ARHGAP35 encoding the RhoGAP p190A is one of the most frequently mutated genes in human cancer, which suggests that p190A may have a tumor-suppressive function. Frank et al. demonstrate that p190A and its paralog p190B mediate contact inhibition of epithelial cell proliferation by repressing YAP-mediated gene transcription.
Abstract
ARHGAP35 encoding p190A RhoGAP is a cancer-associated gene with a mutation spectrum suggestive of a tumor-suppressor function. In this study, we demonstrate that loss of heterozygosity for ARHGAP35 occurs in human tumors. We sought to identify tumor-suppressor capacities for p190A RhoGAP (p190A) and its paralog p190B in epithelial cells. We reveal an essential role for p190A and p190B to promote contact inhibition of cell proliferation (CIP), a function that relies on RhoGAP activity. Unbiased mRNA sequencing analyses establish that p190A and p190B modulate expression of genes associated with the Hippo pathway. Accordingly, we determine that p190A and p190B induce CIP by repressing YAP–TEAD-regulated gene transcription through activation of LATS kinases and inhibition of the Rho–ROCK pathway. Finally, we demonstrate that loss of a single p190 paralog is sufficient to elicit nuclear translocation of YAP and perturb CIP in epithelial cells cultured in Matrigel. Collectively, our data reveal a novel mechanism consistent with a tumor-suppressor function for ARHGAP35.
Introduction
Two recent studies mapping the landscape of somatic mutations in human cancer identified ARHGAP35 as a major cancer gene (Kandoth et al., 2013; Lawrence et al., 2014). These studies demonstrated that ARHGAP35 is mutated in >2% of all tumors and thus ranks among the top ∼30 most significantly mutated genes in human cancer. This discovery was surprising because ARHGAP35 was the only gene with such high frequency of mutations that was not included in the Cancer Gene Census at that time. The mutation rate of ARHGAP35 is particularly high in uterine corpus endometrioid carcinoma, and the gene is also frequently mutated in squamous cell carcinoma and adenocarcinoma of the lung, head and neck cancer, and renal cell carcinoma (Kandoth et al., 2013; Lawrence et al., 2014). In addition, ARHGAP35 is located in a region of chromosome 19 that is focally deleted in numerous carcinomas (Zack et al., 2013).
ARHGAP35 encodes p190A RhoGAP (p190A), a major GTPase-activating protein (GAP) for Rho family proteins (Settleman et al., 1992). p190A exhibits 50% sequence identity and the same overall structure as p190B RhoGAP (p190B), which is encoded by ARHGAP5 (Burbelo et al., 1995). Both p190A and p190B are widely coexpressed, and each is essential for normal mouse development and tissue homeostasis (Brouns et al., 2000; Sordella et al., 2002). p190A and p190B provide spatial and temporal control of Rho activity in response to extracellular signaling (Burbelo et al., 1995; Nakahara et al., 1998; Wildenberg et al., 2006). In this capacity, p190A and p190B exert profound effects on the actin cytoskeleton and cellular processes directly dependent on actin polymerization. In addition, p190A and p190B have been shown to regulate transcriptional responses through TFII-I and CREB, respectively (Sordella et al., 2002; Jiang et al., 2005).
Cancer genome sequencing data support a tumor-suppressor role for ARHGAP35 (Kandoth et al., 2013; Lawrence et al., 2014). However, cellular functions of p190A consistent with such a role have not been determined. p190A plays pivotal roles in proliferative and motile capacities of mammalian cells, but the effects are not consistent with a tumor-suppressor role. Inhibition of p190A function by knockdown or overexpression of GAP-deficient p190A inhibits cell spreading and protrusion, resulting in loss of cell polarity and perturbation of cell migration (Arthur and Burridge, 2001). A recent publication by Binamé et al. (2016) confirms that p190A is required for directional cell motility and that certain p190A mutations found in human cancer perturb directional cell motility. However, loss of directional motility is not a hallmark of cancer (Hanahan and Weinberg, 2011). A role for p190A in cytokinesis has also been established (Su et al., 2003). Overexpression of p190A perturbs cytokinesis, resulting in the emergence of multinucleate cells, and loss of p190A might therefore seem advantageous to cancerous cells. However, endogenous levels of p190A do not affect cytokinesis (Su et al., 2009). Moreover, depletion of p190A inhibits entry into the cell cycle, thereby perturbing cell proliferation (Su et al., 2009). Collectively, the published effects on proliferative and motile capacities associated with loss of p190A function are not consistent with a tumor-suppressor role.
In contrast, we demonstrate in this study that p190A promotes contact inhibition of cell proliferation (CIP). Loss of CIP represents one of the earliest appreciated hallmarks of cancer (Hanahan and Weinberg, 2011). This effect of p190A is shared with p190B. Next, using an unbiased approach, we show that p190A and p190B suppress the transcriptional activity of YAP, an effector of the Hippo pathway and an established modulator of CIP (Zeng and Hong, 2008; McClatchey and Yap, 2012; Gumbiner and Kim, 2014). We show that p190A and p190B signal to prevent translocation of YAP from the cytosol to the nucleus. Furthermore, we establish that p190A and p190B repress YAP-mediated gene transcription by activating large tumor suppressor (LATS) kinases as well as by inhibiting Rho–ROCK signaling. Finally, we show that depletion of a single p190 paralog is sufficient to induce alterations associated with oncogenic transformation when epithelial cells are cultured in Matrigel. Collectively, our data support a tumor-suppressor function for p190A through activation of canonical Hippo signaling and inhibition of mechanotransduction to induce CIP.
Results
Analysis of ARHGAP35 mutations suggests p190A loss of function in epithelial cancers
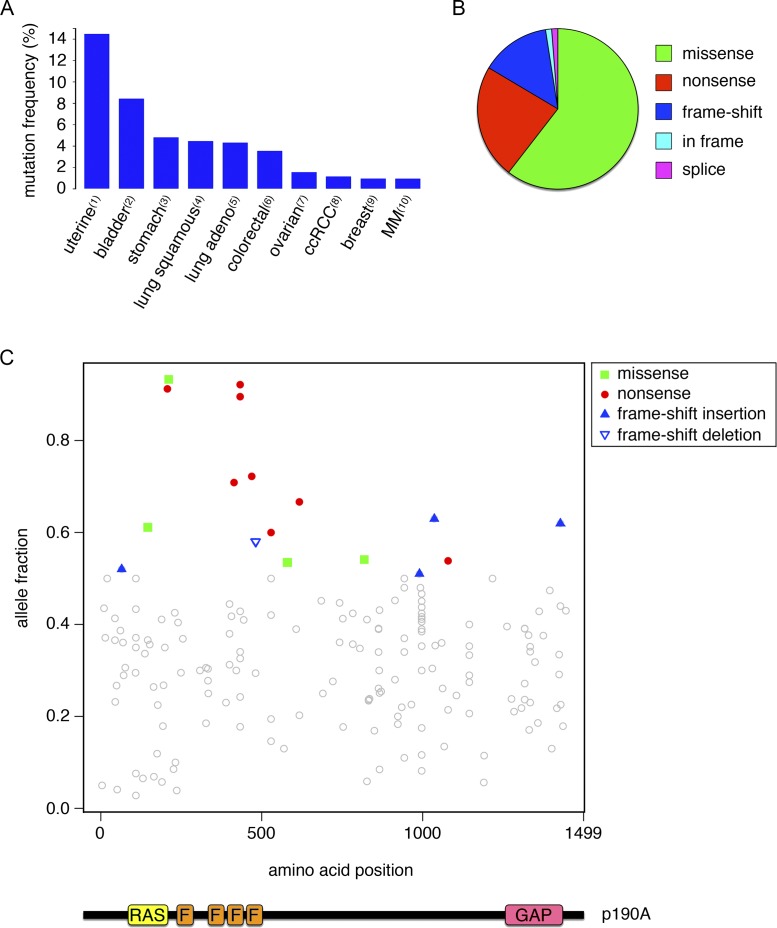
ARHGAP35 mutations occur predominantly in epithelial cancers (Fig. 1 A). Analysis of the spectrum of mutations occurring in ARHGAP35 suggests a role as tumor suppressor. Approximately 40% of nonsynonymous ARHGAP35 mutations are nonsense or frame-shift mutations (Fig. 1 B). These mutations are scattered throughout the coding region, which is difficult to reconcile with a gain-of-function effect (Fig. 1 C). Instead, if ARHGAP35 were to exert tumor-suppressor capacities, one might expect that a subset of tumors would exhibit loss of heterozygosity. To this end, we performed an analysis of somatic ARHGAP35 mutations in the PanCancer mutation dataset derived from ∼5,000 patients. This analysis revealed a wide range of mutant allele frequencies, with several tumors having mutant allele fractions >0.5, thus indicative of loss of heterozygosity (Fig. 1 C). It should be noted that even among tumors with an allele fraction <0.5, many will likely exhibit loss of heterozygosity because tumor samples contain stromal and inflammatory components. A tumor-suppressor function is also supported by the observation that ARHGAP35 is located in a region of chromosome 19, which frequently is focally deleted in carcinomas (Zack et al., 2013).
Figure 1.
Mutations in ARHGAP35 encoding p190A in human cancer. (A) Frequency distribution of ARHGAP35 mutations in different types of human cancer. Numbers refer to the following published studies: (1) Kandoth et al. (2013); (2) Cancer Genome Atlas Research Network (2014); (3) Bass et al. (2014); (4) Collisson et al. (2014); (5) Cancer Genome Atlas Research Network (2012); (6) Cancer Genome Atlas Network (2012); (7) Bell et al. (2011); (8) Cancer Genome Atlas Research Network (2013); (9) Cancer Genome Atlas Network (2012); (10) Lohr et al. (2014). MM, multiple myeloma. (B) Pie chart illustrating relative frequencies of missense, nonsense, and frame-shift mutations as well as in-frame deletions and splice mutations in ARHGAP35 mutations in human cancer. (C) Diagram illustrating the location of p190A mutations and relative frequencies of corresponding mutant ARHGAP35 alleles in tumor samples. Each point in the figure indicates an individual somatic mutation from a dataset derived from ∼5,000 cancer patients. The allele fraction of a given mutation is estimated based on the ratio between read counts of the alternative alleles versus the total read counts including both alternative alleles and reference alleles. We highlight mutations with loss-of-function consequences (missense and nonsense/frameshift indel) with allelic fraction >0.5. RAS, GTPase domain; F, FF motifs; GAP, GAP domain.
Depletion of p190 RhoGAP is sufficient to activate Rho signaling in epithelial cells
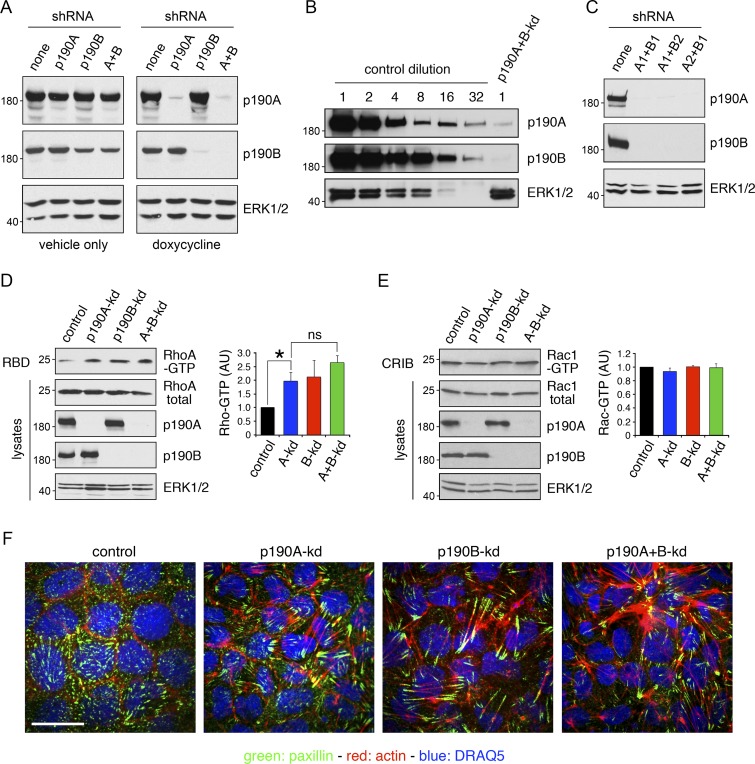
Next, to elucidate the effects of loss of p190A expression in epithelial oncogenesis, we used immortalized, but nontumorigenic, diploid MDCK cells. This model permits a detailed analysis of the function of p190A in epithelial cells without confounding effects resulting from oncogenic transformation. We included the paralog p190B in this analysis because p190B is 50% identical to p190A, structurally highly similar, and coexpressed with p190A in most tissues and cell types. Using previously published methods (Frank et al., 2012), we generated MDCK cells with doxycycline-inducible knockdown of p190A (p190A-kd), p190B (p190B-kd), or p190A combined with p190B (p190A+B-kd; Fig. 2 A). In this system, we achieved almost complete depletion of p190A and p190B (Fig. 2 B). Moreover, combinations of different shRNAs yielded highly effective knockdown of p190A and p190B (Fig. 2 C). Using these cell lines, we first determined the effects of depleting p190A and/or p190B on the activity of endogenous RhoA. To this end, we used the Rhotekin–Ras binding domain (RBD) pulldown assay to detect the active GTP-bound form of RhoA. p190A-kd or p190B-kd significantly increased cellular levels of GTP-bound RhoA, whereas p190A+B-kd yielded a modest additional increase (Fig. 2 D). p190A has also been reported to act as a GAP for Rac proteins in an environment of acidic phospholipids (Ligeti et al., 2004). However, using the PAK3-Cdc42/Rac interactive binding (CRIB) pulldown assay, we observed no changes in Rac-GTP levels upon p190A-kd and/or p190B-kd (Fig. 2 E). Furthermore, depletion of p190A and/or p190B elicited potent actin-stress fiber and focal adhesion formation (Fig. 2 F), which is consistent with activation of RhoA.
Figure 2.
p190A and/or p190B repress Rho signaling in epithelial cells. (A) MDCK TR-T10 cells expressing inducible shRNAs targeting p190A and/or p190B. Cells were incubated with either vehicle or doxycycline (dox) to induce knockdown of gene expression. Cells were cultured for 3 d at sparse density to achieve optimal knockdown followed by 5 d at high density. Whole-cell lysates were then subjected to Western blotting to detect p190A, p190B, or ERK1/2. All other figures show data from doxycycline-treated cells only. (B) Whole-cell lysates from control cells were subjected to serial twofold dilution. These samples, along with undiluted lysates from p190A+B-kd cells, were processed for Western blotting to detect p190A, p190B, and ERK1/2. (C) Knockdown of gene expression using different combinations of shRNAs targeting p190A or p190B. (D) Effects of p190A and/or p190B depletion on cellular RhoA-GTP levels were measured by a Rhotekin–RBD pulldown assay. Data are presented as mean ± SD (n = 3); *, P < 0.05 (n = 3). (E) Rac1-GTP levels in cells with p190A-kd and/or p190B-kd were determined by a PAK3-CRIB pulldown assay. Data are presented as mean ± SD (n = 3); *, P < 0.05 (n = 3). Molecular masses are given as kilodaltons. (F) Control, p190A-kd, p190B-kd, or p190A+B-kd cells were processed to localize the focal adhesion constituent paxillin (green), incubated with Alexa Fluor 594–phalloidin to detect polymerized actin (red), and stained with the DNA-intercalating dye DRAQ5 to show nuclei (blue). Bar, 10 µm.
p190A and p190B are essential for CIP in epithelial cells
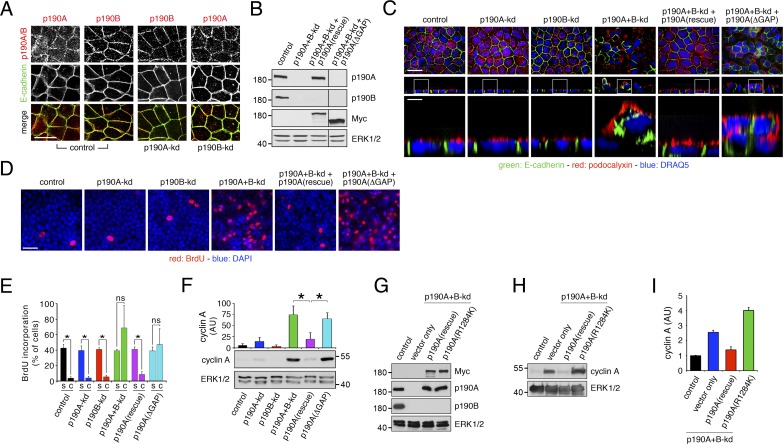
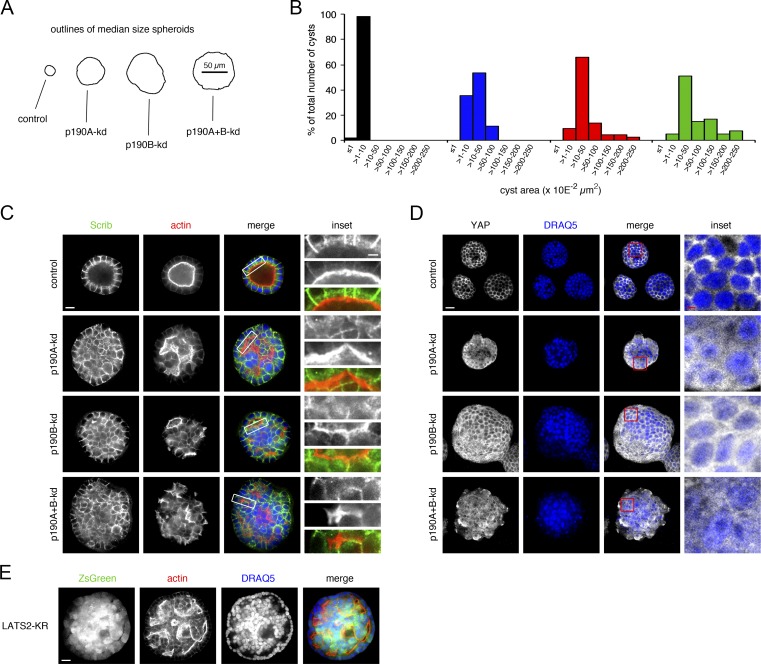
Phase-contrast imaging of confluent p190A+B-kd cell cultures revealed formation of foci-like structures that were not observed with control, p190A-kd, or p190B-kd cells (Fig. S1 A). To elucidate the nature of these structures, we first examined dense cultures of p190A-kd and/or p190B-kd cells grown on permeable supports. Both p190A and p190B localized to adherens junctions, and individual knockdown of p190A or p190B did not affect localization of the other paralog (Fig. 3 A). We next analyzed the effects of p190A and/or p190B depletion on epithelial architecture and included p190A+B-kd cells that had been reconstituted with either Myc-tagged, knockdown-resistant full-length p190A or p190A(ΔGAP), a mutant comprised of amino acids 1–1,228 and thus lacking the C-terminal GAP domain (Fig. 3 B). Knockdown of p190A or p190B individually had no major effects on epithelial architecture (Fig. 3 C). In contrast, p190A+B-kd cultures exhibited abundant areas with cell multilayering, an effect that was rescued by expression of knockdown-resistant full-length p190A but not p190A(ΔGAP) (Figs. 3 C and S1 A). Phalloidin staining revealed that multilayered cells were piled up many layers high and that several foci-like structures of multilayered cells contained lumens (Fig. S2, A–D). Combined knockdown of p190A and p190B also elicited extensive cell sloughing as demonstrated in cytospins of media harvested from cell cultures (Fig. S1 B). Again, these effects were rescued by expression of knockdown-resistant p190A but not p190A(ΔGAP) (Fig. S1 B). The sloughed cells were undergoing apoptosis as determined by TdT dUTP Nick-end labeling (TUNEL) staining (Fig. S1 B). Thus, sloughing may result from multilayered cells receiving insufficient survival signals upon segregation from the cell–substratum interface.
Figure 3.
p190A and p190B are essential for CIP in epithelial cells. (A) p190A and p190B localize in part to adherence junctions in MDCK cells grown on permeable supports. Knockdown of p190A and/or p190B was achieved as described in the legend to Fig. 1 A. Cells were then fixed and labeled to detect E-cadherin (green) and p190A or p190B (red). Bar, 10 µm. (B) Expression of knockdown-resistant Myc-tagged p190A or p190A(ΔGAP), which is missing the C-terminal GAP-domain, in p190A+B-kd cells. Whole-cell lysates were processed for Western blotting to detect p190A, p190B, Myc epitope, and ERK. The lines between lanes 3 and 4 indicate omission of two lanes from the original gel. The omitted lanes contained samples with additional clones expressing knockdown-resistant Myc-tagged p190A or p190A(ΔGAP). (C) Depletion of p190A and p190B together, but not individually, elicits cell multilayering. Exogenous expression of knockdown-resistant Myc-tagged p190A, but not a mutant from which the C-terminal GAP domain is deleted, restores normal epithelial architecture. Cells were labeled to localize E-cadherin (green) and podocalyxin (red); nuclei were stained with DRAQ5 (blue). Bars: (low magnification) 10 µm; (high magnification) 2 µm. (D) BrdU incorporation (red) in dense cultures of control, p190A-kd, p190B-kd, or p190A+B-kd cells as well as in p190A+B-kd cells expressing knockdown-resistant full-length p190A or GAP-deficient p190A(ΔGAP). Samples were counterstained with DAPI (blue) to detect cell nuclei. Bar, 20 µm. (E) Quantification of BrdU incorporation data are presented as mean ± SD (n = 3); *, P < 0.01. s, subconfluent culture; c, confluent culture. (F) Cyclin A expression in dense cultures of control, p190A-kd, p190B-kd, or p190A+B-kd cells with or without expression of knockdown-resistant p190A or p190A(ΔGAP). Whole-cell lysates were processed for Western blotting to detect cyclin A and ERK. Data are presented as mean ± SD (n = 4); *, P < 0.02. (G) Expression of Myc-tagged, knockdown-resistant, WT, or catalytically inactive p190A(R1284K) in MDCK cells depleted of p190A and p190B. Western blotting of whole-cell lysates was performed to detect Myc-epitope, p190A, p190B, and ERK1/2. (H) Cyclin A expression in whole-cell lysates from control and p190A+B-kd cells transduced with empty vector, Myc-tagged WT, or catalytically inactive p190A(R1284K). Molecular masses are given as kilodaltons. (I) Quantification by densitometry of cyclin A levels from the conditions described in H. The histograms show mean ± SD (n = 4).
Collectively, the data above suggest that cells with joint depletion of p190A and p190B fail to undergo CIP. To test this possibility directly, we measured incorporation of BrdU in subconfluent and confluent cultures of MDCK cells. Although control, p190A-kd, and p190B-kd cells underwent quiescence at confluent density, p190A+B-kd cells continued to incorporate BrdU at high levels, similar to subconfluent cultures (Fig. 3, D and E). We confirmed these results independently by analysis of cyclin A levels, which were low in control, p190A-kd, and p190B-kd cells upon reaching saturation densities. In contrast, cyclin A levels remained high in dense cultures of p190A+B-kd cells (Figs. 3 F and S1, C and D). BrdU incorporation and cyclin A levels in p190A+B-kd cells were normalized by expression of knockdown-resistant full-length p190A but not p190A(ΔGAP) (Fig. 3, D–F). A p190A(R1284K) construct containing a function-ablating mutation in the essential arginine residue of the GAP domain, which also occurs in a patient tumor sample (TCGA-CV-7425), similarly failed to restore contact inhibition in cells depleted of p190A and p190B (Fig. 3, G–I). These results establish that p190A and p190B collectively are essential to mediate CIP, at least in part through their function as RhoGAPs. Moreover, the data show that perturbation of CIP in p190A+B-kd cells results in cell multilayering and cell sloughing.
Given that Rho proteins regulate the formation of junctional complexes in epithelial cells (Baum and Georgiou, 2011), we hypothesized that p190A and p190B might exert their effects on CIP through adherens and/or tight junctions. However, cadherin-mediated cell–cell adhesion remained intact after depletion of p190A or p190B alone or together. This was indicated by localization of E-cadherin to membranes engaged in cell–cell contact (Fig. 3 C) as well as by aggregation assays (Fig. S3, A and B). Tight junctions were also intact after knockdown of p190A and/or p190B. This was determined by staining for the tight junction constituent ZO-1 and by measurements of transepithelial resistance (Fig. S3, C and D). Moreover, E-cadherin remained sequestered from the apical protein podocalyxin in p190A+B-kd cells (Fig. S3 E). Collectively, these results indicate that p190A and p190B do not promote CIP through major effects on cell–cell junctions or cell polarity.
p190A and p190B repress transcriptional activation by the Hippo transducer YAP
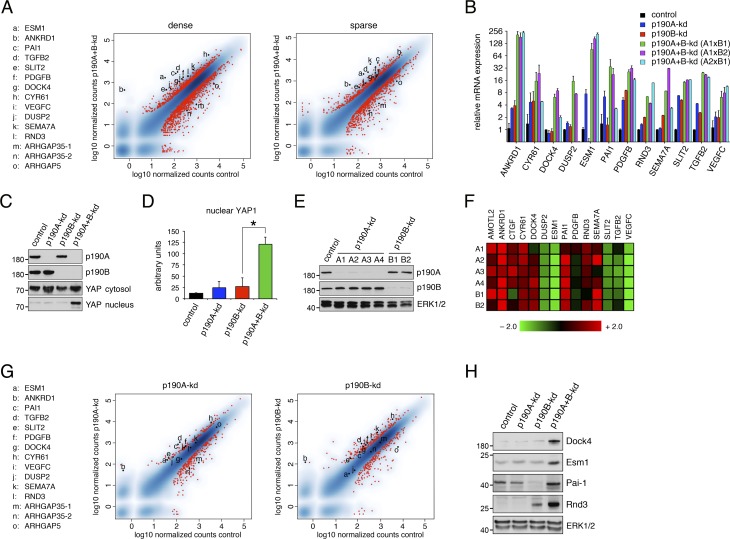
Both p190A and p190B have also been implicated in modulating transcriptional responses (Sordella et al., 2002; Jiang et al., 2005). Therefore, we considered that loss of CIP in p190A+B-kd cells might result from effects on gene expression. To test this possibility, we conducted genome-wide mRNA sequencing (mRNA-seq) analyses on MDCK cells with or without knockdown of p190A and/or p190B. These analyses were performed for both sparse and dense cultures. We focused on genes that exhibited altered expression in p190A+B-kd cells relative to control cells as well as cells with individual knockdown of p190A or p190B. Using criteria detailed in Materials and methods, we determined that 347 and 757 gene transcripts in dense cultures of p190A+B-kd cells were significantly up- and down-regulated, respectively (Tables S1 and S2). In sparse cultures of p190A+B-kd cells, 224 gene transcripts were up-regulated and another 224 gene transcripts down-regulated (Tables S3 and S4). Strikingly, genes that ranked at the top among up-regulated genes in both datasets included several that previously have been associated with Hippo signaling (Aragona et al., 2013; Mohseni et al., 2014). These genes include ANKRD1, PAI1, TGFB2, SLIT2, PDGFB, CYR61, SEMA7A, and RND3, which ranked 2, 3, 4, 8, 11, 13, 18, and 28, respectively, in dense p190A+B-kd cells (Table S1). These and other genes selected for validation are shown in Fig. 4 A. Importantly, the majority of these genes were ranked highly in both dense and sparse cells (Fig. 4 A), consistent with p190A+B-kd cells exhibiting a defect in sensing cell density. Induced expression of top ranked genes in the transcriptome of dense p190A+B-kd cells was validated by quantitative PCR (qPCR; Fig. 4 B).
Figure 4.
p190A and p190B repress expression of genes associated with the Hippo pathway. (A) Genome-wide mRNA-seq analyses from MDCK cells depleted of p190A and p190B expression reveal strong induction of genes associated with Hippo signaling in both dense and sparse cultures. The data are represented as x–y plots with relative gene expression levels in control cells and p190A+B-kd cells on the abscissa and ordinate, respectively. Red dots correspond with individual genes with differential gene expression between p190A+B-kd and control cells using a threshold of log2(fold change) ≥1 and adjusted P value ≤0.05. Genes selected for further validation as well as ARHGAP35 (two splice forms) and ARHGAP5 genes encoding p190A and p190B, respectively, are listed as a–o and marked individually as black dots. (B) Validation of transcriptomes from dense cultures of control, p190A-kd, p190B-kd, and p190A+B-kd cells by qPCR analysis. p190A and p190B depletion was achieved by three distinct combinations of shRNAs targeting p190A and p190B. Data are presented as mean ± SD (n = 3). (C) Knockdown of p190A and p190B promotes nuclear accumulation of YAP as determined by Western blotting to detect YAP1 in the nuclear fractions as well as p190A, p190B, and total YAP1 in the cytosolic fraction. (D) Quantification of nuclear YAP1 levels in dense cultures of control, p190A-kd, p190B-kd, or p190A+B-kd cells. Data are presented as mean ± SD (n = 4); *, P < 0.02. (E) Depletion of p190A or p190B in HEC-1-A uterine carcinoma cells as determined by Western blotting to detect p190A, p190B, and ERK. (F) Heat map representing relative fold change (linear scale) in gene expression levels in HEC-1-A cells depleted of p190A or p190B. Four distinct shRNAs (A1–A4) targeting p190A and two distinct shRNAs (B1 and B2) targeting p190B were used in this experiment. (G) Transcriptomes from MDCK cells depleted of either p190A or p190B expression show modest induction of Hippo-associated genes relative to control cells. The data are presented as described in A. Knockdown of either p190A or p190B expression was induced for 3 d at sparse cell density followed by replating at high cell density and continued culture for another 3 d, after which gene expression analysis was performed. The red dots correspond with individual genes with differential gene expression between p190A-kd or p190B-kd and control cells using a threshold of |log2(fold change)| ≥1 and adjusted P value ≤0.05. Genes are listed as a–o and marked individually as described in A. (H) Validation of gene expression elicited by depletion of p190A and/or p190B by Western blotting using antibodies specific for Dock4, Esm1, Pai-1, and Rnd3 with ERK1/2 as control for equal protein loading. Molecular masses are given as kilodaltons.
To further test whether p190A and p190B modulate Hippo signaling in MDCK cells, we assayed the effects of knocking down p190A and/or p190B on localization of YAP, a transcriptional coactivator that is a substrate of LATS kinases and a key transducer of Hippo signaling. Phosphorylation of YAP prevents its entry into the nucleus and thus inhibits its activity (Zhao et al., 2007). In control cells as well as p190A-kd or p190B-kd cells, we observed low to modest levels of YAP in the nuclear fraction isolated from confluent cell monolayers. In contrast, the amount of YAP was significantly increased in the nuclear fraction from p190A+B-kd cells (Fig. 4, C and D).
Current genome sequencing data do not support that p190A and p190B are simultaneously mutated and/or deleted in cancer. To address the possibility that p190A or p190B individually induces expression of genes modulated by the Hippo pathway, we chose human HEC-1-A uterine carcinoma cells for analysis because ARHGAP35 is highly mutated in uterine carcinoma (Fig. 1 A). Thus, we generated HEC-1-A cells with constitutive knockdown of p190A or p190B (Fig. 4 E). Indeed, depletion of either p190A or p190B was sufficient to augment expression of established Hippo-modulated genes including AMOTL2, ANKRD1, CTGF, CYR61, PAI1, RND3, and SEMA7A (Fig. 4 F). Expression of Hippo pathway–associated genes was also modestly up-regulated in genome-wide analyses of MDCK cells depleted of p190A or p190B alone (Fig. 4 G). Collectively, these results demonstrate a role for p190A and p190B as essential modulators of YAP-regulated gene expression in epithelial cells.
Finally, we were able to validate gene expression data by Western blotting to the extent that antibodies cross-reactive with canine gene products could be obtained (Fig. 4 H). Interestingly, this included strongly induced expression of Rnd3, which interacts directly with p190A and p190B and enhances their GAP activity (Wennerberg et al., 2003). ROCK-phosphorylated Rnd3 has been demonstrated to stabilize adherens junctions by sequestering p190B in the cytosol (Priya et al., 2015). However, Rnd3 evidently cannot exert this function in the absence of p190B, which may explain why adherens junctions appear intact in cells depleted of p190A and/or p190B. Rnd3 has moreover been implicated in promoting CIP (Hernández-Sánchez et al., 2015). Evidently, in spite of strongly induced expression of Rnd3, p190A+B-kd cells fail to reach quiescence at high cell density. Hence, if Rnd3 promotes CIP in MDCK cells, it may execute this function through its established effector proteins p190A and/or p190B.
p190A and p190B promote CIP through inhibition of YAP-mediated gene transcription
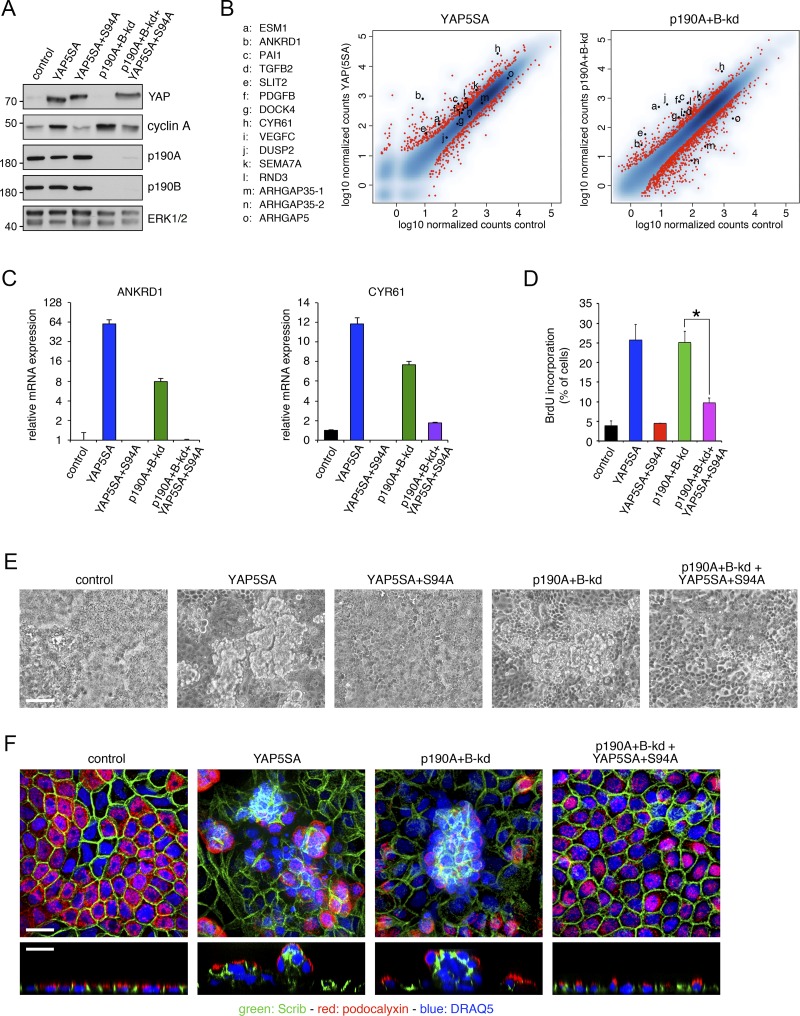
YAP serves as a coactivator for TEAD family transcription factors (Vassilev et al., 2001). Phosphorylation of YAP on five serines, including S127 and S397, prevents its entry into the nucleus and promotes its degradation by the β-TRCP E3-ligase (Zhao et al., 2007, 2010). Mutation of each of these five serines to alanine generates a YAP5SA mutant that is constitutively active (Zhao et al., 2007). Introduction of an additional S94A mutation in YAP5SA yields YAP5SA+S94A, which no longer can bind to TEAD family transcription factors and serves as a dominant inhibitory mutant (Zhao et al., 2008). To better define the extent to which p190A and p190B regulate transcription of Hippo-associated genes, we generated MDCK cells expressing YAP5SA (Fig. 5 A). We then performed mRNA-seq analysis to directly compare gene expression induced by YAP5SA to that of p190A+B-kd in the same experiment. Strikingly, ANKRD1, CYR61, ESM1, PAI1, RND3, SEMA7A, and SLIT2 were all strongly induced in both YAP5SA-expressing and p190A+B-kd cells (Fig. 5 B). To further validate that p190A and p190B repress YAP-mediated gene transcription, we generated MDCK cells expressing YAP5SA+S94A with or without knockdown of p190A and p190B (Fig. 5 A) and assayed the effects on transcript levels of the bona fide YAP target genes ANKRD1 and CYR61. Expression of YAP5SA+S94A strongly reduced expression levels of ANKRD1 and CYR61 transcripts in p190A+B-kd cells (Fig. 5 C).
Figure 5.
p190A and p190B promote CIP by repressing YAP-mediated gene transcription. (A) Expression of constitutively active YAP5SA or dominant-negative YAP5SA+S94A in control and p190A+B-kd MDCK cells. Western blotting of whole-cell lysates was performed to detect YAP1, cyclin A, p190A, p190B, and ERK. Molecular masses are given as kilodaltons. (B) mRNA-seq transcriptomes from dense cultures of MDCK cells expressing constitutively active YAP5SA and from p190A+B-kd cells were derived from samples processed concomitantly to permit direct comparison. The data are represented as described in the legend to Fig. 5 A. (C) qPCR analysis of ANKRD1 and CYR61 expression in control and p190A+B-kd cells with or without expression of YAP5SA or YAP5SA+S94A. Data are presented as mean ± SD (n = 3). (D) BrdU incorporation in dense cultures of control and p190A+B-kd cells with or without expression of YAP5SA or YAP5SA+S94A. Data are presented as mean ± SD (n = 4); *, P < 0.01. (E) Phase-contrast imaging of control, YAP5SA, YAP5SA+S94A, and p190A+B-kd cells as well as p190A+B-kd cells expressing YAP5SA+S94A. Bar, 50 µm. (F) Confocal microscopy of control, YAP5SA, and p190A+B-kd cells as well as p190A+B-kd cells expressing YAP5SA+S94A. Cells were labeled to detect Scrib (green) and podocalyxin (red), and nuclei were stained with DRAQ5 (blue). Bars, 10 µm.
Next, we tested whether the capacity of p190A and p190B to repress YAP-mediated gene transcription is responsible for CIP in epithelial cells. Expression of YAP5SA alone led to loss of CIP as indicated by increased expression of cyclin A and elevated BrdU incorporation at high cell density, effects that mimic those observed in p190A+B-kd cells (Fig. 5, A and D). Moreover, as previously described (Zhao et al., 2011), expression of YAP5SA in MDCK cells elicited extensive cell multilayering (Fig. 5, E and F). In contrast, expression of YAP5SA+S94A in p190A+B-kd cells effectively reduced both cyclin A levels and BrdU incorporation in comparison with p190A+B-kd cells (Fig. 5, A and D). Expression of YAP5SA+S94A moreover attenuated multilayering in cultures of p190A+B-kd cells (Fig. 5, E and F). Collectively, these data establish that p190A and p190B induce CIP by repressing YAP-mediated gene transcription.
Rho–ROCK signaling is required for loss of CIP in MDCK cells depleted of p190A and p190B
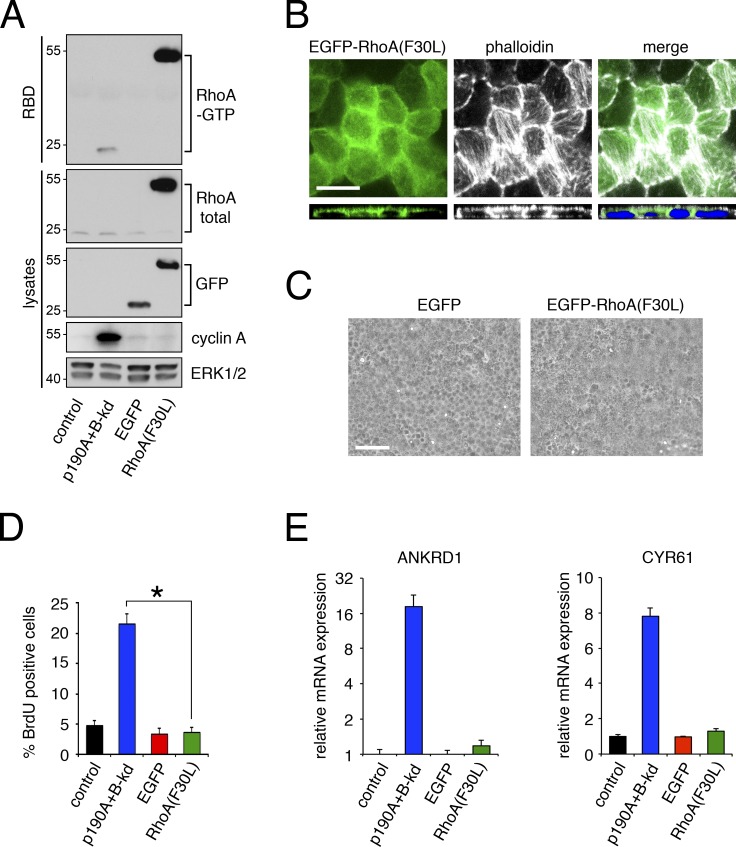
Mechanotransduction by Rho–ROCK-dependent actomyosin contractility has previously been shown to induce expression of YAP-regulated genes (Berenjeno et al., 2007; Dupont et al., 2011). Therefore, we tested whether Rho–ROCK signaling was sufficient and/or required for YAP-mediated gene transcription and loss of CIP in MDCK cells. To mimic the loss of RhoGAP activity, we generated MDCK cells with doxycycline-inducible expression of “fast-cycling” RhoA(F30L) (Fig. 6 A). Expression of RhoA(F30L) led to a major increase in RhoA-GTP levels and potent formation of actin stress fibers (Fig. 6, A and B). However, RhoA(F30L) expression did not perturb CIP as indicated by the absence of cell multilayering, foci-like structures or increased BrdU incorporation (Fig. 6, B–D). In addition, expression of RhoA(F30L) did not induce expression of ANKRD1 or CYR61 (Fig. 6 E). These results are consistent with published studies on MDCK cells, which demonstrate that expression of constitutively active forms of RhoA, G14V, and Q63L do not induce cell multilayering (Takaishi et al., 1997; Jou and Nelson, 1998; Hansen et al., 2000).
Figure 6.
Active RhoA is not sufficient to perturb CIP or induce expression of YAP-regulated genes. (A) RhoA-GTP levels in MDCK cells expressing EGFP-tagged fast-cycling RhoA(F30L) or EGFP only relative to p190A+B-kd or control cells. GTP-bound RhoA was precipitated by a Rhotekin–RBD pulldown assay followed by immunoblotting to detect active and total RhoA as well as GFP, cyclin A, and ERK. Molecular masses are given as kilodaltons. (B) Effects of expressing EGFP-RhoA(F30L) (green) on actin stress fiber formation detected by staining with Alexa Fluor 594–phalloidin (white). Nuclei were labeled with DRAQ5 (blue). Bar, 10 µm. (C) Phase images of cells expressing EGFP-RhoA(F30L) or EGFP only. Bar, 50 µm. (D) BrdU incorporation in dense monolayers of cells expressing EGFP-RhoA(F30L) or EGFP only. p190A+B-kd and control cells were included for comparison. Data are presented as mean ± SD (n = 4); *, P < 0.02. (E) qPCR analysis of ANKRD1 and CYR61 expression in cells expressing EGFP-RhoA(F30L) or EGFP only relative to p190A+B-kd or control cells. Data are presented as mean ± SD (n = 3).
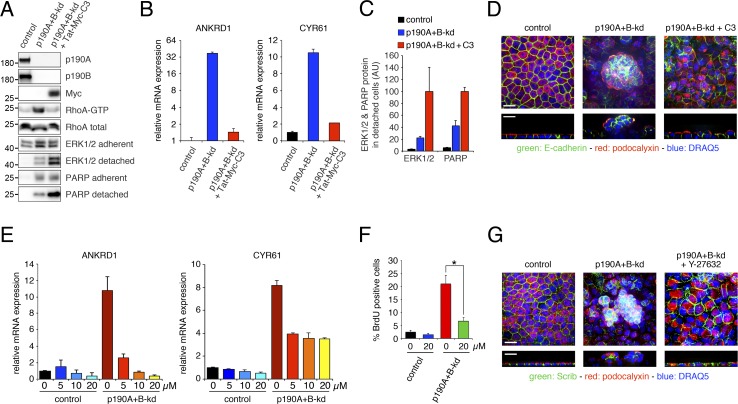
We then sought to determine whether Rho signaling is required for bypassing CIP in confluent cultures of p190A+B-kd cells. Various attempts to achieve stable or inducible expression of dominant negative RhoA in MDCK cells were unsuccessful. Instead, we used Tat-derivatized and Myc-tagged Clostridium botulinum C3 (Tat-Myc-C3) toxin to acutely inactivate endogenous Rho proteins. Upon overnight incubation, Tat-Myc-C3 toxin entered p190A+B-kd cells and efficiently depleted RhoA-GTP levels (Fig. 7 A). Treatment with Tat-Myc-C3 toxin strongly reduced expression of ANKRD1 and CYR61 induced by p190A+B-kd (Fig. 7 B). It moreover led to dramatic sloughing of cells depleted of p190A and p190B, thus demonstrating that Rho signaling is required not only for bypass of CIP but also for survival of multilayered cells. Given that the integrity of sloughed cells is disrupted, we used Western blotting for extracellular signal-related kinase (ERK) protein from harvested cell culture medium as a proxy readout for the amounts of sloughed cells. Treatment with Tat-Myc-C3 toxin led to a significant increase in the amount of cell sloughing in p190A+B-kd cells (Fig. 7 C). Moreover, the sloughed cells were undergoing apoptosis as indicated by a significant increase in the amount of cleaved poly (ADP-ribose) polymerase (PARP) protein in medium harvested from p190A+B-kd cells after incubation with Tat-Myc-C3 toxin (Fig. 7 C). Confocal microscopy demonstrated that cell multilayering was substantially reduced after treatment with Tat-Myc-C3 toxin (Fig. 7 D). Collectively, these results demonstrate that Rho signaling is essential for YAP-regulated gene transcription, protection against apoptosis, and cell multilayering in MDCK cells depleted of p190A and p190B. It should be noted that it was not formally possible to assess CIP in cells treated with C3 toxin because of toxicity upon prolonged exposure.
Figure 7.
Rho–ROCK signaling promotes YAP-mediated gene transcription and loss of CIP in cells depleted of p190A and p190B. (A) Delivery of Tat-Myc-C3 transferase (Tat-Myc-C3) protein to p190A+B-kd cells. Control or p190A+B-kd cells were incubated with estimated equal amount of GST only or cleaved Tat-Myc-C3 for 18 h. Western blotting of whole-cell lysates was performed to detect Tat-Myc-C3 protein as well as p190A, p190B, RhoA, ERK1/2, and cleaved PARP fragment. Lysates of detached cells and cellular fragments were processed for Western blotting to detect ERK1/2 and cleaved PARP fragment. Moreover, RBD pulldown was performed to detect levels of Rho-GTP in parallel cultures. Molecular masses are given as kilodaltons. (B) qPCR analysis of ANKRD1 and CYR61 expression in control and p190A+B-kd cells with or without uptake of Tat-Myc-C3 protein. Data are presented as mean ± SD (n = 3). (C) Quantification by densitometry of relative amounts of ERK1/2 and cleaved PARP protein in medium harvested from control and p190A+B-kd cells incubated with GST only or cleaved Tat-Myc-C3. (D) Confocal microscopy of control and p190A+B-kd cells with or without incubation with Tat-Myc-C3 protein. Cells were labeled to detect E-cadherin (green) and podocalyxin (red), and nuclei were stained with DRAQ5 (blue). (E) qPCR analysis of mRNA levels in control and p190A+B-kd cells treated with 0–20 µM ROCK inhibitor Y-27632 for 24 h. (F) BrdU incorporation in control or p190A+B-kd cells incubated with 0 or 20 µM Y27632 for 3 d at low cell density followed by 5 d at high cell density. Data are presented as mean ± SD (n = 4); *, P < 0.02. (G) Confocal microscopy of control and p190A+B-kd cells as well as p190A+B-kd cells treated with 20 µM Y-27632 during 5 d of culturing on permeable supports. Bars, 10 µm.
Next, we tested a requirement for ROCK signaling in loss of CIP and YAP-mediated gene transcription in MDCK cells depleted of p190A and p190B. Incubation of confluent p190A+B-kd cells with a range of 5–20 µM of the ROCK inhibitor Y-27632 for 24 h yielded substantial concentration-dependent reductions in the expression of ANKRD1 and CYR61 (Fig. 7 E). Furthermore, prolonged treatment of p190A+B-kd cells with 20 µM Y-27632 significantly reduced BrdU incorporation (Fig. 7 F) as well as cyclin A levels (Fig. S4, A and B). In addition, incubation with ROCK inhibitor attenuated multilayering in p190A+B-kd cell cultures (Figs. 7 G and S4 C). In comparison, the effects of inhibiting Rho–ROCK signaling in MDCK cells without depletion of p190A and p190B were relatively subtle (Fig. S4, D–G). Collectively, our results are consistent with the conclusion that p190A and p190B modulate YAP-mediated gene expression and associated cellular effects in a manner for which Rho–ROCK signaling is essential.
Modulation of CIP and YAP-mediated gene expression by p190A and p190B is LATS dependent
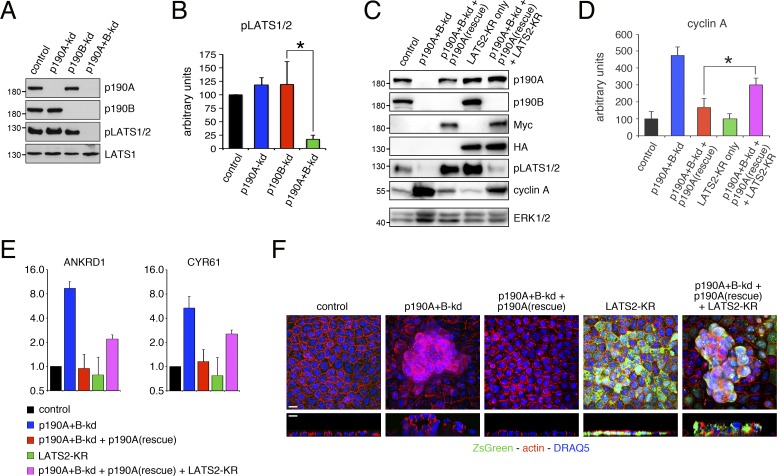
The data presented above demonstrate that Rho–ROCK signaling is required but not sufficient to promote YAP-mediated gene transcription in MDCK cells depleted of p190A and p190B. YAP is a substrate of the AGC family kinases LATS1 and LATS2, and canonical Hippo signaling is defined as LATS1/2 dependent (Meng et al., 2016). To determine whether p190A and p190B modulate Hippo signaling in MDCK cells, we performed Western blots for phospho-S909/S872 in the activation loop of LATS1/2. Depletion of p190A or p190B alone did not elicit major effects on phosphorylation of S909/S872 in LATS1/2 in confluent monolayers. However, combined knockdown of p190A and p190B led to a significant decrease in LATS1/2 S909/S872 phosphorylation (Fig. 8, A and B). Thus, expression of p190A and p190B is required for LATS1/2 phosphorylation in MDCK cells.
Figure 8.
p190A and p190B activate LATS, and p190A promotes CIP in a LATS-dependent manner. (A) Depletion of p190A and p190B inactivates LATS. pLATS1/2 refers to both LATS1(pS909) and LATS2(pS872) because the antibody does not distinguish between the two paralogs. (B) Quantification of phospho-LATS1/2 levels in dense cultures of control, p190A-kd, p190B-kd, or p190A+B-kd cells. (C) Expression of knockdown-resistant p190A in p190A+B-kd cells promotes CIP in a LATS-dependent manner. Molecular masses are given as kilodaltons. (D) Quantification of cyclin A levels in control and p190A+B-kd cells with or without expression of knockdown-resistant p190A and with and without expression of kinase-dead LATS2(K655R) (LATS2-KR). Data are presented as mean ± SD (n = 4); *, P < 0.02. (E) qPCR analysis of ANKRD1 and CYR61 expression in control and p190A+B-kd cells with or without expression of LATS2-KR. Data are presented as mean ± SD (n = 3). (F) Confocal microscopy of control and p190A+B-kd cells without or with expression of knockdown-resistant p190A as well as with and without expression of LATS2-KR. Cells were stained with Alexa Fluor 594–phalloidin and DRAQ5 to detect polymerized actin (red) and nuclei (blue). Cells expressing LATS2-KR coexpress ZsGreen from an IRES cassette (green). Bars, 10 µm.
We then sought to establish whether LATS1/2 activation is necessary for p190A to promote CIP and repress YAP-mediated gene transcription. Specifically, we asked whether the induction of CIP and attenuation of YAP-regulated gene transcription in p190A+B-kd cells by knockdown-resistant p190A is LATS dependent. To this end, we engineered control or p190A+B-kd cells to express HA-tagged kinase-dead murine LATS2(K655R), abbreviated as LATS2-KR, upstream of an IRES-ZsGreen cassette (Fig. 8 C). Expression of LATS2-KR to interfere with endogenous LATS signaling has been devised and validated previously (Zhao et al., 2007). Using this strategy for MDCK cells cultured on permeable supports, we found that expression of LATS2-KR in p190A+B-kd cells perturbed the capacity of exogenous knockdown-resistant p190A to restore CIP and suppress YAP-mediated gene transcription (Fig. 8, C–E). Moreover, in p190A+B-kd cells restored with exogenous knockdown-resistant p190A, expression of LATS2-KR elicited cell multilayering (Fig. 8 F). In contrast, expression of LATS2-KR on its own did not perturb CIP or affect YAP-regulated gene expression in MDCK cells. Thus, neither cyclin A levels nor ANKRD1 or CYR61 transcripts were noticeably altered by LATS2-KR expression alone, which moreover did not impact epithelial architecture detectably (Fig. 8, C–F). The latter result is consistent with mechanotransduction serving a permissive role for LATS signaling to YAP as demonstrated previously (Aragona et al., 2013); i.e., in the absence of strong cytoskeletal tension, inhibition of LATS is insufficient to drive YAP activation. Collectively, the data demonstrate that in addition to modulating mechanotransduction, p190A controls CIP via canonical, i.e., LATS-dependent, Hippo signaling. These results may furthermore explain why Rho signaling, while required for loss of CIP in response to p190A+B-kd, is not sufficient to promote YAP-mediated gene transcription and bypass CIP in cells depleted of p190A and p190B.
Both p190A and p190B are required for epithelial architecture of MDCK cells cultured in Matrigel
Cells with preserved epithelial characteristics such as MDCK cells form cysts when suspended individually and propagated in Matrigel. Hippo signaling is highly context dependent. Therefore, as an alternative to permeable supports, we tested the effects of p190A and/or p190B depletion in MDCK cells cultured in Matrigel. Strikingly, although control cells formed cysts with well-defined lumens, depletion of p190A or p190B alone or combined resulted in a significant increase in spheroid size indicative of perturbation of CIP (Fig. 9, A and B). Moreover, spheroids formed by p190A- and/or p190B-depleted MDCK cells were for the most part solid and thus without well-defined lumens, a trait associated with transformation of epithelial cells (Fig. 9 C). The formation of largely solid spheroids from cells with knockdown of p190A and/or p190B correlated with nuclear translocation of YAP (Fig. 9 D). This was most evident from optical sections sampled from the periphery of spheroids but was also clearly visible when sectioning through the center (Fig. S5 A). The sufficiency of individual p190A or p190B knockdown in eliciting nuclear translocation of YAP and perturbation of CIP in cells grown in Matrigel underscores the established importance of extracellular matrix in modulation of Hippo signaling (Serrano et al., 2013). Consistent with mechanical properties of Matrigel playing a key role in influencing the balance between canonical Hippo signaling and mechanotransduction, expression of LATS2-KR alone in MDCK cells led to formation of mostly solid spheroids with larger-than-normal diameter (Fig. 9 E).
Figure 9.
Both p190 RhoGAP paralogs are required for normal epithelial architecture of MDCK cells propagated in Matrigel. (A) Outlines of median-size spheroids derived from MDCK cells with or without depletion of p190A and/or p190B cultured in Matrigel for 14 d. (B) Size (area) distribution of spheroids with or without p190A and/or p190B knockdown after 14 d of culture in Matrigel. The areas from ≥40 cysts from each condition were measured. (C) Immunolabeling for Scrib (green) and phalloidin staining to detect polymerized actin (red) in spheroids with or without p190A and/or p190B depletion after 10 d of culture in Matrigel. Nuclei were stained with DRAQ5 (blue). Bars: (low magnification) 10 µm; (inset) 2.5 µm. (D) Localization of YAP in MDCK cell spheroids with or without knockdown of p190A and/or p190B after 14 d of culture in Matrigel. The images for YAP (white) were superimposed on the DRAQ5 stain (blue) using the screen function in Photoshop. Optical sections close to the perimeter of spheroids are shown. Bars: (low magnification) 20 µm; (inset) 5 µm. (E) Staining of LATS2-KR–expressing MDCK cells cultured for 14 d in Matrigel with Alexa Fluor 594–phalloidin to detect polymerized actin (red) and DRAQ5 to label nuclei (red). LATS2 and ZsGreen are coexpressed from a bicistronic vector. Bar, 20 µm.
In spite of the striking effects of individual p190A/B knockdown on spheroid architecture in Matrigel, we observed no major effects on adherens junctions consistent with findings for cells grown on permeable supports. Moreover, cells facing mostly small and/or slit-shaped lumens in p190A- and/or p190B-depleted spheroids exhibited strong subluminal phalloidin staining, a characteristic of apical membranes in epithelial cysts (Fig. 9 C). In such cells, Scrib was confined to membranes engaged in cell–cell contact. E-cadherin and β-catenin were similarly localized to membranes engaged in cell–cell contact (Fig. S5 B). Collectively, the data obtained with MDCK cells cultured in Matrigel point to an essential role for p190A and p190B to control CIP by impacting Hippo signaling and are consistent with a tumor-suppressor role for p190A in epithelial cells.
Discussion
Two recent comprehensive studies identified ARHGAP35, which encodes p190A RhoGAP, as a major human cancer gene (Kandoth et al., 2013; Lawrence et al., 2014). This observation was unexpected because ARHGAP35 had not been strongly linked to cancer, and yet it ranked among the top 30 cancer genes. The high frequency of nonsense and frame-shift mutations in ARHGAP35 is indicative of a role for p190A as tumor suppressor. Importantly, the ARHGAP35 locus is found in a region of the human genome that is frequently lost in tumors (Zack et al., 2013). Thus, loss-of-function mutations in one allele of ARHGAP35 may be sufficient to eliminate p190A signaling in such tumors. This hypothesis is validated by our demonstration that loss-of-function mutations occur in human tumors with loss of heterozygosity. Previously, p190A was shown to suppress Ras-induced transformation of NIH3T3 fibroblast cells (Wang et al., 1997) as well as to inhibit PDGF-induced glioma formation in mice (Wolf et al., 2003). Recently, loss-of-function mutations in p190A were also linked to impairment of directional motility (Binamé et al., 2016). However, cellular functions of p190A consistent with a tumor-suppressor role are not well defined.
In this study, we demonstrate that p190A and its paralog p190B play an essential role in CIP, an established barrier for oncogenic transformation (Hanahan and Weinberg, 2011). A detectable fraction of both p190A and p190B localize to lateral membranes of polarized MDCK cells. A simple mechanism whereby p190A and p190B might promote CIP would be to induce and/or enhance RhoA-dependent formation of adherens and/or tight junctions. However, depletion p190A and p190B does not appear to perturb cell–cell junctions or cell polarity. In absence of such effects, we considered the possibility that p190A and p190B might affect gene transcription as there is precedent in the literature for such a role (Sordella et al., 2002; Jiang et al., 2005). We determined that p190A and p190B repress expression of a cassette of genes that are regulated by the transcriptional coactivator YAP, a transducer of the Hippo pathway (Camargo et al., 2007; Dong et al., 2007). Accordingly, depletion of p190A and p190B expression elicits translocation of YAP to the nucleus and activation of a YAP-dependent transcriptional program. We demonstrate that suppression of YAP-regulated gene transcription underlies the capacity of p190A and p190B to induce CIP. This result is consistent with a tumor-suppressor function of p190A as YAP is an oncoprotein that plays a pivotal role in human cancer (Moroishi et al., 2015; Zanconato et al., 2016).
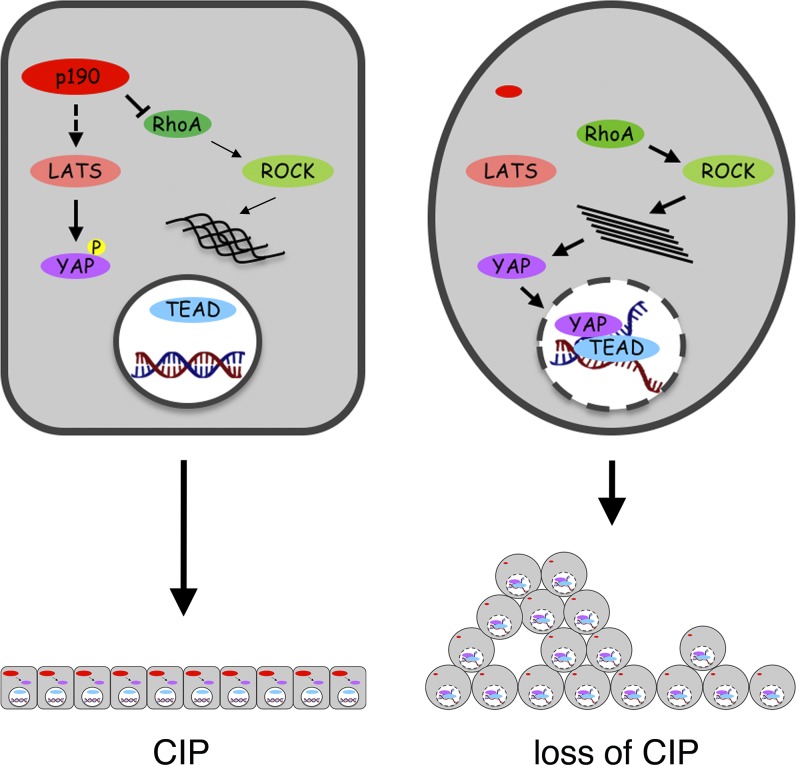
YAP activity is regulated by both the canonical Hippo pathway and by mechanotransduction, which is defined as ROCK dependent (Dupont et al., 2011; Aragona et al., 2013). Our data demonstrate that in MDCK cells, p190A and p190B promote hydrolysis of GTP-bound RhoA but not Rac1. We moreover show that catalytic activity is essential for exogenous p190A to induce CIP in dense epithelial cell cultures depleted of endogenous p190A and p190B. Furthermore, we establish that Rho–ROCK signaling plays a major role in eliciting transcription of YAP-regulated genes upon knockdown of p190A and p190B. However, elevated cellular levels of GTP-bound RhoA alone are not sufficient to bypass CIP as cells expressing highly active fast-cycling RhoA fail to promote transcription of YAP-regulated genes and undergo quiescence at high cell density. Moreover, we and others have not observed loss of CIP upon expression of constitutively active forms of RhoA in MDCK cells (Takaishi et al., 1997; Jou and Nelson, 1998; Hansen et al., 2000). Therefore, in addition to catalytic activity, nonenzymatic functions, i.e., scaffolding activities of p190A and p190B, are required for inducing CIP in dense cultures. Such scaffolding activities likely act upstream of LATS as indicated by our demonstration that p190A and p190B promote LATS activation and that LATS is required for induction of CIP in p190A+B-kd cells reconstituted with knockdown-resistant p190A. Fig. 10 presents a model for how p190 RhoGAPs control CIP based on our current knowledge. Further work is required to define scaffolding functions of p190 RhoGAPs, an arduous undertaking given the sheer size and relative scarcity of defined structural domains.
Figure 10.
Model illustrating the role of p190 RhoGAPs to promote CIP by repressing YAP activity. This cartoon summarizes our findings for how p190A and p190B modulate CIP through effects on YAP-regulated gene transcription in MDCK cells cultured on permeable supports. Left: A cell with normal expression of p190A and p190B. The activities of p190 RhoGAPs in this context are dual. The catalytic function of p190 proteins represses Rho–ROCK signaling to attenuate mechanotransduction signals. Concomitantly, most likely through scaffolding activities, p190 proteins inhibit LATS activation. The net result is repression of YAP activation and YAP–TEAD-regulated gene transcription to promote CIP. Right: A cell depleted of p190A and p190B. In this cell, LATS is inactive, which combined with potent mechanotransduction signaling leads to YAP activation, transcription of YAP–TEAD-regulated genes, and escape from CIP, resulting in cell multilayering at high cell density. Importantly, inhibition of LATS signaling as well as Rho–ROCK activation is required to reach a threshold sufficient to trigger escape from CIP. The relative importance of the dual regulation may be context dependent.
In this work, we relied on a nononcogenically transformed epithelial cell system to study the role of p190A and p190B in CIP. This is essential because carcinoma cells exhibit loss of CIP to varying degrees. The data reveal that p190A and p190B possess similar functions to repress YAP-mediated transcription and induce CIP in epithelial cells. The apparent redundancy may confer an additional level of safeguarding against deregulated growth resulting from loss of a single paralog. However, only ARHGAP35 encoding p190A is significantly mutated or lost in cancer. This raises the question of why loss of p190A but not p190B might initiate, promote, and/or sustain oncogenic transformation. The catalytic activity of p190B is higher than that of p190A (Vincent and Settleman, 1999). Accordingly, p190B may not be dispensable for cancer cell survival. Individual knockdown of p190A or p190B is sufficient to elicit modest increases in transcription of YAP-regulated genes in both MDCK cells and HEC-1-A uterine carcinoma cells. These results suggest that there is a threshold for induced expression of YAP-regulated genes to bypass CIP, which is reached in MDCK cells cultured on permeable supports only upon combined depletion of p190A and p190B but not individual depletion. However, in cells with activating mutations in oncogenes or loss-of-function mutations in tumor-suppressor genes, the threshold for inducing loss of CIP may be lower and loss of p190A expression therefore sufficient.
p190A and p190B exhibit 50% sequence identity but also large stretches of divergent sequence and separate modes of regulation. Another possibility is that in epithelial cells undergoing oncogenic transformation, p190B expression, localization, and/or activity may be altered in a manner that leaves the cell more responsive to loss of p190A. Alternatively, ARHGAP35 mutations may occur at a later stage when cancer cells are surrounded by a more rigid matrix, which lowers the threshold for robust YAP activation through inhibition of the canonical Hippo pathway. Our results obtained with culturing MDCK cells in Matrigel are compatible with this scenario. In this setting, loss of a single p190 paralog is sufficient to elicit architectural changes associated with oncogenic transformation. It is pertinent to direct future studies to better understand the role of ARHGAP35 as a major novel cancer gene and use this information to develop personalized therapeutics for cancer. In this context, the potential to perturb the YAP–TEAD interaction as demonstrated previously (Liu-Chittenden et al., 2012; Jiao et al., 2014) may prove relevant in tumors with loss-of-function mutations in ARHGAP35.
Materials and methods
Reagents
The following reagents were used for this work: BrdU (Sigma-Aldrich); doxycycline (Sigma-Aldrich); DAPI (Thermo Fisher Scientific); DRAQ5 (Biostatus); FluorSave (EMD Millipore); FuGENE6 (Promega); phalloidin Alexa Fluor 594 (Thermo Fisher Scientific); PageRuler prestained protein ladder 10–180 kD (Thermo Fisher Scientific); EDTA-free Pierce protease inhibitor tablets (Thermo Fisher Scientific); and ROCK inhibitor (Y-27632; Sigma-Aldrich).
Antibodies
Antibodies used for this study are listed in Table 1. Secondary antibodies purchased from Invitrogen were as follows: goat anti–mouse/Alexa Fluor 488; goat anti–mouse/Alexa Fluor 555; goat anti–rabbit/Alexa Fluor 488; goat anti–rat/Alexa Fluor 488; goat anti–mouse/HRP; and goat anti–rabbit/HRP.
Table 1. Antibodies used.
| Antigen | Species | Company | Catalog no. (or clone) |
|---|---|---|---|
| β-Catenin | Mouse monoclonal | BD | 610153 |
| BrdU | Mouse monoclonal | EMD Millipore/Sigma-Aldrich | 05-633 |
| Cyclin A | Rabbit polyclonal | Santa Cruz Biotechnology, Inc. | sc-751 |
| DOCK4 | Rabbit polyclonal | Abcam | ab85723 |
| E-cadherin | Rabbit polyclonal | Cell Signaling Technologies | 3195 |
| E-cadherin | Rat monoclonal | EMD Millipore/Sigma-Aldrich | MABT26 |
| ERK1 | Mouse monoclonal | Santa Cruz Biotechnology, Inc. | sc-271269 |
| ERK1/2 | Rabbit polyclonal | Santa Cruz Biotechnology, Inc. | sc-94 |
| ESM1 | Rabbit polyclonal | Abcam | ab103590 |
| HA tag | Mouse monoclonal | Hansen laboratory, Boston Children's Hospital, Boston, MA | clone 12CA5 |
| LATS1 | Rabbit monoclonal | Cell Signaling Technologies | 3477S |
| pLATS1/2 | Rabbit monoclonal | Cell Signaling Technologies | 9157S |
| Myc tag | Mouse monoclonal | Cell Signaling Technologies | 2276 |
| p190A | Mouse monoclonal | BD | 610150 |
| p190A | Rabbit polyclonal | Bethyl Laboratories | A304-549A |
| p190B | Mouse monoclonal | BD | 611613 |
| p190B | Rabbit polyclonal | Bethyl Laboratories | A301-736A |
| PAI1 | Mouse monoclonal | Finsen Laboratory, Rigshospitalet, Copenhagen, Denmark | N/A |
| PARP | Mouse monoclonal | BD | 611039 |
| Rac1 | Mouse monoclonal | BD | 610651 |
| RhoA | Mouse monoclonal | Santa Cruz Biotechnology, Inc. | sc-418 |
| Rnd3 | Mouse monoclonal | EMD Millipore/Sigma-Aldrich | 05-723 |
| Scrib | Rabbit polyclonal | Santa Cruz Biotechnology, Inc. | sc-28737 |
| YAP1 | Mouse monoclonal | Santa Cruz Biotechnology, Inc. | sc-101199 |
| YAP1 | Rabbit polyclonal | Novus Biologicals | NB110-58358 |
| ZO-1 | Mouse monoclonal | Invitrogen | 33-9100 |
Preparation of Tat-Myc-C3 transferase protein
Tat-Myc-C3 transferase was cloned into pGEXTEV vector (gift from K. Orth, University of Texas Southwestern, Dallas, TX). Plasmid was transformed into Rosetta2DE3pLYS bacterial cells, plated on Luria-Bertani agar plates, and incubated at 37°C overnight. The next day, bacteria scraped from the plates were used for a starter culture and incubated for 3–4 h at 37°C. The starter culture was inoculated to larger flasks at a 1:100 volume ratio and grown to OD600 = 0.6–1.0. Protein expression was induced with 0.4 mM IPTG, and the bacteria were grown for 16 h at RT. The next day, the bacteria were spun at 6,000 g for 10 min and harvested and resuspended in 20 mM Tris, pH 8, 150 mM NaCl, 0.5% Triton X-100, and 1 mM PMSF. The bacterial suspension was lysed with Emulsiflex-C3 (Avestin). The lysate was centrifuged at 15,000 g for 10 min, and supernatant was transferred to a tube containing glutathione resin (Goldbio) and incubated at 4°C on a rotating platform for 1 h. The supernatant was removed, and the beads were transferred to an Econo-Pac column (Bio-Rad Laboratories) and washed with 20 mM Tris, pH 8, 150 mM NaCl, and 0.5% Triton X-100. Next, to cleave the GST moiety, His-TEV protease was added and incubated at 4°C overnight. The next day, cobalt agarose resin (Goldbio) was added to remove His-TEV protease, and flow-through containing cleaved Tat-Myc-C3 toxin was collected. The protein was dialyzed to 20 mM Tris, pH 8, and 150 mM NaCl, and aliquots were flash-frozen with liquid nitrogen and stored at −80°C.
Cell culture
TR-T10 MDCK cells permitting conditional knockdown of gene expression have been described previously (Frank et al., 2012). TR-T10 cells as well as retroviral and lentiviral 293 packaging cells were propagated in DMEM supplemented with 10% FBS, and HEC-1-A cells were grown in McCoy’s medium supplemented with 10% FBS. For culture in Matrigel, ∼3,000 cells were mixed thoroughly in 30 µl ice-cold Matrigel and placed onto collagen-coated coverslips for 15–30 min to harden before addition of cell culture medium. Cysts/spheroids were cultured for 10–14 d before fixation, labeling, and imaging.
Plasmid constructs
For RNA interference, complementary 80–87-base oligonucleotides containing BglII and HindIII overhangs were annealed and cloned into tetracycline-inducible pReSI-H1-Puro or pReSI-H1-Hygro retroviral expression vectors described previously (Frank et al., 2012). Sequences of shRNA sense oligonucleotides used for MDCK cells in this work are as follows: canine p190A shRNA 1, 5′-GATCCCGGAACAGCGATTTAAAGCATTGCCCGGGCCGCCCTCAATGCTTTAAATCGCTGTTCCTTTTTTGGAATCGATA-3′; and shRNA 2, 5′-GATCCCGCACCTTAGTGCAACTCATTGGCCCGGGCCGCCCTCCAATGAGTTGCACTAAGGTGCTTTTTTGGAATCGATA-3′; canine p190B shRNA 1, 5′-GATCCCGGACGAGTAGTAAACAATGTTGCTTCCTGTCACAACATTGTTTACTACTCGTCCTTTTTTGGAACTCGAGA-3′; and shRNA 2, 5′-GATCCCGGTAACTGTGGAGTTGGAATTGCTTCCTGTCACAATTCCAACTCCACAGTTACCTTTTTTGGAACTCGAGA-3′; and α-catenin shRNA 1, 5′-GATCCCGGCTAACAGAGACCTGATATTCGGGCCCGGCGGGAGAATATCAGGTCTCTGTTAGCCTTTTTTGGAATCGATA-3′.
Lentiviral pZIP vectors encoding shRNAs targeting human p190A or p190B message and conferring resistance to neomycin were purchased from transOMIC Technologies (TRHS1000-35 [ARHGAP35/p190A] and TRHS1000-394 [ARHGAP5/p190B]). The sequences were as follows: human p190A shRNA 1, 5′-TGCTGTTGACAGTGAGCGCCCTAATCTAGATGAAATAGAATAGTGAAGCCACAGATGTATTCTATTTCATCTAGATTAGGTTGCCTACTGCCTCGGA-3′; shRNA 2, 5′-TGCTGTTGACAGTGAGCGACAGGATTCAGAAGAAGATATATAGTGAAGCCACAGATGTATATATCTTCTTCTGAATCCTGCTGCCTACTGCCTCGGA-3′; shRNA 3, 5′-TGCTGTTGACAGTGAGCGCCCTGTCCATTTTGAAATTACATAGTGAAGCCACAGATGTATGTAATTTCAAAATGGACAGGATGCCTACTGCCTCGGA-3′; and shRNA 4, 5′-TGCTGTTGACAGTGAGCGCAAGTAGGGAACAGCTAACTGATAGTGAAGCCACAGATGTATCAGTTAGCTGTTCCCTACTTATGCCTACTGCCTCGGA-3′; human p190B shRNA 1, 5′-TGCTGTTGACAGTGAGCGCCAGCAGAAAAACTAATGTACATAGTGAAGCCACAGATGTATGTACATTAGTTTTTCTGCTGATGCCTACTGCCTCGGA-3′; and shRNA 2, 5′-TGCTGTTGACAGTGAGCGCACATATAGAACAACTTAAACATAGTGAAGCCACAGATGTATGTTTAAGTTGTTCTATATGTTTGCCTACTGCCTCGGA-3′.
Expression constructs encoding rat WT p190A, p190A(R1284K), or p190A(ΔGAP) comprising amino acids 1–1,228, each resistant to knockdown by p190A shRNA 1, were generated in pWZL-neo that had been modified to encode a double Myc tag on the N terminus. Expression constructs encoding YAP5SA or YAP5SA+S94A in pInducer20 were a gift from F.D. Camargo (Boston Children's Hospital, Boston, MA) and have been described previously (Mori et al., 2014). Expression constructs encoding EGFP-RhoA(F30L) or EGFP only under control of TetR were generated by PCR and cloned into pQCTXN, a derivative of pQCXIN that contains the tetO element upstream of the CMV promoter.
Inducible RNA interference and gene expression
To permit inducible knockdown of p190A and/or p190B expression, TR-T10 cells were transduced with retrovirus encoding p190A shRNA or p190B shRNA, and the cells were then selected with hygromycin (p190A shRNA) or puromycin (p190B shRNA). To generate TR-T10 cells with combined knockdown of p190A and p190B, p190B-kd cells were transduced with retrovirus encoding p190A shRNA, and cells were selected with hygromycin. To induce knockdown of gene expression, cells were plated at low density (4 × 103 cells per cm2) and cultured for 3 d in the presence of 4 µg/ml doxycycline. Cells were then harvested and replated at low (4 × 103 cells per cm2) or high (2 × 105 cells per cm2) density and cultured for another 3–5 d before experimentation.
TR-T10 or p190A+B-kd cells with doxycycline-inducible expression of YAP5SA or YAP5SA+S94A were generated by lentiviral transduction and selected with G418. In cells with p190A+B-kd, expression of YAP5SA or YAP5SA+S94A is linked to knockdown of p190A and p190B as both are under control of the TetR. TR-T10 cells with inducible expression of EGFP or EGFP-RhoA(F30L) were generated by retroviral transduction and selected with G418. Gene expression was induced according to the same protocol as described above for knocking down p190A and p190B expression. Plasmid encoding HA-tagged murine kinase-dead LATS2(K655R) in pcDNA3-HA generated by Zhao et al. (2007) was obtained from Addgene (plasmid 33100). The mammalian expression cassette was amplified by PCR and inserted into pHAGE-CMV-IZsGreen, thus permitting lentiviral transduction of TR-T10 cells with or without inducible p190A and p190B knockdown. High-level-expressing cells were selected by FACS and used for experimentation. HEC-1-A cells with constitutive knockdown of p190A or p190B were generated by lentiviral transduction and selected in G418-containing medium.
RBD and CRIB assays
RBD and CRIB pulldown assays to detect activated RhoA and Rac1, respectively, were performed essentially as previously described (Hansen et al., 2000) except that RBD and CRIB beads were purchased from Cytoskeleton. In brief, control and p190A- and/or p190B-depleted cells with or without treatment with Tat-Myc-C3 toxin were lysed in modified gold lysis buffer (1% Triton X-100, 20 mM Tris, pH 8.0, 500 mM NaCl, 10 mM MgCl2, and 15% glycerol) containing protease inhibitors. Cell lysates were cleared by centrifugation for 2 min at 12,000 rpm at 4°C in a TOMY MX-301 centrifuge. An aliquot of 100 µl lysate was removed and mixed with 2× Laemmli sample buffer for detection of total Rho/Rac. Next, lysates were transferred to tubes containing 20-µl aliquots of RBD or CRIB beads and tumbled end over end at for 20 min at 4°C. The RBD/CRIB beads were then rinsed twice with modified gold lysis buffer with all remaining liquid removed using a Hamilton syringe. The beads were mixed with 30 µl of 2× Laemmli sample buffer and heated to 95°C for 3 min. Finally, samples were electrophoresed on SDS-PAGE gels, and active Rho and Rac were detected by Western blotting. Aliquots of 1% of total cell lysates were run in parallel to determine total Rho and Rac levels.
Confocal fluorescence microscopy
Samples for immunostaining of MDCK cells cultured on permeable supports were processed essentially as detailed previously (Hansen et al., 2000). Samples were rinsed once with PBS and fixed in 3.7% formalin containing 10% methanol for 10 min at RT. After rinsing three times with PBS, samples were incubated for 30 min at RT with blocking buffer (10% normal goat serum, 0.2% fish skin gelatin, and 0.1% Triton X-100 in PBS). Next, samples were incubated with primary antibodies for 2 h at RT, rinsed extensively with PBS containing 0.1% Triton X-100 for 30 min, and then incubated with secondary antibodies for 60 min at RT. After further extensive rinsing for 30 min, samples were stained for 15 min at RT with DRAQ5 to detect nuclei and when relevant, fluorescent phalloidin to visualize polymerized actin. After three additional brief rinses, samples were mounted with FluorSave (EMD Millipore). For MDCK cells cultured in Matrigel, incubation with primary antibody was performed at 4°C overnight, and incubation with secondary antibody for 2 h at RT, but otherwise the process was as described for cells grown on permeable supports. Imaging was performed using line-scanning and spinning-disk confocal microscopes.
TUNEL assay
TUNEL analysis was performed to detect apoptosis in cells shed from MDCK cell monolayers. To this end, medium from cell cultures was harvested, and cells were pelleted on slides using a Cytospin apparatus. The slides were then immersed in 2% formalin for 30 min and rinsed two times with PBS and once with distilled water. Cells were permeabilized with ice-cold 70% (vol/vol) ethanol for 30 min on ice and processed for TUNEL staining according to the manufacturer's instructions (In situ Cell Death Detection kit; Roche). Finally, nuclei were labeled with DAPI and imaged by fluorescence microscopy.
BrdU incorporation
For determining proliferation, cells cultured in 24-well format were incubated with BrdU at a final concentration of 10 µM for 3 h at 37°C. Cells were fixed with 3.7% formalin for 20 min at RT, incubated with 1 N HCl for 1 h, and neutralized by addition of an equal volume of 0.1 M Na2B4O7. Samples were then processed to detect BrdU incorporation by immunofluorescence microscopy as described above, except that nuclei were stained with DAPI to permit visualization by conventional immunofluorescence microscopy. Next, quantification of BrdU incorporation was performed by determining the ratio of BrdU-positive nuclei to total number of nuclei essentially as previously described (Hansen et al., 2000).
Aggregation assay
MDCK cells with and without knockdown of p190A and/or p190B were harvested by trypsinization, and 2 × 106 cells per condition were plated into each well of a 24-well ultralow adhesion cluster plate (Corning). After culture for 8 h in suspension, cell clusters were triturated 10 times through a 200-µl pipette tip. As negative control, we used cells depleted of α-catenin described previously (Frank et al., 2012). Quantification of cell aggregation was performed by determining the ratio of area of cells in aggregates to individual cells using ImageJ (National Institutes of Health), essentially as detailed by others (Qin et al., 2005).
Real-time qPCR analyses
For real-time qPCR, total RNA was extracted from cell pellets using RNeasy Mini and QIAshredder kits (both QIAGEN) according to the manufacturer’s instructions. cDNA was synthetized using iScript Reagents (Bio-Rad Laboratories). Quantitative RT-PCR was performed with the One Step plus Sequence Detection System using Fast SYBR green master mix reagent (Applied Biosystems Ltd.). Gene expression levels were normalized to the mean of at least two housekeeping genes. qPCR primers for canine and human genes are listed in Tables 2 and 3, respectively.
Table 2. qPCR primers for canine genes.
| Gene | Forward sequence (5′–3′) | Reverse sequence (5′–3′) |
|---|---|---|
| ACTB | AGATCTGGCACCACACCTTCTA | CATGATCTGGGTCATCTTCTCA |
| ANKRD1 | GGCTGAACCGTTACAAGATGAT | TGGTTCCATTCTGCCAGTGTAG |
| CYR61 | CAACGAGGACTGCAGCAAAATG | CCATTCTGGTAGATTCGGGAGT |
| DOCK4 | CCCAAATCTCTCTGCACCTGAG | ATCCGACATGTTCTTGGCTTCT |
| DUSP2 | GCCCTGTGGAGATCTTACCCTA | TAGCGGAAAAGGCCCTCAAAG |
| ESM1 | ACCGGCAAGTGTCTGAAATTTC | TTCTCTCACAGCATTGCCATCT |
| HPRT | CTTTGCTGACCTGCTGGATTAT | TTTATGTCCCCTGTTGACTGGT |
| PAI1 | TGACCGACATGTTTAGGCCAAA | TGTGCCACTCTCATTCACTTCA |
| PDGFB | CTCGATCCGCTCCTTTGATGA | ATCGGGTCAAATTCAGGTCCAA |
| RND3 | GCCAGTTTTGAGATCGACACAC | CAAATCAGCACAGCATCCGAAT |
| RPS18 | TGAGAAGTTCCAGCACATTTTG | ATCGATGTCTGCTTTCCTCAAT |
| SEMA7A | GACCTTCCACGTGCTTTACCTA | CATGATGTTGAAGACCAGGCTG |
| SLIT2 | ACATTGCCGTGGAGCTGTATC | AGGGCAAGTAGTTCCACAATGT |
| TGFB2 | AGTTCAGACACTCAGCACAGT | TGGGTGTTTTGCCGATGTAGTA |
| VEGFC | TCAGTGCAAGAACAGAAGAGACT | CCAAACTCCTTCCCCACATCTA |
Table 3. qPCR primers for human genes.
| Gene | Forward sequence (5′–3′) | Reverse sequence (5′–3′) |
|---|---|---|
| AMOTL2 | AGCTTCAATGAGGGTCTGCTC | CTGCTGAAGGACCTTGATCACT |
| ANKRD1 | TGATTATGTATGGCGCGGATCT | GCGAGAGGTCTTGTAGGAGTTC |
| CTGF | GAAGCTGACCTGGAAGAGAACA | CGTCGGTACATACTCCACAGAA |
| CYR61 | CATTCCTCTGTGTCCCCAAGAA | TACTATCCTCGTCACAGACCCA |
| DOCK4 | ATGAGACCATCTTCCAGGCAGA | AGCTTTTGATGTTGTCCGGAAC |
| DUSP2 | TTCCGCTACAAGAGTATCCCTG | TTCACCCAGTCAATGAAGCCTA |
| ESM1 | GCATGACATGGCATCTGGAGA | CTCAGAAATCACAGCCGGGATC |
| HPRT | AGCCAGACTTTGTTGGATTTG | TTTACTGGCGATGTCAATAGG |
| PAI1 | TCTCTGCCCTCACCAACATTC | CGGTCATTCCCAGGTTCTCTAG |
| PDGFB | TCCCGAGGAGCTTTATGAGATG | GGGTCATGTTCAGGTCCAACTC |
| SLIT2 | CTGTGTGGGGAATGACAGTTTC | AGAGTATCAAATGCCCCTGGTG |
| VEGFC | ACCAAACAAGGAGCTGGATGAA | ACACTGGCATGAGTTTCTGTCT |
| RND3 | GGCCAGTTTTGAAATCGACACA | CAAATCAGCACAGCATCCGAAT |
| RPS18 | TTCGGAACTGAGGCCATGAT | TTTCGCTCTGGTCCGTCTTG |
| SEMA7A | AGACGCCATTGTTCCACTCTAA | TGCCCCTGTCTGTAGTTAGGTA |
| TGFB2 | CCCCACATCTCCTGCTAATGTT | AAGTGGACGTAGGCAGCAATTA |
Genome-wide mRNA expression profiling
Total RNA was isolated from 106 MDCK cells using RNeasy kit (QIAGEN). RNA integrity was analyzed on a 2100 Bioanalyzer using the Agilent RNA 6000 Nano kit (Agilent Technologies). Only samples with an RNA integrity number >9.5 was used for further analysis. cDNAs and libraries were prepared using 1 µg total RNA and the TruSeq mRNA sample preparation kit according to the manufacturer’s instructions (Illumina). Library quality was analyzed on a 2100 Bioanalyzer using the Agilent DNA 1000 kit (Agilent Technologies). Library samples were quantified using the Qubit dsDNA BR assay kit (Invitrogen) and diluted to 10 nM before single-read sequencing on an Illumina HiSeq 2000 sequencing platform performed at the National High-throughput DNA Sequencing Centre at the University of Copenhagen.
For read mapping and data analysis, reads were aligned in single-end mode to canine genome (canFam3; http://www.ensembl.org/Canis_familiaris/) using STAR (version 2.3.1n) aligner using default settings (Dobin et al., 2013). Mapped data were converted in gene level counts using HTseq-count 0.6.0 and the ENSEMBL annotation (canFam3.1). Differential expression was evaluated using DESeq2 Bioconductor package 1.4.5 (Love et al., 2014).
To identify genes associated with loss of CIP in p190A+B-kd cells, we first determined differential gene expression between p190A+B-kd and control cells using a threshold of |log2(fold change)| ≥1 and adjusted P value ≤0.05. Next, we determined differential gene expression between p190A+B-kd and p190A-kd as well as p190A+B-kd and p190B-kd cells using a threshold of |log2(fold change)| ≥0.5 and adjusted P value ≤0.05 for both analyses. Genes filtered by these thresholds are listed in Tables S1 and S2.
Determinations of allelic fraction profile of ARHGAP35 somatic mutations
We used a compiled patient-derived PanCancer mutation dataset (Lawrence et al., 2014) to examine loss-of-function mutations in ARHGAP35. A somatic mutation causing protein changes (nonsense/frameshift indel/missense) and allele fraction >0.5 would indicate a complete loss of function of the two normal alleles.
Statistical analyses
Unpaired two-tailed Student t tests were performed as previously detailed (Hansen et al., 1992).
Online supplemental material
Fig. S1 shows phase images of p190A- and/or p190B-depleted MDCK cells by phase microscopy, by TUNEL staining of cells detached from cultures grown on permeable supports with or without knockdown of p190A and/or p190B as well as cyclin A cell proliferation data for adherent cells. Fig. S2 shows a z stack of confocal microscopy images of p190A+B-kd cells along with accompanying x–y and z images. Fig. S3 illustrates effects of p190A and/or p190B knockdown on adherens and tight junctions. Fig. S4 demonstrates effects of C3 toxin and ROCK inhibitor on p190A- and/or p190B-depleted cells. Fig. S5 shows images of YAP, E-cadherin, and β-catenin staining control, p190A-kd, p190B-kd, and p190A+B-kd MCDK cell spheroids cultured in Matrigel. Table S1 shows mRNA-seq analysis of up-regulated dense cultures. Table S2 shows mRNA-seq analysis of down-regulated dense cultures. Table S3 shows mRNA-seq analysis of up-regulated sparse cultures. Table S4 shows mRNA-seq analysis of down-regulated sparse cultures.
Supplementary Material
Acknowledgments
We are grateful to Fernando D. Camargo for YAP expression constructs.
G.G. Galli was supported by an American-Italian Cancer Foundation postdoctoral research fellowship. This work used Harvard University Digestive Diseases Center core facilities and was funded by National Institutes of Health R01 CA205158 and an endowed investigatorship from the Roy and Lynne Frank Foundation (to S.H. Hansen). G. Getz is the inaugural incumbent of the Paul C. Zamecnik Chair in Oncology at the Massachusetts General Hospital Cancer Center.
The authors declare no competing financial interests.
Author contributions: S.R. Frank, C.P. Köllman, P. Luong, M. Frödin, and S.H. Hansen conducted cell-based experiments. C.P. Köllman and G.G. Galli performed qPCR analyses. L. Zou, A. Bernards, and G. Getz generated human tumor data. R.A. Calogero analyzed all mRNA-seq data. S.H. Hansen designed research and wrote the manuscript with input from coauthors.
References
- Aragona M., Panciera T., Manfrin A., Giulitti S., Michielin F., Elvassore N., Dupont S., and Piccolo S.. 2013. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell. 154:1047–1059. 10.1016/j.cell.2013.07.042 [DOI] [PubMed] [Google Scholar]
- Arthur W.T., and Burridge K.. 2001. RhoA inactivation by p190RhoGAP regulates cell spreading and migration by promoting membrane protrusion and polarity. Mol. Biol. Cell. 12:2711–2720. 10.1091/mbc.12.9.2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass A.J., Thorsson V., Shmulevich I., Reynolds S.M., Miller M., Bernard B., Hinoue T., Laird P.W., Curtis C., Shen H., et al. Cancer Genome Atlas Research Network . 2014. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 513:202–209. 10.1038/nature13480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum B., and Georgiou M.. 2011. Dynamics of adherens junctions in epithelial establishment, maintenance, and remodeling. J. Cell Biol. 192:907–917. 10.1083/jcb.201009141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell D., Berchuck A., Birrer M., Chien J., Cramer D.W., Dao F., Dhir R., DiSaia P., Gabra H., Glenn P., et al. Cancer Genome Atlas Research Network . 2011. Integrated genomic analyses of ovarian carcinoma. Nature. 474:609–615. 10.1038/nature10166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berenjeno I.M., Núñez F., and Bustelo X.R.. 2007. Transcriptomal profiling of the cellular transformation induced by Rho subfamily GTPases. Oncogene. 26:4295–4305. 10.1038/sj.onc.1210194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binamé F., Bidaud-Meynard A., Magnan L., Piquet L., Montibus B., Chabadel A., Saltel F., Lagrée V., and Moreau V.. 2016. Cancer-associated mutations in the protrusion-targeting region of p190RhoGAP impact tumor cell migration. J. Cell Biol. 214:859–873. 10.1083/jcb.201601063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouns M.R., Matheson S.F., Hu K.Q., Delalle I., Caviness V.S., Silver J., Bronson R.T., and Settleman J.. 2000. The adhesion signaling molecule p190 RhoGAP is required for morphogenetic processes in neural development. Development. 127:4891–4903. [DOI] [PubMed] [Google Scholar]
- Burbelo P.D., Miyamoto S., Utani A., Brill S., Yamada K.M., Hall A., and Yamada Y.. 1995. p190-B, a new member of the Rho GAP family, and Rho are induced to cluster after integrin cross-linking. J. Biol. Chem. 270:30919–30926. 10.1074/jbc.270.52.30919 [DOI] [PubMed] [Google Scholar]
- Camargo F.D., Gokhale S., Johnnidis J.B., Fu D., Bell G.W., Jaenisch R., and Brummelkamp T.R.. 2007. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr. Biol. 17:2054–2060. 10.1016/j.cub.2007.10.039 [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network 2012. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 487:330–337. 10.1038/nature11252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network 2012. Comprehensive molecular portraits of human breast tumours. Nature. 490:61–70. 10.1038/nature11412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network 2012. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 489:519–525. 10.1038/nature11404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network 2013. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 499:43–49. 10.1038/nature12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network 2014. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 507:315–322. 10.1038/nature12965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collisson E.A., Campbell J.D., Brooks A.N., Berger A.H., Lee W., Chmielecki J., Beer D.G., Cope L., Creighton C.J., Danilova L., et al. Cancer Genome Atlas Research Network . 2014. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 511:543–550. 10.1038/nature13385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Feldmann G., Huang J., Wu S., Zhang N., Comerford S.A., Gayyed M.F., Anders R.A., Maitra A., and Pan D.. 2007. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 130:1120–1133. 10.1016/j.cell.2007.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S., Morsut L., Aragona M., Enzo E., Giulitti S., Cordenonsi M., Zanconato F., Le Digabel J., Forcato M., Bicciato S., et al. . 2011. Role of YAP/TAZ in mechanotransduction. Nature. 474:179–183. 10.1038/nature10137 [DOI] [PubMed] [Google Scholar]
- Frank S.R., Bell J.H., Frödin M., and Hansen S.H.. 2012. A βPIX-PAK2 complex confers protection against Scrib-dependent and cadherin-mediated apoptosis. Curr. Biol. 22:1747–1754. 10.1016/j.cub.2012.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbiner B.M., and Kim N.G.. 2014. The Hippo-YAP signaling pathway and contact inhibition of growth. J. Cell Sci. 127:709–717. 10.1242/jcs.140103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., and Weinberg R.A.. 2011. Hallmarks of cancer: The next generation. Cell. 144:646–674. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Hansen S.H., Sandvig K., and van Deurs B.. 1992. Internalization efficiency of the transferrin receptor. Exp. Cell Res. 199:19–28. 10.1016/0014-4827(92)90457-J [DOI] [PubMed] [Google Scholar]
- Hansen S.H., Zegers M.M., Woodrow M., Rodriguez-Viciana P., Chardin P., Mostov K.E., and McMahon M.. 2000. Induced expression of Rnd3 is associated with transformation of polarized epithelial cells by the Raf-MEK-extracellular signal-regulated kinase pathway. Mol. Cell. Biol. 20:9364–9375. 10.1128/MCB.20.24.9364-9375.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Sánchez M., Poch E., Guasch R.M., Ortega J., López-Almela I., Palmero I., and Pérez-Roger I.. 2015. RhoE is required for contact inhibition and negatively regulates tumor initiation and progression. Oncotarget. 6:17479–17490. 10.18632/oncotarget.4127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W., Sordella R., Chen G.C., Hakre S., Roy A.L., and Settleman J.. 2005. An FF domain-dependent protein interaction mediates a signaling pathway for growth factor-induced gene expression. Mol. Cell. 17:23–35. 10.1016/j.molcel.2004.11.024 [DOI] [PubMed] [Google Scholar]
- Jiao S., Wang H., Shi Z., Dong A., Zhang W., Song X., He F., Wang Y., Zhang Z., Wang W., et al. . 2014. A peptide mimicking VGLL4 function acts as a YAP antagonist therapy against gastric cancer. Cancer Cell. 25:166–180. 10.1016/j.ccr.2014.01.010 [DOI] [PubMed] [Google Scholar]
- Jou T.S., and Nelson W.J.. 1998. Effects of regulated expression of mutant RhoA and Rac1 small GTPases on the development of epithelial (MDCK) cell polarity. J. Cell Biol. 142:85–100. 10.1083/jcb.142.1.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth C., McLellan M.D., Vandin F., Ye K., Niu B., Lu C., Xie M., Zhang Q., McMichael J.F., Wyczalkowski M.A., et al. . 2013. Mutational landscape and significance across 12 major cancer types. Nature. 502:333–339. 10.1038/nature12634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth C., Schultz N., Cherniack A.D., Akbani R., Liu Y., Shen H., Robertson A.G., Pashtan I., Shen R., Benz C.C., et al. Cancer Genome Atlas Research Network . 2013. Integrated genomic characterization of endometrial carcinoma. Nature. 497:67–73. 10.1038/nature12113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M.S., Stojanov P., Mermel C.H., Robinson J.T., Garraway L.A., Golub T.R., Meyerson M., Gabriel S.B., Lander E.S., and Getz G.. 2014. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 505:495–501. 10.1038/nature12912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligeti E., Dagher M.C., Hernandez S.E., Koleske A.J., and Settleman J.. 2004. Phospholipids can switch the GTPase substrate preference of a GTPase-activating protein. J. Biol. Chem. 279:5055–5058. 10.1074/jbc.C300547200 [DOI] [PubMed] [Google Scholar]
- Liu-Chittenden Y., Huang B., Shim J.S., Chen Q., Lee S.J., Anders R.A., Liu J.O., and Pan D.. 2012. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 26:1300–1305. 10.1101/gad.192856.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr J.G., Stojanov P., Carter S.L., Cruz-Gordillo P., Lawrence M.S., Auclair D., Sougnez C., Knoechel B., Gould J., Saksena G., et al. Multiple Myeloma Research Consortium . 2014. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell. 25:91–101. 10.1016/j.ccr.2013.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClatchey A.I., and Yap A.S.. 2012. Contact inhibition (of proliferation) redux. Curr. Opin. Cell Biol. 24:685–694. 10.1016/j.ceb.2012.06.009 [DOI] [PubMed] [Google Scholar]
- Meng Z., Moroishi T., and Guan K.L.. 2016. Mechanisms of Hippo pathway regulation. Genes Dev. 30:1–17. 10.1101/gad.274027.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohseni M., Sun J., Lau A., Curtis S., Goldsmith J., Fox V.L., Wei C., Frazier M., Samson O., Wong K.K., et al. . 2014. A genetic screen identifies an LKB1-MARK signalling axis controlling the Hippo-YAP pathway. Nat. Cell Biol. 16:108–117. 10.1038/ncb2884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori M., Triboulet R., Mohseni M., Schlegelmilch K., Shrestha K., Camargo F.D., and Gregory R.I.. 2014. Hippo signaling regulates microprocessor and links cell-density-dependent miRNA biogenesis to cancer. Cell. 156:893–906. 10.1016/j.cell.2013.12.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroishi T., Hansen C.G., and Guan K.L.. 2015. The emerging roles of YAP and TAZ in cancer. Nat. Rev. Cancer. 15:73–79. 10.1038/nrc3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara H., Mueller S.C., Nomizu M., Yamada Y., Yeh Y., and Chen W.T.. 1998. Activation of beta1 integrin signaling stimulates tyrosine phosphorylation of p190RhoGAP and membrane-protrusive activities at invadopodia. J. Biol. Chem. 273:9–12. 10.1074/jbc.273.1.9 [DOI] [PubMed] [Google Scholar]
- Priya R., Gomez G.A., Budnar S., Verma S., Cox H.L., Hamilton N.A., and Yap A.S.. 2015. Feedback regulation through myosin II confers robustness on RhoA signalling at E-cadherin junctions. Nat. Cell Biol. 17:1282–1293. 10.1038/ncb3239 [DOI] [PubMed] [Google Scholar]
- Qin Y., Capaldo C., Gumbiner B.M., and Macara I.G.. 2005. The mammalian Scribble polarity protein regulates epithelial cell adhesion and migration through E-cadherin. J. Cell Biol. 171:1061–1071. 10.1083/jcb.200506094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano I., McDonald P.C., Lock F., Muller W.J., and Dedhar S.. 2013. Inactivation of the Hippo tumour suppressor pathway by integrin-linked kinase. Nat. Commun. 4:2976 10.1038/ncomms3976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settleman J., Narasimhan V., Foster L.C., and Weinberg R.A.. 1992. Molecular cloning of cDNAs encoding the GAP-associated protein p190: implications for a signaling pathway from ras to the nucleus. Cell. 69:539–549. 10.1016/0092-8674(92)90454-K [DOI] [PubMed] [Google Scholar]
- Sordella R., Classon M., Hu K.Q., Matheson S.F., Brouns M.R., Fine B., Zhang L., Takami H., Yamada Y., and Settleman J.. 2002. Modulation of CREB activity by the Rho GTPase regulates cell and organism size during mouse embryonic development. Dev. Cell. 2:553–565. 10.1016/S1534-5807(02)00162-4 [DOI] [PubMed] [Google Scholar]
- Su L., Agati J.M., and Parsons S.J.. 2003. p190RhoGAP is cell cycle regulated and affects cytokinesis. J. Cell Biol. 163:571–582. 10.1083/jcb.200308007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L., Pertz O., Mikawa M., Hahn K., and Parsons S.J.. 2009. p190RhoGAP negatively regulates Rho activity at the cleavage furrow of mitotic cells. Exp. Cell Res. 315:1347–1359. 10.1016/j.yexcr.2009.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaishi K., Sasaki T., Kotani H., Nishioka H., and Takai Y.. 1997. Regulation of cell–cell adhesion by rac and rho small G proteins in MDCK cells. J. Cell Biol. 139:1047–1059. 10.1083/jcb.139.4.1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev A., Kaneko K.J., Shu H., Zhao Y., and DePamphilis M.L.. 2001. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 15:1229–1241. 10.1101/gad.888601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent S., and Settleman J.. 1999. Inhibition of RhoGAP activity is sufficient for the induction of Rho-mediated actin reorganization. Eur. J. Cell Biol. 78:539–548. 10.1016/S0171-9335(99)80019-3 [DOI] [PubMed] [Google Scholar]
- Wang D.Z., Nur-E-Kamal M.S., Tikoo A., Montague W., and Maruta H.. 1997. The GTPase and Rho GAP domains of p190, a tumor suppressor protein that binds the M(r) 120,000 Ras GAP, independently function as anti-Ras tumor suppressors. Cancer Res. 57:2478–2484. [PubMed] [Google Scholar]
- Wennerberg K., Forget M.A., Ellerbroek S.M., Arthur W.T., Burridge K., Settleman J., Der C.J., and Hansen S.H.. 2003. Rnd proteins function as RhoA antagonists by activating p190 RhoGAP. Curr. Biol. 13:1106–1115. 10.1016/S0960-9822(03)00418-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildenberg G.A., Dohn M.R., Carnahan R.H., Davis M.A., Lobdell N.A., Settleman J., and Reynolds A.B.. 2006. p120-catenin and p190RhoGAP regulate cell-cell adhesion by coordinating antagonism between Rac and Rho. Cell. 127:1027–1039. 10.1016/j.cell.2006.09.046 [DOI] [PubMed] [Google Scholar]
- Wolf R.M., Draghi N., Liang X., Dai C., Uhrbom L., Eklöf C., Westermark B., Holland E.C., and Resh M.D.. 2003. p190RhoGAP can act to inhibit PDGF-induced gliomas in mice: A putative tumor suppressor encoded on human chromosome 19q13.3. Genes Dev. 17:476–487. 10.1101/gad.1040003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack T.I., Schumacher S.E., Carter S.L., Cherniack A.D., Saksena G., Tabak B., Lawrence M.S., Zhsng C.Z., Wala J., Mermel C.H., et al. . 2013. Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 45:1134–1140. 10.1038/ng.2760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanconato F., Cordenonsi M., and Piccolo S.. 2016. YAP/TAZ at the roots of cancer. Cancer Cell. 29:783–803. 10.1016/j.ccell.2016.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Q., and Hong W.. 2008. The emerging role of the hippo pathway in cell contact inhibition, organ size control, and cancer development in mammals. Cancer Cell. 13:188–192. 10.1016/j.ccr.2008.02.011 [DOI] [PubMed] [Google Scholar]
- Zhao B., Wei X., Li W., Udan R.S., Yang Q., Kim J., Xie J., Ikenoue T., Yu J., Li L., et al. . 2007. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 21:2747–2761. 10.1101/gad.1602907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Ye X., Yu J., Li L., Li W., Li S., Yu J., Lin J.D., Wang C.Y., Chinnaiyan A.M., et al. . 2008. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 22:1962–1971. 10.1101/gad.1664408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Li L., Tumaneng K., Wang C.Y., and Guan K.L.. 2010. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP). Genes Dev. 24:72–85. 10.1101/gad.1843810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Li L., Lu Q., Wang L.H., Liu C.Y., Lei Q., and Guan K.L.. 2011. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 25:51–63. 10.1101/gad.2000111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.