Abstract
De novo membrane protein structure determination is often limited by the availability of large crystals and the difficulties in obtaining accurate diffraction data for experimental phasing. Here we present a method that combines in situ serial crystallography with de novo phasing for fast, efficient membrane protein structure determination. The method enables systematic diffraction screening and rapid data collection from hundreds of microcrystals in in meso crystallization wells without the need for direct crystal harvesting. The requisite data quality for experimental phasing is achieved by accumulating diffraction signals from isomorphous crystals identified post-data collection. The method works in all experimental phasing scenarios and is particularly attractive with fragile, weakly diffracting microcrystals. The automated serial data collection approach can be readily adopted at most microfocus macromolecular crystallography beamlines.
Huang et al. report a fully automated method for collecting and merging data from thousands of in meso-grown microcrystals. This in situ approach provides a fast and efficient way to determine the structures of membrane proteins from fragile microcrystals.
Introduction
Membrane proteins perform essential roles in signal and energy transduction, metabolism, and transport and contribute to the structural integrity of cells. They account for close to a third of all cellular proteins and are important drug targets. High-resolution three-dimensional structural information is key to understanding how membrane protein work at a molecular level and can be used to inform structure-based drug design and discovery1,2. The vast majority of membrane protein structure work has been performed using macromolecular crystallography. Recent advances in cryogenic electron microscopy (cryo-EM) and de novo prediction methods will undoubtedly contribute to providing much-needed new structures3,4. However, many membrane proteins are still too small to be imaged at high resolution by cryo-EM and de novo prediction methods, as yet, do not provide atomic-level resolution. Macromolecular crystallography is limited too in that membrane protein crystals of high diffraction quality are difficult to generate. The lipid cubic phase (LCP) or in meso method of crystallization has made important contributions in this regard, in part, because proteins crystallize from within and into a native membrane-like environment5. In recent years, the in meso method accounts for close to 40% of all new unique membrane protein crystal structures (Supplementary Fig. 1).
Another challenge associated with membrane protein structure determination is that most new and interesting targets have novel structures with which the most common phasing method, molecular replacement, is rarely useful. In such cases, de novo structure determination by experimental phasing is required6. Of the 738 published unique membrane protein structures as of May 2018 (http://blanco.biomol.uci.edu/mpstruc/), 617 were solved by crystallography, of which 46% used experimental phasing with single-wavelength anomalous diffraction (SAD) the most popular de novo experimental phasing method (Supplementary Fig. 1). Experimental phasing comes with its own challenges. It requires highly accurate diffraction data, which are difficult to acquire especially when using small, fragile crystals of the type commonly encountered with membrane proteins. Heavy atom experimental phasing of membrane proteins with serial crystallography approaches has been demonstrated using crystals harvested with micro-meshes or standard loops7–9. But harvesting of the large numbers of crystals required for serial data collection is very time-consuming and particularly challenging for crystals grown in meso. This background serves to highlight the requirement for efficient and robust de novo phasing methods for use with membrane protein microcrystals.
Responding to this critical need, we introduce here a fully automated method for collecting, selecting, and merging data from hundreds to thousands of in meso-grown microcrystals. This approach boosts the collective phasing signal of many tiny crystals to a level where de novo phasing of highly challenging membrane protein targets becomes not only possible but also fast and high-throughput. We refer to it as the in meso in situ serial crystallography—experimental phasing (IMISX-EP) method. IMISX-EP has three components: (1) in meso crystallization (and in situ soaking where appropriate), (2) fast, automated grid scanning, and serial data collection in situ, and (3) real-time data processing and selection of data sets followed by structure solution (Fig. 1). IMISX-EP eliminates one of the major bottlenecks subsequent to in meso crystallization—direct crystal harvesting, and enables in situ diffraction data collection by combining in meso crystallization and in situ serial synchrotron crystallography10,11 at cryogenic temperatures. In addition, the method provides convenient and highly effective in situ soaking capabilities (Fig. 2). We demonstrate the utility of the method with four real-life integral membrane proteins using all major anomalous phasing methods, and show that IMISX-EP provides a fast, efficient, and a direct means for de novo structure determination of membrane protein as microcrystals.
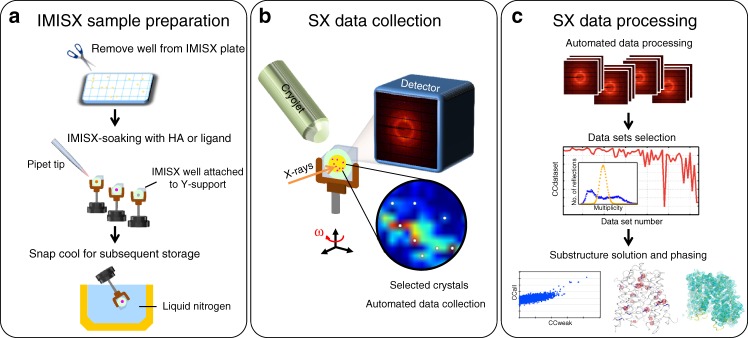
Fig. 1.
Overview of the IMISX-EP workflow for high-throughput in situ de novo phasing of in meso-grown microcrystals of membrane proteins. a IMISX crystallization, well preparation, and post-crystallization treatment, snap-cooling, and storage. b Automated grid scanning and serial data collection on hundreds of microcrystals in situ with a fast frame-rate detector. c Selection of isomorphous data sets based on intensity correlation coefficients (red line), analysis of crystal orientations (inset), and phase determination
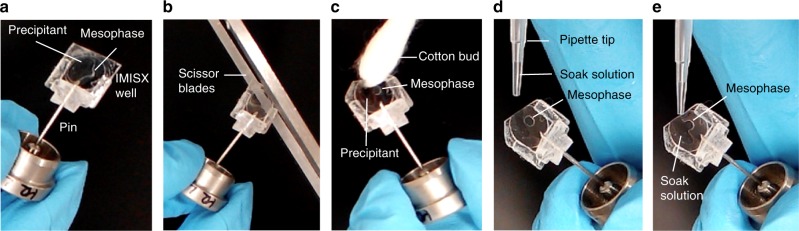
Fig. 2.
Steps involved in the heavy atom soaking of crystals growing in the lipid cubic mesophase inside IMISX wells for use in IMISX-EP. a, b Photographic images of the process of cutting open an IMISX well with a scissor, c wicking away excess precipitant solution with a cotton bud from around the crystal-laden mesophase, and d, e adding heavy atom containing precipitant solution by pipette
Results
The first demonstration of IMISX-EP was carried out using the most popular experimental phasing method, SAD phasing, with the Se-Met labeled proteins, PepTSt and LspA. Instead of harvesting mesophase-containing individual or clusters of crystals from each well by the traditional loop-harvesting method, the entire IMISX well was removed from the plate, mounted on a pin and snap-cooled in liquid nitrogen (Methods, Supplementary Fig. 2). This enabled in situ X-ray diffraction data collection to be carried out subsequently on all crystals in the IMISX well. From two such wells, 210 PepTSt crystals were measured, 145 partial data sets were indexed and processed, and 89 were selected on the basis of diffraction intensity correlation coefficient (CCdataset) (Supplementary Fig. 3a, Table 1 and Table 2) for successful Se-SAD phasing with SHELXD/E12 (Fig. 3a, Table 2, Supplementary Figs. 4a, 5a). The second example of Se-SAD phasing proved to be a particularly challenging case study for serial crystallography because the target protein, LspA, was incompletely Se-Met labelled. The average Se-Met incorporation was estimated at 59% based on the refined Se anomalous scattering signal (Methods). In this instance, the high-throughput feature of the IMISX-EP method proved invaluable enabling diffraction data to be collected from 974 LspA crystals in 32 IMISX wells (Table 1 and Table 2). The final data set derived from 497 crystals (Supplementary Fig. 3b). The substructure was determined by SHELXD and an interpretable map was generated by CRANK213 (Fig. 3b, Table 2, Supplementary Figs. 4b, 5b).
Table 1.
Data collection and refinement statistics*
| Se-PepTSt (6FMR) | Se/S-LspA (6FMS) | Hg-BacA soaking SAD (6FMT) | Hg-BacA soaking SIRAS | BacA Native SIRAS (6FMV) | Hg-BacA Co-crystallization (6FMW) | W-PgpB (6FMX) | S-PepTSt (6FMY) | |
|---|---|---|---|---|---|---|---|---|
| Data collection | ||||||||
| Space group | C2221 | C2 | C222 | C222 | C222 | C222 | I222 | C2221 |
| Cell dimensions | ||||||||
| a, b, c (Å) | 103.7, 110.9, 110.4 | 112.8, 110.1, 86.0 | 112.6, 145.6, 40.3 | 112.6, 145.6, 40.3 | 113.4, 144.4, 40.4 | 114.1, 145.1, 40.0 | 68.8, 76.8, 98.9 | 101.2, 110.2, 111.5 |
| α, β, γ (°) | 90, 90, 90 | 90, 97.1, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Wavelength (Å) | 0.97854 | 0.97858 | 1.9 | 1.9 | 1.0 | 1.9 | 1.21371 | 2.0664 |
| Resolution (Å) | 49.54–2.70 (2.77–2.70) | 46.27–3.00 (3.08–3.00) | 44.58–3.00 (3.08–3.00) | 44.56–3.00 (3.08–3.00) | 40.39–2.30 (2.36–2.30) | 44.45–2.60 (2.67–2.60) | 35.6–1.79 (1.90–1.79) | 49.29–2.70 (2.77–2.70) |
| R meas | 0.44 (3.69) | 0.32 (3.46) | 0.48 (3.93) | 0.38 (2.86) | 0.50 (6.39) | 0.38 (2.00) | 0.10 (1.32) | 0.32 (1.69) |
| I /σ (I) | 8.90 (1.74) | 9.14 (1.45) | 10.62 (1.97) | 10.34 (1.98) | 9.85 (1.56) | 8.95 (1.96) | 7.93 (0.91) | 40.2 (8.8) |
| CC1/2 (%) | 99.4 (45.3) | 99.6 (40.9) | 99.7 (69.9) | 99.8 (72.8) | 99.5 (53.5) | 99.4 (32.5) | 99.7 (50.6) | 100 (96.0) |
| Completeness (%) | 100 (100) | 99.9 (99.8) | 100 (100) | 100 (100) | 100 (100) | 99.9 (99.5) | 91.7 (90.0) | 100 (100) |
| Multiplicity | 25.4 (23.8) | 22.7 (22.7) | 65.4 (60.7) | 49.9 (47.2) | 16.95 (14.09) | 20.7 (15.7) | 2.60 (2.50) | 423.6 (378.1) |
| Refinement** | ||||||||
| Resolution (Å) | 49.55–2.70 | 46.27–3.00 | 44.56–3.00 | – | 40.39–2.30 | 44.45–2.60 | 38.40–1.79 | 49.29–2.70 |
| No. of unique reflections | 33,746/1655 | 41,032/2058 | 12,841/641 | – | 15,224/761 | 19,671/975 | 39,988/2006 | 33,070/1661 |
| Rwork/Rfree | 0.23/0.25 | 0.23/0.27 | 0.24/0.28 | – | 0.19/0.22 | 0.22/0.25 | 0.24/0.26 | 0.21/0.25 |
| No. of atoms | ||||||||
| Protein | 3,649 | 4,990 | 1,896 | 2,081 | 2,061 | 1,587 | 3,448 | |
| Ligand/ion | 244 | 466 | 89 | – | 197 | 57 | 129 | 320 |
| Water | 37 | 10 | 4 | – | 65 | 6 | 93 | 10 |
| B-factor | ||||||||
| Proteins | 53.7 | 80.4 | 60.4 | – | 38.9 | 51.5 | 33.3 | 47.4 |
| Ligand/ion | 62.6 | 87.4 | 69.1 | – | 58.4 | 60.3 | 51.5 | 61.3 |
| Water | 50.2 | 73.9 | 60.7 | – | 42.4 | 44.3 | 38.7 | 46.1 |
| R.m.s. deviations | ||||||||
| Bond lengths (Å) | 0.002 | 0.003 | 0.003 | – | 0.003 | 0.003 | 0.005 | 0.003 |
| Bond angles (°) | 0.443 | 0.525 | 0.528 | – | 0.611 | 0.640 | 0.822 | 0.705 |
*Data processing statistics are reported with Friedel pairs separated. Values in parentheses are for the highest resolution shell
**Refinement statistics are reported with Friedel pairs separated for all cases except BacA native SIRAS
Table 2.
Sample consumption and phasing statistics
| Se-PepTSt (6FMR) | Se/S-LspA (6FMS) | Hg-BacA soaking SAD (6FMT) | Hg-BacA soaking SIRAS | BacA Native SIRAS (6FMV) | Hg-BacA Co-crystallization (6FMW) | W-PgpB (6FMX) | S-PepTSt (6FMY) | |
|---|---|---|---|---|---|---|---|---|
| Sample consumption | ||||||||
| Heavy atom labeling | Se-Met | Se/S-Met | Hg-soaking | Hg-soaking | None | Hg-co-crystallization | W-co-crystallization | Native sulfur |
| Protein consumption (μg) | 0.5 | 7.7 | 1.9 | 1.9 | 1.2 | 0.48 | 0.3 | 14.8 |
| No. of wells | 2 | 32 | 8 | 8 | 5 | 2 | 1 | 59 |
| No. of crystals | 210 | 974 | 968 | 968 | 125 | 66 | 1 | 6,639 |
| No. of processed data sets | 145 | 614 | 742 | 742 | 110 | 64 | 1 | 4,528 |
| No. of merged data sets | 89 | 497 | 360 | 271 | 94 | 55 | 1 | 1,595 |
| Selection rate (%) | 42 | 51 | 37 | 28 | 75 | 83 | 100 | 24 |
| Total data (°) | 1,335 | 4,970 | 3,600 | 2,710 | 940 | 1,170 | 140 | 15,950 |
| Phasing | ||||||||
| Phasing method | Se-SAD | Se-SAD | Hg-SAD | Hg-SIRAS | Hg-SIRAS | Hg-SAD | W-SAD | S-SAD |
| SHELXD resolution range (Å) | 49.54–3.20 | 44.35–4.20 | 44.58–3.30 | 44.56–3.30 | – | 44.45–4.00 | 35.60–1.80 | 49.29–3.50 |
| SHELXD CCall/CCweak (%) | 44.8/21.6 | 41.5/16.5 | 29.4/17.1 | 23.3/16.8 | – | 38.5/24.0 | 39.1/20.1 | 31.0/12.6 |
| Heavy atom sites | 18 Se | 12 Se | 1 Hg | 1 Hg | – | 2 Hg | 1 W | 13 S |
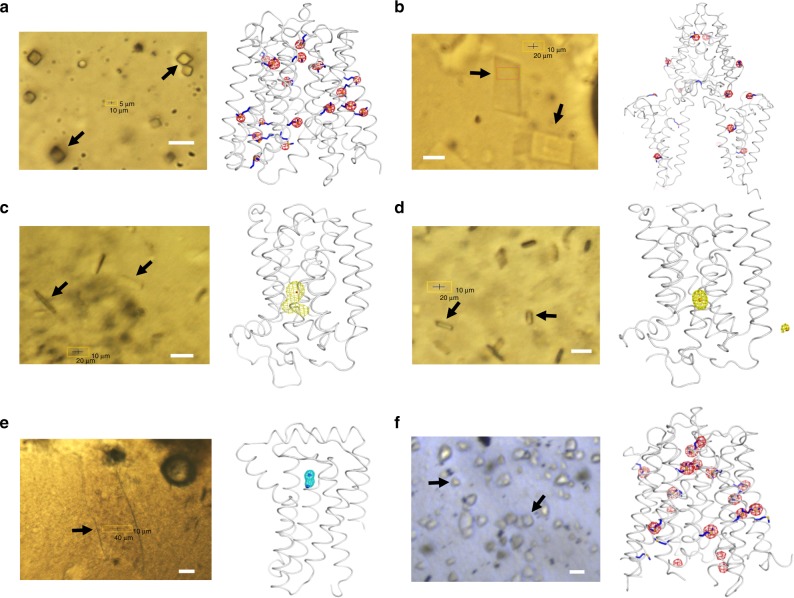
Fig. 3.
Structures of membrane proteins solved by IMISX-EP and the crystals used for data collection. Photographic images of crystals in IMISX wells held in the cryo-stream were recorded with an in-line microscope at beamline X06SA-PXI. a Se-PepTSt, 2.7 Å, 89 crystals, two wells, Se-SAD, 18 Se. b Se/S-LspA, 3.0 Å, 497 crystals, 32 wells, Se-SAD, 12 Se. c Hg-BacA IMISX-soaking, 3.0 Å, 360 crystals, eight wells, Hg-SAD, 1 Hg. d Hg-BacA co-crystallization, 2.6 Å, 55 crystals, two wells, Hg-SAD, 2 Hg. e W-PgpB, 1.8 Å, one crystal, one well, W-SAD, 1 W. f S-PepTSt, 2.7 Å, 1595 crystals, 59 wells, S-SAD, 13 S. Structures are shown in ribbon representation with anomalous sub-structures depicted as spheres and SHELXE anomalous Fourier maps contoured at 5 σ. The SHELXE anomalous Fourier map was calculated using the observed anomalous difference (|FA|) and the phases of the sub-structure determined using SHELXD. The black arrows point to representative crystals in each well. The white scale bar corresponds to 20 μm
When Se-Met labeling compromises expression and/or protein and crystal quality, traditional heavy atom derivatization is an alternative and proven approach for experimental phasing. The heavy atom can be introduced either by co-crystallizing it with the protein or by soaking it into the crystal. We demonstrated the IMISX-EP approach for both heavy atom derivatization methods with two membrane proteins, BacA and PgpB. Indeed, BacA is the first de novo structure solved by the IMISX-EP method14. BacA crystals were derivatized by in situ soaking with HgCl2 in IMISX wells directly (Fig. 2). These wells provide an ideal environment in which to optimize heavy atom concentration and soaking time. Further, labeling benefits from the sticky, viscous, and nanoporous nature of the mesophase in which the crystals are suspended during soaking and buffer exchange (Methods, Fig. 2). From the 968 BacA crystals identified, two groups were created with which to evaluate two phasing methods (Supplementary Fig. 3c and 3d, Table 1 and Table 2). One contained 360, the other 271 data sets. The first group was used successfully for SAD phasing with SHELXD and CRANK2 (Fig. 3c, Supplementary Figs. 4c, 5c). The smaller, second group was combined with a high-resolution native data set for phasing with single isomorphous replacement with anomalous scattering (SIRAS), and the structure was phased readily with SHELXD/E (Table 2, Supplementary Figs. 4d, 5d). This example illustrates convincingly how SIRAS with a relatively weak heavy atom data set can be combined with a higher resolution native data set for experimental phasing in serial crystallography.
IMISX-EP using heavy atom co-crystallization was demonstrated with BacA and PgpB. BacA, was labeled with HgCl2 in solution ahead of setting up trials in IMISX crystallization plates. Of the 66 crystals measured from two IMISX wells, 64 were indexed and integrated, and 55 were selected (Supplementary Fig. 3e, Table 1 and Table 2). Mercury sites were identified with SHELXD, and an interpretable map was obtained by employing 25 cycles of SHELXE density modification with autobuilding (Fig. 3d, Table 2, Supplementary Figs. 4e, 5e). PgpB was pre-incubated with Na2WO4 and subsequently crystallized in the cubic mesophase. The crystal used for data collection was relatively large and measured 10 × 50 × 150 μm3. As the crystal had its flat face oriented approximately parallel to the windows of the well (Fig. 3e), it was possible to record 140° of data using a rotation angle of ± 70° from this one crystal without compromising data quality at high-tilt angles (Supplementary Fig. 6). Because of the strong anomalous signal from tungsten and the high diffraction resolution, the structure was solved with the relatively low redundancy of 2.6 and a completeness of 91% with SHELXD/E (Fig. 3e, Supplementary Figs. 4f, 5f). This example shows that the IMISX-EP approach can be used with large single crystals.
To demonstrate the full potential of IMISX-EP, the most-challenging experimental phasing method, native-SAD using the anomalous signal from light elements (Z ≤ 20) only, was attempted with unlabeled PepTSt crystals. The estimated Bijvoet ratio (ΔF/F) of sulfur (S)-PepTSt is ~ 1/100 (1%) at 6 keV (2.067 Å), which is approximately one-fourth that of Se-PepTSt at the Se absorption edge. Given the success of Se-SAD phasing using 89 Se-PepTSt crystals as described above, one expects that thousands of PepTSt microcrystals would be needed for native-SAD phasing. To expedite data collection on thousands of microcrystals, a high-throughput serial data collection protocol was implemented. It combined 50–100 Hz grid scanning of the entire 1–2 mm diameter LCP bolus with a micrometer-sized X-ray beam and automated data collection from each crystal identified on the basis of the grid scan (Methods, Supplementary Fig. 7). A total of 98,050° of data were recorded from 6639 crystals in 59 IMISX wells using 27 h of beamtime. Of these, 4528 crystals were indexed and processed and the final data derived from 1595 crystals included 15,950° of data (Supplementary Fig. 3f, g, Table 1 and Table 2). SHELXD located 13 of the 18 internal S sites in PepTSt and SHELXE produced an interpretable map (Fig. 3f, Table 2, Supplementary Figs. 4g, 5g).
Discussion
It is clear from the case studies presented that IMISX-EP is a powerful and generally applicable high-throughput method. It combines the advantages of conventional crystallography with those of serial crystallography7–11,15–19 and transforms traditional micro-crystallography20 to an in situ serial crystallography method that is considerably more efficient and that requires miniscule quantities of protein. Specifically, in the most-challenging native-SAD phasing example above, < 20 μg PepTSt was used. Not only does the method extend phasing to microcrystals while minimizing the detrimental effects of radiation damage, it also effectively reduces both random and systematic errors in diffraction data to such an extent that the weak anomalous signal from light elements can be extracted reliably. At first blush, serial crystallography may appear to differ dramatically from conventional crystallography. In fact, the two are similar in that both seek to accumulate sufficient diffraction signal; by exploiting the voluminous diffraction of a large single crystal in conventional crystallography and by combining the diffraction volume of many microcrystals in serial crystallography21,22. Therefore, IMISX-EP very effectively improves the success rate of the phasing experiment by simple signal accumulation using a highly efficient serial crystallography method. It represents a paradigm shift from the conventional, subjective selection of crystals based on appearance, ease of harvesting, available harvesting time, and storage space to an exhaustive measurement of all crystals to the limits of radiation damage from which accurate data can be gleaned based upon a carefully chosen subset of isomorphous crystals.
In this study, the experimental phasing examples used for purposes of demonstration ranged from simple and easy to perform to extremely challenging. The strength of the phasing signal, as estimated by the Bijvoet ratio, ranges from 1% in the case of native-SAD PepTSt to 7% for tungsten phasing of PgpB. The observable anomalous peak height, which determines the success of a SAD experiment, is proportional to the square root of the number of observed reflections23, which in turn is related directly to the diffraction resolution. Therefore, experimental phasing becomes considerably more difficult in going from working with the high-resolution data available for heavy atom-labeled W-PgpB to the mid-resolution data available for native S-PepTSt. This is clearly reflected in the total number of degrees of data used for successful phasing in six cases and is congruent with the results of comparable phasing studies in the literature, which include soluble proteins7,9,11,17 (Supplementary Fig. 8). The weaker the phasing signal, the more critical becomes the process of selecting out and combining isomorphous data sets. Different criteria and clustering methods have been tested and implemented recently24–30. For native-SAD PepTSt data that have a weak signal and a large number of data sets, an iterative procedure based on CCdataset proved most effective. It rejects non-isomorphous data sets, whilst retaining weak but isomorphous data sets, which contribute to providing the needed anomalous signal.
The IMISX method takes advantage of the intrinsic viscosity of the cubic mesophase in which crystals grow. Thus, the entire crystal-laden bolus is contained in a confined space within the IMISX well, which can be cryogenically cooled for safe storage and to extend crystal lifetime in the X-ray beam. Another advantage is the ease with which heavy atom and ligand-soaking experiments can be performed in true in situ fashion without ever touching the crystals, which remain suspended in the viscous mesophase. Wider use of this convenient soaking feature will greatly facilitate mechanistic studies and drug discovery with membrane protein targets.
IMISX-EP has wide ranging applicability. It can be used with all experimental phasing approaches for membrane protein structure determination extending from traditional heavy atom derivatization and Se-Met labeling to the more recent native-SAD6,21,31 and emerging methods that include iodo-detergent labeling32 and fast halide soaking9. IMISX-EP is compatible with different plate and sample holder types19,33–35 and can be performed at most modern synchrotron macromolecular crystallography beamlines. It is likely to prove important in the effective utilization of next generation synchrotron sources such as the Diffraction Limited Storage Ring36. Furthermore, it is anticipated that the entire IMISX well mounting, crystal-laden bolus centering, rastering, and serial data collection process will be carried out unattended once the initial data collection parameters are known. Taken together, IMISX-EP, and serial crystallography in general, will contribute to having membrane protein structures included in the protein structure knowledge base to a level that reflects the frequency with which they are found in the cell. Given the importance of membrane proteins as therapeutic targets, these methods, in turn, will greatly facilitate high-throughput structure-based drug design and discovery.
Methods
Protein purification
Four proteins were used in this study as follows: the peptide transporter, PepTSt, from Streptococcus thermophilus37, the lipoprotein signal peptidase II, LspA, from Pseudomonas aeruginosa (PAO1)38, the undecaprenyl-pyrophosphate phosphatase, BacA, from Escherichia coli (K-12)39, and the phosphatidic acid phosphatase, PgpB, from Bacillus subtilis40. Se-LspA, PgpB, BacA, and native S-PepTSt were produced recombinantly and purified from biomass following published protocols37–40. Se-PepTSt was produced by using E. coli C43 (DE3) (NEB) cells with the pWaldo-dtpT plasmid. Cells were grown in SelenoMet™ Medium (Molecular Dimensions) supplemented with 50 mg/L Se-Met (Sigma) and 50 mg/L kanamycin (Melford) following supplier’s instructions. Se-PepTSt was purified by following a published protocol37. All protein samples were stored at − 80 °C.
Crystallization
Crystals were grown from purified concentrated proteins by the LCP method41 in IMISX plates10. Complexes with globomycin (Sigma), cefadroxil (Sigma), and Ala-Phe (H-Ala-Phe-OH) (BACHEM) were used in the crystallization of Se-LspA, Se-PepTSt, and native S-PepTSt, respectively. Co-crystallization of Hg-derivatized BacA and W-derivatized PgpB was performed by pre-incubating protein solution at 4 °C with 2 mM HgCl2 (Hampton) for 10 min and 100 mM Na2WO4 (Hampton) for 30 min, respectively, before setting up crystallization trials. For Se-Met substituted LspA, Hg-derivatized BacA and W-derivatized PgpB, the mesophase was produced by homogenizing two volumes of protein solution at ~ 12 mg/mL with three volumes of monoolein (9.9 monoacylglycerol, MAG) (NuCheck) in a LCP coupled syringe mixing device at 20 °C42. A similar protocol was used with PepTSt with the following modifications. The hosting lipid, 7.8 MAG (Avanti), was used for native S-PepTSt and Se-PepTSt using equal volumes of lipid and protein solution to make the mesophase, and the concentration of the protein solution was 10 mg/mL. Crystallization trials were set up robotically at 20 °C using 50 nL protein-laden mesophase and 800–1000 nL precipitant solution for Se-PepTSt, Se-LspA, Hg-derivatized BacA, and W-derivatized PgpB. In the case of native S-PepTSt, crystals were grown in syringes. For this purpose, 20 μL protein-laden LCP was injected into a 500 μL-syringe containing 400 μL precipitant solution43. Proteins were crystallized under the following conditions: Se-PepTSt (21–22 %(v/v) PEG- 400, 250–325 mM ammonium dihydrogen phosphate, 100 mM HEPES, pH 7.0, and 10 mM cefadroxil), Se-LspA (35–43 %(v/v) PEG-400, 100 mM MES, pH 5.6–6.0, and 60–100 mM ammonium phosphate monobasic), Hg-derivatized BacA (40 %(v/v) PEG-400, 300–500 mM ammonium citrate dibasic and 100 mM sodium citrate pH 5.0), W-derivatized PgpB (40 %(v/v) PEG-400, 100 mM HEPES pH 7 and 100 mM lithium citrate tribasic tetrahydrate) and native S-PepTSt (21–22 %(v/v) PEG-400, 250–325 mM ammonium dihydrogen phosphate, 100 mM HEPES, pH 7.0, and 10 mM Ala-Phe). Crystals of native-BacA for soaking experiments were grown in the same precipitant as was used for Hg-derivatized BacA crystal production. Se-PepTSt crystals grew as pyramids with an average size of 5 × 10 × 10 μm3. Se-LspA crystals grew as thin plates with an average size of 10 × 20 × 50 μm3. BacA crystals grew as thin plates with an average size of 5 × 10 × 20 μm3. Hg-BacA crystals grew as thin plates with an average size of 3 × 5 × 15 μm3. W-PgpB crystal grew as plates to a size of 10 × 50 × 150 μm3. Native S-PepTSt crystals grew as pyramids with an average size of 10 × 10 × 10 μm3.
IMISX-soaking and sample preparation
All of the samples used for data collection were prepared using the IMISX method as described previously10,11 with two important modifications. First, a Y-shaped holder (Supplementary Fig. 2) was used to secure the flexible IMISX well stably in the cryo-stream for effective raster scanning and data collection (Supplementary Fig. 7). Second, soaking crystals with heavy atoms was performed directly in the IMISX well without touching the hosting mesophase or the crystals therein. For this purpose, the intact well containing the crystal-laden mesophase bolus was removed from the double-sandwich IMISX plate and was mounted on a Y-support. One corner of the well was snipped off with a scissor and precipitant solution was wicked away from around the mesophase bolus using a cotton bud or tissue paper. Heavy atom reagent, dissolved in the same precipitant solution, was pipetted into the well via the open corner and manipulated so as to make contact with and to fully bathe the mesophase bolus. After a period of incubation at 20 °C (Rumed incubator, model 3101), the IMISX well was snap-cooled immediately in liquid nitrogen (Fig. 1). Samples were placed in pucks and shipped in a Dewar to the Swiss Light Source for measurement. Screening for optimum heavy atom labeling conditions was performed directly in IMISX wells in a process that involved different soaking times, heavy atom concentrations and heavy atom types chosen based on BacA sequence analysis in conjunction with the HATODAS II server44. Soaking with 10 mM HgCl2 for 70 min provided sufficient labeling for successful experimental phasing.
Automated serial data collection
X-ray diffraction experiments were carried out on protein crystallography beamlines X06SA-PXI and X10SA-PXII at the Swiss Light Source, Villigen, Switzerland. Data were collected at 100 K using cryo-cooled IMISX wells in a cryo-stream, as described11. Measurements were made in steps of 0.1–0.2° at a speed of 0.1 s/step with either an EIGER 16 M or a PILATUS 6M-F detector operated in continuous/shutterless data collection mode45. Beam size was adjusted according to crystal dimensions to optimize signal/noise for data collection and typically measured 10 × 10 μm2 or 20 × 20 μm2. With the Swiss Light Source data acquisition software (DA+)46 an automated serial data collection protocol (CY+) was developed enabling grid scan rates of 50–100 Hz47. This made it possible to raster scan an entire 2 × 2 mm2 mesophase bolus with X-rays at micrometer resolution in about 8 min. The resulting finely sampled grid map accurately located all crystals in the bolus and provided a ranking of diffraction quality in heat map form. Immediately after completing the grid scan, the automated data collection protocol CY+ was launched for serial data collection on crystals above a defined diffraction quality threshold over a specified rotation range (typically 10–20°) and a defined beam attenuation. Following this protocol, a few hundred crystals were measured in an hour (Supplementary Fig. 7). Radiation damage per crystal was maintained below 5 MGy for all proteins except Se-PepTSt where a dose of 17 MGy per crystal was used.
Data processing, selection, and merging
In serial crystallography, real-time data analysis is indispensable. This was implemented in the current IMISX-EP workflow by combining parallel data processing using XDS with data set selection and merging based on correlation coefficient and diffraction signal strength (Supplementary Fig. 3). Data collection wedges of 10–140° per crystal were indexed and processed using XDS48,49, as described11. For data set selection, all data sets were initially sorted in ascending order based on the averaged Rmeas values calculated from the three lowest resolution shells of each data set. Next, the sorted data sets were scaled and merged with XSCALE to obtain a preliminary scaled and merged data set. Then, the correlation coefficients (CCdataset) based on intensities in resolution shells were calculated between the preliminary merged data and each individual data set. The selection of data sets was carried out on the basis of the CCdataset values of each data set in a chosen resolution shell and the data sets below a certain CCdataset were rejected. This CCdataset selection process is usually carried out in an iterative manner based on a case-by-case basis. With Se-LspA, an additional selection based on the asymptotic < I/σ > ratio (ISa)50, as determined by XSCALE, was used to remove outlier data sets before CCdataset selection. Final data sets were scaled and merged with XSCALE. Data collection and processing statistics are provided in Table 1.
Structure determination and refinement
The SAD method was employed for experimental phasing using anomalous diffraction data sets from crystals of Se-PepTSt, Se/S-LspA, W-PgpB, Hg-BacA (soaking), Hg-BacA (co-crystallization), and native S-PepTSt. The SIRAS method was also used for Hg-BacA (soaking). Heavy atom location, structure phasing, and density modification were performed using the HKL2MAP51 interface of SHELXC/D/E12 for all structures and produced interpretable electron density maps for Se-PepTSt, Hg-BacA (soaking, SIRAS), Hg-BacA (co-crystallization), W-PgpB, and native S-PepTSt. Additional phase improvement and iterative auto-model building were carried out with CRANK213 for Se/S-LspA and Hg-BacA (soaking, SAD). From the experimentally phased maps, BUCCANEER52 and PHENIX AutoBuild53 were used for initial model building and models were completed manually using COOT54. Phenix.refine55 and BUSTER56 were used during the refinement of all structures. Phasing and refinement statistics are reported in Table 1 and Table 2. Figures of molecular structures were generated with PyMOL57.
Refinement of anomalous scattering factor f”
The anomalous scattering factor (f”) of Se in the Se-Met of PepTSt and LspA was refined using phenix.refine55 as follows. First, the atomic coordinates and B-factors were refined with standard PHENIX refinement to converged Rwork/Rfree values. Then, the B-factor of each Se atom was set to the average B-factor of its neighboring carbon atoms (Cγ and Cε) and its occupancy was set to one. Finally, the anomalous scattering factor f”, the atomic scattering factor f’, and atomic coordinates were refined using fixed B-factor and occupancy values for all Se atoms. The refined average values of f” were 5.38 and 3.16 for PepTSt and LspA, respectively. This result indicates a Se-Met substitution value of ~ 59% in LspA using PepTSt as reference with 100% substitution.
Code availability
Custom computer codes for correlation coefficient based data set selection and diffraction data are available at https://github.com/ranganawarshamanage/mxt.
Data availability
All diffraction data and refined models have been deposited in the Protein Data Bank under PDB identifiers 6FMR (Se-PepTSt), 6FMS (Se/S-LspA), 6FMT (Hg-BacA soaking SAD), 6FMV (BacA Native SIRAS), 6FMW (Hg-BacA co-crystallization), 6FMX (W-PgpB), and 6FMY (PepTSt). All data sets generated during the current study are available upon request.
Electronic supplementary material
Acknowledgements
We thank M. El Ghachi and F. Kreff for providing BacA and PgpB protein. The assistance and support of beamline scientists at the Swiss Light Source beamline X06SA and X10SA is acknowledged. C.-Y.H. is partially supported by the European Union’s Horizon 2020 research and innovation program under the Marie-Skłodowska-Curie grant agreement No. 701647. The work was funded in part by Science Foundation Ireland (16/IA/4435).
Author contributions
C.-Y.H., N.H. and L.V. produced and crystallized proteins. C.-Y.H., V.O., N.H., L.V. and E.P. collected X-ray diffraction data. C.-Y.H., V.O., R.W., T.W., S.B., K.D. and M.W. analyzed data. C.-Y.H., V.O., M.C. and M.W. wrote manuscript with contributions from all co-authors.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Chia-Ying Huang, Vincent Olieric.
Contributor Information
Martin Caffrey, Email: martin.caffrey@tcd.ie.
Meitian Wang, Email: meitian.wang@psi.ch.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s42003-018-0123-6.
References
- 1.Hendrickson WA. Atomic-level analysis of membrane-protein structure. Nat. Struct. Mol. Biol. 2016;23:464–467. doi: 10.1038/nsmb.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Santos R, et al. A comprehensive map of molecular drug targets. Nat. Rev. Drug. Discov. 2017;16:19–34. doi: 10.1038/nrd.2016.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiu W, Downing KH. Editorial overview: cryo electron microscopy: exciting advances in CryoEM Herald a new era in structural biology. Curr. Opin. Struct. Biol. 2017;46:iv–viii. doi: 10.1016/j.sbi.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ovchinnikov S, et al. Protein structure determination using metagenome sequence data. Science. 2017;355:294–298. doi: 10.1126/science.aah4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caffrey M. A comprehensive review of the lipid cubic phase or in meso method for crystallizing membrane and soluble proteins and complexes. Acta Crystallogr. Sect. F. 2015;71:3–18. doi: 10.1107/S2053230X14026843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hendrickson WA. Anomalous diffraction in crystallographic phase evaluation. Q. Rev. Biophys. 2014;47:49–93. doi: 10.1017/S0033583514000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zander U, et al. MeshAndCollect: an automated multi-crystal data-collection workflow for synchrotron macromolecular crystallography beamlines. Acta Crystallogr. D Biol. Crystallogr. 2015;71:2328–2343. doi: 10.1107/S1399004715017927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamamoto M, et al. Protein microcrystallography using synchrotron radiation. IUCrJ. 2017;4:529–539. doi: 10.1107/S2052252517008193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Melnikov I, et al. Fast iodide-SAD phasing for high-throughput membrane protein structure determination. Sci. Adv. 2017;3:e1602952. doi: 10.1126/sciadv.1602952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang CY, et al. In meso in situ serial X-ray crystallography of soluble and membrane proteins. Acta Crystallogr. D Biol. Crystallogr. 2015;71:1238–1256. doi: 10.1107/S1399004715005210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang CY, et al. In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures. Acta Crystallogr. D Struct. Biol. 2016;72:93–112. doi: 10.1107/S2059798315021683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheldrick GM. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr. D. 2010;66:479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skubák P, Pannu NS. Automatic protein structure solution from weak X-ray data. Nat. Commun. 2013;4:2777. doi: 10.1038/ncomms3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El Ghachi, M. et al. Structure of undecaprenyl-pyrophosphate phosphatase, BacA: an enzyme with an interdigitated inverted-topology repeat. Nat. Commun. 9, 1078 (2018). [DOI] [PMC free article] [PubMed]
- 15.Gati, C. et al. Serial crystallography on in vivo grown microcrystals using synchrotron radiation. IUCrJ 1, 87–94 (2014). [DOI] [PMC free article] [PubMed]
- 16.Botha S, et al. Room-temperature serial crystallography at synchrotron X-ray sources using slowly flowing free-standing high-viscosity microstreams. Acta Crystallogr. D Biol. Crystallogr. 2015;71:387–397. doi: 10.1107/S1399004714026327. [DOI] [PubMed] [Google Scholar]
- 17.Hasegawa K, et al. Development of a dose-limiting data collection strategy for serial synchrotron rotation crystallography. J. Synchrotron Radiat. 2017;24:29–41. doi: 10.1107/S1600577516016362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diederichs K, Wang M. Serial synchrotron X-ray crystallography (SSX) Methods Mol. Biol. 2017;1607:239–272. doi: 10.1007/978-1-4939-7000-1_10. [DOI] [PubMed] [Google Scholar]
- 19.Zander U, et al. Automated harvesting and processing of protein crystals through laser photoablation. Acta Crystallogr. D Struct. Biol. 2016;72:454–466. doi: 10.1107/S2059798316000954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith JL, Fischetti RF, Yamamoto M. Micro-crystallography comes of age. Curr. Opin. Struct. Biol. 2012;22:602–612. doi: 10.1016/j.sbi.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Q, Hendrickson WA. Crystallographic phasing from weak anomalous signals. Curr. Opin. Struct. Biol. 2015;34:99–107. doi: 10.1016/j.sbi.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karplus PA, Diederichs K. Assessing and maximizing data quality in macromolecular crystallography. Curr. Opin. Struct. Biol. 2015;34:60–68. doi: 10.1016/j.sbi.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Terwilliger TC, et al. Can I solve my structure by SAD phasing? Anomalous signal in SAD phasing. Acta Crystallogr. D Biol. Crystallogr. 2016;72:346–358. doi: 10.1107/S2059798315019269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giordano R, Leal RMF, Bourenkov GP, McSweeney S, Popov AN. The application of hierarchical cluster analysis to the selection of isomorphous crystals. Acta Crystallogr. D Biol. Crystallogr. 2012;68:649–658. doi: 10.1107/S0907444912006841. [DOI] [PubMed] [Google Scholar]
- 25.Assmann G, Brehm W, Diederichs K. Identification of rogue datasets in serial crystallography. J. Appl. Crystallogr. 2016;49:1021–1028. doi: 10.1107/S1600576716005471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diederichs K. Dissecting random and systematic differences between noisy composite data sets. Acta Crystallogr. D Struct. Biol. 2017;73:286–293. doi: 10.1107/S2059798317000699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foadi J, et al. Clustering procedures for the optimal selection of data sets from multiple crystals in macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2013;69:1617–1632. doi: 10.1107/S0907444913012274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zander U, et al. Merging of synchrotron serial crystallographic data by a genetic algorithm. Acta Crystallogr. D Struct. Biol. 2016;72:1026–1035. doi: 10.1107/S2059798316012079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamashita K, Hirata K, Yamamoto M. Acta Crystallogr. D Struct Biol. 2018. KAMO: towards automated data processing for microcrystals; pp. 441–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo G, et al. Sample manipulation and data assembly for robust microcrystal synchrotron crystallography. IUCrJ. 2018;5:238–246. doi: 10.1107/S2052252518005389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weinert T, et al. Fast native-SAD phasing for routine macromolecular structure determination. Nat. Methods. 2015;12:131–133. doi: 10.1038/nmeth.3211. [DOI] [PubMed] [Google Scholar]
- 32.Nakane T, et al. Membrane protein structure determination by SAD, SIR, or SIRAS phasing in serial femtosecond crystallography using an iododetergent. Proc. Natl. Acad. Sci. USA. 2016;113:13039–13044. doi: 10.1073/pnas.1602531113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Axford D, Aller P, Sanchez-Weatherby J, Sandy J. Applications of thin-film sandwich crystallization platforms. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2016;72:313–319. doi: 10.1107/S2053230X16004386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Broecker J, et al. A versatile system for high-throughput in situ X-ray screening and data collection of soluble and membrane-protein crystals. Cryst. Growth Des. 2016;16:6318–6326. doi: 10.1021/acs.cgd.6b00950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Broecker J, et al. High-throughput in situ X-ray screening of and data collection from protein crystals at room temperature and under cryogenic conditions. Nat. Protoc. 2018;13:260–292. doi: 10.1038/nprot.2017.135. [DOI] [PubMed] [Google Scholar]
- 36.Eriksson M, van der Veen JF, Quitmann C. Diffraction-limited storage rings – a window to the science of tomorrow. J. Synchrotron Radiat. 2014;21:837–842. doi: 10.1107/S1600577514019286. [DOI] [PubMed] [Google Scholar]
- 37.Lyons Ja, et al. Structural basis for polyspecificity in the POT family of proton-coupled oligopeptide transporters. EMBO Rep. 2014;15:886–893. doi: 10.15252/embr.201338403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vogeley L, et al. Structural basis of lipoprotein signal peptidase II action and inhibition by the antibiotic globomycin. Science. 2016;351:876–880. doi: 10.1126/science.aad3747. [DOI] [PubMed] [Google Scholar]
- 39.Manat G, et al. Membrane topology and biochemical characterization of the escherichia coli BacA undecaprenyl-pyrophosphate phosphatase. PLoS ONE. 2015;10:e0142870. doi: 10.1371/journal.pone.0142870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghachi ME, et al. Crystal structure and biochemical characterization of the transmembrane PAP2 type phosphatidylglycerol phosphate phosphatase from Bacillus subtilis. Cell Mol. Life Sci. 2017;74:2319–2332. doi: 10.1007/s00018-017-2464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caffrey M, Cherezov V. Crystallizing membrane proteins using lipidic mesophases. Nat. Protoc. 2009;4:706. doi: 10.1038/nprot.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng A, Hummel B, Qiu H, Caffrey M. A simple mechanical mixer for small viscous lipid-containing samples. Chem. Phys. Lipids. 1998;95:11–21. doi: 10.1016/S0009-3084(98)00060-7. [DOI] [PubMed] [Google Scholar]
- 43.Liu W, Ishchenko A, Cherezov V. Preparation of microcrystals in lipidic cubic phase for serial femtosecond crystallography. Nat. Protoc. 2014;9:2123–2134. doi: 10.1038/nprot.2014.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sugahara M, et al. Heavy-atom Database System: a tool for the preparation of heavy-atom derivatives of protein crystals based on amino-acid sequence and crystallization conditions. Acta Crystallogr. D Biol. Crystallogr. 2005;61:1302–1305. doi: 10.1107/S0907444905019670. [DOI] [PubMed] [Google Scholar]
- 45.Casanas A, et al. EIGER detector: application in macromolecular crystallography. Acta Crystallogr. D Struct. Biol. 2016;72:1036–1048. doi: 10.1107/S2059798316012304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wojdyla JA, et al. DA+data acquisition and analysis software at the Swiss Light Source macromolecular crystallography beamlines. J. Synchrotron Radiat. 2018;25:293–303. doi: 10.1107/S1600577517014503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wojdyla JA, et al. Fast two-dimensional grid and transmission X-ray microscopy scanning methods for visualizing and characterizing protein crystals. J. Appl. Crystallogr. 2016;49:944–952. doi: 10.1107/S1600576716006233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kabsch, W. Xds. Acta Crystallogr. D66, 125–132 (2010). [DOI] [PMC free article] [PubMed]
- 49.Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D. 2010;66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Diederichs K. Quantifying instrument errors in macromolecular X-ray data sets. Acta Crystallogr. D Biol. Crystallogr. 2010;66:733–740. doi: 10.1107/S0907444910014836. [DOI] [PubMed] [Google Scholar]
- 51.Pape T, Schneider TR. HKL2MAP: a graphical user interface for macromolecular phasing with SHELX programs. J. Appl. Crystallogr. 2004;37:843–844. doi: 10.1107/S0021889804018047. [DOI] [Google Scholar]
- 52.Cowtan K. Completion of autobuilt protein models using a database of protein fragments. Acta Crystallogr. D Biol. Crystallogr. 2012;68:328–335. doi: 10.1107/S0907444911039655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Terwilliger TC, et al. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 55.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blanc E, et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2210–2221. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- 57.The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All diffraction data and refined models have been deposited in the Protein Data Bank under PDB identifiers 6FMR (Se-PepTSt), 6FMS (Se/S-LspA), 6FMT (Hg-BacA soaking SAD), 6FMV (BacA Native SIRAS), 6FMW (Hg-BacA co-crystallization), 6FMX (W-PgpB), and 6FMY (PepTSt). All data sets generated during the current study are available upon request.