Cheng et al. combine quantitative immunoelectron microscopy and light imaging microscopy to comprehensively analyze LAMP1 subcellular distribution in neurons. The data show that a significant portion of LAMP1-positive organelles are not degradative, illustrating the need for additional lysosomal and endocytic markers to define endolysosomal compartments in the nervous system.
Abstract
Despite widespread distribution of LAMP1 and the heterogeneous nature of LAMP1-labeled compartments, LAMP1 is routinely used as a lysosomal marker, and LAMP1-positive organelles are often referred to as lysosomes. In this study, we use immunoelectron microscopy and confocal imaging to provide quantitative analysis of LAMP1 distribution in various autophagic and endolysosomal organelles in neurons. Our study demonstrates that a significant portion of LAMP1-labeled organelles do not contain detectable lysosomal hydrolases including cathepsins D and B and glucocerebrosidase. A bovine serum albumin–gold pulse–chase assay followed by ultrastructural analysis suggests a heterogeneity of degradative capacity in LAMP1-labeled endolysosomal organelles. Gradient fractionation displays differential distribution patterns of LAMP1/2 and cathepsins D/B in neurons. We further reveal that LAMP1 intensity in familial amyotrophic lateral sclerosis–linked motor neurons does not necessarily reflect lysosomal deficits in vivo. Our study suggests that labeling a set of lysosomal hydrolases combined with various endolysosomal markers would be more accurate than simply relying on LAMP1/2 staining to assess neuronal lysosome distribution, trafficking, and functionality under physiological and pathological conditions.
Introduction
Lysosomes serve as the terminal degradation hubs for autophagic/endocytic components, thus maintaining cellular homeostasis essential for neuronal growth and survival as well as synaptic remodeling (Cai et al., 2010; Farías et al., 2017; Padamsey et al., 2017). Lysosomal hydrolases degrade extracellular materials that have been internalized by endocytosis and intracellular components sequestered by autophagy. In addition, lysosomes play a critical role in nutrient sensing and recycling of the catabolic pathway (Luzio et al., 2007). Endolysosomal trafficking from early endosomes (EEs) to late endosomes (LEs) and finally into mature lysosomes is essential for delivering target materials and maintaining efficient degradation capacity. Newly synthesized precursors of lysosomal hydrolases are transported from the TGN to endosomes and lysosomes (Geuze et al., 1985; Griffiths et al., 1988, 1990; Lobel et al., 1989; Ishidoh and Kominami, 2002). Autophagosomes undergo a stepwise maturation through fusion of autophagosomes with endosomes to form amphisomes or with lysosomes to form degradative autolysosomes (Klionsky and Emr, 2000; Levine and Klionsky, 2004; Maday et al., 2012, 2014; Nixon, 2013; Cheng et al., 2015). Before endocytosed/autophagic organelles mature into lysosomes, they are a continuum of intermediates that continually pass over endocytosed cargos, exchange membrane constituents, and add lysosomal hydrolases. These heterogeneous intermediates, having diverse qualities in morphology, membrane components, hydrolase contents, luminal pH, and distinct cellular functions, represent the different stages of maturation within the endolysosomal pathway (Saftig and Klumperman, 2009). A mature lysosome is thus defined as: (A) a storage organelle for active forms of degradative enzymes with acidic pH optimum; (B) the hydrolysis of substrates occurring within; (C) limiting membranes with specific glycosylated membrane-associated proteins such as LAMP1; and (D) the lack of nonlysosomal proteins such as mannose-6-phosphate receptors (MPRs; Saftig, 2005; Luzio et al., 2007).
Neurons are highly polarized and postmitotic cells with three functionally and structurally different compartments: soma, dendrites, and axons. These unique features add yet another layer of complexity to the distribution and maturation of lysosomes. To achieve maturation, autophagic and endocytic vesicles undergo long-distance transport from distal regions back to the soma, a distance that can span up to 1 m long in peripheral neurons (Overly and Hollenbeck, 1996). Impaired retrograde transport of endolysosomal organelles causes an accumulation of immature lysosomes in neurons, leading to dramatically decreased proteolytic capability (Cai et al., 2010; Lee et al., 2011; Gowrishankar et al., 2015, 2017). Emerging evidence demonstrates that defects in the endolysosomal system, which are often accompanied by impaired endolysosomal trafficking, are associated with the pathogenesis of several major neurodegenerative diseases such as Alzheimer’s disease (AD), Parkinson’s disease, Huntington’s disease, and amyotrophic lateral sclerosis (Levine and Kroemer, 2008; Nixon et al., 2008; Gowrishankar et al., 2015; Wong and Holzbaur, 2015; Xie et al., 2015). In addition, several hereditary lysosomal storage diseases affect the central nervous system (Ballabio and Gieselmann, 2009). With these clinical implications, there is growing interest in understanding the causal relationship between dysregulation of endolysosome systems and progression of neurological diseases. Thus, it is necessary to develop practical guidelines for assessing neuronal lysosome distribution and trafficking, by extension revealing the pathological response of lysosomes to disease conditions in both in vitro and in vivo nervous systems.
Lysosome-associated membrane proteins (LAMPs; mainly LAMP1/2) are type-1 transmembrane proteins with considerable sequence homology. Although LAMP1/2 target to lysosomes, they are not static components of the lysosomal membrane; rather, they are in dynamic equilibrium between lysosomes, endosomes, and the plasma membrane (Deng and Storrie, 1988; Patterson and Lippincott-Schwartz, 2002; Eskelinen et al., 2003). Newly synthesized LAMP1 exits the TGN and enters the plasma membrane and endolysosomal pathway (Cook et al., 2004), where it exists as the most abundant endolysosome membrane protein (Saftig and Klumperman, 2009). In addition, LAMP1 is also detected in AP1-clathrin–positive TGN membranes (Höning et al., 1996; Hunziker and Geuze, 1996; Janvier and Bonifacino, 2005). Recent studies in neurons also suggest that LAMP1-labeled organelles are heterogeneous (Gowrishankar et al., 2015, 2017; Goo et al., 2017) and represent intermediates of endocytic and autophagic pathways and lysosomal biogenesis (Hollenbeck, 1993; Lee et al., 2011; Maday et al., 2012, 2014; Cheng et al., 2015). Despite widespread distribution patterns of LAMP1 and the heterogeneous nature of LAMP1-labeled compartments, LAMP1 continues to be the most routinely used marker for neuronal lysosomes, and many studies refer to LAMP1-positive organelles as degradative lysosomes. This practice could lead to misinterpretation of the actual roles of mature active lysosomes in neuronal growth, function, and survival in healthy brains as well as their pathological impact on neurodegenerative diseases. Thus, there is an urgent need to quantitatively characterize LAMP1-labeled degradative and nondegradative organelles in both in vitro and in vivo nervous systems.
In this study, by using immunotransmission EM (iTEM) and light confocal imaging combined with Airyscan superresolution microscopy, we expand on the limited literature to provide a comprehensive and quantitative analysis of relative LAMP1 distribution in various endolysosomal organelles in neurons. Our study demonstrates that a significant portion of neuronal LAMP1-labeled organelles does not contain three major lysosomal hydrolases including cathepsins D and B and glucocerebrosidase (GCase). We further show that the relative intensity of LAMP1 in familial amyotrophic lateral sclerosis (fALS)-linked spinal motor neurons (sMNs) does not necessarily reflect lysosomal deficits in vivo. Thus, one must be more cautious when interpreting LAMP1-labeled organelles in neurons where LAMP1 intensity, trafficking, and distribution do not necessarily represent degradative lysosomes under physiological and pathological conditions.
Results
Differential distribution patterns of LAMP1 and cathepsin D in adult neurons in vivo
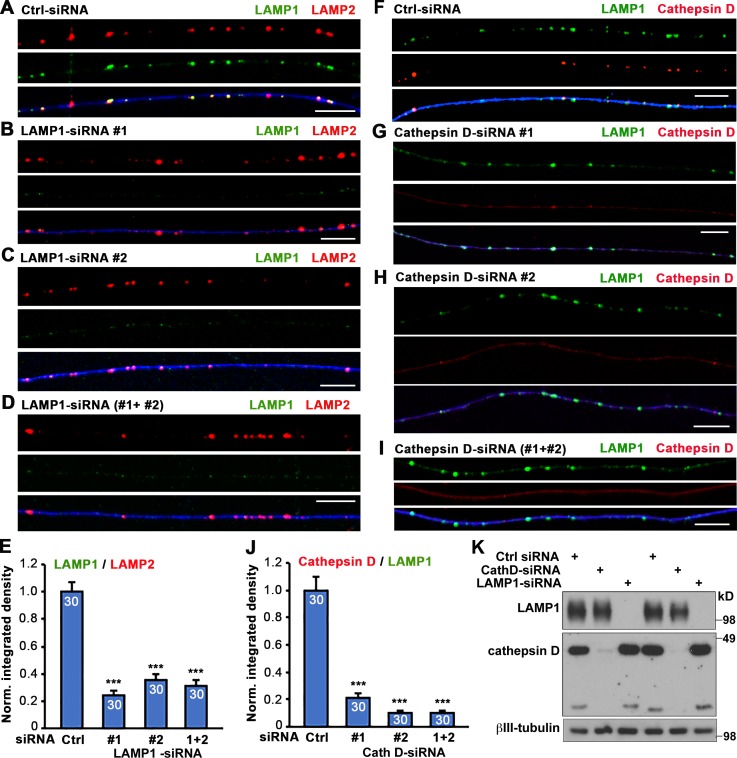
One hallmark of degradative lysosomes is the presence of active hydrolases, among which cathepsins constitute a major category. Cathepsin D, an aspartyl protease that primarily resides within lysosomes, is a prominent member of the cathepsin family. Given the increasing application of LAMP1 as a neuronal lysosomal marker, we sought to investigate whether it can reliably label cathepsin-containing lysosomes in mature neurons in vivo. Application of LAMP1 and cathepsin D antibodies has been widely reported, and their specificity has been repeatedly verified in recent studies (Marwaha et al., 2017; Sathyanarayan et al., 2017). We further confirmed their specificity in neurons by knocking down LAMP1 or cathepsin D via RNAi approaches. Mouse dorsal root ganglion (DRG) neurons at day in vitro (DIV) 0 were transfected with control, LAMP1-, or cathepsin D–targeted siRNAs through nucleofection, followed by immunostaining or immunoblotting at DIV3. Compared with neurons transfected with control siRNA, LAMP1 signals were selectively depressed in neurons transfected with LAMP1-targeted siRNA1-, 2-, or combined 1+2 (Fig. 1, A–D). Similarly, cathepsin D signals were selectively depressed in neurons transfected with cathepsin D–targeted siRNA1-, 2-, or combined 1+2 (Fig. 1, F–I). The normalized integrated density ratio of LAMP1/LAMP2 (Fig. 1 E) or cathepsin D/LAMP1 (Fig. 1 J) was robustly reduced in LAMP1- or cathepsin D–depleted neurons, respectively. Western blotting consistently showed a selective depletion of LAMP1 or cathepsin D by their corresponding siRNAs (Fig. 1 K). These findings verify the specificity of LAMP1 and cathepsin D antibodies in primary neurons, prompting us to characterize colocalization of LAMP1 and cathepsin D signals in vitro and in vivo.
Figure 1.
Verified specificity of LAMP1 and cathepsin D antibodies by knockdown assays in DRG neurons. (A–E) Representative images of DRG neuron axons (A–D) and quantitative analysis (E) showing substantially reduced LAMP1 staining in axons after knockdown by LAMP1-siRNA1 (B), LAMP1-siRNA2 (C), or combined LAMP1-siRNA1+2 (D). Neurons were cotransfected at DIV0 with 30 nM control siRNA (Ctrl) or LAMP1-siRNA oligonucleotides with GFP (pseudocolor blue) and analyzed by coimmunostaining 3 d after transfection using antibodies against LAMP1 and LAMP2. Note that LAMP1-siRNAs abolish LAMP1 staining without reducing LAMP2 signals, thus verifying the specificity of LAMP1 antibody. The integrated density of LAMP1 was normalized to LAMP2 from the same images (E). (F–J) Representative images of DRG neuron axons (F–I) and quantitative analysis (J) showing a substantial reduction in cathepsin D staining along axons after the knockdown by cathepsin D–siRNA1 (G), cathepsin D–siRNA2 (H), or combined cathepsin D–siRNA1+2 (I). Neurons were cotransfected at DIV0 with 30 nM control siRNA or cathepsin D–siRNA oligonucleotides with GFP (pseudocolor blue) as indicated and then were analyzed by coimmunostaining 3 d after transfection using antibodies against LAMP1 and cathepsin D. Note that cathepsin D–siRNAs abolished cathepsin D staining without reducing LAMP1 detection, thus verifying the specificity of cathepsin D antibody. The integrated density of cathepsin D was normalized to LAMP1 from the same images (J). Data were collected from 30 axons in three repeats. Error bars indicate SEM. Bars, 10 µm. ***, P < 0.001. (K) Representative Western blots showing effective knockdown of neuronal LAMP1 and cathepsin D with LAMP1-siRNA1+2 or cathepsin D–siRNA1+2, respectively. DRG neurons were transfected at DIV0 with 30 nM control or targeted siRNA oligonucleotides and analyzed by Western blot 3 d after transfection using antibodies against LAMP1, cathepsin D, and βIII-tubulin.
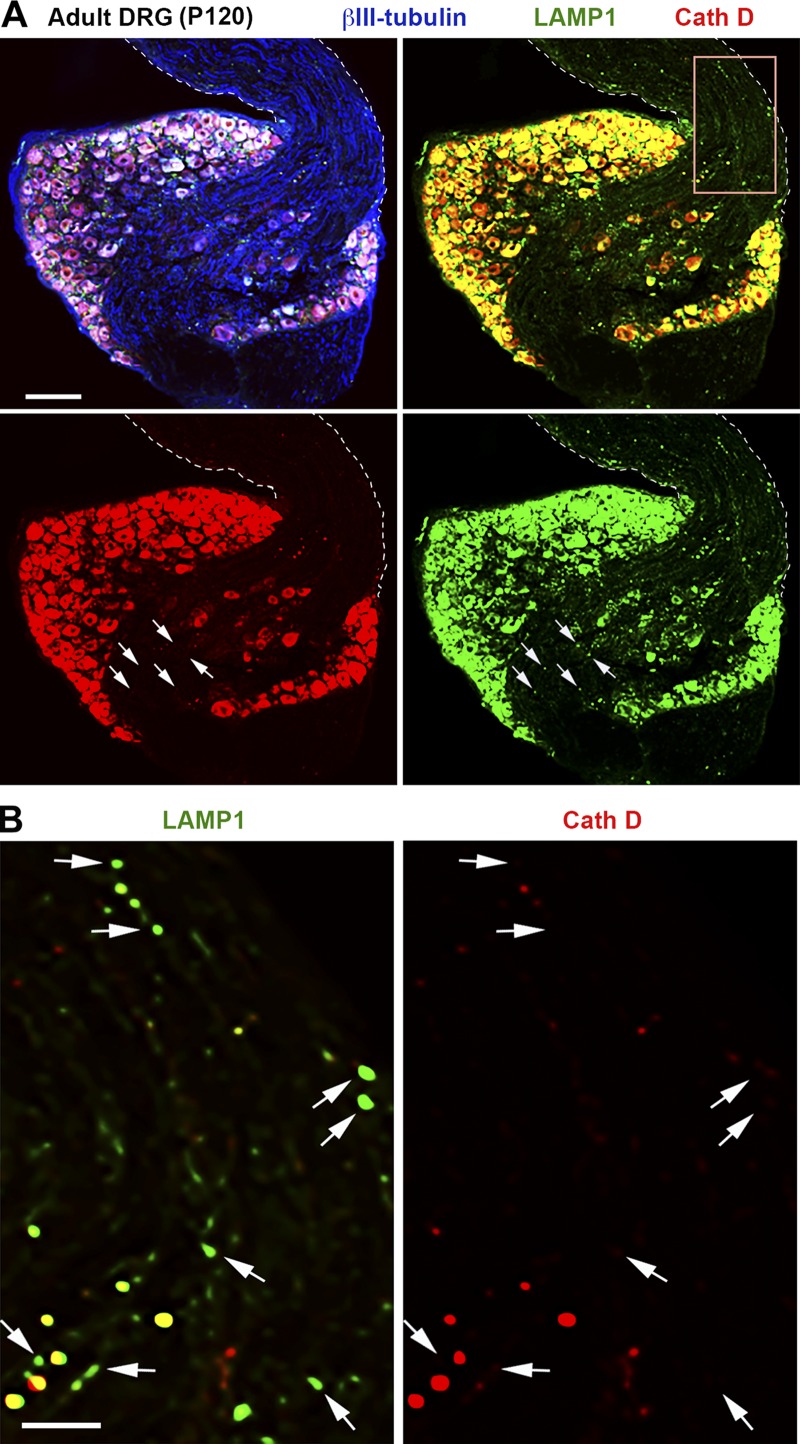
We chose coimmunostaining of adult mouse DRGs, which are composed of sensory neuron cell bodies and pseudounipolar axon bundles extending from the dorsal root toward the periphery and gray matter of the spinal cord (Fig. 2 A). The unique axon-bundled structures within the DRG make it an ideal model for tracing distribution and colocalization of axonal organelles in vivo. DRGs isolated from adult mice at P120 (postnatal day 120) were coimmunostained with antibodies against LAMP1, cathepsin D, and neuron-specific βIII-tubulin. LAMP1 and cathepsin D signals are intensely accumulated in the soma of DRG neurons, prohibiting colocalization analysis. Relative colocalization was assessed by Manders’ colocalization coefficient (MCC)-based analysis and particle-based analysis in axonal bundles. LAMP1 and cathepsin D were only partially colocalized in DRG axon bundles labeled by βIII-tubulin (Fig. 2 A) such that only 16.62 ± 1.39% (MCC-based) or 21.78 ± 2.13% (particle-based) of LAMP1-labeled vesicles contained detectable cathepsin D (Fig. 2 B), indicating their differential distribution patterns in mature adult neurons in vivo.
Figure 2.
Differential distribution patterns of LAMP1 and cathepsin D in adult mature DRG neurons in vivo. (A and B) Representative images (A) showing partial colocalization of LAMP1 and cathepsin D in DRG axon bundles from adult mice at P120. Mice were perfused with 50% Bouin’s solution, and DRGs were isolated for immunohistochemical staining with antibodies against LAMP1 (green), cathepsin D (red), and βIII-tubulin (blue). Dashed lines show the edges of axon bundles labeled by βIII-tubulin. Arrows indicate LAMP1-labeled organelles with no detectable cathepsin D. Note that although LAMP1 signals were detectable within axon bundles as shown in the enlarged boxed area (B), only 16.62 ± 1.39% (MCC based) or 21.78 ± 2.13% (particle based) of them colocalized with detectable cathepsin D. Data were quantified from a total of 15 images of DRG axonal bundles. Bars: (A) 100 µm; (B) 20 µm.
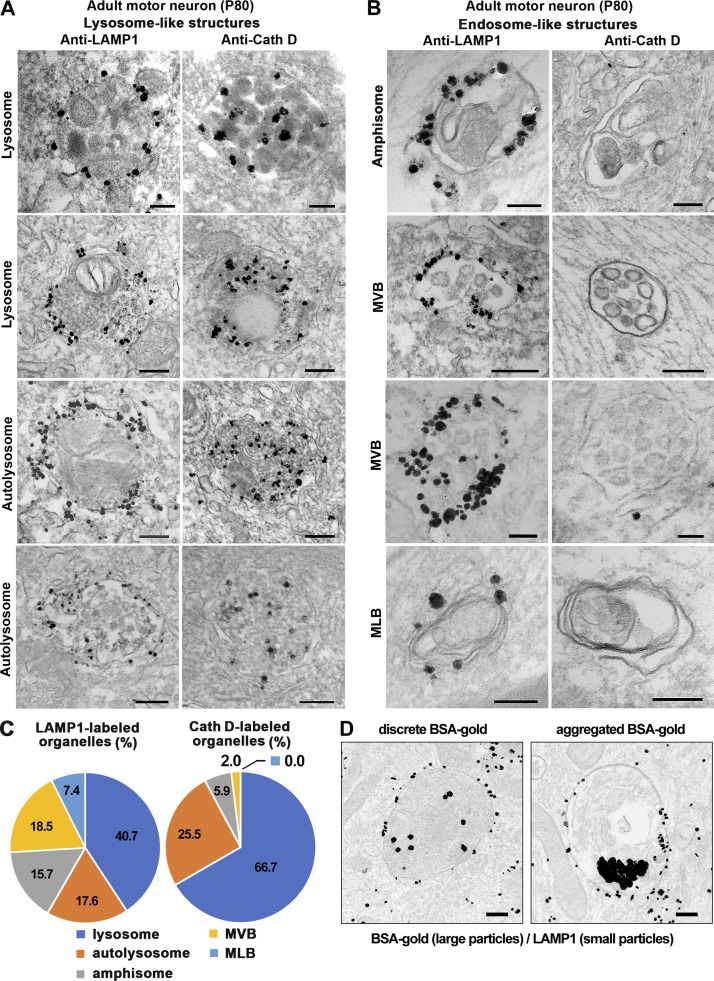
To confirm these findings at the ultrastructural level, we examined mouse sMNs from adult mice at P80 with iTEM. Spinal cord sections were immunostained with an antibody against LAMP1 or cathepsin D followed by incubation with Nanogold-conjugated secondary antibodies and silver enhancement. LAMP1 signals were readily detected around the surface of various endosome- and lysosome-like organelles including mature lysosomes with a high electron density, degradative autolysosomes with double membranes and electron density (Fig. 3 A), immature autophagosomes with double membranes containing nondegraded translucent contents, multivesicular bodies (MVBs) containing multiple internal vesicles, and multilamellar bodies (MLBs) composed of concentric membrane layers (Fig. 3 B). In contrast, cathepsin D signals were only detected in the luminal side of mature lysosomes and autolysosomes (Fig. 3 A) but were rarely found in other types of organelles (Fig. 3 B). The relative frequency of cathepsin D detection in amphisome- and endosome-like organelles (MVB and MLB) is much lower compared with the frequency of LAMP1 immunostaining (Fig. 3 C).
Figure 3.
Ultrastructural analysis showing differential distribution of LAMP1 and cathepsin D in various endolysosomal and autophagic organelles in adult motor and DRG neurons. (A–C) Representative iTEM micrographs (A and B) and quantitative analysis (C) showing differential distribution patterns of LAMP1 and cathepsin D in lysosome- (A) and endosome-like (B) structures. Mouse spinal cords were dissected at P80 and sectioned into 100-µm sections, and then LAMP1 and cathepsin D were labeled with primary antibodies followed by incubation with Nanogold-conjugated secondary antibodies and silver enhancement. Note that LAMP1 signals can be detected around the surface of both autophagic and endolysosome-like organelles including lysosomes with single membranes and high electron density (A), degradative autolysosomes with double membranes and high electron density (A), immature autophagosomes containing nondegraded translucent contents that likely represent amphisomes after fusion with LEs (B), MVBs containing multiple internal vesicles (B), and MLBs composed of concentric membrane layers (B). In contrast, cathepsin D is mainly detected within the luminal side of lysosomes and autolysosomes (A and C) but was rarely found in endosome-like organelles (B and C). The percentages of LAMP1 or cathepsin detected in these organelles are shown in pie charts (C). The data were collected from three adult WT mice, and only those organelles with clear membrane structures were selected for quantification. A total of 108 LAMP1-positive organelles was found from a total of 503 EM micrographs from 12 grids, and 51 cathepsin D–positive organelles were found in 326 EM micrographs from 11 grids. (D) Representative iTEM micrographs of pulse–chase BSA-gold assay showing a heterogeneity of degradative capacity in LAMP1-labeled endolysosomal organelles. Adult DRG neurons at DIV4 were fed with the fluid-phase endocytic marker BSA-gold conjugates (6 nm) for 4 h at 37°C followed by a 4-h chasing period in a conjugate-free medium. Neurons were then fixed for LAMP1 immunogold staining and silver enhancement. LAMP1-gold particles (1.4 nm) were located on the surface of endolysosomal vesicles, whereas the larger BSA-gold particles (6 nm) were internalized into the luminal side. Note that BSA-gold particles appear as flocculation because of BSA degradation (right) or discretion because of lack of BSA degradation (left), indicating a heterogeneity of degradative capacity in these LAMP1-labeled endolysosomal organelles. Bars, 200 nm.
As the presence of degradative capacity is a necessary prerequisite for mature lysosomes, we further conducted a BSA-gold pulse–chase assay in live neurons followed by iTEM to examine whether BSA could be degraded within these LAMP1-labeled organelles. BSA conjugated with colloidal gold (BSA-gold) would be internalized after fluid phase endocytosis, and the gold particles should only flocculate in the lumen of mature lysosomes, where proteolytic degradation of BSA occurs (Griffiths et al., 1988; Bright et al., 1997; Cai et al., 2010). After a 4-h pulse and 4-h chase procedure, we confirmed the fate of internalized BSA-gold in LAMP1-labeled lysosomal compartments at the ultrastructural level by iTEM. Among a total of 431 LAMP1-labeled organelles with internalized gold particles, 354 (82.1%) of them contained flocculated gold particles, whereas 77 (17.9%) showed discrete gold particles, reflecting the heterogeneous degradative capacity among LAMP1-labeled organelles (Fig. 3 D). Given the fact that these BSA-gold–containing LAMP1 organelles represent the late stages of the endocytic pathway after a prolonged pulse–chase in live neurons, it is conceivable that the percentage of degradative LAMP1 organelles (82.1%) is higher than that observed in LAMP1 immunostaining assays (Fig. 4) that detect all LAMP1 organelles including those outside of the endocytic pathway.
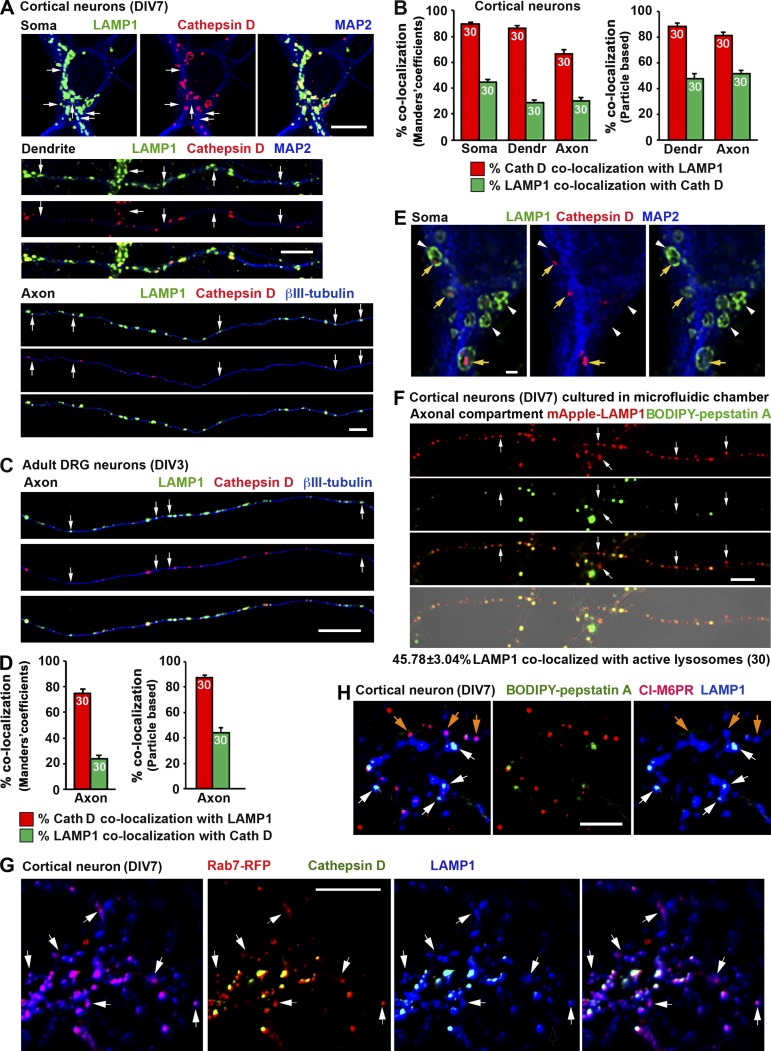
Figure 4.
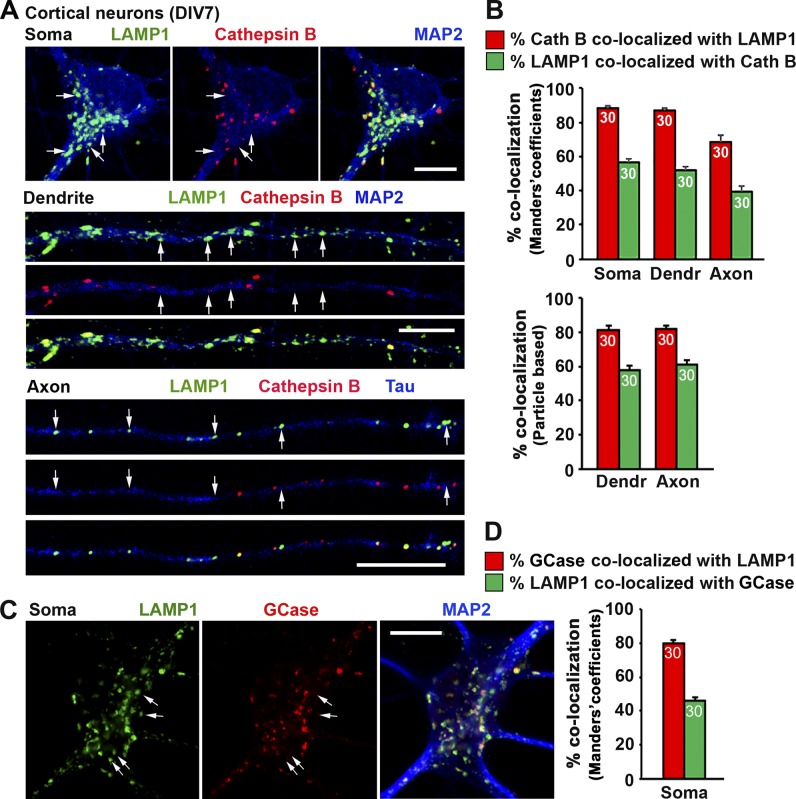
A significant portion of LAMP1-labeled organelles does not contain lysosomal hydrolase cathepsin D in cultured embryonic cortical neurons and adult DRG neurons. (A and B) Representative images (A) and quantitative analysis (B) showing a partial colocalization of LAMP1 and cathepsin D in cortical neurons. Neurons were coimmunostained at DIV7 with antibodies against LAMP1, cathepsin D, and MAP2 or βIII-tubulin. Images were taken from the soma, dendrite, and axon as indicated. Arrows indicate LAMP1-positive organelles lacking cathepsin D. (C and D) Representative images (C) and quantitative analysis (D) showing a large number of LAMP1 organelles negative for cathepsin D in axons of adult DRG neurons. Neurons isolated from adult mice (P30) were coimmunostained at DIV3 with antibodies against LAMP1, cathepsin D, and βIII-tubulin. Images were taken from axon segments. Arrows indicate LAMP1-positive organelles with no detectable cathepsin D. Relative colocalization was assessed by MCC-based analysis (B and D, left) and particle-based analysis (B and D, right). Data were quantified from the total number of neurons indicated in the bar graphs for each group from more than three experiments. Error bars indicate SEM. (E) Representative Airyscan superresolution images (obtained with an LSM 880 microscope) demonstrate ring-like LAMP1 structures containing cathepsin D in their luminal side. Mouse cortical neurons were coimmunostained at DIV7 with antibodies against LAMP1, cathepsin D, and MAP2. Images were taken from the soma region. Yellow arrows indicate LAMP1 ring-like structures containing detectable cathepsin D. White arrowheads indicate LAMP1-positive organelles lacking cathepsin D. Note that only a small fraction of LAMP1 ring-like structures surrounded cathepsin D. (F) Representative images showing a partial colocalization of LAMP1 with the active cathepsin D marker BODIPY–pepstatin A in cortical neuron axons. Neurons were infected with mApple-LAMP1 at DIV0 before plating in a microfluidic chamber. At DIV7, BODIPY–pepstatin A (1 µM) was loaded for 1 h before live imaging of the axon chamber. Arrows indicate LAMP1-positive organelles with nondetectable BODIPY–pepstatin A. Quantitative analysis shows 45.78 ± 3.04% of mApple-LAMP1 organelles colocalized with active cathepsin D as indicated by BODIPY–pepstatin A staining. (G) Representative images showing LAMP1-positive and cathepsin D–negative organelles as LEs in nature. Cortical neurons transfected with Rab7-RFP were fixed at DIV7 with 50% Bouin’s solution and immunostained with antibodies against LAMP1 and cathepsin D. White arrows indicate LAMP1-positive and cathepsin-negative organelles that were colabeled by the late endosomal marker Rab7. (H) Representative images showing the late endosomal nature of LAMP1-positive organelles lacking degradative capacity in cortical neurons. Neurons at DIV7 were stained with BODIPY–pepstatin A (1 µM) for 1 h followed by fixation and immunostaining with antibodies against LAMP1 and CI-M6PR. White arrows indicate LAMP1-labeled organelles with active cathepsin D, orange arrows point to nondegradative LAMP1 organelles that are costained by the late endosomal marker CI-M6PR. Bars: (A, C, F, G, and H) 10 µm; (E) 1 µm.
A significant portion of LAMP1-labeled organelles do not contain cathepsin D
Using cultured cortical neurons as a model, we further dissected the properties of LAMP1-labeled organelles in three distinct neuronal compartments: soma, axons, and dendrites. Bouin’s fixation was previously reported to reliably detect lysosomal luminal proteins by immunofluorescence (Lin and Lobel, 2001; Zhang et al., 2003; Sun et al., 2008; Harrison et al., 2009). In this study, we modified Bouin’s fixation protocol by using 50% Bouin’s solution to allow optimal codetection of both lysosomal hydrolases and LAMP1. This modified protocol significantly enhanced the signal detection of cathepsin D by 2.91 ± 0.26-fold (P < 0.0001) compared with 4% PFA fixation solution in neurons (not depicted). Using this fixation protocol, we first examined whether LAMP1-labeled organelles contain cathepsin D in cortical neurons at DIV7. Relative colocalization was assessed by MCC-based analysis. In the soma region, the majority (89.67 ± 1.33%) of cathepsin D–labeled organelles were also positive for LAMP1. However, only 44.8 ± 1.87% of LAMP1-labeled organelles contained cathepsin D. This number further dropped to 28.6 ± 2.07% in dendrites and 30.31 ± 2.39% in axons (Fig. 4, A and B). We alternatively assessed relative colocalization with particle-based analysis in dendrites and axons. Consistently, although 88.48 ± 2.78% (dendrites) or 81.43 ± 2.28% (axons) of cathepsin D–containing organelles were colabeled by LAMP1, a relatively small population of LAMP1 particles contained lysosome hydrolase cathepsin D (47.71 ± 3.98% in dendrites and 51. 64 ± 2.66% in axons). We next confirmed these observations in axons of DRG neurons isolated from adult mice at P30 because almost all neurites from DRG neurons are tau-positive axons (Perlson et al., 2009). Consistently, only 23.7 ± 2.98% (MCC-based analysis) or 43.93 ± 4.30% (particle-based analysis) of LAMP1 was colocalized with cathepsin D along DRG neuron axons (Fig. 4, C and D).
To illustrate differential LAMP1 and cathepsin D staining patterns in more detail, we applied Airyscan superresolution microscopy with 32 GaAsP detectors (Müller and Enderlein, 2010; Sheppard et al., 2013). LAMP1 clearly delineated the limiting membrane of the organelles, whereas cathepsin D resided inside; a sizable portion of LAMP1 ring-like structures did not contain detectable cathepsin D (Fig. 4 E). These Airyscan images are consistent with iTEM (Fig. 3, A and B) at the ultrastructural level and thus support the notion that approximately half of LAMP1-labeled organelles contain a major lysosome hydrolase.
Maintaining an acidified intraluminal environment is necessary for hydrolase activation and substrate degradation, the defining feature of mature lysosomes (Saftig, 2005). To test whether neuronal LAMP1-labeled organelles represent acidified structures containing activated hydrolases, we used boron-dipyrromethene (BODIPY)-modified full-length (FL)–pepstatin A, a fluorophore-tagged pepstatin that specifically binds active cathepsin D in acidic environments at pH 4.5. Cortical neurons were infected with lenti-mApple-LAMP1 and plated in microfluidic chamber devices in which neuronal cell bodies and dendrites are restricted to the soma chamber, whereas axons grow through long microgrooves (450 µm) into the axon terminal chamber (Zhou et al., 2012, 2016). Neurons at DIV7 were loaded with BODIPY–pepstatin A in both soma and axon chambers; images were taken from the axon chamber to trace the colocalization of mApple-LAMP1 and BODIPY–pepstatin A. Consistent with our cathepsin immunostaining and iTEM data, only 45.78 ± 3.04% of LAMP1-positive organelles colocalized with BODIPY–pepstatin A in axons (Fig. 4 F), further indicating the nondegradative LAMP1-labeled organelles within axons.
To determine the identity of the LAMP1-positive and cathepsin D–negative organelles, we performed triple labeling of LAMP1, cathepsin D, and Rab7 in cortical neurons. Representative images (Fig. 4 G) show that a sizable portion of LAMP1-positive and cathepsin-negative organelles are LEs in nature. To examine whether these LAMP1 organelles have degradative capacity, we stained cortical neurons (DIV7) with BODIPY–pepstatin A (1 µM) for 1 h followed by fixation and immunostaining with antibodies against LAMP1 and cation-independent MPR (CI-M6PR), a membrane protein preferentially located in LEs and immature lysosomes (Griffiths et al., 1988). A majority of LAMP1-positive organelles colabeled with CI-M6PR lacked degradative capacity (Fig. 4 H). The results of triple-color imaging were consistent with our iTEM analysis, where cathepsin D signals were only detected in the luminal side of mature lysosomes and autolysosomes (Fig. 3 A) but were rarely found in endosome-like organelles (Fig. 3 B).
Partial colocalization of LAMP1/2 with other lysosomal hydrolases
To exclude the possibility for a selective deficiency of cathepsin D in LAMP1-labeled organelles, we examined other prominent lysosomal hydrolases. Cathepsin B, a lysosomal cysteine protease, plays an important role in lysosomal proteolysis. Consistently, LAMP1 and cathepsin B are partially colocalized, with 56.76 ± 2.13% of LAMP1 signals colocalizing with cathepsin B in the soma, 51.83 ± 2.02% in dendrites, and 39.14% ± 3.81% in axons (MCC-based analysis). For particle-based analysis, colocalization of LAMP1 with cathepsin B in dendrites and axons are 57.59 ± 2.82% and 60.93 ± 2.64%, respectively (Fig. 5, A and B). To further confirm our findings, we examined a third lysosomal hydrolase: the acid β-glucosidase-GCase, a glucocerebrosidase (GBA)-encoded enzyme involved in sphingolipid catabolism in lysosomes (Grabowski et al., 1990). Homozygous GBA mutations cause Gaucher disease, the most common neuropathic lysosomal storage disorder (Sidransky, 2012). Similarly, in the soma of mouse cortical neurons, only 46.24 ± 2.06% of LAMP1 colocalized with GCase, whereas 79.72 ± 1.93% of GCase colocalized with LAMP1 (Fig. 5, C and D). Thus, our study with three different lysosomal hydrolases consistently supports the notion that although the majority of lysosomal hydrolases colocalize with LAMP1, a significant portion of LAMP1-labeled organelles in neurons do not contain detectable lysosomal hydrolases, representing nondegradative endolysosomal structures.
Figure 5.
A significant portion of LAMP1-labeled organelles does not contain detectable lysosomal hydrolases cathepsin B and GCase in cortical neurons. (A–D) Representative images (A and C) and quantitative analysis (B and D) showing a partial colocalization of LAMP1 with cathepsin B (A and B) or GCase (C and D). Cortical neurons were coimmunostained at DIV7 with antibodies against LAMP1, cathepsin B or GCase, and MAP2 or tau. Images were taken from soma, dendrites, and axons as indicated. Arrows point to LAMP1-positive organelles without detectable cathepsin B (A) or GCase (C). Relative colocalization was assessed by MCC-based analysis and particle-based analysis as indicated in bar graphs. Data were quantified from 30 neurons (n = 30) for each group in three experiments. Error bars indicate SEM. Bars, 10 µm.
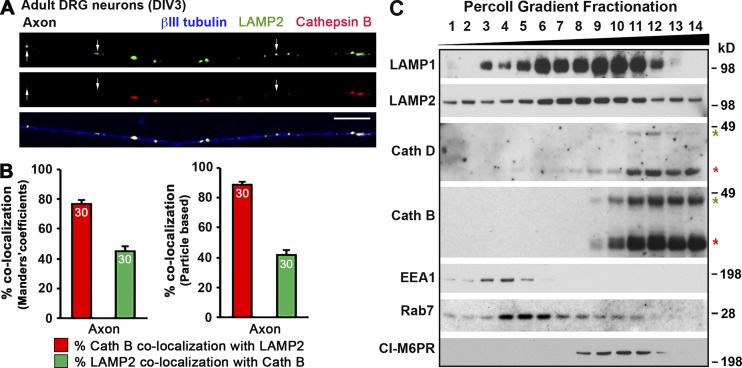
Next, we asked whether LAMP2 shares a similar distribution pattern as LAMP1 by coimmunostaining adult DRG neurons with antibodies against LAMP2, cathepsin B, and βIII tubulin at DIV3. Consistently, LAMP2 and cathepsin B were only partially colocalized, with 44.90 ± 3.28% (MCC based) or 41.55 ± 3.44% (particle based) of LAMP1-labeled organelles colocalizing with cathepsin B in the axons, respectively (Fig. 6, A and B). We alternatively applied biochemical gradient fractionation to confirm our imaging observations. Cultured cortical neurons at DIV7 were homogenized and subjected to subcellular fractionation by Percoll gradient centrifugation. Immunoblotting analysis demonstrates the differential gradient fractionation patterns of LAMP1/2 and cathepsins D/B in neurons; although LAMP1/2 were widely present in most fractions (3–12), mature forms of cathepsins D/B were enriched in the heavier-gradient fractions 10–14 (Fig. 6 C). Importantly, EEA1 (an early endosomal marker) was in the lighter gradient fractions 3–5, and Rab7 and CI-M6PR were mainly in middle fractions (4–10 and 8–11, respectively), indicating effective gradient fractionation.
Figure 6.
Differential distribution and fractionation patterns of LAMP1/2 and lysosomal hydrolases cathepsins D/B in neurons. (A and B) Representative images (A) and quantitative analysis (B) showing a large number of LAMP2 organelles with no detectable cathepsin B in axons of adult DRG neurons. Neurons isolated from adult mice (P30) were coimmunostained at DIV3 with antibodies against LAMP2, cathepsin B, and βIII-tubulin. Images were taken from distal axon segments. Arrows indicate LAMP2-labeled organelles with no detectable cathepsin B. Relative colocalization was assessed by MCC-based analysis (B, left) and particle-based analysis (B, right). Data were quantified from 30 neurons for each group in three experiments. Error bars indicate SEM. Bar, 10 µm. (C) Percoll gradient fractionation showing differential fractionation patterns of LAMP1/2 and cathepsins D/B in neurons. Cultured cortical neurons at DIV7 were homogenized and subjected to subcellular fractionation by Percoll gradient centrifugation. A total of 14 gradient fractions were collected from top (fraction 1) to bottom (fraction 14). All fractions were analyzed by sequential immunoblotting with antibodies against endolysosomal membrane proteins LAMP1/2, lysosomal hydrolases cathepsins D/B, early endosomal marker EEA1, and late endosomal markers Rab7 and CI-M6PR. Red asterisks mark cleaved mature forms of cathepsins D/B, and green asterisks indicate intermediate forms of cathepsins D/B. Note that approximately half of both mature forms of cathepsins D/B are distributed in fractions 10–12, where LAMP1 is also abundantly detected; Rab7 is mainly detected in fractions 4–10, where LAMP1 is predominantly distributed, indicating a considerable gradient codistribution of LAMP1 with cathepsins D/B and Rab7.
Analysis of LAMP1 appears insufficient for characterizing lysosomal deficits in fALS-linked motor neurons
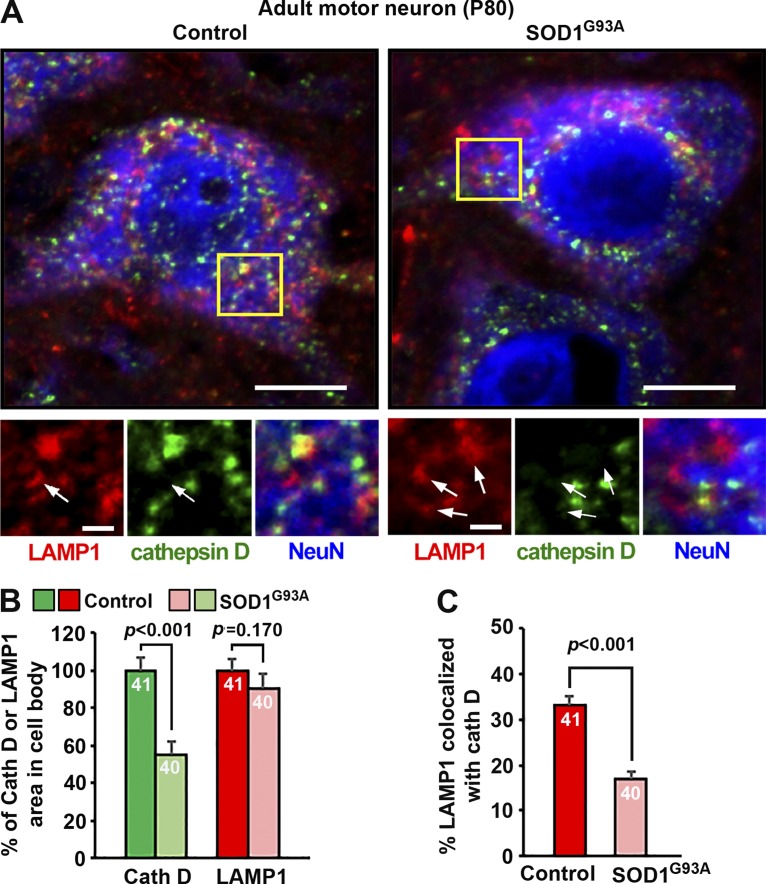
Our previous study revealed progressive lysosomal defects in sMNs of the fALS-linked hSOD1G93A mutant mouse starting at an early asymptomatic stage (P40; Xie et al., 2015). To examine whether LAMP1 is a sensitive marker for assessing lysosomal defects in neurodegenerative diseases, we coimmunostained ventral root motor neurons of both WT and fALS-linked hSOD1G93A mice at P80 with antibodies against LAMP1, cathepsin D, and NeuN, a neuron marker (Fig. 7 A). Compared with age-matched WT controls, cathepsin D mean intensity was dramatically reduced (54.86 ± 7.59%; P < 0.001) in hSOD1G93A motor neuron cell bodies. In contrast, there was no detectable change in LAMP1 mean intensity in the same hSOD1G93A motor neurons compared with WT mice (P = 0.170; Fig. 7 B). In addition, the proportion of lysosomes identified as LAMP1 and cathepsin D double-positive organelles was also reduced from 33.25 ± 1.66% to 16.92 ± 1.66% in hSOD1G93A mutant motor neurons (Fig. 7 C). Such inconsistency has also been demonstrated in AD, where altered protein levels of cathepsins B and D were observed in the hippocampus of postmortem patient brains, whereas LAMP1 remained unchanged (Bordi et al., 2016). Consistently, in AD-linked PS1-knockout (KO)/PS1–familial AD cells where acidification of lysosomes is defective because of reduced targeting of vacuolar-type H+ ATPase, LAMP1/2 staining was unaffected (Lee et al., 2010). Given the fact that not all LAMP1-labeled organelles represent degradative lysosomes, labeling a set of lysosomal hydrolases combined with various endolysosomal markers is more accurate than simply relying on LAMP1 staining to determine any alteration of the endolysosomal pathway in neurodegenerative diseases.
Figure 7.
LAMP1 and cathepsin D respond differently to lysosomal deficits in fALS-linked sMNs. (A–C) Representative images (A) and quantitative analysis (B and C) showing differential changes in the mean intensity and colocalization of LAMP1 (red) and cathepsin D (green) in sMNs (labeled with NeuN; blue) from adult control or fALS-linked mutant hSOD1G93A mice at presymptomatic stages (P80). Arrows indicate LAMP1-positive organelles with no detectable cathepsin D. Consistent with early lysosomal deficits in sMNs of fALS-linked hSOD1G93A mutant mice (Xie et al., 2015), cathepsin D signals were significantly reduced to 54.86 ± 7.59% in the hSOD1G93A motor neurons relative to age-matched control motor neurons (P < 0.001), whereas LAMP1 signals remained unchanged (P = 0.17) at the same disease stage (B). Note that although 33.25 ± 1.66% of LAMP1-positive organelles were colocalized with cathepsin D in control MNs, only 16.92 ± 1.66% (P < 0.001) were colocalized in mutant SOD1G93A motor neurons (C). Relative colocalization was assessed by MCC-based analysis. Data were quantified from total numbers of neurons indicated in the bar graphs and analyzed using the Student’s t test. Error bars indicate SEM. Bars: (main images) 20 µm; (insets) 5 µm.
LAMP1 is distributed among a heterogeneous population of endocytic organelles
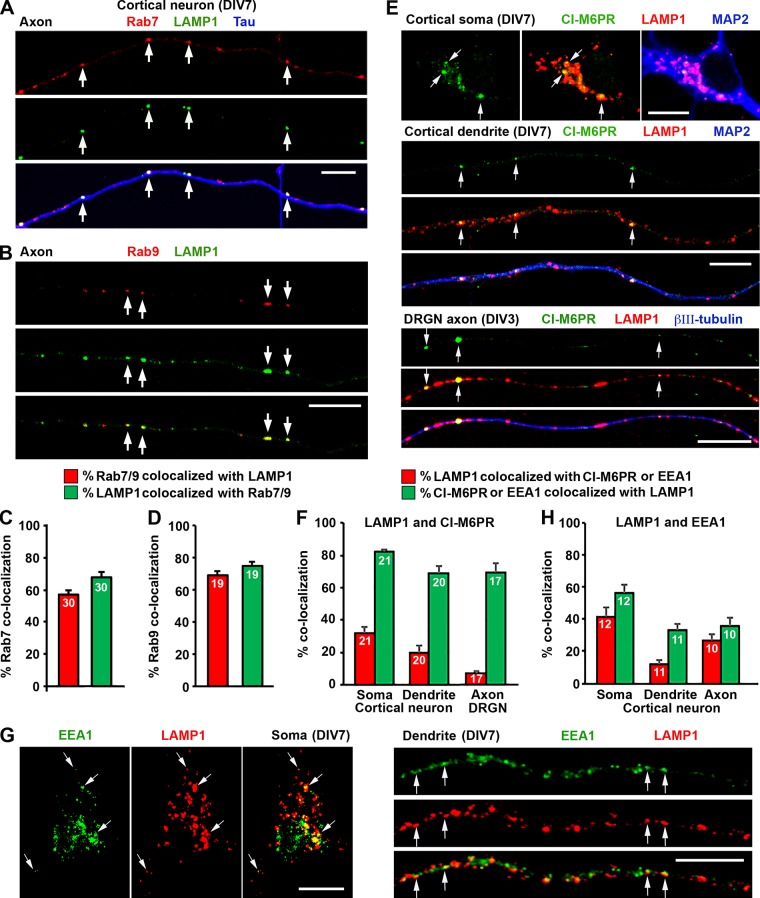
A significant proportion of LAMP1/2 traffics through the endocytic pathway to lysosomes, and their sorting and targeting are regulated by the endocytic machinery (Janvier and Bonifacino, 2005). LAMP1 is primarily located in LEs and lysosomes, with a smaller population in EEs and plasma membranes (Höning et al., 1996; Hunziker and Geuze, 1996; Saftig and Klumperman, 2009). However, quantitative assessment of LAMP1 relative distribution in these endocytic organelles has not been reported in neurons. Thus, we examined the identities of LAMP1-targeted organelles by colabeling other endocytic markers. We first coimmunostained cortical neurons with LAMP1 and the LE marker Rab7. The majority (67.88 ± 3.4%) of axonal LAMP1 signals colocalized with Rab7, indicating LEs/endolysosome in nature (Fig. 8, A and C). We confirmed this by costaining LAMP1 and Rab9, the Ras-like GTPase that shuttles between LEs and Golgi apparatus (Lombardi et al., 1993). 74.67 ± 2.83% of LAMP1-labeled organelles colocalized with Rab9 in axons (Fig. 8, B and D). Next, we costained neurons with LAMP1 and CI-M6PR. We found that 32.38 ± 3.52% of LAMP1-labeled organelles colocalized with CI-M6PR in the soma of cortical neurons, whereas in dendrites, this number dropped to 20.16 ± 4.19% (Fig. 8, E and F). Interestingly, only 6.98 ± 1.67% of LAMP1-labeled organelles colocalized with CI-M6PR in axons of DRG neurons. Furthermore, 41.28 ± 6.02% of LAMP1-labeled organelles colocalized with EEA1 (EE marker) in the soma. This number dropped to 26.77 ± 3.61% and 11.95 ± 2.33% in axons and dendrites, respectively (Fig. 8, G and H). Altogether, our imaging analysis provides quantitative evidence that LAMP1-labeled organelles represent a heterogeneous population of endolysosomal structures within neurons. This conclusion is consistent with our observations in the iTEM study (Fig. 3, B and C) and gradient fractionation study (Fig. 6 C).
Figure 8.
LAMP1 targets a heterogeneous population of endocytic organelles. (A–D) Representative images (A and B) and quantitative analysis (C and D) showing a partial colocalization of LAMP1 with Rab7-positive LEs (A and C) or Rab9-positive LEs/retromers (B and D). Mouse cortical neurons were coimmunostained at DIV7, and images were taken from axon segments. Arrows indicate LAMP1 colocalized with Rab7 or Rab9. (E and F) Representative images (E) and quantitative analysis (F) showing a partial colocalization of LAMP1 with CI-M6PR in cortical and DRG neurons (DRGNs). Embryonic mouse cortical neurons and adult DRG neurons were coimmunostained at DIV7 or 3, respectively, with antibodies against CI-M6PR, LAMP1, and MAP2 or βIII-tubulin. Images were taken from the soma (top), dendrites (middle), and DRG axons (bottom), respectively. Arrows indicate LAMP1-positive organelles that were colabeled with CI-M6PR. (G and H) Representative images (G) and quantitative analysis (H) showing a small portion of LAMP1 colocalized with the EE marker EEA1. Cortical neurons were coimmunostained at DIV7 with antibodies against EEA1 and LAMP1, followed by imaging from the soma (left), dendrites (right), and axons (not depicted). Arrows indicate colocalization of LAMP1 and EEA1 in the soma and along dendrites. Relative colocalization was assessed by MCC-based analysis. Data were quantified from the total number of neurons indicated within the bar graphs for each group from three experiments. Error bars indicate SEM. Bars, 10 µm.
Discussion
Our study using iTEM and light imaging analysis provides quantitative analysis of LAMP1 distribution in various endolysosomal organelles in neurons. We demonstrate that a significant portion of LAMP1/2-labeled organelles does not represent degradative lysosomes in neurons but instead belongs to a wide spectrum of endocytic organelles. Therefore, changes in the distribution, density, and trafficking of LAMP1-labeled organelles do not necessarily reflect lysosome function or deficits under physiological or pathological conditions. We suggest that codetection of LAMP1 with various lysosomal hydrolases will provide a better understanding of how lysosomal function or its deficit contributes to neuronal health and disease progression.
Our quantitative iTEM analysis and BSA-gold pulse–chase assays are consistent with several recent studies showing heterogeneous nature of LAMP1-labeled organelles in various types of cell lines and neurons. In a study by Johnson et al. (2016), 79 ± 2% of LAMP1 was colocalized with saposin C, a glycoprotein thought to be restricted to lysosomes; this number dropped to 64 ± 4% in the periphery, suggesting heterogeneous populations of LAMP1-labeled organelles. The EM ultrastructural study of lysosomal distribution in dendrites of hippocampal neurons using APEX2 technology also showed that the luminal electron density of LAMP1-labeled organelles is heterogeneous (Goo et al., 2017). A third study identified a subpopulation of LAMP1-labeled organelles that are deficient in major cathepsins B/D/L in both distal processes of the hippocampal region of WT control mice and within the swollen axons of amyloid plaques of AD mice (Gowrishankar et al., 2015). The same group further characterized axonal lysosomal trafficking in JIP3 KO neurons and revealed an accumulation of LAMP1-labeled organelles in axon swellings. Further EM analysis indicated that these LAMP1 organelles are morphologically similar to LEs (MLBs or MVBs) and devoid of lysosomal hydrolases (Gowrishankar et al., 2017). Our iTEM ultrastructural observations in adult neurons in vivo are also consistent with the previous iTEM analysis in nonneuronal human HepG2 cells where endogenous LAMP1 localization can vary among distinct endolysosomal intermediates (Saftig and Klumperman, 2009). Our quantitative analysis showing partial colocalization of LAMP1 and major lysosomal hydrolases is supported by a recent study in rat hippocampal neurons where ectopic LAMP1-GFP was coexpressed with cathepsin D–GFP or stained with the fluorogenic cathepsin B substrate Magic red. Consistently, LAMP1-labeled organelles and cathepsin-containing vesicles were not completely colocalized (Farías et al., 2017). Altogether, these studies support our notion that a significant portion of LAMP1-labeled organelles in healthy and diseased neurons do not contain major lysosomal hydrolases. Instead, these LAMP1-positive organelles are likely intermediates of endocytic and autophagic pathways and lysosomal biogenesis (Hollenbeck, 1993; Lee et al., 2011; Maday et al., 2012, 2014; Cheng et al., 2015).
To more sensitively detect lysosomal luminal hydrolases, we modified Bouin’s fixation protocol (Lin and Lobel, 2001; Zhang et al., 2003; Harrison et al., 2009) to allow optimal codetection of both luminal protein cathepsin D and membrane protein LAMP1. This modified protocol significantly enhances the signal detection of cathepsin D by approximately threefold compared with 4% PFA fixation solution in neurons. With this protocol and MCC-based colocalization analysis, we found that 44.8 ± 1.87% of LAMP1-labeled organelles in the soma contain cathepsin D. However, this number drops to 28.6 ± 2.07% (P < 0.0001) in dendrites and 30.31 ± 2.39% (P < 0.0001) in axons (Fig. 4, A and B), suggesting that the percentage of degradative LAMP1 organelles is relatively higher in the soma than in neuronal processes. Using the pH-sensitive chimeric LAMP1 construct, which has a superecliptic pHluorin fused to the luminal side, Farías et al. (2017) also showed a differential acidity of LAMP1-labeled organelles in distal and proximal axons, thus suggesting that LAMP1-positive organelles do not necessarily represent degradative lysosomes in distal axons. Altogether, these studies provide some insights into the differential endolysosomal maturation and degradation activity in these neuronal compartments.
Materials and methods
Mouse line and animal care
C57BL/6J WT and B6.Cg-Tg (SOD1G93A) 1Gur/J mice were purchased from The Jackson Laboratory and maintained on the C57BL/6J genetic background in our animal facility. The Sprague–Dawley rats were purchased from Charles River Laboratory. Animals were housed in a 12-h light/dark cycle and fed with regular diet ad libitum. All animal procedures were performed following National Institutes of Health guidelines and were approved by the National Institute of Neurological Disorders and Stroke/National Institute on Deafness and Other Communication Disorders Animal Care and Use Committee.
Antibodies, constructs, and siRNAs
Sources of antibodies or reagents are as follows: antibodies against LAMP1 (rabbit; ab24170; Abcam; and rat; 1D4B; Developmental Studies Hybridoma Bank), cathepsin D (rat; MAB1029; R&D Systems), cathepsin B (goat; AF965; R&D Systems), βIII-tubulin (mouse; MAB5564; EMD Millipore; and mouse; T8578; Sigma-Aldrich), MAP2 (rabbit; AB5622; EMD Millipore; and mouse; 556320; BD), Tau-1 (mouse; MAB3420; EMD Millipore), NeuN (mouse; MAB377; EMD Millipore), GBA (rabbit; G4171; Sigma-Aldrich), CI-M6PR (mouse; ab2733; Abcam), EEA1 (mouse; 610456; BD), Rab7 (rabbit; 9367P; Cell Signaling Technology), Rab9a (mouse; MA3-067; Thermo Fisher Scientific); BSA-gold tracer (6 nm; Electron Microscopy Sciences); BODIPY FL–pepstatin A (Invitrogen); Alexa Fluor 546– or 633–conjugated secondary antibodies (Invitrogen), and Bouin’s solution (HT10132; Sigma-Aldrich). LAMP1-mApple (promoter, phosphoglycerate kinase; backbone, pLex; lentiviral) was generated in M. Ward’s laboratory (National Institute of Neurological Disorders and Stroke, Bethesda, MD). mRFP-Rab7 (promoter, cytomegalovirus; backbone, pmRFP-C3) was a gift from A. Helenius (14436; Addgene; Vonderheit and Helenius, 2005). The constructs were verified by DNA sequencing. Stealth siRNAs are from Thermo Fisher Scientific (LAMP1-siRNA1, MSS237022; LAMP1-siRNA2, MSS275304; cathepsin D–siRNA1, MSS203352; and cathepsin D–siRNA2, MSS203353).
Immunohistochemistry
Mice were anesthetized and transcardially perfused sequentially with 1× PBS followed by 4% PFA in PBS. Tissues were collected, followed by postfixation in 4% PFA overnight, and then they were soaked in 30% sucrose for 3 d. Spinal cord and DRG sections (10 µm) were obtained with a CM3050S cryostat (Leica Biosystems), rinsed with PBS, and then subjected to antigen retrieval with sodium citrate buffer (10 mM sodium citrate and 0.05% Tween-20, pH 6.0) if necessary. After incubating with primary antibody (4°C overnight) and three washes with 1× PBS, sections were incubated with an appropriate Alexa Fluor–conjugated secondary antibody (1:1,000; Invitrogen) at RT for 1 h. After three washes in PBS, sections were coverslipped with VectaMount (Vector Laboratories) mounting medium.
iTEM
iTEM was performed as previously described (Lin et al., 2017). For preparing mice spinal cords, adult mice were anesthetized and perfused with freshly made EM fixative (4% PFA and 0.05% glutaraldehyde in PBS, pH 7.4). Tissues were dissected and sectioned into 100-µm sections using a VT1200S vibratome (Leica Biosystems). Cultured DRG neurons at DIV4 were fixed with the same EM fixatives for 1 h at RT. DRG neurons or 100-µm tissue sections were washed with PBS and then permeabilized and blocked with 0.1% saponin/5% normal goat serum in PBS for 1 h. Endogenous LAMP1 and cathepsin D were labeled with LAMP1 or cathepsin D antibody for 1 h at RT. Samples were washed with PBS and incubated with secondary antibody conjugated to 1.4-nm Nanogold (Nanoprobes) for 1 h. After the wash with PBS, samples were fixed with 2% glutaraldehyde in PBS for at least 30 min and then sent to the National Institute for Neurological Disorders and Stroke EM facility for processing. All steps were performed at RT unless otherwise indicated. The control for specificity of immunogold labeling is by omitting the primary antibody. Images were acquired using an electron microscope (1200EX; JEOL).
BSA-gold pulse–chase assays
DRG neurons at DIV4 were incubated with mixed 0.25 ml BSA-gold (particle size 6 nm) with 0.75 ml neuron feeding medium at 37°C for 4 h for fluid phase endocytosis followed by a 4-h chase in conjugate-free medium. The cells were fixed with 4% PFA and 0.05% glutaraldehyde in PBS, pH 7.4, for 1 h at RT and then proceeded for immuno-EM with an anti-LAMP1 antibody and a secondary antibody conjugated to 1.4-nm Nanogold as described above.
Microfluidic chamber preparation
For house-made microfluidic devices, a silicon wafer with a pattern made out of SU-8 by photolithography was used to cast the polydimethyl siloxane (PDMS) microfluidic devices. In brief, SYLGARD 184 silicone elastomer base (Corning) was mixed with the curing agent at a ratio of 10:1. The PDMS was then mixed well using a ARF-310 mixer (THINKY) in two steps: mixing at 2,000 rpm for 4 min and defoaming at 2,200 rpm for 4 min. The well-mixed PDMS was poured onto the silicon wafer and then placed in a Bel-Art vacuum desiccator for 3 h to help remove air bubbles from the PDMS. The wafer with PDMS was placed in an 80°C oven for 1–2 h to cure. Once the PDMS was cured, the PDMS was pulled out, and reservoirs were punched out. The PDMS devices were extensively washed and autoclaved before being attached to coated coverslips.
Primary neuron cultures
Cortical neurons were collected from E18–19 rat or mouse embryos as described previously (Kang et al., 2008). In brief, after digestion with papain (Worthington Biochemical Corporation) for 45 min at 37°C and triturating with fire-polished pipettes, the cortical cells were recovered by centrifugation. After preparing a single-cell suspension, for the microfluidic chambers, 2 × 105/20 µl dissociated neurons were infected with a concentrated lentivirus containing mApple-LAMP1 for 10 min and then added into the cell body chamber of the microfluidic devices. After cells were attached, 100 µl of preequilibrated culture medium was added into each well of the device. Axons passed through microgrooves into the axonal terminal chamber at DIV4 as they grew. For regular culture, neurons were plated onto 12-mm coverslips coated with polyornithine (diluted 1:4 in double-distilled H2O; Sigma-Aldrich) and laminin (diluted 1:500 in PBS; Roche). After 24 h of growing neurons in plating medium, half of the plating medium was replaced with the same amount of neuronal feeding medium (2% B27 and 0.5 mM GlutaMAX in neurobasal medium). Cells were fed every 3 d by aspirating half the medium and replacing it with the same amount of neuronal feeding medium.
DRG neurons were isolated from P8–10 Sprague–Dawley rat or P30 mouse spinal cords in HBSS and digested in 2.5 U/ml dispase II (Roche) and 200 U/ml collagenase (Worthington Biochemical Corporation) at 37°C for 30 min followed by 35 min shaking at RT. Neurons were then collected using a 70-µm nylon cell strainer (Falcon). DRG neurons were plated on coverslips coated with polyornithine (diluted 1:4 in double-distilled H2O) and laminin (diluted 1:50 in PBS). Neurons were then maintained in neurobasal medium supplemented with 2% FBS, 2 mM GlutaMAX, and B27 (Invitrogen) for 3 d at 37°C in a 5% CO2 incubator.
Immunocytochemistry
Primary cultures of DRG or cortical neurons were fixed in 50% Bouin’s solution with 4% sucrose in PBS for 30 min if lysosomal hydrolases were costained; for other staining, we used 4% PFA (with 4% sucrose in PBS) for 15 min. Cells were then washed with PBS, incubated with a blocking buffer (0.4% saponin, 1% BSA, and 5% goat or donkey serum in PBS) for 1 h and then incubated with a primary antibody diluted in the incubation buffer (0.1% saponin, 1% BSA, and 5% goat or donkey serum in PBS) overnight at 4°C. After three washes in PBS, the cells were incubated with an appropriate Alexa Fluor–conjugated secondary antibody (1:400; Invitrogen) at RT for 1 h. After three washes in PBS, cells were coverslipped with VectaMount mounting medium.
Image acquisition and quantification
Confocal images were obtained using an LSM 880 oil-immersion 63× 1.4 NA oil objective with sequential acquisition setting (ZEISS). For live-cell imaging, neurons grown in the microfluidic chambers were replaced with prewarmed Hibernate E low-fluorescence medium (BraiBits) supplemented with 2% B27 and 0.5 mM GlutaMAX at the day of imaging. The temperature was maintained at 37°C. For fixed cells, images were taken at RT. Images were acquired using the same settings with no saturation and no bleedthrough and minimized noise at a resolution of 1,024 × 1,024 pixels (16 bit). For colocalization quantification, images were preprocessed with subtraction of a median filter-processed image and background, and then two images were proceeded to the ImageJ plugin JACOP (National Institutes of Health). MCCs were chosen as the colocalization ratio readout. Because the MCC is sensitive to noise and background, true structural pixels were identified by applying a threshold to the images; all pixels with intensities above this threshold were considered to be an object. The threshold value was defined manually after visual inspection (Dunn et al., 2011). For particle-based analysis, the percentage of LAMP1-labeled particles colocalizing with visible cathepsin D/B signals were calculated. Because lysosomal density in the soma is too dense to reveal single vesicles, particle-based analysis was chosen to quantify the colocalization along axons and dendrites. The superresolution images were taken in Airyscan mode. Intensity measurements were also performed using ImageJ. For quantification of SOD1G93A neurons, fluorescence intensity of endogenous LAMP1 staining was measured as mean intensity and expressed in arbitrary units of fluorescence per square area. The mean intensity of LAMP1 was normalized as a percentile ratio relative to the mean intensity of NeuN staining in the same soma of neurons; the values of normalized mean intensity from SOD1G93A neurons were further compared with those from WT controls.
Subcellular fractionation by Percoll gradient centrifugation
Rat cortical neurons in DIV7 were homogenized in homogenization buffer (250 mM sucrose, 20 mM Tris-HCl, pH 7.4, 1 mM EGTA, 1 mM EDTA, and protease inhibitor mixture). After removing the unbroken cells and nuclei by centrifugation at 750 g for 10 min at 4°C, the supernatant (one volume) was laid on the top of nine volumes of 20% (vol/vol) Percoll (GE17-0891; Sigma-Aldrich) with homogenization buffer and centrifuged at 20,000 g for 2.5 h at 4°C. 14 fractions were sequentially collected from top (fraction 1) to bottom (fraction 14). Equal amounts of each fraction were analyzed by sequential immunoblotting with antibodies against LAMP1 (ab24170; Abcam), LAMP2 (NB300-591; Novus Biologicals), cathepsin B (AF965; R&D Systems), cathepsin D (MAB1029; R&D Systems), EEA1 (610456; BD), Rab7 (9367P; Cell Signaling Technology), and M6PR (ab2733; Abcam). The blot membranes were stripped between applications of each antibody.
Statistical analysis
Statistical analysis was performed using Prism (GraphPad Software). Two groups were compared using Student’s t test (sample size n ≥ 30) or Mann-Whitney test (sample size n < 30). Data are expressed as means ± SEM. Differences were considered significant with P < 0.05.
Acknowledgments
We thank members of the Sheng laboratory for technical assistance and constructive discussion, S. Cuddy for mouse cortical neuron culture, M. Ward for lentiviral construct, K. Chamberlain and D. Schoenberg for critical reading, the National Institute of Neurological Disorders and Stroke EM Facility for EM sample handling, and P. Lobel for advising on Bouin’s fixation for lysosomal hydrolase detection.
This work is supported by the Intramural Research Program of National Institute of Neurological Disorders and Stroke and National Institutes of Health grants ZIA NS003029 (to Z.-H. Sheng) and ZIA NS002946 (to Z.-H. Sheng).
The authors declare no competing financial interests.
Author contributions: X.-T. Cheng, Y.-X. Xie, B. Zhou, and N. Huang designed and performed experiments and analyzed and interpreted data; T. Farfel-Becker provided experimental advice on hydrolase staining and lentiviral construct; Z.-H. Sheng is the senior author who conceived and directed the project; X.-T. Cheng, Y.-X. Xie, and Z.-H. Sheng wrote the manuscript.
References
- Ballabio A., and Gieselmann V.. 2009. Lysosomal disorders: from storage to cellular damage. Biochim. Biophys. Acta. 1793:684–696. 10.1016/j.bbamcr.2008.12.001 [DOI] [PubMed] [Google Scholar]
- Bordi M., Berg M.J., Mohan P.S., Peterhoff C.M., Alldred M.J., Che S., Ginsberg S.D., and Nixon R.A.. 2016. Autophagy flux in CA1 neurons of Alzheimer hippocampus: Increased induction overburdens failing lysosomes to propel neuritic dystrophy. Autophagy. 12:2467–2483. 10.1080/15548627.2016.1239003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright N.A., Reaves B.J., Mullock B.M., and Luzio J.P.. 1997. Dense core lysosomes can fuse with late endosomes and are re-formed from the resultant hybrid organelles. J. Cell Sci. 110:2027–2040. [DOI] [PubMed] [Google Scholar]
- Cai Q., Lu L., Tian J.H., Zhu Y.B., Qiao H., and Sheng Z.H.. 2010. Snapin-regulated late endosomal transport is critical for efficient autophagy-lysosomal function in neurons. Neuron. 68:73–86. 10.1016/j.neuron.2010.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X.T., Zhou B., Lin M.Y., Cai Q., and Sheng Z.H.. 2015. Axonal autophagosomes recruit dynein for retrograde transport through fusion with late endosomes. J. Cell Biol. 209:377–386. 10.1083/jcb.201412046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook N.R., Row P.E., and Davidson H.W.. 2004. Lysosome associated membrane protein 1 (Lamp1) traffics directly from the TGN to early endosomes. Traffic. 5:685–699. 10.1111/j.1600-0854.2004.00212.x [DOI] [PubMed] [Google Scholar]
- Deng Y.P., and Storrie B.. 1988. Animal cell lysosomes rapidly exchange membrane proteins. Proc. Natl. Acad. Sci. USA. 85:3860–3864. 10.1073/pnas.85.11.3860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn K.W., Kamocka M.M., and McDonald J.H.. 2011. A practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol. Cell Physiol. 300:C723–C742. 10.1152/ajpcell.00462.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskelinen E.-L., Tanaka Y., and Saftig P.. 2003. At the acidic edge: emerging functions for lysosomal membrane proteins. Trends Cell Biol. 13:137–145. 10.1016/S0962-8924(03)00005-9 [DOI] [PubMed] [Google Scholar]
- Farías G.G., Guardia C.M., De Pace R., Britt D.J., and Bonifacino J.S.. 2017. BORC/kinesin-1 ensemble drives polarized transport of lysosomes into the axon. Proc. Natl. Acad. Sci. USA. 114:E2955–E2964. 10.1073/pnas.1616363114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuze H.J., Slot J.W., Strous G.J., Hasilik A., and von Figura K.. 1985. Possible pathways for lysosomal enzyme delivery. J. Cell Biol. 101:2253–2262. 10.1083/jcb.101.6.2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goo M.S., Sancho L., Slepak N., Boassa D., Deerinck T.J., Ellisman M.H., Bloodgood B.L., and Patrick G.N.. 2017. Activity-dependent trafficking of lysosomes in dendrites and dendritic spines. J. Cell Biol. 216:2499–2513. 10.1083/jcb.201704068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowrishankar S., Yuan P., Wu Y., Schrag M., Paradise S., Grutzendler J., De Camilli P., and Ferguson S.M.. 2015. Massive accumulation of luminal protease-deficient axonal lysosomes at Alzheimer’s disease amyloid plaques. Proc. Natl. Acad. Sci. USA. 112:E3699–E3708. 10.1073/pnas.1510329112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowrishankar S., Wu Y., and Ferguson S.M.. 2017. Impaired JIP3-dependent axonal lysosome transport promotes amyloid plaque pathology. J. Cell Biol. 216:3291–3305. 10.1083/jcb.201612148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowski G.A., Gatt S., and Horowitz M.. 1990. Acid beta-glucosidase: enzymology and molecular biology of Gaucher disease. Crit. Rev. Biochem. Mol. Biol. 25:385–414. 10.3109/10409239009090616 [DOI] [PubMed] [Google Scholar]
- Griffiths G., Hoflack B., Simons K., Mellman I., and Kornfeld S.. 1988. The mannose 6-phosphate receptor and the biogenesis of lysosomes. Cell. 52:329–341. 10.1016/S0092-8674(88)80026-6 [DOI] [PubMed] [Google Scholar]
- Griffiths G., Matteoni R., Back R., and Hoflack B.. 1990. Characterization of the cation-independent mannose 6-phosphate receptor-enriched prelysosomal compartment in NRK cells. J. Cell Sci. 95:441–461. [DOI] [PubMed] [Google Scholar]
- Harrison K.D., Miao R.Q., Fernandez-Hernándo C., Suárez Y., Dávalos A., and Sessa W.C.. 2009. Nogo-B receptor stabilizes Niemann-Pick type C2 protein and regulates intracellular cholesterol trafficking. Cell Metab. 10:208–218. 10.1016/j.cmet.2009.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenbeck P.J. 1993. Products of endocytosis and autophagy are retrieved from axons by regulated retrograde organelle transport. J. Cell Biol. 121:305–315. 10.1083/jcb.121.2.305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höning S., Griffith J., Geuze H.J., and Hunziker W.. 1996. The tyrosine-based lysosomal targeting signal in lamp-1 mediates sorting into Golgi-derived clathrin-coated vesicles. EMBO J. 15:5230–5239. [PMC free article] [PubMed] [Google Scholar]
- Hunziker W., and Geuze H.J.. 1996. Intracellular trafficking of lysosomal membrane proteins. BioEssays. 18:379–389. 10.1002/bies.950180508 [DOI] [PubMed] [Google Scholar]
- Ishidoh K., and Kominami E.. 2002. Processing and activation of lysosomal proteinases. Biol. Chem. 383:1827–1831. 10.1515/BC.2002.206 [DOI] [PubMed] [Google Scholar]
- Janvier K., and Bonifacino J.S.. 2005. Role of the endocytic machinery in the sorting of lysosome-associated membrane proteins. Mol. Biol. Cell. 16:4231–4242. 10.1091/mbc.E05-03-0213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D.E., Ostrowski P., Jaumouillé V., and Grinstein S.. 2016. The position of lysosomes within the cell determines their luminal pH. J. Cell Biol. 212:677–692. 10.1083/jcb.201507112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J.S., Tian J.H., Pan P.Y., Zald P., Li C., Deng C., Sheng Z.H. Docking of axonal mitochondria by syntaphilin controls their mobility and affects short-term. 2008 doi: 10.1016/j.cell.2007.11.024. [DOI] [PMC free article] [PubMed]
- Klionsky D.J., and Emr S.D.. 2000. Autophagy as a regulated pathway of cellular degradation. Science. 290:1717–1721. 10.1126/science.290.5497.1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Yu W.H., Kumar A., Lee S., Mohan P.S., Peterhoff C.M., Wolfe D.M., Martinez-Vicente M., Massey A.C., Sovak G., et al. . 2010. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell. 141:1146–1158. 10.1016/j.cell.2010.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Sato Y., and Nixon R.A.. 2011. Lysosomal proteolysis inhibition selectively disrupts axonal transport of degradative organelles and causes an Alzheimer’s-like axonal dystrophy. J. Neurosci. 31:7817–7830. 10.1523/JNEUROSCI.6412-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., and Klionsky D.J.. 2004. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell. 6:463–477. 10.1016/S1534-5807(04)00099-1 [DOI] [PubMed] [Google Scholar]
- Levine B., and Kroemer G.. 2008. Autophagy in the pathogenesis of disease. Cell. 132:27–42. 10.1016/j.cell.2007.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., and Lobel P.. 2001. Production and characterization of recombinant human CLN2 protein for enzyme-replacement therapy in late infantile neuronal ceroid lipofuscinosis. Biochem. J. 357:49–55. 10.1042/bj3570049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M.Y., Cheng X.T., Tammineni P., Xie Y., Zhou B., Cai Q., and Sheng Z.-H.. 2017. Releasing Syntaphilin Removes Stressed Mitochondria from Axons Independent of Mitophagy under Pathophysiological Conditions. Neuron. 94:595–610.e6. 10.1016/j.neuron.2017.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobel P., Fujimoto K., Ye R.D., Griffiths G., and Kornfeld S.. 1989. Mutations in the cytoplasmic domain of the 275 kd mannose 6-phosphate receptor differentially alter lysosomal enzyme sorting and endocytosis. Cell. 57:787–796. 10.1016/0092-8674(89)90793-9 [DOI] [PubMed] [Google Scholar]
- Lombardi D., Soldati T., Riederer M.A., Goda Y., Zerial M., and Pfeffer S.R.. 1993. Rab9 functions in transport between late endosomes and the trans Golgi network. EMBO J. 12:677–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzio J.P., Pryor P.R., and Bright N.A.. 2007. Lysosomes: fusion and function. Nat. Rev. Mol. Cell Biol. 8:622–632. 10.1038/nrm2217 [DOI] [PubMed] [Google Scholar]
- Maday S., Wallace K.E., and Holzbaur E.L.. 2012. Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J. Cell Biol. 196:407–417. 10.1083/jcb.201106120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maday S., Twelvetrees A.E., Moughamian A.J., and Holzbaur E.L.. 2014. Axonal transport: cargo-specific mechanisms of motility and regulation. Neuron. 84:292–309. 10.1016/j.neuron.2014.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marwaha R., Arya S.B., Jagga D., Kaur H., Tuli A., and Sharma M.. 2017. The Rab7 effector PLEKHM1 binds Arl8b to promote cargo traffic to lysosomes. J. Cell Biol. 216:1051–1070. 10.1083/jcb.201607085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller C.B., and Enderlein J.. 2010. Image scanning microscopy. Phys. Rev. Lett. 104:198101 10.1103/PhysRevLett.104.198101 [DOI] [PubMed] [Google Scholar]
- Nixon R.A. 2013. The role of autophagy in neurodegenerative disease. Nat. Med. 19:983–997. 10.1038/nm.3232 [DOI] [PubMed] [Google Scholar]
- Nixon R.A., Yang D.S., and Lee J.H.. 2008. Neurodegenerative lysosomal disorders: a continuum from development to late age. Autophagy. 4:590–599. 10.4161/auto.6259 [DOI] [PubMed] [Google Scholar]
- Overly C.C., and Hollenbeck P.J.. 1996. Dynamic organization of endocytic pathways in axons of cultured sympathetic neurons. J. Neurosci. 16:6056–6064. 10.1523/JNEUROSCI.16-19-06056.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padamsey Z., McGuinness L., Bardo S.J., Reinhart M., Tong R., Hedegaard A., Hart M.L., and Emptage N.J.. 2017. Activity-Dependent Exocytosis of Lysosomes Regulates the Structural Plasticity of Dendritic Spines. Neuron. 93:132–146. 10.1016/j.neuron.2016.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson G.H., and Lippincott-Schwartz J.. 2002. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 297:1873–1877. 10.1126/science.1074952 [DOI] [PubMed] [Google Scholar]
- Perlson E., Jeong G.B., Ross J.L., Dixit R., Wallace K.E., Kalb R.G., and Holzbaur E.L.. 2009. A switch in retrograde signaling from survival to stress in rapid-onset neurodegeneration. J. Neurosci. 29:9903–9917. 10.1523/JNEUROSCI.0813-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saftig P. 2005. Lysosomes. Springer; New York: 197pp. [Google Scholar]
- Saftig P., and Klumperman J.. 2009. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat. Rev. Mol. Cell Biol. 10:623–635. 10.1038/nrm2745 [DOI] [PubMed] [Google Scholar]
- Sathyanarayan A., Mashek M.T., and Mashek D.G.. 2017. ATGL Promotes Autophagy/Lipophagy via SIRT1 to Control Hepatic Lipid Droplet Catabolism. Cell Reports. 19:1–9. 10.1016/j.celrep.2017.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard C.J.R., Mehta S.B., and Heintzmann R.. 2013. Superresolution by image scanning microscopy using pixel reassignment. Opt. Lett. 38:2889–2892. 10.1364/OL.38.002889 [DOI] [PubMed] [Google Scholar]
- Sidransky E. 2012. Gaucher disease: insights from a rare Mendelian disorder. Discov. Med. 14:273–281. [PMC free article] [PubMed] [Google Scholar]
- Sun P., Sleat D.E., Lecocq M., Hayman A.R., Jadot M., and Lobel P.. 2008. Acid phosphatase 5 is responsible for removing the mannose 6-phosphate recognition marker from lysosomal proteins. Proc. Natl. Acad. Sci. USA. 105:16590–16595. 10.1073/pnas.0807472105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonderheit A, and Helenius A. 2005. Rab7 associates with early endosomes to mediate sorting and transort of Semliki forest virus to late endosomes. Plos Biology. 3:e233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong Y.C., and Holzbaur E.L.. 2015. Temporal dynamics of PARK2/parkin and OPTN/optineurin recruitment during the mitophagy of damaged mitochondria. Autophagy. 11:422–424. 10.1080/15548627.2015.1009792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Zhou B., Lin M.-Y., Wang S., Foust K.D., and Sheng Z.-H.. 2015. Endolysosomal Deficits Augment Mitochondria Pathology in Spinal Motor Neurons of Asymptomatic fALS Mice. Neuron. 87:355–370. 10.1016/j.neuron.2015.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Sun M., Dwyer N.K., Comly M.E., Patel S.C., Sundaram R., Hanover J.A., and Blanchette-Mackie E.J.. 2003. Differential trafficking of the Niemann-Pick C1 and 2 proteins highlights distinct roles in late endocytic lipid trafficking. Acta Paediatr. Suppl. 92:63–73. 10.1111/j.1651-2227.2003.tb00224.x [DOI] [PubMed] [Google Scholar]
- Zhou B., Cai Q., Xie Y., and Sheng Z.H.. 2012. Snapin recruits dynein to BDNF-TrkB signaling endosomes for retrograde axonal transport and is essential for dendrite growth of cortical neurons. Cell Reports. 2:42–51. 10.1016/j.celrep.2012.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Yu P., Lin M.Y., Sun T., Chen Y., and Sheng Z.H.. 2016. Facilitation of axon regeneration by enhancing mitochondrial transport and rescuing energy deficits. J. Cell Biol. 214:103–119. 10.1083/jcb.201605101 [DOI] [PMC free article] [PubMed] [Google Scholar]