Desmosomal mutations result in potentially deadly cardiocutaneous disease caused by electrical conduction defects and disruption of gap junctions. Kam et al. demonstrate a mechanism whereby loss of the intermediate filament anchoring protein desmoplakin stimulates Cx43 turnover by increasing K-Ras expression, marking Cx43 for lysosomal degradation through ERK1/2 phosphorylation.
Abstract
Desmoplakin (DP) is an obligate component of desmosomes, intercellular adhesive junctions that maintain the integrity of the epidermis and myocardium. Mutations in DP can cause cardiac and cutaneous disease, including arrhythmogenic cardiomyopathy (ACM), an inherited disorder that frequently results in deadly arrhythmias. Conduction defects in ACM are linked to the remodeling and functional interference with Cx43-based gap junctions that electrically and chemically couple cells. How DP loss impairs gap junctions is poorly understood. We show that DP prevents lysosomal-mediated degradation of Cx43. DP loss triggered robust activation of ERK1/2–MAPK and increased phosphorylation of S279/282 of Cx43, which signals clathrin-mediated internalization and subsequent lysosomal degradation of Cx43. RNA sequencing revealed Ras-GTPases as candidates for the aberrant activation of ERK1/2 upon loss of DP. Using a novel Ras inhibitor, Ras/Rap1-specific peptidase (RRSP), or K-Ras knockdown, we demonstrate restoration of Cx43 in DP-deficient cardiomyocytes. Collectively, our results reveal a novel mechanism for the regulation of the Cx43 life cycle by DP in cardiocutaneous models.
Introduction
Maintenance of tissue homeostasis and integrity is achieved through highly specialized membrane–cytoskeleton attachment sites. The most prominent of these in the epidermis and myocardium are desmosomes, which are cadherin-based junctional complexes that tether the intermediate filament (IF) cytoskeleton to sites of cell–cell adhesion (Leung et al., 2002; Green and Simpson, 2007). In both epithelial and cardiac cells, desmosomes are structurally and functionally associated with actin-associated adherens junctions, which organize the cortical actin cytoskeleton, and connexin-containing gap junctions, which are pore-like structures that allow for the passage of small molecules and ions between two neighboring cells (Saez et al., 2003; Laird, 2006; Goodenough and Paul, 2009). In cardiomyocytes, close association between junctions is facilitated by intercalated discs (IDs), which are present where cardiomyocytes meet end to end and serve to integrate force transmission and electrical coupling in the heart (Forbes and Sperelakis, 1985). The ID in adult mammals is further specialized to form the area composita, in which adherens junction and desmosome components are intermixed, further stabilizing the junction (Goossens et al., 2007; Pieperhoff and Franke, 2007; Pieperhoff et al., 2008).
The desmosomal component that physically links the transmembrane desmosomal cadherin complex to the IF cytoskeleton is desmoplakin (DP), a requisite mediator of mechanical strength in skin and heart. The importance of DP is underscored by the failure of DP-null murine embryos to survive beyond embryonic day 6.5 (E6.5) because of destabilization of desmosomes leading to egg cylinder elongation defects (Gallicano et al., 1998). Tetraploid analysis and tissue-specific conditional knockouts (KOs) later demonstrated a requirement for DP in vascular development, epidermal integrity, and cardiac function (Gallicano et al., 2001; Vasioukhin et al., 2001; Lyon et al., 2014). In the case of heart, conditional DP KO recapitulated the pathology of the human cardiac disease arrhythmogenic cardiomyopathy (ACM), which has been associated with mutations in desmosomal components (Asimaki and Saffitz, 2014; Lyon et al., 2014). Pathogenic DP mutations are the second most frequently identified in ACM and can be present either with or without cutaneous phenotypes (Alcalai et al., 2003; López-Ayala et al., 2014; Molho-Pessach et al., 2015; Castelletti et al., 2017). In ACM, healthy myocardium is progressively replaced with fibrofatty tissue deposits that can ultimately lead to ventricular arrhythmias and sudden cardiac death (Basso et al., 1996, 2009; Sen-Chowdhry et al., 2007). Although best known for their roles in intercellular adhesion, desmosome components have emerged as regulators of intracellular signaling (Bass-Zubek et al., 2008; Godsel et al., 2010; Wolf et al., 2013; Rötzer et al., 2016). DP in particular has been implicated in the regulation of signaling pathways involved in the adipogenic and fibrogenic aspects of ACM pathogenesis (Garcia-Gras et al., 2006; Dubash et al., 2016; Lombardi et al., 2016). However, the molecular mechanisms by which desmosomal mutations lead to disease pathogenesis are not well understood.
While desmosomal and adherens junctions components of the ID facilitate mechanical coupling between neighboring cardiomyocytes, evidence suggests that they are physically and functionally integrated with gap junctions and sodium channels to facilitate electrical conduction at these nodes (Saez et al., 2003; Laird, 2006; Goodenough and Paul, 2009; Stroemlund et al., 2015). Vertebrate gap junctions are made up of proteins from the 21-gene connexin family, whereby hexamers of connexin proteins (termed connexons) formed on two adjacent cells dock with each other to produce channels that allow for the passage of small molecules and ions (Laird, 2006; Goodenough and Paul, 2009). Of these members, connexin-43 (Cx43) is the most ubiquitously expressed connexin in many tissues and cultured cells (Laird, 2006). We previously showed that DP plays an important role in the delivery of Cx43 to the plasma membrane via an EB1–microtubule-dependent pathway (Patel et al., 2014). In addition, two independent mouse models with cardiac conditional DP KO (DPcKO) reported that DP loss also leads to a significant reduction in Cx43 protein levels (Gomes et al., 2012; Lyon et al., 2014). The loss of Cx43 expression in these models was speculated to contribute to a feature of ACM called the “concealed phase,” in which electrical conduction defects occur before fibrofatty replacement of healthy myocardium (Saffitz et al., 2010; Gomes et al., 2012; Lyon et al., 2014). However, the molecular mechanism by which loss of DP leads to a decrease in Cx43 is unknown. Given the unique organization of cardiac cell junctions, cardiomyocytes provide an ideal model for studying the structural and functional dynamics of intercellular junctions and how interfering with normal integration of intercellular junctions leads to disease (Garcia-Gras et al., 2006; Cerrone et al., 2012; Lyon et al., 2014).
In this study, we characterize a novel mechanism by which DP loss leads to Cx43 protein degradation. Depletion of DP triggers robust activation of the ERK1/2–MAPK pathway, resulting in phosphorylation of residues S279 and 282 within the intracellular C terminus of Cx43. These consensus ERK1/2 phosphorylation sites are known to be phosphorylated in tandem to mark Cx43 for degradation (Fong et al., 2014). We further show that DP loss elevates mRNA, protein, and activity of the canonical upstream activator of ERK1/2, K-Ras, in cardiac cells but not in an epidermal model of DP loss. Inhibition of Ras using either K-Ras knockdown (KD) or a novel peptide inhibitor derived from a toxin secreted by the bacterium Vibrio vulnificus rescued Cx43 expression and localization caused by DP loss. Along with our previous work (Patel et al., 2014), these data support the idea that loss of DP interferes with both transcriptional and posttranslational mechanisms controlling Cx43 expression and localization and provide a new perspective on the molecular events leading to cardiac arrhythmias and other connexin-based disorders in skin and heart.
Results
DP loss impairs the expression and localization of the gap junction protein Cx43
Previous studies showed that DP loss is associated with decreased expression of Cx43 in cardiac tissue (Gomes et al., 2012; Lyon et al., 2014). Toward determining the mechanism by which DP regulates Cx43 protein levels, we used cultured neonatal rat ventricular cardiomyocytes (NRVCMs) and the HL-1 mouse atrial cardiomyocyte line (Claycomb et al., 1998). Previous in vitro analyses comparing primary cardiomyocytes and HL-1 cells with in vivo models of cardiac disease demonstrated the utility of these cell culture models for defining molecular mechanisms underlying desmosome-related cardiac disorders (Garcia-Gras et al., 2006; Cerrone et al., 2012, 2014; Lyon et al., 2014).
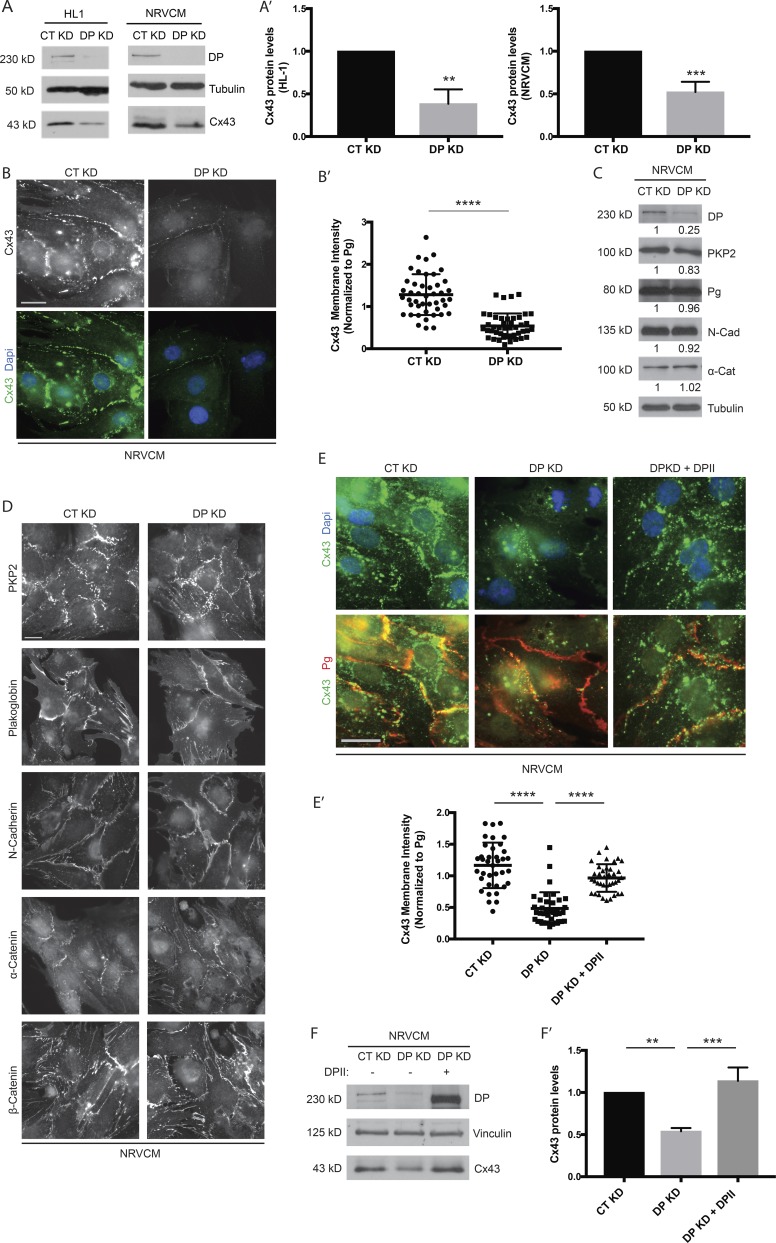
To verify the dependence of Cx43 protein levels on the presence of DP in vitro, freshly isolated NRVCMs were treated with an adenovirus harboring either control (nontargeting) or DP KD shRNA constructs, and HL-1 cells were transfected with either control or DP KD–specific siRNA. In both cardiac cell types, depletion of DP resulted in a substantial decrease in Cx43 protein levels, consistent with that observed in a cardiac restricted mouse model harboring DP deletion (Fig. 1, A and A′; Lyon et al., 2014). Furthermore, the fluorescence intensity of Cx43 staining at cell–cell junctions was substantially reduced in DP KD cardiomyocytes compared with controls (Fig. 1, B and B′). DP loss did not detectably alter the expression or distribution of other cardiac junctional components whose disruption might have contributed to the observed decrease in Cx43, including plakoglobin (Pg), N-cadherin, and α-catenin (Fig. 1, C and D). Although DP KD resulted in a small decrease in plakophilin 2 (PKP2) levels (Fig. 1 C), we previously showed that DP can restore defects resulting from PKP2 loss (Dubash et al., 2016).
Figure 1.
DP KD results in decreased levels of Cx43 in cardiac cells. (A) NRVCMs and HL-1 cells were treated with DP-specific shRNA and siRNA, respectively, and samples were analyzed 72 h after infection/transfection. DP KD leads to a significant reduction of Cx43 protein levels in both cell types (HL-1, n = 4; NRVCM, n = 6. (A′) Densitometry quantification of Cx43 levels normalized to loading control. (B) Control (CT) and DP KD NRVCMs grown on glass coverslips and stained for Cx43. DP KD results in a decrease of Cx43 junctional localization (n = 5). (B′) Quantification of Cx43 membrane intensity (normalized to junctional Pg signal) in control and DP KD NRVCMs. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by two tailed Student’s t test. (C) Control and DP KD NRVCMs were blotted for the following cardiac junctional components: DP, PKP2, Pg, N-cadherin, and α-catenin (n = 3). Densitometry quantification is representative of three independent experiments. (D) Control and DP KD NRVCMs were seeded on glass coverslips and stained for PKP2, Pg, N-cadherin, α-catenin, and β-catenin (n = 3). (E and E′) Re-expression of full-length human DPII rescues Cx43 junctional localization in NRVCMs (n = 3). Quantification of junctional Cx43 (normalized to Pg) shows that DPII expression causes a significant restoration of Cx43 membrane localization. Bars, 20 µm. (F) Re-expression of DPII restores Cx43 protein levels in NRVCMs by Western blot (n = 3). (F′) Densitometry quantification of Cx43 levels normalized to loading control. Error bars represent mean ± SD. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by one-way ANOVA followed by Tukey’s post hoc test.
To test whether the loss of DP is responsible for the observed decrease in Cx43 protein levels, we assessed whether expression of exogenous DP was sufficient to recover Cx43 expression. To achieve this, DPII-GFP was delivered using an adenovirus in the background of DP KD. DPII is a naturally occurring isoform of DPI that is lacking in the rod domain (6.8 vs. 8.6 kb). This construct was used to overcome technical limits in size for adenoviral packaging and transduction. Like DPI, DPII is localized to desmosomes and confers adhesive strength to cell–cell junctions (Angst et al., 1990; Green et al., 1990; Cabral et al., 2012; Patel and Green, 2014). Expression of full-length DPII was sufficient to restore Cx43 localization to cell junctions as assessed by immunofluorescence (Fig. 1, E and E′). Additionally, DPII expression restored Cx43 protein to control levels in the background of DP KD as assessed by Western blot (Fig. 1, F and F′). Collectively, these data demonstrate that DP is essential for the stable expression of Cx43 protein and its incorporation into gap junctions in cardiac cells.
DP loss results in elevated phosphorylation of Cx43 at MAPK-regulated S279/282 sites
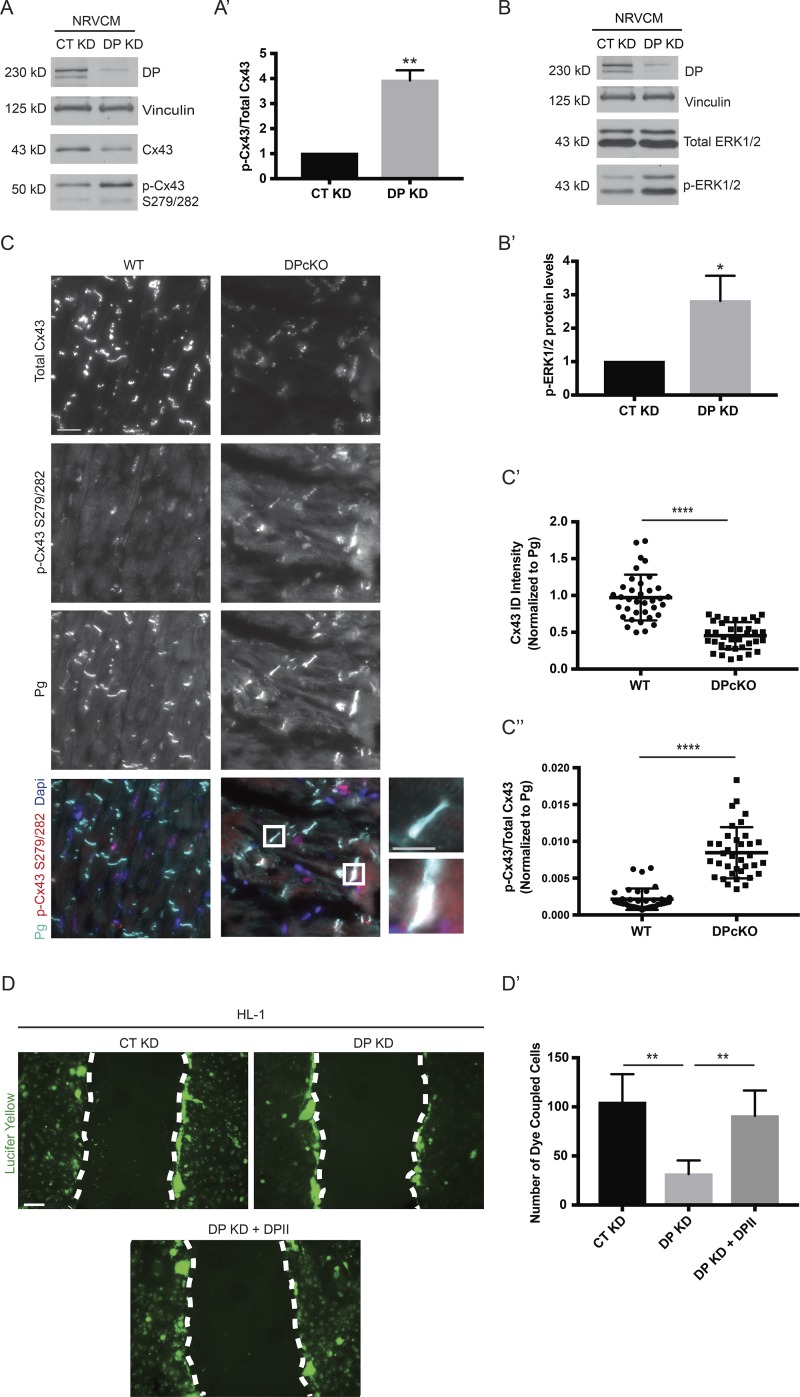
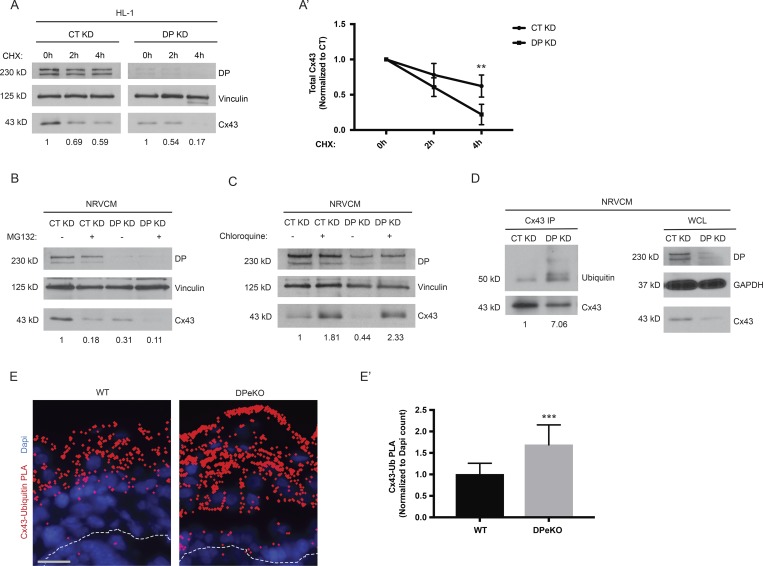
Connexins exhibit short half-lives on the order of 1–5 h compared with other membrane proteins, which typically exhibit half-lives on the order of 24–48 h (Laird et al., 1991; Beardslee et al., 1998). Gap junctions are therefore highly dynamic structures that are constantly undergoing assembly, remodeling, and turnover (Solan and Lampe, 2014). Posttranslational modification of Cx43, particularly phosphorylation, has been extensively studied as a mechanism by which gap junction dynamics are regulated. Of the 26 serines in the intracellular serine-rich C-terminal region of Cx43, at least 19, along with two of six tyrosines, have been identified as phosphorylation sites (Axelsen et al., 2013). Given the importance of phosphorylation for the connexin life cycle, we asked whether DP plays a role in modulating Cx43 phosphorylation. To address this question, control and DP KD NRVCMs were tested with phosphosite-specific Cx43 antibodies (Márquez-Rosado et al., 2012). Interestingly, an increase in phosphorylation of a linked pair of Cx43 sites at S279/282 was detected in response to DP KD (Fig. 2 A). Phosphorylation at this pair has been implicated in down-regulation of gap junction intercellular communication as well as internalization and degradation of Cx43 (Norris et al., 2008; Johnson et al., 2013; Fong et al., 2014). When normalized to total Cx43 protein levels that are reduced in the absence of DP, the ratio of p-Cx43 S279/282 to total Cx43 increased approximately fourfold in DP KD NRVCMs (Fig. 2 A′).
Figure 2.
Loss of DP leads to elevated phosphorylation of Cx43 at S279/282 in cardiac models. (A) NRVCMs treated with control and DP KD shRNA were blotted for total Cx43 and p-Cx43 S279/282 72 h after infection (n = 3). (A′) Densitometry was performed to quantify the ratio of p-Cx43 S279/282 to total Cx43 levels. (B) Control and DP KD NRVCMs were blotted for p-ERK1/2 levels, revealing an increase in p-ERK1/2 levels in DP KD compared with control (n = 4). (B′) Densitometry quantification of p-ERK1/2 levels normalized to loading controls. (C) WT and DPcKO heart sections were stained for total Cx43, p-Cx43 S279/282, and Pg (n = 3). Boxed regions in color overlays are magnified in the panels on the right. Bars: 20 µm (main images); 10 µm (insets). (C′) Quantification of total Cx43 localized at IDs (normalized to Pg). (C″) Quantification of the p-Cx43 S279/282 to total Cx43 ratio localized at IDs. *, P < 0.05; **, P < 0.01; ****, P < 0.0001 by two-tailed Student’s t test. (D and D′) Scrape loading dye transfer assays were performed on CT KD, DP KD, and DP KD HL-1 cells expressing full-length DPII. The total number of cells displaying LY dye transfer was quantified (n = 3). Bar, 100 µm. Error bars represent mean ± SD. **, P < 0.01 by one-way ANOVA followed by Tukey’s post hoc test.
As phosphorylation at Cx43 S279/282 sites is mediated by ERK1/2, we tested whether depletion of DP affects activation of the ERK1/2–MAPK pathway (Warn-Cramer et al., 1996). Indeed, by Western blot, robust activation of p-ERK1/2 levels was observed in response to DP KD in NRVCMs (Fig. 2, B and B′). The original observation that DP loss leads to reduced Cx43 protein expression was made in a cardiac conditional KO mouse model (Lyon et al., 2014). To test whether DP loss leads to increased p-Cx43 S279/282 in cardiac tissue, immunofluorescence analysis was performed on cardiac sections from the aforementioned DPcKO mouse. Cx43 staining intensity at the IDs of DPcKO heart sections was significantly decreased compared with WT (Fig. 2, C and C′). However, DPcKO cardiac sections display elevated levels of p-Cx43 S279/282 staining compared with WT (Fig. 2 C). Quantification of p-Cx43 S279/282 normalized to total Cx43 at IDs (as marked by Pg staining) revealed an approximately fourfold increase in the p-Cx43/total Cx43 ratio in DPcKO versus WT hearts (Fig. 2 C″).
To determine whether the observed loss of Cx43 was associated with a functional loss of gap junction intercellular communication (GJIC), scrape loading dye transfer assays were performed. HL-1 cells grown to a confluent monolayer were scraped and treated with the gap junction permeable dye Lucifer Yellow (LY). GJIC was evaluated by the number of cells displaying positive LY signal, indicative of level of dye transfer between neighboring cells. DP depletion resulted in a significant reduction in LY signal transferred between cells compared with control (Fig. 2, D and D′). Furthermore, reexpression of full-length DPII restored levels of dye transfer to that of control cells (Fig. 2, D and D′). These data demonstrate that the decreased expression and assembly of Cx43 into gap junctions caused by DP loss perturbs gap junction function.
Cx43 phosphorylation is altered in ACM
To determine whether this pathway has relevance in ACM, a blinded experiment was performed using cardiac sections from healthy controls or ACM patients harboring pathogenic mutations in DP or PKP2. All patient samples discussed in this section are unpublished. Immunofluorescence analysis of total Cx43 and p-Cx43 S279/282 was assessed, and in parallel, DP staining was performed to evaluate the consequence of the mutations to DP’s localization at IDs. Healthy control tissue displayed strong total Cx43 signal at IDs (Fig. 3 B and S1 A). Additionally, control sections did not show appreciable ID or cytoplasmic p-Cx43 S279/282 signal (Fig. 3 B). In contrast, cardiac tissue from two of the ACM patients harboring mutations in DP at 1,218+1 G→A and C3337T, respectively, displayed almost complete loss of total Cx43. The only detectable signal was nonspecific lipofuscin autofluorescence (Fig. S1 A, white arrowheads). These data are consistent with the reported loss of Cx43 from IDs of both DP ACM and Carvajal syndrome (cardiocutaneous syndrome caused by DP mutations) patients (Kaplan et al., 2004; Asimaki et al., 2009; Gomes et al., 2012). Assessment of p-Cx43 S279/282 could not be performed in these patients as we could not detect appreciable total Cx43 signal. Both patients also showed severe impairment of DP localization to IDs as marked by Pg staining compared with healthy controls (Fig. S1, B and B′).
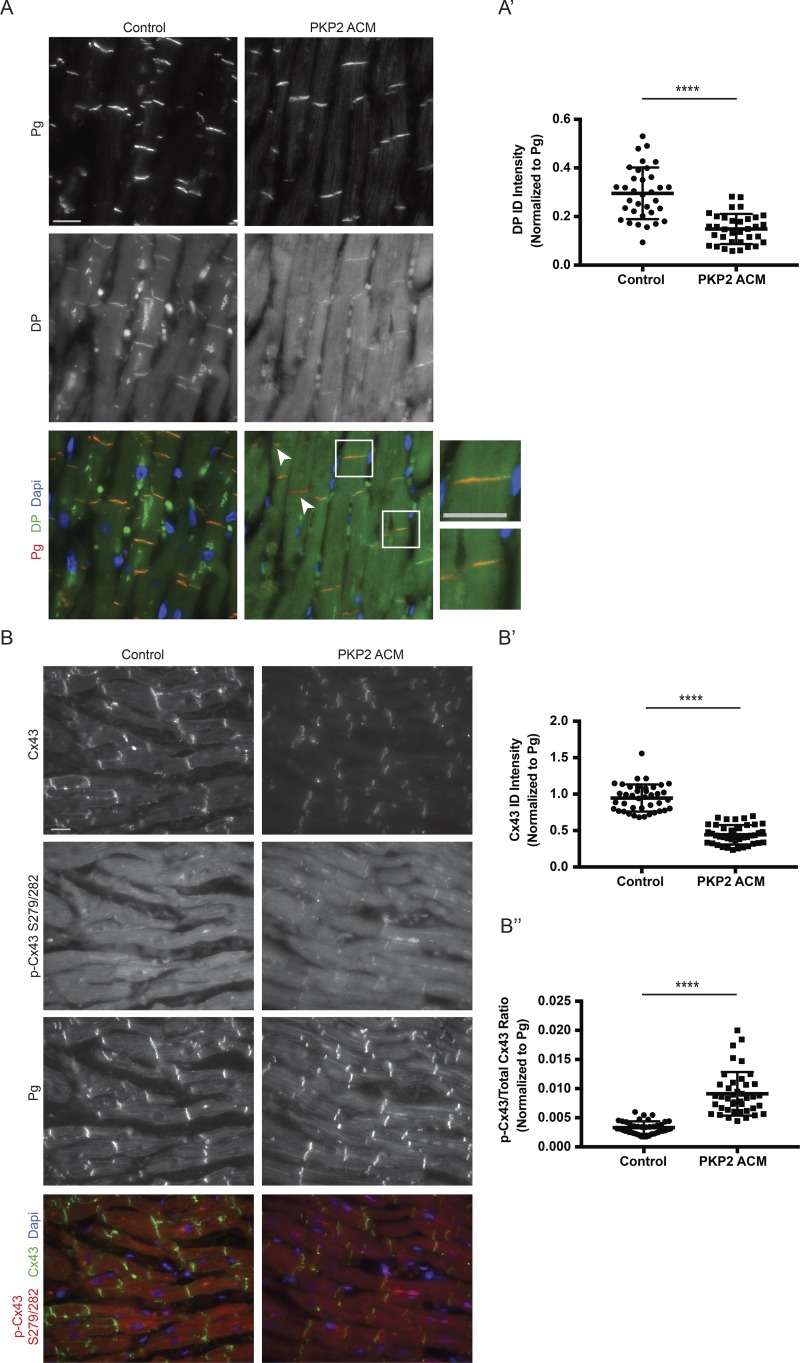
Figure 3.
Increased phosphorylation of Cx43 at S279/282 is conserved in a PKP2 ACM patient sample. (A and A′) Healthy control and PKP2 ACM (971_980del10) human cardiac sections were stained for DP and Pg as a marker of IDs (n = 1). DP ID localization was quantified (normalized to Pg ID intensity). Boxed regions in color overlays are magnified in the panels on the right showing regions with retained DP ID localization. White arrows denote IDs showing positive Pg staining without presence of DP. (B) Control and PKP2 ACM cardiac sections were stained for total Cx43, p-Cx43 S279/282, and Pg (n = 1). Bars: 20 µm (main images); 10 µm (insets). (B′) Quantification of total Cx43 localized at IDs normalized to Pg. (B″) Quantification of the p-Cx43 S279/282 to total Cx43 ratio at IDs normalized to Pg. Error bars represent mean ± SD. ****, P < 0.0001 by two-tailed Student’s t test.
In contrast, analysis of DP ID localization in cardiac tissue from an ACM patient harboring a pathogenic mutation in PKP2 at 971_980del10 showed significantly impaired (Fig. 3 A, white arrowheads) but not complete loss of DP from IDs compared with control (Fig. 3 A [inset] and Fig. 3 A′). The loss of DP resulting from a PKP2 mutation is consistent with our previous work showing that PKP2 loss in cardiac cells promotes the destabilization of DP protein and its subsequent degradation via the proteasome (Dubash et al., 2016). Furthermore, this patient displayed significantly lowered Cx43 ID staining compared with control without complete loss of expression (Fig. 3, B and B′). This patient’s cardiac tissue showed an approximately threefold increase in p-Cx43 S279/282 to total Cx43 (Fig. 3 B″), reminiscent of elevated p-Cx43 S279/282 signal seen in DPcKO murine hearts (Fig. 2 C). The milder Cx43 phenotype observed in this ACM patient compared with those harboring pathogenic DP mutations may have resulted from the presence of residual DP at IDs.
Elevated phosphorylation of Cx43 at S279/282 is conserved in an epidermal restricted DP mouse KO model
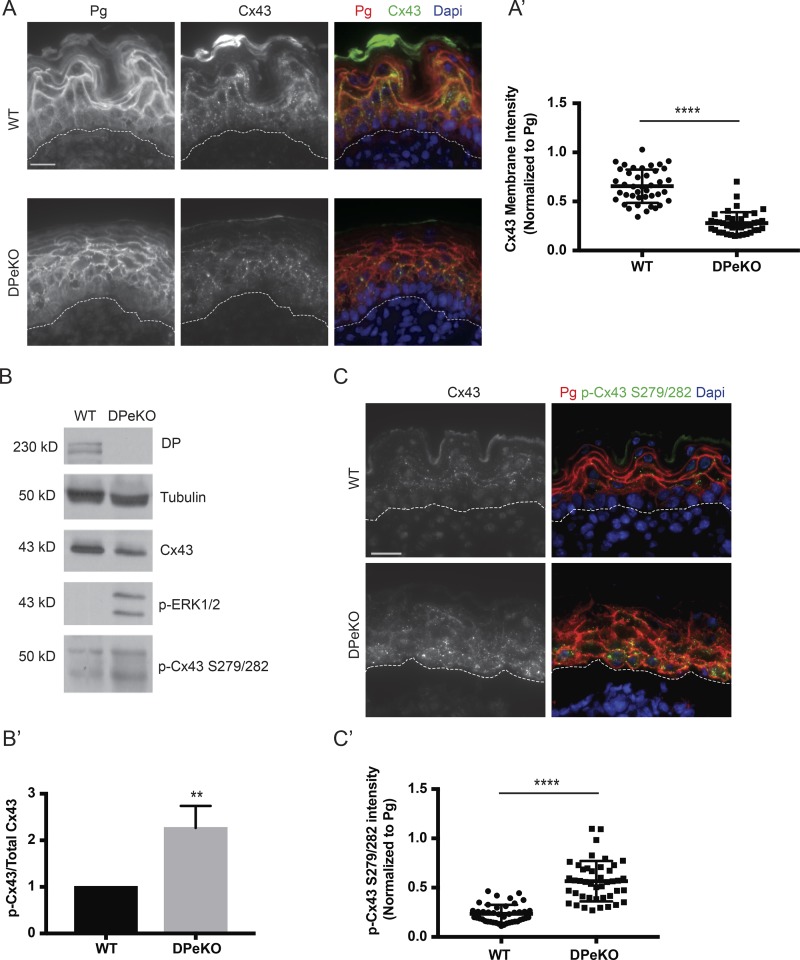
In addition to cardiocutaneous disorders such as Carvajal syndrome, dominant de novo DP mutations have recently been reported to cause erythrokeratodermia–cardiomyopathy syndrome associated with impaired Cx43 membrane localization in the skin (Boyden et al., 2016). Thus, we next addressed whether DP regulates Cx43 levels in the epidermis, where it plays a vital role in epidermal proliferation and differentiation (Norgett et al., 2000; Langlois et al., 2007, 2010), using an epidermal conditional KO mouse model of DP (DPeKO; Vasioukhin et al., 2001). Immunofluorescence analysis demonstrated that the previously reported abnormal morphology in DPeKO epidermis was accompanied by significantly lower Cx43 membrane staining (Fig. 4, A and A′; Vasioukhin et al., 2001). We also observed an increase in p-Cx43 S279/282 levels in DPeKO samples compared with WT by Western blot (Fig. 4 B) and immunofluorescence (Fig. 4, C and C′), paralleling what was observed in cardiac cells. Furthermore, the ratio of p-Cx43 S279/282 to total Cx43 levels revealed an approximately twofold increase in DPeKO samples compared with WT (Fig. 4 B′). Accompanying the observed changes in Cx43, p-ERK1/2 levels increased in DPeKO epidermis compared with WT (Fig. 4 B). Collectively, these data suggest that DP-dependent regulation of Cx43 phosphorylation at the p-ERK1/2–MAPK-specific residues, S279/282, is a conserved mechanism in cardiac and cutaneous models.
Figure 4.
Elevated Cx43 phosphorylation at S279/282 is conserved in an epidermal restricted model of DP loss. (A and A′) WT and DPeKO epidermal sections were stained for Cx43 and Pg. DPeKO sections display significantly lowered Cx43 membrane intensity (n = 3). (B) Western blot analysis performed on WT and DPeKO lysates show decreased total Cx43 and increases in p-ERK1/2 and p-Cx43 S279/282 levels in DPeKO compared with WT (n = 3). (B′) Densitometry was performed to quantify the ratio of p-Cx43 S279/282 to total Cx43 levels. (C and C′) WT and DPeKO epidermis were stained for p-Cx43 S279/282 and Pg. DPeKO show significantly higher levels of p-Cx43 S279/282 staining intensity (n = 3). Bars, 20 µm. Error bars represent mean ± SD. **, P < 0.01; ****, P < 0.0001 by two-tailed Student’s t test.
DP loss leads to elevated ubiquitination and degradation of Cx43
Phosphorylation of Cx43 at S279/282 has been shown to trigger internalization and degradation of Cx43 (Fong et al., 2014). To test whether depletion of DP leads to degradation of Cx43 in cardiac cells, Cx43 protein stability was analyzed in response to DP KD by treating HL-1 cells with cycloheximide for the indicated times. Cx43 protein degraded at a higher rate in DP KD cells compared with control (Fig. 5, A and A′). Quantitative PCR (qPCR) analysis showed that DP KD did not have a significant effect on Cx43 transcript levels in NRVCMs, suggesting that DP-dependent regulation of Cx43 expression does not occur at the mRNA level (Fig. S2). Degradation of Cx43 has been shown to occur via either the proteasomal or lysosomal pathways depending on cell type and degradation stimulus (Laing et al., 1997; Leithe and Rivedal, 2004; Leithe et al., 2006). To identify the machinery responsible for Cx43 degradation in response to DP loss, we used the proteasomal inhibitor MG132 or the lysosomal inhibitor chloroquine in the background of DP KD. Surprisingly, MG132 treatment resulted in a decrease in Cx43 expression in control cells without decreasing DP levels (Fig. 5 B). MG132 treatment further decreased the levels of Cx43 observed upon DP KD (Fig. 5 B). In contrast, chloroquine treatment caused an accumulation Cx43 protein levels in control cells and restored Cx43 expression in DP KD cells. These data are consistent with lysosomal-mediated degradation of Cx43 at steady state, which is exacerbated by loss of DP (Fig. 5 C).
Figure 5.
Loss of DP promotes ubiquitination and degradation of Cx43. (A) Control and DP KD HL-1 cells were treated with cycloheximide for the indicated time points, and samples were blotted for DP and Cx43 (n = 4). (A′) Densitometry quantification was performed to assess rate of Cx43 degradation. (B) MG132 was used to assess role of proteasomal degradation of Cx43 caused by DP loss in NRVCMs (n = 3). (C) The lysosomal inhibitor Chloroquine was able to restore Cx43 expression levels in response to DP KD in NRVCMs (n = 3). (D) Cx43 was immunoprecipitated from control and DP KD NRVCMs. Samples were subsequently blotted for Cx43 and ubiquitin. Whole-cell lysate (WCL) from these samples is to the right (n = 4). (E and E′) WT and DPeKO epidermal sections were subjected to PLA analysis with Cx43 and ubiquitin antibody pairs (n = 3). Bar, 20 µm. Quantification of PLA dots normalized to DAPI count revealed a significant increase in Cx43-Ubiquitin PLA in DPeKO compared with WT skin. Error bars represent mean ± SD. **, P < 0.01; ***, P < 0.001 by two-tailed Student’s t test.
Cx43 is modified by ubiquitination, a tag that is thought to target the connexin for subsequent degradation (Laing and Beyer, 1995; Leithe and Rivedal, 2004; Chen et al., 2012). To determine whether Cx43 is ubiquitinated in response to DP loss, immunoprecipitates of endogenous Cx43 from control and DP KD NRVCMs were immunoblotted for endogenous ubiquitin. Using the mono/polyubiquitin antibody FK2, it was observed that Cx43 is more highly ubiquitinated in DP KD samples compared with control (Fig. 5 D). To determine whether increased ubiquitination of Cx43 is conserved in mouse epidermis lacking DP, proximity ligation assay (PLA) was used to detect ubiquitin modification of Cx43 in mouse tissue sections. PLA allows for the in situ visualization of two antigens that are in close proximity to each other (40–100 nm). This assay is used frequently to detect posttranslational modifications on target proteins (Fredriksson et al., 2002; Söderberg et al., 2006; Ristic et al., 2016). DPeKO epidermis produced significantly more Cx43-Ub PLA signal compared with WT, suggesting that DP-dependent ubiquitination of Cx43 is conserved in the heart and skin (Fig. 5, E and E′).
Blockade of Cx43 phosphorylation is sufficient to rescue Cx43 expression and localization
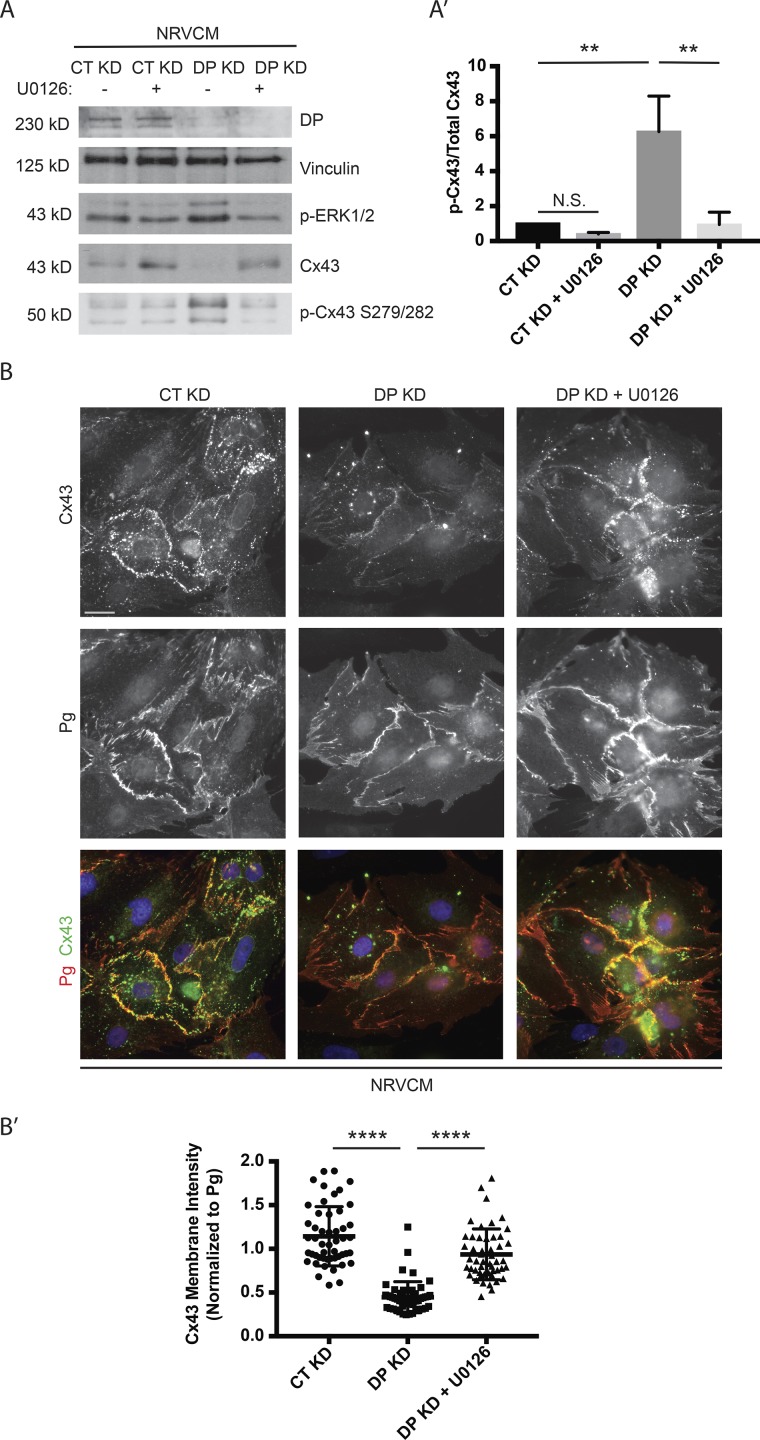
Our data suggest that DP loss activates p-ERK1/2–MAPK, which in turn phosphorylates Cx43 on S279/282 to target Cx43 for degradation. To directly test whether preventing Cx43 phosphorylation blocks degradation and restores junctional localization of Cx43, DP KD NRVCMs were treated with U0126, a small-molecule inhibitor of MEK1/2, the direct upstream activators of p-ERK1/2. U0126 treatment restored p-ERK1/2 and total Cx43 expression levels to those of control (Fig. 6 A). Additionally, when normalized to total Cx43 levels, the p-Cx43 S279/282 signal was also restored to a level comparable with control (Fig. 6 A′).
Figure 6.
Blockade of p-ERK1/2–MAPK activation is sufficient to rescue Cx43 expression and localization. (A) Control, DP KD, and DP KD NRVCMs treated with 5 µM U0126 or vehicle were blotted for DP, p-ERK1/2, Cx43, and p-Cx43 S279/282 (n = 4). (A′) Densitometry quantification shows that U0126 treatment was sufficient to significantly restore the p-Cx43 S279/282 to total Cx43 ratio to control levels. (B) Immunofluorescence analysis of NRVCMs treated with control or DP KD shRNA in the presence or absence of 5 µM U0126 stained for Cx43 and Pg (n = 3). Bar, 20 µm. (B′) Quantification of Cx43 junctional localization normalized to junctional Pg. U0126 treatment led to significant restoration of junctional Cx43. Error bars represent mean ± SD. **, P < 0.01; ****, P < 0.0001 by one-way ANOVA followed by Tukey’s post hoc test.
Immunofluorescence analysis using Pg as a marker of cell junctions revealed that U0126 treatment also partially restores localization of Cx43 to cell junctions (Fig. 6, B and B′). The difference in restoration between protein levels and localization may be explained by our previous demonstration that DP also functions posttranslationally to promote the delivery of Cx43 to cell junctions via microtubules (MTs) in keratinocytes via an association with MT plus end protein EB1 (Patel et al., 2014). These data suggest that blocking the initial phosphorylation of Cx43 by p-ERK1/2 is sufficient to prevent Cx43 loss in DP-deficient conditions and to help restore membrane localization of Cx43, where it can be assembled into gap junction complexes.
DP inhibits the ERK–MAPK pathway via suppression of RAS
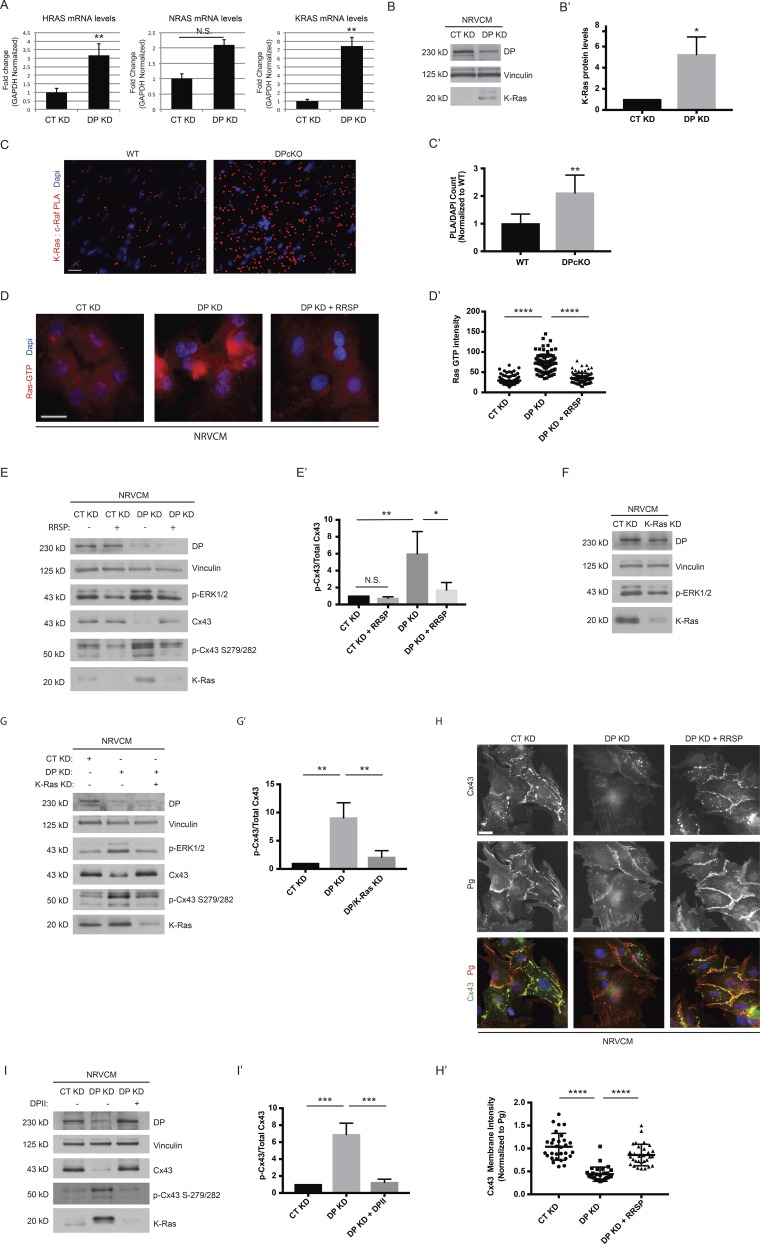
Toward identifying candidate mechanisms by which DP loss activates the ERK1/2–MAPK pathway, we compared control and DPKD NRVCMs using large-scale unbiased RNA sequencing (RNA-seq) analysis. Notably, levels of V-Ki-ras2 Kirsten rat sarcoma viral oncogene homologue (KRAS) RNA were significantly up-regulated in DP KD samples. K-Ras as is one of the three major isoforms of the Ras GTPase family, which include H-Ras and N-Ras (Castellano and Santos, 2011; Hobbs et al., 2016). Mutations in these proteins frequently lead to malignancies, with gain-of-function missense mutations in RAS genes having been found in ∼25% of human cancers (Hobbs et al., 2016). Ras is well known to initiate a signaling cascade involving the subsequent activation of the Raf–MEK–ERK pathway (Roberts and Der, 2007). To validate DP’s regulation of Ras transcript levels, qPCR analysis of the three major Ras isoforms (KRAS, HRAS, and NRAS) was compared in control and DP KD NRVCMs. Depletion of DP resulted in significant increases of K-Ras and H-Ras transcript levels but not of N-Ras (Fig. 7 A).
Figure 7.
DP modulates the ERK1/2–MAPK pathway via regulation of Ras expression. (A) Control and DP KD NRVCMs were analyzed for mRNA levels of KRAS, HRAS, and NRAS (n = 3). (B) NRVCMs treated with control and DP KD shRNA were blotted with a K-Ras antibody (n = 3). (B′) Densitometry quantification of K-ras levels normalized to loading controls. (C and C′) WT and DPcKO cardiac sections were subjected to PLA analysis with K-Ras/c-Raf antibody pairs (n = 3). Quantification of PLA signal normalized to DAPI count shows that K-Ras/c-Raf PLA signal is significantly increased in DPcKO compared with WT hearts. *, P < 0.05; **, P < 0.01 by two-tailed Student’s t test. (D) Immunofluorescence analysis of Ras-GTP staining was performed on control, DP KD, and DP KD NRVCMs treated with 2 nM RRSP (n = 3). (D′) Quantification of Ras GTP staining intensity. (E) CT KD and DP KD were compared with CT KD and DP KD NRVCMs treated with 2 nM RRSP by blotting for DP, p-ERK1/2, p-Cx43 S279/282, and Cx43 (n = 3). (E′) Densitometry quantification of p-Cx43 S279/282 to total Cx43 ratio. (F) Control and K-Ras KD NRVCMs were blotted for DP, p-ERK1/2, and K-Ras protein levels (n = 3). (G and G′) Control, DP KD, and DP/K-Ras double KD NRVCMs were subjected to Western blot analysis for DP, p-ERK1/2, Cx43, p-Cx43 S279/282, and K-Ras levels (n = 3). The ratio of Cx43 to p-Cx43 S279/282 was quantified by densitometry. (H) Control, DP KD, and DP KD NRVCMs treated with 2 nM RRSP were stained for Cx43 and Pg as a marker of cell junctions (n = 3). (H′) Quantification of Cx43 junctional intensity normalized to junctional Pg. Bars, 20 µm. (I and I′) Control, DP KD, and DP KD NRVCMs expressing DPII were blotted for DP, Cx43, p-Cx43 S279/282, and K-Ras. The ratio of Cx43 to p-Cx43 S279/282 was quantified by densitometry. Error bars represent mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by one-way ANOVA followed by Tukey’s post hoc test.
Because the KRAS transcript displayed the most substantial increase (sevenfold), we assessed K-Ras protein levels in response to DP KD. Western blot analysis showed that K-Ras protein expression is elevated in DP KD cells compared with control (Fig. 7, B and B′). The protein kinase c-Raf directly binds to GTP-loaded Ras proteins to propagate the MAPK pathway via the subsequent phosphorylation and activation of MEK, the kinase that in turn phosphorylates and activates ERK1/2 (Roberts and Der, 2007). As such, we exploited PLA to measure Ras–Raf interactions as an indicator of elevated Ras activation in vivo under DP KO conditions. K-Ras/c-Raf PLA signal was significantly increased in DPcKO hearts compared with WT, affirming that DP-dependent Ras activation is conserved in vivo (Fig. 7, C and C′).
Toward determining whether K-Ras is responsible for DP-dependent activation of the ERK1/2–MAPK pathway, we used a recently described bacterial toxin-derived Ras/Rap1-specific peptidase (RRSP; Antic et al., 2015). RRSP’s ability to inhibit Ras activity in mammalian cells is notable as Ras proteins have been notoriously difficult to target pharmacologically (Cox et al., 2014). RRSP is derived from DUF5Vv (domain of unknown function in fifth position), a naturally occurring effector domain from the multifunctional-autoprocessing-repeats-in-toxins (MARTX) toxin of the bacterium V. vulnificus (Antic et al., 2015; Biancucci et al., 2017). This protein represents a new class of endopeptidase that specifically processes all three major Ras isoforms as well as Rap1, blunting their activity and downstream activation of the ERK1/2–MAPK pathway, and which has so far been tested solely on cancer cell lines in an in vitro setting (Antic et al., 2015).
RRSP was functionalized for delivery into cells by fusion of the DUF5Vv domain to the N terminus of anthrax toxin lethal factor (LFNDUF5Vv), followed by introduction of the fusion protein to NRVCMs using anthrax toxin protective agent (PA; Antic et al., 2015). RRSP effectively reduced p-ERK1/2 levels in NRVCMs, consistent with dampened Ras activation (Fig. S3). Having validated RRSP efficacy in cardiomyocytes, we next tested whether RRSP would be able to rescue DP-dependent Ras activation. We used a Ras-GTP antibody that specifically recognizes Ras in its GTP-bound active state. Immunofluorescence analysis revealed that DP KD resulted in a significant up-regulation of Ras-GTP signal, which was restored to control levels with RRSP treatment (Fig. 7, D and D′). RRSP treatment also efficiently inhibited p-ERK1/2 activation that occurred in response to DP KD (Fig. 7 E).
To confirm that RRSP suppresses p-ERK by inhibiting K-Ras, we generated shRNA specific to K-Ras and showed that it is capable of knocking down K-Ras along with a concurrent decrease in p-ERK1/2 in NRVCMs (Fig. 7 F). We next performed a double KD of K-Ras and DP and showed that silencing of K-Ras reduced p-ERK1/2 back to control levels in DP-deficient NRVCMs, indicating that DP-dependent activation of the ERK1/2–MAPK pathway is caused by elevated K-Ras signaling (Fig. 7 G). As robust activation of ERK1/2–MAPK signaling was observed in an epidermal model of DP deletion (Fig. 4 B), we addressed whether DP’s effects on Ras expression are conserved in the skin. qPCR analysis revealed that transcript levels of all three major Ras isoforms are not significantly changed in DPeKO epidermis compared with WT (Fig. S4). This finding is consistent with the idea that ERK1/2–MAPK activation depends on factors other than modulation of Ras mRNA levels in DPeKO epidermis.
To determine whether ERK-dependent reduction of Cx43 depends on K-Ras in DP-deficient cells, we assessed Cx43 levels in RRSP-treated or K-Ras KD NRVCMs. Both RRSP and K-Ras shRNA restored total Cx43 as well as the ratio of p-Cx43 S279/282 to that of control samples (Fig. 7, E, E′, G, and G′). Similar to U0126 treatment, RRSP also significantly restored Cx43 localization to cell junctions in NRVCMs in DP KD cells (Fig. 7, H and H′). Finally, DPII expression in the background of DP KD was sufficient to restore K-Ras, p-ERK1/2, and the ratio of p-Cx43 S279/282 to total Cx43 levels to that of control (Fig. 7, I and I′).
Discussion
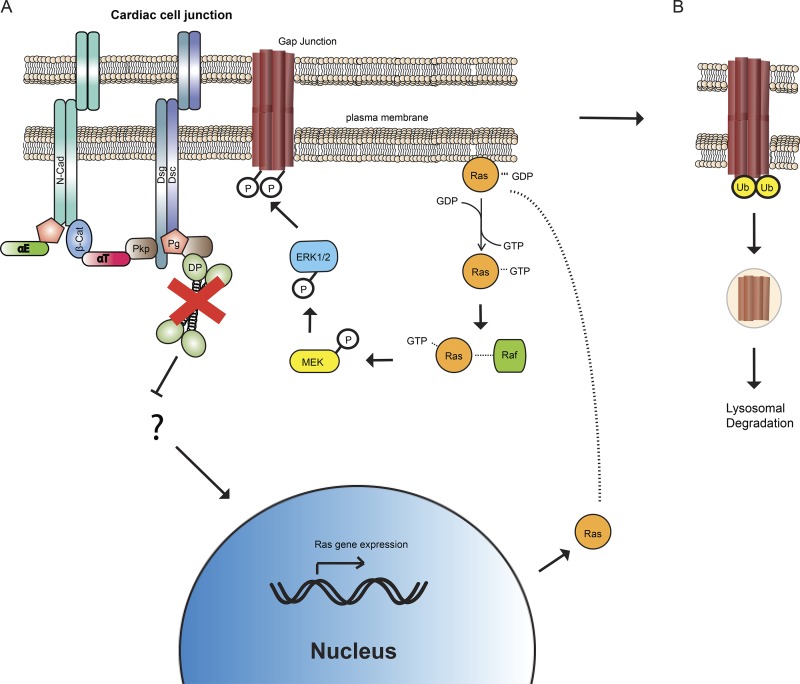
This work reveals a novel mechanism by which DP keeps Ras in check to stabilize Cx43 in the gap junctions of cardiomyocytes. Upon loss of DP either in vitro or in vivo, ERK1/2–MAPK signaling is activated, resulting in phosphorylation of Cx43 at S279/282, an event that signals Cx43 for degradation that is performed via the lysosome (Fig. 8). This finding provides new insight into the role of DP in the maintenance of gap junction stability and provides a potential explanation for the previously observed decrease in Cx43 expression identified in a mouse model of ACM (Lyon et al., 2014). In these mice, diminished Cx43 was associated with cardiac electrical conduction defects before fibrofatty replacement of the myocardium, similar to the concealed phase of ACM (Asimaki and Saffitz, 2012; Lyon et al., 2014). Our data provide a potential explanation for how DP loss could contribute to the concealed phase through regulation of Cx43 protein levels via modulation of Cx43 posttranslational modifications.
Figure 8.
Model of DP dependent regulation of Cx43 phosphorylation and degradation. (A) Loss of DP leads to elevated Ras gene expression that results in elevated Ras activation. Activation of the Ras signaling pathway leads to increased p-ERK1/2–MAPK levels that in turn phosphorylate Cx43 at S279/282. N-Cad, N-Cadherin; β-Cat, β-catenin; αT, αT-catenin; αE, αE-catenin; Dsg, Desmoglein; Dsc, Desmocollin. (B) Loss of DP also results in increased ubiquitination of Cx43. Cx43 is subsequently targeted for degradation via a lysosomal pathway. Ub, Ubiquitin.
We identified Ras as a key link between DP and Cx43 regulation. Inhibition of Ras activity using a novel Ras inhibitor, RRSP, or by silencing with shRNA targeting K-Ras, demonstrated that ERK1/2 activation and associated loss of Cx43 expression was mediated by increased Ras expression and activity in DP-deficient cells. How DP regulates Ras mRNA levels is not yet understood. Although regulatory mechanisms controlling Ras transcription are not well characterized, one possibility is that loss of DP enhances the rate of Ras gene transcription via activation of transcription factors/cofactors. DP-dependent regulation of Ras transcript levels could also be through epigenetic mechanisms, for instance via the action of specific microRNAs that have been shown to be responsible for controlling Ras expression levels in various cell types (Shin et al., 2011; Wang et al., 2014; Zhang et al., 2014). To date, there have been no pharmacological inhibitors of Ras that have made it to the clinic despite massive efforts to target this pathway given its prevalence in human cancers, prompting the assertion that Ras proteins are “undruggable” (Baines et al., 2011; Cox et al., 2014). In addition to its ability to process all three major Ras isoforms, RRSP has also been shown to be effective toward the inhibition of various mutant forms of K-ras that are widely associated with human cancers (Antic et al., 2015). Our work provides additional evidence that RRSP is an effective inhibitor of Ras and establishes a potential use of this novel reagent for clinical purposes.
We also show that DP loss leads to activation of the ERK1/2–MAPK pathway in epidermal and cardiac models of DP deficiency. It is well established that elevated ERK1/2–MAPK signaling in the heart results in cardiac hypertrophy, an abnormal enlargement of the heart caused by increased cardiomyocyte size (Wang, 2007; Lorenz et al., 2009). Thus, it is plausible that abnormal ERK1/2 signaling could contribute to disease pathogenesis including cardiomyocyte hypertrophy in cardiac patients harboring mutations in DP (or factors that regulate its expression or localization). In epithelial cells, both activation of ERK1/2 signaling and loss or impairment of DP can lead to altered proliferation, migration, and differentiation (Matsubayashi et al., 2004; Setzer et al., 2004; Khavari and Rinn, 2007; Yang et al., 2012). That depletion of DP in the HaCaT keratinocyte cell line resulted in increased proliferation associated with elevated p-ERK1/2 levels is consistent with the possibility that DP-dependent regulation of cell behavior is mediated at least in part through its ability to attenuate ERK activity (Wan et al., 2007).
In this work, we use a previously published mouse model harboring cardiomyocyte specific conditional KO of DP using ventricular myosin light chain-2–Cre mice that recapitulates the pathological progression of ACM (Lyon et al., 2014). The authors observed wavefront propagation defects that were associated with loss of Cx43 expression. Our data suggest that DP loss triggers activation of the ERK1/2–MAPK pathway that then signals Cx43 for subsequent degradation by phosphorylation of the putative MAPK sites, S279/282. A conditional KO model whereby DP was deleted from the sinoatrial node (SAN) using the Hcn4-Cre-ERT2 mouse line has also been generated (Mezzano et al., 2016). Loss of DP from the SAN, which houses the pacemaker cells of the heart, resulted in beat-to-beat defects associated with loss of the pacemaker complex connexin, Cx45. Notably, no changes in Cx45 transcript level were detected, raising the possibility that Cx45 protein levels are regulated posttranslationally. Although Cx45 has been demonstrated to be a phosphoprotein with multiple serine residues in its C terminus, the exact role of Cx45 phosphorylation or the specific kinases that may modulate it are not known (Lampe and Lau, 2000; van Veen et al., 2000). Along with the data reported in this study, these observations raise the possibility that DP controls Cx45 expression through modulation of specific Cx45 phosphosites that govern its dynamics.
The importance of Cx43 to the cardiac conduction system was highlighted in a recent study that implicated tissue resident macrophages present in mouse and human atrioventricular nodes in the regulation of cardiac electrical conduction (Hulsmans et al., 2017). The authors demonstrate that macrophages and cardiomyocytes are connected by Cx43-based gap junctions, whereby the resident macrophages function to modulate the electrical conduction of cardiomyocytes (Hulsmans et al., 2017). This finding could have important implications for both cardiac homeostasis and diseases such as ACM, in which increased cytokine expression and inflammation may contribute to disease progression (Corrado et al., 1997; Campian et al., 2010; Sen-Chowdhry et al., 2010; Asimaki et al., 2011).
The physiological relevance of our findings is supported by our analysis of cardiac tissue from a patient harboring a pathogenic PKP2 ACM mutation, which exhibited elevated levels of p-Cx43 S279/282 compared with control heart, reminiscent of that observed in murine DPcKO hearts. This was interesting in light of our previous observation that loss of PKP2 in cardiac cells promotes DP destabilization through proteasome-mediated degradation (Dubash et al., 2016). Although previous studies have shown that depletion of PKP2 resulted in defects in DP localization to cell junctions, effects on its protein expression levels appear to be exclusive to the cardiac system (Bass-Zubek et al., 2008; Godsel et al., 2010). This is likely because PKP2 is the lone isoform expressed in cardiac cells as opposed to epithelial cells that express PKP1 and PKP3 (Bass-Zubek et al., 2009). In the same study, we demonstrated that a profibrotic signaling pathway triggered by the activation of TGF-β in response to PKP2 depletion was caused by the secondary loss of DP. This finding, coupled with the present observation that a PKP2 ACM mutation phenocopies the effects of DP loss, suggests the possibility that a portion of the pathogenic mutations in ACM associated with PKP2 may be corollary to its effects on DP expression and its loss.
Materials and methods
Isolation of NRVCMs, cell culture, and mouse models
NRVCMs were isolated as previously described (Dubash et al., 2016) from 1- to 3-d-old Sprague-Dawley rats (Charles River). Briefly, hearts were dissected from neonatal rats, enzymatically digested, and resuspended in M199 medium (Lonza) containing 10% FBS supplemented with 15 µM vitamin B12. Cells were then plated for 2 h on plastic cell culture dishes to separate the cardiac fibroblasts that adhere more rapidly than cardiomyocytes. The remaining cell suspension was then filtered through a 40-µM filter, spun down, and resuspended in M199 medium containing 10% horse serum, 15 µM vitamin B12, and BrdU. Plating of cells was then performed in six-well dishes or coverslips that had been coated with collagen type IV. Media was changed every 24–48 h, and adenoviral infection was performed 48 h after isolation. These protocols were conducted with the approval of the Northwestern University Institutional Animal Care and Use Committee, and all animal care protocols conform to National Institutes of Health (NIH) guidelines and the recommendations of the Panel on Euthanasia of the American Veterinary Medical Association. The HL-1 cell line was maintained with Claycomb medium (Sigma-Aldrich) supplemented with 10% FBS (Atlanta Biologicals), 0.1 mM norepinephrine, 2 mM l-glutamine, and penicillin/streptomycin solution (Sigma-Aldrich). Cells were grown on plastic dishes precoated with a solution of FN-0.02% gelatin.
To delete the DP gene in the mouse epidermis, conditional gene targeting was used whereby Exon 2 of the DP gene was flanked with LoxP sites and removed by breeding with mice transgenic for Cre recombinase under the control of the keratin 14 promoter (Vasioukhin et al., 2001). The K14-Cre transgenic and the DP-floxed (DSPflox/flox) mice were gifts from E. Fuchs (Rockefeller University, New York, NY). Epidermis obtained from E18.5 embryos were snap-frozen in liquid nitrogen and either processed for Western blotting or sectioned for immunofluorescence analysis. To delete the DP gene in the mouse myocardium, DP-floxed mice were bred with heterozygous ventricular myosin light chain-2 Cre [MLC2v(cre+)] mice. DPcKO mice and their control littermates were on a congenic C56Bl/6 background. Hearts obtained from 8-wk-old mice were snap-frozen in liquid nitrogen and sectioned for immunofluorescence analysis.
DNA, siRNA, and shRNA constructs, adenovirus production, and chemical reagents
The DPII-GFP construct was generated with the QuikChange II XL site-directed mutagenesis kit (Agilent Technologies) whereby nucleotides 3,584–5,380 were spliced out of a GFP-tagged full-length DPI construct (Godsel et al., 2005). Adenoviral DPII was generated using Gateway recombination, whereby DPII-GFP was cloned into the pAd CMV/V5-DEST vector. The EmGFP BLOCK-iT PolIII miR RNAi Expression Vector kit (Thermo Fisher Scientific) was used to design and generate control and rat DP-specific oligonucleotides (target sequences: 5′-AAACCGGAAACATCATCTCTT-3′ and 5′-TGGTAATAGTTGACCCAGAAA-3′). These oligonucleotides where then cloned into the pAd CMV/V5-DEST vector using Gateway recombination. Adenovirus was generated using the ViraPower Adenoviral Expression System (Invitrogen). K-Ras shRNA was generated from a rat K-Ras specific sequence within the lentiviral backbone pLKO (target sequence, 5′-CCGGCTATACATTAGTCCGAGAAATCTCGAGATTTCTCGGACTAATGTATAGTTTTTG-3′; Sigma-Aldrich). Lentiviral particles were generated by the Northwestern University Skin Disease Research Center DNA/RNA Delivery Core. DP-specific murine siRNA oligonucleotides were obtained from GE Healthcare (sense sequences: 5′-GUGCAGAACUUGGUAAACA-3′ and 5′-GCAUCCAGCUUCAGACAAA-3′). Cells were treated with the following reagents at the stated concentrations: 10 µg/ml cycloheximide, 100 µM chloroquine, 10 µM MG132, 5 µM U0126, and 2 nM RRSP.
Transfections
Transient transfections were performed using DharmaFECT (GE Healthcare). Cells plated at 60–70% confluence were incubated with 0.2–5 µg DNA and DharmaFECT reagent premixed in serum-free media. Fresh medium was added 24 h after transfection, and samples were lysed or fixed 72 h after transfection.
Antibodies
The following primary antibodies were used in this study: NW6 rabbit anti-DP directed against the C-terminal domain of DP (Angst et al., 1990), 1407 chicken anti-Pg (Aves Laboratories), mouse anti-PKP2 (651101; Progen), 115F mouse anti-DP (a gift from D. Garrod, University of Manchester, Manchester, UK), 12G10 mouse anti–α-tubulin (Developmental Studies Hybridoma Bank), rabbit anti-Cx43 (AB1728; EMD Millipore), mouse anti-Cx43 (MAB3068; EMD Millipore), IF1 mouse anti-Cx43 (P. Lampe, Fred Hutchinson Research Center, Seattle, WA), mouse anti–N-cadherin (Invitrogen), mouse anti–α-catenin, rabbit anti–β-catenin (C2206; Sigma-Aldrich), mouse anti-Vinculin (V9131; Sigma-Aldrich), rabbit anti–phospho-Cx43 S279/282 (sc-12900-R; Santa Cruz Biotechnology), rabbit anti–phospho-Cx43 S279/282 (P. Lampe), rabbit anti–p-44/42 (9101; Cell Signaling Technologies), rabbit anti–total 44/42 (V1141; Promega), FK2 mouse anti–ubiquitylated proteins (04-263; EMD Millipore), 1296 anti-Myosin VI (S. Polo, FIRC Institute of Molecular Oncology, Milan, Italy), mouse anti–K-ras (WH0003845M1; Sigma-Aldrich), and mouse anti–Ras-GTP (26909; NewEast Biosciences). Peroxidase-conjugated anti–mouse, –rabbit, and –chicken secondary antibodies were used for Western blot analysis (Kiekegaard & Perry Laboratories). Alexa Fluor 488/568/647–conjugated goat anti–mouse, –rabbit, and –chicken secondary antibodies were used for immunofluorescence assays (Invitrogen).
Western blotting and coimmunoprecipitation assays
For analysis of protein expression levels, cells were washed once with PBS and lysed in urea sample buffer (8 M deionized urea, 1% sodium dodecyl sulfate, 10% glycerol, 60 mM Tris, pH 6.8, and 5% β-mercaptoethanol). Total protein concentrations were equalized, and samples were run on 7.5–12% SDS-PAGE gels. Gels were transferred to polyvinylidene fluoride membranes to be probed with primary and secondary antibodies against proteins of interest.
Cells used for coimmunoprecipitation assays were washed twice with PBS and lysed with modified RIPA buffer (500 mM NaCl, 50 mM Tris, pH 7.6, 10 mM MgCl2, 1% Triton X-100, 0.1% SDS, and 0.5% deoxycholate) supplemented with complete protease inhibitor cocktail (Roche). Samples were then incubated with 1 µg antibody against the protein of interest to be pulled down by rotating overnight at 4°C. Subsequently, samples were incubated with Protein A/G PLUS Agarose beads (Santa Cruz Biotechnology) for 30 min at 4°C. Samples were washed and eluted in Laemmli buffer with 5% β-mercaptoethanol, followed by SDS-PAGE and Western blot analysis.
PLA
Reagents required for PLA analysis were purchased from Sigma-Aldrich. Cells were rinsed and fixed as described for conventional immunofluorescence. Samples were then incubated with primary antibodies for the proteins of interest for 1 h at 37°C followed by incubation with secondary PLA probes for 1 h at 37°C. This was followed by a 30-min incubation with PLA ligase for ligation of nucleotides on corresponding probes. Subsequently, PLA polymerase was added for 100 min at 37°C to amplify the PLA signal, which results in red fluorescent dots if the antigens in question are in close proximity. Samples were mounted with PLA mounting medium containing DAPI, and ImageJ software (NIH) was used to quantify the number of PLA dots, which were then normalized to DAPI.
qPCR
Total RNA was isolated from cells using the RNeasy Mini kit (Qiagen) according to the manufacturer’s instructions. Samples were then equalized for total RNA concentration, and cDNA was synthesized using the Superscript III First Strand kit (Invitrogen) according to the manufacturer’s instructions. qPCR was performed using SYBR Green PCR master mix (Applied Biosystems) along with the appropriate primers using a StepOnePlus instrument (Applied Biosystems). Calculations for relative mRNA levels were performed using the ΔΔCt method using levels of GAPDH mRNA as a control. Relative levels were presented as fold-change values compared with control samples. Two-tailed Student’s t test was used to perform statistical analysis, with P values <0.05 considered statistically significant.
Immunofluorescence and quantification of pixel intensity
Cells grown on glass coverslips were washed once with PBS and fixed using 4% PFA or ice-cold methanol. Cells were then permeabilized with 0.2% Triton X-100 in PBS. Primary antibodies were added to coverslips and incubated at 37°C for 1 h followed by multiple washes in PBS. Alexa Fluor secondary antibodies (Thermo Fisher Scientific) were added and incubated for 30 min, followed by PBS washes and mounting of coverslips in polyvinyl alcohol (Sigma-Aldrich). Samples were visualized using a DMR microscope (Leica Microsystems) fitted with 40-Å (NA 1.0, Plan Fluotar) and 63-Å (NA 1.32, Plan Apochromat) objective lenses, and images were captured with an Orca 100 charge-coupled device camera (model C4742-95; Hamamatsu Photonics) and MetaMorph 7.7 imaging software (Molecular Devices). Mean fluorescence pixel intensity was quantified using raw, unsaturated 12-bit images with line or area tracing as stated using ImageJ. For quantification of junctional/ID fluorescence intensity, cell borders as marked by Pg signal using ImageJ line tracing and fluorescence intensity of Pg and the costained protein were assessed from the same line position. Subsequently, signal intensity values of the protein of interest were normalized to Pg intensity.
Immunohistochemistry
Tissue samples were fixed in 10% neutral-buffered formalin and embedded in paraffin. Paraffin-embedded sections were deparaffinized in sequential xylene and ethanol washes. Antigen retrieval for paraffin-embedded sections was performed by heating samples to 95°C in 0.01 M citrate buffer. Sections were blocked in 10% normal goat serum and incubated with primary and secondary antibodies for 1 h at 37°C. After mounting in polyvinyl alcohol, sections were visualized as described above.
Surface biotinylation assay
Cells were washed twice with ice-cold PBS and incubated with 2 mg/ml EZ-Link Sulfo-NHS-SS-biotin (Thermo Fisher Scientific) for 30 min at 4°C. Excess biotin was quenched with 100 mM glycine in PBS by washing three times for 10 min each. Cells were then washed with PBS and lysed in RIPA buffer (10 mM Tris, pH 7.5, 140 mM NaCl, 1% Triton X-100, 0.1% SDS, 0.5% sodium deoxycholate, 5 mM EDTA, 2 mM EGTA, and complete protease inhibitor). Lysates were centrifuged, and cell supernatants were normalized for total protein. 30 µl whole cell lysate was saved for input controls. Streptavidin beads (Thermo Fisher Scientific) were then used to pull down biotinylated proteins by rotating overnight at 4°C. After several washes in lysis buffer, samples were eluted using Laemmli sample buffer at 95°C and analyzed by Western blot as described above.
Scrape loading dye transfer assay
Cells were grown to confluence on glass coverslips and treated with appropriate KD conditions. A surgical blade was used to scrape lines in the confluent monolayer. Medium was aspirated and replaced with cell culture media premixed with 100 µl LY at 12 mg/ml stock (Thermo Fisher Scientific). Cells were incubated at 37°C for 15 min followed by PBS washes. Subsequently, samples were fixed in PFA and processed for immunofluorescence analysis as described above.
Delivery of RRSP into cells
NRVCMs seeded onto either six-well plates or glass coverslips were treated with the appropriate adenoviral constructs. Purification and cellular intoxication with LFNDUF5Vv and PA (referred to simply as RRSP) were conducted as previously described previously (Antic et al., 2015). Before treatment, cells in culture were provided with fresh medium. Next, 4.6 nM PA and 2 nM RRSP were added to the media for overnight incubation. Cells were either lysed in appropriate lysis buffer for Western blot analysis or fixed for image processing.
Statistics
All quantifications are presented as the mean ± SD. One-way ANOVA followed by Tukey’s post hoc analysis or Dunnett’s multiple comparison test was used for multiple comparisons. For data comparing two conditions, statistical differences were analyzed with a two-tailed Student’s t test. P < 0.05 was considered significant. For statistical analysis presented as fold change (bar graphs), the mean of data from control samples was assigned a value of 1, and all other samples were calculated relative to the control. All densitometry quantification was normalized to respective loading controls.
Study approval
For the DPeKO mouse model, all mice were housed in accredited animal facilities, and the use and care of mice were in accordance with Northwestern University’s Institutional Care and Use Committee. For the DPcKO mouse model, all animal procedures were in full compliance with the guidelines approved by the University of California, San Diego, Animal Care and Use Committee. For human samples, informed consent was not required for the experiments performed in this study as all human samples were obtained as deidentified tissue and are covered by Beth Israel Deaconess Medical Center exemption protocols.
Online supplemental material
Fig. S1 shows that tissue sections from two independent DP ACM patients display a complete loss of DP and Cx43 localization at IDs. Fig. S2 demonstrates that depletion of DP in NRVCMs does not result in a significant change in Cx43 transcript levels. Fig. S3 shows that dose-dependent addition of RRSP to NRVCMs results in corresponding inhibition of p-ERK1/2 signal. Fig. S4 demonstrates that DPeKO epidermis does not display a significant change in KRAS, HRAS, or NRAS mRNA levels compared with WT tissue.
Supplementary Material
Acknowledgments
We thank Jeffrey Saffitz (Beth Israel Deaconess Medical Center) for providing ACM patient cardiac samples. We thank Gillian Fitz, Lisa Godsel, and Jennifer Koetsier for their experimental/technical assistance. We also thank Jong Kook Park and Robert Lavker for their guidance in carrying out LY dye transfer assays. Sequencing services were performed at the Northwestern University Genomics Core Facility. Tissue sectioning and lentiviral particle production was performed at the Northwestern University Skin Disease Research Center (supported in part by P30AR057216).
This work was supported by NIH grant R37AR43380 and the J.L. Mayberry Endowment (to K.J. Green). P.D. Lampe is supported by NIH grant GM55632. F. Sheikh is supported by grants from NIH/National Heart, Lung, and Blood Institute (R01 HL09780-06) and the Tobacco-Related Disease Research Program (24RT-022). K.J.F. Satchell is supported by a Translational Research Grant from Robert H. Lurie Comprehensive Cancer Center and the Northwestern Medicine Catalyst Fund. S. Polo is supported by the Associazione Italiana per la Ricerca sul Cancro (IG11627) and Worldwide Cancer Research (11-0051). C.Y. Kam was supported by a American Heart Association predoctoral fellowship (15PRE25560138).
The authors declare no competing financial interests.
Author contributions: C.Y. Kam and K.J. Green designed the study, analyzed the data, and wrote the manuscript. C.Y. Kam performed most of the experiments. A.D. Dubash assisted with the generation of reagents and analysis of data. P. Lampe generated the phosphospecific Cx43 antibodies used for this study. F. Sheikh generated the DPcKO mouse model. K.J.F. Satchell generated RRSP and provided technical assistance for its use. S. Polo and E. Magistrati provided reagents and participated in the preparation of the manuscript. All authors provided critical feedback on the manuscript.
References
- Alcalai R., Metzger S., Rosenheck S., Meiner V., and Chajek-Shaul T.. 2003. A recessive mutation in desmoplakin causes arrhythmogenic right ventricular dysplasia, skin disorder, and woolly hair. J. Am. Coll. Cardiol. 42:319–327. 10.1016/S0735-1097(03)00628-4 [DOI] [PubMed] [Google Scholar]
- Angst B.D., Nilles L.A., and Green K.J.. 1990. Desmoplakin II expression is not restricted to stratified epithelia. J. Cell Sci. 97:247–257. [DOI] [PubMed] [Google Scholar]
- Antic I., Biancucci M., Zhu Y., Gius D.R., and Satchell K.J.. 2015. Site-specific processing of Ras and Rap1 Switch I by a MARTX toxin effector domain. Nat. Commun. 6:7396 10.1038/ncomms8396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asimaki A., and Saffitz J.E.. 2012. Gap junctions and arrhythmogenic cardiomyopathy. Heart Rhythm. 9:992–995. 10.1016/j.hrthm.2011.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asimaki A., and Saffitz J.E.. 2014. Remodeling of cell-cell junctions in arrhythmogenic cardiomyopathy. Cell Commun. Adhes. 21:13–23. 10.3109/15419061.2013.876016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asimaki A., Tandri H., Huang H., Halushka M.K., Gautam S., Basso C., Thiene G., Tsatsopoulou A., Protonotarios N., McKenna W.J., et al. 2009. A new diagnostic test for arrhythmogenic right ventricular cardiomyopathy. N. Engl. J. Med. 360:1075–1084. 10.1056/NEJMoa0808138 [DOI] [PubMed] [Google Scholar]
- Asimaki A., Tandri H., Duffy E.R., Winterfield J.R., Mackey-Bojack S., Picken M.M., Cooper L.T., Wilber D.J., Marcus F.I., Basso C., et al. 2011. Altered desmosomal proteins in granulomatous myocarditis and potential pathogenic links to arrhythmogenic right ventricular cardiomyopathy. Circ Arrhythm Electrophysiol. 4:743–752. 10.1161/CIRCEP.111.964890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsen L.N., Calloe K., Holstein-Rathlou N.H., and Nielsen M.S.. 2013. Managing the complexity of communication: Regulation of gap junctions by post-translational modification. Front. Pharmacol. 4:130 10.3389/fphar.2013.00130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines A.T., Xu D., and Der C.J.. 2011. Inhibition of Ras for cancer treatment: The search continues. Future Med. Chem. 3:1787–1808. 10.4155/fmc.11.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso C., Thiene G., Corrado D., Angelini A., Nava A., and Valente M.. 1996. Arrhythmogenic right ventricular cardiomyopathy. Dysplasia, dystrophy, or myocarditis? Circulation. 94:983–991. 10.1161/01.CIR.94.5.983 [DOI] [PubMed] [Google Scholar]
- Basso C., Corrado D., Marcus F.I., Nava A., and Thiene G.. 2009. Arrhythmogenic right ventricular cardiomyopathy. Lancet. 373:1289–1300. 10.1016/S0140-6736(09)60256-7 [DOI] [PubMed] [Google Scholar]
- Bass-Zubek A.E., Hobbs R.P., Amargo E.V., Garcia N.J., Hsieh S.N., Chen X., Wahl J.K. III, Denning M.F., and Green K.J.. 2008. Plakophilin 2: A critical scaffold for PKCα that regulates intercellular junction assembly. J. Cell Biol. 181:605–613. 10.1083/jcb.200712133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass-Zubek A.E., Godsel L.M., Delmar M., and Green K.J.. 2009. Plakophilins: Multifunctional scaffolds for adhesion and signaling. Curr. Opin. Cell Biol. 21:708–716. 10.1016/j.ceb.2009.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beardslee M.A., Laing J.G., Beyer E.C., and Saffitz J.E.. 1998. Rapid turnover of connexin43 in the adult rat heart. Circ. Res. 83:629–635. 10.1161/01.RES.83.6.629 [DOI] [PubMed] [Google Scholar]
- Biancucci M., Rabideau A.E., Lu Z., Loftis A.R., Pentelute B.L., and Satchell K.J.F.. 2017. Substrate recognition of MARTX Ras/Rap1-specific endopeptidase. Biochemistry. 56:2747–2757. 10.1021/acs.biochem.7b00246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden L.M., Kam C.Y., Hernández-Martín A., Zhou J., Craiglow B.G., Sidbury R., Mathes E.F., Maguiness S.M., Crumrine D.A., Williams M.L., et al. 2016. Dominant de novo DSP mutations cause erythrokeratodermia-cardiomyopathy syndrome. Hum. Mol. Genet. 25:348–357. 10.1093/hmg/ddv481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral R.M., Tattersall D., Patel V., McPhail G.D., Hatzimasoura E., Abrams D.J., South A.P., and Kelsell D.P.. 2012. The DSPII splice variant is crucial for desmosome-mediated adhesion in HaCaT keratinocytes. J. Cell Sci. 125:2853–2861. 10.1242/jcs.084152 [DOI] [PubMed] [Google Scholar]
- Campian M.E., Verberne H.J., Hardziyenka M., de Groot E.A., van Moerkerken A.F., van Eck-Smit B.L., and Tan H.L.. 2010. Assessment of inflammation in patients with arrhythmogenic right ventricular cardiomyopathy/dysplasia. Eur. J. Nucl. Med. Mol. Imaging. 37:2079–2085. 10.1007/s00259-010-1525-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano E., and Santos E.. 2011. Functional specificity of ras isoforms: So similar but so different. Genes Cancer. 2:216–231. 10.1177/1947601911408081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelletti S., Vischer A.S., Syrris P., Crotti L., Spazzolini C., Ghidoni A., Parati G., Jenkins S., Kotta M.C., McKenna W.J., et al. 2017. Desmoplakin missense and non-missense mutations in arrhythmogenic right ventricular cardiomyopathy: Genotype-phenotype correlation. Int. J. Cardiol. 249:268–273. 10.1016/j.ijcard.2017.05.018 [DOI] [PubMed] [Google Scholar]
- Cerrone M., Noorman M., Lin X., Chkourko H., Liang F.X., van der Nagel R., Hund T., Birchmeier W., Mohler P., van Veen T.A., et al. 2012. Sodium current deficit and arrhythmogenesis in a murine model of plakophilin-2 haploinsufficiency. Cardiovasc. Res. 95:460–468. 10.1093/cvr/cvs218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerrone M., Lin X., Zhang M., Agullo-Pascual E., Pfenniger A., Chkourko Gusky H., Novelli V., Kim C., Tirasawadichai T., Judge D.P., et al. 2014. Missense mutations in plakophilin-2 cause sodium current deficit and associate with a Brugada syndrome phenotype. Circulation. 129:1092–1103. 10.1161/CIRCULATIONAHA.113.003077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen V.C., Kristensen A.R., Foster L.J., and Naus C.C.. 2012. Association of connexin43 with E3 ubiquitin ligase TRIM21 reveals a mechanism for gap junction phosphodegron control. J. Proteome Res. 11:6134–6146. 10.1021/pr300790h [DOI] [PubMed] [Google Scholar]
- Claycomb W.C., Lanson N.A. Jr., Stallworth B.S., Egeland D.B., Delcarpio J.B., Bahinski A., and Izzo N.J. Jr. 1998. HL-1 cells: A cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc. Natl. Acad. Sci. USA. 95:2979–2984. 10.1073/pnas.95.6.2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrado D., Basso C., Thiene G., McKenna W.J., Davies M.J., Fontaliran F., Nava A., Silvestri F., Blomstrom-Lundqvist C., Wlodarska E.K., et al. 1997. Spectrum of clinicopathologic manifestations of arrhythmogenic right ventricular cardiomyopathy/dysplasia: A multicenter study. J. Am. Coll. Cardiol. 30:1512–1520. 10.1016/S0735-1097(97)00332-X [DOI] [PubMed] [Google Scholar]
- Cox A.D., Fesik S.W., Kimmelman A.C., Luo J., and Der C.J.. 2014. Drugging the undruggable RAS: Mission possible? Nat. Rev. Drug Discov. 13:828–851. 10.1038/nrd4389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubash A.D., Kam C.Y., Aguado B.A., Patel D.M., Delmar M., Shea L.D., and Green K.J.. 2016. Plakophilin-2 loss promotes TGF-β1/p38 MAPK-dependent fibrotic gene expression in cardiomyocytes. J. Cell Biol. 212:425–438. 10.1083/jcb.201507018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong J.T., Nimlamool W., and Falk M.M.. 2014. EGF induces efficient Cx43 gap junction endocytosis in mouse embryonic stem cell colonies via phosphorylation of Ser262, Ser279/282, and Ser368. FEBS Lett. 588:836–844. 10.1016/j.febslet.2014.01.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes M.S., and Sperelakis N.. 1985. Intercalated discs of mammalian heart: A review of structure and function. Tissue Cell. 17:605–648. 10.1016/0040-8166(85)90001-1 [DOI] [PubMed] [Google Scholar]
- Fredriksson S., Gullberg M., Jarvius J., Olsson C., Pietras K., Gústafsdóttir S.M., Ostman A., and Landegren U.. 2002. Protein detection using proximity-dependent DNA ligation assays. Nat. Biotechnol. 20:473–477. 10.1038/nbt0502-473 [DOI] [PubMed] [Google Scholar]
- Gallicano G.I., Kouklis P., Bauer C., Yin M., Vasioukhin V., Degenstein L., and Fuchs E.. 1998. Desmoplakin is required early in development for assembly of desmosomes and cytoskeletal linkage. J. Cell Biol. 143:2009–2022. 10.1083/jcb.143.7.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallicano G.I., Bauer C., and Fuchs E.. 2001. Rescuing desmoplakin function in extra-embryonic ectoderm reveals the importance of this protein in embryonic heart, neuroepithelium, skin and vasculature. Development. 128:929–941. [DOI] [PubMed] [Google Scholar]
- Garcia-Gras E., Lombardi R., Giocondo M.J., Willerson J.T., Schneider M.D., Khoury D.S., and Marian A.J.. 2006. Suppression of canonical Wnt/beta-catenin signaling by nuclear plakoglobin recapitulates phenotype of arrhythmogenic right ventricular cardiomyopathy. J. Clin. Invest. 116:2012–2021. 10.1172/JCI27751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godsel L.M., Hsieh S.N., Amargo E.V., Bass A.E., Pascoe-McGillicuddy L.T., Huen A.C., Thorne M.E., Gaudry C.A., Park J.K., Myung K., et al. 2005. Desmoplakin assembly dynamics in four dimensions: Multiple phases differentially regulated by intermediate filaments and actin. J. Cell Biol. 171:1045–1059. 10.1083/jcb.200510038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godsel L.M., Dubash A.D., Bass-Zubek A.E., Amargo E.V., Klessner J.L., Hobbs R.P., Chen X., and Green K.J.. 2010. Plakophilin 2 couples actomyosin remodeling to desmosomal plaque assembly via RhoA. Mol. Biol. Cell. 21:2844–2859. 10.1091/mbc.e10-02-0131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes J., Finlay M., Ahmed A.K., Ciaccio E.J., Asimaki A., Saffitz J.E., Quarta G., Nobles M., Syrris P., Chaubey S., et al. 2012. Electrophysiological abnormalities precede overt structural changes in arrhythmogenic right ventricular cardiomyopathy due to mutations in desmoplakin-A combined murine and human study. Eur. Heart J. 33:1942–1953. 10.1093/eurheartj/ehr472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough D.A., and Paul D.L.. 2009. Gap junctions. Cold Spring Harb. Perspect. Biol. 1:a002576 10.1101/cshperspect.a002576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goossens S., Janssens B., Bonné S., De Rycke R., Braet F., van Hengel J., and van Roy F.. 2007. A unique and specific interaction between alphaT-catenin and plakophilin-2 in the area composita, the mixed-type junctional structure of cardiac intercalated discs. J. Cell Sci. 120:2126–2136. 10.1242/jcs.004713 [DOI] [PubMed] [Google Scholar]
- Green K.J., and Simpson C.L.. 2007. Desmosomes: New perspectives on a classic. J. Invest. Dermatol. 127:2499–2515. 10.1038/sj.jid.5701015 [DOI] [PubMed] [Google Scholar]
- Green K.J., Parry D.A., Steinert P.M., Virata M.L., Wagner R.M., Angst B.D., and Nilles L.A.. 1990. Structure of the human desmoplakins. Implications for function in the desmosomal plaque. J. Biol. Chem. 265:2603–2612. [PubMed] [Google Scholar]
- Hobbs G.A., Der C.J., and Rossman K.L.. 2016. RAS isoforms and mutations in cancer at a glance. J. Cell Sci. 129:1287–1292. 10.1242/jcs.182873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsmans M., Clauss S., Xiao L., Aguirre A.D., King K.R., Hanley A., Hucker W.J., Wülfers E.M., Seemann G., Courties G., et al. 2017. Macrophages facilitate electrical conduction in the heart. Cell. 169:510–522.e20. 10.1016/j.cell.2017.03.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K.E., Mitra S., Katoch P., Kelsey L.S., Johnson K.R., and Mehta P.P.. 2013. Phosphorylation on Ser-279 and Ser-282 of connexin43 regulates endocytosis and gap junction assembly in pancreatic cancer cells. Mol. Biol. Cell. 24:715–733. 10.1091/mbc.e12-07-0537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan S.R., Gard J.J., Carvajal-Huerta L., Ruiz-Cabezas J.C., Thiene G., and Saffitz J.E.. 2004. Structural and molecular pathology of the heart in Carvajal syndrome. Cardiovasc. Pathol. 13:26–32. 10.1016/S1054-8807(03)00107-8 [DOI] [PubMed] [Google Scholar]
- Khavari T.A., and Rinn J.. 2007. Ras/Erk MAPK signaling in epidermal homeostasis and neoplasia. Cell Cycle. 6:2928–2931. 10.4161/cc.6.23.4998 [DOI] [PubMed] [Google Scholar]
- Laing J.G., and Beyer E.C.. 1995. The gap junction protein connexin43 is degraded via the ubiquitin proteasome pathway. J. Biol. Chem. 270:26399–26403. 10.1074/jbc.270.44.26399 [DOI] [PubMed] [Google Scholar]
- Laing J.G., Tadros P.N., Westphale E.M., and Beyer E.C.. 1997. Degradation of connexin43 gap junctions involves both the proteasome and the lysosome. Exp. Cell Res. 236:482–492. 10.1006/excr.1997.3747 [DOI] [PubMed] [Google Scholar]
- Laird D.W. 2006. Life cycle of connexins in health and disease. Biochem. J. 394:527–543. 10.1042/BJ20051922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird D.W., Puranam K.L., and Revel J.P.. 1991. Turnover and phosphorylation dynamics of connexin43 gap junction protein in cultured cardiac myocytes. Biochem. J. 273:67–72. 10.1042/bj2730067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampe P.D., and Lau A.F.. 2000. Regulation of gap junctions by phosphorylation of connexins. Arch. Biochem. Biophys. 384:205–215. 10.1006/abbi.2000.2131 [DOI] [PubMed] [Google Scholar]
- Langlois S., Maher A.C., Manias J.L., Shao Q., Kidder G.M., and Laird D.W.. 2007. Connexin levels regulate keratinocyte differentiation in the epidermis. J. Biol. Chem. 282:30171–30180. 10.1074/jbc.M703623200 [DOI] [PubMed] [Google Scholar]
- Langlois S., Cowan K.N., Shao Q., Cowan B.J., and Laird D.W.. 2010. The tumor-suppressive function of Connexin43 in keratinocytes is mediated in part via interaction with caveolin-1. Cancer Res. 70:4222–4232. 10.1158/0008-5472.CAN-09-3281 [DOI] [PubMed] [Google Scholar]
- Leithe E., and Rivedal E.. 2004. Ubiquitination and down-regulation of gap junction protein connexin-43 in response to 12-O-tetradecanoylphorbol 13-acetate treatment. J. Biol. Chem. 279:50089–50096. 10.1074/jbc.M402006200 [DOI] [PubMed] [Google Scholar]
- Leithe E., Brech A., and Rivedal E.. 2006. Endocytic processing of connexin43 gap junctions: A morphological study. Biochem. J. 393:59–67. 10.1042/BJ20050674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung C.L., Green K.J., and Liem R.K.. 2002. Plakins: A family of versatile cytolinker proteins. Trends Cell Biol. 12:37–45. 10.1016/S0962-8924(01)02180-8 [DOI] [PubMed] [Google Scholar]
- Lombardi R., Chen S.N., Ruggiero A., Gurha P., Czernuszewicz G.Z., Willerson J.T., and Marian A.J.. 2016. Cardiac fibro-adipocyte progenitors express desmosome proteins and preferentially differentiate to adipocytes upon deletion of the desmoplakin gene. Circ. Res. 119:41–54. 10.1161/CIRCRESAHA.115.308136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Ayala J.M., Gómez-Milanés I., Sánchez Muñoz J.J., Ruiz-Espejo F., Ortíz M., González-Carrillo J., López-Cuenca D., Oliva-Sandoval M.J., Monserrat L., Valdés M., and Gimeno J.R.. 2014. Desmoplakin truncations and arrhythmogenic left ventricular cardiomyopathy: Characterizing a phenotype. Europace. 16:1838–1846. 10.1093/europace/euu128 [DOI] [PubMed] [Google Scholar]
- Lorenz K., Schmitt J.P., Vidal M., and Lohse M.J.. 2009. Cardiac hypertrophy: Targeting Raf/MEK/ERK1/2-signaling. Int. J. Biochem. Cell Biol. 41:2351–2355. 10.1016/j.biocel.2009.08.002 [DOI] [PubMed] [Google Scholar]
- Lyon R.C., Mezzano V., Wright A.T., Pfeiffer E., Chuang J., Banares K., Castaneda A., Ouyang K., Cui L., Contu R., et al. 2014. Connexin defects underlie arrhythmogenic right ventricular cardiomyopathy in a novel mouse model. Hum. Mol. Genet. 23:1134–1150. 10.1093/hmg/ddt508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Márquez-Rosado L., Solan J.L., Dunn C.A., Norris R.P., and Lampe P.D.. 2012. Connexin43 phosphorylation in brain, cardiac, endothelial and epithelial tissues. Biochim. Biophys. Acta. 1818:1985–1992. 10.1016/j.bbamem.2011.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y., Ebisuya M., Honjoh S., and Nishida E.. 2004. ERK activation propagates in epithelial cell sheets and regulates their migration during wound healing. Curr. Biol. 14:731–735. 10.1016/j.cub.2004.03.060 [DOI] [PubMed] [Google Scholar]
- Mezzano V., Liang Y., Wright A.T., Lyon R.C., Pfeiffer E., Song M.Y., Gu Y., Dalton N.D., Scheinman M., Peterson K.L., et al. 2016. Desmosomal junctions are necessary for adult sinus node function. Cardiovasc. Res. 111:274–286. 10.1093/cvr/cvw083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molho-Pessach V., Sheffer S., Siam R., Tams S., Siam I., Awwad R., Babay S., Golender J., Simanovsky N., Ramot Y., and Zlotogorski A.. 2015. Two novel homozygous desmoplakin mutations in Carvajal syndrome. Pediatr. Dermatol. 32:641–646. 10.1111/pde.12541 [DOI] [PubMed] [Google Scholar]
- Norgett E.E., Hatsell S.J., Carvajal-Huerta L., Cabezas J.C., Common J., Purkis P.E., Whittock N., Leigh I.M., Stevens H.P., and Kelsell D.P.. 2000. Recessive mutation in desmoplakin disrupts desmoplakin-intermediate filament interactions and causes dilated cardiomyopathy, woolly hair and keratoderma. Hum. Mol. Genet. 9:2761–2766. 10.1093/hmg/9.18.2761 [DOI] [PubMed] [Google Scholar]
- Norris R.P., Freudzon M., Mehlmann L.M., Cowan A.E., Simon A.M., Paul D.L., Lampe P.D., and Jaffe L.A.. 2008. Luteinizing hormone causes MAP kinase-dependent phosphorylation and closure of connexin 43 gap junctions in mouse ovarian follicles: One of two paths to meiotic resumption. Development. 135:3229–3238. 10.1242/dev.025494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel D.M., and Green K.J.. 2014. Desmosomes in the heart: A review of clinical and mechanistic analyses. Cell Commun. Adhes. 21:109–128. 10.3109/15419061.2014.906533 [DOI] [PubMed] [Google Scholar]
- Patel D.M., Dubash A.D., Kreitzer G., and Green K.J.. 2014. Disease mutations in desmoplakin inhibit Cx43 membrane targeting mediated by desmoplakin–EB1 interactions. J. Cell Biol. 206:779–797. 10.1083/jcb.201312110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieperhoff S., and Franke W.W.. 2007. The area composita of adhering junctions connecting heart muscle cells of vertebrates - IV: Coalescence and amalgamation of desmosomal and adhaerens junction components—late processes in mammalian heart development. Eur. J. Cell Biol. 86:377–391. 10.1016/j.ejcb.2007.04.001 [DOI] [PubMed] [Google Scholar]
- Pieperhoff S., Schumacher H., and Franke W.W.. 2008. The area composita of adhering junctions connecting heart muscle cells of vertebrates. V. The importance of plakophilin-2 demonstrated by small interference RNA-mediated knockdown in cultured rat cardiomyocytes. Eur. J. Cell Biol. 87:399–411. 10.1016/j.ejcb.2007.12.002 [DOI] [PubMed] [Google Scholar]
- Ristic M., Brockly F., Piechaczyk M., and Bossis G.. 2016. Detection of protein-protein interactions and posttranslational modifications using the proximity ligation assay: Application to the study of the SUMO pathway. Methods Mol. Biol. 1449:279–290. 10.1007/978-1-4939-3756-1_17 [DOI] [PubMed] [Google Scholar]
- Roberts P.J., and Der C.J.. 2007. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 26:3291–3310. 10.1038/sj.onc.1210422 [DOI] [PubMed] [Google Scholar]
- Rötzer V., Hartlieb E., Winkler J., Walter E., Schlipp A., Sardy M., Spindler V., and Waschke J.. 2016. Desmoglein 3-dependent signaling regulates keratinocyte migration and wound healing. J. Invest. Dermatol. 136:301–310. 10.1038/JID.2015.380 [DOI] [PubMed] [Google Scholar]
- Saez J.C., Berthoud V.M., Branes M.C., Martinez A.D., and Beyer E.C.. 2003. Plasma membrane channels formed by connexins: Their regulation and functions. Physiol. Rev. 83:1359–1400. 10.1152/physrev.00007.2003 [DOI] [PubMed] [Google Scholar]
- Saffitz J.E., Asimaki A., and Huang H.. 2010. Arrhythmogenic right ventricular cardiomyopathy: New insights into mechanisms of disease. Cardiovasc. Pathol. 19:166–170. 10.1016/j.carpath.2009.10.006 [DOI] [PubMed] [Google Scholar]
- Sen-Chowdhry S., Syrris P., Ward D., Asimaki A., Sevdalis E., and McKenna W.J.. 2007. Clinical and genetic characterization of families with arrhythmogenic right ventricular dysplasia/cardiomyopathy provides novel insights into patterns of disease expression. Circulation. 115:1710–1720. 10.1161/CIRCULATIONAHA.106.660241 [DOI] [PubMed] [Google Scholar]
- Sen-Chowdhry S., Morgan R.D., Chambers J.C., and McKenna W.J.. 2010. Arrhythmogenic cardiomyopathy: Etiology, diagnosis, and treatment. Annu. Rev. Med. 61:233–253. 10.1146/annurev.med.052208.130419 [DOI] [PubMed] [Google Scholar]
- Setzer S.V., Calkins C.C., Garner J., Summers S., Green K.J., and Kowalczyk A.P.. 2004. Comparative analysis of armadillo family proteins in the regulation of a431 epithelial cell junction assembly, adhesion and migration. J. Invest. Dermatol. 123:426–433. 10.1111/j.0022-202X.2004.23319.x [DOI] [PubMed] [Google Scholar]
- Shin K.H., Bae S.D., Hong H.S., Kim R.H., Kang M.K., and Park N.H.. 2011. miR-181a shows tumor suppressive effect against oral squamous cell carcinoma cells by downregulating K-ras. Biochem. Biophys. Res. Commun. 404:896–902. 10.1016/j.bbrc.2010.12.055 [DOI] [PubMed] [Google Scholar]
- Söderberg O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K.J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L.G., and Landegren U.. 2006. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods. 3:995–1000. 10.1038/nmeth947 [DOI] [PubMed] [Google Scholar]
- Solan J.L., and Lampe P.D.. 2014. Specific Cx43 phosphorylation events regulate gap junction turnover in vivo. FEBS Lett. 588:1423–1429. 10.1016/j.febslet.2014.01.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroemlund L.W., Jensen C.F., Qvortrup K., Delmar M., and Nielsen M.S.. 2015. Gap junctions—Guards of excitability. Biochem. Soc. Trans. 43:508–512. 10.1042/BST20150059 [DOI] [PubMed] [Google Scholar]
- van Veen T.A., van Rijen H.V., and Jongsma H.J.. 2000. Electrical conductance of mouse connexin45 gap junction channels is modulated by phosphorylation. Cardiovasc. Res. 46:496–510. 10.1016/S0008-6363(00)00047-X [DOI] [PubMed] [Google Scholar]
- Vasioukhin V., Bowers E., Bauer C., Degenstein L., and Fuchs E.. 2001. Desmoplakin is essential in epidermal sheet formation. Nat. Cell Biol. 3:1076–1085. 10.1038/ncb1201-1076 [DOI] [PubMed] [Google Scholar]
- Wan H., South A.P., and Hart I.R.. 2007. Increased keratinocyte proliferation initiated through downregulation of desmoplakin by RNA interference. Exp. Cell Res. 313:2336–2344. 10.1016/j.yexcr.2007.01.010 [DOI] [PubMed] [Google Scholar]
- Wang Y. 2007. Mitogen-activated protein kinases in heart development and diseases. Circulation. 116:1413–1423. 10.1161/CIRCULATIONAHA.106.679589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Shi Z.M., Jiang C.F., Liu X., Chen Q.D., Qian X., Li D.M., Ge X., Wang X.F., Liu L.Z., et al. 2014. MiR-143 acts as a tumor suppressor by targeting N-RAS and enhances temozolomide-induced apoptosis in glioma. Oncotarget. 5:5416–5427. 10.18632/oncotarget.2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warn-Cramer B.J., Lampe P.D., Kurata W.E., Kanemitsu M.Y., Loo L.W., Eckhart W., and Lau A.F.. 1996. Characterization of the mitogen-activated protein kinase phosphorylation sites on the connexin-43 gap junction protein. J. Biol. Chem. 271:3779–3786. 10.1074/jbc.271.7.3779 [DOI] [PubMed] [Google Scholar]
- Wolf A., Rietscher K., Glaß M., Hüttelmaier S., Schutkowski M., Ihling C., Sinz A., Wingenfeld A., Mun A., and Hatzfeld M.. 2013. Insulin signaling via Akt2 switches plakophilin 1 function from stabilizing cell adhesion to promoting cell proliferation. J. Cell Sci. 126:1832–1844. 10.1242/jcs.118992 [DOI] [PubMed] [Google Scholar]
- Yang L., Chen Y., Cui T., Knösel T., Zhang Q., Albring K.F., Huber O., and Petersen I.. 2012. Desmoplakin acts as a tumor suppressor by inhibition of the Wnt/β-catenin signaling pathway in human lung cancer. Carcinogenesis. 33:1863–1870. 10.1093/carcin/bgs226 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Kim J., Mueller A.C., Dey B., Yang Y., Lee D.H., Hachmann J., Finderle S., Park D.M., Christensen J., et al. 2014. Multiple receptor tyrosine kinases converge on microRNA-134 to control KRAS, STAT5B, and glioblastoma. Cell Death Differ. 21:720–734. 10.1038/cdd.2013.196 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.