Abstract
Successfully implementing pharmacogenomics into routine clinical practice requires an efficient process to order genetic tests and report the results to clinicians and patients. Lack of standardized approaches and terminology in clinical laboratory processes, ordering of the test and reporting of test results all impede this workflow. Expert groups such as the Association for Molecular Pathology and the Clinical Pharmacogenetics Implementation Consortium have published recommendations for standardizing laboratory genetic testing, reporting and terminology. Other resources such as PharmGKB, ClinVar, ClinGen and PharmVar have established databases of nomenclature for pharmacogenetic alleles and variants. Opportunities remain to develop new standards and further disseminate existing standards which will accelerate the implementation of pharmacogenomics.
Keywords: : CPIC, implementation, pharmacogenetics, pharmacogenomics, PharmGKB, standardization
Over the past two decades, implementation of pharmacogenomics into routine clinical practice has been steadily increasing and moving beyond the limits of a few select genes into a broader and preemptive application of the technology [1–4]. Similar to other advances in healthcare, a lack of standardization across the field represents an ongoing challenge. For successful integration of pharmacogenomics into routine practice, standardization is essential across all implementation steps, including laboratory processes, genetic test ordering and reporting results to clinicians and patients.
Resources such as PharmGKB (pharmgkb.org) and the Clinical Pharmacogenetics Implementation Consortium (cpicpgx.org) have been advancing the goal of translating complex genomic information into actionable phenotypes that are useful for patient care [5–7]. Standardization across all areas of the implementation process will enable and accelerate the use of pharmacogenomics in patient care. The electronic health record (EHR) is vital to implementing pharmacogenomics as it enables consistent use of genetic information at the point of care (e.g., for medication prescribing or dispensing) [8,9]. Standardization allows for efficient development and use of the EHR for pharmacogenomics through consistent clinical decision making, portability, scalability, durability of information and clinician education [10].
Recent efforts to improve standardization include published results from the Centers for Disease Control and Prevention (CDC) PGx workgroup that provided recommendations to improve pharmacogenomic implementation by standardizing the allele and variant nomenclature of test results [11]. Also, CPIC generated consensus for standard phenotype terms through the CPIC term standardization project [12]. Although progress is being made to standardize pharmacogenomics in practice, additional standardization opportunities remain. Limited standards may cause confusion related to the genetic test result, the exchange of structured interpretations between laboratories, and the continuity of genetic information delivered through EHRs. Ultimately, the lack of standardization may hinder the development of coherent reimbursement policies that drive patient access and facilitate increased adoption.
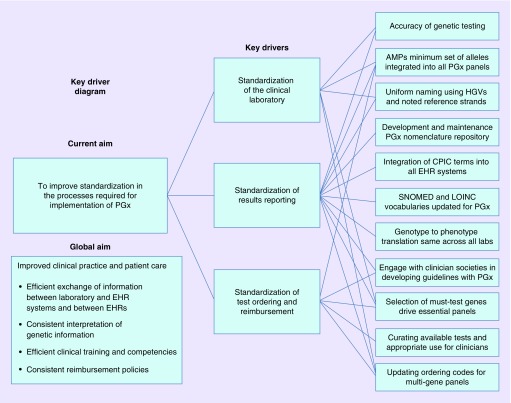
In this selective review, we summarize the state of current standardization efforts in pharmacogenomics. We focus on three processes that impact the eventual use of pharmacogenomics in clinical practice: the clinical laboratory, genetic test ordering and finally reporting test results (Tables 1 & 2 & Figure 1). Ongoing efforts and the remaining gaps in each process are highlighted and discussed.
Table 1. . Current and desired states of standardization of clinical laboratory processes and reporting.
| Process | Current state | Desired state | Ongoing initiatives | Gaps/opportunities |
|---|---|---|---|---|
| Laboratory processes | ||||
| Genetic test platform | Single nucleotide variant, copy number variant or sequence | Accuracy of testing | CDC workgroup recommendations [11] | Increase reference materials to account for genomic variation |
| Alleles/variants tested | Variants interrogated are not standardized across labs and same alleles not always tested | Variants reported and alleles tested should meet a minimum standard (e.g., ‘recommended gene panels for inclusion in all tests’) | European Pharmacogenetic Implementation Consortium – list of PGx alleles for clinical tests. AMP working on project to define must-test alleles for pharmacogenes |
Dissemination and uptake of recommendations |
| Reporting of pharmacogenomic results | ||||
| Reference strand used | Genotype calls from coding or noncoding strands using different guides (HGRA, HapMap project) | Specific indication of accession and version number of the sequence (Use the LRG, RefSeqGene and/or HGRA) | RefSeq project of the NCBI | Dissemination and uptake of recommendations |
| Nomenclature of PGx variants | Variance in systems leads to potential inadequacies (e.g., HGVS, rsID). HGVS does not specify a reference sequence | Uniform system for naming variants used across all labs (conversion of star or other nomenclatures into HGVS nomenclature) | CDC workgroup recommendations [11] ClinVar and Mutalyzer SNP converter tool, Leiden Open Variation database 3.0 PharmVar creating LRGs and combining all nomenclatures into one database |
Dissemination and uptake of standardized nomenclature and recommendations from CDC workgroup |
| Nomenclature of PGx genes/alleles | Summaries of variation have been published and star allele nomenclature assigned, but not systematically maintained (e.g., SLCO1B1; DPYD) | A systematically maintained, centralized PGx data repository | PharmVar – initial focus will be CYP enzymes, with plan to expand to other PGx genes | Dissemination and uptake of standardized nomenclature |
| Variants tested | Laboratory reports only listing genotypes detected in a sample | Reports should also include variants that can be detected by test | CDC workgroup recommendations [11] | Dissemination and uptake of CDC workgroup recommendations |
| Allele function and phenotype nomenclature | Laboratories report a variety of different terms to describe allele function and the corresponding phenotypes | Standardized terms used; integration of these terms into EHRs | CPIC term standardization project [12] | Dissemination and uptake of standardized nomenclature |
| Vocabularies (e.g., SNOMED, LOINC) | Phenotype terms used in EHR systems vary | Vocabularies (e.g., SNOMED, LOINC) | SNOMED and LOINC terms now available | Continued updates and additional terms added as needed |
| Genotype to phenotype translation | Laboratories and clinical practice guidelines do not translate phenotype based on genotype in the same way (e.g., CYP2D6) Reporting of allele function and corresponding phenotype may differ between laboratories |
Standardized translation – allele function and phenotype must match across all laboratories | CPIC continues to disseminate standardized phenotype terms and translation tables (e.g., used in each guideline, added to various medical vocabularies) CYP2D6 genotype to phenotype standardization project ongoing |
Dissemination and uptake of standardized translation method |
AMP: Association for Molecular Pathology; CPIC: Clinical Pharmacogenetics Implementation Consortium; EHR: Electronic health record; HGRA: Human Genome Reference Assembly; HGVS: Human Genome Variation Society; LOINC: Logical Observation Identifier Names and Code; LRG: Locus Reference Genomic; SNOMED: Systematized Nomenclature of Medicine; SNP: Single nucleotide polymorphism.
Table 2. . Current and desired states of standardization of pharmacogenomic test ordering and reimbursement.
| Process | Current state | Desired state | Ongoing initiatives | Gaps/opportunities |
|---|---|---|---|---|
| Gene/drug selections | Genes are selected by laboratory doing the test, often structured around reimbursement and clinicians must navigate these tests after determining the gene to interrogate | Must-test genes drive more consistent panel reimbursement and easier ordering process. Disease-specific guidelines provide recommendations of use of genetic test | CPIC – list of actionable genes and corresponding drugs, uses levels of evidence to support clinical utility (https://cpicpgx.org/genes-drugs/) | List updates needed on ongoing basis Effects of WGS/WES on gene selection/reimbursement |
| Market awareness: available tests and appropriate use | Clinicians under-aware of the types of pharmacogenomic tests available and the clinical utility of each | Userfriendly and intuitive database to provide available tests by medication class and disease state with clinical recommendations for use | GTR (www.ncbi.nlm.nih.gov/gtr/) provides central location for voluntary submission of genetic test information by providers | GTR relies on voluntary submission and could be more complete Resources should be readily understandable by patients |
| Current reimbursement and out of pocket costs (OOP) | No consensus, little reimbursement for testing outside of oncology, and lack of unique CPT codes | Sharp increase in the number of unique CPT codes for expanding number of tests, payer engagement in requirements from laboratories | Approximately 10 new CPT Tier 1 codes available for 2018 Active research and interest among field |
Further understand payer perspectives on pharmacogenomics, especially evidence requirements and decision making processes Develop structures and tools to reuse pharmacogenomic results over time which increases value of testing |
CPT: Current procedural terminology; GTR: Genetic testing registry; WES: Whole exome sequencing; WGS: Whole genome sequencing.
Figure 1. . Pharmacogenomic implementation processes and impact on clinical practice and patient care.
AMP: Association for Molecular Pathology; CPIC: Clinical Pharmacogenetics Implementation Consortium; EHR: Electronic health record; HGVS: Human Genome Variation Society; LOINC: Logical Observation Identifier Names and Code.
Standardization in clinical laboratory processes
Genetic testing platform
Current state
Advancements in genetic testing technology can make decisions on which genetic test to order complex. The use of targeted technologies to detect single nucleotide polymorphism and copy number variation has been the industry standard for decades. The declining costs of next generation or massively parallel sequencing technologies have accelerated their use, especially in the research setting [13]. However, targeted platforms are primarily used in clinical practice [14] as there can be technical difficulties such as with pseudogenes and gene conversions in pharmacogenes that are still challenging by massively parallel sequencing. Additionally, determining functionality of novel variants identified by massively parallel sequencing is still evolving.
Manufacturers of testing platforms include many pharmacogenes and variants. Depending on whether the platform is used clinically or as a discovery tool can determine the variants included on the platform. Clinical Laboratory Improvement Amendments of 1988 (CLIA; 42 CFR 493) is intended to ensure that laboratory results used in patient care are accurate. As required by regulatory agencies, during the analytical validation process for clinical testing, laboratories must validate that each variant tested is accurate and precise and that the test platform has appropriate analytical sensitivity and analytical specificity. There are many guidelines for analytical validation [15,16]. The CDC's Genetic Testing Reference Materials Coordination Program (wwwn.cdc.gov/clia/Resources/GetRM/) uses consensus verification to characterize DNA samples for test development and validation as well as for proficiency testing and quality control.
Research laboratories are not regulated in this manner and thus are not required to rigorously validate a platform, which allows flexibility to allow for innovation and creative exploration. Additionally, non-CLIA research laboratories are not permitted under law to provide clinical results. If results generated in a non-CLIA environment are returned to patients, they should be counseled that these results are not for treatment purposes and additional testing through their regular healthcare providers at CLIA-certified laboratories should be sought. Following this process, the researcher would not be giving results for diagnosis or treatment purposes and therefore would not violate the applicable CLIA regulations [17].
Preferred state & ongoing initiatives
Regardless of the laboratory, there should be adequate quality control in place to ensure accurate clinical and research testing. Due to biological diversity in the human genome, there will probably never be one assay that accurately tests for all types of genomic variation (e.g., from the genome to the metabolome). Reference materials that span the scope of genomic variants can help ensure that any platform accurately detects what it says it can detect.
Genetic variants tested
Current state
Clinical laboratories and genetic test assays interrogate various genes and variants, which can lead to meaningful differences in results. Currently, standards do not exist that define which variants must be tested [18]. The CDC's Genetic Testing Reference Material Coordination Program conducted a study in which nine volunteer clinical, research and commercial laboratories were provided with blinded genomic DNA samples for genotyping for a number of pharmacogenes using a variety of different methods (i.e., single nucleotide variant genotyping, copy number variant assessment and DNA sequence analysis) [18]. None of the laboratories interrogated the same set of variants for any of the pharmacogenes analyzed; therefore, different haplotype calls were reported for the same allele. Though all of the results were consistent among the laboratories based on the variants tested, the implications of these findings are clinically important as it could cause an incorrect assignment of phenotype and ultimately result in different recommended clinical actions. For example, the no function CYP2C19*4 allele has been identified in linkage disequilibrium with the increased function CYP2C19*17 (c.-806C>T) allele in certain ethnic subpopulations (CYP2C19*4B) [19,20]. If rs28399504, which defines the CYP2C19*4 haplotype, is not tested, a CYP2C19 intermediate metabolizer could be misclassified as an ultrarapid or normal metabolizer [11].
Preferred state & current initiatives
Variants reported and alleles tested should meet a minimum standard (e.g., recommended alleles for inclusion in all tests). The Association for Molecular Pathology (AMP) has an effort underway to create recommendations for a minimum set of alleles to include in each clinical pharmacogene genotyping panel. These recommendations are intended to inform clinical laboratory professionals when designing and validating clinical pharmacogenomic assays, to promote standardization of pharmacogenomic testing across different laboratories, and to complement other clinical guidelines such as CPIC. Using criteria such as allele function, population frequency and availability of reference materials, the AMP Pharmacogenomics Working Group has proposed a recommended minimum set of alleles and their defining variants (Tier 1) that should be included in clinical pharmacogenomic tests for CYP2C19 [14]. In addition, alleles that do not currently meet one or more of the criteria for inclusion in Tier 1 are considered optional for clinical testing (Tier 2). These recommendations are intended to facilitate testing by laboratories and to improve genotyping concordance across laboratories. AMP plans to address additional pharmacogenes. Efforts such as those from the AMP to standardize clinical tests allows for comparison of results from different institutions or studies and further provides evidence of the clinical and analytic validity. The Ubiquitous Pharmacogenomics PREPARE (Preemptive Pharmacogenomic Testing for Preventing Adverse Drug Reactions) is one such clinical study where the Ubiquitous Pharmacogenomics consortium is implementing a single standardized platform to evaluate the impact of pharmacogenomic testing on therapy outcomes in seven European clinical centers [21].
Standardizing the reporting of pharmacogenomic results
Reporting pharmacogenomic results to clinicians and patients has many steps, from the initial laboratory results, to the transfer of the laboratory report to the EHR, and to the information presented to the prescribing clinician at the point of care (Table 1 & Figure 1). Standardizing all these processes, while facilitating communication, is crucial to consistent and effective implementation of pharmacogenomics in clinical practice. The CDC PGx workgroup described areas of the laboratory report that are not standardized and summarizes the lack of standardization for nomenclature for sequence variants and alleles [11]. In all, nine recommendations for standardization are outlined including four addressing the naming of sequence variants, one for naming alleles and four recommendations for the test report. However, the CDC PGx workgroup did not provide recommendations for other areas of reporting of pharmacogenomic results such as the translation of genotype to phenotype and nomenclature used to describe allele function and phenotype, but this is addressed by CPIC, PharmGKB and PharmVar [12,22].
Sequence variant & allele nomenclature
Current state
Sequence variant naming uses the Human Genome Variation Society (HGVS) nomenclature. However, because the HGVS nomenclature can be used with any of a number of reference sequences, the same variant could be named using various reference sequences.
The star (*) allele nomenclature is the most common nomenclature for naming alleles for pharmacogenomics. However, some genes, such as VKORC1, have been reported using a variety of nomenclature systems. Furthermore, the naming of the star (*) alleles is generally maintained by a specific nomenclature committee for the specific gene (e.g., TPMT, UGT, NAT, HLA), but each one is not always updated in a systematic and efficient way. For example, the UGT nomenclature database has not been updated since 2007 [23]. For some genes, such as SLCO1B1 and DPYD, formal nomenclature committees do not exist and naming of alleles is not systematically managed.
Preferred state & ongoing initiatives
The CDC workgroup did recommend naming sequence variants by using HGVS nomenclature but also recommended which reference sequences should be utilized (i.e., Locus Reference Genomic, RefSeqGene and/or Human Genome Reference Assembly [11]). Efforts are underway to address the allele nomenclature issues. PharmVar, a rebranding and expansion of the Human Cytochrome P450 Allele Nomenclature Database, aims to improve pharmacogenomics nomenclature by serving as a “centralized ‘Next-Generation’ Pharmacogene Variation data repository” (pharmvar.org). The major focus of PharmVar is to continue the mission of serving as the official and unified allele designation system for CYP450 genes and will expand to other pharmacogenes such as SLCO1B1 and DPYD [22].
Reporting variants tested
Current state
As genotype test vary between laboratories, genotypes (i.e., * alleles) are called based on different variants. Currently, College of American Pathology (CAP) requires a laboratory to list all genotypes tested in the laboratory report; however, they might list only the * allele tested (e.g., CYP2D6*2) but not the defining variants for that allele.
Preferred state & ongoing initiatives
Knowledge of the all variants tested is needed to interpret the genetic test result. Therefore, the CDC workgroup also provided a recommendation for how to report the variants observed as well as listing all variants that can be detected by the test.
Allele function & phenotype nomenclature
Current state
Laboratories report a variety of different terms to describe allele function and the corresponding phenotypes (e.g., normal metabolizer vs extensive metabolizer). Not only are the various terms used to describe phenotype confusing to patients and clinicians, but this also impedes best use practice in the EHR with clinical decision support (CDS) and sharing of pharmacogenomic data across different platforms.
Preferred state & current initiatives
Standardization of allele function and phenotype nomenclature is critical for proper implementation of pharmacogenomics into the EHR and for use in clinical practice. In 2013, the Health Level Seven International (HL7) released an implementation guide for clinical genomics that details how genetic test results should be implemented into the EHR for both sequencing and genotyping-based tests and includes both disease causing and pharmacogenomic variants [24].
In 2014, CPIC led a consensus effort to standardize terms for clinical pharmacogenomic alleles and phenotypes. The goal of the project was to create standardized terms to be used in CPIC guidelines and to have these standardized pharmacogenomic terms adopted broadly by clinical genetic testing laboratories and relevant professional societies and organizations [12]. The Logical Observation Identifier Names and Codes (LOINC) terminology system has become standard for the reporting of laboratory test results and interpretations. CPIC has worked to further facilitate improvements in CDS by obtaining LOINC identifiers for pharmacogenomic interpretation codes, which specify the gene product and its role (metabolize vs function vs risk), and the answer list specifying the level of each interpretation code [12]. CPIC guidelines and PharmGKB now use these standardized terms and are included with pharmacogenomic data submissions to ClinVar. Shortly after in 2015, AMP and CAP (clinical laboratory proficiency testing program; CAP-PGx) formally endorsed the set of standardized terms describing allele function and phenotype [25].
Continuing work in this area, led by the CPIC Informatics Working Group, aims to standardize another widely used medical terminology particularly important to CDS, the Systematized Nomenclature of Medicine – Clinical Terms (SNOMED CT). The working group identified 39 concepts in need of replacement or updating and 18 to be removed from the current SNOMED CT library. A formal request was submitted and subsequently approved. The new concepts are available as of January 2018 (www.snomed.org/snomed-ct).
Genotype to phenotype translation
Current state
The process of translating genotype to phenotype is critical for implementation [8,26]. CPIC and PharmGKB create dedicated tables that provide a comprehensive translation from diplotype to interpreted phenotype. However, this translation is not standardized across laboratories or even pharmacogenomic guidelines. For example, some laboratories and the Dutch Pharmacogenetic Working Group (DPWG) guidelines consider a CYP2D6 activity score of 1.0 (e.g., combination of a normal and no function allele or two decreased function alleles) as a CYP2D6 intermediate metabolizer while the package insert of the obsolete Roche AmpliChip™ and CPIC guideline categorizes this score as an CYP2D6 normal metabolizer [27–29].
Other discrepancies exist between pharmacogenomic guidelines with regards to phenotype assignment and clinical recommendations. A recent collaborative paper between CPIC and the DPWG highlighted differences between variant calls, associated clinical recommendations and the underlying methodologies [30]. Although highly similar, discordances were found for approximately 20% gene–drug pairs in terminology for both allele function and phenotypes, and more concerning, the differences in clinical classification for specific variants. For example, individuals with the CYP2C19 *1/*17 genotype are classified as rapid metabolizers by CPIC and normal metabolizers by DPWG.
Preferred state & current initiatives
Since most pharmacogenomic recommendations are based on phenotype groupings, the assignment of phenotype based on genotype is an important aspect to clinical implementation. Reporting of different inferred phenotypes across laboratories and guidelines has created confusion and inconsistencies in recommendations. To maximize the utility of pharmacogenomic test results, it is desirable to standardize these definitions. PharmGKB, CPIC, American College of Medical Genetics (ACMG) and other groups including representation from ClinVar/ClinGen are working together to establish more unified translation of genotype to phenotype in the future, similar to a system used for disease risk (e.g., benign, pathogenic, etc.) [31]. Furthermore, CPIC and the DPWG are conducting a consensus project among CYP2D6 experts for the definitions used to assign CYP2D6 phenotype based on genotype. Once complete, both the CPIC and DPWG will adopt these definitions. Other discrepancies between guidelines are also being addressed through ongoing collaboration [30].
Standardizing the pharmacogenomic test ordering & coverage policy processes
Pharmacogenomic test selection
Current state
As the number of pharmacogenomic tests available to clinicians continue to increase, ordering the most appropriate and cost–effective test is paramount. Standardized clinical language across various gene panels so that clinicians can easily move between different types of tests will aid adoption (Table 2). The issue of standardizing which genes are included on the panels brings forward financial and market issues. As laboratories vie for market position, more genes with potential clinical utility and gene/drug associations can be included, but this inclusion may not be consistent with the latest evidence such as guidance from CPIC. Some tests offered by different laboratories specify gene panels based on the highly curated work of CPIC and PharmGKB while others extend their reach into lower levels of actionability. CPIC provides a list of gene–drug pairs assigning levels A, B, C and D based on actionability with CPIC level A being the highest level (strong to moderate recommendations) to CPIC level D being the lowest level with no actionability [7]. If whole genome and whole exome sequencing (WGS and WES) testing becomes more common, the focus will shift from a selection of genes for individual panels to one of interpreting the relevant pharmacogenes and actionable variants [13]. Greater availability of sequencing data would disrupt the current reimbursement structure which rewards individual pharmacogenes over broader panel approaches.
Preferred state & ongoing initiatives
The ideal state for pharmacogenomic test ordering is one where genetic results are available to the clinician at the first point of prescribing through preemptive testing (Table 2). Preemptive testing using multigene panels has been shown to be feasible based on experiences at multiple institutions before WGS or WES data are more widely used [9,32,33]. Carefully selected and intuitively designed alerts in the CDS systems will provide the most advanced use of these data. Until this state is realized the most optimal situation would be one where clinicians are aware of the utility behind the pharmacogenomic tests most applicable to their care responsibilities. Institutions implementing pharmacogenomics into practice must invest in informatics resources to design CDS systems with both active and passive alerts for drugs ordered that have pharmacogenomic associations [8,9,33–38]. This, coupled with new, more specific current procedural terminology (CPT) codes, should allow for greater clarity on reimbursement and cost-sharing discussions between patient and provider, as well as budget impact projections for the organization.
Several academic institutions and consortiums have been working on integrating pharmacogenomic information into the EHR and following the actionability of this data [8,9,32,33,36,39]. It is these types of projects that should facilitate research and the subsequent education of clinicians from general practice to more specialized fields on the utility and appropriate application of the variety of tests that are available. A greater understanding of when testing should be applied will also improve the conversations with payers and enable clinicians to advocate for reimbursement on behalf of their patients [38].
Pharmacogenomic testing reimbursement
Establishing consistent reimbursement for new health technologies may serve to make innovations more available to patients. The reimbursement landscape can also dictate the level of investment and guide decision making of laboratories and others relevant to pharmacogenomics implementation. As discussed, some laboratories may pursue larger panels, sacrificing high levels of utility to attract self-pay patients while others depend largely on the work of CPIC to guide gene selection. Payers’ perspectives and future plans can have a significant impact on a laboratories decision to pursue diagnostic development testing beyond what they know is currently reimbursed. Standardization across the pharmacogenomics research community can serve as a strong signal to payers to those that ultimately influence accessibility to pharmacogenomic testing.
Current state
Health insurance reimbursement for pharmacogenomics is limited, especially the preemptive approach [12,40,41]. Opportunities for standardization exist from at least two perspectives that have direct bearing on coverage of pharmacogenomic testing. Payer perspectives on the clinical utility of pharmacogenomics differ between organizations, and few organizations are equipped to lead a standardization in reimbursement. However, a Centers for Medicare and Medicaid Services (CMS) national coverage decision is seen as a potential benchmark for the commercial industry [42]. Thus far, CMS appears skeptical of the utility of pharmacogenomic testing, and in mid-2015 implemented restrictive changes in reimbursement for several drug–gene pairs [43,44]. Although a hierarchy of quality evidence at least informally exists for payers, there is clear variability in how different types of payers make these decisions, especially for pharmacogenomics [42,45].
The other key challenge in reimbursement standardization is more technical. Unique CPT codes for panel testing need to be developed as more codes move from Tier 2 to Tier 1 classifications with Human Genome Organization approved gene symbols through the Coding Change Application system. The lack of more specific CPT codes for panel testing that cover multiple genes and variants leads to claims for multigene panels that are currently filed and reimbursed by CMS and some commercial payers under an ‘unlisted molecular pathology procedure’ CPT code (81479). Most other germline pharmacogenomic testing CPT codes currently available are limited to variants in single enzymes or genes (CYP2C19, 2D6, 2C9 and VKORC1, UGT1A1). Although some payers will only accept CPT code 81479 for panel reimbursement, others allow gene code stacking to account for panel ordering. Use of a generic CPT code for multigene panels precludes efficient implementation of multigene panel testing. Payers are unable to track the appropriate use of tests ordered under this current structure, and this likely leads to additional inefficiencies. Further, CPT codes can be used track healthcare services subsequent outcomes to inform clinical utility, and this is not possible with generic CPT codes.
Preferred state & ongoing initiatives
A deeper understanding of how pharmacogenomics fits into payers evidence hierarchies, and how this evidence should be developed, is needed. Private and public payers should work alongside the pharmacogenomics community to define the acceptable level and type of evidence, from study design to clinical guidelines, which will lead to efficiencies in the mechanisms of this process while remaining economically sustainable.
Notable progress has been made in moving codes to Tier 1, as well as developing new codes for genes such as G6PD. With approximately ten new CPT Tier 1 codes for pharmacogenetic genes becoming usable in 2018, the use of stacked coding, submitting individual, reimbursable codes for multiple genes on a panel test may be a possible, yet cumbersome, way to recoup at least part of the cost. Ultimately, the hope would be that WGS or WES becomes widely accessible and that coding practice follows suit to enable large scale economic studies on the health system and population impacts of certain pharmacogene variants.
Reimbursement procedures for pharmacogenomics must accommodate both physician fee schedules and clinical laboratory fee schedules, which adds complexity. Separate Healthcare Common Procedure Coding System codes have been developed specifically for the interpretation and reporting of results in the clinical setting by the physician. More standardized reimbursement policy from an authoritative source such as a National Coverage Determinations for pharmacogenomics from CMS would provide a benchmark. A reference point like this may adjudicate differences across the Local Coverage Determinations made by Medicare Administrative Contractors throughout the country, as well as among the coverage policies for numerous other payers.
Discussion: impacts on clinical practice & patient care
We have highlighted the current state of standardization in the clinical laboratory, result reporting and test ordering. As standardization develops further, implementation of pharmacogenomics into routine care will become more efficient across healthcare settings. Figure 1 summarizes these processes and how standardization will impact clinical practice and patient care. Ultimately, standardization impacts the transfer of information to and between the EHR, the use of the information to make clinical recommendations, the type and complexity of education of clinicians and the reasonable reimbursement so patients can receive the test in the first place.
Integration into the EHR
Reporting understandable and actionable pharmacogenomics results to clinicians is an essential aspect of effectively implementing pharmacogenomics in practice which makes advances in clinical informatics crucial [8]. Several large academic health centers and national consortia have dedicated considerable resources to learning how best to integrate pharmacogenomics into the EHR for clinical care [2,4,32,33,35,36,39,46–48]. Different models are emerging and the lack of standardization of clinical laboratory processes and reporting complicates many aspects of implementing pharmacogenomics into the EHR, including transferring the initial laboratory results to the EHR as well as alerts presented to the clinician [26]. As pharmacogenomic results can have lifetime implications, results must be standardized to facilitate the exchange of information among different EHRs over time. To effectively implement and maintain new gene/drug pairs into a decision-support system there must consistency of genetic testing performance and reporting using standard nomenclature.
Standard vocabularies and other standardization efforts will support emerging strategies such as Substitutable Medical Applications and Reusable Technologies (SMART) on Fast Health Interoperability Resources (FHIR) which is a promising strategy to provide flexibility and support informatics needs for pharmacogenomics across different EHRs [49–51]. As more software applications per these standards are developed, it is likely fewer local informatics resources will be needed to implement pharmacogenomics into the EHR.
The CDC PGx workgroup, CPIC, PharmGKB and AMP have published projects addressing standardization issues (i.e., test reporting, allele/phenotype terms and variants tested, respectively). However, the uptake of CDC PGx workgroup recommendations has been slow. Bousman et al. compared the coverage and results reporting of CYP2D6 and CYP2C19 for pharmacogenomic testing in psychiatry [52]. They found that most test panels included the major alleles for CYP2D6 (*2, *4, *5, *10, *17) and CYP2C19 (*2, *3, *17), but no two panels contained the same combination and none fulfilled the nine recommendations laid out in the CDC recommendations [52]. The terms developed as part of the CPIC term standardization project have been more widely accepted and included as LOINC identifiers and SNOMED codes. Furthermore, these terms have been adopted by ClinGen and ClinVar and are being used by the CAP proficiency testing programs [12]. Until accrediting organizations like CAP adopts these standards, the recommendations from professional organizations will remain best practices.
Clinical recommendations
Pharmacogenomic recommendations are based on the assigned or inferred phenotype; therefore, consistent interpretation of the genotype result to a phenotype is critical for accurate pharmacogenomic-based prescribing. Inconsistencies in naming sequence variants and alleles, variants tested and diplotype to phenotype translation make interpreting genotype data very difficult and additional expertise is generally needed. Although CPIC and PharmGKB provide gene definition tables and comprehensive genotype to phenotype tables for each guideline to aid in consistent reporting and interpretation of genetic results [12,26], it is ultimately up to the laboratories and implementation leaders to ensure the correct phenotype assignment is made. The clinician, in most cases, is not trained to interpret these results and depends on these interpretations to guide prescribing [8].
Clinician training & competencies
Because initial clinical training does not provide the depth of expertise to interpret pharmacogenomics and interpreting genetic test results can be complex, leaders in implementing pharmacogenomics dedicate trained personnel to interpret genetic test results and educate clinicians [53,54]. Competencies, resources, and models are emerging to train clinicians in pharmacogenomics [55,56]. As genotype interpretation is standardized, clinician education can increasingly focus on how to use the patient's phenotype to optimize medication use instead of the details of interpreting results. It will remain useful for clinicians to understand genotype interpretation, but they can also trust the underlying processes to confidently use pharmacogenomic information. Projects from NHGRI like the Genetics/Genomics Competency Center (G2C2) and Inter-Society Coordinating Committee for Practitioner Education in Genomics (ISCC) provide leadership and educational resources to educate clinicians on genomics, including pharmacogenomics. Better defining expert interpretation roles and clinician roles will simplify clinical implementation and facilitate the uptake of pharmacogenomics into routine clinical practice.
Reimbursement effects
Limited reimbursement in pharmacogenomics will have direct impacts on test development and test ordering. The standardization of coverage policies across public and private payers in the USA remains a challenge. A recent study showed that the ‘absence of out-of-pocket costs to the patient’ was the second most important factor to clinicians when deciding to order a pharmacogenomic test [57]. Overall coverage policy development for genomics is stymied by the staggering number of nearly 75,000 genetic testing units (any orderable combination of analytes) currently being produced by US-based CLIA-certified labs and available on the market [58]. Newly developed pharmacogenomic tests represent are a minority of these tests (just over 10% of net new tests), with more attention and reimbursement focused on other types of genomic testing. The pharmacogenomics community must continue to pursue standardization in pharmacogenomics reimbursement – both the policy decisions and technical coding process. Without changes in the reimbursement structure, the benefits from pharmacogenomic testing may remain available only to patients who are able and willing to pay the full out-of-pocket costs.
Future perspective
Many of the efforts highlighted here are projects funded over a defined period through grant or other support. Currently, NHGRI funds ClinGen, ClinVar and CPIC. Professional organizations such as AMP and ACMG have made important contributions to pool resources to evaluate and share information that also provides standards. If government funding is no longer available, hopefully the pharmacogenomics community will identify other means of support for these critical resources together to support these critical endeavors.
Standardization across the implementation process will enable efficient and successful integration of pharmacogenomics into routine clinical care at a larger scale. Resources and working groups such as PharmGKB, CPIC, the CDC PGx workgroup and others have been working toward this goal [5–7]. Existing standards should be further disseminated. Opportunities remain to develop new standards and facilitate dissemination of existing standards to enable the implementation of pharmacogenomics.
Executive summary.
Importance of standardization across the implementation process
Standardization across the implementation process (i.e., clinical laboratory, genetic test ordering and reporting) will enable efficient and successful integration of pharmacogenomics into routine clinical care and the electronic health record at a larger scale.
Standardization in clinical laboratory processes
Standardization in the genetic testing platform and genetic variants tested will ensure adequate quality control and consistent interpretation of genetic information.
Efforts are underway to create recommendations for a minimum set of alleles to include in each genotyping panels [14].
Standardizing the reporting of pharmacogenomic results
The CDC PGx workgroup addressed many of the key issues of lack of standardization in laboratory reporting but uptake of these recommendations has been slow [11].
Lack of standardized terms to describe allele and phenotypes impedes implementation of pharmacogenetics into the electronic health record and sharing of pharmacogenetic data across platforms. Pharmacogenetic phenotype LOINC and Systematized Nomenclature of Medicine terms have been created based on the CPIC term standardization project [12].
Differences exist between translating genotype to phenotype translation between clinical laboratories and pharmacogenetic clinical guidelines. Projects are underway to standardize these processes.
Standardizing the pharmacogenomic test ordering & coverage policy processes
Lack of consistent coverage policies across public and private payers impedes genetic test development and ordering and remains a challenge.
Clinician education & training
Initial clinical training in pharmacogenetics is inconsistent and impedes use of pharmacogenetics in clinical practice.
As genotype interpretation is standardized, clinician education can be streamlined and focus to optimize medication use using genetic information instead of the details of interpreting results.
Footnotes
Financial & competing interests disclosure
This work was funded by the NIH for CPIC (R24GM115264; KE Caudle, JM Hoffman, TE Klein, M Whirl-Carrillo) and PharmGKB (R24GM61374; TE Klein, M Whirl-Carrillo). VM Pratt is supported by the IGNITE project grant (U01HG007762) and the Indiana University Health – Indiana University School of Medicine Strategic Research Initiative. The Indiana University School of Medicine Pharmacogenomics Laboratory is a fee-for-service clinical laboratory that offers clinical pharmacogenetic testing. ALSAC supports KE Caudle, JM Hoffman and NJ Keeling. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of thismanuscript.
References
- 1.Rosenman MB, Decker B, Levy KD, Holmes AM, Pratt VM, Eadon MT. Lessons learned when introducing pharmacogenomic panel testing into clinical practice. Value Health. 2017;20(1):54–59. doi: 10.1016/j.jval.2016.08.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dunnenberger HM, Crews KR, Hoffman JM, et al. Preemptive clinical pharmacogenetics implementation: current programs in five US medical centers. Annu. Rev. Pharmacol. Toxicol. 2015;55:89–106. doi: 10.1146/annurev-pharmtox-010814-124835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roden DM, Van Driest SL, Mosley JD, et al. Benefit of pre-emptive pharmacogenetic information on clinical outcome. Clin. Pharmacol. Ther. 2018;103(5):787–794. doi: 10.1002/cpt.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Relling MV, Evans WE. Pharmacogenomics in the clinic. Nature. 2015;526(7573):343–350. doi: 10.1038/nature15817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caudle KE, Klein TE, Hoffman JM, et al. Incorporation of pharmacogenomics into routine clinical practice: the Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline development process. Curr. Drug Metab. 2014;15(2):209–217. doi: 10.2174/1389200215666140130124910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whirl-Carrillo M, McDonagh EM, Hebert JM, et al. Pharmacogenomics knowledge for personalized medicine. Clin. Pharm. Ther. 2012;92(4):414–417. doi: 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caudle KE, Gammal RS, Whirl-Carrillo M, Hoffman JM, Relling MV, Klein TE. Evidence and resources to implement pharmacogenetic knowledge for precision medicine. Am. J. Health Sys. Pharm. 2016;73(23):1977–1985. doi: 10.2146/ajhp150977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hicks JK, Dunnenberger HM, Gumpper KF, Haidar CE, Hoffman JM. Integrating pharmacogenomics into electronic health records with clinical decision support. Am. J. Health Sys. Pharm. 2016;73(23):1967–1976. doi: 10.2146/ajhp160030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freimuth RR, Formea CM, Hoffman JM, Matey E, Peterson JF, Boyce RD. Implementing genomic clinical decision support for drug-based precision medicine. CPT Pharmacometrics Syst. Pharmacol. 2017;6(3):153–155. doi: 10.1002/psp4.12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hicks JK, Crews KR, Hoffman JM, et al. A clinician-driven automated system for integration of pharmacogenetic interpretations into an electronic medical record. Clinical Pharm. Ther. 2012;92(5):563–566. doi: 10.1038/clpt.2012.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalman LV, Agundez J, Appell ML, et al. Pharmacogenetic allele nomenclature: international workgroup recommendations for test result reporting. Clin. Pharm. Ther. 2016;99(2):172–185. doi: 10.1002/cpt.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caudle KE, Dunnenberger HM, Freimuth RR, et al. Standardizing terms for clinical pharmacogenetic test results: consensus terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC) Genet. Med. 2017;19(2):215–223. doi: 10.1038/gim.2016.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang W, Wu G, Broeckel U, et al. Comparison of genome sequencing and clinical genotyping for pharmacogenes. Clin. Pharm. Ther. 2016;100(4):380–388. doi: 10.1002/cpt.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pratt VM, Del Tredici AL, Hachad H, et al. Recommendations for cinical CYP2C19 genotyping allele selection: a report of the Association for Molecular Pathology. J. Mol. Diagn. 2018;20(3):269–276. doi: 10.1016/j.jmoldx.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 15.Mattocks CJ, Morris MA, Matthijs G, et al. A standardized framework for the validation and verification of clinical molecular genetic tests. Eur. J. Hum. Genet. 2010;18(12):1276–1288. doi: 10.1038/ejhg.2010.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jennings L, Van Deerlin VM, Gulley ML College of American Pathologists Molecular Pathology Resource Comittee. Recommended principles and practices for validating clinical molecular pathology tests. Arch. Pathol. Lab. Med. 2009;133(5):743–755. doi: 10.5858/133.5.743. [DOI] [PubMed] [Google Scholar]
- 17.HHS. Return of individual results and special consideration of issues arising from amendments of HIPAA and CLIA, 2018(2/27) www.hhs.gov/ohrp/sachrp-committee/recommendations/2015-september-28-attachment-c/index.html
- 18.Pratt VM, Zehnbauer B, Wilson JA, et al. Characterization of 107 genomic DNA reference materials for CYP2D6, CYP2C19, CYP2C9, VKORC1, and UGT1A1: a GeT-RM and Association for Molecular Pathology collaborative project. J. Mol. Diagn. 2010;12(6):835–846. doi: 10.2353/jmoldx.2010.100090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott SA, Sangkuhl K, Gardner EE, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450-2C19 (CYP2C19) genotype and clopidogrel therapy. Clin. Pharm. Ther. 2011;90(2):328–332. doi: 10.1038/clpt.2011.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scott SA, Martis S, Peter I, Kasai Y, Kornreich R, Desnick RJ. Identification of CYP2C19*4B: pharmacogenetic implications for drug metabolism including clopidogrel responsiveness. Pharmacogenomics J. 2012;12(4):297–305. doi: 10.1038/tpj.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cecchin E, Roncato R, Guchelaar HJ, Toffoli G Ubiquitous Pharmacogenomics Consortium. Ubiquitous pharmacogenomics (U-PGx): the time for implementation is now. an Horizon2020 program to drive pharmacogenomics into clinical practice. Curr. Pharm. Biotechnol. 2017;18(3):204–209. doi: 10.2174/1389201018666170103103619. [DOI] [PubMed] [Google Scholar]
- 22.Gaedigk A, Ingelman-Sundberg M, Miller NA, et al. The Pharmacogene Variation (PharmVar) Consortium: incorporation of the human cytochrome P450 (CYP) allele nomenclature database. Clin. Pharm. Ther. 2018;103(3):399–401. doi: 10.1002/cpt.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the pharmacogenomics research network. Clin. Pharm. Ther. 2011;89(3):464–467. doi: 10.1038/clpt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Health Level Seven® INTERNATIONAL. HL7 version 2 implementation guide: clinical genomics; fully LOINC-qualified genetic variation model (US Realm) 2013. www.hl7.org/implement/standards/product_brief.cfm?product_id=23 Implementation Guides.
- 25.AMP. Position Statements & Letters. 2017. www.amp.org/advocacy/position-statements-letters/
- 26.Hoffman JM, Dunnenberger HM, Kevin Hicks J, et al. Developing knowledge resources to support precision medicine: principles from the Clinical Pharmacogenetics Implementation Consortium (CPIC) J. Am. Med. Inform. Assoc. 2016;23(4):796–801. doi: 10.1093/jamia/ocw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monnier N, Kozak-Ribbens G, Krivosic-Horber R, et al. Correlations between genotype and pharmacological, histological, functional, and clinical phenotypes in malignant hyperthermia susceptibility. Hum. Mutat. 2005;26(5):413–425. doi: 10.1002/humu.20231. [DOI] [PubMed] [Google Scholar]
- 28.Crews KR, Gaedigk A, Dunnenberger HM, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy: 2014 update. Clin. Pharm. Ther. 2014;95(4):376–382. doi: 10.1038/clpt.2013.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hicks JK, Bishop JR, Sangkuhl K, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin. Pharm. Ther. 2015;98(2):127–134. doi: 10.1002/cpt.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bank P, Caudle KE, Swen JJ, et al. Comparison of the Guidelines of the Clinical Pharmacogenetics Implementation Consortium and the Dutch Pharmacogenetics Working Group. Clin. Pharm. Ther. 2017;103(4):599–618. doi: 10.1002/cpt.762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoffman JM, Haidar CE, Wilkinson MR, et al. PG4KDS: a model for the clinical implementation of pre-emptive pharmacogenetics. Am. J. Med. Genet. C Semin. Med. Genet. 2014;166C(1):45–55. doi: 10.1002/ajmg.c.31391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Donnell PH, Wadhwa N, Danahey K, et al. Pharmacogenomics-based point-of-care clinical decision support significantly alters drug prescribing. Clin. Pharm. Ther. 2017;102(5):859–869. doi: 10.1002/cpt.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bell GC, Crews KR, Wilkinson MR, et al. Development and use of active clinical decision support for preemptive pharmacogenomics. J. Am. Med. Inform. Assoc. 2014;21(e1):e93–e99. doi: 10.1136/amiajnl-2013-001993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rasmussen-Torvik LJ, Stallings SC, Gordon AS, et al. Design and anticipated outcomes of the eMERGE-PGx project: a multicenter pilot for preemptive pharmacogenomics in electronic health record systems. Clin. Pharm. Ther. 2014;96(4):482–489. doi: 10.1038/clpt.2014.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldspiel BR, Flegel WA, Dipatrizio G, et al. Integrating pharmacogenetic information and clinical decision support into the electronic health record. J. Am. Med. Inform. Assoc. 2014;21(3):522–528. doi: 10.1136/amiajnl-2013-001873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herr TM, Bielinski SJ, Bottinger E, et al. Practical considerations in genomic decision support: the eMERGE experience. J. Pathol. Inform. 2015;6:50. doi: 10.4103/2153-3539.165999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Veenstra DL. The value of routine pharmacogenomic screening – are we there yet? A perspective on the costs and benefits of routine screening-shouldn't everyone have this done? Clin. Pharm. Ther. 2016;99(2):164–166. doi: 10.1002/cpt.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caraballo PJ, Sutton JA, Moyer AM, et al. Technical challenges and opportunities when implementing pharmacogenomics decision support integrated in the electronic health record. Stud. Health Technol. Inform. 2017;245:1255. [PubMed] [Google Scholar]
- 40.Faulkner E, Annemans L, Garrison L, et al. Challenges in the development and reimbursement of personalized medicine-payer and manufacturer perspectives and implications for health economics and outcomes research: a report of the ISPOR personalized medicine special interest group. Value Health. 2012;15(8):1162–1171. doi: 10.1016/j.jval.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 41.Cohen J, Wilson A, Manzolillo K. Clinical and economic challenges facing pharmacogenomics. Pharmacogenomics J. 2013;13(4):378–388. doi: 10.1038/tpj.2011.63. [DOI] [PubMed] [Google Scholar]
- 42.Keeling NJ, Rosenthal MM, West-Strum D, Patel AS, Haidar CE, Hoffman JM. Preemptive pharmacogenetic testing: exploring the knowledge and perspectives of US payers. Genet. Med. 2017 doi: 10.1038/gim.2017.181. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lynch JA, Berse B, Dotson WD, Khoury MJ, Coomer N, Kautter J. Utilization of genetic tests: analysis of gene-specific billing in Medicare claims data. Genet. Med. 2017;19(8):890–899. doi: 10.1038/gim.2016.209. [DOI] [PubMed] [Google Scholar]
- 44.Centers for Medicare & Medicaid Services. MD, USA: 2015. Local Coverage Determination (LCD): CYP2C19, CYP2D6, CYP2C9, and VKORC1 genetic testing (L36312)https://med.noridianmedicare.com/documents/10546/6990983/MolDX+CYP2C19%2C%20CYP2D6%2C%20CYP2C9%2C%20and+VKORC1+Genetic+Testing+LCD/469a28e0-cb6d-41a6-abc3-58d476755277 [Google Scholar]
- 45.Leung MY, Halpern MT, West ND. Pharmaceutical technology assessment: perspectives from payers. J. Manag. Care Pharm. 2012;18(3):256–264. doi: 10.18553/jmcp.2012.18.3.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rohrer Vitek CR, Abul-Husn NS, Connolly JJ, et al. Healthcare provider education to support integration of pharmacogenomics in practice: the eMERGE network experience. Pharmacogenomics. 2017;18(10):1013–1025. doi: 10.2217/pgs-2017-0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shuldiner AR, Relling MV, Peterson JF, et al. The Pharmacogenomics Research Network Translational Pharmacogenetics Program: overcoming challenges of real-world implementation. Clin. Pharma. Ther. 2013;94(2):207–210. doi: 10.1038/clpt.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manzi SF, Fusaro VA, Chadwick L, et al. Creating a scalable clinical pharmacogenomics service with automated interpretation and medical record result integration – experience from a pediatric tertiary care facility. J. Am. Med. Inform. Assoc. 2017;24(1):74–80. doi: 10.1093/jamia/ocw052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alterovitz G, Warner J, Zhang P, et al. SMART on FHIR genomics: facilitating standardized clinico-genomic apps. J. Am. Med. Inform. Assoc. 2015;22(6):1173–1178. doi: 10.1093/jamia/ocv045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mandl KD, Mandel JC, Murphy SN, et al. The SMART platform: early experience enabling substitutable applications for electronic health records. J. Am. Med. Inform. Assoc. 2012;19(4):597–603. doi: 10.1136/amiajnl-2011-000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rasmussen LV, Overby CL, Connolly J, et al. Practical considerations for implementing genomic information resources. Experiences from eMERGE and CSER. Appl. Clin. Inform. 2016;7(3):870–882. doi: 10.4338/ACI-2016-04-RA-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bousman CA, Jaksa P, Pantelis C. Systematic evaluation of commercial pharmacogenetic testing in psychiatry: a focus on CYP2D6 and CYP2C19 allele coverage and results reporting. Pharmacogenet. Genomics. 2017;27(11):387–393. doi: 10.1097/FPC.0000000000000303. [DOI] [PubMed] [Google Scholar]
- 53.Nickola TJ, Green JS, Harralson AF, O'Brien TJ. The current and future state of pharmacogenomics medical education in the USA. Pharmacogenomics. 2012;13(12):1419–1425. doi: 10.2217/pgs.12.113. [DOI] [PubMed] [Google Scholar]
- 54.Murphy JE, Green JS, Adams LA, Squire RB, Kuo GM, McKay A. Pharmacogenomics in the curricula of colleges and schools of pharmacy in the United States. Am. J. Pharm. Educ. 2010;74(1):7. doi: 10.5688/aj740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roederer MW, Kuo GM, Kisor DF, et al. Pharmacogenomics competencies in pharmacy practice: a blueprint for change. J. Am. Pharm. Assoc. 2017;57(1):120–125. doi: 10.1016/j.japh.2016.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adams SM, Anderson KB, Coons JC, et al. Advancing pharmacogenomics education in the core PharmD curriculum through student personal genomic testing. Am. J. Pharm. Educ. 2016;80(1):3. doi: 10.5688/ajpe8013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peterson JF, Field JR, Shi Y, et al. Attitudes of clinicians following large-scale pharmacogenomics implementation. Pharmacogenomics J. 2016;16(4):393–398. doi: 10.1038/tpj.2015.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.The current landscape of genetic testing: market growth, reimbursement trends, challenges and opportunities. 2018. www.concertgenetics.com/resources/2018-current-landscape-genetic-testing/