Mitochondrial β-barrel proteins are imported from the cytosol into the organelle. Jores et al. provide new insights into the early events of this process by describing an array of cytosolic chaperones and cochaperones that associate with newly synthesized β-barrel proteins and assure their optimal biogenesis.
Abstract
Mitochondrial β-barrel proteins are encoded in the nucleus, translated by cytosolic ribosomes, and then imported into the organelle. Recently, a detailed understanding of the intramitochondrial import pathway of β-barrel proteins was obtained. In contrast, it is still completely unclear how newly synthesized β-barrel proteins reach the mitochondrial surface in an import-competent conformation. In this study, we show that cytosolic Hsp70 chaperones and their Hsp40 cochaperones Ydj1 and Sis1 interact with newly synthesized β-barrel proteins. These interactions are highly relevant for proper biogenesis, as inhibiting the activity of the cytosolic Hsp70, preventing its docking to the mitochondrial receptor Tom70, or depleting both Ydj1 and Sis1 resulted in a significant reduction in the import of such substrates into mitochondria. Further experiments demonstrate that the interactions between β-barrel proteins and Hsp70 chaperones and their importance are conserved also in mammalian cells. Collectively, this study outlines a novel mechanism in the early events of the biogenesis of mitochondrial outer membrane β-barrel proteins.
Introduction
In eukaryotes, membrane-embedded β-barrel proteins can be found in the outer membrane (OM) of chloroplasts and mitochondria where they perform many crucial functions, including the transport of ions, small molecules, and nucleic acids. Additionally, some β-barrel proteins have essential roles in the organellar import of cytosolically synthesized proteins and in the exchange of lipids with other compartments.
Mitochondrial β-barrel proteins are translated on cytosolic ribosomes and then imported into their target organelles (Höhr et al., 2015; Ulrich and Rapaport, 2015). Upon reaching the mitochondrial surface, they are translocated across the OM into the intermembrane space (IMS) with the help of the translocase of the mitochondrial OM (TOM) complex (Rapaport and Neupert, 1999; Pfanner et al., 2004). The β-barrel precursor proteins interact with the import receptor Tom20 that can recognize them via their targeting signal, which is composed of a highly hydrophobic β-hairpin motif (Söllner et al., 1989; Rapaport and Neupert, 1999; Schleiff et al., 1999; Krimmer et al., 2001; Yamano et al., 2008; Jores et al., 2016). Next, they are translocated across the OM via a pore formed by Tom40, the core subunit of the TOM complex.
In the IMS, the small translocase of the inner membrane (TIM) chaperone complexes Tim8/13 and Tim9/10 prevent the aggregation of the β-barrel precursor proteins (Hoppins and Nargang, 2004; Wiedemann et al., 2004; Habib et al., 2005). Finally, OM insertion of such proteins is facilitated by the topogenesis of outer membrane β-barrel proteins (TOB) complex (also known as sorting and assembly machinery [SAM]; Paschen et al., 2003; Wiedemann et al., 2003; Chan and Lithgow, 2008).
Although the mitochondrial steps of the β-barrel biogenesis are rather well defined, much less is known about the early cytosolic events of the biogenesis. Importantly, during their transit through the cytosol, the newly synthesized β-barrel proteins must be kept in an import-competent conformation. This is a difficult task, as these proteins are prone to aggregation as a result of their high overall hydrophobicity and their tendency to form β-sheets (Hecht, 1994).
The cell possesses a multitude of different molecular chaperones and cochaperones that should prevent misfolding and aggregation of their client proteins (Kim et al., 2013). The ATP-dependent chaperones of the heat shock protein 70 (Hsp70) family help in the folding of newly synthesized proteins, but can also unfold and disaggregate misfolded proteins (Morano, 2007; Mayer, 2013; Clerico et al., 2015; Nillegoda and Bukau, 2015). Although Hsp70 chaperones can interact with a vast range of proteins, their activity and substrate specificity are often fine-tuned by cochaperones of the Hsp40 family, also known as J proteins (Walsh et al., 2004; Kampinga and Craig, 2010; Clerico et al., 2015). Proteins of the Hsp90 family form a second group of ATP-dependent chaperones that are often involved in the maturation of signaling molecules (Young et al., 2001; Terasawa et al., 2005; Schopf et al., 2017).
Chaperones of the Hsp90 and Hsp70 families, as well as some of their cochaperones, have been implicated in the import of mitochondrial preproteins (Deshaies et al., 1988; Caplan et al., 1992; Endo et al., 1996; Hoseini et al., 2016; Xie et al., 2017) and of carrier proteins of the mitochondrial inner membrane (Young et al., 2003; Bhangoo et al., 2007). Furthermore, the newly synthesized form of the OM protein Mim1 was shown to interact specifically with the Hsp40 protein Djp1 (Papić et al., 2013). In many cases, however, the data were only obtained in in vitro experiments, and the physiological relevance of the chaperone–precursor interaction is unclear. Additionally, the role of the individual chaperones in the biogenesis of the mitochondrial proteins is often ill defined. Similarly unclear is whether the chaperones are solely preventing the aggregation of the newly synthesized proteins or whether they are also involved in the mitochondrial targeting of their substrates. Finally, it is still unknown whether the involvement of molecular chaperones in the biogenesis of mitochondrial precursor proteins is restricted to some mitochondrial proteins or if this is a general principle. For example, so far, β-barrel proteins were not reported to interact with any cytosolic chaperone or cochaperone.
To address these open questions and to better understand the early events in the biogenesis of mitochondrial β-barrel proteins, we characterized their interactions with cytosolic factors and analyzed the importance of these interactions for the biogenesis pathway of these proteins. We found that, in yeast, several chaperones of the Hsp90, Hsp70, and Hsp40 families interact with β-barrel proteins and that such interactions are required for an efficient mitochondrial import in vitro and in vivo. Collectively, the current study sheds new light on the early events in the biogenesis of mitochondrial β-barrel proteins.
Results
Cytosolic chaperones interact in vitro with newly synthesized β-barrel proteins
To remain import competent, newly synthesized mitochondrial β-barrel proteins should avoid aggregation in the cytosol, a task that might require the function of chaperones. To study the role of cytosolic chaperones in the biogenesis of these proteins, we first wanted to identify the chaperones that interact with such proteins. To that end, we performed pull-down experiments with an HA-tagged version of the β-barrel protein Porin translated in vitro in yeast extract. We reasoned that because the yeast extract does not contain mitochondria, the newly synthesized proteins cannot be integrated into a membrane, implying that they should be kept in solution by proteins present in the extract. After pulling down Porin-HA with anti-HA beads, we identified copurified proteins by mass spectrometry. Among the coeluted proteins, we identified molecular chaperones of the Hsp90 and Hsp70 families, as well as Hsp40 family cochaperones (Table 1). Of note, among the hits of the mass spectrometric analysis, we found also ubiquitin and in the pull-down eluates, we could detect a band with a size corresponding to the one expected for an ubiquitinated version of Porin (Fig. S1 A). Thus, it might be that a certain proportion of the expressed Porin molecules is getting ubiquitinated.
Table 1. Proteins that co-purified with in vitro translated Porin.
| Protein name | Gene name | iBAQa |
|---|---|---|
| Ubiquitin | RPS31;RPL40B;RPL40A;UBI4 | 255830000 |
| Porin | POR1 | 234220000 |
| Heat shock protein SSA2 | SSA2 | 138040000 |
| Elongation factor 1-α | TEF1 | 135660000 |
| Mitochondrial protein import protein MAS5 | YDJ1 | 105990000 |
| Heat shock protein 60, mitochondrial | HSP60 | 92565000 |
| Glyceraldehyde-3-phosphate dehydrogenase 3 | TDH3 | 78971000 |
| Tryptophan-tRNA ligase, cytoplasmic | WRS1 | 64375000 |
| Pyruvate kinase 1 | CDC19 | 59277000 |
| Heat shock protein SSB1 | SSB1 | 57047000 |
| ATP-dependent RNA helicase eIF4A | TIF1 | 44960000 |
| Tubulin beta chain | TUB2 | 39876000 |
| Plasma membrane ATPase 1/2 | PMA1;PMA2 | 35671000 |
| Long-chain-fatty-acid-CoA ligase 4 | FAA4 | 32266000 |
| 40S ribosomal protein S3 | RPS3 | 31676000 |
| H/ACA ribonucleoprotein complex subunit 2 | NHP2 | 30161000 |
| ATP-dependent molecular chaperone HSC82 | HSC82 | 27457000 |
| 60S ribosomal protein L27-A/B | RPL27B;RPL27A | 23564000 |
| Acetolactate synthase catalytic subunit, mitochondrial | ILV2 | 21029000 |
| 60S ribosomal protein L17-B | RPL17B | 20275000 |
Only the 20 proteins with the highest iBAQ values are shown. Proteins with a chaperone-like function are highlighted.
Intensity-based absolute quantification.
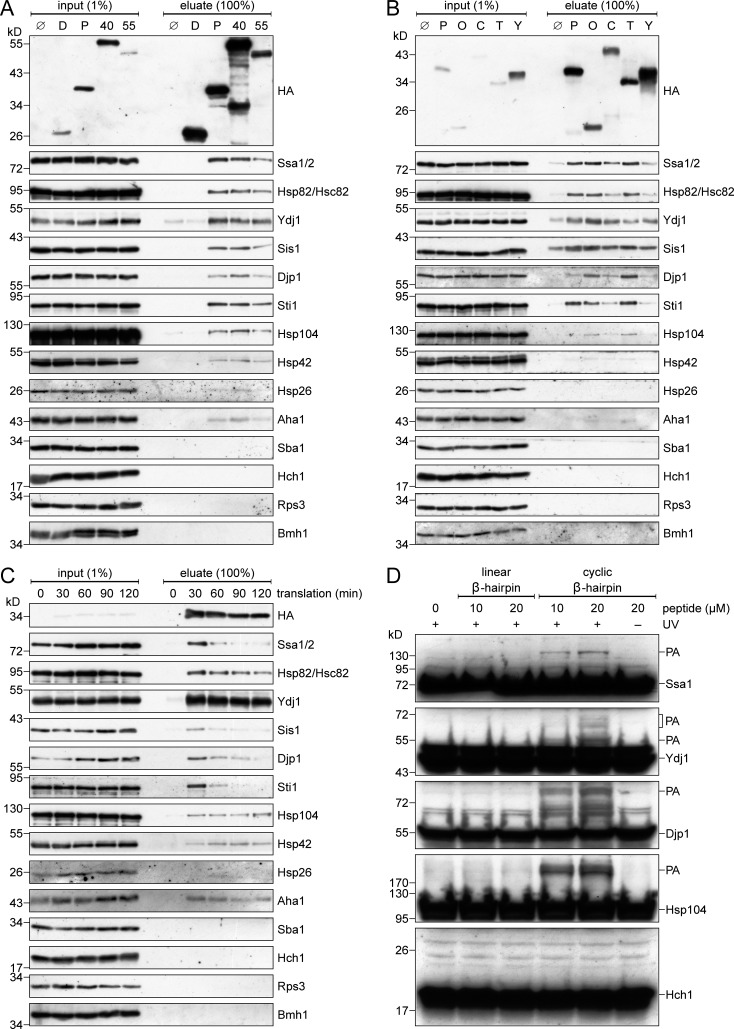
To verify the mass spectrometry analysis and to test whether the chaperones also interact with other β-barrel proteins, we translated in yeast extracts HA-tagged variants of either the β-barrel proteins Porin, Tom40, or Tob55 as well as the control protein, dihydrofolate reductase (DHFR), and analyzed the copurified proteins by immunodecoration with antibodies against several chaperones and cochaperones (Fig. 1 A). To assure that potential interactions are not mediated by the N-terminal POTRA domain of Tob55, we used a truncated version of the protein that is composed only of its β-barrel domain (Pfitzner et al., 2016). We found that all three β-barrel proteins coeluted with the Hsp70 chaperones Ssa1/Ssa2 and the Hsp90 chaperones Hsc82/Hsp82. Of note, the paralogous proteins Ssa1/Ssa2 and Hsc82/Hsp82 are very similar to each other and our antibodies cannot discriminate between the two isoforms. Additionally, the eluates contained the Hsp40 cochaperones Ydj1, Sis1, and Djp1, two cochaperones interacting with Hsp90 proteins, namely Sti1 and Aha1, and two chaperones, Hsp104 and Hsp42, that were previously found to associate with misfolded or aggregated proteins (Bösl et al., 2006; Ungelenk et al., 2016). The cochaperones Sba1 and Hch1, as well as the ribosomal protein Rps3 and the cytosolic 14-3-3 protein Bmh1 were not coeluted with any of the translated proteins (Fig. 1 A). Furthermore, also the cytosolic chaperonin CCT/TRiC did not interact with newly synthesized Porin (Fig. S1 B). Control reactions without any translated protein or with DHFR lead to a negligible copurification, if at all, of the above-mentioned chaperones (Fig. 1 A). These findings indicate interactions of newly synthesized β-barrel proteins with a restricted subset of cytosolic chaperones.
Figure 1.
Cytosolic chaperones interact with newly synthesized β-barrel proteins. (A and B) In vitro translation reactions using yeast extracts without mRNA (Ø) or programmed with mRNA encoding DHFR-HA (D), Porin-HA (P), Tom40-HA (40), Tob55Δ120-HA (55), Om14-HA (O), Cyc3-HA (C), Tim23-HA (T), or Yah1-HA (Y) were subjected to a pull-down with anti-HA beads. Samples from the input and the eluates were analyzed by SDS-PAGE and immunodecoration with the indicated antibodies. (C) Yeast extracts programmed with mRNA encoding for Porin-HA were incubated for the indicated times. Afterward, the reactions were subjected to an anti-HA pull-down and analyzed as in A. (D) Yeast extracts were incubated with the indicated Bpa-containing peptide at the specified concentrations. Some samples were either illuminated with UV light (+UV) to induce cross-linking or were left in darkness (−UV). Then, the samples were analyzed by SDS-PAGE and immunodecoration with the indicated antibodies. PA, specific photoadducts.
To test whether the observed interactions are specific to β-barrel proteins, we performed similar pull-down experiments with other mitochondrial precursor proteins. We found that the α-helical OM protein Om14 and the inner membrane protein Tim23 also associate with Hsp70 and Hsp90 chaperones (Fig. 1 B). However, there are clear differences between the levels of the associated cochaperones Ydj1, Sis1, Djp1, and Sti1 to the helical hydrophobic proteins as compared with their interactions with β-barrel proteins. Of note, the two soluble proteins Cyc3 and Yah1 that are targeted to the IMS and matrix, respectively, displayed only weak interactions with cytosolic chaperones. Collectively, these results indicate that the interactions between precursor proteins and Hsp70 or Hsp90 chaperones are mainly governed by hydrophobicity, whereas the cochaperones are sensitive to more subtle differences in their substrate proteins and are, therefore, able to fine tune the specificity of the chaperone–substrate complexes.
The interactions of chaperones with β-barrel proteins are dynamic
Having established an assay to monitor specific association of cytosolic factors with β-barrel proteins, we next analyzed whether the observed interactions vary with time. When we increased the in vitro translation reactions from 30 to 120 min, we observed that the levels of most associated chaperones were reduced over time, whereas the levels of Hsp104 and Hsp42 were slightly increased in the samples from longer incubation times (Fig. 1 C). Because these proteins associate with misfolded or aggregated proteins, our findings suggest that the Hsp70 and Hsp90 chaperone systems are early interaction partners that can keep the newly synthesized β-barrel proteins in solution for only a limited time. Longer incubation times, therefore, lead to an increased misfolding and possibly to the aggregation of the substrate protein and consequently also to an increased association with Hsp104 and Hsp42.
Next, we tested if the chaperones depend on each other for binding to β-barrel proteins. To this end, we translated Porin in yeast extracts prepared from cells that were lacking the cochaperones Sti1, Ydj1, or Sis1 (Fig. S1, C–E). The absence of Sti1 led to a decreased association of Porin with Hsp82. This is in line with the reported function of Sti1 as mediator of the transfer of substrate proteins from Hsp70 to Hsp90 (Scheufler et al., 2000; Wegele et al., 2003). Additionally, the amount of copurified Ydj1 was increased, whereas the level of Djp1 was reduced in the eluates obtained from the sti1Δ extract (Fig. S1 C). When an extract from ydj1Δ cells was used, the levels of coeluted Sis1 and Djp1 were reduced, whereas Hsp104, Hsp42 and Hsp26 were present in the eluate in higher amounts (Fig. S1 D). These observations substantiate the importance of Ydj1 in preventing misfolding or aggregation of newly synthesized β-barrel proteins.
To prepare a yeast extract without Sis1, we constructed a yeast strain that contains the SIS1 gene under the control of a tetracycline-repressible promoter. Cells were depleted of Sis1 by adding the tetracycline analogue doxycycline to the growth medium and the obtained yeast extract was compared with one from the same cells grown without doxycycline. As expected, Sis1 could not be detected in the Dox-treated cells (Fig. S1 E). Using these extracts, we observed a slightly increased association of Ssa1, Hsp82, Sti1, and Hsp104 with newly synthesized Porin, whereas the levels of copurified Ydj1 and Djp1 were reduced (Fig. S1 E). These findings show that the chaperone system that prevents the aggregation of newly synthesized β-barrel proteins can dynamically adapt to the chaperone composition of the cell. It seems that the association with chaperones is not critically dependent on any single cochaperone, as the lack of Sti1, Ydj1, or Sis1 can be compensated by the remaining chaperones and cochaperones.
A β-hairpin motif is sufficient for the interaction with chaperones
Because we could demonstrate that full-length β-barrel proteins can interact with several (co)chaperones, we asked whether a β-hairpin motif, as the smallest building block of all β-barrel proteins, is sufficient for this interaction. To address this question, we used a linear and a cyclic β-hairpin peptide derived from the human β-barrel protein VDAC1. We previously showed that the cyclic β-hairpin peptide is recognized by the cell as a mitochondrial targeting signal of β-barrel proteins and interacts with various TOM components (Jores et al., 2016). The peptides used in this study harbor the photoreactive amino acid p-benzoyl-l-phenylalanine (Bpa) and can form covalent bonds to nearby proteins upon irradiation with UV light. When we mixed the Bpa-containing peptides with a yeast extract and illuminated the sample with UV light, we observed a dose-dependent formation of photoadducts (PA) of the cyclic, but not the linear, β-hairpin peptide with Ssa1, Ydj1, Djp1, and Hsp104 (Fig. 1 D). For some chaperones we observed the formation of several PAs that were migrating at an apparent molecular mass higher than expected for an adduct of the chaperones with a single peptide molecule. Such multiple bands were observed in previous cross-linking experiments (Plath et al., 1998; Jores et al., 2016; Wu et al., 2018) and might result from multiple peptides binding to the same chaperone molecule and/or from different SDS-PAGE mobility of the various PAs. The observed adducts are specific, as the cochaperone Hch1 did not form any PA with the β-hairpin peptide. We conclude that the β-hairpin motif is sufficient for a direct interaction with cytosolic (co)chaperones.
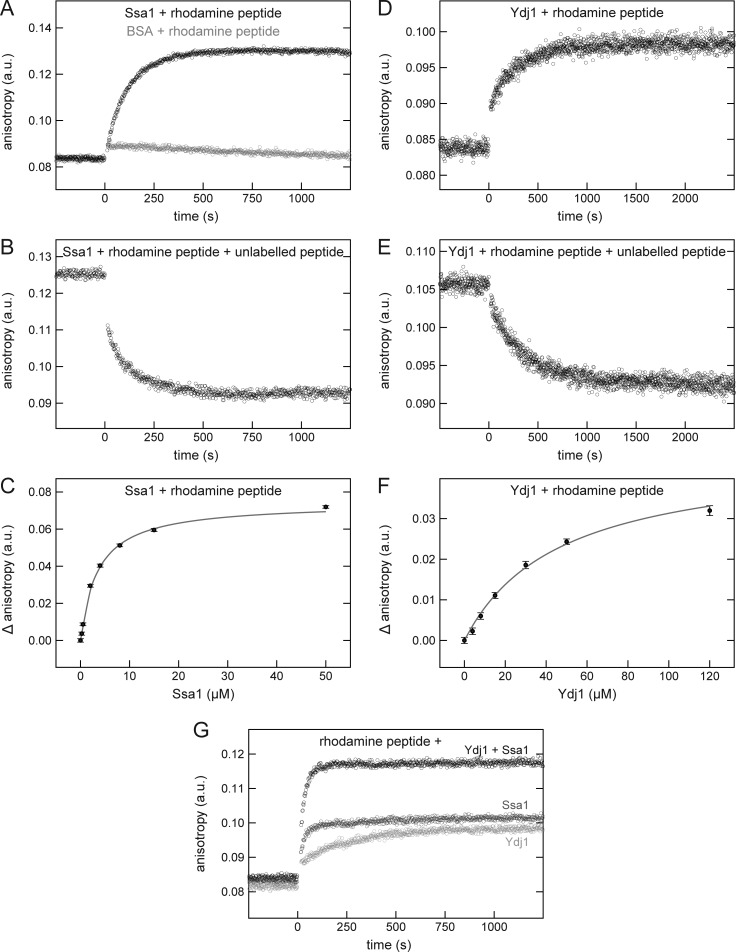
To further characterize the interaction between the β-hairpin peptide and the Hsp70 chaperone Ssa1, we analyzed their binding kinetics and affinity by fluorescence anisotropy. The anisotropy of a rhodamine-labeled cyclic β-hairpin peptide increased upon addition of recombinant Ssa1 indicating the formation of a peptide-Ssa1 complex (Fig. 2 A, black circles). This interaction is specific as the addition of the control protein BSA did not lead to such an increase (Fig. 2 A, gray circles). When the Ssa1/rhodamine-labeled peptide complex was supplemented with an excess of unlabeled cyclic β-hairpin peptide, the labeled peptide dissociated from Ssa1 demonstrating that the chaperone can also interact with the unlabeled peptide (Fig. 2 B). Next, we used titration experiments with increasing concentrations of Ssa1 to determine the affinity between the chaperone and the cyclic β-hairpin peptide (Fig. 2 C). A dissociation constant (Kd) of 3.5 µM was derived from a hyperbolic regression curve fitted to the data. This affinity is in line with previous studies that reported affinities in the low micromolar range for the interaction of Hsp70s with substrate peptides (Schmid et al., 1994; Endo et al., 1996; Pierpaoli et al., 1997; Ricci and Williams, 2008; Schneider et al., 2016).
Figure 2.
A cyclic β-hairpin peptide binds directly to Ssa1 or Ydj1. (A–F) The fluorescence anisotropy of a rhodamine-labeled cyclic β-hairpin peptide was measured in the presence of 10 µM Ssa1 (A–C, black circles), 30 µM BSA (A, gray circles), or 30 µM Ydj1 (D–F). After association of the rhodamine-labeled peptide with the indicated proteins (A and D), the samples were supplemented with a 100-fold excess of the unlabeled peptide and dissociation was recorded (B and E). For affinity determinations, the rhodamine-labeled peptide was mixed with the indicated concentrations of either Ssa1 (C) or Ydj1 (F), and the difference in anisotropy (Δ anisotropy) of the bound and free peptide was plotted against the (co)chaperone concentration. The data were fitted using a hyperbolic regression curve (gray). (G) The fluorescence anisotropy of a rhodamine-labeled cyclic β-hairpin peptide was measured in the presence of 5 µM Ssa1 (dark gray circles), 30 µM Ydj1 (light gray circles), or both proteins (black circles).
Next, we performed similar experiments with a recombinant Ydj1 and observed an association of Ydj1 with both the rhodamine-labeled (Fig. 2 D) and the unlabeled cyclic peptide (Fig. 2 E). Titration experiment revealed the Kd of the Ydj1/peptide interaction to be 46.8 µM (Fig. 2 F). Similar two-digit micromolar affinities have been previously determined for interactions of Hsp40 cochaperones with their substrate peptides (Pierpaoli et al., 1997; Li and Sha, 2004). Collectively, a cyclic β-hairpin peptide can bind directly to both the Hsp70 chaperone Ssa1 and its Hsp40 cochaperone Ydj1.
Finally, we analyzed if Ssa1 and Ydj1 can form a ternary complex with the β-hairpin peptide. To this end, we followed the changes in the anisotropy of the rhodamine-labeled peptide upon addition of either the individual proteins separately or both together. The change in anisotropy was much bigger when both chaperones were present in the reaction, suggesting the formation of a ternary complex composed of Ssa1, Ydj1, and the β-hairpin peptide.
Hsp70 chaperones are required for the biogenesis of β-barrel proteins
Having established that chaperones can bind to newly synthesized β-barrel proteins, we wanted to test if these interactions are required for an efficient mitochondrial import of these proteins. To this end, we aimed to perform in vitro import experiments with β-barrel proteins. First, we analyzed the import efficiency of Porin translated in yeast extract for 30, 60, or 120 min. In line with the observed reduced interactions with Hsp70 and its cochaperones after longer translation times (Fig. 1 C), we observed also reduced import of Porin upon longer translation reactions (Fig. S2 A).
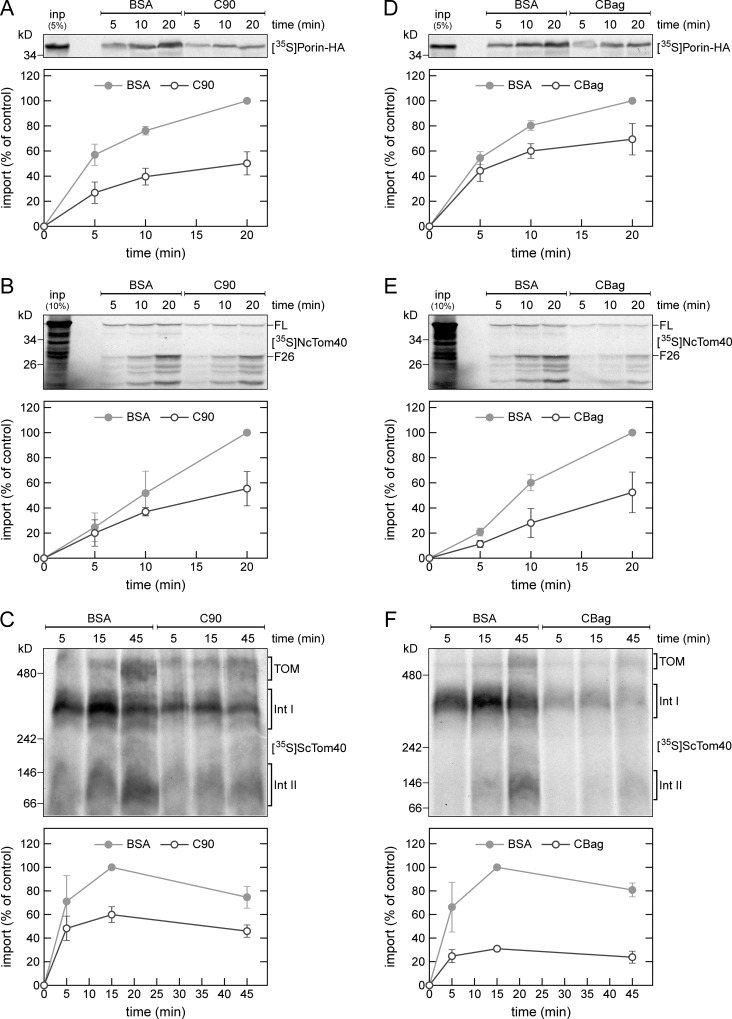
Next, we assessed the requirement for chaperones in the import of β-barrel proteins by testing if the C-terminal domain of human Hsp90 (C90) can inhibit the β-barrel protein import. Previous studies showed that C90 inhibits the import of mitochondrial inner membrane carrier proteins by blocking the chaperone binding site on the import receptor Tom70 (Young et al., 2003; Bhangoo et al., 2007). Importantly, the in vitro import of radiolabeled Porin was reduced by ∼50% in the presence of C90 (Fig. 3 A). Comparable results were obtained when we used Tom40 from Neurospora crassa (NcTom40) as a substrate (Fig. 3 B). The proper membrane integration of NcTom40 was monitored by the formation of a proteinase K–protected fragment of 26 kD (F26; Rapaport and Neupert, 1999). In a third import assay, we tested the effect of C90 on the assembly of yeast Tom40 (ScTom40) into the TOM complex, as monitored by BN-PAGE. On its assembly pathway into the mature TOM complex, Tom40 forms two stable intermediates that are detected by BN-PAGE, a first assembly intermediate with the TOB complex, and a second intermediate composed of Tom40, Tom22, and the small TOM subunits (Rapaport and Neupert, 1999; Model et al., 2001; Wiedemann et al., 2003). We observed that the addition of C90 to the import reaction caused a clear reduction in the formation of the two assembly intermediates and of the mature TOM complex (Fig. 3 C). The impaired formation of the first intermediate indicates that the import of Tom40 is inhibited in an early step before its association with the TOB complex.
Figure 3.
Cytosolic chaperones are required for the in vitro import of β-barrel proteins. (A–F) Top, radiolabeled precursor proteins of Porin-HA (A and D), N. crassa Tom40 (NcTom40; B and E), or yeast Tom40 (ScTom40; C and F) were produced in yeast extract, and subjected to in vitro import reactions using isolated mitochondria. Before the import reaction, the mitochondria were mixed with either C90 or BSA (A–C). Alternatively, the translation reactions were supplemented with either CBag or BSA (D–F). After import for the indicated times, the mitochondria were subjected to carbonate extraction (A and D) or were treated with proteinase K (B and E). The samples were subjected to SDS-PAGE (A, B, D, and E) or BN-PAGE (C and F) and autoradiography. Bottom, intensities of the bands corresponding to Porin-HA, the protease-protected fragment F26 of NcTom40, or assembly intermediate I of Tom40 from three independent experiments were quantified and the mean intensity from the 20-min (A, B, D, and E) or 15-min (C and F) import in the presence of BSA was set to 100%. Error bars represent ± SD. FL, full-length. F26, protease-protected fragment of 26 kD. The migration of the assembled TOM complex and assembly intermediates (Int) I and II of Tom40 are indicated.
To exclude the possibility that the observed import inhibition is caused by a general import deficiency of the C90-treated mitochondria, we performed import experiments using the matrix-destined precursor protein pSu9-DHFR. Supporting a specific effect on β-barrel proteins, the presence of C90 in the import reaction did not affect the import of pSu9-DHFR (Fig. S2 B). Collectively, these results demonstrate that binding of chaperones to Tom70 is required for an efficient import of mitochondrial β-barrel proteins.
To directly analyze the putative involvement of Hsp70 chaperones, we performed in vitro import experiments in the presence of the Hsp70 inhibitor CBag that is composed of the C-terminal Bag domain of human Bag-1M (Young et al., 2003; Bhangoo et al., 2007). The mitochondrial import of Porin and NcTom40 as well as the assembly of ScTom40 were reduced by 30–70% when these proteins were synthesized in CBag-treated yeast extract (Fig. 3, D and F). As a control, the import of the matrix-destined protein pSu9-DHFR was not affected under these conditions (Fig. S2 C). Furthermore, we tested if the Hsp90 inhibitor Radicicol affects the import of β-barrel proteins but could not detect such an effect under the tested conditions (unpublished data). These experiments show that Hsp70 chaperones are required for proper import of mitochondrial β-barrel proteins.
To substantiate these findings, we used Porin molecules that were translated in yeast extract and denatured with urea to remove bound chaperones. We found that the inhibitory effect of C90 on the import of urea-denatured Porin is less pronounced when compared with nondenatured, chaperone-bound Porin (Fig. S2 D). The residual inhibition of the import of urea-denatured Porin by C90 can be explained by the latter sterically hindering Porin from binding to the TOM complex. Supporting this notion is the previous observation that antibodies against Tom70 can inhibit the binding of Tom40 precursor molecules to the mitochondrial surface (Keil et al., 1993). As expected, the Hsp70 inhibitor CBag completely failed to impair the import of urea-denatured Porin (Fig. S2 E). These findings confirm that the inhibitory effects of C90 and CBag result from interactions of chaperones with β-barrel precursor proteins.
Tom70 is involved in the import of β-barrel proteins
We next analyzed if Tom70 is required directly for an efficient biogenesis of β-barrel proteins. To this end, we performed in vitro import experiments using Porin and mitochondria isolated from either WT or cells deleted for TOM70 and its paralog TOM71 (tom70/71Δ). Mitochondria lacking Tom70/71 imported Porin with reduced efficiency as compared with control organelles (Fig. S3 A). To substantiate the role of Tom70 in the import of β-barrel proteins, we tested its ability to directly bind the targeting signal of these proteins. When Bpa-containing linear or cyclic β-hairpin peptides were incubated with fusion proteins composed of GST and the cytosolic domain of Tom70 (GST-Tom70) or Tom20 (GST-Tom20), we observed specific PAs mainly of the cyclic peptide with the receptor proteins but not with GST that served as a control (Fig. S3 B). Collectively, these results demonstrate that Tom70 has a dual function in the biogenesis of β-barrel proteins; a docking site for cytosolic chaperones and an import receptor recognizing the β-barrel protein targeting signal.
Cytosolic chaperones interact in vivo with a β-hairpin peptide
We next asked whether the interactions that we observed in the yeast extract can be detected also in vivo. To this end, we used a hybrid protein composed of the β-hairpin peptide fused to the passenger domain DHFR-HA. We demonstrated previously that this fusion protein is targeted to mitochondria (Jores et al., 2016). As we expected only transient association of the β-hairpin peptide and chaperones, we used a photo cross-linking approach to take a snapshot of the interactions of the β-hairpin peptide. For this aim, we used an amber suppression system to insert in vivo the unnatural amino acid Bpa into the β-hairpin moiety (Chen et al., 2007).
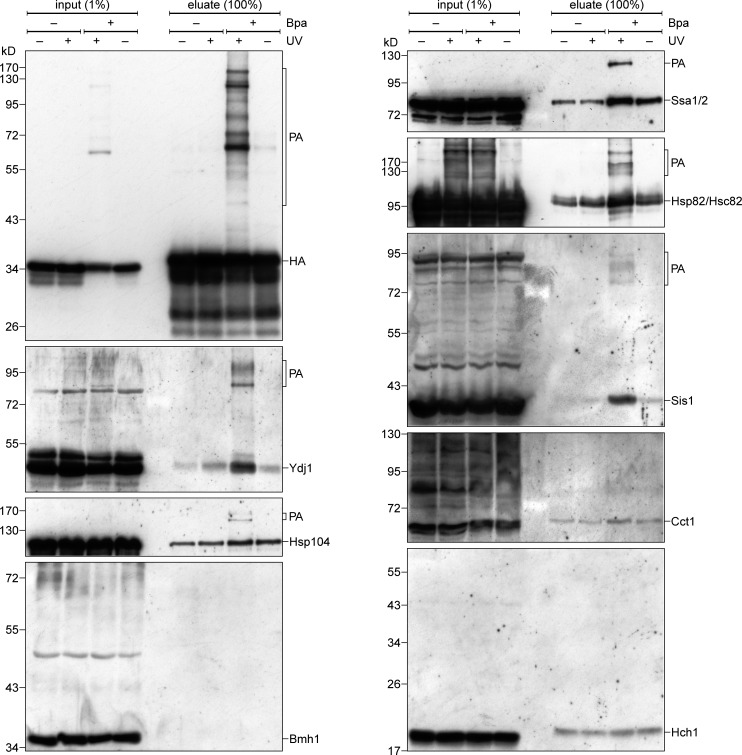
When cells expressing the β-hairpin(Bpa)–DHFR fusion protein were irradiated with UV light, we detected the formation of several PAs (Fig. 4, upper left panel). The specificity of these PAs is demonstrated by their absence when either the cells were not exposed to UV light or upon usage of proteins lacking the Bpa residue (Fig. 4). To analyze if the β-hairpin is cross-linked to chaperones, we first enriched PAs of the fusion protein by a pull-down with anti-HA beads and then probed the samples with antibodies against some of the chaperones that we found to bind in vitro to β-barrel proteins. In this way, we detected specific PAs for Ssa1, Hsp82, Ydj1, Sis1, and Hsp104 (Fig. 4). In all cases, no PAs were observed in the absence of UV irradiation or using constructs without Bpa residue. Furthermore, we could not detect PAs for the control cytosolic proteins Cct1, Bmh1, and Hch1. These results demonstrate that cytosolic (co)chaperones directly interact in vivo with β-barrel proteins.
Figure 4.
Cytosolic chaperones interact in vivo with a β-hairpin peptide. Yeast cells expressing either control hp18(VDAC)-DHFR-HA (−Bpa) or a variant with Bpa inserted in the β-hairpin (+Bpa) were subjected to in vivo photo cross-linking (+UV) or kept in the dark (−UV). Next, the cells were lysed and subjected to a pull-down with anti-HA beads. Samples from the input and the eluates were analyzed by SDS-PAGE and immunodecoration with the indicated antibodies.
Hsp40 cochaperones are required for the biogenesis of β-barrel proteins
We then asked whether the interactions of Ydj1 and Sis1 with β-barrel proteins contribute to the biogenesis of these proteins. Ydj1 was previously implicated with the import of the mitochondrial presequence-containing inner membrane protein F1β and the same study also showed that overexpression of SIS1 can partially complement the growth phenotype of yeast cells harboring a temperature-sensitive allele of YDJ1 (Caplan et al., 1992). However, so far, Sis1 was not reported to be involved in the import of mitochondrial proteins.
Sis1 is an essential protein and yeast cells with a deletion of YDJ1 have a severe growth defect and tend to accumulate suppressors. Therefore, we created strains that express SIS1, YDJ1, or both genes under the control of a tetracycline-repressible promoter. To further speed up the depletion of the corresponding proteins, they were fused at their N termini with ubiquitin followed by a leucine residue. After translation, the ubiquitin part is cleaved off leaving Ydj1 or Sis1 with an N-terminal leucine residue that reduces the half-life of the proteins (Gnanasundram and Koš, 2015).
We used a protocol where cells harboring endogenous Porin-HA were grown for 4 h in the presence or absence of doxycycline, followed by 1 h incubation in medium without methionine. Then, the medium was supplemented with [35S]methionine and the cells were harvested immediately or 5, 15, or 30 min afterward. The cells were lysed, and a crude mitochondrial fraction was obtained, solubilized, and subjected to a pull-down of the HA-tagged Porin. Of note, two populations of Porin-HA molecules can be detected in such experiments: freshly synthesized radiolabeled ones by autoradiography and preexisting ones together with the newly synthesized molecules by immunodecoration with anti-HA antibodies. Initially, as a control, we performed these experiments with WT cells that do not contain tetracycline-repressible promoters. As expected, the levels of radiolabeled Porin-HA from doxycycline-treated or untreated cells were indistinguishable (Fig. S4 A), verifying that the addition of doxycycline does not affect the biogenesis of Porin-HA.
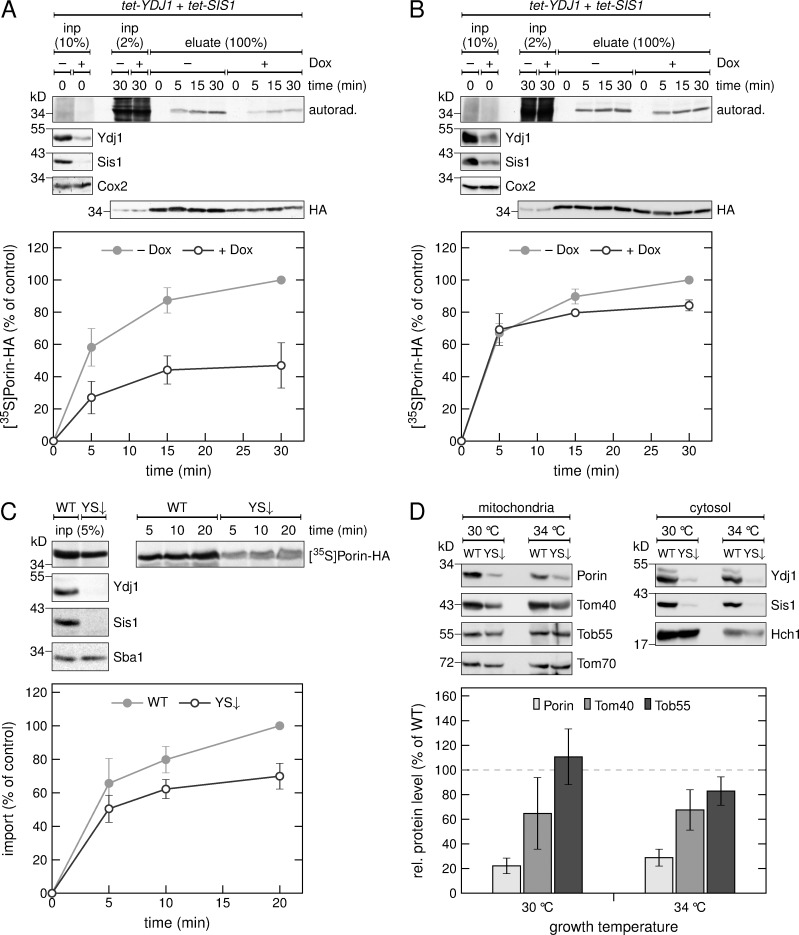
Next, we tested if a depletion of either Ydj1 or Sis1 hampers the biogenesis of Porin-HA and observed only a marginal reduction in the level of the radiolabeled β-barrel protein (Fig. S4, B and C). Because it was reported that Sis1 can partially complement for the absence of Ydj1, we also analyzed the effect of a double depletion of both proteins. Interestingly, the levels of radiolabeled Porin-HA in the mitochondrial fraction was reduced by ∼50% when cells were depleted of both, Ydj1 and Sis1 (Fig. 5 A). In addition, we could also observe a reduction in the overall mitochondrial amounts of Porin-HA as detected by immunodecoration with anti-HA antibody (Fig. 5 A).
Figure 5.
The cochaperones Ydj1 and Sis1 are required for the in vivo biogenesis of Porin. (A and B) HA-tagged Porin was expressed in a strain with tetracycline-repressible promoters controlling the expression of YDJ1 and SIS1 in the absence (−) or presence (+) of doxycycline (Dox) followed by methionine starvation. Synthesis of radiolabeled proteins was initiated by addition of [35S]Met to the medium, and cells were harvested after the indicated time periods. Then, a crude mitochondrial fraction (A) or the whole cell lysate (B) were obtained. The samples were solubilized and subjected to a pull-down with anti-HA beads. Input samples from the whole cell lysate (inp) and the eluates were analyzed by SDS-PAGE, autoradiography (autorad.) and immunodecoration with the indicated antibodies. Bottom, intensities of the bands corresponding to Porin-HA from three independent experiments were quantified, and the mean intensity from the 30-min samples without doxycycline was set to 100%. Error bars represent ± SD. (C) Top, radiolabeled Porin-HA was translated in yeast extract from WT yeast cells or from cells depleted for Ydj1 and Sis1 (YS↓). The translation reactions were subjected to in vitro import reactions using mitochondria isolated from YS↓ cells. After import for the indicated times, the mitochondria were subjected to carbonate extraction. The samples were analyzed by SDS-PAGE, autoradiography ([35S]Porin-HA) and immunodecoration with the indicated antibodies. Bottom, the intensities of the bands corresponding to Porin-HA were quantified and the intensity from import of Porin-HA translated in the WT strain was set to 100%. Error bars represent ± SD. (D) Top, a crude mitochondrial and a cytosolic fraction were isolated from WT or YS↓ yeast cells analyzed by SDS-PAGE and immunodecoration with the indicated antibodies. Bottom, intensities of the bands in the mitochondrial fraction from three independent experiments were quantified and normalized to the level of Tom70. The level of the proteins in the WT strain was set to 100%. Error bars represent ± SD.
To exclude the possibility that the reduced levels of Porin-HA are caused by an altered translation rate, we analyzed the total cellular levels of newly synthesized Porin-HA and observed only mild reduction (Fig. 5 B). Furthermore, this reduction is not seen in the initial time point (5 min), but only after longer times. This observation might indicate that this minor reduction is not caused by a compromised translation but rather by a degradation of nonimported precursor molecules.
Because Ydj1 has previously been shown to be involved in the mitochondrial import of presequence-containing proteins (Caplan et al., 1992; Xie et al., 2017), it might be that the observed import defect of Porin-HA is caused by an accumulation of nonimported preproteins that clog the TOM complex. Therefore, we analyzed the import of Porin-HA in an in vitro import assay where it is the only imported protein. When we translated Porin-HA in a yeast extract from cells depleted for Ydj1 and Sis1, we observed compromised import of the β-barrel protein as compared with extract from control WT cells (Fig. 5 C).
To generalize these findings, we monitored the steady-state levels of the three β-barrel proteins Porin, Tom40, and Tob55 in yeast cells depleted for Ydj1 and Sis1 (Fig. 5 D). We observed strongly reduced levels of Porin at both normal growth temperature (30°C) and at slightly elevated temperatures (34°C) where chaperone actions might be more important. We could also detect reduced levels of Tom40 under these conditions, whereas the levels of Tob55 were not affected at 30°C and only mildly reduced at 34°C (Fig. 5 D). Collectively, these results highlight the role of the Hsp40 cochaperones Ydj1 and Sis1 in the biogenesis of β-barrel proteins. However, the extent to which the different β-barrel proteins rely on these cochaperones varies.
Mammalian Hsp70 is involved in the biogenesis of β-barrel proteins
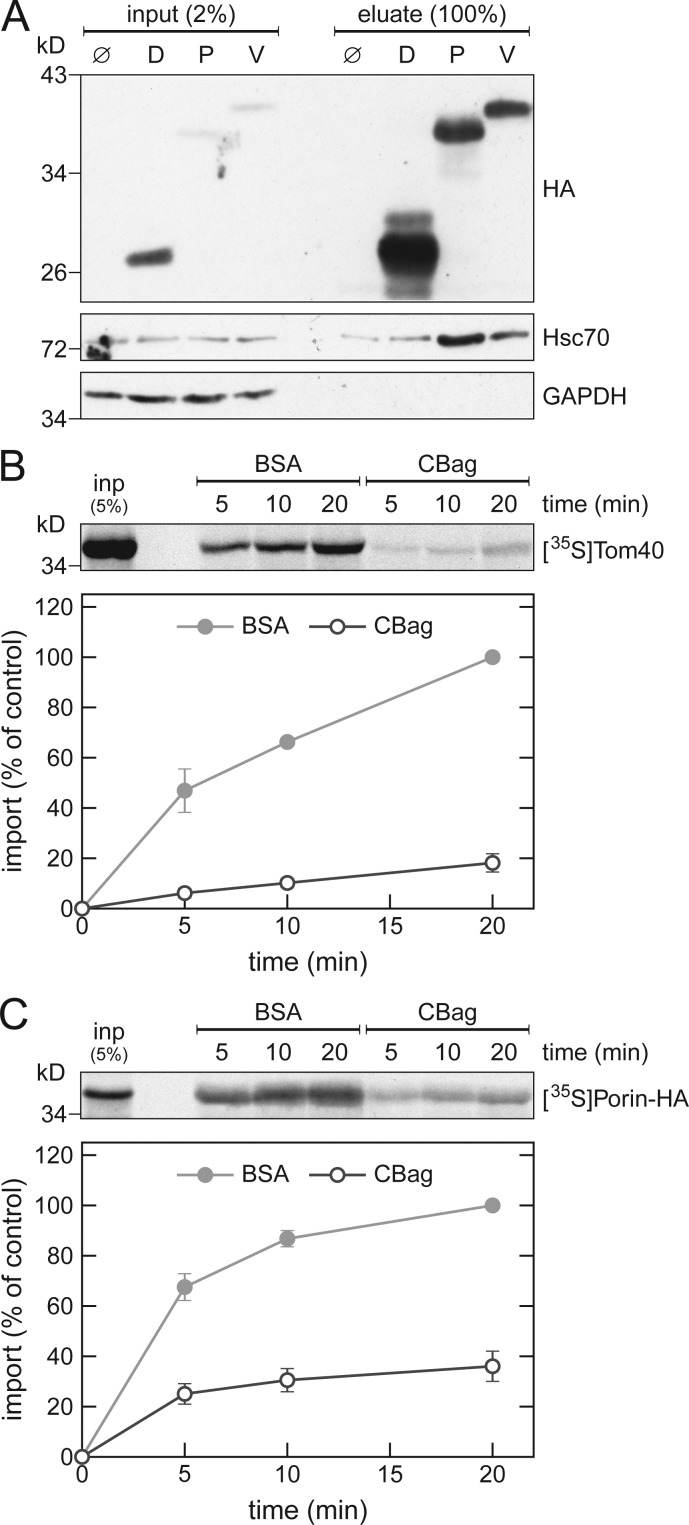
To test whether the involvement of cytosolic Hsp70s in the biogenesis of β-barrel proteins is also conserved in higher eukaryotes, we performed pull-down experiments with β-barrel proteins translated in rabbit reticulocyte lysate. We found that the mammalian Hsp70 chaperone, Hsc70, coelutes with yeast Porin, and its human homologue VDAC1. Only negligible levels were obtained when no protein was synthesized or when the control protein DHFR was used (Fig. 6 A). As a negative control, the cytosolic protein GAPDH did not coelute with any of the tested proteins.
Figure 6.
Mammalian Hsc70 is involved in the biogenesis of β-barrel proteins. (A) In vitro translation reactions using rabbit reticulocyte lysate without mRNA (Ø) or programmed with mRNA encoding DHFR-HA (D), Porin-HA (P), or VDAC1-HA (V) were subjected to a pull-down with anti-HA beads. Samples from the input and the eluates were analyzed by SDS-PAGE and immunodecoration with the indicated antibodies. (B and C) Top, rabbit reticulocyte lysate was used to synthesize radiolabeled human Tom40 (B) or yeast Porin (C). After translation, the lysate was supplemented with either CBag or BSA and subjected to in vitro import reactions using mitochondria isolated from human (B) or yeast (C) cells. After import for the indicated times, the mitochondria were treated with proteinase K (B) or were subjected to carbonate extraction (C), and the samples were analyzed by SDS-PAGE and autoradiography. Bottom, intensities of the bands corresponding to Tom40 or Porin from three independent experiments were quantified and the mean intensity from the 20-min import with BSA was set to 100%. Error bars represent ± SD.
Next, we analyzed whether mammalian Hsc70 is indeed required for import of β-barrel proteins. To this end, we performed in vitro import experiments using mitochondria isolated from human cells and precursor proteins of human Tom40 translated in rabbit reticulocyte lysate that was treated with the Hsp70 inhibitor CBag or, as a control, with BSA. A severe reduction of ∼80% in the import of Tom40 after CBag-treatment was observed (Fig. 6 B). Next, we tested if the mammalian Hsc70 can also function in a heterologous system by performing in vitro import experiments with yeast Porin that was translated in CBag-treated rabbit reticulocyte lysate and then imported into yeast mitochondria. As before, the CBag-treatment led to a drastically reduced import of the precursor protein (Fig. 6 C). Collectively, these results indicate that the interactions of Hsp70 chaperones with β-barrel protein are conserved from yeast to human.
Discussion
Currently, little is known about the mechanisms that assure the safe passage of mitochondrial precursor proteins through the cytosol. To keep these newly synthesized proteins in an import-competent conformation, a tight folding as well as an aggregation of the precursors must be prevented. In this study, we identified several cytosolic chaperones that can bind to newly synthesized β-barrel proteins. We further demonstrate that this binding has a physiological significance, as inhibiting the cytosolic Hsp70 chaperones of the Ssa subfamily or down-regulating the Hsp40 cochaperones Ydj1 and Sis1 hampered the biogenesis of β-barrel proteins. Although pull-down experiments followed by mass spectrometry analysis identified yeast Hsp90 as an interaction partner of β-barrel proteins, subsequent assays with inhibitors of this chaperone failed to detect any effect on the biogenesis of these substrate proteins. Hence, it seems that the binding to Hsp90 is not absolutely required under the tested conditions. This situation is rather similar to the binding preferences in yeast cells of carrier proteins of the mitochondrial inner membrane. These latter proteins require for their proper import in yeast cells cytosolic Hsp70 but not Hsp90 (Young et al., 2003). As Hsp70 and Hsp90 chaperones require ATP for their proper function, their involvement in the biogenesis of β-barrel proteins shed light on previous unexplained observations namely, that the biogenesis of these proteins requires ATP (Pfanner et al., 1988; Hwang and Schatz, 1989; Rapaport and Neupert, 1999; Asai et al., 2004).
Members of the Hsp70 and Hsp90 families serve as multipurpose chaperones and were implicated with the mitochondrial import of inner membrane proteins (Deshaies et al., 1988; Young et al., 2003). Both chaperones have a very broad substrate range and specificity is often generated by interaction with cochaperones (Walsh et al., 2004; Kampinga and Craig, 2010; Röhl et al., 2013). Accordingly, we found that newly synthesized β-barrel proteins are bound by several cochaperones that can also interact with Hsp70 and/or Hsp90 chaperones. Furthermore, when we tested the ability of cytosolic chaperones to bind to different mitochondrial precursor proteins, we observed that Sti1 and the Hsp40 cochaperones Ydj1, Sis1, and Djp1 associated to varying extents with the membrane proteins Porin, Om14, and Tim23, whereas the general Hsp70 and Hsp90 chaperones did not distinguish between these substrates. Notable, the lower affinity of the β-hairpin motif to the cochaperone Ydj1, as compared with its affinity to Hsp70, is in line with the assumption that the substrate might be recognized first by the cochaperone and then transferred to the better binder, the Hsp70 chaperone.
When we analyzed the role that these cochaperones play in vivo in the biogenesis of mitochondrial β-barrel proteins, we found that they have overlapping functions, as none of them is absolutely required for the binding of β-barrel proteins to other chaperones and cochaperones. This multifaceted involvement of cochaperones matches emerging concepts about the function of these proteins as fine-tuners of the ability of Hsp70s to participate in diverse cellular processes (Cyr and Ramos, 2015; Craig and Marszalek, 2017). Furthermore, we could demonstrate that cells depleted of Ydj1 or Sis1 alone imported β-barrel proteins with only slightly reduced efficiency. However, when the cells were depleted of both proteins simultaneously, the biogenesis of mitochondrial β-barrel proteins was reduced. Although Ydj1 and its mammalian orthologue DNAJA1 were reported to be involved in the import of mitochondrial inner membrane proteins (Caplan et al., 1992; Bhangoo et al., 2007), an involvement of Sis1 in mitochondrial protein import was not described so far. Our observation that a certain level of import is still maintained in the double depletion strain might suggest that additional, yet unknown cochaperones can replace Ydj1 and Sis1. Alternatively, the remaining import levels might reflect the basal chaperoning function of Hsp70 without contribution of cochaperones. Our findings regarding the binding of β-barrel proteins to multiple (co)chaperones are in line with a systematic study that found that a given yeast protein can interact with up to 25 different chaperones during its lifetime in the cell (Gong et al., 2009).
Of note, we found that a β-hairpin, which can serve as a targeting signal for mitochondrial β-barrel proteins is sufficient for the binding to cytosolic chaperones. This raises the possibility that the initial binding of newly synthesized β-barrel proteins to chaperones can occur already cotranslationally, namely while the rest of the molecule is still being synthesized. In this way, premature aggregation of the hydrophobic segments of the protein will be prevented already upon their exit from the ribosome.
In this study, we could show that, apart from keeping the β-barrel proteins in an import-competent conformation, the Hsp70 chaperones are also directly involved in the mitochondrial import of their substrates, probably by targeting them to the receptor Tom70. Interestingly, so far, Tom20 was assumed to be the major import receptor for mitochondrial β-barrel proteins (Söllner et al., 1989; Rapaport and Neupert, 1999; Schleiff et al., 1999; Krimmer et al., 2001; Yamano et al., 2008; Jores et al., 2016). Our results, however, demonstrate, that Tom70 is also involved in the biogenesis of these proteins. This involvement can be via direct recognition of the β-barrel precursor protein and/or serving as an anchor for the cytosolic chaperones. Our findings agree with previous studies suggesting that Tom70 is playing a role in the import of β-barrel precursor proteins (Keil et al., 1993; Habib et al., 2005; Jores et al., 2016).
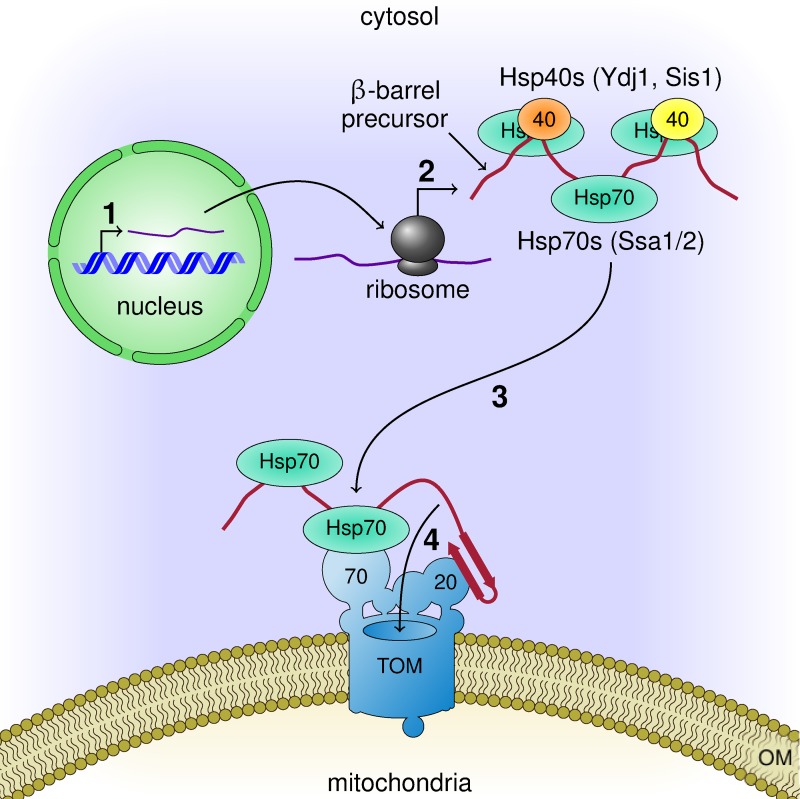
Our current observations together with previous knowledge allow us to suggest a working model for the early stages of the biogenesis of mitochondrial β-barrel proteins (Fig. 7). Upon their synthesis on cytosolic ribosomes, or shortly afterward, these proteins associate with various cochaperones that mediate their association with Hsp70s. This interaction keeps the newly synthesized proteins in an import-competent conformation. The chaperone-substrate complex then docks on the import receptor Tom70, while appropriate β-hairpin elements, which serve as targeting signal, can be recognized by the import receptors Tom20 and Tom70. Both alternatives of initial binding to either Tom20 or Tom70 can be envisaged. The combined action of both receptors increases the fidelity of the recognition, provides a double-check mechanism, and supports productive continuation of the import process by transfer of the substrate to the import pore of the TOM complex. The results obtained with the mammalian components suggest that these principles are conserved also in higher eukaryotes. Collectively, our findings outline a novel mechanism in the early events of the biogenesis of mitochondrial β-barrel proteins.
Figure 7.
Working model for the early events in the biogenesis of β-barrel proteins. Mitochondrial β-barrel proteins are encoded by nuclear genes (1). Upon their synthesis on cytosolic ribosomes (2), they associate with Hsp70 and Hsp40 chaperones that keep them in an import-competent conformation. On the mitochondrial surface, the chaperone–substrate complex can dock to the import receptor Tom70 while the targeting signal in the form of a β-hairpin motif is recognized by the import receptor Tom20 (3). Then, the newly synthesized protein is translocated across the OM through the import pore of the TOM complex (4).
Materials and methods
Yeast strains and growth methods
Standard genetic techniques were used for growth and manipulation of yeast strains. All strains used in this study are listed in Table S1. The tom70/71Δ strain was created by successive knock-outs of TOM70 and TOM71 using a KanMX4 and a NatNT2 cassette, respectively. For strains with genes under the control of a tetracycline-repressible promoter, the strain YMK120 was used (Gnanasundram and Koš, 2015). The tetracycline operator was inserted into the genome by homologous recombination using an insertion cassette amplified from the plasmids pMK632 (Gnanasundram and Koš, 2015), pMK632Kan, or pMK632His. Strains with two genes under the control of the tetracycline operator were obtained by mating of strains with a single tet-regulated gene followed by tetrad dissection. Strains with genomically HA-tagged Porin were created by homologous recombination using an insertion cassette amplified from the plasmid pFA6a-3HA-NatNT2.
Human cell model
Reprogramming of fibroblasts from a healthy individual to induced pluripotent stem cells (iPSCs) was achieved using an episomal transfection method (Takahashi et al., 2007). The healthy iPSCs were available from healthy control, which have been previously characterized (Reinhardt et al., 2013). Small molecule neuroprogenitor cells (smNPCs) were differentiated from the healthy iPSCs according to a previously established method (Reinhardt et al., 2013) and the smNPCs were cultured in two 20 cm2 dishes coated with Matrigel (Sigma Aldrich) in culture medium–containing purmorphic acid (Marrone et al., 2018).
Recombinant DNA techniques
The plasmid pGEM4 (Promega) was amplified by PCR using primers TJ207 and TJ208 to create pGEM4polyA with a poly-A stretch after the multiple cloning site. The plasmid pRS316-Atg32-3HAn was used as template for the PCR-amplification of a triple HA-tag. The amplification product was inserted into either pGEM4polyA or pRS426-TPI using BamHI and SalI restriction sites. The DHFR, POR1, TOM40, TOB55, OM14, CYC3, TIM23, YAH1, NcTOM40, and VDAC1 genes were amplified by PCR from pGEM4 plasmids harboring these genes or from yeast genomic DNA and were then inserted into pGEM4polyA or pGEM4polyA-3HA using suitable restriction sites. The plasmids pGEM4-pSu9-DHFR and pGEM4-VDAC1 were used as templates for the amplification of the pSu9 and hp18(VDAC) gene fragments. The amplification products were cloned into pGEM4polyA-DHFR-3HA using EcoRI and KpnI restriction sites.
The Thr258Bpa mutation within hp18(VDAC) was introduced via site-directed mutagenesis using the QuikChange Site-Directed Mutagenesis kit (Stratagene) according to the manufacturer’s instructions. The hp18(VDAC)-DHFR-3HA constructs were subcloned into pRS426-TPI using EcoRI and SalI restriction sites. The C-terminal triple HA-tag and CYC1 terminator from pRS426-TPI-3HA were amplified by PCR and inserted into the plasmid pFA6A-NatNT2 using HindIII and BamHI restriction sites. For the exchange of the NatMX cassette in pMK632, the plasmid was PCR-amplified using primers TJ203 and TJ204. The HIS3MX and KanMX cassettes were amplified by PCR using the plasmids pFA6a-HIS3MX6 and pFA6a-KanMX4 as templates and TJ201 and TJ202 as primers. The PCR products were combined using a previously described method (Jacobus and Gross, 2015). All primers and plasmids used in this study are listed in Tables S2 and S3, respectively.
Immunoblotting
A list of primary antibodies used in this study is included in Table S4. The secondary antibodies were Horseradish peroxidase-coupled goat anti–rabbit (1721019; BioRad) or goat anti–rat (ab6845; Abcam) that were diluted 1:10,000 or 1:2,000, respectively.
Protein purification
Recombinant CBag and C90 were expressed in BL21 cells from the plasmid pPROEX HTa CBag or pPROEX HTa C90 (gift of F.U. Hartl, Max Planck Institute of Biochemistry, Martinsried, Germany; Young et al., 2003). Expression was induced with 1 mM IPTG for 4 h at 37°C. The cells were harvested, resuspended in French Press buffer (40 mM Hepes, 100 mM KCl, 20 mM imidazole, 2 mM PMSF, and 1× EDTA-free cOmplete protease inhibitor [Roche], pH 7.5) and lysed with an EmulsiFlex-C5 French Press. The cell lysate was subjected to a clarifying spin (15,000 g, 15 min, 4°C) and the recombinant proteins were purified using a 1-ml HisTrap HP column (GE Healthcare) on an ÄKTA Start chromatography system (GE Healthcare). The bound proteins were washed with 20 ml wash buffer (40 mM Hepes, 100 mM KCl, and 50 mM imidazole, pH 7.5) and eluted with elution buffer (40 mM Hepes, 100 mM KCl, and 300 mM imidazole, pH 7.5).
Ssa1 was expressed and purified as described earlier (Schmid et al., 2012). For recombinant Ydj1, BL21-Codon Plus (DE3)-RIL cells were used containing a pET28 vector carrying the full length Ydj1 gene and an N-terminal His6-SUMO-tag. Expression was induced with 1 mM IPTG for 4 h at 30°C. Cells were harvested and resuspended in Ni-NTA buffer A (40 mM NaH2PO4, 500 mM NaCl, 20 mM imidazole, and 2 mM β-mercaptoethanol, pH 8.0) supplemented with EDTA-free protease inhibitor (SERVA) and DNase1. Cells were lysed using a Cell Disruption System (Constant Systems) at 1.8 kbar. After lysate clarification (20,000 g, 1 h, 4°C), the supernatant was loaded on a 5-ml HisTrap HP column (GE Healthcare), washed with 10 column volumes (CV) Ni-NTA buffer A and 10 CV 5% Ni-NTA buffer B (40 mM NaH2PO4, 500 mM NaCl, 500 mM imidazole, and 2 mM β-mercaptoethanol, pH 8.0). The bound proteins were eluted with 100% Ni-NTA buffer B (40 mM NaH2PO4, 500 mM NaCl, 500 mM imidazole, and 2 mM β-mercaptoethanol, pH 8.0). Fractions were diluted 1:10 with Ni-NTA buffer A/H2O (1:1), supplemented with His6-tagged SUMO-protease and incubated overnight at 4°C. The protein solution was then loaded on a 5-ml HisTrap HP column. The flow through was concentrated and loaded on a Superdex 16/60 200 pg SEC column (GE Healthcare). Proteins were eluted with SEC buffer (40 mM Hepes, 150 mM KCl, 5 mM MgCl2, and 1 mM DTT). Purity was checked by SDS-PAGE and the identity of the protein was confirmed by mass spectrometry.
GST, GST-Tom70, and GST-Tom20 were expressed and purified as described earlier (Papić et al., 2013).
Biochemical procedures
For the preparation of human mitochondria, smNPCs were removed from the monolayer of the culture dish using Accutase (Stem Cell Technologies) and the cell pellet was resuspended in 1 ml buffer (70 mM Tris base, 1 mM EDTA, and 0.25 M sucrose, pH 7.4) and kept on ice. The cells were then homogenized using 10 passes of a 20-G needle, 10 passes of a 23-G needle, and then 15 passes of a 30-G needle. The resulting homogenate was cleared by centrifugation (1,000 g, 5 min, 4°C) and the supernatant was centrifuged (10,000 g, 15 min, 4°C) resulting in a crude mitochondrial pellet that was kept on ice before its use in import reactions.
Mitochondria were isolated from yeast cells by differential centrifugation as described (Daum et al., 1982). For the isolation of crude mitochondrial fractions, yeast cells were grown in selective media to logarithmic phase and harvested by centrifugation. The cells were resuspended in SEM buffer (250 mM sucrose, 10 mM MOPS, and 1 mM EDTA, pH 7.4) supplemented with 2 mM PMSF, mixed with glass beads, and lysed with a FastPrep-24 5G homogenizer (MP Biomedicals) for 30 s at 6 m/s. The whole cell lysate was separated from glass beads and cell debris by centrifugation (1,000 g, 3 min, 2°C). Crude mitochondria were isolated from the cell lysate by centrifugation (8,000 g, 10 min, 2°C). A sample from the cytosol-containing supernatant was subjected to chloroform-methanol precipitation. The samples were then subjected to SDS-PAGE followed by Western blotting.
In vitro translation and import of radiolabeled proteins
Yeast extracts for in vitro translation were prepared as described (Wu and Sachs, 2014). For the preparation of yeast extracts from Ydj1- and/or Sis1-depleted cells, they were grown in the presence of 2 µg/ml doxycycline for 8 h before extract preparation. Proteins were synthesized in yeast extract or rabbit reticulocyte lysate (Promega) after in vitro transcription by SP6 polymerase from pGEM4 vectors. Radiolabeled proteins were synthesized in the presence of [35S]methionine (Hartmann Analytik). If not indicated otherwise, the translation times were 30 min for the yeast extract and 60 min for the rabbit reticulocyte lysate.
After translation, the reactions were centrifuged (100,000 g, 45 min, 2°C) to remove ribosomes. The supernatant was diluted with import buffer (250 mM sucrose, 0.25 mg/ml BSA, 80 mM KCl, 5 mM MgCl2, 10 mM MOPS, 2 mM NADH, and 2 mM ATP, pH 7.2) and incubated for 10 min at 25°C. Where indicated, the supernatant was supplemented with 20 µM CBag or, as a control, with an equivalent amount of BSA. To remove aggregated proteins, the supernatant was centrifuged (16,000 g, 10 min). Isolated mitochondria were diluted in import buffer and, where indicated, supplemented with 20 µM of either C90 or, as a control, BSA. The import reactions composed of the precursor proteins mixed with isolated mitochondria were performed by incubation at 25°C for the indicated times. The import reactions were stopped by diluting the samples with SEM-K80 buffer (250 mM sucrose, 80 mM KCl, 10 mM MOPS, and 1 mM EDTA, pH 7.2) and mitochondria were reisolated (13,200 g, 2 min, 2°C).
For urea treatment, the supernatant after the 100,000 g centrifugation was mixed with two volumes of saturated ammonium sulfate solution. After precipitation on ice and centrifugation, the pellet was resuspended in urea buffer (8 M urea and 10 mM MOPS-KOH, pH 7.2). For efficient denaturation, the urea-treated lysate was incubated at 25°C for 30 min before its use in import reactions.
The import of Porin-3HA was analyzed by carbonate extraction. The mitochondria were resuspended in 0.1 M Na2CO3, incubated on ice for 30 min and reisolated by centrifugation (100,000 g, 30 min, 2°C). The pellets were resuspended in 2× Lämmli buffer, incubated for 10 min at 95°C and subjected to SDS-PAGE. The import of pSu9-DHFR-3HA, NcTom40, and human Tom40 was analyzed by proteinase K (PK) treatment (50 µg/ml PK for 30 min on ice). After inhibition of PK with 5 mM PMSF, the samples were boiled at 95°C for 10 min before their analysis by SDS-PAGE. Correct import was analyzed by the formation of typical proteolytic fragments (NcTom40) or PK-resistance of either the mature form of pSu9-DHFR-3HA or the membrane integrated form of human Tom40.
Assembly intermediates of Tom40 were analyzed by BN-PAGE. The mitochondria were solubilized with digitonin buffer (1% digitonin, 20 mM Tris, 0.1 mM EDTA, 50 mM NaCl, and10% glycerol, pH 7.4) for 30 min at 4°C on an overhead shaker (12 rpm). After a clarifying spin (30,000 g, 15 min, 2°C), 10× sample buffer (5% [wt/vol] Coomassie brilliant blue G-250, 100 mM Bis-Tris, and 500 mM 6-aminocaproic acid, pH 7.0) was added and the mixture was analyzed by electrophoresis in a blue native gel containing an 8–13% gradient of acrylamide (Schägger et al., 1994). In all cases, gel separation was followed by Western blotting and autoradiography. For dissipation of membrane potential, isolated mitochondria were mixed with 20 µM CCCP and incubated for 5 min at 4°C before the import reaction.
Pull-down of in vitro translated proteins
Reactions containing proteins translated in either yeast extract or rabbit reticulocyte lysate were loaded onto magnetic anti-HA beads (ThermoFisher) that were previously equilibrated with KHM buffer (110 mM potassium acetate, 20 mM Hepes, and 2 mM MgCl2, pH 7.4). Binding to the beads was performed for 2 h at 4°C on an overhead shaker (12 rpm). The beads were then washed 4 times with KHM buffer. In the third washing step, the beads were transferred to a new reaction tube. Bound proteins were eluted by addition of 2× Lämmli buffer containing 0.05% H2O2 and 5 min incubation at 95°C. The beads were collected and the supernatant was transferred to a new tube, supplemented with 5% β-mercaptoethanol and incubated for another 5 min at 95°C before analysis by SDS-PAGE followed by Western blotting or mass spectrometry.
Mass spectrometry
Eluted proteins were purified by SDS-PAGE and Coomassie-stained gel pieces were digested in gel with trypsin as described previously (Borchert et al., 2010). After desalting using C18 Stage tips, extracted peptides were separated on an EasyLC nano-HPLC (ThermoFisher) coupled to an LTQ Orbitrap Elite (ThermoFisher) as described elsewhere (Franz-Wachtel et al., 2012) with slight modifications: the peptide mixtures were separated using a 127-min segmented gradient from to 5–33% and 50–90% of HPLC solvent B (80% acetonitrile in 0.1% formic acid) in HPLC solvent A (0.1% formic acid) at a flow rate of 200 nL/min. The 15 most intense precursor ions were sequentially fragmented in each scan cycle using collision-induced dissociation (CID). The target values for MS/MS fragmentation were 5,000 charges and 106 charges for the MS scan. Acquired MS spectra were processed with MaxQuant software with integrated Andromeda search engine (Cox et al., 2011). Database search was performed against a target decoy Saccharomyces cerevisiae database obtained from Uniprot, the sequences of the constructs DHFR-3HA and Porin-3HA, and 285 commonly observed contaminants. Initial maximum allowed mass tolerance was set to 4.5 ppm (for the survey scan) and 0.5 D for CID fragment ions. A false discovery rate of 1% was applied at the peptide and protein level.
Metabolic labeling
Yeast cells grown in 25 ml SD medium at 30°C to early logarithmic phase (OD600 ≈ 0.3) were supplemented with 2 µg/ml doxycycline and grown for another 4 h. The cells were harvested, washed with water, and resuspended in SD medium lacking methionine (SD-Met). After 1 h shaking at 30°C, the cells were harvested and resuspended in 2.5 ml SD-Met. After 10 min at 30°C, the cultures were supplemented with 10 µM [35S]methionine and incubated further at 30°C with shaking. Samples (540 µl) were collected after various time periods and mixed with sodium azide and methionine (final concentration 10 mM and 0.003%, respectively). The samples were centrifuged (5,000 g, 1 min, 2°C) and the cell pellets were washed with a 10 mM sodium azide solution.
For isolation of crude mitochondria, the cells were resuspended in 350 µl lysis buffer (10 mM Tris, 1 mM EDTA, and 2 mM PMSF, pH 8.0) mixed with 300 mg glass beads (diameter 0.25–0.5 mm) and lysed by five cycles of vortexing for 1 min with 1-min break on ice in between. The glass beads and cellular debris were removed by centrifugation (1,000 g, 3 min, 2°C) and crude mitochondria were isolated from the supernatant by centrifugation (8,000 g, 10 min, 2°C). The mitochondrial pellet was solubilized in 300 µl solubilization buffer (50 mM Tris, 5 mM EDTA, 150 mM NaCl, and 0.5% Triton X-100, pH 8.0) for 30 min at 4°C on an overhead shaker (12 rpm). After a clarifying spin (30,000 g, 15 min, 2°C), the supernatant was loaded onto magnetic anti-HA beads (ThermoFisher), the beads were washed with buffer containing 50 mM Tris, 5 mM EDTA, 500 mM NaCl, 0.5% Triton X-100, 0.003% methionine, and 5% sucrose, pH 7.4, and further treatment was as described above for the pull-down of in vitro synthesized proteins. For the analysis of proteins in the whole cell lysate, the same protocol was used; however, the centrifugation step to pellet mitochondria was omitted. Instead the whole cell lysate was mixed with an equal volume of 2× solubilization buffer and incubated for 30 min at 4°C on an overhead shaker (12 rpm). The further treatment was as described above.
UV-induced cross-linking
Synthesis of the Bpa-containing linear and cyclic β-hairpin peptides was described before (Jores et al., 2016). In vitro photo cross-linking was performed by mixing yeast extract or recombinant GST-fusion proteins with Bpa-containing β-hairpin peptides in import buffer without BSA. The mixture was incubated for 10 min on ice before UV irradiation for 30 min at 4°C using a Blak-Ray B-100 AP UV lamp at a distance of 10 cm from the samples. After the UV-illumination, the samples were mixed with 4× Lämmli buffer, incubated at 95°C for 5 min, and subjected to analysis by SDS-PAGE followed by Western blotting.
In vivo photo cross-linking was performed according to a previously published protocol (Shiota et al., 2013). Yeast cells were transformed with a plasmid coding for hp18(VDAC)-DHFR-3HA with or without the Thr258Bpa mutation and with the plasmid pBpa2-PGK1+3SUP4-tRNACUA that harbors a suppressor tRNA and an aminoacyl-tRNA synthetase that can charge it with Bpa. The cells were grown at 30°C in 500 ml SD-Ura-Trp medium supplemented with 0.6 mM Bpa (Bachem) to logarithmic phase (OD600 ≈ 1) and harvested by centrifugation (3,000 g, 5 min, RT). For photo cross-linking, the cells were resuspended in 6 ml water, divided into four samples of 1.5 ml each and two samples were transferred to a four-section glass Petri dish. The Petri dish was wrapped with aluminum foil on the bottom and sides and was placed on ice. A UV lamp as above was placed immediately on top of the Petri dish, and the cells were subjected to UV-irradiation for 15 min at 4°C. The remaining two samples were kept in the dark. Irradiated and nonirradiated cells were harvested by centrifugation (3,000 g, 5 min, 2°C) and resuspended in 1.2 ml lysis buffer (10 mM Tris, 150 mM NaCl, 2 mM PMSF, 1× EDTA-free cOmplete protease inhibitor [Roche], and 5 mM EDTA, pH 7.5). The cell suspension was distributed into two 1.5-ml test-tubes and crude mitochondria were isolated as described above. Membranes were solubilized with 1% Triton X-100 for 30 min at 4°C on an overhead shaker (12 rpm). After a clarifying spin (30,000 g, 30 min, 2°C), the supernatant was loaded onto magnetic anti-HA beads (ThermoFisher), the beads were washed with a buffer containing 50 mM Tris, 5 mM EDTA, 500 mM NaCl, and 0.5% Triton X-100, pH 7.5, and further treatment was as described above for the pull-down of in vitro synthesized proteins.
Fluorescence anisotropy measurements
Cyclic β-hairpin peptides were synthesized as described (Jores et al., 2016), coupled to TAMRA and used for the determination of binding kinetics and affinity of peptide/chaperone complexes by fluorescence anisotropy. Measurements were performed at 30°C in a Jasco FP-8500 Fluorospectrometer equipped with polarizers. Excitation and emission wavelengths were set to 554 nm and 579 nm, respectively. Samples containing 2 µM rhodamine-labeled peptide were equilibrated for ∼15 min before 5 or 10 µM Ssa1, 30 µM Ydj1, or 30 µM BSA were added. After reaching steady-state, a 100-fold molar excess of the unlabeled cyclic β-hairpin peptide was added to the cuvette and dissociation was recorded. For affinity measurements, 2 µM rhodamine-labeled peptide were supplemented with the indicated concentrations of Ssa1 or Ydj1 and the difference in anisotropy of bound and free peptide were plotted against the (co)chaperone concentration.
Online supplemental material
Fig. S1 shows additional controls for the pull-down experiments described in Fig. 1 and pull-down experiments from yeast extracts lacking certain cochaperones. Fig. S2 shows control in vitro import experiments. Fig. S3 shows that the mitochondrial import receptor Tom70 is involved in the biogenesis of β-barrel proteins. Fig. S4 shows metabolic labeling experiments performed with WT cells or cells depleted for either Ydj1 or Sis1. Table S1 contains a list of yeast strains used in this study. Table S2 contains a list of primers used in this study. Table S3 contains a list of plasmids used in this study. Table S4 contains a list of antibodies used in this study.
Supplementary Material
Acknowledgments
We thank E. Kracker and G. Hack for technical support and Dr. F.U. Hartl for constructs.
This work was supported by the Deutsche Forschungsgemeinschaft (RA 1028/8-1 and RA 1028/10-1 to D. Rapaort and SFB766 TPs Z1 and B11 to B. Macek and D. Rapaort).
The authors declare no competing financial interests.
Author contributions: T. Jores, J. Lawatscheck, V. Beke, and K. Yunoki, conducted experiments; J.C. Fitzgerald isolated mitochondria, M. Franz-Wachtel, and B. Macek performed mass spectrometry analysis; H. Kalbacher synthesized and purified peptides; T. Jores, T. Endo, J. Buchner, and D. Rapaport designed experiments and analyzed data. T. Jores and D. Rapaport wrote the manuscript.
References
- Asai T., Takahashi T., Esaki M., Nishikawa S., Ohtsuka K., Nakai M., and Endo T.. 2004. Reinvestigation of the requirement of cytosolic ATP for mitochondrial protein import. J. Biol. Chem. 279:19464–19470. 10.1074/jbc.M401291200 [DOI] [PubMed] [Google Scholar]
- Bhangoo M.K., Tzankov S., Fan A.C.Y., Dejgaard K., Thomas D.Y., and Young J.C.. 2007. Multiple 40-kDa heat-shock protein chaperones function in Tom70-dependent mitochondrial import. Mol. Biol. Cell. 18:3414–3428. 10.1091/mbc.e07-01-0088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchert N., Dieterich C., Krug K., Schütz W., Jung S., Nordheim A., Sommer R.J., and Macek B.. 2010. Proteogenomics of Pristionchus pacificus reveals distinct proteome structure of nematode models. Genome Res. 20:837–846. 10.1101/gr.103119.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bösl B., Grimminger V., and Walter S.. 2006. The molecular chaperone Hsp104--a molecular machine for protein disaggregation. J. Struct. Biol. 156:139–148. 10.1016/j.jsb.2006.02.004 [DOI] [PubMed] [Google Scholar]
- Caplan A.J., Cyr D.M., and Douglas M.G.. 1992. YDJ1p facilitates polypeptide translocation across different intracellular membranes by a conserved mechanism. Cell. 71:1143–1155. 10.1016/S0092-8674(05)80063-7 [DOI] [PubMed] [Google Scholar]
- Chan N.C., and Lithgow T.. 2008. The peripheral membrane subunits of the SAM complex function codependently in mitochondrial outer membrane biogenesis. Mol. Biol. Cell. 19:126–136. 10.1091/mbc.e07-08-0796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Schultz P.G., and Brock A.. 2007. An improved system for the generation and analysis of mutant proteins containing unnatural amino acids in Saccharomyces cerevisiae. J. Mol. Biol. 371:112–122. 10.1016/j.jmb.2007.05.017 [DOI] [PubMed] [Google Scholar]
- Clerico E.M., Tilitsky J.M., Meng W., and Gierasch L.M.. 2015. How hsp70 molecular machines interact with their substrates to mediate diverse physiological functions. J. Mol. Biol. 427:1575–1588. 10.1016/j.jmb.2015.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., and Mann M.. 2011. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10:1794–1805. 10.1021/pr101065j [DOI] [PubMed] [Google Scholar]
- Craig E.A., and Marszalek J.. 2017. How Do J-Proteins Get Hsp70 to Do So Many Different Things? Trends Biochem. Sci. 42:355–368. 10.1016/j.tibs.2017.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyr D.M., and Ramos C.H.. 2015. Specification of Hsp70 function by Type I and Type II Hsp40. Subcell. Biochem. 78:91–102. 10.1007/978-3-319-11731-7_4 [DOI] [PubMed] [Google Scholar]
- Daum G., Böhni P.C., and Schatz G.. 1982. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J. Biol. Chem. 257:13028–13033. [PubMed] [Google Scholar]
- Deshaies R.J., Koch B.D., Werner-Washburne M., Craig E.A., and Schekman R.. 1988. A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides. Nature. 332:800–805. 10.1038/332800a0 [DOI] [PubMed] [Google Scholar]
- Endo T., Mitsui S., Nakai M., and Roise D.. 1996. Binding of mitochondrial presequences to yeast cytosolic heat shock protein 70 depends on the amphiphilicity of the presequence. J. Biol. Chem. 271:4161–4167. 10.1074/jbc.271.8.4161 [DOI] [PubMed] [Google Scholar]
- Franz-Wachtel M., Eisler S.A., Krug K., Wahl S., Carpy A., Nordheim A., Pfizenmaier K., Hausser A., and Macek B.. 2012. Global detection of protein kinase D-dependent phosphorylation events in nocodazole-treated human cells. Mol. Cell. Proteomics. 11:160–170. 10.1074/mcp.M111.016014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnanasundram S.V., and Koš M.. 2015. Fast protein-depletion system utilizing tetracycline repressible promoter and N-end rule in yeast. Mol. Biol. Cell. 26:762–768. 10.1091/mbc.e14-07-1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y., Kakihara Y., Krogan N., Greenblatt J., Emili A., Zhang Z., and Houry W.A.. 2009. An atlas of chaperone-protein interactions in Saccharomyces cerevisiae: implications to protein folding pathways in the cell. Mol. Syst. Biol. 5:275 10.1038/msb.2009.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habib S.J., Waizenegger T., Lech M., Neupert W., and Rapaport D.. 2005. Assembly of the TOB complex of mitochondria. J. Biol. Chem. 280:6434–6440. 10.1074/jbc.M411510200 [DOI] [PubMed] [Google Scholar]
- Hecht M.H. 1994. De novo design of beta-sheet proteins. Proc. Natl. Acad. Sci. USA. 91:8729–8730. 10.1073/pnas.91.19.8729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höhr A.I.C., Straub S.P., Warscheid B., Becker T., and Wiedemann N.. 2015. Assembly of β-barrel proteins in the mitochondrial outer membrane. Biochim. Biophys. Acta. 1853:74–88. 10.1016/j.bbamcr.2014.10.006 [DOI] [PubMed] [Google Scholar]
- Hoppins S.C., and Nargang F.E.. 2004. The Tim8-Tim13 complex of Neurospora crassa functions in the assembly of proteins into both mitochondrial membranes. J. Biol. Chem. 279:12396–12405. 10.1074/jbc.M313037200 [DOI] [PubMed] [Google Scholar]
- Hoseini H., Pandey S., Jores T., Schmitt A., Franz-Wachtel M., Macek B., Buchner J., Dimmer K.S., and Rapaport D.. 2016. The cytosolic cochaperone Sti1 is relevant for mitochondrial biogenesis and morphology. FEBS J. 283:3338–3352. 10.1111/febs.13813 [DOI] [PubMed] [Google Scholar]
- Hwang S.T., and Schatz G.. 1989. Translocation of proteins across the mitochondrial inner membrane, but not into the outer membrane, requires nucleoside triphosphates in the matrix. Proc. Natl. Acad. Sci. USA. 86:8432–8436. 10.1073/pnas.86.21.8432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobus A.P., and Gross J.. 2015. Optimal cloning of PCR fragments by homologous recombination in Escherichia coli. PLoS One. 10:e0119221 10.1371/journal.pone.0119221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jores T., Klinger A., Groß L.E., Kawano S., Flinner N., Duchardt-Ferner E., Wöhnert J., Kalbacher H., Endo T., Schleiff E., and Rapaport D.. 2016. Characterization of the targeting signal in mitochondrial β-barrel proteins. Nat. Commun. 7:12036 10.1038/ncomms12036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampinga H.H., and Craig E.A.. 2010. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11:579–592. 10.1038/nrm2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil P., Weinzierl A., Kiebler M., Dietmeier K., Söllner T., and Pfanner N.. 1993. Biogenesis of the mitochondrial receptor complex. Two receptors are required for binding of MOM38 to the outer membrane surface. J. Biol. Chem. 268:19177–19180. [PubMed] [Google Scholar]
- Kim Y.E., Hipp M.S., Bracher A., Hayer-Hartl M., and Hartl F.U.. 2013. Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 82:323–355. 10.1146/annurev-biochem-060208-092442 [DOI] [PubMed] [Google Scholar]
- Krimmer T., Rapaport D., Ryan M.T., Meisinger C., Kassenbrock C.K., Blachly-Dyson E., Forte M., Douglas M.G., Neupert W., Nargang F.E., and Pfanner N.. 2001. Biogenesis of porin of the outer mitochondrial membrane involves an import pathway via receptors and the general import pore of the TOM complex. J. Cell Biol. 152:289–300. 10.1083/jcb.152.2.289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., and Sha B.. 2004. Peptide substrate identification for yeast Hsp40 Ydj1 by screening the phage display library. Biol. Proced. Online. 6:204–208. 10.1251/bpo90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrone L., Bus C., Schöndorf D., Fitzgerald J.C., Kübler M., Schmid B., Reinhardt P., Reinhardt L., Deleidi M., Levin T., et al. 2018. Generation of iPSCs carrying a common LRRK2 risk allele for in vitro modeling of idiopathic Parkinson’s disease. PLoS One. 13:e0192497 10.1371/journal.pone.0192497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer M.P. 2013. Hsp70 chaperone dynamics and molecular mechanism. Trends Biochem. Sci. 38:507–514. 10.1016/j.tibs.2013.08.001 [DOI] [PubMed] [Google Scholar]
- Model K., Meisinger C., Prinz T., Wiedemann N., Truscott K.N., Pfanner N., and Ryan M.T.. 2001. Multistep assembly of the protein import channel of the mitochondrial outer membrane. Nat. Struct. Biol. 8:361–370. 10.1038/86253 [DOI] [PubMed] [Google Scholar]
- Morano K.A. 2007. New tricks for an old dog: the evolving world of Hsp70. Ann. N. Y. Acad. Sci. 1113:1–14. 10.1196/annals.1391.018 [DOI] [PubMed] [Google Scholar]
- Nillegoda N.B., and Bukau B.. 2015. Metazoan Hsp70-based protein disaggregases: emergence and mechanisms. Front. Mol. Biosci. 2:57 10.3389/fmolb.2015.00057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papić D., Elbaz-Alon Y., Koerdt S.N., Leopold K., Worm D., Jung M., Schuldiner M., and Rapaport D.. 2013. The role of Djp1 in import of the mitochondrial protein Mim1 demonstrates specificity between a cochaperone and its substrate protein. Mol. Cell. Biol. 33:4083–4094. 10.1128/MCB.00227-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschen S.A., Waizenegger T., Stan T., Preuss M., Cyrklaff M., Hell K., Rapaport D., and Neupert W.. 2003. Evolutionary conservation of biogenesis of β-barrel membrane proteins. Nature. 426:862–866. 10.1038/nature02208 [DOI] [PubMed] [Google Scholar]
- Pfanner N., Pfaller R., Kleene R., Ito M., Tropschug M., and Neupert W.. 1988. Role of ATP in mitochondrial protein import. Conformational alteration of a precursor protein can substitute for ATP requirement. J. Biol. Chem. 263:4049–4051. [PubMed] [Google Scholar]
- Pfanner N., Wiedemann N., Meisinger C., and Lithgow T.. 2004. Assembling the mitochondrial outer membrane. Nat. Struct. Mol. Biol. 11:1044–1048. 10.1038/nsmb852 [DOI] [PubMed] [Google Scholar]
- Pfitzner A.-K., Steblau N., Ulrich T., Oberhettinger P., Autenrieth I.B., Schütz M., and Rapaport D.. 2016. Mitochondrial-bacterial hybrids of BamA/Tob55 suggest variable requirements for the membrane integration of β-barrel proteins. Sci. Rep. 6:39053 10.1038/srep39053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierpaoli E.V., Sandmeier E., Baici A., Schönfeld H.J., Gisler S., and Christen P.. 1997. The power stroke of the DnaK/DnaJ/GrpE molecular chaperone system. J. Mol. Biol. 269:757–768. 10.1006/jmbi.1997.1072 [DOI] [PubMed] [Google Scholar]
- Plath K., Mothes W., Wilkinson B.M., Stirling C.J., and Rapoport T.A.. 1998. Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell. 94:795–807. 10.1016/S0092-8674(00)81738-9 [DOI] [PubMed] [Google Scholar]
- Rapaport D., and Neupert W.. 1999. Biogenesis of Tom40, core component of the TOM complex of mitochondria. J. Cell Biol. 146:321–331. 10.1083/jcb.146.2.321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt P., Glatza M., Hemmer K., Tsytsyura Y., Thiel C.S., Höing S., Moritz S., Parga J.A., Wagner L., Bruder J.M., et al. 2013. Derivation and expansion using only small molecules of human neural progenitors for neurodegenerative disease modeling. PLoS One. 8:e59252 10.1371/journal.pone.0059252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci L., and Williams K.P.. 2008. Development of fluorescence polarization assays for the molecular chaperone Hsp70 family members: Hsp72 and DnaK. Curr. Chem. Genomics. 2:90–95. 10.2174/1875397300802010090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röhl A., Rohrberg J., and Buchner J.. 2013. The chaperone Hsp90: changing partners for demanding clients. Trends Biochem. Sci. 38:253–262. 10.1016/j.tibs.2013.02.003 [DOI] [PubMed] [Google Scholar]
- Schägger H., Cramer W.A., and von Jagow G.. 1994. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217:220–230. 10.1006/abio.1994.1112 [DOI] [PubMed] [Google Scholar]
- Scheufler C., Brinker A., Bourenkov G., Pegoraro S., Moroder L., Bartunik H., Hartl F.U., and Moarefi I.. 2000. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 101:199–210. 10.1016/S0092-8674(00)80830-2 [DOI] [PubMed] [Google Scholar]
- Schleiff E., Silvius J.R., and Shore G.C.. 1999. Direct membrane insertion of voltage-dependent anion-selective channel protein catalyzed by mitochondrial Tom20. J. Cell Biol. 145:973–978. 10.1083/jcb.145.5.973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid A.B., Lagleder S., Gräwert M.A., Röhl A., Hagn F., Wandinger S.K., Cox M.B., Demmer O., Richter K., Groll M., et al. 2012. The architecture of functional modules in the Hsp90 co-chaperone Sti1/Hop. EMBO J. 31:1506–1517. 10.1038/emboj.2011.472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid D., Baici A., Gehring H., and Christen P.. 1994. Kinetics of molecular chaperone action. Science. 263:971–973. 10.1126/science.8310296 [DOI] [PubMed] [Google Scholar]
- Schneider M., Rosam M., Glaser M., Patronov A., Shah H., Back K.C., Daake M.A., Buchner J., and Antes I.. 2016. BiPPred: Combined sequence- and structure-based prediction of peptide binding to the Hsp70 chaperone BiP. Proteins. 84:1390–1407. 10.1002/prot.25084 [DOI] [PubMed] [Google Scholar]
- Schopf F.H., Biebl M.M., and Buchner J.. 2017. The HSP90 chaperone machinery. Nat. Rev. Mol. Cell Biol. 18:345–360. 10.1038/nrm.2017.20 [DOI] [PubMed] [Google Scholar]
- Shiota T., Nishikawa S., and Endo T.. 2013. Analyses of protein-protein interactions by in vivo photocrosslinking in budding yeast. Methods Mol. Biol. 1033:207–217. 10.1007/978-1-62703-487-6_14 [DOI] [PubMed] [Google Scholar]
- Söllner T., Griffiths G., Pfaller R., Pfanner N., and Neupert W.. 1989. MOM19, an import receptor for mitochondrial precursor proteins. Cell. 59:1061–1070. 10.1016/0092-8674(89)90762-9 [DOI] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., and Yamanaka S.. 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 131:861–872. 10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]
- Terasawa K., Minami M., and Minami Y.. 2005. Constantly updated knowledge of Hsp90. J. Biochem. 137:443–447. 10.1093/jb/mvi056 [DOI] [PubMed] [Google Scholar]
- Ulrich T., and Rapaport D.. 2015. Biogenesis of β-barrel proteins in evolutionary context. Int. J. Med. Microbiol. 305:259–264. 10.1016/j.ijmm.2014.12.009 [DOI] [PubMed] [Google Scholar]
- Ungelenk S., Moayed F., Ho C.-T., Grousl T., Scharf A., Mashaghi A., Tans S., Mayer M.P., Mogk A., and Bukau B.. 2016. Small heat shock proteins sequester misfolding proteins in near-native conformation for cellular protection and efficient refolding. Nat. Commun. 7:13673 10.1038/ncomms13673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh P., Bursać D., Law Y.C., Cyr D., and Lithgow T.. 2004. The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep. 5:567–571. 10.1038/sj.embor.7400172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegele H., Haslbeck M., Reinstein J., and Buchner J.. 2003. Sti1 is a novel activator of the Ssa proteins. J. Biol. Chem. 278:25970–25976. 10.1074/jbc.M301548200 [DOI] [PubMed] [Google Scholar]
- Wiedemann N., Kozjak V., Chacinska A., Schönfisch B., Rospert S., Ryan M.T., Pfanner N., and Meisinger C.. 2003. Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature. 424:565–571. 10.1038/nature01753 [DOI] [PubMed] [Google Scholar]
- Wiedemann N., Truscott K.N., Pfannschmidt S., Guiard B., Meisinger C., and Pfanner N.. 2004. Biogenesis of the protein import channel Tom40 of the mitochondrial outer membrane: intermembrane space components are involved in an early stage of the assembly pathway. J. Biol. Chem. 279:18188–18194. 10.1074/jbc.M400050200 [DOI] [PubMed] [Google Scholar]
- Wu C., and Sachs M.S.. 2014. Preparation of a Saccharomyces cerevisiae cell-free extract for in vitro translation. Methods Enzymol. 539:17–28. 10.1016/B978-0-12-420120-0.00002-5 [DOI] [PubMed] [Google Scholar]
- Wu X., Li L., and Jiang H.. 2018. Mitochondrial inner-membrane protease Yme1 degrades outer-membrane proteins Tom22 and Om45. J. Cell Biol. 217:139–149. 10.1083/jcb.201702125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J.L., Bohovych I., Wong E.O.Y., Lambert J.-P., Gingras A.-C., Khalimonchuk O., Cowen L.E., and Leach M.D.. 2017. Ydj1 governs fungal morphogenesis and stress response, and facilitates mitochondrial protein import via Mas1 and Mas2. Microb. Cell. 4:342–361. 10.15698/mic2017.10.594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano K., Yatsukawa Y., Esaki M., Hobbs A.E.A., Jensen R.E., and Endo T.. 2008. Tom20 and Tom22 share the common signal recognition pathway in mitochondrial protein import. J. Biol. Chem. 283:3799–3807. 10.1074/jbc.M708339200 [DOI] [PubMed] [Google Scholar]
- Young J.C., Moarefi I., and Hartl F.U.. 2001. Hsp90. J. Cell Biol. 154:267–273. 10.1083/jcb.200104079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J.C., Hoogenraad N.J., and Hartl F.U.. 2003. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell. 112:41–50. 10.1016/S0092-8674(02)01250-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.