Sivakumar and Kurpios review how transcription regulates cell shape changes during organ development in embryogenesis.
Abstract
The emerging field of transcriptional regulation of cell shape changes aims to address the critical question of how gene expression programs produce a change in cell shape. Together with cell growth, division, and death, changes in cell shape are essential for organ morphogenesis. Whereas most studies of cell shape focus on posttranslational events involved in protein organization and distribution, cell shape changes can be genetically programmed. This review highlights the essential role of transcriptional regulation of cell shape during morphogenesis of the heart, lungs, gastrointestinal tract, and kidneys. We emphasize the evolutionary conservation of these processes across different model organisms and discuss perspectives on open questions and research avenues that may provide mechanistic insights toward understanding birth defects.
Introduction
Morphogenesis, or the process of shape formation, requires precise spatial coordination of a limited repertoire of cellular behaviors such as oriented cell division, polarized growth, directional migration, differentiation, and cell death (Table 1). Understanding these mechanisms of cell shape changes is therefore fundamental to understanding organ morphogenesis.
Table 1. Changes in epithelial cell shape are central to morphogenesis.
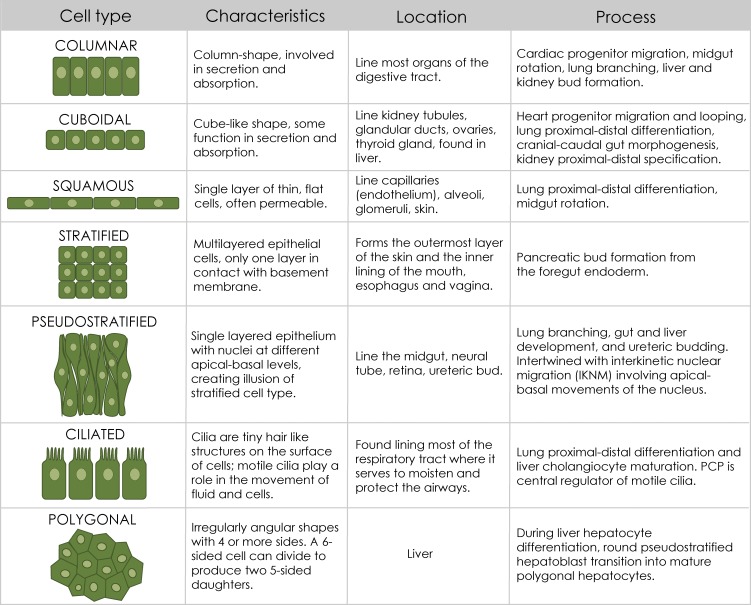 |
Major fundamental cell shapes discussed in this review are depicted. All epithelia have a typical ABP, but their morphologies range from flat or squamous, to cuboidal or columnar. Epithelia can either consist of a single cell layer, referred to as simple epithelia, or host multiple cell layers, known as stratified epithelia. In pseudostratified epithelium, cells exist in a single layer, but their nuclei travel between apical and basal surfaces, a process known as IKNM. In vertebrates, most cells possess single nonmotile primary cilium, which serves as critical regulator of signal transduction during development and homeostasis. Whereas a cell’s shape is defined by a global observation, round, cuboidal, polygonal, etc., this review focuses on the transcriptional mechanisms by which a cell can change its shape to execute its function within a developing organ.
Cell shape in a cluster is the result of the interplay between cell–cell, cell–matrix adhesion, and cortical tension (Vogel and Sheetz, 2006; Lecuit and Lenne, 2007). While cortical tension is an isotropic regulator of cell shape, the distribution of the protein complexes involved in cell–matrix and cell–cell adhesion can be polarized and is primarily governed by the planar cell polarity (PCP) and apical–basal polarity (ABP) pathways. PCP, the orientation and alignment of cells within a sheet, involves proteins encoded by PCP genes that establish geometric states within a cell to orient cellular behaviors along the plane of a cell sheet (reviewed in Karner et al., 2006; Seifert and Mlodzik, 2007; Wallingford, 2012). These behaviors include convergent extension (Keller et al., 2000; Keller, 2006), oriented cell division (Williams and Fuchs, 2013), directional migration (Carmona-Fontaine et al., 2008), and cellular rearrangements such as directed intercalation and polarized ciliary beating (Wallingford, 2010, 2012). The ABP pathway involves evolutionarily conserved asymmetrically localized multiprotein complexes that demarcate the boundary between the apical, lateral, and basal membranes, forming specialized epithelial surfaces (reviewed in Macara, 2004; Mellman and Nelson, 2008; Elsum et al., 2012).
Embryonic organ development is driven by the coordination and alignment of local cellular behaviors with the anteroposterior, dorsoventral, and left–right (LR) axes (Bakkers et al., 2009). Embryonic spatiotemporal patterning is largely conserved across evolution and is governed by tissue-specific gene regulatory networks, which ultimately regulate PCP and ABP. Early studies of cell shape changes provided significant insight on protein trafficking and cytoskeleton rearrangements of the structurally and functionally distinct apical and basal–lateral plasma membrane domains and on the role of extracellular cues in initiating and orienting cellular reorganization (Le Bivic et al., 1990; Matter et al., 1990; Yeaman et al., 1999). However, cell shape changes are also programmed at the level of the genome (Halbleib et al., 2007). Moreover, PCP coordinates morphogenetic behaviors of individual cells and cell populations with global patterning information (Gray et al., 2011). Here we discuss emerging studies of the role of transcriptional regulation of cell shape changes during organ morphogenesis. We review the developmental processes and underlying cell shape changes involved in morphogenesis of the heart, lungs, stomach, intestine, pancreas, liver, and kidneys. Knowledge from different model organisms has been integrated to bridge the link between the transcriptional machinery and cell shape changes driving organ formation.
Transcriptional regulation of cell shape during heart development
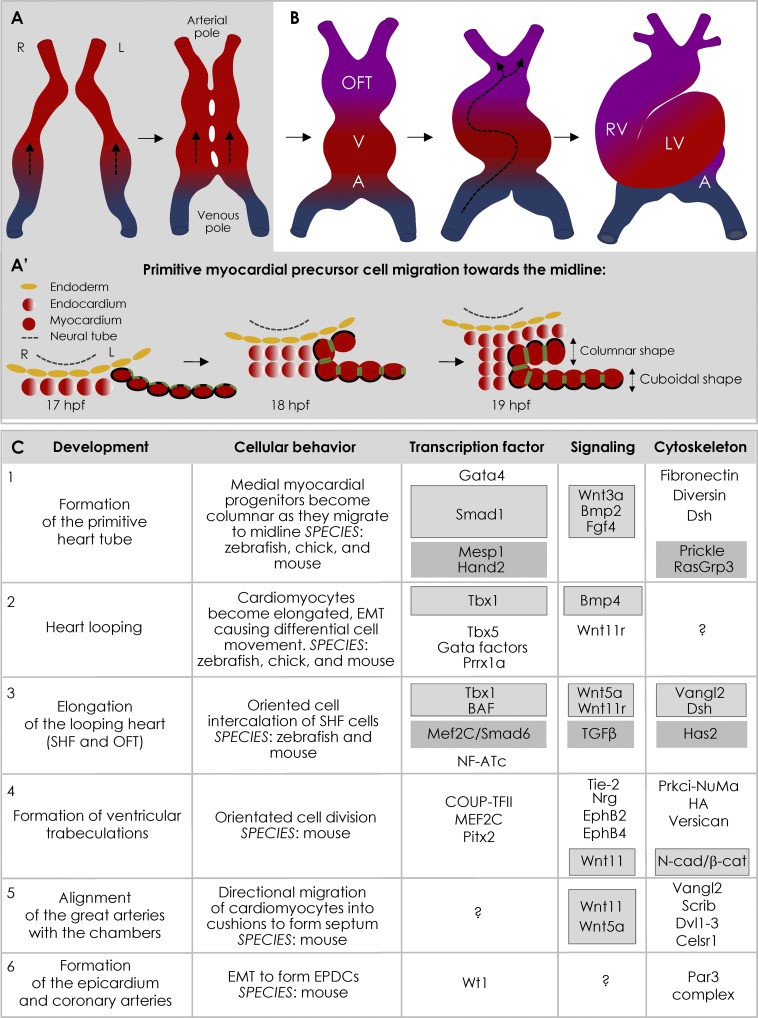
The heart is the first organ to function during vertebrate embryogenesis. The muscular (myocardial) layer and the endothelial (endocardial) layer of the adult heart are derived from bilateral populations of mesodermal cardiac precursor cells in the lateral mesoderm (Stainier, 2001; McFadden and Olson, 2002; Evans et al., 2010; Fig. 1, A and B). These migrate and fuse at the embryonic midline forming the linear primary heart tube, which subsequently transforms into a looped, multichambered, valved organ (Fig. 1, A and B).
Figure 1.
Cellular processes during heart development. (A and B) Migration of the left (L) and right (R) cardiac precursors and their fusion at the midline forms primary heart tube (A). Addition of the SHF cardiac progenitors (purple) transforms the primitive heart tube into a looped, multichambered, and valved organ (B). Red marks arterial pole, blue is venous pole, and arrows represent the direction of blood flow. A, atrium; V, ventricle. (A′) In zebrafish, the migrating myocardial precursors change shape from cuboidal (lateral cells) to columnar (closest to midline). aPKC staining (green) is more prominent in the medial cells and restricted to the apicolateral membranes, while β-catenin (black) localizes to the basolateral membranes. hpf, hours post fertilization. (C) Transcriptional and signaling pathways that regulate the cellular processes in the forming heart. Linked TFs, their downstream pathways, and cytoskeletal regulators have been similarly boxed. In example 3, Tbx1 regulates Wnt5a and Wnt11r, which regulate Vangl2 and Dsh to cause addition of SHF progenitors by oriented intercalation. Genes without any known interactors are left unboxed.
The cell shape changes pertinent to heart development are observed throughout its morphogenesis, with spatial and temporal insights into the critical molecular players obtained from a range of studies in zebrafish, chicken, and mouse.
Cell shape changes during primitive heart tube formation
As the left and right cardiac precursors migrate toward the midline, the medial myocardial progenitor cells become columnar and adopt a polarized epithelial organization, with a basolateral distribution of β-catenin, apical localization of atypical PKC (aPKC) iota, and junctional localization of zonula occludens (ZO-1; Trinh and Stainier, 2004a; Rohr et al., 2006; Fig. 1 A′). In contrast, the lateral myocardial progenitors remain mostly cuboidal. Midline migration defects result in cardia bifida, the formation of two hearts (Garavito-Aguilar et al., 2010).
In chicken and mouse embryos, Wnt3A and the BMP2 pathways converge to regulate the transcription factor (TF) SMAD-1, ensuring proper migration of cardiac progenitors (Yue et al., 2008; Song et al., 2014; Fig. 1 C). The GATA4 TF also regulates cardiac progenitor migration, although its cell-intrinsic roles in regulating shape are unknown (Kuo et al., 1997; Fig. 1 C).
Mesp1, a homeobox TF, acts cell autonomously to coordinate the specification and polarity of myocardial progenitors as they move toward the midline (Chiapparo et al., 2016). Directionality of this movement is regulated by Prickle1, a core PCP component, while migration speed is governed by the Mesp1 target RasGRP3, a critical regulator of cell motility and ERK signaling (Chiapparo et al., 2016; Kelly, 2016; Fig. 1 C).
In zebrafish, the PCP components Diversin and Dishevelled (Dsh), as well as deposition of fibronectin, are the primary downstream regulators of cell shape changes that accompany cardiac precursor migration (Trinh and Stainier, 2004b; Moeller et al., 2006); however, their transcriptional regulation remains unidentified (Fig. 1 C). Similarly, whereas fibronectin deposition to establish the myocardial ABP is driven by the TF Hand2 (Trinh et al., 2005), the downstream signaling events that regulate this polarity are unknown (Fig. 1 C).
Cell shape changes during initiation of heart looping
As the cardiac progenitors migrate to the midline, they fuse to form the cardiac cone (Trinh and Stainier, 2004a; Holtzman et al., 2007). The cardiac cone undergoes a leftward displacement as it extends, known as cardiac “jogging,” after which the linear heart tube bends at the boundary between the ventricle and the atrium to create the S-shaped loop (Wittig and Münsterberg, 2016; Fig. 1).
At the onset of looping, cardiomyocytes change from small and rounded (cuboidal and isotropic) to a flattened and elongated shape in the outer curvature (Taber, 2001; Hosseini et al., 2017). Tbx1, a T-box TF, is the cell-intrinsic master regulator of cardiomyocyte shape during looping (Choudhry and Trede, 2013). In Tbx1−/− zebrafish, cells in the outer and inner curvatures retain isotropic morphology and do not elongate. Tbx1 directly targets Wnt11r to regulate these cellular changes; however, the cellular effectors remain unknown (Choudhry and Trede, 2013; Fig. 1 C).
In chickens and mice, heart looping initiates with the dorsal closure of the heart tube and the simultaneous disappearance of the dorsal mesocardium of the splanchnic mesoderm (Goenezen et al., 2012; Wittig and Münsterberg, 2016). Differential growth (Taber, 2001; Wittig and Münsterberg, 2016; Hosseini et al., 2017) and changes in cardiomyocyte shape (Taber, 2001; Hosseini et al., 2017) as well as external forces from neighboring tissues (Männer, 2000; Loots et al., 2003) are involved in heart looping. In zebrafish, the TF Prrx1a, an inducer of epithelial-to-mesenchymal transition downstream of BMP, was recently identified as the driver of differential cell movement of cardiac progenitors toward the midline, leading to a leftward displacement of the cardiac posterior pole through an actomyosin-dependent process (Ocaña et al., 2017). In the mouse, SNAIL1 is thought to act in a similar manner to the zebrafish Prrx1a, suggesting a conserved mechanism (Ocaña et al., 2017).
Cell shape changes in the second heart field (SHF)
Subsequent morphogenesis of the heart occurs by addition of SHF cardiac progenitor cells, located in the splanchnic mesoderm (Kelly et al., 2001; Mjaatvedt et al., 2001; Waldo et al., 2001; Cai et al., 2003). SHF populations contribute to the formation of right ventricle and outflow tract (OFT) at the arterial pole and atrial myocardium at the venous pole (Kelly et al., 2001; Vincent and Buckingham, 2010; Fig. 1 B). Perturbation to SHF can lead to severe morphological anomalies including OFT malformation, one of the most common congenital heart defects in humans (Šamánek, 2000; Hoffman and Kaplan, 2002; Dyer and Kirby, 2009).
SHF cells constitute an atypical, apicobasally polarized epithelium characterized by apical monocilia and dynamic actin-rich basal filopodia (Francou et al., 2014). Although the dynamic properties of these protrusions remain uncharacterized, cell intercalation was suggested during the incorporation of the SHF progenitors (Sinha et al., 2012). This process is regulated by Wnt5a-activated PCP signaling as both Dvl1/2 and Wnt5a mutants display aberrant cell packing and defective actin polymerization and filopodia formation in SHF cells, leading to OFT shortening defects (Sinha et al., 2012; Fig. 1 C). Thus, a Wnt5a-Dvl PCP signaling cascade may regulate the protrusive cell behavior of SHF progenitors to promote their deployment and OFT lengthening (Sinha et al., 2012). Of note, Wnt5a is expressed in SHF cells, and mutations of the Wnt5a gene or its receptor tyrosine kinase Ror2 have been found in patients with cardiac OFT defects (Cohen et al., 2012).
In mice, Tbx1 is a critical regulator of SHF morphology in addition to regulating proliferation and differentiation of SHF progenitors (Francou et al., 2014). Cell shape changes in Tbx1 mutant embryos include rounder cell shape, a reduced basolateral membrane domain, and impaired filopodial activity associated with elevated aPKC zeta levels. Moreover, manipulating the epithelial properties of SHF progenitors by increasing activated aPKC zeta leads to reduced extension of the myocardial OFT. In humans, Tbx1 is haploinsufficient in DiGeorge syndrome, which is associated with cardiac OFT defects (Scambler, 2010; Choudhry and Trede, 2013). Tbx1 also interacts with the BAF chromatin remodeling complex to regulate the expression of Wnt5a, which interacts with Wnt11, Dsh, and VANGL PCP 2 (Vangl2) to induce oriented cell intercalation (Chen et al., 2012; Fig. 1 C).
Cell shape changes during ventricular trabeculation
Cardiac trabeculation is crucial to the formation of cardiac muscle and initiates when differentiated ventricular cardiomyocytes extrude and expand from the luminal surface of the primitive heart into the surrounding cardiac jelly (Samsa et al., 2013). Many signaling pathways regulate this process (Suri et al., 1996; Morris et al., 1999; Toyofuku et al., 2004; Liu et al., 2010; Chen et al., 2013; Rasouli and Stainier, 2017), with a recent study implicating aPKC iota, a component of the Par polarity complex, in directing cardiomyocyte cell polarization in mice (Passer et al., 2016). Before the onset of trabeculation, aPKC iota (also known as Prkci) and its interacting partner nuclear mitotic apparatus (NuMA) localize asymmetrically in luminal myocardial cells, which then undergo oriented cell division relative to the axis of polarity defined by the heart lumen (Passer et al., 2016; Fig. 1 C). Disruption of the Prkci–NuMA complex results in aberrant mitotic spindle alignment, loss of polarized cardiomyocyte division, and loss of normal myocardial trabeculation. The hyaluronan-rich cardiac jelly is essential for the luminal localization of the Prkci because depletion of hyaluronan results in loss of the luminal localization of the Par complex and randomized orientation of cardiomyocytes (Passer et al., 2016).
Cell shape changes during alignment of the great arteries
The initially single OFT separates into the aorta and the pulmonary artery, which align with their corresponding cardiac chambers (Anderson et al., 2003a,b). The essential role of the PCP signaling pathway in this process can be observed in mouse mutants of Vangl2 (Henderson et al., 2001; Phillips et al., 2005), Scrib (Phillips et al., 2007), Dvl1-3 (Hamblet et al., 2002; Etheridge et al., 2008), and Celsr1 (Curtin et al., 2003) that have defective polarized cardiomyocyte migration and disrupted alignment of great arteries with ventricles (Fig. 1 C). While both Wnt11 (Zhou et al., 2007; Nagy et al., 2010) and Wnt5a (Schleiffarth et al., 2007) mutants have similar defects, the epistatic relationship between Wnts, the PCP components, and the cell-intrinsic transcriptional networks upstream remain to be elucidated (Fig. 1 C).
Cell shape changes during formation of epicardium and coronary vasculature
The maturing heart is enveloped by the apically–basally polarized epicardium, the outer layer of the heart wall (Hirose et al., 2006; Martínez-Estrada et al., 2010). A subset of cells in the epicardium undergoes epithelial-to-mesenchymal transition, producing cardiovascular progenitors known as epicardially derived cells (EPDCs; Wessels and Pérez-Pomares, 2004). In mice, EPDCs contribute to endothelial and smooth muscle cells and to the developing coronary vessels and connective tissue (Wessels and Pérez-Pomares, 2004; Martínez-Estrada et al., 2010).
Par3, a critical ABP component, plays an essential role in the formation of the mouse epicardium, and Par3-null mice develop epithelial cysts, preventing the normal development of the epicardium (Hirose et al., 2006). Moreover, disrupted polarity and EPDC organization results in failure of coronary vessel network formation and a thin ventricular myocardium (Hirose et al., 2006). The regulatory mechanisms upstream of Par3-mediated epicardial polarity remain unknown.
Perspectives on cell shape changes during heart development
While the master transcriptional mechanisms for cardiac progenitor migration appear conserved across species, the molecular mechanisms underlying fusion of the progenitors to form the heart tube remain poorly understood in the mouse. Furthermore, the TFs regulating the differences between migrating lateral and medial cell populations are unknown. The transcriptional control of cell shape changes in the later stages of heart development also needs investigation. Challenges will involve combining our knowledge of the dynamic heart anatomy with the simultaneous SHF morphogenesis and the coordinated repetitive contractions of the differentiated cardiomyocytes (Clowes et al., 2014; Lindsey et al., 2014). Live-imaging techniques using transgenic models coupled with our understanding of the later stages are critical to improving our ability to diagnose debilitating cardiac disorders.
Transcriptional regulation of cell shape during lung development
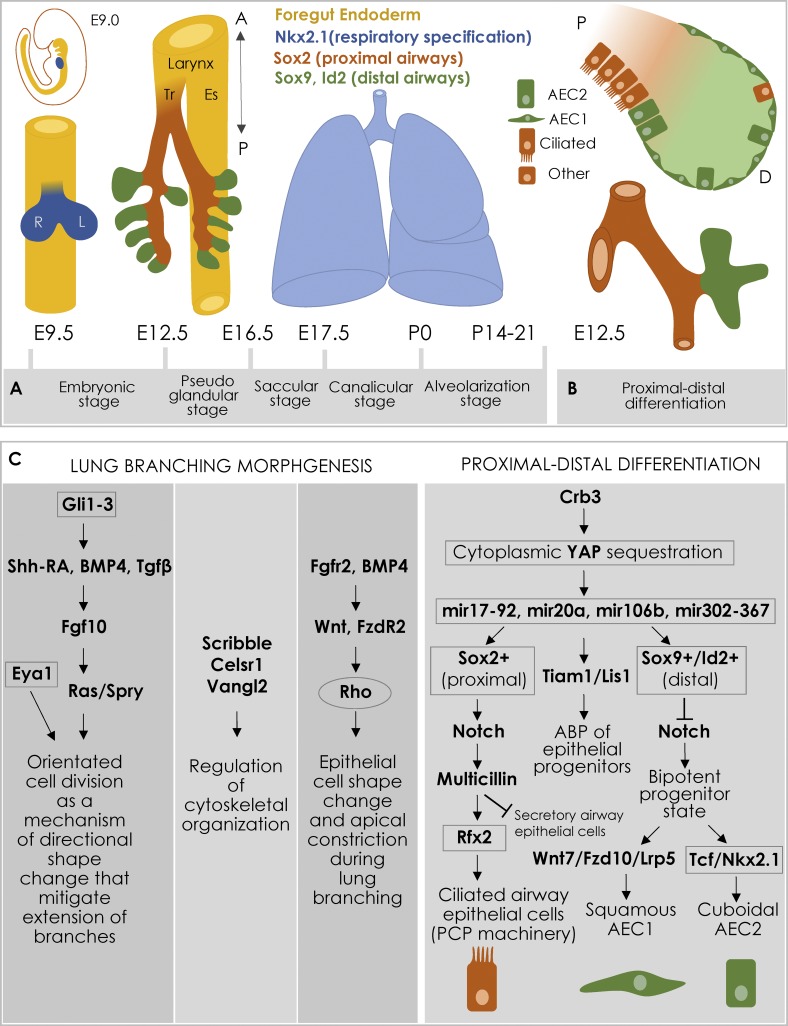
Lung development initiates with the specification of the ventral foregut endoderm into the respiratory endoderm (Lazzaro et al., 1991; Kimura and Deutsch, 2007; Herriges and Morrisey, 2014). This is driven by Nkx2.1 expression in the foregut dependent on signals from the surrounding heterogeneous mesenchyme (Morrisey and Hogan, 2010; Fig. 2 A). At the same time, the foregut separates into two tubes: a dorsal esophagus and a ventral trachea that connects to the lung buds (Rosekrans et al., 2015; Swarr and Morrisey, 2015). Once formed, the primary lung buds extend into the mesenchyme and begin the process of branching morphogenesis, forming a highly conserved network of airways (Metzger et al., 2008; Morrisey and Hogan, 2010; Swarr and Morrisey, 2015; Fig. 2 A). After E16.5, lung development switches from branching morphogenesis to the canalicular and saccular stages during which the terminal branches become narrower and form clusters of epithelial sacs at their terminal ends. These develop into alveoli, which mature during alveolarization following birth (Morrisey and Hogan, 2010; Rock and Hogan, 2011; McCulley et al., 2015; Weibel, 2015; Frank et al., 2016).
Figure 2.
Cellular processes during lung development. (A) Lung endoderm specification begins at E9.0 on the ventral side of the foregut endoderm (yellow) where initiation of Nkx2.1 expression (blue) begins. The trachea (Tr) forms during the embryonic stage and is visible at E9.5–E10; by E12.5 it is separated from the esophagus (Es). The other stages of lung development are displayed. A, anterior; P, posterior. (B) Proximal–distal differentiation (E12.5). The Nkx2.1+ endoderm gives rise to Sox2+ proximal (P; orange) and Sox9/Id2+ distal (D; green) progenitors. The proximal progenitors give rise to ciliated and other cell types (neuroendocrine, secretory, and mucosal). The distal progenitors give rise to AEC1 and AEC2. (C) Summary of the main regulatory pathways, including transcriptional regulators (boxed), signaling molecules, and cytoskeletal effectors (circled), underlying lung branching morphogenesis (left) and proximal–distal differentiation (right).
Cell shape changes during lung branching morphogenesis
Lung branching proceeds in a stereotypical manner by concurrent and repeated use of three major branching mechanisms: domain branching, planar bifurcation, and orthogonal bifurcation (Metzger et al., 2008). Fgf10, secreted by the mesenchyme at the distal end of primary lung buds, is the primary chemoattractive cue directing lung outgrowth (Bellusci et al., 1997; Sekine et al., 1999). Fgf10 expression is regulated via interplay of the retinoic acid, Shh, and BMP4/TGF-β signaling pathways (Lebeche et al., 1999; Warburton et al., 2010; Herriges and Morrisey, 2014). Downstream of Fgf10 signaling, Ras/Spry activity regulates spindle pole orientation, leading to a specific directionality in epithelial cell proliferation during extension of branches (El-Hashash and Warburton, 2011; Fig. 2 C).
Cell shape changes during bud tip generation (Kim et al., 2013; Kadzik et al., 2014; Swarr and Morrisey, 2015) are regulated by the PCP components Scribble, Celsr1, and Vangl2, in response to FGF signaling (Yates et al., 2013). In contrast, the epithelial cell shape and apical constriction during stalk extension and branching (Kadzik et al., 2014) are controlled by Wnt/Frizzled 2 signaling (Kim et al., 2013; Kadzik et al., 2014). Disruption of canonical Wnt signaling by conditional deletion of β-catenin or overexpression of the Wnt inhibitor Dickkopf1 severely impairs lung branching (Rajagopal et al., 2008). This Wnt/Fzd-mediated signaling is regulated at least in part via Fgfr2 and BMP4 in the lung epithelium (Volckaert and De Langhe, 2015; Fig. 2 C).
Cell shape changes during lung differentiation along the proximal-distal axis
The endoderm lining the lung undergoes dramatic morphological changes as it develops, giving rise to specialized cells within branched airways that transport air to gas-exchanging alveoli (Morrisey and Hogan, 2010; Hogan et al., 2014; Fig. 2 B). Expression of the TF Sox2 specifies the proximal endoderm progenitor lineage in the stalk region (reviewed in Herriges and Morrisey, 2014; Fig. 2 B). Due to apical constriction (Kadzik et al., 2014; Fumoto et al., 2017), these progenitor cells are bottle-shaped and give rise to airway neuroendocrine, secretory, ciliated, and mucosal cells (Kadzik et al., 2014). In contrast, the combined expression of Nkx2.1, Sox9, and Id2 marks the distal endoderm progenitor lineage (reviewed in Herriges and Morrisey, 2014; Fig. 2 B). These distal progenitors are short columnar and give rise to type 1 and type 2 alveolar epithelial cells (AEC1 and AEC2) that make up the distal gas exchange region composed of millions of alveoli (Kadzik et al., 2014; Fumoto et al., 2017; Fig. 2 B).
In the mouse airway cells, Sox2 expression requires phosphorylation and nuclear-cytoplasmic distribution of the transcriptional regulator Yes-associated protein (Yap; Szymaniak et al., 2015; Fig. 2 C). The Crb3-mediated ABP in the proximal airway epithelium causes Yap to bind to Lats1/2 kinases present at the apical junctions to promote Yap cytoplasmic sequestration. Thus, the preestablished ABP controls airway cell differentiation (Szymaniak et al., 2015).
In the mouse, the balance of lung endoderm proliferation and differentiation as well as ABP is regulated by expression of the miR302/367 cluster, downstream of Gata6 (Ventura et al., 2008; Fig. 2 C). Increased miR302/367 expression results in the formation of an undifferentiated multilayered lung endoderm. Loss of miR302/367 activity results in decreased proliferation and enhanced differentiation of the lung endoderm. MiR302/367 coordinates the balance between proliferation and differentiation through direct regulation of Rbl2 and Cdkn1a, whereas ABP is controlled by Tiam1 and Lis1 (Ventura et al., 2008; Fig. 2 C). Thus, miR302/367 directs lung endoderm development by coordinating multiple aspects of progenitor cell behavior.
Proximal airway epithelium differentiation is modulated by Notch signaling and is critical to establishing and maintaining the balance between ciliated and secretory cells within the proximal lineage (Rawlins et al., 2009; Fig. 2 C). In ciliated cells, the central target of Notch is the transcriptional cofactor Multicilin, whose targets include regulators of motile ciliogenesis, including multiple Rfx family members, C-Myb, and FoxJ1 (Stubbs et al., 2012; Fig. 2 C).
The differentiation of the distal airway epithelium generates AEC1 and AEC2 cells (Fig. 2 C). AEC1 are squamous epithelial cells that form a thin barrier between the alveolar airspace and blood-filled capillaries (Weibel, 2015). In contrast, AEC2 are cuboidal with defined ABP and serve to produce surfactant proteins (Weibel, 2015; Rozycki and Hendricks-Munos, 2017). AEC1 development requires reactivation of canonical Wnt signaling by the interaction of Wnt7 with Fzd1/10 and Lrp5 (Wang et al., 2005; Mutze et al., 2015). In contrast, differentiation of AEC2 cells requires Wnt-dependent TGF/β-catenin activity and a dosage of Nkx2.1 expression (Wang et al., 2005; Frank et al., 2016; Fig. 2 C).
While the transcriptional networks and signaling pathways critical for the establishment and maintenance of AEC1 and AEC2 are known, the downstream players and cellular mechanisms that drive the ABP of AEC2 remain to be elucidated. Moreover, AEC2 function as stem and progenitor cells in the adult alveoli and can proliferate and differentiate into AEC1, suggesting plasticity within the alveolar epithelium (Kapanci et al., 1969; Evans et al., 1975; Barkauskas et al., 2013). Identifying the signaling pathways that regulate such plasticity and the cell behaviors that maintain and can repair the alveoli is therefore an important research goal.
Perspectives on cell shape regulation during lung morphogenesis
Transcriptional regulation of shape during lung branching morphogenesis has been well studied. In contrast, much less is known about the early morphogenetic events involved in the primary lung bud formation and the trachea–esophagus separation or mechanisms involved in cell shape changes during the later stages. The plasticity within the AEC1 and AEC2 epithelial cells further complicates the ability to directly infer the transcriptional networks responsible for the establishment of particular cell shapes. Further identification of specific genetic markers for lineage tracing of distinct alveolar populations is needed to understand the relationship between cell types and regulation of their shape. Toward this effort, studies using single-cell RNA sequencing may provide additional clues on lung alveolar epithelial heterogeneity (Treutlein et al., 2014).
The mechanisms that regulate the organization of cells in the heterogeneous lung mesenchyme are also poorly understood (Zepp et al., 2017). Recent study involving single-cell RNA sequencing of the mouse lung has generated a spatial and transcriptional map of the lung mesenchyme (Zepp et al., 2017). This paves the way to characterizing the mechanisms that regulate cell division, alignment, adhesion, and mesenchymal motility, all of which involve changes in cell shape.
Transcriptional regulation of cell shape in the digestive system
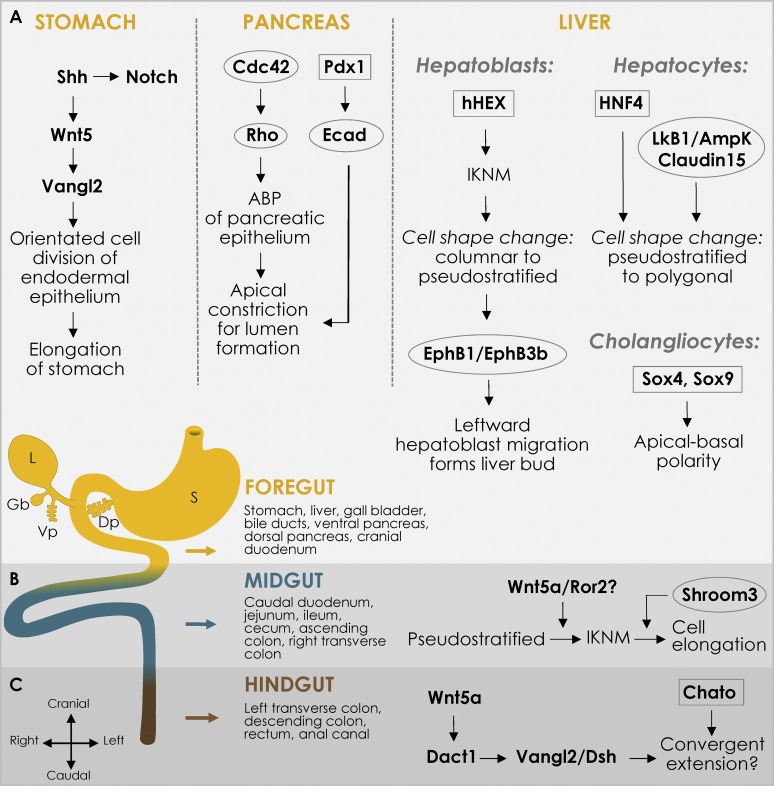
The primitive gut tube consists of endodermally derived epithelial cells surrounded by a cylinder of loose mesenchyme, suspended in the coelomic cavity by the dorsal mesentery (DM). It is divided along the cranial–caudal axis into the foregut (Fig. 3 A), midgut (Fig. 3 B), and hindgut (Roberts, 2000; Fig. 3 C). The gut tube undergoes differential elongation along the cranial–caudal axis with the midgut region elongating disproportionately (McGrath and Wells, 2015). As it lengthens, asymmetries across the LR axis establish the stomach curvature (Raya and Izpisúa Belmonte, 2006; Sadler and Langman, 2012) and trigger midgut rotation and looping morphogenesis (Davis et al., 2008). Concurrently, the gut endoderm undergoes extensive remodeling, forming finger-like villi (E14.5) for nutrient absorption (Mathan et al., 1976; Matsumoto et al., 2002; de Santa Barbara et al., 2003). Each villus is covered by a simple columnar epithelium and contains a mesodermal core with supporting blood vasculature, lymphatic lacteals, enteric nerves, smooth muscle, fibroblasts, myofibroblasts, and immune cells (reviewed in Walton et al., 2018). Key transcriptional players that regulate cell shape changes during gut elongation, LR asymmetry, and epithelial cytodifferentiation along the crypt–villus axis have been described.
Figure 3.
Cellular processes during the craniocaudal morphogenesis of the gastrointestinal tract and its accessory organs. The primitive gut tube (shown here as a cartoon at E11) is divided along the cranial–caudal axis into foregut (yellow), midgut (blue), and hindgut (brown) segments. The liver (L), dorsal and ventral pancreatic buds (Dp and Vp, respectively), and the gall bladder (Gb) form as endodermal diverticula of the foregut, which also forms the stomach (S). The outline of the mechanisms involved in the development of each segment has been depicted including TFs (boxed), signaling molecules, and cytoskeletal effectors (circled).
Regulation of cell shape changes during gut elongation
The early midgut undergoes dramatic elongation (Figs. 3 B and 4 A), a process critical for nutrient absorption and diet adaptations; failure to elongate leads to congenital short bowel syndrome with high associated mortality in humans (Hasosah et al., 2008). The early mouse midgut endoderm was initially characterized as stratified (Mathan et al., 1976; Matsumoto et al., 2002), while intercalation and convergent extension-like movements were proposed to drive midgut and hindgut elongation (Matsumoto et al., 2002; García-García et al., 2008; Cervantes et al., 2009; Matsuyama et al., 2009; Reed et al., 2009; Fig. 3, B and C). However, recent studies established that this endoderm is pseudostratified (Grosse et al., 2011; Yamada et al., 2013; Freddo et al., 2016; Norden, 2017; Table 1) and elongates using interkinetic nuclear migration (IKNM; Sauer, 1935; Meyer et al., 2011). The actin-binding protein Shroom3 (Hildebrand, 2005) is required for maintenance of a single-layered pseudostratified midgut epithelium, and, in its absence, apical surfaces are disorganized and the epithelium temporarily assumes a stratified structure during villus formation (Grosse et al., 2011).
Figure 4.
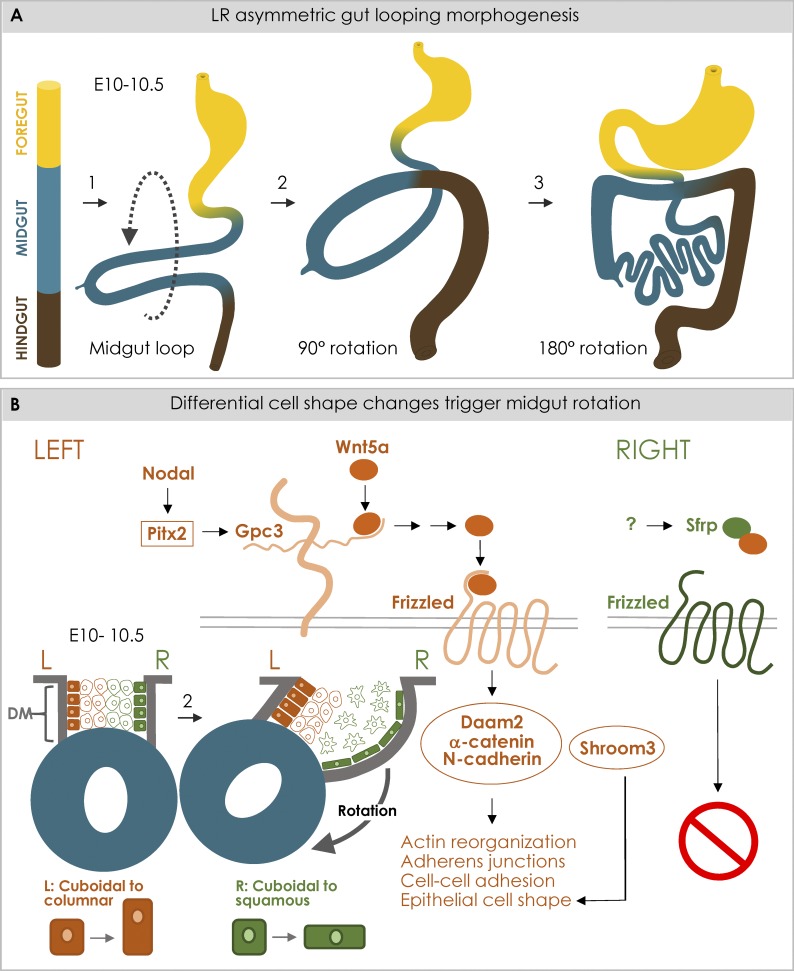
Cellular processes involved in asymmetric midgut rotation. (A) The primitive gut is divided into foregut (yellow), midgut (blue), and hindgut (brown). Primary midgut loop forms due to rapid midgut growth with respect to the embryo (1). (2) The loop rotates 90° counterclockwise (ventral view). (3) Additional loops form as the gut rotates another 180°. (B) The mechanisms that lead to midgut rotation (2 in A). The midgut is suspended by the DM, which comprises four distinct cell compartments, and changes within each cause the DM to deform and tilt the gut tube leftward. Transcriptional regulation of Wnt pathway genes by Pitx2 leads to Daam2 activation, which mediates adhesion at cell junctions by binding α-catenin and N-cadherin. Actin remodeling and lengthening of junctions cause left condensation. Antagonized Wnt signaling causes right mesenchymal cells to remain dispersed. Shroom3 with N-cadherin on the left are also involved in the conversion of cuboidal to columnar cell morphology. Consequently, the left DM shortens, and the right side lengthens, deforming the DM and shifting midgut leftward. Left compartment is orange; right is green. Midgut is blue.
Several mechanistic studies of gut elongation have interpreted experimental data in the context of the stratified model (Saotome et al., 2004; Kim et al., 2007; García-García et al., 2008; Cervantes et al., 2009; Yamada et al., 2010; Fig. 3, B and C). Of interest, Wnt5a−/− and Ror2−/− mouse embryos have significantly shorter midgut with thicker epithelium and disrupted ABP, consistent with disruption of IKNM (Cervantes et al., 2009; Matsuyama et al., 2009). Similarly, secreted frizzled related protein (Sfrp) 1 and 2 control ABP and oriented cell division in the midgut and foregut epithelium by interacting with Vangl2, downstream of Wnt5a signaling (Matsuyama et al., 2009). Additionally, the Hh signaling pathway is required for normal intestine lengthening, but the cellular mechanisms remain unexplored (Mao et al., 2010; Kim et al., 2011). Thus, it will be important to test whether these signaling pathways regulate IKNM and intersect with Shroom3 to drive cell shape changes during gut elongation.
Regulation of cell shape changes across the LR axis
LR asymmetry is fundamental to gut anatomy and function. The first evidence of LR asymmetry involves the emergence of the conserved greater and lesser stomach curvatures, located on the left and right sides, respectively (Chapman, 1997,). In Xenopus laevis and mice, stomach curvature forms due to differential tissue architecture that develops in the left and right foregut (future stomach) walls, a process that is intrinsic to the gut tube itself (Davis et al., 2017). The foregut lumen is initially aligned with symmetric walls along the LR axis. Stomach curvature initiation is accompanied by an outward bend and lengthening, followed by the thinning of the left wall compared with the right, driven by LR asymmetric cell rearrangements and differences in endoderm cell polarity (Davis et al., 2017).
The LR patterning genes including Foxj1, Nodal, and Pitx2 regulate cellular rearrangements within the left wall to give rise to stomach curvature (Bamforth et al., 2004; Shiratori et al., 2006; Davis et al., 2017); however, the downstream cellular effectors that drive physical stomach bending remain to be elucidated.
At the midgut region, a leftward tilting of the gut tube initiates its rotation (Ueda et al., 2016; Fig. 4 A). Unlike stomach asymmetries, the midgut tilt is driven by the DM, an adjacent bridge of mesoderm that suspends the entire gut tube from the dorsal body wall (Hecksher-Sørensen et al., 2004; Davis et al., 2008; Fig. 4 B). In birds and mice, the embryonic DM consists of four juxtaposed cellular compartments: left cuboidal epithelium, left mesenchyme, right mesenchyme, and right cuboidal epithelium (Davis et al., 2008; Fig. 4 B). To initiate gut rotation, the left epithelial cells change shape from cuboidal to columnar, the left mesenchyme compacts, the right mesenchyme expands, and the right epithelial cells become squamous. Consequently, the left DM shortens while the right side lengthens, deforming the shape of the DM and shifting the attached gut tube to the left (Davis et al., 2008; Fig. 4 B). There are no asymmetries in cell number, proliferation, or cell death within the DM; thus, gut rotation is strictly a consequence of changes in cell shape and behavior across the LR axis.
Similar to the stomach, Pitx2 expression regulates the changes in cell behavior within the left DM compartment (Davis et al., 2008; Kurpios et al., 2008). Left-side compaction results from the expression of the Pitx2 effector Gpc3, integrating noncanonical Wnt5a to activate the formin Daam2, to stabilize N-cadherin–dependent cell adhesion (Welsh et al., 2013; Fig. 4 B). Simultaneously, the cytoskeleton of mesenchymal cells reorganizes to polarize and orient to the left (Welsh et al., 2013). These morphogenetic behaviors are spatiotemporally linked with gut arterial and lymphatic development, suggesting that gut rotation is tightly coordinated with gut vasculogenesis (Mahadevan et al., 2014).
Loss of Pitx2 results in a cuboidal instead of columnar epithelium within the left DM (Davis et al., 2008), a double-right phenotype. A similar phenotype is observed in mouse embryos lacking Shroom3 (Hildebrand, 2005; Plageman et al., 2011). Shroom3 expression in the DM is not dependent on Pitx2; rather, Shroom3 functions cooperatively with the Pitx2 target gene N-cadherin to regulate the columnar epithelium (Plageman et al., 2011; Fig. 4 B). Given the established role for Shroom proteins in defining cell shape and remodeling of the midgut (Grosse et al., 2011) and mesenteric epithelium, it will be important to identify which upstream factors drive Shroom3 (and Shroom2) expression in these tissues (Chung et al., 2010; Plageman et al., 2010).
In zebrafish, gut looping results from the asymmetric migration of the neighboring lateral plate mesoderm cells, while compromise of Nodal activity randomizes this migration and gut looping chirality (Horne-Badovinac et al., 2003). Thus, the Nodal-Pitx2 transcriptional regulation of cell shape during LR midgut asymmetry is likely evolutionarily conserved.
Cell shape changes across the crypt-villus axis
The functional epithelium of the adult small intestine is organized into finger-like villi, which project into the gut lumen, and flask-like crypts embedded within the mesenchyme. The crypts are proliferative and contain intestinal stem cells that differentiate into specialized, postmitotic epithelial cells along the crypt–villus axis in the adult intestine. These include the absorptive enterocytes, mucus-secreting goblet cells, hormone-secreting enteroendocrine cells, and Paneth cells with function in innate immunity and antimicrobial defense (van der Flier and Clevers, 2009).
Enterocytes are the major polarized cell type within the intestine. While the transcriptional regulation of their polarization has been challenging to study in vivo, recent pioneering work using human Caco-2 cells, which develop structural and functional polarity similar to enterocytes in vivo, has provided an important genomic portrait of the changes in gene expression patterns during enterocyte cell polarization (Halbleib et al., 2007; Sääf et al., 2007).
The transition from proliferating, nonpolarized Caco-2 cells to postmitotic polarizing cells involves a switch in transcriptional signature resembling structural characteristics of migrating and differentiating intestinal epithelial cells in vivo (Halbleib et al., 2007; Sääf et al., 2007). This result was surprising because the transcriptional changes underlying Caco-2 polarization occurred despite the absence of tissue morphogen gradients in vitro. For example, whereas Wnt target gene expression is robust in mitotic nonpolarized Caco-2 cells, it decreases as cells become postmitotic and polarized, akin to the decreasing gradient of Wnt activity from crypt to villus in vivo (Halbleib et al., 2007; Sääf et al., 2007). After polarization, the decrease of Wnt gene expression is accompanied by an increase of β-catenin levels in the cell nucleus and is associated with TCF4 (Sääf et al., 2007). Furthermore, transcriptional profiling reveals a critical interplay between polarization and the TGFβ, FGF, SHH, and Notch signaling pathways for which cellular details remain to be uncovered (Halbleib et al., 2007). In addition, KLF4, a zinc finger TF, regulates polarity-related genes, which include LKB1, EPHB2, and EPHB3 (Yu et al., 2009). Thus, Caco-2 cells may serve as a powerful in vitro model to study the genetic programing of intestinal progenitor cells when they leave the crypt to become differentiated cell types of the adult intestine.
Regulation of cell shape changes during development of the liver and pancreas
The liver and pancreas form as endodermal diverticula of the foregut (Yin, 2017; Fig. 3 A). Liver bud formation includes three stages (Bort et al., 2006; Ober and Lemaigre, 2018). First, elongation of the ventral endoderm causes the cuboidal epithelia to become columnar, resulting in endodermal thickening. The second stage involves IKNM leading to the formation of a pseudostratified layer of cells called hepatoblasts (Bort et al., 2006). This is regulated by hHex, a homeobox-containing TF of the Antennapedia/Ftz class, expressed in the foregut endoderm (Bort et al., 2006; Ober and Lemaigre, 2018; Fig. 3 A). In the third stage, pseudostratified hepatoblasts delaminate and proliferate. Following repulsive cues provided by EphrinB1/EphB3b signaling in the right lateral plate mesoderm, they become motile and undergo a leftward migration, producing the left-sided liver bud (Bort et al., 2006; Cayuso et al., 2016; Fig. 3 B). Mutants for aPKC iota, Pard6γb, and actin modulators, have been shown to disrupt this asymmetric migration resulting in the formation of a midline liver (Huang et al., 2008).
Liver bud formation is accompanied by remodeling of the adjacent endothelium (Antoniou et al., 2009). Subsequently, hepatoblasts more distal to the portal vein differentiate along the hepatocyte lineage, while those proximal commit to cholangiocyte differentiation (Carpentier et al., 2011). This positions the hepatocytes between the endothelial sinusoids and bile canaliculi for their function in the directional exchange of macromolecules between the sinusoids and canaliculi (Treyer and Müsch, 2013; Ober and Lemaigre, 2018).
Fetal inactivation of HNF4α, a key transcriptional regulator of hepatocyte differentiation, arrests hepatocyte maturation by blocking the transition of round, pseudostratified hepatoblasts into mature polygonal hepatocytes. This arrest is characterized by loss of cell–cell contacts, disrupted expression of cell adhesion factors, and mislocalization of junctional proteins such as E-cadherin and ZO-1 (Parviz et al., 2003). While the exact downstream effectors of HNF4α are unknown, studies in zebrafish and mice have identified the LKB1–AMPk pathway and Claudin15 as critical regulators of hepatocyte polarity (Cheung et al., 2012; Gissen and Arias, 2015; Fig. 3 B). Mice lacking either LKB1 or Claudin15 display loss of hepatocyte polarity, disrupted tight junctions, and dilated hepatic canaliculi (Shackelford and Shaw, 2009; Cheung et al., 2012).
Cholangiocytes are distinguished by their cuboidal morphology and proximity to the portal vein; they also develop primary apical cilia and produce tight junctions that separate their apicolateral from basolateral junctions (Tanimizu et al., 2007). The SRY-related high mobility group box TFs Sox4 and Sox9 cooperatively set up cholangiocyte ABP (Poncy et al., 2015; Fig. 3 B), while the tight junctions are governed by Grainheadlike-2, a TF that regulates Claudins and Rab25 (Tanimizu et al., 2013). The direct molecular mechanisms downstream of Sox4 and Sox9 remain unknown; however, the presence of primary cilia in the apical surface suggests PCP pathway involvement. Further investigations should provide mechanistic insights into the role of aberrant cholangiocyte differentiation in liver pathologies.
Pancreatic development initiates with the evagination of the dorsal gut endoderm of the posterior foregut into the surrounding condensed mesenchyme, producing the dorsal pancreatic bud (Yin, 2017; Fig. 3 B). A similar ventral evagination arises caudal to the hepatic/biliary bud, resulting in the formation of the ventral pancreatic bud (Mastracci and Sussel, 2012; Fig. 3 B).
The epithelial–mesenchymal crosstalk guiding endodermal evagination is modulated by mesenchymal FGF7 signaling (Elghazi et al., 2002). Following bud formation, the stalk elongates and initiates branching morphogenesis of the buds (Gittes, 2009). The initially multilayered stratified epithelium transforms into single-layered polarized epithelial cells, which invade and branch extensively into the underlying mesenchymal compartment (Fujitani, 2017). During this transition, small microlumens form between the epithelial cells, and the rise of apical epithelial polarity results in microlumen expansion, creating independent luminal networks (Kesavan et al., 2009). While the Cdc42-Rho signaling axis directly regulates this ABP, the TF Pdx1 drives E-cadherin to induce the apical constriction necessary for lumen formation and maintenance (Marty-Santos and Cleaver, 2016; Fig. 3 B).
Unlike the liver, much less is known about the transcriptional regulation of cell shape in the pancreas. Future studies may help us understand how perturbation of these processes may lead to various forms of diabetes (Larsen and Grapin-Botton, 2017).
Perspectives on the regulation of cell shape changes in the digestive system
The events shaping the digestive system are relatively understudied compared with the heart and lung, perhaps owing to the complex and dynamic events of gut looping morphogenesis. In addition to differentiation along the crypt–villus axis, such studies are complicated by the variations among derivatives of the midgut, hindgut, and foregut, with further complexity arising from gastrointestinal tract morphological diversity across species. Moreover, until recently, the early mouse gut tube was mischaracterized as a stratified epithelium. It will thus be important that previous and future models of intestinal growth be reexamined in this context. For example, radial intercalation as a mechanism for gut lengthening is not possible in the context of a single-layer, pseudostratified epithelium. Such studies highlight the importance of correctly describing cell shape events driving intestinal morphogenesis. Given that pseudostratification is a hallmark of many organoid systems (Eiraku et al., 2011; Lancaster et al., 2013; Ranga et al., 2016), organoid technology used to model intestinal development and diseases in vitro (Clevers, 2016) may hold promise for interpreting the in vivo molecular and cellular mechanisms driving this tissue arrangement in the context of important human disorders, such as congenital short bowel syndrome.
Transcriptional regulation of cell shape changes during kidney development
Kidney development is initiated when the caudal region of the mesonephric duct (Wolffian duct), an epithelial tube derived from intermediate mesoderm, buds out into the adjacent metanephric mesenchyme (MM) forming the ureteric bud (UB; Saxén, 1987; Fig. 5 A). UB undergoes branching morphogenesis to give rise to the renal collecting duct system, while it induces MM cells to condense into epithelial vesicles (renal vesicles) to form the nephron (Fig. 5 C). UB growth and branching are critical for urogenital development, and their failure leads to a spectrum of birth defects ranging from renal agenesis to reduced kidney size and nephron number (Costantini, 2006). Extensive studies have characterized the signaling pathways that control kidney development and patterning (reviewed in Dressler, 2006, 2009; Michos, 2009; Costantini and Kopan, 2010; Carroll and Das, 2011; Lienkamp et al., 2012; Little and McMahon, 2012; Blake and Rosenblum, 2014; Bertram et al., 2016). Below we focus on the cellular mechanisms that initiate UB budding and branching morphogenesis and the process of nephron development.
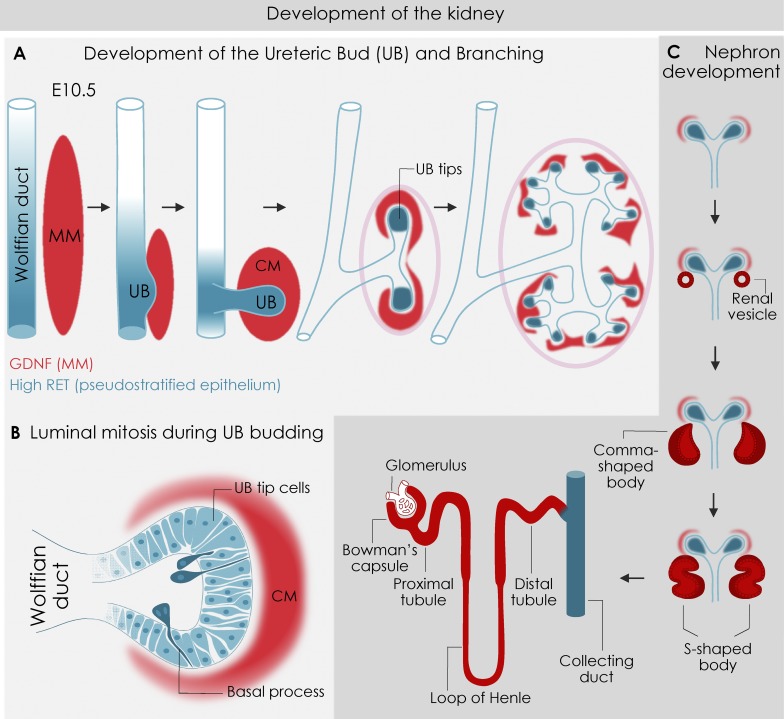
Figure 5.
Kidney development. (A) Kidney is formed via reciprocal interactions between the Wolffian duct and the MM. MM cells condense, forming cap mesenchyme (CM) surrounding the UB tips. MM-derived GDNF (red) signaling via RET in the Wolffian duct (blue) induces UB budding. Ret is initially broadly expressed in the Wolffian duct but becomes restricted to the distal tips of the budding UB. (B) Cross-section view of pseudostratified UB bud (blue) demonstrating mitosis-associated cell dispersal. Dividing cells (darker blue) leave the epithelium for the lumen, retaining a basal process; they later reenter the epithelium. (C) Sequential nephron development: The CM-derived cells (red) form most of the nephron (renal vesicle, then C- and S-shaped bodies, then mature nephron), while the UB-derived cells (blue) form the collecting ducts. Proximal–distal patterning along the nephron gives rise to glomerulus, Bowman’s capsule, and the renal tubules (proximal tubule, loop of Henle, distal tubule, and the connecting tubule, which connects to the UB-derived collecting duct).
UB formation and early branching
The UB arises from the pseudostratified epithelium of the caudal mesonephric duct (Chi et al., 2009; Fig. 5 A). This budding process is driven by extensive cell movements within the duct epithelium regulated by the glial cell line–derived neurotrophic factor (GDNF) signaling through its receptor RET (receptor tyrosine kinase) and coreceptor GFAR1 (Brophy et al., 2001; Takahashi, 2001; Tang et al., 2002; Arighi et al., 2005; Costantini and Kopan, 2010). GDNF is expressed in the underlying MM downstream of Eya1 and Hox11 (Xu et al., 1999; Rocque and Torban, 2015), while RET and GFAR1 are expressed in the Wolffian duct epithelium governed by Gata3 (Gong et al., 2007; Grote et al., 2008). GDNF expression in the MM is also maintained and reinforced by Wnt11 (Majumdar et al., 2003), which is regulated by canonical Wnt/β-catenin signaling downstream of Wnt9b (Karner et al., 2011). The TFs HNFβ1 (Paces-Fessy et al., 2012) and Ap-2 B (Moser et al., 1997) regulate Wnt9b expression during UB formation and outgrowth, following which the UB undergoes branching morphogenesis controlled by the PCP components Fzd4 and Fzd8 (Ye et al., 2011). In addition, Fgf7-10/FgfR2 and the synergistic interaction of the HGF–Met and EGF–EGFR pathways are critical during UB branching morphogenesis (Sakurai et al., 1997).
While GDNF-Ret signaling is critical for kidney development, its specific role in UB branching morphogenesis is unclear. Ret expression defines a population of highly mitotic UB tip cells (Fig. 5, A and B), which are distinct from the differentiating trunk cells behind the tips, and where most of the cell proliferation and growth occurs (Michael and Davies, 2004). The significance of GDNF/RET signaling in the regulation of cell migration and rearrangement during UB budding from the metanephric duct can be observed in chimeric mice, in which cells with higher RET-signaling activity outcompete cells with lower activity and migrate toward the budding site where the initial UB outgrowth occurs (Chi et al., 2009). In contrast, cells lacking RET are excluded from the tips of the branching UB in chimeric kidneys, suggesting that cell sorting regulated by GDNF/RET is the mechanism responsible for UB outgrowth (Chi et al., 2009).
Time-lapse microscopy and genetic strategies have uncovered a unique mitotic behavior in the branching tips of the UB termed “mitosis-associated cell dispersal” or luminal mitosis (Packard et al., 2013; Fig. 5 B). During this process, dividing tip cells delaminate from the epithelium and transiently enter the lumen, retaining a thin connection with the basement membrane (known as the basal process; Fig. 5 B). One daughter cell inherits its mother’s location, whereas the other travels along the lumen then reinserts into the epithelial plane a few cell diameters away. Interestingly, luminal mitosis bears some similarities to the process of IKNM, a hallmark of pseudostratified epithelia, discussed above (Kosodo, 2012; Spear and Erickson, 2012). However, unlike luminal mitosis in the UB, mitosis in pseudostratified epithelia occurs within the confines of the epithelium.
IKNM can be mediated by either microtubules or the actin cytoskeleton (Kosodo, 2012; Spear and Erickson, 2012), and loss of actin depolymerizing factors cofilin1 and destrin disrupts UB branching morphogenesis (Kuure et al., 2010). Whether GDNF/RET signaling is connected to the subcellular mechanisms during luminal migration of UB tip cells and whether the regulators of the actin cytoskeleton are downstream of GDNF/RET remain to be elucidated.
Renal vesicle induction and nephron morphogenesis
As the UB tip cells bifurcate, Wnt4, Fgf8, and Pax8 expression within the MM drive mesenchymal-to-epithelial transition, forming polarized renal vesicles (Schmidt-Ott and Barasch, 2008; Fig. 5 C). Once formed, they elongate and transition into the comma- and S-shaped body. The S-shaped body distal end fuses with the UB, while the remaining proximal end generates the proximal and distal tubules of the nephron (Dressler, 2006).
The Hippo pathway transcriptional regulator Yap regulates distal fusion of the S-shaped body to the UB, a process dependent on Cdc42-driven nuclear localization of YAP (Reginensi et al., 2013). However, the more proximal components of the nephron require Notch signaling to differentiate from the more distal tubules (Cheng and Kopan, 2005). During proximal–distal specification, tubule cuboidal epithelial cells elongate perpendicular to the tubule proximal–distal axis and undergo rosette-based cell intercalation (Lienkamp et al., 2012). The noncanonical PCP pathway downstream of Wnt9b drives this myosin-dependent rosette-based cell intercalation and regulates polarized cell division to cause tubule elongation by convergent extension (Karner et al., 2011; Lienkamp et al., 2012).
Key to proximal tubule development is the differentiation of epithelial precursors at the S-shaped body stage into podocytes, polarized renal glomerular cells with a central role in generating glomerular ultrafiltrate. Accompanying this differentiation is a dynamic epithelial cell shape change thought to occur during the comma body stage when cuboidal epithelial cells transition to columnar morphology (Greka and Mundel, 2012). As the comma body adopts an S shape, the VEGF-dependent intrusion of endothelium and capillary loop formation causes dramatic expansion of the apical epithelial surface (Schell et al., 2014). This expansion is accompanied by repositioning of cell–cell junctions from the apical to the basal pole of the cell followed by restructuring and replacement of tight junctions with sieve-like complexes called slit diaphragms (Grahammer et al., 2013).
Three polarity complexes regulate podocyte differentiation: the apical Crumbs complex (Ebarasi et al., 2009; Pieczynski and Margolis, 2011), the apical Cdc42/Par3/Par6/αPKC complex (Hartleben et al., 2008; Scott et al., 2012), and the Nephrin/Nck2/n-WASP/Ark2/3 pathway (New et al., 2013; Schell et al., 2013). Podocyte differentiation is governed by Pax2 and Wt1 interplay, which regulate transcriptional control during podocyte development (Guo et al., 2002; Kobayashi et al., 2004). However, their downstream effectors of cell polarity remain to be elucidated.
As the proximal and distal components form, they are spatially organized in the corticomedullary axis. The cortex contains the renal corpuscles and the proximal and distal tubules, while the renal medulla consists of the loop of Henle and the collecting duct tubular network (Little et al., 2007; Fig. 5 C). In the medulla, the tubular elongation and the growth of the loop of Henle are regulated by paracrine canonical Wnt7b signaling. In Wnt7b mutant mice, cell division planes of the collecting duct epithelium are biased along the radial versus the longitudinal axis (Yu et al., 2009). A similar loss of the medullary region has been documented in Vangl2 and Fat4 mutant mice in addition to reduced tubular formation (Yates et al., 2010), indicating that components of the core PCP network function alongside canonical Wnt7b to drive tubular genesis and organization in the medulla.
Insights into regulation of cell shape changes during kidney morphogenesis
Epithelial cell shape changes and cell polarity have established critical roles throughout the development and functioning of the nephron, from early stages of morphogenesis to the later stages of differentiation across the proximal–distal axis. For example, perturbation of cell polarity, cell shape, and tubular dilations has been implicated in polycystic kidney disease (Schedl, 2007; Saburi et al., 2008). However, it is presently unclear to what extent the observed defective polarities are causative or secondary consequences of this pathology. Furthermore, the exact roles of cytogenesis genes, implicated in polycystic kidney disease and other kidney injury conditions, during the spatiotemporal morphogenesis of the kidney remain unknown. Effects on kidney development can be investigated using conditional knockout mice with lineage-specific abrogation of polarity. Such models coupled with transgenic fluorescent labeling of specific cell types would facilitate dynamic analyses of cell shape during tubulogenesis and extension and the relationship between cell shape and kidney disease. Finally, while interactions between the mesenchymal stroma and the epithelia are well understood during UB outgrowth, studies investigating later kidney development, such as formation of the renal vesicle, comma-, and S-shaped bodies, have mainly focused on the epithelia. Hence, mechanisms that regulate behavior and differentiation of mesenchymal stromal cells warrant further investigations.
Conclusion
While the importance of transcriptional regulation underlying cell shape changes during organogenesis is apparent, substantial exciting questions remain unanswered. The integration of findings drawn from genetics, cell biology, evolution, computational biology, and high-resolution live imaging will offer a deeper understanding of the generation of cell shape. For example, the combination of forward and reverse genetics with multispecies approaches, and informed by sensitive transcriptomic and proteomic methods, is likely to provide detailed, integrated insights into molecular and cellular mechanisms during organogenesis. A more complete understanding of the regulation of cell shape during morphogenesis is a critical first step toward better diagnostics and ultimately new therapies to address a broad spectrum of organ malformations that can cause life-threatening or debilitating disorders.
Acknowledgments
The authors would like to thank Drs. John Wallingford, Robert Huebner, and members of the Kurpios laboratory for many helpful discussions on this topic and Cindy Westmiller for assisting with reference material.
This work was supported by the National Institutes of Health (R01 DK092776) and March of Dimes (1-FY11-520).
The authors declare no competing financial interests.
References
- Anderson R.H., Webb S., Brown N.A., Lamers W., and Moorman A.. 2003a Development of the heart: (2) Septation of the atriums and ventricles. Heart. 89:949–958. 10.1136/heart.89.8.949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R.H., Webb S., Brown N.A., Lamers W., and Moorman A.. 2003b Development of the heart: (3) Formation of the ventricular outflow tracts, arterial valves, and intrapericardial arterial trunks. Heart. 89:1110–1118. 10.1136/heart.89.9.1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniou A., Raynaud P., Cordi S., Zong Y., Tronche F., Stanger B.Z., Jacquemin P., Pierreux C.E., Clotman F., and Lemaigre F.P.. 2009. Intrahepatic bile ducts develop according to a new mode of tubulogenesis regulated by the transcription factor SOX9. Gastroenterology. 136:2325–2333. 10.1053/j.gastro.2009.02.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arighi E., Borrello M.G., and Sariola H.. 2005. RET tyrosine kinase signaling in development and cancer. Cytokine Growth Factor Rev. 16:441–467. 10.1016/j.cytogfr.2005.05.010 [DOI] [PubMed] [Google Scholar]
- Bakkers J., Verhoeven M.C., and Abdelilah-Seyfried S.. 2009. Shaping the zebrafish heart: From left-right axis specification to epithelial tissue morphogenesis. Dev. Biol. 330:213–220. 10.1016/j.ydbio.2009.04.011 [DOI] [PubMed] [Google Scholar]
- Bamforth S.D., Bragança J., Farthing C.R., Schneider J.E., Broadbent C., Michell A.C., Clarke K., Neubauer S., Norris D., Brown N.A., et al. 2004. Cited2 controls left-right patterning and heart development through a Nodal-Pitx2c pathway. Nat. Genet. 36:1189–1196. 10.1038/ng1446 [DOI] [PubMed] [Google Scholar]
- Barkauskas C.E., Cronce M.J., Rackley C.R., Bowie E.J., Keene D.R., Stripp B.R., Randell S.H., Noble P.W., and Hogan B.L.. 2013. Type 2 alveolar cells are stem cells in adult lung. J. Clin. Invest. 123:3025–3036. 10.1172/JCI68782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellusci S., Grindley J., Emoto H., Itoh N., and Hogan B.L.. 1997. Fibroblast growth factor 10 (FGF10) and branching morphogenesis in the embryonic mouse lung. Development. 124:4867–4878. [DOI] [PubMed] [Google Scholar]
- Bertram J.F., Goldstein S.L., Pape L., Schaefer F., Shroff R.C., and Warady B.A.. 2016. Kidney disease in children: Latest advances and remaining challenges. Nat. Rev. Nephrol. 12:182–191. 10.1038/nrneph.2015.219 [DOI] [PubMed] [Google Scholar]
- Blake J., and Rosenblum N.D.. 2014. Renal branching morphogenesis: Morphogenetic and signaling mechanisms. Semin. Cell Dev. Biol. 36:2–12. 10.1016/j.semcdb.2014.07.011 [DOI] [PubMed] [Google Scholar]
- Bort R., Signore M., Tremblay K., Martinez Barbera J.P., and Zaret K.S.. 2006. Hex homeobox gene controls the transition of the endoderm to a pseudostratified, cell emergent epithelium for liver bud development. Dev. Biol. 290:44–56. 10.1016/j.ydbio.2005.11.006 [DOI] [PubMed] [Google Scholar]
- Brophy P.D., Ostrom L., Lang K.M., and Dressler G.R.. 2001. Regulation of ureteric bud outgrowth by Pax2-dependent activation of the glial derived neurotrophic factor gene. Development. 128:4747–4756. [DOI] [PubMed] [Google Scholar]
- Cai C.L., Liang X., Shi Y., Chu P.H., Pfaff S.L., Chen J., and Evans S.. 2003. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev. Cell. 5:877–889. 10.1016/S1534-5807(03)00363-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona-Fontaine C., Matthews H., and Mayor R.. 2008. Directional cell migration in vivo: Wnt at the crest. Cell Adhes. Migr. 2:240–242. 10.4161/cam.2.4.6747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpentier R., Suñer R.E., van Hul N., Kopp J.L., Beaudry J.B., Cordi S., Antoniou A., Raynaud P., Lepreux S., Jacquemin P., et al. 2011. Embryonic ductal plate cells give rise to cholangiocytes, periportal hepatocytes, and adult liver progenitor cells. Gastroenterology. 141:1432–1438: 1438.e1–1438.e4. 10.1053/j.gastro.2011.06.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll T.J., and Das A.. 2011. Planar cell polarity in kidney development and disease. Organogenesis. 7:180–190. 10.4161/org.7.3.18320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayuso J., Dzementsei A., Fischer J.C., Karemore G., Caviglia S., Bartholdson J., Wright G.J., and Ober E.A.. 2016. EphrinB1/EphB3b coordinate bidirectional epithelial-mesenchymal interactions controlling liver morphogenesis and laterality. Dev. Cell. 39:316–328. 10.1016/j.devcel.2016.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes S., Yamaguchi T.P., and Hebrok M.. 2009. Wnt5a is essential for intestinal elongation in mice. Dev. Biol. 326:285–294. 10.1016/j.ydbio.2008.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman H.W. 1997. Comparative Physiology of the Vertebrate Digestive System. Second edition. Cambridge University Press. 38:576–577. [Google Scholar]
- Chen H., Zhang W., Sun X., Yoshimoto M., Chen Z., Zhu W., Liu J., Shen Y., Yong W., Li D., et al. 2013. Fkbp1a controls ventricular myocardium trabeculation and compaction by regulating endocardial Notch1 activity. Development. 140:1946–1957. 10.1242/dev.089920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Fulcoli F.G., Ferrentino R., Martucciello S., Illingworth E.A., and Baldini A.. 2012. Transcriptional control in cardiac progenitors: Tbx1 interacts with the BAF chromatin remodeling complex and regulates Wnt5a. PLoS Genet. 8:e1002571 10.1371/journal.pgen.1002571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H.T., and Kopan R.. 2005. The role of Notch signaling in specification of podocyte and proximal tubules within the developing mouse kidney. Kidney Int. 68:1951–1952. 10.1111/j.1523-1755.2005.00627.x [DOI] [PubMed] [Google Scholar]
- Cheung I.D., Bagnat M., Ma T.P., Datta A., Evason K., Moore J.C., Lawson N.D., Mostov K.E., Moens C.B., and Stainier D.Y.. 2012. Regulation of intrahepatic biliary duct morphogenesis by Claudin 15-like b. Dev. Biol. 361:68–78. 10.1016/j.ydbio.2011.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Michos O., Shakya R., Riccio P., Enomoto H., Licht J.D., Asai N., Takahashi M., Ohgami N., Kato M., et al. 2009. Ret-dependent cell rearrangements in the Wolffian duct epithelium initiate ureteric bud morphogenesis. Dev. Cell. 17:199–209. 10.1016/j.devcel.2009.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiapparo G., Lin X., Lescroart F., Chabab S., Paulissen C., Pitisci L., Bondue A., and Blanpain C.. 2016. Mesp1 controls the speed, polarity, and directionality of cardiovascular progenitor migration. J. Cell Biol. 213:463–477. 10.1083/jcb.201505082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhry P., and Trede N.S.. 2013. DiGeorge syndrome gene tbx1 functions through wnt11r to regulate heart looping and differentiation. PLoS One. 8:e58145 10.1371/journal.pone.0058145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung M.I., Nascone-Yoder N.M., Grover S.A., Drysdale T.A., and Wallingford J.B.. 2010. Direct activation of Shroom3 transcription by Pitx proteins drives epithelial morphogenesis in the developing gut. Development. 137:1339–1349. 10.1242/dev.044610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. 2016. Modeling development and disease with organoids. Cell. 165:1586–1597. 10.1016/j.cell.2016.05.082 [DOI] [PubMed] [Google Scholar]
- Clowes C., Boylan M.G., Ridge L.A., Barnes E., Wright J.A., and Hentges K.E.. 2014. The functional diversity of essential genes required for mammalian cardiac development. Genesis. 52:713–737. 10.1002/dvg.22794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen E.D., Miller M.F., Wang Z., Moon R.T., and Morrisey E.E.. 2012. Wnt5a and Wnt11 are essential for second heart field progenitor development. Development. 139:1931–1940. 10.1242/dev.069377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini F. 2006. Renal branching morphogenesis: Concepts, questions, and recent advances. Differentiation. 74:402–421. 10.1111/j.1432-0436.2006.00106.x [DOI] [PubMed] [Google Scholar]
- Costantini F., and Kopan R.. 2010. Patterning a complex organ: Branching morphogenesis and nephron segmentation in kidney development. Dev. Cell. 18:698–712. 10.1016/j.devcel.2010.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin J.A., Quint E., Tsipouri V., Arkell R.M., Cattanach B., Copp A.J., Henderson D.J., Spurr N., Stanier P., Fisher E.M., et al. 2003. Mutation of Celsr1 disrupts planar polarity of inner ear hair cells and causes severe neural tube defects in the mouse. Curr. Biol. 13:1129–1133. 10.1016/S0960-9822(03)00374-9 [DOI] [PubMed] [Google Scholar]
- Davis A., Amin N.M., Johnson C., Bagley K., Ghashghaei H.T., and Nascone-Yoder N.. 2017. Stomach curvature is generated by left-right asymmetric gut morphogenesis. Development. 144:1477–1483. 10.1242/dev.143701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis N.M., Kurpios N.A., Sun X., Gros J., Martin J.F., and Tabin C.J.. 2008. The chirality of gut rotation derives from left-right asymmetric changes in the architecture of the dorsal mesentery. Dev. Cell. 15:134–145. 10.1016/j.devcel.2008.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Santa Barbara P., van den Brink G.R., and Roberts D.J.. 2003. Development and differentiation of the intestinal epithelium. Cell. Mol. Life Sci. 60:1322–1332. 10.1007/s00018-003-2289-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressler G.R. 2006. The cellular basis of kidney development. Annu. Rev. Cell Dev. Biol. 22:509–529. 10.1146/annurev.cellbio.22.010305.104340 [DOI] [PubMed] [Google Scholar]
- Dressler G.R. 2009. Advances in early kidney specification, development and patterning. Development. 136:3863–3874. 10.1242/dev.034876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer L.A., and Kirby M.L.. 2009. The role of secondary heart field in cardiac development. Dev. Biol. 336:137–144. 10.1016/j.ydbio.2009.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebarasi L., He L., Hultenby K., Takemoto M., Betsholtz C., Tryggvason K., and Majumdar A.. 2009. A reverse genetic screen in the zebrafish identifies crb2b as a regulator of the glomerular filtration barrier. Dev. Biol. 334:1–9. 10.1016/j.ydbio.2009.04.017 [DOI] [PubMed] [Google Scholar]
- Eiraku M., Takata N., Ishibashi H., Kawada M., Sakakura E., Okuda S., Sekiguchi K., Adachi T., and Sasai Y.. 2011. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature. 472:51–56. 10.1038/nature09941 [DOI] [PubMed] [Google Scholar]
- Elghazi L., Cras-Méneur C., Czernichow P., and Scharfmann R.. 2002. Role for FGFR2IIIb-mediated signals in controlling pancreatic endocrine progenitor cell proliferation. Proc. Natl. Acad. Sci. USA. 99:3884–3889. 10.1073/pnas.062321799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Hashash A.H., and Warburton D.. 2011. Cell polarity and spindle orientation in the distal epithelium of embryonic lung. Dev. Dyn. 240:441–445. 10.1002/dvdy.22551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsum I., Yates L., Humbert P.O., and Richardson H.E.. 2012. The Scribble-Dlg-Lgl polarity module in development and cancer: From flies to man. Essays Biochem. 53:141–168. 10.1042/bse0530141 [DOI] [PubMed] [Google Scholar]
- Etheridge S.L., Ray S., Li S., Hamblet N.S., Lijam N., Tsang M., Greer J., Kardos N., Wang J., Sussman D.J., et al. 2008. Murine dishevelled 3 functions in redundant pathways with dishevelled 1 and 2 in normal cardiac outflow tract, cochlea, and neural tube development. PLoS Genet. 4:e1000259 10.1371/journal.pgen.1000259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M.J., Cabral L.J., Stephens R.J., and Freeman G.. 1975. Transformation of alveolar type 2 cells to type 1 cells following exposure to NO2. Exp. Mol. Pathol. 22:142–150. 10.1016/0014-4800(75)90059-3 [DOI] [PubMed] [Google Scholar]
- Evans S.M., Yelon D., Conlon F.L., and Kirby M.L.. 2010. Myocardial lineage development. Circ. Res. 107:1428–1444. 10.1161/CIRCRESAHA.110.227405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francou A., Saint-Michel E., Mesbah K., and Kelly R.G.. 2014. TBX1 regulates epithelial polarity and dynamic basal filopodia in the second heart field. Development. 141:4320–4331. 10.1242/dev.115022 [DOI] [PubMed] [Google Scholar]
- Frank D.B., Peng T., Zepp J.A., Snitow M., Vincent T.L., Penkala I.J., Cui Z., Herriges M.J., Morley M.P., Zhou S., et al. 2016. Emergence of a wave of Wnt signaling that regulates lung alveologenesis through controlling epithelial self-renewal and differentiation. Cell Reports. 17:2312–2325. 10.1016/j.celrep.2016.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freddo A.M., Shoffner S.K., Shao Y., Taniguchi K., Grosse A.S., Guysinger M.N., Wang S., Rudraraju S., Margolis B., Garikipati K., et al. 2016. Coordination of signaling and tissue mechanics during morphogenesis of murine intestinal villi: A role for mitotic cell rounding. Integr. Biol. 8:918–928. 10.1039/C6IB00046K [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujitani Y. 2017. Transcriptional regulation of pancreas development and β-cell function [Review]. Endocr. J. 64:477–486. 10.1507/endocrj.EJ17-0098 [DOI] [PubMed] [Google Scholar]
- Fumoto K., Takigawa-Imamura H., Sumiyama K., Kaneiwa T., and Kikuchi A.. 2017. Modulation of apical constriction by Wnt signaling is required for lung epithelial shape transition. Development. 144:151–162. 10.1242/dev.141325 [DOI] [PubMed] [Google Scholar]
- Garavito-Aguilar Z.V., Riley H.E., and Yelon D.. 2010. Hand2 ensures an appropriate environment for cardiac fusion by limiting Fibronectin function. Development. 137:3215–3220. 10.1242/dev.052225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-García M.J., Shibata M., and Anderson K.V.. 2008. Chato, a KRAB zinc-finger protein, regulates convergent extension in the mouse embryo. Development. 135:3053–3062. 10.1242/dev.022897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gissen P., and Arias I.M.. 2015. Structural and functional hepatocyte polarity and liver disease. J. Hepatol. 63:1023–1037. 10.1016/j.jhep.2015.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittes G.K. 2009. Developmental biology of the pancreas: A comprehensive review. Dev. Biol. 326:4–35. 10.1016/j.ydbio.2008.10.024 [DOI] [PubMed] [Google Scholar]
- Goenezen S., Rennie M.Y., and Rugonyi S.. 2012. Biomechanics of early cardiac development. Biomech. Model. Mechanobiol. 11:1187–1204. 10.1007/s10237-012-0414-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong K.Q., Yallowitz A.R., Sun H., Dressler G.R., and Wellik D.M.. 2007. A Hox-Eya-Pax complex regulates early kidney developmental gene expression. Mol. Cell. Biol. 27:7661–7668. 10.1128/MCB.00465-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grahammer F., Schell C., and Huber T.B.. 2013. The podocyte slit diaphragm--from a thin grey line to a complex signalling hub. Nat. Rev. Nephrol. 9:587–598. 10.1038/nrneph.2013.169 [DOI] [PubMed] [Google Scholar]
- Gray R.S., Roszko I., and Solnica-Krezel L.. 2011. Planar cell polarity: Coordinating morphogenetic cell behaviors with embryonic polarity. Dev. Cell. 21:120–133. 10.1016/j.devcel.2011.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greka A., and Mundel P.. 2012. Cell biology and pathology of podocytes. Annu. Rev. Physiol. 74:299–323. 10.1146/annurev-physiol-020911-153238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosse A.S., Pressprich M.F., Curley L.B., Hamilton K.L., Margolis B., Hildebrand J.D., and Gumucio D.L.. 2011. Cell dynamics in fetal intestinal epithelium: Implications for intestinal growth and morphogenesis. Development. 138:4423–4432. 10.1242/dev.065789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote D., Boualia S.K., Souabni A., Merkel C., Chi X., Costantini F., Carroll T., and Bouchard M.. 2008. Gata3 acts downstream of beta-catenin signaling to prevent ectopic metanephric kidney induction. PLoS Genet. 4:e1000316 10.1371/journal.pgen.1000316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J.K., Menke A.L., Gubler M.C., Clarke A.R., Harrison D., Hammes A., Hastie N.D., and Schedl A.. 2002. WT1 is a key regulator of podocyte function: Reduced expression levels cause crescentic glomerulonephritis and mesangial sclerosis. Hum. Mol. Genet. 11:651–659. 10.1093/hmg/11.6.651 [DOI] [PubMed] [Google Scholar]
- Halbleib J.M., Sääf A.M., Brown P.O., and Nelson W.J.. 2007. Transcriptional modulation of genes encoding structural characteristics of differentiating enterocytes during development of a polarized epithelium in vitro. Mol. Biol. Cell. 18:4261–4278. 10.1091/mbc.e07-04-0308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamblet N.S., Lijam N., Ruiz-Lozano P., Wang J., Yang Y., Luo Z., Mei L., Chien K.R., Sussman D.J., and Wynshaw-Boris A.. 2002. Dishevelled 2 is essential for cardiac outflow tract development, somite segmentation and neural tube closure. Development. 129:5827–5838. 10.1242/dev.00164 [DOI] [PubMed] [Google Scholar]
- Hartleben B., Schweizer H., Lübben P., Bartram M.P., Möller C.C., Herr R., Wei C., Neumann-Haefelin E., Schermer B., Zentgraf H., et al. 2008. Neph-Nephrin proteins bind the Par3-Par6-atypical protein kinase C (aPKC) complex to regulate podocyte cell polarity. J. Biol. Chem. 283:23033–23038. 10.1074/jbc.M803143200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasosah M., Lemberg D.A., Skarsgard E., and Schreiber R.. 2008. Congenital short bowel syndrome: A case report and review of the literature. Can. J. Gastroenterol. 22:71–74. 10.1155/2008/590143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecksher-Sørensen J., Watson R.P., Lettice L.A., Serup P., Eley L., De Angelis C., Ahlgren U., and Hill R.E.. 2004. The splanchnic mesodermal plate directs spleen and pancreatic laterality, and is regulated by Bapx1/Nkx3.2. Development. 131:4665–4675. 10.1242/dev.01364 [DOI] [PubMed] [Google Scholar]
- Henderson D.J., Conway S.J., Greene N.D., Gerrelli D., Murdoch J.N., Anderson R.H., and Copp A.J.. 2001. Cardiovascular defects associated with abnormalities in midline development in the Loop-tail mouse mutant. Circ. Res. 89:6–12. 10.1161/hh1301.092497 [DOI] [PubMed] [Google Scholar]
- Herriges M., and Morrisey E.E.. 2014. Lung development: Orchestrating the generation and regeneration of a complex organ. Development. 141:502–513. 10.1242/dev.098186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand J.D. 2005. Shroom regulates epithelial cell shape via the apical positioning of an actomyosin network. J. Cell Sci. 118:5191–5203. 10.1242/jcs.02626 [DOI] [PubMed] [Google Scholar]
- Hirose T., Karasawa M., Sugitani Y., Fujisawa M., Akimoto K., Ohno S., and Noda T.. 2006. PAR3 is essential for cyst-mediated epicardial development by establishing apical cortical domains. Development. 133:1389–1398. 10.1242/dev.02294 [DOI] [PubMed] [Google Scholar]
- Hoffman J.I., and Kaplan S.. 2002. The incidence of congenital heart disease. J. Am. Coll. Cardiol. 39:1890–1900. 10.1016/S0735-1097(02)01886-7 [DOI] [PubMed] [Google Scholar]
- Hogan B.L., Barkauskas C.E., Chapman H.A., Epstein J.A., Jain R., Hsia C.C., Niklason L., Calle E., Le A., Randell S.H., et al. 2014. Repair and regeneration of the respiratory system: Complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell. 15:123–138. 10.1016/j.stem.2014.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman N.G., Schoenebeck J.J., Tsai H.J., and Yelon D.. 2007. Endocardium is necessary for cardiomyocyte movement during heart tube assembly. Development. 134:2379–2386. 10.1242/dev.02857 [DOI] [PubMed] [Google Scholar]
- Horne-Badovinac S., Rebagliati M., and Stainier D.Y.. 2003. A cellular framework for gut-looping morphogenesis in zebrafish. Science. 302:662–665. 10.1126/science.1085397 [DOI] [PubMed] [Google Scholar]
- Hosseini H.S., Garcia K.E., and Taber L.A.. 2017. A new hypothesis for foregut and heart tube formation based on differential growth and actomyosin contraction. Development. 144:2381–2391. 10.1242/dev.145193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Ruan H., Aw M.Y., Hussain A., Guo L., Gao C., Qian F., Leung T., Song H., Kimelman D., et al. 2008. Mypt1-mediated spatial positioning of Bmp2-producing cells is essential for liver organogenesis. Development. 135:3209–3218. 10.1242/dev.024406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadzik R.S., Cohen E.D., Morley M.P., Stewart K.M., Lu M.M., and Morrisey E.E.. 2014. Wnt ligand/Frizzled 2 receptor signaling regulates tube shape and branch-point formation in the lung through control of epithelial cell shape. Proc. Natl. Acad. Sci. USA. 111:12444–12449. 10.1073/pnas.1406639111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapanci Y., Weibel E.R., Kaplan H.P., and Robinson F.R.. 1969. Pathogenesis and reversibility of the pulmonary lesions of oxygen toxicity in monkeys. II. Ultrastructural and morphometric studies. Lab. Invest. 20:101–118. [PubMed] [Google Scholar]
- Karner C., Wharton K.A. Jr., and Carroll T.J.. 2006. Planar cell polarity and vertebrate organogenesis. Semin. Cell Dev. Biol. 17:194–203. 10.1016/j.semcdb.2006.05.003 [DOI] [PubMed] [Google Scholar]
- Karner C.M., Das A., Ma Z., Self M., Chen C., Lum L., Oliver G., and Carroll T.J.. 2011. Canonical Wnt9b signaling balances progenitor cell expansion and differentiation during kidney development. Development. 138:1247–1257. 10.1242/dev.057646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller R. 2006. Mechanisms of elongation in embryogenesis. Development. 133:2291–2302. 10.1242/dev.02406 [DOI] [PubMed] [Google Scholar]
- Keller R., Davidson L., Edlund A., Elul T., Ezin M., Shook D., and Skoglund P.. 2000. Mechanisms of convergence and extension by cell intercalation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355:897–922. 10.1098/rstb.2000.0626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly R.G. 2016. How Mesp1 makes a move. J. Cell Biol. 213:411–413. 10.1083/jcb.201604121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly R.G., Brown N.A., and Buckingham M.E.. 2001. The arterial pole of the mouse heart forms from Fgf10-expressing cells in pharyngeal mesoderm. Dev. Cell. 1:435–440. 10.1016/S1534-5807(01)00040-5 [DOI] [PubMed] [Google Scholar]
- Kesavan G., Sand F.W., Greiner T.U., Johansson J.K., Kobberup S., Wu X., Brakebusch C., and Semb H.. 2009. Cdc42-mediated tubulogenesis controls cell specification. Cell. 139:791–801. 10.1016/j.cell.2009.08.049 [DOI] [PubMed] [Google Scholar]
- Kim B.M., Mao J., Taketo M.M., and Shivdasani R.A.. 2007. Phases of canonical Wnt signaling during the development of mouse intestinal epithelium. Gastroenterology. 133:529–538. 10.1053/j.gastro.2007.04.072 [DOI] [PubMed] [Google Scholar]
- Kim H.Y., Varner V.D., and Nelson C.M.. 2013. Apical constriction initiates new bud formation during monopodial branching of the embryonic chicken lung. Development. 140:3146–3155. 10.1242/dev.093682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.H., Kim B.M., Mao J., Rowan S., and Shivdasani R.A.. 2011. Endodermal Hedgehog signals modulate Notch pathway activity in the developing digestive tract mesenchyme. Development. 138:3225–3233. 10.1242/dev.066233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura J., and Deutsch G.H.. 2007. Key mechanisms of early lung development. Pediatr. Dev. Pathol. 10:335–347. 10.2350/07-06-0290.1 [DOI] [PubMed] [Google Scholar]
- Kobayashi N., Gao S.Y., Chen J., Saito K., Miyawaki K., Li C.Y., Pan L., Saito S., Terashita T., and Matsuda S.. 2004. Process formation of the renal glomerular podocyte: Is there common molecular machinery for processes of podocytes and neurons? Anat. Sci. Int. 79:1–10. 10.1111/j.1447-073x.2004.00066.x [DOI] [PubMed] [Google Scholar]
- Kosodo Y. 2012. Interkinetic nuclear migration: Beyond a hallmark of neurogenesis. Cell. Mol. Life Sci. 69:2727–2738. 10.1007/s00018-012-0952-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C.T., Morrisey E.E., Anandappa R., Sigrist K., Lu M.M., Parmacek M.S., Soudais C., and Leiden J.M.. 1997. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 11:1048–1060. 10.1101/gad.11.8.1048 [DOI] [PubMed] [Google Scholar]
- Kurpios N.A., Ibañes M., Davis N.M., Lui W., Katz T., Martin J.F., Izpisúa Belmonte J.C., and Tabin C.J.. 2008. The direction of gut looping is established by changes in the extracellular matrix and in cell:cell adhesion. Proc. Natl. Acad. Sci. USA. 105:8499–8506. 10.1073/pnas.0803578105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuure S., Cebrian C., Machingo Q., Lu B.C., Chi X., Hyink D., D’Agati V., Gurniak C., Witke W., and Costantini F.. 2010. Actin depolymerizing factors cofilin1 and destrin are required for ureteric bud branching morphogenesis. PLoS Genet. 6:e1001176 10.1371/journal.pgen.1001176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M.A., Renner M., Martin C.A., Wenzel D., Bicknell L.S., Hurles M.E., Homfray T., Penninger J.M., Jackson A.P., and Knoblich J.A.. 2013. Cerebral organoids model human brain development and microcephaly. Nature. 501:373–379. 10.1038/nature12517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen H.L., and Grapin-Botton A.. 2017. The molecular and morphogenetic basis of pancreas organogenesis. Semin. Cell Dev. Biol. 66:51–68. 10.1016/j.semcdb.2017.01.005 [DOI] [PubMed] [Google Scholar]
- Lazzaro D., Price M., de Felice M., and Di Lauro R.. 1991. The transcription factor TTF-1 is expressed at the onset of thyroid and lung morphogenesis and in restricted regions of the foetal brain. Development. 113:1093–1104. [DOI] [PubMed] [Google Scholar]
- Lebeche D., Malpel S., and Cardoso W.V.. 1999. Fibroblast growth factor interactions in the developing lung. Mech. Dev. 86:125–136. 10.1016/S0925-4773(99)00124-0 [DOI] [PubMed] [Google Scholar]
- Le Bivic A., Quaroni A., Nichols B., and Rodriguez-Boulan E.. 1990. Biogenetic pathways of plasma membrane proteins in Caco-2, a human intestinal epithelial cell line. J. Cell Biol. 111:1351–1361. 10.1083/jcb.111.4.1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuit T., and Lenne P.F.. 2007. Cell surface mechanics and the control of cell shape, tissue patterns and morphogenesis. Nat. Rev. Mol. Cell Biol. 8:633–644. 10.1038/nrm2222 [DOI] [PubMed] [Google Scholar]
- Lienkamp S.S., Liu K., Karner C.M., Carroll T.J., Ronneberger O., Wallingford J.B., and Walz G.. 2012. Vertebrate kidney tubules elongate using a planar cell polarity-dependent, rosette-based mechanism of convergent extension. Nat. Genet. 44:1382–1387. 10.1038/ng.2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey S.E., Butcher J.T., and Yalcin H.C.. 2014. Mechanical regulation of cardiac development. Front. Physiol. 5:318 10.3389/fphys.2014.00318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little M.H., and McMahon A.P.. 2012. Mammalian kidney development: Principles, progress, and projections. Cold Spring Harb. Perspect. Biol. 4:a008300 10.1101/cshperspect.a008300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little M.H., Brennan J., Georgas K., Davies J.A., Davidson D.R., Baldock R.A., Beverdam A., Bertram J.F., Capel B., Chiu H.S., et al. 2007. A high-resolution anatomical ontology of the developing murine genitourinary tract. Gene Expr. Patterns. 7:680–699. 10.1016/j.modgep.2007.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Bressan M., Hassel D., Huisken J., Staudt D., Kikuchi K., Poss K.D., Mikawa T., and Stainier D.Y.. 2010. A dual role for ErbB2 signaling in cardiac trabeculation. Development. 137:3867–3875. 10.1242/dev.053736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loots E., Hillen B., and Veldman A.E.. 2003. The role of hemodynamics in the development of the outflow tract of the heart. J. Eng. Math. 45:91–104. 10.1023/A:1022029300196 [DOI] [Google Scholar]
- Macara I.G. 2004. Par proteins: Partners in polarization. Curr. Biol. 14:R160–R162. 10.1016/j.cub.2004.01.048 [DOI] [PubMed] [Google Scholar]
- Mahadevan A., Welsh I.C., Sivakumar A., Gludish D.W., Shilvock A.R., Noden D.M., Huss D., Lansford R., and Kurpios N.A.. 2014. The left-right Pitx2 pathway drives organ-specific arterial and lymphatic development in the intestine. Dev. Cell. 31:690–706. 10.1016/j.devcel.2014.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar A., Vainio S., Kispert A., McMahon J., and McMahon A.P.. 2003. Wnt11 and Ret/Gdnf pathways cooperate in regulating ureteric branching during metanephric kidney development. Development. 130:3175–3185. 10.1242/dev.00520 [DOI] [PubMed] [Google Scholar]
- Männer J. 2000. Cardiac looping in the chick embryo: A morphological review with special reference to terminological and biomechanical aspects of the looping process. Anat. Rec. 259:248–262. [DOI] [PubMed] [Google Scholar]
- Mao J., Kim B.M., Rajurkar M., Shivdasani R.A., and McMahon A.P.. 2010. Hedgehog signaling controls mesenchymal growth in the developing mammalian digestive tract. Development. 137:1721–1729. 10.1242/dev.044586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Estrada O.M., Lettice L.A., Essafi A., Guadix J.A., Slight J., Velecela V., Hall E., Reichmann J., Devenney P.S., Hohenstein P., et al. 2010. Wt1 is required for cardiovascular progenitor cell formation through transcriptional control of Snail and E-cadherin. Nat. Genet. 42:89–93. 10.1038/ng.494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty-Santos L., and Cleaver O.. 2016. Pdx1 regulates pancreas tubulogenesis and E-cadherin expression. Development. 143:101–112. 10.1242/dev.126755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastracci T.L., and Sussel L.. 2012. The endocrine pancreas: Insights into development, differentiation, and diabetes. Wiley Interdiscip. Rev. Dev. Biol. 1:609–628. 10.1002/wdev.44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathan M., Moxey P.C., and Trier J.S.. 1976. Morphogenesis of fetal rat duodenal villi. Am. J. Anat. 146:73–92. 10.1002/aja.1001460104 [DOI] [PubMed] [Google Scholar]
- Matsumoto A., Hashimoto K., Yoshioka T., and Otani H.. 2002. Occlusion and subsequent re-canalization in early duodenal development of human embryos: Integrated organogenesis and histogenesis through a possible epithelial-mesenchymal interaction. Anat. Embryol. (Berl.). 205:53–65. 10.1007/s00429-001-0226-5 [DOI] [PubMed] [Google Scholar]
- Matsuyama M., Aizawa S., and Shimono A.. 2009. Sfrp controls apicobasal polarity and oriented cell division in developing gut epithelium. PLoS Genet. 5:e1000427 10.1371/journal.pgen.1000427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matter K., Brauchbar M., Bucher K., and Hauri H.P.. 1990. Sorting of endogenous plasma membrane proteins occurs from two sites in cultured human intestinal epithelial cells (Caco-2). Cell. 60:429–437. 10.1016/0092-8674(90)90594-5 [DOI] [PubMed] [Google Scholar]
- McCulley D., Wienhold M., and Sun X.. 2015. The pulmonary mesenchyme directs lung development. Curr. Opin. Genet. Dev. 32:98–105. 10.1016/j.gde.2015.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden D.G., and Olson E.N.. 2002. Heart development: Learning from mistakes. Curr. Opin. Genet. Dev. 12:328–335. 10.1016/S0959-437X(02)00306-4 [DOI] [PubMed] [Google Scholar]
- McGrath P.S., and Wells J.M.. 2015. SnapShot: GI tract development. Cell. 161:176–176.e1. 10.1016/j.cell.2015.03.014 [DOI] [PubMed] [Google Scholar]
- Mellman I., and Nelson W.J.. 2008. Coordinated protein sorting, targeting and distribution in polarized cells. Nat. Rev. Mol. Cell Biol. 9:833–845. 10.1038/nrm2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger R.J., Klein O.D., Martin G.R., and Krasnow M.A.. 2008. The branching programme of mouse lung development. Nature. 453:745–750. 10.1038/nature07005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer E.J., Ikmi A., and Gibson M.C.. 2011. Interkinetic nuclear migration is a broadly conserved feature of cell division in pseudostratified epithelia. Curr. Biol. 21:485–491. 10.1016/j.cub.2011.02.002 [DOI] [PubMed] [Google Scholar]
- Michael L., and Davies J.A.. 2004. Pattern and regulation of cell proliferation during murine ureteric bud development. J. Anat. 204:241–255. 10.1111/j.0021-8782.2004.00285.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michos O. 2009. Kidney development: From ureteric bud formation to branching morphogenesis. Curr. Opin. Genet. Dev. 19:484–490. 10.1016/j.gde.2009.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mjaatvedt C.H., Nakaoka T., Moreno-Rodriguez R., Norris R.A., Kern M.J., Eisenberg C.A., Turner D., and Markwald R.R.. 2001. The outflow tract of the heart is recruited from a novel heart-forming field. Dev. Biol. 238:97–109. 10.1006/dbio.2001.0409 [DOI] [PubMed] [Google Scholar]
- Moeller H., Jenny A., Schaeffer H.J., Schwarz-Romond T., Mlodzik M., Hammerschmidt M., and Birchmeier W.. 2006. Diversin regulates heart formation and gastrulation movements in development. Proc. Natl. Acad. Sci. USA. 103:15900–15905. 10.1073/pnas.0603808103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J.K., Lin W., Hauser C., Marchuk Y., Getman D., and Lee K.F.. 1999. Rescue of the cardiac defect in ErbB2 mutant mice reveals essential roles of ErbB2 in peripheral nervous system development. Neuron. 23:273–283. 10.1016/S0896-6273(00)80779-5 [DOI] [PubMed] [Google Scholar]
- Morrisey E.E., and Hogan B.L.. 2010. Preparing for the first breath: Genetic and cellular mechanisms in lung development. Dev. Cell. 18:8–23. 10.1016/j.devcel.2009.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser M., Pscherer A., Roth C., Becker J., Mücher G., Zerres K., Dixkens C., Weis J., Guay-Woodford L., Buettner R., and Fässler R.. 1997. Enhanced apoptotic cell death of renal epithelial cells in mice lacking transcription factor AP-2β. Genes Dev. 11:1938–1948. 10.1101/gad.11.15.1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutze K., Vierkotten S., Milosevic J., Eickelberg O., and Königshoff M.. 2015. Enolase 1 (ENO1) and protein disulfide-isomerase associated 3 (PDIA3) regulate Wnt/β-catenin-driven trans-differentiation of murine alveolar epithelial cells. Dis. Model. Mech. 8:877–890. 10.1242/dmm.019117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy I.I., Railo A., Rapila R., Hast T., Sormunen R., Tavi P., Räsänen J., and Vainio S.J.. 2010. Wnt-11 signalling controls ventricular myocardium development by patterning N-cadherin and beta-catenin expression. Cardiovasc. Res. 85:100–109. 10.1093/cvr/cvp254 [DOI] [PubMed] [Google Scholar]
- New L.A., Keyvani Chahi A., and Jones N.. 2013. Direct regulation of nephrin tyrosine phosphorylation by Nck adaptor proteins. J. Biol. Chem. 288:1500–1510. 10.1074/jbc.M112.439463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norden C. 2017. Pseudostratified epithelia - cell biology, diversity and roles in organ formation at a glance. J. Cell Sci. 130:1859–1863. 10.1242/jcs.192997 [DOI] [PubMed] [Google Scholar]
- Ober E.A., and Lemaigre F.P.. 2018. Development of the liver: Insights into organ and tissue morphogenesis. J. Hepatol. 68:1049–1062. 10.1016/j.jhep.2018.01.005 [DOI] [PubMed] [Google Scholar]
- Ocaña O.H., Coskun H., Minguillón C., Murawala P., Tanaka E.M., Galcerán J., Muñoz-Chápuli R., and Nieto M.A.. 2017. A right-handed signalling pathway drives heart looping in vertebrates. Nature. 549:86–90. 10.1038/nature23454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paces-Fessy M., Fabre M., Lesaulnier C., and Cereghini S.. 2012. Hnf1b and Pax2 cooperate to control different pathways in kidney and ureter morphogenesis. Hum. Mol. Genet. 21:3143–3155. 10.1093/hmg/dds141 [DOI] [PubMed] [Google Scholar]
- Packard A., Georgas K., Michos O., Riccio P., Cebrian C., Combes A.N., Ju A., Ferrer-Vaquer A., Hadjantonakis A.K., Zong H., et al. 2013. Luminal mitosis drives epithelial cell dispersal within the branching ureteric bud. Dev. Cell. 27:319–330. 10.1016/j.devcel.2013.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parviz F., Matullo C., Garrison W.D., Savatski L., Adamson J.W., Ning G., Kaestner K.H., Rossi J.M., Zaret K.S., and Duncan S.A.. 2003. Hepatocyte nuclear factor 4α controls the development of a hepatic epithelium and liver morphogenesis. Nat. Genet. 34:292–296. 10.1038/ng1175 [DOI] [PubMed] [Google Scholar]
- Passer D., van de Vrugt A., Atmanli A., and Domian I.J.. 2016. Atypical protein kinase C-dependent polarized cell division is required for myocardial trabeculation. Cell Reports. 14:1662–1672. 10.1016/j.celrep.2016.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips H.M., Murdoch J.N., Chaudhry B., Copp A.J., and Henderson D.J.. 2005. Vangl2 acts via RhoA signaling to regulate polarized cell movements during development of the proximal outflow tract. Circ. Res. 96:292–299. 10.1161/01.RES.0000154912.08695.88 [DOI] [PubMed] [Google Scholar]
- Phillips H.M., Rhee H.J., Murdoch J.N., Hildreth V., Peat J.D., Anderson R.H., Copp A.J., Chaudhry B., and Henderson D.J.. 2007. Disruption of planar cell polarity signaling results in congenital heart defects and cardiomyopathy attributable to early cardiomyocyte disorganization. Circ. Res. 101:137–145. 10.1161/CIRCRESAHA.106.142406 [DOI] [PubMed] [Google Scholar]
- Pieczynski J., and Margolis B.. 2011. Protein complexes that control renal epithelial polarity. Am. J. Physiol. Renal Physiol. 300:F589–F601. 10.1152/ajprenal.00615.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plageman T.F. Jr., Chung M.I., Lou M., Smith A.N., Hildebrand J.D., Wallingford J.B., and Lang R.A.. 2010. Pax6-dependent Shroom3 expression regulates apical constriction during lens placode invagination. Development. 137:405–415. 10.1242/dev.045369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plageman T.F. Jr., Zacharias A.L., Gage P.J., and Lang R.A.. 2011. Shroom3 and a Pitx2-N-cadherin pathway function cooperatively to generate asymmetric cell shape changes during gut morphogenesis. Dev. Biol. 357:227–234. 10.1016/j.ydbio.2011.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poncy A., Antoniou A., Cordi S., Pierreux C.E., Jacquemin P., and Lemaigre F.P.. 2015. Transcription factors SOX4 and SOX9 cooperatively control development of bile ducts. Dev. Biol. 404:136–148. 10.1016/j.ydbio.2015.05.012 [DOI] [PubMed] [Google Scholar]
- Rajagopal J., Carroll T.J., Guseh J.S., Bores S.A., Blank L.J., Anderson W.J., Yu J., Zhou Q., McMahon A.P., and Melton D.A.. 2008. Wnt7b stimulates embryonic lung growth by coordinately increasing the replication of epithelium and mesenchyme. Development. 135:1625–1634. 10.1242/dev.015495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranga A., Girgin M., Meinhardt A., Eberle D., Caiazzo M., Tanaka E.M., and Lutolf M.P.. 2016. Neural tube morphogenesis in synthetic 3D microenvironments. Proc. Natl. Acad. Sci. USA. 113:E6831–E6839. 10.1073/pnas.1603529113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasouli S.J., and Stainier D.Y.R.. 2017. Regulation of cardiomyocyte behavior in zebrafish trabeculation by Neuregulin 2a signaling. Nat. Commun. 8:15281 10.1038/ncomms15281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins E.L., Clark C.P., Xue Y., and Hogan B.L.. 2009. The Id2+ distal tip lung epithelium contains individual multipotent embryonic progenitor cells. Development. 136:3741–3745. 10.1242/dev.037317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raya A., and Izpisúa Belmonte J.C.. 2006. Left-right asymmetry in the vertebrate embryo: from early information to higher-level integration. Nat. Rev. Genet. 7:283–293. 10.1038/nrg1830 [DOI] [PubMed] [Google Scholar]
- Reed R.A., Womble M.A., Dush M.K., Tull R.R., Bloom S.K., Morckel A.R., Devlin E.W., and Nascone-Yoder N.M.. 2009. Morphogenesis of the primitive gut tube is generated by Rho/ROCK/myosin II-mediated endoderm rearrangements. Dev. Dyn. 238:3111–3125. 10.1002/dvdy.22157 [DOI] [PubMed] [Google Scholar]
- Reginensi A., Scott R.P., Gregorieff A., Bagherie-Lachidan M., Chung C., Lim D.S., Pawson T., Wrana J., and McNeill H.. 2013. Yap- and Cdc42-dependent nephrogenesis and morphogenesis during mouse kidney development. PLoS Genet. 9:e1003380 10.1371/journal.pgen.1003380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts D.J. 2000. Molecular mechanisms of development of the gastrointestinal tract. Dev. Dyn. 219:109–120. [DOI] [PubMed] [Google Scholar]
- Rock J.R., and Hogan B.L.. 2011. Epithelial progenitor cells in lung development, maintenance, repair, and disease. Annu. Rev. Cell Dev. Biol. 27:493–512. 10.1146/annurev-cellbio-100109-104040 [DOI] [PubMed] [Google Scholar]
- Rocque B., and Torban E.. 2015. Planar cell polarity pathway in kidney development and function. Adv. Nephrol. 2015:1–15. 10.1155/2015/764682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohr S., Bit-Avragim N., and Abdelilah-Seyfried S.. 2006. Heart and soul/PRKCi and nagie oko/Mpp5 regulate myocardial coherence and remodeling during cardiac morphogenesis. Development. 133:107–115. 10.1242/dev.02182 [DOI] [PubMed] [Google Scholar]
- Rosekrans S.L., Baan B., Muncan V., and van den Brink G.R.. 2015. Esophageal development and epithelial homeostasis. Am. J. Physiol. Gastrointest. Liver Physiol. 309:G216–G228. 10.1152/ajpgi.00088.2015 [DOI] [PubMed] [Google Scholar]
- Rozycki H.J., and Hendricks-Munos K.D.. 2017. Structure and development of alveolar epithelial cells. In Fetal and Neonatal Physiology. Fifth edition Polin R.A., Abman S.H., Rowitch D., Benitz W.E., and Fox W.W., editors. Elsevier; 809–813. 10.1016/B978-0-323-35214-7.00081-0 [DOI] [Google Scholar]
- Sääf A.M., Halbleib J.M., Chen X., Yuen S.T., Leung S.Y., Nelson W.J., and Brown P.O.. 2007. Parallels between global transcriptional programs of polarizing Caco-2 intestinal epithelial cells in vitro and gene expression programs in normal colon and colon cancer. Mol. Biol. Cell. 18:4245–4260. 10.1091/mbc.e07-04-0309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saburi S., Hester I., Fischer E., Pontoglio M., Eremina V., Gessler M., Quaggin S.E., Harrison R., Mount R., and McNeill H.. 2008. Loss of Fat4 disrupts PCP signaling and oriented cell division and leads to cystic kidney disease. Nat. Genet. 40:1010–1015. 10.1038/ng.179 [DOI] [PubMed] [Google Scholar]
- Sadler T.W., and Langman J.. 2012. Langman’s Medical Embryology. Wolters Kluwer Health/Lippincott Williams & Wilkins, Philadelphia. [Google Scholar]
- Sakurai H., Tsukamoto T., Kjelsberg C.A., Cantley L.G., and Nigam S.K.. 1997. EGF receptor ligands are a large fraction of in vitro branching morphogens secreted by embryonic kidney. Am. J. Physiol. 273:F463–F472. [DOI] [PubMed] [Google Scholar]
- Šamánek M. 2000. Congenital heart malformations: Prevalence, severity, survival, and quality of life. Cardiol. Young. 10:179–185. 10.1017/S1047951100009082 [DOI] [PubMed] [Google Scholar]
- Samsa L.A., Yang B., and Liu J.. 2013. Embryonic cardiac chamber maturation: Trabeculation, conduction, and cardiomyocyte proliferation. Am. J. Med. Genet. C. Semin. Med. Genet. 163:157–168. 10.1002/ajmg.c.31366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saotome I., Curto M., and McClatchey A.I.. 2004. Ezrin is essential for epithelial organization and villus morphogenesis in the developing intestine. Dev. Cell. 6:855–864. 10.1016/j.devcel.2004.05.007 [DOI] [PubMed] [Google Scholar]
- Sauer F.C. 1935. Mitosis in the neural tube. J. Comp. Neurol. 62:377–405. 10.1002/cne.900620207 [DOI] [Google Scholar]
- Saxén L. 1987. Organogenesis of the Kidney. Cambridge University Press, Cambridge: 10.1017/CBO9780511565083 [DOI] [Google Scholar]
- Scambler P.J. 2010. 22q11 deletion syndrome: A role for TBX1 in pharyngeal and cardiovascular development. Pediatr. Cardiol. 31:378–390. 10.1007/s00246-009-9613-0 [DOI] [PubMed] [Google Scholar]
- Schedl A. 2007. Renal abnormalities and their developmental origin. Nat. Rev. Genet. 8:791–802. 10.1038/nrg2205 [DOI] [PubMed] [Google Scholar]
- Schell C., Baumhakl L., Salou S., Conzelmann A.C., Meyer C., Helmstädter M., Wrede C., Grahammer F., Eimer S., Kerjaschki D., et al. 2013. N-wasp is required for stabilization of podocyte foot processes. J. Am. Soc. Nephrol. 24:713–721. 10.1681/ASN.2012080844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell C., Wanner N., and Huber T.B.. 2014. Glomerular development--shaping the multi-cellular filtration unit. Semin. Cell Dev. Biol. 36:39–49. 10.1016/j.semcdb.2014.07.016 [DOI] [PubMed] [Google Scholar]
- Schleiffarth J.R., Person A.D., Martinsen B.J., Sukovich D.J., Neumann A., Baker C.V., Lohr J.L., Cornfield D.N., Ekker S.C., and Petryk A.. 2007. Wnt5a is required for cardiac outflow tract septation in mice. Pediatr. Res. 61:386–391. 10.1203/pdr.0b013e3180323810 [DOI] [PubMed] [Google Scholar]
- Schmidt-Ott K.M., and Barasch J.. 2008. WNT/beta-catenin signaling in nephron progenitors and their epithelial progeny. Kidney Int. 74:1004–1008. 10.1038/ki.2008.322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott R.P., Hawley S.P., Ruston J., Du J., Brakebusch C., Jones N., and Pawson T.. 2012. Podocyte-specific loss of Cdc42 leads to congenital nephropathy. J. Am. Soc. Nephrol. 23:1149–1154. 10.1681/ASN.2011121206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert J.R., and Mlodzik M.. 2007. Frizzled/PCP signalling: A conserved mechanism regulating cell polarity and directed motility. Nat. Rev. Genet. 8:126–138. 10.1038/nrg2042 [DOI] [PubMed] [Google Scholar]
- Sekine K., Ohuchi H., Fujiwara M., Yamasaki M., Yoshizawa T., Sato T., Yagishita N., Matsui D., Koga Y., Itoh N., and Kato S.. 1999. Fgf10 is essential for limb and lung formation. Nat. Genet. 21:138–141. 10.1038/5096 [DOI] [PubMed] [Google Scholar]
- Shackelford D.B., and Shaw R.J.. 2009. The LKB1-AMPK pathway: Metabolism and growth control in tumour suppression. Nat. Rev. Cancer. 9:563–575. 10.1038/nrc2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiratori H., Yashiro K., Shen M.M., and Hamada H.. 2006. Conserved regulation and role of Pitx2 in situs-specific morphogenesis of visceral organs. Development. 133:3015–3025. 10.1242/dev.02470 [DOI] [PubMed] [Google Scholar]
- Sinha T., Wang B., Evans S., Wynshaw-Boris A., and Wang J.. 2012. Disheveled mediated planar cell polarity signaling is required in the second heart field lineage for outflow tract morphogenesis. Dev. Biol. 370:135–144. 10.1016/j.ydbio.2012.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., McColl J., Camp E., Kennerley N., Mok G.F., McCormick D., Grocott T., Wheeler G.N., and Münsterberg A.E.. 2014. Smad1 transcription factor integrates BMP2 and Wnt3a signals in migrating cardiac progenitor cells. Proc. Natl. Acad. Sci. USA. 111:7337–7342. 10.1073/pnas.1321764111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear P.C., and Erickson C.A.. 2012. Interkinetic nuclear migration: A mysterious process in search of a function. Dev. Growth Differ. 54:306–316. 10.1111/j.1440-169X.2012.01342.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stainier D.Y. 2001. Zebrafish genetics and vertebrate heart formation. Nat. Rev. Genet. 2:39–48. 10.1038/35047564 [DOI] [PubMed] [Google Scholar]
- Stubbs J.L., Vladar E.K., Axelrod J.D., and Kintner C.. 2012. Multicilin promotes centriole assembly and ciliogenesis during multiciliate cell differentiation. Nat. Cell Biol. 14:140–147. 10.1038/ncb2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suri C., Jones P.F., Patan S., Bartunkova S., Maisonpierre P.C., Davis S., Sato T.N., and Yancopoulos G.D.. 1996. Requisite role of angiopoietin-1, a ligand for the TIE2 receptor, during embryonic angiogenesis. Cell. 87:1171–1180. 10.1016/S0092-8674(00)81813-9 [DOI] [PubMed] [Google Scholar]
- Swarr D.T., and Morrisey E.E.. 2015. Lung endoderm morphogenesis: Gasping for form and function. Annu. Rev. Cell Dev. Biol. 31:553–573. 10.1146/annurev-cellbio-100814-125249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymaniak A.D., Mahoney J.E., Cardoso W.V., and Varelas X.. 2015. Crumbs3-mediated polarity directs airway epithelial cell fate through the Hippo pathway effector Yap. Dev. Cell. 34:283–296. 10.1016/j.devcel.2015.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taber L.A. 2001. Biomechanics of cardiovascular development. Annu. Rev. Biomed. Eng. 3:1–25. 10.1146/annurev.bioeng.3.1.1 [DOI] [PubMed] [Google Scholar]
- Takahashi M. 2001. The GDNF/RET signaling pathway and human diseases. Cytokine Growth Factor Rev. 12:361–373. 10.1016/S1359-6101(01)00012-0 [DOI] [PubMed] [Google Scholar]
- Tang M.J., Cai Y., Tsai S.J., Wang Y.K., and Dressler G.R.. 2002. Ureteric bud outgrowth in response to RET activation is mediated by phosphatidylinositol 3-kinase. Dev. Biol. 243:128–136. 10.1006/dbio.2001.0557 [DOI] [PubMed] [Google Scholar]
- Tanimizu N., Miyajima A., and Mostov K.E.. 2007. Liver progenitor cells develop cholangiocyte-type epithelial polarity in three-dimensional culture. Mol. Biol. Cell. 18:1472–1479. 10.1091/mbc.e06-09-0848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimizu N., Nakamura Y., Ichinohe N., Mizuguchi T., Hirata K., and Mitaka T.. 2013. Hepatic biliary epithelial cells acquire epithelial integrity but lose plasticity to differentiate into hepatocytes in vitro during development. J. Cell Sci. 126:5239–5246. 10.1242/jcs.133082 [DOI] [PubMed] [Google Scholar]
- Toyofuku T., Zhang H., Kumanogoh A., Takegahara N., Yabuki M., Harada K., Hori M., and Kikutani H.. 2004. Guidance of myocardial patterning in cardiac development by Sema6D reverse signalling. Nat. Cell Biol. 6:1204–1211. 10.1038/ncb1193 [DOI] [PubMed] [Google Scholar]
- Treutlein B., Brownfield D.G., Wu A.R., Neff N.F., Mantalas G.L., Espinoza F.H., Desai T.J., Krasnow M.A., and Quake S.R.. 2014. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 509:371–375. 10.1038/nature13173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treyer A., and Müsch A.. 2013. Hepatocyte polarity. Compr. Physiol. 3:243–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh L.A., and Stainier D.Y.. 2004a Cardiac development. Methods Cell Biol. 76:455–473. 10.1016/S0091-679X(04)76020-3 [DOI] [PubMed] [Google Scholar]
- Trinh L.A., and Stainier D.Y.. 2004b Fibronectin regulates epithelial organization during myocardial migration in zebrafish. Dev. Cell. 6:371–382. 10.1016/S1534-5807(04)00063-2 [DOI] [PubMed] [Google Scholar]
- Trinh L.A., Yelon D., and Stainier D.Y.. 2005. Hand2 regulates epithelial formation during myocardial diferentiation. Curr. Biol. 15:441–446. 10.1016/j.cub.2004.12.083 [DOI] [PubMed] [Google Scholar]
- Ueda Y., Yamada S., Uwabe C., Kose K., and Takakuwa T.. 2016. Intestinal rotation and physiological umbilical herniation during the embryonic period. Anat. Rec. (Hoboken). 299:197–206. 10.1002/ar.23296 [DOI] [PubMed] [Google Scholar]
- van der Flier L.G., and Clevers H.. 2009. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu. Rev. Physiol. 71:241–260. 10.1146/annurev.physiol.010908.163145 [DOI] [PubMed] [Google Scholar]
- Ventura A., Young A.G., Winslow M.M., Lintault L., Meissner A., Erkeland S.J., Newman J., Bronson R.T., Crowley D., Stone J.R., et al. 2008. Targeted deletion reveals essential and overlapping functions of the miR-17∼92 family of miRNA clusters. Cell. 132:875–886. 10.1016/j.cell.2008.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent S.D., and Buckingham M.E.. 2010. How to make a heart: The origin and regulation of cardiac progenitor cells. Curr. Top. Dev. Biol. 90:1–41. 10.1016/S0070-2153(10)90001-X [DOI] [PubMed] [Google Scholar]
- Vogel V., and Sheetz M.. 2006. Local force and geometry sensing regulate cell functions. Nat. Rev. Mol. Cell Biol. 7:265–275. 10.1038/nrm1890 [DOI] [PubMed] [Google Scholar]
- Volckaert T., and De Langhe S.P.. 2015. Wnt and FGF mediated epithelial-mesenchymal crosstalk during lung development. Dev. Dyn. 244:342–366. 10.1002/dvdy.24234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldo K.L., Kumiski D.H., Wallis K.T., Stadt H.A., Hutson M.R., Platt D.H., and Kirby M.L.. 2001. Conotruncal myocardium arises from a secondary heart field. Development. 128:3179–3188. [DOI] [PubMed] [Google Scholar]
- Wallingford J.B. 2010. Planar cell polarity signaling, cilia and polarized ciliary beating. Curr. Opin. Cell Biol. 22:597–604. 10.1016/j.ceb.2010.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallingford J.B. 2012. Planar cell polarity and the developmental control of cell behavior in vertebrate embryos. Annu. Rev. Cell Dev. Biol. 28:627–653. 10.1146/annurev-cellbio-092910-154208 [DOI] [PubMed] [Google Scholar]
- Walton K.D., Mishkind D., Riddle M.R., Tabin C.J., and Gumucio D.L.. 2018. Blueprint for an intestinal villus: Species-specific assembly required. Wiley Interdiscip. Rev. Dev. Biol. 7:e317 10.1002/wdev.317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Shu W., Lu M.M., and Morrisey E.E.. 2005. Wnt7b activates canonical signaling in epithelial and vascular smooth muscle cells through interactions with Fzd1, Fzd10, and LRP5. Mol. Cell. Biol. 25:5022–5030. 10.1128/MCB.25.12.5022-5030.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton D., El-Hashash A., Carraro G., Tiozzo C., Sala F., Rogers O., De Langhe S., Kemp P.J., Riccardi D., Torday J., et al. 2010. Lung organogenesis. Curr. Top. Dev. Biol. 90:73–158. 10.1016/S0070-2153(10)90003-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weibel E.R. 2015. On the tricks alveolar epithelial cells play to make a good lung. Am. J. Respir. Crit. Care Med. 191:504–513. 10.1164/rccm.201409-1663OE [DOI] [PubMed] [Google Scholar]
- Welsh I.C., Thomsen M., Gludish D.W., Alfonso-Parra C., Bai Y., Martin J.F., and Kurpios N.A.. 2013. Integration of left-right Pitx2 transcription and Wnt signaling drives asymmetric gut morphogenesis via Daam2. Dev. Cell. 26:629–644. 10.1016/j.devcel.2013.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels A., and Pérez-Pomares J.M.. 2004. The epicardium and epicardially derived cells (EPDCs) as cardiac stem cells. Anat. Rec. A Discov. Mol. Cell. Evol. Biol. 276A:43–57. 10.1002/ar.a.10129 [DOI] [PubMed] [Google Scholar]
- Williams S.E., and Fuchs E.. 2013. Oriented divisions, fate decisions. Curr. Opin. Cell Biol. 25:749–758. 10.1016/j.ceb.2013.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittig J.G., and Münsterberg A.. 2016. The early stages of heart development: Insights from chicken embryos. J. Cardiovasc. Dev. Dis. 3:12 10.3390/jcdd3020012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P.X., Adams J., Peters H., Brown M.C., Heaney S., and Maas R.. 1999. Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat. Genet. 23:113–117. 10.1038/12722 [DOI] [PubMed] [Google Scholar]
- Yamada M., Udagawa J., Matsumoto A., Hashimoto R., Hatta T., Nishita M., Minami Y., and Otani H.. 2010. Ror2 is required for midgut elongation during mouse development. Dev. Dyn. 239:941–953. 10.1002/dvdy.22212 [DOI] [PubMed] [Google Scholar]
- Yamada M., Udagawa J., Hashimoto R., Matsumoto A., Hatta T., and Otani H.. 2013. Interkinetic nuclear migration during early development of midgut and ureteric epithelia. Anat. Sci. Int. 88:31–37. 10.1007/s12565-012-0156-8 [DOI] [PubMed] [Google Scholar]
- Yates L.L., Papakrivopoulou J., Long D.A., Goggolidou P., Connolly J.O., Woolf A.S., and Dean C.H.. 2010. The planar cell polarity gene Vangl2 is required for mammalian kidney-branching morphogenesis and glomerular maturation. Hum. Mol. Genet. 19:4663–4676. 10.1093/hmg/ddq397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates L.L., Schnatwinkel C., Hazelwood L., Chessum L., Paudyal A., Hilton H., Romero M.R., Wilde J., Bogani D., Sanderson J., et al. 2013. Scribble is required for normal epithelial cell-cell contacts and lumen morphogenesis in the mammalian lung. Dev. Biol. 373:267–280. 10.1016/j.ydbio.2012.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X., Wang Y., Rattner A., and Nathans J.. 2011. Genetic mosaic analysis reveals a major role for frizzled 4 and frizzled 8 in controlling ureteric growth in the developing kidney. Development. 138:1161–1172. 10.1242/dev.057620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeaman C., Grindstaff K.K., and Nelson W.J.. 1999. New perspectives on mechanisms involved in generating epithelial cell polarity. Physiol. Rev. 79:73–98. 10.1152/physrev.1999.79.1.73 [DOI] [PubMed] [Google Scholar]
- Yin C. 2017. Molecular mechanisms of Sox transcription factors during the development of liver, bile duct, and pancreas. Semin. Cell Dev. Biol. 63:68–78. 10.1016/j.semcdb.2016.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Carroll T.J., Rajagopal J., Kobayashi A., Ren Q., and McMahon A.P.. 2009. A Wnt7b-dependent pathway regulates the orientation of epithelial cell division and establishes the cortico-medullary axis of the mammalian kidney. Development. 136:161–171. 10.1242/dev.022087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Q., Wagstaff L., Yang X., Weijer C., and Münsterberg A.. 2008. Wnt3a-mediated chemorepulsion controls movement patterns of cardiac progenitors and requires RhoA function. Development. 135:1029–1037. 10.1242/dev.015321 [DOI] [PubMed] [Google Scholar]
- Zepp J.A., Zacharias W.J., Frank D.B., Cavanaugh C.A., Zhou S., Morley M.P., and Morrisey E.E.. 2017. Distinct mesenchymal lineages and niches promote epithelial self-renewal and myofibrogenesis in the lung. Cell. 170:1134–1148.e10. 10.1016/j.cell.2017.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Lin L., Majumdar A., Li X., Zhang X., Liu W., Etheridge L., Shi Y., Martin J., Van de Ven W., et al. 2007. Modulation of morphogenesis by noncanonical Wnt signaling requires ATF/CREB family-mediated transcriptional activation of TGFbeta2. Nat. Genet. 39:1225–1234. 10.1038/ng2112 [DOI] [PMC free article] [PubMed] [Google Scholar]