Abstract
Redundancy and compensation provide robustness to biological systems but may contribute to therapy resistance. Both tumor-associated macrophages (TAMs) and Foxp3+ regulatory T (Treg) cells promote tumor progression by limiting antitumor immunity. Here we show that genetic ablation of CSF1 in colorectal cancer cells reduces the influx of immunosuppressive CSF1R+ TAMs within tumors. This reduction in CSF1-dependent TAMs resulted in increased CD8+ T cell attack on tumors, but its effect on tumor growth was limited by a compensatory increase in Foxp3+ Treg cells. Similarly, disruption of Treg cell activity through their experimental ablation produced moderate effects on tumor growth and was associated with elevated numbers of CSF1R+ TAMs. Importantly, codepletion of CSF1R+ TAMs and Foxp3+ Treg cells resulted in an increased influx of CD8+ T cells, augmentation of their function, and a synergistic reduction in tumor growth. Further, inhibition of Treg cell activity either through systemic pharmacological blockade of PI3Kδ, or its genetic inactivation within Foxp3+ Treg cells, sensitized previously unresponsive solid tumors to CSF1R+ TAM depletion and enhanced the effect of CSF1R blockade. These findings identify CSF1R+ TAMs and PI3Kδ-driven Foxp3+ Treg cells as the dominant compensatory cellular components of the immunosuppressive tumor microenvironment, with implications for the design of combinatorial immunotherapies.
Keywords: Immunology, Oncology
Keywords: Cancer immunotherapy, Macrophages, T cells
Compensatory immunosuppression by CSF1R+ tumor-associated macrophages and PI3Kδ-driven Foxp3+ regulatory T cells mediates resistance to cancer immune monotherapy.
Introduction
Tumors grow in immunocompetent hosts despite the ability of the adaptive immune system to recognize and kill cancer cells. In part this is attributable to the phenomenon of immunosuppression. Tumor-associated macrophages (TAMs) are among the most abundant nontransformed cell types in solid cancers (1–4), and growing evidence suggests that TAMs can promote cancer progression and therapeutic resistance in a wide range of human malignancies (5–15). TAMs can be acutely targeted via the inhibition of colony-stimulating factor-1 (CSF1 or M-CSF) or its receptor, CSF1R (CD115), and in preclinical studies, CSF1/CSF1R inhibitors reduce tumor growth in murine tumor models (15–20). Further, CSF1 inhibitors are currently being tested in clinical trials (21), of which the most advanced is the small-molecule CSF1R inhibitor PLX3397 (22). Although early preclinical data suggested good tolerability and promising effects, CSF1R inhibitors have shown very limited antitumor effects in patients (23). Further, blocking the CSF1 pathway can also cause systemic effects not limited to macrophage depletion, such as rebound hematopoiesis. The current lack of understanding of the mechanism that regulates CSF1-driven tumor progression raises concerns regarding the potential clinical utility of these therapeutic strategies.
Regulatory T cells (Treg cells) are a subpopulation of lymphocytes that are critical in maintaining tolerance to self-antigens and innocuous foreign antigens under physiological conditions, but can also be co-opted by tumors to escape immunity (24). Phosphoinositide 3-kinase δ (PI3Kδ) is abundant in lymphocytes (25) and is activated by antigen, cytokine, and growth factor receptors (26). Recent evidence has shown that genetic inactivation of PI3Kδ in mice protects against a wide range of solid tumors (27). This immunomodulatory effect was due to the inactivation of PI3Kδ in CD4+Foxp3+ Treg cells, unleashing CD8+ cytotoxic T lymphocytes which could then induce tumor regression (27). The PI3Kδ inhibitor, idelalisib (Zydelig, Gilead) has proven highly effective for the treatment of chronic lymphocytic leukemia (26), and exerts its main effect by blocking the interactions between lymphocytic leukemia cells and stromal cells in their lymphoid niche. However, the extent to which PI3Kδ inhibitors may have more widespread utility in the treatment of solid tumors is only beginning to be explored (25).
CSF1R+ TAMs and Foxp3+ Treg cells have been shown to influence tumor growth via a number of different molecular mechanisms, some acting directly on the tumor cells themselves, and some via inhibition of effector cells of the immune system, most notably CD8+ cytotoxic T lymphocytes. However, the extent to which these mechanisms form a partially redundant cellular network contributing robustness to tumor immunosuppression and a compensatory network contributing resistance to immunotherapy is unclear. In this study we provide evidence that the growth of adenocarcinomas in a mouse model of colorectal cancer was partially dependent upon suppression of CD8+ T cells by CSF1R+ macrophages recruited by tumor cell–derived CSF1. However, genetic ablation of CSF1 in the tumor cells, or pharmacological blockade of CSF1R by PLX3397, resulted in an increased accumulation of Foxp3+ Treg cells, limiting an effective CD8+ T cell response against the tumor. Vice versa, depletion of Foxp3+ Treg cells resulted in a large accumulation of CSF1R+ macrophages, suppressive to CD8+ T cell function. Importantly, genetic inactivation of PI3Kδ in Foxp3+ Treg cells hugely sensitized solid tumors to depletion of CSF1R+ TAMs. Further, combined inhibition of CSF1R and PI3Kδ by PLX3397 and idelalisib, respectively, substantially and synergistically inhibited tumor growth. These findings highlight the potential for TAMs and Treg cells to each overcome the individual blockade of the other, and provide a rationale for a new form of combinatorial immunotherapy.
Results
Tumor cell–derived CSF1 promotes solid tumor growth and recruits TAMs.
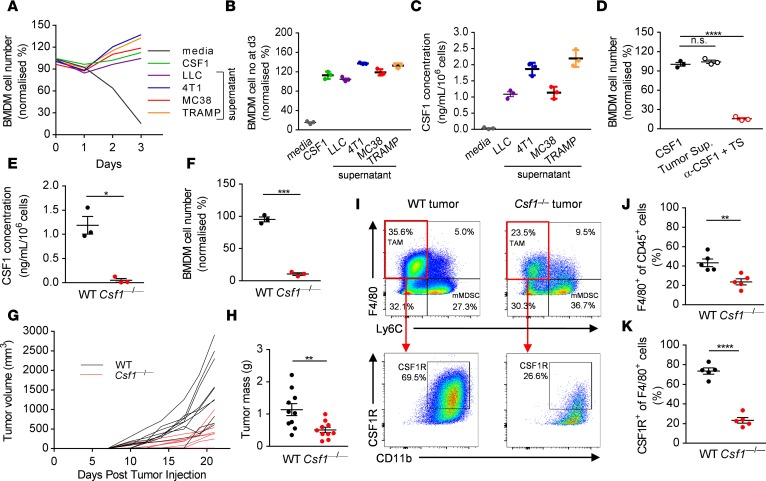
Several different cell types have the potential to produce CSF1 within the tumor microenvironment, such as stromal fibroblasts, immune cells, and tumor cells themselves (28). To more clearly define the extent to which tumor cell–derived CSF1 affects macrophage biology, we first asked whether selected mouse tumor cell lines — the LLC lung carcinoma, 4T1 mammary carcinoma, MC38 colorectal adenocarcinoma, and TRAMP-C1 prostate adenocarcinoma cells — secrete functional CSF1. We cultured bone marrow–derived macrophages (BMDMs) in the presence of supernatant collected from the different tumor cells. Conditioned media derived from all the tested tumor cell lines (LLC, 4T1, MC38, and TRAMP-C1) promoted the survival of primary BMDMs (Figure 1A). We tested tumor supernatants for the presence of different growth factors and cytokines known to affect the survival of BMDMs. ELISA analysis indicated that LLC, 4T1, MC38, and TRAMP-C1 secreted high levels of CSF1 into the culture supernatant in a fashion that correlated with the relative effects of their conditioned medium on BMDM survival (Figure 1, B and C). To determine whether the major survival factor responsible for the observed effect was CSF1, we tested whether neutralization of CSF1 in culture supernatants abrogates the survival of the BMDMs in the presence of MC38 tumor supernatant. Strikingly, CSF1 accounted for the entirety of the macrophage survival effect of MC38 culture supernatants (Figure 1D).
Figure 1. Tumor cell–derived CSF1 promotes tumor growth and recruits CSF1R+ tumor-associated macrophages.
(A and B) Representative graph (A) and quantification at day 3 (B) of the number of BMDMs cultured in the presence of complete media supplemented with recombinant CSF1 or tumor-derived supernatant from LLC (lung), 4T1 (breast), MC38 (colon), and TRAMP (prostate) cancer cell lines. (C) Concentration of CSF1 in the tumor cell supernatants measured by ELISA. (D) Quantification of BMDMs cultured in the presence of MC38 tumor cell–derived supernatant with or without anti-CSF1 neutralizing antibody. (E) Concentration of CSF1 in the WT and CRISPR/Cas9-enginered Csf1–/– MC38 tumor cell supernatants measured by ELISA. (F) Survival of BMDMs cultured in the presence of WT and Csf1–/– MC38 tumor cell–derived supernatant. (G and H) In vivo growth curves (G) and tumor masses (H) of WT and Csf1–/– MC38 tumor cells in WT mice following s.c. injections at day 0. (I–K) Representative dot plots (I) and quantification of the F4/80+ Ly6C– (J) and CSF1R+ of F4/80+ (K) immune cell infiltrates of WT and Csf1–/– MC38 tumors derived from WT mice at day 21. Graphs show mean ± SEM of data from at least 3 independent experiments or 5 to 10 individual mice per group. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; n.s., non-significant by Student’s t test or 2-way ANOVA.
Having found that high levels of CSF1 are secreted into the culture supernatant by a range of mouse solid tumor cell lines, we asked whether tumor cell–derived CSF1 is the predominant factor supporting TAM survival in vivo. To this end, we disrupted the gene encoding CSF1 using CRISPR/Cas9 mutagenesis in MC38 cells. We confirmed that Csf1–/– MC38 cells are not able to secrete CSF1 into the culture supernatant (Figure 1E), and cannot support macrophage survival in vitro (Figure 1F). We detected no significant difference between the growth rates of parental (WT) and Csf1–/– MC38 cells in vitro (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.120631DS1), or when CSF1 was neutralized within WT tumor cell line cultures (Supplemental Figure 1B), arguing against an autocrine role of CSF1 on tumor cells themselves. However, we observed impaired growth of Csf1–/– tumor cells compared with the parental MC38 cells in vivo when implanted into C57BL/6 mice, although tumor rejection was incomplete (Figure 1, G and H). When we isolated and analyzed the immune cell infiltrates of primary tumors removed at day 21 after implantation, we found a significant reduction in the F4/80+ Ly6C– leukocyte population in Csf1–/– tumors compared with WT tumors (Figure 1, I and J). Moreover, the decrease in the number of CSF1R+ cells among the F4/80+ cell population was even more pronounced between the 2 genotypes (Figure 1, I and K). Deletion of CSF1 in tumor cells also resulted in significantly reduced numbers of macrophages expressing CD206 and CD163, markers which are present on M2-polarized TAMs (Supplemental Figure 2, A–C). These results indicate that tumor cell–derived CSF1 has an important role in promoting solid tumor growth and in recruiting CSF1R+ TAMs into the tumor microenvironment but raise the possibility that other factors contribute to residual immunosuppression in the absence of tumor cell–derived CSF1.
Tumor cell–derived CSF1 recruits TAMs, which act through the inhibition of CD8+ T cells.
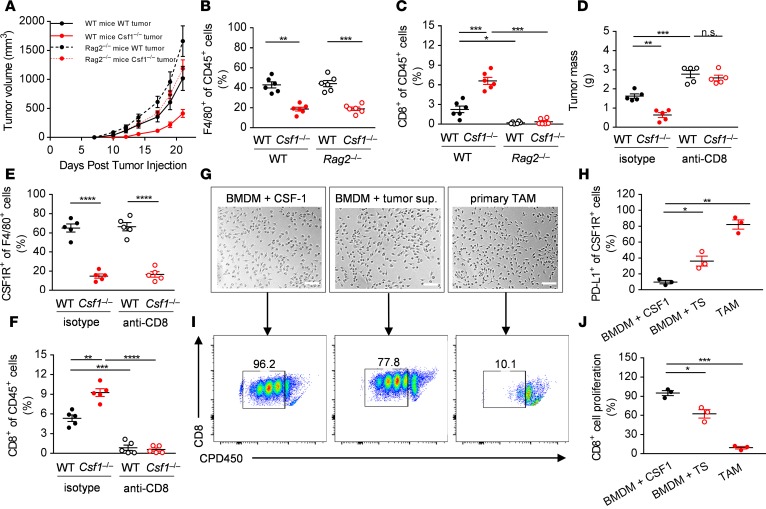
We were able to detect CSF1R+ cells colocalizing with CD8+ cells in the primary MC38 tumors and this observation led us to ask whether such cells might influence CD8+ T cell function. To test this, we implanted Csf1–/– tumor cells into Rag2–/– mice, deficient in T and B cells of the adaptive immune system. Strikingly, growth of WT and Csf1–/– tumors in Rag2–/– animals was similar, in contrast to their growth in immunocompetent hosts (Figure 2A). Furthermore, the growth defect of the Csf1–/– tumors was completely reversed following antibody-mediated depletion of CD8+ T cells (Figure 2D). We confirmed the depletion of F4/80+ and CSF1R+ TAMs in Csf1–/– MC38 tumor–bearing mice and the depletion of CD8+ T cells in Rag2–/– animals or antibody-treated mice by flow cytometry (Figure 2, B, C, E, and F). Although the recruitment of TAMs was unaffected by loss of CD8+ T cells (Figure 2, B and E), we found large increases in CD8+ T cell numbers in the primary Csf1–/– tumors compared with the WT tumors (Figure 2, C and F). These data strongly indicate that tumor cell–derived CSF1 drives TAM-dependent suppression of antitumor CD8+ T cell responses.
Figure 2. CSF1R+ tumor-associated macrophages inhibit CD8+ T cells of the adaptive immune system.
(A) In vivo growth curves of WT and Csf1–/– MC38 tumor cells implanted s.c. at day 0 into WT and Rag2–/– mice. (B and C) Quantification of the F4/80+ (B) and CD8+ (C) immune cells infiltrating the WT and Csf1–/– tumors isolated from the WT and Rag2–/– mice at day 21. (D) WT and Csf1–/– MC38 primary tumor masses removed at day 21 from WT mice treated with anti-CD8 antibody or control antibody. (E and F) Quantification of the CSF1R+ (E) and CD8+ (F) immune cells infiltrating the WT and Csf1–/– tumors isolated from the WT mice treated with anti-CD8 or control antibody at day 21. (G and H) Representative photomicrographs of (G) and PD-L1 expression on (H) BMDMs cultured in the presence of recombinant CSF1 or MC38 tumor supernatant and of primary CSF1R+ TAMs isolated from the MC38 primary tumors. (I and J) Representative flow panels (I) and quantification (J) of the in vitro proliferation of CD8+ lymphocytes co-cultured with BMDMs (as treated above) or CSF1R+ TAMs isolated from the MC38 tumors. Graphs show mean ± SEM of data from at least 3 independent experiments or 5–6 individual mice per group. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; n.s., non-significant by Student’s t test or 2-way ANOVA. Scale bars: 50 μm.
Given the observation that CSF1R+ TAMs limit adaptive immunity, we asked whether the immunosuppressive effect of tumor cell–derived CSF1 was specifically dependent upon expression or secretion of certain molecules by macrophages. To this end, we isolated CSF1R+ TAMs from the primary MC38 tumors using a discontinuous Percoll gradient and CD115-based positive magnetic selection. With this approach, primary TAMs could be cultured for a few days in the presence of tumor-conditioned media and showed similar morphology to primary BMDMs cultured in the presence of tumor supernatant (Figure 2G).
Tumor cells frequently express programmed cell death-ligand 1 (PD-L1), facilitating their escape from the immune system (29). However, little is known about the role of PD-L1 on TAMs, so we next tested the expression of PD-L1 on the primary MC38 tumor-isolated macrophages. As shown on Figure 2H, tumor-derived primary TAMs strongly expressed PD-L1 compared with naive BMDMs. We asked whether this is in part attributable to a factor secreted by tumor cells. Remarkably, culturing BMDMs in the presence of tumor cell–derived conditioned media significantly increased the ratio of cells expressing PD-L1 on their surface (Figure 2H). Furthermore, in the culture supernatants of CSF1R+ TAMs we were also able to detect high amounts of TGF-β1 (1.48 ± 0.14 ng/ml/106 cells) capable of inhibiting lymphocyte proliferation and function. As a consequence, primary TAMs and tumor re-educated BMDMs — but not naive BMDMs — could strongly suppress CD8+ T lymphocyte proliferation in vitro (Figure 2, I and J). Taken together, these data indicate that CSF1R+ TAMs express PD-L1, secrete TGF-β1, and are capable of limiting CD8+ T lymphocyte proliferation ex vivo, but other sources of immunosuppression may contribute to the failure of total tumor rejection with CSF1 ablation alone.
Depletion of CSF1R+ macrophages synergizes with genetic ablation of Foxp3+ Treg cells and with deletion of PI3Kδ specifically in the Foxp3+ Treg compartment.
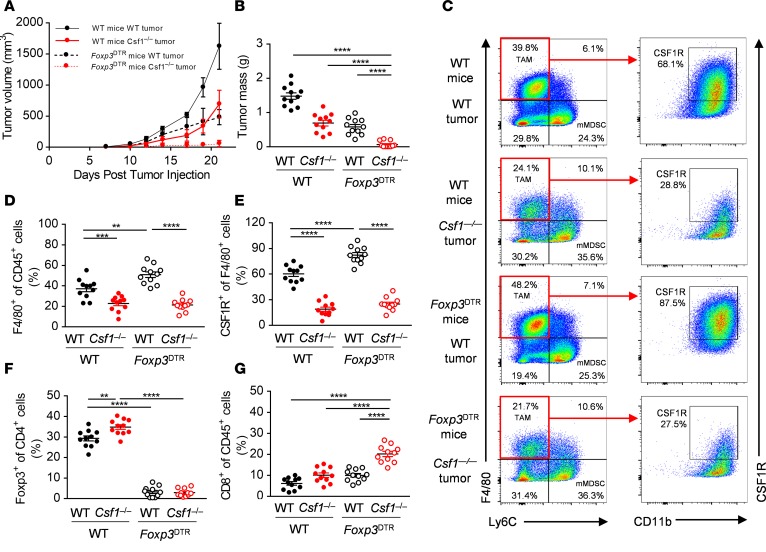
To determine the dependence of MC38 tumors on Treg-mediated immunosuppression, we depleted Treg cells from MC38 tumor-bearing Foxp3DTR mice treated with 25 μg/kg diphtheria toxin by intraperitoneal injection on days 3, 7, 10, 14, and 17 after tumor injection. Treg depletion led to reduced MC38 tumor growth, similar to the effects of deleting MC38-derived CSF1 alone, indicating that Treg cells were also an important component of immunosuppression in the MC38 model (Figure 3, A and B). Strikingly, deletion of both tumor cell–derived CSF1 and Foxp3+ Treg cells caused almost complete tumor rejection (Figure 3, A and B); half of the mice were completely tumor-free. Depletion of Treg cells alone led to significant increases in F4/80+ and CSF1R+ TAMs (Figure 3, C–E) and deletion of tumor-derived CSF1 caused significant increases in intratumoral Foxp3+ cells (Figure 3F). This suggested that each of these 2 immunosuppressive mechanisms might compensate for the loss of the other, and that inhibition of both was required to unleash full CD8+ T cell–mediated attack on the tumor, thereby leading to complete tumor rejection. In support of this idea, we found that combined deletion of tumor-derived CSF1 and Foxp3+ Treg cells produced a supra-additive effect on the number of tumor-associated CD8+ T cells (Figure 3G).
Figure 3. Depletion of CSF1R+ macrophages synergizes with genetic ablation of Foxp3+ Treg cells.
(A and B) In vivo growth curves (A) and primary tumor masses at day 21 (B) of WT and Csf1–/– MC38 tumor cells implanted s.c. at day 0 into WT and Foxp3DTR mice treated with 25 μg/kg diphtheria toxin (DTx) i.p. (C–G) Representative flow panels (C) and quantification of the F4/80+ (D) CSF1R+ (E), Foxp3+ (F), and CD8+ (G) immune cell infiltrates of WT and Csf1–/– MC38 tumors isolated from WT and Foxp3DTR mice treated with DTx. Graphs show mean ± SEM of data from 10 individual mice per group. **P < 0.01; ***P < 0.001; ****P < 0.0001 by 2-way ANOVA.
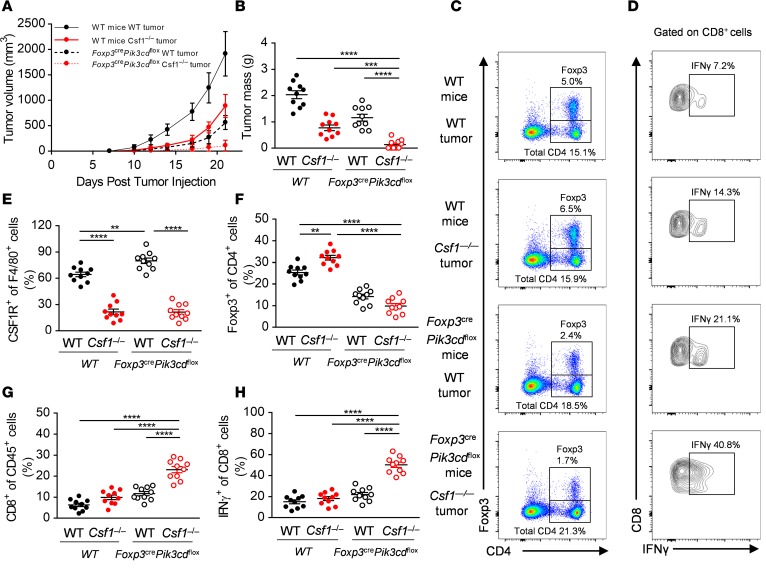
Recent studies indicate that PI3Kδ plays an important role in the maturation of Foxp3+ Treg cells and that this effect can supercede a smaller role for PI3Kδ in CD8+ T cell function, such that tumors relying heavily on Treg -mediated suppression of CD8+ T cells for growth can be inhibited by deletion of PI3Kδ (27). We investigated a potential role for PI3Kδ in the MC38 model using mice with a Treg-specific deletion of PI3Kδ. Foxp3YFP-Cre Pik3cdfl/fl mice had reduced numbers of intratumoral Foxp3+ Treg cells and behaved similarly to the Foxp3DTR mice, both in terms of their partial resistance to tumor growth, a compensatory increase in F4/80+ and CSF1R+ TAMs, and their combinatorial effects with deletion of tumor-derived CSF1 (Figure 4, A–E). Thus, Csf1–/– tumors had greatly reduced volume and mass when grown in Foxp3YFP-Cre Pik3cdfl/fl mice (Figure 4, A and B). Deletion of both tumor-derived CSF1 and PI3Kδ in Treg cells produced supra-additive increases in the intratumoral CD8+ T cell population and a striking synergistic augmentation of their function in terms of IFN-γ production (Figure 4, D, G, and H).
Figure 4. Depletion of CSF1R+ macrophages synergizes with Foxp3+ Treg cell–specific deletion of PI3Kδ.
(A and B) In vivo growth curves (A) and primary tumor masses at day 21 (B) of WT and Csf1–/– MC38 tumor cells implanted into WT and Foxp3YFP-Cre Pik3cdfl/fl mice. (C–H) Representative flow panels (C and D) and quantification of the CSF1R+ (E), Foxp3+ (F), CD8+ (G), and IFN-γ+ of CD8+ (H) immune cell infiltrates of WT and Csf1–/– MC38 tumors isolated from the WT and Foxp3YFP-Cre Pik3cdfl/fl mice. Graphs show mean ± SEM of data from 10 individual mice per group. **P < 0.01; ***P < 0.001; ****P < 0.0001 by 2-way ANOVA.
Combined inhibition of CSF1R and PI3Kδ effectively blocks solid tumor immunosuppression.
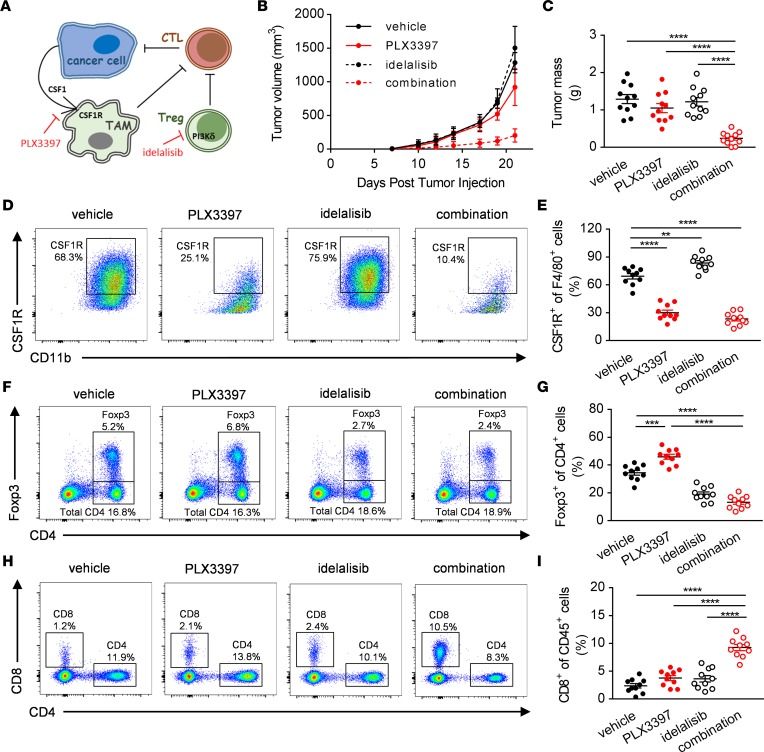
Our observations that combined depletion of Foxp3+ Treg cells and CSF1R+ TAMs can lead to very effective inhibition of tumor growth led us to explore the potential for combined pharmacological inhibition of CSF1R and PI3Kδ, in particular to investigate whether compensatory immunosuppression between CSF1R+ TAMs and Foxp3+ Treg cells might contribute to monotherapy resistance (Figure 5A). To this end, C57BL/6 mice were orally dosed with 40 mg/kg PLX3397 and/or 100 mg/kg idelalisib daily from day 7 after tumor implantation, when the tumors became palpable. Control mice received vehicle (0.5% w/v methylcellulose). Importantly, and consistent with compensatory immunosuppression driving therapy resistance to immune monotherapy, only tumors treated with the combination of PLX3397 and idelalisib showed a statistically significant reduction in tumor growth and primary tumor mass (Figure 5, B and C). By contrast, single-agent PLX3397 or idelalisib-treated tumors grew at the same rates compared with the vehicle controls. As a confirmation of the depletion of TAMs by PLX3397, we detected a reduced proportion of CSF1R+ macrophages among intratumoral myeloid cells by flow cytometry (Figure 5, D and E). Similarly, flow cytometry confirmed the depletion of Treg cells by idelalisib as a reduced proportion of CD4+ Foxp3+ Treg cells among intratumoral lymphocytes (Figure 5, F and G). Consistent with a synergistic reversal of immunosuppression, we detected an increase in CSF1R+ macrophages in idelalisib-treated tumors (Figure 5, D and E), an increase in Foxp3+ Treg cells in PLX3397-treated tumors (Figure 5, F and G), and a significant increase in the proportion of CD8+ T lymphocytes among CD45+ cells in the MC38 tumors treated with the combination of PLX3397 and idelalisib (Figure 5, H and I). Further, we detected similar synergistic effects when combining CSF1 blockade with PI3Kδ inhibition in the B16-F10 tumor model (Supplemental Figure 3, A and B). Collectively, these findings provide evidence of compensatory immunosuppression between CSF1R+ macrophages and PI3Kδ-driven Foxp3+ Treg cells and provide a rationale for combinatorial therapy using CSF1- and PI3Kδ-targeted approaches.
Figure 5. Combined inhibition of CSF1R and PI3Kδ effectively blocks tumor immunosuppression.
(A) Graphical abstract. (B and C) In vivo growth curves (B) and tumor masses at day 21 (C) of WT mice orally treated with vehicle (0.5% methylcellulose) or 40 mg/kg PLX3397 or 100 mg/kg idelalisib or the combination of the 2 from day 7 following s.c. injections of MC38 cells. (D–I) Representative flow cytometry panels (left) and quantification (right) of the CSF1R+ (D and E), Foxp3+ (F and G), and CD8+ (H and I) immune cell infiltrates of WT mice treated with vehicle (0.5% methylcellulose) or 40 mg/kg PLX3397 or 100 mg/kg idelalisib or the combination of the two. Graphs show mean ± SEM of data from 10 individual mice per group. **P < 0.01; ***P < 0.001; ****P < 0.0001 by 2-way ANOVA.
Discussion
TAMs and Treg cells are critical components of the tumor microenvironment, and contribute to every aspect of tumor growth and progression (5, 21, 30). Here, we provide evidence that Foxp3+ Treg cells and CSFR1+ TAMs form a partially redundant cellular network contributing robustness to tumor immunosuppression and a compensatory network contributing resistance to immunotherapy. Consistently, we found that tumor growth in the MC38 model was partially dependent upon CSF1 made by the tumor cells themselves and likely mediated through suppression of CD8+ T cells by CSF1R+ TAMs. However, blocking this pathway alone resulted in increased accumulation of Foxp3+ Treg cells inside the tumor and a modest effect on tumor growth. Conversely, depletion of Foxp3+ Treg cells alone resulted in increased accumulation of CSF1R+ TAMs and, again, a modest effect on tumor growth. These results suggested the presence of compensation between these 2 dominant immunosuppressive populations. Consistent with this hypothesis, deletion of both tumor-derived CSF1 and Foxp3+ Treg cells resulted in a dramatic influx of CD8+ T cells, augmentation of their function and a strikingly synergistic reduction in tumor growth.
Targeting the nontransformed tumor microenvironment is a promising approach to treat cancer, since it presents genetically stable targets for therapy (31, 32). However, the extent to which TAMs and Treg cells use common versus segregated molecular mechanisms to control CD8+ T cells or, indeed, combine with other cell types to regulate tumorigenesis are still unclear. In line with this, clinical trials with PLX3397 used as a monotherapy have shown only limited potential to arrest solid tumor growth, possibly due to the development of additional resistance mechanisms and acute toxicity problems when used close to maximum-tolerated doses (e.g., rebound myelopoiesis) (33). Similarly, idelalisib, capable of disrupting Treg cell function, showed only limited potential to arrest solid tumor growth in preclinical studies when used as a monotherapy. Our results suggest that recruitment of the main 2 immunosuppressive subsets capable of limiting CD8+ T cell immunity, CSF1R+ macrophages and Foxp3+ Treg cells, can be responsible for maintaining significant immunosuppression in the absence of the other. In support of this, we show that PLX3397, at doses predicted to achieve substantial target engagement, had insignificant effects on the growth of tumors in the MC38 model but was remarkably effective in combination with idelalisib, providing a strong rationale for exploring the combined pharmacological inhibition of both TAMs and Treg cells by compounds currently in separate clinical development, and raising the prospect that these drugs might be effectively administered below their maximum-tolerated doses to reduce acute toxicity. Recently, several other groups have shown beneficial effects using combinations of CSF1R inhibitors with various other agents, including paclitaxel (34), radiation therapy (35), adoptive cell transfer (18, 36), checkpoint inhibition therapy (16), and CXCR2 blockade (37). The most appropriate combinatorial approach will likely vary with the precise nature of the tumor but, in principle, combined inhibition of CSF1R+ macrophages and Foxp3+ Treg cells carries the potential for an effective immunotherapy against a range of tumors that rely upon immunosuppression for their survival and development.
Methods
Mice.
C57BL/6 mice were obtained from the breeding colonies in the Biological Services Unit of the Babraham Institute. Foxp3DTR mice, expressing the human diphtheria toxin receptor and an eGFP reporter under the Foxp3 promoter, allowed specific depletion of Foxp3+ cells upon administration of diphtheria toxin (38). Foxp3YFP-Cre mice expressing a YFP-Cre fusion protein reporter from the Foxp3 promoter were crossed with a mouse strain harboring loxP sites flanking alleles 1–9 of the Pik3cd gene (Foxp3FYC-Cre Pik3cdfl/fl mice), allowing deletion of PI3Kδ specifically in Foxp3+ cells (27). Rag2–/– mice were a gift from Marc Veldhoen (Instituto de Medicina Molecular, Lisboa, Portugal). In all experiments, mice were compared with appropriate age- and strain-matched controls. Mice were housed in the Biological Support Unit at the Babraham Institute under specific pathogen–free conditions.
Tumor cells.
4T1 mammary carcinoma, TRAMP-C1 prostate adenocarcinoma, and B16-F10 malignant melanoma cell lines were obtained from ATCC. LLC Lewis lung carcinoma cells were obtained from Matthew Kraman (F-star Biotechnology, Cambridge, United Kingdom). The MC38 colon carcinoma cell line was obtained from Miguel Gaspar (F-star Biotechnology, Cambridge, United Kingdom), with permission from Mark Smyth (QIMR Berghofer, Brisbane, Australia). All tumor cells were maintained in culture in DMEM containing 10% FCS (GIBCO) and dissociated from culture flasks using Cell Dissociation Buffer (GIBCO).
ELISA.
Levels of CSF1 and TGF-β1 present within the supernatants of the cells were measured using the Mouse M-CSF and Human TGF-β1 Quantikine ELISA kits (R&D Systems), in accordance with the manufacturer’s instructions.
Deletion of Csf1.
gRNA sequences directed against exon 1, 4, 5 of the murine Csf1 gene were designed using the CHOPCHOP web tool for genome engineering (39). Analysis of likely off-target genes was performed in silico. No genes directly involved in cell proliferation and differentiation were identified as off-targets. The following CRISPR guide oligonucleotides were ordered: 5′-CACCGGCTGCCCGTATGACCGCGCG-3′ (forward), 5′-AAACCGCGCGGTCATACGGGCAGCC-3′ (reverse), 5′-CACCGGGTGTCCATTCCCAATCATG-3′ (forward), 5′-AAACCATGATTGGGAATGGACACCC-3′ (reverse), 5′-CACCGGAGTTCCTGGAGCCTCTCGG-3′ (forward) 5′-AAACCCGAGAGGCTCCAGGACCTCC-3′ (reverse), and subcloned into the pSpCas9 vector (PX458, Addgene) using the BbsI overhangs. After sequence verification of the inserts, MC38 tumor cells were transfected with the pSpCas9Csf1 and pBABEpuro vectors using Lipofectamine 2000 (ThermoFisher Scientific) transfection reagent. Cells were subsequently selected with 5 μg/ml puromycin for 72 hours and used for single-cell clone generation. Genomic modification of single-cell clones was assessed by Western blotting and ELISA. Five different clones were tested in vitro and 2 independent clones were implanted in in vivo experiments.
Isolation of BMDMs.
Primary macrophages were isolated from long bones (femurs, tibia) of WT mice essentially as described previously (40). Briefly, suspensions of bone marrow cells were cultured for 48 hours in complete medium (RPMI 1640 medium containing L-glutamine supplemented with 10% fetal bovine serum, 1% penicillin and streptomycin, 1 mM sodium pyruvate, 1% nonessential amino acids) in the presence of 10 ng/ml recombinant mouse CSF1 (Peprotech). Nonadherent cells (macrophage precursors) were then plated at 0.5 million per cm2 and cultured in the presence of 10 ng/ml mouse CSF1 for 5 more days with media changes and replacement of the cytokines every 2 to 3 days. Flow cytometric analysis of the cells (to detect CD11b and F4/80 markers) revealed that greater than 97% were mature macrophages. In further experiments, CSF1 was supplied in the form of purified protein (recombinant cytokine) or as a 10% conditioned medium derived from the different tumor cell lines.
Viability assays.
PrestoBlue Cell Viability Reagent (Molecular Probes) was incubated with the cells for 1 hour at 37°C and then fluorescence in the wells was detected on a PHERAstar FSX plate reader (BMG LabTech) using a 575/620 filter, with wells containing no cells set as a ‘blank’ standard according to the manufacturer’s instructions.
Isolation of TAMs.
A maximum of 1 gram of tumor tissue per sample was placed into 2.5 ml of tumor dissociation buffer (Tumor Dissociation Kit, Mouse, Miltenyi Biotec) and cut finely with scalpels. The minced tissue was transferred into gentleMACS C tubes (Miltenyi Biotec) and processed on a gentleMACS Dissociator (Miltenyi Biotec) according to the ‘soft/medium tissue’ protocol from the Tumor Dissociation Kit. Tubes were then placed in a shaking incubator at 37°C for 1 hour, and processed once more on the gentleMACS Dissociator. The digested tumors were filtered through 40-μM cell strainers (Falcon) to remove remaining solid tissue, washed through with PBS, and centrifuged at 500 g for 7 minutes. The cell pellets were then resuspended in PBS and cell counts determined on the CASY cell counter (OMNI Life Sciences). Tumor-infiltrating leukocytes were separated from cancer cells using a discontinuous Percoll (GE Healthcare) gradient of 40% Percoll (0.6 ml 10× PBS, 5.4 ml Percoll, and 9 ml DMEM) and 80% Percoll (1.2 ml 10× PBS, 10.8 ml Percoll, and 3 ml DMEM). Then, TAMs were isolated from other leukocytes by positive selection for CD115 (anti-mouse CD115-PE, clone AFS98, Biolegend) used at a 1:200 dilution in FACS buffer and incubated at 4°C for 30 minutes. After washing, the cells were incubated with anti-PE microbeads (Miltenyi Biotec) at a 1:10 dilution in FACS buffer at 4°C for a further 20 minutes. Washed cells were passed through LS columns (Miltenyi Biotec) mounted on a μMACS separator (Miltenyi Biotec), with the flowthrough containing the unlabeled leukocytes. Magnetically labeled CD115+ macrophages were eluted from the columns by firmly pushing the plunger into the column outside the magnet.
Lymphocyte isolation.
Spleens were pushed through 40-μm cell strainers and washed with PBS. After centrifugation at 500 g for 5 minutes, the cell pellets were resuspended in 1 ml red blood cell lysis buffer (Sigma-Aldrich) and incubated at room temperature for 5 minutes. Cells were then washed and resuspended in PBS, and cell counts were determined on the CASY cell counter. CD8+ T cells were isolated from the spleen suspensions by negative selection against CD4+ T cells, B cells, natural killer (NK) cells, and antigen-presenting cells (APCs), respectively marked by anti-CD4 (clone RM4-5, Biolegend), anti-CD19 (clone 6D5, Biolegend), anti-NK1.1 (clone PK136, Biolegend), and anti-MHC class II (clone M5/114.15-2, Biolegend), all conjugated with FITC. These antibodies were applied to cells at a 1:200 dilution in FACS buffer and incubated at 4°C for 30 minutes. After washing, the cells were incubated with anti-FITC microbeads (Miltenyi Biotec) at a 1:10 dilution in FACS buffer at 4°C for a further 20 minutes. Washed cells were passed through LS columns mounted on the μMACS separator, with the flowthrough collected as CD8+ T cells of 90% to 95% purity.
Lymphocyte proliferation.
CD8+ T cells were resuspended in RPMI 1640 medium (Invitrogen) supplemented with 10% FCS and a final concentration of 10 μM Cell Proliferation Dye 450 (eBioscience) was added to 1-ml aliquots of lymphocytes with immediate vortexing to ensure rapid and homogeneous labeling of cells. Cells were incubated at 37°C for 10 minutes, then washed 3 times with RPMI 1640 supplemented with 10% FCS. Fluorescent dye–labeled lymphocytes were cultured for 3 days in coculture with BMDMs or primary TAMs before antibody staining and analysis by flow cytometry.
Tumor models.
Suspensions of tumor cells were prepared in sterile PBS, and cell counts were determined on a CASY cell counter. MC38 (2 × 106) or B16-F10 (1 × 105) cells were administered in 200 μl by subcutaneous injection in the shaved left flank of isoflurane-anesthetized mice. When tumors became palpable (around day 7–10), tumor growth was monitored by caliper measurements every 2 to 3 days. Tumor volume (mm3) was calculated as length × width2. Mice bearing MC38 tumors were culled at 21 days after implantation. Mice bearing B16-F10 tumors were culled at 19 days after implantation. Any mice bearing tumors that approached or exceeded the terminal size limit (10% of the weight of the mouse at the start of the study) during the course of the study were culled and excluded from the study. Tumors were collected into cold PBS at the end of the study and weights recorded prior to further processing.
Flow cytometry.
Surface stains were carried out in PBS containing 2% FCS (FACS buffer) for 30 minutes at 4°C. All of the following antibodies were used at a 1:200 dilution: anti-mouse CD8-BV711 (clone 53-6.7), CD11b-APC (clone M1/70), CD45-BV650 (clone 30-F11), CD115-PE (clone AFS98), CD274-BV605 (clone 10F.9G2), F4/80-PEcy7 (clone BM8), Ly6C-AF700 (clone HK1.4), IFN-γ-BV510 (clone XMG1.2, all from Biolegend); CD206-PE (clone MR6F3), CD163-PerCP eF710 (clone TNKUPJ), MHC class II-APC (M5/114.15.2, all from eBioscience). Fixation and permeabilization for intracellular staining was carried out with the Foxp3/Transcription Factor Staining Buffer set (eBioscience), with a 15-minute fixation at room temperature, followed by intracellular staining in permeabilization buffer for 30 minutes at 4°C. Unless otherwise stated, in assays where cells are fixed, Fixable Viability Dye eFluor 780 (eBioscience) was added to the surface stain solution at a 1:3,000 dilution. Samples were acquired on the BD Fortessa analyzers in the Babraham Institute Flow Core Facility. Data collected were analyzed using FlowJo software (Tree Star).
In vivo inhibitor and depleting antibody treatments.
PLX3397 and idelalisib (both from Selleckchem) were administered orally once per day at 40 mg/kg and 100 mg/kg, respectively, from day 7 after tumor implantation. Methylcellulose (0.5% w/v) was used as the vehicle control. B16-F10 tumor–bearing mice were treated intraperitoneally (i.p.) with 150 μg InVivoMAb anti-mouse CSF1 antibody (clone 5A1, Bio X Cell) on days 3, 7, 10, 14, and 17 after tumor implantation. The isotype control used in this experiment was rat IgG1 (clone HRPN, Bio X Cell). Diphtheria toxin from Corynebacterium diphtheriae (Sigma-Aldrich) was obtained in lyophilized powder form, and reconstituted in 0.5 ml sterile ddH2O according to the manufacturer’s instructions. Solutions for injection were made up in sterile PBS to a dose of 25 μg/kg, separately calculated based on average weights of male and female mice in each cohort. To achieve transient depletion of Treg cells in Foxp3DTR mice, diphtheria toxin was administered to the peritoneum in a 100 μl volume, with mice under inhalation anesthesia, on days 3, 7, 10, 14, and 17 after tumor implantation. Tumor growth was monitored as described above, and processing of tissues at the end of the experiment followed the same method as detailed in previous sections.
Study approval.
All animal work was performed under Home Office UK Project license PPL 70/8100.
Statistics.
Data were compared using a 2-tailed, unpaired Student’s t test or 2-way, repeated-measures ANOVA with Bonferroni’s post hoc test (GraphPad Prism software). Differences with P values of < 0.05 were considered significant: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Author contributions
DG designed and performed the experiments and wrote the manuscript. RR, PTH, ELL, KO, and LRS reviewed and edited the manuscript. FMG, DS, SJS, and KO provided important reagents.
Supplementary Material
Acknowledgments
We are grateful to the staff from the Biological Services Unit, Flow Cytometry and Imaging Core Facilities of the Babraham Institute for expert assistance. D. Gyori was funded by a research grant from Karus Therapeutics. E.L. Lim was supported by a Yousef Jameel Scholarship (Cambridge Trust). R. Roychoudhuri and K. Okkenhaug received institute support from Biotechnology and Biological Sciences Research Council (BBSRC) (BBS/E/B/000 -C0407, -C0409, -C0427, and -C0428). K. Okkenhaug was also supported by Wellcome Trust grant 095198/Z/10/Z. L.R. Stephens and P.T. Hawkins were supported by an institute grant from the BBSRC (BB/J004456/1). R. Roychoudhuri is supported by the Wellcome Trust/Royal Society (grant 105663/Z/14/Z), the UK Biotechnology and Biological Sciences Research Council (grant BB/N007794/1), and Cancer Research UK (grant C52623/A22597).
Version 1. 06/07/2018
Electronic publication
Footnotes
DG’s present address is: Department of Physiology, Semmelweis University, Budapest, Hungary.
Conflict of interest: DG was funded by a grant from Karus Therapeutics. KO received consultation fees from Karus Therapeutics and Gilead Sciences. LRS and PTH received consultation fees from Karus Therapeutics.
License: This work is licensed under the Creative Commons Attribution 4.0 International License. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
Reference information: JCI Insight. 2018;3(11):e120631. https://doi.org/10.1172/jci.insight.120631.
Contributor Information
David Gyori, Email: gyoridavid@med.semmelweis-univ.hu.
Ee Lyn Lim, Email: eelynlim@ifrec.osaka-u.ac.jp.
Dominik Spensberger, Email: dominik.spensberger@babraham.ac.uk.
Rahul Roychoudhuri, Email: rahul.roychoudhuri@babraham.ac.uk.
Klaus Okkenhaug, Email: ko256@cam.ac.uk.
References
- 1.Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature. 2013;496(7446):445–455. doi: 10.1038/nature12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ginhoux F, Jung S. Monocytes and macrophages: developmental pathways and tissue homeostasis. Nat Rev Immunol. 2014;14(6):392–404. doi: 10.1038/nri3671. [DOI] [PubMed] [Google Scholar]
- 3.McAllister SS, Weinberg RA. The tumour-induced systemic environment as a critical regulator of cancer progression and metastasis. Nat Cell Biol. 2014;16(8):717–727. doi: 10.1038/ncb3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mosely SI, et al. Rational selection of syngeneic preclinical tumor models for immunotherapeutic drug discovery. Cancer Immunol Res. 2017;5(1):29–41. doi: 10.1158/2326-6066.CIR-16-0114. [DOI] [PubMed] [Google Scholar]
- 5.Noy R, Pollard JW. Tumor-associated macrophages: from mechanisms to therapy. Immunity. 2014;41(1):49–61. doi: 10.1016/j.immuni.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol. 2010;11(10):889–896. doi: 10.1038/ni.1937. [DOI] [PubMed] [Google Scholar]
- 7.Kitamura T, Qian BZ, Pollard JW. Immune cell promotion of metastasis. Nat Rev Immunol. 2015;15(2):73–86. doi: 10.1038/nri3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitchem JB, et al. Targeting tumor-infiltrating macrophages decreases tumor-initiating cells, relieves immunosuppression, and improves chemotherapeutic responses. Cancer Res. 2013;73(3):1128–1141. doi: 10.1158/0008-5472.CAN-12-2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franklin RA, et al. The cellular and molecular origin of tumor-associated macrophages. Science. 2014;344(6186):921–925. doi: 10.1126/science.1252510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strachan DC, et al. CSF1R inhibition delays cervical and mammary tumor growth in murine models by attenuating the turnover of tumor-associated macrophages and enhancing infiltration by CD8+ T cells. Oncoimmunology. 2013;2(12):e26968. doi: 10.4161/onci.26968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kübler K, et al. Prognostic significance of tumor-associated macrophages in endometrial adenocarcinoma. Gynecol Oncol. 2014;135(2):176–183. doi: 10.1016/j.ygyno.2014.08.028. [DOI] [PubMed] [Google Scholar]
- 12.Hu W, et al. Alternatively activated macrophages are associated with metastasis and poor prognosis in prostate adenocarcinoma. Oncol Lett. 2015;10(3):1390–1396. doi: 10.3892/ol.2015.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skrzypski M, et al. Main histologic types of non-small-cell lung cancer differ in expression of prognosis-related genes. Clin Lung Cancer. 2013;14(6):666–673.e2. doi: 10.1016/j.cllc.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 14.Yoshikawa K, et al. Impact of tumor-associated macrophages on invasive ductal carcinoma of the pancreas head. Cancer Sci. 2012;103(11):2012–2020. doi: 10.1111/j.1349-7006.2012.02411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pyonteck SM, et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat Med. 2013;19(10):1264–1272. doi: 10.1038/nm.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu Y, et al. CSF1/CSF1R blockade reprograms tumor-infiltrating macrophages and improves response to T-cell checkpoint immunotherapy in pancreatic cancer models. Cancer Res. 2014;74(18):5057–5069. doi: 10.1158/0008-5472.CAN-13-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quail DF, et al. The tumor microenvironment underlies acquired resistance to CSF-1R inhibition in gliomas. Science. 2016;352(6288):aad3018. doi: 10.1126/science.aad3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sluijter M, et al. Inhibition of CSF-1R supports T-cell mediated melanoma therapy. PLoS One. 2014;9(8):e104230. doi: 10.1371/journal.pone.0104230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mok S, et al. Inhibition of CSF-1 receptor improves the antitumor efficacy of adoptive cell transfer immunotherapy. Cancer Res. 2014;74(1):153–161. doi: 10.1158/0008-5472.CAN-13-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ries CH, et al. Targeting tumor-associated macrophages with anti-CSF-1R antibody reveals a strategy for cancer therapy. Cancer Cell. 2014;25(6):846–859. doi: 10.1016/j.ccr.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 21.Ries CH, Hoves S, Cannarile MA, Rüttinger D. CSF-1/CSF-1R targeting agents in clinical development for cancer therapy. Curr Opin Pharmacol. 2015;23:45–51. doi: 10.1016/j.coph.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 22.Butowski N, et al. Orally administered colony stimulating factor 1 receptor inhibitor PLX3397 in recurrent glioblastoma: an Ivy Foundation Early Phase Clinical Trials Consortium phase II study. Neuro-oncology. 2016;18(4):557–564. doi: 10.1093/neuonc/nov245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quail DF, Joyce JA. Molecular pathways: deciphering mechanisms of resistance to macrophage-targeted therapies. Clin Cancer Res. 2017;23(4):876–884. doi: 10.1158/1078-0432.CCR-16-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roychoudhuri R, Eil RL, Restifo NP. The interplay of effector and regulatory T cells in cancer. Curr Opin Immunol. 2015;33:101–111. doi: 10.1016/j.coi.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 25.Okkenhaug K, Graupera M, Vanhaesebroeck B. Targeting PI3K in cancer: impact on tumor cells, their protective stroma, angiogenesis, and immunotherapy. Cancer Discov. 2016;6(10):1090–1105. doi: 10.1158/2159-8290.CD-16-0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okkenhaug K, Burger JA. PI3K signaling in normal B cells and chronic lymphocytic leukemia (CLL) Curr Top Microbiol Immunol. 2016;393:123–142. doi: 10.1007/82_2015_484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ali K, et al. Inactivation of PI(3)K p110δ breaks regulatory T-cell-mediated immune tolerance to cancer. Nature. 2014;510(7505):407–411. doi: 10.1038/nature13444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel S, Player MR. Colony-stimulating factor-1 receptor inhibitors for the treatment of cancer and inflammatory disease. Curr Top Med Chem. 2009;9(7):599–610. doi: 10.2174/156802609789007327. [DOI] [PubMed] [Google Scholar]
- 29.Dong H, et al. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med. 2002;8(8):793–800. doi: 10.1038/nm730. [DOI] [PubMed] [Google Scholar]
- 30.Chitu V, Stanley ER. Colony-stimulating factor-1 in immunity and inflammation. Curr Opin Immunol. 2006;18(1):39–48. doi: 10.1016/j.coi.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 31.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19(11):1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity. 2013;39(1):1–10. doi: 10.1016/j.immuni.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 33.Gyori D, Chessa T, Hawkins PT, Stephens LR. Class (I) phosphoinositide 3-kinases in the tumor microenvironment. Cancers (Basel) 2017;9(3):E24. doi: 10.3390/cancers9030024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeNardo DG, et al. Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy. Cancer Discov. 2011;1(1):54–67. doi: 10.1158/2159-8274.CD-10-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu J, et al. CSF1R signaling blockade stanches tumor-infiltrating myeloid cells and improves the efficacy of radiotherapy in prostate cancer. Cancer Res. 2013;73(9):2782–2794. doi: 10.1158/0008-5472.CAN-12-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mok S, et al. Inhibition of colony stimulating factor-1 receptor improves antitumor efficacy of BRAF inhibition. BMC Cancer. 2015;15:356. doi: 10.1186/s12885-015-1377-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar V, et al. Cancer-associated fibroblasts neutralize the anti-tumor effect of CSF1 receptor blockade by inducing PMN-MDSC infiltration of tumors. Cancer Cell. 2017;32(5):654–668.e5. doi: 10.1016/j.ccell.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim JM, Rasmussen JP, Rudensky AY. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat Immunol. 2007;8(2):191–197. doi: 10.1038/ni1428. [DOI] [PubMed] [Google Scholar]
- 39.Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 2014;42(Web Server issue):W401–W407. doi: 10.1093/nar/gku410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gyori D, et al. The phosphoinositide 3-kinase isoform PI3KB regulates osteoclast-mediated bone resorption in humans and mice. Arthritis Rheumatol. 2014;66(8):2210–2221. doi: 10.1002/art.38660. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.