Abstract
Background
Childhood asthma in inner city populations is a major public health burden and understanding early life immune mechanisms that promote asthma onset is key to disease prevention. Children who develop asthma demonstrate a high prevalence of aeroallergen sensitization and T helper 2 (Th2)-type inflammation, however the early life immune events that lead to Th2 skewing and disease development are unknown.
Objective
We sought to use RNA sequencing of peripheral blood mononuclear cells (PBMCs) collected at age 2 to determine networks of immune responses that occur in children who develop allergy and asthma.
Methods
In a high asthma risk inner city birth cohort, we compared gene expression by RNA sequencing in PBMCs collected at age 2 between children who developed ≥2 aeroallergen sensitizations including dust mite (DM) and/or cockroach (CR) by age 3 and asthma by age 7 (cases) and matched controls who did not develop any aeroallergen sensitization or asthma by age 7.
Results
PBMCs from the cases showed higher levels of expression of natural killer (NK) cell related genes. After CR or DM allergen but not tetanus antigen stimulation, PBMCs from the cases compared to the control group, showed differential expression of 244 genes. This gene set included upregulation of a densely interconnected NK cell-like gene network reflecting a pattern of cell activation and induction of inflammatory signaling molecules including key Th2-type cytokines IL9, IL13, and CCL17 as well as a dendritic cell (DC)-like gene network including upregulation of CD1 lipid antigen presentation molecules. The NK cell-like response was reproducible in an independent group of children with later onset allergic sensitization and asthma, and was found to be specific to only those children that develop both aeroallergen sensitization and asthma.
Conclusion
These findings provide important mechanistic insight into an early life immune pathway involved in Th2 polarization leading to development of allergic asthma.
Introduction
Asthma has emerged as a major public health problem in the United States (US) over the past 25 years. Its incidence has risen over several decades and it now affects over 23 million Americans, including 7 million children, and costs US society $56 billion annually.(1) The prevalence and severity of asthma are particularly high in low-income urban populations in the US.(2, 3) While asthma is a heterogeneous disease consisting of multiple endotypes, a substantial subset of asthma begins in childhood and the immune changes leading to disease occur early in life.(4–6) Currently no prevention strategies exist, so better understanding of the early life immune changes that contribute to development of disease is critical.
During childhood, allergic sensitization and associated Th2 cell immune elements are key risk factors for airway inflammation, low lung function, airway hyperresponsiveness and asthma.(7–10) Developing sensitization to multiple allergens in the first few years of life is especially important in light of its poor prognosis with respect to persistent disease, increased exacerbations, and low lung function.(11–13) While some genetic and environmental risk factors for Th2 inflammation and asthma development are known(14, 15), the early life immune pathways that drive the imbalance towards a Th2 immune response in humans are not well understood(6). Furthermore, not all individuals who develop allergen specific Th2 inflammation will develop asthma and the systemic immunologic changes that specifically predispose to allergic asthma are unclear.
Several innate immune cells including innate lymphoid cells type 2 (ILC2), NK cells, invariant natural killer T cells (iNKTs), γδ T cells, dendritic cells (DCs), monocytes, and others can contribute to increased Th2-type cytokine signaling and these cells are all key early responders to exogenous antigens and infections.(16–19) Studies in mouse models have demonstrated the importance of innate immune cells in the development of allergic airway responses (20–24), and differences in ILC2, NK, and iNKT cell populations have been observed in adults with established asthma.(25–28) However, demonstration of the innate immune mechanisms that can initiate Th2-asthma in humans is lacking.
To identify immunologic antecedents of allergic sensitization and asthma in early childhood, we studied children enrolled in an ongoing high-risk asthma birth cohort study, Urban Environment and Childhood Asthma (URECA).(29) Enrolled participants all have a parental history of allergic diseases or asthma and live in low-income census tracts. Per study design, the entire cohort had PBMCs collected at age 2 and stimulated with dust mite (DM), cockroach (CR) extracts, tetanus toxoid (TT), or medium alone (non-stimulated; NS). We used an unbiased whole genome RNA-sequencing approach coupled with cell deconvolution to investigate PBMC gene expression and stimulation responses, and here we report immunologic differences that can be detected at age 2 and correspond with the development of aeroallergen sensitization and asthma.
Methods
URECA population
URECA is a birth cohort study initiated in 2005 in inner city Baltimore, Boston, New York City and St. Louis, and details of the study design have been described elsewhere.(29) In brief, pregnant women age 18 or older were recruited with selection criteria including a history of asthma, allergic rhinitis, or eczema in the mother or father. Between February 2005 and March 2007, 1850 families were screened, 776 met eligibility criteria, and 560 newborns were enrolled at birth. Informed consent was obtained from the parent or legal guardian of the infant.
Maternal questionnaires were administered prenatally and postnatally, every three months through age 7 years, to ascertain wheezing illnesses and rhinitis symptoms. Allergen specific IgE (ImmunoCAP, Phadia, Uppsala Sweden) for milk, egg, peanut, and German cockroach was measured yearly until age 7 years. Specific IgE for dust mites (Dermatophagoides farinae, Dermatophagoides pteronyssinus), dog, cat, mouse, and Alternaria was added for ages 2–7 years. The lower limit of detection for the specific IgEs was 0.10 kU/L and we manually set all values that were below this to be 0.09 kU/L. Prick skin testing was performed at 3, 5, and 7 years of age for 14 common aeroallergens. Allergen sensitization was defined as either a wheal ≥ 3mm larger than the saline control on prick skin testing or specific IgE ≥ 0.35 kU/L. Asthma at age 7 was defined using a prespecified algorithm that included parent-reported physician asthma diagnosis, asthma symptoms, health care use for asthma, use of asthma medications in the previous year, spirometry with reversibility, and bronchial hyperresponsiveness assessed by methacholine inhalation challenge. Home visits three months after birth included house dust collection to assay for the allergens Bla g 1 (cockroach), Can f 1 (dog), Fel d 1 (cat), Der f 1 and Der p 1 (house dust mites), and Mus m 1 (mouse) by ELISA (Indoor Biotechnologies, Charlottesville, VA).
PBMC processing and stimulation studies
Peripheral blood samples were collected at age 2 and processed as previously described.(29) In short, at each URECA research center, within 16 hours of blood collection, 1.0x10^6 mononuclear cells were separated and incubated (48 hours, 37C, 5% CO2) in the presence of German cockroach extract (10mg/mL) (Greer Laboratories, Lenoir NC), dust mite extract (Dermatophagoides pteronyssinus) (10mg/mL) (Greer Laboratories, Lenoir NC), tetanus toxoid (5mg/mL) (MassBiologics, Mattapan MA), or medium alone (AIM-V media with human serum albumin (Invitrogen #12055-091)). Following incubation, cells were lysed using TRIzol (Tri-Reagent LS, Fisher Scientific), and maintained at −80C pending extraction of total cellular RNA.
Study Design
An overview of the study design and analytical flow is presented in Fig. S1. For the initial case-control cohort (cohort 1), cases (n=21) were URECA participants who met the following inclusion criteria: a) ≥2 aeroallergen sensitizations at age 3, 5, and 7 including sensitization to DM and/or CR at each age b) a diagnosis of asthma at age 7, and c) had PBMC RNA samples collected at age 2. Controls (n=30) were URECA participants with: a) no aeroallergen sensitization through age 7, b) did not have asthma at age 7, and c) had PBMC RNA samples collected at age 2. Controls were selected to match to cases on at least a bivariate level according to the following variables: gender, race, URECA site, birth season, and levels of Bla g 1, Der f 1, and Def p 1 measured in the home 3 months after birth. All available NS, CR, DM, and TT stimulation PBMC samples from these 51 individuals were selected for RNA-sequencing.
For cohort 2, seven additional URECA participants were selected who met the following inclusion criteria: a) ≥1 aeroallergen sensitization by age 7 including CR sensitization, b) a diagnosis of asthma at age 7, and c) PBMC RNA samples collected at age 2. Fourteen additional controls were selected to match these cases by the same criteria as the controls of cohort 1.
Furthermore all URECA participants with PBMC RNA samples collected at age 2 and who had sufficient year 7 outcome data for group classification (full cohort) were added to the sequencing run to allow a 4 group comparison based on CR sensitization (age 3, yes/no) and asthma (age 7, yes/no). For the latter analyses, only NS and CR stimulation PBMC samples were used for RNA-sequencing.
RNA sequencing
RNA was extracted from TRIzol following the manufacturer’s protocol. RNA quality was assessed using RNA electrophoresis (Agilent). All samples had RIN values ≥7. Sequencing libraries were constructed from total RNA using TruSeq RNA Sample Preparation Kits v2 (Illumina) and clustered onto a flowcell using a cBOT amplification system with a HiSeq SR v4 Cluster Kit (Illumina). Single-read sequencing was carried out on a HiSeq2500 sequencer (Illumina), using a HiSeq SBS v4 Kit to generate 58-base reads, with a target of approximately 10 million reads per sample.
Samples were processed in two separate batches. Cohort 1 was processed as a single batch. All additional samples (cohort 2 and full cohort) were processed as a second batch. One half of the samples from cohort 1 had sequencing library preparation and RNA-sequencing repeated during the second batch to control for batch effects.
Bioinformatic analysis
FASTQ files were downloaded from https://basespace.illumina.com. Libraries were processed via Galaxy.(30) Libraries were aligned via TopHat (v1.4.1)(31) to the human reference genome, Ensembl’s Homo sapiens GRCh38 version 77 (GRCh38.77.gtf). The single-paired flag was set to “single,” while all other TopHat parameters were set to defaults. HTSeq-count(32) was used to generate gene counts with mode as “Intersection (nonempty)” and minimum alignment quality set to 0 and otherwise set to default parameters.
Statistics
Genes with a trimmed mean of M values (TMM) normalized(33) count of at least 1 in 20% of libraries and a gene biotype(34) of protein coding went into further analysis, leaving 13,867 Ensembl IDs that went into further analysis. Count data were converted into log2-counts per million, while weighting the observations to estimate the mean-variance relationship and incorporating the correlation between samples from the same individual, using the voom function(35) in limma(36).
Normalized voom counts were then used to test case vs. control in NS samples using linear modeling including batch effects of site, gender and blood draw year. To test differences between cases and controls in response to CR stimulation, a linear model was built containing group (case vs. control), stimulation (non-stimulation and CR stimulation), site, gender, blood draw year and home allergen levels (Bla g 1 in bedroom and living room) and a random effect for individual. The equivalent was done for DM stimulation but using NS, DM stimulation, and Der f 1 and Der p 1 levels in bedroom and living room. Where cell specific gene sets were tested rather than individual genes, the geometric mean of all the genes within the set was used as a summary expression for the mixed effect linear model. Multiple testing correction was by the Benjamini–Hochberg procedure controlling the false discovery rate (FDR) at level 0.05.
Cell deconvolution to assess which cell types significantly impact gene expression was performed using the Immune Response In Silico (IRIS) data set(37) as the reference data set to determine cell specific markers genes.(38) Marker genes for B cell, DC, monocyte, NK cell, CD4+ T cell, and CD8+ T cell were used. Surrogate cell proportions were calculated for our samples using the digital sorting algorithm (DSA).(39) Finally, genes that were significant for case vs. control in response to stimulation (CR or DM) were assigned to cell type using the Subset Prediction from Enrichment Correlation (SPEC) algorithm.(40) The stability of the gene assignment to cell type was assessed through sub-sampling the marker genes through 100 iterations. Gene set correlation p-values were calculated by simulating random gene sets of equivalent size over 10,000 simulations.
To assess for correlations among gene sets and clinical data, a general linear model was used with a gamma distribution and log link function to calculate a parameter estimate and p-value. For comparisons of clinical and demographic data among groups, p-values are calculated as appropriate for data – t-test for normally distributed, continuous data, Kruskal-Wallis test for non-normally distributed, continuous data, and either chi-squared or Fisher’s exact tests for categorical data.
Pathway Analysis
Differentially expressed genes and gene sets were investigated using Database for Annotation Visualization and Integrated Discovery version 6.7.(41, 42) This database uses a modified Fisher exact test to identify specific biological/functional categories that are overrepresented in gene sets in comparison with a reference set (default human genome was used as the reference set). Gene Ontology biological processes (GO BP), canonical pathways from Kyoto Encyclopedia of Genes and Genomes (KEGG), and pathways from BioCarta that were associated with the differentially expressed genes were used as databases.
STRING version 10.0, which is a database of known and predicted protein-protein interactions was used to determine interaction networks of cell specific gene sets. Default parameters were used, including all interaction sources and using a minimum required interaction score or 0.40 to be included in the network.(43) Cytoscape version 3.2.1 was used to draw interaction networks according to a prefuse force directed layout using the combined interaction score exported from STRING.(44) Unconnected nodes were excluded.
Results
Increased NK cell-type gene expression is associated with development of allergic asthma
We initially focused on two groups of children defined by clinical outcome in a nested case-control design (cohort 1). The case group was composed of all URECA participants who had ≥2 aeroallergen sensitizations including DM and/or CR at age 3, 5, and 7 and had a diagnosis of asthma at age 7. We defined this group as early onset multiple sensitizations and asthma (“Early”, n=21). The control group (“Neither”, n=30) was composed of URECA participants who had no aeroallergen sensitization at age 3, 5, or 7 and did not have asthma at age 7. These individuals were matched to the Early group according to demographic variables and allergen exposure in the home. RNA-sequencing was performed on the year 2 NS, CR, DM, and TT samples from these 51 individuals.
The Early group differed clinically from the Neither group with a higher incidence of wheezing illnesses and higher eczema area and severity index (EASI) scores in the first year of life, which are known risk factors for development of asthma. At age 2, the time of blood draw, they had higher total IgE levels and higher specific IgEs to food and airborne allergens, though most specific IgEs were below the cutoff for definition of sensitization (≤0.35 kU/L). By age 7 these same clinical differences were more pronounced and the IgE levels markedly increased and demonstrated multiple specific sensitizations (Table S1).
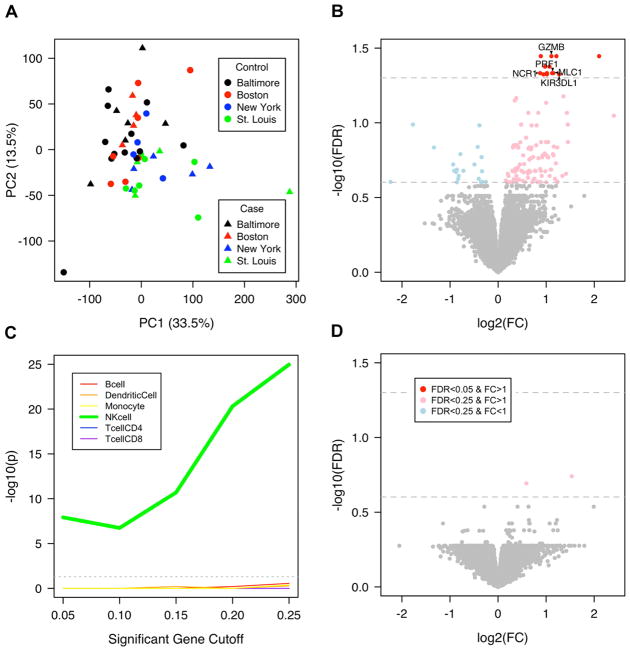
Principal component analysis (PCA) of gene expression of unstimulated PBMCs from these two groups showed similar global patterns among individuals (Fig. 1A). The primary source of variability in gene expression was city of residence (site); all four sites were equally represented in both groups. Assessment for differentially expressed genes comparing Early versus Neither while controlling for site demonstrated a small set of differentially expressed genes (13 genes at FDR<0.05, 131 genes at FDR<0.25) between groups, the majority of which (13/13; 110/131) were expressed at a higher level in the Early group (Fig. 1B). The upregulated genes were highly enriched for NK cell specific genes and no other cell type from the IRIS reference dataset(37) (Fig. 1C) as well as the KEGG (45, 46) pathway NK cell mediated cytotoxicity (FDR=2.7E-6).
Fig. 1. Differential expression of NK cell genes is associated with allergic asthma.
(A) Site, not case-control assignment, accounted for the greatest source of gene expression variability in unstimulated PBMCs by PCA. (B) The set of differentially expressed genes between Early and Neither in unstimulated PBMC samples (NS linear model with fixed effects for site, gender, blood draw year; nEarly = 19, nNeither = 30). 13 genes were significant at FDR<0.05 and FC>1, indicated by a red dot. Labelled genes (arrowheads) are NK cell marker genes in the IRIS database that have an FDR<0.05. Genes with a semi-transparent red dot have a FC>1 and FDR<0.25. Genes with a semi-transparent blue dot have a FC<1 and FDR<0.25. (C) Genes that showed higher expression in Early compared to Neither were highly enriched for IRIS NK cell marker genes and no other cell type by Fisher’s exact test of overlap between significant genes and IRIS marker genes. (D) There were no differentially expressed genes with FDR<0.05 between Early and Neither after tetanus toxoid stimulation (tetanus stimulation linear model with fixed effects for site, gender, blood draw year; nEarly = 19, nNeither = 30).
Gene expression of PBMCs from the same individuals after stimulation with the control antigen TT (to which all children had been vaccinated) demonstrated robust gene expression changes due to stimulation (Fig. S2A) and no significant differences in gene expression between groups after stimulation (Fig. 1D), documenting a similar peripheral immune response to this common antigen to which children have uniform exposure. These results demonstrate that children who will develop early onset multiple sensitizations and asthma have higher gene expression of NK cell-related genes in resting PBMCs at age 2 relative to matched controls, but show similar responses to stimulation with a common vaccine antigen.
Allergen stimulation causes NK cell and Th2-like gene activation
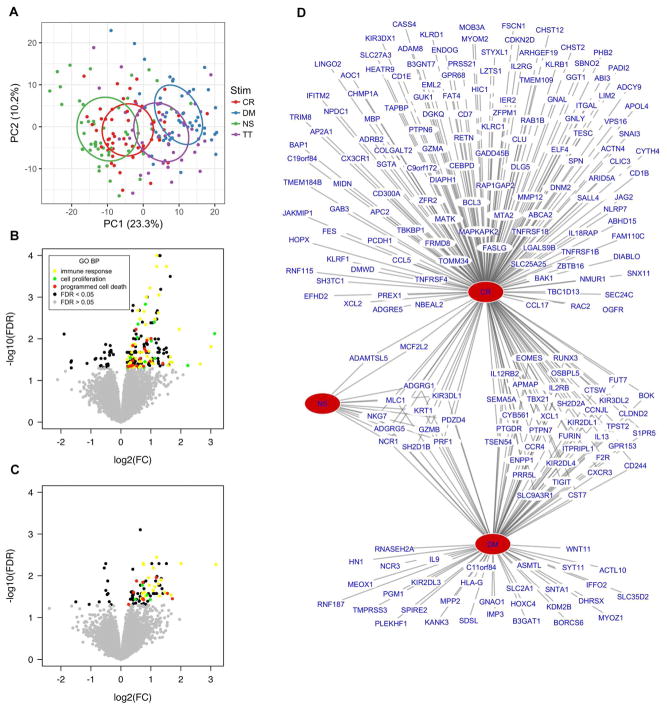
We next examined the CR and DM stimulated samples in cohort 1. Among all individuals, each allergen stimulation showed robust global gene expression relative to unstimulated samples (Fig. S2B, S2C). The type of stimulation accounted for the greatest amount of variability among these samples and each allergen stimulation showed a relatively distinct pattern of global gene expression (Fig. 2A).
Fig. 2. Allergen stimulation of PBMCs leads to robust gene expression changes.
(A) Stimulus accounts for the greatest source of gene expression variability in PBMCs by PCA. Ellipses represent 95% confidence intervals. (B) CR-Early individuals demonstrated significant differences in expression of 206 genes in response to CR stimulation compared to Neither (FDR<0.05). (C) DM-Early individuals demonstrated significant differences in expression of 91 genes in response to DM stimulation compared to Neither (FDR<0.05) (Stimulation linear modeling with fixed effects for site, gender, blood draw year, home allergen levels, and stimulation and random effect for individual; nCR-Early = 16, nDM-Early = 13, nNeither = 30). Differentially expressed genes in both comparisons are enriched for similar GO BP pathways. (D) Significantly upregulated genes in the Early group in NS, CR, and DM stimulated samples show a high degree of overlap as depicted in a bipartite network (P=3.6E-75; Fisher’s exact test, CR and DM upregulated genes).
Of the 21 Early individuals, the 16 who developed CR sensitization by age 3 (CR-Early) demonstrated significant differences in expression of 206 genes in response to CR stimulation compared to the Neither group (FDR<0.05) (Fig. 2B). Similarly the 13 Early who were DM sensitized by age 3 (DM-Early) demonstrated significantly different expression of 91 genes in response to DM stimulation compared to Neither (FDR<0.05) (Fig. 2C). No genes were differentially expressed among the samples in association with total or allergen specific IgE levels at age 2 in linear models. Furthermore a similar pattern of gene expression differences was observed between children in the Early group stratified by allergen sensitization at age 2, demonstrating these findings are independent of whether the Early individuals were already sensitized at the time of blood draw.
The CR and DM-induced gene lists showed enrichment for the same GO biologic processes: immune response, cell proliferation, and programmed cell death. Genes in each of these pathways showed increased expression with both allergen stimulations in Early relative to Neither (Fig. 2B, 2C). These two gene lists, which are derived from different subsets of individuals and are induced by different allergen stimulations show highly significant overlap (Fig. 2D; p=3.6E-75; Fisher’s exact test) suggesting the observed responses represent a common molecular response to allergen stimulation in susceptible individuals. Of all the 244 genes differentially expressed in the combined gene list, we focused on the 213 that showed increased expression in Early relative to Neither with both allergen stimulations (Table S2). None of these 213 genes were differentially expressed between groups after TT stimulation.
The set of allergen-upregulated genes specific to the Early group contains the same 13 genes that showed higher expression in the unstimulated samples (Fig. 2D); all these genes were further upregulated by allergen stimulation in Early (Table S2). These 213 genes include numerous additional NK cell-related genes (FDR=6.9E-21 for IRIS NK cell marker genes; FDR=2.8E-6 for KEGG natural killer cell-mediated cytotoxicity) as well as several cytokine signaling genes (FDR=7.5E-4 for KEGG cytokine-cytokine receptor interaction) including multiple signaling genes implicated in Th2-type inflammation such as IL9, IL13, CCL17, CCL5, and CCR4.
NK cells and DCs represent a primary source of response to allergen
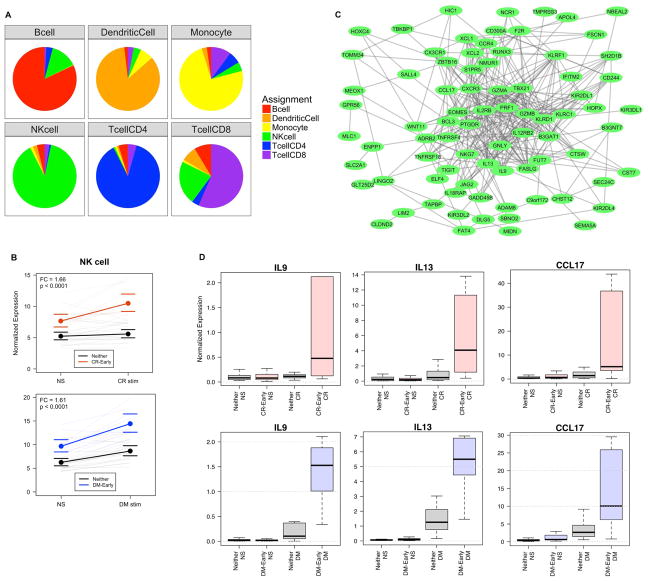
The primary cellular source of the 244 differentially expressed gene transcripts was assessed by computational deconvolution using established markers for 6 major cell types contained in PBMCs (Table S3).(40, 47) 108 of these 244 differentially expressed genes could be assigned to NK cells and were not associated with other cell types available for deconvolution (Fig. 3A, Fig. S3). All of these NK cell assigned genes were increased in expression after CR or DM stimulation in Early compared to Neither. These NK cell-associated genes show a high level of gene-gene correlation (r=0.31 p<0.0001) and as a gene set show a coherent increase in expression level with both CR and DM stimulation in Early but not Neither (Fig. 3B).
Fig. 3. Allergen stimulation of PBMCs cause NK cell activation and Th2-like inflammation.
(A) The 244 genes could each be assigned to 1 of the 6 cell subsets using marker gene cell deconvolution. Sensitivity analysis showed that the gene assignments were relatively stable over 100 bootstrap simulations (SPEC sensitivity analysis). (B) The NK cell gene set shows significantly higher baseline expression in Early compared to Neither and increases significantly with allergen stimulation in both the CR-Early and DM-Early groups; bounds represent 95% confidence intervals (Stimulation linear models with fixed effects for site, gender, blood draw year, home allergen levels, and stimulation; random effect for individual; p-value and FC represent group term controlling for stimulation). (C) NK cell genes represent a densely connected network of known gene-gene interactions. (D) Boxplot of normalized expression of IL9, IL13, and CCL17 showing group difference with allergen stimulation (Pink=CR-Early, Blue=DM-Early, Grey=Neither).
These NK cell-associated genes represent a densely clustered network of known gene-gene interactions and associations (Fig. 3C) and are enriched for the KEGG pathways NK cell mediated cytotoxicity (FDR=2.0E-5), cytokine-cytokine receptor interaction (FDR=3.4E-5), antigen processing and presentation (FDR=3.9E-5), and chemokine signaling pathway (FDR=5.3E-2). The NK cell-type response includes both Th1 and Th2 type inflammation genes, but shows particularly high elevation of critical Th2-inflammatory cytokines IL9, IL13, and CCL17 (Fig. 3D) suggesting that NK cell derived Th2 cytokines may be a principal source of skewing towards Th2 inflammation at this developmental stage of the immune system. In contrast, CD4+ T cell assigned genes were predominantly decreased in expression by allergen stimulation (Table S3) and were not correlated with these Th2-inflammatory cytokines.
A separate set of 52 of the 244 differentially expressed genes showed an expression pattern highly correlated with DC marker genes and not with other cell types (Fig. 3A, Fig. S3). These DC-associated genes show a high level of gene-gene correlation (r=0.53 p<0.0001) and a moderate increase in response to CR allergen stimulation specific to CR-Early and not seen in Neither (Fig. S4A). In contrast, the DC gene response to DM stimulation was not significant in the DM-Early individuals (Fig. S4B). These DC-associated genes form a smaller network of known gene-gene interactions (Fig. S4C) but do not show significant pathway specific enrichment. The observed DC gene set includes the important lipid antigen presenting genes CD1B and CD1E, both of which were highly induced by CR stimulation in the CR-Early group and also show increase with DM stimulation in the DM-Early group (Fig. S4D).
The remaining differentially expressed genes show correlation with B cell, CD4+ T cell, CD8+ T cell, and monocyte marker genes, but they form smaller gene sets and do not show enrichment for known molecular pathways. As gene sets, they show relatively modest differences between Early and Neither with stimulation. The relevance of these differences is less evident and could either represent changes in cell composition, uncharacterized molecular pathways, or functional pathways this study is underpowered to detect.
NK cell-type response is specific to development of allergy and asthma
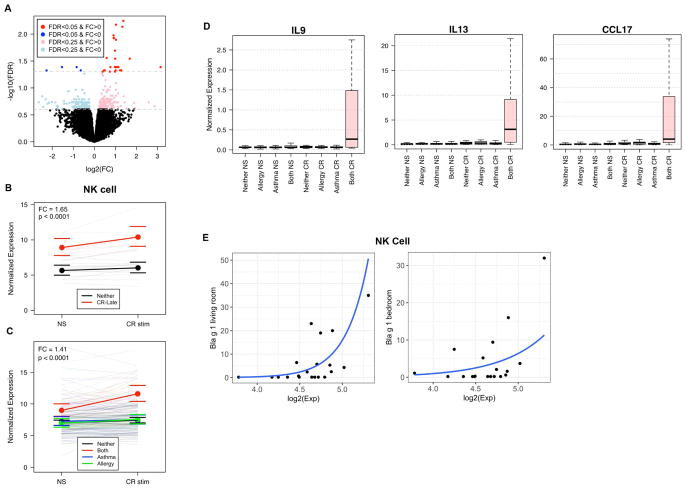
To assess the reproducibility of our findings and the specificity to the development of both allergy and asthma, we investigated the response to stimulation in an independently selected group of URECA participants (cohort 2). Cohort 1 had focused on individuals with the most severe phenotype of interest – specifically those with multiple sensitizations by age 3 and asthma by age 7. There were no additional URECA participants who met this selection criteria, however there were participants who developed CR sensitization between ages 3 and 7 years and asthma by age 7. Seven such individuals were identified, 3 became CR sensitized by age 5 and the other 4 became CR sensitized by age 7; 6/7 had multiple allergic sensitizations at age 7. This group was considered “Late” allergy and asthma onset and was compared to 14 newly selected and matched “Neither” individuals who did not develop allergic sensitization or asthma by age 7. Despite the later onset of allergen sensitization, a similar difference in gene expression was observed in PBMCs collected at age 2. In Late compared to Neither, unstimulated PBMCs and those stimulated with CR extract overexpressed the same NK cell gene set that was exhibited by the Early group (Fig. 4A, 4B). A difference in the DC gene set was not observed in this cohort.
Fig. 4. NK cell gene activation is specific to allergic asthma.
(A) The Late group demonstrated significant differences in expression of similar genes in response to CR stimulation compared to controls (31 genes at FDR<0.05, 326 genes at FDR<0.25). (Stimulation linear model with fixed effects for site, gender, blood draw year, home allergen levels, and stimulation, and random effect for individual; nLate = 7, nNeither = 14). (B) The NK cell gene set showed significantly higher baseline expression in Late compared to Neither, and expression increased significantly with CR stimulation in Late (p-value and FC represent group term controlling for stimulation; number of individuals in each comparison is represented in parentheses; bounds represent 95% confidence intervals) (C) The NK cell gene set shows significantly higher baseline expression in all cases (Red; “Both” = Early and Late groups, n=20) compared to Neither (Black, n=102), asthma only (Blue; “Asthma”, n=40), and CR allergy only (Green; “Allergy”, n=35) groups and increases with CR stimulation only in the Both group. (D) Boxplot of normalized expression of IL9, IL13, and CCL17 showing group difference with allergen stimulation (Pink=CR-Early, Blue=DM-Early, Grey=Neither). (E) The average expression value of the NK cell gene set showed correlation with Bla g 1 levels measured in the living room (p<0.05) and bedroom (p=0.08) in the Both group.
We next tested whether NK cell expression was related to allergy, asthma, or both. Children from the CR-Early and Late groups were combined to a single group (n=20), and their NS and CR stimulation gene expression data was compared to other subgroups of URECA participants classified by outcomes at age 7: those who developed CR sensitization without asthma (n=35), those who developed asthma without any allergic sensitization (n=40), and those who developed neither allergic sensitization nor asthma (n=102) (Table S4). Increased NK cell gene expression at age 2 was found only in children who developed both CR sensitization and asthma by age 7 (Fig. 4C, 4D). Notably, similar results were found for increased expression of the DC gene set after CR stimulation (Fig. S4E). The other 3 groups were indistinguishable from one another in expression of NK and DC gene sets.
To better understand the etiology of this NK cell gene set response among the Early and Late groups, we investigated the correlation of multiple clinical variables with NK cell gene set expression. The demographic variable that most highly correlated with expression level of the NK cell gene set was CR allergen levels in the home measured in the first year of life. Among these 20 individuals, a higher level of CR exposure correlated with a higher level of NK cell gene set expression in response to CR stimulation (Fig. 4E). Home allergen levels did not correlate with NK cell gene expression within the other 3 groups. This finding suggests these disease susceptible individuals may develop an NK response directly influenced by higher levels of allergen exposure early in life.
Discussion
Children who develop multiple allergies in early life have an increased risk for wheezing and asthma. To identify immunologic correlates of early multiple allergic sensitization and asthma in high-risk urban children participating in the URECA study, we analyzed patterns of allergen-specific gene expression at age 2 years and compared these findings to longitudinal patterns of allergic sensitization and asthma assessed through age 7 years. Our results reveal differential gene expression in unstimulated and allergen stimulated PBMCs collected at age 2 in children who had early multiple sensitization followed by asthma. Affected children exhibited increased expression of NK cell-related genes in unstimulated PBMCs, and differential expression of these NK cell-related genes was augmented by stimulation with CR and DM allergen but not tetanus toxoid. The largest set of differentially expressed genes after allergen stimulation was attributed to NK cells through cell marker deconvolution. This gene set describes a densely interconnected gene-gene interaction network that likely represents expansion of NK cells or a similar population of cytotoxic lymphocytes, as well as activation of these cells leading to transcription of key immune signaling molecules including the canonical Th2 cytokines IL9, IL13, and CCL17. We also observed with CR stimulation an upregulation of DC-related genes including CD1 lipid presentation molecules.
Our results suggest that NK cell-like innate immune responses in early life contribute to development of aeroallergen sensitization driven asthma. Mouse models have demonstrated a key role for NK cells in allergy and asthma development and lack of NK cells attenuates airway inflammation in allergen sensitized mice.(23, 24, 48, 49) Furthermore, children and adults with asthma have previously been observed to have higher levels of NK cells than nonasthmatics after disease onset(28, 50–52) and NK cells are a known source of Th2 cytokines(53, 54). Specifically subpopulations of NK cells from adults with asthma have been shown to produce Th2 cytokines (55) and also to directly regulate T cell activation(56). Our results are among the first evidence that NK cell-type responses in early life appear to play a direct role in the development of allergic asthma in children.
We have also found evidence of upregulation of lipid antigen presentation by DCs in response to CR, which is coincident with the observed NK cell response. Bidirectional NK-DC cross-talk can modify the differentiation of memory CD4+ T cell responses.(57–59) In mouse models, NK cells can also induce maturation of DCs and regulate their participation in allergen presentation and allergic sensitization.(23, 60–62) Our data implicate immune responses related to both NK cells and DCs in the early life immunopathogenesis leading to allergic asthma.
Having observed the NK cell and DC type responses within this nested case-control cohort, we were able to demonstrate the same NK cell finding in a second group of children who developed allergic sensitization after age 3. Furthermore, we showed the specificity of these findings to children who develop aeroallergen sensitization with asthma compared to those who develop only one of the conditions. Thus our results suggest that increased expression of NK-related genes occurs before allergic sensitization, and predisposes specifically to asthma associated with allergic sensitization.
NK cells and DCs may differ intrinsically in children who develop allergic sensitization and asthma, or may be modulated early in life by environmental factors. Given that these children tend to have more wheezing illnesses and respiratory viral infections in early life as observed here and in previous work (63) one possible mechanism is a recruitment of NK cells from the periphery to the lung during those infections as suggested by findings in adult asthma (64), where they could skew towards type 2 inflammation. However, viral infection alone seems insufficient to drive this NK signature since it was lacking in the group that developed asthma without allergic sensitization despite those individuals also having a higher incidence of early life wheezing illnesses (Table S4). We did observe that among children who develop disease, the magnitude of the NK cell-type response correlated with CR allergen levels in house dust suggesting allergen or associated exposures may play a key role in this immune response. Since cockroach allergen is also a marker of specific microbial exposures, and the cockroach extract used in this study contains endotoxins and other bacterial components, it is possible that the microbial environment of these children also contributes to the observed immune response in light of known associations between the microbiome and disease development in this population (65, 66). Whether NK cell responses are shaped by environmental, genetic, or epigenetic differences is under investigation. We have also yet to determine how NK cell responses change longitudinally during immune development and manifestation of clinical disease among these children. Assessing each of these factors will be critical to better understand the mechanisms by which NK cells can contribute to disease development, improving identification of at risk children early in life, and ultimately identifying disease prevention strategies.
We used computational cell deconvolution to assess the source of the gene expression differences observed. A distinct advantage to assessing immune responses by this method in a cell mixture is the ability to observe global expression patterns along with cell specificity and to infer potential cell-cell interactions.(47) However, while the gene sets described show high correlation with NK and DC cell marker genes, this assignment is probabilistic and limited by the ability to resolve cell types by patterns of marker gene expression in a complex mixture. Cells with highly similar transcriptional profiles including for example NK cells, iNKT cells, and γδ T cells could each be contributing in part to the observed NK cell-type gene expression. In this study only bulk PBMC RNA had been stored, but in future studies cell sorting followed by population or single cell RNA-sequencing would help to resolve the source of the signal with greater specificity.
We conclude that children prone to aeroallergen sensitization and asthma display an increased expression of NK cell-related genes in PBMCs in early life that is markedly upregulated upon stimulation with allergen but not tetanus toxoid. This gene expression pattern includes transcription of IL9, IL13, CCL17, CCL5, and CCR4 that likely promote skewing towards Th2 polarization. These findings are further supported by observed upregulation of DC genes after allergen stimulation including those involved in lipid antigen presentation. These findings provide evidence for a mechanistic link for susceptibility to allergic asthma in high-risk children and suggest potential methods for early disease risk identification and disease prevention. Our findings also have the potential to identify key biomarkers for allergic asthma development, which will be necessary to conduct population targeted asthma prevention studies.
Supplementary Material
Key Messages.
-
Inner city children who develop allergy and asthma by age 7 already show differences in PBMC gene expression at age 2 with:
1) Higher expression of cytotoxic lymphocyte genes in resting PBMCs
2) Upregulation of two distinct gene networks related to NK cells and dendritic cells that include multiple Th2-type cytokines
These findings are specific to the development of allergic asthma and not observed in children who develop only allergy or non-allergic asthma.
Acknowledgments
We are grateful to the entire URECA collaboration which consists of the following institutions and investigators (principal investigators are indicated by asterisks): Johns Hopkins University School of Medicine, Baltimore, MD – R. Wood*, S. Leimenstoll, R. Spellman; Boston University School of Medicine, Boston, MA, – G. O’Connor*, B. Caldwell, R. Cohen, E. Collins, S. Dorans, A. Florea, L. Gagalis, E. Gjerasi, D. Kretschman, M. Macharia, C. Sakow, S. Steinbach, A. Strakus, B. West; Columbia University Medical Center, New York, NY – M. Kattan*, E. Arteago-Solis, A. Bello, Y. Fernandez-Pau, C. Lamm, S. Lovinsky-Desir, D. Perlaza, L. Peters, M. Pierce, C. Sanabia, S. Tsang, A. Valones, N. Whitney, P. Yaniv; Washington University School of Medicine and St Louis Children’s Hospital, St. Louis, Missouri – L. Bacharier*, A. Beigelman, G. Bloomberg, E. Tesson, A. Freie; National Institute of Allergy and Infectious Diseases, Bethesda, MD – P. Gergen, A. Togias, E. Smartt, C. Czarniecki; University of Wisconsin School of Medicine and Public Health, Madison, WI – J. Gern, W. Busse*, C. Sorkness, S. Doyle, R. Burton, P. Heinritz, K. Spring, A. Dresen, S. Ramratnam; Rho Inc. Chapel Hill, NC – S. Arbes*, C. Visness*, A. Calatroni, K. Jaffee, M. Yaeger, and H. Mitchell. We are grateful to all the families who have participated in the URECA study. We would like to thank D. Chaussabel and G. Nepom for their advice and discussions.
Funding: This project has been funded in whole or in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, under Contract numbers NO1-AI-25496, NO1-AI-25482, HHSN272200900052C, HHSN272201000052I, 1UM1AI114271-01 and UM2AI117870. Additional support was provided by the National Center for Research Resources, National Institutes of Health, under grants RR00052, M01RR00533, 1UL1RR025771, M01RR00071, 1UL1RR024156, UL1TR000040, UL1TR001079 and 5UL1RR024992-02.
Abbreviations
- CR
Cockroach
- DC
Dendritic Cell
- DM
Dust Mite
- DSA
Digital Sorting Algorithm
- EASI
Eczema Area and Severity Index
- FC
Fold Change
- FDR
False Discovery Rate
- GO BP
Gene Ontology Biological Process
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- IgE
Immunoglobulin E
- ILC2
Innate Lymphoid Cells type 2
- iNKT
Invariant Natural Killer T cell
- IRIS
Immune Response In Silico
- NK
Natural Killer
- PBMC
Peripheral Blood Mononuclear Cell
- PCA
Principal Component Analysis
- RNA
Ribonucleic Acid
- SPEC
Subset Prediction from Enrichment Correlation
- Th2
T helper 2
- TMM
Trimmed Mean of M values
- TT
Tetanus Toxoid
- URECA
Urban Environment and Childhood Asthma
- US
United States
Footnotes
Author contributions: RNA sequencing data were generated at the Benaroya Research Institute. M.C.A., S.P., W.W.B., and J.E.G. designed the study. M.C.A., E.W., and K.J. conducted the analyses. G.T.O, L.B.B., G.R.B., M.K., R.A.W., are the URECA site PIs responsible for sample collection. C.M.V. and P.L. coordinated sample and data management. M.C.A. and J.E.G. wrote the manuscript.
Competing interests: The authors declare no conflict of interest.
Data and material availability: RNA-sequencing data were deposited to the National Center for Biotechnology Information Gene Expression Omnibus with accession number GSE96783.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Barnett SBL, Nurmagambetov TA. Costs of asthma in the United States: 2002–2007. Journal of Allergy and Clinical Immunology. 2011;127(1):145–52. doi: 10.1016/j.jaci.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 2.Weiss KB, Gergen PJ, Crain EF. Inner-city asthma. The epidemiology of an emerging US public health concern. Chest. 1992;101(6):362S–7. doi: 10.1378/chest.101.6.362s. [DOI] [PubMed] [Google Scholar]
- 3.Crain EF, Weiss KB, Bijur PE, Hersh M, Westbrook L, Stein RE. An estimate of the prevalence of asthma and wheezing among inner-city children. Pediatrics. 1994;94(3):356–62. [PubMed] [Google Scholar]
- 4.Macaubas C, de Klerk NH, Holt BJ, Wee C, Kendall G, Firth M, et al. Association between antenatal cytokine production and the development of atopy and asthma at age 6 years. The Lancet. 2003;362(9391):1192–7. doi: 10.1016/s0140-6736(03)14542-4. [DOI] [PubMed] [Google Scholar]
- 5.Stern DA, Guerra S, Halonen M, Wright AL, Martinez FD. Low IFN-γ production in the first year of life as a predictor of wheeze during childhood. Journal of Allergy and Clinical Immunology. 2007;120(4):835–41. doi: 10.1016/j.jaci.2007.05.050. [DOI] [PubMed] [Google Scholar]
- 6.Bousquet J, Gern JE, Martinez FD, Anto JM, Johnson CC, Holt PG, et al. Birth cohorts in asthma and allergic diseases: report of a NIAID/NHLBI/MeDALL joint workshop. J Allergy Clin Immunol. 2014;133(6):1535–46. doi: 10.1016/j.jaci.2014.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robinson DS, Hamid Q, Ying S, Tsicopoulos A, Barkans J, Bentley AM, et al. Predominant T H2 -like Bronchoalveolar T-Lymphocyte Population in Atopic Asthma. New England Journal of Medicine. 1992;326(5):298–304. doi: 10.1056/NEJM199201303260504. [DOI] [PubMed] [Google Scholar]
- 8.Grunig G, Warnock M, Wakil AE, Venkayya R, Brombacher F, Rennick DM, et al. Requirement for IL-13 independently of IL-4 in experimental asthma. Science. 1998;282(5397):2261–3. doi: 10.1126/science.282.5397.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wills-Karp M. Interleukin-13: Central Mediator of Allergic Asthma. Science. 1998;282(5397):2258–61. doi: 10.1126/science.282.5397.2258. [DOI] [PubMed] [Google Scholar]
- 10.Zhang D-H, Yang L, Cohn L, Parkyn L, Homer R, Ray P, et al. Inhibition of Allergic Inflammation in a Murine Model of Asthma by Expression of a Dominant-Negative Mutant of GATA-3. Immunity. 1999;11(4):473–82. doi: 10.1016/s1074-7613(00)80122-3. [DOI] [PubMed] [Google Scholar]
- 11.Simpson A, Tan VY, Winn J, Svensen M, Bishop CM, Heckerman DE, et al. Beyond atopy: multiple patterns of sensitization in relation to asthma in a birth cohort study. Am J Respir Crit Care Med. 2010;181(11):1200–6. doi: 10.1164/rccm.200907-1101OC. [DOI] [PubMed] [Google Scholar]
- 12.Havstad S, Johnson CC, Kim H, Levin AM, Zoratti EM, Joseph CL, et al. Atopic phenotypes identified with latent class analyses at age 2 years. J Allergy Clin Immunol. 2014;134(3):722–7e2. doi: 10.1016/j.jaci.2014.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belgrave DC, Buchan I, Bishop C, Lowe L, Simpson A, Custovic A. Trajectories of lung function during childhood. Am J Respir Crit Care Med. 2014;189(9):1101–9. doi: 10.1164/rccm.201309-1700OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ober C, Yao TC. The genetics of asthma and allergic disease: a 21st century perspective. Immunol Rev. 2011;242(1):10–30. doi: 10.1111/j.1600-065X.2011.01029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raedler D, Schaub B. Immune mechanisms and development of childhood asthma. The Lancet Respiratory Medicine. 2014;2(8):647–56. doi: 10.1016/S2213-2600(14)70129-8. [DOI] [PubMed] [Google Scholar]
- 16.Deniz G, van de Veen W, Akdis M. Natural killer cells in patients with allergic diseases. Journal of Allergy and Clinical Immunology. 2013;132(3):527–35. doi: 10.1016/j.jaci.2013.07.030. [DOI] [PubMed] [Google Scholar]
- 17.Walker JA, Barlow JL, McKenzie ANJ. Innate lymphoid cells — how did we miss them? Nat Rev Immunol. 2013;13(2):75–87. doi: 10.1038/nri3349. [DOI] [PubMed] [Google Scholar]
- 18.Holt PG, Sly PD. Interaction Between Adaptive and Innate Immune Pathways in the Pathogenesis of Atopic Asthma. Chest. 2011;139(5):1165–71. doi: 10.1378/chest.10-2397. [DOI] [PubMed] [Google Scholar]
- 19.Subrata LS, Bizzintino J, Mamessier E, Bosco A, McKenna KL, Wikstrom ME, et al. Interactions between Innate Antiviral and Atopic Immunoinflammatory Pathways Precipitate and Sustain Asthma Exacerbations in Children. The Journal of Immunology. 2009;183(4):2793–800. doi: 10.4049/jimmunol.0900695. [DOI] [PubMed] [Google Scholar]
- 20.Lambrecht BN, Hammad H. Taking our breath away: dendritic cells in the pathogenesis of asthma. Nat Rev Immunol. 2003;3(12):994–1003. doi: 10.1038/nri1249. [DOI] [PubMed] [Google Scholar]
- 21.Chang YJ, Kim HY, Albacker LA, Baumgarth N, McKenzie AN, Smith DE, et al. Innate lymphoid cells mediate influenza-induced airway hyper-reactivity independently of adaptive immunity. Nat Immunol. 2011;12(7):631–8. doi: 10.1038/ni.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim HY, Chang YJ, Subramanian S, Lee HH, Albacker LA, Matangkasombut P, et al. Innate lymphoid cells responding to IL-33 mediate airway hyperreactivity independently of adaptive immunity. J Allergy Clin Immunol. 2012;129(1):216–27. e1–6. doi: 10.1016/j.jaci.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mathias CB, Guernsey LA, Zammit D, Brammer C, Wu CA, Thrall RS, et al. Pro-inflammatory role of natural killer cells in the development of allergic airway disease. Clinical & Experimental Allergy. 2014;44(4):589–601. doi: 10.1111/cea.12271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ple C, Barrier M, Amniai L, Marquillies P, Bertout J, Tsicopoulos A, et al. Natural Killer Cells Accumulate in Lung-Draining Lymph Nodes and Regulate Airway Eosinophilia in a Murine Model of Asthma. Scandinavian Journal of Immunology. 2010;72(2):118–27. doi: 10.1111/j.1365-3083.2010.02419.x. [DOI] [PubMed] [Google Scholar]
- 25.Akbari O, Faul JL, Hoyte EG, Berry GJ, Wahlström J, Kronenberg M, et al. CD4+ Invariant T-Cell–Receptor+ Natural Killer T Cells in Bronchial Asthma. New England Journal of Medicine. 2006;354(11):1117–29. doi: 10.1056/NEJMoa053614. [DOI] [PubMed] [Google Scholar]
- 26.Shim J-U, Koh Y-I. Increased Th2-like Invariant Natural Killer T cells in Peripheral Blood From Patients With Asthma. Allergy Asthma Immunol Res. 2014;6(5):444. doi: 10.4168/aair.2014.6.5.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barnig C, Cernadas M, Dutile S, Liu X, Perrella MA, Kazani S, et al. Lipoxin A4 Regulates Natural Killer Cell and Type 2 Innate Lymphoid Cell Activation in Asthma. Science Translational Medicine. 2013;5(174):174ra26–ra26. doi: 10.1126/scitranslmed.3004812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duvall MG, Barnig C, Cernadas M, Ricklefs I, Krishnamoorthy N, Grossman NL, et al. Natural killer cell-mediated inflammation resolution is disabled in severe asthma. Sci Immunol. 2017;2(9) doi: 10.1126/sciimmunol.aam5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gern JE, Visness CM, Gergen PJ, Wood RA, Bloomberg GR, O'Connor GT, et al. The Urban Environment and Childhood Asthma (URECA) birth cohort study: design, methods, and study population. BMC Pulm Med. 2009;9:17. doi: 10.1186/1471-2466-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Èech M, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016;44(W1):W3–W10. doi: 10.1093/nar/gkw343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–78. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anders S, Pyl PT, Huber W. HTSeq - A Python framework to work with high-throughput sequencing data. Vol. 2014. Cold Spring Harbor Laboratory Press; 2014. Feb 20, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smedley D, Haider S, Durinck S, Pandini L, Provero P, Allen J, et al. The BioMart community portal: an innovative alternative to large, centralized data repositories. Nucleic Acids Res. 2015;43(W1):W589–98. doi: 10.1093/nar/gkv350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu R, Holik AZ, Su S, Jansz N, Chen K, Leong HS, et al. Why weight? Modelling sample and observational level variability improves power in RNA-seq analyses. Nucleic Acids Res. 2015;43(15):e97. doi: 10.1093/nar/gkv412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abbas AR, Baldwin D, Ma Y, Ouyang W, Gurney A, Martin F, et al. Immune response in silico (IRIS): immune-specific genes identified from a compendium of microarray expression data. Genes Immun. 2005;6(4):319–31. doi: 10.1038/sj.gene.6364173. [DOI] [PubMed] [Google Scholar]
- 38.Chikina M, Zaslavsky E, Sealfon SC. CellCODE: a robust latent variable approach to differential expression analysis for heterogeneous cell populations. Bioinformatics. 2015;31(10):1584–91. doi: 10.1093/bioinformatics/btv015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhong Y, Wan YW, Pang K, Chow LM, Liu Z. Digital sorting of complex tissues for cell type-specific gene expression profiles. BMC Bioinformatics. 2013;14:89. doi: 10.1186/1471-2105-14-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bolen CR, Uduman M, Kleinstein SH. Cell subset prediction for blood genomic studies. BMC Bioinformatics. 2011;12:258. doi: 10.1186/1471-2105-12-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2008;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 42.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research. 2008;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Research. 2014;43(D1):D447–D52. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shannon P. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Research. 2003;13(11):2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44(D1):D457–62. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen-Orr SS, Gaujoux R. Computational deconvolution: extracting cell type-specific information from heterogeneous samples. Curr Opin Immunol. 2013;25(5):571–8. doi: 10.1016/j.coi.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Korsgren M, Persson CGA, Sundler F, Bjerke T, Hansson T, Chambers BJ, et al. Natural Killer Cells Determine Development of Allergen-induced Eosinophilic Airway Inflammation in Mice. The Journal of Experimental Medicine. 1999;189(3):553–62. doi: 10.1084/jem.189.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Manners S, Alam R, Schwartz DA, Gorska MM. A mouse model links asthma susceptibility to prenatal exposure to diesel exhaust. J Allergy Clin Immunol. 2014;134(1):63–72. doi: 10.1016/j.jaci.2013.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krejsek J, Král B, Vokurková D, Derner V, Toušková M, Paráková Z, et al. Decreased peripheral blood gamma delta T cells in patients with bronchial asthma. Allergy. 1998;53(1):73–7. doi: 10.1111/j.1398-9995.1998.tb03776.x. [DOI] [PubMed] [Google Scholar]
- 51.Lin SJ, Chang LY, Yan DC, Huang YJ, Lin TJ, Lin TY. Decreased intercellular adhesion molecule-1 (CD54) and L-selectin (CD62L) expression on peripheral blood natural killer cells in asthmatic children with acute exacerbation. Allergy. 2003;58(1):67–71. doi: 10.1034/j.1398-9995.2003.t01-1-23697.x. [DOI] [PubMed] [Google Scholar]
- 52.Timonen T, Stenius-Aarniala B. Natural killer cell activity in asthma. Clin Exp Immunol. 1985;59(1):85–90. [PMC free article] [PubMed] [Google Scholar]
- 53.Warren HS, Kinnear BF, Phillips JH, Lanier LL. Production of IL-5 by human NK cells and regulation of IL-5 secretion by IL-4, IL-10, and IL-12. J Immunol. 1995;154(10):5144–52. [PubMed] [Google Scholar]
- 54.Loza MJ, Zamai L, Azzoni L, Rosati E, Perussia B. Expression of type 1 (interferon gamma) and type 2 (interleukin-13, interleukin-5) cytokines at distinct stages of natural killer cell differentiation from progenitor cells. Blood. 2002;99(4):1273–81. doi: 10.1182/blood.v99.4.1273. [DOI] [PubMed] [Google Scholar]
- 55.Wei H, Zhang J, Xiao W, Feng J, Sun R, Tian Z. Involvement of human natural killer cells in asthma pathogenesis: natural killer 2 cells in type 2 cytokine predominance. J Allergy Clin Immunol. 2005;115(4):841–7. doi: 10.1016/j.jaci.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 56.Wingett D, Nielson CP. Divergence in NK cell and cyclic AMP regulation of T cell CD40L expression in asthmatic subjects. J Leukoc Biol. 2003;74(4):531–41. doi: 10.1189/jlb.0303103. [DOI] [PubMed] [Google Scholar]
- 57.Jacobs B, Ullrich E. The Interaction of NK Cells and Dendritic Cells in the Tumor Environment: How to Enforce NK Cell & DC Action Under Immunosuppressive Conditions? CMC. 2012;19(12):1771–9. doi: 10.2174/092986712800099857. [DOI] [PubMed] [Google Scholar]
- 58.Van Elssen CHMJ, Vanderlocht J, Oth T, Senden-Gijsbers BLMG, Germeraad WTV, Bos GMJ. Inflammation restraining effects of prostaglandin E2 on natural killer-dendritic cell (NK-DC) interaction are imprinted during DC maturation. Blood. 2011;118(9):2473–82. doi: 10.1182/blood-2010-09-307835. [DOI] [PubMed] [Google Scholar]
- 59.Harizi H. Reciprocal crosstalk between dendritic cells and natural killer cells under the effects of PGE2 in immunity and immunopathology. Cell Mol Immunol. 2013;10(3):213–21. doi: 10.1038/cmi.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deniz G, Akdis M, Aktas E, Blaser K, Akdis CA. Human NK1 and NK2 subsets determined by purification of IFN-γ-secreting and IFN-γ-nonsecreting NK cells. European Journal of Immunology. 2002;32(3):879. doi: 10.1002/1521-4141(200203)32:3<879::AID-IMMU879>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 61.Di Santo JP. Functionally distinct NK-cell subsets: Developmental origins and biological implications. European Journal of Immunology. 2008;38(11):2948–51. doi: 10.1002/eji.200838830. [DOI] [PubMed] [Google Scholar]
- 62.Hoshino T, Winkler-Pickett RT, Mason AT, Ortaldo JR, Young HA. IL-13 production by NK cells: IL-13-producing NK and T cells are present in vivo in the absence of IFN-gamma. J Immunol. 1999;162(1):51–9. [PubMed] [Google Scholar]
- 63.Jackson DJ, Gangnon RE, Evans MD, Roberg KA, Anderson EL, Pappas TE, et al. Wheezing rhinovirus illnesses in early life predict asthma development in high-risk children. Am J Respir Crit Care Med. 2008;178(7):667–72. doi: 10.1164/rccm.200802-309OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Message SD, Laza-Stanca V, Mallia P, Parker HL, Zhu J, Kebadze T, et al. Rhinovirus-induced lower respiratory illness is increased in asthma and related to virus load and Th1/2 cytokine and IL-10 production. Proc Natl Acad Sci U S A. 2008;105(36):13562–7. doi: 10.1073/pnas.0804181105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lynch SV, Wood RA, Boushey H, Bacharier LB, Bloomberg GR, Kattan M, et al. Effects of early-life exposure to allergens and bacteria on recurrent wheeze and atopy in urban children. J Allergy Clin Immunol. 2014;134(3):593–601. e12. doi: 10.1016/j.jaci.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.O'Connor GT, Lynch SV, Bloomberg GR, Kattan M, Wood RA, Gergen PJ, et al. Early-life home environment and risk of asthma among inner-city children. J Allergy Clin Immunol. 2017 doi: 10.1016/j.jaci.2017.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.