Using an in vitro wound healing assay, De Pascalis et al. show that intermediate filaments (IFs) participate in the dynamics of the acto-myosin network and its association with adhesions in astrocytes during collective migration. Glial IFs control the distribution of forces and the interactions between neighboring cells, ultimately determining the speed and direction of collective migration.
Abstract
Mesenchymal cell migration relies on the coordinated regulation of the actin and microtubule networks that participate in polarized cell protrusion, adhesion, and contraction. During collective migration, most of the traction forces are generated by the acto-myosin network linked to focal adhesions at the front of leader cells, which transmit these pulling forces to the followers. Here, using an in vitro wound healing assay to induce polarization and collective directed migration of primary astrocytes, we show that the intermediate filament (IF) network composed of vimentin, glial fibrillary acidic protein, and nestin contributes to directed collective movement by controlling the distribution of forces in the migrating cell monolayer. Together with the cytoskeletal linker plectin, these IFs control the organization and dynamics of the acto-myosin network, promoting the actin-driven treadmilling of adherens junctions, thereby facilitating the polarization of leader cells. Independently of their effect on adherens junctions, IFs influence the dynamics and localization of focal adhesions and limit their mechanical coupling to the acto-myosin network. We thus conclude that IFs promote collective directed migration in astrocytes by restricting the generation of traction forces to the front of leader cells, preventing aberrant tractions in the followers, and by contributing to the maintenance of lateral cell–cell interactions.
Introduction
During morphogenesis, tissue repair, and cancer, cells frequently migrate in a collective manner as groups, chains, or sheets (Haeger et al., 2015; Mayor and Etienne-Manneville, 2016). Collectively migrating cells move with a similar speed and direction (Etienne-Manneville, 2014). The cytoskeleton, composed of actin microfilaments, microtubules, and intermediate filaments (IFs), plays a key role in single and collective cell migration. The roles of the actin and microtubule network in cell migration have been well characterized (Gardel et al., 2010; Etienne-Manneville, 2013). Similar to actin and microtubules, IFs affect cell migration (Leduc and Etienne-Manneville, 2015). During oncogenesis, changes in IF protein expression, in particular increased vimentin levels, have been associated with cell invasion and tumor spreading (Chung et al., 2013; Leduc and Etienne-Manneville, 2015). However, the exact functions of IFs during migration are still not well understood.
Migration of cell sheets is characterized by specific mechanical features (De Pascalis and Etienne-Manneville, 2017). High tractions generated at the front of leader cells (du Roure et al., 2005; Trepat et al., 2009) are transmitted to the rest of the monolayer (Tambe et al., 2011; Serra-Picamal, 2012). Because of their structural characteristics, IFs are hypothesized to be key players in cell mechanics, helping maintain cell and tissue integrity (Kreplak and Fudge, 2007; Herrmann et al., 2009). Keratin IFs have recently been shown to control traction forces during collective migration of epithelial cells (Sonavane et al., 2017). Moreover, alterations of the vimentin network in endothelial cells perturb acto-myosin contractility and cell mechanical resilience (Osmanagic-Myers et al., 2015), suggesting that nonkeratin IFs may also contribute to the mechanical properties of collectively migrating cells.
In this study, we used primary astrocytes to investigate the role of IFs in collective migration. Astrocytes are major glial cells that mainly express the three IF proteins glial fibrillary acidic protein (GFAP), nestin, and vimentin, which assemble together in cytoplasmic IFs (Leduc and Etienne-Manneville, 2017). Increased levels of these IF proteins have been reported in glioblastomas, which are highly invasive primary glial tumors (Ma et al., 2008; Matsuda et al., 2015; Lin et al., 2016; Lv et al., 2017). Astrocytes migrate in a collective manner during development (Gnanaguru et al., 2013; Liu et al., 2015). In the adult brain, reactive astrocytes, which express higher levels of GFAP, can polarize to eventually migrate in the direction of inflammatory sites (Burda et al., 2016; Pekny et al., 2016). The collective migration of reactive astrocytes can be recapitulated in vitro in a wound healing assay that induces the polarization of wound edge cells and the collective movement of the cell sheet in a timely controlled manner (Etienne-Manneville and Hall, 2001; Etienne-Manneville, 2006). Using this assay, we have investigated the role of vimentin, GFAP, and nestin in collective migration. We show that these three IF proteins participate in the dynamics of the acto-myosin network and its association with focal adhesions (FAs) and adherens junctions (AJs). Glial IFs thus control the distribution of forces in the migrating monolayer and the interactions between neighboring cells, ultimately determining the speed and direction of collective migration.
Results and discussion
IFs control the distribution and strength of traction forces in a migrating monolayer
We used previously described siRNAs (si triple IF) to specifically decrease the expression of each of the three main glial cytoplasmic IF proteins (GFAP, vimentin, and nestin) expressed in primary rat astrocytes (Dupin et al., 2011). A second independent set of siRNAs was also used to reduce the amount of IFs in astrocytes (Fig. S1, A and B). Upon IF protein depletion, cell area and size during migration was increased, as previously reported (Middeldorp and Hol, 2011; Fig. S1, C and D). In wound healing assays, IF-depleted cells (si triple IF) were slower than control cells (Fig. 1, A and C; Fig. S1 E; and Video 1), confirming that IFs are involved in astrocyte migration as previously observed in vitro and in vivo (Lepekhin et al., 2001; Dupin et al., 2011; Sakamoto et al., 2013). Reduction of IFs also significantly decreased the persistence and directionality of migration of primary astrocytes (Fig. 1, B and C; and Fig S1 E) and centrosome reorientation, an indicator of the front-rear polarization of the leader cells (Fig. S1 F). Depletion of either of the three IF proteins present in astrocytes had similar although less pronounced effects on migration than the concomitant depletion of the three proteins, suggesting that they all participate in the functions of the glial IF network (Fig. 1 C).
Figure 1.
IFs control collective astrocyte migration by regulating traction forces. (A) Phase-contrast images of astrocyte (shown in Video 1) wound healing after 24 h of migration. Black lines represent the initial position (0 h), and white lines show the final position (24 h), of the leading edge. (B) Simplified method for calculating persistence and directionality of migration of a cell with nuclear tracking. For more detailed formulas, see Materials and methods. (C) Graphs of cell velocity, directionality, and persistence measured by manual nuclear tracking of leader cells after 24 h of migration. (D) Simplified protocol for plating cells into PDMS rectangular stamps onto hydrogels. The black dotted arrows show the main directions of migration. The central image shows a monolayer of cells migrating on a hydrogel embedded with fluorescent beads (green dots) with representative tractions (T, blue arrows). The last image represents schematically the components of tractions (Tx and Ty) analyzed with TFM. (E) Phase-contrast images of astrocytes migrating on a 9-kPa collagen-coated polyacrylamide hydrogel at different time points. The white dotted line represents the leading edge, and the arrows show the direction of migration. See also Video 2. (F) Tractions in the x direction (Tx) at indicated time points. See also Video 3. (G) Representative kymographs of total tractions (|T|). (H) Graphs of tractions in the x direction (Tx), mean values (left) and values at the edge of the monolayer (middle) and at the center (right). (I) Graph showing the ratio of the tractions at the edge over tractions at the center, plotted as a function of time of migration. Data are from n = 3 independent experiments. The sample size for each repeat is: si ctl 50, 50, 50; si triple IF 50, 50, 50; si GFAP 50, 50, 50; si vimentin 50, 50, 49; si nestin 50, 50, 50 cells for C; two, one, and four videos of si ctl and two, three, and three videos of si triple IF for E–I. Graphs show mean ± SEM. *, P < 0.05; **, P < 0.01; and ***, P < 0.001. Bars, 50 µm.
To analyze the effect of IF depletion on traction forces in a migrating astrocyte monolayer, we performed traction force microscopy (TFM; Trepat et al., 2009; Fig. 1, D–F; and Video 2). Cells were plated on collagen-coated polyacrylamide substrates inside a polydimethylsiloxane (PDMS) stamp of rectangular shape. Removal of the PDMS stamp allows the cells to migrate outward (Fig. 1, D and E; and Video 2). Analysis of traction forces in control cells showed that most traction forces were exerted at the edges of the monolayer, whereas follower cells produced weaker forces (Fig. 1, F–I; and Video 3). Although in these conditions cell migration speed was not significantly reduced by IF depletion, possibly indicating that IF effects on velocity are sensitive to substrate rigidity, si triple IF cells showed stronger tractions at all time points (Fig. 1, F–I). Moreover, in IF-depleted astrocytes, tractions were generated not only by leaders but also by followers. In fact, most of the increase in traction forces corresponded to increased forces in the follower cells, resulting in the generation of similar traction forces at the edge and in the center of the monolayer (Fig. 1, F–I). These results show that IFs restrict the generation of traction forces to the leader cells and prevent the accumulation of traction forces within the monolayer.
IFs control the organization and dynamics of the acto-myosin network
In single cells, IFs have been shown to influence the organization of the actin cytoskeleton as well as acto-myosin contractility (Gregor et al., 2014; Jiu et al., 2015, 2017; Costigliola et al., 2017), leading us to investigate how IFs influence the acto-myosin network in migrating monolayers. During collective migration, astrocytes showed two main types of actin fibers (Figs. 2 A and S2 A). In addition to the longitudinal stress fibers oriented in the direction of migration and associated with FAs at the cell front, collectively migrating astrocytes display interjunctional transverse arcs (ITAs), which are anchored at AJs on the lateral sides of adjacent cells (Peglion et al., 2014). IF-depleted leader cells often completely lacked visible ITAs and only showed longitudinal stress fibers (Fig. 2, A and B; and Fig. S2, A and B). Silencing any of the three IF proteins similarly favored the generation of longitudinal stress fibers at the expense of ITAs (Fig. S2 C). The general orientation of the actin cables was also affected in follower cells just behind the leaders (Figs. 2 C and S2 D). Although control cells showed actin fibers both perpendicular and parallel to the wound, si triple IF cells mainly displayed stress fibers oriented perpendicular to the wound in both leaders and followers (Figs. 2 C and S2 D).
Figure 2.
IFs control the organization and dynamics of the actin network. (A) Immunofluorescence images of actin fibers in migrating astrocytes stained for nuclei (cyan) and actin (phalloidin, gray). (B) Graph showing the percentage of leader cells that present ITAs in si ctl and si triple IF cells. (C) Rose plot showing the frequency of angle distribution of actin fibers analyzed in follower cells (Fig. S3 D). Fibers with a 0° angle are perpendicular to the wound, and fibers with an angle close to 90° are parallel to the wound. (D) Schematics showing the position of the kymographs acquired in E–G. (E) Frames of migrating si ctl and si triple IF astrocytes transfected with LifeAct-mCherry. The white dotted line represents the wound, and the pink dotted lines represent the positions in which the kymographs were calculated. Kymographs (fire LUT) show the retrograde flow of actin on cell–cell junctions over a time period of 4 h. The graph shows the mean retrograde flow speed of actin cables measured at the level of the cell–cell junctions. (F) Frames from Video 4 of migrating astrocytes transfected with GFP-MRLC. The thick white dotted line represents the outline of nearby cells. The kymographs were obtained along the pink dotted lines. f and b on the side of the kymograph indicate the front and the rear of the line. Kymographs (fire LUT) show the retrograde flow of myosin at the front and at the rear of myosin longitudinal fibers over a time period of 15 min. The graph shows the mean speed of the myosin retrograde flow at the cell front and at the rear of the longitudinal fiber calculated from the kymographs. The white dotted lines indicate the position of the wound. (G) Immunofluorescence images from Video 5 showing GFP-N-cadherin–expressing astrocytes. Insets show the corresponding phase-contrast image. N, nucleus. The kymographs were obtained along the pink dotted lines. f and b on the side of the kymographs indicate the front and the rear of the line. Kymographs (fire LUT) show the retrograde flow of N-cadherin over a time period of 2 h. The graph shows the mean retrograde flow speed of the N-cadherin flow in si ctl and si triple IF migrating astrocytes. (H) Staining for N-cadherin (gray) and nuclei (cyan) in migrating astrocytes nucleofected with the indicated siRNAs. Histogram shows the mean percentage ± SEM of continuous junctions between adjacent leader cells. White dotted line indicates the position of the wound. Data are from n = 3 independent experiments. The sample size for each repeat: si ctl 64, 67, 92 and si triple IF 44, 96, 122 for B; si ctl 11, 10, eight stacks and si triple IF 12, 10, eight stacks for C; si ctl 30, 40, 32 and si triple IF 18, 33, 30 for E; si triple IF f 22, 27, 18 and si triple IF b 22, 24, 20 for F; si ctl 14, 38, 28 and si triple IF 38, 49, 14 for G; si ctl 104, 120, 178, si GFAP 134, 137, 125, si vimentin 113, 180, 133, si nestin 132, 190, 123, and si triple IF 122, 225, 112 for H. ***, P < 0.00. Bars: (main images) 10 µm; (kymographs and insets of H) 5 µm.
The dynamics of the acto-myosin network is one of the main factors responsible for the generation of traction forces exerted on the substrate. We used LifeAct-mCherry–expressing astrocytes to analyze the dynamics of the actin network in leader cells. In wound edge cells, actin ITAs undergo a continuous retrograde flow coupled to the retrograde flow of associated AJs (Peglion et al., 2014). The retrograde flow of ITAs was visible in cells transfected with control siRNA (si ctl; Fig. 2, D and E). In the few si triple IF cells showing ITAs, the retrograde flow was much slower (Fig. 2 E). To analyze the retrograde flow in the longitudinal stress fibers, we used cells expressing GFP-labeled myosin regulatory light chain (GFP-MRLC; Fig. 2, D and F). Kymograph analysis at the front and rear ends of these longitudinal fibers showed that retrograde flow of myosin was slower in IF-depleted cells than in control cells (Fig. 2, D and F; and Video 4). Moreover, although myosin flow was higher at the front than at the rear of the fibers in control cells, this difference was abolished in si triple IF cells (Fig. 2 F). Altogether, these results show that IFs play a key role in the organization and dynamics of the acto-myosin network during collective cell migration.
IFs are required for actin-driven treadmilling of AJs and maintenance of cell–cell interactions
The coordination of the actin-driven retrograde flow between adjacent leader cells drives the treadmilling of cadherin-based Ajs, which participates in the maintenance of AJs and thereby controls centrosome orientation and direction of migration in leader cells (Dupin et al., 2011; Camand et al., 2012; Peglion et al., 2014). Using GFP-N-cadherin–expressing cells, we observed that depletion of IFs strongly reduced AJ retrograde flow, which was sometimes absent or in the opposite direction in some cells (Fig. 2, D and G; and Video 5). As previously described (Peglion et al., 2014), the reduction of AJ retrograde flow by si triple IF or single siRNAs was associated with the altered morphology of AJs, which appeared more discontinuous than in control cells (Fig. 2 H and Fig. S2, E and F). The alteration of the lateral AJs was confirmed by the increase of nonadherent regions between neighboring wound-edge cells (Fig. S2 F). Altogether, these results show that IFs facilitate the actin-driven dynamics of AJs between adjacent leader cells. The reduction of AJ retrograde flow in IF-depleted cells along with the strong decrease of ITA cables probably contributes to the decrease of cell polarity and the loss of coordination between leader cells.
IFs and plectin interact with FAs and control their dynamics and distribution
The impact of IFs on longitudinal stress fibers anchored at FAs (Fig. S2 A) led us to investigate the relationship between IFs and FAs in migrating astrocytes expressing GFP-vimentin and paxillin-Orange (or RFP-vimentin and GFP-paxillin). Live superresolution imaging (3D-structured illumination microscopy [SIM]) showed that in astrocytes, as in other cell types (Gonzales et al., 2001; Burgstaller et al., 2010; Mendez et al., 2010; Lynch et al., 2013), IFs come in very close proximity to FAs (Fig. 3 A). It also illustrated the highly dynamic behavior of IFs near FAs (Videos 6 and 7).
Figure 3.
IFs and plectin regulate FA localization and dynamics. (A) Fluorescence SIM-3D images of a migrating astrocytes transfected with GFP-vimentin and paxillin-Orange shown in Video 6 (see also Video 7). The orthogonal projections show that vimentin and paxillin are found in the same focal plane. (B) Immunofluorescence images of migrating astrocytes stained for nuclei (cyan) and paxillin (green). The white dotted lines indicate the position of the wound. Insets show enlarged images of FAs at the leading edge and in a central region of the cell. (C) The top left graph shows the mean number of FAs per cell, and the bottom left graph, the mean area of FAs. Adhesions were projected on the nucleus-tip axis (see schematics). The central top graph shows the normalized distance to the nucleus center of each FA. The central bottom graph shows the distribution of adhesions along the nucleus-tip axis. (D) Lifetime of GFP-paxillin–positive adhesions in migrating leader astrocytes (top). Duration time of assembly and disassembly of these adhesions (bottom). (E) Lifetime of GFP-paxillin positive adhesions in migrating astrocytes (left) of the second and third rows. Duration time of assembly and disassembly of these adhesions (right). (F) Immunofluorescence images of astrocytes in the migrating monolayer stained for nuclei (cyan) and paxillin (green). Data are from n = 3 independent experiments. The sample size for each repeat: si ctl 49, 50, 50, si triple IF 50, 50, 50, and si plectin 50, 50, 55 for C; si ctl 26, 40, 40 and si triple IF 26, 40, 40 for D; si ctl 16, 16, 35 and si triple IF 8, 36, 37 for E. n.s., P > 0.05; *, P < 0.05; **, P < 0.01; and ***, P < 0.001. Bars: (orthogonal projections of A) 1 µm; (A and insets of B) 5 µm; (B top and F) 10 µm.
We thus asked whether IFs could affect FAs, as previously reported in vimentin-expressing cells (Burgstaller et al., 2010; Gregor et al., 2014). In a migrating monolayer of control astrocytes, FAs are strongly concentrated at the leading edge of the leader cells (Fig. 3, B and C), whereas only a few small FAs are visible further back in the protrusion or at the rear of leader cells and in followers (Fig. 3, B–D). Depletion of IF proteins increased the number and size of FAs in leader cells (Fig. 3, B and C; and Fig. S3 A). In this condition, FAs appeared more dispersed between the front edge and the nucleus (Fig. 3, B [insets] and C). Depletion of any single IF protein had a similar effect (Fig. S3 B). Using GFP-paxillin–expressing astrocytes, we observed that in leader cells as well as in follower cells of the second or third rows, the lifetime of FAs was longer in si triple IF cells than in control cells (Fig. 3, D and E; and Video 8). This reduced FA turnover resulted from both a slower assembly and a slower disassembly rate (Fig. 3, D and E). Further away from the wound, the turnover of FAs was too slow to be analyzed; however, we noticed that IF depletion also led to a strong increase of FAs (Fig. 3 F), which is in agreement with the higher tractions seen in the center of the monolayer of si triple IF cells (Fig. 1, F–I). Because microtubules, which are tightly associated with IFs, can affect the dynamics of FAs (Ezratty et al., 2005; Akhmanova et al., 2009; Gan et al., 2016), we analyzed the impact of the si triple IF on microtubule organization and dynamics. In si triple IF cells, the microtubule network was intact and globally oriented along the axis of migration as in control cells (Fig. S3 C). We analyzed microtubule dynamics by tracking the plus end protein end-binding protein 3 (EB3). We observed that IF depletion slightly but significantly modified the speed and directionality of EB3 comets (Fig. S3 D and Video 9), indicating that IFs impact microtubule dynamic instability. This also confirms that loss of IFs has a slight effect on microtubule orientation, as previously reported (Gan et al., 2016). Thus, we conclude that the effect of IF depletion on FA dynamics does not result from major changes in the microtubule network but may involve alterations of microtubule dynamics and the control of microtubule-associated proteins (Chang et al., 2008; Jiu et al., 2017).
In migrating astrocytes, the cytoskeletal cross-linker plectin colocalizes with IFs and paxillin at the distal end of FAs (Fig. 4 A). Although we could not distinguish between the different plectin isoforms, plectin 1f is the most likely candidate, as it has been shown to anchor vimentin to FAs and control FA turnover in fibroblasts (Burgstaller et al., 2010). IF interaction with plectin and FAs was confirmed by coimmunoprecipitations. Vimentin and GFAP also coimmunoprecipitated with plectin, talin, and vinculin but were not found in control immunoprecipitations performed with irrelevant IgGs (Fig. 4 B). Comparison of immunoprecipitations performed on nonmigrating and migrating cells (4 h after wounding) showed that IF association with plectin and FA proteins strongly increased during wound-induced migration (Fig. 4 B).
Figure 4.
Plectin knockdown phenocopies IF depletion. (A) Immunofluorescence images of migrating astrocytes stained for nuclei (cyan), paxillin (green), and plectin (magenta). (B) Western blot analysis using indicated antibodies of total astrocyte lysates (left) and proteins immunoprecipitated with antibodies against plectin, talin, or vinculin and control antibodies (IgG Ms) before and 4 h after wounding. Input lysate corresponds to 2% of the total lysate used for the IP, with a higher exposition shown for plectin and talin bands. (C) Immunofluorescence images of migrating astrocytes stained for nuclei (cyan) and actin (phalloidin, gray) compared with si ctl (Fig. 2 A). The white dotted line represents the position of the wound. The quantification shows the percentage of cells that present ITAs. (D) Immunofluorescence images of migrating astrocytes stained for the AJ marker α-E-catenin (gray) and nuclei (cyan). The white dotted line represents the position of the wound. (E) Quantification of nuclear tracking of migrating astrocytes after 24 h of migration. The graphs show cell velocity, directionality, and persistence of migration of astrocytes nucleofected with si ctl or si plectin. (F) Immunofluorescence images of centrosome orientation in plectin-depleted astrocytes stained for nuclei (cyan) and centrin (red) compared with si ctl (Fig. S1 F). White arrowheads were scored as polarized centrosomes, and yellow ones were scored as nonpolarized centrosomes. The graphs show the percentage of cells with the centrosome located in the wound-facing quadrant in front of the nucleus. Data are from n = 3 independent experiments. The sample size for each repeat: si ctl 262, 247, 150 and si plectin 384, 245, 155 for C; si ctl 40, 50, 39 and si plectin 50, 50, 49 for E; si ctl 92, 222, 83 and si plectin 101, 210, 123 for F. ***, P < 0.001. Histograms show mean ± SEM. Bar, 10 µm.
We then investigated the role of plectin in astrocyte migration, using siRNA to reduce plectin expression in primary astrocytes (Fig. S3, E and F). Plectin depletion strongly affected the distribution of FAs, which are not confined only to the cell tip (Fig. 3, B and C). It also affected actin organization (presence of stress fibers but not ITAs; Fig. 4 C) and AJs (loss of junctions; Fig. 4 D), mimicking the effects observed after IF protein depletion and confirming observations made on plectin-depleted endothelial cells (Osmanagic-Myers et al., 2015). Plectin depletion reduced the speed, directionality, and persistence of migration (Fig. 4 E), as well as centrosome orientation (Fig. 4 F), further supporting the idea that plectin and IFs act together to control AJ and FA dynamics, actin organization, and collective cell migration (Leduc and Etienne-Manneville, 2015; Leube et al., 2015; Seltmann et al., 2015; Wiche et al., 2015).
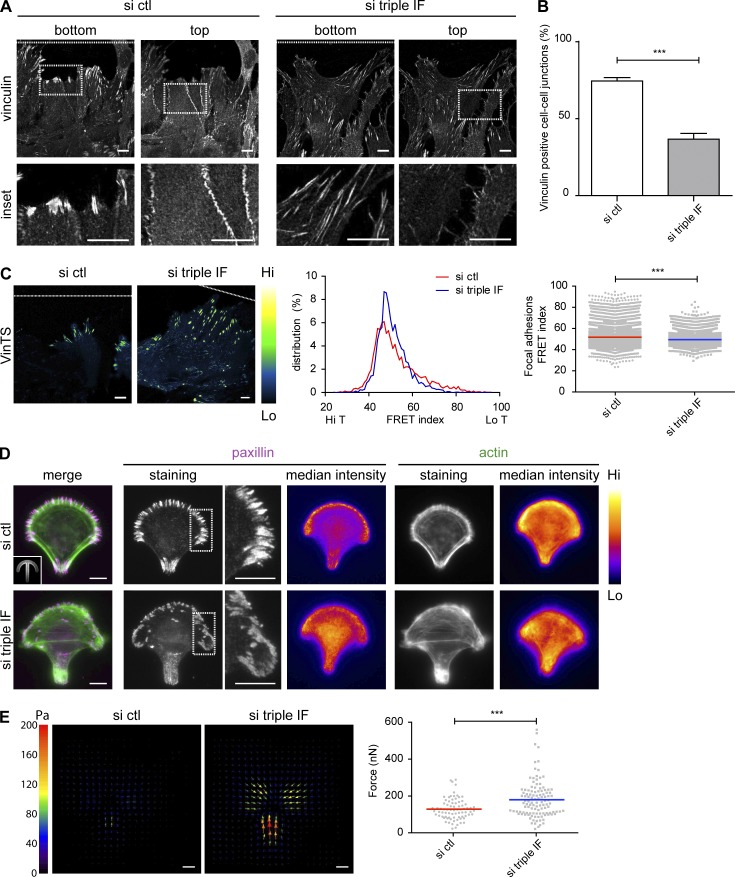
IFs control vinculin-mediated traction forces at FAs
Because IFs strongly influence actin organization and the generation of traction forces in the migrating monolayer, we asked whether IF depletion may affect the localization of vinculin. Vinculin is a key mechanosensor protein controlling the interaction of actin with both FAs and AJs (Gregor et al., 2014; De Pascalis and Etienne-Manneville, 2017). In most control cells, vinculin was found at both FAs and AJs (Fig. 5, A and B). In contrast, in a large majority of IF-depleted cells, vinculin was found in FAs but was frequently absent from AJs (Fig. 5, A and B). Vinculin exists in a closed conformation that opens upon stretching when actin fibers pull on the carboxy-terminal domain of talin-bound vinculin (Bakolitsa et al., 2004). We took advantage of the vinculin fluorescence resonance energy transfer (FRET) tension sensor (Grashoff et al., 2010) to quantify the tension exerted on vinculin during cell migration. The FRET index distribution in FAs was shifted to statistically lower values in si triple IF leader cells than in control leader cells (Fig. 5 C), suggesting that vinculin-mediated tension was increased in IF-depleted cells.
Figure 5.
IFs limit the generation of traction forces at FAs. (A) Immunofluorescence confocal images in two distinct focus planes of astrocytes stained for vinculin. The white dotted line indicates the position of the wound. (B) Graph showing the percentage of junctions showing positive vinculin staining. (C) Fluorescence images of migrating astrocytes expressing VinTS (Green Fire Blue LUT). Representative frequency distribution of frequencies of FRET index (central graph) and quantification of the mean FRET index (inversely related to vinculin tension) of vinculin in FAs (right graph). Each dot represents a FA. The white dotted line indicates the position of the wound. (D) Immunofluorescence and intensity projection (fire LUT) of paxillin (magenta) and actin (green) on micropatterned cells. Inset shows the crossbow-shaped micropattern. Projections are calculated on one experiment representative of three. (E) Left: Tractions fields of astrocytes plated on collagen-coated crossbows. Arrows indicate the orientation, color, and length of the local magnitude of the force in Pa. Right: Quantification of traction forces in single micropatterned cells. Data are from n = 3 independent experiments. Histograms show mean ± SEM. The sample size for each repeat: si ctl 137, 247, 150 and si triple IF 222, 200, 151 for B; si ctl 2,537, 3,312, 1,473 adhesions and si triple IF 1,447, 2,548, 2,081 adhesions for C; si ctl 32, 21, 25 and si triple IF 30, 46, 38 for E. ***, P < 0.001. Bar, 10 µm.
Because AJs have been shown to locally inhibit FA formation (Camand et al., 2012), the effect of IFs on FAs may result from the alteration of AJs, leading to a redistribution of FAs and actin. To test this hypothesis, we plated single cells on adhesive micropatterns, which allowed us to study the role of IFs on FAs and traction forces independently of cell migration and cell–cell interactions. On crossbow-shaped micropatterns, FAs reproducibly concentrated on the outer edge of the arc and on the rear side of the bar forming the pattern, whereas the actin cable systematically formed on the two nonadhesive edges of the cell (Fig. 5 D). IF depletion led to globally wider distribution of FAs, which appeared on the inner region of the adhesive stripes, whereas actin cables formed in more random direction across the cell body (Fig. 5 D). We then used TFM to analyze the orientation and strength of traction forces of single cells plated on crossbow-shaped micropatterns. IF-depleted cells exerted significantly stronger traction forces than control cells (Fig. 5 E). These results show that IFs affect FAs, actin organization, and traction forces independently of their effect on AJs and cell migration.
In conclusion, our results show that glial IFs increase migration speed, direction, and persistence during astrocyte collective movement. Keratin depletion has an opposite effect on keratinocyte migration (Osmanagic-Myers et al., 2006), strongly suggesting that IF functions on cell migration depend on their composition. Although the silencing of IF protein is only partial, it strongly affects collective migration, showing that limited perturbations of the IF network as observed in IF-related diseases or oncogenesis are sufficient to alter cell functions. We show that glial IFs together with plectin control the distribution of FAs and the organization of the actin cytoskeleton in the migrating cell sheet as well as on the single cell. The glial IF network controls the distribution of traction forces generated in the migrating cell sheet, preventing the accumulation of such forces within the monolayer. It is thus likely that IF-depleted followers are less pulled by the leader cells, and instead essentially migrate in response to the tractions they exert on the substrate, leading to an alteration of the collective behavior. This could be partially because IF-depleted glial cells cannot interact with their neighbors, as a similar effect was observed for keratin 8 morphants of Xenopus laevis mesoendoderm explants (Sonavane et al., 2017). In agreement with this hypothesis, we show here that IFs promote the formation of ITAs, which connect the lateral sides of the adjacent leaders and control the treadmilling of AJs. Such actin-driven AJ dynamics is required for the maintenance of cell–cell contacts, which control the directionality and the persistence of migration (Camand et al., 2012; Peglion et al., 2014). However, experiments of single cells indicate that the control of AJs by IFs is not solely responsible for the impact of IFs on FAs and traction forces. The IF-associated protein, plectin, interacts with IFs and localizes at FAs, and its depletion mimics most of the effects observed after IF depletion. IFs may act via plectin to control vinculin recruitment and the mechanical coupling between FAs and actin and thereby traction forces and/or act indirectly by controlling the acto-myosin network (Costigliola et al., 2017) or regulating FA dynamics through microtubules (Ezratty et al., 2005; Akhmanova et al., 2009; Gan et al., 2016).
Altogether, these observations provide new insights into how higher IF levels may contribute to the invasive phenotype of glioblastoma, suggesting that IFs may serve as an interesting therapeutic target to reduce the invasive capacity of these tumors.
Materials and methods
Cell culture
Primary astrocytes were obtained from E17 rat embryos (Etienne-Manneville, 2006). The striatum of pup brains devoid of meninges was dissected and mechanically dissociated with Pasteur pipettes. The cell suspension was plated on P100 poly-l-ornithine (Sigma-Aldrich)–coated dishes and incubated for 7 d, after which the non-astrocytic cells were eliminated by several washes with PBS. 3 d later, the cell culture containing at least 98% GFAP-positive cells could be used. Use of these animals is in compliance with ethical regulations and has been approved by the Prefecture de Police and Direction départementale des services vétérinaires de Paris. Astrocytes were grown in 1 g/l glucose DMEM supplemented with 10% FBS (Invitrogen), 1% penicillin-streptomycin (Gibco), and 1% amphotericin B (Gibco) at 5% CO2 and 37°C.
Transfection
Astrocytes were transfected with Lonza glial transfection solution and electroporated with a Nucleofector machine (Lonza). Cells were then plated on appropriate supports previously coated with poly-l-ornithine. Experiments were performed 3 or 4 d posttransfection, and comparable protein silencing was observed. siRNAs were used at 1 nmol, and DNA was used at 5 µg (or 2.5 µg for pLifeAct-mCherry). siRNA sequences used were as follows: Luciferase (control) 5′-UAAGGCUAUGAAGAGAUAC-3′, GFAP rat 5′-GAGUGGUAUCGGUCCAAGU-3′, GFAP rat set B 5′-CAACCUGGCUGUGUACAGA-3′, vimentin rat 5′-UGAAGAAGCUGCACGAUGA-3′, vimentin rat set B 5′-CGAGCUCAGCACAUAACAA-3′, nestin rat 5′-GUUCCAGCUGGCUGUGGAA-3′, nestin rat set B 5′-AUAAGAGCCUUCUAGAAGA-3′, and plectin rat 5′-AGAGCGAGCUGGAGCGACA-3′. DNA plasmids used were pEGFP-paxillin (gift from E.M. Vallés, Institut Curie, Paris, France), pPaxillin psmOrange (31923; Addgene), pPaxillin-mCherry cloned from pPaxillin psmOrange with BamHI and NotI, pEGFP-N3-vimentin (Sakamoto et al., 2013), RFP-N3-vimentin cloned from pEGFP-N3-vimentin with NotI and EcoRI, pVimentin psmOrange (31922; Addgene), pLifeAct-mCherry (gift from M. Piel, Institut Pierre-Gilles de Gennes, Paris, France), pEGFP-N-cadherin (gift from C. Gauthier-Rouvière, Cell Biology Research Center, Montpellier, France), eGFP-RLC Myosin II (gift from B. Latge and B. Goud, Institut Curie), vinculin tension sensor (Vin TS; 26019; Addgene; Grashoff et al., 2010), and mEB3-FL-pEGFP-N1 (gift from N. Morin, Cell Biology Research Center, Montpellier, France).
2D wound healing assay
Cells were plated on appropriate supports (dishes, plates, coverslips, or glass-bottom; MatTek) and allowed to grow to confluence. Fresh medium was added the day after transfection and the day before the experiment. The cell monolayer was then scratched with a p200 pipette tip to induce migration.
Immunofluorescence
Cells migrating for 8 h (unless otherwise stated) were fixed with cold methanol for 3–5 min at −20°C or 4% warm PFA for 10 min and permeabilized for 10 min with Triton X-100 0.2%. Coverslips were blocked for 30 min with 4% BSA in PBS. The same solution was used for primary and secondary antibody incubation for 1 h. Nuclei were stained with DAPI or Hoechst and mounted with Mowiol. Epifluorescence images were acquired with a Leica DM6000 microscope equipped with 40× 1.25-NA or 63× 1.4-NA oil objectives and recorded on a CCD Leica DFC350 FX camera with Leica software. Confocal images were acquired with a Zeiss LSM 700 inverted microscope equipped with a 63× 1.3-NA oil objective with Zen software. Super-resolution 3D-SIM images were acquired with a Zeiss LSM780 ELYRA with 63× 1.4-NA or 100× 1.46-NA oil objectives and recorded on an EMCCD camera Andor Ixon 887 1K with Zen software.
Primary antibodies used were anti–α-E-catenin (32635, rabbit polyclonal, lot 4 of 04/2016; Abcam), anti–α-tubulin (MCA77G YL1/2, rat monoclonal; Bio-Rad), anti-GFAP (sc-6170, goat polyclonal, clone C-19, lots F0313 and H189; Santa Cruz; and Z0334, rabbit polyclonal, lot 20035993; Dako), anti–N-cadherin (ab12221, rabbit polyclonal, lot GR139340-26, and sc-31030, goat polyclonal, clone K-20, lot B2014; Abcam), anti-nestin (MAB353, mouse monoclonal, lot 2780475; Millipore), anti-paxillin (610051, mouse monoclonal, lot 5246880; BD; and ab32084, rabbit monoclonal, clone Y133 and lot GR215998-1; Abcam), anti-Phalloidin (176759, lot GR278180-3; Abcam), anti-pericentrin (PRB 432-C, rabbit polyclonal; Covance), anti-plectin (P9318, mouse monoclonal, clone 7A8; Sigma-Aldrich), anti-talin (T3287, mouse monoclonal, clone 8D4, lot 035M4805V; Sigma-Aldrich), anti-vimentin (sc-7557R, rabbit polyclonal, clone C-20, lot E3113, and 7557, goat polyclonal, clone C-20, lots E0313 and B1510; Santa Cruz; and V6630, mouse monoclonal lot 10M4831; Sigma-Aldrich), anti-vinculin (V9131, mouse monoclonal, lot 036M4797V; Sigma-Aldrich). Secondary antibodies were Alexa Fluor 488 donkey anti–rabbit (711-545-152), Rhodamine (TRITC) donkey anti–rabbit (711-025-152), Alexa Fluor 647 donkey anti–rabbit (711-695-152), Alexa Fluor 488 donkey anti–mouse (715-545-151), Rhodamine (TRITC) donkey anti–mouse (715-025-151), Alexa Fluor 647 donkey anti–goat (705-605-147), and Alexa Fluor 488 donkey anti–rat (712-545-153); all from Jackson ImmunoResearch.
Live imaging
For phase-contrast wound healing assays, cells were wounded in 12-well plates with normal astrocyte medium. Hepes was added to the medium, and paraffin was used to cover the well to prevent medium evaporation. Acquisition started 30 min after wounding. Videos were acquired with a Zeiss Axiovert 200M equipped with a thermostatic humid chamber with 5% CO2 and 37°C. All images were acquired with a 10× 0.45-NA air objective and an EMCCD pco.edge sCMOS camera with Metamorph software. Images were acquired every 15 min for 24 h.
Nuclei of leader cells were manually tracked with Fiji software (Manual Tracking plugin). Calculations for 24 h of migration were normalized by pixel size and time with the following formulas (n is the number of time points acquired and ϑ is velocity):
mean velocity,
persistence,
and directionality,
For fluorescence live imaging, cells on appropriate dishes (Ibidi or MatTek) were wounded and allowed to migrate for 4 h before image acquisition. Hepes and antioxidants were added to the medium before acquisition. Epifluorescence experiments investigating paxillin turnover and N-cadherin and actin retrograde flow were performed with a Nikon BioStation IM-Q (40× air objective) at a rate of 1 frame/5 min for 3–5 h with 5% CO2 and 37°C in normal astrocyte medium. Confocal experiments for MRLC and EB3 were performed on a PerkinElmer spinning disk Ultraview Vox confocal microscope equipped with an EMCCD Hamamatsu camera and a 63× 1.4-NA oil objective and Volocity software with 5% CO2 and 37°C in normal astrocyte medium. Superresolution experiments were acquired with the Zeiss LSM780 ELYRA PS.1 with an EMCCD Andor Ixon 887 1K camera and a 63× 1.4-NA oil objective at 37°C in normal astrocyte medium. Images were acquired and automatically processed for structured illumination with Zen software.
FRET tension sensor experiments
Cells were transfected with VinTS. The day before the experiment, medium was changed to phenol-free (FluoroBrite DMEM, A18967; Gibco). Cells were wounded for 4 h, and Hepes was added before acquisition. Images were acquired with a Zeiss LSM780 with a 63× 1.4-NA oil objective and Definite Focus with Zen software at 5% CO2 and 37°C in phenol-free medium. The donor was excited at 458 nm, and the emission spectrum of the sensor was in 13 contiguous channels from 465 to 545 nm, to include both the donor and acceptor emission wavelengths. Images were analyzed through the Fiji plugin PixFret. After background correction, the FRET index was calculated on thresholded images to delineate the FAs. PixFret calculates the FRET index by measuring the ratio of intensity IFRET/(IDonor + IFRET) between the donor channel and the FRET channel (channels 3 and 6).
FACS
Migrating and nonmigrating cells in 100-mm dishes were trypsinized and fixed in 4% warm PFA for 10 min, and 10,000 cells were analyzed on a BD LSRFortessa II flow cytometer applying a live cell gate based on forward scatter and side scatter.
Micropatterns
Coverslips were plasma-cleaned for 45 s and incubated immediately with 0.1 mg/ml poly-l-lysine/polyethylene glycol (PLL-PEG) diluted in 10 mM Hepes for 30 min at room temperature. Excess of PLL-PEG was allowed to slide off, and the coverslips were dried before printing. Micropatterns were printed for 4 min with specifically designed chrome masks and coated for 1 h at 37°C with collagen (100 µg/ml) and fluorescent fibrinogen (Alexa Fluor 647, 20 µg/ml) diluted in fresh NaHCO3, pH 8.3, 100 mM. Micropatterned coverslips were washed twice in PBS and used immediately for the experiments. Plated cells were allowed to adhere for 16 h before fixation. Crossbow patterns have a diameter of 40 µm, with an area of ∼1,000 µm2.
TFM experiments on micropatterns
After printing, micropatterns were coated with collagen and fluorescent fibrinogen for 1 h. A thin layer of polyacrylamide solution (52.5 µl acrylamide, 50.25 µl bis-acrylamide, 381 µl water, 10 µl of 0.2-µm-diameter fluorescent beads [FluoSpheres; Molecular Probes], 5 µl of 10% APS, and 3 µl Temed for a 40-kPa gel), previously vacuumed, was allowed to polymerize on the collagen for 20 min. Gels were then washed twice in NaHCO3, pH 8.3, 100 mM, and cells were plated immediately. 16 h later, stacks of single cells were acquired before and after trypsin treatment. Acquisitions were performed with a Nikon Eclipse Ti-E inverted microscope, with a 40× 0.6 NA air objective equipped with an ORCA-Flash 4.0 Hamamatsu CMOS camera with Micromanager software at 5% CO2 and 37°C in normal astrocyte medium. Only single cells, checked through Hoechst staining, were analyzed. Analysis of TFM on micropatterns was performed with a custom-designed macro in Fiji based on work by Tseng et al. (2012). The topmost planes of beads before and after trypsinization were selected and aligned using a normalized cross-correlation algorithm (Align Slices in Stack plugin). The displacement field was computed from bead movements using particle image velocimetry (PIV). Parameters for the PIV were three interrogation windows of 128, 96, and 64 pixels with a correlation of 0.60. Traction forces were calculated from the displacement field using Fourier transform traction cytometry and a Young’s modulus of 40 kPa, a regularization factor of 10−9, and a Poisson ratio of 0.5.
TFM experiments on migrating cells
The protocol for these experiments has been adapted from Trepat et al. (2009), Tambe et al. (2011), and Serra-Picamal et al. (2015). Hydrogels with a Young’s modulus of 9 kPa containing fluorescent beads (375.3 µl of 10 mM Hepes, 93.75 µl of 40% acrylamide, 25 µl of 2% bis-acrylamide, 3.2 µl of 0.2-µm-diameter FluoSpheres, 2.5 µl APS, and 0.25 µl Temed) were prepared on a six-well MatTek plate. Gel surface was activated with a solution of 2 mg/ml sulfo-SANPAH (22589; Thermo Scientific) for 5 min under UV light (wavelength of 365 nm at a distance of 5 cm). After three washes to remove the excess sulfo-SANPAH, gels were coated with a solution of rat tail type I collagen (100 µg/ml) overnight at 4°C. At this point, gels were ready to seed cells.
Monolayer patterning was achieved by using PDMS membranes with rectangular openings. In brief, SU8 masters with the desired shape were produced by soft lithography. After master silanization (trichloro(1H,1H,2H,2H-perfluoro-octyl)silane; Sigma-Aldrich), PDMS was spin-coated on top at 4,200 rpm for 1 min and 15 s and allowed to polymerize at 80°C for 2 h. For handling purposes, thick borders of PDMS were added to the membranes. To avoid cell attachment, PDMS membranes were passivated using a 2% solution of Pluronic F-127 (Sigma-Aldrich) in PBS for 1 h, washed, and allowed to dry. Afterward, membranes were carefully placed on top of coated polyacrylamide gels, and a 15–20-µl drop containing 500,000 cells was seeded right on the PDMS opening. Thus, cells only attached in the desired rectangular configuration. After at least 2 h, PDMS was removed, allowing the monolayer to spread, and imaging started. Images of the cells and the fluorescent beads were acquired every 15 min for 24 h on an inverted epifluorescence Nikon Eclipse Ti microscope with an ORCA-Flash 4.0 CMOS IMCD camera, 10× 0.3-NA air objective with PFS using Metamorph software at 5% CO2 and 37°C in normal astrocyte medium. At the end of the experiment, cells were trypsinized to image the gel in a relaxed configuration to be used as a reference. Analysis was performed every 1 h for the 24-h time course with custom-made Matlab software by comparing the beads at every time point to the reference image. First, the two images were aligned using a custom-made 2D cross-correlation algorithm. Then, PIV analysis was performed using a window of 32 × 32 pixels and an overlap of 0.5, thus obtaining a displacement field for every time point. Tractions were computed from displacements using Fourier transform traction microscopy with finite substrate thickness (Trepat et al., 2009; Tambe et al., 2011; Serra-Picamal et al., 2015).
Electrophoresis and Western blot
Cells lysates were obtained with Laemmli buffer composed of 60 mM Tris-HCl, pH 6.8, 10% glycerol, 2% SDS, and 50 mM DTT with the addition of antiprotease (cOmplete cocktail, 11 873 588 001; Roche), 1 mM glycerol phosphate, 1 mM Na orthovanadate, and 1 mM Na fluoride. Samples were boiled for 5 min at 95°C before loading on polyacrylamide gels. Transfer occurred at 0.2 or 0.3 A overnight on nitrocellulose membranes. Membranes were blotted with TBST (0.2% Tween) and 5% milk and incubated for 1 h with the primary antibody and for 1 h with HRP-conjugated secondary antibody. Bands were revealed with ECL chemiluminescent substrate (Thermo Scientific).
Primary antibodies used were anti–β-catenin (610154, mouse monoclonal, lot 3137536; BD), anti-GFAP (sc-6170, goat polyclonal, clone C-19, lots F0313 and H189; Santa Cruz), anti-GAPDH (MAB374, mouse monoclonal, lot 2689153; Millipore), anti-nestin (MAB353, mouse monoclonal, lot 2780475; Millipore), anti-plectin (mouse monoclonal, clone 7A8; Sigma-Aldrich), anti-talin (T3286, mouse monoclonal, clone 8D4, lot 035M4805V; Sigma-Aldrich), anti-vimentin (V6630, mouse monoclonal, lot 10M4831; Sigma-Aldrich), and anti-vinculin (V9131, mouse monoclonal, lot 036M4797V; Sigma-Aldrich). Secondary HRP-conjugated antibodies used were peroxidase donkey anti–rabbit (711-035-152), peroxidase donkey anti–mouse (715-035-151), and peroxidase donkey anti–goat (705-035-147); all from Jackson ImmunoResearch.
Immunoprecipitation
Confluent 100-mm-diameter dishes of WT astrocytes were washed with cold PBS and lysed with IP B buffer (25 mM Tris HCl, pH 7.5, 1 mM EDTA, pH 8, 150 mM NaCl, 1 mM MgCl2, and 0.5% Triton X-100) with the addition of fresh anti-protease (cOmplete cocktail). Lysates were centrifuged at 13,000 rpm for 2.5 min at 4°C. 15 µl (2% of the total lysate) of supernatant was stored at −20°C with the addition of Laemmli buffer, and the rest of the supernatant was incubated with 50 µl protein G (previously washed in PBS) and 1 µg primary antibody or control antibody for 2 h at 4°C on a spinning wheel. Beads were washed at least eight times with the IP B buffer before loading in Laemmli buffer on precast gels (Invitrogen).
Quantification of immunofluorescent images
All images were analyzed with Fiji software (Schindelin et al., 2012). For actin distribution and orientation, cells in the first row were scored manually as positive when containing both ITAs and stress fibers or negative when they possessed only stress fibers and no ITAs. To measure actin fiber orientation in the followers, phalloidin z-stacks were projected for maximum intensity onto one image. After filtering with a specific kernel and despeckling, they were analyzed with a specific Fiji directionality plugin. Vinculin localization on FAs or cell–cell junctions was manually scored in the first row of cells. Polarity was also scored by counting the percentage of cells with the centrosome (centrin staining) oriented in the 90° quadrant toward the wound.
To quantify retrograde flow, kymographs (width, 5 pixels) were drawn on the cell–cell junctions (front part) or on the central axis (tip and behind) of the cell for myosin. They were plotted with the Multi Kymograph plugin, from which three slopes were measured to obtain a mean value for each cell. To quantify FAs, background-subtracted images of FA-stained cells were thresholded with the same value for all images. Cell contours were drawn manually to count the number and size of FAs. The relative projected distance between FAs and the nucleus was defined to characterize the distribution of FAs by projecting the FA position onto a line drawn between the centroid of the nucleus and the tip of the cell (see schematics in Fig. 3 C). To quantify paxillin FA turnover, integrated fluorescent intensity was analyzed. Turnover was defined as the time elapsed between their birth (first frame) and their death (last frame). Assembly was defined as the time between birth and the peak of fluorescent intensity, and disassembly was defined as the time between the highest peak of intensity and death of the FA.
EB3 tracking was performed with Fiji TrackMate Plugin with the following settings: LoG detector, estimated blob diameter 1 pixel, simple LAP tracker (1 pixel linking max distance, 1.5 pixel gap-closing max distance, and 3 gap-closing max frame gap), and filters for duration of track (>4) and track displacement (>2.5). Velocity, persistence, and directionality toward the wound were calculated with the formulas previously described for cell migration (in the x direction) with a custom-designed Fiji macro.
Statistical analysis
All data are presented as the mean in dot plots or mean ± SEM in histograms of at least three independent experiments. Statistical analysis was obtained with Student’s t test for Figs. 2 (E and G), 3 (E and F), 4 E, and 5 (C and E) and Figs. S1 (A and C–E) and 3 (D and E). ANOVA followed by Bonferroni post hoc test for Figs. 1 C, 2 F, and 3 (C, E, and F) and Fig. S2 F or χ2 test for contingency on the raw data for Figs. 2 (B and H), 4 (C and F), and 5 B and Figs. S1 F, 2 C, and 3D. Analysis was performed with GraphPad Prism 5.0. P-values are reported as not significant (n.s.) for P > 0.05, *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
Online supplemental material
Fig. S1 shows the levels of silencing of IFs on migrating astrocytes (Western blots and immunofluorescence) and their effects on collective migration and centrosome polarity. Fig. S2 shows the effect of single IF siRNA on the actin cytoskeleton and AJs. Fig. S3 shows the effect of single IF siRNA on FAs, the effects of si triple IF on microtubules and their dynamics, and the levels of silencing of si plectin (Western blot and immunofluorescence). Video 1 shows a wound healing assay of primary astrocytes transfected with si ctl or si triple IF. Videos 2 and 3 show migration of primary astrocytes transfected with si ctl and si triple IF and their tractions (Tx), respectively. Video 4 shows the effects of IFs on myosin retrograde flow during collective migration. Video 5 shows the effects of IFs on N-cadherin retrograde flow during collective migration. Videos 6 and 7 show the dynamics of vimentin IFs near FAs. Video 8 shows the effect of IFs on FA turnover during collective migration. Video 9 shows EB3 dynamics of si ctl and si triple IF astrocytes during collective migration.
Supplementary Material
Acknowledgments
We thank members of the Etienne-Manneville, Trepat, and Théry laboratories for support and discussion, as well as J.B. Manneville and C. Alibert for stimulating discussion and careful reading of the manuscript. We gratefully thank N. Festuccia for support and help with FACS. We gratefully acknowledge J.Y. Tinevez and A. Salles and the Imagopole of Institut Pasteur (Paris, France), as well as the France-BioImaging infrastructure network for the use of the Elyra microscope. The authors gratefully acknowledge the Nikon Imaging Centre, Institut Curie, and the ImagoSeine core facility of the Institut Jacques Monod.
This work was supported by La Ligue Contre le Cancer (EL2017.LNCC) and the Institut Pasteur. C. De Pascalis was a scholar in the Pasteur–Paris University International PhD program and received a stipend from Fondation pour la Recherche Médicale, Institut Pasteur, and a short-term European Molecular Biology Organization fellowship. C. Pérez-González acknowledges support from Fundación Caixa. The France-BioImaging infrastructure network is supported by the French Agence Nationale de la Recherche (ANR10-INBS-04; Investments for the Future) and the Région Ile-de-France (program Domaine d’Intérêt Majeur-Malinf). The Nikon Imaging Centre is a member of the Agence Nationale de la Recherche Infrastructure France-BioImaging (ANR10-INBS-04), and the Institut Jacques Monod is a member of Infrastructures en Biologie Santé et Agronomie and Agence Nationale de la Recherche France-BioImaging (ANR10-INBS-04).
The authors declare no competing financial interests.
Author contributions: C. De Pascalis designed, performed experiments, and wrote the paper; C. Pérez-González assisted in the setup of TFM and analyzed results; S. Seetharaman performed part of revision experiments; B. Boëda assisted with biochemistry and discussions; B. Vianay assisted with the setup of TFM on micropatterns and designed a custom macro for analysis of TFM and EB3; M. Burute assisted with the setup of TFM on micropatterns; C. Leduc designed the macro for FA analysis and assisted with SRM images; N. Borghi assisted with FRET microscopy analysis; X. Trepat assisted with TFM and discussions; and S. Etienne-Manneville supervised the project, analyzed the results, and wrote the paper.
References
- Akhmanova A., Stehbens S.J., and Yap A.S.. 2009. Touch, grasp, deliver and control: Functional cross-talk between microtubules and cell adhesions. Traffic. 10:268–274. 10.1111/j.1600-0854.2008.00869.x [DOI] [PubMed] [Google Scholar]
- Bakolitsa C., Cohen D.M., Bankston L.A., Bobkov A.A., Cadwell G.W., Jennings L., Critchley D.R., Craig S.W., and Liddington R.C.. 2004. Structural basis for vinculin activation at sites of cell adhesion. Nature. 430:583–586. 10.1038/nature02610 [DOI] [PubMed] [Google Scholar]
- Burda J.E., Bernstein A.M., and Sofroniew M.V.. 2016. Astrocyte roles in traumatic brain injury. Exp. Neurol. 275:305–315. 10.1016/j.expneurol.2015.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgstaller G., Gregor M., Winter L., and Wiche G.. 2010. Keeping the vimentin network under control: Cell-matrix adhesion-associated plectin 1f affects cell shape and polarity of fibroblasts. Mol. Biol. Cell. 21:3362–3375. 10.1091/mbc.e10-02-0094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camand E., Peglion F., Osmani N., Sanson M., and Etienne-Manneville S.. 2012. N-cadherin expression level modulates integrin-mediated polarity and strongly impacts on the speed and directionality of glial cell migration. J. Cell Sci. 125:844–857. 10.1242/jcs.087668 [DOI] [PubMed] [Google Scholar]
- Chang Y.C., Nalbant P., Birkenfeld J., Chang Z.F., and Bokoch G.M.. 2008. GEF-H1 couples nocodazole-induced microtubule disassembly to cell contractility via RhoA. Mol. Biol. Cell. 19:2147–2153. 10.1091/mbc.e07-12-1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung B.M., Rotty J.D., and Coulombe P.A.. 2013. Networking galore: Intermediate filaments and cell migration. Curr. Opin. Cell Biol. 25:600–612. 10.1016/j.ceb.2013.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costigliola N., Ding L., Burckhardt C.J., Han S.J., Gutierrez E., Mota A., Groisman A., Mitchison T.J., and Danuser G.. 2017. Vimentin fibers orient traction stress. Proc. Natl. Acad. Sci. USA. 114:5195–5200. 10.1073/pnas.1614610114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pascalis C., and Etienne-Manneville S.. 2017. Single and collective cell migration: The mechanics of adhesions. Mol. Biol. Cell. 28:1833–1846. 10.1091/mbc.e17-03-0134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupin I., Sakamoto Y., and Etienne-Manneville S.. 2011. Cytoplasmic intermediate filaments mediate actin-driven positioning of the nucleus. J. Cell Sci. 124:865–872. 10.1242/jcs.076356 [DOI] [PubMed] [Google Scholar]
- du Roure O., Saez A., Buguin A., Austin R.H., Chavrier P., Silberzan P., and Ladoux B.. 2005. Force mapping in epithelial cell migration. Proc. Natl. Acad. Sci. USA. 102:2390–2395. 10.1073/pnas.0408482102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville S. 2006. In vitro assay of primary astrocyte migration as a tool to study Rho GTPase function in cell polarization. Methods Enzymol. 406:565–578. 10.1016/S0076-6879(06)06044-7 [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S. 2013. Microtubules in cell migration. Annu. Rev. Cell Dev. Biol. 29:471–499. 10.1146/annurev-cellbio-101011-155711 [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S. 2014. Neighborly relations during collective migration. Curr. Opin. Cell Biol. 30:51–59. 10.1016/j.ceb.2014.06.004 [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S., and Hall A.. 2001. Integrin-mediated activation of Cdc42 controls cell polarity in migrating astrocytes through PKCzeta. Cell. 106:489–498. 10.1016/S0092-8674(01)00471-8 [DOI] [PubMed] [Google Scholar]
- Ezratty E.J., Partridge M.A., and Gundersen G.G.. 2005. Microtubule-induced focal adhesion disassembly is mediated by dynamin and focal adhesion kinase. Nat. Cell Biol. 7:581–590. 10.1038/ncb1262 [DOI] [PubMed] [Google Scholar]
- Gan Z., Ding L., Burckhardt C.J., Lowery J., Zaritsky A., Sitterley K., Mota A., Costigliola N., Starker C.G., Voytas D.F., et al. 2016. Vimentin intermediate filaments template microtubule networks to enhance persistence in cell polarity and directed migration. Cell Syst. 3:500–501. 10.1016/j.cels.2016.11.011 [DOI] [PubMed] [Google Scholar]
- Gardel M.L., Schneider I.C., Aratyn-Schaus Y., and Waterman C.M.. 2010. Mechanical integration of actin and adhesion dynamics in cell migration. Annu. Rev. Cell Dev. Biol. 26:315–333. 10.1146/annurev.cellbio.011209.122036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnanaguru G., Bachay G., Biswas S., Pinzón-Duarte G., Hunter D.D., and Brunken W.J.. 2013. Laminins containing the β2 and γ3 chains regulate astrocyte migration and angiogenesis in the retina. Development. 140:2050–2060. 10.1242/dev.087817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales M., Weksler B., Tsuruta D., Goldman R.D., Yoon K.J., Hopkinson S.B., Flitney F.W., and Jones J.C.. 2001. Structure and function of a vimentin-associated matrix adhesion in endothelial cells. Mol. Biol. Cell. 12:85–100. 10.1091/mbc.12.1.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grashoff C., Hoffman B.D., Brenner M.D., Zhou R., Parsons M., Yang M.T., McLean M.A., Sligar S.G., Chen C.S., Ha T., and Schwartz M.A.. 2010. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 466:263–266. 10.1038/nature09198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor M., Osmanagic-Myers S., Burgstaller G., Wolfram M., Fischer I., Walko G., Resch G.P., Jörgl A., Herrmann H., and Wiche G.. 2014. Mechanosensing through focal adhesion-anchored intermediate filaments. FASEB J. 28:715–729. 10.1096/fj.13-231829 [DOI] [PubMed] [Google Scholar]
- Haeger A., Wolf K., Zegers M.M., and Friedl P.. 2015. Collective cell migration: Guidance principles and hierarchies. Trends Cell Biol. 25:556–566. 10.1016/j.tcb.2015.06.003 [DOI] [PubMed] [Google Scholar]
- Herrmann H., Strelkov S.V., Burkhard P., and Aebi U.. 2009. Intermediate filaments: Primary determinants of cell architecture and plasticity. J. Clin. Invest. 119:1772–1783. 10.1172/JCI38214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiu Y., Lehtimäki J., Tojkander S., Cheng F., Jäälinoja H., Liu X., Varjosalo M., Eriksson J.E., and Lappalainen P.. 2015. Bidirectional interplay between vimentin intermediate filaments and contractile actin stress fibers. Cell Reports. 11:1511–1518. 10.1016/j.celrep.2015.05.008 [DOI] [PubMed] [Google Scholar]
- Jiu Y., Peränen J., Schaible N., Cheng F., Eriksson J.E., Krishnan R., and Lappalainen P.. 2017. Vimentin intermediate filaments control actin stress fiber assembly through GEF-H1 and RhoA. J. Cell Sci. 130:892–902. 10.1242/jcs.196881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreplak L., and Fudge D.. 2007. Biomechanical properties of intermediate filaments: From tissues to single filaments and back. BioEssays. 29:26–35. 10.1002/bies.20514 [DOI] [PubMed] [Google Scholar]
- Leduc C., and Etienne-Manneville S.. 2015. Intermediate filaments in cell migration and invasion: The unusual suspects. Curr. Opin. Cell Biol. 32:102–112. 10.1016/j.ceb.2015.01.005 [DOI] [PubMed] [Google Scholar]
- Leduc C., and Etienne-Manneville S.. 2017. Regulation of microtubule-associated motors drives intermediate filament network polarization. J. Cell Biol. 216:1689–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepekhin E.A., Eliasson C., Berthold C.H., Berezin V., Bock E., and Pekny M.. 2001. Intermediate filaments regulate astrocyte motility. J. Neurochem. 79:617–625. 10.1046/j.1471-4159.2001.00595.x [DOI] [PubMed] [Google Scholar]
- Leube R.E., Moch M., and Windoffer R.. 2015. Intermediate filaments and the regulation of focal adhesion. Curr. Opin. Cell Biol. 32:13–20. 10.1016/j.ceb.2014.09.011 [DOI] [PubMed] [Google Scholar]
- Lin L., Wang G., Ming J., Meng X., Han B., Sun B., Cai J., and Jiang C.. 2016. Analysis of expression and prognostic significance of vimentin and the response to temozolomide in glioma patients. Tumour Biol. 37:15333–15339. 10.1007/s13277-016-5462-7 [DOI] [PubMed] [Google Scholar]
- Liu C.Y., Lin H.H., Tang M.J., and Wang Y.K.. 2015. Vimentin contributes to epithelial-mesenchymal transition cancer cell mechanics by mediating cytoskeletal organization and focal adhesion maturation. Oncotarget. 6:15966–15983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv D., Lu L., Hu Z., Fei Z., Liu M., Wei L., and Xu J.. 2017. Nestin expression is associated with poor clinicopathological features and prognosis in glioma patients: An association study and meta-analysis. Mol. Neurobiol. 54:727–735. 10.1007/s12035-016-9689-5 [DOI] [PubMed] [Google Scholar]
- Lynch C.D., Lazar A.M., Iskratsch T., Zhang X., and Sheetz M.P.. 2013. Endoplasmic spreading requires coalescence of vimentin intermediate filaments at force-bearing adhesions. Mol. Biol. Cell. 24:21–30. 10.1091/mbc.e12-05-0377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y.H., Mentlein R., Knerlich F., Kruse M.L., Mehdorn H.M., and Held-Feindt J.. 2008. Expression of stem cell markers in human astrocytomas of different WHO grades. J. Neurooncol. 86:31–45. 10.1007/s11060-007-9439-7 [DOI] [PubMed] [Google Scholar]
- Matsuda Y., Ishiwata T., Yoshimura H., Hagio M., and Arai T.. 2015. Inhibition of nestin suppresses stem cell phenotype of glioblastomas through the alteration of post-translational modification of heat shock protein HSPA8/HSC71. Cancer Lett. 357:602–611. 10.1016/j.canlet.2014.12.030 [DOI] [PubMed] [Google Scholar]
- Mayor R., and Etienne-Manneville S.. 2016. The front and rear of collective cell migration. Nat. Rev. Mol. Cell Biol. 17:97–109. 10.1038/nrm.2015.14 [DOI] [PubMed] [Google Scholar]
- Mendez M.G., Kojima S., and Goldman R.D.. 2010. Vimentin induces changes in cell shape, motility, and adhesion during the epithelial to mesenchymal transition. FASEB J. 24:1838–1851. 10.1096/fj.09-151639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middeldorp J., and Hol E.M.. 2011. GFAP in health and disease. Prog. Neurobiol. 93:421–443. 10.1016/j.pneurobio.2011.01.005 [DOI] [PubMed] [Google Scholar]
- Osmanagic-Myers S., Gregor M., Walko G., Burgstaller G., Reipert S., and Wiche G.. 2006. Plectin-controlled keratin cytoarchitecture affects MAP kinases involved in cellular stress response and migration. J. Cell Biol. 174:557–568. 10.1083/jcb.200605172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmanagic-Myers S., Rus S., Wolfram M., Brunner D., Goldmann W.H., Bonakdar N., Fischer I., Reipert S., Zuzuarregui A., Walko G., and Wiche G.. 2015. Plectin reinforces vascular integrity by mediating crosstalk between the vimentin and the actin networks. J. Cell Sci. 128:4138–4150. 10.1242/jcs.172056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peglion F., Llense F., and Etienne-Manneville S.. 2014. Adherens junction treadmilling during collective migration. Nat. Cell Biol. 16:639–651. 10.1038/ncb2985 [DOI] [PubMed] [Google Scholar]
- Pekny M., Pekna M., Messing A., Steinhäuser C., Lee J.M., Parpura V., Hol E.M., Sofroniew M.V., and Verkhratsky A.. 2016. Astrocytes: A central element in neurological diseases. Acta Neuropathol. 131:323–345. 10.1007/s00401-015-1513-1 [DOI] [PubMed] [Google Scholar]
- Sakamoto Y., Boëda B., and Etienne-Manneville S.. 2013. APC binds intermediate filaments and is required for their reorganization during cell migration. J. Cell Biol. 200:249–258. 10.1083/jcb.201206010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: An open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seltmann K., Cheng F., Wiche G., Eriksson J.E., and Magin T.M.. 2015. Keratins stabilize hemidesmosomes through regulation of β4-integrin turnover. J. Invest. Dermatol. 135:1609–1620. 10.1038/jid.2015.46 [DOI] [PubMed] [Google Scholar]
- Serra-Picamal X. 2012. Mechanical waves during tissue expansion. Nat. Phys. 8:628–634. 10.1038/nphys2355 [DOI] [Google Scholar]
- Serra-Picamal X., Conte V., Sunyer R., Muñoz J.J., and Trepat X.. 2015. Mapping forces and kinematics during collective cell migration. Methods Cell Biol. 125:309–330. 10.1016/bs.mcb.2014.11.003 [DOI] [PubMed] [Google Scholar]
- Sonavane P.R., Wang C., Dzamba B., Weber G.F., Periasamy A., and DeSimone D.W.. 2017. Mechanical and signaling roles for keratin intermediate filaments in the assembly and morphogenesis of Xenopus mesendoderm tissue at gastrulation. Development. 144:4363–4376. 10.1242/dev.155200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tambe D.T., Hardin C.C., Angelini T.E., Rajendran K., Park C.Y., Serra-Picamal X., Zhou E.H., Zaman M.H., Butler J.P., Weitz D.A., et al. 2011. Collective cell guidance by cooperative intercellular forces. Nat. Mater. 10:469–475. 10.1038/nmat3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trepat X., Wasserman M.R., Angelini T.E., Millet E., Weitz D.A., Butler J.P., and Fredberg J.J.. 2009. Physical forces during collective cell migration. Nat. Phys. 5:426–430. 10.1038/nphys1269 [DOI] [Google Scholar]
- Tseng Q., Duchemin-Pelletier E., Deshiere A., Balland M., Guillou H., Filhol O., and Théry M.. 2012. Spatial organization of the extracellular matrix regulates cell–cell junction positioning. Proc. Natl. Acad. Sci. USA. 109:1506–1511. 10.1073/pnas.1106377109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiche G., Osmanagic-Myers S., and Castañón M.J.. 2015. Networking and anchoring through plectin: A key to IF functionality and mechanotransduction. Curr. Opin. Cell Biol. 32:21–29. 10.1016/j.ceb.2014.10.002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.