207.5 ± 24.0 signals/cell), whereas cells coexpressing the hCLR(V205del) variant showed a complete abatement of PLA signal (1.4 ± 0.8 signals/cell; Fig. 4 B). Collectively, these data demonstrate that V205 in hCLR is a critical residue for the formation of the hCLR:hRAMP2 heterodimer, which is furthermore required for its normal subcellular localization and signaling.
Figure 4.
hCLR(V205del) does not interact with RAMPs. (A) Representative saturation isotherms from BRET assays of transiently transfected HEK293T cells with hCLR (top graph) or hCLR(V205) (bottom graph; fused to renilla luciferase) and increasing amounts of hRAMP2 (fused to eYFP). Experiments were performed at least three times and a representative experiment is shown. (B) PLA in transiently transfected HEK293T cells. Red indicates positive PLA signal, blue is nuclear DAPI stain, green is plasma membrane β-catenin. Negative controls included no transfection of hRAMP2. Graph depicts quantitation of signal, with asterisks denoting significant differences compared with control; ***, P < 0.001. n = 2 independent experiments, in duplicate, with 20 cells quantitated per condition. Bar, 10 µm.
Lymphatic Calcrl is essential for murine survival and lymphatic vascular development
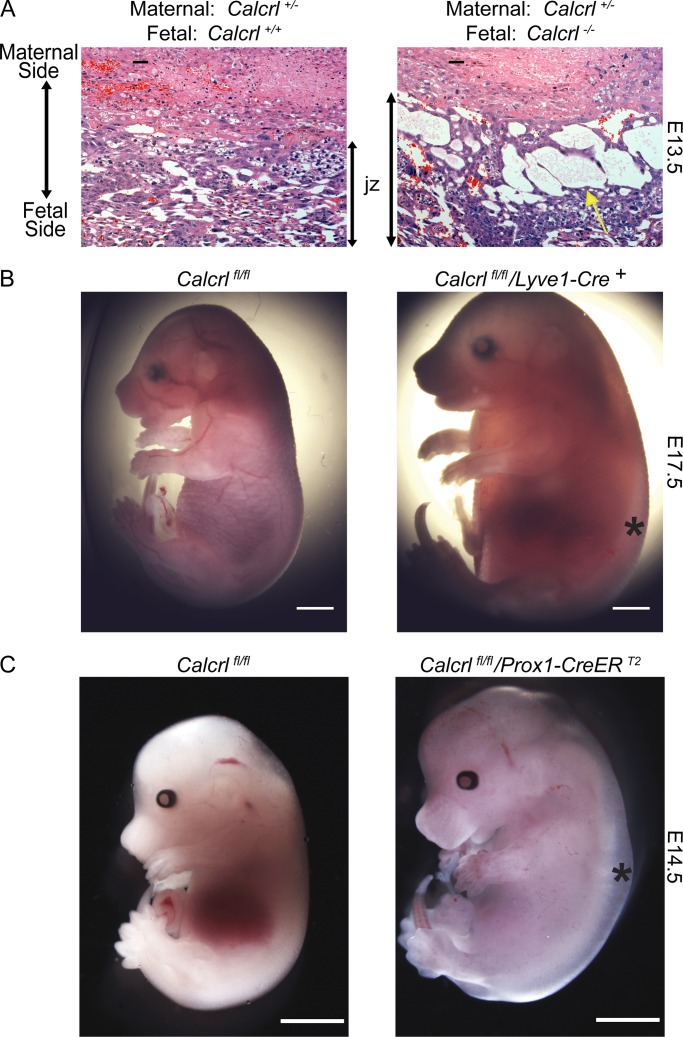
CLR is a broadly expressed receptor with abundant expression in cardiovascular tissues and the peripheral nervous system. Similar to the family case report, global deletion of the Calcrl in mice results in mid-gestation embryonic lethality with profound interstitial edema due to arrested lymphangiogenesis, an HF phenotype that is fully recapitulated in both Adm−/− and Ramp2−/− mice (Caron and Smithies, 2001; Dackor et al., 2006; Fritz-Six et al., 2008; Kechele et al., 2016). In addition, we found that placentas from Calcrl−/− fetuses also exhibited marked interstitial edema within the labyrinth and enlarged junctional zone (Fig. 5 A), reflecting the pathology observed in the human condition.
Figure 5.
Placental edema, HF, and fetal demise in mice lacking Calcrl. (A) Placenta from E13.5 Calcrl+/+ and edematous Calcrl−/− littermates, revealing significant interstitial edema within an expanded junctional zone (jz). Yellow arrows highlight fluid filled spaces in the jz. (B) Representative images of E17.5 Calcrlfl/fl and Calcrlfl/fl/Lyve1-Cre littermates. Asterisks indicate edema. n = 5–7 animals in each group. (C) Representative images of E14.5 Calcrlfl/fl and Calcrlfl/fl; Prox1-CreERT2 littermates from tamoxifen treated dams. Asterisks indicate edema. n = 8–10 animals in each group. Bars, 2 mm.
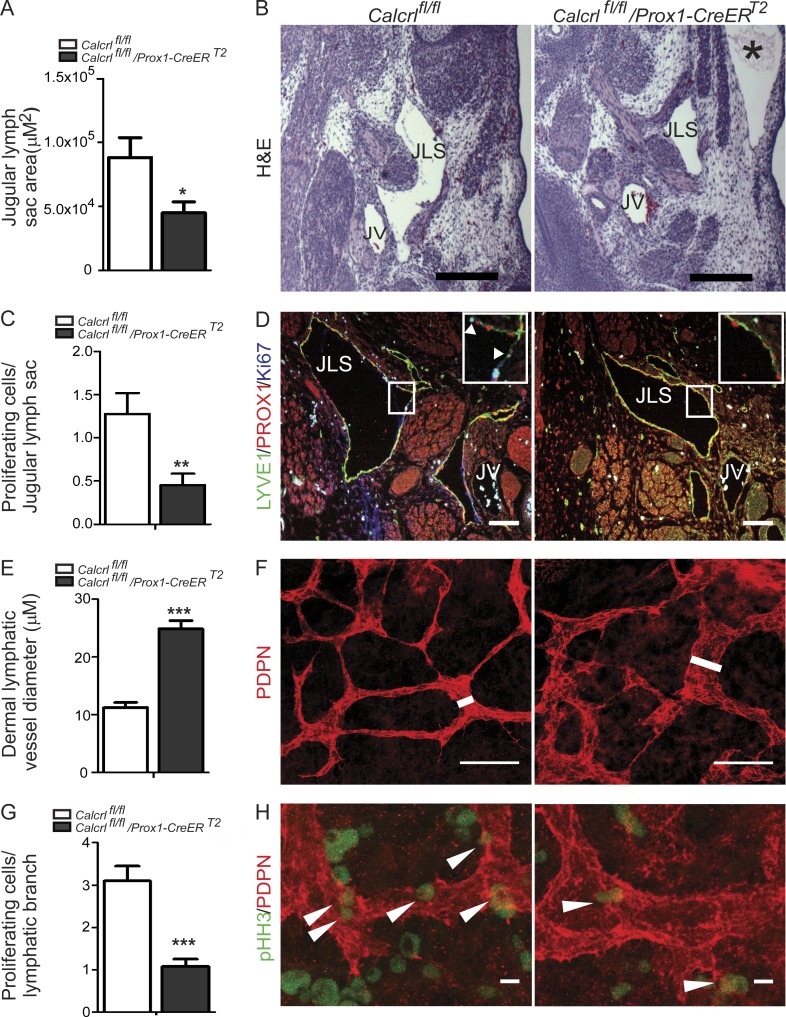
Therefore, to gain insights into the functional consequences of lymphatic endothelial Calcrl loss during development, we generated lymphatic-specific Calcrl null animals using two independent Cre-driver lines: constitutively expressed, Lyve1-Cre (Pham et al., 2010), and the inducible Prox1-CreERT2 (Srinivasan et al., 2007). All Calcrlfl/fl;Lyve1-Cre animals died in late gestation (embryonic day 16.5 [E16.5]–17.5) with profound interstitial edema (Fig. 5 B). Similarly, tamoxifen injection of pregnant Calcrlfl/fl dams mated with Calcrlfl/fl;Prox1-CreERT2 males resulted in excision of the Calcrl gene and mid-gestation lethality with HF between E14.5 and 16.5 of Calcrlfl/fl;Prox1-CreERT2 embryos (Fig. 5 C; Fig. S3, A–D). Morphometric analysis of the developing jugular lymph vessels of Calcrlfl/fl;Prox1-CreERT2 embryos revealed subcutaneous edema and significantly smaller lymphatic vessels (Fig. 6, A and B) that were developmentally arrested, as indicated by a >50% reduction in Ki67-positive lymphatic endothelial cells within the jugular lymph sacs (Fig. 6, C and D) compared with littermate Calcrlfl/fl embryos. Analysis of dermal lymphatic vessels within the embryonic skin of Calcrlfl/fl;Prox1-CreERT2 embryos also revealed the presence of developmentally arrested lymphatic capillaries that appeared significantly dilated and malformed within the environment of subcutaneous edema compared with the dermal lymphatic capillaries of normal Calcrlfl/fl embryos (Fig. 6, E–H).
Figure 6.
Lymphatic deletion of Calcrl leads to developmentally arrested lymphatics, hypoplastic jugular lymph sacs, and dilated dermal lymphatic vessels. (A) Measurement of jugular lymph sac area in E14.5 Calcrlfl/fl and Calcrlfl/fl/Prox1-CreERT2 embryos. Quantitative data are represented as mean and SEM for area. n = 6–7 animals per group. (B) Representative images of H&E-stained sections of E14.5 Calcrlfl/fl and Calcrlfl/fl/Prox1-CreERT2 embryos showing jugular lymph sac (JLS), jugular vein (JV), and severe edema (*). Bars, 100 µm. (C) Number of Ki67-positive cells per jugular lymph sac in Calcrlfl/fl and Calcrlfl/fl/Prox1-CreERT2 embryos. Quantitative data are represented as mean and SEM for number of Ki67-positive cells. n = 6 animals per group. (D) Representative images of LYVE1 (green), PROX1 (red), and Ki67 (blue) stained sections of E14.5 Calcrlfl/fl and Calcrlfl/fl/Prox1-CreERT2 embryos showing proliferating cells in jugular lymph sac (JLS). Boxed regions are shown as magnified insets. Arrowheads highlight Ki67-positive lymphatic endothelial cells. Bars, 100 µm. (E) Dermal lymphatic vessel diameter of Calcrlfl/fl and Calcrlfl/fl/Prox1-CreERT2 embryos. n = 3–5 animals per group. (F) Podoplanin (PDPN, red) stained dermal lymphatic vessels from E13.5 Calcrlfl/fl and Calcrlfl/fl/Prox1-CreERT2 embryos. White line highlights width of the lymphatic vessel. Bars, 25 µm. (G) Quantification of phosphohistone H3 (pHH3)–positive cells in E13.5 Calcrlfl/fl and Calcrlfl/fl/Prox1-CreERT2 dermal lymphatic vessels. Data are represented as mean and SEM for number of pHH3-positive cells per branch. n = 3–5 animals per group. (H) Representative images of PDPN (red) and pHH3 (green) stained dermal lymphatics of E13.5 Calcrlfl/fl and Calcrlfl/fl/Prox1-CreERT2 embryos. Arrowheads indicate pHH3-positive cells. Bars, 1 µm. Quantitative data are represented as mean and SEM. In all panels, significance was determined by Student t test (tail = 2, type = 2) with *, P < 0.05; **, P < 0.005; ***, P < 0.0001.
Since abnormal lymphovenous valve formation during development has recently been linked to GLD and NIHF in both mice and humans (Martin-Almedina et al., 2016), we carefully examined the lymphovenous valves of Calcrlfl/fl;Prox1-CreERT2 embryos and observed normal structural morphology, appropriate compartmentalization of blood cells from lymph and high levels of Prox1 and Lyve1 staining (Fig. S4). Moreover, because Prox1 is expressed in myocardial cells and global loss of Calcrl results in reduced myocardial proliferation in mice (Dackor et al., 2006), we carefully evaluated the structure and development of the hearts of Calcrlfl/fl;Prox1-CreERT2 embryos and found them comparable in size and structure to wild-type littermates (Fig. S5).
Collectively, these results establish that loss of functional Calcrl within the lymphatic vasculature causes a mid-gestational proliferative arrest in lymphangiogenesis resulting in profound HF with associated placental edema, which are phenotypes entirely consistent with the clinical presentation of NIHF in humans with the homozygous hCLR(V205del) variant.
Loss of RAMP2-mediated CLR signaling in mice recapitulates the hCLR(V205del) haplotype
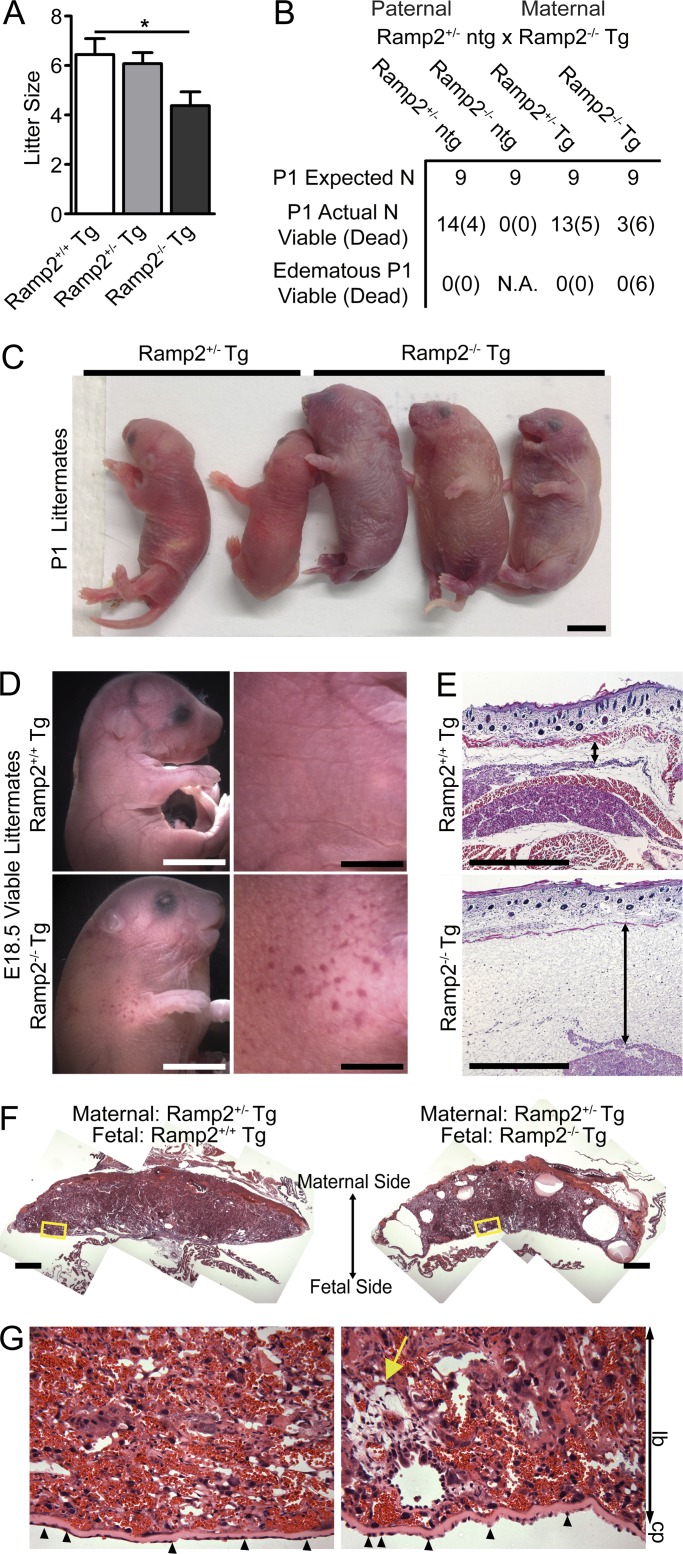
CLR serves as the receptor for both AM and CGRP ligands, depending on which RAMP the receptor is heterodimerized with. Thus, patients with the hCLR(V205del) variant are likely to be functionally deficient for both AM/RAMP2/3-mediated and CGRP/RAMP1-mediated signaling. We have previously demonstrated that Ramp1 and Ramp3 null mice survive to adulthood with little to no overt phenotypes, consistent with a lack of overt phenotypes in mice deficit for CGRP (Dackor et al., 2007). Therefore, to more carefully parse the relative contributions of RAMP2-mediated signaling, we characterized the phenotypes of a unique Ramp2 transgenic line in which endothelial restoration of Ramp2 using a vascular endothelial-specific promoter to drive a Ramp2 transgene (Tg) is sufficient to partially rescue the edematous phenotype and embryonic lethality of Ramp2−/− mice (Kechele et al., 2016). Similar to the previously reported Ramp2+/− subfertility phenotype (Kadmiel et al., 2011), Ramp2−/−-Tg dams undergo implantation and maintain pregnancy to term, but they have significantly smaller litter sizes than Ramp2+/+-Tg or Ramp2+/−-Tg females (Fig. 7 A), further supporting the observation that haploinsufficiency for CLR-RAMP2 signaling in females, both in mice and in humans, is associated with subfertility (Kadmiel et al., 2011; Li et al., 2013). While Ramp2−/−-Tg pups were born at nearly Mendelian ratios (Fig. 7 B), ∼60–70% died perinatally with marked fluid accumulation, small focal hemorrhages in the skin, and HF (Fig. 7, C and D), likely due to the low level of VE-cadherin–mediated Ramp2 reconstitution in lymphatic vessels. Histology confirmed substantial amounts of interstitial edema within a thickened subcutaneous layer of the skin in Ramp2−/−-Tg embryos compared with littermate Ramp2+/+-Tg controls (Fig. 7 E). Furthermore, the placentas from edematous Ramp2−/−-Tg embryos showed marked fluid-filled cavities within the labyrinth and junctional zones of the fetal placenta (Fig. 7, F and G). Collectively, the morphological phenotypes observed in the majority of Ramp2−/−-Tg embryos, including HF, thickened skin, placental edema, and embryonic demise are fully consistent with the clinical characteristics of NIHF in humans and further establishes the CLR–RAMP2–AM signaling axis as the molecular basis for NIHF and female subfertility observed in humans with a hCLR(V205del) variant.
Figure 7.
Genetic loss of Ramp2 leads to subfertility, perinatal HF, and lethality. (A) Litter sizes from Ramp2+/− ntg males crossed with Ramp2+/+Tg, Ramp2+/−Tg, and Ramp2−/−Tg females. n = 8–12 litters per genotype. Significance was determined by one-way ANOVA with Tukey’s Multiple Comparison test. (B) Breeding results, both expected and observed Mendelian ratios from Ramp2+/− ntg male crossed to Ramp2−/−Tg female. n = 45 from 8 litters. (C and D) Representative images of P1 (C) and E18.5 Ramp2−/−Tg (D) mice compared with littermate controls. Bars, 5 mm. (E and F) Representative histological images of skin (E) and placenta (F) from E18.5 Ramp2+/+Tg and edematous Ramp2−/−Tg littermates. Double-sided arrow indicates subcutaneous edema. Bars, 500 µm. (G) Higher magnification of boxed regions from panel F. Yellow arrow points to fluid filled edematous regions. Small arrowheads indicate abnormally shaped trophoblast cells lining the chorionic plate. lb, labyrinth; cp, chorionic plate. Bars, 100 µm.
Discussion
In summary, an in-frame deletion of a highly conserved valine reside in extracellular loop 1 of CLR, has been identified in six members of a consanguineous family, resulting in autosomal recessive GLD, with NIHF in homozygous carriers and female subfertility in heterozygous carriers. The mutation imparts structural changes within the receptor that diminishes its interaction with the requisite accessory chaperone protein RAMP, resulting in reduction of receptor at the cell surface and significant impairment of ligand-mediated signaling. In animal models, we show that the expression of the CLR–RAMP2–AM signaling axis in lymphatic endothelial cells is both necessary and sufficient for mammalian survival and normal female reproductive function, providing a cellular and molecular foundation for the clinical manifestations observed in humans with the hCLR(V205del) variant. The impact and relevance of these findings can be extrapolated to at least three different areas.
First, these findings provide, to our knowledge, the third case of genetically inherited NIHF associated with GLD, thereby increasing the repertoire of candidate factors for genetic testing and screening beyond the very recently identified disease-associated PIEZO1 and EPHB4 genes. Moreover, based on deductions from animal model studies, as well as the extensive, four-generation pedigree, we also conclude that haploinsufficiency for the CLR–RAMP2–AM axis is associated with clinical subfertility in female carriers. Collectively, this work provides additional diagnostic criteria for causes of NIHF and female subfertility, as well as three additional genes for candidate genetic testing: CALCRL, ADM, and RAMP2.
Second, the remarkable conservation of phenotype between the human patients and the genetic animal models provides compelling evidence for the evolutionarily conserved functions of the AM–CLR–RAMP2 signaling axis in embryonic vascular development and female fertility. In contrast, the hCLR(V205del) variant appeared to have little consequence on CGRP-mediated physiology, which is further supported by the lack of overt phenotypic effects in either CGRPΑ−/−, CGRPΒ−/−, Ramp1−/−, or Ramp3−/− mice (Dackor et al., 2007). This may help explain why antibody-mediated CGRP antagonist therapies have recently shown tremendous promise, with little side effects, in several large-scale phase III trials for the treatment of migraine (Goadsby et al., 2017; Silberstein et al., 2017).
Finally, using molecular dynamic modeling and biochemical approaches we have demonstrated the requisite functions imparted by a GPCR–RAMP interaction for the normal trafficking, signaling, and physiology of a family B GPCR. These findings are particularly timely, since the molecular crystallization of three family B GPCRs, CRF, GLP-1, and glucagon receptors, have recently been elucidated (Hollenstein et al., 2013; Song et al., 2017; Zhang et al., 2017). Notably, Zhang et al. demonstrated that the ECL1 region of the glucagon receptor, which also interacts with RAMPs (Weston et al., 2015), is critical for forming the peptide binding interface. Together, these findings demonstrate that ECL1 is a functionally critical structure of family B GPCRs, and imparts physiological relevance to this motif in humans. Future cocrystallization of active-state family B GPCRs in oligomeric complex with RAMPs will likely reveal novel insights into the structure–function relationships imparted by these protein–protein interactions. Indeed, while the current work was focused on the canonical RAMP-associated receptor, CLR, it is worth emphasizing that over a dozen GPCRs have been shown to interact and function through RAMPs, including calcitonin (Armour et al., 1999), parathyroid hormone, and vasointestinal peptide receptors (Christopoulos et al., 2003) as well as other nonfamily B GPCRs such as GPR30 (Lenhart et al., 2013), glucagon (Weston et al., 2015), and calcium sensing receptors (Bouschet et al., 2005). Thus, the physiological implications of aberrant GPCR-RAMP oligomerization may be vastly underappreciated. Furthermore, these studies open potential avenues for the development of therapeutic pharmacological chaperones to enhance GPCR–RAMP interaction as a means to promote receptor trafficking, as has successfully been accomplished for other disease-causing plasma membrane protein variants such as V2 vasopressin receptor (Morello et al., 2000), CFTR (Drumm et al., 1991), and aquaporin-2 (Tamarappoo and Verkman, 1998).
Materials and methods
Targeted capture and parallel sequencing
Genomic DNA was extracted from the fetal cord blood of family member IV.3 and from blood samples of family members II.3, III.3, III.4, III.6, III.10, and IV.1. Whole-exome sequencing was performed on exon targets isolated from patient IV.3 by capture using nextSeq Exome sequencing Platform (Illumina) at CentoGene and an end-to-end bioinformatics pipeline was applied. This included base calling, primary filtering of low quality reads, and probable artifacts and annotation of variants. For medical evaluation purposes, all disease causing variants reported in HGMD, in ClinVar (class 1 and 0), and all variants with minor allele frequencies of >1% in the ExAC database were considered. All variants that could possibly impair the protein sequence, i.e., disruption of conserved splice sites, missense, nonsense, read-throughs, or small insertion/deletion were prioritized. The percent coverage at 10× for patient IV.3 was 95.64%, while the coverage for the mother III.4 and father III.3 was 96.59% and 97.10%, respectively. The average fold coverage for patient IV.3 was 117.57%, while for the mother III.4 and father III.3 was 133.36% and 140.96%, respectively.
Molecular modeling of hCLR, hCLR(V205del), and RAMP interaction
Two hCLR and hCLR(V205del) GPCR homology models were constructed. The hCLR and hCLR(V205del) sequences (UniProtKB: Q16602 and Q16602 with valine 205 deleted) were used as the targets. The GPCRs were aligned against human corticotropin-releasing factor receptor 1 (PDB no. 4K5Y; Hollenstein et al., 2013) using SWISS-MODEL automated mode (Biasini et al., 2014). These models resulted in a Model-Template Range of 138–389 amino acids. The top homology models were chosen using the QMEAN score (Benkert et al., 2009a,b, 2011). In brief, QMEAN is a composite scoring function for the estimation of the global and local model quality. It consists of four structural descriptors: the local geometry is analyzed by a torsion angle potential over three consecutive amino acids. Two pairwise distance-dependent potentials are used to assess all-atom and C-β interactions. A solvation potential describes the burial status of the residues. Furthermore, the Corticotropin-releasing factor receptor 1 was use as the target GPCR for generation of the homology model because it is a member of the secretin receptor family and the same subfamily, B1, as hCLR. Once the models were generated they were exported for use in PyMOL. In PyMOL, both GPCR structures were probed for changes in polar contacts involving side chains centered on the site of the valine 205 deletion (magenta). The labeled residues indicate the changes in predicted hydrogen bonding, large positional changes, cysteine bonds, or the site of the amino acid deletion.
Homology models of hCLR were generated using the recently published cryo-electron microscopy structure of 5VAI and the x-ray crystallographic of 3N7P as described previously (Woolley et al., 2017). The position of the N-terminal region of hRAMP1 was aided by superimposing the crystal structure of 4RWG to the homology model of hCLR. After which the hCLR component (leaving the RAMP region intact) was removed and the complex minimized using MODELLER. The transmembrane component of hRAMP1 was generated using an in house script and docked into the hCLR + N’ terminal hRAMP model using GRAMM in low resolution helix docking mode. Each of the 10,000 docked positions of hRAMP1 TM against the hCLR-RAMP1 N’ terminal model was refined using ROSETTA. The top 1% of the best scoring structures were clustered using a cutoff of 2A. Three clusters emerged and the middle structure of each cluster was used to generate three complete homology models of the hCLR-hRAMP1 complex. REMC simulations were used to refine each model (16 temperatures spanning 300–500 K following a Boltzmann distribution), after which the final structures were scored using ROSETTA and the best structure was used for the subsequent study. The deletion of V205 was performed using MODELLER. Parallel REMC simulations of the wild-type hCLR–hRAMP1 complex as well as the deletion were performed and compared. To access the research data supporting the molecular dynamic simulations in this publication, see Poyner and Simms, 2018.
Generation of hCLR and hRAMP2 expression constructs
The hCLR mammalian expression plasmid Myc-hCLR-phRLucN3 (hCLR-rLuc) were donated by the laboratory of M. Bouvier (Université de Montréal, Montreal, Canada) and used as the template for site-directed mutagenesis using the QuikChange II XL Site-Directed Mutagenesis kit (Agilent Genomics) to generate the hCLR(V205del) mutant. Mutagenesis primers were designed using QuikChange Primer Design Tool (Agilent Genomics). Myc-hCLR and Myc-hCLR(V205del) were further subcloned into pcDNA3.1(+) to remove the rLuc moiety for additional experiments. The hRAMP2 mammalian expression plasmid hRAMP2/pcDNA3.1 was purchased from the cDNA Resource Center (Bloomsburg University, Bloomsburg, PA). The hRAMP2 gene was subcloned into the pEYFP-N1 vector to generate hRAMP2-eYFP. All constructs were verified by DNA sequencing.
BRET
HEK293T cells were maintained in standard conditions. Transfections were performed using Lipofectamine 2000 Reagent (Thermo Fisher) following the manufacturer’s specifications. In brief, 96-well white, clear bottom plates (Corning) were coated with poly-d-lysine. 50,000 cells were seeded into each well and grown overnight in 37°C, 5% CO2. The next day, cells were transfected with a serial dilution of hRAMP2-eYFP DNA (15–0 µg) with a constant concentration of hCLR-rLuc or hCLR(V205del) to generate the saturation isotherm. On day 3, the media was aspirated and replaced with 90 µl 1× PBS and 10 µl of 10× coelenterazine, incubated for 10 min, and the plates were read on a Mithras LB 940 Multimode Microplate Reader (Berthold Technologies).
cAMP EPAC biosensor assay
HEK293T cells were transfected using the calcium phosphate transfection protocol; see reference above. In brief, 10-cm dishes were seeded with four million cells and grown overnight in standard conditions. The next day, each dish was transfected with 4 µg EPAC biosensor, 2.5 µg of hRAMP2, and 2.5 µg of either hCLR or hCLR(V205) or empty vector. After 18 h, cells are harvest and seeded at 100,000 cells per well into a white 96-well poly-d-lysine coated plate in phenol-free reduced serum DMEM (supplemented with 2% FBS, 1% Hepes, and 750 µl of Gentamicin; Sigma). On the day of the experiment, fresh AM stock was made at 1 mM, and a half-log dilution series was generated with a final concentration in the assay of 10 µM to 31.6 pM. Media was aspirated from each well and replace with 80 µl of PBS and 10 µl of 10× coelenterazine and incubated 10 min. Next, 10 µl of AM was added to each well and the plate was incubated for 45 min in the dark at room temperature. All assays were read on a Mithras LB 940 Multimode Microplate Reader.
Fluorogen activating protein (FAP) assay
Fluorogen was obtained from the laboratory of A. Waggoner (Carnegie Mellon University Technology Center of Networks and Pathways, Pittsburgh, PA). Gateway cloning technology (Thermo Fisher) was use to generate the FAP-RAMP2 fusion protein. To investigate the hRAMP2 chaperone activity all experiments were performed using poly-d-lysine–coated, 24-well clear plates. 175,000 HEK293T cells were seeded into each well and grown overnight. The following day, each well was transfected by calcium phosphate transfection with 250 ng of FAP-hRAMP2 and 375 ng of hCLR, hCLR(V205del), β2ADR, or pcDNA3.1. Cells were grown for 18 h in transfection media and assayed for FAP expression. Media was aspirated and replaced with 250 µl of room-temperature PBS with a 1:6,000 dilution of fluorogen SCi1 and incubated for 10 min. Next, 250 µl of PBS without fluorogen and each plate was read on an Odyssey CLx imaging system (Li-COR Biosciences).
PLA
HEK293T cells were seeded into poly-d-lysine coated MatTek plates and transfected with hCLR or hCLR(V205) or hβ2 adrenergic receptor and hRAMP2 or pcDNA3.1 using calcium phosphate. At 24 h after transfection, cells were washed with room temperature PBS and fixed onto plates with fresh 4% paraformaldehyde in PBS. Cells were then probed by PLA using the Duolink In Situ Red kit Goat/Rabbit (Sigma) in a humidity chamber according to manufacturer’s specifications. In brief, cells were blocked with 4% BSA in PBS, incubated with appropriate primary antibodies and then incubated with PLA probes, anti-Goat and anti-Rabbit secondary antibodies conjugated to unique oligonucleotides. Each sample was treated with a ligation solution which allows the oligo pairs in close proximity for form a close loop. Next, rolling-circle amplification was initiated by the addition of an amplification solution that contains polymerase and fluorescently labeled oligonucleotides, which produces discrete fluorescent dots. After the PLA protocol, cells were counterstained with a mouse anti–β-catenin primary antibody (610153; BD Biosciences) and donkey anti-mouse Alexa Fluor 488 secondary antibody to visualize the plasma membrane and Hoechst to mark the nucleus. Cells were analyzed by confocal laser scanning microscopy. A minimum of 25 cells were imaged for analysis, and each red dot was scored as a single interaction using BlobFinder software (Allalou and Wählby, 2009).
Animal studies
All animal procedures were approved by the University of North Carolina Chapel Hill Institutional Animal Care and Use Committee.
Calcrlfl/fl/Prox1-CreERT2 mice were generated by crossing Calcrlfl/fl mice that were on a N7-10 on C57BL/6 background to Prox1-CreERT2 NMRI line (Srinivasan et al., 2007) a gift from G. Oliver (Northwestern University, Chicago, IL). Cre-mediated recombination was induced by administering tamoxifen (Tam; T5648; Sigma) dissolved in corn oil and ethanol to pregnant female mice aged 4–6 mo for three consecutive days from E8.5 to10.5 at a dose of 5 mg/40 g intraperitoneally and their embryos harvested at various stages during gestation. Calcrlfl/fl/Lyve1-Cre mice were generated by crossing the aforementioned Calcrlfl/fl line to Lyve-1 Cre+ line obtained from Jackson Lab (JAX stock 012601; Pham et al., 2010). Ramp2+/− were crossed to Tg(Cdh5-Ramp2)1Car hemizygous mice and maintained on an isogenic C57BL/6 background as previously reported (Dackor et al., 2007; Kechele et al., 2016). Ramp2+/− males were crossed to Ramp2+/−; Tg(Cdh5-Ramp2) dames, and viable progeny were harvested during late gestation. Littermate controls were used for all animal studies. Primers used for genotyping were Calcrl wild-type allele, 5′-GCGGAGCATATTCAATCACAAG-3′ and 5′-GAAATGTGCTGTATGTTCAAGC-3′; Calcrl floxed allele, 5′GCGGAGCATATTCAATCACAAG-3′ and 5′-GACGAGTTCTTCTGAGGGGA-3′; Prox1-CreERT2 allele, 5′-CGAGCTCTTTCTCTCTACAGTTCAACA-3′ and 5′-GGCCAGTAACGTTAGGGAGAGG-3′; Lyve-1 Cre+ allele, 5′-TGCCACCTGAAGTCTCTCCT-3′ and 5′-TGAGCCACAGAAGGGTTAGG-3′ and 5′-GAGGATGGGGACTGAAACTG-3′; excised Calcrl allele, 5′GCGGAGCATATTCAATCACAAG-3′ and 5′-GAATAAGTTGAGCTGGGCAG-3′; Ramp2 wild-type allele, 5′-FAAGTCAGGCAGTCAGGGTTG-3′ and 5′-TCTGTCTGGATGCTGCCTTGC-3′; Ramp2 null allele, 5′-GACGAGTTCTTCTGAGGGGA-3′ and 5′-TCTGTCTGGATGCTGCCTTGC-3′; and the Tg(Cdh5-Ramp2) allele, 5′-GACTACAAAGACGATGACGACAAGC-3′ and 5′-GTCCATGCAACTCTTGTACTCATACC-3′.
Tissue preparation, histology, and immunofluorescence
For cellular staining, 500,000 HEK293T cells were seeded into 35-mm MatTek glass-bottom dishes coated with poly-d-lysine and grown overnight. Cells were transfected with 150 ng of hRAMP2 and 150 ng of either hCLR or hCLR(V205del) or pcDNA3.1 using calcium phosphate. The following day, cells were washed with room temperature PBS and fixed onto plates with fresh 4% paraformaldehyde in PBS.
Viable mouse embryos were collected and fixed in 4% paraformaldehyde in PBS for 24 h at 4°C. Whole mount back skin was collected and stained as described in Li and Mukouyama (2011). In brief, limb skins were peeled after dehydrating in 100% MeOH followed by rehydration and antibody staining. Embryos were paraffin embedded, transversely sectioned, and stained with either H&E or Masson’s Trichrome according to standard protocols. Tissues and cells were washed with PBS and permeabilized with 0.1% Triton X-100 in PBS, washed, and blocked for 1 h at room temperature with 5% normal donkey serum or 4% BSA in PBS. Samples were probed with primary antibodies directed toward rabbit anti-LYVE1 (1:250; Fitzgerald), rat anti-Ki67 (1:150; VP-RM04; Vector Labs), rabbit anti-pHH3 (1:200; 6-570; Cell Signaling) human anti-Prox1 (1:100; AF2727; R&D Systems), Syrian hamster-anti-podoplanin (1:100; DSHB8.1.1), rat anti-PECAM (1:100; 550274; BD PharMingen), rabbit anti-myc (1:500; ab9106; Abcam), and goat anti-HA (1:250; ab9134; Abcam) overnight at 4°C. Samples were washed with 0.1% Triton X-100 in PBS and probed with donkey anti-rabbit Cy2 (1:200; Jackson ImmunoResearch) and donkey anti-goat Cy3 (1:400; Jackson ImmunoResearch) conjugated secondary antibodies for 1.5 h at room temperature in the dark. Bisbenzimide H 33258 (Hoechst; 1:1000; B1155; Sigma) was used to label nuclei. Samples were washed with PBS and imaged.
Microscopy and image analysis
Fixed whole mount mouse embryos were imaged using a MZ16FA dissecting stereoscope (Leica) with a QImaging Micropublisher 5.0 color camera using QCapture imaging software (QImaging). H&E- and Masson’s Trichrome–stained tissues were imaged using on a Dialux 20 microscope (Leitz) using similar camera and software. Fluorescently stained mouse tissue was imaged using either a Nikon E800 fluorescence microscope with a Hamamatsu Orca camera and MetaMorph software (Molecular Devices Corporation) or a Zeiss LSM 700 confocal laser-scanning microscope and ZEN 2011 software. Stained HEK293T cells were visualized with a Zeiss 880 scanning confocal microscope mounted on an Inverted Axio Observer Z1 equipped with Definite Focus. All images were psuedocolored and analyzed using ImageJ (National Institutes of Health); Fiji and R-values were calculated using Coloc2 (Schindelin et al., 2012).
Real-time qPCR
Mouse embryonic tissue was collected and stored in RNAlater (AM7021; Ambion). RNA was extracted and converted to cDNA as per Davis et al. (2017). HEK293T cells were seeded at 2.5 million cells per 10-cm dish and grown overnight in standard conditions (DMEM supplemented with 10% FBS, 1% Hepes, and 750 µl of Gentamicin; Sigma). Transfections were performed using calcium phosphate transfection as outlined by Kingston et al. (Kingston et al., 2003a,b). In brief, 1 ml of transfection mixture containing 10 µg of Myc-hCLR, Myc-hCLR(V205del), or pcDNA3.1 was added to each dish and grown overnight. RNA from tissues and cells was harvested using TRIzol reagent (15596026; Ambion) followed by DNase (M6101; Promega) treatment, and cDNA was prepared using iScript (170-8890; BioRad).
Real time-qPCR was performed using single-tube Assay on Demand (Thermo Fisher Scientific/Applied Biosystems) for human CALCRL (Hs00907738_m1), mouse Calcrl (Mm00516986_m1), human GAPDH (4310884E), and mouse Gapdh (Mm99999915_g1) on a StepOnePlus Real-Time PCR system (Applied Biosystems). Comparative ΔΔCT method was used to analyze relative gene expression normalized to GAPDH housekeeping control.
Statistical analysis
Statistical analysis was determined using GraphPad 5.0, and the data are represented as the mean ± SEM. Significant differences were determined by either Student’s t test or one-way ANOVA with Tukey’s Multiple Comparison test and represented as *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
Online supplemental material
Fig. S1 shows conservation of hCLR Valine205 residue across species. Fig. S2 shows the establishment and optimization of the IRFAP assay. Fig. S3 shows spatiotemporal deletion of Calcrl in developing lymphatic vessels. Fig. S4 shows that lymphovenous valves develop normally in Calcrlfl/fl;Prox1-CreERT2 embryos. Fig. S5 shows that heart structure appears normal in Calcrlfl/fl;Prox1-CreERT2 embryos. Video 1 shows the dynamics of CLR–RAMP2–AM complexes (hCLR(V205del) and wild-type hCLR) following REMC simulations at 300 K.
Supplementary Material
Acknowledgments
We are indebted to the family for their time and collaboration. This work is dedicated to our former colleague and mentor, Dr. Oliver Smithies (1925-2017), who personally inspired the translational of these genetic animal studies to human disease. We thank the University of North Carolina Histology Research Core, the Center of Gastrointestinal Biology and Disease Histology Core (National Institutes of Health [NIH] grant P30-DK34987), the Michael Hooker Imaging Core, and the Microscopy Service Laboratory. We also thank Dr. Alan Waggoner, Carnegie Mellon University, Pittsburgh, and Dr. Michel Bouvier, Université de Montréal for providing reagents, as well as members of the Caron laboratory for helpful insights, advice and discussion.
Funding comes from NIH grants RO1-DK099156, RO1-HD060860, and RO1-HL129086 to K.M. Caron.; American Heart Association Innovator Award 16IRG27260077 to K.M. Caron; NIH grant F32-HL134279 to D.I. Mackie.; American Heart Association grant 15POST25270006 to R.B. Davis; NIH grant F31-CA174194 to D.O. Kechele, Biotechnology and Biological Sciences Research Council grant BB/M007529/1 to D.R. Poyner and J. Simms. NIH NCI K12 Training Grant 5K12-CA100639-10 and NIH NCI grant 1K22CA212058-01 were awarded to J.C. Snyder. National Institute of Drug Abuse grant 5P30-DA029925-06 to was awarded to M.G. Caron.
The authors declare no competing financial interests.
Author contributions: D.I. Mackie designed research studies, conducted experiments, and wrote the manuscript. F. Al Mutairi designed research studies, conducted experiments, and edited the manuscript. R.B. Davis designed research studies, conducted experiments, and wrote the manuscript. D.O. Kechele conducted experiments and wrote the manuscript. N.R. Nielsen conducted experiments, analyzed data, and edited the manuscript. J.C. Snyder designed research studies, provided intellectual support, and edited the manuscript. M.G. Caron provided intellectual support and edited the manuscript. H.J. Kliman provided placental samples, analyzed data, provided intellectual support, and edited the manuscript. J.S. Berg analyzed research results, provided intellectual support, and edited the manuscript. J. Simms and D.R. Poyner designed research studies, conducted experiments, analyzed results, provided intellectual support, and edited the manuscript. K.M. Caron designed research studies, provided intellectual support, and wrote the manuscript.
References
- Achen, M.G., Jeltsch M., Kukk E., Mäkinen T., Vitali A., Wilks A.F., Alitalo K., and Stacker S.A.. 1998. Vascular endothelial growth factor D (VEGF-D) is a ligand for the tyrosine kinases VEGF receptor 2 (Flk1) and VEGF receptor 3 (Flt4). Proc. Natl. Acad. Sci. USA. 95:548–553. 10.1073/pnas.95.2.548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alders, M., Hogan B.M., Gjini E., Salehi F., Al-Gazali L., Hennekam E.A., Holmberg E.E., Mannens M.M., Mulder M.F., Offerhaus G.J., et al. . 2009. Mutations in CCBE1 cause generalized lymph vessel dysplasia in humans. Nat. Genet. 41:1272–1274. 10.1038/ng.484 [DOI] [PubMed] [Google Scholar]
- Alders, M., Al-Gazali L., Cordeiro I., Dallapiccola B., Garavelli L., Tuysuz B., Salehi F., Haagmans M.A., Mook O.R., Majoie C.B., et al. . 2014. Hennekam syndrome can be caused by FAT4 mutations and be allelic to Van Maldergem syndrome. Hum. Genet. 133:1161–1167. 10.1007/s00439-014-1456-y [DOI] [PubMed] [Google Scholar]
- Allalou, A., and Wählby C.. 2009. BlobFinder, a tool for fluorescence microscopy image cytometry. Comput. Methods Programs Biomed. 94:58–65. 10.1016/j.cmpb.2008.08.006 [DOI] [PubMed] [Google Scholar]
- Apkon, M. 1995. Pathophysiology of hydrops fetalis. Semin. Perinatol. 19:437–446. 10.1016/S0146-0005(05)80051-5 [DOI] [PubMed] [Google Scholar]
- Armour, S.L., Foord S., Kenakin T., and Chen W.J.. 1999. Pharmacological characterization of receptor-activity-modifying proteins (RAMPs) and the human calcitonin receptor. J. Pharmacol. Toxicol. Methods. 42:217–224. 10.1016/S1056-8719(00)00074-5 [DOI] [PubMed] [Google Scholar]
- Arnold, K., Bordoli L., Kopp J., and Schwede T.. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 22:195–201. 10.1093/bioinformatics/bti770 [DOI] [PubMed] [Google Scholar]
- Barak, L.S., Salahpour A., Zhang X., Masri B., Sotnikova T.D., Ramsey A.J., Violin J.D., Lefkowitz R.J., Caron M.G., and Gainetdinov R.R.. 2008. Pharmacological characterization of membrane-expressed human trace amine-associated receptor 1 (TAAR1) by a bioluminescence resonance energy transfer cAMP biosensor. Mol. Pharmacol. 74:585–594. 10.1124/mol.108.048884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth, P., Schonbrun J., and Baker D.. 2007. Toward high-resolution prediction and design of transmembrane helical protein structures. Proc. Natl. Acad. Sci. USA. 104:15682–15687. 10.1073/pnas.0702515104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barwell, J., Conner A., and Poyner D.R.. 2011. Extracellular loops 1 and 3 and their associated transmembrane regions of the calcitonin receptor-like receptor are needed for CGRP receptor function. Biochim. Biophys. Acta. 1813:1906–1916. 10.1016/j.bbamcr.2011.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barwell, J., Woolley M.J., Wheatley M., Conner A.C., and Poyner D.R.. 2012. The role of the extracellular loops of the CGRP receptor, a family B GPCR. Biochem. Soc. Trans. 40:433–437. 10.1042/BST20110726 [DOI] [PubMed] [Google Scholar]
- Bellini, C., and Hennekam R.C.. 2012. Non-immune hydrops fetalis: a short review of etiology and pathophysiology. Am. J. Med. Genet. A. 158A:597–605. 10.1002/ajmg.a.34438 [DOI] [PubMed] [Google Scholar]
- Bellini, C., Hennekam R.C., Boccardo F., Campisi C., Serra G., and Bonioli E.. 2006. Nonimmune idiopathic hydrops fetalis and congenital lymphatic dysplasia. Am. J. Med. Genet. A. 140:678–684. 10.1002/ajmg.a.31100 [DOI] [PubMed] [Google Scholar]
- Bellini, C., Hennekam R.C., Fulcheri E., Rutigliani M., Morcaldi G., Boccardo F., and Bonioli E.. 2009. Etiology of nonimmune hydrops fetalis: a systematic review. Am. J. Med. Genet. A. 149A:844–851. 10.1002/ajmg.a.32655 [DOI] [PubMed] [Google Scholar]
- Bellini, C., Donarini G., Paladini D., Calevo M.G., Bellini T., Ramenghi L.A., and Hennekam R.C.. 2015. Etiology of non-immune hydrops fetalis: An update. Am. J. Med. Genet. A. 167A:1082–1088. 10.1002/ajmg.a.36988 [DOI] [PubMed] [Google Scholar]
- Benkert, P., Schwede T., and Tosatto S.C.. 2009a. QMEANclust: estimation of protein model quality by combining a composite scoring function with structural density information. BMC Struct. Biol. 9:35. 10.1186/1472-6807-9-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkert, P., Tosatto S.C., and Schwede T.. 2009b. Global and local model quality estimation at CASP8 using the scoring functions QMEAN and QMEANclust. Proteins. 77(S9, Suppl 9):173–180. 10.1002/prot.22532 [DOI] [PubMed] [Google Scholar]
- Benkert, P., Biasini M., and Schwede T.. 2011. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. 27:343–350. 10.1093/bioinformatics/btq662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biasini, M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Gallo Cassarino T., Bertoni M., Bordoli L., and Schwede T.. 2014. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 42(W1):W252-258. 10.1093/nar/gku340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouschet, T., Martin S., and Henley J.M.. 2005. Receptor-activity-modifying proteins are required for forward trafficking of the calcium-sensing receptor to the plasma membrane. J. Cell Sci. 118:4709–4720. 10.1242/jcs.02598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brace, R.A. 1989. Effects of outflow pressure on fetal lymph flow. Am. J. Obstet. Gynecol. 160:494–497. 10.1016/0002-9378(89)90479-1 [DOI] [PubMed] [Google Scholar]
- Brace, R.A., and Valenzuela G.J.. 1990. Effects of outflow pressure and vascular volume loading on thoracic duct lymph flow in adult sheep. Am. J. Physiol. 258:R240–R244. [DOI] [PubMed] [Google Scholar]
- Brouillard, P., Dupont L., Helaers R., Coulie R., Tiller G.E., Peeden J., Colige A., and Vikkula M.. 2017. Loss of ADAMTS3 activity causes Hennekam lymphangiectasia-lymphedema syndrome 3. Hum. Mol. Genet. 26:4095–4104. 10.1093/hmg/ddx297 [DOI] [PubMed] [Google Scholar]
- Bukowski, R., and Saade G.R.. 2000. Hydrops fetalis. Clin. Perinatol. 27:1007–1031. 10.1016/S0095-5108(05)70061-0 [DOI] [PubMed] [Google Scholar]
- Caron, K.M., and Smithies O.. 2001. Extreme hydrops fetalis and cardiovascular abnormalities in mice lacking a functional Adrenomedullin gene. Proc. Natl. Acad. Sci. USA. 98:615–619. 10.1073/pnas.98.2.615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan, M., Yallampalli U., Banadakappa M., and Yallampalli C.. 2015. Involvement of Receptor Activity-Modifying Protein 3 (RAMP3) in the Vascular Actions of Adrenomedullin in Rat Mesenteric Artery Smooth Muscle Cells. Biol. Reprod. 93:116. 10.1095/biolreprod.115.134585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopoulos, A., Christopoulos G., Morfis M., Udawela M., Laburthe M., Couvineau A., Kuwasako K., Tilakaratne N., and Sexton P.M.. 2003. Novel receptor partners and function of receptor activity-modifying proteins. J. Biol. Chem. 278:3293–3297. 10.1074/jbc.C200629200 [DOI] [PubMed] [Google Scholar]
- Connell, F., Kalidas K., Ostergaard P., Brice G., Homfray T., Roberts L., Bunyan D.J., Mitton S., Mansour S., Mortimer P., and Jeffery S.. Lymphoedema Consortium . 2010. Linkage and sequence analysis indicate that CCBE1 is mutated in recessively inherited generalised lymphatic dysplasia. Hum. Genet. 127:231–241. 10.1007/s00439-009-0766-y [DOI] [PubMed] [Google Scholar]
- Connell, F.C., Gordon K., Brice G., Keeley V., Jeffery S., Mortimer P.S., Mansour S., and Ostergaard P.. 2013. The classification and diagnostic algorithm for primary lymphatic dysplasia: an update from 2010 to include molecular findings. Clin. Genet. 84:303–314. 10.1111/cge.12173 [DOI] [PubMed] [Google Scholar]
- Dackor, R.T., Fritz-Six K., Dunworth W.P., Gibbons C.L., Smithies O., and Caron K.M.. 2006. Hydrops fetalis, cardiovascular defects, and embryonic lethality in mice lacking the calcitonin receptor-like receptor gene. Mol. Cell. Biol. 26:2511–2518. 10.1128/MCB.26.7.2511-2518.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dackor, R., Fritz-Six K., Smithies O., and Caron K.. 2007. Receptor activity-modifying proteins 2 and 3 have distinct physiological functions from embryogenesis to old age. J. Biol. Chem. 282:18094–18099. 10.1074/jbc.M703544200 [DOI] [PubMed] [Google Scholar]
- Davis, R.B., Kechele D.O., Blakeney E.S., Pawlak J.B., and Caron K.M.. 2017. Lymphatic deletion of calcitonin receptor-like receptor exacerbates intestinal inflammation. JCI Insight. 2:e92465. 10.1172/jci.insight.92465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drumm, M.L., Wilkinson D.J., Smit L.S., Worrell R.T., Strong T.V., Frizzell R.A., Dawson D.C., and Collins F.S.. 1991. Chloride conductance expressed by delta F508 and other mutant CFTRs in Xenopus oocytes. Science. 254:1797–1799. 10.1126/science.1722350 [DOI] [PubMed] [Google Scholar]
- Fang, J., Dagenais S.L., Erickson R.P., Arlt M.F., Glynn M.W., Gorski J.L., Seaver L.H., and Glover T.W.. 2000. Mutations in FOXC2 (MFH-1), a forkhead family transcription factor, are responsible for the hereditary lymphedema-distichiasis syndrome. Am. J. Hum. Genet. 67:1382–1388. 10.1086/316915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, G.W., Fuhrman M.H., Adler S.A., Szent-Gyorgyi C., Waggoner A.S., and Jarvik J.W.. 2014. Self-Checking Cell-Based Assays for GPCR Desensitization and Resensitization. J. Biomol. Screen. 19:1220–1226. 10.1177/1087057114534299 [DOI] [PubMed] [Google Scholar]
- Fotiou, E., Martin-Almedina S., Simpson M.A., Lin S., Gordon K., Brice G., Atton G., Jeffery I., Rees D.C., Mignot C., et al. . 2015. Novel mutations in PIEZO1 cause an autosomal recessive generalized lymphatic dysplasia with non-immune hydrops fetalis. Nat. Commun. 6:8085. 10.1038/ncomms9085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- François, M., Caprini A., Hosking B., Orsenigo F., Wilhelm D., Browne C., Paavonen K., Karnezis T., Shayan R., Downes M., et al. . 2008. Sox18 induces development of the lymphatic vasculature in mice. Nature. 456:643–647. 10.1038/nature07391 [DOI] [PubMed] [Google Scholar]
- Fritz-Six, K.L., Dunworth W.P., Li M., and Caron K.M.. 2008. Adrenomedullin signaling is necessary for murine lymphatic vascular development. J. Clin. Invest. 118:40–50. 10.1172/JCI33302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goadsby, P.J., Reuter U., Hallström Y., Broessner G., Bonner J.H., Zhang F., Sapra S., Picard H., Mikol D.D., and Lenz R.A.. 2017. A Controlled Trial of Erenumab for Episodic Migraine. N. Engl. J. Med. 377:2123–2132. 10.1056/NEJMoa1705848 [DOI] [PubMed] [Google Scholar]
- Gordon, K., Spiden S.L., Connell F.C., Brice G., Cottrell S., Short J., Taylor R., Jeffery S., Mortimer P.S., Mansour S., and Ostergaard P.. 2013. FLT4/VEGFR3 and Milroy disease: novel mutations, a review of published variants and database update. Hum. Mutat. 34:23–31. 10.1002/humu.22223 [DOI] [PubMed] [Google Scholar]
- Guex, N., Peitsch M.C., and Schwede T.. 2009. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis. 30(S1, Suppl 1):S162–S173. 10.1002/elps.200900140 [DOI] [PubMed] [Google Scholar]
- Hay, D.L., Howitt S.G., Conner A.C., Schindler M., Smith D.M., and Poyner D.R.. 2003. CL/RAMP2 and CL/RAMP3 produce pharmacologically distinct adrenomedullin receptors: a comparison of effects of adrenomedullin22-52, CGRP8-37 and BIBN4096BS. Br. J. Pharmacol. 140:477–486. 10.1038/sj.bjp.0705472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay, D.L., Poyner D.R., and Sexton P.M.. 2006. GPCR modulation by RAMPs. Pharmacol. Ther. 109:173–197. 10.1016/j.pharmthera.2005.06.015 [DOI] [PubMed] [Google Scholar]
- Hollenstein, K., Kean J., Bortolato A., Cheng R.K., Doré A.S., Jazayeri A., Cooke R.M., Weir M., and Marshall F.H.. 2013. Structure of class B GPCR corticotropin-releasing factor receptor 1. Nature. 499:438–443. 10.1038/nature12357 [DOI] [PubMed] [Google Scholar]
- Ichikawa-Shindo, Y., Sakurai T., Kamiyoshi A., Kawate H., Iinuma N., Yoshizawa T., Koyama T., Fukuchi J., Iimuro S., Moriyama N., et al. . 2008. The GPCR modulator protein RAMP2 is essential for angiogenesis and vascular integrity. J. Clin. Invest. 118:29–39. 10.1172/JCI33022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joukov, V., Pajusola K., Kaipainen A., Chilov D., Lahtinen I., Kukk E., Saksela O., Kalkkinen N., and Alitalo K.. 1996. A novel vascular endothelial growth factor, VEGF-C, is a ligand for the Flt4 (VEGFR-3) and KDR (VEGFR-2) receptor tyrosine kinases. EMBO J. 15:1751. [PMC free article] [PubMed] [Google Scholar]
- Kadmiel, M., Fritz-Six K., Pacharne S., Richards G.O., Li M., Skerry T.M., and Caron K.M.. 2011. Research resource: Haploinsufficiency of receptor activity-modifying protein-2 (RAMP2) causes reduced fertility, hyperprolactinemia, skeletal abnormalities, and endocrine dysfunction in mice. Mol. Endocrinol. 25:1244–1253. 10.1210/me.2010-0400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitani, S., Asakawa M., Shimekake Y., Kuwasako K., Nakahara K., and Sakata T.. 1999. The RAMP2/CRLR complex is a functional adrenomedullin receptor in human endothelial and vascular smooth muscle cells. FEBS Lett. 448:111–114. 10.1016/S0014-5793(99)00358-0 [DOI] [PubMed] [Google Scholar]
- Kattner, E., Schäfer A., and Harzer K.. 1997. Hydrops fetalis: manifestation in lysosomal storage diseases including Farber disease. Eur. J. Pediatr. 156:292–295. 10.1007/s004310050603 [DOI] [PubMed] [Google Scholar]
- Kechele, D.O., Dunworth W.P., Trincot C.E., Wetzel-Strong S.E., Li M., Ma H., Liu J., and Caron K.M.. 2016. Endothelial Restoration of Receptor Activity-Modifying Protein 2 Is Sufficient to Rescue Lethality, but Survivors Develop Dilated Cardiomyopathy. Hypertension. 68:667–677. 10.1161/HYPERTENSIONAHA.116.07191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer, F., Arnold K., Künzli M., Bordoli L., and Schwede T.. 2009. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 37(Database):D387–D392. 10.1093/nar/gkn750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston, R.E., Chen C.A., and Okayama H.. 2003a. Calcium phosphate transfection. Curr. Protoc. Cell Biol. Chapter 20.3. [DOI] [PubMed] [Google Scholar]
- Kingston, R.E., Chen C.A., and Rose J.K.. 2003b. Calcium phosphate transfection. Curr. Protoc. Mol. Biol. Chapter 9.1. [DOI] [PubMed] [Google Scholar]
- Koyama, T., Ochoa-Callejero L., Sakurai T., Kamiyoshi A., Ichikawa-Shindo Y., Iinuma N., Arai T., Yoshizawa T., Iesato Y., Lei Y., et al. . 2013. Vascular endothelial adrenomedullin-RAMP2 system is essential for vascular integrity and organ homeostasis. Circulation. 127:842–853. 10.1161/CIRCULATIONAHA.112.000756 [DOI] [PubMed] [Google Scholar]
- Kuwasako, K., Kitamura K., Nagata S., Hikosaka T., Takei Y., and Kato J.. 2011. Shared and separate functions of the RAMP-based adrenomedullin receptors. Peptides. 32:1540–1550. 10.1016/j.peptides.2011.05.022 [DOI] [PubMed] [Google Scholar]
- Lek, M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., et al. Exome Aggregation Consortium . 2016. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 536:285–291. 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhart, P.M., Broselid S., Barrick C.J., Leeb-Lundberg L.M., and Caron K.M.. 2013. G-protein-coupled receptor 30 interacts with receptor activity-modifying protein 3 and confers sex-dependent cardioprotection. J. Mol. Endocrinol. 51:191–202. 10.1530/JME-13-0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., and Mukouyama Y.S.. 2011. Whole-mount immunohistochemical analysis for embryonic limb skin vasculature: a model system to study vascular branching morphogenesis in embryo. J. Vis. Exp. 51:2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M., Yee D., Magnuson T.R., Smithies O., and Caron K.M.. 2006. Reduced maternal expression of adrenomedullin disrupts fertility, placentation, and fetal growth in mice. J. Clin. Invest. 116:2653–2662. 10.1172/JCI28462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M., Wu Y., and Caron K.M.. 2008. Haploinsufficiency for adrenomedullin reduces pinopodes and diminishes uterine receptivity in mice. Biol. Reprod. 79:1169–1175. 10.1095/biolreprod.108.069336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M., Schwerbrock N.M., Lenhart P.M., Fritz-Six K.L., Kadmiel M., Christine K.S., Kraus D.M., Espenschied S.T., Willcockson H.H., Mack C.P., and Caron K.M.. 2013. Fetal-derived adrenomedullin mediates the innate immune milieu of the placenta. J. Clin. Invest. 123:2408–2420. 10.1172/JCI67039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., Cowley A., Uludag M., Gur T., McWilliam H., Squizzato S., Park Y.M., Buso N., and Lopez R.. 2015. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 43(W1):W580-584. 10.1093/nar/gkv279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacs, V., Mathur J., Mao R., Bayrak-Toydemir P., Procter M., Cahalan S.M., Kim H.J., Bandell M., Longo N., Day R.W., et al. . 2015. Impaired PIEZO1 function in patients with a novel autosomal recessive congenital lymphatic dysplasia. Nat. Commun. 6:8329. 10.1038/ncomms9329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Almedina, S., Martinez-Corral I., Holdhus R., Vicente A., Fotiou E., Lin S., Petersen K., Simpson M.A., Hoischen A., Gilissen C., et al. . 2016. EPHB4 kinase-inactivating mutations cause autosomal dominant lymphatic-related hydrops fetalis. J. Clin. Invest. 126:3080–3088. 10.1172/JCI85794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLatchie, L.M., Fraser N.J., Main M.J., Wise A., Brown J., Thompson N., Solari R., Lee M.G., and Foord S.M.. 1998. RAMPs regulate the transport and ligand specificity of the calcitonin-receptor-like receptor. Nature. 393:333–339. 10.1038/30666 [DOI] [PubMed] [Google Scholar]
- McWilliam, H., Li W., Uludag M., Squizzato S., Park Y.M., Buso N., Cowley A.P., and Lopez R.. 2013. Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res. 41(W1):W597-600. 10.1093/nar/gkt376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morello, J.P., Salahpour A., Laperrière A., Bernier V., Arthus M.F., Lonergan M., Petäjä-Repo U., Angers S., Morin D., Bichet D.G., and Bouvier M.. 2000. Pharmacological chaperones rescue cell-surface expression and function of misfolded V2 vasopressin receptor mutants. J. Clin. Invest. 105:887–895. 10.1172/JCI8688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham, T.H., Baluk P., Xu Y., Grigorova I., Bankovich A.J., Pappu R., Coughlin S.R., McDonald D.M., Schwab S.R., and Cyster J.G.. 2010. Lymphatic endothelial cell sphingosine kinase activity is required for lymphocyte egress and lymphatic patterning. J. Exp. Med. 207:17–27. 10.1084/jem.20091619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyner, D., and Simms J.. 2018. Molecular dynamic simulations of CLR/RAMP1 and CLR/RAMP2 heterodimers, with and without deletions of V205 in CLR. 10.17036/researchdata.aston.ac.uk.00000369 [DOI]
- Randenberg, A.L. 2010a. Nonimmune hydrops fetalis part I: etiology and pathophysiology. Neonatal Netw. 29:281–295. 10.1891/0730-0832.29.5.281 [DOI] [PubMed] [Google Scholar]
- Randenberg, A.L. 2010b. Nonimmune hydrops fetalis part II: does etiology influence mortality? Neonatal Netw. 29:367–380. 10.1891/0730-0832.29.6.367 [DOI] [PubMed] [Google Scholar]
- Sabine, A., Agalarov Y., Maby-El Hajjami H., Jaquet M., Hägerling R., Pollmann C., Bebber D., Pfenniger A., Miura N., Dormond O., et al. . 2012. Mechanotransduction, PROX1, and FOXC2 cooperate to control connexin37 and calcineurin during lymphatic-valve formation. Dev. Cell. 22:430–445. 10.1016/j.devcel.2011.12.020 [DOI] [PubMed] [Google Scholar]
- Salahpour, A., Espinoza S., Masri B., Lam V., Barak L.S., and Gainetdinov R.R.. 2012. BRET biosensors to study GPCR biology, pharmacology, and signal transduction. Front. Endocrinol. (Lausanne). 3:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šali, A., and Blundell T.L.. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779–815. 10.1006/jmbi.1993.1626 [DOI] [PubMed] [Google Scholar]
- Schindelin, J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. . 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers, F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., et al. . 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberstein, S.D., Dodick D.W., Bigal M.E., Yeung P.P., Goadsby P.J., Blankenbiller T., Grozinski-Wolff M., Yang R., Ma Y., and Aycardi E.. 2017. Fremanezumab for the Preventive Treatment of Chronic Migraine. N. Engl. J. Med. 377:2113–2122. 10.1056/NEJMoa1709038 [DOI] [PubMed] [Google Scholar]
- Snyder, J.C., Pack T.F., Rochelle L.K., Chakraborty S.K., Zhang M., Eaton A.W., Bai Y., Ernst L.A., Barak L.S., Waggoner A.S., and Caron M.G.. 2015. A rapid and affordable screening platform for membrane protein trafficking. BMC Biol. 13:107. 10.1186/s12915-015-0216-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder, J.C., Rochelle L.K., Ray C., Pack T.F., Bock C.B., Lubkov V., Lyerly H.K., Waggoner A.S., Barak L.S., and Caron M.G.. 2017. Inhibiting clathrin-mediated endocytosis of the leucine-rich G protein-coupled receptor-5 diminishes cell fitness. J. Biol. Chem. 292:7208–7222. 10.1074/jbc.M116.756635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, G., Yang D., Wang Y., de Graaf C., Zhou Q., Jiang S., Liu K., Cai X., Dai A., Lin G., et al. . 2017. Human GLP-1 receptor transmembrane domain structure in complex with allosteric modulators. Nature. 546:312–315. 10.1038/nature22378 [DOI] [PubMed] [Google Scholar]
- Srinivasan, R.S., Dillard M.E., Lagutin O.V., Lin F.J., Tsai S., Tsai M.J., Samokhvalov I.M., and Oliver G.. 2007. Lineage tracing demonstrates the venous origin of the mammalian lymphatic vasculature. Genes Dev. 21:2422–2432. 10.1101/gad.1588407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szent-Gyorgyi, C., Schmidt B.F., Creeger Y., Fisher G.W., Zakel K.L., Adler S., Fitzpatrick J.A., Woolford C.A., Yan Q., Vasilev K.V., et al. . 2008. Fluorogen-activating single-chain antibodies for imaging cell surface proteins. Nat. Biotechnol. 26:235–240. 10.1038/nbt1368 [DOI] [PubMed] [Google Scholar]
- Tamarappoo, B.K., and Verkman A.S.. 1998. Defective aquaporin-2 trafficking in nephrogenic diabetes insipidus and correction by chemical chaperones. J. Clin. Invest. 101:2257–2267. 10.1172/JCI2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins, H.A., Chakravarthy M., Abhayawardana R.S., Gingell J.J., Garelja M., Pardamwar M., McElhinney J.M., Lathbridge A., Constantine A., Harris P.W., et al. . 2016. Receptor Activity-modifying Proteins 2 and 3 Generate Adrenomedullin Receptor Subtypes with Distinct Molecular Properties. J. Biol. Chem. 291:11657–11675. 10.1074/jbc.M115.688218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston, C., Lu J., Li N., Barkan K., Richards G.O., Roberts D.J., Skerry T.M., Poyner D., Pardamwar M., Reynolds C.A., et al. . 2015. Modulation of Glucagon Receptor Pharmacology by Receptor Activity-modifying Protein-2 (RAMP2). J. Biol. Chem. 290:23009–23022. 10.1074/jbc.M114.624601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolley, M.J., and Conner A.C.. 2013. Comparing the molecular pharmacology of CGRP and adrenomedullin. Curr. Protein Pept. Sci. 14:358–374. 10.2174/13892037113149990053 [DOI] [PubMed] [Google Scholar]
- Woolley, M.J., Simms J., Uddin S., Poyner D.R., and Conner A.C.. 2017. Relative Antagonism of Mutants of the CGRP Receptor Extracellular Loop 2 Domain (ECL2) Using a Truncated Competitive Antagonist (CGRP8-37): Evidence for the Dual Involvement of ECL2 in the Two-Domain Binding Model. Biochemistry. 56:3877–3880. 10.1021/acs.biochem.7b00077 [DOI] [PubMed] [Google Scholar]
- Zhang, H., Qiao A., Yang D., Yang L., Dai A., de Graaf C., Reedtz-Runge S., Dharmarajan V., Zhang H., Han G.W., et al. . 2017. Structure of the full-length glucagon class B G-protein-coupled receptor. Nature. 546:259–264. 10.1038/nature22363 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.