Abstract
Objective: To compare three potential sources of controlled clinical terminology (READ codes version 3.1, SNOMED International, and Unified Medical Language System (UMLS) version 1.6) relative to attributes of completeness, clinical taxonomy, administrative mapping, term definitions and clarity (duplicate coding rate).
Methods: The authors assembled 1929 source concept records from a variety of clinical information taken from four medical centers across the United States. The source data included medical as well as ample nursing terminology. The source records were coded in each scheme by an investigator and checked by the coding scheme owner. The codings were then scored by an independent panel of clinicians for acceptability. Codes were checked for definitions provided with the scheme. Codes for a random sample of source records were analyzed by an investigator for “parent” and “child” codes within the scheme. Parent and child pairs were scored by an independent panel of medical informatics specialists for clinical acceptability. Administrative and billing code mapping from the published scheme were reviewed for all coded records and analyzed by independent reviewers for accuracy. The investigator for each scheme exhaustively searched a sample of coded records for duplications.
Results: SNOMED was judged to be significantly more complete in coding the source material than the other schemes (SNOMED* 70%; READ 57%; UMLS 50%; *p <.00001). SNOMED also had a richer clinical taxonomy judged by the number of acceptable first-degree relatives per coded concept (SNOMED* 4.56; UMLS 3.17; READ 2.14, *p <.005). Only the UMLS provided any definitions; these were found for 49% of records which had a coding assignment. READ and UMLS had better administrative mappings (composite score: READ* 40.6%; UMLS* 36.1%; SNOMED 20.7%, *p <. 00001), and SNOMED had substantially more duplications of coding assignments (duplication rate: READ 0%; UMLS 4.2%; SNOMED* 13.9%, *p <. 004) associated with a loss of clarity.
Conclusion: No major terminology source can lay claim to being the ideal resource for a computer-based patient record. However, based upon this analysis of releases for April 1995, SNOMED International is considerably more complete, has a compositional nature and a richer taxonomy. It suffers from less clarity, resulting from a lack of syntax and evolutionary changes in its coding scheme. READ has greater clarity and better mapping to administrative schemes (ICD-10 and OPCS-4), is rapidly changing and is less complete. UMLS is a rich lexical resource, with mappings to many source vocabularies. It provides definitions for many of its terms. However, due to the varying granularities and purposes of its source schemes, it has limitations for representation of clinical concepts within a computer-based patient record.
A computer-based patient record (CPR) is an electronic patient record that supports users by providing accessibility to complete and accurate data, alerts, reminders, clinical decision support and links to medical knowledge.1 It is supported by a system of programs which provide useful access to the data, allow entry and collation of information, and secure storage. In 1992 the Institute of Medicine treatise on the CPR1 clearly pointed to the importance of a central data dictionary and industry coding standards as key elements. Methods for achieving those goals have remained illusory, in part because of historical emphasis by software developers on billing and epidemiology rather than patient care, and also due to intense debate as to the merits of standardized schemes for classification. There has been no general agreement on the attributes of such systems, much less whether a particular scheme does the job.
Although some will argue the pragmatics of individual attributes, building upon the work of others within the informatics community,2,3 we maintain that a classification scheme for implementation within a CPR should have the following features:
Complete and comprehensive2—The classification scheme should cover the entire clinical spectrum, including all component disciplines involved in patient care at sufficient granularity (depth and level of detail) to depict the care process.
Clarity (clear and non-redundant)2—A code is an assignment of an identifier within the scheme to represent a concept from clinical practice. A term4 is a natural language phrase associated with and representing that concept in clinical parlance. Although synonym terms are desirable since they allow a clinician reasonable variation in expression, a concept should be neither vague nor ambiguous, and should not have overlapping meanings within the scheme. This means that a single concept should not have many code representations within the scheme.
Mapping (administrative cross references)—To be useful, a clinical scheme must point to related entities in widely used administrative and epidemiologic reporting systems.
-
Atomic and compositional character—Clinical classifications that break findings and events (concepts) into basic component pieces, have substantial practical advantages by avoiding an explosion of terms with the additional of new knowledge. Such a scheme is multiaxial and compositional, as opposed ato a precoordinated scheme wherein each concept—no matter how complex—has a single representational code. To illustrate, contrast the two representations of the concept “back pain”:
UMLS
SNOMED International
C0004604 BACK PAIN
T-D2100 BACK,
F-A26000 PAIN
Synonyms2—The scheme must support alternate terminology as required by the clinicians
Attributes—The scheme should support a mechanism to modify or qualify meaning of the core term
Uncertainty—The scheme must support a graduated record of certainty for findings and assessments.
Hierarchies and inheritance2—A hierarchical organization of concepts, linking logically more general and more specific terms, facilitates the use of classifications within an “intelligent” record by supporting inductive reasoning. A single term should be allowed many parents or children as clinically appropriate.
Context-free identifiers—The codes themselves must be devoid of meaning to avoid assignment conflicts as the body of clinical knowledge evolves.
Unique identifiers—A code must not be re-used when it is declared obsolete.
Definitions—Concepts should be associated with concise explanations of their meaning. Publication of definitions does not guarantee clarity, but it promotes the development of a clear schema.
Language independence—The scheme should be freely translated across the human languages in use by patients and caregivers.
Syntax and grammar—Compositional schemes must be accompanied by a set of rules that define logical and clinically relevant constructions of the codes. Pre-coordinated schemes do not have this requirement.
In an earlier publication5 we studied seven major systems for the attribute of completeness. This work drew on other projects which had previously evaluated one or a few systems in limited application areas.6,7,8,9,10,11,12,13,14 Our conclusion was that no scheme available in the English-speaking world was sufficiently comprehensive to be readily implemented within a CPR. However, three candidate systems were substantially better than competitors: the READ classification15,16,4 of the National Health Service of Great Britain; SNOMED International,17,18,19 published by the College of American Pathologists (CAP); and the Unified Medical Language System (Metathesaurus)20,21,22,23,24 of the National Library of Medicine (NLM). Our study was limited in scope, especially within the domain of nursing practice. Furthermore, coding staff scored their own evaluations and we did not study any attributes other than completeness. It is our purpose in this paper to extend our initial work, and evaluate the 1995 release of these three schemes relative to the attributes of completeness, clarity, definitions, inheritance and administrative mapping.
Methods
Evaluation Set
As part of an initial examination of coding schemes previously reported,5 we took source material from clinical records found in four medical centers across the United States. Our first report details the methods whereby source material was prepared for analysis. The data originated with both textual and flowcharted information found in active clinical charts. Dr. Chute reviewed this material for concepts and formed an initial evaluation set of 3061 records. These records were hierarchically linked constructions of sometimes complex conceptual entities. Retrospectively analyzing our evaluation set, we identified two limitations of the source material: (1) comprehensiveness and (2) granularity. In particular, we determined that nursing care was one portion of the clinical record that was underrepresented. Furthermore, the level of detail of the evaluation records differed from item to item. We therefore revised our initial evaluation set. We first expanded our phase one material with additional concepts taken from nursing care plans, flowcharts and notes. We used the same methods to gather and prepare this data as we had previously described.
In order ot identify the universe of conceptual information to be found in the CPR, and to better define its granular nature (i.e., level of detail), we organized a consensus discussion within the Computer-based Patient Record Institute (CPRI) workgroup on codes. From this discussion, we formulated a list of the conceptual domains of the CPR. This is included in abbreviated form in the Appendix. We used the domain definitions from this discussion and reorganized the original evaluation records so that they followed this categorization. We eliminated all duplicates, since some records appeared more than once, and assigned a domain to each record based upon the clinical context of the material from its source.
By way of example, the source record detailed in our earlier paper5 was modified as follows:
Source text: “... it was identified as a superficial spreading melanoma Clark's level 2, with depth of invasion 0.84 mm.”
Phase I Evaluation Records:
<Diagnosis>:melanoma
<Extent>:Clark's level 2
<Quantitative>:0.184 mm [depth of invasion]
<Mode>:superficial spreading
Phase II Evaluation Records:
<6.1.1|Medical diagnosis>:superficial spreading melanoma
<3.|Attributes>:Clark's level 2
<3.|Attributes>:depth of invasion [0.84 mm]
In this particular case, the concept “superficial spreading melanoma” was judged to fit within the definition of a medical diagnosis more appropriately than an attribute/diagnosis pair. Examples of material added to the evaluation set from nursing documentation included:
Phase II Nursing Data:
<6.2.1|Nursing diagnosis>:ineffective individual coping
<4.4|Educational intervention>:explain lab values to patient
<5.1.1|Symptom>:fear of suffocation
Completeness
We gave the evaluation records to three coding team leaders: Drs. Sneiderman (READ), Warren (SNOMED), and Cohn (UMLS). We asked the owners of the three systems to give us copies that would be current in April, 1995. We received copies of the three schemes as follows:
SNOMED International version 3.1; publication April 1995; delivered March 1995
READ Version 3.1; publication May 1995; delivered August 1995
UMLS Version 1.6; publication January 1995; delivered July 1995
Each coding team leader commented upon problems with coding of the source records using public or commercial browsing tools. This necessitated review by the scheme owner to assure fairness. The READ browser, published by Computer Aided Medical Systems, frequently missed terms—many times due to cultural differences in phrasing and spelling. It did not employ translation techniques to assist with differences characterized by the two terms: “NEVUS” and “NAEVUS.” The COACH browser distributed by the National Library of Medicine often buried the term which happened to match the source term uniquely at the bottom of a list of hundreds of marginal choices. In an effort to be linguistically complete, it was often misleading or troublesome to use. SNOMED did not have an adequate public-domain browser and we were forced to purchase a tool from an independent developer (Medsight Informatique Inc., 1801 McTavish St-Bruno, Quebec, Canada) that was functionally superior to many others but not well suited to high volumes of coding.
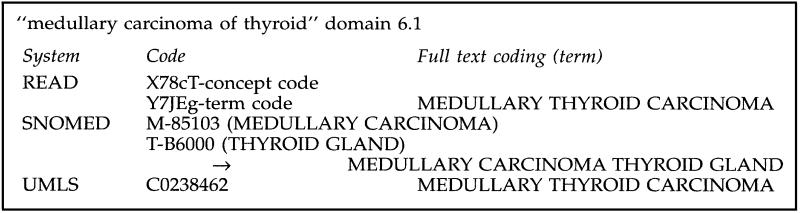
Using these tools, each coding team leader led the effort to classify the source record from the phase II evaluation set within the scheme provided. The best mapping was recorded by code and full text coding record. The full text record was stripped of technical verbiage such as “NOS” and “NEC” for readability by a clinician whose assignment would be to judge meaning and relevance, not totality of the scheme. We took this action so as not to bias the scorers (described below—a team of clinician experts) at a time when they would have no knowledge of the source scheme. Take for example our full text coding of the record shown in Figure 1.
Figure 1.
Sample full-text codings for clinical scoring.
We made every effort when using the compositional features of SNOMED to employ sensible and logical constructions since some “nonsense” constructions were possible. The mapping was reviewed by a second author and then passed to the publisher of the scheme for comment and correction. Based upon this response, a fraction of the original coding was revised. Most changes occurred in the READ scheme, where unfamiliarity with differences in culture and administrative systems caused some confusion.
The scoring team leader, Dr. Carpenter, assembled a team of nine clinicians (six physicians and three nurses) from the Mayo Clinic for scoring of the matched sets. Each scorer had a minimum of ten years of staff clinical experience. The nurses had primarily inpatient experience. Physicians were trained in the disciplines of Endocrinology, Internal Medicine, Orthopedic Surgery, Cardiology and Pediatrics and practiced within a variety of settings. Five individuals from this group (three physicians and two nurses) scored each set of records on a fivepoint Likert scale, rating the acceptability of the match between source concept and coded result. (All evaluation steps used the same Likert rating: 1 = strongly disagree, 3 = neutral, 5 = strongly agree.) Only the full text coding was reviewed by the clinical scoring team. In cases where their assessment score was 3 or less, we also asked them to rate whether the offered term was too specific, too general, or was unrelated to the source term. We scored exact lexical matches, excluding issues of number and minor changes in word order, as six points without clinician review. We scored all records without a match as zero. Scoring sheets assembled by the scoring team leader were collated, checked for accuracy, and entered into a SAS database for analysis. 4% of records were found to be in error (for example, the scoring sheet contained the wrong candidate term) and were reanalyzed by two team members and scored using the same Likert scale. We used an average of the scores for the final analyses.
We used the SAS procedure25 FREQ to calculate descriptive statistics of the clinician scoring by scheme. For calculation of a completeness score, we accepted only code matches with a Likert score of 4 or higher. Using this categorization, we computed the Chi-Square statistic to analyze for statistical differences between the schemes in aggregate. To compare coding schemes by information domain, we employed the SAS procedure GLM and used Bonferroni adjusted t-tests to compare mean scores by domain and compute confidence intervals.
In order to assess variability in rater scoring, we randomly selected 85 cases from all source material which had coding matches in all three schemes, but exact lexical matches in none of them. We did this several months after the original scoring step. These cases were prepared on a new scoring sheet in which the matches for the three schemes were displayed side-by-side, but still blinded as to source. We then asked three of the original clinician-scorers to rate this new set, and compared their scores to the original ratings using simple descriptive statistics. A sample record from the scoring sheet is shown in Figure 2.
Figure 2.
Sample from clinical scoring sheet.
Definitions
Neither SNOMED nor READ provides definitions apart from that implied by the code term. In the case of UMLS which provides a separate file (MRDEF) of definitions, we searched the file for definitions of all the UMLS codes selected by the coding team. A raw score was computed for the frequency of availability of definitions.
Taxonomy
From the source data covering the domains 4.X-Interventions, 5.X-Findings, 6.X-Diagnoses and impressions, 10.X-Anatomy and 11.X-Etiology, we selected a pragmatic sample of 10% of source codes at random. We used the computer files provided by the owner of each scheme to analyze the codes within this sample of records for all “parents” and “children.” This was subject to some interpretation in the case of UMLS, and we chose to analyze only the “context relationships” from data file MRCXT. This file lists hierarchical relationships between UMLS concepts as taken from the UMLS source vocabularies. UMLS also has a semantic network which defines a set of hierarchical relationships but we did not analyze these.
We compiled only the first-generation relatives for codes from all schemes and arranged them in a pair-wise fashion for analysis. Six clinician-informatics specialists were recruited and generously volunteered their time for the analysis. These reviewers are all experienced practicing clinicians, who are also active in systems development at their home institutions. All are active members of the American Medical Informatics Association (AMIA). Via Internet mail, they reviewed random samples of 125 parent-child pairs for each coding scheme. They used a five point Likert scale (1 = extremely dissatisfied with pairing, 3 = neutral, 5 = extremely satisfied with pairing) to rate the clinical utility of each pairing. Each reviewer was blinded to the source scheme they were rating. We asked them to score the pair following these directions:
“We ask you to rate the clinical and programmatic utility of this pairing by choosing a score for each pair based upon your judgement of their appropriateness as a CLINICALLY USEFUL classification hierarchy. We ask you to judge this pair based upon the following criteria:
the pairing is clinically relevant and sound for support of clinical reminders and other features of the computerized record
the pairing is sensible and appropriate when viewed as an informatics structural element of the computerized record”
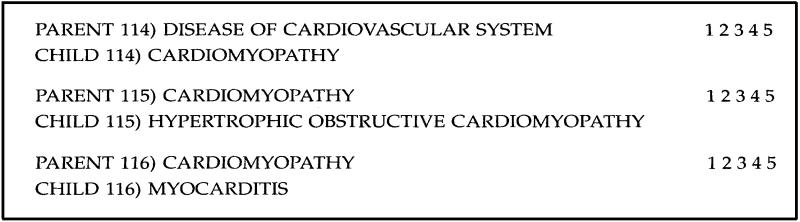
A sample from the coding sheet for SNOMED appears in Figure 3.
Figure 3.
Sample taxonomic scoring sheet.
Mapping
Using the computer files provided by the scheme owners, we evaluated the mapping of codes we found to the billing and administrative schemes in use in the host countries, whenever these cross references were supplied in the published scheme. This meant that we reviewed codes for records from source domains 6.X (diagnoses) and 5.1.X (symptoms and reports) for mappings to ICD-9-CM for SNOMED and UMLS, and to ICD-10 for READ. For procedures, we also studied records from domains 4.X (procedures) excluding domain 4.2.X (medications) for mappings to CPT or ICD-9-CM for SNOMED and UMLS, and to OPCS-4 for READ. At two separate sites, we assembled a team of medical encoding specialists who reviewed the mapped codes for accuracy.
Clarity (Coding Duplications)
Each coding team leader further analyzed the 10% random sample of records mentioned above to search the scheme for duplicate coding. In this analysis, we specifically looked for clinical concepts that had more than one coding assignment (ignoring the differences in terms that might be available). For this sample of 190 records, the team leaders did an exhaustive analysis of the published scheme using the browser, looking for additional coding representations of the same (source) concept. We assembled all possible representations and then a second member of the study team evaluated them to judge if they were true duplications of meaning or were representational variants. A duplication was judged to be present if the coding scheme had a second identifier for the source concept. For precoordinated schemes, any second coding would be a duplication. For SNOMED, the issue is more complex and we looked carefully at the semantics (coding axes and code combinations) of the coding assignment. In the case of SNOMED, we judged second representations to be duplicates only if they used the same semantic structure. For SNOMED, we classified additional code representations as a representational variant if the duplicate code had different semantics. For example,
[T-D9510 RIGHT ANKLE] and [T-D9500, G-A100 ANKLE, RIGHT]
was judged to be a duplication since both codings have root in the topology (T-axis) semantic. However, for the source record “S2-second heart sound,”
[F-35040 SECOND HEART SOUND] and [G-A702 SECOND; T-32000 HEART; A-25100 SOUND]
were judged to be representational variants. Although variants might be confusing or misleading to the untrained user, an experienced developer could possibly exploit such richness to their advantage. We summarized the frequency of both events as an indication of duplications in the published schema.
Results
Evaluation Set
Once we edited the source material for content, added nursing data, and eliminated duplications, the evaluation set consisted of 1,929 records. Table 1 summarizes the number of records by information domain and clinical source. Nursing documents, history and physicals and progress notes were taken from both inpatient and outpatient care environments. The majority of source records were classified as attributes, interventions, findings and diagnoses. Within each of these major categories, the subsets most heavily represented were laboratory tests, therapeutic procedures, symptoms, physical findings, medical diagnoses and body parts or organs. The distribution of records by information domain was driven entirely by clinical record content and therefore represented the concerns and focus of the health care providers who recorded the source clinical documents.
Table 1.
Phase II Source Records: Text Source by Information Domain
| Source Documents | 2 Admin | 3 Demo | 4 Intrvtn | 5 Finding | 6 Diagns | 7 Plans | 8 Equip | 9 Events | 10 Anatmy | 11 Etiol | 14 Agents | Totals |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Consults | 4 | 23 | 13 | 29 | 21 | 4 | 1 | 1 | 11 | 0 | 0 | 107 |
| Discharge summaries | 5 | 30 | 111 | 94 | 46 | 11 | 3 | 2 | 19 | 1 | 0 | 322 |
| Nursing documents | 2 | 5 | 90 | 130 | 65 | 95 | 2 | 0 | 1 | 0 | 0 | 390 |
| History and physicals | 7 | 90 | 129 | 248 | 102 | 9 | 6 | 0 | 42 | 3 | 4 | 640 |
| Operative notes | 0 | 18 | 85 | 29 | 22 | 2 | 17 | 0 | 54 | 0 | 0 | 227 |
| Progress notes | 0 | 20 | 24 | 37 | 16 | 2 | 1 | 0 | 10 | 3 | 0 | 113 |
| X-ray reports | 0 | 20 | 13 | 46 | 17 | 2 | 0 | 0 | 31 | 1 | 0 | 130 |
| Totals | 18 | 206 | 465 | 613 | 289 | 125 | 30 | 3 | 168 | 8 | 4 | 1929 |
Completeness
Table 2 summarizes the scoring results of the best coding matches by scheme. The upper portion of the table summarizes scoring frequency and mean scores for each scheme. At the bottom, we used as definition of success a minimum score of four (4 = agree, 5 = strongly agree or 6 = lexical match). From this definition, the completeness scores by scheme were READ—57.0%, SNOMED—69.7% and UMLS—49.6% (χ2 = 164, 2 df, p <.00001). We found that coding assignments scored at 3 or less were judged similarly for specificity across all schemes. In particular, when a rater was dissatisfied with the coding match, they rated it as too general approximately 80% of the time, too specific 10% of the time, and as unrelated 10% of the time. This varied only slightly between READ, SNOMED and UMLS.
Table 2.
Average Scoring Frequency by Scheme: Number of Scores (Percent of total)
| Score | READ | SNOMED | UMLS |
|---|---|---|---|
| 0 (no match) | 80 (4.1) | 103 (5.3) | 312 (16.2) |
| 1-1.99 | 51 (2.6) | 23 (1.2) | 65 (3.4) |
| 2-2.99 | 378 (19.6) | 215 (11.1) | 375 (19.4) |
| 3-3.99 | 321 (16.6) | 245 (12.7) | 221 (11.5) |
| 4-4.99 | 406 (21.0) | 383 (19.9) | 238 (12.3) |
| 5 | 202 (10.5) | 530 (27.5) | 140 (7.3) |
| 6 (exact match) | 491 (25.5) | 430 (22.3) | 578 (30.0) |
| Mean score | 3.96* (95% CI 3.89-4.04) | 4.31* (4.24-4.38) | 3.63* (3.54-3.73) |
| Completeness (score 4 or greater) | 57.0%** | 69.7%** | 49.6%** |
P <.05 for each pair-wise comparison of mean scores.
χ2 = 164 df = 2 for comparison of completeness scores, p <.00001.
Table 3 lists the average score between systems by information domain. Mean scores are accompanied by Bonferroni confidence intervals for the thirty-three subsets analyzed. For readability, the final column summarizes statistically significant differences between systems within the domain.
Table 3.
Average Score by Domain and Scheme (Confidence Intervals Employing Correction)
| Domain | N | READ | SNOMED | UMLS | Statistical Summary |
|---|---|---|---|---|---|
| Demographics | 18 | 3.70 (2.38-5.02) | 3.39 (2.07-4.71) | 3.02 (1.70-4.34) | R = S = U |
| Attributes | 206 | 3.43 (3.04-3.81) | 3.97 (3.59-4.36) | 2.52 (2.13-2.91) | S > R > U |
| Interventions | 465 | 3.91 (3.65-4.17) | 4.36 (4.10-4.62) | 4.06 (3.80-4.31) | S > U, R |
| Findings | 613 | 3.79 (3.56-4.02) | 4.31 (4.08-4.53) | 3.44 (3.22-3.67) | S > R > U |
| Diagnoses/impressions | 289 | 4.04 (3.70-4.36) | 4.62 (4.29-4.95) | 4.15 (3.82-4.48) | S > U, R |
| Plans | 125 | 2.93 (2.43-3.43) | 3.58 (3.08-4.08) | 2.88 (2.38-3.38) | S > R, U |
| Equipment/devices | 30 | 2.49 (1.47-3.51) | 3.25 (2.23-4.27) | 3.39 (2.37-4.41) | U, S > R |
| Events | 3 | 2.93 (-.29-6.16) | 3.2 (-.03-6.43) | 2.53 (-.69-5.76) | S = R = U |
| Human anatomy | 168 | 4.60 (4.17-5.03) | 4.88 (4.44-5.31) | 4.33 (3.90-4.76) | S, R > U |
| Etiologic agents | 8 | 3.93 (1.95-5.90) | 4.78 (2.80-6.75) | 4.55 (2.57-6.53) | S = U = R |
| Agents | 4 | 3.05 (.25-5.85) | 4.05 (1.25-6.84) | 1.55 (-1.25-4.35) | S = R = U |
Intrarater Variability
When we compared the average scores of the three raters for the 85 cases, 67% of cases were assigned the same rating, 15.5% were rated up one category, 0.4% were rated up two categories, 13.9% were rated down one category and 3.2% were rated down two categories. We analyzed this across all three coding schemes and found a tendency to rate SNOMED records downward, and an opposing change upward for UMLS. Mean scores for this set were: ▶
Table 4.
| SCHEME | Mean | SD | Differential from original score |
|---|---|---|---|
| READ | 3.28. | .91 | (-.18) |
| SNOMED | 3.48 | .86 | (-.74) |
| UMLS | 3.57 | .80 | (+.20) |
Reviewing the source records and the repeat scoring, there seemed to be a tendency to assign a “blanket” score on the cross-record rating. In fact, when linear regression was used to evaluate interdependency of scores for the validation set, 50-60% of variance in the score for one scheme could be explained based upon either of the other sets of scores. Therefore the differential observed between SNOMED and UMLS may represent a testing sequence bias, but seemed more likely to represent a regression towards the mean based upon the high degree of correlation among scores in this small validation set.
Definitions
Definitions were published in the UMLS source file MRDEF for 49.1% of the unique concept codes identified during the completeness analysis.
Taxonomy
The sampling procedure identified 165 records for taxonomic analysis. Table 4 summarizes the results of that examination. Column one lists the total number of records for which a match was found within the scheme. Column two lists the total number of first degree relatives (immediate parents or children) found for the terms that had a match. Columns three and four summarize the scoring of the relatives by domain experts, using an average score of four or better as acceptable. Finally, column five lists the average number of relatives scored as acceptable, indexed against the number of codes with matches from column one.
Table 4.
Summary of Taxonomy Analysis for 165 Source Records
| System | Source Matches | Number of First-Degree Relatives | Score 1-3 (%) | Score 4-5 (%) | Average Acceptable Relatives per Code |
|---|---|---|---|---|---|
| READ | 158 | 516 | 34.6 | 65.4 | 2.14* |
| SNOMED | 158 | 1065 | 32.3 | 67.7 | 4.56* |
| UMLS | 144 | 616 | 25.8 | 74.2 | 3.17* |
Pair-wise comparison of number of acceptable relatives shows significance for all pairings:
READ-SNOMED χ2 = 34.51, df = 2, p <.00001
SNOMED-UMLS χ2 = 7.87, df = 2, p <.005
UMLS-READ χ2 = 8.49, df = 2, p <.004.
Mapping
Table 5 summarizes the analysis of administrative cross references published with each scheme. The READ publication consistently did the best job of recognizing that mapping from a clinical scheme to an epidemiologic system may not always be one-to-one. In cases of ambiguity or overlap, they published a list of related codes and indicated where manual review would be necessary based upon context. Unfortunately, the best coding matches were not always listed first or flagged as primary.
Table 5.
Summary of Administrative Mapping: Symptoms (Domains 5.1.X); Diagnoses (Domains 6.X); Procedures (Domains 4.X excluding 4.2.X)
| System | Symptoms | Diagnoses | Procedures | Composite Score |
|---|---|---|---|---|
| READ (ICD-10, OPCS-4) | 35/207 | 138/290 | 107/192 | 40.6% |
| SNOMED (ICD-9-CM) | 42/207 | 79/290 | 22/192 | 20.7%* |
| UMLS (ICD-9-CM, CPT-IV) | 66/207 | 117/290 | 66/192 (ICD-9-CM) | 36.1% |
| 5/192 (CPT-IV) |
Pair-wise comparison with READ and UMLS shows significant difference, χ2 = 64.02, 43.59 respectively, df = 2, p <.00001
Because of difficulty in obtaining a copy of OPCS-4 for our reviewing team, procedural cross mapping for READ is listed as unreviewed raw scores. Otherwise, Table 5 lists only cross reference mappings found acceptable by reviewers. Subjectively, SNOMED had a higher error rate for ICD-9-CM mapping and seemed to be out of date in greater proportion, suggesting that editorial work may not be current in this area. In particular, both SNOMED and UMLS did not reflect many recent five digit changes to ICD-9-CM. UMLS of course, because of its origin and design, had many cross references to other schemes not analyzed in this paper. In general, Table 5 demonstrates that use of any of these three schemes will require substantial manual editing before they might be used to represent core clinical content and still accomplish administrative reporting for billing or epidemiologic purposes.
Clarity (Coding Duplications)
Table 6 summarizes the results of the search for duplication of coding within each scheme. Of the 165 records randomly selected for review, column one lists the number of records for which a match was found in the scheme. This serves as a denominator for the frequency calculations in columns two and three. Column two lists the number of duplicate codes found by reviewers and the relative frequency. Column three provides a similar tally of representational (semantic) variants—which were only relevant to the case of SNOMED.
Table 6.
Summary of Duplications Found in 165 Random Records
| System | Records with Matches | Duplications (Rate) | Representational Variants (Rate) |
|---|---|---|---|
| READ | 158 | 0.00* | N/A |
| SNOMED | 158 | 22 (13.9%)* | 8 (5.1%) |
| UMLS | 144 | 6 (4.2%)* | N/A |
Two-by-two comparison of duplications shows significance for all pairings:
READ-SNOMED χ2 = 23.65, df = 2 p <.00001
SNOMED-UMLS χ2 = 8.53, df = 2 p <.004
UMLS-READ χ2 = 6.72, df = 2 p <.01
Discussion
Methods and Study Limitations
Reviewing the conduct of this study, we have attempted to create a set of protocols for evaluating a clinical classification scheme and for providing feed-back to scheme developers. Earlier projects6,7,8,9,10,11,12,13,14 have studied a single feature—usually completeness, and have generally employed highly focused evaluation sets. Limitations of those projects have grown out of finite resources, confusion regarding study objectives, and a lack of documented and validated methods. We believe that we have addressed some, but not all, of these concerns.
In particular, we have begun our project with a proposal for the attributes of an ideal classification scheme. We do this as an outgrowth of a discussion within CPRI and on the basis of our own personal study, but also because the discussion of these features must begin in earnest in a broader forum. We hope that this paper will help to fuel and focus that discussion. Although some of the attributes we propose rest on common sense alone, others clearly require CPR systems research for validation of utility.
To address the nature of completeness, we have begun by expanding our source material to include previously unstudied areas—especially in nursing. In a sense, a complete evaluation set cannot yet be collected since we do not yet know for certain which aspects of the CPR will benefit objectively from coding. Also the use of material from an earlier project5 exposes our methods to criticisms of contamination (allowing code vendors to `tailor' their schemes). Furthermore, biases in selection of source material by the authors can create unfair comparisons of systems. To address these concerns we can only note once more5 that we selected documents across geographical regions and from many clinical viewpoints. The segregation of the source material into study records clearly represents the editorial opinion of the authors (JRC and CGC) and cannot be considered universal or standardized. In an effort to be fair, we believe that we more often overstated systems capabilities than denied them. The greatest concern of our investigator team in this regard is that limited browser function may have presented the greatest challenge to validity of our data since every tool we used had some limitations.
The bias (or error) created by the browser that we employed was a serious concern relative to an accurate estimate of duplication in the coding schemes. We are not aware of any earlier studies that have defined this problem or evaluated this issue in a quantitative manner. We are concerned that “exhaustive search” is an imprecise and non-reproducible proposal for study methods but lack an acceptable alternative at this time. We also found that our goals for duplication analysis were less well-defined in the setting of a compositional system such as SNOMED. This aside, we believe that the quality (if not precision) of the information presented does present real issues for implementation of clinical coding, since in a certain sense duplication rate (lack of clarity) can be a greater limitation than lack of completeness when using a system clinically.
From our study of rater team scoring validity, we are concerned that we may have introduced a “testing-sequence” bias. We cannot completely resolve this question with our current data set. Our data does not include rater identification that would permit analysis of variance—a better method for testing the question of bias. A better study plan would have mixed codes from the three source vocabularies when presenting them to the clinical judges, thus eliminating any tendency for scoring “drift” to impact upon scores by scheme. Since we received material from the scheme publishers over a six month period, and since all coding was done with donated time, this was not technically possible without delaying the whole project substantially. From experience, we observe that studies such as this require some haste in order to make observations that are not immediately discounted by vendors for being out-of-date.
As we have tested new study methods, we have also raised questions regarding alternative strategies. In particular, in our study of taxonomy we studied “the nearest neighbors” of each randomly selected code element for clinical utility of the taxonomic link. This method ignores “taxonomic depth” (Is the granularity of the hierarchy and depth of the hierarchy best?) and may not properly measure multiple inheritance as a scheme feature. An alternative method would ask domain experts to rate every member of the full-depth taxonomy surrounding a randomly selected code, and possibly to score the frequency and types of semantic links to the code.
Classification Systems: Observations
The structure and utility of each coding system that we studied fell short of the ideals that we propose, but we also encountered many strengths that offer potential use for the CPR system developer. A summary view of the three schemes we studied is provided in Table 7, referencing the features of an ideal scheme that we introduced above.
Table 7.
Summary of System Features by Scheme
| Feature | READ | SNOMED | UMLS |
|---|---|---|---|
| Complete | .57 | .70 | .50 |
| Clear1 | 1.00 | .86 | .96 |
| Mapping2 | .41 | .21 | .36 |
| Compositional | Partial | Yes | No |
| Synonyms | Yes | Yes | Yes |
| Attributes and uncertainty | Yes (qualifiers) | Yes (modifier axis) | No |
| Taxonomy3 | 2.14 | 4.56 | 3.17 |
| Meaningless identifiers | Yes | No | Yes |
| Unique identifiers | Yes | Yes | Yes |
| Definitions4 | 0.00 | 0.00 | .49 |
| Language independence | English only | English, French, Chinese and Portuguese | (English, French, German, Portuguese, Spanish5) |
| Syntax/grammar | Yes (qualifier mapping) | No | N/A (precoordinated) |
Score for clarity = (1-duplication rate)
Fraction of candidate concepts with administrative mapping
Number of first-degree relatives per concept
Fraction of concepts with definitions provided
MeSH terms only
We delayed our study of READ for release of the version 3.0, which includes nursing terms and attempts to coordinate efforts at clinical classification beyond primary care. The National Health Service (NHS) of Great Britain has numerous clinical teams revising READ, with many differing clinical emphases. Comparing versions 2 and 3 we note progress toward greater completeness. However, in the course of our efforts we encountered codes marked as “obsolete” which often seemed to represent sound clinical terms. These terms represent less than 3% of all records coded. Publications from NHS for April 1995 did not discuss the purpose of such `obsolete' markings, although NHS personnel insist that these codes are permissible for use. They emphasize that no READ code is ever eliminated from the scheme for reasons of historical release management. Based upon this feedback from NHS, all codes flagged as `obsolete' were included in the study report since a sensitivity analysis demonstrated that their inclusion did not materially affect the results.
READ has excellent clarity based upon our limited sampling. The administrative mapping of READ was superior to others we studied, and the coding of a source term into READ was generally straightforward when compared with SNOMED, although ICD-10 and OPCS-4 may have limited utility for investigators in the United States. Synonymy is well supported; the READ scheme employs multiple inheritance, which is superior to SNOMED.
READ has changed structure with the release of version 3, and it now includes meaningless concept identifiers. It also employs a limited compositional scheme featuring “qualifiers” which are linked in an organized list to parent concepts. “Qualifiers” generally provide quantification, anatomical location or specification for the terms to which they are linked. Although READ cannot be described as a fully compositional scheme on this basis, this change is clearly a decisive step in that direction. Furthermore, it has a clearer structure than the SNOMED compositional model since it has a defined syntax for permissible combinations. READ provides no definitions, commenting in their release notes that the preferred term for each code defines the concept; and is available only in English.
Our initial analysis5 of coding schemes suggested that SNOMED was the most complete classification system available today. This second phase analysis continues to support that observation, documenting the steady clinical evolution of SNOMED. The compositional design of SNOMED is the best we have evaluated of those proposed for clinical medicine. Although SNOMED poses implementation problems for the CPR developer, it offers clear advantages for exhaustively describing a rapidly evolving clinical care environment. READ has recognized the soundness of a compositional structure in its latest version, but still lags well behind SNOMED in that regard.
A problem with SNOMED that is clearly highlighted by our analysis is a high rate of duplicate codes—it lacks clarity. This problem is created in part by the compositional nature of the scheme, coupled with an evolution that has not proffered definitions, sometimes ignores orthogonality (non-overlapping construction), and has not developed a coding syntax. Investigators have previously suggested procedures for such steps2,26 and reports indicate that the SNOMED editorial board is considering these options. Ultimately, a compositional scheme must develop these features in order to achieve its full potential and utility.
Contrasting READ and UMLS to SNOMED relative to the features of completeness and clarity highlights the potential—and the pitfalls—of a compositional approach to clinical coding. To create an extreme example, random assembly of entries from a very large dictionary could include completely all terms in use for clinical care. This proposal would also introduce all the ambiguity and duplication that controlled vocabularies are intended to minimize. By defining component features of Diagnoses, Function, Topography, Morphology, Living Organisms, Procedures etc., SNOMED has attempted to organize clinical practice into a set of characteristics that define an element while they also dissect out its nature. Thus in a real sense, better completeness for SNOMED might be intuitive to some. The question emerges from this analysis however, whether such an approach can also be definitive and non-ambiguous. The pre-coordinated scheme formulated by READ clearly enhances the latter attributes. Challenges remain both for the systems designer and the scheme developer to creatively exploit and manage these issues.
Mapping within SNOMED is weak—if the systems implementor wishes to develop links to billing systems, additional work will be required. It supports ample synonyms within the publication and the addition of a “General Modifier” axis has created a structure to handle uncertainty and quantification. SNOMED has done more than any other publication to become an international scheme. It now supports four major languages: English, French, Chinese and Portuguese.
SNOMED falls short by continued ties to a rigid hierarchy in which identifiers are also taxonomic links. Clinical taxonomies must support multiple inheritance to be useful for decision logic.2 For example rheumatoid arthritis is both an arthritis and an auto-immune disease. The scheme currently employed by SNOMED ties the classification to a numerical assignment scheme that makes this impossible.
The UMLS Metathesaurus is not a classification system by design; rather, it is an inter-lingua or translation tool primarily designed for information retrieval. In that sense, we wish to make it clear that this study is not a proper evaluation of UMLS relative to the principles for which it was designed. Nonetheless, we included the Metathesaurus in our study because of the substantial national investment in this product and the clinical classification interest in UMLS voiced by CPR developers.7,10,27 Furthermore, for translating from the clinical arena to the medical literature, investigators have maintained that the UMLS is critical to workstation development. In this sense, one can argue that the Metathesaurus should be at least as complete as that clinical environment it proposes to serve.
However, based upon our observations, the Metathesaurus is not sufficiently complete nor organized in such a way to serve as a controlled terminology within a CPR. Comparing acceptability scores across schemes, it is notable that UMLS was much more dichotomous (a clean hit or a clean miss) than SNOMED with substantially less completeness—due in large part to its precoordinated paradigm. It publishes terms from both compositional and precoordinated schemes that may overlap without a definition of a canonical or preferred concept. It remains focused on the content of the source vocabularies that it connects and that material is not chosen primarily for clinical descriptive purposes. Although on the one hand NLM plans to subsume the entirety of SNOMED, it does not exploit the richness of composition that make SNOMED as complete as we found it. In the process of including SNOMED elements (for example), UMLS does not publish the minimal syntactical and compositional guidelines provided by CAP in SNOMED.
In order to exploit such richness from its source vocabularies, UMLS would have to include such compositional guidelines and promote their use in some fashion within its publication.
UMLS is well organized and clear with only rare duplications. Mapping is a fundamental purpose of the publication, and it includes links to some thirty-four different source schemes. Some investigators have reported upon its utility for translation between coding schemes.28,29 In keeping with the general results of that work, the CPR developer will find that creation of direct links to ICD-9-CM and CPT-V will require additional translation steps.
UMLS has an abundant lexicon of terms from which to choose, rich synonymy, and more definitions than any other scheme—suggesting that UMLS may aid the CPR developer most in creation of a local lexicon that will serve as the language interface with the clinician. Furthermore, the additional semantic and lexical features of UMLS make it a good resource for natural language analysis and building of the human interface.
Although UMLS maintains both a semantic network and the inheritance of terms from its source vocabularies, these taxonomies are clinical only in part. In fact, the UMLS taxonomies are incomplete from a clinical standpoint because the component vocabularies are of different granularity, they differ in their capacity to support multiple inheritance, and they are derived from different compositional semantics. The UMLS take attribute terms from its source schemes, but offers no guidelines for composing complex elements, and therefore we analyzed it as a pre-coordinated (one concept = one code) system. It maintains a system of unique identifiers for its concepts and terms. The Unified Medical Language System also supports five languages—but only for Medical Subject Heading (MeSH) terminology.
In conclusion, we cannot claim any universal solution to the coding problem of the CPR based upon our data. We do believe that the analyses we have made suggest strategies that are reasonable for CPR developers in the United States and Europe and we welcome the consideration of those questions in the critical light of our data. We also believe that our material points out important priorities for classification scheme developers to pursue in order to improve their product. On the national scene, the board of directors of AMIA30 has suggested coding sets for use in selected domains—especially medications and observations. This analysis suggests the prudence of some of their recommendations and perhaps urges action in other areas. We welcome the debate that will inevitably follow the publication of this paper.
Acknowledgments
We thank the clinicians of the Mayo Foundation who donated tremendous amounts of their time to score coding assignments: Sheryl Ness, RN, Julia Behrenbeck, MS, RN, Donald C. Purnell, MD, Lawrence H. Lee, MD, Robert C. Northcutt, MD, Vahab Fatourechi, MD, Sean F. Dinneen, MD, Maria Collazo-Clavell, MD, Robert A. Narotzky, MD.
We owe special thanks to the members of the informatics community who evaluated and scored hierarchical relationships: James J. Cimino, MD, Columbia University, New York NY; George Hripcsak, MD, Columbia University, New York NY; J. Marc Overhage, MD, PhD, Regenstrief Institute, Indianapolis IN; Thomas Payne, MD, Group Health Cooperative of Puget Sound, Seattle WA; William Hersh, MD, Oregon Health Sciences University, Portland OR; Patricia Flatley Brennan, RN, PhD, University of Wisconsin, Madison WI.
Members of the Codes and Structures Workgroup of the Computer-based Patient Record Institute donated time and their thoughts to development of the domains descriptions. We thank them.
Kathy Brouch organized a team of coding specialists to review administrative mapping data. We thank them for their assistance.
Thomas G. Tape, MD, kindly provided guidance with statistical analysis. We thank him.
Papers authored under the auspices of CPRI represent the consensus of the membership, but do not necessarily represent the views of individual CPRI member organizations.
Appendix
Concept Domains of the CPR
-
NAME: Administrative Concepts
DEFINITION: Administrative concepts are attributes of the CPR that are properties of the health care system (the environment and circumstances of the health care delivery process) and are necessary data elements within the CPR.
- 1.1 NAME: Facilities and institutions
- 1.2 NAME: Practitioners and care givers
- 1.3 NAME: Patients and clients
- 1.4 NAME: Payers and reimbursement sources; financial information
-
NAME: Demographics
-
DEFINITION: Demographic attributes/concepts are descriptors of living situations, major ethnic/racial categories, social or behavioral characteristics, or other properties of health care clients that identify them as individuals or quantify clinical risk.
- 2.1 NAME: Address
- 2.2 NAME: Telephone
- 2.3 NAME: Ethnicity
- 2.4 NAME: Religion
- 2.5 NAME: Occupation
- 2.6 NAME: Date of birth
- 2.7 NAME: Marital status
- 2.8 NAME: Race
- 2.9 NAME: Language
- 2.10 NAME: Educational level
-
-
NAME: Attributes
- DEFINITION: Attributes are features that change the meaning or enhance the description of an event or concept.
- EXAMPLE: Classes of attributes might include: Topography (excludes anatomy), Site, Negation, Severity, Stage, Clinical scoring, Disease activity, Time, Interval, Baseline, Trend
-
NAME: Interventions
-
DEFINITION: Activities used to alter, modify or enhance the condition of a patient in order to achieve a goal of better health, cure of disease, or optimal life style.
- 4.1 NAME: Diagnostic Procedure
- 4.1.1 NAME: Laboratory procedure or test
- 4.1.2 NAME: Procedure for functional testing or assessment
- 4.1.3 NAME: Radiographic procedure or test
- 4.2 NAME: Therapeutic interventions
- 4.2.1 NAME: Medication
- 4.2.2 NAME: Therapeutic procedure
- 4.2.3 NAME: Physical therapeutic intervention
- 4.3 NAME: Environmental interventions
- 4.4 NAME: Educational interventions
- 4.5 NAME: Behavioral and perceptual interventions
-
-
NAME: Finding
-
DEFINITION: A finding is an observation regarding a patient. It may be discovered by inquiry of a patient or patient's close contact. It may be directly measured by observing the patient during function or by stimulating the patient and assessing response. It may be the measurement of an attribute of the patient, a physical feature, or determination of a bodily function.
- 5.1 NAME: History and patient centered reports
- 5.1.1 NAME: Symptoms (and reports of disease or abnormal function)
- 5.1.2 NAME: Personal habits and functional reports
- 5.1.3 NAME: Reason for Encounter
- 5.1.4 NAME: Family history
- 5.2 NAME: Physical Exam
- 5.3 NAME: Laboratory and Testing Result
- 5.4 NAME: Educational or Psychiatric Testing Result
- 5.5 NAME: Functional Assessment Result
-
-
NAME: Diagnoses and Impressions
-
DEFINITION: A diagnosis is the determination or description of the nature of a problem or disease; a concise technical description of the cause, nature, or manifestations of a condition, situation or problem.
- 6.1 NAME: Disease-focused diagnosis
- 6.1.1 NAME: Medical diagnosis
- 6.1.2 NAME: Testing diagnosis
- 6.2 NAME: Function-focused diagnosis
- 6.2.1 NAME: Nursing diagnosis
- 6.2.2 NAME: Disability assessment
-
-
NAME: Plans
-
DEFINITION: A method or proposed procedure, documented in the CPR, for achieving a patient/client goal or outcome.
- 7.1 NAME: Referrals
- 7.2 NAME: Patient intervention contracts
- 7.3 NAME: Order
- 7.4 NAME: Appointments
- 7.5 NAME: Disposition
- 7.6 NAME: Nursing Intervention
-
-
NAME: Equipment and devices
-
DEFINITION: Objects used by providers or client/patients during the provision of health care services, in the pursuit of wellness, or to educate and instruct
- 8.1 NAME: Medical Device
- 8.2 NAME: Biomedical or Dental Material
- 8.2.1 NAME: Biomedical supply
-
-
NAME: Event
-
DEFINITION: A broad attribute type used for grouping activities, processes and states into recognizable associations. (UMLS) A noteworthy occurrence or happening (Webster 3rd Int Dict).
- 9.1 NAME: Encounter
- 9.2 NAME: Patient life event
- 9.3 NAME: Episode of care
-
-
NAME: Human anatomy
-
DEFINITION: A set of concepts relating to components or regions of the human body, used in the description of procedures, findings, and diagnoses.
- 10.1 NAME: Body Location or Region
- 10.2 NAME: Body Part, Organ, or Organ Component
- 10.3 NAME: Body Space or Junction
- 10.4 NAME: Body Substance
- 10.5 NAME: Body System
- 10.6 NAME: Hormone
-
-
NAME: Etiologic agents
-
DEFINITION: Forces, situations, occurrences, living organisms, or other elements that may be instrumental or causative in the pathogenesis of human illness or suffering.
- 11.1 NAME: Infectious Agent
- 11.2 NAME: Trauma
-
-
NAME: Documents
- DEFINITION: A writing, as a book, report or letter, conveying information about a patient, event, or procedure.
-
NAME: Legal agreements
- DEFINITION: Contractual and other legal documents, made by or on behalf of the patient/client, in order to document patient wishes, enforce or empower patient priorities, or to assure legal resolution of issues in a manner in keeping with the patient's personal choices.
-
NAME: Agents
DEFINITION: Agents are other individuals, possibly themselves clients or patients, who must be referenced in the CPR because of important family or personal relationships to the client/patient.
References
- 1.Dick RS, Steen EB. The computer-based patient record: An essential technology for health care. Washington, DC: National Academy Press, 1991. [PubMed]
- 2.Cimino JJ, Clayton PD, Hripcsak G, Johnson S. Knowledge-based approaches to the maintenance of a large controlled medical terminology. JAMIA. 1994;1: 35-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.International Standards Organization. International Standard ISO 1087: Terminology—Vocabulary. Geneva, Switzerland: International Standards Organization, 1990.
- 4.Read JD, Sanderson HF, Drennan YM. Terming, encoding and grouping. MEDINFO 1995: Proceedings of the Eighth World Congress on Medical Informatics. Greenes RA (ed). 1995; North Holland Publishing, pp. 56-59. [PubMed]
- 5.Chute C, Cohn S, Campbell K, Oliver D, Campbell JR. The content coverage of clinical classifications. JAMIA. 1996;3: 224-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell JR, Payne TH. A comparison of four schemes for codification of problem lists. Proc Ann Symp Comput Appl Med Care. 1994; 201-5. [PMC free article] [PubMed]
- 7.Rosenberg KM, Coultas DB. Acceptability of Unified Medical Language System terms as substitute for natural language general medicine clinic diagnoses. Proc Annu Symp Comput Appl Med Care. 1994; 193-7. [PMC free article] [PubMed]
- 8.Henry SB, Campbell KE, Holzemer WL. Representation of nursing terms for the description of patient problems using SNOMED III. Proc Ann Symp Comp Appl Med Care. 1993; 700-4. [PMC free article] [PubMed]
- 9.Campbell JR, Kallenberg GA, Sherrick RC. The Clinical Utility of META: an analysis for hypertension. Proc Ann Symp Comput App Med Care. 1992; 397-401. [PMC free article] [PubMed]
- 10.Payne TH, Martin DR. How useful is the UMLS metathesaurus in developing a controlled vocabulary for an automated problem list? Proc Ann Symp Comput App Med Care. 1993; 705-9. [PMC free article] [PubMed]
- 11.Payne TH, Murphy GR, Salazar AA. How well does ICD9 represent phrases used in the medical record problem list? Proc Ann Symp Comput App Med Care. 1992; 205-9. [PMC free article] [PubMed]
- 12.Friedman C. The UMLS coverage of clinical radiology. Proc Ann Symp Comput App Med Care. 1992; 309-13. [PMC free article] [PubMed]
- 13.Henry SB, Holzemer WL, Reilly CA, Campbell KE. Terms used by nurses to describe patient problems. JAMIA. 1994; 1: 61-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zielstorf RD, Cimino C, Barnett GO, Hassan L, Blewett DR. Representation of nursing terminology in the UMLS metathesaurus; a pilot study. Proc Ann Symp Comp App Med Care. 1992; 392-6. [PMC free article] [PubMed]
- 15.Chisolm J. The Read Classification. BMJ. 1990;300: 1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Read Codes File Structure Version 3: Overview and Technical description. Woodgate, Leicestershire, UK: NHS Centre for Coding and Classification, 1994.
- 17.Cote RA, Rothwell DJ, Palotay JL, Beckett RS, Brochu L, eds. The Systematized Nomenclature of Human and Veterinary Medicine: SNOMED International. College of American Pathologists, 1993.
- 18.Cad RA, Robboy S. Progress in medical information management. Systematized nomenclature of medicine (SNOMED). JAMA. 1980;243: 756-62. [DOI] [PubMed] [Google Scholar]
- 19.Evans DA, Rothwell DJ, Monarch IA, et al. Toward representations for medical concepts. Med Decision Making. 1991;11(4 Suppl): S102-8. [PubMed] [Google Scholar]
- 20.Lindberg DA, Humphreys BL, McCray AT. The Unified Medical Language System. Meth Inf Med. 1993;32: 281-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindberg DAB, Humphreys BL, McCray AT. Yearbook of Medical Informatics. “The Unified Medical Language System.” 1993; 41-51. [DOI] [PMC free article] [PubMed]
- 22.Humphreys BL, Lindberg DA. The UMLS project: making the conceptual connection between users and the information they need. Bull Med Libr Assoc. 1993;81: 170-7. [PMC free article] [PubMed] [Google Scholar]
- 23.McCray AT, Razi A. The UMLS knowledge source server. MEDINFO. 1995; 144-7. [PubMed]
- 24.McCray AT, Divita G. ASN.1 Defining a Grammar for the UMLS Knowledge Sources. Proc Ann Symp Comp App Med Care. 1995; 868-72. [PMC free article] [PubMed]
- 25.Overview of the SAS System. SAS Institute Inc. Cary, North Carolina. 1990.
- 26.Campbell KE, Das AK, Musen MA. A logical foundation for the representation of clinical data. JAMIA. 1994; 218-32. [DOI] [PMC free article] [PubMed]
- 27.Stitt FW. A standards-based clinical information system for HIV/AIDS. Med Info. 1995;8 Pt 1: 402. [PubMed] [Google Scholar]
- 28.Cimino JJ. From ICD-9-CM to MESH using the UMLS: a how-to-guide. Proc Ann Symp Comput Appl Med Care. 1993: 730-4. [PMC free article] [PubMed]
- 29.Cimino JJ. Representation of clinical laboratory terminology in the Unified Medical Language System. Proc Annu Symp Comput Appl Med Care. 1991; 199-203. [PMC free article] [PubMed]
- 30.Board of Directors of the American Medical Informatics Association. Standards for Medical Identifiers, Codes and Messages Needed to Create an Efficient Computer-stored Medical Record. JAMIA. 1994;1: 1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]