Abstract
A recently reported family of soluble cyanobacterial carotenoproteins, homologs of the C-terminal domain (CTDH) of the photoprotective Orange Carotenoid Protein, is suggested to mediate carotenoid transfer from the thylakoid membrane to the Helical Carotenoid Proteins, which are paralogs of the N-terminal domain of the OCP. Here we present the three-dimensional structure of a carotenoid-free CTDH variant from Anabaena (Nostoc) PCC 7120. This CTDH contains a cysteine residue at position 103. Two dimer-forming interfaces were identified, one stabilized by a disulfide bond between monomers and the second between each monomer’s β-sheets, both compatible with small-angle X-ray scattering data and likely representing intermediates of carotenoid transfer processes. The crystal structure revealed a major positional change of the C-terminal tail. Further mutational analysis revealed the importance of the C-terminal tail in both carotenoid uptake and delivery. These results have allowed us to suggest a detailed model for carotenoid transfer via these soluble proteins.
Dvir Harris et al. present the structure of a homolog of the orange carotenoid protein (OCP) C-terminal domain, elaborating on this protein family’s carotenoid transfer mechanism. They observed major structural shifts in the homolog compared to that of the OCP C-terminal domain, with a strong positive impact on carotenoid uptake and delivery.
Introduction
Photosynthetic organisms, including cyanobacteria, utilize the energy of the sun to sustain themselves in a wide range of static and dynamically changing conditions. The photosynthetic process is initiated by light absorption by antenna complexes, which are the phycobilisomes1,2 in cyanobacteria. Excess absorbed energy can become deleterious without proper regulation as overexcitation may cause damage to the photochemical reaction centers, leading to cell stress and eventually cell death3. Cyanobacteria have evolved several protection mechanisms including non-photochemical quenching, induced by the Orange Carotenoid Protein (OCP)4,5. OCP is a water-soluble, carotenoid-binding, 35 kDa protein found to induce dissipation of phycobilisome excitation energy minimizing energy flux toward the reaction centers5–7. OCP can also directly protect against the presence of singlet oxygen8,9. OCP is comprised of two domains: the N-terminal domain (NTD) and the C-terminal domain (CTD) connected through an inter-domain flexible linker loop9. In darkness and low light, OCP is in its stable inactive orange state (OCPO). Upon strong illumination, changes in the carotenoid and the protein lead to the formation of the metastable, active red state (OCPR) in which CTD and NTD are completely separated6. Only the OCPR state interacts with the phycobilisome to induce fluorescence quenching through its NTD10,11. The CTD also interacts with the fluorescence recovery protein, which supports the reorganization of OCPR to OCPO12–14. The structure of OCPO (PDB code: 5UI2, Supplementary Figure 1) demonstrated that the carotenoid is situated in a hydrophobic cavity formed by both the CTD and the NTD9,15–18.
Although the structure of the OCPR state remains elusive to date, the structure of the isolated NTD with bound carotenoid (holo-NTD) was determined (PDB code: 4XB4)16. This holo-NTD structure revealed that, upon photoactivation, the carotenoid becomes more planar and is translocated by 12 Å into the NTD cavity16. The crystal structure of a carotenoid-free CTD (apo-CTD) has not yet been determined (Supplementary Figure 1). Phylogenetic studies discovered that cyanobacteria contain genes that encode for homologs of both the NTD and CTD19–21. NTD homologs have been termed Helical Carotenoid Proteins (HCPs) for their all α-helical structure19,22. Anabaena (Nostoc) PCC 7120 (hereafter Anabaena) possesses four variants of HCPs (out of nine total clades identified in cyanobacteria19). The holo-HCP1 was isolated from Anabaena and its structure was solved, proving HCPs to be carotenoid-binding proteins19. Its structure exhibited high similarity to the NTD of OCP, which can individually exist as a proteolytic cleavage product of OCP10. Previous studies have shown that each of the HCP clades demonstrates different features22. Unlike the wide variety of HCPs, the CTD homologs (CTDH) fall only into two clades, of which only one is present in Anabaena (a clade 2 CTDH). The CTDH gene in Anabaena is located adjacent to the HCP4 gene, therefore it was suggested that HCP4 and CTDH are the progenitors of OCP19,23,24.
Recently, it was revealed that isolated CTD of Synechocystis OCP and CTDHs from Anabaena and Thermosynechococcus elongatus (a clade 1 CTDH) are also capable of carotenoid binding25,26. Both CTD and CTDH were shown to mediate carotenoid transfer to apo-OCP, apo-NTD, or apo-HCP (from clade 4)25,26. OCP and CTDH are also capable of taking up a carotenoid molecule from membranes while HCP4 and isolated NTD are unable to do so25. Thus one likely role of CTDHs is to serve as intermediate transfer molecules required for delivery of carotenoids from membranes to the HCPs. Anabaena CTDH was shown to form a disulfide-linked dimer in the presence or absence of carotenoid under oxidizing conditions. By contrast, the CTDH from T. elongatus (hereafter TeCTDH) that lacks Cys103 forms a dimer only in the presence of a carotenoid that is shared by the two monomers25. When stabilized by the presence of the disulfide bond, the CTDH dimer could not donate its carotenoid to an acceptor molecule; the reduced form of holo-AnaCTDH was able to transfer its carotenoid to HCP. This suggests that the disulfide bond containing clade 2 CTDH dimer has additional regulation and may be controlled by changes in the redox state in the cell, functioning upon stress induction.
We present here three important findings on the carotenoid transfer mechanism by CTDH. First, there is a major structural shift of the C-terminal tail of the CTDH (with respect to its position in the holo-form of CTD-OCP), bringing it into close proximity to the carotenoid-binding pocket. Second, we show that the C-terminal tail has a strong positive impact on both the carotenoid uptake as well as the carotenoid delivery by the CTDH; when absent, the rates of these processes are considerably reduced. Finally, the role of CTDH as carotenoid donor to HCPs was confirmed. Surprisingly, apo-AnaCTDH was shown to be also capable of receiving a carotenoid molecule from HCP1, suggesting an increased complexity for carotenoid transfer between different partners.
Results
The AnaCTDH can form a large homogeneous oligomer
Anabaena apo-CTDH was prepared for crystallization as described in Muzzopappa et al. (2017)25. Native-polyacrylamide gel electrophoresis (native-PAGE) indicated that the major assembly is a dimeric form, suggesting the presence of a disulfide bond between the CTDH monomers (Supplementary Figure 2A). Under reducing conditions, the CTDH appeared as a monomer (Supplementary Figure 2B). Size exclusion chromatography revealed that the CTDH protein in solution was mostly in a higher oligomeric state (Supplementary Figure 3, dashed bold line) with additional, smaller oligomeric states in solution. This discrepancy concerning AnaCTDH oligomeric state between native-PAGE and size exclusion chromatography has previously been published25.
Crystallization trials were attempted on this apparent mixture of forms and large crystals were obtained (Supplementary Figure 4A). Using a laboratory X-ray source, the crystals diffracted to 2.9 Å (Supplementary Figure 4B). Indexing revealed a rather large unit cell (Table 1) with respect to the molecular weight (MW) of the AnaCTDH dimer (34 kDa). The calculated Matthews coefficient indicated that the asymmetric unit could contain 10–24 AnaCTDH monomers.
Table 1.
Data-collection and final refinement statistics
| Native | 2 M urea | |
|---|---|---|
| Data collection | ||
| Space group | P212121 | P6522 |
| Cell dimensions | ||
| a, b, c (Å) | 65.65, 112.28, 318.54 | 81.08, 81.08, 162.94 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 120.0 |
| Resolution (Å) | 47.99–2.90 (3.06–2.90)a | 53.19–2.75 (2.85–2.75)a |
| R merge | 0.06 (0.42)a | 0.07 (0.33)a |
| 〈I/σ(I)〉 | 16.7 (3.3)a | 11.7 (5.0)a |
| Completeness (%) | 99.9 (100)a | 99.8 (100)a |
| Redundancy | 3.7 (3.9)a | 5.6 (6.1)a |
| Refinement | ||
| Resolution (Å) | 53.19–2.75 | |
| No. of reflections | 8742 | |
| Rwork/Rfree | 0.241/0.292 | |
| Completeness (%) | 99.58 | |
| No. of non-hydrogen atoms | 1855 | |
| Protein | 1828 | |
| Ligands | 12 | |
| Water | 15 | |
| B factors (Å2) | ||
| Protein | 67.31 | |
| Ligands | 78.36 | |
| Water | 45.16 | |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.012 | |
| Bond angles (°) | 1.47 | |
Values in parentheses are for the outer shell
Whether this large oligomerization state has any biological significance as a quaternary assembly has to be further investigated. The fact that it was successfully crystallized and diffracted well suggests that this state is not a random aggregation and that different faces of interaction between the monomers (or between dimers) must exist. The presence of high MW oligomers was also detected in carotenoid-containing versions of AnaCTDH, TeCTDH, and Synechocystis CTD-OCP preparations obtained in Escherichia coli cells25, suggesting that oligomerization is a more general characteristic of CTD-like proteins. Nevertheless, these large oligomers were not detected in size exclusion chromatography when the holo-proteins were isolated from Synechocystis cells or when AnaCTDH was isolated under reducing conditions or in a AnaCTDH Cys103Phe mutant25.
Crystallization of apo-AnaCTDH dimer
In order to obtain a more homogeneous preparation of apo-AnaCTDH dimers, we added 2 M urea to the solution27,28, which caused a large proportion of the high oligomeric states to disassemble, resulting in predominantly dimeric CTDH (Supplementary Figure 3, solid line). Crystals (Supplementary Figure 5A) were obtained in the presence of urea (see Methods). The apo-AnaCTDH dimer crystals were found to diffract to 2.43 Å with synchrotron radiation (Supplementary Figure 5B, and Table 1).
Determination of the urea-treated dimeric apo-AnaCTDH structure
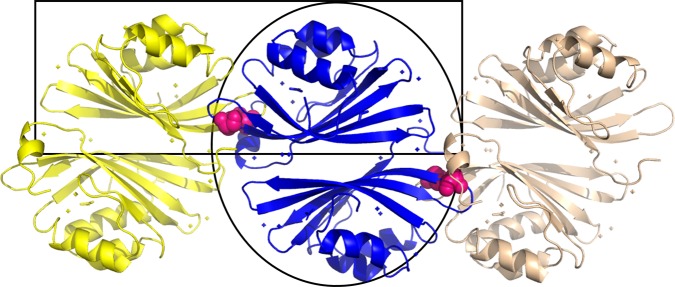
The X-ray crystallographic structure of apo-AnaCTDH was determined by molecular replacement (MR) to 2.75 Å resolution using the OCPO-CTD as the search model (from the 5UI2 structure), with two monomers in the asymmetric unit. Somewhat surprisingly, the disulfide bond between Cys103 residues of two monomers mentioned above was found to be between two different asymmetric units and not between the two monomers of one asymmetric unit. The asymmetric unit dimer (hence called A-type or back-to-back) was formed via an interaction plane between the β-sheets of two monomers, quite distant from Cys103 (Fig. 1). The presence of the oxidized disulfide bond was confirmed by the presence of continuous electron density between the two Cys103 residues in the Fo-Fc omit map (Supplementary Figure 6).
Fig. 1.
Two dimeric interfaces are revealed in the CTDH structure. Three adjacent asymmetric units are depicted in yellow, blue, and light pink. The Cys103 residues forming the disulfide bonds are presented as pink spheres. The asymmetric unit contains a back-to-back dimer interface (called A-type; black oval). The disulfide bonds link the head-to-head functional dimer (F-type; black rectangle)
Analysis using the PISA macromolecular interaction tool29 showed that both the A-type dimer and the functional disulfide-containing dimer (hence called an F-type or head-to-head) exhibited negative calculated ΔG values, predicted to be stable. PISA suggested that only the F-type dimer has biological significance, (ΔiGP value of 0.067 for the F-type dimer interface). This finding agrees with the suggestion proposed by Moldenhauer et al. (2017)26 and Muzzopappa et al. (2017)25 that the interaction plane between the CTD(H) dimer should be at the same site as the CTD and NTD in OCPO. In contrast, a ΔiGP value of 0.518 in the case of the A-type dimer interface implies possibly crystal-stabilized interface (as a ΔiGP value <0.5 is considered necessary for biological significance29). However, this interface might still be important for the assembly into large oligomers in solution, which was shown to be condition-dependent25 as well as a possible different type of dimeric interaction under some specific conditions.
Analysis of the dimeric interfaces in solution
The presence of the disulfide bond forces the formation of the F-type dimer of apo-AnaCTDH. However, upon the reduction of cysteines or in their absence (in clade 1 CTDHs or in CTD-OCP), the A-type dimerization can become possible. The heterogeneous nature of the apo-AnaCTDH solution (Supplementary Figure 3) and this protein’s propensity to monomerize in the absence of carotenoid25 impedes structural analysis of its non-covalently bound dimers. In contrast, individual apo-CTDs from Synechocystis OCP (carrying a Phe instead of Cys at residue position 103) efficiently dimerize without forming higher-order oligomers26, making apo-CTD dimers suitable for structural characterization in solution by using small-angle X-ray scattering (SAXS).
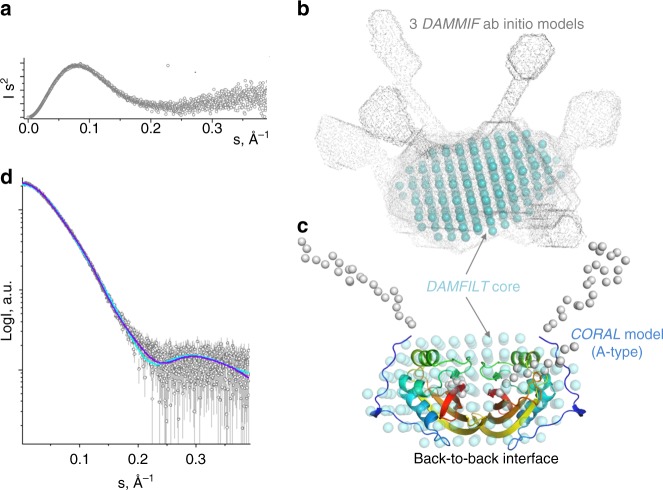
At 270 µM, the 18.7 kDa apo-CTD construct gave particles with a radius of gyration (Rg) of 2.7 nm and the maximum particle dimension (Dmax) of 9.5 nm. The bell-shaped Kratky plot with the gradual rise of the curve at high angles (Fig. 2a) suggested particles with a compact globular core and moderate flexibility. The Porod volume of 56 nm3 suggests that the corresponding MW estimate (35 kDa) is close to that expected for apo-CTD dimers (37 kDa), in line with their known dimerization at concentrations above 150 µM26,30. Ab initio molecular shape reconstruction using DAMMIF31 resulted in globular models with pairs of protrusions at variable positions (Fig. 2b), consistent with the Kratky plot. Averaging of the DAMMIF models with DAMAVER32 revealed a core, common for the generated models (DAMFILT core; shown by cyan spheres in Fig. 2b). Given the similarity of the primary structure and fold of apo-CTD from Synechocystis and apo-AnaCTDH (Cα r.m.s.d. <1 Å), the two types of dimeric cores found in the crystal lattice (Fig. 1) were built by spatial overlay, yielding either the F-type or the A-type dimer. To account for local differences between the apo-CTD and apo-AnaCTDH structures, the A-type dimer of apo-CTD was further refined by local protein–protein docking approach using RosettaDock server33, which resulted in a more connected, realistic structural model free from steric clashes (Supplementary Figure 7A). This model, matching the DAMFILT-derived globular core (Fig. 2c), was used as a template to model the unstructured terminal regions (including the C-terminal tails) using CORAL (Fig. 2c). Likewise, the F-type of the apo-CTD dimer was slightly corrected to eliminate apparent clashes and then used to build a CORAL model with flexible termini. CORAL-derived structural models for both types of dimers described the experimental SAXS data reasonably well; however, statistical analysis including χ2 and the correlation map P value (CorMap)34 showed much better fits in the case of A-type dimer (Fig. 2d). This favored fit for the A-type apo-CTD dimer may indicate that, in the absence of the Cys–Cys bond, the back-to-back interface might be more favorable in solution. It is likely that the compromised ability of apo-AnaCTDH to form non-covalent dimers, which presumably occurs only at high protein concentrations, is associated with the absence of the functionally important phenylalanine residues in the external side of the β-sheet (in positions 299 and 300 of Synechocystis OCP), which in AnaCTDH are occupied by Ile-His residues (see Supplementary Figures 7B and 7C). This hypothesis is in line with the reported inability of apo-TeCTDH lacking a Phe in position homologous to 299 in the CTD of Synechocystis OCP to form stable dimers25. Attempts to fit the SAXS data with either a monomer or a trimer of apo-CTD gave significantly poorer fits (χ2 ~ 19 for a monomer, χ2 ~ 5 for a trimer).
Fig. 2.
Analysis of the solution conformation of Synechocystis apo-CTD dimers using SAXS. a Kratky plot showing that apo-CTD is represented by particles with a globular core and flexible termini/loops. b Ab initio shape reconstruction using DAMMIF. Three best-fitting DAMMIF models are shown superimposed to reveal the common core (DAMFILT core, cyan spheres). c CORAL-derived structural model of the A-type apo-CTD dimer obtained upon modeling of the flexible N- (23 residues) and C-terminal (12 residues) tails in order to minimize the discrepancy between the experimental and theoretical SAXS curves calculated from the model. The CORAL model overlaid with the DAMFILT core (cyan spheres) from b is shown by ribbons colored by gradient from blue (N-terminus) to red (C-terminus), with the flexible termini represented by Cα atoms (gray spheres). d Fitting of the experimental SAXS profile for apo-CTD by the best-fitting CORAL-derived models corresponding to either A-type (dark blue; χ2 = 1.14, CorMap = 0.16) or F-type (light blue; χ2 = 2.51, CorMap = 0.00) dimers
These observations indicate that transitions between the two types of dimers may occur, which may be relevant for the carotenoid transfer mechanism. Owing to the steric constraints, the F-type crystallographic apo-AnaCTDH dimer is not capable of performing carotenoid uptake from the membranes, as the carotenoid channel is partially blocked by the C-terminal tail and filled with the Cys103-containing loop (hereafter β5/β6 loop), while in the A-type dimer, the carotenoid cavity is open. Notably, the carotenoid transfer is suppressed when the dimeric holoform is stabilized by the disulfide bond25. Furthermore, the A-type conformation may represent one of the intermediates during carotenoid transfer into the HCPs. One may speculate that, during the transfer, the F-type dimer disassembles, exposing one side of the carotenoid, and yields an intermediate A-type dimer with the carotenoid accessible in one holo-monomer, which then forms a new F-type-like interface with an apo-HCP counterpart.
Structural changes related to the shortening of CTDH relative to CTD
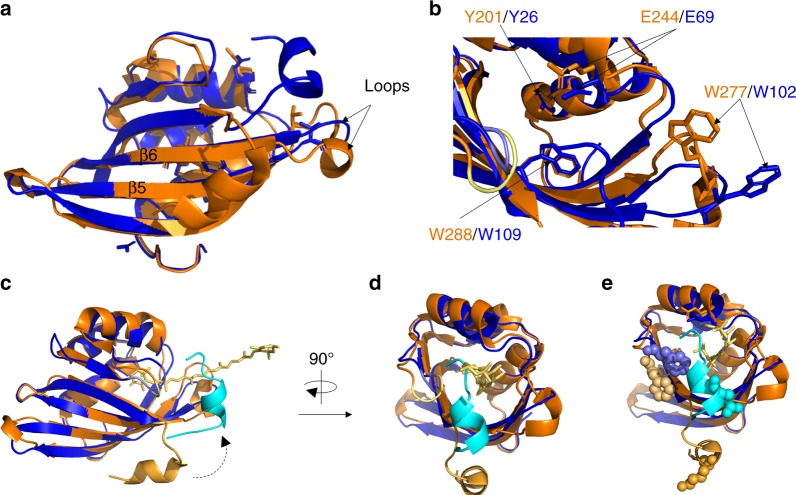
To date, the only high-resolution structural information available on the CTD is when it is in association with the NTD in the OCPO state of the full-length OCP protein (closed carotenoid-associated conformation–PDB codes: 5UI2, 4XB5, 5TUX, 3MG1, etc.). As opposed to the CTDs of OCPs, all CTDHs lack the four amino acids that immediately follow residue 103 (residue 278 in Synechocystis OCP) in the CTD (Supplementary Figure 7C). Therefore, tertiary structure differences were expected to be present between the proteins25. Homology modeling suggested that, in the CTDH, the gap would result in a shortening of the β6 strand, while the loop that connects this strand with β5 would become longer forcing it into a different position25. The resolution of the three-dimensional structure of apo-AnaCTDH presented in this study shows that the lack of these four amino acids causes structural changes, different than those predicted by modeling. The β6 strand is longer and the loop between strands β6 and β5 is shorter (Fig. 3a). These structural changes lead to a change in the position of Glu244 and Trp277 (Fig. 3b), which play an essential role in the interactions between the CTD and NTD in the OCPO state15. These changes in positions can affect assembly processes, which in turn result in diverse activities by different protein pairs. More specifically, when holo-CTD and apo-NTD interact, an OCP-like protein is formed, in which the carotenoid is shared by the CTD and NTD (similarly to OCPO); however, when holo-CTDH and apo-HCP interact, the carotenoid is not shared by both domains since full carotenoid transfer occurs (from CTDH to HCP)25. In contrast to the residues noted above, Tyr201 and Trp288 in Synechocystis CTD (Tyr26 and Trp109 in AnaCTDH), which are found in interaction with the carotenoid in OCPO, are in almost identical positions in OCP and in the apo-AnaCTDH structure presented here, suggesting that the carotenoid can interact with these amino acids in CTDH dimers.
Fig. 3.
Crystal structure comparison between apo-AnaCTDH and CTD OCPO (PDB code: 5UI2). a CTD-OCP (PDB code: 5UI2, orange) and apo-AnaCTDH (blue) structures are superimposed. Both elongation of β6 strand and shortening of the loop connecting β5 and β6 strands are observed for apo-AnaCTDH. b These differences lead to spatial changes in both E244/E69 and W277/W102. In contrast, Y201/Y26 and W288/W109 overlap well in both structures. c Cartoon representation of CTDH (blue shades) super-positioned with CTD of OCPO (orange shades). The C-terminal tail changes its orientation (indicated by dashed arrow) substantially from an external position (tail in orange) to an internal position for apo-AnaCTDH (tail in light blue). The position of the carotenoid molecule (yellow stick representation) is preserved from the 5UI2 structure. d Same as c but rotated clockwise by 90°. e Same as b but with amino acids known to be substantially perturbed in their solvent accessibility upon holo-to-apo transition in CTD-OCP
In an engineered AnaCTDH mutant in which the four amino acids missing in the sequence (Supplementary Figure 7C) were reintroduced and the Cys103 was changed to Phe, the holo-dimer was less stable and easily lost the carotenoid25. This suggests that the structural change induced by the lack of the four amino acids is important for carotenoid stabilization in the CTDH dimer. Here we show that the addition of the 4 amino acids also hinders carotenoid uptake. Holo-HCP4, -HCP3, and -HCP2 were unable to give the carotenoid to apo-AnaCTDH-C103F mutant protein (which cannot form the critical disulfide bond, thus making carotenoid transfer possible) (Supplementary Figure 8A, 8B, and 8C).
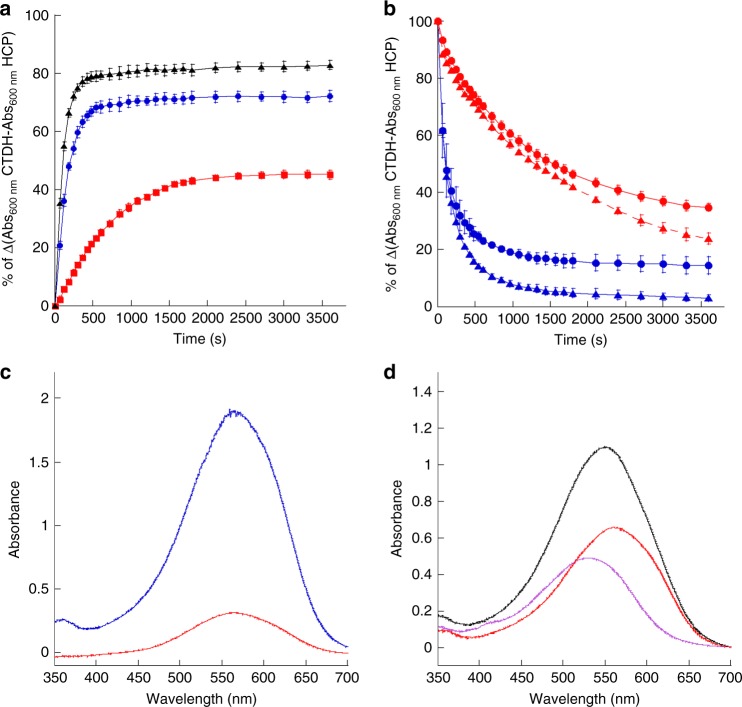
Surprisingly, HCP1 largely transferred its carotenoid to apo-AnaCTDH-C103F (Fig. 4a, blue and black spectra, and Supplementary Figure 9A). The carotenoid transfer was slower from HCP1 to apo-AnaCTDH-C103F+4aa (Fig. 4a, red spectrum), indicating that this CTDH mutant is hindered in the uptake and stabilization of the carotenoid. In conclusion, in CTDHs, the structural changes generated by the absence of the four amino acids following Cys103 are critical for carotenoid uptake, binding, and stabilization of the dimer.
Fig. 4.
Carotenoid transfer between HCPs and CTDHs and carotenoid uptake from membranes. a Kinetics of carotenoid transfer from holo-HCP1 to apo-CTDH-C103F-Cter-Histag (blue), apo-CTDH-C103F-Nter-Histag (black), and apo-CTDH-C103F+4aa (red) followed by an increase of absorbance at 600 nm. b Kinetics of carotenoid transfer from holo-CTDH-C103F (blue) and from holo-CTDH-C103F-∆C-terminal tail (∆C-terminal tail abbreviated to CTT on spectra, red) to apo-HCP1 (triangles) and apo-HCP4 (circles) followed by the decrease of absorbance at 600 nm. The CTDHs used in these experiments have the His-tag in the C-terminus. c Absorption spectra of the supernatant after 1 h incubation of canthaxanthin containing E. coli membranes with apo-CTDH-C103F (blue) and Apo-CTDH-C103-∆C-terminal tail (red) following membranes removal. 100% of apo-CTDH-C103F was converted to holo-protein while only 20(±5)% of apo-CTDHC103F-ΔC was converted to holo-protein. d Absorption spectrum of the supernatant after 1 h incubation of canthaxanthin containing E. coli membranes with apo-CTDH-C103F and apo-HCP1 (black). The membranes were removed before spectrum recording. Deconvolution of the graph into HCP1 (46%) and CTDH (54%) components is also shown. The ratio CTDH to HCP1 was 1 during the incubation. The experiments were done at least three times and protein concentrations were similar
The position of the C-terminal tail in the CTDH structure
The most striking difference between the apo-AnaCTDH structure presented here and the CTD-OCPO structure is a major shift in the position of the C-terminal tail (Fig. 3c). In the CTD-OCPO structure, the C-terminal tail is in an external position interacting with the β-sheets and possibly with the N-terminal extension (based on their proximity, as demonstrated in 5UI2 structure). In contrast, in the apo-AnaCTDH structure, the C-terminal tail adopts an internal position, coming closer to the F-type interface between the CTDH monomers (CTD-NTD interface in OCP), partially covering the opening of the designated carotenoid-accommodating cavity in the apo-AnaCTDH structure (Fig. 3d).
This dramatic shift in the position of the C-terminal tail is in good agreement with the literature—the structural changes taking place upon OCPO to OCPR photoconversion were studied previously17 by examining amino acid solvent accessibility. It was demonstrated that residues Pro225-Pro226-Phe227 (Phe227 is shown as shaded wheat-orange spheres in Fig. 3e) exhibit the second highest decrease in accessibility in the CTD upon phototransformation. It was postulated that this PPF patch serves as a cap for the carotenoid cavity in the CTD upon translocation of the carotenoid in the OCPR state. However, Lys310 (bottom-most light orange spheres in Fig. 3e) exhibited the highest decrease in accessibility, yet no explanation was suggested for this observation. The apo-AnaCTDH structure provides a more comprehensive interpretation of these results. Upon loss of the carotenoid to the NTD, the C-terminal tail, which points away from the OCPO globular structure and was highly accessible, now rotates by nearly 180° and turns into the carotenoid cavity, capping it. This capping function covers the PPF patch completely while internalizing Gln132, the structurally homologous CTDH residue of Lys310 in the CTD (Fig. 3e).
Role of the C-terminal tail
In order to assess the role of the C-terminal tail, two AnaCTDH-C103F mutants lacking the C-terminal tail were constructed. Both mutated proteins lacked 11 C-terminal amino acids from Leu126 to Leu136. One of the proteins had the His6-tag at the C-terminus just after Lys125 while the second mutant had the His6-tag at the N-terminus (see Methods). Regardless of the position of the His-tag, the mutated CTDH-C103F-ΔC-terminal tail failed to receive a carotenoid from holo-HCP1 (Supplementary Figures 8E and 8F). The carotenoid uptake from the membranes was also largely reduced when the C-terminal tail was absent (only 20% of holo-protein was produced (Fig. 4c).
The importance of the C-terminal tail was further examined for the carotenoid delivery by the holo-CTDH. For both apo-HCP1 and apo-HCP4 as recipients, holo-CTDH was able to transfer its carotenoid with or without the C-terminal tail (Supplementary Figures 9b and 9c), yet the absence of the C-terminal tail from the holo-CTDH decreased the rate of carotenoid transfer (Fig. 4b).
These results demonstrate that the internal, cavity-blocking position of the C-terminal tail in the apo-form does not hinder carotenoid uptake. In fact, it facilitates its uptake from both the membrane and, surprisingly, from holo-HCP1. Moreover, the C-terminal tail facilitates transfer the carotenoid to HCPs, possibly starting from its external position (as in OCPO), then moving closer to the carotenoid to expedite the process. Therefore, the C-terminal tail, which starts with a relatively hydrophobic patch and ends in a relatively hydrophilic patch (Supplementary Figure 7c), may serve as a facilitator for carotenoid uptake and delivery. For uptake, the hydrophilic end may bind to the hydrophilic lipid head groups followed by forming an interaction between the hydrophobic patch and the membrane-embedded carotenoid molecule. For delivery, the hydrophilic patch would bind to the appropriate site on the HCP, followed by transfer of the carotenoid to the HCP-binding cavity, facilitated by the hydrophobic patch.
Possible structural changes anticipated in CTDH following carotenoid uptake
The apo-CTDH structure is expected to change upon carotenoid binding, so we attempted to understand the structural effects of canthaxanthin binding on apo-AnaCTDH. Computational docking of a carotenoid molecule to the apo-AnaCTDH structure was executed using the Swissdock server35. When the F-type dimer was used as the host (with or without the C-terminal tails), not a single 1 of the 34 docking clusters predicted a carotenoid positioned inside the cavity formed by the dimer (Supplementary Figures 10A and 10B). Manual positioning of the carotenoid molecule was also unsuccessful. Thus the F-type dimer cannot accommodate the carotenoid unless structural changes occur. Indeed, size exclusion chromatography and native-PAGE showed different volumes for holo- and apo-AnaCTDH25. The same situation seems to be valid for CTD-OCP, where the holo-dimer was reported to have a larger apparent size than the apo-dimer26.
By contrast, when an apo-AnaCTDH monomer was used as the host (with or without the C-terminal tail), 31 out of the top 34 clusters suggested that the carotenoid would be positioned in the pocket that has been shown to bind the carotenoid in OCPO (Supplementary Figures 10C and 10D). In both docking trials, the outward-facing region of the C-terminal tail was found to be a good docking site for the carotenoid molecule (red circles, Supplementary Figures 10B and 10D), suggesting that there could be an interaction between the two (as proposed above).
Two possible structural obstacles for carotenoid accommodation in the F-type CTDH dimer based on the apo-AnaCTDH structure can be suggested. First, the β5/β6 loops (Gln100-Gly106, see Supplementary Figure 11), from both monomers, fill the opposing cavities (Supplementary Figure 11), which together are suspected to host the carotenoid molecule in the holo-AnaCTDH dimer form as inferred from the OCPO structure. When using an apo-AnaCTDH monomer as a docking target, the β5/β6 loop from the other monomer is absent in the cavity, thus possibly enabling the carotenoid molecule to be docked in the monomeric target. Based on the scenario described above, we suggest that, upon carotenoid association, the position of these loops is altered, facilitating the binding of the carotenoid.
In addition, as already noted, the volume of holo-AnaCTDH is larger than that of apo-AnaCTDH. This could be related to a slight separation of monomers to yield a sufficiently sized cavity, long enough to accommodate the carotenoid. The distances between Trp110–Trp110’ and Tyr27–Tyr27’ in the F-type dimer (~25 and ~30 Å, respectively) are too small for the accommodation of a canthaxanthin molecule (~30 Å), especially if these amino acids should be allowed to form H-bonds with the carotenoid carbonyls. The Trp288Ala mutant of CTD-OCP was shown to transfer the carotenoid more easily than the wild type (WT)30. This implied that the Trp stabilizes the binding of the carotenoid in CTD(H)s as in full-size OCPs.
It is important to note that carotenoid-containing monomeric forms of CTD(H)s were detected previously25,30. We thus hypothesized that a monomer can temporarily accommodate the carotenoid taken up from a membrane. Carotenoid binding could occur during CTDH synthesis, although uptake into apo-AnaCTDH has been shown experimentally25. The question arises about how this is possible with the position of the C-terminal tail in apo-AnaCTDH acting as an intermediate cap of the carotenoid-binding site. As discussed, we observed that not only apo-AnaCTDH can take the carotenoid from the membrane but also from HCP1. These results suggest that the C-terminal tail moves rather easily. Moreover, our results indicate that the C-terminal tail facilitates carotenoid uptake and delivery. It was suggested that the position of the carotenoid in the monomer (completely buried) must be different to that in the dimer in which it is shared by two monomers25,30. We propose that, first, the C-terminal tail supports the uptake of the carotenoid and that structural changes (i.e., β5/β6 loop movement) assist to better host the carotenoid molecule within the first monomer as a moiety of an A-type CTDH dimer. Second, after a local energetic minimum is reached, reorganization of the monomers yields the F-type dimer while undergoing another structural change in the second monomer cavity to facilitate carotenoid binding.
CTDH role and carotenoid transfer directionality
In a previous publication, we concluded that the role of CTDH is to help the assembly of holo-HCPs25. We proposed that the carotenoid transfer process could be unidirectional, i.e., from membranes to CTDH to HCPs. The fact that holo-AnaHCP4, -HCP3, or -HCP2 cannot transfer its carotenoid to the apo-AnaCTDH supported our hypothesis, suggesting it to be a possible in vivo scenario. Here it is shown that holo-AnaCTDH can supply the carotenoid to apo-HCP1 and apo-HCP4 with similar efficiency (Fig. 4b). In addition, holo-HCP1 is formed only in the presence of CTDH since HCP1 (like HCP4) is unable to extract the carotenoid from the membrane (at least in vitro) (Fig. 4d and Muzzopappa et al. (2017)25). Thus the presence of a CTDH is essential for the formation of all HCPs25.
Nevertheless, here we showed that, at least in vitro, HCP1 (but not HCP2, HCP3, and HCP4) can transfer its carotenoid to the apo-AnaCTDH. The question arises whether this process can occur in vivo as well. It is important to note that HCP1 was proposed to be a carotenoid carrier rather than a 1O2 or phosphate-buffered saline fluorescence quencher. Thus the fact that it can supply the carotenoid to the CTDHs supports this hypothesis. This result also suggests that the structure of HCP1, which is similar to the one of the NTD-OCP19, must be slightly different from that of other HCPs, at least concerning the interface with the CTDH, since it is the only HCP that is able to transfer the carotenoid to the various CTDH isoforms.
The results described here indicate that primary and tertiary structures of both CTDH (and CTD) and HCPs (and NTD) determine the possibility and/or the directionality of carotenoid transfer from one to the other. Slight structural modifications generated during evolution most probably appeared to suppress disadvantageous carotenoid transfer pathways and to add more effective and adaptive means of regulation.
Structural position of holo-AnaCTDH subunits yields absorption differences compared to OCPO
When comparing the OCPO structure and the F-type apo-AnaCTDH dimer, the NTD-equivalent, second CTDH monomer is positioned differently in space with respect to the first (CTD super-positioned) CTDH monomer, as can be seen in Supplementary Figure 12. While the carotenoid cavity in OCPO is substantially tilted between the two domains, it is relatively straight in the apo-AnaCTDH dimer. Indeed, the carotenoid in OCPO adopts a bent conformation, while it has a planar conformation in the holo-NTD variant16. The increased chromophore planarity and rotation of the terminal rings are known to induce the red shift of absorption peak due to elongation of the effective conjugation length36,37. While some structural alterations are anticipated upon carotenoid uptake by the apo-AnaCTDH, the overall axial positions of the two monomers are not expected to be dramatically changed. Thus the carotenoid cavity in the CTDH dimer will allow a planar conformation of the carotenoid. This should be clarified by structural studies using holo-CTD(H) dimer.
CTDH carotenoid uptake and delivery cycle
In chloroplasts and cyanobacteria, various redox-activation processes of enzymes in the stroma exist. These activations occur via ferredoxin, ferredoxin-thioredoxin reductase, and thioredoxin (Trx)38,39. Trx can also be reduced by NADPH-Trx reductases. Trx also acts as an electron donor for antioxidant defense systems (e.g., peroxiredoxins, catalases)40. From the known functions of Trx, it can be clearly inferred that proteins with oxidized disulfide bonds exist in cyanobacterial cytoplasm under various conditions. The CTDH dimer therefore could also be a Trx-regulated protein (and/or other regulatory agent).
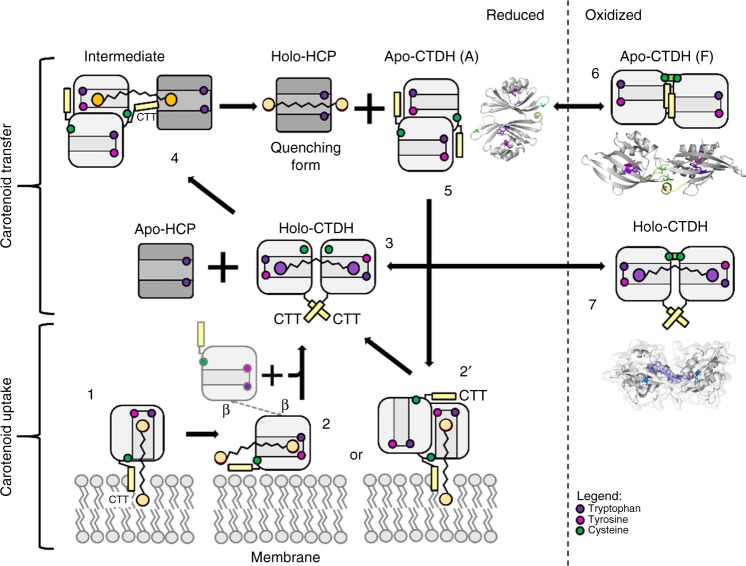
Based on these data and previous work of our laboratories, we can suggest a mechanism (Fig. 5) describing carotenoid delivery and uptake by a clade 2 CTDH. In vivo, in low light conditions or darkness (where no photoprotection is needed), Trx is more oxidized owing to low photosynthetic activity, and thus holo- and apo-CTDH dimers are stabilized by the disulfide bond between monomers that inhibits carotenoid uptake and delivery (Fig. 5, steps 6 and 7). Under conditions where photoprotection against oxidative stress is needed (reducing conditions, high light, low CO2), the Trx pool is largely reduced which in turn reduces both the apo- and holo-CTDH dimer to facilitate carotenoid transfer (Fig. 5, step 3) and uptake (Fig. 5, step 5). When reduced, and with the aid of HCP, the holo F-type dimer is converted to an A-type dimer, leaving the carotenoid bound to one monomer, bringing it one step closer to be transferred to the HCP. Then, by the assistance of the C-terminal tail, the carotenoid can be transferred directly to HCPs (Fig. 5, step 4).
Fig. 5.
CTDH carotenoid uptake and delivery cycle. apo-AnaCTDH monomer (1) or a dimer in its A-type state (2′) may extract a carotenoid molecule from the membrane with the assistance of the C-terminal tail (labeled as CTT in the figure). Then a holo-dimer is formed (3) in an F-type state (with association of another monomer; 2) Then the holo-CTDH can either be redox regulated in its F-type state between the reduced (3) and oxidized states (7) or interact with apo-HCP to allow carotenoid transfer through the A-type state with the assistance of the C-terminal tail (4). This yields the holo-HCP and the apo-CTDH dimer that can undergo A-to-F type transition and either be in its reduced state (5) and thus can take another carotenoid molecule or be modified to its oxidized states (6). Different carotenoid molecule colors represent different binding modes and thus different spectroscopic characteristics
After carotenoid transfer, the C-terminal tail remains near the carotenoid cavity and the CTDH can dimerize to yield the F-type apo-CTDH shown in this work. This apo-dimer is stable and will be able to receive a new carotenoid molecule only upon reduction of the disulfide bond. The uptake of the carotenoid from the membrane (and from HCP1) can be done by either a monomer (Fig. 5, step 1) or a reduced dimer (Fig. 5, step 2) in its A-type state; however, in either case the extension of the C-terminal tail toward the carotenoid site facilitates its translocation toward the designated cavity in the CTDH. A second monomer (Fig. 5, step 2′) is then assembled in order to co-encapsulate the carotenoid and form the F-type holo-CTDH (Fig. 5, step 3), which under regular conditions undergoes an oxidation event to yield the carotenoid-locked holo-CTDH (Fig. 5, step 7). We believe that F-type apo-dimer can be a result of two possible scenarios, which are not necessarily mutually exclusive. First, it is possible that carotenoid uptake is already completed during or immediately after CTDH synthesis before the formation of the dimer and disulfide bond. In this case, the F-type apo-dimer will be an intermediate stage along the CTDH carotenoid cycle. Nevertheless, it is also possible that the protein dimer scaffold can be formed prior to carotenoid uptake, and thus interchange between redox states can occur. In this case, F-type apo-dimer would be a preliminary stage of the cycle. These alternatives require further investigation.
Methods
Construction of plasmids containing Synechocystis ctd-ocp and Anabaena ctdh and hcp genes for expression in E. coli
The construction of the plasmid containing the Synechocystis 6803 CTD used for SAXS experiments was previously described26. The construction of plasmids containing Anabaena hcp1, hcp2, hcp3, and hcp4 genes were previously described22 and of those containing the Anabaena WT ctdh and mutated ctdh-C103F genes (pCDFCTDHAna-4940Ctag and pCDF-CTDHAna-C103F, respectively) were previously described25. The C-terminal tail was deleted by mutagenesis using the plasmid pCDF-CTDHAna-C103F and the primers F-4940-ΔC-terminal tail- (5′-GATCATCCACACCCAAATTAAACACCACCACCACCACCACTAGTCTTG-3′) and R-4940-ΔC-terminal tail (5′-TTTAATTTGGGTGTGGATGATCTGTTTTTCTTGGTTAAGAG-3′) to create the pCDF-CTDHAna-C103F-ΔC-terminal tail. In all these CTDH plasmids, the sequence coding for the His-tag were in the 3′ terminus of the gene (C-terminus of the protein).
To obtain N-terminal His-tagged CTDHs, the primers F-4940-BamHI (5′-CACACGGATCCGAAAGCTGCTGAGTCTCTCCC-3′) and R-4940-NotI (5′-CATTATGCGGCCGCCTTATTGATGACAGCGCCCC-3′) were used to amplify the ctdh gene from genomic DNA of Anabaena PCC 7120 and cloned in pCDFDuet-1 digested by BamHI and NotI. The point mutation C103F (see Muzzopappa et al. (2017)25 for primers) and the deletion of the C-terminal tail (using F-4940-ΔC-terminal tail (5′-GATCATCCACACCCAAATTAAATAGTCTTGATTAATTACTTTAAC-3′) and R-4940-ΔC-terminal tail) were introduced by mutagenesis. Then a part of the N-terminal prolongation present in these plasmids was excised by mutagenesis using the primers F-4940-ΔBamHI (5′-CATCACCATCATCACCACAAAGCTGCTGAGTCTCTCCC-3′) and R-ΔDuet (5′-GTGGTGATGATGGTGATGCATGGTATATCTCCT-3′) to create the plasmids pCDF-NHISCTDHAna-C103F and pCDF-NHISCTDHAna-C103F-ΔC-terminal tail. The proteins obtained using these plasmids contained only six His after the first Met and then the CTDH primary sequence.
HCPs and apo-CTDH production, isolation, and purification
The production and isolation of apo and holo-HCPs (Lopez-Igual et al.22) and apo- and holo-AnaCTDH were previously described25. For the apo-Ana-CTDH isolation used to crystallization, lysis buffer (40 mM Tris pH 8, 10% glycerol, 300 mM NaCl, 1 mM EDTA, 1 mM PMSF, 1 mM caproic acid, 1 mM benzamidic acid, 50 µg mL−1 DNAse) was then passed through a French Press machine. Following removal of membranes, the supernatant was further purified using a nickel affinity column (Ni-Probond resin, Invitrogen). Proteins were eluted with 250 mM imidazole and followed by dialysis with 40 mM Tris-HCl pH 8. To further disassemble AnaCTDH oligomers into smaller assemblies, the isolated protein was treated with 2 M urea and put for an overnight incubation prior to size exclusion chromatography.
Sodium dodecyl sulfate (SDS)–PAGE and Native-PAGE
Crude apo-AnaCTDH sample was analyzed as previously described25. SDS–PAGE was performed on a 15% polyacrylamide/2 M Urea in TRIS/MES system41. Non-denaturing gel electrophoresis was done to examine the oligomeric state of the apo-AnaCTDH. Following purification, CTDH was loaded onto 15% native polyacrylamide gels (pH 8), and electrophoresis was conducted in a buffer at pH 8 (25 mM Tris-HCl, 192 mM glycine). Carbonic anhydrase (29 kDa) and α-Lactalbumin (14 kDa) were used as markers.
Size exclusion chromatography
The crude apo-AnaCTDH oligomeric mixture was applied onto an analytical size exclusion chromatography (BioSEC-5, 1000 Å, Agilent, CA, USA) with 40 mM Tris-HCl (pH 8) used as elution buffer. To further purify AnaCTDH dimers, a 2 M urea-treated oligomeric mixture was applied onto a preparative size exclusion chromatography and eluted with a 40 mM Tris-HCl buffer (pH 8) containing 2 M urea. Ultraviolet detection was conducted at λabs = 225 nm with a flow rate of 0.5 mL min−1. Weight calibration was executed using the following markers: Thyroglobulin, bovine (669 kDa); Apoferritin, horse spleen (443 kDa); Albumin, bovine serum (66 kDa); Carbonic anhydrase, Erythrocyte, bovine (29 kDa), and Cytochrome C, horse heart (12.4 kDa).
Crystallization and data collection
AnaCTDH oligomer crystals diffracting to 2.9 Å were obtained at 20 °C by hanging drop diffusion method when grown in 1.26 M NaH2PO4 and 0.14 M K2HPO4, pH 5 (being an optimized condition following INDEX HT screen, Hampton research). Large football-shaped crystals were obtained after several days and data sets were collected at Technion Center for Structural Biology (TCSB). AnaCTDH dimers in 2 M urea were crystallized at 15 °C by hanging drop vapor diffusion method (final urea concentration was 1 M). AnaCTDH crystals diffracting to a resolution of 2.43 Å were obtained by growth in 0.1 M citric acid, 25% w/v PEG 3350, pH 4.2 (optimized following INDEX HT screen, Hampton research). Smaller crystals were obtained after several days, and data sets were collected at the European Synchrotron Radiation Facility (ESRF) on beamline ID 30-A1 using MXPressE automatic data collection service. The data sets for both oligomeric states were scaled and merged using MOSFLM42 and SCALA43, respectively. Molecular replacement runs were carried out using Phaser44. While the AnaCTDH oligomer structure was failed to be solved using molecular replacement, the structure of the dimeric AnaCTDH was solved and then refined using both NCSref and Phenix.refine45. Structural solution inspections and manual modifications were made using Coot46. PDBREDO server47 was utilized to minimize errors prior to deposition to the PDB. The structure was then examined and compared to other structures using PyMoL48. A section of the composite Fo-Fc omit map calculated using the Phenix protocol and visualized in Pymol is shown in Supplementary Figure 6. The overall B-factors of the CTDH structure are higher in the peripheral surfaces of the protein, perhaps due to the presence of 1 M urea in the crystallization liquor (Table 1). Three urea molecules were modeled into densities too large to be modeled as solvent and not corresponding to bound ionic species. The B-factors of the urea molecules are higher than the average protein B-factors, further indicating that the urea molecules required to avoid oligomerization are weakly bound to the protein elements.
Homology modeling
A homology-based model of the AnaCTDH structure was built using Swiss-model49, utilizing the CTD structure from OCP (PDB code: 5UI2), to serve as a template for MR attempts.
Docking
Docking simulations were executed using Swissdock server35, with the structure of the monomer, dimer, and monomer/dimer without C-terminal tail as the rigid body and canthaxanthin as the ligand to fit.
SAXS of apo-OCP-CTD
Apo-CTD from Synechocystis (residues 165–317), carrying an uncleavable N-terminal hexahistidine tag26, was analyzed in 20 mM Tris-HCl buffer (pH 7.6) containing 150 mM NaCl, 0.1 mM EDTA, 2 mM dithiothreitol, and 3% glycerol by synchrotron SAXS at the EMBL P12 beam line (PETRA III, DESY Hamburg, Germany)50. SAXS curves collected in a batch mode (1 s exposure time, collected as 20 × 50 ms frames) at different protein concentrations showed substantial concentration dependence, in line with the previous observations using size exclusion chromatography26. Since no extrapolation was possible, to ensure the predominance of the dimeric species, the SAXS data at the highest protein concentration (270 µM per monomer) were analyzed and used for modeling. The averaged SAXS curve for the sample was buffer subtracted in PRIMUS51. No radiation damage was detected by inspection of the time course of the scattering for protein frames. The Guinier region was linear and was used to determine experimental radius of gyration, Rg. Pairwise distance distribution, P(r), was calculated by GNOM52 at s ≤ 0.28 Å−1 to determine the maximum particle dimension, Dmax, and the Porod volume. Ab initio shape reconstruction was performed using DAMMIF31 and three best-fitting models were averaged using DAMAVER32 to reveal the average core common among the models (DAMFILT). Theoretical SAXS curves and fitting to the experimental data were calculated using CRYSOL53. The core models of the apo-CTD dimers were built by superposition of the Synechocystis OCP-CTDs (PDB code: 4XB5; residues 173–305 out of 317) onto the crystallographic Anabaena apo-CTDH dimers in order to preserve the subunit interfaces. Few clashes in the amino acid side chains and flexible loops were relieved manually in Coot46 in the case of the F-type apo-CTD dimer preserving the head-to-head orientation. In the case of A-type apo-CTD dimer, the local protein–protein docking of the two subunits was required to account for local differences between apo-CTD and apo-CTDH sequences and to relieve clashes. The RosettaDock server33 was used with the default set of parameters, which resulted in a more connected and realistic top-scoring model (Supplementary Figure 7), devoid of steric clashes but preserving the back-to-back subunit orientation. On the basis of the fixed core dimers (either A or F) thus obtained, the unstructured parts of the protein (23 residues in the N-terminal tail and 12 residues in the C-terminal tail) were modeled using CORAL54 to minimize the difference between the model-derived and experimental SAXS curves. The modeling procedure was repeated ten times for each scenario to verify that a stable solution is found and the data corresponding to the best-fitting solution are presented along with the statistical analysis using χ2 and correlation map P value34. To assess the robustness of the solution, the hypothetical modeling by either apo-CTD monomer or trimer (supplemented with the flexible termini) was also performed but yielded much worse fits to the SAXS data.
Absorbance measurements and experiments of carotenoid transfer
Absorbance spectra and kinetics of carotenoid transfer between CTDH and HCPs were measured in a Specord S600 spectrophotometer (Analytic Jena) at 23 °C. To study the carotenoid transfer from holo-proteins to apo-proteins, holo-HCPs to apo-CTDHs (1 holo-HCP per 2.5 apo-CTDHs molar ratio) and holo-CTDHs to apo-HCPs (1 holo-CTDH to 5 apo-HCP molar ratio) protein mixtures were incubated in 40 mM Tris-HCl buffer (pH 8) at 23 °C for 1 h in darkness. A triplicate of absorbance spectra were recorded for 1 h and carotenoid transfer was followed by changes in absorbance at 600 nm. To determine the percentage of carotenoid transferred, a spectral deconvolution was performed using Excel to fit the data to the sum of the reference spectra of the holo-proteins involved in the experiment (described in ref. 26). To study the carotenoid transfer from membranes to HCPs and CTDHs, 12 µM apo-dimers were incubated with an E. coli canthaxanthin-containing membrane suspension (48 µM canthaxanthin, measured by acetone extraction) at 33 °C for 1 h in darkness. Holo-protein formation was measured by absorbance spectroscopy after precipitation of membranes. The percentage of holo-protein formed was determined by comparing the spectra of 100% holo-proteins (at 12 µM) to those of the supernatant.
Data availability
The data sets generated during the current study are available in the Protein Data Bank repository, [https://www.rcsb.org/] under accession code 6FEJ. All other data sets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Electronic supplementary material
Acknowledgements
This work was supported by the Israel Science Foundation founded by the Israel Academy of Sciences and Humanities (843/16), grants from the Agence Nationale de la Recherche RECYFUEL [ANR-16-CE05-0026]), and by the European Union’s Horizon 2020 research and innovation program under grant agreement no. 675006 (SE2B). F.M.’s salary is financed by SE2B. The research is also supported by the Centre National de la Recherche Scientifique and the Commissariat à l’Energie Atomique. We gratefully thank the staff of the ESRF (beamline ID30A-1/MASSIF-1) for provision of synchrotron radiation facilities and assistance in data collection. We would also like to thank Dr. Yael Pazy-Benhar and the Technion Center for Structural Biology (TCSB) for assisting with radiation facilities and data collection. N.N.S. is grateful to Yury B. Slonimskiy (A.N. Bach Institute of Biochemistry) for help with protein purification. T.F. acknowledges support by the German Federal Ministry of Education and Research (WTZ-RUS grant 01DJ15007) and the German Research Foundation (Cluster of Excellence “Unifying Concepts in Catalysis” and FR 1276/5-1). N.N.S. acknowledges the Russian Foundation for Basic Research grant (18-04-00691). This study was supported by Russian Scientific Foundation project No. 18-44-04002.
Author contributions
N.A., D.K. and E.G.M. conceived the idea. D.H., D.K., E.G.M. and N.A. designed the experiments. D.H., A.W., F.M. and N.N.S. performed all of the structural and biochemical experiments. D.H., D.K., E.G.M., T.F. and N.A. analyzed the data. D.H., N.N.S., T.F., E.G.M., D.K. and N.A. wrote the paper.
Competing interests
The authors declare no competing Interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Diana Kirilovsky, Email: diana.kirilovsky@cea.fr.
Noam Adir, Email: noam@ch.technion.ac.il.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s42003-018-0132-5.
References
- 1.Blankenship RE. Origin and early evolution of photosynthesis. Photosynth. Res. 1992;33:91–111. doi: 10.1007/BF00039173. [DOI] [PubMed] [Google Scholar]
- 2.Adir N. Elucidation of the molecular structures of components of the phycobilisome: reconstructing a giant. Photosynth. Res. 2005;85:15–32. doi: 10.1007/s11120-004-2143-y. [DOI] [PubMed] [Google Scholar]
- 3.Krieger-Liszkay A, Fufezan C, Trebst A. Singlet oxygen production in photosystem II and related protection mechanism. Photosynth. Res. 2008;98:551–564. doi: 10.1007/s11120-008-9349-3. [DOI] [PubMed] [Google Scholar]
- 4.Niyogi KK, Truong TB. Evolution of flexible non-photochemical quenching mechanisms that regulate light harvesting in oxygenic photosynthesis. Curr. Opin. Plant Biol. 2013;16:307–314. doi: 10.1016/j.pbi.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 5.Wilson A, et al. A soluble carotenoid protein involved in phycobilisome-related energy dissipation in cyanobacteria. Plant Cell. 2006;18:992–1007. doi: 10.1105/tpc.105.040121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson A, et al. A photoactive carotenoid protein acting as light intensity sensor. Proc. Natl. Acad. Sci. USA. 2008;105:12075–12080. doi: 10.1073/pnas.0804636105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gwizdala M, Wilson A, Omairi-Nasser A, Kirilovsky D. Characterization of the synechocystis PCC 6803 fluorescence recovery protein involved in photoprotection. Biochim. Biophys. Acta. 2013;1827:348–354. doi: 10.1016/j.bbabio.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 8.Sedoud A, et al. The cyanobacterial photoactive orange carotenoid protein is an excellent singlet oxygen quencher. Plant Cell. 2014;26:1781–1791. doi: 10.1105/tpc.114.123802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kerfeld CA, et al. The crystal structure of a cyanobacterial water-soluble carotenoid binding protein. Structure. 2003;11:55–65. doi: 10.1016/S0969-2126(02)00936-X. [DOI] [PubMed] [Google Scholar]
- 10.Leverenz RL, et al. Structural and functional modularity of the orange carotenoid protein: distinct roles for the N- and C-terminal domains in cyanobacterial photoprotection. Plant Cell. 2014;26:426–437. doi: 10.1105/tpc.113.118588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gwizdala M, Wilson A, Kirilovsky D. In vitro reconstitution of the cyanobacterial photoprotective mechanism mediated by the Orange Carotenoid Protein in Synechocystis PCC 6803. Plant Cell. 2011;23:2631–2643. doi: 10.1105/tpc.111.086884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sutter M, et al. Crystal structure of the FRP and identification of the active site for modulation of OCP-mediated photoprotection in cyanobacteria. Proc. Natl. Acad. Sci. USA. 2013;110:10022–10027. doi: 10.1073/pnas.1303673110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boulay C, Wilson A, D’Haene S, Kirilovsky D. Identification of a protein required for recovery of full antenna capacity in OCP-related photoprotective mechanism in cyanobacteria. Proc. Natl. Acad. Sci. 2010;107:11620–11625. doi: 10.1073/pnas.1002912107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sluchanko NN, et al. The purple Trp288Ala mutant of synechocystis OCP persistently quenches phycobilisome fluorescence and tightly interacts with FRP. Biochim. Biophys. Acta. 2017;1858:1–11. doi: 10.1016/j.bbabio.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 15.Wilson A, et al. Structural determinants underlying photoprotection in the photoactive orange carotenoid protein of cyanobacteria. J. Biol. Chem. 2010;285:18364–18375. doi: 10.1074/jbc.M110.115709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leverenz RLL, et al. A 12 A carotenoid translocation in a photoswitch associated with cyanobacterial photoprotection. Science. 2015;348:1463–1466. doi: 10.1126/science.aaa7234. [DOI] [PubMed] [Google Scholar]
- 17.Gupta S, et al. Local and global structural drivers for the photoactivation of the orange carotenoid protein. Proc. Natl. Acad. Sci. 2015;112:E5567–E5574. doi: 10.1073/pnas.1512240112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirilovsky D, Kerfeld CA. Cyanobacterial photoprotection by the orange carotenoid protein. Nat. Plants. 2016;2:16180. doi: 10.1038/nplants.2016.180. [DOI] [PubMed] [Google Scholar]
- 19.Melnicki MR, et al. Structure, diversity, and evolution of a new family of soluble carotenoid-binding proteins in cyanobacteria. Mol. Plant. 2016;9:1379–1394. doi: 10.1016/j.molp.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 20.Kerfeld CA. Water-soluble carotenoid proteins of cyanobacteria. Arch. Biochem. Biophys. 2004;430:2–9. doi: 10.1016/j.abb.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 21.Kirilovsky D, Kerfeld CA. The orange carotenoid protein in photoprotection of photosystem II in cyanobacteria. Biochim. Biophys. Acta. 2012;1817:158–166. doi: 10.1016/j.bbabio.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 22.Lopez-Igual R, et al. Different functions of the paralogs to the N-terminal domain of the Orange Carotenoid Protein in the cyanobacterium Anabaena sp. PCC 7120. Plant Physiol. 2016;171:1852–1866. doi: 10.1104/pp.16.00502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bao H, Melnicki MR, Kerfeld CA. Structure and functions of Orange Carotenoid Protein homologs in cyanobacteria. Curr. Opin. Plant Biol. 2017;37:1–9. doi: 10.1016/j.pbi.2017.03.010. [DOI] [PubMed] [Google Scholar]
- 24.Lechno-Yossef S, Melnicki MR, Bao H, Montgomery BL, Kerfeld CA. Synthetic OCP heterodimers are photoactive and recapitulate the fusion of two primitive carotenoproteins in the evolution of cyanobacterial photoprotection. Plant J. 2017;91:646–656. doi: 10.1111/tpj.13593. [DOI] [PubMed] [Google Scholar]
- 25.Muzzopappa F, et al. The paralogs to the C-terminal domain of the cyanobacterial OCP are carotenoid donors to HCPs. Plant Physiol. 2017;175:1283–1303. doi: 10.1104/pp.17.01040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moldenhauer M, et al. Assembly of photoactive orange carotenoid protein from its domains unravels a carotenoid shuttle mechanism. Photosynth. Res. 2017;133:327–341. doi: 10.1007/s11120-017-0353-3. [DOI] [PubMed] [Google Scholar]
- 27.Dines M, Sendersky E, David L, Schwarz R, Adir N. Structural, functional, and mutational analysis of the NblA protein provides insight into possible modes of interaction with the phycobilisome. J. Biol. Chem. 2008;283:30330–30340. doi: 10.1074/jbc.M804241200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dines M, Sendersky E, Schwarz R, Adir N. Crystallization of sparingly soluble stress-related proteins from cyanobacteria by controlled urea solublization. J. Struct. Biol. 2007;158:116–121. doi: 10.1016/j.jsb.2006.10.021. [DOI] [PubMed] [Google Scholar]
- 29.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 30.Maksimov EG, et al. The unique protein-to-protein carotenoid transfer mechanism. Biophys. J. 2017;113:402–414. doi: 10.1016/j.bpj.2017.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2003;36:860–864. doi: 10.1107/S0021889803000268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lyskov S, Gray JJ. The RosettaDock server for local protein–protein docking. Nucleic Acids Res. 2008;36:W233–W238. doi: 10.1093/nar/gkn216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Franke D, Jeffries CM, Svergun DI. Correlation map, a goodness-of-fit test for one-dimensional X-ray scattering spectra. Nat. Methods. 2015;12:419–422. doi: 10.1038/nmeth.3358. [DOI] [PubMed] [Google Scholar]
- 35.Grosdidier A, Zoete V, Michielin O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011;39:W270–W277. doi: 10.1093/nar/gkr366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cianci M, et al. The molecular basis of the coloration mechanism in lobster shell: β-crustacyanin at 3.2-Å resolution. Proc. Natl. Acad. Sci. 2002;99:9795–9800. doi: 10.1073/pnas.152088999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng PP, et al. The structure of allophycocyanin B from Synechocystis PCC 6803 reveals the structural basis for the extreme redshift of the terminal emitter in phycobilisomes. Acta Crystallogr. D. Biol. Crystallogr. 2014;70:2558–2569. doi: 10.1107/S1399004714015776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindahl M, Kieselbach T. Disulphide proteomes and interactions with thioredoxin on the track towards understanding redox regulation in chloroplasts and cyanobacteria. J. Proteomics. 2009;72:416–438. doi: 10.1016/j.jprot.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 39.Florencio FJ, Pérez-Pérez ME, López-Maury L, Mata-Cabana A, Lindahl M. The diversity and complexity of the cyanobacterial thioredoxin systems. Photosynth. Res. 2006;89:157–171. doi: 10.1007/s11120-006-9093-5. [DOI] [PubMed] [Google Scholar]
- 40.Lindahl M, Florencio FJ. Thioredoxin-linked processes in cyanobacteria are as numerous as in chloroplasts, but targets are different. Proc. Natl. Acad. Sci. 2003;100:16107–16112. doi: 10.1073/pnas.2534397100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kashino Y, Koike H, Satoh K. An improved sodium dodecyl sulfate‐polyacrylamide gel electrophoresis system for the analysis of membrane protein complexes. Electrophoresis. 2001;22:1004–1007. doi: 10.1002/1522-2683()22:6<1004::AID-ELPS1004>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 42.Battye, T. G. G., Kontogiannis, L., Johnson, O., Powell, H. R. & Leslie, A. G. W. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Cryst. 67, 271-281 (2011). [DOI] [PMC free article] [PubMed]
- 43.Evans P. Scaling and assessment of data quality. Acta Crystallogr. Sect. D. Biol. Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 44.McCoy AJ, et al. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix. refine. Acta Crystallogr. Sect. D. Biol. Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 47.Joosten RP, Long F, Murshudov GN, Perrakis A. The PDB_REDO server for macromolecular structure model optimization. IUCrJ. 2014;1:213–220. doi: 10.1107/S2052252514009324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DeLano, W. L. The PyMOL Molecular Graphics System. http://www.pymol.org (2002).
- 49.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 50.Blanchet CE, et al. Versatile sample environments and automation for biological solution X-ray scattering experiments at the P12 beamline (PETRA III, DESY) J. Appl. Crystallogr. 2015;48:431–443. doi: 10.1107/S160057671500254X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003;36:1277–1282. doi: 10.1107/S0021889803012779. [DOI] [Google Scholar]
- 52.Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992;25:495–503. doi: 10.1107/S0021889892001663. [DOI] [Google Scholar]
- 53.Svergun D, Barberato C, Koch MHJ. CRYSOL–a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 1995;28:768–773. doi: 10.1107/S0021889895007047. [DOI] [Google Scholar]
- 54.Petoukhov MV, et al. New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 2012;45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets generated during the current study are available in the Protein Data Bank repository, [https://www.rcsb.org/] under accession code 6FEJ. All other data sets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.