Unlike man-made wheels that are centered and rotated via an axle, the mitotic spindle of a human cell is rotated by external cortical pulling mechanisms. Zulkipli et al. identify MARK2’s role in equatorial spindle centering and astral microtubule length, which in turn control spindle rotation.
Abstract
The plane of cell division is defined by the final position of the mitotic spindle. The spindle is pulled and rotated to the correct position by cortical dynein. However, it is unclear how the spindle’s rotational center is maintained and what the consequences of an equatorially off centered spindle are in human cells. We analyzed spindle movements in 100s of cells exposed to protein depletions or drug treatments and uncovered a novel role for MARK2 in maintaining the spindle at the cell’s geometric center. Following MARK2 depletion, spindles glide along the cell cortex, leading to a failure in identifying the correct division plane. Surprisingly, spindle off centering in MARK2-depleted cells is not caused by excessive pull by dynein. We show that MARK2 modulates mitotic microtubule growth and length and that codepleting mitotic centromere-associated protein (MCAK), a microtubule destabilizer, rescues spindle off centering in MARK2-depleted cells. Thus, we provide the first insight into a spindle-centering mechanism needed for proper spindle rotation and, in turn, the correct division plane in human cells.
Introduction
Loss of tissue organization is a hallmark of aggressive carcinomas. In epithelial tissues, during cell division, the position of the mitotic spindle defines the plane of division, and in turn, the position of daughter cells within the growing and stratifying epithelial tissue (Kulukian and Fuchs, 2013; Chin et al., 2014; Macara et al., 2014). The spindle is brought to the correct position by cortical dynein-mediated forces that pull and rotate the spindle; how these pulling forces are counteracted to maintain the spindle’s center of rotation is an intriguing physical and biological problem. Spindle centering forces were recently measured in worm embryos (Garzon-Coral et al., 2016) that are 10 times larger than human cells. Master regulators that sense and control spindle centering are not known in human cells, although changes in microtubule dynamics can alter spindle centering (Draviam et al., 2006), suggesting the existence of a centering mechanism in human cells as well.
Unlike equatorial spindle-centering mechanisms (in the x–y plane), spindle orientation mechanisms (in the z-plane) have been explored in detail in human cells. Proper 3D orientation of the spindle requires the interactions of astral microtubules with cytoplasmic and cortical force generators (O’Connell and Wang, 2000; Wühr et al., 2010; Kimura and Kimura, 2011; Markus and Lee, 2011; Collins et al., 2012; Kiyomitsu and Cheeseman, 2012). In cell cultures, dynein is required to rotate and orient the spindle along a predetermined axis: the interphase long axis of the cell (O’Connell and Wang, 2000; Corrigan et al., 2013). Importantly, two pathways that influence cortical dynein, LGN–NuMA–Gαi pathway (Kotak et al., 2012) and CHICA-dependent dynein signaling pathway (Dunsch et al., 2012), orient the spindle parallel to the substratum, and excessive dynein activity can cause spindle tumbling with respect to the substratum (Samora et al., 2011; Kotak et al., 2012). Thus, cortical dynein-mediated pull is currently considered to be the primary force-generating pathway for powering spindle movements in human cells. In contrast, in the yeast Saccharomyces cerevisiae, dynein is dispensable for early spindle rotation or alignment and is essential only for the later translocation of the spindle into the bud neck (Sheeman et al., 2003; Huisman and Segal, 2005; Ten Hoopen et al., 2012). Whether similar mechanisms independent of cortical dynein operate in human cells to dynamically control spindle positions is not known.
MARK2Par1b/EMK1 (hereafter referred as MARK2) of the MAP–microtubule affinity regulating kinase (MARK) family of kinases is evolutionarily conserved from yeasts to humans. MARK2-null mice are dwarf, with most tissues proportionately smaller, hypofertile, lean, and resistant to high-fat diet–induced weight gain (Bessone et al., 1999; Hurov and Piwnica-Worms, 2007). In nematodes, mutations in par-1 disrupt spindle positioning, leading to disorganized embryos that lack germ cells (Guo and Kemphues, 1995). In flies, mutations in par-1 disrupt the oocyte microtubule network, leading to defects in posterior patterning of the embryo (Shulman et al., 2000). In mouse hepatocytes, MARK2 is required for the asymmetric inheritance of apical domains through cell division (Slim et al., 2013). However, the precise role of MARK2 during the process of spindle orientation in human cells is not clear.
MARK2’s role in the regulation of interphase microtubule dynamics has been characterized in several cell lines (Cohen et al., 2004; Mandelkow et al., 2004; Schaar et al., 2004; Hayashi et al., 2011; Nishimura et al., 2012; Sato et al., 2013, 2014). Whether MARK2 controls mitotic microtubule dynamics is not known.
To study spindle rotation and centering mechanisms, we used time-lapse microscopy to track changes in spindle positions in 100s of dividing human epithelial cells. We used our quantitative single-cell methodology to link a variety of interphase cell shapes to dynamically changing spindle positions in cultures that retain cell–cell contacts (Corrigan et al., 2013) so that we can systematically compare the roles of proteins in centering, rotating, and oscillating movements of the mitotic spindle. We report that the spindles of MARK2-depleted cells are equatorially off centered and that spindles undergo gliding instead of rotational movements. In the absence of MARK2, even spindles born close to the geometric center of the cell undergo dramatic off centering and remain equatorially off centered until anaphase. Importantly, equatorially off-centered spindles of MARK2-depleted cells fail to rotate properly toward the correct final position, predetermined by interphase cell shape. This shows the importance of equatorial centering mechanisms in controlling spindle rotation and determining the correct division plane. To identify the underlying molecular cause for off-centered spindles in MARK2-depleted cells, we studied the behavior of cortical dynein and astral microtubules. MARK2-depleted cells display longer astral microtubules, increased mitotic microtubule growth rates, and diffuse cortical dynein along the cell cortex. Equatorial off centering in MARK2-depleted cells could not be simply explained by excessive pulling by cortical dynein because the codepletion of LGN leads to loss of cortical dynein and spindle rotation as expected, but spindles remain equatorially off centered. In contrast, disrupting microtubule dynamics through mitotic centromere-associated protein (MCAK) depletion rescues the spindle off-centering phenotype in MARK2-depleted cells. We propose a model where cortical dynein–mediated pulling forces are actively counteracted by a MARK2-mediated spindle-centering mechanism to ensure the successful rotation of the spindle toward its predetermined final position. Thus, this study uncovers the role and regulation of equatorial spindle centering in determining the correct plane of division in human cells.
Results
Equatorial centering of the spindle is an early mitotic event
Using our single-cell method developed to track the spindle’s biased rotation to its final position determined by interphase cell shape (Corrigan et al., 2013, 2015), we studied mitotic spindle movements from nuclear envelope breakdown (NEBD) to anaphase onset in epithelial cell cultures that retained cell–cell and cell–substrate contacts. To study the range of spindle centering and off-centering events, we tracked spindle movements, once every 4 min, in a spindle reporter cell line HeLaHis2B-GFP;mCherry-tubulin that constitutively expresses low levels of mCherry-tubulin and histone2B-GFP (Corrigan et al., 2013). We assessed the extent of spindle centering along two axes: (1) the equatorial axis by measuring the nearest distance between spindle equator and the cortex (for equatorial centering) and (2) the pole-to-pole axis by measuring the nearest distance between the spindle pole and cortex (for longitudinal centering; Fig. 1 a). Analysis of mCherry-tubulin signals in time-lapse videos showed that at the onset of mitosis (marked by NEBD), spindles born in the geometric center are retained in the center, whereas spindles born away from the geometric center are actively brought to the center within the first 12 min of NEBD (Fig. 1, a and b). We observed that equatorial spindle centering did not require a mature spindle structure as equatorial centering was observed even in immature and bent spindles (compare Fig. 1 b and Fig. 1 c). In contrast and as reported earlier (Kiyomitsu and Cheeseman, 2012; Kotak et al., 2012; Corrigan et al., 2013), centering along the spindle’s longitudinal pole-to-pole axis was dislodged by spindle oscillations (Fig. 1 a). Consequently, longitudinally off-centered spindles arise as part of the normal spindle oscillatory movement until late metaphase (Fig. 1 a). We conclude that the equatorial centering of the spindle apparatus is accomplished very early on during mitosis.
Figure 1.
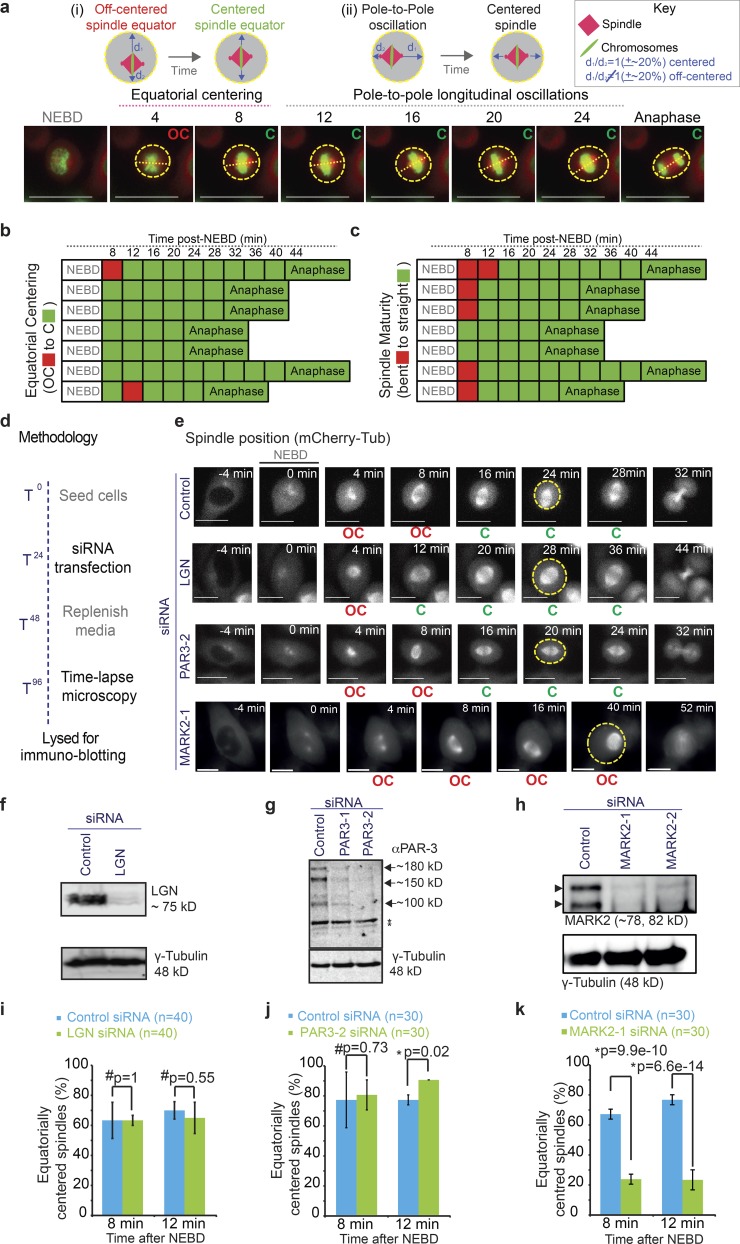
Equatorial spindle centering is an early mitotic event that is dependent on MARK2, but not LGN or PAR3. (a) Top: Cartoons of mitotic spindle in either equatorially (i) or longitudinally (ii) off-centered (OC; left) versus centered (C; right) positions. Spindles were scored as off centered when the two distances (d1 and d2; marked by arrows) were unequal by >20%. Bottom: Time-lapse images of control siRNA–transfected HeLa His2B-GFP;mCherry-Tub cells acquired once every 4 min showing normal equatorial spindle centering before longitudinal centering and oscillation along the pole-to-pole axis. Outline of cell cortex identified using cytoplasmic signal of mCherry-tubulin is marked in yellow; pole-to-pole axis marked with a dashed line. Bars, 15 µm (b and c) Temporal evolution of spindle centering along the equatorial axis (b) and corresponding spindle maturation (from bent to straight bipolar spindle; c) in seven representative cells from videos acquired as shown in a. Boxes corresponding with sequential time frames in the video are colored either in red to indicate off-centered (b), or immature (c) spindles or in green to indicate centered (b) or mature (c) spindles. (d) Experimental regimen showing timeline of siRNA treatment followed by microscopy and lysate collection for immunoblotting to register protein depletion extent. (e) Time-lapse images of HeLa His2B-GFP;mCherry-Tub cells treated with control or LGN siRNA showing normal spindle centering but no oscillation of the mitotic spindle or Par3-2 siRNA showing normal centering of the mitotic spindle or MARK2-1 siRNA showing equatorially off centering of the mitotic spindle. Bars, 10 µm. (f–h) Blot showing depletion extent of LGN (f), PAR3 (g), or MARK2 (h) using one or two different siRNA oligonucleotides. (i–k) Graph of percentage of equatorially centered spindles at 8 or 12 min after NEBD in control or LGN siRNA–treated cells (i), control or PAR3-2 siRNA–treated cells (j), and control or MARK2-1 siRNA–treated cells (k) as assessed from time-lapse videos of HeLaHis2B-GFP;mCherry-Tub cells as shown in e. Note that post-siRNA transfection time-matched control siRNA data are used for graphs and immunoblots. Error bars refer to SEM values from three independent experiments. P-values showing significant (*) or insignificant (#) differences were calculated using a proportion test.
Equatorial centering of spindles fail in the absence of MARK2
To identify the molecular players required for the equatorial centering of the spindle, we used time-lapse microscopy and followed spindle movements in HeLaHis2B-GFP;mCherry-tubulin cells depleted of LGN, MARK2Par1b or PAR3, using multiple siRNA oligonucleotides. To confirm the extent of protein depletion in the cells studied using time-lapse microscopy, we collected cell lysates after each time-lapse imaging session (Fig. 1 d). Fluorescent immunoblots of lysates showed that >90% of protein depletion was achieved after respective siRNA treatments (Fig. 1, f–h).
Analysis of time-lapse videos of HeLaHis2B-GFP;mCherry-tubulin cells showed normal equatorial centering of the spindle in cells depleted of LGN or PAR3 (Fig. 1, e, i, and j). In contrast, equatorial centering of spindles was severely disrupted in MARK2-depleted cells compared with control or LGN- or PAR3-depleted cells (Fig. 1, e and i–k), revealing MARK2 as the first kinase with a role in the equatorial centering of the human mitotic spindle.
MARK2Par1b, but not MARK3 Par1c, is required for equatorial spindle centering
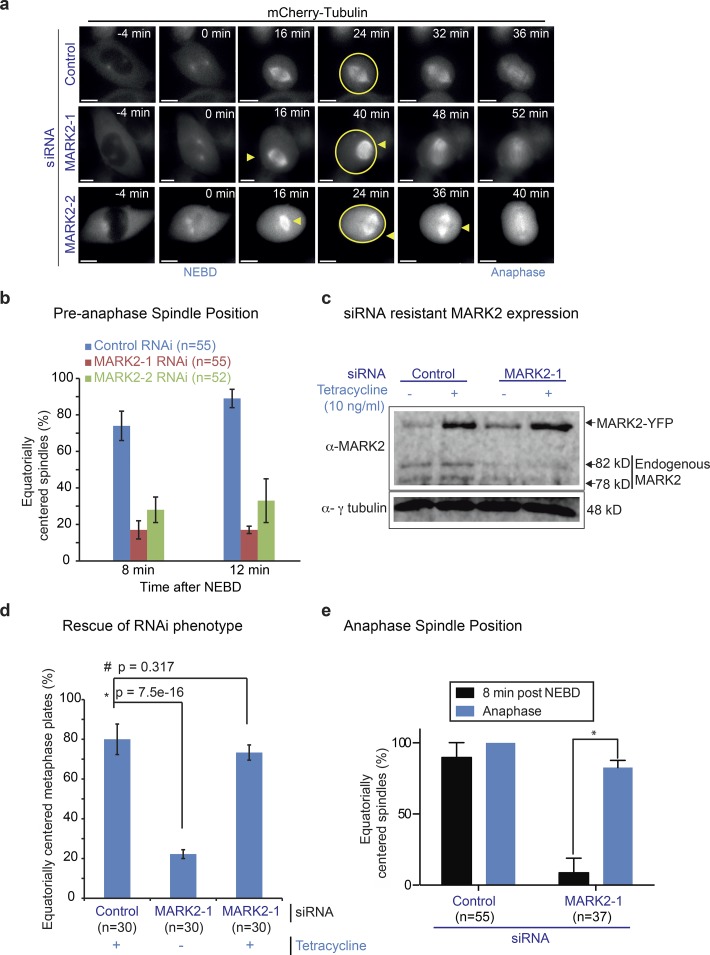
To confirm that MARK2 siRNA–induced spindle-centering defects are specific to MARK2 depletion and not an off-target effect, we used two strategies. First, we confirmed that two different siRNA oligonucleotides against MARK2 caused the spindle off-centering defect. Time-lapse videos of HeLaHis2B-GFP;mCherry-tubulin cells showed equatorially off-centered mitotic spindles after treatment with either MARK2-1 or MARK2-2 siRNA but not control siRNA (Figs. 2 a and S1 a). Quantitative analysis showed that 12 min after NEBD, equatorially centered spindles were found only in 28% and 38% of MARK2-1 and MARK2-2 siRNA–treated cells, respectively, compared with 90% of control siRNA–treated cells (Fig. 2 b). The severity of the spindle off-centering phenotype (Figs. 2 a and S1 a) correlated well with the extent of MARK2 depletion (Fig. 1 h). Thus, two different siRNA-mediated depletions of MARK2 perturb the equatorial centering of mitotic spindles.
Figure 2.
MARK2 is required for equatorial spindle centering up to anaphase. (a) Representative single-plane deconvolved images of z stack videos of HeLaHis2B-GFP;mCherry-tubulin cells treated with MARK2 or control siRNA oligonucleotides as indicated. Only mCherry-tubulin signals are shown for clarity. Bars, 5 µm. Arrowheads point to off-centered spindles. Circles highlight the cell cortex. Note that the cell showed in MARK2-1 row is shown in Fig 1 e. (b) Graph of the percentage of mitotic cells with equatorially centered spindles at 8 or 12 min after NEBD. (c) Uncropped immunoblot showing the expression of siRNA-resistant MARK2-siRes-GFP in cells treated with control or MARK2 siRNA. Lysates of HeLaMARK2-siRes-GFP cells treated with siRNA and exposed to tetracycline as indicated were immunoblotted with antibody against hMARK2; anti–γ-tubulin antibody was used as loading control. Endogenous MARK2 and Tet-inducible siRNA-resistant MARK fused with YFP are shown with arrows. (d) Bar graph showing the percentage of mitotic HeLaMARK2-siRes-GFP cells with equatorially centered spindles. Control or MARK2-1 siRNA–treated HeLaMARK2-siRes-GFP cells in the presence or absence of tetracycline (as indicated) were treated with MG132 to arrest them at metaphase prior during live-cell imaging. Cell expressing (green; +) or not expressing (nongreen, –) the siRNA-resistant form of MARK2-siRes-GFP were imaged, and chromosome positions were ascertained using differential interference contrast, which was used to determine equatorially centered or off-centered metaphase plates. (e) Bar graph comparing the extent of equatorial centering of spindles at 8 min after NEBD versus anaphase onset in control or MARK2-1 siRNA–treated cells as assessed from time-lapse videos as shown in a. Error bars represent SEM across three experimental repeats. P-values were obtained using a proportion test on percentage values * and # indicate significant and insignificant difference, respectively.
As a second strategy, to fully confirm that the equatorial centering defect observed in MARK2 siRNA–treated cells is specifically caused by MARK2 depletion and not an artifact caused by off-target depletion, we generated siRNA-resistant form of human MARK2 fused to YFP (hMARK2siRes-YFP) and expressed it in low levels using a Tet-inducible system (Fig. 2 c). After MARK2 siRNA treatment, hMARK2siRes-YFP–expressing cells (green cells) showed normally centered metaphase chromosomes, whereas nongreen cells showed equatorially off-centered chromosome positions (Figs. 2 d and S1 b). Thus, MARK2 depletion–induced spindle centering can be rescued by Tet-inducible expression of human MARK2siRes-YFP. We conclude that MARK2 is required for the equatorial centering of mitotic spindle.
Furthermore, in cells treated with two different siRNA against MARK3Par1c, a close family member of MARK2, we observed normal equatorial centering of spindles, similar to control siRNA–treated cells (Fig. S1, c and d). We conclude that MARK2 but not its close relative MARK3 is required for equatorial centering of the spindle.
Equatorial spindle centering is regulated differently in early and late mitosis
Biased rotation of the spindle occurs in early mitosis (Corrigan et al., 2013). We were intrigued to find that equatorially off-centered spindles of MARK2-depleted cells were observed primarily in early mitosis and these off-centered spindles were frequently restored back to the center in anaphase (Fig. S1 a). To confirm this, we quantitatively compared early and late mitotic spindle positions in MARK2-depleted cells and found that spindles were equatorially centered normally in anaphase but not early mitosis (Fig. 2 e). We conclude that at least two equatorial spindle-centering mechanisms operate in human cells, which are mitotic phase dependent. MARK2 is required for spindle centering in early mitosis when the spindle undergoes rotation.
Equatorial centering is required for proper spindle rotation, oscillation, and biased division plane
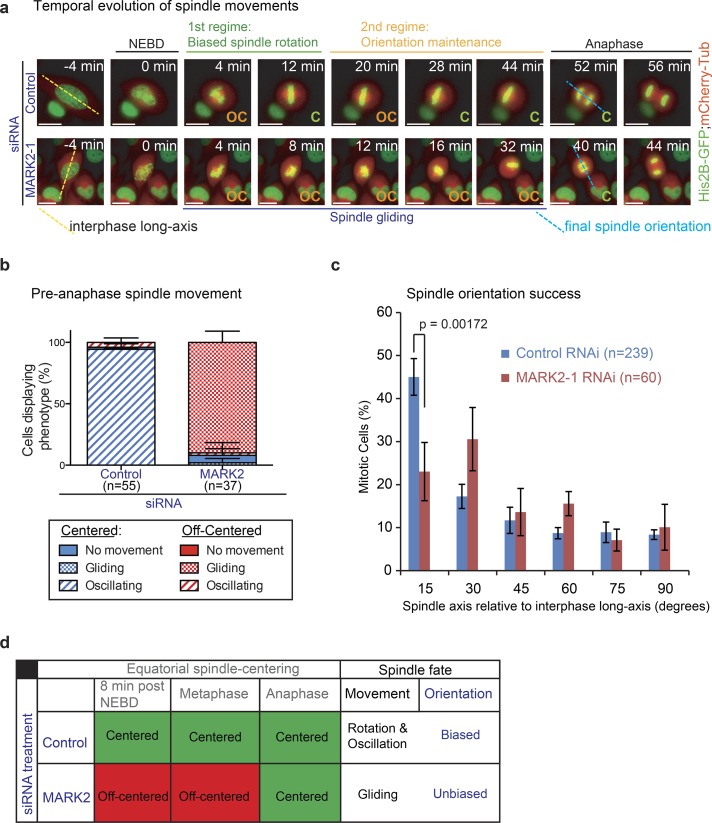
The plane of epithelial cell division is predetermined by interphase cell shape and cell–cell or cell–substrate contact sites (Chin et al., 2014). We reported previously that the mitotic spindle first rotates with a directional bias toward the interphase cell’s long axis (during prometaphase, when chromosomes congress) and then remains oriented along the long axis, where it undergoes spindle oscillations (until anaphase, when chromosomes segregate; Corrigan et al., 2013, 2015). Whether any of these key mitotic events are perturbed in cells that fail to equatorially center their spindles is not known. Hence, we studied whether MARK2-depleted cells displayed normal chromosome congression, chromosome segregation, spindle rotation, spindle oscillation, and spindle orientation along the long axis of the cell. Time-lapse videos of HeLaHis2B-GFP;mCherry-tubulin cells treated with MARK2 siRNA showed no significant difference in chromosome congression and segregation times compared with control cells (Fig. S2, a–d). However, the analysis of spindle movements indicated a striking reduction in spindle rotation or oscillatory pole-to-pole movements in MARK2-depleted cells compared with controls (Figs. 2 a and 3, a and b; Videos 1, 2, and 3). Importantly, despite the absence of pole-to-pole oscillations, spindles in MARK2-depleted cells were dynamic and displayed gliding movements along the cell cortex (Figs. 2 a and 3, a and b). Unlike the equatorially centered spindles in control cells that displayed biased rotation toward the interphase long axis (4–12 min; Fig. 3 a), the off-centered spindles in MARK2-depleted cells showed a prolonged duration of gliding motion along the cell cortex; thus, the absence of spindle centering disrupted normal spindle rotation. The gliding motion and lack of rotation was most obvious in MARK2-depleted cells when congression was occasionally delayed (Video 3). In summary, MARK2 depletion disrupts equatorial spindle centering, spindle rotation, and oscillations (Fig. 3 b), which are events that precede anaphase.
Figure 3.
Off-centered spindles in MARK2-depleted cells fail to rotate properly toward the predetermined spindle position at anaphase. (a) Representative time-lapse images of HeLaHis2B-GFP;mCherry-Tub cells treated with siRNA as indicated. Final spindle (pole-to-pole) axis at anaphase onset (blue dashed line) and interphase long axis (yellow dashed line) are shown. Biased spindle rotation (green bar) and spindle orientation maintenance and oscillation (orange bar) regimes, off-centered (OC) spindle gliding proximal to the cortex (blue bar) are all indicated. Bars, 10 μm. (b) Graph showing percentage of cells with centered (C) or off-centered spindles that displayed no movements (gliding or oscillatory) as assessed from time-lapse videos of HeLaHis2B-GFP;mCherry-Tub cells treated with control or MARK2-1 siRNA as indicated. (c) Distribution of final spindle orientation angles relative to the interphase long axis in cells as assessed from time-lapse videos of HeLaHis2B-GFP;mCherry-Tub cells treated with control or MARK2-1 siRNA as in (a). P-value was obtained using a proportion test on percentage values. Error bars are SEM values across three experimental repeats. (d) Table summarizing spindle position (i; equatorial centering success), spindle movement (ii; oscillatory, rotational, or gliding), and anaphase spindle orientation bias along interphase long axis (iii) in MARK2 or control siRNA–treated cells.
Because spindle centering, rotation, and oscillations were all disrupted in MARK2-depleted cells, we tested whether spindles could define the plane of cell division normally. To determine the efficiency with which cells could properly rotate their spindles and arrive at the final predefined position along the interphase long axis, we performed semiautomated analysis of our videos using our Spindle3D software (Corrigan et al., 2013). Analysis of final spindle orientation angles at the metaphase–anaphase transition showed a statistically significant reduction in the percentage of cells that correctly aligned the spindle along the interphase long axis after MARK2 depletion compared with control depletion (Fig. 3, c and d). Thus, MARK2 depletion induced spindle off centering is coincident with severe defects in both spindle rotation and identifying the correct plane of cell division (Fig. 3 d).
MARK2 depletion delays, but does not abrogate, mitotic cell rounding
Compared with control-depleted cells, MARK2-depleted cells showed a delay in mitotic cell rounding (Fig. S2 e). However, mitotic cell rounding was not completely abrogated as the vast majority of MARK2-depleted cells had completed mitotic rounding in late prometaphase (at least 8 min before anaphase onset; Fig. S2 e). In contrast, equatorial spindle centering remained severely compromised in late prometaphase MARK2-depleted cells (Fig. S2 f); at this stage, spindles were bipolar and normally oriented parallel to the substratum as assessed by spindle-pole positions (Fig. S2 g). Based on these analyses, we conclude that equatorial spindle off centering in MARK2-depleted cells is not directly caused by the delay in mitotic cell rounding.
MARK2 localizes to centrosomes and cell cortex, and its depletion alters mitotic microtubule growth and function
To understand the underlying reason for spindle off centering in MARK2-depleted cells, we next studied the localization of MARK2 in HeLa cells using YFP-tagged MARK2. YFP-MARK2 localized to both interphase and mitotic centrosomes independent of microtubules (Fig. S3). In mitotic cells, MARK2 distinctly localized to the cell cortex and faintly associated with the mitotic spindle in a microtubule-dependent manner (Figs. 3 d and S3, b and d).
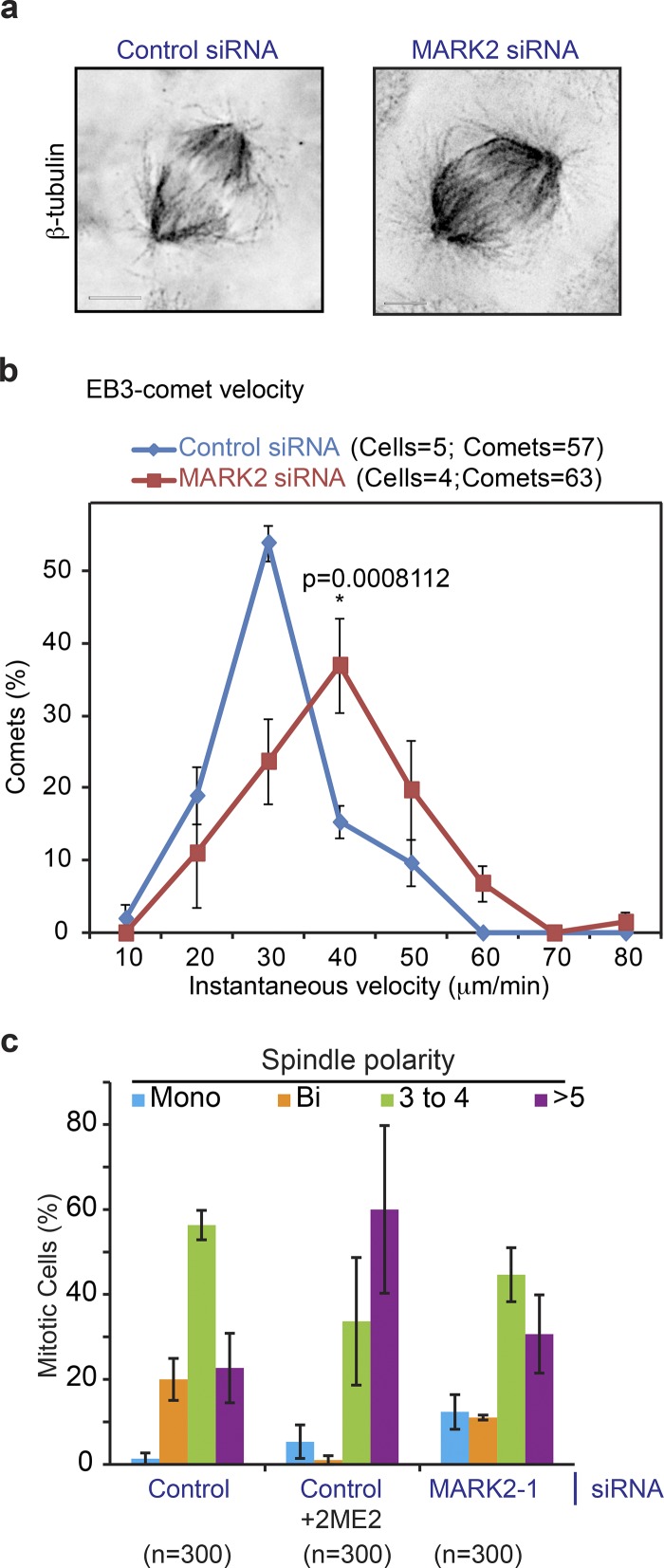
We next investigated whether depletion of MARK2 altered the distribution of astral microtubules in mitosis. After a brief exposure to ice-cold methanol for 60 s, we immunostained siRNA-treated cells using tubulin antibodies to assess the status of cold stable astral microtubules (Fig. S4 a). Compared with control siRNA–treated cells, MARK2 siRNA–treated cells showed a noticeable increase in astral microtubule length and density near poles (Figs. 4 a and S4 b). Mean lengths of cold-stable astral microtubules were 1.69 (SD = 0.61) μm (n = 6 cells) after control siRNA treatment and 3.27 (SD = 1.25) μm (n = 10 cells) after MARK2 siRNA treatment. Mean densities of cold-stable astral microtubules near the spindle pole area were ∼2 microtubules/23 μm2 (n = 10 cells) after control siRNA treatment and ∼6 microtubules/23 μm2 (n = 10 cells) after MARK2 siRNA treatment. These findings in mitotic cells are consistent with the reported interphase role of MARK2 as an enhancer of microtubule destabilization by controlling microtubule lifetime and growth rate (Nishimura et al., 2012).
Figure 4.
MARK2 regulates mitotic microtubule growth rate and function. (a) Single plane deconvolved images of cells immunostained with antibodies against anti–β-tubulin after control or MARK2-1 siRNA treatment. Bars, 5 µm. Grayscale image signals are inverted to highlight astral microtubule length and density. (b) Graph comparing the distribution of instantaneous velocities of EB3 comets in cells expressing EB3-mKate and treated with either MARK2 or control siRNA. Values were obtained manually by clicking spots using SoftWoRx software. Nonoverlapping peak values between control and MARK2 siRNA–treated cells signify statistically significant differences (*, P < 0.01) using the proportion test. Error bars are SEM values across cells from two independent repeats. (c) Bar graph shows frequency distribution of spindle polarity (based on the number of spindle poles: one [mono], two [bi], three to four, or more than five). Cells were treated with siRNA as indicated, exposed to 1.7 µM nocodazole for 3 h to depolymerize all microtubules, recovered for 10 min in nocodazole-free medium to reassemble spindles, and immunostained using α-tubulin antibody. 100 nM 2ME2 was added in the recovery medium of control siRNA–treated cells. Error bars represent SEM from three independent experiments.
Whether MARK2 regulates mitotic microtubule dynamics is not known. To address this, we depleted MARK2 in cells expressing EB3-TdTomato (HeLa EB3-TdTomato), a routine marker for growing mitotic microtubule ends (Shrestha et al., 2014; Iorio et al., 2015; Nakai et al., 2015; Tamura et al., 2015), and measured instantaneous velocity of growing microtubules in the metaphase spindle. We found a small but significant increase in mitotic microtubule growth rates in MARK2-depleted cells compared with control-depleted cells (Fig. 4 b). Because of the modest changes in microtubule growth rates after MARK2 depletion, we used a sensitive mitotic microtubule function assay to confirm the role of MARK2 in regulating mitotic microtubules. We treated cells with nocodazole to fully disassemble mitotic spindle microtubules and then washed off the drug to study the rate of spindle reassembly, which is dependent on microtubule function. As a positive control, nocodazole-washed-off cells were exposed to 2-methyl estradiol (2ME2), a microtubule-pausing drug (Corrigan et al., 2013) that is expected to delay bipolar spindle reassembly. To study bipolar spindle reassembly efficiency in the presence and absence of MARK2, we fixed and immunostained cells at various time points after nocodazole wash off. As expected, 10 min after nocodazole wash off, control cells form multipolar spindles that rapidly coalesce into a single bipolar spindle (Tulu et al., 2006). In contrast, MARK2 siRNA– or 2ME2-treated cells were delayed in coalescing multipolar spindles into bipolar ones compared with control siRNA–treated cells (Fig. 4 c), providing further evidence for the role of MARK2 in regulating mitotic microtubule function. We conclude that MARK2 kinase controls microtubule plus end growth rates, astral microtubule length, and microtubule function during mitosis.
Cortical dynein is diffuse, but spindle movement rates are unperturbed in MARK2-depleted cells
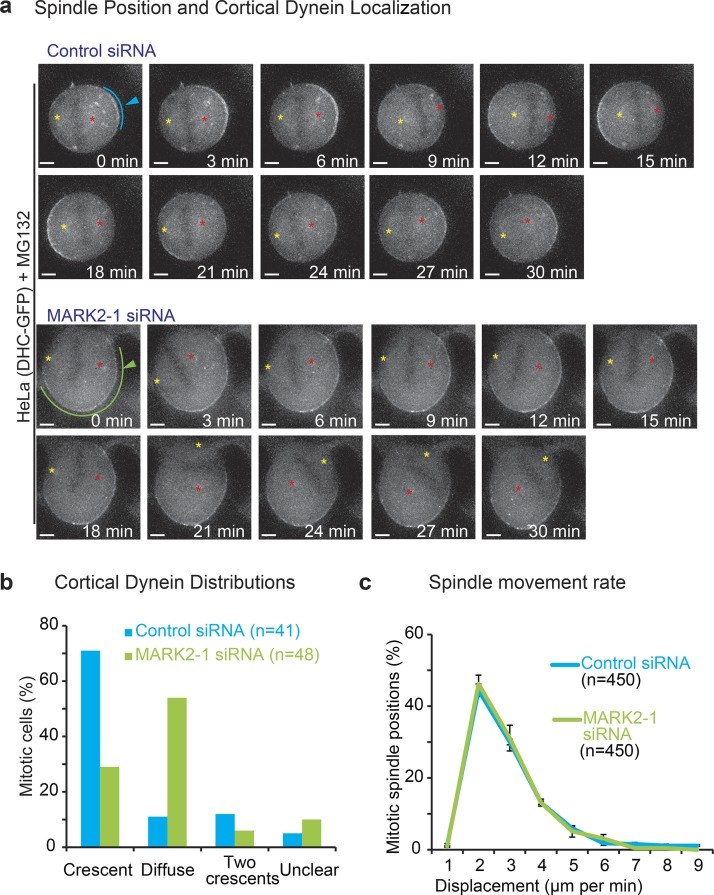
Cortical dynein is required for a stable 3D positioning of the spindle parallel to the substratum and also for spindle oscillations along the longitudinal pole-to-pole axis (Kiyomitsu and Cheeseman, 2012; Kotak et al., 2012; Corrigan et al., 2013). We therefore tested whether cortical dynein localization is perturbed after MARK2 depletion using a HeLaDHC1-GFP cell line expressing GFP-fused to mouse dynein heavy chain 1 (DHC1) under the control of its endogenous promoter (Poser et al., 2008).
Using DHC1-GFP localization at spindle poles, we once again confirmed equatorially off-centered spindles in HeLaDHC1-GFP cells treated with MARK2 siRNA (Fig. 5 a). As expected (Kiyomitsu and Cheeseman, 2012; Tame et al., 2014), in control siRNA–treated cells, DHC1-GFP signal at the cortex was asymmetrically enriched in a small region (Fig. 5 a, blue arrowhead; and Fig. 5 b, crescent) and was mostly excluded in regions proximal to the spindle pole (Fig. 5 a). However, in MARK2 siRNA–treated cells, DHC1-GFP signal at the cortex was distributed along a wider region compared with controls and lacked the crescent-like enrichment (Fig. 5 a, green arrowhead; and Fig. 5 b, diffuse), although DHC1-GFP was normally excluded at the cell cortex proximal to the spindle pole (Fig. 5 a). This normal exclusion of DHC1-GFP signal from cortical areas close to the spindle pole and chromosomes suggest that Plk1- and Ran-dependent mechanisms that exclude DHC-GFP from the cortex (Kiyomitsu and Cheeseman, 2012) are unlikely to be perturbed after MARK2 depletion (Fig. 5 a). Because chromosome and spindle pole positions can influence the position of cortical dynein localization (Kiyomitsu and Cheeseman, 2012), the diffuse cortical dynein localization could be a consequence of off-centered spindles in MARK2-depleted cells.
Figure 5.
Cortical dynein distributions, but not spindle movement rates, are perturbed after MARK2 depletion. (a) Representative images of DHC-GFP signals in HeLaDHC-GFP cells treated with the indicated siRNA from time-lapse videos acquired once every minute. Cells were arrested at metaphase with MG132 treatment immediately before imaging. Spindle poles are marked with yellow and red asterisks to follow spindle movements. Blue and green arrowheads refer to crescent or diffuse DHC-GFP signals, respectively. Bars, 3 µm. (b) Bar graph shows frequency distributions of DHC-GFP localizations in cell treated with the indicated siRNA oligonucleotides. Data collated over three independent experiments. (c) Graph shows frequency distribution of spindle pole displacement in HeLa cells treated with MG132 immediately before imaging. Values are obtained from time-lapse videos obtained in a. n refers to spindle positions acquired from at least 15 cells for each RNAi condition from three independent experiments. Distances are measured from the spindle pole furthest away from the cortex in the first frame, with the distance measured each subsequent frame for that specific pole. Error bars show SEM.
To test whether the diffuse cortical localization of dynein can impair the extent of spindle movement, we measured spindle displacement toward the cortex in control and MARK2-depleted HeLaDHC1-GFP cells using DHC-GFP as a spindle pole marker. Comparing the distribution of spindle pole displacement between control and MARK2-depleted cells showed no significant difference (Fig. 5 c). Thus, despite the equatorial off centering of spindles and diffuse localization of cortical dynein, spindle displacement extent by itself is not noticeably impaired in MARK2-depleted cells, suggestive of comparable pulling toward the cortex in the presence and absence of MARK2.
Spindle off centering in MARK2-depleted cells is not caused by excessive pulling by cortical dynein
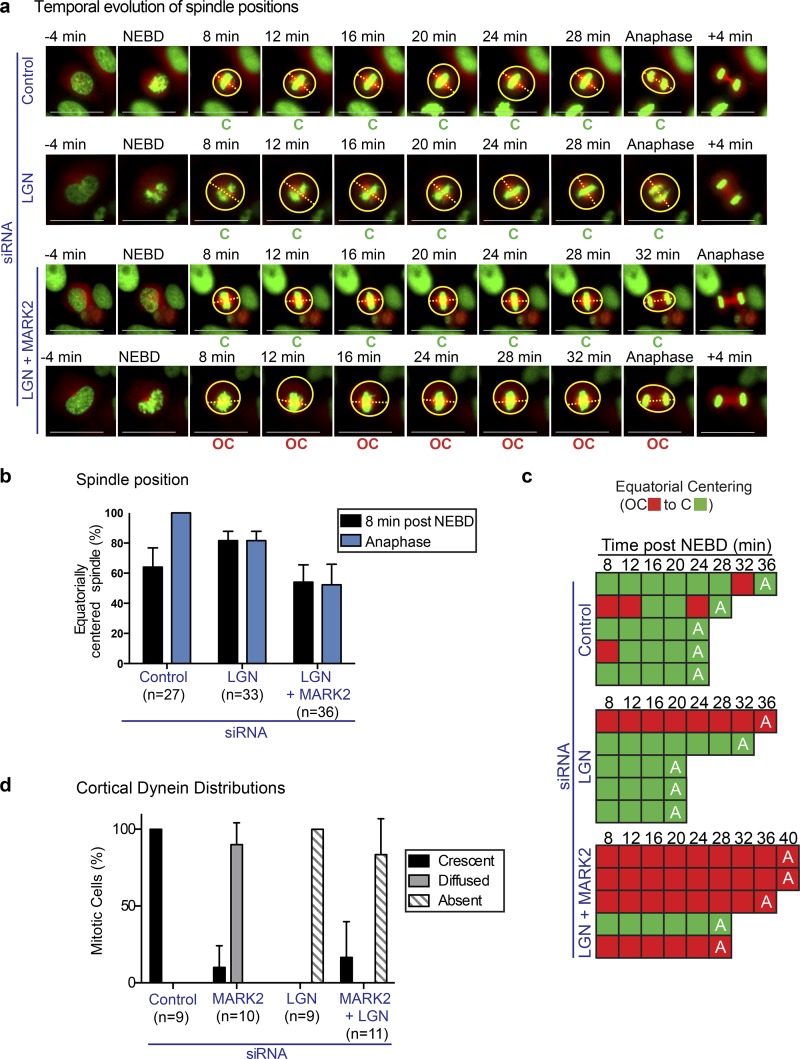
Longer astral microtubules in MARK2-depleted cells compared with control cells (Fig. 4 a) raised the possibility that spindle off-centering may arise from prolonged pulling of microtubules by cortical dynein, which may have been missed in our spindle displacement research. Therefore, we codepleted LGN (cortical dynein platform) and MARK2 and used time-lapse microscopy to assess equatorial centering of spindles in the absence of cortical dynein. We first confirmed that cortical dynein is absent in LGN and MARK2–codepleted HeLaDHC1-GFP cells (Fig. 6 d).
Figure 6.
Equatorial spindle off centering after MARK2 depletion is not caused by excessive pull by cortical dynein. (a) Representative time-lapse images of HeLaHis2B-GFP;mCherry-Tub cells treated with siRNA as indicated. Equatorially centered (C) and off-centered (OC) spindles are marked. mCherry-tubulin signal is used to outline cell cortex (circles) and pole-to-pole axis is marked with a dashed line. Bars, 15 μm. (b) Graph of percentage of equatorially centered spindles in cells treated with control or LGN with MARK2-1 siRNA as assessed from time-lapse videos as in a. Spindle positions 8 min after NEBD or at anaphase are reported. Error bars are SEM values across three experimental repeats. (c) Temporal evolution of spindle positions in five representative cells, treated with siRNA as indicated assessed from time-lapse videos as in a. A, anaphase. Red and green boxes indicate equatorially off-centered and centered spindle states, respectively. (d) Bar graph shows frequency distributions of DHC-GFP localizations in HeLaDHC-GFP cells treated with the indicated siRNA oligonucleotides as assessed from time-lapse videos acquired once every minute. Error bars are SEM values from data collated over three independent experiments.
As expected from a loss of cortical dynein phenotype, the spindles of LGN and MARK2–codepleted cells did not display much movement throughout mitosis (Fig. 6 a). Importantly, comparing the extent of spindle centering in LGN versus LGN and MARK2–codepleted HeLa cellsHis2B-GFP;mCherry-tubulin showed increased spindle off centering in codepleted cells (Fig. 6, a–c), confirming MARK2’s role in spindle centering. Because LGN depletion did not fully rescue the equatorial centering defect seen after MARK2 depletion (Fig. 6, a–c), we conclude that spindle off centering in MARK2-depleted cells is not simply caused by excessive pull by cortical dynein.
Importantly, those spindles born at the geometric center remained centered in MARK2 and LGN–codepleted cells, unlike MARK2-depleted cells, where spindles born at the geometric center become actively off centered (compare Fig. 6 a with Fig. 2 a). These data suggest that MARK2 activity plays an important unrecognized role in resisting an outwardly pull of the spindle by cortical dynein in specifically in early mitosis.
In summary, the persistence of off-centered spindles in cells codepleted of LGN and MARK2 demonstrate a role for MARK2 in equatorial centering of spindles independent of the LGN pathway. We conclude that equatorial off centering of spindles in MARK2-depleted cells is not caused by excessive pull by cortical dynein.
MARK2 loss induced spindle off centering can be rescued by MCAK depletion
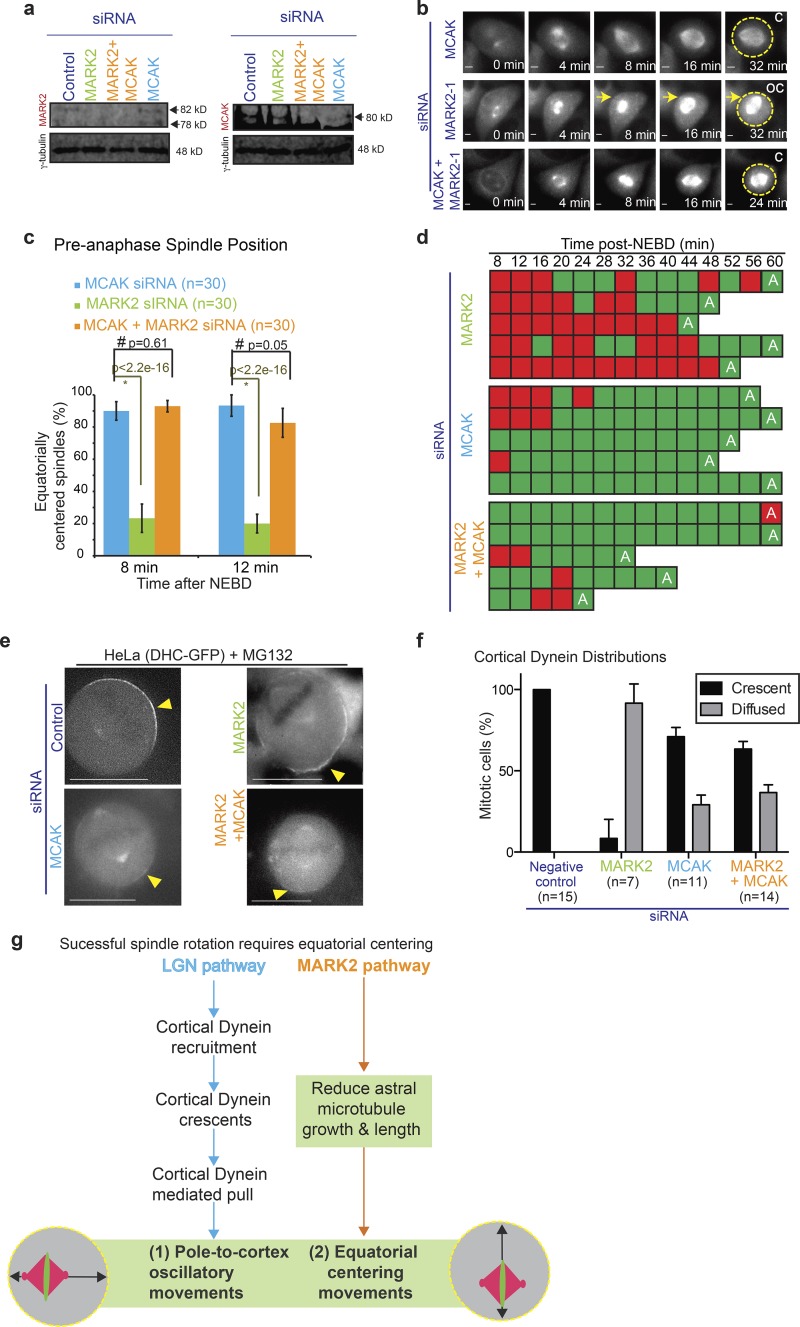
MARK2 depletion induced an increase in microtubule plus end growth rate. Therefore, we tested whether reducing plus end dynamics could rescue MARK2 depletion induced spindle off centering. For this purpose, we chose to deplete MCAK, a microtubule destabilizer that tracks with microtubule tips during mitosis (Domnitz et al., 2012). MCAK depletion causes an increase in astral microtubule stability (Rizk et al., 2009) and reduction in microtubule growth velocities (Braun et al., 2014). We had previously shown that MCAK is not required for spindle orientation success or rotational bias of spindles (Corrigan et al., 2013), but whether it is important for equatorial centering of the spindle was not known. We first analyzed spindle-centering success after MCAK depletion in HeLaHis2B-GFP;mCherry-tubulin cells. We observed no significant difference in equatorial spindle-centering extent between MCAK- and control-depleted cells (compare Fig. 7 b and Fig. 2 a, control), although as reported previously (Kline-Smith et al., 2004), chromosome congression time was prolonged and lagging chromosomes were seen in MCAK-depleted cells (mean NEBD to anaphase onset time: control siRNA, 30 min [n = 66 cells]; MCAK siRNA, 41 min [n = 117 cells]). We conclude MCAK is dispensable for the equatorial centering of mitotic spindles.
Figure 7.
Aberrant equatorial centering and dynein distribution after MARK2 depletion can be rescued by MCAK depletion. (a) Immunoblots show depletion extent of MARK2 and MCAK in lysates of HeLaHis2B-GFP;mCherry-Tub cells after siRNA treatment with siRNA oligonucleotides as indicated. γ-Tubulin is used as loading control. (b) Representative time-lapse images of HeLaHis2B-GFP;mCherry-Tub cells treated with siRNA against MARK2 or MCAK alone or both MCAK and MARK2. Equatorially centered (C) and off-centered (OC) spindles are marked. mCherry-tubulin signal alone is shown and the signal was used to outline the cell cortex (circles). Bars, 2 μm. (c) Graph of percentage of equatorially centered spindles assessed from time-lapse videos as in b. Error bars are SEM values across three experimental repeats. (d) Temporal evolution of spindle positions in five representative cells treated with indicated siRNA showing normal recentering of equatorially off-centered spindles in cells treated with siRNA against either MCAK alone or both MCAK and MARK2 but not MARK2 alone. A, anaphase. Red and green boxes indicate equatorially off-centered and centered spindles, respectively. (e) Representative live-cell images of HeLaDHC-GFP cells treated with the indicated siRNA oligonucleotides. Yellow arrowheads mark cortical DHC-GFP localization. Bars, 15 μm. (f) Bar graph shows percentage of HeLaDHC-GFP cells with crescent or diffused cortical dynein localization after treatments with the indicated siRNA oligonucleotides. Error bars are SEM values across data collated over three independent experiments. (g) Successful spindle rotation requires equatorial centering: Cartoon shows two separate pathways governing equatorial and longitudinal spindle positions: (i) LGN directly controls cortical dynein recruitment and, in turn, cortical dynein–mediated pull that powers spindle displacement (blue arrows). (ii) In contrast, MARK2/Par1b controls astral microtubule length and dynamics and ensures equatorial spindle centering (orange arrows). Both equatorial centering mediated by MARK2 and cortical pulling powered by dynein are needed for successful spindle rotation. Equatorial off centering and longitudinal oscillations have an indirect impact on cortical dynein crescent formation.
To test whether MCAK depletion could rescue MARK2 depletion–induced off centering of spindles, we analyzed spindle positions in time-lapse videos of cells codepleted of MCAK and MARK2. The percentage of cells with equatorially centered spindles at 8 and 12 min after NEBD was significantly increased after the codepletion of MCAK and MARK2 compared with MARK2 depletion alone, indicating a rescue of the spindle off-centering phenotype induced by MARK2 depletion (Fig. 7, a–d).
Although the spindle off-centering phenotype was rescued, the mitotic cell rounding delay remained until late prometaphase in cells codepleted of MARK2 and MCAK (Fig. S4 f). These findings show that the depletion of MCAK specifically rescues the spindle-centering defect but not the mitotic cell rounding delay in MARK2-depleted cells.
We investigated whether MCAK depletion rescued spindle off centering in MARK2-depleted cells simply by further increasing the length of astral microtubules. For this purpose, we briefly exposed cells to cold and immunostained siRNA-treated cells using tubulin antibodies to assess the status of cold stable astral microtubules (Fig. S4 a). Analyzing the length of astral microtubules relative to pole–cortex distance in centered and off-centered spindles showed a comparable increase in astral microtubule occupancy in cells depleted of MARK2, MCAK, or both compared with controls (Fig. S4, b–d). These findings show that the rescue of spindle centering is not simply caused by a dramatic increase in the lengths or densities of astral microtubules in MCAK and MARK2–codepleted cells. Moreover, using immunostaining, we did not observe any obvious change in colocalization between MCAK and EB3 (a microtubule plus end marker) after MARK2 depletion (Fig. S4 e), ruling out a direct role for MARK2 in altering MCAK localization or function.
Detailed analysis of spindle positions once every 4 min after NEBD (Fig. 7 d) showed that the rescue of spindle centering is dynamic; in conditions where MCAK alone or both MCAK and MARK2 were depleted, off-centered spindles were rapidly restored back to the center, unlike in MARK2-depleted cells. These data show that MARK2 depletion–induced equatorial off centering of spindles is being dynamically rescued by codepleting the microtubule depolymerizer MCAK. Importantly, MCAK codepletion rescued the diffuse cortical dynein localization observed in MARK2-depleted cells (Fig. 7, e and f), showing that cortical dynein crescents are reliant on equatorial spindle-centering status. We conclude MARK2 depletion–induced equatorial spindle off centering is dynamically rescued by MCAK depletion, highlighting the importance of MARK2-mediated modulation of microtubule dynamics in the equatorial centering of spindles.
Discussion
By tracking the temporal evolution of spindle positions, we demonstrate that proper rotation of the mitotic spindle requires two closely related mechanisms: MARK2-mediated spindle centering and cortical dynein–mediated spindle pulling. Although dynein-mediated pull powers the movement of spindles, we show that MARK2 is needed to restore an off-centered spindle back to the center of the cell; a loss of either of these pathways will disrupt spindle rotation, leading to an incorrect division plane. By showing that MARK2’s centering role is dependent on microtubule dynamics, our findings reveal a previously unrecognized spindle-centering mechanism that is crucial for successful spindle rotation. We present a dynamic model wherein the rotation of mitotic spindle to a predetermined position is dependent on both cortical dynein–mediated spindle movement and MARK2-mediated spindle-centering mechanisms.
In vitro research of microtubule asters devoid of rotation shows that although dynein can support centration, it is not needed for centering the asters (Laan et al., 2012). In worm embryos, cortical force generation in fact antagonizes metaphase spindle centration (Garzon-Coral et al., 2016). In agreement, our in vivo research of LGN-depleted cells show that cortical dynein is dispensable for spindle centering, in metaphase, along both the equatorial and longitudinal axes. Therefore, cortical dynein is not required for equatorial spindle centering in human cells but is instead required for dislodging the spindle from its position as part of the spindle rotation process (Fig. 7 g). Thus, by using MARK2 depletion as a molecular probe, we have uncovered an essential role for equatorial spindle centering in forming cortical dynein crescents and, in turn, mediating proper spindle rotation.
We find that cold-stable astral microtubule length and density are increased in MARK2-depleted cells, similarly to cells depleted of MCAK, a potent microtubule destabilizer (Tamura and Draviam, 2012) revealing MARK2’s significant role in regulating astral microtubules. During interphase, MARK2 regulates the levels of MAP2 and Tau proteins on the microtubule (Drewes et al., 1995, 1997; Illenberger et al., 1996), whereas the C terminus of MCAK by itself regulates its binding to the ends of microtubules (Talapatra et al., 2015). Consistent with this difference in regulation, we find that MARK2 depletion increases microtubule growth speed, whereas MCAK depletion reduces microtubule growth velocities (Braun et al., 2014). These two microtubule regulatory pathways are likely to cross talk as the codepletion of MCAK dynamically rescues both the off-centered spindles and loss of cortical dynein crescents in MARK2-depleted cells.
We hypothesize that microtubule ends are sufficient, given certain physical constraints, to drive cell-shape–associated spindle centering in human cells. A similar conclusion has been drawn for nuclear centering (Zhao et al., 2012) and centrosome centering in interphase cells (Zhu et al., 2010) and spindle centering in worm embryos (Garzon-Coral et al., 2016). In this context, our discovery of MARK2’s role in controlling mitotic microtubule dynamics is important. The MARK family of kinases was originally identified in a screen for kinases that phosphorylate microtubule-associated proteins (Tau, MAP2, and MAP4) and trigger microtubule disruption in interphase (Drewes et al., 1995, 1997; Illenberger et al., 1996). Subsequently, MARK-mediated regulation of microtubule dynamics during interphase was characterized in several cell lines (Cohen et al., 2004; Mandelkow et al., 2004; Schaar et al., 2004; Hayashi et al., 2011; Nishimura et al., 2012). Our findings extend this interphase role of MARK2 to mitosis for the first time. Previously, it was not known whether the MARK family played any role in dividing human epithelial cells, whether through its regulation of microtubule dynamics or otherwise.
Finally, we provide the first evidence for a cell cycle–dependent role for MARK2 in spindle centering during early mitosis, when spindle rotation occurs. Several microtubule-associated proteins (EB1 and APC), actin meshwork regulators (myosin-X, Abl1, PI(3)K, PAK2, LIMK1, β1 integrin, and Cdc42) are known to be important for spindle orientation (Draviam et al., 2006; Toyoshima and Nishida, 2007; Toyoshima et al., 2007; Kaji et al., 2008; Mitsushima et al., 2009; Samora et al., 2011; Dunsch et al., 2012; Kiyomitsu and Cheeseman, 2012; Kotak et al., 2012; Matsumura et al., 2012), but it is not clear whether these are mitotic phase–specific roles. In addition, for previously reported spindle-centering regulators (moesin, LIMK1, and the microtubule-end–binding proteins EB1 and APC; Draviam et al., 2006; Carreno et al., 2008; Kaji et al., 2008; Kunda et al., 2008; Roubinet et al., 2011), it is not clear whether disrupting their role leads to spindle gliding instead of spindle rotation in a cell cycle–regulated manner. However, it is clear that several proteins that occupy microtubule ends are mitotic phase dependent (Syred et al., 2013; Tamura et al., 2015). Our study shows that depletion of MARK2Par1b alone but not LGN, PAR3, MARK3Par1c, or MCAK severely disrupts the equatorial centering in pre-anaphase spindles. In contrast, codepletion of LGN and MARK2 disrupts equatorial centering in post-anaphase spindles as well, showing the temporally separable roles of LGN and MARK2. How MARK2 regulates mitotic microtubule dynamics and restores spindle centering in a cell cycle–specific manner are important questions for the future.
MARK2 loss does not perturb chromosome congression or segregation, despite impairing spindle centering and spindle rotation. This shows that spindle-centering mechanisms can be exquisitely sensitive to microtubule dynamics compared with other microtubule-driven mitotic events. A wider and important biomedical implication of this study is that spindle centering, and hence cell fate decisions and in turn tissue organization (Chin et al., 2014; Patel et al., 2016), might be more readily disrupted relative to chromosomal stability after antimicrotubule cancer therapies.
Materials and methods
Cell culture and synchronization
HeLa cells were cultured in DMEM (Tergaonkar et al., 1997) supplemented with 10% FCS and antibiotics (penicillin and streptomycin) and plated onto glass-bottomed dishes (LabTek) or 13-mm round coverslips for imaging. For inhibition experiments, cells were treated with 10 µM MG132 (1748; Tocris). Cells were synchronized using a single 1 µg/ml aphidicolin block for 24 h and then released for 7 h before filming.
Cell line generation
The HeLahMARK2-siRES-YFP cell line was generated by transfecting a Tet-inducible expression vector encoding siRNA-resistant YFP-hMARK2 and followed by colony picking. Cell line generation procedures were followed according to the FRT/TO system protocol (Invitrogen).
Plasmid and siRNA transfections
HeLa cells were transfected with siRNAs or plasmid vectors as described previously (Shrestha et al., 2014). siRNA oligonucleotides used against MARK2 (5′-CCUCCAGAAACUAUUCCGCGAAGUA-3′ for MARK2-1 and 5′-UCUUGGAUGCUGAUAUGAACAUCAA-3′ for MARK2-2), LGN (Corrigan et al., 2013), MARK3 (5′-AUAUGUUGCGGUUCGCCGUUCCCGG-3′ for MARK3-2 and 5′-GCGGUAAACUCGACACGUU-3′ for MARK3-3), and PAR-3 (5′-CAACAGCUGGCUUCCUCAAGCAGAA-3′ for PAR3-1 and 5′-GCAAGAGGCUUAAUAUCCGACUUAA-3′ for PAR3-2) were purchased from GE Healthcare. Sequences of all plasmids are available upon request.
Live-cell time-lapse imaging and analysis
Cells were transfected with siRNA oligonucleotides or plasmid vectors, 48 or 24 h, respectively, before imaging and transferred to Leibovitz L15 medium (Invitrogen) for imaging at 37°C. To observe chromosome and spindle movements, images were acquired with exposures of 0.1 s from at least three Z planes 3 μm apart every 4 min for 5 h using a 40× 0.75 NA objective on an DeltaVision Core microscope (Applied Precision Ltd.) equipped with a Cascade2 camera under EM mode. For imaging plus end dynamics in HeLa EB3-TdTomato (gift from A. McAinsh and A. Straube, University of Warwick, Coventry, England, UK), images were acquired with exposures of 0.04 s from at least 10 Z planes 0.1 μm apart every 10 s for 5 min using a 100× 1.2 NA objective on the microscope described above. For imaging DHC-GFP signals, images were acquired with exposures of 0.04 s from at least 10 Z planes 0.1 μm apart every 1 min for 10 min using a 100× 1.2 NA objective on the microscope described above. For MARK2 colocalization studies, the centrosome marker (a fragment of pericentrin-tagged to RFP) was used (kind gift from J. Pines, University of Cambridge, Cambridge, England, UK). Time-lapse videos were analyzed manually using SoftWoRx. Spindles in HeLaHis-GFP;mCherry-Tub cells were visually scored as equatorially off centered when unequal distances were observed between the cell cortex and the two opposing edges of metaphase plate (histone-GFP signal) or the cell cortex and the two walls of the spindle at the equator (mCherry-tubulin signal). In HeLaDHC-GFP cells, the pole-to-pole axis and spindle walls (DHC-GFP signal) were used to similarly assess spindle centering at the equator.
Immunofluorescence and immunoblotting
For immunofluorescence, antibodies against GFP (1:1,000, 1181446001; Roche), MARK2 (1:500, H00002011-M01; Novus Biologicals), MARK3 (1:1,000, 9311; Cell Signaling Technology), PAR3 (1:1,000, 07–330; EMD Millipore), α-tubulin (1:500, Ab6160; Abcam), EB3 (Abcam), MCAK (Andrews et al., 2004), and β-tubulin (1:1,000, T4026; Sigma-Aldrich) were used. Images of immunostained cells were acquired using a 100× 1.2 NA objective on a DeltaVision Core microscope equipped with CoolSnap HQ Camera (Photometrics). For immunoblotting, antibodies against γ-tubulin (T6793; Sigma-Aldrich) and others indicated above were used. Immunoblots were developed using fluorescent secondary antibodies (LI-COR Biosciences), and fluorescent immunoblots were quantified using the Odyssey (LI-COR Biosciences) software.
Nocodazole wash-off assay
Cells were exposed to 1.7 µM nocodazole (Thermo Fisher Scientific) for 3 h to depolymerize microtubules. Nocodazole was removed by washing cells three times with warm PBS, and cells were allowed to recover for 10 min in nocodazole-free complete DMEM. Cells were fixed and immunostained using antitubulin antibody.
Statistical analysis
Error bars show SEM values obtained across experiments, cells, or kinetochores as indicated in legend. P-values representing significance were obtained using Mann-Whitney U test, proportion test, or paired sample t test.
Online supplemental material
Fig. S1 shows that MARK2 but not MARK3 is required for equatorial spindle centering. Fig. S2 shows normal chromosome congression and segregation in MARK2-depleted cells. Fig. S3 shows MARK2 localization in interphase and mitosis. Fig. S4 shows the rescue of MARK2 depletion–induced spindle off centering by codepleting MCAK. Videos 1, 2, and 3 show time-lapse deconvolution microscopy of spindle rotation or gliding movements in cells treated with control or MARK2 siRNA, respectively.
Supplementary Material
Acknowledgments
We thank the Pines group for the plasmid vector encoding pericentrin fragment and the McAinsh and Straube groups for the HeLa cell line expressing EB3-TdTomato. We thank Viviane Boilot and Nicholas Levin for support with image analysis.
This work was supported by a Cancer Research UK Career Development Award (C28598/A9787), Biotechnology and Biological Sciences Research Council Project grant (BB/R01003X/1), and a Queen Mary University of London Laboratory startup grant to V.M. Draviam, a Universiti Brunei Darussalam PhD studentship to I. Zulkipli, a Queen Mary University of London PhD studentship to M. Hart, a London Interdisciplinary Biosciences Consortium Biotechnology and Biological Sciences Research Council–Doctoral Training Partnerships PhD studentship to D. Dang (cosupervised by V.M. Draviam and N. Sastry; BB/M009513/1), and an Islamic Development Bank PhD studentship to P. Gul.
The authors declare no competing financial interests.
Author contributions: I. Zulkipli, J. Clark, M. Hart, and R.L. Shrestha designed and performed experiments and analyzed and interpreted data. P. Gul, T. Kasichiwin, I. Zulkipli, and D. Dang analyzed data. N. Sastry and V.M. Draviam cosupervised D. Dang. V.M. Draviam planned the study, discussed the experimental design, analyzed and interpreted data, and wrote the manuscript. V.M. Draviam and M. Hart edited the manuscript.
References
- Andrews P.D., Ovechkina Y., Morrice N., Wagenbach M., Duncan K., Wordeman L., and Swedlow J.R.. 2004. Aurora B regulates MCAK at the mitotic centromere. Dev. Cell. 6:253–268. 10.1016/S1534-5807(04)00025-5 [DOI] [PubMed] [Google Scholar]
- Bessone S., Vidal F., Le Bouc Y., Epelbaum J., Bluet-Pajot M.-T., and Darmon M.. 1999. EMK protein kinase-null mice: dwarfism and hypofertility associated with alterations in the somatotrope and prolactin pathways. Dev. Biol. 214:87–101. 10.1006/dbio.1999.9379 [DOI] [PubMed] [Google Scholar]
- Braun A., Dang K., Buslig F., Baird M.A., Davidson M.W., Waterman C.M., and Myers K.A.. 2014. Rac1 and Aurora A regulate MCAK to polarize microtubule growth in migrating endothelial cells. J. Cell Biol. 206:97–112. 10.1083/jcb.201401063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreno S., Kouranti I., Glusman E.S., Fuller M.T., Echard A., and Payre F.. 2008. Moesin and its activating kinase Slik are required for cortical stability and microtubule organization in mitotic cells. J. Cell Biol. 180:739–746. 10.1083/jcb.200709161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin H.M.S., Nandra K., Clark J., and Draviam V.M.. 2014. Need for multi-scale systems to identify spindle orientation regulators relevant to tissue disorganization in solid cancers. Front. Physiol. 5:278 10.3389/fphys.2014.00278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen D., Brennwald P.J., Rodriguez-Boulan E., and Müsch A.. 2004. Mammalian PAR-1 determines epithelial lumen polarity by organizing the microtubule cytoskeleton. J. Cell Biol. 164:717–727. 10.1083/jcb.200308104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins E.S., Balchand S.K., Faraci J.L., Wadsworth P., and Lee W.L.. 2012. Cell cycle-regulated cortical dynein/dynactin promotes symmetric cell division by differential pole motion in anaphase. Mol. Biol. Cell. 23:3380–3390. 10.1091/mbc.e12-02-0109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan A.M., Shrestha R.L., Zulkipli I., Hiroi N., Liu Y., Tamura N., Yang B., Patel J., Funahashi A., Donald A., and Draviam V.M.. 2013. Automated tracking of mitotic spindle pole positions shows that LGN is required for spindle rotation but not orientation maintenance. Cell Cycle. 12:2643–2655. 10.4161/cc.25671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan A.M., Shrestha R., Draviam V.M., and Donald A.M.. 2015. Modeling of Noisy Spindle Dynamics Reveals Separable Contributions to Achieving Correct Orientation. Biophys. J. 109:1398–1409. 10.1016/j.bpj.2015.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domnitz S.B., Wagenbach M., Decarreau J., and Wordeman L.. 2012. MCAK activity at microtubule tips regulates spindle microtubule length to promote robust kinetochore attachment. J. Cell Biol. 197:231–237. 10.1083/jcb.201108147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draviam V.M., Shapiro I., Aldridge B., and Sorger P.K.. 2006. Misorientation and reduced stretching of aligned sister kinetochores promote chromosome missegregation in EB1- or APC-depleted cells. EMBO J. 25:2814–2827. 10.1038/sj.emboj.7601168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewes G., Trinczek B., Illenberger S., Biernat J., Schmitt-Ulms G., Meyer H.E., Mandelkow E.M., and Mandelkow E.. 1995. Microtubule-associated protein/microtubule affinity-regulating kinase (p110mark): A novel protein kinase that regulates tau-microtubule interactions and dynamic instability by phosphorylation at the Alzheimer-specific site serine 262. J. Biol. Chem. 270:7679–7688. 10.1074/jbc.270.13.7679 [DOI] [PubMed] [Google Scholar]
- Drewes G., Ebneth A., Preuss U., Mandelkow E.M., and Mandelkow E.. 1997. MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell. 89:297–308. 10.1016/S0092-8674(00)80208-1 [DOI] [PubMed] [Google Scholar]
- Dunsch A.K., Hammond D., Lloyd J., Schermelleh L., Gruneberg U., and Barr F.A.. 2012. Dynein light chain 1 and a spindle-associated adaptor promote dynein asymmetry and spindle orientation. J. Cell Biol. 198:1039–1054. 10.1083/jcb.201202112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon-Coral C., Fantana H.A., and Howard J.. 2016. A force-generating machinery maintains the spindle at the cell center during mitosis. Science. 352:1124–1127. 10.1126/science.aad9745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S., and Kemphues K.J.. 1995. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell. 81:611–620. 10.1016/0092-8674(95)90082-9 [DOI] [PubMed] [Google Scholar]
- Hayashi K., Suzuki A., Hirai S., Kurihara Y., Hoogenraad C.C., and Ohno S.. 2011. Maintenance of dendritic spine morphology by partitioning-defective 1b through regulation of microtubule growth. J. Neurosci. 31:12094–12103. 10.1523/JNEUROSCI.0751-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman S.M., and Segal M.. 2005. Cortical capture of microtubules and spindle polarity in budding yeast - where’s the catch? J. Cell Sci. 118:463–471. 10.1242/jcs.01650 [DOI] [PubMed] [Google Scholar]
- Hurov J., and Piwnica-Worms H.. 2007. The Par-1/MARK family of protein kinases: from polarity to metabolism. Cell Cycle. 6:1966–1969. 10.4161/cc.6.16.4576 [DOI] [PubMed] [Google Scholar]
- Illenberger S., Drewes G., Trinczek B., Biernat J., Meyer H.E., Olmsted J.B., Mandelkow E.M., and Mandelkow E.. 1996. Phosphorylation of microtubule-associated proteins MAP2 and MAP4 by the protein kinase p110mark. Phosphorylation sites and regulation of microtubule dynamics. J. Biol. Chem. 271:10834–10843. 10.1074/jbc.271.18.10834 [DOI] [PubMed] [Google Scholar]
- Iorio F., Shrestha R.L., Levin N., Boilot V., Garnett M.J., Saez-Rodriguez J., and Draviam V.M.. 2015. A Semi-Supervised Approach for Refining Transcriptional Signatures of Drug Response and Repositioning Predictions. PLoS One. 10:e0139446 10.1371/journal.pone.0139446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji N., Muramoto A., and Mizuno K.. 2008. LIM kinase-mediated cofilin phosphorylation during mitosis is required for precise spindle positioning. J. Biol. Chem. 283:4983–4992. 10.1074/jbc.M708644200 [DOI] [PubMed] [Google Scholar]
- Kimura K., and Kimura A.. 2011. A novel mechanism of microtubule length-dependent force to pull centrosomes toward the cell center. Bioarchitecture. 1:74–79. 10.4161/bioa.1.2.15549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T., and Cheeseman I.M.. 2012. Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat. Cell Biol. 14:311–317. 10.1038/ncb2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline-Smith S.L., Khodjakov A., Hergert P., and Walczak C.E.. 2004. Depletion of centromeric MCAK leads to chromosome congression and segregation defects due to improper kinetochore attachments. Mol. Biol. Cell. 15:1146–1159. 10.1091/mbc.e03-08-0581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotak S., Busso C., and Gönczy P.. 2012. Cortical dynein is critical for proper spindle positioning in human cells. J. Cell Biol. 199:97–110. 10.1083/jcb.201203166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulukian A., and Fuchs E.. 2013. Spindle orientation and epidermal morphogenesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368:20130016 10.1098/rstb.2013.0016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunda P., Pelling A.E., Liu T., and Baum B.. 2008. Moesin controls cortical rigidity, cell rounding, and spindle morphogenesis during mitosis. Curr. Biol. 18:91–101. 10.1016/j.cub.2007.12.051 [DOI] [PubMed] [Google Scholar]
- Laan L., Pavin N., Husson J., Romet-Lemonne G., van Duijn M., López M.P., Vale R.D., Jülicher F., Reck-Peterson S.L., and Dogterom M.. 2012. Cortical dynein controls microtubule dynamics to generate pulling forces that position microtubule asters. Cell. 148:502–514. 10.1016/j.cell.2012.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macara I.G., Guyer R., Richardson G., Huo Y., and Ahmed S.M.. 2014. Epithelial homeostasis. Curr. Biol. 24:R815–R825. 10.1016/j.cub.2014.06.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelkow E.M., Thies E., Trinczek B., Biernat J., and Mandelkow E.. 2004. MARK/PAR1 kinase is a regulator of microtubule-dependent transport in axons. J. Cell Biol. 167:99–110. 10.1083/jcb.200401085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus S.M., and Lee W.L.. 2011. Regulated offloading of cytoplasmic dynein from microtubule plus ends to the cortex. Dev. Cell. 20:639–651. 10.1016/j.devcel.2011.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura S., Hamasaki M., Yamamoto T., Ebisuya M., Sato M., Nishida E., and Toyoshima F.. 2012. ABL1 regulates spindle orientation in adherent cells and mammalian skin. Nat. Commun. 3:626 10.1038/ncomms1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsushima M., Toyoshima F., and Nishida E.. 2009. Dual role of Cdc42 in spindle orientation control of adherent cells. Mol. Cell. Biol. 29:2816–2827. 10.1128/MCB.01713-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai Y., Ozeki M., Hiraiwa T., Tanimoto R., Funahashi A., Hiroi N., Taniguchi A., Nonaka S., Boilot V., Shrestha R., et al. . 2015. High-speed microscopy with an electrically tunable lens to image the dynamics of in vivo molecular complexes. Rev. Sci. Instrum. 86:013707 10.1063/1.4905330 [DOI] [PubMed] [Google Scholar]
- Nishimura Y., Applegate K., Davidson M.W., Danuser G., and Waterman C.M.. 2012. Automated screening of microtubule growth dynamics identifies MARK2 as a regulator of leading edge microtubules downstream of Rac1 in migrating cells. PLoS One. 7:e41413 10.1371/journal.pone.0041413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell C.B., and Wang Y.L.. 2000. Mammalian spindle orientation and position respond to changes in cell shape in a dynein-dependent fashion. Mol. Biol. Cell. 11:1765–1774. 10.1091/mbc.11.5.1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel H., Stavrou I., Shrestha R.L., Draviam V., Frame M.C., and Brunton V.G.. 2016. Kindlin1 regulates microtubule function to ensure normal mitosis. J. Mol. Cell Biol. 8:338–348. 10.1093/jmcb/mjw009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poser I., Sarov M., Hutchins J.R.A., Hériché J.-K., Toyoda Y., Pozniakovsky A., Weigl D., Nitzsche A., Hegemann B., Bird A.W., et al. . 2008. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat. Methods. 5:409–415. 10.1038/nmeth.1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizk R.S., Bohannon K.P., Wetzel L.A., Powers J., Shaw S.L., and Walczak C.E.. 2009. MCAK and paclitaxel have differential effects on spindle microtubule organization and dynamics. Mol. Biol. Cell. 20:1639–1651. 10.1091/mbc.e08-09-0985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roubinet C., Decelle B., Chicanne G., Dorn J.F., Payrastre B., Payre F., and Carreno S.. 2011. Molecular networks linked by Moesin drive remodeling of the cell cortex during mitosis. J. Cell Biol. 195:99–112. 10.1083/jcb.201106048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samora C.P., Mogessie B., Conway L., Ross J.L., Straube A., and McAinsh A.D.. 2011. MAP4 and CLASP1 operate as a safety mechanism to maintain a stable spindle position in mitosis. Nat. Cell Biol. 13:1040–1050. 10.1038/ncb2297 [DOI] [PubMed] [Google Scholar]
- Sato Y., Akitsu M., Amano Y., Yamashita K., Ide M., Shimada K., Yamashita A., Hirano H., Arakawa N., Maki T., et al. . 2013. The novel PAR-1-binding protein MTCL1 has crucial roles in organizing microtubules in polarizing epithelial cells. J. Cell Sci. 126:4671–4683. 10.1242/jcs.127845 [DOI] [PubMed] [Google Scholar]
- Sato Y., Hayashi K., Amano Y., Takahashi M., Yonemura S., Hayashi I., Hirose H., Ohno S., and Suzuki A.. 2014. MTCL1 crosslinks and stabilizes non-centrosomal microtubules on the Golgi membrane. Nat. Commun. 5:5266 10.1038/ncomms6266 [DOI] [PubMed] [Google Scholar]
- Schaar B.T., Kinoshita K., and McConnell S.K.. 2004. Doublecortin microtubule affinity is regulated by a balance of kinase and phosphatase activity at the leading edge of migrating neurons. Neuron. 41:203–213. 10.1016/S0896-6273(03)00843-2 [DOI] [PubMed] [Google Scholar]
- Sheeman B., Carvalho P., Sagot I., Geiser J., Kho D., Hoyt M.A., and Pellman D.. 2003. Determinants of S. cerevisiae dynein localization and activation: implications for the mechanism of spindle positioning. Curr. Biol. 13:364–372. 10.1016/S0960-9822(03)00013-7 [DOI] [PubMed] [Google Scholar]
- Shrestha R.L., Tamura N., Fries A., Levin N., Clark J., and Draviam V.M.. 2014. TAO1 kinase maintains chromosomal stability by facilitating proper congression of chromosomes. Open Biol. 4:130108 10.1098/rsob.130108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman J.M., Benton R., and St Johnston D.. 2000. The Drosophila homolog of C. elegans PAR-1 organizes the oocyte cytoskeleton and directs oskar mRNA localization to the posterior pole. Cell. 101:377–388. 10.1016/S0092-8674(00)80848-X [DOI] [PubMed] [Google Scholar]
- Slim C.L., Lázaro-Diéguez F., Bijlard M., Toussaint M.J.M., de Bruin A., Du Q., Müsch A., and van Ijzendoorn S.C.D.. 2013. Par1b induces asymmetric inheritance of plasma membrane domains via LGN-dependent mitotic spindle orientation in proliferating hepatocytes. PLoS Biol. 11:e1001739 10.1371/journal.pbio.1001739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syred H.M., Welburn J., Rappsilber J., and Ohkura H.. 2013. Cell cycle regulation of microtubule interactomes: multi-layered regulation is critical for the interphase/mitosis transition. Mol. Cell. Proteomics. 12:3135–3147. 10.1074/mcp.M113.028563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talapatra S., Harker B., and Welburn J.. 2015. The C-terminal region of the motor protein MCAK controls its structure and activity through a conformational switch. eLife. 4:e06421 10.7554/eLife.06421.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tame M.A., Raaijmakers J.A., van den Broek B., Lindqvist A., Jalink K., and Medema R.H.. 2014. Astral microtubules control redistribution of dynein at the cell cortex to facilitate spindle positioning. Cell Cycle. 13:1162–1170. 10.4161/cc.28031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura N., and Draviam V.M.. 2012. Microtubule plus-ends within a mitotic cell are ‘moving platforms’ with anchoring, signalling and force-coupling roles. Open Biol. 2:120132 10.1098/rsob.120132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura N., Simon J.E., Nayak A., Shenoy R., Hiroi N., Boilot V., Funahashi A., and Draviam V.M.. 2015. A proteomic study of mitotic phase-specific interactors of EB1 reveals a role for SXIP-mediated protein interactions in anaphase onset. Biol. Open. 4:155–169. 10.1242/bio.201410413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ten Hoopen R., Cepeda-García C., Fernández-Arruti R., Juanes M.A., Delgehyr N., and Segal M.. 2012. Mechanism for astral microtubule capture by cortical Bud6p priming spindle polarity in S. cerevisiae. Curr. Biol. 22:1075–1083. 10.1016/j.cub.2012.04.059 [DOI] [PubMed] [Google Scholar]
- Tergaonkar V., Mythily D.V., and Krishna S.. 1997. Cytokeratin patterns of expression in human epithelial cell lines correlate with transcriptional activity of the human papillomavirus type 16 upstream regulatory region. J. Gen. Virol. 78:2601–2606. 10.1099/0022-1317-78-10-2601 [DOI] [PubMed] [Google Scholar]
- Toyoshima F., and Nishida E.. 2007. Integrin-mediated adhesion orients the spindle parallel to the substratum in an EB1- and myosin X-dependent manner. EMBO J. 26:1487–1498. 10.1038/sj.emboj.7601599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima F., Matsumura S., Morimoto H., Mitsushima M., and Nishida E.. 2007. PtdIns(3,4,5)P3 regulates spindle orientation in adherent cells. Dev. Cell. 13:796–811. 10.1016/j.devcel.2007.10.014 [DOI] [PubMed] [Google Scholar]
- Tulu U.S., Fagerstrom C., Ferenz N.P., and Wadsworth P.. 2006. Molecular requirements for kinetochore-associated microtubule formation in mammalian cells. Curr. Biol. 16:536–541. 10.1016/j.cub.2006.01.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wühr M., Tan E.S., Parker S.K., Detrich H.W. III, and Mitchison T.J.. 2010. A model for cleavage plane determination in early amphibian and fish embryos. Curr. Biol. 20:2040–2045. 10.1016/j.cub.2010.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T., Graham O.S., Raposo A., and St Johnston D.. 2012. Growing microtubules push the oocyte nucleus to polarize the Drosophila dorsal-ventral axis. Science. 336:999–1003. 10.1126/science.1219147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Burakov A., Rodionov V., and Mogilner A.. 2010. Finding the cell center by a balance of dynein and myosin pulling and microtubule pushing: a computational study. Mol. Biol. Cell. 21:4418–4427. 10.1091/mbc.e10-07-0627 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.