Abstract
At high latitudes, climatic shifts hypothetically initiate recurrent episodes of divergence by isolating populations in glacial refugia—ice-free regions that enable terrestrial species persistence. Upon glacial recession, populations subsequently expand and often come into contact with other independently diverging populations, resulting in gene flow. To understand how recurrent periods of isolation and contact may have impacted evolution at high latitudes, we investigated introgression dynamics in the stoat (Mustela erminea), a Holarctic mammalian carnivore, using whole-genome sequences. We identify two spatio-temporally distinct episodes of introgression coincident with large-scale climatic shifts: contemporary introgression in a mainland contact zone and ancient contact ~200 km south of the contemporary zone, in the archipelagos along North America’s North Pacific Coast. Repeated episodes of gene flow highlight the central role of cyclic climates in structuring high-latitude diversity, through refugial divergence and introgressive hybridization. When introgression is followed by allopatric isolation (e.g., insularization) it may ultimately expedite divergence.
Jocelyn Colella et al. report whole-genome sequences of 10 stoats (Mustela erminea) from four regions of glacial refugia. They find evidence for two past introgressive events between lineages that coincide with interglacial periods, a pattern that may extend to other high–latitude species.
Introduction
Glacial refugia are fragmented pockets of unglaciated land where terrestrial species have persisted and diverged through ice ages, and which have significantly structured biological diversity at high latitudes1–3. Three primary macro-refugial locations are hypothesized to have enabled the persistence of temperate terrestrial North American species through Pleistocene glacial cycles (>24 glacial/interglacial cycles; 2.6 Mya–11.7 kya). These include an expansive Northern Beringian refugium shared with Asia4 and two Southern refugia, separated into East and West by the Rocky Mountains or Great Plains/Mississippi drainage5,6. A fourth, smaller refugium (or series of refugia), hypothesized to exist on areas of exposed continental shelf during the last glacial maximum near contemporary North Pacific Coastal archipelagos7–10, may have provided another isolated sanctuary for long-term coastal persistence of terrestrial species. Although this refugium has received varying degrees of empirical support, its existence is critical to understanding dynamics of intercontinental biotic exchange and it may have played a primary role in the early peopling of the Americas11.
Phylogeography of high-latitude Mustela erminea, the stoat or ermine (Family: Mustelidae) delineates four genetically distinct mitochondrial lineages geographically corresponding to these four refugia12. Although glacial isolation appears to have impacted the long-term evolution of this mammalian meso-carnivore, the effects of glacial recession and subsequent secondary contact between refugial lineages remain unexplored. Therefore, we investigated the consequences of cyclic climates and pulsed introgression on genomic diversity in this circumboreal species complex. During inter-glacial warming, divergent lineages expanded from refugial centers until they reached environmental or biological barriers13. In some cases, admixture occurred upon secondary contact with other lineages. If M. erminea experienced serial bouts of climate-mediated introgression, a repeated signature of introgressive hybridization should be evident in the genomes of refugial descendants14.
Contact zones are windows into evolutionary processes15 now being explored on a genomic scale. We define introgressive hybridization as interbreeding and the movement of alleles between two genetically distinct lineages. Although often difficult to parse introgression from incomplete lineage sorting in closely related taxa or lineages16, investigations of divergent lineages, such as those within the M. erminea complex, provide insight into the genomic consequences of introgression. For example, genetic swamping or homogenization can prevent divergence between mixing lineages, while Bateson–Dobzhansky–Muller17–19 and mitonuclear20 incompatibilities can lead to hybrid breakdown, thereby promoting divergence through reinforcement21,22. Although our understanding of hybridization emerges largely from research on plants23,24, the genomic era has uncovered substantial reticulate evolution (e.g., introgressive hybridization) in a growing number of animal species25,26, including humans27. As histories of divergence and speciation-with-gene flow continue to be discovered (e.g.,28,29), the implications of hybridization for the conservation of evolutionarily distinct units appear profound30.
To test whether genomic structure in stoats reflects refugial origins and to explore the consequences of contact among lineages, we generated ten whole-genome sequences of representatives from each refugial clade. With expanded genomic coverage, we test for signatures of refugial divergence and introgression, characterize the timing of introgression events relative to climatic oscillations, and infer the impact of introgressive hybridization on divergence across the complex landscape of northwestern North America.
Results
Refugial origins and phylogeography
To test our hypothesis that the four mitochondrial stoat lineages originated as the result of isolation and divergence in each of the four major North American refugia, we generated whole-genome sequences for ten M. erminea drawn from each refugial lineage and two spatially disjunct contact zones (the interior border of Alaska-Yukon Territory and the NPC; Fig. 1). We contrasted mitochondrial and nuclear phylogenies against geographic clade distributions, estimated pairwise diversity metrics (pairwise mitochondrial and nuclear divergence, FST31, relatedness32), and used ADMIXTURE33 analyses to define populations.
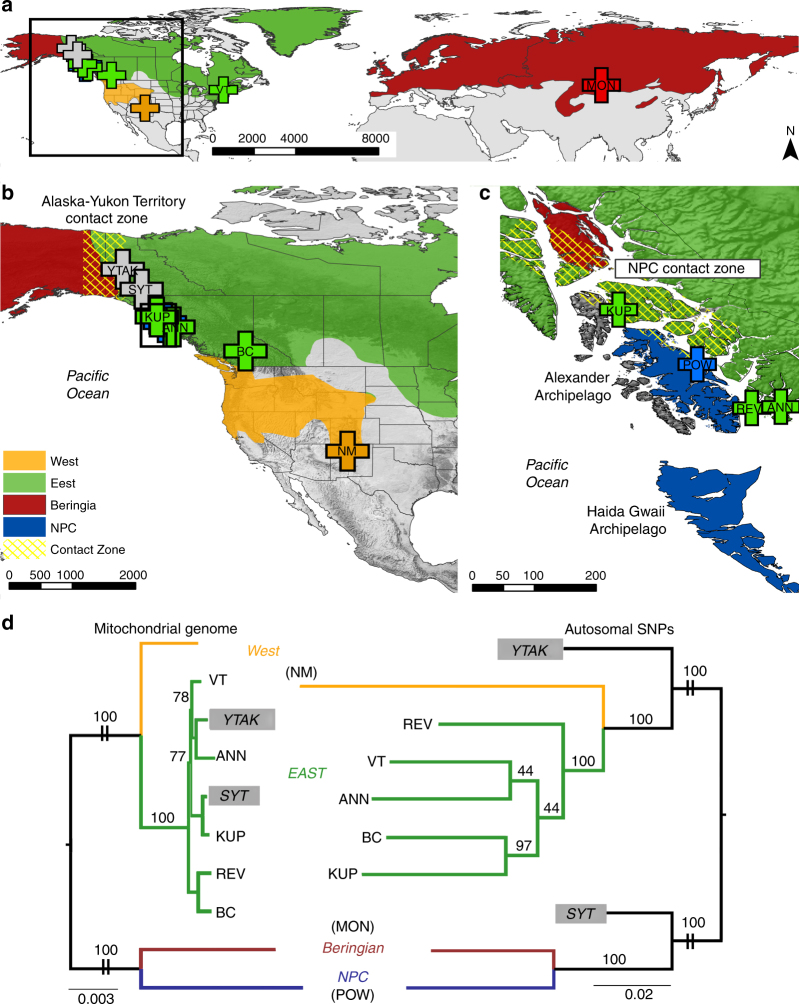
Fig. 1.
Mustela erminea clade geographic distributions and phylogenetic relationships. a–c Stoat clade distributions (clade names reflect refugial origins) based on amplicon mitochondrial haplotype distributions12 and IUCN (International Union for Conservation of Nature, www.iucn.org) range information. Genomic samples are labeled with locality abbreviations: ANN (Annette Island, Alaska, USA), BC (southern British Columbia, Canada), KUP (Kupreanof Island, Alaska, USA), MON (Mongolia), NM (New Mexico, USA), POW (Prince of Wales Island, Alaska, USA), REV (Revillagigedo Island, Alaska, USA), SYT (Southern Yukon Territory, Canada), YTAK (northern Yukon-Alaska border), VT (Vermont, USA). Contact zones are denoted with yellow hatching, based on the presence of mixed mitochondrial haplotypes12. Scale bars are in kilometers. a Global sampling scheme. b North American sampling localities. c Sampling within the northern North Pacific Coast (NPC) archipelagos. d Mid-point rooted maximum-likelihood phylogenies, scaled by genetic distance. d (left) Complete mitochondrial genomes and d (right) autosomal SNPs for ten M. erminea individuals. Phylogenies demonstrate strong support for four major refugial clades and mitonuclear discordance for hybrid samples (SYT, YTAK). Deep divergence between the NPC Island (POW) and Beringia (MON) clades is evident in the long branches in both phylogenies
Our results showed strong mitochondrial (Fig. 1; Supplementary Fig. 1) and nuclear (Figs. 1–3; Supplementary Fig. 2; Supplementary Table 1) support for four stoat clades geographically coincident with North American refugia. We identified deep divergence of a highly distinctive West clade, a group previously lacking strong nuclear support12 (Figs. 1, 2a and c; Supplementary Fig. 2; Supplementary Tables 1 and 2). We detected substantial genetic divergence between mainland stoat lineages (e.g., 21% average pairwise difference across >3 million autosomal SNPs between Beringia and East clades), comparable to estimates between Beringia stoats and the outgroup species, M. putorius (domestic ferret, 20%), calling into question whether this widespread mustelid is a single species (Supplementary Table 1). Centrality of the Vermont sample is consistent with widespread expansion of the East lineage from a single eastern refugium followed by intra-clade divergence (Fig. 2b). Similar refugial signatures have been detected in other North American species (e.g., martens34 and black bears35). Additional structure within the East clade (Fig. 2b) may reflect microrefugial diversification36. Geographic proximity (Fig. 1) is not predictive of genetic similarity (Fig. 2b; e.g., REV and BC), consistent with patterns also uncovered in primary stoat prey species such as voles37 and lemmings38.
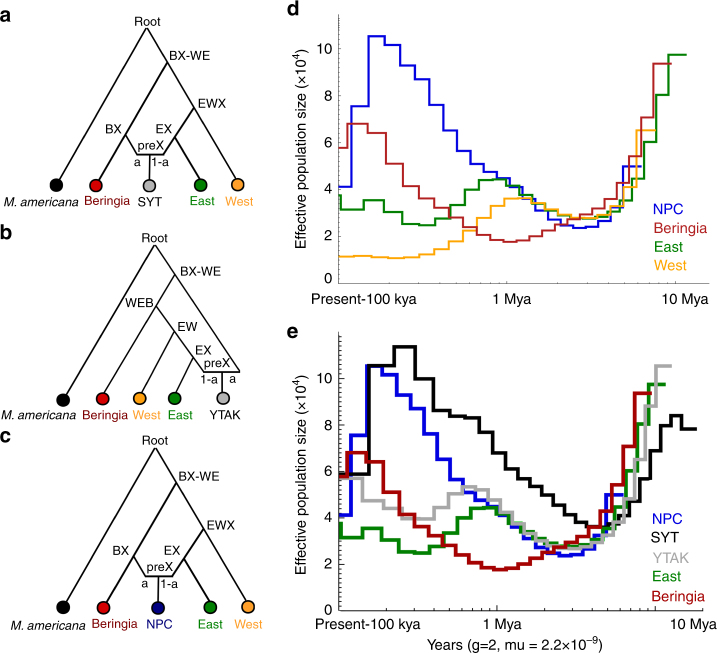
Fig. 3.
Phylogenetic placement of admixed samples and historical demography. a–c Optimal phylogenetic placement of hybrid samples onto our non-admixed RAxML topology (see also Supplementary Fig. 5) based on f4-statistics fitting in Admixture Graph. a SYT originates from ancestors of Beringia and East populations; b YTAK is admixed from East and an unsampled ancestor of all M. erminea lineages; c NPC stems from ancestors of Beringia and East clades. (d distributions of effective population sizes (Ne) through time from pairwise sequential Markovian coalescent (PSMC) analysis scaled by a general mammalian mutation rate (2.2 × 10−9) and a 2-year generation time. d PSMC plots for a representative from each refugial population (East, West, Beringia, NPC). e PSMC plots for hybrid samples (YTAK, SYT, NPC) and their source populations (Beringia, East). Hybrid distributions mimic that of their most-backcrossed source (f4) with artefactual Ne inflation, as expected for admixed demographic histories
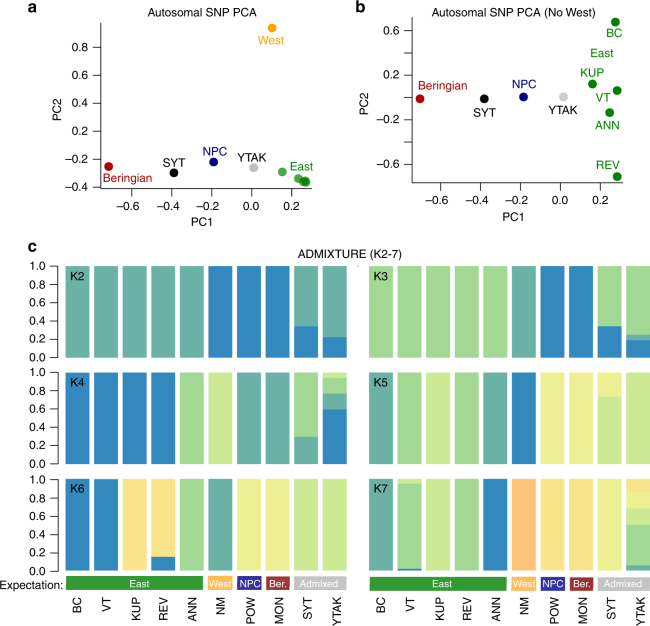
Fig. 2.
Principal component analysis and ADMIXTURE results. a, b SNP principal component analysis (PCA). All samples identified as admixed by f3-statistics (NPC, SYT, YTAK) lie intermediate to Beringia and East clades, their purported source populations. a PCA with all samples demonstrates substantial nuclear genetic differentiation of the West clade. PC1 explains 20.7% of the total variation, and PC2 explains 14.1%. b PCA with the West population removed demonstrates additional structure within the East clade, centered on Vermont (VT). Hybrid samples are intermediate to their source populations. PC1 explains 29.8% of the total variation and PC2 11.8%. c ADMIXTURE plots for K2–7. K = 3 best matches our expectation based on the geographic distributions of refugial clades and is consistent with admixed ancestry for SYT and YTAK samples: (1) East samples as a distinct group, (2) a West population, and (3) a combined Beringia-NPC population with two hybrids admixed from these groups. K = 6 has the lowest cross-validation score for our data and shows an identical pattern to K3 but with additional substructure within the East clade, indicative of microrefugial substructure near Appalachia and consistent with PCA results suggesting East expansion from a single refugium
The sister relationship between NPC and Beringia stoats, combined with substantial genetic divergence (nuclear >10%; mitochondrial >4%; Supplementary Table 1) between these clades and their current geographic distributions, are consistent with the Coastal Refugium Hypothesis7–10. Ancestry estimation33 identified autonomous East and West populations but a single combined Beringia/NPC population (K = 3; Fig. 2c), again highlighting the paleoendemic ancestry of the NPC Island lineage. Pairwise mitochondrial divergence is also particularly elevated for the NPC Island lineage, bordering the species-level divergence threshold suggested for distinct mammal species39.
Detecting introgression and determining source populations
Evidence of recurrent introgressive hybridization, whether as a single or recurrent event, should be evident in the genomes of refugial descendants. We hypothesized that signatures of introgression should be present in individuals collected within the Alaska-Yukon Border hybrid zone and expected that stoats from Beringia and East clades are the most likely source populations based on contemporary clade distributions. In addition to contrasting mitochondrial and nuclear phylogenies for discordance, we performed a series of tests for introgression including ADMIXTURE33 and f3-statistics (Supplementary Table 3)40,41 to assess shared genetic drift among all possible combinations of two source populations42.
We identified mitonuclear discordance for two individuals collected near the Alaska-Yukon border (herein SYT and YTAK, representing Southern and Northern localities along the contact zone, respectively), which can indicate a history of genomic introgression or incomplete lineage sorting. ADMIXTURE identified the same two individuals as admixed between East (K3: SYT 66%, YTAK 74%) and Beringia (K3: SYT 34%, YTAK 19%) clades or as a single, but distinct population, consistent with shared source populations (Fig. 2c). Both Alaska-Yukon samples were positioned between their hypothesized source populations in our PCA results (Fig. 2a and b), with SYT closer to Beringia and YTAK closer to the East clade, further supporting their mixed ancestry and suggested asymmetric backcrossing distributions. Lastly, f2 statistics, a measure of distance between populations, were also lowest between the putative Alaska-Yukon hybrids and their suspected source populations, further supporting their derivation from the same source populations (Supplementary Table 4).
Surprisingly, NPC Island stoats also tested positive for admixture, with Beringia-East parentage (Table 1; Fig. 2a and b). These island stoats have been isolated off the NPC (Prince of Wales Island and the neighboring Haida Gwaii archipelago of British Columbia, Canada [not sequenced here]) for at least 10,000 years12; therefore, we suspect they are derived from an ancient hybridization event that occurred during an earlier interglacial period, prior to the contemporary insularization of this clade. We found no evidence of admixture, however, in the Kupreanof Island sample.
Table 1.
Source populations and admixture date estimates for each hybrid sample
| Target | Source1 | Source2 | f3-statistics | MixMapper date estimates | ||||
|---|---|---|---|---|---|---|---|---|
| f3 | Err | Z | α | Mixed drift | Years (Ne = 375k) | |||
| NPC | East | Beringian | −0.05 | 0.009 | −5.28 | 0.48–0.59 | 0.00–0.00 | 0 |
| SYT | East | Beringian | −0.17 | 0.003 | −48.66 | 0.52–0.78 | 0.00–0.00 | 0 |
| YTAK | East | Beringian | −0.15 | 0.003 | −47.41 | 0.48-0.73 | 0.00–0.23 | 0–393,996.5 |
Admixture date estimates in years based on an effective population size (Ne) of 375k (ref. 12) for the North Pacific Coast island (NPC) and the two Alaska-Yukon hybrids (SYT from Southern Yukon and YTAK from the northern Alaska-Yukon border). Err indicates the standard error and Z indicates the Z-score
Testing for asymmetric introgression
Biased or asymmetric introgression may indicate a selective advantage, the existence of genetic incompatibilities, or a demographic imbalance. Alternatively, equal contributions from both parental source populations may suggest that there are minimal genetic incompatibilities between hybridizing stoat clades, as we would expect within a single species. For the NPC Island clade, biased allelic overlap with one source population may shed light on interglacial colonization dynamics, as the island may have been colonized by both Beringia and East ancestors who subsequently interbred. We used F4-statistics, similar to D-statistics43,44 and ABBA/BABA45, to highlight differential backcrossing histories among lineages and used admixture graph fitting46 to estimate the topological placement of admixed samples relative to their potential sources.
First, our f4 results highlight the influence of repeated glaciation in North America as reflected in multiple episodes of gene flow between lineages originating in independent refugia. Allele sharing (f4) reflected phylogenetic relationships, with stepwise genetic distance from the ancestral Beringia clade increasing with greater geographic distance from Beringia (Fig. 1; Supplementary Tables 5 and 6), a pattern consistent with more frequent or prolonged contact between geographically proximal refugial lineages.
Admixture graph fitting identified SYT as the result of Beringia and East admixture, while YTAK was produced from an ancestral admixture event between East and a shared ancestor of all stoat lineages (Fig. 3a and b; Supplementary Table 7). The vast majority of f4 comparisons (Supplementary Tables 5 and 6) were highly significant (Z-score >5), reflecting substantial intra-specific divergence within M. erminea. Biased allelic overlap suggested that YTAK shares more alleles with the East source, whereas SYT is more backcrossed with the Beringia source (Table 2). Differentially backcrossed hybrid offspring within this broad, mainland contact zone suggests that hybridization between East and Beringia lineages may have minimal negative fitness consequences (e.g., reduced fertility, viability)47. These preliminary inferences require additional sequencing of hybrids from the interior Alaska-Yukon hybrid zone to characterize the specific genomic regions that may be crossing semi-permeable lineage boundaries and to rigorously test for asymmetric introgression, which could be driven by selection (e.g, genetic incompatibilities, Darwin’s corollary to Haldane’s rule48), drift, or demographic parameters. If gene flow is neutral, extended periods of contact may promote fusion between East and Beringian lineages. However, the contemporary persistence of both Old World Beringian and New World East lineages in North America suggests that gene flow during brief periods of interglacial contact through the Pleistocene were insufficient to homogenize refugial divergence.
Table 2.
f4-statistics for hybrid samples (X) with M. putorius outgroup (W)
| W | X | Y | Z | f4 | Z-scores |
|---|---|---|---|---|---|
| OG | NPC | East | Beringia | 0.30 | 31.79 |
| OG | NPC | West | East | 0.32 | 76.99 |
| OG | NPC | West | Beringia | 0.51 | 70.42 |
| OG | SYT | East | Beringia | 0.13 | 9.29 |
| OG | SYT | West | East | 0.37 | 86.48 |
| OG | SYT | West | Beringia | 0.41 | 42.17 |
| OG | YTAK | Beringia | East | 0.28 | 27.12 |
| OG | YTAK | West | East | 0.40 | 90.67 |
| OG | YTAK | West | Beringia | 0.08 | 10.18 |
F4(W,X:Y,Z) results for each hybrid sample (NPC North Pacific Coast island, SYT Southern Alaska-Yukon border, and YTAK northern Alaska-Yukon border) relative to all possible parental source populations (East, West, Beringia)
All Z-scores are positive for ease of interpretation and significant (>5). Rotating the order of a pair (e.g., YZ to ZY) switches the sign +/− of both the f4-statistic and the Z-score but does not change the relationships
Biased allele sharing between NPC and Beringia (Table 2) is consistent with phylogenetic relationships (Fig. 1d) and three possible evolutionary histories: (1) differential introgression (selection) favoring Beringia alleles early in the NPC Island clade’s origin; (2) extended or more frequent connection between Beringia and NPC refugia throughout the Pleistocene; or (3) stochasticity, potentially compounded by founder effects or bottlenecks.
Timing introgressive events
Given the pattern of North American glaciation49, the current geographic isolation of the NPC Island clade, and the limited dispersal abilities of insular M. erminea50, we suspect that hybridization is not ongoing on NPC islands. In contrast, based on the current distribution of mitochondrial diversity12 and our understanding of postglacial population expansion, we expect that stoats from East and Beringia clades are actively interbreeding along the Alaska-Yukon border. To approximate the timing of admixture events, we converted drift unit branch lengths (D), output from MixMapper51, to absolute time (years) using the formula D ≈ 1−e−t/2Ne(solved for t generations)52.
The estimated time of admixture for Alaska-Yukon hybrids did not differ from zero (Table 1), suggesting that the Alaska-Yukon Territory border is an active hybrid zone between the leading-edge of Beringia and East stoat clades. In contrast, NPC admixture is estimated at up to 394 kya (Table 1; Supplementary Table 8), prior to the Wisconsin glacial (30–80kya53,54) and Sangamonian interglacial (80–140kya53,54). The weaker NPC hybrid signal relative to contemporary Alaska-Yukon hybrids (f3 Z-score), the insular isolation of this clade for at least the past 10,000 years12,55, and the short generation time of stoats (2 years56) suggest that NPC Island admixture is more ancient than that occurring in the mainland Alaska-Yukon zone. We hypothesize that the hybrid origin of the NPC clade predated a period of glaciation and refugial isolation, but this point requires further exploration with expanded sampling and tests of linkage disequilibrium. If NPC stoats continue to evolve in isolation, our results suggest that continued divergence in allopatry may ultimately lead to speciation.
Based on our results, NPC islands were colonized either by admixed individuals or independent invasions of both parental lineages (East, Beringia) that subsequently hybridized. Introgressive hybridization, preceding divergence in isolation, may have disproportionately elevated the evolutionary distance between NPC Island stoats and other lineages (Figs. 1, 2, and 3d, e; Supplementary Tables 1–4). Isolation of a genetically recombinant population, at least through the Last Glacial Maximum, in a coastal refugium reinforced an evolutionary trajectory independent from either source population (Supplementary Fig. 3). Further, low amplicon variation in NPC stoats12 suggests that island founder effects or a bottleneck5 occurred among NPC stoats in the coastal refugium57. Drift may impact small populations and may have further accelerated NPC stoat differentiation.
Historical demography
We expected glacial cycling (periods of isolation/contact) to have impacted the historical demography of each refugial stoat clade differentially. Further, we hypothesized that individuals with a history of introgression will exhibit elevated effective population sizes (Ne), with a distribution most similar to that of their most-backcrossed source population (East for YTAK, and Beringia for SYT). Last, we expected cladogenesis to coincide with refugial isolation during glacial periods and demographic troughs, while Ne is expected to rise during interglacials as a consequence of population expansion.
Stoat demographic histories (PSMC58; Fig. 3d, e) showed clade-level responses to cyclic glaciation, highlighting the central role of large-scale climatic shifts in shaping diversity at high latitudes through episodic isolation and contact. Overall low, but relatively stable Ne in East and West clades reflected similar demographic responses to New World glacial cycling, where the West refugium was likely smaller than the East, as reflected in smaller Ne (Fig. 3d). Unlike Nearctic populations, Western Beringia experienced substantial expansion historically, corresponding to reduced glaciation in eastern Eurasia and the larger size of Beringia compared to the NPC refugium. Dates of cladogenesis mirrored estimates from the calibrated mitochondrial genome phylogeny (Supplementary Fig. 1), with inter-clade divergence beginning around 2 Mya when Beringia split from the other stoat lineages, followed by West (1.3 Mya) divergence, and most recently Island/East (0.9 Mya) cladogenesis (Fig. 3d). However, the hybrid ancestry of the NPC island sample suggest that cladogenesis estimates based on Ne may be unreliable. As expected, when hybrid distributions are juxtaposed against their parental populations (Fig. 3e), they mirror that of their most-backcrossed source population, but with an upward shifted Ne59. If the increase in hybrid Ne occurs at the time of hybridization between parental populations59, SYT and YTAK admixture may be older than predicted based on admixture date estimates alone.
Combined with diversity metrics (Supplementary Fig. 2; Supplementary Tables 1–4), highly distinct demographic histories suggest taxonomic revision of M. erminea may be warranted, pending wider geographic sampling. This is a particularly compelling example of cryptic diversification in carnivores when contrasted against weakly differentiated PSMC demographic histories uncovered in some canid60 (wolf, dingo, domestic dog) and felid61 (tiger, leopard, cheetah) genomes.
Discussion
We document multiple temporally and spatially offset bouts of introgressive contact between divergent stoat lineages, corresponding to distinct phases of glacial cycling. Therefore, we predict that other higher latitude species with shared biogeographic histories may also exhibit genomic signatures of repeated introgression corresponding to cyclic climates. This process of recurrent introgressive hybridization between geographically proximal lineages, followed by refugial or insular isolation and divergence, may contribute to elevated levels of island endemism, particularly for continental archipelagos adjacent to glacially dynamic mainland areas.
When backcrossing occurs into both parental populations, as in the Alaska-Yukon hybrid zone, introgression may be stochastic or selectively neutral. Consequently, extended periods of contact may lead to homogenization18,62 and result in a net loss of regional diversity63, such that the divergent lineages evident today (East, Beringia) may be ephemeral64. The narrow Alaska-Yukon hybrid zone and strong East-Beringia genomic differentiation, however, suggests that such peripheral hybrid populations are ephemeral and minimally contribute to long-term evolution of high latitude diversity. If introgression events are temporary, genomic evidence of serial hybridization offers a model for detecting refugia and tracking colonization routes among high latitude species. Here, we provide yet another example of introgressive hybridization in mammals, highlighting both the prevalence and varied role of introgression in mammalian evolution. Our results demonstrate that, for geographically widespread species, the long-term evolutionary role of introgression often may be inconsequential relative to larger biogeographical processes, such as island vicariance, refugial isolation, or rapid environmental change.
In some situations, however, a population bottleneck in a small refugium or in response to insularization can enable the temporary persistence of less fit recombinant genotypes65. Even a temporary reduction in fitness can enable traversal between fitness-landscape optima and promulgate alternative evolutionary trajectories, which can permanently impact the genomic structure of high latitude populations and potentially lead to speciation65,66. Genomic evidence of hybridization, followed by divergence in geographic isolation, is consistent with a hybrid origin for the NPC lineage. Though still incipient in this case, the process of introgression followed by allopatry may lead to hybrid speciation67. Hybridization can catalyze the evolution of reproductive barriers68 or expedite divergence through the rapid generation of novel genetic recombinants. Although the mechanisms underlying this special case of divergence with gene flow remain elusive, a plausible precondition for speciation is the separation of lineages into allopatric ranges, which may enhance the genetic divergence of a recombinant population, even if introgression is not directly implicated in speciation. Therefore, NPC stoats potentially highlight the integral role of post-hybridization allopatric divergence in the evolutionary histories of island and refugial taxa, but expanded insular sampling is required to fully test this mechanism of divergence.
It has long been argued that pre-zygotic isolation is reinforced when diverging groups return to parapatry and hybridize18,20, although this idea is not without controversy. Under this scenario, hybridization between incipient species can result in unfit hybrids and natural selection might act against hybridization, reinforcing pre-zygotic isolation mechanisms, eventually leading to speciation and avoiding maladaptive hybridization18,69. In this fashion, continuing and progressive reinforcement of pre-zygotic mechanisms would ultimately result in speciation and complete isolation between lineages18,70, if hybridization and selective pressures on both lineages are symmetrical71. With an active and plausibly ongoing hybrid zone, along with more ancient island admixture, stoats provide a unique opportunity to investigate the impact of symmetrical versus asymmetrical introgression using genomics.
With multiple waves of mainland colonizers corresponding to climatic cycles and potentially adding novel genetic elements to insular genomic architectures, the process of repeated, climate-mediated hybridization may have played a role in diversification across other land-bridge island systems. Irish stoats, for example, exhibit elevated genetic diversity compared to neighboring populations in Great Britain, despite persisting on a smaller island farther from a continental source13,72. In this parallel system, the English Channel separating Britain from the European mainland has been serially exposed through Pleistocene glacial cycles73, effectively homogenizing populations across the Channel. On the other hand, the Irish Sea, nearly twice the depth of the English Channel, further separates Ireland from Britain, presenting a formidable barrier to gene flow that allowed only occasional influx of novel alleles from mainland or British immigrants during extreme glacial events. This mechanism of post-hybridization allopatric divergence is not restricted to land bridge or continental archipelagos. Oceanic island populations of Darwin’s storied finches on the Galapagos Islands also were impacted by climate-mediated pulsed hybridization that was coincident with changes in sea level due to glaciation74.
Admixture in this insular endemic (NPC Island stoats, subspecies: M. e. haidarum) poses a particular challenge for the U.S. Endangered Species Act, which currently denies protection for hybrid taxa30,75. Mustela erminea haidarum is federally protected in Canada on the Haida Gwaii Islands of British Columbia56, but also occurs on Prince of Wales Island, Alaska, where it has no protection. This study is congruent with previous work noting that this island-restricted stoat subspecies is genetically12,76 and morphologically77 distinctive, but levels of management protection at the federal level differ profoundly between Canada and the United States.
Genomic signatures of repeated admixture and refugial isolation among North American stoats highlight the episodic role of Late Quaternary climate cycles in generating and structuring high latitude diversity. We demonstrate two instances of gene flow between stoat lineages coincident with interglacial periods: one contemporaneous along the interior Alaska-Yukon border and a second, more ancient event, occurring along the NPC that left a signature that persisted through multiple glacial cycles. Each hybridization event exemplifies an antagonistic outcome of hybridization (e.g., homogenization versus diversity generation) within a single species complex, highlighting the multifaceted role of introgression in mammalian evolution and the varied temporal and geographic scales at which this process unfolds. At high latitudes, where dramatic range shifts in response to glacial cycling afforded recurring opportunities for contact and isolation, ephemeral hybridization has repeatedly shaped biological diversity2,78,79.
Methods
Sequencing, assembly and post-processing
Contact zones and populations were defined based on expanded mitochondrial sampling12. Ten whole-genome sequences for representatives from each refugial stoat clade, and with additional sampling in suspected contact zones (Fig. 1), were generated on an Illumina HiSeq 2000 (paired-end reads [100 bp]), three samples in four Illumina lanes and the remaining samples in one lane each (Supplementary Table 9). Liver subsamples were obtained post-mortem from the University of New Mexico’s Museum of Southwestern Biology in compliance with ethical regulations (IACUC #16-200526-MC). We used a DNeasy Blood and Tissue Kit (QIAGEN, USA) extraction. Next-generation sequencing libraries were prepared using the Illumina TruSeq DNA Sample Prep Kit. Our genome assembly pipeline followed Lan et al.79 with reads examined using FastQC80 and adapter sequences and sex chromosomes removed (Trimmomatic v0.33 (ref. 81)). Quality scales were compared across samples and rescaled to Illumina 1.8 + phred + 33 quality score, when necessary, to maintain consistency (Picard v1.9: http://picard.sourceforge.net; SAMTools82). Reads were mapped to the domestic ferret genome83 (Mustela putorius furo) using the Burrows-Wheeler aligner84 (BWA). An additional BWA iteration extracted mitochondrial genomes. Final depth of coverage ranged from 10 to 61× (Supplementary Table 9). PCR duplicates were removed using the MarkDuplicates tools from the Picard suite. Nuclear and mitochondrial genome consensus sequences were called using mpileup in SAMtools. SNPs were called using HaplotypeCaller in the Genomic Analysis Toolkit85 (GATK) for all stoats and again against each of two outgroups (M. putorius and Martes americana) and filtered by minimum depth (minDP = 2), genotype quality (minGQ = 30), minimum minor allele frequency (MAF = 0.1), and scaffold size and location (1 Mb), and then private alleles and indels were removed using VCFtools86 (Supplementary Table 10). We intentionally selected an MAF of 0.1 to remove singletons (e.g., individual-specific, rare mutations), and thereby any potential sequencing errors, which are more common in low-coverage genomes. Although singletons can be informative when analyzing within population variation, our analyses focus on shared alleles (e.g., f-statistics) to identify introgression and backcrossing histories, therefore singletons, since they are not shared, do not provide information about allelic overlap among populations. Our minDP (2) was set as one-third of the coverage of our lowest coverage samples (e.g., roughly one-third of 8×) as recommended by the PSMC manual. Also, 2 is the minimum requirement for detecting heterozygotes. Format conversions (vcf, ped, bed) were conducted using PLINK87. Missing data were removed (--max-missing) based on analysis specifications. SNPs were spaced (1 per 50 bp window) to account for linkage disequilibrium (e.g., 25) and sorted into 46 ‘pseudo-chromosomes’ to enable the application of human-specific analyses on a non-model system with 40 chromosomes.
Phylogenetic methods
jModeltest88,89 estimated the most appropriate model of evolution for mitochondrial genomes and autosomal SNPs, with and without outgroup sequences (M. putorius and Martes americana). We generated phylogenies for the entire mitochondrial genome (calibrated/uncalibrated) and autosomal nuclear SNPs (uncalibrated) using RAxML90 (GTRCAT model, 10,000 generations, random starting seed) and Beast291 (1 million generations, 4 chains, 2 runs, sampling frequency of 1000). It should be noted that phylogeny inference using highly variable data (e.g., SNPs) can induce acquisition bias resulting in longer branch lengths92, however, other divergence metrics (e.g., PCA, relatedness, FST) provide additional evidence of substantial divergence between refugial stoat clades. The oldest known M. erminea fossil93 (1.8 My) was used as a log-normal fossil calibration (mean = 5.16, sd = 0.25, offset = 180) to estimate divergence times in Beast2 (Supplementary Fig. 1) following the TreeThinkers tutorial (http://treethinkers.org/tutorials/divergence-time-estimation-using-beast/). A log-normal distribution places the highest probability on ages that are somewhat older than the fossil date, with non-zero probability to infinity. Log-normal distributional priors were selected to place 1.8 Mya at the median and shape priors encompassed late Pliocene to early Pleistocene, similar to Harding and Smith94. Fossil-calibrated divergence estimates separate two pairs of stoat clades (Beringia-NPC and East–West) at 4.47 Mya (±1.21 My, Pliocene), with the Beringia-NPC split dated within the early Pleistocene at 2.1 Mya (±0.6 My), over 1 million years before East–West cladogenesis (1 Mya ± 0.32; Supplementary Fig. 1). Phylogenies were visualized in FigTree v1.2.2 (http://tree.bio.ed.ac.uk/software/figtree/).
Traditional phylogenetic approaches were contrasted against TreeMix ancestral graphs modeling historic gene flow (migration events) among populations95. To accommodate genetic exchange between populations of the same species, TreeMix uses genome-wide allele frequency data to infer historical relationships among populations to maximize the composite likelihood of the sample covariance matrix (What), while modeling both cladogenesis and migration. TreeMix iterates over a standard bifurcating topology to identify a series of migration edges that most increase the likelihood score. TreeMix was run without default sample size corrections (-noss), because multiple populations were comprised of a single individual. We limited the number of migration events (-m) to three, to examine only the most influential events among the few groups examined. TreeMix simulations with M. putorius as the outgroup identified ancient geneflow between Beringia stoat ancestors and M. putorius. Simulations were re-run with M. americana as an outgroup to assess whether long-branch attraction between distantly diverged M. erminea and M. americana (>11Mya96) impacts migration inference in TreeMix, but results were consistent, regardless of the outgroup used. After TreeMix identified gene flow between M. erminea and M. putorius (Supplementary Fig. 4), we only show Admixture Graph results with M. americana as outgroup.
PCAs were generated in SNPrelate97. Diversity statistics (relatedness2, Supplementary Fig. 2; nucleotide diversity, Supplementary Table 11; FST, Supplementary Table 2) were calculated using VCFtools. For ADMIXTURE33 analysis, sites with >80% missing data were removed and the lowest cross-validation (cv = 10- and 5-fold) score identified the most appropriate number of populations (K = 6) by iteratively leaving a sample out and reexamining the partitioning of genetic structure among the remaining samples.
Introgression analyses
F-statistics were run in AdmixTools41 and MixMapper51 using M. americana as an outgroup after TreeMix identified ancient gene flow between M. erminea and M. putorius (Supplementary Fig. 4). f2 statistics [f2(Pop1, Pop2)] quantify drift through summary statistics (e.g., allele frequencies, heterozygosity, covariance, and the probability of two lineages coalescing), with larger values indicating greater divergence42 (Supplementary Table 4). f2 statistics were generated from the compute_moment_stats and compute_most_additive_trees functions in MixMapper with 1000 bootstraps and SNP blocks of 50 (1 per 50 bp). f3-statistics explicitly test for admixture (AdmixTools, 3PopTest) and considered all permutations where Source1, Source2, and the Target samples came from different populations. Populations (e.g., clades) were defined by phylogenetic relationships (Fig. 1) and ADMIXTURE results (Fig. 2c). A positive f3 value does not necessarily indicate the absence of admixture42. Small sample sizes and the absence of a linkage map for M. erminea prevents linkage disequilibrium-based estimates of Ne and refined dating of admixture events.
To parse the backcrossing history of each hybrid sample, f4-statistics were used. We used block-jackknifing to accommodate non-independence between loci42. Although f-statistics alone cannot deduce the direction of gene flow in a system, admixture graph fitting can test whether a proposed evolutionary model fits the data well45,51. Admixture Graph46 fit hybrid individuals into a non-admixed tree topology (Supplementary Fig. 5a) based on f4 results (Supplementary Tables 5 and 6). We ran an additional Admixture Graph simulation on the optimal (lowest minimal error) 4-taxa non-admixed backbone topology identified by Admixture Graph (Supplementary Fig. 5b; Supplementary Table 12) and again for Alaska-Yukon hybrids onto a 5-taxon backbone (Supplementary Fig. 5c) to evaluate how inferred relationships changed (Supplementary Fig. 6). We compared individuals from each population against members of alternative populations (e.g., f4 (Outgroup, NPC; East, Beringia)). We also compared hybrid individuals (as identified by f3-statistics) against individuals from ‘pure’ populations (e.g., f4(Outgroup, Hybrid; East, Beringia)) and admixed populations (e.g., f4 (East, SYT; Beringia; YTAK)) to decipher the backcrossing histories of hybrid samples and characterize patterns of gene flow across populations. f4-statistics are negative (Z-score ≤−5) if there is more allelic overlap between X and Y than between X and Z since the evolutionary split between Y and Z, and positive (Z-score ≥5) if there has been more recent gene flow between X and Z than between X and Y. f4 results are in Table 2, Supplementary Tables 5 and 6, and Supplementary Fig. 7. Dawson et al.’s12 Ne Model 2 using multi-locus Approximate Bayesian Computation approximates mean clade Ne at: 250k for Beringia, 50k for NPC, 125k for East, and 75k for West; therefore, the combined Ne for NPC, East, and Beringia populations is 375k. D is tightly correlated with Ne and additional efforts to refine historical population sizes (Pleistocene, Pliocene) should improve the resolution of our admixture date estimates. Generational output was converted to years using a M. erminea generation time of 2 years56.
Historical demography
PSMC58 was used on consensus genomic sequence data (using both complete and down-sampled sequence data) to characterize historical demography (Fig. 3d, e) by examining heterozygosity densities in a 100 bp sliding windows across the genome. PSMC was run twice for each individual, once utilizing all mapped sequence data (coverage 10–61×) and again on data down-sampled to 10× coverage (matching the lowest coverage sample) using the DownsampleSam tool (Picard). Results were scaled by a general mammalian mutation rate (2.2 × 10−9 per base pair per year98) and stoat generation time, resulting in distributions of Ne through time (Fig. 3d, e). One hundred PSMC bootstrap replicates were performed and plotted for both full-coverage and down-sampled data to confirm consistent distributional shapes and enable comparison across individuals, as PSMC is sensitive to variation in coverage depth (ideal coverage 18× (ref. 99)).
Code availability
Our custom python spacing script is available on GitHub at https://github.com/jpcolella/Ermine_WGS_2018.
Data Availability
Genomic reads are available in NCBI’s Sequence Read Archive, accession: SRP138989. Assembled FASTA files are available on Dryad (10.5061/dryad.bv2g720).
Electronic supplementary material
Acknowledgements
We thank the Cook Lab (University of New Mexico), Enrique Lessa (Universidad de la República, Uruguay), Webb Miller (Penn State University), Aakrosh Ratan (University of Virginia School of Medicine), Kalle Leppälä (University of Aarhus), and Victor Albert (University at Buffalo; UB) for help with data analyses and thoughtful independent reviews; Tilottama Roy and Clark Driscoll (UB) for assistance in the lab; Kimberley Farr (UB) for fruitful scientific discussion; Tom Jung, Batsaikhan Nyamsuren, Natalie Dawson, and Ray Slayton for facilitating specimen acquisition; and the Museum of Southwestern Biology for providing tissue samples. This work was supported by the National Science Foundation (grant number NSF 1258010). Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Author contributions
J.P.C., S.L.T., J.A.C., and C.L conceived the project. J.P.C. assembled the genomes, conducted all analyses, and wrote initial manuscript draft with scientific and editorial input from T.L., S.L.T., J.A.C., and C.L. T.L. designed the bioinformatics pipeline and provided computational support throughout the analysis process. S.C.S. conducted whole-genome sequencing at Penn State University’s Huck Institutes of the Life Sciences. S.L.T. provided substantial conceptual and editorial support. J.A.C. collected and provided tissue samples from the field via the Museum of Southwestern Biology. J.A.C guided sampling efforts based on previous research and provided substantial conceptual support in manuscript preparation and revisions. C.L. led the team and provided access to computational resources.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jocelyn P. Colella, Email: jcolella.jc@gmail.com
Charlotte Lindqvist, Email: cl243@buffalo.edu.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s42003-018-0058-y.
References
- 1.Hewitt GM. The genetic legacy of the Quaternary ice ages. Nature. 2000;405:907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- 2.Hewitt GM. Genetic consequences of climatic oscillations in the Quaternary. Proc. R. Soc. Lond. Ser. B Biol. Sci. 2004;359:183–195. doi: 10.1098/rstb.2003.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Provan J, Bennett KD. Phylogeographic insights into cryptic glacial refugia. Trends Ecol. Evol. 2008;23:564–571. doi: 10.1016/j.tree.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 4.Hultén, E. Outline of the History of Arctic and Boreal Biota During the Quaternary Period: Their Evolution During and After the Glacial Period as Indicated by the Equiformal Progressive Areas of Present Plant Species (J. Cramer, Leuter Shausen, Germany, 1972).
- 5.Avise, J. C. Phylogeography: The History and Formation of Species (Harvard University Press, Cambridge, MA, 2000).
- 6.Swenson NG, Howard DJ. Clustering of contact zones, hybrid zones, and phylogeographic breaks in North America. Am. Nat. 2005;166:581–591. doi: 10.1086/491688. [DOI] [PubMed] [Google Scholar]
- 7.Heusser, C. J. in The Outer Shores, 327 (eds G. G. E. Scudder & N. Gessler, University of British Columbia, Queen Charlotte Islands Museum Press, Skidgate, British Columbia, UK, 1989).
- 8.Josenhans HW, Fedje DW, Conway KW, Barrie JV. Post glacial sea levels on the Western Canadian continental shelf: evidence for rapid change, extensive subaerial exposure, and early human habitation. Mar. Geol. 1995;125:73–94. doi: 10.1016/0025-3227(95)00024-S. [DOI] [Google Scholar]
- 9.Carrara P, Ager TA, Baichtal JF. Possible refugia in the Alexander Archipelago of southeastern Alaska during the late Wisconsin glaciation. Can. J. Earth. Sci. 2007;44:229–244. doi: 10.1139/e06-081. [DOI] [Google Scholar]
- 10.Foster, J. B. The Evolution of the Mammals of the Queen Charlotte Islands, British Columbia. B. C. Prov. Mus. Occas. Pap. No 14, 1–130 (1965).
- 11.Llamas B, et al. Ancient mitochondrial DNA provides high-resolution time scale of the peopling of the Americas. Sci. Adv. 2016;2:e1501385. doi: 10.1126/sciadv.1501385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dawson NG, Hope AG, Talbot SL, Cook JA. A multilocus evaluation of ermine (Mustela erminea) across the Holarctic, testing hypotheses of Pleistocene diversification in response to climate change. J. Biogeogr. 2014;41:464–475. doi: 10.1111/jbi.12221. [DOI] [Google Scholar]
- 13.Martinkova N, McDonald RA, Searle JB. Stoats (Mustela erminea) provide evidence of natural overland colonization of Ireland. Proc. Biol. Sci. 2007;274:1387–1393. doi: 10.1098/rspb.2007.0334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrison RG, Larson EL. Hybridization, introgression, and the nature of species boundaries. J. Hered. 2014;105:795–809. doi: 10.1093/jhered/esu033. [DOI] [PubMed] [Google Scholar]
- 15.Harrison RG. Hybrid zones: windows on evolutionary process. Oxf. Surv. Evolut. Biol. 1990;7:69–128. [Google Scholar]
- 16.Nichols R. Gene trees and species trees are not the same. Trends Ecol. Evol. 2001;16:358–364. doi: 10.1016/S0169-5347(01)02203-0. [DOI] [PubMed] [Google Scholar]
- 17.Bateson, W. in Darwin and Modern Science, 85–101 (eds A. C. Seward, Cambridge University Press, Cambridge, United Kingdom, 1909).
- 18.Dobzhansky, T. H. Genetics and the Origin of Species (Columbia University Press, New York City, NY, 1937).
- 19.Muller HJ. Isolating mechanisms, evolution, and temperature. Biol. Symp. 1942;6:71–125. [Google Scholar]
- 20.Coyne, J. A. & Orr, H. A. Speciation (Sinauer Associates, Sunderland, MA, 2004).
- 21.Burke J, Arnold ML. Genetics and the fitness of hybrids. Annu. Rev. Genet. 2001;35:31–52. doi: 10.1146/annurev.genet.35.102401.085719. [DOI] [PubMed] [Google Scholar]
- 22.Burton RS, Barreto FS. A disproportionate role for mtDNA and Dobzhansky-Muller incompatibilities? Mol. Ecol. 2012;21:4942–4957. doi: 10.1111/mec.12006. [DOI] [PubMed] [Google Scholar]
- 23.Arnold, M. L. Evolution through Genetic Exchange (Oxford University Press, Oxford, United Kingdom, 2007).
- 24.Folk, R. A., Soltis, P. S., Soltis, D. E. & Guralnick, R. New prospects in the detection and comparative analysis of hybridization in the tree of life. Am. J. Bot. 105, 1–12 (2018). [DOI] [PubMed]
- 25.Miller W, et al. Polar and brown bear genomes reveal ancient admixture and demographic footprints of past climate change. Proc. Natl. Acad. Sci. 2012;109:E2382–E2390. doi: 10.1073/pnas.1210506109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edwards SV, Potter S, Schmitt CJ, Bragg JG, Moritz C. Reticulation, divergence, and the phylogeography-phylogenetics continuum. Proc. Natl. Acad. Sci. USA. 2016;113:8025–8032. doi: 10.1073/pnas.1601066113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lazaridis I, et al. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature. 2014;513:409–413. doi: 10.1038/nature13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feder JL, Egan SP, Nosil P. The genomics of speciation-with-gene-flow. Trends Genet. 2012;28:342–350. doi: 10.1016/j.tig.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 29.Sullivan J, et al. Divergence with gene flow within the recent chipmunk radiation (Tamias) Heredity. 2014;113:185–194. doi: 10.1038/hdy.2014.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haig, S. M. & Allendorf, F. W. in The Endangered Species Act at Thirty, Vol. 2: Conserving Biodiversity in Human-Dominated Landscapes Vol. 708, 150–163 (eds Scott, J. M., Goble, D. D. & Davis, F. W.) (Island Press, Washington, 2006).
- 31.Wright S. The genetical structure of populations. Ann. Hum. Genet. 1949;15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- 32.Manichaikul A, et al. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stone KD, Flynn RW, Cook JA. Post-glacial colonization of northwestern North America by the forest-associated American marten (Martes americana, Mammalia: Carnivora: Mustelidae) Mol. Ecol. 2002;11:2049–2063. doi: 10.1046/j.1365-294X.2002.01596.x. [DOI] [PubMed] [Google Scholar]
- 35.Wooding S, Ward R. Phylogeography and Pleistocene evolution in the North American black bear. Mol. Biol. Evol. 1997;14:1096–1105. doi: 10.1093/oxfordjournals.molbev.a025719. [DOI] [PubMed] [Google Scholar]
- 36.Rull V. Microrefugia. J. Biogeogr. 2009;36:481–484. doi: 10.1111/j.1365-2699.2008.02023.x. [DOI] [Google Scholar]
- 37.Sawyer YE, Cook JA. Phylogeographic structure in long-tailed voles (Rodentia: Arvicolinae) belies the complex Pleistocene history of isolation, divergence, and recolonization of Northwest North America’s fauna. Ecol. Evol. 2016;6:6633–6647. doi: 10.1002/ece3.2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fedorov VB, Stenseth NC. Multiple glacial refugia in the North American Arctic: inference from phylogeography of the collared lemming (Dicrostonyx groenlandicus) Proc. R. Soc. Lond. Ser. B Biol. Sci. 2002;269:2071–2077. doi: 10.1098/rspb.2002.2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baker RJ, Bradley RD. Speciation in mammals and the genetic species concept. J. Mammal. 2006;87:643–662. doi: 10.1644/06-MAMM-F-038R2.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature. 2009;461:489–494. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patterson N, et al. Ancient admixture in human history. Genetics. 2012;192:1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peter BM. Admixture, population structure, F-statistics. Genetics. 2016;202:1485–1501. doi: 10.1534/genetics.115.183913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Green RE, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Durand EY, Patterson N, Reich D, Slatkin M. Testing for ancient admixture between closely related populations. Mol. Biol. Evol. 2011;28:2239–2252. doi: 10.1093/molbev/msr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin SH, Davey JW, Jiggins CD. Evaluating the use of ABBA-BABA statistics to locate introgressed loci. Mol. Biol. Evol. 2015;32:244–257. doi: 10.1093/molbev/msu269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mailund T, Leppälä K, Nielsen S. AdmixtureGraph: admixture graph manipulation and fitting. Bioinformatics. 2017;33:1738–1740. doi: 10.1093/bioinformatics/btx048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Orr HA. Haldane’s Rule. Annu. Rev. Ecol. Syst. 1997;28:195–218. doi: 10.1146/annurev.ecolsys.28.1.195. [DOI] [Google Scholar]
- 48.Brandvain Y, Pauly GB, May MR, Turelli M. Explaining Darwin’s corollary to Haldane’s Rule: the role of mitonuclear interactions in asymmetric postzygotic isolation among toads. Genetics. 2014;197:743–747. doi: 10.1534/genetics.113.161133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dyke, A. S., Moore, A. & Robertson, L. Deglaciation of North America, Ottawa Open File 1574 (Natural Resources Canada, 2003).
- 50.King CM, McMillan CD. Population structure and dispersal of peak-year cohorts of stoats (Mustela erminea) in two New Zealand forests, with especial reference to control. NZ J. Ecol. 1982;5:59–66. [Google Scholar]
- 51.Lipson M, et al. Efficient moment-based inference of admixture parameters and sources of gene-flow. Mol. Biol. Evol. 2013;30:1788–1802. doi: 10.1093/molbev/mst099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Puckett EE, Etter PD, Johnson EA, Eggert LS. Phylogeographic analyses of American black bears (Ursus americanus) suggest four glacial refugia and complex patterns of postglacial admixture. Mol. Biol. Evol. 2015;32:2338–2350. doi: 10.1093/molbev/msv114. [DOI] [PubMed] [Google Scholar]
- 53.De Jong J. Climate variability during the last three million years, as indicated by vegetational evolution in northwest Europe and with emphasis on data from The Netherlands. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1988;318:603–617. doi: 10.1098/rstb.1988.0025. [DOI] [Google Scholar]
- 54.Gibbard PL, et al. Early and middle Pleistocene correlations in the southern North Sea basin. Quat. Sci. Rev. 1991;10:23–52. doi: 10.1016/0277-3791(91)90029-T. [DOI] [Google Scholar]
- 55.Carrara, P., Ager, T. A., Baichtal, J. F. & VanSistine, D. P. Map of glacial limits and possible refugia in the southern Alexander Archipelago, Alaska during the Late Wisconsin Glaciation. U.S. Geological Survey, Miscellaneous Field Studies Map MP-2424, 1–13 (2003).
- 56.COSEWIC. Assessment and status report on the Ermine haidarum subspecies Mustela erminea haidarum in Canada (Committee on the Status of Endangered Wildlife in Canada, 2009).
- 57.Heaton TH, Talbot SL, Shields GF. An ice age refugium for large mammals in the Alexander Archipelago, Southeastern Alaska. Quat. Res. 1996;46:186–192. doi: 10.1006/qres.1996.0058. [DOI] [Google Scholar]
- 58.Li H, Durbin R. Inference of human population history from whole genome sequence of a single individual. Nature. 2011;475:493–496. doi: 10.1038/nature10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cahill JA, Soares AER, Green RE, Shapiro B. Inferring species divergence times using pairwise sequential Markovian coalescent modelling and low-coverage genomic dataPlease provide journal name, volume and page number in ref. 60. Journal Name: PLoS GenetVolume (Issue): 10(1)Page number: e1004016. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2016;371:20150138. doi: 10.1098/rstb.2015.0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Freedman, A. H. et al. Genome sequencing highlights the dynamic early history of dogs. PLoS Genet.10, e1004016. [DOI] [PMC free article] [PubMed]
- 61.Kim, S. et al. Comparison of carnivore, omnivore, and herbivore mammalian genomes with a new leopard assembly. Genome. Biol. 17, 211 (2016). [DOI] [PMC free article] [PubMed]
- 62.Fitzpatrick BM, Fordyce J, Gavrilets S. What, if anything, is sympatric speciation? J. Evol. Biol. 2008;21:1452–1459. doi: 10.1111/j.1420-9101.2008.01611.x. [DOI] [PubMed] [Google Scholar]
- 63.Rhymer JM, Simberloff D. Extinction by hybridization and introgression. Annu. Rev. Ecol. Syst. 1996;27:83–109. doi: 10.1146/annurev.ecolsys.27.1.83. [DOI] [Google Scholar]
- 64.Rosenblum EB, et al. Goldilocks meets Santa Rosalia: an ephemeral speciation model explains patterns of diversification across time scales. Evol. Biol. 2012;39:255–261. doi: 10.1007/s11692-012-9171-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gavrilets, S. Fitness Landscapes and the Origin of Species (Princeton University Press, Princeton, NJ, 2004).
- 66.Barton NH. Fitness landscapes and the origin of species. Evolution. 2005;59:246–248. [Google Scholar]
- 67.Schumer M, Rosenthal GG, Andolfatto P. How common is homoploid hybrid speciation? Evolution. 2014;68:1553–1560. doi: 10.1111/evo.12399. [DOI] [PubMed] [Google Scholar]
- 68.Meier JI, et al. Ancient hybridization fuels rapid cichlid fish adaptive radiations. Nat. Commun. 2017;8:14363. doi: 10.1038/ncomms14363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Serevdio MR. 2004. The what and why of research on reinforcement. PLoS Biol. 2004;2:e420. doi: 10.1371/journal.pbio.0020420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Turelli M, Moyle LC. Asymmetric postmating isolation: Darwin’s corollary to Haldane’s rule. Genetics. 2007;176:1059–1088. doi: 10.1534/genetics.106.065979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yukilevich R. Asymmetrical patterns of speciation uniquely support reinforcement in Drosophila. Evolution. 2012;66:1430–1446. doi: 10.1111/j.1558-5646.2011.01534.x. [DOI] [PubMed] [Google Scholar]
- 72.MacArthur, R. H. & Wilson, E. O. The Theory of Island Biogeography (Princeton University Press, Princeton, NJ, 1967).
- 73.Preece, R. C. Island Britain: A Quaternary perspective. Geological Society Special Publication No. 96, 1995.
- 74.Lamichhaney S, et al. Evolution of Darwin’s finches and their beaks revealed by genome sequencing. Nature. 2015;518:371–375. doi: 10.1038/nature14181. [DOI] [PubMed] [Google Scholar]
- 75.Wayne RK, Shaffer HB. Hybridization and endangered species protection in the molecular era. Mol. Ecol. 2016;25:2680–2589. doi: 10.1111/mec.13642. [DOI] [PubMed] [Google Scholar]
- 76.Fleming MA, Cook JA. Phylogeography of endemic ermine (Mustela erminea) in southeast Alaska. Mol. Ecol. 2002;11:795–897. doi: 10.1046/j.1365-294X.2002.01472.x. [DOI] [PubMed] [Google Scholar]
- 77.Hall, E. R. American Weasels (University of Kansas Publications, Museum of Natural History, Lawrence, KS, 1952).
- 78.Hewitt GM. Quaternary phylogeography: the roots of hybrid zones. Genetica. 2011;139:617–638. doi: 10.1007/s10709-011-9547-3. [DOI] [PubMed] [Google Scholar]
- 79.Lan, T. et al. Genome-wide evidence for a hybrid origin of modern polar bears. BioRxiv 047498. Preprint at https://www.biorxiv.org/content/early/2016/04/06/047498 (2016).
- 80.Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc (2010).
- 81.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Peng X, et al. The draft genome sequence of the ferret (Mustela putorius furo) facilitates study of human respiratory disease. Nat. Biotechnol. 2014;32:1250–1255. doi: 10.1038/nbt.3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McKenna A, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genom. Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Danecek P, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Purcell S, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Guindon S, Gascuel O. A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 90.Stamatakis A. RAxML Version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bouckaert R, et al. BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2014;10:e1003537. doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Leaché, A. D., Banbury, B. L., Felsenstein, J. & Stamatakis, A. Short tree, long tree, right tree, wrong tree: new acquisition bias corrections for inferring SNP phylogenies. Syst. Biol. 64, 1032–1047 (2015). [DOI] [PMC free article] [PubMed]
- 93.King, C. M. Mustela erminea. Mammalian Species No. 195, 1–8 (1983).
- 94.Harding KE, Smith FA. Mustela or vison? Evidence for the taxonomic status of the American mink and a distinct biogeographic radiation of American weasels. Mol. Phylogenet. Evol. 2009;52:632–634. doi: 10.1016/j.ympev.2009.05.036. [DOI] [PubMed] [Google Scholar]
- 95.Pickrell JK, Pritchard JK. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012;8:e1992967. doi: 10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Koepfli KP, et al. Multigene phylogeny of the mustelidae: resolving relationships, tempo, and biogeographic history of a mammalian adaptive radiation. BMC Biol. 2008;6:10. doi: 10.1186/1741-7007-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zheng X, et al. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics. 2012;28:3326–3328. doi: 10.1093/bioinformatics/bts606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kumar S, Subramanian S. Mutation rates in mammalian genomes. Proc. Natl. Acad. Sci. USA. 2002;99:803–808. doi: 10.1073/pnas.022629899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nadachowska-Brzyska K, Burri R, Smeds L, Ellegren H. PSMC analysis of effective population sizes in molecular ecology and its application to black-and-white Ficedula flycatchers. Mol. Ecol. 2016;25:1058–1072. doi: 10.1111/mec.13540. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genomic reads are available in NCBI’s Sequence Read Archive, accession: SRP138989. Assembled FASTA files are available on Dryad (10.5061/dryad.bv2g720).