Actin polymerization is important to generate primary cilia. Drummond et al. show that upstream actin regulators are necessary for this process by controlling aPKC and Src kinase activity to promote Hedgehog signaling and restrict primary cilia.
Abstract
Primary cilia are polarized organelles that allow detection of extracellular signals such as Hedgehog (Hh). How the cytoskeleton supporting the cilium generates and maintains a structure that finely tunes cellular response remains unclear. Here, we find that regulation of actin polymerization controls primary cilia and Hh signaling. Disrupting actin polymerization, or knockdown of N-WASp/Arp3, increases ciliation frequency, axoneme length, and Hh signaling. Cdc42, a potent actin regulator, recruits both atypical protein pinase C iota/lambda (aPKC) and Missing-in-Metastasis (MIM) to the basal body to maintain actin polymerization and restrict axoneme length. Transcriptome analysis implicates the Src pathway as a major aPKC effector. aPKC promotes whereas MIM antagonizes Src activity to maintain proper levels of primary cilia, actin polymerization, and Hh signaling. Hh pathway activation requires Smoothened-, Gli-, and Gli1-specific activation by aPKC. Surprisingly, longer axonemes can amplify Hh signaling, except when aPKC is disrupted, reinforcing the importance of the Cdc42–aPKC–Gli axis in actin-dependent regulation of primary cilia signaling.
Introduction
A key unresolved issue in cell biology is how the actin cytoskeleton regulates microtubule-based structures and signaling during development and disease progression. Primary cilia are polarized microtubule-based signaling organelles found on most vertebrate cells (Kim and Dynlacht, 2013). They form during G0/G1 when the mother centriole (now termed basal body) docks at the apical membrane in epithelia or near the nucleus in fibroblasts. This docking event clears away cortical actin and nucleates a membrane-enclosed microtubule shaft called an axoneme (Sorokin, 1968; Francis et al., 2011). Several signaling pathways use primary cilia to transduce signal, with Hedgehog (Hh) being the most well known (Goetz and Anderson, 2010). Deficits in ciliogenesis or cilia-mediated signaling can result in cancer or developmental abnormalities collectively termed ciliopathies that affect many parts of the body that rely on cilia for normal function (Atwood et al., 2012; Yuan and Sun, 2013).
After basal body docking, axoneme extension occurs at the plus ends of microtubules, eventually reaching a state of dynamic stability (Avasthi and Marshall, 2012). Transcriptional control, activity of intraflagellar transport complexes, transport of cargo by a complex of proteins called the Bardet-Biedl syndrome network, intracellular signaling components, and the actin cytoskeleton have all been shown to affect axoneme length (Avasthi and Marshall, 2012; Keeling et al., 2016), yet how the actin cytoskeleton in particular influences axoneme length and signaling remains unclear. Focal adhesion complexes connect the basal body to the surrounding actin cytoskeletal meshwork, and their down-regulation leads to similar defects as disrupting actin polymerization (Antoniades et al., 2014), pointing to a mechanical force that may regulate dynamic stability of the axoneme. Actin and Myosin6 mediate the scission of ectosomes from primary cilia tips, which is required for Hh signaling (Nager et al., 2017), indicating that vesicle release or docking may regulate cilia-specific signaling. Additionally, the ciliary membrane is enriched with actin-binding proteins that further accumulate when actin polymerization is disrupted (Kohli et al., 2017), suggesting that a balance of actin polymerization is needed for proper cilia-specific functions.
Polarity proteins such as the serine/threonine kinase atypical PKC iota/lamda (aPKC) direct polarizing events in many cell types and tissues to generate the architecture of organs to ensure proper development of organisms (Rodriguez-Boulan and Macara, 2014). Typically, aPKC is a constitutive partner of the PDZ domain containing protein Par6, and the GTPase Cdc42 localizes the aPKC-Par6 cassette to specific locations within the cell. The PDZ protein Par3 is an aPKC substrate and transient member of the polarity complex that can influence complex activity and localization under certain contexts (Wirtz-Peitz et al., 2008; Morais-de-Sá et al., 2010). Although polarity proteins can localize to the basal body or ciliary shaft depending on the cell type and protein (Fan et al., 2004; Atwood et al., 2013), how they influence ciliogenesis has not been fully elucidated. The transmembrane protein Crumbs interacts with the polarity proteins and the microtubule motor Kif3 to connect ciliary membrane proteins to anterograde transport (Fan et al., 2004; Sfakianos et al., 2007). Additionally, Par6 and Cdc42 can interact with Sec10, a component of the exocyst complex, at the cilium to potentially target and dock protein-containing vesicles that may be useful for ciliogenesis (Zuo et al., 2011; Choi et al., 2013). aPKC also drives Hh signaling by phosphorylating and activating the Hh transcription factor Gli1 (Atwood et al., 2013), providing a crucial functional link between protein localization and activity.
Here, we report the connection between actin regulation and polarity proteins during ciliogenesis and Hh signaling. We find that Cdc42 recruits distinct regulators of actin dynamics, aPKC and Missing-in-Metastasis (MIM), to the basal body to restrict both the frequency of ciliated cells and axoneme length. aPKC activity is necessary for these functions, and transcriptome analysis of cells undergoing pharmacological aPKC inhibition implicates the Src pathway as a major effector. The aPKC and MIM complexes differentially regulate Src activity to maintain homeostatic levels of filamentous actin. Loss of the Cdc42 substrates N-WASp-Arp2/3 or the Cdc42 effector Toca1 also deregulates cilia frequency and axoneme length, providing a direct link between actin dynamics and ciliogenesis. Surprisingly, axoneme length directly regulates Hh signaling independently of cilia frequency and is dependent on canonical Hh pathway activation through the Smoothened (Smo)–Gli–aPKC axis, indicating a functional role for axoneme length regulation of primary cilia-mediated signaling.
Results
Axoneme length control regulates Hh response
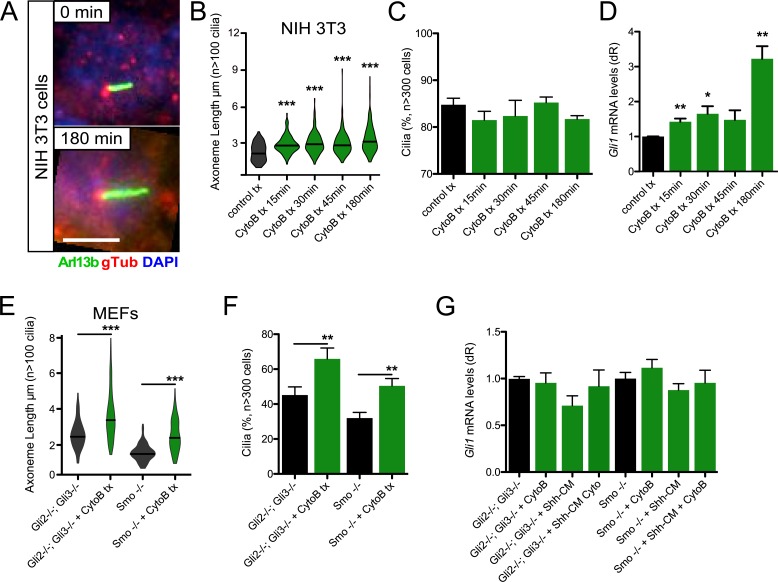
One of the major functions of the primary cilium is to transduce the Hh signal, although how disrupting axoneme length influences signal transduction is unclear. Increases in ciliogenesis and axoneme length are seen when actin regulation is disrupted using genetic or pharmacological means in immortalized human retinal pigmented epithelial (htRPE) cells (Kim et al., 2010). Using cytochalasin B to block actin monomer addition to growing actin polymers, we also observed significant increases in axoneme length over 3 h of treatment with confluent Hh-inducible NIH 3T3 mouse embryonic fibroblast (MEF) cell line (Fig. 1, A and B). We observed similar changes in axoneme length in primary mouse dermal cells (mDCs), where length changes are reversible when cytochalasin B is washed away (Fig. S1). Significant increases in Hh pathway activation coupled with axoneme length were seen during 3-h cytochalasin B treatment in the presence of Sonic Hh–conditioned media (Shh-CM), as determined by mRNA levels of Hh target gene Gli1, despite no significant changes in the percentage of ciliated cells (Fig. 1, C and D). This suggests that axoneme length control can influence primary cilia–mediated Hh signaling.
Figure 1.
Cytochalasin B induces axoneme length changes to regulate Hh signaling. (A) Immunofluorescence of NIH 3T3 cells treated with or without 10 µM cytochalasin B (CytoB) for the indicated times and stained for Arl13b, γ-tubulin (gTub), and DNA (DAPI). Bar, 5 µm. (B and C) Violin plot of axoneme lengths from confluent NIH 3T3 cells (B) and percentage of cells displaying primary cilia (C) after treatment with 10 µM CytoB for the indicated amounts of time (n = 3 experiments). tx, treatment. (D) Gli1 mRNA levels of confluent NIH 3T3 cells treated with Shh-CM and 10 µM CytoB for the indicated amounts of time (n = 3 experiments). dR, delta reporter signal normalized to passive reference dye. (E and F) Violin plot of axoneme lengths from subconfluent Gli2−/−; Gli3−/− and Smo−/− MEFs (E) and percentage of cells displaying primary cilia (F) after 3-h treatment with 10 µM CytoB (n = 3 experiments). (G) Gli1 mRNA levels of Gli2−/−; Gli3−/− and Smo−/− MEFs treated with or without Shh-CM and/or 3 h of 10 µM CytoB (n = 3 experiments). CM, conditioned media. Error bars represent SEM. Significance was determined by unpaired two-tailed t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
What mediates the increases in Hh signal in longer axonemes is unknown. Previous studies in mouse basal cell carcinoma and medulloblastoma have suggested that primary cilia are necessary for tumors driven by the canonical Hh pathway activator Smo but repress tumor growth driven by a constitutively active Gli2 transcription factor (Han et al., 2009; Wong et al., 2009). Both studies demonstrate that primary cilia are necessary to cleave Gli3 into a repressor form, whose absence may help facilitate the enhanced Hh pathway activation and tumor growth seen in this background.
To begin to parse the relationship between axoneme length and Hh signal transduction, we used Gli2−/−; Gli3−/− MEFs (Lipinski et al., 2008), which are deficient in the transcription factors necessary to initiate and repress Hh target gene induction but still contain the transcriptional amplifier Gli1. These cells show significant increases in axoneme length and percentage of ciliated cells upon cytochalasin B treatment (Fig. 1, E and F) but do not display increases in Hh pathway activation in the presence of Shh-CM or cytochalasin B (Fig. 1 G), suggesting that Gli2 and Gli3 are necessary for the increases in Hh target gene induction and that Gli1 is not sufficient for Hh activation in this context. We next used Smo−/− MEFs (Varjosalo et al., 2006), which are deficient in the canonical Hh pathway activator Smo but leave the three Gli transcription factors untouched. These cells also show significant increases in axoneme length and percentage of ciliated cells with cytochalasin B treatment (Fig. 1, E and F). However, the Hh pathway was not activated in the presence of Shh-CM or cytochalasin B (Fig. 1 G). Altogether, these data suggest canonical Smo-mediated and not noncanonical Gli-mediated activation is necessary for increased Hh signaling when actin is disrupted and that actin’s effect on axoneme length is separable from Hh signaling.
Actin polymerizing proteins restrict primary cilia and Hh response
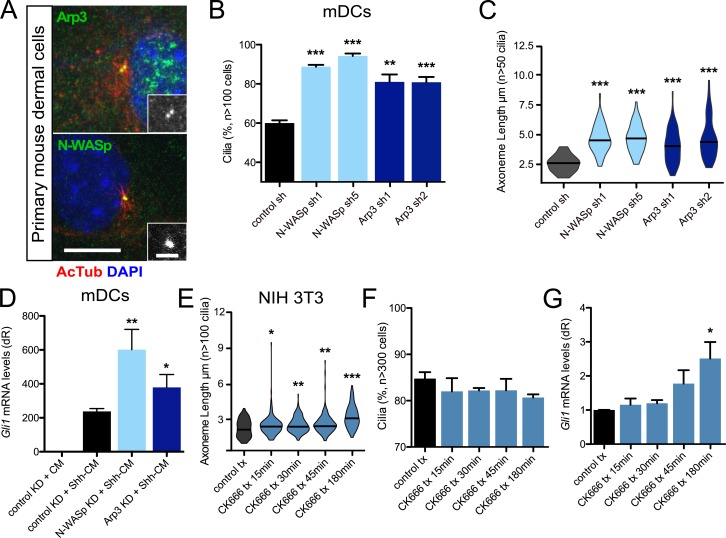
Because cytochalasin B blocks actin polymerization by preventing monomer addition to a growing filament, we next looked at proteins responsible for actin polymerization and their effects on ciliogenesis and Hh signaling. Both the branched actin nucleator Arp3 of the Arp2/3 complex and actin nucleator N-WASp localized to the basal body at the base of the primary cilium in mDCs (Fig. 2 A). Loss of Arp3 has been previously shown to increase axoneme length in htRPE cells (Kim et al., 2010), and we observed a similar increase in axoneme length and percentage of ciliated cells when Arp3 protein was knocked down using two distinct shRNAs targeting Arp3 in mDCs (Fig. 2, B and C; and Fig. S2). We also observed significant increases in axoneme length and ciliated cells when N-WASp was knocked down using two distinct shRNAs (Fig. 2, B and C; and Fig. S2). Loss of either Arp3 or N-WASp resulted in increases in Hh pathway activation in the presence of Shh-CM, similar to cytochalasin B treatment (Fig. 2 D). The increases in Hh signaling at least partly stem from lengthening of the axoneme, which was determined using an Arp2/3 small-molecule inhibitor (CK666) that suppresses actin filament binding (Hetrick et al., 2013). We found that acute treatment of confluent cells altered axoneme length but did not significantly alter cilia frequency, providing an assay to monitor signaling events while only changing one parameter. CK666 treatment of confluent NIH 3T3 cells produced significant increases in axoneme length and Hh pathway activation but no change in the percentage of ciliated cells over the course of 3 h (Fig. 2, E–G). These data suggest that elevated Hh pathway activation is not a consequence of off-target effects of cytochalasin B treatment but is a genuine consequence of primary cilia alterations mediated through disrupting actin polymerization.
Figure 2.
Actin nucleators restrict axoneme length and Hh signaling. (A) Immunofluorescence of mDCs stained for Arp3 or N-WASp with acetylated-tubulin (AcTub)–positive primary cilia. Immunoreactivity around the basal body is highlighted in the lower right of each panel. Bars: 10 µm; (inset) 2 µm. (B and C) Percentage of subconfluent mDCs displaying primary cilia (B) and violin plot of axoneme lengths (C) after shRNA knockdown of control (n = 4 experiments), N-WASp (n = 3 experiments), or Arp3 (n = 4 experiments). sh, short hairpin. (D) Gli1 mRNA levels of confluent mDCs after shRNA knockdown of control (n = 9 experiments), N-WASp (n = 5 experiments), or Arp3 (n = 3 experiments) and treated with control-conditioned media or Shh-CM. CM, conditioned media; KD, knockdown. dR, delta reporter signal normalized to passive reference dye. (E and F) Violin plot of axoneme lengths from confluent NIH 3T3 cells (E) and percentage of cells displaying primary cilia (F) after treatment with 20 µM CK666 for the indicated amounts of time (n = 3 experiments). tx, treatment. (G) Gli1 mRNA levels of confluent NIH 3T3 cells treated with Shh-CM and 20 µM CK666 for the indicated amounts of time (n = 3 experiments). Error bars represent SEM. Significance determined by unpaired two-tailed t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
The Cdc42–Par6–aPKC complex restricts primary cilia
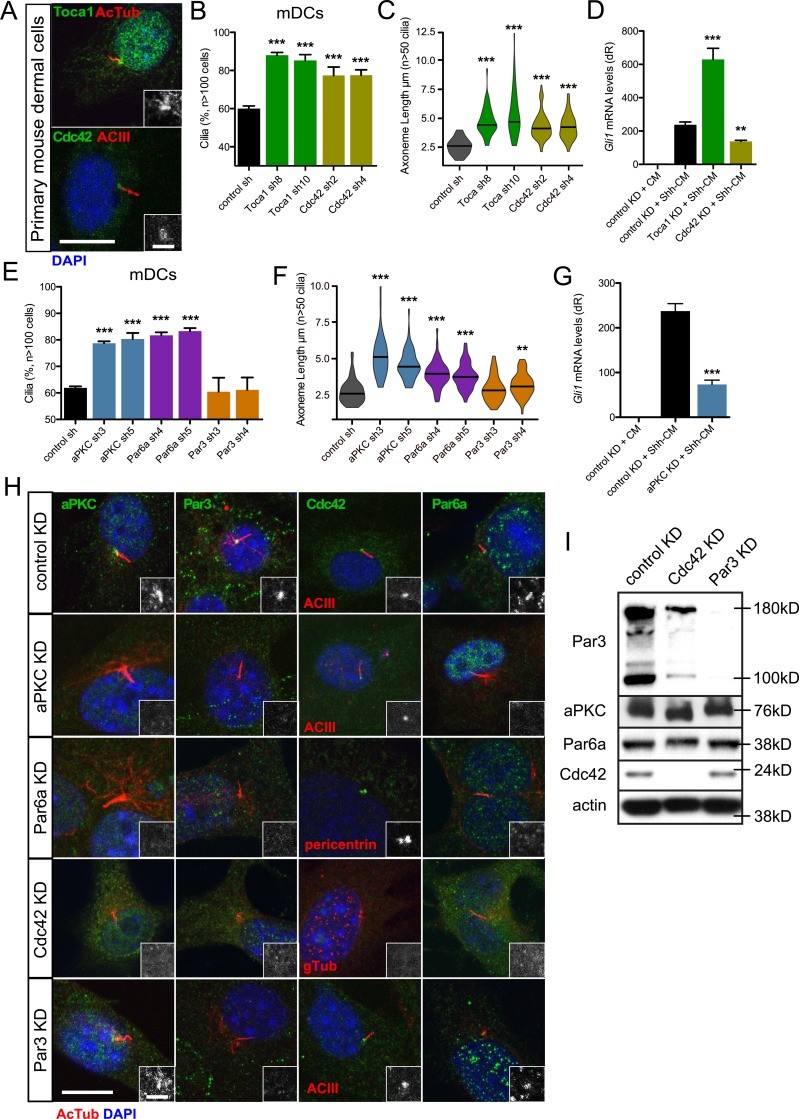
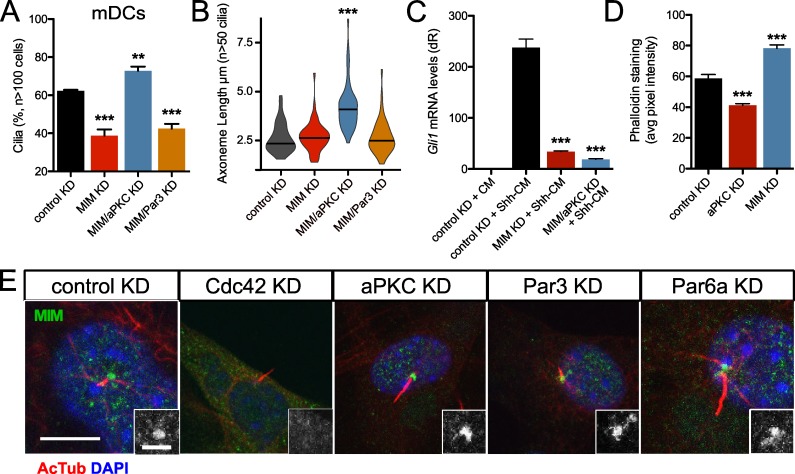
How actin-binding proteins are regulated at the primary cilium remains unclear. One potent actin regulator, Cdc42, has been shown to promote primary cilia formation in MDCK cells and in kidney collecting duct cells from Hoxb7-cre; Cdc42 fl/fl mice (Zuo et al., 2011; Elias et al., 2015). Cdc42 also localizes to the basal body in mDCs, mouse keratinocytes, and mouse basal cell carcinoma cells (Atwood et al., 2013). Cdc42 directly promotes N-WASp-Arp2/3–mediated actin nucleation and indirectly affects N-WASp function through Toca-1 (Rohatgi et al., 1999; Ho et al., 2004), suggesting that Cdc42 is a strong candidate for regulating actin nucleators at the basal body. We detect both Cdc42 and Toca-1 at the basal body in mDCs (Fig. 3 A). Knockdown of either protein using two distinct shRNAs results in significant increases in axoneme length and ciliated cells (Fig. 3, B and C; and Fig. S2), mimicking knockdown of Arp3 and N-WASp (Fig. 2, B and C; and Fig. S2) but in direct contrast to the Cdc42-deficient phenotype observed in kidney cells (Zuo et al., 2011; Elias et al., 2015). As expected, loss of Toca-1 results in significant increases in Hh pathway activation in the presence of Shh-CM; however, loss of Cdc42 suppressed Hh signaling, despite the increases in primary cilia frequency and length (Fig. 3 D). This disparity between primary cilia and Hh signaling when Cdc42 is lost is similar to our previous results obtained after we knocked down aPKC in mDCs and observed increases in the percentage of ciliated cells but significant suppression of Hh pathway activation (Atwood et al., 2013).
Figure 3.
Cdc42 regulates primary cilia and Hh signaling by recruiting aPKC–Par6a to the basal body. (A) Immunofluorescence of mDCs stained for Toca1 or Cdc42 with acetylated-tubulin (AcTub) – or adenyl cyclase III (ACIII)–positive primary cilia. Immunoreactivity around the basal body is highlighted in the lower right of each panel. Bars: 10 µm; (inset) 2 µm. (B and C) Percentage of subconfluent mDCs displaying primary cilia (B) and violin plot of axoneme lengths (C) after shRNA knockdown of control (n = 4 experiments), Toca1 (n = 3 experiments), or Cdc42 (n = 4 experiments). sh, short hairpin. (D) Gli1 mRNA levels of confluent mDCs after shRNA knockdown of control (n = 9 experiments), Toca1 (n = 2 experiments), or Cdc42 (n = 3 experiments) and treated with control-conditioned media or Shh-CM. CM, conditioned media. dR, delta reporter signal normalized to passive reference dye. (E and F) Percentage of subconfluent mDCs displaying primary cilia (E) and violin plot of axoneme lengths (F) after shRNA knockdown of control (n = 4 experiments), aPKC (n = 4 experiments), Par6a (n = 4 experiments), or Par3 (n = 4 experiments). sh, short hairpin. (G) Gli1 mRNA levels of confluent mDCs after shRNA knockdown of control (n = 9 experiments) or aPKC (n = 6 experiments) and treated with control-conditioned media or Shh-CM. KD, knockdown. Error bars represent SEM. Significance determined by unpaired two-tailed t test (**, P < 0.01; ***, P < 0.001). (H) Immunofluorescence of control, aPKC, Par6a, Cdc42, or Par3 knockdown mDCs stained for indicated proteins and AcTub/ACIII–positive primary cilia or pericentrin/γ-tubulin (gtub)–positive basal bodies. Immunoreactivity around the basal body is highlighted in the lower right of each panel. Bars: 10 µm; (inset) 2 µm. (I) Western blot of control, Cdc42, or Par3 knockdown mDCs that are probed for polarity proteins and actin.
Indeed, when we replicated our work by knocking down aPKC using two distinct shRNAs specific to the iota/lambda isoform, we observed not only the expected increase in the frequency of ciliated cells and loss of Hh signaling but also increases in axoneme length, which we did not previously appreciate (Fig. 3, E–G; and Fig. S2). Because aPKC is typically part of a larger complex of polarity proteins that regulate cell and tissue polarity across metazoa, we analyzed the role of other complex members Par6a and Par3 in primary cilia formation. Knockdown of Par6a expression phenocopied the gain in primary cilia frequency and axoneme length (Fig. 3, E and F; and Fig. S2). Surprisingly, knockdown of Par3 did not substantially alter cilia frequency or axoneme length despite its prominent role in aPKC-Par6 function in other cell types, possibly because of Par3’s transient participation in the complex (Fig. 3, E and F; and Fig. S2; Rodriguez-Boulan and Macara, 2014).
Because Cdc42 recruits the Par6–aPKC complex to subcellular locales in other contexts (Hutterer et al., 2004; Atwood et al., 2007), we wanted to determine how these polarity proteins localize to the basal body. Cdc42, aPKC, Par6a, and Par3 localize to the basal body or centrosome in multiple cell types with or without a primary cilium (Atwood et al., 2013). As expected, we observed an enrichment of polarity proteins at the basal body in mDCs and at the centrosomes during mitosis in mDCs and mouse keratinocytes, as seen with aPKC immunostaining (Figs. 3 H and S3). When we knocked down Cdc42, we observed loss of aPKC, Par6a, and Par3 basal body localization, positioning Cdc42 as the master regulator of polarity protein localization. Knockdown of aPKC or Par6a disrupted their basal body localization, but Cdc42 remained at the basal body. Interestingly, Par3 knockdown did not disturb the basal body enrichment of Cdc42, Par6a, or aPKC. This is in sharp contrast to Par3’s role in directing apical recruitment of the polarity proteins in epithelia (Rodriguez-Boulan and Macara, 2014). Although Par6a and aPKC recruit Par3 to the basal body, we observed no discernable Par3 function during ciliogenesis in mesenchymal cells, despite its reported role in epithelia (Sfakianos et al., 2007). Consistent with this idea, biochemical analysis in Par3 knockdown cells showed no changes in aPKC, Par6a, or Cdc42 protein, whereas Cdc42 knockdown altered aPKC and Par6a mobility and Par3 stability, which may stem from loss of posttranslational modifications from improper localization (Fig. 3 I).
aPKC kinase activity is critical to restrict primary cilia
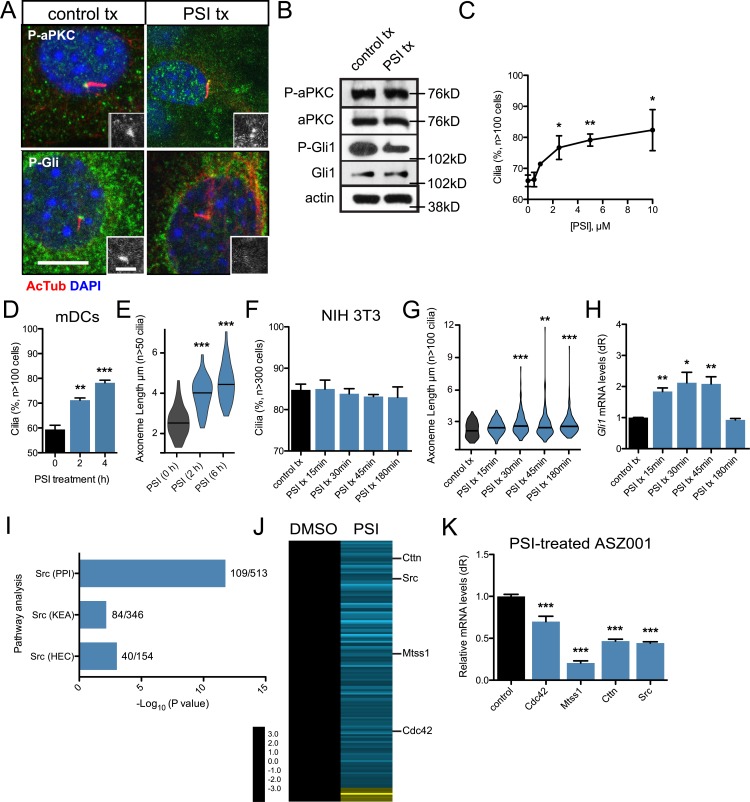
An essential function of Cdc42 during polarity establishment is to recruit and activate aPKC’s kinase activity at specific cellular locations to stimulate downstream signaling events (Drummond and Prehoda, 2016). To determine if aPKC kinase activity is essential at the primary cilium, we used the aPKC psuedosubstrate peptide inhibitor (PSI; Atwood et al., 2013) to block aPKC kinase activity in mDCs. The peptide inhibitor broadly inhibits both full-length aPKC isoforms (iota/lambda and zeta) but has little to no effect on catalytic-domain–only isoforms that do not contain the amino-terminal regulatory regions (Wu-Zhang et al., 2012; Tsai et al., 2015). Inhibiting kinase activity did not disrupt aPKC basal body localization, protein expression, or protein maturation (Fig. 4, A and B), reinforcing the idea that aPKC is not regulated by autophosphorylation (Tobias et al., 2016). However, PSI treatment did inhibit aPKC-mediated phosphorylation of Gli1, which is necessary for robust Gli1 activation (Atwood et al., 2013), at the basal body, as determined by Western blot and an in-house–generated phospho-specific antibody that recognizes the aPKC-specific Gli1 T304 phosphosite (Fig. 4, A and B), suggesting that loss of active phospho-Gli1 may be the root cause of Hh pathway inhibition when aPKC is inhibited.
Figure 4.
aPKC activity is necessary to regulate primary cilia frequency and axoneme length. (A) Immunofluorescence of control-treated or 10 µM PSI–treated mDCs stained for phosphorylated aPKC (P-aPKC), phosphorylated Gli1 (P-Gli), and primary cilia (AcTub). Immunoreactivity around the basal body is highlighted in the lower right of each panel. tx, treatment. Bars: 10 µm; (inset) 2 µm. (B) Western blot of control or PSI-treated mDCs that are probed for aPKC, P-aPKC, Gli1, P-Gli1, and actin. (C and D) Percentage of subconfluent mDCs with primary cilia upon dose-dependent (C; n = 2 experiments) or temporal addition of 10 µM PSI (D; n = 3 experiments). (E) Violin plot of axoneme length upon temporal addition of 10 µM PSI. (F and G) Percentage of cells displaying primary cilia (F) and violin plot of axoneme lengths (G) from confluent NIH 3T3 cells after treatment with 10 µM PSI for the indicated amounts of time (n = 3 experiments). (H) Gli1 mRNA levels of confluent NIH 3T3 cells treated with Shh-CM and 10 µM PSI for the indicated amounts of time (n = 3 experiments). dR, delta reporter signal normalized to passive reference dye. (I) Pathway analysis of down-regulated transcripts in PSI-treated ASZ001 cells compared with DMSO control. Number of genes in dataset compared with total number of genes displayed as a fraction to the right of each bar. KEA, kinase enrichment analysis; PPI, protein–protein interaction database. Human endogenous complexome, HEC. (J) Heat map of significantly changed transcripts in the Src pathway in PSI-treated ASZ001 cells compared with DMSO control. (K) Quantitative reverse-transcription PCR validation of selected components of the Src pathway in PSI-treated ASZ001 cells compared with DMSO control (n = 3 experiments). Error bars represent SEM. Significance determined by unpaired two-tailed t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Recapitulating the aPKC knockdown phenotype, the percentage of cells with primary cilia increased in a dose-dependent manner in cells treated with PSI (Fig. 4 C). PSI’s effect on primary cilia are at a concentration that does not affect the in vitro kinase activity of other PKC isoforms such as PKCalpha (Bogard and Tavalin, 2015), further reinforcing the role of aPKC kinase activity in this process. This effect was relatively rapid, as PSI stimulated more primary cilia with longer axonemes in as little as 2 h (Fig. 4, D and E). We observed increased frequencies of primary cilia with longer axonemes in the presence of PSI in multiple cell type types, suggesting aPKC kinase activity may serve as a general mechanism to restrict primary cilia frequency and axoneme length (Fig. S3). Interestingly, Hh pathway activation initially increases before shutting down over the course of 3-h PSI treatment in confluent NIH 3T3 cells despite increasing axoneme lengths and no change in ciliated cell frequency (Fig. 4, F–H), suggesting that aPKC’s effects on axoneme length are rapid and can indirectly influence Hh signaling before aPKC’s direct effects on Gli1 are seen. This may be caused by a delay in phospho-Gli1 degradation that allows higher transient Hh pathway activation coupled to longer axonemes before the phospho-Gli1 signal is lost.
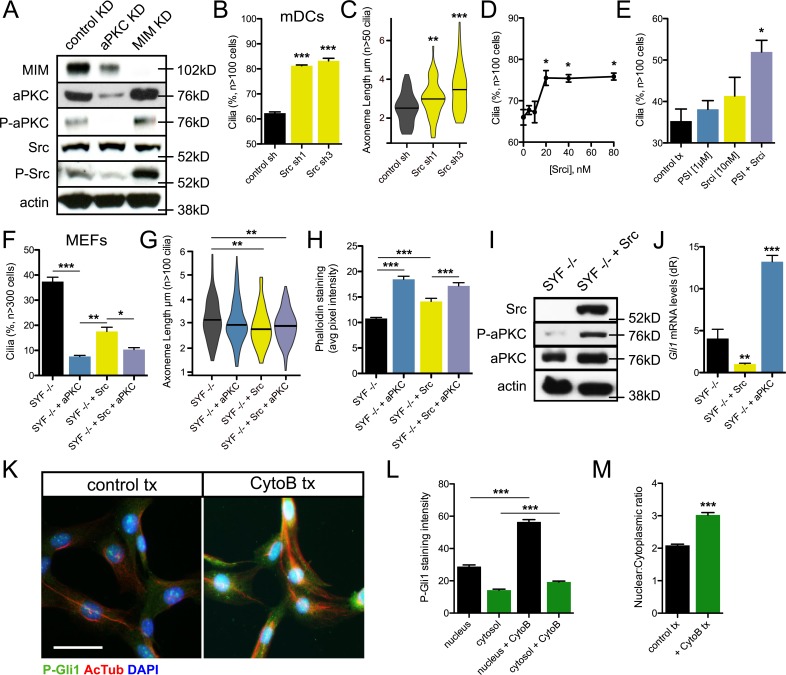
To narrow down the molecular events that aPKC regulates to restrict primary cilia, we reanalyzed our 3′-end polyadenylated RNA sequencing dataset from PSI-treated keratinocyte-derived mouse cell line ASZ001 (Atwood et al., 2013). We performed differential analysis using DE-seq, an R package to analyze count data from RNA-sequencing, with a minimum 100-count cutoff and found that expression levels of 2,781 PSI-dependent transcripts changed twofold or more when compared with control-treated cells. Pathway analysis of down-regulated transcripts indicated the Src pathway was significantly altered in our dataset (Fig. 4 I and Tables S1, S2, and S3). This was intriguing, as Src also restricts primary cilia number and axoneme length in mDCs (Bershteyn et al., 2010). In addition to Src, Cortactin, Mtss1 (MIM), and Cdc42 were down-regulated in our dataset (Fig. 4 J). We verified expression levels using quantitative RT-PCR in ASZ001 cells and found that the relative values from our dataset closely mirror actual mRNA levels, suggesting that aPKC may cooperate with the Src pathway to restrict primary cilia number and axoneme length (Fig. 4 K).
aPKC antagonizes MIM’s ciliary function downstream of Cdc42
MIM is a scaffold protein that promotes primary ciliogenesis in part by restricting the activities of Src and actin polymerization at the basal body (Bershteyn et al., 2010). Although aPKC interacts with MIM (Atwood et al., 2013), the functional effect of this interaction on ciliogenesis is unknown. As expected, MIM shRNA-mediated knockdown resulted in a loss of primary cilia in mDCs but did not result in shorter primary cilia (Fig. 5, A and B). Concomitant shRNA-mediated reduction in aPKC, but not Par3, rescued the MIM knockdown phenotype and resulted in longer axonemes (Fig. 5, A and B), suggesting aPKC’s role in restricting primary cilia is epistatic to MIM. MIM knockdown also suppressed Hh signaling as expected from loss of cilia, but aPKC-MIM double knockdown did not rescue the Hh signaling deficits. Linking these results to the alterations in actin polymerization, aPKC knockdown cells displayed significantly less phalloidin staining, whereas MIM knockdown cells displayed significantly more phalloidin staining than control cells (Figs. 5 D and S4 A), suggesting that their respective roles in regulating primary cilia frequency and axoneme length are actin dependent.
Figure 5.
aPKC antagonizes MIM downstream of Cdc42. (A and B) Percentage of subconfluent mDCs displaying primary cilia (A) and violin plot of axoneme length (B) after shRNA knockdown (KD) of control (n = 4 experiments), MIM (n = 7 experiments), aPKC and MIM (n = 7 experiments), or Par3 and MIM (n = 3 experiments). (C) Gli1 mRNA levels of confluent mDCs after shRNA knockdown of control (n = 9 experiments), MIM (n = 3 experiments), or aPKC and MIM (n = 3 experiments) and treated with control-conditioned media or Shh-CM. CM, conditioned media. dR, delta reporter signal normalized to passive reference dye. (D) Phalloidin staining quantification of subconfluent mDCs after shRNA knockdown of control, aPKC, or MIM (n > 50 cells). Error bars represent SEM. Significance was determined by unpaired two-tailed t test (**, P < 0.01; ***, P < 0.001). (E) Immunofluorescence of control, aPKC, Par6a, Cdc42, or Par3 knockdown mDCs stained for MIM and acetylated-tubulin (AcTub)–positive primary cilia. MIM staining around the basal body is highlighted in the lower right of each panel. Bars: 10 µm; (inset) 2 µm.
To determine whether aPKC and MIM operate in the same or parallel pathways, we knocked down MIM and the polarity proteins to assess how depletion of one factor affects the basal body localization of the other proteins. aPKC, Cdc42, Par6a, and Par3 all localized to the basal body in the absence of MIM, indicating MIM does not influence the Cdc42-aPKC complex localization (Fig. S5 A). However, MIM is lost at the basal body upon Cdc42 knockdown, but not knockdown of the other polarity proteins, suggesting Cdc42 recruits both aPKC and MIM to the basal body to regulate actin polymerization, ciliogenesis, Hh signaling, and axoneme length (Fig. 5 E).
aPKC promotes Src activity to restrict primary cilia
Next, we sought to determine how aPKC antagonism of MIM regulates their function at the primary cilium. Because MIM is known to regulate ciliogenesis through antagonism of Src activity, we hypothesized that aPKC might promote Src activity. We assayed protein levels after shRNA knockdown of either aPKC or MIM and found that MIM and aPKC regulate each other, as loss of MIM increases aPKC protein levels and loss of aPKC decreases MIM levels (Figs. 6 A and S5 B). We also found an increase in phosphorylated Src protein, a readout of Src activity, after MIM knockdown without increasing total Src levels. aPKC knockdown decreased phosphorylated Src levels, suggesting regulation of Src activity may serve as a nexus between MIM and aPKC to regulate actin polymerization and primary cilia. shRNA-mediated knockdown of Src phenocopied loss of aPKC, displaying a greater frequency of ciliated cells and longer axonemes (Fig. 6, B and C). In addition, pharmacological inhibition of Src activity increased ciliogenesis in a dose-dependent manner (Fig. 6 D). As both aPKC and Src restrict primary cilia, we asked if they work in concert to perform their functions. Subthreshold concentrations of PSI or Src inhibitor (Srci) had modest effects on ciliogenesis in subconfluent cells (Fig. 6 E). However, inhibiting both proteins resulted in a significant gain in primary cilia, suggesting aPKC and Src likely target similar pathways.
Figure 6.
aPKC and Src positively regulate each other to promote Gli1 activity. (A) Western blot of control, aPKC, or MIM knockdown mDCs that are probed for MIM, total and activated forms of aPKC and Src, and actin. (B and C) Percentage of subconfluent mDCs displaying primary cilia (B) and violin plot of axoneme length (C) after shRNA knockdown of control (n = 2 experiments) or Src (n = 2 experiments for each short hairpin). (D) Percentage of mDCs with primary cilia upon dose-dependent addition of Src inhibitor I (Srci; n = 2 experiments). (E) Percentage of subconfluent mDCs with primary cilia in control treatment (tx) or subthreshold concentrations of PSI, Srci, or both (n = 3 experiments). (F–H) Percentage of cells displaying primary cilia (F), violin plot of axoneme lengths (G), and phalloidin staining quantification (H; n > 100 cells) from subconfluent Src/Fyn/Yes (SYF)−/− MEFs with or without stable transfection of aPKC and/or Src (n = 3 experiments). (I) Western blot of SYF−/− MEFs with or without addition of Src and probed for total and activated forms of aPKC and total Src. (J) Gli1 mRNA levels of SYF−/− MEFs with or without addition of Src or aPKC (n = 4 experiments). dR, delta reporter signal normalized to passive reference dye. (K) Immunofluorescence of NIH 3T3 cells treated with or without 2.5 µM cytochalasin B (CytoB) for 3 h and stained for the indicated proteins. Bar, 30 µm. (L and M) Quantification (L) and nuclear-to-cytoplasmic ratio (M) of the aPKC-specific phosphorylated-Gli1 immunostain (n > 100 cells). Error bars represent SEM. Significance was determined by unpaired two-tailed t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
To further examine the aPKC–Src interaction, we used Src-Yes-Fyn−/− (SYF−/−) MEFs (Klinghoffer et al., 1999) to delineate their respective contributions. Overexpression of aPKC robustly inhibited ciliogenesis in SYF−/− MEFs to a greater extent than overexpression of Src, whereas overexpression of both proteins mimicked the aPKC-only phenotype (Fig. 6 F). Although no significant differences in axoneme length were observed in these cells (Fig. 6 G), addition of either aPKC or Src, or both proteins, resulted in significant increases in phalloidin staining (Figs. 6 H and S4 B). These results are consistent biochemically, as we observed a loss in active, phosphorylated aPKC protein that was rescued upon Src addition, suggesting Src and aPKC positively regulate each other’s activity (Fig. 6, A and I; and Fig. S5C). We also observe a significant inhibition of Hh signaling in SYF−/− MEFs upon the addition of Src, whereas aPKC addition significantly increases Hh signaling despite the lower frequency of ciliated cells, suggesting that their Hh signaling functions are separable in contrast to their roles in ciliogenesis and actin polymerization (Fig. 6, F and J). As aPKC activity consistently increases Hh signaling, we assayed whether disrupting actin increases aPKC activity. Using cytochalasin B, we observe significant increases in aPKC-specific Gli1 phosphorylation in both the nucleus and cytosol, where cytochalasin B–treated cells showed a higher nuclear-to-cytoplasmic ratio, suggesting that aPKC-mediated activation of Gli1 is responsible for the elevated Hh pathway activation when actin is disrupted (Fig. 6, K–M; and Fig. S5 D).
Discussion
Ciliogenesis represents a dynamic process where actin, microtubule, and membrane dynamics work in concert to generate a signaling organelle. Here, we show that Cdc42 localizes to the basal body to regulate Hh signaling and actin dynamics, restricting the frequency of ciliated cells and axoneme length. Cdc42 recruits both the aPKC–Par6a complex and MIM to the basal body, where they regulate the activity of Src to maintain appropriate levels of actin polymerization, ciliogenesis, axoneme length, and Hh signaling (Fig. S5 E). How Cdc42 recruits two genetically distinct complexes to the basal body may stem from complex heterogeneity where one molecule of Cdc42 can only interact with one Par6a–aPKC or one MIM. Alternatively, Cdc42 may recruit one large complex as aPKC and MIM interact at the basal body in mDCs (Atwood et al., 2013).
Actin dynamics orchestrate several processes that are necessary for ciliogenesis. Transporting the mother centriole (future basal body) to the appropriate cortex of the cell is an actin-dependent process (Euteneuer and Schliwa, 1985). Once there, focal adhesion proteins anchor the basal body to the underlying actin cytoskeleton (Antoniades et al., 2014). Local clearing of the actin cytoskeleton commences (Francis et al., 2011) and the axoneme elongates through microtubule nucleation. Once the basal body docks, a reduction in cortical actin would presumably promote more ciliated cells, as there would be no physical barrier to prevent axoneme elongation. How actin regulates axoneme length is less clear. One hypothesis is that cortical actin reinforces the ciliary necklace to help provide a diffusion barrier and prevent vesicle trafficking and anterograde transport to grow the axoneme shaft. This is particularly attractive as the polarity proteins are implicated in anterograde transport at the primary cilium (Fan et al., 2004; Sfakianos et al., 2007). Alternatively, actin may serve as a mechanical force that prevents the axoneme from growing too long, akin to twisting balloons. Relaxation of the force or widening of the actin anchor surrounding the basal body would provide more slack to grow the axoneme, whereas constricting the actin anchor would result in the opposite effect.
Primary cilia are necessary to amplify Hh signal and loss of the cilium results in a host of Hh-deficient abnormalities (Briscoe and Thérond, 2013). How mutants that disrupt actin regulation and promote primary cilia alter Hh signaling is less clear. We show that disruption of actin through loss of direct actin nucleators amplify Hh target gene expression more than simply adding cells with primary cilia. In support of this idea, we show that axoneme length regulation is functionally important as longer primary cilia can transduce more Hh signal. This signaling requires canonical Hh pathway activation through Smo and Gli2/Gli3. Proteome mapping using APEX2 tagging has revealed an abundance of actin-binding and other actin-related proteins enriched at the primary cilium when actin is depolymerized using cytochalasin D in htRPE cells (Kohli et al., 2017). For instance, Myo5a volume at the primary cilium was increased upon cytochalasin D treatment, but not the intensity of staining, suggesting that a longer axoneme can hold more protein such as the positive Hh regulator SMO that could lead to more Hh signaling. Additionally, many actin-related proteins enrich at the basal body upon actin disruption (Kohli et al., 2017), which could sequester these proteins away from other subcellular locations. This could be a mechanism to sequester the negative HH regulator ACTRT1, which binds to the GLI1 promoter in humans to suppress HH signaling (Bal et al., 2017). Although disrupting actin has pleiotropic effects, we show that Hh pathway activation is not necessary for axoneme elongation, suggesting that actin’s effect on cilia and Hh signaling are separable.
aPKC activity is necessary for Hh pathway activation and functions by phosphorylating and activating Gli1. Intriguingly, our data also support a role for Cdc42 in Hh pathway activation, presumably by recruiting and activating aPKC at the basal body, which is consistent with the holoprosencephaly phenotype seen when Cdc42 is lost (Chen et al., 2006). The authors thought the defect was independent of the Hh pathway, but they only examined Shh expression and not downstream targets such as Gli1 or Ptch1, leaving open the possibility that the defect is Hh dependent. Separately, aPKC and MIM antagonism controls ciliogenesis and axoneme length through regulation of Src activity. We provide several lines of evidence to support this model. First, aPKC promotes MIM protein levels, whereas MIM suppresses aPKC levels. Second, aPKC and Src positively regulate each other’s activity, whereas MIM suppresses both aPKC and Src activity, in line with previous observations that show interactions between aPKC and Src (Wooten et al., 2001) and MIM and Src (Bershteyn et al., 2010). Third, aPKC knockdown rescues the cilia defect in MIM knockdown cells. Finally, Src knockdown, or pharmacological inhibition, phenocopies loss of aPKC. These data point to a signaling nexus that centers on appropriate control of Src activity at the basal body. In summary, our results open up new avenues of inquiry into primary cilia length control and suggest that reanalyzing previously published cilia work may be beneficial in understanding cell and tissue-level Hh regulation.
Materials and methods
Cell culture, drug treatments, and quantitative PCR
Primary mDCs were isolated as previously described (Woo et al., 2013). In brief, skin was dissected from 1- to 3-d-old pups and incubated in 2.5 mg/ml dispase in HBSS overnight at 4°C. The dermis was separated from the epidermis and minced, and dermal pieces were incubated with 0.25% (wt/vol) collagenase type I in DMEM at 37°C for 1 h to isolate single cells. mDCs were grown in Amniomax media containing supplement and penicillin/streptomycin (Life Technologies). ASZ001 cells were grown in 154CF media containing 2% chelated FBS, Human Keratinocyte Growth Supplement, and penicillin/streptomycin (Life Technologies). NIH 3T3, 10T1/2 (C3H10T1/2), SYF−/− MEFs, Smo−/− MEFs, and Gli1−/−; Gli2−/− MEFs were grown in DMEM media containing 10% FBS and penicillin/streptomycin (Life Technologies). MDCK cells were grown in Eagle’s Minimum Essential Medium containing 10% FBS and penicillin/streptomycin (Life Technologies). Cells were serum-starved or serum-starved and treated with or without control-conditioned media or Shh-CM for 24 h to induce ciliogenesis before drug treatments unless stipulated otherwise. Drug treatments were performed with 10 µM myristoylated PSI (Atwood et al., 2013), 20 nM Srci (Src inhibitor I; Calbiochem), 10 µM cytochalasin B (Sigma), or 20 µM CK666 (Calbiochem) unless otherwise specified. RNA was harvested using the RNeasy Minikit (QIAGEN). Quantitative RT-PCR was performed using the Brilliant II SYBR Green QRT-PCR Master Mix kit (Agilent Technologies) on a Mx3000P qPCR System (Agilent Technologies) or the StepOnePlus System (Invitrogen) using primers to Gli1 (forward: 5′-GCAGGTGTGAGGCCAGGTAGTGACGATG-3′, reverse: 5′-CGCGGGCAGCACTGAGGACTTGTC-3′), Cdc42 (forward: 5′-GCAGGGCAAGAGGATTATGAC-3′, reverse: 5′-TCTCAGGCACCCACTTTTC-3′), Mtss1 (forward: 5′-GAAGCTGCAGAAGAAGGC-3′, reverse: 5′-GAGCGCTGTCCAACTGAGGC-3′), Cttn (forward: 5′-ATTTGGTGTTCAGTCCGAGAG-3′, reverse: 5′-CTTGTCCATCCGATCCTTCTG-3′), Src (forward: 5′-GACCGAGCTCACCACTAAGG-3′, reverse: 5′-CTGTGGCTCAGTGGACGTAA-3′), Gapdh (forward: 5′-AATGAATACGGCTACAGCAACAGGGTG-3′, reverse: 5′-AATTGTGAGGGAGATGCTCAGTGTTGGG-3′). Fold change in mRNA expression was measured using ΔΔCt analysis with Gapdh as an internal control gene.
Lentiviral knockdown
Lentiviral pLKO.1 vector containing shRNAs (Open Biosystems) to Prkci (sh3: 5′-CCGTTCACCATGAAATGGATA-3′, sh5: 5′-CCAGACAGAAAGCAGGTTGTT-3′; Atwood et al., 2013), Pard6a (sh4: 5′-AAACTGTCATCGTTGGTGAGG-3′, sh5: 5′-AAGGATCTCATCACTGACCGC-3′), Cdc42 (sh2: 5′-AAGAAAGGAGTCTTTGGACAG-3′, sh4: 5′-TGTCTGTGGATAACTTAGCGG-3′), Pard3 (sh3: 5′-ATTTGCCTGCTAAATCTACTC-3′, sh4: 5′-TATGTTCCCATTATCCTGCTC-3′), Src (sh1: 5′-ATTCCCGTCTAGTGATCTTGC-3′, sh3: 5′-TTATTGACAATCTGCAGCCGC-3′), Fnbp1l (sh8: 5′-TTAAGTTCATCAATGCGCTGC-3′, sh10: 5′-TGATAGTCCCATCAGAAATGG-3′), Was (sh1: 5′-TATGAGAGGTGAAAGGTGACG-3′, sh5: 5′-TCTCTCCTCATTGATTGGTGC-3′), Actr3 (sh1: 5′-TTAGCTCTCTTCTACATCTGC-3′, sh2: 5′-AAAGTAATGATCTTCAGGTTC-3′); or pSicoR-puro vector containing shRNAs to Mtss1 (5′-GCAAGGCACTCATCGAAGA-3′; Bershteyn et al., 2010) were used. Lentiviral infection was performed and cells assayed between 2 and 4 d depending on the efficiency of knockdown, as determined by protein levels on Western blot or immunofluorescence.
Antibodies, immunofluorescence, and Western blots
Cells were fixed with either 4% paraformaldehyde or 100% methanol for 10 min in 1% normal horse serum, and 0.1% Triton X-100 in PBS was used for blocking. The following antibodies were used: rabbit anti-aPKC (1:500, sc-216; Santa Cruz Biotechnology), rabbit anti–p-aPKC T410 (1:100, sc-12894; Santa Cruz Biotechnology), rabbit anti-Pard6a (1:100, sc-25525; Santa Cruz Biotechnology), mouse anti-Pard6a (1:100, sc-365323; Santa Cruz Biotechnology), mouse anti-Pard3 (1:500, MABF28; Millipore), mouse anti-Cdc42 (1:500, sc-8401; Santa Cruz Biotechnology), rabbit anti-MIM (1:1,000; Bershteyn et al., 2010), rabbit anti-Src (1:1,000, 2108; Cell Signaling), rabbit anti–p-Src Y416 (1:1,000, 2101; Cell Signaling), rabbit anti-Fnbp1l (1:500, ab67310; Abcam), rabbit anti–N-WASp (1:100, sc-20770; Santa Cruz Biotechnology), mouse anti-Arp3 (1:500, ab49671; Abcam), rabbit anti–γ-tubulin (1:500, SAB4503045; Sigma), mouse anti–γ-tubulin (1:500, ab11316; Abcam), mouse anti–acetylated tubulin (1:2,000, T7451; Sigma), goat anti–adenyl cyclase III (1:50, sc-32113; Santa Cruz Biotechnology), mouse anti-Arl13b (1:2,000, 75–287; Antibodies Inc.) mouse anti–β actin (1:5,000, A2228; Sigma), rabbit anti-Gli1 (1:1,000, AF3455; R&D Systems), and rabbit anti–P-T304 Gli1 (1:500). Secondary antibodies included Alexa Fluor 488, 546, and 647 (Jackson ImmunoResearch) and Alexa Fluor 488 phalloidin (Life Technologies). Slides were mounted with Prolong Diamond Antifade Mountant from Molecular Probes. Confocal images were acquired at room temperature on a Leica SP2 AOBS laser scanning microscope with HCX PL APO 40× and 63× oil immersion objectives or a Zeiss LSM700 laser scanning microscope with Plan-Apochromat 40× and 63× oil immersion objectives. Fluorescent images were acquired at room temperature on an EVOS FL Color Imaging System with Plan Fluorite 40× objective. Images were arranged with ImageJ, Affinity Photo, and Affinity Designer.
ImageJ was used to estimate the percentage of cells with cilia (total cells were counted by nuclear DAPI stain and compared with the number of acetylated tubulin– or Arl13b-positive cells) and axoneme length. For Western blots, cells were lysed with 2× SDS sample buffer (100 mM Tris HCl 6.8, 200 nM DTT, 4% SDS, 0.2% bromophenol blue, and 20% glycerol) and boiled at 100°C for 10 min. Samples were resolved on a 4–12% polyacrylamide gradient gel and transferred to nitrocellulose membrane by a semidry transfer apparatus. Membranes were blocked with 5% milk in TBS with 0.05% Tween-20 before sequential addition of primary and secondary antibodies. Membranes were either imaged using (a) HRP-conjugated secondary antibodies, ECL Western blotting substrate (Pierce), and x-ray film, or (b) Alexa Fluor secondary antibodies and the LI-COR Odyssey imaging system. Bands were quantified using ImageJ.
Pathway analysis
Total read counts from aligned 3′ end RNA sequencing of PSI-treated ASZ001 cells (Atwood et al., 2013) were generated using DE-seq. Genes from differentially expressed transcripts with a 100-count minimum threshold and at least a 50% reduction from control-treated samples were analyzed using Enrichr (Chen et al., 2013; Kuleshov et al., 2016). Src pathway signatures were extracted from protein–protein interaction hub analysis, kinase enrichment analysis, and NURSA human endogenous complexome analysis. All differentially expressed Src pathway signature genes were hierarchically clustered using the Cluster program and visualized in Java Treeview.
Statistics
Statistical analyses were done using two-tailed t tests and compared with control cells using GraphPad Prism.
Online supplemental material
Fig. S1 shows cytochalasin B washout restores primary cilium length. Fig. S2 shows Western blot analysis of protein knockdown. Fig. S3 shows aPKC controls primary cilia and Hh signaling. Fig. S4 shows phalloidin staining is altered upon disruption of upstream regulators of actin nucleators. Fig. S5 shows MIM and phospho-Gli1 staining. Table S1 shows protein–protein interaction hub proteins. Table S2 shows kinase enrichment analysis proteins. Table S3 shows human endogenous complexome proteins.
Supplementary Material
Acknowledgments
We wish to thank K. Blake for help with statistics, as well as M. Bershteyn, J. Nelson, J. Allard, and members of the S.X. Atwood and A.E. Oro laboratories for help and guidance.
This work was funded by a National Science Foundation Graduate Research Fellowship (T.T.L. Nguyen), National Institutes of Health Award R01ARO52785 (A.E. Oro), National Institutes of Health Pathway to Independence Award R00CA176847 (S.X. Atwood), and University California, Irvine, startup funds (S.X. Atwood).
The authors declare no competing financial interests.
Author contributions: S.X. Atwood and A.E. Oro conceived the project. S.X. Atwood, M.L. Drummond, and M. Li performed most of the experiments. E. Tarapore performed SYF−/− experiments. T.T.L. Nguyen performed the PSI treatment experiments shown in Fig. S3. B.J. Barouni, S. Cruz, and K.C. Tan imaged and quantified cilia time-course and knockout MEF data. S.X. Atwood, M. Drummond, and A.E. Oro wrote the manuscript. All authors analyzed and discussed the results and commented on the manuscript.
References
- Antoniades I., Stylianou P., and Skourides P.A.. 2014. Making the connection: ciliary adhesion complexes anchor basal bodies to the actin cytoskeleton. Dev. Cell. 28:70–80. 10.1016/j.devcel.2013.12.003 [DOI] [PubMed] [Google Scholar]
- Atwood S.X., Chabu C., Penkert R.R., Doe C.Q., and Prehoda K.E.. 2007. Cdc42 acts downstream of Bazooka to regulate neuroblast polarity through Par-6 aPKC. J. Cell Sci. 120:3200–3206. 10.1242/jcs.014902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwood S.X., Chang A.L.S., and Oro A.E.. 2012. Hedgehog pathway inhibition and the race against tumor evolution. J. Cell Biol. 199:193–197. 10.1083/jcb.201207140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwood S.X., Li M., Lee A., Tang J.Y., and Oro A.E.. 2013. GLI activation by atypical protein kinase C ι/λ regulates the growth of basal cell carcinomas. Nature. 494:484–488. 10.1038/nature11889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avasthi P., and Marshall W.F.. 2012. Stages of ciliogenesis and regulation of ciliary length. Differentiation. 83:S30–S42. 10.1016/j.diff.2011.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bal E., Park H.-S., Belaid-Choucair Z., Kayserili H., Naville M., Madrange M., Chiticariu E., Hadj-Rabia S., Cagnard N., Kuonen F., et al. . 2017. Mutations in ACTRT1 and its enhancer RNA elements lead to aberrant activation of Hedgehog signaling in inherited and sporadic basal cell carcinomas. Nat. Med. 23:1226–1233. 10.1038/nm.4368 [DOI] [PubMed] [Google Scholar]
- Bershteyn M., Atwood S.X., Woo W.-M., Li M., and Oro A.E.. 2010. MIM and cortactin antagonism regulates ciliogenesis and hedgehog signaling. Dev. Cell. 19:270–283. 10.1016/j.devcel.2010.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogard A.S., and Tavalin S.J.. 2015. Protein Kinase C (PKC)ζ Pseudosubstrate Inhibitor Peptide Promiscuously Binds PKC Family Isoforms and Disrupts Conventional PKC Targeting and Translocation. Mol. Pharmacol. 88:728–735. 10.1124/mol.115.099457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J., and Thérond P.P.. 2013. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 14:416–429. 10.1038/nrm3598 [DOI] [PubMed] [Google Scholar]
- Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., and Ma’ayan A.. 2013. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 14:128 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Liao G., Yang L., Campbell K., Nakafuku M., Kuan C.-Y., and Zheng Y.. 2006. Cdc42 deficiency causes Sonic hedgehog-independent holoprosencephaly. Proc. Natl. Acad. Sci. USA. 103:16520–16525. 10.1073/pnas.0603533103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S.Y., Chacon-Heszele M.F., Huang L., McKenna S., Wilson F.P., Zuo X., and Lipschutz J.H.. 2013. Cdc42 deficiency causes ciliary abnormalities and cystic kidneys. J. Am. Soc. Nephrol. 24:1435–1450. 10.1681/ASN.2012121236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond M.L., and Prehoda K.E.. 2016. Molecular Control of Atypical Protein Kinase C: Tipping the Balance between Self-Renewal and Differentiation. J. Mol. Biol. 428:1455–1464. 10.1016/j.jmb.2016.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias B.C., Das A., Parekh D.V., Mernaugh G., Adams R., Yang Z., Brakebusch C., Pozzi A., Marciano D.K., Carroll T.J., and Zent R.. 2015. Cdc42 regulates epithelial cell polarity and cytoskeletal function during kidney tubule development. J. Cell Sci. 128:4293–4305. 10.1242/jcs.164509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euteneuer U., and Schliwa M.. 1985. Evidence for an involvement of actin in the positioning and motility of centrosomes. J. Cell Biol. 101:96–103. 10.1083/jcb.101.1.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan S., Hurd T.W., Liu C.-J., Straight S.W., Weimbs T., Hurd E.A., Domino S.E., and Margolis B.. 2004. Polarity proteins control ciliogenesis via kinesin motor interactions. Curr. Biol. 14:1451–1461. 10.1016/j.cub.2004.08.025 [DOI] [PubMed] [Google Scholar]
- Francis S.S., Sfakianos J., Lo B., and Mellman I.. 2011. A hierarchy of signals regulates entry of membrane proteins into the ciliary membrane domain in epithelial cells. J. Cell Biol. 193:219–233. 10.1083/jcb.201009001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz S.C., and Anderson K.V.. 2010. The primary cilium: a signalling centre during vertebrate development. Nat. Rev. Genet. 11:331–344. 10.1038/nrg2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y.-G., Kim H.J., Dlugosz A.A., Ellison D.W., Gilbertson R.J., and Alvarez-Buylla A.. 2009. Dual and opposing roles of primary cilia in medulloblastoma development. Nat. Med. 15:1062–1065. 10.1038/nm.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetrick B., Han M.S., Helgeson L.A., and Nolen B.J.. 2013. Small molecules CK-666 and CK-869 inhibit actin-related protein 2/3 complex by blocking an activating conformational change. Chem. Biol. 20:701–712. 10.1016/j.chembiol.2013.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho H.-Y.H., Rohatgi R., Lebensohn A.M., Le Ma J., Li S.P., Gygi, and Kirschner M.W.. 2004. Toca-1 mediates Cdc42-dependent actin nucleation by activating the N-WASP-WIP complex. Cell. 118:203–216. 10.1016/j.cell.2004.06.027 [DOI] [PubMed] [Google Scholar]
- Hutterer A., Betschinger J., Petronczki M., and Knoblich J.A.. 2004. Sequential roles of Cdc42, Par-6, aPKC, and Lgl in the establishment of epithelial polarity during Drosophila embryogenesis. Dev. Cell. 6:845–854. 10.1016/j.devcel.2004.05.003 [DOI] [PubMed] [Google Scholar]
- Keeling J., Tsiokas L., and Maskey D.. 2016. Cellular Mechanisms of Ciliary Length Control. Cells. 5:E6 10.3390/cells5010006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., and Dynlacht B.D.. 2013. Assembling a primary cilium. Curr. Opin. Cell Biol. 25:506–511. 10.1016/j.ceb.2013.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Lee J.E., Heynen-Genel S., Suyama E., Ono K., Lee K., Ideker T., Aza-Blanc P., and Gleeson J.G.. 2010. Functional genomic screen for modulators of ciliogenesis and cilium length. Nature. 464:1048–1051. 10.1038/nature08895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinghoffer R.A., Sachsenmaier C., Cooper J.A., and Soriano P.. 1999. Src family kinases are required for integrin but not PDGFR signal transduction. EMBO J. 18:2459–2471. 10.1093/emboj/18.9.2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli P., Höhne M., Jüngst C., Bertsch S., Ebert L.K., Schauss A.C., Benzing T., Rinschen M.M., and Schermer B.. 2017. The ciliary membrane-associated proteome reveals actin-binding proteins as key components of cilia. EMBO Rep. 18:1521–1535. 10.15252/embr.201643846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., et al. . 2016. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44(W1):W90–W97. 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski R.J., Bijlsma M.F., Gipp J.J., Podhaizer D.J., and Bushman W.. 2008. Establishment and characterization of immortalized Gli-null mouse embryonic fibroblast cell lines. BMC Cell Biol. 9:49 10.1186/1471-2121-9-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morais-de-Sá E., Mirouse V., and St Johnston D.. 2010. aPKC phosphorylation of Bazooka defines the apical/lateral border in Drosophila epithelial cells. Cell. 141:509–523. 10.1016/j.cell.2010.02.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nager A.R., Goldstein J.S., Herranz-Pérez V., Portran D., Ye F., Garcia-Verdugo J.M., and Nachury M.V.. 2017. An Actin Network Dispatches Ciliary GPCRs into Extracellular Vesicles to Modulate Signaling. Cell. 168:252–263.e14. 10.1016/j.cell.2016.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Boulan E., and Macara I.G.. 2014. Organization and execution of the epithelial polarity programme. Nat. Rev. Mol. Cell Biol. 15:225–242. 10.1038/nrm3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi R., Ma L., Miki H., Lopez M., Kirchhausen T., Takenawa T., and Kirschner M.W.. 1999. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 97:221–231. 10.1016/S0092-8674(00)80732-1 [DOI] [PubMed] [Google Scholar]
- Sfakianos J., Togawa A., Maday S., Hull M., Pypaert M., Cantley L., Toomre D., and Mellman I.. 2007. Par3 functions in the biogenesis of the primary cilium in polarized epithelial cells. J. Cell Biol. 179:1133–1140. 10.1083/jcb.200709111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin S.P. 1968. Reconstructions of centriole formation and ciliogenesis in mammalian lungs. J. Cell Sci. 3:207–230. [DOI] [PubMed] [Google Scholar]
- Tobias I.S., Kaulich M., Kim P.K., Simon N., Jacinto E., Dowdy S.F., King C.C., and Newton A.C.. 2016. Protein kinase Cζ exhibits constitutive phosphorylation and phosphatidylinositol-3,4,5-triphosphate-independent regulation. Biochem. J. 473:509–523. 10.1042/BJ20151013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai L.-C.L., Xie L., Dore K., Xie L., Del Rio J.C., King C.C., Martinez-Ariza G., Hulme C., Malinow R., Bourne P.E., and Newton A.C.. 2015. Zeta Inhibitory Peptide Disrupts Electrostatic Interactions That Maintain Atypical Protein Kinase C in Its Active Conformation on the Scaffold p62. J. Biol. Chem. 290:21845–21856. 10.1074/jbc.M115.676221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varjosalo M., Li S.-P., and Taipale J.. 2006. Divergence of hedgehog signal transduction mechanism between Drosophila and mammals. Dev. Cell. 10:177–186. 10.1016/j.devcel.2005.12.014 [DOI] [PubMed] [Google Scholar]
- Wirtz-Peitz F., Nishimura T., and Knoblich J.A.. 2008. Linking cell cycle to asymmetric division: Aurora-A phosphorylates the Par complex to regulate Numb localization. Cell. 135:161–173. 10.1016/j.cell.2008.07.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S.Y., Seol A.D., So P.-L., Ermilov A.N., Bichakjian C.K., Epstein E.H. Jr., Dlugosz A.A., and Reiter J.F.. 2009. Primary cilia can both mediate and suppress Hedgehog pathway-dependent tumorigenesis. Nat. Med. 15:1055–1061. 10.1038/nm.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo W.-M., Atwood S.X., Zhen H.H., and Oro A.E.. 2013. Rapid genetic analysis of epithelial-mesenchymal signaling during hair regeneration. J. Vis. Exp. 72:e4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooten M.W., Vandenplas M.L., Seibenhener M.L., Geetha T., and Diaz-Meco M.T.. 2001. Nerve growth factor stimulates multisite tyrosine phosphorylation and activation of the atypical protein kinase C’s via a src kinase pathway. Mol. Cell. Biol. 21:8414–8427. 10.1128/MCB.21.24.8414-8427.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu-Zhang A.X., Schramm C.L., Nabavi S., Malinow R., and Newton A.C.. 2012. Cellular pharmacology of protein kinase Mζ (PKMζ) contrasts with its in vitro profile: implications for PKMζ as a mediator of memory. J. Biol. Chem. 287:12879–12885. 10.1074/jbc.M112.357244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S., and Sun Z.. 2013. Expanding horizons: ciliary proteins reach beyond cilia. Annu. Rev. Genet. 47:353–376. 10.1146/annurev-genet-111212-133243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X., Fogelgren B., and Lipschutz J.H.. 2011. The small GTPase Cdc42 is necessary for primary ciliogenesis in renal tubular epithelial cells. J. Biol. Chem. 286:22469–22477. 10.1074/jbc.M111.238469 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.