Abstract
Successful outcomes of surgical cancer resection necessitate negative, cancer-free surgical margins. Currently, tissue samples are sent to pathology for diagnostic confirmation. Hyperspectral imaging (HSI) is an emerging, non-contact optical imaging technique. A reliable optical method could serve to diagnose and biopsy specimens in real-time. Using convolutional neural networks (CNNs) as a tissue classifier, we developed a method to use HSI to perform an optical biopsy of ex-vivo surgical specimens, collected from 21 patients undergoing surgical cancer resection. Training and testing on samples from different patients, the CNN can distinguish squamous cell carcinoma (SCCa) from normal aerodigestive tract tissues with an area under the curve (AUC) of 0.82, 81% accuracy, 81% sensitivity, and 80% specificity. Additionally, normal oral tissues can be sub-classified into epithelium, muscle, and glandular mucosa using a decision tree method, with an average AUC of 0.94, 90% accuracy, 93% sensitivity, and 89% specificity. After separately training on thyroid tissue, the CNN differentiates between thyroid carcinoma and normal thyroid with an AUC of 0.95, 92% accuracy, 92% sensitivity, and 92% specificity. Moreover, the CNN can discriminate medullary thyroid carcinoma from benign multi-nodular goiter (MNG) with an AUC of 0.93, 87% accuracy, 88% sensitivity, and 85% specificity. Classical-type papillary thyroid carcinoma is differentiated from benign MNG with an AUC of 0.91, 86% accuracy, 86% sensitivity, and 86% specificity. Our preliminary results demonstrate that an HSI-based optical biopsy method using CNNs can provide multi-category diagnostic information for normal head-and-neck tissue, SCCa, and thyroid carcinomas. More patient data are needed in order to fully investigate the proposed technique to establish reliability and generalizability of the work.
Keywords: Hyperspectral imaging, convolutional neural network, deep learning, optical biopsy, intraoperative imaging, head and neck surgery, head and neck cancer
1. INTRODUCTION
Estimated cancer incidence world-wide in 2012 for cancers of the lip, oral cavity, nasopharynx, other pharynx, larynx, and thyroid combined was 1.4 million people newly diagnosed. This corresponds with an age-standardized incidence rate of about 25 men out of 100,000 and 14 women out of 100,000 diagnosed in 2012 alone with these types of cancer.1 Approximately 90% of cancer at sites including the lips, gums, mouth, plate, and anterior two-thirds of the tongue are squamous cell carcinoma (SCCa).2 There were over 800,000 cancer deaths in 2012 from these types of cancer.1 The procedure for diagnosing these cancers is a tissue biopsy, and cases usually require surgical resection. Surgical resection is required for treatment of these cancers in order to prevent disease recurrence.3
Hyperspectral imaging (HSI) is a non-contact optical imaging modality capable of acquiring hundreds of images at multiple wavelengths. Preliminary research demonstrates that HSI may contain potential for providing diagnostic information for a myriad of diseases.4 Convolutional neural networks (CNNs), a type of machine learning and artificial intelligence algorithm, have demonstrated astonishing ability at image classification tasks.5,6 This study aims to investigate the ability of HSI to classify tissues from the thyroid, aerodigestive tract, and oral cavity, using convolutional neural networks. Both normal and cancerous tissue classification performance is investigated. If proven reliable, this method could help provide diagnostic information, perhaps as a computer-aided diagnostic tool for physicians diagnosing and treating these types of cancer.
2. METHODS
To investigate the ability of HSI to perform optical biopsy, we recruited human cancer patients into our study, acquired and processed gross-level HSI of freshly excised tissue specimens, trained our convolutional neural network, and evaluated system performance.
2.1. Experimental Design
In collaboration with the Otolaryngology Department and the Department of Pathology and Laboratory Medicine at Emory University Hospital Midtown, 21 head and neck cancer patients who were electing to undergo surgical cancer resection were recruited for our study to evaluate the efficacy of using HSI for optical biopsy.7,8 From these 21 patients, a total of 60 excised tissue samples were collected. Three tissue samples were collected from each patient, i.e., a sample of the tumor, a normal tissue sample, and a sample at the tumor-normal interface. The collected tissues were kept in cold PBS during transport to the imaging laboratory where the specimens were scanned with a hyperspectral imaging system.9,10
Two regions of interest were used for this study: firstly, the upper aerodigestive tract sites, including tongue, larynx, pharynx, and mandible; and secondly, the thyroid and associated carcinomas. Head and neck squamous cell carcinoma (HNSCCa) of the aerodigestive tract represented the first group, comprised of 7 patients. Normal tissue was obtained from all patients in the HNSCCa group, and SCCa was obtained from 6 of these patients. The thyroid group consisted of 14 patients total and included one benign neoplasm and three malignant neoplasms: benign multi-nodular goiter (MNG, 3 patients), classical-type papillary thyroid carcinoma (cPTC, 4 patients), follicular-type papillary thyroid carcinoma (fPTC, 4 patients), and medullary thyroid carcinoma (MTC, 3 patients), respectively.
After imaging with HSI, tissues were fixed in formalin, underwent haemotoxylin and eosin (H&E) staining, paraffin embedded, sectioned, and digitized. A head and neck specialized, certified pathologist confirmed the diagnoses of the ex-vivo tissues using the digitized histology slides in Aperio ImageScope (Leica Biosystems Inc, Buffalo Grove, IL, USA). The histological images serve as the ground truth for the experiment.
2.2. Hyperspectral Imaging and Preprocessing
The hyperspectral images were acquired using a CRI Maestro imaging system (Perkin Elmer Inc., Waltham, Massachusetts), which is comprised of a Xenon white-light illumination source, a liquid crystal tunable filter, and a 16-bit charge-coupled device (CCD) camera capturing images at a resolution of 1040 by 1,392 pixels and a spatial resolution of 25 μm per pixel.7,9,11,12 The hypercube contains 91 spectral bands, ranging from 450 to 900 nm with a 5 nm spectral sampling interval.
The hyperspectral data were normalized at each wavelength (λ) sampled for all pixels (i, j) by subtracting the inherent dark current (captured by imaging with a closed camera shutter) and dividing by a white reference disk according to the following equation.8,12
| (1) |
Specular glare is created on the tissue surfaces due to wet surfaces completely reflecting incident light. Glare pixels do not contain useful spectral information for tissue classification and are hence removed from each HSI by converting the RGB composite image of the hypercube to grayscale and experimentally setting an intensity threshold that sufficiently removes the glare pixels, assessed by visual inspection.
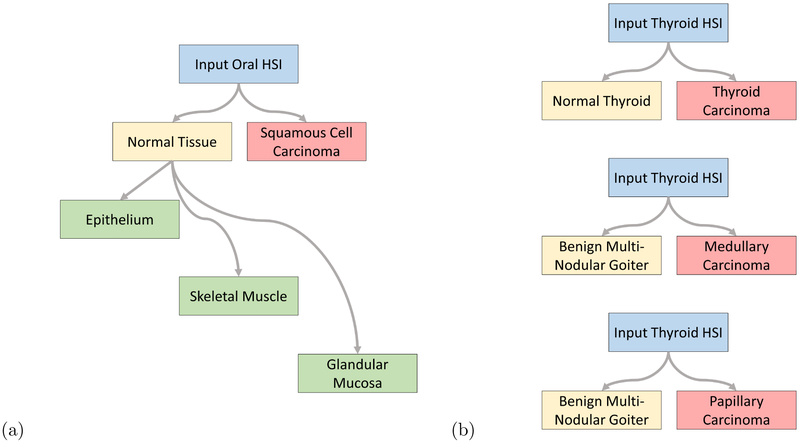
A schematic of the classification scheme is shown in Figure 1. For binary cancer classification, the classes used are normal aerodigestive tissue versus SCCa, and medullary and papillary thyroid carcinoma versus normal thyroid tissue. For multi-class classification of oral and aerodigestive tract tissue, epithelium, skeletal muscle, and gland are used. In addition, for multi-class sub-classification, the number of normal samples were augmented by 90, 180, and 270 degree rotations and vertical and horizontal reflections, to produce six times the number of samples. For multi-class classification of thyroid cancer, classical-type papillary thyroid carcinoma, medullary thyroid carcinoma, and multi-nodular thyroid goiter tissue are used.
Figure 1:
Tissue classification scheme.
For training and testing the CNN, each patient HSI needs to be divided into patches. Patches are produced from each HSI after normalization and glare removal to create 25×25×91 non-overlapping patches that do not include any ‘black-holes’ where pixels have been removed due to specular glare, see Table 1. Using the binary mask created from the gold-standard, the areas of normal tissue were investigated under histology to extract regions of interest.
Table 1:
Patch-based data for CNN classification.
| Class | No. Patients | Total Patches | |
|---|---|---|---|
| Thyroid | Normal Thyroid | 11 | 14,491 |
| Benign MNG | 3 | 9,778 | |
| MTC | 3 | 10,334 | |
| Classical PTC | 4 | 6,836 | |
| Follicular PTC | 4 | 13,200 | |
| HNSCCa | Epithelium | 4 | 6,366 |
| Skeletal Muscle | 3 | 5,238 | |
| Mucosal Gland | 4 | 5,316 | |
| SCCa | 6 | 4,008 | |
2.3. Convolutional Neural Network
To classify thyroid tissues, a 3D-CNN based on AlexNet, an ImageNet classification model, was implemented using TensorFlow.5,13 The model consisted of six convolutional layers with 50, 45, 40, 35, 30, and 25 convolutional filters, respectively. Convolutions were performed with a convolutional kernel of 5×5×9, which correspond to the x-y-λ dimensions. Following the convolutional layers were two fully connected layers of 400 and 100 neurons each. A drop-out rate of 80% was applied after each layer. Convolutional units were activated using rectified linear units (ReLu) with Xavier convolutional initializer and a 0.1 constant initial neuron bias.14 Step-wise training was done in batches of 10 patches for each step. Every one thousand steps the validation performance was evaluated and the training data were randomly shuffled for improved training. Training was done using the AdaDelta, adaptive learning, optimizer for reducing the cross-entropy loss with an epsilon of 1×10−8 and rho of 0.90.15 For thyroid normal versus carcinoma, the training was performed at a learning rate of 0.1 for two to six thousand steps depending on the group. For MNG versus MTC and for MNG versus cPTC, the training was done at a learning rate of 0.005 for exactly two thousand steps for all groups.
To classify oral cavity tissues, the AlexNet CNN architecture was expanded to include an adapted version of the inception module that does not include max-pools and uses larger convolutional kernels, implemented using TensorFlow.5,6,13 As shown in Figure 2, the modified inception module simultaneously performs a series of convolutions with different kernel sizes: a 1×1 convolution; and convolutions with 3×3, 5×5, and 7×7 kernels following a 1×1 convolution. The model consisted of two consecutive inception modules, followed by a traditional convolutional layer with a 9×9 kernel, followed by a final inception module. After the convolutional layers were two consecutive fully connected layers, followed by a final soft-max layer equal to the number of classes. A dropout rate of 60% was applied after each layer. For binary classification, the number of convolutional filters were 355, 350, 75, and 350, and the fully connected layers had 256 and 218 neurons. For multi-class classification, the number of convolutional filters were 496, 464, 36, and 464, and the fully connected layers had 1024 and 512 neurons. Convolutional units were activated using rectified linear units (ReLu) with Xavier convolutional initializer and a 0.1 constant initial neuron bias.14 Step-wise training was done in batches of 10 (for binary) or 15 (for multi-class) patches for each step. Every one thousand steps the validation performance was evaluated and the training data were randomly shuffled for improved training. Training was done using the AdaDelta, adaptive learning, optimizer for reducing the cross-entropy loss with an epsilon of 1×10−8 (for binary) or 1×10−9 (for multi-class) and rho of 0.8 (for binary) or 0.95 (for multi-class).15 For normal oral tissue versus SCCa binary classification, the training was done at a learning rate of 0.05 for five to fifteen thousand steps depending on the patient-held-out iteration. For multi-class sub-classification of normal oral tissues, the training was done at a learning rate of 0.01 for three to five thousand steps depending on the patient-held-out iteration.
Figure 2:
Modified inception module.
2.4. Validation
The final layer of the CNN labels each test case as the class with the highest probability overall, so each test patch has exactly one label. In addition, the probabilities of each test patch belonging to all classes are output from the network. The class probabilities for all patches of a test patient case are used to construct receiver operator characteristic (ROC) curves using MATLAB (MathWorks Inc, Natick, MA, USA). For binary classification, only one ROC curve is created per patient test case, but for multi-class classification, each class is used to generate a respective ROC curve; true positive rate and false positive rate are calculated as that class against all others.
The CNN classification performance was evaluated using leave-one-patient-out external-validation to calculate the sensitivity, specificity, and accuracy, defined below, using the optimal operating point of each patient’s ROC curve.8,10
| (2) |
| (3) |
| (4) |
Where TP, TN, FP, and FN represent the number of true positives, true negatives, false positives, and false negatives, respectively. The area under the curve (AUC) for the ROC curves is calculated as well and averaged across patients.
3. RESULTS
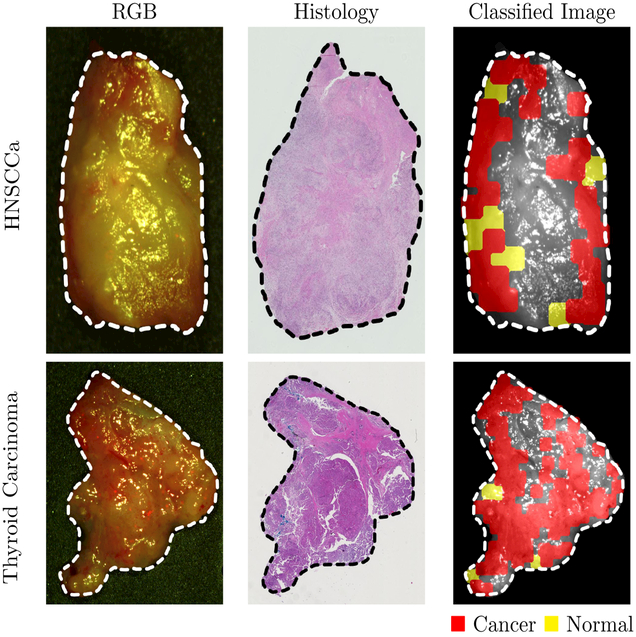
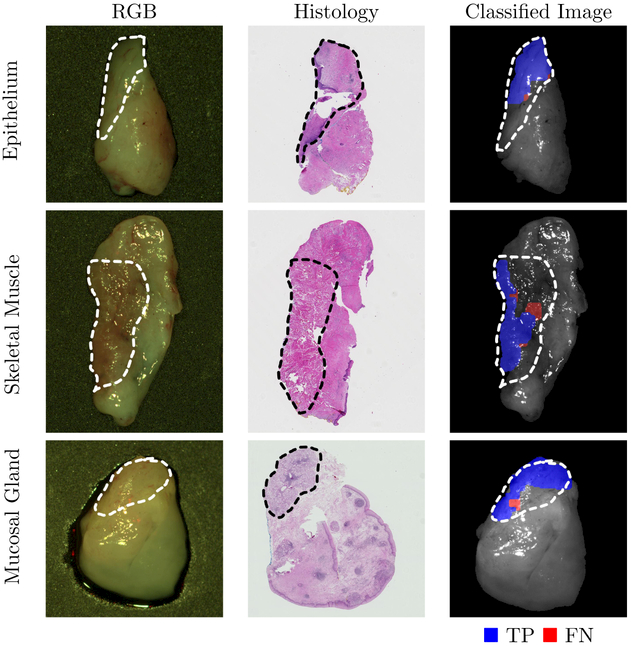
Training and testing on samples from different patients, the CNN can distinguish squamous cell carcinoma (SCCa) from normal oral tissues with an AUC of 0.82, 81% accuracy, 81% sensitivity, and 80% specificity. Representative HNSCCa classification results are shown in Figure 3. Additionally, normal oral tissues can be sub-classified into epithelium, muscle, and glandular mucosa using a decision tree method, with an average AUC of 0.94, 90% accuracy, 93% sensitivity, and 89% specificity. See Table 2 for the full results. Representative normal sub-classification results are shown in Figure 4 and full results are in Table 3.
Figure 3:
Representative results of binary cancer classification. Left: HSI-RGB composite images with cancer ROI outlined. Center: Respective histological gold standard with corresponding ROI outlined. Right: Artificially colored CNN classification results. True positive results representing correct cancer identification are visualized in red, and false negatives representing incorrect normal identification are shown in yellow. Tissue shown in grayscale represents tissue that is not classified due to the tissue surface containing glare pixels causing insufficient area to produce the necessary patch-size for classification.
Table 2:
Results of inter-patient CNN classification (leave-one-patient-out validation).
| No. Patients | AUC | Accuracy | Sensitivity | Specificity | ||
|---|---|---|---|---|---|---|
| Thyroid | Normal vs. Carcinoma | 11 | 0.95 ± 0.07 | 92 ± 9% | 92 ± 8% | 92 ± 10% |
| PTC vs MNG | 7 | 0.91 ± 0.10 | 86 ± 13% | 86 ± 14% | 86 ± 13% | |
| MTC vs MNG | 6 | 0.93 ± 0.04 | 87 ± 5% | 88 ± 4% | 85 ± 7% | |
| HNSCCa | Normal vs. SCCa | 6 | 0.82 ± 0.13 | 81 ± 11% | 81 ± 15% | 80 ± 16% |
| Multi-Class | 7 | 0.94 ± 0.08 | 90 ± 9% | 93 ± 6% | 89 ± 13% | |
Figure 4:
Representative results of sub-classification of normal oral tissues. Left: HSI-RGB composites are shown with ROI of the tissue type outlined. Center: Respective histological gold standard with corresponding ROI outlined. Right: Artificially colored CNN classification results of the ROI only. True positive results representing correct tissue-subtype are visualized in blue, and false negatives are shown in red. Tissue within the ROI that is shown in grayscale represents tissue that is not classified due to glare pixels or insufficient area to produce the necessary patch-size.
Table 3:
Results of inter-patient CNN classification of sub-classified normal oral tissues.
| No. Patients | AUC | Accuracy | Sensitivity | Specificity | |
|---|---|---|---|---|---|
| Epithelium | 4 | 0.94 ± 0.06 | 90 ± 5% | 91 ± 3% | 91 ± 7% |
| Skeletal Muscle | 3 | 0.99 ± 0.01 | 98 ± 3% | 98 ± 3% | 97 ± 4% |
| Mucosal Gland | 4 | 0.89 ± 0.10 | 83 ± 13% | 90 ± 8% | 79 ± 18% |
After separately training on thyroid tissue, the CNN differentiates between thyroid carcinoma and normal thyroid with an AUC of 0.95, 92 accuracy, 92% sensitivity, and 92% specificity. Representative thyroid carcinoma classification results are shown in Figure 3. Moreover, the CNN can discriminate MTC from benign MNG with an AUC of 0.93, 87% accuracy, 88% sensitivity, and 85% specificity. Classical-type papillary thyroid carcinoma is differentiated from MNG with an AUC of 0.91, 86% accuracy, 86% sensitivity, and 86% specificity.
4. CONCLUSION
We developed a deep learning based classification method for hyperspectral images of fresh surgical specimens. The study demonstrated the ability of HSI and convolutional neural networks for discriminating between normal tissue and carcinoma. The novel results of normal tissue sub-classification into categories of epithelium, skeletal muscle, and glandular mucosa demonstrate that there is further classification potential for HSI.
In this study, the limited patient dataset reduces the generalizability of the results. In addition, the ROI technique for outlining tissues of interest for normal multi-class sub-classification creates the potential to introduce error into the experiment. Both of these issues could be resolved by utilizing a large number of patient data. By acquiring and processing more patient HSI data, the artificially intelligent technique will allow studies of more tissue types and potentially produce results with a more universal application. Further work involves investigating the multiple pre-processing approaches and refining the proposed deep learning architectures.
ACKNOWLEDGMENTS
This research is supported in part by NIH grants CA176684, CA156775 and CA204254, Georgia Cancer Coalition Distinguished Clinicians and Scientists Award, and a pilot project fund from the Winship Cancer Institute of Emory University under the award number P30CA138292. The authors would like to thank the surgical pathology team at Emory University Hospital Midtown including Andrew Balicki, Jacqueline Ernst, Tara Meade, Dana Uesry, and Mark Mainiero, for their help in collecting fresh tissue specimens.
Footnotes
DISCLOSURES
The authors have no relevant financial interests in this article and no potential conflicts of interest to disclose. Informed consent was obtained from all patients in accordance with Emory Institutional Review Board policies under the Head and Neck Satellite Tissue Bank protocol.
REFERENCES
- [1].Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin D, Forman D, and Bray F, “GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11.” http://globocan.iarc.fr (2013). [DOI] [PubMed] [Google Scholar]
- [2].Joseph LJ, Goodman M, Higgins K, Pilai R, Ramalingam SS, Magliocca K, Patel MR, El-Deiry M, Wadsworth JT, Owonikoko TK, Beitler JJ, Khuri FR, Shin DM, and Saba NF, “Racial disparities in squamous cell carcinoma of the oral tongue among women: A SEER data analysis,” Oral Oncology 51(6), 586–592 (2015). [DOI] [PubMed] [Google Scholar]
- [3].Kim BY, Choi JE, Lee E, Son YI, Baek CH, Kim SW, and Chung MK, “Prognostic factors for recurrence of locally advanced differentiated thyroid cancer,” Journal of Surgical Oncology (2017). [DOI] [PubMed] [Google Scholar]
- [4].Lu G and Fei B, “Medical hyperspectral imaging: a review,” Journal of Biomedical Optics 19(1) (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Krizhevsky A, Sutskever I, and Hinton GE, “Imagenet classification with deep convolutional neural networks,” Proceedings of the 25th International Conference on Neural Information Processing Systems 1, 1097–1105 (2012). [Google Scholar]
- [6].Szegedy C, Liu W, Jia Y, Sermanet P, Reed SE, Anguelov D, Erhan D, Vanhoucke V, and Rabinovich A, “Going deeper with convolutions,” 2015 IEEE Conference on Computer Vision and Pattern Recognition (2015). [Google Scholar]
- [7].Fei B, Lu G, Wang X, Zhang H, Little JV, Patel MR, Griffith CC, El-Diery MW, and Chen AY, “Label-free reflectance hyperspectral imaging for tumor margin assessment: a pilot study on surgical specimens of cancer patients,” Journal of Biomedical Optics 22(8) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Fei B, Lu G, Halicek MT, Wang X, Zhang H, Little JV, Magliocca KR, Patel M, Griffith CC, El-Deiry MW, and Chen AY, “Label-free hyperspectral imaging and quantification methods for surgical margin assessment of tissue specimens of cancer patients,” in [2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC)], 4041–4045 (July 2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Halicek M, Lu G, Little JV, Wang X, Patel M, Griffith CC, El-Deiry MW, Chen AY, and Fei B, “Deep convolutional neural networks for classifying head and neck cancer using hyperspectral imaging,” Journal of Biomedical Optics 22(6) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lu G, Little JV, Wang X, Zhang H, Patel M, Griffith CC, El-Deiry M, Chen AY, and Fei B, “Detection of head and neck cancer in surgical specimens using quantitative hyperspectral imaging,” Clinical Cancer Research (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lu G, Wang D, Qin X, Muller S, Wang X, Chen AY, Chen ZG, and Fei B, “Detection and delineation of squamous neoplasia with hyperspectral imaging in a mouse model of tongue carcinogenesis,” Journal of Biophotonics (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lu G, Wang D, Qin X, Halig L, Muller S, Zhang H, Chen A, Pogue BW, Chen Z, and Fei B, “Framework for hyperspectral image processing and quantification for cancer detection during animal tumor surgery,” Journal of Biomedical Optics 20(12) (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, Corrado GS, Davis A, Dean J, Devin M, Ghemawat S, Goodfellow I, Harp A, Irving G, Isard M, Jia Y, Jozefowicz R, Kaiser L, Kudlur M, Levenberg J, Mané D, Monga R, Moore S, Murray D, Olah C, Schuster M, Shlens J, Steiner B, Sutskever I, Talwar K, Tucker P, Vanhoucke V, Vasudevan V, Viégas F, Vinyals O, Warden P, Wattenberg M, Wicke M, Yu Y, and Zheng X, “TensorFlow: Large-scale machine learning on heterogeneous systems,” (2015). Software available from tensorflow.org. [Google Scholar]
- [14].Glorot X and Bengio Y, “Understanding the difficulty of training deep feedforward neural networks,” in [Proceedings of the Thirteenth International Conference on Artificial Intelligence and Statistics], Teh YW and Titterington M, eds., Proceedings of Machine Learning Research 9, 249–256, PMLR, Chia Laguna Resort, Sardinia, Italy: (13–15 May 2010). [Google Scholar]
- [15].Zeiler MD, “ADADELTA: an adaptive learning rate method,” CoRR (2012). [Google Scholar]