Abstract
Adenosine-to-inosine (A-to-I) RNA editing, a process mediated by adenosine deaminases that act on the RNA (ADAR) gene family, is a recently discovered epigenetic modification dysregulated in human cancers. However, the clinical significance and the functional role of RNA editing in colorectal cancer (CRC) remain unclear. We have systematically and comprehensively investigated the significance of the expression status of ADAR1 and of the RNA editing levels of antizyme inhibitor 1 (AZIN1), one of the most frequently edited genes in cancers, in 392 colorectal tissues from multiple independent CRC patient cohorts. Both ADAR1 expression and AZIN1 RNA editing levels were significantly elevated in CRC tissues when compared with corresponding normal mucosa. High levels of AZIN1 RNA editing emerged as a prognostic factor for overall survival and disease-free survival and were an independent risk factor for lymph node and distant metastasis. Furthermore, elevated AZIN1 editing identified high-risk stage II CRC patients. Mechanistically, edited AZIN1 enhances stemness and appears to drive the metastatic processes. We have demonstrated that edited AZIN1 functions as an oncogene and a potential therapeutic target in CRC. Moreover, AZIN1 RNA editing status could be used as a clinically relevant prognostic indicator in CRC patients.
Keywords: Gastroenterology, Oncology
Keywords: Colorectal cancer, Epigenetics, RNA processing
Antizyme inhibitor 1 (AZIN1) functions as an oncogene, and drives stemness and the metastatic process in human colorectal cancer.
Introduction
Colorectal cancer (CRC) is one of the most frequently occurring malignancies worldwide, and it remains the second leading cause of cancer-related deaths in Western countries (1). CRC pathogenesis is strongly associated with lifestyle choices, such as diet, obesity, and smoking (2). Emerging evidence indicates that these factors have a profound effect on epigenetic alterations, including histone modification (3), DNA methylation (4), and regulation of noncoding RNAs (5). These epigenetic modifications can directly influence key cellular mechanisms, such as gene expression and enhancer activity, to maintain cellular homeostasis and prevent various diseases, including cancer (6).
RNA editing is a recently identified epigenetic alteration that is involved in the posttranscriptional regulation of key genes associated with human cancers (7). Specifically, the conversion of adenosine to inosine (A-to-I), where splicing and translational machineries recognize the inosine residues as guanosines, is emerging as the most frequent type of RNA editing process in humans. Such A-to-I RNA editing is primarily catalyzed by enzymes encoded by the family of adenosine deaminases that act on the RNA (ADAR) genes (ADAR1, ADAR2, and ADAR3) (8). Intriguingly, several recent studies have shown that A-to-I RNA editing by ADAR1 was prominent in several cancers, including hepatocellular carcinoma (HCC), esophageal cancer, and gastric cancer (9–11). Antizyme inhibitor 1 (AZIN1) was identified as one of the most frequently occurring A-to-I RNA alterations in HCC (9). Mechanistically, edited AZIN1 conferred the gain-of-function phenotype associated with aggressive tumors and promoted ornithine decarboxylase (ODC) and polyamines accumulation (9, 10). In spite of the growing evidence for A-to-I RNA editing in tumorigenesis, the clinical significance and the functional role of AZIN1 RNA editing in CRC remains unexplored.
Herein, we demonstrate for the first time to our knowledge that AZIN1 RNA editing levels and ADAR1 expression are significantly increased in CRC and edited AZIN1 is an independent prognostic factor for overall survival (OS) and disease-free survival (DFS) in patients with this malignancy. Furthermore, edited AZIN1 appears to function as an oncogene; enhances cellular proliferation, invasion, and migration capabilities; and promotes cancer stem-like cell features. Collectively, we illustrate that RNA editing is dysregulated in CRC and edited AZIN1 acts as an oncogene in CRC.
Results
The RNA editing gene, ADAR1, and AZIN1 RNA editing levels are frequently dysregulated in CRC.
With the hypothesis to determine whether RNA editing levels are dysregulated in CRC, we first assessed the expression levels of ADAR1 and ADAR2 in training (n = 24) and validation cohorts (n = 50) of patients by analyzing their CRC and matched adjacent normal mucosa tissues (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.99976DS1). Although 3 ADARs (ADAR1, ADAR2, and ADAR3) have been identified as members of the ADAR family in vertebrates, since ADAR3 is primarily expressed in brain cells (12), its expression was excluded in our CRC cohort. We found that ADAR1 expression was significantly upregulated in CRC tissues in both cohorts (P = 0.0002 in the training cohort, P < 0.0001 in the validation cohort; Figure 1A), while the expression of ADAR2 was significantly downregulated in CRC tissues when compared with normal mucosa (P < 0.0001 in both cohorts; Supplemental Figure 2). To further validate the overexpression of ADAR1 in CRC, we used immunohistochemical analysis and observed a strong staining for ADAR1 in CRC cells, which was primarily confined to the cytoplasm, while very limited expression was detected in adjacent normal mucosa, which is consistent with previous reports in other human cancers (10, 13) (Supplemental Figure 3, A and B).
Figure 1. RNA editing gene ADAR1 is deregulated along with AZIN1 RNA editing levels in CRC.
(A) ADAR1 expression levels in CRC tissues compared with that in normal mucosa in the training cohort and validation cohort (Wilcoxon’s signed-rank test). (B) AZIN1 RNA editing levels in CRC tissues compared to that of normal mucosa in the training cohort and validation cohort (Wilcoxon’s signed-rank test). (C) Correlation between AZIN1 RNA editing and ADAR1 expression levels in the validation cohort and the clinical evaluation cohort (Spearman’s rank correlation analysis). ***P < 0.001.
Considering that we have previously identified AZIN1 as one of the most frequently edited genes in esophageal cancer and HCC (9, 10), we asked whether the dysregulation of ADAR1 corresponds to the alteration of edited AZIN1 levels in the CRC clinical cohorts. To quantify AZIN1 RNA editing levels, we employed RNA editing site-specific quantitative PCR (RESSq-PCR), as described previously (14). Consistent with the outcomes of ADAR1 dysregulation, AZIN1 RNA editing levels were significantly increased in neoplastic tissues when compared with normal mucosa (P < 0.0001 in both cohorts; Figure 1B).
Since AZIN1 RNA is edited by ADAR1 and not by ADAR2 (9), we assessed whether AZIN1 RNA editing levels were associated with ADAR1 expression in CRC cohorts. As expected, edited AZIN1 positively correlated with ADAR1 expression in both clinical cohorts (P < 0.0001, ρ = 0.5 in both cohorts; Figure 1C), suggesting that the dysregulation of ADAR1 might contribute to increased levels of AZIN1 RNA editing in CRC, highlighting that the dysregulation of RNA editing is a prominent feature in CRC patients.
Edited AZIN1 and aberrant expression of ADAR1 associate with disease progression, recurrence, and prognosis in CRC patients.
Next, we evaluated the clinical significance of edited AZIN1 and the expression level of ADAR1 in a large clinical evaluation cohort (Supplemental Table 1). Surprisingly, although the expression status of ADAR1 was not significantly different across tumor stages, AZIN1 RNA editing levels were higher in stage IV CRCs compared with stage I CRCs (P < 0.05; Figure 2A). Additionally, a time-to-event analysis revealed that highly edited AZIN1 and a high expression of ADAR1 resulted in poor OS (ADAR1, P = 0.004; AZIN1 RNA editing, P = 0.0003; Figure 2B). Furthermore, elevated editing levels of AZIN1 also correlated with poor DFS (ADAR1, P = 0.076; AZIN1 RNA editing, P = 0.027; Figure 2C). The prognostic significant of the results was analyzed by normalizing AZIN1 expression results with both β-actin as well as GAPDH, and the results were quite similar.
Figure 2. High AZIN1 RNA editing levels and high expression of ADAR1 correlate with poor prognosis of OS and DFS in the clinical evaluation cohort.
(A) TNM stage-dependent expression status of ADAR1 and AZIN1 RNA editing levels in CRC tissues (Kruskal-Wallis test). (B) Kaplan-Meier survival curves for OS in CRC patients in the clinical evaluation cohort (n = 220) sorted into low and high expression levels of ADAR1 and low and high AZIN1 RNA editing status (log-rank test). (C) Kaplan-Meier survival curves for DFS in CRC patients in the clinical evaluation cohort (n = 220) were sorted into low and high expression levels of ADAR1 and low and high AZIN1 RNA editing status (log-rank test).
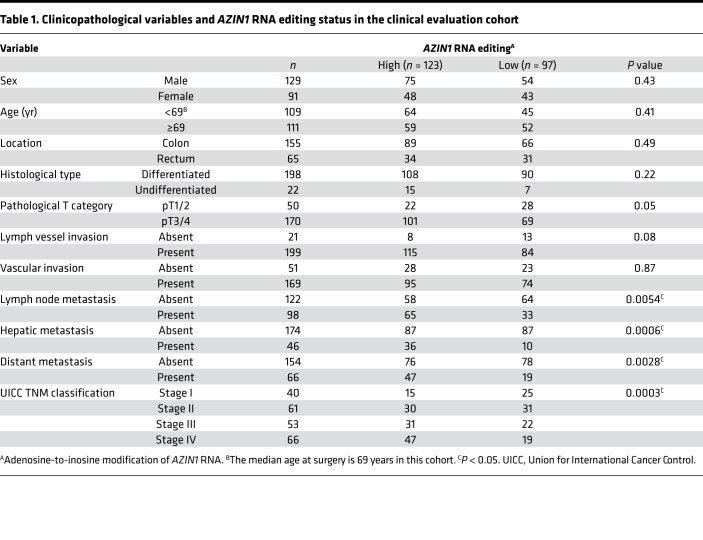
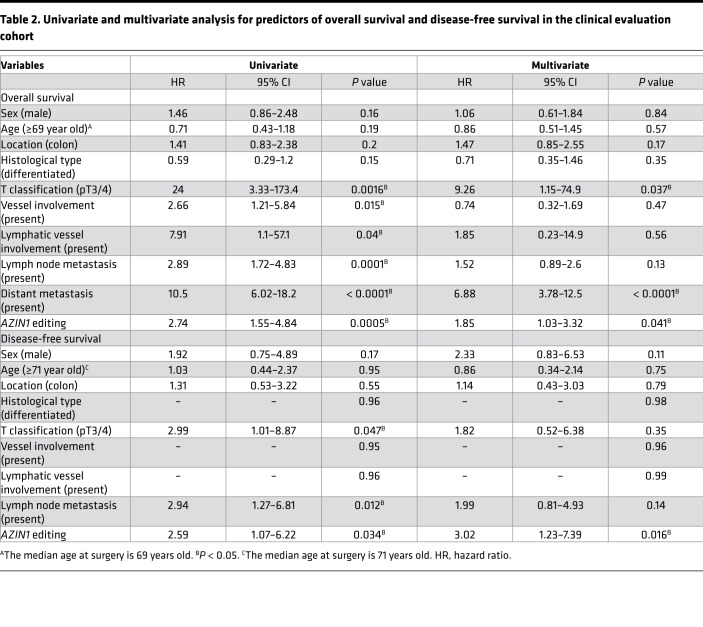
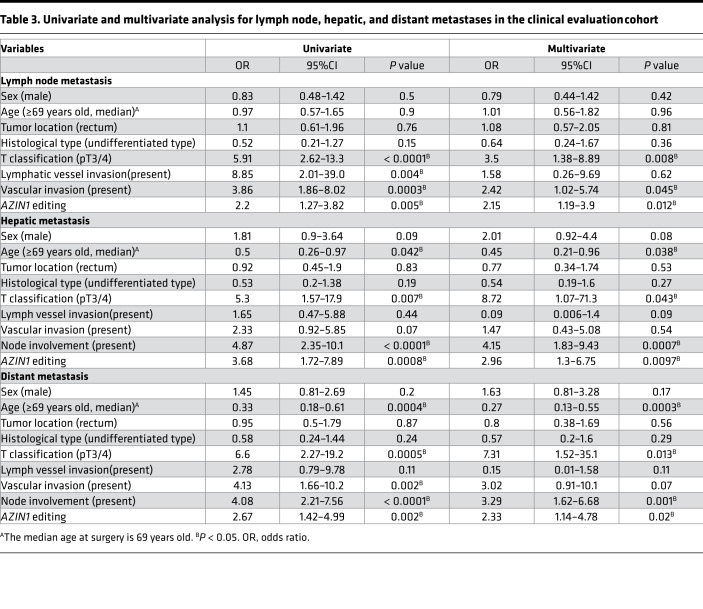
In order to determine the clinical significance of edited AZIN1 and ADAR1 expression levels, we assessed whether the expression of these genes associated with various clinicopathological factors (Table 1 and Supplemental Table 3). Quantitative profiling analyses revealed that increased AZIN1 RNA editing correlated significantly with the presence of lymph node metastasis (P = 0.0054), hepatic metastasis (P = 0.0006), distant metastasis (P = 0.0028), and progression of tumor node metastasis (TNM) staging (P = 0.0003) in CRC patients (Table 1). Likewise, the overexpression of ADAR1 was associated with male sex (P = 0.016), hepatic metastasis (P = 0.0095), distant metastasis (P = 0.016), and progression of TNM staging (P = 0.025; Supplemental Table 3). To further evaluate the prognostic biomarker potential of AZIN1 RNA editing, we performed a multivariate Cox regression analysis. In addition to the presence of distant metastasis, high levels of edited AZIN1 emerged as an independent prognostic factor for OS (hazard ratio [HR], 1.85; 95% CI, 1.03–3.32, P = 0.041; Table 2) and DFS in CRC patients (HR, 3.02; 95% CI, 1.23–7.39, P = 0.016; Table 2). Moreover, a multivariate analysis revealed that high levels of AZIN1 editing were an independent predictive factor for lymph node metastasis (odds ratio [OR] 2.15, 95% CI 1.19–3.9, P = 0.012), hepatic metastasis (OR 2.96, 95% CI 1.3–6.75, P = 0.0097), and distant metastasis (OR 2.33, 95% CI 1.14–4.78, P = 0.02; Table 3), suggesting that the dysregulation of edited AZIN1 may also be involved in metastatic disease progression.
Table 1. Clinicopathological variables and AZIN1 RNA editing status in the clinical evaluation cohort.
Table 2. Univariate and multivariate analysis for predictors of overall survival and disease-free survival in the clinical evaluation cohort.
Table 3. Univariate and multivariate analysis for lymph node, hepatic, and distant metastases in the clinical evaluation cohort.
Next, we examined whether AZIN1 RNA editing levels and the expression status of ADAR1 could be used as predictive biomarkers of recurrence and prognosis in stage II CRC patients. High levels of ADAR expression and edited AZIN1 were both associated with poor DFS in CRC patients in the clinical evaluation cohort (P = 0.007 and P = 0.03; Supplemental Figure 4). Furthermore, elevated edited AZIN1 in CRC tumors was significantly associated with poor OS in stage II disease (P = 0.016), suggesting that edited AZIN1 RNA status could be used to identify high-risk stage II CRC patients.
AZIN1 RNA editing levels are increased in colorectal adenomas.
Bearing in mind that edited AZIN1 was elevated in CRCs, we then assessed the levels of edited AZIN1 in colorectal adenomas compared to those of matched normal mucosa to determine whether this epigenetic modification has a role in the multistep cascade of progression from adenoma to carcinoma. To our surprise, edited AZIN1 levels were also elevated in the colorectal adenomas compared with adjacent normal mucosa (P = 0.016), suggesting that AZIN1 RNA editing is an early event and that it may play a key role in cancer initiation (Supplemental Figure 5).
AZIN1 RNA editing promotes cellular proliferation, invasion, and migration in CRC.
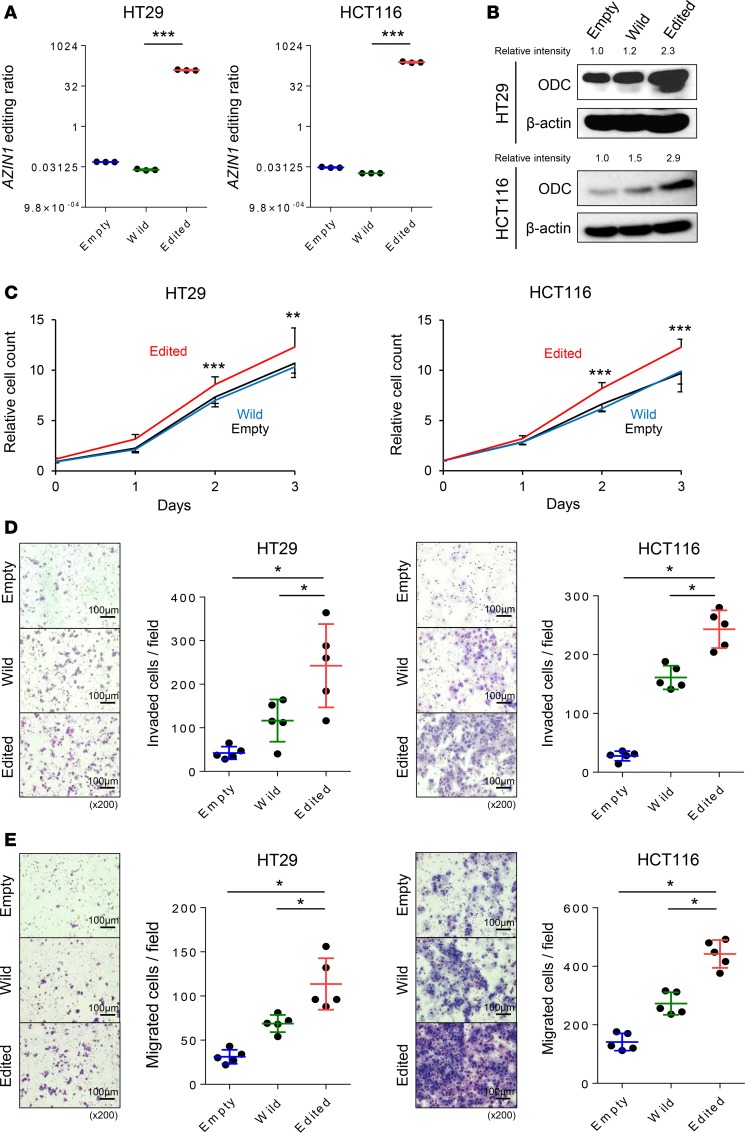
To gain insights into the biological relevance of edited AZIN1 in CRC, we overexpressed either wild-type or edited AZIN1 in HT29 (microsatellite stable) and HCT116 (microsatellite unstable) cell lines (P < 0.001 in both cell lines; Figure 3A). Consistent with a previous study (9), immunofluorescence staining revealed localization of wild-type AZIN1 within the cytoplasm, while edited AZIN1 was present in both the nucleus and the cytoplasm of the transfected CRC cells (Supplemental Figure 6). We have previously demonstrated that the oncogene ODC is a downstream target of edited AZIN1 (9). The overexpression of edited AZIN1 resulted in upregulation of ODC protein in both cell lines, confirming that edited AZIN1 stabilizes ODC more effectively than wild-type AZIN1 (Figure 3B). Next, we assessed whether edited AZIN1 enhances cellular proliferation in CRC using an MTT assay. Overexpression of edited AZIN1 significantly increased cellular proliferation in both CRC cell lines when compared with wild-type AZIN1 (P < 0.01 in HT29, P < 0.001 in HCT116; Figure 3C).
Figure 3. AZIN1 RNA editing promotes cellular proliferation, invasion, and migration.
(A) Edited to wild-type AZIN1 RNA ratios in HT29 and HCT116 cell lines transfected with empty, wild-type, or edited AZIN1-containing plasmids, as determined by RESSq-PCR (n = 3) (Steel test). (B) ODC protein expression levels in the transfected HT29 and HCT116 cell lines analyzed by Western blot. (C) Effect on cellular proliferation of HT29 and HCT116 cells after transfection resulting in overexpression of wild-type or edited AZIN1 (n = 16) (Wilcoxon’s rank-sum test). (D) Effect of overexpression of wild-type or edited AZIN1 on invasiveness of HT29 and HCT116 cells (n = 5) (Steel test). (E) Effect of overexpression of wild-type or edited AZIN1 on migration of HT29 and HCT116 cells (n = 5) (Steel test). *P < 0.05, **P < 0.01, ***P < 0.001. Scale bar: 100 μm; original magnification, ×200.
We next performed invasion and migration assays to determine whether edited AZIN1 promotes the invasive and migratory potential of CRC cells. Overexpression of edited AZIN1 enhanced invasiveness relative to both empty vector (P < 0.05 in both cell lines) and wild-type AZIN1 (P < 0.05 in both cell lines; Figure 3D). Similarly, the overexpression of edited AZIN1 increased migration levels compared with both the controls (P < 0.05 in both cell lines) and wild-type AZIN1-overexpressed cells (P < 0.05 in both cell lines; Figure 3E). Collectively, these results suggest that AZIN1 RNA editing plays a critical role in CRC pathogenesis.
AZIN1 RNA editing promotes stemness in CRC cells.
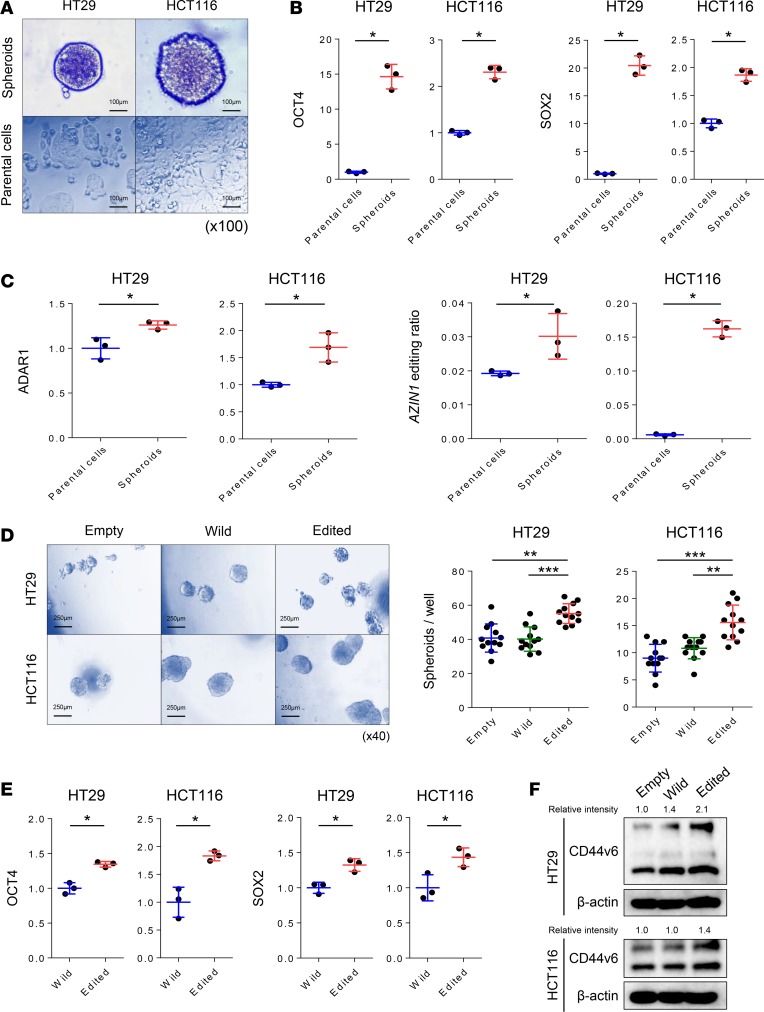
Cancer stem cells (CSCs) have been hypothesized as one of the underlying causes of metastasis and disease recurrence (15). Therefore, we assessed whether AZIN1 editing influences CRC stemness. First, we used spheroids to enrich CSCs in HT29 and HCT116 cell lines (Figure 4A). To validate the enrichment of CSCs in spheroids, we first confirmed overexpression of OCT4 and SOX2, stemness-associated genes, in spheroids in comparison to parental cells (P < 0.05; Figure 4B). We thereafter analyzed the expression of ADAR1 between spheroids and parental cells to determine whether ADAR1 activity is higher in CSCs. Interestingly, we found that ADAR1 expression was overexpressed in spheroids when compared with parental cells in both cell lines (P < 0.05 in both cell lines; Figure 4C). Consistently, edited AZIN1 was also overexpressed in the spheroids (P < 0.05 in both cell lines; Figure 4C). These results suggest that the overexpression of ADAR1 in CSCs may have resulted in the overexpression of edited AZIN1. In order to determine whether edited AZIN1 drives stemness or whether it is a passenger, we assessed the spheroid-forming capacity of cells overexpressing edited AZIN1. CRC cells overexpressing edited AZIN1 possessed significantly greater spheroid-forming capacity when compared with parental or wild-type AZIN1-overexpressing cells (HT29, P < 0.01 and HCT116, P < 0.001 vs. empty vector and HT29, P < 0.001, and HCT116, P < 0.01 vs. wild-type AZIN1, Figure 4D). Moreover, OCT4 and SOX2 levels were significantly higher in spheroids overexpressing edited AZIN1 when compared with the overexpressed wild-type AZIN1 in HT29 (P < 0.05) and HCT116 cells (P < 0.05; Figure 4E). Recently, a variant isoform of CD44, CD44v6, has been identified as a key CRC CSC marker that is typically overexpressed in cancer tissues (16, 17). Elevated levels of CD44v6 expression are associated with poor patient prognosis in various cancers, including CRC (15, 18–22). Substantial overexpression of CD44v6 was observed in CRC cells overexpressing edited AZIN1 in comparison with wild-type AZIN1-overexpressing cells (Figure 4F), providing more indication that AZIN1 RNA editing is involved in driving cancer stemness in CRC.
Figure 4. AZIN1 RNA editing promotes stemness in CRC cells.
(A) Spheroids established from HT29 and HCT116 cells (scale bar: 100 μm; original magnification, ×100). (B) OCT4 and SOX2 RNA expression levels in parental cells and spheroids derived from HT29 or HCT116 cell lines (n = 3) (Wilcoxon’s rank-sum test). (C) AZIN1 RNA editing levels and ADAR1 expression levels in parental cells and spheroids derived from HT29 and HCT116 cell lines (n = 3) (Wilcoxon’s rank-sum test). (D) Comparison of spheroid formation by HT29 and HCT116 cells overexpressing wild-type or edited AZIN1 with cells transfected with empty vector (n = 12) (Steel test) (scale bar: 250 μm; original magnification, ×40). (E) OCT4 and SOX2 RNA expression levels in spheroids derived from either wild-type or edited AZIN1 overexpressing HT29 and HCT116 cells (n = 3) (Wilcoxon’s rank-sum test). (F) CD44v6 protein expression in HT29 and HCT116 cells transfected with empty, wild-type, or edited AZIN1-containing plasmids. *P < 0.05, **P < 0.01, ***P < 0.001.
AZIN1 RNA editing promotes tumor growth in a xenograft animal model.
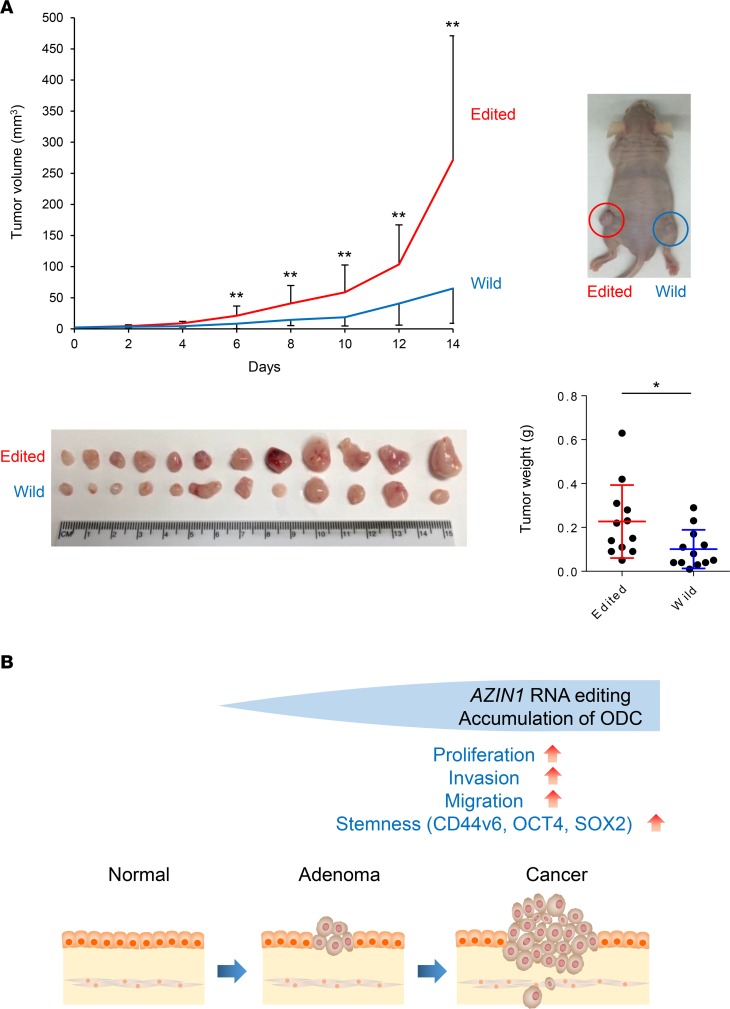
To confirm our in vitro findings, we established xenograft tumors using HCT116 cells that were transfected with vectors containing either wild-type or edited AZIN1. Mice injected with edited AZIN1-transfected cells showed accelerated tumor growth when compared with the wild-type AZIN1-transfected group. After 2 weeks of tumor growth, the mice implanted with edited AZIN1-transfected cells had both larger tumor volume and greater tumor weight than those implanted with wild-type AZIN1-transfected cells (tumor volume, P < 0.01; tumor weight, P = 0.019), further supporting the oncogenic role of edited AZIN1 in CRC (Figure 5A). Collectively, our findings suggest that AZIN1 RNA editing may perform a crucial function in various stages of CRC tumorigenesis (Figure 5B).
Figure 5. AZIN1 RNA editing promotes tumor growth in a xenograft animal model.
(A) Development of xenograft tumors was established by injecting mice with HCT116 cells that were transfected with either wild-type or edited AZIN1-containing plasmids (Wilcoxon’s rank-sum test). (B) AZIN1 RNA editing can promote proliferation, invasion, migration, and stemness in CRC. In combination with our findings from the clinical study, AZIN1 RNA editing may have a crucial role in the shift from the preneoplastic step to the advanced metastatic step in CRC tumorigenesis. *P < 0.05, **P < 0.01.
Discussion
CRC arises through stepwise, sequential accumulation of genetic and epigenetic alterations in colorectal tissues. RNA editing is a recently discovered epigenetic modification that appears to be frequently dysregulated in various cancers. Although RNA editing was initially considered to be a rare event, limited specifically to coding exons within genes, high-throughput sequencing data have now revealed that it occurs more prevalently (23). In particular, A-to-I RNA editing mediated by ADARs is the most prominent form of RNA editing in humans, and various studies have recently demonstrated cancer-specific dysregulation of ADAR as well as subsequent site-specific RNA editing in several human cancers (9–11, 24). More specifically, AZIN1 has been identified as one of the most frequently edited genes in HCC and esophageal cancers (9, 10). However, whether RNA editing holds a significant biological purpose in CRC is unknown. In this study, we demonstrated the dysregulation of ADAR1 in CRC, with corresponding AZIN1 editing alteration using multiple independent CRC cohorts. AZIN1 RNA editing levels showed strong association with various metastasis-associated parameters. Mechanistically, we used a series of functional validation studies in CRC cell lines and xenograft animal models to demonstrate that edited AZIN1 acquires oncogenic properties, including enhancement of stem-like characteristics.
In the present study, we demonstrated for the first time to our knowledge that RNA editing is dysregulated in CRC, a finding which is in consonance with previous reports in other cancers (9–11, 25, 26). We showed the dysregulation of ADAR1, with a corresponding increase in AZIN1 RNA editing levels in cancer tissues when compared with normal mucosa in multiple CRC cohorts. Intriguingly, the enhancement of edited AZIN1 was also observed in premalignant adenomas, suggesting that aberrant expression of ADAR1 with a concomitant increase in edited AZIN1 RNA levels may be an important event in the early initiation steps of colorectal carcinogenesis. Furthermore, we showed that high AZIN1 RNA editing levels, as well as the expression of ADAR1 in CRC tissues, were potential predictors for recurrence and poor prognosis in CRC patients. AZIN1 RNA edited status showed a significant correlation with metastasis-associated clinical factors, including lymph node, hepatic, and distant metastases in CRC patients. Additionally, high AZIN1 RNA editing status associated with disease recurrence and poor survival in stage II CRC patients, an important clinical finding considering that a significant proportion of stage II CRC patients (25%–30%) develop recurrence. Accordingly, AZIN1 RNA editing could be used as a prognostic biomarker for the identification of high-risk stage II CRC patients who could truly benefit from adjuvant chemotherapy (27–29). Collectively, our data indicate that AZIN1 RNA editing could be used as a prognostic biomarker for disease progression, especially for the metastatic process in CRC.
Using a series of experiments, we interrogated the functional role of edited AZIN1 in CRC. We demonstrated that AZIN1 RNA editing promoted the accumulation of ODC and subsequently enhanced the malignant potential of cells through increased proliferation, invasion, and migration. ODC is a key protein that catalyzes the rate-limiting step in polyamine synthesis and controls the rate of the cell cycle (30). AZIN1 is an ODC homolog, which controls ODC accumulation through the inhibition of antizyme, a negative regulator of ODC (31). Hence, AZIN1 prevents proteolytic degradation of ODC by sequestering antizyme from ODC. This stabilization of ODC leads to the accumulation of polyamine and increases cellular proliferation. Interestingly, recent studies show that edited AZIN1 may confer even greater antizyme-binding affinity than standard AZIN1, thereby augmenting the effect on cellular proliferation (32, 33). Our results corroborate some of the previous studies reporting the oncogenic potential of edited AZIN1 in HCC and esophageal cancer (9, 10). Additionally, ODC is known to promote self-renewal of embryonic stem cells via accumulation of polyamine (34). We showed that AZIN1 RNA editing levels are increased in spheroid-derived cancer stem-like cells and edited AZIN1 significantly enhanced spheroid formation, with a corresponding increase in stemness markers. Considering that CSCs are thought be involved in metastatic processes, the enhancement of stemness could be a pivotal link to our clinical findings.
One of the limitations of our study is that we did not determine whether edited AZIN1 may enhance metastatic potential of CRC cells in vivo. We are currently in the process of establishing such an animal model for analyzing the metastatic potential of edited AZIN1-transfected cancer cells. Furthermore, considering that we have identified prognostic potential of edited AZIN1 and demonstrated its involvement in cancer stemness, it would be interesting to determine whether edited AZIN1 expression is associated with drug resistance.
In summary, our study provides evidence we believe to be novel for the oncogenic role of edited AZIN1 in CRC. Our study highlights the clinical and biological significance of AZIN1 RNA editing in CRC, including the enhancement of cancer stem-like features. Therefore, we propose that AZIN1 RNA editing status could be used as a prognostic indicator in CRC patients and as a potential therapeutic target in CRC.
Methods
Patients and sample collection.
In this study, we examined a total of 392 tissue specimens, which included 294 fresh-frozen primary CRCs, 12 adenomas, and 86 normal mucosae collected from 4 independent patient cohorts. These cohorts were enrolled at the National Cancer Center Hospital (training cohort), Okayama University Hospital (validation cohort), Mie University Hospital (clinical evaluation cohort), and Tokushima University Hospital (adenoma cohort), as described in Supplemental Table 1.
The diagnosis of CRC was confirmed for all enrolled patients based on clinicopathological findings. The TNM staging system from the American Joint Committee on Cancer was used for the pathological tumor staging of CRCs. All CRC patients who underwent surgery were followed up for tumor recurrence at regular intervals for up to 5 years. During each annual hospital visit, all patients underwent a chest x-ray, colonoscopy, and abdominal computed tomography. Patients treated with radiotherapy or chemotherapy before surgery were excluded from this study. All patients with stage III/IV disease received 5-fluorouracil–based chemotherapy, whereas no adjuvant chemotherapy was given to stage I and II patients.
RNA extraction and cDNA synthesis.
Fresh-frozen surgical specimens were homogenized with a Mixer Mill MM 300 homogenizer (QIAGEN). The total RNA from tissues and cell lines was isolated using the RNeasy Mini kits (QIAGEN) according to the manufacturer’s instructions. The cDNA was synthesized from 1.0 μg total RNA using the Advantage RT PCR kit (Clontech Laboratories Inc.).
RESSq-PCR.
RNA editing of AZIN1 was analyzed using the RESSq-PCR method published previously (14). In brief, specific primers for the wild-type and edited AZIN1 sequences were designed (Supplemental Figure 1A). Based on the difference in the Ct values, the ratios between the edited and wild-type AZIN1 were calculated using the formula 2–(Ct edited – Ct wild-type). Primer sequences for the PCRs are shown in Supplemental Table 2. To confirm the reliability of this method for the assessment of AZIN1 RNA editing levels, we first examined the levels of predetermined mixtures of oligonucleotides derived from wild-type or edited AZIN1 sequences, ranging from 0% to 100%, and whether PCR can accurately quantify these oligonucleotides. We found that RESSq-PCR could reliably identify the two variants in the mixture, and the observed differences in the Ct values were adequate to generate ratios for edited and wild-type AZIN1 (Supplemental Figure 1, B and C).
Real-time quantitative PCR analyses for ADAR1, ADAR2, OCT4, and SOX2.
Real-time quantitative PCR was performed for gene expression analysis using the StepOne Real Time PCR System and Power SYBR Green Master Mix (Life Technologies), as previously described (35). GAPDH was used as a normalization control. The relative expression of each mRNA was determined using the ΔΔCt method. Primer sequences are shown in Supplemental Table 2.
Cell lines.
The HT29 (microsatellite stable) and HCT116 (microsatellite unstable) CRC cell lines were purchased from ATCC. All cell lines were cultured according to the manufacturer’s specifications. All cell lines were tested and authenticated every few months using a panel of established genetic markers. All experiments were performed using cells that did not exceed 15–20 passages.
Immunohistochemical analysis.
Paraffin-embedded sections were deparaffinized using xylene and ethanol, and endogenous peroxidase activity was eliminated with H2O2. Following antigen retrieval by autoclaving the tissues at 121°C for 15 minutes, slides were incubated with an anti-ADAR1 antibody at a 1:100 dilution (Abcam) overnight. The color development was achieved using the EnVision + Dual Link Kit (DAKO), and slides were counterstained with hematoxylin. Negative controls were run in parallel. The level of ADAR1 staining was evaluated using the Allred proportion score (0, none; 1, 1%; 2, 1%–10%; 3, 10%–33%; 4, 33%–67%; and 5, >67% positive cells), measured 3 times by 2 independent investigators who were blinded to the nature of the specimens and antibodies used.
Immunofluorescence analysis.
Following fixation by methanol, cultured cells were stained with an anti-AZIN1 antibody at a 1:200 dilution (Abcam) overnight, followed by Alexa Fluor 488–conjugated goat anti-mouse IgG (H+L) secondary antibody at a 1:200 dilution (A-11001, Thermo Fisher Scientific). Immunofluorescence was examined using an upright fluorescence microscope from the Olympus Laboratories.
Wild-type and edited AZIN1 overexpression assays.
Plasmids bearing wild-type or edited AZIN1 cDNA sequences were used to overexpress AZIN1 (9). These plasmids were ligated into the pLenti6/V5-TOPO vector (9, 10), and each AZIN1 expression construct was transfected into HT29 or HCT116 cells (2,000 ng per 1 million cells) using Lipofectamine 2,000 (Invitrogen). For all transfections, empty pLenti6/V5-TOPO vector was used as the control vector. Forty-eight hours after transfection, the transfected cells were collected for further experiments.
Western immunoblotting.
Western immunoblotting experiments were performed as described previously (36). Anti-ADAR1 (1:2,000 dilution; ab88574, Abcam), anti-ODC (1:2,000 dilution; ab66067, Abcam), and anti-CD44v6 (1:1,000 dilution; ab78960, Abcam) antibodies were used to detect target proteins, and an anti–β-actin antibody (1:5,000 dilution; A5441, MilliporeSigma) was used as the loading control.
Cell proliferation assays.
The MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay (MilliporeSigma) was performed to measure the effect on cell proliferation following the overexpression of wild-type or edited AZIN1 using the method described previously (35).
Invasion/migration assays.
The invasiveness of cancer cells was evaluated using BioCoat Matrigel Invasion Chambers (Corning Life Sciences) as described previously (35).
Establishment of spheroid-derived CSCs.
Spheroid-derived CSCs were generated from HT29 and HCT116 cells in serum-free DMEM/F12 medium containing B27, N2 supplements (Gibco), 10 ng/ml human recombinant basic fibroblast growth factor (Gibco), and 20 ng/ml epidermal growth factor (MilliporeSigma) and cultured in Costar ultra-low attachment flasks (Corning) as described previously (37).
Spheroid formation assays.
HT29 and HCT116 cells were cultured in serum-free DMEM/F12 medium, including B27 and N2 supplements, 10 ng/ml human recombinant basic fibroblast growth factor (Gibco), and 20 ng/ml epidermal growth factor (MilliporeSigma), using ultra-low-attachment 96-well plates (Corning). The number of spheroids was counted using a microscope.
Xenograft studies.
Male athymic nude mice were obtained from Harlan Laboratories at 5 weeks of age and kept under controlled conditions (12-hour light/dark cycles), with food and water ad libitum. To establish a xenograft tumor model, HCT116 cell lines transfected with wild-type or edited AZIN1 cDNA sequence plasmids were subcutaneously injected in the left and right flanks of 12 mice (3 × 106 cells/injection site) with 50 μl Matrigel (Corning). Mice were monitored for 14 days following the injection, and subcutaneous tumors were measured every second day (1/2 length × width × height). At 2 weeks after injection, all animals were sacrificed.
Statistics.
Results are expressed as mean ± SD. JMP software (version 10.0, SAS Institute Inc.) and MedCalc (version 16.8.4, MedCalc Software) were used to perform statistical analyses. Differences between groups were estimated by Wilcoxon’s signed-rank test, Wilcoxon’s rank-sum test, the χ2 test, Steel test, Kruskal-Wallis test, and 1-way ANOVA analysis, as appropriate. The correlation of two groups was estimated by Spearman’s rank correlation analysis. For time-to-event analyses, survival estimates were calculated using the Kaplan-Meier analysis, and groups were compared with the log-rank test. Receiver operating characteristic curves were established to determine the cutoff values for analyzing risk factors for prognosis and each metastasis type by Youden’s index. OS was measured from the date patients underwent surgery to the date of death, resulting from any cause, or the last known follow-up for patients that were still alive. DFS analysis was measured from the date the patient underwent curative surgery to the date of disease recurrence, death from any cause, or until last contact with the patient.
The Cox’s proportional hazards models were used to estimate HRs for recurrence or death. Assumptions of proportionality were confirmed for the Cox proportional hazards analyses by generating Kaplan-Meier survival curves (e.g., high vs. low expression groups) and by ensuring that the two curves did not intersect each other. Multivariate logistic regression models were used to predict factors influencing lymph node metastasis, hepatic metastasis, and distant metastasis. Forced-entry regression was used to include these variables in all multivariable equations in order to analyze whether each of the predictors affected the outcome after adjusting for known confounders. All P values were 2 sided, and those less than 0.05 were considered statistically significant.
Study approval.
Written informed consent was obtained from each patient, and the institutional review boards of Baylor University Medical Center; Okayama University Graduate School of Medicine, Dentistry, and Pharmaceutical Sciences; Mie University Graduate School of Medicine; University of Tokushima; National Cancer Center Hospital; and National University of Singapore approved this study. The animal protocol was approved by the Institutional Animal Care and Use Committee of the Baylor Scott & White Research Institute.
Author contributions
KS, YO, ST, JM, and AG conceived and designed experiments. KS, YO, and ST performed experiments. KS, YO, ST, JM, and AG analyzed data. KS, YT, TN, MK, NT, TT, YY, TF, and LC contributed reagents, materials, and other analytical tools. KS, YO, ST, and AG wrote the manuscript.
Supplementary Material
Acknowledgments
The present work was supported by grants CA72851, CA181572, CA184792, CA187956, and CA202797 from the National Cancer Institute, National Institutes of Health; grant RP140784 from the Cancer Prevention Research Institute of Texas; and grants from the Sammons Cancer Center and Baylor Foundation as well as funds from the Baylor Scott & White Research Institute awarded to AG. This work was also supported by a grant from the Uehara Memorial Foundation to KS and YO.
Version 1. 06/21/2018
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2018;3(12):e99976. https://doi.org/10.1172/jci.insight.99976.
Contributor Information
Kunitoshi Shigeyasu, Email: gmd421045@s.okayama-u.ac.jp.
Yoshinaga Okugawa, Email: yoshinaga.okugawa@gmail.com.
Shusuke Toden, Email: shusuke.toden@gmail.com.
Jinsei Miyoshi, Email: jinsei03442000@gmail.com.
Yuji Toiyama, Email: ytoi0725@clin.medic.mie-u.ac.jp.
Takeshi Nagasaka, Email: nagasakatahino@gmail.com.
Naoki Takahashi, Email: naoki19800623@gmail.com.
Masato Kusunoki, Email: kusunoki@clin.medic.mie-u.ac.jp.
Tetsuji Takayama, Email: takayama@tokushima-u.ac.jp.
Yasuhide Yamada, Email: yamada0304@aol.com.
Toshiyoshi Fujiwara, Email: toshi_f@md.okayama-u.ac.jp.
Ajay Goel, Email: Ajay.Goel@baylorhealth.edu.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Center MM, Jemal A, Smith RA, Ward E. Worldwide variations in colorectal cancer. CA Cancer J Clin. 2009;59(6):366–378. doi: 10.3322/caac.20038. [DOI] [PubMed] [Google Scholar]
- 3.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 4.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4(2):143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 5.Mattick JS, Makunin Non-coding RNA. Hum Mol Genet. 2006;15 Spec No 1:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 6.Okugawa Y, Grady WM, Goel A. Epigenetic alterations in colorectal cancer: emerging biomarkers. Gastroenterology. 2015;149(5):1204–1225.e12. doi: 10.1053/j.gastro.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qi L, Chan TH, Tenen DG, Chen L. RNA editome imbalance in hepatocellular carcinoma. Cancer Res. 2014;74(5):1301–1306. doi: 10.1158/0008-5472.CAN-13-3485. [DOI] [PubMed] [Google Scholar]
- 8.Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen L, et al. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat Med. 2013;19(2):209–216. doi: 10.1038/nm.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qin YR, et al. Adenosine-to-inosine RNA editing mediated by ADARs in esophageal squamous cell carcinoma. Cancer Res. 2014;74(3):840–851. doi: 10.1158/0008-5472.CAN-13-2545. [DOI] [PubMed] [Google Scholar]
- 11.Chan TH, et al. ADAR-mediated rna editing predicts progression and prognosis of gastric cancer. Gastroenterology. 2016;151(4):637–650.e10. doi: 10.1053/j.gastro.2016.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Avesson L, Barry G. The emerging role of RNA and DNA editing in cancer. Biochim Biophys Acta. 2014;1845(2):308–316. doi: 10.1016/j.bbcan.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 13.Chan TH, et al. A disrupted RNA editing balance mediated by ADARs (Adenosine DeAminases that act on RNA) in human hepatocellular carcinoma. Gut. 2014;63(5):832–843. doi: 10.1136/gutjnl-2012-304037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crews LA, et al. An RNA editing fingerprint of cancer stem cell reprogramming. J Transl Med. 2015;13:52. doi: 10.1186/s12967-014-0370-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang JC. Cancer stem cells: Role in tumor growth, recurrence, metastasis, and treatment resistance. Medicine (Baltimore) 2016;95(1 Suppl 1):S20–S25. doi: 10.1097/MD.0000000000004766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heider KH, Kuthan H, Stehle G, Munzert G. CD44v6: a target for antibody-based cancer therapy. Cancer Immunol Immunother. 2004;53(7):567–579. doi: 10.1007/s00262-003-0494-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jung T, Gross W, Zöller M. CD44v6 coordinates tumor matrix-triggered motility and apoptosis resistance. J Biol Chem. 2011;286(18):15862–15874. doi: 10.1074/jbc.M110.208421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J, et al. Clinical and prognostic significance of HIF-1α, PTEN, CD44v6, and survivin for gastric cancer: a meta-analysis. PLoS One. 2014;9(3):e91842. doi: 10.1371/journal.pone.0091842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, et al. Prognostic significance of CD44V6 expression in osteosarcoma: a meta-analysis. J Orthop Surg Res. 2015;10:187. doi: 10.1186/s13018-015-0328-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang H, Zhao W, Shao W. Prognostic value of CD44 and CD44v6 expression in patients with non-small cell lung cancer: meta-analysis. Tumour Biol. 2014;35(8):7383–7389. doi: 10.1007/s13277-014-2150-3. [DOI] [PubMed] [Google Scholar]
- 21.Hu B, Luo W, Hu RT, Zhou Y, Qin SY, Jiang HX. Meta-analysis of prognostic and clinical significance of CD44v6 in esophageal cancer. Medicine (Baltimore) 2015;94(31):e1238. doi: 10.1097/MD.0000000000001238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang JL, et al. CD44v6 overexpression related to metastasis and poor prognosis of colorectal cancer: A meta-analysis. Oncotarget. 2017;8(8):12866–12876. doi: 10.18632/oncotarget.14163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baysal BE, Sharma S, Hashemikhabir S, Janga SC. RNA Editing in pathogenesis of cancer. Cancer Res. 2017;77(14):3733–3739. doi: 10.1158/0008-5472.CAN-17-0520. [DOI] [PubMed] [Google Scholar]
- 24.Shah SP, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461(7265):809–813. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- 25.Anadón C, et al. Gene amplification-associated overexpression of the RNA editing enzyme ADAR1 enhances human lung tumorigenesis. Oncogene. 2016;35(33):4407–4413. doi: 10.1038/onc.2015.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olsen RR, Zetter BR. Evidence of a role for antizyme and antizyme inhibitor as regulators of human cancer. Mol Cancer Res. 2011;9(10):1285–1293. doi: 10.1158/1541-7786.MCR-11-0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benson AB, et al. American Society of Clinical Oncology recommendations on adjuvant chemotherapy for stage II colon cancer. J Clin Oncol. 2004;22(16):3408–3419. doi: 10.1200/JCO.2004.05.063. [DOI] [PubMed] [Google Scholar]
- 28.Quasar Collaborative Group. et al. Adjuvant chemotherapy versus observation in patients with colorectal cancer: a randomised study. Lancet. 2007;370(9604):2020–2029. doi: 10.1016/S0140-6736(07)61866-2. [DOI] [PubMed] [Google Scholar]
- 29.Baddi L, Benson A. Adjuvant therapy in stage II colon cancer: current approaches. Oncologist. 2005;10(5):325–331. doi: 10.1634/theoncologist.10-5-325. [DOI] [PubMed] [Google Scholar]
- 30.Wallace HM, Fraser AV, Hughes A. A perspective of polyamine metabolism. Biochem J. 2003;376(Pt 1):1–14. doi: 10.1042/BJ20031327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujita K, Murakami Y, Hayashi S. A macromolecular inhibitor of the antizyme to ornithine decarboxylase. Biochem J. 1982;204(3):647–652. doi: 10.1042/bj2040647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morgan DO. Principles of CDK regulation. Nature. 1995;374(6518):131–134. doi: 10.1038/374131a0. [DOI] [PubMed] [Google Scholar]
- 33.Pyronnet S, Pradayrol L, Sonenberg N. A cell cycle-dependent internal ribosome entry site. Mol Cell. 2000;5(4):607–616. doi: 10.1016/S1097-2765(00)80240-3. [DOI] [PubMed] [Google Scholar]
- 34.Zhao T, Goh KJ, Ng HH, Vardy LA. A role for polyamine regulators in ESC self-renewal. Cell Cycle. 2012;11(24):4517–4523. doi: 10.4161/cc.22772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okugawa Y, et al. Metastasis-associated long non-coding RNA drives gastric cancer development and promotes peritoneal metastasis. Carcinogenesis. 2014;35(12):2731–2739. doi: 10.1093/carcin/bgu200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toden S, et al. Novel evidence for curcumin and boswellic acid-induced chemoprevention through regulation of miR-34a and miR-27a in colorectal cancer. Cancer Prev Res (Phila) 2015;8(5):431–443. doi: 10.1158/1940-6207.CAPR-14-0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toden S, Tran HM, Tovar-Camargo OA, Okugawa Y, Goel A. Epigallocatechin-3-gallate targets cancer stem-like cells and enhances 5-fluorouracil chemosensitivity in colorectal cancer. Oncotarget. 2016;7(13):16158–16171. doi: 10.18632/oncotarget.7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.