Abstract
Site-1 protease (S1P), encoded by MBTPS1, is a serine protease in the Golgi. S1P regulates lipogenesis, endoplasmic reticulum (ER) function, and lysosome biogenesis in mice and in cultured cells. However, how S1P differentially regulates these diverse functions in humans has been unclear. In addition, no human disease with S1P deficiency has been identified. Here, we report a pediatric patient with an amorphic and a severely hypomorphic mutation in MBTPS1. The unique combination of these mutations results in a frequency of functional MBTPS1 transcripts of approximately 1%, a finding that is associated with skeletal dysplasia and elevated blood lysosomal enzymes. We found that the residually expressed S1P is sufficient for lipid homeostasis but not for ER and lysosomal functions, especially in chondrocytes. The defective S1P function specifically impairs activation of the ER stress transducer BBF2H7, leading to ER retention of collagen in chondrocytes. S1P deficiency also causes abnormal secretion of lysosomal enzymes due to partial impairment of mannose-6-phosphate–dependent delivery to lysosomes. Collectively, these abnormalities lead to apoptosis of chondrocytes and lysosomal enzyme–mediated degradation of the bone matrix. Correction of an MBTPS1 variant or reduction of ER stress mitigated collagen-trafficking defects. These results define a new congenital human skeletal disorder and, more importantly, reveal that S1P is particularly required for skeletal development in humans. Our findings may also lead to new therapies for other genetic skeletal diseases, as ER dysfunction is common in these disorders.
Keywords: Cell Biology, Genetics
Keywords: Genetic diseases, Glycobiology, Proteases
Site-1 protease (S1P) deficiency causes human skeletal dysplasia due to defective S1P-mediated ER-to-Golgi and Golgi-to-lysosome inter-organelle protein trafficking.
Introduction
S1P belongs to the proprotein convertase subtilisin/kexin (PCSK) family and is also known as PCSK8 and SKI-1 (1–3). It is encoded by membrane-bound transcription factor peptidase, site 1 (MBTPS1). Site-1 protease (S1P) is ubiquitously expressed in the Golgi where it functions sequentially with the site-2 protease (S2P), encoded by the membrane-bound transcription factor peptidase, site 2 (MBTPS2), to proteolytically activate unique membrane-bound latent transcription factors. A well-studied representative of this type of transcription factor is the SREBPs (SREBP-1a, -1c, and -2), which are key regulators of cholesterol and fatty acid biosynthesis and uptake (2). SREBPs are synthesized as integral membrane proteins in the endoplasmic reticulum (ER). When intracellular cholesterol levels drop, the SREBPs translocate to the Golgi, where they are sequentially cleaved by S1P and S2P to become active. Activated SREBPs then translocate to the nucleus to bind to cis-acting sterol-responsive elements required for cholesterol biosynthesis, thereby restoring cholesterol homeostasis. Similarly, S1P and S2P also process membrane-bound activating transcription factor 6 (ATF6) during ER stress response (3). When unfolded proteins accumulate in the ER, ATF6 is sequentially cleaved by S1P and S2P in the Golgi to release its cytoplasmic domain, which enters the nucleus to activate the transcription of genes encoding ER chaperones (3).
S1P may also function independently of S2P. Recently, in vitro studies showed that S1P activates GlcNAc-1-phosphotransferase (GPT) by cleaving its α/β subunit precursor (4), which is required for the mannose-6-phosphate (M6P) modification of lysosomal enzymes in the Golgi for their targeted transport to the lysosome through specific M6P receptors (5). Defective M6P modification causes the lysosomal storage diseases ML-II/III (6). However, the in vivo function of S1P in inter-organelle lysosomal enzyme trafficking is unclear.
Mice with global constitutive deficiency of S1P die prior to implantation during embryogenesis (7). Mice homozygous for floxed S1P alleles bred with mice expressing Cre recombinase under the control of an interferon-inducible MX1 promoter exhibit 70%–90% liver-specific deletion of floxed MBTPS1 alleles after induction. These mice are viable and have reduced biosynthesis of cholesterol and fatty acids (7). The role of S1P in skeletal development was first reported in the zebrafish gonzo mutant (8). Mice with osteoblast- or chondrocyte-specific deletion of S1P died during or shortly after birth, likely due to respiratory defects due to chondrodysplasia (9, 10). These lethal phenotypes indicate essential roles for S1P during development. However, how diverse functions of S1P are integrated in vivo after birth remains unclear. In addition, no human disease with S1P deficiency has been identified to date.
Here, we report a pediatric patient with skeletal dysplasia and elevated blood lysosomal enzymes. Whole-exome sequencing identified novel biallelic variants in MBTPS1. The maternal MBTPS1 variant, through creation of a rare dominant splice donor site in exon 9, resulted in a major form of loss of function and a minor form of functional MBTPS1 transcripts. Combined with the paternal nonsense mutation, the patient had about 1% of the expected functional MBTPS1 transcripts, which was sufficient for the patient to bypass intrauterine lethality. Therefore, the combination of an amorphic and a severely hypomorphic MBTPS1 mutation make this patient an excellent and rare model to study how the function of S1P is differentially required in humans. Using patient fibroblasts, patient-derived induced pluripotent stem cells (iPSCs), and in vivo models, our studies reveal insights into differential requirement of S1P in lysosomal enzyme trafficking, lipid biosynthesis, and skeletal development in humans.
Results
Unique amorphic and severely hypomorphic mutations in S1P gene were identified in a patient with skeletal dysplasia and elevated blood lysosomal enzymes.
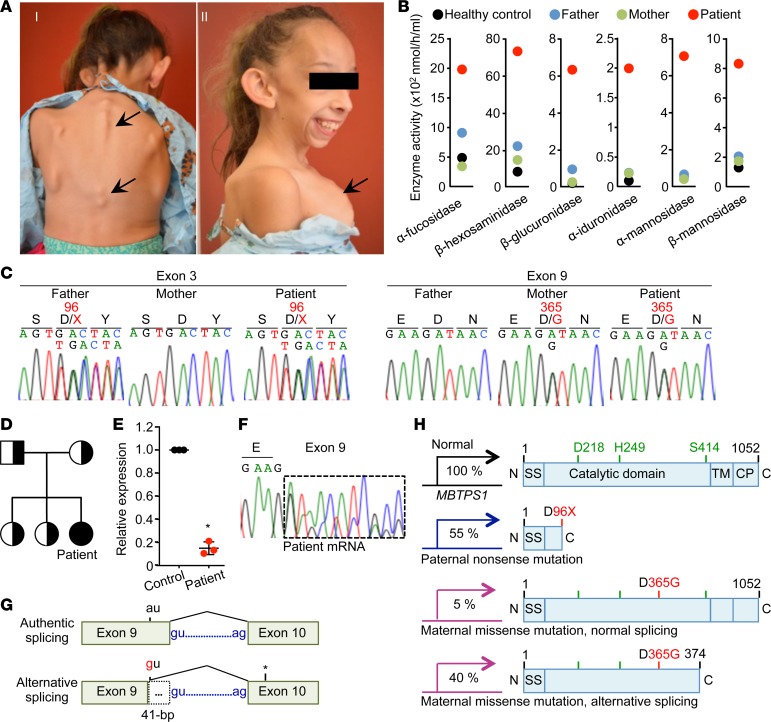
The patient was born with birth weight of 2.07 kg and no gross abnormalities. At 6 months of age, her length and weight were at 5% compared with healthy controls, after which growth in height and weight slowed down (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.121596DS1). At 2 years of age, attainment of gross motor milestones was delayed, but speech and cognitive development were normal. At 3 years of age, she was thought to have Russell-Silver syndrome and, therefore, growth hormone replacement therapy was started but discontinued after 1 year due to limited response. She had elective bilateral inguinal hernia repair at age 3.5 years. At 6 years of age, she had spondyloepiphyseal dysplasia with associated kyphosis as well as dysmorphic facial features, with prominent forehead, prominent cheekbones, and large ears (Figure 1A). Her primary clinical presentations are retarded growth with skeletal abnormalities, including a bone mineral density of approximately 60% relative to healthy controls. At age 8 years, she was found to have normal serum chemistry, liver function, and blood cell counts but displayed markedly elevated plasma levels of lysosomal enzymes compared with those of her parents and a healthy control (Figure 1B). Her parents and two sisters are healthy. Based on her clinical presentations and abnormal blood chemistry (Figure 1, A and B, and Supplemental Figure 1A), she was given a provisional diagnosis of mucolipidosis III (ML-III), a lysosomal storage disease that is caused by mutations in the genes GNPTAB or GNPTG that encode GPT subunits (11, 12). Of note, however, she lacked common features of ML-II/III (cognitive and hearing impairment or cardiac valvular defects) (4, 11–13).
Figure 1. Identification of MBTPS1 compound heterozygote variants in a patient with skeletal dysplasia and elevated circulating levels of lysosomal enzymes.
(A) The patient exhibits skeletal dysplasia with kyphoscoliosis (I) and skeletal dysplasia with dysmorphic facial features, with large ears and pectus carinatum (II). (B) Serum lysosomal enzyme activities. (C) DNA sequences of MBTPS1 exon 3 and exon 9. Letters and numbers in red indicate mutated residues and sites in the S1P protein sequence. (D) Pedigree of the family. The black circle indicates the patient; half-black square or circles indicate heterozygotes. (E) Relative MBTPS1 expression in patient B cells by qRT-PCR normalized with control B cells. A forward primer designed in exon 19 and a reverse primer designed in exon 21 of MBTPS1 were used. Mean ± SEM; n = 3, *P < 0.01, Student’s t test. Patient cells exhibited 20% MBTPS1 expression compared with irrelevant control cells. (F) MBTPS1 cDNA sequence. The dashed box indicates an aberrant transcript. (G) Diagram showing that the maternal variant creates an alternative splice donor site, resulting in the 41-bp deletion in exon 9. The asterisk indicates the premature termination created by alternative splicing causing the 41-bp deletion in exon 9. (H) Schematic domain of S1P in the patient. Green, catalytic triad; red, mutated residues. SS, signal sequence; TM, transmembrane domain; CP, cytoplasmic domain. The three different S1P variants are expressed in the patient.
Unexpectedly, Sanger sequencing of GNPTAB and GNPTG did not reveal findings consistent with ML-II/III; instead, using whole-exome sequencing, biallelic variants in MBTPS1 were identified. The paternal variant was a nucleotide duplication (NM_003791.3: c.285dupT) in exon 3, predicted to create a nonsense change (p.D96X) (Figure 1C). The maternal variant was a nucleotide substitution (NM_003791.3: c.1094A>G) in exon 9, predicted to result in a missense variant substituting aspartic acid for glycine (p.D365G) (Figure 1C).
The paternal variant (p.D96X) is expected to encode an S1P that lacks the entire catalytic domain (Figure 1, D and H). The maternal variant (p.D365G) was not located within the S1P catalytic triad (Figure 1, D and H, and Supplemental Figure 1B), and in vitro site-directed mutagenesis of MBTPS1 cDNA with the maternal c.1094A>G variant indicated that S1P harboring the D365G (S1PD365G) had normal catalytic activity (Supplemental Figure 1C). However, qRT-PCR showed an 80% reduction of MBTPS1 expression in B cells from the patient compared with a normal control (Figure 1E). To determine how the maternal c.1094A>G variant impairs MBTPS1 expression, we individually cloned and sequenced RT-PCR amplicons from the patient MBTPS1 exons 7–10 and found that the maternal c.1094A>G variant created a dominant splice donor site in exon 9, resulting in an alternatively spliced transcript with a 41-bp deletion of exon 9, leading to a loss of S414 in the catalytic triad (Figure 1, F and G, and Supplemental Figure 1D). Treatment of parental B cells with a nonsense-mediated mRNA decay (NMD) inhibitor (cycloheximide) stabilized mutant MBTPS1 transcripts, indicating that this reduced MBTPS1 expression in the patient was caused by NMD (14) (Supplemental Figure 1E). Together, the paternal and maternal variants generate only approximately 1% of the normally spliced, functional MBTPS1 (p.D365G) transcripts in the patient compared with healthy control (Supplemental Figure 1F).
S1P deficiency causes a partially defective M6P-dependent Golgi-to-lysosome transport of lysosomal enzymes.
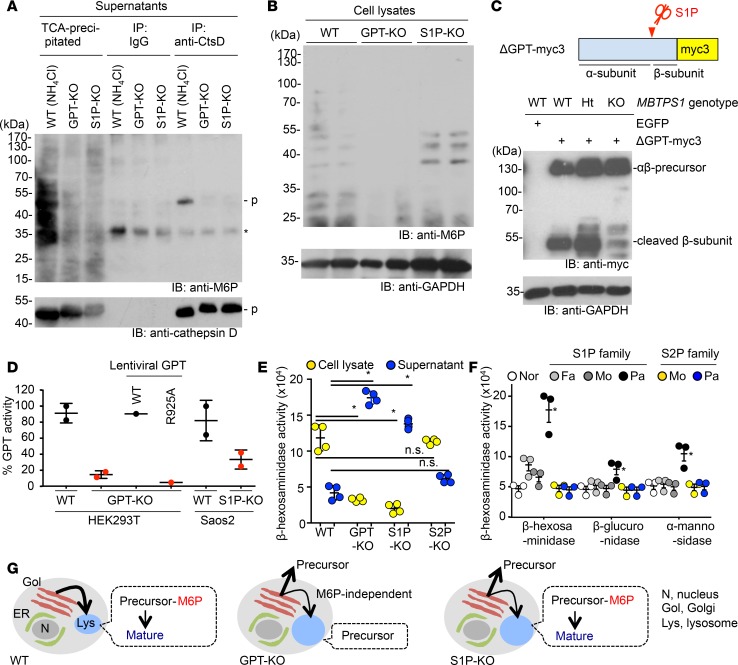
Previous in vitro studies indicate that S1P activates GPT by cleaving its α/β subunit precursor (4), which is required for the M6P modification of lysosomal enzymes in the Golgi apparatus for their targeted transport to the lysosome (5). Defective M6P modification causes the lysosomal storage diseases ML-II/III (6). Consistent with this, the patient exhibited elevated circulating lysosomal enzyme activities. We therefore evaluated patient fibroblasts for the lysosomal abnormalities typically observed in ML-II/III patient cells, such as inclusion bodies, enlarged lysosomes, increased levels of lysosomal proteins, and diffused distribution or increased secretion of lysosomal enzymes (11–13). Surprisingly, we detected neither defective M6P modification nor any of the aforementioned lysosomal abnormalities in the patient fibroblasts (Figure 2A and Supplemental Figure 2, A and B). However, once the maternal and patient fibroblasts were treated with S1P inhibitor PF-429242, elevated secretion of lysosomal enzymes and lysosomal hypertrophy were found in both cells, indicating that residually expressed S1P is sufficient to maintain lysosomal function in patient fibroblasts (Supplemental Figure 2, C and D). Given that the patient has skeletal dysplasia (Supplemental Figure 1A), we reasoned that secretory cells, such as osteoblasts or chondrocytes, might be more susceptible than fibroblasts to decreased S1P expression. To test this, we generated a Saos2 osteosarcoma cell line lacking S1P (S1P-KO cells) (15) (Supplemental Figure 3, A–E). Saos2 cells lacking GPT (GPT-KO cells) were generated as the positive control. Culture supernatants from S1P-KO and GPT-KO clones showed increased activity of the M6P-dependent lysosomal enzyme β-hexosaminidase compared with supernatants from WT cells, indicating that S1P functions in targeting lysosomal enzymes to lysosomes (Figure 2B). Cathepsin D, which is synthesized as a precursor form, is one of the M6P-modified lysosomal enzymes. Once cathepsin D precursor was transported from the Golgi apparatus to the lysosome, it was processed into its mature form (Figure 2C). Relative to WT cells, both S1P-KO and GPT-KO cells secreted high levels of the lysosomal enzyme cathepsin D precursor (55 kDa, Figure 2D, middle), supporting a default secretory pathway. The precursor form of cathepsin D was also detected in patient sera but not in maternal sera (Figure 2E). M6P-marked cathepsin D was detected in supernatants from WT cells treated with NH4Cl, which induces direct secretion of lysosomal enzymes from the Golgi apparatus, but not in untreated S1P-KO or GPT-KO cells (Figure 3A). These data indicate that the excessive secretion of cathepsin D from S1P-KO cells is due to defective M6P modification as in GPT-KO cells.
Figure 2. S1P deficiency causes a partially defective M6P-dependent Golgi-to-lysosome transport of lysosomal enzymes.
(A) Representative immunofluorescence images of fibroblasts from the patient and her parents. ML-II and ML-IIIa cells are positive controls derived from patients with these conditions. Cathepsin B, lysosome enzyme; LAMP1, lysosome marker; TO-PRO, nuclear counterstaining. Scale bar: 10 μm. The number of N indicates the exact number of samples. (B) M6P-dependent lysosomal enzyme β-hexosaminidase activity from Saos2 lysates or supernatants, as indicated. Data represent mean ± SEM; n = 3. *P < 0.05, 1-way ANOVA followed by Student’s t test. (C) Inter-organelle trafficking of cathepsin D. Without M6P modification, cathepsin D precursor is directly secreted into extracellular space. Ionomycin triggers the release of lysosomal contents through inducing the lysosomal exocytosis of matured cathepsin D. (D) Western blotting of precursor (p), intermediate (i), and mature (m) forms of cathepsin D from Saos2 lysates or supernatants, with or without ionomycin treatment. The precursor form of cathepsin D was detected in supernatant from GPT-KO and S1P-KO cells. However, upon ionomycin treatment, the mature form of cathepsin D was detected in supernatant from S1P-KO cells, indicating correct targeting of cathepsin D in lysosome in S1P-KO cells. (E) Sera from the patient (pat) and her mother (mom) were analyzed by Western blotting (top). Lysates from Saos2 cells were used as a control. The same membrane was stained with Ponceau S to confirm equivalent loading of serum proteins (bottom). (F) Immunofluorescence images of WT and mutant Saos2 cells. Insets show high-magnification images of lysosomes. Scale bars: 5 μm; 125 nm (insets). Enlarged lysosomes were found in GPT-KO and S1P-KO cells. Unlike GPT-KO cells in which cathepsin D is absent in lysosomes, cathepsin D was detected in lysosomes in S1P-KO cells. (G) Phase-contrast images of WT and mutant Saos2 cells. Arrowheads indicate inclusion bodies. Scale bar: 10 μm.
Figure 3. S1P is important but is not the sole enzyme that activates GPT.
(A) Supernatants from WT and mutant Saos2 cells were immunoprecipitated with anti-cathepsin D antibody (anti-CtsD) or normal goat IgG or precipitated with trichloroacetic acid (TCA)/acetone (input). M6P moieties were detected with anti-M6P. WT Saos2 cells were treated with NH4Cl to mimic ML-II/III cells (positive control). p, cathepsin D precursor. Asterisks indicate nonspecific bands. As expected, M6P moiety was detected on secreted cathepsin D from WT cells induced by NH4Cl treatment but not on secreted cathepsin D from untreated GPT-KO and S1P-KO cells. (B) Western blotting of M6P modification of lysates. Anti-GAPDH was used as loading control. Unlike GPT-KO cells, M6P bands were detected in lysates from S1P-KO cells. (C) Established clones were transfected with myc3-tagged GPT reporter (top). Cleavage of the reporter substrate in S1P-KO cells was considerably impaired but a small amount of cleaved reporter substrate was detected, supporting an alternative S1P-independent GPT-activating mechanism (bottom). (D) GPT activity in membrane fractions of WT cells, mutant HEK293T cells, or WT or S1P-KO Saos2 cells. Compared with WT cells, GPT-KO cells showed a substantial reduction in GPT activity, which was rescued by WT cells but not by the cleavage resistant GPT-R925A construct, which has missense mutation in the cleavage site by S1P to endow the resistance to the cleavage. Residual GPT activity was observed in S1P-KO cells, suggesting an alternative S1P-independent GPT-activating mechanism. (E) β-Hexosaminidase activity in lysates and culture supernatants from WT and mutant Saos2 cells. S2P-KO Saos2 cells do not show excessive secretion of β-hexosaminidase. Mean ± SEM for 4 independent experiments are shown. *P < 0.05, 1-way ANOVA followed by Student’s t test. (F) Serum lysosomal enzyme activities. Nor, normal individual; Fa, father; Mo, mother; Pa, patient. S2P patient does not have lysosomal abnormality. Mean ± SEM for 3 independent experiments are shown. *P < 0.05, 1-way ANOVA followed by Student’s t test. (G) Models of lysosomal enzyme trafficking in WT and mutant cells.
S1P is not the sole GPT-activating enzyme.
Unexpectedly, the mature form (28 kDa) of cathepsin D was detected in the supernatant of S1P-KO cells but not in the supernatant of GPT-KO cells upon ionomycin-induced lysosomal exocytosis, indicating that cathepsin D was partially targeted to the lysosome in S1P-KO cells (Figure 2D, right). Further, S1P-KO cells had cathepsin D in lysosomes and did not have inclusion bodies, which is similar to WT cells (Figure 2, F and G). In addition, we found that M6P modification in lysates from S1P-KO cells was similar to that from WT cells (Figure 3B), consistent with residual lysosomal targeting of M6P-dependent enzymes in S1P-KO cells. We monitored cleavage of a myc3-tagged GPT reporter substrate, which is an artificial substrate of S1P, and observed some, albeit reduced, cleavage of myc3-tagged GPT reporter upon loss of S1P, suggesting an unknown S1P-independent mechanism for residual GPT activation (Figure 3C). We also measured GPT activity in those cells, and we found that S1P-KO cells showed 30% activity compared with WT control cells (Figure 3D). In summary, our data show that S1P-KO cells had less severe lysosomal phenotypes than in GPT-KO cells, indicating that S1P is important but not the sole enzyme to proteolytically activate GPT.
S1P regulates lysosomal enzyme trafficking in vivo, and chondrocytes are a major source of circulating lysosomal enzymes in the absence of S1P.
To investigate the physiological requirements for S1P, we analyzed mice with an inducible global (Mbtps1fl/fl;CagCreERTM) or chondrocyte-specific (Mbtps1fl/fl;Col2a1CreERT2) deficiency of S1P (7, 10). Both mouse lines displayed elevated plasma lysosomal enzyme activities and bone defects, similar to the patient, whereas these phenotypes were not observed in their littermate controls. However, Mbtps1fl/fl;CagCreERTM mice, similar to the patient, did not have neurological defects, a common sign of ML-II/III (Supplemental Figure 4, A–E), which also supports a S1P-independent mechanism for GPT activation, because GPT-deficient mice are reported to have inclusion bodies in neural cells (16). The elevated plasma lysosomal enzyme activities in Mbtps1fl/fl;Col2a1CreERT2 mice indicate that chondrocytes are a major source of circulating lysosomal enzymes.
The patient has no obvious metabolic abnormalities.
In the Golgi membrane, S1P, acting sequentially with S2P encoded by MBTPS2, cleaves membrane-bound transcription factors in response to ER stress or decreased sterol metabolites. These substrates include SREBPs, which are critical for cholesterol biosynthesis, and ATF6, which is important for regulating ER stress responses (1, 7). Our data show that the patient did not have any obvious metabolic abnormalities and her lipid levels were only slightly lower compared with controls under physiological conditions (Supplemental Figure 5A). Of note, the patient has a relatively low level of HDL, suggesting an important role of S1P in regulating HDL.
We also identified a patient with an S2P mutation (Supplemental Figure 5D). Both patient study and in vitro experiments showed that the S2P pathway is not required for lysosomal enzyme trafficking (Figure 3, E–G, and Supplemental Figure 5, B and C).
S1P deficiency causes defective UPR and subsequent cell death, owing to collagen accumulation in the ER of the patient chondrocytes.
Skeletal dysplasia was the major clinical presentation of the S1P patient (Figure 4A and Supplemental Figure 1A), and it was also observed in Mbtps1-deficient mice (Supplemental Figure 4, C and D). S1P regulates ER stress responses (1, 3). We hypothesized that skeletal dysplasia was due to an impaired unfolded protein response (UPR), which is typically triggered by physiological ER stress during normal skeletal development to increase protein folding and secretory capacities, especially in secretory cells such as osteoblasts and chondrocytes (17, 18). To test this, we generated iPSCs from primary fibroblasts from the patient and her mother (Figure 4B). The patient iPSCs displayed defective osteoblast differentiation when compared with the maternal iPSCs and had impaired induced expression of UPR-related genes, such as HSPA5 and DDIT3 (19) (Supplemental Figure 6A). We used these patient- and maternal-derived iPSCs to generate teratomas (Figure 4, C and D), which contain multiple tissues, including cartilages. Chondrocytes in patient iPSC-derived teratomas displayed increased lysosomes, enlarged ER, an abnormal ER retention of collagens, and increased apoptosis compared with those in maternal iPSC-derived teratomas (Figure 4, E–G, and Supplemental Figure 6, B–D). These phenotypes were recapitulated in patient fibroblasts treated with TGF-β1, which induces ER stress (Figure 5A) (20). In addition, the size of the ER in patient fibroblasts was considerably enlarged compared with that of maternal fibroblasts (Figure 5B). These data supported the hypothesis that S1P deficiency causes defective UPR and subsequent cell death, owing to collagen accumulation in the ER.
Figure 4. S1P deficiency causes defective UPR and subsequent cell death, owing to collagen accumulation in the ER of patient chondrocytes.
(A) Hand radiographs. Arrows mark shortening in tubular bones and delayed ossification of epiphyses and carpal bones in the patient compared with a healthy girl. (B) Flow cytometric profiles of iPSCs derived from fibroblasts of the patient and her parents using antibodies against the pluripotent stem cell markers TRA-1-60 and SSEA4. Normal fibroblasts were used as negative controls. (C) Scheme of generating iPSC-derived teratomas in immune-deficient NRG mice. (D) Images of H&E-stained teratomas. Scale bar: 100 μm. (E) Representative images of teratoma tissue sections. Scale bar: 5 μm. Identical chondrocytes in the patient teratoma have enlarged calnexin+ ER and enlarged Lamp1+ lysosomes compared with maternal chondrocytes. Arrowheads indicate enlarged ER. (F) Immunofluorescence images of cartilage in teratomas. TO-PRO, nuclear staining; yellow, merged. Arrowheads indicate collagen II retained in calnexin+ ER in patient-derived cartilage. Scale bar: 10 μm. (G) Images of TUNEL staining of maternal and patient-derived teratomas. Scale bar: 100 μm. TUNEL+ apoptotic cells were detected in cartilages but not in noncartilaginous tissues in the patient teratoma. (H) Expression of COP-II vesicle-related genes in maternal and patient-derived teratoma, as measured by qRT-PCR. Each dot represents a teratoma-bearing mouse. (I) Confocal images of cartilage in teratomas. Scale bar: 20 μm; 6.67 μm (insets). Tango1 and Hsp47 were decreased in patient cartilages compared with maternal cartilages. All data are presented as mean ± SEM of at least 3 independent experiments. *P < 0.05; **P < 0.01, Student’s t test.
Figure 5. Impaired S1P-BBF2H7-Tango1 axis causes abnormal collagen retention in the ER.
(A) Quantification of mean fluorescent intensity (MFI) of immunofluorescence images of collagen I in fibroblasts treated with or without TGF-β1 treatment for 48 hours. Regardless of TGF-β1 treatment, the patient fibroblasts showed accumulation of collagen-I. (B) Transmission electron microscopy (TEM) images of maternal and patient fibroblasts. Arrowheads indicate rough ER. Enlarged rough ER was found in patient fibroblasts compared with maternal fibroblasts. Scale bar: 500 nm. (C) Expression of COP-II mega vesicle-related genes in maternal and patient-derived fibroblasts, as measured by qRT-PCR, normalized to maternal fibroblasts. (D) Expression of collagen I (COL1A1), ER stress-related genes (HSPA5 and DDIT3), and COP-II component (SEC23A) genes in maternal and patient fibroblasts, by qRT-PCR normalized to maternal fibroblasts treated with or without TGF-β1. Compared with maternal cells, patient cells showed defective induction of UPR triggered by TGF-β1 treatment. (E) Western blotting of Sec23a, Tango1, and Hsp47 in fibroblasts. Mo., maternal fibroblasts; PF, PF-429242 (S1P inhibitor). (F) Secreted procollagen I from maternal and patient fibroblasts, treated as indicated, was measured by ELISA. PF, PF-429242. S1P-dependent collagen-I secretion is shown. (G) Schematic protein domain structures of BBF2H7 and the constitutively active BBF2H7 p60 mutant. S1P cleaves the luminal domain of BBF2H7 followed by cleavage of the transmembrane domain by S2P in the Golgi apparatus, resulting in translocation of the cytoplasmic domain of BBF2H7 (BBF2H7 p60) to the nucleus to promote gene expression. (H) Expression of COL1A1, HSPA5, DDIT3, OASIS (CREB3L1), SEC23A, and COP-II–enlarging proteins (Sedlin, Tango1, and Hsp47) in BBF2H7 p60-expressing patient fibroblasts compared with uninfected cells as analyzed by quantitative RT-PCR. All data are from 3 independent experiments. *P < 0.05; **P < 0.01, Student’s t test.
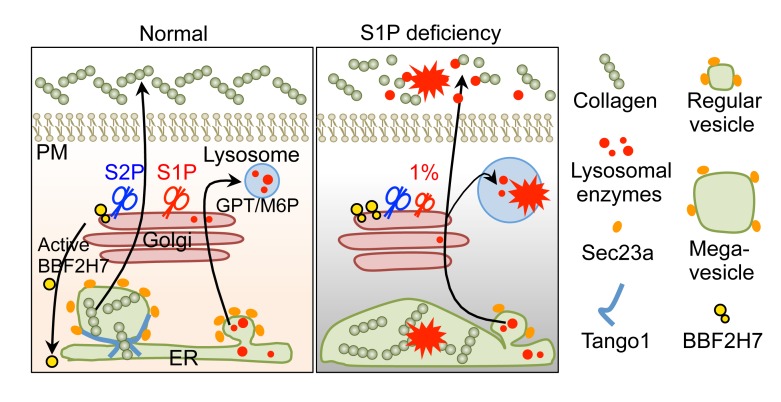
Impaired S1P-BBF2H7-Tango1 axis causes impaired mega COP-II vesicle formation essential for collagen trafficking between the ER and Golgi.
Extracellular matrix (ECM) proteins are essential for skeletal development. ECM proteins are biosynthesized in the ER and transported by coat protein complex II (COP-II) vesicles to the Golgi apparatus en route to the extracellular spaces. The COP-II vesicles are typically approximately 80 nanometers in diameter. However, collagens, which are major ECM proteins for skeletal development, are over several hundred nanometers long. Therefore, specially enlarged COP-II vesicles (mega vesicles), which are regulated by the UPR, are required to export collagens to the Golgi apparatus for secretion (10, 21). S1P proteolytically activates membrane-bound transcription factor BBF2H7, a key UPR transducer, to regulate expression of a set of genes (Sec23a, Tango1, Sedlin, and Hsp47) essential for the formation of mega vesicles in ECM-secreting cells (8, 18, 21–25). These genes were downregulated in patient teratomas and patient fibroblasts compared with controls (Figure 4H and Figure 5, C and D). As the patient fibroblasts express a residual amount of MBTPS1, we treated the cells with an S1P inhibitor, which further decreased Tango1 and Hsp47 in patient fibroblasts and impaired collagen I secretion (Figure 5, E and F). Importantly, Tango1 and Hsp47 were also decreased in cartilage in patient teratomas (Figure 4I), indicating a critical role of S1P in UPR-regulated mega vesicle formation. Overexpression of a constitutively active form of BBF2H7 (p60), which is only composed of cytoplasmic domain of BBF2H7 and, thus, cleavage by S1P is not required for the translocation of p60 into the nucleus, significantly increased the expression of genes essential for the formation of mega vesicles in patient fibroblasts (Figure 5, G and H), indicating that active BBF2H7 positively regulates these genes. Thus, impaired S1P-BBF2H7-Tango1-ER-axis-regulated mega vesicle formation underlies collagen retention in patient cells (Supplemental Figure 6E).
Abnormally secreted lysosomal enzyme–mediated ECM degradation contributes to skeletal dysplasia in the patient.
Upon ER stress, the proteolytic activation of BBF2H7 requires sequential actions of S1P and S2P in the Golgi membrane. Patients with deficiency of S2P have been reported (26). To differentiate our S1P-deficinet patient from the S2P-deficient patient, we identified a new S2P-deficient patient with a short stature (Supplemental Table 2) (26), suggesting that the skeletal dysplasia resembles the S1P-deficient patient. However, the S2P-deficient patient had no elevated levels of serum lysosomal enzymes and urine N-telopeptides, a collagen degradation product (Figure 3F and Supplemental Figure 5E). In contrast, the S1P patient has increased levels of urine N-telopeptides, suggesting abnormal ECM degradation due to abnormally secreted lysosomal enzymes with collagenase activity in the absence of S1P. Thus, these data indicate that the defective S1P-GPT-lysosome axis plays an additional contribution to the skeletal dysplasia in this patient (Figure 6F) (27).
Figure 6. Improving ER function or antisense morpholino oligo splicing therapy mitigates collagen retention in the ER of MBTPS1 mutant cells.
(A) Structure of MBTPS1 pre-mRNA in abnormal and corrected maternal transcripts. The antisense morpholino oligo (AMO) was designed to block the maternal alternative MBTPS1 splicing site. The red box indicates AMO designed to block cryptic splice site. The asterisk indicates the premature termination. AMO hampers pathogenic alternative splicing and promote correct mRNA splicing. (B) Maternal (blue) and patient (red) fibroblasts were treated with AMO for 12, 24, and 48 hours, and MBTPS1 expression was analyzed by qRT-PCR normalized to HPRT expression. Numbers indicate fold change compared with time 0. For total MBTPS1, a forward primer designed in exon 19 and a reverse primer designed in exon 21 of MBTPS1 were used. For MBTPS1 with Δ41 bp, a forward primer designed in the boundary of exon 7–8 and a reverse primer designed in the boundary of exon 9–10 of MBTPS1 were used. (C) Western blotting of endogenous BBF2H7 in AMO or control oligo-treated patient fibroblasts. PF, S1P inhibitor PF-429242. Defective S1P-dependent cleavage of BBF2H7 and improved cleavage by AMO treatment in patient fibroblasts. (D) Secreted procollagen I from maternal and patient fibroblasts, treated as indicated, was measured by ELISA. AMO treatment increased collagen I secretion in patient fibroblasts. (E) Immunofluorescence images of collagen I in fibroblasts treated with or without the PF-429242 or the PBA for 48 hours. TO-PRO, nuclear staining. Scale bar: 5 μm. Quantification of mean fluorescent intensity (MFI) of intracellular collagen I staining. A dramatic decrease in collagen I in PBA-treated patient fibroblasts was seen. (F) A model showing how S1P deficiency causes compound defects in the ER and lysosome, leading to skeletal dysplasia. All data are from at least 3 independent experiments. Mean ± SEM. *P < 0.05; **P < 0.01, Student’s t test.
Blocking the pathogenic splicing site by AMO restored ER function in patient cells.
To validate that the maternal alternative splicing site is pathogenic, we treated patient fibroblasts with an antisense morpholino oligonucleotide (AMO) (Figure 6A) (28). AMO treatment resulted in a 20-fold increase in properly spliced MBTPS1 transcripts and a substantial decrease in the 41-bp–deleted pathogenic splice variant in the patient cells compared with control oligo (Figure 6B and Supplemental Figure 7A). AMO treatment also increased the S1P-dependent activation of BBF2H7 as well as the expression of UPR- and COP-II–related genes in patient fibroblasts (Figure 6C and Supplemental Figure 7B). Importantly, AMO treatment increased secretion of collagen I in patient fibroblasts (Figure 6D). These results not only validate the disease-causing variants of MBTPS1, but also reveal a potential individualized gene therapy for this patient.
Improving ER function mitigates collagen accumulation in the ER of patient cells.
To investigate whether improving ER function mitigates the S1P-deficient cellular abnormalities, we tested the FDA-approved histone deacetylase inhibitor named PBA (sodium phenylbutyrate) (29). Our results indicate that PBA treatment diminished collagen I accumulation in patient fibroblasts (Figure 6E), suggesting a potential therapy to improve the skeletal developmental defects.
Discussion
In this paper, we report an interesting pediatric patient with skeletal dysplasia and increased circulating lysosomal enzymes resulting from biallelic MBTPS1 pathogenic variants that cause S1P deficiency. Although S1P is a critical protease for activating various membrane-bound transcription factors needed to induce physiological ER stress responses that is required for normal organ development, the skeletal dysplasia present in this patient is by far the most obvious clinical manifestation. This indicates that S1P function is especially required for skeletal development.
S1P is the only enzyme to proteolytically activate GPT based on reported in vitro studies (4). We found that S1P deficiency impaired proteolytic activation of GPT, with resultant extracellular secretion of lysosomal enzymes in cultured cells as well as in mice. However, cells with S1P deficiency had less severe lysosomal phenotypes than GPT-deficient cells. In addition, M6P modification in lysates from S1P-deficient cells was similar to those from WT cells. These data demonstrate that, although S1P plays an important role in proteolytically activating GPT (Figure 3C), it is not the sole enzyme that has this function. Thus, future studies are necessary to identify additional new protease(s).
S1P was discovered as a critical regulator for synthesis of cholesterol and fatty acids in cultured cells and in mice (1, 3). Previous studies in mice suggest that it is a potential target for lipid-lowering drugs (7), but whether humans respond to S1P deficiency in the same way as mice has been unclear, because cholesterol metabolism is different between humans and mice. In addition, the early embryonic lethality phenotype observed in S1P-deficient mice suggests potential detrimental complications of S1P blockade. Our data show that the patient did not have any obvious metabolic abnormalities and that her lipid levels were only slightly lower compared with those of controls without challenge, suggesting that the residual S1P activity is sufficient to maintain lipid homeostasis under physiological conditions.
Sec23a has been considered the target gene regulated by BBF2H7 for the formation of both regular and mega vesicles in mice (Supplemental Figure 6B) (18). In contract, Tango1 and Hsp47, regulated by BBF2H7, are known to be important in mega vesicle formation (17, 30, 31). Our results show that collagens but not regular cargo proteins, such as lysosomal enzymes, had defective ER-to-Golgi trafficking and secretion in the ER of the patient cells, suggesting that mega vesicle but not regular COP-II vesicle-mediated inter-organelle trafficking is impaired. These results demonstrate that reduction of BBF2H7-regulated critical targets, such as Tango1 and Hsp47, causes the impaired ER exporting of collagen, leading to defective skeletal development in the patient.
During ER stress, the proteolytic activation of S1P substrates, including BBF2H7, requires sequential actions of S1P and S2P in the Golgi membrane (1). The S2P-deficient patient we identified has short stature, as previously reported (Supplemental Table 2) (26), suggesting skeletal dysplasia similar to that of the S1P-deficient patient. However, the S2P-deficient patient had no elevated levels of circulating lysosomal enzymes and urine N-telopeptides, a collagen degradation product (Supplemental 5E and Supplemental Table 2), indicating that S2P function is not required for the targeted intracellular transport of lysosomal enzymes. In contrast, the S1P patient has increased levels of urine N-telopeptides, indicating an additional contribution of abnormal ECM degradation to the skeletal dysplasia due to abnormally secreted lysosomal enzymes with bone matrix–degrading activity (32). Although most of the substrates of S1P are shared with S2P, mutations in the two genes result in widely different clinical manifestations, indicating differential function of these regulatory proteases.
In summary, our data reveal a compound defect in the S1P-BBF2H7-Tango1-ER axis and S1P-GPT-lysosome axis for the pathogenesis of a new skeletal dysplasia upon loss of S1P (Supplemental Table 2). Importantly, this study provides insights into differential requirements of S1P functions in humans and suggests personalized therapies for this patient. Genetic skeletal diseases constitute a large and diverse group (33). Each individual disease is rare. Collectively, they are common (1 per 4,000 children) and have very few therapeutic options. ER dysfunction is common in these patients (33). Our findings may lead to new therapies for these genetic skeletal diseases.
Methods
Additional details are available in the Supplemental Methods.
Experimental design
The study design was centered on the functional characterization of the pathogenic variants in MBTPS1 in the patient we identified. These include in vitro and in vivo approaches to determine how the MBTPS1 pathogenic variants causes S1P deficiency as well as how S1P deficiency results in skeletal dysplasia in the patient. Patient fibroblasts, fibroblast-derived iPSCs, and mouse models were used in this study.
Cell culture
Primary fibroblasts from ML-II/IIIa patients were purchased from Coriell Cell Repositories (GM02013/ML-II, GM00113/ML-IIIa). Saos2, a human osteosarcoma cell line purchased from the ATCC collection, was grown in McCoy’s 5A supplemented with 15% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. Human primary fibroblasts were isolated from patient tissue samples and immortalized by human telomerase reverse transcriptase gene through lentiviral infection. Fibroblasts were cultured in DMEM supplemented with 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. Epstein-Barr virus–transformed (EBV-transformed) human immortalized B cells were grown in RPMI1640 supplemented with 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin.
Measurement of lysosomal enzyme activities
A fixed volume of plasma from mice and human or culture supernatant from Saos2 cells was incubated with an enzyme-specific 4-methylumbelliferone–conjugated (4-MU–conjugated) substrate for a specified length of time (27). The fluorescence released during the enzymatic reaction was measured using a FLUOstar Omega microplate reader (BMG Labtech). As a positive control for β-hexosaminidase activity in the supernatant of cultured cells, ionomycin was used to stimulate suspended Saos2 cells for 10 minutes. After centrifugation with 500 g for 3 minutes, supernatant from Saos2 cells was collected for the assay.
Prediction of the catalytic domain structure of human S1P
The predicted catalytic domain of human S1P was built using homology-based modeling in SWISS-MODEL (https://swissmodel.expasy.org/). The molecular visualization system PyMOL (open-source software published by DeLano Scientific) was used to depict the structures.
Genetic mutation analysis in this family
Direct sequencing of PCR amplicons from genomic DNA extracted from EBV-mediated immortalized human B cells was performed to confirm germline mutation of MBTPS1 using an ABI 3730 capillary sequencer (Applied Biosystems). The sequences were analyzed using MacVector software (MacVector Inc.). For analysis of alternative splicing, PCR amplicons containing exon 9 from cDNA synthesized from patient-derived immortalized B cells were subcloned into pcDNA3.1(+), and then individual clones were sequenced.
Gene expression analyses with qPCR
Gene expression levels in the patient and irrelevant healthy control immortalized B cells or immortalized fibroblasts were analyzed by qPCR using primers listed in Supplemental Table 3. Total RNA extracted from the cells using Trizol was converted to cDNA using the M-MLV Reverse Transcriptase system. PCR analysis of cDNA was performed with the SYBR Green qPCR system using a CFX96 Real-time system instrument (Bio-Rad). To test for NMD, treatment with cycloheximide (100 μM) was performed by incubating patient’s parental immortalized B cells for 6 hours, and then transcripts were analyzed by direct sequencing.
Plasmids
The C-terminal myc-FLAG–tagged human S1P in the expression vector pCMV6 was purchased from OriGene. S414A- and D365G-carrying S1P expression vectors and R925A-carrying a GPT α/β subunit precursor were generated by site-directed mutagenesis. A truncated α/β subunit precursor lacking amino acids 431–819, designated as myc3-tagged GPT reporter, was generated using the original GPT α/β-subunit precursor tagged with C-terminal myc3 as a template for inverse PCR (4). C-terminal myc3-tagged BBF2H7 was subcloned into pcDNA3.1(+). C-terminal myc-FLAG–tagged human S1P, C-terminal myc3-tagged α/β subunit precursor, and C-terminal myc3 tagged BBF2H7 p60 (residue 1–377 amino acids) were subcloned into pHIV-ZsGreen for lentivirus transduction system (18) (a gift from Bryam Welm, University of Utah, Salt Lake City, Utah, USA).
Transfection
CHO-7 and SRD-12B cells were transfected with myc3-tagged GPT reporter and with each S1P expression vector at a ratio of 100:1 using Fugene6 transfection reagent. After 48 hours, transfection efficiency was determined by EGFP expression with flow cytometry. Infection-competent lentivirus was generated from HEK293T cells transfected with expression vectors pLV-HELP and pMD2.G using PEImax transfection reagent (Polysciences Inc.). After 48 hours, lentivirus-containing culture supernatant was collected, and then cells of interest were infected and sorted based on ZsGreen expression.
Western blotting
Protein extracts from cultured cells or tissues were prepared using a 1% Triton X-100–based cell lysis buffer. Lysates were applied to an 8%–12 % SDS-PAGE gel. Separated proteins were transferred onto an Immobilon-P membrane (Millipore). After blots were blocked, the membranes were incubated with antibodies and were then developed with ECL systems. Exposed X-ray films were scanned and analyzed with ImageJ software (NIH). For protein extraction from cartilage from mouse ribs, proteins from frozen tissues were extracted into 8 M urea lysis buffer using Bead-beater (34) (Biospec Products Inc.). For detecting endogenous GPT in immortalized B cells, 1.5 × 107 cells were suspended in hypotonic lysis buffer and then ultrasonicated for 30 seconds. After removing debris by spinning at 1,000 g for 3 minutes, supernatant was further ultracentrifuged at 100,000 g for 2 hours. The membrane fractions were solubilized with 3% Triton X-100–containing lysis buffer. 150 μg protein was analyzed by Western blotting.
Immunoprecipitation
The supernatants collected from Saos2 cells cultured in serum-free conditions for 2 days were concentrated 10-fold using Amicon filters, mixed with SDS (final concentration of 2.86%), and then boiled for 5 minutes. After a 10-fold dilution with RIPA buffer, goat anti-cathepsin D (4 μg) and protein-G beads were added for the immunoprecipitation. Immunoprecipitates were eluted with SDS sample buffer and were then analyzed with a nonreducing SDS/PAGE gel. Trichloroacetic acid (TCA)/acetone precipitation was performed to prepare the input positive control. Briefly, 100 μl 100% TCA was added to 900 μl culture medium, and the sample was then incubated for 1 hour on ice. After centrifugation at 15,000 g for 20 minutes, pellets were washed once with cold acetone (Fisher Scientific). Finally, dried washed pellets were solubilized in 1× SDS sample buffer using ultrasonication.
CRISPR/Cas9
S1P-KO, GPT-KO, and S2P-KO cells were generated with CRISPR/Cas9 technology. Briefly, sgRNA oligonucleotides targeting the coding region of individual genes were designed using CHOPCHOP (http://chopchop.cbu.uib.no) and were subcloned into the Bbs1 site of the CRISPR plasmid pSpCas9 (BB)-2A-GFP (Addgene, plasmid 48138) (15). After transfection into Saos2 and HEK293T cells, GFP-positive cells were sorted into a 96-well plate at a density of 1 cell per well with a MoFLO cell sorter (Beckman Coulter). After the colonies had expanded to 50% confluency, the genomic DNA was extracted to perform genotyping PCR followed by direct DNA sequencing. Once S1P-KO clones were established based on genotyping, a cholesterol auxotrophy assay for survival and a reporter assay based on transfecting myc3-tagged GPT reporter were performed to confirm S1P deficiency (3, 35). Established S1P-KO and S2P-KO clones were confirmed to be cholesterol auxotrophic. Briefly, cells were cultured in serum-free OPTI-MEM1 medium with or without 50 μM sodium mevalonate, 20 μM sodium oleate, 5 μg/ml cholesterol, and 1% fatty acid-free BSA for 3 days followed by staining with crystal violet. GPT deficiency was validated by Western blotting of the membrane fraction using an anti–α subunit specific antibody (catalog sc-107561, Santa Cruz Biotech).
Generation of iPSCs
Primary skin fibroblasts derived from the patient and her parents were transfected with the Human iPS Cell Reprogramming Episomal Plasmid (ALSTEM) (36) according to the manufacturer’s protocol using a Nucleofector II instrument (Lonza). The single colonies were isolated, expanded, and then validated as iPSCs by staining with antibodies against SSEA4 (catalog MC-813-70, STEMCELL Technologies) and TRA-1-60 (catalog TRA-1-60R, STEMCELL Technologies).
Immunofluorescence
Cells plated on a glass coverslip were treated with 10 ng/ml TGF-β1 (Peprotech), 5–10 μM PF-429242, or 10 mM PBA for 48 hours. Then, cells were fixed with cold 100% methanol (Fisher Scientific) for 5 minutes at –20°C. After blocking with 1% BSA in PBS-T, the cells were incubated with primary antibodies for 2 hours and then with fluorescence-conjugated secondary antibodies for 1 hour. Counterstaining was performed using DAPI or TO-PRO3. Tissue sections were incubated with primary antibodies overnight at 4°C. Secondary antibodies were added for 1 hour at room temperature (37). Primary antibodies were against cathepsin D (catalog sc-6488), cathepsin B (catalog sc-13985), calnexin (catalog sc-23954), Sec31a (catalog sc-376587), Hsp47 (catalog sc-5293), FLAG (catalog sc-166355), myc (catalog sc-789) and BBF2H7 (catalog sc-69372, Santa Cruz Biotech); LAMP1 (catalog 328602, Biolegend); collagen-II (catalog MA5-13026, Thermo Fisher Scientific); calnexin (catalog GTX109669) and Sec23a (catalog GTX109488, GeneTex Inc.); Tango1 (catalog HPA055922, MilliporeSigma); and collagen I (catalog ab34710) and collagen X (catalog ab58632, Abcam). Secondary antibodies were conjugated with DyLight 488, Alexa Fluor 555, or DyLight 649 (Jackson ImmunoResearch). The TUNEL-positive apoptotic cells were stained using the In Situ Cell Death Detection Kit, Fluorescein (MilliporeSigma). Samples were analyzed by confocal laser scanning microscopy using a Nikon C1 scanning head mounted on a Nikon ECLIPSE 2000U inverted microscope (Plan Apochromat dry objective lens, ×20, NA 0.75; Nikon Instruments Inc.) or a superresolution microscope (DeltaVision, OMX-SR).
Microscopy
For histology, femurs and brains from mice and human teratomas were fixed in 4% paraformaldehyde overnight at 4°C and were then washed and embedded in paraffin. Sections (5 μm) were cut using a microtome and then stained with H&E. For confocal microscopy of cryosections, human teratomas were fixed in 4% paraformaldehyde overnight at 4°C, washed in PBS, cryoprotected in 20% sucrose in PBS at 4°C overnight, embedded in 50% tissue freezing medium/50% OCT and cryosectioned (20- to 30-μm sections).
In vitro differentiation of osteoblasts from human iPSCs
Maternal and patient-derived iPSCs were seeded onto Matrigel-coated 6-well plates in the presence of Y-27632 (Cayman Chemical) and incubated at 37°C in a 5% CO2 incubator. The next day, mTeSR1 medium was replaced with fresh 10% FBS-containing D-MEM medium supplemented with 50 μg/ml ascorbic acid, 10 mM β-glycerophosphate (Santa Cruz Biotech), and 100 nM dexamethasone. This osteogenic medium was changed everyday without harvesting and reseeding. After 2 weeks, the cells were either stained with Alizarin Red S or the RNA was extracted for qRT-PCR (17).
AMO
The AMO (5′-AGAAAAGCGGGCGATGTTAcCTTCA-3′ [the lowercase “c” represents the mutated base compared to healthy control]) against exon 9 in the maternal mutant transcript containing cryptic splice donor site was designed (Gene Tools Inc.) (38). Forty thousand fibroblasts were seeded onto a 24-well plate. Introduction of AMO (10 μM) in maternal and patient fibroblasts was carried out using the Endo-Porter (6 μM) peptide delivery system (Gene Tools Inc.). After 12, 24, and 48 hours, RNA was extracted for quantitative RT-PCR as well as cloning of MBTPS1 cDNA. The following primer pair was used to quantify a MBTPS1 transcript with 41-bp deletion in exon 9: forward primer targeting the boundary exons 7 and 8, 5′-CCGTTTGTTGACAAGGTGTG-3′; reverse primer targeting exon 10, 5′-CTGGTAGCTCCTTCAAAGTC-3′.
Measurement of secreted procollagen I
Supernatant from cultured fibroblasts was collected at 48 hours after treatment with AMO and was then diluted 500-fold with PBS (39). Secreted procollagen 1a in diluted samples was measured with an ELISA kit according to the manufacturer’s instructions (R&D Systems).
Other reagents
4-MU N-acetyl-β-D-glucosaminide, cycloheximide, trichloroacetic acid, PBA, BSA, mevalonate, oleate, cholesterol, fatty acid–free BSA, crystal violet, alizarin red S, ascorbic acid, and dexamethasone were purchased from MilliporeSigma. Trizol and protein G magnetic Dynabeads were purchased from Invitrogen. The M-MLV Reverse Transcriptase system, ECL systems, DAPI, and TO-PRO3 were purchased from Thermo Fisher Scientific. The SYBR Green qPCR system was purchased from Bio-Rad. Fugene6 transfection reagent was purchased from Promega. A myc-His–tagged, single-chain variable fragment (scFv) against M6P residues (scFv M6P-1) was purchased from Ascenion GmbH.
Animal models
Teratoma formation assay.
Human iPSCs derived from the patient and her parents were grown to near confluence, treated with Accutase (STEMCELL Technologies), harvested in PBS, and then resuspended in 30% Matrigel-containing DMEM-F12 (Corning). Immune-deficient NRG mice (either sex, 6–8 weeks of age) were anesthetized and then subcutaneously injected with approximately 1 × 106 to 5 × 106 cells in 200 μl/injection site. Mice were injected in the dorsolateral area into the subcutaneous space on both sides. After 7–10 weeks, mice were euthanized by CO2 exposure, and then the teratomas were dissected.
S1P-deficient mice.
To generate mice with global inducible deletion of S1P (Mbtps1f/f;CagCreERTM), Mbtps1f/f mice (provided by Jay Horton, University of Texas Southwestern Medical Center, Dallas, Texas, USA) (7) were crossed with the CagCreERTM-transgenic mice (B6.Cg-Tg(CAG-cre/Esr1*)5Amc/J, Jackson Laboratories). To induce postnatal deletion of S1P, tamoxifen (MP Biomedicals) was dissolved in ethanol/sunflower oil (1:9) and administered orally (p.o.) (20 μg per day) to pups from P1 to P7. Adult deletion was accomplished by administering tamoxifen p.o. (500 μg per day) every other day for 4 times. WT littermates (Mbtps1f/w;CagCre or Mbtps1f/f) treated with the same regimen were used as controls. Mice deficient for S1P in chondrocytes (Mbtps1f/f;Col2a1CreERT2) were generated by crossing Mbtps1f/f mice with Col2a1CreERT2-transgenic mice. All mice were maintained on a C57BL/6J background. Mice were bred and maintained in specific pathogen–free AAALAC-accredited animal facilities. Both sexes (6–12 weeks of age) were used for experiments. Mice were genotyped routinely by PCR assay on genomic DNA isolated from tail clips.
Statistics
Statistical tests were performed using Prism software (GraphPad). Two-sided, two-tailed Student’s t tests were performed to assess the statistical significance of differences between 2 groups after the data were confirmed to fulfill the criteria of normal distribution and equal variance. One-way analysis of variance was used to analyze the significance of differences among 3 or more groups. Differences were considered statistically significant at P < 0.05.
Study approval
Animal studies were conducted with protocols approved by the Institutional Animal Care and Use Committee of the Oklahoma Medical Research Foundation. The patient study was approved by Institutional Review Boards of the University of Oklahoma Health Sciences Center and the Oklahoma Medical Research Foundation. Written informed consent was received from the patient’s parents prior to inclusion in the study, and written informed consent was provided for use of photographs of the patient in the manuscript.
Author contributions
KJW performed medical evaluations of the patients. KJW and PMG discovered the MBTPS1 mutations. YK, JF, HW, CH, JMM, JS, RS, SC, LP, TY, GW, S. Macwana, S. McGee, JG, and DP performed experiments. YK, JF, HW, KW, PMG, and LX designed the research and analyzed data. SL, CG, KF, JAJ, CR, and RPM provided advice, reagents, or technical expertise. YK and LX wrote the manuscript with input from the coauthors.
Supplementary Material
Acknowledgments
We thank Joseph Goldstein, Jay Horton, Linda Sandell, and Thomas Braulke for providing reagents, cell lines, or mice; Jeffery Esko, Paul Kincade, Christopher West, and Manu Nair for advice and support; and Thomas Lewis, Sowmya Krishnan, and Golder Wilson for patient care. This work was funded by grants from the NIH (GM114731, HL128390, HD083418, AR066590, GM110766, AR063124, and AR056360), Oklahoma Applied Research Support (AR16-019), an Investigator-Initiated Research Award from Pfizer (study ID WI204780), the Japanese Society for the Promotion of Science Postdoctoral Fellowships for Research Abroad, the National Natural Science Foundation of China (81520108005, 81470825, 81370617), the Jiangsu Provincial Special Program of Medical Science (BL2012005), Jiangsu Province’s Key Medical Center (ZX201102), and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Version 1. 07/26/2018
Electronic publication
Footnotes
KF’s present address is: Department of Biomedical Sciences, Chubu University College of Life and Health Sciences, Kasugai, Aichi, Japan.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2018;3(14):e121596. https://doi.org/10.1172/jci.insight.121596.
Contributor Information
Yuji Kondo, Email: yuji-kondo@omrf.org.
Jianxin Fu, Email: jianxin-fu@omrf.org.
Hua Wang, Email: Hua-wang@ouhsc.edu.
Christopher Hoover, Email: Christopher-Hoover@omrf.org.
Richard Steet, Email: rsteet@ccrc.uga.edu.
Debabrata Patra, Email: patrad@wudosis.wustl.edu.
Jianhua Song, Email: songj@omrf.org.
Laura Pollard, Email: lpollard@ggc.org.
Sara Cathey, Email: scathey@ggc.org.
Tadayuki Yago, Email: tadayuki-yago@omrf.org.
Graham Wiley, Email: Graham-Wiley@omrf.org.
Samuel McGee, Email: sam-mcgee@omrf.org.
Shibo Li, Email: Shibo-Li@ouhsc.edu.
Courtney Griffin, Email: courtney-griffin@omrf.org.
Koichi Furukawa, Email: koichi@med.nagoya-u.ac.jp.
Judith A. James, Email: Judith-James@omrf.org.
Changgeng Ruan, Email: ruanchanggeng@suda.edu.cn.
Rodger P. McEver, Email: rodger-mcever@omrf.org.
Patrick M. Gaffney, Email: gaffneyp@omrf.org.
Lijun Xia, Email: lijun-xia@omrf.org.
References
- 1.Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89(3):331–340. doi: 10.1016/S0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 2.Wang X, Sato R, Brown MS, Hua X, Goldstein JL. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell. 1994;77(1):53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- 3.Ye J, et al. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6(6):1355–1364. doi: 10.1016/S1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 4.Marschner K, Kollmann K, Schweizer M, Braulke T, Pohl S. A key enzyme in the biogenesis of lysosomes is a protease that regulates cholesterol metabolism. Science. 2011;333(6038):87–90. doi: 10.1126/science.1205677. [DOI] [PubMed] [Google Scholar]
- 5.Baranski TJ, Faust PL, Kornfeld S. Generation of a lysosomal enzyme targeting signal in the secretory protein pepsinogen. Cell. 1990;63(2):281–291. doi: 10.1016/0092-8674(90)90161-7. [DOI] [PubMed] [Google Scholar]
- 6.Steet RA, et al. A splicing mutation in the alpha/beta GlcNAc-1-phosphotransferase gene results in an adult onset form of mucolipidosis III associated with sensory neuropathy and cardiomyopathy. Am J Med Genet A. 2005;132A(4):369–375. doi: 10.1002/ajmg.a.30498. [DOI] [PubMed] [Google Scholar]
- 7.Yang J, Goldstein JL, Hammer RE, Moon YA, Brown MS, Horton JD. Decreased lipid synthesis in livers of mice with disrupted site-1 protease gene. Proc Natl Acad Sci USA. 2001;98(24):13607–13612. doi: 10.1073/pnas.201524598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schlombs K, Wagner T, Scheel J. Site-1 protease is required for cartilage development in zebrafish. Proc Natl Acad Sci USA. 2003;100(24):14024–14029. doi: 10.1073/pnas.2331794100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Achilleos A, et al. MBTPS1/SKI-1/S1P proprotein convertase is required for ECM signaling and axial elongation during somitogenesis and vertebral development†. Hum Mol Genet. 2015;24(10):2884–2898. doi: 10.1093/hmg/ddv050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patra D, et al. Site-1 protease is essential for endochondral bone formation in mice. J Cell Biol. 2007;179(4):687–700. doi: 10.1083/jcb.200708092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kudo M, Brem MS, Canfield WM. Mucolipidosis II (I-cell disease) and mucolipidosis IIIA (classical pseudo-hurler polydystrophy) are caused by mutations in the GlcNAc-phosphotransferase alpha / beta -subunits precursor gene. Am J Hum Genet. 2006;78(3):451–463. doi: 10.1086/500849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tiede S, et al. Mucolipidosis II is caused by mutations in GNPTA encoding the alpha/beta GlcNAc-1-phosphotransferase. Nat Med. 2005;11(10):1109–1112. doi: 10.1038/nm1305. [DOI] [PubMed] [Google Scholar]
- 13.Tiede S, Muschol N, Reutter G, Cantz M, Ullrich K, Braulke T. Missense mutations in N-acetylglucosamine-1-phosphotransferase alpha/beta subunit gene in a patient with mucolipidosis III and a mild clinical phenotype. Am J Med Genet A. 2005;137A(3):235–240. doi: 10.1002/ajmg.a.30868. [DOI] [PubMed] [Google Scholar]
- 14.Rodríguez-Pascau L, Coll MJ, Vilageliu L, Grinberg D. Antisense oligonucleotide treatment for a pseudoexon-generating mutation in the NPC1 gene causing Niemann-Pick type C disease. Hum Mutat. 2009;30(11):E993–E1001. doi: 10.1002/humu.21119. [DOI] [PubMed] [Google Scholar]
- 15.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kollmann K, et al. Lysosomal dysfunction causes neurodegeneration in mucolipidosis II ‘knock-in’ mice. Brain. 2012;135(Pt 9):2661–2675. doi: 10.1093/brain/aws209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishikawa T, et al. UPR transducer BBF2H7 allows export of type II collagen in a cargo- and developmental stage-specific manner. J Cell Biol. 2017;216(6):1761–1774. doi: 10.1083/jcb.201609100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saito A, et al. Regulation of endoplasmic reticulum stress response by a BBF2H7-mediated Sec23a pathway is essential for chondrogenesis. Nat Cell Biol. 2009;11(10):1197–1204. doi: 10.1038/ncb1962. [DOI] [PubMed] [Google Scholar]
- 19.Yamamoto K, et al. Direct conversion of human fibroblasts into functional osteoblasts by defined factors. Proc Natl Acad Sci USA. 2015;112(19):6152–6157. doi: 10.1073/pnas.1420713112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim SI, Na HJ, Ding Y, Wang Z, Lee SJ, Choi ME. Autophagy promotes intracellular degradation of type I collagen induced by transforming growth factor (TGF)-β1. J Biol Chem. 2012;287(15):11677–11688. doi: 10.1074/jbc.M111.308460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishikawa Y, Ito S, Nagata K, Sakai LY, Bächinger HP. Intracellular mechanisms of molecular recognition and sorting for transport of large extracellular matrix molecules. Proc Natl Acad Sci USA. 2016;113(41):E6036–E6044. doi: 10.1073/pnas.1609571113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Venditti R, et al. Sedlin controls the ER export of procollagen by regulating the Sar1 cycle. Science. 2012;337(6102):1668–1672. doi: 10.1126/science.1224947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kondo S, et al. BBF2H7, a novel transmembrane bZIP transcription factor, is a new type of endoplasmic reticulum stress transducer. Mol Cell Biol. 2007;27(5):1716–1729. doi: 10.1128/MCB.01552-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masago Y, et al. The molecular chaperone Hsp47 is essential for cartilage and endochondral bone formation. J Cell Sci. 2012;125(Pt 5):1118–1128. doi: 10.1242/jcs.089748. [DOI] [PubMed] [Google Scholar]
- 25.Wilson DG, et al. Global defects in collagen secretion in a Mia3/TANGO1 knockout mouse. J Cell Biol. 2011;193(5):935–951. doi: 10.1083/jcb.201007162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindert U, et al. MBTPS2 mutations cause defective regulated intramembrane proteolysis in X-linked osteogenesis imperfecta. Nat Commun. 2016;7:11920. doi: 10.1038/ncomms11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petrey AC, et al. Excessive activity of cathepsin K is associated with cartilage defects in a zebrafish model of mucolipidosis II. Dis Model Mech. 2012;5(2):177–190. doi: 10.1242/dmm.008219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Osorio FG, et al. Splicing-directed therapy in a new mouse model of human accelerated aging. Sci Transl Med. 2011;3(106):106ra107. doi: 10.1126/scitranslmed.3002847. [DOI] [PubMed] [Google Scholar]
- 29.Rishikof DC, Ricupero DA, Liu H, Goldstein RH. Phenylbutyrate decreases type I collagen production in human lung fibroblasts. J Cell Biochem. 2004;91(4):740–748. doi: 10.1002/jcb.10742. [DOI] [PubMed] [Google Scholar]
- 30.Maeda M, Saito K, Katada T. Distinct isoform-specific complexes of TANGO1 cooperatively facilitate collagen secretion from the endoplasmic reticulum. Mol Biol Cell. 2016;27(17):2688–2696. doi: 10.1091/mbc.e16-03-0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saito K, et al. TANGO1 facilitates cargo loading at endoplasmic reticulum exit sites. Cell. 2009;136(5):891–902. doi: 10.1016/j.cell.2008.12.025. [DOI] [PubMed] [Google Scholar]
- 32.Kollmann K, et al. Decreased bone formation and increased osteoclastogenesis cause bone loss in mucolipidosis II. EMBO Mol Med. 2013;5(12):1871–1886. doi: 10.1002/emmm.201302979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Briggs MD, Bell PA, Wright MJ, Pirog KA. New therapeutic targets in rare genetic skeletal diseases. Expert Opin Orphan Drugs. 2015;3(10):1137–1154. doi: 10.1517/21678707.2015.1083853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bergstrom K, et al. Defective intestinal mucin-type O-glycosylation causes spontaneous colitis-associated cancer in mice. Gastroenterology. 2016;151(1):152–164.e11. doi: 10.1053/j.gastro.2016.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rawson RB, DeBose-Boyd R, Goldstein JL, Brown MS. Failure to cleave sterol regulatory element-binding proteins (SREBPs) causes cholesterol auxotrophy in Chinese hamster ovary cells with genetic absence of SREBP cleavage-activating protein. J Biol Chem. 1999;274(40):28549–28556. doi: 10.1074/jbc.274.40.28549. [DOI] [PubMed] [Google Scholar]
- 36.Okita K, et al. A more efficient method to generate integration-free human iPS cells. Nat Methods. 2011;8(5):409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- 37.Herzog BH, et al. Podoplanin maintains high endothelial venule integrity by interacting with platelet CLEC-2. Nature. 2013;502(7469):105–109. doi: 10.1038/nature12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Du L, Pollard JM, Gatti RA. Correction of prototypic ATM splicing mutations and aberrant ATM function with antisense morpholino oligonucleotides. Proc Natl Acad Sci USA. 2007;104(14):6007–6012. doi: 10.1073/pnas.0608616104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Delitto D, et al. Human pancreatic cancer cells induce a MyD88-dependent stromal response to promote a tumor-tolerant immune microenvironment. Cancer Res. 2017;77(3):672–683. doi: 10.1158/0008-5472.CAN-16-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.