Abstract
The nasal mucosa is an important component of mucosal immunity. Immunogenic particles in inspired air are known to activate the local nasal mucosal immune system and can lead to sinonasal inflammation; however, little is known about the effect of this activation on the lung immune environment. Here, we showed that nasal inoculation of murine coronavirus (CoV) in the absence of direct lung infection primes the lung immune environment by recruiting activated monocytes (Ly6C+ inflammatory monocytes) and NK cells into the lungs. Unlike infiltration of these cells into directly infected lungs, a process that requires type I IFN signaling, nasally induced infiltration of Ly6C+ inflammatory monocytes into the lungs is IFN-I independent. These activated macrophages ingested antigen and migrated to pulmonary lymph nodes, and enhanced both innate and adaptive immunity after heterologous virus infection. Clinically, such nasal-only inoculation of MHV-1 failed to cause pneumonia but significantly reduced mortality and morbidity of lethal pneumonia caused by severe acute respiratory syndrome CoV (SARS-CoV) or influenza A virus. Together, the data indicate that the nose and upper airway remotely prime the lung immunity to protect the lungs from direct viral infections.
Keywords: Immunology, Virology
Keywords: Innate immunity
Nasal priming with a pathogen in the absence of any lung infection protects against lethal heterologous virus challenge.
Introduction
Epidemic and pandemic lethal viral pneumonia periodically occur, resulting in substantial morbidity and mortality. Effective treatment against lethal viral pneumonia remains lacking. A major physiological role of the human nose is to protect the relatively vulnerable lungs (1). As the first point of contact of the respiratory tract with the environment, the nose has many unique histologic and anatomic features that allow it to effectively filter, humidify, and warm inhaled air to minimize detrimental stimulation of the lower airway and lungs. While these physical-type protective mechanisms are well recognized and studied, little is known about whether the nose can prime immunity in the lungs, and thereby protect them.
The human nose is an important branch of the mucosal immune system. Mucosal surfaces that cover the gastrointestinal (GI), respiratory, urinary, and genital tracts function as immunologic sensors to detect dangerous signals including pathogens (2, 3). Recent studies have shown that activation of the GI mucosal immune system not only contributes to the local immune response, but also regulates immunity and function of distant organs, including the lungs and the brain (4–6). For instance, interaction between the GI mucosa and microbiota can alter both T cell and B cell responses to influenza A virus (IAV) infections (7). Similar to the GI system, the human nose is covered by type I mucosa that comprises a single layer of epithelial cells, abundant antigen-presenting cells (APCs), goblet cells, and specialized mucosal lymphoid organs (Waldeyer’s ring). The human nose and upper airway effectively capture the majority of common immunogenic particles and droplets that contain pathogens in inhaled air. While these foreign antigens, bacteria, and viruses can activate the local mucosal immune system to cause sino-nasal inflammatory diseases, little is known about the immunologic effect on tissues distal to the nose and upper airway, especially the lungs.
In order to examine such effects, we intranasally infected BALB/c mice with a pneumotropic murine coronavirus (CoV) delivered in a very small volume (1 μl in each nostril, 2% of normal inoculate volume) to avoid aspiration. We failed to recover any virus in the lungs during the course of the experiment using either real-time quantitative RT-PCR (qRT-PCR) or virus titration, and lung disease was absent. Strikingly, such limited local nasal infection significantly reduced the mortality rates after challenge with 2 heterologous pathogenic respiratory viruses, SARS-CoV (from 100% to 0%) and IAV (from 100% to 60%) and reduced clinical disease. We observed a significant increase in numbers of Ly6C+ inflammatory monocytes (IMs) in the lungs, 2 days after nasal infection. Moreover, Ly6C+ IM recruited by low-volume nasal infection were less activated and proinflammatory compared with those detected after direct lung infection. Our results suggest that these monocytes dramatically enhanced the innate and virus-specific CD4+ and CD8+ T cell responses after heterologous virus challenge. These findings indicate that activating the nasal innate immune response may be very useful for the prevention and treatment of lethal viral pneumonia.
Results
Nasal administration of MHV-1 dramatically reduced mortality and morbidity after heterologous virus infection.
We delivered mouse hepatitis virus type 1 (MHV-1; 104 PFU) intranasally in a small volume (1 μl in each nostril) to generate nasal-only inoculation (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.99025DS1). Using this amount of inoculum, we detected viruses in the nasal lavage fluid (NLF) only on day 1 after infection (p.i.), but did not detect virus in the lungs during the course of the experiment (Supplemental Figure 1C). No virus was detected in the lungs by plaque assay or qRT-PCR. Clinically, mice with such nasal inoculum did not lose weight (Supplemental Figure 1D) or display any signs of MHV-1 pneumonia. Cell numbers (Supplemental Figure 1E) and differential in bronchoalveolar fluids (BALFs) from mice after intranasal MHV-1 were indistinguishable when compared with control mice intranasally treated with vehicle. The major cell type in BALFs from both groups was alveolar macrophages, which were defined by high autofluorescence and CD11c, SiglecF, and F40/80 expression as described previously (8). Both groups had very few T cells (CD3+), B cells (CD19+), neutrophils (Ly6G+CD11b+), or NK cells (NKp46+CD3–) cells in the BALF (data not shown). Additionally, nasal infection by MHV-1 caused significant lymphocyte expansion in the cervical lymph nodes but not in the MLNs, consistent with the presence of virus in the upper airway but not in the lungs (Supplemental Figure 1F). Since we failed to observe any signs of direct virus infection of the lungs after intranasal inoculation with 104 PFU MHV-1 in 2 μl, we used this volume/dose in all subsequent experiments (designated as “nasal-only inoculation”). We next analyzed the early immune response in the nasal-associated lymphoid tissue (NALT; Supplemental Figure 1G) and superficial and deep cervical lymph nodes within 24 hours after nasal-only inoculation. We observed that upregulated B cell expression of Ly6C (Supplemental Figure 1H) was one of the earliest events in the NALT after nasal-only MHV-1 inoculation, suggestive of B cell activation. Further, the B/T cell ratio, B cell frequency, and numbers in the NALT but not the cervical lymph nodes were significantly reduced within 12–24 hours p.i. (Supplemental Figure 1, I–L), suggesting that B cell egress from the NALT contributed to the immune response in a distant organ, the lungs.
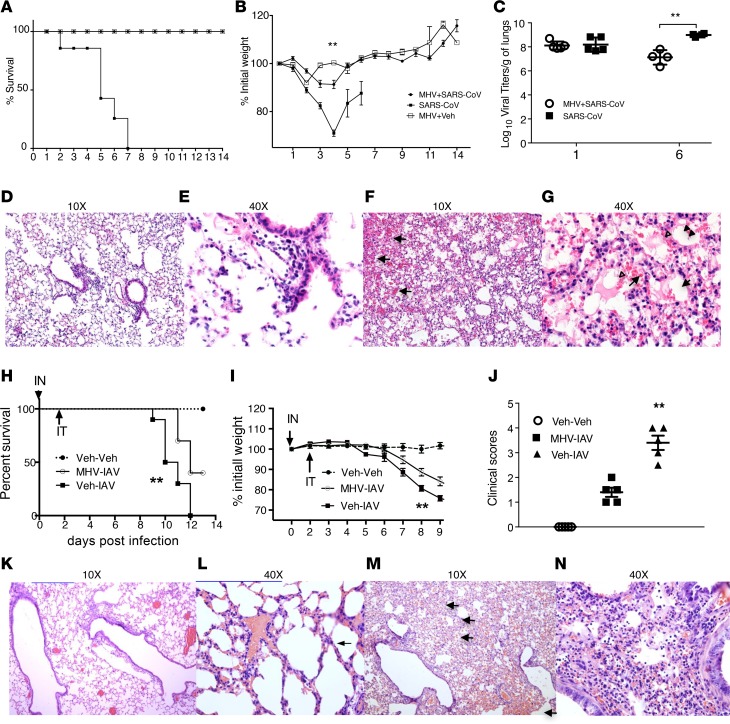
We next examined whether nasal-only inoculation with MHV-1 could modulate the mortality and morbidity of lethal pneumonia by SARS-CoV and IAV. BALB/c mice were intranasally treated with MHV-1 (104 PFU in 2 μl). Two days later, mice were intratracheally (IT) infected with unrelated viruses, IAV or SARS-CoV, at a lethal dose. Prior nasal MHV-1 inoculation dramatically reduced the mortality rate of SARS-CoV pneumonia from 100% to 0% (Figure 1A). Morbidity, represented by weight loss, was significantly reduced (Figure 1B), whereas the kinetics of SARS-CoV clearance was significantly enhanced (>10 fold) in the lungs of mice with prior nasal MHV-1 inoculation (MHV+SARS-CoV) (Figure 1C). Histologically, mice without prior nasal MHV-1 inoculation exhibited evidence of extensive alveolar damage characterized by alveolar edema with inflammation and vascular congestion/hemorrhage, while in mice with prior nasal MHV inoculum, only minor interstitial inflammation and minor vascular congestion were observed (Figure 1, D–G). To extend these results to a non-CoV infection, we infected mice with mouse-adapted IAV (PR-8 strain, 1,160 tissue culture infectious units [TCIU] in 50 μl per mouse via IT instillation) following MHV-1 priming. Similar to SARS-CoV infection, prior nasal MHV-1 inoculation significantly reduced IAV pneumonia–induced mortality (100% to 60%) (Figure 1H), weight loss (Figure 1I), and clinical scores (Figure 1J). Lung histological examination revealed less severe alveolar wall damage in the group with prior MHV-1 inoculation (Figure 1, K–N). Taken together, these data indicate that nasal-only inoculation with MHV-1 generated robust protection against lethal pneumonia caused by heterologous viruses.
Figure 1. Nasal administration of MHV-1 dramatically reduced mortality and morbidity of lethal pneumonia by heterologous viruses.
BALB/c mice were intranasally infected with MHV-1 (2 μl, 104 PFU) or vehicle. Two days later, mice were infected with SARS-CoV (104 PFU) or IAV (PR-8 strain, 1,160 TCIU) (50 μl per mouse via IT instillation). The negative controls were mice that only received vehicle. Mortality and morbidity were then monitored daily. Mice that lost >30% of their initial weight were euthanized per institutional IACUC protocols. (A) Survival rates of SARS-CoV–infected mice. P < 0.0001, SARS (n = 10) vs. MHV+SARS (n = 10) and MHV+vehicle (MHV+Veh) (n = 5). No difference was found between MHV+SARS and MHV+Veh, using both log-rank (Mantel-Cox) test and Gehan-Breslow-Wilcoxon test. (B) Weight is expressed as percentage of original weight. **P < 0.01, among MHV-SARS (n = 10), SARS (n = 10), and MHV+Veh (n = 5) using repeated-measures ANOVA; P < 0.01, MHV-SARS vs. SARS, using LSD. (C) Viral titers at 6 days p.i. **P < 0.001, MHV-SARS (n = 5) vs. SARS (n = 5). (D–G) Lung histology: D (×10) and E (×40), representative from MHV-SARS group; F (×10) and G (×40), representative from SARS group. Hemorrhage (arrows), hyalinization (arrowheads), and extensive infiltration of inflammatory cells in both alveolar space and lung parenchyma are illustrated. (H) Survival rate. **P < 0.01, both infected groups (n = 10 for each) vs. controls (n = 6); and MHV-IAV (n = 10) vs. Veh-IAV (n = 10). (I) Weight was expressed as percentage of original weight. **P < 0.01, among MHV-IAV (n = 10), Veh-IAV (n = 10), and controls (n = 6). (J) Clinical scores on day 9 p.i. **P < 0.01, Veh-IAV vs. MHV-IAV, and both Veh-IAV and MHV-IAV vs. Veh-Veh. n = 5 for each group. (K–N) lung histology. K (10×) and L (40×), representative from MHV-IAV–infected mice; M (10×) and N (40×), representative from IAV-infected mice. Hemorrhage (arrows) and extensive infiltration of inflammatory cells (N) in both alveolar space and lung parenchyma are illustrated. Modest amounts of congestion and inflammatory infiltration in the alveolar septum was found in mice with MHV-IAV infections (thin arrow). Data are from at least 2 independent experiments. IN, intranasal; Inf Mon, inflammatory monocytes.
Nasal-only inoculation of MHV-1 remotely primes lung innate immunity by recruiting Ly6C+ IMs.
To determine whether this MHV-1 protective effect was accompanied by changes in infiltrating cell composition, we analyzed mouse lungs on days 0, 1, 2, 4, and 20. While total cell numbers in the lungs did not chang after nasal-only inoculation (Figure 2A), the cellular composition was significantly altered, with decreased frequencies of both T and B cells, and increased frequency of Ly6C+ IMs (Ly6C+CD11b+CD11cintF4/80intCD3–CD19–SiglecF–Ly6G–) (Supplemental Figure 2A; Figure 2, B–D) and NK cells (Supplemental Figure 2B) at day 2 and 4 p.i. These results were further supported by nasal-only inoculation of MHV-JHM, a neurotropic coronavirus (9, 10), which also caused infiltration of Ly6C+ IMs in the lungs (3,000 PFU, 1 μl ×2 for intranasal delivery; Supplemental Figure 2, C and D). After nasal-only inoculation, MHV-JHM–infected mice developed no signs of clinical disease.
Figure 2. Nasal administration of MHV-1 remotely recruits Ly6C+ IMs into the lungs.
(A) Total cell numbers in the lungs after nasal-only MHV-1 infection. n = 3–7 per group, pooled from 2 independent experiments. No differences were observed. (B and C) Ly6C+CD11b+ cell infiltration in the lungs after nasal infection. Data in B are expressed as percentage of CD45+ cells. **P < 0.01 vs. dpi 0. Gating strategy is shown in C. n = 3–12, pooled from 3 different experiments. (D) Phenotypic analysis of Ly6ChiCD11b+ cells. Blue lines represent the Ly6ChiCD11b+ cells; red lines represent negative or positive controls in each small panel as indicated. Blue-filled represents F4/80 negative control. Ly6ChiCD11b+ cells were Ly6G–CD19–CD3-SiglecF–NKp46–F4/80int. Neut, neutrophils; T, T cells; B, B cells; NK, NK cells; aM, alveolar macrophages. (E–G) Localization of Ly6C+ IMs in the lungs. BALB/c mice were intranasally infected with MHV-1 (104 PFU in 2 μl MEM, 1 μl/nostril) or vehicle, and were then sacrificed at 2 dpi. The left lungs were clamped, and the right pulmonary vessels were then exclusively perfused via the right ventricle (E). The frequencies of Ly6C+ IMs in both left and right lungs were then analyzed and expressed as percentage of CD45+ singlet cells (F and G). **P < 0.01 vs. controls. n = 3 per group.
Ly6C+ IMs that accumulated after nasal-only inoculation could be present in the lung parenchyma, alveolar spaces, or blood vessels. To distinguish these possibilities, we performed unilateral perfusion of the pulmonary vessels and examined whether such intervention changed the frequency of these cells in comparison to the non-perfused side in the same animals (Figure 2E). As shown in Figure 2, F and G, the frequency of these cells was not changed by perfusion of the pulmonary circulation, indicating that they were not circulating in pulmonary blood vessels, although they could still be adherent to vessel walls (11) or in the lymphatic vessels. The lack of change in cell number (Supplemental Figure 1E) or differential in BALFs after nasal-only inoculation suggests that the Ly6C+ IMs are situated primarily in the lung parenchyma but not in the pulmonary vessels or alveolar spaces.
Nasal-only inoculation–induced infiltration of Ly6C+ inflammatory monocytes into the lungs is IFN-I independent.
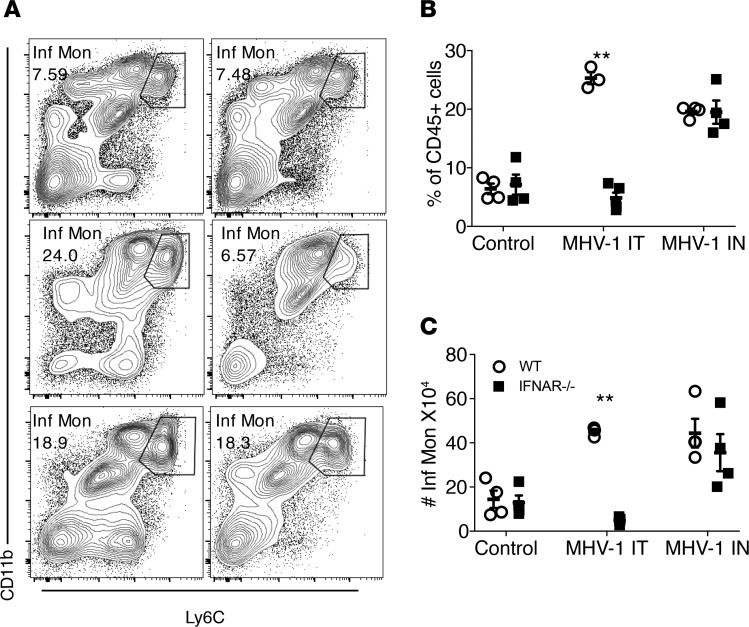
Previous studies have shown that the recruitment of Ly6C+ IMs to sites of virus infection in the lung is IFN-I dependent (12–14) . In order to determine whether IM infiltration into the lungs after nasal-only inoculation was also IFN-I dependent, we infected mice lacking IFN-I receptors (BALB/c IFNAR–/–) and BALB/c controls with MHV-1 intranasally (2 μl, 104 PFU) or IT (50 μl 104 PFU), or with vehicle. Lungs were then analyzed on day 2 p.i. (Figure 3). As expected, direct lung infection with 104 PFU MHV-1 significantly increased the accumulation of Ly6C+ IMs in the lungs of WT mice but not IFNAR–/– mice. In marked contrast, in mice intranasally primed with MHV-1, Ly6C+ IMs equivalently infiltrated the lungs of IFNAR–/– and WT mice. Recent studies have demonstrated an important role for the vagus nerve in the regulation of excessive immune responses in the GI tract and lungs (15–17). Next, to assess whether the vagus nerve has a similar role after nasal-only MHV-1 inoculation, we performed unilateral cervical vagotomy (18). The contralateral side was sham-treated without vagotomy. Mice were then intranasally treated with MHV-1 (2 μl, 104 PFU) 14 days later. As shown in Supplemental Figure 3, there was nearly identical Ly6C+ IM infiltration in the lungs in vagotomized and sham-treated sides of the same animals. Taken together, these data indicate that Ly6C+ IM recruitment in the lungs after nasal MHV-1 priming is IFN-I– and vagus nerve–independent, and thus mechanistically different from that caused by direct lung infection.
Figure 3. Ly6C+ IM recruitment by nasal MHV-1 infection is not IFN-I dependent.
IFNAR–/– and BALB/c mice were treated with MHV-1 intranasally (IN, 2 μl, 104 PFU) or IT (50 μl, 104 PFU) or vehicle. The mice were sacrificed at 2 days p.i. The frequency and numbers of Ly6C+ IMs in the lungs were then determined. (A) Flow cytometric plots of representative mice from each group. WT denotes BALB/c mice. (B and C) Frequency and numbers of Ly6C+ IMs in the lungs. **P < 0.01 vs. IFNAR–/– in MHV-1 IT group. n = 4–5 per group.
Ly6C+ IMs recruited by nasal-only inoculation are less activated/mature than those recruited by IT infection.
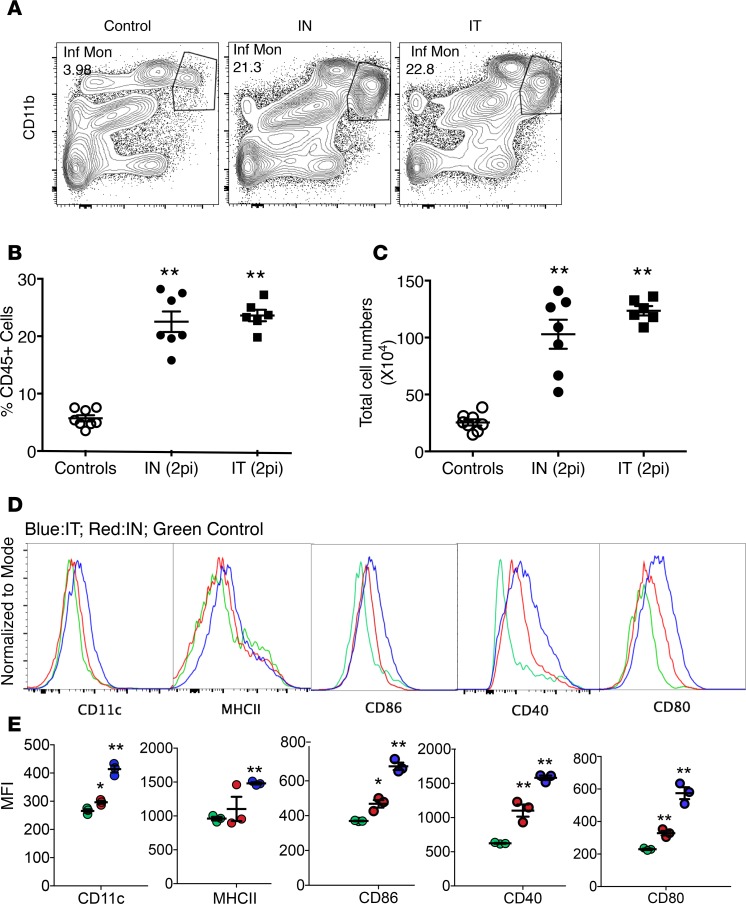
We next compared the expression levels of activation/maturation markers including CD11c, MHC class II (MHCII), CCR7, CD86, CD80, and CD40 on Ly6C+ IMs recruited by nasal-only as opposed to IT MHV-1 infection. As shown in Figure 4, A–C, nasal-only and IT infection MHV-1 resulted in nearly identical frequencies and numbers of Ly6C+ IMs in the lungs on day 2 p.i. However, in mice with nasal-only inoculation, the expression levels of CD11c, MHCII, CD86, CD80, and CD40 on Ly6C+ IMs were increased to a lesser degree than after IT infection (Figure 4D), indicating the cells were less activated/mature than those recruited by direct lung infection.
Figure 4. Activation markers on Ly6C+ IMs in mice after nasal-only and IT MHV-1 infection.
BALB/c mice were treated with MHV-1 intranasally (2 μl, 104 PFU) or IT (50 μl 104 PFU), or with vehicle. Their lungs were then analyzed at 2 days p.i. by flow cytometry. (A) Flow cytometric plots of representative mice from control, IN, and IT groups. (B and C) Frequencies and numbers of Ly6C+ IMs. **P < 0.01 vs. controls. No differences in either frequencies or numbers between IN and IT groups was detected. n = 6–8 per group. (D and E) Histogram and mean fluorescence intensity (MFI) of activation markers (representative of two independent experiments). Blue lines, IT; red lines, IN; green lines, control. *P < 0.05, **P < 0.01, vs. controls in each panel. n = 3 per group.
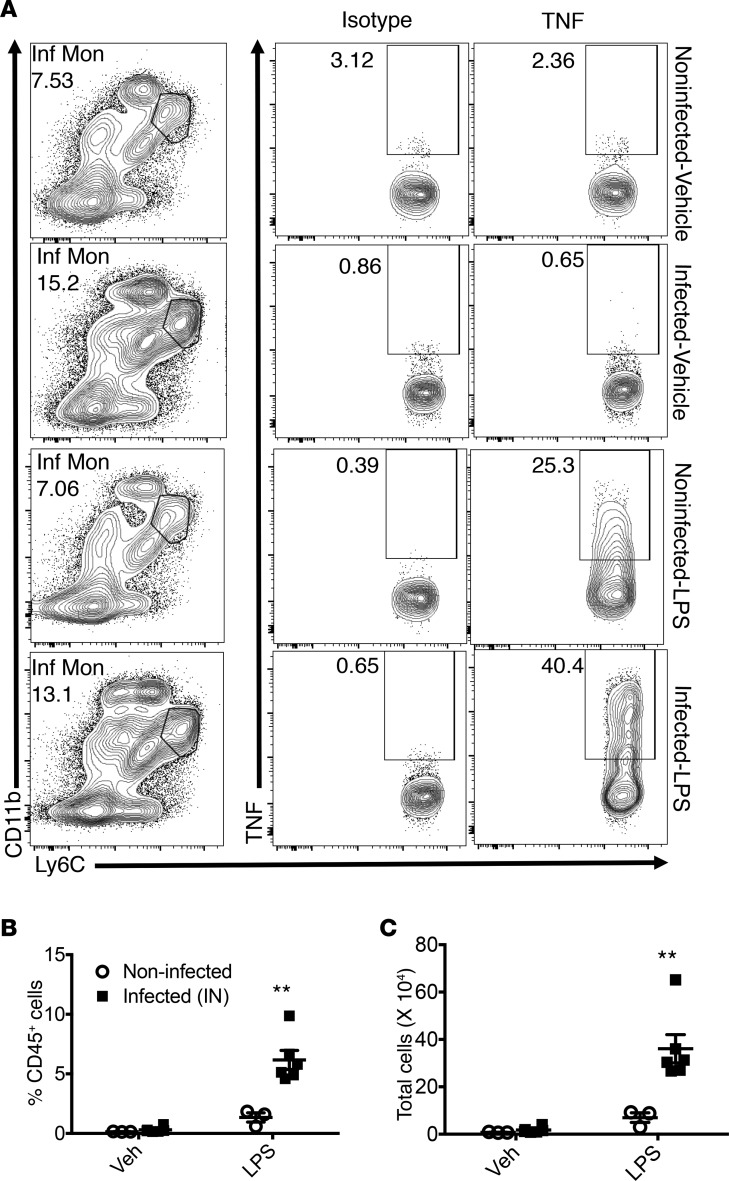
Ly6C+ IMs recruited by nasal priming can produce TNF.
Previous studies have shown that secretion of TNF is one major antiviral mechanism employed by Ly6C+ IMs (13, 19, 20). To assess whether Ly6C+ IMs recruited by nasal MHV-1 priming produced TNF, we harvested lung-derived cells on day 2 p.i. and treated them with LPS (1 ng/μl) or vehicle (RP10) for 6 hours directly ex vivo, followed by intracellular staining for TNF. In the absence of LPS treatment, Ly6C+ IMs from both noninfected and MHV-1–primed mice did not produce TNF, as there was no difference between isotype-stained cells and TNF antibody–treated cells in the 2 groups (Figure 5A). After LPS stimulation, mice with nasal-only inoculation exhibited significantly more TNF-secreting Ly6C+ IMs in the lungs, in comparison to noninfected controls (Figure 5, A–C). There was no difference in the amount of TNF expressed per cells, however, based on measurements of MFI (data not shown). These data showing increased LPS-induced TNF production by Ly6C+ IMs recruited to the lungs following nasal-only inoculation with MHV-1 suggest that nasal exposure to virus-primed immune cells in the lung generates a more robust innate immune response to pathogens.
Figure 5. Intracellular expression of TNF by IMs.
BALB/c mice were treated with MHV-1 intranasally (2 μl, 104 PFU, infected, n = 6 per group) or vehicle (noninfected, n = 3 per group). Their lungs were harvested to prepare single-cell suspension at 2 dpi. The cells were then treated with LPS (1 ng/μl) or vehicle (RP10) in the presence of GolgiPlug for 6 hours at 37°C. Intracellular staining for TNF was then performed. Isotype was used for all samples for optimal gating. (A) Representative flow cytometric plots from each group. (B and C) Frequency and number of TNF-producing Ly6C+ IMs. **P < 0.01. Representative of 2 independent experiments.
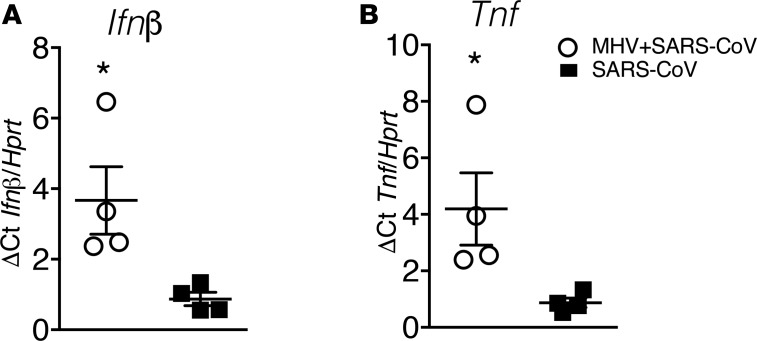
Increased TNF and INF-β in mice with nasal-only inoculation after SARS-CoV infection.
To assess whether prior nasal-only MHV-1 inoculation generated a more robust innate immune response after SARS-CoV challenge, were infected mice intranasally with MHV-1 and challenged them with SARS-CoV at 2 days p.i. As shown in Figure 6, A and B, both TNF and IFN-β levels were dramatically increased at 24 hours after SARS-CoV infection in the MHV-1–primed mice, further supporting the notion that nasal-only inoculation enhanced the lung innate immune response after lethal virus infection.
Figure 6. INF-β and TNF expression in vivo 24 hours after SARS-CoV infection.
BALB/c mice were intranasally infected with MHV-1, followed by IT infection with SARS-CoV. (A) IFN-β and (B) TNF levels 24 hours after SARS-CoV infection were determined using qRT-PCR. *P < 0.05, MHV+SARS vs. SARS. n = 4 per group.
Migration of lung Ly6C+ IMs in mice after nasal-only inoculation.
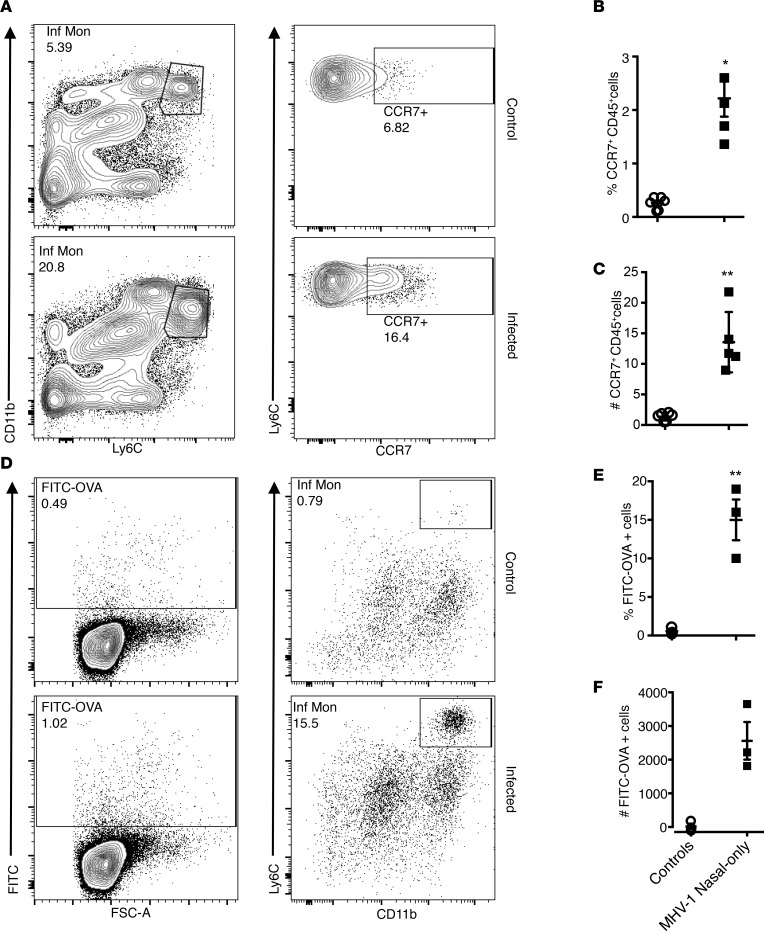
Since infiltrating Ly6C+ IMs may function as APCs to orchestrate adaptive immunity (21–24), we next examined whether intranasal MHV-1 priming also augmented APC numbers and function. After nasal-only inoculation, not only was the number of Ly6C+ IMs in the lungs increased vs. controls, but the fraction expressing CCR7 also increased (Figure 7, A–C). Since CCR7 expression is required for APC migration to draining lymph nodes, we next instilled OVA-FITC IT into the lungs of mice and analyzed mediastinal lymph nodes (MLNs) 18 hours later for FITC+ cell migration. Mice with nasal infection but not controls exhibited more FITC+ Ly6C+CD11b+ IMs in MLNs (Figure 7, D–F), demonstrating that Ly6C+ IMs recruited by nasal MHV-1 inoculation were primed for both antigen uptake and migration to the MLNs.
Figure 7. Migration of Ly6C+ IMs.
(A–C) CCR7 expression on Ly6C+ IMs. (A) Flow cytometric plots showing increased CCR7+Ly6+ IM levels in mice after nasal-only inoculation. (B and C) Frequencies of CCR7+Ly6+ IMs in the lungs of mice with and without nasal infection. **P < 0.01; *P < 0.05. n = 5–6 per group. (D) Migration of Ly6+ IMs into the MLNs. BALB/c mice were intranasally infected with MHV-1 (2 μl, 104 PFU) or vehicle. Two days later, FITC-OVA was instilled IT. MLNs were then collected to evaluate cell migration. Data are representative of 2 independent experiments. (D) Flow cytometric plots showing increased frequency of FITC-OVA+Ly6C+ IMs in the MLNs. (E and F) Frequencies and numbers of FITC-OVA+Ly6C+ IMs in the MLNs collected from mice with and without intranasal infection. **P < 0.01 vs. controls. n = 3 per group.
Enhanced adaptive immune response against heterologous virus challenge in the lungs after nasal infection.
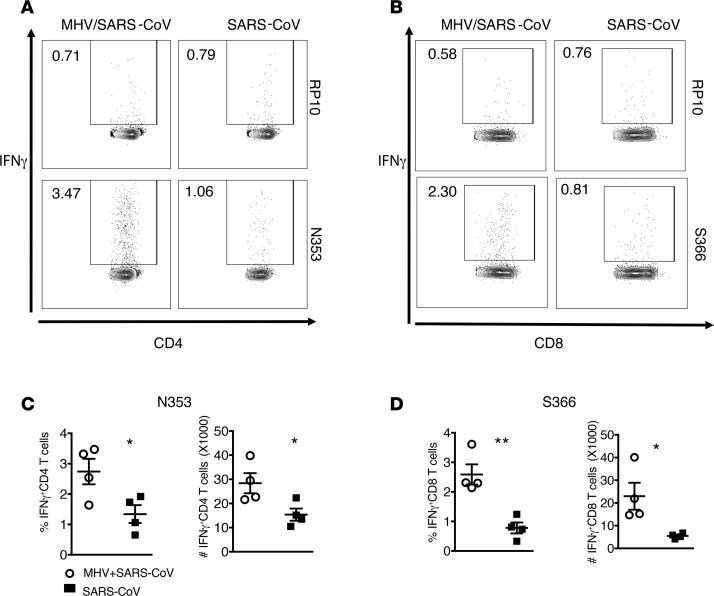
To assess whether enhanced Ly6C+ CD11b+ IM migration to the MLN resulted in a stronger T cell response in the lungs of MHV-1–primed mice, were treated mice intranasally with MHV-1 2 days prior to challenge with a lethal dose of SARS-CoV. The SARS-CoV–specific T cell response was then investigated using an intracellular cytokine staining assay after stimulation with virus-specific peptides (N353 for CD4+ T cells; S366 for CD8+ T cells) (25). As shown in Figure 8, prior nasal inoculation of MHV-1 significantly enhanced the SARS-CoV–specific CD4+ and CD8+ cell responses in the lungs, suggesting that enhancement of the T cell response resulting from nasal priming by MHV-1 is one possible mechanism underlying the observed protection against viral pneumonia.
Figure 8. T cell responses after SARS-CoV infection in mice with or without prior nasal-only MHV-1 inoculation.
SARS-CoV–infected mice with or without prior nasal-only MHV-1 inoculation were analyzed for virus-specific T cell responses in the lungs. (A and B) Representative flow plots showing frequency of virus-specific CD8 (S366-specific) and CD4 (N353-specific) T cell responses on day 6 after infection with 104 PFU SARS-CoV. RP10 medium without specific peptide. (C and D) Summary data for frequency and total number of virus-specific T cells are shown in the bar graphs. *P < 0.05; **P < 0.01. n = 4 per group.
Discussion
In this report, we demonstrated that intranasal exposure to low volumes of pneumotropic or neurotropic murine CoV remotely primes the immune environment in the lungs by increasing the infiltration of Ly6C+CD11b+ IMs in the absence of direct pulmonary infection. These cells, in comparison to those recruited by direct viral infection in the lungs, were less activated and thus less proinflammatory, as they expressed lower levels of CD86/80, CD40, MHCII, and CD11c. However, after LPS stimulation, these cells were able to secrete TNF in the lungs, suggestive of a role in the initiation or enhancement of the innate immune response. They also exhibited the ability to ingest antigen, migrate to MLNs, and enhance the T cell response to heterologous viral infection in the lungs. As a result, nasal priming by MHV-1 dramatically reduced the mortality and morbidity resulting from subsequent lethal infection by both SARS-CoV and IAV. A major physiological role of the human nose is to protect the lungs. Our results reveal that in addition to warming, humidifying and filtering inspired air, immunological priming is another lung-protective mechanism initiated in the nose and upper airway.
Immune priming of the lung by prior pulmonary infection has been previously described (26). For instance, IAV infection of mice diminished the clinical and pathologic consequences of subsequent respiratory syncytial virus infection (26, 27). Further, pulmonary treatment with immunogenic substances such as poly(I:C) and CpG generated protection against subsequent lethal pneumonia caused by unrelated pathogens including IAV, SARS-CoV, and Middle East respiratory syndrome CoV (MERS-CoV) (28, 29). In this study, we show that nasal-only exposure to a mouse CoV can prime the lung environment. This priming phenomenon appears to be not only restricted to MHV-1, as MHV-JHM, a neurotropic virus that primarily causes encephalitis but not pneumonia, also recruits antiviral monocytes to the lungs. In contrast to previous reports where direct inoculation of the lungs occurred through aspiration, our data indicate that indirect remote priming of the lung environment by viral exposure of the nose can prepare the lung for more robust infections. We rigorously excluded the possibility of lung exposure to virus, as we failed to detect any MHV-1 in the lungs using either qRT-PCR or viral titration, and failed to find any clinical evidence of direct lung infection.
Interactions between upper and lower airways have long been observed. For instance, allergic rhinitis is the single most important risk factor for asthma (30). Nasal allergen provocation in patients with allergic rhinitis upregulates the expression of ICAM-1, VCAM-1, and E-selectin in the bronchial tissues (31). The common cold is a very frequent cause of asthma exacerbation (32). However, the mechanism by which inflammation in the nose impacts the lungs remains poorly understood. Recently, neuroimmune interactions were postulated to play an important role in the regulation of innate immunity in response to infectious and other environmental pathogens (15–17). Kradin et al. (17) showed that pulmonary DCs are normally located in immediate proximity to unmyelinated nerves. Capsaicin, a neurotoxic protein, effectively inhibited the accumulation of DCs and other inflammatory cells around small pulmonary vessels after exposure to inhaled antigens. Further, the nose and lungs are connected through the nasobronchial reflex (33), which consists of afferent signals carried through the trigeminal nerve in the nose and efferent signals carried through the vagus nerve to the lungs. However, we were unable to provide experimental data supporting a role for priming via the nasobronchial reflex, since unilateral cervical vagotomy did not diminish Ly6C+ IM infiltration into the lungs after nasal-only inoculation (Supplemental Figure 3, A and B).
Cross talk between 2 distinct and distant mucosal sites has been previously reported. Schuijt et al. (34) showed that the GI mucosal system plays a critical role in regulation of lung immunity, since depletion of the gut microbiota was associated with increased bacterial dissemination, decreased TNF and IL-10 secretion, and increased organ damage and mortality during pneumococcal pneumonia. These phenotypic alterations in gut microbiota–depleted mice were completely reversed by fecal microbiota transplantation. Other groups have also reported similar cross-mucosal interaction (34). However, the underlying mechanism remains poorly understood but may involve B cell activation. Johansen et al. showed that activated B cells from human tonsils preferentially migrate to distal mucosal sites in the lower airway, lacrimal glands, salivary glands, and vagina (35). Heidegger (36) et al. showed that viral infection in the gut also caused early reduction in B cell numbers in Peyer’s patches but not mesenteric lymph nodes. Such reduction was abolished in mice pretreated with FTY720, a compound that inhibits B cell egress from secondary lymphoid organs (36) . Other studies showed roles of B cells in innate immunity. For instance, Harvey et al. (37) showed that B cells transfer antigen to other APCs, especially macrophages. Bao et al. (38) reported that IFN-γ–producing B cells can directly stimulate macrophages and play an important role in innate immunity. B cells also are responsible for the formation of mucosa-associated lymphoid tissue (MALT) in the stomach after H. pylori infection (39) . Our data demonstrated early activation and possible egress of B cells in the NALT after nasal-only MHV-1 inoculation. However, whether B cells from the NALT act as a messenger between the nose and lungs, and thereby prime lung immunity, requires further investigation (Supplemental Figure 1, G–L).
Mounting evidence has demonstrated protective and pathogenic roles for Ly6C+ IMs in the host response to different viral diseases, including respiratory syncytial virus (RSV) pneumonia, HSV-2 vaginitis, IAV pneumonia, HSV-1 keratoconjunctivitis, and cytomegalovirus hepatitis (12–14, 22, 40). Goritzka et al. (13) reported that RSV infection recruited protective Ly6C+ IMs via alveolar macrophage–derived type I IFN. In mice with HSV-2–induced vaginitis, the absence of CCR2, a chemokine receptor required for Ly6C+ IMs egress from the bone marrow and entry into sites of inflammation, resulted in significantly higher viral titers in the vagina and dramatically higher mortality rates compared with CCR2+ controls (22). In another study, depletion of Ly6C+ IMs but not DCs or other myeloid cells, resulted in higher susceptibility to HSV-2 infection (41). In contrast, a robust immunopathogenic role for Ly6C+ IMs in mice infected with SARS-CoV was demonstrated (25), resulting in increased lethality. Several hypotheses have been proposed for the differing effects of these cells. First, Ly6C+ IM migration is IFN-I dependent, and the timing of IFN-I induction may influence the extent of Ly6C+ IM accumulation. Delayed IFN-I signaling was associated with accumulation of pathogenic Ly6C+ IMs in the lungs after SARS-CoV infection (25). Second, the amount of infiltration of Ly6C+ IMs may influence outcomes. Aldridge et al. (42) showed that moderate infiltration of Ly6C+ IMs in the lungs protected mice from lethal IAV pneumonia; however, both excessive infiltration and elimination of these cells were detrimental due to associated immunopathology and reduced CD8+ T cell responses, respectively. The quality of infiltrating Ly6C+ IMs may also contribute to outcomes. In our study, nasal-only inoculation and IT instillation caused comparable infiltration of Ly6C+ IMs in the lungs, but Ly6C+IMs driven by nasal-only inoculation were less mature and proinflammatory compared with those driven by IT instillation.
How Ly6C+ IMs prevent disease in low-dose MHV-1–infected mice is not yet understood (13). Ly6C+ IMs enhanced the apoptosis of lung epithelial cells after IAV infection through a TRAIL-DR5–mediated (TNF-related apoptosis-inducing ligand-death-inducing receptor 5–mediated pathway) (19, 20) and were also implicated in the development of IAV-specific CD8+ T cell responses (42). Ly6C+ IMs were also shown to activate NK cells via an IL-18–dependent mechanism in a mouse model of HSV-2 vaginitis (41). We observed increased production of TNF by Ly6C+ IMs and early infiltration and activation of NK cells in the lungs of mice after nasal exposure to MHV-1 (Supplemental Figure 2B). We also observed increased expression of CCR7 on Ly6C+ IMs, increased ability to ingest antigens and migrate to the lung draining lymph nodes, and significantly enhanced T cell responses in the lungs to heterologous virus infections.
In summary, we demonstrate that nasal exposure to coronavirus remotely primes lung immunity in the absence of direct lung infection, and that such priming provides effective protection against subsequent lethal viral infection in the lungs. Our data reveal that early exposure to pathogens in the nose and upper airways can immunologically prepare the lungs for more rapid initiation of innate and adaptive immune response against homologous and heterologous infection. While clinical and epidemiological evidence supports the concept of a “unified airway,” the underlying pathobiology remains poorly understood. Our data implicate crosstalk between distant mucosal sites in the nose and lung, suggesting a common mucosal immune system as a mechanism connecting the nose and lungs. These findings may have important therapeutic implications for the use of nasal immunotherapy to prevent or reduce the severity of viral pneumonia, which presently causes significant morbidity and mortality and have limited therapeutic options for treatment. Use of nasal priming will need to be carefully tailored to individual patients. For example, if TNF is key for nasal priming, this approach would not be useful in patients treated with infliximab or etanercept, which inhibit TNF signaling. Further investigation not will only advance our understanding of airway mucosal immunology, but may also help open novel directions for management of lower airway diseases and to define the limitation of such nasal immunotherapy.
Methods
Mice.
BALB/c mice were purchased from the National Cancer Institute (NCI) and housed and bred in a specific pathogen–free facility at the University of Iowa, with a 12-hour light cycle. The mice used in the study were 7- to 11-week-old females. Food and water were provided ad libitum for all mice throughout the duration of the study. IFNAR–/– BALB/c mice were provided by Joan Durbin at Rutgers New Jersey Medical School, Newark, New Jersey, USA.
Viral infection.
Mouse coronavirus MHV-1 and JHM strains were propagated as previously described (9). Mouse adapted influenza virus (IAV, PR-8) was provided by Kevin Legge of the University of Iowa and prepared as described previously(43). Mouse-adapted SARS-CoV (MA15) was obtained from Kanta Subbarao (NIH, Bethesda, Maryland, USA). For intranasal infection, we first determined a volume that did not cause aspiration. BALB/c mice were intranasally treated with 1% Evans blue in PBS at the indicated volume (under light anesthesia using isoflurane) prior to euthanization 10 minutes later. BALF was obtained and assayed for Evan’s blue using a spectrophotometer. In a separate experiment, radiolabeled sulfur colloid solution (5 μl per nostril) was intranasally administered to mice under light anesthesia using isoflurane. The radioactivity was determined scintigraphically 20 minutes later. Based on the results, we used 2 μl (1 μl per nostril) containing 104 PFU MHV-1 for nasal-only inoculation after light anesthesia with isoflurane. For IT instillation, BALB/c mice were anesthetized with isoflurane and hung vertically through the upper incisors on a tower, and the tongue was gently retracted. A 50-μl viral solution was then pipetted into the hypopharynx.
Viral titration.
MHV-1 viral titers were determined by a standard plaque assay using HeLa cells stably transfected with the MHV receptor as described previously (44). SARS-CoV titers were determined using Vero E6 cells as described previously (45). Viral titers are expressed as PFU/g.
qRT-PCR.
RNA was isolated from whole mouse lung homogenized in TRIzol using a standard protocol (9). cDNA was created by normalizing each sample to 2 μg RNA. qRT-PCR was performed on a Applied Biosystems 7300 system using SYBR Green ROX qPCR Mastermix (QIAGEN). Previously described primers (9), located within Nsp12, specific for MHV-1 genomic RNA (Fwd: 5′-AGGGAGTTTGACCTTGTTCAG-3′, Rev: 5′-ATAATGCACCTGTCATCCTCG-3′) were used to measure expression of genomic RNA. Samples with Ct values at or below those from mock-infected mice were considered negative for viral genomic RNA. For cytokine measurements, whole lungs of infection and controls were used.
Antibodies and flow cytometry.
The following antibodies were used in this study: CD45 (PE/Cy7 and PerCP-Cy5.5) (cl. 30-F11, BioLegend), Ly6C (PE and FITC) (clone AL-21, BD Biosciences), CD11b (eFluor 450 and PE) (clone M1/70, eBioscience), CD3 (PE) (clone 17A2, BioLegend), CD19 (FITC and PE) (clone 6D5, BioLegend), Ly6G (PE and allophycocyanin) (clone IA8, BioLegend), SiglecF (PE) (clone E50-2440, BD Biosciences), NKp46 (PE/Cy7) (clone 29A1.4, BioLegend), F4/80 (PE), (clone BM8, BioLegend), MHCII (PerCP-Cy5.5) (M5-114.15.2, eBioscience), CD40 (allophycocyanin) (cl. 3/23, BioLegend), CD80 (allophycocyanin) (clone 2D10, BioLegend), CD86 (FITC) (clone GL1, BioLegend), CD11c (PE) (clone HL3, BD Biosciences), TNF (PE) (cl. MP6-XT22, BioLegend), CD4 (PE) (clone RM4-5, BioLegend), CD8 (FITC) (cl.53-6.7, BioLegend), IFN-γ (allophycocyanin) (clone XMG1.2, BioLegend), and CCR7 (allophycocyanin) (4B12, BioLegend). Cells were analyzed using a FACSVerse (BD). For antigen-specific T cells following SARS-CoV infection, 1 × 106 cells were cultured in a 96-well round-bottom plate in the presence of Brefeldin A and peptides corresponding to a CD8+ T cell epitope (S566) or a CD4+ T cell epitope (N353) (Bio-Synthesis).
Whole lung lavage, nasal lavage, unilateral vagotomy, single-cell suspension, and isolation of cervical lymph nodes, MLNs, and NALT.
Whole lung lavage was performed as described previously (46). For NLFs, mice were decapitated after euthanasia and exsanguination. The mandible was then removed to expose the posterior opening of the nasopharyngeal tube in the oropharynx. A 20-gauge catheter was inserted into the nasopharynx. One milliliter PBS was used to lavage the nasal cavity. Unilateral vagotomy was performed as described previously under general anesthesia (18). The NALT, cervical lymph nodes, and MLNs were isolated in standard fashion microscopically (45, 47). Lung single-cell suspensions were generated per standard protocols (48). Briefly, lungs were minced and digested in HBSS containing 0.1 mg/ml of DNase (Roche), 1 mg/ml collagenase D (Roche), 2% FCS, and 25 mM HEPES at room temperature for 30 minutes. Digested tissues were then gently dispersed and passed through a 70-μm nylon filter to prepare single-cell suspension.
Intracellular cytokine staining.
To detect TNF production by Ly6C+ IMs, mice were infected intranasally with MHV-1 (2 μl, 104 PFU) or vehicle. Two days later, mice were sacrificed, and single-cell suspensions of the lungs were prepared. The cells were treated with LPS or vehicle in the presence of GolgiPlug (BD, 1 μl/ml) for 6 hours. They were then surface stained (CD45, Ly6C, and CD11b), followed by cell fixation and permeabilization (BD Cytofix/Cytoperm). Intracellular staining was then performed using anti-TNF antibody (PE, BD Biosciences). Isotype was used for gating for each sample.
Cell migration to the draining lymph nodes.
To evaluate the migration of Ly6C+ IMs in the lungs to the MLNs, BALB/c mice were infected intranasally with MHV-1 (2 μl, 104 PFU) or vehicle. Two days later, OVA-FITC (200 μg/ 50 μl) was IT instilled into the lungs. OVA-FITC–positive Ly6C+ IMs in the MLNs were then assayed 18 hours later.
Survival, clinical scores, and lung histology.
After viral infection, mice were monitored daily for morbidity and mortality. Mice with greater than 30% weight loss were euthanized per institutional IACUC protocols. Clinical illness was determined blindly using a grading system as follows: 0, healthy; 0.5, huddling; 0.5, reduced activity; 1, inactive; 1, ruffled fur; 2, emaciated; 6, dead (49). Lung histology was performed as described previously (46).
Statistics.
Data are expressed as mean ± SEM. Two-tailed unpaired Student’s t tests were used to analyze differences in mean values between 2 groups. ANOVA was used for comparison among 3 groups. Weight loss was analyzed using repeated-measures ANOVA. Least significant difference (LSD) was used for multiple comparisons. Log-rank (Mantel-Cox) tests and Gehan-Breslow-Wilcoxon test were used to analyze differences in survival. P < 0.05 were considered significant.
Study approval.
All animal care and experimental procedures used in this study were approved by the IACUC of the University of Iowa.
Author contributions
XH, SP, and NP conceived the idea; XH, ST, RV, JA, and SP designed experiments; XH, RV, JA, and RC performed experiments; DKM performed histology analysis; XH and SP wrote the manuscript.
Supplementary Material
Acknowledgments
The authors would like to thank all members of the Perlman laboratory for discussion and technical assistance and Paul McCray for reviewing the manuscript. This work was supported in part by funding from NIH grants NIDCD 2T32DC000040-21 (XH/PI, B.J. Gantz), AAO-HNS/Cook Medical CORE grant 314113 (XH), NIAID 1R21AI096139-01A1 (XH and ST), NIH ES025198 (ST), and 1R01 NS36592 (SP).
Version 1. 06/07/2018
Electronic publication
Version 2. 06/25/2018
Rudragouda Channappanavar's name was spelled incorrectly.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2018;3(11):e99025. https://doi.org/10.1172/jci.insight.99025.
Contributor Information
Xiaoyang Hua, Email: Xiaoyang-hua@uiowa.edu.
Rahul Vijay, Email: rahul-vijay@uiowa.edu.
Jeremiah Athmer, Email: athmerjeremiah@gmail.com.
David K. Meyerholz, Email: david-meyerholz@uiowa.edu.
Nitin Pagedar, Email: nitin-pagedar@uiowa.edu.
Stephen Tilley, Email: stephen_tilley@med.unc.edu.
Stanley Perlman, Email: stanley-perlman@uiowa.edu.
References
- 1.Hens G, Hellings PW. The nose: gatekeeper and trigger of bronchial disease. Rhinology. 2006;44(3):179–187. [PubMed] [Google Scholar]
- 2.Neutra MR, Kozlowski PA. Mucosal vaccines: the promise and the challenge. Nat Rev Immunol. 2006;6(2):148–158. doi: 10.1038/nri1777. [DOI] [PubMed] [Google Scholar]
- 3.Davis SS. Nasal vaccines. Adv Drug Deliv Rev. 2001;51(1-3):21–42. doi: 10.1016/S0169-409X(01)00162-4. [DOI] [PubMed] [Google Scholar]
- 4.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157(1):121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foster JA, McVey Neufeld KA. Gut-brain axis: how the microbiome influences anxiety and depression. Trends Neurosci. 2013;36(5):305–312. doi: 10.1016/j.tins.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 6.Luna RA, Foster JA. Gut brain axis: diet microbiota interactions and implications for modulation of anxiety and depression. Curr Opin Biotechnol. 2015;32:35–41. doi: 10.1016/j.copbio.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 7.Ichinohe T, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci USA. 2011;108(13):5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith KA, Harcus Y, Garbi N, Hämmerling GJ, MacDonald AS, Maizels RM. Type 2 innate immunity in helminth infection is induced redundantly and acts autonomously following CD11c(+) cell depletion. Infect Immun. 2012;80(10):3481–3489. doi: 10.1128/IAI.00436-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fehr AR, Athmer J, Channappanavar R, Phillips JM, Meyerholz DK, Perlman S. The nsp3 macrodomain promotes virulence in mice with coronavirus-induced encephalitis. J Virol. 2015;89(3):1523–1536. doi: 10.1128/JVI.02596-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bergmann CC, Lane TE, Stohlman SA. Coronavirus infection of the central nervous system: host-virus stand-off. Nat Rev Microbiol. 2006;4(2):121–132. doi: 10.1038/nrmicro1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson KG, et al. Cutting edge: intravascular staining redefines lung CD8 T cell responses. J Immunol. 2012;189(6):2702–2706. doi: 10.4049/jimmunol.1201682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seo SU, et al. Type I interferon signaling regulates Ly6C(hi) monocytes and neutrophils during acute viral pneumonia in mice. PLoS Pathog. 2011;7(2):e1001304. doi: 10.1371/journal.ppat.1001304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goritzka M, et al. Alveolar macrophage-derived type I interferons orchestrate innate immunity to RSV through recruitment of antiviral monocytes. J Exp Med. 2015;212(5):699–714. doi: 10.1084/jem.20140825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conrady CD, Zheng M, Mandal NA, van Rooijen N, Carr DJ. IFN-α-driven CCL2 production recruits inflammatory monocytes to infection site in mice. Mucosal Immunol. 2013;6(1):45–55. doi: 10.1038/mi.2012.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olofsson PS, Rosas-Ballina M, Levine YA, Tracey KJ. Rethinking inflammation: neural circuits in the regulation of immunity. Immunol Rev. 2012;248(1):188–204. doi: 10.1111/j.1600-065X.2012.01138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mirakaj V, Dalli J, Granja T, Rosenberger P, Serhan CN. Vagus nerve controls resolution and pro-resolving mediators of inflammation. J Exp Med. 2014;211(6):1037–1048. doi: 10.1084/jem.20132103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kradin R, MacLean J, Duckett S, Schneeberger EE, Waeber C, Pinto C. Pulmonary response to inhaled antigen: neuroimmune interactions promote the recruitment of dendritic cells to the lung and the cellular immune response to inhaled antigen. Am J Pathol. 1997;150(5):1735–1743. [PMC free article] [PubMed] [Google Scholar]
- 18.Erin N, Akdas Barkan G, Harms JF, Clawson GA. Vagotomy enhances experimental metastases of 4THMpc breast cancer cells and alters substance P level. Regul Pept. 2008;151(1-3):35–42. doi: 10.1016/j.regpep.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 19.Davidson S, Crotta S, McCabe TM, Wack A. Pathogenic potential of interferon αβ in acute influenza infection. Nat Commun. 2014;5:3864. doi: 10.1038/ncomms4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herold S, et al. Lung epithelial apoptosis in influenza virus pneumonia: the role of macrophage-expressed TNF-related apoptosis-inducing ligand. J Exp Med. 2008;205(13):3065–3077. doi: 10.1084/jem.20080201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samstein M, Schreiber HA, Leiner IM, Susac B, Glickman MS, Pamer EG. Essential yet limited role for CCR2+ inflammatory monocytes during Mycobacterium tuberculosis-specific T cell priming. Elife. 2013;2:e01086. doi: 10.7554/eLife.01086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iijima N, Mattei LM, Iwasaki A. Recruited inflammatory monocytes stimulate antiviral Th1 immunity in infected tissue. Proc Natl Acad Sci USA. 2011;108(1):284–289. doi: 10.1073/pnas.1005201108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammad H, et al. Inflammatory dendritic cells — not basophils — are necessary and sufficient for induction of Th2 immunity to inhaled house dust mite allergen. J Exp Med. 2010;207(10):2097–2111. doi: 10.1084/jem.20101563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plantinga M, et al. Conventional and monocyte-derived CD11b(+) dendritic cells initiate and maintain T helper 2 cell-mediated immunity to house dust mite allergen. Immunity. 2013;38(2):322–335. doi: 10.1016/j.immuni.2012.10.016. [DOI] [PubMed] [Google Scholar]
- 25.Channappanavar R, et al. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe. 2016;19(2):181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wissinger E, Goulding J, Hussell T. Immune homeostasis in the respiratory tract and its impact on heterologous infection. Semin Immunol. 2009;21(3):147–155. doi: 10.1016/j.smim.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 27.Walzl G, Tafuro S, Moss P, Openshaw PJ, Hussell T. Influenza virus lung infection protects from respiratory syncytial virus-induced immunopathology. J Exp Med. 2000;192(9):1317–1326. doi: 10.1084/jem.192.9.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao J, et al. Intranasal treatment with poly(I•C) protects aged mice from lethal respiratory virus infections. J Virol. 2012;86(21):11416–11424. doi: 10.1128/JVI.01410-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao J, et al. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc Natl Acad Sci USA. 2014;111(13):4970–4975. doi: 10.1073/pnas.1323279111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Togias A. Rhinitis and asthma: evidence for respiratory system integration. J Allergy Clin Immunol. 2003;111(6):1171–1183. doi: 10.1067/mai.2003.1592. [DOI] [PubMed] [Google Scholar]
- 31.Braunstahl GJ, Overbeek SE, Kleinjan A, Prins JB, Hoogsteden HC, Fokkens WJ. Nasal allergen provocation induces adhesion molecule expression and tissue eosinophilia in upper and lower airways. J Allergy Clin Immunol. 2001;107(3):469–476. doi: 10.1067/mai.2001.113046. [DOI] [PubMed] [Google Scholar]
- 32.Tan WC. Viruses in asthma exacerbations. Curr Opin Pulm Med. 2005;11(1):21–26. doi: 10.1097/01.mcp.0000146781.11092.0d. [DOI] [PubMed] [Google Scholar]
- 33.Togias A. Mechanisms of nose-lung interaction. Allergy. 1999;54 Suppl 57:94–105. doi: 10.1111/j.1398-9995.1999.tb04410.x. [DOI] [PubMed] [Google Scholar]
- 34.Schuijt TJ, et al. The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut. 2016;65(4):575–583. doi: 10.1136/gutjnl-2015-309728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johansen FE, Baekkevold ES, Carlsen HS, Farstad IN, Soler D, Brandtzaeg P. Regional induction of adhesion molecules and chemokine receptors explains disparate homing of human B cells to systemic and mucosal effector sites: dispersion from tonsils. Blood. 2005;106(2):593–600. doi: 10.1182/blood-2004-12-4630. [DOI] [PubMed] [Google Scholar]
- 36.Heidegger S, et al. Virus-associated activation of innate immunity induces rapid disruption of Peyer’s patches in mice. Blood. 2013;122(15):2591–2599. doi: 10.1182/blood-2013-01-479311. [DOI] [PubMed] [Google Scholar]
- 37.Harvey BP, Gee RJ, Haberman AM, Shlomchik MJ, Mamula MJ. Antigen presentation and transfer between B cells and macrophages. Eur J Immunol. 2007;37(7):1739–1751. doi: 10.1002/eji.200636452. [DOI] [PubMed] [Google Scholar]
- 38.Bao Y, et al. Identification of IFN-γ-producing innate B cells. Cell Res. 2014;24(2):161–176. doi: 10.1038/cr.2013.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang L, et al. Interferon-γ-producing B cells induce the formation of gastric lymphoid follicles after Helicobacter suis infection. Mucosal Immunol. 2015;8(2):279–295. doi: 10.1038/mi.2014.66. [DOI] [PubMed] [Google Scholar]
- 40.Crane MJ, Hokeness-Antonelli KL, Salazar-Mather TP. Regulation of inflammatory monocyte/macrophage recruitment from the bone marrow during murine cytomegalovirus infection: role for type I interferons in localized induction of CCR2 ligands. J Immunol. 2009;183(4):2810–2817. doi: 10.4049/jimmunol.0900205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee AJ, et al. Inflammatory monocytes require type I interferon receptor signaling to activate NK cells via IL-18 during a mucosal viral infection. J Exp Med. 2017;214(4):1153–1167. doi: 10.1084/jem.20160880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aldridge JR, et al. TNF/iNOS-producing dendritic cells are the necessary evil of lethal influenza virus infection. Proc Natl Acad Sci USA. 2009;106(13):5306–5311. doi: 10.1073/pnas.0900655106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Legge KL, Braciale TJ. Lymph node dendritic cells control CD8+ T cell responses through regulated FasL expression. Immunity. 2005;23(6):649–659. doi: 10.1016/j.immuni.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 44.Khanolkar A, et al. Protective and pathologic roles of the immune response to mouse hepatitis virus type 1: implications for severe acute respiratory syndrome. J Virol. 2009;83(18):9258–9272. doi: 10.1128/JVI.00355-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vijay R, et al. Critical role of phospholipase A2 group IID in age-related susceptibility to severe acute respiratory syndrome-CoV infection. J Exp Med. 2015;212(11):1851–1868. doi: 10.1084/jem.20150632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tilley SL, et al. Retinoid-related orphan receptor gamma controls immunoglobulin production and Th1/Th2 cytokine balance in the adaptive immune response to allergen. J Immunol. 2007;178(5):3208–3218. doi: 10.4049/jimmunol.178.5.3208. [DOI] [PubMed] [Google Scholar]
- 47.Asanuma H, et al. Isolation and characterization of mouse nasal-associated lymphoid tissue. J Immunol Methods. 1997;202(2):123–131. doi: 10.1016/S0022-1759(96)00243-8. [DOI] [PubMed] [Google Scholar]
- 48.Channappanavar R, Fett C, Zhao J, Meyerholz DK, Perlman S. Virus-specific memory CD8 T cells provide substantial protection from lethal severe acute respiratory syndrome coronavirus infection. J Virol. 2014;88(19):11034–11044. doi: 10.1128/JVI.01505-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Darniot M, Pitoiset C, Petrella T, Aho S, Pothier P, Manoha C. Age-associated aggravation of clinical disease after primary metapneumovirus infection of BALB/c mice. J Virol. 2009;83(7):3323–3332. doi: 10.1128/JVI.02198-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.