The functional importance of N 6 -methyladenosine (m 6 A) modification in human health and disease has recently entered the research spotlight. In a recent paper published in Cell Research , Li et al. report a remarkable finding that deficiency in m 6 A reader Ythdf2 results in a striking expansion of mouse and human hematopoietic stem cells.
RNA is made up of four nucleotides: A, C, G and U. Information on RNA molecules relies on their core sequence, and also their chemical modifications. RNA chemical modifications play important roles in various steps of RNA processing, degradation, translation, and thus in the regulation of gene expression. Amongst the more than 100 different chemical modifications on RNA identified, N6-methyladenosine (m6A) is the most abundant epigenetic mark on eukaryotic messenger RNAs (mRNAs). The m6A modification has been implicated in regulating a range of biological processes, and dysregulation of m6A modification has been associated with many human diseases including cancer and obesity.1
Recent research has identified enzymes specifically involved in adjusting m6A methylation levels and their functional outcomes; this has been considered another layer of epigenetic control similar to DNA and histone modifications, and has increased our knowledge of epitranscriptomics.2 The m6A methyltransferase complex, or the “writer”, consists of Methyl transferase-like 3 and 14 (METTL3 and METTL14) with the cofactor Wilms tumor 1 associated protein (WTAP). METTL3 and METTL14 play critical roles in both normal hematopoiesis and leukemogenesis.3 On the other hand, fat mass and obesity associated protein (FTO) and ALKBH5 act as demethylases to erase m6A modifications, and thus maintain a dynamic balance of m6A levels. The function of FTO has been associated with body fat homeostasis, and recently with oncogenesis of acute myeloid leukemia.3
The m6A modification is recognized by its readers, YTH domain family proteins, amongst which YTHDF2 destabilizes m6A-modified mRNAs after binding within the consensus sequences.2 A recent study reports that YTHDF2 deficiency results in female infertility in mice, and YTHDF2 is maternally required for oocyte maturation.4 However, the physiological role of YTHDF2 in hematopoietic stem cell (HSC) biology was still unknown. In the present study, Li et al. observe a significant increase in numbers of HSCs in Ythdf2 conditional knockout mice compared to wild-type littermates.5 The Ythdf2-knockout mice showed no difference in numbers of hematopoietic progenitors and mature lineage cells, suggesting that Ythdf2 deficiency in hematopoietic lineages specifically increases HSC numbers. Limiting dilution in vivo engraftment assay revealed a 2.2-fold increase in competitive repopulating units (a measure of the numbers of functionally active HSCs) in Ythdf2-knockout, compared with control, bone marrow cells. Importantly, Ythdf2-knockout HSCs manifested normal lineage differentiation, with no apparent signs of hematological malignancies.
Another interesting observation is that the m6A modifications are enriched in mRNAs of several transcriptional factors that are crucial for self-renewal of HSCs, such as Gata2, Tal1, Etv6 and Stat55. Ythdf2 deficiency resulted in extended stability of these m6A-marked mRNAs and elevated expression of these transcriptional factors (Fig. 1). Decapping of mRNA is an important step in mRNA decay, and Dcp1a encodes an mRNA decapping enzyme. Using fluorescence in situ hybridization, Li et al. were able to demonstrate co-localization of Tal1 mRNA, Dcp1a and Ythdf2 in wild-type cells but not in Ythdf2-knockout cells. They also showed that knockdown of Tal1 suppressed the increased engraftment of Ythdf2-knockout HSCs, supporting their conclusion that upregulation of HSC self-renewal factors accounted for the HSC expansion phenotype in Ythdf2-knockout mice.5
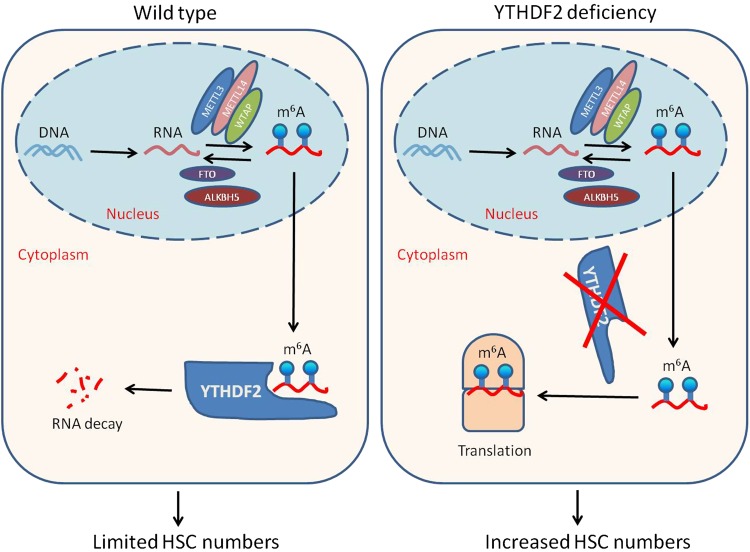
Fig. 1.
In wild-type cells, m6A reader Ythdf2 recognizes m6A-modified RNAs and thus targets these RNAs for degradation (left). In the present study, Li et al. demonstrate that Ythdf2 deficiency results in increased stability of m6A-modified RNAs, leading to elevated expression of HSC self-renewal factors and increased HSC numbers (right)
Human cord blood (CB) is an alternative source of HSCs for transplantation in clinical applications.6,7 However, limited numbers of HSCs in single CB collections have hampered wider use of CB in patients with malignant and nonmalignant disorders. While progress has been made to expand CB HSC numbers ex vivo,7 additional means are still needed to meet clinical demands. In an attempt to reveal whether the function of m6A and Ythdf2 in HSCs would be conserved in human cells, Li et al. first performed m6A RNA-seq in human CB CD34+ cells and found that many transcriptional factors involved in HSC self-renewal were marked by m6A modifications. They were then able to show that knockdown of YTHDF2 resulted in increased HSC numbers with long-term engraftment capability in vivo in immune-deficient mice. Thus, the exciting work by Li et al. adds to our understanding of m6A and YTHDF2 function in HSC expansion in both mouse and human (Fig. 1).
Recent progress has shed new light on enhancing CB transplantation efficacy by several approaches: collecting and processing HSCs at physiological low oxygen levels to prevent their differentiation and increase numbers;8 modifying membrane expression of a homing receptor, CXCR4;9 modulating epigenetic regulations to promote the HSC homing process;10,11 and by manipulating the cellular metabolism of HSCs to ex vivo expand them.12 Thus, it would be interesting to see whether the effects will be even greater by combining several different approaches for both enhanced HSC homing and ex vivo expansion with YTHDF2 inhibition.
To conclude, Li et al. present important information towards further understanding the physiological impact of m6A modification and HSC biology. Looking to the future, the new findings that HSC expansion can be achieved by Ythdf2 suppression open up a new and potent direction to simultaneously target multiple key transcriptional factors for HSC self-renewal, and to engineer HSCs for therapeutic advantage, effects that may be relevant beyond the enhancement of hematopoiesis. Identifying specific small-molecule inhibitors for Ythdf2 may provide a promising way to enhance HSC transplantation, especially with CB cells, and to also benefit HSC-based gene therapy.
References
- 1.Fu Y, et al. Nat. Rev. Genet. 2014;15:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- 2.Roundtree IA, et al. Cell. 2017;169:1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deng X, et al. Cell Res. 2018;28:507–517. doi: 10.1038/s41422-018-0034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ivanova I, et al. Mol. Cell. 2017;67:1059–1067. doi: 10.1016/j.molcel.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Zhenrui, Qian Pengxu, Shao Wanqing, Shi Hailing, He Xi C., Gogol Madelaine, Yu Zulin, Wang Yongfu, Qi Meijie, Zhu Yunfei, Perry John M., Zhang Kai, Tao Fang, Zhou Kun, Hu Deqing, Han Yingli, Zhao Chongbei, Alexander Richard, Xu Hanzhang, Chen Shiyuan, Peak Allison, Hall Kathyrn, Peterson Michael, Perera Anoja, Haug Jeffrey S., Parmely Tari, Li Hua, Shen Bin, Zeitlinger Julia, He Chuan, Li Linheng. Suppression of m6A reader Ythdf2 promotes hematopoietic stem cell expansion. Cell Research. 2018;28(9):904–917. doi: 10.1038/s41422-018-0072-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ballen KK, Gluckman E, Broxmeyer HE. Blood. 2013;122:491–498. doi: 10.1182/blood-2013-02-453175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broxmeyer HE. Transfus. Apher. Sci. 2016;54:364–372. doi: 10.1016/j.transci.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mantel CR, et al. Cell. 2015;161:1553–1565. doi: 10.1016/j.cell.2015.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Capitano ML, et al. Stem Cells. 2015;33:1975–1984. doi: 10.1002/stem.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang X, et al. Nat. Commun. 2018;9:2741. doi: 10.1038/s41467-018-05178-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo B, et al. Nat. Med. 2017;23:424–428. doi: 10.1038/nm.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo B, et al. Nat. Med. 2018;24:360–367. doi: 10.1038/nm.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]