Abstract
Kinesins are versatile nano‐machines that utilize variable non‐motor domains to tune specific motor microtubule encounters. During plant cytokinesis, the kinesin‐12 orthologs, PHRAGMOPLAST ORIENTING KINESIN (POK)1 and POK2, are essential for rapid centrifugal expansion of the cytokinetic apparatus, the phragmoplast, toward a pre‐selected cell plate fusion site at the cell cortex. Here, we report on the spatio‐temporal localization pattern of POK2, mediated by distinct protein domains. Functional dissection of POK2 domains revealed the association of POK2 with the site of the future cell division plane and with the phragmoplast during cytokinesis. Accumulation of POK2 at the phragmoplast midzone depends on its functional POK2 motor domain and is fine‐tuned by its carboxy‐terminal region that also directs POK2 to the division site. Furthermore, POK2 likely stabilizes the phragmoplast midzone via interaction with the conserved microtubule‐associated protein MAP65‐3/PLEIADE, a well‐established microtubule cross‐linker. Collectively, our results suggest that dual localized POK2 plays multiple roles during plant cell division.
Keywords: cytokinesis, cytoskeleton, division plane, phragmoplast, preprophase band
Subject Categories: Cell Adhesion, Polarity & Cytoskeleton; Cell Cycle; Plant Biology
Introduction
During cytokinesis, the physical partitioning of all cellular content, plant cells separate by an inward‐to‐outward directed division mode of centrifugal cell plate assembly, as opposed to a centripetal furrowing predominant in metazoan cytokinesis 1, 2, 3. The plant cytokinetic apparatus, called phragmoplast, aids in cell plate biosynthesis. Here, we adopt the current nomenclature of plant cytokinesis structures and functions 4. The phragmoplast is a bipolar array of highly dynamic microtubules, actin filaments, and endomembranes that coordinates the delivery of cell plate biosynthetic vesicles for subsequent fusion within the plane of cell division 4, 5. The majority of microtubules are organized anti‐parallel with their minus ends pointing toward the daughter nuclei (distal phragmoplast) and their plus ends adjoining the plane of cell division (phragmoplast midzone). This overall, bipolar organization guides secretory vesicle trafficking toward the division plane from both daughter cells 6, 7.
The conserved microtubule cross‐linkers MAP65 are integral components of the phragmoplast. Their homologs in yeast, Ase1 and human, PRC1, maintain anti‐parallel microtubule overlap regions in the spindle and the midbody, the animal analog of the phragmoplast 8, 9, 10. In plants, the microtubule cross‐linking function of MAP65‐3/PLEIADE (PLE), member of a protein family of nine in Arabidopsis, is in particular required for phragmoplast integrity and efficient cell plate formation 11, 12, 13, 14, 15. Loss of MAP65‐3/PLE results in a functionally compromised phragmoplast due to a wider midzone than wild type, causing incomplete cell plate formation featuring stubs and gaps 11, 12. Several other members of the MAP65 family act redundantly with MAP65‐3/PLE and localize at the entire phragmoplast or its midzone 15, 16, 17, 18. Spatial confinement to the phragmoplast midzone and MAP65 activity during cytokinesis is regulated by phosphorylation 19, 20. Aurora kinase and mitogen‐activated protein kinases (MAPK) deactivate MAP65 and release the anti‐parallel microtubule overlap 18, 19, 21.
Subsequently, microtubules depolymerize in the central zone of the phragmoplast, where cell plate assembly initiated, while new microtubules polymerize at the phragmoplast leading zone, promoting its centrifugal expansion 4, 6. Phragmoplast expansion is tightly coordinated with cell plate growth by the addition of newly arriving vesicles at its margins 22, 23. The initial disk phragmoplast turns into a ring that expands radially, concurrent with the centrifugally evolving cell plate 24, 25. Finally, the fusion of the cell plate with the parental plasma membrane terminates cytokinesis, but the subcellular location of cell plate fusion is not random.
A poorly understood guidance mechanism directs the radial expansion of the phragmoplast toward a pre‐determined division site that is marked in prophase by the plant‐specific cytoskeletal preprophase band 26, 27. Essentially, the preprophase band delineates the periphery of the division plane at the cell cortex and facilitates the recruitment of proteins that remain there, serving as fiducial markers of the division plane throughout cell division 4, 24, 28, 29, 30, 31, 32. Perturbations of the division site or the guidance mechanism that ensures centrifugal phragmoplast expansion toward this site lead to oblique cell plate insertions with dramatic consequences on growth performance 30, 31, 33, 34, 35.
Previous work established a closely related pair of Arabidopsis kinesin‐12 motor proteins, PHRAGMOPLAST ORIENTING KINESIN (POK) 1, and its homolog POK2 as essential contributors of cytokinesis 31. Based on the persistent presence of POK1 at the division site and the requirement for both POK1 and its homolog POK2 to retain division site resident proteins beyond prophase, these kinesins are regarded as pivotal factors to identify and maintain the division site 28, 29, 31, 36. In pok1 pok2 double mutants, the distinct subcellular localization of division site resident proteins is lost from the division site upon metaphase, suggesting a scaffolding function for POKs at the division site 29, 31. In these mutants, co‐alignment of preprophase band, phragmoplast and cell plate fusion site, is disrupted, due to a notable slant of the phragmoplast 31, 34. Furthermore, in pok1 pok2 cytokinetic cells, the rate of phragmoplast expansion is slower compared to wild type 31, implicating an additional function of POKs in phragmoplast dynamics. Here, we report a novel spatio‐temporal localization pattern of POK2 that requires distinct protein domains. In addition to its anticipated localization at the cortical division zone, the unexpected accumulation of POK2 at the phragmoplast midzone accounts for the phragmoplast expansion delay observed in the pok1 pok2 double mutant. POK2 localization at the phragmoplast midzone requires motility and the microtubule cross‐linker MAP65‐3/PLE. Surprisingly, two separate POK2 regions bind to MAP65 proteins with distinct specificities. We propose that POK2 interaction further enhances MAP65‐3‐mediated stability of the midzone allowing rapid centrifugal expansion of phragmoplast.
Results
POK2 facilitates timely phragmoplast expansion
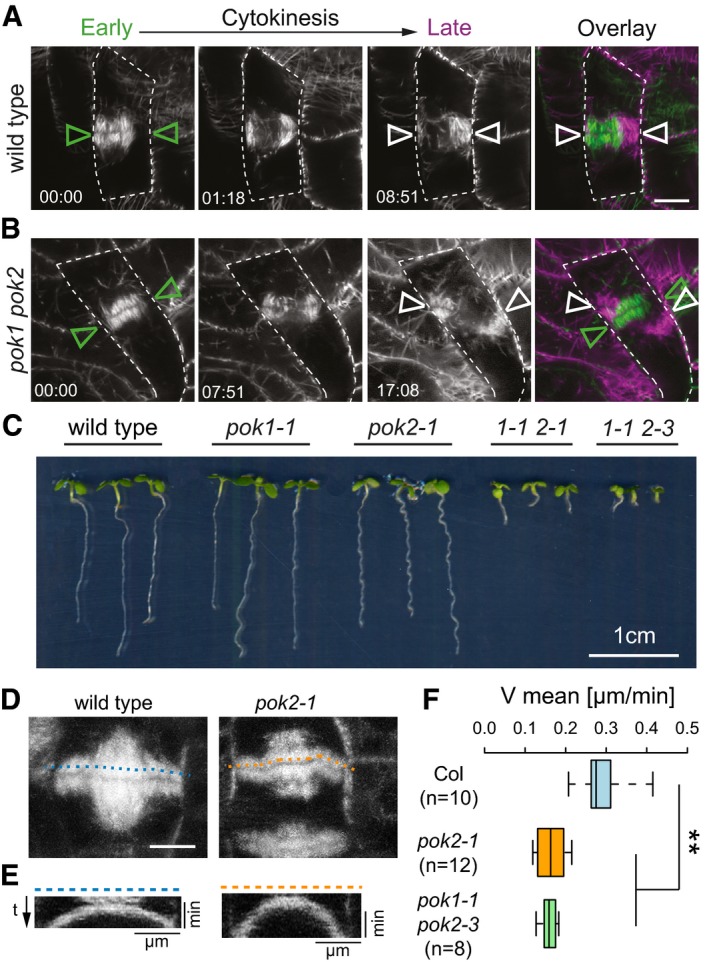
Previously, we reported on the cellular phenotype of pok1 pok2 double mutants. Phragmoplast guidance is compromised, slowing down cytokinesis and causing oblique insertion of cell plates at high frequency, consequently affecting meristem organization and growth (Fig 1A–C) 31. However, POK1 localizes exclusively at the division site, supporting its role in division site maintenance, but this localization pattern does not offer an immediate explanation for the puzzling reduction in the phragmoplast expansion rate that we observed in the pok1 pok2 double mutant 31. We suspected that the yet uncharacterized ortholog of POK1, POK2 performs distinct functions in phragmoplast expansion. Therefore, we investigated the expansion rate in pok2‐1 single mutants using kymograph analysis 37. Phragmoplasts of pok2‐1 mutants expand at a mean velocity (0.16 μm/min ± 0.04) similar to the pok1 pok2 double mutant (0.15 μm/min ± 0.02), while wild‐type phragmoplasts expand about twice as fast (0.32 μm/min ± 0.07), consistent with our hypothesis that POK2 is involved in phragmoplast dynamics (Fig 1D–F) 31.
Figure 1. Comparative analysis of phragmoplast expansion.
-
A, BThree representative time points (in min) of phragmoplast expansion in (A) wild‐type and (B) pok1 pok2 double mutant are depicted. Microtubules are visualized with fluorescent reporters. (A) In wild type, the principal orientation of the phragmoplast does not alter between early (green triangles) and late cytokinesis (white triangles). (B) pok1 pok2 double mutant, note the mis‐alignment of phragmoplast orientation in early (green triangles) and late cytokinesis (white triangles). Overlays show merges of early and late cytokinesis time points. Dashed line traces cell outlines. Scale bar indicates 5 μm. Images were taken on Zeiss LSM880 with the Airyscan detector.
-
CComparison of growth of 6‐day‐old seedlings from different genetic background.
-
DTime projection of cell division in wild‐type (Col‐0) and pok2‐1 single mutants. Scale bar indicates 5 μm.
-
EKymographs of the microtubule reporter signal, using line selections corresponding to the dashed lines in (D).
-
FBox plot depicting velocities of phragmoplast expansion deduced from kymograph analysis. Wild‐type (Col‐0) phragmoplasts expand faster (0.32 μm/min ± 0.07, mean ± SD) than pok2‐1 single (0.16 μm/min ± 0.04, mean ± SD, **P = 0.001) and pok1 pok2 double mutants (0.15 μm/min ± 0.02, mean ± SD, **P = 0.001). Box ranges represent median and 25th and 75th percentiles, respectively. Whiskers extend to minimum and maximum values. (n) indicates the number of phragmoplasts analyzed. Significance value (**P) was determined using one‐way ANOVA with Tukey HSD post hoc test.
Dual localization of POK2 at the division site and the phragmoplast midzone
To investigate POK2 involvement in phragmoplast expansion further, we generated transgenic plant lines expressing a green fluorescent protein (GFP)‐POK2 fusion protein (Figs 2A and EV1A). Our attempts to propagate full‐length cDNA or full genomic clones and in Agrobacterium failed. However, we succeeded to generate a full‐length clone consisting of POK2 cDNA and genomic DNA (see Materials and Methods) driven by the p35S promoter. Polymerase chain reaction of wild‐type, transgenic, and rescue line cDNA yielded identical fragments sizes, indicating correct splicing of the transgene (Fig EV1B). Introgression of p35S:GFP‐POK2 into the pok1 pok2 mutant restored the phenotypic defects, confirming the functionality of GFP‐POK2 transgene (Fig 2B, Appendix Fig S1A–G). Subsequently, we determined the cell cycle‐specific, subcellular localization of GFP‐POK2 in plant lines that co‐express the microtubule reporter RFP‐MBD (Figs 2C and EV1C and D). We observed that POK2 displays a dual localization pattern. Consistent with our expectation and reminiscent of POK1 localization 31, GFP‐POK2 marks the division site throughout cell division, forming a continuous equatorial ring at the plasma membrane (Fig 2C–F, Movies EV1 and EV2). This circular GFP‐POK2 assembly accumulates at the preprophase band and remains associated with the underlying plasma membrane region, designated cortical division zone, beyond the disassembly of the preprophase band (Figs 2C and EV1C) 4. The initially broad GFP‐POK2 rings narrow during cytokinesis, resembling the narrowing of POK1, TAN, and RanGAP1 at the cell plate fusion site (Fig 2E) 4, 28, 29, 31. However, in addition to POK2 at the cortical division zone and cell plate fusion site, we observe GFP‐POK2 at the midzone of the early disk phragmoplast, where it remains present throughout radial phragmoplast expansion (ring phragmoplast) (Figs 2E and F, and EV1D, Table 1). Ultimately, the phragmoplast resident POK2 population merge with the division site resident POK2 as the phragmoplast leading edge approaches the division site upon cell plate fusion. The merge does not occur simultaneously at all sites, but is particularly lagging behind only in cell corners (Fig 2F). The association of GFP‐POK2 with the phragmoplast midzone strongly supports the requirement for POK2 in phragmoplast expansion. We cannot exclude that the reported localization pattern deviates from the endogenous POK2. However, the fusion protein is regulated in a cell cycle‐dependent manner and shows the anticipated localization pattern and dynamics, based on POK2 functional analysis.
Figure 2. GFP‐POK2 localization depends on the cell cycle stage.
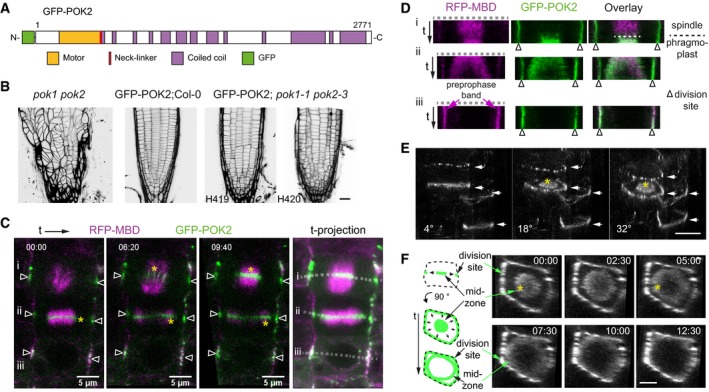
- Protein domain organization of POK2.
- Propidium iodide stained root meristems of pok1 pok2 double mutant, wild type (Col‐0) expressing p35S:GFP‐POK2 (GFP‐POK2) and two examples (H419, H420) of rescue lines expressing p35S:GFP‐POK2 (GFP‐POK2) in pok1 pok2 (inverted gray scale). Scale bar indicates 20 μm.
- Three time points of a time series showing GFP‐POK2 and microtubules (RFP‐MBD) during cell division. Three cells, at different cell cycle stages (i anaphase, ii early cytokinesis, iii prophase), were recorded. Triangles indicate the division site. Yellow asterisks indicate the enrichment of GFP‐POK2 at the phragmoplast midzone. Dotted lines in each stage indicate line selection used for kymographs (space‐time plots) depicted in (D).
- Single‐channel images and overlay of kymographs along line selection (dashed line) in (C). Note the differences in GFP and RFP signal distribution. Triangles indicate the division site. Relates to Appendix Fig S1 and Movie EV1.
- Rotational views (angle is indicated) of 3‐D reconstructed GFP‐POK2 localization patterns, displaying rings (arrows) and a disk‐like distribution at the phragmoplast midzone (asterisk). Note the variety of ring morphologies (filamentous, diffuse, and pearl‐string). Scale bar indicates 10 μm.
- Top view (90° rotation) and a graphical representation of a cytokinetic cell displaying GFP‐POK2 at the phragmoplast midzone (disk) and at the division site (ring). Asterisk in time point 00:00 min indicates GFP‐POK2 association with disk phragmoplast and at 05:00 min with ring phragmoplast. At 12:30, the merge of midzone GFP‐POK2 and POK2 at division site is almost completed. Scale bar indicates 5 μm.
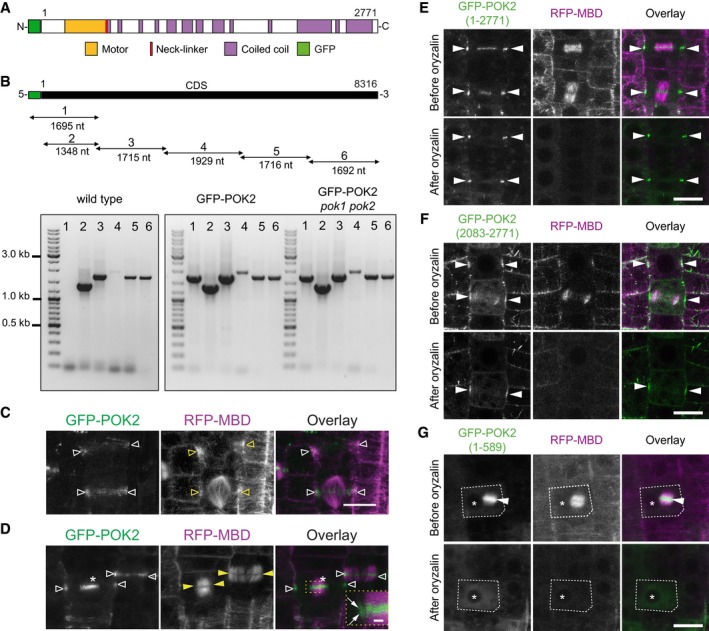
Figure EV1. POK2 expression and microtubule dependencies of POK2 protein domains.
-
AGFP‐POK2 protein organization.
-
BGraphical representation of transgene coding sequence (CDS) and relative position of PCR fragments (consecutive numbers and fragment sizes are indicated) amplified from cDNA derived from wild type, transgenic line (GFP‐POK2), and rescue line (GFP‐POK2 pok1 pok2). Primer combinations are given in Appendix Table S2.
-
C, DCo‐localization of GFP‐POK2 and microtubule marker RFP‐MBD. (C) Prophase cells displaying preprophase bands and pro‐spindle (yellow triangles), respectively, and filamentous GFP‐POK2 (white triangles). (D) Telophase cells displaying phragmoplasts (yellow arrowheads) and GFP‐POK2 at the division site (white triangles) and at the phragmoplast midzone (asterisk). The inset shows the leading edge of the phragmoplast (arrows) at a higher magnification. Images are Z‐projections. Scale bar 10 μm. Relates to Fig 1 and Movies EV1 and EV2.
-
E–GRoot meristem cells expressing (E) GFP‐POK2 (1–2,771), (F) GFP‐POK2 (2,083–2,771), or (G) GFP‐POK2 (1–589) along with the microtubule marker RFP‐MBD are shown before (upper row) and after (lower row) treatment with 10 μM oryzalin. Images of the same cells are shown in each case. Arrow heads point to GFP signal at the division site (E and F) and to the phragmoplast midzone (G, dashed line indicates cell outline). The asterisks mark a conspicuous vacuole present before and after treatment. Note the persistence of GFP signal at the division site in (E) and (F), despite the depolymerization of phragmoplast microtubules and cytosolic GFP in (G). Images are maximum Z‐projections. Scale bars indicate 10 μm. Relates to Figs 1, 2, 3.
Table 1.
Differential subcellular distribution of POK2 fusion proteins during cell division
| Prophase | Meta‐/Anaphase | Cytokinesis | |||
|---|---|---|---|---|---|
| PPB | Spindle/CDZ | Phragmoplast midzone only | Phragmoplast midzone/CDZ | Cell plate fusion site only | |
| POK2 (1–2,771) (n = 12 seedlings) | 98% (n = 49) | 100% (n = 14) | 4% | 96% (n = 79) | 0% |
| POK2 (2,083–2,771) (n = 34 seedlings) | 100% (n = 120) | 89% (n = 37) | 8% | 64% (n = 78) | 21% |
| POK2 (1–589) (n = 28 seedlings) | 16% (n = 55) | 9% (n = 13) | 99% | 0% (n = 104) | 0% |
| POK2 (Δ589–2,771) (n = 18 seedlings) | 100% (n = 36) | 25% (n = 4) | 9% | 59% (n = 22) | 32% |
To determine whether POK2 localization depends on microtubules, we treated seedlings with the microtubule depolymerizing drug oryzalin. GFP‐POK2 persists at the cortical division zone, even after the complete depolymerization of microtubules, similar to POK1 31. In contrast, GFP‐POK2 at the phragmoplast vanished upon microtubule depolymerization (Fig EV1E and Table 2). Consequently, once POK2 tethers to the division site, it is independent of microtubules, while its association with the phragmoplast midzone depends on the intact microtubule cytoskeleton, indicating that distinct mechanisms mediate division site and phragmoplast association of POK2.
Table 2.
Signal distribution of GFP‐POK2 fusion proteins before and after microtubule depolymerization with oryzalin
| Prophase | Meta‐/Anaphase | Cytokinesis | ||||||
|---|---|---|---|---|---|---|---|---|
| PPB | Spindle/cortical division zone | Phragmoplast midzone | Cell plate fusion site | |||||
| GFP | MT | GFP | MT | GFP | MT | GFP | MT | |
| POK2 (1–2,771) (n = 6 roots) | ||||||||
| Before oryzalin | 16 | 16 | 6 | 6 | 25 | 27 | 27 | 0 |
| After oryzalin | 4 | 0 | 5 | 0 | 0 | 0 | 25a | 0 |
| POK2 (2,083–2,771) (n = 6 roots) | ||||||||
| Before oryzalin | 21 | 21 | 3 | 3 | 6 | 10 | 10 | 0 |
| After oryzalin | 8 | 0 | 3 | 0 | 0 | 0 | 7b | 0 |
| POK2 (1–589) (n = 6 roots) | ||||||||
| Before oryzalin | 1 | 13 | 2 | 5 | 30 | 31 | 0 | 0 |
| After oryzalin | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Green fluorescent protein (GFP) and red fluorescent protein (microtubule reporter, MT) signal distribution was examined before treatment with oryzalin, and after depolymerization of microtubules. Cell cycle stages are indicated, according to mitotic microtubule arrays. We scored cells for presence of GFP signal at PPB, cortical division zone, phragmoplast midzone, and cell plate fusion site. Relates to Fig EV1.
a n = 2, b n = 3; Cells without GFP signal after treatment due to completion of cytokinesis during incubation with oryzalin.
To examine how individual protein domains contribute to and establish the dual localization pattern and function of POK2, we decided to analyze the localization of POK2‐domain deletion mutants.
POK2 carboxy‐terminal domain is sufficient to mark the division site
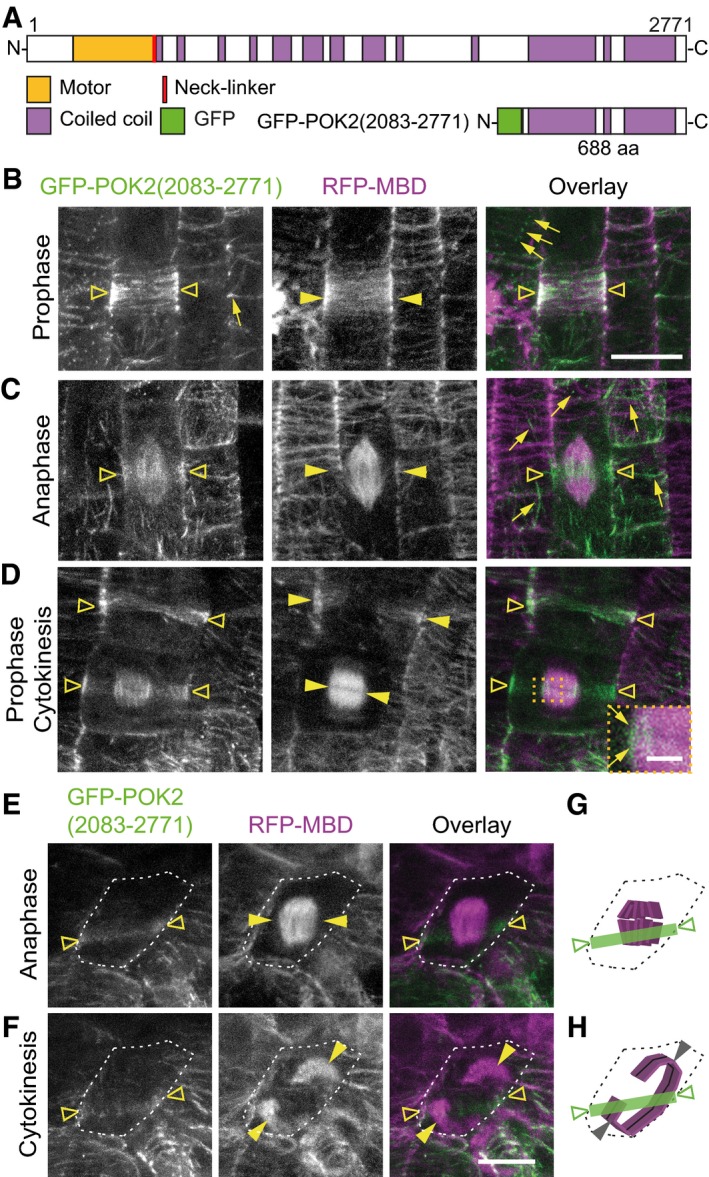
First, we determined the localization of the POK2 C‐terminal region that corresponds to the respective POK1 region responsible for division site targeting (Figs 3A and EV2A) 31, 34. GFP‐POK2 (2,083–2,771) decorates filaments, reminiscent of cortical microtubules and puncta or clusters close to the plasma membrane in interphase (Fig 3B). In mitotic cells, GFP‐POK2 (2,083–2,771) co‐localizes with the preprophase band and disperses on spindle and phragmoplast, but it distinctly decorates the division site throughout mitosis and cytokinesis, demonstrating that the C‐terminal region is sufficient for division site targeting (Fig 3C–E, Table 1). De‐polymerization of microtubules using 10 μM oryzalin abolishes filamentous GFP‐POK2 (2,083–2,771), indicating its dependency on the microtubule cytoskeleton, but GFP‐POK2 (2,083–2,771) that is already present at the division site persists throughout the treatment, revealing POK2 microtubule‐independent maintenance at the division site (Fig EV1F and Table 2). To exclude a contribution of endogenous POK2 to the observed localization pattern, we examined GFP‐POK2 (2,083–2,771) distribution in the pok1 pok2 double mutant (Fig EV2B–D). Compared to wild‐type mitotic cells that display intact GFP‐POK2 (2,083–2,771) rings, in pok1 pok2 mutants about 30% of GFP‐POK2 (2,083–2,771) rings are discontinuous, unveiling a reduction in the efficacy of cortical division zone targeting or retention (Fig 3E–H, Table 1). However, the majority of pok1‐1 pok2‐1 double mutant cells forms continuous rings at the position imposed by the preprophase band 34. Yet, the expanding phragmoplasts disregard the spatial information provided by the cortical GFP‐POK2 (2,083–2,771) rings resulting in inappropriate cell plate positioning (Figs 3E and F, and EV2C and D). Therefore, demarcation of the division site by GFP‐POK2 (2,083–2,771) alone is not sufficient to guide the phragmoplast.
Figure 3. The carboxy‐terminal domain POK2 (2,083–2,771) localizes to the division site in mitosis.
-
AProtein domain organization of POK2 and GFP‐POK2 (2,083–2,771).
-
B–DPOK2 (2,083–2,771) localizes to the cortical division zone (triangles) throughout mitosis. (B) POK2 (2,083–2,771) (triangles) co‐localizes with preprophase band (arrow heads). (C) Anaphase cell exhibiting GFP‐POK2 signal (triangles) at the cortical division zone and the spindle and (D) prophase and cytokinetic cell displaying preprophase band and phragmoplast (arrow heads) and GFP‐POK2 (2,083–2,771) (triangles) at the preprophase band, phragmoplast and cortical division zone. Inset shows magnification of boxed area; arrows point to GFP‐POK2 (2,083–2,771) at the phragmoplast leading zone. Examples of GFP‐POK2 (2,083–2,771) labeled interphase microtubules are indicated by arrows in (B) and (C).
-
E, FCell of pok1 pok2 double mutant with intact GFP‐POK2 (2,083–2,771) cortical division zone localization (triangle). (E) Upper and (F) lower panels show the same cell (cell shape is indicated by dashed line in ana‐ and telophase). Note the mismatch between the GFP‐POK2 (2,083–2,771) cortical division zone signal at the plasma membrane (triangles) and direction of the phragmoplast expansion (arrow heads). Microtubules are visualized by RFP‐MBD (magenta).
-
G, HIllustration of cell outlines (dashed line), microtubules (magenta), and GFP‐POK2 (2,083–2,771) signal distribution (green) of cell depicted in (E and F).
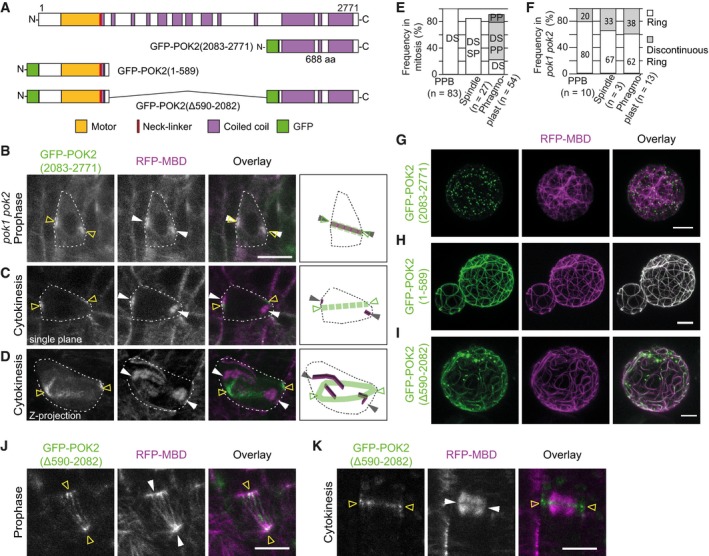
Figure EV2. Localization patterns of different POK2 fusion proteins in transgenic lines and transiently expressed in protoplasts.
-
AGraphical representation of protein domain organization of POK2, GFP‐POK2 (2,083–2,771), GFP‐POK2 (1–589) and GFP‐POK2 (Δ590‐2,082).
-
B–D(B) GFP‐POK2 (2,083–2,771) co‐localizes with the preprophase band (arrow heads) (C and D) Mis‐alignment of GFP‐POK2 (2,083–2,771) (triangles) and phragmoplasts (arrow heads). (B and C) are single plane images. (D) shows a maximum Z‐projection. Cartoons illustrate microtubule (magenta) and GFP‐POK2 (2,083–2,771) subcellular distribution in cells, outlined by dashed lines.
-
EFrequency (%) of mitotic cells displaying GFP‐POK2 (2,083–2,771) in wild type. Numbers indicate the percentage of cells that showed GFP‐POK2 (2,083–2,771) association with preprophase band (PPB), spindle (SP), phragmoplast (PP), and division site (DS).
-
FFrequency (%) of complete and discontinuous rings at the division site of pok1 pok2 mutants during cell division.
-
G–ITransient co‐expression of (G) GFP‐POK2 (2,083–2,771), (H) GFP‐POK2 (1–589), and (I) GFP‐POK2 (Δ590–2,082) with the microtubule reporter RFP‐MBD in Arabidopsis protoplasts. Note the complete co‐localization of GFP‐POK2 (1–589) and microtubules.
-
J, KLocalization of GFP‐POK2 (Δ590–2,082) (triangles) in (J) prophase cell with a preprophase band (arrow heads) and (K) telophase cell with a phragmoplast (arrow heads).
In Arabidopsis interphase protoplasts, transiently expressed GFP‐POK2 (2,083–2,771) forms punctate clusters similar to the ones recorded in plants, but lack the microtubule association (Fig EV2G), suggesting that a cell cycle‐specific binding partner facilitates microtubule interaction in planta. Hence, in planta, the POK2 C‐terminal domain associates with microtubules either directly, and/or through interaction partners. Since POK2 C‐terminal domain is sufficient to identify the division site, but fails to complement the mutant phenotype, these observations denote a critical contribution of POK2 motor domain for POK function at the division site.
The motor domain targets POK2 to the phragmoplast midzone
Next, we investigated the localization of the N‐terminal POK2 motor domain by expression of GFP‐POK2 (1–589). This fusion protein contains the first coiled‐coil domain of POK2 to facilitate anticipated dimerization (Fig 4A). As expected for a kinesin motor, GFP‐POK2 (1–589) associates with cortical microtubules in Arabidopsis interphase protoplasts (Fig EV2H). However, in Arabidopsis seedlings, we rarely observe GFP‐POK2 (1–589) association with cortical microtubules in interphase cells and only one‐fifth of preprophase bands are decorated with GFP‐POK2 (1–589) before it disappears upon preprophase band disassembly, in contrast to the C‐terminal domain POK2 (2,083–2,771) (Table 1, Appendix Fig S2A). In meta‐ and anaphase, GFP‐POK2 (1–589) remains cytosolic but, as the cells proceed to telophase, GFP‐POK2 (1–589) progressively associates with the midzone of the evolving phragmoplast and subsequently persists at the phragmoplast midzone until completion of cytokinesis (Fig 4B, Movie EV3). In pok1 pok2 mutants, we observe the same topology, confirming that the N‐terminal motor domain directs POK2 toward the phragmoplast midzone, but this is not sufficient to rescue the mutant phenotype (Fig 4C). Moreover, GFP‐POK2 (1–589) association with the phragmoplast disappears upon microtubule de‐polymerization with oryzalin, demonstrating its microtubule dependency (Fig EV1G and Table 2). Taken together, these results show that POK2 N‐terminal domain mediates the interaction with microtubules. However, the distinct localization of GFP‐POK2 (1–589) at the preprophase band and the phragmoplast midzone, together with its absence from the spindle, support the existence of a regulatory mechanism underlying its cell cycle stage‐specific accumulation.
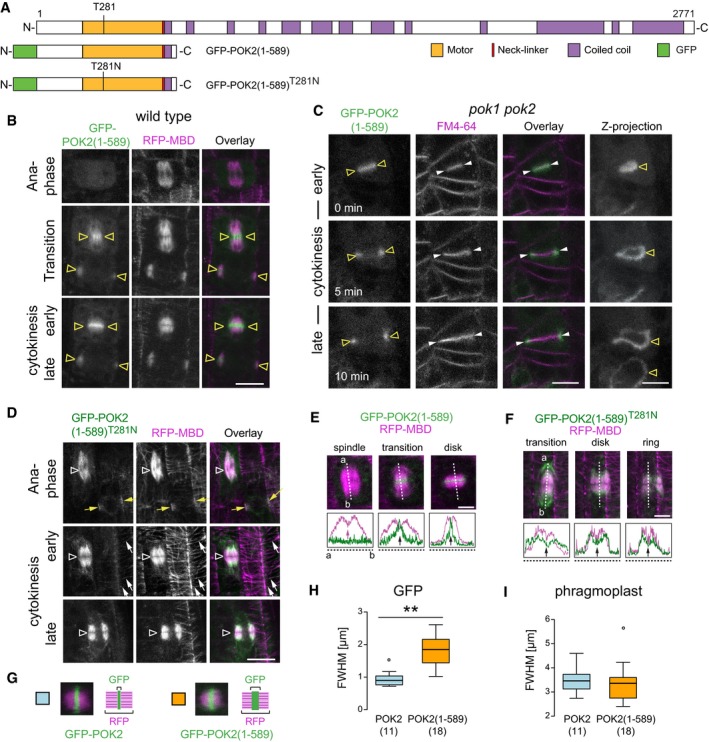
Figure 4. POK2 motor domain, POK2 (1–589), is sufficient for phragmoplast midzone targeting during cytokinesis.
-
ADomain organization and overview of fusion proteins. N‐terminal domain POK2 (1–589) including predicted disordered regions (white), motor domain (yellow), neck‐linker (red), and coiled coil. The position of conserved threonine (T) 281 in the ATP‐binding site and mutation of T281 to asparagine (N) in GFP‐POK2 (1–589)T281N rigor mutant is indicated.
-
BGFP‐POK2 (1–589) accumulates at the phragmoplast midzone (triangles) during the transition from ana‐ to telophase and remains restricted to the midzone.
-
CSingle plane images of a time series during cytokinesis in the pok1 pok2 double mutant. GFP‐POK2 (1–589) co‐localizes with the early cell plate (triangles), stained with FM4‐64 (arrow heads), and associates with the cell plate margins throughout cytokinesis. Z‐projections illustrate the ring‐shaped localization of GFP‐POK2 (1–589). Note that the POK2 motor domain does not localize to the division site and does not rescue the pok1 pok2 mutant phenotype.
-
DThe rigor mutant GFP‐POK2 (1–589)T281N localizes to mitotic and cortical microtubules (white arrows). Yellow arrows indicate a preprophase band in the upper panel; triangles indicate the phragmoplast midzone region.
-
E, FIndividual images of a time series, showing (E) GFP‐POK2 (1–589) and (GFP‐POK2 (1–589)T281N signal in cytokinesis. Plot profiles illustrate GFP (green) and RFP (magenta) signal distribution along the dashed lines in the respective figure panels above. Arrows in plot profiles point to intensity maxima (peaks) or local minima (valleys).
-
G–IComparison between signal width of GFP‐POK2 and GFP‐POK2 (1–589) in cytokinesis. (H and I) Full width at half maximum (FWHM) of (H) GFP‐POK2 (blue, 0.94 ± 0.25 μm, mean ± SD, n = 11) and GFP‐POK2 (1–589) (yellow, 1.81 ± 0.47 μm, mean ± SD, n = 18) differs significantly (**P = 0.001, one‐way ANOVA and Tukey HSD) while FWHMs of (I) phragmoplast are comparable (3.45 ± 0.52 μm, mean ± SD, n = 11 and 3.37 ± 0.80 μm, mean ± SD, n = 18, respectively). Box ranges represent median and 25th and 75th percentiles, respectively. Tukey–whiskers extend to data points that are < 1.5 × IQR away from 25th/75th quartile, outlier is indicated by circles.
ATP‐hydrolysis is a prerequisite for POK2 phragmoplast midzone targeting
The exclusive localization of GFP‐POK2 (1–589) at the phragmoplast midzone where the majority of microtubule plus ends convene suggests microtubule plus end‐directed motility, consistent with the prediction for N‐terminal motor domains 38. Consequently, we asked whether motility is a requirement for phragmoplast midzone association of GFP‐POK2 (1–589). Since ATP‐hydrolysis is essential for motor motility, we generated a hydrolysis‐deficient mutant GFP‐POK2 (1–589)T281N by replacing the conserved threonine with asparagine at position 281 of the ATP‐binding site within the motor domain (Fig 4A). Such rigor mutants bind to microtubules tightly, but are incapable of stepping along them 39. Contrary to GFP‐POK2 (1–589), the rigor mutant GFP‐POK2 (1–589)T281N indeed associates with the entire length of microtubules and lacks the distinct accumulation at the phragmoplast midzone (Fig 4D–F, Appendix Fig S2B). In support of POK2 motility, GFP‐POK2 (1–589) traces linear trajectories with a mean velocity of 4.19 ± 1.03 μm/min (Appendix Fig S2I) in interphase cells of the root meristem (Appendix Fig S2C–E, Movie EV4). When co‐expressed with RFP‐End Binding (EB) 1b, a bona fide microtubule plus end tracking protein, their co‐localization strongly supports the view that the POK2 motor domain is motile and moves toward microtubule plus ends (Appendix Fig S2F–H, Movie EV5). Collectively, these data suggest that microtubule plus end‐directed motility is a prerequisite for POK2 phragmoplast midzone targeting.
POK2 midzone association requires fine‐tuning
Comparing the signal distribution with GFP‐POK2, we noticed that the signal of GFP‐POK2 (1–589) at the phragmoplast midzone appeared wider (Figs 4G and EV1E and G, Appendix Fig S2A). Therefore, we quantified the average full width half maximum (FWHM) 40 of both signals at the midzone and confirmed that the distribution of GFP‐POK2 (1–589) is considerably broader than GFP‐POK2, while the phragmoplast size remains equal (Fig 4H and I). This implies that POK2 restriction to the midzone is fine‐tuned by POK2 domains that are absent from GFP‐POK2 (1–589).
In contrast to the C‐terminal POK2 (2,083–2,771), the N‐terminal GFP‐POK2 (1–589) does not localize to the division site, but is responsible for POK2 association with the phragmoplast midzone. Thus, the dual localization pattern of POK2 at the division site and at the phragmoplast midzone is mediated independently, utilizing distinct protein domains and is regulated in a cell cycle‐dependent manner, likely by specific binding partners, by post‐translational protein‐modifications and/or by intramolecular inhibition. While POK2 at the plasma membrane marks/maintains the division site together with POK1, POK2 at the phragmoplast assists its timely expansion toward the division site. During the final stages of cytokinesis, the seemingly separate populations merge at the division site (Fig 2F).
Investigation of the POK2 central domain, lacking motor domain and C‐terminal domain, was hampered by our unsuccessful cloning attempts. However, deletion of the central domain in GFP‐POK2(Δ590–2,082) did not interfere with microtubule association (Fig EV2I) or its localization pattern during mitosis, although it appears less abundant compared to POK2 (1–2,771) (Fig EV2J and K, Table 1). This indicates that the central domain is dispensable for POK2 localization, but it might contribute to protein stability.
MAP65‐3/PLE retains POK2 at the phragmoplast midzone
The striking localization of POK2 at the phragmoplast midzone is reminiscent of MAP65‐3/PLE, an effective microtubule cross‐linker of anti‐parallel microtubules that is essential for phragmoplast integrity 11, 13. Impairment of MAP65‐3/PLE function causes a widening of the phragmoplast at the midzone that is hampering efficient cell plate assembly, featuring multinucleate cells, cell plate stubs and gaps (Fig 5, Appendix Fig S3) 11, 12. The similarity of POK2 and MAP65‐3/PLE localization in the phragmoplast midzone prompted us to investigate the potential interaction between the two proteins. We examined the phragmoplast midzone localization of GFP‐POK2 in the MAP65‐3/PLE mutant pleiade (ple)‐2 (Fig 5) 23, 41. While GFP‐POK2 signal at the division site is readily detectable, abundance of GFP‐POK2 is diminished at the midzone of ple‐2 mutants compared to wild type (Fig 5A and B). Furthermore, in ple‐2 cytokinetic cells, the motor domain GFP‐POK2 (1–589) lacks the confinement to the phragmoplast midzone; instead, GFP‐POK2 (1–589) frequently decorates the entire length of phragmoplast microtubules, revealing the involvement of MAP65‐3/PLE in restricting POK2 to the phragmoplast midzone (Fig 5C, Appendix Fig S3B). As for the localization of the C‐terminal domain POK2 (2,083–2,771), we did not observe an altered localization in ple‐2 mutants. Like in wild type, GFP‐POK2 (2,083–2,771) localizes to the division site and scarcely decorates phragmoplast microtubules (Fig 5D, Appendix Fig S3A, C and D). In contrast to the GFP‐POK2 (1–589) motors that move toward the microtubule plus end‐enriched midzone where they are retained by MAP65‐3/PLE, the C‐terminal GFP‐POK2 (2,083–2,771) does not even reach the midzone due to the lack of motility, however, it might bind to other interaction partners along the phragmoplast.
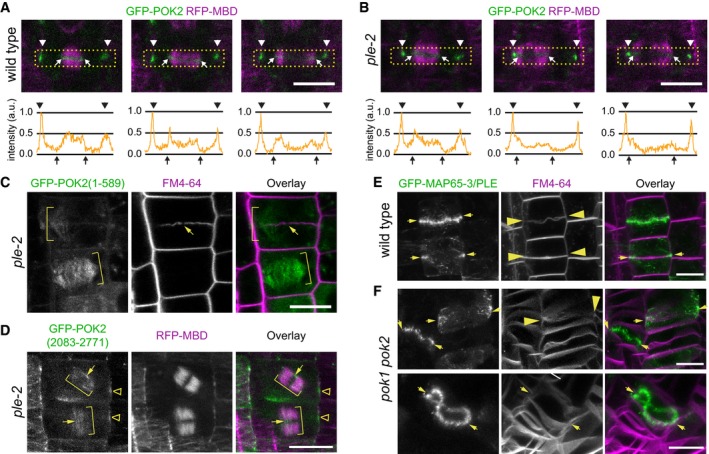
Figure 5. POK2 MAP65‐3/PLEIADE interaction at the phragmoplast midzone requires an N‐terminal binding site.
-
A, BComparison of GFP‐POK2 localization in (A) wild‐type and (B) pleiade (ple)‐2 mutant at three consecutive stages of cytokinesis. Arrow heads indicate the division site. Arrows point to the midzone margins. Lower panels show intensity profile plots of GFP‐POK2 in the rectangular selection indicated in the corresponding confocal image above. Images are single confocal sections depicting median planes recorded with identical settings. Normalized intensities are shown in arbitrary units (a.u.).
-
CLocalization of GFP‐POK2. Localization of GFP‐POK2 (1–589) motor domain along phragmoplast microtubules (brackets) in the ple‐2 mutant. Note that GFP‐POK2 (1–589) in wild type localizes to the phragmoplast midzone (Fig 4). Plasma membranes are stained with FM4‐64. Arrow indicates a characteristic gap in the incomplete cell plate.
-
DCo‐expression of GFP‐POK2 (2,083–2,771) and microtubule reporter (RFP‐MBD) in ple‐2 mutant. Note the increased phragmoplast size (brackets) and the wider than normal midzone. GFP‐POK2 (2,083–2,771) faintly decorates phragmoplast microtubules, also across the midzone (arrows) and accumulates at the cortical division zone (triangles). Note that the localization is not altered compared to wild‐type (Fig 3).
-
E, FGFP‐MAP65‐3/PLE localizes at the midzone (arrows) in (E) wild‐type and (F) pok1 pok2 mutant cytokinetic cells. FM4‐64 staining indicates the plasma membranes. Arrows indicate cell plate fusion sites. Images are maximum Z‐projections. Note the extreme midzone undulations in pok1 pok2 mutants (lower panel).
Next, we investigated whether loss of POK1 and POK2 interferes with the localization pattern of MAP65‐3/PLE in root meristematic cells 42. In pok1 pok2 mutants, as in wild type, GFP‐MAP65‐3/PLE is directed to the phragmoplast midzone, indicating that POK2 functions downstream of MAP65‐3/PLE (Fig 5E and F, Appendix Fig S3E and F). However, the phragmoplast midzone in pok1 pok2 mutants displays pronounced undulations hinting to a lack of stability (Fig 5E and F, Appendix Fig S3E and F), consistent with the reduction in phragmoplast expansion rate due to the loss of pok2. Taking these data together, we conclude that MAP65‐3/PLE acts upstream of POK2.
POK2 interacts with MAP65‐3/PLE via two distinct binding sites
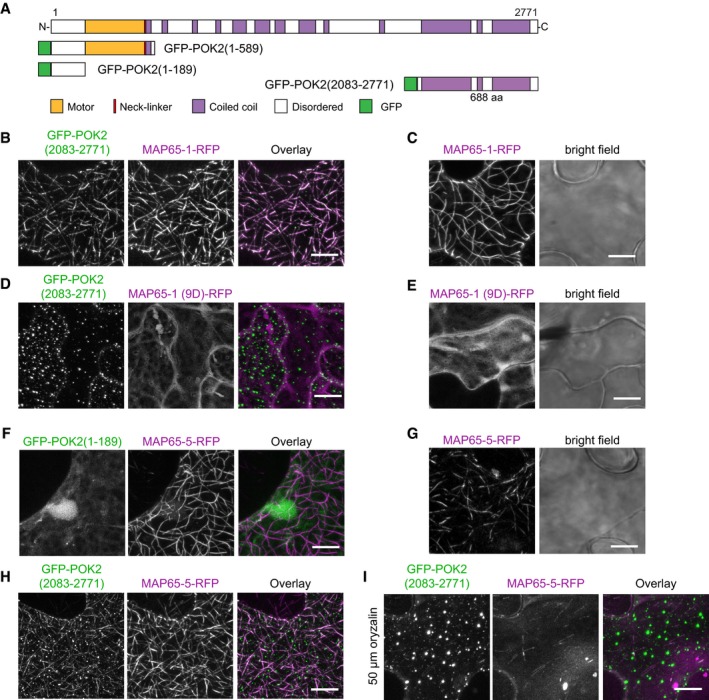
The genetic interaction between POK2 and MAP65 might be indirect. Therefore, we investigated the interaction between POK2 and MAP65‐3/PLE further by transient co‐expression of GFP‐POK2 domains and MAP65‐3/PLE‐RFP in tobacco leaves (Fig 6A). Notably, in tobacco, there is no expression of a MAP65‐3 homolog 17. Co‐expression of the N‐terminal GFP‐POK2 (1–589) with MAP65‐3 showed the anticipated co‐localization along microtubules, as each protein when expressed alone decorates microtubules (Fig 6B–D). However, MAP65‐3/PLE, which specifically cross‐links anti‐parallel microtubules, labeled fewer microtubules than GFP‐POK2 (1–589). Since the motor domain POK2 (1–589) is mis‐localized in the ple‐2 mutants (Fig 5C, Appendix Fig S3B), we wondered whether the disordered region upstream of the motor domain might be responsible for interaction with MAP65‐3/PLE. So, we removed the motor domain and created GFP‐POK2 (1–189). This fusion protein does not bind to microtubules in tobacco pavement cells, but remains cytosolic (Fig 6E). However, when co‐expressed with MAP65‐3/PLE, we observe co‐localization of the short fragment GFP‐POK2 (1–189) with MAP65‐3/PLE (Fig 6F), but not with MAP65‐5 which also localizes to the phragmoplast midzone (Fig EV3F and G) 17, suggesting that this disordered POK2 domain is sufficient to facilitate interaction with the midzone resident MAP65‐3/PLE.
Figure 6. Tobacco abaxial leaf epidermal cells transiently expressing fusion proteins.
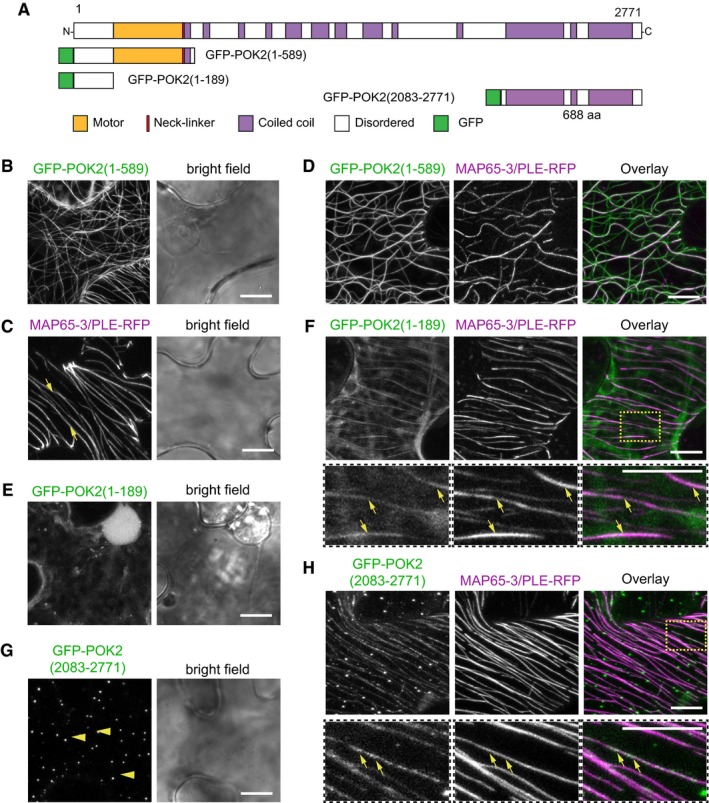
- Domain organization and overview of fusion proteins.
- Single expression of GFP‐POK2 (1–589). Note the association with microtubules.
- MAP65‐3/PLE‐RFP labels microtubules (individual examples indicated by arrows).
- Co‐expression of GFP‐POK2 (1–589) and MAP65‐3/PLE‐RFP. Overlay shows partial co‐localization of GFP‐POK2 (1–589) and MAP65‐3/PLE‐RFP.
- Single expression of GFP‐POK2 (1–189). Note the cytosolic distribution.
- Co‐expression of GFP‐POK2 (1–189) and MAP65‐3/PLE‐RFP. Overlay shows partial co‐localization of GFP‐POK2 (1–189) and MAP65‐3/PLE‐RFP (individual examples indicated by arrows). Lower panels show threefold magnification of boxed area in (F overlay).
- Single expression of GFP‐POK2 (2,083–2,771). Note that GFP‐POK2 (2,083–2,771) alone does not associate with microtubules, but forms punctate clusters (examples indicated by arrow heads).
- Co‐expression of GFP‐POK2 (2,083–2,771) and MAP65‐3/PLE‐RFP. Overlay shows partial co‐localization of GFP‐POK2 (2,083–2,771) and MAP65‐3/PLE‐RFP. Lower panels show threefold magnification of boxed area in (H overlay). Arrows indicate GFP‐POK2 (2,083–2,771) along MAP65‐3/PLE‐RFP labeled microtubules.
Figure EV3. Transient co‐expression of fusion proteins in tobacco abaxial leaf epidermal cells.
-
ADomain organization and overview of fusion proteins.
-
B–ITransient co‐expression of GFP‐POK2 (2,083–2,771) and (B) MAP65‐1‐RFP, (D) MAP65‐1(9D)‐RFP, (H and I) MAP65‐5‐RFP in tobacco abaxial leaf epidermal cells. Note that microtubules are labeled by both fusion proteins in cells co‐expressing GFP‐POK2 (2,083–2,771) and MAP65‐1‐RFP, and MAP65‐5‐RFP. In cells expressing MAP65‐1(9D), which does not bind microtubules, and MAP65‐5‐RFP treated with oryzalin to depolymerize microtubules, GFP‐POK2 (2,083–2,771) displays punctate clusters, similar to the pattern observed when expressed alone (Fig 6G). Single expression of (C) MAP65‐1‐RFP, (E) MAP65‐1(9D)‐RFP, and (G) MAP65‐5‐RFP. Note that MAP65‐1(9D)‐RFP is cytosolic. (F) Co‐expression of GFP‐POK2 (1–189) with MAP65‐5 RFP. Scale bar indicates 10 μm.
Considering that the C‐terminal domain POK2 (2,083–2,771) might fine‐tune POK2 localization at the midzone, we also investigated the interaction between POK2 (2,083–2,771) and MAP65‐3/PLE. In tobacco pavement cells, GFP‐POK2 (2,083–2,771) accumulates in punctate clusters, similar to the pattern observed in Arabidopsis interphase protoplasts (Figs 6G and EV2G). When we co‐expressed with MAP65‐3/PLE‐RFP, we observe substantial overlap of GFP‐POK2 (2,083–2,771) and MAP65‐3/PLE‐RFP signal suggesting MAP65‐3/PLE recruits the POK2 C‐terminal domain to microtubules (Fig 6H). A mating‐based split‐ubiquitin assay in yeast further corroborates direct interaction between both the N‐terminal GFP‐POK2 (1–189) (Fig 7A–C) and the C‐terminal POK2 (2,083–2,771) (Fig 7C) with MAP65‐3/PLE. Expression of interactors was confirmed by immuno‐blotting (Fig 7D–G). Together, these observations suggest that POK2 uses two distinct domains to interact with MAP65‐3/PLE. We wondered about the specificity of the observed interactions and tested additional MAP65 family members, reported to associate with the phragmoplast, for co‐localization with C‐terminal POK2 (2,083–2,771) in tobacco transient expression. In addition to MAP65‐3/PLE, POK2 C‐terminal domain co‐localized with MAP65‐1 and MAP65‐5 (Fig EV3B, C, H and G), indicating that the POK2 C‐terminal binding site was not specific for MAP65‐3/PLE. Interestingly, GFP‐POK2 (2,083–2,771) only co‐localizes with MAP65‐1‐RFP along microtubules, but it is not recruited to the cytosol by MAP65‐1(9D) mutant that fails to bind microtubules in tobacco (Fig EV3D and E) 21, suggesting that interaction of POK2 and MAP65 requires the presence of microtubules. Along these lines, upon treatment with oryzalin, C‐terminal POK2 (2,083–2,771) reforms clusters close to the plasma membrane while MAP65‐5 diffuses into the cytosol (Fig EV3I). Together, the results support the view that POK2 interacts with microtubule bound‐MAP65 isoforms.
Figure 7. The yeast mating‐based Split‐Ubiquitin Cyto‐SUS interaction assay.
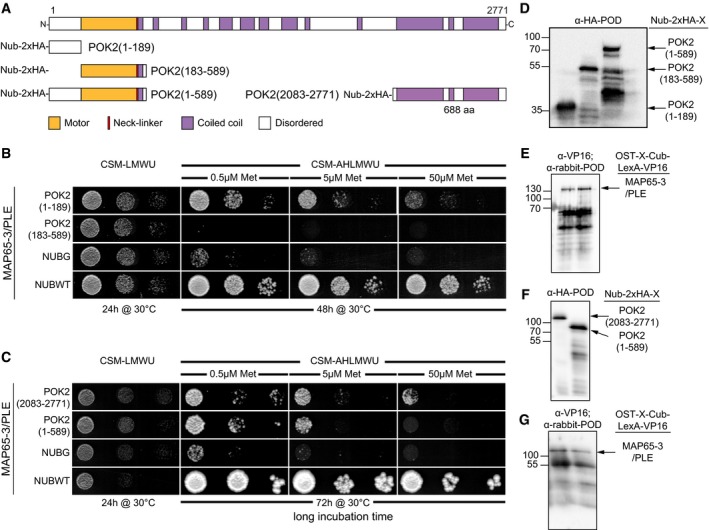
-
ADomain organization and overview of fusion proteins.
-
B, CInteraction of OST‐MAP65‐3/PLE‐Cub and NubG–POK2x fusion as indicated, with controls (NubG, negative; NubWT (wild type), positive). Yeast diploids were spotted (from left to right, dropped at 1.0, 0.1, and 0.01 A600 in each case) on CSM‐Leu‐, Met‐, Trp‐, Ura‐ (CSM‐LMWU) medium to verify mating, and on CSM‐Ade‐, His‐, Leu‐, Met‐, Trp‐, Ura‐ (CSM‐AHLMWU) with the addition of different methionine concentrations to assay for interaction.
-
D–GImmunoblot‐detection of fusion proteins expressed in yeast. (D) Nub‐2xHA‐POK2 (1–189) (31 kDa) and Nub‐2xHA‐POK2 (183–589) (57 kDa), corresponding to (B), and (F) Nub‐2xHA‐POK2 (1–589) (75 kDa) and Nub‐2xHA‐POK2 (2,083–2,771) (91 kDa) corresponding to (C), using an anti‐HA antibody conjugated with peroxidase (POD). (E, G) Immunoblot‐detection of MAP65‐3/PLE‐Cub‐VP16 (137 kDa) in yeast protein extracts, using an anti‐VPS16 as a primary antibody and anti‐rabbit‐POD as a secondary antibody. (E) corresponds to (B), (G) corresponds to (C). Relates to Fig 6.
In summary, we show that two distinct protein domains are responsible for the dual localization of POK2 during cell division. The C‐terminal domain is required for accurate localization at the division site, and the motor domain directs POK2 toward the phragmoplast midzone, where it contributes to phragmoplast stability in a MAP65‐3/PLE‐dependent manner. ATP‐dependent motor motility ensures microtubule plus end‐directed translocation of POK2 toward phragmoplast midzone, where it is retained by MAP65‐3/PLE through its interaction with the intrinsically disordered N‐terminus of POK2 and further fine‐tuned by the C‐terminal POK2 domain.
Discussion
In this study, we clarified the cause of the phragmoplast expansion phenotype in the pok1 pok2 double mutant phenotype. We investigated the role of the kinesin‐12 POK2 in phragmoplast expansion and identified its dual localization pattern. POK2 resides at the division site aiding in its maintenance, whereas the unexpected association with the phragmoplast assists radial expansion. Furthermore, we identified two potential MAP65‐3/PLE binding sites in POK2 that likely differ in their specificity for MAP65 binding.
POK2 acts redundantly with POK1 in division site maintenance
Our previous work determined the homologous genes POK1 and POK2 as essential for division plane maintenance. Their joint effort identifies and preserves the division site by retaining division site resident proteins 29, 31, 36. Reminiscent of POK1 localization 31, also POK2, tethers to the division site and displays comparable cell cycle and microtubule dependencies, suggesting redundancy of POK1 and POK2 activities. Of the six kinesin‐12 in Arabidopsis, so far, only POK1 and POK2 are present to the division site 43, 44, 45. Beyond their function in division site maintenance, we previously proposed POKs active involvement in phragmoplast guidance during late cytokinesis, mediated by peripheral microtubules that emanate from the phragmoplast leading zone 31. In tobacco bright yellow (BY)‐2 cells, peripheral microtubules of opposing phragmoplast halves polymerize from extant microtubules at shallow angles, some being cross‐linked in the midzone by MAP65 46. In this study, in addition to its anticipated localization at the division site, we report POK2 at the phragmoplast midzone. Thus, POK2 localizes to these entities that merge upon cell plate fusion. This novel finding identifies POK2 as realistic candidate for interaction with peripheral microtubules, bridging the phragmoplast and the division site. Strikingly, in the moss Physcomitrella patens, the actin‐dependent motor protein MYOSIN (MYO) 8A moves toward the phragmoplast midzone along peripheral microtubules and localizes to the division site 47. Genetic impairment of all five moss MYO8 genes causes disorganized cell wall insertion 47, together, pointing to possible mechanistic analogies between MYO8 and POK2 in Physcomitrella and Arabidopsis, respectively. Nonetheless, how the expanding phragmoplast and the division site at the cell cortex communicate exactly is still unresolved.
Regarding the exact mode of POK2 function at the division site and at the phragmoplast midzone, only hypotheses can be made. Evidence concerning how kinesin‐12 interacts with microtubules from plants is still missing. However, in metazoan, kinesin‐12 displays microtubule bundling and sliding activity. HsKif15 regulates spindle dynamics although the exact mechanism is currently debated 48, 49, 50, 51. In vitro, dimeric HsKif15 promotes microtubule sliding of higher order microtubules utilizing a motile motor in concert with a non‐motor microtubule binding domain 50. Other in vitro studies report tetrameric HsKif15 switching between microtubule tracks and HsKif15 motor collectives that display processive movement at steady velocity 52. Recently, formation of tetrameric HsKif15 was reported also in vivo 53. Whether POK2 molecules form di‐ or tetramers and how these interact with microtubules requires further scrutiny. Moreover, the dual localization of POK2 might complicate matters. POK2, at the division site, tethered by its C‐terminal domain is likely dimeric and independent of microtubules. On the other hand, POK2 at the phragmoplast midzone depends on motility along microtubules and might be capable of forming tetramers. Therefore, the mode of interaction of POK2 with microtubules might vary depending on its subcellular localization. Given the enormous size of POK2 and the different subcellular activities, intramolecular inactivation must be deemed likely and might be regulated post‐translationally.
POK2 phragmoplast midzone targeting requires motility and accelerates phragmoplast expansion
The specific localization of POK2 at the phragmoplast midzone indicates a site‐specific function. In pok2 single and pok1 pok2 double mutants alike, the mean phragmoplast expansion velocity is significantly slower than wild type, indicating POK2 is a major contributor to the timely expansion of the phragmoplast. Nevertheless, neither does the slower expansion rate interfere with the phragmoplast guidance mechanism toward the division site, since pok2 single mutants do not show division plane defects 34, nor is the rapid phragmoplast expansion required for cell plate biosynthesis, as cell wall stubs, gaps, and multinucleate cells, characteristics of failing cell plate biosynthesis, are absent from pok2 single and pok1 pok2 double mutants 31, 34. In contrast, another pair of kinesin‐12 in Arabidopsis, KINESIN‐12A, and KINESIN‐12B exclusively localize to the phragmoplast in a MAP65‐3/PLE‐dependent manner and together these kinesins are essential for proper cell plate biosynthesis during male gametogenesis 13, 43, 54, 55. Unless unknown redundancies are unresolved, rapid phragmoplast expansion is not compulsory for successful phragmoplast guidance or for normal cell plate biosynthesis.
Our results suggest that motility is the prerequisite for POK2 midzone accumulation. The N‐terminal region of POK2, containing the motor domain, localizes to the phragmoplast midzone in a microtubule‐dependent manner, demonstrating that this region is sufficient for POK2 midzone targeting. However, compared to full‐length POK2 at the midzone, the accumulation of the motor domain is slightly wider, suggesting the existence of a fine‐tuning mechanism that involves additional regions of POK2. In contrast, the catalytically‐inactive, motility‐impaired mutant POK2 (1–589)T281N fails to localize to the midzone. This finding implies that POK2 moves toward microtubule plus ends to reach the division plane, where it likely contributes to microtubule plus end stabilization, as observed for high concentrations of tetrameric hKif15 which disables catastrophe at microtubule plus ends in vitro 51. Consistent with the notion that POK2 might serve auxiliary function in microtubules plus end stabilization, phragmoplasts are often twisted and bend in pok1 pok2 mutants.
Kinesin‐12 at the phragmoplast interact with MAP65
Although their roles in cytokinesis apparently differ, POK2 and kinesin‐12A/B share the specific localization at the phragmoplast midzone, suggesting they utilize a similar targeting and/or retention mechanism possibly via conserved interactions with a specific binding partner, such as the phragmoplast‐specific MAP65‐3/PLE. MAP65 proteins were proposed to serve as local platforms for the recruitment of kinesins 56, 57. Indeed, kinesin‐12A/B are completely abolished in the MAP65‐3/PLE mutant dyc283, while PAKRP2 kinesin becomes evenly distributed along phragmoplast microtubules 12, similar to the N‐terminal GFP‐POK2 (1–589) in the ple‐2 allele. On the other hand, MAP65‐3/PLE localization is not impaired in pok1 pok2 mutants, placing POK2 function downstream of MAP65‐3/PLE.
Together with the genetic data, our co‐localization and interaction assays demonstrate that MAP65‐3/PLE may interact directly with both POK2 motor domain and C‐terminal domain, recruiting them to microtubules. This provides compelling evidence for interaction of POK2 and MAP65 at the phragmoplast midzone. In tobacco, the N‐terminal binding site in POK2 (1–189) interacts specifically with MAP65‐3, but not with phragmoplast midzone resident MAP65‐5, while the C‐terminal binding site in POK2 (2,083–2,771) interacts with all MAP65 isoforms examined. This result suggests that the POK2 C‐terminal domain may also associate with other paralogs of MAP65‐3/PLE at the phragmoplast, such as MAP65‐1/2, MAP65‐5, and MAP65‐4 15, 17, 18. While MAP65‐1 decorates the entire phragmoplast, MAP65‐5 is restricted to the midzone, similar to MAP65‐3/PLE 16. We propose that the interaction of the C‐terminal GFP‐POK2 (2,083–2,771) with MAP65‐3/PLE might reflect a general affinity of POK2 to MAP65 proteins, which also explains why POK2 recruitment to the midzone is not prohibited, but it is less efficient in ple‐2 mutants. Hence, microtubule association of POK2 C‐terminal domain with the phragmoplast and cortical microtubules, which has never been observed in the case of POK1 C‐terminal domain when constitutively expressed 31, is likely mediated by MAP65 proteins.
We summarized the possible mechanism regarding potential POK2 functions in cytokinesis in a schematic model (Fig EV4). We show that motile POK2 moves toward microtubule plus ends and arrives at the midzone utilizing its motor domain (Fig EV4). Both the motor domain and the C‐terminal domain bind MAP65‐3/PLE at anti‐parallel microtubule overlaps, thereby sequestering POK2 at the phragmoplast midzone. We hypothesize that, in analogy to hKif15, POK2 might interact with microtubules, in the immediate vicinity of MAP65‐3/PLE, which in turn might increase the affinity of POK2 C‐terminal domain for microtubule interaction. Potentially, like HsKif15, POK2 motor collectives impede microtubule plus end catastrophe, thereby conferring further stability to the expanding phragmoplast. Besides, at the division site, tethered POK2 motors might stabilize microtubule plus ends of peripheral phragmoplast microtubules 31, 46, forming stable, transient connections at final stages of cytokinesis.
Figure EV4. Putative model for POK2 activities in cytokinesis.
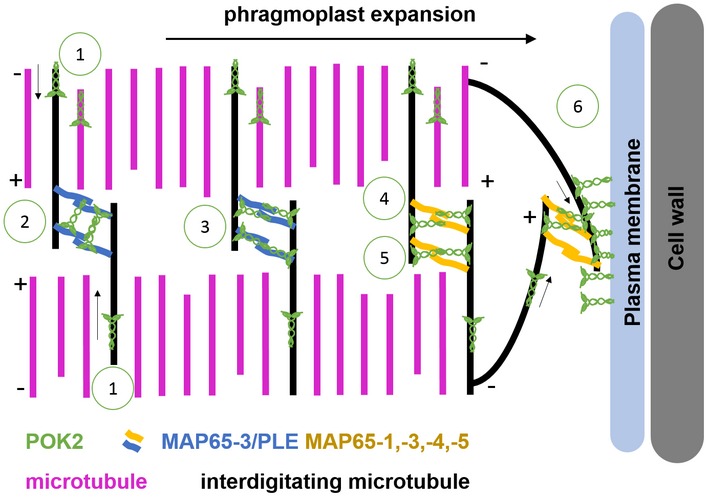
Schematic representation of phragmoplast expansion during cytokinesis. Based on our data and available literature, we propose the following scenarios for POK2 activities during centrifugal phragmoplast expansion: (1) POK2 moves toward the phragmoplast midzone where it associates with MAP65/PLE and other MAP65 dimers. In accordance with HsKif15‐microtubule encounters, POK2 forms dimers and/or tetramers and potentially prevents microtubule plus end catastrophe. POK2 interacts with MAP65 isoforms via distinct protein domains. Interaction with MAP65‐3/PLE (2, 3) is mediated by both the disordered region (1–189) and the C‐terminal domain (2,083–2,771). POK2 might bind between two MAP65‐3/PLE dimers (2) or parallel to the axis of one MAP65‐3 dimer (3). For interaction with other MAP65 isoforms, dimeric (4) or tetrameric (5) POK2 solely utilizes the C‐terminal region. POK2 at the division site (6) is likely dimeric, due to the tethering via its C‐terminal region. With their motor domains, POKs might interact with peripheral microtubules directly. Microtubule plus ends (+), microtubule minus ends (−). Arrows indicate direction of POK2 motility.
The dual localization of POK2 essentially represents the intersection of known plant kinesin‐12 localization patterns. Its closest relative POK1 resides exclusively at the division site, although we must not categorically dismiss POK1 abundance at the phragmoplast below the detection limit of our imaging system 31. The more distant relatives kinesin‐12A and kinesin‐12B exclusively localize to the phragmoplast midzone 54. Animal kinesin‐12 serves in spindle assembly, suggesting that microtubule sliding, bundling, and stabilizing activity is its core function that might contribute to phragmoplast midzone stability in concert with MAP65‐mediated phragmoplast organization. Therefore, POK localization at the division site likely mediates a derived plant‐specific function, while the presence at the phragmoplast might reflect an ancestral function. Thorough phylogenetic analysis and domain swap experiments might clarify whether POK2 incarnates an evolutionary intermediate between kinesin‐12A/B and POK1.
Materials and Methods
Plant material
In the present study, wild‐type, transgenic, and mutant plants of the Arabidopsis thaliana accession Columbia (Col‐0) were utilized. Mutants pok1 pok2 and pleiade‐2 and GFP‐MAP65‐3/PLE were described previously 23, 31, 41, 42. The allele combination pok1‐1 pok2‐3 was used throughout the study unless otherwise indicated.
Growth conditions
Arabidopsis seeds were surface sterilized with 6% (v/v) sodium hypochlorite (Roth, Cat. 9062.3) and sown on plates containing ½ × Murashige and Skoog medium (Duchefa‐Biochemie, MO221.0005) in 1% (w/v) agar (Serva, 11396.03). Seeds were stratified for at least 24 h at 4°C in darkness. Subsequently, seedlings were grown at standard conditions (22°C, 16 h light/8 h darkness cycle). For crossing or reproduction, 2‐week‐old seedlings were transferred to soil and grown in a plant growth chamber under the above‐listed conditions.
Imaging
Imaging of seedlings was performed using either a Leica TCS‐SP8, or a Zeiss LSM 880, both equipped with Argon/Krypton mixed gas laser source, proper filters and detectors and water immersion objective lens 40× or 63× with a numerical aperture of 1.10 or 1.20, respectively. Fluorescence signal in both microscopes was detected either with conventional photomultipliers or using Leica hybrid detectors and Zeiss GaAsP or the Airyscan detectors. GFP was exited with a 488 nm laser line, whereas the detection wavelength range was 500–550 nm. For excitation of RFP, propidium iodide, and FM4‐64, a 561 solid state laser line was used and the detection wavelength range was adjusted (570–650 nm and 570–720, respectively). Imaging was performed at constant room temperature of 22°C.
Localization patterns of POK2 fusion proteins
Dividing cells from seedlings co‐expressing either of the fusion proteins along with the microtubule reporter RFP‐MBD 31 were classified into individual cell cycle stages, based on mitotic microtubule array organization. For determining the presence or absence of GFP signal at a distinct subcellular location, only cells exhibiting RFP‐MBD were taken into consideration.
Full width half maximum (FWHM) analysis
In ImageJ, single image planes of midzone/phragmoplasts were rotated (bicubic interpolation) to align the midzone with the y‐axis. A plot profile of a rectangular selection of the phragmoplast was generated for each channel and the FWHM of the resulting plot profiles was determined in Excel. Measurements were averaged and Box plots were created using BoxPlotR (http://boxplot.tyerslab.com/). The Tukey–whiskers extend to data points that are < 1.5 × IQR away from 1st/3rd quartile. The significance values P were determined using one‐way ANOVA with the post hoc Tukey HSD.
Treatment with oryzalin
Young seedlings, co‐expressing either of the GFP fusion proteins and RFP‐MBD, were treated with 10 μΜ oryzalin (Supelco, PS‐410). Confocal images of the same root meristem region were obtained before and after treatment. An aqueous solution of 10 μΜ oryzalin was added at one edge of the microscope slide, and simultaneously, the mounting medium was removed from the other edge using filter paper. During treatment, z‐stacks were taken at 3–5 min intervals. Depolymerization of microtubules was confirmed by the disappearance of RFP signal.
Image processing
Raw images were processed by ImageJ/Fiji 58, http://rsb.info.nih.gov/ij/. To correct for drift of single‐channel and multichannel stacks were appropriate, the “StackReg” plugin was applied. Time laps movies were processed in ImageJ v1.51k. To reduce noise, “Subtract Background” was performed with using Rolling Ball Radius 100.0. Kymographs were created in ImageJ v1.51k, using “Multiple Kymograph” plugin. Color merges were carried out with ImageJ v.1.48s, ImageJ v1.51k, or Adobe Photoshop CS5 v12.0.4 (Adobe Systems). Only linear adjustments were applied. Figures were assembled in Adobe Illustrator CS5 v15.0.2.
Generating cDNA, amplification of PCR products and ligation
cDNA was essentially generated as described previously 34, using Superscript Reverse Transcriptase II (Invitrogen, 18064‐022). PCR products for cloning purposes were amplified with Phusion DNA Polymerase (New England Biolabs, M0530L) (Appendix Table S2). Standard cloning was performed using Quick Ligation Kit (New England Biolabs, M2200L).
Generating entry clones
pENTR:POK2 (2,083–2,771): The corresponding coding sequence was amplified from flower cDNA, digested with BamHI/XbaI and ligated with pENTR3C, digested with BamHI/XbaI.
pDONR:POK2 (2,083–2,771): Coding sequence was amplified from pENTR:POK2 (2,083–2,771) using primers flanked with minimal attB sites (Appendix Table S1). The PCR product was amplified with full‐length attB1F and attB2R (Appendix Table S1) and cloned into pDONR221 via a Gateway BP reaction (Invitrogen, 11789‐020).
pENTR:POK2 (1–589): Corresponding coding sequence was amplified from flower cDNA and inserted into pOCC10 sub‐cloning vector via NotI/AscI. Subsequently, the insert digested with AscI, blunted and digested with BamHI. Simultaneously, pENTR3C was digested with BamHI/EcoRV. Afterwards, fragments were ligated.
pENTR:POK2 (1–589)T281N: Point mutations in the ATP‐binding site (T281N) were introduced by amplification from pENTR3C_POK2 (1–589) using mismatch primers (Appendix Table S1). The two overlapping amplicons were fused via extension PCR. The resulting fragment was cloned into the pENTR:POK2 (1–589) via NotI/BstEII digest to replace the respective wild‐type fragment.
pENTR:POK2 (1–189) and pENTR:POK2 (183–589): PCR products corresponding to fragments POK2 (1–189) or POK2 (183–589) were amplified with appropriate primers (Appendix Table S1), digested with NotI/XbaI, and ligated into NotI/XbaI sites of pENTR:POK2 (1–589).
pENTR:POK2 (Δ590–2,082): POK2 (1–589) and POK2 (2,083–2,771) were amplified from respective pENTR clones and subsequently combined by fusion PCR introducing a short linker sequence (Appendix Table S1).
The PCR product and pENTR:POK2 (1–589) were digested with NcoI/XbaI and ligated, resulting in pENTR3C:POK2 (Δ590–2,082).
pENTR:POK2 (1–2,771): The pENTR:POK2(Δ590‐2082) vector and PCR product PCRI (Appendix Table S1) were digested with BstEII/XhoI and ligated. The resulting vector was linearized with BstEII and a 3,654 bp BstEII fragment, resulting from a digest of PCR product PCRII (Appendix Table S1), was inserted. The final full‐length clone represents a hybrid of cDNA and genomic DNA.
pENTR:MAP65‐3/PLE: The coding sequence, without stop codon, was amplified (Appendix Table S1) from seedling cDNA and cloned into pENTR3C via KpnI/NotI.
pENTR:EB1b: The coding sequence of EB1b was amplified from seedling cDNA and cloned in to the pGEM T‐easy vector (Promega, A1360). From there, it was cloned into pENTR2B via EcoRI/XhoI digest and subsequent ligation.
Generating XFP‐expression clones
GFP‐POK2 (2,083–2,771): pDONR221: POK2 (2,083–2,771) was used in a Gateway LR reaction (Invitrogen, 11791‐020) with pK7WGF2 to create the N‐terminal GFP fusion protein 59.
GFP‐POK2 (1–589): Subcloning vector pOCC10:POK2 (1–589) was sequentially digested with SgsI, blunted and digested with XbaI. Simultaneously, binary vector pFK241 pGreenIIS was sequentially digested with BsrgI, blunted and digested with XbaI. The resulting fragments were ligated.
GFP‐POK2 (1–2,771): pENTR:POK2 (1–2,771) was recombined with pFK241 pGreenIIS in a Gateway LR reaction.
MAP65‐X‐RFP: pENTR:MAP65‐3/PLE, pDONR207: MAP65‐1 16, pDONR207: MAP65‐1‐(9D) 21 and pDONR207: MAP65‐5 16 were used in a Gateway LR reaction (Invitrogen, 11791‐020) with pB7RWG2 to create the C‐terminal RFP‐fusion proteins 59.
Plant transformation
Transgene integration into the plant genome was accomplished by the Agrobacterium tumefaciens transformation 60, using strain GV3101. Screening for resistant transformants was performed on appropriate selective medium.
Rescue lines
To examine the rescue ability of GFP‐POK2 (1–2,771), a selected transgenic T2 line was crossed with the pok1‐1 pok2‐3 double mutant. F2 plants were examined for the presence of the GFP‐POK2 (1–2,771) transgene (resistance to kanamycin) and the pok1‐1 pok2‐3 mutant phenotypes, which was reduced to 9% (n = 137) compared to the expected 20% usually obtained for pok1‐1 pok2‐3 mutant 31. F2 plants were genotyped for the presence of the T‐DNA insertion in pok1‐1 pok2‐3 31. Phenotypes of selected F3 plants (87 > n < 102) homozygous for both were further examined. Phenotypic defects, characteristic of pok1‐1 pok2‐3 double mutant (number of cotyledons and angle between them, reduced seed germination), and root development are repressed in the F3 lines compared to pok1‐1 pok2‐3 mutants and GFP‐POK2 (1–2,771) transgenic line, implying that GFP‐POK2 (1–2,771) is sufficient to rescue the mutant phenotype. All F3 plants were genotyped and confirmed to be pok1‐1 pok2‐3.
Staining with fluorescent dyes
To visualize cell plate formation and the root architecture, 4‐ to 6‐day‐old seedlings were mounted in aqueous solution of either FM4‐64 (1:1,000; Invitrogen Cat. F34653) or propidium iodide (10 μg/ml; Sigma‐Aldrich, 1002395778). FM4‐64 stains the plasma membrane as well as the developing cell plate, and propidium iodide labels the cell walls of living plant cells.
Mating‐based Cyto‐SUS assays
The Mating Based Cyto‐SUS Assay was performed as described previously 36. In single Gateway LR reactions, pENTR:MAP65‐3/PLE was cloned into the OST‐Cub destination vector, whereas pENTR:POK2 (1–189), pENTR:POK2 (183–589), pENTR:POK2 (1–589), and pENTR:POK2 (2,083–2,771) were cloned into the pNX35‐Dest destination vector 61, 62. Expression clones were transformed into yeast, mated and dropped on selection plates. To confirm fusion protein expression, yeast cells were harvested before mating and proteins were extracted. To detect fusion proteins, we performed Western blot analyses using anti‐HA antibody, coupled with peroxidase (1:10,000; Roche, Cat. 12013819001) for the detection of NubG‐2xHA‐X fusion proteins; for the detection of OST‐MAP65‐3/PLE‐LexA fused to Cub, anti‐VP16 (1:1,000; GeneTex, Cat. GTX30776) antibody was used as a first antibody and an anti‐rabbit‐POD as a second antibody (1:10,000; Merck‐Millipore, Cat. AP307P, BM Chemiluminescence Blotting Substrate POD, Roche 11500708001).
Transient expression of fusion proteins in Tobacco leaves
Wild‐type tobacco (Nicotiana benthamiana) plants were grown in soil at 26°C on standard day and night conditions (16 h light/8 h darkness cycle). Up to three fully expanded leaves of 4‐ to 5‐week‐old tobacco plants were infiltrated with 2 ml of Agrobacterium tumefaciens GV3101 cultures carrying respective plasmids. Before infiltration, cultures, grown to OD600 1.0, were pelleted and washed twice in dH2O, then adjusted to OD600 0.8. For co‐expression of fusion proteins, Agrobacterium cultures carrying different plasmids were mixed before infiltration.
Bioinformatic analyses
Motor domain prediction was extracted from Uniprot (http://www.uniprot.org/uniprot/A0A178VJB1) 63. Coiled‐coil domains were predicted by Pair Coil (http://cb.csail.mit.edu/cb/paircoil2/) 64.
Data availability
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: POK2 (AT3G19050), MAP65‐3/PLEIADE (AT5G51600), MAP65‐1 (AT5G55230), MAP65‐5 (AT2G38720), EB1b (AT5G62500).
Author contributions
EL, AH, and PL performed experiments and analyzed data. AG, DVD, and M‐TH provided material. SM analyzed data, conceptualized research, and wrote the initial manuscript. All authors contributed to editing of the final manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Movie EV1
Movie EV2
Movie EV3
Movie EV4
Movie EV5
Review Process File
Acknowledgements
We acknowledge the ABRC and NASC for distribution of seeds used in this study. We appreciate the help of Bettina Alber, Jens Reich, Leander Rohr, Steffi Zimmermann, and Luise Zühl with data collection. We thank Frank Küttner for sharing the plasmid pFK241 pGreenIIS. We gratefully acknowledge support from the University of Tübingen and funding from the Deutsche Forschungsgemeinschaft to SM (MU 3133/1‐1 and MU 3133/5‐1).
EMBO Reports (2018) 19: e46085
References
- 1. Rappaport R (1986) Establishment of the mechanism of cytokinesis in animal cells. Int Rev Cytol 105: 245–281 [DOI] [PubMed] [Google Scholar]
- 2. Jurgens G (2005) Cytokinesis in higher plants. Annu Rev Plant Biol 56: 281–299 [DOI] [PubMed] [Google Scholar]
- 3. Lipka E, Herrmann A, Mueller S (2015) Mechanisms of plant cell division. WIREs Dev Biol 4: 391–405 [DOI] [PubMed] [Google Scholar]
- 4. Smertenko A, Assaad F, Baluska F, Bezanilla M, Buschmann H, Van Damme D, Drakakaki G, Hauser M‐T, Janson M, Mineyuki Y et al (2017) Plant cytokinesis: terminology for structures and processes. Trends Cell Biol 27: 885–894 [DOI] [PubMed] [Google Scholar]
- 5. Park M, Touihri S, Muller I, Mayer U, Jurgens G (2012) Sec1/Munc18 protein stabilizes fusion‐competent syntaxin for membrane fusion in Arabidopsis cytokinesis. Dev Cell 22: 989–1000 [DOI] [PubMed] [Google Scholar]
- 6. Smertenko AP, Piette B, Hussey PJ (2011) The origin of phragmoplast asymmetry. Curr Biol 22: 1924–1930 [DOI] [PubMed] [Google Scholar]
- 7. Richter S, Kientz M, Brumm S, Nielsen ME, Park M, Gavidia R, Krause C, Voss U, Beckmann H, Mayer U et al (2014) Delivery of endocytosed proteins to the cell‐division plane requires change of pathway from recycling to secretion. Elife 3: e02131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Otegui MS, Verbrugghe KJ, Skop AR (2005) Midbodies and phragmoplasts: analogous structures involved in cytokinesis. Trends Cell Biol 15: 404–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yamashita A, Sato M, Fujita A, Yamamoto M, Toda T (2005) The roles of fission yeast ASE1 in mitotic cell division, meiotic nuclear oscillation, and cytokinesis checkpoint signaling. Mol Biol Cell 16: 1378–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Subramanian R, Wilson‐Kubalek EM, Arthur CP, Bick MJ, Campbell EA, Darst SA, Milligan RA, Kapoor TM (2010) Insights into antiparallel microtubule crosslinking by PRC1, a conserved nonmotor microtubule binding protein. Cell 142: 433–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Müller S, Smertenko A, Wagner V, Heinrich M, Hussey PJ, Hauser MT (2004) The plant microtubule‐associated protein AtMAP65‐3/PLE is essential for cytokinetic phragmoplast function. Curr Biol 14: 412–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ho C‐MK, Hotta T, Guo F, Roberson RW, Lee Y‐RJ, Liu B (2011) Interaction of antiparallel microtubules in the phragmoplast is mediated by the microtubule‐associated protein MAP65‐3 in Arabidopsis . Plant Cell 23: 2909–2923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ho C‐MK, Lee Y‐RJ, Kiyama LD, Dinesh‐Kumar SP, Liu B (2012) Arabidopsis microtubule‐associated protein MAP65‐3 cross‐links antiparallel microtubules toward their plus ends in the phragmoplast via its distinct C‐terminal microtubule binding domain. Plant Cell 24: 2071–2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kosetsu K, de Keijzer J, Janson ME, Goshima G (2013) Microtubule‐associated protein65 is essential for maintenance of phragmoplast bipolarity and formation of the cell plate in Physcomitrella patens . Plant Cell 25: 4479–4492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li H, Sun B, Sasabe M, Deng X, Machida Y, Lin H, Julie Lee YR, Liu B (2017) Arabidopsis MAP65‐4 plays a role in phragmoplast microtubule organization and marks the cortical cell division site. New Phytol 215: 187–201 [DOI] [PubMed] [Google Scholar]
- 16. Van Damme D, Van Poucke K, Boutant E, Ritzenthaler C, Inzé D, Geelen D (2004) In vivo dynamics and differential microtubule‐binding activities of MAP65 proteins. Plant Physiol 136: 3956–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Smertenko AP, Kaloriti D, Chang H‐Y, Fiserova J, Opatrny Z, Hussey PJ (2008) The C‐terminal variable region specifies the dynamic properties of Arabidopsis microtubule‐associated protein MAP65 isotypes. Plant Cell 20: 3346–3358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sasabe M, Kosetsu K, Hidaka M, Murase A, Machida Y (2011) Arabidopsis thaliana MAP65‐1 and MAP65‐2 function redundantly with MAP65‐3/PLEIADE in cytokinesis downstream of MPK4. Plant Signal Behav 6: 743–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sasabe M, Machida Y (2006) MAP65: a bridge linking a MAP kinase to microtubule turnover. Curr Opin Plant Biol 9: 563–570 [DOI] [PubMed] [Google Scholar]
- 20. Sasabe M, Soyano T, Takahashi Y, Sonobe S, Igarashi H, Itoh TJ, Hidaka M, Machida Y (2006) Phosphorylation of NtMAP65‐1 by a MAP kinase down‐regulates its activity of microtubule bundling and stimulates progression of cytokinesis of tobacco cells. Genes Dev 20: 1004–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Boruc J, Weimer AK, Stoppin‐Mellet V, Mylle E, Kosetsu K, Cedeño C, Jaquinod M, Njo M, De Milde L, Tompa P et al (2017) Phosphorylation of MAP65‐1 by Arabidopsis aurora kinases is required for efficient cell cycle progression. Plant Physiol 173: 582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Boruc J, Van Damme D (2015) Endomembrane trafficking overarching cell plate formation. Curr Opin Plant Biol 28: 92–98 [DOI] [PubMed] [Google Scholar]
- 23. Steiner A, Rybak K, Altmann M, McFarlane HE, Klaeger S, Nguyen N, Facher E, Ivakov A, Wanner G, Kuster B et al (2016) Cell cycle‐regulated PLEIADE/AtMAP65‐3 links membrane and microtubule dynamics during plant cytokinesis. Plant J 88: 531–541 [DOI] [PubMed] [Google Scholar]
- 24. Müller S, Jürgens G (2016) Plant cytokinesis—No ring, no constriction but centrifugal construction of the partitioning membrane. Semin Cell Dev Biol 53: 10–18 [DOI] [PubMed] [Google Scholar]
- 25. Smertenko A, Hewitt SL, Jacques CN, Kacprzyk R, Liu Y, Marcec MJ, Moyo L, Ogden A, Oung HM, Schmidt S et al (2018) Phragmoplast microtubule dynamics ‐ a game of zones. J Cell Sci 131: jcs203331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wick SM, Duniec J (1983) Immunofluorescence microscopy of tubulin and microtubule arrays in plant cells. I. Preprophase band development and concomitant appearance of nuclear envelope‐associated tubulin. J Cell Biol 97: 235–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mineyuki Y (1999) The preprophase band of microtubules: its function as a cytokinetic apparatus in higher plants In International review of cytology, Kwang WJ. (ed.), pp 1–49. Cambridge, MA: Academic Press; [Google Scholar]
- 28. Walker KL, Muller S, Moss D, Ehrhardt DW, Smith LG (2007) Arabidopsis tangled identifies the division plane throughout mitosis and cytokinesis. Curr Biol 17: 1827–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xu XM, Zhao Q, Rodrigo‐Peiris T, Brkljacic J, He CS, Muller S, Meier I (2008) RanGAP1 is a continuous marker of the Arabidopsis cell division plane. Proc Natl Acad Sci USA 105: 18637–18642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Martinez P, Luo A, Sylvester A, Rasmussen CG (2017) Proper division plane orientation and mitotic progression together allow normal growth of maize. Proc Natl Acad Sci USA 114: 2759–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lipka E, Gadeyne A, Stöckle D, Zimmermann S, De Jaeger G, Ehrhardt DW, Kirik V, Van Damme D, Müller S (2014) The phragmoplast‐orienting kinesin‐12 class proteins translate the positional information of the preprophase band to establish the cortical division zone in Arabidopsis thaliana . Plant Cell 26: 2617–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Buschmann H, Dols J, Kopischke S, Peña EJ, Andrade‐Navarro MA, Heinlein M, Szymanski DB, Zachgo S, Doonan JH, Lloyd CW (2015) Arabidopsis KCBP interacts with AIR9 but stays in the cortical division zone throughout mitosis via its MyTH4‐FERM domain. J Cell Sci 128: 2033–2046 [DOI] [PubMed] [Google Scholar]
- 33. Smith LG, Hake S, Sylvester AW (1996) The tangled‐1 mutation alters cell division orientations throughout maize leaf development without altering leaf shape. Development 122: 481–489 [DOI] [PubMed] [Google Scholar]
- 34. Müller S, Han S, Smith LG (2006) Two kinesins are involved in the spatial control of cytokinesis in Arabidopsis thaliana . Curr Biol 16: 888–894 [DOI] [PubMed] [Google Scholar]
- 35. Mir R, Morris VH, Buschmann H, Rasmussen CG (2018) Division plane orientation defects revealed by a synthetic double mutant phenotype. Plant Physiol 176: 418–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stöckle D, Herrmann A, Lipka E, Lauster T, Gavidia R, Zimmermann S, Müller S (2016) Putative RopGAPs impact division plane selection and interact with kinesin‐12 POK1. Nat Plants 2: 16120 [DOI] [PubMed] [Google Scholar]
- 37. Livanos P, Chugh M, Müller S (2017) Analysis of phragmoplast kinetics during plant cytokinesis In Plant protein secretion: methods and protocols, Jiang L. (ed.), pp 137–150. New York: Springer; [DOI] [PubMed] [Google Scholar]
- 38. Endow SA, Kull FJ, Liu H (2010) Kinesins at a glance. J Cell Sci 123: 3420–3424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nakata T, Hirokawa N (1995) Point mutation of adenosine triphosphate‐binding motif generated rigor kinesin that selectively blocks anterograde lysosome membrane transport. J Cell Biol 131: 1039–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. de Keijzer J, Kieft H, Ketelaar T, Goshima G, Janson ME (2017) Shortening of microtubule overlap regions defines membrane delivery sites during plant cytokinesis. Curr Biol 27: 514–520 [DOI] [PubMed] [Google Scholar]
- 41. Müller S, Fuchs E, Ovecka M, Wysocka‐Diller J, Benfey PN, Hauser MT (2002) Two new loci, PLEIADE and HYADE, implicate organ‐specific regulation of cytokinesis in Arabidopsis . Plant Physiol 130: 312–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Steiner A, Muller L, Rybak K, Vodermaier V, Facher E, Thellmann M, Ravikumar R, Wanner G, Hauser MT, Assaad FF (2016) The membrane‐associated Sec1/Munc18 KEULE is required for phragmoplast microtubule reorganization during cytokinesis in Arabidopsis . Mol Plant 9: 528–540 [DOI] [PubMed] [Google Scholar]
- 43. Lee YR, Li Y, Liu B (2007) Two Arabidopsis phragmoplast‐associated kinesins play a critical role in cytokinesis during male gametogenesis. Plant Cell 19: 2595–2605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lipka E, Müller S (2012) Potential roles for Kinesins at the cortical division site. Front Plant Sci 3: 158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhu C, Dixit R (2012) Functions of the Arabidopsis kinesin superfamily of microtubule‐based motor proteins. Protoplasma 249: 887–899 [DOI] [PubMed] [Google Scholar]
- 46. Murata T, Sano T, Sasabe M, Nonaka S, Higashiyama T, Hasezawa S, Machida Y, Hasebe M (2013) Mechanism of microtubule array expansion in the cytokinetic phragmoplast. Nat Commun 4: 1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wu S‐Z, Bezanilla M (2014) Myosin VIII associates with microtubule ends and together with actin plays a role in guiding plant cell division. Elife 3: e03498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Boleti H, Karsenti E, Vernos I (1996) Xklp2, a novel xenopus centrosomal kinesin‐like protein required for centrosome separation during mitosis. Cell 84: 49–59 [DOI] [PubMed] [Google Scholar]
- 49. Hancock William O (2014) Mitotic kinesins: a reason to delve into kinesin‐12. Curr Biol 24: R968–R970 [DOI] [PubMed] [Google Scholar]
- 50. Sturgill Emma G, Das Dibyendu K, Takizawa Y, Shin Y, Collier Scott E, Ohi Melanie D, Hwang W, Lang Matthew J, Ohi R (2014) Kinesin‐12 Kif15 targets kinetochore fibers through an intrinsic two‐step mechanism. Curr Biol 24: 2307–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Drechsler H, McAinsh AD (2016) Kinesin‐12 motors cooperate to suppress microtubule catastrophes and drive the formation of parallel microtubule bundles. Proc Natl Acad Sci USA 113: E1635–E1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Drechsler H, McHugh T, Singleton MR, Carter NJ, McAinsh AD (2014) The Kinesin‐12 Kif15 is a processive track‐switching tetramer. Elife 3: e01724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mann BJ, Balchand SK, Wadsworth P (2017) Regulation of Kif15 localization and motility by the C‐terminus of TPX2 and microtubule dynamics. Mol Biol Cell 28: 65–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lee YR, Liu B (2000) Identification of a phragmoplast‐associated kinesin‐related protein in higher plants. Curr Biol 10: 797–800 [DOI] [PubMed] [Google Scholar]
- 55. Pan R, Lee YR, Liu B (2004) Localization of two homologous Arabidopsis kinesin‐related proteins in the phragmoplast. Planta 220: 156–164 [DOI] [PubMed] [Google Scholar]
- 56. Lee Y‐RJ, Liu B (2013) The rise and fall of the phragmoplast microtubule array. Curr Opin Plant Biol 16: 757–763 [DOI] [PubMed] [Google Scholar]
- 57. Walczak CE, Shaw SL (2010) A MAP for bundling microtubules. Cell 142: 364–367 [DOI] [PubMed] [Google Scholar]
- 58. Rasband WS (1997. ‐2014) ImageJ. Bethesda, MD: US National Institutes of Health; [Google Scholar]
- 59. Karimi M, Inzé D, Depicker A (2002) GATEWAY™ vectors for Agrobacterium‐mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- 60. Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- 61. Grefen C, Blatt MR (2012) Do calcineurin B‐like proteins interact independently of the serine threonine kinase CIPK23 with the K+ channel AKT1? Lessons learned from a ménage à trois. Plant Physiol 159: 915–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Karnik R, Zhang B, Waghmare S, Aderhold C, Grefen C, Blatt MR (2015) Binding of SEC11 indicates its role in SNARE recycling after vesicle fusion and identifies two pathways for vesicular traffic to the plasma membrane. Plant Cell 27: 675–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Consortium TU (2017) UniProt: the universal protein knowledgebase. Nucleic Acids Res 45: D158–D169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. McDonnell AV, Jiang T, Keating AE, Berger B (2006) Paircoil2: improved prediction of coiled coils from sequence. Bioinformatics 22: 356–358 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Movie EV1
Movie EV2
Movie EV3
Movie EV4
Movie EV5
Review Process File
Data Availability Statement
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: POK2 (AT3G19050), MAP65‐3/PLEIADE (AT5G51600), MAP65‐1 (AT5G55230), MAP65‐5 (AT2G38720), EB1b (AT5G62500).


