Abstract
Germline SAMD9 and SAMD9L mutations cause a spectrum of multisystem disorders that carry a markedly increased risk of developing myeloid malignancies with somatic monosomy 7. Here, we describe 16 siblings, the majority of which were phenotypically normal, from 5 families diagnosed with myelodysplasia and leukemia syndrome with monosomy 7 (MLSM7; OMIM 252270) who primarily had onset of hematologic abnormalities during the first decade of life. Molecular analyses uncovered germline SAMD9L (n = 4) or SAMD9 (n = 1) mutations in these families. Affected individuals had a highly variable clinical course that ranged from mild and transient dyspoietic changes in the bone marrow to a rapid progression of myelodysplastic syndrome (MDS) or acute myeloid leukemia (AML) with monosomy 7. Expression of these gain-of-function SAMD9 and SAMD9L mutations reduces cell cycle progression, and deep sequencing demonstrated selective pressure favoring the outgrowth of clones that have either lost the mutant allele or acquired revertant mutations. The myeloid malignancies of affected siblings acquired cooperating mutations in genes that are also altered in sporadic cases of AML characterized by monosomy 7. These data have implications for understanding how SAMD9 and SAMD9L mutations contribute to myeloid transformation and for recognizing, counseling, and treating affected families.
Keywords: Hematology
Keywords: Leukemias
Germline mutations in SAMD9 or SAMD9L define familial myelodysplasia and leukemia syndrome with monosomy 7, and result in a variety of hematologic disorders.
Introduction
In 2016, Narumi and colleagues (1) identified heterozygous germline missense SAMD9 mutations as the cause of MIRAGE syndrome (OMIM 617053), a multisystem developmental disorder characterized by an elevated risk of early-onset myelodysplastic syndrome (MDS) with somatic monosomy 7 (Mo7). Enforced expression of these mutant SAMD9 proteins in cultured cell lines revealed gain-of-function growth-suppressive activities and impaired ERK activation (1). Remarkably, germline SAMD9 mutations were invariably absent in the Mo7 bone marrow (BM) cells of affected patients, indicating selective pressure for loss of the chromosome 7 homolog harboring the mutant allele during leukemic outgrowth. In keeping with its previous use to describe stress-related aneuploidy in hepatocytes (2), the authors referred to this novel mechanism of myeloid transformation as “adaptation by aneuploidy” (1).
This landmark discovery stimulated additional studies that revealed germline SAMD9 mutations in additional MIRAGE syndrome patients (3), as well as SAMD9L mutations in patients with ataxia pancytopenia syndrome (ATXPC; OMIM 159550) and in other children with BM Mo7, MDS, aplastic anemia, and/or neurologic abnormalities (4–6). We recently identified a germline SAMD9 mutation in siblings that presented with MDS without developmental or neurologic abnormalities (7). Our sequencing of Mo7 BM samples from this family, as well as other recent reports, revealed complex patterns of clonal evolution with the acquisition of copy-neutral loss of heterozygosity (CN-LOH) as a result of somatic uniparental disomy (UPD) for chromosome 7q and revertant mosaic SAMD9 or SAMD9L mutant alleles (3, 5, 7, 8). We also discovered germline SAMD9 or SAMD9L variants in 8 of 46 (17%) unselected primary pediatric MDS cases seen at a single institution (9). Together, these data collectively identified SAMD9 and SAMD9L as strong candidate genes in other familial hematologic disorders associated with MDS, including myelodysplasia and leukemia syndrome with Mo7 (MLSM7; OMIM 252270) (10, 11). MLSM7 was first reported in phenotypically normal siblings who developed MDS or acute myeloid leukemia (AML) associated with Mo7 during the first 2 decades of life (10, 11). Interestingly, Southern blot analysis identified DNA segments from different parental chromosome 7 homologs in the 7q22-q34 interval in MDS and AML samples (11). These results are inconsistent with the Knudson paradigm of recessive tumor suppressor genes, but compatible with adaptation by aneuploidy.
Here, we describe molecular analysis of blood and/or BM samples from 5 MLSM7 families that revealed germline mutation in either SAMD9L or SAMD9 in 16 siblings who were evaluated for hematologic abnormalities, MDS, or AML during the first decade of life. We observed remarkable clinical and hematologic variability in different families, including spontaneous improvement with loss of Mo7 cells in some individuals, and early progression to AML in others. Deep sequencing and single nucleotide polymorphism (SNP) array analyses uncovered multiple acquired revertant alterations that presumably remove or counteract the growth-suppressive germline allele, and we also observed discrete secondary mutations upon progression to MDS or AML within different members of the same family. In addition to further illuminating the pathogenesis of MLSM7, these data have implications for diagnosis and clinical management.
Results
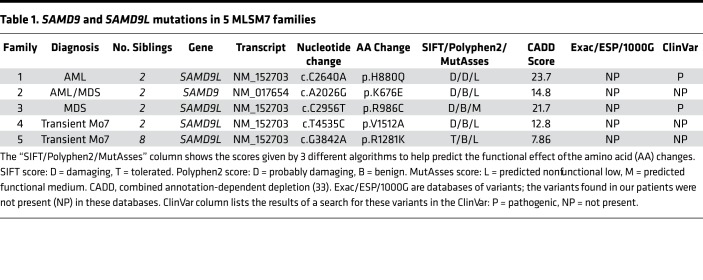
Table 1 and Supplemental Table 1 (supplemental material available online with this article; https://doi.org/10.1172/jci.insight.121086DS1) present selected clinical, pathologic, and molecular findings from 5 families with a germline mutation in SAMD9 (n = 1) or SAMD9L (n = 4), and the mutational data are summarized in Figure 1 (12). Families 1 and 2 were included in the original description of MLSM7 (10, 11). Individual pedigrees for families 1, 2, and 5 are shown in Figures 2–4, and in the supplemental materials for families 3 and 4 (Supplemental Figures 1 and 2).
Table 1. SAMD9 and SAMD9L mutations in 5 MLSM7 families.
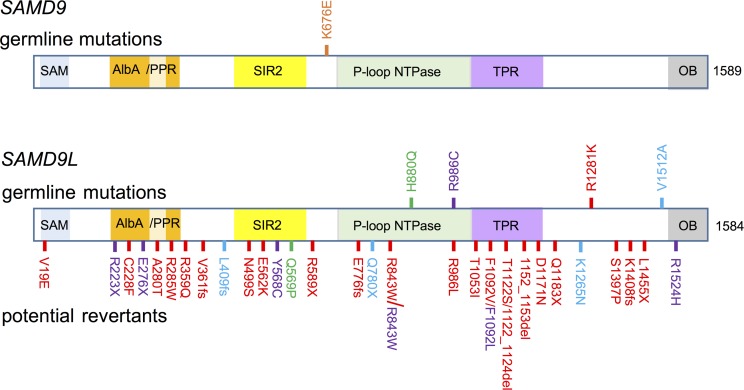
Figure 1. SAMD9 and SAMD9L variants identified in 5 MLSM7 families.
Schematic diagram of the locations of germline mutations and potential revertants with respect to the predicted domains of SAMD9 (NM_017654) and SAMD9L (NM_152703) proteins based on Mekhedov et al. (12). Germline mutations (top) and potential revertants (bottom) are shown. Mutations and revertants are color coded for each family (1: green, 2: orange, 3: purple, 4: blue, 5: red). SAM, sterile α motif domain; AlbA, acetylation lowers binding affinity; DNA-binding domain; PPR, pentatricopeptide repeat; SIR2, silent mating–type information regulation 2; P-loop NTPase, P-loop–containing nucleoside triphosphate hydrolase; TPR, tetratricopeptide repeat–containing domain; OB, oligonucleotide/oligosaccharide–binding fold domain. Figure not drawn to scale.
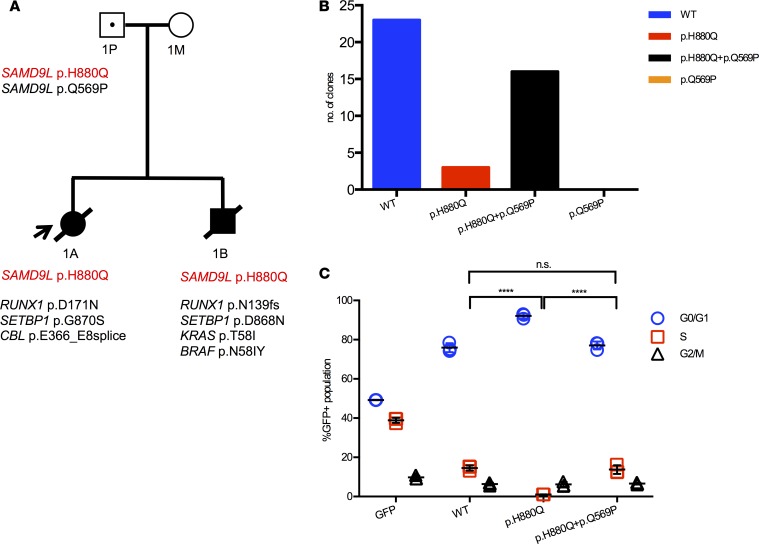
Figure 2. SAMD9L H880Q mutation in MLSM7 family 1.
(A) Pedigree showing the germline SAMD9L mutation (red font) in the father (1P) and patients 1A and 1B, as well as secondary somatic mutations identified in their AML bone marrows (black font). Squares indicate male family members, circles female members. Unaffected individuals are indicated with open symbols, unaffected mutation carriers are denoted with black dots, affected individuals with a clinical history of monosomy 7 are shown with filled symbols, and symbols with a slash denote deceased individuals. Arrow indicates the proband. (B) Sequencing data from the father (1P) showing the number of individual amplicons with normal sequence (left), those with the p.H880Q mutation only (middle), and those with the p.H880Q and p.Q569P substitutions in cis (right). The p.Q569P second-site mutation was only detected in the context of the p.H880Q variant and accounted for over 80% of mutant transcripts. (C) EdU cell cycle results showing that 293T cells expressing SAMD9L p.H880Q exhibit a cell cycle arrest phenotype compared with cells expressing the normal SAMD9L protein. This defect is partially corrected by the second-site p.Q569P substitution detected in 1P. Percentages of cells in the S phase of the cell division cycle were compared using 1-way ANOVAs with repeated measures followed by Tukey’s post hoc multiple-comparisons test; significance was based on α = 0.05 (n = 3). ****P < 0.0001. n.s., not significant. Individual data points, and mean ± SD are shown. Data representative of 3 experiments completed in triplicate.
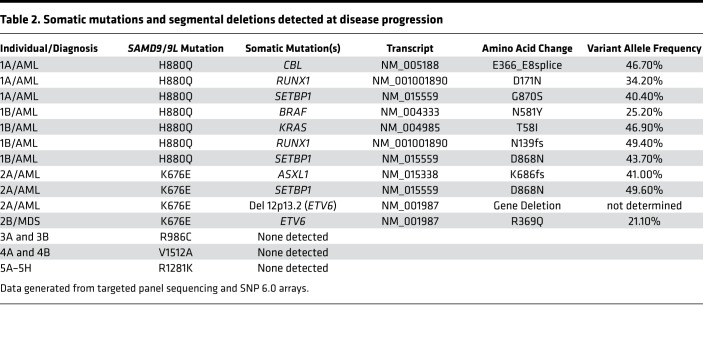
As previously reported (10, 11), the proband in family 1 presented with AML. Review of the published restriction fragment length polymorphism (RFLP) data showed that each patient’s BM deleted the chromosome 7 homolog inherited from the father (Supplemental Figure 3). Consistent with these data, both siblings inherited a mutation in SAMD9L (p.H880Q) from their asymptomatic father (Figure 2A and Supplemental Table 2). This SAMD9L p.H880Q mutation has been previously reported in families with MDS and ATXPC (4, 8). Deep sequencing showed that the mutation was present at a variant allele frequency (VAF) of less than 5% in the BM of both children, consistent with a high percentage of Mo7 cells detected by cytogenetic and SNP array analyses (Supplemental Table 3). By contrast, this mutation was present at a VAF of 43.17% in the healthy father. In addition, we detected a second-site p.Q569P mutation in cis at a VAF of 41.72% in the same DNA specimen from the father (Figure 2B and Supplemental Table 4) that was not observed in either sibling. We reasoned that this represented a somatic mutation that might confer a fitness advantage in cells. Consistent with this idea, 293T cells transiently expressing the p.H880Q variant showed a reduced fraction of cells in S phase compared with controls, which was partially rescued by the p.Q569P mutation when in cis with p.H880Q (Figure 2C). The AML cells of the brother and sister in family 1 acquired different pathogenic RUNX1 and SETBP1 mutations, both of which were present at high VAFs (Table 2 and Supplemental Table 5). The AML clone of patient 1B also acquired mutations in BRAF (p.N581Y), and KRAS (p.T58I) — the latter is a recurrent mutation in sporadic AML (13–16). Patient 1A died with refractory AML, while his sibling (patient 1B) died from infection after hematopoietic stem cell transplantation (HSCT).
Table 2. Somatic mutations and segmental deletions detected at disease progression.
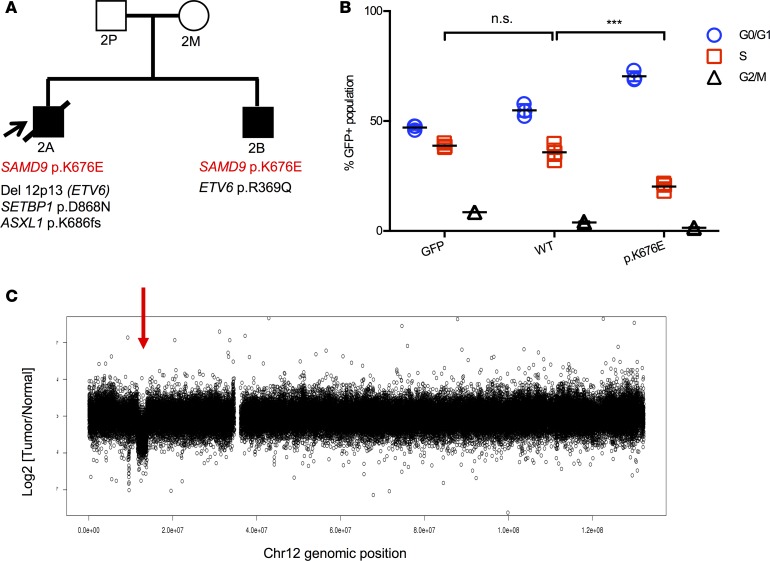
The siblings in family 2 (patients 2A and 2B) presented with AML and MDS, respectively (Table 1 and Supplemental Table 1). Molecular analysis uncovered a previously unidentified germline mutation in SAMD9 (p.K676E) that was present at a greatly reduced VAF in BM samples from both children with a high percentage of Mo7 cells (Supplemental Tables 2 and 3). In contrast to family 1, this variant was not detected in either parent, despite the fact that previous RFLP analysis by Southern blot demonstrated loss of the paternal chromosome 7 homolog in both siblings (Supplemental Figure 3B). This p.K676E SAMD9 variant suppressed cell cycle progression in 293T cells (Figure 3B). The AML cells of patient 2A acquired a SETBP1 mutation, as well as heterozygous deletion of chromosome band 12p13, which contains the ETV6 gene (Figure 3C, Table 2, and Supplemental Tables 3 and 5). Remarkably, his brother’s MDS cells retained both ETV6 alleles, but acquired a heterozygous p.R369Q mutation. Patient 2A died of sepsis during induction, while patient 2B recovered following HSCT.
Figure 3. SAMD9 K676E mutation in MLSM7 family 2.
(A) Pedigree of family 2 with a germline SAMD9 p.K676E mutation (red font). The cooperating mutations found in the bone marrows of patients 2A (AML) and 2B (MDS) are shown in black text. Squares indicate male family members, circles female members. Unaffected individuals are indicated with open symbols, unaffected mutation carriers are denoted with black dots, affected individuals with a clinical history of monosomy 7 are shown with filled symbols, and symbols with a slash denote deceased individuals. Arrow indicates the proband. (B) EdU cell cycle results from 293T cells transfected with an empty GFP vector, a cDNA encoding WT SAMD9, or a SAMD9 p.K676E cDNA. Percentages of cells in the S phase of the cell division cycle were compared using 1-way ANOVAs with repeated measures followed by Tukey’s post hoc multiple-comparisons test; significance was based on α = 0.05 (n = 3). ***P < 0.001. n.s., not significant. Individual data points, and mean ± SD are shown. Data representative of 2 experiments completed in triplicate. (C) SNP array analysis showing a focal 12p13.2 deletion that includes the ETV6 gene (red arrow) in the AML sample of patient 2A.
MLSM7 families 3–5 also harbored germline SAMD9L mutations, all of which caused growth arrest in 293T cells (Figure 4 and Supplemental Figures 1 and 2). However, in contrast to families 1 and 2 above, mutation carriers exhibited remarkably less severe hematologic diseases. Family 3 was defined by a p.R986C mutation in SAMD9L. The proband (patient 3A) presented with MDS and Mo7. Cytogenetic analysis of the BM of her older sister (patient 3B), who had no overt hematologic abnormalities, revealed a focal somatic deletion of 7q11-36, which contains the SAMD9L locus, in 7 of 27 metaphase cells (Supplemental Figure 1D). Interestingly, she also harbored numerous revertant mutations in SAMD9L that abrogated the growth-suppressive properties of the p.R986C substitution (Supplemental Figure 1, A–C, and Supplemental Table 4). This variant was inherited from the patient’s mother, who also had a p.R1524H substitution in SAMD9L at a VAF of approximately 50%. Interestingly, this variant did not rescue the growth-suppressive effects of the p.R986C mutation in 293T cells (Supplemental Figure 1E).
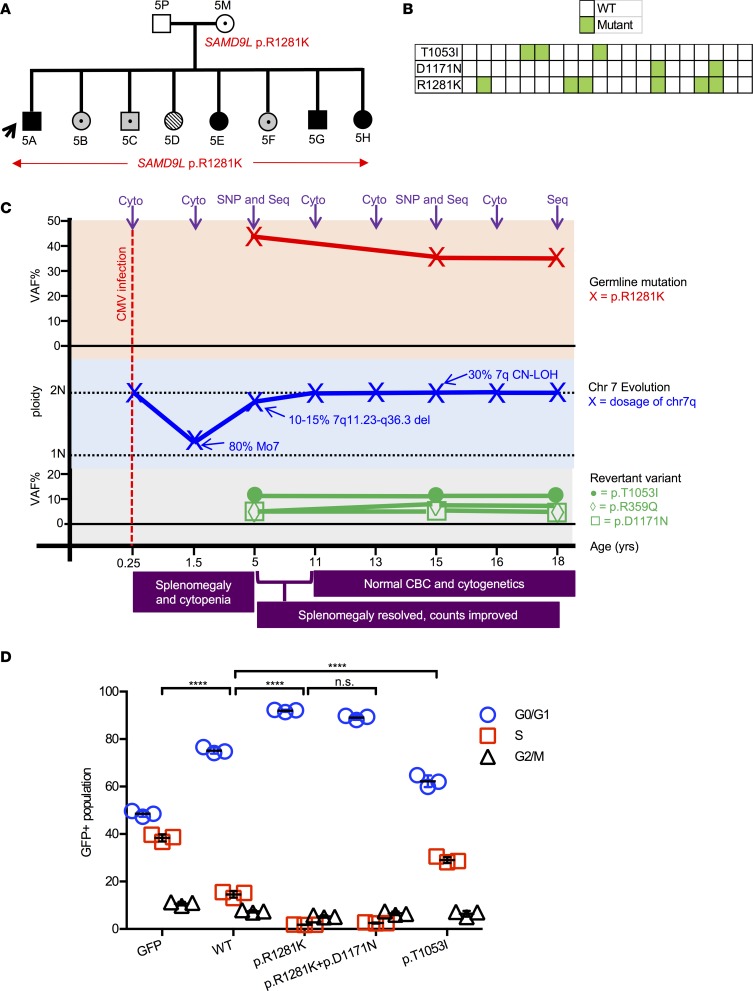
Figure 4. SAMD9L R1281K mutation in MLSM7 family 5.
(A) Pedigree showing the germline SAMD9L p.R1281K mutation (in red) in the healthy carrier (5M) and all 8 children. Squares indicate male family members and circles female members. Arrow indicates the proband. Unaffected individuals are indicated with open symbols, mutation carriers are denoted with black dots, individuals with a clinical history of BM Mo7 are shown with filled symbols, hatched symbol indicates CN-LOH involving chromosome 7q, carriers with dysplastic BM changes are shown with symbols filled with gray in addition to a black dot. (B) Sequencing results of individual PCR products from the amplified genomic DNA of patient 5A at the age of 15. The green boxes indicate individual missense mutations and the white boxes depict the normal sequence. Each column is an individual clone. Note that the p.D1171N mutation occur in cis with the germline p.R1281K mutation, while the p.T1053I does not. (C) Clonal evolution of BM cells from patient 5A (proband) over 18 years. Purple arrows indicate the time points at which cytogenetics (Cyto), SNP array (SNP), or SAMD9L sequencing (Seq) was performed. (D) EdU cell cycle in 293T cells transfected with vectors encoding GFP (empty vector), wild-type SAMD9L, SAMD9L p.R1281K, SAMD9L p.R1281K+p.D1171N, or SAMD9L p.T1053I. Percentages of cells in the S phase of the cell division cycle were compared using 1-way ANOVAs with repeated measures followed by Tukey’s post hoc multiple-comparisons test; significance was based on α = 0.05 (n = 3). ****P < 0.0001. n.s., not significant. Individual data points, and mean ± SD are shown. Data representative of 2 experiments completed in triplicate.
The proband in family 4 (patient 4A) presented when she was 3 years old with pneumonia, oral candidiasis, pancytopenia, and Mo7 in 100% of BM metaphase cells. She was treated with antibiotics and her blood counts recovered. At age 7, cytogenetic analysis of her BM was normal. Her younger sister (patient 4B) was 1.5 years old and in good health when blood tests and BM biopsies were performed. Although her blood counts were normal, cytogenetic analysis revealed Mo7 in 9 of 10 BM metaphase cells. By age 4, her BM cytogenetics were normal. We performed molecular analyses on samples obtained from patient 4A when she was almost 5 years old, and from patient 4B when she was almost 3. Both had a germline SAMD9L p.V1512A variant. We also detected CN-LOH at 7q11.21-q36.3 in the proband (patient 4A; see Supplemental Figure 2C). No somatic mutations in genes frequently altered in MDS or AML were identified in families 3–5.
In family 5, the mother and all 8 children harbor a p.R1281K SAMD9L mutation (Figure 4A). The index case (patient 5A) presented with anemia, thrombocytopenia, and splenomegaly at age 4 months, and he was diagnosed with a cytomegalovirus (CMV) infection. BM cytogenetics were normal at that time despite morphologic dyspoiesis. At age 14 months, he continued to have mild thrombocytopenia and neutropenia with slightly macrocytic red cell indices despite no evidence of persistent CMV infection. A BM aspirate was performed at age 17 months, which revealed Mo7 in 28 of 35 cells analyzed. His blood counts and BM Mo7 resolved over time and he is now 26 years old. Similarly, his siblings all had hypocellular marrows and dyspoietic changes in their erythroid and megakaryocytic cells, but with variable peripheral blood cell counts. Unlike families 1–4, some siblings have extrahematopoietic phenotypes, including camptodactyly (patient 5B) and a sensory and motor peripheral neuropathy (patient 5E), although the majority are phenotypically normal. Three siblings have been diagnosed with BM Mo7 (patients 5E, 5G, and 5H), which resolved without treatment. Consistent with the absence of Mo7 after hematologic recovery, deep sequencing of blood and BM samples from the members of this family revealed a mutant VAF of approximately 50% in the mother (patient 5M) and 5 of the siblings (patients 5B, 5C, 5E, 5F, and 5H) (Supplemental Tables 1 and 2). The remaining 3 siblings either had subclonal 7q deletions (patient 5A; age 5) or varying degrees of 7q CN-LOH involving the SAMD9/9L locus (patient 5A; age 15, and patients 5D and 5G) with a corresponding decrease in the p.R1281K VAF (Supplemental Figure 4A and Supplemental Table 2). Most notably, patient 5D had a p.R1281K VAF of 7.87% and SNP array analysis revealed segmental somatic CN-LOH for the father’s normal chromosome 7 homolog from 7q11.21-qtel (Supplemental Table 2 and Supplemental Figures 4A and 5).
We performed additional sequencing on samples from patient 5A that were collected at 3 time points. At age 5, he had a 7q heterozygous deletion in approximately 10%–15% of his BM cells involving 7q11.23-q36.3, four different potential revertant mutations (p.R70C, p.R359Q, p.T1053I, and p.D1171N), and a p.R1281K VAF of 43% (Figures 4, B and C, and Supplemental Tables 2, 3, and 4). While p.D1171N is in cis with p.R1281K, the 3 clones with the p.T1053I mutation did not contain the p.R1281K germline variant. A sample collected at 15 years of age exhibited clonal evolution, now with CN-LOH involving 7q11.23-7q36.3 in approximately 30% of cells and a lower p.R1281K VAF (35%) (Supplemental Tables 2 and 3). All 4 acquired variants previously detected at age 5 persisted, and 2 additional variants were present at low allelic frequencies (p.A280T and p.F1092V) (Supplemental Table 4). Little VAF change was noted between the samples collected at ages 15 and 18.
Based on a study in which Tesi and colleagues (5) demonstrated a germline loss-of-function SAMD9L mutation in trans that likely modulated the clinical effects of a causative gain-of-function (GOF) SAMD9L allele, we reasoned that the somatic p.T1053I mutation in the BM of patient 5A might behave similarly. Indeed, expressing the SAMD9L p.T1053I mutation in 293T cells resulted in an increase in the S-phase fraction (Figure 4D), suggesting that this is a loss-of-function allele that could counteract the growth-suppressive effects of the p.R1281K mutation when expressed in the same cell.
Deep sequencing uncovered additional potential somatic revertant mutations in other members of family 5 (Supplemental Figure 4B and Supplemental Table 4). Clonal evolution with respect to the percentage of BM cells with Mo7, CN-LOH, and the presence of variant mutations was not unique to patient 5A, but was also evident in patient 5G (Supplemental Figure 4C and Supplemental Tables 3 and 4). Despite the benign clinical course to date in the members of family 5, the p.R1281K variant induced a cell cycle arrest phenotype in transduced 293T cells similar to that of other germline SAMD9L mutations (Supplemental Figure 6).
Discussion
We discovered and functionally characterized germline SAMD9 or SAMD9L mutations in 16 children from 5 MLSM7 families that presented with a range of hematologic abnormalities (Supplemental Figure 7). Together, along with other recent reports (6–9), this study has implications for diagnosing, managing, and counseling families of phenotypically normal children with BM Mo7 regardless of previous history. The recent identification of patients with isolated hematologic disease and of healthy adult SAMD9 or SAMD9L mutation carriers (5, 6, 8) underscores the substantial heterogeneity in disease presentation and clinical course, and supports viewing germline SAMD9 or SAMD9L mutations as the underlying causes of a diverse spectrum of disorders that perturb the same growth-control pathway in multiple tissues. This is reminiscent of how alterations that converge on the Ras/Raf/MEK/ERK signaling pathway underlie the Rasopathies, which include a spectrum of diseases including neurofibromatosis type 1, Noonan syndrome, and other developmental disorders that all have distinct clinical phenotypes (17).
Inflammatory molecules such as tumor necrosis factor α (TNF-α) and interferon α (IFN-α) that are produced in the BM microenvironment during viral infections and in response to other stress signals induce SAMD9 and SAMD9L expression (5, 18). SAMD9 and SAMD9L proteins exhibit GOF biochemical properties that reduce Raf/MEK/ERK signaling and inhibit the growth of cultured cell lines (1, 19), and this likely results in selective pressure favoring the outgrowth of hematopoietic stem and progenitors (HSPCs) with loss of the chromosome 7 homolog harboring the mutant allele (i.e., adaptation by aneuploidy). This idea is consistent with the acquisition of a p.Q569P substitution in cis with the p.H880Q germline variant in the father from family 1 and other second-site mutations that abrogate the GOF activities of mutant SAMD9 and SAMD9L proteins, and with the observation of somatic CN-LOH events that result in the nonrandom loss of the mutant SAMD9 or SAMD9L alleles in the BM cells of some affected individuals with normal BM cytogenetics in our cohort and in previous studies (5, 8).
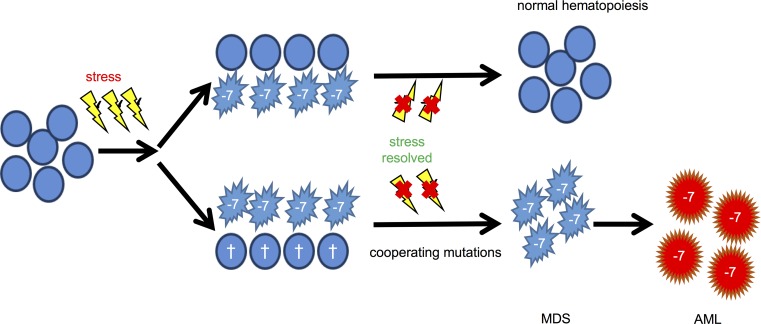
Molecular analysis of BM specimens from the 2 index MLSM7 families supports the idea that the acquisition of somatic oncogenic mutations is a major determinant of progression to MDS/AML. Remarkably, the myeloid malignancies of different patients independently acquired secondary mutations in ETV6, RUNX1, and SETBP1. These genes are also recurrently mutated in other cases of MDS or AML characterized by Mo7, including some that are associated with known inherited or environmental risk factors (20–25). Figure 5 incorporates these new observations into the existing model of clonal evolution and hematologic outcomes in patients with germline SAMD9 and SAMD9L mutations. Specifically, our studies of MLSM7 patients who developed MDS/AML support the existence of an evolutionary bottleneck that strongly predisposes a susceptible HSPC to acquire a limited spectrum of secondary mutations that drive clonal outgrowth and leukemic transformation. We speculate that this bottleneck is a consequence of extensive damage to the HSPC compartment, and is determined by the severity and duration of the inciting inflammatory insult and the specific germline SAMD9 or SAMD9L mutation. Accordingly, patients that retain a large reservoir of non-Mo7 HSPCs after the inciting inflammatory stress resolves are likely to recover normal blood cell counts through a variety of genetic and nongenetic mechanisms (Figure 5, top). This is associated with loss of the Mo7 clone in some cases. In contrast, we propose that severe attrition of non-Mo7 HSPCs facilitates persistence of the Mo7 clone, which is then strongly predisposed to acquire cooperating mutations that drive progression to MDS and AML (Figure 5, bottom). Alternatively, non-Mo7 HSPCs may acquire specific oncogenic mutations that are able to confer a fitness advantage during inflammatory stress, and ultimately give rise to MDS and AML with subsequent loss of chromosome 7.
Figure 5. Proposed roles of SAMD9/9L haploinsufficiency in BM Mo7 and subsequent hematologic outcomes.
In accordance with the model proposed by Tesi and colleagues (5), inflammatory stress in the BM microenvironment (yellow thunderbolt) provides selective pressure favoring the outgrowth of Mo7 clones (–7) in carriers of SAMD9/9L GOF mutations. Hematopoietic recovery occurs in many patients due to outgrowth of a residual population of HSPCs without Mo7, which also frequently exhibit revertant SAMD9/9L mutations or somatic UPD (top pathway). This pattern was seen in the members of families 3–5. Our analysis of families 1 and 2, in which both siblings progressed to MDS/AML, supports the existence of an alternative pathway that involves irreversible stress-induced damage to the non-Mo7 HSPC compartment (indicated as crosses). This creates a genetic bottleneck and results in persistence of the Mo7 clone that has a high probability of acquiring a limited spectrum of cooperating mutations that drive progression to leukemia.
Samd9l-deficient mice (mice lack a Samd9 gene) spontaneously develop MDS with incomplete penetrance, and the MOL4070LTR retrovirus cooperates with Samd9l haploinsufficiency to drive progression to AML (19). In addition, Samd9l-deficient HSPCs exhibited a growth advantage over wild-type HSPCs in competitive repopulation experiments, which was suggested to be due, at least in part, to hypersensitivity to growth factors. In light of these data, it is unclear why durable hematologic recovery with loss of the Mo7 clone occurs spontaneously in some children with germline SAMD9 and SAMD9L mutations. For example, Mo7 clones disappeared without treatment in multiple members of family 5, and all 9 carriers are now well without hematologic disease (Supplemental Figure 8). It is possible that HSPCs that have deleted one chromosome 7 homolog (versus isolated loss of one allele of Samd9l in the mouse model) are at a competitive growth advantage only during stress signaling within the BM microenvironment. Our serial genetic analysis of patients 5A and 5G is intriguing, as the Mo7 clone regressed in both patients over time and this was associated with the emergence of different clones that either lose a germline SAMD9/SAMD9L mutation or attenuate its effects through an alternative genetic mechanism. While these data support the idea that some mutant SAMD9L alleles are compatible with effective hematopoiesis for many years, they also indicate the presence of ongoing selective pressure favoring the growth of HSPCs with revertant mutations. Although the mutant SAMD9 and SAMD9L alleles characterized to date all had similar functional effects on the cell cycle when expressed exogenously in 293T cells (Supplemental Figure 6 and ref. 9), the clinical variability in MLSM7 families underscores the importance of performing additional functional analyses in a more physiologic context.
Alternatively, spontaneous regression of Mo7 clones may represent developmental involution of a fetal HSPC population, as is thought to occur in the self-limited myeloproliferative disorders observed in infants and young children with Down and Noonan syndromes (26, 27). Along these lines, it is striking that we and others have identified multiple healthy adult carriers of SAMD9 and SAMD9L mutations (5, 8). While these observations provide indirect evidence that young children with germline SAMD9 or SAMD9L mutations are particularly susceptible to myeloid transformation, further longitudinal and prospective studies are needed to fully evaluate the impact of these mutations over time and during aging, particularly as a 42-year-old male with MDS and a germline SAMD9L mutation has been reported (8). Notably, clinical records from family 4 indicated that a paternal aunt died from AML with Mo7. Although her tissues were not available for molecular analysis, these findings suggest that some adults with germline SAMD9/SAMD9L mutations are at elevated risk of developing hematologic cancers. Monitoring family 5 and other kindreds with multiple affected individuals will likely be particularly informative.
In contrast to many children with germline SAMD9 and SAMD9L mutations reported to date, the 16 patients with MLSM7 in this study, as well as 5 children from 2 families previously reported by our group (7, 9), predominantly presented with isolated hematologic disease. These observations have implications for diagnosis, molecular testing, and clinical management. First and foremost, SAMD9 and SAMD9L mutation testing should be part of the standard laboratory assessment of children who present with pancytopenia (6) or MDS during the first decade of life, and for all pediatric patients with MDS, or AML associated with BM Mo7. Clinical sequencing for acquired mutations in genes implicated in MDS and AML, such as ETV6, SETBP1, KRAS, and RUNX1 may identify patients at high risk for leukemic transformation who would likely benefit most from HSCT. In the absence of known cooperating mutations, careful follow-up with serial physical examinations and regular laboratory testing seems warranted. In addition, sequencing of the SAMD9/SAMD9L locus can provide important information regarding somatic revertant mutations or somatic CN-LOH events that may influence clinical management. The choice of treatment will likely also be influenced by any extrahematopoietic phenotypes in children with SAMD9/SAMD9L germline mutations, as recent studies showed a poor outcome after HSCT for patients with a severe MIRAGE syndrome phenotype (28). By contrast, 2 of the MLSM7 patients with MDS reported here (2B and 3A) underwent successful HSCT with 100% donor chimerism and long-term survival (Supplemental Figure 9, B and C). A third patient with AML (1B) died of sepsis after receiving a haploidentical HSCT (Supplemental Figure 9A). Finally, the families of children with germline SAMD9 or SAMD9L mutations should be counseled regarding the implications of this finding and offered appropriate genetic testing.
Methods
SNP arrays.
DNA isolated from patients was analyzed using Affymetrix SNP 6.0 arrays. SNP array data were analyzed using dChip software (29) with a reference normalization algorithm (30) and a circular binary segmentation algorithm was used to identify copy number abnormalities as previously described (31–34). B-allele frequency information from SNP markers was used to identify CN-LOH and to confirm copy number gains and losses.
Haloplex.
A targeted sequencing panel using the Haloplex platform (Agilent) was designed to interrogate 56 genes recurrently mutated in myeloid disorders (Supplemental Table 6). Following sequencing on an Illumina MiSeq, samples were aligned and SNVs were called using SureCall software (Agilent) (Supplemental Table 7). The software merges duplicate sequencing reads into single reads using a molecular bar code system and the number of merged reads is recorded in the resulting bam file. Using these numbers, absolute read count coverage for every SNV was calculated. In some instances, reads collapsed from a single barcode (reads that were not duplicated) were excluded from the analysis.
Amplicon sequencing.
Both SAMD9 and SAMD9L were sequenced in at least one sample from each individual (parents and siblings) by amplifying approximately 1.5-kb regions (3 per gene) with more than 50 overlapping bases. The amplicons were submitted to the Hartwell Center for Nextera XT library preparation, per the manufacturer’s protocol, and sequenced on a MiSeq with a 500cycle nano kit, as previously described (9) (Supplemental Table 8). When necessary, short amplicons were used to validate the known SAMD9/SAMD9L germline variants in each family (Supplemental Table 9). Oligonucleotides were designed to amplify an approximately 350-bp fragment surrounding each variant and these oligonucleotides included the following Illumina adapters: TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG-[forward primer] and GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG-[reverse primer]. The amplicons were then purified with Ampure XP beads, PCR-amplified (5 cycles) to attach indices and adapters, followed by an additional purification with Ampure XP beads and quality assessment on a LabChip GX. The samples were run on a MiSeq with a 500cycle nano kit. VarScan 2 was used to call variants (35). Somatic revertants at a VAF of greater than 2% were reported, unless multiple samples were available for that patient. In those cases, a VAF greater than 1% was reported if the variant was observed in more than one sample. Only variants either not present in the ExAC database or at an allele frequency of ≤0.01% were reported.
Functional assessment of SAMD9/9L mutations.
The pCMV6-Entry[SAMD9] (RC219076) and pCMV6-Entry[SAMD9L] (RC207886) vectors were purchased from Origene. SAMD9 and SAMD9L were PCR amplified such that the resultant amplicons contained a 5′-CACC overhang to allow for direct subcloning into the Gateway entry vector, pENTR/D-TOPO. Patient-specific mutations were introduced into the wild-type SAMD9/SAMD9L–containing pENTR/D-TOPO vectors using the GENEART Site-Directed Mutagenesis System (Invitrogen) kit and the mutagenesis primers found in Supplemental Table 10. SAMD9/SAMD9L–GFP fusions were created by using the Gateway LR clonase II reaction with the pcDNA6.2/N-EmGFP-DEST Gateway destination vector, which was purchased from Lifetech (V35620). The pcDNA6.2/N-EmGFP/GW/CAT vector was used as the GFP-empty vector control. HEK-293T cells (ATCC) were treated with 10 μM EdU for 2 hours starting 24–48 hours after transfection with mutant or wild-type SAMD9/SAMD9L. Following the EdU incubation, cells were harvested and fixed with 4% paraformaldehyde. After fixing, the Click-It reaction (Invitrogen, C10635) was performed according to the manufacturer’s protocol. Following the Click-It reaction, total DNA was labeled with FxCycle (Invitrogen, F10347), and analyzed by flow cytometry.
Colony PCR assays.
Genomic patient DNA was amplified and the PCR products were subcloned into PCR-Blunt II-TOPO (Invitrogen, 45-0245) and sequenced to confirm mutational status of the SAMD9L/SAMD9 locus.
Statistics.
Statistical significance between S phases was established by performing 1-way ANOVAs with repeated measures followed by Tukey’s post hoc multiple-comparisons test; significance was based on α = 0.05. A P value of less than 0.05 was considered significant. Quantitative data are presented as dot plots showing the individual data points, the mean, and error bars shown as the standard deviation.
Data availability and accession codes.
Genomic data have been deposited in the European Genome-phenome Archive (EGA), which is hosted by the European Bioinformatics Institute (EBI), under accession EGAS00001002202. All other remaining data are available within this article and supplementary files, or available from the authors upon request.
Study approval.
BM and peripheral blood samples from parents, patients, and siblings were banked for research after informed consent was obtained (Supplemental Table 11). All studies were approved by the Institutional Review Board at UCSF and at St. Jude Children’s Research Hospital.
Author contributions
JCW designed the study, performed research studies, analyzed data, and wrote the manuscript. VB assisted with experimental design, performed research studies, analyzed data, and wrote the manuscript. TL, JS, and MDPA performed research studies. JM and MW performed bioinformatic analyses. CGM, MLL, and JRD reviewed the data and edited the manuscript. RR, WLC, JD, SG, PCR, SI, and RY identified patients and contributed clinical data. KS and JMK designed the study, analyzed data, and wrote the manuscript.
Supplementary Material
Acknowledgments
We thank all the patients and their parents for contributing material for this study, as well as the numerous clinicians and supporting staff at the collaborating hospitals that have helped to identify these patients. We thank the Hartwell Center for Bioinformatics and Biotechnology of St. Jude Children’s Research Hospital for assistance with sequencing. Jinghui Zhang and the Department of Computational Biology graciously provided previously established bioinformatic pipelines and support. This work was supported by the American Lebanese and Syrian Associated Charities of St. Jude Children’s Research Hospital, by awards to K. Shannon from the St. Baldrick’s Foundation and the Evans MDS Initiative, and by grants from the US National Institutes of Health: P30 CA021765, Cancer Center Support Grant to J.M. Klco; K08 HL116605 to J.M. Klco; R50 CA211452 to J.C. Wong; R35 CA197695-01A1 to C.G. Mullighan; R01 CA216352 to K. Shannon; and P30 CA082103 Cancer Center Support Grant to the Helen Diller Family Comprehensive Cancer Center at UCSF. J.M. Klco holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund and a Scholar Award from the V Foundation. K. Shannon is an American Cancer Society Research Professor.
Version 1. 07/25/2018
Electronic publication
Funding Statement
JMK
JMK
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2018;3(14):e121086. https://doi.org/10.1172/jci.insight.121086.
Contributor Information
Jasmine C. Wong, Email: WongJa@peds.ucsf.edu.
Victoria Bryant, Email: victoria.bryant@stjude.org.
Tamara Lamprecht, Email: Tamara.Lamprecht@StJude.org.
Jing Ma, Email: jing.ma@stjude.org.
Maria del pilar Alzamora, Email: Pilar.Alzamora@ucsf.edu.
Charles G. Mullighan, Email: charles.mullighan@stjude.org.
Mignon L. Loh, Email: mignon.loh@ucsf.edu.
James R. Downing, Email: James.Downing@STJUDE.ORG.
William L. Carroll, Email: william.carroll@nyumc.org.
Stuart Gold, Email: stuart_gold@med.unc.edu.
Paul C. Rogers, Email: progers@cw.bc.ca.
Rochelle Yanofsky, Email: ryanofsk@gmail.com.
Kevin Shannon, Email: shannonk@peds.ucsf.edu.
References
- 1.Narumi S, et al. SAMD9 mutations cause a novel multisystem disorder, MIRAGE syndrome, and are associated with loss of chromosome 7. Nat Genet. 2016;48(7):792–797. doi: 10.1038/ng.3569. [DOI] [PubMed] [Google Scholar]
- 2.Duncan AW, et al. Aneuploidy as a mechanism for stress-induced liver adaptation. J Clin Invest. 2012;122(9):3307–3315. doi: 10.1172/JCI64026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buonocore F, et al. Somatic mutations and progressive monosomy modify SAMD9-related phenotypes in humans. J Clin Invest. 2017;127(5):1700–1713. doi: 10.1172/JCI91913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen DH, et al. Ataxia-pancytopenia syndrome is caused by missense mutations in SAMD9L. Am J Hum Genet. 2016;98(6):1146–1158. doi: 10.1016/j.ajhg.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tesi B, et al. Gain-of-function SAMD9L mutations cause a syndrome of cytopenia, immunodeficiency, MDS, and neurological symptoms. Blood. 2017;129(16):2266–2279. doi: 10.1182/blood-2016-10-743302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bluteau O, et al. A landscape of germ line mutations in a cohort of inherited bone marrow failure patients. Blood. 2018;131(7):717–732. doi: 10.1182/blood-2017-09-806489. [DOI] [PubMed] [Google Scholar]
- 7.Schwartz JR, et al. Germline SAMD9 mutation in siblings with monosomy 7 and myelodysplastic syndrome. Leukemia. 2017;31(8):1827–1830. doi: 10.1038/leu.2017.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pastor VB, et al. Constitutional SAMD9L mutations cause familial myelodysplastic syndrome and transient monosomy 7. Haematologica. 2018;103(3):427–437. doi: 10.3324/haematol.2017.180778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwartz JR, et al. The genomic landscape of pediatric myelodysplastic syndromes. Nat Commun. 2017;8(1):1557. doi: 10.1038/s41467-017-01590-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carroll WL, Morgan R, Glader BE. Childhood bone marrow monosomy 7 syndrome: a familial disorder? J Pediatr. 1985;107(4):578–580. doi: 10.1016/S0022-3476(85)80027-5. [DOI] [PubMed] [Google Scholar]
- 11.Shannon KM, et al. Familial bone marrow monosomy 7. Evidence that the predisposing locus is not on the long arm of chromosome 7. J Clin Invest. 1989;84(3):984–989. doi: 10.1172/JCI114262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mekhedov SL, Makarova KS, Koonin EV. The complex domain architecture of SAMD9 family proteins, predicted STAND-like NTPases, suggests new links to inflammation and apoptosis. Biol Direct. 2017;12(1):13. doi: 10.1186/s13062-017-0185-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Papaemmanuil E, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374(23):2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thol F, et al. Acute myeloid leukemia derived from lympho-myeloid clonal hematopoiesis. Leukemia. 2017;31(6):1286–1295. doi: 10.1038/leu.2016.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunlap J, et al. Multiplex high-throughput gene mutation analysis in acute myeloid leukemia. Hum Pathol. 2012;43(12):2167–2176. doi: 10.1016/j.humpath.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 16.Metzeler KH, et al. Spectrum and prognostic relevance of driver gene mutations in acute myeloid leukemia. Blood. 2016;128(5):686–698. doi: 10.1182/blood-2016-01-693879. [DOI] [PubMed] [Google Scholar]
- 17.Simanshu DK, Nissley DV, McCormick F. RAS proteins and their regulators in human disease. Cell. 2017;170(1):17–33. doi: 10.1016/j.cell.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hershkovitz D, et al. Functional characterization of SAMD9, a protein deficient in normophosphatemic familial tumoral calcinosis. J Invest Dermatol. 2011;131(3):662–669. doi: 10.1038/jid.2010.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagamachi A, et al. Haploinsufficiency of SAMD9L, an endosome fusion facilitator, causes myeloid malignancies in mice mimicking human diseases with monosomy 7. Cancer Cell. 2013;24(3):305–317. doi: 10.1016/j.ccr.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 20.Wall M, Rayeroux KC, MacKinnon RN, Zordan A, Campbell LJ. ETV6 deletion is a common additional abnormality in patients with myelodysplastic syndromes or acute myeloid leukemia and monosomy 7. Haematologica. 2012;97(12):1933–1936. doi: 10.3324/haematol.2012.069716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Makishima H, et al. Somatic SETBP1 mutations in myeloid malignancies. Nat Genet. 2013;45(8):942–946. doi: 10.1038/ng.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hasle H, et al. Monosomy 7 and deletion 7q in children and adolescents with acute myeloid leukemia: an international retrospective study. Blood. 2007;109(11):4641–4647. doi: 10.1182/blood-2006-10-051342. [DOI] [PubMed] [Google Scholar]
- 23.Feurstein S, Godley LA. Germline ETV6 mutations and predisposition to hematological malignancies. Int J Hematol. 2017;106(2):189–195. doi: 10.1007/s12185-017-2259-4. [DOI] [PubMed] [Google Scholar]
- 24.Osato M. Point mutations in the RUNX1/AML1 gene: another actor in RUNX leukemia. Oncogene. 2004;23(24):4284–4296. doi: 10.1038/sj.onc.1207779. [DOI] [PubMed] [Google Scholar]
- 25.Coccaro N, Tota G, Zagaria A, Anelli L, Specchia G, Albano F. SETBP1 dysregulation in congenital disorders and myeloid neoplasms. Oncotarget. 2017;8(31):51920–51935. doi: 10.18632/oncotarget.17231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roberts I, Izraeli S. Haematopoietic development and leukaemia in Down syndrome. Br J Haematol. 2014;167(5):587–599. doi: 10.1111/bjh.13096. [DOI] [PubMed] [Google Scholar]
- 27.Romano AA, et al. Noonan syndrome: clinical features, diagnosis, and management guidelines. Pediatrics. 2010;126(4):746–759. doi: 10.1542/peds.2009-3207. [DOI] [PubMed] [Google Scholar]
- 28.Sarthy J, et al. Poor outcome with hematopoietic stem cell transplantation for bone marrow failure and MDS with severe MIRAGE syndrome phenotype. Blood Adv. 2018;2(2):120–125. doi: 10.1182/bloodadvances.2017012682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin M, Wei LJ, Sellers WR, Lieberfarb M, Wong WH, Li C. dChipSNP: significance curve and clustering of SNP-array-based loss-of-heterozygosity data. Bioinformatics. 2004;20(8):1233–1240. doi: 10.1093/bioinformatics/bth069. [DOI] [PubMed] [Google Scholar]
- 30.Pounds S, Cheng C, Mullighan C, Raimondi SC, Shurtleff S, Downing JR. Reference alignment of SNP microarray signals for copy number analysis of tumors. Bioinformatics. 2009;25(3):315–321. doi: 10.1093/bioinformatics/btn624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mullighan CG, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446(7137):758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 32.Mullighan CG, et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science. 2008;322(5906):1377–1380. doi: 10.1126/science.1164266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mullighan CG, et al. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 2008;453(7191):110–114. doi: 10.1038/nature06866. [DOI] [PubMed] [Google Scholar]
- 34.Mullighan CG, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009;360(5):470–480. doi: 10.1056/NEJMoa0808253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koboldt DC, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22(3):568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genomic data have been deposited in the European Genome-phenome Archive (EGA), which is hosted by the European Bioinformatics Institute (EBI), under accession EGAS00001002202. All other remaining data are available within this article and supplementary files, or available from the authors upon request.