Abstract
Developmental dysplasia of the hip (DDH) is the most common skeletal developmental disease. However, its genetic architecture is poorly understood. We conduct the largest DDH genome-wide association study to date and replicate our findings in independent cohorts. We find the heritable component of DDH attributable to common genetic variants to be 55% and distributed equally across the autosomal and X-chromosomes. We identify replicating evidence for association between GDF5 promoter variation and DDH (rs143384, effect allele A, odds ratio 1.44, 95% confidence interval 1.34–1.56, P = 3.55 × 10−22). Gene-based analysis implicates GDF5 (P = 9.24 × 10−12), UQCC1 (P = 1.86 × 10−10), MMP24 (P = 3.18 × 10−9), RETSAT (P = 3.70 × 10−8) and PDRG1 (P = 1.06 × 10−7) in DDH susceptibility. We find shared genetic architecture between DDH and hip osteoarthritis, but no predictive power of osteoarthritis polygenic risk score on DDH status, underscoring the complex nature of the two traits. We report a scalable, time-efficient recruitment strategy and establish for the first time to our knowledge a robust DDH genetic association locus at GDF5.
Konstantinos Hatzikotoulas et al. report the largest genome-wide association study to date for developmental dysplasia of the hip using national clinical audit data from the UK. They find a significant association with the GDF5 locus and evidence for shared genetic architecture with hip osteoarthritis.
Introduction
Developmental dysplasia of the hip (DDH) is a disorder characterised by abnormal development of the hip joint and presents with varying severity from mild uncovering of the femoral head to complete dislocation of the joint1. It is the most common developmental musculoskeletal anomaly, with population-weighted average incidence that ranges strongly with ethnic background from 0.06 per 1000 live births in Black Africans to 76.1 per 1000 live births in Native Americans2, and with an incidence in the UK European population of 3.6 per 1000. DDH is a complex disorder, with known associations including female sex, first-born, breech presentation, and family history2. There is a seven-fold increase in the incidence between siblings and a 10-fold increase in the parents of probands compared to the general population3, and a concordance rate of 41% between identical twins versus 3% in dizygotic twins4.
While DDH is heritable, its genetic architecture remains poorly characterised. Several linkage scans and candidate gene studies have implicated possible associated genetic variants, including in GDF55, but to date no replicated loci of genome-wide significance have been identified2,6–9. A recent genome-wide association study (GWAS) of 386 patients and 558 controls in the Han Chinese population suggested an association with variation in UQCC (odds ratio (OR) 1.35, P = 3.63 × 10-6), a gene adjacent to GDF510. Morphological abnormalities of the hip such as DDH are also recognised as risk factors for the development of secondary degenerative change of the hip11,12, and commonly result in hip replacement in adult life12,13. Idiopathic hip osteoarthritis is also very common and has a substantial heritable component14, and may share common genetic aetiology with DDH2,7,15.
Systematic examination of genome-wide variation in DDH in larger sample sizes is necessary to clarify its heritable biology and inform mechanism-driven preventative strategies. However, sample collection for uncommon diseases can be challenging and time-consuming, as attested by the relative paucity of sample sets for genetic studies of DDH. Routinely collected national clinical audit datasets present an opportunity for efficient case ascertainment in genetic epidemiology. The National Joint Registry for England, Wales, Northern Ireland, and the Isle of Man (NJR, http://www.njrcentre.org.uk/) was established in 2003 to collect audit data on all hip and knee replacement surgery in these regions, for which it has a completeness rate of 97% (http://www.njrreports.org.uk/Data-Completeness-and-quality). As at 31 December 2014, the dataset held information on 711,765 primary hip replacement procedures, including data on the diagnostic indication for surgery.
We use the NJR as a case-ascertainment tool to conduct a nationwide genome-wide association scan to characterise the genetic architecture of DDH. We examine the heritable contribution to DDH, and use fine-mapping and gene-based enrichment approaches to identify likely causal variants and genes. Finally, we use independent osteoarthritis datasets to examine the potential for shared genetic aetiology between DDH and hip osteoarthritis. We report a scalable, time-efficient recruitment strategy. We find the heritable component of DDH attributable to common variants to be 55% with the lead replicating signal at genome-wide significance being rs143384 within the GDF5 promoter. We also find shared genetic architecture between DDH and hip osteoarthritis.
Results
Recruitment strategy
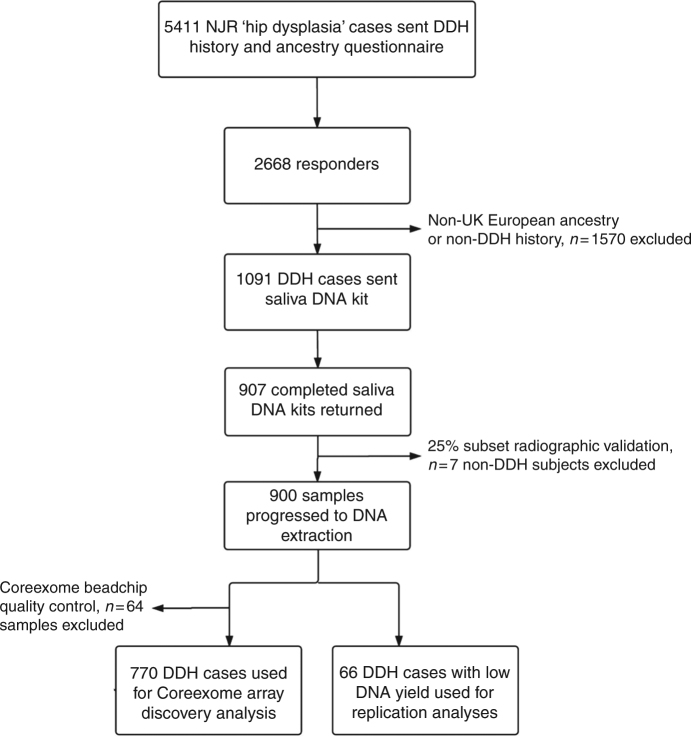
At the time of recruitment, the NJR had “hip dysplasia” recorded as the indication for hip replacement surgery in 5411 patients from the total database of 711,765 procedures (0.76%). All of these individuals were invited to participate in the postal screening phase of the study (Fig. 1). Following screening, 1091 consenting individuals with self-confirmed UK European ancestry and idiopathic DDH diagnosed in childhood were invited to submit a saliva sample. Of the 907 who returned a sample, 834 provided sufficient DNA to proceed to genome-wide genotyping. After quality control, 64 more individuals were excluded, leading the final number of DDH cases used in the genome-wide association analysis to 770. The sex distribution of the responding subjects was similar to that of the DDH population in the UK4. The diagnosis of DDH was independently validated by assessment of pelvic radiographs in a convenience sample of 25% of the subjects completing saliva return. The age and sex distribution of the radiographic validation subset was similar to the 907 that returned a saliva sample and radiographic analysis confirmed DDH in 224/231 (97%) subjects. Non-DDH cases were excluded from further analysis. The time taken from first patient mailshot to completion of genome-wide association analysis was approximately 18 months.
Fig. 1.
Recruitment flowchart
Controls were drawn from the United Kingdom Household Longitudinal Study (UKHLS), also known as Understanding Society (http://www.understandingsociety.ac.uk/). The UKHLS is an annual longitudinal panel survey of over 40,000 UK households from England, Scotland, Wales and Northern Ireland beginning in 2009–2010. Participants are surveyed annually and the study includes a wide range of sociodemographic, phenotypic, self-reported medical history and medication use data. Biomedical measures and blood samples were collected during a single nurse visit and DNA has been extracted and stored for genetic analyses. In total, 10,480 samples were genotyped on the Illumina HumanCoreExome-12v1-0_A chip at the Wellcome Trust Sanger Institute. After quality control (QC), genotype data for 9961 individuals were available. We further excluded all participants with any musculoskeletal disorder and the final dataset comprised 8016 individuals. A case:control ratio of ~1:4 was selected to be used in both discovery and replication stage to guard against case:control imbalance causing association tests to miscalculate for low frequency variants16.
The heritable component of DDH
Using genetic complex trait analysis (GCTA)17 across 770 cases and 3364 controls in the discovery GWAS, we found that common-frequency autosomal single-nucleotide polymorphisms (SNPs) explain 55% (±se = 6%, P < 0.0001) of the liability-scale heritability of DDH (Supplementary Table 1). The heritability estimate was similar (54.7 ± 5.8%) when the analysis was repeated using sex as a covariate. When the heritability analysis was stratified by chromosome, we found that the X chromosome contributed a similar amount to overall DDH heritability as each of the autosomes (Supplementary Fig. 1), and we identified no individual X-chromosome signals.
Discovery GWAS
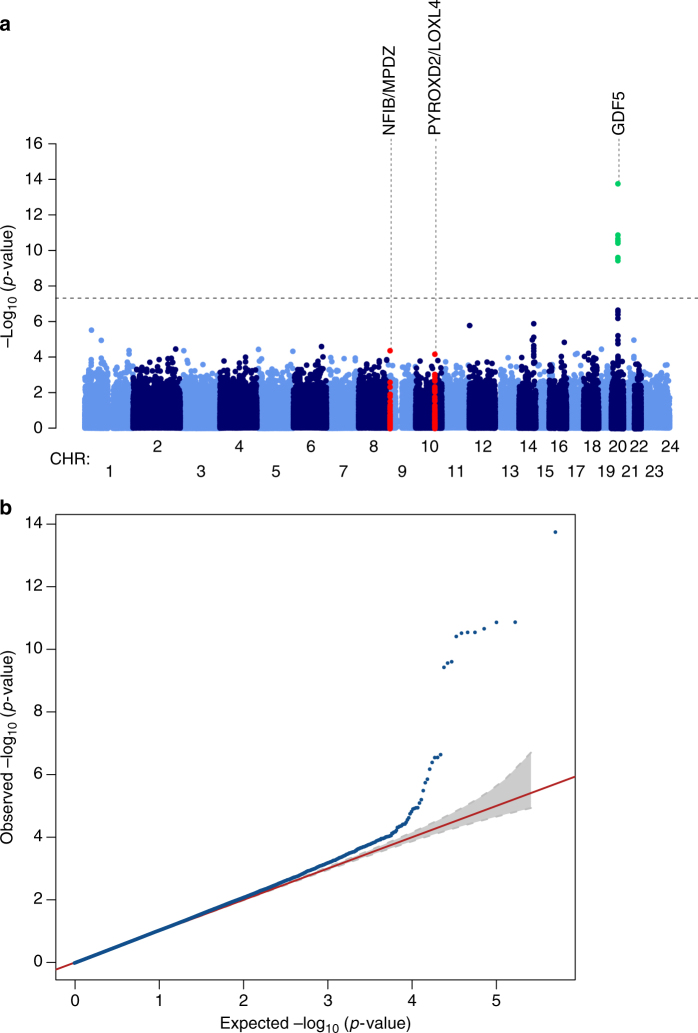
Genome-wide likelihood ratio test under an additive genetic model for association with DDH showed an excess of signals in the discovery sample set (Fig. 2a, b). Data were pruned for linkage disequilibrium using the clumping function in PLINK and 53 SNPs comprising 25 independent signals showed suggestive evidence for association with DDH with P < 1 × 10−4 compared to three independent variants expected under the null hypothesis of no association (binomial P = 9.57 × 10−17, Supplementary Table 2). Eleven correlated variants reached genome-wide significance (P < 5.0 × 10−8) and all reside in the same region. The lead variant, rs143384 (effect allele A, effect allele frequency (EAF) 0.60, OR [95% CI] 1.57 [1.3–1.77], P = 1.72 × 10−14), is located in the 5′ untranslated region (UTR) of GDF5 (20q11.22). We performed a conditional analysis specifically for rs143384 and rs143383 (effect allele A, EAF 0.64, OR [95% CI] 1.51 [1.33–1.70], P = 1.29 × 10−11) as they have both previously shown suggestive association with DDH. When rs143383 is conditioned on rs143384, the P-value becomes insignificant ((effect allele A, EAF 0.64, OR [95% CI] 1.51 [1.33–1.70], P = 0.61), suggesting the association observed with rs143383 is not an independent signal. Since DDH is more prevalent in women, we also repeated the analysis using sex as a covariate and found no qualitative difference in the results (Supplementary Table 3).
Fig. 2.
Manhattan and quantile–quantile plot of the DDH genome-wide association scan. a The dashed line indicates the genome-wide significance threshold (P = 5.0 × 10-8). Green dots represent variants for which P-values reached the genome-wide significance threshold and red dots illustrate the other two replicating signals. Chromosomes X and pseudo-autosomal regions on the chromosome X are represented by number 23 and 24, respectively. b Quantile–quantile plot of the data used in the GWAS
Replication
Independent association signals were taken forward to replication in three DDH cohorts of UK European ancestry, totalling 1129 cases and 4652 UKHLS controls. Following QC, one of the variants was excluded from further analysis due to low call rate. Five SNPs showed evidence for nominally significant (P < 0.05) association with the same direction of effect as the discovery cohort, compared with 1.35 under the null expectation (binomial test P = 0.01; Supplementary Table 4). rs143384 reached genome-wide significance in the replication dataset alone (effect allele A, EAF 0.61, OR [95% CI] 1.37 [1.24–1.51], P = 1.33 × 10−10). When the replication analysis was performed excluding the 66 NJR subjects that failed genome-wide genotyping QC, the findings were similar.
Meta-analysis
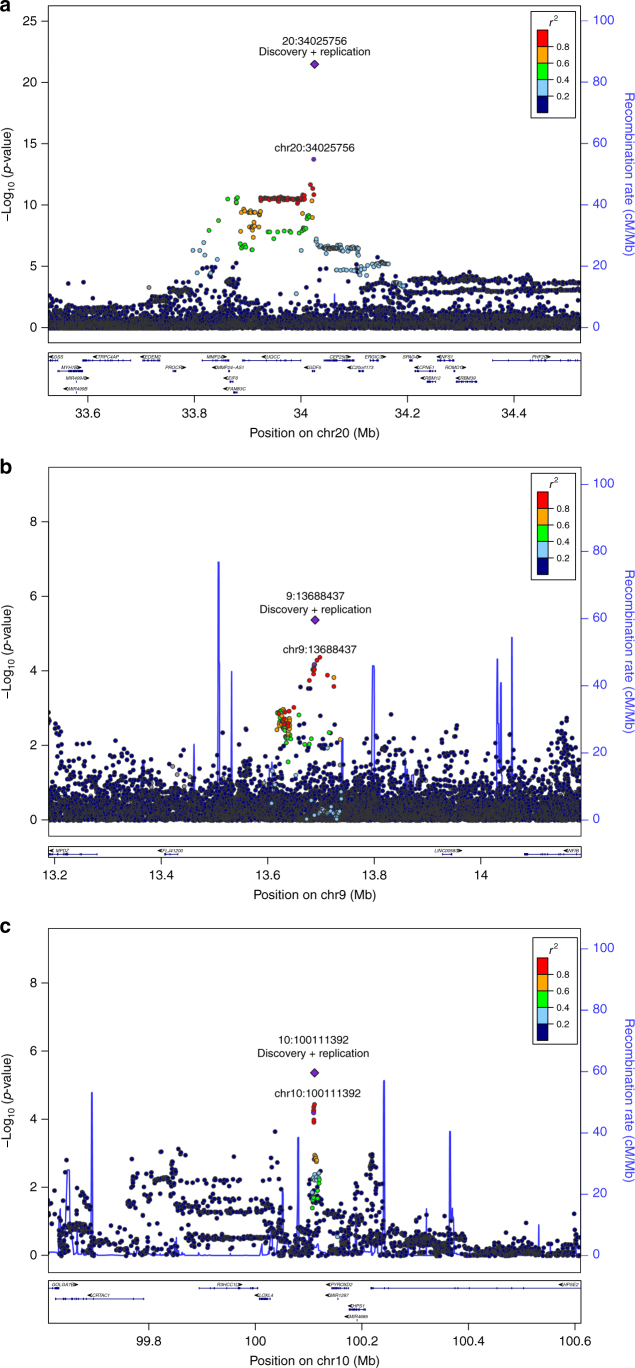
Twenty SNPs had the same direction of effect in both the discovery and replication analysis. Upon meta-analysis, three SNPs in the same region of chromosome 20 showed association with DDH at genome-wide significance: rs143384 in GDF5 (effect allele A, OR [95% CI] 1.44 [1.34–1.56], P = 3.55 × 10−22; Fig. 3a) rs12479765 in MMP24 (effect allele G, OR [95% CI] 1.33 [1.20–1.47], P = 3.18 × 10−8), and rs2050729 in RMB39 (effect allele G, OR [95% CI] 1.41 [1.25–1.58], P = 1.15 × 10−8). Conditional analysis of rs12479765 and rs2050729 on the lead variant rs143384 attenuated their association with DDH. Although the linkage disequilibrium (LD) r2 correlations among these three variants are lower than 0.16 (Supplementary Table 5), the P-values after conditioning on rs143384 were P = 0.004 and P = 0.045 for rs2050729 and rs12479765, respectively, suggesting residual nominal independent association of these variants with DDH. Rs143384 explained 0.96% of DDH variance on the liability scale (h2L) and the sibling relative risk ratio (λs) attributable to this variant was λs = 1.04 (Supplementary Fig. 2).
Fig. 3.
Regional association plots of the three replicating signals. Regional association plot of rs143384 (20:34025756) (a), rs4740554 (9:13688437) (b) and rs4919218 (10:100111392) (c). Each filled circle represents the P-value of analysed variants (as −log10 P-values) plotted against their physical position (NCBI Build 37). The P-value at the discovery stage and combined discovery and replication cohorts is represented by a purple circle and diamond, respectively. The other variants in the region are coloured depending on their degree of correlation (r2) with the index variant according to a scale from r2 = 0 (blue) to r2 = 1 (red). Gene location is annotated based on the UCSC genome browser
Two further independent signals showed evidence of replicating association with DDH, although they did not reach genome-wide significance at meta-analysis: rs4740554 (effect allele C, EAF 0.10, OR [95% CI] 1.30 [1.16–1.45], P = 4.44 × 10−6) and rs4919218 (effect allele C, EAF 0.31, OR [95% CI] 1.19 [1.10–1.28], P = 4.38 × 10−6; Fig. 3b, c; Table 1). rs4740554 and rs4919218 explained 0.36% and 0.24% of phenotypic variance (λs = 1.02 and 1.01), respectively (Supplementary Fig. 2).
Table 1.
Replicating variants associated with DDH
| SNP ID (chromosome: position) | Effect allele/other allele | Effect allele frequency (discovery stage) | Discovery odds ratio (95% CI) | Discovery P-value | Replication odds ratio (95% CI) | Replication P-value | Meta-analysis odds ratio (95% CI) | Meta-analysis P-value |
|---|---|---|---|---|---|---|---|---|
| rs143384 (20:34025756) | A/G | 0.60 | 1.57 (1.40–1.77) | 1.72 × 10−14 | 1.37 (1.24–1.51) | 1.33 × 10−10 | 1.44 (1.34–1.56) | 3.55 × 10−22 |
| rs4740554 (9:13688437) | C/A | 0.10 | 1.44 (1.22–1.71) | 4.09 × 10−05 | 1.20 (1.03–1.39) | 0.01 | 1.30 (1.16–1.45) | 4.44 × 10−6 |
| rs4919218 (10:100111392) | C/T | 0.69 | 1.27 (1.13–1.43) | 6.48 × 10−05 | 1.14 (1.03–1.25) | 0.0089 | 1.19 (1.10–1.28) | 4.38 × 10−6 |
Fine-mapping
For the GDF5 signal, the sum of probabilities of causality for five variants in the fine-mapped region was ≥0.95. Among these, both rs143384 and rs143383 have high epigenomic scores as they are located in the 5′ UTR of GDF5, have high nucleotide sequence conservation, and overlap DNase hypersensitivity and activating histone marks. Considering annotations and association P-values together, rs143384 had a >99% likelihood of being the causal variant, assuming only one variant as causal. For the other two replicating signals it was not possible to narrow down the location of the likely causal variants, as the chr10 and chr9 locus credible sets contain 483 and 618 variants, respectively.
Gene-based analyses
We performed gene-based analysis using MAGMA v1.03 (ref. 18) to identify genes which contain multiple variants that contribute to DDH. Prior to gene analysis, MAGMA applies an internal SNP QC process. MAGMA uses all remaining SNPs in a gene to compute gene P-values while accounting for LD and after adjusting for the family-wise error rate (FWER). We found five genes that were significantly associated with DDH susceptibility: GDF5, UQCC1, MMP24, RETSAT and PDRG1 (P = 9.24 × 10−12, P = 1.86 × 10−10, P = 3.18 × 10−9, P = 3.70 × 10−8 and P = 1.06 × 10−7, respectively). Following QC, 13, 18, 17, 20 and 6 SNPs were included in the analysis for GDF5, UQCC1, MMP24, RETSAT and PDRG1, respectively (Supplementary Fig. 3). To exclude association through GDF5, we repeated the analysis by removing GDF5 and all genes remained significantly associated with DDH. We further looked up the functionality of the variants that were used in the analysis for each gene to better understand whether their effects are local to the gene itself, or may have more distant regulatory effects. Over 59% of these remaining variants in each gene analysed by MAGMA are assumed to have high (disruptive) impact in the protein (e.g. missense, stop codon) (Supplementary Fig. 3) and none of the significant genes share variants that are in high LD with each other across genes (Supplementary Table 6). Fourteen variants are expression quantitative trait loci (eQTL) that regulate various genes nearby (Supplementary Data 1).
Genetic overlap with hip osteoarthritis
We calculated polygenic risk scores to detect shared genetic aetiology between DDH and hip osteoarthritis. We found no evidence that hip osteoarthritis polygenic risk score predicts DDH status. The best-fit polygenic risk score for hip osteoarthritis in the arcOGEN19 and in the UK Biobank (UKBB) International Classification of Diseases (ICD) 10 coded hip osteoarthritis datasets explained ~0.05% and 0.35% of variation in DDH susceptibility, respectively (FWER P > 0.05; Supplementary Fig. 4A, B). To further investigate the genetic overlap with hip osteoarthritis, we utilised LD score regression (LDSC)20 to estimate the genetic correlation between DDH and UKBB hip osteoarthritis datasets. DDH showed a nominally significant positive genetic correlation (rg) with hip osteoarthritis (rg = 0.5839, s.e. = 0.2068, P = 0.0047). However, this finding does not distinguish between shared genetic causes and a genetic causal relationship between the two traits. We also calculated the genetic correlations between DDH and 235 other traits and diseases using GWAS summary statistics and LD score regression implemented in the online software LD Hub21. After applying the false discovery rate (5% FDR) procedure to correct for multiple testing, none of the traits/diseases had a significant genetic correlation with DDH (Supplementary Data 2). We found a nominally significant genetic correlation with citrate (rg = 0.5934, s.e. = 0.2211, P = 0.0073), acetate (rg = −0.6778, s.e. = 0.2545, P = 0.0077), birth weight (rg = 0.2399, s.e. = 0.1104, P = 0.0298), infant head circumference (rg = 0.3932, s.e. = 0.1826, P = 0.0313) and child birth weight (rg = 0.3576, s.e. = 0.1815, P = 0.0488) which did not pass multiple-testing correction.
Discussion
We used case-ascertainment by national clinical audit database search and postal recruitment to facilitate the largest genome-wide association scan for DDH to date. We find that the heritable component of DDH due to common autosomal variants is approximately 55%, consistent with the complex nature of the disease, and find evidence for genetic correlation but a lack of predictive power of osteoarthritis polygenic risk scores on DDH. We establish variation within GDF5 on chromosome 20 as robustly associated with DDH susceptibility with rs143384 as the causal signal by fine-mapping, although GDF5 variation makes only a small contribution to overall DDH heritability. Through gene-based analyses we identify GDF5, UQCC1, MMP24, RETSAT and PDRG1 to be associated with DDH susceptibility.
The patients recruited in the discovery cohort were adults with a diagnosis of hip dysplasia ascertained via a national clinical audit database for hip replacement. We employed a stepwise filtration approach to mitigate misclassification bias in case ascertainment, using indication for surgery recorded in the NJR followed by self-reported questionnaires for phenotype confirmation. The discovery cohort studied here may reflect individuals with either untreated DDH or who have had treated disease eventually leading to the need for hip arthroplasty, resulting in potential selection bias. In mitigation against this, the female to male case ratio in our final NJR DDH cohort was similar to that expected in the general DDH population in the UK2, and contrasts with the sex ratio found in the population with idiopathic osteoarthritis of the hip22. The case filtration approach used was also validated with high concordance by examination of plain radiographic images acquired through the National Data Sharing Network. Ancestry screening by questionnaire in this population also resulted in only a small percentage of outliers requiring exclusion by genotyping. Finally, our replication cohorts mainly comprised independent populations of children recruited prospectively with a firm diagnosis of DDH, and in whom the EAF and odds ratio of association for the primary variant signal was almost identical as that found in the NJR discovery sample. DDH is a risk factor for degenerative disease at the hip11,12,23, and osteoarthritis susceptibility genes influence the association between hip morphology and osteoarthritis15,24,25. We therefore also examined whether we were simply detecting osteoarthritis-associated genetic loci in our DDH discovery cohort, as case identification required NJR registration for a hip replacement. Polygenic risk scores derived using independent hip osteoarthritis cohorts predicted only a small amount of the genetic variability in the DDH discovery population. However, using LDSC we found a 58% positive genetic correlation between the two diseases. The main difference between the two analyses is that PRS uses variants under a specific P-value threshold where LDSC uses genome-wide summary statistics. Although LDSC is a method to estimate the degree to which genetic risk factors are shared between two diseases or traits, it does not ascribe causation. Thus the observed correlation might be attributed either to shared genetic effects on both traits or because of causal pathways between traits. Moreover, it’s not clear if one variable is causal to the other or if causation is mediated by an independent factor. Future studies using Mendelian randomisation analyses may determine the causal relationships between DDH and hip osteoarthritis.
Previous studies examining the epidemiology of DDH within families using linkage approaches have demonstrated that heritable factors contribute between 50% and 85% of the total risk or liability of disease3,26,27, depending on population ancestry. Here, we used dense genome-wide genotyping data to estimate the heritable risk of DDH among unrelated individuals of UK European ancestry in the general English population. Our estimate of DDH heritability attributable to variation within common autosomal chromosomes was broadly similar to the estimates derived from linkage analyses, and confirms the complex nature of the disease2. Although DDH shows strong sexual dimorphism, the heritability estimate was almost identical (54.7 ± 5.8%) when the analysis was repeated using sex as a covariate. When the heritability analysis was stratified by chromosome, we found that the X-chromosome contributed a similar amount (adjusted for length) to overall DDH heritability as the autosomes (Supplementary Fig. 1), and we identified no individual chromosome X signals. We also conducted the discovery association analyses both with and without adjustment for sex (Supplementary Tables 2 and 3), and this made no qualitative difference to the results. Taken together, these data suggest that although DDH is a strongly sex-linked condition, this is not substantially due to variation in genomic DNA.
The gene GDF5 encodes growth differentiation factor 5 (GDF5), belonging to the transforming growth factor beta superfamily28. GDF5 is required for normal bone and joint development by promoting cartilage condensation and increasing the size of the skeletal elements through proliferation within epiphyseal cartilage29. Two GDF5 variants, rs143383 and rs143384, have previously shown suggestive association with DDH in unreplicated candidate gene studies in Asians and in Europeans5,30. Here we validate and extend these findings at the genome-wide level and with robust replication. These variants are in high linkage disequilibrium (r2 = 0.82, D′′ = 0.99), and were both directly genotyped in this study. Our data implicate rs143384 as the lead variant, with the rs143383 association with DDH being two orders of magnitude weaker. Conditional analyses substantiate this observation.
We identify an extended region of 16 DDH-associated variants at genome-wide significance and in high LD extending across GDF5 and UQCC, with rs143384 as the lead signal. Interrogation of the GTEx database indicates that rs143384 is an eQTL for multiple genes across various tissues (Supplementary Table 7)31,32. Recently, Chen et al.33 have shown that the GDF5 locus contains many separate regulatory elements that control expression of the gene at different joint sites, and that these flanking regions are large. The same group have since described a novel enhancer region GROW1 in an extended downstream regulatory region of GDF534. The lead variant in this 5′ enhancer is rs4911178 (Chr:Pos 20:33952620), a G > A substitution in which the variant allele A occurs at high frequency in Eurasian populations, and in which DDH and osteoarthritis are also common34. This variant was also directly typed in our discovery cohort, and identified in our extended association region on chromosome 20 (Supplementary Table 3). The effect allele G (OR 0.67, 95% CI 0.59–0.76, P = 2.98 × 10−11) was in strong LD (r2 = 0.8) with rs143384, consistent with the findings of Capellini et al.34. This annotated function provides novel opportunities for investigation of the role of GDF5 as a candidate target for DDH prevention.
Gene-based enrichment analysis can boost power to identify contributing loci by combining information across multiple SNPs co-localised at the gene level. We found significant associations between DDH and the GDF5, UQCC1, MMP24, RETSAT and PDRG1 genes. The results of functional look up, LD calculations and after removing the GDF5 locus from the analysis, tie the association to the above genes. However, these results refer only to the variants used in the gene-based analysis and further investigation through functional experiments is warranted to asses if these signals are truly independent. UQCC1 gene encodes a trans-membrane protein ubiquinol-cytochrome-c reductase complex chaperone, which is structurally similar to the mouse basic fibroblast growth factor repressed ZIC-binding protein. UQCC is expressed in differentiating chondrocytes and regulates growth control in mouse35,36. In humans, polymorphisms in this gene are associated with DDH10, bone size37, height38 and hip axis length39. MMP24 encodes a member of the peptidase M10 family of matrix metalloproteinases (MMPs) which are involved in the breakdown of extracellular matrix in normal physiological processes, such as embryonic development and tissue remodelling40. Polymorphisms within MMP24 have also been associated with height variation in childhood41,42. RETSAT codes for retinol saturase, an enzyme centrally involved in the metabolism of vitamin A43. Retinoic acid signalling is essential for normal limb bud development, including bone and cartilage formation44. The RETSAT pathways provides a further candidate therapeutic target for the prevention of DDH.
In conclusion, this largest study of DDH to date provides a comprehensive picture of its complex genetic architecture, the contribution of common variants to its heritability, and establishes the first robust DDH genetic locus to our knowledge. We demonstrate proof of principle for the utility of national clinical audit-based case ascertainment and recruitment for genetics and genomics studies using a strategy that is readily transferrable to other complex diseases in which large-scale datasets are held.
Methods
Study oversight
The study was approved by the National Research Ethics Service in England (NRES 12/YH/0390, 30 October 2012). Initial contact was made by the NJR data controller and all subjects provided written informed consent prior to participation.
Study populations
National Joint Registry (NJR) cohort: The NJR hip dysplasia base population comprised 5411 adult individuals (4095 females) living in England who had undergone a hip replacement recorded in the NJR by the operating surgeon as being for the indication of hip dysplasia and who had given written permission for re-contact for research purposes. All were invited to participate in the postal screening phase of the study between January 2013 and April 2014 (Fig. 1, recruitment flowchart). Screening for UK European descent was made using self-declared ancestry as: White European, Black African, Black Caribbean, Asian, Arabic, Chinese/Oriental, Mixed, or Other; and supplemented by country of birth, mother’s country of birth, and father’s country of birth. A further check for ethnicity outliers was made at genotyping, where DNA samples proceeded to analysis. DDH history was confirmed by self-completed questionnaire (Supplementary Table 8). Inclusion required a positive response to both question 1 and 2. Subjects responding positively to any items within questions 6, 7 or 10 were excluded from further participation. One thousand and ninety-one individuals confirming both UK European ancestry and idiopathic DDH diagnosed in childhood were invited to return by post a saliva sample for DNA extraction and genotyping. Nine hundred and seven subjects (mean age 51.6 ± 12.4 years; 803 (88%) females) returned a saliva sample for DNA processing. The sex distribution of the subjects returning a sample was similar to that of the DDH population in the UK2. In 231 subjects (mean age 51.6 ± 12.4 years; 211 (91%) females) operated in hospitals from which >10 cases were recruited and independent radiographic validation of the DDH phenotype was made following retrieval of the pelvic radiograph preceding hip replacement using the National Data Sharing Network (http://www.image-exchange.co.uk/). The images were reviewed independently by two experienced orthopaedic surgeons for the presence and grade of DDH using the Hartofilakidis45 classification. A third observer made the final judgement if there was disagreement. This analysis confirmed DDH in 224 (97%) of subjects. Eighty had radiographic evidence of a previous corrective pelvic and/or femoral osteotomy that precluded DDH grade classification beyond confirmation of the DDH phenotype. Of the remaining 151 subjects, 77 showed Hartofilakidis grade A dysplasia (femoral head contained within the original acetabulum despite the degree of subluxation), 48 were of grade B (low dislocation, femoral head articulates with a false acetabulum that partially covers the true acetabulum to a varying degree), and 31 were of grade C (high dislocation, femoral head is completely out of the true acetabulum and migrated superiorly and posteriorly to a varying degree). Seven subjects were excluded from further analysis, and all had a radiographic phenotype of idiopathic osteoarthritis. This distribution of disease grades was consistent with expectation of an adult patient cohort with a history of childhood DDH2,46,47. The mean age of the 770 subjects (693, 90% females) participating in the discovery GWAS was 50 ± 12 years.
DDH case control cohort: This population comprised 838 children (725 female) mean age 7 ± 5 years recruited by face-to-face contact at 1 of 18 participating hospitals (see related manuscript file) and undergoing hospital treatment for DDH, or currently under follow-up for hospital-treated DDH. These subjects had been previously diagnosed with DDH by patient history and clinical examination in conjunction with radiography or ultrasonography. Subjects with borderline forms of DDH, such as a brief episode of harness treatment for mild instability, were excluded. None of the participants in the DDH case control cohort have undergone hip replacement and are therefore independent of the participants in the NJR cohort.
Royal National Orthopaedic Hospital (RNOH) cohort: The subjects within this cohort comprised 225 adults (220 female), mean age 32 ± 16 years participating in the RNOH DDH biobank, with a clinician-confirmed childhood history of idiopathic DDH by patient history and clinical examination in conjunction with radiography or ultrasonography, and undergoing osteotomy for residual dysplasia and/or degenerative changes. Subject level identification checks between the participants in the RNOH versus NJR adult recruitment cohorts was used to exclude any subject overlap between the cohorts.
The replication cohort comprised the 838 children from the UK DDH case control consortium (DDH CCC) and the 225 adults with DDH recruited in the RNOH DDH biobank. In addition, 66 cases (59 female) from the NJR discovery cohort that were of insufficient DNA yield for genome-wide genotyping were also included in the replication cohort. The mean age and sex distribution of the 66 NJR subjects participating in the replication cohort was similar (age 50 ± 14 years, 59 (89%) females) to that participating in the NJR discovery cohort. Subject overlap between the discovery and replication populations, and between the replication cohorts was excluded using individual patient identifiers and by cross-referencing to the NJR dataset. Case duplication was excluded in the discovery cohort at both subject ID and genotype quality control stages.
The control population comprised 8016 participants from the UKHLS. A sample of 3364 UKHLS controls were included in the discovery stage and an independent sample of 4652 UKHLS controls was used in the replication stage at a case:control ratio of ~1:4. The proportion of female controls in the discovery stage was selected to reflect the distribution in cases. All replication and control participants were of UK European ancestry.
arcOGEN cohort: The arcOGEN dataset comprises 7410 unrelated men and women with osteoarthritis and 11008 unrelated controls from the UK19. The osteoarthritis hip strata comprises 3266 cases with symptomatic hip osteoarthritis and a radiographic KL score ≥2. The arcOGEN study used two different types of controls: population-based, unrelated UK controls which came from five distinct sources: the 1958 Birth Cohort (58BC) and the UK Blood Donor Service (UKBS) from the Wellcome Trust Case Control Consortium 2 (WTCCC2) study, the 1958 Birth Cohort from the Type 1 Diabetes Genetics Consortium (T1DGC) study, the Avon Longitudinal Study of Parents and Children (ALSPAC) and the People of the British Isles (PoBI) study (ST1); and unrelated, osteoarthritis-free controls (females only) from the TwinsUK cohort which comprises twins ascertained to study the heritability and genetics of age-related diseases48. These unselected twins were recruited from the general population through national media campaigns in the UK and shown to be comparable to age-matched population singletons in terms of disease-related and lifestyle characteristics49.
UK Biobank cohort: The UK Biobank is a prospective cohort of 500,000 men and women aged between 40 and 69 years at recruitment50. Hospital episode statistics were used to define case status for osteoarthritis in the UKBB sample. Inclusion and exclusion criteria were based on the International Statistical Classification of Diseases and Related Health Problems (ICD) 9 or ICD-10 codes. In total, 2396 cases were defined with a code for hip osteoarthritis, and no inflammatory arthritis syndromes or other musculoskeletal disorders. Age-matched controls were selected on the condition that they did not have any osteoarthritis-related ICD-9 or ICD-10 codes, or self-reported musculoskeletal disorders or symptoms.
Genotyping, quality control, and association analyses
DNA from 834 NJR DDH cases was genotyped using the Illumina HumanCoreExome-24 BeadChip (Illumina, San Diego, USA). Genotypes were called using Illumina Genome Studio Gencall. The UKHLS were genotyped on the Illumina HumanCoreExome-12v1-0_A chip and had previously been called using the same algorithm. After removing samples and variants with call rate <90%, samples underwent standard quality-control procedures, with exclusion criteria as follows: (i) call rate <98%, (ii) gender discrepancy, (iii) excess heterozygosity, (iv) duplicates and/or related, (v) ethnicity outliers, (vi) Fluidigm concordance (this identity check looks at sample concordance between Illumina and Fluidigm genotypes) (Supplementary Table 9) and variants with (i) call rate <98%, (ii) Hardy–Weinberg Equilibrium (HWE) P < 1 × 10−4, (iii) cluster separation score <0.4, (iv) minor allele frequency (MAF) <0.01, (v) minor allele count (MAC) <4 in cases and controls separately.
Data were pruned for linkage disequilibrium using the clumping function in PLINK51. Parameters used: (a) significance threshold for index single single-nucleotide polymorphism (SNP): 1e-4, (b) LD threshold for clumping: 0.20, and (c) physical distance threshold for clumping: 500 kb.
Following quality-control exclusions, the discovery cohort comprised 770 (693 females) DDH cases and 3364 (3048 females) controls. Power was calculated using Quanto v1.2.452, assuming a population mean of 0 and a standard deviation (SD) of 1. We calculated power (at the genome-wide significance threshold P = 5 × 10-8) to detect an effect size of 1 SD by fixing the sample size to the post-QC discovery sample, which gave >80% power to detect common variants with moderate effect size (Supplementary Fig. 5). Following QC checks at both the sample and SNP level, 256,867 overlapping variants were tested for association with DDH using SNPTEST v253 under an additive model and using a maximum likelihood ratio test. Association testing on the X chromosome assumes a model of full X inactivation where pseudo-autosomal regions on the X were treated like autosomes. The Y chromosome was not included in the analysis. Cluster plots of all prioritised variants were examined in cases and controls separately to minimise the possibility of spurious association due to genotyping error. Independent variants with P < 1 × 10−4 in the discovery set were selected for replication using the clumping function in PLINK.
De novo genotyping (prioritised variants genotyped in independent cohorts) in the 1129 replication cases was conducted using the iPLEX® Assay and the MassARRAY® System (Agena Bioscience, Inc). Variants with poor Agena design metrics were replaced with highly correlated proxies. Sample exclusions were based on sex inconsistencies and a sample call rate <80%. Variants with a call rate <90% and exact HWE P < 1 × 10−6 were also excluded. Case control association analysis in the replication cohort of 1129 (1004 females) cases and 4652 (2527 females) controls was performed under an additive genetic model using SNPTEST v2. When the replication analysis was performed excluding the 66 NJR subjects who failed genome-wide genotyping QC, the findings were similar (Supplementary Table 10). Finally, a fixed-effects meta-analysis across the discovery and replication datasets was conducted using GWAMA54, comprising a total of 1899 cases and 8016 controls. Genome-wide significance was defined as P < 5.0 × 10−8. Conditional single-variant association analyses were performed in SNPTEST v2 and were used to identify statistical independence. SNPTEST performs conditioning of a signal by adding any potentially dependent SNP as a covariate in the analysis. It then tests for a genetic effect over and above the phenotypic variability that is explained by the covariate(s). A variant was considered independent of the index SNP if the pre- and post-conditioning P-value difference was lower than two orders of magnitude. We repeated the analysis by adjusting for the first 10 principal components to guard against population stratification. We observed no qualitative difference and report the results of the unadjusted analysis. Since DDH prevalence varies widely between populations, we applied multidimensional scaling analyses to characterise our samples (cases and controls) in the context of broader global diversity and further ensure the homogeneity of the tested cohort. As shown in Supplementary Fig. 6 our cohort formed a well-defined cluster with GBR population of 1000 Genomes Project.
Estimation of heritability
We applied the genome-wide complex trait analysis (GCTA) approach to estimate the proportion of phenotypic variance in DDH susceptibility that is explained by genome-wide autosomal SNPs17, assuming a DDH prevalence of 3.6 per 100 live births in the UK2, and using an MAF cut-off of 0.01. GCTA uses a genetic relationship matrix (GRM) of pairs of samples as input for the restricted maximum likelihood (REML) analysis to estimate narrow sense heritability (h2). GCTA also converts h2 to liability-scale heritability by applying the transformation described by Lee et al.55, to make the estimates comparable with those for quantitative traits. An alternative approach based on phenotype correlation–genotype correlation (PCGC)56 regression has been proposed to avoid biases in REML estimation of heritability in the context of ascertained case–control studies. Here, we applied both GCTA and PCGC methodologies in order to validate our results and ensure that the heritability estimates did not suffer from case–control ascertainment bias. The sibling relative risk ratio and liability-scale variance explained were calculated according to the methods described by Morris et al.57. To investigate if the DDH strongly sex-linked condition is due to variation in genomic DNA, we further partitioned the genetic variance onto individual chromosomes and then compared them to heritability estimates of chromosome X. In this analysis, GCTA uses a joint analysis (all the chromosomes are fitted in one model) to protect against inter-chromosomal correlations and report the estimates of variance explained by different chromosomes independently of each other. GCTA contains built-in options for GRM and heritability estimation for the X chromosome and assumes a model of full X inactivation58.
Fine-mapping
In order to fine-map the replicating association signals, we used association statistics from imputed genotypes in determining the credible set, since an assumption of fine-mapping is that the causal variant is among the variants tested. All variants within 500 kb of the three replicating lead SNPs were pre-phased in SHAPEITv259 and the resulting haplotypes were imputed with IMPUTE260 using 1000 Genomes Project61 combined with UK10K Project62 data as the reference panel. Post-imputation quality control (QC) criteria for SNP exclusion were HWE P-value <1.0 × 10−4 and imputation info score <0.4. We then applied a method that assigns a relative “probability of regulatory function” (PRF) score among candidate causal variants, reweighting association statistics based on epigenomic annotations, and delineating the 95% probability set of likely causal variants. We collected a set of 70 genomic and epigenomic annotations, primarily Gencode gene annotations (v19), FANTOM transcription start sites and enhancers63,64, and imputed Roadmap Epigenomics histone marks, DNase hypersensitivity, and ChromHMM genome segmentations for the lymphoblastoid cell line epigenome (GM12878)65,66. We used the fgwas software67 to train a Bayesian hierarchical model to compute enrichment of eQTLs in these annotations based on summary statistics from the Geuvadis RNA sequencing project68. We used forward stepwise selection followed by cross-validation to arrive at a combined model with 37 annotations and their associated enrichments. The respective annotations from 119 Roadmap epigenomes were used to compute PRF scores for each variant at the new loci in each of the 119 epigenomes. At each locus we defined the credible set of causal variants using the WTCCC method with approximate Bayes factors (BF)69, where SNP k’s posterior probability of being causal is . Beginning with the most associated SNP, SNPs are sequentially added to the set until the combined probability is ≥95%. We then selected the top four epigenomes based on the maximum regulatory score among variants in the 95% credible set, and examined the regulatory annotations for variants in the credible set.
Gene-based analyses
In order to quantify the degree of association each gene has with the phenotype and understand the potential functional effects of the identified variants, we used the GWAS results to perform gene-based enrichment analyses in MAGMA v1.0318 to identify the joint association of all markers in a given gene with the phenotype. We used all 19,427 protein-coding genes from the NCBI 37.3 gene definitions and each gene was assigned the SNPs located between the gene’s start and stop sites. We used all variants that passed QC and after SNP annotation there were 17,516 genes that were covered by at least one SNP. Gene-based analysis was then carried out based on a multiple principal components regression model that accounts for linkage disequilibrium between SNPs, using an F-test to compute the gene-based P-value (self-contained P-value). P-values generated by MAGMA are not corrected for multiple testing. Since Bonferroni correction is conservative when variables (genes) are correlated, MAGMA provides a built-in FWER method which uses a permutation procedure to obtain P-values corrected for multiple testing. In all, 10,000 permutations were used in the analysis and the significance threshold was set at α = 0.05 (FWER threshold = 2.29−07).
Genetic overlap with hip osteoarthritis
Polygenic risks scores were created for all genotyped participants to test for shared genetic aetiology with idiopathic hip osteoarthritis. Polygenic risk scores were calculated as the summation of an individual’s genotype across many genetic loci using PRSice70, and weighted by the effect size estimated from two independent UK-based cohorts: arcOGEN hip osteoarthritis cases versus controls and UK Biobank hip osteoarthritis cases versus controls, as outlined in the Study Populations section above. Age-matched controls were selected on the condition that they did not have a hospital diagnosed (ICD-9 or ICD-10) or self-reported musculoskeletal disorders or symptoms. Before creating the scores, the analysis was restricted to autosomal SNPs and clumping was used to obtain SNPs in linkage equilibrium with an r2 < 0.1 within a 250-bp window. Several polygenic scores were created containing SNPs according to the significance of their association with the hip osteoarthritis. Logistic regression models were used to test the associations between the polygenic profiles of hip osteoarthritis and DDH and the first two PCs were used to adjust for population structure. Nagelkerke’s pseudo-R2 calculation as a measure of the variance explained and the most predictive threshold are shown in Supplementary Fig. 4A, B. The genetic correlation between DDH and hip osteoarthritis was estimated using LD score regression by Bulik-Sullivan et al.20. This method exploits the LD structure of SNPs across the genome and uses the cross-products of summary test statistics provided from two GWASs at each SNP and then regresses the cross-product on the LD score to calculate the genetic correlations between traits/diseases. To test whether DDH has a shared genetic aetiology with 235 other diseases, we used LD score regression as implemented in the online software LDHub21 (http://ldsc.broadinstitute.org/). For both analyses we used summary statistics of the imputed DDH dataset. Genotype imputation was performed on the Michigan Imputation Server (http://www.haplotype-reference-consortium.org/) using the updated Haplotype Reference Consortium71. The combined UK10K/1000 Genomes Project haplotype reference panel was used to impute UKBB dataset centrally (http://www.ukbiobank.ac.uk/wp-content/uploads/2014/04/imputation_documentation_May2015.pdf).
Data availability
Genome-wide genotype data of the DDH cases and UKHLS controls have been deposited to the European Genome-Phenome Archive (www.ebi.ac.uk/ega/home, DDH accession numbers: EGAD00010000766 and EGAS00001000916; UKHLS accession numbers: EGAD00010000891 and EGAS00001001232) and of the DDH NJR cases to the NJR data archive (www.njrcentre.org.uk).
Electronic supplementary material
Description of Additional Supplementary Files
Acknowledgements
We thank the patients and staff of all the hospitals who have contributed data to the National Joint Registry. We are grateful to the Healthcare Quality Improvement Partnership (HQIP), the National Joint Registry Steering Committee (NJRSC), and staff at the NJR Centre for facilitating this work. The views expressed represent those of the authors and do not necessarily reflect those of the NJRSC or HQIP who do not vouch for how the information is presented. This research has been conducted using the UK Biobank Resource under Application Number 9979. The study also utilised summary statistics data from arcOGEN (http://www.arcogen.org.uk/) funded by a special purpose grant from Arthritis Research UK (grant 18030). The study was funded by the Healthcare Quality Improvement Partnership, the National Institute for Health Research, The Wellcome Trust (098051), Medical Research Council and Arthritis Research UK as part of the MRC-Arthritis Research UK Centre for Integrated Research into Musculoskeletal Ageing (CIMA), The European Union’s Seventh Framework Program for research, technological development and demonstration (D-BOARD grant #305815), The Great Ormond Street Hospital National Institute for Health Research Biomedical Research Centre, and The Special Trustees of the Royal National Orthopaedic Hospital. The UK Household Longitudinal Study is led by the Institute for Social and Economic Research at the University of Essex and funded by the Economic and Social Research Council. The survey was conducted by NatCen and the genome-wide scan data were analysed and deposited by the Wellcome Trust Sanger Institute. Information on how to access the data can be found on the Understanding Society website https://www.understandingsociety.ac.uk/.
Author contributions
Study concept and design: J.M.W. and E.Z; subject recruitment: J.M.W., A.R., and DDH CCC, D.M.E.; data analysis and interpretation: K.H., J.S., E. Zengini, J.M.W., E.Z.; drafting and revising of manuscript and approval of the final version to be published: all authors; agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolve: K.H., A.R., J.M.W. and E.Z.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Konstantinos Hatzikotoulas, Andreas Roposch, Eleftheria Zeggini and J. Mark Wilkinson.
Contributor Information
J. Mark Wilkinson, Phone: +44 114 2714705, Email: j.m.wilkinson@sheffield.ac.uk.
The DDH Case Control Consortium:
Andrew Wainwright, Tim Theologis, Nicholas M. P. Clarke, Jonathan S. M. Dwyer, Aresh Hashemi-Nejad, Nigel Kiely, Marcos Katchburian, Nicolas Nicolaou, Johnathan Page, Martin Gargan, Colin Bruce, Anish Sanghrajka, Paul Marshall, Mark Flowers, Olivia Malaga-Shaw, Piers Mitchell, Ben Holroyd, and Manoj Ramachandran
Electronic supplementary material
Supplementary Information accompanies this paper at 10.1038/s42003-018-0052-4.
References
- 1.Harcke HT. Developmental dysplasia of the hip: a spectrum of abnormality. Pediatrics. 1999;103:152. doi: 10.1542/peds.103.1.152. [DOI] [PubMed] [Google Scholar]
- 2.Loder RT, Skopelja EN. The epidemiology and demographics of hip dysplasia. ISRN Orthop. 2011;2011:238607. doi: 10.5402/2011/238607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bjerkreim I, van der Hagen CB. Congenital dislocation of the hip joint in Norway. V. Evaluation of genetic and environmental factors. Clin. Genet. 1974;5:433–448. doi: 10.1111/j.1399-0004.1974.tb01717.x. [DOI] [PubMed] [Google Scholar]
- 4.Idelberger KH. [Orthopedic genetics and family counseling (proceedings)] Z. Orthop. Ihre Grenzgeb. 1978;116:552–554. [PubMed] [Google Scholar]
- 5.Rouault K, et al. Evidence of association between GDF5 polymorphisms and congenital dislocation of the hip in a Caucasian population. Osteoarthritis Cartilage. 2010;18:1144–1149. doi: 10.1016/j.joca.2010.05.018. [DOI] [PubMed] [Google Scholar]
- 6.Cilliers HJ, Beighton P. Beukes familial hip dysplasia: an autosomal dominant entity. Am. J. Med. Genet. 1990;36:386–390. doi: 10.1002/ajmg.1320360403. [DOI] [PubMed] [Google Scholar]
- 7.Mabuchi A, Nakamura S, Takatori Y, Ikegawa S. Familial osteoarthritis of the hip joint associated with acetabular dysplasia maps to chromosome 13q. Am. J. Hum. Genet. 2006;79:163–168. doi: 10.1086/505088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li LY, et al. [Gene mapping of developmental dysplasia of the hip in chromosome 17q21 region] Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2010;27:620–625. doi: 10.3760/cma.j.issn.1003-9406.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 9.Feldman G, et al. The Otto Aufranc Award: identification of a 4 Mb region on chromosome 17q21 linked to developmental dysplasia of the hip in one 18-member, multigeneration family. Clin. Orthop. Relat. Res. 2010;468:337–344. doi: 10.1007/s11999-009-1073-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun Y, et al. A common variant of ubiquinol-cytochrome c reductase complex is associated with DDH. PLoS ONE. 2015;10:e0120212. doi: 10.1371/journal.pone.0120212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murray RO. The aetiology of primary osteoarthritis of the hip. Br. J. Radiol. 1965;38:810–824. doi: 10.1259/0007-1285-38-455-810. [DOI] [PubMed] [Google Scholar]
- 12.Solomon L. Patterns of osteoarthritis of the hip. J. Bone Jt. Surg. Br. 1976;58:176–183. doi: 10.1302/0301-620X.58B2.932079. [DOI] [PubMed] [Google Scholar]
- 13.Murphy SB, Ganz R, Muller ME. The prognosis in untreated dysplasia of the hip. A study of radiographic factors that predict the outcome. J. Bone Jt. Surg. Am. 1995;77:985–989. doi: 10.2106/00004623-199507000-00002. [DOI] [PubMed] [Google Scholar]
- 14.Spector TD, MacGregor AJ. Risk factors for osteoarthritis: genetics. Osteoarthritis Cartilage. 2004;12(Suppl A):S39–S44. doi: 10.1016/j.joca.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 15.Pollard TC, et al. The hereditary predisposition to hip osteoarthritis and its association with abnormal joint morphology. Osteoarthritis Cartilage. 2013;21:314–321. doi: 10.1016/j.joca.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 16.Ma C, Blackwell T, Boehnke M, Scott LJ, Go TDi. Recommended joint and meta-analysis strategies for case-control association testing of single low-count variants. Genet. Epidemiol. 2013;37:539–550. doi: 10.1002/gepi.21742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 2015;11:e1004219. doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.arcOGEN Consortium; arcOGEN Collaborators. Zeggini E, et al. Identification of new susceptibility loci for osteoarthritis (arcOGEN): a genome-wide association study. Lancet. 2012;380:815–823. doi: 10.1016/S0140-6736(12)60681-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bulik-Sullivan BK, et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng J, et al. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2017;33:272–279. doi: 10.1093/bioinformatics/btw613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Felson DT, et al. Evidence for a Mendelian gene in a segregation analysis of generalized radiographic osteoarthritis: the Framingham Study [see comments] Arthritis Rheum. 1998;41:1064–1071. doi: 10.1002/1529-0131(199806)41:6<1064::AID-ART13>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 23.McWilliams DF, et al. Mild acetabular dysplasia and risk of osteoarthritis of the hip: a case-control study. Ann. Rheum. Dis. 2010;69:1774–1778. doi: 10.1136/ard.2009.127076. [DOI] [PubMed] [Google Scholar]
- 24.Waarsing JH, et al. Osteoarthritis susceptibility genes influence the association between hip morphology and osteoarthritis. Arthritis Rheum. 2011;63:1349–1354. doi: 10.1002/art.30288. [DOI] [PubMed] [Google Scholar]
- 25.Agricola R, et al. Validation of statistical shape modelling to predict hip osteoarthritis in females: data from two prospective cohort studies (Cohort Hip and Cohort Knee and Chingford) Rheumatology (Oxford) 2015;54:2033–2041. doi: 10.1093/rheumatology/kev232. [DOI] [PubMed] [Google Scholar]
- 26.Atasu M, Akkoyunlu U, Tokgozoglu N, Say B. The heritability of liability to congenital dislocation of the hip. Turk. J. Pediatr. 1972;14:23–26. [PubMed] [Google Scholar]
- 27.Li L, et al. Heritability and sibling recurrent risk of developmental dysplasia of the hip in Chinese population. Eur. J. Clin. Invest. 2013;43:589–594. doi: 10.1111/eci.12084. [DOI] [PubMed] [Google Scholar]
- 28.Hotten G, Neidhardt H, Jacobowsky B, Pohl J. Cloning and expression of recombinant human growth/differentiation factor 5. Biochem. Biophys. Res. Commun. 1994;204:646–652. doi: 10.1006/bbrc.1994.2508. [DOI] [PubMed] [Google Scholar]
- 29.Francis-West PH, et al. Mechanisms of GDF-5 action during skeletal development. Development. 1999;126:1305–1315. doi: 10.1242/dev.126.6.1305. [DOI] [PubMed] [Google Scholar]
- 30.Dai J, et al. Association of a single nucleotide polymorphism in growth differentiate factor 5 with congenital dysplasia of the hip: a case-control study. Arthritis Res. Ther. 2008;10:R126. doi: 10.1186/ar2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Consortium GT. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Consortium GT. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen H, et al. Heads, shoulders, elbows, knees, and toes: modular Gdf5 enhancers control different joints in the vertebrate skeleton. PLoS Genet. 2016;12:e1006454. doi: 10.1371/journal.pgen.1006454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Capellini TD, et al. Ancient selection for derived alleles at a GDF5 enhancer influencing human growth and osteoarthritis risk. Nat. Genet. 2017;49:1202–1210. doi: 10.1038/ng.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Imabayashi H, et al. Redifferentiation of dedifferentiated chondrocytes and chondrogenesis of human bone marrow stromal cells via chondrosphere formation with expression profiling by large-scale cDNA analysis. Exp. Cell Res. 2003;288:35–50. doi: 10.1016/S0014-4827(03)00130-7. [DOI] [PubMed] [Google Scholar]
- 36.Vetter K, Wurst W. Expression of a novel mouse gene ‘mbFZb’ in distinct regions of the developing nervous system and the adult brain. Mech. Dev. 2001;100:123–125. doi: 10.1016/S0925-4773(00)00511-6. [DOI] [PubMed] [Google Scholar]
- 37.Deng FY, et al. Genome-wide association study identified UQCC locus for spine bone size in humans. Bone. 2013;53:129–133. doi: 10.1016/j.bone.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanna S, et al. Common variants in the GDF5-UQCC region are associated with variation in human height. Nat. Genet. 2008;40:198–203. doi: 10.1038/ng.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soranzo N, et al. Meta-analysis of genome-wide scans for human adult stature identifies novel loci and associations with measures of skeletal frame size. PLoS Genet. 2009;5:e1000445. doi: 10.1371/journal.pgen.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Borkakoti N. Matrix metalloproteases: variations on a theme. Prog. Biophys. Mol. Biol. 1998;70:73–94. doi: 10.1016/S0079-6107(98)00003-0. [DOI] [PubMed] [Google Scholar]
- 41.Fontenele EG, et al. Association study of GWAS-derived loci with height in Brazilian children: importance of MAP3K3, MMP24 and IGF1R polymorphisms for height variation. Horm. Res. Paediatr. 2015;84:248–253. doi: 10.1159/000437324. [DOI] [PubMed] [Google Scholar]
- 42.Zhao J, et al. The role of height-associated loci identified in genome wide association studies in the determination of pediatric stature. BMC Med. Genet. 2010;11:96. doi: 10.1186/1471-2350-11-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moise AR, Kuksa V, Imanishi Y, Palczewski K. Identification of all-trans-retinol:all-trans-13,14-dihydroretinol saturase. J. Biol. Chem. 2004;279:50230–50242. doi: 10.1074/jbc.M409130200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ross SA, McCaffery PJ, Drager UC, De Luca LM. Retinoids in embryonal development. Physiol. Rev. 2000;80:1021–1054. doi: 10.1152/physrev.2000.80.3.1021. [DOI] [PubMed] [Google Scholar]
- 45.Hartofilakidis G, Stamos K, Karachalios T, Ioannidis TT, Zacharakis N. Congenital hip disease in adults. Classification of acetabular deficiencies and operative treatment with acetabuloplasty combined with total hip arthroplasty. J. Bone Jt. Surg. Am. 1996;78:683–692. doi: 10.2106/00004623-199605000-00007. [DOI] [PubMed] [Google Scholar]
- 46.Engesaeter LB, Furnes O, Havelin LI. Developmental dysplasia of the hip--good results of later total hip arthroplasty: 7135 primary total hip arthroplasties after developmental dysplasia of the hip compared with 59774 total hip arthroplasties in idiopathic coxarthrosis followed for 0 to 15 years in the Norwegian Arthroplasty Register. J. Arthroplast. 2008;23:235–240. doi: 10.1016/j.arth.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 47.Engesaeter IO, et al. Prevalence of radiographic findings associated with hip dysplasia in a population-based cohort of 2081 19-year-old Norwegians. Bone Jt. J. 2013;95-B:279–285. doi: 10.1302/0301-620X.95B2.30744. [DOI] [PubMed] [Google Scholar]
- 48.TwinsUK-website. TwinsUK. Vol. 2010.
- 49.Andrew T, et al. Are twins and singletons comparable? A study of disease-related and lifestyle characteristics in adult women. Twin Res. 2001;4:464–477. doi: 10.1375/twin.4.6.464. [DOI] [PubMed] [Google Scholar]
- 50.Sudlow C, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gauderman, W. M. J. QUANTO 1.1: A Computer Program for Power and Sample Size Calculations for Genetic Epidemiology Studies. http://biostats.usc.edu/Quanto.html (2006).
- 53.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 54.Magi R, Morris AP. GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics. 2010;11:288. doi: 10.1186/1471-2105-11-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating missing heritability for disease from genome-wide association studies. Am. J. Hum. Genet. 2011;88:294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Golan D, Lander ES, Rosset S. Measuring missing heritability: inferring the contribution of common variants. Proc. Natl. Acad. Sci. USA. 2014;111:E5272–E5281. doi: 10.1073/pnas.1419064111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morris AP, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang J, et al. Genome partitioning of genetic variation for complex traits using common SNPs. Nat. Genet. 2011;43:519–525. doi: 10.1038/ng.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods. 2013;10:5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 60.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abecasis GR, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Consortium UK, et al. The UK10K project identifies rare variants in health and disease. Nature. 2015;526:82–90. doi: 10.1038/nature14962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Consortium F, et al. A promoter-level mammalian expression atlas. Nature. 2014;507:462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Andersson R, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roadmap Epigenomics C, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ernst J, Kellis M. Large-scale imputation of epigenomic datasets for systematic annotation of diverse human tissues. Nat. Biotechnol. 2015;33:364–376. doi: 10.1038/nbt.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pickrell JK. Joint analysis of functional genomic data and genome-wide association studies of 18 human traits. Am. J. Hum. Genet. 2014;94:559–573. doi: 10.1016/j.ajhg.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lappalainen T, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wellcome Trust Case Control, C.. et al. Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat. Genet. 2012;44:1294–1301. doi: 10.1038/ng.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Euesden J, Lewis CM, O’Reilly PF. PRSice: Polygenic Risk Score software. Bioinformatics. 2015;31:1466–1468. doi: 10.1093/bioinformatics/btu848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McCarthy S, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Genome-wide genotype data of the DDH cases and UKHLS controls have been deposited to the European Genome-Phenome Archive (www.ebi.ac.uk/ega/home, DDH accession numbers: EGAD00010000766 and EGAS00001000916; UKHLS accession numbers: EGAD00010000891 and EGAS00001001232) and of the DDH NJR cases to the NJR data archive (www.njrcentre.org.uk).