Drosophila melanogaster neural stem cells (neuroblasts) divide asymmetrically by differentially segregating protein determinants into their daughter cells. Wissel et al. use time-resolved transcriptional profiling to identify a v-ATPase/Notch regulatory loop that acts in multiple stem cell lineages both during nervous system development and in the adult gut.
Abstract
Drosophila melanogaster neural stem cells (neuroblasts [NBs]) divide asymmetrically by differentially segregating protein determinants into their daughter cells. Although the machinery for asymmetric protein segregation is well understood, the events that reprogram one of the two daughter cells toward terminal differentiation are less clear. In this study, we use time-resolved transcriptional profiling to identify the earliest transcriptional differences between the daughter cells on their way toward distinct fates. By screening for coregulated protein complexes, we identify vacuolar-type H+–ATPase (v-ATPase) among the first and most significantly down-regulated complexes in differentiating daughter cells. We show that v-ATPase is essential for NB growth and persistent activity of the Notch signaling pathway. Our data suggest that v-ATPase and Notch form a regulatory loop that acts in multiple stem cell lineages both during nervous system development and in the adult gut. We provide a unique resource for investigating neural stem cell biology and demonstrate that cell fate changes can be induced by transcriptional regulation of basic, cell-essential pathways.
Introduction
Stem cells must balance self-renewal and differentiation during development and tissue homeostasis. Understanding how different cell fates are established and maintained is critically important for both developmental biology and cancer research as disruption of this unique balance can result in tumorigenesis or tissue degeneration (Morrison and Kimble, 2006). Generation of different cell fates after a stem cell division can be achieved either stochastically or through an asymmetric cell division (Horvitz and Herskowitz, 1992). When stem cells divide asymmetrically, one daughter cell reproducibly maintains stem cell identity while the other commits to differentiation (Simons and Clevers, 2011). Asymmetric cell division can be attained intrinsically whereby the stem cell segregates cell fate determinants into only one of the two daughter cells. Alternatively, the mitotic spindle of the stem cell is oriented so that after division only one of the two daughter cells continues to receive self-renewal factors released by the stem cell niche (Knoblich, 2008). Ultimately, differential exposure to niche factors or unequal concentrations of segregating determinants need to be translated into distinct and stable cell fates by instructing or repressing particular transcriptional programs. These programs are implemented through very dynamic gene regulatory networks (Gloss et al., 2017). As most of our knowledge about transcriptional changes is based on end-point analysis, a time-resolved overview of these transitional states is essential to fully understand the molecular mechanisms shaping and maintaining the distinct fates of the two daughter cells. In this study, we fill this knowledge gap by establishing high-resolution time-course transcriptome datasets that extend our current understanding of the events occurring after stem cell division.
Drosophila melanogaster larval neuroblasts (NBs) are a well-established model system to study stem cell biology (Doe, 2008; Homem and Knoblich, 2012; Homem et al., 2015). Several types of NBs can be distinguished in the central larval brain based on their division mode (Bello et al., 2008; Boone and Doe, 2008; Bowman et al., 2008). Type I NBs divide into a larger cell that retains NB characteristics and a smaller ganglion mother cell (GMC) that gives rise to two postmitotic neurons or glial cells (see Fig. 1 a). Type II NBs also divide asymmetrically, generating an NB and a smaller intermediate neural progenitor (INP) cell. Newly formed INPs go through defined maturation steps to become transit-amplifying INPs, which undergo three to six asymmetric divisions generating one INP and one GMC that also divides into two neurons or glial cells (Bello et al., 2008; Boone and Doe, 2008; Bowman et al., 2008).
Figure 1.
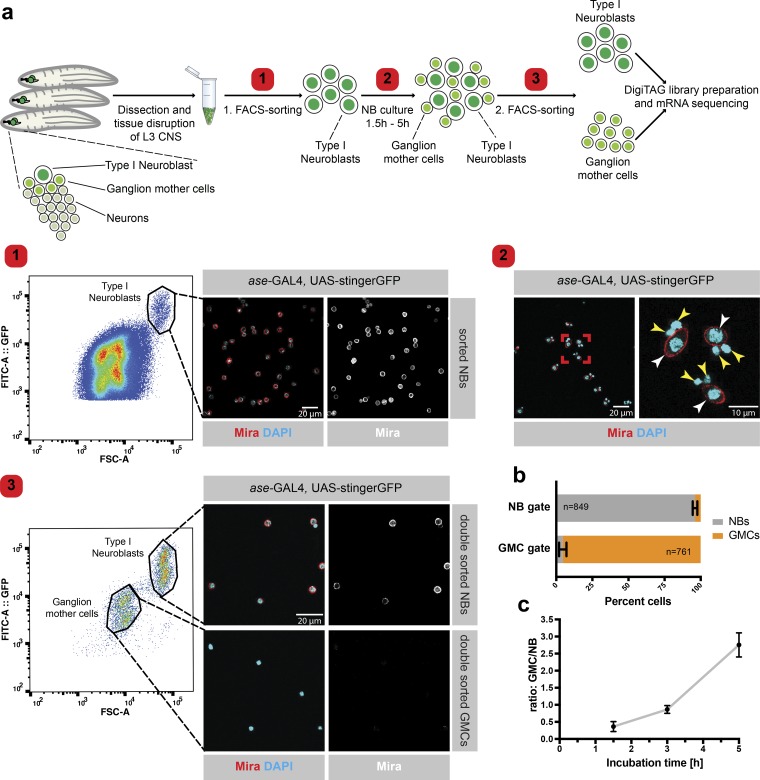
Pure populations of larval NBs and GMCs of different ages can be obtained by FACS. (a) Larval central nervous systems (CNS) expressing a nuclear GFP in a type I NB–specific manner (ase-GAL4 and UAS-stingerGFP) were dissected and the tissue was disturbed into a single-cell solution. NBs were sorted according to cell size and GFP intensity (step 1) and were cultured in Schneider’s medium for 1.5, 3, or 5 h (step 2); white arrowheads indicate NBs, and yellow arrowheads indicate GMCs. The mixture of NBs and newly formed GMCs was subjected to a second FACS sort, displaying two clearly distinct cell populations in the FACS plot. The population containing large cells with a high GFP intensity signal comprises NBs (Mira+), whereas the small cells with the low GFP signal intensity are GMCs. The FACS plot is a representative example of the second FACS sort after 3 h of NB culture (step 3). By using a GAL4 strain harboring both heterozygous and homozygous GAL4 and UAS-stingerGFP, we were able to separate two NB and two GMC populations when plotting the strength of the GFP signal to the size of the cells (forward scatter [FSC]-A). Both subpopulations are equal in size but different in the strength of the GFP signal as heterozygous insertions result in weaker GFP signals than homozygous insertions. (b) Essentially pure populations of NBs and GMCs were obtained after the second FACS sort. n (NB gate) = 849 cells, n (GMC gate) = 761 cells. (c) Increased incubation time between the two consecutive FACS sorts resulted in an increased GMC/NB ratio. n ≥ 3 Experiments. Error bars represent mean ± SD.
NBs and INPs divide asymmetrically in an intrinsic manner through the differential localization of cell fate determinants. Brat, Numb, and Prospero (Pros) are segregated into the GMC to drive a differentiation program. Pros is a transcription factor that activates proneural genes and inhibits cell cycle genes (Choksi et al., 2006), whereas Brat acts as a translational repressor (Sonoda and Wharton, 2001) and Numb inhibits Notch signaling in the GMC by promoting endocytosis of the Notch receptor (Schweisguth, 2004; Couturier et al., 2012). Loss of these cell fate determinants disturbs the balance between differentiation and self-renewal. For example, in a brat mutant, type II NB–generated INPs fail to mature and revert into NB-like cells giving rise to transplantable tumors (Caussinus and Gonzalez, 2005; Bello et al., 2006; Betschinger et al., 2006; Lee et al., 2006; Bowman et al., 2008).
Generation of different cell fates after asymmetric cell division implies several fundamental differences in the biology of the two daughter cells, including their proliferation and cell growth potential. Larval NBs regrow after each cell division to their original size before they continue dividing, whereas GMCs do not alter their cell volume (Homem et al., 2013). NBs and GMCs also differ in their cell cycle times: type I NBs require 1.3 h for each cell division compared with 4.2 h for the final division of GMCs (Bowman et al., 2008; Homem et al., 2013). The molecular machinery that regulates the asymmetric segregation of the cell fate determinants during mitosis is well understood. However, how the asymmetric distribution of the cell fate determinants translates into these fundamental differences between the two daughter cells is less clear (Doe, 2008; Reichert, 2011; Homem and Knoblich, 2012). A regulatory network including deadpan, klumpfuss, worniu, and E(spl)mγ that controls self-renewal in NBs has been identified (Slack et al., 2006; Neumüller et al., 2011; Berger et al., 2012). Nevertheless, the dynamic molecular events stabilizing the distinct fates after asymmetric distribution of the cell fate determinants are poorly understood. To fill this gap, we performed time-resolved transcriptome analysis of NBs and maturing GMCs.
For this, we established a method to purify type I NBs and GMCs at three different time points after their asymmetric cell division. This is possible as cell cycle, cell growth, and maturation times in NBs are highly synchronous, allowing us to derive high-quality transcriptomes by pooling NBs and GMCs isolated at defined times after mitosis. We performed complex enrichment analysis using the Complex Enrichment Analysis Tool (COMPLEAT; Vinayagam et al., 2013) on the transcriptome data to determine protein complexes whose members undergo similar transcriptional changes in NBs and GMCs over time. This analysis highlighted a progressive down-regulation of the vacuolar-type H+–ATPase (v-ATPase) complex in GMCs over time. v-ATPase is a multisubunit proton pump found in the endomembrane system of all eukaryotic cells, where it is essential for the acidification of intracellular organelles, including endosomes and lysosomes. Consequently, v-ATPase plays a crucial role in protein trafficking and degradation and was only recently found to modulate several signaling transduction cascades (Forgac, 2007; Sun-Wada and Wada, 2015; Oot et al., 2017). We show that the v-ATPase complex is required for efficient NB self-renewal and acts in a regulatory loop involving the Notch signaling pathway. Our data indicate that this regulatory loop acts across multiple stem cell lineages at different times of the Drosophila life cycle. In addition, our data provide a resource for investigating neural developmental biology with high temporal resolution and may shed light on the mechanisms that establish the fundamental differences between stem cells and their differentiating sibling.
Results
Pure populations of larval NBs and GMCs of different ages can be obtained by FACS
To understand the temporal changes in NBs and GMCs, we retrieved transcriptomes from cells of different ages. To this end, we cultured FACS-sorted NBs from dissociated brains (Berger et al., 2012; Harzer et al., 2013; Homem et al., 2013) for 1.5, 3, or 5 h as they continued to divide asymmetrically (Fig. 1 a). To collect nearly pure populations of both NBs and GMCs, we subjected the cell suspension containing NBs and newly formed GMCs to a second FACS sort (Fig. 1, a and b; and Fig. S1). In each experiment, we obtained ∼1,000 NBs and, depending on the incubation time, up to 3,000 GMCs with a maximum age of 1.5, 3, or 5 h (Fig. 1 c). Low-input sequencing was achieved by combining transposon-mediated library preparation with molecular barcoding. Adding a unique identifier barcode to the 3′ end of each original cDNA molecule allowed us to extensively amplify our sequencing library without risking misrepresentation by PCR amplification artifacts (Landskron et al., 2018). In sum, the combination of our two-step FACS strategy and low-input sequencing technique allowed us to obtain the transcriptome of NBs and differently matured GMCs.
To assess how the short culture period affected NB gene expression, we performed a differential expression analysis of our 1.5-, 3-, and 5-h NB datasets and identified 180 differentially expressed genes (false discovery rate [FDR] = 0.05, P < 0.05; log2 fold change [log2fc] >2; Fig. S2 a). However, the vast majority of these genes are lowly expressed (Fig. S2, b and c) and more than one third of them have an reads per million mapped reads (RPM) value of <10 at all three time points (Fig. S2 b). Thus, only minor transcriptional changes affecting lowly expressed genes occur in NBs during the culture time, while most moderately or highly expressed genes remain unchanged. Hierarchical clustering of the log2RPM values of the 180 differently expressed genes showed that four out of five clusters were not enriched for any gene ontology (GO) term, indicating that NB culture does not affect a specific biological process within these clusters (Fig. S2 a). Cluster 4 contained predominantly genes that are lowly expressed in 1.5- and 3-h NBs but are up-regulated at the 5-h time point. Genes in this cluster were slightly enriched for the GO terms synaptic signaling and neuron development, indicating that longer culture might induce the expression of neuronal genes in NBs. Altogether, short-term NB culture has modest effects on the NB transcriptome. To avoid artifacts arising from those changes, however, we only compare NB and GMC datasets from identical culture times in the following analyses.
Time-resolved transcriptomics demonstrates gradual fate commitment
2,438 genes were differentially expressed between NBs and GMCs isolated 1.5 h after asymmetric division (FDR = 0.05, P < 0.05; log2fc >1). Over time, the total number of genes expressed unequally between GMCs and NBs increased, reflecting their continuous move toward differentiation (Fig. 2 a and Table S1). 1,028 genes were deregulated between NBs and GMCs at all time points (Fig. 2 b), including the known NB markers deadpan, worniu, and string as well as the known differentiation/neuronal markers dacapo, embryonic lethal abnormal vision, and nerfin-1, confirming the reliability of our datasets (Fig. S2 e). Pros, Inscutable, Cherub, and Staufen RNAs are known to be asymmetrically inherited between NBs and GMCs (Li et al., 1997; Hughes et al., 2004; Landskron et al., 2018). Except for Staufen, all RNAs follow the expected trend, further verifying the high quality of our datasets (Fig. S2 d). Furthermore, we observed higher overlap of deregulated genes between the 1.5–3-h and 3–5-h time points compared with the 1.5–5-h time points, suggesting continuous and dynamic transcriptional changes over the lifetime of a GMC (Fig. 2 b).
Figure 2.
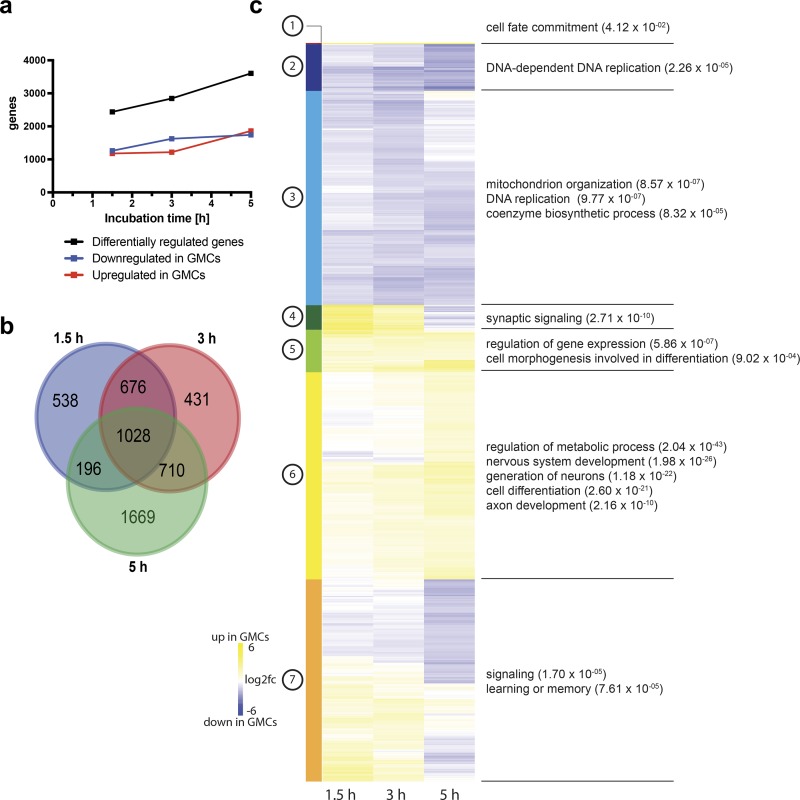
Time-resolved transcriptomics demonstrates gradual fate commitment. (a) The number of differentially expressed genes between NBs and GMCs increases over time. (b) Venn diagram representation of genes deregulated between NBs and GMCs at 1.5-, 3-, and 5-h time points and their respective overlap. (c) Unsupervised hierarchical clustering analysis of gene log2fc between NBs and GMCs at the indicated time point with subsequent GO term analysis of each cluster.
We used unsupervised hierarchical clustering (Fig. 2 c and Table S2) to identify a set of genes whose expression in GMCs compared with NBs was increased throughout the time-course (clusters 1 and 5) and a set of genes expressed equally in NB and GMCs at the 1.5-h time point but up-regulated in GMCs at the later time points (clusters 6). As expected, genes in these clusters were enriched for the GO terms differentiation and neurogenesis. We also identified genes that were either permanently or progressively down-regulated in GMCs compared with NBs (clusters 2 and 3). These clusters were enriched for DNA replication and metabolic processes, consistent with the slower cell cycle and reduced proliferation capacity of GMCs. Other sets (clusters 4 and 7) contained genes expressed more highly in 1.5- and 3-h GMCs and less in 5-h GMCs compared with NBs. Surprisingly, these clusters scored for the GO terms synaptic signaling and learning and could include interesting candidates for further studies of neural differentiation processes. Taken together, the GMC fate program is not completely established immediately after asymmetric cell division but instead is a gradual process that occurs over the entire lifespan of the cell.
Complex enrichment analysis reveals the gradual transcriptional down-regulation of v-ATPase in GMCs
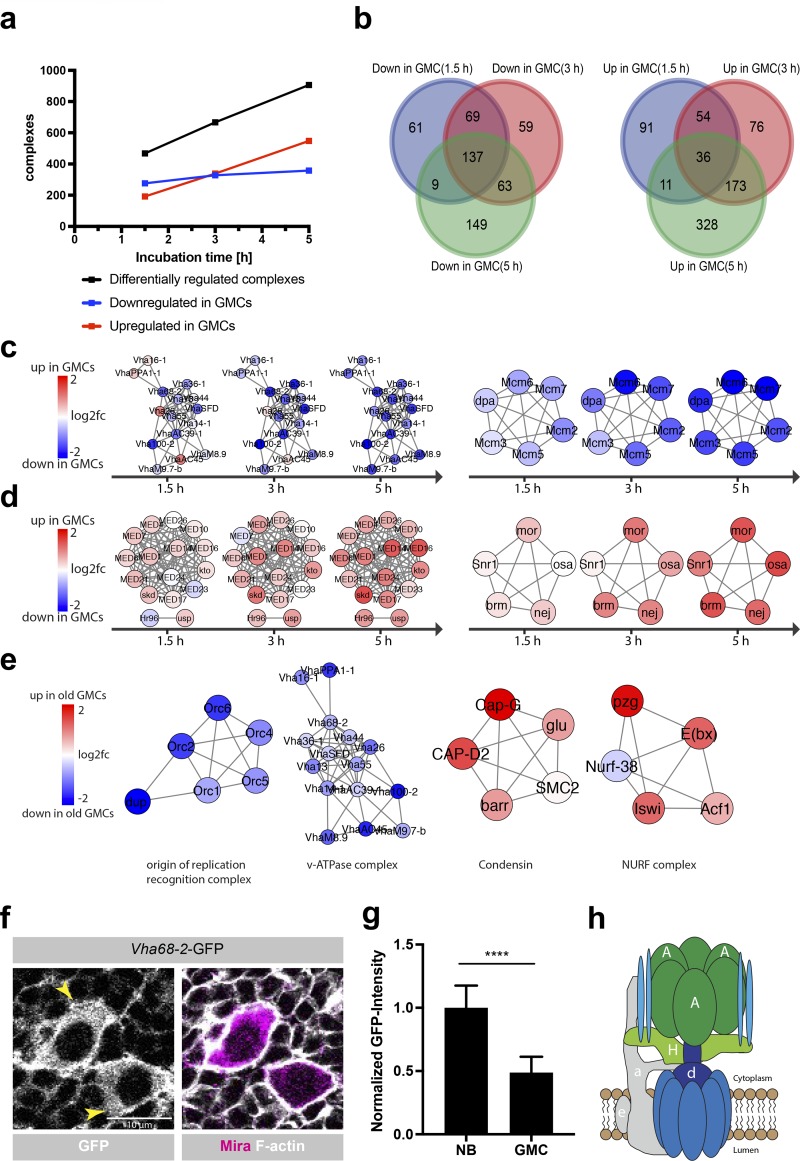
To identify coregulated genes involved in similar molecular processes, we performed a protein complex enrichment analysis using COMPLEAT (Vinayagam et al., 2013). This revealed an increasing number of protein complexes differentially regulated between NBs and GMCs over time (Fig. 3 a). 137 complexes were down-regulated in GMCs compared with NBs at all three time points (Fig. 3 b). This included protein complexes required for DNA replication (e.g., Mcm2–7, GINS, and DNA polymerases), DNA repair (e.g., MSH2/6–BLM–p53–RAD51, PCNA–MutS–α–MutL–α–DNA complex, and BASC complex), regulation of the mitotic cell cycle progression and metabolic processes (e.g., β-oxidation, inosine monophosphate biosynthesis, and gluconeogenesis; Fig. 3 c and Table S3). Interestingly, only 36 complexes were up-regulated at all three time points in GMCs compared with NBs. The highest overlap of up-regulated complexes in GMCs versus NBs was detectable between 3 and 5 h, suggesting that GMCs first down-regulate complexes required for proliferation and subsequently up-regulate new sets of complexes promoting differentiation and their final division (Fig. 3 b). Complexes up-regulated in 3- and 5-h GMCs included the Notch corepressor complex spen–CtBP–Su(H) (Oswald et al., 2005), potentially reflecting an interesting cross talk between transcriptional regulation and post-transcriptional inhibition of Notch by Numb (Schweisguth, 2004; Couturier et al., 2012). In line with the emerging evidence for cell fate control by alternative splicing (Chen et al., 2015; Abramczuk et al., 2017), we found several RNA splicing complexes (e.g., commitment complex and C complex spliceosome) up-regulated in GMCs compared with NBs. Finally, comparing mature GMCs and NBs revealed the up-regulation of several chromatin remodeling complexes (p300–CBP–p270–SWI/SNF and ING2 complex) and the Mediator–DRIP complex in GMCs, which have been shown to act as master coordinators of cell fate determination (Fig. 3 d and Table S3; Eroglu et al., 2014; Homem et al., 2014; Yin and Wang, 2014). Altogether, we identified a core set of protein complexes deregulated between NBs and GMCs that might provide a framework for further analysis of cell fate establishment and maintenance.
Figure 3.
Complex enrichment analysis reveals the gradual transcriptional down-regulation of v-ATPase in GMCs. (a) The number of significantly differentially regulated complexes between NBs and GMCs increases over time. (b) Venn diagram representation of complexes that are down- or up-regulated in GMCs compared with NBs at a specific time point and their respective overlap. (c) Examples of protein complexes that were progressively down-regulated in GMCs compared with NB at all three time points (v-ATPase, left; Mcm2–7 complex, right). (d) Examples of protein complexes that were progressively up-regulated in GMCs compared with NBs (DRIP–Mediator complex, left; p300–CBP–p270–SWI/SNF complex, right). (e) Examples of protein complexes that were either down- (nuclear origin of replication recognition complex; v-ATPase) or up-regulated (Condensin; NURF complex) between young (1.5 h) and old (5 h) GMCs. (f) Endogenous Vha68-2-GFP protein expression. Vha68-2-GFP shows a strong signal in the cytoplasm and at the membrane of NBs, whereas a reduced signal was detectable in GMCs. Larval brains were stained for F-actin and Mira; arrowheads indicate NBs. (g) Quantification of Vha68-2-GFP protein expression levels in NBs and GMCs. n (NB) = 104, n (GMC) = 205 cells. Error bars represent mean ± SD. ****, P < 0.0001; unpaired two-sided Student’s t test. (h) Schematic of the v-ATPase complex (adapted from Forgac, 2007). A, Vha68-2; H, VhaSFD; a, Vha100-2; e, VhaM9.7-b; d, VhaAC39-1.
To identify protein complexes that are deregulated in the process of GMC maturation, we compared the transcriptome of mature (5 h) and young (1.5 h) GMCs. The 343 protein complexes down-regulated in old compared with young GMCs included DNA replication complexes (Mcm2–7, nuclear origin of replication recognition complex, and DNA synthesome complex), the large and small ribosome subunits and complexes of the respiratory chain (Fig. 3 e and Table S3). This is expected as GMCs do not grow in cell size and slow down their cell cycle after asymmetric cell division. Complexes up-regulated during the maturation of GMCs included the Mediator–DRIP complex, chromatin remodeler (p300–CBP–p270–SWI/SNF complex and nucleosome remodeling factor [NURF] complex), the 13S condensin complex and genes related to the mitotic cell cycle G2/M transition DNA damage checkpoint (Fig. 3 e and Table S3). We propose that the increased expression of the 13S condensin complex and genes required for the G2/M transition relates to the final cell division of the GMC into two neurons/glia cells. Additionally, the NURF complex was shown to be essential for the differentiation of melanocyte stem cells (Koludrovic et al., 2015), suggesting the intriguing possibility that NURF could play a similar role in GMC differentiation.
Among the protein complexes we identified, the v-ATPase complex stood out as it is one of the few complexes gradually down-regulated in GMCs over time, both when compared with NBs as well as when compared with each other (Fig. 3, c, e, and h; and Fig. S3, a and b). Asymmetric inheritance of RNAs between NBs and GMCs has been reported previously (Li et al., 1997; Hughes et al., 2004; Landskron et al., 2018). Therefore, we performed FISH for the catalytic v-ATPase subunit Vha68-2. During NB division, no asymmetric mRNA localization was detectable, suggesting that Vha68-2 mRNA is not asymmetrically segregated between NBs and GMCs and that different mechanisms are responsible for v-ATPase down-regulation in GMCs (Fig. S3, c and d). To confirm the transcriptome data, we generated a fly strain where Vha68-2 was tagged with GFP. Vha68-2-GFP was strongly expressed in a punctate manner in the cytoplasm and at the cell membrane of NBs, whereas a significantly reduced Vha68-2-GFP signal was detectable in GMCs (Fig. 3, f and g). Thus, consistent with the transcriptome data, the v-ATPase complex is down-regulated in GMCs after asymmetric cell division.
v-ATPase is required for NB regrowth after asymmetric cell division
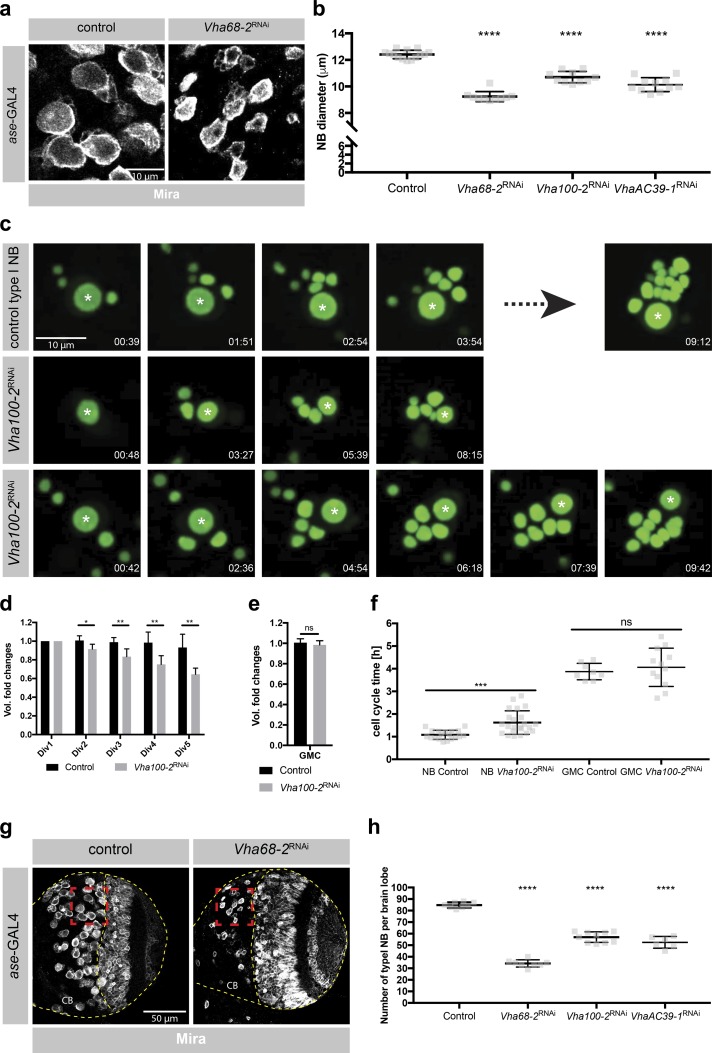
To understand the function of v-ATPase, we inhibited its components by RNAi in type I NBs. Both depletion of the catalytic subunit (Fig. 4, a and b) and inhibition of several other v-ATPase subunits (Fig. 4 b) resulted in a prominent reduction in NB size, suggesting that v-ATPase might be required for cell growth. To further understand this loss of function phenotype, we performed ex vivo live imaging of NBs labeled by nuclear GFP (Homem et al., 2013). It is well established that larval NBs regrow after each cell division to their original size before they continue dividing, while GMCs do not alter their cell volume (Homem et al., 2013). In line with that ex vivo control NBs regrew to their original size after each cell division (Fig. 4, c and d; and Video 1). v-ATPase deficient NBs still divided into two daughter cells of unequal size but did not grow to the same extent as control NBs before the next division (Fig. 4, c and d; and Videos 2 and 3). GMCs generated by control or v-ATPase deficient NBs, instead did not show volume alterations during their lifespan (Fig. 4 e). In addition, cell cycle time in NBs but not in GMCs was prolonged upon v-ATPase knockdown (Fig. 4 f). Thus, v-ATPase is required for cell growth in NBs but not in GMCs, consistent with its down-regulation in GMCs. We also observed a significant decrease in NB number upon v-ATPase knockdown (Fig. 4, g and h). To exclude apoptosis as a potential explanation for the loss of v-ATPaseRNAi NBs, we performed TdT dUTP Nick-End labeling (TUNEL) assays and did not observe any TUNEL-positive NBs upon v-ATPase depletion (Fig. S3 e). This confirms previous research showing that NBs, like other stem cells, can only maintain their stemness if they grow at a minimum rate (Song and Lu, 2011). Thus, v-ATPase promotes NB self-renewal by driving cell cycle progression and NB regrowth after asymmetric cell division.
Figure 4.
The v-ATPase is required for NB regrowth after asymmetric cell division. (a) Vha68-2 inhibition by RNAi caused a reduction in NB size. Images are magnifications of the red boxes in g. Larval brains were stained for the stem cell marker Mira. (b) Quantification of NB diameter upon knockdown of Vha68-2, Vha100-2, or VhaAC39-1. Each data point represents the average type I NB diameter per brain lobe. n (control) = 14, n(Vha68-2RNAi) = 10, n (Vha100-2RNAi) = 10, n (VhaAC39-1RNAi) = 11. (c) Vha100-2 knockdown by RNAi resulted in a defect in NB regrowth after asymmetric cell division and in a prolonged NB cell cycle time. GMC cell cycle times were not altered. Single frames from time-lapse videos of cultured NBs expressing UAS-stingerGFP under the control of ase-GAL4. Asterisks label NBs. Time in hours:minutes. (d) Automated quantification of volume variation with each cell division of control or Vha100-2RNAi NBs. n (control) = 5, n (Vha100-2RNAi) = 5. Div, cell division. (e) Automated quantification of volume variation within the lifespan of GMCs originating either from control or from Vha100-2RNAi NBs. n (control) = 5, n (Vha100-2RNAi) = 5. (f) Quantification of cell cycle times of control or Vha100-2RNAi NBs and their siblings. Each data point represents the duration of one cell cycle. n (NB control) = 19, n (NB Vha100-2RNAi) = 23, n (GMC control) = 8, n (GMC Vha100-2RNAi) = 12. (g) Vha68-2 depletion by RNAi caused a reduction in NB number. Larval brains were stained for Mira. CB, central brain. (h) Quantification of NB number per brain lobe upon knockdown of Vha68-2, Vha100-2, or VhaAC39-1. Each data point represents the number of type I NBs per brain lobe. n (control) = 8, n (Vha68-2RNAi) = 8, n (Vha100-2RNAi) = 10, n (VhaAC39-1RNAi) = 6. Error bars represent mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; unpaired two-sided Student’s t test.
v-ATPase is required for Notch signaling in NBs
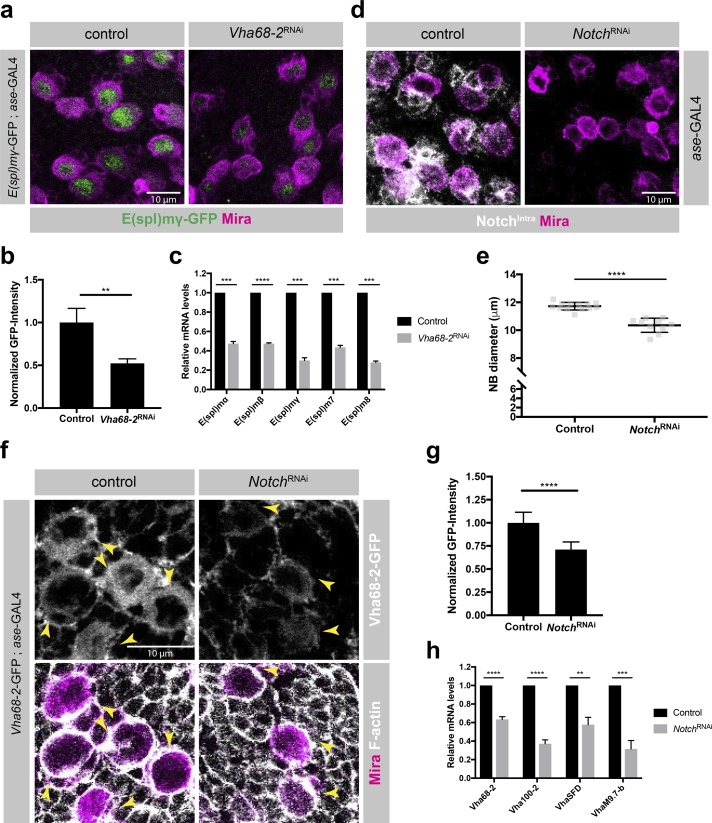
The interplay between v-ATPase and several signaling pathways including TOR, Wnt, Notch, and JNK signaling has been reported previously (Yan et al., 2009; Buechling et al., 2010; Cruciat et al., 2010; Vaccari et al., 2010; Zoncu et al., 2011; Petzoldt et al., 2013; Gleixner et al., 2014). Because our transcriptome datasets showed down-regulation of Notch target gene expression in GMCs compared with NBs (Table S1) and because Notch signaling is essential for NB self-renewal (San-Juán and Baonza, 2011; Song and Lu, 2011), we investigated a potential interaction between v-ATPase and the Notch signaling pathway. Loss of v-ATPase resulted in reduced expression levels of the Notch reporter E(spl)mγ-GFP (Almeida and Bray, 2005), suggesting that v-ATPase is required for an active Notch signaling pathway in NBs (Fig. 5, a and b). Indeed, the mRNA levels for E(spl)mγas well as other Notch targets were down-regulated in FACS-purified Vha68-2RNAi NBs (Fig. 5 c). Furthermore, depletion of Notch by RNAi caused reduced NB diameter, phenocopying the effect of v-ATPase inhibition by RNAi (Fig. 5, d and e). Therefore, v-ATPase is required for an active Notch signaling pathway in NBs.
Figure 5.
Regulatory loop between Notch signaling and v-ATPase orchestrates NB growth and self-renewal. (a) Depletion of Vha68-2 by RNAi resulted in reduced protein expression of the Notch reporter E(spl)mγ-GFP. Larval brains were stained for Mira. (b) Quantification of E(spl)mγ-GFP signal intensity in control and Vha68-2RNAi NBs. n (control) =3, n (Vha68-2RNAi) = 3 brain lobes. (c) Expression levels of Notch target genes in sorted NBs assessed by quantitative RT-PCR upon depletion of Vha68-2 by RNAi. n = 3 experiments. (d) Knockdown of Notch in NBs resulted in decreased NB size. Larval brains were stained for NotchIntra and Mira. (e) Quantification of NB diameter upon Notch depletion by RNAi. Each data point represents the mean type I NB diameter per brain lobe. n (control) = 13, n (NotchRNAi) = 10. (f) Down-regulation of Vha68-2-GFP protein expression upon NotchRNAi. Larval brains were stained for F-actin and Mira. Arrowheads indicate NBs. (g) Quantification of Vha68-2-GFP signal intensity in control and NotchRNAi NBs. n (control) = 14, n (NotchRNAi) = 14 brain lobes. (h) Expression levels of v-ATPase subunits in FACS-purified NBs assessed by quantitative RT-PCR upon Notch depletion by RNAi. n = 4 experiments. Error bars represent mean ± SD. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; unpaired two-sided Student’s t test.
v-ATPase and the Notch signaling pathway form a regulatory loop
Because Notch signaling is active in NBs but not in GMCs, we next asked whether v-ATPase expression in NBs might be driven through a regulatory loop involving the Notch signaling pathway. Interestingly, Vha68-2–GFP protein expression levels were significantly reduced upon Notch depletion (Fig. 5, f and g). Furthermore, sorted Notch-deficient NBs showed transcriptional down-regulation of all probed v-ATPase subunits, suggesting that the Notch singling pathway is either directly or indirectly driving v-ATPase expression in NBs (Fig. 5 h). Altogether, our results suggest that a regulatory loop between the Notch signaling pathway and v-ATPase ensures NB self-renewal.
v-ATPase is required for brat tumor progression
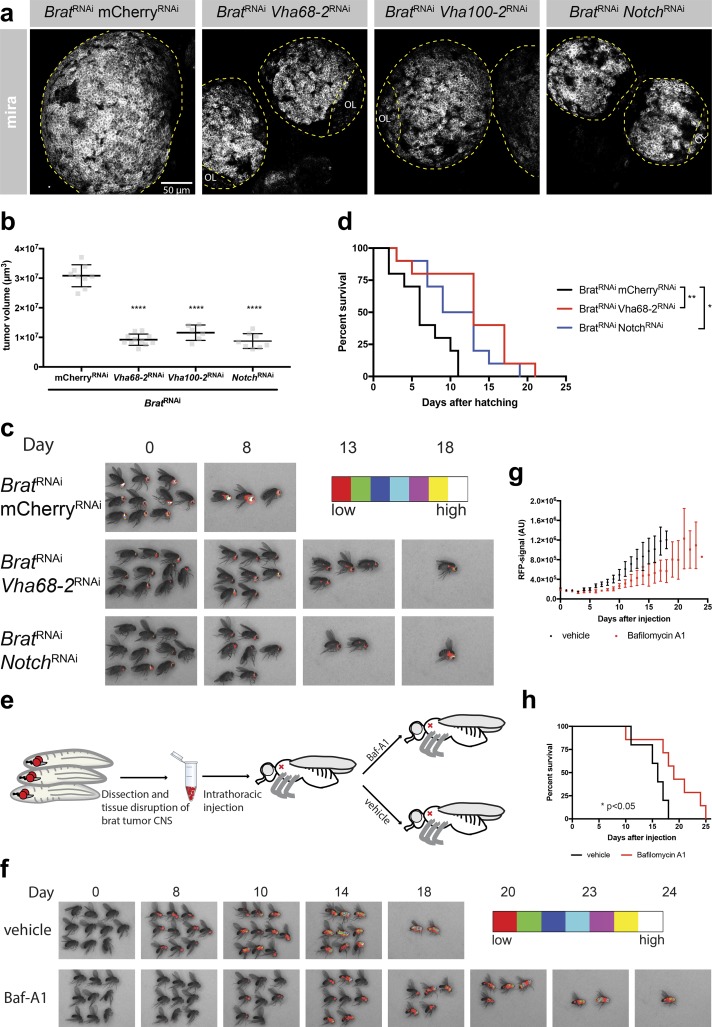
As the Notch pathway is one of the most common signaling pathways in cancer (Yuan et al., 2015) and v-ATPase is considered a potential anticancer target (Stransky et al., 2016), we tested the significance of the connection between v-ATPase and the Notch signaling pathway for tumor development. It is well established that mutations affecting asymmetric NB division (e.g., brat mutations) can result in persistent proliferation of both daughter cells and the formation of lethal, transplantable brain tumors (Caussinus and Gonzalez, 2005; Bello et al., 2006; Betschinger et al., 2006; Bowman et al., 2008; Homem et al., 2015). To test whether deregulation of the Notch/v-ATPase regulatory loop might be implicated in the formation of those tumors, we inhibited both Notch and v-ATPase using RNAi. As has been shown for NotchRNAi (Song and Lu, 2011), v-ATPaseRNAi significantly reduced the brain tumor volume in larvae (Fig. 6, a and b). Like Notch signaling, v-ATPase is required for brat tumor progression in adult flies as it delays the increase of an RFP signal driven by a tumor NB-specific promoter (Fig. 6 c). Consistently, the lifespan of bratRNAi tumor bearing flies is significantly increased upon simultaneous Notch or v-ATPase inhibition (Fig. 6 d). We also tested whether pharmacological inhibition of v-ATPase could rescue tumor progression. For this, we transplanted 1,000 brat tumor cells each into WT host flies and then injected the v-ATPase inhibitor bafilomycin A1 (Baf-A1; Fig. 6 e). Flies treated with a single dose of Baf-A1 showed delayed tumor progression (Fig. 6, f and g), resulting in a significant delay in metastasis-induced lethality (Fig. 6 h). These results indicate that v-ATPase is required for Notch-dependent brat tumor progression.
Figure 6.
v-ATPase is required for brat tumor progression. (a) Knockdown of either the v-ATPase or Notch in a bratRNAi background results in reduced larval brain tumor volume compared with mCherryRNAi control. Larval brains were stained for Mira. OL, optic lobe. (b) Quantification of the brain tumor volume upon knockdown of brat in the combination with mCherryRNAi, Vha68-2RNAi, Vha100-2RNAi, or NotchRNAi. Each data point represents the volume of one brain lobe. n (mCherryRNAi) = 9, n (Vha68-2RNAi) = 11, n (Vha100-2RNAi) = 6, n (NotchRNAi) = 8. Error bars represent mean ± SD. ****, P < 0.0001; unpaired two-sided Student’s t test. (c) Vha68-2RNAi or NotchRNAi slowed down brat tumor progression and increased the survival of adult flies compared with bratRNAi, mCherryRNAi control. mCherryRNAi does not target the RFP that we used to label tumor cells. Pictures of real-time tumor metastasis burden; RFP signal is displayed in false colors (BRGBCMYW). (d) Kaplan–Meier plot showing the increased survival of adult flies upon Vha68-2RNAi or NotchRNAi compared with mCherryRNAi in a bratRNAi background. n = 10 flies per genotype. (e) Cartoon depicting transplantation of brat tumors into adult host flies and subsequent treatment of these flies. CNS, central nervous system. (f) Single injection of Baf-A1 slows down brat tumor progression and prolongs fly survival compared with vehicle control. Pictures of real-time tumor metastasis burden; RFP signal is displayed in false colors (BRGBCMYW). First detectable metastasis appeared 3 d later in Baf-A1–treated flies in comparison with nontreated flies. (g) RFP signal intensity quantification of whole flies; RFP signals of all flies on one picture were averaged. n = 3 experiments. (h) Kaplan–Meier plot showing the increased survival of Baf-A1–treated flies compared with vehicle-treated flies. n ≥ 9 flies per genotype. *, P < 0.05; **, P < 0.01; Mantel–Cox test.
To directly test the significance of the regulatory loop between v-ATPase and the Notch signaling pathway for tumor development, we expressed a constitutively active Notch in NBs, which resulted in brain tumor formation as previously reported (Wang et al., 2006; Bowman et al., 2008). Additional loss of v-ATPase resulted in significantly reduced brain tumor volume (Fig. S4, a and b), suggesting that v-ATPase is required for Notch-driven tumor progression.
v-ATPase depletion induces overproliferation of intestinal stem cells (ISCs)
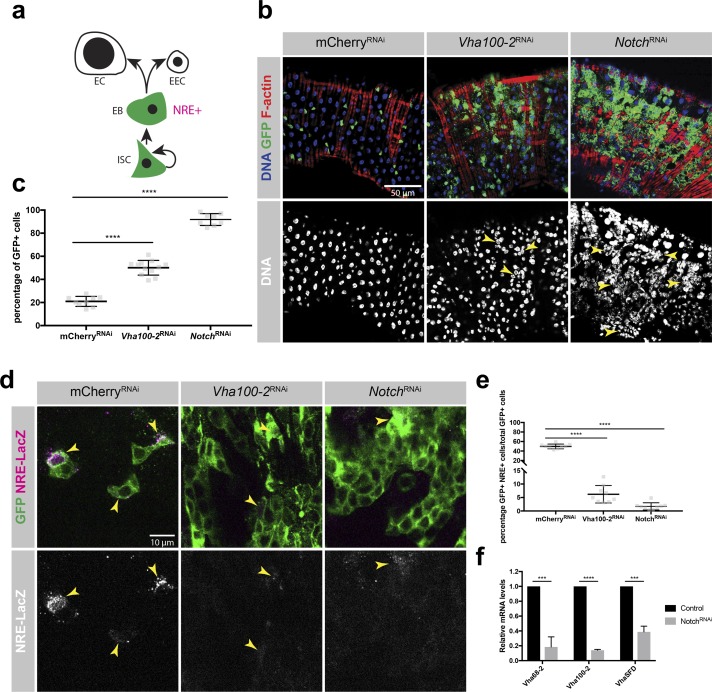
v-ATPase could be required for brat tumor progression either because of its basic cell-essential housekeeping function (Allan et al., 2005; Blomen et al., 2015) in protein trafficking and degradation or more specifically because of its role in Notch signaling. To distinguish those possibilities, we investigated the role of v-ATPase in Drosophila ISCs. ISCs maintain the adult Drosophila midgut by repeated asymmetric cell divisions (Nászai et al., 2015). ISCs self-renew and give rise to enteroblasts (EBs) that differentiate into either polyploid enterocytes (ECs) or enteroendocrine cells (Fig. 7 a). Unlike in NBs, where NotchRNAi causes underproliferation, loss of Notch in ISC lineages of the posterior midgut (NotchRNAi driven by esg-GAL4) results in the overproliferation of ISC/EB-like cells (Ohlstein and Spradling, 2006). Interestingly, depletion of v-ATPase in ISCs and EBs caused overproliferation rather than loss of esg+ ISC/EB-like cells at the expense of ECs in the posterior midgut (Fig. 7, b and c). Loss of either v-ATPase or Notch resulted in significantly reduced expression levels of the Notch reporter NRE-LacZ (Fig. 7, d and e; Furriols and Bray, 2001; Lucchetta and Ohlstein, 2017). Thus, v-ATPase promotes Notch signaling in stem cell lineages, resulting in distinct consequences for intestinal and neural stem cells. Furthermore, sorted Notch-deficient esg+ ISC/EB-like cells showed transcriptional down-regulation of probed v-ATPase subunits, indicating that the Notch signaling pathway drives v-ATPase expression in ISCs (Fig. 7 f).
Figure 7.
v-ATPaseRNAi induces ISC/EB overproliferation. (a) Drosophila ISCs divide asymmetrically to self-renew and generate EBs. EBs differentiate to either ECs or enteroendocrine cells (EECs). Esg is expressed in ISCs and EBs; the Notch reporter NRE marks EBs. (b) Depletion of either Vha100-2 or Notch in ISCs by esg-GAL4 induced overproliferation of esg+ ISC/EB-like cells in the expanse of ECs in the posterior midgut (arrowheads). v-ATPase seems to promote Notch signaling in stem cell lineages, resulting in distinct consequences for ISCs and neural stem cells. Green represents esg-GAL4, UAS-GFP visualized by anti-GFP staining. (c) Percentage of esg+ ISC/EB-like cells per posterior midgut image. n (mCherryRNAi) = 9, n (Vha100-2RNAi)=12, n (NotchRNAi) = 8. (d) Depletion of either Vha100-2 or Notch by esg-GAL4 results in reduced expression levels of the Notch reporter NRE-LacZ. NRE+ cells are marked by arrowheads. Green represents esg-GAL4, UAS-GFP visualized by anti-GFP staining. (e) Quantification of the percentage of NRE+ cells among esg+ cells per midgut image. n (mCherryRNAi) = 9, n (Vha100-2RNAi) = 9, n (NotchRNAi) = 9. (f) Expression levels of v-ATPase subunits in sorted esg+ ISC/EB-like cells assessed by qRT-PCR upon depletion of Notch. n = 3 experiments. Error bars represent mean ± SD. ***, P < 0.001; ****, P < 0.0001; unpaired two-sided Student’s t test.
Taken together, our data indicate that the interplay between v-ATPase and the Notch signaling pathway accounts for reduced brat tumor progression upon v-ATPase knockdown. Furthermore, we demonstrated that the regulatory loop between v-ATPase and Notch is not exclusive to the brain but acts in other stem cells as well.
Discussion
Our results establish a method for quantifying the transcriptional changes occurring in the two daughter cells of an asymmetric stem cell division over time. We purified NBs and GMCs at different time points during their development, which enabled us to follow transcriptional changes that occur during the process of differentiation upon asymmetric cell division with high temporal resolution.
Previous studies have successfully described transcriptome dynamics during differentiation in several mammalian stem cell systems, including neural, cardiac, and pancreatic tissue (Wu et al., 2010; Fathi et al., 2011; Piccini et al., 2016; Huang et al., 2017). These studies followed the differentiation of stem cells into specified cell types over days or weeks, which allowed the identification of optimal cell culture conditions for differentiation or to determine sets of genes or pathways required for the fate commitment of specific cell types. Our study complements these experiments by providing a highly time-resolved transcriptome analysis that assesses the fast changes occurring in differentiating daughter cells after asymmetric cell division. The combination of our time-resolved transcriptomes and the protein complex enrichment analysis lays the foundation for a more targeted search for factors and protein complexes that are required to stabilize the NB or GMC fate. Multiple protein complexes known for their role in fate regulation in several distinct species were enriched in our dataset, providing the potential applicability of this knowledge to other stem cell systems.
We identified the v-ATPase complex as one of the strongest and most consistently differently expressed complexes between NBs and GMCs over time. Our study reveals a regulatory loop between v-ATPase and the Notch signaling pathway in NBs. This interplay ensures the balance between self-renewal and differentiation and therefore constitutes an additional and novel layer of Notch signal pathway regulation in NB lineage development. The requirement for Notch signaling during G1/S transition in neural stem cells (Borghese et al., 2010) could explain the prolonged NB cell cycle time observed upon v-ATPase knockdown. v-ATPase expression by the Notch signaling pathway is further supported by transcriptome data showing that the expression of all v-ATPase subunits are more highly expressed in the Notch-active ECs compared with ISCs (Dutta et al., 2015). Our experiments in the adult ISC system are also conceptually surprising: reducing the expression of a protein complex that was initially considered as a “housekeeping” complex was able to induce an overproliferation of stem-like cells. This highlights the role of the v-ATPase in cellular signaling and tissue homeostasis.
Previous studies suggested that v-ATPase acts in endosomes to create an acidic environment required for efficient Notch S3 cleavage (Yan et al., 2009; Vaccari et al., 2010; Lange et al., 2011). The S3 cleavage process generates the free Notch intracellular domain that translocates to the nucleus to drive target gene expression (Schweisguth, 2004). Although it is likely that the v-ATPase plays a similar role during Notch signaling in NBs, it is less clear what drives v-ATPase expression in NBs but not in GMCs. In other tissues, either Rbcn-3 or the transcription factor Mitf can induce the expression of v-ATPase subunits (Yan et al., 2009; Zhang et al., 2015; Bouché et al., 2016; Tognon et al., 2016). In Drosophila NBs, however, neither loss nor overexpression of Mitf nor loss of Rbcn-3 had any effect on self-renewal capacity (unpublished data). Our results indicate that the Notch pathway is driving v-ATPase expression in NBs. This effect seems to be indirect, as v-ATPase is not among the targets suggested by chromatin immunoprecipitation analysis of the Notch-binding protein Su(H) in hyperplastic Drosophila larval brains (Zacharioudaki et al., 2015). Consistently, prominent Notch targets such as E(spl) act as transcriptional repressors (Paroush et al., 1994) that could indirectly influence v-ATPase in NBs and GMCs.
Interestingly, reduced v-ATPase expression in NBs caused them to acquire GMC-specific characteristics including a prolonged cell cycle time and a reduced proliferation and cell growth potential. This raises the exciting possibility that overexpression of the v-ATPase complex might revert GMCs into NB-like cells as does overexpression of E(spl)mγ (Berger et al., 2012). As the v-ATPase complex contains 15 core subunits (many of which are gradually down-regulated), such a gain-of-function study would be technically very challenging. We nonetheless generated transgenic Drosophila lines driving the overexpression of the three most strongly down-regulated v-ATPase subunits. However, even upon simultaneous overexpression of those three transgenes in both type I and type II NBs and GMCs, self-renewal and differentiation behavior was not detectably changed (unpublished data). This result could be explained by an uneven stoichiometry of all the functional v-ATPase complex subunits. Alternatively, additional factors not present in GMCs could be required for v-ATPase to be active; v-ATPase activity was shown to be regulated by a reversible assembling processes of the two major domains V0 and V1 depending on nutrients response, cell maturation, hormones, and growth factors (Oot et al., 2017).
The Notch pathway is one of the most commonly activated signaling pathways in cancer (Yuan et al., 2015). Our data show that inhibition of the v-ATPase either genetically or chemically impairs proliferation capacity of the Notch signaling-dependent brat tumors. Consistently, cell proliferation can be reduced in Notch-addicted breast tumor cell lines upon v-ATPase inhibition (Kobia et al., 2014). Based on this and other research, v-ATPase is considered as a potential novel anticancer target (Stransky et al., 2016). We demonstrated that reduction of v-ATPase expression in ISCs and EBs primarily affects Notch signaling, suggesting that the regulatory loop involving v-ATPase and the Notch pathway might also be critical for brat tumor progression. Investigating whether a similar pathway operates in human tumors as well might result in important mechanistic insight into the mechanism of action for v-ATPase inhibiting antitumor compounds.
In conclusion, we provide a time-resolved transcriptome dataset that represents a significant resource for investigating new mechanisms of neural stem cell biology and might help to explain how the fundamental differences between stem cells and their differentiating siblings are established.
Materials and methods
Fly strains and RNAi analysis
The following Drosophila stocks were used: UAS-Vha68-2RNAi (34390; Vienna Drosophila Resource Center [VDRC] and BL34582; Bloomington Drosophila Stock Center), UAS-Vha100-2RNAi (30297; VDRC), UAS-VhaAC39-1RNAi (20950; VDRC), E(spl)mγ-GFP (Almeida and Bray, 2005), NRE-LacZ; esg-GAL4, UAS-GFP, tubulin-GAL80ts (Lucchetta and Ohlstein, 2017), UAS-NotchRNAi (100002; VDRC), UAS-NotchΔECD (Larkin et al., 1996), UAS-bratRNAi (31333; VDRC), and UAS-mCherryRNAi (BL35785). The following GAL4 driver lines were used: UAS-dcr2; ase-GAL4, UAS-stingerGFP (Barolo et al., 2000), ase-GAL4 (Zhu et al., 2006), UAS-dcr2; wor-GAL4, ase-GAL80 (Neumüller et al., 2011), UAS-stingerRFP (Homem et al., 2014), and esg-GAL4, UAS-mCD8::GFP, tubulin-GAL80ts (Goulas et al., 2012).
Stock generated in this study was Vha68-2-GFP. gRNA was cloned into pU6-BbsI-chiRNA (45946; 100 ng/μl; Addgene) and coinjected with the donor plasmid (250 ng/μl) into act>Cas9 embryos. The gRNA used was GGAGGACTAGAGACCGCGC.
Fly crosses were set up at 25°C for 24 h and then shifted to 29°C. For experiments in Fig. 7 (b–f), fly crosses were set up and reared at 18°C. 3-d-old female flies were shifted to 29°C for 6 d. For the brat rescue, double RNAi crosses were set up and reared at 29°C. Flies were collected 2 d after eclosion and kept at 29°C.
Antibodies and immunohistochemistry
Larval brains were dissected in PBS and fixed at RT in 5% PFA in PBS for 20 min. Brains were washed three times with 0.1% Triton X-100 in PBS (PBST) and incubated 1 h in blocking solution (1% normal goat serum in PBST). Incubation with primary antibodies in blocking solution was performed overnight at 4°C. Brains were washed three times with PBST, incubated for 2 h at RT with secondary antibody, washed five times with PBST, and mounted in Vectashield mounting medium (containing DAPI; Vector Labs).
Dissected guts were fixed in PBS containing 4% PFA for 30 min at RT. Guts were washed with PBS and blocked in PBS containing 0.05% of Tween-20 and 5% of normal goat serum (blocking solution) for 1 h. Guts were then incubated with primary antibodies (contained in blocking solution) overnight at 4°C. Guts were then washed three times for 15 min in PBS containing 0.05% of Tween-20 (washing solution) and labeled with secondary antibodies (contained in blocking solution) overnight at 4°C. Finally, guts were washed three times for 15 min in washing solution and mounted in a solution of Vectashield/DAPI (Vector Labs).
Antibodies used in this study were rabbit anti-Mira (1:250; Betschinger et al., 2006), guinea pig anti-Mira (1:200; Eroglu et al., 2014), chicken anti-GFP (1:500; ab13970; Abcam), mouse anti-Notchintra (1:1,000; C17.9C6; Developmental Studies Hybridoma Bank), mouse anti–β-galactosidase (1:100; Z378B; Promega), mouse anti-Dacapo (1:100; AB_10805540; Developmental Studies Hybridoma Bank), rat anti-Elav (1:100; 7E8A10; Developmental Studies Hybridoma Bank), guinea pig anti-Dpn (1:1,000; Eroglu et al., 2014), mouse anti-Pros (1:100; Developmental Studies Hybridoma Bank), and Alexa Fluor 568 phalloidin (A12380; Invitrogen).
Secondary antibodies were coupled to Alexa Fluor 405, Alexa Fluor 488, Alexa Fluor 568, and Alexa Fluor 647 (Invitrogen; all used in 1:500 dilution). An in situ cell death detection kit (TMR red; 12156792910; Roche) was used for the TUNEL assay.
Microscopy and in vitro live imaging
Immunofluorescence images were acquired at room temperature using ZEN 2011 software (Zeiss) on an LSM780 microscope (Zeiss) equipped with a GaAsP detector and 25×/0.8 Plan Apochromat or 40×/1.4 enhanced chemiluminescence Plan Apochromat oil differential interference contrast or 40×/1.3 enhanced chemiluminescence Plan-Neofluar oil differential interference contrast objectives (Zeiss).
In vitro live imaging of cultured cells was performed using an UltraView Vox spinning-disk confocal system (PerkinElmer) installed on an AxioObserver Z1 microscope (Zeiss). Images were recorded with an Hamamatsu electron-multiplying charge-coupled device 9100-13 camera using 40×/1.3 enhanced chemiluminescence Plan Neofluar lens (Zeiss). Acquisition of video sequences was done with the Volocity 3D image software (PerkinElmer). Multiple positions were acquired simultaneously. At each position, z stacks were captured every 3 min. Collected images were deconvolved using Huygens deconvolution suite (SVI). Nuclei volumes and cell cycle times were automatically analyzed using Definiens as described previously (Homem et al., 2013, 2014).
FISH
FISH was performed as previously described (Landskron et al., 2018) with minor changes. Briefly, L3 brains were fixed at RT for 40 min in 5% PFA. Brains were washed three times with PBST and then permeabilized in 70% ethanol overnight at 4°C. Ethanol was removed, and brains were incubated at 37°C for 5 min in 400 µl washing buffer (2× SSC and 10% formamide). Upon removal of the washing buffer, brains were incubated in the dark in 100 µl hybridization buffer (1 mg/ml Escherichia coli tRNA, 2 mM vanadyl ribonucleoside complex, 200 nM BSA, 2× SSC, 10% formamide, and 100 mg/ml dextran sulfate) including the FISH probes at 37°C for 6 h. Brains were washed twice with washing buffer, each time for 30 min at 30°C and mounted in 2× SSC. Vha68-2 FISH probes were labeled with Quasar 570 dye and designed using the Stellaris probe designer (Stellaris Biosearch Technologies).
Against Vha68-2 mRNA (labeled with Quasar 570 dye), we used 5′-CCAACTCGTACATAGCTGAT-3′, 5′-ACCGGATCTCCGACAGTTAC-3′, 5′-GAAAGAGGCTTGCCGGTACG-3′, 5′-AAAGATGCTGCCCATGATAC-3′, 5′-AGCTCGTTAATGTCCTTCAG-3′, 5′-TGGGGATGTAGATGGATTCG-3′, 5′-GACAAACTGGGCACGTTCAC-3′, 5′-TTGACGTTCAGGGGGTTGAA-3′, 5′-TTGACCAGAGTGTTCTCATG-3′, 5′-GGGGTTCACAATCATCTTGT-3′, 5′-TGATCTCTCCATCGAACTCG-3′, 5′-CACACCTGCAACATGGTGTG-3′, 5′-GAAGAGCGAGTCGAGCACAC-3′, 5′-AACCGAAAGCTCCGGGAATG-3′, 5′-CTGCGAGATCACAGTCTTGC-3′, 5′-ATCGGAGTTGGAGTACTTGG-3′, 5′-CGCAACCGACGTAGATGATG-3′, 5′-TACCTCAGACATCTCGTTAC-3′, 5′-TTCATGATGGACTCGGTGAC-3′, 5′-CAGGCATGTTGGAGGTGTTG-3′, 5′-GATACCAGTGTAGATGGAGG-3′, 5′-ATCACGGAAGTATTCGGACA-3′, 5′-TCATGGACACGTTGTAACCC-3′, 5′-CAACGGGAGGTGGAATCAGC-3′, 5′-AATTTCACGAAGAGCCTCAG-3′, 5′-AGGCATCTCAGCGAGACGAC-3′, 5′-TCGTAGAAGGAGGCCAGACG-3′, 5′-GGTTACCCAAGCACTTAACG-3′, 5′-GAGACACAGCTCCGACAATG-3′, 5′-CACGGGATCGGAGAAGTCAC-3′, 5′-CAGAACACCTGCACGATACC-3′, 5′-GGCCAACTTCTTGTCGAGAC-3′, 5′-CAGTTGATCGAGGGGAAGTG-3′, 5′-GCATGTACTTCGAGTAGGAG-3′, 5′-TCATAGAAGTCATCCAGAGC-3′, 5′-CACGAATTCGGGGAAGTTCT-3′, 5′-GATCTCCTTGACCTTGGTAC-3′, 5′-TGCACGATCTCAGACAGATC-3′, 5′-AGCGTGATCTTGTCGGTTTC-3′, 5′-AGGAAATCGTCCTTCAGCAG-3′, 5′-GATCGTACGAGGAGTAGGAG-3′, 5′-GTCTTGTAGAAGGGGCAGAA-3′, 5′-GATGATGTTCCTCAACATGC-3′, 5′-GACGGGCCATGTCGTAGAAG-3′, 5′-TTCTCAGACTGAGCCGTAGA-3′, 5′-GAATCACGTTCCAGGTGATC-3′, 5′-TACATAATGTTGCCCATTGC-3′, and 5′-GAACTTCATGGATGACAGCT-3′.
Cell dissociation, FACS, sample preparation, and RNA sequencing
Type I NBs were sorted as described previously with minor modifications (Berger et al., 2012; Harzer et al., 2013). Briefly, third-instar larva were washed in PBS, brains were dissected in Schneider’s medium (21720024; Gibco) and collected in Rinaldini’s solution at room temperature. Brains were dissociated in a dissociation solution (collagenase I [C0130; Sigma-Aldrich] and papain [P4762; Sigma-Aldrich]; final concentration 1 mg/ml in Rinaldini’s solution) for 20 min at 25°C. Brains were washed twice in Rinaldini’s solution and disrupted manually in supplemented Schneider’s medium. NBs were sorted into supplemented Schneider’s medium with a FACSAriaIII machine (BD) according to cell size and GFP intensity and incubated for respective times (1.5, 3, or 5 h) at 25°C. 20 min before the end of the incubation time, collagenase I and papain (Sigma-Aldrich) solutions were added (final concentration 1 mg/ml), and cells were pipetted up and down to dissociate NBs and newly formed GMCs. NBs and GMCs were sorted according to cell size and GFP intensity.
For cell staining, cells were sorted on coated glass-bottomed dishes and stained as described previously (Berger et al., 2012). For RNA isolation, NBs and GMCs were sorted directly in TRIzol LS (10296010; Invitrogen).
Sample preparation and RNA sequencing (DigiTAG)
Per experiment, total RNA from 1,000 to 3,000 FACS-sorted NBs and GMCs was isolated by TRIzol purification. We analyzed three independent biological replicates for each NB time point and two replicates for each GMC time point (Gene Expression Omnibus accession number GSE104049). Reverse transcription and enrichment for mRNAs was performed by incubating total RNA with 50 U Superscript III Reverse Transcriptase (18080044; Invitrogen), oligo(dT)20, 10 U RNaseOUT (Invitrogen), dNTPs, first strand buffer, MgCl2, and DTT for 50 min at 50°C and subsequently for 5 min at 85°C. To generate the second-strand cDNA, second-strand buffer (Invitrogen), dNTPs, RNase H, and DNA polymerase I (Invitrogen) were added to the first-strand cDNA and incubated for 2 h at 16°C. Double-stranded cDNA was purified using magnetic beads (AMPure XP beads; Beckman Coulter). Purified cDNA was simultaneously fragmented and tagged with adapter sequences by using the Nextera DNA Library Preparation Kit (Illumina) for 5 min at 55°C. After another purification step (AMPure XP beads; Beckman Coulter), tagmented cDNA was PCR amplified by Phusion HF master mix (Thermo Fisher Scientific), 20× Eva Green (Biotium), Nextera primers mix, Index 2 primers (N501-N506 for multiplexing), and modified Index 1 primers. The latter contains a random 8-mer molecular barcode that allowed us to extensively amplify our sequencing library without running into the risk of misrepresentations by PCR amplification artifacts. Purified libraries were subjected to 50-bp single-end sequencing on a Hiseq 2000 platform (Illumina).
Transcriptome data analysis
Alignment
Unstranded reads were screened for ribosomal RNA by aligning with BWA (v0.7.12; Li and Durbin, 2009) against known rRNA sequences (RefSeq). The rRNA subtracted reads were aligned with TopHat (v2.1.1; Kim et al., 2013) against the Drosophila genome (FlyBase r6.12). Introns between 20 and 150,000 bp are allowed, which is based on FlyBase statistics. Microexon-search was enabled. Additionally, a gene model was provided as GTF (FlyBase r6.12).
Deduplication
Reads arising from duplication events are marked as such in the alignment (SAM/BAM files) as follows. The different tags are counted at each genomic position. Thereafter, the diversity of tags at each position is examined. First, tags are sorted descending by their count. If several tags have the same occurrence, they are further sorted alphanumerically. Reads sharing the same tag are sorted by the mean PHRED quality. Again, if several reads have the same quality, they are further sorted alphanumerically. Now the tags are cycled through by their counts. Within one tag, the read with the highest mean PHRED quality is the unique correct read and all subsequent reads with the same tag are marked as duplicates. Furthermore, all reads that have tags with one mismatch difference compared the pool of valid read tags are also marked as duplicates.
Summarization
Small nuclear RNA, rRNA, tRNA, small nucleolar RNA, and pseudogenes are masked from the GTF (FlyBase r6.12) with subtractBed from bedtools (v2.26.0; Quinlan and Hall, 2010). The aligned reads were counted with HTSeq (v0.6.1; intersection-nonempty), and genes were subjected to differential expression analysis with DESeq2 (v1.12.4; Love et al., 2014).
Hierarchical clustering analysis
Genes are filtered by the indicated log2fc and an adjusted P value < 0.05 in at least one pairwise comparison. In addition, a minimal expression of 5 RPM in at least one condition was required. The log2RPM (Fig. S2 a) or log2fc (Fig. 2 c) are hierarchically clustered (“euclidean” and “complete”) and the tree cut into five (Fig. S2 a) or seven (Fig. 2 c) clusters (different cluster numbers were tested; Kolde, 2015). GO analysis was performed with FlyMine (Lyne et al., 2007).
FACS sorting of esg+ ISCs/EBs
FACS of esg+ cells was performed as previously described (Dutta et al., 2013). Briefly, after dissection of Drosophila midguts, the tissue was disrupted by elastase treatment (final concentration of 1 mg/ml for 1 h at 27°C). Guts were washed with PBS and pipetted up and down to dissociate the tissue. After centrifugation (20 min at 300g and 4°C), cell pellet was resuspended in PBS and sorted into TRIzol LS (Invitrogen) based on GFP intensity and cell size.
Quantitative RT-PCR analysis
First-strand cDNA was generated using random primers on TRIzol-extracted total cell RNA. Quantitative PCR was done using IQ SYBR Green Supermix on a CFX96 cycler (Bio-Rad Laboratories). Expression of each gene was normalized to Act5c (NBs) or RpL32 (ISCs/EBs), and relative levels were calculated using the 2-ΔΔCT method (Livak and Schmittgen, 2001). The following primer pairs were used: Act5c, 5′-AGTGGTGGAAGTTTGGAGTG-3′ and 5′-GATAATGATGATGGTGTGCAGG-3′; RpL32, 5′-CGCTTCAAGGGACAGTATCTG-3′ and 5′-AAACGCGGTTCTGCATGAG-3′; E(spl)mα, 5′-CTATGCCGAGATCGATGAGAAC-3′ and 5′-GAAGCGAACTGGGACATAGAC-3′; E(spl)mβ, 5′-GTGACCATAGCTTGATCCTCTAC-3′ and 5′-TTGGCCATCGTCTCAACTAC-3′; E(spl)mγ, 5′-CTGGACGAGCTAAAGGATCTTATG-3′ and 5′-GTTCCAGGATATCGGCTTTCTC-3′; E(spl)m7, 5′-CGTCAACACTCCACTCAGTATC-3′ and 5′-TCGTTGTCGCTGGCATATC-3′; E(spl)m8, 5′-TGGCTCAGGAAGAACAATCC-3′ and 5′-CAGGTGAGTCATCACCGATTT-3′; Vha68-2, 5′-TGTCCGAATACTTCCGTGATATG-3′ and 5′-CAGCGAGACGACCAGAAAT-3′; Vha100-2, 5′-GGACAGTCGTTGTGTTGTAGT-3′ and 5′-TCTGCTGATGGGTCTGTTTG-3′; VhaSFD, 5′-TGTGGCGTACTCATCAAAGG-3′ and 5′-CTTCGACGACGAGGACATTAC-3′; and VhaM9.7-b, 5′-TATCCGAGTGGGTGGCA-3′ and 5′-GTCCTCGGGCGAAGAAG-3′.
COMPLEAT complex enrichment analysis
The transcriptomics data were preprocessed using custom-made Python scripts. Log2fc gene expression data between indicated time points and cell types (e.g., NB vs. GMC) were calculated and enriched protein complexes were subsequently identified using the COMPLEAT algorithm (Vinayagam et al., 2013). Deregulated complexes were visualized using the Cytoscape platform (Shannon et al., 2003). Interaction data were obtained from the BioGRID interaction database (Stark et al., 2006).
Brain tumor transplantation, Baf-A1 treatment and metastasis quantification
L3 brains from UAS-dcr2/UAS-bratRNAi; wor-GAL4, ase-GAL80/+; UAS-stingerRFP were dissected and disrupted (see the Cell dissociation, FACS, sample preparation, and RNA sequencing section). Concentrations of RFP+ brat tumor cells were quantified on a Neubauer cell counter, and ∼1,000 cells were intrathoracically injected into 3–6-d-old adult females with a Nanoject II (Drummond). After a recovery phase of 4 h, flies were injected with either the v-ATPase inhibitor Baf-A1 (9.2 nL of 50 nM Baf-A1; EMD Millipore) or vehicle (DMSO in PBS). Brightfield, GFP, and RFP pictures of living flies were taken on a SteREO Lumar.V12 (Zeiss) with a Pursuit-XS monochrome camera (Diagnostic Instruments) every 24 h after transplantation. GFP autofluorescence signal was subtracted from tumor-specific RFP signal and displayed in false colors black-red-green-blue-cyan-magenta-yellow-white (BRGBCMYW) and subsequently merged with the corresponding brightfield image. The RFP-specific signal was quantified on the outlined area of whole flies after GFP autofluorescence subtraction on ImageJ. The mean intensity of all flies in one picture was plotted.
Statistics
Statistical analyses were performed with Prism 7 (GraphPad Software). Unpaired two-sided Student’s t test was used to assess statistical significance between two genotypes/conditions. Mantel-Cox test was used to assess statistical significance in the survival of two conditions. No statistical methods were used to predetermine the sample size. Sample sizes for experiments were estimated based on previous experience with a similar setup that showed significance. Data distribution was assumed to be normal, but this was not formally tested. Experiments were not randomized, and the investigator was not blinded.
Online supplemental material
Fig. S1 shows that sorted NBs divide asymmetrically and that both daughter cells express the expected cell markers (corresponds to Fig. 1 a, step 2). Fig. S2 shows that NB culture has modest effects on the NB transcriptome. Fig. S3 shows the transcriptional down-regulation of the v-ATPase in GMCs over time and that Vha68-2 mRNA is not asymmetrically localized during NB cell division. Fig. S4 shows that the v-ATPase is required for Notch-driven tumor progression. Video 1 shows a control type I NB that divides asymmetrically multiple times. After each division, the NB regrows to its original size before it continues dividing. (Video 1 corresponds to Fig. 4 c.) Videos 2 and 3 show Vha100-2RNAi NBs that divide asymmetrically multiple times. Regrowth potential upon cell division is impaired, resulting in decreasing NB volume. (Videos 2 and 3 correspond to Fig. 4 c.) Table S1 contains a complete list of genes differentially expressed in NBs and GMCs at 1.5, 3, and 5 h with a FDR of <0.05. Table S2 contains a complete list of genes for all seven clusters of the hierarchical clustering analysis and all enriched GO terms for each gene cluster (corresponds to Fig. 2 c). Table S3 includes a complete list of enriched protein complexes (based on COMPLEAT analysis) for the following time points and cell types: 1.5 h GMC versus 1.5 h NB, 3 h GMC versus 3 h NB, 5 h GMC versus 5 h NB, and 5 h GMC versus 1.5 h GMC (corresponds to Fig. 3, a–e).
Supplementary Material
Acknowledgments
We are grateful to members of the J.A. Knoblich laboratory for feedback. We also thank the BioOptics facility (notably T. Lendl, M. Weninger, G. Schmauss, and P. Pasierbek),the fly house (notably P. Duchek, J.F. Gokcezade, and V. Steinmann), M.D. Abdusselamoglu for technical assistance, the VDRC, the Bloomington Drosophila Stock Center, and the Vienna Biocenter Core Facilities Next-Generation Sequencing unit.
Work in the J.A. Knoblich laboratory is supported by the Austrian Federal Ministry of Education, Science and Research, the Austrian Academy of Sciences, the Austrian Science Fund (Z_153_B09), and the City of Vienna. This project has received funding from the European Research Council under the European Union’s Horizon 2020 research and innovation programme (grants 695642 and 693184) and under the FP7 programme (grant 250342). S. Wissel is supported by a Boehringer Ingelheim Fonds PhD fellowship. F. Bonnay was supported by a European Molecular Biology Organization Long-Term Fellowship (LTF 1280-2014).
The authors declare no competing financial interests.
Author contributions: S. Wissel, H. Harzer, and J.A. Knoblich designed experiments. S. Wissel and J.A. Knoblich interpreted experiments. S. Wissel performed experiments. H. Harzer performed 5-h time-course experiments. F. Bonnay and S. Wissel performed Drosophila gut experiments and Baf-A1 experiments. T.R. Burkard performed RNA sequencing data analysis. R.A. Neumüller performed complex enrichment analysis. S. Wissel and J.A. Knoblich wrote the manuscript with input from all authors.
References
- Abramczuk M.K., Burkard T.R., Rolland V., Steinmann V., Duchek P., Jiang Y., Wissel S., Reichert H., and Knoblich J.A.. 2017. The splicing co-factor Barricade/Tat-SF1, is required for cell cycle and lineage progression in Drosophila neural stem cells. Development. 144:3932–3945. 10.1242/dev.152199 [DOI] [PubMed] [Google Scholar]
- Allan A.K., Du J., Davies S.A., and Dow J.A.T.. 2005. Genome-wide survey of V-ATPase genes in Drosophila reveals a conserved renal phenotype for lethal alleles. Physiol. Genomics. 22:128–138. 10.1152/physiolgenomics.00233.2004 [DOI] [PubMed] [Google Scholar]
- Almeida M.S., and Bray S.J.. 2005. Regulation of post-embryonic neuroblasts by Drosophila Grainyhead. Mech. Dev. 122:1282–1293. 10.1016/j.mod.2005.08.004 [DOI] [PubMed] [Google Scholar]
- Barolo S., Carver L.A., and Posakony J.W.. 2000. GFP and beta-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. Biotechniques. 29:726–732. [DOI] [PubMed] [Google Scholar]
- Bello B., Reichert H., and Hirth F.. 2006. The brain tumor gene negatively regulates neural progenitor cell proliferation in the larval central brain of Drosophila. Development. 133:2639–2648. 10.1242/dev.02429 [DOI] [PubMed] [Google Scholar]
- Bello B.C., Izergina N., Caussinus E., and Reichert H.. 2008. Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural Dev. 3:5 10.1186/1749-8104-3-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger C., Harzer H., Burkard T.R., Steinmann J., van der Horst S., Laurenson A.S., Novatchkova M., Reichert H., and Knoblich J.A.. 2012. FACS purification and transcriptome analysis of drosophila neural stem cells reveals a role for Klumpfuss in self-renewal. Cell Reports. 2:407–418. 10.1016/j.celrep.2012.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betschinger J., Mechtler K., and Knoblich J.A.. 2006. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell. 124:1241–1253. 10.1016/j.cell.2006.01.038 [DOI] [PubMed] [Google Scholar]
- Blomen V.A., Májek P., Jae L.T., Bigenzahn J.W., Nieuwenhuis J., Staring J., Sacco R., van Diemen F.R., Olk N., Stukalov A., et al. 2015. Gene essentiality and synthetic lethality in haploid human cells. Science. 350:1092–1096. 10.1126/science.aac7557 [DOI] [PubMed] [Google Scholar]
- Boone J.Q., and Doe C.Q.. 2008. Identification of Drosophila type II neuroblast lineages containing transit amplifying ganglion mother cells. Dev. Neurobiol. 68:1185–1195. 10.1002/dneu.20648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghese L., Dolezalova D., Opitz T., Haupt S., Leinhaas A., Steinfarz B., Koch P., Edenhofer F., Hampl A., and Brüstle O.. 2010. Inhibition of notch signaling in human embryonic stem cell-derived neural stem cells delays G1/S phase transition and accelerates neuronal differentiation in vitro and in vivo. Stem Cells. 28:955–964. 10.1002/stem.408 [DOI] [PubMed] [Google Scholar]
- Bouché V., Perez Espinosa A., Leone L., Sardiello M., Ballabio A., and Botas J.. 2016. Drosophila Mitf regulates the V-ATPase and the lysosomal-autophagic pathway. Autophagy. 12:484–498. 10.1080/15548627.2015.1134081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman S.K., Rolland V., Betschinger J., Kinsey K.A., Emery G., and Knoblich J.A.. 2008. The tumor suppressors Brat and Numb regulate transit-amplifying neuroblast lineages in Drosophila. Dev. Cell. 14:535–546. 10.1016/j.devcel.2008.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buechling T., Bartscherer K., Ohkawara B., Chaudhary V., Spirohn K., Niehrs C., and Boutros M.. 2010. Wnt/Frizzled signaling requires dPRR, the Drosophila homolog of the prorenin receptor. Curr. Biol. 20:1263–1268. 10.1016/j.cub.2010.05.028 [DOI] [PubMed] [Google Scholar]
- Caussinus E., and Gonzalez C.. 2005. Induction of tumor growth by altered stem-cell asymmetric division in Drosophila melanogaster. Nat. Genet. 37:1125–1129. 10.1038/ng1632 [DOI] [PubMed] [Google Scholar]
- Chen K., Dai X., and Wu J.. 2015. Alternative splicing: An important mechanism in stem cell biology. World J. Stem Cells. 7:1–10. 10.4252/wjsc.v7.i1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choksi S.P., Southall T.D., Bossing T., Edoff K., de Wit E., Fischer B.E., van Steensel B., Micklem G., and Brand A.H.. 2006. Prospero acts as a binary switch between self-renewal and differentiation in Drosophila neural stem cells. Dev. Cell. 11:775–789. 10.1016/j.devcel.2006.09.015 [DOI] [PubMed] [Google Scholar]
- Couturier L., Vodovar N., and Schweisguth F.. 2012. Endocytosis by Numb breaks Notch symmetry at cytokinesis. Nat. Cell Biol. 14:131–139. 10.1038/ncb2419 [DOI] [PubMed] [Google Scholar]
- Cruciat C.-M., Ohkawara B., Acebron S.P., Karaulanov E., Reinhard C., Ingelfinger D., Boutros M., and Niehrs C.. 2010. Requirement of prorenin receptor and vacuolar H+-ATPase-mediated acidification for Wnt signaling. Science. 327:459–463. 10.1126/science.1179802 [DOI] [PubMed] [Google Scholar]
- Doe C.Q. 2008. Neural stem cells: balancing self-renewal with differentiation. Development. 135:1575–1587. 10.1242/dev.014977 [DOI] [PubMed] [Google Scholar]
- Dutta D., Xiang J., and Edgar B.A.. 2013. RNA expression profiling from FACS-isolated cells of the Drosophila intestine. Curr Protoc Stem Cell Biol. 27. Unit. 2F:2 10.1002/9780470151808.sc02f02s27 [DOI] [PubMed] [Google Scholar]
- Dutta D., Dobson A.J., Houtz P.L., Gläßer C., Revah J., Korzelius J., Patel P.H., Edgar B.A., and Buchon N.. 2015. Regional Cell-Specific Transcriptome Mapping Reveals Regulatory Complexity in the Adult Drosophila Midgut. Cell Reports. 12:346–358. 10.1016/j.celrep.2015.06.009 [DOI] [PubMed] [Google Scholar]
- Eroglu E., Burkard T.R., Jiang Y., Saini N., Homem C.C.F., Reichert H., and Knoblich J.A.. 2014. SWI/SNF complex prevents lineage reversion and induces temporal patterning in neural stem cells. Cell. 156:1259–1273. 10.1016/j.cell.2014.01.053 [DOI] [PubMed] [Google Scholar]
- Fathi A., Hatami M., Hajihosseini V., Fattahi F., Kiani S., Baharvand H., and Salekdeh G.H.. 2011. Comprehensive gene expression analysis of human embryonic stem cells during differentiation into neural cells. PLoS One. 6:e22856 10.1371/journal.pone.0022856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forgac M. 2007. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 8:917–929. 10.1038/nrm2272 [DOI] [PubMed] [Google Scholar]
- Furriols M., and Bray S.. 2001. A model Notch response element detects Suppressor of Hairless-dependent molecular switch. Curr. Biol. 11:60–64. 10.1016/S0960-9822(00)00044-0 [DOI] [PubMed] [Google Scholar]
- Gleixner E.M., Canaud G., Hermle T., Guida M.C., Kretz O., Helmstädter M., Huber T.B., Eimer S., Terzi F., and Simons M.. 2014. V-ATPase/mTOR signaling regulates megalin-mediated apical endocytosis. Cell Reports. 8:10–19. 10.1016/j.celrep.2014.05.035 [DOI] [PubMed] [Google Scholar]
- Gloss B.S., Signal B., Cheetham S.W., Gruhl F., Kaczorowski D.C., Perkins A.C., and Dinger M.E.. 2017. High resolution temporal transcriptomics of mouse embryoid body development reveals complex expression dynamics of coding and noncoding loci. Sci. Rep. 7:6731 10.1038/s41598-017-06110-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulas S., Conder R., and Knoblich J.A.. 2012. The Par complex and integrins direct asymmetric cell division in adult intestinal stem cells. Cell Stem Cell. 11:529–540. 10.1016/j.stem.2012.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harzer H., Berger C., Conder R., Schmauss G., and Knoblich J.A.. 2013. FACS purification of Drosophila larval neuroblasts for next-generation sequencing. Nat. Protoc. 8:1088–1099. 10.1038/nprot.2013.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homem C.C.F., and Knoblich J.A.. 2012. Drosophila neuroblasts: a model for stem cell biology. Development. 139:4297–4310. 10.1242/dev.080515 [DOI] [PubMed] [Google Scholar]
- Homem C.C.F., Reichardt I., Berger C., Lendl T., and Knoblich J.A.. 2013. Long-term live cell imaging and automated 4D analysis of drosophila neuroblast lineages. PLoS One. 8:e79588 10.1371/journal.pone.0079588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homem C.C.F., Steinmann V., Burkard T.R., Jais A., Esterbauer H., and Knoblich J.A.. 2014. Ecdysone and mediator change energy metabolism to terminate proliferation in Drosophila neural stem cells. Cell. 158:874–888. 10.1016/j.cell.2014.06.024 [DOI] [PubMed] [Google Scholar]
- Homem C.C.F., Repic M., and Knoblich J.A.. 2015. Proliferation control in neural stem and progenitor cells. Nat. Rev. Neurosci. 16:647–659. 10.1038/nrn4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvitz H.R., and Herskowitz I.. 1992. Mechanisms of asymmetric cell division: two Bs or not two Bs, that is the question. Cell. 68:237–255. 10.1016/0092-8674(92)90468-R [DOI] [PubMed] [Google Scholar]
- Huang Y., Wan J., Guo Y., Zhu S., Wang Y., Wang L., Guo Q., Lu Y., and Wang Z.. 2017. Transcriptome analysis of the induced pluripotent stem cell (iPSC)-derived pancreatic β-like cell differentiation. Cell Transplant. 26:1380–1391. 10.1177/0963689717720281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J.R., Bullock S.L., and Ish-Horowicz D.. 2004. Inscuteable mRNA localization is dynein-dependent and regulates apicobasal polarity and spindle length in Drosophila neuroblasts. Curr. Biol. 14:1950–1956. 10.1016/j.cub.2004.10.022 [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., and Salzberg S.L.. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich J.A. 2008. Mechanisms of asymmetric stem cell division. Cell. 132:583–597. 10.1016/j.cell.2008.02.007 [DOI] [PubMed] [Google Scholar]
- Kobia F., Duchi S., Deflorian G., and Vaccari T.. 2014. Pharmacologic inhibition of vacuolar H+ ATPase reduces physiologic and oncogenic Notch signaling. Mol. Oncol. 8:207–220. 10.1016/j.molonc.2013.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolde R.2015. pheatmap: Pretty heatmaps. R package version 1.0. 8. https://CRAN.R-project.org/package=pheatmap
- Koludrovic D., Laurette P., Strub T., Keime C., Le Coz M., Coassolo S., Mengus G., Larue L., and Davidson I.. 2015. Chromatin-Remodelling Complex NURF Is Essential for Differentiation of Adult Melanocyte Stem Cells. PLoS Genet. 11:e1005555 10.1371/journal.pgen.1005555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landskron L., Steinmann V., Bonnay F., Burkard T.R., Steinmann J., Reichardt I., Harzer H., Laurenson A.-S., Reichert H., and Knoblich J.A.. 2018. The asymmetrically segregating lncRNA cherub is required for transforming stem cells into malignant cells. eLife. 7:e31347 10.7554/eLife.31347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange C., Prenninger S., Knuckles P., Taylor V., Levin M., and Calegari F.. 2011. The H(+) vacuolar ATPase maintains neural stem cells in the developing mouse cortex. Stem Cells Dev. 20:843–850. 10.1089/scd.2010.0484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M.K., Holder K., Yost C., Giniger E., and Ruohola-Baker H.. 1996. Expression of constitutively active Notch arrests follicle cells at a precursor stage during Drosophila oogenesis and disrupts the anterior-posterior axis of the oocyte. Development. 122:3639–3650. [DOI] [PubMed] [Google Scholar]
- Lee C.-Y., Robinson K.J., and Doe C.Q.. 2006. Lgl, Pins and aPKC regulate neuroblast self-renewal versus differentiation. Nature. 439:594–598. 10.1038/nature04299 [DOI] [PubMed] [Google Scholar]
- Li H., and Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25:1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Yang X., Wasser M., Cai Y., and Chia W.. 1997. Inscuteable and Staufen mediate asymmetric localization and segregation of prospero RNA during Drosophila neuroblast cell divisions. Cell. 90:437–447. 10.1016/S0092-8674(00)80504-8 [DOI] [PubMed] [Google Scholar]
- Livak K.J., and Schmittgen T.D.. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) Method. Methods. 25:402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchetta E.M., and Ohlstein B.. 2017. Amitosis of Polyploid Cells Regenerates Functional Stem Cells in the Drosophila Intestine. Cell Stem Cell. 20:609–620.e6. 10.1016/j.stem.2017.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyne R., Smith R., Rutherford K., Wakeling M., Varley A., Guillier F., Janssens H., Ji W., Mclaren P., North P., et al. 2007. FlyMine: an integrated database for Drosophila and Anopheles genomics. Genome Biol. 8:R129 10.1186/gb-2007-8-7-r129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison S.J., and Kimble J.. 2006. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 441:1068–1074. 10.1038/nature04956 [DOI] [PubMed] [Google Scholar]
- Nászai M., Carroll L.R., and Cordero J.B.. 2015. Intestinal stem cell proliferation and epithelial homeostasis in the adult Drosophila midgut. Insect Biochem. Mol. Biol. 67:9–14. 10.1016/j.ibmb.2015.05.016 [DOI] [PubMed] [Google Scholar]
- Neumüller R.A., Richter C., Fischer A., Novatchkova M., Neumüller K.G., and Knoblich J.A.. 2011. Genome-wide analysis of self-renewal in Drosophila neural stem cells by transgenic RNAi. Cell Stem Cell. 8:580–593. 10.1016/j.stem.2011.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein B., and Spradling A.. 2006. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature. 439:470–474. 10.1038/nature04333 [DOI] [PubMed] [Google Scholar]
- Oot R.A., Couoh-Cardel S., Sharma S., Stam N.J., and Wilkens S.. 2017. Breaking up and making up: The secret life of the vacuolar H+ -ATPase. Protein Sci. 26:896–909. 10.1002/pro.3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald F., Winkler M., Cao Y., Astrahantseff K., Bourteele S., Knöchel W., and Borggrefe T.. 2005. RBP-Jkappa/SHARP recruits CtIP/CtBP corepressors to silence Notch target genes. Mol. Cell. Biol. 25:10379–10390. 10.1128/MCB.25.23.10379-10390.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paroush Z., Finley R.L. Jr., Kidd T., Wainwright S.M., Ingham P.W., Brent R., and Ish-Horowicz D.. 1994. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell. 79:805–815. 10.1016/0092-8674(94)90070-1 [DOI] [PubMed] [Google Scholar]
- Petzoldt A.G., Gleixner E.M., Fumagalli A., Vaccari T., and Simons M.. 2013. Elevated expression of the V-ATPase C subunit triggers JNK-dependent cell invasion and overgrowth in a Drosophila epithelium. Dis. Model. Mech. 6:689–700. 10.1242/dmm.010660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccini I., Araúzo-Bravo M., Seebohm G., and Greber B.. 2016. Functional high-resolution time-course expression analysis of human embryonic stem cells undergoing cardiac induction. Genom. Data. 10:71–74. 10.1016/j.gdata.2016.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A.R., and Hall I.M.. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 26:841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichert H. 2011. Drosophila neural stem cells: cell cycle control of self-renewal, differentiation, and termination in brain development. Results Probl. Cell Differ. 53:529–546. 10.1007/978-3-642-19065-0_21 [DOI] [PubMed] [Google Scholar]
- San-Juán B.P., and Baonza A.. 2011. The bHLH factor deadpan is a direct target of Notch signaling and regulates neuroblast self-renewal in Drosophila. Dev. Biol. 352:70–82. 10.1016/j.ydbio.2011.01.019 [DOI] [PubMed] [Google Scholar]
- Schweisguth F. 2004. Regulation of notch signaling activity. Curr. Biol. 14:R129–R138. 10.1016/j.cub.2004.01.023 [DOI] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., and Ideker T.. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13:2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons B.D., and Clevers H.. 2011. Strategies for homeostatic stem cell self-renewal in adult tissues. Cell. 145:851–862. 10.1016/j.cell.2011.05.033 [DOI] [PubMed] [Google Scholar]
- Slack C., Somers W.G., Sousa-Nunes R., Chia W., and Overton P.M.. 2006. A mosaic genetic screen for novel mutations affecting Drosophila neuroblast divisions. BMC Genet. 7:33 10.1186/1471-2156-7-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., and Lu B.. 2011. Regulation of cell growth by Notch signaling and its differential requirement in normal vs. tumor-forming stem cells in Drosophila. Genes Dev. 25:2644–2658. 10.1101/gad.171959.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda J., and Wharton R.P.. 2001. Drosophila Brain Tumor is a translational repressor. Genes Dev. 15:762–773. 10.1101/gad.870801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C., Breitkreutz B.-J., Reguly T., Boucher L., Breitkreutz A., and Tyers M.. 2006. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 34:D535–D539. 10.1093/nar/gkj109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stransky L., Cotter K., and Forgac M.. 2016. The Function of V-ATPases in Cancer. Physiol. Rev. 96:1071–1091. 10.1152/physrev.00035.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun-Wada G.-H., and Wada Y.. 2015. Role of vacuolar-type proton ATPase in signal transduction. Biochim. Biophys. Acta. 1847:1166–1172. 10.1016/j.bbabio.2015.06.010 [DOI] [PubMed] [Google Scholar]
- Tognon E., Kobia F., Busi I., Fumagalli A., De Masi F., and Vaccari T.. 2016. Control of lysosomal biogenesis and Notch-dependent tissue patterning by components of the TFEB-V-ATPase axis in Drosophila melanogaster. Autophagy. 12:499–514. 10.1080/15548627.2015.1134080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccari T., Duchi S., Cortese K., Tacchetti C., and Bilder D.. 2010. The vacuolar ATPase is required for physiological as well as pathological activation of the Notch receptor. Development. 137:1825–1832. 10.1242/dev.045484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinayagam A., Hu Y., Kulkarni M., Roesel C., Sopko R., Mohr S.E., and Perrimon N.. 2013. Protein complex-based analysis framework for high-throughput data sets. Sci. Signal. 6:rs5 10.1126/scisignal.2003629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Somers G.W., Bashirullah A., Heberlein U., Yu F., and Chia W.. 2006. Aurora-A acts as a tumor suppressor and regulates self-renewal of Drosophila neuroblasts. Genes Dev. 20:3453–3463. 10.1101/gad.1487506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.Q., Habegger L., Noisa P., Szekely A., Qiu C., Hutchison S., Raha D., Egholm M., Lin H., Weissman S., et al. 2010. Dynamic transcriptomes during neural differentiation of human embryonic stem cells revealed by short, long, and paired-end sequencing. Proc. Natl. Acad. Sci. USA. 107:5254–5259. 10.1073/pnas.0914114107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Denef N., and Schüpbach T.. 2009. The vacuolar proton pump, V-ATPase, is required for notch signaling and endosomal trafficking in Drosophila. Dev. Cell. 17:387–402. 10.1016/j.devcel.2009.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J.-W., and Wang G.. 2014. The Mediator complex: a master coordinator of transcription and cell lineage development. Development. 141:977–987. 10.1242/dev.098392 [DOI] [PubMed] [Google Scholar]
- Yuan X., Wu H., Xu H., Xiong H., Chu Q., Yu S., Wu G.S., and Wu K.. 2015. Notch signaling: an emerging therapeutic target for cancer treatment. Cancer Lett. 369:20–27. 10.1016/j.canlet.2015.07.048 [DOI] [PubMed] [Google Scholar]
- Zacharioudaki E., Housden B.E., Garinis G., Stojnic R., Delidakis C., and Bray S.. 2015. Genes implicated in stem-cell identity and temporal-program are directly targeted by Notch in neuroblast tumours. Development. 10.1242/dev.126326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Zhou Q., Ogmundsdottir M.H., Möller K., Siddaway R., Larue L., Hsing M., Kong S.W., Goding C., Palsson A., et al. 2015. Mitf is a master regulator of the v-ATPase, forming a control module for cellular homeostasis with v-ATPase and TORC1. J. Cell Sci. 128:2938–2950. 10.1242/jcs.173807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S., Lin S., Kao C.-F., Awasaki T., Chiang A.-S., and Lee T.. 2006. Gradients of the Drosophila Chinmo BTB-zinc finger protein govern neuronal temporal identity. Cell. 127:409–422. 10.1016/j.cell.2006.08.045 [DOI] [PubMed] [Google Scholar]
- Zoncu R., Bar-Peled L., Efeyan A., Wang S., Sancak Y., and Sabatini D.M.. 2011. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 334:678–683. 10.1126/science.1207056 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.