Abstract
Background
Neuroanatomical asymmetries have recently been associated with the progression of Alzheimer’s disease (AD) but the biological basis of asymmetric brain changes in disease remains unknown.
Methods
We investigated genetic influences on brain asymmetry by identifying associations between MRI-derived measures of asymmetry and candidate single-nucleotide polymorphisms (SNPs) that have previously been identified in genome-wide association studies (GWAS) for AD diagnosis and for brain subcortical volumes. For the longitudinal neuroimaging data (1,241 individuals; 6,395 scans), we use a mixed effects model with interaction between genotype and diagnosis.
Results
We found significant associations between asymmetry of amygdala, hippocampus, and putamen and SNPs in the genes BIN1, CD2AP, ZCWPW1, ABCA7, TNKS, and DLG2. For AD candidate SNPs, we demonstrated an asymmetric effect on subcortical brain structures.
Conclusions
The associations between SNPs in the genes TNKS and DLG2 and AD-related increases in shape asymmetry are of particular interest; these SNPs have previously been associated with subcortical volumes of amygdala and putamen but have not yet been associated with Alzheimer’s pathology. This provides novel evidence about the biological underpinnings of brain asymmetry as a disease marker. Contralateral brain structures represent a unique, within-patient, reference element for disease and asymmetries can provide a personalized measure of the accumulation of past disease processes.
Keywords: Imaging, Genetics, Alzheimer’s, Asymmetry, Shape, Longitudinal
Introduction
Alzheimer’s disease (AD) is a progressive and irreversible brain disorder characterized by a gradual degradation of cognitive functions over many years that includes a long preclinical phase (1). Because brain atrophy as measured with magnetic resonance imaging (MRI) correlates with neuron loss (2), longitudinal in vivo neuroimaging has become invaluable for studying trajectories of pathophysiological change in AD. Repeated volumetric measurements of brain volumes in the same individuals have shown accelerated rates of atrophy in patients with AD compared to healthy controls, even in preclinical stages (3).
Volume measurements are, however, only a crude simplification of the complex anatomical change that occurs in aging and AD and often ignore the fact that atrophy is not uniform across a brain structure, e.g. the hippocampus, (4–6). In contrast, shape descriptors are sensitive to such changes as they retain more geometrical information (7). Indeed, a recent study revealed that subtle preclinical changes in the shape asymmetry of subcortical brain structures could predict the conversion from mild cognitive impairment to dementia more accurately than volumetric asymmetry (8). These asymmetries are undirectional, i.e., they do not have a consistent hemispheric effect and therefore refer to the magnitude of asymmetry independent of direction.
While structural asymmetries could serve as imaging biomarker for the early pre-symptomatic classification and prediction of AD, possible biological mechanisms that underlie asymmetric manifestation of AD pathology are unclear. Here, we investigate the genetic influence on shape asymmetry in AD in an imaging quantitative trait loci (QTL) analysis including healthy controls, MCI stable, MCI progressor and AD patients. While previous genome-wide association studies (GWAS) studies have revealed single-nucleotide polymorphisms (SNP) that are related to AD diagnosis, the mechanism through which they affect the disease remains largely unknown. Relating these same SNPs to imaging markers helps our understanding of how common genetic variants alter specific structures and pathways in the living human brain (9). Because prior research is inconsistent with respect to heritability of brain structural asymmetry (10–12), we model both main effects of SNP as well as interactions between diagnosis groups and SNP on brain asymmetry. Whereas main effects speak to heritability per se, a significant interaction would reveal genetic influences on brain shape asymmetry that are magnified in AD patients and thus can be interpreted to reflect the development of disease (13).
Imaging QTL studies may have several potential advantages over case-control studies; including increased power (14). Imaging endophenotypes of disease in QTL studies can separate diseased and normal subjects more accurately and therefore limit the confound of including asymptomatic subjects in the control group, which is particularly important for the long clinically silent prodromal phase of AD (13,15–21). These previous imaging QTL studies on AD risk variants and MRI measures employed a cross-sectional design and focused on volume of brain structures and cortical thickness. Here, we use a longitudinal model to study the genetics of shape asymmetry, yielding increased power for detecting genetic associations (22), although this may also depend on the genetic architecture of the specific traits being studied.
The current study utilizes longitudinal imaging data from over 6,000 MRI scans and genetic data from 1,241 individuals in the Alzheimer’s Disease Neuroimaging Initiative (ADNI). We focus on four brain structures (hippocampus, amygdala, putamen, caudate), selected a priori based on previous reports of increased volume and shape asymmetries in AD (8,23). Shape asymmetry is computed with the Mahalanobis distance of lateralized brain structures within a subject. We include 31 SNPs, selected a priori based on previous GWAS results. These SNPs are primarily composed of 21 candidate SNPs that have been associated to late-onset AD in GWAS (24–26). If these genetic risk variants for AD also influence shape asymmetry in AD the results could suggest mechanisms or pathways through which these genes might be exerting their influence in AD. In a more exploratory analysis, 10 additional SNPS are included that have recently been associated to subcortical volume in large-scale GWAS (27), but not to AD per se. Here, the motivation is to identify possible genetic predispositions that render the brain vulnerable to shape asymmetry in disease. For example, genes that are associated with smaller hippocampi might render the hippocampus more vulnerable to AD pathology even if the genes are not directly implicated in AD pathology.
Methods
Data
We analyzed data from the ADNI, which was launched in 2003 as a public-private partnership, led by Principal Investigator Michael W. Weiner, MD. The primary goal of ADNI has been to test whether serial MRI, PET, other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early AD. For up-to-date information, see www.adni-info.org. We select all subjects with genetic information and at least three longitudinal MRI scans from the ADNI cohort, yielding N=1,241 individuals and 6,395 scans with summary statistics listed in supplementary table 1. Supplementary figure 1 shows a histogram of the number of scans per subjects.
Analysis of brain structure shape
We compute the brain structure shape based on the brain descriptor BrainPrint, which relies on the automated segmentation with FreeSurfer (28–31). BrainPrint and its application to study shape asymmetry has previously been described in detail (7,8). Briefly, after image segmentation, geometric representations are extracted for the identified subcortical structures via the marching cubes algorithm. shapeDNA (32) is used as the shape descriptor of the individual structures in the BrainPrint, which performs among the best in a comparison of methods for non-rigid 3D shape retrieval (33). shapeDNA is based on the eigenvalues of the Laplace-Beltrami operator and, therefore, isometry invariant. Eigenvalues of the Laplace-Beltrami operator Δ can be computed via finite element analysis by solving the Laplacian eigenvalue problem (Helmholtz equation) on the given shape
The solution consists of eigenvalue λi ∈ ℝ and eigenfunction fi pairs. The first l non-zero eigenvalues form the descriptor: λ̄ = (λ1, …, λl), where we set l = 50 (7). A key property of the eigenvalues is their isometry invariance, i.e., length-preserving deformations will not change the spectrum. Isometry invariance includes rigid body motion as well as reflections, and, therefore, permits to directly compare shapes across individuals without any registration. The collection of shape descriptors from cortical and subcortical structures forms the BrainPrint, which has recently shown high potential for the automated diagnosis of dementia (34,35).
Brain Asymmetry from BrainPrint
Asymmetry of a lateralized brain structure s is measured by directly computing the Mahalanobis distance between the descriptors
where we use a diagonal covariance matrix Σ with the i-th element Σii = i2 to reduce the impact of higher eigenvalues on the distance (7). The asymmetry computation completely avoids lateral processing bias as it works on both hemispheres independently. The asymmetry measure presents a within-subject measure that can identify directional and undirectional asymmetry. In addition, it allows for quantifying localized asymmetries potentially induced by morphometric changes in subnuclei, which can be crucial for tracking the progression of dementia (4,5).
Genetic data
In ADNI, GWAS genotyping was performed using three different Illumina platforms, Illumina Human610-Quad, Illumina HumanOmni Express, and Ilumina Omni2.5M BeadChips (36). The APOE ε4 allele defining SNPs (rs429358, rs7412) were separately obtained using standard methods (36). Standard quality control procedures for genetic markers and subjects are performed as described previously: 1) for SNP, SNP call rate < 95%, Hardy-Weinberg equilibrium test p<1 × 10−6, and minor allele frequency (MAF) < 1%; 2) for subject, subject sex and identity check and subject call rate < 95% (37). Due to the impact of population stratification on association analysis, we select only non-Hispanic Caucasian participants using multidimensional scale analysis and HapMap GWAS genotypes (38). As the ADNI used different genotyping platforms, we impute ungenotyped SNPs separately in each platform using MACH (39) with the reference panel of the Haplotype Reference Consortium (HRC). After the imputation, we impose an r2 = 0.30 as the threshold to accept the imputed genotypes. From the imputed data, we select 21 candidate AD SNPs (25) and 10 SNPs that have been associated with subcortical brain structures (27) based on a cut-off of p<1 × 10−7, listed in the Appendix.
Statistical Analysis
We use linear mixed effects models (40,41) to study the association between longitudinal change in brain asymmetry (i.e. the lateral shape distance) and genetics. Genotypes are coded as 0, 1, and 2 representing the number of minor alleles in the genotype, following an additive genetic model. We denote age at baseline for individual i with Bi, years-from-baseline at follow-up scan j with Xij, diagnosis with Di, and the additive encoding of the SNP with Si. To establish whether an association between SNP and asymmetry differs between diagnosis groups, the effect of interest in the model is the interaction term SNP × Diagnosis. The linear model for asymmetry Yij as dependent variable is
| (1) |
where β0, β1, β2, β3 are fixed effects regression coefficients and b0i, b1i are random effects regression coefficients. The random effects enable modeling individual-specific intercept and slope with respect to the time from the baseline. The fixed effect coefficient β2 models the longitudinal change on a population level. The statistical model is an adaptation of previous longitudinal models on the ADNI (8,40). We further evaluate a simplified model without an interaction between SNP and diagnosis. The following additional parameters are included as fixed effects (not shown in Eq. (1)): years of education, sex, intracranial volume (ICV), and the number of APOE ε4 risk alleles.
For the diagnosis, we differentiate between control subjects, MCI subjects that remain stable, MCI subjects that progress to AD, and AD subjects. In our analysis, we encode the diagnosis once as a quantitative variable and once as a categorical variable with four levels. The continuous coding of diagnosis has for instance been used in (17,42) that also investigated the interaction of SNP with diagnosis. In their cross-sectional analyses, they have not differentiated between MCI progressor and stable. In a post-hoc analysis, we include the interaction SNP x years-from-baseline to the model to evaluate whether SNPs might have time-varied effects on asymmetry. We use false discovery rate (FDR) with q=0.05 (43) to control for multiple hypothesis testing across SNPs similar to (19), but also discuss Bonferroni correction in the result’s section. The appendix reports details on the implementation of the models.
Results
In the following, we describe the results of the SNP-asymmetry analyses, where we use models with and without the interaction of SNP and diagnosis together with a quantitative and categorical coding of the diagnosis. Table 1 summarizes all the SNPs that showed significant associations in the different models together with their closest genes, location, major/minor alleles, minor allele frequency, genotype count, population-attributable fractions (PAF) or preventive fractions.
Table 1.
Summary of SNPs with significant associations to brain asymmetry. SNPs included in the study were selected from Alzheimer’s GWAS (25) and from subcortical brain GWAS (27).
| SNP | Chr. | Position | Closest gene | Major/minor alleles | GWASa | Asymmetryb | Effectc | PAF Typed | PAF (%) | Genotype Counte | MAFf |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs6733839 | 2 | 127892810 | BIN1 | C/T | AD | Putamen | SNP x Disease | risk | 8.1 | 2382/2977/1036 | T=0.3950 |
| rs10948363 | 6 | 47487762 | CD2AP | A/G | AD | Amygdala | SNP | risk | 2.3 | 3375/2549/471 | G=0.1881 |
| rs4147929 | 19 | 1063443 | ABCA7 | G/A | AD | Hippocampus | SNP x Disease | risk | 2.8 | 3337/2503/555 | C=0.2115 |
| rs1476679 | 7 | 100004446 | ZCWPW1 | T/C | AD | Hippocampus | SNP x Disease | preventive | 3.2 | 4369/1828/198 | A=0.1751 |
| rs117253277 | 8 | 9304257 | TNKS | C/A | Amygdala | Amygdala | SNP x Disease | 6320/75/0 | A=0.0264 | ||
| rs683250 | 11 | 83276168 | DLG2 | G/A | Putamen | Putamen | SNP, SNP x Disease | 2173/3318/904 | T=0.3950 |
GWAS identification of SNPs. Brain structures for subcortical GWAS.
Association of SNP to brain asymmetry identified in this work
Significant main or interaction effect in our model
Population-attributable fractions (PAF) type for AD risk SNPs (from Supplementary Table 6 in (25))
Genotype count for (0/1/2) for image scans
Minor allele frequency (MAF) based on the 1000 Genomes project
In the following, significant interactions are reported in more detail, grouped by whether they were identified with a quantitative coding of disease (0=CN, 1=MCI-Stable, 2=MCI-Progressor, 3=AD; Table 2) or categorical coding (Table 3). We show standardized regression coefficients and p-values for the main effects, and in addition adjusted p-values after FDR correction for the interaction. Results are only included in the tables 2 and 3 when the adjusted p-value of the interaction is below 0.05.
Table 2.
Standardized regression coefficients and p-values for the analysis of asymmetry with genetic loci for the linear mixed effects model with interactions. Adjusted p-values for the interaction are presented, where we only show significant associations after FDR correction. p-values are rounded to five decimal places. The diagnosis is modeled as continuous variable.
| SNP | GWAS | β3 (SNP) | β4 (Diagnosis) | β5 (SNP x Diagnosis) | ||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Beta | P-value | Beta | P-value | Beta | P-value | Adj. P-value | ||
| Amygdala asymmetry | ||||||||
| rs117253277 | Amygdala | −0.539 | 0.04136 | 0.225 | 0.00000 | 0.585 | 0.00129 | 0.03738 |
| Putamen asymmetry | ||||||||
| rs683250 | Putamen | −0.015 | 0.76344 | 0.026 | 0.43458 | 0.097 | 0.00124 | 0.03833 |
| rs6733839 | AD | 0.060 | 0.18796 | 0.180 | 0.00000 | −0.086 | 0.00314 | 0.04864 |
Table 3.
Standardized regression coefficients and p-values for the analysis of lateral asymmetry with genetic loci for the linear mixed effects model with interactions. Adjusted p-values for the interaction are presented, where we only show significant associations after FDR correction. p-values are rounded to five decimal places. The diagnosis is modeled as categorical variable, where we only report difference between CN and AD. The other factors were not significant.
| SNP | GWAS | β3 (SNP) | β4 (Diagnosis) | β5 (SNP x Diagnosis) CN -> AD | ||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Beta | P-value | Beta | P-value | Beta | P-value | Adj. P-value | ||
| Hippocampus asymmetry | ||||||||
| rs1476679 | AD | −0.037 | 0.42071 | 0.600 | 0.00000 | 0.255 | 0.00148 | 0.02213 |
| rs4147929 | AD | 0.060 | 0.29610 | 0.787 | 0.00000 | −0.309 | 0.00147 | 0.02213 |
| Amygdala asymmetry | ||||||||
| rs117253277 | Amygdala | −0.268 | 0.38490 | 0.638 | 0.00000 | 2.360 | 0.00006 | 0.00160 |
| Putamen asymmetry | ||||||||
| rs6733839 | AD | 0.044 | 0.41800 | 0.607 | 0.00000 | −0.323 | 0.00051 | 0.01363 |
Interactions with a quantitative coding of disease reveal genetic variants that are associated to shape asymmetry in a stage-dependent manner. Significant associations exist between amygdala asymmetry and rs117253277, as well as between putamen asymmetry and rs683250 and rs6733839 (Table 2). The main SNP effect is not significant for any of these associations after FDR correction. The main diagnosis effect is highly significant for rs117253277 and rs6733839, but not for rs683250. Notably, all regression coefficients for diagnosis are positive, which indicates an increase in asymmetry with the progression of dementia, consistent with our previous results (8). rs117253277shows a negative coefficient for SNP (−0.539), which means that the presence of a minor allele A decreases the asymmetry. Importantly, the positive interaction (coefficient estimate = 0.585) signifies that asymmetry increases with the number of minor alleles for demented subjects. For rs683250, the pattern is inverted, with minor alleles yielding an increase in asymmetry in controls but a decrease in the demented population. The SNPs (rs117253277 and rs683250) were identified in the subcortical GWAS for amygdala and putamen, respectively, which is consistent with the structures in which we observe disease-dependent associations to asymmetry; rs6733839 was identified in the AD GWAS.
Table 3 reports the results for the categorical coding of diagnosis. The categorical coding is less hypothesis-driven than the continuous coding because it allows for non-linear interactions that are driven by only two groups (CN -> MCI-s, CN -> MCI-p, and CN -> AD). That said, this analysis only revealed significant coefficients for the factor CN -> AD. Factors that distinguish MCI groups from CN do not show significant results for the interaction. The main SNP effect is not significant for any of these associations, where the main diagnosis effect is highly significant for all. As for the continuous model, the interactions of diagnosis with rs117253277 and rs6733839 are significant. In addition, SNP x diagnosis interactions are obtained for hippocampal asymmetry for rs1476679 and rs4147929. Both of these SNPs have been reported in AD GWAS. Interestingly, they have an inverted effect on asymmetry, with a positive interaction coefficient for rs1476679 (estimate = 0.255) and a negative coefficient for rs4147929 (estimate = −0.309). This is consistent with their respective role in AD, as reported in Table 1: rs4147929 is a risk locus, whereas rs1476679 is a preventive locus (25). The minor allele frequency and the genotype count of rs117253277 are low, which may bias the results. For confirmation, we created random samples of similar sample size that matched the diagnostic distribution. The estimates for the interaction SNP x diagnosis over 50 repetitions are plotted in supplementary figure 2. The median of 2.08 and the mean of 2.01 are close to the estimate of the original model (2.36).
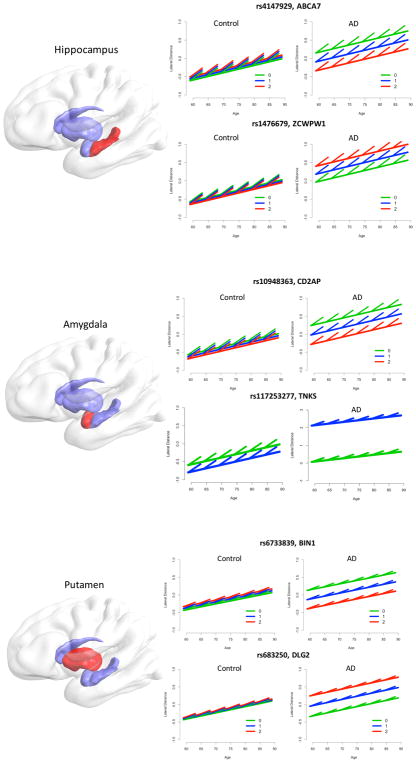
Figure 1 displays the estimated intra- and inter-individual change of the lateral shape asymmetry for hippocampus, amygdala and putamen with the associated loci. We show the genotype for control and AD. Solid lines depict the global age effect, where the offset in intercept is determined by the genotype. Short line ticks depict the longitudinal intra-individual effect. The common pattern, except for rs117253277, is that the genotype has limited effect on the asymmetry of control subjects but a strong effect for AD patients. For rs117253277, the number of minor alleles also influences the asymmetry of control subjects, which illustrates the strong main effect of SNP (−0.268) in Table 3. For hippocampus and amygdala, we observe a higher intra-individual increase in asymmetry compared to the inter-individual increase (i.e., the age effect), as previously reported (8). Note that cross-sectional and longitudinal effects can vary substantially in Figure 1, which may result from positive selection bias for very old adults in cross-sectional studies.
Figure 1.
Longitudinal analysis of lateral asymmetry measures of the hippocampus, amygdala, and putamen with significantly associated SNPs. Lines and ticks illustrate estimates of the different linear mixed effects models with categorical coding of diagnosis. The global age effect is depicted by the slope of the long solid lines, short line ticks depict longitudinal slopes. Plots are shown for control subjects and AD patients. The number of minor alleles can be related to higher or lower asymmetry in AD, depending on the SNP and its biological function.
Table 4 provides statistical detail for the model with main effect of SNP only for quantitative coding (see supplementary table 2 for categorical coding). The association of rs683250 to putamen asymmetry is consistent with the interaction. A new association with amygdala asymmetry is found for the AD candidate SNP rs10948363.
Table 4.
Standardized regression coefficients and p-values for the analysis of lateral asymmetry with genetic loci for the linear mixed effects model without interactions. Adjusted p-values for the main effect SNP are presented, where we only show significant associations after FDR correction. p-values are rounded to five decimal places. The diagnosis is modeled as continuous variable.
| SNP | GWAS | β3 (SNP) | β4 (Diagnosis) | |||
|---|---|---|---|---|---|---|
|
| ||||||
| Beta | P-value | Adj. P-value | Beta | P-value | ||
| Amygdala asymmetry | ||||||
| rs10948363 | AD | −0.112 | 0.00033 | 0.01034 | 0.235 | 0.00000 |
| Putamen asymmetry | ||||||
| rs683250 | Putamen | 0.108 | 0.00069 | 0.02127 | 0.107 | 0.00000 |
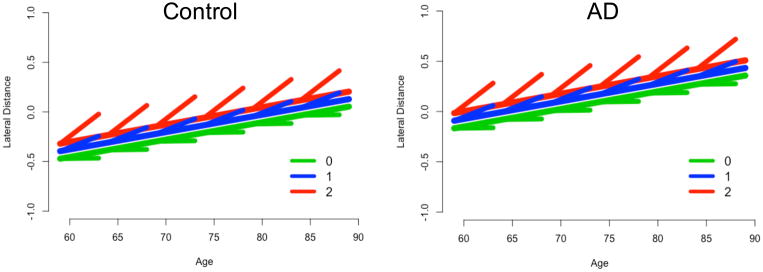
In a post-hoc analysis, we added the interaction SNP x years-from-baseline to the models and evaluated whether the interaction was significant for the above identified pairings of asymmetry and SNP. In the model without SNP x diagnosis interaction, we found that the interaction SNP x years-from-baseline was significant for rs683250 and putamen asymmetry (beta=0.043, p=0.00127). Figure 2 illustrates the intra- and inter-individual change by genotype, which shows that minor alleles were associated with a steeper increase in asymmetry over time.
Figure 2.
Longitudinal analysis for the SNP rs683250 and putamen asymmetry with interaction SNP x years-from-baseline. Plots are shown for controls and AD subjects for different genotype counts. A larger number of minor alleles corresponds to a steeper longitudinal increase.
In all the presented analyses, we included the number of APOE4 risk alleles as a covariate. In an additional analysis, we removed it from covariates and consider it as the SNP of interest. Across all the models, with and without interaction, as well as the different coding of diagnosis, there were no significant associations between asymmetry and APOE4.
We used FDR correction to control for multiple testing but almost all results would also be significant with the conservative Bonferroni correction (p-value threshold of 0.00161). The only exception is the interaction of rs6733839 with diagnosis for putamen asymmetry in the quantitative coding, although the interaction would still be significant for the categorical coding. Note that we do not correct for multiple comparisons across models.
Discussion
In a series of linear mixed effects models of longitudinal neuroanatomical change, we have identified genetic risk variants associated with an increase in brain shape asymmetry in AD. The closest genes associated with significant SNPs include BIN1 (rs6733839), CD2AP (rs10948363) and ABCA7 (rs4147929), which code for proteins involved in amyloid generation, secretion and clearance and are thus likely directly implicated in the accumulation of AD pathology. Another gene, ZCWPW1 (rs1476679) has previously been identified to have a preventative effect in AD. Interestingly, we also identified SNPs in the genes TNKS and DLG2 as risk variants for AD-related increases in shape asymmetry. These are SNPs that have previously been associated to subcortical volumes of amygdala and putamen, respectively (27). Here, we show that these same SNPs also convey risk for AD pathology in these same structures. Previous reports have found several associations between neuroimaging measures and APOE4 (19). In contrast, we found no significant associations between neuroanatomical asymmetry and APOE4, suggesting that the association between APOE4 and atrophy is global, i.e., not symmetric. Below, we review the specifics of each significant SNP before discussing our results more generally.
The interaction of rs117253277 (TNKS) with diagnosis showed the most significant association in our study (p=6 × 10−6). The SNP was identified in the subcortical GWAS as a common variant associated with differences in amygdala volume. Our results show that the SNP also influences amygdala asymmetry in the context of AD pathology, which provides novel evidence that inherent differences in amygdala volumes make the brain more vulnerable to AD-related patterns of atrophy (i.e. increases in shape asymmetry). Importantly also, the interaction between SNP and disease was significant both when coding disease categorically and continuously, which shows that the SNP promotes increases in shape asymmetry already in the preclinical stages of AD in a dose-dependent response. Additional experiments were performed on matched samples to confirm the reliable estimate despite the low MAF of the SNP. The closest gene is TNKS (tankyrase) that catalyzes the ADP-ribosylation of target proteins.
rs683250 is an intronic locus within DLG2 and was identified in the subcortical GWAS. It seems to predispose the brain to undergo asymmetric shape atrophy in the progression to AD. For rs683250, however, we also identified a significant main effect of SNP on shape asymmetry, suggesting that the related gene has an effect on putamen asymmetry per se that is magnified in disease. Moreover, we found in the post-hoc analysis that there was a significant interaction between SNP and years-from-baseline, indicating that the SNP affects change in putamen asymmetry over time. Genetic variants in DLG2 affect learning and cognitive flexibility (44) and are associated with schizophrenia (45). The link to schizophrenia of the SNP is interesting, as abnormal asymmetries in subcortical structures have previously been reported for schizophrenia (46,47).
rs6733839 is within the bridging integrator 1 (BIN1) gene and the most important genetic susceptibility locus after APOE4 for individuals of European ancestry (48). As rs117253277, the SNP was significant for categorical and continuous coding. GWAS studies with MRI measures found association of BIN1 with atrophy in hippocampus (16), entorhinal and temporal pole cortex (15), left parahippocampal and right inferior parietal cortex (19). Functions of the BIN1 gene include the production and clearance of amyloid–beta (Aβ) and cellular signaling; it increases the risk for AD by modulating tau pathology and is also involved in endocytosis, inflammation, calcium homeostasis, and apoptosis (49). Unlike the two AD-related SNPs related to hippocampus asymmetry, reviewed below, the SNP x disease interaction for rs6733839 was significant for the continuous coding of disease groups, suggesting a linear increase in shape asymmetry with disease progression that includes preclinical stages.
For hippocampal asymmetry, two SNPs, rs1476679 and rs4147929, showed significant interaction with diagnosis. They have an inverse effect on asymmetry, which is consistent with their different roles as preventative or risk locus in AD. rs1476679 is intronic in the ZCWPW1 (encoding zinc finger) gene, is a histone modification reader and is involved in epigenetic regulation (50). The preventative effect of rs1476679, reducing the risk of AD, was reported in Caucasians (25), a Spanish sample (52), and Han Chinese (53).
rs4147929 is within the ATP-binding cassette transporter A7 (ABCA7) gene. It is a transmembrane protein that influences neuronal cholesterol efflux and Aβ secretion (54). The gene is strongly expressed in hippocampus subfield CA1 (55) and associated with amyloid plaque burden (56). The minor allele of ABCA7 increases the risk of AD, as shown in an autopsy-confirmed research cohort (57). ABCA7 showed a significant association with hippocampal atrophy (20) and gray matter density (13). The expression of the gene in the subfield CA1 and the association with hippocampal atrophy are supportive of our results, as we have shown in previous work that the increase in asymmetry in hippocampus is not uniform but localized; one of the reasons for the improved results with shape descriptors compared to volumetric analyses.
There was only one SNP, rs10948363 (CD2AP), which showed a significant association with shape asymmetry independent of disease status. This confirms previous reports that brain structural asymmetry per se is not strongly heritable (11), but also shows that QTL studies can reveal subtle genetic influences that are not detectable in twin studies. CD2AP rs10948363 potentially contributes to amyloid precursor protein (APP) metabolism and subsequent Aβ generation through its regulation of clathrin-mediated endocytosis (58). It is probably linked to modulating Aβ clearance and tau neurotoxicity (56) and was associated to FDG PET metabolism (13). Our results suggest its influence on amygdala asymmetry, which showed one of the strongest associations to dementia in our previous work (8).
Supplementary data from the UK Brain Expression Consortium on gene expression QTLs from postmortem healthy human brains (http://www.braineac.org/), revealed that homozygotes of the minor allele type for rs683250 show greater expression of DLG2 in putamen of healthy individuals as compared to individuals carrying at least one major allele (supplementary figure 3). The results of the cis-eQTL mapping analysis are consistent with our results of the main effect of this SNP on putamen asymmetry.
Collectively, our results provide novel evidence for genes that may drive asymmetric accumulation of AD pathology and also suggests that sequence variants may act through their influence on neuroanatomical asymmetry. It is important to note that brain structural asymmetry in AD, or disease more generally, is different from the lateralization of language and motor function, where higher asymmetry tends to relate to higher functioning. Instead, higher asymmetry in AD is associated with the progression of preclinical and prodromal stages of disease, and reflects an asymmetric effect of pathologic processes on brain morphology. The core genetic mechanisms of lateralized human brain development are unknown (59). Future work may be directed at studying the relation between functional asymmetry in development and asymmetric disease manifestation.
Several strengths and limitations of our work are worth noting. Working with the ADNI dataset is a strength as it is a large-scale, publicly available dataset that includes a rigorous clinical and genetic examination. However, ADNI was designed to simulate clinical trials and therefore uses more stringent inclusion and exclusion criteria, which necessitates a replication on an independent sample in the general population. Another limitation of the study may be the a priori selection of candidate genes. A limitation of working with a global shape descriptor is that we cannot visualize the localized changes in shape asymmetry. A strength is that genetic associations of asymmetry are studied in a longitudinal design, where baseline asymmetry and change during the study period is modeled. This allowed us to not only model controls and AD patients, but further differentiate between MCI subjects that remain stable and those that progress to AD.
Supplementary Material
Supplementary Figure S1: Histogram of the number of scans per subject. Most subjects have between four and five scans.
Supplementary Figure S2: Boxplot of estimates for the interaction SNP x diagnosis for SNP rs117253277 and amygdala asymmetry with categorical coding. Matched samples for genotypes CC and CA were created to have a similar number of images per diagnostic group. We used random sampling to generate the matched samples and repeated this procedure 50 times. The estimate of the original model based on unequal sample size is shown as red dot. The median 2.08 and the mean 2.01 are close to the estimate of the original model (2.36).
Supplementary Figure S3: We studied gene expression quantitative trait loci (eQTL) of significant SNPs using gene expression data from postmortem healthy human brains from the UK Brain Expression Consortium (http://www.braineac.org/). The project analyzed up to 10 brain regions for 134 individuals: cerebellar cortex (CRBL), frontal cortex (FCTX), hippocampus (HIPP), medulla (specifically inferior olivary nucleus, MEDU), occipital cortex (specifically primary visual cortex, OCTX), putamen (PUTM), substantia nigra (SNIG), thalamus (THAL), temporal cortex (TCTX) and intralobular white matter (WHMT).
The plots show the gene expression levels stratified by genotypes of the corresponding SNP. Most of our findings were for SNP x diagnosis interactions; only rs10948363 and rs683250 showed a significant main effect SNP. Since expression data was collected on healthy subjects, we only expect differences for SNPs with significant main effect. rs10948363 was associated to amygdala asymmetry, but amygdala was not included in the eQTL mapping analysis. This only leaves SNP rs683250, which was associated to putamen asymmetry. In plot (a), we observe higher expression levels of DLG2 for GG than for GA or AA in putamen (red box). For completeness, we also include results for cis-eQTL mapping analysis for the other SNPs (b) – (e), except for rs117253277, which was not part of the Braineac database.
Supplementary Table S1: Diagnostic and demographic information of the ADNI sample used for the experiments.
Supplementary Table S2: Standardized regression coefficients and p-values for the analysis of lateral asymmetry with genetic loci for the linear mixed effects model without interactions. Adjusted p-values for the main effect SNP are presented, where we only show significant associations after FDR correction. p-values are rounded to five decimal places. The diagnosis is modeled as categorical variable, where we only report difference between CN and AD. The other factors were not significant.
Acknowledgments
Support for this research was provided in part by the Bavarian State Ministry of Education, Science and the Arts in the framework of the Centre Digitisation.Bavaria (ZD.B). Additional support for data analysis was provided by NLM R01 LM012535 and NIA grants R03 AG054936, P30 AG010133 and R01 AG019771. Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare;; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Footnotes
Financial Disclosures
All authors report no biomedical financial interests or potential conflicts of interest.
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wpcontent/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Christian Wachinger, Lab for Artificial Intelligence in Medical Imaging (AI-Med), Klinikum der Universität München, Ludwig-Maximilians-Universität München, Waltherstr. 23, 80337 München, Germany.
Kwangsik Nho, Center for Neuroimaging and Indiana Alzheimer Disease Center, Department of Radiology and Imaging Sciences, Indiana University School of Medicine, Indianapolis, IN, 46202, USA.
Andrew J. Saykin, Center for Neuroimaging and Indiana Alzheimer Disease Center, Department of Radiology and Imaging Sciences, Indiana University School of Medicine, Indianapolis, IN, 46202, USA
Martin Reuter, A.A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, 149 Thirteenth Street, Suite 2301, Charlestown, MA 02129, USA. Deutsches Zentrum für Neurodegenerative Erkrankungen (DZNE), Bonn, Germany.
Anna Rieckmann, Department of Radiation Sciences, Umea University, 3A Norrlands universitetssjukhus, 901 87 Umeå, Sweden.
References
- 1.Sperling RA, Aisen PS, Beckett LA, Bennett DA, Craft S, Fagan AM, et al. Toward defining the preclinical stages of Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7(3):280–92. doi: 10.1016/j.jalz.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jack CR, Knopman DS, Jagust WJ, Petersen RC, Weiner MW, Aisen PS, et al. Tracking pathophysiological processes in Alzheimer’s disease: an updated hypothetical model of dynamic biomarkers. Lancet Neurol. 2013;12(2):207–16. doi: 10.1016/S1474-4422(12)70291-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jack CR, Weigand SD, Shiung MM, Przybelski SA, O’Brien PC, Gunter JL, et al. Atrophy rates accelerate in amnestic mild cognitive impairment. Neurology. 2008;70(19 Part 2):1740–52. doi: 10.1212/01.wnl.0000281688.77598.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jagust W. Vulnerable neural systems and the borderland of brain aging and neurodegeneration. Neuron. 2013;77(2):219–234. doi: 10.1016/j.neuron.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pievani M, Galluzzi S, Thompson PM, Rasser PE, Bonetti M, Frisoni GB. APOE4 is associated with greater atrophy of the hippocampal formation in Alzheimer’s disease. Neuroimage. 2011;55(3):909–19. doi: 10.1016/j.neuroimage.2010.12.081. [DOI] [PubMed] [Google Scholar]
- 6.Woolard AA, Heckers S. Anatomical and functional correlates of human hippocampal volume asymmetry. Psychiatry Res Neuroimaging. 2012;201(1):48–53. doi: 10.1016/j.pscychresns.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wachinger C, Golland P, Kremen W, Fischl B, Reuter M. BrainPrint: A discriminative characterization of brain morphology. NeuroImage. 2015 Apr 1;109:232–48. doi: 10.1016/j.neuroimage.2015.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wachinger C, Salat DH, Weiner M, Reuter M, Initiative ADN. Whole-brain analysis reveals increased neuroanatomical asymmetries in dementia for hippocampus and amygdala. Brain. 2016;139(12):3253–66. doi: 10.1093/brain/aww243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Medland SE, Jahanshad N, Neale BM, Thompson PM. Whole-genome analyses of whole-brain data: working within an expanded search space. Nat Neurosci. 2014;17(6):791–800. doi: 10.1038/nn.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bishop DV. Cerebral asymmetry and language development: cause, correlate, or consequence? Science. 2013;340(6138):1230531. doi: 10.1126/science.1230531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eyler LT, Vuoksimaa E, Panizzon MS, Fennema-Notestine C, Neale MC, Chen C-H, et al. Conceptual and data-based investigation of genetic influences and brain asymmetry: a twin study of multiple structural phenotypes. J Cogn Neurosci. 2014;26(5):1100–17. doi: 10.1162/jocn_a_00531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guadalupe T, Mathias SR, Theo GM, Whelan CD, Zwiers MP, Abe Y, et al. Human subcortical brain asymmetries in 15,847 people worldwide reveal effects of age and sex. Brain Imaging Behav. 2017;11(5):1497–514. doi: 10.1007/s11682-016-9629-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stage E, Duran T, Risacher SL, Goukasian N, Do TM, West JD, et al. The effect of the top 20 Alzheimer disease risk genes on gray-matter density and FDG PET brain metabolism. Alzheimers Dement Diagn Assess Dis Monit. 2016;5:53–66. doi: 10.1016/j.dadm.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Potkin SG, Guffanti G, Lakatos A, Turner JA, Kruggel F, Fallon JH, et al. Hippocampal atrophy as a quantitative trait in a genome-wide association study identifying novel susceptibility genes for Alzheimer’s disease. PloS One. 2009;4(8):e6501. doi: 10.1371/journal.pone.0006501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biffi A, Anderson CD, Desikan RS, Sabuncu M, Cortellini L, Schmansky N, et al. Genetic variation and neuroimaging measures in Alzheimer disease. Arch Neurol. 2010;67(6):677–85. doi: 10.1001/archneurol.2010.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chauhan G, Adams HH, Bis JC, Weinstein G, Yu L, Töglhofer AM, et al. Association of Alzheimer’s disease GWAS loci with MRI markers of brain aging. Neurobiol Aging. 2015;36(4):1765e7–1765. e16. doi: 10.1016/j.neurobiolaging.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furney SJ, Simmons A, Breen G, Pedroso I, Lunnon K, Proitsi P, et al. Genome-wide association with MRI atrophy measures as a quantitative trait locus for Alzheimer’s disease. Mol Psychiatry. 2011;16(11):1130–8. doi: 10.1038/mp.2010.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohannim O, Hua X, Rajagopalan P, Hibar DP, Jahanshad N, Grill JD, et al. Multilocus genetic profiling to empower drug trials and predict brain atrophy. NeuroImage Clin. 2013;2:827–35. doi: 10.1016/j.nicl.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J-Q, Wang H-F, Zhu X-C, Sun F-R, Tan M-S, Tan C-C, et al. GWAS-Linked Loci and Neuroimaging Measures in Alzheimer’s Disease. Mol Neurobiol. 2017;54(1):146– 53. doi: 10.1007/s12035-015-9669-1. [DOI] [PubMed] [Google Scholar]
- 20.Ramirez LM, Goukasian N, Porat S, Hwang KS, Eastman JA, Hurtz S, et al. Common variants in ABCA7 and MS4A6A are associated with cortical and hippocampal atrophy. Neurobiol Aging. 2016;39:82–9. doi: 10.1016/j.neurobiolaging.2015.10.037. [DOI] [PubMed] [Google Scholar]
- 21.Shen L, Kim S, Risacher SL, Nho K, Swaminathan S, West JD, et al. Whole genome association study of brain-wide imaging phenotypes for identifying quantitative trait loci in MCI and AD: A study of the ADNI cohort. Neuroimage. 2010;53(3):1051–63. doi: 10.1016/j.neuroimage.2010.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Z, Shen X, Pan W, Initiative ADN. Longitudinal analysis is more powerful than cross-sectional analysis in detecting genetic association with neuroimaging phenotypes. PloS One. 2014;9(8):e102312. doi: 10.1371/journal.pone.0102312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fox NC, Warrington EK, Freeborough PA, Hartikainen P, Kennedy AM, Stevens JM, et al. Presymptomatic hippocampal atrophy in Alzheimer’s disease: A longitudinal MRI study. Brain. 1996;119(6):2001–7. doi: 10.1093/brain/119.6.2001. [DOI] [PubMed] [Google Scholar]
- 24.Hollingworth P, Harold D, Sims R, Gerrish A, Lambert J-C, Carrasquillo MM, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet. 2011;43(5):429–35. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lambert J-C, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45(12):1452–8. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naj AC, Jun G, Beecham GW, Wang L-S, Vardarajan BN, Buros J, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet. 2011;43(5):436–41. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hibar DP, Stein JL, Renteria ME, Arias-Vasquez A, Desrivières S, Jahanshad N, et al. Common genetic variants influence human subcortical brain structures. Nature. 2015 doi: 10.1038/nature14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis: I. Segmentation and surface reconstruction. Neuroimage. 1999;9(2):179–94. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- 29.Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33(3):341–355. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- 30.Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis: II: inflation, flattening, and a surface-based coordinate system. Neuroimage. 1999;9(2):195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- 31.Fischl B, Sereno MI, Tootell RB, Dale AM. High-resolution intersubject averaging and a coordinate system for the cortical surface. Hum Brain Mapp. 1999;8(4):272–84. doi: 10.1002/(SICI)1097-0193(1999)8:4<272::AID-HBM10>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reuter M, Wolter F-E, Peinecke N. Laplace-Beltrami spectra as “Shape-DNA” of surfaces and solids. Comput-Aided Des. 2006;38(4):342–366. [Google Scholar]
- 33.Lian Z, Godil A, Bustos B, Daoudi M, Hermans J, Kawamura S, et al. A comparison of methods for non-rigid 3D shape retrieval. Pattern Recognit. 2013;46(1):449–61. [Google Scholar]
- 34.Bron EE, Smits M, van der Flier WM, Vrenken H, Barkhof F, Scheltens P, et al. Standardized evaluation of algorithms for computer-aided diagnosis of dementia based on structural MRI: The CADDementia challenge. NeuroImage. 2015 May 1;111:562–79. doi: 10.1016/j.neuroimage.2015.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wachinger C, Reuter M, Initiative ADN. Domain adaptation for Alzheimer’s disease diagnostics. Neuroimage. 2016;139:470–9. doi: 10.1016/j.neuroimage.2016.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saykin AJ, Shen L, Yao X, Kim S, Nho K, Risacher SL, et al. Genetic studies of quantitative MCI and AD phenotypes in ADNI: Progress, opportunities, and plans. Alzheimers Dement. 2015;11(7):792–814. doi: 10.1016/j.jalz.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nho K, Corneveaux JJ, Kim S, Lin H, Risacher SL, Shen L, et al. Whole-exome sequencing and imaging genetics identify functional variants for rate of change in hippocampal volume in mild cognitive impairment. Mol Psychiatry. 2013;18(7):781. doi: 10.1038/mp.2013.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nho K, Kim S, Risacher SL, Shen L, Corneveaux JJ, Swaminathan S, et al. Protective variant for hippocampal atrophy identified by whole exome sequencing. Ann Neurol. 2015;77(3):547–52. doi: 10.1002/ana.24349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34(8):816–34. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson WK, Hallmayer J, O’Hara R. Design considerations for characterizing psychiatric trajectories across the lifespan: application to effects of APOE-ε4 on cerebral cortical thickness in Alzheimer’s disease. Am J Psychiatry. 2011;168(9):894– 903. doi: 10.1176/appi.ajp.2011.10111690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verbeke G, Molenberghs G. Linear mixed models for longitudinal data. Springer Science & Business Media; 2009. [Google Scholar]
- 42.Khondoker M, Newhouse S, Westman E, Muehlboeck J, Mecocci P, Vellas B, et al. Linking genetics of brain changes to Alzheimer’s disease: sparse whole genome association scan of regional MRI volumes in the ADNI and AddNeuroMed cohorts. J Alzheimers Dis. 2015;45(3):851–64. doi: 10.3233/JAD-142214. [DOI] [PubMed] [Google Scholar]
- 43.Benjamini Y, Hochberg Y. On the adaptive control of the false discovery rate in multiple testing with independent statistics. J Educ Behav Stat. 2000;25(1):60–83. [Google Scholar]
- 44.Nithianantharajah J, Komiyama NH, McKechanie A, Johnstone M, Blackwood DH, St Clair D, et al. Synaptic scaffold evolution generated components of vertebrate cognitive complexity. Nat Neurosci. 2013;16(1):16–24. doi: 10.1038/nn.3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kirov G, Pocklington AJ, Holmans P, Ivanov D, Ikeda M, Ruderfer D, et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry. 2012;17(2):142–53. doi: 10.1038/mp.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oertel-Knöchel V, Linden DE. Cerebral asymmetry in schizophrenia. The Neuroscientist. 2011;17(5):456–67. doi: 10.1177/1073858410386493. [DOI] [PubMed] [Google Scholar]
- 47.Okada N, Fukunaga M, Yamashita F, Koshiyama D, Yamamori H, Ohi K, et al. Abnormal asymmetries in subcortical brain volume in schizophrenia. Mol Psychiatry. 2016;21(10):1460–6. doi: 10.1038/mp.2015.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chapuis J, Hansmannel F, Gistelinck M, Mounier A, Van Cauwenberghe C, Kolen KV, et al. Increased expression of BIN1 mediates Alzheimer genetic risk by modulating tau pathology. Mol Psychiatry. 2013;18(11):1225–34. doi: 10.1038/mp.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tan M-S, Yu J-T, Tan L. Bridging integrator 1 (BIN1): form, function, and Alzheimer’s disease. Trends Mol Med. 2013;19(10):594–603. doi: 10.1016/j.molmed.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 50.He F, Umehara T, Saito K, Harada T, Watanabe S, Yabuki T, et al. Structural insight into the zinc finger CW domain as a histone modification reader. Structure. 2010;18(9):1127–39. doi: 10.1016/j.str.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 51.Rosenthal SL, Barmada MM, Wang X, Demirci FY, Kamboh MI. Connecting the dots: potential of data integration to identify regulatory SNPs in late-onset Alzheimer’s disease GWAS findings. PloS One. 2014;9(4):e95152. doi: 10.1371/journal.pone.0095152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruiz A, Heilmann S, Becker T, Hernández I, Wagner H, Thelen M, et al. Follow-up of loci from the International Genomics of Alzheimer’s Disease Project identifies TRIP4 as a novel susceptibility gene. Transl Psychiatry. 2014;4(2):e358. doi: 10.1038/tp.2014.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao Y, Tan M-S, Wang H-F, Zhang W, Wang Z-X, Jiang T, et al. ZCWPW1 is associated with late-onset Alzheimer’s disease in Han Chinese: a replication study and meta-analyses. Oncotarget. 2016;7(15):20305. doi: 10.18632/oncotarget.7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chan SL, Kim WS, Kwok JB, Hill AF, Cappai R, Rye K-A, et al. ATP-binding cassette transporter A7 regulates processing of amyloid precursor protein in vitro. J Neurochem. 2008;106(2):793–804. doi: 10.1111/j.1471-4159.2008.05433.x. [DOI] [PubMed] [Google Scholar]
- 55.Rosenthal SL, Kamboh MI. Late-onset Alzheimer’s disease genes and the potentially implicated pathways. Curr Genet Med Rep. 2014;2(2):85–101. doi: 10.1007/s40142-014-0034-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shulman JM, Chen K, Keenan BT, Chibnik LB, Fleisher A, Thiyyagura P, et al. Genetic susceptibility for Alzheimer disease neuritic plaque pathology. JAMA Neurol. 2013;70(9):1150–7. doi: 10.1001/jamaneurol.2013.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Monsell SE, Mock C, Fardo DW, Bertelsen S, Cairns NJ, Roe CM, et al. Genetic differences between symptomatic and asymptomatic persons with Alzheimer’s disease neuropathologic change. Alzheimers Dement J Alzheimers Assoc. 2015;11(7):P767. [Google Scholar]
- 58.Dunstan ML, Gerrish A, Morgan T, Owens H, Badarinarayan N, Thomas RS, et al. THE ROLE OF CD2AP IN APP PROCESSING. Alzheimers Dement J Alzheimers Assoc. 2016;12(7):P458–9. [Google Scholar]
- 59.Francks C. Exploring human brain lateralization with molecular genetics and genomics. Ann N Y Acad Sci. 2015;1359(1):1–13. doi: 10.1111/nyas.12770. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1: Histogram of the number of scans per subject. Most subjects have between four and five scans.
Supplementary Figure S2: Boxplot of estimates for the interaction SNP x diagnosis for SNP rs117253277 and amygdala asymmetry with categorical coding. Matched samples for genotypes CC and CA were created to have a similar number of images per diagnostic group. We used random sampling to generate the matched samples and repeated this procedure 50 times. The estimate of the original model based on unequal sample size is shown as red dot. The median 2.08 and the mean 2.01 are close to the estimate of the original model (2.36).
Supplementary Figure S3: We studied gene expression quantitative trait loci (eQTL) of significant SNPs using gene expression data from postmortem healthy human brains from the UK Brain Expression Consortium (http://www.braineac.org/). The project analyzed up to 10 brain regions for 134 individuals: cerebellar cortex (CRBL), frontal cortex (FCTX), hippocampus (HIPP), medulla (specifically inferior olivary nucleus, MEDU), occipital cortex (specifically primary visual cortex, OCTX), putamen (PUTM), substantia nigra (SNIG), thalamus (THAL), temporal cortex (TCTX) and intralobular white matter (WHMT).
The plots show the gene expression levels stratified by genotypes of the corresponding SNP. Most of our findings were for SNP x diagnosis interactions; only rs10948363 and rs683250 showed a significant main effect SNP. Since expression data was collected on healthy subjects, we only expect differences for SNPs with significant main effect. rs10948363 was associated to amygdala asymmetry, but amygdala was not included in the eQTL mapping analysis. This only leaves SNP rs683250, which was associated to putamen asymmetry. In plot (a), we observe higher expression levels of DLG2 for GG than for GA or AA in putamen (red box). For completeness, we also include results for cis-eQTL mapping analysis for the other SNPs (b) – (e), except for rs117253277, which was not part of the Braineac database.
Supplementary Table S1: Diagnostic and demographic information of the ADNI sample used for the experiments.
Supplementary Table S2: Standardized regression coefficients and p-values for the analysis of lateral asymmetry with genetic loci for the linear mixed effects model without interactions. Adjusted p-values for the main effect SNP are presented, where we only show significant associations after FDR correction. p-values are rounded to five decimal places. The diagnosis is modeled as categorical variable, where we only report difference between CN and AD. The other factors were not significant.