Abstract
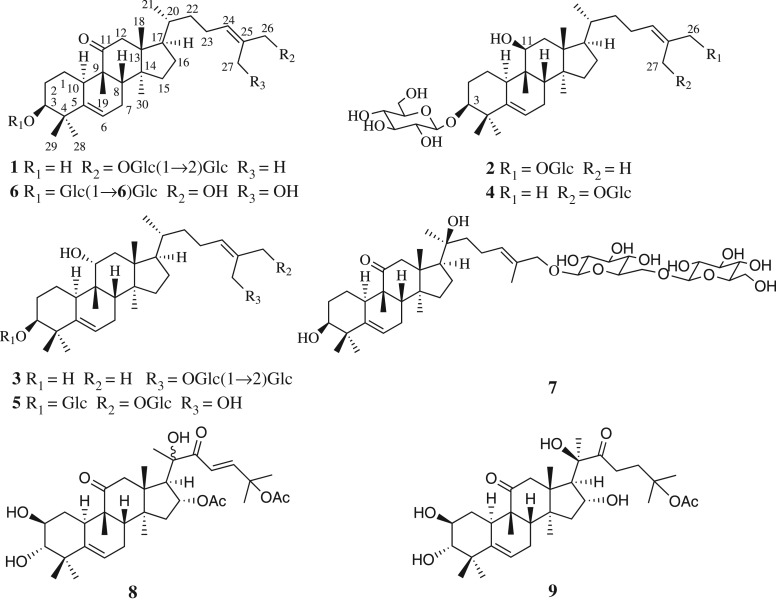
Seven new cucurbitane glucosides, hemslepensides J-P (1–7), and two known compounds, 16,25-O-diacetyl-cucurbitane F (8) and 25-O-acetyl-23,24-dihydrocucurbitacin F (9), were isolated from the tubers of Hemsleya pengxianensis var. jinfushanensis. The structures of 1–7 were elucidated using infrared absorption spectroscopy, nuclear magnetic resonance spectroscopy and high-resolution electrospray ionization mass spectrometry. The treatment of HT29 cells, human colon cancer cells, with compounds 8 and 9 inhibited cell proliferation. Further study demonstrated that compounds 8 and 9 induced F-actin aggregation, G2/M phase cell cycle arrest and cell apoptosis in HT29 cells. In summary, the present study enriched the chemical composition research of H. pengxianensis, and suggested that the compounds 8/9 treatment may be a potentially useful therapeutic option for colon cancer.
Keywords: Cucurbitaceae, Hemsleya pengxianensis, cucurbitane triterpenoid, anti-cancer
1. Introduction
Tetracyclic triterpenes are a diverse group of natural products consisting of four rings and 30 carbon atoms. Extracted from the rhizomes, roots or the juice of various plants such as Siraitia grosvenorii (Cucurbitaceae) or Momordica Linn. (Cucurbitaceae), tetracyclic triterpenes have been reported with multiple biological activities including anti-inflammatory function, cytotoxicity and anti-cancer, preventive and curative effects against CCl4-induced hepatotoxicity and anti-fertility activities [1–3]. The genus Hemsleya (Cucurbitaceae) includes approximately 30 species, distributed in the subtropical to temperate regions in Asia, eastern India and northern Vietnam. In China, it is mainly distributed in the southwest to south central regions. Tetracyclic triterpenes were identified as the main chemical constituents in the tubers of Hemsleya pengxianensis var. jinfushanensis [4–8].
As previously reported, our studies had identified 16 new compounds with cytotoxic activity against the human cancer cell lines [6–8]. Recently, further investigation of H. pengxianensis var. jinfushanensis resulted in the discovery of another seven new cucurbitane-type triterpenoids, named hemslepensides J-P (1–7), together with two known aglycones (8–9) (figure 1). In this paper, we elucidated the isolation and structure of seven new saponins. Moreover, based on our research results and previous published data [4–8], the relationships between the nuclear magnetic resonance (NMR) characteristics and the common substituents of cucurbitane tetracyclic triterpenes were discussed.
Figure 1.
The structures of compounds 1–9.
Compound 8 was first isolated in our laboratory and its cytotoxic activity has been screened in H460, SW620 and DU145 cell lines [6]. As the main component of H. pengxianensis var. jinfushanensis, compound 9 has been shown to inhibit human lung adenocarcinoma cell line A549 growth both in vitro and in vivo [9,10]. Although compounds 8 and 9 exhibit potent cytotoxicity, the underlying mechanism in colon cancer cells remains to be elucidated. In previous experiments, we screened the cytotoxicity of compounds 8 and 9. The results revealed that these two compounds were more effective than cisplatin against colon cancer HT29 cells. So, the anti-colon cancer mechanism of compounds 8 and 9 was studied targeting the HT29 cell line.
2. Experimental procedure
2.1. Material and methods
The NMR spectra were acquired using a Bruker 600 Ultrashield NMR spectrometer (Bruker Biospin, Rheinstetten, Germany). The infrared (IR) spectra were measured using a Thermo Nicolet NEXUS 670 FTIR spectrometer (GMI, Ramsey, USA). The optical rotations were run on a JASCO P-2000 polarimeter (Jasco, Tokyo, Japan). High-resolution electrospray ionization mass spectrometry (HRESIMS) was measured on Thermo Scientific Accela PDA and LTQ-Orbitrap XL mass spectrometers (Thermo Fisher Scientific, Inc., Waltham, MA, USA). Preparative HPLC was run on an LC-6AD Shimadzu liquid chromatograph equipped with an SPD-20A UV/Vis detector (Shimadzu Corporation, Kyoto, Japan) and an ODS column (YMC-Pack ODS-A column, 250 × 20 mm, 5 µm, 12 nm, YMC Co., Ltd, Tokyo, Japan). The data of cell cycle and apoptosis were acquired using BD FACSCalibur flow cytometer (Becton, Dickinson and Company, NJ, USA). The optical density (OD) value was measured using a Victor 1420 multifunctional-counter instrument (Perkin Elmer, Waltham, MA, USA). Morphological changes were analysed under an FV3000 confocal laser scanning microscope (Olympus, Tokyo, Japan). Column chromatography (CC) was performed using an ODS C18 column (50 µm, YMC Co., Ltd, Tokyo, Japan) and silica gel (300–400 mesh, Qingdao Marine Chemistry Ltd, Qingdao, China). RPMI-1640, newborn calf serum and phosphate-buffered saline (PBS) were purchased from Hyclone (Logan, UT, USA). The trypsin/EDTA and Hoechst 33342 were purchased from Solarbio (Beijing Solarbio Science and Technology, Beijing, China). Thiazolyl blue tetrazolium bromide (MTT), d-glucose and l-glucose were purchased from Sigma-Aldrich (St Louis, USA). Propidium iodide (PI) cell cycle kit, Annexin V/PI apoptosis kit and Actin-Tracker Green kit were purchased from Beyotime Biotechnology (Shanghai, China).
2.2. Plant material
The rhizomes of H. pengxianensis were collected in September 2012 in Chongqing, China, and were identified by Dr Sirong Yi from the Chongqing Institute of Pharmaceutical Plants. Its voucher specimen (HA201209) was deposited in the Institute of Materia Medica of the Shandong Academy of Medical Science in China.
2.3. Extraction and isolation
The air-dried rhizomes (8.95 kg) of H. pengxianensis were decocted with 95% ethanol under refluxing each for 2 h three times. The total extraction was suspended in H2O and partitioned with EtOAc five times. The EtOAc fraction (650 g) was treated with silica gel CC (200–300 mesh, 10 × 120 cm, 2 kg) using a successive CH2Cl2–MeOH eluent (100 : 0→0 : 100, v/v) to give 10 fractions (A–J). Further isolation of Fr.H (35 g) was achieved via silica gel CC (CH2Cl2–MeOH, 99 : 1→0 : 100), repeated ODS gel CC (MeOH–H2O, 60 : 40 → 100 : 0) and prep-HPLC (MeOH–H2O, 80 : 20, 6.0 ml min−1, 210 nm) to give compound 1 (20 mg). Fr.I (71 g) was applied to silica gel CC (CH2Cl2–MeOH, 98 : 2 → 0 : 100) to yield subfractions (I1–I10). Fr.I5 (3.2 g), which was further purified by repeated ODS gel CC (MeOH–H2O, 50 : 50 → 100 : 0) followed by prep-HPLC (MeOH–H2O, 80 : 20, 73 : 27, respectively) to yield compounds 3 (15 mg), 4 (8 mg) and 2 (7.5 mg). The same methods with different proportions of eluent (prep-HPLC, MeOH–H2O, 68:32) were applied to Fr.I8 (6.5 g) to obtain compounds 5 (18 mg) and 6 (8 mg). Fr.I10 (2.7 g) was subjected to repeated ODS gel CC (MeOH–H2O, 40 : 60 → 100 : 0) followed by prep-HPLC (MeOH–H2O, 63 : 37) to give compound 7 (15 mg). Fr.B (236 g) was separated by silica gel CC using CH2Cl2–MeOH gradient mixtures (100 : 0 → 70 : 30) to afford six fractions (B1–B6). Fr.B3 (21.8 g) was repeated purified via ODS gel CC (MeOH–H2O, 45 : 55 → 70 : 30) to give compound 8 (4.1 g). Fr.B4 (155.2 g) was repeated crystallized in EtOAc to obtain compound 9 (121.3 g).
2.3.1. Hemslepenside J (1)
C42H68O13, white amorphous powder; [α]20D +96.95 (c 0.1, MeOH : H2O = 1:1); IR (KBr) νmax 3378, 2937, 2874, 1697, 1462, 1374 and 1070 cm−1; 1H NMR and 13C NMR spectral data, see tables 1 and 2; HRESIMS m/z 798.5005 [M+NH4]+ (calcd. for C42H68O13NH4, 798.5004).
Table 1.
The 1H NMR spectroscopic data of compounds 1–7 (δ in ppm, J in Hz).
| no. | 1a | 2b | 3b | 4b | 5b | 6b | 7b |
|---|---|---|---|---|---|---|---|
| 1 | 1.06–1.09 (1H, m) | 1.51–1.55 (1H, m) | 1.53–1.57 (1H, m) | 1.51–1.55 (1H, m) | 1.31–1.35 (1H, m) | 1.24–1.27 (1H, m) | 1.22–1.25 (1H, m) |
| 1.42–1.49 (1H, m) | 1.62–1.67 (1H, m) | 2.19–2.25 (1H, m) | 1.61–1.68 (1H, m) | 2.17–2.25 (1H, m) | 1.51–1.54 (1H, m) | 1.52–1.57 (1H, m) | |
| 2 | 1.42–1.49 (1H, m) | 1.78–1.84 (1H, m) | 1.42–1.45 (1H, m) | 1.81–1.84 (1H, m) | 1.93–1.94 (1H, m) | 1.32–1.37 (1H, m) | 1.57–1.63 (1H, m) |
| 1.70–1.75 (1H, m) | 2.05–2.09 (1H, m) | 1.50–1.53 (1H, m) | 2.05–2.10 (1H, m) | 2.21–2.25 (1H, m) | 1.98–2.02 (1H, m) | 1.79–1.84 (1H, m) | |
| 3 | 3.28 (1H, br. s) | 3.42 (1H, br. s) | 3.41 (1H, br. s) | 3.42 (1H, br. s) | 3.42 (1H, br. s) | 3.46 (1H, br. s) | 3.43 (1H, br. s) |
| 6 | 5.45–5.48 (1H, m) | 5.54 (1H, dd, 5.8, 2.1) | 5.49 (1H, dt, 6.5, 1.7) | 5.54 (1H, dd, 6.2, 1.8) | 5.49 (1H, d, 6.1) | 5.61 (1H, dt, 4.3, 2.0) | 5.63 (1H, d, 5.6) |
| 7 | 1.84–1.86 (1H, m) | 1.84–1.88 (1H, m) | 1.82–1.84 (1H, m) | 1.84–1.88 (1H, m) | 1.81–1.83 (1H, m) | 1.93–1.96 (1H, m) | 1.91–1.95 (1H, m) |
| 2.24–2.31 (1H, m) | 2.31–2.38 (1H, m) | 2.39–2.45 (1H, m) | 2.31–2.38 (1H, m) | 2.35–2.42 (1H, m) | 2.33–2.37 (1H, m) | 2.37–2.40 (1H, m) | |
| 8 | 1.82–1.84 (1H, m) | 1.93–1.94 (1H, m) | 1.67 (1H, d, 7.5) | 1.93–1.95 (1H, m) | 1.67 (1H, d, 7.5) | 1.93–1.96 (1H, m) | 1.95–1.99 (1H, m) |
| 10 | 2.33–2.39 (1H, m) | 2.02–2.05 (1H, m) | 2.49 (1H, dd, 12.2, 3.4) | 2.02–2.05 (1H, m) | 2.48 (1H, d, 12.3) | 1.44–1.48 (1H, m) | 2.40–2.44 (1H, m) |
| 11 | 3.88 (1H, dd, 4.2, 2.0) | 3.85 (1H, dd, 11.5, 5.5) | 3.88 (1H, dd, 4.4, 2.1) | 3.84 (1H, dd, 11.4, 5.1) | |||
| 12 | 2.21 (1H, d, 14.2) | 1.97–2.02 (2H, m) | 1.77–1.82 (2H, m) | 1.97–2.02 (2H, m) | 1.83–1.89 (2H, m) | 2.38 (1H, d, 14.2) | 2.47 (1H, d, 14.3) |
| 3.09 (1H, d, 14.2) | 3.07 (1H, d, 14.2) | 3.09 (1H, d, 14.3) | |||||
| 15 | 1.19–1.24 (1H, m) | 1.21–1.26 (2H, m) | 1.16–1.26 (2H, m) | 1.21–1.26 (2H, m) | 1.12–1.14 (1H, m) | 1.28–1.32 (1H, m) | 1.33–1.42 (2H, m) |
| 1.31–1.34 (1H, m) | 1.20–1.23 (1H, m) | 1.40–1.42 (1H, m) | |||||
| 16 | 1.28–1.31 (1H, m) | 1.28–1.30 (1H, m) | 1.27–1.33 (1H, m) | 1.26–1.31 (1H, m) | 1.27–1.30 (1H, m) | 1.38–1.40 (1H, m) | 1.84–1.91 (2H, m) |
| 1.92–1.99 (1H, m) | 1.88–1.91 (1H, m) | 1.96–1.99 (1H, m) | 1.88–1.91 (1H, m) | 1.94–1.96 (1H, m) | 1.74–1.77 (1H, m) | ||
| 17 | 1.65–1.70 (1H, m) | 1.49–1.51 (1H, m) | 1.57–1.63 (1H, m) | 1.48–1.50 (1H, m) | 1.61 (1H, q, 9.0) | 1.77–1.79 (1H, m) | 2.11 (1H, t, 9.5) |
| 18 | 0.65 (3H, s) | 1.02 (3H, s) | 0.91 (3H, s) | 1.02 (3H, s) | 0.91 (3H, s) | 0.75 (3H, s) | 0.91 (3H, s) |
| 19 | 0.96 (3H, s) | 0.97 (3H, s) | 1.15 (3H, s) | 0.97 (3H, s) | 1.11 (3H, s) | 1.06 (3H, s) | 1.10 (3H, s) |
| 20 | 1.35–1.41 (1H, m) | 1.46–1.48 (1H, m) | 1.45–1.47 (1H, m) | 1.45–1.47 (1H, m) | 1.49–1.51 (1H, m) | 2.40–2.45 (1H, m) | |
| 21 | 0.87 (3H, d, 6.4) | 0.98 (3H, d, 6.7) | 0.97 (3H, d, 6.4) | 0.98 (3H, d, 4.7) | 0.98 (3H, d, 6.3) | 0.94 (3H, d, 6.5) | 1.26 (3H, s) |
| 22 | 1.35–1.41 (1H, m) | 1.04–1.14 (2H, m) | 1.00–1.04 (1H, m) | 1.03–1.13 (2H, m) | 1.14–1.18 (1H, m) | 1.10–1.14 (1H, m) | 1.42–1.52 (2H, m) |
| 1.05–1.06 (1H, m) | 1.06–1.09 (1H, m) | 1.51–1.54 (1H, m) | 1.48–1.51 (1H, m) | ||||
| 23 | 1.86–1.89 (1H, m) | 1.95–1.97 (1H, m) | 1.99–2.05 (1H, m) | 1.95–1.97 (1H, m) | 1.76–1.81 (1H, m) | 2.02–2.09 (1H, m) | 2.06 (2H, q, 7.9) |
| 2.00–2.07 (1H, m) | 2.09–2.15 (1H, m) | 2.11–2.19 (1H, m) | 2.12–2.19 (1H, m) | 2.02–2.10 (1H, m) | 2.17–2.24 (1H, m) | ||
| 24 | 5.40 (1H, t, 6.8) | 5.47 (1H, t, 7.1) | 5.37 (1H, t, 7.0) | 5.38 (1H, t, 7.0) | 5.63 (1H, t, 7.4) | 5.55 (1H, t, 7.3) | 5.49 (1H, t, 6.7) |
| 26 | 3.89 (1H, d, 11.7) | 4.04 (1H, d, 11.5) | 1.79 (3H, s) | 1.77 (3H, s) | 4.16 (1H, d, 11.7) | 4.08 (2H, s) | 4.01 (1H, d, 11.5) |
| 4.11 (1H, d, 11.7) | 4.20 (1H, d, 11.5) | 4.38 (1H, d, 11.7) | 4.20 (1H, d, 11.5) | ||||
| 27 | 1.60 (3H, s) | 1.68 (3H, s) | 4.27 (1H, d, 11.6) | 4.19 (1H, d, 11.5) | 4.17 (1H, d, 12.3) | 4.16 (2H, s) | 1.68 (3H, s) |
| 4.31 (1H, d, 11.6) | 4.33 (1H, d, 11.5) | 4.20 (1H, d, 12.3) | |||||
| 28 | 0.95 (3H, s) | 1.01 (3H, s) | 1.10 (3H, s) | 1.01 (3H, s) | 1.06 (3H, s) | 1.06 (3H, s) | 1.04 (3H, s) |
| 29 | 1.04 (3H, s) | 1.20 (3H, s) | 1.05 (3H, s) | 1.20 (3H, s) | 1.19 (3H, s) | 1.23 (3H, s) | 1.14 (3H, s) |
| 30 | 1.00 (3H, s) | 0.84 (3H, s) | 0.86 (3H, s) | 0.84 (3H, s) | 0.87 (3H, s) | 1.09 (3H,s) | 1.11 (3H, s) |
| Glc′ | 26-Glc (inner) | 3-Glc | 27-Glc (inner) | 3-Glc | 3-Glc | 3-Glc (inner) | 26-Glc (inner) |
|---|---|---|---|---|---|---|---|
| 1′ | 4.26 (1H, d, 7.8) | 4.28 (1H, d, 7.8) | 4.36 (1H, d, 7.7) | 4.28 (1H, d, 7.8) | 4.27 (1H, d, 7.8) | 4.28 (1H, d, 7.8) | 4.24 (1H, d, 7.9) |
| 2′ | 3.24–3.27 (1H, m) | 3.31–3.33 (1H, m) | 3.45–3.49 (1H, m) | 3.31–3.32 (1H, m) | 3.19–3.20 (1H, m) | 3.35–3.37 (1H, m) | 3.18–3.21 (1H, m) |
| 3′ | 3.14–3.18 (1H, m) | 3.33–3.35 (1H, m) | 3.52–3.56 (1H, m) | 3.33–3.34 (1H, m) | 3.31–3.33 (1H, m) | 3.19–3.21 (1H, m) | 3.38–3.42 (1H, m) |
| 4′ | 3.07–3.11 (1H, m) | 3.25–3.28 (1H, m) | 3.35–3.37 (1H, m) | 3.26–3.29 (1H, m) | 3.25–3.30 (1H, m) | 3.26–3.28 (1H, m) | 3.34–3.35 (1H, m) |
| 5′ | 3.11–3.14 (1H, m) | 3.20–3.23 (1H, m) | 3.21–3.23 (1H, m) | 3.20–3.24 (1H, m) | 3.25–3.30 (1H, m) | 3.29–3.30 (1H, m) | 3.21–3.27 (1H, m) |
| 6′ | 3.63–3.68 (1H, m) | 3.65–3.68 (1H, m) | 3.67–3.70 (1H, m) | 3.67–3.71 (1H,m) | 3.65–3.68 (1H, m) | 3.78 (1H, dd, 12.0, 6.0) | 3.78 (1H, dd, 12.1, 5.0) |
| 3.45–3.50 (1H, m) | 3.84–3.87 (1H, m) | 3.83–3.85 (1H, m) | 3.84–3.87 (1H, m) | 3.83–3.88 (1H, m) | 4.07 (1H, dd, 12.0, 1.9) | 4.13 (1H, dd, 12.1, 1.9) |
| Glc″ | 26-Glc (terminal) | 26-Glc | 27-Glc (terminal) | 27-Glc | 26-Glc | 3-Glc (terminal) | 26-Glc (terminal) |
|---|---|---|---|---|---|---|---|
| 1″ | 4.40 (1H, d, 7.8) | 4.24 (1H, d, 7.9) | 4.62 (1H, d, 7.8) | 4.21 (1H, d, 7.8) | 4.28 (1H, d, 7.8) | 4.41 (1H, d, 7.8) | 4.39 (1H, d, 7.8) |
| 2″ | 2.97–3.01 (1H, m) | 3.16–3.18 (1H, m) | 3.24–3.26 (1H, m) | 3.16–3.20 (1H,m) | 3.17–3.19 (1H, m) | 3.17–3.19 (1H, m) | 3.32–3.34 (1H, m) |
| 3″ | 3.02–3.05 (1H, m) | 3.28–3.30 (1H, m) | 3.23–3.24 (1H, m) | 3.31–3.34 (1H,m) | 3.31–3.33 (1H, m) | 3.39–3.43 (1H, m) | 3.35–3.38 (1H, m) |
| 4″ | 3.07–3.11 (1H, m) | 3.18–3.20 (1H, m) | 3.27–3.30 (1H, m) | 3.20–3.24 (1H,m) | 3.21–3.25 (1H, m) | 3.32–3.34 (1H, m) | 3.28 (1H, d, 8.2) |
| 5″ | 3.35–3.37 (1H, m) | 3.20–3.23 (1H, m) | 3.31–3.34 (1H, m) | 3.16–3.20 (1H,m) | 3.20–3.21 (1H, m) | 3.28–3.29 (1H, m) | 3.21–3.27 (1H, m) |
| 6″ | 3.58–3.63 (1H, m) | 3.63–3.65 (1H, m) | 3.63–3.67 (1H, m) | 3.63–3.67 (1H,m) | 3.63–3.65 (1H, m) | 3.67 (1H, dd, 11.9, 5.2) | 3.66 (1H, dd, 11.9, 5.5) |
| 3.40–3.45 (1H, m) | 3.81–3.84 (1H, m) | 3.80–3.83 (1H, m) | 3.81–3.84 (1H, m) | 3.79–3.83 (1H, m) | 3.87 (1H, dd, 11.9, 2.1) | 3.86 (1H, dd, 11.9, 2.1) |
a1H NMR data measured in DMSO-d6 at 600 MHz.
b1H NMR data measured in MeOH-d4 at 600 MHz.
Table 2.
The 13C NMR (150 MHz) spectroscopic data for compounds 1–7.
| no. | 1a | 2b | 3b | 4b | 5b | 6b | 7b |
|---|---|---|---|---|---|---|---|
| δc, type | δc, type | δc, type | δc, type | δc, type | δc, type | δc, type | |
| 1 | 20.2, CH2 | 24.4, CH2 | 26.6, CH2 | 24.4, CH2 | 27.4, CH2 | 22.7, CH2 | 21.8, CH2 |
| 2 | 28.5, CH2 | 29.7, CH2 | 30.6, CH2 | 29.7, CH2 | 29.7, CH2 | 29.1, CH2 | 29.9, CH2 |
| 3 | 74.2, CH | 88.6, CH | 78.0, CH | 88.6, CH | 88.7, CH | 87.8, CH | 77.1, CH |
| 4 | 41.0, C | 42.6, C | 42.8, C | 42.6, C | 43.0, C | 42.7, C | 42.4, C |
| 5 | 140.9, C | 143.8, C | 144.1, C | 143.8, C | 145.1, C | 142.2, C | 141.5, C |
| 6 | 117.5, CH | 120.5, CH | 120.7, CH | 120.5, CH | 119.8, CH | 119.8, CH | 120.6, CH |
| 7 | 23.5, CH2 | 25.4, CH2 | 25.2, CH2 | 25.4, CH2 | 25.2, CH2 | 25.0, CH2 | 24.9, CH2 |
| 8 | 43.2, CH | 43.0, CH | 44.9, CH | 43.0, CH | 44.8, CH | 45.5, CH | 44.8, CH |
| 9 | 48.1, C | 41.4, C | 41.0, C | 41.4, C | 41.2, C | 50.5, C | 50.3, C |
| 10 | 34.5, CH | 41.0, CH | 37.2, CH | 41.0, CH | 37.4, CH | 37.0, CH | 36.8, CH |
| 11 | 213.5, C | 74.0, CH | 79.4, CH | 74.0, CH | 79.5, CH | 217.9, C | 217.9, C |
| 12 | 48.3, CH2 | 39.5, CH2 | 41.2, CH2 | 39.5, CH2 | 41.0, CH2 | 49.7, CH2 | 50.1, CH2 |
| 13 | 48.4, C | 46.4, C | 48.4, C | 46.4, C | 48.4, C | 50.2, C | 51.3, C |
| 14 | 49.0, C | 50.8, C | 50.7, C | 50.8, C | 50.7, C | 50.9, C | 50.3, C |
| 15 | 33.9, CH2 | 36.1, CH2 | 35.5, CH2 | 36.1, CH2 | 35.5, CH2 | 35.6, CH2 | 35.1, CH2 |
| 16 | 27.3, CH2 | 29.0, CH2 | 29.3, CH2 | 29.0, CH2 | 29.2, CH2 | 29.0, CH2 | 22.9, CH2 |
| 17 | 48.7, CH | 52.3, CH | 51.8, CH | 52.3, CH | 51.7, CH | 50.7, CH | 52.2, CH |
| 18 | 16.6, CH3 | 18.1, CH3 | 17.3, CH3 | 18.0, CH3 | 17.3, CH3 | 17.4, CH3 | 19.4, CH3 |
| 19 | 19.6, CH3 | 22.6, CH3 | 26.4, CH3 | 22.6, CH3 | 26.3, CH3 | 20.4, CH3 | 20.4, CH3 |
| 20 | 35.1, CH | 37.3, CH | 37.3, CH | 37.3, CH | 37.2, CH | 37.0, CH | 75.8, C |
| 21 | 18.2, CH3 | 19.2, CH3 | 19.4, CH3 | 19.3, CH3 | 19.2, CH3 | 19.0, CH3 | 26.1, CH3 |
| 22 | 35.5, CH2 | 37.3, CH2 | 37.9, CH2 | 38.0, CH2 | 37.5, CH2 | 37.4, CH2 | 44.9, CH2 |
| 23 | 23.7, CH2 | 25.7, CH2 | 25.8, CH2 | 25.7, CH2 | 25.4, CH2 | 25.2, CH2 | 23.8, CH2 |
| 24 | 127.0, CH | 130.9, CH | 132.0, CH | 132.1, CH | 134.6, CH | 130.9, CH | 130.2, CH |
| 25 | 131.5, C | 132.7, C | 132.5, C | 132.4, C | 136.2, C | 139.2, C | 132.9, C |
| 26 | 73.5, CH2 | 76.1, CH2 | 22.1, CH3 | 22.0, CH3 | 73.2, CH2 | 65.7, CH2 | 76.2, CH2 |
| 27 | 13.7, CH3 | 14.3, CH3 | 68.4, CH2 | 68.0, CH2 | 58.5, CH2 | 58.4, CH2 | 14.3, CH3 |
| 28 | 27.5, CH3 | 28.6, CH3 | 27.6, CH3 | 28.7, CH3 | 28.0, CH3 | 28.8, CH3 | 28.5, CH3 |
| 29 | 25.7, CH3 | 26.1, CH3 | 26.6, CH3 | 26.1, CH3 | 26.5, CH3 | 26.0, CH3 | 26.2, CH3 |
| 30 | 17.7, CH3 | 18.7, CH3 | 20.0, CH3 | 18.7, CH3 | 19.9, CH3 | 19.0, CH3 | 19.2, CH3 |
| Glc′ | 26-Glc (inner) | 3-Glc | 27-Glc (inner) | 3-Glc | 3-Glc | 3-Glc (inner) | 26-Glc (inner) |
|---|---|---|---|---|---|---|---|
| 1′ | 100.2, CH | 106.8, CH | 101.4, CH | 106.8, CH | 106.7, CH | 106.5, CH | 102.9, CH |
| 2′ | 82.0, CH | 75.7, CH | 82.1, CH | 75.7, CH | 75.7, CH | 75.7, CH | 75.2, CH |
| 3′ | 76.1, CH | 78.4, CH | 78.4, CH | 78.4, CH | 78.4, CH | 78.1, CH | 77.2, CH |
| 4′ | 69.8, CH | 71.8, CH | 71.7, CH | 71.8, CH | 71.8, CH | 71.8, CH | 71.6, CH |
| 5′ | 76.7, CH | 78.0, CH | 77.9, CH | 78.0, CH | 78.1, CH | 78.3, CH | 78.2, CH |
| 6′ | 60.8, CH2 | 63.0, CH2 | 62.8, CH2 | 63.0, CH2 | 63.0, CH2 | 70.0, CH2 | 69.8, CH2 |
| Glc″ | 26-Glc (terminal) | 26-Glc | 27-Glc (terminal) | 27-Glc | 26-Glc | 3-Glc (terminal) | 26-Glc (terminal) |
|---|---|---|---|---|---|---|---|
| 1″ | 104.1, CH | 102.6, CH | 104.9, CH | 102.6, CH | 102.9, CH | 105.0, CH | 104.9, CH |
| 2″ | 75.0, CH | 75.2, CH | 76.0, CH | 75.2, CH | 75.2, CH | 75.3, CH | 75.1, CH |
| 3″ | 76.9, CH | 78.3, CH | 78.3, CH | 78.3, CH | 78.3, CH | 77.3, CH | 78.1, CH |
| 4″ | 69.7, CH | 71.8, CH | 71.5, CH | 71.7, CH | 71.8, CH | 71.7, CH | 71.7, CH |
| 5″ | 76.2, CH | 77.8, CH | 77.7, CH | 77.8, CH | 77.8, CH | 78.1, CH | 78.2, CH |
| 6″ | 60.9, CH2 | 62.9, CH2 | 62.9, CH2 | 62.8, CH2 | 62.9, CH2 | 62.9, CH2 | 62.9, CH2 |
a13C NMR data measured in DMSO-d6 at 150 MHz.
b13C NMR data measured in MeOH-d4 at 150 MHz.
2.3.2. Hemslepenside K (2)
C42H70O13, white amorphous powder; [α]20D −21.77 (c 0.1, MeOH); IR (KBr) νmax 3417, 2926, 2866, 1075 and 1019 cm−1; 1H NMR and 13C NMR spectral data, see tables 1 and 2; HRESIMS m/z 805.4705 [M+Na]+ (calcd. for C42H70O13Na, 805.4714).
2.3.3. Hemslepenside L (3)
C42H70O13, white amorphous powder; [α]20D +32.33 (c 0.1, MeOH); IR (KBr) νmax 3417, 2932, 2871, 1076 and 1025 cm−1; 1H NMR and 13C NMR spectral data, see tables 1 and 2; HRESIMS m/z 805.4714 [M+Na]+ (calcd. for C42H70O13Na, 805.4714).
2.3.4. Hemslepenside M (4)
C42H70O13, white amorphous powder; [α]20D +19.89 (c 0.1, MeOH); IR (KBr) νmax 3385, 2935, 2866, 1074 and 1020 cm−1; 1H NMR and 13C NMR spectral data, see tables 1 and 2; HRESIMS m/z 805.4710 [M+Na]+ (calcd. for C42H70O13Na, 805.4714).
2.3.5. Hemslepenside N (5)
C42H70O14, white amorphous powder; [α]20D +17.35 (c 0.1, MeOH); IR (KBr) νmax 3384, 2941, 2866, 1076 and 1029 cm−1; 1H NMR and 13C NMR spectral data, see tables 1 and 2; HRESIMS m/z 799.4846 [M+H]+ (calcd. for C42H71O14, 799.4844).
2.3.6. Hemslepenside O (6)
C42H68O14, white amorphous powder; [α]20D +93.49 (c 0.1, MeOH); IR (KBr) νmax 3386, 2930, 2866, 1691, 1075 and 1033 cm−1; 1H NMR and 13C NMR spectral data, see tables 1 and 2; HRESIMS m/z 814.4963 [M+NH4]+ (calcd. for C42H68O14NH4, 814.4953).
2.3.7. Hemslepenside P (7)
C42H68O14, white amorphous powder; [α]20D +44.43 (c 0.1, MeOH); IR (KBr) νmax 3419, 2929, 2877, 1686, 1076 and 1032 cm−1; 1H NMR and 13C NMR spectral data, see tables 1 and 2; HRESIMS m/z 814.4964 [M+NH4]+ (calcd. for C42H68O14NH4, 814.4953).
2.4. Enzymatic hydrolysis of 1–7 and determination of the absolute configuration of the monosaccharides
To each solution of compounds 1–7 (4 mg) in H2O (4 ml) was added cellase (each 20 mg) and stirred at 40°C for 120 h. The reaction mixture was partitioned between EtOAc and H2O and the EtOAc layer was separated. The aqueous layer was evaporated under reduced press to afford solid saccharide mixture. The authentic samples of d-(+)-glucose and l-(−)-glucose and the saccharide mixture were dissolved in 1 ml of H2O and mixed with 1 ml of EtOH to which (S)-(−)-α-methylbenzylamine (7 µl) and NaBH3CN (6.75 mg) were added, respectively. The mixture was stirred at 40°C for 4 h followed by the addition of glacial acetic acid (0.2 ml) and evaporated under reduced pressure to afford solid mixture. To the solid mixture was added acetic anhydride (0.3 ml) in pyridine (0.3 ml) for acetylation for 24 h at room temperature. H2O (1 ml) was added to the reaction mixture and evaporated under reduced pressure to remove residual pyridine (repeated three times). The residue was suspended in H2O (0.5 ml) and subjected to a Cleanert C18-N column (Agela) eluented with H2O, 20% CH3CN and 50% CH3CN (15, 15 and 5 ml) successively in order to obtain 50% CH3CN fraction. The 50% CH3CN fraction was monitored at 210 nm by HPLC under the following conditions: Agilent SB-C18 column (4.6 × 250 mm, 5 µm); 40% CH3CN/H2O; 0.8 ml min−1; 30°C. The composition sugar of compounds 1–7 was determined to be d-(+)-glucose by comparing the retention time tR (min) of their 1-[(S)-N-acetyl-a-methylbenzylamino]-1-deoxyglucitol acetate derivatives with those of authentics: 20.8 min (derivative of d-glucose) and 19.4 min (derivative of l-glucose).
2.5. Cell culture and cytotoxicity assay
The HT29 cell lines were purchased from the Institute of Materia Medica of the Chinese Academy of Medical Sciences. The cells were cultured in RPMI-1640 containing 10% newborn calf serum at 37°C in a humidified 5% CO2 air. The cells were detached using a solution of 0.25% trypsin/EDTA. Approximately 6 × 103 cells per well were plated in 96-well plates. After incubating for 24 h, cisplatin, DMSO (2‰) and isolated compounds (100, 10, 1, 0.1, 0.01 and 0 µM) were added for another 48 h, respectively. Then, the cells per well were treated with 10 µl of MTT solution (5 mg ml−1 in PBS) and reincubated for 4 h at 37°C. The medium was removed and 150 µl well−1 DMSO was replaced. The plates were oscillated gently for 5–10 min to dissolve fully in a 96-well plate oscillator. The OD value of each well was measured at 570 nm using the Victor 1420 instrument, and the half-maximal inhibitory concentration (IC50) values were calculated using the SPSS statistical software. Cisplatin served as a positive control. At least, three independent experiments were performed.
2.6. Flow cytometric analysis for cell cycle distribution
The 3 × 105 well−1 HT29 cells were seeded into six-well plates in 2 ml of medium and incubated at 37°C overnight. Then, the HT29 cells were treated with DMSO (2‰), compounds 8 and 9 (0, 0.1 or 1 µM, respectively) for 24 h. Afterwards, the cells were collected by trypsinization and fixed in 70% ice-cold ethanol at 4°C for another 24 h. The cells were then washed twice with PBS and stained with fluorochrome solution including PI and RNAse in the dark for 30 min at 37°C. The DNA content was analysed by flow cytometer. At least three independent experiments were performed.
2.7. Cell apoptosis detection
The HT29 cells were plated at a concentration of 3 × 105 cells per well in 2 ml of medium in six-well plates overnight. Then, the cells were treated with either 0.1 or 1 µM compounds 8 and 9, respectively, for 24 h. Following harvesting by trypsinization, the cells were washed once with PBS and then were resuspended in 195 µl of Annexin V-FITC binding buffer. Afterwards, 5 µl of Annexin V-FITC and 10 µl of PI were added in turn followed by incubating in the dark for 15 min at room temperature (20–25°C). The cells were analysed using flow cytometer. At least three independent experiments were performed.
2.8. Fluorescence microscopy
Approximately 5 × 104 HT29 cells per well were grown on sterilized glass coverslips. After the treatment with either 0.1 or 1 µM compounds 8 and 9, respectively, for 24 h, the cells were fixed with beyotime P0098 fixative (15 min) and then washed three times with beyotime P0106 clearing solution (5 min each time). The actin-tracker green staining solution (200 µl well−1) was added to the coverslips in the dark and incubated for 60 min at room temperature. The stained cells were rinsed three times with beyotime P0106 clearing solution (5 min each time) followed by staining using the Hoechst 33342 for 5 min and washed three times (3 min each time) in PBS. Morphological changes were analysed under an FV3000 confocal laser scanning microscope.
2.9. Statistical analysis
All statistical comparisons were made by Statistical Product and Service Solutions (SPSS, v. 20.0). The differences were described as statistically significant if p < 0.05.
3. Results and discussion
3.1. The structure elucidation of seven new saponins
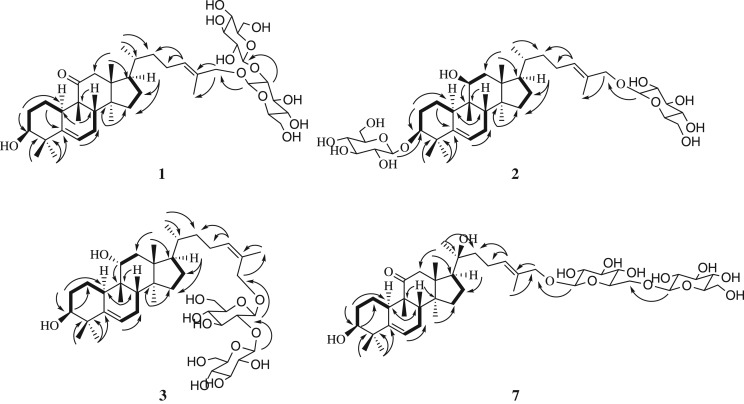
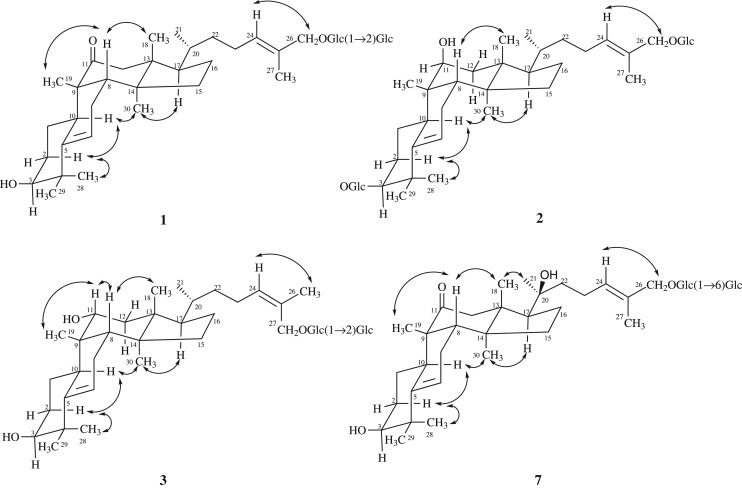
Hemslepenside J (1) was obtained as a white solid. The molecular formula was determined as C42H68O13 on the basis of the HRESIMS data analysis (m/z 798.5005 [M+NH4]+, calcd. for 798.5004). The IR absorption bands at 3378, 2937 and 1697 cm−1 indicated the presence of hydroxyl, methyl, methylene and carbonyl groups. Comparison of the NMR data of 1 (tables 1 and 2) with those of delavanoside A indicated that these two compounds have the same aglycone and they just differ in the attachment point of the sugar residue [11]. The sugar residue was attached to C-26 (δC 73.5) in 1, which was confirmed by the HMBC correlations from δH 4.26 (1H, d, J = 7.8 Hz, H-1′) to δC 73.5 (C-26) (figure 2). The equatorial configuration of H-3 could be deduced from its couple constant J3eq, 2ax = J3eq, 2eq and an obvious broad singlet of oxygenated methine proton at δH 3.28 (H-3). The relative configuration of 1 was determined via NOESY data (figure 3), in which correlations of H-28/H-10, H-10/H-30 and H-30/H-17 indicated the α-orientation of H-10, H-17 and CH3-30. Meanwhile, the NOESY correlations of H-19/H-8 and H-8/H-18 indicated the β-orientation of H-8, CH3-18 and CH3-19. Enzymatic hydrolysis of 1 afforded d-glucose as the composition sugar via the HPLC analysis of its 1-[(S)-N-acetyl-α-methylbenzylamino]-1-deoxyglucitol acetate derivatives. The large coupling constant value 7.8 Hz (JH-1′,H-2′ and JH-1″,H-2″) of anomeric protons showed the presence of two β-glucopyranosyl moieties. The trans configuration of Δ24 was confirmed by the NOESY correlation of H-24/H-26 (figure 3). Thus, compound 1 was determined to be 3β,26-dihydroxycucurbita-5,24(E)-diene-11-one-26-O-β-d-glucopyranosyl(1 → 2)-β-d-glucopyranoside.
Figure 2.
Key 1H-1H COSY (bold bonds) and HMBC (arrows) correlations for compounds 1–3 and 7.
Figure 3.
Key NOESY correlations of compounds 1–3 and 7.
Hemslepenside K (2) was isolated as a white solid. The molecular formula was determined as C42H70O13 from the molecular ion peaks [M+Na]+ at m/z 805.4705 (calcd. for C42H70O13Na, 805.4714) in the positive-mode HRESIMS. The NMR data of 2 (tables 1 and 2) were almost identical with that of hemslepenside H except for C-11 which could be ascribed to the α-orientation hydroxyl of C-11 in hemslepenside H replaced by β-orientation hydroxyl of C-11 in 2 [8]. Above deduction was confirmed by the two small coupling constants 4.2 Hz (JH-11,Ha-12) and 2.0 Hz (JH-11,Hb-12) of δH 3.88 (1H, dd, H-11). In addition, the trans configuration of the Δ24 was determined via the NOESY correlations from δH 5.47 (1H, t, J = 7.1 Hz, H-24) to δH 4.04 (1H, d, J = 11.5 Hz, Ha-26) and 4.20 (1H, d, J = 11.5 Hz, Hb-26). Thus, the structure of 2 was determined to be 3-O-β-d-glucopyranosyl-3β,11β,26-trihydroxycucurbita-5,24(E)-diene-26-O-β-d-glucopyranoside.
Hemslepenside L (3) was isolated as a white solid. The molecular formula C42H70O13 was determined via its HRESIMS data (m/z 805.4714 [M+Na]+, calcd. for 805.4714), which suggested that 3 and hemslepenside H were a pair of isomers [8]. The 13C NMR data of 3 (table 2) and hemslepenside H were almost identical with the exception of the β-d-glucose link location. Two β-d-glucoses were linked to C-3 and C-26, respectively, in hemslepenside H, whereas two β-d-glucoses composed a disaccharide unit, then linked to C-27 in 3, which could be confirmed by HMBC correlations from δH 4.36 (1H, d, J = 7.7 Hz, H-1′) to δC 68.4 (C-27), from δH 4.62 (1H, d, J = 7.8 Hz, H-1″) to δC 82.1 (C-2′). The relative configuration of 3 was deduced from NOESY data (figure 3), especially the NOESY correlations from δH 5.37 (1H, t, J = 7.0 Hz, H-24) to δH 1.79 (3H, s, H-26) showed that the terminal double bond was the cis configuration. Thus, the structure of 3 was determined to be 3β,11α,27-trihydroxycucurbita-5,24(Z)-diene-27-O-β-d-glucopyranosyl(1 → 2)-β-d-glucopyranoside.
Hemslepenside M (4) was isolated as a white solid and an isomer of compound 2 according to the HRESIMS data (m/z 805.4710 [M+Na]+, calcd. for 805.4714). The 1H and 13C NMR spectra (tables 1 and 2) displayed one β-orientation hydroxyl at C-11 (δC 74.0), and two β-d-glucoses linked at C-3 (δC 88.6) and C-27 (δC 68.0), respectively. The above deduction was confirmed by the two small coupling constants 4.4 Hz (JH-11,Ha-12) and 2.1 Hz (JH-11,Hb-12) of δH 3.88 (H-11), and the HMBC correlations from H-1′ (δH 4.28, 1H, d, J = 7.8 Hz) to δC 88.6 (C-3) and from H-1″ (δH 4.21, 1H, d, J = 7.8 Hz) to δC 68.0 (C-27). The structure and relative configuration of 4 were almost identical to those of 2 by comparing the NMR and NOESY data (figure 3). The significant differences, the NOESY correlation between δH 5.38 (1H, t, J = 7.0 Hz, H-24) and 1.77 (3H, s, H-26) in 4, emerged at C-26, which further confirmed the cis double bond at C-24 in 4 instead of the trans one in 2. Thus, the structure of 4 was determined to be 3-O-β-d-glucopyranosyl-3β,11β,27-trihydroxycucurbita-5,24(Z)-diene-27-O-β-d-glucopyranoside.
Hemslepenside N (5) was obtained as a white solid. It showed a positive ion peak at m/z 799.4846 [M+H]+ (calcd. for 799.4844), which determined the molecular formula of 5 as C42H70O14. The molecular weight of 5 was 16 mass units more than that of hemslepenside H [8], which implied an additional hydroxyl unit in 5. The significant difference was a hydroxymethyl signal at δC 58.5 (C-27) in 5 but a methyl signal at δC 15.0 (C-27) in hemslepenside H, which was consistent with the results of molecular weight analysis. In addition, the 24(E)-ene was deduced from the NOESY correlation of H-24/H-26. Thus, the structure of 5 was determined to be 3-O-β-d-glucopyranosyl-3β,11α,26,27-tetrahydroxycucurbita-5,24(E)-diene-26-O-β-d-glucopyranoside.
Hemslepenside O (6) was isolated as a white amorphous powder and had a molecular formula of C42H68O14 according to positive ion HRESIMS (m/z 814.4963 [M+NH4]+, calcd. for 814.4953). The molecular weight of 6 was 162 mass units more than that of jinfushanoside B [4], which suggested an additional hexose unit existed in 6. The additional β-d-glucose was connected to C-6′, which could be deduced from the HMBC correlations from δH 4.41 (1H, d, J = 7.8 Hz, H-1″) to δC 70.0 (C-6′). The same NOESY and HMBC correlations for the 26,27-dihydroxycucurbita-11-one skeleton as those in jinfushanoside B were found. Thus, the structure of 6 was assigned as 3β,26,27-trihydroxycucurbita-5,24-diene-11-one-3-O-β-D-glucopyranosyl(1 → 6)-β-D-glucopyranoside.
Hemslepenside P (7) was isolated as a white amorphous powder. The elemental formula for 7 was assigned as C42H68O14 from the ion peak [M+NH4]+ at m/z 814.4964 (calcd. for 814.4953) in the positive-mode HRESIMS. In the 13C NMR and DEPT spectra, the significant difference between 7 and carnosifloside I came from the great downfield of C-20 (from δC 35.9 in carnosifloside I to 75.8 in 7), which indicated the attachment of a hydroxyl to C-20 in 7 [12]. The above deduction was confirmed by its 16 mass units more than that of carnosifloside I and supported by HMBC correlations from δH 1.26 (3H, s, H-21) to δC 75.8 (C-20), 44.9 (C-22) and 52.2 (C-17). The NOESY correlations from δH 0.91 (3H, s, H-18) to 1.26 (3H, s, H-21) indicated the S-configuration of C-20. Thus, the structure of 7 was established as 3β,20S,26-trihydroxycucurbita-5,24(E)-diene-11-one-26-O-β-d-glucopyranosyl(1→6)-β-d-glucopyranoside.
3.2. Cell experiments
3.2.1. Growth inhibitory properties of compounds 8 and 9 in HT29 cells
Seven new compounds (1–7) and two known compounds (8, 9) were evaluated for cytotoxicity in HT29 cell line. HT29 cells were treated with different concentrations (0, 0.01, 0.1, 1, 10 or 100 µM) of 1–9 for 48 h, or treated with DMSO (2‰) as solvent control. After 48 h of treatment, their IC50 values are shown in table 3. Compounds 8 and 9 displayed significant cytotoxicity; however, except compound 2, the other six new compounds had no cytotoxicity. The results suggest that the cytotoxicity of cucurbitane tetracyclic triterpenes sapogenins is generally more potent than that of saponins based on previous reports of our group [6–8].
Table 3.
The IC50 values of compounds 1–9 against HT29 cell line.
| compound code | HT29 cell line |
|---|---|
| 1 | >100 |
| 2 | 17.71 ± 0.95 |
| 3 | >100 |
| 4 | >100 |
| 5 | >100 |
| 6 | >100 |
| 7 | >100 |
| 8 | 0.69 ± 0.06 |
| 9 | 0.37 ± 0.025 |
| cisplatin | 4.87 ± 0.38 |
3.2.2. The effects of compounds 8 and 9 on the G2/M phase of the cell cycle in HT29 cells
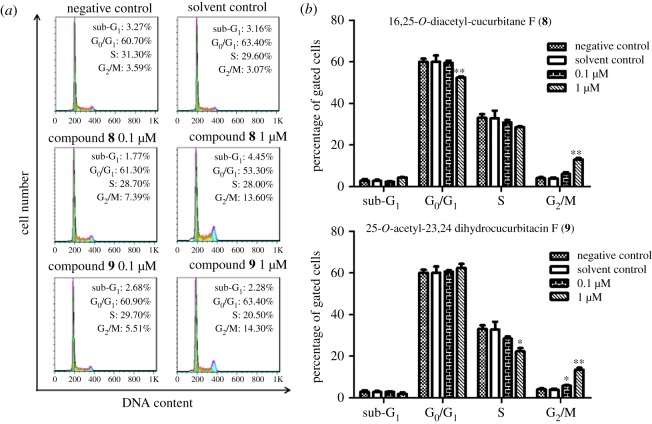
To investigate the underlying mechanism of the proliferation inhibitory effects of compounds 8 and 9 in HT29 cells, the cell cycle check points were examined by flow cytometry. HT29 cells were treated with 0.1 and 1 µM of compounds 8 and 9 for 24 h in the experimental group, while the control group was treated with the DMSO (2‰). As shown in figure 4, treatment with DMSO (2‰) had no effect on the distribution of cell cycle. Compared with the control group, the percentage of cells in the G2/M phase increased significantly with the increasing dose of 8 and 9. After treatment with 1 µM compound 8 for 24 h, the percentage of cells in the G2/M phase increased from 4.1% to 13.1%. Compound 9 showed similar effects to compound 8. These results suggested that induction of G2/M cell cycle arrest may contribute to the inhibitory effects of compounds 8 and 9 on cell proliferation.
Figure 4.
Effect of 16,25-O-diacetyl-cucurbitane F (8) and 25-O-acetyl-23,24-dihydrocucurbitacin F (9) on cell cycle distribution. (a) Representative DNA histograms of cell cycle analysis in HT29 cells. The cells were treated with either 8 (0, 0.1 and 1.0 µM), 9 (0.1 and 1.0 µM) or DMSO (2‰) for 24 h, stained with PI and analysed by flow cytometry, respectively. (b) Graphical representation of the cell cycle phase distribution. The results show the mean ± s.e.m. of three independent experiments. Significantly different (*p < 0.05, **p < 0.01) compared with negative control (0 µM-treated).
3.2.3. The effects of compounds 8 and 9 on apoptotic induction in HT29 cells
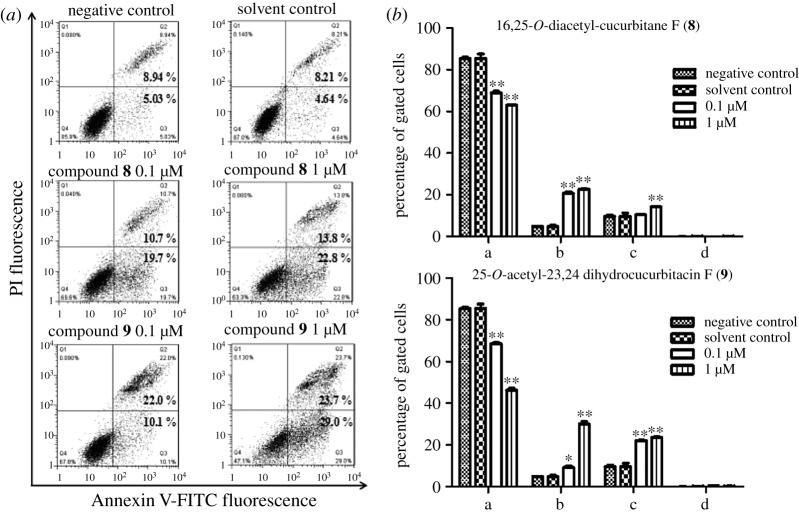
To determine if apoptosis is involved in the two compounds' induced inhibition of cell proliferation, we employed Annexin V/PI double staining to examine the effect of compounds 8 and 9 on apoptosis. The results indicated that treating with 0.1 and 1 µM compounds 8 or 9 for 24 h induced a significant increase in the percentage of apoptotic and necrotic cells when compared with the control group. As shown in figure 5, DMSO (2‰) did not induce cell apoptosis, while compounds 8 and 9 significantly increased the percentage of apoptotic and necrotic cells in a dose-dependent manner. These results suggested that induction of cell apoptosis may be involved in the inhibitory effects of compounds 8 and 9 on cell proliferation in HT29 cells.
Figure 5.
Effect of 16,25-O-diacetyl-cucurbitane F (8) and 25-O-acetyl-23,24-dihydrocucurbitacin F (9) on cell apoptosis. (a) Density plots showing the percentage distribution of compounds 8 and 9 treated cells. The HT29 cells were stained with Annexin V/PI and analysed by flow cytometry. Upper left quadrant: Annexin V(−)/PI(+) are labelled as damaged cells. Upper right quadrant: Annexin V(+)/PI(+) are labelled as late apoptotic/secondary necrotic cells. Lower right quadrant: Annexin V(+)/PI(−) are labelled as early apoptotic cells. Lower left quadrant: Annexin V(−)/PI(−) are labelled as live cells. (b) Graphical representation of the cell apoptosis phase distribution. The results show the mean ± s.e.m. of three independent experiments. Significantly different (*p < 0.05, **p < 0.01) compared with negative control (0 µM-treated). The abscissa ‘a, b, c and d’ represents in turn ‘live cells, early apoptotic cells, late apoptotic/secondary necrotic cells, and damaged cells'.
3.2.4. Induction of cell morphological changes by compounds 8 and 9 in HT29 cells
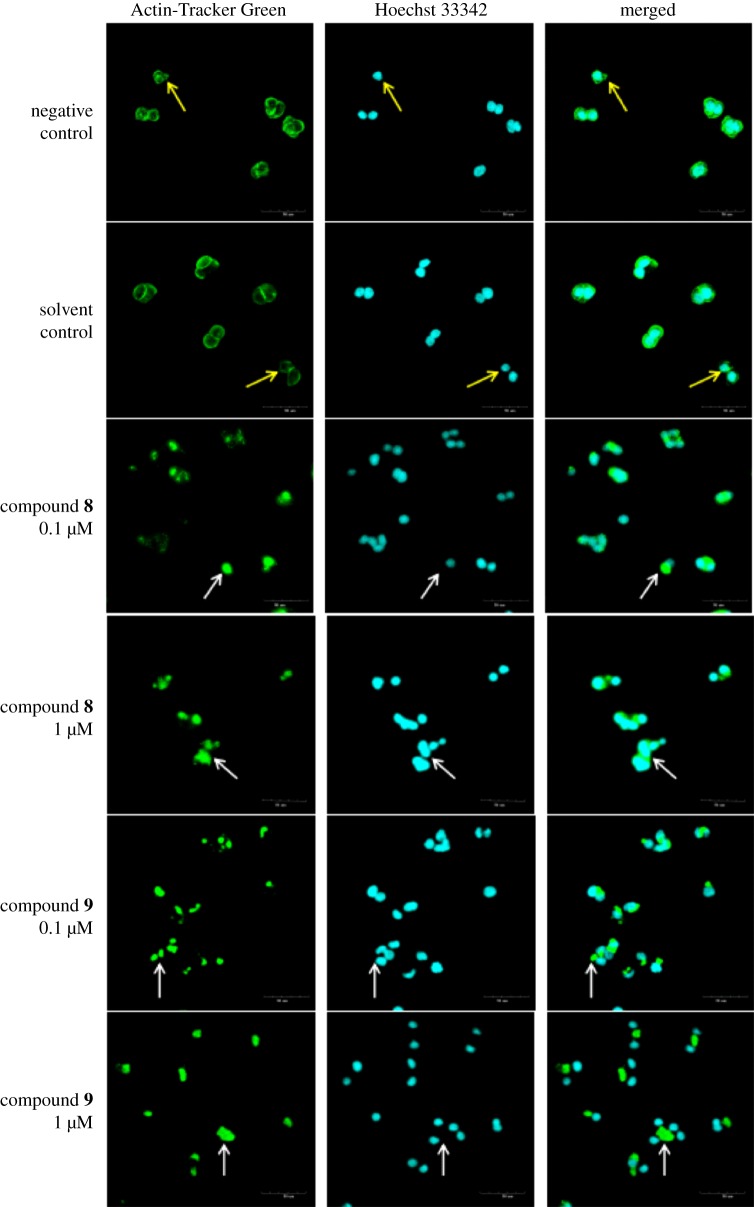
As the cytoskeleton is very important for maintaining the native morphology and function of cells, we investigated that the microfilaments change by staining F-actin after the treatment with compounds 8 and 9. Negative control HT29 cells exhibited F-actin that mainly surrounded the edge of cells, especially in the contact area of cells (figure 6). Meanwhile, the formation of lamellipodia was observed at the leading edge (indicated by yellow arrows in figure 6). The microfilaments in solvent control cells (2‰ DMSO) were as intact as that of negative control, whereas destruction of cytoskeleton was observed in cells treated with compounds 8 and 9. The distribution of F-actin was diminished significantly in area near the surface of the cells (figure 6). Overall, the F-actin aggregation (indicated by white arrows in figure 6) was increased with increasing concentration of compounds 8 and 9. Aggregated F-actin completely detached from the nucleus after 24 h treatment with 1 µM compound 9 (figure 6). The results indicated that compound 9 is more potent than compound 8 in inducing F-actin aggregation.
Figure 6.
16,25-O-diacetyl-cucurbitane F (8) and 25-O-acetyl-23,24-dihydrocucurbitacin F (9) induced the morphological changes in the HT29 cells. Fluorescence photomicrographic images of HT29 cells stained with Actin-Tracker Green (actin, green) fluorescent probe and Hoechst 33342 (DNA, blue). The HT29 cells were incubated with either DMSO (2‰), compound 8 (0.1, 1 µM, respectively) or compound 9 (0.1, 1 µM, respectively) for 24 h. The negative control was without treatment. Scale bar, 50 µm.
4. Conclusion
13C NMR data play an important role in inferring the structure of triterpenoids. Based on previous reports and our data in this paper, the 13C NMR spectral characteristics of common substituents of 5,24-diene cucurbitane tetracyclic triterpenes were summarized [4–8,11–13]. In the A ring, with a broad singlet signal (H-3) in 1H NMR, the 3β-OH was often connected with sugar, the obvious characteristic is that the δC-3 75–81 (CH) was replaced by 85–90 (CH). In the B ring, the presence of δC 140–146 (C-5, C) and 116–122 (C-6, CH) indicated that there exists a double bond. In the C ring, the big changes often occur at δC-11 211–219 (C, carbonyl), 77–80 (CH, α-OH) with 1H (dd, J = 10–12, 4–6 Hz), 72–74 (CH, β-OH) with 1H (dd, J = 3–5, 1–3 Hz) and 26–32 (CH2). In the side chain, it was δC-20 73–82 (C) instead of 35–38 (CH) that revealed the appearance of OH (C-20). In addition, the substituents of terminal Δ24, such as δC 64–68 (C-26, CH2OH), 58–62 (C-27, CH2OH), 21–23 (C-26, CH3) and 13–15 (C-27, CH3), have been identified. These data could help the interpreter to infer the characteristic groups quickly.
We compared the structures of cucurbitane triterpenoids with anti-cancer activity, which could provide direction for structural modification of triterpenoids. Cucurbitacins with carbonyl group at C-11, such as cucurbitacin B, D, E, I and compounds 8 and 9, exhibited strong anti-cancer activity. The α-hydroxy-ketone at C-21 and C-22 might be another important active group. And it could be deduced that a hydroxyl at C-16 would enhance cytotoxicity from the comparison results of compounds 8 and 9 in this paper.
Moreover, we not only investigated the effects of compounds 1–9 on HT29 cell proliferation, but also preliminarily explored the mechanism of compounds 8 and 9 on anti-proliferative effect in colon cancer cells. Cucurbitacins are a group of highly oxidized tetracyclic triterpenoids, and the screening study of these on the cytotoxic activity indicated that cucurbitacins possess strong anti-cancer activity, especially cucurbitacin B, D, E, I and compound 9 [3,10,14–17]. The report pointed out that cucurbitacin-I induced G2/M cell cycle arrest in SW480 cells accompanied by the downregulation of cyclin A, cyclin B1, CDK1 and CDC25C in vitro and in vivo [18]. However, cucurbitacin B induces cell cycle G2 arrest in SW480 cells without downregulation of CDK1 expression [19]. Besides cell cycle arrest, apoptosis is another mechanism for anti-proliferative effect of cancer cell. Cucurbitacin-I induced apoptosis in SW480 cells via downregulation of Bcl-2 expression and increasing the expression of apoptosis-related proteins (cleaved caspases-3, -7, -8 and -9, and PARP) [18]. But cucurbitacin B-induced apoptosis did not alter the expression of anti-apoptotic proteins such as Bcl-2 and Bcl-xL [19]. Thus, there are controversies about the anti-cancer mechanisms of different cucurbitacins even for the same cells. Certainly, there are also differences in anti-tumour mechanisms of same cucurbitacin on different cell lines [20,21]. In addition, recent studies have reported that cucurbitacins have an inhibitory effect on the proliferation of cancer cells, as well as on the polymerization and permeability of actin [22–24]. Therefore, we need further investigation to clarify their mechanisms.
In conclusion, our studies enriched the chemical composition research of H. pengxianensis and provided scientific basis for the development of 16,25-O-diacetyl-cucurbitane F (8) and 25-O-acetyl-23,24-dihydrocucurbitacin F (9) as a chemotherapeutic agent against colon cancer.
Acknowledgements
Thanks very much for support from the Innovation Project of Shandong Academy of Medical Sciences, PR China. We are also grateful to two anonymous reviewers and editors, who provided comments that substantially improved the manuscript.
Ethics
In this study, we do not experiment with animals. Informed consent was obtained from all participants.
Data accessibility
This article does not contain any additional data.
Authors' contributions
Q.Y. and Y.M. conceived of the study, designed the study and coordinated the study. W.W. and H.Y. performed the data analysis, participated in the design of the study and drafted the manuscript. Yi.L. and Z.Z. contributed significantly to revise the manuscript. Yo.L. and H.W. helped perform the analysis with constructive discussions. All authors gave final approval for publication.
Competing interests
We declare we have no competing interests.
Funding
Q.Y. and Y.M. are grateful for support from the Innovation Project of Shandong Academy of Medical Sciences.
References
- 1.Chen JC, Chiu MH, Nie RL, Cordell GA, Qiu SX. 2005. Cucurbitacins and cucurbitane glycosides: structures and biological activities. Nat. Prod. Rep. 22, 386–399. ( 10.1039/b418841c) [DOI] [PubMed] [Google Scholar]
- 2.Passos GF, Medeiros R, Marcon R, Nascimento AF, Calixto JB, Pianowski LF. 2013. The role of PKC/ERK1/2 signaling in the anti-inflammatory effect of tetracyclic triterpene euphol on TPA-induced skin inflammation in mice. Eur. J. Pharmacol. 698, 413–420. ( 10.1016/j.ejphar.2012.10.019) [DOI] [PubMed] [Google Scholar]
- 3.Gao Y, Islam MS, Tian J, Lui VW, Xiao D. 2014. Inactivation of ATP citrate lyase by Cucurbitacin B: a bioactive compound from cucumber, inhibits prostate cancer growth. Cancer Lett. 349, 15–25. ( 10.1016/j.canlet.2014.03.015) [DOI] [PubMed] [Google Scholar]
- 4.Chen JC, Niu XM, Li ZR, Qiu MH. 2005. Four new cucurbitane glycosides from Hemsleya jinfushanensis. Planta Med. 71, 983–986. ( 10.1055/s-2005-873110) [DOI] [PubMed] [Google Scholar]
- 5.Chen JC, Zhou L, Wang YH, Tian RR, Yan YX, Nian Y, Sun Y, Zheng YT, Qiu MH. 2012. Cucurbitane triterpenoids from Hemsleya penxianensis. Nat. Prod. Bioprospect. 2, 138–144. ( 10.1007/s13659-011-0044-2) [DOI] [Google Scholar]
- 6.Li Y, Zheng Z, Zhou L, Liu Y, Wang H, Li L, Yao Q. 2015. Five new cucurbitane triterpenoids with cytotoxic activity from Hemsleya jinfushanensis. Phytochem. Lett. 14, 239–244. ( 10.1016/j.phytol.2015.10.019) [DOI] [Google Scholar]
- 7.Xu X, Bai H, Zhou L, Deng Z, Zhong H, Wu Z, Yao Q. 2014. Three new cucurbitane triterpenoids from Hemsleya penxianensis and their cytotoxic activities. Bioorg. Med. Chem. Lett. 24, 2159–2162. ( 10.1016/j.bmcl.2014.03.027) [DOI] [PubMed] [Google Scholar]
- 8.Li Y, Wang WX, Zheng ZF, Mu YL, Liu YJ, Wang HY, Li L, Yao QQ. 2018. Eight new cucurbitane triterpenoids from ‘Xue Dan,’ the roots of Hemsleya pengxianensis. J. Asian Nat. Prod. Res. 20, 36–48. ( 10.1080/10286020.2017.1355363) [DOI] [PubMed] [Google Scholar]
- 9.Wu J, Wu YJ, Yang BB. 2002. Anticancer activity of Hemsleya amabilis extract. Life Sci. 71, 2161–2170. ( 10.1016/S0024-3205(02)02013-1) [DOI] [PubMed] [Google Scholar]
- 10.Boykin C, Zhang G, Chen YH, Zhang RW, Fan XE, Yang WM, Lu Q. 2011. Cucurbitacin IIa: a novel class of anti-cancer drug inducing non-reversible actin aggregation and inhibiting survivin independent of JAK2/STAT3 phosphorylation. Br. J. Cancer 104, 781–789. ( 10.1038/bjc.2011.10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen JC, Zhang ZQ, Qiu MH. 2007. Chemical constituents from the tubers of Hemsleya delavayi. Acta Chim. Sin. 65, 1679–1684. [Google Scholar]
- 12.Kasai R, Matsumoto K, Nie RL, Morita T, Awazu A, Zhou J, Tanaka O. 1987. Sweet and bitter cucurbitane glycosides from Hemsleya carnosiflora. Phytochemistry 26, 1371–1376. ( 10.1016/S0031-9422(00)81815-4) [DOI] [Google Scholar]
- 13.Li P, et al. 2017. New cucurbitane triterpenoids with cytotoxic activities from Hemsleya penxianensis. Fitoterapia 120, 158–163. ( 10.1016/j.fitote.2017.06.009) [DOI] [PubMed] [Google Scholar]
- 14.Cai Y, Fang X, He C, Li P, Xiao F, Wang Y, Chen M. 2015. Cucurbitacins: a systematic review of the phytochemistry and anticancer activity. Am. J. Chin. Med. 43, 1331–1350. ( 10.1142/S0192415X15500755) [DOI] [PubMed] [Google Scholar]
- 15.Ishii T, Kira N, Yoshida T, Narahara H. 2013. Cucurbitacin D induces growth inhibition, cell cycle arrest, and apoptosis in human endometrial and ovarian cancer cells. Tumour Biol. 34, 285–291. ( 10.1007/s13277-012-0549-2) [DOI] [PubMed] [Google Scholar]
- 16.Hung CM, Chang CC, Lin CW, Ko SY, Hsu YC. 2013. Cucurbitacin E as inducer of cell death and apoptosis in human oral squamous cell carcinoma cell line SAS. Int. J. Mol. Sci. 14, 17 147–17 156. ( 10.3390/ijms140817147) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopez-Haber C, Kazanietz MG. 2013. Cucurbitacin I inhibits Rac1 activation in breast cancer cells by a Ros-mediated mechanism and independently of Jak2 and P-Rex1. Mol. Pharmacol. 83, 1141–1154. ( 10.1124/mol.112.084293) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim HJ, Park JH, Kim JK. 2014. Cucurbitacin-I, a natural cell-permeable triterpenoid isolated from Cucurbitaceae, exerts potent anticancer effect in colon cancer. Chem. Biol. Interact. 219, 1–8. ( 10.1016/j.cbi.2014.05.005) [DOI] [PubMed] [Google Scholar]
- 19.Yasuda S, Yogosawa S, Izutani Y, Nakamura Y, Watanabe H, Sakai T. 2010. Cucurbitacin B induces G2 arrest and apoptosis via a reactive oxygen species-dependent mechanism in human colon adenocarcinoma SW480 cells. Mol. Nutr. Food Res. 54, 559–565. ( 10.1002/mnfr.200900165) [DOI] [PubMed] [Google Scholar]
- 20.Zheng Q, et al. 2014. Cucurbitacin B inhibits growth and induces apoptosis through the JAK2/STAT3 and MAPK pathways in SH-SY5Y human neuroblastoma cells. Mol. Med. Rep. 10, 89–94. ( 10.3892/mmr.2014.2175) [DOI] [PubMed] [Google Scholar]
- 21.Zhang T, et al. 2012. Cucurbitacin induces autophagy through mitochondrial ROS production which counteracts to limit caspase-dependent apoptosis. Autophagy 8, 559–576. ( 10.4161/auto.18867) [DOI] [PubMed] [Google Scholar]
- 22.Gupta P, Srivastava SK. 2014. Inhibition of HER2-integrin signaling by Cucurbitacin B leads to in vitro and in vivo breast tumor growth suppression. Oncotarget 5, 1812–1828. (doi:10.18632/oncotarget.1743) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kausar H, Munagala R, Bansal SS, Aqil F, Vadhanam MV, Gupta RC. 2013. Cucurbitacin B potently suppresses non-small-cell lung cancer growth: identification of intracellular thiols as critical targets. Cancer Lett. 332, 35–45. ( 10.1016/j.canlet.2013.01.008) [DOI] [PubMed] [Google Scholar]
- 24.Gabrielsen M, et al. 2013. Cucurbitacin covalent bonding to cysteine thiols: the filamentous-actin severing protein Cofilin1 as an exemplary target. Cell Commun. Signal. 11, 58–68. ( 10.1186/1478-811X-11-58) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article does not contain any additional data.