Abstract
Pseudohyphal growth is a multicellular phenotype naturally performed by wild budding yeast cells in response to stress. Unicellular yeast cells undergo gross changes in their gene regulation and elongate to form branched filament structures consisting of connected cells. Here, we construct synthetic gene regulation systems to enable external induction of pseudohyphal growth in Saccharomyces cerevisiae. By controlling the expression of the natural PHD1 and FLO8 genes we are able to trigger pseudohyphal growth in both diploid and haploid yeast, even in different types of rich media. Using this system, we also investigate how members of the BUD gene family control filamentation in haploid cells. Finally, we employ a synthetic genetic timer network to control pseudohyphal growth and further explore the reversibility of differentiation. Our work demonstrates that synthetic regulation can exert control over a complex multigene phenotype and offers opportunities for rationally modifying the resulting multicellular structure.
Georgios Pothoulakis and Tom Ellis report the construction of a synthetic gene regulation system for inducing pseudohyphal growth in Saccharomyces cerevisiae. This multicellular yeast phenotype can now be switched on and off via external control in a variety of conditions.
Introduction
As biological research and efforts in synthetic biology accelerate the incorporation of ever-more complex systems into organisms, they are also shifting from tasks performed in individual cells to tasks achieved by more complex multicellular systems such as pattern formation. Formation of multicellular patterns is a widely observed characteristic found in a large number of organisms and is the result of intricate arrangements of differentiated cells under the control of development and differentiation mechanisms. These arrangements enable the formation of structures that serve specialist roles, such as tissues and organs1.
While multicellular phenotypes are expressed commonly by multicellular organisms, they can also emerge in organisms traditionally considered unicellular. A classic example are biofilms, which are microbial cell aggregations that secrete extracellular polymeric substances forming a protective matrix around a colony population2. Unicellular organisms offer the advantage of being reliable and easy to genetically manipulate, while also not naturally displaying multicellular phenotypes in rich growth media. One such organism is Saccharomyces cerevisiae yeast, which can display a variety of characteristics normally associated with multicellularity, specialization, and pattern formation. S. cerevisiae is able to take multicellular growth forms such as flocs, flors, biofilms, and filaments in response to environmental condition by changing the way that the cells adhere to each other and to biotic or abiotic substrates using proteins called adhesins or flocculins3,4.
Filamentation is an important differentiation mechanism for many fungal species as a response to external stimuli such as nitrogen starvation while, for some yeast species like Candida albicans, the phenotype it is also connected to pathogenicity5,6. There is great variety in the morphology among the different filamenting fungal species and in the case of S. cerevisiae yeast cells are forming pseudohyphae: chains of elongated cells fully-separated by cytokinesis but attached through adhesion proteins5. Pseudohyphal growth is a highly complex phenotype that involves a large number of processes including substrate adhesion, bud site selection and cell morphogenesis7. Both haploid and diploid yeast cells create kinds of pseudohyphae formation as a stress response although the underlying genetic and morphological differences between the two cell types during the expression of this response leads to a distinction. The term pseudohyphal growth is usually reserved for diploid cells, while the term invasive growth is used for haploid cells due to the characteristic invasion of the colonies into agar substrate5. Extensive research has elucidated many of the genetic and biochemical mechanisms behind pseudohyphal growth, a complex multi-faceted phenotype regulated by four highly characterized signaling pathways—the Ras2/cAMP-PKA pathway, the protein kinase Snf1 pathway, the TOR (target of rapamycin) pathway and the MAPK pathway5,8. Interestingly, only diploid MATa/MATα yeast strains can form pseudohyphae, as pseudohyphal growth is a characteristic of the bipolar budding pattern. However, haploid cells lacking BUD4, which encodes a protein responsible for the conversion of the bipolar budding pattern to an axial budding pattern, are known to grow following the bipolar budding pattern and so offer potential to enable filament formation from haploid cells9. While pseudohyphal growth is in general a natural response to nutrient starvation, i.e., nitrogen stress or glucose deprivation, it can also be induced independently by manipulating the expression of specific gene targets10. Two such targets are the genes PHD1 and the FLO8, which were previously investigated based on their ability to induce pseudohyphal growth when overexpressed in diploid yeast10,11.
In previous work, we showed that the simplest multicellular phenotype that is normally triggered by stress in S. cerevisiae—flocculation—can be put under the control of synthetic gene networks in standard lab strains12. Here we take this a step further, using synthetic gene networks introduced into the genome of S288C-derived yeast in order to enable the more complex pseudohyphal growth multicellular phenotype to be externally triggered in rich growth media in both haploid and diploid strains. This is achieved by using synthetic genetic regulation to control the filamentous growth master regulator genes and in two different designs, we describe versions that work in galactose-based media and glucose-based media with different chemical inducers. By monitoring the growth characteristics directed by these networks in strains with BUD gene deletions, we are able to observe the roles of BUD8 and BUD9 in defining the multicellular growth patterns. Finally, we explore the applicability of established yeast synthetic network designs in the context of growth phenotype control by using multigene networks to control the transition back from pseudohyphal to normal colony growth over a set amount of time12. This work demonstrates the use of synthetic regulation to control and reverse a complex multigene phenotype in yeast that directs a multicellular growth morphology from normally unicellular cells.
Results
A synthetic gene network for inducible pseudohyphal growth
To construct a synthetic gene network to independently induce filamentation in rich media we chose to target the PHD1 and FLO8 master regulator genes. Both the Phd1 and Flo8 proteins are involved in the pseudohyphal growth cascade of the cAMP-PKA pathway and act as transcriptional regulators of the MUC1/FLO11 gene that encodes for a flocculin that plays a central role in pseudohyphal growth and in other yeast multicellular phenotypes (Fig. 1)5,13. The MUC1/FLO11 gene is regulated by a large promoter bound by multiple transcription factors from several signaling pathways and its expression can have dramatic effects on cell adherence with other substrates5. In nutrient rich conditions, many of these transcription factors, including Phd1, are down-regulated thus prohibiting pseudohyphal growth11. Past studies have shown that overexpression of PHD1 induces pseudohyphal growth in Σ1278b diploid strains even in nutrient-rich conditions, although deletion of this gene appears to have no negative effect in filamentation8,10. Gimeno and Fink have also shown that over-expressing PHD1 in specific BUD4 negative Σ1278b-based haploid strains causes the pseudohyphal growth pattern during nutrient starvation10. However, even in nitrogen-limiting conditions, expression of pseudohyphal growth is impossible without proper expression of the FLO8 gene11,14.
Fig. 1.
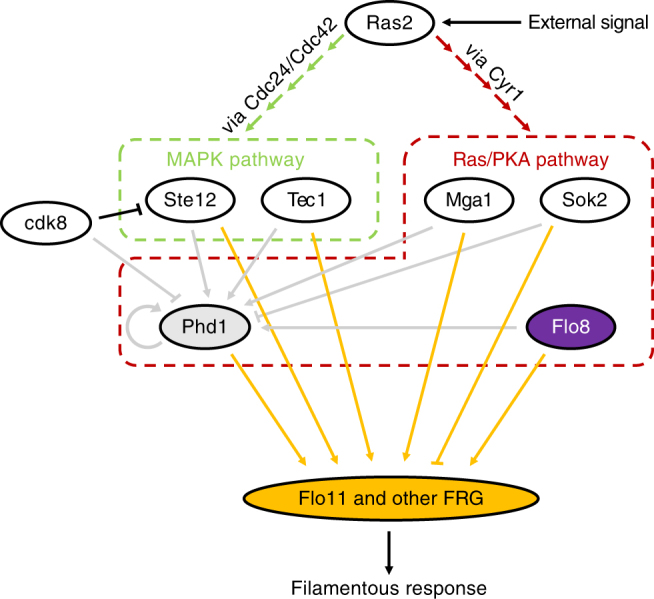
Regulation of Muc1/Flo11 protein. External signals such as nitrogen starvation activate Ras2, which in turn triggers both MAP kinase and Ras/PKA pathways through a cascade involving Cdc24/Cdc42 and Cyr1, respectively8. Several factors from both pathways have been shown to control Phd1 expression including Ste12 and Tec1 of the MAP kinase pathway and Mga1, Sok2, and Flo8 from the Ras/PKA pathway and Phd1 itself11. In addition, Cdk8 which is expressed in normally growing cells has been shown to cause Phd1 and Ste12 protein destabilization11. Ultimately, several filamentous response genes (FRG) including Flo11 are regulated by Phd1, Flo8, Ste12, Tec1, Mga1, and Sok2 and direct the phenotype. Arrows indicate upregulation while lines with bars indicate inhibition5, 11
Most commonly-used lab and industrial yeasts such as those derived from the S288C strain carry a mutation in the FLO8 gene that leads to the expression of a truncated version of the protein (flo8-1) making pseudohyphal growth impossible14–16. This mutation is not present in the Σ1278b strain used to study yeast multicellular phenotypes. Removing this mutation to enable proper Flo8 protein expression in S288C cells restores its filamentation capabilities14. Raithatha et al.11 have shown that co-expression of the un-truncated version of the FLO8 gene with a version of the PHD1 gene that carries a natural polymorphism on the 92nd codon (S92F) found in the Σ1278b strain, enhances filamentation of the S288C strain. This polymorphism eliminates a Cdk8-dependent phosphorylation site, thus further stabilizing the Phd1 protein.
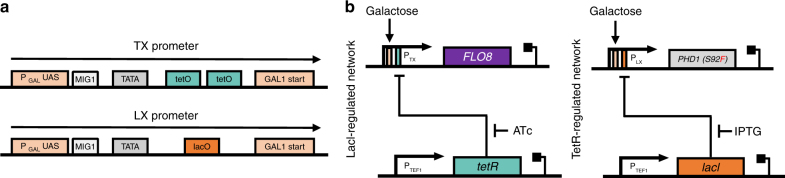
Two previously described synthetic promoters were used to generate an initial synthetic regulation network that triggers pseudohyphal growth when yeast are grown in galactose media and given external chemical inducers (Fig. 2a). The two promoters used, TX and LX, are repressible by the bacterial TetR and LacI repressors, respectively, when these transcription factors are co-expressed12,17. Promoter repression from the TetR and LacI proteins can be relieved by the addition of anhydrotetracycline (ATc) and Isopropyl β-D-1-thiogalactopyranoside (IPTG), respectively12. Since both TX and LX promoters are based on GAL1, a natural yeast promoter repressed in the presence of glucose and induced by galactose, both promoters are only active in galactose-containing media18,19. And since both the tetR and lacI genes originate from E. coli, for inhibition to occur both need to be added to the constructs20,21. Thus, combining the TX and LX promoters with genomically integrated cassettes that constitutively express tetR and lacI, results in the complete synthetic gene network. Initially, a network with two genes under independent regulation was constructed, so that the growth morphology of cells overexpressing the modified PHD1 gene (S92F mutant) and the fully-functioning FLO8 gene could be independently assessed (Fig. 2b).
Fig. 2.
Designing synthetic regulatory networks for pseudohyphal growth induction. a Diagrams of the TX and LX synthetic promoters. Both promoters contain the GAL1 upstream activating sequence (UAS), which is required for galactose activation but is normally repressed in glucose, a MIG1 protein repressor recognition site that inhibits activation in glucose, a TATA box and either tetO2 or lacO operator sites. Each promoter is represented by a black arrow containing all critical sites shown as colored rectangles. Transcription from each promoter occurs at the defined ‘start’ sites. b Diagram of the synthetic gene networks for controlling pseudohyphal growth. PHD1(S92F) is under the control of the GAL1-based LX promoter carrying a Lac operator (lacO) site and induced by IPTG, while FLO8 is under the control of the GAL1-based TX promoter carrying a Tet operator (tetO2) site and controlled by ATc. Both lacI and tetR genes are expressed from the constitutive TEF1 promoter. Genes are shown as colored boxes, promoters as arrows. Regulation is shown as rectangles inside the promoters (orange for LAC, green for TET, and pink for GAL1)
As pseudohyphal growth is a characteristic of the bipolar budding pattern normally only observed in diploid cells, these two gene networks were first incorporated into the BY4743 diploid yeast strain. Rather than making any modifications to the natural, inactivated PHD1 and FLO8 genomic loci in BY4743, the synthetic gene networks were integrated at the well-characterized URA3 landing-pad locus elsewhere in the genome. Since we also desired to test these networks in haploid cells we repeated integration of the constructs as in the diploid but now in a BY4741 haploid with a bud4 gene knockout (strain Y02569). Knockout of this gene is known to force the haploid cells to grow in a bipolar budding pattern and thus may aid in establishing pseudohyphal growth22.
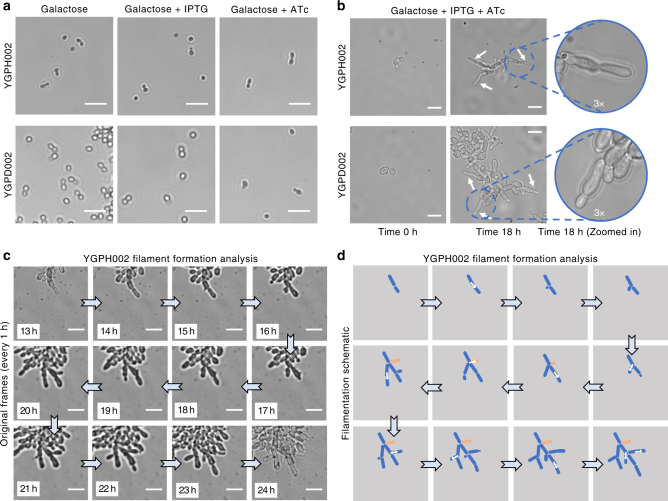
The two resulting strains, YGPH002 (haploid) and YGPD002 (diploid), were first assessed for their ability to grow in filamentous form in induction conditions. In the absence of either or both inducer chemicals in galactose growth media, no filamentous growth is seen (Fig. 3a). This demonstrates that overexpression of just one of the two genes (PHD1(S92F) or FLO8) is not enough to induce pseudohyphal growth. However, both strains shown signs of flocculation when grown in galactose media, even in the absence of IPTG and ATc. This is likely indicative of some leaky expression from the synthetic networks possibly as a result imperfect repression of the TetR and LacI-regulated promoters and cells being sensitive to even the smallest amounts of the Phd1 or Flo8 proteins. As Flo8 is directly involved in the flocculation phenotype it is most likely leaky expression of this gene that leads to a degree of flocculation.
Fig. 3.
Implementing inducible synthetic regulatory networks for pseudohyphal growth induction. a Inverted microscope images of haploid YGPH002 and diploid YGPD002 strains in different induction conditions. Images were taken after 24 h of growth in galactose media or galactose media supplemented with either 1 mM IPTG or 250 ng/ml ATc. Brightfield images were acquired using a 20× objective. Small white lines are scales which correspond to 20 μm. b Images of the haploid YGPH002 and diploid YGPD002 strains taken at time 0 h or after 18 h of growth inside the ONIX microfluidic platform in galactose media with 250 ng/ml of ATc and 1 mM IPTG supplemented. Areas of the 18 h time point frames have been enlarged by 3× to highlight filament formation. Brightfield images were acquired using a 60× objective. White arrows are pointing to newly formed filaments. Small white lines are scales which correspond to 10 μm. c Time-lapse microscopy and (d) filament formation analysis of the haploid YGPH002 strain when pseudohyphal growth is induced. Cells were induced for 24 h. Here, frames from 1 h intervals starting from the 13 h time point were selected and magnified in order to highlight the generation of branched formations from cells following a unipolar pattern. White arrows highlight growth of new branches from mother cells. Brightfield images were taken using a 60× objective. The white lines correspond to scales with a length of 10 μm. The cell shown in orange is not clearly visible after the 8th frame since it’s covered by other cells
In galactose media with both ATc and IPTG inducers present, both the haploid and diploid trains showed clear morphological differences, visibly forming filaments as they grow (Fig. 3b). This filamentous growth was imaged on the ONIX microfluidics platform with images of cells taken using an inverted microscope at 0 and 18 h post induction. Cells appeared considerably elongated and attached to their mother cells, validating the anticipated phenotype.
Following verification of the desired phenotype, we next generated a time-lapse to track the haploid YGPH002 cells as they transition from normal to pseudohyphal growth and generate filaments (Fig. 3c, d). The results confirmed that upon induction haploid cells form filaments that branch off as new daughter cells appear. They also confirmed that upon pseudohyphal growth induction, cells turn from a bipolar to a unipolar budding pattern where new buds form on the pole opposite of the birth scar in an effort to branch away from the center of the colony. A further observation that affirms successful induction of pseudohyphal growth is the visible “waiting” of the mother cells to further divide while their daughter cells are elongating. It is clear that daughter buds appear only after the previous daughter cells have reached maximum size. It was proposed by Kron et al.23 that during pseudohyphal growth, mitosis in the mother cell is delayed by a G2 checkpoint while the daughter elongates, and this results in synchronization of budding between mother and daughter cells, something not seen during normal budding growth.
The above results verify that it is possible to design and construct synthetic regulatory networks capable of inducing the pseudohyphal growth phenotype and subsequent filament formation in S. cerevisiae, even when these cells are not stressed by external factors. Importantly, this work shows that the synthetic networks can induce filamentation in haploid lab yeast.
Inducible pseudohyphal growth independent of carbon source
The initial two synthetic gene networks enable stressor-free externally inducible pseudohyphal growth in haploid and diploid S. cerevisiae, however this system is only functional in galactose-rich growth media. Therefore, we next turned to the previously developed Z3EV-mediated gene induction system described by McIsaac et al.24 to obtain external induction in the absence of galactose in the media. Z3EV is an artificial transcription factor which is a fusion of the mouse Zif268 DNA binding domain, the ligand binding domain of the human estrogen receptor and VP16 (viral protein 16). In the presence of β-estradiol this selectively binds to promoters containing Zif268 binding motifs and activates their expression25.
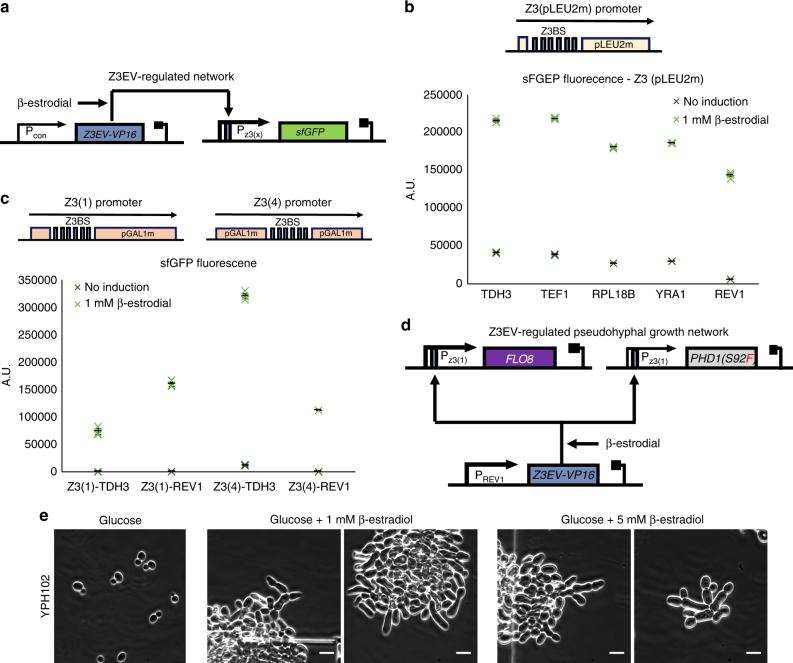
Following the approach described by McIsaac et al., we constructed our own Z3EV-activated promoter by placing 6 Zif268 binding sites (from now on referred to as Z3BS) upstream of a minimal LEU2 promoter to create promoter Z3(pLEU2m) (Fig. 4a). In previous uses, the Z3EV transcription factor has been constitutively expressed from the strong ADH1 promoter, however as we required promoter leakiness to be minimized, we opted to reduce Z3EV expression to the minimum required for good expression25. Thus, we created a library of constructs carrying five constitutive promoters of various strengths driving Z3EV expression. To characterize these, the Z3(pLEU2m) promoter was placed upstream of a sfGFP gene. In the presence of the β-estradiol inducer, the Z3EV transcription factor binds to the Z3(pLEU2) promoter and activates sfGFP transcription which can be quantified by measuring the green fluorescence of cells using flow cytometry.
Fig. 4.
Extending external inducible pseudohyphal growth to glucose-rich media. a Diagram of the Z3 promoter characterization system. The yEGFP fluorescent marker is under the control of one of the three Z3 promoter derivatives, activated by the Z3EV-VP16 transcription factor in the presence of β-estradiol. Z3EV-VP16 is expressed from a constitutive promoter. Genes are shown as colored boxes, promoters as arrows. Z3EV-VP16 regulation sites are shown as blue rectangles within the promoters. b Characterization of the Z3(pLEU2) promoter when paired with different combinations of constitutive promoters driving Z3EV-VP16 expression (TDH3, TEF1, RPL18B, YRA1, or REV1). The yEGFP fluorescent output of five different constructs is tested when induced with 1 mM of β-estradiol for 5 h. Data shows fluorescence values from three repeats (colored X markers) as determined by flow cytometry, black bars indicate mean values, error bars indicate standard error. c Characterization of the Z3(1) and Z3(4) promoters when paired with either the TDH3 or REV1 constitutive promoters driving Z3EV-VP16 expression. The green fluorescence output of the four different constructs is tested when induced with 1 mM of β-estradiol for 5 h. Data shows fluorescence values from three repeats (colored X markers) as determined by flow cytometry, black bars indicate mean fluorescence values, error bars indicate standard error. d Diagram of the final Z3EV-regulated pseudohyphal growth network. Both the FLO8 and PHD1(S92F) genes are under the control of the Z3(1) promoter while Z3EV-VP16 is expressed from the constitutive REV1 promoter. Genes are shown as colored boxes while promoters as arrows. Z3EV-VP16 regulation sites are shown as blue rectangles within promoters. e Inverted microscope images of the haploid YPH102 strain taken after 28 h of growth inside the ONIX microfluidic platform in glucose media without β-estradiol or with either 1 or 5 mM of β-estradiol supplemented. Brightfield images were acquired using a 60× objective. Small white lines are scales and correspond to 10 μm
All five constructs were transformed into Y02569 haploid yeast cells and induced with 1 mM of β-estradiol for 5 h and measured compared to their uninduced counterparts (Fig. 4b). The mean fluorescence values with the five different promoters for Z3EV expression reveal that the relative activation levels of the Z3(pLEU2m) promoter are comparable. This therefore allows us to eliminate the use of strong expression for this transcription factor. Furthermore, there is a notable fold-increase in expression upon induction in all cases and this is most prominent for the REV1 promoter, the weakest of those tested here. The construct with this promoter, has the most desirable on/off characteristics needed for the synthetic gene network but still shows some leaky expression from the Z3(pLEU2m) promoter when no inducer is present.
In order to improve on this, we replaced the Z3(pLEU2m) promoter with two designs containing the GAL1 core promoter that more closely match the P1 and P4 promoters described by McIsaac et al. These two new promoters, named Z3(1) and Z3(4) were tested as before in cells where Z3EV expression is driven by the strongest (TDH1) and weakest (REV1) of the five constitutive promoters. Performance of their expression as determined by flow cytometry, is shown in Fig. 4c. The previously observed leaky expression from both TDH1 and REV1 promoters has now been drastically decreased, and in particular for the REV1-containing constructs the leakage is practically eliminated.
Using these optimized promoter combinations, a final design with Z3EV expression from the REV1 promoter and PHD1(S92F) and FLO8 expression from the Z3(1) promoter was selected for induction of pseudohyphal growth in glucose-containing media (Fig. 4d). This synthetic regulatory network, designed to be dependent only on the presence of β-estradiol, was introduced into haploid Y02569 cells to create strain YGPH102. This strain was then grown in glucose, and induced with varying concentrations of β-estradiol for 28 h while under observation in the ONIX microfluidics platform (Fig. 4e). The resulting images show that cell morphology indeed changes considerably due to the induction of pseudohyphal growth with external inducer, with cells elongating and visibly staying attached to each other in the growing colony. In contrast when no inducer is added, cells follow normal growth patterns.
Altering the inducible pseudohyphal morphology in haploids
We next asked whether growth morphology could be further perturbed through programmed genetic changes. We focused on the BUD gene family that is known to affect budding patterns during growth by controlling the cell sites where new buds appear during division and are also known to be involved in regulating the filamentous growth MAPK pathway26,27. Two proteins in particular play major roles in bud site selection in bipolar budding diploid cells; Bud8 and Bud9. The Bud8 protein localizes primarily to the distal pole, forcing distal pole site selection, while the Bud9 protein localizes primarily to the proximal pole to give the reverse outcome28. Previously, it has been shown that creating gene knockouts of the BUD8 gene in diploid cells forces cells to bud almost exclusively from the proximal pole, while knocking out the BUD9 gene forces cells to bud almost exclusively from the distal pole29. In addition, Harkins et al.28 has shown that similar results can be taken by controlling the relative levels of expression of the BUD8 and BUD9 genes rather than via gene deletion. In normal haploid cells, deletion of these genes does not have any effects on the axial budding pattern and does not affect cell viability or growth rates, although they do play a role during haploid invasive growth where the Bud8 protein promotes this phenotype while the Bud9 impedes it28,30. Therefore, we chose to investigate the knockout of these two genes and their effects on morphology using our inducible pseudohyphal growth haploid strain.
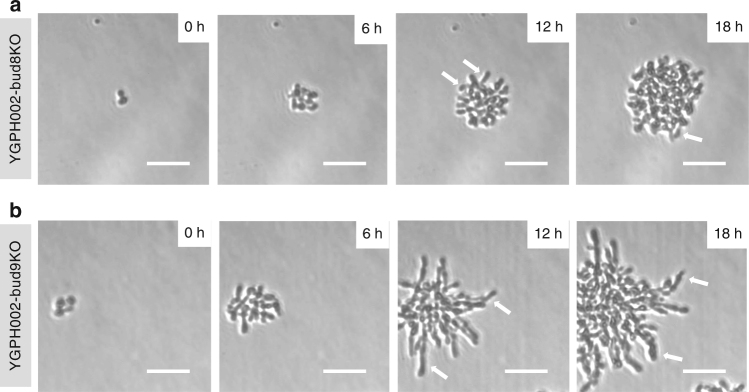
The BUD8 and BUD9 genes were individually deleted from our haploid YGPH002 strain, in each case by transforming and recombining the linear K. lactis URA3 marker into the relevant chromosomal loci to disrupt the gene. In both cases, many viable colonies were seen post transformation, confirming that knockouts of these genes do not affect cell viability28,31. The two new strains were externally induced as before and their pseudohyphal growth capabilities were assessed using an inverted microscope and growth in galactose media. Cells were grown on agarose pads to enable generation of time-lapse images (Fig. 5, Supplementary Movies 1 and 2). These images clearly show that the -bud8 strain (YGPH002-bud8KO) has difficulty creating filaments that extend outwards from the colony, although elongated cells are seen. Instead, the colony appears to be concentrated at the center with new buds developing inwards, a strong indication of proximal budding. In contrast, the -bud9 strain (YGPH002-bud9KO), clearly exhibits a classic pseudohyphal growth phenotype with extensive filaments forming and always extending outwards of the center of the colony.
Fig. 5.
Induced pseudohyphal growth in haploid cells with BUD8 or BUD9 gene deletion. a YGPH002-bud8KO time-lapse. b YGPH002-bud9KO time-lapse. Arrows highlight some of the filaments formed as a result of the induced pseudohyphal growth phenotype. Cells are grown for 18 h on 1.2% agarose pads made with 2% synthetic complete galactose media, supplemented with 400 ng/ml ATc and 1 mM IPTG. Brightfield images were captured using a 20× objective and a Phase 1 contrast filter every 10 min but only selected frames are shown here. White arrows point to cells exhibiting filamentous-like morphology. White lines represent scale bars with a length of 50 μm
Synthetic regulation for phenotype reversal during growth
The orthogonal, non-native transcription factors used within our inducible synthetic gene networks allow the switch to pseudohyphal growth to be connected to inputs other than external induction. In the absence of inducer chemicals, filamentous growth can be activated by any regulation that triggers Z3EV expression (in the glucose media system) or that inhibits expression of LacI and TetR (in the galactose media system). This design theoretically enables the change in growth phenotype to be linked to a wide variety of inputs that change gene expression in yeast, such a light-based induction, cell-to-cell signaling and environmental sensing32–34. It also enables the growth phenotype to be connected to existing synthetic gene networks that have been developed previously in work by the yeast synthetic biology field35.
To demonstrate this feature, we modified our galactose-media pseudohyphal growth induction system to link it to a synthetic gene network motif previously described as a genetic ‘timer’. This motif, consisting of mutual inhibition of LacI and TetR, delays the return of an initial gene expression state for several generations after an initial pulse of chemical induction, in this case with ATc12. It enables a population to grow performing strong expression of one or more genes for multiple generations, before switching these off and reverting back to expression of other genes as before the induction. When linked to FLO8 and PHD1 expression, the timer motif is designed initiate a population to differentiate and grow for several generations in pseudohyphal mode before the cells within this then begin reverse their differentiation and revert back to normal budding unicellular growth. This can be used to create a population on a plate that initially forms expansive filaments, before then budding to form a full colony.
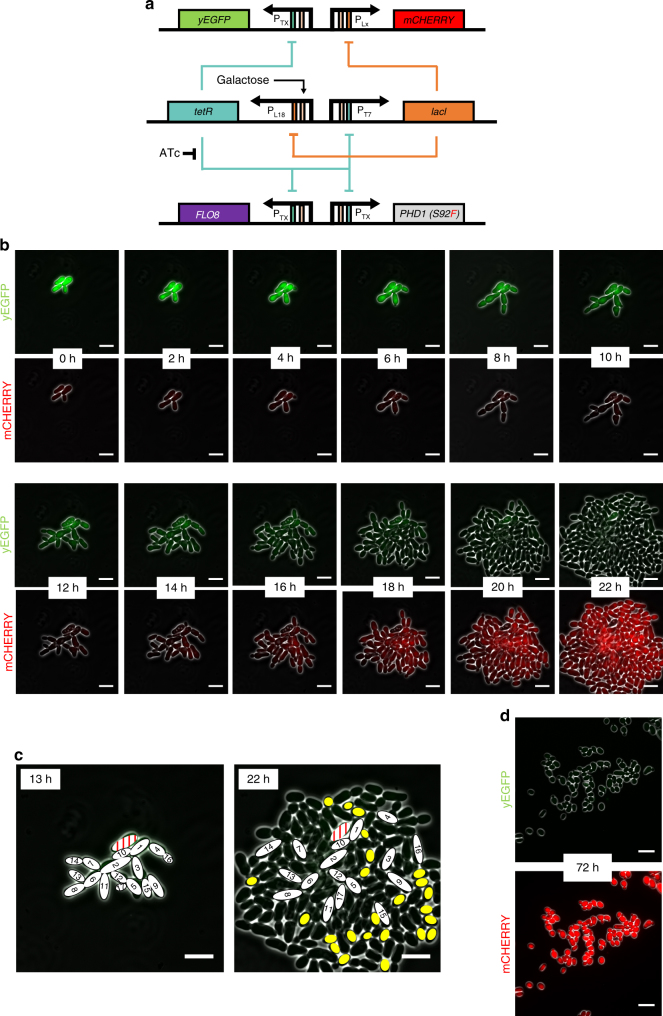
For the genetic timer motif (Fig. 6a) we modified the previously described T7-L18 genetic timer which uses mutated versions of the TX and LX promoters with reduced maximum expressions to ensure a long reset time after initial induction is removed12. Since the LACI gene originates from E. coli, we encoded a strong c-Myc nuclear localization signal (NLS, amino acid sequence PAAKRVKLD) into the C-terminal of the protein to ensure this regulator gave strong repression in S. cerevisiae36. Both the PHD1(S92F) and FLO8 genes necessary for pseudohyphal growth induction were placed downstream of TX promoters making them co-regulated by the TetR levels in the yeast. By default, the T7-L18 timer rests at the tetR ON state and so in this configuration, the filamentation genes are repressed when cells grow normally in galactose-rich media. Upon induction with ATc, tetR regulation is supressed, the network switches to the lacI ON state (pseudohyphal growth) and stays there as long as the inducer is present. After removal of the inducer the network slowly reverts back to the tetR ON state at a rate dependent on the relative strengths of the opposing T7 and L18 promoters. For characterization purposes, this synthetic gene network also included two further genes, the yEGFP and mCHERRY genes that encode green and red fluorescent proteins, respectively. These were expressed from further copies of the TX and LX promoters and used to monitor transition of the timer motifs from one state to another in a manner similar to that used before by Wu et al.37.
Fig. 6.
Using a genetic timer network to temporally control pseudohyphal growth within a growing colony. a Diagram of the T7-L18 synthetic timer network interfaced with expression of pseudohyphal growth genes and green and red fluorescent proteins. The lacI, PHD1, FLO8, and yEGFP genes are under the control of the TX promoter or its derivative, T7. The mCHERRY gene is expressed from the LX promoter while its derivative L18 promoter is controlling the tetR gene. Genes are shown as colored boxes, promoters as arrows. Regulation sites are shown as rectangles inside promoters (orange for lac, green for tet, and pink for galactose). b Time-lapse fluorescence microscopy of the YGPTIMER strain carrying the T7-L18 genetic timer. Cells pre-treated with ATc for 24 h were grown inside the ONIX microfluidics platform in synthetic complete galactose media after ATc had been removed by washing. Brightfield, green (yEGFP) and red (mCherry) fluorescence images are taken every 10 min. Here, selected frames from 2 h intervals are shown. c Analysis of cell position and shape for the YGPTIMER strain carrying the T7-L18 genetic timer. The exact same cells present at 13 h after the removal of ATc are shown in the final colony image at 22 h after ATc removal. Cells are numbered based on the order of appearance. One cell that arrests after emerging and never gives a daughter cell appears in red. Yellow ovals represent cells that appear oval after reaching full growth and are also detached from their mother cells. d Inverted microscope images of the YGPTIMERX strain taken after 72 h of growth in synthetic complete galactose media after ATc had been removed by washing. The green (top) and red (bottom) fluorescence channels are each combined with the brightfield channel to enhance cell visibility. Images are taken using the 60× objective and the Phase 3 contrast filter. White lines represent scale bars with a length of 10 μm
The complete six gene network was constructed as three cassettes and integrated into the haploid Y02569 strain to create a strain called YGPTIMER (Fig. 6a, Supplementary Movies 3 and 4). Cells from this strain were then pre-treated with ATc to induce filamentation and then washed to remove this inducer before being transferred into the ONIX microfluidics platform for live-cell imaging over 22 h (Fig. 6b). The initial cells of the colony grew in filamentous form to expand from the center for the first 12 h. During this time, they exhibited a strong yEGFP signal associated with low levels of TetR and high levels of LacI in each cell. Between 12 and 18 h this signal decreased and mCherry expression began, signifying a switch to lower LacI levels, back to the tetR-ON state of the timers. Accompanying this, the number of cells in the colony now begins to increase more rapidly and outward exploration with new filaments starts to cease. By 22 h most of the new cells within the colony are now red in fluorescence rather than green, and exhibit a more oval morphology compared to the initial filamenting cells, suggesting that budding unicellular growth starts appearing in the colony. To get a better understanding on how the colony behaves after the initial induction of filamentation and the resulting cell placement when the system has fully reset, the localization of the filamenting cells from the 13 h and 22 h time points was analyzed (Fig. 6c). This revealed that the cells that had been undergoing pseudohyphal growth in the first 13 h still individually looked filamentous after 22 h, but were now no longer adhered to one another, instead being broken apart by the rapid growth of the new budding cells. Many of new cells in the later timepoints are oval shaped and virtually all cells on the outer edge of the colony show detachment from their mother cells. Yet there is also a clear lag between the switching of fluorescent protein production from green to red and the disappearance of the various features of pseudohyphal growth. As the phenotype is reversed, we first see increased occurrence of detachment and then the new cells gradually grow less elongated, although not all new cells can be said to be oval. During the experiment, all cells continue to bud exclusively from the distal pole (a sign of unipolarity) suggesting that this is the slowest differentiation feature to revert. Finally, to confirm the full reversion of the system, we imaged a timer strain left to grow in liquid phase for 72 h after removal of the inducer and observed cells with standard budding morphology exhibiting high red fluorescence and no green fluorescence (Fig. 6d).
Discussion
This work utilized synthetic gene regulatory networks to activate yeast pseudohyphal growth even in nitrogen rich media. Initially two separate inducible synthetic regulatory networks based on the TetR and LacI-regulated TX and LX promoters, respectively, were shown to work in both haploid and diploid cells that grow in galactose media (Fig. 3). To our understanding, this is the first time where pseudohyphal growth has been induced in a haploid strain in rich media. Previously, PHD1 overexpression has been shown to induce pseudohyphal growth in bud4 mutant haploid cells only in nitrogen limited (SLAD) media10. In addition, it has been shown that a constitutive pseudohyphal growth phenotype is observed in elm1Δ fus3Δ haploid cells38. Importantly, both strains formed filaments without the presence of external stressors. A similar design chosen to be independent of the carbon source was implemented with Z3-based regulation and upon induction of this the haploid strain showed altered growth morphology but with a certain number of cells remaining spherical, and the overall filamentation characteristics appearing less strong than in galactose-grown strains (Fig. 4). A likely explanation for this is that the Z3(1) promoter is weaker than the TX and LX promoters and thus upon induction leads to lower PHD1 and FLO8 expression. Taken together this work highlights the use of promoter-based synthetic regulatory networks for controlling complex phenotypes. As the regulated promoters used are modular and orthogonal it is possible that these networks could be implemented in other species. Indeed, related TetR-regulated promoters have been used for gene expression control in many organisms, including Candida albicans12,39.
Enabling control over the induction of the pseudohyphal growth in cells grown in glucose and nitrogen-rich conditions potentially leads to filaments with different metabolic behavior from cells that traditionally form filaments under nitrogen starvation or glucose deprivation. Central metabolic functions in particular, are very sensitive to glucose availability and there is a general decrease in cytoplasmic ribosomal mRNA availability when there is very low glucose availability40. In addition, the absence of glucose can lead to general inhibition of translation in S. cerevisiae and also acts as a trigger of invasive growth in wild-type haploid strains that follow the axial budding pattern41,42. The limited biomass formation caused by filamentation in combination with normal glucose-enabled metabolism could potentially be a route towards higher yields of biochemical products from metabolically engineered cells that also have the morphology control system described here43.
Previously it has been proposed that mutations in the BUD8 gene eliminate pseudohyphal growth and based on our experiments of triggering PHD1 and FLO8 overexpression when Bud8 is absent, that does appear to be the case and cells do exhibit a proximal pole bias (Fig. 5)7. Having said that, cell morphology appears to change and elongated cells become apparent. Since bud site selection is affected by the relative levels of the Bud8 and Bud9 proteins, further exploration could be achieved through coordinated overexpression or repression of these two genes rather than via direct gene knockouts. This could enable additional possibilities for rational synthetic regulation of colony morphology, colony size and the pattern and directions of multicellular growth.
An important goal of synthetic regulatory network design is the ability to construct and implement predictable and mathematically-described systems that are able to achieve complex phenotypical results and here that was attempted by linking our synthetic pseudohyphal switch to a previously-described timer network (Fig. 6). The intentional slow-reversion of the phenotype in this experiment yields insights into how the cells are behaving when their morphology is under the control of synthetic mutual inhibition network. Overall, we observe an initial expansion of the area of the colony due to pseudohyphal growth but at the cost of slow cell growth. Eventually, as newly-formed cells start switching to a more unicellular growth mode and appearing less elongated, growth rate accelerates and filament formation brakes as a result of both mechanical forces of newly formed cells that push old ones and elimination of adhesion. The unexpected speed with which the unicellular-growing yeast takes over the colony is not surprising as they can divide much faster in rich media. Less obvious is why the genetic timer system used here only enforced the pseudohyphal phenotype for 4 or so generations over the first 20 h. When previously described in liquid growth experiments, the T7-L18 timer used here took five times as long to reset back after initial induction12. It is clear that there are major differences in the intracellular environment and gene expression in filamenting cells compared to in unicellular yeast.
Overall, this work describes how synthetic gene networks can be used to control the growth of S. cerevisiae yeast into filamented multicellular colonies, both in haploid and diploid standard lab strains and in rich growth media too. Bottom-up engineered synthetic multicellularity from unicellular yeast or using other organisms offers opportunities for exciting applications of synthetic gene networks, where populations of cells can be repurposed for new tasks (e.g., to grow patterned materials) or can be used to investigate the principles of natural pattern formation (e.g., the roles of cell-to-cell signaling, feedback and regulation). Control over pseudohyphal growth as described here represents a major initial step towards achieving predicable multicellular patterned growth from unicellular cells. With further genetic engineering, it could presumably be possible to gain even greater control of the subsequent colony morphology. Synthetic gene networks could be employed to provide additional control over budding localization along filaments, and differential gene expression through the colony (e.g., altered expression of genes in cells at the edge of the filaments vs. those in the center of the colony). These networks could employ quorum sensing mechanisms, cell-to-cell signaling or mother- or daughter-cell specific promoters in order to have differential regulation between the cells. Theoretically, genetic programs could be written that define when and where fractal-like filamentous growth patterns occur and how ‘branched’ the filaments are (by controlling budding frequency and location). These synthetic multicellular patterns could be utilized in scenarios where increased surface area and substrate removal from a colony of yeast is beneficial.
Ultimately, this work aims to accelerate the use of synthetic biology in the exploration and generation of genetically-encoded multicellular pattern formation programs, and therefore it is especially valuable that this work has been achieved in one of the most widely-used model organisms and works in standard lab growth conditions. As such this work offers a toolkit towards programming synthetic multicellular morphologies within growing 2-dimensional yeast colonies.
Methods
Strains and cultures
All engineered strains we derived either from the haploid S. cerevisiae Y02569 (BY4741; MATa; ura3Δ0; leu2Δ0; his3Δ1; met15Δ0; YJR092w::kanMX4) strain provided by EUROSCARF or from the diploid S. cerevisiae BY4743 (MATa/α; his3Δ1/his3Δ1; leu2Δ0/leu2Δ0; LYS2/lys2Δ0; met15Δ0/MET15; ura3Δ0/ura3Δ0) strain, also provided by EUROSCARF. The YGPH002 and YGPD002 strains were created by transforming both the pGPY002 and pGPY003 plasmids into the Y02569 and BY4743 strains respectively. The YGPH078, YGPH079, YGPH080, YGPH081, YGPH082, YGPH092, YGPH093, YGPH094, and YGPH095 strains were created by transforming the corresponding pGPY078-82 and pGPY092-95 plasmids into the Y02569 strain. The YGPH102 strain was created by transforming the pGPY102 plasmid into the Y02569 strain. The YGPH002-bud8KO and YGPH002-bud9KO knockout haploid strains were created through the insertion of the K. lactis URA3 gene in place of the BUD8 or BUD9 gene respectively thus replacing the corresponding open reading frame (ORF) in its entirety and part of the upstream promoter sequence in YGPH002 cells. The YGPTIMER and YGPTIMERX strains were created by subsequently transforming the pGPY006, pT7L18 and pGPY062 plasmids in the Y02569 strain. An NLS sequence (amino acid sequence PAAKRVKLD) was added to YGPTIMER through CRISPR editing. Cells are cultured in synthetic complete drop-out glucose media (SC-Glu) for transformation and general proliferation and in synthetic complete drop-out galactose media (SC-Gal) media with either IPTG, ATc or both supplemented. During the time-lapse and ONIX experiments, cells were grown at 30 °C inside the microscope. For all other cultures, cells were grown in liquid cultures at 30 °C with shaking at 225 rpm.
Plasmid construction
The pGPY002 plasmid carrying the PHD1(S92F) gene under the LX promoter and the lacI gene under the TEF1 promoter and the pGPY003 plasmid carrying the FLO8 gene under the TX promoter and the tetR gene under the TEF1 promoter were based on the pLVGI and pTVGI plasmids respectively (both based on pRS4D1 plasmid) described in Ellis et al.12. The PHD1 and flo8-1 genes were acquired from the YPH500 yeast strain through colony PCRs that also introduce restriction sites compatible with the pLVGI and pTVGI plasmids thus replacing the yEGFP gene. The PHD1 gene inside the pGPY002 plasmid was altered on the 92nd codon so that it encodes Phenylalaline (TTT codon) instead of Serine (TCT codon) while the flo8-1 gene of the pGPY003 plasmid was altered to remove the early stop codon (TAG-142nd codon) and replace it with TGG so that it creates the fully functional FLO8 gene. To make the new plasmids compatible with the Y02569 and BY4743 strains, the TRP1 markers of both the LVGI and TVGI plasmids were replaced with the HIS3 and LEU2 markers respectively taken from pRS based plasmids through simple restriction digestion and ligation. For the Z3 promoter experiments all plasmids were assembled using the MoClo method of modular assembly in combination with the Yeast ToolKit created by the Dueber lab44. All necessary promoter, open reading frame (ORF) and terminator parts were amplified accordingly and stored in parts plasmids compatible with the rest of the YTK kit. The Z3BS, Z3(1), Z3(4), and Z3EV parts were created using sequences obtained from the pRS416-yZ3EV-Z3pr-yEGFP (RB3579) which was a gift from David Botstein (Addgene plasmid # 69100). From there, cassette plasmids containing the whole genes of interest were created and finally assembled together using the pYTK096 URA3 integrating plasmid as a backbone thus generating the pGPY078-82, pGPY092-95 and pGPY102 plasmids. The pGPY006 plasmid was based on the pGPY002 plasmid. The LX promoter of the PHD1(S92F) gene and the lacI gene along with the TEF1 promoter were removed completely and replaced with a TX promoter and the FLO8 gene controlled by TX as well. The pT7L18 plasmid carrying the tetR and lacI genes controlled by the L18 and T7 promoters respectively is described in Ellis et al.12. The pGPY062 plasmid carrying the yEGFP gene under the TX promoter and the mCHERRY gene under the LX promoter is a modified version of the p714 plasmid described in the same study with its TRP marker replaced by URA3 to make it compatible with the Y02569 strain12. Supplementary Data 1 contains sequences of all plasmids used in this study in commented Genbank format.
Linear BUD gene family replacing fragment generation
In order to generate the linear fragments that enabled the creation of the BUD8 and BUD9 knockouts the K. lactis URA3 gene was PCR amplified from the pMirage plasmid provided by Dr Ben Blount (Imperial College London) using three different primer pairs that introduced the necessary overlaps for homologous recombination to directly replace the BUD8 or BUD9 genes at their genomic loci and a part of their upstream regions.
Inverted microscope image capture
Images were taken through 20x/0.45 or 60x/1.40 CPI60 objectives mounted on a Nikon Eclipse Ti inverted microscope with live cells imaged on plain slides, 1.2% SC-Gal agarose pad slides or using the ONIX microfluidic platform. To visualize the samples a Phase filter 1 or 3 is used to enhance contrast and Brightfield illumination. For fluorescence capture, excitation, emission filters, and exposures were respectively 480 nm, 535 nm, 1000 ms for the GFP channel (yEGFP) and 532 nm, 590 nm, 2000 ms for Cy3 channel (mCherry). During time-lapse experiments, the software autofocus function of the microscope is used to adjust for any potential movement of the cells during growth in order to keep clear track of the samples. The microscope experiment of Fig. 2b, the time-lapse experiment of Fig. 2c and the time-lapse experiment of Fig. 3c, d were carried out using the CellASIC ONIX Microfluidic platform (Merck Millipore). For cell growth, in most cases flow rate was adjusted between Psi = 2 and Psi = 4 which was determined to be good for yeast cells. For haploid yeast cells the Y04C-02-5PK plates were used. The time-lapse microscopy experiments shown on Fig. 3 were carried out on 1.2% SC-Gal agarose pad slides including 400 ng/ml ATc and 1 mM IPTG which were prepared following a protocol adapted from Rines et al.,45. NIS-Elements Microscope Imaging Software (Nikon) is used for capturing and ImageJ (National Institutes of Health) is used for image presentation.
Flow cytometry analysis
Flow cytometry assays were performed using the Attune NxT flow cytometer with the Attune NxT autosampler attachment from ThermoFisher Scientific. A 488 nm laser was used for excitation of green fluorescence detecting through a 530 nm band-pass filter (BL1). The voltages of the FSC, SSC and BL1 channels for the promoter characterization experiments were 200, 320 and 480 respectively. A threshold of 3.0 × 103 A.U. was applied to the forward (FSC) scatter to minimize non yeast events. Data analysis was performed using FlowJo software (Tree Star), gating samples for forward scatter and side scatter to exclude non-yeast events and obtaining fluorescence values from BL1-H (height) channels.
Data availability
Time-lapse movies of the experiments shown in Figs. 5a, b and 6a, b are provided in Supplementary Movies 1–4. High resolution versions of all microscopy images shown in the manuscript are available via Figshare at DOI: 10.6084/m9.figshare.566017946. Maps of new plasmids generated in this study are provided in Supplementary Data 1 in commented GenBank format and all plasmids will be made available via Addgene. All data generated or analyzed during this study that are not included in this published article and its Supporting Information files are available upon request.
Electronic supplementary material
Description of Additional Supplementary Files
Acknowledgements
The authors wish to thank William Shaw (Imperial College London) for his assistance on the implementation of the Yeast ToolKit and providing certain YTK part plasmids. We also thank Marios Tomazou, Felix Jonas and all other helpful members of CSynBI at Imperial College London. This work was funded by The Leverhulme Trust under project grant RPG-2012-562 and by the BBSRC via grant BB/K006290/1.
Author contributions
G.P. and T.E. designed the study and wrote the manuscript. G.P. analyzed the data and performed the experiments.
Competing interests
The authors declare no competing financial interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information is available for this paper at 10.1038/s42003-017-0008-0.
References
- 1.Salazar-Ciudad I, Jernvall J, Newman SA. Mechanisms of pattern formation in development and evolution. Development. 2003;130:2027–2037. doi: 10.1242/dev.00425. [DOI] [PubMed] [Google Scholar]
- 2.Sharma G, et al. Escherichia coli biofilm: development and therapeutic strategies. J. Appl. Microbiol. 2016;121:309–319. doi: 10.1111/jam.13078. [DOI] [PubMed] [Google Scholar]
- 3.Dranginis AM, Rauceo JM, Coronado JE, Lipke PN. A biochemical guide to yeast adhesins: glycoproteins for social and antisocial occasions. Microbiol. Mol. Biol. Rev. 2007;71:282–294. doi: 10.1128/MMBR.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruckner S, Mosch HU. Choosing the right lifestyle: adhesion and development in Saccharomyces cerevisiae. Fems. Microbiol. Rev. 2012;36:25–58. doi: 10.1111/j.1574-6976.2011.00275.x. [DOI] [PubMed] [Google Scholar]
- 5.Cullen PJ, Sprague GF., Jr. The regulation of filamentous growth in yeast. Genetics. 2012;190:23–49. doi: 10.1534/genetics.111.127456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumamoto CA. A contact-activated kinase signals Candida albicans invasive growth and biofilm development. Proc. Natl Acad. Sci. USA. 2005;102:5576–5581. doi: 10.1073/pnas.0407097102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mosch HU, Fink GR. Dissection of filamentous growth by transposon mutagenesis in Saccharomyces cerevisiae. Genetics. 1997;145:671–684. doi: 10.1093/genetics/145.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gancedo JM. Control of pseudohyphae formation in Saccharomyces cerevisiae. Fems. Microbiol. Rev. 2001;25:107–123. doi: 10.1111/j.1574-6976.2001.tb00573.x. [DOI] [PubMed] [Google Scholar]
- 9.Sanders SL, Herskowitz I. The Bud4 protein of yeast, required for axial budding, is localized to the mother/bud neck in a cell cycle-dependent manner. J. Cell. Biol. 1996;134:413–427. doi: 10.1083/jcb.134.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gimeno CJ, Fink GR. Induction of pseudohyphal growth by overexpression of PHD1, a Saccharomyces cerevisiae gene related to transcriptional regulators of fungal development. Mol. Cell. Biol. 1994;14:2100–2112. doi: 10.1128/MCB.14.3.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raithatha S, Su TC, Lourenco P, Goto S, Sadowski I. Cdk8 regulates stability of the transcription factor Phd1 to control pseudohyphal differentiation of Saccharomyces cerevisiae. Mol. Cell. Biol. 2012;32:664–674. doi: 10.1128/MCB.05420-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellis T, Wang X, Collins JJ. Diversity-based, model-guided construction of synthetic gene networks with predicted functions. Nat. Biotechnol. 2009;27:465–471. doi: 10.1038/nbt.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Douglas LM, Li L, Yang Y, Dranginis AM. Expression and characterization of the flocculin Flo11/Muc1, a Saccharomyces cerevisiae mannoprotein with homotypic properties of adhesion. Eukaryot. Cell. 2007;6:2214–2221. doi: 10.1128/EC.00284-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu HP, Styles CA, Fink GR. Saccharomyces cerevisiae S288C has a mutation in FL08 a gene required for filamentous growth. Genetics. 1996;144:967–978. doi: 10.1093/genetics/144.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi O, Suda H, Ohtani T, Sone H. Molecular cloning and analysis of the dominant flocculation gene FLO8 from Saccharomyces cerevisiae. Mol. Gen. Genet. 1996;251:707–715. doi: 10.1007/BF02174120. [DOI] [PubMed] [Google Scholar]
- 16.Ryan O, et al. Global gene deletion analysis exploring yeast filamentous growth. Science. 2012;337:1353–1356. doi: 10.1126/science.1224339. [DOI] [PubMed] [Google Scholar]
- 17.Murphy KF, Balazsi G, Collins JJ. Combinatorial promoter design for engineering noisy gene expression. Proc. Natl Acad. Sci. USA. 2007;104:12726–12731. doi: 10.1073/pnas.0608451104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nehlin JO, Carlberg M, Ronne H. Control of yeast GAL genes by MIG1 repressor: a transcriptional cascade in the glucose response. Embo. J. 1991;10:3373–3377. doi: 10.1002/j.1460-2075.1991.tb04901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flick JS, Johnston M. Two systems of glucose repression of the GAL1 promoter in Saccharomyces cerevisiae. Mol. Cell. Biol. 1990;10:4757–4769. doi: 10.1128/MCB.10.9.4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramos JL, et al. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 2005;69:326–356. doi: 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farabaugh PJ. Sequence of the lacI gene. Nature. 1978;274:765–769. doi: 10.1038/274765a0. [DOI] [PubMed] [Google Scholar]
- 22.Voth WP, Olsen AE, Sbia M, Freedman KH, Stillman DJ. ACE2, CBK1, and BUD4 in budding and cell separation. Eukaryot. Cell. 2005;4:1018–1028. doi: 10.1128/EC.4.6.1018-1028.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kron SJ, Styles CA, Fink GR. Symmetric cell division in pseudohyphae of the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 1994;5:1003–1022. doi: 10.1091/mbc.5.9.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McIsaac RS, et al. Synthetic gene expression perturbation systems with rapid, tunable, single-gene specificity in yeast. Nucleic Acids Res. 2013;41:e57. doi: 10.1093/nar/gks1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McIsaac RS, Gibney PA, Chandran SS, Benjamin KR, Botstein D. Synthetic biology tools for programming gene expression without nutritional perturbations in Saccharomyces cerevisiae. Nucleic Acids Res. 2014;42:e48. doi: 10.1093/nar/gkt1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Casamayor A, Snyder M. Bud-site selection and cell polarity in budding yeast. Curr. Opin. Microbiol. 2002;5:179–186. doi: 10.1016/S1369-5274(02)00300-4. [DOI] [PubMed] [Google Scholar]
- 27.Basu S, et al. Spatial landmarks regulate a Cdc42-dependent MAPK pathway to control differentiation and the response to positional compromise. Proc. Natl Acad. Sci. USA. 2016;113:E2019–E2028. doi: 10.1073/pnas.1522679113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harkins HA, et al. Bud8p and Bud9p, proteins that may mark the sites for bipolar budding in yeast. Mol. Biol. Cell. 2001;12:2497–2518. doi: 10.1091/mbc.12.8.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zahner JE, Harkins HA, Pringle JR. Genetic analysis of the bipolar pattern of bud site selection in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:1857–1870. doi: 10.1128/MCB.16.4.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cullen PJ, Sprague GF., Jr. The roles of bud-site-selection proteins during haploid invasive growth in yeast. Mol. Biol. Cell. 2002;13:2990–3004. doi: 10.1091/mbc.E02-03-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ni L, Snyder M. A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae. Mol. Biol. Cell. 2001;12:2147–2170. doi: 10.1091/mbc.12.7.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen MT, Weiss R. Artificial cell-cell communication in yeast Saccharomyces cerevisiae using signaling elements from Arabidopsis thaliana. Nat. Biotechnol. 2005;23:1551–1555. doi: 10.1038/nbt1162. [DOI] [PubMed] [Google Scholar]
- 33.Hughes RM, Bolger S, Tapadia H, Tucker CL. Light-mediated control of DNA transcription in yeast. Methods. 2012;58:385–391. doi: 10.1016/j.ymeth.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bahn YS, et al. Sensing the environment: lessons from fungi. Nat. Rev. Microbiol. 2007;5:57–69. doi: 10.1038/nrmicro1578. [DOI] [PubMed] [Google Scholar]
- 35.Blount BA, Weenink T, Ellis T. Construction of synthetic regulatory networks in yeast. FEBS Lett. 2012;586:2112–2121. doi: 10.1016/j.febslet.2012.01.053. [DOI] [PubMed] [Google Scholar]
- 36.Dang CV, Lee WM. Identification of the human c-myc protein nuclear translocation signal. Mol. Cell. Biol. 1988;8:4048–4054. doi: 10.1128/MCB.8.10.4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu M, et al. Engineering of regulated stochastic cell fate determination. PNAS. 2013;110:10610–10615. doi: 10.1073/pnas.1305423110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim J, Rose MD. Stable pseudohyphal growth in budding yeast induced by synergism between septin defects and altered MAP-kinase signaling. PLoS Genet. 2015;11:e1005684. doi: 10.1371/journal.pgen.1005684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park YN, Morschhauser J. Tetracycline-inducible gene expression and gene deletion in Candida albicans. Eukaryot. Cell. 2005;4:1328–1342. doi: 10.1128/EC.4.8.1328-1342.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yin Z, et al. Glucose triggers different global responses in yeast, depending on the strength of the signal, and transiently stabilizes ribosomal protein mRNAs. Mol. Microbiol. 2003;48:713–724. doi: 10.1046/j.1365-2958.2003.03478.x. [DOI] [PubMed] [Google Scholar]
- 41.Ashe MP, De Long SK, Sachs AB. Glucose depletion rapidly inhibits translation initiation in yeast. Mol. Biol. Cell. 2000;11:833–848. doi: 10.1091/mbc.11.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cullen PJ, Sprague GF., Jr. Glucose depletion causes haploid invasive growth in yeast. Proc. Natl Acad. Sci. USA. 2000;97:13619–13624. doi: 10.1073/pnas.240345197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Albers E, Larsson C, Andlid T, Walsh MC, Gustafsson L. Effect of nutrient starvation on the cellular composition and metabolic capacity of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2007;73:4839–4848. doi: 10.1128/AEM.00425-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee ME, DeLoache WC, Cervantes B, Dueber JE. A highly characterized yeast toolkit for modular, multipart assembly. ACS Synth. Biol. 2015;4:975–986. doi: 10.1021/sb500366v. [DOI] [PubMed] [Google Scholar]
- 45.Rines, D. R., Thomann, D., Dorn, J. F., Goodwin, P. & Sorger, P. K. Live cell imaging of yeast. Cold Spring Harbor Protoc.2011, 1026–1041 (2011). [DOI] [PubMed]
- 46.Pothoulakis, G. & Ellis, T. Supporting Material. Figshare. 10.6084/m9.figshare.5660179.v1. Accessed 3 Dec 2017 (2017).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
Time-lapse movies of the experiments shown in Figs. 5a, b and 6a, b are provided in Supplementary Movies 1–4. High resolution versions of all microscopy images shown in the manuscript are available via Figshare at DOI: 10.6084/m9.figshare.566017946. Maps of new plasmids generated in this study are provided in Supplementary Data 1 in commented GenBank format and all plasmids will be made available via Addgene. All data generated or analyzed during this study that are not included in this published article and its Supporting Information files are available upon request.