Abstract
Background:
The allergenicity of dust mite exposure might be dependent on variants in the gene for IL-10 (IL10).
Objectives:
To evaluate whether dust mite exposure modifies the effect of single nucleotide polymorphisms (SNPs) in IL10 on allergy and asthma exacerbations.
Methods:
We genotyped 6 SNPs in IL10 in 417 Costa Rican children and 503 white children in the Childhood Asthma Management Program (CAMP) with asthma and their parents. We used family-based and population-based approaches to test for interactions between IL10 SNPs and dust mite allergen on serum IgE to dust mite in Costa Rica and on asthma exacerbations in Costa Rica and CAMP.
Results:
Dust mite exposure significantly modified the relation between 3 SNPs in IL10 (rs1800896, rs3024492, and rs3024496) and IgE to dust mite in Costa Rica (P for interaction, .0004 for SNP rs1800896). For each of these SNPs, homozygosity for the minor allele was associated with increased levels of IgE to dust mite with increased dust mite exposure. Homozygosity for the minor allele of each of the 3 SNPs was associated with increased risk of occurrence (~3-fold to 39-fold increase) and frequency of asthma exacerbations among children exposed to ≥10 μg/g dust mite allergen in Costa Rica. Similar results were obtained for 2 of these SNPs (rs1800896 and rs3024496) among white children in CAMP.
Conclusion:
Our findings suggest that dust mite allergen levels modify the effect of IL10 SNPs on allergy and asthma exacerbations and may partly explain conflicting findings in this field. (J Allergy Clin Immunol 2008;122:93–8.)
Keywords: IL10, asthma, IgE, dust mite, interaction, exacerbations, Costa Rica, CAMP
IL-10 is a pleiotropic immunoregulatory cytokine produced by T-regulatory cells, B cells, monocytes, alveolar macrophages, mast cells, and pulmonary dendritic cells.1–4 IL-10 plays a central role in the induction of antigen-specific tolerance in human beings 2 and protects mice from antigen-induced airway inflam- mation,5 a hallmark of asthma.
In human beings, the gene for IL-10 (IL10) is on chromosome 1q31–32, a genomic region linked to asthma and related pheno-types.6 Most,7,8 but not all,9 association studies of polymorphisms in the promoter region of IL10 and asthma-related phenotypes have been positive. Although these polymorphisms have been shown to influence levels of IL10 expression, there have been conflicting findings with regard to the direction of these associations.10,11 A potential reason for these inconsistent results is failure to model gene-by-environment interactions appropriately.12
Dust mite allergen is an environmental exposure known to influence allergic sensitization,13,14 asthma exacerbations,15 and, importantly, IL-10 production16 in human beings. We hypothesized that dust mite allergen exposure would modify the relationship between polymorphisms in IL10 and both allergen-specific immune responses and disease exacerbations in children with asthma. To test this hypothesis, we first assessed whether levels of IgE to the house dust mite Dermatophagoides pteronyssinus (Der p 1) and asthma exacerbations were influenced by single nucleotide polymorphisms (SNPs) in IL10 among children in the Genetics of Asthma in Costa Rica Study. We further assessed whether levels of Der p 1 in house dust modified the relation between IL10 SNPs and IgE to dust mite and asthma exacerbations in Costa Rican children. Finally, we attempted to replicate significant findings for asthma exacerbations (because specific IgE to dust mite was not available) in white (non-Hispanic) children with asthma in the Childhood Asthma Management Program (CAMP).
METHODS
Study populations
The protocols for subject recruitment and data collection for the Genetics of Asthma in Costa Rica Study have been previously described in detail and are included along with a detailed Methods section in the Online Repository at www.jacionline.org.17 The population of the Central Valley of Costa Rica is a genetic isolate18,19 of mixed Spanish and Amerindian ancestry with a prevalence of asthma that ranks among the highest in the world.20 Children included in the study had asthma (defined as physician-diagnosed asthma and ≥2 respiratory symptoms or asthma attacks in the previous year, and either airway responsiveness [an FEV1 decline of at least 20% from the best FEV1 value after inhalation of methacholine ≤16.81 mmol], or bronchodilator responsiveness [an increment of at least 12% and at least 200 mL in baseline FEV1 all loci. In Illumina, 5 samples were repeated on each of15 plates, with no discordant runs. The genotypic pass rate was >99% for all Illumina loci. after administration of albuterol]) and high probability of having ≥6 great-grandparents born in the Central Valley. Of the 439 participating children, 426 had DNA that passed quality control and were included in this analysis along with their parents. Study participants completed a protocol that included questionnaires, house dust collection, and collection of blood samples. Written parental consent was obtained for participating children, from whom written assent was also obtained. The study was approved by the Institutional Review Boards of the Hospital Nacional de Ninos (San Jose, Costa Rica) and Brigham and Women’s Hospital (Boston, Mass).
CAMP was a multicenter clinical trial of the effects of anti-inflammatory medications in children with mild to moderate asthma. Participating children had asthma defined by symptoms greater than 2 times per week, the use of an inhaled bronchodilator at least twice weekly or the use of daily medication for asthma, and increased airway responsiveness to methacholine (PC20 ≤ 12.5 mg/mL).21 Of the 1041 children enrolled in the original clinical trial, 968 children and 1518 of their parents contributed DNA samples. This analysis was restricted to 483 families of white children. Questionnaire data was collected at baseline and during the course of the 4-year clinical trial, and blood samples and house dust samples were collected at baseline.21 Written informed consent was obtained from parents of participating children. CAMP was approved by the Institutional Review Boards of Brigham and Women’s Hospital and the other participating centers.
Measurement of allergen-specific IgE
In Costa Rican children, serum IgE to Der p 1 was measured by using the UniCAP 250 system (Pharmacia & Upjohn, Kalamazoo, Mich), with samples measured in duplicate. IgE to dust mite was not measured in CAMP.
Statistical analysis
In Costa Rica, level of IgE to Der p 1 was treated as a continuous variable, and asthma exacerbations were defined as having at least 1 hospitalization for asthma in the previous year. For CAMP, we present results for exacerbations at 2 time points. In CAMP, asthma exacerbations were defined as a having at least 1 emergency department visit or hospitalization for asthma. Levels of Der plin house dust were treated as continuous in analyses of specific IgE to Der p 1 and were dichotomized at ≥10 μg/g in models of asthma exacerbations for ease of exposition and because of previous evidence that this level influences the risk of asthma exacerbations.27,28 Hardy-Weinberg equilibrium was tested in parental data by a χ2 goodness-of-fit test, and deviations from mendelian inheritance were tested with PedCheck (http://watson.hgen.pitt.edu/register/docs/pedcheck.html).29 Estimates of measures of linkage disequilibrium (LD) -D’ and r2 were obtained from Haploview v3.11 (http://www.broad.mit.edu/mpg/haploview/).30 All analyses were performed assuming an additive genetic model. Family-based association analyses were performed with the family-based association test (FBAT) statistic implemented in HelixTree with PBAT v5 (Golden Helix, Bozeman, Mont).31 Gene-by-environment interactions were then tested by using the family-based association tests of interaction (FBATIs) implemented in PBAT.32 SNPs with evidence of interaction (nominal P values <.05) were carried forward to a population-based analysis of association (linear regression) in index children by using SASv9.1.2 (SAS Institute, Cary, NC). Logistic regression was used to examine gene-by-environment interactions on asthma exacerbations. The Fisher method was used to obtain P values for both studies (Costa Rica and CAMP) combined, as follows:
RESULTS
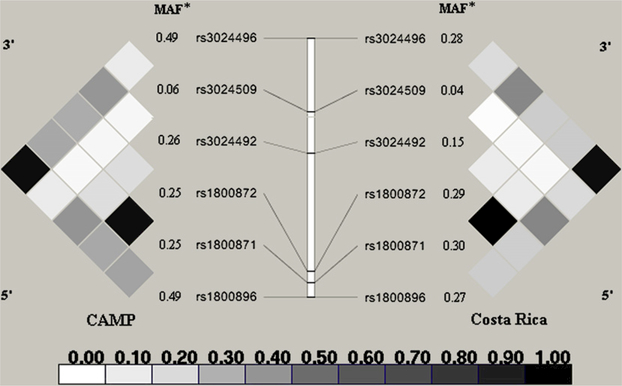
Of the 426 participating families in Costa Rica, 9 were excluded from this analysis because of mendelian inconsistencies, leaving 417 children and their parents. Of the 483 families of white children in CAMP, 13 were removed from this analysis because of mendelian inconsistencies, leaving 470 families (and 503 children). Parental genotypes were in Hardy-Weinberg equilibrium for all SNPs in Costa Rica and CAMP (P > .05 in all cases). Although the minor allele frequencies differed, the LD pattern (r2) across IL10 was similar in both Costa Rica and CAMP (Fig 1; see this article’s Table E1 in the Online Repository at www.jacionline.org). Baseline characteristics for children with asthma in Costa Rica and white children with asthma in CAMP are presented in Table I. Exposure to high levels of dust mite allergen (whether ≥2 μg/g or ≥10 μg/g house dust) was more common in Costa Rican children than in white children in CAMP.
FIG 1.
Pairwise (r2) LD for IL10 in Costa Rican and white (non-Hispanic) CAMP parents. *MAF.
Table E1.
Genotypic data for IL10 polymorphisms in children from Costa Rica and CAMP
| Chromosome 1 position | Alleles | Location | Costa Rica |
CAMP |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| H/H* | H/h† | h/h‡ | missing | H/H* | H/h† | h/h‡ | missing | |||
| rs1800896 203335292 | A>G | Promoter | 219 | 154 | 33 | 11 | 126 | 238 | 117 | 22 |
| rs1800871 203335029 | C>T | Promoter | 204 | 164 | 39 | 10 | 259 | 168 | 31 | 45 |
| rs1800872 203334802 | C>A | Promoter | 206 | 165 | 39 | 7 | 274 | 174 | 32 | 23 |
| rs3024492 203332507 | A>T | Intron | 300 | 94 | 12 | 11 | 272 | 195 | 34 | 2 |
| rs3024509 203331692 | T>C | Intron (boundary) | 381 | 29 | 0 | 7 | 439 | 59 | 1 | 4 |
| rs3024496 203330259 | T>C | Exon | 214 | 159 | 32 | 12 | 129 | 254 | 119 | 1 |
Homozygotes for the major allele
Heterozygotes.
Homozygotes for the minor allele.
Table I.
Baseline characteristics of children with asthma in Costa Rica and white children with asthma in CAMP
| Median (interquartile range) or count(%) |
||
|---|---|---|
| Costa Rica | CAMP (white) | |
| Variable | (n = 417) | (n = 503) |
| Age (y) | 8.7 (7.7–10.4) | 8.6 (7.0–10.5) |
| Sex, female | 157 (38%) | 192 (38%) |
| Total serum IgE (IU/mL)* | 414 (117–962) | 402 (157–1068) |
| Specific IgE to Der p 1 | 14.1 (0.5–75) | Not measured |
| Der p 1 ≥2 0μg/g in house dust* | 382 (94%) | 108 (23%) |
| Der p 1 ≥10 μs/s in house dust* | 203 (44%) | 48 (10%) |
| ≥ 1 Asthma exacerbation† | 21 (5%) | 153 (31%) |
Numbers and percentages may reflect missing data on some subjects. Information missing on some subjects in Costa Rica measurements of Der p 1 in house dust (n 510) and asthma exacerbations (n = 1). Information missing on some subjects in CAMP for total IgE (n = 7) and measurements of Der p 1 in house dust (n = 27).
In Costa Rica, at least 1 hospitalization for asthma in the previous year. In CAMP, at least 1 emergency department visit or hospitalization for asthma during the first 4 years of the trial.
Family-based association analysis of IgE to dust mite in Costa Rica
We found no significant association between SNPs in IL10 and IgE to dust mite among Costa Rican children in models that did not consider the possibility of effect modification (FBAT results in Table II). The results of the family-based analysis of association between SNPs in IL10 and IgE to dust mite in Costa Rican children, assuming a genotype-by-dust-mite allergen interaction, are shown in Table II (FBATI column). We found significant evidence of such an interaction on IgE to dust mite allergen for 3 SNPs in IL10 (rs1800896, rs3024492, and rs3024496). Two of these SNPs (rs1800896 and rs3024496) were in strong LD (r2 = 0.94).
Table II.
Family-based analysis of association between IL10 polymorphisms and IgE to Der p 1 and interaction between IL10 polymorphisms and exposure to Der p 1 on serum IgE to Der p 1 among children in Costa Rica
| Chromosome 1 position | Alleles | Location | MAF | No.* | FBAT† P value | FBATI‡ P value |
|---|---|---|---|---|---|---|
| rs1800896 203335292 | A>G | Promoter | 0.27 | 264 | .38 | .04 |
| rs1800871 203335029 | C>T | Promoter | 0.30 | 266 | .64 | .75 |
| rs1800872 203334802 | C>A | Promoter | 0.30 | 271 | .68 | .82 |
| rs3024492 203332507 | A>T | Intron | 0.15 | 176 | .40 | .003 |
| rs3024509 203331692 | T>C | Intron (boundary) | 0.04 | 74 | .54 | .17 |
| rs3024496 203330259 | T>C | Exon | 0.28 | 269 | .35 | .02 |
Number of informative families.
All models were adjusted for age and sex. Family-based analysis of association between IL10 polymorphisms and measurements of serum IgE to Der p 1 in a model not accounting for IL10 SNP-by-dust mite allergen interaction.
All models were adjusted for age and sex. Family-based analysis of interaction modeling the significance of IL10 SNP-by-Der p 1 allergen interaction in the prediction of specific IgE to Der p 1.
House dust collection
In both Costa Rica and CAMP, a Douglas vacuum cleaner (model 6735, Douglas, Walnut Ridge, Ark) was used to collect a global dust sample. Dust samples collected in Costa Rica were mailed to the Allergy and Immunology Reference Laboratory of Johns Hopkins Hospital, where the dust was weighed, sifted, and aliquoted for measurement of Der p 1 allergen by 2-site mAb ELISA assays.22 In CAMP, dust samples (collected ~6 months after randomization) were analyzed for dust mite allergen by using standardized mAb-based immunoenzymetric assays at a central laboratory.21
Genotyping
Markers chosen for genotyping were selected on the basis of previous literature, functional data (rs1800871, rs1800872, and rs1800896),11,23 validation in dbSNP (Single Nucleotide Polymorphism Database, www.ncbi.nlm.nih.gov/projects/SNP), and a haplotype-tagging algorithm in CAMP based on previous results of gene sequencing.24 The 6 SNPs genotyped capture ≥90% of the HapMap SNPs in IL10 in Centre d’etude du polymorphisme hu-main trios at an r2 ≥0.8. In Costa Rica, 5 SNPs were genotyped by using Gold-enGate assays on the Illumina BeadStation 500 system (San Diego, Calif), and 1 SNP (rs1800872) was genotyped with aTaqman genotyping assay (Applied Biosystems, Foster City, Calif). In CAMP, all SNPs were genotyped by using the SnaPshot Multiplex Kit (Applied Biosystems).26 The quality of the genotypic data was assessed by several methods. Duplicate genotyping was performed on approximately 5% of the samples to assess genotype reproducibility. In samples run with SnaPshot or Taqman, no discordant genotypes were detected, with genotype completion rates between 95% and 99% for all loci. In Illumina, 5 samples were repeated on each of 15 plates, with no discordant runs. The genotypic pass rate was >99% for all Illumina loci.
Population-based association analysis of IgE to dust mite in Costa Rica
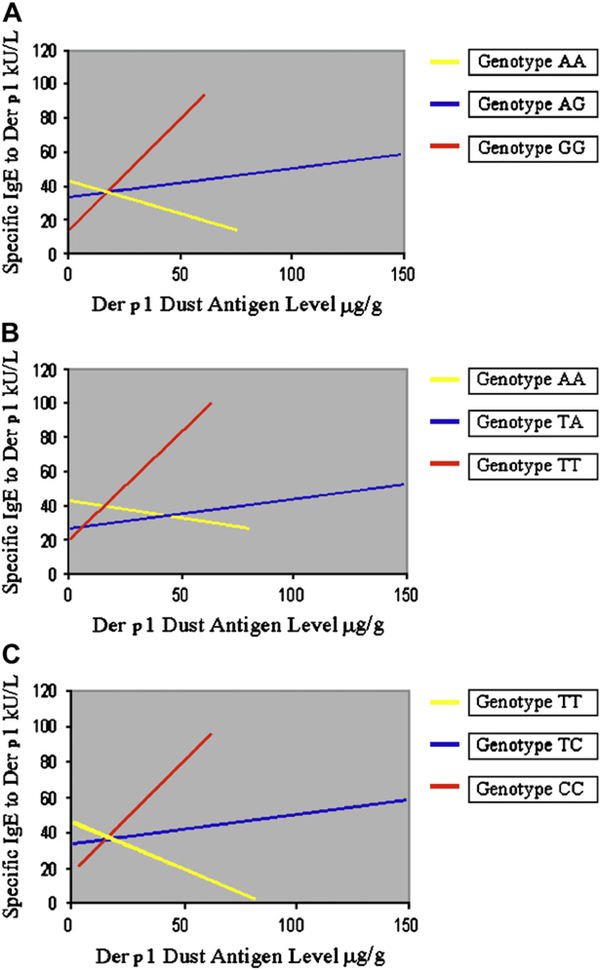
The results for the analysis of association between SNPs in IL10 and IgE to dust mite are presented in Table III. Similar to the results of our family-based analysis, no IL10 SNP was significantly associated with IgE to dust mite in models not accounting for genotype-by-allergen interactions (model 1). Including such interactions in our models resulted in significant effects for genotypes, Der p 1 levels (except for the model including SNP rs3024492), and genotype-by-dust-mite allergen interactions for all 3 SNPs (model 2). Fig 2 shows the results of our generalized linear models. For each SNP (rs1800896, rs3024492, and rs3024496 in A, B, and C, respectively), children homozygous for the minor allele (shown in red) had higher levels of IgE to dust mite than children homozygous for the major allele (shown in yellow) at high levels of dust mite allergen exposure; heterozygote children (shown in blue) had intermediate effects. In contrast, opposite effects are noted at low levels of dust mite allergen exposure. Bootstrapped regression resulted in smaller CIs for the interaction terms in our models for 2 SNPs (rs1800896 and rs3024496; model 3 inTable III). Models accounting for the possibility of right-censoring in the data for children whose specific IgE to dust mite reached the upper limit of detection (100 kU/L) also show significant results for genotype-by-allergen interaction for all SNPs except rs3024492 (model 4 in Table III).
Table III.
Population-based analysis of interaction between IL10 polymorphisms and exposure to Der p 1 on serum IgE to Der p 1 among children in Costa Rica
| Adjusted models | Model 1 |
Model 2 |
Model 3 |
Model 4 |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Baseline (no interaction) |
Interaction |
Bootstrap |
Life-table regression |
||||||
| Estimate | SE | P value | Estimate | SE | P value | P value | P value | ||
| A. | rs1800896* | -1.89 | 3.17 | .55 | -12.51 | 4.29 | .004 | .006 | .02 |
| Der p 1 (μg/g) | 0.04 | 0.11 | .72 | -0.46 | 0.17 | .008 | .004 | .02 | |
| Interaction* | 0.68 | 0.19 | .0004 | .0002 | .005 | ||||
| B. | rs3024492* | - 3.76 | 3.95 | .34 | -12.44 | 5.16 | .02 | .02 | .08 |
| Der p 1 (μg/g) | 0.04 | 0.11 | .70 | -0.24 | 0.15 | .11 | .11 | .40 | |
| Interaction* | 0.50 | 0.19 | .01 | .02 | .08 | ||||
| C. | rs3024496* | -1.12 | 3.21 | .73 | -13.69 | 4.38 | .002 | .001 | .002 |
| Der p 1 (μg/g) | 0.03 | 0.11 | .80 | -0.57 | 0.19 | .002 | <.0001 | .003 | |
| Interaction* | 0.79 | 0.19 | <.0001 | <.0001 | <.0001 | ||||
Effect estimates are in reference to increasing content of the minor allele.
FIG 2.
Regression lines by genotype for SNPs rs1800896 (A), rs3024492 (B), and rs3024496 (C) comparing the specific IgE response to Der p 1 antigen by measured Der p 1 antigen level. Yellow, Homozygotes for the major allele; blue, heterozygotes; red, homozygotes for the minor allele.
Analysis of asthma exacerbations
The results of logistic regression models for asthma exacerbations are presented in Table IV. After adjusting for age and sex, there was no significant association between any of the 3 SNPs of interest in IL10 (rs1800896, rs3024492, and rs3024496) and asthma exacerbations in Costa Rica or CAMP (results not shown). On the other hand, there were significant interactions between genotypes in the IL10 SNPs and dust mite allergen exposure on asthma exacerbations (Table IV). Among Costa Rican children who were homozygous for the minor allele of each of the 3 IL10 SNPs, those exposed to high dust mite allergen levels had significantly increased odds of asthma hospitalizations (~3-fold to 39-fold increment). The effect of high levels of dust mite allergen was the opposite for children homozygous for the major allele of each of the 3 SNPs (~81% to 86% decrease), with intermediate effects on heterozygous children.
Table IV.
Analysis of the interaction between IL10 polymorphisms and Der p 1 exposure on the occurrence of asthma exacerbations among children in Costa Rica and CAMP
| Logistic regression | Costa Rican children with asthma |
White children with asthma in CAMP |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Model 1 |
Model 2 |
Model 3 |
Combined |
||||||||
| Year before enrollment |
Year 2 CAMP |
Year 4 CAMP |
|||||||||
| Estimate | SE | P value | Estimate | SE | P value | Estimate | SE | P value | P value† | ||
| A. | rs1800896* | -2.20 | 1.14 | .05 | -1.68 | 0.84 | .05 | -1.23 | 0.72 | .09 | (.02-.03) |
| Der p 1 >10 (μg/g) | -2.00 | 0.78 | .01 | - 2.47 | 1.15 | .03 | -2.08 | 0.90 | .02 | (.002-.003) | |
| Interaction* | 1.62 | 0.76 | .03 | 1.55 | 0.80 | .05 | 1.19 | 0.67 | .07 | (.01-.02) | |
| B | rs3024492* | -4.34 | 2.14 | .04 | -0.74 | 0.86 | .39 | -0.62 | 0.71 | .40 | (.08-.08) |
| Der p 1 >10 (μg/g) | -1.68 | 0.63 | .008 | -1.14 | 0.58 | .05 | -1.20 | 0.51 | .02 | (.002-.003) | |
| Interaction* | 2.67 | 1.19 | .03 | 0.51 | 0.80 | .52 | 0.48 | 0.68 | .47 | (.07-.08) | |
| C. | rs3024496* | -2.09 | 1.15 | .07 | -1.26 | 0.80 | .12 | -0.95 | 0.70 | .17 | (.05-.06) |
| Der p 1 >10 (μg/g) | -1.97 | 0.78 | .01 | -2.06 | 0.98 | .04 | -1.90 | 0.81 | .02 | (.002-.003) | |
| Interaction* | 1.52 | 0.77 | .05 | 1.16 | 0.75 | .12 | 0.96 | 0.65 | .14 | (.04-.04) | |
Effect estimates are in reference to increasing content of the minor allele.
The Fisher method has been used to obtain combined P values from Costa Rica with those from CAMP at years 2 (presented first in the parenthesis) and 4 of the trial.
Although attenuated, the results for 2 common SNPs in IL10 (rs1800896 and rs3024496, both with minor allele frequency [MAF] of 0.49 in CAMP; Fig 1) similarly modified the effect of Der p 1 allergen on asthma exacerbations among children at years 2(P value for interaction [rs1800896] = .05; [rs3024496] = .12) and 4 (P value for interaction [rs1800896] = .07; [rs3024496] = .14) of the CAMP trial. Among children homozygous for the minor allele of each of the 2 IL10 SNPs, those exposed to high levels of dust mite allergen had increased odds of asthma hospitalizations (~1.3-fold to 1.9-fold increase at 2 years, and a ~1.02- fold to 1.3-fold increase at 4 years). The effect of high levels of dust mite allergen was the opposite for children homozygous for the major allele of each of the 2 SNPs (~87% to 92% decrease at 2 years, and a ~85% to 88% decrease at 4 years), with intermediate effects on heterozygous children. Although consistent in direction to results obtained in Costa Rica, results for SNP rs3024492 (MAF, 0.26 in CAMP) were not significant. Results from the combined analysis of Costa Rica and CAMP (Table IV) demonstrate that for 2 SNPs (rs1800896 and rs3024496), genotype-by-allergen interactions were significant in the prediction asthma exacerbations.
Results similar to those from logistic regression were obtained for rs1800896 in analyses of the counted numbers of asthma exacerbations in Costa Rica and CAMP (see Table E2 in the Online Repository at www.jacionline.org).
Table E2.
Analysis of the interaction between IL10 polymorphisms and Der p 1 among children in Costa Rica and CAMP exposure on the number of asthma exacerbations
| Costa Rica |
CAMP |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Negative binomial regression |
Poisson regression |
|||||||||
| Model 1 |
Model 2 |
Model 3 |
||||||||
| Year before enrollment |
Year 2 CAMP |
Year 4 CAMP |
Combined |
|||||||
| Estimate | SE | P value | Estimate | SE | P value | Estimate | SE | P value | P values† | |
| A. rs1800896* | -0.37 | 0.23 | .11 | -1.55 | 0.68 | .02 | -1.03 | 0.49 | .04 | (.02-.03) |
| Der p 1 >10 (μg/g) | -0.11 | 0.12 | .37 | - 3.03 | 1.10 | .006 | -2.26 | 0.70 | .001 | (.003-.02) |
| Interaction* | 0.28 | 0.15 | .06 | 1.61 | 0.66 | .02 | 1.07 | 0.47 | .02 | (.009-.009) |
| B rs3024492* | -0.09 | 0.29 | .75 | -1.74 | 0.60 | .004 | -0.92 | 0.50 | .07 | (.02-.21) |
| Der p 1 >10 (μg/g) | 0.03 | 0.11 | .78 | -1.68 | 0.49 | .001 | -1.45 | 0.37 | <.0001 | NA‡ |
| Interaction* | -0.10 | 0.19 | .60 | 1.18 | 0.55 | .03 | 0.63 | 0.47 | .18 | NA‡ |
| C. rs3024496* | -0.33 | 0.24 | .17 | -1.13 | 0.60 | .06 | -0.84 | 0.48 | .07 | (.06-.07) |
| Der p 1 >10 (μg/g) | -0.10 | 0.13 | .43 | -2.32 | 0.83 | .005 | -2.02 | 0.61 | .001 | (.004-.02) |
| Interaction* | 0.24 | 0.15 | .18 | 1.12 | 0.58 | .05 | 0.86 | 0.45 | .06 | (.05-.06) |
Effect estimates are in reference to increasing content of the minor allele.
Fisher’s method has been used to obtain combined P values . The range presented reflects the result of combining the P values obtained from Costa Rica with CAMP at both 2 and 4 years of the trial
Not applicable. These P values are not combined because the effect estimates are in opposite directions.
DISCUSSION
Among children with asthma in Costa Rica, dust mite allergen exposure significantly modified the effect of polymorphisms in the promoter of IL10 on production of serum IgE to dust mite. Among children in 2 independent populations (Costa Rica and CAMP), dust mite allergen exposure significantly modified the effect of 2 IL10 SNPs (rs1800896 and rs3024496) on asthma exacerbations. To our knowledge, this is the first report of replication of an interaction between a genetic variant and an objectively measured environmental exposure on asthma-related phenotypes in 2 populations.
Specific IgE responses to Der p 1 (dust mite) exposure are critically dependent on baseline IL-10 production and TH2-predominant immune responses in a time-dependent fashion.33,34 Baseline elevation of IL-10 can markedly reduce specific-IgE production to Der p 1 in TH2-stimulated PBMCs.34 In contrast, increased levels of IL-10 after the induction of a specific-IgE response can lead to further increases in specific-IgE.34 Thus IL-10 is a plausible and necessary intermediary between dust mite exposure and dust mite-specific immune responses.
Of note, the most consistently replicated genotype-by-dust mite allergen interaction (rs1800896-by-dust antigen level) in our study involves a well described functional variant in IL10. Although many studies have shown decreased IL-10 production associated with the common A allele compared with the G allele of rs1800896,35,36 others have demonstrated the opposite effect, and 1 study showed no effect.37 On the basis of these previous studies and our current findings, we speculate that the effect of functional promoter polymorphisms in IL10 on IL-10 transcription by may be complex and linked to the degree of exposure to specific antigenic stimuli (as would be predicted in Fig 1). Assuming that elevated IL-10 levels are associated with decreased specific IgE responses at a given level of exposure,34 we hypothesize that the previously described functional variant in IL10 (rs1800896) is associated with both increased IL-10 expression at low levels of antigenic exposure and decreased IL-10 expression at high levels of antigenic exposure.
Of interest, there was a difference between Costa Rica and CAMP in the magnitude of the modification of the effect of IL10 SNPs on asthma exacerbations by dust mite allergen levels. Possible explanations include regression to the mean, differences in phenotypic ascertainment of asthma exacerbations, time from exposure measurement, differences in the MAF of some of the SNPs of interest (likely because of the relative isolation of the Costa Rican population), and (most importantly) differences in Der p 1 levels in house dust. Although the Costa Rican and CAMP studies ascertained children on similarly strict definitions of asthma17,21 and used similar dust collection protocols, it is remarkable (given the differences noted) that a significant modification of the effect of IL10 SNPs by Der p 1 allergen was found in both populations.
Although it is generally assumed that level of exposure is directly correlated with sensitization to dust mite allergen, this assertion has been challenged.38 Der p 1 levels have been associated with Der p 1 sensitization in a number of cohorts,13,14,39–41 many of which have included subjects recruited on the basis of atopy14,40 or a strong family history of atopy.13,39 Other studies have found no association between exposure and sensitization to Der p 1,29,42,43 and some have demonstrated an inverse association.44 In longitudinal cohort studies, Der p 1 exposure has been shown to be a risk factor for Der p 1 sensitization40 in atopic children45 or in cohorts enriched with children at high risk for atopy,40 whereas 1 study collecting longitudinal data on 3 independent population-based cohorts found no association.38 Our findings are consistent with those of cross-sectional and longitudinal studies suggesting that increased exposure leads to increased sensitization to dust mite in children with parental history of asthma and allergies (if those high-risk children had susceptibility variants for allergen sensitization).39,46 In a study with repeated skin test measurements over a 3-year period, newly diagnosed sensitization to Dermatophagoides pteronyssinus was dependent on a previous history of atopy, exposure to Der p 1 allergen, and parental history of atopy.45 Our results suggest that inconsistent findings for dust mite allergen and asthma phenotypes may be a result of untested gene-by-environment interactions.
Our study has several limitations. First, we may have had limited statistical power to detect modest interactions between some IL10 SNPs and dust mite allergen exposure. Second, dust mite allergen levels were measured only once in CAMP. Our finding of a modification of the effect of IL10 SNPs by dust mite allergen levels on asthma exacerbations in CAMP assumes that indoor exposure to dust mite is relatively stable over time and/or that a singular level of exposure can confer longitudinal risk. Although we cannot comment on the latter suggestion, a recent study from The Netherlands suggests that a single home-allergen measurement of Der p 1 adequately represents levels of exposure over a period of 4 years, and we obtained similar results for asthma exacerbations after 2 and 4 years of follow-up (Table IV). Finally, we could not test for an interaction between IL10 SNPs and dust mite allergen exposure on IgE to dust mite in CAMP, and thus our finding for dust mite sensitization in Costa Rica must be interpreted with caution pending replication in other populations.
In summary, our findings suggest that dust mite allergen exposure significantly modifies the effect of polymorphisms in IL10 on sensitization to dust mite and asthma exacerbations in childhood. In addition, our results suggest that failing to account for biologically appropriate gene-by-environment interactions may obscure important genetic effects. Our study highlights an important variable that may help to determine which group with asthma will derive the greatest benefit from reducing or avoiding dust mite allergen exposure.
METHODS
Study populations
The protocols for subject recruitment and data collection for the Genetics of Asthma in Costa Rica Study have been previously described in detail.E1,E2 The population of the Central Valley of Costa Rica is a genetic isolateE3,E4 of mixed Spanish and Amerindian ancestry with a prevalence of asthma that ranks among the highest in the world.E5 Children included in the study had asthma (defined as physician-diagnosed asthma and ≥2 respiratory symptoms or asthma attacks in the previous year, and either airway responsiveness [an FEV1 decline of at least 20% from the best FEV1 value after inhalation of meth-acholine ≤16.81 mmol], or bronchodilator responsiveness [an increment of at least 12% and at least 200 mL in baseline FEV1 after administration of albuterol]) and high probability of having ≥6 great-grandparents born in the Central Valley (confirmed by our study genealogist in 416 (95%) of 439 participating children). The latter criterion was required to increase the likelihood that children would be descendants of the founder population of the Central Valley.E6 Of the 439 participating children, 426 had DNA that passed quality control and were included in this analysis along with their parents. Families identified for removal on the basis of quality control checks include those that consistently demonstrate mendelian inconsistencies (because of either nonpaternity or sample mislabeling) or have an insufficient concentration of DNA available for analysis. Study participants completed a protocol that included questionnaires, house dust collection, and collection of blood samples. Written parental consent was obtained for participating children, from whom written assent was also obtained. The study was approved by the Institutional Review Boards of the Hospital Nacional de Niños (San Jose, Costa Rica) and Brigham and Women’s Hospital (Boston, Mass).
The CAMP was a multicenter clinical trial of the effects of anti-inflammatory medications in children with mild to moderate asthma. All recruited children had asthma defined by symptoms greater than 2 times per week, the use of an inhaled bronchodilator at least twice weekly or the use of daily medication for asthma, and airway responsiveness to methacholine ≤12.5 mg/ mL.E7,E8 Children with severe asthma or other clinically significant conditions were excluded.E7 Of the 1041 children enrolled in the original clinical trial, 968 children and 1518 of their parents contributed DNA samples. This analysis was restricted to 483 families of white (non-Hispanic) children. Questionnaire data were collected at baseline and during the course of the 4-year clinical trial, and blood samples and house dust samples were collected approximately 6 months after randomization.E7 Written informed consent was obtained from parents of participating children. CAMP was approved by the Institutional Review Boards of Brigham and Women’s Hospital and the other participating centers.
Measurement of allergen-specific IgE
In Costa Rican children, serum IgE to Der p 1 was measured by using the UniCAP 250 system (Pharmacia & Upjohn, Kalamazoo, Mich), with samples measured in duplicate. IgE to dust mite was not measured in CAMP.
House dust collection
In both Costa Rica and CAMP, a Douglas vacuum cleaner (model 6735, Douglas, Walnut Ridge, Ark) was used to collect a global dust sample from 5 areas of the child’s household: the upper mattress surface of the child’s bed, an upholstered chair or sofa in the family room or the living room, and floor samples from the child’s bedroom, the family room or living room, and the kitchen. Dust samples collected in Costa Rica were mailed to the Allergy and Immunology Reference Laboratory of Johns Hopkins Hospital, where the dust was weighed, sifted, and aliquoted for measurement of Der p 1 allergen by 2-site mAb ELISA assays.E9 In CAMP, dust samples were analyzed for dust mite allergen by using standardized mAb-based immunoenzymetric assays at a central laboratory.E7
Genotyping
Protocol details, SNP flanking sequence, and primer data are available at http://innateimmunity.net. Markers chosen for genotyping were selected on the basis of previous literature, functional data (rs1800871, rs1800872, and rs1800896)E10,E11 validation in dbSNP (Single Nucleotide Polymorphism Database, www.ncbi.nlm.nih.gov/projects/SNP), and a haplotype-tagging algorithm in CAMP based on previous results of gene sequencing.E12 We genotyped these 6 SNPs in IL10 in Costa Rican children with asthma and their parents and in families of white children with asthma in CAMP. The 6 SNPs genotyped capture ≥90% of the HapMap SNPs in IL10 in Centre d’etude du polymorphisme humain trios at an r2 ≥0.8. In Costa Rica, 5 SNPs were genotyped by using GoldenGate assays on the Illumina BeadStation 500 system (San Diego, Calif),E13 and 1 SNP (rs1800872) was genotyped with a Taqman genotyping assay (Applied Biosystems, Foster City, Calif). In CAMP, all SNPs were genotyped by using the SnaPshot Multiplex Kit (Applied Biosys-tems).E14 The quality of the genotypic data was assessed by several methods. Duplicate genotyping was performed on approximately 5% of the samples to assess genotype reproducibility. In samples run with SnaPshot or Taqman, no discordant genotypes were detected, with genotype completion rates between 95% and 99% for all loci. In Illumina, 5 samples were repeated on each of 15 plates, with no discordant runs. The genotypic pass rate was >99% for all Il- lumina loci.
Statistical analysis
In Costa Rica, level of IgE to Der p 1 was treated as a continuous variable, and asthma exacerbations were defined as binary (at least 1 hospitalization for asthma in the previous year) and continuous (number of unscheduled doctor visits for asthma in the previous year) variables. For CAMP, we present results for exacerbations at 2 time points (2 and 4 years) to provide estimates close to the date of dust sample measurement (2 years), and estimates that extend to the reported limit of acceptability for a singular dust sample measurement (up to 4 years; the within-home variance is less than the between-home variance for repeated dust sample measurements).E15 In CAMP, asthma exacerbations were defined as binary (at least 1 emergency department visit or hospitalization for asthma) and continuous (total number of emergency department visits or hospitalizations for asthma) variables. Levels of Der p 1 in house dust were treated as continuous in analyses of specific IgE to Der p 1 and were dichotomized at 10 μg/g in models of asthma exacerbations for ease of exposition and because of previous evidence that this level influences the risk of asthma exacerbations.E16–E18
Hardy-Weinberg equilibrium was tested in parental data by a χ2 goodness-of-fit test, and deviations from mendelian inheritance were tested with PedCheck (http://watson.hgen.pitt.edu/register/docs/pedcheck.html).E19 Genotypes of families with mendelian inconsistencies were set to missing. Estimates of measures of LD -D’ and r2 were obtained from Haploview v3.11 (http://www.broad.mit.edu/mpg/haploview/).E20 All analyses were performed assuming an additive genetic model. Family-based association analyses were performed with the FBAT statistic implemented in HelixTree with PBAT v5 (Golden Helix, Bozeman, Mont).E21 Gene-by-environment interactions were then tested by using the FBATI implemented in PBAT.E22 The FBATI statistic is generated by using a causal inference regression approach in which the interaction term is not dependent on the main genetic effect. SNPs with evidence of interaction (nominal P values <.05) were carried forward to a population-based analysis of association (linear regression) in index children by using SASv9.1.2 (SAS Institute, Cary, NC). We generated 1000 bootstrapped regression samples to recalculate CIs and P values to assess for violation of parametric assumptions and to deal with the potential influence of outliers. The Proc Lifereg procedure (T distribution) in SAS was used to assess the possible effects of right censoring in our data for serum IgE to dust mite (with an upper limit of detection at 100 kU/L). Logistic, Poisson (for the counted numbers of asthma exacerbations in CAMP), and negative binomial (to address overdispersion in our count data for asthma exacerbations in Costa Rica) regressions were used to examine gene-by-environment interactions on asthma exacerbations. All models used to examine gene-by-environment interactions on the outcomes of interest included the following: age, sex, IL10 genotype, dust levels of Der p 1, and an interaction term for IL10 genotype and Der p 1 level. Model fitness for regression models was assessed as following: F statistics were evaluated for linear regression, Hosmer-Lemeshow tests were used for logistic regression, and deviance/degrees of freedom were evaluated for Poisson regression (and negative binomial regression).
RESULTS
Of the 426 participating families in Costa Rica, 9 were excluded from this analysis because of mendelian inconsistencies, leaving 417 children and their parents. Of the 483 families of white children in CAMP, 13 were removed from this analysis because of Mendelian inconsistencies, leaving 470 families (and 503 children). Parental genotypes were in Hardy-Weinberg equilibrium for all SNPs in Costa Rica and CAMP (P > .05 in all cases). Although the minor allele frequencies differed, the LD pattern (r2) across IL10 was similar in both Costa Rica and CAMP (Fig 1; Table E1). Baseline characteristics for children with asthma in Costa Rica and white children with asthma in CAMP are presented in Table I. Exposure to high levels of dust mite allergen (whether ≥ 2 μg/g or ≥ 10 μg/g house dust) was more common in Costa Rican children than in white children in CAMP.
Family-based association analysis of IgE to dust mite in Costa Rica
We found no significant association between SNPs in 1L10 and IgE to dust mite among Costa Rican children in models that did not consider the possibility of effect modification (FBAT results in Table II). The results of the family-based analysis of association between SNPs in 1L10 and IgE to dust mite in Costa Rican children, assuming a genotype-by-dust-mite allergen interaction, are shown in Table II (FBATI column). We found significant evidence of such an interaction on IgE to dust mite allergen for 3 SNPs in 1L10 (rs1800896, rs3024492, and rs3024496). Two of these SNPs (rs1800896 and rs3024496) were in strong LD (r2 = 0.94).
Population-based association analysis of IgE to dust mite in Costa Rica
The results for the analysis of association between SNPs in 1L10 and IgE to dust mite are presented in Table III. Similar to the results of our family-based analysis, no 1L10 SNP was significantly associated with IgE to dust mite in models not accounting for genotype-by-allergen interactions (model 1). Including such interactions in our models resulted in significant effects for genotypes, Der p 1 levels (except for the model including SNP rs3024492), and genotype-by-dust-mite allergen interactions for all 3 SNPs (model 2). Fig 2 shows the results of our generalized linear models. For each SNP (rs1800896, rs3024492, and rs3024496 in A, B, and C, respectively), children homozygous for the minor allele (shown in red) had higher levels of IgE to dust mite than children homozygous for the major allele (shown in yellow) at high levels of dust mite allergen exposure; heterozygote children (shown in blue) had intermediate effects. In contrast, opposite effects are noted at low levels of dust mite allergen exposure. Bootstrapped regression resulted in smaller CIs for the interaction terms in our models for 2 SNPs (rs1800896 and rs3024496; model 3 in Table III). Models accounting for the possibility of right-censoring in the data for children whose specific IgE to dust mite reached the upper limit of detection (100 kU/L) also show significant results for genotype-by-allergen interaction for all SNPs except rs3024492 (model 4 in Table III). P values for F statistics were examined to assess model fitness. Comparable to our findings for individual predictors, models were not adequately fit (P values for model 1, A-C, are all >.4) without the inclusion of interaction terms (P values for model 2, A-C, are all <.05).
Analysis of asthma exacerbations
The results of logistic regression models for asthma exacerbations are presented in Table IV . After adjusting for age and sex, there was no significant association between any of the 3 SNPs of interest in 1L10 (rs1800896, rs3024492, and rs3024496) and asthma exacerbations in Costa Rica or CAMP (results not shown). On the other hand, there were significant interactions between genotypes in the IL10 SNPs and dust mite allergen exposure on asthma exacerbations (Table IV). Among Costa Rican children who were homozygous for the minor allele of each of the 3 1L10 SNPs, those exposed to high dust mite allergen levels had significantly increased odds of asthma hospitalizations (~3-fold to 39-fold increment). The effect of high levels of dust mite allergen was the opposite for children homozygous for the major allele of each of the 3 SNPs (~81% to 86% decrease), with intermediate effects on heterozygous children.
While attenuated, the results for 2 common SNPs in IL10 (rs1800896 and rs3024496, both with MAF of 0.49 in CAMP; Fig 1) similarly modified the effect of Der p 1 allergen on asthma exacerbations among children at years 2 (P value for interaction [rs1800896] = .05; [rs3024496] = .12) and 4 (P value for interaction [rs1800896] = .07; [rs3024496] = .14) of the CAMP trial. Among children homozygous for the minor alleles of each of the 2 1L10 SNPs, those exposed to high levels of dust mite allergen had increased odds of asthma hospitalizations (~1.3-fold to 1.9-fold increase at 2 years, and a ~1.02-fold to 1.3-fold increase at 4 years). The effect of high levels of dust mite allergen was the opposite for children homozygous for the major allele of each of the 2 SNPs (~87% to 92% decrease at 2 years, and a ~85% to 88% decrease at 4 years), with intermediate effects on heterozygous children. Although consistent in direction with results obtained in Costa Rica, results for SNP rs3024492 (MAF, 0.26 in CAMP) were not significant. Consistent with these findings, result from the combined analysis of Costa Rica and CAMP (Table IV) demonstrate that for 2 SNPs (rs1800896 and rs3024496), genotype-by-allergen interactions were significant in the prediction asthma exacerbations. P values for the Hosmer-Lemeshow statistic ranged between 0.4 and 0.8 for all logistic models presented, consistent with adequate model fitness.
Poisson regression models were originally used to analyze counted numbers of asthma exacerbations in Costa Rica and CAMP. Deviance/degrees of freedom were assessed to evaluate the adequacy of fit for our models. Although overdispersion did not appear to be a significant factor in the fit of our Poisson regression models in CAMP (deviance/degrees of freedom ranged between 1 and 2), our results in Costa Rica were strongly suggestive of overdispersion (deviance/degrees of freedom ~4). To account for this overdispersion, we reanalyzed our data for Costa Rica by using negative binomial models (deviance/degrees of freedom in negative binomial models from Costa Rica were all ~1). Results from our Poisson and negative binomial regression models (Table E2) suggest that dust mite allergen exposure modified the effect of 1 SNP in 1L10 (rs1800896) on the number of asthma exacerbations in Costa Rica and in CAMP. Among Costa Rican children who were homozygous for the minor allele of each of these 2 SNPs, those exposed to high levels of dust mite allergen had an increase (75%) in the count of asthma exacerbations; however, this result was of marginal significance (P = .06). Similar results were obtained in CAMP. Among white children in CAMP who were homozygous for the minor allele of SNP rs1800896, those exposed to high levels of dust mite allergen had a significantly increased number of asthma exacerbations in years 2 and 4. Results for SNP rs3024496 were similar to results of SNP rs1800896 in both Costa Rica and CAMP; however, most of these results were of marginal statistical significance. Significant evidence for modification of the effect of SNP rs3024492 on the number of asthma exacerbations by dust mite allergen exposure was demonstrated in CAMP at year 2 but not in CAMP at year 4 or in the Costa Rican cohort. Consistent with these findings and those of logistic regression, the results from the combined analysis of Costa Rica and CAMP (Table E2) demonstrate that SNP rs1800896-by-allergen interactions were significantly predictive of asthma exacerbations. These findings were less compelling for SNPs rs3024492 and rs3024496.
Acknowledgments
We thank all families for their invaluable participation in the Genetics of Asthma in Costa Rica and CAMP studies. We acknowledge the CAMP investigators and research team, supported by the National Heart, Lung, and Blood Institute, for collection of CAMP Genetic Ancillary Study data. All work on data collected from the Genetics of Asthma in Costa Rica and the CAMP Genetic Ancillary Study was conducted at the Channing Laboratory of Brigham and Women’s Hospital under appropriate CAMP policies and human subjects protections.
The Genetics of Asthma in Costa Rica Study is supported by National Institutes of Health grants HL66289, HL04370, and HL074193. The Childhood Asthma Management Program Genetics Ancillary Study is supported by the National Heart, Lung, and Blood Institute, grants N01-HR-16049, U01HL075419, U01HL65899, P01HL083069, R01HL086601, and T32HL07427. G.M.H. is the recipient of an Individual National Research Service Award (1F32HL083634).
Abbreviations used
- CAMP
Childhood Asthma Management Program
- FBAT
Family-based association test
- FBATI
Family-based test of interaction
- LD
Linkage disequilibrium
- MAF
Minor allele frequency
- SNP
Single nucleotide polymorphism
Footnotes
Disclosure of potential conflict of interest: D. R. Gold has given talks on indoor allergens for Indoor Biotechnologies Ltd. S. T. Weiss has consulting arrangements with Schering-Plough, Genentech, Variagenics, Genome Therapeutics, and Roche Pharmaceuticals and has received research support from AstraZeneca, Millennium Pharmaceuticals, Pfizer, Boehringer Ingelheim, and Glaxo Wellcome. The rest of the authors have declared that they have no conflict of interest.
REFERENCES
- 1.Taylor A, Verhagen J, Blaser K, Akdis M, Akdis CA. Mechanisms of immune suppression by interleukin-10 and transforming growth factor-beta: the role of T regulatory cells. Immunology 2006;117:433–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akbari O, DeKruyff RH, Umetsu DT. Pulmonary dendritic cells producing IL-10 mediate tolerance induced by respiratory exposure to antigen. Nat Immunol 2001;2:725–31. [DOI] [PubMed] [Google Scholar]
- 3.Lim S, Caramori G, Tomita K, Jazrawi E, Oates T, Chung KF, et al. Differential expression ofIL-10receptor by epithelial cells and alveolar macrophages. Allergy 2004;59:505–14. [DOI] [PubMed] [Google Scholar]
- 4.Matsumura Y, Byrne SN, Nghiem DX, Miyahara Y, Ullrich SE. A role for inflammatory mediators in the induction of immunoregulatory B cells. J Immunol 2006;177:4810–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oh JW, Seroogy CM, Meyer EH, Akbari O, Berry G, Fathman CG, et al. CD4 T-helper cells engineered to produce IL-10 prevent allergen-induced airway hyperreactivity and inflammation. J Allergy Clin Immunol 2002;110:460–8. [DOI] [PubMed] [Google Scholar]
- 6.Kurz T, Strauch K, Heinzmann A, Braun S, Jung M, Ruschendorf F, et al. A European study on the genetics of mite sensitization. J Allergy Clin Immunol 2000;106:925–32. [DOI] [PubMed] [Google Scholar]
- 7.Lyon H, Lange C, Lake S, Silverman EK, Randolph AG, Kwiatkowski D, et al. IL10 gene polymorphisms are associated with asthma phenotypes in children. Genet Epidemiol 2004;26:155–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Steen K, McQueen MB, Herbert A, Raby B, Lyon H, Demeo DL, et al. Genomic screening and replication using the same data set in family-based association testing. Nat Genet 2005;37:683–91. [DOI] [PubMed] [Google Scholar]
- 9.Hakonarson H, Bjornsdottir US, Ostermann E, Arnason T, Adalsteinsdottir AE, Halapi E, et al. Allelic frequencies and patterns of single-nucleotide polymorphisms in candidate genes for asthma and atopy in Iceland. Am J Respir Crit Care Med 2001;164:2036–44. [DOI] [PubMed] [Google Scholar]
- 10.Eskdale J, Gallagher G, Verweij CL, Keijsers V, Westendorp RG, Huizinga TW. Interleukin 10 secretion in relation to human IL-10 locus haplotypes. Proc Natl Acad SciUSA 1998;95:9465–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibson AW, Edberg JC, Wu J, Westendorp RG, Huizinga TW, Kimberly RP. Novel single nucleotide polymorphisms in the distal IL-10promoter affect IL-10production and enhance the risk of systemic lupus erythematosus. J Immunol 2001;166:3915–22. [DOI] [PubMed] [Google Scholar]
- 12.Martinez FD. Gene-environment interactions in asthma: with apologies to William of Ockham. Proc Am Thorac Soc 2007;4:26–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Price JA, Pollock I, Little SA, Longbottom JL, Warner JO. Measurement of airborne mite antigen in homes of asthmatic children. Lancet 1990;336:895–7. [DOI] [PubMed] [Google Scholar]
- 14.Celedon JC, Milton DK, Ramsey CD, Litonjua AA, Ryan L, Platts-Mills TA, et al. Exposure to dust mite allergen and endotoxin in early life and asthma and atopy in childhood. J Allergy Clin Immunol 2007;120:144–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green RM, Custovic A, Sanderson G, Hunter J, Johnston SL, Woodcock A. Synergism between allergens and viruses and risk of hospital admission with asthma: case-control study. BMJ 2002;324:763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong CK, Li ML, Wang CB, Ip WK, Tian YP, Lam CW. House dust mite allergen Der p 1 elevates the release of inflammatory cytokines and expression of adhesion molecules in co-culture of human eosinophils and bronchial epithelial cells. Int Immunol 2006;18:1327–35. [DOI] [PubMed] [Google Scholar]
- 17.Hunninghake GM, Soto-Quiros ME, Avila L, Ly NP, Liang C, Sylvia JS, et al. Sensitization to Ascaris lumbricoides and severity of childhood asthma in Costa Rica. J Allergy Clin Immunol 2007;119:654–61. [DOI] [PubMed] [Google Scholar]
- 18.Carvajal-Carmona LG, Ophoff RA, Service S, Hartiala J, Molina J, Leon P, et al. Genetic demography of Antioquia (Colombia) and the Central Valley of Costa Rica. Hum Genet 2003;112:534–41. [DOI] [PubMed] [Google Scholar]
- 19.Escamilla MA, Spensy M, Reus VI, Gallegos A, Meza L, Molina J, et al. Use of linkage disequilibrium approaches to map genes for bipolar disorder in the Costa Rican population. Am J Med Genet 1996;67:244–53. [DOI] [PubMed] [Google Scholar]
- 20.Pearce N, Ait-Khaled N, Beasley R, Mallol J, Keil U, Mitchell E, et al. Worldwide trends in the prevalence of asthma symptoms: phase III of the International Study of Asthma and Allergies in Childhood (ISAAC). Thorax 2007;62:758–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.The Childhood Asthma Management Program (CAMP): design, rationale, and methods. Childhood Asthma Management Program Research Group. Control Clin Trials 1999;20:91–120. [PubMed] [Google Scholar]
- 22.Luczynska CM, Arruda LK, Platts-Mills TA, Miller JD, Lopez M, Chapman MD. A two-site monoclonal antibody ELISA for the quantification of the major Dermatopha- goides spp. allergens, Derp I and Der f I. J Immunol Methods 1989;118:227–35. [DOI] [PubMed] [Google Scholar]
- 23.Kurreeman FA, Schonkeren JJ, Heijmans BT, Toes RE, Huizinga TW. Transcription of the IL10 gene reveals allele-specific regulation at the mRNA level. Hum Mol Genet 2004;13:1755–62. [DOI] [PubMed] [Google Scholar]
- 24.Lazarus R, Klimecki WT, Palmer LJ, Kwiatkowski DJ, Silverman EK, Brown A, et al. Single-nucleotide polymorphisms in the interleukin-10 gene: differences in frequencies, linkage disequilibrium patterns, and haplotypes in three United States ethnic groups. Genomics 2002;80:223–8. [DOI] [PubMed] [Google Scholar]
- 25.Gunderson KL, Steemers FJ, Lee G, Mendoza LG, Chee MS. A genome-wide scalable SNP genotyping assay using microarray technology. Nat Genet 2005;37:549–54. [DOI] [PubMed] [Google Scholar]
- 26.Jungerius BJ, Veenendaal A, Van Oost BA, Te Pas MF, Groenen MA. Typing single-nucleotide polymorphisms using a gel-based sequencer: a new data analysis tool and suggestions for improved efficiency. Mol Biotechnol 2003;25:283–8. [DOI] [PubMed] [Google Scholar]
- 27.Platts-Mills TA, Hayden ML, Chapman MD, Wilkins SR. Seasonal variation in dust mite and grass-pollen allergens in dust from the houses of patients with asthma. J Allergy Clin Immunol 1987;79:781–91. [DOI] [PubMed] [Google Scholar]
- 28.Sporik R, Platts-Mills TA, Cogswell JJ. Exposure to house dust mite allergen of children admitted to hospital with asthma. Clin Exp Allergy 1993;23:740–6. [DOI] [PubMed] [Google Scholar]
- 29.O’Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 1998;63:259–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005;21:263–5. [DOI] [PubMed] [Google Scholar]
- 31.Lange C, DeMeo D, Silverman EK, Weiss ST, Laird NM. PBAT: tools for family-based association studies. Am J Hum Genet 2004;74:367–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lake SL, Laird NM. Tests of gene-environment interaction for case-parent triads with general environmental exposures. Ann Hum Genet 2004;68:55–64. [DOI] [PubMed] [Google Scholar]
- 33.Cosmi L, Santarlasci V, Angeli R, Liotta F, Maggi L, Frosali F, et al. Sublingual immunotherapy with Dermatophagoides monomeric allergoid down-regulates allergen-specific immunoglobulin E and increases both interferon-gamma- and interleukin-10-production. Clin Exp Allergy 2006;36:261–72. [DOI] [PubMed] [Google Scholar]
- 34.Jeannin P, Lecoanet S, Delneste Y, Gauchat JF, Bonnefoy JY. IgE versus IgG4 production can be differentially regulated by IL-10. J Immunol 1998;160:3555–61. [PubMed] [Google Scholar]
- 35.Chung EY, Liu J, Zhang Y, Ma X. Differential expression in lupus-associated IL-10 promoter single-nucleotide polymorphisms is mediated by poly(ADP-ribose) polymerase-1. Genes Immun 2007;8:577–89. [DOI] [PubMed] [Google Scholar]
- 36.Demeo DL, Campbell EJ, Barker AF, Brantly ML, Eden E, McElvaney NG, et al. IL10 polymorphisms are associated with airflow obstruction in severe alpha1-anti-trypsin deficiency. Am J Respir Cell Mol Biol 2008;38:114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoffjan S, Ostrovnaja I, Nicolae D, Newman DL, Nicolae R, Gangnon R, et al. Genetic variation in immunoregulatory pathways and atopic phenotypes in infancy. J Allergy Clin Immunol 2004;113:511–8. [DOI] [PubMed] [Google Scholar]
- 38.Torrent M, Sunyer J, Munoz L, Cullinan P, Iturriaga MV, Figueroa C, et al. Early- life domestic aeroallergen exposure and IgE sensitization at age 4 years. J Allergy Clin Immunol 2006;118:742–8. [DOI] [PubMed] [Google Scholar]
- 39.Wahn U, Lau S, Bergmann R, Kulig M, Forster J, Bergmann K, et al. Indoor allergen exposure is a risk factor for sensitization during the first three years of life. J Allergy Clin Immunol 1997;99:763–9. [DOI] [PubMed] [Google Scholar]
- 40.Lau S, Illi S, Sommerfeld C, Niggemann B, Bergmann R, von Mutius E, et al. Early exposure to house-dust mite and cat allergens and development of childhood asthma: a cohort study. Multicentre Allergy Study Group. Lancet 2000;356:1392–7. [DOI] [PubMed] [Google Scholar]
- 41.Huss K, Adkinson NF Jr, Eggleston PA, Dawson C, Van Natta ML, Hamilton RG. House dust mite and cockroach exposure are strong risk factors for positive allergy skin test responses in the Childhood Asthma Management Program. J Allergy Clin Immunol 2001;107:48–54. [DOI] [PubMed] [Google Scholar]
- 42.Custovic A, Taggart SC, Francis HC, Chapman MD, Woodcock A. Exposure to house dust mite allergens and the clinical activity of asthma. J Allergy Clin Immunol 1996;98:64–72. [DOI] [PubMed] [Google Scholar]
- 43.Marks GB, Tovey ER, Toelle BG, Wachinger S, Peat JK, Woolcock AJ. Mite allergen (Der p 1) concentration in houses and its relation to the presence and severity of asthma in a population of Sydney schoolchildren. J Allergy Clin Immunol 1995;96:441–8. [DOI] [PubMed] [Google Scholar]
- 44.Dharmage S, Bailey M, Raven J, Mitakakis T, Cheng A, Guest D, et al. Current indoor allergen levels of fungi and cats, butnot house dustmites, influence allergy and asthmain adults with high dust mite exposure. Am J Respir Crit Care Med 2001;164:65–71. [DOI] [PubMed] [Google Scholar]
- 45.Kuehr J, Frischer T, Meinert R, Barth R, Forster J, Schraub S,et al. Mite allergen exposure is arisk for the incidence of specific sensitization. J Allergy Clin Immunol 1994;94:44–52. [DOI] [PubMed] [Google Scholar]
- 46.Cole Johnson C, Ownby DR, Havstad SL, Peterson EL. Family history, dust mite exposure in early childhood, and risk for pediatric atopy and asthma. J Allergy Clin Immunol 2004;114:105–10. [DOI] [PubMed] [Google Scholar]
References
- E1.Hunninghake GM, Soto-Quiros ME, Avila L, Ly NP, Liang C, Sylvia JS, et al. Sensitization to Ascaris lumbricoides and severity of childhood asthma in Costa Rica. J Allergy Clin Immunol 2007;119:654–61. [DOI] [PubMed] [Google Scholar]
- E2.Hersh CP, Raby BA, Soto-Quiros ME, Murphy AJ, Avila L, Lasky-Su J, et al. Comprehensive testing of positionally cloned asthma genes in two populations. Am J Respir Crit Care Med 2007;176:849–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E3.Service S, Deyoung J, Karayiorgou M, Roos JL, Pretorious H, Bedoya G, et al. Magnitude and distribution of linkage disequilibrium in population isolates and implications for genome-wide association studies. Nat Genet 2006;38:556–60. [DOI] [PubMed] [Google Scholar]
- E4.Carvajal-Carmona LG, Ophoff RA, Service S, Hartiala J, Molina J, Leon P, et al. Genetic demography of Antioquia (Colombia) and the Central Valley of Costa Rica. Hum Genet 2003;112:534–41. [DOI] [PubMed] [Google Scholar]
- E5.Pearce N, Ait-Khaled N, Beasley R, Mallol J, Keil U, Mitchell E, et al. Worldwide trends in the prevalence of asthma symptoms: phase III of the International Study of Asthma and Allergies in Childhood (ISAAC). Thorax 2007;62:758–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E6.Escamilla MA, Spensy M, Reus VI, Gallegos A, Meza L, Molina J, et al. Use of linkage disequilibrium approaches to map genes for bipolar disorder in the Costa Rican population. Am J Med Genet 1996;67:244–53. [DOI] [PubMed] [Google Scholar]
- E7.The Childhood Asthma Management Program (CAMP): design, rationale, and methods. Childhood Asthma Management Program Research Group. Control Clin Trials 1999;20:91–120. [PubMed] [Google Scholar]
- E8.Fleming DM, Cross KW, Sunderland R, Ross AM. Comparison of the seasonal patterns of asthma identified in general practitioner episodes, hospital admissions, and deaths. Thorax 2000;55:662–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E9.Luczynska CM, Arruda LK, Platts-Mills TA, Miller JD, Lopez M, Chapman MD. A two-site monoclonal antibody ELISA for the quantification of the major Dermato-phagoides spp. allergens, Der p I and Der f I. JImmunol Methods 1989;118:227–35. [DOI] [PubMed] [Google Scholar]
- E10.Kurreeman FA, Schonkeren JJ, Heijmans BT, Toes RE, Huizinga TW. Transcription of the IL10 gene reveals allele-specific regulation at the mRNA level. Hum Mol Genet 2004;13:1755–62. [DOI] [PubMed] [Google Scholar]
- E11.Gibson AW, Edberg JC, Wu J, Westendorp RG, Huizinga TW, Kimberly RP. Novel single nucleotide polymorphisms in the distal IL-10 promoter affect IL- 10 production and enhance the risk of systemic lupus erythematosus. J Immunol 2001;166:3915–22. [DOI] [PubMed] [Google Scholar]
- E12.Lazarus R, Klimecki WT, Palmer LJ, Kwiatkowski DJ, Silverman EK, Brown A, et al. Single-nucleotide polymorphisms in the interleukin-10 gene: differences in frequencies, linkage disequilibrium patterns, and haplotypes in three United States ethnic groups. Genomics 2002;80:223–8. [DOI] [PubMed] [Google Scholar]
- E13.Gunderson KL, Steemers FJ, Lee G, Mendoza LG, Chee MS. A genome-wide scalable SNP genotyping assay using microarray technology. Nat Genet 2005;37:549–54. [DOI] [PubMed] [Google Scholar]
- E14.Jungerius BJ, Veenendaal A, Van Oost BA, Te Pas MF, Groenen MA. Typing single-nucleotide polymorphisms using a gel-based sequencer: a new data analysis tool and suggestions for improved efficiency. Mol Biotechnol 2003;25:283–8. [DOI] [PubMed] [Google Scholar]
- E15.Antens CJ, Oldenwening M, Wolse A, Gehring U, Smit HA, Aalberse RC, et al. Repeated measurements of mite and pet allergen levels in house dust over a time period of 8 years. Clin Exp Allergy 2006;36:1525–31. [DOI] [PubMed] [Google Scholar]
- E16.Platts-Mills TA, Hayden ML, Chapman MD, Wilkins SR. Seasonal variation in dust mite and grass-pollen allergens in dust from the houses of patients with asthma. J Allergy Clin Immunol 1987;79:781–91. [DOI] [PubMed] [Google Scholar]
- E17.Sporik R, Holgate ST, Platts-Mills TA, Cogswell JJ. Exposure to house-dust mite allergen (Der p I) and the development of asthma in childhood: a prospective study. N Engl J Med 1990;323:502–7. [DOI] [PubMed] [Google Scholar]
- E18.Sporik R, Platts-Mills TA, Cogswell JJ. Exposure to house dust mite allergen of children admitted to hospital with asthma. Clin Exp Allergy 1993;23:740–6. [DOI] [PubMed] [Google Scholar]
- E19.O’Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 1998;63:259–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E20.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005;21:263–5. [DOI] [PubMed] [Google Scholar]
- E21.Lange C, DeMeo D, Silverman EK, Weiss ST, Laird NM. PBAT: tools for family-based association studies. Am J Hum Genet 2004;74:367–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- E22.Vansteelandt S, Demeo DL, Lasky-Su J, Smoller JW, Murphy AJ, McQueen M, et al. Testing and estimating gene-environment interactions in family-based association studies. Biometrics 2007; October 26 [epub ahead of print]. [DOI] [PubMed] [Google Scholar]