Abstract
Metabolic labeling is one of the most powerful methods to label the live cell for in vitro and in vivo tracking. However, the cellular mechanisms by modified glycosylation due to metabolic agents are not fully understood. Therefore, metabolic labeling has not yet been widely used in EPC tracking and labeling. In this study, cell functional properties such as proliferation, migration and permeability and gene expression patterns of metabolic labeling agent-treated hUCB-EPCs were analyzed to demonstrate cellular effects of metabolic labeling agents. As the results, 10 μM Ac4ManNAz treatment had no effects on cellular function or gene regulations, however, higher concentration of Ac4ManNAz (>20 μM) led to the inhibition of functional properties (proliferation rate, viability and rate of endocytosis) and down-regulation of genes related to cell adhesion, PI3K/AKT, FGF and EGFR signaling pathways. Interestingly, the new blood vessel formation and angiogenic potential of hUCB-EPCs were not affected by Ac4ManNAz concentration. Based on our results, we suggest 10 μM as the optimal concentration of Ac4ManNAz for in vivo hUCB-EPC labeling and tracking. Additionally, we expect that our approach can be used for understanding the efficacy and safety of stem cell-based therapy in vivo.
Introduction
Human umbilical cord blood-derived endothelial progenitor cells (hUCB-EPCs) have a great potential therapeutic impact in clinical trials of acute nervous system disorders, myocardial infarction and stroke1–3. hUCB-EPCs have been also used to investigate the repair of injured vessels and neovascularization or regeneration of ischemic tissues and have been used for therapeutic re-endothelialization of vein graft because of their ability to induce neovascularization under ischemic conditions4–6. Moreover, hUCB-EPC can be obtained without extensive surgical procedure7 and are immediately available, there are no risks to the donor and there is a low risk of transmitting infectious diseases8. For these reasons, currently, hUCB-EPCs are the preferred tool for animal- and patient-based studies than other forms of pluripotent hematopoietic stem cells and mesenchymal stem cells for transplantation9–11. However, many unknown factors, including their regenerative property, the fate of transplanted hUCB-EPCs, in vivo migration to the site of injury and in situ differentiation have yet to be exploited in more efficient ways to treat various diseases12–14.
For understanding biological mechanisms and the therapeutic effects of inoculated cells in vivo, cell labeling and tracking are very useful processes15. Although direct labeling with specific dyes or indirect labeling with genetic cell modifications and reporter genes16 have been used to observe unknown factors of transplanted hUCB-EPCs. These labeling methods have several safety and technical issues, such as photobleaching, quenching, sensitivity to pH changes and multiple labeling steps17. Recently, various techniques, including fluorescence, bioluminescence, positron emission tomography (PET), single photon emission computed tomography (SPECT) and magnetic resonance imaging (MRI), have been developed and used for hUCB-EPC labeling and tracking18–21. Many studies have used MRI methods with iron oxide- and 19F-based probes22–25. Wang et al. and Willenbrock et al. reported that magnetically labeled hUCB-EPCs in a mouse model were detectable for more than 7 days post-transplantation and have many advantages such as a simple and low-cost labeling, an effective imaging window and good signal intensity26,27. However, MRI labeling is limited by the persistence of the in vivo signal even after death of the transplanted cells, such that the MRI signal does not correlate with the viability of transplanted cells28.
Metabolic labeling is the preferred labeling technique for tracking live cells because it has many advantages such as low background, correlation of cell survival and simple steps for cell labeling29,30. In addition, the reaction produces very few toxic and non-toxic byproducts and therefore, metabolic labeling has advantages that apply to in vivo studies. Till now, the metabolic labeling technique in stem cells has been mostly used for detecting glycoprotein markers of mesenchymal stem cell differentiation31, for identifying or isolating live colon cancer stem cells32 and for quantifying protein abundance in embryonic stem cells (ESCs)33. However, the cellular mechanisms by which modified glycosylation due to metabolic agents are not completely understood34. Recently, Lee. et al. reported that the metabolic labeling technique can stably label stem cells, but this study only focused on efficient labeling of stem cells35.
In this study, to validate the safety and optimize the metabolic labeling method, we analyze cell functional properties, such as proliferation, migration and permeability and gene expression patterns of metabolic labeling agents-treated hUCB-EPCs. First, we screened the azido sugars for more efficient labeling of hUCB-EPCs. And then, we observed changes in basic cellular events including cell growth, migration, permeability and mitochondrial function. We analyzed transcriptomic changes by RNA-seq, functional characterization of hUCB-EPCs by observation of tube formation and marker gene expression using flow cytometric analysis.
Results
Screening of metabolic labeling agent for efficient labeling of hUCB-EPCs
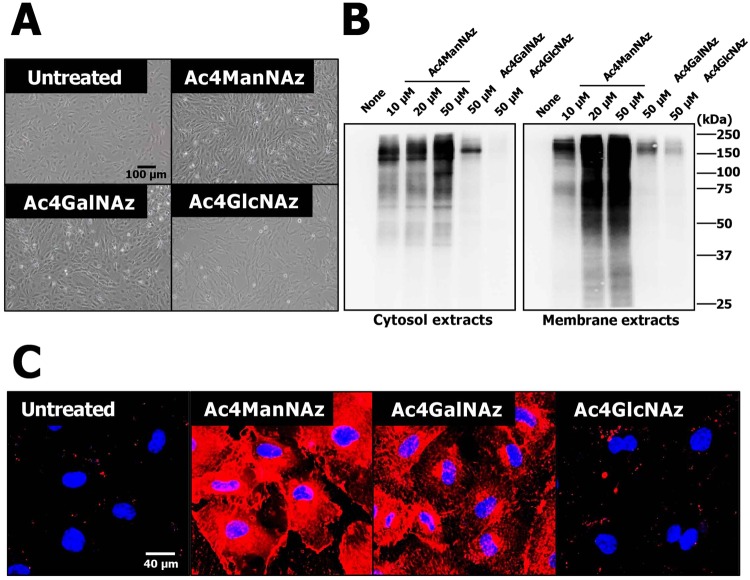
Metabolic labeling method was described as the introduction of subtle modifications into monosaccharide precursors, such as in the case of introducing an azido group from azide-functionalized monosaccharides (tetra-acetylated N-azidoacetylmannosamine (Ac4ManNAz), tetra-acetylated N-azidoacetylgalactosamine (Ac4GalNAz), or tetra-acetylated N-azidoacetylglucosamine (Ac4GlcNAz)) via post-translational modification (PTM)36. Once the azido group was introduced into cellular glycans, the glycosylation position and type of three azide-functionalized monosaccharides differed based on each sugar type37. In detail, the Ac4ManNAz was metabolically converted to azido sialic acid derivative, which is used for N-linked glycosylation of cell surface proteins38, whereas Ac4GlcNAz and Ac4GalNAz were predominantly used for O-linked glycosylation as a substitute for O-GlcNAc and O-GalNAc, which are attached to serine and threonine side chain of numerous intracellular proteins39. When using metabolic labeling reagents, the choice of reagents was found to be very important for efficiently labeling the target cells. Thus, we first screened the metabolic labeling reagents for efficient labeling hUCB-EPCs.
Isolated hUCB-EPCs were treated with 10, 20, or 50 μM Ac4ManNAz, Ac4GlcNAz, or Ac4GalNAz, which can easily access the modified surface and intracellular proteins with the azido group via PTM. hUCB-EPCs treated with three azido sugars did not show changes or differences in cell morphology (Fig. 1A). To analyze the incorporation of azido groups, Western blot analysis was performed as described in materials and methods. Interestingly, Ac4ManNAz treatment resulted in a higher generation efficiency of the azido group than Ac4GalNAz and Ac4GlcNAz treatment (Fig. 1B). Azido groups were incorporated into cytosolic and membrane proteins on hUCB-EPCs treated with Ac4ManNAz and the rate of incorporation of labeled proteins gradually increased relative to Ac4ManNAz concentration. However, hUCB-EPCs treated with 50 μM Ac4GalNAz or Ac4GlcNAz generated only the modified proteins and Ac4GlcNAz treatment labeled membrane proteins. In addition, the generation of azido groups was determined by biorthogonal copper-free click chemistry with dye-labeled dibenzyl cyclooctyne (DBCO-Cy5) (Fig. 1C). Ac4ManNAz treatment resulted in higher labeling efficiency than Ac4GalNAz and Ac4GlcNAz, which was similar to results from the Western blot analysis. Thus, we finally selected Ac4ManNAz for metabolic labeling of hUCB-EPCs.
Figure 1.
Screening metabolic labeling agents for hUCB-EPC labeling and tracking. (A) Morphological properties of hUCB-EPCs treated with three metabolic labeling agents (Ac4ManNAz, Ac4GlcNAz and Ac4GalNAz) were analyzed using microscopic observation. (B) Western blot analysis of metabolic labeling agent-treated hUCB-EPCs showing total proteins and generated azide groups. In the figure indicated full-length expression and all gels were run under the same experimental conditions while images of western blots displayed. (C) Visualization of metabolic labeling agent-treated hUCB-EPCs using DBCO-cy5 (n = 3).
Bio-physiological effects of Ac4ManNAz-treated hUCB-EPCs
When hUCB-EPCs were treated with Ac4ManNAz, an abiotic azido group was introduced to native proteins by PTM. Additionally, Ac4ManNAz, which is an analog of ManNAc, was used as a carbon source40. Generally, the carbon source concentration can impair many cellular functions such as cell proliferation, viability and permeability41,42. Additionally, glycosylation of native proteins regulates their functions and kinetics and changes in glycosylation can alter cellular functions such as host cell surface interactions and modulation of cell signaling and gene expression43. Thus, Ac4ManNAz labeling of hUCB-EPCs still introduces the possibility of altering cellular physiology.
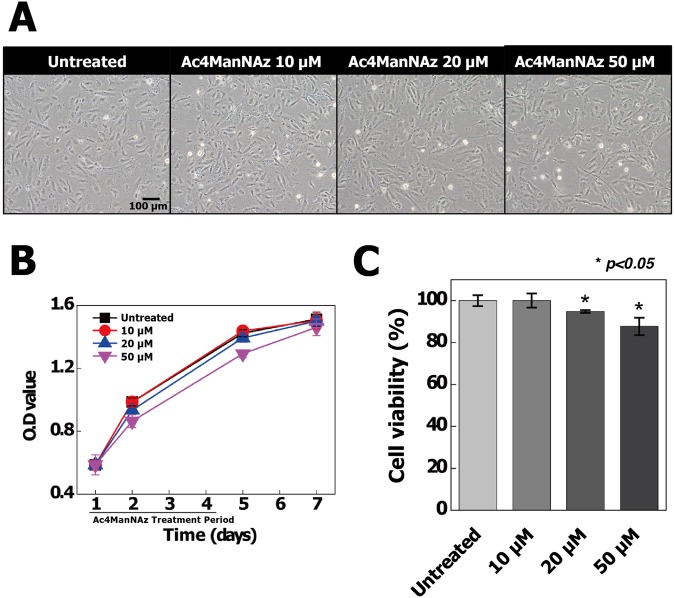
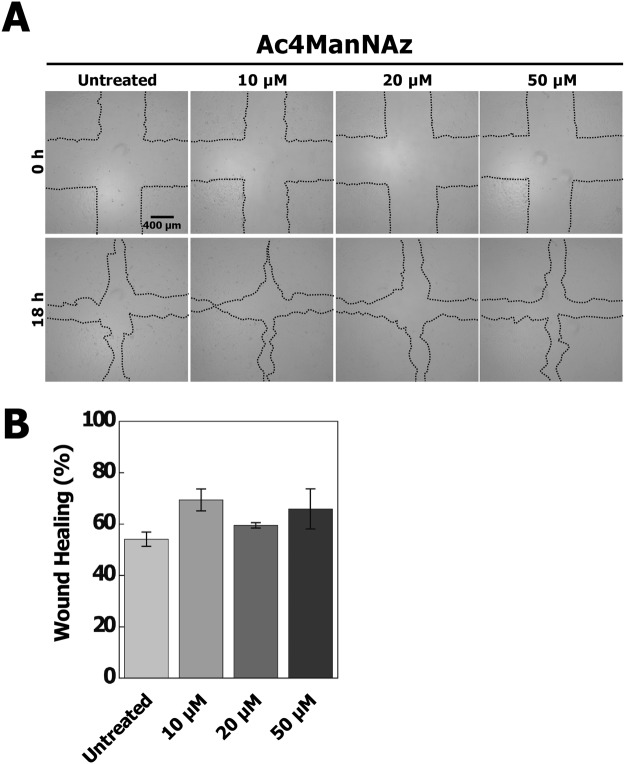
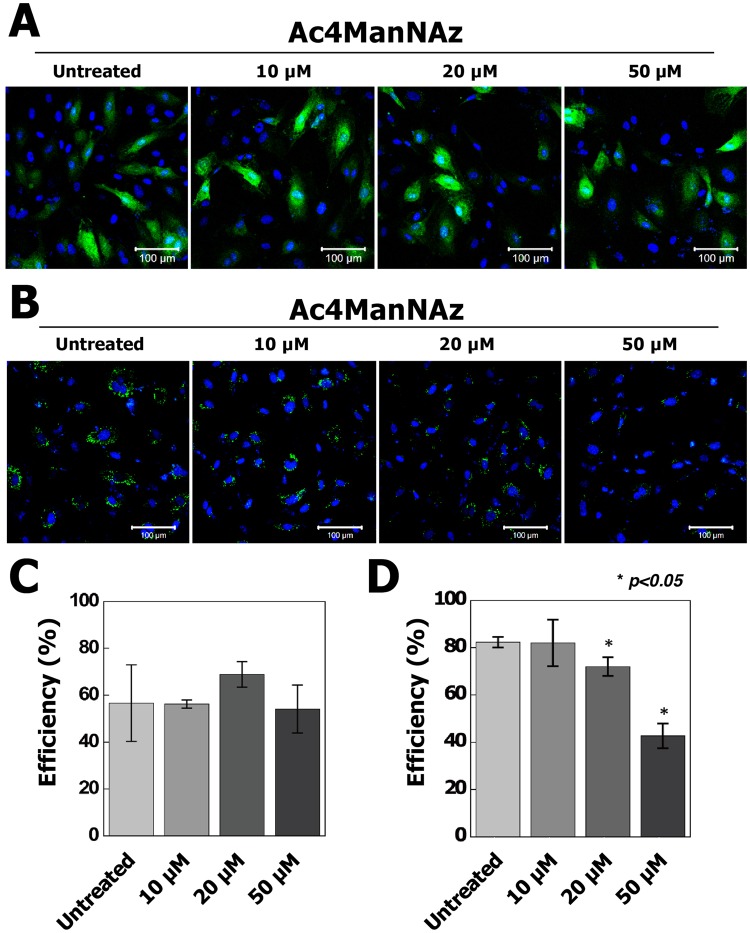
To validate the influence of Ac4ManNAz on hUCB-EPCs, we firstly performed cytotoxic tests using CCK-8 assay and cell morphology assessment. Morphological study suggests that treatment of Ac4ManNAz did not affect significantly on cell morphology (Fig. 2A). However, the growth rate gradually decreased in hUCB-EPCs treated with >20 µM Ac4ManNAz (Fig. 2B) and cell viability data showed that treatment of >20 µM Ac4ManNAz significantly decreased cell viability, approximately 6.2% and 12.3% (*p < 0.05) at the concentrations 20 µM and 50 µM of Ac4ManNAz as compared to control (Fig. 2C). We furthermore examined the effects of Ac4ManNAz using in vitro wound healing assay. The scratch wounds were almost the same size in each experimental group at 0 h; after 18 h, the difference in the reduction in wound size was not statistically significant in any Ac4ManNAz (10, 20 and 50 µM) treatment group (Fig. 3). In addition, we analyzed the permeability of hUCB-EPCs treated with Ac4ManNAz using liposome-mediated transfection of a pcDNA3-eGFP plasmid and Qdot 525 probe. The results of the permeability test using eGFP showed no significant differences between the treated and untreated conditions (Fig. 4A,C). However, results from the Qdot analysis revealed a gradual decrease in the rate of endocytosis of hUCB-EPCs treated with >20 µM Ac4ManNAz (Fig. 4B,D). These results suggested that the membrane fusion was not changed; however, the rate of endocytosis was reduced in cells treated with >20 µM Ac4ManNAz.
Figure 2.
Analysis of morphological properties, proliferation ability and viability of Ac4ManNAz-treated hUCB-EPCs. All hUCB-EPCs were incubated with various concentrations of Ac4ManNAz (0 to 50 µM). (A) Ac4ManNAz concentration-dependent morphological changes were analyzed by microscopic observation. Cell growth rate (B) and viability (C) were analyzed by CCK-8 and manual microscopic counting.
Figure 3.
Wound healing assay in Ac4ManNAz-treated hUCB-EPCs. Wound healing assay was performed to assess the effect of Ac4ManNAz on the migration of hUCB-EPCs. The assay was repeated three times and representative images (A) and quantification (B) are shown.
Figure 4.
Analysis of cell permeability via transfection and Qdot 525 labeling in Ac4ManNAz-treated hUCB-EPCs. (A) Ac4ManNAz-treated hUCB-EPCs were transfected with pcDNA3-eGFP and GFP fluorescence and analyzed using fluorescence microscopy. (B) Confocal fluorescence images of live hUCB-EPC labeling with a Qdot 525 probe (green). Quantification of the transfection (C) and Qdot labeling (D) efficiency was conducted.
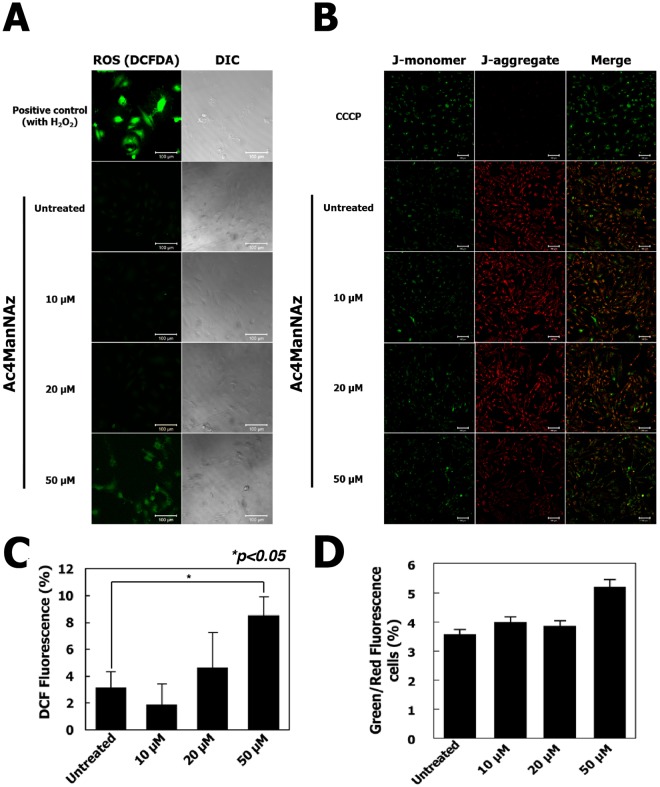
Additionally, we conducted the reactive oxygen species (ROS) generation assay and assessment of mitochondrial membrane potential (ΔΨm) to analyze the apoptosis induction by Ac4ManNAz treatments. The Fig. 5A suggests that hUCB-EPCs treated with 50 µM Ac4ManNAz significantly increased ROS intensity as compared to control. The quantitative measurement of ROS intensity was sustained increasingly to approximately 5.4% (*p < 0.05) at the concentrations 50 µM of Ac4ManNAz (Fig. 5C). The increased and decreased florescent intensity of red and green florescent caused by JC-1 indicates the change in mitochondrial membrane potential (ΔΨm). Result indicated that hUCB-EPCs treated with 50 µM of Ac4ManNAz reduced a red fluorescence (Fig. 5B). As shown from quantitative data, the Green/Red-fluorescence cells ratio were increased to 1.5% at 50 µM of Ac4ManNAz treatment (Fig. 5D). Although increased ROS generation and reduced of JC-1 red fluorescence indicated potent apoptotic activity of treatment of 50 µM Ac4ManNAz, these changes are not clear which of them are directly associated with hUCB-EPCs apoptosis. To prove this, we additionally analyzed the apoptotic effects of Ac4ManNAz by Annexin V staining (Fig. S1). As the results, apoptosis rates of hUCB-EPCs treated with higher concentration (more than 20 μM) of Ac4ManNAz were not changed compared with non-treated hUCB-EPCs. These results described that high concentration of Ac4ManNAz slightly modulated the generation of ROS and mitochondrial membrane potential, but these changes were not induced apoptosis.
Figure 5.
Analysis of reactive oxygen species (ROS) production and mitochondrial function in hUCB-EPCs treated with Ac4ManNAz. (A) hUCB-EPCs were treated with 0, 10, 20, or 50 μM Ac4ManNAz, and intracellular ROS levels were then measured using the fluorescent probe 2′, 7′-dichlorfluorescein-diacetate (DCFH-DA). (B) Changes in mitochondrial membrane potential (ΔΨm) were measured in hUCB-EPCs using the fluorescent probe JC-1. (C) Quantification of ROS generation are expressed as the percentage of fluorescence intensity relative to the control. (D) Value of mitochondrial membrane potential were expressed as % Green/Red fluorescence cells.
Transcriptomic change in Ac4ManNAz-treated hUCB-EPCs
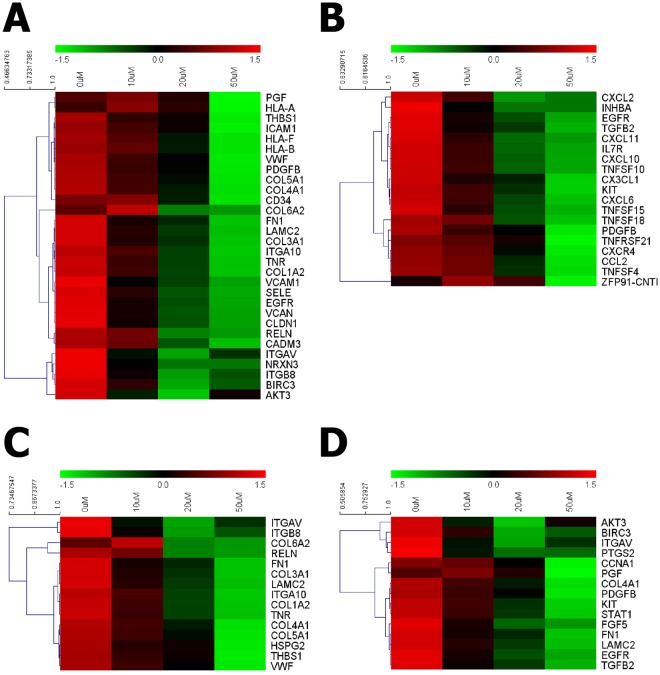
To additionally address the metabolic labeling effects, we explored the transcriptome patterns of hUCB-EPCs treated with various concentrations of Ac4ManNAz. mRNA from hUCB-EPCs treated with 10, 20 and 50 μM of Ac4ManNAz and untreated hUCB-EPCs was extracted and analyzed using the HiSeq2000 platform. Next, based on the expression levels of known genes, we identified 615 genes that were changed by 2-fold or more with a threshold of “p < 0.05 and FDR < 5”. And then, to identify cellular functional process-related genes, we carried out GO term analysis. Interestingly, in accordance with the increase in Ac4ManNAz concentration on hUCB-EPCs, 468 genes related to cell adhesion, cytokine-cytokine receptor interaction, extracellular matrix (ECM)-receptor interaction and cancer pathway were down-regulated (Fig. 6).
Figure 6.
Transcriptomic analysis of effects of Ac4ManNAz treatment on hUCB-EPCs. Expression profiles of 0, 10, 20, and 50 μM Ac4ManNAz-treated hUCB-EPCs are shown. Heat map representation of mapped reads corresponds to protein-coding genes and the bio-functional pathway. (A) Cell adhesion, (B) cytokine-cytokine receptor interaction, (C) ECM-receptor interaction and (D) cancer-related pathways.
Of note, treatment with 20 and 50 µM Ac4ManNAz resulted in considerable down-regulation of many genes in the cell functional pathways of hUCB-EPCs. In cell adhesion, genes related to primary interactions with the cell matrix (LAMC3, ITGA5, ITGA10, ITGB8, TNR, EGFR and RELN) and integrin (CAMs) (ICAM1 and VCAM1), primary roles in cell-cell interactions (SELE), cell-cell interaction-mediated MHC (HLA-B, HLA-F and CD34) and collagen proteins (COL1A1, COL3A1, COL4A1, COL5A1 and COL6A2) were significantly down-regulated (Fig. 6A). In addition, genes related to cytokine-cytokine receptor interaction and ECM-receptor interaction were also down-regulated. In detail, gene expression levels of chemotactic cytokines (chemokines) (CCL2, CX3CL1, CXCL2, CXCL6, CXCL10, CXCL11 and CXCR4), tumor necrosis factors (TNFs) (TNFSF4, TNFSF10, TNFSF15, TNFSF18 and TNFRSF21), integrins (ITGA5, ITGA10, ITGB8) and ECM proteins (FN1, TNR and THBS1) were reduced by increasing the Ac4ManNAz concentration (Fig. 6B,C). Additionally, genes related to the regulation of cell cycle (CCNA1), cell growth, proliferation, apoptosis and the immune response (AKT3, STAT1, BIRC3, PTGS2, KIT, EGFR) were down-regulated (Fig. 6D). These results showed that Ac4ManNAz regulated a wide range of cell adhesion pathways and cell physiological pathways. In particular, these results also suggest that treatment >20 µM Ac4ManNAz induces the immune response and inhibits cell cycle progression, cell proliferation and cell adhesion. Interestingly, contrary to previous reports, Ac4ManNAz modulated cell biological functions.
Analysis of hUCB-EPC functions after Ac4ManNAz treatment
In our results regarding the effects of metabolic labeling on hUCB-EPCs, higher concentrations of Ac4ManNAz negatively affected cell growth, proliferation, adhesion and rate of endocytosis and induction of ROS generation and mitochondrial depolarization. Of note, results from the gene expression analysis revealed the down-regulated expression of integrins, which are important for stem cell proliferation and self-renewal regulated via PI3K and focal adhesion kinase (FAK) signaling pathways44. Additionally, AKT3 gene, which is downstream of the PI3K and FAK signaling pathways45, was down-regulated. However, interestingly, there was no significant change in the gene expression of cadherin, which is one of the most important molecules in stem cell pluripotency and stemness46,47. Stem cells are known to have two important characteristics that distinguish them from other types of cells48. First, all stem cells are unspecialized. Second, under certain biochemical cues, stem cells can be induced to differentiate. Thus, as it was unclear whether Ac4ManNAz affects hUCB-EPC functions, we conducted the tubule formation assay and immunophenotyping of marker genes to analyze these important characteristics of EPCs.
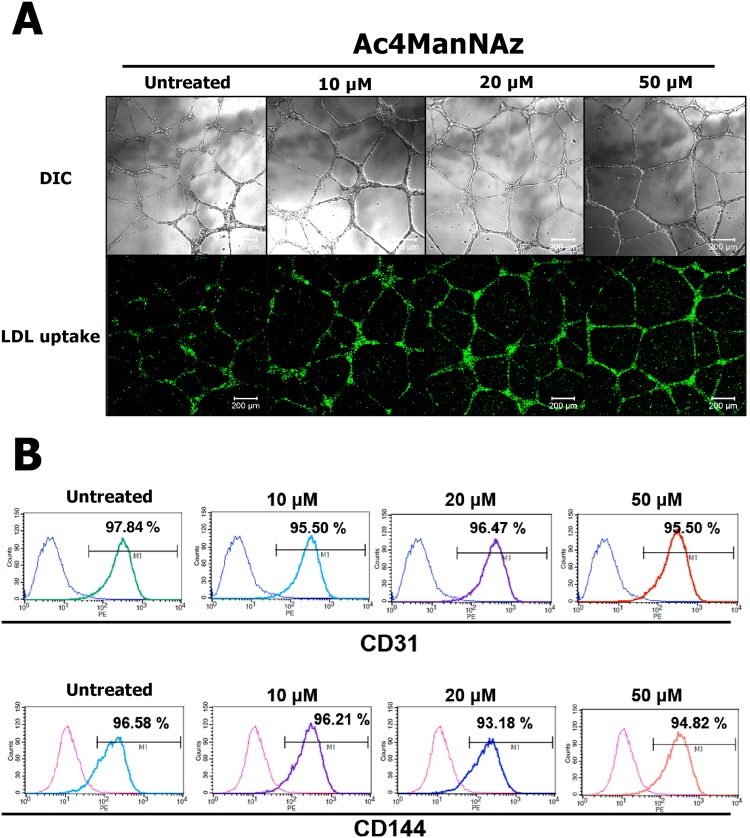
To define the effects of Ac4ManNAz on hUCB-EPC functions, we carried out a tube formation assay on Matrigel to determine the angiogenic potential. We observed tube-like structures 4 days after treating hUCB-EPCs with 0, 10, 20 or 50 µM Ac4ManNAz. The upper of Fig. 7A shows typical tube formation in all samples. One of the hallmarks of hUCB-EPCs is their ability to internalize ac-LDL via the “scavenger cell pathway” of LDL metabolism49. At the endpoints of each protocol, we incubated our endothelial cultures with 1 μg/mL ac-LDL conjugated with a fluorescent dye (Fig. 7A lower). In addition, the expression of specific endothelial cell surface markers CD31 (PECAM-1) and CD144 (VE-cadherin) of hUCB-EPCs were analyzed by flow cytometry (Fig. 7B). Interestingly, these results showed that Ac4ManNAz does not affect the natural characteristics of hUCB-EPCs.
Figure 7.
Analysis of tube formation and LDL uptake assay of Ac4ManNAz-treated hUCB-EPCs. (A) hUCB-EPCSs treated with 0, 10, 20, or 50 μM Ac4ManNAz were transferred to Matrigel-coated plates and cell rearrangement and tube structure formation were captured by a light microscope equipped with digital charge-coupled device camera after 48 h. Images of ac-LDL uptake were captured by fluorescence microscopy. (B) Representative FACS analysis demonstrated the function of hUCB-EPC by their markers CD31 and CD144.
Discussion
Stem cell-based therapy holds great promise for repairing damage and injury of the human body; however, the safety and efficacy of stem cell in vivo conditions have not been fully confirmed50. Additionally, although hUCB-EPCs have great potential as therapeutics for cardiovascular diseases including coronary artery disease and stroke, the roles, behavior and fate of hUCB-EPCs in vivo are not completely understood51. Many cell labeling and tracking methods have been used and developed for hUCB-EPCs safety and efficacy in vivo17. Metabolic labeling is one of the most powerful methods for cell labeling and tracking and has proven to be superior to other imaging approaches29,30. However, metabolic labeling has not yet been widely used in stem cell tracking, because the introduction of abiotic azido groups in native proteins via PTM can potentially affect cell signaling pathways, cell adhesion, cell proliferation and the immune response. We previously reported the effects of metabolic labeling agents on cancer cells, showing that a higher concentration of Ac4ManNAz, which was recommended to achieve the best labeling efficiency, led to inhibition of basic cellular properties and down-regulation of many genes related to cell biological functions52. Therefore, to successfully apply the metabolic labeling technique to EPC tracking and labeling, the effects of metabolic labeling agents must be demonstrated. Thus, in this study, we reported for the first time the effects and optimized conditions of a metabolic labeling agent in hUCB-EPCs.
Metabolic labeling agents are known to have three type of azido sugars (Ac4ManNAz, Ac4GalNAz and Ac4GlcNAz), which introduce the azido group onto a native protein using different glycosylation processes. Thus, we tested all azido sugars to screen for optimal metabolic labeling agents for hUCB-EPCs. As a result, treatments with Ac4GalNAz and Ac4GlcNAz showed weaker metabolic labeling efficiency than Ac4ManNAz. In addition, Ac4GlcNAz had very low labeling efficiency. Generally, O-glycosylated proteins are highly abundant in mammalian cells53 and Ac4GalNAz and Ac4GlcNAz use the endogenous salvage pathway to introduce the azido group via O-linked glycosylation, which synthesizes UDP-GalNAc and GlcNAc from GalNAc and GlcNAc. Although these modification pathways involving numerous proteins, including various enzymes and nuclear pore proteins, are well-defined, conjugating an azido group on GalNAc and GlcNAc could be affected by enzyme function54. Indeed, the Bertozzi research group has shown that AGX1 encodes UDP-GlcNAc pyrophosphorylase, which is the final enzyme in UDP-GalNAc and GlcNAc biosynthesis and has the greatest loss of catalytic efficiency with azide-functionalized substrate analogs55,56. However, our study was focused on the optimization of hUCB-EPC labeling using the metabolic labeling technique. Thus, we finally selected Ac4ManNAz.
In the manufacturer’s procedure, the treatment of 50 μM Ac4ManNAz as maximum level was recommended for the highest labeling efficiency. Also, many studies of cell or virus labeling and tracking was normally used in Ac4ManNAz concentration of 50 μM or more57,58. However, in our results, the decreased proliferation rate, viability, rate of endocytosis and induction of ROS generation and mitochondrial depolarization were caused by high concentration of Ac4ManNAz. The metabolic labeling agent Ac4ManNAz is composed of a sugar molecule (ManNAc) and an azido group, which are incorporated into cellular proteins. Therefore, Ac4ManNAz has the potential to affect metabolic flux and cellular mechanisms by addition of sugar molecules. Regarding this, in previous reports, a high concentration (1 mM) of ManNAc treatment in mouse ESCs induced switching of epigenetic factors from Sirt1/Ogt to Mgea5 at the Hcrt gene locus59. A high concentration (>1,000 µM) of Ac4ManNAc, which is a ManNAc analog without the azido group, dramatically increased cytotoxicity by inducing the accumulation of acetic acid in cells60. Additionally, we previously analyzed the effects of ManNAc treatment on the A549 cancer cell line, ManNAc treatment at the same concentration of metabolic labeling agents does not affect cellular function or proliferation52. Also, the carbon energy source in the EGM-2-MV BulletKit Medium is the 5 mM. Ac4ManNAz were used less than 50 μM, which is a lower concentration by 100-fold. So, the addition of Ac4ManNAz as carbon energy source shown the negligible effects. These reports suggested that, changes of cellular physiology and mechanisms on hUCB-EPCs were caused by introduction of the azido group derived from Ac4ManNAz onto a native protein.
Non-toxic labeling method of azide group was the strain-promoted azide alkyne click chemistry (SPAAC) reaction, which rely on the use of cyclooctynes derivatives such as bicyclo[6.1.0]nonyne (BCN), difluorooctyne (DIFO), dibenzylcyclooctyne (DIBO) and DBCO61. These cyclooctynes derivatives have high selectivity of azide-functionalized biomolecules. However, several groups have recently reported that thiol-reactive group (-SH) can be conjugated with various cyclooctyne derivatives in the presence of high thiol concentrations as thiol-yne reaction, which can lead to issues associated with nonspecific background labeling and affected the efficiency of biomolecules62–64. Nevertheless, in mammalian proteins, the occurrence frequency of cysteine targets, which present the thiol group, is 3.3% and SPAAC reaction have the significantly higher reaction rates compared to thiol-yne reactions. For example, the BCN–azide reaction (10−1 m−1 s−1) is approximately three orders of magnitude greater than the rate constant for the BCN–thiol reaction (10−4 m−1 s−1)64–66. In addition, Chin Fen Teo et al. optimized the ratio of the reaction between azido-DBCO functionalities to the formation of thiol-DBCO product. These results shown that the reaction to proceed longer than 1 hour leads to increase the non-specific thiol-DBCO reaction and azido-DBCO specific reaction reaches completion within 1 hour67. Based on these results, in our experiments, hUCB-EPCs were treated with metabolic labeling agents supplemented medium (10, 20 and 50 µM) for 72 hours and all metabolic labeling agents-treated hUCB-EPCs were incubated with DBCO-Cy5 (10 µM, final concentration) for 1 hour at 37 °C. Thus, our optimized labeling procedures can be provided when highly specific labeling of hUCB-EPCs is required, thereby reducing the artifacts.
Our results reveal that the highly modified glycosylation by Ac4ManNAz treatments in hUCB-EPCs lead to a change in the physiological and biochemical properties of these cells. Notably, treatments with >20 µM Ac4ManNAz resulted in decreased growth rate, viability and rate of endocytosis and induction of ROS generation and mitochondrial depolarization. These results were consistent with a change in the expression of genes related to cell adhesion. Higher concentrations of Ac4ManNAz down-regulated the expression of integrin, collagen and fibronectin. Generally, integrins play an important role in various cell-signaling events including PI3K/AKT and FAK signaling pathways68, which help to regulate stem cell proliferation and self-renewal69. Accordingly, genes related to the PI3K/AKT, FGF and EGFR signaling pathways, which regulated cell proliferation, apoptosis, survival and immune response, were down-regulated46,70. Interestingly, expression of cadherins, which are cell adhesion molecules that actively contribute to cell death, survival and proliferation, was not inhibited by Ac4ManNAz treatments. The characterization of hUCB-EPCs were also not affected. Although treatments with >20 µM Ac4ManNAz led to negative outcomes in hUCB-EPCs, the effects of treatments with control or 10 µM Ac4ManNAz were not remarkably different. Additionally, it appears that 10 µM Ac4ManNAz provides sufficient labeling for tracking and monitoring hUCB-EPCs (Fig. S2). Thus, we emphasize the treatment of 10 µM Ac4ManNAz for labeling and tracking hUCB-EPCs as the optimal concentration because this concentration optimizes the proliferative capacity and functional property that are important for maximizing the use of EPCs as therapeutic agents.
In conclusion, this study described the optimal condition for the metabolic labeling technique for efficient labeling and tracking of hUCB-EPCs and the effects of metabolic labeling agents. Although high concentrations of Ac4ManNAz negatively affected hUCB-EPC properties, cell adhesion and cellular signaling pathways, 10 μM Ac4ManNAz showed the least adverse effects when compared to untreated hUCB-EPCs and this concentration provided sufficient labeling efficiency for cell labeling and tracking. Based on our results, we suggest 10 μM as the optimal concentration of Ac4ManNAz for in vivo cell labeling and tracking of hUCB-EPCs. Additionally, we expect that our approach can be used for understanding the efficacy and safety of stem cell-based therapy in vivo and to help determine the utility of stem cells in downstream experiments.
Methods
Ethic statement
All experiments were conducted in compliance with the relevant laws and institutional guidelines of the Korea Institute of Toxicology. The protocol was approved by the Committee on Biological Research of Korea Institute of Toxicology and Institutional Review Board (P01-201509-41-001).
hUCB-EPCs culture
hUCB-EPCs were purchased from AllCells (Alameda, CA, USA). hUCB-EPCs culture were performed as previously described71,72. Briefly, mononuclear cells (MNCs) were first isolated from fresh hUCB-EPCs by density gradient centrifugation using Ficoll reagent (GE Healthcare, Piscataway, NJ, USA). MNCs were plated on fibronectin-coated tissue culture plates at a density of 3 ~ 6 × 106 cells/6 wells in EGM-2-MV BulletKit Medium (Lonza, Walkersville, MD, USA) and cells were maintained for 5 ~ 7 days and used as an enriched EPC population.
In vitro cell labeling and imaging
hUCB-EPCs (5 × 104 cells/35 mm glass-bottom dishes) were treated with Ac4ManNAz, Ac4GalNAz, or Ac4GlcNAz (Invitrogen, Carlsbad, CA, USA) supplemented medium (50 µM, final concentration of each) for 72 h. Cells were washed twice with Dulbecco’s phosphate-buffered saline (DPBS) and subsequently incubated with DBCO-Cy5 (10 µM, final concentration) for 1 h at 37 °C. Cells were then washed and fixed with 4% paraformaldehyde for 15 min. After fixation, nuclei were stained with DAPI solution (Invitrogen, Carlsbad, CA, USA). All cell images were obtained using a confocal laser scanning microscope (Leica Microsystems, Mannheim, Germany) equipped with a 405 diode (405 nm) and HeNe-Red (633 nm) lasers.
Western blot analysis
To confirm the introduction of azide (−N3) in the hUCB-EPCs, each 10–50 µM of Ac4ManNAz-, Ac4GalNAz-, or Ac4GlcNAz-treated hUCB-EPCs (2 × 106 cells/100 mm dish) were prepared. Cells were washed twice with DPBS, pH 7.4 and harvested to extract cellular protein. A subcellular protein fractionation kit (Thermo Fisher Scientific, Rockford, IL, USA) was used according to the manufacturer’s instructions for segregating proteins from different cellular compartments including cytosol, membrane, nucleus, chromatin and cytoskeletons. Protein lysate concentrations were individually determined by bicinchoninic acid (BCA) protein assay (Thermo Fisher Scientific, Rockford, IL, USA) and protein concentrations were adjusted to 1 mg/mL. Then, 100 µL of lysate was incubated with 5 mM phosphine-PEG3-biotin in DPBS for 6 h at 37 °C to assess specific interactions between phosphine and an azide group of the cellular protein. Samples were boiled with SDS loading buffer and resolved by 10% SPS-PAGE and proteins were transferred to Hybond-P membrane (Amersham, St. Albans, UK). After blocking with 5% bovine serum albumin in TBST (50 mM Tris∙HCl, 150 mM NaCl, 0.1% Tween-20, pH 7.4), the membrane was incubated in streptavidin-HRP (diluted 1:10,000 in TBST) overnight at 4 °C. Then, the membrane was washed three times with TBST and developed using ECL Western Blotting Substrate (Thermo Fisher Scientific, Rockford, IL, USA).
Cell viability and wound healing assay
To measure cell viability, hUCB-EPCs were seeded in 96-well plates (5 × 103 cells/well) and incubated for 1 day. Cells were incubated with various concentrations of Ac4MAnNAz (0 to 50 µM) for 3 days at 37 °C. Cell Counting Kit-8 solution (10 µL) (Dojindo Molecular Technologies Inc., Kumamoto, Japan) was then added to each well. After further incubation for 2 h at 37 °C, the absorbance of each well was measured at 450 nm using a microplate reader (VersaMaxTM, Molecular Devices Corp., Sunnyvale, CA, USA). For wound healing, a sterile pipette tip was used to clear a small area across the diameter of 10 cm dishes with confluent monolayers of untreated or Ac4MAnNAz-treated hUCB-EPCs. Cell migration was measured and photographed from the wound/scratch edge after 18 h.
Analysis of reactive oxygen species (ROS) generation and mitochondrial membrane potential
Microscopic fluorescence imaging was used to study reactive oxygen species (ROS) generation in hUCB-EPCs after treatments to different concentrations of Ac4ManNAz. Cells (1 × 104 per well) were seeded and were then treated to 0 µM, 10 µM, 20 µM and 50 µM concentrations of Ac4ManNAz for 3 days at 37 °C. Cells were incubated with 2,7-Dichlorodihydrofluorescein diacetate (DCF-DA) (10 mM) for 30 min at 37 °C. The reaction mixture was aspirated and replaced by 200 µl of phosphate-buffered saline (PBS) in each well. The plate was kept on a shaker for 10 min at room temperature in the dark. An inverted fluorescent microscope was used to visualize intracellular fluorescence of cells and to capture images. Mitochondrial membrane potential was measured using JC-1 dye (5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolyl-carbocyanine iodide; Life Technologies, Eugen, Oregon, USA) according to the manufacturer’s instructions. Briefly, hUCB-EPCs were grown in 24-well plate and treated with different concentrations of Ac4ManNAz. Ac4MAnNAz-treated or untreated hUCB-EPCs were washed with PBS and stained with 10 µg/mL JC-1 dye for 15 min at 37 °C and fluorescence images were acquired.
Transfection and Qdot 525 labeling
hUCB-EPCs were transfected with 100 ng of pcDNA3-EGFP using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. In addition, 1 μM Qdot 525 (Thermo Fisher Scientific, Rockford, IL, USA) in medium was incubated for 4–6 h at 37 °C and hUCB-EPC samples were diluted in DPBS immediately before measurement. Then, we analyzed samples using fluorescence microscopy.
RNA-seq analysis
hUCB-EPCs were cultured with 0, 10 or 50 μM Ac4ManNAz and harvested. Total RNA was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and the quantity and quality of total RNA were evaluated using a NanoDrop spectrophotometer (NanoDrop Technologies, Montchanin, DE, USA) and a 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). An RNA sequencing library was generated using TruSeq RNA Library Preparation Kit (Illumina, San Diego, CA, USA) according to the user’s instruction manual. Briefly, mRNA was separated from total RNA using Oligo (dT) beads and chemically fragmented. After double-strand cDNA synthesis of the fragmented mRNA, end-repair, adenylation of the 3′-end and sequencing adapter ligation were performed and followed by DNA purification with magnetic beads and PCR amplification. Finally, the amplified library was purified, quantified and then applied for template preparation. A HiSeq2000 platform was used to generate 99-bp paired-end sequencing reads (Illumina, San Diego, CA, USA). All 99-bp paired-end sequence reads were mapped to the human genome using TopHat version 2.0.4. Finally, we identified differentially expressed genes (DEgenes). To characterize the biological pathways related to differentially expressed sequences and transcription factors, representative pathways were analyzed in the context of several databases such as KEGG (http://www.genome.ad.jp), BioCarta (http://www.biocarta.com) and Reactome (http://www.reactome.org), as suggested by MsigDB v4.0. Additionally, we used Fisher’s exact test and FDR to examine mapping pathways (filtering options: p < 0.05 and FDR < 5). To identify cellular functional process-related genes, the DEgenes were subjected to gene ontology (GO) analysis using the Database for Annotation, Visualization and Integrated Discovery (DAVID), KEGG, BIOCARTA, REACTOME and Pathway Interaction Database (PID). We used a Gene Set Enrichment Analysis technique to identify statistically up- and down-regulated gene set.
In vitro tube formation assay
Functional characterization of hUCB-EPCs was performed using vascular tube formation and acetylated low-density lipoprotein (ac-LDL; Invitrogen, Carlsbad, CA, USA) uptake assays. For the vascular tube formation assay, 50 μL of Matrigel Basement Membrane Matrix (BD Biosciences, San Diego, CA, USA) was added to a 48-well plate and allowed to solidify at 37 °C for 30 min; 5 × 104 hUCB-EPCs were suspended in 100 μL of culture medium and plated onto the Matrigel layer. After 24 h, the medium was removed and the formation of vascular tube-like structures was assessed with an inverted microscope (Eclipse TS100; Nikon, Tokyo, Japan) and a digital camera system for imaging (Digital SLR Camera D300; Nikon, Tokyo, Japan). For the ac-LDL uptake assay, hUCB-EPCs were seeded onto glass coverslips in 24-well plates at a density of 5 × 104 cells/well in MV2 medium. When cells reached ∼80% confluence, cultures were serum-deprived overnight in Iscove’s Modified Dulbecco’s Medium (IMDM, Sigma-Aldrich, St. Louis, MO, USA) supplemented with 2% lipoprotein-deficient serum from human plasma (Sigma-Aldrich, St. Louis, MO, USA) and then, the medium was replaced with IMDM supplemented with 100 μg/mL human ac-LDL. After 24 h, cells on coverslips were stained with Nile Red (Sigma-Aldrich, St. Louis, MO, USA) and examined by fluorescence microscopy with an inverted microscope (Eclipse TS100; Nikon, Tokyo, Japan) and a digital camera system for imaging (Digital SLR Camera D300; Nikon, Tokyo, Japan).
Flow Cytometry
To analyze hUCB-EPC marker genes, immunophenotyping was performed using the following monoclonal antibodies: anti-CD31-FITC (1:25, BD Biosciences, San Jose, CA, USA) and anti-CD144-PE (1:10, Beckman Coulter, Fullerton, CA, USA). Antibodies and matched isotype control (Beckman Coulter, Fullerton, CA, USA) were incubated for 30 min at 4 °C. Data were acquired and analyzed on a five-parameter flow cytometer (FACSCalibur, Becton Dickinson, San Jose, CA) with Weasel software (WEHI, Melbourne, Australia).
Statistical analysis
Experimental data are presented as a mean ± standard deviation and were analyzed using Analysis of Variance (ANOVA) tests. A value of p < 0.05 was considered statistically significant.
Electronic supplementary material
Acknowledgements
This work supported by grants (NRF-2017M3A9C 7065685 and NRF-2016M3A9B4919616) from National Research Foundation funded by the Ministry of Science and ICT, Korea, and the Research Center for High Quality Livestock Products through Agriculture, Ministry of Agriculture, Food and Rural Affairs (Grant no. 715003-07). We thank Eun Hee Han (Korea Basic Science Institute) and Yong-Jin Kim (AMOREFPACIFIC) for technical support in analyses.
Author Contributions
S.-S.H. and H.-E.S. conceived the study and designed most experiments. H.-E.S. and S.-J.P. carried out isolation and in-vitro culture of hUCB-EPCs. S.-S.H. and H.-E.S. performed in-vitro cell labeling and an analysis of basic cellular properties. S.-S.H. and B.-C.K. analyzed the gene expression patterns of metabolic labeled hUCB-EPCs. D.-E.L. carried out the cell permeability analysis. S.-J.P., H.-M.C. and S.-W.M. analyzed the flow cytometry analysis. All authors analyzed the data and contributed the manuscript preparation. S.-S.H., S.-H.M. and S.-W.K. wrote the manuscript, which was reviewed and edited by the other co-authors.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Sang-Soo Han and Hye-Eun Shim contributed equally.
Contributor Information
Sung-Hwan Moon, Email: sunghwanmoon@kku.ac.kr.
Sun-Woong Kang, Email: swkang@kitox.re.kr.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-31594-0.
References
- 1.Asahara T, et al. Isolation of putative progenitor endothelial cells for angiogenesis. Science. 1997;275:964–967. doi: 10.1126/science.275.5302.964. [DOI] [PubMed] [Google Scholar]
- 2.Sukmawati D, Tanaka R. Introduction to next generation of endothelial progenitor cell therapy: a promise in vascular medicine. Am J Transl Res. 2015;7:411–421. [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao YH, et al. Endothelial Progenitor Cells: Therapeutic Perspective for Ischemic Stroke. Cns Neurosci Ther. 2013;19:67–75. doi: 10.1111/cns.12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitchell KE, et al. Matrix cells from Wharton’s jelly form neurons and glia. Stem Cells. 2003;21:50–60. doi: 10.1634/stemcells.21-1-50. [DOI] [PubMed] [Google Scholar]
- 5.Schmidt D, et al. Umbilical cord blood derived endothelial progenitor cells for tissue engineering of vascular grafts. Ann Thorac Surg. 2004;78:2094–2098. doi: 10.1016/j.athoracsur.2004.06.052. [DOI] [PubMed] [Google Scholar]
- 6.Yang HM, et al. Therapeutic Efficacy of Human Embryonic Stem Cell-Derived Endothelial Cells in Humanized Mouse Models Harboring a Human Immune System. Arterioscl Throm Vas. 2013;33:2839–2849. doi: 10.1161/ATVBAHA.113.302462. [DOI] [PubMed] [Google Scholar]
- 7.Ingram DA, et al. Identification of a novel hierarchy of endothelial progenitor cells using human peripheral and umbilical cord blood. Blood. 2004;104:2752–2760. doi: 10.1182/blood-2004-04-1396. [DOI] [PubMed] [Google Scholar]
- 8.Rocha V, Gluckman E. Improving outcomes of cord blood transplantation: HLA matching, cell dose and other graft- and transplantation-related factors. British journal of haematology. 2009;147:262–274. doi: 10.1111/j.1365-2141.2009.07883.x. [DOI] [PubMed] [Google Scholar]
- 9.Madlambayan G, Rogers I. Umbilical cord-derived stem cells for tissue therapy: current and future uses. Regenerative medicine. 2006;1:777–787. doi: 10.2217/17460751.1.6.777. [DOI] [PubMed] [Google Scholar]
- 10.Bliss T, Guzman R, Daadi M, Steinberg GK. Cell transplantation therapy for stroke. Stroke. 2007;38:817–826. doi: 10.1161/01.STR.0000247888.25985.62. [DOI] [PubMed] [Google Scholar]
- 11.Laughlin MJ, et al. Outcomes after transplantation of cord blood or bone marrow from unrelated donors in adults with leukemia. New Engl J Med. 2004;351:2265–2275. doi: 10.1056/NEJMoa041276. [DOI] [PubMed] [Google Scholar]
- 12.Li Y-F, et al. Endothelial progenitor cells in ischemic stroke: an exploration from hypothesis to therapy. Journal of Hematology & Oncology. 2015;8:33. doi: 10.1186/s13045-015-0130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song E, Lu C-W, Fang L-J, Yang W. Culture and identification of endothelial progenitor cells from human umbilical cord blood. International Journal of Ophthalmology. 2010;3:49–53. doi: 10.3980/j.issn.2222-3959.2010.01.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sudchada S, et al. CD14−/CD34+ is the founding population of umbilical cord blood-derived endothelial progenitor cells and angiogenin1 is an important factor promoting the colony formation. Annals of Hematology. 2012;91:321–329. doi: 10.1007/s00277-011-1303-3. [DOI] [PubMed] [Google Scholar]
- 15.Wang J, Jokerst JV. Stem Cell Imaging: Tools to Improve Cell Delivery and Viability. Stem Cells International. 2016;2016:16. doi: 10.1155/2016/9240652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kircher, M. F., Gambhir, S. S. & Grimm, J. Noninvasive cell-tracking methods. Nat Rev Clin Oncol8, 677–688, http://www.nature.com/nrclinonc/journal/v8/n11/suppinfo/nrclinonc.2011.141_S1.html (2011). [DOI] [PubMed]
- 17.Janic B, Arbab AS. Cord blood endothelial progenitor cells as therapeutic and imaging probes. Imaging in medicine. 2012;4:477–490. doi: 10.2217/iim.12.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higuchi T, et al. Reporter gene PET for monitoring survival of transplanted endothelial progenitor cells in the rat heart after pretreatment with VEGF and atorvastatin. J Nucl Med. 2009;50:1881–1886. doi: 10.2967/jnumed.109.067801. [DOI] [PubMed] [Google Scholar]
- 19.Varma, N. R. S. et al. Endothelial Progenitor Cells (EPCs) as Gene Carrier System for Rat Model of Human Glioma. Plos One7, 10.1371/journal.pone.0030310 (2012). [DOI] [PMC free article] [PubMed]
- 20.Schlechta B, et al. Ex-Vivo Expanded Umbilical Cord Blood Stem Cells Retain Capacity for Myocardial Regeneration. Circ J. 2010;74:188–194. doi: 10.1253/circj.CJ-09-0409. [DOI] [PubMed] [Google Scholar]
- 21.Shi H, et al. Tracking of CFSE-labeled endothelial progenitor cells in laser-injured mouse retina. Chinese Med J-Peking. 2011;124:751–757. [PubMed] [Google Scholar]
- 22.Arbab AS, et al. Labeling of cells with ferumoxides-protamine sulfate complexes does not inhibit function or differentiation capacity of hematopoietic or mesenchymal stem cells. NMR in biomedicine. 2005;18:553–559. doi: 10.1002/nbm.991. [DOI] [PubMed] [Google Scholar]
- 23.Ju S, et al. In vitro labeling and MRI of mesenchymal stem cells from human umbilical cord blood. Magnetic resonance imaging. 2006;24:611–617. doi: 10.1016/j.mri.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 24.Siow TY, Chen CC, Lin CY, Chen JY, Chang C. MR phase imaging: sensitive and contrast-enhancing visualization in cellular imaging. Magnetic resonance imaging. 2012;30:247–253. doi: 10.1016/j.mri.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 25.Hu SL, et al. In vivo magnetic resonance imaging tracking of SPIO-labeled human umbilical cord mesenchymal stem cells. Journal of cellular biochemistry. 2012;113:1005–1012. doi: 10.1002/jcb.23432. [DOI] [PubMed] [Google Scholar]
- 26.Wang S, et al. Magnetic resonance imaging targeting of intracranial glioma xenografts by Resovist-labeled endothelial progenitor cells. Journal of neuro-oncology. 2011;105:67–75. doi: 10.1007/s11060-011-0569-6. [DOI] [PubMed] [Google Scholar]
- 27.Willenbrock S, et al. In vivo MRI of intraspinally injected SPIO-labelled human CD34+ cells in a transgenic mouse model of ALS. In vivo (Athens, Greece) 2012;26:31–38. [PubMed] [Google Scholar]
- 28.Mahmoudi, M. et al. Novel MRI Contrast Agent from Magnetotactic Bacteria Enables In Vivo Tracking of iPSC-derived Cardiomyocytes. Sci Rep-Uk6, 10.1038/srep26960 (2016). [DOI] [PMC free article] [PubMed]
- 29.Diaz, S. & Varki, A. Metabolic radiolabeling of animal cell glycoconjugates. Curr Protoc Protein Sci Chapter 12, Unit12 12 12 12 11–55, 10.1002/0471140864.ps1202s57 (2009). [DOI] [PubMed]
- 30.Lee SY, et al. Non-invasive stem cell tracking in hindlimb ischemia animal model using bio-orthogonal copper-free click chemistry. Biochem Bioph Res Co. 2016;479:779–786. doi: 10.1016/j.bbrc.2016.09.132. [DOI] [PubMed] [Google Scholar]
- 31.Hart C, Chase LG, Hajivandi M, Agnew B. Metabolic labeling and click chemistry detection of glycoprotein markers of mesenchymal stem cell differentiation. Methods Mol Biol. 2011;698:459–484. doi: 10.1007/978-1-60761-999-4_33. [DOI] [PubMed] [Google Scholar]
- 32.Sun L, Fu H, Li Y, Duan X, Li Z. Rapid Recognition and Isolation of Live Colon Cancer Stem Cells by Using Metabolic Labeling of Azido Sugar and Magnetic Beads. Analytical Chemistry. 2016;88:3953–3958. doi: 10.1021/acs.analchem.6b00154. [DOI] [PubMed] [Google Scholar]
- 33.Vogt JA, et al. Protein abundance quantification in embryonic stem cells using incomplete metabolic labelling with 15N amino acids, matrix-assisted laser desorption/ionisation time-of-flight mass spectrometry and analysis of relative isotopologue abundances of peptides. Rapid communications in mass spectrometry: RCM. 2003;17:1273–1282. doi: 10.1002/rcm.1045. [DOI] [PubMed] [Google Scholar]
- 34.Kang K, et al. Tissue-based metabolic labeling of polysialic acids in living primary hippocampal neurons. Proceedings of the National Academy of Sciences. 2015;112:E241–E248. doi: 10.1073/pnas.1419683112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee S, et al. In vivo stem cell tracking with imageable nanoparticles that bind bioorthogonal chemical receptors on the stem cell surface. Biomaterials. 2017;139:12–29. doi: 10.1016/j.biomaterials.2017.05.050. [DOI] [PubMed] [Google Scholar]
- 36.Agnew B, Hart C, Nyberg T, Lakshmanaswamy R. In vivo metabolic labeling and detection of specific glycoprotein subclasses in a mouse breast cancer model. Cancer Research. 2007;67:2465–2465. [Google Scholar]
- 37.Laughlin ST, Bertozzi CR. Metabolic labeling of glycans with azido sugars and subsequent glycan-profiling and visualization via Staudinger ligation. Nat. Protocols. 2007;2:2930–2944. doi: 10.1038/nprot.2007.422. [DOI] [PubMed] [Google Scholar]
- 38.Tian Y, Zhang H. Characterization of disease-associated N-linked glycoproteins. Proteomics. 2013;13:504–511. doi: 10.1002/pmic.201200333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dube DH, Prescher JA, Quang CN, Bertozzi CR. Probing mucin-type O-linked glycosylation in living animals. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:4819–4824. doi: 10.1073/pnas.0506855103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim EJ, et al. Characterization of the Metabolic Flux and Apoptotic Effects of O-Hydroxyl- and N-Acyl-modified N-Acetylmannosamine Analogs in Jurkat Cells. Journal of Biological Chemistry. 2004;279:18342–18352. doi: 10.1074/jbc.M400205200. [DOI] [PubMed] [Google Scholar]
- 41.Saki N, Jalalifar MA, Soleimani M, Hajizamani S, Rahim F. Adverse Effect of High Glucose Concentration on Stem Cell Therapy. International Journal of Hematology-Oncology and Stem Cell Research. 2013;7:34–40. [PMC free article] [PubMed] [Google Scholar]
- 42.Follmar KE, et al. Effects of glutamine, glucose and oxygen concentration on the metabolism and proliferation of rabbit adipose-derived stem cells. Tissue engineering. 2006;12:3525–3533. doi: 10.1089/ten.2006.12.3525. [DOI] [PubMed] [Google Scholar]
- 43.Purcell MK, et al. Identification of the major capsid protein of erythrocytic necrosis virus (ENV) and development of quantitative real-time PCR assays for quantification of ENV DNA. J Vet Diagn Invest. 2016;28:382–391. doi: 10.1177/1040638716646411. [DOI] [PubMed] [Google Scholar]
- 44.Rowland TJ, et al. Roles of integrins in human induced pluripotent stem cell growth on Matrigel and vitronectin. Stem cells and development. 2010;19:1231–1240. doi: 10.1089/scd.2009.0328. [DOI] [PubMed] [Google Scholar]
- 45.Rodgers, S. J., Ferguson, D. T., Mitchell, C. A. & Ooms, L. M. Regulation of PI3K effector signalling in cancer by the phosphoinositide phosphatases. Bioscience Rep37, 10.1042/Bsr20160432 (2017). [DOI] [PMC free article] [PubMed]
- 46.Soncin F, Ward CM. The Function of E-Cadherin in Stem Cell Pluripotency and Self-Renewal. Genes. 2011;2:229–259. doi: 10.3390/genes2010229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li L, Bennett SAL, Wang L. Role of E-cadherin and other cell adhesion molecules in survival and differentiation of human pluripotent stem cells. Cell Adhesion & Migration. 2012;6:59–70. doi: 10.4161/cam.19583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bindu, H. & Srilatha, B. Potency of various types of stem cells and their transplantation. J Stem Cell Res Ther1 (2011).
- 49.Voyta JC, Via DP, Butterfield CE, Zetter BR. Identification and isolation of endothelial cells based on their increased uptake of acetylated-low density lipoprotein. The Journal of cell biology. 1984;99:2034–2040. doi: 10.1083/jcb.99.6.2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marks PW, Witten CM, Califf RM. Clarifying Stem-Cell Therapy’s Benefits and Risks. New Engl J Med. 2017;376:1007–1009. doi: 10.1056/NEJMp1613723. [DOI] [PubMed] [Google Scholar]
- 51.Adamiak, M., Madeja, Z. & Zuba-Surma, E. K. In Adult Stem Cell Therapies: Alternatives to Plasticity (ed Mariusz Z. Ratajczak) 35–51 (Springer New York, 2014).
- 52.Han S-S, et al. Physiological Effects of Ac4ManNAz and Optimization of Metabolic Labeling for Cell Tracking. Theranostics. 2017;7:1164–1176. doi: 10.7150/thno.17711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roth Z, Yehezkel G, Khalaila I. Identification and Quantification of Protein Glycosylation. International Journal of Carbohydrate Chemistry. 2012;2012:10. doi: 10.1155/2012/640923. [DOI] [Google Scholar]
- 54.Comer FI, Hart GW. O-Glycosylation of Nuclear and Cytosolic Proteins: DYNAMIC INTERPLAY BETWEEN O-GlcNAc ANDO-PHOSPHATE. Journal of Biological Chemistry. 2000;275:29179–29182. doi: 10.1074/jbc.R000010200. [DOI] [PubMed] [Google Scholar]
- 55.Hang HC, Yu C, Kato DL, Bertozzi CR. A metabolic labeling approach toward proteomic analysis of mucin-type O-linked glycosylation. Proceedings of the National Academy of Sciences. 2003;100:14846–14851. doi: 10.1073/pnas.2335201100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vocadlo DJ, Hang HC, Kim EJ, Hanover JA, Bertozzi CR. A chemical approach for identifying O-GlcNAc-modified proteins in cells. Proc Natl Acad Sci USA. 2003;100:9116–9121. doi: 10.1073/pnas.1632821100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chang PV, et al. Metabolic labeling of sialic acids in living animals with alkynyl sugars. Angew Chem Int Ed Engl. 2009;48:4030–4033. doi: 10.1002/anie.200806319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao X, et al. Labeling of Enveloped Virus via Metabolic Incorporation of Azido Sugars. Bioconjug Chem. 2015;26:1868–1872. doi: 10.1021/acs.bioconjchem.5b00310. [DOI] [PubMed] [Google Scholar]
- 59.Hayakawa K, et al. Epigenetic switching by the metabolism-sensing factors in the generation of orexin neurons from mouse embryonic stem cells. The Journal of biological chemistry. 2013;288:17099–17110. doi: 10.1074/jbc.M113.455899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen L, Liang JF. Metabolic Monosaccharides Altered Cell Responses to Anticancer Drugs. European journal of pharmaceutics and biopharmaceutics: official journal of Arbeitsgemeinschaft fur Pharmazeutische Verfahrenstechnik e.V. 2012;81:339–345. doi: 10.1016/j.ejpb.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baskin JM, Bertozzi CR. Bioorthogonal click chemistry: covalent labeling in living systems. QSAR & Combinatorial Science. 2007;26:1211–1219. doi: 10.1002/qsar.200740086. [DOI] [Google Scholar]
- 62.van Geel R, Pruijn GJ, van Delft FL, Boelens WC. Preventing thiol-yne addition improves the specificity of strain-promoted azide-alkyne cycloaddition. Bioconjugate chemistry. 2012;23:392–398. doi: 10.1021/bc200365k. [DOI] [PubMed] [Google Scholar]
- 63.Jewett JC, Sletten EM, Bertozzi CR. Rapid Cu-Free Click Chemistry with Readily Synthesized Biarylazacyclooctynones. Journal of the American Chemical Society. 2010;132:3688–3690. doi: 10.1021/ja100014q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tian C, et al. Use of steric encumbrance to develop conjugated nanoporous polymers for metal-free catalytic hydrogenation. Chemical Communications. 2016;52:11919–11922. doi: 10.1039/C6CC06372A. [DOI] [PubMed] [Google Scholar]
- 65.Dommerholt J, et al. Readily accessible bicyclononynes for bioorthogonal labeling and three-dimensional imaging of living cells. Angewandte Chemie (International ed. in English) 2010;49:9422–9425. doi: 10.1002/anie.201003761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Madl CM, Heilshorn SC. Bioorthogonal Strategies for Engineering Extracellular Matrices. Advanced Functional Materials. 2018;28:1706046. doi: 10.1002/adfm.201706046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Teo CF, Wells L. Monitoring protein O-linked β-N-acetylglucosamine status via metabolic labeling and copper-free click chemistry. Analytical biochemistry. 2014;464:70–72. doi: 10.1016/j.ab.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Akiyama SK. Integrins in cell adhesion and signaling. Human cell. 1996;9:181–186. [PubMed] [Google Scholar]
- 69.Ellis SJ, Tanentzapf G. Integrin-mediated adhesion and stem-cell-niche interactions. Cell and Tissue Research. 2009;339:121. doi: 10.1007/s00441-009-0828-4. [DOI] [PubMed] [Google Scholar]
- 70.Redmer T, et al. E-cadherin is crucial for embryonic stem cell pluripotency and can replace OCT4 during somatic cell reprogramming. EMBO Reports. 2011;12:720–726. doi: 10.1038/embor.2011.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Finney MR, et al. Direct comparison of umbilical cord blood versus bone marrow-derived endothelial precursor cells in mediating neovascularization in response to vascular ischemia. Biology of blood and marrow transplantation: journal of the American Society for Blood and Marrow Transplantation. 2006;12:585–593. doi: 10.1016/j.bbmt.2005.12.037. [DOI] [PubMed] [Google Scholar]
- 72.Lee MJ, et al. Enhancement of wound healing by secretory factors of endothelial precursor cells derived from human embryonic stem cells. Cytotherapy. 2011;13:165–178. doi: 10.3109/14653249.2010.512632. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.