Dear Editor,
N6-methyladenosine (m6A) is the prominent dynamic mRNA modification, governed by methyltransferase complex (“writers”), demethylases (“erasers”) and RNA-binding proteins (‘readers’).1 m6A modification directs mRNAs to distinct fates by grouping them for differential processing, translation and decay in the processes such as cell differentiation, embryonic development and stress responses.1,2 Owing to a deeper understanding of this modification and the technological advance, functional characterizations of m6A in gene regulation have become a hot topic that warrants further dissection.3,4
Autophagy is a fundamental cellular process in eukaryotic cells, which sequesters cytoplasmic material within double-membraned autophagosomes and leads to degradation and recycling of the engulfed substrates.5,6 Unc-51-like kinase 1 (ULK1) serves as a protein kinase that is activated upon autophagy stimulation and is critical for the recruitment of other autophagy-related proteins to the autophagosome formation site.5 To date, the regulation of autophagy initiation mainly focused on the post-translational modification (PTM) and transactivation of autophagy-related genes.5,6 Whether m6A modification could regulate the mRNA processing of autophagy-related genes and affect autophagy remains unknown.
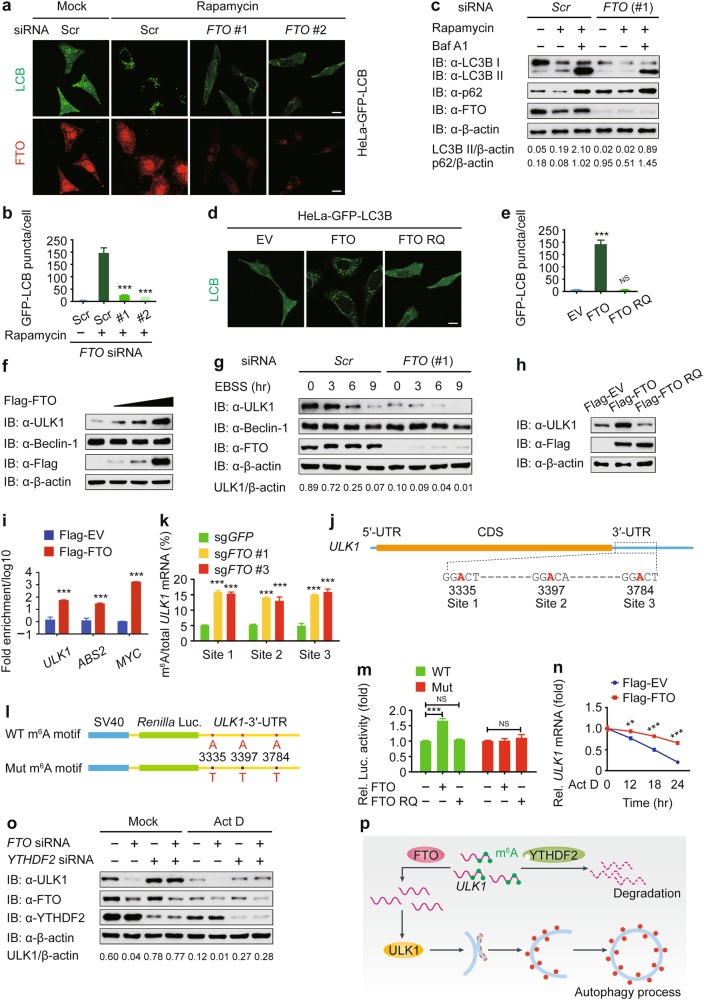
To investigate the role of m6A modification in autophagy, we performed a screen by using small interfering RNAs (siRNAs) targeting genes encoding the “writers”, “erasers”, and “readers” and identified fat mass and obesity associated (FTO) protein as a positive regulator of autophagy (Supplementary information, Figure S1a). We first confirmed the knockdown efficiency of FTO-specific siRNAs (Supplementary information, Figure S1b). We then examined the puncta formation of autophagy marker light chain 3B (LC3B). Under both basal and rapamycin-induced autophagy conditions, depletion of FTO significantly reduced the formation of GFP-LC3B puncta (Fig. 1a, b). We further monitored the autophagic flux by detecting LC3B in the presence of bafilomycin A1 (Baf A1), a H+-ATPase inhibitor that can block the autophagosome turnover. We observed that knockdown of FTO suppressed the accumulation of LC3B II. Consistently, the level of p62/SQSTM1 (an autophagy substrate) was higher in FTO-knockdown cells than that in control cells (Fig. 1c).
Fig. 1.
FTO-mediated demethylation of ULK1 transcripts promotes autophagy flux by upregulating ULK1 protein abundance. a Representative images of GFP-LC3B puncta in HeLa-GFP-LC3B cells transfected with scrambled siRNA or FTO-specific siRNA after rapamycin (250 nM) treatment for 18 h. Scale bar, 20 μm. b Quantification of GFP-LC3B puncta in HeLa-GFP-LC3B cells transfected with scrambled siRNA or FTO-specific siRNA. Bars represent mean ± SEM of triplicate samples (20 cells per sample). ***P < 0.001. c HEK293T cells transfected with control or FTO-specific siRNA were treated with rapamycin (250 nM) for 18 h. The lysates were subjected to immunoblot analysis with the indicated antibodies. d HeLa cells stably expressing GFP-LC3B were transfected with pcDNA3.1 empty vector or plasmids expressing wild-type or the mutant form of FTO (RQ, short for R316Q). Scale bar, 20 μm. e Average GFP-LC3B puncta per cell were calculated. Bars represent mean ± SEM of triplicate samples (20 cells per sample). ***P < 0.001; NS, not significant. f Immunoblot analysis of extracts of 293T cells transfected with the increasing doses of Flag-FTO expression vector (wedge). g 293T cells were transfected with control or FTO-specific siRNA, followed by EBSS treatment for the indicated times. The protein expression levels of Beclin-1 and ULK1 were detected by immunoblot. h 293T cells were transfected with plasmids encoding Flag-FTO or Flag-FTO RQ, and the cellular extracts were analyzed by immunoblot. i Confirmation of FTO binding to ULK1 mRNA by RNA immunoprecipitation. ABS2 and MYC were used as the positive control. Data are normalized to negative control that is set to a value of 1. The relative Flag-FTO/Flag-EV ratios (log10) are plotted. Data are shown as means ± SEM of three independent experiments. ***P < 0.001. j Schematic representation of the position of m6A motifs within ULK1 transcript. k Abundance of ULK1 transcripts among mRNA immunoprecipitated with anti-m6A antibody from sgGFP-expressing and sgFTO-expressing cells. Data are expressed as means ± SEM of three independent experiments. ***P < 0.001. l Wild-type or m6A consensus sequence mutant (A-to-T mutation) ULK1-3′-UTR was fused with Renilla luciferase reporter. m Relative luciferase activity of ULK1-3′-UTR with wild-type or mutated m6A sites after co-transfection with empty vector, FTO, or FTO RQ into 293T cell. Renilla luciferase activity was measured and normalized to firefly luciferase activity. n 293T cells transfected with plasmids encoding Flag-FTO were treated with Act D (5 μg/mL) for the indicated times. The expression of ULK1 was analyzed by real-time PCR. Data are expressed as means ± SEM of three independent experiments. **P < 0.01 and ***P < 0.001. o 293T cells were co-transfected with control or FTO-specific siRNA and YTHDF2-specific siRNA, followed by Act D (5 μg/mL) treatment for 18 h. The lysates were subjected to immunoblot analysis with the indicated antibodies. p A proposed working model to illustrate how FTO mediates the m6A modification of ULK1 transcripts and positively regulates autophagy
To further confirm the role of FTO in autophagy, we ectopically expressed Flag-tagged FTO and found that overexpression of FTO remarkably enhanced LC3B II accumulation and p62 consumption (Supplementary information, Figures S1c and d). Since FTO is a demethylase, we next generated catalytically inactive FTO mutant RQ (R316Q) and observed that cells expressing wild-type (WT) FTO, rather than FTO RQ mutant, displayed an increased number of GFP-LC3B puncta (Fig. 1d, e). Together, our findings reveal that FTO positively regulates autophagy in an enzyme activity-dependent manner.
To reveal the molecular mechanisms underpinning promotion of autophagy by FTO, we overexpressed FTO and observed that ULK1 protein level positively correlated with FTO protein level (Fig. 1f). In addition, FTO-mediated ULK1 stabilization could be potentiated by treatment of Earle’s balanced salt solution (EBSS; an autophagy agonist) (Fig. 1g and Supplementary information, Figure S1e). Moreover, we found that FTO RQ mutant could not stabilize ULK1 (Fig. 1h). These results suggest that FTO-mediated ULK1 stabilization depends on the demethylase activity of FTO. To determine whether the ULK1 transcripts are bound by FTO, we performed anti-Flag-based RNA immunoprecipitation (IP) coupled to real-time PCR (RIP-PCR) with gene-specific primers. We first verified the efficiency of this immunoprecipitation assay by detecting the expression of Flag-tagged FTO (Supplementary information, Figure S1f). The IP samples from Flag-FTO cells revealed a strong signal for ULK1 mRNA (Fig. 1i), while ASB2 and MYC were chosen as the positive control.7 Collectively, these results indicate that FTO targets the ULK1 transcripts and positively regulates the abundance of ULK1 protein.
As FTO is an m6A “eraser” of RNA methylation, we next investigated the m6A modification of ULK1 transcripts. N6-methyladenosine-sequencing (m6A-seq) is an immunocapturing approach for the unbiased transcriptome-wide localization of m6A in high resolution.8,9 To assess the distribution of m6A within ULK1 transcripts, we performed m6A-seq and found three distributions of m6A in the 3′-untranslated region (3′-UTR) of ULK1 transcript (Fig. 1j). To elucidate the function of FTO in the m6A modification of ULK1 mRNA, we generated an FTO-low-expressing (sgFTO) HEK293T cell line by using the CRISPR/Cas9 genome-editing tool (Supplementary information, Figure S1g). We immunoprecipitated the total mRNA with anti-m6A antibody and found that mRNA isolated from sgFTO cells showed a much higher enrichment of anti-m6A-bound EEF1A1 gene than that from control (sgGFP) cells (Supplementary information, Figure S1h), indicating that EEF1A1 gene can be m6A modified as described by the instructions of Magna MeRIPTM m6A Kit (Merck Millipore, Cat# 17-10499) and FTO could demethylate it. We further used the same samples for real-time PCR analysis to detect the potential ULK1 m6A-bound fraction using gene-specific primers and observed that the anti-m6A-bound fraction showed greater enrichment for ULK1 at three sites in sgFTO cells (Fig. 1k). Furthermore, we performed luciferase reporter assay and found that WT FTO could not promote the luciferase activity of the reporter construct bearing ULK1-3′-UTR with the mutations at these three m6A sites (Fig. 1l, m). Together, these results confirm that the m6A sites on ULK1 transcripts are the direct substrates of FTO-catalyzed demethylation.
Increasing evidence suggests that m6A modification affects many aspects of gene expression, including nuclear export, mRNA decay, alternative pre-mRNA splicing, 3′-end processing and translation.4 We investigated the effect of FTO on the intracellular distribution of the ULK1 transcripts and found that FTO did not affect the translocation of ULK1 mRNA from the nucleus to the cytoplasm (Supplementary information, Figures S1i and j). We then investigated whether FTO-mediated demethylation is implicated in ULK1 mRNA decay. We observed that the decay rate of ULK1 mRNA was much slower in FTO-overexpressing cells, compared to that in control cells, when the transcription was halted with actinomycin D (Act D; Fig. 1n). Consistently, the protein level of ULK1 was also higher in FTO-overexpressing cells than in control cells (Supplementary information, Figure S1k). Furthermore, we found that the decay of ULK1 transcripts was enhanced in 293T cells transfected with FTO-specific siRNA than that with scramble siRNA (Supplementary information, Figure S1l). As YTHDF2 is the major m6A reader for the decay of m6A-modified mRNA, we next determined whether YTHDF2 targets cellular m6A-bound ULK1 transcripts and found that the reduction of ULK1 protein by FTO impairment was almost abolished in YTHDF2-knockdown cells (Fig. 1o and Supplementary information, Figure S1m). Together, our data suggest that the m6A changes caused by FTO influence the stability of ULK1 transcripts, likely through a YTHDF2-dependent manner.
Our results sheds light on how FTO specifically upregulates the ULK1 protein abundance, thereby promoting the initiation of autophagy. ULK1 mRNA undergoes m6A modification in the 3′-UTR and the m6A-marked ULK1 transcripts can further be targeted for degradation by YTHDF2. FTO reverses the m6A mRNA modification of ULK1 transcripts, thus prolonging the half-life time of ULK1 transcripts (Fig. 1p). Actually, since a great number of genes are involved in autophagy, we cannot exclude the possibility that m6A modification regulates autophagy by targeting other autophagic genes. As previous studies indicated that FTO functioned as an oncogenic m6A demethylase in leukemia and brain tumor,7,10 considering the importance of autophagy in tumorigenesis, we should not ignore the oncogenic roles of FTO via autophagy. Our study for the first time reveals a crosstalk of mRNA m6A modification and autophagy, which broadens the multilayer regulation of autophagy and expands our understanding of such interplays that are essential for therapeutic application.
Electronic supplementary material
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31700760, 31522018, 91629101, 91753137, 31771462, 31471252, 31500813), the National Key Basic Research Program of China (2015CB859800, 2014CB910800), Science and Technology Planning Project of Guangzhou, China (201804010385, 201604020003, 201604046001), and China Postdoctoral Science Foundation (2017M622862).
Competing interests
The authors declare no competing interests.
Contributor Information
Jian Ren, Email: renjian.sysu@gmail.com.
Jun Cui, Email: cuij5@mail.sysu.edu.cn.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41422-018-0069-8.
References
- 1.Roignant JY, Soller M. Trends Genet. 2017;33:380–390. doi: 10.1016/j.tig.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 2.Gilbert WV, Bell TA, Schaening C. Science. 2016;352:1408–1412. doi: 10.1126/science.aad8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jia GF, et al. Nat. Chem. Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roundtree IA, et al. Cell. 2017;169:1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morel E, et al. Annu. Rev. Pharmacol. Toxicol. 2017;57:375–398. doi: 10.1146/annurev-pharmtox-010716-104936. [DOI] [PubMed] [Google Scholar]
- 6.Jin SH, et al. EMBO J. 2016;35:866–880. doi: 10.15252/embj.201593596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Su R, et al. Cell. 2018;172:90–105. doi: 10.1016/j.cell.2017.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dominissini D, et al. Nature. 2012;485:201–284. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 9.Dominissini D, et al. Nat. Protoc. 2013;8:176–189. doi: 10.1038/nprot.2012.148. [DOI] [PubMed] [Google Scholar]
- 10.Cui Q, et al. Cell Rep. 2017;18:2622–2634. doi: 10.1016/j.celrep.2017.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.