How the endocytic protein NUMB modulates mesenchymal migration is unclear. Zobel et al. show that NUMB limits the formation of apically restricted, actin-based protrusions called circular dorsal ruffles (CDRs). NUMB activates the ARF6 guanine nucleotide exchange factor EFA6B and controls the trafficking rate of ARF6-dependent cargos relevant for CDR formation, such as RAC1, and negatively regulates HGF-induced cell migration and invasion.
Abstract
The endocytic protein NUMB has been implicated in the control of various polarized cellular processes, including the acquisition of mesenchymal migratory traits through molecular mechanisms that have only been partially defined. Here, we report that NUMB is a negative regulator of a specialized set of understudied, apically restricted, actin-based protrusions, the circular dorsal ruffles (CDRs), induced by either PDGF or HGF stimulation. Through its PTB domain, NUMB binds directly to an N-terminal NPLF motif of the ARF6 guanine nucleotide exchange factor, EFA6B, and promotes its exchange activity in vitro. In cells, a NUMB–EFA6B–ARF6 axis regulates the recycling of the actin regulatory cargo RAC1 and is critical for the formation of CDRs that mark the acquisition of a mesenchymal mode of motility. Consistently, loss of NUMB promotes HGF-induced cell migration and invasion. Thus, NUMB negatively controls membrane protrusions and the acquisition of mesenchymal migratory traits by modulating EFA6B–ARF6 activity.
Introduction
A key hallmark of mesenchymal motility is the acquisition of a prototypical front–back polarity driven by the extension of polarized, actin-rich membrane protrusions. These protrusions include finger-like filopodia and flat lamellipodia, sausage-like lobopodia adopted by cell moving in 3D, blebs, and a set of understudied, apically restricted, circular dorsal ruffles (CDRs; Ridley, 2011). These latter structures have recently attracted attention, as they display features of an oscillatory and recurrent process (Hoon et al., 2012; Sero et al., 2012; Bernitt et al., 2015, 2017; Corallino et al., 2018) whose formation is typically induced and biased following stimulation with certain types of growth factors, including PDGFs in fibroblasts and hepatocyte growth factors (HGFs) in epithelial cells. In keeping with these observations, CDRs have been suggested to steer cells along chemotactic gradients and to mark the acquisition of an elongated, mesenchymal mode of motility (Lanzetti et al., 2004; Palamidessi et al., 2008; Gu et al., 2011; Zeng et al., 2011; Sero et al., 2012). These properties, together with the fact that CDRs are an “easy-to-follow read out” through phase-contrast time-lapse microscopy (Palamidessi et al., 2008), have prompted their use for the identification of novel signaling molecules, pathways, and cellular processes involved in their formation and important for directional, mesenchymal motility. CDRs are also endocytic structures through which large molecular weight protein and fluids can be internalized via micropinocytosis (Hoon et al., 2012). This entry route is important for the uptake of proteinaceous materials that sustain cell growth under limited availability of nutrients (Commisso et al., 2013; Palm et al., 2015; Kim et al., 2016). Additionally, CDRs promote the internalization of growth factors, actin regulatory GTPases, and integrins that are subsequently delivered back to the plasma membrane (PM) through recycling (Orth et al., 2006; Gu et al., 2011; Hoon et al., 2012). In turn, their formation is controlled by endocytic/trafficking pathways. Consistently, perturbing early endosomal functions by deregulated expression of RAB5 or its GTPase-activating protein (GAP), RN-tre (also called USP6NL), impairs these structures (Spaargaren and Bos, 1999; Lanzetti et al., 2004; Topp et al., 2004; Kunita et al., 2007; Mojica-Vázquez et al., 2017). Similarly, CDRs strictly require the functional activity of ARF6, which acts by controlling the recycling of a specialized set of cargos, including RAC1 (Palamidessi et al., 2008; Donaldson et al., 2009; Schweitzer et al., 2011). Notably, the activation of ARF6 must be followed by its rapid deactivation for these cargos to be efficiently delivered to the PM (Allaire et al., 2013; Dutta and Donaldson, 2015; Kobayashi et al., 2015; Loskutov et al., 2015), indicating that tight temporal control of ARF6 cycles and, specifically, of the duration of its activation might be crucial also for the formation of CDRs. Here, we set out to identify novel critical molecular determinants of CDR formation and further characterize the molecular and trafficking routes onto which they act. We focused on the endocytic molecule NUMB (Santolini et al., 2000).
NUMB is a multifunctional adaptor protein, originally identified as a cell-autonomous fate determinant (Uemura et al., 1989), involved in several aspects of membrane dynamics (Pece et al., 2011). Its role as an endocytic adaptor is supported by its localization to endocytic organelles, cotrafficking with internalizing receptors, and interaction with the α-adaptin subunit of the major clathrin adaptor AP2 (Santolini et al., 2000; Dho et al., 2006; Wei et al., 2014). In addition, it also localizes, in Caenorhabditis elegans and Drosophila melanogaster, to recycling endosomes and regulates the delivery of cargos back to the PM (Nilsson et al., 2008, 2011; Cotton et al., 2013; Couturier et al., 2013), a function conserved in mammals where NUMB regulates postendocytic sorting of cargos such as NOTCHs and ERBB2 (Smith et al., 2004; McGill et al., 2009; Hirai et al., 2017).
One relevant feature of NUMB is that it acts as a tumor suppressor in breast cancer (Pece et al., 2004; Colaluca et al., 2008, 2018; Tosoni et al., 2017), and likely in other cancers as well (Maiorano et al., 2007; Westhoff et al., 2009; Hong et al., 2014). This property has been linked to the ability of NUMB to suppress the oncogenic NOTCH pathway and to enhance p53-dependent signaling (Colaluca et al., 2017; Pece et al., 2004; Colaluca et al., 2008; Westhoff et al., 2009; Tosoni et al., 2017). The tumor suppressor activity of NUMB might, however, extend beyond these circuitries. In the mammary gland compartment, for instance, NUMB plays a dual role in the stem cell (SC) and in progenitors (Tosoni et al., 2015). In SCs, NUMB imparts an asymmetric mode of cellular division by asymmetrically partitioning at mitosis in the daughter that retains the SC identity, where it sustains high p53 activity. In progenitors, however, loss of NUMB correlates with epithelial-to-mesenchymal transition (EMT) and the reacquisition of stemness (Tosoni et al., 2015). Consistent with this latter finding and the notion that it might regulate the acquisition of mesenchymal features, NUMB loss was shown to perturb epithelial polarity and cell–cell adhesion and promote EMT (Wang et al., 2009; Pece et al., 2011; Zhang et al., 2016). The molecular and cellular mechanisms through which NUMB exerts its function in the maintenance of an epithelial phenotype and, vice versa, how its loss leads to a mesenchymal morphological transition that precedes gene rewiring typical of EMT, is still elusive.
Here, we show that NUMB is a negative regulator of CDR formation and controls mesenchymal motility. It does so by physically interacting with the guanine nucleotide exchange factor (GEF) EFA6B and by activating its exchange activity in vitro. In cells, a NUMB–EFA6B–ARF6 axis operates in the control of recycling back to the PM of cargos, including RAC1, which is ultimately critical for the formation of CDRs and the acquisition of the mesenchymal mode of motility.
Results
NUMB is a negative regulator of CDR formation downstream of receptor tyrosine kinase (RTK) activation
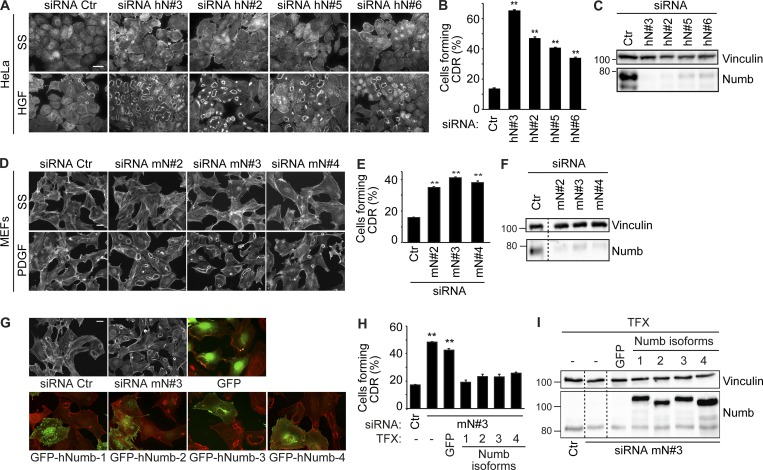
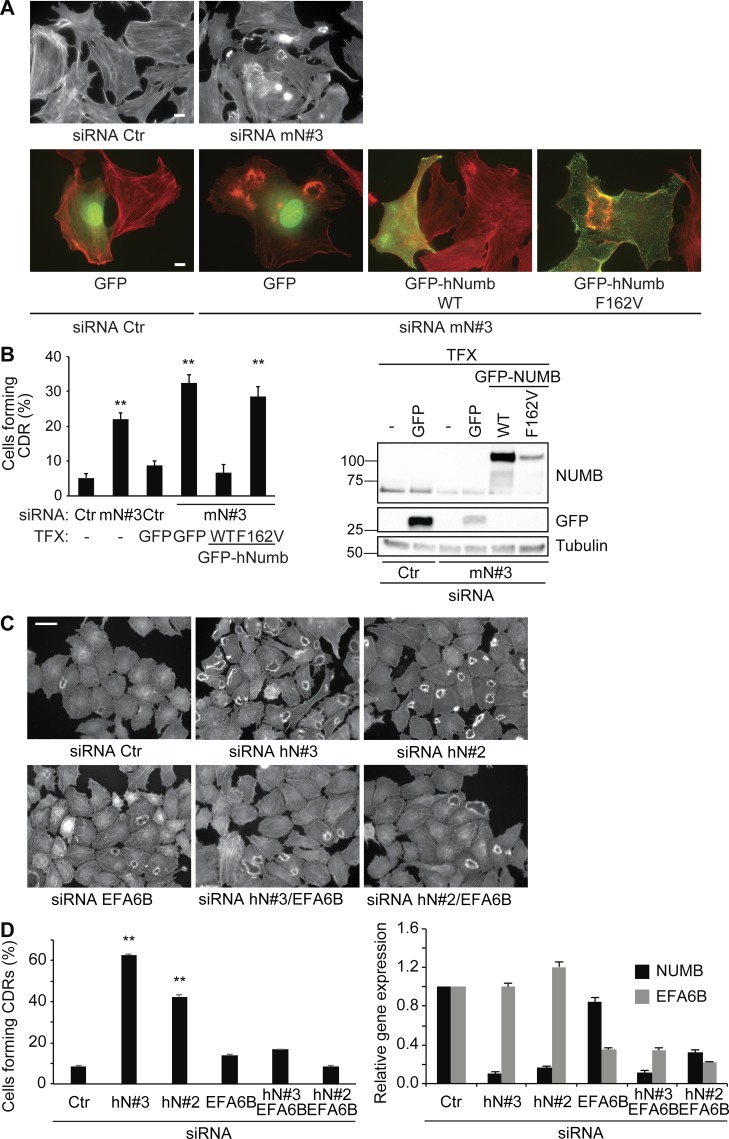
To assess the impact of endocytic/trafficking proteins on CDR formation and mesenchymal motility, we exploited an RNAi screening of a selected set of endocytic regulators using CDR formation as read-out markers (Palamidessi et al., 2008; Frittoli et al., 2014). Among CDR regulatory candidates, we focused for further analysis on the endocytic adaptor NUMB that robustly increased CDR formation (Table S1). We corroborated our initial findings using four independent siRNAs against NUMB, which were all effective in significantly inhibiting protein expression and promoting CDR formation induced by HGF stimulation in HeLa cells (Fig. 1, A–C). We further verified the general requirement of NUMB in the process by assessing whether its silencing with another set of independent oligos also impacted the formation of CDRs in mouse embryo fibroblasts (MEFs) stimulated with PDGF, a known potent inducer of these structures (Buccione et al., 2004). NUMB ablation also significantly increased the number of cells forming CDRs upon PDGF stimulation, specifically at suboptimal concentrations of the ligand (Fig. 1, D–F). Under the latter conditions, the number of control cells forming CDRs is reduced, but removal of NUMB robustly increases this response.
Figure 1.
NUMB is a negative regulator of growth factor–induced CDR formation. (A) HeLa cells seeded on Matrigel-coated coverslips were transfected with four different siRNAs (hN#3, hN#2, hN#5, and hN#6) against human NUMB or scrambled oligos (siRNA Ctr, control). Cells were serum starved (SS) for 2 h, stimulated with 100 ng/ml HGF for 4 min, and stained to detect F-actin. Bar, 40 μm. (B) Number of cells forming CDRs over the total number of cells per field. (C) The expression levels of NUMB and vinculin were analyzed by immunoblotting (IB). (D) MEF cells transfected with three different siRNAs (m#2, mN#3, and mN#4) against murine NUMB or siRNA Ctr as control were seeded on gelatin-coated coverslips. Cells were serum starved for 2 h, stimulated with 1 ng/ml PDGF for 8 min, and stained to detect F-actin. Bar, 40 µm. (E) The number of cells forming CDRs over the total number of cells per field. (B and E) The data are the mean ± SEM (n > 100 cells/condition in 20 fields/each condition out of three independent experiments). (F) The expression level of NUMB and vinculin was analyzed by IB. (G) MEF cells were transfected with siRNA Ctr or with siRNA against murine NUMB (mN#3, upper panels), either alone or in combination with GFP empty vector (upper panels, right) or GFP-hNUMB isoform 1, 2, 3, or 4 (lower panels). Cells were serum starved for 2 h and stimulated with 1 ng/ml PDGF for 8 min, fixed and processed for epifluorescence to visualize GFP or GFP-NUMB (green, where indicated), and stained to detect F-actin (red). Bar, 40 µm; 20 µm in GFP or GFP-NUMB panels. (H) The number of cells forming CDRs over the total number of cells per field was counted. Representative images are shown. The data are the mean ± SEM (n > 100 cells/condition in 20 fields/each condition out of four independent experiments). (I) The expression levels of NUMB and vinculin were analyzed by IB. (C, F, and H) Molecular weight (MW) markers are shown on the left in kilodaltons. TFX, transfected. A Student’s t test was used to calculate the P values: **, P < 0.01. Dashed lines indicate that the intervening lanes have been spliced out.
There are four distinct, ubiquitously expressed NUMB splice isoforms (Dho et al., 1999; Verdi et al., 1999). To investigate their involvement in CDR formation and provide genetic evidence for the role of NUMB in the process, we reconstituted NUMB expression by transfecting each of the four isoforms in siRNA-treated MEFs. The expression of all four human NUMB isoforms restored CDR formation to a level similar to the one observed in control NUMB-proficient cells, therefore ruling out spurious effects exerted by siRNA (Fig. 1, G–I). Collectively, these findings indicate that NUMB is a negative regulator of CDR formation downstream of either activated C-Met or PDGF receptors.
NUMB controls CDR formation through the ARF6 trafficking pathway
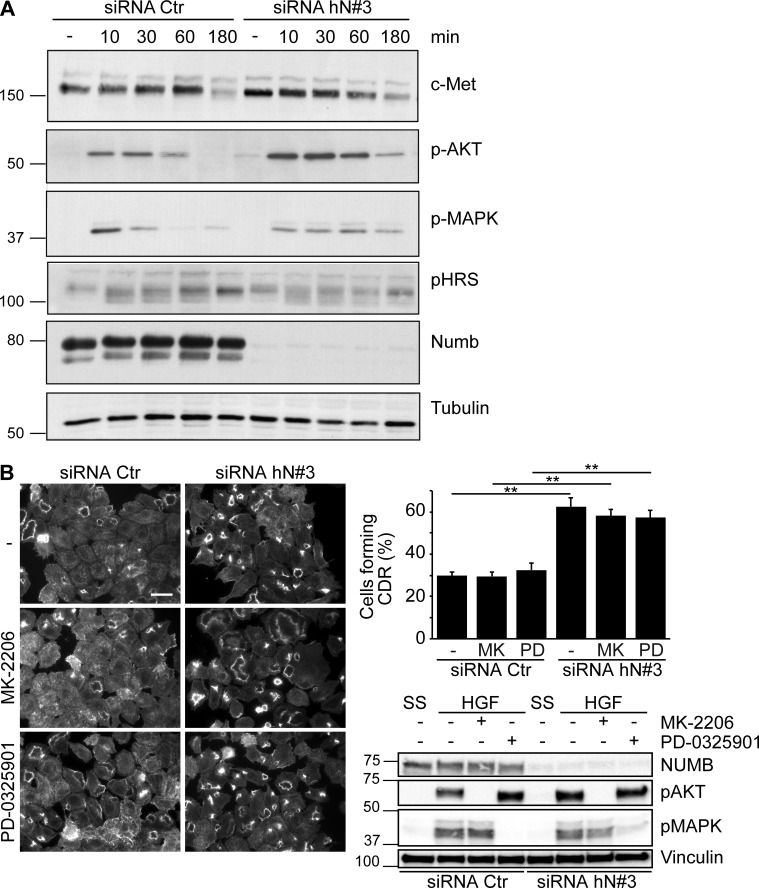
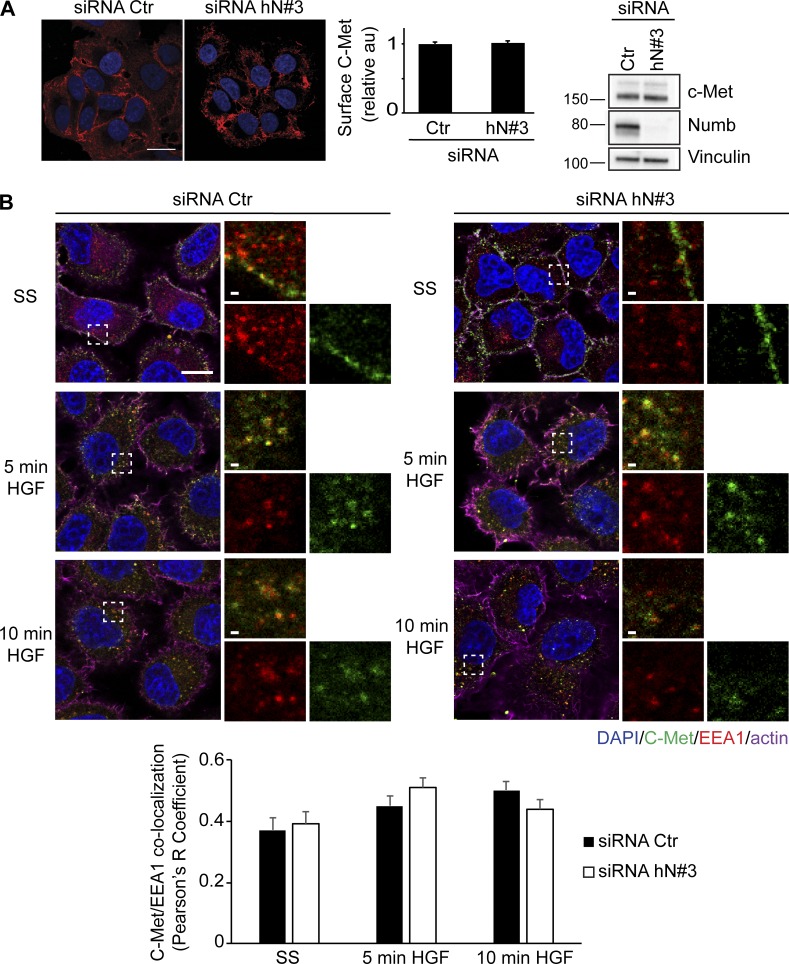
Next, we set out to identify the signaling axis through which NUMB controls CDR formation. CDRs are strictly dependent on stimulation with a subset of growth factors that activate specific RTKs; thus, one obvious possibility is that NUMB silencing causes aberrant RTK signaling or trafficking. Focusing on the HGF–C-Met axis, we found, however, that loss of NUMB had no effect on total C-Met levels and stability, but slightly elevated and prolonged the phosphorylation of ERK1/2 and AKT (Fig. 2 A). However, neither of these signaling axes contributed significantly to CDR formation since pharmacological inhibition of the activity of ERK1/2 with PD-0325901, a selective and non–ATP-competitive MAPK/ERK kinase inhibitor (Henderson et al., 2010), or AKT with MK-2206, an allosteric inhibitor (Yap et al., 2011), had no effect on CDRs (Fig. 2 B). These findings are consistent with a recent set of results showing that inhibition of AKT has no effect on CDR formation nor on macropinocytic-dependent dextran internalization (Palm et al., 2017; Corallino et al., 2018), which nearly invariably accompanies the enclosure of these apical, actin-rich structures. We also detected no alterations in C-Met surface levels (Fig. 3 A) or in its localization in early endosomes induced by HGF stimulation in control versus NUMB KD (Fig. 3 B), suggesting that NUMB loss had no major impact on receptor trafficking. This is consistent with data showing that NUMB loss did not alter the internalization (not shown) or recycling kinetics of EGF receptor (EGFR) and transferrin (Tf; Fig. S1 A), either in the absence or the presence of HGF (Fig. S1 B).
Figure 2.
Slightly elevated and prolonged phosphorylation of ERK1/2 and AKT caused by NUMB loss has no impact on CDR formation. (A) HeLa cells transfected with an siRNA (siRNA hN#3) against NUMB or scrambled oligo (siRNA Ctr) were serum starved (SS) for 2 h (−) and stimulated with 100 ng/ml HGF for the indicated time. The expression levels of the indicated total and phospho-proteins (p-) were analyzed by IB. (B) Left panels: HeLa cells were seeded on Matrigel-coated coverslips and transfected with siRNA against human NUMB (hN#3) or siRNA Ctr as control. Cells treated with DMSO (–), MK-2206 inhibitor (1 μM, 24 h), or PD-0325901 inhibitor (1 μM, 1 h) were serum starved for 2 h and stimulated with 100 ng/ml HGF for 4 min before staining with phalloidin to detect F-actin. Bar, 40 μm. Upper right graph: the number of cells forming CDRs/the total number of cells per field. Representative images are shown. The data are the mean ± SEM (n > 100 cells/condition out of 10 fields/condition in three independent experiments). Lower right panel: the expression levels of indicated proteins were analyzed by IB. MW markers are shown on the left in kilodaltons. A Student’s t test was used to calculate the P values: **, P < 0.01.
Figure 3.
Loss of NUMB does not affect trafficking of C-Met. (A) Left panels: confocal analysis of HeLa cells transfected with an siRNA (siRNA hN#3) against NUMB or scrambled oligo (siRNA Ctr). Cells were stained with specific antibody to detect cell surface C-Met (red; without permeabilization) or with DAPI (blue) to visualize nuclei. Bar, 20 μm. Central graph: integrated density of C-Met signaling (defined using ImageJ) normalized over the number of cells and expressed as fold increase with respect to siCTR scrambled control cells. Representative images are shown in the middle panel. The data are the mean ± SEM (n > 100 cells/condition out of 20 fields/condition in three independent experiments). Right panel: expression levels of indicated proteins were analyzed by IB. MW markers are shown on the left in kilodaltons. (B) Confocal analysis of HeLa cells transfected with siRNA against NUMB (siRNA hN#3) or scrambled oligo as control (siRNA Ctr). Cells, seeded on glass coverslips, were serum starved (SS) for 2 h and stimulated with 100 ng/ml HGF at different time points. Cells were stained for C-Met (green), EEA1 (red), DAPI (blue), and actin (magenta). Magnification of the boxed, selected areas is shown. Bars, 10 μm. Lower graph: Pearson’s R correlation value between C-Met and EEA1 (see Materials and methods for details). The data are the mean ± SEM (n > 50 cells/condition in two independent experiments). Representative images are shown in the upper panel.
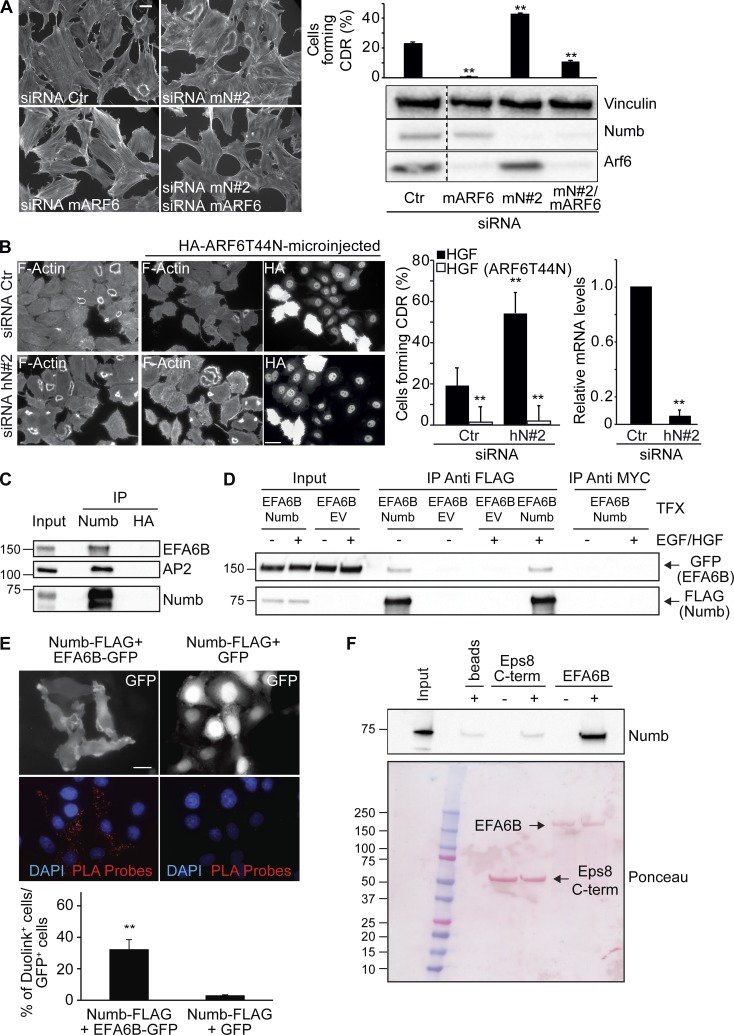
Trafficking routes through early RAB5-positive and ARF6-recycling endosomes have been implicated in CDR formation (Buccione et al., 2004; Palamidessi et al., 2008). For instance, we have previously shown that stimulation of RTKs promotes RAB5 activation and instigates the association with endosomes of the small GTPase RAC1, which is found on these structures together with its GEF, TIAM1 (Palamidessi et al., 2008; Viaud et al., 2014; Bouchet et al., 2016). Subsequent delivery of activated RAC1 back to the PM is brought about by a specific ARF6-dependent recycling route and is required for CDR formation (Palamidessi et al., 2008). We found that endogenous or GFP-tagged NUMB is enriched both on enlarged early endosomes induced by RAB5 ectopic expression (Fig. 4 A) as well as on enlarged recycling endosomes induced by dominant active ARF6 (ARF6-Q67L; Fig. 4 A). Notably, endogenous NUMB endosomal localization is not significantly affected by HGF treatment, which reduces, instead, its cell surface distribution (Fig. 4 B). Thus, NUMB may be implicated in RAB5 or ARF6-endosomal trafficking routes. Consistently, and more relevantly, the ablation of ARF6 expression reduced CDR formation induced by NUMB loss (Fig. 5 A). Similar results were also obtained by expressing a dominant-negative ARF6T44N mutant, indicating that the cycling of the GTPase is required for CDR formation (Fig. 5 B).
Figure 4.
NUMB localizes on RAB5- and ARF6-induced endosomes. (A) Confocal analysis of HeLa cells transfected with RAB5A or HA-ARF6-Q67L either alone (right panels) or in combination with GFP-NUMB (left panels). Cells were processed for epifluorescence (GFP) or stained to detect NUMB, EEA1, HA-ARF6-Q67L, or F-actin, and the nuclei were stained with DAPI. Cells with enlarged EEA1 endosomes and F-actin vesicles induced by RAB5A and ARF6-Q67L, respectively, are marked by asterisks. Bars, 20 μm. Lower images: 4× magnification of the boxed areas. Bars, 5 μm. (B) Confocal analysis of HeLa cells serum starved (SS) for 2 h and stimulated with 100 ng/ml HGF at the indicated time points. Cells were fixed and stained for NUMB (green), EEA1 (red), and DAPI (blue). Bars, 20 μm. Right graph: Pearson’s R correlation value between NUMB and EEA1 (see Materials and methods for details). The data are the mean ± SEM (n > 20 cells/condition in three independent experiments).
Figure 5.
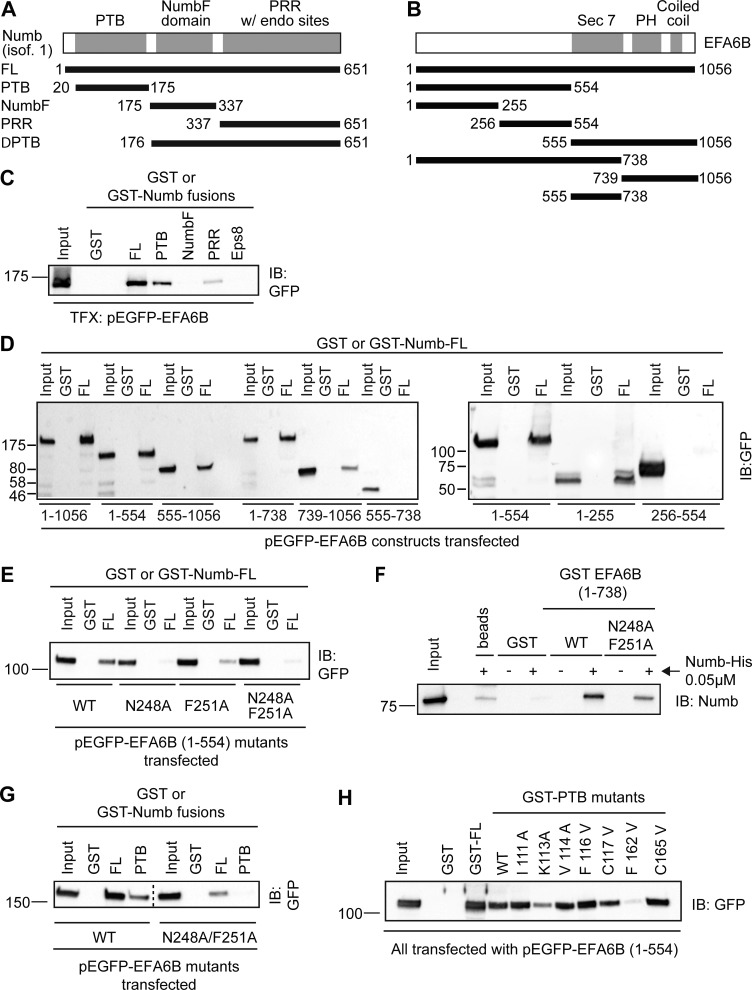
Functional and physical interaction of NUMB with EFA6B, a GEF for ARF6. (A) MEFs transfected with control (siRNA Ctr) or the indicated siRNAs against mNUMB (mN#2) or mARF6 (siRNA mARF6) were serum starved, stimulated with PDGF (1 ng/ml) for 8 min, and stained to detect F-actin. Bar, 40 μm. The percentage of cells forming CDRs with respect to the total is on the right. Data are the mean ± SD (n = 100 cells/condition in two independent experiments). The expression levels of indicated proteins were analyzed by IB. MW markers are shown on the right in kilodaltons. Dashed line indicates that the intervening lanes have been spliced out. (B) HeLa cells transfected with control (siRNA Ctr) or the siRNAs against hNUMB (hN#2) were microinjected with HA-ARF6T44N. Cells were then serum starved, stimulated with HGF for 4 min, and stained with phalloidin to detect F-actin and with anti-HA to detect microinjected cells. Bar, 40 μm. The percentage of cells forming CDRs with respect to the total is on the right. Data are the mean ± SD (n = 100 cells/condition in three independent experiments). The mRNA levels of NUMB were assessed by qRT-PCR, expressed relative to control after normalizing to GAPDH. The data are the relative level of gene expression compared with control cells (siCTR) expressed as mean ± SD (n = 3 independent experiments). A Student’s t test was used to calculate the P values: **, P < 0.01. (C) Equal amounts of total HeLa cell lysate (3.0 mg) were immunoprecipitated with anti-NUMB, anti-HA, or an unrelated IgG. Inputs (0.5%) and immunoprecipitates (IP) were analyzed by IB with indicated antibodies. The known interactor of NUMB, AP2, was used as positive control for the immunoprecipitates. A representative experiment of at least three independent experiments is shown. (D) Phoenix cells cotransfected with GFP-EFA6B and NUMB-FLAG or an empty FLAG-containing vector, EV (control), were serum starved for 4 h (−) and stimulated for 5 min with 100 ng/ml HGF and EGF. Equal amounts of cell lysate (1.5 mg) were immunoprecipitated with anti-FLAG M2 affinity beads (two rounds with 15 μl of bead for each round) or anti-MYC as control. Inputs (1%) and immunoprecipitates were analyzed by IB with indicated antibodies. A representative experiment of at least three independent experiments is shown. (E) Upper panels: HeLa cells cotransfected with NUMB-FLAG and EFA6B-GFP or GFP alone (control) were stained with anti-FLAG monoclonal and anti-GFP polyclonal antibodies. The signals were detected by Duolink 30 Detection Kit 613 (red), and the nuclei were counterstained with DAPI (blue). Red dots represent the positive protein–protein interaction Duolink signal. Lower graph: percentage of Duolink-positive cells over the total number of GFP-positive cells. The data are the mean ± SEM (at least 100 cells/condition in three independent experiments). Bar, 20 μm. A Student’s t test was used to calculate the P values: **, P < 0.01. (F) Purified His-tagged NUMB-His (0.5 μM) was incubated with 0.5 μM immobilized MBP-EFA6 or MBP C-terminal (586–821 aa) actin-binding region of EPS8 (EPS8 C-term), used as negative control, or empty beads. Input (1/5) and bound material were analyzed by IB with anti-NUMB to visualize NUMB or by Ponceau staining to detect MBP recombinant protein. (C, D, and F) MW markers are shown on the left in kilodaltons.
NUMB interacts with EFA6B, a GEF for ARF6
Like all small GTPases, ARF6 cycles between its active GTP-bound and its inactive GDP-bound conformations (Donaldson and Jackson, 2011). GEFs activate ARF6 by promoting the exchange of GTP for GDP, whereas GAPs down-regulate ARF6 activity through hydrolysis of GTP to GDP (Donaldson and Jackson, 2011). Since GEFs are the primary regulators of small GTPase nucleotide status (Donaldson and Jackson, 2011), we investigated whether NUMB interacted with any of the eight identified ARF6 GEFs that belong to three distinct families: cytohesin, EFA6, and BRAG (Casanova, 2007). Among the latter molecules, we focused our attention on EFA6A-D, the cytohesin family that has been reported to control actin remodeling (Casanova, 2007; Gillingham and Munro, 2007), which is required for the formation of CDRs and cell migration (Hoon et al., 2012), and BRAG2. In our experimental model systems, HeLa and MEF cells, ARNO (a cytohesin family member), EFA6B and EFA6D, and BRAG2 were expressed (Fig. S2 A). By GST pull-down assays, however, NUMB could only interact with EFA6B, but not with EFA6D or ARNO (Fig. S2 B). Similarly, in coimmunoprecipitation experiments, we detected no interaction with BRAG2, which is the only BRAG family member possessing putative motives interacting with phosphotyrosine-binding (PTB) domains, similar to EFA6B (see below; Fig. S2 C). While we did not test the interaction with cytohesin and Grp1 (the other two cytohesin family members), the results above point to EFA6B as the relevant interactor.
We further characterized the interaction between EFA6B and NUMB by showing that the two proteins coimmunoprecipitated both under endogenous (Fig. 5 C) or upon overexpression conditions in a growth factor–independent fashion (Fig. 5 D) and by in situ proximity ligation experiments (Fig. 5 E). Finally, we also showed that recombinant, purified NUMB and EFA6B interacted directly (Fig. 5 F). Collectively, these results indicate that NUMB may impinge on ARF6 activity by binding and possibly modulating its GEF, EFA6B.
The N-terminal NPLF motif of EFA6B is required for binding to the PTB domain of NUMB
To characterize the minimal regions of NUMB and EFA6B required for the interaction, we performed a series of pull-down assays with different deletion constructs of both proteins (Fig. 6, A and B). We found that (i) the NUMB PTB domain, but not its C-terminal region, was required for binding to EFA6B (Fig. 6 C and Fig. S3, A and B); and (ii) the N-terminal domain of EFA6B encompassing the first 554 amino acids bound to NUMB at levels comparable to those of the full-length protein (Fig. 6 D and Fig. S3 C). The C-terminal region (555–1,056 aa) of EFA6B displayed, instead, a much weaker interaction (Figs. 6 D and S3 C). Of note, the isolated Sec7 domain (555–738 aa), responsible for the guanine nucleotide exchange activity on ARF6, did not bind to NUMB (Fig. 6 D and Fig. S3 C), whereas the pleckstrin homology domain and the coiled-coil region (739–1,056 aa) weakly associated with NUMB (Fig. 6 D and Fig. S3 C). We concluded that the major determinants mediating the interaction with NUMB reside in the N-terminal region of EFA6B.
Figure 6.
The N-terminal NPLF motif of EFA6B is required for binding to NUMB via its PTB domain. (A and B) Scheme of NUMB (A) and EFA6B (B) domain organization and of the fragments used to characterize the interaction between the two proteins. Numbers correspond to amino acid boundaries. (C) Lysates (2.5 mg) of Phoenix cells transfected with pEGFP-EFA6B were incubated with 0.5 μM GST or GST-fused to NUMB of the indicated NUMB fragments (PTB, 20–175 aa; NUMBF, 175–337 aa; NUMB-PRR, 337–651 aa) or GST-PTB of EPS8, used as negative control. Input lysates (1%) and bound material were analyzed by IB with anti-GFP to visualize EFA6B or by Ponceau staining to detect recombinant GST-fusion proteins (shown in Fig. S3 A). MW markers are shown on the left in kilodaltons. TFX, transfected. (D) Lysates (1.5 mg) of Phoenix cells transfected with either the full-length pEGFP-EFA6B or the indicated GFP-fused EFA6B fragments (amino acid boundaries indicated at the bottom) were incubated with 0.5 μM GST or GST-NUMB immobilized on beads. Input lysates (1% of the total) and bound material were analyzed by IB with anti-GFP to visualize EFA6B and its deletion mutants, or by Ponceau staining to detect the recombinant GST proteins (see Fig. S3 C). MW markers are shown on the left in kilodaltons. (E) Lysates (750 μg) of Phoenix cells transfected with pEGFP-EFA6B 1–554 either WT or carrying N248A, F251A point mutations, alone or in combination, were incubated with 0.5 μM GST or GST-NUMB immobilized on beads. Input lysates (2%) and bound material were analyzed by IB with anti-GFP to visualize EFA6B and its point mutants, or by Ponceau staining to detect recombinant GST proteins (see Fig. S3 D). MW markers are on the left in kilodaltons. (F) Purified, His-tagged His-NUMB (0.05 μM) was incubated with 0.5 μM GST-EFA6B 1–738 either WT or carrying double N248A/F251A point mutations, or with GST alone (control) or in the absence of any protein (beads) to rule out unspecific signals of the antibodies. Input (1/5 of total) and bound material were analyzed by IB with anti-NUMB to visualize NUMB. The Ponceau staining to detect GST recombinant proteins is in Fig. S3 F. (G) Lysates (2 mg) of Phoenix cells transfected with pEGFP-EFA6B WT or pEGFP-EFA6B N248A-F251A were incubated with 0.5 μM GST, GST-NUMB, or GST-PTB immobilized on beads. Input lysates (2%) and bound material were analyzed by IB with anti-GFP to visualize EFA6B. Ponceau staining to detect the recombinant GST proteins is in Fig. S3 H. Dashed line indicates that the intervening lanes have been spliced out. (H) Lysates (2 mg) of Phoenix cells transfected with pEGFP-EFA6B 1–554 were incubated with 0.5 μM GST, GST-NUMB, GST-PTB, or the indicated GST-PTB point mutants immobilized on beads. Input lysates (2%) and bound material were analyzed by IB with anti-GFP to visualize EFA6B. Ponceau staining to detect the recombinant GST proteins is in Fig. S3 J. (F–H) MW markers are shown on the left in kilodaltons.
The inspection of the amino acid sequence of this latter region revealed the presence of an NPLF (248–251 aa) motif, which corresponds to a consensus NPxF sequence for binding to the NUMB PTB domain (Qin et al., 2004; Bogdanović et al., 2012). Indeed, a fragment encompassing the first 255 aa of EFA6B, which includes the NPLF motif, bound to NUMB to a similar extent as the longer N-terminal region (1–554 aa) in a pull-down assay (Fig. 6 D and Fig. S3 C). The critical importance of the NPLF motif was further corroborated by the generation of single point N248A and F251A mutants, introduced either alone or in combination in the context of the EFA6B N-terminal region (1–554 aa). These mutants display either reduced or undetectable binding to NUMB (Fig. 6 E and Fig. S3 D) or its PTB domain (Fig. S3 E). We confirmed this finding in a set of pull-down assays using recombinant purified proteins. Under these conditions, a construct of EFA6B (1–738 aa) carrying the N248A and F251A mutations in the NPLF motif showed markedly reduced binding to recombinant purified NUMB (Fig. 6 F and Fig. S3 F) or its isolated PTB domain (Fig. S3 G), as compared with WT GST-EFA6B. Similarly, these mutations severely impaired the interaction between full-length EFA6B and full-length NUMB (Fig. 6 G and Fig. S3 H), while they displayed no association with the isolated and purified PTB domain (Fig. 6 G and Fig. S3, H and I).
The binding between the NUMB PTB domain and the consensus NPxF motif is mediated by a set of conserved residues within the PTB domain (Zwahlen et al., 2000). We exploited these observations and generated a set of single point mutants (Fig. 6 H and Fig. S3 J). The mutants K113A and F162V in the NUMB PTB domain, predicted to impair the association with NPxF-containing peptides, severely impaired the binding to the N-terminus of EFA6B, albeit to a different extent (Fig. 6 H and Fig. S3 J). All together, these experiments indicate that the 248-NPLF-251 motif of EFA6B and the canonical peptide binding groove of the NUMB PTB are the major interaction surfaces. Consistently, the expression of NUMB mutants (either in a single amino acid, NUMB-F162V [Fig. 7, A and B], or lacking the entire PTB domain [Fig. S4]), which are impaired in binding to a number of NUMB interactors, including EFA6B, failed in rescuing CDR formation.
Figure 7.
EFA6B acts downstream of NUMB in controlling CDR formation. (A) MEF cells were transfected with a scrambled oligo (siRNA Ctr) as control or with siRNA against murine NUMB (mN#3), either alone or in combination with GFP empty vector, GFP-hNUMB WT, or F162V mutant (as indicated). Cells seeded on gelatin-coated coverslips were serum starved for 2 h, stimulated with 1 ng/ml PDGF for 8 min, and processed for epifluorescence to visualize GFP or GFP-NUMB (green, where indicated) or stained to detect F-actin (red). Bars, 20 μm; 10 μm in GFP or GFP-NUMB panels. (B) Left graph: percentage of cells forming CDRs over the total number of cells per field. The data are the mean ± SEM (n > 100 cells/condition in two independent experiments). Right panel: the expression levels of indicated proteins were analyzed by IB. MW markers are shown on the left in kilodaltons. TFX, transfected. (C) HeLa cells transfected with siRNA against NUMB (siRNA hN#3, hN#2) or EFA6B (siRNA hEFA6B) alone or in combination with NUMB (siRNA hN#3/hEFA6B or siRNA hN#2/hEFA6B) siRNA or scrambled oligo (siRNA Ctr, control) were seeded on Matrigel-coated coverslips. Cells were serum starved for 2 h and stimulated with 100 ng/ml HGF for 4 min, stained to detect F-actin antibody. Bar, 40 μm. (D) Left graph: the number of cells forming CDRs over the total number of cells per field. The data are the mean ± SEM (n > 100 cells/condition in three independent experiments). Right graph: NUMB and EFA6B mRNA levels were analyzed by RT-PCR. The data are the relative level of gene expression compared with control cells (siCTR) expressed as mean ± SD (n = 3 independent experiments). A Student’s t test was used to calculate the P values: **, P < 0.01.
EFA6B acts downstream of NUMB in controlling CDR formation
Since EFA6B is a GEF for ARF6 (Donaldson and Jackson, 2011), we analyzed its subcellular localization with respect to the ARF6 recycling compartment (Fig. S5, A and B). The expression of dominant active ARF6-Q67L induced, as expected, the formation of enlarged endosomes, where EFA6B, as well as NUMB (Fig. 4 A), prominently localized (Fig. S5 B). The enrichment of these proteins in the limited membrane of ARF6-positive endosomes prompted us to investigate whether active ARF6 promoted their interaction. Consistently, we found that the expression of ARF6-Q67L significantly enhanced the coimmunoprecipitation between NUMB and EFA6B (Fig. S5 C).
NUMB may regulate ARF6-dependent recycling by interacting and possibly by modulating its GEF, EFA6B. If this hypothesis were correct, the down-regulation of EFA6B should abrogate the increase in growth factor–induced CDRs caused by NUMB ablation. Consistently, the concomitant silencing of NUMB and EFA6B reverted the increase in CDR formation caused by the sole NUMB loss, while the individual silencing of EFA6B had no effect on CDRs (Fig. 7, C and D). Thus, NUMB is epistatic to EFA6B during CDR formation.
NUMB is an activator of EFA6B
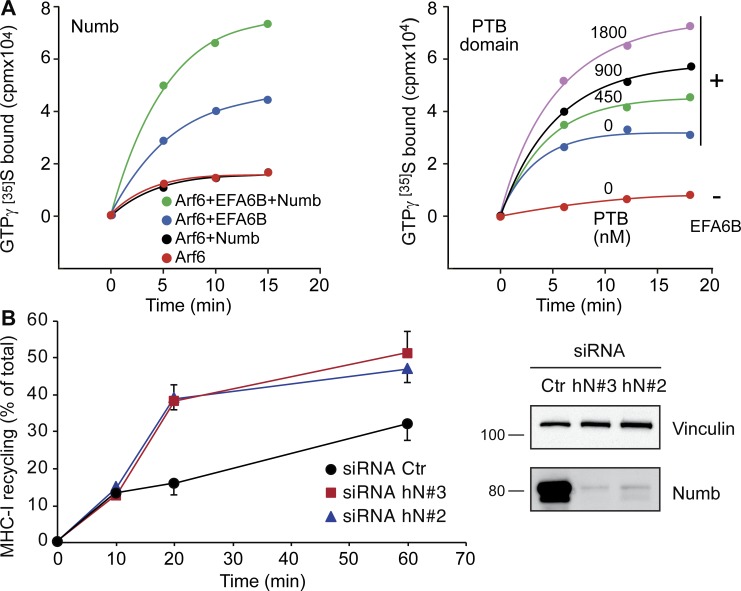
To directly analyze the biochemical consequence of the interaction between NUMB and EFA6B, we performed a series of GEF assays using purified proteins and ARF6 as substrate. As expected, EFA6B significantly accelerated the exchange of nucleotides on myristoylated ARF6 in the presence of liposomes. The addition of NUMB or its isolated PTB domain had no effect on ARF6 intrinsic nucleotide exchange activity but robustly increased the EFA6B-dependent catalysis (Fig. 8 A). Thus, NUMB acts as a positive regulator of EFA6B nucleotide exchange activity on ARF6 in vitro.
Figure 8.
NUMB activates EFA6B and modulates ARF6 recycling. (A) Left graph: guanine nucleotide exchange activity of ARF6 was assessed by measuring the kinetic of binding of [35S]GTPγS to myristoylated ARF6 (ARF6) in the presence of phospholipid vesicles and in the absence (red line) or presence of purified MBP-tagged EFA6B (200 nM; blue line) and recombinant His-tagged NUMB (1.7 µM; green line). The addition of recombinant NUMB to ARF6 had no effect on the kinetic of binding of [35S]GTPγS (black line). Right graph: kinetic of binding of [35S]GTPγS to myristoylated ARF6 (ARF6) in the absence (PTB 0 nm, −EFA6B) or presence of purified recombinant EFA6B (250 nM) and increasing concentrations of purified recombinant NUMB PTB domain. Data are the mean of duplicate measurements and are representative of at least three independent experiments. (B) Left graph: MHC I recycling kinetics were determined by FACS analysis. HeLa cells transfected with siRNA against NUMB (siRNA hN#3, hN#2) or scrambled oligo (siRNA Ctr) were incubated with MHC I–specific antibody for 3 h at 16°C to allow internalization while blocking recycling. Cells were then acid washed with 0.5 M NaCl and 0.5% acetic acid, pH 3, to strip surface-bound MHC I antibodies. Next, cells were pulsed with complete medium and incubated at 37°C for 0, 10, 20, and 60 min (chase) to allow recycling of MHC I to the cell surface. After each time point, cells were washed, fixed with 2% PFA, and analyzed by FACS using MHC I–specific antibodies. Data are the mean ± SD (n = 3 independent experiments). Right panels: the expression levels of NUMB and vinculin were by IB. MW markers are shown on the left in kilodaltons.
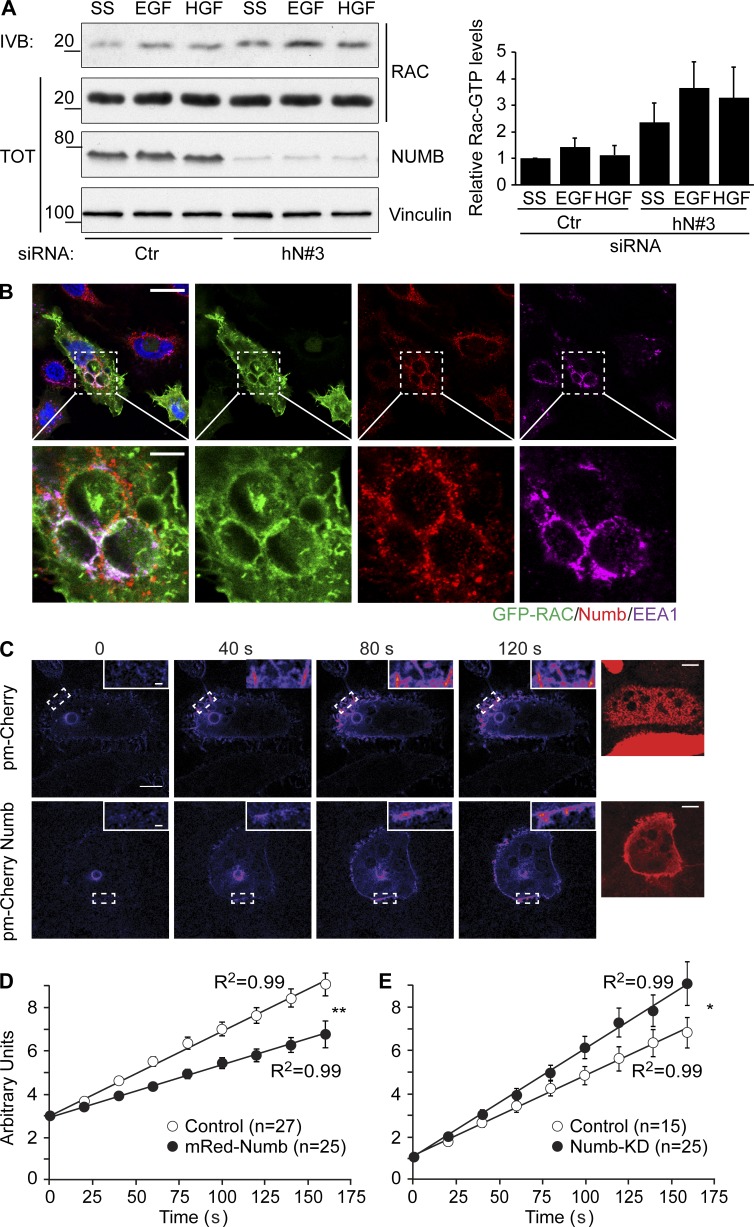
How can the increased NUMB-dependent, EFA6B-mediated activation of ARF6 lead to modulation of recycling and regulation of CDRs? At first glance, since CDR formation strictly requires ARF6 and its recycling, and since the loss of NUMB robustly enhances their appearance, one might have expected NUMB to be a negative regulator of EFA6B activity. A number of recent findings, however, suggest that ARF6 must undergo rapid cycles of activation and deactivation to execute its function in recycling (Allaire et al., 2013; Dutta and Donaldson, 2015; Kobayashi et al., 2015; Loskutov et al., 2015). More importantly, it has been shown that the deactivation of ARF6 is absolutely required to promote cargo recycling (Radhakrishna and Donaldson, 1997). We therefore hypothesized that in the absence of NUMB, EFA6B activity is reduced, limiting ARF6 activation and facilitating its subsequent deactivation for recycling to proceed. If this were the case, one would predict that loss of NUMB should lead not only to enhanced CDR formation (as shown herein), but also to increased recycling of specific cargo delivered back to the PM by ARF6-dependent routes (Schweitzer et al., 2011), including the canonical major histocompatibility complex (MHC) class I (Schweitzer et al., 2011) and RAC1 (Palamidessi et al., 2008). We directly tested these predictions experimentally. First, we set up MHC class I recycling assays using an antibody that recognizes the extracellular portion of the receptor. We found that depletion of NUMB significantly increased the rate of recycling of this cargo (Fig. 8 B). Next, we focused on RAC1. First, we showed that removal of NUMB slightly increased RAC1-GTP levels even in the absence of growth factor stimulation (Fig. 9 A). Additionally, consistent with previous reports (Palamidessi et al., 2008; Viaud et al., 2014; Bouchet et al., 2016), we found GFP-RAC1 localized on the limited membrane of EEA1-positive, enlarged endosomes where endogenous NUMB also resides (Fig. 9 B), suggesting that NUMB may regulate RAC1 trafficking. To test this possibility directly, we measured the delivery of photoactivatable RAC1 from endosomes back to the PM. We used two-photon illumination to ensure that only the RAC1 molecules present in the 3D limited space of the endosomal membrane were hit by photons and activated, and measured the relative amount of photoactivated RAC1 that reached the PM (Palamidessi et al., 2008, 2010). The ectopic expression of NUMB significantly decreased the rate of RAC1 recycling versus control cells (Fig. 9, C and D). The opposite effect was detected upon NUMB removal (Fig. 9 E). Together, these data show that NUMB controls the rate of recycling of cargos relevant for CDR formation, likely through the regulation of ARF6 GTP/GDP cycles. Notably, NUMB appears to act specifically on the ARF6 trafficking route, as its removal has no effects on the recycling of cargos, such as EGFR and Tf receptors (TfRs), that do not use this pathway (Fig. S1).
Figure 9.
Loss of NUMB increases RAC-GTP levels and RAC1 recycling. (A) HeLa cells transfected with siRNA against human NUMB (hN#3) or scrambled oligo (Ctr) as control were serum starved (SS) for 2 h and stimulated with either 100 ng/ml EGF (8 min) or 100 ng/ml HGF (7 min) as indicated. Equal amounts of cell lysate were immunoprecipitated with GST-CRIB. Total lysates (TOT) and bound material (IVB) were analyzed by IB with the indicated antibody. MW markers are on the left in kilodaltons. Right graph: quantification of the relative RAC1-GTP levels. The relative intensity of each RAC1-GTP band was normalized to the total amount of RAC1. Data are the mean ± SD of three independent experiments. (B) Confocal analysis of HeLa cells cotransfected with GFP-RAC1 and RAB5, which induces the formation of enlarged endosomes. Cells were fixed and processed for epifluorescence analysis to visualize GFP-RAC (green) or stained with specific antibodies or with DAPI to visualize NUMB (red), EEA1 (magenta), or nuclei (blue), respectively. Bar, 20 μm. Lower panels: magnifications of the indicated regions. Bar, 5 μm. (C) HeLa cells were cotransfected with photoactivatable paGFP RAC and pmCherry-NUMB or empty vector. paGFP barely absorbs 488 nm light in its native form, thereby exhibiting a weak fluorescence emission. Focal plane–restricted, multiphoton-induced photoactivation of paGFP-RAC was achieved by exciting the area corresponding to a vesicle. Images were then collected every 20 s (exciting fluorescence at 488 nm). Insets, 3× magnifications of the indicated areas. Right panels: NUMB Red epifluorescence images are shown. Bars, 10 μm. (D) The increase in mean fluorescence over time in a target region of the PM adjacent to the photoactivatable vesicle. The data are the mean ± SEM of 27 and 25 events counted for the empty vector (control) and NUMB (pmCherry-NUMB) transfected cells, respectively. (E) The increase in mean fluorescence over time in a target region of the PM adjacent to the photoactivatable vesicle. The data are the mean ± SEM of 15 and 18 events counted for cControl and NUMB KD cells, respectively. An ANOVA F-test was used to calculate P values of the different slope: **, P < 0.005; *, P < 0.05.
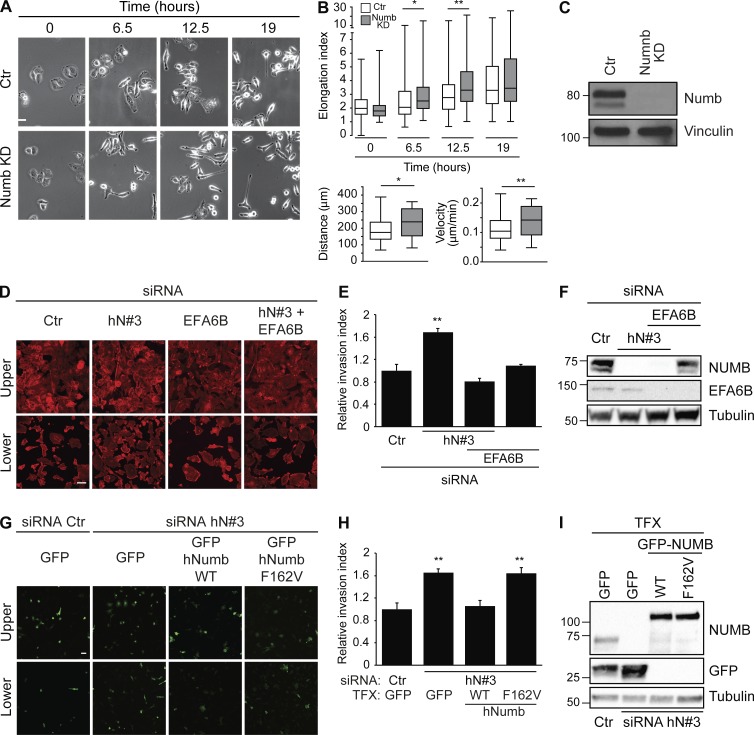
NUMB is a negative regulator of HGF-induced cell migration and invasion
CDR formation invariably marks the acquisition of a mesenchymal, elongated mode of cell migration (Palamidessi et al., 2008). Hence, we tested whether NUMB perturbation impacted cell migration in 2D and 3D cell cultures upon activation of RTKs. Loss of NUMB caused a significant increase in the cell elongation index (the ratio between the major axis and the minor axis), which directly correlates with mesenchymal cell locomotion (Palamidessi et al., 2008) and was accompanied by a concomitant increase in migration velocity (Fig. 10, A–C). We next investigated the role of NUMB in cell motility in a 3D matrix. Matrigel-coated Boyden chambers were employed, and HGF was used as a chemoattractant. We found that removal of NUMB by siRNA promoted cell invasion toward HGF (Fig. 10, D–I), consistent with increased CDR formation and motility. Additionally, and more relevantly, the concomitant silencing of NUMB and EFA6B reverted the increased chemoinvasiveness caused by the sole NUMB loss (Fig. 10, D–F). Importantly, the individual silencing of EFA6B had no effect on invasion (Fig. 10, D–F). Using a similar experimental setting, we also showed that the expression of siRNA-resistant variants of NUMB WT but not of the NUMB-F162V mutant, which no longer binds to EFA6B, reverted the increased chemotactic invasion caused by the loss of endogenous NUMB (Fig. 10, G–I).
Figure 10.
NUMB is a negative regulator of HGF-induced cell migration and invasion. (A) HeLa cells transfected with an siRNA against NUMB (NUMB KD) or scrambled oligo (Ctr) as control were serum starved for 2 h, stimulated with 100 ng/ml HGF, and analyzed by time-lapse microscopy. Representative images for each condition at specific time points (0 h, 6 h 30 min, 12 h 30 min, and 19 h) are shown. Bar, 20 μm. (B) Upper graph: elongation index (the ratio between the major and minor axis of a cell) was quantified for each condition at the indicated time in at least 30 single cells/experiment of three independent experiments. Lower graphs: total accumulated distance of cells (left graph) and mean velocity (right panel) were measured in at least 30 single cells/experiment of three independent experiments. The data are presented as the mean ± SEM. (C) The expression levels of NUMB and vinculin were analyzed by IB. (D) HeLa cells were transfected with a scrambled oligo (siRNA Ctr) as control, or with siRNA against human NUMB (hN#3) and EFA6 either alone or in combination (as indicated). Cells were seeded on Matrigel-coated chambers. 100 ng/ml HGF was added in the lower chamber to generate a chemoattractive gradient. After 48 h, cells were fixed, stained to detect F-actin, and analyzed by confocal microscopy. Representative images of upper, noninvasive cells (upper panels) and lower, invasive cells (lower panels) are shown. Bar, 10 μm. (E) Relative invasion index. Quantification of the number of cells passing through the semipermeable membrane is shown as fold increase compared with control cells. At least 10 fields were analyzed for each condition. The data are the mean ± SEM (n > 100 cells/condition in two independent experiments). (F) The expression levels of NUMB, EFA6, and tubulin were analyzed by IB. MW markers are shown on the left in kilodaltons. (G) HeLa cells were transfected with a scrambled oligo (siRNA Ctr) as control, siRNA against human NUMB (hN#3) either alone or in combination with GFP empty vector, siRNA-resistant GFP-hNUMB WT, or F162V mutant (as indicated). Cells were seeded on Matrigel-coated chambers. 100 ng/ml HGF was added in the lower chamber to generate a chemoattractive gradient. Cells were fixed and GFP-positive cells were analyzed by confocal microscopy. Representative images of upper, noninvasive cells (upper panels) and lower, invasive cells (lower panel) are shown. Bar, 20 µm. (H) Relative invasion index of cells passing through the semipermeable membrane (invasion index = ratio between GFP invasive cells/GFP noninvasive cells/field) is shown as fold increase compared with control cells (siRNA Ctr-GFP). The data are the mean ± SEM (n > 100 cells/condition in two independent experiments). (I) The expression levels of NUMB, GFP, GFP NUMB-WT, or NUMB-F162V and tubulin were analyzed by WB. (C and H) MW markers are shown on the left in kilodaltons. TFX, transfected. A Student’s t test was used to calculate the P values: *, P < 0.05; **, P < 0.01.
Collectively, these observations support the notion that NUMB, by controlling CDR formation, acts as a negative regulator of HGF-induced mesenchymal motility and invasive programs.
Discussion
The endocytic adaptor NUMB is at the center of diverse cellular phenotypes, including cell fate developmental decisions, maintenance of SC compartments, regulation of cell polarity, adhesion, and migration, which all impact epithelial cell states (Pece et al., 2011). Molecularly, NUMB controls the trafficking of various cargos, including E-cadherin and integrin β1, in addition to being implicated in a variety of signaling pathways, including NOTCH, Hedgehog, and TP53 (Colaluca et al., 2017; Zhong et al., 1997; Di Marcotullio et al., 2006; Nishimura and Kaibuchi, 2007; Gold et al., 2009; Wang et al., 2009; Pece et al., 2011; Qi et al., 2015; Zhang et al., 2016; Sheng et al., 2017; Tosoni et al., 2017). Our findings uncover a novel facet of the pleiotropic role of NUMB by providing in vitro and in vivo evidence that it acts by directly interacting and modulating the enzymatic activity of EFA6B. We show that this interaction is necessary to tune the activity of ARF6 and ARF6-dependent recycling routes needed, in turn, for the efficient delivery of cargos to the PM. This axis is critical for the control of polarized membrane protrusions that mark the acquisition of a mesenchymal mode of migration in response to PDGF and HGF stimulation. Notably, the latter growth factor has been shown to be a potent inducer of a transition from an epithelial to a mesenchymal state, a process that is accelerated following NUMB loss (Wang and Li, 2010; Lau and McGlade, 2011). A similar role has also been reported for EFA6B, which is needed for the maintenance of tight junctions in MDCK (Théard et al., 2010), while in breast cancer lines, EFA6B opposes TGF-β induction of EMT (Zangari et al., 2014), suggesting that a NUMB–EFA6–ARF6 axis might also be relevant in control of the latter phenotypes. Mechanistically, the positive activity exerted by NUMB via EFA6B on ARF6 in vitro appears in contrast with its negative role in the regulation of ARF6-dependent recycling and biological activity. However, in order to function properly, ARF6 must be kept under tight temporal and spatial control (Allaire et al., 2013; Dutta and Donaldson, 2015; Kobayashi et al., 2015; Loskutov et al., 2015), as both too little or too much (or persistent) ARF6-GTP levels have detrimental biological effects. In addition, the activity of EFA6B is regulated through a negative feedback loop by GTP-loaded ARF6 that binds to an allosteric site on the pleckstrin homology–C-terminal region (Padovani et al., 2014). Within this context, the NUMB–EFA6B axis, herein identified, is likely part of this self-regulated molecular network that might also include GAPs and effectors of ARF6, whose action must be finely tuned and coordinated to ensure appropriate spatiotemporal control of the activity of this GTPase. In keeping with the latter possibility, in a set of preliminary experiments, we also found that NUMB, in addition to binding to EFA6B, also coimmunoprecipitated with ACAP1, a GAP for ARF6 (Jackson et al., 2000; Dai et al., 2004). Thus, NUMB may indeed act as a core factor for the assembly of a multiprotein complex regulating ARF6 activity in a spatiotemporally defined manner. In this respect, it is noteworthy that while ARF6 predominantly localizes to the PM and endosomal compartments, it plays important roles in a variety of cellular processes, including endocytosis of the PM, exocytosis, endosomal recycling, cytokinesis, and actin cytoskeleton reorganization (Hongu and Kanaho, 2014). In these processes, ARF6 is likely regulated through distinct molecular machineries. Not surprisingly, there are at least eight GEFs belonging to three distinct families (cytohesin, EFA6, and BRAG families) that have been demonstrated to promote GTP/GDP exchange on ARF6 (Casanova, 2007). These proteins display variable distribution and functions, acting in a site-specific fashion (Hongu and Kanaho, 2014). Our data suggest that the NUMB–EFA6B axes specifically impact ARF6 endosomal function controlling the recycling of the specialized set of cargos important for CDR formation. Whether this control is exerted directly on endosomal vesicles or at the PM where the two proteins reside remains to be firmly established. Future work will also be needed to unveil all the components of the NUMB–EFA6B network, which we predict will play a pivotal role in the control of epithelial identity and in the transition to a mesenchymal state, with obvious relevance to cancer.
Materials and methods
Antibodies and reagents
The antibodies used were anti-NUMB (Pece et al., 2004); mouse monoclonal anti-vinculin (Sigma) and rabbit polyclonal anti-GFP for immunofluorescence (a gift from Jan Faix, Hannover Medical School Institute of Biophysical Chemistry, Hannover, Germany); rabbit polyclonal GFP for Western blotting (WB; Sigma); mouse anti-FLAG (Sigma); rabbit anti-FLAG (Sigma); mouse monoclonal anti-His (sc-8036; Santa Cruz Biotechnology); rabbit polyclonal anti-RAB5A (sc-19; Santa Cruz Biotechnology); rabbit polyclonal anti-HA (sc-805; Santa Cruz Biotechnology); goat polyclonal anti-EEA1 (sc-6415; Santa Cruz Biotechnology); mouse monoclonal anti-EEA1 (a gift from Mrino Zerial, Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany); mouse monoclonal anti–MHC class I W6/32 (sc-32235; Santa Cruz Biotechnology); rabbit monoclonal anti-ARF6 (5740; Cell Signaling); rabbit polyclonal anti–C-Met for WB (4560S; Cell Signaling); goat polyclonal anti–C-Met immunofluorescence (AF276; R&D Systems); rabbit polyclonal anti-pAKT (9271; Cell Signaling); rabbit polyclonal anti–phospho p44/42 MAPK (ERK1/2) (4370S; Cell Signaling); rabbit polyclonal anti–β-adaptin 2 (PA1-1066; Thermo Fisher); rabbit polyclonal anti-EFA6B (HPA034722; Sigma); and rabbit monoclonal anti-NUMB (2756; Cell Signaling).
Recombinant human HGF (used at 100 ng/ml) was from R&D Systems. Recombinant human PDGF-BB (used at 1 ng/ml) was from Immunological Sciences. Recombinant EGF was from Vinci Biochem.
Plasmids
NUMB isoforms were cloned into pEGFP-C1 (Clontech) vector as a HindIII/Sal1 fragment, as described previously (Colaluca et al., 2008). NUMBΔPTB (176–651) was cloned by BglII/SalI digestion in pEGFP-N1 vector. FL NUMB FLAG, NUMB PTB (20–175) FLAG, and NUMBΔPTB (176–651) FLAG were cloned by NheI/EcoRI digestion in pLL3.7 vector also carrying short hairpin targeting endogenous NUMB, as described in Colaluca et al. (2008). FL NUMB, NUMB PTB (20–175), NUMB PRR1 (175–337), and NUMB PRR2 (337–651) were cloned by BamHI/SalI digestion in modified pGEX6p1 (GE Healthcare) for GST fusion protein expression, described in Colaluca et al. (2017), with the GST at the N-terminus. NUMB His and NUMB PTB were expressed in a pET vector, described in Colaluca et al. (2017). GFP NUMB and GST NUMB PTB point mutations were obtained by the QuikChange Mutagenesis Kit (Stratagene) according to the manufacturer’s instructions. pEGFC3 EFA6B, pEGFPC3 EFA6A, and pEYFP ARNO were described in Derrien et al. (2002) and Franco et al. (1999). pCMV-RAB5A, paGFP-RAC, and pCDNA-ARF6-QL were described in Palamidessi et al. (2008). pBAC-His-MBP-TEV vector was a gift from S. Pasqualato (European Institute of Oncology, Milan, Italy). pEGFPC1-EFA6B deletion mutants and Myelin basic protein (MBP)–EFA6B were obtained by PCR.
Generation of EFA6B deletion constructs
EFA6B deletion constructs containing different fragments of the C-terminal region of the protein (648–821 aa) were obtained through PCR, using as a template the pEGFP_C3 vector containing the cDNA sequence of human EFA6B.
To clone the different constructs, specific primers (Table S2) were designed, flanked by BglII and SalI sites. DNA fragments were then subcloned either in an empty pGEFP_C1 vector after BglII-SalI double digestion, in an empty pGEX6p1 vector after BamHI-SalI double digestion, or in an empty pBAC_His-MBP-TEV vector after BamHI-SalI double digestion and subsequent ligation.
Generation of EFA6B point mutants
The different EFA6B mutants were generated by performing site-directed mutagenesis using the QuikChange Mutagenesis Kit according to the manufacturer’s instructions. Specific primers were designed (see Table S2).
The pGEX EFA6B 1–554 aa N248A, F251A, the pGEX EFA6B 1–739 aa N248A, F251A, and the pBAC EFA6B N248A, F251A, were generated by GenScript. Then the EFA6B 1–554 aa N248A, F251A fragment, and the EFA6B N248A, F251A, were amplified by PCR reaction with primers flanked by BglII and SalI sites. DNA fragments were then subcloned in an empty pGEFP_C1 vector after BglII-SalI double digestion and subsequent ligation.
Cell culture
HeLa cells were grown in MEM (Invitrogen) supplemented with 10% South American serum (EuroClone), 1% nonessential amino acids, and 1% sodium pyruvate. Immortalized MEFs (Scita et al., 1999) were grown in DMEM (Lonza) supplemented with 10% South American serum and 1% L-glutamine (EuroClone). Phoenix cells were grown in DMEM, supplemented with South American serum and 1% L-glutamine. Cells were grown at 37°C in 10% CO2. The identity of the cells used was verified routinely by finger printing.
Transfection
Transfections were performed using the calcium phosphate method, electroporation, Lipofectamine (Invitrogen), or FuGene HD (Promega).
Phoenix cells were transfected using the calcium phosphate procedure. In this case, DNA (20 μg for a 15-cm plate) was diluted in 878 μl of double-distilled H2O, and 122 μl of 2 M CaCl2 was added. This solution was added, dropwise, to 1 ml of HBS twice (50 mM Hepes, pH 7.5, 10 mM KCl, 12 mM dextrose, 280 mM NaCl, and 1.5 mM Na2HPO4). Then, the precipitate was added to the cells (plated on 15-cm dishes) and removed after 12–16 h.
Electroporation was used to transfect MEF cells for CDR rescue experiment using the Micro-Porator system (Digital Bio Technology) according to the manufacturer’s instructions. Cells were microporated with one pulse of 20 ms at 1,650 V, with a cell density of 5 × 106 cells.
Lipofectamine or FuGene HD was used to transfect HeLa cells for immunofluorescence experiments according to the manufacturer’s instructions.
siRNA experiments
siRNA delivery was achieved by mixing 5–50 nM specific siRNAs with Opti-MEM and Lipofectamine RNAiMAX Transfection Reagent (Life Technologies) by two cycles of transfection, according to the manufacturer’s instructions. For each RNAi experiment, negative control was performed with the same amounts of scrambled siRNAs (siCTR). KD efficiency was controlled by WB or quantitative RT-PCR (qRT-PCR).
Four different siRNAs were used for hNUMB KD, three different siRNAs were used for mNUMB KD, and two different siRNAs were used for EFA6B KD, with comparable results.
Oligo details are as follows: hNUMB oligo #3: 5′-TACACACCTCTTCTAACCATCGGTC-3′ (Invitrogen); hNUMB oligo #2: 5′-GGACCTCATAGTTGACCAGTT-3′ (NUMB Nishimura; Thermo Fisher); hNUMB oligo #5: 5′-AAATGTAGCTTCCCGGTTA-3′ (ON-TARGETplus NUMB siRNA J-015902-05; Dharmacon); hNUMB oligo #6: 5′-TAAGATAGTCGTTGGTTCA-3′ (ON-TARGETplus NUMB siRNA J-015902-06; Dharmacon); mNUMB oligo #2: 5′-AGAAAGAAAGACCTTT-3′ (Riboox); mNUMB oligo #3: 5′-CGGGAAAGAAAGCAG (Riboox); mNUMB oligo #4: 5′-ATCTGTCATTGTTTC-3′ (Riboox); hEFA6B: 5′-GAACCGCAATCAGCTGTGA-3′ (ON-TARGETplus PSD4 siRNA J-019959-06; Thermo Fisher); and mARF6: 5′-GCAAGACAACGAUCCUGUA-3′ (s62577; Ambion). For all siRNA experiments, the appropriate scrambled oligos were used as control siRNAs (siCTR).
qRT-PCR detection of mRNAs
Gene expression was analyzed using TaqMan gene expression assay (Applied Biosystems). In detail, 0.1 ng cDNA was amplified, in triplicate, in a reaction volume of 25 µl with 10 pmol of each gene-specific primer and the SYBR-green PCR MasterMix (Applied Biosystems). Real-time PCR was performed on the 14 ABI/Prism 7700 Sequence Detector System (PerkinElmer/Applied Biosystems) using a pre-PCR step of 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 60 s at 60°C. Specificity of the amplified products was confirmed by melting curve analysis (Dissociation Curve TM; PerkinElmer/Applied Biosystems) and by 6% PAGE. Samples were amplified with primers for each gene (listed in Table S3). The Ct values were normalized to the GAPDH curve. Results were quantified using the 2−ΔΔCT method. PCR experiments were performed in triplicate.
Imaging techniques
Immunofluorescence
Cells were plated on glass coverslips (preincubated with 0.5% gelatin in PBS at 37°C for 30 min, in the case of MEFs). After 24 h, cells were processed for epifluorescence or indirect immunofluorescence microscopy. Cells were fixed in 4% PFA for 10 min, washed with PBS, and permeabilized in PBS and 0.1% Triton X-100 for 10 min at RT. To prevent nonspecific binding of the antibodies, cells were then incubated with PBS in the presence of 1% BSA for 10 min.
The coverslips were then gently deposited, face down, on 50 μl of primary antibody diluted in PBS and 1% BSA, spotted on Parafilm. After 40 min of incubation at RT, coverslips were transferred into 12-well plates and washed three times with PBS. Cells were then incubated for 40 min at RT with the appropriate secondary antibody. F-actin was detected by staining with FITC- or TRITC-conjugated phalloidin (Molecular Probes) at a concentration of 6.7 U/ml.
After three washes in PBS, coverslips were transferred into 12-well plates and incubated in PBS containing DAPI (1:3,000) for 5 min at RT. Coverslips were washed three times in PBS, mounted in Mowiol (20% Mowiol [Sigma], 5% glycerol, 2.5% DABCO [Molecular Probes], and 0.02% NaN3 in PBS), examined by fluorescent optical microscopy or in a 90% glycerol solution containing diazabicyclo-(2.2.2)octane antifade (Sigma), and examined under a Bio-Rad MRC 1024 confocal microscope equipped with a 20-mW Kr-Ar laser for colocalization analysis. Confocal image acquisition was performed in sequential mode to limit channel cross-talk and corrected for residual fluorescence bleed-through. Green fluorescence was collected through a 520 ± 35–nm band-pass filter, while the red channel filter was collected through a 605 ± 20–nm band-pass. Images were further processed with the ImageJ software (Adobe).
For two-photon fluorescence microscopy, a Chameleon-XR (Coherent) Ti:sapphire laser source was directly coupled to the scanning head of a Leica TCS SP5 AOBS confocal microscope using an infrared port. Two-photon activation was performed by irradiating a specific vesicle at λ = 790 nm with a pixel dwell time of 4.9 μs and an activation average power < p > = 10 mW. The targeted region was subjected to pulses of high energy–density infrared light (λ = 790 nm) to induce photoactivation. Each pulse had a 1-s duration, and a time interval of 20 s between pulses was set to avoid photobleaching of the activated molecules. Imaging of the activated proteins was obtained using the 488-nm line of a 20-mW Argon ion laser using a 100× oil NA = 1.4 objective HCX PL APO (Leica Microsystems). The spectral window used for collecting fluorescence was 500–600 nm.
Microscope image acquisition
For each of the images in our figures, we employed the following conditions of acquisition. Please refer to Table S4 for microscopes and settings, objectives, cameras, and acquisition/processing software. Details and types of operations employed in processing the images, using the ImageJ software, are specified as follows and are in the indicated Materials and methods sections: Fig. 1 A: Olympus BX 51 FL; objective, 60× oil, 0.5 zoom camera; fluorochrome, TRITC-phalloidin. Fig. 1, D and G: Olympus BX 51 FL; objective, 40× oil, 0.5 zoom camera; fluorochromes, TRITC-phalloidin and GFP. Fig. 2 B: Olympus BX 51 FL; objective, 60× oil, 0.5 zoom camera; fluorochrome, TRITC-phalloidin. Fig. 3 A: Leica SP5; objective, 63× oil immersion; fluorochromes, DAPI and Cy3-donkey anti-goat. Fig. 3 B: Leica SP5; objective, 63× oil immersion; fluorochromes, Alexa 647–donkey anti-goat, Alexa 555–goat anti-human, FITC-phalloidin, and DAPI. Fig. 4 A: Leica TCS SP2 AOBS; objective, 63× oil immersion; fluorochromes (left panels), GFP, Cy3-donkey anti-goat, Cy3-donkey anti-rabbit, and DAPI; fluorochromes (right panels), Alexa 488–donkey anti-mouse, TRITC-phalloidin, Cy3-donkey anti-goat, and DAPI. Fig. 4 B: Leica TCS SP2 AOBS; objective, 63× oil immersion; fluorochromes, Alexa 488–donkey anti-mouse, Cy3-donkey anti-goat, and DAPI. Fig. 5 A: Olympus BX 51 FL; objective, 40× oil, 0.5 zoom camera; fluorochrome, FITC-phalloidin. Fig. 5 B: Olympus BX 51 FL; objective, 60× oil, 0.5 zoom camera; fluorochromes, FITC-phalloidin and CY3-donkey anti-rabbit. Fig. 5 E: Olympus BX 51 FL; objective, 60× oil; fluorochromes, GFP, PLA-probe (Texas Red), and DAPI. Fig. 7 A: Olympus BX 51 FL; objective, 60× oil, 0.5 zoom camera; fluorochromes, GFP and TRITC-phalloidin. Fig. 7 C: Olympus BX 51 FL; objective, 60× oil, 0.5 zoom camera; fluorochrome, TRITC-phalloidin. Fig. 9 B: SARA Leica SP5; objective, 63× oil immersion; fluorochromes, GFP, Cy3-donkey anti-mouse, Alexa 647–donkey anti-goat, and DAPI. Fig. 9 C: Two-photon fluorescence microscopy; described in detail in the Immunofluorescence section. Fig. 10 A: Described in detail in the Single cell migration assay section. Fig. 10 D: Leica SP8; objective, 40× oil; fluorochrome, TRITC-phalloidin. Fig. 10 G: Leica SP8; objective, 20×; fluorochrome, GFP. Fig. S4: Olympus BX 51 FL; objective, 40× oil, 0.5 zoom camera; fluorochromes, GFP and TRITC-phalloidin. Fig. S5 A: Leica TCS SP2 AOBS; objective, 63× oil immersion; fluorochromes, GFP, Cy3-donkey anti-goat, and DAPI. Fig. S5 B: Leica TCS SP2 AOBS; objective, 63× oil immersion; fluorochromes, GFP, TRITC-phalloidin, and DAPI.
siRNA-based screening
A candidate siRNA-based screening was performed to identify endocytic molecules specifically required for the formation of HGF-induced CDRs. HeLa cells were transfected with scrambled control (siCTR) or the indicated specific siRNA and stimulated with HGF (100 ng/ml) and/or EGF (100 ng/ml) for 10 min before fixing and staining with phalloidin. The identification of F-actin–positive apical CDRs was facilitated by 3D reconstruction of confocal serial z sections. The fraction of siRNA-treated HeLa cells forming CDRs or peripheral ruffle structures relative to that of control, HGF-stimulated cells was calculated. Three independent experiments were performed and 40 cells/experiment were analyzed.
CDR formation assay
To evaluate CDR formation, HeLa cells were seeded on Matrigel-coated coverslips. To perform the coating, Matrigel was diluted at 0.5 mg/ml in cold 50% DMEM/F12 (without serum) on ice. Then a refrigerated 6-well plate with coverslips was placed on ice, and 1 ml of solution was added and homogeneously distributed. The plate was incubated for 1 h at RT. After two washes with PBS, 75,000–100,000 HeLa cells were seeded. After 48 h, cells were serum starved for 2 h and stimulated with HGF.
Single-cell migration assay
Single-cell migration was monitored as follows. Briefly, siCTR and siNUMB HeLa cells, upon 24 h of interference, were seeded sparsely in a 6-well plate (2 × 104 cells/well) in complete medium. After 24 h, cells were serum starved for 2 h and stimulated with 100 ng/ml HGF. Random cell motility was monitored over a 19-h period. Pictures were taken every 5 min from 10 positions/condition using a motorized Olympus ScanR inverted microscope with 40× objective. All experiments were performed using an environmental microscope incubator set to 37°C and 5% CO2 perfusion. Single cells were manually tracked using the Manual Tracking Tool ImageJ software plugin. Elongation index was calculated as the ratio between the major and the minor axis. Distance and velocity were obtained by Chemotaxis and the Migration Tool ImageJ software plugin.
Invasion assay
Cell invasion assays were performed using a BD BioCoat GFR Matrigel Invasion Chamber (BD Biosciences) composed of a polycarbonate membrane, containing 8-μm pores, and covered with a thin layer of GFR Matrigel Basement Membrane Matrix. An equal number of cells (2.5 × 104) was seeded into the upper chamber of the Transwell and allowed to migrate for 48 h. The lower chamber additionally contained HGF (100 ng/ml). Cells were fixed with 4% PFA for 10 min, washed with PBS, and stained with TRITC-phalloidin or processed for epifluorescence analysis to visualize GFP-positive cells. Cells were analyzed by serial confocal z sections and taken from the top of the Matrigel to the bottom of the supporting plastic porous filter. The number of cells was counted in four randomly chosen fields. Data are expressed as the mean ± SEM of two independent experiments.
C-Met surface level quantification
siCTR or siNUMB cells, upon 72 h of interference, were washed twice with PBS and fixed with PFA. Cells were stained with C-Met–specific antibody in the absence of permeabilization to visualize only cell surface receptor, and with DAPI (blue) to visualize nuclei. To quantify the amount of C-Met receptor on the cell surface, integrated density of C-Met signaling (defined using ImageJ) normalized over the number of cells was quantified in the different conditions. Values were normalized over the siCTR scrambled control cells and shown as fold increase. At least 20 fields of view were analyzed in each condition, and three independent experiments were performed. The data are the mean ± SEM. P values were obtained using Student’s t test.
C-Met internalization by immunofluorescence
48 h after interference, siCTR or siNUMB (hN#3) HeLa cells were serum starved for 2 h, stimulated with 100 ng/ml HGF for 5 and 10 min, and fixed with PFA. Cells were stained with C-Met antibody, hEEA1 antibody, phalloidin, and DAPI to visualize the nuclei. Images were acquired with an SP5 Leica confocal system (63× magnification, 2.5× zoom).
For C-Met/EEA1 colocalization analysis, a mask was generated around EEA1 vesicles, and the colocalization was calculated as Pearson’s R value using the ImageJ Coloc2 tool, analyzing at least 50 cells for each condition.
NUMB/EEA1 colocalization analysis
A mask was generated around EEA1 vesicles, and the colocalization was calculated as Pearson’s R value using the ImageJ Coloc2 tool, analyzing at least 20 cells for each condition.
EGF recycling assay
Recycling assays of 125I-EGF were performed as described in Sigismund et al. (2008). In brief, cells were serum starved and then incubated with 125I-EGF (20 ng/ml) for 15 min at 37°C, followed by mild acid/salt treatment to remove bound EGF. Cells were then chased at 37°C in a medium containing 4 μg/ml EGF for the indicated times to allow recycling. At the end of each chase time, the medium was collected and subjected to TCA precipitation to determine the amount of intact (TCA precipitable) and degraded (TCA soluble) 125I-EGF present in it. Surface-bound 125I-EGF was extracted by acid treatment (0.2 M acetic acid, pH 2.8, and 0.5 M NaCl).
Finally, cells were lysed in 1 N NaOH to determine intracellular 125I-EGF. Data are expressed as the fraction of intact 125I-EGF in the medium with respect to the total (total medium + total surface + total intracellular). Nonspecific counts were measured for each time point in the presence of a 200-fold excess of cold ligand and were never >3–10% of the total counts.
Tf recycling assay
Recycling assays of the TfR were performed as described by Tosoni et al. (2005). In brief, cells were incubated for 1 h at 37°C in serum-free L-15 medium (Gibco) with 0.2% BSA, followed by incubation for 1 h at 16°C with 2 μg/ml human Tf (400 ng/ml 125I-Tf and 1,600 ng/ml cold Tf) in serum-free L-15 with 0.2% BSA. Cells were then washed twice with ice-cold PBS, and surface-bound Tf was removed by incubation for 15 min at 4°C in 1 ml of 150 mM NaCl, 1 mM CaCl2, and 20 mM sodium acetate buffer (pH 4.6), followed by two washes with ice-cold PBS. Cells were then incubated in serum-free MEM supplemented with 0.2% BSA, 20 mM Hepes (pH 7.4), and 100 µg/ml cold human Tf (in the absence or presence of 100 ng/ml HGF when indicated) for 5 min at 4°C and then warmed to 37°C to allow recycling of internalized Tf. At various times, incubations were stopped by placing the culture dishes on ice. The medium was collected (medium), and surface-bound Tf was removed by stripping for 5 min at 4°C, with 300 µl of 0.5 M NaCl and 0.5 M acetic acid (pH 2.5; bound). Finally, cells were solubilized in 1 M NaOH (internalized). For each time point, Tf recycling is expressed as the ratio of medium + bound (recycled)/internalized + medium + bound (total).
MHC class I recycling
siCTR and siNUMB HeLa cells, upon 48 h of interference, were washed twice with PBS and incubated at 16°C for 3 h in CO2-independent L-15 medium (Leibovitz; Gibco), supplemented with 10% FBS and containing 10 µg/ml of mouse MHC I antibody (sc-32235). Under these conditions, recycling is blocked, whereas internalization proceeds, albeit at a reduced rate (Punnonen et al., 1998). The cells were washed twice with ice-cold PBS and three times with acid wash buffer (0.5 M NaCl and 0.5 M acetic acid, pH 2.5), followed by successive rinses with PBS to remove surface antibody, and were incubated in complete medium. The cells were then transferred to 37°C to allow recycling of internalized MHC I. At various time points, cells were harvested on ice in PBS and 10 mM EDTA, transferred in a clean tube, and washed once in PBS and 1% BSA. Pellets were incubated with secondary antibody resuspended in PBS and 1% BSA. Cells were washed three times with PBS and 1% BSA. Cells were fixed in 2% formaldehyde for 20 min on ice. Cells were washed once in PBS and 1% BSA, resuspended in PBS, and kept at 4°C until FACS analysis. The amount of MHC class I recycled at different time points was calculated as the percentage of total MHC class I internalized at time 0 (t0).
In situ proximity ligation assay
We detected the association in situ between NUMB-FLAG and GFP-EFA6B with a Duolink II Detection Kit (Olink Bioscience), according to the manufacturer’s instructions. Upon fixation in 4% PFA, cells were incubated for 10 min with blocking solution (PBS, 0.1% Tween 20, and 1% BSA). Primary antibodies against FLAG and GFP were incubated in the presence of blocking solution (PBS, 0.05% Tween 20, and 1% BSA). This was followed by incubation with secondary antibodies conjugated to oligonucleotides that are ligated to form a closed circle in the presence of Duolink Ligation Solution (Söderberg et al., 2006). In the final step, we added DNA polymerase in order to amplify ligated oligos, which were detected using complementary, fluorescently labeled oligonucleotides.
Biochemical procedures
Coimmunoprecipitation assay
Lysates prepared in JS buffer (50 mM Hepes, pH 7.5, 50 mM NaCl, 1% glycerol, 1% Triton X-100, 1.5 mM MgCl2, and 5 mM EGTA) were incubated in the presence of the anti-FLAG M2 affinity gel (a purified murine IgG1 monoclonal antibody covalently attached to agarose; Sigma) for two cycles of 1 h each at 4°C with rocking.
Immunoprecipitates were then washed three times in JS buffer. After washing, beads were resuspended in a 1:1 volume of 2× SDS-PAGE sample buffer, boiled for 10 min at 95°C, centrifuged for 1 min, and then loaded onto polyacrylamide gels.
Pull-down assay
Lysates prepared in JS buffer were incubated in the presence of 0.5 µM GST-tagged purified proteins immobilized on beads for 1 h at 4°C under mild agitation. Beads were then washed three times in JS buffer. After washing, beads were resuspended in a 1:1 volume of 2× SDS-PAGE sample buffer, boiled for 10 min at 95°C, centrifuged for 1 min, and then loaded onto polyacrylamide gels.
In vitro binding assay
0.5 µM purified proteins immobilized on beads were incubated with the indicated amount of purified protein in JS buffer for 1 h at 4°C under mild agitation. Beads were then washed three times in JS buffer. After washing, beads were resuspended in a 1:1 volume of 2× SDS-PAGE sample buffer, boiled for 10 min at 95°C, centrifuged for 1 min, and then loaded onto polyacrylamide gels.
Production of proteins
All the GST and His fusion proteins used were produced in bacteria using Escherichia coli BL21 Rosetta (DE3) competent cells transformed with the pGEX6P1 or pET30 vector in which the desired construct had been cloned.
E. coli BL21 Rosetta (DE3) cells picked from individual colonies and transformed with the indicated GST-fusion or His-fusion constructs were used to inoculate 200 ml of lysogeny broth (LB) medium (containing ampicillin at 50 μg/ml) and were grown overnight at 37°C. Between 10 and 100 ml of the overnight culture was diluted in 1 liter of LB and was grown at 37°C (240 rpm, shaking) until it reached approximately OD = 0.4–0.6. Then, IPTG was used to induce the protein production using different conditions depending on the protein (Table S5). After this, the induction cells were pelleted down at 6,000 rpm for 15 min at 4°C, and pellets were used immediately or conserved at −80°C after washing in PBS once.
Pellets were suspended in GST-lysis buffer (2× TBS, 0.5 mM EDTA, 10% glycerol, protease inhibitor cocktail [Roche; freshly added], and 1 mM DTT [freshly added]) or His-lysis buffer (2× TBS, 2 mM β-mercaptoethanol, 10% glycerol, protease inhibitor cocktail [Roche; freshly added], and 20 mM imidazole) based on the tag (15 ml for 1-liter culture). Samples were sonicated on ice three times for 30 s each and were pelleted down at 13,200 rpm for 30 min at 4°C using a JA-20 Beckman rotor or at 40,000 rpm for 45 min at 4°C using a 55.2 Ti Beckman rotor. 1 ml of glutathione-Sepharose beads (Amersham Bioscience) or 5 ml of Ni-NTA (Qiagen), previously washed three times with GST-lysis buffer or His-lysis buffer, respectively, was added to the supernatant, and samples were incubated for 1–2 h at 4°C while rocking. Beads were then washed three times (with 5 min of incubation at 4°C each) in the GST- or His-lysis solution. GST proteins were resuspended in 50% slurry in the GST-lysis solution. The quantification was achieved in an SDS-PAGE gel using a titration curve with BSA.
Elution of His-tagged NUMB protein
Beads were washed once in the washing solution (1× TBS, 50 mM imidazole, and 10% glycerol) and then subjected to six cycles of elution in the elution buffer (1× TBS, 200 mM imidazole, and 10% glycerol) in Poly-Prep Chromatography columns (Bio-Rad). Eluted proteins were collected in clean tubes containing 1 mM EDTA and 1 mM DTT. The best fractions identified in SDS-PAGE gel were collected together and dialyzed overnight in dialysis buffer (20 mM Tris, pH 7.1, 10% glycerol, 0.5 mM EDTA, 1 mM DTT, and 160 mM NaCl). After dialysis, precipitates were removed by centrifugation. The supernatant was diluted to finally have 40 mM NaCl, and was purified through ion exchange chromatography using a Resource Q column (GE Healthcare Life Science) on an AKTA purifier system (Amersham Bioscience). The fractions in the area under the curve were collected together, concentrated with Vivaspin concentrators (Vivaspin 20 MWCO 100000; Sartorius Stedim Biotech), flash frozen using liquid nitrogen, and stored at −80°C. The storage buffer used was 20 mM Tris pH 7.1, 10% glycerol, 0.5 mM EDTA, 1 mM DTT, and 160 mM NaCl.
Elution of His-tagged NUMB PTB protein
Beads were washed once in the washing solution (1× TBS and 50 mM imidazole) and then subjected to six cycles of elution in the elution buffer (1× TBS and 200 mM imidazole) in Poly-Prep Chromatography columns (Bio-Rad). Eluted proteins were collected in clean tubes containing 1 mM EDTA and 1 mM DTT. The best fractions identified in SDS-PAGE gel were collected together and dialyzed overnight in dialysis buffer (1× TBS, 0.5 mM EDTA, and 1 mM DTT). After dialysis, precipitates were removed by centrifugation, and the supernatant was concentrated in Vivaspin concentrators (Vivaspin 20 MWCO 3000; GE Healthcare Life Sciences), purified through gel filtration using a Superdex 200 10/30 column (GE Healthcare Life Sciences), and equilibrated with storage buffer on an AKTA purifier system (Amersham Bioscience). Proteins were loaded in a 500-μl volume using a Hamilton syringe. Typical flow rate was 0.5 ml/min. Finally, the fractions in the area under the curve were collected together, concentrated with Vivaspin concentrators, flash frozen using liquid nitrogen, and stored at −80°C. The storage buffer used was 20 mM Tris, pH 7.6, 5% glycerol, 10 mM NaCl, 0.5 mM EDTA, and 1 mM DTT.
MBP-EFA6B protein production
The MBP-EFA6B protein used was produced in a baculovirus system. EFA6B was cloned in pBAC_His-MBP-TEV vector.
Transposition
Transposition was made using DH10MultiBac-YFP electrocompetent cells (a gift from A. Musacchio, Max Planck Institute of Molecular Physiology, Dortmund, Germany) that were electroporated in the presence of 1 μl of DNA from average-yield miniprep.
Selection of the recombinant bacteria was made after plating on LB plates containing 50 μg/ml kanamycin, 7 μg/ml gentamycin, 10 μg/ml tetracycline, 200 μg/ml X-gal, and 40 µg/ml IPTG for at least 48 h.
White recombinant proteins were selected, and bacmid DNA was isolated using the Promega Wizard Plus SV Minipreps kit with a slight protocol modification that included precipitation with isopropanol instead of elution from the column kit.
Virus production
DNA was used to transfect Sf21 insect cells (Invitrogen) in order to produce the virus. Sf21 cells were plated at 0.5 × 106/ml in a 6-well plate and transfected using Insectogene reagent (Biontex Laboratories) according to the manufacturer’s instructions. Cell transfection was monitored by looking at the appearance of YFP-positive cells. After 72 h, the supernatant was collected (V0, primary stock virus production). The virus was amplified in H5 cells (Invitrogen). 25 ml of H5 cells at 0.5 × 106/ml was infected using 1.5 ml of the V0 virus. Cell infection was monitored by looking at the appearance of YFP-positive cells and at the cell growth arrest caused by the viral infection. After 72 h from the cell growth arrest, the supernatant was collected (V1 generation). The virus was amplified a second time using 200 ml of H5 cells infected with 2.5 ml of the V1 virus, following the same protocol.
Proteins were finally produced from 500 ml of H5 cells seeded at a concentration of 0.5 × 106 cells/ml and infected with 12.5 ml of V2 virus.
For protein purification, the cells were harvested 3 d after infection. After 15 min of centrifugation at 1,200 rpm, pellets were suspended in lysis buffer (2× TBS, 0.5 mM EDTA, 10% glycerol, protease inhibitor cocktail [Roche; freshly added], and 1 mM DTT [freshly added]; 25 ml for 1-liter culture) or conserved at −80°C after washing in PBS once. Samples were sonicated on ice three times for 30 s each and were pelleted down at 45,000 rpm for 1 h at 4°C using a 55.2 Ti Beckman rotor. 1 ml of amylose beads (NEB), previously washed three times with lysis buffer, was added to the supernatant, and samples were incubated for 1–2 h at 4°C while rocking. Beads were then washed three times (with 5 min of incubation at 4°C each) with lysis solution.
MBP-EFA6B protein elution
Beads were washed once in the lysis solution and then subjected to six cycles of elution in the elution buffer (2× TBS, 10% glycerol, 10 mM amylose, 0.5 mM EDTA, and 1 mM DTT, freshly added) in Poly-Prep Chromatography columns (Bio-Rad). Eluted proteins were collected in clean tubes. The best fractions identified in SDS-PAGE gel were collected together and dialyzed overnight in dialysis buffer (1× TBS, 10% glycerol, 0.5 mM EDTA, and 1 mM DTT). After dialysis, precipitates were removed by centrifugation, and the supernatant was concentrated in Vivaspin, flash frozen using liquid nitrogen, and stored at –80°C.
Statistical analysis
In the CDR formation assay, 20 fields were analyzed for each sample, and the percentage of cells forming CDRs (number of cells with CDRs/total number of cells in the field) was scored. In case of reinfection with GFP constructs, only GFP-positive cells were considered for the analysis. All data are presented as the mean ± SEM from at least three independent experiments. A Student’s t test was used to calculate the P values. *, P < 0.05; **, P < 0.01.
Online supplemental material
Table S1 contains a summary of the effects on CDR of oligos silencing a set of selected endocytic regulators. Tables S2–S5 contain details of the primers, ID assays, and microscope acquisition settings used. Fig. S1 shows that loss of NUMB has no or marginal effect on EGFR and TfR trafficking. Fig. S2 shows that NUMB interacts with EFA6B, but not with other ARF6 GEFs, including EFA6A, ARNO, and BRAG2. Fig. S3 shows additional experiments characterizing the interaction between NUMB and EFA6B. Fig. S4 shows that a mutant of NUMB devoid of the PTB domain is no longer able to restore CDR phenotypes in NUMB-silenced cells. Fig. S5 shows the localization of EFA6B on ARF6-induced enlarged endosomes. Fig. S6 contains all uncropped gels.
Supplementary Material
Acknowledgments
We thank Sebastiano Pasqualato and Silvia Monzani of the Crystallization Facility of European Institute of Oncology for technical assistance with the protein purification.
This work has been supported by grants from the Associazione Italiana per la Ricerca sul Cancro (AIRC) to G. Scita (IG-18621), P.P. Di Fiore (IG-18988 and MCO 10.000), and S. Pece (IG-11904); the Italian Ministry of University and Scientific Research to P.P. Di Fiore and S. Pece; the International Association for Cancer Research (AICR-14-0335) to G. Scita; the European Research Council (Advanced-ERC#268836) to G. Scita; and the Italian Ministry of Health (RF-2013-02358446) to G. Scita. S. Bisi was supported by an AIRC fellowship.
The authors declare no competing financial interests.
Author contributions: M. Zobel and A. Disanza performed, analyzed, and wrote the results; F. Senic-Matuglia, I.N. Colaluca, S. Bisi, E. Barbieri, G. Caldieri, and S. Sigismund performed some of the experiments in cell biology and biochemistry and analyzed data; M. Franco performed the biochemical enzymatic assays; S. Confalonieri provided all bioinformatics and statistical support; S. Pece and P. Chavrier provided key reagents; and P.P. Di Fiore and G. Scita conceived the work and wrote the manuscript.
References
- Allaire P.D., Seyed Sadr M., Chaineau M., Seyed Sadr E., Konefal S., Fotouhi M., Maret D., Ritter B., Del Maestro R.F., and McPherson P.S.. 2013. Interplay between Rab35 and Arf6 controls cargo recycling to coordinate cell adhesion and migration. J. Cell Sci. 126:722–731. 10.1242/jcs.112375 [DOI] [PubMed] [Google Scholar]
- Bernitt E., Koh C.G., Gov N., and Döbereiner H.G.. 2015. Dynamics of actin waves on patterned substrates: a quantitative analysis of circular dorsal ruffles. PLoS One. 10:e0115857 10.1371/journal.pone.0115857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernitt E., Döbereiner H.G., Gov N.S., and Yochelis A.. 2017. Fronts and waves of actin polymerization in a bistability-based mechanism of circular dorsal ruffles. Nat. Commun. 8:15863 10.1038/ncomms15863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanović O., Delfino-Machín M., Nicolás-Pérez M., Gavilán M.P., Gago-Rodrigues I., Fernández-Miñán A., Lillo C., Ríos R.M., Wittbrodt J., and Martínez-Morales J.R.. 2012. Numb/Numbl-Opo antagonism controls retinal epithelium morphogenesis by regulating integrin endocytosis. Dev. Cell. 23:782–795. 10.1016/j.devcel.2012.09.004 [DOI] [PubMed] [Google Scholar]
- Bouchet J., Del Río-Iñiguez I., Lasserre R., Agüera-Gonzalez S., Cuche C., Danckaert A., McCaffrey M.W., Di Bartolo V., and Alcover A.. 2016. Rac1-Rab11-FIP3 regulatory hub coordinates vesicle traffic with actin remodeling and T-cell activation. EMBO J. 35:1160–1174. 10.15252/embj.201593274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buccione R., Orth J.D., and McNiven M.A.. 2004. Foot and mouth: podosomes, invadopodia and circular dorsal ruffles. Nat. Rev. Mol. Cell Biol. 5:647–657. 10.1038/nrm1436 [DOI] [PubMed] [Google Scholar]
- Casanova J.E. 2007. Regulation of Arf activation: the Sec7 family of guanine nucleotide exchange factors. Traffic. 8:1476–1485. 10.1111/j.1600-0854.2007.00634.x [DOI] [PubMed] [Google Scholar]
- Colaluca I.N., Tosoni D., Nuciforo P., Senic-Matuglia F., Galimberti V., Viale G., Pece S., and Di Fiore P.P.. 2008. NUMB controls p53 tumour suppressor activity. Nature. 451:76–80. 10.1038/nature06412 [DOI] [PubMed] [Google Scholar]
- Colaluca I.N., Basile A., Freiburger L., D’Uva V., Disalvatore D., Vecchi M., Confalonieri S., Tosoni D., Cecatiello V., Malabarba M.G., et al. . 2018. A Numb-Mdm2 fuzzy complex reveals an isoform-specific involvement of Numb in breast cancer. J. Cell Biol. 217:745–762. 10.1083/jcb.201709092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commisso C., Davidson S.M., Soydaner-Azeloglu R.G., Parker S.J., Kamphorst J.J., Hackett S., Grabocka E., Nofal M., Drebin J.A., Thompson C.B., et al. . 2013. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 497:633–637. 10.1038/nature12138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corallino S., Malinverno C., Neumann B., Tischer C., Palamidessi A., Frittoli E., Panagiotakopoulou M., Disanza A., Malet-Engra G., Nastaly P., et al. . 2018. A RAB35-p85/PI3K axis controls oscillatory apical protrusions required for efficient chemotactic migration. Nat. Commun. 9:1475 10.1038/s41467-018-03571-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton M., Benhra N., and Le Borgne R.. 2013. Numb inhibits the recycling of Sanpodo in Drosophila sensory organ precursor. Curr. Biol. 23:581–587. 10.1016/j.cub.2013.02.020 [DOI] [PubMed] [Google Scholar]
- Couturier L., Mazouni K., and Schweisguth F.. 2013. Numb localizes at endosomes and controls the endosomal sorting of notch after asymmetric division in Drosophila. Curr. Biol. 23:588–593. 10.1016/j.cub.2013.03.002 [DOI] [PubMed] [Google Scholar]
- Dai J., Li J., Bos E., Porcionatto M., Premont R.T., Bourgoin S., Peters P.J., and Hsu V.W.. 2004. ACAP1 promotes endocytic recycling by recognizing recycling sorting signals. Dev. Cell. 7:771–776. 10.1016/j.devcel.2004.10.002 [DOI] [PubMed] [Google Scholar]
- Derrien V., Couillault C., Franco M., Martineau S., Montcourrier P., Houlgatte R., and Chavrier P.. 2002. A conserved C-terminal domain of EFA6-family ARF6-guanine nucleotide exchange factors induces lengthening of microvilli-like membrane protrusions. J. Cell Sci. 115:2867–2879. [DOI] [PubMed] [Google Scholar]
- Dho S.E., French M.B., Woods S.A., and McGlade C.J.. 1999. Characterization of four mammalian numb protein isoforms. Identification of cytoplasmic and membrane-associated variants of the phosphotyrosine binding domain. J. Biol. Chem. 274:33097–33104. 10.1074/jbc.274.46.33097 [DOI] [PubMed] [Google Scholar]
- Dho S.E., Trejo J., Siderovski D.P., and McGlade C.J.. 2006. Dynamic regulation of mammalian numb by G protein-coupled receptors and protein kinase C activation: Structural determinants of numb association with the cortical membrane. Mol. Biol. Cell. 17:4142–4155. 10.1091/mbc.e06-02-0097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Marcotullio L., Ferretti E., Greco A., De Smaele E., Po A., Sico M.A., Alimandi M., Giannini G., Maroder M., Screpanti I., and Gulino A.. 2006. Numb is a suppressor of Hedgehog signalling and targets Gli1 for Itch-dependent ubiquitination. Nat. Cell Biol. 8:1415–1423. 10.1038/ncb1510 [DOI] [PubMed] [Google Scholar]
- Donaldson J.G., and Jackson C.L.. 2011. ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat. Rev. Mol. Cell Biol. 12:362–375. 10.1038/nrm3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson J.G., Porat-Shliom N., and Cohen L.A.. 2009. Clathrin-independent endocytosis: a unique platform for cell signaling and PM remodeling. Cell. Signal. 21:1–6. 10.1016/j.cellsig.2008.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta D., and Donaldson J.G.. 2015. Sorting of Clathrin-Independent Cargo Proteins Depends on Rab35 Delivered by Clathrin-Mediated Endocytosis. Traffic. 16:994–1009. 10.1111/tra.12302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco M., Peters P.J., Boretto J., van Donselaar E., Neri A., D’Souza-Schorey C., and Chavrier P.. 1999. EFA6, a sec7 domain-containing exchange factor for ARF6, coordinates membrane recycling and actin cytoskeleton organization. EMBO J. 18:1480–1491. 10.1093/emboj/18.6.1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frittoli E., Palamidessi A., Marighetti P., Confalonieri S., Bianchi F., Malinverno C., Mazzarol G., Viale G., Martin-Padura I., Garré M., et al. . 2014. A RAB5/RAB4 recycling circuitry induces a proteolytic invasive program and promotes tumor dissemination. J. Cell Biol. 206:307–328. 10.1083/jcb.201403127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham A.K., and Munro S.. 2007. The small G proteins of the Arf family and their regulators. Annu. Rev. Cell Dev. Biol. 23:579–611. 10.1146/annurev.cellbio.23.090506.123209 [DOI] [PubMed] [Google Scholar]
- Gold K., Cotton J.A., and Stollewerk A.. 2009. The role of Notch signalling and numb function in mechanosensory organ formation in the spider Cupiennius salei. Dev. Biol. 327:121–131. 10.1016/j.ydbio.2008.12.004 [DOI] [PubMed] [Google Scholar]
- Gu Z., Noss E.H., Hsu V.W., and Brenner M.B.. 2011. Integrins traffic rapidly via circular dorsal ruffles and macropinocytosis during stimulated cell migration. J. Cell Biol. 193:61–70. 10.1083/jcb.201007003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson Y.C., Chen Y., Frederick M.J., Lai S.Y., and Clayman G.L.. 2010. MEK inhibitor PD0325901 significantly reduces the growth of papillary thyroid carcinoma cells in vitro and in vivo. Mol. Cancer Ther. 9:1968–1976. 10.1158/1535-7163.MCT-10-0062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai M., Arita Y., McGlade C.J., Lee K.F., Chen J., and Evans S.M.. 2017. Adaptor proteins NUMB and NUMBL promote cell cycle withdrawal by targeting ERBB2 for degradation. J. Clin. Invest. 127:569–582. 10.1172/JCI91081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J., Liu Z., Zhu H., Zhang X., Liang Y., Yao S., Wang F., Xie X., Zhang B., Tan T., et al. . 2014. The tumor suppressive role of NUMB isoform 1 in esophageal squamous cell carcinoma. Oncotarget. 5:5602–5614. 10.18632/oncotarget.2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hongu T., and Kanaho Y.. 2014. Activation machinery of the small GTPase Arf6. Adv. Biol. Regul. 54:59–66. 10.1016/j.jbior.2013.09.014 [DOI] [PubMed] [Google Scholar]
- Hoon J.L., Wong W.K., and Koh C.G.. 2012. Functions and regulation of circular dorsal ruffles. Mol. Cell. Biol. 32:4246–4257. 10.1128/MCB.00551-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson T.R., Brown F.D., Nie Z., Miura K., Foroni L., Sun J., Hsu V.W., Donaldson J.G., and Randazzo P.A.. 2000. ACAPs are arf6 GTPase-activating proteins that function in the cell periphery. J. Cell Biol. 151:627–638. 10.1083/jcb.151.3.627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.M., Roy S.G., Chen B., Nguyen T.M., McMonigle R.J., McCracken A.N., Zhang Y., Kofuji S., Hou J., Selwan E., et al. . 2016. Targeting cancer metabolism by simultaneously disrupting parallel nutrient access pathways. J. Clin. Invest. 126:4088–4102. 10.1172/JCI87148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H., Etoh K., Marubashi S., Ohbayashi N., and Fukuda M.. 2015. Measurement of Rab35 activity with the GTP-Rab35 trapper RBD35. Methods Mol. Biol. 1298:207–216. 10.1007/978-1-4939-2569-8_18 [DOI] [PubMed] [Google Scholar]
- Kunita R., Otomo A., Mizumura H., Suzuki-Utsunomiya K., Hadano S., and Ikeda J.E.. 2007. The Rab5 activator ALS2/alsin acts as a novel Rac1 effector through Rac1-activated endocytosis. J. Biol. Chem. 282:16599–16611. 10.1074/jbc.M610682200 [DOI] [PubMed] [Google Scholar]
- Lanzetti L., Palamidessi A., Areces L., Scita G., and Di Fiore P.P.. 2004. Rab5 is a signalling GTPase involved in actin remodelling by receptor tyrosine kinases. Nature. 429:309–314. 10.1038/nature02542 [DOI] [PubMed] [Google Scholar]
- Lau K.M., and McGlade C.J.. 2011. Numb is a negative regulator of HGF dependent cell scattering and Rac1 activation. Exp. Cell Res. 317:539–551. 10.1016/j.yexcr.2010.12.005 [DOI] [PubMed] [Google Scholar]
- Loskutov Y.V., Kozyulina P.Y., Kozyreva V.K., Ice R.J., Jones B.C., Roston T.J., Smolkin M.B., Ivanov A.V., Wysolmerski R.B., and Pugacheva E.N.. 2015. NEDD9/Arf6-dependent endocytic trafficking of matrix metalloproteinase 14: a novel mechanism for blocking mesenchymal cell invasion and metastasis of breast cancer. Oncogene. 34:3662–3675. 10.1038/onc.2014.297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiorano E., Favia G., Pece S., Resta L., Maisonneuve P., Di Fiore P.P., Capodiferro S., Urbani U., and Viale G.. 2007. Prognostic implications of NUMB immunoreactivity in salivary gland carcinomas. Int. J. Immunopathol. Pharmacol. 20:779–789. 10.1177/039463200702000414 [DOI] [PubMed] [Google Scholar]
- McGill M.A., Dho S.E., Weinmaster G., and McGlade C.J.. 2009. Numb regulates post-endocytic trafficking and degradation of notch1. J. Biol. Chem. 284:26427–26438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica-Vázquez L.H., Benetah M.H., Baanannou A., Bernat-Fabre S., Deplancke B., Cribbs D.L., Bourbon H.M., and Boube M.. 2017. Tissue-specific enhancer repression through molecular integration of cell signaling inputs. PLoS Genet. 13:e1006718 10.1371/journal.pgen.1006718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson L., Conradt B., Ruaud A.F., Chen C.C., Hatzold J., Bessereau J.L., Grant B.D., and Tuck S.. 2008. Caenorhabditis elegans num-1 negatively regulates endocytic recycling. Genetics. 179:375–387. 10.1534/genetics.108.087247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson L., Jonsson E., and Tuck S.. 2011. Caenorhabditis elegans numb inhibits endocytic recycling by binding TAT-1 aminophospholipid translocase. Traffic. 12:1839–1849. 10.1111/j.1600-0854.2011.01271.x [DOI] [PubMed] [Google Scholar]
- Nishimura T., and Kaibuchi K.. 2007. Numb controls integrin endocytosis for directional cell migration with aPKC and PAR-3. Dev. Cell. 13:15–28. 10.1016/j.devcel.2007.05.003 [DOI] [PubMed] [Google Scholar]
- Orth J.D., Krueger E.W., Weller S.G., and McNiven M.A.. 2006. A novel endocytic mechanism of epidermal growth factor receptor sequestration and internalization. Cancer Res. 66:3603–3610. 10.1158/0008-5472.CAN-05-2916 [DOI] [PubMed] [Google Scholar]
- Padovani D., Folly-Klan M., Labarde A., Boulakirba S., Campanacci V., Franco M., Zeghouf M., and Cherfils J.. 2014. EFA6 controls Arf1 and Arf6 activation through a negative feedback loop. Proc. Natl. Acad. Sci. USA. 111:12378–12383. 10.1073/pnas.1409832111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palamidessi A., Frittoli E., Garré M., Faretta M., Mione M., Testa I., Diaspro A., Lanzetti L., Scita G., and Di Fiore P.P.. 2008. Endocytic trafficking of Rac is required for the spatial restriction of signaling in cell migration. Cell. 134:135–147. 10.1016/j.cell.2008.05.034 [DOI] [PubMed] [Google Scholar]
- Palamidessi A., Testa I., Frittoli E., Barozzi S., Garrè M., Mazza D., Di Fiore P.P., Diaspro A., Scita G., and Faretta M.. 2010. Understanding biological dynamics: following cells and molecules to track functions and mechanisms. Eur. Biophys. J. 39:947–957. 10.1007/s00249-009-0461-x [DOI] [PubMed] [Google Scholar]
- Palm W., Park Y., Wright K., Pavlova N.N., Tuveson D.A., and Thompson C.B.. 2015. The Utilization of Extracellular Proteins as Nutrients Is Suppressed by mTORC1. Cell. 162:259–270. 10.1016/j.cell.2015.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W., Araki J., King B., DeMatteo R.G., and Thompson C.B.. 2017. Critical role for PI3-kinase in regulating the use of proteins as an amino acid source. Proc. Natl. Acad. Sci. USA. 114:E8628–E8636. 10.1073/pnas.1712726114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pece S., Serresi M., Santolini E., Capra M., Hulleman E., Galimberti V., Zurrida S., Maisonneuve P., Viale G., and Di Fiore P.P.. 2004. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J. Cell Biol. 167:215–221. 10.1083/jcb.200406140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pece S., Confalonieri S., R Romano P., and Di Fiore P.P.. 2011. NUMB-ing down cancer by more than just a NOTCH. Biochim. Biophys. Acta. 1815:26–43. [DOI] [PubMed] [Google Scholar]
- Punnonen E.L., Ryhänen K., and Marjomäki V.S.. 1998. At reduced temperature, endocytic membrane traffic is blocked in multivesicular carrier endosomes in rat cardiac myocytes. Eur. J. Cell Biol. 75:344–352. 10.1016/S0171-9335(98)80067-8 [DOI] [PubMed] [Google Scholar]
- Qi S., Zhao X., Li M., Zhang X., Lu Z., Yang C., Zhang C., Zhang H., and Zhang N.. 2015. Aberrant expression of Notch1/numb/snail signaling, an epithelial mesenchymal transition related pathway, in adenomyosis. Reprod. Biol. Endocrinol. 13:96 10.1186/s12958-015-0084-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H., Percival-Smith A., Li C., Jia C.Y., Gloor G., and Li S.S.. 2004. A novel transmembrane protein recruits numb to the plasma membrane during asymmetric cell division. J. Biol. Chem. 279:11304–11312. 10.1074/jbc.M311733200 [DOI] [PubMed] [Google Scholar]
- Radhakrishna H., and Donaldson J.G.. 1997. ADP-ribosylation factor 6 regulates a novel plasma membrane recycling pathway. J. Cell Biol. 139:49–61. 10.1083/jcb.139.1.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley A.J. 2011. Life at the leading edge. Cell. 145:1012–1022. 10.1016/j.cell.2011.06.010 [DOI] [PubMed] [Google Scholar]
- Santolini E., Puri C., Salcini A.E., Gagliani M.C., Pelicci P.G., Tacchetti C., and Di Fiore P.P.. 2000. Numb is an endocytic protein. J. Cell Biol. 151:1345–1352. 10.1083/jcb.151.6.1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer J.K., Sedgwick A.E., and D’Souza-Schorey C.. 2011. ARF6-mediated endocytic recycling impacts cell movement, cell division and lipid homeostasis. Semin. Cell Dev. Biol. 22:39–47. 10.1016/j.semcdb.2010.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scita G., Nordstrom J., Carbone R., Tenca P., Giardina G., Gutkind S., Bjarnegård M., Betsholtz C., and Di Fiore P.P.. 1999. EPS8 and E3B1 transduce signals from Ras to Rac. Nature. 401:290–293. 10.1038/45822 [DOI] [PubMed] [Google Scholar]
- Sero J.E., German A.E., Mammoto A., and Ingber D.E.. 2012. Paxillin controls directional cell motility in response to physical cues. Cell Adhes. Migr. 6:502–508. 10.4161/cam.21672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng W., Dong M., Chen C., Wang Z., Li Y., Wang K., Li Y., and Zhou J.. 2017. Cooperation of Musashi-2, Numb, MDM2, and P53 in drug resistance and malignant biology of pancreatic cancer. FASEB J. 31:2429–2438. 10.1096/fj.201601240R [DOI] [PubMed] [Google Scholar]
- Sigismund S., Argenzio E., Tosoni D., Cavallaro E., Polo S., and Di Fiore P.P.. 2008. Clathrin-mediated internalization is essential for sustained EGFR signaling but dispensable for degradation. Dev. Cell. 15:209–219. 10.1016/j.devcel.2008.06.012 [DOI] [PubMed] [Google Scholar]
- Smith C.A., Dho S.E., Donaldson J., Tepass U., and McGlade C.J.. 2004. The cell fate determinant numb interacts with EHD/Rme-1 family proteins and has a role in endocytic recycling. Mol. Biol. Cell. 15:3698–3708. 10.1091/mbc.e04-01-0026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderberg O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K.J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L.G., and Landegren U.. 2006. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods. 3:995–1000. 10.1038/nmeth947 [DOI] [PubMed] [Google Scholar]
- Spaargaren M., and Bos J.L.. 1999. Rab5 induces Rac-independent lamellipodia formation and cell migration. Mol. Biol. Cell. 10:3239–3250. 10.1091/mbc.10.10.3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théard D., Labarrade F., Partisani M., Milanini J., Sakagami H., Fon E.A., Wood S.A., Franco M., and Luton F.. 2010. USP9x-mediated deubiquitination of EFA6 regulates de novo tight junction assembly. EMBO J. 29:1499–1509. 10.1038/emboj.2010.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topp J.D., Gray N.W., Gerard R.D., and Horazdovsky B.F.. 2004. Alsin is a Rab5 and Rac1 guanine nucleotide exchange factor. J. Biol. Chem. 279:24612–24623. 10.1074/jbc.M313504200 [DOI] [PubMed] [Google Scholar]
- Tosoni D., Puri C., Confalonieri S., Salcini A.E., De Camilli P., Tacchetti C., and Di Fiore P.P.. 2005. TTP specifically regulates the internalization of the transferrin receptor. Cell. 123:875–888. 10.1016/j.cell.2005.10.021 [DOI] [PubMed] [Google Scholar]
- Tosoni D., Zecchini S., Coazzoli M., Colaluca I., Mazzarol G., Rubio A., Caccia M., Villa E., Zilian O., Di Fiore P.P., and Pece S.. 2015. The Numb/p53 circuitry couples replicative self-renewal and tumor suppression in mammary epithelial cells. J. Cell Biol. 211:845–862. 10.1083/jcb.201505037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosoni D., Pambianco S., Ekalle Soppo B., Zecchini S., Bertalot G., Pruneri G., Viale G., Di Fiore P.P., and Pece S.. 2017. Pre-clinical validation of a selective anti-cancer stem cell therapy for Numb-deficient human breast cancers. EMBO Mol. Med. 9:655–671. 10.15252/emmm.201606940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T., Shepherd S., Ackerman L., Jan L.Y., and Jan Y.N.. 1989. numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell. 58:349–360. 10.1016/0092-8674(89)90849-0 [DOI] [PubMed] [Google Scholar]
- Verdi J.M., Bashirullah A., Goldhawk D.E., Kubu C.J., Jamali M., Meakin S.O., and Lipshitz H.D.. 1999. Distinct human NUMB isoforms regulate differentiation vs. proliferation in the neuronal lineage. Proc. Natl. Acad. Sci. USA. 96:10472–10476. 10.1073/pnas.96.18.10472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viaud J., Lagarrigue F., Ramel D., Allart S., Chicanne G., Ceccato L., Courilleau D., Xuereb J.M., Pertz O., Payrastre B., and Gaits-Iacovoni F.. 2014. Phosphatidylinositol 5-phosphate regulates invasion through binding and activation of Tiam1. Nat. Commun. 5:4080 10.1038/ncomms5080 [DOI] [PubMed] [Google Scholar]
- Wang Z., and Li S.S.. 2010. Numb: A new player in EMT. Cell Adhes. Migr. 4:176–179. 10.4161/cam.4.2.10690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Sandiford S., Wu C., and Li S.S.. 2009. Numb regulates cell-cell adhesion and polarity in response to tyrosine kinase signalling. EMBO J. 28:2360–2373. 10.1038/emboj.2009.190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J., Fu Z.Y., Li P.S., Miao H.H., Li B.L., Ma Y.T., and Song B.L.. 2014. The clathrin adaptor proteins ARH, Dab2, and numb play distinct roles in Niemann-Pick C1-Like 1 versus low density lipoprotein receptor-mediated cholesterol uptake. J. Biol. Chem. 289:33689–33700. 10.1074/jbc.M114.593764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhoff B., Colaluca I.N., D’Ario G., Donzelli M., Tosoni D., Volorio S., Pelosi G., Spaggiari L., Mazzarol G., Viale G., et al. . 2009. Alterations of the Notch pathway in lung cancer. Proc. Natl. Acad. Sci. USA. 106:22293–22298. 10.1073/pnas.0907781106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap T.A., Yan L., Patnaik A., Fearen I., Olmos D., Papadopoulos K., Baird R.D., Delgado L., Taylor A., Lupinacci L., et al. . 2011. First-in-man clinical trial of the oral pan-AKT inhibitor MK-2206 in patients with advanced solid tumors. J. Clin. Oncol. 29:4688–4695. 10.1200/JCO.2011.35.5263 [DOI] [PubMed] [Google Scholar]
- Zangari J., Partisani M., Bertucci F., Milanini J., Bidaut G., Berruyer-Pouyet C., Finetti P., Long E., Brau F., Cabaud O., et al. . 2014. EFA6B antagonizes breast cancer. Cancer Res. 74:5493–5506. 10.1158/0008-5472.CAN-14-0298 [DOI] [PubMed] [Google Scholar]
- Zeng Y., Lai T., Koh C.G., LeDuc P.R., and Chiam K.H.. 2011. Investigating circular dorsal ruffles through varying substrate stiffness and mathematical modeling. Biophys. J. 101:2122–2130. 10.1016/j.bpj.2011.09.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Li F., Song Y., Sheng X., Ren F., Xiong K., Chen L., Zhang H., Liu D., Lengner C.J., et al. . 2016. Numb and Numbl act to determine mammary myoepithelial cell fate, maintain epithelial identity, and support lactogenesis. FASEB J. 30:3474–3488. 10.1096/fj.201600387R [DOI] [PubMed] [Google Scholar]
- Zhong W., Jiang M.M., Weinmaster G., Jan L.Y., and Jan Y.N.. 1997. Differential expression of mammalian Numb, Numblike and Notch1 suggests distinct roles during mouse cortical neurogenesis. Development. 124:1887–1897. [DOI] [PubMed] [Google Scholar]
- Zwahlen C., Li S.C., Kay L.E., Pawson T., and Forman-Kay J.D.. 2000. Multiple modes of peptide recognition by the PTB domain of the cell fate determinant Numb. EMBO J. 19:1505–1515. 10.1093/emboj/19.7.1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.