Dear Editor,
γ-aminobutyric acid type A (GABAA) receptors mediate rapid inhibitory neurotransmission by opening a chloride selective pore in response to binding of γ-aminobutyric acid (GABA), and thus are vital for controlling excitability in the brain.1 Dysfunctional GABAA receptors are directly involved in the pathogenesis of many neurologic diseases and psychiatric disorders.2 Moreover, GABAA receptors are modulated, directly activated or inhibited by over hundreds of pharmacologically and clinically important compounds of different structural classes.3 As members of Cys loop-type ligand-gated ion channel superfamily that also includes nicotinic acetylcholine receptors, glycine receptors and serotonin type 3 receptor, human GABAA receptors are typically heteropentamers assembled from a repertoire of 19 different subunits (α, β, γ, δ, ε, θ, π, and ρ subunits), giving rise to a spectrum of GABAA receptor subtypes with different subunit compositions and arrangements, as well as distinct biophysical and pharmacological properties.1,4 Although the subunit stoichiometries and arrangements of functional GABAA receptor subtypes have been intensively investigated in the last two decades, the assembling principles of these receptor subtypes remain unknown.4 The unique property that keeps GABAA receptors apart from other members of the Cys-loop superfamily is the activating ligand GABA. Early studies with reconstituted recombinant receptors have revealed that robust GABA-activated channel formation occurred with combinations of α and β subunits.5 All Cys-loop receptors share a similar neurotransmitter binding pocket formed at the extracellular interface between two adjacent subunits by three loops from the principle (+) and three loops/strands from the complementary (−) subunits; and the pocket at the extracellular β(+)/α(−) interface is a GABA-binding site.3 However, how GABA selectively binds at the interface, and how the binding signal is transmitted quickly and efficiently to open an integral ion channel remains elusive, significantly limiting our understanding of the ligand-gating mechanism of the GABAA receptors.
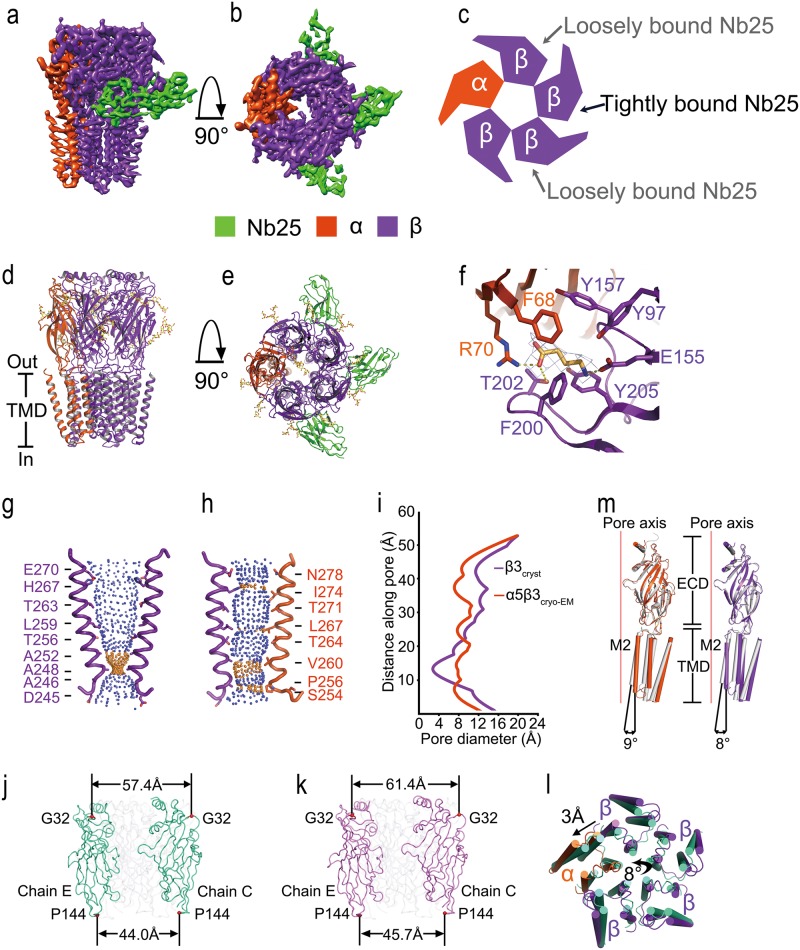
To elucidate the assembly principle and the ligand-gating mechanism of GABAA receptors containing both α and β subunits, we overexpressed and purified human α5β3 GABAA receptor from HEK293S-GnTI− cells. To eliminate the possibility that contaminated proteins interact with GABAA receptor through an intracellular loop between transmembrane helices 3 and 4 with very high affinity, we substituted this loop by a short linker. Similar approach has previously been applied to GABAA receptors6–11 and other Cys-loop receptor members. The α5 and β3 subunits share ∼35% amino acid sequence identity and likely adopt near identical backbone conformations, which makes the two different subunits indistinguishable at low resolution, and thus challenges the cryo-EM analysis, especially at particle alignment stage. We took advantage of a nanobody (Nb25) that specifically binds to the β3 extracellular domain (ECD),6 to facilitate the 2D and 3D classifications and to distinguish the two different subunits. The GABAA receptor-Nb25 complex was co-purified in the presence of 1 mM GABA and was used to prepare sample grids for cryo-EM. By combining 161,455 particle images from 3724 electron-counting movies recorded in a TF20 and a Titan Krios cryo-electron microscope, we obtained a cryo-EM structure of 3.51 Å by single-particle analysis (Supplementary Information, Data S1, Figure S5, S6). The cryo-EM density map reveals that five well-resolved subunits form a cylinder-shaped central ion channel in a pseudo-symmetrical arrangement, and three Nb25s bind between adjacent ECD interfaces of two subunits (Fig. 1a–e). Each subunit contains a large N-terminal ECD surrounding an extracellular vestibule, and a C-terminal four-helix bundle transmembrane domain (TMD) forming a funnel-shaped transmembrane channel (Fig. 1d, e). Two side Nb25s bind loosely, as judged by the slightly lower densities (Fig. 1b, c).
Fig. 1.
Cryo-EM structure of the human α5β3 GABAA receptor. a, b Surface views parallel to the plasma membrane (a) or from the extracellular space down the five-fold pseudo-symmetry axis (b) of the cryo-EM density map of the human α5β3 GABAA receptor in complex with Nb25 reveals a distinct assembly of one α (red) and four β subunits (purple), and three bound Nb25s (green). c Schematic top-down view of the α5β3 GABAA receptor indicating two different interfaces bound by Nb25s. d, e α5β3 GABAA receptor viewed parallel to the plasma membrane (d) or from the extracellular space down the five-fold pseudo-symmetry axis with three Nb25s bound (e). N-linked glycans are shown in ball-and stick representation. f GABA-binding site at β( + )/α(−) interface with density at 3σ contour level. Dashed links indicate salt bridges or hydrogen bond. The residues in β( + ), α(−) and GABA are depicted in sticks. g, h Solvent contours of the human β3 GABAA receptor pore (PDB ID: 4COF) (g) or of the human α5β3 GABAA receptor pore (this study) (h) showing the M2 helices of subunits and the side chains of pore-lining residues, labeled according to protein sequence. Small blue spheres define a radius of >4.0 Å and yellow spheres represent a radius of 1.6–4.0 Å. i Illustration of the pore radii as a function of distance along the pore axis of desensitized (purple) and open (red) GABAA receptor pores. Pore radii in panels E-G were calculated using the computer program Hole. j, k The extracellular domains of homopentameric β3 GABAA receptor in the desensitized state (j) and that of heteropentameric α5β3 GABAA receptor in the open state (k) showing that the extracellular domains ‘open up’ in a way akin to petals opening on a flower. The distances between Gly 32 and that between Pro 144 in two opposing subunits are shown. l Superposition of the transmembrane domains of homopentameric β3 GABAA receptor in the desensitized state (green) and that of heteropentameric α5β3 GABAA receptor in the open state (α subunit in red color and β subunits in purple color) showing that they undergo an anticlockwise rotation of ∼8° relative to the pore axis, with the tilting of the M2 helix ∼3 Å away from the five-fold axis. m Superpositions of the extracellular domains showing conformational changes within GABAA subunits. Left, α5 subunit in the open state (red) to β3 subunit in the desensitized state (grey). Right, β3 subunit in the open (purple) to desensitized (grey) state
The structure of the α5β3 GABAA receptor reveals an unexpected subunit stoichiometry of one α and four β subunits. In theory, a huge number of GABAA receptor subtypes may be assembled from a repertoire of 19 different subunits.1 However, very few combinations have been conclusively identified. A GABAA receptor containing α, β and γ subunits seems to have defined subunit stoichiometry and arrangement,12 which has been directly visualized recently.9–11 Of note, accumulating evidence, especially that from single channel electrophysiological recordings,13 supports the existence of αβ GABAA receptors at extrasynaptic locations. A pentameric arrangement containing two α and three β subunits,12 or three α and two β subunits,14 have been proposed. However, the observation of three Nb25s binding to a α5β3 GABAA receptor suggests an existence of four continuous β3 subunits, since Nb25 has been previously observed to bind the ECDs between two adjacent β subunits.7 The quality of the density map is excellent, allowing for visualization of many medium and large side chains, as well as glycosylation of Asn side chains. We observed clear densities corresponding to eight N-linked glycans located on the external surface of the cylinder on four subunits at two sites (Asn80 and Asn149) (Fig. 1d, e, Supplementary Information, Figure S1), characteristic of β3 subunit as previously reported7 (Supplementary Information, Figure S2), indicating the existence of only one α5 subunit in the α5β3 assembly. The α5 subunit identity was further substantiated by clear densities of two N-linked glycans, one of which located in the extracellular vestibule at Asn114 (Fig. 1d, e, Supplementary Information, Figure S3), whereas the other one located on the external surface of the cylinder at Asn205 (Fig. 1d, e, Supplementary Information, Figure S4), two potential glycosylation sites based on sequence analysis.15 Previous findings suggest that a glycosylation site within the extracellular vestibule blocks the formation of pentamers with more than two α subunits,10 thus excluding the possibility that the GABAA receptor contains three α and two β subunits.14
To understand the molecular recognition of GABA, the α5β3 GABAA receptor structure was determined in the presence of saturating GABA. Among the canonical neurotransmitter-binding site, we observed significant density for GABA at the β(+)/α(−) interface of the α5β3 GABAA receptor (Fig. 1f). Three loops from each side of the interface contribute to GABA binding, A, B and C from the (+) side, and D, E and F from the (−) side (Supplementary Information, Figure S2). Aromatic residues from loops A–E, including Tyr 97, Tyr 157, Phe 200, and Tyr 205 from the β subunit, Phe 68 from the α subunit, form a tightly packed aromatic cage surrounding a GABA molecule wedging between the β(+)/α(−) interface with its amino group interacting with the β(+) subunit and its carboxyl tail mainly with the α(−) subunit. The amino group of GABA forms cation–π interaction with Tyr205, and a salt bridge with the carboxylate group of Glu155 in β-strand 7. Whereas the carboxylate group of GABA forms a salt bridge with the guanidinium moiety of Arg 70, and a hydrogen bond with the hydroxy group of Thr 202. It is worth noting that, we did not observe similar density at the α(+)/β(−) interface in α5β3 GABAA receptor structure, as compared with α1β1γ2 GABAA receptor structure.10
The structure of the heteropentameric α5β3 GABAA receptor is clearly distinct to that of the benzamidine-bound, homopentameric β3 GABAA receptor in a desensitized state.7 In the β3 receptor, the pore-lining M2 helices become gradually narrower at the intracellular end with the narrowest region at Ala 248 (Ala -2′ on the M2, pore-lining helix), restricting the pore to ∼3.15 Å in diameter, too small for the conduction of chloride ions (Fig. 1g). Whereas in the GABA-bound α5β3 receptor, the M2 helices are nearly parallel to the pore axis with four narrow regions at Ile 274, Leu 267, Val 260 and Pro 256 of α5, and His 267, Leu 259, Ala 252 and Ala 248 of β3 subunits (Fig. 1h). The pore is most constricted at Pro 256 of α5, and Ala 248 of β3 subunits, yielding a pore of ∼7 Å in diameter (Fig. 1h, i). Given that chloride has a Pauling radius of 1.8 Å, and the Cl···H-O (water) hydrogen bond can be as short as ∼2.5 Å, Cl− can snugly pass through the pore with one layer of bound water molecules. It is then reasonable to hypothesize that the α5β3 GABAA receptor structure represents an open state.
The two GABAA receptor structures, in the desensitized and open states, differ in two main respects. First, the extracellular domains ‘open up’ in a way akin to petals opening on a flower, i.e., the upper part of the extracellular domains expands during the transition from the desensitized to the open state. In the desensitized state the extracellular domains are separated by ∼57.4 Å as measured at Gly 32 in two opposing subunits (Fig. 1j). This distance expands to ∼ 61.4 Å in the open state (Fig. 1k). By contrast, the bottom part of the extracellular domains only slightly expands. The distance between Pro 144 in two opposing subunits is ∼44.0 Å in the desensitized state (Fig. 1j), and is ∼45.7 Å in the open state (Fig. 1k). Second, in the open state, the transmembrane domains undergo an anticlockwise rotation of ∼8° relative to the pore axis, ‘splaying open’ with the tilting of the M2 helix ∼3 Å away from the five-fold axis and opening of the pore (Fig. 1l).
Superpositions of individual subunits from the desensitized and open states reveal that the extracellular and transmembrane domains undergo mainly rigid body movements relative to one another (Fig. 1m). Superimposing the ECD from the same β3 subunit in the two states give rise to a rotated transmembrane domain. Transition from the desensitized to the open state therefore involves tilting of the pore lining M2 helix ∼8° away from the ion channel (Fig. 1m). Superimposing the ECD from an α5 subunit in open state to a β3 subunit in the other state shows a slightly larger rotation with tilting of the pore lining M2 helix ∼9° away from the ion channel (Fig. 1m).
In summary, we have used single particle cryo-EM coupled with nanobody to determine the structure of a heteropentameric α5β3 GABAA receptor. The structure shows an unexpected subunit stoichiometry of one α and four β subunits. In agreement with the observation of a GABA binding at a canonical ligand-binding “aromatic cage” at the β(+)/α(−) interface, the receptor adopts a conductive, open channel conformation. Structural comparisons reveal a quaternary activation mechanism arising from rigid-body movements between the extracellular and transmembrane domains. The α5β3 receptor contains only one GABA-binding site, represents the simplest heteropentameric GABAA receptor, and provides us a unique opportunity for further biophysical analysis of the channel gating mechanism.
Electronic supplementary material
Acknowledgements
We acknowledge the use of cryoEM instruments and computing resources in Center for Integrative Imaging of Hefei National Laboratory for Physical Sciences at the Microscale of University of Science and Technology of China, Center of Cryo-Electron Microscopy, Zhejiang University and the Electron Imaging Center for Nanomachines at University of California, Los Angeles. We thank the Bioinformatics Center of the University of Science and Technology of China, School of Life Science, for providing supercomputing resources for this project. This work was supported in part by funds from the Ministry of Science and Technology (Awards 2016YFA0500404 and 2014CB910300 to S.Y. and 2016YFA0501102 to X.Z.), the National Natural Science Foundation of China (Awards 31525001 and 31430019 to S.Y., 31600606 to X.Z. and 31630030 to G.Q.B.), and the Fundamental Research Funds for the Central Universities (to S.Y.). The Electron Imaging Center for Nanomachines at University of California, Los Angeles is supported by grants from NIH (GM071940, S10RR23057, S10OD018111 and U24GM116792) and NSF (DBI-1338135 and DMR-1548924). Structure coordinates and cryo-EM density maps have been deposited in the protein data bank under accession number 6A96 and EMD-6998.
Competing interests
The authors declare no competing interests.
Contributor Information
Z. Hong Zhou, Email: hong.zhou@ucla.edu.
Xiaokang Zhang, Email: xzhang1965@zju.edu.cn.
Sheng Ye, Email: sye@zju.edu.cn.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41422-018-0077-8.
References
- 1.Sigel E, Steinmann ME. J. Biol. Chem. 2012;287:40224–40231. doi: 10.1074/jbc.R112.386664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rudolph U, Mohler H. Annu. Rev. Pharmacol. Toxicol. 2014;54:483–507. doi: 10.1146/annurev-pharmtox-011613-135947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chua HC, Chebib M. Adv. Pharmacol. 2017;79:1–34. doi: 10.1016/bs.apha.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Olsen RW, Sieghart W. Pharmacol. Rev. 2008;60:243–260. doi: 10.1124/pr.108.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Angelotti TP, Macdonald RL. J. Neurosci. 1993;13:1429–1440. doi: 10.1523/JNEUROSCI.13-04-01429.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller PS, et al. Nat. Struct. Mol. Biol. 2017;24:986–992. doi: 10.1038/nsmb.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller PS, Aricescu AR. Nature. 2014;512:270–275. doi: 10.1038/nature13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laverty D, et al. Nat. Struct. Mol. Biol. 2017;24:977–985. doi: 10.1038/nsmb.3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller P, et al. bioRxiv. 2018 doi: 10.1101/338343. [DOI] [Google Scholar]
- 10.Phulera S, Zhu H, Yu J, Claxton DP, Yoder N, Yoshioka C, Gouaux E. Cryo-EM structure of the benzodiazepine-sensitive α1β1γ2S tri-heteromeric GABAA receptor in complex with GABA. Elife. Jul 25;7. pii: e39383. 10.7554/eLife.39383 (2018). [DOI] [PMC free article] [PubMed]
- 11.Zhu S, et al. Structure of a human synaptic GABAA receptor. Nature. 2018;559:67–72. doi: 10.1038/s41586-018-0255-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tretter V, Ehya N, Fuchs K, Sieghart W. J. Neurosci. 1997;17:2728–2737. doi: 10.1523/JNEUROSCI.17-08-02728.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mortensen M, Smart TG. J. Physiol. 2006;577:841–856. doi: 10.1113/jphysiol.2006.117952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Im WB, Pregenzer JF, Binder JA, Dillon GH, Alberts GL. J. Biol. Chem. 1995;270:26063–26066. doi: 10.1074/jbc.270.44.26063. [DOI] [PubMed] [Google Scholar]
- 15.Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S. Proteomics. 2004;4:1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.