Abstract
Background:
Success of infection treatment depends on the availability of accurate, reliable, and comprehensive data, information, and knowledge at the point of therapeutic decision-making. The identification of a national minimum data set will support the development and implementation of an effective surveillance system. The goal of this study was to develop a national antimicrobial resistance surveillance minimum data set.
Methods:
In this cross-sectional and descriptive study, data were collected from selected pioneering countries and organizations which have national or international antimicrobial resistance surveillance systems. A minimum data set checklist was extracted and validated. The ultimate data elements of the minimum data set were determined by applying the Delphi technique.
Results:
Through the Delphi technique, we obtained 80 data elements in 8 axes. The resistance data categories comprised basic, clinical, electronic reporting, infection control, microbiology, pharmacy, World Health Organization-derived, and expert-recommended data. Relevance coding was extracted based on the Iranian electronic health record coding system.
Conclusion:
This study provides a set of data elements and a schematic framework for the implementation of an antimicrobial resistance surveillance system. A uniform minimum data set was created based on key informants’ opinions to cover essential needs in the early implementation of a global antimicrobial resistance surveillance system in Iran.
Keywords: Drug resistance , Microbial , Dataset , Biosurveillance , Global health , Iran
What’s Known
Demand to survey and predict antimicrobial resistance (AMR) has resulted in the design and implementation of surveillance systems. Minimum data sets are necessary to establish national surveillance systems with consistent core data elements covering all aspects of the domain problem.
What’s New
Current study provides a set of data elements and a schematic framework for the implementation of an AMR surveillance system, both nationally and globally. Suggested minimum data set is compatible with the WHO GLASS standards and international ontologies and coding systems.
Introduction
For many years, antimicrobial resistance (AMR) has been a growing threat to the effective treatment and management of a range of infections caused by bacterial pathogens.1 If no effective plans to combat this resistance are implemented, it is forecast that it will lead to an annual 10 million deaths by 2050.2 In developing countries, the risk of AMR is growing rapidly.3 Iran has one of the highest global antibiotic consumptions, and there are concerns that the irresponsible use of antimicrobial agents will result in a high level of AMR.4,5 Iran is facing severe antibiotic misuse and overprescription, which is creating a large economic burden. According to a report, antibiotics were used inappropriately in 98% of all procedures.6 Success of treatment depends on the availability of accurate, reliable, and comprehensive data and knowledge at the point of therapeutic decision-making to identify the origin, spread, and patterns of AMR.7 The demand to survey and predict such resistance has resulted in the design and implementation of surveillance systems at all geographical levels.8,9 A comprehensive and up-to-date surveillance system is a critical requirement in monitoring and controlling AMR trends.10 Despite the steps taken by countries and organizations to boost monitoring and surveillance, data gaps remain because interoperability between regional levels has yet to be materialized.11 These gaps, together with a lack of unified standards for data sharing at national and global levels, are hampering efforts to produce knowledge at a global level with a view to enabling a comprehensive surveillance of the occurrence and trends of resistance worldwide.12 To meet such interoperability challenges, researchers have recommended various AMR control strategies such as the establishment of local, national, and global strategies, surveillance systems, and networks based on harmonized and well-informed efforts across all nations.13 For the effective and early implementation of antimicrobial resistance surveillance systems (AMRSSs) at a national level, a stepwise approach should be taken in the design and development of software modules. Database design is the first important step in the life-cycle of the information system, and it should be performed in accordance with a proper plan.14 Minimum data sets (MDSs) facilitate the establishment of national databases with consistent core data elements covering all aspects of the domain problem.15 Numerous scientific committees that aim to facilitate multidisciplinary and multi-institutional studies have latterly been established. However, irrespective of this aim, a standard method for the collection, analysis, distribution, and reporting of data to other participants is required.16 Management of AMR data at a national level requires the storage, retrieval, and exchange standards in the MDS of our AMRSS17 because the identification of an MDS is the most basic task with regard to the data collection of a surveillance system.18
Unfortunately, there is no national AMRSS in Iran. The Iranian Ministry of Health and Medical Education (MOHME), the Iranian Food and Drug Administration, and public health agencies have emphasized the need for a national AMRSS to improve preventive actions against this increased threat. The identification of a national MDS will support the development and implementation of an effective surveillance system and will provide conceptual interoperability at local, national, and global levels by covering data and standardization gaps based on novel interoperability strategies such as the Global Antimicrobial Resistance Surveillance System (GLASS). Thus, the present study was conducted to provide a comprehensive MDS as a template for establishing an AMRSS.
Related Works
There is a paucity of research on the development of an AMRSS MDS due to the short history of AMRSSs. The World Health Organization (WHO) recommended the development and implementation of nationwide surveillance systems to collect, analyze, and report AMR data on behalf of upper regional and global levels, and in 2015 it announced its GLASS to enable countries to create a worldwide picture of AMR by collecting data regarding major pathogens and antimicrobial resistant organisms from their respective national AMRSS.19 The WHO suggested 7 categories, including identification number, names, gender, dates, hospitalization data, and specimen data.12
In 2014, the European Antimicrobial Resistance Surveillance Network (EARS-Net) introduced its own MDS to AMRSS in a supra-national manner. The recommended MDS incorporated 53 data elements in 8 categories, consisting of laboratory, patient, date, specimen, technical, and epidemiological data elements.20 The United States and Australia have a similar approach; they recommended their own MDS in the first phase of their AMRSS development. The United States filtered demographic data due to confidentiality and privacy challenges. Australia introduced 2 distinct MDSs by first developing an AMRSS MDS and then going a step beyond by recommending through the Drug Utilization Sub-Committee a surveillance MDS for antimicrobial drug therapy and management. Now many hospitals contribute data on a voluntary basis to antimicrobial utilization programs.21 Recently, some other countries have collaborated in developing AMRSSs.22,23
Materials and Methods
Our cross-sectional, descriptive study was performed in 2016 at Tehran University of Medical Sciences (TUMS) in Iran. Data were collected from selected pioneering countries and organizations, including the United States,24 Australia,25 the WHO,26 and the European Union,27 all of which have national or international AMRSSs in place. Related material was retrieved via a literature review. The material encompassed papers, guidelines, strategies, gray documents, reports, types of software, and webpages available on general and specific search engines. Relevant studies were identified via keywords comprising the following: minimum data set, MDS, antimicrobial resistance, standard data, data element, AMR data elements, WHO, EARS-Net, National Antimicrobial Resistance Monitoring System (NARMS), and Australia. Google, Google Scholar, PubMed, ScienceDirect, and Scopus were searched for related keywords. All resources were confirmed from 2000 to May 2016. In this step, no sampling method was used, and all of the literature was evaluated on the basis of the inclusion criteria. The literature review was limited to the English and Persian languages during this period, in the form of full-texts and their valid sources. Materials without full-text access via the TUMS central library were excluded from the study. This process continued until data saturation was achieved. To determine the data saturation point, we conducted a simple interview with 2 key informants: the curator of the National Reference Laboratory and an expert from the Centers for Disease Control and Prevention (CDC). They validated the data elements in a quick and simple process. The collected data elements were classified into basic, clinical, laboratory, electronic reporting, infection control, microbiology, pharmacy, WHO-derived, and other (expert-recommended) data. These data elements were combined with the selected countries and organizations’ recommendations in order to construct the final version of the MDS checklist. A questionnaire was designed on the basis of the extracted checklist; it was composed of 3 columns with “yes” and “no” alongside each data element. Expert recommendation data elements were recorded in a blank row. The content validity of the questionnaire was assessed by 6 experts: 2 infectious diseases clinicians, 2 bacteriology specialists, and 2 health information management experts. The inclusion criteria were comprised of having been an expert with national responsibility in AMR stewardship in the preceding 3 years, having a major role in the local Antimicrobial Stewardship Program (ASP), full professorship or associate professorship, and active collaboration in the WHO meetings and programs (e.g., the WHO focal points in AMR surveillance programs). Uninterested experts and nonacademic experts were excluded from the study. Many references have recommended that an ASP team be comprised of 6 to 9 experts with different disciplines related to surveillance (as is the case with the selected team with national responsibilities in Iran). All guidelines on ASP activities have limited the number of experts in the design and development of AMRSS strategies.28
In the present study, after an initial coordination by telephone, 6 targeted e-mails were sent to the selected experts, who returned a completed checklist to us within a week. The results were aggregated, and the reliability of the questionnaire was evaluated using test-retest at a week’s interval by resending a new e-mail in SPSS software. A Spearman correlation coefficient of 78% was achieved.
The ultimate data elements of the AMRSS MDS were determined by using the Delphi technique with the participation of available informants, who were 6 experts from the MOHME, Tehran University of Medical Sciences, Tabriz University of Medical Sciences, and Iran University of Medical Sciences. The demographic features of the informants are shown in table 1.
Table 1.
Demographic features of the experts in the Delphi technique
| Specialty | Age | Gender | Academic Level | Job | Experience |
|---|---|---|---|---|---|
| Pediatrics infectious disease fellowship | 52 | Male | PhD | MOHME CDC expert | >15 y |
| Infectious disease medicine | 40 | Male | PhD | University professor | >7 y |
| Bacteriology expert | 46 | Male | PhD | University professor | >10 y |
| Bacteriology expert | 65 | Male | PhD | University professor | >25 y |
| Health information management expert | Female | PhD | University professor | >30 y | |
| Health information management expert | 35 | Male | PhD | University professor | >7 y |
MOHME: Iranian ministry of health and medical education; CDC: Centers for disease control and prevention
The inclusion and exclusion criteria were the same as those in the previous step. We used a 5-point Likert scale checklist, which allows the respondents to express to what extent they agree or disagree with a particular data element. Data elements with agreement levels of under 50% were excluded, and those with agreement levels of over 75% were accepted as required data elements. Data elements between 50% and 75% were reassessed using the checklist in the second round in a similar manner and elements with greater than 75% agreements were added to the final data elements. The final checklist was distributed by visiting the experts in their personal office.
In the next step, the recommended data elements were mapped to the common international coding systems used in Iran. Finally, adaptation of the recommended MDS to that of other countries was evaluated in the form of a comparative table.
Results
The AMRSS data elements were classified into 8 categories. The total number of the ultimate data elements was 144. Table 2 shows the result of the Delphi technique on the data elements.
Table 2.
Result of the Delphi technique on the extracted data elements
| Categories | Number of Data Elements | Delphi First Round | Delphi Second Round | Result | ||||
|---|---|---|---|---|---|---|---|---|
| <50% | 50-75% | >75% | <50% | 50-75% | >75% | |||
| Basic data | 29 | 6 | 7 | 16 | 3 | 0 | 4 | 20 |
| Clinical data | 11 | 1 | 3 | 7 | 1 | 0 | 2 | 9 |
| EARS-Net data | 5 | 5 | 0 | 0 | 0 | 0 | 0 | 0 |
| Electronic reporting data | 19 | 12 | 5 | 2 | 2 | 0 | 3 | 5 |
| Nutrition data | 17 | 13 | 4 | 0 | 4 | 0 | 0 | 0 |
| Infection control data | 8 | 0 | 3 | 5 | 0 | 0 | 3 | 8 |
| Microbiology data | 19 | 3 | 7 | 9 | 4 | 0 | 3 | 12 |
| Drug data | 7 | 4 | 0 | 3 | 4 | 0 | 0 | 3 |
| Specimen data | 5 | 4 | 1 | 0 | 1 | 0 | 0 | 0 |
| WHO-derived data | 24 | 6 | 8 | 10 | 5 | 0 | 3 | 13 |
| Total | 144 | 54 | 38 | 52 | 24 | 0 | 18 | 80 |
EARS Net data: European antimicrobial resistance surveillance network; WHO: World health organization
Through the Delphi technique, we obtained 80 data elements in 8 axes (table 3).
Table 3.
AMRSS data categories and the elements for Iran and related MAKSA coding
| No | Categories | Data Elements | With an Equivalent Field in GLASS | MAKSA Coding |
|---|---|---|---|---|
| 1 | Basic data | Country | * | ISO_3166-1 |
| 2 | Laboratory | String | ||
| 3 | Identification number (general unique ID, e.g., national code) | * | String | |
| 4 | Date of birth | Do _Date | ||
| 5 | Age | * | Derived element - integer | |
| 6 | Gender | * | ||
| 7 | WHO age category | * | WHO standard international age classification | |
| 8 | Specimen number | String | ||
| 9 | Specimen date | * | Do _Date | |
| 10 | Specimen type | * | thritaEHR. Specimen type | |
| 11 | Specimen start and end date | Do _Date | ||
| 12 | Reason (unresponsive to treatment, etc.) | SCTID: 408773008(SNOMED-CT) | ||
| 13 | Isolate number | String | ||
| 14 | Organism | * | SCTID: 264395009(SNOMED-CT) | |
| 15 | Organism type (subgroups) | SCTID: 264395009 (SNOMED-CT) | ||
| 16 | Beta-lactamase | SCTID: 79744009 (SNOMED-CT) | ||
| 17 | ESBL | SCTID: 409799006(SNOMED-CT) | ||
| 18 | Carbapenem | String | ||
| 19 | MRSA screening test | CPT 901617 | ||
| 20 | Serotype (in detailed reports) | SCTID: 260820006(SNOMED-CT) | ||
| 21 | Clinical data | Date of admission | Do _Date | |
| 22 | Date of discharge | Do _Date | ||
| 23 | Date of operation | Do _Date | ||
| 24 | Date of ward admission | Do _Date | ||
| 25 | Diagnosis | ICD-10 | ||
| 26 | Operation | ICD-10-CM | ||
| 27 | Prior antibiotic therapy | SCTID: 281789004(SNOMED-CT) | ||
| 28 | Response to treatment | SCTID: 182985004(SNOMED-CT) | ||
| 29 | Origin (hospital- or community-acquired) | * | SCTID: 255396000(SNOMED-CT) | |
| 30 | Electronic reporting data (death data) | Date of death | Do _Date | |
| 31 | Time of death | Do _Time | ||
| 32 | Death speed (diagnosis to death, shows disease severity) | ICD-10 | ||
| 33 | Patient address (street, city, and province) or only geographical region | Country divisions | ||
| 34 | Specimen site | SCTID: 430304001(SNOMED-CT) | ||
| 35 | Infection control | Bacteremia | SCTID: 5758002(SNOMED-CT) | |
| 36 | Central catheter | SCTID: 52124006(SNOMED-CT) | ||
| 37 | Urine catheter | SCTID: 20568009(SNOMED-CT) | ||
| 38 | Ventilator | SCTID: 706172005(SNOMED-CT) | ||
| 39 | Nosocomial infection | SCTID: 19168005(SNOMED-CT) | ||
| 40 | Peripheral catheter | SCTID: 423954007(SNOMED-CT) | ||
| 41 | Surgical site infection | SCTID: 423954007(SNOMED-CT) | ||
| 42 | Urinary tract infection | SCTID: 68566005(SNOMED-CT) | ||
| 43 | Microbiology data | ESBL producer | SCTID: 409799006(SNOMED-CT) | |
| 44 | Gram stain | SCTID: 62777006(SNOMED-CT) | ||
| 45 | H antigen phase 1 | SCTID: 115744003(SNOMED-CT) | ||
| 46 | H antigen phase 2 | “ | ||
| 47 | Hodge test | SCTID: 336769001(SNOMED-CT) | ||
| 48 | Microscopy | SCTID: 117259009(SNOMED-CT) | ||
| 49 | O antigen | SCTID: 23434000 (SNOMED-CT) | ||
| 50 | Oxacillin screen plate | SCTID: 67466005 (SNOMED-CT) | ||
| 51 | Urine colony count | SCTID: 104193001 (SNOMED-CT) | ||
| 52 | Vancomycin screen plate | String | ||
| 53 | VRE | SCTID: 113727004 (SNOMED-CT) | ||
| 54 | BLNAR | SCTID: 712679007 (SNOMED-CT) | ||
| 55 | Pharmacy data | Antibiotic treatment | SCTID: 281789004 (SNOMED-CT) | |
| 56 | Start date | Do _Date | ||
| 57 | End date | Do _Date | ||
| 58 | WHO-derived data | District | * | Country Divisions |
| 59 | Epid No (Unique ID) | * | String | |
| 60 | Date sent to ref lab | * | Do _Date | |
| 61 | Date received in ref lab | * | Do _Date | |
| 62 | Date result to the district | * | Do _Date | |
| 63 | PMN% | SCTID: 43657008(SNOMED-CT) | ||
| 64 | Lymphocyte% | SCTID: 409599009 (SNOMED-CT) | ||
| 65 | Monocyte% | SCTID: 401100006(SNOMED-CT) | ||
| 66 | Eosinophil% | SCTID: 71960002(SNOMED-CT) | ||
| 67 | Leucocytes% | SCTID: 106200001(SNOMED-CT) | ||
| 68 | RBC | SCTID: 14089001(SNOMED-CT) | ||
| 69 | Parasites | SCTID: 252400008(SNOMED-CT) | ||
| 70 | User Information | * | string | |
| 71 | Expert recommended data | Underlying disease | ICD-10 | |
| 72 | Immunodeficiency | SCTID: 234532001(SNOMED-CT) | ||
| 73 | History of travel | SCTID: 443846001(SNOMED-CT) | ||
| 74 | Destination in travel | String | ||
| 75 | Length of stay in destination | Integer | ||
| 76 | Infection exposure history in 2 weeks or more | ICD-10 | ||
| 77 | Type of antimicrobial dosage (intravenous or oral) | SCTID: 18629005(SNOMED-CT) | ||
| 78 | Cancer complication | ICD-10 | ||
| 79 | Transfusion history | SCTID: 161664006(SNOMED-CT) | ||
| 80 | Vaccination history | SCTID: 425457005(SNOMED-CT) | ||
AMRSS: Antimicrobial resistance surveillance system; GLASS: Global antimicrobial resistance surveillance system; MAKSA: Iranian electronic health record coding system
The AMR data categories were as follows: basic, clinical, electronic reporting, infection control, microbiology, pharmacy, WHO-derived, and expert-recommended data. The required data elements of the WHO GLASS aggregate reports were indicated. Relevance coding was extracted based on the Iranian electronic health record coding system (MAKSA) to facilitate data exchange between various systems in SEPAS middleware.
As is shown in table 4, the WHO recommended an MDS for each pathogen (2002).29 .
Table 4.
WHO-required data elements in the national reporting of each participant country
| Pathogens | Recommended Minimum Data Set | Recommended on GLASS | Iran National AMRSS MDS | NARMS | EARS-Net | AMRU |
|---|---|---|---|---|---|---|
| E. coli | Unique identifier | + | + | + (O157) | + | + |
| Age or date of birth | + | + | + (O157) | + | + | |
| Gender | + | + | + (O157) | + | + | |
| Health-care facility | - | + | + (O157) | + | + | |
| Care group | + | + | + (O157) | + | + | |
| Date of admission | + | + | + (O157) | + | - | |
| Type of specimen | + | + | + (O157) | + | + | |
| Test result (R/I/S) | + | + | + (O157) | + | + | |
| K. pneumonia | Unique identifier | + | + | - | + | + |
| Age or date of birth | + | + | - | + | + | |
| Gender | + | + | - | + | + | |
| Health-care facility | - | + | - | + | + | |
| Care group | + | + | - | + | + | |
| Date of admission | + | + | - | + | - | |
| Type of specimen | + | + | - | + | + | |
| Test result (R/I/S) | + | + | - | + | + | |
| Presenting symptoms | - | + | - | - | - | |
| Predisposing factors (e.g., cancer, travel, catheter, etc.) | - | + | - | - | - | |
| A. baumannii | Not mentioned | - | + | - | + | + |
| S. aureus | Unique identifier | + | + | - | + | + |
| Age or date of birth | + | + | - | + | + | |
| Gender | + | + | - | + | + | |
| Health-care facility | - | + | + | |||
| Care group | + | + | - | + | + | |
| Date of admission | + | + | - | + | - | |
| Type of specimen | + | + | - | + | + | |
| Test result (R/I/S) | + | + | - | + | + | |
| Presenting symptoms | - | + | - | - | - | |
| Predisposing factors (e.g., surgery, trauma, etc.) | - | - | - | |||
| Method of identification | - | + | - | - | - | |
| Shigella | Unique identifier | + | + | + | - | + |
| Age or date of birth | + | + | + | - | + | |
| Gender | + | + | + | - | + | |
| Health-care facility | - | + | + | - | + | |
| Care group | + | + | + | - | + | |
| Date of admission | + | + | + | - | - | |
| Type of specimen | + | + | + | - | + | |
| Test result (R/I/S) | + | + | + | - | + | |
| Outcome (death or recovery) | - | + | + | - | - | |
| S. pneumonia | As for Shigella | “ | “ | - | + | + |
| Vaccination history | - | + | - | - | - | |
| Salmonella spp. | As for Shigella | “ | “ | + | - | + |
| N. gonorrhoeae | Unique identifier | + | + | - | - | + |
| Age or date of birth | + | + | - | - | + | |
| Gender | + | + | - | - | + | |
| Risk behaviors | - | - | - | - | - | |
| Date of onset symptoms | - | + | - | - | - | |
| Date of specimen | + | + | - | - | + | |
| Anatomic site sampled | - | + | - | - | - | |
| Test result (R/I/S) | + | + | - | - | + | |
+Positive result; -Negative result or not surveyed; WHO: World Health Organization; AMRSS: Antimicrobial resistance surveillance system; MDS: Minimal data set; NARMS: National Antimicrobial Resistance Monitoring System; EARS Net data: European Antimicrobial Resistance Surveillance Network
The WHO GLASS-required data elements (2015),12 EARS-Net data set, NARMS data elements, Australian AMRU (JETACAR), and Iran national AMRSS MDS were compared to assess the compatibility of the selected systems with the WHO standards and GLASS perquisites and to ensure that there were no missing data elements at both national and global levels. These data elements constituted the global report of each participant to the WHO.
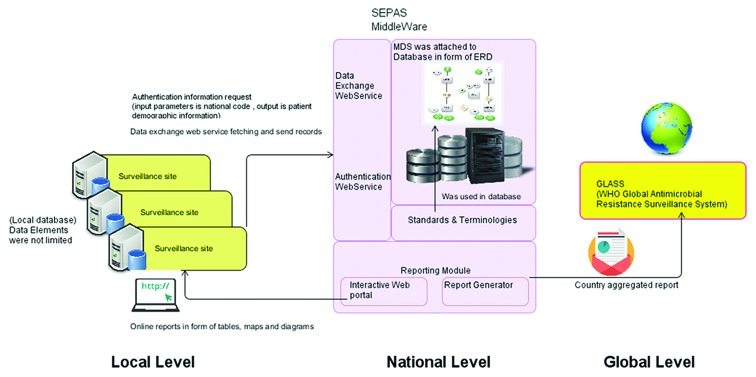
The basic data category included some essential identical data, dates, numbers, and organisms, as well as specimens and their types for collection and analysis. If any confidential piece of information was required for policy considerations, it could be accessed by queries across SEPAS authentication web services.
The clinical data category included important dates, diagnosis, operations, details of prior treatments, and responses to those treatments. We obtained free access to the coding systems and imposed no limit on the range of codes. The electronic reporting data category incorporated some important data regarding patient deaths such as date, time, reason, geographical information, and specimen site.
The infection control category consisted of infection source data such as bacteremia and catheter-related infections. These data were extracted from the WHO documents and platforms for AMRSS. Another important category was that of microbiologic data. This category contained specific data relating to the AMR test, as well as detailed qualitative and quantitative relevance data. The pharmacy data category encompassed antibiotic treatment, dosage, dosage interval, start date, and end date, while the WHO-derived data included some data applicable to rural and under-served areas whereby district-wide test samples were forwarded to a reference laboratory with information such as date sent, date received, and date reported. The district data and other data elements such as some laboratory parameters to satisfy the global objectives pursued by the WHO were also included.
The final category included expert recommendations for Iran. They emphasized underlying diseases, immunodeficiency problems, history and destination of travel, and length of stay in neighboring countries. Infection exposure history (whether it was 2 weeks or more), antimicrobial dosage type (intravenous or oral), cancer complications, transfusion history, and vaccination history were the recommended information. Where possible, MAKSA codes were assigned to each data element to meet interoperability requirements and to reduce data redundancy.
Discussion
Given the status of antibiotic use in Iran and the lack of effective surveillance, the current study determined the national AMRSS MDS, containing 80 data elements, to collect, analyze, and report AMR indicators. Each data element was mapped to common coding standards and terminologies to facilitate interoperability between various health systems at local, national, and global levels. Another important outcome of the current study was fulfillment of the WHO GLASS guideline in the data infrastructure of the Iranian AMRSS by applying cross-mapping of data elements. These achievements are essential to implement a GLASS-oriented AMRSS.
Kalankesh et al.18 reported that the identification of an MDS is the most basic task in the data collection of a surveillance system. Our literature review yielded no standard and uniform data collection formats in Iran. The absence of such a format has thwarted efforts to establish an AMRSS and precluded the participation of Iran in the WHO GLASS. For an effective collaboration in international efforts, the data should be uniform. Therefore, it is critical to develop a national AMRSS MDS (linked with the WHO GLASS) as a database infrastructure for collecting, analyzing, sharing, and reporting AMR data in Iran. It was imperative that our study meet a standard set of data elements at local and national levels.
Many countries and organizations have underscored the identification of an MDS in AMR data collection as a key area in a national program.30 Eurostat is responsible for providing statistical information to the institutions of the European Union and for promoting harmonization of statistical methods across its members. In addition, it seeks to furnish information to policy-makers with 2 fundamental objectives: 1) defining an MDS and 2) developing efficient and effective data collection and analysis methods.31 About 80% of countries have focused on determining electronic health record-related MDSs. The result of the present study displayed that our suggested MDS was compatible with international frameworks, especially the WHO standards, more than other countries and organizations. The current study is the first research on the implementation of the GLASS since the publication of GLASS documents.32
MDS definition and implementation is intended to improve the consistency of reporting methods and the interpretability of the reported results, allowing for a better comparison of the published AMR reports and articles, provision of guidance for future research, and fostering of a generation of on-demand knowledge.33 Our study encompassed these 2 key areas, particularly the identification of an MDS.
SEPAS is a national project that aims to integrate health-related data for each individual to improve data accessibility and efficiency across Iran. In order to manage AMR data at a national level, it was necessary to meet SEPAS storage, retrieval, and exchange standards in our study.17 SEPAS utilizes some standards and terminology such as ICD-10, SNOMED, ISO13606, SOA, and MAKSA. All software systems should implement these standards to provide interoperability with SEPAS.34 As an early step in the Iranian national AMRSS, we met these standards at the required level. A schematic view of the application of the MDS in the Iranian National Health System is proposed in figure 1.
Figure1.
Shows schematic view of the application of a minimum data set (MDS) in the Iranian National Health System.
Documentation of data elements at the point of care (when the patient is transferred to another ward, hospital, or city for any reason) would support health-care workers in making decisions concerning patient transfer, health center equipment, and management of the available resources with a view to augmenting interoperability.35,36 SEPAS provides this advantage to physicians, health workers, and policy-makers anytime, anywhere. In order to implement conceptual interoperability, health information managers should identify important data elements and create a data dictionary to generate consistency between health-care providers in various disciplines.34 SEPAS provides this advantage to physicians, health workers, and policy-makers anytime, anywhere. In order to implement conceptual interoperability, health information managers should identify important data elements and create a data dictionary to generate consistency between health-care providers in various disciplines.34 Accordingly, identification of a national MDS should be compatible with SEPAS.37 In our study, the equivalent codes of each data element were extracted on the basis of the reference health-coding of Iran.
The WHO emphasizes that, for the successful establishment of AMRSSs, participation at the country level be made as simple as possible. Thus, systems should collect a minimum set of data needed for useful AMR surveillance and follow an evidence-based approach to public health interventions.29 In the current study, Iranian CDC experts were incorporated in the evaluation group.
Information technology staff is critical to integrating AMR surveillance standards into existing antimicrobial stewardship programs. Facilitating the collection and reporting of antibiotic data in a standard manner is an important task in policy making vis-à-vis stewardship programs.38 Consequently, integration of a defined MDS in an antimicrobial stewardship program is a task critical to the implementation of an effective data collection, analysis, and reporting process.
The present study has 2 limitations. First, we aimed to determine a national MDS without enhanced surveillance capabilities; thus, a selected team was enough for the defined objectives. To conduct an enhanced surveillance, it is recommended that clinical domain data sets be combined. Definition of an enhanced surveillance MDS requires that a large sample size of domain experts be drawn upon. Second, we were faced with a lack of comparative studies on the axes of the WHO GLASS and related business intelligence capabilities. We seek to overcome this limitation in our next 2 national studies in the future.
Conclusion
Creation of an effective antimicrobial stewardship program requires the sound collection, analysis, and reporting of accurate and reliable AMR data. To obtain a clear national picture of AMR statutes in Iran, we need a uniform and consistent data model that satisfies concept interoperability. Definition of a national MDS should meet these requirements as a first step in the establishment of a surveillance database. The present study provides a set of data elements and a schematic framework for the implementation of an AMRSS, both nationally and globally. For the early implementation of the GLASS in Iran, we recommend the development of conceptual models of surveillance systems and the identification of priority values and scales for each data element such as priority pathogens, antimicrobial agents, surveillance methods, and key performance indicators as the next step.
Acknowledgement
This study is a part of a PhD dissertation financially supported by Tehran University of Medical Sciences (Grant #280/6/MI/11).
Conflict of Interest:None declared.
References
- 1.Organization WH. Antimicrobial resistance: global report on surveillance. Geneva: World Health Organization; 2014. [Google Scholar]
- 2.Hoffman SJ, Outterson K, Rottingen JA, Cars O, Clift C, Rizvi Z, et al. An international legal framework to address antimicrobial resistance. Bull World Health Organ. 2015;93:66. doi: 10.2471/BLT.15.152710. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alsan M, Schoemaker L, Eggleston K, Kammili N, Kolli P, Bhattacharya J. Out-of-pocket health expenditures and antimicrobial resistance in low-income and middle-income countries: an economic analysis. Lancet Infect Dis. 2015;15:1203–10. doi: 10.1016/S1473-3099(15)00149-8. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soltani J, Poorabbas B, Miri N, Mardaneh J. Health care associated infections, antibiotic resistance and clinical outcome: A surveillance study from Sanandaj, Iran. World J Clin Cases. 2016;4:63–70. doi: 10.12998/wjcc.v4.i3.63. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mansouri S, Abbasi S. Prevalence of multiple drug resistant clinical isolates of extended-spectrum beta-lactamase producing Enterobacteriaceae in Southeast Iran. Iran J Med Sci. 2010;35:101–8. [Google Scholar]
- 6.Hatam N, Askarian M, Moravveji AR, Assadian O. Economic burden of inappropriate antibiotic use for prophylactic purpose in shiraz, iran. Iran Red Crescent Med J. 2011;13:234–8. [ PMC Free Article] [PMC free article] [PubMed] [Google Scholar]
- 7.Amábile-Cuevas CF. Antimicrobial resistance in bacteria. Poole: Horizon Scientific Press; 2007. [Google Scholar]
- 8.Shakiba M, Haghdoost A, Majdzadeh S. The Application of Geographical Information System in Explaining Spatial Distribution of Low Birth Weight; a Case Study in North of Iran. Iran J Med Sci. 2008;33:220–5. [Google Scholar]
- 9.de Kraker ME, Jarlier V, Monen JC, Heuer OE, van de Sande N, Grundmann H. The changing epidemiology of bacteraemias in Europe: trends from the European Antimicrobial Resistance Surveillance System. Clin Microbiol Infect. 2013;19:860–8. doi: 10.1111/1469-0691.12028. [DOI] [PubMed] [Google Scholar]
- 10.Lewis DA, Lukehart SA. Antimicrobial resistance in Neisseria gonorrhoeae and Treponema pallidum: evolution, therapeutic challenges and the need to strengthen global surveillance. Sex Transm Infect. 2011;87:ii39–43. doi: 10.1136/sti.2010.047712. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crosse M. Antibiotic Resistance (AR): Data Gaps Will Remain Despite HHS Taking Steps to Improve Monitoring. Collingdale: Diane Publishing Company; 2011. [Google Scholar]
- 12.Organization WH. Global antimicrobial resistance surveillance system (GLASS): technical meeting on the early implementation phase: 22-23 October 2015: WHO Regional Office for Europe Copenhagen, Denmark: meeting report. 2016. [Google Scholar]
- 13.Ghosh TK, Kundu R, Kalra A, Ganguly N, Mitra M. Infectious diseases in children and newer vaccines. Infectious diseases in children and newer vaccines. 2007. [Google Scholar]
- 14.Mitea AC. An optimization algorithm for physical database design. WSEAS Transactions on Information Science and Applications. 2007;4:122–7. [Google Scholar]
- 15. Health Professions Minimum Data Set US [Internet] Department of Health and Human Services: Health Resource and Services Administration; [cited 2016 Mar 3] Available from: [http://bhw.hrsa.gov/healthworkforce/data/minimumdataset/index.html. ]
- 16.Ghaneie M, Rezaie A, Ghorbani NR, Heidari R, Arjomandi M, Zare M. Designing a minimum data set for breast cancer: a starting point for breast cancer registration in iran. Iran J Public Health. 2013;42:66–73. [ PMC Free Article] [PMC free article] [PubMed] [Google Scholar]
- 17.Niakan Kalhori SR, Tayefi B, Noori A, Mearaji M, Rahimzade S, Zandian E, et al. Inpatient data, inevitable need for policy making at national and sub-national levels: a lesson learned from NASBOD. Arch Iran Med. 2014;17:16–21. [PubMed] [Google Scholar]
- 18.Kalankesh LR, Dastgiri S, Rafeey M, Rasouli N, Vahedi L. Minimum data set for cystic fibrosis registry: a case study in iran. Acta Inform Med. 2015;23:18–21. doi: 10.5455/aim.2015.23.18-21. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Organization WH. Call for participation: Global Antimicrobial Resistance Surveillance System 2015 [cited June 2016] Available from: [http://www.who.int/drugresistance/surveillance/en. ] [Google Scholar]
- 20.Cornaglia G, Hryniewicz W, Jarlier V, Kahlmeter G, Mittermayer H, Stratchounski L, et al. European recommendations for antimicrobial resistance surveillance. Clin Microbiol Infect. 2004;10:349–83. doi: 10.1111/j.1198-743X.2004.00887.x. [DOI] [PubMed] [Google Scholar]
- 21.Cairns KA, Roberts JA, Cotta MO, Cheng AC. Antimicrobial Stewardship in Australian Hospitals and Other Settings. Infect Dis Ther. 2015;4:27–38. doi: 10.1007/s40121-015-0083-9. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Organization WH. Antimicrobial resistance in the Western Pacific Region: a review of surveillance and health systems response. Manila: WHO Regional Office for the Western Pacific; 2015. [Google Scholar]
- 23. Pan-Canadian Public Health Network. Antimicrobial resistance surveillance data requirements for priority organisms. The Communicable and Infectious Disease Steering Committee Antimicrobial Resistance Surveillance Task Group. Ottawa: Public Health Network Council; 2016. [Google Scholar]
- 24.Prevention CfDCa. National Antimicrobial Resistance Monitoring System for Enteric Bacteria (NARMS): Human Isolates Final Report, 2013. Atlanta, Georgia: US Department of Health and Human Services, CDC ; 2013. [Google Scholar]
- 25.Shaban R, Simon G, Trott D, Turnidge J, Jordan D [Internet] Surveillance and reporting of antimicrobial resistance and antibiotic usage in animals and agriculture in Australia. c2014- [cited 2016 March 12] Available from: [https://research-repository.griffith.edu.au/bitstream/handle/10072/65151/98828_1.pdf?sequence=1. ]
- 26.Organization WH. Global antimicrobial resistance surveillance system: Manual for early implementation. Geneva: World Health Organization; 2015. [Google Scholar]
- 27.System EARS, [Internet] Summary of latest data on antibiotic resistance in the European Union 2010. c2010- [cited 2016 July 03] . Available from: [http://www.ecdc.europa.eu/en/activities/surveillance/EARS-Net/Pages/index. aspx. ]
- 28.Griffith M, Postelnick M, Scheetz M. Antimicrobial stewardship programs: methods of operation and suggested outcomes. Expert Rev Anti Infect Ther. 2012;10:63–73. doi: 10.1586/eri.11.153. [DOI] [PubMed] [Google Scholar]
- 29.Organization WH. Surveillance standards for antimicrobial resistance. Geneva: World Health Organization; 2002. [Google Scholar]
- 30.Nathwani D, Christie P. The Scottish approach to enhancing antimicrobial stewardship. J Antimicrob Chemother. 2007;60:i69–71. doi: 10.1093/jac/dkm162. [DOI] [PubMed] [Google Scholar]
- 31.Basys CI, Credes I. Defning a minimum data set and related indicators for use with the system of health accounts in the European Union. Luxembourg: IGSS. 2004;128 [Google Scholar]
- 32.Walker DS, Visger JM, Levi A. Midwifery data collection: options and opportunities. J Midwifery Womens Health. 2008;53:421–9. doi: 10.1016/j.jmwh.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 33.Jason LA, Unger ER, Dimitrakoff JD, Fagin AP, Houghton M, Cook DB, et al. Minimum data elements for research reports on CFS. Brain Behav Immun. 2012;26:401–6. doi: 10.1016/j.bbi.2012.01.014. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Asadi F, Moghaddasi H, Rabiei R, Rahimi F, Mirshekarlou SJ. The Evaluation of SEPAS National Project Based on Electronic Health Record System (EHRS) Coordinates in Iran. Acta Inform Med. 2015;23:369–73. doi: 10.5455/aim.2015.23.369-373. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahmadi M, Mohammadi A, Chraghbaigi R, Fathi T, Shojaee Baghini M. Developing a minimum data set of the information management system for orthopedic injuries in iran. Iran Red Crescent Med J. 2014;16:e17020. doi: 10.5812/ircmj.17020. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hachesu PR, Ahmadi M, Rezapoor A, Salahzadeh Z, Farahnaz S, Maroufi N. Clinical care improvement with use of health information technology focusing on evidence based medicine. Healthc Inform Res. 2012;18:164–70. doi: 10.4258/hir.2012.18.3.164. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mirnia K, Samad Soltani T, Rezaei M, Heidarzadeh M, Piri Z. Design and evaluation of electronic briefs of neonatal intensive care unit in Taleghani hospital, Tabriz, Iran. Glob J Health Sci. 2014;6:125–31. doi: 10.5539/gjhs.v6n5p125. [ PMC Free Article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Centers for Disease Control and Prevention [Internet] Core Elements of Hospital Antibiotic Stewardship Programs 2015. [cited 2016 April 03] Available from: [http://www.cdc.gov/getsmart/healthcare/implementation/core-elements.html. ]