Abstract
Caveolae are plasma membrane invaginations involved in transport, signalling and mechanical membrane sensing in metazoans. Their formation depends upon multiple interactions between membrane‐embedded caveolins, lipids and cytosolic cavin proteins. Of the four cavin family members, only cavin1 is strictly required for caveola formation. Here, we demonstrate that an eleven residue (undecad) repeat sequence (UC1) exclusive to cavin1 is essential for caveolar localization and promotes membrane remodelling through binding to phosphatidylserine. In the notochord of mechanically stimulated zebrafish embryos, the UC1 domain is required for caveolar stability and resistance to membrane stress. The number of undecad repeats in the cavin1 UC1 domain varies throughout evolution, and we find that an increased number also correlates with increased caveolar stability. Lastly, we show that the cavin1 UC1 domain induces dramatic remodelling of the plasma membrane when grafted into cavin2 suggesting an important role in membrane sculpting. Overall, our work defines a novel conserved cavin1 modular domain that controls caveolar assembly and stability.
Keywords: caveolae, cavin, coiled‐coil, undecad repeat
Subject Categories: Membrane & Intracellular Transport
Introduction
Caveolae are bulb‐shaped invaginations of the cell surface that play central roles in intracellular signalling, trafficking, lipid metabolism and membrane tension responses 1. These characteristic 50–60 nm structures are formed by co‐operative interactions between two families of proteins called caveolins and cavins. Caveolins are integral membrane proteins and were the first components shown to be essential for caveolae formation. Caveolin1 and caveolin2 (CAV1, CAV2) 2, 3 are ubiquitously expressed in all tissues while caveolin3 (CAV3) is a muscle‐specific isoform 4. CAV1 and CAV3 are essential for caveolae formation in respective cell types, 5 and CAV1 knockout mice show a complete loss of morphological caveolae 6. Over the past decade, a second family of cytosolic peripheral membrane proteins have been identified, the cavins, that specifically associate with caveolae in heteromeric complexes 7. In humans, there are four family members (cavin1‐4), with cavin4 being highly muscle‐specific. Cavin1 (also called PTRF; polymerase 1 transcript and release factor) is a central component of the cavin coat assembly; cavin1 knockout mice lack caveolae, similar to knockout of CAV1, cavin1 is sufficient to generate caveolae in the absence of other cavins, and cavin1 is required for heteromeric association of cavin2 and cavin3 with caveolins 8, 9, 10. Cavin2 plays a specific role in caveola formation in lung endothelial cells 11, while cavin3 plays a role in endocytosis through caveolae 12 and is implicated in maintaining circadian rhythm and ERK/AKT signalling through caveolae 13, 14. Caveolae also act as membrane reservoirs, flattening out in response to mechanical stress and releasing cavin proteins to the cytosol 15. Genetic mutations in both caveolins and cavins are implicated in development of human pathologies such as lipodystrophy and muscular dystrophy 16, 17, 18, 19, 20.
Recent studies have begun to characterize the structural and functional properties of cavin proteins. Cavins were initially shown to form high molecular weight (60S) complexes on sucrose gradients, suggesting the formation of oligomeric assemblies 21. Subsequently, it was shown that cavin1 could assemble into homo and hetero‐oligomeric complexes of cavin1‐cavin1, cavin1‐cavin2 and cavin1‐cavin3 on individual caveola 10, 22. The cavin proteins have a distinctive architecture at the primary and secondary structure levels, possessing N‐ and C‐terminal α‐helical regions (HR1 and HR2) alternating with three disordered regions (DR1, DR2 and DR3; Fig 1A). The X‐ray crystallographic structures of the cavin1 and cavin4 HR1 domains showed a highly extended trimeric coiled‐coil domain that is required for homo‐ and hetero‐interactions of the cavin proteins in vitro 23. HR1 domains possess a positively charged surface that binds preferentially to phosphatidylinositol phospholipids [phosphoinositides (PIs)]. The trimerization of HR1 further promotes the folding of HR2 into an α‐helical structure that binds preferentially to phosphatidylserine (PS) 23. Phosphoinositide‐binding lysine residues within the cavin1 HR1 domain are also ubiquitinated, promoting degradation in response to mechanical stress 24. Although progress has been made in understanding the molecular mechanisms that underpin caveola formation and disassembly, many questions remain unanswered, such as what properties of cavin1 lead to its prime importance in caveola assembly, and how is the disassembly of caveolae regulated?
Figure 1. Role of cavin1 HR2 domain in caveolar localization.

- GFP‐tagged truncations of cavin1 are indicated schematically.
- GFP‐tagged cavin1 truncations were expressed in A431 cell line (green) and assessed for co‐localization with CAV1 in caveolae (red). Scale bars = 10 μm.
- The PLA signal between co‐expressed GFP‐tagged constructs in PC3 cell line and endogenous CAV1 was quantitated using ImageJ as number of PLA dots per cell is plotted, N = 2 (independent biological replicates), n = 15–25 (sample size). Data are presented as mean, and error bars indicate standard deviation. ****P < 0.0001, ns = not significant; one‐way ANOVA followed by Bonferroni's multiple comparisons.
In this study, we show that cavin1 contains a unique repeated sequence [an undecad or hendecad repeat we term UC1 (undecad of cavin1)] that is predicted to form a right‐handed coiled‐coil and that this domain confers binding to phosphatidylserine. Deletion of the UC1 domain in cavin1 disrupts the formation of caveolae, while mutation of potential membrane binding residues in the UC1 domain reduces the affinity for phosphatidylserine‐containing membranes. We then tested the importance of these repeats in vivo using zebrafish deleted for the notochord‐specific paralog zcavin1b 25. Here, we show for the first time that notochord lesions caused by excessive mechanical stress are rescued by expressing the wild‐type zcavin1b but not by a mutant lacking the zcavin1b UC1 domain. Finally, using model cell systems we show that the progressive deletion of undecad units from UC1 domain modulates stability of caveolae. This work outlines fundamental characteristics of cavin1 essential for function of caveola membrane domains.
Results and Discussion
Model cell lines and animal model used for analysing cavins and caveola formation
In this work, we have employed three human cell lines (PC3, MCF7 and A431) that differ in their expression of caveolar protein components. The PC3 prostate cancer cell line does not express any cavin family proteins, but expresses CAV1 that is diffusely localized throughout the plasma membrane 8. They lack morphological caveolae, but heterologous expression of cavin1‐GFP induces caveola formation 8, 26. The MCF7 breast cancer cell line does not express either cavin or caveolin family proteins, and exogenous cavin proteins show a cytosolic distribution 24, 27. The A431 cell line expresses CAV1 as well as cavin1 and forms morphological caveolae at the plasma membrane 24, 28, 29. Genome‐edited zebrafish used in this study lack the notochord‐specific isoform of cavin1b and show prominent notochord lesions, reduced locomotor capacity and reduced caveola density 25.
Mutational analysis of the cavin1 HR2 domain and its role in caveola association
Our previous work showed that both HR1 and HR2 domains are essential for cavin1 recruitment to caveolae 23. In order to determine the specific region in the cavin1 HR2 domain that facilitates caveola localization, we designed truncation mutants guided by amino acid sequence alignment and secondary structure prediction spanning the length of the HR2 domain (Figs 1A and EV1A). Subcellular localization of each mutant was analysed based on their co‐localization with endogenous CAV1 in A431 cells, which also express endogenous full‐length cavin1 (Fig 1B). Similar to cavin1 HR1 domain, cavin1 truncation mutants containing amino acid residues from 45 to 240 did not associate with CAV1‐containing puncta by confocal microscopy (Fig 1B). However, in clear contrast, addition of cavin1 sequences beyond residue 250 fully restored the co‐localization with CAV1 at the plasma membrane.
Figure EV1. Amino acid sequence alignment of cavin family proteins and PLA analysis of cavin1 truncation mutants.

- Amino acid sequence alignment of cavin family proteins from mouse and zebrafish species highlighting helical regions (HR1 and HR2) and disordered regions (DR1, DR2 and DR3). Highly conserved residues are boxed in red.
- Each GFP‐tagged construct (green) was co‐expressed with cavin1‐mCherry (red) in PC3 line, and PLA was performed as per manufacturer's protocol (described in Materials and Methods) between GFP and CAV1. Greyscale images shows PLA signal as white dots. Scale bars = 10 μm.
- Representative EM micrograph corresponding to Fig 1C showing caveolar morphology of PC3 cells transfected with GFP + cavin1‐mCherry, GFP‐cavin1 (45–230) + Cavin1‐mCherry and GFP‐cavin1 (45–280) + cavin1‐mCherry. GFP fusions and cavin1‐mCherry were co‐expressed in PC3 cells and processed for ruthenium red staining. Scale bar = 100 nm.
We next used the proximity ligation assay (PLA) method to quantitate the association between GFP‐tagged cavin1 truncation mutants and CAV1 in PC3 cells co‐expressing full‐length cavin1‐mCherry (Figs 1C and EV1B). Increased interaction between two proteins is indicated by a higher PLA signal in the form of a greater number of puncta. Control experiments with cavin1 FL‐GFP showed a high PLA signal, whereas the low PLA signal with GFP alone was considered to be the background (Figs 1C and EV1B). In close agreement with the co‐localization experiments (Fig 1B), while C‐terminal truncations beyond residue 240 showed a low PLA signal similar to GFP, the cavin1 (45–250), (45–280) and (45–300) constructs showed a robust PLA signal similar to the full‐length cavin1 protein. Electron microscopy analysis shows that the caveolae generated in the presence of these truncated proteins are morphologically similar to those generated by full‐length cavin1 (Fig EV1C). These results indicate that amino acid residues from 250 onwards are essential for cavin1 localization and generation of caveola structures.
Identification and characterization of undecad repeat (UC1) domain unique to cavin1
We recently identified a repeated sequence of eleven amino acids (undecad or hendecad repeats) within the cavin1 HR2 domain (residues 240–269 in mouse cavin1) that is absent from other cavin family proteins 7. These sequences, we term the UC1 for undecad repeat of cavin1, are highly conserved but are variable in the number of repeats that are present across species (Figs 2A and EV2). Detailed analysis of cavin1 sequences among metazoans highlights the HR1 and HR2 regions as the most conserved structural domains, with the UC1 domain insert in the HR2 region as the primary distinguishing feature from other cavins. Terrestrial vertebrates only possess two copies of the UC1 repeat sequence, with the exception of chicken, mallard (duck) and coelacanth. However, teleost species in the clade Actinopterygii possess two isoforms of cavin1, cavin1a and cavin1b, with three and five copies of the UC1 domain, respectively (Fig 2A). Notably, the great blue mudskipper cavin1b contains six copies of the UC1 domain (Figs 2A and EV2). Domain duplication has been proposed to be important for the evolution of complex functions in vertebrates, although no common genetic mechanism for domain repetition could be discerned 30. The UC1 repeats are always inserted directly between the highly conserved amino acid sequences “KEK” and “LEKTR” pointing to the repetition of the UC1 coding sequence at a specific genomic location within exon 2 (Fig EV2).
Figure 2. Identification and bioinformatics analysis of UC1 domain of cavin1.

- A cladogram was constructed using the sequences shown in Fig EV2, and red numbers at nodes show branch support values. Marked in blue are species in the clade Sarcopterygii; pink (cavin1b) and green (cavin1a) colour represent species in the clade Actinopterygii. The number of UC1 domain repeats in cavin1 is shown in black circles with exceptions marked in squares.
- Amino acid sequence alignment of mouse cavin1 and zebrafish cavin1b HR2 domains highlighting conserved inserts in the HR2 domain.
- The cavin1 HR2 insert is composed of an eleven residue (undecad) repeat of the sequence [ML]EKT[KR]x[KR]T[KR]EN. Mouse (and human) cavin1 contains two repeats, while zebrafish cavin1b contains five.
- Model of human and zebrafish cavin1 undecad repeats based on the undecad repeats of the trimeric autotransporter adhesin EIBD (PDB 2XQH).
Figure EV2. Alignment of cavin1 protein sequences from various species.
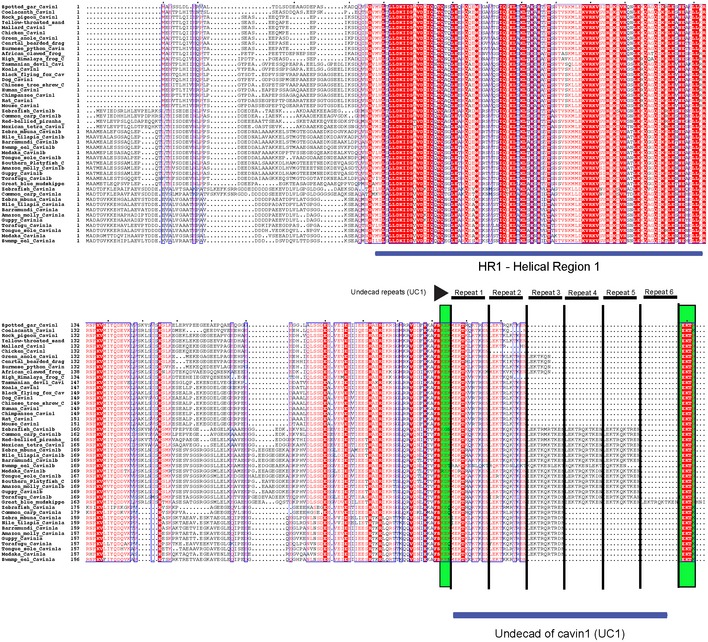
Multiple protein sequence alignment of cavin1 was performed using Multialign program. Repeating units of the UC1 domain are marked with solid lines and the highly conserved amino acid sequences “KEK” and “LEKTR” are marked in light green.
Undecad repeats (with residues labelled abcdefghijk) typically possess hydrophobic residues at a, d and h positions, and are predicted to fold into straight or slightly right‐handed coiled‐coil trimers or tetramers (Fig 2B and C) 31. As the cavin1 N‐terminal HR1 domain forms a trimeric coiled‐coil 23, this suggests a trimeric assembly of the cavin1 UC1 domain is the most probable arrangement within full‐length cavin1. We generated models of this region for both human cavin1 (2 copies of UC1) and zebrafish cavin1b (five copies of UC1) based on the undecad repeats of the trimeric autotransporter adhesin EIBD (PDB 2XQH; Fig 2D). One notable feature of these mouse cavin1 and zebrafish cavin1b models is the presence of alternating acidic and basic ridges along their surfaces. When expressed and purified in E. coli zebrafish cavin1b UC1 domain (zC1bUC1) does not form oligomers in solution (Fig EV3A), and circular dichroism (CD) spectroscopy showed that zC1bUC1 domain is largely random coil in isolation precluding further high‐resolution structural analysis (Fig EV3B). We then generated a chimeric construct [mC1HR1‐3CPL‐zC1bUC1] containing mouse cavin1 HR1 domain (mC1HR1) fused to the UC1 domain of zebrafish cavin1b using a 13 residue‐long flexible linker with a 3C protease cleavage site and rich in glycine/serine (Fig EV3C). Secondary structure comparisons using CD spectroscopy showed a 93% measured α‐helical content for mC1HR1 (Fig EV3D) as expected from its crystal structure 23 and 87% α‐helical content for the chimeric construct mC1HR1‐3CPL‐zC1bUC1 (Fig EV3D). The high percentage of α‐helicity for the chimeric protein infers that the zebrafish cavin1b undecad repeats are also forming significant α‐helical structure in this context. The total predicted α‐helicity of mCHR1‐3CPL‐zC1bUC1 would be < 55% if zebrafish undecad repeats were disordered. The ratio of molar ellipticity values at 222 nm and 208 nm for mC1HR1 [(MRE)222/(MRE)208 – 1.04] and mC1HR1‐3CPL‐zC1bUC1 [(MRE)222/(MRE)208 – 1.13] is higher than 1 indicating the presence of coiled‐coils in both domains, which is consistent with our structural predictions. Consistent with formation of an extended structure, the chimeric mC1HR1‐3CPL‐zC1bUC1 protein also showed a higher stability than mC1HR1 in the presence of guanidine hydrochloride denaturant (Fig EV3E).
Figure EV3. In vitro biochemical analysis of zebrafish cavin1b UC1 domain.

- Size exclusion chromatography of zebrafish cavin1b UC1 domain expressed and affinity purified in E. coli analysed by SDS–PAGE and Coomassie Blue stain for protein. Molecular weight markers on the top with arrows represent size exclusion chromatography elution positions for standard globular proteins, while markers on the left indicate SDS–PAGE denatured protein markers.
- Circular dichroism spectroscopy analysis of zebrafish cavin1b UC1 domain showing spectra typical to largely random coil protein.
- Schematic representation of fusion construct used in (D) and (E).
- Circular dichroism spectroscopy analysis of mouse cavin1 HR1 domain and fusion construct mC1HR1‐3CPL‐zC1bUC1 with measured α‐helical content using K2D3 server.
- Plot of fraction of folded protein for mC1HR1 and mC1HR1‐3CPL‐zC1b UC1 fusion using guanidine hydrochloride denaturant.
Our truncation analyses highlight the UC1 domain as being required for caveola association of cavin1 (Fig 1). To confirm the role of the UC1 domain of cavin1 in caveolar localization, we generated a series of mutants as shown in Fig 3A and analysed their subcellular localization in A431 cells. In comparison with the full‐length protein's punctate co‐localization with CAV1, the Δ2UC1 cavin1‐GFP deletion mutant and the slightly longer deletion ΔHR2 cavin1‐GFP both showed a cytoplasmic distribution with a reduced cavin1 puncta at the plasma membrane (Figs 3B and EV4A). Δ2UC1 cavin1‐GFP and ΔHR2 cavin1‐GFP over‐expression in A431 cells also caused a significant reduction in CAV1 puncta at the plasma membrane (Fig EV4B), suggesting that mutants lacking the HR2 domain or UC1 may associate with endogenous cavin1 and cause dominant negative inhibition of its ability to form caveolae. These data are also in agreement with studies of cavin1 mutants in HeLa cells 32. Overall, these studies highlight the importance of the UC1 domain in caveola formation.
Figure 3. The UC1 domain of cavin1 is important for caveola function in vivo .

- Schematic representation of UC1 and HR2 domain deletions in cavin1.
- GFP‐tagged WT cavin1, Δ2 UC1 cavin1, ΔHR2 cavin1 were expressed in A431 cells and co‐stained with CAV1 antibody (red), scale bar = 10 μm.
- GFP‐tagged cavin1b full‐length and ∆5 UC1 cavin1b expression in 60 and 90 hpf live cavin1b −/− embryos. Scale bar = 100 μm.
- Live bright field images of uninjected cavin1b −/− zebrafish, and cavin1b −/− zebrafish injected with zcavin1b and ∆5 UC1 zcavin1b. 60 hpf and 90 hpf timepoints shown. Embryos at 60 hpf were electrically stimulated (10 min) and imaged at 90 hpf. Injected WT zebrafish presented no observable lesions even upon electrical stimulation (n = 75, clutch‐N = 3). Arrowheads indicate lesions. “S” next to arrowhead denotes a severe lesion. Scale bar = 100 μm.
- Plot of lesion number for 60 hpf embryos, 90 hpf embryos and 90 hpf embryos electrically stimulated at 60 hpf for 10 min. For 60 hpf, 90 hpf and 90 hpf stimulated; fish number (uninjected) = 84, 43, 39 respectively, fish number (zcavin1b‐GFP) = 41, 18, 24 respectively, fish number (∆5 UC1 zcavin1b‐GFP) = 52, 22, 28, respectively. Ordinary one‐way ANOVA, ns = not significant, ****P < 0.0001. Data are presented as mean, and error bars indicate standard deviation.
Figure EV4. Quantitative analysis (related to Fig 3B and E).

- Quantitation of cavin1‐GFP punctae in A431 cells (related to Fig 3B). Data are presented as mean, and error bars indicate standard deviation. N = 2 (independent biological replicates), n = 10–15 (sample size) ***P < 0.001; one‐way ANOVA followed by Bonferroni's multiple comparisons.
- Quantitation of CAV1 punctae in A431 cells (related to Fig 3B) after expression of GFP‐tagged WT cavin1, Δ2 UC1 cavin1 and ΔHR2 cavin1. Data are presented as mean, and error bars indicate standard deviation. N = 2 (independent biological replicates), n = 10–15 (sample size) ***P < 0.001, **P < 0.01; one‐way ANOVA followed by Bonferroni's multiple comparisons.
- Plot of lesion severity index for 60 hpf embryos, 90 hpf embryos and 90 hpf embryos electrically stimulated at 60 hpf for 10 min. For 60 hpf, 90 hpf and 90 hpf stim; fish number (uninjected) = 84, 43, 39, respectively, fish number (zcavin1b‐GFP) = 41, 18, 24, respectively, fish number (∆5 UC1 zcavin1b‐GFP) = 52, 22, 28, respectively. Data are presented as mean and error bars indicate standard deviation. ns = not significant, ****P < 0.0001; one‐way ANOVA followed by Bonferroni's multiple comparisons.
Importance of the UC1 domain in vivo
We, and others, recently showed that genetic ablation of notochord‐specific cavin1b in zebrafish using CRISPR/Cas9 results in the formation of characteristic lesions in vacuolated notochord cells during development 25, 33. The zebrafish notochord plays an important role in facilitating locomotion by providing a stiff but flexible midline structure for bracing muscle contractions and is constantly exposed to mechanical stress. The phenotypic readout of the cavin1b −/− mutant is to measure lesion numbers and severity. In initial experiments, we confirmed that re‐expressing zebrafish cavin1b (zcavin1b) with a C‐terminal GFP tag was able to significantly rescue the lesion phenotype, reducing both the number and severity of lesions (Figs 3C and D, and EV4C). Constructs were microinjected into embryos at the one‐cell stage, and embryos with ubiquitous GFP fluorescence were selected at 24 hpf and screened at 60 and 90 hpf to confirm similarity in expression levels across groups (Fig 3C). In contrast, despite similar expression levels, zcavin1b lacking the UC1 domain (∆5UC1; deletion of all five undecad repeats, amino acids from 252–317) was unable to restore the protective activity of zcavin1b in the notochord (Figs 3D and E, and EV4C).
We then used constant voltage electrical stimulation to exaggerate the mechanical stress by inducing muscle contraction on the notochord of cavin1b −/− zebrafish. At 60 hpf, cavin1b −/− embryos injected with wild‐type or mutant zcavin1b were subjected to electrical stimulation for 10 min and lesions were scored 30 h later. Cavin1b −/− zebrafish injected with wild‐type zcavin1b appeared to be protected from mechanical stress due to stimulation, as there were no significant differences between stimulated and unstimulated groups (Figs 3D and E, and EV4C). However, the cavin1b −/− zebrafish expressing the ∆5UC1 mutant showed a significant increase in lesion number and severity after stimulation (38.8% ± 11.7) similar to uninjected cavin1b −/− zebrafish (40.8% ± 12.6; Fig 3E). These experiments confirm the importance of the UC1 domain in modulating the mechanical properties of zebrafish notochord membrane and provide a valuable insight into the function of the UC1 domain in vivo.
The UC1 domain of cavin1 is important for membrane binding and the stability of caveolae
Our structural model of the human UC1 domain (analogous to mouse cavin1) highlights multiple surface exposed lysine/arginine residues that may potentially be involved in lipid binding during caveola biogenesis (Fig 2D). To test this, we mutated six basic residues (K245E, K247E, R249D, K256E, R258D, K260E; 6KR/ED) in the UC1 region of mouse cavin1 and analysed its subcellular localization in A431 cells. The 6KR/ED cavin1‐GFP mutant showed a punctate membrane distribution similar to wild‐type cavin1 in A431 cells most likely due to the presence of endogenous cavin1 (Fig 4A). In the absence of endogenous cavin1 in PC3 cells, 6KR/ED mutant showed a moderate reduction in cavin1 puncta formation compared to WT cavin1 (Fig EV5A). This indicates that the potential lipid binding residues of the UC1 domain are important for caveola recruitment and formation. We next performed liposome binding assays with GFP‐tagged cavin1 purified from HEK293 cells (Fig 4B). Varying the molar percentage (mol%) of phosphatidylinositol(4,5)bisphosphate [PI(4,5)P 2] and phosphatidylserine (PS) in liposomes was used to test whether mutating the UC1 domain altered the relative affinities for either of these negatively charged lipids. While wild‐type cavin1 showed a clear increase in membrane binding with increasing concentrations of both lipids, the 6KR/ED cavin1 mutant showed a clearly reduced affinity for PS, while not affecting PI(4,5)P 2 interactions. This suggests that these basic residues in the UC1 domain are involved in co‐operative PS binding. In line with previous work 23, this supports a model whereby the HR1 domain contributes primarily to PI(4,5)P2 binding while the UC1 domain preferentially associates with PS. Thus, cavin1 contains two distinct positively charged regions with differential lipid binding preferences. The functional implications of these differences are yet to be fully understood, but it has been postulated that cavins could potentially act as lipid organizers during caveola formation while controlling the release of specific lipids into non‐caveolae membrane upon disassembly 7. We thus propose that caveola formation involves multiple co‐operative cavin1‐membrane interactions where the unique UC1 domain of cavin1 is important for efficient caveola formation.
Figure 4. The UC1 domain of cavin1 modulates the stability of caveolae.
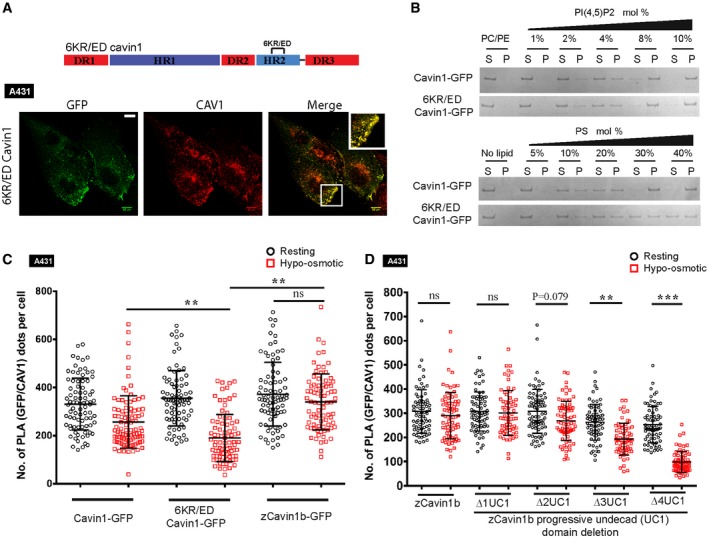
-
ASchematic representation of undecad region mutation in cavin1. GFP‐tagged 6KR/ED Cavin1 was expressed in A431 cells and co‐stained with CAV1 antibody (red), scale bar = 10 μm.
-
BLiposome binding and SDS–PAGE analysis with varying mol% of either PI(4,5)P2 or PS and purified GFP‐tagged cavin1 proteins. (S) indicates supernatant, and (P) indicates liposome pellet bound fraction.
-
C, DA431 cells expressing GFP‐tagged cavin1, 6KR/ED cavin1, zebrafish cavin1b (C) and zebrafish cavin1b mutants (D) with progressive deletion of undecad repeat units (denoted as Δ1, Δ2, Δ3 and Δ4 UC1) were subjected to osmotic swelling assay followed by in situ PLA using antibodies specific to CAV1 and cavin1. The PLA signal between GFP‐tagged cavin constructs and endogenous CAV1 before (resting—black circle) and after (hypotonic—red square) osmotic swelling was quantitated using ImageJ and plotted as number of PLA dots per cell, N = 3 (independent biological replicates), n = 25–30 (sample size). Data are presented as mean, and error bars indicate standard deviation. ***P < 0.001, **P < 0.01, ns = not significant; one‐way ANOVA followed by Bonferroni's multiple comparisons.
Figure EV5. Representative images related to Fig 4C.

- Quantitation of GFP‐positive puncta in PC3 cells after expression of GFP‐tagged constructs as shown. N = 2 (independent biological replicates), n = 10–15 (sample size). Data are presented as mean, and error bars indicate standard deviation. Scale bars = 10 μm.
- Representative images of PLA analysis presented in Fig 4C. Scale bars = 10 μm.
To characterize the functional importance of the UC1 domain of cavin1, we used an established cell swelling method to study the disassembly of caveolae under mechanical force. Recent studies have shown that mechanical stress causes caveolar disassembly and release of caveolar components into the cytosol including an increase in the cytosolic pool of cavin1 15, 34, 35. We first performed PLA to assess the changes in Cavin1/CAV1 association in A431 cells subjected to membrane stretching by exposure to hypo‐osmotic medium (Figs 4C and EV5B). A431 cells expressing cavin1‐GFP showed an expected reduction in PLA signals upon cell treatment, consistent with reduced caveola localization. The 6KR/ED cavin1‐GFP mutant protein showed an even greater reduction in PLA signal with CAV1 (46.5% decrease versus 22% for the wild‐type protein; Fig 4C), which supports a role for membrane association by the UC1 domain in stabilizing caveolae against membrane tension. Zebrafish cavin1b in contrast, which contains three additional UC1 sequences, showed only a marginal 8.7% decrease in its association with CAV1 in response to hypo‐osmotic treatment (Fig 4C). This raised the possibility that increased numbers of undecad repeats might enhance caveola stability. To test this, we performed systematic truncations of the zcavin1b undecad repeats. The complete deletion of all five undecad units from zebrafish cavin1b increases the cytosolic pool of protein with a marked reduction in cavin1 puncta (Fig EV5A) upon expression in PC3 cells. As such this protein was not included in caveola stability studies. Compared to full‐length zcavin1b, the deletion of two C‐terminal undecad units has a marginal effect on the stability of caveolae. However, deletion of three or four undecad repeats caused significantly reduced caveola stability in response to hypo‐osmotic treatment; association of the cavin mutants with CAV1 was not significantly different from WT CAV1 under control conditions but significantly reduced upon hypo‐osmotic treatment (Fig 4D). These results suggest that the presence of multiple copies of the UC1 sequences, such as in zebrafish cavin1b, can enhance the resistance of caveolae to disassembly under mechanical stress.
The UC1 domain promotes membrane remodelling
We next asked whether the UC1 domain might be sufficient to promote a similar caveola biogenesis activity when inserted into cavin2, which is unable to make caveolae in PC3 cells that lack endogenous cavin1. Mouse cavin1 amino acid residues 240–269 were inserted into the cavin2 HR2 domain (following equivalent cavin2 amino acid residue 246), and we compared the subcellular distribution of cavin1‐GFP and cavin2‐GFP with the UC1‐containing cavin2‐GFP mutant in PC3 cells. Cavin1‐GFP expression in PC3 cells induces caveola formation and shows a typical punctate distribution co‐localizing with endogenous CAV1 (Fig 5A). Cavin2‐GFP shows a predominantly cytosolic distribution when expressed alone (Fig 5A), with no co‐localization with CAV1 (note that a small proportion of very highly expressing cells showed membrane tubules decorated with cavin2‐GFP, consistent with previous studies 36). In contrast to cavin2, UC1 cavin2‐GFP expressed alone showed a striking redistribution to the plasma membrane, forming extensive tubules that partially co‐localized with CAV1 but did not result in formation of typical caveola puncta (Fig 5A). Electron microscopy of cells using APEX2 fused to a GFP‐binding protein 37 showed that APEX2 reaction product was predominantly observed in the cytosol of PC3 cells expressing only cavin2‐GFP, but was clearly evident on patches of the plasma membrane as well as extensive tubular structures when UC1 cavin2‐GFP was expressed (Fig 5B). Liposome binding analysis of cavin2‐GFP purified from HEK293 cells showed an increase in PS binding by the addition of the UC1 domain to cavin2 with little effect on PI(4,5)P2 binding (Fig 5C). While these experiments show that insertion of the UC1 domain into cavin2 does not confer normal caveola‐forming activity on its own, it does confer a remarkable ability to bind and tubulate the plasma membrane. We thus propose that the UC1 domain of cavin1 is essential for the unique ability of cavin1 to organize membrane domains that co‐localize with CAV1 and to remodel the plasma membrane by providing an additional PS binding interface.
Figure 5. Inserting the UC1 domain into cavin2 induces extensive plasma membrane tubulation.
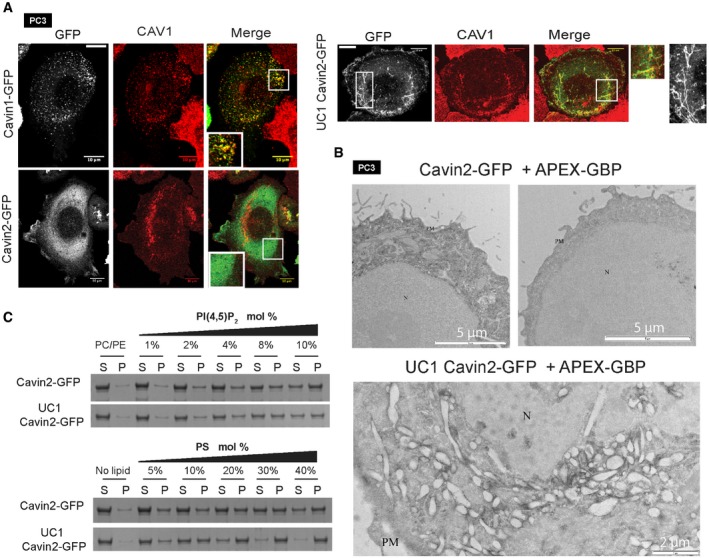
- Expression of GFP‐tagged WT cavin1, WT cavin2 and UC1 cavin2 in PC3 cell line and co‐localization with CAV1. Tubule morphologies are highlighted in the right‐hand greyscale image with expanded representative regions shown. Scale bars = 10 μm and 1 μm (insets).
- GFP‐tagged WT cavin2 and UC1 cavin2 were co‐expressed in PC3 cells with APEX‐GBP, fixed in 2.5% gluteraldehyde and processed for DAB reaction. The WT cavin2 shows only diffuse staining, while cells expressing UC1 cavin2‐GFP show a large accumulation of membrane tubules coated by the cavin2‐GFP mutant protein. N—nucleus, PM—plasma membrane
- Liposome binding and SDS–PAGE analysis with varying mol% of either PI(4,5)P2 or PS and purified GFP‐tagged cavin2 proteins as shown. (S) indicates supernatant, and (P) indicates liposome pellet bound fraction.
In summary, we propose that the caveolar coat is built with multiple homo‐ and hetero‐oligomeric species of cavin1, cavin1‐cavin2 and cavin1‐cavin3, as well as through relatively low‐affinity interactions with CAV1, other membrane proteins 29 and specific membrane lipids including phosphoinositides via the HR1 domain 23 and PS via the UC1 domain (this study). How the extended UC1 domain combines with the trimeric HR1 coiled‐coil domain to form the extended rod‐like structure of the core cavin assembly remains to be determined. The curvature of the membrane and the unique lipid environment promoted by caveolin may also be a key factor in the assembly of caveolae. We propose that the multiple low‐affinity interactions involved in building the cavin coat are also crucial for caveola disassembly, a rapid energy‐independent process which occurs in response to increased membrane tension 10, 15. Our data show that the caveolar disassembly is counter‐balanced by the stabilizing effect of the UC1 domain of cavin1. Recent studies have characterized the role of other caveolar coat components such as EHD proteins that contribute to the mechano‐protective activity of caveolae 38 and our studies now suggest that caveolar stability in different organisms/tissues has been further tuned by the number of repeats in the cavin1 UC1 domain. Defining the roles of specific caveolar proteins and domains in membrane association, membrane remodelling, protein oligomerization, and the cycles of caveola disassembly and reassembly will be essential for understanding the multiple functions controlled by caveolae.
Materials and Methods
Cell lines maintenance and materials
PC3 cells were maintained in RPMI medium (Gibco® Life technologies), while A431 and MCF7 cell lines were maintained in DMEM medium (Gibco® Life technologies) supplemented with 10% foetal bovine serum (FBS) and penicillin/streptomycin. For all experiments, 2 × 105 PC3 or MCF7 cells or 1 × 105 A431 cells were plated in either 6‐well culture dishes (Nunc™, Cat. No. 140675, culture area—9.6 cm2) or 35‐mm tissue culture dishes (TPP® 93040, culture area—9.2 cm2). Antibodies used were as follows: rabbit polyclonal anti‐caveolin1 (BD Transduction Laboratories, Cat. No. 610060), mouse monoclonal anti‐GFP (Roche Diagnostics Cat. No. 11814460001, Dilution 1:1,000), Donkey anti‐Rabbit IgG (H+L) Secondary Antibody Alexa Fluor® 647 conjugate.
Zebrafish line and maintenance
Zebrafish were raised, fed and maintained according to institutional guidelines (University of Queensland). Zebrafish embryos were obtained from paired matings of adult zebrafish and raised at 28.5°C in standard E3 media (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2 and 0.33 mM MgSO4). The following zebrafish strains were used: wild‐type (TAB) and cavin1b −/−uq7rp 25. All animal experiments were approved by The University of Queensland Animal Ethics Committee.
Molecular cloning and plasmids
All mouse cavin1 truncations were cloned in pEGFP‐C1 vector at the EcoRI site using an overlap extension PCR strategy, to be expressed with an N‐terminal GFP fusion. All cavin domain mutants, cavin2 undecad insertions, cavin1 undecad and HR2 deletion were cloned in pEGFP‐C1 vector at EcoRI site using circular polymerase extension cloning (CPEC) method 39. Cavin1‐mCherry 26 and APEX‐GBP 37 are described previously. Zebrafish Cavin1b, mutant of zcavin1b lacking UC1 domain (∆5UC1; deletion of amino acids from 252–317) and sequential undecad repeats deletion mutants were cloned in pCS2+GFP at EcoRI site using overlap extension strategy with C‐terminal GFP tag.
RNA synthesis
pCS2+GFP vector containing zcavin1b or ∆5UC1 zcavin1b was linearized and purified using the QIAquick PCR Purification Kit (Qiagen), and products were checked on a 3% TBE agarose gel. Purified products were then transcribed using the mESSAGE mMACHINE SP6 Transcription Kit (Ambion) and purified using Zymo Research RNA Clean & Concentrator Kit.
Zebrafish stimulation assay
Zebrafish microinjection and electrical stimulation were carried out as previously described with modifications 25. Briefly, yolk microinjection (Purified zC1b or ∆5UC1 zC1b, 400 μg/ml with phenol red as an indicator) was carried out on one‐cell stage zebrafish embryos. Embryos ubiquitously expressing green fluorescence were selected at 1 dpf, and screened again at 60 hpf and 90 hpf to confirm similarity in expression levels. At 60 hpf, embryos were dechorionated by incubation in 1 mg/ml pronase solution at RT for 5 min and then washed three times in E3 media. 60 hpf notochord lesion analysis was then carried out as described below, before dividing embryos into unstimulated control and stimulated groups. 60 hpf embryos in stimulated groups were stimulated using a constant voltage electrical stimulator (Square Pulse Stimulator S44, Grass Instruments) with the following settings: stimulation rate: 4 pps × 0.1, delay: 9 ms × 0.1, duration: 0 ms × 0.1, voltage: 12 V × 10 for 10 min. Zebrafish were then rested at 28°C before 90 hpf notochord lesion analysis.
Zebrafish notochord lesion analysis
For notochord lesion analysis, zebrafish were first anesthetized in tricaine (0.16 mg/ml in E3 media) before lesion number and severity was measured and analysed 25. Lesion number represents mean number of lesions counted per notochord of each embryo. Lesion severity uses a severity index scoring system, which scores mild (slightly delaminated notochord cells), moderate (more cell delamination compared to mild lesions with occasional fragmented vacuoles) and severe lesions (dense cellular regions flanked by, and/or consist of fragmented vacuoles) within one notochord with the arbitrary values of 2, 3 and 4, respectively. Statistical analyses were carried out using ordinary one‐way ANOVA with post hoc Tukey's multiple‐comparison test.
Cavin1 bioinformatics
The undecad FASTA sequence with query subranges 1–55 (zebrafish cavin1b—five repeats) or 1–66 (mudskipper cavin1b—six repeats; see Appendix) was searched using BLASTP (NCBI) via the nr database with additional search sets including and excluding organisms teleostei,(taxid:32443), sarcopterygii (taxid:8287) and actinopterygii (taxid:7898). Refseq sequences corresponding to selected taxonomic groups as appeared in resulting BLAST Taxonomy Report pages were aligned using the T‐Coffee program (Myers‐Miller mode) via the MacVector software package to identify paralog proteins. Phylogenetic reconstruction was carried out using Phylogeny.fr web service 40.
Protein purification from mammalian cells
HEK 293 cells were maintained in DMEM supplemented with 10% FBS and plated in 100‐mm cell culture dishes (Corning® 430176). Transfections of GFP‐tagged constructs were performed using polyethylenimine (PEI) transfection reagent (Sigma‐Aldrich Cat. No. 408727) with 1:4 w/w ratio (DNA:PEI), and cells were harvested 24 h post‐transfection. Cell lysis was performed in 20 mM Hepes‐KOH pH 7.6, 500 mM NaCl buffer containing 1% Triton X‐100 with three times 3‐s sonication pulse at output power 10. Lysate was then centrifuged at 5,000× g for 10 min, and supernatant fraction was incubated with purified GFP nanobody tagged with MBP for 30 min at 4°C. Finally, lysate nanobody mixture was incubated with amylose resin (NEB Cat No. E8021L) for 2 h at 4°C. Amylose resin was then extensively washed with 20 mM Hepes‐KOH pH 7.6, 500 mM NaCl buffer containing 1% Triton X‐100, and elution was performed in 20 mM Hepes‐KOH pH 7.6, 500 mM NaCl buffer containing 20 mM Maltose (Sigma‐Aldrich Cat. No. M2250). Elute was concentrated with Amicon® Ultra‐4 10kD cut‐off (Merck‐Millipore Cat. No. UFC801024) and finally loaded on Superose 6 10/30GL size exclusion column equilibrated in 20 mM Hepes‐KOH pH 7.6, 150 mM NaCl. Concentration of gel filtration elute was avoided as it leads to precipitation in lower salt content.
Recombinant protein expression and purification
E.coli strain Rosetta™(DE3)pLysS (Novagen) was used for protein expression and purification. Briefly, E. coli cells expressing respective construct were propagated in TB media for 16 h at 20°C. Cells were disrupted in 20 mM Hepes‐KOH pH 7.6, 500 mM NaCl with subsequent addition of Triton X‐100 followed by high‐speed centrifugation to remove cell debris. Purification of His‐ubiquitin tagged proteins was done using HisTrap HP cartridges (GE Healthcare, Cat. No. 17‐5247‐01) by the addition of 500 mM imidazole to elution buffer. Removal of ubiquitin tag was done using de‐ubiquitination protease prepared as described previously 23. Protein samples after tag cleavage were loaded on Superdex 75 10/30GL gel filtration column. Size exclusion chromatography analysis was done in gel filtration buffer (20 mM Hepes‐KOH pH 7.6, 500 mM NaCl).
Circular Dichroism (CD) spectroscopy
CD measurements were performed on a Jasco J‐810 spectropolarimeter (JASCO Corp) in 20 mM Hepes‐KOH pH 7.6, 500 mM NaCl using a 0.1 mm path‐length cuvette at 25°C. Data were collected from 250 to 200 nm with 1 mn resolution at 50 nm/min, and two acquisitions were averaged per sample. Protein concentration was kept at 50 μM for all measurements. Estimation of helical content was done using K2D3 server (http://cbdm-01.zdv.uni-mainz.de/~andrade/k2d3/). Guanidine hydrochloride (GdnHCl) induced denaturation of protein was done in 20 mM Hepes‐KOH pH 7.6, 500 mM NaCl buffer at room temperature by adding increasing amount of GdnHCl to a fixed protein concentration (50 μM). The molar fraction of folded protein was calculated from equation f = ([q]−[q]u)/([q]n−[q]u) where [q] is the observed mean residue ellipticity at any particular denaturant concentration. [q]n and [q]u is mean residue ellipticities of the native folded and denatured states.
Liposome preparation and binding analysis
Liposomes were prepared by rehydration of lipid films composed of phosphatidylcholine (PC), phosphatidylethanol (PE) with increasing mole percentage of either PI(4,5)P2 (1, 2, 4, 8, 10%) or phosphatidylserine (PS; 5, 10, 20, 30, 40%) in 20 mM Hepes‐KOH pH 7. 6, 150 mM NaCl buffer followed by 10 freeze/thaw cycles and vigorous vortexing to generate multilamellar vesicles. Liposome binding analysis was done by mixing purified proteins and lipids in specific protein : lipid ratio (1:25 for cavin1 and 1:10 for cavin2), followed by 10‐min incubation at room temperature and subsequent centrifugation at 60,000× g at 22°C for 10 min.
Osmotic swelling assay
Cells were incubated in pre‐warmed hypo‐osmotic medium containing 3:7 ratio of DMEM with 10% FBS/sterile water (constituting 70% hypo‐osmotic medium) for 15 min.
Immunofluorescence and proximity ligations assay (PLA)
A431 and PC3 cells were grown at 40–60% confluency in DMEM or RPMI 1640 medium supplemented with 10% FBS. Cells were then transfected with each domain construct using Lipofectamine 2000 (Invitrogen) as per manufacturer's instructions. Cells were fixed 24 h post‐transfection with 4% paraformaldehyde in phosphate‐buffered saline (PBS) at 4°C and subsequently permeabilized with 0.1% Triton X‐100 in PBS for 7 min. Cells were probed with CAV1 antibody (dilution 1:800) and anti‐rabbit secondary antibody (2 μg/ml). Confocal images were acquired on LSM 510 Meta (Carl Zeiss, Inc) equipped with 63× oil immersion objective, NA 1.4. Images were acquired at different laser power for GFP‐tagged truncation mutants and detector gain settings in order to avoid oversaturation of pixels. This was done because all cavin domain constructs express different levels of protein. All images were processed for background correction and brightness/contrast using ImageJ.
For PLA, A431 and PC3 cells were fixed in 4% paraformaldehyde in PBS at 4°C and subsequent permeabilization with 0.1% Triton X‐100 in PBS (7 min) was performed. Cells were then incubated with primary antibodies for GFP and CAV1 for 1 h (1:1,000 dilution‐ PC3 and 1:800 dilution A431). Afterwards, in situ PLA was performed as per manufacturer's instructions using Duolink anti‐Mouse MINUS and anti‐Rabbit PLUS In Situ PLA probes and the Duolink In Situ Detection Reagents Red. Images were then acquired on Zeiss LSM 710 confocal microscope (Carl Zeiss, Inc) equipped with 63× oil immersion objective, and quantitation of PLA dots per cell was performed using find maxima function in ImageJ with offset of 50 (A431) and 40 (PC3). Quantitation of cavin1/CAV1 puncta for A431 cell line was also done using find maxima function in ImageJ with offset of 35 (cavin1) and 45 (CAV1).
Electron microscopy
PC3 cells were plated onto 30‐mm tissue culture dishes. Cells were allowed to adhere to dishes for 48 h prior to transfection. Cells were then co‐transfected with APEX‐GBP 37 and wild‐type cavin2‐GFP or undecad cavin2‐GFP, fixed in 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (cacodylate; pH 7.4) for 1 h. DAB (3,3′diaminobenzidine tetrahydrochloride, Sigma‐Aldrich Cat. No. D5905) reaction was then performed as follows. Briefly, cells were treated with DAB/cacodylate mixture (DAB final concentration—1 mg/ml) + 5.88 mM H2O2 (hydrogen peroxide, Sigma‐Aldrich Cat. No. H1009) for 2 min. Cells were then washed with 0.1 M sodium cacodylate buffer and fixed with 1% osmium tetroxide for 2 min. Cells were then embedded in LX112 resin, and thin sections were cut as described previously 37. Images were acquired on JEOL 1011 electron microscope at 80 kV. Caveola morphology was analysed by fixing PC3 cells in the presence of ruthenium red and processed as described previously 8.
Statistical analyses
Statistical analysis and P‐value calculations were performed by one‐way ANOVA (unless stated otherwise) followed by Bonferroni's multiple comparisons using GraphPad Prism software.
Author contributions
RGP and BMC conceived the project and study. VAT performed molecular cloning, biochemical and cellular assay. Y‐WL performed all animal (zebrafish) studies with the help of HPL and TEH. OK and BMC explored the undecad repeats concept. CF performed cellular processing for electron microscopy and RGP acquired images and interpreted the data. MB and K‐AM assisted in cellular assays. SM and KA assisted in reagent preparation and data analysis. All authors commented on the manuscript. VAT, Y‐WL, BMC and RGP wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Review Process File
Acknowledgements
This work was supported by grants from the National Health and Medical Research Council of Australia (NHMRC; to RGP Grant Number APP569542 and APP1037320) and the Australian Research Council (ARC; to BMC, Grant Number DP120101298). RGP is supported by an NHMRC Senior Principal Research Fellowship from the NHMRC (APP1058565) and by the Australian Research Council Centre of Excellence in Convergent Bio‐Nano Science and Technology (R.G. Parton). BMC is supported by NHMRC Career Development Fellowship (APP1061574). Confocal microscopy was performed at the Australian Cancer Research Foundation (ACRF)/Institute for Molecular Bioscience (IMB) Dynamic Imaging Facility for Cancer Biology, established with funding from the ACRF.
EMBO Reports (2018) 19: e45775
Contributor Information
Brett M Collins, Email: b.collins@imb.uq.edu.au.
Robert G Parton, Email: r.parton@imb.uq.edu.au.
References
- 1. Parton RG, del Pozo MA (2013) Caveolae as plasma membrane sensors, protectors and organizers. Nat Rev Mol Cell Biol 14: 98–112 [DOI] [PubMed] [Google Scholar]
- 2. Rothberg KG, Heuser JE, Donzell WC, Ying YS, Glenney JR, Anderson RG (1992) Caveolin, a protein component of caveolae membrane coats. Cell 68: 673–682 [DOI] [PubMed] [Google Scholar]
- 3. Scherer PE, Okamoto T, Chun M, Nishimoto I, Lodish HF, Lisanti MP (1996) Identification, sequence, and expression of caveolin‐2 defines a caveolin gene family. Proc Natl Acad Sci USA 93: 131–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Way M, Parton RG (1995) M‐caveolin, a muscle‐specific caveolin‐related protein. FEBS Lett 376: 108–112 [DOI] [PubMed] [Google Scholar]
- 5. Drab M, Verkade P, Elger M, Kasper M, Lohn M, Lauterbach B, Menne J, Lindschau C, Mende F, Luft FC et al (2001) Loss of caveolae, vascular dysfunction, and pulmonary defects in caveolin‐1 gene‐disrupted mice. Science 293: 2449–2452 [DOI] [PubMed] [Google Scholar]
- 6. Razani B, Combs TP, Wang XB, Frank PG, Park DS, Russell RG, Li M, Tang B, Jelicks LA, Scherer PE et al (2002) Caveolin‐1‐deficient mice are lean, resistant to diet‐induced obesity, and show hypertriglyceridemia with adipocyte abnormalities. J Biol Chem 277: 8635–8647 [DOI] [PubMed] [Google Scholar]
- 7. Kovtun O, Tillu VA, Ariotti N, Parton RG, Collins BM (2015) Cavin family proteins and the assembly of caveolae. J Cell Sci 128: 1269–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hill MM, Bastiani M, Luetterforst R, Kirkham M, Kirkham A, Nixon SJ, Walser P, Abankwa D, Oorschot VM, Martin S et al (2008) PTRF‐Cavin, a conserved cytoplasmic protein required for caveola formation and function. Cell 132: 113–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu L, Brown D, McKee M, Lebrasseur NK, Yang D, Albrecht KH, Ravid K, Pilch PF (2008) Deletion of Cavin/PTRF causes global loss of caveolae, dyslipidemia, and glucose intolerance. Cell Metab 8: 310–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gambin Y, Ariotti N, McMahon KA, Bastiani M, Sierecki E, Kovtun O, Polinkovsky ME, Magenau A, Jung W, Okano S et al (2014) Single‐molecule analysis reveals self assembly and nanoscale segregation of two distinct cavin subcomplexes on caveolae. Elife 3: e01434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hansen CG, Shvets E, Howard G, Riento K, Nichols BJ (2013) Deletion of cavin genes reveals tissue‐specific mechanisms for morphogenesis of endothelial caveolae. Nat Commun 4: 1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McMahon KA, Zajicek H, Li WP, Peyton MJ, Minna JD, Hernandez VJ, Luby‐Phelps K, Anderson RG (2009) SRBC/cavin‐3 is a caveolin adapter protein that regulates caveolae function. EMBO J 28: 1001–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hernandez VJ, Weng J, Ly P, Pompey S, Dong H, Mishra L, Schwarz M, Anderson RG, Michaely P (2013) Cavin‐3 dictates the balance between ERK and Akt signaling. Elife 2: e00905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schneider K, Köcher T, Andersin T, Kurzchalia T, Schibler U, Gatfield D (2012) CAVIN‐3 regulates circadian period length and PER:CRY protein abundance and interactions. EMBO Rep 13: 1138–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sinha B, Koster D, Ruez R, Gonnord P, Bastiani M, Abankwa D, Stan RV, Butler‐Browne G, Vedie B, Johannes L et al (2011) Cells respond to mechanical stress by rapid disassembly of caveolae. Cell 144: 402–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hayashi YK, Matsuda C, Ogawa M, Goto K, Tominaga K, Mitsuhashi S, Park YE, Nonaka I, Hino‐Fukuyo N, Haginoya K et al (2009) Human PTRF mutations cause secondary deficiency of caveolins resulting in muscular dystrophy with generalized lipodystrophy. J Clin Invest 119: 2623–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rajab A, Straub V, McCann LJ, Seelow D, Varon R, Barresi R, Schulze A, Lucke B, Lutzkendorf S, Karbasiyan M et al (2010) Fatal cardiac arrhythmia and long‐QT syndrome in a new form of congenital generalized lipodystrophy with muscle rippling (CGL4) due to PTRF‐CAVIN mutations. PLoS Genet 6: e1000874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim CA, Delepine M, Boutet E, El Mourabit H, Le Lay S, Meier M, Nemani M, Bridel E, Leite CC, Bertola DR et al (2008) Association of a homozygous nonsense caveolin‐1 mutation with Berardinelli‐Seip congenital lipodystrophy. J Clin Endocrinol Metab 93: 1129–1134 [DOI] [PubMed] [Google Scholar]
- 19. Gazzerro E, Bonetto A, Minetti C (2011) Caveolinopathies: translational implications of caveolin‐3 in skeletal and cardiac muscle disorders. Handb Clin Neurol 101: 135–142 [DOI] [PubMed] [Google Scholar]
- 20. Garg A, Agarwal AK (2008) Caveolin‐1: a new locus for human lipodystrophy. J Clin Endocrinol Metab 93: 1183–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hayer A, Stoeber M, Bissig C, Helenius A (2010) Biogenesis of caveolae: stepwise assembly of large caveolin and cavin complexes. Traffic 11: 361–382 [DOI] [PubMed] [Google Scholar]
- 22. Ludwig A, Howard G, Mendoza‐Topaz C, Deerinck T, Mackey M, Sandin S, Ellisman MH, Nichols BJ (2013) Molecular composition and ultrastructure of the caveolar coat complex. PLoS Biol 11: e1001640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kovtun O, Tillu VA, Jung W, Leneva N, Ariotti N, Chaudhary N, Mandyam RA, Ferguson C, Morgan GP, Johnston WA et al (2014) Structural insights into the organization of the cavin membrane coat complex. Dev Cell 31: 405–419 [DOI] [PubMed] [Google Scholar]
- 24. Tillu VA, Kovtun O, McMahon K‐A, Collins BM, Parton RG (2015) A phosphoinositide‐binding cluster in cavin1 acts as a molecular sensor for cavin1 degradation. Mol Biol Cell 26: 3561–3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lim Y‐W, Lo HP, Ferguson C, Martel N, Giacomotto J, Gomez GA, Yap AS, Hall TE, Parton RG (2017) Caveolae protect notochord cells against catastrophic mechanical failure during development. Curr Biol 27: 1968–1981.e1967 [DOI] [PubMed] [Google Scholar]
- 26. Bastiani M, Liu L, Hill MM, Jedrychowski MP, Nixon SJ, Lo HP, Abankwa D, Luetterforst R, Fernandez‐Rojo M, Breen MR et al (2009) MURC/Cavin‐4 and cavin family members form tissue‐specific caveolar complexes. J Cell Biol 185: 1259–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lavie Y, Fiucci G, Liscovitch M (1998) Up‐regulation of caveolae and caveolar constituents in multidrug‐resistant cancer cells. J Biol Chem 273: 32380–32383 [DOI] [PubMed] [Google Scholar]
- 28. Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A et al (2015) Tissue‐based map of the human proteome. Science 347: 385–394 [DOI] [PubMed] [Google Scholar]
- 29. Yamaguchi T, Lu C, Ida L, Yanagisawa K, Usukura J, Cheng J, Hotta N, Shimada Y, Isomura H, Suzuki M et al (2016) ROR1 sustains caveolae and survival signalling as a scaffold of cavin‐1 and caveolin‐1. Nat Commun 7: 10060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Björklund ÅK, Ekman D, Elofsson A (2006) Expansion of protein domain repeats. PLoS Comput Biol 2: e114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Burkhard P, Stetefeld J, Strelkov SV (2001) Coiled coils: a highly versatile protein folding motif. Trends Cell Biol 11: 82–88 [DOI] [PubMed] [Google Scholar]
- 32. Stoeber M, Schellenberger P, Siebert CA, Leyrat C, Helenius A, Grunewald K (2016) Model for the architecture of caveolae based on a flexible, net‐like assembly of Cavin1 and Caveolin discs. Proc Natl Acad Sci USA 113: E8069–E8078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Garcia J, Bagwell J, Njaine B, Norman J, Levic DS, Wopat S, Miller SE, Liu X, Locasale JW, Stainier DYR et al (2017) Sheath cell invasion and trans‐differentiation repair mechanical damage caused by loss of caveolae in the zebrafish notochord. Curr Biol 27: 1982–1989.e1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lo HP, Nixon SJ, Hall TE, Cowling BS, Ferguson C, Morgan GP, Schieber NL, Fernandez‐Rojo MA, Bastiani M, Floetenmeyer M et al (2015) The caveolin–cavin system plays a conserved and critical role in mechanoprotection of skeletal muscle. J Cell Biol 210: 833–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cheng JPX, Mendoza‐Topaz C, Howard G, Chadwick J, Shvets E, Cowburn AS, Dunmore BJ, Crosby A, Morrell NW, Nichols BJ (2015) Caveolae protect endothelial cells from membrane rupture during increased cardiac output. J Cell Biol 211: 53–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hansen CG, Bright NA, Howard G, Nichols BJ (2009) SDPR induces membrane curvature and functions in the formation of caveolae. Nat Cell Biol 11: 807–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ariotti N, Hall Thomas E, Rae J, Ferguson C, McMahon K‐A, Martel N, Webb Robyn E, Webb Richard I, Teasdale Rohan D, Parton Robert G (2015) Modular detection of GFP‐labeled proteins for rapid screening by electron microscopy in cells and organisms. Dev Cell 35: 513–525 [DOI] [PubMed] [Google Scholar]
- 38. Yeow I, Howard G, Chadwick J, Mendoza‐Topaz C, Hansen CG, Nichols BJ, Shvets E (2017) EHD proteins cooperate to generate caveolar clusters and to maintain caveolae during repeated mechanical stress. Curr Biol 27: 2951–2962.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Quan J, Tian J (2011) Circular polymerase extension cloning for high‐throughput cloning of complex and combinatorial DNA libraries. Nat Protoc 6: 242–251 [DOI] [PubMed] [Google Scholar]
- 40. Dereeper A, Audic S, Claverie J‐M, Blanc G (2010) BLAST‐EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol Biol 10: 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Review Process File


