Abstract
We evaluated genetics of hyperuricemia and gout, their interaction with kidney function and medication intake in chronic kidney disease (CKD) patients. Genome-wide association studies (GWAS) of urate and gout were performed in 4941 CKD patients in the German Chronic Kidney Disease (GCKD) study. Effect estimates of 26 known urate-associated population-based single nucleotide polymorphisms (SNPs) were examined. Interactions of urate-associated variants with urate-altering medications and clinical characteristics of gout were evaluated. Genome-wide significant associations with serum urate and gout were identified for known loci at SLC2A9 and ABCG2, but not for novel loci. Effects of the 26 known SNPs were of similar magnitude in CKD patients compared to population-based individuals, except for SNPs at ABCG2 that showed greater effects in CKD. Gene-medication interactions were not significant when accounting for multiple testing. Associations with gout in specific joints were significant for SLC2A9 rs12498742 in wrists and midfoot joints. Known genetic variants in SLC2A9 and ABCG2 were associated with urate and gout in a CKD cohort, with effect sizes for ABCG2 significantly greater in CKD compared to the general population. CKD patients are at high risk of gout due to reduced kidney function, diuretics intake and genetic predisposition, making treatment to target challenging.
Introduction
Gout is a progressive painful debilitating disease, and the most common inflammatory arthritis in many Western countries1. Population-based studies have identified genetic variants in multiple genes2 including SLC2A93–5 and ABCG25,6 associated with serum urate concentrations. Individuals with chronic kidney disease (CKD) represent a high-risk population for hyperuricemia and gout due to decreased renal clearance of urate and consecutive increase in serum urate concentrations. About ~25% of CKD patients from the prospective German Chronic Kidney Disease (GCKD) study reported a physician diagnosis of gout at study baseline and two thirds of patients were hyperuricemic7. This high prevalence of gout is most relevant given that CKD affects about 10% of the adult population in many countries8.
Despite the importance of CKD as a risk factor for gout, knowledge about genetic determinants of serum urate in the setting of CKD is limited. One candidate gene study that focused on 11 urate transporters reported that the strength of association - as quantified by the association p-value - between genetic variants in ABCG2 was stronger in patients with CKD compared to 481 population-based individuals, while the opposite was observed for variants in SLC2A99. While the authors hypothesized that this could be interpreted by a compensatory role of the ABCG2 transporter in the intestine in the setting of reduced kidney function, they did not formally compare effect sizes or test for differences in effect. Additional aspects of urate genetics in CKD that have not been addressed are interactions between genetic risk variants for gout and medication intake as well as clinical characteristics of gout attacks.
We aimed to evaluate the genetic underpinnings of hyperuricemia and gout in a large cohort of CKD patients, by carrying out genome-wide association studies (GWAS) of serum urate concentrations and gout. Effect sizes of known urate-associated variants detected in the general population were formally compared to their counterparts among CKD patients. Interactions between medications influencing serum urate concentrations and commonly prescribed in CKD were evaluated, and associations between genetic risk variants and clinical characteristics of gout were examined.
Results
Study population
Baseline clinical characteristics of 4,941 GCKD study participants with complete clinical information required for the GWAS are summarized in Table 1. A quarter of the patients reported diagnosis of gout at study baseline. Participants with gout compared to those without gout were significantly (p-value < 0.05) more likely to be male, older, have a higher BMI, be hypertensive, diabetic, have higher triglyceride levels and have lower eGFR. There were no significant differences in serum urate concentrations between the two groups, but patients with gout were significantly more likely to be treated with allopurinol compared to those without gout7. The most pronounced significant differences were observed for eGFR, age, gender, BMI, and diuretic and gout medication intake.
Table 1.
Study sample characteristics of GCKD participants with complete urate, gout and genotype information.
| Characteristic | Overall (n = 4941) | No gout (n = 3724) | Gout (n = 1217) | P-value |
|---|---|---|---|---|
| eGFR, ml/min/1.73 m2 | 49.5 ± 18.1 | 51.0 ± 18.9 | 44.9 ± 14.8 | <2.2e-16 |
| Age | 60.1 ± 12.0 | 59.1 ± 12.6 | 63.1 ± 9.1 | <2.2e-16 |
| Female, n (%) | 39.8% | 44.5% | 25.4% | <2.2e-16 |
| BMI, kg/m2 | 29.8 ± 6.0 | 29.3 ± 5.8 | 31.6 ± 6.1 | <2.2e-16 |
| Hypertension, n (%) | 96.2% | 95.4% | 98.6% | 4.9E-07 |
| Systolic BP, mmHg | 139.4 ± 20.4 | 138.8 ± 20.1 | 141.2 ± 21.1 | 5.2E-04 |
| Cholesterol, mg/dL | 211.3 ± 53.0 | 213.0 ± 53.3 | 206.1 ± 51.8 | 6.0E-05 |
| Triglycerides, mg/dL | 168.1 (118.2, 239.0) | 161.2 (113.8, 229.3) | 193.6 (133.4, 270.4) | 6.3E-15 |
| CHD, n (%) | 19.9% | 17.5% | 27.0% | 6.4E-13 |
| DM 2, n (%) | 24.3% | 21.8% | 32.1% | 4.3E-13 |
| Serum urate, mg/dL | 7.21 ± 1.92 | 7.20 ± 1.91 | 7.23 ± 1.96 | 7.3E-01 |
| Hyperuricemia, n (%) | 61.0% | 57.2% | 62.3% | 1.8E-03 |
| Alcohol intake, moderate to large amount | 19.0% | 17.3% | 24.1% | 9.4E-07 |
| Diuretic intake, n (%) | 61.0% | 56.9% | 73.7% | <2.2e-16 |
| Gout medication intake, n (%) | 32.8% | 20.8% | 69.5% | <2.2e-16 |
Continuous variables are mean (SD) unless otherwise noted. N = 4941, except for systolic blood pressure (n = 4913), cholesterol (n = 4935), triglycerides (n = 4933). Estimated glomerular filtration rate (eGFR), body mass index (BMI), systolic blood pressure (systolic BP), coronary heart disease (CHD), diabetes mellitus type 2 (DM 2); p-value for comparison of each characteristic in individuals with and without gout.
Genome-wide association analyses
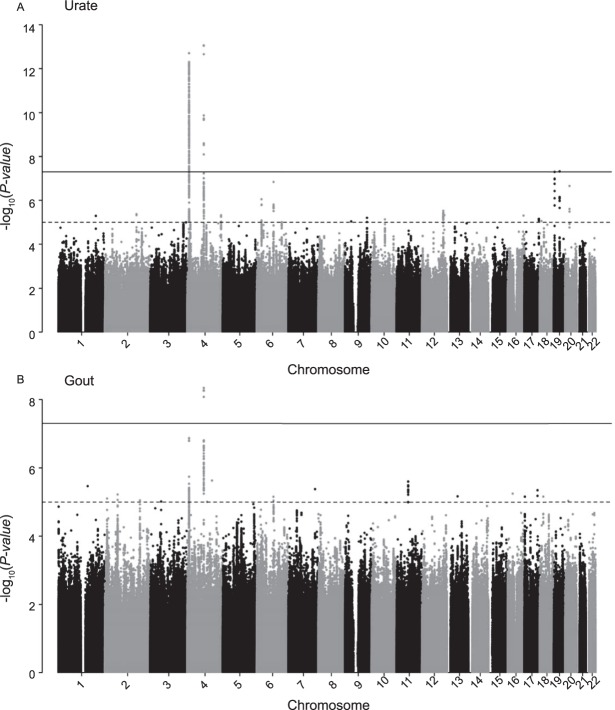
We conducted GWAS for serum urate concentrations and gout using 9,281,895 autosomal SNPs. A quantile-quantile plot comparing observed and expected p-values from the serum urate GWAS showed SNPs with very low observed p-values for serum urate and gout. There was no inflation of median test statistics, consistent with the absence of systematic errors (Supplemental Fig. 1). In the serum urate GWAS, three loci showed genome-wide significant associations (p-value < 5.0E-08, Fig. 1). Variants with the lowest association p-values mapped into the previously reported genes SLC2A9 (rs13111638) and ABCG2 (rs2231142). Per each minor allele copy, serum urate concentrations were 0.40 mg/dl (23.79 μmol/L) higher for ABCG2 rs2231142 and 0.31 mg/dl (18.44 μmol/L) lower for SLC2A9 rs13111638 (Table 2). A third locus, URI1, had not been detected in prior population-based studies. Because CKD cohorts with gout information were not available for replication, and because our findings supported the presence of similar associated loci in population-based and CKD patient studies, the index SNP at this locus was investigated in the well-powered summary results from GWAS meta-analysis of serum urate of >140,000 participants in the GUGC Consortium2. The association was not significant and therefore not followed up on. Odds Ratios (OR) for the associations with gout from the GWAS of gout were 1.54 (95% CI: 1.34; 1.78) for ABCG2 rs2231142 and 0.77 (95% CI: 0.68; 0.86) for SLC2A9 rs13111638 (Table 2). Leading SNPs with association p-values of p < 1.0E-06 (suggestive significance) are listed in Supplemental Tables 1 and 2.
Figure 1.
Manhattan plot for GWAS of serum urate (A) and gout (B) from the regression models. Y-axis: association p-values; x-axis: plotted SNPs by chromosomal position. Continuous line: genome-wide significance level (5E-8). Dashed line: suggestive significance level (1E-5). Data from 1000 Genomes imputation-based GWAS in CKD subjects of European-ancestry.
Table 2.
Association estimates for index SNPs associated at genome-wide significance with serum urate, and their corresponding association with gout.
| SNP | Funct. | Closest Genes | Chr | Position | A1 | A2 | Freq.A1 | Urate | Gout | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| effect | SE | p-value | OR | 95% CI | p-value | ||||||||
| rs2231142 | exonic | ABCG2 | 4 | 89052323 | T | G | 0.11 | 0.40 | 0.05 | 8.8E-14 | 1.54 | 1.34, 1.78 | 5.3E-09 |
| rs13111638 | intronic | SLC2A9 | 4 | 9996890 | T | C | 0.19 | −0.31 | 0.04 | 2.0E-13 | 0.77 | 0.68, 0.86 | 2.9E-05 |
SNP indicates the SNP with the lowest p-value for urate; the index SNP at SLC2A9 for gout was rs59834205 (p-value 1.36E-07, LD with rs13111638: r2 = 0.60, D′ = 0.79, using 1000 G phase 3); the index SNP at ABCG2 for gout was rs4148155 (p-value 4.54E-09; LD with rs2231142: r2 = 0.99, D′ = 1, using 1000 G phase 3). Estimates for rs4148155 and rs59834205 are shown in Supplemental Table 2. Abbreviations: Funct., function; Chr, chromosome; A1, coded allele; A2, non-coded allele; Freq.A1, the frequency of the coded allele; effect, effect of A1; SE, standard error; OR, odds ratio; 95% CI, 95% confidence interval. Associations with urate were adjusted for age, sex, log(eGFR), BMI, significant principal components (p < 0.05), intake of diuretics and gout medication, while associations with gout were adjusted for age, sex, log(eGFR), BMI, and intake of diuretics. Associations were filtered for plausibility and quality.
Comparison of genetic effects to the general population
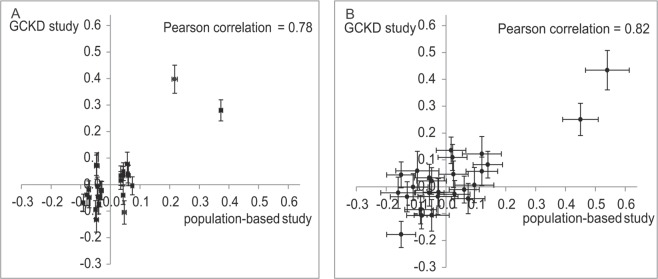
Next we compared the effect sizes of 26 SNPs known to be associated with serum urate in the general population2 to the results from the GWAS in the GCKD study. Of the 26 SNPs discovered among approximately 140,000 individuals, seven SNPs were nominally associated (p-value < 0.05) with serum urate among CKD patients in the GCKD study (Supplemental Table 3) and two SNPs, at ABCG2 and at SLC2A9, were significant after adjusting for multiple testing (p-value < 1.9E-03, 0.05/26). Effect sizes of the genetic associations on serum urate were of similar magnitude in the GCKD study and the population-based meta-analysis for most SNPs, with high correlations across all risk variants observed for both urate (Pearson correlation coefficient 0.78 (Fig. 2, Panel A)) and gout (correlation 0.82 (Fig. 2, Panel B)). The two SNPs that were significantly associated in the GCKD study, at SLC2A9 and ABCG2, were tested for differences in genetic effect sizes on serum urate. The association for rs2231142 in ABCG2 was significantly higher in CKD patients compared to the results from the general population (effect 0.4 mg/dl in CKD vs. 0.22 mg/dl in population-based studies, p-value for difference = 1.5E-03; Supplemental Table 4).
Figure 2.
Scatter plot comparing effect sizes of 26 known urate-associated SNPs from a meta-analysis of population-based studies (X-axis) to those of CKD patients (Y-axis). Upper panel shows the association with urate (Panel A), lower panel shows the associations with gout (Panel B). Gout effects are presented on the log odds scale.
SNP-kidney-function interaction
Although genetic effects of most variants on serum urate discovered in the general population were similar among CKD patients, the effect of individual variants on serum urate may change as eGFR declines. We therefore compared associations of the two genome-wide significant urate-associated SNPs across different categories of eGFR. When stratified by eGFR, the minor T allele of rs2231142 in ABCG2 was significantly associated with serum urate concentrations in each stratum and showed a trend for larger effects at eGFR values <30 ml/min/1.73 m2 (Supplemental Table 5). A statistical test for interaction across the entire eGFR range was not significant. The SNP in SLC2A9 did not show any clear effect size differences or interactions with categories of eGFR.
Gene-environment interaction of candidate variants and medications
Several medications commonly prescribed to CKD patients are known to affect serum urate concentrations. We therefore assessed potential effect modifications of these medications on the associations of urate-associated SNPs mapping into the interacting transport proteins with serum urate and gout. The intake of allopurinol, thiazide diuretics, loop diuretics, potassium sparing diuretics and losartan was evaluated for interaction with known urate-associated SNPs mapping into the loci for SLC2A9, ABCG2, SLC17A1, and SLC22A12 (p < 2.5E-03, 0.05/(4 * 5)). Table 3 shows results for the interaction between all SNPs and medications that showed at least a nominally significant (p < 0.05) SNP* medication interaction, namely thiazides and loop diuretics. Two models were evaluated because of polypharmacy in patients of moderate and advanced CKD. One compared the effect between patients taking the respective medication to those not taking it, and one compared the effect between patients who were only taking this medication to patients not taking any of the evaluated medications. Three of the interactions achieved nominal significance in at least one comparison and had similar effect size differences in both types of comparisons, but none of these interactions were statistically significant after correcting for multiple testing. A detailed list of all tested gene-medication interactions can be found in Supplemental Table 6.
Table 3.
Selected interactions between known urate-associated risk alleles in urate transporters and medications known to affect serum urate concentrations.
| SNP | Medication | With this medication | Without this medication | All (SNP × medication) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| effect | SE | p-value | n | effect | SE | p-value | n | effect | SE | p-value | n | ||
| SLC2A9 rs12498742 (G) | thiazide | −0.23 | 0.08 | 2.7E-03 | 1570 | −0.33 | 0.05 | 7.2E-13 | 3371 | 0.11 | 0.08 | 2.0E-01 | 4941 |
| loop diuretic | −0.37 | 0.07 | 1.0E-07 | 1900 | −0.25 | 0.05 | 1.5E-07 | 3041 | −0.08 | 0.08 | 2.9E-01 | 4941 | |
| ABCG2 rs2231142 (T) | thiazide | 0.37 | 0.10 | 3.1E-04 | 1570 | 0.43 | 0.06 | 3.0E-12 | 3371 | −0.06 | 0.11 | 6.0E-01 | 4941 |
| loop diuretic | 0.58 | 0.10 | 2.1E-09 | 1900 | 0.30 | 0.06 | 9.3E-07 | 3041 | 0.22 | 0.11 | 4.4E-02 | 4941 | |
| SLC17A1 rs1165151 (G) | thiazide | 0.14 | 0.06 | 2.5E-02 | 1570 | 0.03 | 0.04 | 4.4E-01 | 3371 | 0.10 | 0.07 | 1.5E-01 | 4941 |
| loop diuretic | −0.03 | 0.06 | 6.2E-01 | 1900 | 0.11 | 0.04 | 3.1E-03 | 3041 | −0.16 | 0.07 | 2.0E-02 | 4941 | |
| NRXN2/SLC22A12 rs478607 (A) | thiazide | −0.13 | 0.09 | 1.5E-01 | 1570 | −0.12 | 0.05 | 2.1E-02 | 3371 | −0.02 | 0.10 | 8.7E-01 | 4941 |
| loop diuretic | −0.29 | 0.08 | 4.5E-04 | 1900 | −0.01 | 0.05 | 7.9E-01 | 3041 | −0.27 | 0.09 | 3.6E-03 | 4941 | |
| SNP | Medication | Only this medication | No medication** | All (SNP × medication) | |||||||||
| effect | SE | p-value | n | effect | SE | p-value | n | effect | SE | p-value | n | ||
| SLC2A9 rs12498742 (G) | thiazide | −0.06 | 0.12 | 5.9E-01 | 604 | −0.29 | 0.06 | 3.9E-06 | 1503 | 0.23 | 0.12 | 6.3E-02 | 2107 |
| loop diuretic | −0.44 | 0.11 | 6.3E-05 | 589 | −0.29 | 0.06 | 3.9E-06 | 1503 | −0.17 | 0.12 | 1.6E-01 | 2092 | |
| ABCG2 rs2231142 (T) | thiazide | 0.35 | 0.16 | 3.3E-02 | 604 | 0.27 | 0.09 | 1.9E-03 | 1503 | 0.11 | 0.17 | 5.1E-01 | 2107 |
| loop diuretic | 0.66 | 0.17 | 1.6E-04 | 589 | 0.27 | 0.09 | 1.9E-03 | 1503 | 0.40 | 0.18 | 2.4E-02 | 2092 | |
| SLC17A1 rs1165151 (G) | thiazide | 0.24 | 0.10 | 2.1E-02 | 604 | 0.02 | 0.05 | 6.7E-01 | 1503 | 0.21 | 0.11 | 4.6E-02 | 2107 |
| loop diuretic | −0.02 | 0.10 | 8.8E-01 | 589 | 0.02 | 0.05 | 6.7E-01 | 1503 | −0.06 | 0.10 | 5.9E-01 | 2092 | |
| NRXN2/SLC22A12 rs478607 (A) | thiazide | 0.04 | 0.14 | 7.5E-01 | 604 | −0.03 | 0.07 | 6.9E-01 | 1503 | 0.08 | 0.14 | 5.8E-01 | 2107 |
| loop diuretic | −0.23 | 0.13 | 9.0E-02 | 589 | −0.03 | 0.07 | 6.9E-01 | 1503 | −0.21 | 0.14 | 1.4E-01 | 2092 | |
**No medication refers to not taking gout medications, diuretics and losartan. Associations were adjusted for age, sex, log(eGFR), BMI, and medications (gout medication, diuretics, losartan) other than the category being evaluated. The p-value for interaction adjusted for multiple testing was p < 2.5E-03. Rs12498742 (SLC2A9) is the reported index SNP from the GUGC Consortium and is in LD with rs13111638 (r2 = 0.74, D′ = 0.98).
Association with clinical characteristics of gout
Hyperuricemia can lead to monosodium urate crystal formation, influenced by factors such as temperature, pH, salt concentration, and cartilage matrix components. Processes of crystal formation and initiation of joint inflammation are incompletely understood and more underlying factors seem to be at play. We investigated whether genetic effects were more clearly discernible in joints across which temperature, weight pressure or usage strain may differ for the genome-wide significant urate-associated index SNPs at ABCG2 and SLC2A9. The SNP rs12498742 in SLC2A9 was significantly (p < 3.1E-03, 0.05/(2 * 8)) associated with gout affection in wrist and midfoot joints (Table 4). The strongest effect was observed with gout affection of the wrists (OR 0.35, 95% CI 0.18–0.60, p = 4.7E-04), with each copy of the G allele associated with 65% lower odds of gout affection of the wrists. Because of the lower minor allele frequency, subgroups for rs2231142 in ABCG2 were small, and only one joint localization showed a nominally significant (p < 0.05) association (ankles, OR = 1.47, 95% CI 1.02–2.08, p = 3.2E-02). The possibility to study a clear pattern of joint affection in relation to genetic risk variants therefore requires study in even larger samples.
Table 4.
Association between genome-wide significant SNPs and clinical characteristics of gout.
| Affected joints | Controls | Cases | % of female cases | SLC2A9 rs12498742 (G) | ABCG2 rs2231142 (T) | ||
|---|---|---|---|---|---|---|---|
| OR (95%CI) | p-value | OR (95%CI) | p-value | ||||
| shoulder | 2007 | 19 | 26.3% | 0.67 (0.25–1.47) | 3.6E-01 | 0.38 (0.06–1.27) | 1.9E-01 |
| elbow | 1988 | 38 | 10.5% | 0.36 (0.15–0.73) | 9.7E-03 | 0.92 (0.42–1.77) | 8.2E-01 |
| wrist | 1959 | 67 | 23.9% | 0.35 (0.18–0.60) | 4.7E-04 | 1.18 (0.70–1.89) | 5.1E-01 |
| finger | 1929 | 97 | 35.1% | 0.73 (0.49–1.04) | 9.7E-02 | 0.64 (0.37–1.04) | 9.3E-02 |
| knee | 1928 | 98 | 7.1% | 0.59 (0.39–0.87) | 1.0E-02 | 1.41 (0.94–2.07) | 8.7E-02 |
| ankle | 1903 | 123 | 14.6% | 0.67 (0.47–0.93) | 2.3E-02 | 1.47 (1.02–2.08) | 3.2E-02 |
| toe | 1504 | 522 | 20.5% | 0.92 (0.77–1.09) | 3.3E-01 | 1.15 (0.93–1.43) | 1.9E-01 |
| midfoot | 1816 | 210 | 21.4% | 0.61 (0.46–0.80) | 4.2E-04 | 1.31 (0.97–1.73) | 7.0E-02 |
Covariates included age, sex and log (eGFR). OR: Odds Ratio, 95% CI: 95% Confidence interval. For SLC2A9, rs12498742 is the previously reported index SNP from the GUGC Consortium; it is in LD with rs13111638 (r2 = 0.74, D′ = 0.98), the index SNP for SLC2A9 in the GCKD study. The p-value adjusted for multiple testing was p < 3.1E-03 (in bold).
Discussion
In this study of serum urate and gout genetics in CKD, we found genome-wide significant associations with SNPs in two previously identified loci, ABCG2 and SLC2A9. The magnitude of genetic effects identified in previous population-based studies was generally similar except for ABCG2, where effects were larger in CKD patients. Effect modification of risk variants in ABCG2 and SLC2A9 by eGFR categories or intake of urate-altering medications was not significant after accounting for multiple testing, but there were several nominally significant gene-by-medication interactions. Genetic associations with detailed information about the affected joint localization showed significant associations for the SLC2A9 risk allele with gout of the wrists and midfoot joints.
SLC2A9 and ABCG2 are the two most prominent players in gout explaining ~5% of variance in serum urate levels10. GLUT9, affecting uric acid transport through urate reabsorption at the basolateral membrane11 of renal proximal tubules, is encoded by SLC2A9. The ABCG2 transporter, encoded by ABCG2 and localized to the brush border of renal proximal tubules and intestinal cells, functions to extrude urate in an ATP-dependent fashion2,5,12–18.
Similar effect sizes of the lead SNP in SLC2A9 were found when comparing our results to those from population-based studies2,4,5 and one previous study in individuals with CKD stage 3. The previous study of CKD patients by Bhatnagar and colleagues evaluated a bi-racial study population with fewer and younger patients overall and smaller patient numbers in the European ancestry subgroup9,19. Other differences include a genome-wide screen in our study, allowing for the comparison of effect sizes outside of the 11 transporter genes they studied, and the detailed clinical characterization of gout in our study. While Bhatnagar and colleagues9 reported that the index variant in ABCG2 in CKD patients was associated with serum urate with similar effect sizes but at lower p-values when compared to population-based studies2,4,5, we found that the effect sizes were almost twice as large in CKD patients compared to population-based samples. Differences between the studies, most importantly differences in ethnic composition and the proportion of individuals with diabetes, but also in age may account for this difference, but random variation should also be taken into consideration.
Besides renal excretion, recent findings demonstrated physiological and pathophysiological roles of ABCG2 on intestinal urate excretion in humans9,20, where the degree of intestinal ABCG2 dysfunction showed strong associations with the severity of hyperuricemia in the setting of end-stage renal disease20. We further tested whether the risk conferred by rs2231142 in ABCG2 was altered in the setting of reduced eGFR using stratified and interaction analyses. A trend for higher genetic effects on serum urate levels in individuals with eGFR <30 ml/min/1.73 m2 lends some support to the importance of intestinal ABCG2 function, but results from our interaction analysis with eGFR did not support the hypothesis that genetic effects of ABCG2 on serum urate become larger i.e. more important in individuals with CKD compared to the general population in a linear fashion.
To our knowledge, this is the first study to quantify the interaction between gout risk genes and sub-classes of diuretics or allopurinol. Diuretics are widely used for CKD management, hypertension and heart failure21, the latter being frequent comorbidities in patients of the GCKD study. Allopurinol is the most frequently prescribed urate-lowering therapy in the GCKD study despite its side effects and a reduced dose recommendation in CKD. Our findings are suggestive of a greater impact of ABCG2 risk alleles among those using loop diuretics. The observation that adjustment for allopurinol intake was important in detecting genetic effects on gout in this study implies that allopurinol is a well working drug regardless of the observed SLC2A9 and ABCG2 genotype.
Gene-environment interaction-studies have shown conflicting results for the role of genetic risk factors in gout patients using diuretics22–24, likely related to polypharmacy in CKD and limited sample sizes. Such sample sizes have to be several times larger than the ones needed to detect main gene effects, highlighting the need for future, bigger cohort studies to explore gene-environment interactions. Worldwide rates of initiation and continuation of urate-lowering therapy are low and achievement of serum urate targets therefore infrequent7, although gout in combination with CKD7 contributes to poor health-related quality of life in these patients25.
Our results on joint location of acute gout flares do not clearly answer the hypothesis that genetic effects are more important in joints with higher weight pressure and usage strain26. However, sample sizes of each subgroup were small, highlighting the need of large gout research cohorts.
Strengths of our study are being one of the largest CKD cohorts world-wide, a detailed data collection, including medication intake data and information from a validated gout questionnaire27–29 based on ACR criteria to survey gout, the availability of genome-wide genetic data for all patients allowing for an unbiased search for gout risk loci in the setting of CKD and a comprehensive examination of known genetic risk loci. Knowledge generated from CKD cohorts such as the GCKD study may inform future interventional trials that focus on reducing the burden of hyperuricemia and its associated progression of CKD and CVD30–34.
The present study has some limitations. Gout status was based on self-reported physician diagnosis of gout, possibly introducing some misclassification, but good sensitivity and reliability on self-reported gout has been reported previously7,35. Our analyses were based on a Caucasian patient population of mostly CKD stage 3, potentially compromising generalizability. Sample sizes were small for interaction analyses, although the GCKD study is one of the largest observational cohorts of CKD patients world-wide. Pooled samples from different CKD cohorts with available information on serum urate concentrations and gout under the framework of CKD consortia around the world could address this limitation36.
In summary, the known genetic associations at ABCG2 and SLC2A9 with serum urate and gout were replicated in CKD patients with higher effect sizes for ABCG2 in patients with CKD compared to individuals from the general population. Interactions with medications and effect modification by eGFR categories were not significant after correcting for multiple testing. Affected joint localizations showed significant associations for SLC2A9 with wrists and midfoot joints. Patients with CKD are at risk for gout due to reduced kidney function, diuretics intake and genetics7. This large and complex patient group will become even more relevant in ageing populations and with increasing polypharmacy, and yet is still understudied.
Methods
Study population
The GCKD study is a multi-centric, prospective observational study of 5,217 CKD patients of European ancestry. Patients undergo regular, standardized study visits including questionnaires, physical examinations and biosampling by trained personnel. The GCKD study has implemented a data protection concept according to the data protection recommendations of the platform for technology and methods for networked medical research (TMF; www.tmf-ev.de), supported by the German Ministry of Education and Research (www.bmbf.de). The study protocol was approved by local institutional review boards (1 ethics committee of the RWTH Aachen University, Aachen, Germany; 2 ethics committee of the Charité – University-Medicine, Berlin, Germany; 3 ethics committee of the Friedrich-Alexander University, Erlangen, Germany; 4 ethics committee of the Albert-Ludwigs-University, Freiburg, Germany; 5 ethics committee of the Friedrich-Schiller University, Jena, Germany; 6 ethics committee of the Hannover Medical School, Hannover, Germany; 7 ethics committee of the medical faculty, Ruprecht-Karls University, Heidelberg, Germany; 8 ethics committee of the medical faculty, Ludwig-Maximilians-University, Munich, Germany; 9 ethics committee of the Julius-Maximilians-University, Würzburg, Germany) at each participating academic institution, the data protection concept was reviewed by the data protection officer of the State of Hessen37 and the study was registered in the national registry of clinical studies (DRKS 0003971). All methods were carried out in accordance with relevant guidelines and regulations. Written informed consent was obtained for all participants. Study procedures and main baseline findings have been reported19,37. Prevalence and correlates of gout at study baseline were published elsewhere7.
A comprehensive gout questionnaire, implemented due to the high prevalence of gout observed at study baseline, designed to meet the American College of Rheumatology (ACR) criteria used to survey gout and validated in previous observational studies of gout27–29, was used to assess clinical characteristics of patients, reporting a physician diagnosis of gout. The questionnaire covered physician diagnosis of gout, age of onset, annual number of attacks, attack duration, appearance of the affected joints, affected joint number and localization, presence of tophi, presence of crystals from fine-needle aspiration, presence of hyperuricemia, and family history of gout.
Genotyping, quality control and imputation
DNA was isolated from peripheral blood leukocytes and genotyped for 5,123 patients using the Illumina HumanOmni2.5-8 v1.2 BeadChip (Illumina, GenomeStudio, Genotyping Module Version 1.9.4) at the Helmholtz Center Munich. Standardized protocols were used for data cleaning38. Custom written scripts (R, Perl) and plink1.939 software was used for quality control. Quality control steps included call rate, sex check, mean heterozygosity, genetic ancestry and cryptic relatedness, leading to the exclusion of 89 samples. Single nucleotide polymorphism (SNP) exclusion criteria prior to imputation were a call rate of <0.96, deviation from Hardy-Weinberg equilibrium (p < 1.0E-05), and duplicate positions, yielding clean genotype data for 2,337,794 autosomal SNPs of the Omni 2.5 component from 5,034 patients. Genotypes were then imputed by pre-phasing40 and imputation with Impute2 (v2.3.1)41 using the 1000 Genomes Phase 3 version 5 ALL as reference panel. After genotype imputation, 9,281,895 SNPs of high imputation quality (info score > 0.8) and a minor allele frequency of >0.01 were kept for association analyses to generate robust association statistics.
Statistical analyses
Patient characteristics were compared using t-tests for continuous and Chi-square tests for categorical variables.
Genome-wide association analyses
Single variant genome-wide association analyses were conducted using linear regression (urate) and logistic regression (gout), assuming an additive genetic model using genotype dosages to account for imputation uncertainty. For urate as the outcome, the regression model was adjusted for age, sex, estimated glomerular filtration rate (log(eGFR)), body mass index (BMI), intake of diuretics, and intake of gout medication. For gout as the outcome, the models was adjusted for age, sex, log(eGFR), BMI, and intake of diuretics. In addition, principal components (PCs) were included as covariates when significantly (p < 0.05) associated with evaluated outcomes of the respective models.
Association analyses were performed using SNPTEST v2.542. Functional annotation of variants was conducted using ANNOVAR43.
Comparison of genetic effects to the general population
Twenty-six SNPs previously reported as associated with serum urate and replicated in a meta-analysis of mostly population-based cohorts in the Global Urate Genetics Consortium (GUGC)2 were used as candidate variants comparing their effect estimates in general populations with CKD patients in the GCKD study. Differences in effect estimates were computed for significantly associated SNPs (in the GCKD study) after adjusting for multiple testing (p-value < 1.9E-03, 0.05/26) between population-based results and CKD patients using a 2-sample t-test. SNP associations were compared for minor allele frequencies, direction and magnitude and evaluated visually.
SNP-kidney-function interaction
Interactions were evaluated for SNPs showing genome-wide significant associations with serum urate in the GCKD study. According to the Kidney Disease: Improving Global Outcomes CKD guidelines44, eGFR was categorized into stages G1/2 (≥60 ml/min/1.73 m2), G3a (45–59 ml/min/1.73 m2), G3b (30–44 ml/min/1.73 m2) and G4/5 (˂30 ml/min/1.73 m2). For each of the four eGFR categories, the multivariable adjusted odds ratios (OR) and 95% confidence intervals (95% CI; all covariates as above except for eGFR) were compared. Interactions between genotype and eGFR category were evaluated by adding a SNP*eGFR category term to the model and evaluating its statistical significance.
Gene-environment interaction of candidate variants and medications
Gene environment-interactions were assessed for known urate-associated variants2 mapping into or near transporter genes reported to interact with medications (SLC2A9, ABCG2, NRXN2/SLC22A12 and SLC17A1)11 and medications known to affect urate concentration and commonly prescribed in CKD patients: allopurinol, thiazide diuretics, loop diuretics, potassium sparing diuretics and losartan. Allele dosages of imputed candidate SNPs were converted to the most likely genotypes. Current medication intake was assessed for all GCKD patients and coded using Anatomical Therapeutic Chemical (ATC) codes. Allopurinol, thiazide diuretics, loop diuretics, potassium sparing diuretics and losartan intake were represented by codes M04AA01/M04AA51, C03A/C03B C03C, C03D and C09CA01.
For each SNP and medication combination, two types of stratified and interaction analyses were performed adjusted for age, sex, log(eGFR), BMI, intake of medications other than the evaluated class and PCs significantly associated with serum urate. Firstly, the stratified analyses by intake of the specific medication under evaluation and the analysis including an interaction term (SNP × specific medication) were assessed among all individuals. Secondly, the stratified analyses by intake of the specific medication under evaluation and the analysis including an interaction term (SNP × specific medication) were assessed excluding individuals taking any of the other classes of medication under evaluation. Statistical significance for multiple testing (p < 2.5E-03) was obtained by dividing the significance threshold of 0.05 by the four tested SNPs multiplied with the five evaluated medications.
Association with clinical characteristics of gout
SNPs associated with serum urate at genome-wide significance were characterized for their associations with clinical characteristics of gout. Case numbers were only sufficient for a comparison of affected joint localizations across genotypes. Because of the limited sample size, only the most important covariates, age, sex and log(eGFR), were included. Statistical significance for multiple testing (p < 3.1E-03) was obtained by dividing the significance threshold of 0.05 by the two tested SNPs multiplied with the eight evaluated locations of gout attacks. All candidate variant analyses were conducted using R version 3.3.0.
Electronic supplementary material
Acknowledgements
The work of UTS was supported by a scholarship from the Else Kroener Fresenius Foundation (NAKSYS). The work of J.J. was supported by the Chinese Scholarship Council. The work of AK was supported by the German Research Foundation (CRC 1140, 992 and KO 3598/3-1). This research was conducted with support from Astra Zeneca and Grünenthal. Genotyping was supported by Bayer Pharma Aktiengesellschaft (AG). The GCKD study is funded by grants from the German Ministry of Education and Research (BMBF, Grant Number 01ER0804) and the KfH Foundation for Preventive Medicine. We are grateful for the willingness of the patients to participate in the GCKD study. The enormous effort of the study personnel of the various regional centres is highly appreciated. We thank the large number of nephrologists who provide routine care for the patients and collaborate with the GCKD study. A list of nephrologists currently collaborating with the GCKD study is available at http://www.gckd.org. Current GCKD investigators are: University of Erlangen-Nürnberg: Kai-Uwe Eckardt, Heike Meiselbach, Markus Schneider, Thomas Dienemann, Hans-Ulrich Prokosch, Barbara Bärthlein, André Reis, Arif B. Ekici, Susanne Avendaño, Dinah Becker-Grosspitsch, Birgit Hausknecht, Rita Zitzmann, Anke Weigel, Andreas Beck, Thomas Ganslandt, Sabine Knispel. University of Freiburg: Gerd Walz, Anna Köttgen, Ulla Schultheiß, Fruzsina Kotsis, Simone Meder, Erna Mitsch, Ursula Reinhard. RWTH Aachen University: Jürgen Floege, Georg Schlieper, Turgay Saritas, Sabine Ernst, Astrid Blasius, Kerstin Schmitz. Charité, University Medicine Berlin: Elke Schaeffner, Seema Baid-Agrawal, Kerstin Theisen. Hannover Medical School: Hermann Haller, Jan Menne, Elisabeth Bahlmann. University of Heidelberg: Martin Zeier, Claudia Sommerer, Claudia Föllinger. University of Jena: Gunter Wolf, Martin Busch, Rainer Fuß. Ludwig-Maximilians University of München: Thomas Sitter, Claudia Blank. University of Würzburg: Christoph Wanner, Vera Krane, Karina Schönowsky, Antje Börner-Klein. Medical University of Innsbruck, Division of Genetic Epidemiology: Florian Kronenberg, Julia Raschenberger, Barbara Kollerits, Lukas Forer, Sebastian Schönherr, Hansi Weissensteiner. University of Regensburg, Institute of Functional Genomics: Peter Oefner, Wolfram Gronwald, Helena Zacharias; Department of Medical Biometry, Informatics and Epidemiology (IMBIE), University of Bonn: Matthias Schmid, Jennifer Nadal. The article processing charge was funded by the German Research Foundation (DFG) and the University of Freiburg in the funding programme Open Access Publishing.
Author Contributions
Authors involved in study conception are: J.J., K.-U.E., Y.L., A.K.; authors involved in the study design are: J.J., K.-U.E., Y.L., F.K., A.K., U.T.S.; authors involved in data acquisition are: A.B.E., T.S., E.S., K.-U.E., Y.L., F.K., A.K., U.T.S.; authors involved in data analysis are: J.J., Y.L., A.K.; authors involved in data interpretation are: K.-U.E., Y.L., F.K., A.K., U.T.S.; authors involved in giving critical intellectual input are: A.B.E., T.S., E.S., K.-U.E., Y.L., F.K., A.K., U.T.S.; authors involved in drafting the article are: J.J., A.K., U.T.S.; and authors involved in the critical revision of the article are: A.B.E., T.S., E.S., K.-U.E., Y.L., F.K., A.K., U.T.S.
Data availability
The data that supports the findings of this study is available for authors upon reasonable request and with permission of the Steering Committee of the GCKD study http://www.gckd.org.
Competing Interests
J.J., A.B.E., T.S., K.-U.E., E.S., Y.L., F.K., and U.T.S. declare no competing financial interests. A.K. received research support from Astra Zeneca, Aries and Grünenthal. E.S. declares as non-financial interest the membership of the extended board of the German Society of Nephrology (DGfN); J.J., A.B.E., T.S., K.-U.E., Y.L., F.K., A.K., U.T.S. declare no competing non-financial interests. The results presented in this paper have not been published previously in whole or part, except in abstract format.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-31282-z.
References
- 1.Lim, S. Y. et al. Trends in Gout and Rheumatoid Arthritis Hospitalizations in the United States, 1993–2011. JAMA315, 2345–2347, 10.1001/jama.2016.3517 (2016). [DOI] [PMC free article] [PubMed]
- 2.Kottgen, A. et al. Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat Genet45, 145–154, 10.1038/ng.2500 (2012). [DOI] [PMC free article] [PubMed]
- 3.Li, S. et al. The GLUT9 gene is associated with serum uric acid levels in Sardinia and Chianti cohorts. Plos Genet3, e194, https://doi.org/07-PLGE-RA-0574 (2007). [DOI] [PMC free article] [PubMed]
- 4.Doring, A. et al. SLC2A9 influences uric acid concentrations with pronounced sex-specific effects. Nat Genet40, 430–436, 10.1038/ng.107 (2008). [DOI] [PubMed]
- 5.Vitart, V. et al. SLC2A9 is a newly identified urate transporter influencing serum urate concentration, urate excretion and gout. Nat Genet40, 437–442, 10.1038/ng.106 (2008). [DOI] [PubMed]
- 6.Dehghan, A. et al. Association of three genetic loci with uric acid concentration and risk of gout: a genome-wide association study. Lancet372, 1953–1961, 10.1016/S0140-6736(08)61343-4 (2008). [DOI] [PMC free article] [PubMed]
- 7.Jing, J. et al. Prevalence and correlates of gout in a large cohort of patients with chronic kidney disease: the German Chronic Kidney Disease (GCKD) study. Nephrol Dial Transplant30, 613–621, 10.1093/ndt/gfu352 (2014). [DOI] [PubMed]
- 8.Eckardt, K. U. et al. Evolving importance of kidney disease: from subspecialty to global health burden. Lancet382, 158–169, 10.1016/S0140-6736(13)60439-0 (2013). [DOI] [PubMed]
- 9.Bhatnagar, V. et al. Analysis of ABCG2 and other urate transporters in uric acid homeostasis in chronic kidney disease: potential role of remote sensing and signaling. Clin Kidney J9, 444–453, 10.1093/ckj/sfw010 (2016). [DOI] [PMC free article] [PubMed]
- 10.Kuo, C. F., Grainge, M. J., Zhang, W. & Doherty, M. Global epidemiology of gout: prevalence, incidence and risk factors. Nat Rev Rheumatol11, 649–662, 10.1038/nrrheum.2015.91 (2015). [DOI] [PubMed]
- 11.Bobulescu, I. A. & Moe, O. W. Renal transport of uric acid: evolving concepts and uncertainties. Adv Chronic Kidney Dis19, 358–371, 10.1053/j.ackd.2012.07.009 (2012). [DOI] [PMC free article] [PubMed]
- 12.Kolz, M. et al. Meta-analysis of 28,141 individuals identifies common variants within five new loci that influence uric acid concentrations. Plos Genet5, e1000504, 10.1371/journal.pgen.1000504 (2009). [DOI] [PMC free article] [PubMed]
- 13.Tin, A. et al. Genome-wide association study for serum urate concentrations and gout among African Americans identifies genomic risk loci and a novel URAT1 loss-of-function allele. Hum Mol Genet20, 4056–4068, 10.1093/hmg/ddr307 (2011). [DOI] [PMC free article] [PubMed]
- 14.Hosomi, A., Nakanishi, T., Fujita, T. & Tamai, I. Extra-renal elimination of uric acid via intestinal efflux transporter BCRP/ABCG2. Plos One7, e30456, 10.1371/journal.pone.0030456 (2012). [DOI] [PMC free article] [PubMed]
- 15.Matsuo, H. et al. Common defects of ABCG2, a high-capacity urate exporter, cause gout: a function-based genetic analysis in a Japanese population. Sci Transl Med1, 5ra11, 10.1126/scitranslmed.3000237 (2009). [DOI] [PubMed]
- 16.Matsuo, H. et al. ABCG2 dysfunction increases the risk of renal overload hyperuricemia. Nucleosides Nucleotides Nucleic Acids33, 266–274, 10.1080/15257770.2013.866679 (2014). [DOI] [PubMed]
- 17.Hu, M. & Tomlinson, B. Gender-dependent associations of uric acid levels with a polymorphism in SLC2A9 in Han Chinese patients. Scand J Rheumatol41, 161–163, 10.3109/03009742.2011.637952 (2011). [DOI] [PubMed]
- 18.Nakayama, A. et al. ABCG2 is a high-capacity urate transporter and its genetic impairment increases serum uric acid levels in humans. Nucleosides Nucleotides Nucleic Acids30, 1091–1097, 10.1080/15257770.2011.633953 (2011). [DOI] [PubMed]
- 19.Titze, S. et al. Disease burden and risk profile in referred patients with moderate chronic kidney disease: composition of the German Chronic Kidney Disease (GCKD) cohort. Nephrol Dial Transplant30, 441–451, 10.1093/ndt/gfu294 (2014). [DOI] [PubMed]
- 20.Matsuo, H. et al. Hyperuricemia in acute gastroenteritis is caused by decreased urate excretion via ABCG2. Sci Rep6, 31003, 10.1038/srep31003 (2016). [DOI] [PMC free article] [PubMed]
- 21.Beck, H. et al. Heart failure in a cohort of patients with chronic kidney disease: the GCKD study. Plos One10, e0122552, 10.1371/journal.pone.0122552 (2015). [DOI] [PMC free article] [PubMed]
- 22.Bao, Y. et al. Lack of gene-diuretic interactions on the risk of incident gout: the Nurses’ Health Study and Health Professionals Follow-up Study. Ann Rheum Dis74, 1394–1398, 10.1136/annrheumdis-2014-206534 (2014). [DOI] [PMC free article] [PubMed]
- 23.Mitnala, S. et al. Clinical and genetic features of diuretic-associated gout: a case-control study. Rheumatology (Oxford) 55, 1172–1176, 10.1093/rheumatology/kew018 (2016). [DOI] [PubMed]
- 24.McAdams-DeMarco, M. A. et al. A urate gene-by-diuretic interaction and gout risk in participants with hypertension: results from the ARIC study. Ann Rheum Dis72, 701–706, 10.1136/annrheumdis-2011-201186 (2011). [DOI] [PMC free article] [PubMed]
- 25.Chandratre, P. et al. Health-related quality of life in gout: a systematic review. Rheumatology (Oxford) 52, 2031–2040, 10.1093/rheumatology/ket265 (2013). [DOI] [PMC free article] [PubMed]
- 26.Dalbeth, N. & Stamp, L. Hyperuricaemia and gout: time for a new staging system? Ann Rheum Dis73, 1598–1600, 10.1136/annrheumdis-2014-205304 (2014). [DOI] [PubMed]
- 27.Choi, H. K., Atkinson, K., Karlson, E. W., Willett, W. & Curhan, G. Purine-rich foods, dairy and protein intake, and the risk of gout in men. N Engl J Med350, 1093–1103, 10.1056/NEJMoa035700 (2004). [DOI] [PubMed]
- 28.Choi, H. K., Atkinson, K., Karlson, E. W., Willett, W. & Curhan, G. Alcohol intake and risk of incident gout in men: a prospective study. Lancet363, 1277–1281, 10.1016/S0140-6736(04)16000-5 (2004). [DOI] [PubMed]
- 29.Choi, H. K. & Curhan, G. Coffee consumption and risk of incident gout in women: the Nurses’ Health Study. Am J Clin Nutr92, 922–927, 10.3945/ajcn.2010.29565 (2010). [DOI] [PMC free article] [PubMed]
- 30.Johnson, R. J. et al. Uric acid and chronic kidney disease: which is chasing which? Nephrol Dial Transplant28, 2221–2228, 10.1093/ndt/gft029 (2013). [DOI] [PMC free article] [PubMed]
- 31.Miyaoka, T., Mochizuki, T., Takei, T., Tsuchiya, K. & Nitta, K. Serum uric acid levels and long-term outcomes in chronic kidney disease. Heart Vessels29, 504–512, 10.1007/s00380-013-0396-0 (2013). [DOI] [PubMed]
- 32.Rodenbach, K. E. et al. Hyperuricemia and Progression of CKD in Children and Adolescents: The Chronic Kidney Disease in Children (CKiD) Cohort Study. Am J Kidney Dis66, 984–992, 10.1053/j.ajkd.2015.06.015 (2015). [DOI] [PMC free article] [PubMed]
- 33.Feig, D. I., Kang, D. H. & Johnson, R. J. Uric acid and cardiovascular risk. N Engl J Med359, 1811–1821, 10.1056/NEJMra0800885 (2008). [DOI] [PMC free article] [PubMed]
- 34.Choi, H. K. & Curhan, G. Independent impact of gout on mortality and risk for coronary heart disease. Circulation116, 894–900, https://doi.org/CIRCULATIONAHA.107.703389 (2007). [DOI] [PubMed]
- 35.McAdams, M. A. et al. Reliability and sensitivity of the self-report of physician-diagnosed gout in the campaign against cancer and heart disease and the atherosclerosis risk in the community cohorts. J Rheumatol38, 135–141, 10.3899/jrheum.100418 (2011). [DOI] [PMC free article] [PubMed]
- 36.Dienemann, T. et al. International Network of Chronic Kidney Disease cohort studies (iNET-CKD): a global network of chronic kidney disease cohorts. BMC Nephrol17, 121, 10.1186/s12882-016-0335-2 (2016). [DOI] [PMC free article] [PubMed]
- 37.Eckardt, K. U. et al. The German Chronic Kidney Disease (GCKD) study: design and methods. Nephrol Dial Transplant27, 1454–1460, 10.1093/ndt/gfr456 (2011). [DOI] [PubMed]
- 38.Anderson, C. A. et al. Data quality control in genetic case-control association studies. Nat Protoc5, 1564–1573, 10.1038/nprot.2010.116 (2010). [DOI] [PMC free article] [PubMed]
- 39.Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet81, 559–575, https://doi.org/S0002-9297(07)61352-4 (2007). [DOI] [PMC free article] [PubMed]
- 40.Delaneau, O., Zagury, J. F. & Marchini, J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods10, 5–6, 10.1038/nmeth.2307 (2013). [DOI] [PubMed]
- 41.Howie, B., Fuchsberger, C., Stephens, M., Marchini, J. & Abecasis, G. R. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet44, 955–959, 10.1038/ng.2354 (2012). [DOI] [PMC free article] [PubMed]
- 42.Marchini, J. & Howie, B. Genotype imputation for genome-wide association studies. Nat Rev Genet11, 499–511, 10.1038/nrg2796 (2010). [DOI] [PubMed]
- 43.Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res38, e164, 10.1093/nar/gkq603 (2010). [DOI] [PMC free article] [PubMed]
- 44.KDIGO clinical practice guideline for the evaluation and management of chronic kidney disease. Kidney Int Suppl3, 1–150, 10.1038/kisup.2012.77 (2012).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that supports the findings of this study is available for authors upon reasonable request and with permission of the Steering Committee of the GCKD study http://www.gckd.org.